Abstract
Viroporins are indispensable for viral replication. As intracellular ion channels they disturb pH gradients of organelles and allow Ca2+ flux across ER membranes. Viroporins interact with numerous intracellular proteins and pathways and can trigger inflammatory responses. Thus, they are relevant targets in the search for antiviral drugs. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) underlies the world-wide pandemic of COVID-19, where an effective therapy is still lacking despite impressive progress in the development of vaccines and vaccination campaigns. Among the 29 proteins of SARS-CoV-2, the E- and ORF3a proteins have been identified as viroporins that contribute to the massive release of inflammatory cytokines observed in COVID-19. Here, we describe structure and function of viroporins and their role in inflammasome activation and cellular processes during the virus replication cycle. Techniques to study viroporin function are presented, with a focus on cellular and electrophysiological assays. Contributions of SARS-CoV-2 viroporins to the viral life cycle are discussed with respect to their structure, channel function, binding partners, and their role in viral infection and virus replication. Viroporin sequences of new variants of concern (α–ο) of SARS-CoV-2 are briefly reviewed as they harbour changes in E and 3a proteins that may affect their function.
Keywords: Viroporins, Viroporin activity, Structure-function relations, Viroporin assays, SARSCoV-2, Review
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has caused the ongoing pandemic of Coronavirus Disease 2019 (COVID-19). SARS-CoV-2 belongs to the genus Beta-coronavirus of the Coronaviridae family, which includes SARS-CoV and Middle East respiratory syndrome coronavirus (MERS CoV) (de Wit et al., 2016). Structural proteins S, M, and E are part of the virus envelope and mediate virus – host cell attachment and entry. Coronavirus-encoded accessory proteins play critical roles in virus–host interactions and modulate host immune responses, thereby participating in coronaviral pathogenicity (Lim et al., 2016). One group of accessory proteins that have found interest as potential pharmaceutical targets are viroporins. Viroporins are small, highly hydrophobic, virus-encoded proteins that interact with membranes modifying the cell’s permeability to ions or other small molecules. The name viroporin was introduced after the discovery of proteins from several different types of viruses that share common characteristics, such as binding to intracellular proteins and activity as ion channels (Carrasco et al., 1993). Ion channel activity of viroporins has been described earlier, when enhanced membrane permeability was found in several virus-related cell systems (Carrasco, 1978, Gonzalez and Carrasco, 2003, McClenaghan et al., 2020). The main contribution of viroporins to the viral life cycle involves virion assembly and release from infected cells (Iwatsuki-Horimoto et al., 2006, Lu et al., 2006, Steinmann et al., 2007, Ruiz et al., 2010) as shown by viroporin defective viruses that were unable to accomplish proper folding and viral release (Terwilliger et al., 1989, Iwatsuki-Horimoto et al., 2006, Lu et al., 2006, Jones et al., 2007, Steinmann et al., 2007). Viroporins have also been suggested to be involved in virus-induced apoptosis (O'Brien, 1998).
2. Definition of viroporins
A viroporin is a transmembrane hydrophilic pore including a selectivity filter, possessing defined charge selectivity and translocation efficiency that allows ions or small solutes to pass the host cell's membranes along their electrochemical gradient. Viroporins have been described in numerous viruses ( Table 1), and they classically participate in the viral replication cycle by locating to membranes connecting the ER lumen and cytosol. Presence of viroporins on the plasma membrane has been observed in some studies and not found in others. To date, the question remains whether viroporins allow random passage of their permeant ions, or if they are controlled by a gating mechanism similar to ligand- or voltage-gated neuronal ion channels.
Table 1.
Properties of a selection of well-characterized viroporins.
| Viroporin Class |
Virus Family |
Virusname | Viroporin | proposed oligomerization | Size [aa] |
Ion selectivity | References |
|
|---|---|---|---|---|---|---|---|---|
| Inflammasome activation | Structure | |||||||
| IA | Coronaviridae | SARS CoV | S1-E | 2, 3, 5 (Torres et al., 2005) | 76 | K+/Na+ Ca2+ |
(Nieto-Torres et al., 2015) | (Pervushin et al., 2009) (Li et al., 2014),(Surya et al., 2018) |
| SARS CoV-2 | S2-E | 5 (Mandala et al., 2020) | 75 | K+/Na+ >Ca2+/Mg2+; H+ |
– | (Mandala et al., 2020), (Xia et al., 2021) | ||
| Orthomyxo-viridae | Influenza A Virus | M2 | 4 (Salom et al., 2000), (Nieva et al., 2012)1 | 97 | H+ | (Ichinohe et al., 2010) | (Schnell and Chou, 2008, Stouffer et al., 2008) (Acharya et al., 2010), (Wang et al., 2013) | |
| Retroviridae | HIV-1 | Vpu | 5 (Hussain et al., 2007), (Padhi et al., 2013)* | 81 | K+/Na+ | (Triantafilou et al., 2021) | (Park et al., 2003) | |
| IB | Paramyxoviridae | RSV | SH | 5 (Gan et al., 2012) | 64–65 | K+/Na+cations | (Triantafilou et al., 2013) | modeling only§ |
| IIA | Flaviviridae | Hepatitis C virus | p7 | 6 (Luik et al., 2009) 4–7 (Chandler et al., 2012) |
63 | H+ cations |
(Shrivastava et al., 2013, Farag et al., 2017) | (Montserret et al., 2010), (Cook et al., 2013) |
| IIB | Picornaviridae | Poliovirus | 2B | 2, 4 (Gonzalez and Carrasco, 2003, Clarke et al., 2006) | 97 | Ca2+ | – | – |
| III | Coronaviridae | SARS CoV | S1–3a | 275 | K+ | (Chen et al., 2019) | – | |
| SARS CoV-2 | S2–3a | 2, 4 (Kern et al., 2021) | 287 | Ca2+ > K+ > Na+ | – | (Kern et al., 2021) | ||
All selected viruses are enveloped (exception: picornaviridae), single-stranded +RNA viruses except HIV-1 (reverse transcribing RNA virus). Ion selectivities have been adapted from a previous review (Scott and Griffin, 2015) except HCV p7 (Montserret et al., 2010), and SARS CoV-2 E protein (Xia et al., 2021) and ORF 3a (Kern et al., 2021) proteins. §only modeled structures available **Rhinolophus sinicus (Chinese rufous horseshoe bat); Rhinolophus ferrumequinum (Greater horseshoe bat).
3. General architecture and function of viroporins
Viroporins have been found in numerous RNA viruses, including E viroporins of SARS-CoV, mouse hepatitis virus, and infectious bronchitis virus, 6 K protein from togaviruses, the small hydrophobic protein (SH) from paramyxoviruses, non-structural protein 4 (NSP4) from rotaviruses, p10 from avian orthoreovirus, p7 from pestiviruses and flaviviridae including human hepatitis C virus, and Kcv from Paramecium bursaria. Viroporins and membrane proteins with putative channel function were also found on DNA viruses such as the agnoprotein of JC polyomavirus, viral protein 4 of Simian virus 40 (SV40), and E5 of human papillomavirus type 16. A number of excellent reviews on channel proteins of viruses are available (Nieva et al., 2012, Gonzalez and Carrasco, 2003, Scott and Griffin, 2015). Properties of the best-characterized viroporins are summarized in Table 1.
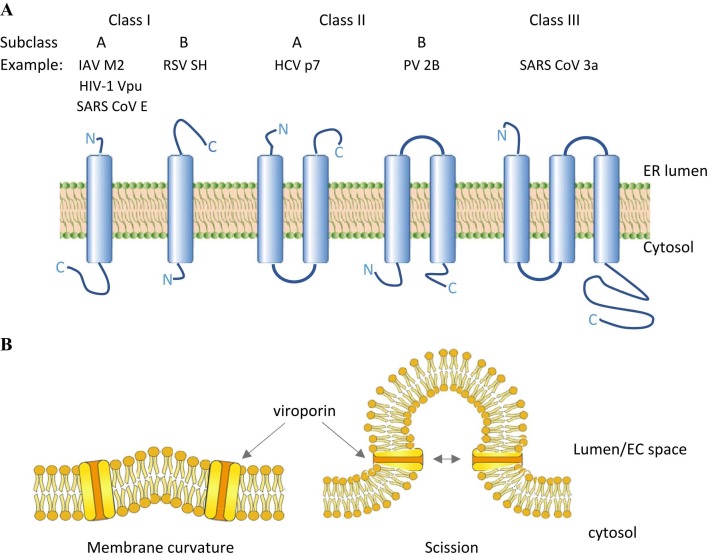
Viroporins can be classified into classes I to III, named after their number of transmembrane domains. The position of the amino- and carboxyl-terminal domains can be luminal or cytosolic which determines their subgroup (Nieva et al., 2012). Class IA viroporins contain a short luminal amino- (N) terminal domain (< 25 amino acids) and a larger carboxyl-(C) terminal domain of around 50 amino acids. Class IB viroporins possess a short cytosolic amino (N)-terminal domain while its longer carboxyl (C)-terminal part is located in the endoplasmic reticulum (ER) lumen. The third class has so far only one member, namely the SARS CoV open reading frame (ORF) 3a. Here, the short N-terminal domain points to the ER lumen, followed by three transmembrane domains, hence the longer C-terminal domain is situated in the cytosol ( Fig. 1A).
Fig. 1.
Classes and subclasses of viroporins and their transmembrane topology. (A) Transmembrane topology. Schemes represent one subunit only, while in vivo all viroporin assemble into dimers up to pentamers (heptamers have been reported for HCV p7 channels) to form the ion channel. IAV = Influenza A virus; HIV-1 = human immunodeficiency virus type 1; RSV = respiratory syncytial virus; SH = small hydrophobic; HCV = hepatitis C virus; PV = poliovirus. (B) Postulated viroporin activity in membrane curvature needed during budding and scission.
Proposed mechanism is adapted from IAV M2 protein studies (Rossman et al., 2010)
3.1. Viroporins and virus glycoproteins modify membrane permeability
In addition to viroporins, several other virus-associated glycoproteins were described to increase cell membrane permeability (Chernomordik et al., 1994, Arroyo et al., 1995, Newton et al., 1997, Ciccaglione et al., 1998). Some viral glycoproteins are able to oligomerize, thereby forming pore-like structures that allow transmembrane permeation of ions, as viroporins do. In addition, domains adjacent to the transmembrane region could act to destabilize membrane structure. Indeed, viroporin activity is fully replaced in the HIV-2 virus that does not express typical viroporins (Bour and Strebel, 1996). Taken together, in viruses lacking the typical viroporins their function could be replaced by pore-forming glycoproteins, while other viruses are in need of ion channel activity (Bour and Strebel, 1996, Gonzalez and Carrasco, 2003). It was proposed that pore-forming glycoproteins play key roles during virus entry and in some cases during virus budding, while viroporins are essential in virus assembly and release (Gonzalez and Carrasco, 2003). It should be noted that transmembrane channel activity is essential for virus replication and proliferation, whether provided by viroporins or other pore-forming proteins. It has been suggested that viroporins help to create the negative membrane curvature that is needed for closure and release of virus-containing particles (Fig. 1B).
3.2. Viroporin channel activity modulate cellular homeostasis
The maintenance of membrane gradients and the specific ionic composition within defined cellular compartments and organelles is essential for cellular homeostasis. Therefore, destruction of these finely balanced systems by viroporin activity will affect multiple cellular processes including trafficking, signaling and the induction of cell death by apoptosis (Scott and Griffin, 2015). Calcium homeostasis is disturbed when Ca2+ leaks from intracellular stores such as mitochondria, ER and Golgi complexes, as is commonly observed after viroporin expression. Examples are poliovirus protein 2B (Aldabe et al., 1997), picornavirus 2B proteins (de Jong et al., 2008) and rotavirus NSP4 viroporin (Diaz et al., 2008, Hyser et al., 2010). It was further shown that the envelope proteins (E) of both, SARS-CoV, and SARS-CoV-2 form calcium selective channels, thereby triggering the activation of the NLRP3 inflammasome, leading to IL-1β overproduction (Nieto-Torres et al., 2015), (Xia et al., 2021). Ca2+ acts as an intracellular messenger and regulator of numerous cellular processes, so changes in calcium levels affect these processes inside both, host cells or the virus (Zhou et al., 2009). Host cell apoptosis is often observed after viroporin expression (Hajnoczky et al., 2003, Aweya et al., 2013, Breitinger et al., 2021), and can also be induced by high calcium levels (Madan et al., 2008). In addition, increased calcium levels can indirectly alter processes like gene expression, viral maturation, or release (Zhou et al., 2009, Nieva et al., 2012). Several viroporins were described as pH-gated, and H+ conducting channels, thereby they would be able to change the pH of the cytosol and organelles. It has been proposed that viroporins action might lead to an increase of cytosolic pH by de-acidification of the ER-Golgi intermediate compartment (ERGIC). Use of this mechanism was suggested for IAV M2 (Ciampor et al., 1992, Ciampor et al., 1992, Sakaguchi et al., 1996), HCV p7 (Wozniak et al., 2010, Breitinger et al., 2016), infectious Bronchitis CoV (IBV) envelope protein (Westerbeck and Machamer, 2019) and SARS CoV-2 E protein (Nieto-Torres et al., 2015, Cabrera-Garcia et al., 2021, Trobec, 2021). In all these cases, shunting of pH may help to protect acid-labile proteins or particles during viral release. In case of the IBV envelope protein, it is proposed, that the altered pH in the Golgi protects the IBV spike protein and helps in releasing virus particles (Westerbeck and Machamer, 2019). Notably, the cytosol constitutes the largest cellular fraction by volume (54%), while rough and smooth ER together occupy 15%, and lysosomes and endosomes only make 2% of eukaryotic cell volume (Alberts et al., 2002). Thus, it would be expected that proton channels in ER, ERGIC, endosomes and lysosomes could change cytosolic pH only locally, and to a small extent, while the pH of these organelles and compartments themselves would be strongly altered in the presence of active viroporins.
