Abstract
It is widely accepted that narcotic use during pregnancy and specific environmental factors (e.g., maternal immune activation and chronic stress) may increase risk of neuropsychiatric illness in offspring. However, little progress has been made in defining human-specific in utero neurodevelopmental pathology due to ethical and technical challenges associated with accessing human prenatal brain tissue. Here we utilized human induced pluripotent stem cells (hiPSCs) to generate reproducible organoids that recapitulate dorsal forebrain development including early corticogenesis. We systemically exposed organoid samples to chemically defined “enviromimetic” compounds to examine the developmental effects of various narcotic and neuropsychiatric-related risk factors within tissue of human origin. In tandem experiments conducted in parallel, we modeled exposure to opiates (μ-opioid agonist endomorphin), cannabinoids (WIN 55,212-2), alcohol (ethanol), smoking (nicotine), chronic stress (human cortisol), and maternal immune activation (human Interleukin-17a; IL17a). Human-derived dorsal forebrain organoids were consequently analyzed via an array of unbiased and high-throughput analytical approaches, including state-of-the-art TMT-16plex liquid chromatography/mass-spectrometry (LC/MS) proteomics, hybrid MS metabolomics, and flow cytometry panels to determine cell-cycle dynamics and rates of cell death. This pipeline subsequently revealed both common and unique proteome, reactome, and metabolome alterations as a consequence of enviromimetic modeling of narcotic use and neuropsychiatric-related risk factors in tissue of human origin. However, of our 6 treatment groups, human-derived organoids treated with the cannabinoid agonist WIN 55,212-2 exhibited the least convergence of all groups. Single-cell analysis revealed that WIN 55,212-2 increased DNA fragmentation, an indicator of apoptosis, in human-derived dorsal forebrain organoids. We subsequently confirmed induction of DNA damage and apoptosis by WIN 55,212-2 within 3D human-derived dorsal forebrain organoids. Lastly, in a BrdU pulse-chase neocortical neurogenesis paradigm, we identified that WIN 55,212-2 was the only enviromimetic treatment to disrupt newborn neuron numbers within human-derived dorsal forebrain organoids. Cumulatively this study serves as both a resource and foundation from which human 3D biologics can be used to resolve the non-genomic effects of neuropsychiatric risk factors under controlled laboratory conditions. While synthetic cannabinoids can differ from naturally occurring compounds in their effects, our data nonetheless suggests that exposure to WIN 55,212-2 elicits neurotoxicity within human-derived developing forebrain tissue. These human-derived data therefore support the long-standing belief that maternal use of cannabinoids may require caution so to avoid any potential neurodevelopmental effects upon developing offspring in utero.
Subject terms: Schizophrenia, Neuroscience
Introduction
Cortical development is regulated by numerous mechanisms that discretely ensure that a series of temporally ordered events unfold in the correct order, generate the correct cell-types, and ultimately generate morphologically patterned tissue. This requires intact programming that emerges from a wide berth of biological processes, including the expression of specific genes [1] and molecules [2], patterns of sustained metabolic activity [3], the prevention of DNA damage [4], ongoing cell-cycle dynamics [5], regulation of cell survival mechanisms [6, 7], as well as orchestrated cell fate decision making [8]. Should any of these processes become altered during in utero cortical development, neocortical neurogenesis may become attenuated and this may yield developmental disorders, disruptions and/or delays. Thus, in utero brain development remains a critical period of risk for numerous neurodevelopmental disorders [9], including autism [10–12], and schizophrenia [13–16].
While risk for highly penetrant cases of neuropsychiatric illness are considered to predominantly arise from latent genetic risk, epidemiological evidence indicates that environmental factors also contribute risk to neurodevelopmental aberrations that may be linked with disease [9, 11–14]. This includes in utero narcotic and/or substance use, maternal immune activation [9, 13–15], as well as other risk factors such as chronic stress [11, 16]. The effects on the brain of most commonly abused drugs of abuse (e.g., cannabinoids or opiods) and bioactive substances (e.g., nicotine in cigarettes [17]) found in consumer products have typically been studied in adolescent and/or adult systems, models, or participants. However, many of these findings are likely to remain relevant to fetal neurodevelopment. For example, nicotine binds cognate nicotinic acetylcholine receptors which are known to affect neural activity (e.g., spike-timing dependent plasticity [18, 19]) and neuronal survival in rodents [20]. In addition, prenatal nicotine exposure has been associated with spine and other, broader, neuroanatomical changes in rats [21]. Similarly, ethanol (as a proxy for alcohol exposure) modulates cortical neuronal excitability [22–24], progenitor cell proliferation [25–27], cortical neuron migration [28], spine density [29, 30], and can cause Fetal Alcohol Syndrome (FAS) [31]. Similarly, opioids have been suggested to alter prenatal neural proliferation [32], induce alterations in growth factor expression (e.g., BDNF [33]), as well as modulate neurogenesis [33, 34]. Notably, cognate receptors for opiates (for e.g., both µ‐ and κ‐opioid receptors) are expressed by neural stem cells and progenitors [35–38]. Due to increasing opioid abuse within communities, there have been both concomitant increases in methadone treatment of opiate dependence during pregnancy as well as rising cases of Neonatal Abstinence Syndrome (NAS) in offspring [38]. Marijuana has become the most commonly abused drug during pregnancy within the United States [39]. Since the endogenous endocannabinoid system is known to be important for both prenatal and postnatal brain development [40], there is particular urgency in understanding how cannabinoids may regulate neurodevelopment. However, due to both increasing recreational use and legalization efforts, it is important to generate appropriate datasets that provide evidence-based guidance regarding public health practice so not to unnecessarily stigmatize recreational marijuana use where legal. The primary psychoactive and molecular constituent of marijuana is Δ9-tetrahydrocannabinol, which functions via activation of cannabinoid type 1 (CB1) receptors [41]. Indeed, CB1 receptors are found during early phases of brain development [42, 43], and are functional as shown by their activation in response to treatment with the cannabinoid CB1 agonist and mimetic WIN 55,212-2 [44]. Consequently cannabinoid receptors have been implicated in numerous biological pathways essential for fetal brain development such as proliferation, migration, and neuronal synaptogenesis (for comprehensive reviews, see [42, 43]). Likewise, synthetic cannabinoid exposure is also a rising issue amongst pregnant women and may also result in prenatal brain alterations [45]. Therefore, understanding the effects of prenatal drug use in a systematic format specifically within human tissue remains an issue of fundamental public health importance.
Beyond substance use/abuse during pregnancy, there is also a rich literature which indicates that environmental risk factors may modulate neurodevelopment and increase risk of specific disorders [9] including autism [10–12] and schizophrenia [13–16]. The Maternal Immune Activation (MIA) model has consequently become a leading hypothesis for autism and schizophrenia that transects brain development, neurodevelopmental disorders, and psychoneuroimmunology [46, 47]. The mechanisms responsible for neurodevelopmental alterations induced by MIA are diverse and likely involves numerous mechanistic intermediaries [48] that may be both time-specific (i.e., dependent upon the neurodevelopmental timing of insult [49–52]) as well as maturation-dependent (i.e., an age-dependent emergence of a phenotype [53]). Recent work has shown that Interleukin 17a (IL17a) principally mediates the neurodevelopmental effects of MIA on the developing cortex [54]. Specifically, IL17a mediates alterations in cortical neuron numbers and organization as well as autism-related behavior in offspring, which could be prevented via attenuation of IL17a in vivo [54]. Another environment-related risk factor for the developing brain is prenatal stress exposure, which is both common and has been associated with various fetal neurodevelopmental and birth outcomes [54]. Pivotal to this is the ability of stress/trauma to potentially modify the expression of neurodevelopment-related genes [55] including growth factors essential for brain assembly such as BDNF [56]. Indeed, in utero exposure to dexamethasone has been shown to disrupt the radial migration of neurons within the developing cortex [57]. Antenatal glucocorticoid therapy (e.g., with betamethasone) also results in lower whole cortex convolution and smaller brain surface area relative to age-matched infant controls [58]. Glucocorticoid receptors have also been shown to potentially underlie critical periods of stress vulnerability during postnatal cortical development and maturation [56, 59, 60]. Indeed, prenatal stress experiences may modify behavior [61] as well as elevate risk of autism [11], depression [62, 63], anxiety [61, 64], and schizophrenia [16, 65]. However, the signatures associated with in utero stress exposure within human tissue still remain largely unknown due to a longstanding inability to ethically access and manipulate developing human neural tissue.
Human-derived Induced Pluripotent Stem Cell (iPSC) technology now offers the potential to study human neurodevelopmental phenotypes via the generation of self-developing and self-organizing neural tissue [66]. These three-dimensional (3D) cultures, known as organoids, mimic the developing cerebral cortex and are approximate models of trimester one of pregnancy [66]. Importantly, a variety of studies have shown that various 3D organoid models faithfully recapitulate aspects of transcriptional [67], epigenetic [68, 69], and proteome programing [70] of fetal brain development. They also exhibit morphology (e.g., ventricles and ventricular zones, as well as developing cortical plates) [66] and cell-types/cellular diversity that is consistent with early corticogenesis [71–73]. Organoids are also typically enriched for various neuronal progenitor and early-born neuron populations that are consistent with early cortical development [71–73]. Organoids therefore provide a promising platform to ethically study developing neural tissue of human origin under controlled laboratory conditions [74]. As such, 3D human-derived models of the developing brain have become a viable model [75] from which to devolve human-specific mechanisms of brain development [76], evolution [77], and diseases [78] such as autism [79–81] and schizophrenia [70, 82–84]. Therefore, human-derived organoids now provide a platform and method from which to experimentally disentangle the effects of neuropsychiatric risk factors, including drug use, in developing neural tissue of human origin.
Here, we sought to determine the prenatal effects of various drug and neuropsychiatric risk factors on early corticogenesis within human-derived tissue. To do this, we generated 3D dorsal forebrain (cortical) organoids (see Methods and [71]) from human iPSCs, and systematically treated samples with various enviromimetic agents. In tandem experiments conducted in parallel, we modeled exposure to opiates (μ-opioid agonist endomorphin), cannabinoids (WIN 55,212-2), alcohol (ethanol), smoking (nicotine), chronic stress (human cortisol), and maternal immune activation (human IL17a) by chronically treating human-derived organoids for 7 Days In Vitro (DIV) before conducting a range of high-throughput analytical assays. This included 16-Plex Tandem Mass Tag (TMT) Liquid-Chromatography/Mass-Spectrometry (LC/MS) proteomics, state-of-the-art hybrid MS metabolomics, single-cell DNA content analysis for cell-cycle determination, a multi-panel flow cytometry assay for cell death and DNA damage, and a pulse-chase neocortical neurogenesis assay. These analyses revealed both convergent and divergent signatures within human-derived dorsal forebrain organoids between enviromimetic treatment groups, and unbiasedly identified that the cannabinoid agonist WIN 55,212-2 is a particularly noxious agent during cortical development.
Results
Generating 3D forebrain organoids for enviromimetic modeling of narcotic and neuropsychiatry-related risk factors
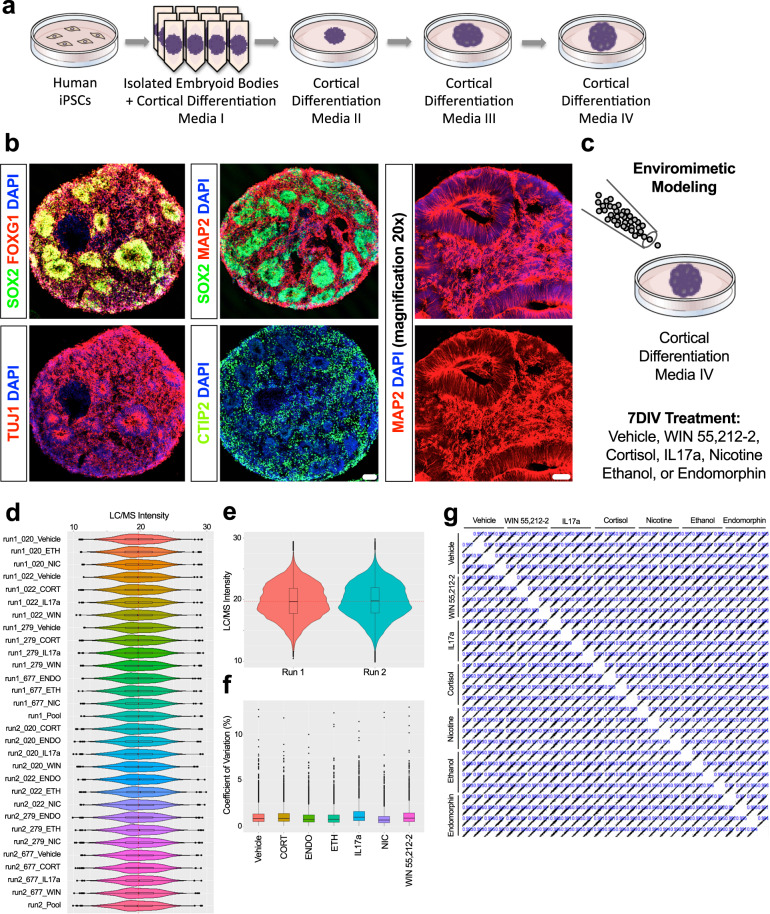
To model human cortical development, we generated self-assembling, self-organizing, and self-maturing 3D organoids that are designed to acquire a dorsal forebrain (i.e., cortical) fate. To do this, we adapted an expedited version [70] of a nascent organoid protocol [71] that reportedly exhibits greater reproducibility than preceding models (e.g., [66]). In rolling pseudorandom quality control assessments of organoids, samples from all donors exhibited evidence of early corticogenesis. This included morphological features such as ventricles, ventricular zones, and evidence of developing cortical fields. Immunostainings consequently confirmed robust neural induction (expression of SOX2 + neural stem cells as well as MAP2 + and TUJ1 + neurons) as well as the acquisition of a forebrain-specific fate (expression of forebrain-specific FOXG1 + progenitors and CTIP2 + early-born cortical neurons). Prototypical morphological features, expected cell-types, and the induction of forebrain-specific cortical transcription factors are shown via whole-organoid Immunostainings in Fig. 1b.
Fig. 1. Enviromimetic forebrain organoids exhibit robust reproducibility.
a Schematic of Human-Derived Dorsal Forebrain Organoid Culturing Pipeline. To generate dorsal forebrain organoids, iPSCs from healthy adult donors were expanded, dissociated into a single-cell suspension, and transferred to ultra-low adherence V-bottom plates. This allowed dissociated iPSCs to proliferate in-suspension to yield highly uniformed (i.e., size constricted) embryoid bodies that were morphologically consistent across wells, plates, and biological donors. In subsequent culturing stages, embryoid bodies were transferred to ultra-low adhesion 6 cm dishes and sequentially transferred through a series of cortical differentiation media (CDM2–4). b Human-Derived Organoids Exhibit Robust Neural Induction and Prototypical Forebrain Developmental Markers Consistent with Early Corticogenesis. As a quality control checkpoint, a pseudorandom assortment of human-derived dorsal forebrain organoids were sampled, drop-fixed, cryosectioned, and immunostained for prototypical forebrain developmental markers. Given the pre-established construct validity of this model, we focused our quality control assessments on the expression of neural stem cells (SOX2 + ; green), forebrain-specific progenitors (FOXG1 + ; red), pan-neuronal markers (TUJ1 + and MAP2 + ; both red in relevant panels), and forebrain-specific early-born neurons (CTIP2 + ; green). This evaluation revealed the expected morphologies of forebrain-specific organoids (e.g., the presence of ventricles, ventricular zones, and developing cortical plates). In addition, all neural-related and forebrain-specific antigens were observed in cultures from all biological donors, indicating the successful assumption of a restricted dorsal forebrain fate. c Schematic of Enviromimetic Treatment Regime. To model exposure to our various narcotic and neuropsychiatric-related enviromimetic risk factors, we chronically treated human-derived dorsal forebrain organoids with a variety of widely-utilized compounds used to study the acute and/or developmental effects of drug exposure and/or environmental risk factors related with mental illnesses. More specifically, this comprised a 7DIV exposure to the cannabinoid receptor agonist WIN 55,212-2, the maternal risk factor IL17a, the human stress hormone cortisol, nicotine, ethanol, and the μ-opioid agonist endomorphin. To ensure consistency across conditions and biological donors, treatments were simultaneously administered to parallel batches of organoids that had been generated from the same originating pool of iPSCs for each human donor. Likewise, all human donor samples were cultured simultaneously and maintained in parallel. d Box plots of Raw TMT-LC/MS Intensities Indicates Robust Reproducibility of 3D Organoid Cultures Across Biological Donors and Replicates. To unbiasedly establish the reproducibility of our culturing pipeline, we adapted cutting-edge Tandem Mass Tag (TMT) chemistry and high-performance liquid-chromatography/mass-spectrometry (LC/MS) to computationally map the molecular composition of human-derived 3D dorsal forebrain organoids from an isobarically-barcoded condensed pool. This real-time detection strategy thus reduces technical noise and variation, and enables rapid high-content sampling of our various treatment conditions. Our TMT-LC/MS dataset identified 40,452 peptides that could be mapped to 5,120 proteins. Based on statistical thresholds and expression levels, 4857 of these proteins could be quantified. Here we present raw TMT-LC/MS intensities for all individual human donors including all possible permutations relating to our enviromimetic treatment conditions. As shown, the distribution of TMT-LC/MS intensities was extremely similar across all groups and biological donors, indicating robust reproducibility of our 3D dorsal forebrain organoids across all biologics and conditions. e Violin Plots of Raw TMT-LC/MS Intensities Reveal No Evidence of Batch Variance. Because of the large number of samples studied here (n = 30 samples in total including pools/internal references, comprising n = 28 human samples for analysis), our TMT-LC/MS analysis had to be split into two 16-plex runs. We therefore sought to determine the reproducibility of our raw TMT-LC/MS datasets across distinct batches of organoids, donors, and treatment conditions. Splitting raw TMT-LC/MS intensities as a function of batch revealed that raw TMT-LC/MS intensities exhibited remarkably similar distributions as denoted in violin plots provided in (1e). f Comparable Coefficients of Variation Across Protein Intensities of All Groups. We next sought to compute coefficients of variation for protein intensities to examine the relative variability within and between our various narcotic and neuropsychiatric-related enviromimetic treatment groups. This analysis once more revealed that all groups exhibited similar coefficients of variation in protein expression, further indicating that our 3D dorsal forebrain cultures remained reproducible even after accounting for divergences in treatment allocation. g Correlation Matrix of All Donors and Groups Confirm Sample Reproducibility. Last, generation of a stringent correlation matrix revealed that there was robust correlation within treatment groups (oftentimes, r2 > 0.99) as expected. In sum, this unbiased statistical analysis confirmed that our culturing pipeline yielded reproducible 3D dorsal forebrain tissue across donors, conditions, and independent batches. For all panels, each donor sample was treated with all experimental compounds. This yielded n = 4 iPSC donors x n = 7 treatment groups for a total n of = 28 experimental samples for LC/MS analysis. Scale bar in b = 100 μm for whole organoid images and 60 μm for ×20 magnification of MAP2 immunostaining.
