Abstract
In budding yeast, the Rho-type GTPase Cdc42p is essential for cell division and regulates pseudohyphal development and invasive growth. Here, we isolated novel Cdc42p mutant proteins with single-amino-acid substitutions that are sufficient to uncouple functions of Cdc42p essential for cell division from regulatory functions required for pseudohyphal development and invasive growth. In haploid cells, Cdc42p is able to regulate invasive growth dependent on and independent of FLO11 gene expression. In diploid cells, Cdc42p regulates pseudohyphal development by controlling pseudohyphal cell (PH cell) morphogenesis and invasive growth. Several of the Cdc42p mutants isolated here block PH cell morphogenesis in response to nitrogen starvation without affecting morphology or polarity of yeast form cells in nutrient-rich conditions, indicating that these proteins are impaired for certain signaling functions. Interaction studies between development-specific Cdc42p mutants and known effector proteins indicate that in addition to the p21-activated (PAK)-like protein kinase Ste20p, the Cdc42p/Rac-interactive-binding domain containing Gic1p and Gic2p proteins and the PAK-like protein kinase Skm1p might be further effectors of Cdc42p that regulate pseudohyphal and invasive growth.
The Rho-type GTPase Cdc42p is a member of the Ras superfamily of small GTP-binding proteins that play an essential role in regulating proliferation and differentiation in all eukaryotes (reviewed in references 24 and 35). In Saccharomyces cerevisiae, Cdc42p has been implicated in the regulation of diverse processes essential for both cell division and cellular development, and a great number of genetic and biochemical studies have shown that the major roles of Cdc42p in these processes are regulation of actin rearrangements and modulation of protein kinase cascades (reviewed in reference 24).
Cdc42 proteins act as molecular switches that, by exchange of GTP for GDP, are placed in an activated, signaling state, which is terminated by the hydrolysis of GTP to GDP (35). Cdc42p must interact with a variety of regulators and downstream effector proteins to constitute a functional GTPase signaling module (24). Regulation is mediated by guanine nucleotide exchange factors, which in S. cerevisiae are represented by Cdc24p (58, 64), and by GTPase-activating proteins that in S. cerevisiae comprise Bem3p and Rga1p (7, 59, 64). A growing number of Cdc42p downstream effector proteins are known that interact with the GTP-bound (activated) form and thereby mediate numerous downstream events. In S. cerevisiae, Cdc42p effectors include the p21-activated (PAK) family protein kinases Ste20p, Cla4p, and Skm1p (2, 9, 11, 27, 36, 47, 57, 63), the formin homology proteins Bni1p and Bnr1p (13, 15, 16, 22, 23, 60, 62), the Wiskott-Aldrich syndrome protein family member Bee1p/Las17p (28, 42), the IQGAP homologue Iqg1p/Cyk1p (12, 30, 45, 55), and the novel Gic1p and Gic2p proteins, which share the conserved Cdc42p/Rac-interactive-binding (CRIB) domain (4, 6).
The molecular mechanisms by which Cdc42p temporally and spatially discriminates between the distinct effectors are largely unknown. Helpful tools for dissecting the distinct functions of Cdc42p are effector mutant proteins that have lost the ability to bind and activate certain effectors but not others. So far, only a few such alleles have been uncovered (29, 46, 49). Most Cdc42p mutants that have been studied to date display defects in functions that are essential for cell division, based on their lethal or temperature-sensitive growth phenotypes (10, 24, 25, 37, 49, 65). No alleles of CDC42 have been described that separate the functions of Cdc42p required for cell division from developmental functions, e.g., appropriate morphological and signaling responses to changes in the environment.
