Summary
Body mass index (BMI) is a complex disease risk factor known to be influenced by genes acting via both metabolic pathways and appetite regulation. In this study, we aimed to gain insight into the phenotypic consequences of BMI-associated genetic variants, which may be mediated by their expression in different tissues. First, we harnessed meta-analyzed gene expression datasets derived from subcutaneous adipose (n = 1257) and brain (n = 1194) tissue to identify 86 and 140 loci, respectively, which provided evidence of genetic colocalization with BMI. These two sets of tissue-partitioned loci had differential effects with respect to waist-to-hip ratio, suggesting that the way they influence fat distribution might vary despite their having very similar average magnitudes of effect on BMI itself (adipose = 0.0148 and brain = 0.0149 standard deviation change in BMI per effect allele). For instance, BMI-associated variants colocalized with TBX15 expression in adipose tissue (posterior probability [PPA] = 0.97), but not when we used TBX15 expression data derived from brain tissue (PPA = 0.04) This gene putatively influences BMI via its role in skeletal development. Conversely, there were loci where BMI-associated variants provided evidence of colocalization with gene expression in brain tissue (e.g., NEGR1, PPA = 0.93), but not when we used data derived from adipose tissue, suggesting that these genes might be more likely to influence BMI via energy balance. Leveraging these tissue-partitioned variant sets through a multivariable Mendelian randomization framework provided strong evidence that the brain-tissue-derived variants are predominantly responsible for driving the genetically predicted effects of BMI on cardiovascular-disease endpoints (e.g., coronary artery disease: odds ratio = 1.05, 95% confidence interval = 1.04–1.07, p = 4.67 × 10−14). In contrast, our analyses suggested that the adipose tissue variants might predominantly be responsible for the underlying relationship between BMI and measures of cardiac function, such as left ventricular stroke volume (beta = 0.21, 95% confidence interval = 0.09–0.32, p = 6.43 × 10−4).
Keywords: adiposity, fat distribution, genetic epidemiology, transcriptome-wide, tissue specificity, genetic colocalization, Mendelian randomization, cardiovascular disease, complex-trait genetics
Introduction
Obesity is a major risk factor for several of the world’s most prevalent diseases, including coronary artery disease (CAD) and type 2 diabetes (T2D).1 A body mass index (BMI) greater than 30 kg/m2 is typically used to classify obesity, which is a complex trait known to have a substantial genetic component.2, 3, 4 Large-scale genome-wide association studies (GWASs) have been successful in identifying genetic variants robustly associated with BMI across the human genome, and approximately 900 independent loci have been uncovered to date.5 However, the functional mechanisms explaining the associations at these loci are for the most part poorly understood. Furthermore, although BMI is commonly used in investigations of adiposity, it is renowned for being heterogeneous,6 suggesting that it is a surrogate measure of various phenotypes. For example, BMI on its own cannot distinguish between fat and lean mass and will also be influenced by traits, such as bone mineral density, that are unrelated to adiposity.7 As such the genetic variants robustly associated with BMI likely exert their effects on this composite trait via alternate biological pathways.
Efforts to obtain insight into BMI-associated loci identified by GWAS have shown that the putatively responsible genes are predominantly expressed in neural tissue.8 Further studies have implicated multiple brain regions in obesity susceptibility,9,10 extending influences beyond the central role implicated for the hypothalamus in appetite regulation in monogenic and rare forms of severe obesity.11, 12, 13 Taken together, these findings establish an important role for the central nervous system in regulating overall body composition and obesity. However, despite this, previous evidence has suggested that the mean effect size of BMI-associated variants on T2D and CAD risk does not drastically differ when clustered by brain-derived tissue types (n = 114 to 205) and other randomly selected sets of BMI SNPs.10
Additionally, recent studies have provided evidence of a link between BMI-increasing alleles, higher fat mass, and lower risk of cardiometabolic disease for a small proportion of variants, consistent with a protective effect mediated via adipose storage capacity and, potentially, site of storage.14,15 In this scenario, a protective effect has been attributed to specific adiposity-increasing alleles on lipid and cardiometabolic traits. This has been shown to be influenced through an effect on fat distribution whereby the adiposity-increasing allele is associated with an increased capacity to store fat subcutaneously as opposed to viscerally (see Loos and Kilpeläinen16 for a review). As such, the extent to which BMI-associated SNPs relate to fat distribution could be important in evaluation of the relationship between excess adiposity (indicated by BMI) and disease.
These findings highlight divergent mechanisms by which BMI-increasing alleles might influence metabolically “favorable” adiposity as opposed to the typically “unfavorable” adiposity leading to obesity and adverse cardiovascular outcomes. The parsimonious explanation for these effects could be that BMI-influencing genes expressed in brain tissue might be more likely to do so via appetite regulation, whereas adipose-expressed genes might have a greater effect in specific fat depots and pathways related to muscle development.16,17 Understanding the relationship that BMI variants and their distinct effects in individual tissue types have with the development of obesity and body composition might prove critical in establishing effective preventative interventions for related co-morbidities.
In this study, we sought to develop insight into the causal pathways by which genetic variants exert their effects on BMI variation by using meta-analyzed gene expression quantitative trait loci (eQTL) datasets derived from subcutaneous adipose and brain-tissue samples. We analyzed these datasets by using Bayesian colocalization to investigate whether the causal variant for BMI variation at each of the known ∼900 genome-wide loci was also the causal eQTL for a proximal gene’s expression in either adipose or brain tissue. Given the amount of heterogeneity detected in conventional Mendelian randomization (MR) analyses of BMI,18 we next reasoned that partitioning BMI variants by predominantly brain and adipose colocalization profiles might provide novel insight into the tissue-specific effects on BMI and their individual contribution to disease risk. To this end, we employed a multivariable MR framework to estimate the independent BMI effects putatively mediated by gene expression in subcutaneous adipose and brain gene expression separately on disease outcomes and measures of cardiac structure and function for which BMI has a known causal effect.19, 20, 21, 22, 23
Material and methods
Overview
A flowchart providing an overview of the analytical pipeline applied in this study can be found in Figure S1.
BMI data
We obtained summary-level data for 2,336,260 SNPs from a meta-analysis of BMI data from the Genetic Investigation of Anthropometric Traits (GIANT) consortium and the UK Biobank (UKB).5 This meta-analysis involved BMI data from the UK Biobank (UKB) study and the GIANT consortium (which did not include data from the UKB cohort), which the original authors conclude as having negligible sample overlap. To ensure that the highest SNP coverage available was implemented for colocalization analyses, we combined these data with summary statistics from a BMI GWAS involving participants of European ancestry from the UK Biobank only (n = 463,005) to obtain summary statistics for SNPs not included in the meta-analysis with GIANT. Further description of the QC information gathered in these GWAS efforts can be found in Document S1, supplementary note 1. We identified independent SNPs robustly associated with BMI (on the basis of p < 5 × 10−8) by using the PLINK softwaretool24 to apply disequilibrium (LD) clumping. The clumping method retains SNPs that have the strongest independent association with BMI across the genome to identify the number of loci where genetic variation influences this trait. Although all SNPs correlated with these top hits are removed during clumping, we then reintroduce all SNPs in the region prior to colocalization analyses (as described below) to test whether there is any evidence of a shared common variant at each locus with gene expression in either brain or adipose tissue. 915 independent SNPs were identified on the basis of a reference panel comprising data on 10,000 unrelated individuals (r2 < 0.01) in the UKB of European ancestry.25
Tissue-specific gene expression data
Meta-analyzed brain eQTL data were obtained from a study conducted by Qi et al.26 This study included data from the GTEx consortium v7,27 the CommonMind Consortium (CMC),28 and the Religious Orders Study and Memory and Aging Project (ROSMAP),29 n = 1,194. The neural tissue included in these studies was derived from 10 brain regions (anterior cingulate cortex, caudate basal ganglia, cerebellar hemisphere, cerebellum, cortex, frontal cortex BA9, hippocampus, hypothalamus, nucleus accumbens basal ganglia, and putamen basal ganglia).26 To ensure that adipose eQTL data of a comparable sample size were included in our study, we performed a meta-analysis of subcutaneous adipose eQTL data by using summary data from two publicly available resources; the MuTHER study (n = 766)30 and individuals of European ancestry in the GTEx consortium v831 (n = 491) (n = 1,257). Consistent with the BMI and brain eQTL data, all adipose eQTL variant data were mapped to the hg19 genome build via the GRCH37 reference assembly. The MuTHER and GTEx eQTL probes were harmonized via Ensembl Gene mappings prior to meta-analysis. Non-autosome and non-protein coding genes as defined by Ensembl were omitted from downstream analyses. Summary-level adipose eQTL data were converted to SMR (summary Mendelian randomization) input format with the “gwas-summary” function in the SMR software package (v. 1.03).32 We applied the SMR package MeCS method, which has been described previously,26 to meta-analyze the harmonized adipose eQTL datasets. A summary of the primary data resources used in this work are summarized in Table S1.