Enviromimetic modeling of narcotic and neuropsychiatric-related risk factors
To model exposure to our various narcotic and neuropsychiatric-related risk factors, we chronically treated human-derived dorsal forebrain organoids with an array of enviromimetic treatments commonly utilized within preclinical neuropsychiatry research. More specifically, after culturing organoids for 30–35 DIV, human-derived dorsal forebrain organoids were exposed to one of a variety of enviromimetic treatments that was added to CDM4 media for an additional 7 DIV (see schematic in Fig. 1c). This comprised exposure to the cannabinoid receptor agonist WIN 55,212-2, the maternal risk factor IL17a, the human stress hormone cortisol, nicotine, ethanol, or the μ-opioid agonist endomorphin (see “Methods” for more information). After this 7 DIV exposure period, organoids were processed and prepared for one of several streams of analysis including liquid-chromatography/mass spectrometry (LC/MS) proteomic profiling, hybrid MS-based metabolomic profiling, high-throughput flow cytometry for unbiased assessments of apoptosis and DNA-damage, single-cell DNA content analysis for cell-cycle and DNA fragmentation analysis, and/or immunohistochemistry for neuronal quantifications. This approach therefore allowed us to ethically examine the neurodevelopmental effects of narcotic and neuropsychiatric-related risk factors in 3D human-derived forebrain tissue under controlled laboratory conditions.
Establishing the reproducibility of 3D human-derived dorsal forebrain organoids
Before conducting experiments, we first sought to ensure that our human-derived dorsal forebrain organoids exhibited sufficient reproducibility across all treatment conditions and donors. In two separate TMT 16-plex pools, a total of 30 samples (comprising n = 28 experimental samples and n = 2 internal references/pools for data normalization) were barcoded with TMT reagents, condensed into a single sample, and subjected to simultaneous detection via LC/MS. In total, this approach identified 40,452 peptides with average sequence coverage of 20.8%. These peptides were subsequently mapped to 5120 proteins of which 4857 could be quantified (94.86% of proteins identified were quantifiable). Of these proteins, 3711 were common to all donor and treatment samples and required no further imputation for statistical analysis.
To ensure that our forebrain enviromimetic organoid model exhibited sufficient reproducibility across treatment conditions, we first conducted a statistical analysis of sample variation. Visual inspection of all raw data points per individual sample revealed that all samples exhibited similar LC/MS intensity distributions for detected proteins (Fig. 1d). To evaluate batch effects data were also split by each TMT-LC/MS run and graphed as violin plots. No evidence of technical batch variation was detected when examining raw TMT-LC/MS intensities for all 28 experimental samples (Fig. 1e). An analysis of treatment-group variation also revealed no evidence of large deviations in coefficients of variation when all data points were examined and mapped as boxplots split by conditions (Fig. 1f). Coefficients of proteome variation revealed that sample variance was less than 1% of the median within treatment groups, and exhibited a <10% range. In addition, generation of a stringent correlation matrix revealed that there was robust similarity within treatment groups (r2 > 0.99) as expected (Fig. 1g). In sum, this statistical analysis confirmed that our culturing pipeline yielded reproducible 3D dorsal forebrain tissue across donors, conditions, and independent batches.
Developmental alterations within the proteome of human-derived organoids following treatment with narcotic and neuropsychiatry-related enviromimetics
We next sought to statistically identify novel molecular alterations between and within our 6 different narcotic and neuropsychiatric risk factor treatment groups. To do this, we first clustered our samples based on a structural equation modeling approach that yielded a principal components solution based on the expression of 3,711 proteins. To unbiasedly identify an effect of treatment group on protein expression, we utilized this dataset for all further downstream analyses for novel factor identification.
A global analysis of all TMT-LC/MS intensities, which did not stratify individual group for a priori hypothesized effects, revealed that 175 proteins differed in at least one group within our total dataset (Fig. 2a; see also Supplementary Table 1). Of these 175 proteins, a stratified analysis revealed that only 41 proteins were specifically different in our 6 enviromimetic treatment groups relative to our vehicle-treated control group (Fig. 2b). In this total pool, WIN 55,212-2 exhibited the most distinct expression profile and did not cluster further with the other narcotic treatment groups. Contrary to this, our neuropsychiatry enviromimetics (both IL17a and cortisol) exhibited similar overall proteome differences and clustered together, whereas nicotine, ethanol, and endomorphin were more similar than any other combination of groups (Fig. 2a-b). Further examination of the 41 differentially expressed proteins identified in the total data pool yielded several notable observations. This list revealed enrichment for factors involved in hypoxia (e.g., hypoxia up-regulated protein 1, or HYOU1), cellular stress responses (e.g., the heatshock proteins HSPA13 and HSPA9), and amyloid-related proteins. Notably, this included amyloid-like protein 2 (APLP2) and the amyloid beta precursor protein (APP). There was also enrichment for a novel growth factor (i.e., mesencephalic astrocyte-derived neurotrophic, or MANF) and proteins localized to, or involved in the functioning of, mitochondria and/or regulation of other cellular metabolic functions (see Table 1). A complete list of these 41 proteins identified in our global/unbiased analysis are listed within Table 1 and, for brevity, a summary of common targets is provided for each individual group in Table 2. Notably, the cannabinoid agonist WIN 55,212-2 and the μ-opioid agonist endomorphin tended to exhibit a more similar overlap in proteome targets, whereas our nicotine, ethanol and cortisol treatment groups tended to be more similar in their combination of proteins targeted (see Table 2).
Fig. 2. Proteome and reactome pathway alterations in human-derived forebrain organoids across narcotic and neuropsychiatric-risk enviromimetic treatment groups.
a–c Statistically unsupervised analysis of TMT-LC/MS proteomics comprising all samples. All 28 of our TMT-LC/MS samples were first subjected to a statistically unbiased analysis that did not consider a priori hypotheses or the quasi-independent nature of each of our treatment conditions. This yielded a clustering solution (a) that identified differential expression of 41 specific proteomic targets that differed from vehicle controls (b). These proteins were correspondingly mapped to their Reactome pathways of origin (c). Note, the ordering of groups for presentation purposes are assigned based on the results of statistically unsupervised clustering. d–i Reactome pathway analysis split by narcotic and neuropsychiatric-related treatment. Analysis of the Reactome pathways split by our individual treatments revealed novel enrichment patterns for each narcotic compound and enviromimetic treatment conditions. As described in-text, WIN 55,212-2, IL17a, and cortisol exhibited the most distinct Reactome pathway patterning, indicating a greater degree of divergence in pathway alterations. Contrary to this, our nicotine, ethanol, and endomorphin treatment groups tended to exhibit a closer degree of convergence. Specifically, these groups tended to exhibit greater enrichment for neurodevelopment-related Reactome pathways including factors mapped to “nervous system development”, “ROBO receptor signaling”, and/or “axon guidance”. However, each of these groups still maintained unique patterns of Reactome pathway enrichment, which perhaps indicates a common role in their ability to alter neurodevelopmental factors albeit via divergent intermediaries. For all panels, each donor sample was treated with all experimental compounds (4 iPSC donors x 7 groups = 28 total samples for analysis).
Table 1.
Top differentially expressed proteins (relative to Ctrls) detected via TMT-LC/MS.
| Short protein name | Long protein name | Uniprot accession | ANOVA p value |
|---|---|---|---|
| NDUFAF2 | NADH Dehydrogenase (Ubiquinone) 1 Alpha Subcomplex Assembly Factor 2 | Q8N183 | 0.000481025 |
| TMEM50B | Transmembrane Protein 50B | P56557 | 0.000548505 |
| IMPA1 | Inositol Monophosphatase 1 | P29218 | 0.001902383 |
| ARMCX1 | Armadillo Repeat-Containing X-Linked Protein 1 | Q9P291 | 0.002196252 |
| GBF1 | Golgi-Specific Brefeldin A-Resistance Guanine Nucleotide Exchange Factor 1 | Q92538-3 | 0.002520107 |
| HDHD5 | Haloacid Dehalogenase-Like Hydrolase Domain-Containing 5 | Q9BXW7-2 | 0.00307041 |
| GTPBP4 | Nucleolar GTP-Binding Protein 1 | Q9BZE4 | 0.00324609 |
| ARF4 | ADP-Ribosylation Factor 4 | P18085 | 0.003766362 |
| GRPEL1 | GrpE Protein Homolog 1, Mitochondrial | Q9HAV7 | 0.004413199 |
| ACAT2 | Acetyl-CoA Acetyltransferase, Cytosolic | Q9BWD1 | 0.004730022 |
| MANF | Mesencephalic Astrocyte-Derived Neurotrophic Factor | P55145 | 0.005179981 |
| DOCK4 | Dedicator of Cytokinesis Protein 4 | Q8N1I0–2 | 0.00538423 |
| TXNRD1 | Thioredoxin-Disulfide Reductase | E9PIR7 | 0.005572821 |
| DIABLO | Diablo Homolog, Mitochondrial | Q9NR28-2 | 0.00627476 |
| HSPA13 | Heat Shock 70 kDa Protein 13 | P48723 | 0.00627476 |
| TFCP2 | Alpha-Globin Transcription Factor CP2 | Q12800-2 | 0.00639284 |
| DYNLT1 | Dynein Light Chain Tctex-Type 1 | P63172 | 0.00671083 |
| HDGF | Hepatoma-Derived Growth Factor | P51858 | 0.007032222 |
| PSMB7 | Proteasome Subunit Beta Type-7 | Q99436 | 0.007289525 |
| UNK | RING Finger Protein Unkempt Homolog | Q9C0B0 | 0.008045913 |
| HYOU1 | Hypoxia Up-Regulated Protein 1 | Q9Y4L1 | 0.008687815 |
| SEC62 | Translocation Protein SEC62 | Q99442 | 0.010325685 |
| APLP2 | Amyloid-Like Protein 2 | Q06481-4 | 0.013408141 |
| USP7 | Ubiquitin Carboxyl-Terminal Hydrolase 7 | Q93009-3 | 0.015504379 |
| KATNAL1 | Katanin p60 ATPase-Containing Subunit A-Like 1 | Q9BW62 | 0.015622147 |
| HSPA5 | Endoplasmic Reticulum Chaperone BiP | P11021 | 0.015742232 |
| DYNC1LI1 | Cytoplasmic Dynein 1 Light Intermediate Chain 1 | Q9Y6G9 | 0.016475164 |
| ABCF1 | ATP-Binding Cassette Sub-Family F Member 1 | Q8NE71-2 | 0.016611257 |
| SHMT2 | Serine Hydroxymethyltransferase, Mitochondrial | P34897-3 | 0.018482781 |
| DHTKD1 | Probable 2-Oxoglutarate Dehydrogenase E1 Component DHKTD1, Mitochonondrial | Q96HY7 | 0.018748869 |
| SLC7A1 | High Affinity Cationic Amino Acid Transporter 1 | P30825 | 0.021172737 |
| PIK3C2B | Phosphatidylinositol 4-Phosphate 3-Kinase C2 Domain-Containing Subunit Beta | O00750 | 0.021899148 |
| SORBS1 | Sorbin and SH3 Domain-Containing Protein 1 | Q9BX66-4 | 0.024998617 |
| HSPA9 | Stress-70 Protein, Mitochondrial | P38646 | 0.027136132 |
| CCDC47 | Coiled-Coil domain-Containing Protein 47 | Q96A33 | 0.028644274 |
| ALG2 | Alpha-1,3/1,6-Mannosyltransferase ALG2 | Q9H553 | 0.030668953 |
| APP | Amyloid-Beta Precursor Protein | P05067-11 | 0.031024857 |
| SUPT6H | Transcription Elongation Factor SPT6 | Q7KZ85 | 0.033263913 |
| RRP12 | RRP12-Like Protein | Q5JTH9-2 | 0.033924326 |
| MLF2 | Myeloid Leukemia Factor 2 | Q15773 | 0.043882525 |
| RPS13 | 40 s Ribosomal Protein S13 | P62277 | 0.047438594 |
Table 2.
Common proteome alterations detected via global analysis of TMT-LC/MS data.
| Enviromimetic drug/risk factor treatment group | |||||||
|---|---|---|---|---|---|---|---|
| Protein name | Uniprot accession | WIN | IL17a | CORT | Nicotine | Ethanol | Endomorphin |
| ADD1 | P35611 | ↓ | ↑ | ||||
| POLR3C | Q9BUI4 | ↑ | ↑ | ||||
| TMEM50B | P56557 | ↑ | ↓ | ||||
| NELFCD | Q8IXH7 | ↑ | ↑ | ||||
| KPNA6 | O60684 | ↑ | ↑ | ||||
| MTX2 | Q75431 | ↑ | ↑ | ||||
| CLPTM1 | Q96005 | ↑ | ↑ | ||||
| APP | P05067 | ↑ | ↑ | ||||
| FXR2 | P51116 | ↑ | ↑ | ↑ | ↑ | ||
| TFCP2 | Q12800 | ↑ | ↑ | ↑ | ↑ | ↑ | |
| SUPT6H | Q7KZ85 | ↑ | ↑ | ||||
| APLP2 | Q06481 | ↑ | ↑ | ↑ | |||
| USP7 | Q93009 | ↑ | ↑ | ↑ | ↑ | ||
| TIMM21 | Q9BVV7 | ↑ | ↑ | ||||
| APMAP | Q9HDC9 | ↑ | ↑ | ↑ | ↑ | ||
| SRP19 | P09132 | ↑ | ↓ | ||||
| RPS27L | Q71UM5 | ↓ | ↓ | ||||
| LSM3 | P62310 | ↑ | ↑ | ↑ | ↑ | ||
| TNPO3 | Q9Y5L0 | ↑ | ↑ | ↑ | ↑ | ||
| SAP30BP | Q9UHR5 | ↑ | ↑ | ↑ | |||
| APLP2 | Q06481 | ↑ | ↑ | ↑ | |||
| TMEM209 | Q96SK2 | ↑ | ↑ | ↑ | |||
| RPL36 | Q9Y3U8 | ↓ | ↓ | ↓ | ↓ | ||
| RPS13 | P62277 | ↓ | ↓ | ↓ | ↓ | ||
| CDC42BPB | Q9Y5S2 | ↑ | ↑ | ↑ | ↑ | ||
| FMNL2 | Q96PY5 | ↑ | ↑ | ↑ | ↑ | ||
| HDHD5 | Q9BXW7 | ↑ | ↑ | ↑ | ↑ | ||
| ZFYVE1 | Q9HBF4 | ↑ | ↑ | ↑ | ↑ | ||
| ARMCX1 | Q9P291 | ↑ | ↑ | ↑ | ↑ | ||
| DHTKD1 | Q96HY7 | ↑ | ↑ | ↑ | ↑ | ||
Because each of our treatment groups represent quasi-independent studies, the proteins identified in our global/unbiased analysis of the dorsal forebrain organoid proteome did not comprise specific pairwise comparisons between individual groups and vehicle-treated controls. This is of note, as this was the primary aim of the current study. To address this, we conducted a further analysis of our TMT-LC/MS intensities stratified specifically by each of our individual treatment groups. In fact, this further analysis of group-by-group differences led us to identify a greater number of differentially expressed proteins (see also Supplementary Table 2). Specifically, we identified 422 distinct proteins that significantly differed in their expression levels across various treatment conditions when each was independently compared against our vehicle-treated control organoids (see also Supplementary Table 3). Broadly, we found that our narcotic mimetic treatments tended to yield a broader spectrum of protein expression changes (endomorphin: n = 196 proteins, nicotine: n = 131 proteins, ethanol: n = 149 proteins, and WIN 55,212-2: n = 84 proteins, respectively). Contrary to this, our “environmental” neuropsychiatric risk factor treatments (cortisol: n = 49 proteins, and IL17a n = 19 proteins, respectively) tended to exhibit fewer overall protein alterations within human-derived dorsal forebrain organoids. A list of differentially expressed proteins in group-segregated pairwise analysis is provided in Table 3. Many of these proteins were differentially expressed across two or more treatment groups, and these common alterations are further summarized in Supplementary Table 4. However, each of our narcotic and neuropsychiatric-related treatments also resulted in unique, non-overlapping, proteome alterations (see Table 3 and Supplementary Table 4).
Table 3.
Group-specific protein alterations via pairwise analysis of TMT-LC/MS data.
| Enviromimetic drug/risk factor treatment group | ||||||
|---|---|---|---|---|---|---|
| WIN 55,212-2 | IL17A | Cortisol | Nicotine | Ethanol | Endomorphin | |
| ↑ TXNRD1 | ↑ GPR89A | ↑ NDUFB9 | ↑ ROBO1 | ↑ ELOF1 | ↑ MTHFD1L | ↑ EFNB3 |
| ↑ UBA6 | ↑ SUB1 | ↑ PBDC1 | ↑ FSD1L | ↑ MPP2 | ↑ MAP4K4 | ↑ NEDD8 |
| ↑ AT2C1 | ↑ DR1 | ↑ RBM28 | ↑ ELAVL2 | ↑ PHF14 | ↑ MYO18A | ↑ TBC1D9B |
| ↑ TRMT10A | ↑ GATD1 | ↑ FLYWCH2 | ↑ CAMK2G | ↑ NELFB | ↑ TSEN34 | ↑ SLC27A4 |
| ↑ TSPAN3 | ↓ NEBL | ↑ ROCK2 | ↑ CNTN2 | ↑ PAK1 | ↑ GATAD2B | ↑ EDC4 |
| ↑ YIPF4 | ↓ SUPT4H1 | ↑ NME7 | ↑ CHCHD3 | ↑ PPP4R2 | ↑ EIPR1 | ↑ LARP4 |
| ↑ TIMM17B | ↓ DIS3 | ↑ RIF1 | ↑ NDRG3 | ↑ NRCAM | ↑ PPIL3 | ↑ TAOK1 |
| ↑ CST3 | ↑ SLC16A2 | ↑ GGA3 | ↑ CLASP2 | ↑ AARSD1 | ↑ BPHL | |
| ↑ GSPT1 | ↑ ABCB6 | ↑ DCAKD | ↑ DCTN6 | ↑ MRPL39 | ↑ SBF2 | |
| ↑ CPE | ↑ ZNF579 | ↑ PRKRA | ↑ CTNND2 | ↑ ARVCF | ↑ ZC3H7A | |
| ↑ ARF4 | ↑ PDLIM3 | ↑ GNAI3 | ↑ CFL1 | ↑ NEFM | ↑ PTGR2 | |
| ↑ MMUT | ↑ GCC2 | ↑ RBM25 | ↑ HS1BP3 | ↑ THY1 | ↑ UBA3 | |
| ↑ SHMT2 | ↑ NDUFA2 | ↑ EIF2B2 | ↑ YLPM1 | ↑ TSG101 | ↑ FAM172A | |
| ↑ NAA10 | ↓ ARMC9 | ↑ MCAM | ↑ MACF1 | ↑ PRKAG1 | ↑ UBTD2 | |
| ↑ TUFM | ↓ INTS9 | ↑ NOVA1 | ↑ MPRIP | ↑ LRRC57 | ↑ RTF1 | |
| ↑ SLC25A3 | ↓ VIRMA | ↑ EIF5 | ↑ DOHH | ↑ EPS15L1 | ↑ CLCC1 | |
| ↑ CPSF1 | ↓ NUP210 | ↑ CDK16 | ↑ DAZAP1 | ↑ AGRN | ↑ TRIM2 | |
| ↑ TRIP12 | ↑ INA | ↑ DPYSL4 | ↑ PSMD14 | ↑ EGLN1 | ||
| ↑ SART3 | ↑ CIAPIN1 | ↑ SPTBN2 | ↑ SDCBP | ↑ FAM234A | ||
| ↑ WDR43 | ↑ DHX57 | ↑ KIF5C | ↑ TRAFD1 | ↑ WDR13 | ||
| ↑ DHCR24 | ↑ UBE2R2 | ↑ RTN3 | ↑ UQCRQ | ↑ DPH5 | ||
| ↑ PCID2 | ↑ TRMT10C | ↑ CTPS1 | ↑ PLXNB2 | ↑ MYG1 | ||
| ↑ METTL3 | ↑ NIT1 | ↑ CTNNA2 | ↑ ZW10 | ↑ HMG20A | ||
| ↑ NDUFAF2 | ↑ TTC5 | ↑ ABCD3 | ↑ NIPSNAP2 | ↑ PHPT1 | ||
| ↑ RDH11 | ↑ TNPO1 | ↑ CARS1 | ↑ PDCD6 | ↑ SH3BP4 | ||
| ↑ GBF1 | ↑ RSF1 | ↑ PEX3 | ↑ GCAT | ↑ UQCR10 | ||
| ↑ EXOC4 | ↑ GAN | ↑ TBL3 | ↑ ATRN | ↑ VPS28 | ||
| ↑ GTPBP4 | ↑ ACTR10 | ↑ DBN1 | ↑ TTC4 | ↑ VPS4A | ||
| ↑ NIF3L1 | ↑ SACS | ↑ TBCEL | ↑ GNAI2 | ↑ PACSIN2 | ||
| ↑ GHITM | ↑ COPG2 | ↑ CADM2 | ↑ EIF2S1 | ↑ PSMD13 | ||
| ↑ EXD2 | ↑ CNOT11 | ↑ AMER2 | ↑ NEFL | ↑ RABGAP1 | ||
| ↑ BPNT2 | ↑ MAGED1 | ↑ GDAP1 | ↑ GALT | ↓ SELENOH | ||
| ↑ YARS2 | ↓ TTC28 | ↑ DTD1 | ↑ CNP | ↓ EIF1AD | ||
| ↑ RRP15 | ↓ MTAP | ↑ CLIP2 | ↑ G6PD | ↓ RPL28 | ||
| ↑ AUP1 | ↓ DOCK4 | ↑ UBQLN2 | ↑ FDPS | ↓ DVL2 | ||
| ↓ DOCK1 | ↓ SYNCRIP | ↑ DCTN4 | ↑ EIF2AK2 | ↓ RPL19 | ||
| ↓ ACSS3 | ↓ PDIA6 | ↑ NOVA2 | ↑ RAB6A | ↓ RPL4 | ||
| ↓ MEX3A | ↓ PDCD4 | ↓ RPS9 | ↑ ATP2B4 | ↓ ARL1 | ||
| ↓ ENSA | ↓ DIPK2A | ↓ SENP3 | ↑ OTX1 | ↓ RPL5 | ||
| ↓ FAM171A2 | ↓ NAA50 | ↑ CRAT | ↓ RPL34 | |||
| ↓ PDCD10 | ↓ RPL15 | ↑ CRK | ↓ NUP107 | |||
| ↓ ATOX1 | ↓ PLOD2 | ↑ CRKL | ↓ RPL23A | |||
| ↓ SORBS2 | ↓ MEIS2 | ↑ BRCC3 | ↓ PPP2CA | |||
| ↓ STX8 | ↓ ATP1A1 | ↑ DHPS | ↓ HNRNPD | |||
| ↓ TLK2 | ↓ S100A6 | ↑ IST1 | ↓ NUFIP2 | |||
| ↓ MIF | ↓ FBL | ↑ CACNA2D1 | ↓ BTF3L4 | |||
| ↓ PPT1 | ↓ FKBP2 | ↑ RAB5B | ↓ API5 | |||
| ↓ CNN3 | ↓ NMT1 | ↑ COPS2 | ↓ LSM6 | |||
| ↓ HMGN3 | ↓ TSFM | ↑ NCALD | ↓ LAS1L | |||
| ↓ PKN2 | ↓ TPMT | ↑ SNRPF | ||||
| ↓ LGALSL | ↓ CASP7 | ↑ RAC1 | ||||
| ↓ ZC3H13 | ↓ NUTF2 | ↑ VAC14 | ||||
| ↓ MTDH | ↓ RPL11 | ↑ TRAF2 | ||||
| ↓ LSM2 | ↓ DDI2 | ↑ IFIT5 | ||||
| ↓ ISOC1 | ↑ HNRNPD | |||||
| ↓ SFXN2 | ↑ UBE4A | |||||
| ↑ ELAVL3 | ||||||
| ↑ LAGE3 | ||||||
| ↑ CRYM | ||||||
In sum, only a modest number of proteins within the developing cortical proteome were found to be robustly responsive to our various narcotic and neuropsychiatric-related treatments. In addition, we discovered that there were various degrees of overlap between treatment conditions in human-derived dorsal forebrain organoids. This implicates that the differential expression of a common ensemble of proteome factors may partially underscore some of the molecular dysfunction induced by our enviromimetic treatments during early human brain development.
