Abstract
Clostridioides difficile is the causative agent of antibiotic-associated diarrhea and is the leading cause of nosocomial infection in developed countries. An increasing number of C. difficile infections are attributed to epidemic strains that produce more toxins and spores. C. difficile spores are the major factor for the transmission and persistence of the organism. Previous studies have identified global regulators that influence sporulation in C. difficile. This study discovers that PdcB, a phosphodiesterase, enhances sporulation in C. difficile strain UK1. Through genetic and biochemical assays, we show that phase-variable expression of pdcB results in hypo- and hyper-sporulation phenotypes. In the “ON” orientation, the identified promotor is in the right orientation to drive the expression of pdcB. Production of the PdcB phosphodiesterase reduces the intracellular cyclic-di-GMP concentration, resulting in a hyper-sporulation phenotype. Loss of PdcB due to the pdcB promoter being in the OFF orientation or mutation of pdcB results in increased cyclic-di-GMP levels and a hypo-sporulation phenotype. Additionally, we demonstrate that CodY binds to the upstream region of pdcB. DNA inversion reorients the CodY binding site so that in the OFF orientation, CodY binds a site that is upstream of the pdcB promoter and can further repress gene expression.
Keywords: Clostridioides difficile, C. difficile, phase variation, gene regulation
Graphic abstract.
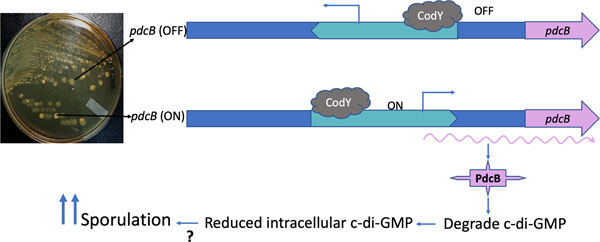
In UK_T translucent cells (pdcB-OFF), the predicted promoter region of pdcB is in the inverted orientation. The binding of CodY downstream further blocks any transcription of the pdcB. In the UK_O opaque cells (pdcB-ON), because of the DNA inversion, the predicted promoter is in the right orientation and drives the expression of pdcB, and is also not affected by the CodY binding to the upstream. Increased production of PdcB results in decreased c-di-GMP, which in turn influences the sporulation rate.
Abbreviated Summary
In the C. difficile UK1 strain, phase variation resulted in two distinct colony morphologies: transparent UK T cells and opaque UK O cells. Whole-genome sequencing of UK_T and UK_O revealed that they differ in the orientation of the DNA sequence upstream of pdcB, which codes for a phosphodiesterase. In UK_T cells, the predicted promoter region of pdcB is in the inverted orientation. The binding of CodY downstream further blocks any transcription of pdcB. In UK O cells, the predicted promoter is in an orientation that promotes the expression of pdcB and is unaffected by CodY binding to the upstream region. Increased production of PdcB results in decreased c-di-GMP, which in turn influences sporulation through a mechanism yet to be discovered.
Introduction
Phase variation is a phenomenon where two distinct phenotypes are established in a reversible manner within a clonal bacterial population. This phenomenon in bacteria long recognized and has been well studied in various bacterial pathogens. Heterogeneity within a bacterial population is readily visible as colony variation and is often associated with pathogenicity (Silverman and Simon, 1980; Jiang et al., 2019; Li and Zhang, 2019; Reyes Ruiz et al., 2020). Genes that are regulated by phase variation are often involved in pathogenesis; these include genes encoding fimbriae, flagella, cell surface protein, and those involved in the synthesis of capsular polysaccharides. These variable bacterial components are responsible for establishing interactions with the host, giving the bacteria an advantage against the challenging environment of the host, aiding in colonization, establishing an infection, and/or evading from host immune response (Li and Zhang, 2019; Reyes Ruiz et al., 2020). Phase variation has been reported in several Gram-negative bacterial pathogens, including Salmonella (Silverman et al., 1979; Kutsukake and Iino, 1980a; Silverman and Simon, 1980; Kutsukake and Iino, 1980b; Scott and Simon, 1982; Smith and Selander, 1990; Salehi et al., 2017), Escherichia coli (Klemm, 1986; Eisenstein, 1988; McClain et al., 1991; McClain et al., 1993), Klebsiella (Struve et al., 2008), Neisseria (van der Ende et al., 2000; Tauseef et al., 2013) and Campylobacter jejuni (Bacon et al., 2001; Holst Sørensen et al., 2012) where it has been employed to evade host immune system, increase adhesion, colonization, and virulence. While there are ample reports on phase-variable genes and their role in the pathogenesis of Gram-negative bacteria, little is known about the role of phase variation in Gram-positive pathogens. Streptococcus pneumoniae utilizes phase variation to switch colony morphology and to express pili (Li and Zhang, 2019). Phase variation has been reported more recently in C. difficile, where two distinct colony morphologies: smooth- circular and rough-filamentous, were observed and were found to be associated with differences in motility and cell shapes (Anjuwon-Foster and Tamayo, 2017; Garrett et al., 2019). In this study, we investigated the molecular mechanisms involved in the phase-variable sporulating (UK1_O) and non-sporulating (UK1_T) colonies observed in the C. difficile UK1 strain.
C. difficile is an anaerobic pathogen that lives in the mammalian GI tract (Edwards and McBride, 2014). C. difficile’s growth in the GI tract is kept in check by the commensal gut microbiota. But during antibiotic treatment, a healthy bacterial population in the gut is disrupted, thereby creating an environment that is favorable for C. difficile to proliferate and cause infection (Edwards and McBride, 2014). C. difficile infection is caused primarily by the toxin produced by the bacteria, which causes inflammation of gut epithelium leading to severe diarrhea, commonly known as antibiotic-associated diarrhea, and ultimately causing fatal pseudomembranous colitis Mullish and Williams, 2018; Prete et al., 2019). Each year >200,000 C. difficile infections (CDI) and >12,000 deaths due to CDI are reported in the United States (CDC, 2020). Due to the increased number of infections and a massive cost of health care of ~ 4.2 billion dollars, CDI has been categorized by the CDC as an urgent threat to public health (Lessa et al., 2015). The increasing number of C. difficile infections has been partly attributed to the emergence of epidemic C. difficile strains that produce more toxins and spores (Denève et al., 2009). C. difficile infections often occur as a result of spore transmission. Ingested spores make their way to the intestine, where they germinate with the help of bile salts into active vegetative cells that secrete cytotoxins, causing tissue damage and inflammation (Paredes-Sabja et al., 2014). Since they are the root source of transmission, it is important to understand the regulation of sporulation in C. difficile. In this study, by characterizing the phase variation involving sporulating phenotypes, we have uncovered a new link between intracellular cyclic-di-GMP (c-di-GMP) concentrations and sporulation.
C-di-GMP is a common bacterial second messenger found in many bacterial species. Two classes of enzymes are important for c-di-GMP homeostasis in a bacterial cell, diguanylate cyclases (DGCs) that synthesize c-di-GMP from 2 GTP molecules and phosphodiesterases (PDEs) that degrade c-di-GMP. In C. difficile, c-di-GMP has been shown to negatively regulate toxin production (McKee et al., 2013; Bordeleau and Burrus, 2015; Purcell et al., 2017; Hall and Lee, 2018; McKee et al., 2018b) and flagella-mediated swimming motility (Purcell et al., 2012; Bordeleau and Burrus, 2015; Hall and Lee, 2018; McKee et al., 2018b), whereas c-di-GMP positively regulates PilA-mediated swarming motility (Bordeleau et al., 2015; Bordeleau and Burrus, 2015; Purcell et al., 2016; Hall and Lee, 2018; McKee et al., 2018b; McKee et al., 2018a), biofilm formation (Purcell et al., 2017; McKee et al., 2018b), cell aggregation (Purcell et al., 2012; Bordeleau et al., 2015) and expression of cell wall anchor structures (Peltier et al., 2015; Arato et al., 2019). While c-di-GMP regulates a plethora of physiological processes, its influence on sporulation has not been investigated. In this study, we discovered a link between low intracellular c-di-GMP levels and a hyper-sporulation phenotype in C. difficile UK1. We find that decreasing c-di-GMP levels by over-expressing pdcB, which encodes a phosphodiesterase, increases sporulation. We also show that a pdcB mutant is hypo-sporulating and overexpression of the pdcB-EAL domain complements the sporulation phenotype. Our data shows that pdcB is expressed during the mid-exponential growth phase and a DNA inversion controls its expression. We locate CodY binding sites near the promoter region of pcdB gene and show that a DNA inversion re-orients the predicted promoter and moves the CodY binding site to its upstream of the promoter. The rearrangement results in the expression of pdcB, which in turn positively influences the sporulation in C. difficile UK1 strain.
Results
C. difficile UK1 strain exhibits phenotypic heterogeneity with two distinguishable colony morphologies
Earlier studies have shown that C. difficile strain R20291 belonging to ribotype 027, associated with epidemic infections, have phenotypic heterogeneity, thereby exhibiting two distinct colony morphologies in vitro (Anjuwon-Foster and Tamayo, 2017; Garrett et al., 2019). Consistent with these studies, we observed two distinct colony types in the UK1 strain during routine plating on TY agar (Fig. 1A). One colony type has a smooth edge and is round, and is termed “opaque” (UK1_O), while the other is irregular with a rough edge and is termed as “translucent” (UK1_T). Under the phase-contrast microscope (Fig.1B), UK1_T cells were longer with an average length of 9.8 µm while the UK1_O cells are significantly shorter with an average length of 4.9 µm (Fig. 1C). To measure the distribution of UK_O and UK_T in a population, we prepared spores from the UK_O colonies and plated them on TY agar. We did not use any germinant in the TY agar plates, since we did not know the effect of germinants on phase variation. We observed that nearly 78% of the colonies appeared after overnight (~20h) incubation were UK_O phenotype, while the remainder were UK_T type colonies. To check whether each phenotype is heritable, we isolated UK1_T and UK1_O colonies and streaked them on TY agar plates. When a single opaque colony was streaked in TY agar and incubated overnight, 100% of the colonies formed were opaque in nature. Upon prolonged incubation (> 72h) however, nearly 70% of the opaque colonies in the TY plate started exhibiting rough edges with the pale yellowish center over time suggesting that opaque colonies could give rise to translucent colonies. When a single UK1_T colony was streaked on to TY agar and incubated overnight, the majority of the colonies remained translucent, but about 2% of the colonies were opaque (Fig. S1A). Nearly 20% of the UK_T colonies gave rise to UK_O colonies after 72h of incubation. Similar observations were made with the broth cultures. Cells from opaque colonies grown in TY broth overnight stayed shorter, but cells from translucent colonies grown in TY broth overnight (~16–20h) produced primarily long cells with some short cells. Serial dilutions made from broth cultures were plated on TY agar to check for colony morphologies upon overnight incubation. UK_T broth cultures produced ~7% opaque colonies, while UK_O broth culture resulted in 100% opaque colonies (Fig. S1B). After 72h, UK_T broth cultures gave rise to ~70% opaque colonies and UK_O broth culture produced nearly 75% opaque colonies. These results together suggest that the phase variation phenotypes are heritable and are stable at least for 20 hours. It also suggests that bacteria in broth culture favor opaque morphology over the translucent phenotype.
Figure 1. Colony and cell morphologies of UK1_T and UK1_O strains.
