Abstract
Parabambusicolaceae is a well-studied family in Massarineae, Pleosporales, comprising nine genera and approximately 16 species. The family was introduced to accommodate saprobic bambusicola-like species in both freshwater and terrestrial environments that mostly occur on bamboos and grasses but are also found on different host substrates. In the present study, we surveyed and collected ascomycetes from bamboo and submerged grass across Yunnan Province, China. A biphasic approach based on morphological characteristics and multigene phylogeny demonstrated five new taxa in Parabambusicolaceae. A novel genus Scolecohyalosporium is introduced as a monotypic genus to accommodate S. submersum sp. nov., collected from dead culms of grass submerged in a freshwater stream. The genus is unique in forming filiform ascospores, which differ from other known genera in Parabambusicolaceae. Multigene phylogeny showed that the genus has a close relationship with Multiseptospora. Moreover, the novel monotypic genus Neomultiseptospora, isolated from bamboo, was introduced to accommodate N. yunnanensis sp. nov. Neomultiseptospora yunnanensis formed a separated branch basal to Scolecohyalosporium submersum and Multiseptospora thailandica with high support (100% ML, 1.00 PP). Furthermore, the newly introduced species, Parabambusicola hongheensis sp. nov. was also isolated from bamboo in terrestrial habitats. Parabambusicola hongheensis clustered with the other three described Parabambusicola species and has a close relationship with P. bambusina with significant support (88% ML, 1.00 PP). Parabambusicola hongheensis was reported as the fourth species in this genus. Detailed description, illustration, and updated phylogeny of Parabambusicolaceae were provided.
Keywords: bambusicolous fungi, freshwater fungi, Neomultiseptospora, Parabambusicola, Scolecohyalosporium, taxonomy
1. Introduction
Parabambusicolaceae is a family in the suborder Massarineae, order Pleosporales, class Dothideomycetes [1,2]. The family was introduced by Tanaka et al. [1] to accommodate taxa resembling the genus Bambusicola in Bambusicolaceae. The family is characterized by scattered or in group, immersed to erumpent, globose to subglobose, or hemispherical papillate ascomata, thin- to thick-walled peridium, composed of brown to dark brown pseudoparenchymatous cells, with heavily melanized cells towards the ostiolar neck. Asci are fissitunicate, broadly cylindrical to clavate asci, embedded in an anastomosed, narrow, branched pseudoparaphyses. Ascospores are clavate to fusiform or ellipsoidal, hyaline, frequently multi-septate [1,3].
The asexual morphs of Parabambusicolaceae are known as phoma-like coelomycetes [4] and monodictys-like or trimmatostroma-like hyphomycetes [1,5,6,7]. The coelomycetous asexual morph was reported by Phukhamsakda et al. [4] for Neoaquastroma bauhiniae and N. krabiense, which were found as saprobes on Bauhinia variegata L. (Fabaceae) and Barringtonia acutangula (Lecythidaceae) in Thailand, respectively. The coelomycetous asexual morph is characterized by pycnidial, dark brown to black, globose, uniloculate, ostiolate conidiomata, enteroblastic, phialidic, integrated conidiogenous cells, and oblong to ellipsoidal or ovoid, hyaline, aseptate, smooth-walled conidia [4]. Moreover, Tanaka et al. [1] also reported the spermatium of Parabambusicola bambusina from the culture of isolate KT 2637, characterized by ellipsoidal or ovoid, hyaline, aseptate, smooth-walled conidia.
The hyphomycete asexual genera in Parabambusicolaceae were referred to as Paramonodictys, Paratrimmatostroma, and Pseudomonodictys [5,6,7]. Paramonodictys and Pseudomonodictys are somewhat similar to Monodictys in having pigmented, dictyosporous conidia. However, these two genera differ from Monodictys due to Paramonodictys forming conidia from stroma [7], while the conidia of Pseudomonodictys form granular contents and the culture also produces red pigments on PDA, which has never been reported for Monodictys [5]. Paramonodictys is characterized by erumpent, erect, subcylindrical, or truncated-cone-form stroma, lacking conidiophores, monoblastic conidiogenous cells, and solitary, muriform, globose to subglobose, olivaceous brown to dark brown conidia [7]. Pseudomonodictys is characterized by semi-macronematous to macronematous, septate, branched, pale brown to brown conidiophores, holoblastic, doliiform, integrated, terminal or intercalary, indeterminate, smooth to verrucose, pale brown conidiogenous cells, and solitary, muriform, top-shaped, reddish-brown to dark brown conidia [5]. Paratrimmatostroma is characterized by effuse or confluent sporodochia, macronematous, or semi-macronematous, mononematous, prostrate, or erect, short, oblong to cylindrical, pale brown conidiophores, arising as lateral branches from creeping hyphae, bearing holoblastic, mono- to polyblastic, integrated, terminal conidiogenous cells and acropleurogenous, dark brown, helicoid, cylindrical, sigmoid, or reniform conidia [6].
The sexual genera in Parabambusicolaceae have similar morphological features. However, these genera can be distinguished based on multigene phylogeny. Tanaka et al. [1] accepted Aquastroma, Multiseptospora, Parabambusicola as well as the other two “Monodictys” species in Parabambusicolaceae. Li et al. [8] included the monotypic genus Multilocularia and Wanasinghe et al. [9] introduced Neoaquastroma to this family. Consequently, Phukhamsakda et al. [4] introduced two novel species in Neoaquastroma and included Pseudomonodictys in Parabambusicolaceae based on multigene phylogeny. Recently, three other monotypic genera viz. Lonicericola, Paratrimmatostroma, and Paramonodictys were phylogenetically affiliated in this family [6,7]. Therefore, nine genera are currently accepted in Parabambusicolaceae based on the morpho-molecular approach [3].
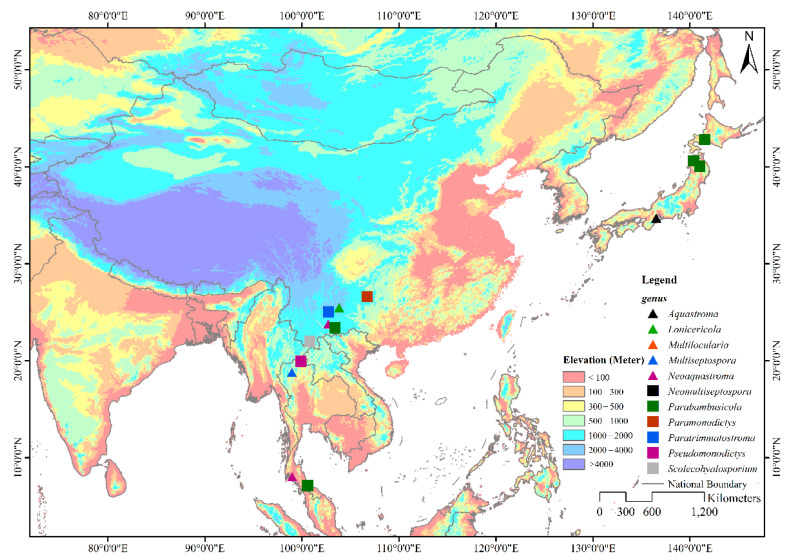
Members of Parabambusicolaceae have been frequently found as saprobes in both aquatic and terrestrial habitats. Species of Parabambusicolaceae occurred on a wide range of hosts, including dicots and monocots, as well as flowerless plants like ferns. However, species in this family are only restricted to Asian countries viz. China (Guizhou and Yunnan), Japan, and Thailand (Figure 1) [1,4,5,6,7,8,10,11,12,13]. Although genera in Parabambusicolaceae are well studied, most of the genera were introduced to accommodate a single species [1,5,6,7,8]. Hence, only 16 species were affiliated in this family under the assumption that more species would be introduced. In the present study, we aim to introduce two monotypic genera and a novel species in Parabambusicolaceae based on multigene phylogeny coupled with morphological distinctiveness.
Figure 1.
Geographical distribution of genera in Parabambusicolaceae. The eleven genera in Parabambusicolaceae that distribute in Asian countries are shown with different symbols on the map.