Reactome modeling reveals biological pathways altered by enviromimetic treatments in human-derived organoids
To gain insight into the specific pathways and biological functions associated with our narcotic and neuropsychiatric-related treatments, we next applied Reactome pathway analysis to this list of candidate differentially expressed factors identified via TMT-LC/MS proteomics. This global analysis revealed that these proteins mapped to biological functions comprising IP2/IP and Ca2+ regulation, cellular responses to heat shock stress, cargo transport pathways including trafficking in and out of the Golgi apparatus, as well as pathways related to protein metabolism and mitochondrial-related (e.g., protein import) functionality (Fig. 2c). However, broadly speaking, a more targeted approach was necessary to statistically parse the individual contributions of specific treatment conditions.
As expected, parsing these differentially regulated proteins for alterations in Reactome pathways differences between our various narcotic and neuropsychiatric enviromimetic treatments revealed group-specific effects. A complete summary of differentially regulated biological processes and pathways between groups is visualized in Fig. 2d-i. Examination of top differentially enriched pathways revealed that human-derived dorsal forebrain organoids treated with the cannabinoid receptor agonist WIN 55,212-2 exhibited altered golgi apparatus VxPx cargo-targeting and trafficking to the cilium and periciliary membrane (FDR = 0.005 and 0.08, respectively; see Fig. 2d). The cilium functions as its own compartmentalized organelle, and is a region where membrane proteins become natively concentrated within the cell [85] that is also required for Sonic Hedgehog (Shh) signal transduction [86]. Not unexpectedly, cilium-related function and transport of membrane proteins has been implicated in both neurodevelopment and disease [86]. Contrary to this, the human maternal immune risk factor IL17a (Fig. 2e) was notably defined by enrichment for RAS processing (FDR = 0.002), which is important for regulation of the brain’s angiotensin system [87]. In addition, IL17a treatment resulted in enrichment for estrogen-stimulated signaling through Protein Kinase C (PKC) Zeta (FDR = 0.002; see Fig. 2e). PKCZ has a suggested role in regulating cell polarity during migration within developing neuroblasts [88] and anterior-posterior axon guidance derived from WNT and Pl3K signaling [89], indicating this pathway may influence normative neurodevelopmental processes within the developing forebrain. Chronic cortisol treatment (Fig. 2f) in human-derived dorsal forebrain organoids yielded enrichment for SRP-mediated protein translation within the endoplasmic reticulum as well as the recruitment of mitotic centrosome proteins and complexes. While the entity p values for these pathways remained low (p = 0.001 and 0.005, respectively) the FDR scores for these cortisol-related pathways were higher than for alterations observed in other groups (FDR = 0.133 for both pathways). Treatment of organoids with nicotine (Fig. 2g) yielded enrichment for numerous canonical pathways essential for normal cortical development, including axon guidance (FDR = 0.009), regulation of RHO/RAS Guanosine Triphosphatases (GTPases; FDR = 0.009), ROBO signaling (FDR = 0.009), and the initiation of eukaryotic protein translation (FDR = 0.01). Similarly, treatment of human-derived organoids with ethanol (Fig. 2h) also revealed enrichment for ROBO receptor signaling (FDR = 6.25E–05), which is important for cortical neurogenesis [90] and axonal guidance [91], as well as an array (~26 proteins) that have been broadly mapped as “nervous system development” factors (FDR = 3.61E–05). Ethanol also exhibited enrichment for cellular stress response factors (FDR = 0.002), Nonsense-Mediated mRNA Decay (NMD; FDR = 0.004), developmental factors (FDR = 0.01), and the regulation of apoptosis (FDR = 0.03). Last, treatment of human-derived dorsal forebrain organoids with the μ-opioid agonist endomorphin (Fig. 2i) also revealed enrichment for ROBO receptor signaling (FDR = 3.34E–05), RNA metabolism factors (FDR = 7.04E–05), exon-enhanced NMD of mRNA transcripts (FDR = 5.05E–04), cellular stress response factors (FDR = 7.02E–04), central nervous system development factors (FDR = 0.0027), and axon guidance factors (FDR = 0.003).
Similar to our analysis of individual proteomic targets, a comparison of Reactome pathway enrichment between our various treatment groups also revealed several noteworthy similarities. Namely, our ethanol, endomorphin, and WIN 55,212-2 treatment groups exhibited enrichment for various mRNA stability and degradation pathways (notably, NMD and 5ʹ to 3ʹ exoribonuclease mRNA degradation pathway factors). Similarly, treatment of human-derived dorsal forebrain organoids with ethanol, endomorphin, cortisol, and nicotine yielded enrichment for rRNA processing factors within the nucleolus and cytosol. Several groups also exhibited differential recruitment of pathways related to protein translation (e.g., nicotine, cortisol), as well as central nervous system development, ROBO receptor signaling, and/or axon guidance (e.g., ethanol, endomorphin, and nicotine). It was also not uncommon for groups to exhibit various degrees of enrichment for pathways involved in mitosis or cell cycle checkpoint activity (e.g., ethanol, endomorphin, and cortisol) as well as pathways involved in cellular stress, apoptosis, or hypoxia (e.g., ethanol, endomorphin cortisol, and nicotine). Of our 7 groups, the most distinct profile belonged to human-derived organoids treated with the cannabinoid agonist WIN 55,212-2, followed by organoids treated with human IL17a and cortisol. Contrary to this, our ethanol, endomorphin, and nicotine groups exhibited a greater number of similarities than any other combination of groups, indicating partially convergent molecular effectors and potentially similar pathways of action within 3D human-derived forebrain organoids.
Metabolomic signatures of narcotic and neuropsychiatric enviromimetic treatments within human-derived forebrain organoids
Across our various proteomic datasets, many of our treatment groups exhibited enrichment for individual protein factors and/or Reactome pathways that have known roles in cellular metabolic and/or bioenergetic function. Therefore, to provide further unbiased systems level analysis of our enviromimetic treatments in human-derived dorsal forebrain organoids, we adapted a targeted hybrid metabolomics panel to map the developing organoid metabolome.
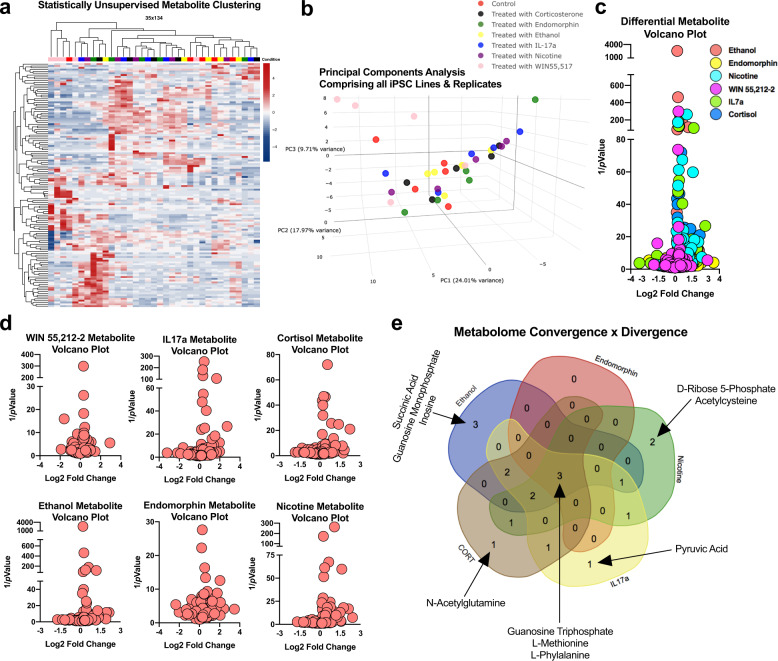
First, imputed sample values were clustered via statistically unsupervised analysis to yield an unbiased visualization of drug and enviromimetic treatment effects (Fig. 3a). Clustering was subsequently cross-validated via another statistically unsupervised analysis that segregated samples into principal components within the broader dataset (Fig. 3b). Both of these unbiased analyses revealed that the metabolome was subtly altered relative to vehicle-treated controls. In subsequent group analyses, data were imputed against metabolite expression patterns in control samples, and analyzed for individual group differences based on Log2 fold change and p values (Fig. 3c–e). A summary of Log2Fold differences and p values, as well as similarities and differences in differentially expressed metabolites, are provided in Tables 4–5. Our analysis consequently led us to identify both the convergence of enviromimetic organoids upon alterations in certain metabolites, as well as unique divergences between groups. Typically speaking, there were 10–11 significantly altered metabolites in most groups. The exception was our endomorphin and WIN 55–212,2 treatment groups, which exhibited an alteration in just 3 factors which were also commonly altered in all other groups (i.e., these treatments did not exhibit any unique differentially expressed metabolites). This included significant alterations in the expression of amino acids (WIN 55–212,2: L-Tyrosine & L-Valine; Endomorphin: L-Methionine) and, in both groups, L-Phenylalanine.
Fig. 3. Hybrid metabolomics reveals novel metabolite alterations within enviromimetic forebrain organoids.
a, b Statistically unsupervised analysis of treatment-induced metabolites. The metabolome of dorsal forebrain organoids exposed to our narcotic and neuropsychiatric-related treatments were first analyzed and in a statistically unsupervised format. This involved unbiased clustering of the metabolomic profiles of all samples (a). In addition, a statistically unsupervised principal components analysis was also conducted to visualize the distinctness of our various treatments within human-derived dorsal forebrain organoids (b). Note, the ordering of groups for presentation purposes are presented differ here due to the hierarchical relationships present between samples in these statistically-unsupervised clustering/principal components analyses. c, d Metabolomic Log2FC alterations visualized as split-axis volcano plots. Metabolic factors were next analyzed as a function of both their Log2FC and statistical significance. This yielded a global metabolite profile for all treatment conditions (see global volcano plot in (c)). Differential metabolite expression was subsequently split as a function of treatment group and was visualized as group-specific volcano plots that had been statistically compared against the metabolite profile of vehicle-treated controls (d). e Metabolome convergence and divergence across narcotic and neuropsychiatric-related risk factors in dorsal forebrain organoids. Further analysis of metabolomes revealed both common and unique metabolite alterations between human-derived dorsal forebrain organoids exposed to our different treatment conditions. This included differential expression of L-Phenylalanine in all treatment groups. The neurodevelopment-related metabolite Guanosine Triphosphate (GTP) was also differentially expressed in all treatment groups except those exposed to WIN 55,212-2. Because WIN 55,212-2 exhibited no further commonalities with our other groups, only the remaining 5 narcotic and neuropsychiatric-related treatment conditions were visualized (e). As discussed in-text, further analysis revealed that treatment of human-derived dorsal forebrain organoids with IL17a altered pyruvic acid expression, cortisol altered N-Acetylglutamine, nicotine altered D-Ribose 5-Phosphate and Acetylcysteine, and ethanol treatment yielded altered succinic acid, guanosine monophosphate, and inosine. Further discussion of metabolite data is provided in-text. Note that (a) and (b) share the same group legend, which is presented in (b). For all panels, each donor sample was treated with all experimental compounds. This yielded n = 5 iPSC donors x n = 7 treatment groups for a total n of = 35 experimental samples for LC/MS analysis.
Table 4.
Differentially expressed metabolites in human enviromimetic organoids.
| Enviromimetic drug/Risk factor treatment group | ||||||
|---|---|---|---|---|---|---|
| WIN | IL17a | CORT | Nicotine | Ethanol | Endomorphin | |
| 3-Phosphoglyceric Acid | ↓ | ↓ | ||||
| Acetylcysteine | ↓ | |||||
| Allantoin | ↓ | ↓ | ||||
| Creatinine | ↓ | ↓ | ↓ | ↓ | ||
| D-Ribose 5-Phosphate | ↓ | |||||
| Guanosine Monophosphate | ↓ | |||||
| Guanosine Triphosphate | ↑ | ↑ | ↑ | ↑ | ↑ | |
| Inosine | ↓ | |||||
| L-Leucine | ↓ | ↓ | ↓ | |||
| L-Methionine | ↓ | ↓ | ↓ | ↓ | ↓ | |
| L-Phenylalanine | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ |
| L-Tyrosine | ↓ | ↓ | ↓ | ↓ | ↓ | |
| L-Valine | ↓ | ↓ | ↓ | ↓ | ||
| N-Acetylglutamine | ↓ | |||||
| N-Acetylornithine | ↓ | ↓ | ↓ | |||
| Pyruvic Acid | ↓ | |||||
| S-Adenosylhomocysteine | ↓ | ↓ | ||||
| Succinic Acid | ↓ | |||||
Table 5.
Statistics for metabolites altered across human-derived organoid groups.
| Enviromimetic Drug/Risk factor treatment group | ||||||
|---|---|---|---|---|---|---|
| WIN | IL17a | CORT | Nicotine | Ethanol | Endomorphin | |
| Log2FC, p Value | Log2FC, p Value | Log2FC, p Value | Log2FC, p Value | Log2FC, p Value | Log2FC, p Value | |
| 3-Phosphoglyceric Acid | −2.72,0.03 | −2.23, 0.04 | ||||
| Acetylcysteine | −0.74,0.04 | |||||
| Allantoin | −0.66,0.02 | −0.65,0.01 | ||||
| Creatinine | −0.36,0.01 | −0.22, 0.03 | −0.27,0.01 | −0.23, 0.02 | ||
| D-Ribose 5-Phosphate | −1,06, 0.003 | |||||
| Guanosine Monophosphate | −0.61, 0.005 | |||||
| Guanosine Triphosphate | Inf,0.04 | Inf,0.04 | Inf,0.04 | Inf,0.04 | Inf,0.04 | |
| Inosine | −1.19, 0.008 | |||||
| L-Leucine | −0.26,0.02 | −0.17, 0.02 | −0.20, 0.01 | |||
| L-Methionine | −0.45,0.003 | −0.39, 0.02 | −0.33,0.01 | −0.38, 0.007 | −0.27,0.03 | |
| L-Phenylalanine | −0.30, 0.003 | −0.32,0.007 | −0.27, 0.02 | −0.25, 0.005 | −0.26, 0.002 | −0.21,0.04 |
| L-Tyrosine | −0.27, 0.013 | −0.30,0.005 | −0.22, 0.02 | −0.22,0.03 | −0.19, 0.0004 | |
| L-Valine | −0.31, 0.03 | −0.19,0.04 | −0.13,0.04 | −0.15, 0.02 | ||
| N-Acetylglutamine | −0.85, 0.03 | |||||
| N-Acetylornithine | −0.51,0.03 | −0.56, 0.01 | −0.47, 0.04 | |||
| Pyruvic Acid | −1.68,0.009 | |||||
| S-Adenosylhomocysteine | −1.39, 0.04 | −1.61,0.01 | ||||
| Succinic Acid | −0.43, 0.03 | |||||
∞/Inf = infinite LC/MS intensity range due to non-detection in the vehicle control group.
All other enviromimetic treatments subsequently exhibited at least 1 specific, and thus unique, metabolome alteration. Ethanol treatment yielded a significant alteration in Succinic Acid (Log2FC = −0.44, p = 0.039), Guanosine Monophosphate (Log2FC = −0.62, p = 0.0056), and Inosine (Log2FC = −1.2, p = 0.0087). Nicotine treatment led to the specific alteration of D-Ribose 5-Phosphate (Log2FC = −1.1, p = 0.0037) and Acetylcysteine (Log2FC = −0.75, p = 0.04). Both IL17a and cortisol treatment led to the identification of only 1 unique metabolome alteration in each group. Notably, IL17a treatment increased Pyruvic Acid (Log2FC = −1.68, p = 0.009), while Cortisol yielded a group specific elevation in N-Acetylglutamine (Log2FC = −0.85, p = 0.038). These data thus indicate that, with the exception of WIN 55–212,2 and endomorphin, there were subtle yet selective metabolome alterations present in all other treatment groups.