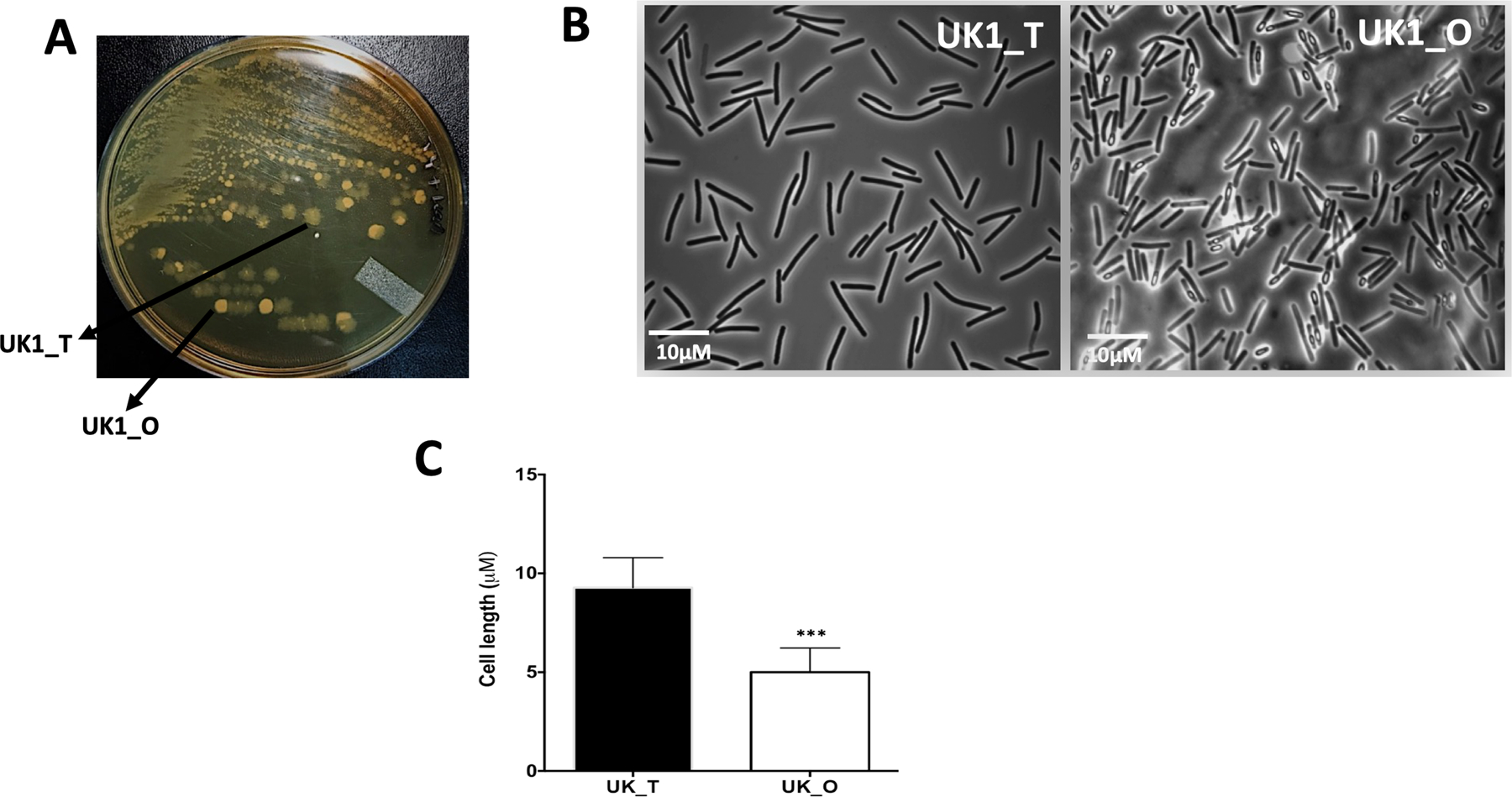
A. UK1 strain when grown in TY medium gives rise to phenotypically distinguishable Translucent (UK1_T) and Opaque (UK1_O) colony morphotypes. B. Phase-contrast microscopic images of UK1_T and UK1_O colony morphotypes. C. Average cell length showing UK1_T cells are significantly longer than UK1_O cells. Twenty-five individual cells per image were measured using ImageJ software. At least four images from two replicates were used (n=100). Data was analyzed using unpaired t-test with Welch’s correction where *** indicates p<0.005.
UK1_T and UK1_O colony morphologies exhibit distinct sporulation and surface motility phenotypes
In order to determine the physiological characteristics of the two colony types, we assayed UK1_T and UK1_O colony types for sporulation, toxin, and motility. To determine the sporulation phenotype, we streaked the isolated UK1_T and UK1_O colonies on solid TY medium, and we found that UK1_O colonies produced significantly more spores compared to UK1_T at 20 h (Fig. 1B, Fig. 2A). We also found that UK1_T and UK1_O produce similar level of cytoplasmic toxins at 16 h of growth (Fig. S2B). While UK1_T and UK1_O exhibited similar swimming motility even after 72h in 0.5% agar TY medium (Fig. S1A), we found that UK1_T exhibited enhanced surface motility compared to the UK1_O when grown in TY medium with 1.8% agar at 72h (Fig. 2B). Similarly, UK1_T formed thick biofilms compared to UK1_O (Fig. 2C). Together, these data demonstrate that UK1_T and UK1_O are phenotypically different in sporulation, swarming motility, and biofilm formation. While the swarming motility and the biofilm formations were previously reported to be associated with phase variation in C. difficile, sporulation was not (Garrett et al., 2019).
Figure 2. Phenotypic characterization of UK1_T and UK1_O strains.
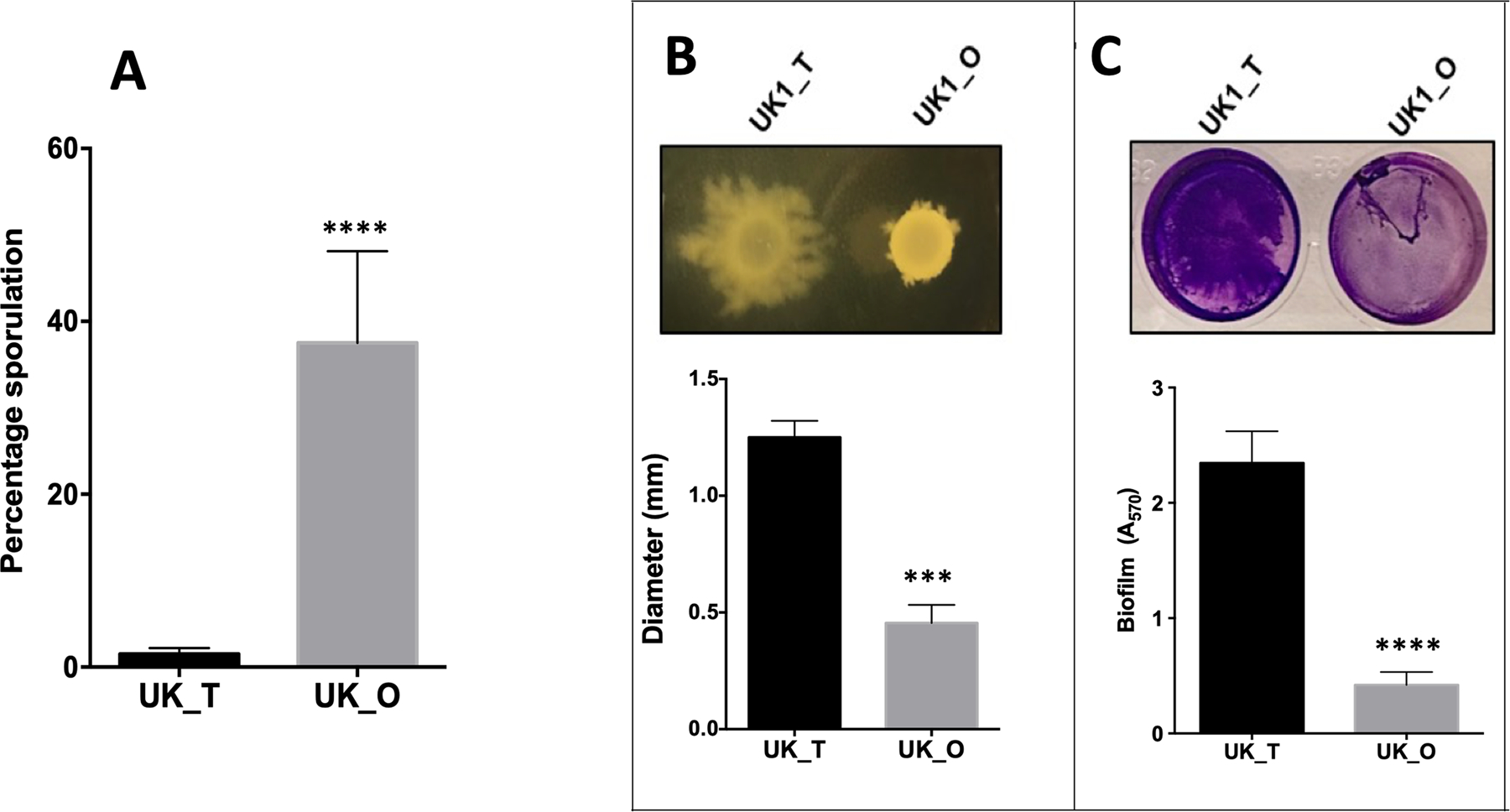
A. Percentage sporulation of UK1_T and UK1_O cells showing hyper sporulation phenotype in UK1_O morphotype. B. Swarming motility exhibited by UK1_T and UK1_O in TY+1.8% agar. C. Biofilm formation of UK1_T and UK1_O morphotypes. OD570 nm was measured to quantify biofilm formation. Three biological replicates were used to carry out the experiments, and data were analyzed by two-tailed unpaired t-test with Welch’s correction where **** indicates p<0.0005 and *** is p<0.005
UK1_O strain undergoes phase variation by DNA inversion
To determine the genetic basis for the phenotypic heterogeneity that gave rise to translucent and opaque colonies in the UK1 strain, we extracted genomic DNA from UK1_T and UK1_O cells harvested from the overnight cultures grown in TY agar (supplementary methods) and performed whole-genome sequencing at Tufts University Core Facility Genomic core. The Sequenced genome was assembled against the closely related and annotated R20291 genome (Stabler et al., 2009). Contrary to our expectation, the sequencing result did not show mutations in any of the known sporulation-associated genes. However, the intergenic region between CDR20291_0684 and CDR20291_0685 genes was listed among unassigned new junction evidence.
The intergenic region between CDR20291_0684 and CDR20291_0685 carried inverted repeat sequences that are known to undergo inversion (Sekulovic et al., 2018). To determine the orientation of this sequence in UK_T and UK_O, we amplified and cloned ~1.5 kb intergenic region in a cloning vector and sent 10 clones from each for sequencing. We aligned the sequences of UK1_T and UK_O to the R20291 genome (Stabler et al., 2009) and analyzed with NCBI BLASTn. The results suggested that all clones from UK1_T perfectly aligned with the published sequence of the R20291. In contrast, all the clones from UK1_O had mismatches in the ~200 bp segment of the intergenic region (Fig. S3), showing DNA inversion in the region.
