Abstract
We have investigated the ability of dexamethasone to regulate interleukin-1β (IL-1β)-induced gene expression, histone acetyltransferase (HAT) and histone deacetylase (HDAC) activity. Low concentrations of dexamethasone (10−10 M) repress IL-1β-stimulated granulocyte-macrophage colony-stimulating factor (GM-CSF) expression and fail to stimulate secretory leukocyte proteinase inhibitor expression. Dexamethasone (10−7 M) and IL-1β (1 ng/ml) both stimulated HAT activity but showed a different pattern of histone H4 acetylation. Dexamethasone targeted lysines K5 and K16, whereas IL-1β targeted K8 and K12. Low concentrations of dexamethasone (10−10 M), which do not transactivate, repressed IL-1β-stimulated K8 and K12 acetylation. Using chromatin immunoprecipitation assays, we show that dexamethasone inhibits IL-1β-enhanced acetylated K8-associated GM-CSF promoter enrichment in a concentration-dependent manner. Neither IL-1β nor dexamethasone elicited any GM-CSF promoter association at acetylated K5 residues. Furthermore, we show that GR acts both as a direct inhibitor of CREB binding protein (CBP)-associated HAT activity and also by recruiting HDAC2 to the p65-CBP HAT complex. This action does not involve de novo synthesis of HDAC protein or altered expression of CBP or p300/CBP-associated factor. This mechanism for glucocorticoid repression is novel and establishes that inhibition of histone acetylation is an additional level of control of inflammatory gene expression. This further suggests that pharmacological manipulation of of specific histone acetylation status is a potentially useful approach for the treatment of inflammatory diseases.
In the resting cell, DNA is tightly compacted to prevent transcription factor accessibility. During activation of the cell this compact inaccessible DNA is made available to DNA-binding proteins, thus allowing the induction of gene transcription (3, 42). DNA is packaged into chromatin, a highly organized and dynamic protein-DNA complex. The fundamental subunit of chromatin, the nucleosome, is composed of an octomer of four core histones, an H3-H4 tetramer and two H2A-H2B dimers, surrounded by 146 bp of DNA (3, 4). The packaging of DNA into nucleosomes acts as a barrier to the initiation of transcription by preventing the access of transcription factors, and RNA polymerase II, to their cognate recognition sequences (43). The N-terminal tails of the core histones contain highly conserved lysines that are sites for posttranscriptional acetylation. In addition, core histones may be modified by phosphorylation, methylation, ADP-ribosylation, or ubiquitinization of specific amino acid residues (44). Histone acetylation is thought to be a dynamic process which occurs on actively transcribed chromatin only (27).
There is compelling evidence that increased gene transcription is associated with an increase in histone acetylation, whereas hypoacetylation is correlated with reduced transcription or gene silencing (40, 42). Targeted acetylation of histone H4 tails plays an important role in allowing regulatory proteins to access DNA and is likely to be a major factor in the regulation of gene transcription (20, 24, 31).
Glucocorticoids are the most effective therapy for the treatment of inflammatory diseases such as asthma, a chronic inflammatory disease of the airway (1). Functionally, they act partly by inducing some anti-inflammatory genes, such as secretory leukocyte proteinase inhibitor (SLPI) (32), Lipocortin-1 (9), and interleukin-1 (IL-1) receptor antagonist (21), but mainly by repression of inflammatory genes, such as cytokines, adhesion molecules, inflammatory enzymes, and receptors (1). They act by binding to a cytosolic glucocorticoid receptor (GR), which upon binding is activated and rapidly translocates to the nucleus. Within the nucleus, GR either induces gene transcription by binding to specific DNA elements in the promoter-enhancer regions of responsive genes or reduces gene transcription by transrepression (37). GR reduces gene transcription by interaction with proinflammatory transcription factors such as AP-1 (Fos-Jun heterodimers) and NF-κB (p65-p50 heterodimers) (2, 29, 37). Both AP-1 and NF-κB and GR mutually repress each other's ability to activate transcription (15) and require the coactivator CREB binding protein (CBP) for maximal activity (11, 16, 26). This suggests that reduction of gene expression by GR may involve interference with transactivation mediated by coactivators such as CBP (34), possibly due to competition (squelching) for limiting amounts of the CBP (16). These studies also suggest that alterations in chromatin structure may be important in modulating glucocorticoid actions. Indeed, it has previously been reported that sodium butyrate, a histone deacetylase (HDAC) inhibitor, interferes with GR-activated transcription (28).
Many of these studies have relied on the overexpression of components of these pathways, which could lead to problems in interpretation. We have therefore examined the role of CBP and associated factors in the regulation of glucocorticoid functions in nontransfected cells. We have investigated the ability of dexamethasone to suppress expression of the inflammatory cytokine granulocyte-macrophage colony-stimulating factor (GM-CSF), to induce SLPI, and to regulate histone acetylation and deacetylation in A549 cells. We demonstrate that dexamethasone shows a different pattern of histone H4 acetylation from that seen with IL-1β and at low concentrations (10−10 M) represses IL-1β-stimulated histone acetylation. This does not involve induction of HDAC protein or activity or squelching of CBP. The mechanism of GR repression of IL-1β-stimulated histone H4 K8 and K12 acetylation was by direct inhibition of CBP-associated histone acetyltransferase (HAT) activity and by active recruitment of a histone deacetylase complex (HDAC2).
MATERIALS AND METHODS
Cell culture.
A549 cells were grown to 50% confluence in Dulbecco modified Eagle medium containing 10% fetal calf serum (FCS) before incubation for 48 to 72 h in serum-free medium. Cells were stimulated by IL-1β (1 ng/ml) in the presence or absence of dexamethasone, and the effects of the HDAC inhibitor trichostatin A (TSA) (Sigma, Poole, United Kingdom) (46) on baseline and IL-1β-stimulated expression of GM-CSF and SLPI release were measured.
GM-CSF, SLPI, and acetylated histone ELISAs.
Determination of GM-CSF expression was measured by sandwich ELISA (enzyme-linked immunosorbent assay; Pharmingen, Lugano, Switzerland) according to the manufacturer's instructions. For immunoassay of SLPI and acetylated histone, polystyrene microtiter plates were coated overnight at 4°C with sample diluted with hydroxy carbonate (pH 9.6). Plates were blocked for 2 h with 5% ovalbumin in phosphate-buffered saline (PBS). Antibodies against SLPI (R&D Systems Europe, Abingdon, United Kingdom), K5, K8, K12, and K16 acetylated histone 4 (Serotec, Oxford, United Kingdom) (39) were diluted 1:300 to 1:1,000 and added to each plate. After 1 h at room temperature, the plates were washed sequentially with 0.1% Tween 20–PBS and incubated with biotinylated goat anti-rabbit antibody (Dako, Cambridge, United Kingdom) for 1 h. Detection was performed using horseradish peroxidase-streptavidin according to Pharmingen instructions. Recombinant human SLPI (R&D Systems Europe) was used as a standard. As a standard for acetylated histone, crude extracted histone from A549 cells incubated with TSA (100 ng/ml) for 6 h was used, and the value was calculated in units, where 1 U is equivalent to the absorbance of 50 ng of TSA-treated hyperacetylated histone after subtraction of bovine serum albumin (BSA)-induced histone acetylation.
Direct histone extraction.
Histones were extracted from nuclei overnight using HCl and H2SO4 at 4°C using a method modified from that as described by Turner and fellows (38) Yoshida et al. (38). Cells were microfuged for 5 min, and the cell pellets were extracted with ice-cold lysis buffer (10 mM Tris-HCl, 50 mM sodium bisulfite, 1% Triton X-100, 10 mM MgCl2, 8.6% sucrose, complete protease inhibitor cocktail [Boehringer-Mannheim, Lewes, United Kingdom]) for 20 min at 4°C. The pellet was repeatedly washed in buffer until the supernatant was clear (centrifuged at 8,000 rpm for 5 min after each wash), and the nuclear pellet was washed in nuclear wash buffer (10 mM Tris-HCl, 13 mM EDTA) and resuspended in 50 μl of 0.2 N HCl and 0.4 N H2SO4 in distilled water. The nuclei were extracted overnight at 4°C, and the residue was microfuged for 10 min. The supernatant was mixed with 1 ml ice-cold acetone and left overnight at −20°C. The sample was microfuged for 10 min, washed with acetone, dried, and diluted in distilled water. Protein concentrations of the histone-containing supernatant were determined by Bradford protein assay kit (Bio-Rad, Hemel Hempstead, United Kingdom).
Western blotting.
Immunoprecipitates, whole-cell extractions, or isolated histones were measured by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis using enhanced chemiluminescence (ECL; Amersham, Amersham, United Kingdom). Proteins were size fractionated by SDS-PAGE and transferred to Hybond-ECL membranes. Immunoreactive bands were detected by ECL.
Immunocytochemistry.