Of particular note, dorsal forebrain organoids treated with our various enviromimetic treatments tended to exhibit a convergence in differentially regulated metabolites. For example, all treatment groups exhibited a significant differential expression in the expression of L-Phenylalanine in dorsal forebrain organoids. However, L-phenylalanine was not the only common alteration detected between groups. We also detected a significant alteration in the expression of GTP in ethanol, endomorphin, nicotine, IL17a, and cortisol treated organoids. Regulators of GTP and GTP-related signaling were also detected in our Reactome pathway analysis (Fig. 2), and were most notably enriched in nicotine treated human-derived dorsal forebrain organoids (Fig. 2g). A schematic summary of metabolomic convergence and divergence between between groups is schematically provided in Fig. 3e. These data therefore indicate that L-Phenylalanine and GTP expression within ethanol, endomorphin, nicotine, IL17a, and cortisol-treated organoids may reflect a commonly noxious role of these compounds within dorsal forebrain-restricted organoids.
Single-cell DNA content analysis of enviromimetic treatments within human-derived forebrain organoids
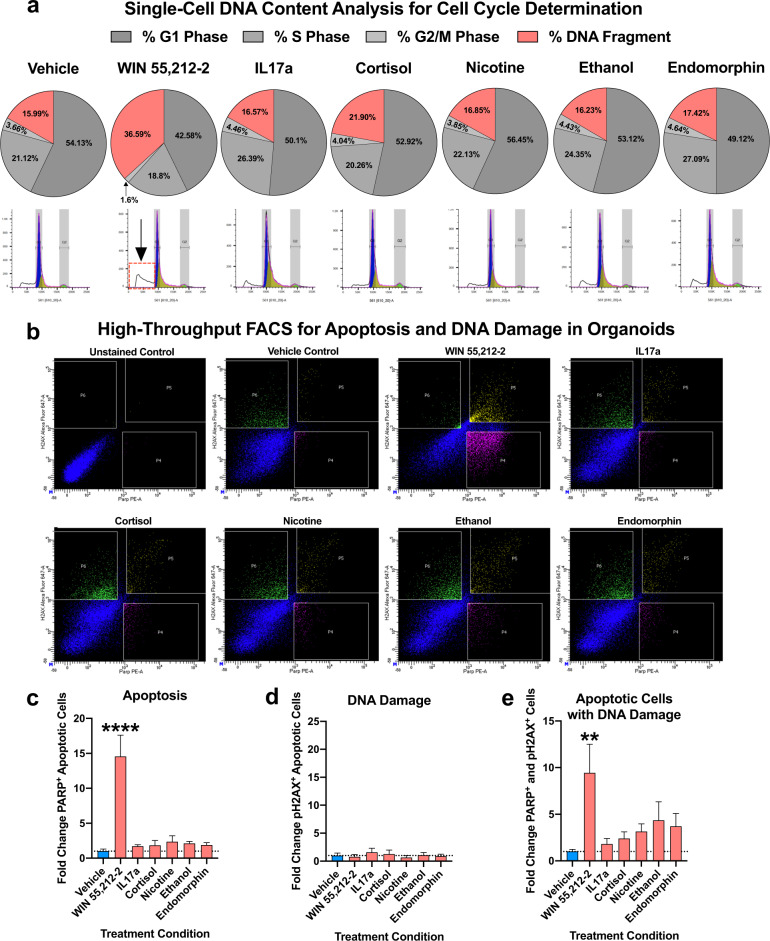
We next considered whether our enviromimetic treatments might elicit an alteration in the proliferative activity and/or cell cycle progression of cells within developing forebrain organoids. Given that our cortical cultures were comprised of ventricular zones enriched for SOX2+ neural stem cells and forebrain-specific FOXG1+ neuronal progenitors (see Fig. 1), our dorsal forebrain organoid cultures are representative of the early stages of corticogenesis whereby ventricular progenitors undergo expansion and amplification for the purpose of supporting neurogenesis and an expanding cortical plate. To yield an unbiased evaluation of cell cycle stage at the whole organoid level, we adapted an established single-cell DNA content analysis [92] that could distinguish cells in G1, S, and G2/M phases. Cells undergoing mitosis characteristically contain increased DNA content due to the DNA replication that occurs as they progress through mitotic cycles. Conversely, cells undergoing apoptosis exhibit fragmented DNA due to the activity of endonucleases that cleave and fragment chromatin into nucleosomal units [93]. Thus, cells undergoing cell division within dorsal forebrain organoids could be identified based on their DNA content [92].
Analysis of single-cell DNA content revealed that there were no substantial alterations in the proportion of G1, S, and G2/M phase cells within human-derived dorsal forebrain organoids that had been treated with IL17a, cortisol, nicotine, ethanol, or endomorphin. However, human-derived dorsal forebrain organoids treated with the cannabinoid agonist WIN 55,212-2 exhibited evidence of increased cell death. Namely, there was a substantial (~2.29 fold) increase in DNA fragmentation within this group, which was co-defined by generally decreased proportions of cells within all other detectable phases of the cell cycle (G1, S, and G2/M phases). Therefore, while we did not identify any alterations in mitotic cell cycle dynamics within our various treatment groups, we did identify a potentially neurotoxic role of the cannabinoid agonist WIN 55,212-2 within human-derived dorsal forebrain organoids.
Induction of apoptosis and DNA damage by narcotic and neuropsychiatry-related enviromimetic treatments within human-derived forebrain organoids
To further investigate the potential for our narcotic and neuropsychiatric-related enviromimetic treatments to induce apoptosis, we adapted a high-throughput FACS DNA damage and cell death panel to corroborate the results our single-cell DNA content analysis.
To do this, we pseudorandomly selected pools of ~10 organoids per line and condition, and dissociated pools to a single-cell suspension. Cell suspensions were next fixed, permeabilized, resuspended in refixation buffer, and consequently labeled with a PE-conjugated cleaved PARP antibody and Alexa647-conjugated phosphorylated H2AX antibody. Of note, an analysis of unlabeled and labeled samples yielded the unbiased capture and quantification of cells exhibiting cell death and DNA damage within human-derived dorsal forebrain organoids without any background (see unstained flow panel in Fig. 4b). In addition, vehicle-treated controls also exhibited exceptionally low rates of PARP+ apoptotic cells (1.58% of all cells), of which only a fraction exhibited evidence of H2AX+ DNA damage (0.4% of all cells were double-positive; see top left panel in Fig. 4b). This indicates that dorsal forebrain organoid cultures were broadly healthy, and did not exhibit any evidence of generalized cell death nor DNA damage at baseline.
Fig. 4. Analysis if DNA content, DNA damage, and apoptosis induction within enviromimetic human-derived forebrain organoids.
a Increased DNA fragmentation in organoids treated with WIN 55,212-2. To examine if our narcotic and neuropsychiatric-related treatments altered the cell cycle of proliferating cells within human-derived dorsal forebrain organoids, we adapted a single-cell DNA content analysis. Briefly, whole organoids were dissociated to a single-cell suspension and labeled with the fluorescent DNA-intercalating agent Propidium Iodide (PI). DNA content below our G1 peak serves as a proxy of cell death due to DNA fragmentation/digestion by endonucleases [93]. No major alterations in the proportion of proliferating cells or DNA fragmentation were detected in our IL17a, cortisol, nicotine, ethanol, and endomorphin treatment groups. However, a robust effect of WIN 55,212-2 treatment was detected for the DNA content, whereby WIN 55,212-2 selectively increased DNA fragmentation in our human-derived dorsal forebrain organoids. This indicated that, of our various narcotic and neuropsychiatric-related enviromimetic treatments, the cannabinoid agonist WIN 55,212-2 may selectively induce neurotoxicity and thus cell death within developing human-derived dorsal forebrain organoids. DNA histograms are provided for all treatment groups underneath their respective pie-charts, which delineate the proportion of cells in G1 phase, S phase, G2/M phase, and those which exhibit DNA fragmentation (i.e., dead and/or dying cells). For reference, a red-box and arrow identifies the DNA fragmentation peak in dorsal forebrain organoids treated with WIN 55,212-2. b–e, Validation of death and DNA damage in organoids treated with WIN 55,212-2. To provide orthogonal validation of our DNA content analysis, we adapted a multi-panel FACS assay to provide a higher-resolution analysis of cell death and DNA damage. Following a series of fixation, permeabilization, and re-fixation steps (designed to gain access to the nucleus), cells were labeled with an Alexa647-conjugated phosphorylated H2AX antibody to detect the presence of DNA damage machinery and a PE-conjugated cleaved PARP antibody for detection of cell death induction. As shown in bar graphs, the endocannabinoid receptor agonist WIN 55,212-2 was the only treatment to significantly increase cell death (PARP-PE + cells, or gate P4 in flow charts) within human-derived dorsal forebrain organoids. There was no increase in P2HAX + healthy cells (i.e., gate P6 in flow charts) that exhibited no evidence of apoptosis amongst any of our treatment groups. However, there was a significant increase in the proportion of DNA damaged apoptotic cells (H2AX + and PARP + double-positive cells, depicted by gate P5 in flow charts) selectively within human-derived dorsal forebrain organoids treated with WIN 55,212-2. For all panels, n = 4–6 iPSC donors x n = 7 treatment groups for a total n of = 34–41 experimental samples for flow cytometry analysis across experiments. All gates/quadrants in flow charts were pseudorandomly labeled. **** denotes p < 0.00001 and ** denotes p < 0.01. Bar graphs represent mean ± SEM.
Analysis of our various enviromimetic dorsal forebrain organoids revealed that all treatment conditions tended to increase cell death. Relative to vehicle-treated controls, the fold change in PARP+ cells exhibiting induction of cell death ranged from 1.71 to 14.56 across groups (see Fig. 4a-b). However, ANOVA test statistics indicated that while there was a significant treatment effect, the only drug treatment to significantly elevate apoptosis in dorsal forebrain organoids was the cannabinoid receptor agonist WIN 55,212-2 (p < 0.0001). An analysis of the fold change in pH2AX+ cells that exhibit DNA damage with or without evidence of apoptosis revealed no significant differences between groups. However, an analysis of the fold change in the proportion of PARP+ and pH2AX+ cells (which selectively identified DNA damaged cells also undergoing death) revealed a similar outcome to our first analysis. Namely, while all treatment groups tended to exhibit increased proportions of DNA damaged dying cells (fold change ranged from 1.80 to 11.13 relative to controls, across groups), post hoc testing revealed that only WIN 55,212-2 exhibited a statistically significant alteration relative to controls (p < 0.01; see Fig. 4d). In sum, only WIN 55,212-2 robustly induced (1) DNA fragmentation, (2) the induction of PARP-mediated cell death, and (3) DNA damage within cells that had already committed to apoptosis.
Neurogenesis within the developing cortical plate of human-derived forebrain organoids treated with narcotic and neuropsychiatry-related enviromimetics
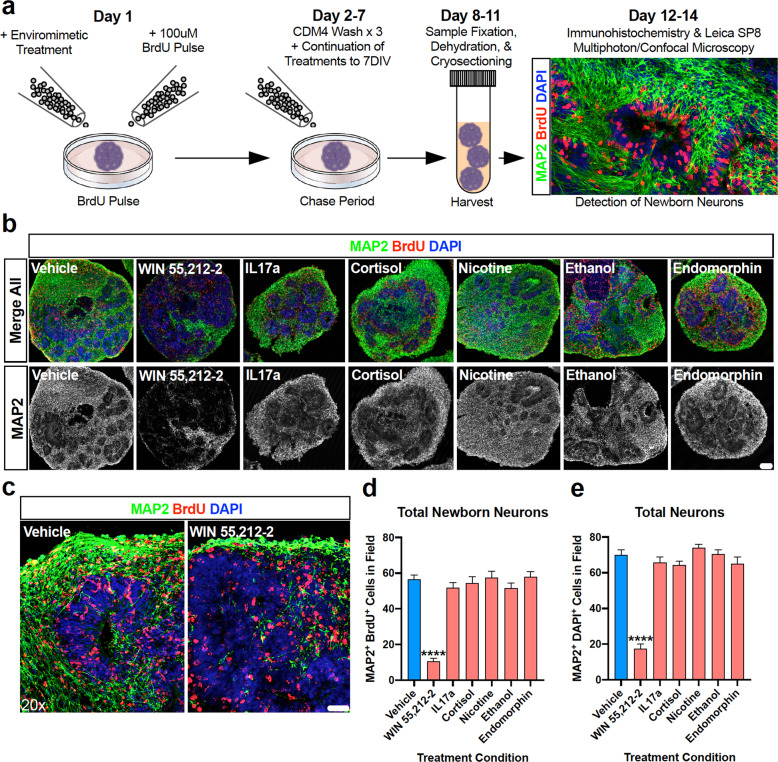
During early forebrain development, ventricular progenitors proliferate, differentiate into newborn neurons, and begin their migration from the ventricular zone into the developing cortical plate [94]. This process is both present and conserved within our 3D human-derived dorsal forebrain organoids (see Fig. 1b). Given validation that the cannabinoid agonist WIN 55,212-2 selectively exerted neurotoxic effects (Fig. 4) in human-derived dorsal forebrain organoids (Fig. 4), we next sought to examine whether WIN 55,212-2 and our other narcotic and neuropsychiatric-related treatments altered neuron numbers within our human-derived dorsal forebrain organoid system. To do this, we adapted a BrdU pulse-chase assay to examine neocortical neurogenesis within organoids [70]. Briefly, at the commencement of drug treatment, organoids were pulsed with 100 μM BrdU for 24 h before continuing their compound exposure routines for a 7 DIV chase period (see Fig. 5a for schematic). This protocol therefore enabled us to label progenitors that were specifically differentiating at the time of drug exposure and track the total quantities of newborn neurons that were subsequently generated. This approach therefore controls for baseline differences in neuron numbers within each organoid, as well as ensures that a specific effect of each drug compound can be identified and assessed for potential effects upon neurogenesis. Analysis revealed that treatment with IL17a, cortisol, nicotine, ethanol, and endomorphin elicited a lack of effect on newborn and total neuron numbers within the developing cortical fields of human-derived dorsal forebrain organoids. Contrary to this, organoids treated with WIN 55,212-2 exhibited a robust depletion of both newborn neurons (MAP2 + neurons with BrdU+ nuclei) and total neurons (MAP2 + neurons with DAPI + nuclei). This indicates that the cannabinoid CB1 agonist WIN 55,212-2 interferes with neocortical neurogenesis within developing human-derived forebrain tissue. This confirms that WIN 55,212-2, and ergo cannabinoids, are potentially noxious compounds during early human fetal brain development.
Fig. 5. The endocannabinoid agonist WIN 55,212-2 selectively depletes human-derived dorsal forebrain organoids of neurons.
a Schematic of neocortical neurogenesis pulse-chase assay. Neocortical neurogenesis was evaluated within human-derived dorsal forebrain organoids via the adaptation of a BrdU pulse-chase assay [70]. Briefly, at the time of treatment commencement, dorsal forebrain organoids were pulsed for 24 h with BrdU. Following this, BrdU was removed and washed out and cultures underwent continuation of their allotted treatment to their endpoints. Organoids were subsequently processed (fixed, dehydrated, cryosectioned, mounted) before being immunostained and imaged for newborn (BrdU + ; red) neurons (MAP2 + ; green) via laser-scanning confocal microscopy. b Representative whole organoid images of enviromimetic treated organoids. Representative whole organoid images for each treatment group are shown as insets. Note that in all groups there was widespread and robust expression of both BrdU (red) and MAP2 (green) across the entire organoid, except those treated with WIN 55,212-2. To emphasize this, MAP2-isolated channels are presented in gray scale below merged images. c–e Depletion of newborn and total neurons in WIN 55,212-2 treated organoids. To determine if our various treatments influenced neuron numbers, we quantified the number of newborn neurons MAP2+ green cells with BrdU+ red nuclei, see (d) and total neurons MAP2+ green cells with DAPI+ blue nuclei, see (e) within cortical fields. The only group to exhibit a significant difference in both newborn and total neuron numbers within the developing cortical plates of dorsal forebrain organoids was the cannabinoid agonist WIN 55,212-2. More specifically, we identified that WIN 55,212-2 treatment reduced newborn neurons by 81.23% and total neurons by 75%. When combined with our single-cell DNA content and DNA-damage cell death flow cytometry panels presented in Fig. 4, this data cumulatively confirms that WIN 55,212-2 is acutely neurotoxic and developmentally disruptive within human-derived dorsal forebrain organoids. For all panels, n = 3 iPSC donors x n = 7 treatment groups x n = 3–4 organoid replicates per donor/condition yielded a total n of = 18–22 cortical fields per condition. **** denotes p < 0.0001. Bar graphs represent mean ± SEM. Scale bar in b = 100 μm and c = 40 μm.
Discussion
The current study sought to determine prenatal signatures related to narcotic use and mental illnesses within 3D human-derived forebrain tissue. In experiments conducted in parallel, we modeled exposure to opiates (μ-opioid agonist endomorphin), cannabinoids (WIN 55,212-2), alcohol (ethanol), smoking (nicotine), chronic stress (human cortisol), and maternal immune activation (human IL17a). Following a range of high-throughput assays, we identified both convergent and divergent signatures between enviromimetic treatment groups, and unbiasedly identified the cannabinoid agonist WIN 55,212-2 as a particularly noxious agent that severely impacts normative cortical development within 3D human-derived dorsal forebrain organoids.
Our first goal was to establish the macromolecular effects of our enviromimetic treatments within human-derived dorsal forebrain organoids. We identified that the molecular composition of enviromimetic-treated organoids were relatively similar, however mapping differentially expressed proteins into their respective Reactome pathways provided additional clarity. Notably, IL17a identified alterations in signaling by ERBB2 factors, which is a pathway of particular relevance to schizophrenia given its interaction with neuregulin-1 to mediate cell adhesion [95], its potential role in schizophrenia risk [96], as well as antipsychotic response/treatment [97]. This result is also of note as MIA experiments in rodents have also identified neuregulin-1 and EGF-related (notably, ERBB4) differences in offspring [98, 99]. Similarly, we also identified WNT signaling dysfunction in our nicotine treated organoids, which is broadly important in the context of neurodevelopmental programming [100, 101], autism risk [102, 103], and schizophrenia too [104–106]. Recent work has shown that nicotine alters embryonic stem cell proliferation [107], and it has been suggested that nicotine may thus also alter neural progenitors as well as neurogenesis [108]. Given the prominence of WNT signaling in the developing brain, these data only reinforce the idea that prenatal exposure to nicotine may have the potential to elicit neurodevelopmental alterations. Nicotine and ethanol treated organoids also mutually exhibited enriched pathways for brain development, including factors related to axon guidance, ROBO receptor signaling, as well as mRNA metabolism and mRNA regulatory pathways. This is prominent as ROBO receptors play an evolutionarily conserved role in both short- and long-distance axon path finding and guidance [109–111]. Similar to nicotine and ethanol, organoids treated with WIN 55,212-2 and endomorphin treatment also exhibited enrichment for mRNA regulatory processes including degradation by 5ʹ to 3ʹ endonucleases and EJC-mediated Nonsense Mediated Decay (NMD), respectively. These pathways, such as NMD, function as important quality and quantity control mechanisms of gene expression [112], and their enrichment within our datasets collectively suggest that there may be disparate, yet similar, responses of these drug treatments upon mRNA degradation and RNA regulatory pathways within the developing brain. However, little work has been conducted on how these fundamental mRNA degradation pathways may regulate neurodevelopment; although it is likely that they are to play an essential role during neural development [113]. The NMD machinery has been implicated in both neural progenitor activity [113] and neuronal differentiation [114]. Notably, NMD has also been shown to specifically regulate axonal guidance in ex vivo open book preparations [115]. Furthermore, in recent work, we have shown that the NMD machinery is expressed in neurons and is operational within their dendrites, where NMD functions to regulate GLUR1 expression, LTP, as well as learning and memory [116]. Variants within the NMD machinery, notably UPF3, have also been associated with developmental delay [117], mental retardation [118], and childhood onset schizophrenia [119]. Therefore, data from our nicotine, ethanol, endomorphin, and WIN 55,212-2 groups indicates that RNA regulation is likely to play a role in the pathogenic effects of these treatments within developing human tissue.