Pseudohyphal development is a process in which S. cerevisiae alters its morphology in response to nutritional signals. When starved for nitrogen, diploid S. cerevisiae strains undergo a developmental transition from growth as single yeast form (YF) cells to a multicellular form consisting of filaments of pseudohyphal (PH) cells (17). This dimorphic switch, referred to as pseudohyphal development, is a composite of genetically dissectable cellular changes, including alterations in the budding pattern, cell morphology, and invasive growth behavior (17, 38). A related phenomenon, invasive growth, occurs in haploid cells (51). Pseudohyphal development and invasive growth are under the control of at least two signaling pathways. One of the routes involves the GTP-binding proteins Ras2p and Gpa2p and the cyclic AMP (cAMP)-dependent protein kinase (26, 34, 61). Activation of the cAMP pathway stimulates expression of FLO11, encoding a cell wall protein required for invasive growth and pseudohyphal development (32, 54). A second pathway that regulates pseudohyphal development and invasive growth involves Cdc42p (40, 41, 52). In this pathway, Ras2p is thought to signal via Cdc42p and Ste20p to the Kss1p mitogen-activated protein kinase (MAPK) cascade that shares several components with the MAPK cascade required for mating (31). The role of Cdc42p in this signaling pathway was deduced from several studies. Dominant activated Cdc42G12V and Cdc42Q61L proteins not only induce pseudohyphal development, but also stimulate expression of genes controlled by the Kss1p MAPK cascade (41). Expression of the dominant negative Cdc42D118A mutant protein inhibits Ras2p-dependent activation of pseudohyphal development, and expression of Cdc42G12Vp or Cdc42Q61Lp rescues defective invasive growth of haploid ras2Δ mutant strains, placing Cdc42p downstream of Ras2p (40). Dominant active Cdc42G12Vp and Cdc42Q61Lp require Ste20p for activation of the Kss1p MAPK pathway, and Cdc42p-Ste20p interactions depend on the CRIB domain of Ste20p, placing Cdc42p upstream of Ste20p (41, 47). However, all studies investigating the function of Cdc42p in pseudohyphal development involved dominant active or inactive variants that also affect functions essential for cell division.
In this study, we isolated novel mutant alleles of CDC42 with the goal of separating functions of Cdc42p required for pseudohyphal and invasive growth from those required for cell division. We find that single-amino-acid substitutions within Cdc42p are sufficient to uncouple functions required for cell division from those regulating cellular development. Several of the Cdc42p mutants isolated here block PH cell morphogenesis in response to nitrogen starvation but do not affect morphology or polarity of the yeast form in nutrient-rich conditions, indicating that these proteins are signaling rather than general morphology mutants. Interaction studies between these development-specific Cdc42p mutants and an array of known effectors indicate that in addition to Ste20p, the Gic1p and Gic2p proteins and the protein kinase Skm1p might be further effectors of Cdc42p that are important for pseudohyphal and invasive growth.
MATERIALS AND METHODS
Yeast strains and growth conditions.
All yeast strains used in this study are congenic to the Σ1278b genetic background (Table 1). The cdc42Δ::HIS3 deletion mutation was introduced using deletion plasmid pME1758 (Table 2). RH2197 and RH2442 are segregants of RH2441, and RH2199 was obtained by mating RH2197 with RH2442. Standard methods for genetic crosses and transformation were used, and standard yeast culture medium was prepared essentially as described (20). Synthetic complete medium (SC) lacking appropriate supplements was used for scoring invasive growth and for β-galactosidase assays. Invasive growth tests were performed as described previously using solid SC medium lacking appropriate supplements (51). Low-ammonium medium (SLAD) was prepared as described (17). When required, uracil was added to SLAD medium to a final concentration of 0.2 mM to make SLAD+Ura.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Plasmid | Reference |
|---|---|---|---|
| RH2441 | MATa/MATα cdc42Δ::HIS3/CDC42 ura3-52/ura3-52 his3::hisG/his3::hisG leu2::hisG/LEU2 trp1::hisG/TRP1 | pTF27-CDC42 | This study |
| RH2197 | MATα cdc42Δ::HIS3 ura3-52 his3::hisG leu2::hisG | pTF27-CDC42 | This study |
| RH2442 | MATa cdc42Δ::HIS3 ura3-52 his3::hisG leu2::hisG | pTF27-CDC42 | This study |