Genetic colocalization
Genetic colocalization is a statistical approach that tests the hypothesis that the same causal variant at a locus is responsible for both a GWAS and a gene-expression-association signal. We systematically applied the Bayesian method “coloc”33 by using default parameters to evaluate the posterior probability (PPA) for colocalization between the 915 independent BMI GWAS SNPs based on LD clumping and the expression of proximal genes within a 200 kb window. The “coloc” method estimates the posterior probability of five competing hypotheses: no association with either trait (PPA0); association with one trait (PPA1, PPA2); association with both traits but with distinct causal variants (PPA3); and association with both traits with a common causal variant (PPA4).33
Colocalization analyses were conducted twice at each locus, first with eQTL data derived from our adipose tissue meta-analysis and then repeated separately with the eQTL data from brain tissue. A PPA4 > 0.8 was considered to be strong evidence of colocalization, as recommended by the authors of the method. Variants within the MHC region (chr6: 25,000,000–35,000,000) were excluded from analyses. Although evidence of genetic colocalization might indicate that changes to a gene’s expression reside along the causal pathway to BMI in this study, there is currently no robust method to rule out horizontal pleiotropy as a possible explanation for these findings.34
Locuszoom plots were generated with code adapted from the “gassocplot” R package. The estimated LD matrix was based on a reference panel comprising 10,000 unrelated individuals in the UKB, as described above.
Characterization of SNPs implemented as instrumental variables
We investigated the association between the BMI SNPs identified in the colocalization analysis and various adiposity traits derived from publicly available GWAS summary statistics. These traits included body-fat percentage; hip circumference; leg-fat percentage (left and right); trunk-fat percentage; subcutaneous adipose tissue (SAT) volume; visceral adipose tissue (VAT) volume; subcutaneous adipose tissue attenuation; visceral adipose tissue attenuation; ratio of visceral-to-subcutaneous adipose tissue volume; and waist-to-hip ratio (WHR). We did this by calculating Pearson correlation coefficients to estimate the correlation of each set of SNPs’ estimates for BMI (calculated as Z scores (i.e., beta/standard error)) with each adiposity trait in turn. A heatmap illustrating the SNP:trait correlations based on hierarchical k-means clustering was generated with the “pheatmap” package in R. A summary of the GWAS data implemented in the analysis is provided in Table S2.
Pathway-enrichment and gene-ontology analyses were carried out with the ConsensusPathDB-human web application.35 The approach mines publicly available data from 32 databases to perform over-representation analyses with cellular-interaction networks. Enrichment analyses were also undertaken with gene expression data from GTEx version 8.31 We first leveraged these data to evaluate whether our brain-tissue-partitioned set of variants was enriched in 13 specific areas of the brain by using the FUMA tool36. We next assessed whether our variants were also eQTLs in all other tissue types from GTEx v8 after excluding adipose- and brain-related tissue types (on the basis of a false discovery rate < 0.05 according to results from GTEx).
Mendelian randomization
We estimated the genetically predicted effect of BMI on six cardiovascular disease outcomes (coronary heart disease, type 2 diabetes, atrial fibrillation, heart failure, peripheral artery disease, and stroke) and four measures of cardiac structure and function (left-ventricular end-diastolic volume, left-ventricular end-systolic volume, stroke volume, and left-ventricular ejection fraction) by applying univariable Mendelian randomization (MR).37,38 We achieved this by using individual-level data from the UKB study by generating a genetic risk score (GRS) with all 915 BMI-associated SNPs weighted by their effect estimates in up to 334,398 unrelated individuals of European descent. This sample size was determined after the removal of individuals who had withdrawn consent, who had evidence of genetic relatedness, or who were not of “white European ancestry” on the basis of a K-means clustering (K = 4). Full details are as described previously.39 MR estimates were then calculated with either linear or logistic regression on all 10 outcomes and adjusted for age, sex, the top 10 principal components, and a binary variable indicating genotype chip. We calculated the type 1 error rate attributed to the proportion of overlap between our GRS derivation dataset and the number of affected individals and controls analyzed in the UKB sample with the “sample overlap”40 web application reported in the Web Resources section. This suggested that, on the basis of the F statistics of our instruments, the bias introduced in our analyses as a result of overlapping samples is likely to be very minimal (type 1 error rate < 0.05).
Next, we repeated the analysis above but used the sets of adipose and brain eQTLs that colocalized with BMI on the basis of PPA4 > 0.8 to derive two separate weighted GRSs (i.e. one based on BMI-associated SNPs with evidence of colocalization in adipose tisue and the other with SNPs with evidence of colocalization in brain tissue). In analyses using the score consisting of adipose-colocalized eQTLs, we refer to this exposure as “adipose-tissue-instrumented BMI” hereafter, whereas when using the GRSs derived from brain-colocalized eQTLs we refer to this exposure as “brain-tissu-instrumented BMI.” We then applied an approach known as multivariable MR, which is used for estimating the independent effects of multiple exposures on an outcome by simultaneously estimating their effects in the same model.41,42 Multivariable MR has previously been applied to separate the effects of extremely correlated traits, such as LDL cholesterol and apolipoprotein B.43 We applied this approach in this study to investigate the genetically predicted effects of adipose- and brain-tissue-instrumented BMI independent of each other on the 10 outcomes in turn. We achieved this by simultaneously modeling the adipose- and brain-tissue-derived GRSs together. An overview of this approach is depicted in Figure S2. Effect estimates are interpretable as a 1-standard-deviation BMI change instrumented with variants that colocalize in one tissue while the contribution of BMI instrumented via variants in the other tissue is accounted for.
In the form of sensitivity analyses, we repeated univariable MR analyses in a two-sample setting by using various techniques developed for investigating whether findings were robust to assumptions. This included the inverse-variance weighted (IVW),44 MR Egger, and weighted median methods and allowed us to leverage findings from large-scale GWASs on the 10 outcomes investigated.45, 46, 47, 48, 49 Global heterogeneity among instrumental variables was calculated via Cochran’s Q statistic. A calculated Q value greater than L-1 (where L is equal to the number of instrumental variables) is indicative of heterogeneity between instrumental variables in the analysis.50 These analyses were all conducted with the “TwoSampleMR” R package. We quantified an evaluation of weak instrument bias by calculating the F statistics of genetic instruments and using the conventional threshold of F > 10 to indicate that our instruments were not prone to this source of bias.51 We additionally applied the MR-Clust approach to evaluate whether our adipose- and brain-tissue-derived instrument sets overlapped with the clusters identified by this approach.52 Finally, we investigated the potential issue whereby SNPs used as instruments could influence BMI prior to their effects on gene expression, which in theory could introduce collider bias into analyses.53 For this reason, we applied the “Steiger” method to remove SNPs more strongly correlated with BMI than with tissue-derived gene expression. All outcome data sources analyzed with both one- and two-sample MR are described in Table S3.
Results
Systematically applying genetic colocalization to highlight loci where BMI and adipose- or brain-derived gene expression share a causal variant
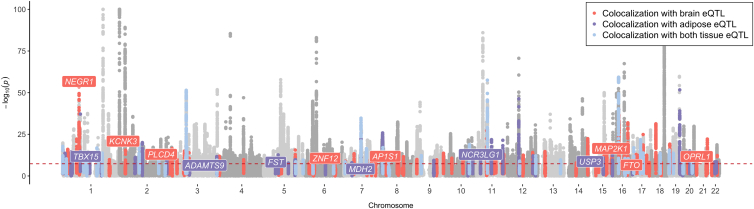
Applying genetic colocalization identified 86 loci where BMI-associated variants colocalized with proximal adipose-derived gene expression (Table S4) and 140 where such variants colocalized with proximal brain-derived gene expression (on the basis of PPA4 > 0.8) (Table S5). In total, 43 variants colocalized with proximal gene expression in both adipose and brain tissues. A subset of candidate loci that are distributed across the genome and provide evidence of colocalization with BMI-associated variants and gene expression in either adipose or brain tissue are highlighted in Figure 1 as an exemplar.