DNA inversion regulates the expression of the downstream gene CDR20291_0685
DNA inversion is known to regulate the expression of the downstream gene as it gives rise to two orientations where the “ON” orientation enhances, and the “OFF” orientation represses the expression of the downstream gene or operon (Silverman and Simon, 1980; Zieg and Simon, 1980; Van de Putte and Goosen, 1992; Jiang et al., 2019). We performed a reporter fusion assay to determine the effect of DNA inversion on the expression of the downstream CDR20291_0685 gene. We fused ~1.5 kb of the upstream DNA segment of CDR20291_0685, containing the invertible region from both the UK1_T and UK1_O cells, with a gusA reporter gene coding beta-glucuronidase, to create plasmid constructs pBA042 and pBA045 (Table S1). Using site-directed mutagenesis, we mutated the inverted repeat sequences that flanked the invertible region to lock the upstream region either in the translucent (pBA043) or opaque (pBA046) orientation. The plasmid constructs were then introduced into the UK1 undifferentiated parent strain by conjugation. The strains were grown anaerobically in TY broth medium at 37°C and were harvested at different time points to perform the reporter gene expression assay. We observed significantly higher beta-glucuronidase activity at each time point with the reporter fusion that was locked in opaque orientation (Fig. 3A). Very minimal reporter activity was recorded with the reporter fusion that was locked in the translucent orientation. This observation is consistent with our qRT-PCR results, where we detected elevated levels of CDR20291_0685 transcript in UK1_O when compared to that from UK1_T (Fig. 3B). Taken together, these results suggest that DNA inversion in the upstream region of the CDR20291_0685 gene resulted in upregulation of its expression in the opaque UK1_O colonies. CDR20291_0685 is the orthologous gene of CD630_07570 in strain CD630. CD630_07570 hydrolyzes c-di-GMP to pGpG in vitro and therefore is a phosphodiesterase (Bordeleau et al., 2011). Therefore, we named CDR20291_0685 as PdcB (Phosphodiesterase of C. difficile B). To emphasize the pdcB’s ON and OFF orientations, we refer to UK1_O as pdcB (ON) and UK1_T as pdcB (OFF), here after.
Figure 3. DNA inversion upregulates the expression of CDR20291_0685 in UK_O cells.
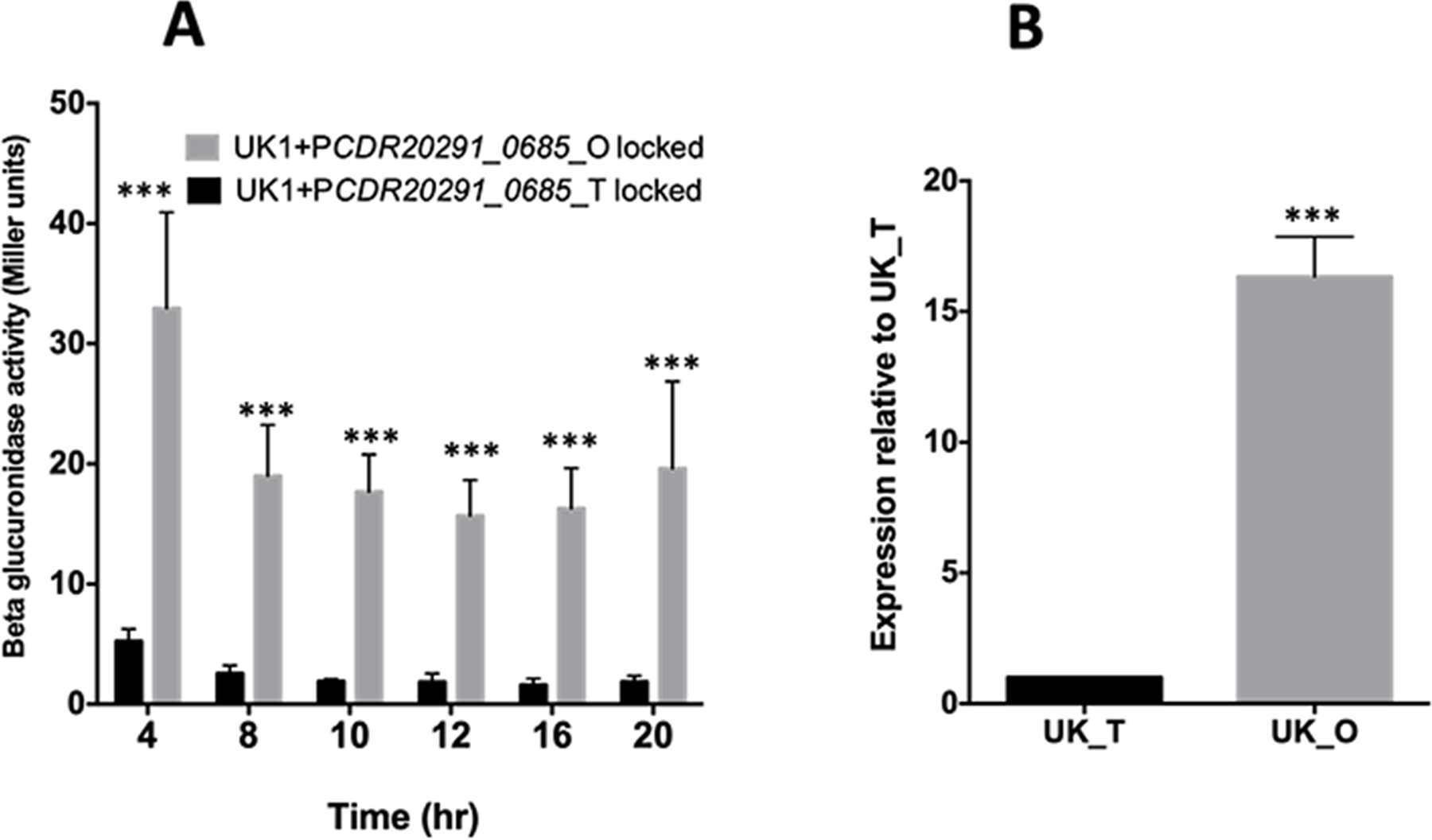
A. The beta-glucuronidase activity of the PCDR20291_0685 upstream-gusA fusions locked in UK1_T and UK1_O orientation in the UK1 parent strain. The data represent the results from three biological replicates. Data were analyzed using 2-way ANOVA (Sidak’s multiple comparisons test) comparing the mean of UK1_T and UK1_O at each time point. *** indicates p<0.005. B. qRT-PCR results showing overexpression of CDR20291_0685 in UK1_O strain at 16h time point. The representative results from three independent experiments are shown. Data were analyzed by a two-tailed unpaired t-test with Welch’s correction where *** is p<0.005.
Promoter of pdcB lies within the invertible region
The mechanism by which DNA inversion regulates the expression of the gene depends upon its location. If DNA inversion occurs in the region that contains the promoter, it will disrupt the promoter, and the gene is not expressed (Zieg and Simon, 1980; Eisenstein, 1988). If the promoter is upstream of the switch, then an intrinsic terminator can be formed to terminate the transcription of the gene (Sekulovic et al., 2018). To determine the mechanism by which this inversion regulates the gene expression, we sought to identify the promoter of pdcB. We hypothesized that the promoter of pdcB is within the invertible region. We extracted total RNA from a 4h culture of pdcB (ON) cells and synthesized pdcB specific cDNA. We carried out PCR using this cDNA as a template, with several primers spanning the inverted repeat sequence (Fig. 4A). Only the primer pairs ORG921/ORG853, and ORG922/ORG853 gave amplification products, suggesting that the promoter region is located around the region spanned by the primer ORG921 (Fig. 4B). Consistent with this observation, we mapped the transcription start of pdcB using 5’RACE. Transcription is initiated 874-bp upstream of the pdcB translational initiation codon and is located next to the right repeat sequence (Fig. 4A). Hexamers −10 (TATTT) and −35 (TTGATA) separated by an −17-bp spacer are found upstream of the transcriptional start site.
Figure 4. The promoter of pdcB is located within the invertible region.
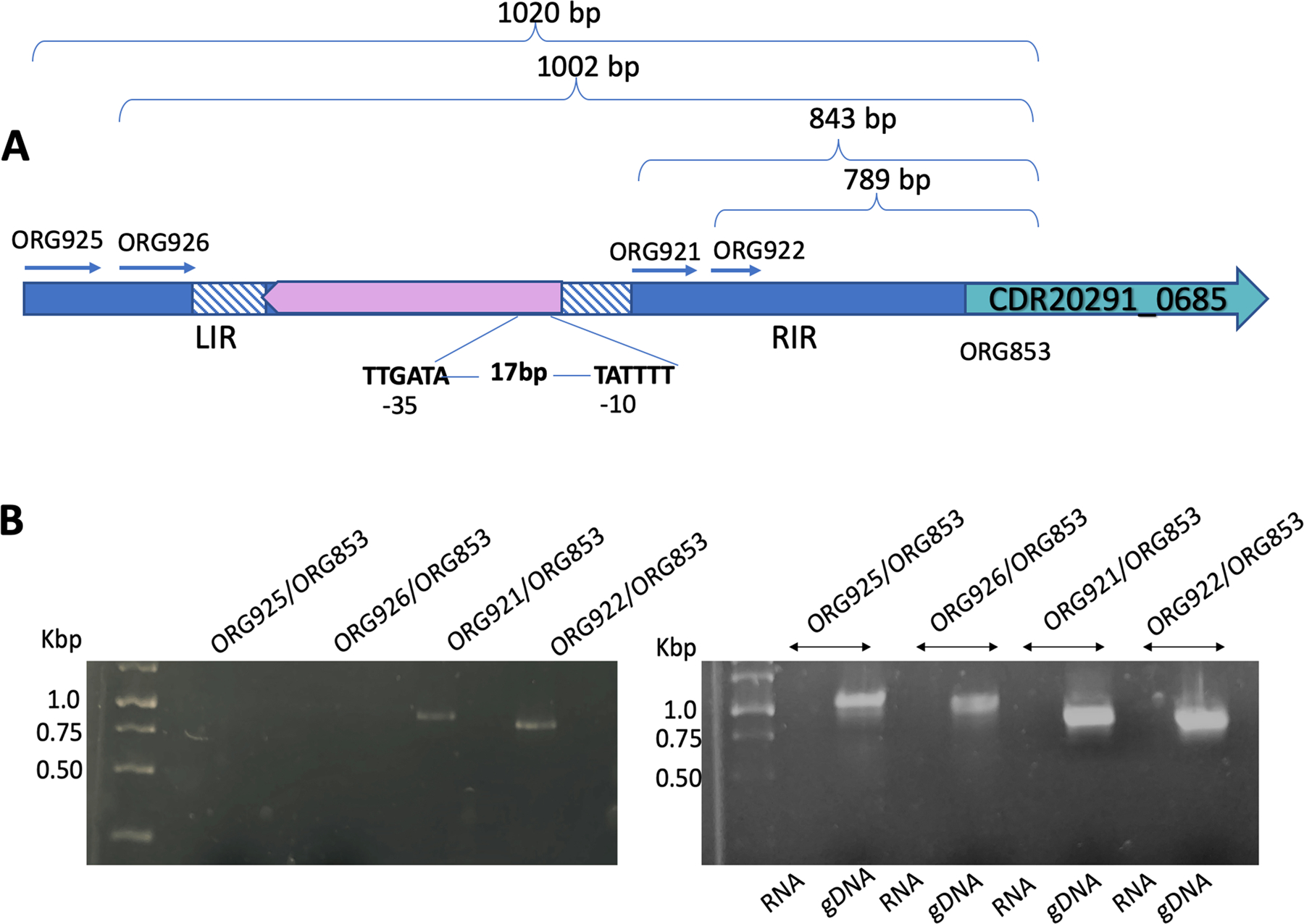
A. Schematic of the upstream region of pdcB from pdcB (ON) cells (the orientation in which pdcB is expressed) depicting the invertible region flanked by the right and left inverted repeats (RIR and LIR). The forward and the reverse primers designed along the length of the invertible region are shown. The transcription initiation site was found near the RIR, and −35 and −10 sites are marked. B. RT PCR detecting the transcripts of pdcB. Genomic DNA (gDNA) and RNA were used as positive and negative controls, respectively. Only the primer pairs ORG921/ORG853, and ORG922/ORG853 gave amplification products of size 843 bp and 789 bp, respectively.