| RH2199 | MATa/MATα cdc42Δ::HIS3/cdc42Δ::HIS3 ura3-52/ura3-52 his3::hisG/his3::hisG leu2::hisG/leu2::hisG | pTF27-CDC42 | This study |
| RH2488 | MATa/MATα bni1::LEU2/bni1::LEU2 ura3-52/ura3-52 leu2::hisG/leu2::hisG trp1::hisG/TRP1 | 38 |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pME1758 | Cassette for full deletion of CDC42 | This study |
| pME1533 | Derivative of pRS316 with mutated CEN region | J. Hegemann |
| pME1534 | CDC42 in pTF27 | This study |
| YCp(CDC42Sc) | CDC42 in pRS315 | 65 |
| pME1552 | CDC42N26I in pRS315 | This study |
| pME1555 | CDC42A30T in pRS315 | This study |
| pME1538 | CDC42I46M in pRS315 | This study |
| pME1557 | CDC42D65N in pRS315 | This study |
| pME1548 | CDC42R68S in pRS315 | This study |
| pME1536 | CDC42S71P in pRS315 | This study |
| pME1560 | CDC42N92D in pRS315 | This study |
| pME1559 | CDC42E95K in pRS315 | This study |
| pME1551 | CDC42E100G in pRS315 | This study |
| pME1545 | CDC42S158T in pRS315 | This study |
| pME1546 | CDC42L160P in pRS315 | This study |
| pME1759 | PSTE20-GFP-STE20 fusion in pRS316 | This study |
| pME1760 | PSTE20-GFP-STE20 fusion in pRS426 | This study |
| B3782 | FLO11-lacZ reporter construct | 54 |
| pEG202 | Vector for construction of LexA fusion proteins | 21 |
| pME1913 | LexA-CDC42C188S in pEG202 | 49 |
| pME1914 | LexA-CDC42N26I,C188S in pEG202 | This study |
| pME1915 | LexA-CDC42A30T,C188S in pEG202 | This study |
| pME1916 | LexA-CDC42I46M,C188S in pEG202 | This study |
| pME1917 | LexA-CDC42D65N,C188S in pEG202 | This study |
| pME1918 | LexA-CDC42R68S,C188S in pEG202 | This study |
| pME1919 | LexA-CDC42S71P,C188S in pEG202 | This study |
| pME1920 | LexA-CDC42N92D,C188S in pEG202 | This study |
| pME1921 | LexA-CDC42E95K,C188S in pEG202 | This study |
| pME1922 | LexA-CDC42E100G,C188S in pEG202 | This study |
| pME1923 | LexA-CDC42S158T,C188S in pEG202 | This study |
| pME1924 | LexA-CDC42L160P,C188S in pEG202 | This study |
| pJG4-5 | Vector for construction of B42 transcription activation domain (B42AD) fusions | 21 |
| pJG4-5(GIC1) | B42AD-GIC1 | 6 |
| pJG4-5(GIC2) | B42AD-GIC2 | 6 |
| pJG4-5(BNI1 1-1214 aa) | B42AD-BNI1(1-1214 aa) | 13 |
| pJG4-5(STE20) | B42AD-STE20 | This study |
| pJG4-5(CLA4) | B42AD-CLA4 | 9 |
| pJG4-5(SKM1) | B42AD-SKM1 | 49 |
Plasmid constructions.
Plasmid pME1758 was created by replacement of the CDC42 coding sequence with the HIS3 selectable marker using a PCR-based three-step cloning strategy. Plasmid pME1534 was constructed by subcloning a 1.7-kb CDC42 BamHI-HindIII fragment from YCp(CDC42Sc) (65) into plasmid pME1533 (pTF27; from J. Hegemann, Heinrich-Heine University, Düsseldorf, Germany). Plasmid pME1533 is a derivative of pRS316 (56) and contains a 24-bp deletion in the CEN6 region that decreases the mitotic stability of the plasmid (Fiedler and Hegemann, unpublished). Plasmids pME1759 and pME1760, both expressing green fluorescent protein (GFP)-Ste20p from the STE20 promoter, were constructed as follows. (i) A 4.9-kb KpnI-NotI fragment carrying STE20 was subcloned from pRS426-STE20 (41) into pRS316 to yield plasmid pRS316-STE20. (ii) A 750-bp DNA fragment containing part of the STE20 promoter was amplified from pRS316-STE20 by PCR, introducing a BglII site after the ATG translational start site of STE20, and cloned into the EcoRV site of pBluescriptKS (Stratagene). (iii) A 510-bp fragment coding for the N-terminal portion of Ste20p was amplified by PCR, introducing a BglII site in front of the second codon of STE20, and inserted as a BglII-XbaI fragment into the BglII and XbaI sites of the construct (ii). (iv) The 375-bp BamHI-SphI fragment from step iii was then exchanged for the corresponding BamHI-SphI fragments in both pRS316-STE20 and pRS426-STE20, yielding plasmids pRS316-STE20-BglII and pRS426-STE20-BglII, respectively, each containing a single BglII site between the ATG translational start site and the second codon of STE20. (v) A 750-bp fragment containing the GFPuv variant of GFP was amplified from plasmid pBAD-GFPuv (Clontech) by PCR, introducing BglII sites before the ATG translational start and before the translational stop codon of GFPuv. After restriction digestion with BglII, the amplified GFPuv fragment was inserted into the BglII site of both plasmids pRS316-STE20-BglII and pRS426-STE20-BglII, yielding plasmids pME1759 and pME1760, respectively. Plasmid pJG4-5(STE20) was constructed by (i) introducing a BglII site into the multiple cloning site of pJG4-5 and (ii) inserting a 3.1-kb BglII-KpnI STE20 fragment from plasmid pRS426-STE20-BglII into the modified version of pJG4-5 from step i.