2. Materials and Methods
2.1. Collection, Examination, Isolation, and Culture Preservation
Dead branches and culms of bamboo and grass were collected from Yunnan Province, China in 2021. The samples were stored in plastic Ziploc bags and returned to the laboratory for observation and examination. Fungal fruiting bodies on the host substrates were observed under an Olympus SZ61 series stereo microscope. Squash-mount slides were prepared to examine micro-morphologies and captured under a Nikon ECLIPSE Ni compound microscope connected with a Nikon DS-Ri2 camera. Indian ink was stained to check the mucilaginous sheath covered the ascospores, while congo red was stained to observe the centrum. The permanent slides contained important morphological features, were prepared by adding Lacto glycerol, and sealed using nail polish. Measurements of morphological features such as ascomata, peridium, pseudoparaphyses, asci, and ascospores were done via NIS-Elements BR 3.2 software v. 5.11.01. Photographic plates were edited and provided by Adobe Photoshop CS6 software (Adobe Systems Inc., San Jose, CA, USA).
Pure cultures were obtained from single spore isolation based on the guidelines of Senanayake et al. [14] and cultivated on potato dextrose agar (PDA) under normal light at 20–25 °C. Culture characteristics were recorded after one week and four weeks. The type specimens were deposited with permanent slides in the Herbarium of Cryptogams Kunming Institute of Botany Academia Sinica (KUN-HKAS), China, and the ex-type living culture was deposited in the Culture Collection of Kunming Institute of Botany (KUMCC). Index Fungorum numbers are provided for the newly described taxa [15].
2.2. DNA Extraction, Amplification, and Sequencing
The generated fungal genomic DNA was extracted from fresh fungal mycelia using Biospin Fungus Genomic DNA Extraction Kit (BioFlux®, Hangzhou, China) following manufacturer protocols (Hangzhou, China). The duplicated strain was also extracted from different fruiting bodies to verify the correctness of DNA sequence data using Forensic DNA Kit (Omega®, Norcross, GA, USA). The generated fungal genomic DNA was stored at 4 °C for PCR amplification and duplicated at −20 °C for long-term storage.
DNA amplification was conducted by polymerase chain reaction (PCR) using four phylogenetic markers viz. the internal transcribed spacers (ITS: ITS1-5.8S-ITS2), 28S large subunit rDNA (LSU), 18S small subunit rDNA (SSU), and the translation elongation factor 1-alpha (TEF1-α). Primer pairs: ITS5 and ITS4 [16], LR0R and LR5 [17], NS1 and NS4 [16], and EF1-983F and EF1-2218R [18] were used to amplify the PCR fragments of these gene regions. The final volume of the PCR reaction was 25 μL containing 2 μL of DNA template, 1 μL of each forward and reverse primers, 12.5 µL of 2× Power Taq PCR Master Mix (mixture of EasyTaqTM DNA Polymerase, dNTPs, and optimized buffer, Beijing Bio Teke Corporation (Bio Teke), Wuxi, China), and 8.5 µL of double-distilled water (ddH2O). The PCR thermal cycle program for ITS, LSU, SSU, and TEF1-α gene regions was set up in the following condition described by Jiang et al. [19]. PCR fragments were purified and sequenced by TsingKe Biological Technology Co., Ltd., Beijing, China. The consensus sequences of the newly generated strains are deposited in GenBank (Table 1).
Table 1.
Taxa used in this study and their GenBank accession numbers. The ex-type and ex-epitype strains are indicated by superscript “T”. The newly generated sequences are indicated in bold.
| Species Name | Strain No. | GenBank Accession Numbers | |||
|---|---|---|---|---|---|
| ITS | LSU | SSU | TEF1-α | ||
| Aquastroma magniostiolata T | HHUF 30122 | NR_153583 | NG_056936 | NG_061000 | AB808486 |
| Bambusicola loculata T | MFLUCC 13-0856 | NR_153609 | NG_069267 | NG_065061 | KP761724 |
| Bambusicola massarinia T | MFLUCC 11-0389 | NR_121548 | NG_058658 | NG_061198 | KP761725 |
| Camarographium koreanum T | CBS 117159 | JQ044432 | JQ044451 | n/a | n/a |
| Dendryphiella fasciculata T | MFLUCC 17-1074 | NR_154044 | NG_059177 | n/a | n/a |
| Dictyocheirospora aquatica T | KUMCC 15-0305 | NR_154030 | KY320513 | n/a | n/a |
| Dictyosporium elegans | NBRC 32502 | DQ018087 | DQ018100 | DQ018079 | n/a |
| Didymocrea sadasivanii | CBS 438.65 | MH858658 | DQ384103 | DQ384074 | n/a |
| Didymosphaeria rubi-ulmifolii T | MFLUCC 14-0023 | n/a | KJ436586 | NG_063557 | n/a |
| Falciformispora lignatilis | BCC 21117 | KF432942 | GU371826 | GU371834 | GU371819 |
| Katumotoa bambusicola T | HHUF 28661 | NR_154103 | NG_059386 | NG_060989 | AB539108 |
| Lonicericola fuyuanensis T | MFLU 19-2850 | NR_172419 | NG_073809 | NG_070329 | MN938324 |
| Lonicericola hyaloseptispora T | KUMCC 18-0149 | NR_164294 | NG_066434 | NG_067680 | n/a |
| Lonicericola hyaloseptispora T | KUMCC 18-0150 | MK098194 | MK098200 | MK098206 | MK098210 |
| Macrodiplodiopsis desmazieri T | CBS 140062 | NR_132924 | NG_058182 | n/a | n/a |
| Magnicamarosporium iriomotense T | HHUF 30125 | NR_153445 | NG_059389 | NG_060999 | AB808485 |
| Melanomma pulvis-pyrius T | CBS 124080 | MH863349 | MH874873 | GU456302 | GU456265 |
| Monodictys sp. | JO 10 | n/a | AB807552 | AB797262 | AB808528 |
| Multilocularia bambusae T | MFLUCC 11-0180 | NR_148099 | NG_059654 | NG_061229 | KU705656 |
| Multiseptospora thailandica T | MFLUCC 11-0183 | NR_148080 | NG_059554 | KP753955 | KU705657 |
| Multiseptospora thailandica | MFLUCC 11-0204 | KU693447 | KU693440 | KU693444 | KU705659 |
| Multiseptospora thailandica | MFLUCC 12-0006 | KU693448 | KU693441 | KU693445 | KU705660 |
| Multiseptospora thysanolaenae T | MFLUCC 11-0202 | n/a | NG_059655 | NG_063600 | KU705658 |
| Neoaquastroma bauhiniae T | MFLUCC 16-0398 | NR_165217 | NG_067814 | NG_070696 | MH028247 |
| Neoaquastroma bauhiniae | MFLUCC 17-2205 | MH025953 | MH023320 | MH023316 | MH028248 |
| Neoaquastroma guttulatum T | MFLUCC 14-0917 | KX949739 | KX949740 | KX949741 | KX949742 |
| Neoaquastroma krabiense T | MFLUCC 16-0419 | NR_165218 | NG_067815 | NG_067670 | MH028249 |
| Neobambusicola strelitziae T | CBS 138869 | NR_137945 | NG_058125 | n/a | MG976037 |
| Neomultiseptospora yunnanensis T | KUMCC 21-0411 | OL898884 | OL898925 | OL898890 | OL964282 |
| Neomultiseptospora yunnanensis T | KUN-HKAS 122240 | OL898885 | OL898926 | OL898891 | OL964283 |
| Palmiascoma gregariascomum T | MFLUCC 11-0175 | NR_154316 | NG_059557 | KP753958 | n/a |
| Parabambusicola aquatica T | MFLUCC 18-1140 | NR_171877 | NG_073791 | n/a | n/a |