To provide further systems-level analysis of our enviromimetic treatments in human-derived dorsal forebrain organoids, we adapted hybrid MS to globally map the developing organoid metabolome. This led to the observation that several factors exhibited convergent alterations in all groups, notably L-Phenylalanine. Prior work has indicated that Phylalanine adversely affects the developing mammalian brain [120] via a variety of mechanisms. For instance, phenylalanine has been shown to induce neuronal death [121], potentiate oxidative stress in the developing cerebral cortex of rats [122], and alter the acid-soluble pool of particular amino acids which modulates their incorporation into proteins within the rat brain [123]. Consistent with this, in Phenylketonuria—a disease characterized by elevated levels of pheylalanine in plasma and cerebrospinal fluid of patients—there are delays in brain development that yields profound insufficiencies in cognitive performance [121]. Thus, the differential expression of L-Phenylalanine in all treatment groups relative to controls indicates a potentially noxious role of all compounds studied within developing dorsal forebrain organoids. This is consistent with discrete alterations in amino acid expression across groups (L-Tyrosine and L-Valine in WIN 55,212-2 treated organoids, and in all other groups L-Methionine).
Excluding WIN, 55,212-2, all other treatment groups also exhibited an alteration in the expression of GTP, which is essential for normative cortical development. Similar to phenylalanine, metabolomic alterations in GTP are also directly linked to neurological disease; namely Segawa disease, which is characterized by deficient dopamine content and dopa-responsive dystonia [124]. However, GTP is broadly more important within the brain, as hydrolysis of GTP into 7,8-DHNP-3’-TP is required for the biosynthesis of numerous monoamine neurotransmitters. Therefore, an intrinsic alteration in GTP within our enviromimetic dorsal forebrain organoids is also likely to hold important implications for healthy corticogenesis within the developing forebrain of our human-derived organoids. In support of this, neuronal GTPase activators are ubiquitously expressed during cortical development [125]. In addition, Rho GTPase proteins [126] and signaling [127] are also known to specifically regulate progenitor proliferation and survival (e.g., RAC1) [128], cell fate of neural progenitors (e.g., CDC42) [129], neuronal migration [126, 127], and axonal development [130]. Therefore, GTP is likely to be important during all phases of brain development and maturation as GTPase factors regulate neural progenitors as well as neuronal differentiation and maturation. Therefore, similar to L-Phenylalanine, significant differences in GTP expression within ethanol, endomorphin, nicotine, IL17a, and cortisol-treated organoids likely reflects a commonly noxious nature of these compounds during human corticogenesis.
Aside from these common metabolic signatures, we also identified a number of highly specific group differences. First, we identified that pyruvic acid was selectively altered in IL17a treated organoids. Interestingly, a prior study has shown that targeting of purine pathway metabolites may be effective in treating autism-related phenotypes in a MIA mouse model [131]. This is paralleled in work that is indicative that pyruvic acid may be abnormal in autism spectrum disorders and may serve as a potential biomarker of the disease and/or a potential therapeutic target [132, 133]. In addition, purine/pyramidine metabolites have been broadly associated with a range of neuropsychiatric disorders that are also defined by neurodevelopmental risk [134]. This further supports the idea that this factor may be a useful biomarker for neuropsychiatric sequelae that may arise from maternal immune activation. Likewise, we identified N-acetylglutamine—which is the acetylated analog of glutamine—as a specific marker of chronic CORT exposure in human-derived dorsal forebrain organoids. This reflects a novel association that does not appear to have been reported in prior works, and little is known of N-acetylglutamine in the developing brain in general. However, given the role of N-acetylglutamine and glutamine in the synthesis of numerous essential units within the cell (e.g., proteins) [135], this result is likely to hold functional importance and will, accordingly, require further investigation. In our ethanol treated organoids, two unique metabolites were identified. The first, succinic acid, may be reflective of generalized alcohol exposure due to the formation of this metabolite during alcoholic fermentation [136], but is a relatively novel hit considering a lack of studies on the topic. The second, Guanosine Monophosphate Inosine, has been identified in a pathway analysis screening of metabolite dysfunction in an animal model of alcoholic liver disease [137] but otherwise also remains a novel hit. Further work on these two potential corollaries may therefore unveil their biological importance in mediating the effects of prenatal alcohol exposure on the developing brain. Last, our analysis of nicotine treated organoids returned group-specific alterations in D-Ribose 5-Phosphate and Acetylcysteine. D-Ribose 5-Phosphate has been previously identified within tobacco leaves [138] but has been scarcely studied, especially in the context of it being a metabolite marker of neuropathology in progenitors, neurons, and brain development. On the other hand, Acetylcysteine is amongst the most widely studied metabolite factors in neuropsychiatry due to its antioxidant properties, and has been extensively studied for its use in the management of substance use disorders [139]. This includes cue-induced nicotine seeking as an endophenotype of smoking behavior in rodents [140–142] well as nicotine-dependence and tobacco use disorder within humans [143, 144]. Acetylcysteine has also been extensively studied and shown to have potential therapeutic benefits in both patient-derived iPSC [145] and a glutathione-deficient rodent model of Schizophrenia [146]. As an estimated ~10.7% of mothers whom smoke will continue to do so during pregnancy, and considering evidence that nicotine may elicit prenatal effects on the developing brain (see [21]), it is imperative that further studies follow-up the metabolite markers here for their role as potential biomarkers as well as their utility as potential therapeutic targets.
We also identified that a variety of our narcotic and enviromimetic organoids also exhibited Reactome pathway alterations related to cell-cycle checkpoints/regulation, mitosis, cellular stress response pathways, as well as apoptosis and the apoptosome (see Fig. 2). Given the importance of progenitor proliferation and survival in the developing brain, these data consequently led us to next consider the impact of our treatments on cell-cycle activity and cell death. This revealed that there were no major differences in the proportion of cells in G1, S, or G2/M phases of the cell cycle between groups. The exception to this was human-derived organoids treated with the cannabinoid agonist WIN 55,212-2, which exhibited alterations due to a dramatic increase in DNA fragmentation (Fig. 3a). DNA fragmentation occurs when endonucleases cleave chromatin into nucleosomal units, and is therefore a marker of cells undergoing apoptosis [93]. Consistent with this, in a follow-up flow cytometry panel we identified that organoids treated with WIN 55,212-2 were selectively enriched for cells exhibiting both apoptosis (cleaved PARP) and DNA damage machinery (pH2AX, see Fig. 3b–e). Importantly, cells which exhibit minor DNA damage will attempt to repair such, whereas those that exhibit more DNA damage will be targeted for apoptosis [147]. The increase in both apoptotic cells, and the proportion of apoptotic cells exhibiting DNA damage, in WIN 55,212-2 treated forebrain organoids is therefore indicative of acute neurotoxicity. Furthermore, in our neocortical neurogenesis pulse-chase assay, WIN 55,212-2 was also the only mimetic condition to elicit a robust depletion in both total neuron and newborn neuron numbers (Fig. 5). This cross-validation therefore confirms that WIN 55,212-2 is acutely noxious in human-derived dorsal forebrain organoids. This result is important, as it addresses and contributes to a major point of contention within the literature. Notably, while there is consensus that prenatal cannabinoid exposure likely alters human neuron development [148] and function [149] in a manner that likely induces protracted risk to behavioral alterations [150, 151], there is conflicting data regarding the specificity of cannabinoid use/abuse/stimulation/modeling upon cell death. For example, synthetic cannabinoids (the accessibility and abuse of which have become a major public health concern [148]) have been shown to elicit cytotoxic effects via the CB1 receptor in mouse forebrain cultures [152]. However, there is also a rich literature which indicates that cannabinoids may also be able to elicit neuroprotective effects by preventing apoptosis [153]. Contributing further ambiguity, at least one study reported that the treatment of pregnant dames with WIN 55,212-2 did not alter cell death within the prenatal mouse cerebral cortex [154]. Indeed, these conflicting data are well described in the literature, and it has been proposed that cannabinoids may elicit both neuroprotective and pro-apoptotic effects in a non-binary and therefore sophisticated manner [155]. Cannabinoid effects on cell death may therefore be dependent upon the cell-type target and the developmental stage at which exposure occurred [156]. Yet, in our prenatal model of the developing forebrain, we observed reproducible evidence of cell death in multiple single-cell analyses (Fig. 4) that were accompanied by a dramatic decrease in neocortical neurogenesis (Fig. 5). These data may therefore be indicative of a potential ‘species-of-origin’ effect, or indicate that other experimental considerations may mediate neurotoxic effects.
It is therefore important to emphasize that the mechanisms of action of synthetic cannabinoids may differ from plant-derived compounds, and that differing concentrations, frequency of use, and timing of consumption may all elicit differential effects. A limitation of this panel study is therefore the fact that only one concentration of each factor was studied, at one particular early time point of development that coincided with early corticogenesis approximately equivalent to trimester one. Effects may therefore scale with increasing doses, longer durations of exposure, or later developmental time points. In addition, it is important to note that while cerebral organoids reproduce aspects of fetal brain transcription [67], epigenetic regulation [68, 69], and proteomic programing [70], this model is not a perfect substitute for the developing brain and yet other factors, phenotypes, or mechanisms may exist outside of those identified in the current study. However, given rising rates of cannabinoid use (both plant-based and synthetic derivatives), such experimental considerations and limitations are important to explicate so not to unnecessarily stigmatize recreational users in jurisdictions where legalization has occurred and/or is currently unfolding. Nonetheless, our work still provides an important contribution to the literature by indicating that chronic WIN 55,212-2 exposure elicits a robust and reproducible neurotoxic effect within human-derived developing forebrain tissue that signals a need for caution. This work therefore emphasizes the need for yet more cannabinoid effects to be reevaluated specifically within human-derived cells and 3D tissue systems, as well as warrants independent replication and further study by other groups.
Following from the work presented here, it will be important for a number of future directions to be explored in more extensive, as well as more specific and topical, investigations. For example, it is possible that other subtle phenotypes may exist in WIN 55,212-2, as well as other, treatment groups. For example, it is of specific note that the endocannabinoid system has been shown to have a potential role in astroglial cell phenotypes, including their differentiation [157]. Indeed, cerebral organoids have already been used to study glial cell diversity and methamphetamine-induced neuroinflammation [158], indicating that similar work could be completed by substituting cannabinoids as well as other agents. Therefore, a future direction following from this work may therefore be to consider differences in the production and proportion of glia, such as astrocytes, in future drug panel organoid studies. In addition, it will be equally important to determine how different types of neurons may be impacted by the enviromimetic conditions examined here. Likewise, it will also be important to continue this line of investigation by examining neuronal dynamics and activity differences. Therefore, a fruitful avenue of further research may be to design a physiological panel to examine how our various treatment groups alter neuronal electrophysiology via high-throughput approaches. Conceptually, due to the overlapping nature of comorbidities and the fact that numerous substances may be simultaneously abused, future studies may wish to consider the compounding effects of simultaneous risk factor exposure within this dorsal forebrain organoid system. Lastly, arguably the most important future direction to emerge from this work would be for other groups to independently replicate our findings, and to adapt the specific controlled and scheduled substances that were modeled here with established mimetic compounds.
In closing, the current study supports the idea that prenatal exposure to a variety of factors can alter the proteome, metabolome, and other discrete cellular processes that are essential for normative cortical development in 3D human-derived developing forebrain tissue. This multi-omic pipeline therefore provides important insight into the dynamics of prenatal brain development, and serves as a valuable human tissue resource that can be screened and cross-referenced by preclinical and clinical researchers alike to identify factors that may mediate the prenatal effects of various drug and neuropsychiatric risk factors in their own datasets.
Materials and methods
Human induced pluripotent stem cells
Human Induced Pluripotent Stem Cells (iPSCs) were maintained as previously described (see [70]). Briefly, iPSCs were maintained in vitronectin-coated plates and passaged using Accutase (Sigma, Material#: A6964) for progression to cerebral organoid generation or with EDTA (prepared in house) for expansion and maintenance. Cells were fed every 24–48 h, as necessary, with mTeSR Plus media (Stem Cell Technologies, Material#: 05825). Human iPSC lines were cultured simultaneously to control for idiosyncratic culturing conditions. All human iPSCs were acquired from deposits made to the National Institute of Mental Health (NIMH) Repository & Genomics Resource center at Rutger’s University (lines beginning with prefix MH, found below) or from the Coriell Institute for Medical Research (lines beginning with prefix GM, also found below). Thus, lines had typically undergone extensive, standardized, testing for common iPSC factors such as pluripotency, viability, and karyotypes. In total, 6 different iPSC lines were sampled across experiments included in the manuscript (MH0159019, MH0159020, MH0159022, MH0174677, GM23279, GM25256) with tissue originating from apparently healthy individuals without any neuropsychiatric or neurological diagnosis or evidence of family history of such. The patients are otherwise listed as healthy donors and devoid of any conflicting or confounding diagnoses. Healthy patient donors comprised 4 males (MH0159020, MH0159022, MH0174677, GM25256) and 2 females (MH0159019 and GM23279). No sex difference in phenotype was observed between iPSC lines. All but one of the donors were adults at the time of biopsy (range: 9, 29, 30, 36, 46, and 58 years of age), alas no age-mediated differences between the iPSC lines were identified in any experimental assay or in quality control assessments.
Generation of human-derived dorsal forebrain organoids
Dorsal forebrain organoids were derived via a directed differentiation protocol from Paola Arlotta’s laboratory at Harvard [71] that we have amended for an expedited/shorter timeline as previously described [70]. Briefly, we cultured undifferentiated iPSCs into colonies before dissociating these into a single cell suspension via Accutase exposure (see above). Suspensions were correspondingly transferred and cultured within ultra-low attachment Aggrewell V-Bottom Plates (Stem Cell Tech, CAT#: 34815) so that 3D stem cell aggregates known as embryoid bodies could be formed. Because of the dimensions and shape of wells within Aggrewell plates, embryoid bodies were reproducibly formed into spherical geometries of a consistent size both within and between iPSC lines and batches. From this point, 3D tissue samples were cycled through successive Cortical Differentiation Media (CDM1–4) every ~7–10 days for a final timeline that yielded early cortical organoids by ~30 DIV. CDM chemical components are described at length within [71]. By the end of our culturing period, all organoids exhibited robust induction of expected morphologies (e.g., ventricles surrounded by ventricular zones) as well as forebrain-specific markers (e.g., FOXG1 + cells) and cortical-specific neurons (e.g. CTIP2 + cells; see Fig. 1). For more extensive protocol details, please refer to [71] for reagents and [70] for timing and other design considerations.
Treatment regime
Human-derived dorsal forebrain organoids were systemically exposed to a barrage of widely accepted drug and environmental mimetics, in tandem, for a total of 7 Days In Vitro (7 DIV). This ensured that all control and treatment conditions were cultured in parallel simultaneously under exacted laboratory conditions (e.g., media batches, feeding regimes etc.). Concentrations were utilized that were known to be phenotypic but not noxious to cultures and/or animals. Consequently, all concentrations were derived from prior studies that generally did not report major neurotoxic effects of each compound, thus avoiding potential ceiling effects on phenotypes. Thus, concentrations were adapted principally from prior basic mechanism studies (and, in our own testing pre-experiment testing, organoid survival) as this allowed a degree of methodological standardization and precision across our 7 different groups that would have not been otherwise achievable. To model alcohol exposure, we utilized a 100 mM [159] of molecular biology grade ethanol (Fisher Scientific, Material#: BP2818100). This concentration is consistent with concentrations reported for ethanol to effect normative brain function in naïve and occasional users [160]. To model chronic stress, we utilized a 10 μg/mL concentration [161] of the human stress hormone cortisol (CORT). We specifically utilized a water-soluble version of CORT, namely hydrocortisone-hemisuccinate (Sigma, Material#: H2270). Similar to ethanol, CORT treatment is a common method for modeling chronic stress under controlled laboratory conditions in both slice [161] and animal studies [60, 162–164]. Cannabis exposure was modeled using a 10 μM concentration [165] of the cannabinoid receptor agonist WIN 55,212-2 (Sigma, Material#: W102), which has also been widely used in cell [166] brain slice [165, 167] studies. To model maternal immune activation, we treated developing dorsal forebrain organoids with 10 ng/mL [168–170] of the human cytokine IL17a (Sigma, Material#: H7791). Importantly, IL17a is downstream of IL6 and was recently shown to mediate the brain-specific effects of maternal immune activation [54, 171]. Therefore, IL17a maintains the necessary construct and predictive validity required to model immune exposure in tissue that otherwise lacks an endogenous immune system. Opiate use was modeled utilizing the μ-opioid receptor agonist endomorphin-1 (1 μM, Abcam, Material#: ab1240411) as utilized in prior studies [172–174]. Last, to model prenatal exposure to smoking, we treated organoids with the nicotinic acetylcholine receptor agonist (-)-nicotine ditartrate (10 μM, Abcam, Material#: ab120562) which was similar to concentrations utilized in prior studies too [175–177]. All concentrations adapted were screened to ensure that no overt evidence of organoid deterioration was induced via chronic exposure for 7 DIV, thus avoiding any manifest ceiling effects upon cell viability and/or death.
Unbiased flow cytometry of apoptosis & DNA damage
To derive an unbiased assessment of DNA damage, cell death, and DNA-damaged cells undergoing apoptosis following drug treatment, we adapted a well validated and widely utilized a Fluorescent-Activated Flow Cytometry (FACS) kit (BD Pharmingen, Material#: 562253). Briefly, pseudorandomly selected organoids were dissociated to a single-cell suspension via a 20-minute exposure to accutase followed by tricheration and serial filtering through 70 → 30 μm pores. Cells were resuspended and incubated in in Cytofix/Cytoperm solution for fixation and initial permeabilization for 30-minutes at room temperature. Cells were consequently washed, resuspended in BD “Plus” permeabilization buffer for 10-minutes on ice, and re-exposed to Cytofix/Cytoperm solution for 5 additional minutes to achieve refixation. Cells were consequently washed, and resuspended in 30 μg of DNAse for 60-minutes at 37 °C. Cells were consequently washed and labeled with PE-Cleaved PARP (BD Pharmingen, Material#: 51–9007684) to label cells committed to apoptosis and Alexa647-H2AX (BD Pharmingen, Material#: 51–9007683) for cells exhibiting DNA damage. Double positive cells represented apoptotic cells exhibiting DNA damage. Per manufacturer instructions, antibodies were diluted at a 1:25 ratio and incubated for 20-minutes at room temperature. Cells were washed and resuspended in staining solution. Labeled suspensions were analyzed utilizing a BD Aria II (Becton Dickinson) cell sorter to acquire multiparameter data files. Data were presented as fold-change of % of cleaved PARP + apoptotic cells, % of H2AX + DNA damaged cells, and % of PARP + H2AX + double-positive cells.
Cellular DNA content & cell-cycle analysis
Organoids were prepared to a single-cell suspension as described above and in [70], and washed successively with calcium/magnesium free PBS at 4 °C to remove residual peptides in solution. Samples were consequently centrifuged, supernatant removed, and pelleted cells resuspended in PBS. Cells were EtOH fixed (100%, at 4 °C) while being gently vortexed. Cells were rehydrated, and incubated with Triton-X with DNAse added for 5 min. Cells were pelleted, Triton-X removed, and resuspended in 200ul of calcium/magnesium free PBS at 4 °C. Immediately prior to flow cytometry, suspensions were incubated with 2 μL of Propidium Iodide (PI; Thermofisher, Material#: P3566), and analyzed for single-cell DNA content utilizing a Sony MA900 cell analysis cytometer. Cell-cycle was subsequently modeled post hoc in the FloJo cytometry analysis package (Becton Dickinson), which allowed cells in G1, S, and G2/M phases to be distinguished based on the DNA content of each individual captured cell.