Overexpression of pdcB results in reduced c-di-GMP levels in UK1
To determine if overexpression of pdcB alters the levels of intracellular c-di-GMP in C. difficile, we first created UK1::pdcB mutant strain using ClosTron mutagenesis (Fig. S4) and extracted and measured the levels of intracellular c-di-GMP from pdcB (OFF), pdcB (ON) and UK1::pdcB strains. Our data suggest that pdcB (OFF) and UK1::pdcB strains have higher levels of c-di-GMP while pdcB (ON) cells has a significantly reduced c-di-GMP level (Fig. 5B). This result indicates that DNA inversion leads to overexpression of the pdcB gene, which encodes an enzyme that cleaves c-di-GMP and reduces the global intracellular concentration of c-di-GMP in the pdcB (ON) cells. Similar to pdcB (OFF) cells, UK1::pdcB mutant also produced translucent colonies and longer cells (Fig 5A), suggesting that these phenotypes are directly related to intracellular levels of c-di-GMP.
Figure 5. Intracellular c-di-GMP levels influence sporulation in UK1 strain.
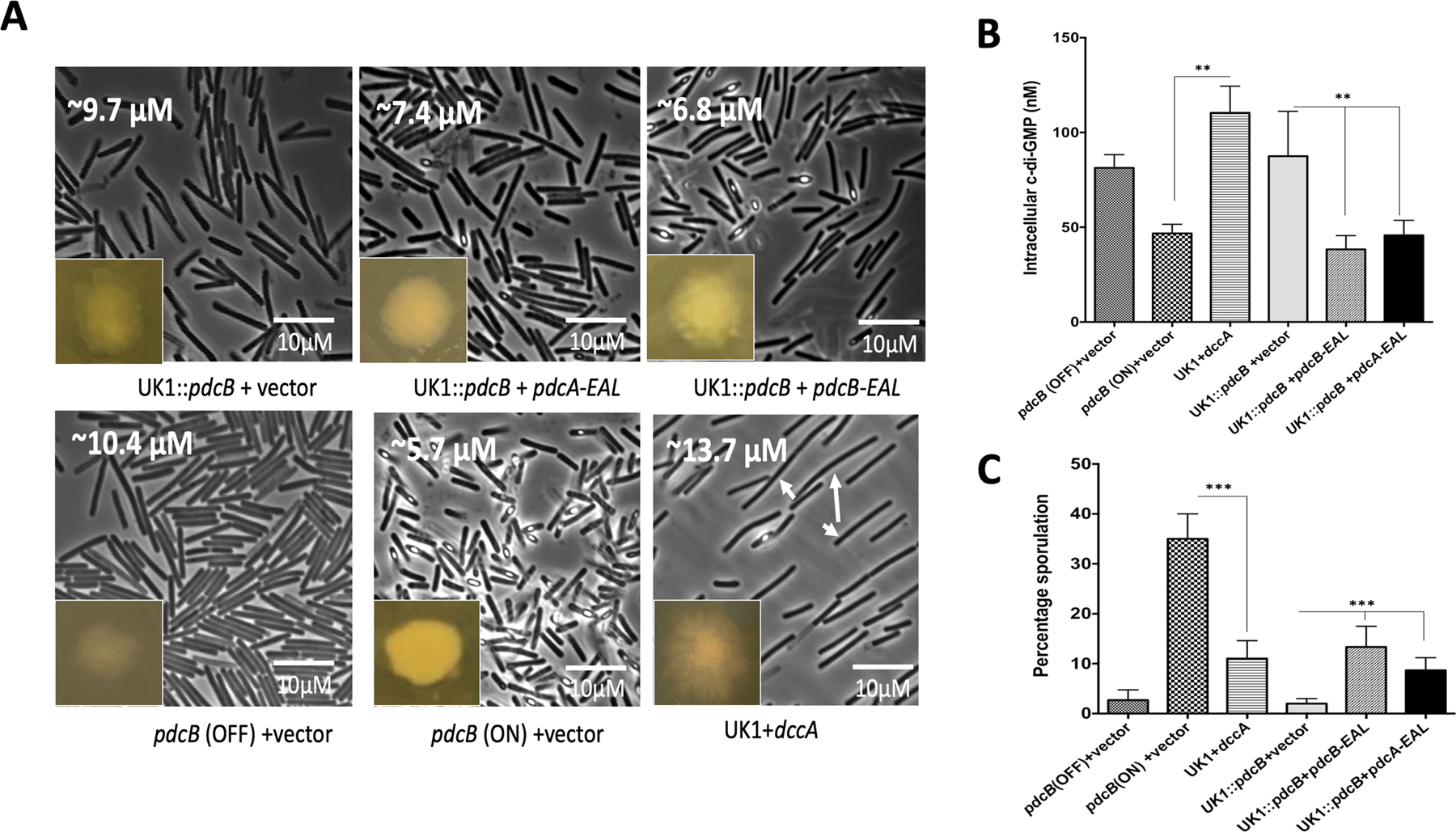
A. Phase-contrast microscopic images along with colony morphology of pdcB (OFF), pdcB (ON), UK1 expressing dccA, UK1::pdcB mutant complemented with pdcB-EAL/ pdcA-EAL. Average cell length is marked at the upper right corner of each image. Using ImageJ software, at least three images from two replicates were used for measuring the cell length (n=100). Long cells appeared upon the induction of dccA were marked with white arrows. The intracellular levels of c-di-GMP (B) and sporulation percentage (C) were measured in these strains. To induce the expression of dccA, pdcB-EAL, pdcA-EAL 100 ngs/ml of ATc was included in the TY agar plates. Three biological replicates were used and data were analyzed by one-way analysis of variance (ANOVA) where ** indicates p<0.05, and *** indicates p<0.005.
Sporulation in C. difficile UK1 is correlated with the intracellular levels of c-di-GMP
Intracellular c-di-GMP homeostasis is maintained by two classes of enzymes, diguanylate cyclases that synthesize c-di-GMP and phosphodiesterases that hydrolyze c-di-GMP. Phosphodiesterases consist of either the EAL domain or an inactive GGDEF and active EAL domain (Hall and Lee, 2018). The PdcB phosphodiesterase contains both the GGDEF and EAL domains, and in vitro assays have shown PdcB can cleave c-di-GMP (Bordeleau et al., 2011). Our results so far showed that DNA inversion upstream of pdcB regulated its expression and hence controlled the intracellular levels of c-di-GMP in C. difficile UK1, which in turn resulted in two phenotypic variants, translucent and opaque colonies. We also observed a significant difference in the frequency of sporulation between the variants. To determine if the intracellular levels of c-di-GMP are associated with a sporulation phenotype, we quantified the percentage sporulation in pdcB (OFF), pdcB (ON), UK1::pdcB mutant, and UK1::pdcB mutant complemented with plasmid-encoded pdcB-EAL under a tetracycline-inducible promoter. Despite multiple attempts, we were not able to complement the UK1::pdcB mutant strain with a full-length plasmid-encoded pdcB. Other studies have shown that complementation with just the EAL domain with the phosphodiesterase activity of pdcA is sufficient to rescue the associated motility phenotypes (Purcell et al., 2017). Since our objective was to determine the role of intracellular levels of c-di-GMP in regulating sporulation in C. difficile UK1 strain, we performed the sporulation assays with the mutant strain complemented with the plasmid that encodes just the PdcB-EAL domain. Percentage sporulation data showed that the hypo-sporulating phenotype in UK1::pdcB strain was partly complemented when the PdcB-EAL was produced in the mutant strain (Fig. 5C). To further confirm that the sporulation phenotype is associated with intracellular levels of c-di-GMP, we complemented the UK1::pdcB mutant with the plasmid-borne PdcA-EAL domain. PdcA is another well-characterized phosphodiesterase of C. difficile, and the conserved GGDEF and EAL domains of PdcB are homologous with those of PdcA (Purcell et al., 2017). We observed that the sporulation phenotype was complemented with the PdcA-EAL domain as well. We also expressed dccA coding for the diguanylate cyclase from the tet inducible promoter to artificially increase the concentration of c-di-GMP and monitored the sporulation percentage upon the induction of dccA. As expected, the production of DccA significantly reduced the sporulation in the UK1 strain (Fig. 5C). Measuring c-di-GMP in strains producing PdcA-EAL or PdcB-EAL or DccA confirmed their enzymatic effect on c-di-GMP concentration (Fig. 5B). The intracellular level of c-di-GMP is known to positively regulate surface motility in C. difficile (Bordeleau et al., 2015). Consistent with previous studies, we observed increased surface motility in the pdcB (OFF) strain, which has increased intracellular c-di-GMP levels compared with the pdcB (ON) strain. So, to further confirm that complementation of the UK1::pdcB mutant with the PdcB-EAL and PdcA-EAL domains reduces the intracellular c-di-GMP, we carried out a surface motility test assay with these strains (Fig. S6). As expected, we observed decreased surface motility in the complemented strains similar to the pdcB (ON) strain, which has reduced intracellular c-di-GMP levels. It is also interesting to note that the UK1::pdcB mutants producing either PdcB-EAL and PdcA-EAL were more opaque, less translucent and produced fewer longer cells (Fig. 5A). In addition to these experiments, we also silenced the pdcB gene using the CRISPRi system ((Müh et al., 2019), where the pdcB specific gRNA was expressed from a xylose-inducible promoter and checked its effect on sporulation. Silencing the pdcB gene significantly reduced sporulation in the UK1 strain (Fig. S5). While this manuscript is under revision, a preprint also reported that increased concentration of c-di-GMP leading to decreased sporulation in C. difficile R20291 and 630erm strains (Edwards et. al., 2021). Taken together, these data strongly suggest that the intracellular levels of c-di-GMP influence sporulation in C. difficile.
CodY binds to the upstream region and represses the expression of pdcB
Several studies have shown CodY-mediated regulation of c-di-GMP signaling proteins. Microarray analysis in the CD630 strain has identified two cyclic di-GMP signaling proteins, CD630_07570 (pdcB) and CD1476, among the 140 genes that were regulated by CodY (Dineen et al., 2010). The study demonstrated enhanced expression of both of these proteins in the codY mutant and identified a consensus CodY binding site in the upstream regions of CD1476 and CD630_07570 (pdcB). Another study has shown that CodY binds directly to the promoter region and represses the expression of pdcA, a phosphodiesterase that cleaves c-di-GMP in the JIR8094 strain (Purcell et al., 2017). To determine if the expression of pdcB is controlled by CodY, we first determined whether a CodY binding consensus sequence is present in the upstream region of pdcB. The two potential CodY binding sites upstream of pdcB are TATTTATTGAAAATTT and ACTTTTCTAAAATTA within the region that undergoes inversion (Fig. 6A). CodY binding site in the opaque orientation is upstream of the predicted promoter region in the pdcB (ON) orientation, which can explain the higher expression of pdcB in this phenotype. We hypothesized that CodY binding would completely shut down even the low-level expression of pdcB in the pdcB (OFF) strain. DNA inversion in the pdcB (ON) strain would shift the CodY binding site to the upstream region of the predicted promoter, leading to the expression of pdcB.
Figure 6. CodY represses the expression of pdcB, which is partially relieved by DNA inversion.