Figure 1.
Loci that provided evidence of genetic colocalization between BMI-associated variants and gene expression derived from brain, adipose tissue, or both
A Manhattan plot illustrating loci and their association with BMI, which showed evidence for colocalization with gene expression in brain tissue (red), adipose tissue (purple), or both (blue). The −log10 of the p value reflecting associations between genetic variants and BMI are plotted on the y axis, and the genomic locations of variants are plotted along the x axis. A subset of loci are annotated here as an exemplar. The complete list of loci that had evidence for colocalization are reported in Tables S4–S6. Gene annotations are based on the gene with the highest PPA4 in the results of the colocalization analysis.
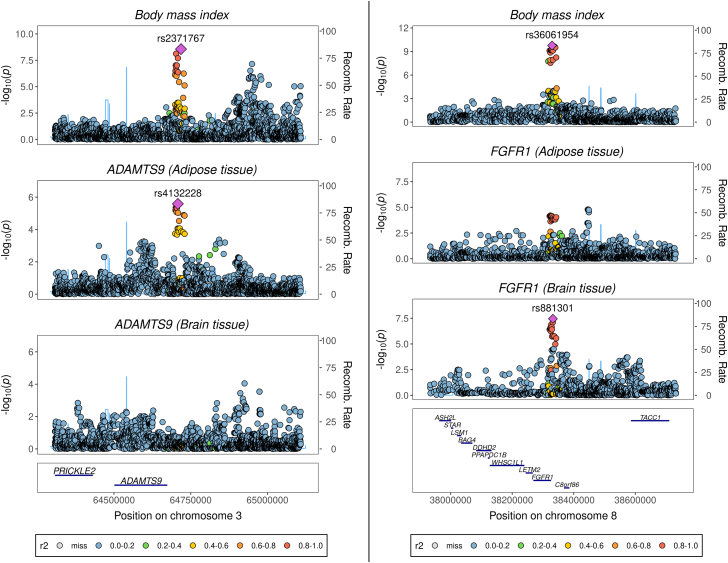
There were many instances where variation in BMI at a locus colocalized with gene expression derived from either subcutaneous adipose or brain-derived tissue but not the other. For example, the ADAMTS9 (a disintegrin-like and metalloprotease with thrombospondin type-1 motif-9) and TBX15 (T-box transcription factor 15) loci provide examples where stronger evidence of colocalization was detected between BMI and their expression in adipose tissue (both PPA4 = 0.97) but not in brain tissue (PPA4 = 0.03 and PPA4 = 0.04, respectively) (Table S6). A locuszoom plot illustrating this contrast at the ADAMTS9 locus is shown in Figure 2A. ADAMTS9 is a secreted metalloproteinase whose expression has been previously linked with decreased insulin sensitivity and signaling in human skeletal muscle,54 whereas TBX15 plays an important role in skeletal development.55
Figure 2.
LocusZoom plots illustrating the association between variants at the ADAMTS9 and FGFR1 loci with body mass index, and each of these genes’ expression in adipose and brain tissue
Variants are plotted according to their chromosomal location at the ADAMTS9 (A) and FGFR1 (B) loci along the x axis, as indicated by the gene track. The strength of their association with each trait is indicated by −log10(p) on the y axis. Recombination rate is calculated from the linkage disequilibrium (LD) structure in the region on the basis of a reference panel of 10,000 individuals of European descent from the UK Biobank (see Methods). LD with respective lead variants in the region is indicated by the color scheme portrayed in the figure legends.
Conversely, NEGR1 (PPA4 = 0.93) and KCNK3 (PPA4 = 0.97) provided strong evidence for colocalization between BMI-associated variants and gene expression when analyses were performed with brain tissue but not when they were performed with gene expression data from adipose tissue (PPA4 = 0.04 and PPA4 = 0.18, respectively) (Table S6). NEGR1 encodes neuronal growth regulator 1, which is involved in synapse formation and neural development,56,57 whereas KCNK3 (potassium two pore domain channel subfamily K member 3) has been previously reported to play a role in taste signaling.58 We also identified loci that provided evidence of colocalization with BMI-associated variants and eQTLs from both adipose and brain tissue; for example, FGFR1, which encodes fibroblast growth factor receptor 1, has an essential role in embryonic development.59 Results at this locus provided evidence of colocalization when we analyzed data from both brain (PPA4 = 0.97) and adipose (PPA4 = 0.92) tissue. The association signal with FGFR1 expression was stronger in the region when we used data derived from brain tissue (p = 3.53 × 10−8) than when we used data derived from adipose tissue (p = 1.64 × 10−5), as depicted in Figure 2B (pcomparison = 0.08). A list of the loci that provided borderline evidence of colocalization (on the basis of PPA4 > 0.7) can be found in Table S7.
Characterization of variants that colocalized with adipose and brain tissue
To investigate how the BMI variants carried forward from the colocalization analysis relate to different measures of anthropometry, we comprehensively compared the correlation between the SNP-BMI associations and 12 GWAS traits representing various aspects of adiposity and body composition. Overall, the SNPs that colocalized with adipose- and brain-tissue BMI shared similar correlation relationships with the majority of adiposity traits and were broadly represented within the two clusters identified by hierarchical k-means clustering (Figure S3). The only exception to this was in relation to WHR and VAT; effect estimates for SNPs that colocalized with BMI-associated variants in brain tissue were more strongly correlated with WHR (r = 0.733) and VAT (r = 0.554) than were estimates for SNPs that colocalized with BMI-associated variants in adipose tissue (WHR: r = 0.445; VAT: r = 0.254) (pcomparison = 0.001 and pcomparison = 0.0088, respectively) (Table S8). This suggests that BMI-associated variants that colocalized with brain-derived gene expression are more likely to play a role in abdominal fat deposition than are variants that colocalized with adipose-derived gene expression. This is particularly noteworthy given that the average effect size of the adipose- and brain-derived variants on BMI were comparable (mean absolute standard deviation change in BMI per effect allele for variants in the adipose set = 0.0148 and brain set = 0.0149). Estimates were similarly comparable after the removal of variants common to both adipose- and brain-tissue-colocalized sets; the adipose variants had a marginally smaller mean magnitude of effect than those specific to brain (adipose = 0.0141 and brain = 0.0146 standard deviation change in BMI).
As an additional sensitivity analysis, we applied this approach on data relating to WHR adjusted for BMI (WHRadjBMI), which found that our adipose subset of SNPs were typically more strongly correlated with this trait (r = −0.423) than were those in the brain subset (r = −0.191) (pcomparison = 0.06). However, we note that caution is required when one interprets these findings because adjusting WHR for BMI might have induced collider bias into these GWAS results.
Pathway analyses provided several examples of overrepresented biological pathways highlighted in each dataset, which could be reflective of important tissue-specific processes related to BMI. For example, genes at adipose-tissue-colocalized loci were enriched among several metabolic pathways, including the malate-aspartate shuttle pathway, which is integral to glycolysis (p = 5.22 × 10−7) (Table S9). Similarly, the fibroblast growth factor (FGF) pathway was enriched for genes at brain-tissue-colocalized loci (p = 3.45 x 10−5); proteins in this pathway constitute a dominant family of signaling molecules in the brain and play important roles in the development and function of the hypothalamus and neuroendocrine system.60,61
Enrichment analyses across 13 individual brain regions for our partitioned sets of SNPs found that the expression of our brain-tissue-colocalized variants tended to be over or underrepresented in certain brain regions compared to others, although overall the evidence of enrichment in these analyses was not particularly strong (full results available in Table S10). Furthermore, after removing SNPs that colocalized in both adipose- and brain tissue, we also found that our sets of variants were on average expressed in various other types of tissues by using data from GTEx v8. In brief, brain-tissue-colocalized variants were eQTLs in a mean of 15.2 (standard deviation [SD] = 10.9) other tissue types (excluding adipose- and brain-related tissues), whereas our adipose partitioned SNPs were eQTLs in a mean of 14.3 (SD = 9.9) other tissues (full results in Table S11).
