Abstract
Hepatocellular carcinoma (HCC) is the most common primary liver malignancy and the third cause of cancer-related death worldwide. Potential microRNAs have been reported as biomarkers for early detection of HCC as well as novel molecular targets for HCC treatment. Various tissue expression profiles of miRNAs using three microarray datasets from groups in Asia (2), Europe, America (GSE147892, GSE21362, GSE74618, GSE40744) and multiple bioinformatics tools were integrated to determine the most significant miRNA groups to assist in the diagnosis of HCC. Statistical analyses identified at least 30 miRNAs with 17 up-regulated and 13 down-regulated in HCC-related tumor tissues. All the miRNAs also showed relevance to the hallmarks of cancer such as cell proliferation, invasion, metastasis, angiogenesis, metabolism, epithelial-mesenchymal transition and apoptosis. Expression levels of miRNAs observed in the European group showed up-regulation at 5–37% compared to both Asian and American groups. Interestingly, four miRNAs divided into two groups as miR-182-5p/miR-1269a and miR-199a/miR-422a were the most promising for diagnosis of HCC patients from healthy controls, with AUC values of 0.902 and 0.892, respectively. Results provided evidence of the correlation between potential miRNAs and HCC that could be useful for disease diagnosis based on in-depth analyses of large case numbers and cohort studies.
Keywords: HCC, MicroRNA, Expression, Bioinformatics tools, Diagnosis, Cancer
HCC; MicroRNA; Expression; Bioinformatics tools; Diagnosis; Cancer.
1. Introduction
Hepatocellular carcinoma (HCC) accounts for 80–90% of primary liver cancer cases as the sixth most common cancer and the third leading cause of cancer deaths worldwide (GLOBOCAN, 2020) (Sung et al., 2021). Hepatitis B virus (HBV), hepatitis C virus (HCV), aflatoxin exposure, excessive alcohol intake and nonalcoholic fatty liver disease (NAFLD) are the most common causes resulting in cirrhosis and the largest risk factors for development of HCC (Dhanasekaran et al., 2012; Herbst and Reddy, 2012). Diagnosis of HCC is typically very late, leading to survival time of less than 2 years with a few cases of 2–6 years (McGlynn et al., 2015). HCC is also a “silent” killer due to non-typical appearance in people with no known liver disease, resulting in late diagnosis and treatment. Until now, no ideal screening modality has been developed for the prognosis of HCC. A biopsy is essential for the diagnosis of HCC and also most types of cancer, with ongoing debate about the ethical issues due to many risks for the patients (Doyle and Sherman, 2017), while other approaches such as alpha-fetoprotein (AFP), abdominal ultrasound (US), CT or MRI have drawbacks due to inadequate sensitivity for proper surveillance and diagnosis. HCC can also be diagnosed using multidisciplinary approaches including clinical, radiological and laboratory modalities with or without liver biopsy (Attwa and El-Etrby, 2015). Other methods such as radiofrequency ablation (RFA), microwave and percutaneous ethanol ablations, transarterial chemoembolization (TACE), systemic chemotherapy and molecularly targeted therapies have also been reported as suitable approaches for the treatment of patients (Attwa and El-Etrby, 2015; Forner et al., 2012; Shariff et al., 2009; Di Bisceglie, 2005).
Recently, the development of non-invasive biomarkers has provided high diagnostic accuracy as effective treatment and surveillance for early detection of HCC. Aberrant miRNA expression has been demonstrated as a potential biomarker in a variety of human cancers including HCC (Visone and Croce, 2009; Wang and Wu, 2009; Calin and Croce, 2006). Various up and down-regulated miRNAs such as miR-122, miR-221, miR-222, miR-224, miR-96, miR-155, miR-81, miR-331 and miR-346 were identified and reported as having diagnostic potential for HCC (Gramantieri et al., 2007; Burchard et al., 2010; Mizuguchi et al., 2011; Pineau et al., 2010; Wang et al., 2008). Application of “omics” technologies also plays an important role in detection of aberrant miRNAs that are small non-coding RNAs causing either translational repression or mRNA degradation (Iorio and Croce, 2009). The occurrence and stability of miRNAs in blood have been reported in previous studies (Jin et al., 2019), while evaluation of expression profiles of miRNAs using microarray and NGS approaches has shown improved understanding of the characteristics of hepatocellular carcinoma (HCC) to determine the significant potential miRNA signatures for use in non-invasive diagnosis of HCC (Jin et al., 2019; Murakami et al., 2014). Here, miRNA expression profiles for HCC and non-tumorous tissue were integrated and compared using multiple bioinformatics tools with three microarray datasets representing groups in Asia, Europe and America. Groups of significant miRNAs and their target genes were investigated to explore the statistical significance of specificity and sensitivity of potential miRNAs in hepatocarcinogenesis.
2. Materials and methods
2.1. Microarray datasets search and analysis
A systematic search of microarray datasets was conducted in Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) (Barrett et al., 2013). Using the keywords “Human Hepatocellular carcinoma”, “miRNA” and “Non coding RNA profiling by array”, 171 datasets were found. The sample count was set to more than 70 samples and the number of miRNA should be greater than 850 for statistical analyses. Finally, four datasets including two in Asia GSE21362 and GSE147892 (the combination of GSE147887 + GSE147889), one in Europe (GSE74618) and one in America (GSE40744) were selected.