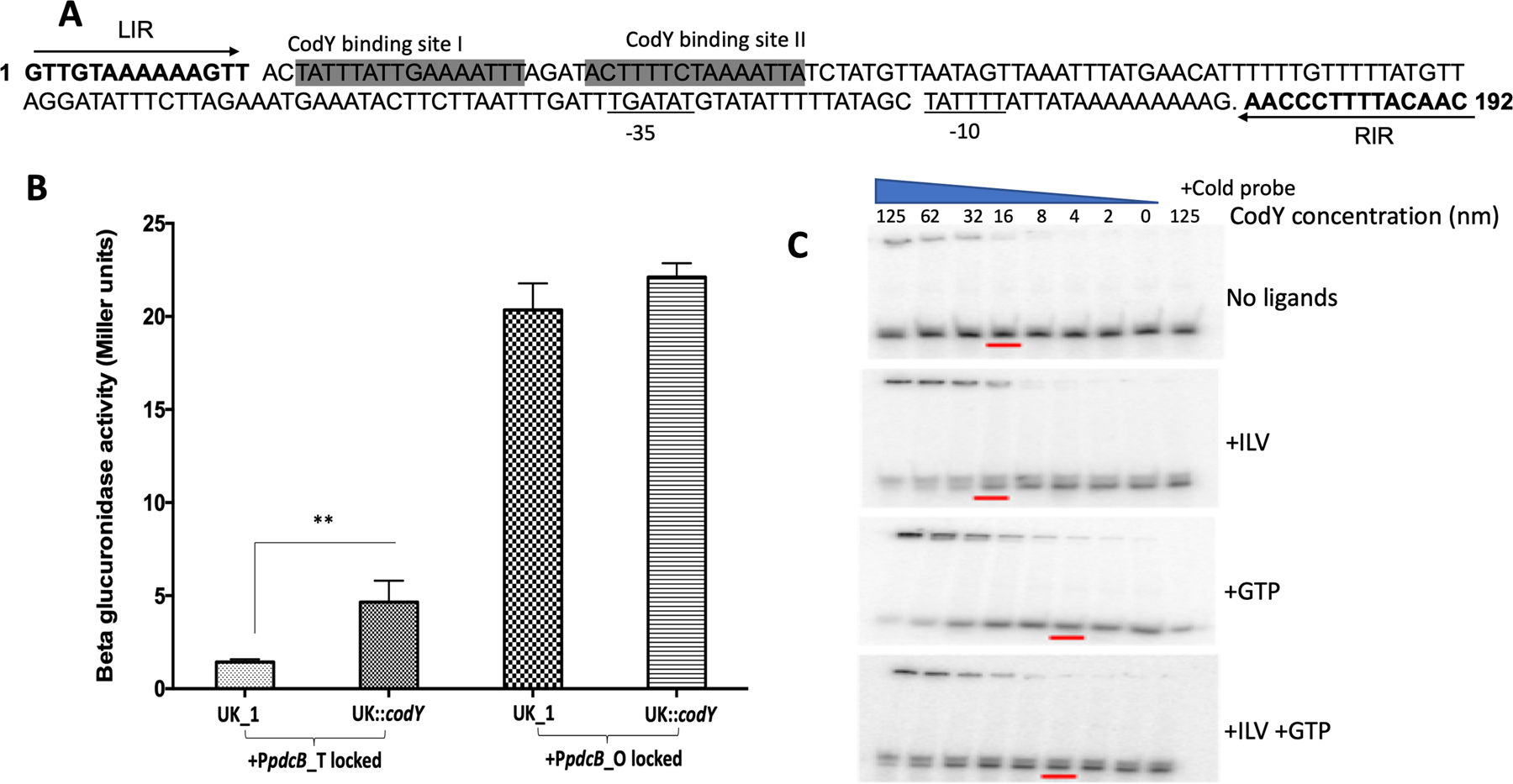
A. Schematic showing the potential CodY binding sites that lie within the invertible region upstream of pdcB. LIR and RIR are in bold and marked with arrows B. Beta-glucuronidase activity of PpdcB-gusA fusions locked in either translucent or opaque orientation in the parent UK1 and UK1::codY mutant. The data shown are the standard errors of the mean of three biological replicates. Statistical analysis was performed using two-way ANOVA.** indicates p<0.05.C. Binding of purified CodY to the upstream region of pdcB. Binding was increased with increasing concentration of CodY. The presence of GTP and ILV further enhanced CodY binding. Ten-fold excess of the cold probe was added with 125 ng of purified CodY in a control reaction to rule out non-specific binding.
To determine if CodY regulates the expression of pdcB, we carried out the reporter fusion assay. We used the same reporter fusions (pBA040, pBA043, pBA046) that were used earlier in this study. Each of the constructs was introduced into the UK1 undifferentiated parent strain and UK1::codY mutant strain by conjugation. The strains were grown anaerobically in TY broth medium at 37°C, and cells were harvested at different time points to perform the reporter assay. We observed increased beta-glucuronidase activity at each time point when it was expressed from the pdcB upstream locked in opaque orientation, with higher expression at mid-exponential phase from 4–12 h for both UK1 parent and UK1::codY mutant strains (Fig. 6B). Minimal reporter activity was observed with the upstream locked in translucent orientation. However, in the absence of CodY, an approximately 1.5-fold increase in the expression could be seen from the promoter locked in the translucent orientation, suggesting the presence of a second but much weaker promoter upstream of the CodY binding site. It is also possible that the purpose of DNA inversion is not only to position the promoter in the right orientation for expression, but also to move the potential CodY binding site upstream to prevent the CodY-mediated repression. Since, the left inverted repeat (LIR) in pdcB upstream is next to the CodY binding site, CodY may also interfere with the binding of the recombinase from recognizing the repeat and can block inversion. This hypothesis, however needs further experimental evidence.
To further confirm the interaction of CodY with pdcB upstream DNA, we carried out an Electrophoretic Mobility Shift Assay (EMSA) with purified CodY protein. We synthesized the DNA fragment of 47 bp containing the potential CodY binding site and radio-labeled with γ- 32 P. The fragment was bound with increasing concentrations of purified CodY, both in the absence and presence of its cofactors ILV (Branched-chain amino acids isoleucine, leucine, and valine) and GTP (Fig. 6C). The shift in the bands at higher concentrations of CodY protein suggests that purified CodY binds to the DNA fragment, and the binding was enhanced in the presence of its cofactors GTP and ILVs.
Discussion
The major objective of this study was to determine the role of phase variation in the physiology of C. difficile UK1 strain. Genome-wide analysis had previously identified seven different types of invertible sites in the C. difficile genome, designated as Cdi1 to Cdi7 (Sekulovic et al., 2018). The Cdi2 site is upstream of pdcB, which encodes a c-di-GMP phosphodiesterase (PDE) (Sekulovic et al., 2018). In this work, we have shown the role of Cdi2 in regulating the expression of pdcB. Our study identified an “ON” orientation to the pdcB promoter region that induces pdcB transcription; over-expression of pdcB reduces c-di-GMP levels, whereas decreased pdcB expression due to the “OFF” promoter orientation increases c-di-GMP levels. Notably, even though C. difficile has 18 predicted PDEs (Bordeleau et al., 2011), expression of pdcB is sufficient to affect intracellular c-di-GMP levels. While c-di-GMP regulates a plethora of physiological processes in C. difficile, its role in sporulation had not been previously described. Only a few studies have investigated the role of c-di-GMP in sporulation. Recently, the relationship between c-di-GMP and sporulation has been explored in Streptomyces venezuelae, a Gram-positive soil bacterium, where low levels of c-di-GMP resulted in hyper sporulation phenotype (Hengst et al., 2010; Tschowri et al., 2014; Al-Bassam et al., 2018; Latoscha et al., 2019; Liu et al., 2019; Gallagher et al., 2020) . Gallagher et al. reported that c-di-GMP bridges the binding of anti-sigma factor RsiG with sigma factor WhiG, strengthening their interaction and preventing sigma factor WhiG-dependent transcription of late sporulation genes (Gallagher et al., 2020). Similarly, another study demonstrated that c-di-GMP is required to control the transition from vegetative to reproductive cells (Bush et al., 2015). It was reported that c-di-GMP was needed to form the active dimeric form of BldB, the master regulator of cell development, and represses the development of reproductive hyphae and keeps Streptomyces in their vegetative state (Hengst et al., 2010; Bush et al., 2015; Al-Bassam et al., 2018; Gallagher et al., 2020). These studies suggest that high c-di-GMP level traps Streptomyces in vegetative growth, and low c-di-GMP levels cause precocious hyper sporulation. It is however, important to note that sporulation in Streptomyces is different from endospore formation in Bacillus and Clostridia.
More recently, proteins involved in c-di-GMP turnover were localized as part of spore proteomes and were found to play a role in spore germination in Bacillus anthracis (Hermanas et al., 2021). Single-cell microscopy studies carried out in B. subtilis have shown a positive correlation of high c-di-GMP levels with sporulation (Weiss et al., 2019). Our current work is the first study to directly correlate intracellular c-di-GMP concentration to sporulation in C. difficile. Hyper sporulation was observed when the intracellular c-di-GMP concentration was reduced by ~1.5 fold in pdcB (ON) compared to the level in pdcB (OFF) cells grown in vitro. It is noteworthy that, in our previous study, R20291::sinRR’ mutant strain, which is asporogenic, had elevated levels of c-di-GMP compared with the R20291 parent strain (Girinathan et al., 2018), corroborating our current observation. By analyzing percentage sporulation in the UK1::pdcB mutant strain and PdcB-EAL and PdcA-EAL producing UK1::pdcB strains, we have shown that production of a phosphodiesterase that reduces c-di-GMP concentration is positively associated with sporulation phenotype. However, overexpression of EAL domains of both PdcB and PdcA only partially complement the sporulation phenotype. This could be because of the overexpression of the PdcB-EAL domain alone without the associated GGDEF domain is not sufficient. Although the GGDEF domain of PdcB-EAL is catalytically inactive (Sekulovic et al., 2018), it might be necessary to enhance the activity of the EAL domain to fully complement sporulation. In homologous proteins which contain both GGDEF and EAL domains, catalytically inactive GGDEF domains are capable of binding to GTP, thus enhancing the PDE activity of the neighboring EAL domain (Christen et al., 2005).
Regulation of C. difficile sporulation can occur either at the transcriptional level by altering the expression of spo0A or at the post-translational level by interfering with the phosphorylation of Spo0A (Edwards and McBride, 2014). There are two known mechanisms by which c-di-GMP is known to function. First, c-di-GMP mediated regulation can occur through RNA-based effectors, which includes direct binding of c-di-GMP to riboswitches present in the 5’ UTR of the target gene transcript leading to premature termination of the transcript. Second, c-di-GMP mediated regulation can occur through protein effectors containing the PilZ domain, GMP binding domain, diguanylate cyclases containing I-sites, GIL proteins, MshEN domains, and the Cle subfamily of CheY proteins which sense and respond to the changes in the intracellular c-di-GMP concentration (McKee et al., 2018b). Whether c-di-GMP binding riboswitches or c-di-GMP binding domains are present in the histidine kinases that phosphorylate Spo0A, and whether c-di-GMP regulates their expression or activity, thus regulating sporulation needs to be further investigated. On the other hand, c-di-GMP could indirectly influence sporulation by affecting the expression of regulators known to affect the expression of spo0A.
Previous studies have shown that the C. difficile UK1::codY mutant exhibits a hyper-sporulation phenotype suggesting that CodY represses sporulation (Nawrocki et al., 2016). The exact mechanism by which CodY regulates sporulation is not understood. However, studies have shown that CodY directly regulates the expression of sinRI (Girinathan et al., 2018) and opp and app (Edwards et al., 2014; Nawrocki et al., 2016), which are positive regulators of sporulation. Results in this study suggest that phosphodiesterases can play a role in regulating sporulation. This could be indirectly by controlling CodY activity, which is directly regulated by intracellular GTP concentration.