3.3. Consequence of disrupted homeostasis: viroporins activate the inflammasome
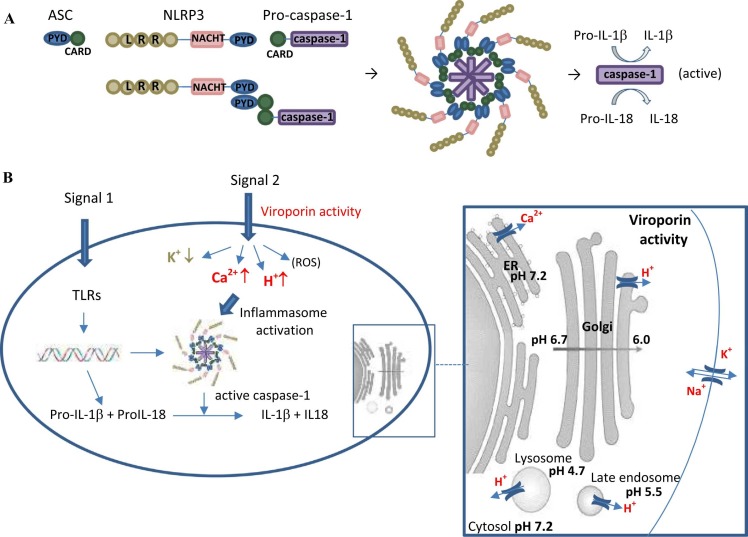
The activation of the inflammasome by viroporins was reviewed before (Farag et al., 2020). Inflammasomes are activated upon cellular infection or stress that trigger the maturation of proinflammatory cytokines such as IL-1β to activate innate immune defenses (Latz et al., 2013). These multi-protein complexes are made up of a sensor protein - the adaptor protein ASC (apoptosis-associated speck-like protein containing CARD) - and the cellular protease caspase-1 (Schroder and Tschopp, 2010). In general, caspases are cysteine proteases that initiate or execute cellular programs, leading to inflammation and/or cell death; inflammasomes activate a specific class called inflammatory caspases (Martinon et al., 2007). Mammalian inflammatory caspases contain a CARD domain followed by a catalytic cysteine residue-containing domain (Boatright and Salvesen, 2003). Caspase-1 is synthesized as an inactive zymogen (pro-caspase-1) that undergoes autocatalytic processing upon appropriate stimulus (Boatright and Salvesen, 2003). Within the inflammasome, caspase-1 is activated upon interaction with the ASC adapter protein bridging the protease with NOD-like receptors (NLRs) (Nadiri et al., 2006). Several pro-inflammatory cytokines of the IL-1 family, mostly IL-1β and IL-18, play an important role in antimicrobial host defense (Dinarello, 1984). IL-1β activates the release of other proinflammatory cytokines such as TNF and IL-6 and is responsible for the acute phase response, which includes fever, acute protein synthesis, anorexia and sleepiness (Martinon et al., 2009). IL-18 is produced during chronic inflammation, in autoimmune diseases, in a variety of cancers, and is connected to several infectious diseases. It induces the production of multiple cytokines including IFN-γ, IL-13, IL-4, IL-8 as well as both Th1 and Th2 lymphokines (Gracie et al., 2003). Proinflammatory stimuli induce expression of the inactive IL-1β and IL-18 pro-forms, while cytokine maturation and release are controlled by inflammasomes (Martinon, Mayor et al., 2009). It is generally accepted that activation and release of IL-1β requires two distinct signals, yet the nature of these signals in the in-vivo situation during inflammation is not completely defined (Negash et al., 2013) (Latz, Xiao et al., 2013). In-vitro studies characterized the first signal to be triggered by various PAMPs (pathogen-associated molecular patterns) and DAMPs (damage-associated molecular patterns), followed by Toll-like receptor (TLR) activation, which induces pro IL-1β synthesis (Negash et al., 2013). The second signal is provoked by different DAMPS and PAMPS promoting NLRP3 inflammasome assembly and caspase-1-mediated activation of pro IL-1β and pro IL-18. The requirement of a second signal might provide a safety-net mechanism to ensure activation of potent inflammatory responses to happen exclusively in the presence of real inflammatory threats, such as pathogen infection and/or tissue injury (Christgen and Kanneganti, 2020). By disruption of the ionic balance of Ca2+ and H+ in the ER/Golgi compartment, several viroporins provide the second signal needed to activate host inflammasomes in processing and releasing of pro-inflammatory cytokines ( Fig. 2). Viroporins triggering inflammasome activation include influenza virus M2 channel (Ichinohe et al., 2010), the respiratory syncytial virus SH protein (Triantafilou et al., 2013), the encephalomyocarditis virus (EMCV) 2B (Ito et al., 2012), hepatitis C virus p7 protein (Shrivastava et al., 2013, Farag et al., 2017), human immunodeficiency virus type 1 (HIV-1) vpu protein (Triantafilou, Ward et al., 2021), classical swine fever virus p7 protein (Lin et al., 2014) and the SARS CoV envelope protein (Nieto-Torres et al., 2014, Nieto-Torres et al., 2015), as well as SARS CoV 3a (Kanzawa et al., 2006, Chen et al., 2019, Siu et al., 2019).
Fig. 2.
Inflammasome and involvement of viroporins. (A) ASC, NLRP3 and Pro-caspase-1 interact via PYD and CARD domains to build the inflammasome. The inflammasome needs to be activated to change pro-caspase-1 into active caspase-1. (B) Two signals (DAMPs/ PAMPs) are needed to induce inflammatory reaction. The first signal activates Toll-like receptors (TLR), which induces synthesis of pro-IL-1β / pro-IL-18, which are converted into active IL-1β / IL-18 only in the presence of an inflammasome complex with active caspase-1. Pro-caspase-1 is transformed into active caspase-1 by a second independent signal (different DAMP or PAMP). Viroporin activity can act as this second signal by increasing cytosolic Ca2 + concentrations (E- or 3a activity in the ER), or by lowering the cytosolic pH due to ion channel activity in the acidic compartments like trans Golgi (E protein and 3a), lysosomes (3a only) and late endosomes (3a only).
3.4. Viroporins and their role in the virus life cycle
Enveloped viruses enter the host cell via endocytosis, and are then released into the cytosol directly at the plasma membrane, or follow the endocytic pathway and enter the cytoplasm through early endosomes or late endosomes depending on their signals to trigger and support fusion (White and Whittaker, 2016). Transcription takes place in the cytosol (SARS CoV, HCV, RSV, PV) or in the nucleus after entering through the nuclear core complex (IAV, HIV-1). Although some viroporins participate in different steps of the viral life cycle, such as cell entry and replication (Castano-Rodriguez et al., 2018) , their main activity concentrates on virion assembly and release from infected cells as shown by the fact that viruses lacking viroporins that fail to accomplish proper assembly and release (Lu et al., 2006, Steinmann et al., 2007, Ruiz et al., 2010). Release of enveloped viruses from infected host cells follows a general mechanism which comprises several stages: (1) preparation and assembly of virion (2) budding, (3) scission and (4) viral release.
Assembly depends on expression, transport, and accumulation of viral structural proteins to cellular membranes where budding occurs. Depending on the specific virus, the zone of budding forms at the plasma membrane (IAV, HIV-1) or on cytosolic organelle membranes, for example the ERGIC (SARS CoV) or ER (HCV), or inside the infected host cell (Kien et al., 2013). Proteins, that are essentially involved in budding-zone formation are HA and neuraminidase for IAV (Chen et al., 2007, Chen et al., 2007), M protein for CoVs, (de Haan et al., 1998, de Haan et al., 2000), the core protein in HCV (Hourioux et al., 2007) and gag for HIV-1 (Gheysen et al., 1989). The next stage in viral release is the formation of the viral bud. Even though budding is mainly triggered by molecular interactions of viral components interacting with core viral proteins (M1 for IAV, N protein for SARS CoV, gag for HIV and core protein for HCV) (Chen et al., 2007, Chen et al., 2007) membrane flexibility is necessary to enable positive membrane curvature. Integration of viroporins inside the appropriate membrane play an important role in this process leading to the dissipation of membrane potential. It has been suggested that M2 protein from IAV could be responsible for the curvature of cellular membranes (Rossman et al., 2010). Next stage in viral release comprises bud closure and scission of the viral envelope from the cellular membrane (Chen et al., 2007, Chen et al., 2007). It is assumed that viroporins are involved in building the negative membrane curvature necessary for closure preceding scission and release (Fig. 1B). Best studied viroporin in this context is the M2 protein of IAV (Rossman et al., 2010). Some viruses like HIV-1 (Garrus et al., 2001), HCV (Ariumi et al., 2011) and herpes simplex virus type 1 (Pawliczek and Crump, 2009), but not IAV (Rossman et al., 2010), use the cellular endosomal sorting complex required for transport (ESCRT) in viral release. Specific cellular receptors or other cellular restricting factors are proposed to help with exocytosis at the end phase of viral release.
4. Viroporin activity assays
One approach towards treatment of virus-induced diseases is the identification of highly specific inhibitors. This can be achieved by testing activity against the entire virus using virus-related assays such as plaque-forming assays. A disadvantage of whole-virus assays is that the target protein itself is not detected, furthermore, high levels of biosafety are required for in vivo studies. If the search is focused on specific proteins – such as viroporins – recombinant expression of the gene of interest is another option. Here, no information on viral activity in vivo is obtained, but the specific target protein can be studied in isolation. In this case, one needs a good, fast, and reliable testing system to guarantee high specificity for a safe preliminary selection of drug candidates. A number of assays for the study of viroporin activity have been described in the literature, each of them having their strengths and limitations. (I) Assays following recombinant expression of viroporins in mammalian cellular systems were applied to test for viroporin activity and the action of inhibitors. (II) A second approach is bacterial overexpression of the target protein (for example in E. coli), followed by purification and incorporation into artificial membranes to study channel function, conductance, and ion selectivity as well as viroporin inhibition. (III) The third approach entails recombinant expression of the target protein in mammalian cell lines followed by direct electrophysiological recording on viroporin expressing cells.
4.1. Cellular hemadsorption assay
The appearance of hemadsorption is dependent on the attachment of red blood cells (RBC) to the surface of cells infected with enveloped, hemagglutinin (HA) – producing viruses, such as influenza, measles, or mumps. Originally developed to detect HA-virus interaction, the assay was adapted for viroporins by using an intracellular HA, which only gets translocated to the cell surface when the active viroporin disrupts vesicular pH. On this base an activity assay was developed (Shelokov et al., 1958, Sakaguchi et al., 1996) and used for activity testing of the IAV M2 channel (Medeiros et al., 2001) and the HCV p7 protein (Griffin et al., 2004, Breitinger et al., 2016). Since the recombinant expression of viroporins induces apoptosis in the host cells, transfected cells – i.e. those that bind RBCs and generate the recorded signal are weakened or destroyed. This, together with the need of numerous washing steps render the assay challenging and ineffective (Breitinger et al., 2021). Washing also breaks weak erythrocyte binding.
4.2. Fluorescence based cellular assays
IAV M2 and HCV p7 were described as mainly H+ selective channels. They target host compartments including the ER, Golgi, mitochondria and lysosomes (Nieva et al., 2012). By integrating viroporins into the membranes of acidic organelles, channel activity results in de-acidification, thereby equilibrating the pH of the cytosol and the acidic compartments. Commercially available acid-sensitive fluorescent dyes show strong fluorescence in acidic environments which is reduced upon increasing pH. Application of these dyes to untransfected control cells will give rise to emission of fluorescence from the intracellular acidic compartments. Viroporin transfected test cells will have the pH of these compartments shunted, leading to a reduced fluorescence signal. Fluorescence based cellular assays were used for HCV p7 channels after recombinant expression on different cells (Wozniak et al., 2010, Breitinger et al., 2016, Breitinger et al., 2021). The test can be used with living cells as well as after fixation of cells with paraformaldehyde (Wozniak et al., 2010).
4.3. Cell viability assays
Recently, a simple cell viability assay was introduced for activity testing on HEK293 cells expressing viroporins. Recombinant expression of active viroporin will generally weaken cells and lead to an increase in the fraction of dead cells by inducing apoptosis (Aweya et al., 2013). This overall loss of living cells can be detected using cellular viability assays. The method has been adapted to assay the viroporin activity (reduced cell viability) as well as function of viroporin inhibitors (cell viability restored) and can also identify cytotoxicity of potential viroporin inhibitors (reduced viability in control culture) (Breitinger et al., 2021, Breitinger et al., 2021).
4.4. Assays using viroporin protein incorporated into artificial lipid membranes
4.4.1. Preparation of lipid vesicles
Commercial membrane-forming lipids such as Phosphatidic acid (PA), phosphatidylcholine acid (PC) and phosphatidylethanolamine are commonly used. Lipids are dried under argon to form a film, then buffers are used to re-hydrate the lipids and form (nano)vesicles. Fluorescent dyes of varying molecular weights can be incorporated into the vesicles for later analysis. Uni-lamellar liposomes can be produced by extrusion through appropriate membrane filters. Liposomes need to be purified by centrifugation and finally resuspended in appropriate buffer (StGelais et al., 2007).
4.4.2. Liposome dye-release assay
Permeability induced by purified, tagged viroporins can be assessed by incubating the target protein with liposomes. Loss of vesicular fluorescence indicates efflux of dye through viroporin activity. Real-time measurement of dye release is performed, using the bee venom peptide melittin as a positive control. Rates of dye efflux, expressed as change in arbitrary fluorescence units per unit time are used to calculate the degree of permeabilization. For inhibition assays, rates of efflux are determined in the presence of addition antivirals compounds (StGelais et al., 2007). Using liposome dye assays several viroporin were shown to conduct small molecules, e.g. the antibiotic hygromycin B, as well as inorganic cations (Lama and Carrasco, 1992, Aldabe et al., 1996, Liao et al., 2006, Carter et al., 2010). The method was used for a screen of potential HCV p7 small-molecule inhibitors (Gervais et al., 2011).
4.4.3. Electrophysiological measurements on artificial membranes
Solvent-free planar lipid bilayers are prepared following specific protocols and sandwiched between two half glass cells. In an example procedure (Montserret et al., 2010), phosphatidylcholine, dissolved in hexane (0.5%), is spread on the top of an electrolyte solution (500 mM KCl, 10 mM HEPES, pH 7.4) in both compartments. Bilayer formation is achieved by lowering and raising the level in one or both compartments and monitoring capacity responses. A transmembrane voltage clamp is applied through an electrode in the cis-side, with grounding on the trans-side. Then, purified peptides are added in nM concentrations. Ion selectivity (Hodgkin and Huxley, 1947) and conductance can be determined using buffers of different concentrations of permeant ions on the cis- and trans side. The method was used for the electrophysiological characterization of several viroporins (Wilson et al., 2004, Torres et al., 2007, Whitfield et al., 2011, Pham et al., 2017, Dey et al., 2019).
