Abstract
Parkinson’s disease (PD) is a neurodegenerative disorder responsible for shaking, rigidity, and trouble in walking and patients’ coordination ability and physical stability deteriorate day by day. Bipolar disorder (BD) is a psychiatric disorder which is the reason behind extreme shiftiness in mood, and frequent mood inversion may reach too high called mania. People with BD have a greater chance of developing PD during the follow-up period. A lot of work has been done to understand the key factors for developing these 2 diseases. But the molecular functionalities that trigger the development of PD in people with BD are not clear yet. In our study, we are intended to identify the molecular biomarkers and pathways shared between BD and PD. We have investigated the RNA-Seq gene expression data sets of PD and BD. A total of 45 common unique genes (32 up-regulated and 13 down-regulated) abnormally expressed in both PD and BD were identified by applying statistical methods on the GEO data sets. Gene ontology (GO) and BioCarta, KEGG, and Reactome pathways analysis of these 45 common dysregulated genes identified numerous altered molecular pathways such as mineral absorption, Epstein-Barr virus infection, HTLV-I infection, antigen processing, and presentation. Analysis of protein-protein interactions revealed 9 significant hub-proteins, namely RPL21, RPL34, CKS2, B2M, TNFRSF10A, DTX2, HLA-B, ATP2A3, and TAPBP. Significant transcription factors (IRF8, SPI1, RUNX1, and FOXA1) and posttranscriptional regulator microRNAs (hsa-miR-491-3p and hsa-miR-1246) are also found by analyzing gene-transcription factors and gene-miRNAs interactions, respectively. Protein-drug interaction analysis revealed hub-protein B2M’s interaction with molecular drug candidates like N-formylmethionine, 3-indolebutyric acid, and doxycycline. Finally, a link between pathological processes of PD and BD is identified at transcriptional level. This study may help us to predict the development of PD among the people suffering from BD and gives some clue to understand significant pathological mechanisms.
Keywords: Parkinson’s disease, bipolar disorder, drug target, differential expressed gene, protein-protein interaction, protein-drug interaction, biomarkers
Introduction
Parkinson’s disease (PD) is an endless and steady degenerative disorder which chiefly invades the motor neuron of the principal nervous system and is perhaps the most widely recognized neurodegenerative disorder after Alzheimer’s disease everywhere around the world.1,2 The early symptoms like shaking, rigidity, slowness of movement, and walking complications may appear very slowly over the long run. Complexities to walk, talk, or even completion of the patient’s daily activities become difficult to do. Sleeping and passionate problems may occur frequently in PD, and in the long run, it may lead to dementia. 3 Roughly 60 000 are diagnosed to have the PD every year in the United States and expanding routinely. In excess of 10 million individuals are living with the PD around the globe. Incidence of PD increases with age; however, around 4% of individuals with PD are diagnosed before the age of 50. 4 Bipolar disorder (BD) is a psychiatric disorder involving episodes of severe mood disturbance, neuropsychological deficiencies, physiological and immunological changes, and even disturbances in functioning. It is now one of the most notable causes of disability around the world and is associated with high rates of premature death from both suicide as well as medical comorbidities. 5 With affecting about 45 million people globally, it is assessed that around 5.7 million adult Americans are struggling with BD each year that is about 2.6% of the adult population from which around 4.4% will develop this disorder eventually in their lives.6,7 Having a previous diagnosis of BD (at a very young age) may increase the probability of developing sporadic (idiopathic and nongenetic) PD with an odds ratio of 3.35. 8 A study published in neurology inspected a case-control study from a Swedish accomplice and found an odds ratio of 3.2 for the being diagnosed with PD within the first year of depression. 9 In another study, it was found that people with BD had a greater chance of developing PD during the follow-up period than control sample. 10 Recently, a study revealed that BD is a premonitory symptom of PD. 11 Day-by-day proof is expanding to support that early life BD may incline people, or be an indicator, of PD development in future life. The reason behind this might be because of common pathways, for example, dopaminergic dysfunction, that can lead both to develop autonomously; however, the significance of these is of now unsure. There are few reports of perspective biomarkers identification using transcriptomic data sets and microarray data that are available in literature.12-16 However, components which are frequently dysregulated in these 2 diseases are not well studied at molecular level until now.
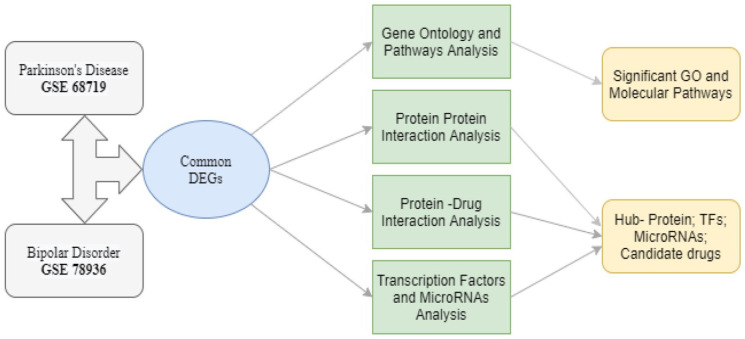
In our study, a systems biology approach has been applied to identify differentially expressed genes (DEGs) and related molecular pathways shared between PD and BD. These DEGs common to PD and BD were subjected to interaction networks utilizing the following tactics: (1) gene enrichment analysis to identify molecular function, cellular components, and biologically processes a gene participates in, (2) protein-protein interaction (PPI) network analysis to find hub genes, (3) transcriptional and posttranscriptional regulatory components (TFs and miRNAs) identification the common DEGs interact with, (4) potential drugs screening using protein-drug interaction networks. The scientific outcomes of this study will help to reveal effective biomarkers and molecular targets for drugs to be used against PD and BD. Overall strategies are summarized in Figure 1.