Library of CDC42 mutants.
CDC42 was mutagenized by PCR amplification of a 1.7-kb fragment from YCp(CDC42Sc) (65) using Taq DNA polymerase and primers T3 and T7 in the presence of 0.24 mM MnCl2. The resulting DNA was digested with BamHI and HindIII and exchanged for the 1.7-kb BamHI-HindIII fragment carrying the wild-type version of CDC42 in plasmid YCp(CDC42Sc) (65). A library of approximately 130,000 recombinants was obtained. Following identification of mutants (see below), both DNA strands of the CDC42 coding sequences were sequenced using the ABI Prism Big Dye terminator sequencing kit and an ABI 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems GmbH, Weiterstadt, Germany).
Screen for CDC42 alleles.
For isolation of haploid invasive growth mutants, strain RH2197 was transformed with the CDC42 mutant library described above. A pool of approximately 26,000 transformants was obtained by growth selection on SC medium without Leu (SC−Leu medium) and subsequent growth on medium containing 0.1% 5-fluoroorotic acid to remove plasmid pME1534 carrying the wild-type CDC42 gene by counterselection. This pool was plated on YPD medium at a density of ∼500 colonies per plate, and mutants were identified by an invasive growth test (51). Phenotypes were confirmed by isolating the CDC42-containing plasmids and reintroducing them into the parental strain by plasmid shuffling. For isolation of pseudohyphal development mutants, strain RH2199 was transformed with the CDC42 mutant library, and a pool of approximately 60,000 transformants was obtained as described above. This pool was plated on SLAD+Ura. Pseudohyphal mutants were identified by the appearance of colonies, visualized with a dissecting microscope. Phenotypes were confirmed by isolating the CDC42-containing plasmids and reintroducing them into the parental strain by plasmid shuffling using either centromere-based or integrative versions.
Pseudohyphal development assays.
Qualitative and quantitative assays for pseudohyphal development, including determination of substrate invasion and cell shape as well as determination of bud site selection patterns, were performed as described previously (38). Pseudohyphal colonies were viewed with a Zeiss axiovert microscope and photographed using a digital camera.
β-Galactosidase assays. (i) FLO11-lacZ activity.
Strains carrying the FLO11-lacZ plasmid B3782 were grown to the exponential growth phase, and extracts were prepared and assayed for β-galactosidase activity as described previously (41). Specific β-galactosidase activity was normalized to the total protein in each extract and equals [the optical density at 420 nm (OD420) × 1.7]/[0.0045 × protein concentration × extract volume × time]. Assays were performed on at least three independent transformants, and the mean value is presented. Standard deviations did not exceed 15%.
Northern blot analysis.
Total RNA was prepared from cultures grown on solid medium for 30 h according to the method described earlier (8). Total RNA was separated on a 1.4% agarose gel containing 3% formaldehyde and transferred onto nylon membranes as described earlier (39). CDC42 and ACT1 transcripts were detected using gene-specific 32P-radiolabeled DNA probes. Hybridizing signals were quantified using a BAS-1500 Phosphor-Imaging scanner (Fuji, Tokyo, Japan).