While we were working on characterizing pdcB role in UK1 phase variation, another study reported the role of cmrRST on phase variation in C. difficile R20291 strain (Garrett et al., 2019). Similar to the pdcB gene, the cmrRST operon carries a DNA inversion element belonging to cdi6 group. CmrR and CmrS make up a two-component signal transduction system coding for a response regulator and a histidine kinase, respectively. To check any connection between cmrRST and phase variation in the UK1 strain, we checked the orientation of the cmrRST upstream region in the pdcB (ON) and pdcB (OFF) cells. In the R20291 strain, the rough colonies (equivalent to the translucent colonies of UK1 where pdcB is in the OFF orientation), the cmrRST operon is in the ON orientation, and the smooth colonies (equivalent to the opaque colonies of UK1 where pdcB is in ON orientation) resulted when the cmrRST is in the OFF orientation (Garrett et al., 2019). In the UK1 strain, however, we observed the opposite. The cmrRST operon was in the ON orientation in pdcB (ON) colonies. In the pdcB (OFF) colonies, the PCR product indicating the cmrRST OFF orientation amplified at higher frequency compared to the product indicating ON orientation (Fig. S7). Even though the frequency of cmrRST being ON/OFF in rough and smooth R20291 cells differs from that of the UK1_O and UK_T cells, there are similarities between the UK1 and R20291 phase-variable colonies. In the cmrRST study, the authors reported that the rough colonies in R20291 (equivalent to translucent pdcB (OFF)) had the Cdi2 region (i.e. upstream of pdcB) in the OFF orientation. Greater levels of intracellular c-di-GMP in the R20291 strain significantly increased cell length and resulted in the formation of rough colonies, similar to what we noticed in pdcB (OFF) cells with higher levels of c-di-GMP. Thus, both the cmrRST study and ours reveal that c-di-GMP plays a role in creating these phase-variable C. difficile colonies.
Site specific recombinases that carry out DNA inversion might be associated with the invertible site or encoded elsewhere in the genome (van de Putte et al., 1980). Prior works with C. dfficile R20291 strain has shown that RecV, the tyrosine recombinase, catalyzes the inversion of the Cdi1, Cdi4, Cdi6 switches, partial inversion of Cdi2, Cdi3, Cdi5 and no effect on Cdi7 inversion (Sekulovic et al., 2018). To determine if RecV has a role in inversion of Cdi2 in UK1 strain we constructed a UK1::recV mutant using ClostTron mutagenesis (Fig. S8AB). If RecV had a role in DNA inversion of Cdi2, we would expect UK1::recV mutant to exist in both the colony UK1_T and UK1_O colony types that are locked in their respective morphotypes in the absence of RecV. However, phase-variable colonies could not be distinguished in UK1::recV mutant (Fig. S8C). The orientation PCR using UK1::recV colonies detected pdcB in both ON and OFF orientation (Fig. S8D). This finding suggested that RecV may be only partially involved in the inversion of the region, which can result in bacterial cells with both ON and OFF orientation to present in a single colony. It is also plausible that a site-specific recombinase other than RecV is involved in the upstream inversion of pdcB in the UK1 strain.
In summary, our study has shown phase variation-mediated regulation of intracellular c-di-GMP homeostasis and its effect on C. difficile physiology. Although both translucent and opaque cells are shown to produce similar levels of toxins, they differ drastically in cell length and sporulation. Based on the swarming motility and biofilm formation phenotypes, it can be suggested that during initiation of infection, UK1 might prefer to exist as the translucent colony cells so that they can better colonize the host and cause infection. Under the selective pressure exerted in the intestinal tract and host immune responses, the UK1 opaque colony type might predominate. This might result in the production of more spores allowing C. difficile to evade the host immune response and persist in the host, to cause recurrent infection, or in the environment, once it is released outside of the infected individual. Understanding the importance of phase-variable expression of pdcB and its associated phenotypes in an in vivo infection model is a priority for future investigation and will shed light on the importance of pdcB in C. difficile pathogenesis.
Material and Methods
Bacterial strains and growth conditions
C. difficile UK1 strains (Table S1) were grown in TY (tryptose and yeast extract) medium agar or broth culture in an anaerobic chamber maintained at 10% H2, 10% CO2, and 80% N2. Transconjugants were selected and grown in TY agar with lincomycin (Linc 20µg/ml) or thiamphenicol (Thio; 15 µg/ml) or both. An E. coli strain optimized for conjugation S17–1 was used for conjugation (Teng et al., 1998), and the E. coli DH5α strain was used for cloning. E. coli strains were cultured aerobically in LB (Luria-Bertani) supplemented with ampicillin (100 µg/ml) or chloramphenicol (25 µg/ml) when necessary.
Phase-contrast microscopy
C. difficile strains were grown in TY medium as described above. One ml of overnight culture was centrifuged at 17,000g for 1 min and washed with 30μl of sterile PBS. The resulting pellets were fixed for 2 hours at room temperature using 2% paraformaldehyde in 1X PBS. A thin layer of 0.7% agarose was applied to the surface of a microscopic slide, and 2μl of concentrated culture was placed on it. The cells were imaged using Zeiss LSM-5 PASCAL (objective lens 100x/1.4 oil). The LSM 5 service pack was used to acquire the images of at least three fields for each strain.
Sporulation assay
Cells grown in TY agar overnight were used for sporulation assay. Bacterial strains carrying plasmids were grown in the presence of thiamphenicol (15 μg/ml). ATc at a concentration of 100 ngs/ml was used to induce the expression of genes cloned under the tet inducible promoter. To enumerate the number of viable spores, cells were harvested using 1 ml of TY broth, and 500μl of the samples from each culture were mixed 1:1 with 95% ethanol and incubated for 30 minutes at room temperature to kill all the vegetative cells. The ethanol-treated samples and the untreated samples were then serially diluted, plated on TY agar with 0.1% taurocholate, and incubated at 37°C for 24 to 48 hours. The percentage of ethanol-resistant spores was calculated by dividing the number of CFU from spores by the total number of CFU and multiplying the value by 100. The results were based on a minimum of three biological replicates.
RNA isolation, reverse transcriptase (RT-PCR) and quantitative reverse transcriptase PCR (qRT-PCR).
C. difficile cultures were grown in TY medium, and 6 ml of cells were harvested at different time points by centrifugation at 3000g for 30 min at 4°C. Total RNA was extracted from the harvested cells following the protocol described previously (El Meouche et al., 2013). Total RNA was treated with DNase (Turbo; Ambion) for 2 hours at 37°C. The cDNA was synthesized using 5μg of total RNA at 42°C for 2 h using avian myeloblastosis virus (AMV) reverse transcriptase (Promega). A 20 µL reaction containing 10 ng or 10 pg (for 16S rRNA) of cDNA, 400 nM gene-specific primers, and 12.75 µL of SYBR PCR master mix (BioRad) was used to perform real-time quantitative PCR using iQPCR instrument (BioRad). Amplification and detection were performed as described previously (El Meouche et al., 2013). The quantity of cDNA of a gene in each sample was normalized to the quantity of C. difficile 16S rRNA gene, and the ratio of normalized target concentrations (threshold cycle [2−ΔΔCt] method) (Saujet et al., 2011; El Meouche et al., 2013) gives the relative change in gene expression. The cDNA prepared was also used as a template to perform reverse transcriptase PCR to detect pdcB transcripts using forward primers ORG925, ORG926, ORG921, ORG922, along with the reverse primer ORG 853 (Table S2).
5’RACE
5′RACE (rapid amplification of cDNA ends) assays were performed with total RNA extracted from pdcB (ON) using a 5′RACE System kit (Sigma Aldrich), according to the instructions provided by the manufacturer. Gene-specific RT primer (SP1) and PCR primer (SP2) were designed as recommended. The 5’-RACE product was used as a template for a nested PCR reaction using a second primer (SP3). The sequences of primers used in the RACE analysis are shown in Table S2. PCR products were cloned in pGEM-T vector and were sequenced.
Mutagenesis of CDR20291_0685 upstream region
Quick Change Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) was used to carry out site-directed mutagenesis whereby the Left Inverted Repeat (LIR) TAGTTGTAAAAGGGTT that flanked the DNA region that undergoes inversion was converted to TACACATGCGAGGGTT. The mutagenic oligonucleotide primers used are listed in Table S2.
Construction of reporter plasmids and beta-glucuronidase assay
The 1.5 kb upstream DNA regions of CDR20291_0685 were amplified by PCR using specific primers with KpnI and SacI (Table S2) recognition sequences. UK1_T and UK1_O chromosomal DNA were used as a template to amplify the upstream region in respective orientations. The upstream region was cloned in the pRPF185 shuttle vector using standard cloning procedures. Plasmid pRPF185 carries a gusA gene for beta-glucuronidase under the tetracycline-inducible (tet) promoter (Fagan and Fairweather, 2011). The tet promoter was removed using KpnI and SacI digestion and was replaced with the CDR20291_0685 upstream region to create plasmids pBA042 and pBA045 (Table S1). The inverted repeats in the upstream region were mutagenized as described earlier to create plasmid pBA043 containing the upstream region locked in translucent orientation and plasmid pBA046 containing the upstream region locked in opaque orientation. The control plasmid pBA040 with promoter-less gusA was created by digesting with KpnI and SacI to remove the tet promoter and then self-ligating after creating blunt ends. Plasmids were introduced into C. difficile strains through conjugation. The transconjugants were grown overnight in TY medium in the presence of thiamphenicol (15 µg/ml). Overnight cultures were used as inoculum at a 1:100 dilution to start a new culture. Bacterial cultures were harvested at different time points of growth, and the amount of beta-glucuronidase activity was assessed as described elsewhere (Mani et al., 2002). Briefly, the cells were washed and resuspended in 1 ml of Z-buffer (60 mM Na2HPO4.7H20 pH 7.0, 40 mM NaH2PO4.H2O, 10 mM KCl, 1mM MgSO4.7H2O, and 50mM β-ME), and lysed by homogenization. The lysate was mixed with 160 µl of 6mM p-nitrophenyl β-D-glucuronide (Sigma) and incubated at 37°C. The reaction was stopped by the addition of 0.4 ml of 1.0 M NaCO3. β-Glucuronidase activity was calculated as described earlier (Dupuy and Sonenshein, 1998; Mani et al., 2002).
Electrophoretic mobility shift assay (EMSAs)
For the CodY binding experiment, the upstream region of the pdcB with the predicted CodY binding sequence 5’ CATAGATAATTTTTAGAAAAGTATCTAAATTTTCAATAAATAGTAAC 3’ was synthesized. It was labeled with [γ- 32 P] dATP-6000 Ci/mmol (PerkinElmer Life Sciences) using T4 polynucleotide kinase. It was then annealed with the complementary oligo to generate a double-stranded DNA probe. The DNA-protein binding reactions were carried out at room temperature for 30 min in 10μl volume containing 1x binding buffer [10mM Tris pH 7.5, 50mM KCl, 50μg BSA, 0.05% NP40, 10% glycerol, 10 mM GTP and 2mM ILV, 100 μg/ml poly dI-dC and 800nM of DNA probe with varying concentration of purified CodY protein. DNA probe in reaction buffer was incubated for 10 min at room temperature before adding purified CodY-6His protein. The reaction was stopped by adding 5µl of gel loading buffer and electrophoresed at 100V for 1.5 h using 6% 1XTBE gel in 0.5X TBE buffer containing 10 mM ILV. Gels were then dried, and autoradiography was performed with Molecular Dynamics Phosphor-Imager technology.