Figure 1.
Overall strategies employed in this study. Differentially expressed genes for BD and PD were identified and then shared dysregulated genes between BD and PD were selected by statistical methods. Gene enrichment study was performed to find significant common pathways and GO terms. Analysis of PPI was done to find hub-proteins, TFs, and miRNAs that regulate those hub-proteins. Finally, prospective drug candidates were identified based on protein-drug interaction.
Materials and Methods
Differential gene expression analysis of PD and BD
The RNA-Seq gene expression data sets for PD- and BD-affected tissues were obtained from the NCBI-GEO (www.ncbi.nlm.nih.gov/geo/) database. The accession number of PD data set was GSE68719, which was generated using human postmortem Brodmann area 9 (BA9), a part of the frontal cortex, 29 brain tissue for PD and 44 neurologically normal individuals. The GEO accession number of BD data set was GSE78936, where RNA sequencing of 7 brain tissue samples from BA9 for BD and 6 neurologically normal individuals were considered. To identify the significant DEGs of interest for each data set, a P value < .05 and absolute log2-fold change ⩾ 1 was considered as threshold criteria. Differentially expressed genes common to both PD and BD were identified using Venny tool (www.bioinfogp.cnb.csic.es/tools/venny/).
Gene ontology and pathway enrichment of DEGs
Gene enrichment analyses were executed to gain insights of the DEGs. Biological processes, molecular functions, and cellular components a gene contributes were obtained from Enrichr (www.maayanlab.cloud/Enrichr/) using a P value < .05 and also common pathways between PD and BD were identified thorough KEGG, Reactome, and BioCarta pathways (www.maayanlab.cloud/Enrichr/). A depiction of significant molecular pathways contributed by DEGs were obtained using NetworkAnalyst (www.networkanalyst.ca/) tool.
PPI network study
Protein-protein interaction network was generated with the help of STRING protein interaction database using NetworkAnalyst online resource by considering a confidence score of 700 (www.networkanalyst.ca/).
Analysis of DEGs interaction with transcriptional and posttranscriptional regulators
Prominent TFs and microRNAs that regulate DEGs of our interest at the transcriptional and posttranscriptional levels were identified using TRANSFAC and miRTarBase databases, respectively with a P value < .05(www.maayanlab.cloud/Enrichr/).
Analysis of protein-drug interactions
Potential drug candidates to be proposed for both PD and BD were identified by analyzing protein-drug interaction using NetworkAnalyst (www.networkanalyst.ca/) tool with the help of DrugBank database (version 5.0). 17
Results
Identification of DEGs shared between PD and BD
Gene expression RNA sequencing data sets were studied for both PD and BD data sets and noteworthy DEGs were identified utilizing statistical methods (Supplementary File 1 and 2). It was found that a total of 45 unique genes was shared between the PD DEGs and BD DEGs of which 32 were up-regulated (RPL21, TNFRSF10A, TMC4, TM4SF1, SRGN, SNORD3A, RPL34, PLAC8, MT1M, MT1G, MT1 F, MT1E, MT1A, MS4A6A, LST1, LOC554223, LILRB1, IL18R1, HLA-DPA1, HLA-B, HCG25, FGF23, DTX2, CYSLTR2, CLIC1, CLEC2B, CKS2, CCDC102A, CASP4, CARD16, BRDT, and B2M). The remaining 13 genes were down-regulated (VEPH1, TAPBP, PBX2, MOG, MIR4516, MDC1, LOC107985075, LOC100507091, KRTAP5-AS1, GRID2IP, DNAH10, CYP2D6, and ATP2A3) (Figures 2 and 3). To know the biological process, cellular component, and molecular function a gene contributes, identified 45 genes were considered to gene set enrichment studies. The enriched biological processes were negative regulation of growth, regulation of growth, negative regulation of multicellular organismal process, negative regulation of secretion, inorganic anion transport, and others as shown in Table 1 (Supplementary File 3).
Figure 2.
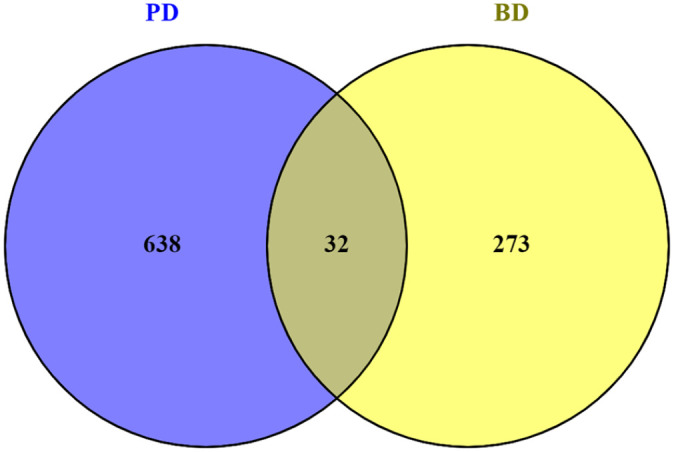
Genes commonly up regulated in PD and BD identified by Venny tool (www.bioinfogp.cnb.csic.es/tools/venny/). Result showing that a number of 32 genes were commonly up-regulated in both PD and BD.