4.5. Electrophysiology measurements on recombinantly expressing mammalian cells
Two sets of experiments have been used: (I) measurement of current traces from single voltage-clamped cells in the whole-cell mode that are lifted and positioned in front of a perfusion device. Inhibitor, or buffers of varying pH conditions are then applied directly onto the cell (Chizhmakov et al., 2003, Breitinger et al., 2016, Breitinger et al., 2021). (II) The second approach is to apply a current-voltage ramp to cells in the whole-cell configuration. In this scenario, the cell can be studied without lifting. Since viroporin induced currents are very small and cells usually are weakened due to cell damage induced by viroporin activity, single trace measurements are challenging, especially since the procedure of detaching the cell and lifting it to the delivery system takes time and stresses the cell further, often resulting in increasing leak currents that overlay the small signal. In case of voltage ramps, currents can be recorded directly after patching and transferring cells into whole cell configuration. Even then, HEK293 cell currents under identical conditions can have a large cell to cell variability (Premkumar et al., 2004, Schwarz et al., 2014, Breitinger et al., 2021, Breitinger et al., 2021, Cabrera-Garcia et al., 2021). Thus, the method requires signals from many cells under each testing condition to be averaged since no direct comparison on single-cell basis is possible. If carefully done, these methods allow the direct study of viroporin channel behaviour under in-vivo like conditions.
5. Coronavirus families
The Coronaviridae family comprises four groups, named α-, β-, γ-, and δ- Coronavirus (CoV). α- and β-CoVs are mainly found in mammals, while γ-CoV and δ-CoV are found in birds. Recombination events have been described in several coronaviruses infecting humans and animals (Vakulenko et al., 2021). Phylogenetic studies suggest that bats are the major source for α-CoV and β-CoV infection of other mammals. Analysis of bat CoVs showed that α-CoV change hosts more frequently than β-CoV. However, β-CoV is more relevant as a source of new pathogenic human viruses, including SARS-CoV-2. Wild birds are the source for highly diversified γ- and δ-CoV (Vakulenko et al., 2021). Table 3 gives some examples, including all coronavirus species discussed here.
Table 3.
SARS-CoV-2 related coronaviruses from different groups and species.
| Group | Name | Species |
|---|---|---|
| alpha | Canine CoV (CCoV) | canine |
| Human CoV 229E (HCoV-229E) | infects humans and bats | |
| beta | Bat SARS-like CoV-WIV1 | Rhinolophus sinicus, Kunming, Yunnan |
| Bat SARS-like CoV YNLF_31C | Rhinolophus ferrumequinum, Lufeng, Yunnan | |
| Bat SARS CoV HKU3 | Rhinolophus sinicus, Hong Kong | |
| Bat SARS related CoV BtRs-HuB2013 | Rhinolophus sinicus, China | |
| Murine hepatitis virus (MHV) | murine | |
| Human CoV HKU1 (HCoV-HKU1) | human, Origin Hong Kong | |
| SARS-CoV | human | |
| MERS-CoV | human | |
| gamma | Avian infectious bronchitis virus (IBV) | avian |
| Turkey CoV (TCoV) | avian | |
| delta | bulbul CoV HKU11 (BuCoV HKU11) | avian: family Pycnonotidae |
| thrush COV12 | avian: family Turdidae |
6. Viroporins from SARS CoV and SARS CoV-2: Sequence comparisons
6.1. Envelope protein
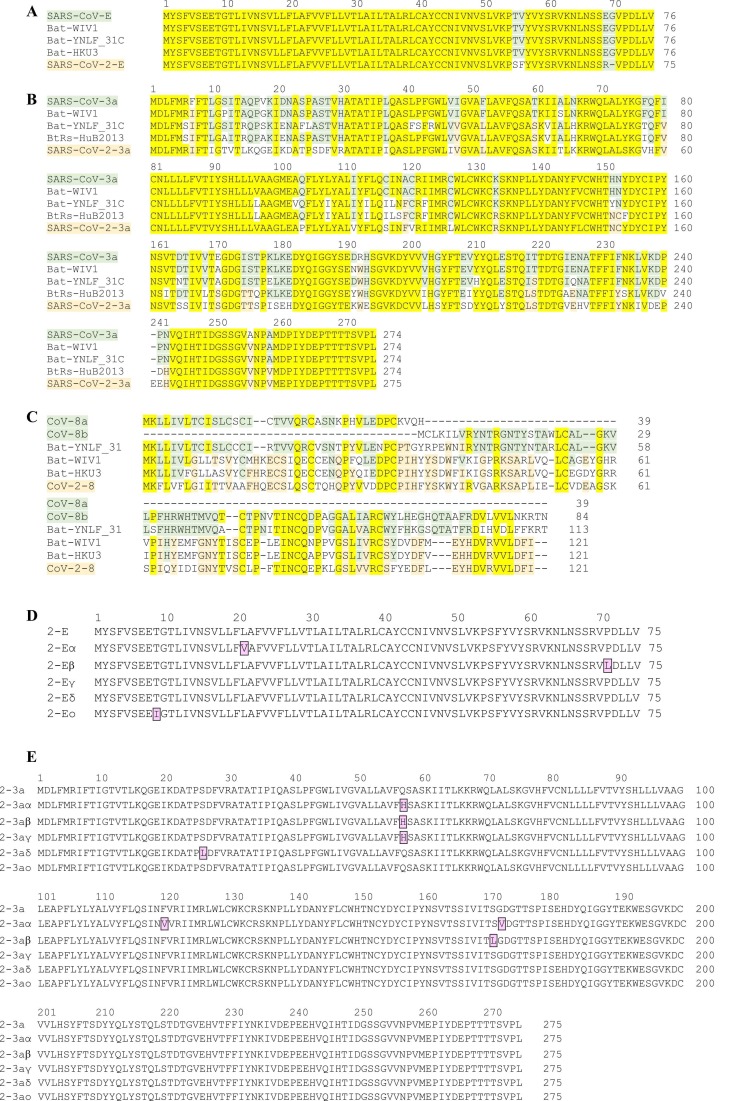
E proteins are well conserved within the different groups of CoVs. In SARS CoV, the 75–76 residue containing ion channel mediates viral assembly and release. When comparing the E protein of SARS CoV to that of SARS-CoV-2, only four amino acid exchanges at the C-terminal domain are found. At position 55/56 threonine and valine in SARS CoV are exchanged to serine and phenylalanine in CoV-2, while at position 69/70 glutamate and glycine in CoV are replaced by a single arginine residue in the 2019 SARS-CoV-2 variant ( Figs. 3A, 4A). Among the new SARS-CoV-2 variants of concern, only very few single mutations were described (Figs. 3D, 4A). The exchange L21V in the α variant as well as T9I in the recent ο strain may affect channel function, while the exchange P71L observed in the β variant is directly preceding the PALS1-binding motif 72DLLV75 (FIG: 3D) and may have an effect on intracellular interactions mediated by the E protein.
Fig. 3.
Viroporin sequence alignments. (A-C) Comparison of viroporin sequences from human (SARS CoV, CoV-2) and bat species CoVs (compare Table 2). Conserved residues are highlighted in yellow, differences between human SARS-CoV and SARS-CoV-2 are highlighted in light green, and differences between human and bat CoVs are highlighted in light orange color. (A) The E protein is 100% conserved between human SARS-CoV and bat variants; SARS-CoV-2 has only 4 amino acid exchanges in the C-terminal region (94.7% identity). (B) ORF3a is less conserved, with CoV 3a has highest similarity to 'Bat-WIV1' (95.3% identity) and ' Bat-YNLF_31C' (86.1% identity), while SARS-CoV-2 has highest similarity to 'BtRs-HuB2013' (75.3% identity). SARS-CoV and SARS-CoV-2 show low similarities between each other and to the bat variants. (C) SARS CoV ORF8a and ORF8b show highest similarity to 'Bat-YNLF_31' (82.9% identity and 76.9%, respectively) while CoV-2 ORF8 has highest similarity with Bat-WIV1 (57.9% identity) and Bat-HKU3 (57.0% identity). (D,E) Sequence comparison of new variants of SARS-CoV-2 that were classified by the WHO as ‘variants of concern’ (SARS-CoV-2 α, β, γ, δ, ο). New mutations are highlighted in purple. Among the SARS-CoV-2 variants, E protein (D) and 3a protein (E) show similar tendency towards developing new mutations. Three mutations were detected in E protein variants on a length of 75 amino acids, while six new mutations are described for 3a variants in a protein of 275 amino acids. Relative to length of the protein, the probability of mutation is even higher in the E protein than in 3a.
Fig. 4.
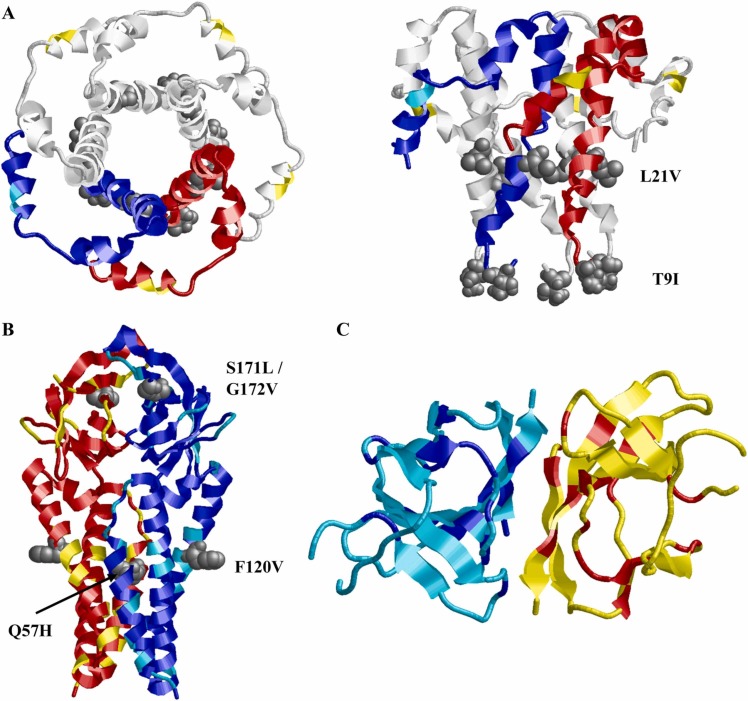
Structures of SARS-CoV-2 Viroporins. Subunits are colored in red/blue and residues that are variable between SARS-CoV, bat species CoVs and SARS-CoV-2 are highlighted in yellow/cyan; the sites of mutation in variants of concern of SARS-CoV-2 are indicated in grey colour and spacefill and indicated in the structures. See Fig. 3 for the respective sequence alignments and Table 1 for literature references. (A) E-protein (based on pdb 5 ×29). Left panel: top view, right panel: side view. Amino acid exchanges between SARS-CoV and SARS-CoV-2 are exclusively located in the C-terminal domain, distant from the channel pore. Mutations found in new strains of SARS-CoV-2 are located in the channel pore region and may potentially affect viroporin function. For clarity, the red/blue subunit coloring scheme was only used for two of the five subunits. Note that exchange P71L is located outside the resolved structure of the protein. (B) 3a protein (based on pdb 6xdc), two subunits (red/blue) are shown. Approximately 26% of residues are exchanged between SARS-CoV and SARS-CoV-2 (yellow/cyan), exchanges found in variants of concern of SARS-CoV-2 are located at or near the pore domains and might affect channel function. Exchange S26L is located outside the resolved structure of the protein. (C) ORF 8 protein (based on pdb 7jtl). Two subunits (red/blue) are shown. The structure shows no similarity correlation to known channel-forming proteins. The low number of conserved (red/blue) compared to nonconserved (yellow/cyan) residues reflects that ORF 8 protein sequences differ significantly between SARS-CoV, bat CoVs, and SARS-CoV-2.
6.2. ORF3a
Among all open reading frames (ORFs) of SARS CoV, ORF3a is the largest; it encodes a transmembrane protein – the 3a protein – of 274 amino acids. While there are large differences in the sequences between SARS-CoV and SARS-CoV-2 (identity of 72.4%) (Figs. 3B, 4B), only minor exchanges are noted among the new, highly infectious SARS-CoV-2 variants (Figs. 3E, 4B). The new variants of concern contain the following mutations: α: Q57H, F120V, G172V; β: Q57H, S171L; γ: Q57H; δ: S26L (Fig. 3E). Of these exchanges, Q→H results in a moderate local increase in basicity, while G→V increases side chain size. Replacement S→L will alter side chain size and polarity. It remains to be seen whether these exchanges result in a pronounced change of protein function and contribution to the viral life cycle.
6.3. ORF8
The intact ORF8 – present in animal (bat) and some early human isolates – encodes a 122-amino-acid polypeptide, 8ab(+), while human SARS-CoV has a deletion of 29 nucleotides, within ORF8a that results in the generation of two proteins, ORF8a and ORF8b, polypeptides of 39 and 84 residues, respectively, that appear without function (Oostra et al., 2007, Muth et al., 2018). It has been shown that the 29 bp deletion causes a dramatic reduction of replication of bat SARS-CoV (Oostra et al., 2007, Muth et al., 2018). Recent SARS-CoV-2 variants possess again the complete protein from the intact ORF8. The ORF8 sequence is changing fast and uncontrolled, leading to a sequence identity of less than 20% between SARS-CoV and SARS-CoV-2, this may be related to some unknown viral infection strategies (Neches et al., 2021). The small ORF8a protein – only present in SARS-CoV – contains a single transmembrane domain which was shown to form active ion channels in artificial lipid bilayers (Chen et al., 2011). ORF8b appeared not to be expressed in SARS-CoV-infected cells. Only after placing the gene immediately behind the T7 promoter, expression was observed, resulting in a soluble monomeric protein found in the cytoplasm, which was highly unstable and rapidly degraded (Oostra et al., 2007). ORF8 of SARS-CoV-2 is a 121-amino acid protein consisting of an N-terminal signal sequence for endoplasmic reticulum import. In contrast to the envelope protein E, ORF3a and ORF8a, ORF8 was lacking any membrane spanning α helices in a structure determined by X-ray crystallography ( Table 2, Figs. 3C, 4C). Instead, the structure revealed that the ORF8 core consisted of two antiparallel β-sheets (Flower, Buffalo et al., 2021), which questions the role of ORF8 and derived proteins as ion channel proteins. Indeed, it has been shown that absence of ORF8a, but not of E- and ORF3a proteins is tolerated in SARS-CoV (Castano-Rodriguez et al., 2018).
Table 2.