Immunohistochemistry and laser-scanning confocal microscopy
Immunohistochemistry of organoids was conducted as previously described [70]. Briefly, organoids were drop-fixed in 4% paraformaldehyde, dehydrated in 30% sucrose, embedded in (Tissue Tek, Material#: 4583) using biopsy molds, and cryosectioned at 30 µm. All sections underwent antigen retrieval in citrate buffer, and were incubated in primary overnight. Primary antibodies comprised SOX2 (1:1000; R&D Systems, Material#: MAB2018-SP), TUJ1/β-tubulin III (1:1000; Abcam, Material#: AB41489), MAP2 (1:1000, Abcam, Material#: AB11267; 1:1000, Abcam, Material#: AB32454), BrdU (1:1000, BD Pharmingen, Material#: 555627), CTIP2 (1:300; Abcam, Material#: AB18465), and FOXG1 (1:500, Abcam, Material#: ab18259). Secondary antibodies were incubated for 2 h at room temperature, and comprised antibodies for rabbit (Fluor 488 Material#: A11008; Fluor 546 Material#: A11035; & Fluor 633 Material#: A21070), mouse (Fluor 488 Material#: A11001; Fluor 546 Material#: A11003; & Fluor 633 Material#: A21052) and chicken (Fluor 546 Material#: A11040). All secondary antibodies were used at a 1:2000 dilution, and sourced from Life Technologies. Microscopy was completed on an Olympus IX81 Laser-Scanning Confocal Microscope or Leica SP8 Multiphoton/Confocal microscope. Images were typically acquired at 1200 × 1200 resolution with optical Z slices (step sizes) ranging from 0.5 to 10 µm depending on the unit of analysis.
Neocortical neurogenesis pulse-chase paradigm
To examine neurogenesis, we conducted a 7DIV BrdU pulse-chase experiment in dorsal forebrain organoids as previously described [70]. Briefly, organoids were pulsed with 10 μM BrdU for 24 h time-locked to when drug treatment commenced. This yielded widespread BrdU incorporation into all proliferating and/or differentiating cells within organoids over a 24 h window. Following this, organoids were maintained for 7DIV with or without treatment, at which point organoids were drop fixed in 4% PFA, dehydrated via sucrose incubation, embedded using Tissue-Tek OCT (CAT#: 4583) compound at −80 °C, and subsequently cryosectioned. Analysis involved immunohistochemistry for new-born cells that exhibit BrdU + (1:1000, BD Pharmingen, CAT#: 555627) nuclei within MAP2 + cell bodies. This subsequently allowed interpretation of the relative degree to which cells that were proliferating at the commencement of our enviromimetic treatments underwent terminal differentiation into neurons (BrdU+ MAP2+ double-positive cells).
Proteomics, barcoding chemistry, & liquid-chromatography/mass-spectrometry
Tandem Mass Tag (TMT) Liquid-Chromatography/Mass-Spectrometry (LC/MS) proteomics was completed as previously described in [70] and [178]. Briefly, organoids were reduced with dithiotreitol and underwent alkylation with iodoacetamide before tryptic digestion at 37 °C overnight. Peptide suspensions were desalted using C18 stage-tips prior to Liquid Chromatography-Mass Spectometry (LC-MS) analysis. An EASY-nLC 1200, which was coupled to a Fusion Lumos mass spectrometer, (ThermoFisher Scientific) was utilized. Buffer A (0.1% FA in water) and buffer B (0.1% FA in 80% ACN) were used as mobile phases for gradient separation [178]. A 75 µm I.D. column (ReproSil-Pur C18-AQ, 3 µm, Dr. Maisch GmbH, German) was packed in-house for separating peptides. A separation gradient of 5–10% buffer B over 1 min, 10–35% buffer B over 229 min, and 35–100% B over 5 min at a flow rate of 300nL/min was adapted. Data dependent mode was selected during operation of the Fusion Lumos mass spectrometer. An Orbitrap mass analyzer acquired Full MS scans over a range of 350–1500 m/z with resolution 120,000 at m/z 200. The top 20 most-abundant precursors were selected with an isolation window of 0.7 Thomsons and fragmented by higher-energy collisional dissociation with normalized collision energy of 40. The Orbitrap mass analyzer was also used to acquire MS/MS scans. The automatic gain control target value was 1e6 for full scans and 5e4 for MS/MS scans respectively, and the maximum ion injection time was 54 ms for both. For TMT chemistry, we adapted the recently released TMTpro 16Plex labeling reagents from ThermoFisher Scientific. This allowed us to run 28 total samples for MS detection of proteins, not including pools for internal standardization, allowing us to sample all 7 group conditions from organoids generated from a maximum of 4 independent iPSC lines (iPSCs ending in 020, 022, 279, and 677 were randomly selected for TMT-LC/MS).
Bioinformatics for proteomics
MS raw files were analyzed using the MaxQuant software [179] and peptide lists were searched against the human Uniprot FASTA database with the Andromeda search engine [180]. A contaminants database was employed and cysteine carbamidomethylation was set as a fixed modification and N-terminal acetylation and methionine oxidations as variable modifications. Further modifications included TMT tags on peptide N termini/lysine residues (+229.16293 Da) set as static modifications. False discovery rate (FDR) was 0.01 for both the protein and peptide level with a minimum length of 7 amino acids for peptides and this FDR was determined by searching a reverse sequence database. Enzyme specificity was set as C-terminal to arginine and lysine as expected using trypsin protease, and a maximum of two missed cleavages were allowed. Peptides were identified with an initial precursor mass deviation of up to 7 ppm and a fragment mass deviation of 20 ppm. Protein identification required at least one unique or razor peptide per protein group. Contaminants, and reverse identification were excluded from further data analysis. Protein intensities were log2 transformed and normalized using quantile normalization from R package preprocessCore. The histogram of the precursor intensity distribution and the boxplot of correlation covariance were visualized using R package ggplot2. Proteins with no missing values were subjected to downstream visualization and statistical analysis using Perseus software of the MaxQuant computational platform [181]. Proteins were subjected to one-way ANOVA test followed by post-hoc test. Proteins with p < 0.05 in one-way ANOVA and differentially regulated versus vehicle controls in post-hoc test were included in downstream functional enrichment analysis. STRING and Reactome databases were utilized for functional enrichment and pathway analysis with significance at p < 0.05.
Hybrid metabolomics liquid-chromatography/mass-spectrometry
Randomly selected samples (organoids generated from iPSCs ending in 019, 020, 022, 256, and 677, each weighing ~20 mg) were subjected to an LC/MS analysis to detect and quantify known peaks. A metabolite extraction was carried out on each sample based on a previously described method [182]. The LC column was a MilliporeTM ZIC-pHILIC (2.1 ×150 mm, 5 μm) coupled to a Dionex Ultimate 3000TM system and the column oven temperature was set to 25oC for the gradient elution. A flow rate of 100 μL/min was used with the following buffers; (A) 10 mM ammonium carbonate in water, pH 9.0, and (B) neat acetonitrile. The gradient profile was as follows; 80–20%B (0–30 min), 20–80%B (30–31 min), 80–80%B (31–42 min). Injection volume was set to 2 μL for all analyses (42 min total run time per injection). MS analyses were carried out by coupling the LC system to a Thermo Q Exactive HFTM mass spectrometer operating in heated electrospray ionization mode (HESI). Method duration was 30 min with a polarity switching data-dependent Top 5 method for both positive and negative modes. Spray voltage for both positive and negative modes was 3.5 kV and capillary temperature was set to 320oC with a sheath gas rate of 35, aux gas of 10, and max spray current of 100 μA. The full MS scan for both polarities utilized 120,000 resolution with an AGC target of 3e6 and a maximum IT of 100 ms, and the scan range was from 67–1000 m/z. Tandem MS spectra for both positive and negative mode used a resolution of 15,000, AGC target of 1e5, maximum IT of 50 ms, isolation window of 0.4 m/z, isolation offset of 0.1 m/z, fixed first mass of 50 m/z, and 3-way multiplexed normalized collision energies (nCE) of 10, 35, 80. The minimum AGC target was 1e4 with an intensity threshold of 2e5. All data were acquired in profile mode.
Bioinformatics for metabolomics
The resulting ThermoTM RAW files were converted to mzXML format using ReAdW.exe version 4.3.1 to enable peak detection and quantification. The centroided data were searched using an in-house python script Mighty_skeleton version 0.0.2 and peak heights were extracted from the mzXML files based on a previously established library of metabolite retention times and accurate masses adapted from the Whitehead Institute [183], and verified with authentic standards and/or high resolution MS/MS spectral manually curated against the NIST14MS/MS [184] and METLIN [185] tandem mass spectral libraries. Metabolite peaks were extracted based on the theoretical m/z of the expected ion type e.g., [M + H] + , with a ± 5 part-per-million (ppm) tolerance, and a ± 7.5 s peak apex retention time tolerance within an initial retention time search window of ± 0.5 min across the study samples. The resulting data matrix of metabolite intensities for all samples and blank controls was processed with an in-house statistical pipeline Metabolyze version 1.0 and final peak detection was calculated based on a signal to noise ratio (S/N) of 3X compared to blank controls, with a floor of 10,000 (arbitrary units). For samples where the peak intensity was lower than the blank threshold, metabolites were annotated as not detected, and the threshold value was imputed for any statistical comparisons to enable an estimate of the fold change as applicable. The resulting blank corrected data matrix was then used for all group-wise comparisons, and t-tests were performed with the Python SciPy (1.1.0) [186] library to test for differences and generate statistics for downstream analyses. Any metabolite with p value < 0.05 was considered significantly regulated (up or down). Heatmaps were generated with hierarchical clustering performed on the imputed matrix values utilizing the R library Pheatmap (1.0.12) [187]. Volcano plots were generated utilizing the R library, Manhattanly (0.2.0). In order to adjust for significant covariate effects (as applicable) in the experimental design the R package, DESeq2 (1.24.0) [188] was used to test for significant differences. Data processing for this correction required the blank corrected matrix to be imputed with zeroes for non-detected values instead of the blank threshold to avoid false positives. This corrected matrix was then analyzed utilizing DESeq2 to calculate the adjusted p value in the covariate model. Final graphing for volcano plots was performed in Graphpad Prism v6.0.
Design & statistical analysis
GraphPad Prism v6.0 was used for all statistical analysis and graphing. All data shown in the manuscript represents Mean ± Standard Error of the Mean (SEM). ANOVAs were the principal statistical test employed to control for the number of group comparisons being completed, as well as correct them when necessary. For comparisons that specifically involved post hoc comparisons or a specific comparison of just two groups, a t test was utilized. Cohen’s d was adapted for effect size estimation when evaluated. Significance was generally set at p < 0.05 per Fisher’s tables, tailed according to statistical-directionality guidelines and corrected for multiple-comparisons. To temper the potential influence of extraneous variance, we adapted unbiased, high-throughput, and analytical methodology where possible (e.g., TMT-LC/MS proteomics, metabolomics, and multiple flow cytometry experiments). This precluded the possibility extraneous variance arising from experimenter bias within these experiments. We also adapted a high-content approach that was concordant with principles of sampling theory. Specifically, we generated, observed, and analyzed as many units of analysis as possible. This resulted in hundreds of organoids being generated and randomly selected for experiments, that were each generated from multiple independent batches. Individual group numbers have been provided throughout the manuscript for clarity. Cellular quantifications were typically conducted in 5000 µm2 Regions of Interest (ROI) that typically extended 0–50 µm and 51–100 µm radially from ventricular zones. Flow cytometry data was analyzed and presented both as a global density %, normalized to total number of cellular singlet events for cell-cycle analysis and as fold-change for apoptosis and DNA damage analyses. Computational analyses were completed, and corrected for false discovery rates, as described above.
Supplementary information
Supplementary Table 1. Global Analysis of TMT-LC:MS Proteomics by ANOVA
Supplementary Table 2. Global Pairwise Analysis of all TMT-LC:MS Quantifiable Proteins
Supplementary Table 3. Summary of Significant TMT-LC:MS Pairwise Comparisons
Supplementary Table 4. Distinct and Common Proteome Alterations Across Groups
Acknowledgements
MN was the recipient of a NHMRC CJ Martin Grant/Fellowship. We acknowledge NYU Langone Health’s Metabolomics Laboratory for its help in acquiring and analyzing the metabolomic data presented, which is partially supported by Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center of NYU. We would also like to thank the WCMC Flow Cytometry, particularly Jason McCormick, and the Proteomics core for technical assistance.
Author contributions
MN and DC conceived the project and designed experiments. MN conducted all experiments, analyzed data, and wrote the manuscript with input from DC. AL provided assistance in maintaining all iPSC lines and generating organoid cultures, as well as provided technical support for experiments. CF provided assistance in generating organoids specifically for metabolomics. EA contributed to single-cell DNA content/cell-cycle analysis. DJ and TR oversaw metabolomics experiments. DG and HF conducted proteomic bioinformatics.
Data availability
The MS-based proteomics data have been deposited in PeptideAtlas repository and are available with identifier PASS01187.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41380-021-01189-9.
References
- 1.Thompson CL, Ng L, Menon V, Martinez S, Lee C-K, Glattfelder K, et al. A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron. 2014;83:309–23. doi: 10.1016/j.neuron.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuurmans C, Guillemot F. Molecular mechanisms underlying cell fate specification in the developing telencephalon. Curr Opin Neurbiol. 2002;12:26–34. doi: 10.1016/s0959-4388(02)00286-6. [DOI] [PubMed] [Google Scholar]
- 3.Kuzawa CW, Chugani HT, Grossman LI, Lipovich L, Muzik O, Hof PR, et al. Metabolic costs and evolutionary implications of human brain development. Proc Nat Acad Sci USA. 2014;111:13010–5. doi: 10.1073/pnas.1323099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Driscoll M, Jeggo PA. The role of the DNA damage response pathways in brain development and microcephaly: insight from human disorders. DNA Rep. 2008;7:1039–50. doi: 10.1016/j.dnarep.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Kosodo Y, Suetsugu T, Suda M, Mimori‐Kiyosue Y, Toida K, Baba SA, et al. Regulation of interkinetic nuclear migration by cell cycle‐coupled active and passive mechanisms in the developing brain. EMBO J. 2011;30:1690–704. doi: 10.1038/emboj.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuan C-Y, Roth KA, Flavell RA, Rakic P. Mechanisms of programmed cell death in the developing brain. Trends Neurosci. 2000;23:291–7. doi: 10.1016/s0166-2236(00)01581-2. [DOI] [PubMed] [Google Scholar]
- 7.Fu W, Killen M, Culmsee C, Dhar S, Pandita TK, Mattson MP. The catalytic subunit of telomerase is expressed in developing brain neurons and serves a cell survival-promoting function. J Mol Neurosci. 2000;14:3–15. doi: 10.1385/JMN:14:1-2:003. [DOI] [PubMed] [Google Scholar]
- 8.Weng Q, Wang J, Wang J, He D, Cheng Z, Zhang F, et al. Single-cell transcriptomics uncovers glial progenitor diversity and cell fate determinants during development and gliomagenesis. Cell Stem Cell. 2019;24:707–23. e8. doi: 10.1016/j.stem.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown AS. Epidemiologic studies of exposure to prenatal infection and risk of schizophrenia and autism. Dev Neurobiol. 2012;72:1272–6. doi: 10.1002/dneu.22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardener H, Spiegelman D, Buka SL. Prenatal risk factors for autism: comprehensive meta-analysis. Br J Neuropsychiatry. 2009;195:7–14. doi: 10.1192/bjp.bp.108.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinney DK, Munir KM, Crowley DJ, Miller AM. Prenatal stress and risk for autism. Neurosci Biobehav Rev. 2008;32:1519–32. doi: 10.1016/j.neubiorev.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolevzon A, Gross R, Reichenberg A. Prenatal and perinatal risk factors for autism: a review and integration of findings. Arch Pediatr Adolesc Med. 2007;161:326–33. doi: 10.1001/archpedi.161.4.326. [DOI] [PubMed] [Google Scholar]
- 13.Brown AS. Prenatal infection as a risk factor for schizophrenia. Schizophr Bull. 2006;32:200–2. doi: 10.1093/schbul/sbj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown AS. Exposure to prenatal infection and risk of schizophrenia. Front Psychiatry. 2011;2:63. doi: 10.3389/fpsyt.2011.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown AS, Begg MD, Gravenstein S, Schaefer CA, Wyatt RJ, Bresnahan M, et al. Serologic evidence of prenatal influenza in the etiology of schizophrenia. Arch Gen Psychiatry. 2004;61:774–80. doi: 10.1001/archpsyc.61.8.774. [DOI] [PubMed] [Google Scholar]
- 16.Kinney DK. Prenatal stress and risk for schizophrenia. Int J Ment Health. 2000;29:62–72. [Google Scholar]
- 17.Dwyer JB, McQuown SC, Leslie FM. The dynamic effects of nicotine on the developing brain. Pharm Ther. 2009;122:125–39. doi: 10.1016/j.pharmthera.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couey JJ, Meredith RM, Spijker S, Poorthuis RB, Smit AB, Brussaard AB, et al. Distributed network actions by nicotine increase the threshold for spike-timing-dependent plasticity in prefrontal cortex. Neuron. 2007;54:73–87. doi: 10.1016/j.neuron.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Goriounova NA, Mansvelder HD. Nicotine exposure during adolescence leads to short-and long-term changes in spike timing-dependent plasticity in rat prefrontal cortex. J Neurosci. 2012;32:10484–93. doi: 10.1523/JNEUROSCI.5502-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miwa JM, Stevens TR, King SL, Caldarone BJ, Ibanez-Tallon I, Xiao C, et al. The prototoxin lynx1 acts on nicotinic acetylcholine receptors to balance neuronal activity and survival in vivo. Neuron. 2006;51:587–600. doi: 10.1016/j.neuron.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 21.Muhammad A, Mychasiuk R, Nakahashi A, Hossain SR, Gibb R, Kolb B. Prenatal nicotine exposure alters neuroanatomical organization of the developing brain. Synapse. 2012;66:950–4. doi: 10.1002/syn.21589. [DOI] [PubMed] [Google Scholar]
- 22.Badanich KA, Mulholland PJ, Beckley JT, Trantham-Davidson H, Woodward JJ. Ethanol reduces neuronal excitability of lateral orbitofrontal cortex neurons via a glycine receptor dependent mechanism. Neuropsychopharmacol. 2013;38:1176–88. doi: 10.1038/npp.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nimitvilai S, Lopez MF, Mulholland PJ, Woodward JJ. Chronic intermittent ethanol exposure enhances the excitability and synaptic plasticity of lateral orbitofrontal cortex neurons and induces a tolerance to the acute inhibitory actions of ethanol. Neuropsychopharmacol. 2016;41:1112–27. doi: 10.1038/npp.2015.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pleil KE, Lowery-Gionta EG, Crowley NA, Li C, Marcinkiewcz CA, Rose JH, et al. Effects of chronic ethanol exposure on neuronal function in the prefrontal cortex and extended amygdala. Neuropharmacol. 2015;99:735–49. doi: 10.1016/j.neuropharm.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sathyan P, Golden HB, Miranda RC. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci. 2007;27:8546–57. doi: 10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice AC, Bullock MR, Shelton KL. Chronic ethanol consumption transiently reduces adult neural progenitor cell proliferation. Brain Res. 2004;1011:94–98. doi: 10.1016/j.brainres.2004.01.091. [DOI] [PubMed] [Google Scholar]
- 27.Crews FT, Nixon K, Wilkie ME. Exercise reverses ethanol inhibition of neural stem cell proliferation. Alcohol. 2004;33:63–71. doi: 10.1016/j.alcohol.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Miller MW. Migration of cortical neurons is altered by gestational exposure to ethanol. Alcohol Clin Exp Res. 1993;17:304–14. doi: 10.1111/j.1530-0277.1993.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 29.King MA, Hunter BE, Walker DW. Alterations and recovery of dendritic spine density in rat hippocampus following long-term ethanol ingestion. Brain Res. 1988;459:381–5. doi: 10.1016/0006-8993(88)90656-7. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence RC, Otero NK, Kelly SJ. Selective effects of perinatal ethanol exposure in medial prefrontal cortex and nucleus accumbens. Neurotoxicol Teratol. 2012;34:128–35. doi: 10.1016/j.ntt.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Streissguth AP, Aase JM, Clarren SK, Randels SP, LaDue RA, Smith DF. Fetal alcohol syndrome in adolescents and adults. JAMA. 1991;265:1961–7. [PubMed] [Google Scholar]
- 32.Zagon IS, MacLaughlin PJ. Endogenous opioid systems regulate cell proliferation in the developing rat brain. Brain Res. 1987;412:68–72. doi: 10.1016/0006-8993(87)91440-5. [DOI] [PubMed] [Google Scholar]
- 33.Wu C-C, Hung C-J, Shen C-H, Chen W-Y, Chang C-Y, Pan H-C, et al. Prenatal buprenorphine exposure decreases neurogenesis in rats. Toxicol Lett. 2014;225:92–101. doi: 10.1016/j.toxlet.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Persson AI, Thorlin T, Bull C, Zarnegar P, Ekman R, Terenius L, et al. Mu‐and delta‐opioid receptor antagonists decrease proliferation and increase neurogenesis in cultures of rat adult hippocampal progenitors. Eur J Neurosci. 2003;17:1159–72. doi: 10.1046/j.1460-9568.2003.02538.x. [DOI] [PubMed] [Google Scholar]
- 35.Kim E, Clark AL, Kiss A, Hahn JW, Wesselschmidt R, Coscia CJ, et al. μ-and κ-opioids induce the differentiation of embryonic stem cells to neural progenitors. J Biol Chem. 2006;281:33749–60. doi: 10.1074/jbc.M603862200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheng WS, Hu S, Herr G, Ni HT, Rock RB, Gekker G, et al. Human neural precursor cells express functional κ-opioid receptors. J Pharm Exp Ther. 2007;322:957–63. doi: 10.1124/jpet.107.121988. [DOI] [PubMed] [Google Scholar]
- 37.Tan KZ, Cunningham AM, Joshi A, Oei JL, Ward MC. Expression of kappa opioid receptors in developing rat brain–implications for perinatal buprenorphine exposure. Reprod Toxicol. 2018;78:81–89. doi: 10.1016/j.reprotox.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Boggess, T & WC Risher, Clinical and basic research investigations into the long‐term effects of prenatal opioid exposure on brain development. J Neurosci Res. 2020;00:1–14. [DOI] [PubMed]
- 39.SAMHSA, Results from the 2005 national survey on drug use and health: national findings. http://www.oas.samhsa.gov/nsduh/2k5nsduh/2k5Results.pdf, 2006.