Figure 3.
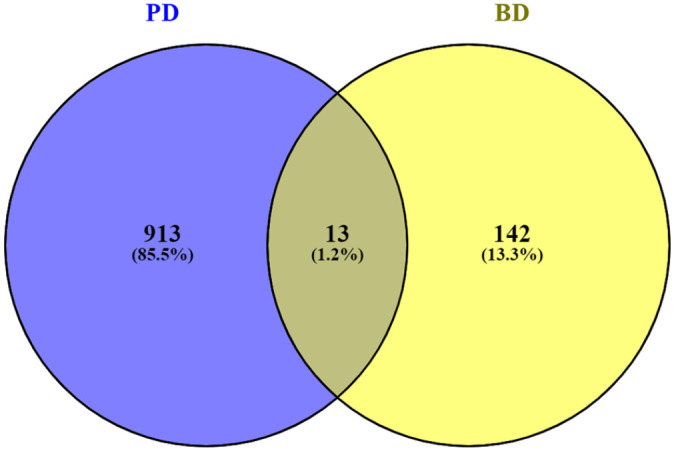
Genes commonly down-regulated in PD and BD identified by Venny tool (www.bioinfogp.cnb.csic.es/tools/venny/). Result showing that a number of 13 genes were commonly down-regulated in both PD and BD.
Table 1.
Significant GO terms related to common differentially expressed genes in BD and PD.
| Group | GO term | P value |
|---|---|---|
| Biological process | Negative regulation of growth | .000004 |
| Regulation of growth | .00120 | |
| Negative regulation of multicellular organismal process | .00202 | |
| Negative regulation of secretion | .00335 | |
| Inorganic anion transport | .00898 | |
| Cellular component | Perinuclear region of cytoplasm | .0002 |
| Integral to endoplasmic reticulum membrane | .0016 | |
| Intrinsic to endoplasmic reticulum membrane | .0023 | |
| Transport vesicle | .0037 | |
| Coated vesicle membrane | .0039 | |
| Molecular function | Copper ion binding | .0003 |
| Hydrolase activity, acting on acid anhydrides, and catalyzing transmembrane movement of substances | .0267 | |
| ATPase activity, coupled to movement of substances | .0275 | |
| Primary active transmembrane transporter activity | .0292 | |
| Structural constituent of ribosome | .0494 |
The important molecular function of DEGs shared by PD and BD were copper ion binding, hydrolase activity, catalyzing transmembrane movement of substances, ATPase activity, primary active transmembrane transporter activity, and structural constituent of ribosome. The important cellular components were found as perinuclear region of cytoplasm, integral to endoplasmic reticulum membrane, intrinsic to endoplasmic reticulum membrane, transport vesicle, and coated vesicle membrane (Table 1).
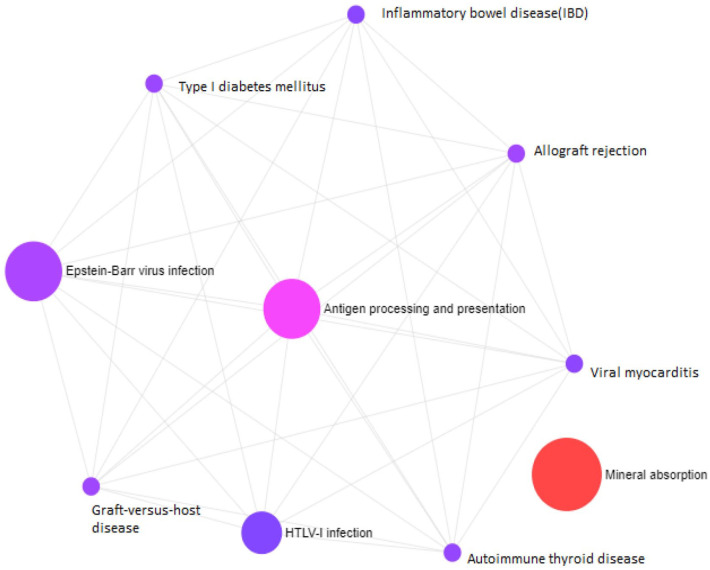
The transformed molecular pathways found as mineral absorption, Epstein-Barr virus infection, HTLV-I infection, and antigen processing and presentation (Figure 4). Common pathway enriched by KEGG. BioCarta, and Reactome are summarized in Table 2 (Supplementary File 4, 5, and 6).
Figure 4.
Important pathways enriched by the shared DEGs between PD and BD. Statistically significant (adj P value < .05) molecular pathways: mineral absorption; antigen processing and presentation.
Table 2.