2.2. Detection of differentially expressed miRNAs (DEMs)
The DEMs were grouped and analyzed to determine differences in expression between cancerous and benign tissues using GraphPad Prism version 8.4.3 (GraphPad Software, Inc., San Diego, CA) with t-test and two-way ANOVA analyses. Determination of miRNA structure and classification was conducted based on the miRbase database (http://www.mirbase.org/) to remove errors in analysis. Comparison of DEMs between HCC and the normal groups was identified based on p value <0.05. Significantly expressed DEMs in each study were listed.
2.3. Combination of the data
Overlapping DEMs were found in all datasets using a Venn diagram creator tool in Bioinformatics and Evolutionary Genomic source (http://bioinformatics.psb.ugent.be/webtools/Venn/). Herein, the list of each dataset contained only one DEM per row with an unlimited number of rows. To avoid missing critical genes, DEMs were selected that overlapped at least two of the datasets in Asia except for Europe and America. The output of this tool was a list of DEMs overlapping between datasets and a visualized Venn diagram plot.
2.4. Area under curve (AUC) analysis
Receiver-operating characteristic (ROC) curves and area under the ROC curve (AUC) were conducted by importing the expression values of all overlapping DEMs into GraphPad Prism version 8.4.3 (GraphPad Software, Inc., San Diego, CA). The detection ability of each miRNA in discriminating HCC patients from the control group based on the sensitivity and specificity of each DEM was assessed, following the method of Wilson and Brown with confidence level set at 95% (Brown et al., 2001).
2.5. Hierarchical clustering analysis
Expression values of significantly up/down-regulated DEMs in HCC (19 DEMs) of 2 datasets with the same platform array were aggregated to create a new data format that was logarithm transformed for the hierarchical clustering analysis using online heat-mapper tool (http://www.heatmapper.ca/). The selected clustering algorithm included the average linkage method and the distance measurement known as “Pearson correlation”. The clustering algorithm was applied to all rows that matched the format of the data, and the dendrogram was shown along with the heatmap.
2.6. MiRNA-mRNA interaction
The list of DEMs was used to include the microRNA query in MirDIP (https://ophid.utoronto.ca/mirDIP/) to synthesize predicted target gene lists of each considered miRNA from 30 different resources (Tokar et al., 2018). The class score was set to very high to determine the list of target genes of the DEMs, and the list was analyzed for function, gene ontology, disease and phenotype ontology using g:Profiler (https://biit.cs.ut.ee/gprofiler/gost) with the option of human analysis and threshold of significance according to the fast discovery rate (FDR) method of Benjamini-Hochberg (Haynes 2013). The miRNA-target network construction analysis was executed in Mirnet (http://www.mirnet.ca). The clear cut pathway related to HCC was selected based on p value ≤0.01. The UCSC Xena gene discovery tool (https://xenabrowser.net/) uses cancer data from TCGA and human genome data to visualize interactions between miR and target genes.
2.7. Logistic regression miR expression
Expression value of each miRNA associated with HCC was examined to differentiate between HCC and healthy groups and described as mean and standard deviation (SD). The HCC group was coded to 0, while the normal group was coded to 1. A binary linear regression equation was used to build up a data series of variables (DEMs) for appropriate groups. Analyses of the area under the ROC curve (AUC) to determine diagnostic levels relative to individual miRNAs or cluster values were conducted using GraphPad Prism version 8.4.3 (GraphPad Software, Inc., San Diego, CA, USA). As described previously, an AUC value of 0.5 can be considered as no discrimination and vice versa, with an AUC value of 1 as perfect discrimination (Shams et al., 2020). Results were set as statistically significant by p < 0.05 levels.
3. Results
3.1. Microarray datasets and overlapped differently expressed miRNAs (DEMs)
In this study, the datasets containing a large number of HCC tissues and healthy controls were selected from the GEO database. As shown in Table 1, GSE147892 is a combination between GSE14887 and GSE14889 datasets of which 97 HCC tissues and 97 normal samples were found and identified (Umezu et al., 2020). GSE21362 included 73 HCC tissues and 73 healthy controls (Sato et al., 2011). GSE74618 consisted of 250 samples collected in Spain and was classified into 4 groups (HCC, HCC cell line, HC and healthy samples) (Martinez-Quetglas et al., 2016). Finally, 76 samples of GSE40744 including HCC, HCV, HBV and healthy controls were also considered as a validation set (Diaz et al., 2013).
Table 1.
Microarray datasets collected from the GEO database.