Using Mendelian randomization instrumented with adipose and brain-regulatory variants to disentangle the putative effects of BMI on disease endpoints
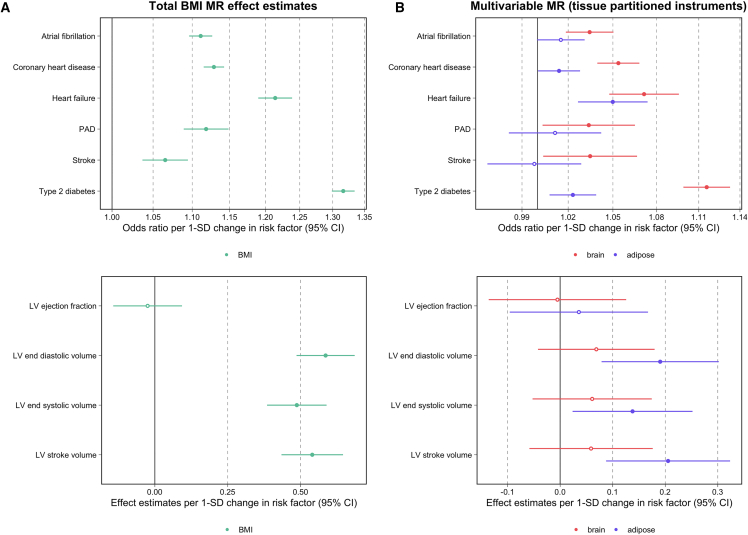
Univariable MR analyses provided strong evidence of a genetically predicted effect between BMI and the cardiovascular endpoints as assessed with a weighted GRS consisting of all 915 SNPs, as well as on the measures of cardiac structure and function with the exception of left ventricular (LV) ejection fracture (beta: −0.025, 95% confidence interval [CI]: −0.143–0.093, p = 0.06) (Figure 3A, Table S12). Repeating analyses in a two-sample analysis via the IVW method supported these findings (Table S13). The estimated Q statistics generated in these analyses (all p < 2.44 × 10−14) highlight the large amount of global heterogeneity associated with the total set of 915 BMI SNPs when these are analyzed against each outcome, suggesting that there are most likely multiple pathways underlying the relationship between these instruments, BMI, and phenotypic endpoints (Table S14). Furthermore, we applied the MR-Clust52 approach, which identified many clusters of BMI variants with comparable effect estimates on outcomes, although none of these clusters substantially overlapped with our adipose- and brain-tissue-expression-partitioned sets (Figure S4, Table S15).
Figure 3.
Forest plots illustrating the Mendelian randomization results
Summary of Mendelian randomization results for BMI on six disease outcomes and four left-ventricular cardiac phenotypes on the basis of (A) univariable analyses using the total set of BMI variants and (B) analyses instrumented in a multivariable setting with tissue-partitioned variants. Forest plots illustrating the odds ratios or effect estimates per standard deviation (SD) change in risk factor and 95% confidence intervals (CIs) for each disease outcome analyzed by MR are shown. The effect estimates of BMI instrumented with all 915 BMI SNPs is illustrated in (A), and the independent effect estimates of BMI instrumented by adipose- (purple) and brain (red)-tissue-derived instruments in the multivariable MR model are illustrated in (B). Circles representing central estimates are filled in when confidence intervals, as illustrated by lines, do not overlap with the null. Abbreviations are as follows: PAD, peripheral artery disease; and LV, left ventricular.
We next investigated the effects of “adipose-“ and “brain-tissue-instrumented BMI” on each outcome by using GRS derived from our subsets of 86 adipose- and 140 brain-tissue variants, respectively. A summary of the adipose- and brain-tissue instruments used, and their effect estimates on BMI, can be found in Table S16. Plots depicting the effect estimates of tissue-partitioned instruments in relation to T2D can be found in Figures S5 and S6 as an exemplar. Broadly, both adipose- and brain-tissue-instrumented BMI provided evidence of an effect on increased risk of cardiovascular disease and increased measures of cardiac structure, with the exception of stroke and peripheral arterial disease, where adipose-tissue-instrumented BMI did not provide strong evidence of an effect (Table S17). Estimates derived in a two-sample setting based on the weighted-median and MR-Egger methods are summarized in Table S18. Repeating analyses with genetic instruments where evidence of colocalization was identified only in adipose or brain tissue (but not the other tissue, on the basis of PPA4 > 0.8) did not drastically alter findings (Table S19). We also applied the Steiger method to filter out instruments that might influence gene expression as a result of their initial effects on BMI variation (Tables S20 and S21).53 This analysis removed two instruments from both the adipose- and brain-tissue sets, although this did not alter overall findings (Table S22).
Next, to separate the effects of adipose- and brain-tissue-instrumented BMI on each outcome, we estimated their independent effects from one another by using multivariable MR. For each of the 6 disease outcomes assessed, the independent effect of adipose-tissue-instrumented BMI was shown to attenuate upon our accounting for the effect of brain-tissue-instrumented BMI in the multivariable MR. For example, the total effect of adipose-tissue-instrumented BMI on coronary heart disease (CHD) risk provided strong evidence of an effect (odds ratio [OR] = 1.04; 95% CI = 1.03–1.05; p = 7.45 × 10−10), although when adipose-tissue-instrumented BMI was analyzed simultaneously with brain-tissue-instrumented BMI in the multivariable model effect estimates attenuated (OR = 1.01; 95% CI = 1.00–1.03; p = 0.04).
In contrast, the multivariable MR estimates for brain-tissue-instrumented BMI were consistent with a strong increasing effect on CHD risk (OR = 1.05; 95% CI = 1.04–1.07; p = 4.67 × 10−14) independent of the effect of BMI instrumented with adipose-tissue SNPs. Similarly, the results of the multivariable MR analysis provided strong evidence of an effect between brain-tissue instrumented BMI and type 2 diabetes (T2D) risk (OR = 1.12; 95% CI = 1.09–1.13; p = 7.16 × 10−46), consistent with the total effect derived in the univariable model (OR = 1.13; 95% CI = 1.11–1.14; p = 8.13 × 10−69). In contrast, evidence of an independent effect for adipose-tissue-instrumented BMI on T2D attenuated in comparison to evidence from the univariable analysis (OR = 1.02; 95% CI = 1.00–1.04; p = 0.003). For detailed results of MR analyses, see Table S17. Figure 3B illustrates similar findings for the other cardiovascular disease endpoints.
In contrast to the analysis above on disease outcomes, applying this approach to the measures of cardiac structure and function revealed that adipose-tissue-instrumented BMI typically predominated in the multivariable model. For example, the univariable MR estimate for adipose-tissue-instrumented BMI on LV stroke volume (beta = 0.23, 95% CI = 0.13–0.34; p = 1.42 × 10−5) remained robust when we accounted for the effect of brain-tissue-instrumented BMI (beta = 0.21, 95% CI = 0.09–0.32, p = 6.43 × 10−4). Conversely, univariable estimates for brain-tissue-instrumented BMI (beta = 0.15, 95% CI = 0.05–0.26, p = 0.004) attenuated to include the null in the multivariable analysis (beta = 0.06, 95% CI = −0.06–0.18, p = 0.33). All multivariable MR estimates from this analysis are also depicted in Figure 3B. A comparison of all univariable and multivariable estimates from the analyses involving tissue-partitioned instruments can be found in Figure S7.
Discussion
We have performed extensive genetic colocalization analyses to gain insights into the distinct contribution of genetic effects on BMI variation putatively mediated by gene expression in adipose and brain tissue. Our findings demonstrate that BMI-associated variants clustered by evidence of colocalization with adipose- and brain-tissue gene expression show differential effects on WHR and VAT, as well as enrichments among biological pathways. This finding is consistent with earlier studies that have suggested that distinct molecular processes and metabolic mechanisms might contribute differentially to fat distribution.8,62,63 Finally, we have undertaken a novel analysis involving these partitioned sets of BMI variants harnessed as genetic instruments by using a multivariable MR framework. Our results suggest that selecting genetic instruments for MR on the basis of their tissue-dependent effects can help elucidate the biological pathways by which an exposure influences disease susceptibility and phenotypic traits.