Different molecular pathway contributed by the differentially expressed genes shared between BD and PD (P value < .05).
| Pathways | P value | Genes involved in pathways |
|---|---|---|
| KEGG | ||
| Mineral absorption | 9.97E−08 | MT1A; MT1M; MT1F; MT1G; MT1E |
| Antigen processing and presentation | 2.68E−05 | HLA-B; B2M; HLA-DPA1; TAPBP |
| Epstein-Barr virus infection | .001069 | HLA-B; B2M; HLA-DPA1; TAPBP |
| Allograft rejection | .003305 | HLA-B; HLA-DPA1 |
| Graft-versus-host disease | .003839 | HLA-B; HLA-DPA1 |
| Type-I diabetes mellitus | .004215 | HLA-B; HLA-DPA1 |
| Autoimmune thyroid disease | .006342 | HLA-B; HLA-DPA1 |
| Viral myocarditis | .007807 | HLA-B; HLA-DPA1 |
| Inflammatory bowel disease (IBD) | .00941 | IL18R1; HLA-DPA1 |
| Human immunodeficiency virus 1 infection | .012015 | HLA-B; B2M; TAPBP |
| Human T-cell leukemia virus 1 infection | .013108 | HLA-B; B2M; HLA-DPA1 |
| Human cytomegalo virus infection | .014088 | HLA-B; B2M; TAPBP |
| Herpes simplex virus 1 infection | .024355 | HLA-B; B2M; HLA-DPA1; TAPBP |
| Natural killer cell-mediated cytotoxicity | .035091 | HLA-B; TNFRSF10A |
| BioCarta | ||
| Antigen processing and presentation homo sapiens h MHC pathway | .026675 | B2M |
| IL12 and Stat4-dependent signaling pathway in Th1 development homo sapiens h IL12 pathway | .033235 | IL18R1 |
| Internal ribosome entry pathway homo sapiens h IRES pathway | .039751 | CASP4 |
| Ras-Independent pathway in NK cell-mediated cytotoxicity homo sapiens h NK cells pathway | .048372 | B2M |
| Reactome | ||
| Response to metal ions homo sapiens R-HSA-5660526 | 2.10E−11 | MT1A; MT1M; MT1F; MT1G; MT1E |
| Metallothioneins bind metals homo sapiens R-HSA-5661231 | 2.10E−11 | MT1A; MT1M; MT1F; MT1G; MT1E |
| Antigen presentation: folding, assembly and peptide loading of class-I MHC homo sapiens R-HSA-983170 | 2.36E−05 | HLA-B; B2M; TAPBP |
| Endosomal/vacuolar pathway homo sapiens R-HSA-1236977 | 3.22E−04 | HLA-B; B2M |
| ER-phagosome pathway homo sapiens R-HSA-1236974 | 4.22E−04 | HLA-B; B2M; TAPBP |
| Immunoregulatory interactions between a Lymphoid and a nonlymphoid cell Homo sapiens R-HSA-198933 | 4.24E−04 | CLEC2B; HLA-B; LILRB1; B2M |
| Antigen processing cross-presentation homo sapiens R-HSA-1236975 | 8.33E−04 | HLA-B; B2M; TAPBP |
Hub-proteins identification from PPI network
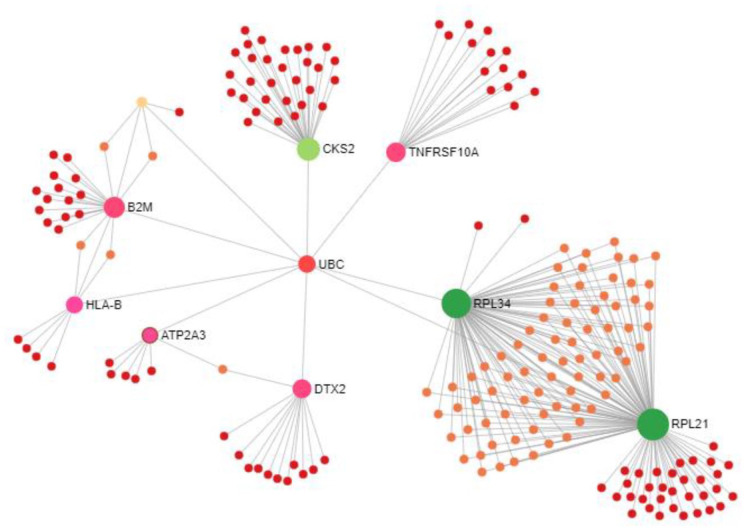
The PPI network created by STRING consists of 205 nodes (9 hub nodes from shared DEGs) and 288 edges (Figure 5) that uncovered 9 hub-proteins, namely RPL21, RPL34, CKS2, B2M, TNFRSF10A, DTX2, HLA-B, ATP2A3, and TAPBP by considering maximum degree of interaction using Cytoscape (https://cytoscape.org/) (Table 3).
Figure 5.
PPI network of shared differentially expressed genes with additional genes for PD and BD samples obtained from STRING database. This network contains 205 nodes (9 hub nodes from shared DEGs) and 288 edges.
Table 3.