| Parabambusicola bambusina | H 4321 | n/a | AB807536 | AB797246 | AB808511 |
| Parabambusicola bambusina | KH 139 | n/a | AB807537 | AB797247 | AB808512 |
| Parabambusicola bambusina | KT 2637 | n/a | AB807538 | AB797248 | AB808513 |
| Parabambusicola honghensis T | KUMCC 21-0410 | OL898880 | OL898921 | OL898886 | n/a |
| Parabambusicola thysanolaenae T | KUMCC 18-0147 | NR_164044 | NG_066435 | NG_067681 | MK098209 |
| Parabambusicola thysanolaenae T | KUMCC 18-0148 | MK098193 | MK098198 | MK098202 | MK098211 |
| Paraconiothyrium estuarinum T | CBS 109850 | NR_166007 | MH874432 | AY642522 | n/a |
| Paraphaeosphaeria michotii T | MFLUCC 13-0349 | NR_155640 | NG_059522 | KJ939285 | n/a |
| Paramonodictys solitarius T | GZCC 20-0007 | MN901152 | MN897835 | MN901118 | MT023012 |
| Paramonodictys solitarius | MFLUCC 17-2353 | MT627707 | MN913703 | MT864299 | MT954397 |
| Paramonodictys solitarius | KH 331 | n/a | AB807553 | AB797263 | AB808529 |
| Paratrimmatostroma kunmingensis T | KUN-HKAS 102224A | MK098192 | MK098196 | MK098204 | MK098208 |
| Paratrimmatostroma kunmingensis T | KUN-HKAS 102224B | MK098195 | MK098201 | MK098207 | n/a |
| Poaceascoma helicoides T | MFLUCC 11-0136 | NR_154317 | NG_059565 | NG_061205 | KP998461 |
| Pseudochaetosphaeronema larense T | CBS 640.73 | NR_132038 | NG_057978 | NG_061147 | KF015684 |
| Pseudocoleophoma calamagrostidis T | HHUF 30450 | NR_154375 | NG_059804 | NG_061264 | LC014614 |
| Pseudomonodictys tectonae T | MFLUCC 12-0552 | n/a | NG_059590 | NG_061213 | KT285571 |
| Scolecohyalosporium submersum T | KUMCC 21-0412 | OL898883 | OL898924 | OL898889 | OL964281 |
| Scolecohyalosporium submersum T | KUMCC 21-0413 | OL898881 | OL898922 | OL898887 | OL964279 |
| Scolecohyalosporium submersum T | KUN-HKAS 122242 | OL898882 | OL898923 | OL898888 | OL964280 |
| Setoseptoria phragmitis T | CBS 114802 | KF251249 | KF251752 | n/a | KF253199 |
| Spegazzinia tessarthra | SH 287 | n/a | AB807584 | AB797294 | AB808560 |
| Sulcatispora acerina T | KT 2982 | LC014597 | LC014610 | LC014605 | LC014615 |
| Sulcatispora berchemiae T | HHUF 29097 | NR_153444 | NG_059390 | NG_064843 | AB808509 |
| Tingoldiago graminicola T | KH 68 | n/a | AB521743 | AB521726 | AB808561 |
| Trematosphaeria grisea T | CBS 332.50 | NR_132039 | NG_057979 | NG_062930 | KF015698 |
| Trematosphaeria pertusa T | CBS 122368 | NR_132040 | NG_057809 | n/a | n/a |
2.3. Sequence Alignment and Phylogenetic Analyses
The consensus ITS, LSU, SSU, and TEF1-α sequences of the newly generated strains were blasted in the nucleotide BLAST search tool via the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 25 November 2021) for searching the similar taxa. The nucleotide BLAST searches showed that the newly generated sequences had the closest similarity with species in Parabambusicolaceae. Hence, the newly generated strains were aligned with representative taxa in Parabambusicolaceae, which were retrieved from GenBank (Table 1). Individual gene alignments were aligned with MAFFT v. 7.475 [20], and the ambiguous sites were trimmed by TrimAl v. 1.3 via the web server phylemon 2 (http://phylemon.bioinfo.cipf.es/utilities.html, accessed on 25 November 2021) and improved manually where necessary using BioEdit v. 6.0.7 [21]. Improved individual gene alignments were prior analyzed by randomized axelerated maximum likelihood (RAxML) analysis via RAxML-HPC v.8 on the XSEDE (8.2.12) tool in the online web portal CIPRES Science Gateway v. 3.3 [22] using default settings but following adjustments described by Jiang et al. [19]. After checking the tree topologies of every individual gene alignment for incongruities, the combined ITS, LSU, SSU, and TEF1-α sequence dataset were then analyzed based on maximum likelihood (RAxML) and Bayesian inference (BI) analyses following commands described by Jiang et al. [19]. Phylogenetic trees were visualized on FigTree v. 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 25 November 2021) and edited by using Microsoft Office PowerPoint 2016 (Microsoft Inc., Redmond, WA, USA). The final sequence dataset and tree were submitted in TreeBASE (https://www.treebase.org/, accessed on 25 November 2021, submission ID: 29072).
3. Results
3.1. Phylogenetic Analyses
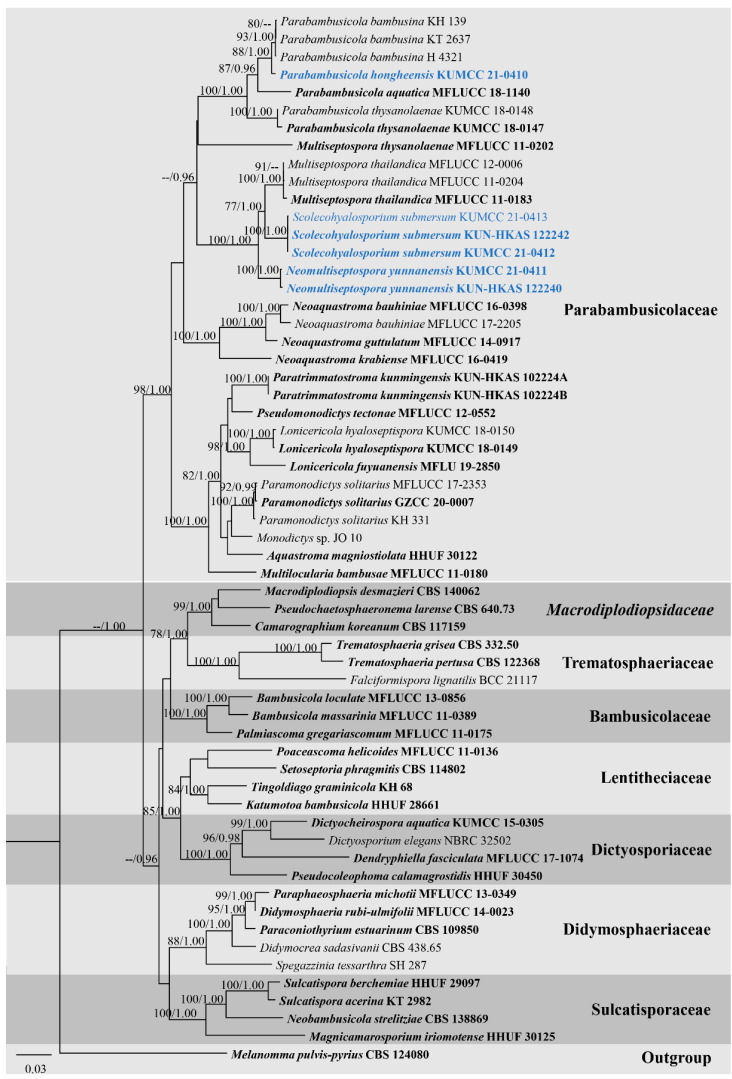
Primarily phylogenetic analyses based on individual genes of ITS, LSU, SSU, and TEF1-α sequence dataset from Table 1 indicated that ITS and TEF1-α are good phylogenetic markers to clarify phylogenetic resolutions of taxa in Parabambusicolaceae. In contrast, LSU and SSU have scarce phylogenetic information to evaluate the phylogenetic status of taxa, and most internal nodes lack significant support (data not shown). Thus, the combined ITS, LSU, SSU, and TEF1-α sequence dataset was conducted to clarify phylogenetic status of taxa in Parabambusicolaceae with other related families. The combined ITS, LSU, SSU, and TEF1-α sequence dataset comprises 59 strains of 46 representative species in Parabambusicolaceae and related families in Massarineae. The dataset consists of 3821 total characters, including gaps (ITS: 1–600 bp, LSU: 601–1486 bp, SSU: 1487–2889 bp, TEF1-α: 2890–3821 bp). The best scoring RAxML tree was selected to represent the phylogenetic relationships of the novel taxa with other representative taxa in Parabambusicolaceae (Figure 2), with a final ML optimization likelihood value of −23,900.440712 (ln). All free model parameters were estimated by the GTRGAMMA model, with 1354 distinct alignment patterns and 28.22% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.236175, C = 0.253070, G = 0.272964, T = 0.237791, with substitution rates AC = 1.158716, AG = 2.682148, AT = 1.418647, CG = 1.002102, CT = 6.123283, GT = 1.000000. The gamma distribution shape parameter alpha = 0.192280 and the Tree-Length = 2.440143. The final average standard deviation of split frequencies at the end of total MCMC generations was calculated as 0.008507 in BI analysis.
Figure 2.
RAxML tree based on ITS, LSU, SSU, and TEF1-α sequence matrix representing the phylogenetic relationships of taxa in Parabambusicolaceae and related families in Massarineae. The tree is rooted in Melanomma pulvis-pyrius (CBS 124080). Bootstrap support values for ML equal to or greater than 70% and the Bayesian posterior probabilities equal to or higher than 0.95 PP are indicated above the nodes as ML/PP. Ex-type strains are in bold and the new taxa are indicated in blue.