Structures of SARS-CoV-2 viroporins.
| Year | Protein | Technique | Residues resolved | PDB code (resolution) | Description | Ref |
|---|---|---|---|---|---|---|
| 2009 | S1-E | Solution NMR in DPC micelles | E8 – R38 | n/a | regular α-helices that form a pentameric left-handed parallel bundle | (Pervushin et al.. 2009) |
| 2014 | S1-E | NMR of ETR in SDS micelles | E8-L65 * | 2MM4 | longer α-helix (Res 15–45) crossing the TM, connected to a shorter C-terminal α-helix (Res 55–65) by a flexible linker (Res 46–54) | (Li et al., 2014) |
| 2018 | S1-E |
NMR in LMPG micelles | E8-L65 * | 5 × 29 | the ETR pentamer is a right- handed α-helical bundle where the C-terminal tails coil around each other (modeling study) | (Surya et al., 2018) |
| 2020 | S1-E or S2-E |
solid-state NMR spectroscopy in ERGIC-mimetic lipid bilayers | E8 – R38 | 7K3G | well-defined, tight 5-helix bundle for Res V14-L34; N-terminal part (Res E8-I13) is dynamic at high temperature but is mostly α-helical, the C-terminal segment (Res T35-R38) is conformationally plastic | (Mandala et al., 2020) |
| 2021 | S2–3a | Cryo-EM in lipid nanodiscs | S40 - D238 | 6XDC (2.9 Å) 7KJR (2.1 Å) |
The TM region connects to the CD through a turn-helix-turn motif following TM3. Each protomer chain forms a pair of opposing β-sheets packed against one another in an eight stranded β-sandwich | (Kern et al., 2021) |
| 2020 | S2-ORF8 |
X-ray crystallography | Q18 – I121 | 7JTL (2.04 Å) | ORF8 adopts an Ig-like fold, it forms a disulfide-linked homodimer containing an intermolecular parallel β-sheet | (Flower et al., 2021) |
Table 2: Published high-resolution structures of SARS CoV viroporins. *All three Cys residues were exchanged with Ala; DPC = dodecyl-phosphatidylcholine; LMPG = lyso-myristoyl phosphatidylglycerol; Res = residues; ETR = truncated E protein; ETM = E protein transmembrane; ERGIC-mimetic lipid bilayer = mixture of phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, phosphatidylserine and cholesterol; CD = cytosolic domain; S1-E = SARS CoV E protein; S2-E = SARS CoV-2 E protein.
It is noteworthy that there are considerable differences between the sequence of the ORF3a proteins of SARS-CoV and SARS-CoV-2 (identity 72.4%), while the E-protein between the two viruses is highly conserved (94.7% identity). Within the new variants of SARS-CoV-2, both viroporins, E and ORF3a, show only minimal variation, on the level of single amino acid exchanges. The reason for this observation is in need of more investigation. It appears that the E protein sequence was already optimized in the earlier stages of evolution of SARS-CoV, while the 3a protein changed considerably from SARS-CoV to SARS-CoV-2. The sequence of the 3a protein in SARS-CoV-2 appears to be improved, since only minor changes are observed in the newer variants of the virus ( Table 3).
7. Function and characteristics of SARS CoV viroporins
7.1. Envelope protein (E)
The first experiments on the E protein of different coronaviruses confirmed that its expression in mammalian cells alters membrane permeability (Liao et al., 2004, Liao et al., 2006). Furthermore, the E protein was found to be located mainly in the endoplasmic reticulum (ER) and Golgi apparatus where its ion channel activity increases the pH of the affected compartments (Nal et al., 2005, Liao et al., 2006, Nieto-Torres et al., 2011, Cabrera-Garcia et al., 2021). Before E protein structures were available, computational secondary structure predictions of the β- and γ-CoV E proteins suggested that a conserved proline residue at position 54, near the C-terminus was centered in a β-coil-β motif (Cohen et al., 2011). This motif was considered as a putative Golgi-complex targeting signal as exchange of this proline with alanine was sufficient to disrupt the localization of a mutant chimeric protein to the Golgi complex, directing it to the plasma membrane instead (Cohen et al., 2011). A reciprocal chimera consisting of the N-terminal and hydrophobic domains from SARS-CoV E fused to the C-terminal tail of vesicular stomatitis virus glycoprotein (VSV-G) was expressed and analyzed by immunofluorescence microscopy, indicating its localization inside the Golgi complex. This suggests that a second Golgi localization tag might be present within the N-terminal region of the E protein (Cohen et al., 2011).
7.1.1. Ion channel characteristics
The topology of the E protein was studied in microsomal membranes and mammalian cells, consistently indicating a single-spanning membrane protein. In the ER the N-terminus is being translocated across the membrane to the luminal side, while the C-terminus remains exposed to the cytoplasmic side of the ER (Duart et al., 2020).
When the ion channel activity of E protein was first studied, it was described as a cation-selective ion channel (Wilson et al., 2004) composed of five subunits (Fig. 4A). Further studies confirmed the E protein to be a functional ion channel in artificial membranes and in mammalian cells after recombinant expression (Pervushin et al., 2009, Breitinger et al., 2021, Xia et al., 2021). Xia et al. investigated the cation selectivity in more detail using purified SARS-CoV-2 E channels, reconstituted in bilayer lipid membranes. Studies of the reversal potentials in solutions with varying concentrations of different cations and Cl– as permeant ions showed that the channel is permeable to K+ and Na+ with similar efficiency. The channel is also permeable to divalent cations Ca2+ and Mg2+, yet their permeability is lower than that of the monovalent ions. The permeability ratio shows a distinct selectivity pattern that is also known for classic voltage-dependent channels (Xia et al., 2021). The Ca2+ permeability of the E protein channel was further studied on the E proteins of both, SARS-CoV (Nieto-Torres, et al., 2015) and SARS-CoV-2 (Verdia-Baguena et al., 2021). Ca2+ permeability is especially relevant, as Ca2+ is an intracellular messenger, and calcium transport is a main trigger for the activation of the NLRP3 inflammasome, leading to IL-1β overproduction (Nieto-Torres et al., 2015).
Notably, while studies of SARS-CoV-2 E protein reconstituted in bilayer lipid membranes had shown an ion preference of monovalent cations over divalent cations (Xia et al., 2021), studies of the E protein of SARS-CoV-2, reconstituted in planar ERGIC-mimic membranes, which include lipids with intrinsic negative curvature, showed enhanced permeability of pure Ca2+ or K+ solutions, and a preferential transport of Ca2+ in mixed KCl-CaCl2 solutions (Verdia-Baguena et al., 2021). Since the putative pore region of the E-protein channel is identical in SARS-CoV and SARS-CoV-2, a likely rationalization is that the ion channel properties of the E protein depend on charge and composition of the lipid membrane as well as on charges on the protein. This was confirmed in a study of SARS-CoV E protein, reconstituted in neutral or negatively charged artificial membranes (Verdia-Baguena et al., 2013). In non-charged phosphatidylcholine membranes, the E protein acts as a non-selective neutral pore, and conductance changes linearly with bulk solution conductivity. In negatively charged phosphatidylserine membranes the influence of the membrane charge became apparent. This was taken as demonstration that even if the conformation of the protein channel is unchanged, protein and membrane charge both determine channel conductance (Verdia-Baguena et al., 2013).
7.1.2. Inhibitors of the E-protein channel
E protein channel activity, like that of other viroporins, can be blocked by various channel blockers or inhibitors. Hexamethylene amiloride (HMA) was first described to inhibit currents induced in lipid bilayers by the E proteins of HCoV-229E and mouse hepatitis virus (MHV) (Wilson et al., 2006). Plaque assays verified HMA action by inhibiting replication of the parent CoVs in cultured cells. Consistent with a specific activity against the E protein, HMA had no antiviral effect on a recombinant MHV that had the entire coding region of E protein deleted (Wilson et al., 2006). Further electrophysiology studies of recombinantly expressed E protein in mammalian cells confirmed the activity of HMA as E protein channel inhibitor (Park et al., 2003, Pervushin, et al., 2009, Breitinger, Ali et al., 2021). NMR studies on the transmembrane domain of E protein (ETM) in dodecyl-phosphatidylcholine micelles identified two binding sites for HMA inside the lumen of the channel, one near the N-terminal, the other one close to the C-terminal end of the pore (Pervushin et al., 2009). A later NMR study performed in lipid bilayers mimicking the ERGIC membrane, confirmed HMA binding to two sites inside the channel pore, with the dominant site near the polar amino-terminal end. The site at the C-terminal part of the pore was affected by acidic pH (Mandala et al., 2020), which agrees with the expected influence of protein and membrane charge on the channel pore (Verdia-Baguena et al., 2013). Amantadine, another classic viroporin inhibitor, was investigated after reconstitution into lipid bilayers and showed only moderate inhibitory activity on E protein of SARS-CoV (Torres et al., 2007). In a further study, amantadine and rimantadine were investigated for their inhibitory ability after recombinant E protein expression in HEK293 cells. Both substances reduced E protein activity in cell viability assay and electrophysiological measurements, with rimantadine inhibition being five-fold more potent than that of amantadine (Breitinger et al., 2021). Genistein and Kaempferol, two flavonoids, were described as viroporin inhibitors before (Sauter et al., 2014, Schwarz et al., 2014). Genistein inhibited the HIV-1 viral protein U (vpu) (Sauter et al., 2014) while Kaempferol derivatives were shown to act as inhibitor against SARS CoV ORF 3a protein (Schwarz et al., 2014) when both ion channels were expressed in Xenopus oocytes and the activity assessed using electrophysiological techniques. When different flavonoids were tested as putative E protein antagonists, Epigallocatechin and quercetin exhibited inhibitory properties at effective concentrations similar to that of rimantadine as shown for SARS-CoV E protein-expressing HEK293 cells using both, cell viability assays and patch-clamp electrophysiological analysis (Breitinger et al., 2021).
7.1.3. Search for new inhibitors using molecular docking
The flavonoid rutin (quercetin-3-O-rutinoside) was identified as putative binding partner of SARS-CoV-2 E protein in a molecular docking study of membrane, envelope and nucleocapsid proteins of the SARS-CoV-2 (Bhowmik et al., 2020). Further inhibitors, that were identified using molecular docking techniques are Tretinoin (Dey et al., 2020); Sinapic Acid showed an in vitro inhibition of -SARS CoV-2 activity, was further studied by molecular dynamic simulations which revealed the E-protein as its major pharmacological target (Orfali et al., 2021). A study based on computational simulation approaches identified Belachinal, Macaflavanone E, and Vibsanol B as putative E protein inhibitors. According to this study, binding of these compounds reduces the random motion of the SARS-CoV-2 E protein, which results in inhibition of its activity (Gupta et al., 2021).
7.1.4. Role of the E protein in assembly, budding and virus release
Despite its role in efficient production and release of new virions, the E protein is incorporated at low levels within the viral envelope. SARS CoVs bud inside the lumen of the ERGIC followed by transport and release through the secretory pathway (Goldsmith et al., 2004). Co-expression experiments in mammalian cells producing virus-like-particles (VLPs) gave dissenting results. While co-expression of SARS-CoV E protein with M and N proteins induced efficient production and release of VLPs, co-expression of E and M without N protein gave very low VLP levels (Siu, Yuen et al., 2019). Assembly of MHV (murine β-CoV) VLPs was independent of N protein when M and E proteins were co-expressed in mammalian OST7–1 cells (Vennema et al., 1996). In contrast, when HEK293 cells were used, co-expression of SARS-CoV M and N structural proteins was sufficient to trigger budding of VLPs even in absence of the E protein (Huang et al., 2004, Hatakeyama et al., 2008). However, secretion of these VLPs was inefficient (Hatakeyama et al., 2008). A recombinant SARS-CoV lacking the E protein showed reduced virus expression and the process of virus release was less efficient than for the wild-type analogue (DeDiego et al., 2007). Budding and exocytosis of virus requires bending of membranes to form vesicles. A potential role of the E protein in inducing membrane curvature has been proposed but still needs further investigation. The SARS-CoV E protein was suggested to build a transmembrane helical hairpin after incorporation into dimyristoylphosphocholine (DMPC) lipid vesicles which was assumed to deform lipid bilayers leading to an increase in their curvature (Arbely et al., 2004). However, this helical transmembrane hairpin has not been confirmed in NMR structures (Pervushin et al., 2009, Li et al., 2014, Surya et al., 2018).
7.1.5. Interaction of E protein with other virus or host proteins
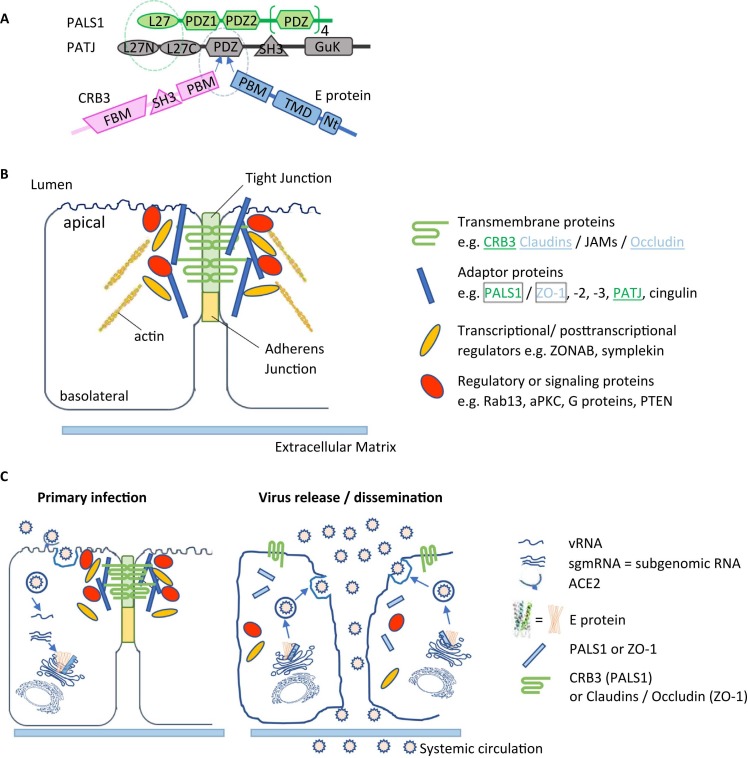
Several interactions between SARS CoV E protein and other virus proteins like membrane (M), non-structural protein 3 (nsp3), nucleocapsid (N) and spike (S) protein have been proposed to be important for different steps in the virus life cycle (Chen et al., 2009, Alvarez et al., 2010, Boson et al., 2021). In addition to communication between viral proteins, several interactions directed towards host proteins are known ( Fig. 5). Best studied interaction site is the PDZ-binding motif (PBM), comprised of the last four carboxy-terminal amino acids (DLLV) of the E protein. PDZ domains are common protein interaction modules recognizing short amino acid motifs at the C-termini of target proteins (Caillet-Saguy et al., 2021). One of the main interactors of the E protein of SARS-CoV-2 is the PDZ containing protein PALS1, a key component of the Crumbs polarity complex (Protein associated with Lin-7 One) (Teoh et al., 2010, Toto et al., 2020, Chai et al., 2021, Javorsky et al., 2021). PALS1 is a tight junction-associated protein that plays a crucial role in establishing and maintaining epithelial polarity in various organisms. It was suggested that the interaction of PALS1 with the SARS-CoV E protein is involved in the disruption of the lung epithelium in SARS patients (Teoh et al., 2010) (Fig. 5). Equilibrium and kinetic binding experiments comparing the affinity of E peptides from SARS-CoV and SARS-CoV-2 for the PDZ domain of PALS1 detected an increased affinity of SARS CoV-2 E protein that could explain the increased virulence of SARS-CoV-2 in relation to CoV (Toto et al., 2020).