- 40.Chadwick B, Miller ML, Hurd YL. Cannabis use during adolescent development: susceptibility to psychiatric illness. Front Psychiatry. 2013;4:129. doi: 10.3389/fpsyt.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Salas-Quiroga A, Díaz-Alonso J, García-Rincón D, Remmers F, Vega D, Gómez-Cañas M, et al. Prenatal exposure to cannabinoids evokes long-lasting functional alterations by targeting CB1 receptors on developing cortical neurons. Proc Nat Acad Sci USA. 2015;112:13693–8. doi: 10.1073/pnas.1514962112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jutras-Aswad D, DiNieri JA, Harkany T, Hurd YL. Neurobiological consequences of maternal cannabis on human fetal development and its neuropsychiatric outcome. Eur Arch Psych Clin Neurosci. 2009;259:395–412. doi: 10.1007/s00406-009-0027-z. [DOI] [PubMed] [Google Scholar]
- 43.Harkany T, Keimpema E, Barabás K, Mulder J. Endocannabinoid functions controlling neuronal specification during brain development. Mol Cell Endocrinol. 2008;286:S84–90. doi: 10.1016/j.mce.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Berrendero F, Garcia-Gil L, Hernandez M, Romero J, Cebeira M, De Miguel R, et al. Localization of mRNA expression and activation of signal transduction mechanisms for cannabinoid receptor in rat brain during fetal development. Dev. 1998;125:3179–88. doi: 10.1242/dev.125.16.3179. [DOI] [PubMed] [Google Scholar]
- 45.Raghunathan R, Liu CH, Kouka A, Singh M, Miranda RC, Larin KV. Assessing the acute effects of prenatal synthetic cannabinoid exposure on murine fetal brain vasculature using optical coherence tomography. J Biophoton. 2019;12:e201900050. doi: 10.1002/jbio.201900050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Estes ML, McAllister AK. Maternal immune activation: implications for neuropsychiatric disorders. Science. 2016;353:772–7. doi: 10.1126/science.aag3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knuesel I, Chicha L, Britschgi M, Schobel SA, Bodmer M, Hellings JA, et al. Maternal immune activation and abnormal brain development across CNS disorders. Nat Rev Neurol. 2014;10:643. doi: 10.1038/nrneurol.2014.187. [DOI] [PubMed] [Google Scholar]
- 48.Richetto J, Chesters R, Cattaneo A, Labouesse MA, Gutierrez AMC, Wood TC, et al. Genome-wide transcriptional profiling and structural magnetic resonance imaging in the maternal immune activation model of neurodevelopmental disorders. Cereb Cortex. 2017;27:3397–413. doi: 10.1093/cercor/bhw320. [DOI] [PubMed] [Google Scholar]
- 49.Haddad FL, Lu L, Baines KJ, Schmid S. Sensory filtering disruption caused by poly I: C-Timing of exposure and other experimental considerations. Brain Behav Immun Health. 2020;9:100156. doi: 10.1016/j.bbih.2020.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giovanoli S, Notter T, Richetto J, Labouesse MA, Vuillermot S, Riva MA, et al. Late prenatal immune activation causes hippocampal deficits in the absence of persistent inflammation across aging. J Neuroinflamm. 2015;12:1–18. doi: 10.1186/s12974-015-0437-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hill R, Nakamura J, Sundram S. Touchscreen cognitive performance following maternal immune activation targeting early and late prenatal neurodevelopmental windows. Biol Psychiatry. 2020;87:S235–236. [Google Scholar]
- 52.Nakamura JP, Schroeder A, Hudson M, Jones N, Gillespie B, Du X, et al. The maternal immune activation model uncovers a role for the Arx gene in GABAergic dysfunction in schizophrenia. Brain Behav Immun. 2019;81:161–71. doi: 10.1016/j.bbi.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 53.Richetto J, Calabrese F, Riva MA, Meyer U. Prenatal immune activation induces maturation-dependent alterations in the prefrontal GABAergic transcriptome. Schizophr Bull. 2014;40:351–61. doi: 10.1093/schbul/sbs195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, et al. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–9. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W, Li Q, Deyssenroth M, Lambertini L, Finik J, Ham J, et al. Timing of prenatal exposure to trauma and altered placental expressions of hpa-axis genes and genes driving neurodevelopment. J Neuroendocrinol. 2018;30:e12581. doi: 10.1111/jne.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Notaras M, van den Buuse M. Neurobiology of BDNF in fear memory, sensitivity to stress, and stress-related disorders. Mol Psychiatry. 2020;25:2251–74. doi: 10.1038/s41380-019-0639-2. [DOI] [PubMed] [Google Scholar]
- 57.Fukumoto K, Morita T, Mayanagi T, Tanokashira D, Yoshida T, Sakai A, et al. Detrimental effects of glucocorticoids on neuronal migration during brain development. Mol Psychiatry. 2009;14:1119–31. doi: 10.1038/mp.2009.60. [DOI] [PubMed] [Google Scholar]
- 58.Modi N, Lewis H, Al-Naqeeb N, Ajayi-Obe M, Doré CJ, Rutherford M. The effects of repeated antenatal glucocorticoid therapy on the developing brain. Ped Res. 2001;50:581–5. doi: 10.1203/00006450-200111000-00008. [DOI] [PubMed] [Google Scholar]
- 59.Sinclair D, Webster MJ, Wong J, Weickert CS. Dynamic molecular and anatomical changes in the glucocorticoid receptor in human cortical development. Mol Psychiatry. 2011;16:504–15. doi: 10.1038/mp.2010.28. [DOI] [PubMed] [Google Scholar]
- 60.Notaras M, Hill R, Gogos J, van den Buuse M. BDNF Val66Met genotype determines hippocampus-dependent behavior via sensitivity to glucocorticoid signaling. Mol Psychiatry. 2016;21:730–2. doi: 10.1038/mp.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinstock M. The long-term behavioural consequences of prenatal stress. Neurosci Biobehav Rev. 2008;32:1073–86. doi: 10.1016/j.neubiorev.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 62.Abe H, Hidaka N, Kawagoe C, Odagiri K, Watanabe Y, Ikeda T, et al. Prenatal psychological stress causes higher emotionality, depression-like behavior, and elevated activity in the hypothalamo-pituitary-adrenal axis. Neurosci Res. 2007;59:145–51. doi: 10.1016/j.neures.2007.06.1465. [DOI] [PubMed] [Google Scholar]
- 63.Markham JA, Koenig JI. Prenatal stress: role in psychotic and depressive diseases. Psychopharmacol. 2011;214:89–106. doi: 10.1007/s00213-010-2035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patin V, Lordi B, Vincent A, Caston J. Effects of prenatal stress on anxiety and social interactions in adult rats. Dev Brain Res. 2005;160:265–74. doi: 10.1016/j.devbrainres.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 65.Van Os,J, Selten J-P. Prenatal exposure to maternal stress and subsequent schizophrenia. Br J Psychiatry. 1998;172:324–6. doi: 10.1192/bjp.172.4.324. [DOI] [PubMed] [Google Scholar]
- 66.Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Nat Acad Sci USA. 2015;112:15672–7. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR. Cerebral organoids recapitulate epigenomic signatures of the human fetal brain. Cell Rep. 2016;17:3369–84. doi: 10.1016/j.celrep.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ziffra, RS, CN Kim, A Wilfert, TN Turner, M Haeussler, AM Casella, et al., Single cell epigenomic atlas of the developing human brain and organoids. bioRxiv, 2020: p. 2019.12.30.891549.
- 70.Notaras, M, A Lodhi, F Dundar, P Collier, M Allen, N Sayles, et al. Multiple neurodevelopmental mechanisms of schizophrenia in patient-derived cerebral organoids. Biological Psychiatry 2021;89:9-S100. [DOI] [PMC free article] [PubMed]
- 71.Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–7. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arlotta P, Paşca SP. Cell diversity in the human cerebral cortex: from the embryo to brain organoids. Curr Op Neurobiol. 2019;56:194–8. doi: 10.1016/j.conb.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 73.Tanaka Y, Cakir B, Xiang Y, Sullivan GJ, Park I-H. Synthetic analyses of single-cell transcriptomes from multiple brain organoids and fetal brain. Cell Rep. 2020;30:1682–9.e3. doi: 10.1016/j.celrep.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prytkova I, Brennand KJ. Prospects for modeling abnormal neuronal function in schizophrenia using human induced pluripotent stem cells. Front Cell Neurosci. 2017;11:360. doi: 10.3389/fncel.2017.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arlotta P. Organoids required! A new path to understanding human brain development and disease. Nat Method. 2018;15:27–29. doi: 10.1038/nmeth.4557. [DOI] [PubMed] [Google Scholar]
- 76.Li Y, Muffat J, Omer A, Bosch I, Lancaster MA, Sur M, et al. Induction of expansion and folding in human cerebral organoids. Cell Stem Cell. 2017;20:385–96. e3. doi: 10.1016/j.stem.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell. 2019;176:743–56.e17. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18:246–54. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 79.Wang P, Mokhtari R, Pedrosa E, Kirschenbaum M, Bayrak C, Zheng D, et al. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol Autism. 2017;8:11. doi: 10.1186/s13229-017-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang W, Ma L, Yang M, Shao Q, Xu J, Lu Z, et al. Cerebral organoid and mouse models reveal a RAB39b–PI3K–mTOR pathway-dependent dysregulation of cortical development leading to macrocephaly/autism phenotypes. Gene Dev. 2020;34:580–97. doi: 10.1101/gad.332494.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ilieva M, Svenningsen ÅF, Thorsen M, Michel TM. Psychiatry in a dish: stem cells and brain organoids modeling autism spectrum disorders. Biol Psychiatry. 2018;83:558–68. doi: 10.1016/j.biopsych.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 82.Stachowiak E, Benson C, Narla S, Dimitri A, Chuye LB, Dhiman S, et al. Cerebral organoids reveal early cortical maldevelopment in schizophrenia—computational anatomy and genomics, role of FGFR1. Transl Psychiatry. 2017;7:1–24. doi: 10.1038/s41398-017-0054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sawada T, Chater TE, Sasagawa Y, Yoshimura M, Fujimori-Tonou N, Tanaka K, et al. Developmental excitation-inhibition imbalance underlying psychoses revealed by single-cell analyses of discordant twins-derived cerebral organoids. Mol Psychiatry. 2020;25:2695–711. doi: 10.1038/s41380-020-0844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kathuria A, Lopez-Lengowski K, Jagtap SS, McPhie D, Perlis RH, Cohen BM, et al. Transcriptomic Landscape and Functional Characterization of Induced Pluripotent Stem Cell–Derived Cerebral Organoids in Schizophrenia. JAMA Psychiatry. 2020;77:745–54. doi: 10.1001/jamapsychiatry.2020.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mukhopadhyay S, Badgandi HB, Hwang S-h, Somatilaka B, Shimada IS, Pal K. Trafficking to the primary cilium membrane. Mol Biol Cell. 2017;28:233–9. doi: 10.1091/mbc.E16-07-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Louvi A, Grove EA. Cilia in the CNS: the quiet organelle claims center stage. Neuron. 2011;69:1046–60. doi: 10.1016/j.neuron.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wright JW, Harding JW. The brain angiotensin system and extracellular matrix molecules in neural plasticity, learning, and memory. Prog Neurobiol. 2004;72:263–93. doi: 10.1016/j.pneurobio.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 88.Kaneko N, Sawada M, Sawamoto K. Mechanisms of neuronal migration in the adult brain. J Neurochem. 2017;141:835–47. doi: 10.1111/jnc.14002. [DOI] [PubMed] [Google Scholar]
- 89.Wolf AM, Lyuksyutova AI, Fenstermaker AG, Shafer B, Lo CG, Zou Y. Phosphatidylinositol-3-kinase–atypical protein kinase C signaling is required for Wnt attraction and anterior–posterior axon guidance. J Neurosci. 2008;28:3456–67. doi: 10.1523/JNEUROSCI.0029-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cárdenas A, Villalba A, de Juan Romero C, Picó E, Kyrousi C, Tzika AC, et al. Evolution of cortical neurogenesis in amniotes controlled by robo signaling levels. Cell. 2018;174:590–606.e21. doi: 10.1016/j.cell.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blockus H, Chédotal A. The multifaceted roles of Slits and Robos in cortical circuits: from proliferation to axon guidance and neurological diseases. Curr Op Neurbiol. 2014;27:82–88. doi: 10.1016/j.conb.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 92.Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc. 2006;1:1458–61. doi: 10.1038/nprot.2006.238. [DOI] [PubMed] [Google Scholar]
- 93.Matassov, D, T Kagan, J Leblanc, M Sikorska, & Z Zakeri, Measurement of apoptosis by DNA fragmentation, in Apoptosis Methods and Protocols. 2004, Springer. p. 1-17. [DOI] [PubMed]
- 94.Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, Van de A, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–7. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 95.Kanakry CG, Li Z, Nakai Y, Sei Y, Weinberger DR. Neuregulin-1 regulates cell adhesion via an ErbB2/phosphoinositide-3 kinase/Akt-dependent pathway: potential implications for schizophrenia and cancer. PloS one. 2007;2:e1369. doi: 10.1371/journal.pone.0001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benzel I, Bansal A, Browning BL, Galwey NW, Maycox PR, McGinnis R, et al. Interactions among genes in the ErbB-Neuregulin signalling network are associated with increased susceptibility to schizophrenia. Behav Brain Func. 2007;3:1–11. doi: 10.1186/1744-9081-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X-D, Su Y-A, Guo C-M, Yang Y, Si T-M. Chronic antipsychotic drug administration alters the expression of neuregulin 1 β, ErbB2, ErbB3, and ErbB4 in the rat prefrontal cortex and hippocampus. Int J Neuropsychopharmacol. 2008;11:553–61. doi: 10.1017/S1461145707008371. [DOI] [PubMed] [Google Scholar]
- 98.Dabbah-Assadi F, Alon D, Golani I, Doron R, Kremer I, Beloosesky R, et al. The influence of immune activation at early vs late gestation on fetal NRG1-ErbB4 expression and behavior in juvenile and adult mice offspring. Brain Behav Immun. 2019;79:207–15. doi: 10.1016/j.bbi.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 99.Hemmerle AM, Ahlbrand R, Bronson SL, Lundgren KH, Richtand NM, Seroogy KB. Modulation of schizophrenia-related genes in the forebrain of adolescent and adult rats exposed to maternal immune activation. Schizophr Res. 2015;168:411–20. doi: 10.1016/j.schres.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Patapoutian A, Reichardt LF. Roles of Wnt proteins in neural development and maintenance. Cur Op Neurobiol. 2000;10:392–9. doi: 10.1016/s0959-4388(00)00100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mulligan KA, Cheyette BN. Neurodevelopmental perspectives on Wnt signaling in psychiatry. Mol Neuropsychiatry. 2016;2:219–46. doi: 10.1159/000453266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kwan V, Unda BK, Singh KK. Wnt signaling networks in autism spectrum disorder and intellectual disability. J Neurodev Dis. 2016;8:1–10. doi: 10.1186/s11689-016-9176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kalkman HO. A review of the evidence for the canonical Wnt pathway in autism spectrum disorders. Mol Autism. 2012;3:1–12. doi: 10.1186/2040-2392-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Topol A, Zhu S, Tran N, Simone A, Fang G, Brennand KJ. Altered WNT signaling in human induced pluripotent stem cell neural progenitor cells derived from four schizophrenia patients. Biol Psychiatry. 2015;78:e29–e34. doi: 10.1016/j.biopsych.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miyaoka T, Seno H, Ishino H. Increased expression of Wnt-1 in schizophrenic brains. Schizophr Res. 1999;38:1–6. doi: 10.1016/s0920-9964(98)00179-0. [DOI] [PubMed] [Google Scholar]
- 106.Cotter D, Kerwin R, Al-Sarraji S, Brion JP, Chadwich A, Lovestone S, et al. Abnormalities of Wnt signalling in schizophrenia–evidence for neurodevelopmental abnormality. Neuroreport. 1998;9:1379–83. doi: 10.1097/00001756-199805110-00024. [DOI] [PubMed] [Google Scholar]
- 107.Qu Q, Zhang F, Zhang X, Yin W. Bidirectional regulation of mouse embryonic stem cell proliferation by nicotine is mediated through Wnt signaling pathway. Dose Response. 2017;15:1559325817739760. doi: 10.1177/1559325817739760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brooks AC, Henderson BJ. Systematic review of nicotine exposure’s effects on neural stem and progenitor cells. Brain Sci. 2021;11:172. doi: 10.3390/brainsci11020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, et al. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 110.Simpson JH, Bland KS, Fetter RD, Goodman CS. Short-range and long-range guidance by Slit and its Robo receptors: a combinatorial code of Robo receptors controls lateral position. Cell. 2000;103:1019–32. doi: 10.1016/s0092-8674(00)00206-3. [DOI] [PubMed] [Google Scholar]
- 111.Dickson BJ, Gilestro GF. Regulation of commissural axon pathfinding by slit and its Robo receptors. Annu Rev Cell Dev Biol. 2006;22:651–75. doi: 10.1146/annurev.cellbio.21.090704.151234. [DOI] [PubMed] [Google Scholar]
- 112.Kurosaki T, Popp MW, Maquat LE. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol. 2019;20:406–20. doi: 10.1038/s41580-019-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet. 2013;22:4673–87. doi: 10.1093/hmg/ddt315. [DOI] [PubMed] [Google Scholar]
- 114.Alrahbeni T, Sartor F, Anderson J, Miedzybrodzka Z, McCaig C, Müller B. Full UPF3B function is critical for neuronal differentiation of neural stem cells. Mol Brain. 2015;8:1–15. doi: 10.1186/s13041-015-0122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Colak D, Ji S-J, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013;153:1252–65. doi: 10.1016/j.cell.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Notaras, M, M Allen, F Longo, N Volk, M Toth, NL Jeon, et al., UPF2 leads to degradation of dendritically targeted mRNAs to regulate synaptic plasticity and cognitive function. Mol Psychiatry, 2019;25:1–20. [DOI] [PMC free article] [PubMed]