Construction and complementation of C. difficile UK1::pdcB mutant strains
The UK1::pdcB mutant in the UK1 strain was created using the ClosTron gene knockout system as described previously (Heap et al., 2010). Briefly, for pdcB disruption, the group II intron insertion site between nucleotides 840 and 841 in the pdcB gene in the sense orientation was selected using a web-based design tool called the Perutka algorithm. The designed retargeted intron was cloned into pMTL007-CE5 as described previously (Govind and Dupuy, 2012; Girinathan et al., 2016). The resulting plasmid pMTL007-CE5::pdcB-840–841s was transferred into C. difficile UK1 cells by conjugation. The potential Ll.ltrB insertions within the target genes in the C. difficile chromosome were conferred by the selection of lincomycin-resistant transconjugants in 20 μg/ ml lincomycin plates. PCR using gene-specific primers (Table S2) in combination with the EBS-U universal was performed to identify putative C. difficile mutants. C. difficile pdcB mutants were complemented by introducing pBA048, which contains the PdcB-EAL domain, and pBA050, which contains the PdcA-EAL domain under the tet inducible promoter, through conjugation.
Cyclic-di-GMP measurement
c-di-GMP was measured as described previously (Girinathan et al., 2017), with a few modifications. Briefly, C. difficile strains were grown in 50 ml TY medium for 16 h. OD600 was recorded and dilution plating was carried out to determine the number of CFU. Cells were centrifuged and pellets were washed once with TE buffer (10 mM Tris [pH 7.5], 1 mM EDTA, pH 8). Pellets were resuspended in extraction solvent consisting of methanol: acetonitrile: distilled water in 40:40:20 ratio plus 0.1 N formic acid. The mixture was incubated in −20°C for 30 min. The extract was harvested by centrifugation. Samples were sent to Kansas University Biochemistry Core facility to be analyzed by high-pressure liquid chromatography (HPLC)-tandem mass spectrometry (MS/MS). To determine the amount of intracellular c-di-GMP, a standard curve was first obtained by analyzing the serial dilution of pure c-di-GMP (Sigma-Aldrich). The peak intensity of each sample was fitted to the standard curve and the value was divided by the total intracellular volume of bacteria extracted. To determine the total intracellular volume of bacteria, the volume of one cell was multiplied by the number of cells extracted, which was based on CFU counts. The volume of one cell was estimated as a cylinder and was determined by measuring the radius and length of the bacterial cell from the electron micrographs.
Supplementary Material
Figure S1. Phenotypes of UK1_T and UK1_O cells. Single UK_T and UK_O colonies were streaked/ inoculated on TY agar (A)/ TY broth (B) and were observed for phase variation. Few UK_T colonies gave rise to UK_O (marked with black arrows) within 20h of incubation, while the UK_O colonies maintained the opaque phenotype. Upon longer incubation (72h), both UK_T and UK_O colonies gave rise to the opaque and translucent colonies, respectively. Few of the UK_O colonies giving to translucent cells are marked with white arrows. At least 6 plates were tested and observed.
Figure S2. Swimming motility and toxin production of UK1_T and UK1_O strains.
A. Swimming motility exhibited by UK1_T and UK1_O in TY+0.5% agar. B. Cytoplasmic toxin levels produced by UK1_T and UK1_O morphotypes during the stationary phase of growth. Three biological replicates were used to carry out the experiments, and data were analyzed by a two-tailed unpaired t-test with Welch’s correction (NS indicates not significant).
Figure S3. UK1_O undergoes DNA inversion upstream of CDR20291_0685.
Alignment of 192 bp segment of the upstream region of CDR20291_0685 from UK1_T and UK_O with the published R20291 sequence. Inverted repeat sequence bordering the sequence is marked in bold.
Figure S4. Construction and confirmation of the pdcB mutation in C. difficile UK1 strain.
A. Schematic representation of ClostTron (group II intron) mediated insertional inactivation of pdcB gene in C. difficile. B. PCR verification of the intron insertion in pdcB in UK1 strain, conducted with intron-specific primer EBS universal [EBS(U)] with pdcB specific primers ORG 771 and ORG 772.
Figure S5. CRISPRi knock-down of pdcB in UK1. Transconjugants carrying pdcB-CRISPRi plasmid were streaked on to TY+Thio plates with or without 1% xylose. Phase contrast microscopic images (A) and sporulation percentage (B) of pdcB-sgRNA induced and uninduced cultures. These are representative results from two independent experiments. Data were analyzed by two-tailed unpaired t-test with Welch’s correction where ***=p<0.005
Figure S6. Increased c-di-GMP is associated with increased swarming motility phenotype in UK1_T strain.
A. Representative swarming motility plates of pdcB (OFF), pdcB (ON), UK1::pdcB mutant, UK1::pdcB + PdcB- EAL and UK1::pdcB + PdcA- EAL complemented strains after 72h of incubation at 37°C.
Figure S7. Orientation of pdcB and cmrRST upstreams in UK_T and UK_O cells. A.PCR amplification of the upstream region of pdcB using primer mixture containing primers ORG886/887/888. Amplification with ORG886/888 results in a product size of 325 bp, while amplification with ORG 887/888 results in a product size of 215 bp. B. PCR amplification of the upstream region of cmrRST using primer mixture containing primers ORG879/880/881. Amplification with ORG879/880 results in a product size of 410 bp, while ORG 880/881 results in 540 bp PCR product. Single bacterial colony of a specified phenotype was resuspended in 50µl TE buffer and was boiled for 10 minutes. The boiled lysate was then centrifuged and 2µl of the supernatant was used a template to perform PCR. At least ten colonies from each phenotype was tested.
Figure S8. Analysis of UK1::recV mutant. A. Schematic representation of ClostTron (group II intron) mediated insertional inactivation of recV gene in C. difficile. B. PCR verification of the intron insertion in recV in UK1 strain, conducted with intron-specific EBS primer with recV specific primers ORG 788 and ORG 789. C. Phase contrast microscopy image of UK1::recV mutant strain. D. Orientation PCR to check the ON and OFF position of pdcB.
Acknowledgment
We thank following investigators for sharing their lab resources: Nigel Minton, University of Nottingham, for the plasmid pMTL007C-E5; Robert Fagan, University of Sheffield for the vector pRPF185; Craig Ellermeier, University of Iowa for the CRISPi plasmid pIA33. We thank Yusuf Ciftci for technical assistance throughout the study, Ruth Welti and Nick Wallace for editing the manuscript.
Footnotes
Data Sharing statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Conflict of Interests
Authors have no conflicts of interests to declare.
References
- Al-Bassam MM, Haist J, Neumann SA, Lindenberg S, and Tschowri N (2018) Expression patterns, genomic conservation and input into developmental regulation of the GGDEF/EAL/HD-GYP domain proteins in Streptomyces. Front Microbiol 9: 2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjuwon-Foster BR, and Tamayo R (2017) A genetic switch controls the production of flagella and toxins in Clostridium difficile. PLoS Genet 13: e1006701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arato V, Gasperini G, Giusti F, Ferlenghi I, Scarselli M, and Leuzzi R (2019) Dual role of the colonization factor CD2831 in Clostridium difficile pathogenesis. Sci Rep 9: 5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon DJ, Szymanski CM, Burr DH, Silver RP, Alm RA, and Guerry P (2001) A phase-variable capsule is involved in virulence of Campylobacter jejuni 81–176. Molecular Microbiology 40: 769–777. [DOI] [PubMed] [Google Scholar]
- Bordeleau E, and Burrus V (2015) Cyclic-di-GMP signaling in the Gram-positive pathogen Clostridium difficile. Curr Genet 61: 497–502. [DOI] [PubMed] [Google Scholar]
- Bordeleau E, Fortier L-C, Malouin F, and Burrus V (2011) C-di-GMP turn-Over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLOS Genetics 7: e1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau E, Purcell EB, Lafontaine DA, Fortier L-C, Tamayo R, and Burrus V (2015) Cyclic Di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile. J Bacteriol 197: 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush MJ, Tschowri N, Schlimpert S, Flärdh K, and Buttner MJ (2015) C-di-GMP signaling and the regulation of developmental transitions in Streptomycetes. Nat Rev Microbiol 13: 749–760. [DOI] [PubMed] [Google Scholar]
- CDC (2020) Antibiotic-resistant Germs: New Threats Centers for Disease Control and Prevention; https://www.cdc.gov/drugresistance/biggest-threats.html. Accessed April 8, 2020. [Google Scholar]
- Christen M, Christen B, Folcher M, Schauerte A, and Jenal U (2005) Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280: 30829–30837. [DOI] [PubMed] [Google Scholar]
- Denève C, Janoir C, Poilane I, Fantinato C, and Collignon A (2009) New trends in Clostridium difficile virulence and pathogenesis. International Journal of Antimicrobial Agents 33: S24–S28. [DOI] [PubMed] [Google Scholar]
- Dineen SS, McBride SM, and Sonenshein AL (2010) integration of metabolism and virulence by Clostridium difficile CodY. Journal of Bacteriology 192: 5350–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy B, and Sonenshein AL (1998) Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 27: 107–120. [DOI] [PubMed] [Google Scholar]
- Edwards AN, and McBride SM (2014) Initiation of sporulation in Clostridium difficile : a twist on the classic model. FEMS Microbiol Lett 358: 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, Nawrocki KL, and McBride SM (2014) Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infect Immun 82: 4276–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, Willams CL, Pareek N, McBride MS, Tamayo R c-di-GMP inhibits early sporulation in Clostridioides difficile. (2021). bioRxiv 2021.06.24.449855; doi: 10.1101/2021.06.24.449855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein BI (1988) Type 1 fimbriae of Escherichia coli: genetic regulation, morphogenesis, and role in pathogenesis. Rev Infect Dis 10 Suppl 2: S341–344. [DOI] [PubMed] [Google Scholar]
- El Meouche I, Peltier J, Monot M, Soutourina O, Pestel-Caron M, Dupuy B, and Pons J-L (2013) Characterization of the SigD regulon of c. Difficile and its positive control of toxin production through the regulation of tcdR. PLoS ONE 8: e83748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ende A. van der, Hopman CTP, and Dankert J (2000) Multiple mechanisms of phase variation of PorA in Neisseria meningitidis. Infection and Immunity 68: 6685–6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan RP, and Fairweather NF (2011) Clostridium difficile has two parallel and essential sec secretion systems. J Biol Chem 286: 27483–27493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KA, Schumacher MA, Bush MJ, Bibb MJ, Chandra G, Holmes NA, et al. (2020) C-di-GMP arms an anti-σ to control progression of multicellular differentiation in Streptomyces. Molecular Cell 77: 586–599.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett EM, Sekulovic O, Wetzel D, Jones JB, Edwards AN, Vargas-Cuebas G, et al. (2019) Phase variation of a signal transduction system controls Clostridioides difficile colony morphology, motility, and virulence. PLoS Biol 17: e3000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girinathan BP, Braun S, Sirigireddy AR, Lopez JE, and Govind R (2016) Importance of Glutamate Dehydrogenase (GDH) in Clostridium difficile colonization in vivo. PLOS ONE 11: e0160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girinathan BP, Monot M, Boyle D, McAllister KN, Sorg JA, Dupuy B, and Govind R (2017) Effect of tcdR mutation on sporulation in the epidemic Clostridium difficile strain R20291. mSphere 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girinathan BP, Ou J, Dupuy B, and Govind R (2018) Pleiotropic roles of Clostridium difficile sin locus. PLoS Pathog 14: e1006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind R, and Dupuy B (2012) Secretion of Clostridium difficile Toxins A and B requires the Holin-like protein TcdE. PLoS Pathog 8: e1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CL, and Lee VT (2018) Cyclic-di-GMP regulation of virulence in bacterial pathogens. Wiley Interdiscip Rev RNA 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, and Minton NP (2010) The ClosTron: Mutagenesis in Clostridium refined and streamlined. J Microbiol Methods 80: 49–55. [DOI] [PubMed] [Google Scholar]