List of identified common biomarker candidates (Proteins and TFs) with their biological roles.
| Biomarker candidate | Full form | Role of biomarkers |
|---|---|---|
| Hub-proteins | ||
| RPL21 | Ribosomal Protein L21 | Diseases associated with RPL21 include hypotrichosis 12 and hypotrichosis simplex |
| RPL34 | Ribosomal Protein L34 | Diseases associated with RPL34 include bone structure disease and Cauda Equina syndrome |
| CKS2 | Cyclin-dependent kinases regulatory subunit 2 | Diseases associated with CKS2 include Coffin-Siris syndrome 1 |
| B2M | Beta-2-microglobulin | Immunodeficiency 43 and amyloidosis, familial visceral are associated with B2M |
| TNFRSF10A | Tumor necrosis factor receptor superfamily member 10A | Diseases associated with TNFRSF10A include Hirata disease and temporal arteritis |
| DTX2 | Deltex E3 ubiquitin ligase 2 | Diseases associated with DTX2 include ependymoblastoma and pthirus pubis infestation |
| HLA-B | Major histocompatibility complex, class I, B | Severe cutaneous adverse reaction and spondyloarthropathy 1 are associated with HLA-B |
| ATP2A3 | ATPase sarcoplasmic/endoplasmic reticulum Ca2 + transporting 3 | Diseases associated with ATP2A3 include Darier-White disease and atrophic muscular disease |
| TAPBP | TAP-binding protein | Diseases associated with TAPBP include Bare lymphocyte syndrome, type-I and immunodeficiency by defective expression Of MHC Class-I |
| Transcription factors; P value < .05 | ||
| IRF8 | Interferon regulatory factor 8 | Diseases associated with IRF8 include immunodeficiency 32A and immunodeficiency 32B |
| SPI1 | Spi-1 proto-oncogene | Diseases associated with SPI1 include inflammatory diarrhea and pulmonary alveolar proteinosis |
| FOXA1 | Forkhead Box A1 | Diseases associated with FOXA1 include estrogen-receptor-negative breast cancer and estrogen-receptor-positive breast cancer |
| RUNX1 | RUNX family transcription factor 1 | RUNX1-associated diseases are platelet disorder, familial, with associated myeloid malignancy and leukemia, acute myeloid |
Transcriptional and posttranscriptional regulators identification that interact with DEGs common to PD and BD
As the expression of genes are regulated at transcriptional and posttranscriptional stages, significant TFs and miRNAs that regulate that expression of shared DEGs between PD and BD were identified. We found significant TFs IRF8 (targeting DEGs MDC1; MS4A6A; ATP2A3; CKS2; DTX2; B2M; TM4SF1; and BRDT), SPI1 (targeting DEGs PLAC8; CYSLTR2; CYP2D6; VEPH1; LST1; MT1G; LILRB1; and BRDT), FOXA1 (targeting DEGs LST1; CASP4; DTX2; CCDC102A; CLIC1; TMC4; BRDT; and GRID2IP), and RUNX1 (targeting DEGs MDC1; MOG; MT1M; LST1; HLA-B; PBX2; LILRB1; PLAC8; MT1A; CLEC2B; VEPH1; MT1F; CARD16; and CLIC1) (Table 3). Besides, top-10 significant miRNAs identified namely, mmu-miR-1965; mmu-miR-1946a; hsa-miR-509-3p; hsa-miR-1246; hsa-miR-4774-3p; hsa-miR-491-3p; hsa-miR-4804-3p; hsa-miR-1285-5p; mmu-miR-1935; and mmu-miR-298-5p are depicted in Table 4.
Table 4.
Top 10 miRNAs that interact with hub genes obtained from miRTarBase 2017 database sorted by P value.
| miRNA name | P value | Associated genes |
|---|---|---|
| mmu-miR-1965 | .002208 | MDC1 |
| mmu-miR-1946a | .003305 | MDC1; TNFRSF10A |
| hsa-miR-509-3p | .003839 | PLAC8; TM4SF1 |
| hsa-miR-1246 | .004215 | CKS2; TAPBP |
| hsa-miR-4774-3p | .006576 | TNFRSF10A; TAPBP |
| hsa-miR-491-3p | .007553 | FGF23; B2M |
| hsa-miR-4804-3p | .009134 | FGF23; B2M |
| hsa-miR-1285-5p | .009691 | PLAC8; TM4SF1 |
| mmu-miR-1935 | .010833 | MDC1; TNFRSF10A; TAPBP |
| mmu-miR-298-5p | .013108 | MDC1; TNFRSF10A; TAPBP |
Drug candidate’s identification using protein-drug interaction network
There are few reports of β2-microglobulin (B2M) which is a part of major histocompatibility complex class 1 (MHC I) molecules involvement in impairing cognitive functions with age and B2M content is seen to be up-regulated in blood and hippocampus of older mice and blood of aged human with impairing cognitive function and neurogenesis. Younger mice treated with B2M injection showed abnormal cognitive function and neurogenesis.18,19 Our hypothesis was to prevent or downregulate this B2M content upon binding with proposed drug compounds. By analyzing protein-drug interactions using NetworkAnalyst tool (www.networkanalyst.ca/), it was found that B2M protein has some interactions with 3 known compounds, namely N-formylmethionine, 3-indolebutyric acid, and doxycycline (Table 5 and Figure 6).
Table 5.
Hub-protein B2M interaction with known 3 drug compounds of drug bank.
| Id | Label | Degree | Betweenness |
|---|---|---|---|
| 567 | B2M | 3 | 3 |
| DB00254 | Doxycycline | 1 | 0 |
| DB02740 | 3-Indolebutyric Acid | 1 | 0 |
| DB04464 | N-Formylmethionine | 1 | 0 |
Figure 6.