A549 cells (0.5 × 106) were cultured in eight-well slide chambers with IL-1β (1 ng/ml) in the presence of dexamethasone. Cells were washed with Hanks balanced salt solution and air dried for 30 min at room temperature. Cells were then fixed in ice-cold acetone-methanol (50:50 [wt/wt]) at −20°C for 10 min. Slides were air dried and incubated with blocking buffer (20% normal swine serum in PBS plus 0.1% saponin) (Dako) for 20 min, followed by a 1-h incubation with primary antibody solution (PBS, 0.1% saponin, 1% BSA). Antibodies against pan-acetylated H4, H4-K5, H4-K8, H4-K12, and H4-K16 (Serotec) (39) were used at a 1:100 to 1:300 dilution. Slides were washed twice and incubated with biotinylated swine anti-rabbit immunoglobulin G (IgG; Dako) (1:200) for 45 min. Slides were washed again before incubation with fluorescein isothiocyante-conjugated streptavidine (1:100) for 45 min. The slides were washed twice more before counterstaining them with 20% hematoxylin and then mounting them. Stained cells were observed by confocal microscopy. Confocal scanning laser microscopy images were collected with a Leica confocal microscope, equipped with a 488- and 514-nm dual-band argon ion laser. An oil immersion objective lens was used, and images were collected using TCSNT software.
Histone acetylation activity.
Cells were plated at a density of 0.25 × 106 cells/ml and exposed to 0.05 mCi/ml of [3H]acetate (Amersham). After incubation for 10 min at 37°C cells were stimulated for 6 h. Histones were isolated and separated by SDS–16% PAGE. Gels were stained with Coomassie brilliant blue, and the core histones (H2A, H2B, H3, and H4) were excised. The radioactivity in extracted core histones was determined by liquid scintillation counting and normalized to protein level.
Histone deacetylation activity.
Radiolabeled histones were prepared from A549 cells following incubation with TSA (100 ng/ml, 6 h) in the presence of 0.1 mCi of [3H]acetate per ml. Histones were dried and resuspended in distilled water. Crude HDAC preparations were extracted from total cellular homogenates with Tris-based buffer (10 mM Tris-HCl, pH 8.0; 500 mM NaCl; 0.25 mM EDTA; 10 mM 2-mercaptoethanol) as previously reported (18). The crude HDAC preparation or immunoprecipitates were incubated with 3H-labeled histone for 30 min at 30°C before the reaction was stopped by the addition of 1 N HCl–0.4 N acetic acid. Released 3H-labeled acetic acid was extracted by ethyl acetate, and the radioactivity of the supernatant was determined by liquid scintillation counting.
IP.
Extracts were prepared using 100 μl of stringent immunoprecipitation (IP) buffer (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 1.0% Triton X-100; 0.5% NP-40; 0.1% SDS; 0.5% deoxycholate; complete protease inhibitor cocktail [Boehringer-Mannheim]) or mild IP buffer (10 mM Tris-HCl, pH 8.0; 150 mM NaCl; 0.5% NP-40; complete protease inhibitor cocktail [Boehringer-Mannheim]). The lysis mixture was incubated on ice for 15 min and microfuged for 10 min at 4°C. Extracts were precleared with 20 μl of A/G agarose (a 50:50 mix; Santa Cruz, Santa Cruz, Calif.) and 2 μg of normal IgG. After microcentrifugation, 20 μl of A/G agarose conjugated with 5 μg of antibody was used to precipitate CBP, p300/CBP-associated factor (PCAF), or GR overnight at 4°C or for 4 h at 4°C for p65 with rotation. The immune complexes were pelleted by gentle centrifugation and washed three times with 1 ml of IP buffer. For the HAT or HDAC assay, immunoprecipitates were washed twice with IP-HAT buffer, and for Western blotting, after a final wash with IP buffer, the buffer was aspirated completely and resuspended in Laemmli buffer.
Purification of GR.
GR was purified from 5 × 109 A549 cells. Total cellular proteins were isolated, and GR was immunoprecipitated as described above using a mouse anti-GR antibody (Serotec). The immunoprecipitate was separated by SDS–8% PAGE, and GR was purified from the excised gel by electroelution according to the manufacturer's instructions (Bio-Rad model 422) and used at a concentration of 10 ng/ml.
IP-HAT assay.
IP-HAT assays were performed using a modified method of Ogryzko et al. (25). Immune complexes with resin were resuspended in 150 μl of HAT buffer (50 mM Tris-HCl, pH 8.0; 10% glycerol; 1 mM dithiothreitol; 0.1 mM EDTA, complete protease inhibitor cocktail). Typically, 20 μl of free core histone solution extracted from A549 cells (final amount, 10 μg) and 30 μl of immunoprecipitate were incubated. Reactions were initiated by the addition of 0.25 μCi of [3H]acetyl coenzyme A (5 Ci/mmol) (Amersham) and were performed for 45 min at 30°C. After incubation, the reaction mixture was spotted onto Whatman p81 phosphocellulose filter paper (Whatman), washed for 30 min with 0.2 M sodium carbonate buffer (pH 9.2) at room temperature with two to three changes of the buffer, and then washed briefly with acetone. The dried filters were counted in a liquid scintillation counter.
Metabolic labeling.
For 32P labeling, cells were cultured in FCS-free medium for 2 days before incubation in a phosphate-free medium for 2 h. Cells were incubated in a phosphate-free medium containing 3 mCi of [32P]orthophosphate (40 μCi/ml; Amersham) for 30 min and then stimulated with IL-1β (1 ng/ml). The cultures were incubated for 6 h at 37°C in an atmosphere of 5% CO2. Cells were collected and lysed with mild IP buffer. Immunoprecipitates of anti-CBP antibody were separated by SDS-7% PAGE and either visualized by using an autoradiograph or quantified by counting the excised radioactive bands.
Chromatin IP (ChIP) assay.
A-549 cells were treated with IL-1β (1 ng/ml) in the presence of dexamethasone as described above. After a 4-h incubation, protein-DNA complexes were fixed by formaldehyde (1%, final concentration) and treated as previously described (13). Cells were resuspended in 200 μl of SDS lysis buffer (50 mM Tris, pH 8.1; 1% SDS; 5 mM EDTA; complete proteinase inhibitor cocktail) and sonicated (three 10-s pulses) on ice. Sonicated samples were centrifuged to spin down cell debris, and the soluble chromatin was immunoprecipitated using sonicated salmon sperm DNA agarose A slurry (Upstate Biotechnology, Buckingham, United Kingdom) as described by Chen et al. (7). Protein-bound immunoprecipitated DNA was washed with LiCl wash buffer and Tris-EDTA (TE), and immune complexes were eluted by adding elution buffer (1% SDS, 0.1 M NaHCO3). The elution was treated successively for 4 h at 65°C in 200 mM NaCl–1% SDS to reverse cross-links and then incubated for 1 h at 45°C with 70 μg of proteinase K (Sigma) per ml, DNA extracted with phenol-chloroform, precipitated with ethanol–0.3 M NaHCOOH–20 μg of glycogen, and resuspended in 50 μl of TE. Quantitative PCR was performed with 10 μl of DNA sample and 30 cycles. Primer pairs of GM-CSF and SLPI were as follows: GM-CSF, forward, 5-CTGACCACCTAGGGAAAAGGC-3; and GM-CSF, reverse, 5-CAGCCACATCCTCCTCCAGAGAAC-3; and SLPI, forward, 5-TCATAGCCTTACCTGGCATAG-3; and SLPI, reverse, 5-TGGACTTCATGGTGAAGGCAG-3. PCR products were resolved by using 3% agarose gel and visualized with ethidium bromide.
Statistics.
Results are expressed as means ± the standard error of the mean (SEM). A multiple comparison was made between the mean of the control and the means from each individual treatment group by Dunnett's test using SAS STAT software (SAS Institute, Inc., Cary, N.C.). All statistical testing was performed using a two-sided 5% level of significance. The concentrations of dexamethasone or trichostatin A producing 50% inhibition (IC50) were calculated from concentration-response curves by linear regression.
RESULTS
Evidence for a role of histone acetylation in IL-1β and dexamethasone-induced gene expression.
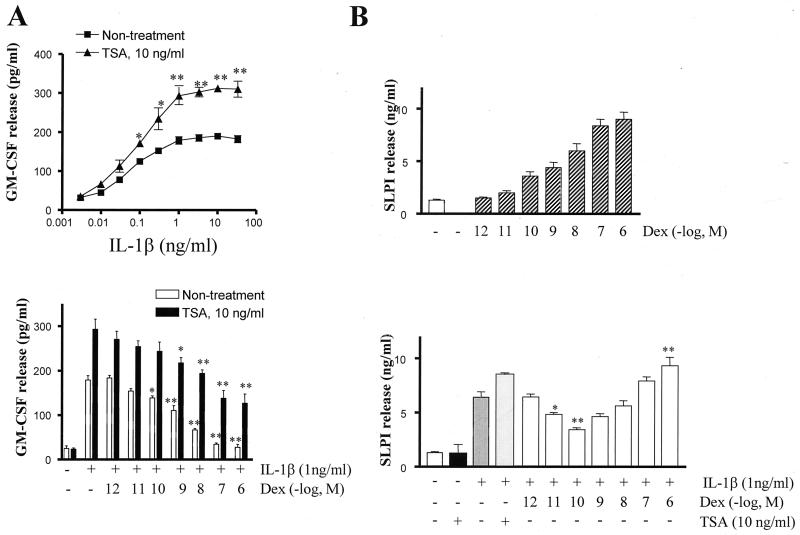
IL-1β (1 ng/ml) stimulated GM-CSF production in A549 cells after 6 h (179 ± 10 versus 25 ± 6 pg/ml). No induction of GM-CSF release was seen before 4 h and a maximum was reached at 24 h. The HDAC inhibitor, TSA gave a concentration-dependent decrease in HDAC activity with an IC50 similar to that previously reported (1.1 pg/ml) (23). This was associated with a marked increase in histone acetylation as measured by [3H]acetate incorporation and by Western blotting analysis (data not shown). In addition, TSA (10 ng/ml) enhanced IL-1β-induced GM-CSF release (293 ± 23 versus 179 ± 10 pg/ml) (Fig. 1A).