Fig. 5.
Cellular binding partners and interactions mediated by viroporins. (A) Interaction at the tight junction: PALS1 and PATJ (two adaptor proteins at the tight junction) interact through their L27 domain. CRB3 and - in case of SARS COV infection - the E protein bind to the PALS1 PDZ domain through their PDZ binding motif (PBM). (B) Composition of lung epithelial tight junctions (TJs). Transmembrane, adaptor and regulatory proteins as well as transcriptional and post-transcriptional regulators are involved in TJ maintenance. Some examples of each group are listed. Many tight junction proteins interact with actin filaments. PALS1 interacts with the tight junction transmembrane protein CRB3 and a second adaptor protein PATJ (green), and also interacts with SARS E protein. ZO-1, a further adaptor protein at the TJ was shown to interact with the S2-E protein, its physiological binding partners are the transmembrane proteins claudins and occludin. (C) Model of the potential loss of the TJ interactions and the consequences on membrane polarity of alveolar epithelial cells after SARS CoV infection. The left scheme illustrates the primary infection of alveolar epithelial cells by incoming viruses. After translation, S, M, N and E proteins target for the ERGIC compartment for virus assembling, budding and exocytosis. In this process, the E protein could bind to PALS1 and ZO-1, hindering them from trafficking to TJ. The right scheme describes possible virus dissemination after TJ disruption. Loss of the TJ transmembrane proteins PALS1 and ZO-1 lead to a progressive decomposition of the TJ and a sub-sequent leakage between neighbouring epithelial cells. As a further consequence, barrier function is lost, and SARS CoV virions can leak into underlying tissues by systemic circulate-on. This process could be the cause of severe alveolar damages observed in SARS CoV-infected patients (Teoh et al., 2010)
The C-terminal sequences of SARS-CoV and SARS-CoV-2 E proteins are notably different (Figs. 3A, 4A), as there is an inversion of charge in position 70 (E for R). Crystal structures of the PDZ – E-protein-DLLV motif interaction of the E proteins of SARS-CoV and SARS-CoV-2 (Javorsky et al., 2021) suggest a subtle difference in the vicinity of the DLLV motif of the two E proteins that might give rise to the better binding of the DLLV motif to PALS1 that was observed for SARS-CoV-2.
Additional interactions of the E protein PBM with the PDZ-domain-2 of the host tight junction protein Zona Occludens-1 (ZO1) were discovered and could contribute to the epithelial barrier damage and pathogenesis (Shepley-McTaggart et al., 2021). An association of the cellular protein syntenin to the PBM of the SARS CoV E protein leads to activation of p38 MAPK (the p38 pathway is the third major signaling cassette of the mitogen-activated protein kinase signaling pathway) and the subsequent overexpression of inflammatory cytokines (Jimenez-Guardeno et al., 2014).
Toll-like receptors (TLRs) are mediators of inflammatory responses and cytokine release. It was shown that infection with SARS-CoV-2 resulted in an upregulation of Myd88, a TLR adaptor protein, and TLRs 1, 2, 4 5, and 8 (Zheng et al., 2021). Expression of these mediators increased with clinical severity of SARS (Zheng et al., 2021). Independent of the PBM – E protein interactions, TLR2 was identified as a direct E protein ligand, and blocking of the TLR2 pathway resulted in protection against SARS infections (Zheng et al., 2021). Expression of the SARS-CoV E protein induces apoptosis in transfected Jurkat T-cells which is inhibited by overexpressed antiapoptotic protein Bcl-x, and an interaction between SARS-CoV E protein and Bcl-xL was indeed demonstrated in vitro and in vivo (Yang et al., 2005). Two further SARS-CoV E protein interaction partners were identified by mass spectrometry: Na+/K+-ATPase, the main cellular ion pump involved in ion homeostasis control and stomatin, an ion channel regulator (Nieto-Torres et al., 2011). In the search for new physiological targets for antiviral drugs, a screen for interactions between 26 SARS-CoV-2 proteins and human host cells suggested a total of 6 interactions for the E protein (Gordon et al., 2020). These included Bromodomain and Extra-terminal domain (BET) proteins BRD2 and BRD4, which control the expression of angiotensin converting enzyme 2 (ACE2) that represents the docking site for SARS CoVs on host cells. A second proposed interaction partner is the beta subunit of the adaptor-related protein complex (AP3B1), a protein involved in protein sorting in the late Golgi network. Other putative binding partners include the cyclophilin CWC27 as well as ZC3H18, involved in mRNA biogenesis, and SLC44A2 which regulates choline transport into mitochondria, thus affecting platelet aggregation exerting an influence on thrombosis (Gordon et al., 2020).
7.1.6. Posttranslational modification of the E protein
Post-translational modification of coronaviruses was reviewed in 2018 with consideration of different postulated membrane topologies. Minor N-linked glycosylation is described for residue N66 of the E protein in a membrane topology where the C-terminus is exposed to the luminal side of the ER (Fung and Liu, 2018). However, the widely accepted topology of the E protein, which is in agreement with NMR structures shows the N-terminus located at the luminal side, while the C-terminus – including N66 – is directed towards the cytosol, this membrane topology would not be amenable to glycosylation. The three cysteine residues C40, C43 and C44 in SARS-CoV E protein are modified by palmitoylation (Liao et al., 2006, Fung and Liu, 2018), which might play a role in its subcellular trafficking and association with lipid rafts. When plasmid DNAs encoding mouse hepatitis CoV (MHV) S, E, M, and N was introduced in HEK293 cells and compared to experiments where E protein residues C40, C44 and C47 were mutated to alanine, VLP formation was significantly reduced (Boscarino et al., 2008). Lopez et al. exchanged the same cysteine residues with alanine in MHV E protein which resulted in crippled virus morphology and reduced yields, as well as faster E protein degradation (Lopez et al., 2008). Taken together, palmitoylation of MHV E protein seems to stabilize the protein and its biological activity during assembly of mature virions. Surprisingly, in E and N protein co-expression experiments, palmitoylation of SARS-CoV E protein had no effect on its association with N protein or subsequent VLP production, and thus is not required for SARS-CoV assembly, and the contributions of E protein palmitoylation to virus packaging and VLP production appear to be independent of each other (Tseng et al., 2014). Molecular dynamics simulations of SARS CoV-2 E protein indicate that the stability of the palmitoylated E protein structure is increased, while loss of palmitoylation causes reduction of the pore radius, and even collapse of the pore”(Sun et al., 2021).
7.1.7. Immunization related studies
Deletion of the E protein in general represses virus growth in culture and abrogates virulence in animals due to its reduced inflammatory reaction. In a murine model, infection with mouse-adapted, mutated SARS-CoV where E protein function was repressed or deleted resulted in very mild or almost absent clinical symptoms while production of anti-inflammatory cytokines and T-cell responses were still robust (Regla-Nava et al., 2015). Several studies performed in mice concentrated on SARS-CoV-ΔE approaches with the goal to find successful vaccine candidates (Netland et al., 2010, Fett et al., 2013, Regla-Nava et al., 2015), and the E protein is considered a promising target for vaccine development.
7.2. ORF 3a protein
ORF 3a is the largest unique open reading frame in the SARS CoV genome, encoding a protein of 274 amino acids. The ORF 3a protein forms an ion channel (Lu et al., 2006), induces vesicle formation (Freundt et al., 2010) and modulates virus release (Lu et al., 2006). The 3a protein belongs among group 3 of viroporins (Fig. 1B) possessing three transmembrane domains with the N-terminus located at the luminal side of the ER and the C-terminal domain directed to the cytosol (Fig. 4B). After the E protein, the 3a protein of SARS-CoV was shown to be involved in membrane rearrangement (Yuan et al., 2005, Freundt et al., 2010) and acting as an ion channel (Lu et al., 2006). Expression of SARS-CoV 3a protein in Xenopus oocytes resulted in a homotetrameric complex, stabilized by disulfide bonds between subunits that conducted K+ ions in two-electrode voltage clamp measurements (Lu et al., 2006). Studies on ion selectivity of SARS-CoV-2 3a channels reconstituted in proteo-liposomes revealed nonselective cation channel activity with a permeability order of Ca2+ > K+ > Na+ > NMDG+ (Kern et al., 2021). Staining and 45Ca overlay studies on an E. coli expressed cytoplasmic domain of SARS-CoV 3a showed that calcium binding introduced a change in conformation which was detected using fluorescence spectroscopy and circular dichroism (Minakshi et al., 2014). Expression of the 3a protein in various in vitro systems indicates that it mainly localizes in the Golgi region (Padhan et al., 2007, Yuan et al., 2007), but was also detected in the ER (Law et al., 2005), late endosomes (Freundt et al., 2010, Yue et al., 2018, Miao et al., 2021), lysosomes (Yue et al., 2018, Zhang et al., 2021), and the trans-Golgi network (Yue et al., 2018).
To date, only few reports on inhibition of ORF 3a ion channel function have been published. Electrophysiological data on heterologously expressed 3a protein in Xenopus oocytes revealed inhibition of 3a-induced currents by Kaempferol derivatives (Schwarz et al., 2014) and Emodin (Schwarz et al., 2011). A molecular docking study on 23 compounds and subsequent experimental testing using fluorescence and UV-Vis spectroscopy identified chlorin, iron(III) protoporphyrin and protoporphyrin as a putative binding partners of ORF 3a (Lebedeva et al., 2021). In a drug repurposing approach, bacteria-based functional assays that measured growth retardation, K+ uptake, and change of cytoplasmic pH of E. coli that overexpressed the 3a protein of SARS-CoV-2 were tested (Tomar et al., 2021). Using these screens, a library of drugs approved for human use an approved human drugs was screened, identifying several potential channel blockers of the 3a channel, namely Capreomycin, Pentamidine, Spectinomycin, Kasugamycin, Plerixafor, Flumatinib, Litronesib, Darapladib, Floxuridine and Fludarabine (Tomar et al., 2021).
7.2.1. Apoptosis and autophagy
The general involvement of the SARS-CoV ORF3a (S1–3a) in the induction of pro-apoptotic reactions was described in several cell lines andshown to be dependent upon its channel function (Law et al., 2005, Chan et al., 2009, Freundt et al., 2010, Ren et al., 2020). Several approaches were followed to better explain the role of S1–3a in pro-apoptotic function. It was shown that the cytoplasmic domain of S1–3a activates the mitochondrial p38 MAP kinase death pathway, which could be repressed by a p38 MAPK inhibitor (Padhan et al., 2008). ER stress in general can cause modulation by different pathways of the unfolded protein response (UPR). Studies on S1–3a-expressing cells show that only one pathway is activated by 3a, namely the PKR-like ER kinase (PERK) pathway (Minakshi et al., 2009). Involvement of 3a protein in inflammatory responses was shown as the 3a protein contributes to assembly of the NLRP3 inflammasome (Chen et al., 2019). Oligomerization of 3a is mediated by Receptor Interacting Protein 3 (Rip3), which leads to lysosomal damage, caspase-1 activation, and subsequent necrotic cell death (Yue et al., 2018). Another aspect of 3a-induced inhibition of cell proliferation could be the observation that S1–3a protein reduces cyclin D3 expression, thereby inhibiting Rb phosphorylation, which subsequently leads to an arrest in the G1 phase of the cell cycle (Yuan et al., 2007).
In general, autophagy is one of the major defense mechanisms of cells in the fight against pathogens (Deretic et al., 2013). However, viruses have developed mechanisms to interfere with the autophagic process, even including autophagy for their own replication. Coronaviruses were suggested to interact with the autophagy pathway (Carmona-Gutierrez et al., 2020) and involvement of S2–3a protein in this process was investigated. Fusion of autophagosomes with lysosomes is prevented by S2–3a, located in the late endosome, blocking interaction of the homotypic fusion and protein sorting (HOPS) complex with the SNARE complex and RAB7 which is required for autolysosome formation, thereby inhibiting fusion of autophagosomes with lysosomes (Miao et al., 2021, Zhang et al., 2021). A further study confirms that 3a of SARS-CoV-2 (S2-3a) alone is able to induce incomplete autophagy in infected cells. ORF3a interacts with autophagy regulator UVRAG, inducing PI3KC3-C1 activity, and at the same time inhibits PI3KC3-C2 (Qu et al., 2021). Surprisingly, despite the high similarity of 72.2% between S1–3a and S2–3a, the cellular autophagy response could only be observed for S2–3a (Qu et al., 2021).
7.2.2. Interaction of 3a protein with other virus or host proteins
ORF3a colocalizes with the M protein in the Golgi compartment, the E protein is also mainly localized in the ERGIC compartment. However, aside of this localization in the same compartment, there is no strong evidence that these virus proteins interact with each other. Some interactions of ORF 3a with proteins involved in autophagy were described in the section 'Apoptosis and autophagy'. Using biochemical, biophysical and genetic techniques, an interaction of the S1–3a with caveolin-1 was identified (Padhan et al., 2007). Caveolins (1−3) are 21–24 kDa proteins building the major structural component of caveolae, a special type of lipid raft responsible in the uptake of small molecules through glycosylphosphatidylinositol (GPI)-anchored receptors; additionally, caveolae play a role in neurotransmitter signaling (Anderson, 1998).
7.2.3. Posttranslational modifications
Pulse-chase analysis on the SARS-CoV 3a protein points to its posttranslational modification through O-glycosylation which was confirmed by an in-situ O-glycosylation assay and the absence of O-linked sugars after substitution of serine and threonine residues in the S1–3a ectodomain (Oostra et al., 2006).