Overall tree topologies based on maximum likelihood (ML), and Bayesian inference (BI) analyses were similar and not significantly different. Three novel species formed well-resolved subclades within Parabambusicolaceae. Parabambusicola hongheensis (KUMCC 21-0410) formed a well-resolved clade with other Parabambusicola species and has a close relationship with P. bambusina (KH 139, KT 2637, H 4321) with significant support (88% ML, 1.00 PP; Figure 2). Two strains of Neomultiseptospora yunnanensis (KUMCC 21-0411, KUN-HKAS 122240) formed a robust clade (100% ML, 1.00 PP) basal to the novel monotypic genus Scolecohyalosporium and the genus Multiseptospora with high support (100% ML, 1.00 PP). Three representative strains of Scolecohyalosporium submersum sp. nov. (KUMCC 21-0412, KUMCC 21-0413, KUN-HKAS 122242) formed a robust clade (100% ML, 1.00 PP), sister to Multiseptospora thailandica (MFLUCC 11-0184, MFLUCC 11-0204, MFLUCC 12-0006) with significant support (77% ML, 1.00 PP).
3.2. Taxonomy
Neomultiseptospora N. Xie, Phookamsak & Hongsanan, gen. nov.
Index Fungorum number: IF 559389
Etymology: Referring to the taxon has a close phylogenetic relationship with the genus Multiseptospora.
Saprobic on bamboo. Sexual morph: Ascomata solitary, scattered, immersed, visible as raised, black dome-shaped on host surface, uni-loculate, hemispherical to subconical, with wedge-shaped at sides, glabrous, ostiole central or aside, with apapillate. Peridium unequally thick-walled, thick at the sides, poorly developed at the base, composed of several layers of brown to dark brown pseudoparenchymatous cells, arranged in textura angularis to textura prismatica, with palisade-like cells at sides, outer layers intermixed with host cortex. Hamathecium composed of dense, filamentous, branched, septate, cellular pseudoparaphyses, slightly constricted at the septa, anastomosed above the asci, embedded in a hyaline gelatinous matrix. Asci 8-spored, bitunicate, fissitunicate, clavate, shortly pedicellate, apically rounded, with the well-developed ocular chamber. Ascospores overlapping 1–2-seriate, hyaline, fusiform to ellipsoidal, or oblong, slightly curved, septate, smooth-walled, surrounded by a distinct, thick, mucilaginous sheath. Asexual morph: Undetermined.
Type species: Neomultiseptospora yunnanensis Phookamsak, Hongsanan and N. Xie
Notes: The novel genus Neomultiseptospora is introduced herein to accommodate the single species N. yunnanensis collected from dead branches of bamboo in Yunnan Province, China. Multigene phylogeny demonstrated that Neomultiseptospora clustered with the genera Multiseptospora and Scolecohyalosporium (Figure 2). Morphologically, Neomultiseptospora resembles Multiseptospora in having phragmosporous, hyaline, multi-septate ascospores but differs in forming raised, hemispherical to subconical ascomata, while Multiseptospora has globose to subglobose ascomata, covered by dark, hair-like hyphae and embedded in host tissue [10]. Additionally, Neomultiseptospora has a close phylogenetic relationship with Scolecohyalosporium but the genus is distinct from Scolecohyalosporium in having phragmosporous ascospores while Scolecohyalosporium has scolecosporous ascospores. Unfortunately, Neomultiseptospora is morphologically similar to the generic type of Parabambusicola in having hemispherical to subconical ascomata and hyaline, fusiform, 5-septate ascospores and occurring on bamboo (Sasa spp.) [1]. However, multigene phylogenetic analyses based on a combined ITS, LSU, SSU, and TEF1-α sequence dataset indicated the distinctiveness between these two genera.
Neomultiseptospora yunnanensis Phookamsak, Hongsanan and N. Xie, sp. nov.
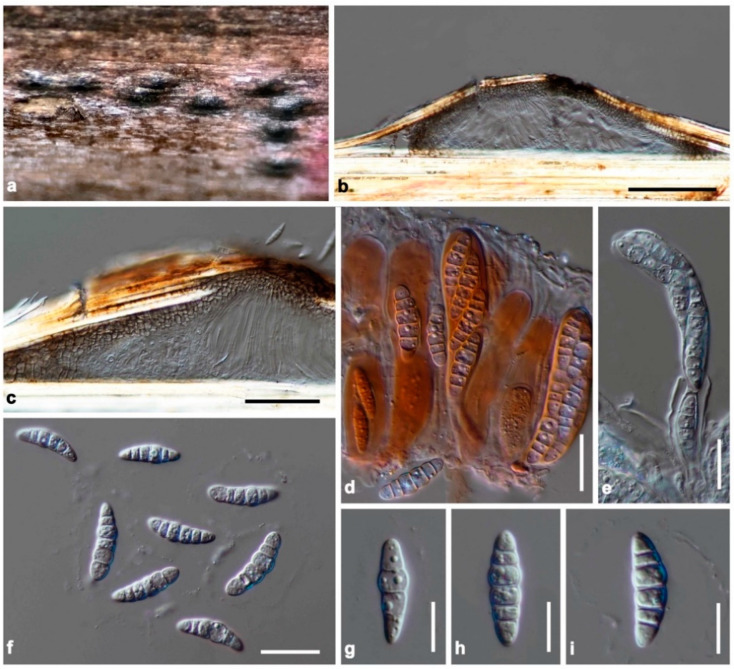
Index Fungorum number: IF 559390, Figure 3.
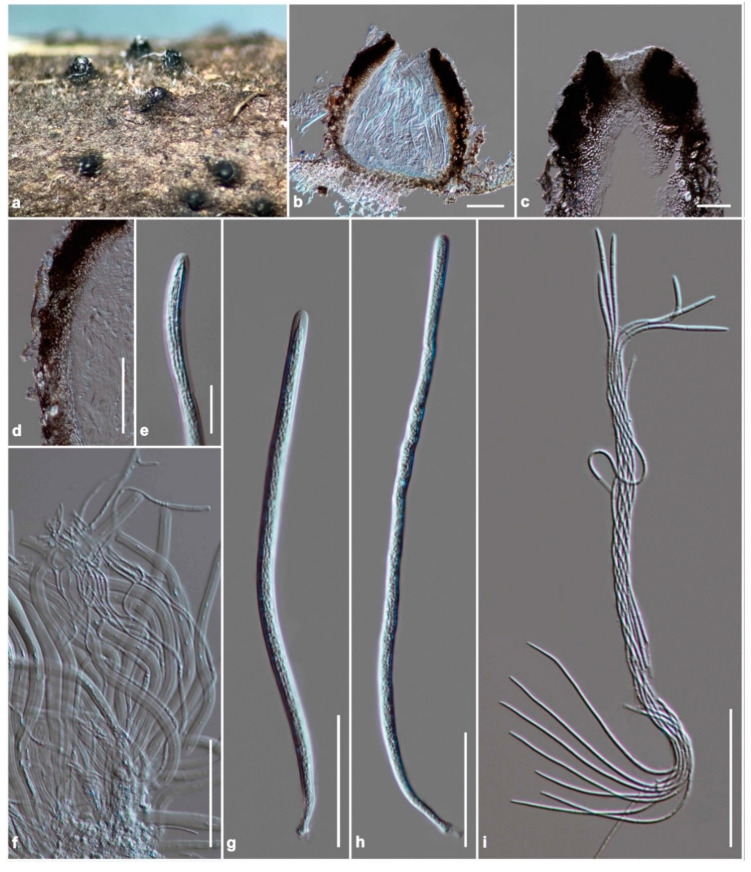
Figure 3.
Neomultiseptospora yunnanensis (KUN-HKAS 122240, holotype). (a) The appearance of ascomata on host substrate; (b) vertical section of ascoma; (c) peridium; (d) asci embedded in cellular pseudoparaphyses stained with congo red; (e) fissitunicate ascus; (f–i) ascospores. Scale bars: (b) = 100 μm, (c) = 50 μm, (d–f) = 20 μm, (g–i) = 10 μm.
Etymology: Referring to the locality, Yunnan Province, China, where the species was collected.