FIG. 1.
Histone acetylation is associated with IL-1β- and dexamethasone (Dex)-induced gene expression. (A) Effect of TSA (10 ng/ml) on IL-1β-stimulated GM-CSF release (upper panel). Cells were stimulated with IL-1β (1 ng/ml) for 6 h. Supernatants were collected and assayed for GM-CSF by ELISA. ∗, P < 0.05; ∗∗, P < 0.01 versus nontreatment. (Lower panel) Effect of 30 min of preincubation with dexamethasone on IL-1β stimulated GM-CSF release. The effect of TSA (10 ng/ml) on dexamethasone inhibition of IL-1β stimulated GM-CSF release was also measured. Results are expressed as mean ± the SEM (n = at least three independent experiments; ∗, P < 0.05, or ∗∗, P < 0.01, versus IL-1β alone). (B) Effects of dexamethasone and IL-1β on SLPI production. Cells were treated with dexamethasone alone (upper panel) or preincubated with dexamethasone for 30 min before incubation with IL-1β (1 ng/ml) for 6 h (lower panel). Supernatants were assayed by ELISA. The effects of TSA (10 ng/ml) on IL-1β-stimulated SLPI release were also measured. Results are expressed as mean ± the SEM (n = at least three independent experiments; ∗, P < 0.05, and ∗∗, P < 0.01, versus IL-1β alone).
IL-1β (1 ng/ml) also increased SLPI production (6.4 ± 0.5 versus 1.3 ± 0.1 ng/ml) (Fig. 1B). This effect was further enhanced by pretreatment with TSA (10 ng/ml) (8.6 ± 0.6 versus 6.4 ± 0.5 ng/ml), whereas TSA alone had no effect.
Role of histone acetylation in dexamethasone-mediated actions.
We next investigated the effect of dexamethasone on IL-1β-stimulated mediator release. Dexamethasone produced a concentration-dependent inhibition of IL-1β-stimulated GM-CSF release, which was maximal at 10−6 M (IC50 of 1.0 × 10−9 M) (Fig. 1A). The inhibitory effect of dexamethasone on IL-1β-induced GM-CSF production was shifted 78-fold to the right in the presence of TSA (10 ng/ml) (IC50 of 7.8 × 10−8 versus 1.0 × 10−9 M), suggesting an involvement of HDACs in the inhibitory actions of dexamethasone (Fig. 1A). In addition, the maximal inhibition achieved was 62%. These results suggest a possible role for histone acetylation and/or deacetylation in the regulation of GM-CSF expression by dexamethasone.
IL-1β (1 ng/ml) and dexamethasone were able to stimulate SLPI production. Dexamethasone caused a concentration-dependent induction of SLPI which reached a maximum at 10−6 M (EC50 = 0.9 × 10−8 M) (Fig. 1B). In contrast, dexamethasone had a biphasic effect on IL-1β-stimulated SLPI production. Low concentrations of dexamethasone (10−10 M) inhibited IL-1β-stimulated SLPI production, whereas higher concentrations of dexamethasone, acting through a glucocorticoid response element (GRE), overcomes this repression and stimulates SLPI release (Fig. 1B). These data suggest that the ability of dexamethasone to inhibit IL-1β-stimulated gene transcription (GM-CSF) occurs at lower concentrations than those required to stimulate gene transcription (SLPI).
Chromatin acetylation is associated with transcriptional activation by IL-1β and dexamethasone.
IL-1β caused both a time- and concentration-dependent four- to fivefold increase in total cellular histone acetylation which preceded GM-CSF production by IL-1β (data not shown). This induction was maximal at 1 ng/ml (137 ± 15 versus 25 ± 3 dpm/μg of protein) and was detectable 30 min after IL-1β stimulation (41 ± 6 versus 18 ± 4 dpm/μg of protein). The stimulation peaked between 4 and 8 h and returned to control levels after 24 h. TSA (1 ng/ml) enhanced both basal (162 ± 21 versus 50 ± 5 dpm/μg of protein) and IL-1β-stimulated (1,543 ± 143 versus 137 ± 15 dpm/μg of protein) histone acetylation. Dexamethasone also produced a time- and concentration-dependent increase in histone acetylation, with a maximum induction at between 4 and 8 h at concentrations of ≥10−8 M (data not shown). TSA enhanced the basal (162 ± 21 versus 20 ± 5 dpm/μg of protein) and dexamethasone-induced (984 ± 50 versus 71 ± 9 dpm/μg of protein) histone acetylation. In subsequent experiments, histone acetylation was measured at 6 h following IL-1β stimulation in the presence of dexamethasone.
Immunofluorescence and confocal microscopy confirmed these results (data not shown). This analysis also showed that IL-1β, but not dexamethasone or TSA, caused nuclear translocation of p65, while dexamethasone, but not IL-1β or TSA, enhanced GR nuclear translocation.
Specific targeting of histone H4 lysine residues by IL-1β and dexamethasone.
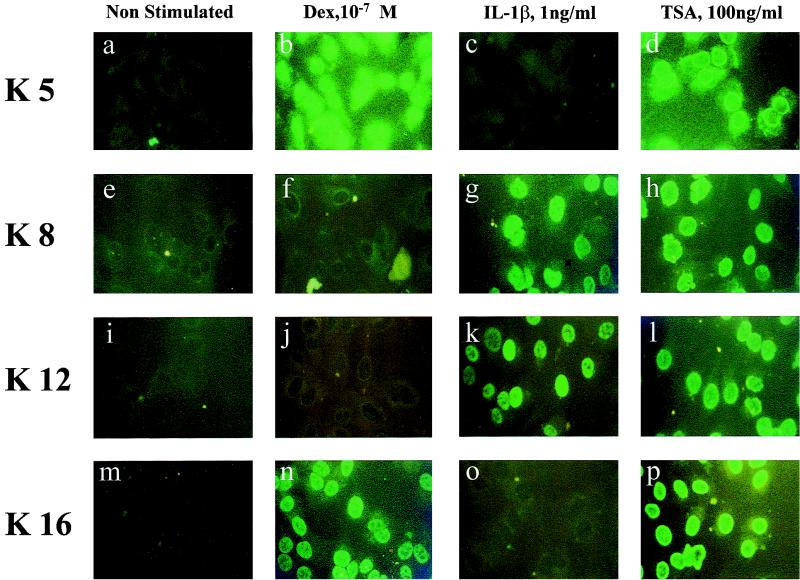
We determined the pattern of lysine acetylation following IL-1β and dexamethasone stimulation. Dexamethasone targeted acetylation on histone H4 lysines K5 and K16 (53 ± 9% versus 36 ± 16% positive nuclei), while IL-1β induced K8 and K12 acetylation (42 ± 15% versus 37 ± 4% positive nuclei). IL-1β also produced a much weaker nuclear staining for acetylated K5 than that seen with dexamethasone (Fig. 2).
FIG. 2.
IL-1β and dexamethasone (Dex) acetylate specific and distinct lysine residues. Immunocytochemical staining for specific histone H4 acetylated lysine residues. Cells were incubated with dexamethasone (10−7 M) (b, f, j, and n), IL-1β (1 ng/ml) (c, g, k, and o), or TSA (100 ng/ml) (d, h, l, and p) for 6 h before staining for acetylated forms of histone H4 lysine residues K5 (a to d), K8 (e to h), K12 (i to l), and K16 (m to p). Results are representative of four independent experiments.
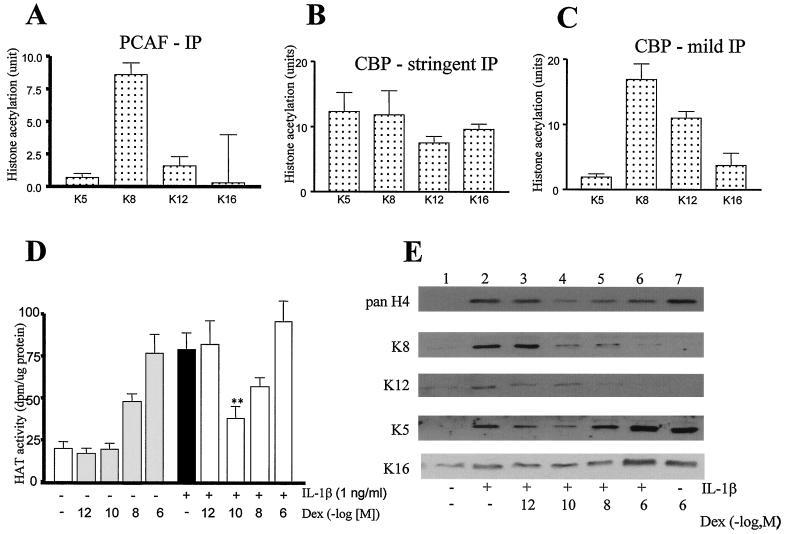
Acetylation of specific lysine residues is mediated through the HAT activities of coactivator molecules, including CBP and PCAF. We therefore examined the possible role of CBP and PCAF in mediating IL-1β-stimulated acetylation of specific histone H4 lysine residues. PCAF was immunoprecipitated from IL-1β-stimulated cells and incubated with histones and acetyl coenzyme A (acetyl-CoA), and acetylated K residues were detected by immunoassay. These results showed that PCAF stimulated K8 acetylation (Fig. 3A), confirming data from Schiltz et al. (33). CBP isolated under stringent IP conditions was able to acetylate all histone H4 lysines (Fig. 3B). In contrast, CBP complexes isolated using mild IP conditions predominantly acetylated K8 and K12 (Fig. 3C), confirming the immunocytochemistry results. This suggests that IL-1β may stimulate K8 and K12 acetylation through a CBP-associated HAT rather than directly through CBP alone.