- 117.Lynch SA, Nguyen LS, Ng LY, Waldron M, McDonald D, Gecz J. Broadening the phenotype associated with mutations in UPF3B: two further cases with renal dysplasia and variable developmental delay. Eur J Med Genet. 2012;55:476–9. doi: 10.1016/j.ejmg.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 118.Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–33. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Addington A, Gauthier J, Piton A, Hamdan F, Raymond A, Gogtay N, et al. A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol Psychiatry. 2011;16:238–9. doi: 10.1038/mp.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Swaiman KF, Wu SR. Phenylalanine and phenylacetate adversely affect developing mammalian brain neurons. Neurol. 1984;34:1246. doi: 10.1212/wnl.34.9.1246. [DOI] [PubMed] [Google Scholar]
- 121.Li D, Gu X, Lu L, Liang L. Effects of phenylalanine on the survival and neurite outgrowth of rat cortical neurons in primary cultures: possible involvement of brain-derived neurotrophic factor. Mol Cell Biochem. 2010;339:1–7. doi: 10.1007/s11010-009-0364-2. [DOI] [PubMed] [Google Scholar]
- 122.Fernandes CG, Leipnitz G, Seminotti B, Amaral AU, Zanatta Â, Vargas CR, et al. Experimental evidence that phenylalanine provokes oxidative stress in hippocampus and cerebral cortex of developing rats. Cell Mol Neurobiol. 2010;30:317–26. doi: 10.1007/s10571-009-9455-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Agrawal H, Bone A, Davison A. Effect of phenylalanine on protein synthesis in the developing rat brain. Biochem J. 1970;117:325–31. doi: 10.1042/bj1170325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Segawa M, Nomura Y, Nishiyama N. Autosomal dominant guanosine triphosphate cyclohydrolase I deficiency (Segawa disease) Ann Neurol. 2003;54:S32–45. doi: 10.1002/ana.10630. [DOI] [PubMed] [Google Scholar]
- 125.Daston MM, Ratner N. Neurofibromin, a predominantly neuronal GTPase activating protein in the adult, is ubiquitously expressed during development. Dev Dynamics. 1992;195:216–26. doi: 10.1002/aja.1001950307. [DOI] [PubMed] [Google Scholar]
- 126.Govek EE, Hatten ME, Van L. Aelst, The role of Rho GTPase proteins in CNS neuronal migration. Dev Neurobiol. 2011;71:528–53. doi: 10.1002/dneu.20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ridley AJ. Rho GTPase signalling in cell migration. Cur Opin Cell Biol. 2015;36:103–12. doi: 10.1016/j.ceb.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leone DP, Srinivasan K, Brakebusch C, McConnell SK. The rho GTPase Rac1 is required for proliferation and survival of progenitors in the developing forebrain. Dev Neurobiol. 2010;70:659–78. doi: 10.1002/dneu.20804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Bräuninger M, et al. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci. 2006;9:1099–107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- 130.Montenegro-Venegas C, Tortosa E, Rosso S, Peretti D, Bollati F, Bisbal M, et al. MAP1B regulates axonal development by modulating Rho-GTPase Rac1 activity. Mol Biol Cell. 2010;21:3518–28. doi: 10.1091/mbc.E09-08-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Naviaux J, Schuchbauer M, Li K, Wang L, Risbrough V, Powell S, et al. Reversal of autism-like behaviors and metabolism in adult mice with single-dose antipurinergic therapy. Transl Psychiatry. 2014;4:e400. doi: 10.1038/tp.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Frye, RE Mitochondrial dysfunction in autism spectrum disorder: Unique abnormalities and targeted treatments. in Sem Pediatr Neurol. 2020. Elsevier. [DOI] [PubMed]
- 133.Shen, L, Y Zhao, H Zhang, C Feng, Y Gao, D Zhao, et al., Advances in biomarker studies in autism spectrum disorders. Reviews on Biomarker Studies in Psychiatric and Neurodegenerative Disorders. 2019. 207-33. [DOI] [PubMed]
- 134.Su S-Y, Hogrefe-Phi CE, Asara JM, Turck CW, Golub MS. Peripheral fibroblast metabolic pathway alterations in juvenile rhesus monkeys undergoing long-term fluoxetine administration. Eur Neuropsychopharmacol. 2016;26:1110–8. doi: 10.1016/j.euroneuro.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bourdon AK, Spano GM, Marshall W, Bellesi M, Tononi G, Serra PA, et al. Metabolomic analysis of mouse prefrontal cortex reveals upregulated analytes during wakefulness compared to sleep. Sci Reps. 2018;8:1–17. doi: 10.1038/s41598-018-29511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fumagalli, C & U.b. Staff, Succinic acid and succinic anhydride. Kirk‐Othmer Encyclopedia of Chemical Technology, 2000.
- 137.Ma T, Li Y, Zhu Y, Jiang S, Cheng C, Peng Z, et al. Differential metabolic pathways and metabolites in a C57BL/6J mouse model of alcoholic liver disease. Med Sci Mon. 2020;26:e924602–1. doi: 10.12659/MSM.924602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kawashima N, Tanabe Y. Purification and properties of ribose phosphate isomerase from tobacco leaves. Plant Cell Physiol. 1976;17:757–64. [Google Scholar]
- 139.McClure EA, Gipson CD, Malcolm RJ, Kalivas PW, Gray KM. Potential role of N-acetylcysteine in the management of substance use disorders. CNS Drugs. 2014;28:95–106. doi: 10.1007/s40263-014-0142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Powell GL, Leyrer‐Jackson JM, Goenaga J, Namba MD, Piña J, Spencer S, et al. Chronic treatment with N‐acetylcysteine decreases extinction responding and reduces cue‐induced nicotine‐seeking. Physiol Rep. 2019;7:e13958. doi: 10.14814/phy2.13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ramirez-Niño AM, D’Souza MS, Markou A. N-acetylcysteine decreased nicotine self-administration and cue-induced reinstatement of nicotine seeking in rats: comparison with the effects of N-acetylcysteine on food responding and food seeking. Psychopharmacol. 2013;225:473–82. doi: 10.1007/s00213-012-2837-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bowers M, Jackson A, Maldoon P, Damaj M. N-acetylcysteine decreased nicotine reward-like properties and withdrawal in mice. Psychopharmacol. 2016;233:995–1003. doi: 10.1007/s00213-015-4179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schmaal L, Berk L, Hulstijn KP, Cousijn J, Wiers RW, van den Brink W. Efficacy of N-acetylcysteine in the treatment of nicotine dependence: a double-blind placebo-controlled pilot study. Eur Add Res. 2011;17:211–6. doi: 10.1159/000327682. [DOI] [PubMed] [Google Scholar]
- 144.Prado E, Maes M, Piccoli LG, Baracat M, Barbosa DS, Franco O, et al. N-acetylcysteine for therapy-resistant tobacco use disorder: a pilot study. Redox Rep. 2015;20:215–22. doi: 10.1179/1351000215Y.0000000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kathuria A, Lopez-Lengowski K, Watmuff B, McPhie D, Cohen BM, Karmacharya R. Synaptic deficits in iPSC-derived cortical interneurons in schizophrenia are mediated by NLGN2 and rescued by N-acetylcysteine. Transl Psychiatry. 2019;9:1–13. doi: 10.1038/s41398-019-0660-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.das Neves Duarte JM, Kulak A, Gholam-Razaee MM, Cuenod M, Gruetter R, et al. N-acetylcysteine normalizes neurochemical changes in the glutathione-deficient schizophrenia mouse model during development. Biol Psychiatry. 2012;71:1006–14. doi: 10.1016/j.biopsych.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 147.Ershova E, Jestkova E, Chestkov I, Porokhovnik L, Izevskaya V, Kutsev S, et al. Quantification of cell-free DNA in blood plasma and DNA damage degree in lymphocytes to evaluate dysregulation of apoptosis in schizophrenia patients. J Psychiatry Res. 2017;87:15–22. doi: 10.1016/j.jpsychires.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 148.Alexandre J, Carmo H, Carvalho F, Silva JP. Synthetic cannabinoids and their impact on neurodevelopmental processes. Addict Biol. 2020;25:e12824. doi: 10.1111/adb.12824. [DOI] [PubMed] [Google Scholar]
- 149.Miranda CC, Barata T, Vaz SH, Ferreira C, Quintas A, Bekman EP. hiPSC-based model of prenatal exposure to cannabinoids: effect on neuronal differentiation. Front Mol Neurosci. 2020;13:119. doi: 10.3389/fnmol.2020.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fernández-Ruiz J, Gómez M, Hernández M, de Miguel R, Ramos JA. Cannabinoids and gene expression during brain development. Neurotox Res. 2004;6:389–401. doi: 10.1007/BF03033314. [DOI] [PubMed] [Google Scholar]
- 151.Wu C-S, Jew CP, Lu H-C. Lasting impacts of prenatal cannabis exposure and the role of endogenous cannabinoids in the developing brain. Fut Neurol. 2011;6:459–80. doi: 10.2217/fnl.11.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tomiyama K-i, Funada M. Cytotoxicity of synthetic cannabinoids on primary neuronal cells of the forebrain: the involvement of cannabinoid CB1 receptors and apoptotic cell death. Toxicol Appl Pharm. 2014;274:17–23. doi: 10.1016/j.taap.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 153.Fernández-Ruiz J, Pazos MR, García-Arencibia M, Sagredo O, Ramos JA. Role of CB2 receptors in neuroprotective effects of cannabinoids. Mol Cel Endocrinol. 2008;286:S91–96. doi: 10.1016/j.mce.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 154.Saez TM, Aronne MP, Caltana L, Brusco AH. Prenatal exposure to the CB 1 and CB 2 cannabinoid receptor agonist WIN 55,212‐2 alters migration of early‐born glutamatergic neurons and GABA ergic interneurons in the rat cerebral cortex. J Neurochem. 2014;129:637–48. doi: 10.1111/jnc.12636. [DOI] [PubMed] [Google Scholar]
- 155.Fernández-Ruiz J, Romero J, Velasco G, Tolón RM, Ramos JA, Guzmán M. Cannabinoid CB2 receptor: a new target for controlling neural cell survival? Trend Pharm Sci. 2007;28:39–45. doi: 10.1016/j.tips.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 156.Guzman, M. Effects on cell viability. Cannabinoids, 2005: p. 627-42. [DOI] [PubMed]
- 157.Aguado T, Palazuelos J, Monory K, Stella N, Cravatt B, Lutz B, et al. The endocannabinoid system promotes astroglial differentiation by acting on neural progenitor cells. J Neurosci. 2006;26:1551–61. doi: 10.1523/JNEUROSCI.3101-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dang, J, SK Tiwari, K Agrawal, H Hui, Y Qin, & TM Rana, Glial cell diversity and methamphetamine-induced neuroinflammation in human cerebral organoids. Mol Psychiatry, 2020;168:1–14. [DOI] [PMC free article] [PubMed]
- 159.Hoffman EJ, Mintz CD, Wang S, McNickle DG, Salton SR, Benson DL. Effects of ethanol on axon outgrowth and branching in developing rat cortical neurons. Neurosci. 2008;157:556–65. doi: 10.1016/j.neuroscience.2008.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Abrahao KP, Salinas AG, Lovinger DM. Alcohol and the brain: neuronal molecular targets, synapses, and circuits. Neuron. 2017;96:1223–38. doi: 10.1016/j.neuron.2017.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kurek A, Kucharczyk M, Detka J, Ślusarczyk J, Trojan E, Głombik K, et al. Pro-apoptotic action of corticosterone in hippocampal organotypic cultures. Neurotox Res. 2016;30:225–38. doi: 10.1007/s12640-016-9630-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dufour BD, McBride JL. Corticosterone dysregulation exacerbates disease progression in the R6/2 transgenic mouse model of Huntington’s disease. Exp Neurol. 2016;283:308–17. doi: 10.1016/j.expneurol.2016.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Notaras M, Du X, Gogos J, Van Den Buuse M, Hill R. The BDNF Val66Met polymorphism regulates glucocorticoid-induced corticohippocampal remodeling and behavioral despair. Transl Psychiatry. 2017;7:e1233. doi: 10.1038/tp.2017.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Notaras MJ, Hill RA, Gogos JA, van den Buuse M. BDNF Val66Met genotype interacts with a history of simulated stress exposure to regulate sensorimotor gating and startle reactivity. Schizophr Bull. 2017;43:665–72. doi: 10.1093/schbul/sbw077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Koch M, Kreutz S, Böttger C, Grabiec U, Ghadban C, Korf HW, et al. The cannabinoid WIN 55,212‐2‐mediated protection of dentate gyrus granule cells is driven by CB1 receptors and modulated by TRPA1 and Cav2. 2 channels. Hippocampus. 2011;21:554–64. doi: 10.1002/hipo.20772. [DOI] [PubMed] [Google Scholar]
- 166.Aguirre-Rueda D, Guerra-Ojeda S, Aldasoro M, Iradi A, Obrador E, Mauricio MD, et al. WIN 55,212-2, agonist of cannabinoid receptors, prevents amyloid β1-42 effects on astrocytes in primary culture. PLoS One. 2015;10:e0122843. doi: 10.1371/journal.pone.0122843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Németh B, Ledent C, Freund TF, Hájos N. CB1 receptor-dependent and-independent inhibition of excitatory postsynaptic currents in the hippocampus by WIN 55,212-2. Neuropharmacol. 2008;54:51–57. doi: 10.1016/j.neuropharm.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Colombo E, Di Dario M, Capitolo E, Chaabane L, Newcombe J, Martino G, et al. Fingolimod may support neuroprotection via blockade of astrocyte nitric oxide. Ann Neurol. 2014;76:325–37. doi: 10.1002/ana.24217. [DOI] [PubMed] [Google Scholar]
- 169.Sarma, JD, B Ciric, R Marek, S Sadhukhan, J Shafagh, DC Fitzgerald, et al., Functional interleukin-17 receptor A is expressed in the central nervous system and upregulated in experimental autoimmune encephalomyelitisa. https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.589.9069&rep=rep1&type=pdf. [DOI] [PMC free article] [PubMed]
- 170.Chen, S, D Shen, NA Popp, AJ Ogilvy, J Tuo, M Abu-Asab, et al., Responses of multipotent retinal stem cells to IL-1β, IL-18, or IL-17. J Opthalmol, 2015;2015:1–9. [DOI] [PMC free article] [PubMed]
- 171.Gumusoglu SB, Hing BWQ, Chilukuri ASS, Dewitt JJ, Scroggins SM, Stevens HE. Chronic maternal interleukin-17 and autism-related cortical gene expression. Neurobiol, Behav Neuropsychopharmacol. 2020;45:1008–17. doi: 10.1038/s41386-020-0640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Irnaten M, Aicher S, Wang J, Venkatesan P, Evans C, Baxi S, et al. μ-Opioid receptors are located postsynaptically and endomorphin-1 inhibits voltage-gated calcium currents in premotor cardiac parasympathetic neurons in the rat nucleus ambiguus. Neurosci. 2003;116:573–82. doi: 10.1016/s0306-4522(02)00657-7. [DOI] [PubMed] [Google Scholar]
- 173.McConalogue K, Grady E, Minnis J, Balestra B, Tonini M, Brecha N, et al. Activation and internalization of the μ-opioid receptor by the newly discovered endogenous agonists, endomorphin-1 and endomorphin-2. Neurosci. 1999;90:1051–9. doi: 10.1016/s0306-4522(98)00514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Seseña E, Vega R, Soto E. Activation of μ-opioid receptors inhibits calcium-currents in the vestibular afferent neurons of the rat through a cAMP dependent mechanism. Front Cell Neurosci. 2014;8:90. doi: 10.3389/fncel.2014.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McGehee DS, Heath M, Gelber S, Devay P, Role LW. Nicotine enhancement of fast excitatory synaptic transmission in CNS by presynaptic receptors. Science. 1995;269:1692–6. doi: 10.1126/science.7569895. [DOI] [PubMed] [Google Scholar]
- 176.Garnier M, Lamacz M, Tonon MC, Vaudry H. Functional characterization of a nonclassical nicotine receptor associated with inositolphospholipid breakdown and mobilization of intracellular calcium pools. Proc Nat Acad Sci USA. 1994;91:11743–7. doi: 10.1073/pnas.91.24.11743. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 177.Corcoran JJ, Kirshner N. Effects of manganese and other divalent cations on calcium uptake and catecholamine secretion by primary cultures of bovine adrenal medulla cells. Cell Calcium. 1983;4:127–37. doi: 10.1016/0143-4160(83)90029-5. [DOI] [PubMed] [Google Scholar]
- 178.Cheng Z, Teo G, Krueger S, Rock TM, Koh HW, Choi H, et al. Differential dynamics of the mammalian mRNA and protein expression response to misfolding stress. Mol Syst. Biology. 2016;12:855. doi: 10.15252/msb.20156423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat Biotech. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 180.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 181.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Method. 2016;13:731. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 182.Pacold ME, Brimacombe KR, Chan SH, Rohde JM, Lewis CA, Swier LJ, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12:452–8. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chen WW, Freinkman E, Wang T, Birsoy K, Sabatini DM. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell. 2016;166:1324–37.e11. doi: 10.1016/j.cell.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Simon-Manso Y, Lowenthal MS, Kilpatrick LE, Sampson ML, Telu KH, Rudnick PA, et al. Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Anal Chem. 2013;85:11725–31. doi: 10.1021/ac402503m. [DOI] [PubMed] [Google Scholar]
- 185.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 186.Jones, E, E Oliphant, & PEA Peterson. SciPy: Open Source Scientific Tools for Python. 2001.
- 187.Kolde, R Pheatmap: Pretty Heatmaps R Package Version 1.0.8. https://CRAN.R-project.org/package=pheatmap. 2015.
- 188.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Global Analysis of TMT-LC:MS Proteomics by ANOVA
Supplementary Table 2. Global Pairwise Analysis of all TMT-LC:MS Quantifiable Proteins
Supplementary Table 3. Summary of Significant TMT-LC:MS Pairwise Comparisons
Supplementary Table 4. Distinct and Common Proteome Alterations Across Groups
Data Availability Statement
The MS-based proteomics data have been deposited in PeptideAtlas repository and are available with identifier PASS01187.