7.3. ORF 8a - 8b and 8ab proteins
In animal isolates and early human isolates, the SARS-CoV encoded a single protein 8ab. However, in later human isolates during the peak of 2003 SARS-CoV epidemic, a 29-nt deletion in the center split ORF8 into two smaller ORFs, encoding proteins 8a and 8b (Oostra et al., 2007, Muth et al., 2018). During the SARS CoV-2 epidemic, however, ORF8ab instead of 8a and 8b are expressed. The intact ORF8 contains a cleavable signal sequence, directing the precursor to the ER and mediating its translocation into the lumen. The cleaved protein is N-glycosylated, and remains stable in the ER (Oostra et al., 2007). ORF8 activates IL-17 signaling pathway and promotes the expression of pro-inflammatory factors thereby contributing to the observed cytokine storm (Lin et al., 2021), independent on viroporin activity. The structure of ORF8 adopts an Ig-like fold, and forms a disulfide-linked homodimer containing an intermolecular parallel β-sheet (Flower et al., 2021).
The 8a polypeptide (39 amino acids) of SARS-CoV was detected in mitochondria, and its overexpression results in increased mitochondrial transmembrane potential, production of reactive oxygen species, increased caspase 3 activity, and cellular apoptosis (Chen et al., 2007, Chen et al., 2007). When reconstituted into artificial lipid bilayers 8a forms cation-selective ion channels, and computational modelling studies in a hydrated POPC lipid bilayer on a truncated 22 amino acid transmembrane helix predicted a putative homo-oligomeric helical bundle model (Chen et al., 2011). In case of SARS-CoV, it was shown that full-length E and 3a proteins were required for maximal virus replication and virulence, while viroporin 8a had only a minor impact on virus viability (Castano-Rodriguez et al., 2018). Thus, ORF 8 proteins of SARS-CoV may be able to form viable ion channels, while ORF 8ab in SARS-CoV-2 does not have a structure that is compatible with an ion channel.
8. Conclusion
The genome of SARS-CoV-2 encodes for 29 proteins, of which the E- and ORF3a protein have been identified as viroporins that participate in viral replication through their channel activity and through interaction with intracellular proteins and pathways. Viroporin activity is indispensable for virus replication. Techniques for the study of activity as well as inhibition of viroporins have been developed making them a highly relevant object of study and a promising target for the development of antiviral drugs.
Acknowledgements
Support by Science and Technology Development Fund (STDF) Egypt grant No 45420 to NS, UB and HGB is gratefully acknowledged.
Competing Interests
The authors report no conflict of interest regarding this work.
CRediT authorship contribution statement
Ulrike Breitinger: Conception, Draft, Writing, Proofreading. Noha S. Farag: Draft, Proofreading. Heinrich Sticht: Conception, Draft, Writing, Proofreading. Hans-Georg Breitinger: Conception, Draft, Writing, Proofreading.
References
- Acharya R., Carnevale V., Fiorin G., Levine B.G., Polishchuk A.L., Balannik V., Samish I., Lamb R.A., Pinto L.H., DeGrado W.F., Klein M.L. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc. Natl. Acad. Sci. U.S.A. 2010;107(34):15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. 4th. Garland; New York: 2002. Molecular Biology of the Cell. [Google Scholar]
- Aldabe R., Barco A., Carrasco L. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 1996;271(38):23134–23137. doi: 10.1074/jbc.271.38.23134. [DOI] [PubMed] [Google Scholar]
- Aldabe R., Irurzun A., Carrasco L. Poliovirus protein 2BC increases cytosolic free calcium concentrations. J. Virol. 1997;71(8):6214–6217. doi: 10.1128/jvi.71.8.6214-6217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez E., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Marcos-Villar L., Enjuanes L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated. Virology. 2010;402(2):281–291. doi: 10.1016/j.virol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.G. The caveolae membrane system. Annu. Rev. Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- Arbely E., Khattari Z., Brotons G., Akkawi M., Salditt T., Arkin I.T. A highly unusual palindromic transmembrane helical hairpin formed by SARS coronavirus E protein. J. Mol. Biol. 2004;341(3):769–779. doi: 10.1016/j.jmb.2004.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y., Kuroki M., Maki M., Ikeda M., Dansako H., Wakita T., Kato N. The ESCRT system is required for hepatitis C virus production. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0014517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo J., Boceta M., Gonzalez M.E., Michel M., Carrasco L. Membrane permeabilization by different regions of the human immunodeficiency virus type 1 transmembrane glycoprotein gp41. J. Virol. 1995;69(7):4095–4102. doi: 10.1128/jvi.69.7.4095-4102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aweya J.J., Mak T.M., Lim S.G., Tan Y.J. The p7 protein of the hepatitis C virus induces cell death differently from the influenza A virus viroporin M2. Virus Res. 2013;172(1–2):24–34. doi: 10.1016/j.virusres.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmik D., Nandi R., Jagadeesan R., Kumar N., Prakash A., Kumar D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infect. Genet. Evol. 2020;84 doi: 10.1016/j.meegid.2020.104451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright K.M., Salvesen G.S. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 2003;15(6):725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Boscarino J.A., Logan H.L., Lacny J.J., Gallagher T.M. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J. Virol. 2008;82(6):2989–2999. doi: 10.1128/JVI.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boson B., Legros V., Zhou B., Siret E., Mathieu C., Cosset F.L., Lavillette D., Denolly S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.016175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour S., Strebel K. The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J. Virol. 1996;70(12):8285–8300. doi: 10.1128/jvi.70.12.8285-8300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitinger U., Ali N.K.M., Sticht H., Breitinger H.G. Inhibition of SARS CoV envelope protein by flavonoids and classical viroporin inhibitors. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.692423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitinger U., Farag N.S., Ali N.K., Breitinger H.G. Patch-clamp study of hepatitis C p7 channels reveals genotype-specific sensitivity to inhibitors. Biophys. J. 2016;110(11):2419–2429. doi: 10.1016/j.bpj.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitinger U., Farag N.S., Ali N.K.M., Ahmed M., El-Azizi M., Breitinger H.G. Cell viability assay as a tool to study activity and inhibition of hepatitis C p7 channels. J. Gen. Virol. 2021;102(3) doi: 10.1099/jgv.0.001571. [DOI] [PubMed] [Google Scholar]
- Cabrera-Garcia D., Bekdash R., Abbott G.W., Yazawa M., Harrison N.L. The envelope protein of SARS-CoV-2 increases intra-Golgi pH and forms a cation channel that is regulated by pH. J. Physiol. 2021;599(11):2851–2868. doi: 10.1113/JP281037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillet-Saguy C., Durbesson F., Rezelj V.V., Gogl G., Tran Q.D., Twizere J.C., Vignuzzi M., Vincentelli R., Wolff N. Host PDZ-containing proteins targeted by SARS-CoV-2. FEBS J. 2021;288(17):5148–5162. doi: 10.1111/febs.15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Gutierrez D., Bauer M.A., Zimmermann A., Kainz K., Hofer S.J., Kroemer G., Madeo F. Digesting the crisis: autophagy and coronaviruses. Micro Cell. 2020;7(5):119–128. doi: 10.15698/mic2020.05.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco L. "Membrane leakiness after viral infection and a new approach to the development of antiviral agents.". Nature. 1978;272(5655):694–699. doi: 10.1038/272694a0. [DOI] [PubMed] [Google Scholar]
- Carrasco L., Perez L., Irurzun A., Lama J., Martinez-Abarca F., Rodrigez P., Guinea R., Castrillo J.L., Sanz M.A., Ayala M.J. In: Regulation of Gene Expression in Animal Viruses. Carrasco L., Sonenberg N., Wimmer E., editors. Plenum; New York: 1993. [Google Scholar]
- Carter S.D., Dent K.C., Atkins E., Foster T.L., Verow M., Gorny P., Harris M., Hiscox J.A., Ranson N.A., Griffin S., Barr J.N. Direct visualization of the small hydrophobic protein of human respiratory syncytial virus reveals the structural basis for membrane permeability. FEBS Lett. 2010;584(13):2786–2790. doi: 10.1016/j.febslet.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano-Rodriguez C., Honrubia J.M., Gutierrez-Alvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Baguena C., Queralt-Martin M., Kochan G., Perlman S., Aguilella V.M., Sola I., Enjuanes L. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018;9:3. doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J., Cai Y., Pang C., Wang L., McSweeney S., Shanklin J., Liu Q. Structural basis for SARS-CoV-2 envelope protein recognition of human cell junction protein PALS1. Nat. Commun. 2021;12(1):3433. doi: 10.1038/s41467-021-23533-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.M., Tsoi H., Chan W.M., Zhai S., Wong C.O., Yao X., Chan W.Y., Tsui S.K., Chan H.Y. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int J. Biochem. Cell Biol. 2009;41(11):2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler D.E., Penin F., Schulten K., Chipot C. The p7 protein of hepatitis C virus forms structurally plastic, minimalist ion channels. PLoS Comput. Biol. 2012;8(9) doi: 10.1371/journal.pcbi.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.J., Leser G.P., Morita E., Lamb R.A. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 2007;81(13):7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.C., Kruger J., Sramala I., Hsu H.J., Henklein P., Chen Y.M., Fischer W.B. ORF8a of SARS-CoV forms an ion channel: experiments and molecular dynamics simulations. Biochim. Biophys. Acta. 2011;1808(2):572–579. doi: 10.1016/j.bbamem.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Ping Y.H., Lee H.C., Chen K.H., Lee Y.M., Chan Y.J., Lien T.C., Jap T.S., Lin C.H., Kao L.S., Chen Y.M. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J. Infect. Dis. 2007;196(3):405–415. doi: 10.1086/519166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.Y., Moriyama M., Chang M.F., Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.C., Lo S.Y., Ma H.C., Li H.C. Expression and membrane integration of SARS-CoV E protein and its interaction with M protein. Virus Genes. 2009;38(3):365–371. doi: 10.1007/s11262-009-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L., Chanturiya A.N., Suss-Toby E., Nora E., Zimmerberg J. An amphipathic peptide from the C-terminal region of the human immunodeficiency virus envelope glycoprotein causes pore formation in membranes. J. Virol. 1994;68(11):7115–7123. doi: 10.1128/jvi.68.11.7115-7123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhmakov I.V., Ogden D.C., Geraghty F.M., Hayhurst A., Skinner A., Betakova T., Hay A.J. Differences in conductance of M2 proton channels of two influenza viruses at low and high pH. J. Physiol. 2003;546(2):427–438. doi: 10.1113/jphysiol.2002.028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christgen S., Kanneganti T.D. Inflammasomes and the fine line between defense and disease. Curr. Opin. Immunol. 2020;62:39–44. doi: 10.1016/j.coi.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampor F., Bayley P.M., Nermut M.V., Hirst E.M., Sugrue R.J., Hay A.J. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus hemagglutinin to the low pH conformation occurs in an acidic trans Golgi compartment. Virology. 1992;188(1):14–24. doi: 10.1016/0042-6822(92)90730-d. [DOI] [PubMed] [Google Scholar]
- Ciampor F., Thompson C.A., Grambas S., Hay A.J. Regulation of pH by the M2 protein of influenza A viruses. Virus Res. 1992;22(3):247–258. doi: 10.1016/0168-1702(92)90056-f. [DOI] [PubMed] [Google Scholar]
- Ciccaglione A.R., Marcantonio C., Costantino A., Equestre M., Geraci A., Rapicetta M. Hepatitis C virus E1 protein induces modification of membrane permeability in E. coli cells. Virology. 1998;250(1):1–8. doi: 10.1006/viro.1998.9380. [DOI] [PubMed] [Google Scholar]
- Clarke D., Griffin S., Beales L., Gelais C.S., Burgess S., Harris M., Rowlands D. Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 2006;281(48):37057–37068. doi: 10.1074/jbc.M602434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J.R., Lin L.D., Machamer C.E. Identification of a Golgi complex-targeting signal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus envelope protein. J. Virol. 2011;85(12):5794–5803. doi: 10.1128/JVI.00060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook G.A., Dawson L.A., Tian Y., Opella S.J. Three-dimensional structure and interaction studies of hepatitis C virus p7 in 1,2-dihexanoyl-sn-glycero-3-phosphocholine by solution nuclear magnetic resonance. Biochemistry. 2013;52(31):5295–5303. doi: 10.1021/bi4006623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Kuo L., Masters P.S., Vennema H., Rottier P.J. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72(8):6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Vennema H., Rottier P.J. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74(11):4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong A.S., de Mattia F., Van Dommelen M.M., Lanke K., Melchers W.J., Willems P.H., van Kuppeveld F.J. Functional analysis of picornavirus 2B proteins: effects on calcium homeostasis and intracellular protein trafficking. J. Virol. 2008;82(7):3782–3790. doi: 10.1128/JVI.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Alvarez E., Almazan F., Rejas M.T., Lamirande E., Roberts A., Shieh W.J., Zaki S.R., Subbarao K., Enjuanes L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007;81(4):1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Saitoh T., Akira S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013;13(10):722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey D., Borkotoky S., Banerjee M. In silico identification of Tretinoin as a SARS-CoV-2 envelope (E) protein ion channel inhibitor. Comput. Biol. Med. 2020;127 doi: 10.1016/j.compbiomed.2020.104063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey D., Siddiqui S.I., Mamidi P., Ghosh S., Kumar C.S., Chattopadhyay S., Ghosh S., Banerjee M. The effect of amantadine on an ion channel protein from Chikungunya virus. PLoS Negl. Trop. Dis. 2019;13(7) doi: 10.1371/journal.pntd.0007548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Y., Chemello M.E., Pena F., Aristimuno O.C., Zambrano J.L., Rojas H., Bartoli F., Salazar L., Chwetzoff S., Sapin C., Trugnan G., Michelangeli F., Ruiz M.C. Expression of nonstructural rotavirus protein NSP4 mimics Ca2+ homeostasis changes induced by rotavirus infection in cultured cells. J. Virol. 2008;82(22):11331–11343. doi: 10.1128/JVI.00577-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C.A. Interleukin-1 and the pathogenesis of the acute-phase response. New Engl. J. Med. 1984;311(22):1413–1418. doi: 10.1056/NEJM198411293112205. [DOI] [PubMed] [Google Scholar]