FIG. 3.
Effects of dexamethasone (Dex) on IL-1β-induced histone acetylation. (A) PCAF acetylates specific histone residues. Cells were treated with IL-1β (1 ng/ml) for 1 h before total cellular proteins were extracted. PCAF was immunoprecipitated under stringent IP conditions and incubated with histones and acetyl-CoA (see Materials and Methods). Using antibodies against specific acetylated lysine residues the level of histone acetylation in the IP sample was measured by immunoassay. Histone acetylation at each lysine residue is expressed in units (1 U is equivalent to the absorbance produced by 50 ng of TSA-treated hyperacetylated histone). Results are expressed as mean ± the SEM (n = at least three independent experiments). (B) CBP acetylation of histone H4 lysine residues. Cells were treated with IL-1β (1 ng/ml) for 1 h before total cellular proteins were extracted. CBP was immunoprecipitated under stringent IP conditions and incubated with histones and acetyl-CoA (see Materials and Methods). Using antibodies against specific acetylated lysine residues the level of histone acetylation in the IP sample was measured by immunoassay. (C) CBP-associated proteins acetylate specific lysine residues. Cells were treated with IL-1β (1 ng/ml) for 1 h before total cellular proteins were extracted. CBP was immunoprecipitated under mild IP conditions and incubated with histones and acetyl-CoA (see Materials and Methods). Using antibodies against specific acetylated lysine residues the level of histone acetylation in the IP sample was measured by immunoassay. (D) Dexamethasone inhibits IL-1β-induced histone acetylation in total cell extracts. Cells were pretreated with dexamethasone for 30 min before incubation with IL-1β (1 ng/ml) for 1 h in the presence of 0.05 mCi of [3H]acetate. Histones were isolated and separated by SDS-PAGE, and [3H]acetate incorporated histones were counted and normalized to the protein level. The data represent the means ± the SEM of three independent experiments. ∗∗, P < 0.01. (E) Western blot analysis of dexamethasone actions on IL-1β-stimulated histone acetylation. Cells were incubated with IL-1β (1 ng/ml) for 6 h in the presence of increasing concentrations of dexamethasone. Protein extracts were obtained and examined for pan-acetylated histone H4 lysine residues and for specific K5, K8, K12, and K16 acetylation by Western blotting. Lanes: control (lane 1); IL-1β stimulation (lane 2); IL-1β stimulation in the presence of dexamethasone at 10−12 M (lane 3), 10−10 M (lane 4), 10−8 M (lane 5), and 10−6 M (lane 6); and dexamethasone, 10−6 M alone (lane 7). The results are representative of three independent experiments.
Dexamethasone targets IL-1β-stimulated acetylation of histone H4 K8, and K12.
We next examined whether IL-1β-stimulated K8 and K12 acetylation was a target for dexamethasone actions. Initial experiments were performed in whole-cell extracts from cells treated with IL-1β and or dexamethasone. IL-1β induced a fourfold increase in histone acetylation (Fig. 3D). Dexamethasone alone had no effect on basal histone acetylation. Dexamethasone had a biphasic effect on IL-1β-stimulated histone acetylation (Fig. 3D). Low concentrations of dexamethasone (10−10 M) inhibited IL-1β-stimulated histone acetylation, while higher concentrations of dexamethasone (10−8 and 10−6 M) returned [3H]acetate incorporation to levels seen with IL-1β alone (Fig. 3D). TSA (100 ng/ml) caused a marked elevation of IL-1β-stimulated histone acetylation (1,543 ± 143 versus 71 ± 9 dpm/μg of protein). In addition, IL-1β plus dexamethasone (10−10 M) stimulated histone acetylation to levels much greater than that seen with IL-1β treatment alone (435 ± 28 versus 71 ± 9 dpm/μg of protein).
Western analysis of specific acetylated lysines showed that dexamethasone inhibited IL-1β-stimulated K8 and K12 acetylation (Fig. 3E, lane 4). In addition, the small induction of K5 acetylation by IL-1β was also suppressed at low (10−12 and 10−10 M) concentrations of dexamethasone (Fig. 3E, lanes 1 to 4), whereas at higher concentrations (10−8 and 10−6 M) marked acetylation of K5 occurred (Fig. 3E, lanes 5 and 6). Dexamethasone also enhanced K16 acetylation at higher concentrations (10−8 and 10−6 M). These data suggest that dexamethasone at low concentrations can inhibit histone acetylation induced by IL-1β, whereas at higher concentrations dexamethasone can itself induce histone acetylation at specific target lysine residues.
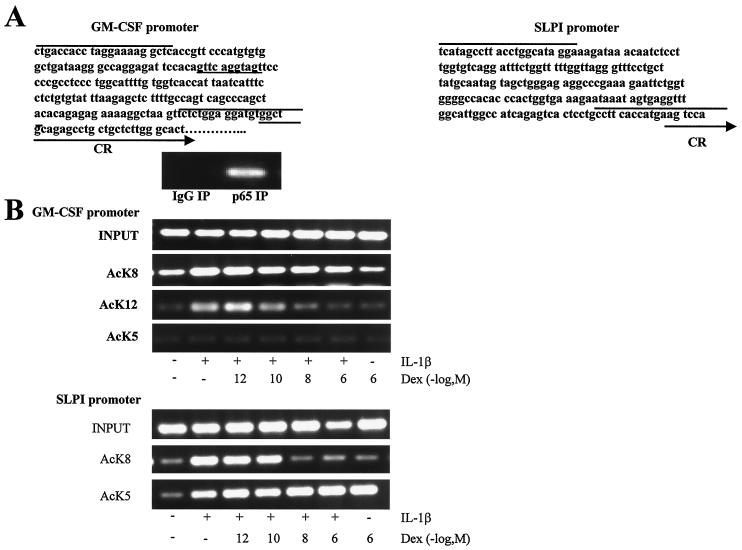
IL-1β increases K8 and K12 acetylation associated with the GM-CSF promoter.
The previous data examined gross histone acetylation. It was essential, therefore, to determine whether the interaction of the p65-activated HAT complex with GR occurs specifically on the GM-CSF and SLPI promoters. We analyzed the nucleosomal events involved in GM-CSF transactivation by semiquantitative ChIP. Two different genomic sites were investigated: the GM-CSF (−191 to +10) and the SLPI (−170 to +32) promoters (Fig. 4A). PCR amplifications were carried out on a fixed amount of immunoprecipitated DNA, followed by 30 cycles of PCR with the appropriate primer pairs. After IL-1β treatment, p65 immunoprecipitates showed a marked enrichment of GM-CSF promoter DNA (Fig. 4A). IP with an antibody against acetylated K8 or K12 resulted in the enrichment for the DNA segments encompassing the GM-CSF promoter following IL-1β treatment (Fig. 4B). These data demonstrate that p65-mediated activation of the GM-CSF promoter in vitro is concomitant with the acetylation of histone H4 K8 and K12 residues. Increasing concentrations of dexamethasone caused a reduction in the enrichment of acetylated K8- and K12-associated GM-CSF promoter fragments (Fig. 4B). This effect correlated well with dexamethasone repression of GM-CSF release. In contrast, acetylated K5 residues were not associated with the GM-CSF promoter segment either at baseline or following IL-1β treatment (Fig. 4B). IP with an antibody against acetylated K8 resulted in the enrichment for the DNA segments encompassing the SLPI promoter following IL-1β treatment. IL-1β stimulation of cells had no effect on K5-associated SLPI promoter DNA. In contrast, dexamethasone caused a concentration-dependent increase in K5-associated DNA enrichment in both basal and IL-1β-treated cells (Fig. 4B). These data indicate that histone acetylation induced by IL-1β or dexamethasone occurs on specific lysine residues associated with distinct pro- and anti-inflammatory genes.
FIG. 4.
Association of specific acetylated lysine residues with GM-CSF and SLPI gene promoters. (A) GM-CSF and SLPI promoter regions. The sequence of the GM-CSF (−191 to +10) and SLPI (−170 to +32) promoter regions amplified by PCR primer pairs. Primers are indicated by overlined sequences. The NF-κB response element in the GM-CSF promoter is underlined. The coding region (CR) of each gene is indicated by an arrow. An enrichment of the GM-CSF promoter DNA is shown following PCR amplification of IP of p65-associated DNA from cells treated with IL-1β (1 ng/ml) for 1 h. (B) Specific lysine residue acetylation at the GM-CSF and SLPI promoters. Cells were incubated with IL-1β (1 ng/ml) in the presence of dexamethasone. Proteins and DNA were cross-linked by formaldehyde treatment, and chromatin pellets were extracted. Following sonication, acetylated histone H4 lysine residues (AcK5, AcK8, and AcK12) were immunoprecipitated, and the associated DNA was amplified by PCR. The results are representative of three independent experiments.
Effect of dexamethasone on p65-induced histone acetylation and deacetylation.