| Accession ID | Platform | Country | No. of samples | Sample groups | Published year | Ref. |
|---|---|---|---|---|---|---|
| GSE74618 | GPL14613 | Spain | 250 | HCC/HCC cell line/HC/Healthy | 2016 | Shen et al. (2010) |
| GSE40744 | GPL14613 | America | 76 | HCC/HCV/HBV/Healthy | 2013 | Sung et al. (2021) |
| GSE147892 (the combination of GSE14887 and GSE14889 datasets | GPL21263 | Japan | 554 | HCV/HCC/sHCC | 2020 | Shams et al. (2020) |
| GSE21362 | GPL10312 | Japan | 146 | Tumor/Non tumor | 2021 | Shariff et al. (2009) |
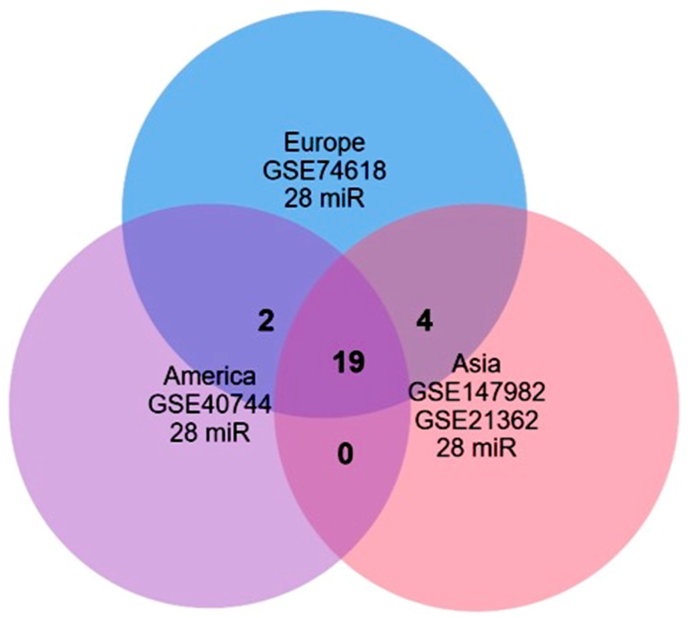
A total of 161, 235, 445 and 227 samples showed significant up/down regulation in GSE74618, GSE40744, GSE147892 and GSE21362 microarray datasets. After integration of the above results, 605 miRNAs that were common in Asia were captured in comparison with 161 and 235 miRNAs that were captured in Europe and America, respectively by using two-way ANOVA analysis (Figure S1). Interestingly, we also found that 28 miRNAs were common among all analyzed datasets in Asia, Europe and America (Figure S1; Table S1).
3.2. Identification of significant DEMs among the three regions
The differences between 28 common DEMs in Asian, European and American regions were also evaluated, based on fold change values to examine up/down regulations of the most significant DEMs. Results indicated statistically significant differences in up/down regulation of 19 DEMs between HCC and normal tissues in the three regions (Figure 1, Table S2).
Figure 1.
Venn diagram representing the most significant DEMs that are common between Asian, European and American regions.
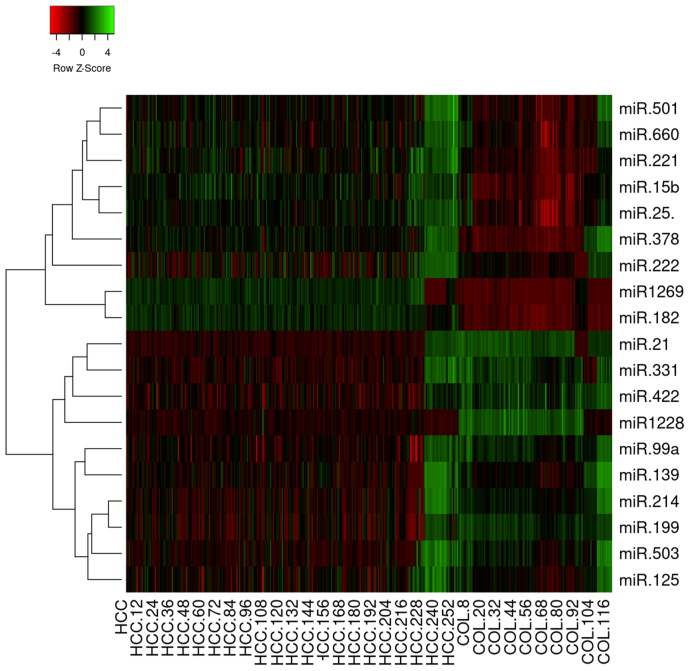
Based on the expression values of all 19 captured DEMs, results of the hierarchically clustered heatmap indicated that ten and nine miRNAs showed their down and up regulation respectively in HCC patients compared with normal subjects (Figure 2). Interestingly, expression profiles of these miRNAs also showed differences among the three regions. Some miRNAs such as miR-224, miR-150 and miR-146b-5 showed up-regulation in European and American populations but down-regulation in the Asian group. By contrast, miR-100, miR-10a, miR-223 and miR-375 showed down-regulation in the European and Asian populations but up-regulation in the American group (Table 2).
Figure 2.
Hierarchically clustered heatmap built up using expression values of all 19 captured DEMs. The heatmap was divided into two groups including up-regulated miRNAs (miR-501, miR-660, miR-15b, miR-25, miR-221, miR-378, miR-222, miR-182 and miR-1269) and down-regulated miRNAs (miR-21, miR-331, miR-422a, miR-1228, miR-99a, miR-139, miR-214, miR-199a, miR-503, miR-125).
Table 2.
Expression profiles of different miRs in the Asian, European and American regions.