- Duart G., Garcia-Murria M.J., Grau B., Acosta-Caceres J.M., Martinez-Gil L., Mingarro I. SARS-CoV-2 envelope protein topology in eukaryotic membranes. Open Biol. 2020;10(9) doi: 10.1098/rsob.200209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag N.S., Breitinger U., Breitinger H.G., Azizi M.A. El. Viroporins and inflammasomes: a key to understand virus-induced inflammation. Int. J. Biochem. Cell Biol. 2020;122 doi: 10.1016/j.biocel.2020.105738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag N.S., Breitinger U., El-Azizi M., Breitinger H.G. The p7 viroporin of the hepatitis C virus contributes to liver inflammation by stimulating production of Interleukin-1β. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863(3):712–720. doi: 10.1016/j.bbadis.2016.12.006. [DOI] [PubMed] [Google Scholar]
- Fett C., DeDiego M.L., Regla-Nava J.A., Enjuanes L., Perlman S. Complete protection against severe acute respiratory syndrome coronavirus-mediated lethal respiratory disease in aged mice by immunization with a mouse-adapted virus lacking E protein. J. Virol. 2013;87(12):6551–6559. doi: 10.1128/JVI.00087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower T.G., Buffalo C.Z., Hooy R.M., Allaire M., Ren X., Hurley J.H. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc. Natl. Acad. Sci. U.S.A. 2021;118:2. doi: 10.1073/pnas.2021785118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt E.C., Yu L., Goldsmith C.S., Welsh S., Cheng A., Yount B., Liu W., Frieman M.B., Buchholz U.J., Screaton G.R., Lippincott-Schwartz J., Zaki S.R., Xu X.N., Baric R.S., Subbarao K., Lenardo M.J. The open reading frame 3a protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death. J. Virol. 2010;84(2):1097–1109. doi: 10.1128/JVI.01662-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13(6):405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S.W., Tan E., Lin X., Yu D., Wang J., Tan G.M., Vararattanavech A., Yeo C.Y., Soon C.H., Soong T.W., Pervushin K., Torres J. The small hydrophobic protein of the human respiratory syncytial virus forms pentameric ion channels. J. Biol. Chem. 2012;287(29):24671–24689. doi: 10.1074/jbc.M111.332791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus J.E., von Schwedler U.K., Pornillos O.W., Morham S.G., Zavitz K.H., Wang H.E., Wettstein D.A., Stray K.M., Cote M., Rich R.L., Myszka D.G., Sundquist W.I. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107(1):55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gervais C., Do F., Cantin A., Kukolj G., White P.W., Gauthier A., Vaillancourt F.H. Development and validation of a high-throughput screening assay for the hepatitis C virus p7 viroporin. J. Biomol. Screen. 2011;16(3):363–369. doi: 10.1177/1087057110396215. [DOI] [PubMed] [Google Scholar]
- Gheysen D., Jacobs E., de Foresta F., Thiriart C., Francotte M., Thines D., Wilde M.De. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59(1):103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- Goldsmith C.S., Tatti K.M., Ksiazek T.G., Rollin P.E., Comer J.A., Lee W.W., Rota P.A., Bankamp B., Bellini W.J., Zaki S.R. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 2004;10(2):320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M.E., Carrasco L. Viroporins. FEBS Lett. 2003;552(1):28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracie J.A., Robertson S.E., McInnes I.B. Interleukin-18. J. Leukoc. Biol. 2003;73(2):213–224. doi: 10.1189/jlb.0602313. [DOI] [PubMed] [Google Scholar]
- Griffin S.D.C., Harvey R., Clarke D.S., Barclay W.S., Harris M., Rowlands D.J. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J. Gen. Virol. 2004;85(Pt 2):451–461. doi: 10.1099/vir.0.19634-0. [DOI] [PubMed] [Google Scholar]
- Gupta M.K., Vemula S., Donde R., Gouda G., Behera L., Vadde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J. Biomol. Struct. Dyn. 2021;39(7):2617–2627. doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnoczky G., Davies E., Madesh M. Calcium signaling and apoptosis. Biochem. Biophys. Res. Commun. 2003;304(3):445–454. doi: 10.1016/s0006-291x(03)00616-8. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., Matsuoka Y., Ueshiba H., Komatsu N., Itoh K., Shichijo S., Kanai T., Fukushi M., Ishida I., Kirikae T., Sasazuki T., Miyoshi-Akiyama T. Dissection and identification of regions required to form pseudoparticles by the interaction between the nucleocapsid (N) and membrane (M) proteins of SARS coronavirus. Virology. 2008;380(1):99–108. doi: 10.1016/j.virol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin A.L., Huxley A.F. Potassium leakage from an active nerve fibre. J. Physiol. 1947;106(3):341–367. doi: 10.1113/jphysiol.1947.sp004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourioux C., Ait-Goughoulte M., Patient R., Fouquenet D., Arcanger-Doudet F., Brand D., Martin A., Roingeard P. Core protein domains involved in hepatitis C virus-like particle assembly and budding at the endoplasmic reticulum membrane. Cell Microbiol. 2007;9(4):1014–1027. doi: 10.1111/j.1462-5822.2006.00848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang Z.Y., Kong W.P., Nabel G.J. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 2004;78(22):12557–12565. doi: 10.1128/JVI.78.22.12557-12565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A., Das S.R., Tanwar C., Jameel S. Oligomerization of the human immunodeficiency virus type 1 (HIV-1) Vpu protein--a genetic, biochemical and biophysical analysis. Virol. J. 2007;4:81. doi: 10.1186/1743-422X-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyser J.M., Collinson-Pautz M.R., Utama B., Estes M.K. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. mBio. 2010;1:5. doi: 10.1128/mBio.00265-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T., Pang I.K., Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 2010;11(5):404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Yanagi Y., Ichinohe T. Encephalomyocarditis virus viroporin 2B activates NLRP3 inflammasome. PLoS Pathog. 2012;8(8) doi: 10.1371/journal.ppat.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki-Horimoto K., Horimoto T., Noda T., Kiso M., Maeda J., Watanabe S., Muramoto Y., Fujii K., Kawaoka Y. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J. Virol. 2006;80(11):5233–5240. doi: 10.1128/JVI.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javorsky A., Humbert P.O., Kvansakul M. Structural basis of coronavirus E protein interactions with human PALS1 PDZ domain. Commun. Biol. 2021;4(1):724. doi: 10.1038/s42003-021-02250-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Guardeno J.M., Nieto-Torres J.L., DeDiego M.L., Regla-Nava J.A., Fernandez-Delgado R., Castano-Rodriguez C., Enjuanes L. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10(8) doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.T., Murray C.L., Eastman D.K., Tassello J., Rice C.M. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 2007;81(16):8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa N., Nishigaki K., Hayashi T., Ishii Y., Furukawa S., Niiro A., Yasui F., Kohara M., Morita K., Matsushima K., Le M.Q., Masuda T., Kannagi M. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-kappaB activation. FEBS Lett. 2006;580(30):6807–6812. doi: 10.1016/j.febslet.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern D.M., Sorum B., Mali S.S., Hoel C.M., Sridharan S., Remis J.P., Toso D.B., Kotecha A., Bautista D.M., Brohawn S.G. Cryo-EM structure of the SARS-CoV-2 3a ion channel in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021;28:573–582. doi: 10.1038/s41594-021-00619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kien, F., Ma, H., Gaisenband, S.D., Nal, B., 2013. Viroporins: Differential Functions at Late Stages of Viral Life Cycles. In: Microbial Pathogenesis: Infection and Immunity. Kishore, U., Nayak, A., ed's, Springer Science+Business Media, LLC, 2013, pp. 38-62.
- Lama J., Carrasco L. Expression of poliovirus nonstructural proteins in Escherichia coli cells. Modification of membrane permeability induced by 2B and 3A. J. Biol. Chem. 1992;267(22):15932–15937. [PubMed] [Google Scholar]
- Latz E., Xiao T.S., Stutz A. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 2013;13(6):397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law P.T.W., Wong C.H., Au T.C.C., Chuck C.P., Kong S.K., Chan P.K.S., To K.F., Lo A.W.I., Chan J.Y.W., Suen Y.K., Chan H.Y.E., Fung K.P., Waye M.M.Y., Sung J.J.Y., Lo Y.M.D., Tsui S.K.W. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005;86(7):1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- Lebedeva N.S., Gubarev Y.A., Mamardashvili G.M., Zaitceva S.V., Zdanovich S.A., Malyasova A.S., Romanenko J.V., Koifman M.O., Koifman O.I. Theoretical and experimental study of interaction of macroheterocyclic compounds with ORF3a of SARS-CoV-2. Sci. Rep. 2021;11(1):19481. doi: 10.1038/s41598-021-99072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Surya W., Claudine S., Torres J. Structure of a conserved Golgi complex-targeting signal in coronavirus envelope proteins. J. Biol. Chem. 2014;289(18):12535–12549. doi: 10.1074/jbc.M114.560094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Lescar J., Tam J.P., Liu D.X. Expression of SARS-coronavirus envelope protein in Escherichia coli cells alters membrane permeability. Biochem. Biophys. Res. Commun. 2004;325(1):374–380. doi: 10.1016/j.bbrc.2004.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Tam J.P., Liu D.X. Viroporin activity of SARS-CoV E protein. Adv. Exp. Med. Biol. 2006;581:199–202. doi: 10.1007/978-0-387-33012-9_34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Yuan Q., Torres J., Tam J.P., Liu D.X. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology. 2006;349(2):264–275. doi: 10.1016/j.virol.2006.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4:3. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Fu B., Yin S., Li Z., Liu H., Zhang H., Xing N., Wang Y., Xue W., Xiong Y., Zhang S., Zhao Q., Xu S., Zhang J., Wang P., Nian W., Wang X., Wu H. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. iScience. 2021;24(4) doi: 10.1016/j.isci.2021.102293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Liang W., Kang K., Li H., Cao Z., Zhang Y. Classical swine fever virus and p7 protein induce secretion of IL-1beta in macrophages. J. Gen. Virol. 2014;95(12):2693–2699. doi: 10.1099/vir.0.068502-0. [DOI] [PubMed] [Google Scholar]
- Lopez L.A., Riffle A.J., Pike S.L., Gardner D., Hogue B.G. Importance of conserved cysteine residues in the coronavirus envelope protein. J. Virol. 2008;82(6):3000–3010. doi: 10.1128/JVI.01914-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. U.S.A. 2006;103(33):12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik P., Chew C., Aittoniemi J., Chang J., Wentworth P., Jr., Dwek R.A., Biggin P.C., Venien-Bryan C., Zitzmann N. The 3-dimensional structure of a hepatitis C virus p7 ion channel by electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 2009;106(31):12712–12716. doi: 10.1073/pnas.0905966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V., Castello A., Carrasco L. Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell Microbiol. 2008;10(2):437–451. doi: 10.1111/j.1462-5822.2007.01057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandala V.S., McKay M.J., Shcherbakov A.A., Dregni A.J., Kolocouris A., Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein in phospholipid bilayers. Nat Struct Mol Biol. 2020;27:1202–1208. doi: 10.1038/s41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F., Gaide O., Petrilli V., Mayor A., Tschopp J. NALP inflammasomes: a central role in innate immunity. Semin. Immunopathol. 2007;29(3):213–229. doi: 10.1007/s00281-007-0079-y. [DOI] [PubMed] [Google Scholar]
- Martinon F., Mayor A., Tschopp J. The inflammasomes: guardians of the body. Annu. Rev. Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- McClenaghan C., Hanson A., Lee S.J., Nichols C.G. Coronavirus proteins as ion channels: current and potential research. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.573339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros R., Escriou N., Naffakh N., Manuguerra J.C., van der Werf S. Hemagglutinin residues of recent human A(H3N2) influenza viruses that contribute to the inability to agglutinate chicken erythrocytes. Virology. 2001;289(1):74–85. doi: 10.1006/viro.2001.1121. [DOI] [PubMed] [Google Scholar]
- Miao G., Zhao H., Li Y., Ji M., Chen Y., Shi Y., Bi Y., Wang P., Zhang H. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell. 2021;56(4):427–442. doi: 10.1016/j.devcel.2020.12.010. e425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4(12) doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rehman S., Hassan M.I., Ahmad F. The SARS Coronavirus 3a protein binds calcium in its cytoplasmic domain. Virus Res. 2014;191:180–183. doi: 10.1016/j.virusres.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montserret R., Saint N., Vanbelle C., Salvay A.G., Simorre J.P., Ebel C., Sapay N., Renisio J.G., Bockmann A., Steinmann E., Pietschmann T., Dubuisson J., Chipot C., Penin F. NMR structure and ion channel activity of the p7 protein from hepatitis C virus. J. Biol. Chem. 2010;285(41):31446–31461. doi: 10.1074/jbc.M110.122895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth D., Corman V.M., Roth H., Binger T., Dijkman R., Gottula L.T., Gloza-Rausch F., Balboni A., Battilani M., Rihtaric D., Toplak I., Ameneiros R.S., Pfeifer A., Thiel V., Drexler J.F., Muller M.A., Drosten C. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8(1):15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadiri A., Wolinski M.K., Saleh M. The inflammatory caspases: key players in the host response to pathogenic invasion and sepsis. J. Immunol. 2006;177(7):4239–4245. doi: 10.4049/jimmunol.177.7.4239. [DOI] [PubMed] [Google Scholar]
- Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., van der Werf S., Yuen K.-Y., Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86(5):1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- Neches R.Y., Kyrpides N.C., Ouzounis C.A. Atypical divergence of SARS-CoV-2 Orf8 from Orf7a within the coronavirus lineage suggests potential stealthy viral strategies in immune evasion. mBio. 2021;12(1):e03014–20. doi: 10.1128/mBio.03014-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negash A.A., Ramos H.J., Crochet N., Lau D.T., Doehle B., Papic N., Delker D.A., Jo J., Bertoletti A., Hagedorn C.H., Gale M., Jr. IL-1beta production through the NLRP3 inflammasome by hepatic macrophages links hepatitis C virus infection with liver inflammation and disease. PLoS Pathog. 2013;9(4) doi: 10.1371/journal.ppat.1003330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netland J., DeDiego M.L., Zhao J., Fett C., Alvarez E., Nieto-Torres J.L., Enjuanes L., Perlman S. Immunization with an attenuated severe acute respiratory syndrome coronavirus deleted in E protein protects against lethal respiratory disease. Virology. 2010;399(1):120–128. doi: 10.1016/j.virol.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K., Meyer J.C., Bellamy A.R., Taylor J.A. Rotavirus nonstructural glycoprotein NSP4 alters plasma membrane permeability in mammalian cells. J. Virol. 1997;71(12):9458–9465. doi: 10.1128/jvi.71.12.9458-9465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., Dediego M.L., Alvarez E., Jimenez-Guardeno J.M., Regla-Nava J.A., Llorente M., Kremer L., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., DeDiego M.L., Verdia-Baguena C., Jimenez-Guardeno J.M., Regla-Nava J.A., Fernandez-Delgado R., Castano-Rodriguez C., Alcaraz A., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10(5) doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., Verdia-Baguena C., Jimenez-Guardeno J.M., Regla-Nava J.A., Castano-Rodriguez C., Fernandez-Delgado R., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J.L., Madan V., Carrasco L. Viroporins: structure and biological functions. Nat. Rev. Microbiol. 2012;10(8):563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien V. Viruses and apoptosis. J. Gen. Virol. 1998;79(Pt 8):1833–1845. doi: 10.1099/0022-1317-79-8-1833. [DOI] [PubMed] [Google Scholar]