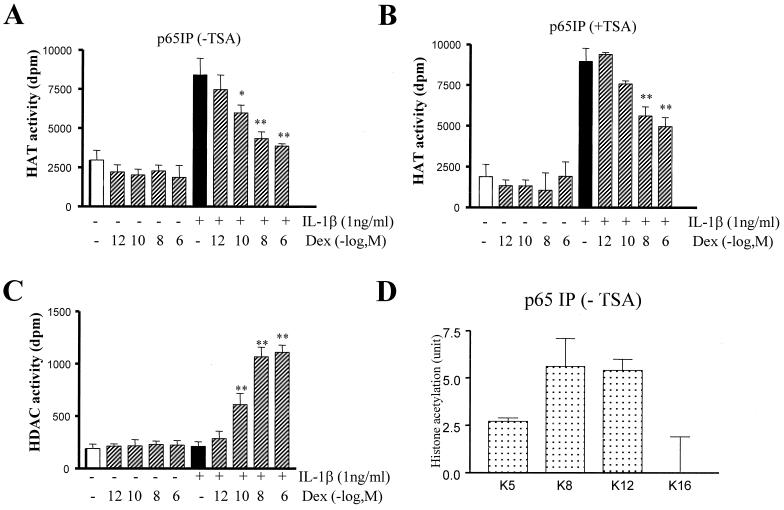
In order to clarify the inhibitory mechanism of dexamethasone on histone acetylation, we investigated p65-associated histone acetylation and deacetylation in IL-1β- and/or dexamethasone-stimulated cells. In some experiments the role of HDACs on dexamethasone action was examined by pretreating the cells with TSA (100 ng/ml). Whole-cell lysates were made, and p65 immunoprecipitates were isolated under mild IP conditions and examined for associated histone acetylation and deacetylation activity (Fig. 5). In these p65 IP experiments, histone acetylation was increased threefold following IL-1β stimulation (Fig. 5A). Dexamethasone inhibited p65-associated IL-1β-induced histone acetylation in a concentration-dependent manner (IC50 of 3.7 × 10−10 M). Dexamethasone alone produced no change in p65-associated histone acetylation from that seen in control untreated samples (Fig. 5A). Control experiments with anti-p65 antibody blocking peptide showed no histone acetylation (221 ± 122 dpm/μg of protein). TSA (100 ng/ml) caused a 50-fold shift in the dexamethasone concentration-response curve, suggesting that the inhibitory effects of dexamethasone require some HDAC involvement (IC50 of 1.9 × 10−8 versus 3.7 × 10−10 M) (Fig. 5B). In the same immunoprecipitates, dexamethasone enhanced histone deacetylation in a concentration-dependent manner (Fig. 5C). To confirm that the p65-IPs were acetylating the same lysine residues as IL-1β, p65-IPs were examined for specific forms of acetylated histone H4 lysines by ELISA. The p65-IPs targeted mainly K8 and K12 acetylation, with a smaller effect on K5 acetylation (Fig. 5D). These data confirmed the results seen by immunocytochemistry and CBP immunoprecipitates isolated under mild IP conditions (see Fig. 2 and Fig. 3C).
FIG. 5.
Dexamethasone (Dex) inhibits p65-associated histone acetylation: a role for HDAC. (A) Dexamethasone inhibits IL-1β-induced p65-immunoprecipitated histone acetylation. Cells were preincubated with dexamethasone for 30 min before IL-1β (1 ng/ml) treatment for a further 1 h. Total cellular proteins were isolated, and p65 was immunoprecipitated under stringent conditions. The associated histone acetylation activity was measured following incubation of the p65-immunoprecipitated extract with 10 μg of free core histones and 0.25 mCi of [3H]acetyl-CoA for 45 min. Radiolabeled histones were counted, and the results are presented as the mean ± the SEM of at least three independent experiments. ∗, P < 0.05; ∗∗, P < 0.01. (B) TSA represses dexamethasone inhibition of p65-associated histone acetylation. Histone acetylation experiments were performed as in panel A in the presence of TSA (100 ng/ml). This resulted in a reduced ability of dexamethasone to suppress p65-associated histone acetylation. Results are presented as the mean ± the SEM of at least three independent experiments. ∗∗, P < 0.01. (C) Effect of IL-1β and dexamethasone on p65-associated histone deacetylation. Using the same immunoprecipitates as in panel A, HDAC activity was measured by incubation of extracts with 3H-labeled histones for 30 min. Free 3H-labeled acetic acid was extracted and counted. The results are presented as the mean ± the SEM of at least three independent experiments. ∗∗, P < 0.01. (D) Specific lysine acetylation by p65. Cells were treated with IL-1β, and total cellular proteins were extracted. p65 was immunoprecipitated under stringent IP conditions and incubated with histones and acetyl-CoA. Using antibodies against specific acetylated lysine residues, the level of histone acetylation in the IP sample was measured by immunoassay. Histone acetylation at each lysine residue is expressed in units (1 U is equivalent to the absorbance produced by 50 ng of TSA-treated hyperacetylated histone). The results are expressed as the mean ± the SEM (n = at least three independent experiments).
Effect of dexamethasone on coactivator expression, association with p65, and phosphorylation.
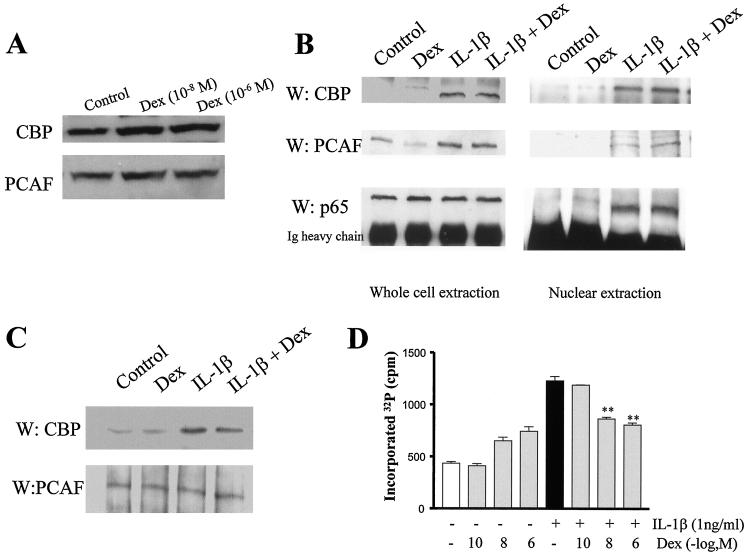
A number of coactivators may be involved in IL-1β-stimulated induction of histone acetylation and its subsequent amelioration by dexamethasone (10, 16, 26, 34). Initially, we examined the effect of dexamethasone on CBP and PCAF expression. Dexamethasone (10−8 and 10−6 M, 6 h) had no effect on CBP or PCAF expression, thus ruling out a reduction in CBP or PCAF expression as a mechanism for inhibiting IL-1β-stimulated histone acetylation (Fig. 6A). An alternative mechanism of dexamethasone action could be to reduce the interaction between the IL-1β-stimulated NF-κB p65 subunit and CBP or PCAF. Using p65 IP, followed by Western blotting, no difference was found in the ability of IL-1β to enhance p65-CBP or p65-PCAF interactions within whole-cell lysates or nuclear extracts following dexamethasone (10−6 M) cotreatment (Fig. 6B). Furthermore, dexamethasone did not inhibit p65 translocation (Fig. 5B) or IL-1β-induced CBP-PCAF association (Fig. 6C).
FIG. 6.
Effect of dexamethasone (Dex) on p65-associated coactivators and GR recruitment. (A) Effect of dexamethasone on CBP and PCAF expression. Cells were incubated with vehicle (control) or increasing concentrations of dexamethasone for 6 h. The results are representative of three independent experiments. (B) Effect of dexamethasone on CBP-p65 interaction and PCAF-p65 interaction. Cells were preincubated with vehicle (control), IL-1β (1 ng/ml), IL-1β and dexamethasone (10−6 M), or dexamethasone (10−6 M) alone for 1 h before total cellular or nuclear proteins were extracted. p65 IP was performed in mild IP buffer. Immunoprecipitates were separated by SDS-PAGE and detected by Western blotting. The results are representative of three independent experiments. (C) Effect of dexamethasone on CBP-PCAF interaction. Cells were preincubated with vehicle (control), IL-1β, or IL-1β and dexamethasone (10−6 M) before protein extraction and PCAF immunoprecipitation under mild IP conditions. The results are representative of three independent experiments. (D) Effect of dexamethasone on CBP phosphorylation. Cells were incubated with [32P]orthophosphate for 30 min before stimulation with IL-1β for 6 h in the presence of dexamethasone. Radioactive bands were excised and counted. The results are expressed as the mean ± the SEM (n = 3; ∗∗, P < 0.01).
Inhibition of phosphorylation by mitogen-activated protein kinase (MAPK) pathways by dexamethasone has been proposed to play an important role in glucocorticoid actions (6, 30, 36). These pathways may also regulate CBP activation by transcription factor phosphorylation or a direct effect on CBP, potentially altering histone acetylation and transactivation capabilities (8). IL-1β significantly induced CBP phosphorylation that was inhibited by dexamethasone (Fig. 6D). However, the concentrations of dexamethasone that repressed IL-1β-stimulated gene expression and histone acetylation had no effect on CBP phosphorylation. This suggests that although higher concentrations of dexamethasone can indeed inhibit CBP phosphorylation, this effect does not account for the repression of histone acetylation by dexamethasone. Direct acetylation has been shown to be important in the activity of some transcription factors and coactivators (5, 12, 14). However, there was no acetylation of CBP or PCAF in these cells following either IL-1β or dexamethasone treatment (data not shown).