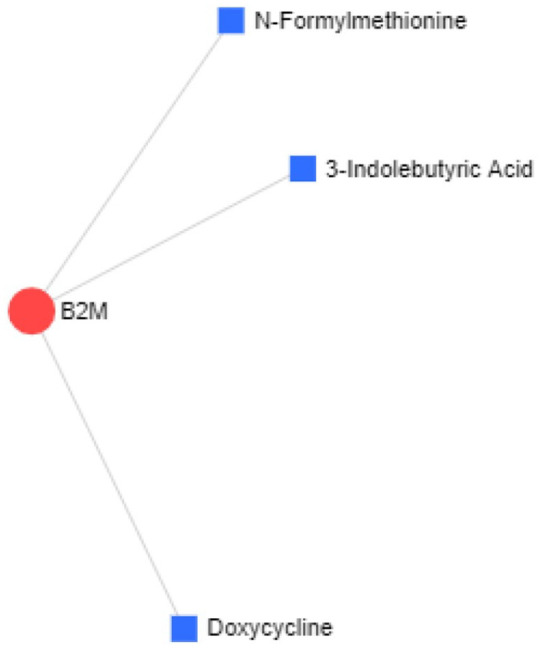
The protein-drug interactions network between hub-protein B2M and proposed drugs obtained with the help of NetworkAnalyst tool where the degree of interaction is represented by the area of the nodes.
N-formyl methionine is effective in initiation of protein synthesis in prokaryotes and mitochondria of eukaryotes while 3-indolebutyric acid is a naturally present plant hormone. Doxycycline, an antibiotic, is used to treat various bacterial infections caused by gram-positive and gram-negative bacteria. As N-formyl methionine, 3-indolebutyric acid can cross the blood brain barrier are capable of donating hydrogen to an acceptor and doxycycline is currently being used, these compounds or modification of it with good absorption, solubility properties upon binding with B2M protein may play role in prevention of amyloid formation.
Discussions
In this study, publicly accessible RNA-seq data of PD (GSE68719) and BD (GSE78936) patients were exploited to identify prospective shared biomarkers and molecular targets in both diseases. Initially, those 2 data sets were statistically investigated to classify DEGs in both samples and then the shared DEGs of both data sets were considered for additional analysis. Gene enrichment analysis revealed that the identified 45 DEGs were mostly responsible for regulation of growth, copper ion binding, hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances, ATPase activity, coupled to movement of substances, primary active transmembrane transporter activity, mineral absorption, antigen processing and presentation, Epstein-Barr virus infection, and so on. (Tables 1 and 2, Figure 4). Further investigation of those DEGs using PPI network revealed 9 hub-proteins, namely RPL21, RPL34, CKS2, B2M, TNFRSF10A, DTX2, HLA-B, ATP2A3, and TAPBP (Figure 5 and Table 3). Pathway analysis of the hub-proteins shows the direct association of B2M gene with neurological disorder due to its role in amyloidosis formation.20,21 According to gene cards database (www.genecards.org/) RPL21 gene is associated with hypotrichosis 12 and hypotrichosis simplex. 22 Diseases associated with RPL34 are bone structure disease, cauda equina syndrome, and hilar cholangiocarcinoma (www.genecards.org/). 23 CKS2 gene is linked with Coffin-Siris syndrome 1 (www.genecards.org/). Mutation in TNFRSF10A is associated with hirata disease and temporal arteritis, whereas DTX2 is linked to ependymoblastoma and pthirus pubis infestation. Diseases associated with HLA-B include severe cutaneous adverse reaction and spondyloarthropathy 1 and gene ATP2A3 is associated with Darier-White disease and atrophic muscular disease (www.genecards.org/). Bare lymphocyte syndrome, type-I and immunodeficiency by defective expression Of MHC class-I are all associated with TAPBP gene regulation. 24
As the instruction of gene expression govern by the TFs and miRNAs at transcriptional and posttranscriptional stages, fundamental evidence for the dysregulation of gene expression can be found from the variations in these biomolecules. So in this study, we examined common DEGs-TFs and DEGs-miRNAs interactions (Tables 3 and 4). Four transcription factors, namely IRF8, FOXA1, SPI1, and RUNX1 are further analyzed for finding their disease association. According to gene cards database (www.genecards.org/) IRF8 is associated with immunodeficiency 32A and immunodeficiency 32B.25,26 Diseases associated with SPI1 includes inflammatory diarrhea and pulmonary alveolar proteinosis while FOXA1 is associated with estrogen-receptor-negative breast cancer and estrogen-receptor-positive breast cancer (www.genecards.org/). Diseases associated with RUNX1 includes platelet disorder, familial, associated myeloid malignancy and leukemia, and acute myeloid.27-29
Among the miRNAs (mmu-miR-1965; mmu-miR-1946a; hsa-miR-509-3p; hsa-miR-1246; hsa-miR-4774-3p; hsa-miR-491-3p; hsa-miR-4804-3p; hsa-miR-1285-5p; mmu-miR-1935; and mmu-miR-298-5p) hsa-miR-1246 and hsa-miR-491-3p may have role in several neurological disorders like amyotrophic lateral sclerosis, attention-deficit hyperactivity disorder (ADHD), and BD.30-34 Dopamine is used as a messenger to transmit signal between nerve cells by interacting with dopamine receptor. Identified biomarker mir-491 is found to be involved in negative regulation of dopamine transporter expression indicating its role in neurodegenerative disorder. 33
To identify drugs which may have a potential influence on PD and BD, we finally studied the known protein-drug interactions network that revealed 3 possible drug compounds namely N-formyl methionine, 3-indolebutyric acid, and doxycycline that interact with hub-protein B2M (Figure 6). There are several reports of involvement of this B2M protein in impairing cognitive function and neurogenesis18,35 The possible drug compound namely N-formyl methionine is a small molecule found to be involved in initiation of protein synthesis in prokaryotic organisms and organelles of eukaryotic organisms. According to chEMBL, this compound is being tested to interact with dopamine receptor and acetylcholinesterase, resulting in negative interaction. Indole-3-butyric acid is a small molecule is shown to interact with acetylcholinesterase, a neurotransmitter, in Torpedo californica, but not in human. These 2 compounds mentioned above have good absorption, solubility values, and can cross the brain-blood barrier. Doxycycline is used to treat infections caused by several gram-positive and gram-negative bacteria. Further justification can determine their interaction, potency, and safety as drug candidate against these neurodegenerative diseases.