- Oostra M., de Haan C.A., de Groot R.J., Rottier P.J. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 2006;80(5):2326–2336. doi: 10.1128/JVI.80.5.2326-2336.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., de Haan C.A., Rottier P.J. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 2007;81(24):13876–13888. doi: 10.1128/JVI.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orfali R., Rateb M.E., Hassan H.M., Alonazi M., Gomaa M.R., Mahrous N., GabAllah M., Kandeil A., Perveen S., Abdelmohsen U.R., Sayed A.M. Sinapic acid suppresses SARS CoV-2 replication by targeting its envelope protein. Antibiotics. 2021;10(4):420. doi: 10.3390/antibiotics10040420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhan K., Minakshi R., Towheed M.A.B., Jameel S. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J. Gen. Virol. 2008;89(8):1960–1969. doi: 10.1099/vir.0.83665-0. [DOI] [PubMed] [Google Scholar]
- Padhan K., Tanwar C., Hussain A., Hui P.Y., Lee M.Y., Cheung C.Y., Peiris J.S.M., Jameel S. Severe acute respiratory syndrome coronavirus Orf3a protein interacts with caveolin. J. Gen. Virol. 2007;88(Pt 11):3067–3077. doi: 10.1099/vir.0.82856-0. [DOI] [PubMed] [Google Scholar]
- Padhi S., Khan N., Jameel S., Priyakumar U.D. Molecular dynamics simulations reveal the HIV-1 Vpu transmembrane protein to form stable pentamers. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0079779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Mrse A.A., Nevzorov A.A., Mesleh M.F., Oblatt-Montal M., Montal M., Opella S.J. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J. Mol. Biol. 2003;333(2):409–424. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- Pawliczek T., Crump C.M. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 2009;83(21):11254–11264. doi: 10.1128/JVI.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K., Tan E., Parthasarathy K., Lin X., Jiang F.L., Yu D., Vararattanavech A., Soong T.W., Liu D.X., Torres J. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5(7) doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T., Perry J.L., Dosey T.L., Delcour A.H., Hyser J.M. The rotavirus NSP4 viroporin domain is a calcium-conducting ion channel. Sci. Rep. 2017;7:43487. doi: 10.1038/srep43487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar A., Wilson L., Ewart G.D., Gage P.W. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 2004;557(1–3):99–103. doi: 10.1016/s0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- Qu Y., Wang X., Zhu Y., Wang W., Wang Y., Hu G., Liu C., Li J., Ren S., Xiao M.Z.X., Liu Z., Wang C., Fu J., Zhang Y., Li P., Zhang R., Liang Q. ORF3a-mediated incomplete autophagy facilitates severe acute respiratory syndrome coronavirus-2 replication. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.716208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regla-Nava J.A., Nieto-Torres J.L., Jimenez-Guardeno J.M., Fernandez-Delgado R., Fett C., Castano-Rodriguez C., Perlman S., Enjuanes L., DeDiego M.L. Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates. J. Virol. 2015;89(7):3870–3887. doi: 10.1128/JVI.03566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.Y., Zhou W., Qiu Y., Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol. Immunol. 2020;17(8):881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman J.S., Jing X., Leser G.P., Lamb R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142(6):902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A., Guatelli J.C., Stephens E.B. The Vpu protein: new concepts in virus release and CD4 down-modulation. Curr. HIV Res. 2010;8(3):240–252. doi: 10.2174/157016210791111124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi T., Leser G.P., Lamb R.A. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J. Cell Biol. 1996;133(4):733–747. doi: 10.1083/jcb.133.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salom D., Hill B.R., Lear J.D., DeGrado W.F. pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry. 2000;39(46):14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D., Schwarz S., Wang K., Zhang R., Sun B., Schwarz W. Genistein as antiviral drug against HIV ion channel. Planta Med. 2014;80(8–9):682–687. doi: 10.1055/s-0034-1368583. [DOI] [PubMed] [Google Scholar]
- Schnell J.R., Chou J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451(7178):591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K., Tschopp J. The inflammasomes. Cell. 2010;140(6):821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Schwarz S., Sauter D., Wang K., Zhang R., Sun B., Karioti A., Bilia A.R., Efferth T., Schwarz W. Kaempferol derivatives as antiviral drugs against the 3a channel protein of coronavirus. Planta Med. 2014;80(2–3):177–182. doi: 10.1055/s-0033-1360277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S., Wang K., Yu W., Sun B., Schwarz W. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antivir. Res. 2011;90(1):64–69. doi: 10.1016/j.antiviral.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C., Griffin S. Viroporins: structure, function and potential as antiviral targets. J. Gen. Virol. 2015;96(8):2000–2027. doi: 10.1099/vir.0.000201. [DOI] [PubMed] [Google Scholar]
- Shelokov A., Vogel J.E., Chi L. Hemadsorption (adsorption-hemagglutination) test for viral agents in tissue culture with special reference to influenza. Proc. Soc. Exp. Biol. Med. 1958;97(4):802–809. doi: 10.3181/00379727-97-23884. [DOI] [PubMed] [Google Scholar]
- Shepley-McTaggart A., Sagum C.A., Oliva I., Rybakovsky E., DiGuilio K., Liang J., Bedford M.T., Cassel J., Sudol M., Mullin J.M., Harty R.N. SARS-CoV-2 envelope (E) protein interacts with PDZ-domain-2 of host tight junction protein ZO1. PLoS One. 2021;16(6):e0251955. doi: 10.1371/journal.pone.0251955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S., Mukherjee A., Ray R., Ray R.B. Hepatitis C virus induces interleukin-1beta (IL-1beta)/IL-18 in circulatory and resident liver macrophages. J. Virol. 2013;87(22):12284–12290. doi: 10.1128/JVI.01962-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.L., Yuen K.S., Castano-Rodriguez C., Ye Z.W., Yeung M.L., Fung S.Y., Yuan S., Chan C.P., Yuen K.Y., Enjuanes L., Jin D.Y. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33(8):8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann E., Penin F., Kallis S., Patel A.H., Bartenschlager R., Pietschmann T. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007;3(7) doi: 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- StGelais C., Tuthill T.J., Clarke D.S., Rowlands D.J., Harris M., Griffin S. Inhibition of hepatitis C virus p7 membrane channels in a liposome-based assay system. Antivir. Res. 2007;76(1):48–58. doi: 10.1016/j.antiviral.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer A.L., Acharya R., Salom D., Levine A.S., Di Costanzo L., Soto C.S., Tereshko V., Nanda V., Stayrook S., DeGrado W.F. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451(7178):596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Karki C., Aguilera J., Lopez Hernandez A.E., Sun J., Li L. Computational study on the function of palmitoylation on the envelope protein in SARS-CoV-2. J. Chem. Theory Comput. 2021;17(10):6483–6490. doi: 10.1021/acs.jctc.1c00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surya W., Li Y., Torres J. Structural model of the SARS coronavirus E channel in LMPG micelles. Biochim Biophys. Acta Biomembr. 2018;1860(6):1309–1317. doi: 10.1016/j.bbamem.2018.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh K.T., Siu Y.L., Chan W.L., Schluter M.A., Liu C.J., Peiris J.S., Bruzzone R., Margolis B., Nal B. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol. Biol. Cell. 2010;21(22):3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger E.F., Cohen E.A., Lu Y.C., Sodroski J.G., Haseltine W.A. Functional role of human immunodeficiency virus type 1 vpu. Proc. Natl. Acad. Sci. U.S.A. 1989;86(13):5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar P.P.S., Krugliak M., Arkin I.T. Blockers of the SARS-CoV-2 3a channel identified by targeted drug repurposing. Viruses. 2021;13(3):532. doi: 10.3390/v13030532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J., Maheswari U., Parthasarathy K., Ng L., Liu D.X., Gong X. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007;16(9):2065–2071. doi: 10.1110/ps.062730007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J., Wang J., Parthasarathy K., Liu D.X. The transmembrane oligomers of coronavirus protein E. Biophys. J. 2005;88(2):1283–1290. doi: 10.1529/biophysj.104.051730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toto A., Ma S., Malagrino F., Visconti L., Pagano L., Stromgaard K., Gianni S. Comparing the binding properties of peptides mimicking the envelope protein of SARS-CoV and SARS-CoV-2 to the PDZ domain of the tight junction-associated PALS1 protein. Protein Sci. 2020;29(10):2038–2042. doi: 10.1002/pro.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K., Kar S., Vakakis E., Kotecha S., Triantafilou M. Human respiratory syncytial virus viroporin SH: a viral recognition pathway used by the host to signal inflammasome activation. Thorax. 2013;68(1):66–75. doi: 10.1136/thoraxjnl-2012-202182. [DOI] [PubMed] [Google Scholar]
- Triantafilou K., Ward C.J.K., Czubala M., Ferris R.G., Koppe E., Haffner C., Piguet V., Patel V.K., Amrine-Madsen H., Modis L.K., Masters S.L., Triantafilou M. Differential recognition of HIV-stimulated IL-1beta and IL-18 secretion through NLR and NAIP signalling in monocyte-derived macrophages. PLoS Pathog. 2021;17(4) doi: 10.1371/journal.ppat.1009417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobec T. The role of the SARS-CoV-2 envelope protein as a pH-dependent cation channel. J. Physiol. 2021;599(14):3435–3436. doi: 10.1113/JP281785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y.T., Wang S.M., Huang K.J., Wang C.T. SARS-CoV envelope protein palmitoylation or nucleocapid association is not required for promoting virus-like particle production. J. Biomed. Sci. 2014;21:34. doi: 10.1186/1423-0127-21-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulenko Y., Deviatkin A., Drexler J.F., Lukashev A. Modular evolution of coronavirus genomes. Viruses. 2021;13(7):1270. doi: 10.3390/v13071270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15(8):2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdia-Baguena C., Aguilella V.M., Queralt-Martin M., Alcaraz A. Transport mechanisms of SARS-CoV-E viroporin in calcium solutions: lipid-dependent anomalous mole fraction effect and regulation of pore conductance. Biochim. Biophys. Acta Biomembr. 2021;1863(6) doi: 10.1016/j.bbamem.2021.183590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdia-Baguena C., Nieto-Torres J.L., Alcaraz A., Dediego M.L., Enjuanes L., Aguilella V.M. Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochim. Biophys. Acta. 2013;1828(9):2026–2031. doi: 10.1016/j.bbamem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wu Y., Ma C., Fiorin G., Wang J., Pinto L.H., Lamb R.A., Klein M.L., Degrado W.F. Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus. Proc. Natl. Acad. Sci. U.S.A. 2013;110(4):1315–1320. doi: 10.1073/pnas.1216526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbeck J.W., Machamer C.E. The infectious bronchitis coronavirus envelope protein alters golgi pH To protect the spike protein and promote the release of infectious virus. J. Virol. 2019;93(11):e00015–19. doi: 10.1128/JVI.00015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Whittaker G.R. Fusion of enveloped viruses in endosomes. Traffic. 2016;17(6):593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield T., Miles A.J., Scheinost J.C., Offer J., Wentworth P., Jr., Dwek R.A., Wallace B.A., Biggin P.C., Zitzmann N. The influence of different lipid environments on the structure and function of the hepatitis C virus p7 ion channel protein. Mol. Membr. Biol. 2011;28(5):254–264. doi: 10.3109/09687688.2011.581253. [DOI] [PubMed] [Google Scholar]
- Wilson L., Gage P., Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353(2):294–306. doi: 10.1016/j.virol.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., McKinlay C., Gage P., Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330(1):322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak A.L., Griffin S., Rowlands D., Harris M., Yi M., Lemon S.M., Weinman S.A. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 2010;6(9) doi: 10.1371/journal.ppat.1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B., Shen X., He Y., Pan X., Liu F.L., Wang Y., Yang F., Fang S., Wu Y., Duan Z., Zuo X., Xie Z., Jiang X., Xu L., Chi H., Li S., Meng Q., Zhou H., Zhou Y., Cheng X., Xin X., Jin L., Zhang H.L., Yu D.D., Li M.H., Feng X.L., Chen J., Jiang H., Xiao G., Zheng Y.T., Zhang L.K., Shen J., Li J., Gao Z. SARS-CoV-2 envelope protein causes acute respiratory distress syndrome (ARDS)-like pathological damages and constitutes an antiviral target. Cell Res. 2021;31(8):847–860. doi: 10.1038/s41422-021-00519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Xiong Z., Zhang S., Yan Y., Nguyen J., Ng B., Lu H., Brendese J., Yang F., Wang H., Yang X.F. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem. J. 2005;392(1):135–143. doi: 10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Li J., Shan Y., Yang Z., Zhao Z., Chen B., Yao Z., Dong B., Wang S., Chen J., Cong Y. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005;109(2):191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Yao Z., Wu J., Zhou Y., Shan Y., Dong B., Zhao Z., Hua P., Chen J., Cong Y. G1 phase cell cycle arrest induced by SARS-CoV 3a protein via the cyclin D3/pRb pathway. Am. J. Respir. Cell Mol. Biol. 2007;37(1):9–19. doi: 10.1165/rcmb.2005-0345RC. [DOI] [PubMed] [Google Scholar]
- Yue Y., Nabar N.R., Shi C.S., Kamenyeva O., Xiao X., Hwang I.Y., Wang M., Kehrl J.H. SARS-coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9(9):904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sun H., Pei R., Mao B., Zhao Z., Li H., Lin Y., Lu K. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 2021;7(1):31. doi: 10.1038/s41421-021-00268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Karki R., Williams E.P., Yang D., Fitzpatrick E., Vogel P., Jonsson C.B., Kanneganti T.D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021;22(7):829–838. doi: 10.1038/s41590-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Frey T.K., Yang J.J. Viral calciomics: interplays between Ca2+ and virus. Cell Calcium. 2009;46(1):1–17. doi: 10.1016/j.ceca.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]