Effect of dexamethasone on coactivator-associated histone acetylation.
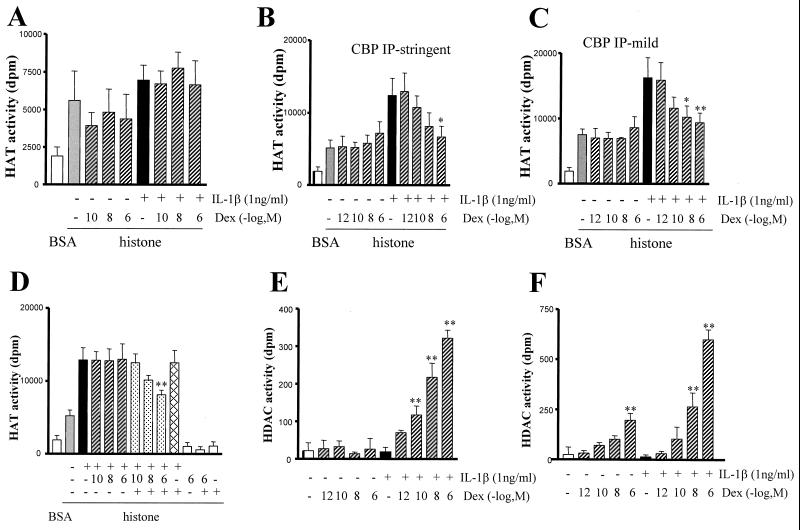
It has previously been shown that PCAF acetylates H4 K8 only (see Fig. 3A and reference 33), and our data showing IL-1β-induced acetylation of K8 and K12 suggest that PCAF alone is unlikely to mediate IL-1β-induced histone acetylation. Further evidence for a lack of a role for PCAF was suggested by a failure of cells treated with IL-1β and/or dexamethasone to show enhanced PCAF histone acetylase activity or for dexamethasone to modify PCAF activity (Fig. 7A).
FIG. 7.
Effect of dexamethasone (Dex) on IL-1β-stimulated CBP-associated histone acetylation and deacetylation activity. (A) No effect of IL-1β and dexamethasone on PCAF IP histone acetylation. Cells were preincubated with dexamethasone (30 min) before IL-1β treatment for 6 h. Total cellular proteins were extracted and PCAF was immunoprecipitated under stringent IP conditions. The associated histone acetylation activity was measured following incubation of the PCAF IP extract with 10 μg of free core histones and 0.25 mCi of [3H]acetyl-CoA for 45 min. Radiolabeled histones were counted, and the results are presented as the mean ± the SEM of at least three independent experiments. (B) Effect of dexamethasone on IL-1β-stimulated CBP immunoprecipitated histone acetylation. Cells were treated as in panel A, and CBP was immunoprecipitated under stringent IP conditions. CBP-associated HAT activity is presented as the mean ± the SEM of at least three independent experiments (∗, P < 0.05). (C) Effect of dexamethasone on IL-1β-stimulated CBP-associated histone acetylation. Cells were treated as in panel A, and CBP was immunoprecipitated under mild IP conditions. CBP-HAT activity is presented as the mean ± the SEM of at least three independent experiments (∗, P < 0.05). (D) Dexamethasone suppression of IL-1β-induced CBP-associated HAT activity requires GR. Cells were treated with IL-1β for 6 h, cellular proteins were extracted, and CBP was immunoprecipitated under stringent IP conditions. CBP IP was incubated with dexamethasone alone or with dexamethasone and GR together with [3H]acetyl-CoA for 45 min in the presence of TSA (100 ng/ml). Results are presented as the mean ± the SEM of at least three independent experiments (∗, P < 0.05). (E) Effect of IL-1β and dexamethasone on histone deacetylation. Using the same immunoprecipitates as in panel C, HDAC activity was measured by incubation of CBP IP extracts with 3H-labeled histones for 30 min. Free 3H-labeled acetic acid was extracted and measured by liquid scintillation counting. The results are presented as the mean ± the SEM of at least three independent experiments (∗∗, P < 0.01). (F) Effect of IL-1β and dexamethasone on GR-mediated histone deacetylation. Cells were treated as in panel A, and total cellular proteins were immunoprecipitated using an anti-GR antibody under stringent IP conditions. The results are presented as the mean ± the SEM of at least three independent experiments (∗∗, P < 0.01).
We have earlier shown that IL-1β stimulated CBP-associated HAT activity. We wanted to investigate whether this CBP-associated activity was a target for dexamethasone activity. Cells were stimulated with IL-1β and/or dexamethasone for 6 h, and CBP was immunoprecipitated under mild or stringent conditions. Histone acetylation assays were performed after the addition of exogenous histones. IL-1β caused an elevation in CBP-dependent histone acetylation under both stringent and mild IP conditions (Fig. 7B and C). This activity peaked at 4 h and returned to baseline by 24 h (data not shown). Dexamethasone caused a concentration-dependent reduction in IL-1β-stimulated CBP-associated histone acetylation (IC50 of 6.5 × 10−9 M) (Fig. 7B). Dexamethasone alone did not inhibit basal-CBP-associated histone acetylation (Fig. 7B).
Under stringent IP conditions IL-1β causes acetylation of all histone H4 lysine residues in contrast to the K8 and K12 pattern seen under mild IP conditions. Using mild IP conditions, IL-1β-induced elevation in CBP-associated histone acetylation was inhibited by dexamethasone (Fig. 7C). CBP isolated under these conditions was more sensitive to the inhibitory effects of dexamethasone than those seen with CBP isolated using more-stringent IP conditions (IC50 of 1 × 10−10 versus 6.5 × 10−9 M). Again, dexamethasone alone did not inhibit basal-CBP-associated histone acetylation. These results suggest that although repression of CBP may account for some of the repressive effect of dexamethasone on IL-1β-stimulated histone acetylation, it is not responsible alone for the repression of histone acetylation by dexamethasone and that CBP-associated cofactors are more sensitive to dexamethasone repression. Additionally, failure of CBP to induce histone acetylation at the higher concentrations of dexamethasone suggests that CBP in isolation does not mediate dexamethasone-induced histone acetylation.
In order to confirm that this inhibitory action of dexamethasone was mediated via GR, we performed HAT assays using immunoprecipitated CBP from cells treated with IL-1β alone and purified GR (10 ng/ml). These experiments were conducted in the presence of TSA (100 ng/ml) in order to inhibit endogenous HDAC activity. IL-1β caused a marked increase in histone acetylation (Fig. 7D). Dexamethasone alone, in the absence of GR, had no direct effect on IL-1β-stimulated histone acetylation. In addition, GR showed no histone acetylation activity in the presence or absence of CBP immunoprecipitate (Fig. 7D). The dexamethasone-GR complex directly inhibited IL-1β-stimulated CBP-mediated histone acetylation in a concentration-dependent manner with a maximal repression of 62% (Fig. 7D). These data suggest that in the absence of HDAC activity, dexamethasone, acting through GR, is able to suppress CBP-associated histone acetylation.
The CBP-associated complex immunoprecipitated under mild IP conditions showed no increase in HDAC activity after IL-1β treatment alone (Fig. 7E). However, with increasing concentrations of dexamethasone the levels of HDAC activity were markedly enhanced, reflecting either induction of HDAC or recruitment of HDAC to the CBP immunoprecipitated complex (Fig. 7E). Dexamethasone alone had no direct effect on HDAC activity. GR immunoprecipitates from both nonstimulated and IL-1β-stimulated cells did not show any histone deacetylation activity (Fig. 7F). In contrast, treatment with dexamethasone induced a concentration-dependent increase in histone deacetylation (Fig. 7F). These experiments showed that GR was associated with a HDAC activity that was induced in a concentration-dependent manner by dexamethasone. This induction reached significant levels at the concentrations that inhibited GM-CSF release and histone acetylation (Fig. 7F, Fig. 1C, and Fig. 3D).
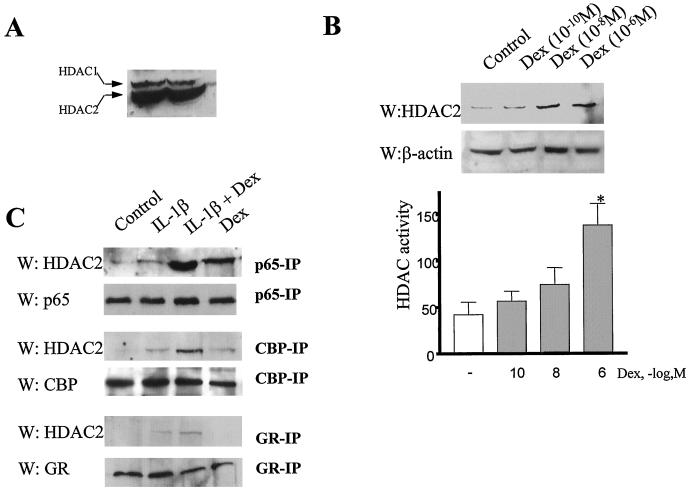
Effect of dexamethasone on HDAC expression, activity, and recruitment.