| miRNAs | Regulation |
||
|---|---|---|---|
| Asia | America | Europe | |
| hsa-miR-150_st | Down | Up | Up |
| hsa-miR-146b-5p | Down | Up | Up |
| hsa-miR-224_st | Down | Up | Up |
| hsa-miR-100_st | Down | Up | Down |
| hsa-miR-10a_st | Down | Up | Down |
| hsa-miR-223_st | Down | Up | Down |
| hsa-miR-375_st | Down | Up | Down |
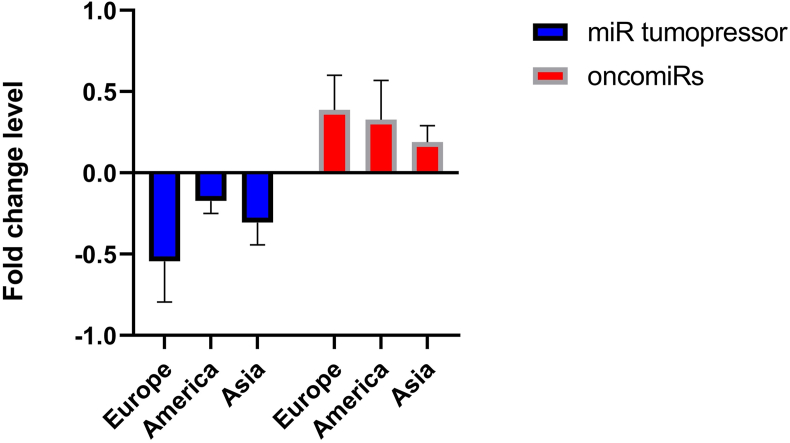
Difference between DEMs was also observed in the three different regions. MiRNAs in the European group showed an increased level of expression compared with the Asian and American groups, while European oncomiRs increased by 5.9% against the American and 19.8 % against the Asian groups with fold change mean values of 0.387/0.328/0.189 in Europe/America/Asia, respectively (Figure 3). Meanwhile, the miR tumorpressor expression in the European group increased 37.01% and 23.09% against the American and Asian groups, respectively with fold change mean values of -0.544, -0.173 and -0.306, respectively (Figure 3).
Figure 3.
Expression profiles of oncomiRs and tumor suppressor miRs in Europe, America and Asia.
3.3. AUC analysis
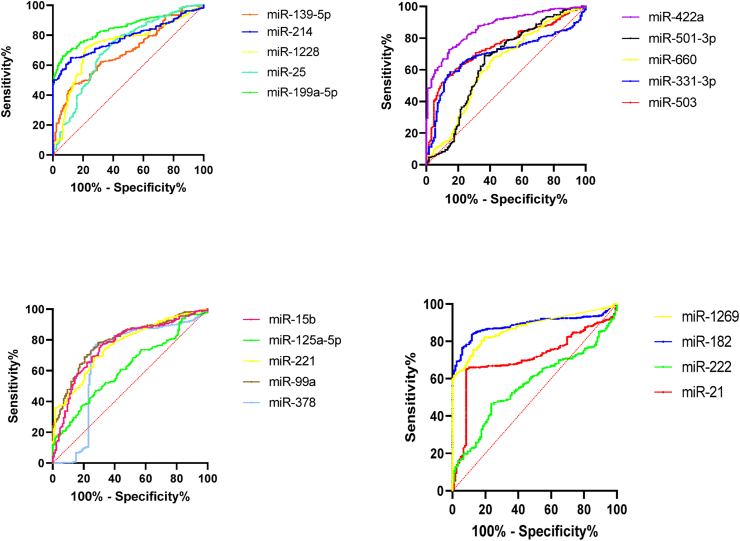
To prove the most reliable miRNAs in discriminating HCC from healthy controls, AUC analysis was performed and the ROC curves for all 19 miRNA were prepared. Four DEMS showed AUC scores greater than 80% (Table S3), with their ROC curves presented in Figure 4. DEMS with AUC greater than 80% are also listed in Table S3. ROC curve analyses showed that the AUC value for miRNA 182 was 0.887 (95% CI, 0.86–0.92) with mir-1269a 0.872 (95% CI, 0.84–0.90). Based on the results, miR-182, miR-1269, miR-422a and miR-199a-5p were the strongest individual signatures for differentiating HCC patients from the healthy control.
Figure 4.
Receiver-operating characteristic (ROC) curves of 19 captured DEMs in Asian, American and European regions.
3.4. Target genes of miRNAs and functional enrichment analysis
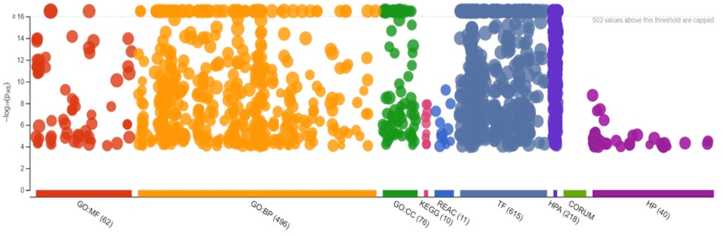
Overall, 4264 target genes for all 19 DEMs were predicted. Enrichment of the identified target genes was performed through g:Profiler online tool and GO and KEGG pathways. Results indicated that the target genes of 19 miRs involved 62 gene ontology (GO) molecular functions (MF), 496 GO biological processes (BP), 76 GO cellular components (CC), 21 biological pathways (KEGG and REACTOME), 615 Transfactors, 215 HPA and 40 HP (Figure 5). Gene ontology, molecular process analysis showed that the target genes were mostly involved in the regulation of cellular metabolic processes, while KEGG analysis indicated that the target genes were mainly implicated in cancer-related pathways including proteoglycans cancer and oncogenic MAPK signaling (Figure 6, Table S4).
Figure 5.
Enrichment analysis and functional annotation of DEMs.