The genes identified by the colocalization analysis highlight different biological features that might reside along the causal pathway to overall body size. Several candidate genes identified by our analysis converge on processes underlying appetite and feeding behavior. For example, NEGR1 has been implicated in conferring risk of obesity in a number of studies64, 65, 66 and is highly expressed in the hypothalamus, where it is known to affect the central regulation of energy balance.67,68 Our results provide further evidence linking the functional effect of NEGR1 expression variants in the brain to phenotypic variation in BMI. Interestingly, eQTLs in the FGFR1 locus showed evidence for colocalization with BMI in both adipose and brain tissue, but the association with FGFR1 expression was stronger in brain tissue at this region, which might be indicative of its functional role with respect to adiposity. Inhibition of FGFR1 has been linked to appetite suppression, which is most likely mediated by hypothalamic FGFR1 signaling cascades underlying energy intake.69,70 Additionally, FGFR1 activation by FGF21 in peripheral tissues has been found to be important for glucose homeostasis and relaying information on nutritional state.71, 72, 73 Importantly, the endocrine signaling cascade affects the central nervous system by enacting a change in preference for carbohydrate consumption, and this has been shown to directly affect weight regulation.74, 75, 76 Some genes highlighted by our analysis have been implicated in rare forms of monogenic obesity. For example, BDNF (encoding brain-derived neurotrophic factor) has been implicated in a rare case of monogenic obesity most likely arising from impaired BDNF expression during hypothalamic development.77 BDNF is an important effector immediately downstream of MC4R (melanocortin-4) in the melanocortin pathway regulating energy balance.78 MC4R is one of the most common genes implicated in monogenic obesity,79 although evidence of colocalization between BMI-associated variants and brain-tissue expression at this locus narrowly missed out on the heuristic threshold applied in this study (PPA = 0.71). Taken together, these results suggest that genetic effects influencing feeding behaviors and the central regulation of body composition play an important role in overall adiposity variation.
Additionally, our colocalization analysis highlighted examples where genetic effects on body composition are most likely mediated by pathways independent to the central regulation of appetite or energy homeostasis in light of evidence of colocalization of BMI-associated variants with gene expression in adipose but not brain tissue. For example, the mesodermal development gene TBX1555,80,81 is differentially expressed within distinct fat depots82, 83, 84 and has been shown to affect adipocyte differentiation, metabolism, and triglyceride storage.85,86 The evidence presented here for colocalization between TBX15 expression in subcutaneous adipose and BMI-associated variants provides further support for the functional relationship between the pathways driving heterogeneity in adipogenesis and fat distribution.87,88 Similarly, ADAMTS9 has been implicated in insulin secretion in peripheral tissues,54,89 which highlights the putative role that this gene might have in insulin metabolism within adipose tissue and fat composition.62
As a result of the distinct sets of BMI-partitioned variants identified by our colocalization analysis, along with their differential effects on WHR and VAT, we hypothesized that the separate biological pathways to which these variants contribute might have differential effects on disease risk. The results of our multivariable MR analysis suggest that BMI instrumented with brain-tissue eQTLs has an independent effect on increased risk for cardiovascular disease endpoints when one accounts for the contribution of BMI instrumented with adipose-tissue eQTLs. One such mechanism by which these SNPs exert their effects on BMI is via appetite regulatory and energy expenditure pathways, highlighting the critical importance of neurogenic adiposity as a risk factor for all-cause mortality.
Conversely, the results of our multivariable MR analysis suggest that the expression of BMI-associated genes in subcutaneous adipose tissue might underlie the relationship between BMI and measures of cardiac structure and function. Previous studies focusing on fat distribution rather than BMI have found that lean body mass is more strongly related to left ventricular traits than BMI or fat mass.90 Given that the enrichment analyses conducted in this study provided evidence that variants colocalizing with adipose-tissue gene expression are more likely to be involved in fat distribution than variants that colocalized with brain-derived gene expression, our results provide further support for the hypothesis that therelationship between body composition and variation in left ventricular remodeling phenotypes is of prognostic importance.91,92
The findings in this study provide insight into the complexity of the genetics of BMI and propose an innovative method of differentiating between the effects of adiposity- and anthropometry-increasing alleles in specific phenotypic contexts. Therefore, although our study was not focused on identifying effects consistent with favorable adiposity, it could be of interest to triangulate our results from alternative approaches in this paradigm to further investigate BMI-associated loci throughout the genome. However, although our approach might prove valuable in terms of developing insight into disease mechanisms, it should be noted that the overall effect of adiposity on disease risk might be the same regardless of which tissue or pathway it results from (as described by the gene-environment equivalence assumption in MR93). In the present context, if the associations of the two sets of instruments with all aspects of body composition across life were known, then their differential associations with disease outcomes could be shown to be entirely due to these differences in body composition.
A limitation of this work is that cell-type-specific effects within bulk adipose-tissue biopsies or brain transcriptomic datasets have not been accounted for. Integrating cell-type data from large samples when they become available is therefore particularly warranted. For example, adipose tissue is comprised of adipocytes, endothelial cells, and multiple immune cell subtypes, which vary proportionally within population data and might contribute differentially to functional changes in tissue composition.94 Although the inclusion of meta-analyzed gene expression data from multiple datasets provided larger statistical power for this study, the development of methods to deconvolute tissue heterogeneity is a growing area of research that could yield higher resolution into molecular and cell-regulatory changes on disease risk.94,95 The findings presented in this study are based on data derived from individuals of European descent, which is primarily due to the lack of available eQTL datasets in the field in non-European participants. However, the approach presented in this manuscript should be worthwhile applying in non-European samples once these data are available in sufficient sample sizes. Additionally, sex-dependent differences in adiposity will also be an important area for future research given that adipose deposition is known to vary between males and females.96 The work presented here focused on non-sex-stratified analyses to initially demonstrate the novel methodology we have developed and has therefore harnessed the most highly powered datasets available. Moreover, the current lack of publicly available tissue-specific eQTL datasets poses a challenge to sex-stratified analyses, as well as sex-differential participation bias in the UK Biobank study.97
Among our adipose- and brain-tissue-derived sets of instruments, we found that there was still a substantial amount of heterogeneity on the basis of Cochran’s Q statistics. We postulate that the most parsimonious explanation for this is that, although integrating tissue-specific data can help bring us closer to understanding the underlying biology of trait-associated variants, there most likely exist various granular-level mechanisms which require further investigation. For instance, we believe it is unlikely that all 140 variants that colocalized with brain-derived gene expression influence BMI via appetite regulation, but additionally via other types of regulatory pathway. We also note that, although adipose and brain tissues were the focus of this study as a result of sample-size availability and their biological relevance to BMI,98 they might not necessarily be the primary tissue by which our colocalized sets of variants influence this trait. Therefore, although our MR framework can exploit the shared tissue specificity among these instrument sets, further research using datasets derived using other tissues is required for comprehensive characterization of each of these BMI SNPs on a case-by-case basis (once these data are accessible in sufficient samples).
It is also important to note that the adipose-tissue-expressed instruments in this study were derived from subcutaneous tissue, and therefore interpretation of these results might not extend to other adipose-related tissues such as visceral fat (the intra-abdominal adipose adjacent to internal organs) and other fat deposits. Furthermore, although in this study we applied the weighted median and MR-Egger methods, which are typically regarded to be more robust to horizontal pleiotropy than the IVW approach, they cannot rule out extensive correlated pleiotropy.99 Finally, as discussed previously, we acknowledge that genetic correlation is necessary, but not sufficient, for causality.34
The results of this study demonstrate that genetic variants underlying complex traits such as BMI, when partitioned according to tissue-specific molecular data, can yield insight into causal pathways and disease etiology via genetic colocalization and MR. Future studies adopting a similar approach should prove valuable in elucidating the distinct contributions of modifiable exposures to phenotypic variation and disease risk.
Acknowledgments
We are extremely grateful to the researchers who made their summary-level data available for the purposes of this study and the participants of the UK Biobank study (application #15825). Funding for this research was provided by the British Heart Foundation (BHF) (grant FS/17/60/33474). G.D.S. works for the Integrative Epidemiology Unit supported by the Medical Research Council (grant MC_UU_00011/1) at the University of Bristol.
Declaration of interests
T.G.R. is a part-time employee of Novo Nordisk outside of the work presented in this manuscript. All other co-authors declare no competing interests.
Published: January 27, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.12.013.
Contributor Information
Genevieve M. Leyden, Email: rj18633@bristol.ac.u.
Tom G. Richardson, Email: tom.g.richardson@bristol.ac.uk.
Data and code availability
GWAS summary statistics on adipose- and brain-tissue-derived gene expression were obtained from the resources in the URL section above. BMI and WHR GWAS summary data were obtained from the GIANT consortium via the link also presented in the URL section. Summary statistics regarding measures of visceral and subcutaneous adipose-tissue volumes were accessed via the NIH GRASP download portal. All other summary-level data analyzed in this study are publicly available from the OpenGWAS Project. Software used in this study can be accessed from the SMR homepage (see web resources). All other code used in this study was taken from various R packages as referenced throughout the manuscript.