Pathways and other network analyses could deliver useful insights in finding of diagnostic and therapeutic interventions. The biomarkers and drug candidate molecules identified could be explored for possible drug targets and activity, respectively, aiming to combat both BD and PD.
Conclusion
Parkinson’s disease and BD, the 2 major forms of neurological diseases, affect large amount of people around the globe. Sufficient diagnosis and effective treatment options are currently not available. In this study, we investigated RNA-seq gene expression profiles utilizing computational biology approaches to identify prospective biomarkers which may play a role in understanding patho-biological mechanisms of PD and BD. The identified biomolecules may be regarded as system biomarkers at the transcription and posttranscription levels. The identified compounds from protein-drug interaction networks may need further investigation to verify their potency and safety as drug candidate against PD and BD.
Supplemental Material
Supplemental material, sj-xlsx-1-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-2-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-3-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-4-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-5-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-6-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights
Acknowledgments
The authors acknowledge the Department of Information and Communication Engineering, Noakhali Science and Technology University for providing support to conduct the research work.
Footnotes
Author Contributions: M.K.I. conceived and designed the experiments, made critical revisions, and approved the final version. M.B.H. and M.K.I. analyzed the data and wrote the first draft of the manuscript. M.K.I., M.B.H, M.Z.I., and A.A reviewed the analysis and contributed to the preparation of the manuscript. All authors reviewed and approved the final manuscript
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iDs: Md. Kobirul Islam  https://orcid.org/0000-0003-0722-5520
https://orcid.org/0000-0003-0722-5520
Apurba Adhikary  https://orcid.org/0000-0003-3970-1878
https://orcid.org/0000-0003-3970-1878
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Tysnes O-B, Storstein A. Epidemiology of Parkinson’s disease. J Neural Transm (Vienna). 2017;124:901-905. 10.1007/s00702-017-1686-y. [DOI] [PubMed] [Google Scholar]
- 2. Hsu W-Y, Lane H-Y, Lin C-H. Medications used for cognitive enhancement in patients with schizophrenia, bipolar disorder, Alzheimer’s disease, and Parkinson’s disease. Front Psychiatry. 2018;9:91. 10.3389/fpsyt.2018.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sveinbjornsdottir S. The clinical symptoms of Parkinson’s disease. J Neurochem. 2016;139:318-324. 10.1111/jnc.13691. [DOI] [PubMed] [Google Scholar]
- 4. Understanding Parkinson’s. https://www.parkinson.org/Understanding-Parkinsons/Statistics. Accessed March 13, 2021.
- 5. Rowland TA, Marwaha S. Epidemiology and risk factors for bipolar disorder. Ther Adv Psychopharmacol. 2018;8:251-269. https://dx.doi.org/10.1177%2F2045125318769235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bipolar Disorder Statistics. https://www.dbsalliance.org/education/bipolar-disorder/bipolar-disorder-statistics/. Accessed March 13, 2021.
- 7. James SL, Abate D, Hassen Abate K, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789-1858. https://www.sciencedirect.com/science/article/pii/S2468125319303334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Faustino PR, Duarte GS, Chendo I, et al. Risk of developing Parkinson disease in bipolar disorder: a systematic review and meta-analysis. JAMA Neurol. 2020;77:192-198. 10.1001/jamaneurol.2019.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pontone GM, Koch G. An association between bipolar disorder and Parkinson disease: when mood makes you move. Neurology. 2019;92:1125-1126. 10.1212/WNL.0000000000007641. [DOI] [PubMed] [Google Scholar]
- 10. Huang M-H, Cheng C-M, Huang K-L, et al. Bipolar disorder and risk of Parkinson disease: a nationwide longitudinal study. Neurology. 2019;92:e2735-e2742. 10.1212/WNL.0000000000007649. [DOI] [PubMed] [Google Scholar]
- 11. Onofrj M, Di Iorio A, Carrarini C, et al. Preexisting bipolar disorder influences the subsequent phenotype of Parkinson’s disease. Mov Disord. 2021;36:2840-2852. https://movementdisorders.onlinelibrary.wiley.com/doi/epdf/10.1002/mds.28745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Emamzadeh FN, Surguchov A. Parkinson’s disease: biomarkers, treatment, and risk factors. Front Neurosci. 2018;12:612. 10.3389/fnins.2018.00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang F, Wu Q, Sun S, Bi G, Guo L. Identification of potential diagnostic biomarkers for Parkinson’s disease. FEBS Open Bio. 2019;9:1460-1468. 10.1002/2211-5463.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krokidis MG. Identification of biomarkers associated with Parkinson’s disease by gene expression profiling studies and bioinformatics analysis. AIMS Neurosci. 