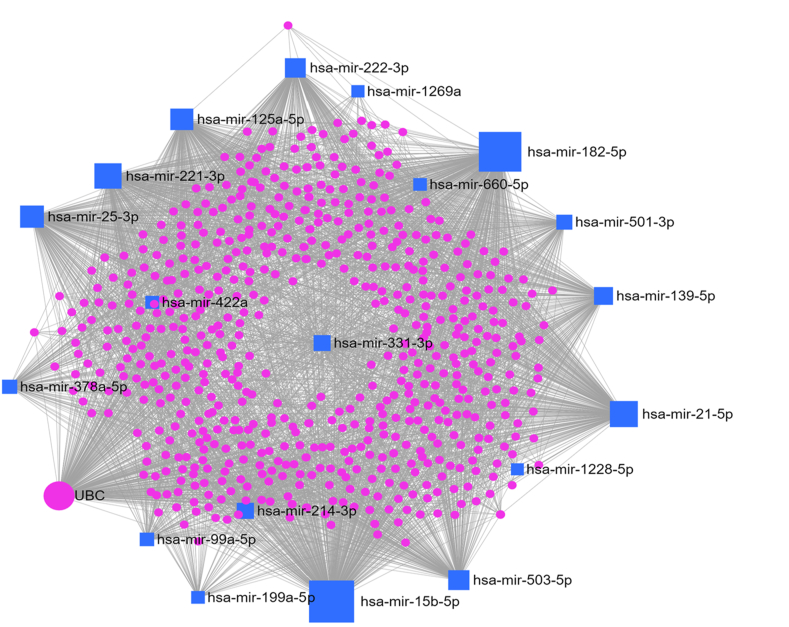
Figure 6.
The miRNA-mRNA interactions among all 19 considered DEMs. The pink circles represent all predicted target genes and the yellow ones represent the genes implicated in cancer related pathways. The network was built bases on string database with target genes from miRTarBase with confident score = 800 and cut off = 6. The network included 3883 nodes and 52455 edges in which miR-182 and miR-15b could play an important role in cancer.
To build an effective diagnostic combination, 4 miRs were selected out of 19 that expressed differently in all 4 datasets with the same prediction of target genes on miRDB, and reported experimentally on NCBI. MiR-182-5p and miR-1269a were identified to act together on the FOXO gene group (FOXO1, FOXO3a) (Cao et al., 2018; Yang et al., 2014). Results obtained by the UCSC Xena tool showed that out of 438 HCC samples collected from the TCGA database, both miR-182-5p and miR-1269a acted as oncomiRs and increased expression resulted in the decrease in expression of the target FOXO gene (Figure S2A). In total, out of 543 target genes of miR-199a-5p and 289 target genes of miR-422a predicted by miRDB, we identified 6 mRNAs that could be regulated by both miR-199a-5p and miR-422a with a score greater than 90 (Table 3). After screening and correlating the expression levels of 6 mRNAs with miR-199a-5p and miR-422a, we found that the expression correlation of 5 mRNAs including RSPH4A, ZBTB20, CDCA7L, SULF1 and ZNF124 was not clear with miR-199a-5p (data not shown), while no analytical data on expression correlation of these mRNAs with miR-422a were available. The reduced expression of only TMEM245 mRNA was observed with up-regulation of miR-199a-5p and vice versa (Figure S2B).
Table 3.
Common potential target mRNAs of miR-422a and miR-199a.
| mRNA | miR-199a-TS | miR-199a-TR | miR-422a-TS | miR-422a-TR |
|---|---|---|---|---|
| TMEM245 | 90 | 77 | 68 | 110 |
| RSPH4A | 73 | 253 | 98 | 71 |
| ZBTB20 | 95 | 27 | 50 | 299 |
| CDCA7L | 96 | 20 | 54 | 241 |
| SULF1 | 97 | 13 | 86 | 21 |
| ZNF124 | 71 | 277 | 92 | 6 |
TS: Target score, TR: Target rank.
3.4.1. Combined diagnosis of miR group and AUC value
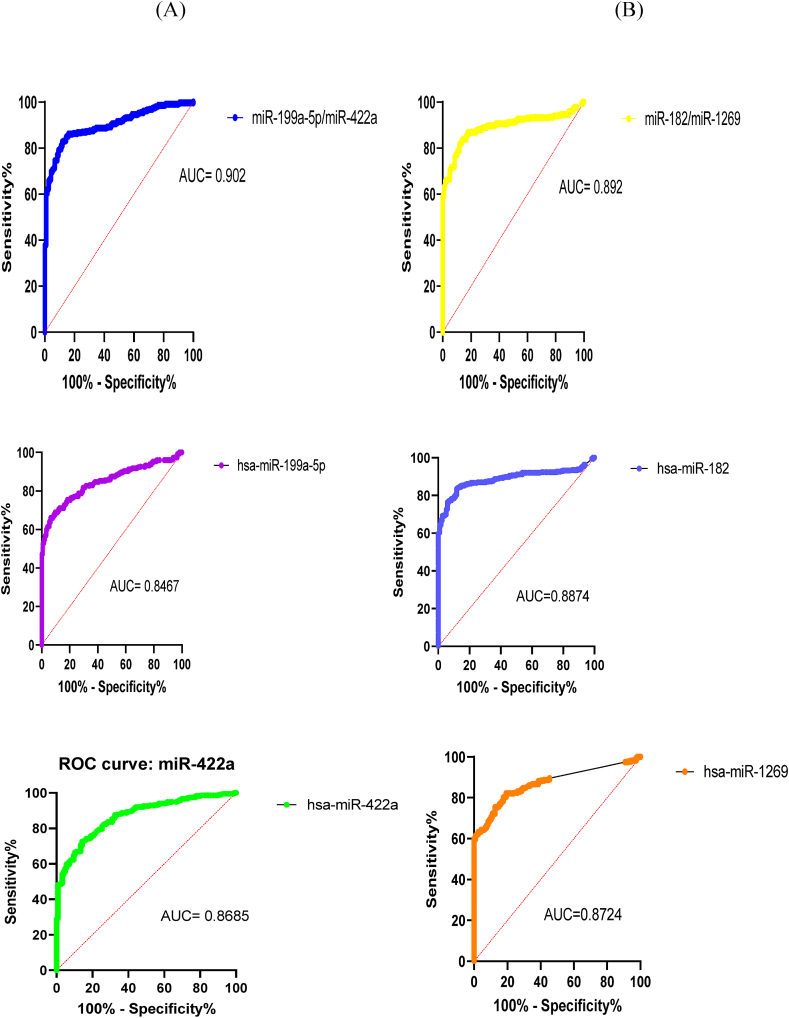
Four miRs were selected to determine AUC diagnostic values. Step-by-step linear regression equations were established and a diagnostic model was built based on cancer appearance in the samples. MiR-199a-5p and miR-422a were combined with the equation y = 0.049 (miR-199a-5p) + 0.092 (miR-422a) - 0.667, while the AUC values of the combination compared to the individual miRs were 0.902, 0.847 and 0.869, respectively (Figure 7A). Similarly, miR-182-5p and miR-1269a were combined according to the equation y = -0.071(miR182-5p) - 0.026(miR1269a) + 0.811, with AUC values of the combination compared to the individual miRs as 0.892, 0.887 and 0.872 (Figure 7B). The diagnostic value of the combination of miR-199a-5p/miR-422a/miR-182-5p/miR-1269a was not recorded when interaction between miRNAs and the symptoms was not significant.