- Hengst CDD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, and Buttner MJ (2010) Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Molecular Microbiology 78: 361–379. [DOI] [PubMed] [Google Scholar]
- Hermanas TM, Subramanian S, Dann CE, and Stewart GC (2021) Spore-sssociated proteins involved in C-di-GMP synthesis and degradation of Bacillus anthracis. J Bacteriol 203: e0013521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst Sørensen MC, Alphen L.B. van, Fodor C, Crowley SM, Christensen BB, Szymanski CM, and Brøndsted L (2012) Phase variable expression of capsular polysaccharide modifications allows Campylobacter jejuni to avoid bacteriophage infection in chickens. Front Cell Infect Microbiol 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Hall AB, Arthur TD, Plichta DR, Covington CT, Poyet M, et al. (2019) Invertible promoters mediate bacterial phase variation, antibiotic resistance, and host adaptation in the gut. Science 363: 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P (1986) Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. The EMBO Journal 5: 1389–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K, and Iino T (1980a) A trans- acting factor mediates inversion of a specific DNA segment in flagellar phase variation of Salmonella. 284: 3. [DOI] [PubMed] [Google Scholar]
- Kutsukake K, and Iino T (1980b) Inversions of specific DNA segments in flagellar phase variation of Salmonella and inversion systems of bacteriophages P1 and Mu. Proc Natl Acad Sci U S A 77: 7338–7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latoscha A, Wörmann ME, and Tschowri N (2019) Nucleotide second messengers in Streptomyces. Microbiology 165: 1153–1165. [DOI] [PubMed] [Google Scholar]
- Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, et al. (2015) Burden of Clostridium difficile Infection in the United States. New England Journal of Medicine 372: 825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, and Zhang J-R (2019) Phase variation of Streptococcus pneumoniae. Microbiol Spectr 7. (1). doi: 10.1128/microbiolspec.GPP3-0005-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovsek S, Leitinger G, and Rupnik M (2013) Ultrastructure of Clostridium difficile colonies. Anaerobe 24: 66–70. [DOI] [PubMed] [Google Scholar]
- Liu X, Zheng G, Wang G, Jiang W, Li L, and Lu Y (2019) Overexpression of the diguanylate cyclase CdgD blocks developmental transitions and antibiotic biosynthesis in Streptomyces coelicolor. Sci China Life Sci 62: 1492–1505. [DOI] [PubMed] [Google Scholar]
- Mani N, Lyras D, Barroso L, Howarth P, Wilkins T, Rood JI, et al. (2002) Environmental Response and Autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. Journal of Bacteriology 184: 5971–5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain MS, Blomfield IC, Eberhardt KJ, and Eisenstein BI (1993) Inversion-independent phase variation of type 1 fimbriae in Escherichia coli. Journal of Bacteriology 175: 4335–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain MS, Blomfield IC, and Eisenstein BI (1991) Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. Journal of Bacteriology 173: 5308–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee RW, Aleksanyan N, Garrett EM, and Tamayo R (2018a) Type IV pili promote Clostridium difficile adherence and persistence in a mouse model of infection. Infect Immun 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee RW, Harvest CK, and Tamayo R (2018b) Cyclic diguanylate regulates virulence factor genes via multiple riboswitches in Clostridium difficile. mSphere 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee RW, Mangalea MR, Purcell EB, Borchardt EK, and Tamayo R (2013) The second messenger Cyclic Di-GMP regulates Clostridium difficile toxin production by controlling the expression of sigD. Journal of Bacteriology 195: 5174–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müh U, Pannullo AG, Weiss DS, and Ellermeier CD (2019) A xylose-inducible expression system and a CRISPR interference plasmid for targeted knockdown of gene expression in Clostridioides difficile. J Bacteriol 201: e00711–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki KL, Edwards AN, Daou N, Bouillaut L, and McBride SM (2016) CodY-dependent regulation of sporulation in Clostridium difficile. J Bacteriol 198: 2113–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes-Sabja D, Shen A, and Sorg JA (2014) Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol 22: 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J, Shaw HA, Couchman EC, Dawson LF, Yu L, Choudhary JS, et al. (2015) Cyclic di-GMP regulates production of sortase substrates of Clostridium difficile and their surface exposure through zmpi protease-mediated cleavage. J Biol Chem 290: 24453–24469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EB, McKee RW, Bordeleau E, Burrus V, and Tamayo R (2016) Regulation of Type IV Pili contributes to surface behaviors of historical and epidemic strains of Clostridium difficile. J Bacteriol 198: 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EB, McKee RW, Courson DS, Garrett EM, McBride SM, Cheney RE, and Tamayo R (2017) A nutrient-regulated cyclic diguanylate phosphodiesterase controls Clostridium difficile Biofilm and toxin production during stationary phase. Infect Immun 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EB, McKee RW, McBride SM, Waters CM, and Tamayo R (2012) Cyclic Diguanylate inversely regulates motility and aggregation in Clostridium difficile. Journal of Bacteriology 194: 3307–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putte P. van de, Cramer S, and Giphart-Gassler M (1980) Invertible DNA determines host specificity of bacteriophage Mu. Nature 286: 218–222. [DOI] [PubMed] [Google Scholar]
- Reyes Ruiz LM., Williams CL, Tamayo R (2020) Enhancing bacterial survival through phenotypic heterogeneity. PLoS Pathog 16: e1008439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi S, Howe K, Lawrence ML, Brooks JP, Bailey RH, and Karsi A (2017) Salmonella enterica Serovar kentucky flagella are required for broiler skin adhesion and caco-2 cell invasion. Appl Environ Microbiol 83: e02115–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saujet L, Monot M, Dupuy B, Soutourina O, and Martin-Verstraete I (2011) The Key Sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. Journal of Bacteriology 193: 3186–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott TN, and Simon MI (1982) Genetic analysis of the mechanism of the Salmonella phase variation site specific recombination system. Mol Gen Genet 188: 313–321. [DOI] [PubMed] [Google Scholar]
- Sekulovic O, Mathias Garrett E, Bourgeois J, Tamayo R, Shen A, and Camilli A (2018) Genome-wide detection of conservative site-specific recombination in bacteria. PLoS Genet 14: e1007332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M, and Simon M (1980) Phase variation: genetic analysis of switching mutants. Cell 19: 845–854. [DOI] [PubMed] [Google Scholar]
- Silverman M, Zieg J, Hilmen M, and Simon M (1979) Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc Natl Acad Sci U S A 76: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NH, and Selander RK (1990) Sequence invariance of the antigen-coding central region of the phase 1 flagellar filament gene (fliC) among strains of Salmonella typhimurium. Journal of Bacteriology 172: 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, et al. (2009) Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struve C, Bojer M, and Krogfelt KA (2008) Characterization of Klebsiella pneumoniae Type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infection and Immunity 76: 4055–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauseef I, Ali YM, and Bayliss CD (2013) Phase variation of PorA, a major outer membrane protein, mediates escape of bactericidal antibodies by Neisseria meningitidis. Infect Immun 81: 1374–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Murray BE, and Weinstock GM (1998) Conjugal Transfer of Plasmid DNA from Escherichia coli to Enterococci: a method to make insertion mutations. Plasmid 39: 182–186. [DOI] [PubMed] [Google Scholar]
- Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, and Buttner MJ (2014) Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158: 1136–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Putte P, and Goosen N (1992) DNA inversions in phages and bacteria. Trends in Genetics 8: 457–462. [DOI] [PubMed] [Google Scholar]
- Weiss CA, Hoberg JA, Liu K, Tu BP, and Winkler WC (2019) Single-cell microscopy reveals that levels of Cyclic di-GMP vary among Bacillus subtilis subpopulations. J Bacteriol 201: e00247–19, /jb/201/16/JB.00247–19.atom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieg J, and Simon M (1980) Analysis of the nucleotide sequence of an invertible controlling element. Proc Natl Acad Sci U S A 77: 4196–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phenotypes of UK1_T and UK1_O cells. Single UK_T and UK_O colonies were streaked/ inoculated on TY agar (A)/ TY broth (B) and were observed for phase variation. Few UK_T colonies gave rise to UK_O (marked with black arrows) within 20h of incubation, while the UK_O colonies maintained the opaque phenotype. Upon longer incubation (72h), both UK_T and UK_O colonies gave rise to the opaque and translucent colonies, respectively. Few of the UK_O colonies giving to translucent cells are marked with white arrows. At least 6 plates were tested and observed.
Figure S2. Swimming motility and toxin production of UK1_T and UK1_O strains.
A. Swimming motility exhibited by UK1_T and UK1_O in TY+0.5% agar. B. Cytoplasmic toxin levels produced by UK1_T and UK1_O morphotypes during the stationary phase of growth. Three biological replicates were used to carry out the experiments, and data were analyzed by a two-tailed unpaired t-test with Welch’s correction (NS indicates not significant).
Figure S3. UK1_O undergoes DNA inversion upstream of CDR20291_0685.
Alignment of 192 bp segment of the upstream region of CDR20291_0685 from UK1_T and UK_O with the published R20291 sequence. Inverted repeat sequence bordering the sequence is marked in bold.
Figure S4. Construction and confirmation of the pdcB mutation in C. difficile UK1 strain.
A. Schematic representation of ClostTron (group II intron) mediated insertional inactivation of pdcB gene in C. difficile. B. PCR verification of the intron insertion in pdcB in UK1 strain, conducted with intron-specific primer EBS universal [EBS(U)] with pdcB specific primers ORG 771 and ORG 772.
Figure S5. CRISPRi knock-down of pdcB in UK1. Transconjugants carrying pdcB-CRISPRi plasmid were streaked on to TY+Thio plates with or without 1% xylose. Phase contrast microscopic images (A) and sporulation percentage (B) of pdcB-sgRNA induced and uninduced cultures. These are representative results from two independent experiments. Data were analyzed by two-tailed unpaired t-test with Welch’s correction where ***=p<0.005
Figure S6. Increased c-di-GMP is associated with increased swarming motility phenotype in UK1_T strain.
A. Representative swarming motility plates of pdcB (OFF), pdcB (ON), UK1::pdcB mutant, UK1::pdcB + PdcB- EAL and UK1::pdcB + PdcA- EAL complemented strains after 72h of incubation at 37°C.
Figure S7. Orientation of pdcB and cmrRST upstreams in UK_T and UK_O cells. A.PCR amplification of the upstream region of pdcB using primer mixture containing primers ORG886/887/888. Amplification with ORG886/888 results in a product size of 325 bp, while amplification with ORG 887/888 results in a product size of 215 bp. B. PCR amplification of the upstream region of cmrRST using primer mixture containing primers ORG879/880/881. Amplification with ORG879/880 results in a product size of 410 bp, while ORG 880/881 results in 540 bp PCR product. Single bacterial colony of a specified phenotype was resuspended in 50µl TE buffer and was boiled for 10 minutes. The boiled lysate was then centrifuged and 2µl of the supernatant was used a template to perform PCR. At least ten colonies from each phenotype was tested.
Figure S8. Analysis of UK1::recV mutant. A. Schematic representation of ClostTron (group II intron) mediated insertional inactivation of recV gene in C. difficile. B. PCR verification of the intron insertion in recV in UK1 strain, conducted with intron-specific EBS primer with recV specific primers ORG 788 and ORG 789. C. Phase contrast microscopy image of UK1::recV mutant strain. D. Orientation PCR to check the ON and OFF position of pdcB.