Web resources
Brain expression meta-analysis data, https://cnsgenomics.com/software/smr/#DataResource
Coloc, https://cran.r-project.org/web/packages/coloc/coloc.pdf
COJO/GCTA, https://cnsgenomics.com/software/gcta/#COJO
ConsensusPath-DB, http://cpdb.molgen.mpg.de/
Locuszoom, https://github.com/jrs95/gassocplot
MuTHER adipose data, http://www.muther.ac.uk/Data.html.
NIH GRASP, https://grasp.nhlbi.nih.gov/FullResults.aspx
OpenGWAS Project, https://gwas.mrcieu.ac.uk/
Sample overlap and type 1 error rate, https://sb452.shinyapps.io/overlap/
Supplemental information
References
- 1.World Health Organization (2020). WHO Obesity and overweight Fact sheet No. 311. https://www.who.int/en/news-room/ fact-sheets/detail/obesity-and-overweight (date last accessed October 27, 2020)
- 2.Elks C.E., den Hoed M., Zhao J.H., Sharp S.J., Wareham N.J., Loos R.J., Ong K.K. Variability in the heritability of body mass index: a systematic review and meta-regression. Front. Endocrinol. (Lausanne) 2012;3:29. doi: 10.3389/fendo.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rokholm B., Silventoinen K., Tynelius P., Gamborg M., Sørensen T.I., Rasmussen F. Increasing genetic variance of body mass index during the Swedish obesity epidemic. PLoS ONE. 2011;6:e27135. doi: 10.1371/journal.pone.0027135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silventoinen K., Jelenkovic A., Sund R., Yokoyama Y., Hur Y.M., Cozen W., Hwang A.E., Mack T.M., Honda C., Inui F., et al. Differences in genetic and environmental variation in adult BMI by sex, age, time period, and region: an individual-based pooled analysis of 40 twin cohorts. Am. J. Clin. Nutr. 2017;106:457–466. doi: 10.3945/ajcn.117.153643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yengo L., Sidorenko J., Kemper K.E., Zheng Z., Wood A.R., Weedon M.N., Frayling T.M., Hirschhorn J., Yang J., Visscher P.M., GIANT Consortium Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sulc J., Winkler T.W., Heid I.M., Kutalik Z. Heterogeneity in Obesity: Genetic Basis and Metabolic Consequences. Curr. Diab. Rep. 2020;20:1. doi: 10.1007/s11892-020-1285-4. [DOI] [PubMed] [Google Scholar]
- 7.Ahima R.S., Lazar M.A. Physiology. The health risk of obesity--better metrics imperative. Science. 2013;341:856–858. doi: 10.1126/science.1241244. [DOI] [PubMed] [Google Scholar]
- 8.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., et al. LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timshel P.N., Thompson J.J., Pers T.H. Genetic mapping of etiologic brain cell types for obesity. eLife. 2020;9:e55851. doi: 10.7554/eLife.55851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verkouter I., de Mutsert R., Smit R.A.J., Trompet S., Rosendaal F.R., van Heemst D., Willems van Dijk K., Noordam R. The contribution of tissue-grouped BMI-associated gene sets to cardiometabolic-disease risk: a Mendelian randomization study. Int. J. Epidemiol. 2020;49:1246–1256. doi: 10.1093/ije/dyaa070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaisse C., Clement K., Guy-Grand B., Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat. Genet. 1998;20:113–114. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 12.Montague C.T., Farooqi I.S., Whitehead J.P., Soos M.A., Rau H., Wareham N.J., Sewter C.P., Digby J.E., Mohammed S.N., Hurst J.A., et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387:903–908. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- 13.van der Klaauw A.A., Farooqi I.S. The hunger genes: pathways to obesity. Cell. 2015;161:119–132. doi: 10.1016/j.cell.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Yaghootkar H., Lotta L.A., Tyrrell J., Smit R.A., Jones S.E., Donnelly L., Beaumont R., Campbell A., Tuke M.A., Hayward C., et al. Genetic Evidence for a Link Between Favorable Adiposity and Lower Risk of Type 2 Diabetes, Hypertension, and Heart Disease. Diabetes. 2016;65:2448–2460. doi: 10.2337/db15-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L.O., Rauch A., Mazzaferro E., Preuss M., Carobbio S., Bayrak C.S., Chami N., Wang Z., Schick U.M., Yang N., et al. Genome-wide discovery of genetic loci that uncouple excess adiposity from its comorbidities. Nat Metab. 2021;3:228–243. doi: 10.1038/s42255-021-00346-2. [DOI] [PubMed] [Google Scholar]
- 16.Loos R.J.F., Kilpeläinen T.O. Genes that make you fat, but keep you healthy. J. Intern. Med. 2018;284:450–463. doi: 10.1111/joim.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gómez-Hernández A., Beneit N., Díaz-Castroverde S., Escribano Ó. Differential Role of Adipose Tissues in Obesity and Related Metabolic and Vascular Complications. Int. J. Endocrinol. 2016;2016:1216783. doi: 10.1155/2016/1216783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rees J.M.B., Wood A.M., Dudbridge F., Burgess S. Robust methods in Mendelian randomization via penalization of heterogeneous causal estimates. PLoS ONE. 2019;14:e0222362. doi: 10.1371/journal.pone.0222362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes M.V., Lange L.A., Palmer T., Lanktree M.B., North K.E., Almoguera B., Buxbaum S., Chandrupatla H.R., Elbers C.C., Guo Y., et al. Causal effects of body mass index on cardiometabolic traits and events: a Mendelian randomization analysis. Am. J. Hum. Genet. 2014;94:198–208. doi: 10.1016/j.ajhg.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wade K.H., Chiesa S.T., Hughes A.D., Chaturvedi N., Charakida M., Rapala A., Muthurangu V., Khan T., Finer N., Sattar N., et al. Assessing the causal role of body mass index on cardiovascular health in young adults: Mendelian randomization and recall-by-genotype analyses. Circulation. 2018;138:2187–2201. doi: 10.1161/CIRCULATIONAHA.117.033278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsson S.C., Bäck M., Rees J.M.B., Mason A.M., Burgess S. Body mass index and body composition in relation to 14 cardiovascular conditions in UK Biobank: a Mendelian randomization study. Eur. Heart J. 2020;41:221–226. doi: 10.1093/eurheartj/ehz388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richardson T.G., Sanderson E., Elsworth B., Tilling K., Davey Smith G. Use of genetic variation to separate the effects of early and later life adiposity on disease risk: mendelian randomisation study. BMJ. 2020;369:m1203. doi: 10.1136/bmj.m1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Funck-Brentano T., Nethander M., Movérare-Skrtic S., Richette P., Ohlsson C. Causal Factors for Knee, Hip, and Hand Osteoarthritis: A Mendelian Randomization Study in the UK Biobank. Arthritis Rheumatol. 2019;71:1634–1641. doi: 10.1002/art.40928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kibinge N.K., Relton C.L., Gaunt T.R., Richardson T.G. Characterizing the Causal Pathway for Genetic Variants Associated with Neurological Phenotypes Using Human Brain-Derived Proteome Data. Am. J. Hum. Genet. 2020;106:885–892. doi: 10.1016/j.ajhg.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi T., Wu Y., Zeng J., Zhang F., Xue A., Jiang L., Zhu Z., Kemper K., Yengo L., Zheng Z., et al. eQTLGen Consortium Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun. 2018;9:2282. doi: 10.1038/s41467-018-04558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration & Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis & Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 28.Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M., Ruderfer D.M., Oh E.C., Topol A., Shah H.R., et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng B., White C.C., Klein H.U., Sieberts S.K., McCabe C., Patrick E., Xu J., Yu L., Gaiteri C., Bennett D.A., et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 2017;20:1418–1426. doi: 10.1038/nn.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grundberg E., Small K.S., Hedman Å.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.P., Meduri E., Barrett A., et al. Multiple Tissue Human Expression Resource (MuTHER) Consortium Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium G., GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 33.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson T.G., Haycock P.C., Zheng J., Timpson N.J., Gaunt T.R., Davey Smith G., Relton C.L., Hemani G. Systematic Mendelian randomization framework elucidates hundreds of CpG sites which may mediate the influence of genetic variants on disease. Hum. Mol. Genet. 