2019;6:333-345. https://dx.doi.org/10.3934%2FNeuroscience.2019.4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sakharkar MK, Singh SKK, Rajamanickam K, et al. A systems biology approach towards the identification of candidate therapeutic genes and potential biomarkers for Parkinson’s disease. PLoS ONE. 2019;14:e0220995. 10.1371/journal.pone.0220995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry. 2004;9:406-416. 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- 17. Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc. 2015;10:823-844. 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- 18. Smith LK, He Y, Park J-S, et al. β2-microglobulin is a systemic pro-aging factor that impairs cognitive function and neurogenesis. Nat Med. 2015;21:932-937. https://www.nature.com/articles/nm.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Staats KA, Schönefeldt S, Van Rillaer M, et al. Beta-2 microglobulin is important for disease progression in a murine model for amyotrophic lateral sclerosis. Front Cell Neurosci. 2013;7:249. https://www.frontiersin.org/articles/10.3389/fncel.2013.00249/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wani MA, Haynes LD, Kim J, et al. Familial hypercatabolic hypoproteinemia caused by deficiency of the neonatal Fc receptor, FcRn, due to a mutant β2-microglobulin gene. Proc Natl Acad Sci U S A. 2006;103:5084-5089. 10.1073/pnas.0600548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Valleix S, Gillmore JD, Bridoux F, et al. Hereditary systemic amyloidosis due to Asp76Asn variant β2-microglobulin. N Engl J Med. 2012;366:2276-2283. 10.1056/NEJMoa1201356. [DOI] [PubMed] [Google Scholar]
- 22. Zhou C, Zang D, Jin Y, et al. Mutation in ribosomal protein L21 underlies hereditary hypotrichosis simplex. Hum Mutat. 2011;32:710-714. 10.1002/humu.21503. [DOI] [PubMed] [Google Scholar]
- 23. Qian J, Xu L, Yu W, et al. Ribosomal protein L34 is a potential prognostic biomarker and therapeutic target in hilar cholangiocarcinoma. Cell Biosci. 2020;10:100. 10.1186/s13578-020-00463-7. [DOI] [Google Scholar]
- 24. Casanova J-L, Conley ME, Seligman SJ, Abel L, Notarangelo LD. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J Exp Med. 2014;211:2137-2149. 10.1084/jem.20140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fleisher G, Starr S, Koven N, et al. A non-x-linked syndrome with susceptibility to severe Epstein-Barr virus infections. J Pediatr. 1982;100:727-730. 10.1016/S0022-3476(82)80572-6. [DOI] [PubMed] [Google Scholar]
- 26. Hambleton S, Salem S, Bustamante J, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365:127-138. 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Béri-Dexheimer M, Latger-Cannard V, Philippe C, et al. Clinical phenotype of germline RUNX1 haploinsufficiency: from point mutations to large genomic deletions. Eur J Hum Genet. 2008;16:1014-1018. 10.1038/ejhg.2008.89. [DOI] [PubMed] [Google Scholar]
- 28. Michaud J, Wu F, Osato M, et al. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood. 2002;99:1364-1372. 10.1182/blood.V99.4.1364. [DOI] [PubMed] [Google Scholar]
- 29. Preudhomme C, Renneville A, Bourdon V, et al. High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood. 2009;113:5583-5587. 10.1182/blood-2008-07-168260. [DOI] [PubMed] [Google Scholar]
- 30. MIR1246 micRNA 1246 [Homo sapiens]. https://www.ncbi.nlm.nih.gov/gene/100302142. Accessed March 13, 2021.
- 31. MicRNA 491—Gene. https://www.genecards.org/cgi-bin/carddisp.pl?gene=MIR491. Accessed March 13, 2021.
- 32. Saucier D, Wajnberg G, Roy J, et al. Identification of a circulating miRNA signature in extracellular vesicles collected from amyotrophic lateral sclerosis patients. Brain Res. 2019;1708:100-108. https://www.sciencedirect.com/science/article/abs/pii/S0006899318306322. [DOI] [PubMed] [Google Scholar]
- 33. Jia X, Wang F, Han Y, et al. miR-137 and miR-491 negatively regulate dopamine transporter expression and function in neural cells. Neurosci Bull. 2016;32:512-522. https://link.springer.com/article/10.1007/s12264-016-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Greenwood TA, Alexander M, Keck PE, et al. Evidence for linkage disequilibrium between the dopamine transporter and bipolar disorder. Am J Med Genet. 2001;105:145-151. https://onlinelibrary.wiley.com/doi/abs/10.1002/1096-8628(2001)9999. [DOI] [PubMed] [Google Scholar]
- 35. Constantinescu R, Andreasson U, Li S, et al. Proteomic profiling of cerebrospinal fluid in parkinsonian disorders. Parkinsonism Relat Disord. 2010;16:545-549. https://www.sciencedirect.com/science/article/abs/pii/S1353802010001434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-2-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-3-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-4-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-5-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-6-bbi-10.1177_11779322221079232 for Bioinformatics Approach to Identify Significant Biomarkers, Drug Targets Shared Between Parkinson’s Disease and Bipolar Disorder: A Pilot Study by Md. Bipul Hossain, Md. Kobirul Islam, Apurba Adhikary, Abidur Rahaman and Md. Zahidul Islam in Bioinformatics and Biology Insights