Figure 7.
ROC analysis of potential groups and individuals of microRNA for HCC diagnosis. AUC values of the combination compared to the individual miRs in the groups of miR-199a-5p and miR-422a (A) and miR-182-5p and miR-1269a (B).
4. Discussion
MiRNAs have been considered as most interesting in cancer diagnostics; however, most recent studies focused on the expression patterns of individual miRNAs along with target genes but not on global microRNA expression patterns and their association with microRNA clusters. In this study, significant differences in the expression of various miRNAs involving HCC were observed in patients living in different worldwide regions, suggesting the impact of external factors such as environments and lifestyle on epigenetic mechanisms such as DNA methylation, histone acetylation and microRNA expression (Alegria-Torres et al., 2011). From more than 2,500 miRs analyzed, the expression of miRs associated with HCC was extremely large, indicating that the effects of miRs are systemic and interactive. Through the analyses of HCC signaling pathways by KEGG and their genes on miRDB, all of the miRs identified were involved in all HCC processes such as proliferation, apoptosis, invasion, metastasis, EMT, angiogenesis, drug resistance and autophagy. Interestingly, we also identified two pairs of miR-199a-5p/miR-422a and miR-182/miR-1269 that were effective for HCC diagnosis based on their interaction with target genes and analysis of AUC values.
The roles of miR-182 and miR1269a have been known as oncomiRs in regulating the development of hepatocellular carcinoma (HCC), together with their overexpression results in the suppression of FOXO3a and FOXO1 (Cao et al., 2018; Yang et al., 2014). Inhibition of FOXO3a and FOXO1 can activate the AKT/FOXO3a pathway and dysregulate p21, cyclin D1, phosphorylated Rb and Ki67 leading to the promotion of HCC proliferation and enhancement of motility and invasive ability of HCC cells, respectively (Cao et al., 2018; Yang et al., 2014). Notably, miR-182 was also reported to be involved in the HCC cancer formation process through direct attachment to the 3′UTR tip of ephrinA5, thereby reducing cell proliferation and migration (Wang et al., 2016), while, SNP re73239138 in the sequence of miR-1269 with GA or AA genotype reduced the incidence of HCC compared with the GG genotype, indicating that it is a protective factor that can suppress tumor growth in HCC by preventing binding to 3′UTR of the SOX6 gene (Xiong et al., 2015). By contrast, miR-182 and miR-1269a and other pairs of miR-199a-5p and miR-422a were demonstrated to be tumor suppressors in HCC that could serve as useful therapeutic agents for miRNA-based HCC therapy (Huang et al., 2017; Zhang et al., 2015). Increase of target genes such as CLTC and DDR1 induced by down-regulation of miR-199a-5p contributed to increased HCC cell invasion and growth of tumors (Huang et al., 2017; Shen et al., 2010). Similarly, the expression level of miR-422a negatively correlated with tumor growth, recurrence and liver metastasis because it can target FOXG1, FOXQ1 and FOXE1 genes and functions involved in significant inhibition of tumor cell proliferation and migration (Zhang et al., 2015). Until now, no evidence has been shown for expression correlation between the TMEM245 gene and miR-199-5p/miR-422a. This should be further investigated even though TMEM245 is not prognostic in liver cancer when using web-based platforms such as the Human Protein Atlas. Thus, both microRNAs could be used as new potential candidates for HCC therapy.