Holotype: KUN-HKAS 122240
Saprobic on dead branches of bamboo. Sexual morph: Ascomata 75–100 μm high, 340–450 μm diam, solitary, scattered, immersed, visible as raised, black, dome-shaped on host surface, uni-loculate, hemispherical to subconical, with wedge-shaped at sides, glabrous, ostiole central or aside, with apapillate. Peridium unequally thick-walled, 15–35 μm wide at the apex, 60–100 μm wide at sides, poorly developed at the base, composed of several layers of brown to dark brown pseudoparenchymatous cells, arranged in textura angularis to textura prismatica, with palisade-like cells at sides, outer layers intermixed with host cortex. Hamathecium composed of dense, 1–2 μm wide, filamentous, branched, septate, cellular pseudoparaphyses, slightly constricted at the septa, anastomosed above the asci, embedded in a hyaline gelatinous matrix. Asci (50–)60–90(–98) × 15–20 μm (x̅ = 76.1 × 17.5 μm, n = 30), 8-spored, bitunicate, fissitunicate, clavate, shortly pedicellate, apically rounded, with the well-developed ocular chamber. Ascospores (22–)23–27 × 5–8.5 μm (x̅ = 25 × 7.1 μm, n = 30), overlapping 1–2-seriate, hyaline, fusiform to ellipsoidal, or oblong, with rounded ends, slightly curved, (4–)5-septate, constricted at the central septum, less constricted at the other septa, smooth-walled, with small guttules, surrounded by a distinct, thick, mucilaginous sheath. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinated on PDA within 24 h. Colonies on PDA reaching 27–30 mm diam after 2 weeks at room temperature (15–20 °C). Colonies dense, irregular in shape, convex to umbonate, surface smooth with an undulate edge, floccose to cottony; from above pale grey to white-grey at the margin, grey at the middle towards the center; from below white-brown at the margin, dark brown to black at the middle towards the center; not producing pigmentation on PDA.
Material examined: China, Yunnan Province, Honghe Autonomous Prefecture, Honghe County, Honghe Hani Rice Terraces (23°5′35″ N, 102°46′47″ E, 1432 + 6 msl), on the dead stem of bamboo in a terrestrial environment, 26 January 2021, R. Phookamsak, BN09A (KUN-HKAS 122240, holotype), ex-type living culture, KUMCC 21-0411.
Notes: The nucleotide BLAST search of ITS sequence showed that the closest similarity of Neomultiseptospora yunnanensis (KUMCC 21-0411) is “Pleosporales sp. strain 1192” (95.26% similarity, Identities = 402/422, with 1 gap) and is identical to Multiseptospora thailandica strain MFLUCC 11-0204 with 94.07% similarity (Identities = 428/455, with no gap), strain MFLUCC 12-0006 with 94.07% similarity (Identities = 428/455, with no gap), and strain MFLUCC 11-0183 with 94.04% similarity (Identities = 426/453, with no gap). Based on the nucleotide BLAST search of LSU sequence, N. yunnanensis is similar to M. thailandica strain MFLUCC 12-0006 (98.10% similarity, Identities = 824/840, with 3 gaps), strain MFLUCC 11-0204 (98.18% similarity, Identities = 808/823, with 2 gaps), and strain MFLUCC 11-0183 (97.91% similarity, Identities = 749/765 with 2 gaps). The closest hits based on TEF1-α sequence are M. thailandica strain MFLUCC 12-0006 (96.12% similarity, Identities = 817/850 with no gap), Neoophiosphaerella sasicola strain KT 1706 (93.16% similarity, Identities = 858/921 with no gap), and Wettsteinina lacustris isolate AFTOL-ID 1592 (93.12% similarity, Identities = 866/930 with no gap).
Based on a nucleotide pairwise comparison, Neomultiseptospora yunnanensis differs from Multiseptospora thailandica (MFLUCC 11-0183, type strain) in 24/416 bp of ITS (5.77%), 15/833 bp of LSU (1.8%), and 39/913 bp of TEF1-α (4.27%). Moreover, N. yunnanensis differs from Scolecohyalosporium submersum (KUMCC 21-0412) in 27/417 bp of ITS (6.47%), 16/850 bp of LSU (1.88%), and 35/923 bp of TEF1-α (4.27%). Neomultiseptospora yunnanensis can be distinguished from M. thailandica in having fusiform to ellipsoidal, or oblong, (4–)5-septate ascospores, whereas M. thailandica has typical fusiform to vermiform, 10–11-septate ascospores [10]. Neomultiseptospora yunnanenis clearly distinct from S. submersum due to S. submersum having filiform, multi-septate (up to 20 septa) ascospores.
Parabambusicola hongheensis H.B. Jiang and Phookamsak, sp. nov.
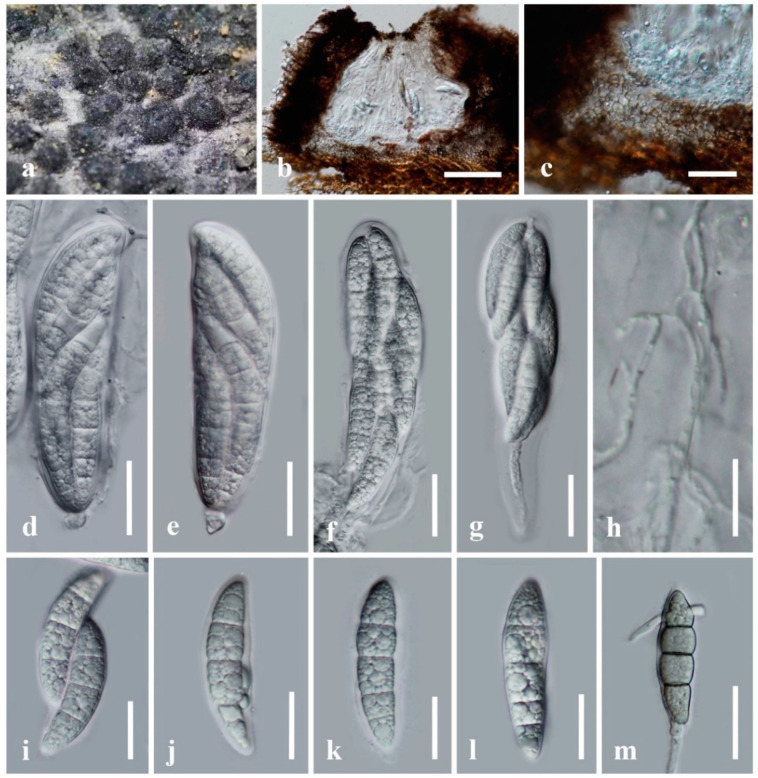
Index Fungorum number: IF 559391, Figure 4.
Figure 4.
Parabambusicola hongheensis (KUN-HKAS 122607, holotype). (a) The appearance of ascomata on host substrate; (b) vertical section of ascoma; (c) peridium; (d–g) asci; (h) pseudoparaphyses; (i–l) ascospores; (m) germinated ascospore. Scale bars: (b) = 100 μm, (c–g,m) = 30 μm, (i–l) = 20 μm, (h) = 15 μm.
Etymology: The specific epithet “hongheensis” refers to the location, Honghe, Yunnan Province of China, where the new species was collected.
Holotype: KUN-HKAS 122607
Saprobic on dead bamboo culms in a terrestrial environment. Sexual morph: Ascomata 200–300 μm high, 340–500 μm diam, gregarious, immersed under host epidermis, raised, dark brown to black, coriaceous, subglobose in surface view, hemispherical or conical with a flattened base in cross-section, uni-loculate, ostiolate, with a pore-like opening at the top. Peridium 20–75 μm wide, comprising dark brown to pale brown cells arranged in textura angularis, intermixed with host tissue. Hamathecium composed of 1.3–2.5 μm wide, filamentous, septate, anastomosed pseudoparaphyses, embedded in a hyaline gelatinous matrix. Asci 115–140 × 30–35 μm (x̅ = 128 × 32.5 μm, n = 20), 8-spored, bitunicate, fissitunicate, broadly cylindrical to clavate, with a swollen end, apically rounded, with an inconspicuous ocular chamber. Ascospores 50–55 × 12–15 μm (x̅ = 53 × 13.5 μm, n = 20), overlapping 2–3-seriate, subhyaline to hyaline, fusiform to vermiform, narrower towards the lower part, enlarged at the 2nd cell from the apex, slightly curved, 3–4-septate, slightly constricted at the septa, smooth-walled, with small, multi-guttulate, surrounded by a thin entire sheath. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinating on PDA within 24 h and germ tubes produced from both cells of ascospores. Colonies on PDA reaching 30 mm diam after 3 weeks at room temperature (10–20 °C). Colonies dense, circular, flattened, floccose; from above pale brown at the middle, pale white at the margin; from below dark brown to black; not producing pigmentation on agar medium. Mycelium superficial to immersed in media, with branched pale grey to black, septate, smooth hyphae.