We have shown that dexamethasone induced histone deacetylation in GR, p65, and CBP immunoprecipitates. Furthermore, TSA decreased the inhibitory effect of dexamethasone on IL-1β-induced GM-CSF production, histone acetylation, and p65-associated histone acetylation. These results suggest that HDACs are involved in the inhibitory effects of dexamethasone. We therefore determined the effect of dexamethasone on HDAC expression, HDAC activity, and p65-HDAC association. A549 cells expressed mainly HDAC2 and very little HDAC1 (Fig. 8A). Dexamethasone induced both HDAC2 expression and histone deacetylation (Fig. 8B and C), but the concentration at which dexamethasone induced these effects (10−6 M) was greater than that which repressed IL-1β-stimulated histone acetylation (10−10 M) (see Fig. 3D). This suggests that dexamethasone repression of IL-1β-stimulated histone acetylation was not due to induction of newly synthesized HDAC protein or activity. We therefore examined HDAC2 association with the activated HAT complexes following incubation of cells with IL-1β and low doses of dexamethasone. Western blot analysis of p65 immunoprecipitates showed a recruitment of HDAC2 to the p65 immunoprecipitated complex following treatment of cells with IL-1β and a low concentration (10−10 M) of dexamethasone (Fig. 8D), suggesting a role for HDAC2 in the suppressive actions of dexamethasone. Similarly, Western blot analysis of CBP and GR immunoprecipitates also showed a recruitment of HDAC2 to the GR IP complexes (Fig. 8D). Dexamethasone (10−10 M) alone had no effect on HDAC2 recruitment to p65, CBP, or GR.
FIG. 8.
Effect of dexamethasone (Dex) on HDAC protein expression, HDAC activity, and HDAC recruitment to the p65 complex. (A) Relative expression of HDAC1 and HDAC2 in A549 cells. A 30-μg portion of protein was size fractionated by SDS–10% PAGE, and Western blot analysis was performed. The results are representative of three independent observations. (B) Effect of dexamethasone on HDAC2 protein expression and HDAC activity. Cells were incubated increasing concentrations of dexamethasone (10−10 to 10−6 M) for 6 h. Western blot analysis of HDAC2 expression is shown in the upper panel, and total cellular HDAC activity is shown in the lower panel. The results are expressed as the mean ± the SEM of three separate experiments (∗, P < 0.05). (C) Recruitment of HDAC2 to p65, CBP, and GR IP complexes. Cells were incubated with IL-1β in the presence of dexamethasone (10−10 M) for 6 h. Total cellular proteins were isolated and immunoprecipitated with anti-p65, anti-CBP, or anti-GR antibodies using mild IP conditions. HDAC2 expression in the IP complexes was measured by Western blotting. p65, CBP, and GR expression in the same samples is shown as a control for protein loading. The results are representative of three separate experiments.
DISCUSSION
IL-1β caused a concentration-dependent increase in GM-CSF expression which was inhibited by dexamethasone at concentrations 5- to 10-fold lower than those which caused transactivation of SLPI. The effect of the HDAC inhibitor TSA suggested that histone acetylation status may play a role in the regulation of GM-CSF and SLPI release and in dexamethasone actions. Increased gene expression by both IL-1β and dexamethasone were associated with increases in histone H4 acetylation status. IL-1β specifically caused acetylation of histone H4 K8 and K12, whereas dexamethasone markedly acetylated K5 and K16, with no effect on K8 and K12. Dexamethasone repressed IL-1β-induced GM-CSF expression and K8 and K12 acetylation at 5- to 10-fold-lower concentrations than that which induced histone acetylation or deacetylation or SLPI induction. Using chromatin IP assays, we confirmed that the differential acetylation of lysine residues by IL-1β and dexamethasone did not occur purely at the gross histone level but also occurred at both the GM-CSF and SLPI promoters.
Previous studies have shown a role for CBP in mediating NF-κB-driven gene transcription (11), and more-recent studies have shown that overexpression of CBP can modulate GR cross-talk with NF-κB (26, 34). The patterns of histone acetylation induced by CBP/p300 and PCAF are distinct, both from each other and from those found in the present study following stimulation by IL-1β or dexamethasone (33). CBP is able to acetylate all the relevant lysine residues of histone H4 (17), suggesting that CBP is the most likely target for competition between GR and p65 or indeed other transactivating proteins in these cells. CBP has several transactivating domains, and the specific domain used varies from one promoter to another and may direct acetylation of specific histone residues (22). CBP regulates the lysine residues acetylated by both IL-1β and dexamethasone; however, the targeting of specific lysine residues requires the association of additional coactivators but not p300 or PCAF, which modulate CBP-mediated histone acetylation.
Under conditions of maximal histone H4 acetylation following TSA treatment, IL-1β (i.e., NF-κB)-mediated GM-CSF activation is increased. This suggests that the GM-CSF promoter is regulated at the NF-κB site by an equilibrium of a CBP-associated coactivator complex interacting with NF-κB and a corepressor complex containing HDACs. The HDAC(s) involved in this complex is not clear in these cells but may include HDAC2 since some HDAC2 is recruited the CBP complex after IL-1β stimulation, albeit to a much lower extent than for IL-1β plus dexamethasone. The recruitment of HDAC to CBP by IL-1β may be part of this process and may account for the effect of TSA on IL-1β-stimulated GM-CSF production. This effect is similar to that reported for tumor necrosis factor alpha-induced IL-6 production through an NF-κB site and for p21CIP/WAF induction through SP1 sites (35, 41).
We show that 50% of glucocorticoid actions in repressing IL-1β-stimulated cytokine release is mediated by HDACs. Thus, the IC50 for GM-CSF repression is inhibited 80-fold by TSA, a finding which correlates with the 50-fold repression in the dexamethasone inhibition of p65-associated HAT activity. In addition, the TSA presence allows dexamethasone to suppress GM-CSF release, p65-associated HAT activity, and CBP-associated HAT activity by only 50 to 60% of the maximal stimulation even at the highest concentrations used.
Our results suggest that the site of cross-talk between p65 and GR occurs at the level of regulation of histone H4 acetylation by CBP and HDAC2. Previous data have suggested a role for CBP and SRC-1 in the nuclear integration of NF-κB and GR actions (34). In this model it was proposed that competition for limiting amounts of CBP, or other coactivators, resulted in an inhibition of NF-κB-driven gene transcription by GR. These studies used overexpression of CBP in order to overcome the actions of GR on NF-κB-mediated gene transcription. In contrast, our data shows no evidence for squelching as a mechanism for GR inhibition of IL-1β actions at least during the short (6-h) time course of these experiments (10, 16, 26, 34). However, exposure of cells for longer periods of time (24 to 48 h) to budesonide, a glucocorticoid agonist, indicates a time- and concentration-dependent reduction in CBP and RNA polymerase II expression (I. M. Adcock and Y. Nasuhara, unpublished observations).
Other studies have suggested that binding of GR to CBP disrupts the CBP-PCAF coactivation complexes (19). We found no evidence that dexamethasone blocked IL-1β-stimulated p65-CBP and p65-PCAF association or the association between CBP and PCAF. Furthermore, our results fail to indicate a major role for PCAF in mediating IL-1β-dependent acetylation of lysines. In contrast, we have shown a direct effect of GR on inhibiting IL-1β-induced CBP complex-mediated histone acetylation. The histone acetylation of CBP immunoprecipitates extracted under mild IP conditions, in which a large number of other proteins were coimmunoprecipitated, was repressed by low concentrations of dexamethasone and was specific to K8 and K12. Our results in which the CBP-associated complex, but not CBP alone, showed specifically for K8 and K12 indicates that other HATs, as well as CBP, are likely to be involved. Alternatively, HATs may interact with one another within a complex to modify the histone target lysines of each specific HAT. As these inhibitory effects of dexamethasone were decreased in the presence of TSA, HDACs were also indicated as playing a role in dexamethasone repression. However, this was not related to the induction of newly synthesized HDAC protein and activity but reflected recruitment of HDAC2 to a p65-CBP complex by GR.
Inhibition of MAPK phosphorylation by dexamethasone has been suggested to play an important role in glucocorticoid actions (6, 30, 36). These pathways also play a role in CBP activation by phosphorylation of transcription factors, such as NF-κB and AP-1 and may also directly phosphorylate CBP, thereby altering transactivation. Although we demonstrated that IL-1β induced phosphorylation of CBP and that this could be inhibited by dexamethasone. The concentration of dexamethasone at which this reduction occurs (10−6 M) is greater than that which inhibited histone acetylation and inflammatory gene expression, indicating that this mechanism of glucocorticoid action is less important for the anti-inflammatory actions of dexamethasone. In addition, we found no evidence for acetylation of CBP by either IL-1β or dexamethasone in these studies.
In summary, we have shown that both dexamethasone and IL-1β stimulated histone acetylation, but each showed a different pattern of histone H4 acetylation. Low concentrations of dexamethasone (10−10 M) which repress IL-1β-stimulated GM-CSF expression also repress IL-1β-stimulated CBP-associated histone acetylation at the GM-CSF promoter. Our data suggest that the activated GR complex inhibits acetylation of K8 and K12 by acting both as a direct inhibitor of CBP-associated histone acetylation and by recruiting HDAC2 to the p65-CBP HAT complex. This action does not involve de novo synthesis of HDAC protein or activity or increased expression of CBP or PCAF. Thus, we found that both HAT and HDAC activities coexist within same complex in the presence of p65 and GR and that they can each act independently without competing with each other (Fig. 9). This mechanism for glucocorticoid repression is novel and establishes that inhibition of histone acetylation is an additional level of control of inflammatory gene expression. This further suggests that pharmacological manipulation of specific histone acetylation status is a potentially useful approach for the treatment of inflammatory diseases. Identification of the precise mechanism by which activated GR recruits HDAC2 may reveal new targets for the development of drugs that may dissociate the anti-inflammatory actions of glucocorticoids from their side effects which are largely due to gene induction.