Using logistic regression, miRNA clusters as potential biomarkers for HCC diagnosis were successfully built up based on microarray datasets and bioinformatics analyses. We found an increase in fold change of miR-183/miR-1269 expression from 0.148 to 1.222 between HCC and normal tissues of patients in Asia, Europe and America, while the fold change of miR-199a-5p and miR-422a down-regulation was from -0.17 to -0.773 (Table 4). Differences in fold change between miR-199a-5p and miR-422a expression were five times, and twice for the groups in Asia and America (Table 4). Interestingly, we also found that AUC values of two pairs of miR-199a-5p/miR-422a and miR-182/miR-1269 (0.9-0.89) were higher than those of AFP, ALT, AST and NLR with AUC values of 0.78, 0.5, 0.66 and 0.74, respectively (Hu et al., 2018). Furthermore, when comparing with other non-invasive diagnostic indicators of HCC such as serum GPC3, GP73 and even the combination of AFP-L3/GP72, the similarity of AUC values was also observed in two pairs of HCC microRNAs identified in this study (Jing et al., 2017; Xu et al., 2014). Results indicated that diagnostic values of miR-199a-5p/miR-422a and miR-182/miR-1269 could be very useful data for the development of diagnostic tools for HCC cancer.
Table 4.
Differential expression levels and expression threshold values of up and down-regulated miRs.
| microRNA | Fold change (FC) |
Q1 (25%) | Q3 (75%) | Expression |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Asia | P value | Europe | P value | America | P value | Mean | Up | Down | |||
| hsa-miR1269 | 0.58 | 0.006 | 0.73 | <0.0001 | 0.53 | <0.0001 | 0.62 | 1.32 ± 0.32 | 4.13 ± 1.34 | ✓ | |
| hsa-miR183 | 0.15 | <0.0001 | 1.1 | <0.0001 | 1.22 | <0.0001 | 0.81 | 3.86 ± 1.94 | 6.89 ± 0.53 | ✓ | |
| hsa-miR199a-5p | -0.48 | <0.0001 | -0.77 | <0.0001 | -0.14 | 0.003 | -0.46 | 6.29 ± 3.29 | 9.48 ± 1.59 | ✓ | |
| hsa-miR422a | -0.17 | <0.0001 | -0.64 | <0.0001 | -0.29 | <0.0001 | -0.37 | 5.87 ± 1.26 | 7.37 ± 1.26 | ✓ | |
5. Conclusions
In summary, we introduced two diagnostic models consisting of different combinations of microRNA molecules belonging to the oncomiR and tumor suppressor groups, based on their key expression algorithms and other functional properties, using bioinformatics tools and microarray datasets in NCBI. The aim of this study was to identify appropriate miRNA biomarkers in tissue samples that could differentiate HCC from healthy individuals. Our results do not support immediate clinical use. The best groups of miRNAs should be selected for more rigorous trials in a large cohort and case-control studies. To date, none of these miRNAs have been tested in experimental studies and they require further experimentation. Furthermore, since miRNA molecules have been shown to be present and stable in serum and plasma in human (Mitchell et al., 2008; He et al., 2015), the expression profiles of two groups of miRNAs for the diagnosis and prognosis of HCC should be evaluated against these samples as a non-invasive approach, as required by medical ethics in clinical diagnosis.
Declarations
Author contribution statement
Vo Hoang Xuan Dat: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Bui Thi Huyen Nhung; Nguyen Bao Quoc: Performed the experiments; Analyzed and interpreted the data.
Nguyen Ngoc Bao Chau: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Pham Hung Cuong; Vo Duc Hieu; Nguyen Thi Minh Linh: Analyzed and interpreted the data.
Funding statement
This work was supported by a grant from the Ministry of Education and Training, Vietnam (B2020-NLS-06).
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Alegria-Torres J.A., Baccarelli A., Bollati V. Epigenetics and lifestyle. Epigenomics. 2011;3(3):267–277. doi: 10.2217/epi.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwa M.H., El-Etrby S.A. Guide for diagnosis and treatment of hepatocellular carcinoma. World J. Hepatol. 2015;7(12):1632–1651. doi: 10.4254/wjh.v7.i12.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Wilhite S.E., et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L., Cai T., DasGupta A. Interval estimation for a binomial proportion. Stat. Sci. 2001;16(2):101–133. [Google Scholar]
- Burchard J., Zhang C., et al. microRNA-122 as a regulator of mitochondrial metabolic gene network un hepatocellular carcinoma. Mol. Syst. Biol. 2010;24(6):402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66(15):7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Cao M.Q., You A.B., Zhu X.D., et al. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J. Hematol. Oncol. 2018;11:12. doi: 10.1186/s13045-018-0555-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran R., Limaye A., Cabrera Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis, and therapeutics. Hepat. Med. 2012;4:19–37. doi: 10.2147/HMER.S16316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bisceglie A.M. Pretransplant treatments for hepatocellular carcinoma: do they improve outcomes? Liver Transplant. 2005;11(S2):S10–S13. doi: 10.1002/lt.20598. [DOI] [PubMed] [Google Scholar]
- Diaz G., Melis M., Tice A., Kleiner D.E., et al. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int. J. Cancer. 2013;133(4):816–824. doi: 10.1002/ijc.28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A., Sherman M. Liver biopsy for hepatocellular carcinoma (HCC): should this be a routine? Hep. Canc. 2017;16:45–60. [Google Scholar]
- Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- Gramantieri L., Ferracin M., et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67(13):6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- Haynes W. In: Encyclopedia of Systems Biology. Dubitzky W., Wolkenhauer O., Cho K.H., Yokota H., editors. Springer; New York, NY: 2013. Benjamini–hochberg method. [Google Scholar]