Material examined: China, Yunnan Province, Honghe Autonomous Prefecture, Honghe County, Baohua Village (23°15′19″ N, 102°21′31.56″ E, altitude 1816.52 msl), on dead culms of bamboo in a terrestrial environment, 28 October 2020, H.B. Jiang, HONGHE015 (KUN-HKAS 122607, holotype), ex-type living culture, KUMCC 21-0410.
Notes: The NCBI nucleotide BLAST search of ITS sequence indicated that Parabambusicola hongheensis (KUMCC 21-0410) is similar to P. bambusina strain KT 2637 with 98.30% similarity (Identities = 521/530, with 3 gaps), and P. bambusina strain H4321 with 98.11% similarity (Identities = 520/530, with 4 gaps). Nucleotide BLAST results of LSU sequence showed that P. hongheensis is similar to “Parabambusicola sp. strain GZCC19-0505” (100% similarity, Identities = 783/783, with no gap), P. bambusina strain H4321 (99.64% similarity, Identities = 840/843, with 1 gap), and P. thysanolaenae strain KUMCC 18-0147 (99.53% similarity, Identities = 839/843, with 1 gap).
A nucleotide pairwise comparison of ITS and LSU sequences indicated that Parabambusicola hongheensis differs from P. bambusina (H 4321, KT 2637, KH 139) in 9/530 bp (1.7%), and 2/843 bp (0.2%), respectively. Parabambusicola hongheensis differs from P. aquatica (MLFUCC 18-1140, type strain) in 40/444 bp of ITS (9%), and 9/836 bp of LSU (1.1%). The species also differs from P. thysanolaenae (KUMCC 18-0147, type strain) in 44/446 bp of ITS (9.9%), and 4/843 bp of LSU (0.4%). Parabambusicola hongheensis typically morphological resembles species in Parabambusicola in forming gregarious, immersed to erumpent, raised, hemispherical ascomata, with flattened base, broadly cylindrical to clavate asci, with subsessile to short pedicel and phragmosporous, hyaline, fusiform ascospores [1,6,12]. Parabambusicola hongheensis can be distinguished from the other three Parabambusicola species in having 3–4-septate ascospores, while the other Parabambusicola species have 5–(6–7)-septate ascospores [1,6,12].
Scolecohyalosporium N. Xie, Phookamsak and Hongsanan, gen. nov.
Index Fungorum number: IF 559392
Etymology: Referring to the taxon forming hyaline scolecosporous ascospores on grass (Poaceae).
Saprobic on stems of unidentified grass submerged in a small stream. Sexual morph: Ascomata solitary, scattered. sometimes clustered, erumpent through host cortex, becoming superficial, uni-loculate, conical to ovoid, glabrous, rough-walled, ostiole central, with truncate apex. Peridium thick-walled, of unequal thickness, thick at sides towards the apex, thin at the base, composed of several layers of small, dark brown to black, pseudoparenchymatous cells, arranged in a textura angularis, with carbonaceous cells at the ostiole, outer layers intermixed with the host tissues. Hamathecium composed of dense, filamentous, branched, septate, cellular pseudoparaphyses, constricted at the septa, anastomosed among the asci, embedded in a gelatinous matrix. Asci 8-spored, bitunicate, fissitunicate, long cylindrical, shortly pedicellate, apically rounded with the indistinct ocular chamber. Ascospores spirally arranged within the ascus, hyaline, filiform, narrower towards the end cells, multi-septate, not constricted at the septa, smooth-walled. Asexual morph: Undetermined.
Type species: Scolecohyalosporium submersum Phookamsak, Hongsanan and N. Xie
Notes: Scolecohyalosporium is introduced herein as a monotypic genus to accommodate S. submersum sp. nov. The species was isolated from dead stems of grass submerged in a freshwater stream in Yunnan, China. Scolecohyalosporium has a unique character that can be distinguished from other sexual genera in Parabambusicolaceae in forming scolecosporous ascospores. Scolecohyalosporium morphological resembles genera Neoophiosphaerella and Poaceascoma in Lentitheciaceae. However, Neoophiosphaerella can be distinguished from Scolecohyalosporium in having subglobose to hemispherical ascomata, with thin-walled, brown cells of peridium, and broader cylindrical asci [1], while the type of Poaceascoma formed setose ascomata [23].
Scolecohyalosporium submersum Phookamsak, Hongsanan and N. Xie, sp. nov.
Index Fungorum number: IF 559393, Figure 5.
Figure 5.
Scolecohyalosporium submersum (KUN-HKAS 122242, holotype). (a) The appearance of ascomata on host substrate; (b) vertical section of ascoma; (c) section through ostiole; (d) peridium; (e) fissitunicate at the apex of the ascus; (f) pseudoparaphyses; (g,h) asci; (i) ascospores. Scale bars: (b,d) = 100 μm, (c,f–i) = 50 μm, (e) = 20 μm.
Etymology: Referring to the habitat of the taxon, submerged in a small freshwater stream.
Holotype: KUN-HKAS 122242
Saprobic on stems of unidentified grass submerged in a small stream. Sexual morph: Ascomata 370–480 μm high, 380–600 μm diam, solitary, scattered. sometimes clustered, erumpent through host cortex, becoming superficial, uni-loculate, conical to ovoid, glabrous, rough-walled, ostiole central, with truncate apex. Peridium 50–90 μm wide at sides, 20–40 μm wide at the base, thick-walled of unequal thickness, thick at sides towards the apex, thin at the base, composed of several layers of small, dark brown to black, pseudoparenchymatous cells, arranged in a textura angularis, with carbonaceous cells at the ostiole, outer layers intermixed with the host tissues. Hamathecium composed of dense, 1.8–3.6 μm wide, filamentous, tapering towards the apex, branched, septate, cellular pseudoparaphyses, constricted at the septa, anastomosed among the asci, embedded in a gelatinous matrix. Asci (200–)250–300(–370) × 7–9(–11) μm (x̅ = 274.6 × 8.2 μm, n = 30), 8-spored, bitunicate, fissitunicate, long cylindrical, shortly pedicellate, apically rounded with the indistinct ocular chamber. Ascospores (230–)260–285(–315) × 1.5–2.2 μm (x̅ = 267.5 × 1.9 μm, n = 30), spirally arranged within the ascus, hyaline, filiform, narrower towards the end cells, multi-septate, up to 20 septa, not constricted at the septa, smooth-walled. Asexual morph: Undetermined.
Culture characteristics: Ascospores germinated on PDA within 24 h. Colonies on PDA reaching 18 mm diam after 2 weeks at room temperature (15–20 °C). Colonies dense, circular, low convex, surface smooth with an entire edge, floccose to cottony; from above pale grey at the margin, grey to dark grey at the center; from below white at the margin, black at the middle, pale grey at the center; not producing pigmentation on PDA.
Material examined: China, Yunnan Province, Xishuangbanna Dai Autonomous Prefecture, Mengla County, Bubeng (21°36′30.13″ N, 101°35′52.54″ E, 664 ± 5 msl), on stems of unidentified grass submerged in a freshwater stream, 27 April 2021, R. Phookamsak, BB21-006 (KUN-HKAS 122242, holotype), ex-type living culture, KUMCC 21-0412, KUMCC 21-0413.
Notes: The NCBI nucleotide BLAST search of ITS sequence indicated that Scolecohyalosporium submersum (KUMCC 21-0412) is similar to “Pleosporales sp. strain 1192” with 96.43% similarity (Identities = 432/448, with 2 gaps), Parabambusicola aquatica strain MFLUCC 18-1140 with 95.58% similarity (Identities = 238/249, with 2 gaps), and Neoaquastroma krabiense strains MFLUCC 16-0419 with 93.65% similarity (Identities = 236/252, with 2 gaps). The nucleotide BLAST search of LSU sequence indicated that the closest hit of S. submersum (KUMCC 21-0412) is Multiseptospora thailandica strain MFLUCC 11-0204 (98.80% similarity, Identities = 823/833, with 1 gap), strain MFLUCC 12-0006 (98.71% similarity, Identities = 839/850, with 2 gaps), and strain MFLUCC 11-0183 (98.56% similarity, Identities = 754/765 with 1 gap). The most similar taxa based on NCBI BLAST search of TEF1-α sequence are M. thailandica strain MFLUCC 12-0006 (96.59% similarity, Identities = 821/850 with no gap), Wettsteinina lacustris isolate AFTOL-ID 1592 (93.44% similarity, Identities = 869/930 with no gap), and “Monodictys sp. strain JO 10 EF45a” (93.20% similarity, Identities = 863/926 with 1 gap). According to Tanaka et al. [1], Wettsteinina lacustris isolate AFTOL-ID 1592 was treated in Lentitheciaceae. Therefore, the isolate was not included in the present phylogenetic analyses. Based on a nucleotide pairwise comparison of ITS, LSU, and TEF1-α sequences, S. submersum differs from M. thailandica (MFLUCC 11-0183, type strain) in 25/419 bp (6%), 11/851 bp (1.3%), 32/895 bp (3.6%), respectively.