2018;27:3293–3304. doi: 10.1093/hmg/ddy210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamburov A., Pentchev K., Galicka H., Wierling C., Lehrach H., Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davey Smith G., Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 38.Davey Smith G., Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson T.G., Harrison S., Hemani G., Davey Smith G. An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. eLife. 2019;8:e43657. doi: 10.7554/eLife.43657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burgess Stephen, Davies Neil M., Thompson Simon G. Bias due to participant overlap in two-sample Mendelian randomization. Genetic Epidemiology. 2016;40(7):597–608. doi: 10.1002/gepi.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanderson E., Davey Smith G., Windmeijer F., Bowden J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int. J. Epidemiol. 2019;48:713–727. doi: 10.1093/ije/dyy262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgess S., Thompson D.J., Rees J.M.B., Day F.R., Perry J.R., Ong K.K. Dissecting Causal Pathways Using Mendelian Randomization with Summarized Genetic Data: Application to Age at Menarche and Risk of Breast Cancer. Genetics. 2017;207:481–487. doi: 10.1534/genetics.117.300191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson T.G., Sanderson E., Palmer T.M., Ala-Korpela M., Ference B.A., Davey Smith G., Holmes M.V. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med. 2020;17:e1003062. doi: 10.1371/journal.pmed.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C., et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., et al. Wellcome Trust Case Control Consortium. Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators. Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen J.B., Thorolfsdottir R.B., Fritsche L.G., Zhou W., Skov M.W., Graham S.E., Herron T.J., McCarthy S., Schmidt E.M., Sveinbjornsson G., et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018;50:1234–1239. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah S., Henry A., Roselli C., Lin H., Sveinbjörnsson G., Fatemifar G., Hedman Å.K., Wilk J.B., Morley M.P., Chaffin M.D., et al. Regeneron Genetics Center Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat. Commun. 2020;11:163. doi: 10.1038/s41467-019-13690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pirruccello J.P., Bick A., Wang M., Chaffin M., Friedman S., Yao J., Guo X., Venkatesh B.A., Taylor K.D., Post W.S., et al. Analysis of cardiac magnetic resonance imaging in 36,000 individuals yields genetic insights into dilated cardiomyopathy. Nat. Commun. 2020;11:2254. doi: 10.1038/s41467-020-15823-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowden J., Hemani G., Davey Smith G. Invited Commentary: Detecting Individual and Global Horizontal Pleiotropy in Mendelian Randomization-A Job for the Humble Heterogeneity Statistic? Am. J. Epidemiol. 2018;187:2681–2685. doi: 10.1093/aje/kwy185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgess S., Thompson S.G., CRP CHD Genetics Collaboration Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011;40:755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 52.Foley C.N., Mason A.M., Kirk P.D.W., Burgess S. MR-Clust: clustering of genetic variants in Mendelian randomization with similar causal estimates. Bioinformatics. 2021;37:531–541. doi: 10.1093/bioinformatics/btaa778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hemani G., Tilling K., Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graae A.S., Grarup N., Ribel-Madsen R., Lystbæk S.H., Boesgaard T., Staiger H., Fritsche A., Wellner N., Sulek K., Kjolby M., et al. ADAMTS9 Regulates Skeletal Muscle Insulin Sensitivity Through Extracellular Matrix Alterations. Diabetes. 2019;68:502–514. doi: 10.2337/db18-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh M.K., Petry M., Haenig B., Lescher B., Leitges M., Kispert A. The T-box transcription factor Tbx15 is required for skeletal development. Mech. Dev. 2005;122:131–144. doi: 10.1016/j.mod.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 56.Hashimoto T., Yamada M., Maekawa S., Nakashima T., Miyata S. IgLON cell adhesion molecule Kilon is a crucial modulator for synapse number in hippocampal neurons. Brain Res. 2008;1224:1–11. doi: 10.1016/j.brainres.2008.05.069. [DOI] [PubMed] [Google Scholar]
- 57.Sanz R., Ferraro G.B., Fournier A.E. IgLON cell adhesion molecules are shed from the cell surface of cortical neurons to promote neuronal growth. J. Biol. Chem. 2015;290:4330–4342. doi: 10.1074/jbc.M114.628438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bachmanov A.A., Beauchamp G.K. Taste receptor genes. Annu. Rev. Nutr. 2007;27:389–414. doi: 10.1146/annurev.nutr.26.061505.111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamaguchi T.P., Harpal K., Henkemeyer M., Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 60.Tsai P.S., Brooks L.R., Rochester J.R., Kavanaugh S.I., Chung W.C. Fibroblast growth factor signaling in the developing neuroendocrine hypothalamus. Front. Neuroendocrinol. 2011;32:95–107. doi: 10.1016/j.yfrne.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaminskas B., Goodman T., Hagan A., Bellusci S., Ornitz D.M., Hajihosseini M.K. Characterisation of endogenous players in fibroblast growth factor-regulated functions of hypothalamic tanycytes and energy-balance nuclei. J. Neuroendocrinol. 2019;31:e12750. doi: 10.1111/jne.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E., et al. ADIPOGen Consortium. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GEFOS Consortium. GENIE Consortium. GLGC. ICBP. International Endogene Consortium. LifeLines Cohort Study. MAGIC Investigators. MuTHER Consortium. PAGE Consortium. ReproGen Consortium New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winkler T.W., Günther F., Höllerer S., Zimmermann M., Loos R.J., Kutalik Z., Heid I.M. A joint view on genetic variants for adiposity differentiates subtypes with distinct metabolic implications. Nat. Commun. 2018;9:1946. doi: 10.1038/s41467-018-04124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thorleifsson G., Walters G.B., Gudbjartsson D.F., Steinthorsdottir V., Sulem P., Helgadottir A., Styrkarsdottir U., Gretarsdottir S., Thorlacius S., Jonsdottir I., et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 65.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C., et al. Wellcome Trust Case Control Consortium. Genetic Investigation of ANthropometric Traits Consortium Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Mägi R., et al. MAGIC. Procardis Consortium Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boender A.J., van Rozen A.J., Adan R.A. Nutritional state affects the expression of the obesity-associated genes Etv5, Faim2, Fto, and Negr1. Obesity (Silver Spring) 2012;20:2420–2425. doi: 10.1038/oby.2012.128. [DOI] [PubMed] [Google Scholar]
- 68.Lee A.W., Hengstler H., Schwald K., Berriel-Diaz M., Loreth D., Kirsch M., Kretz O., Haas C.A., de Angelis M.H., Herzig S., et al. Functional inactivation of the genome-wide association study obesity gene neuronal growth regulator 1 in mice causes a body mass phenotype. PLoS ONE. 2012;7:e41537. doi: 10.1371/journal.pone.0041537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jain V.K., Turner N.C. Challenges and opportunities in the targeting of fibroblast growth factor receptors in breast cancer. Breast Cancer Res. 2012;14:208. doi: 10.1186/bcr3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun H.D., Malabunga M., Tonra J.R., DiRenzo R., Carrick F.E., Zheng H., Berthoud H.R., McGuinness O.P., Shen J., Bohlen P., et al. Monoclonal antibody antagonists of hypothalamic FGFR1 cause potent but reversible hypophagia and weight loss in rodents and monkeys. Am. J. Physiol. Endocrinol. Metab. 2007;292:E964–E976. doi: 10.1152/ajpendo.00089.2006. [DOI] [PubMed] [Google Scholar]
- 71.Adams A.C., Yang C., Coskun T., Cheng C.C., Gimeno R.E., Luo Y., Kharitonenkov A. The breadth of FGF21's metabolic actions are governed by FGFR1 in adipose tissue. Mol. Metab. 2012;2:31–37. doi: 10.1016/j.molmet.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hill C.M., Berthoud H.R., Münzberg H., Morrison C.D. Homeostatic sensing of dietary protein restriction: A case for FGF21. Front. Neuroendocrinol. 2018;51:125–131. doi: 10.1016/j.yfrne.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fisher F.M., Maratos-Flier E. Understanding the Physiology of FGF21. Annu. Rev. Physiol. 2016;78:223–241. doi: 10.1146/annurev-physiol-021115-105339. [DOI] [PubMed] [Google Scholar]