- He Y., Lin J., Kong D., et al. Current state of circulating microRNAs as cancer biomarkers. Clin. Chem. 2015;61:1138–1155. doi: 10.1373/clinchem.2015.241190. [DOI] [PubMed] [Google Scholar]
- Herbst D.A., Reddy K.R. Risk factors for hepatocellular carcinoma. Clin. Liver Dis. 2012;1(6):180–182. doi: 10.1002/cld.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wang N., Yang Y., Ma L., et al. Diagnostic value of alpha-fetoprotein combined with neutrophil to lymphocyte ratio for hepatocellular carcinoma. BMC Gastroenterol. 2018;18:186. doi: 10.1186/s12876-018-0908-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.H., Shan H., Li D., Zhou B., Pang P.F. Mir-199a-5p suppresses tumorigenesis by targeting clathrin heavy chain in hepatocellular carcinoma. Cell Biochem. Funct. 2017;35(2):98–104. doi: 10.1002/cbf.3252. [DOI] [PubMed] [Google Scholar]
- Iorio M.V., Croce C.M. MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 2009;27(34):5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Wong S.W., et al. Circulating microRNA as potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Sci. Rep. 2019;9(1):10464. doi: 10.1038/s41598-019-46872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing J.S., Ye W., Jiang Y.K., et al. The value of GPC3 and GP73 in clinical diagnosis of hepatocellular carcinoma. Clin. Lab. 2017;63(11):1903–1909. doi: 10.7754/Clin.Lab.2017.170712. [DOI] [PubMed] [Google Scholar]
- Martinez-Quetglas I., Pinyol R., Dauch D., Torrecilla S., et al. IGF2 is up-regulated by epigenetic mechanisms in hepatocellular carcinomas and is an actionable oncogene product in experimental models. Gastroenterology. 2016;151(6):1192–1205. doi: 10.1053/j.gastro.2016.09.001. [DOI] [PubMed] [Google Scholar]
- McGlynn K.A., Petrick J.L., London W.T. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clin. Liver Dis. 2015;19(2):223–238. doi: 10.1016/j.cld.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P.S., Parkin R.K., Kroth E.M., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi Y., Mishima T., et al. Sequencing and bioinformatics-based analyses of the microRNA transcriptome in hepatitis B-related hepatocellular carcinoma. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0015304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Tanahashi T., Okada R., et al. Comparison of hepatocellular carcinoma miRNA expression profiling as evaluated by next generation sequencing and microarray. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0106314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau P., Volinia S., et al. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 2010;107(1):264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F., Hatano E., Kitamura K., Myomoto A., et al. MicroRNA profile predicts recurrence after resection in patients with hepatocellular carcinoma within the Milan Criteria. PLoS One. 2011;6(1) doi: 10.1371/journal.pone.0016435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shams R., Saberi S., Zali M., et al. Identification of potential microRNA panels for pancreatic cancer diagnosis using microarray datasets and bioinformatics methods. Sci. Rep. 2020;10:7559. doi: 10.1038/s41598-020-64569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariff M.I.F., Cox I.J., Gomaa A.I., Khan S.A., Gedroyc W., Taylor-Robinson S.D. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expet Rev. Gastroenterol. Hepatol. 2009;3(4):353–367. doi: 10.1586/egh.09.35. [DOI] [PubMed] [Google Scholar]
- Shen Q., Cicinnati V.R., et al. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mol. Cancer. 2010;9:227. doi: 10.1186/1476-4598-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram S., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Tokar T., Pastrello C., et al. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46(D1):D360–D370. doi: 10.1093/nar/gkx1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu T., Tsuneyama K., et al. Comprehensive analysis of liver and blood miRNA in precancerous conditions. Sci. Rep. 2020;10:21766. doi: 10.1038/s41598-020-78500-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R., Croce C.M. miRNAs and cancer. Am. J. Pathol. 2009;174(4):1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lee A.T.C., Ma J.Z.I., et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J. Biol. Chem. 2008;283(19):13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- Wang V., Wu W. MicroRNA-based therapeutics for cancer. BioDrugs. 2009;23:15–23. doi: 10.2165/00063030-200923010-00002. [DOI] [PubMed] [Google Scholar]
- Wang T.H., Yeh C.T., Ho J.Y., Ng K.F., Chen T.C. Oncomir miR-96 and miR-182 promote cell proliferation and invasion through targeting ephrinA5 in hepatocellular carcinoma. Mol. Carcinog. 2016;55(4):366–375. doi: 10.1002/mc.22286. [DOI] [PubMed] [Google Scholar]
- Xiong G., Wang Y., Ding Q., Yang L. Hsa-mir-1269 genetic variant contributes to hepatocellular carcinoma susceptibility through affecting SOX6. Am. J. Transil. Res. 2015;7(10):2091–2098. [PMC free article] [PubMed] [Google Scholar]
- Xu W.J., Guo B.L., Han Y.G., Shi L., Ma W.S. Diagnostic value of alpha-fetoprotein-L3 and Golgi protein 73 in hepatocellular carcinomas with low AFP levels. Tumour Biol. 2014;35(12):12069–12074. doi: 10.1007/s13277-014-2506-8. [DOI] [PubMed] [Google Scholar]
- Yang X.W., Shen G.Z., Cao L.Q., et al. MicroRNA-1269 promotes proliferation in human and hepatocellular carcinoma via downregulation of FOXO1. BMC Cancer. 2014;14:909. doi: 10.1186/1471-2407-14-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yang Y., Yang T., et al. Double-negative feedback loop between microRNA-422a and forkhead box (FOX) G1/Q1/E1 regulates hepatocellular carcinoma tumor growth and metastasis. Hepatology. 2015;61(2):561–573. doi: 10.1002/hep.27491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.