4. Discussion
Parabambusicolaceae is a well-studied family based upon the morphological characterization and molecular phylogeny. Even though the family was introduced in 2015 by K. Tanaka and his colleagues to initially accommodate two novel monotypic genera in Massarineae, the number of genera and species in Parabambusicolaceae has continually increased in recent years [4,6,7,8,9,11,12,13]. In this study, we establish the other two novel genera, Neomultiseptospora and Scolecohyalosporium as well as the novel species Parabambusicola hongheensis in Parabambusicolaceae based on multigene phylogeny coupled with morphological differences. Hence, there are 11 genera and 19 phylogenetic species in this family.
Most genera in Parabambusicolaceae were introduced as monotypic genera, and many genera are currently accommodated as putative species, including Aquastroma, Multilocularia, Neomultiseptospora, Paramonodictys, Paratrimmatostroma, Pseudomonodictys, and Scolecohyalosporium. Aquastroma, Multilocularia, Neomultiseptospora, and Scolecohyalosporium are only represented by the sexual morph, while Paramonodictys, Paratrimmatostroma, and Pseudomonodictys are represented by the hyphomycetous asexual morph. Due to these genera being represented by a single species, the phylogenetic relationships with the related genera are sometimes not well-resolved. This may be because of a few genetic information and taxon sampling.
Multigene phylogenetic analyses demonstrated that Lonicericola, Multilocularia, Neoaquastroma, Neomultiseptospora, Parabambusicola, Paramonodictys, Paratrimmatostroma, and Scolecohyalosporium form well-resolved subclade (up to 70% ML and 0.95 PP support; Figure 2) in this study. However, the phylogenetic relationships among Lonicericola, Paratrimmatostroma, and Pseudomonodictys are not well-resolved. These three genera clade together with low supported values in this study that concurred with Hyde et al. [7]. Paratrimmatostroma (trimmatostroma-like), and Pseudomonodictys (monodictys-like) are represented by their hyphomycetous asexual morph, whereas Lonicericola is represented by massarina-like sexual morph. Hence, the morphological features of Lonicericola cannot be compared with Paratrimmatostroma, and Pseudomonodictys, while Paratrimmatostroma, and Pseudomonodictys have morphological differences. Moreover, Aquastroma (sexual morph) also clusters with Paramonodictys (asexual morph), and “Monodictys sp. JO 10” with low support. Therefore, more described holomorph species are necessary in these genera to support their generic status.
Multiseptospora was introduced as a monotypic genus by Liu et al. [10] to accommodate M. thailandica, isolated from the dead stem of grass (Thysanolaena maxima) in Thailand. Subsequently, Li et al. [8] introduced the second species, M. thysanolaenae, to the genus. Multiseptospora thysanolaenae morphological resembles the type species M. thailandica but differs in forming brown ascospores [8]. Phylogenetic relationships between M. thysanolaenae and M. thailandica were not well-resolved in Li et al. [8] and the species always formed a separated branch basal to Parabambusicola in subsequent studies [4,6,7,9,11]. Due to most sexual genera in Parabambusicolaceae having similar morphology, M. thysanolaenae may not be congeneric with M. thailandica based on phylogenetic evidence. However, the genetic information of M. thysanolaenae is not well-verified (R. Phookamsak, pers. comm.). The ex-type strain of this species needs to be re-sequenced for further evaluating its generic status.
Most sexual genera in Parabambusicolaceae have similar morphology of the ascospores. However, these genera can be distinguished from each other based on the other morphological features such as the shape of ascomata, peridial structure, host occurrences, and habitats, and the affinities of these genera were also supported by phylogenetic evidence. We thus provide the keys to genera in Parabambusicolaceae for better clarifying the generic resolution.
Key to genera of Parabambusicolaceae
1. Asexual morph ................................................................................................................ 2
1. Sexual morph ................................................................................................................... 5
2. Hyphomycetous asexual morph ................................................................................... 3
2. Coelomycetous asexual morph, the sexual morph having obovoid to clavate asci, occurred on dicots .................................................................................. Neoaquastroma
3. Monodictys-like ............................................................................................................... 4
3. Trimmatostroma-like, with varied in shape, brown conidia, occurred on a ferns ........................................................................................................ Paratrimmatostroma
4. Conidia globose or subglobose, with multi cells, arising from subcylindrical or truncated-cone-form stroma, lacking conidiophores ............................... Paramonodictys
4. Conidia acropleurogenous, subglobose to ellipsoidal, with 1–2 cells, irregular in shapes with several cells, arising from semi-macronematous to macronematous conidiophores ...................................................................................... Pseudomonodictys
5. Ascospores phragmosporous, fusiform to ellipsoidal or vermiform ..................... 6
5. Ascospores scolecosporous, filiform ........................................ Scolecohyalosporium
6. Ascomata solitary to gregarious, uni-loculate ............................................................ 7
6. Ascomata immersed in the stroma, multi-loculate, clavate asci with short to long pedicel and hyaline, ellipsoidal, (1–)3(–4)-septate ascospores ........... Multilocularia
7. Ascomata globose to subglobose .................................................................................. 8
7. Ascomata subconical to hemispherical ...................................................................... 10
8. Asci broadly cylindrical to cylindric-clavate, with subsessile to short pedicel ...... 9
8. Asci clavate, with short pedicel, clavate to fusiform, 6–8-septate ascospores, occurred on submerged twigs of dicot .......................................................... Aquastroma
9. Ascomata glabrous, with minute papillate, (8–)9-septate ascospores, occurred on dicots .............................................................................................................. Lonicericola
9. Ascomata setose, covered by tufts of hyphae, apapillate, 10–11-septate ascospores, occurred on monocots ......................................................................... Multiseptospora
10. Ascomata hemispherical to subconical, poorly-developed at the base, fusiform to ellipsoidal, or oblong ascospores, with rounded end cells, slightly constricted at the central septum, less constricted at the other septa .................. Neomultiseptospora
10. Ascomata subglobose to hemispherical or subconical, fusiform to vermiform ascospores with acute ends, frequently not constricted at the septa, covered by a thin entire sheath ....................................................................................... Parabambusiccola
Acknowledgments
The authors express their sincere thanks to the Biology Experimental Center, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences for providing the facilities of the molecular laboratory. Shaun Pennycook at Manaaki Whenua—Landcare Research, New Zealand, is thanked for assistance in naming the novel taxa in Parabambusicolaceae. Chunfang Liao at School of Science, Mae Fah Luang University, Thailand, is thanked for assisting in fungal isolation. Austin G. Smith at World Agroforestry (ICRAF), Kunming Institute of Botany, China, is thanked for editing English grammar. Jian-Bo Yang and Sheng Xu at Honghe Center for Mountain Futures, Kunming Institute of Botany, Chinese Academy of Sciences, Honghe County, China are thanked for providing the distribution map of genera in Parabambusicolaceae. H.-B.J. would like to thank Mae Fah Luang University, Thailand for his Ph.D. scholarship. Chiang Mai University, Thailand is thanked for partial research financial support. We also thank Qi Sun at Central Research Facilities, College of Life Sciences and Oceanography, Shenzhen University for general assistance.