- 74.Lan T., Morgan D.A., Rahmouni K., Sonoda J., Fu X., Burgess S.C., Holland W.L., Kliewer S.A., Mangelsdorf D.J. FGF19, FGF21, and an FGFR1/β-Klotho-Activating Antibody Act on the Nervous System to Regulate Body Weight and Glycemia. Cell Metab. 2017;26:709–718.e3. doi: 10.1016/j.cmet.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jensen-Cody S.O., Flippo K.H., Claflin K.E., Yavuz Y., Sapouckey S.A., Walters G.C., Usachev Y.M., Atasoy D., Gillum M.P., Potthoff M.J. FGF21 Signals to Glutamatergic Neurons in the Ventromedial Hypothalamus to Suppress Carbohydrate Intake. Cell Metab. 2020;32:273–286.e6. doi: 10.1016/j.cmet.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baruch A., Wong C., Chinn L.W., Vaze A., Sonoda J., Gelzleichter T., Chen S., Lewin-Koh N., Morrow L., Dheerendra S., et al. Antibody-mediated activation of the FGFR1/Klothoβ complex corrects metabolic dysfunction and alters food preference in obese humans. Proc. Natl. Acad. Sci. USA. 2020;117:28992–29000. doi: 10.1073/pnas.2012073117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gray J., Yeo G.S., Cox J.J., Morton J., Adlam A.L., Keogh J.M., Yanovski J.A., El Gharbawy A., Han J.C., Tung Y.C., et al. Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) gene. Diabetes. 2006;55:3366–3371. doi: 10.2337/db06-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nicholson J.R., Peter J.C., Lecourt A.C., Barde Y.A., Hofbauer K.G. Melanocortin-4 receptor activation stimulates hypothalamic brain-derived neurotrophic factor release to regulate food intake, body temperature and cardiovascular function. J. Neuroendocrinol. 2007;19:974–982. doi: 10.1111/j.1365-2826.2007.01610.x. [DOI] [PubMed] [Google Scholar]
- 79.Farooqi I.S., Keogh J.M., Yeo G.S., Lank E.J., Cheetham T., O’Rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N. Engl. J. Med. 2003;348:1085–1095. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- 80.Lausch E., Hermanns P., Farin H.F., Alanay Y., Unger S., Nikkel S., Steinwender C., Scherer G., Spranger J., Zabel B., et al. TBX15 mutations cause craniofacial dysmorphism, hypoplasia of scapula and pelvis, and short stature in Cousin syndrome. Am. J. Hum. Genet. 2008;83:649–655. doi: 10.1016/j.ajhg.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee K.Y., Singh M.K., Ussar S., Wetzel P., Hirshman M.F., Goodyear L.J., Kispert A., Kahn C.R. Tbx15 controls skeletal muscle fibre-type determination and muscle metabolism. Nat. Commun. 2015;6:8054. doi: 10.1038/ncomms9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee K.Y., Sharma R., Gase G., Ussar S., Li Y., Welch L., Berryman D.E., Kispert A., Bluher M., Kahn C.R. Tbx15 Defines a Glycolytic Subpopulation and White Adipocyte Heterogeneity. Diabetes. 2017;66:2822–2829. doi: 10.2337/db17-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gesta S., Blüher M., Yamamoto Y., Norris A.W., Berndt J., Kralisch S., Boucher J., Lewis C., Kahn C.R. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc. Natl. Acad. Sci. USA. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schleinitz D., Klöting N., Lindgren C.M., Breitfeld J., Dietrich A., Schön M.R., Lohmann T., Dreßler M., Stumvoll M., McCarthy M.I., et al. Fat depot-specific mRNA expression of novel loci associated with waist-hip ratio. Int. J. Obes. 2014;38:120–125. doi: 10.1038/ijo.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun W., Zhao X., Wang Z., Chu Y., Mao L., Lin S., Gao X., Song Y., Hui X., Jia S., et al. Tbx15 is required for adipocyte browning induced by adrenergic signaling pathway. Mol. Metab. 2019;28:48–57. doi: 10.1016/j.molmet.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gesta S., Bezy O., Mori M.A., Macotela Y., Lee K.Y., Kahn C.R. Mesodermal developmental gene Tbx15 impairs adipocyte differentiation and mitochondrial respiration. Proc. Natl. Acad. Sci. USA. 2011;108:2771–2776. doi: 10.1073/pnas.1019704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heid I.M., Jackson A.U., Randall J.C., Winkler T.W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M.C., Speliotes E.K., Mägi R., et al. MAGIC Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schleinitz D., Böttcher Y., Blüher M., Kovacs P. The genetics of fat distribution. Diabetologia. 2014;57:1276–1286. doi: 10.1007/s00125-014-3214-z. [DOI] [PubMed] [Google Scholar]
- 89.Boesgaard T.W., Gjesing A.P., Grarup N., Rutanen J., Jansson P.A., Hribal M.L., Sesti G., Fritsche A., Stefan N., Staiger H., et al. EUGENE2 Consortium Variant near ADAMTS9 known to associate with type 2 diabetes is related to insulin resistance in offspring of type 2 diabetes patients--EUGENE2 study. PLoS ONE. 2009;4:e7236. doi: 10.1371/journal.pone.0007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bella J.N., Devereux R.B., Roman M.J., O’Grady M.J., Welty T.K., Lee E.T., Fabsitz R.R., Howard B.V., The Strong Heart Study Investigators Relations of left ventricular mass to fat-free and adipose body mass: the strong heart study. Circulation. 1998;98:2538–2544. doi: 10.1161/01.cir.98.23.2538. [DOI] [PubMed] [Google Scholar]
- 91.Neeland I.J., Gupta S., Ayers C.R., Turer A.T., Rame J.E., Das S.R., Berry J.D., Khera A., McGuire D.K., Vega G.L., et al. Relation of regional fat distribution to left ventricular structure and function. Circ Cardiovasc Imaging. 2013;6:800–807. doi: 10.1161/CIRCIMAGING.113.000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aung N., Vargas J.D., Yang C., Cabrera C.P., Warren H.R., Fung K., Tzanis E., Barnes M.R., Rotter J.I., Taylor K.D., et al. Genome-Wide Analysis of Left Ventricular Image-Derived Phenotypes Identifies Fourteen Loci Associated With Cardiac Morphogenesis and Heart Failure Development. Circulation. 2019;140:1318–1330. doi: 10.1161/CIRCULATIONAHA.119.041161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ebrahim S., Davey Smith G. Mendelian randomization: can genetic epidemiology help redress the failures of observational epidemiology? Hum. Genet. 2008;123:15–33. doi: 10.1007/s00439-007-0448-6. [DOI] [PubMed] [Google Scholar]
- 94.Glastonbury C.A., Couto Alves A., El-Sayed Moustafa J.S., Small K.S. Cell-Type Heterogeneity in Adipose Tissue Is Associated with Complex Traits and Reveals Disease-Relevant Cell-Specific eQTLs. Am. J. Hum. Genet. 2019;104:1013–1024. doi: 10.1016/j.ajhg.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Donovan M.K.R., D’Antonio-Chronowska A., D’Antonio M., Frazer K.A. Cellular deconvolution of GTEx tissues powers discovery of disease and cell-type associated regulatory variants. Nat. Commun. 2020;11:955. doi: 10.1038/s41467-020-14561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Randall J.C., Winkler T.W., Kutalik Z., Berndt S.I., Jackson A.U., Monda K.L., Kilpeläinen T.O., Esko T., Mägi R., Li S., et al. DIAGRAM Consortium. MAGIC Investigators Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9:e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pirastu N., Cordioli M., Nandakumar P., Mignogna G., Abdellaoui A., Hollis B., Kanai M., Rajagopal V.M., Parolo P.D.B., Baya N., et al. FinnGen Study. 23andMe Research Team. iPSYCH Consortium Genetic analyses identify widespread sex-differential participation bias. Nat. Genet. 2021;53:663–671. doi: 10.1038/s41588-021-00846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sobreira D.R., Joslin A.C., Zhang Q., Williamson I., Hansen G.T., Farris K.M., Sakabe N.J., Sinnott-Armstrong N., Bozek G., Jensen-Cody S.O., et al. Extensive pleiotropism and allelic heterogeneity mediate metabolic effects of IRX3 and IRX5. Science. 2021;372:1085–1091. doi: 10.1126/science.abf1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morrison J., Knoblauch N., Marcus J.H., Stephens M., He X. Mendelian randomization accounting for correlated and uncorrelated pleiotropic effects using genome-wide summary statistics. Nat. Genet. 2020;52:740–747. doi: 10.1038/s41588-020-0631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GWAS summary statistics on adipose- and brain-tissue-derived gene expression were obtained from the resources in the URL section above. BMI and WHR GWAS summary data were obtained from the GIANT consortium via the link also presented in the URL section. Summary statistics regarding measures of visceral and subcutaneous adipose-tissue volumes were accessed via the NIH GRASP download portal. All other summary-level data analyzed in this study are publicly available from the OpenGWAS Project. Software used in this study can be accessed from the SMR homepage (see web resources). All other code used in this study was taken from various R packages as referenced throughout the manuscript.