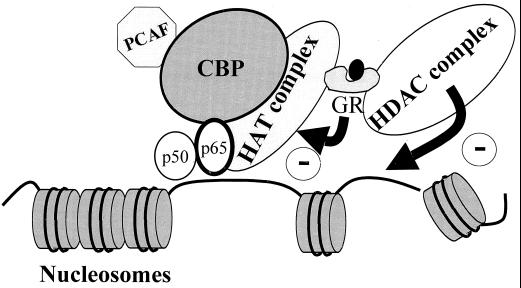
FIG. 9.
Proposed model for dexamethasone-GR complex inhibition of IL-1β-stimulated histone acetylation. DNA bound p65 induces histone acetylation via activation of CBP and a CBP-associated HAT complex. This results in local unwinding of DNA and increased gene transcription. GR, possibly acting as a monomer, interacts with CBP, causing an inhibition of CBP-associated HAT activity. In addition, GR also recruits HDAC2 to the activated p65-CBP complex, further reducing local HAT activity and leading to enhanced nucleosome compaction and repression of transcription.
ACKNOWLEDGMENTS
This work was funded by Glaxo-Wellcome and the Clinical Research Committee (Brompton Hospital).
REFERENCES
- 1.Barnes P J. Anti-inflammatory mechanisms of glucocorticoids. Biochem Soc Trans. 1995;23:940–945. doi: 10.1042/bst0230940. [DOI] [PubMed] [Google Scholar]
- 2.Barnes P J, Adcock I M. Transcription factors and asthma. Eur Respir J. 1998;12:221–234. doi: 10.1183/09031936.98.12010221. [DOI] [PubMed] [Google Scholar]
- 3.Beato M. Chromatin structure and the regulation of gene expression: remodeling at the MMTV promoter. J Mol Med. 1996;74:711–724. doi: 10.1007/s001090050076. [DOI] [PubMed] [Google Scholar]
- 4.Beato M, Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25:3559–3563. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 6.Caelles C, Gonzalez-Sancho J M, Munoz A. Nuclear hormone receptor antagonism with AP-1 by inhibition of the JNK pathway. Genes Dev. 1997;11:3351–3364. doi: 10.1101/gad.11.24.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 8.Espinos E, le Pomi V T, Weber M J. Cooperation between phosphorylation and acetylation processes in transcriptional control. Mol Cell Biol. 1999;19:3474–3484. doi: 10.1128/mcb.19.5.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flower R J, Rothwell N J. Lipocortin-1: cellular mechanisms and clinical relevance. Trends Pharmacol Sci. 1994;15:71–76. doi: 10.1016/0165-6147(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 10.Fontes J D, Kanazawa S, Jean D, Peterlin B M. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol Cell Biol. 1999;19:941–947. doi: 10.1128/mcb.19.1.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 13.Hecht A, Grunstein M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- 14.Imhof A, Wolffe A P. Transcription: gene control by targeted histone acetylation. Curr Biol. 1998;8:R422–R424. doi: 10.1016/s0960-9822(98)70268-4. [DOI] [PubMed] [Google Scholar]
- 15.Jonat C, Rahmsdorf H J, Park K K, Cato A C, Gebel S, Ponta H, Herrlich P. Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell. 1990;62:1189–1204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- 16.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 17.Kimura A, Horikoshi M. How do histone acetyltransferases select lysine residues in core histones? FEBS Lett. 1998;431:131–133. doi: 10.1016/s0014-5793(98)00752-2. [DOI] [PubMed] [Google Scholar]
- 18.Kolle D, Brosch G, Lechner T, Lusser A, Loidl P. Biochemical methods for analysis of histone deacetylases. Methods. 1998;15:323–331. doi: 10.1006/meth.1998.0636. [DOI] [PubMed] [Google Scholar]
- 19.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 20.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 21.Levine S J, Benfield T, Shelhamer J H. Corticosteroids induce intracellular interleukin-1 receptor antagonist type I expression by a human airway epithelial cell line. Am J Respir Cell Mol Biol. 1996;15:245–251. doi: 10.1165/ajrcmb.15.2.8703481. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima H, Kim Y B, Terano H, Yoshida M, Horinouchi S. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp Cell Res. 1998;241:126–133. doi: 10.1006/excr.1998.4027. [DOI] [PubMed] [Google Scholar]
- 24.Nightingale K P, Wellinger R E, Sogo J M, Becker P B. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J. 1998;17:2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 26.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 27.Perry M, Chalkley R. Histone acetylation increases the solubility of chromatin and occurs sequentially over most of the chromatin. A novel model for the biological role of histone acetylation. J Biol Chem. 1982;257:7336–7347. [PubMed] [Google Scholar]
- 28.Plesko M M, Hargrove J L, Granner D K, Chalkley R. Inhibition by sodium butyrate of enzyme induction by glucocorticoids and dibutyryl cyclic AMP. A role for the rapid form of histone acetylation. J Biol Chem. 1983;258:13738–13744. [PubMed] [Google Scholar]
- 29.Ray A, Prefontaine K E. Physical association and functional antagonism between the p65 subunit of transcription factor NF-kappa B and the glucocorticoid receptor. Proc Natl Acad Sci USA. 1994;91:752–756. doi: 10.1073/pnas.91.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rider L G, Hirasawa N, Santini F, Beaven M A. Activation of the mitogen-activated protein kinase cascade is suppressed by low concentrations of dexamethasone in mast cells. J Immunol. 1996;157:2374–2380. [PubMed] [Google Scholar]
- 31.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 32.Sallenave J M, Shulmann J, Crossley J, Jordana M, Gauldie J. Regulation of secretory leukocyte proteinase inhibitor (SLPI) and elastase-specific inhibitor (ESI/elafin) in human airway epithelial cells by cytokines and neutrophilic enzymes. Am J Respir Cell Mol Biol. 1994;11:733–741. doi: 10.1165/ajrcmb.11.6.7946401. [DOI] [PubMed] [Google Scholar]
- 33.Schiltz R L, Mizzen C A, Vassilev A, Cook R G, Allis C D, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 34.Sheppard K A, Phelps K M, Williams A J, Thanos D, Glass C K, Rosenfeld M G, Gerritsen M E, Collins T. Nuclear integration of glucocorticoid receptor and nuclear factor-kappaB signaling by CREB-binding protein and steroid receptor coactivator-1. J Biol Chem. 1998;273:29291–29294. doi: 10.1074/jbc.273.45.29291. [DOI] [PubMed] [Google Scholar]
- 35.Sowa Y, Orita T, Hiranabe-Minamikawa S, Nakano K, Mizuno T, Nomura H, Sakai T. Histone deacetylase inhibitor activates the p21/WAF1/Cip1 gene promoter through the Sp1 sites. Ann N Y Acad Sci. 1999;886:195–199. doi: 10.1111/j.1749-6632.1999.tb09415.x. [DOI] [PubMed] [Google Scholar]
- 36.Swantek J L, Cobb M H, Geppert T D. Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) is required for lipopolysaccharide stimulation of tumor necrosis factor alpha (TNF-alpha) translation: glucocorticoids inhibit TNF-alpha translation by blocking JNK/SAPK. Mol Cell Biol. 1997;17:6274–6282. doi: 10.1128/mcb.17.11.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Truss M, Beato M. Steroid hormone receptors: interaction with deoxyribonucleic acid and transcription factors. Endocrinol Rev. 1993;14:459–479. doi: 10.1210/edrv-14-4-459. [DOI] [PubMed] [Google Scholar]
- 38.Turner B M, Fellows G. Specific antibodies reveal ordered and cell-cycle-related use of histone-H4 acetylation sites in mammalian cells. Eur J Biochem. 1989;179:131–139. doi: 10.1111/j.1432-1033.1989.tb14530.x. [DOI] [PubMed] [Google Scholar]
- 39.Turner B M, O'Neill L P, Allan I M. Histone H4 acetylation in human cells. Frequency of acetylation at different sites defined by immunolabeling with site-specific antibodies. FEBS Lett. 1989;253:141–145. doi: 10.1016/0014-5793(89)80947-0. [DOI] [PubMed] [Google Scholar]
- 40.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe A P. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanden Berghe W, De Bosscher K, Boone E, Plaisance S, Haegeman G. The nuclear factor-kappaB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J Biol Chem. 1999;274:32091–32098. doi: 10.1074/jbc.274.45.32091. [DOI] [PubMed] [Google Scholar]
- 42.Wolffe A P. Transcriptional control. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 43.Workman J L, Buchman A R. Multiple functions of nucleosomes and regulatory factors in transcription. Trends Biochem Sci. 1993;18:90–95. doi: 10.1016/0968-0004(93)90160-o. [DOI] [PubMed] [Google Scholar]
- 44.Wu R S, Panusz H T, Hatch C L, Bonner W M. Histones and their modifications. CRC Crit Rev Biochem. 1986;20:201–263. doi: 10.3109/10409238609083735. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]