Author Contributions
Conceptualization, N.X., R.P., H.J. and S.H.; methodology, N.X., R.P., H.J. and S.H.; resources, R.P. and H.J.; writing—original draft preparation, N.X., R.P. and H.J.; writing—review and editing, N.X., R.P., Y.-J.Z., H.Z. and S.H.; supervision, S.L., J.X., F.X. and S.H.; funding acquisition, N.X., H.Z., S.L. and J.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Key R&D Program of China (2021YFA0910800), Basic and Applied Basic Research Fund of Guangdong Province (2121A1515012166), Stability Support project for Universities in Shenzhen (20200812173625001), and Project of DEGP (2019KTSCX150), the National Natural Foundation of China (grant no. 31861143002), the Department of Science and Technology of Yunnan, China (grant no. 202101AS070045).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data availability was mentioned in the manuscript. The novel taxa were registered in Index Fungorum (http://www.indexfungorum.org/Names/Names.asp, accessed on 18 December 2021) including Index Fungorum numbers IF 559389, IF 559390, IF 559391, IF 559392 and IF 559393. Final alignment and phylogenetic tree were deposited in TreeBase (https://www.treebase.org/ (accessed on 25 November 2021) with submission ID: 29072) and the newly generated sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/submit/, accessed on 25 November 2021) followed as ITS: OL898884, OL898885, OL898880, OL898883, OL898881, OL898882; LSU: OL898925, OL898926, OL898921, OL898924, OL898922, OL898923; SSU: OL898890, OL898891, OL898886, OL898889, OL898887, OL898888; TEF1-α: OL964282, OL964283, OL964281, OL964279, OL964280.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tanaka K., Hirayama K., Yonezawa H., Sato G., Toriyabe A., Kudo H., Hashimoto A., Matsumura M., Harada Y., Kurihara Y., et al. Revision of the massarineae (Pleosporales, Dothideomycetes) Stud. Mycol. 2015;82:75–136. doi: 10.1016/j.simyco.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wijayawardene N.N., Hyde K.D., Al-Ani L.K.T., Tedersoo L., Haelewaters D., Kunhiraman C., Rajeshkumar K.C., Aptroot A., Leontyev V.D., Saxena R.K., et al. Outline of Fungi and fungi-like taxa. Mycosphere. 2020;11:1060–1456. doi: 10.5943/mycosphere/11/1/8. [DOI] [Google Scholar]
- 3.Hongsanan S., Hyde K.D., Phookamsak R., Wanasinghe D.N., McKenzie E.H.C., Sarma V.V., Boonmee S., Lücking R., Bhat D.J., Liu N.G., et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere. 2020;11:1553–2107. doi: 10.5943/mycosphere/11/1/13. [DOI] [Google Scholar]
- 4.Phukhamsakda C., Bhat D.J., Hongsanan S., Xu J.C., Stadler M., Hyde K.D. Two novel species of (Parabambusicolaceae, Pleosporales) with their phoma-like asexual morphs. MycoKeys. 2018;34:47–62. doi: 10.3897/mycokeys.34.25124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ariyawansa H.A., Hyde K.D., Jayasiri S.C., Buyck B., Kandawatte W.T.C., Cui Y.Y., Dai D.Q., Dai Y.C., Daranagama D.A., Jayawardena R., et al. Fungal diversity notes 111–252: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015;75:27–274. doi: 10.1007/s13225-015-0346-5. [DOI] [Google Scholar]
- 6.Phookamsak R., Hyde K.D., Jeewon R., Bhat D.J., Jones E.B.G., Maharachchikumbura S.S.N., Raspé O., Karunarathna S.C., Wanasinghe D., Hongsanan S., et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019;95:1–273. doi: 10.1007/s13225-019-00421-w. [DOI] [Google Scholar]
- 7.Hyde K.D., Dong Y., Phookamsak R., Jeewon R., Bhat D.J., Jones E.B.G., Liu N.-G., Abeywickrama P.D., Mapook A., Wei D., et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020;100:5–277. doi: 10.1007/s13225-020-00439-5. [DOI] [Google Scholar]
- 8.Li G.J., Hyde K.D., Zhao R.L., Hongsanan S., Abdel-Aziz F.A., Abdel-Wahab M.A., Alvarado P., Alves-Silva G., Ammirati J.F., Ariyawansa H.A., et al. Fungal diversity notes 253–366: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;78:1–237. doi: 10.1007/s13225-016-0366-9. [DOI] [Google Scholar]
- 9.Wanasinghe D.N., Hyde K.D., Konta S., To-Anun C., Jones E.B.G. Saprobic Dothideomycetes in Thailand: Neoaquastroma gen. nov. (Parabambusicolaceae) introduced based on morphological and molecular data. Phytotaxa. 2017;302:133–144. doi: 10.11646/phytotaxa.302.2.3. [DOI] [Google Scholar]
- 10.Liu J.K., Hyde K.D., Jones E.B.G., Ariyawansa H.A., Bhat D.J., Boonmee S., Maharachchikumbura S.S.N., McKenzie E.H.C., Phookamsak R., Phukhamsakda C., et al. Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015;72:1–197. doi: 10.1007/s13225-015-0324-y. [DOI] [Google Scholar]
- 11.Samarakoon M.C., Wanasinghe D.N., Liu J.K., Hyde K.D., Promputtha I. The genus Neoaquastroma is widely distributed; a taxonomic novelty, N. cylindricum sp. nov. (Parabambusicolaceae, Pleosporales) from Guizhou, China. AJOM. 2019;2:235–244. doi: 10.5943/ajom/2/1/14. [DOI] [Google Scholar]
- 12.Dong W., Wang B., Hyde K.D., McKenzie E.H.C., Raja H.A., Tanaka K., Abdel-Wahab M.A., Doilom M., Phookamsak R., Hongsanan S., et al. Freshwater Dothideomycetes. Fungal Divers. 2020;105:319–575. doi: 10.1007/s13225-020-00463-5. [DOI] [Google Scholar]
- 13.Yasanthika E., Dissanayake L.S., Wanasinghe D.N., Karunarathna S.C., Mortimer P.E., Samarakoon B.C., Monkai J., Hyde K.D. Lonicericola fuyuanensis (Parabambusicolaceae) a new terrestrial pleosporalean ascomycete from Yunnan Province, China. Phytotaxa. 2020;446:103–113. doi: 10.11646/phytotaxa.446.2.3. [DOI] [Google Scholar]
- 14.Senanayake I.C., Rathnayaka A.R., Marasinghe D.S., Calabon M.S., Gentekaki E., Lee H.B., Xiang M.M., Hurdeal V.G., Pem D., Dissanayake L.S., et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere. 2020;11:2678–2754. doi: 10.5943/mycosphere/11/1/20. [DOI] [Google Scholar]
- 15.Index Fungorum. [(accessed on 18 December 2021)]. Available online: http://www.indexfungorum.org/names/IndexFungorumRegisterName.asp.
- 16.White T.J., Bruns T., Lee S., Taylor J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego, CA, USA: 1990. pp. 315–322. [Google Scholar]
- 17.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehner S. Primers for Elongation Factor 1-Alpha (EF1-alpha) 2001. [(accessed on 15 December 2020)]. Available online: http://ocid.NACSE.ORG/research/deephyphae/EF1primer.pdf.
- 19.Jiang H.B., Phookamsak R., Hyde K.D., Mortimer P.E., Xu J.C., Kakumyan P., Karunarathna S.C., Kumla J.A. A taxonomic appraisal of bambusicolous fungi in Occultibambusaceae (Pleosporales, Dothideomycetes) with new collections from Yunnan Province, China. Life. 2021;11:932. doi: 10.3390/life11090932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh K., Rozewicki J., Yamada K.D. Mafft online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall T. Bioedit Version 6.0.7. [(accessed on 1 March 2021)]. Available online: http://www.mbio.ncsu.edu/bioedit/bioedit.html.
- 22.Miller M.A., Pfeiffer W., Schwartz T. Creating the cipres science gateway for inference of large phylogenetic trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; New Orleans, LA, USA: IEEE; pp. 1–8. [DOI] [Google Scholar]
- 23.Phookamsak R., Manamgoda D.S., Li W.J., Dai D.Q., Singtripop C., Hyde K.D. Poaceascoma helicoides gen et sp. nov., a new genus with scolecospores in Lentitheciaceae. Cryptogam Mycol. 2015;36:225–236. doi: 10.7872/crym/v36.iss2.2015.225. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data availability was mentioned in the manuscript. The novel taxa were registered in Index Fungorum (http://www.indexfungorum.org/Names/Names.asp, accessed on 18 December 2021) including Index Fungorum numbers IF 559389, IF 559390, IF 559391, IF 559392 and IF 559393. Final alignment and phylogenetic tree were deposited in TreeBase (https://www.treebase.org/ (accessed on 25 November 2021) with submission ID: 29072) and the newly generated sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/submit/, accessed on 25 November 2021) followed as ITS: OL898884, OL898885, OL898880, OL898883, OL898881, OL898882; LSU: OL898925, OL898926, OL898921, OL898924, OL898922, OL898923; SSU: OL898890, OL898891, OL898886, OL898889, OL898887, OL898888; TEF1-α: OL964282, OL964283, OL964281, OL964279, OL964280.