Cell extracts and Western blot analysis.
Preparation of total cell extracts and subsequent Western blot analysis were performed essentially as described (52). Cdc42p and Cdc28p proteins were detected using enhanced chemiluminescence technology (Amersham, Buckinghamshire, United Kingdom) after incubation of nitrocellulose membranes with polyclonal anti-Cdc42p (Santa Cruz Biotechnology, Santa Cruz, Calif.) or anti-Cdc28p antibodies and a peroxidase-coupled goat anti-rabbit immunoglobulin G secondary antibody (Dianova, Hamburg, Germany). Cdc42p and Cdc28p signals were quantified using Molecular Analyst software (Bio-Rad, Munich, Germany).
Photomicroscopy.
Staining of bud site chitin rings and actin cytoskeleton was performed essentially as described (1, 48). Briefly, cells were grown to mid-log phase in liquid YNB+Ura medium, fixed in 3.7% formaldehyde for 1 h, and stained in the dark with Calcofluor (Fluorescent Brightener 28; catalog no. F-3543; Sigma) or with rhodamine-phalloidin (R-415; Molecular Probes). Cells and stained bud scars or actin cytoskeleton were visualized on a Zeiss axiovert microscope by either differential interference contrast (DIC) microscopy or fluorescence microscopy using appropriate filter sets. Cells harboring plasmids encoding GFP-Ste20p were grown to exponential growth phase and immediately viewed in vivo using Nomarski optics (DIC) or a GFP filter set (AHF Analysentechnik AG, Tübingen, Germany). All cells shown were photographed using a Xillix Microimager digital camera and the Improvision Openlab software (Improvision, Coventry, England).
Two-hybrid protein interactions.
All CDC42 alleles tested in two-hybrid protein interaction assays were subjected to site-directed PCR mutagenesis in order to introduce the C188S mutations, avoiding membrane localization of the proteins. CDC42 alleles were amplified by PCR and checked by sequence analysis before introduction into vector pEG202 as described (49). The methods for performing two-hybrid analysis have been described before (21). Reporter strain EGY48-p1840 was cotransformed pairwise with the various pEG202(CDC42) and pJG4-5 constructs (Table 2), and transformants were selected on SC−His−Trp medium. Transformants were grown at 23°C in liquid SC−His−Trp medium containing 2% galactose and 2% raffinose to an OD600 of between 1 and 2, and β-galactosidase liquid assays were performed as described previously (18). β-Galactosidase activities were normalized to the activities obtained for Cdc42C188Sp (wild type) and the different effectors, with values set to 100. Absolute values for Cdc42C188Sp were 383 Miller units when interacting with Gic1p, 323 U for Gic2p, 227 U for Bni1p, 39 U for Ste20p, 78 U for Cla4p, and 22 U for Skm1p. All assays were performed in triplicate on at least four independent transformants for each combination of plasmids.
Computer modeling.
A three-dimensional structure model of S. cerevisiae Cdc42p was obtained by homology modeling of the primary structure of S. cerevisiae Cdc42p using the SWISS-MODEL service (19) and the WebLab Viewer software (Molecular Simulations Inc., San Diego, Calif.).
RESULTS
Functions of Cdc42p required for cell division and cellular development can be uncoupled by single-amino-acid substitutions.
We tested whether the different functions of Cdc42p for regulation of distinct processes can be separated by specific amino acid substitutions within the Cdc42p protein. Specifically, we wanted to uncouple functions of Cdc42p required for cell division from those regulating pseudohyphal and invasive growth development. We created a library of PCR-mutagenized CDC42 genes and introduced them on a centromeric vector into both a haploid cdc42Δ mutant and a diploid cdc42Δ/cdc42Δ null strain. Because genomic cdc42Δ null mutations are lethal, CDC42 mutant genes were introduced by plasmid shuffling (20). Mutants of CDC42 causing defective haploid invasive growth on rich medium were isolated from the haploid cdc42Δ background. Mutant alleles of CDC42 leading to both reduced (nonfilamentous) and enhanced (hyperfilamentous) pseudohyphal development on low-ammonium medium were identified from the diploid pool. In all cases, one or more amino acid exchanges in Cdc42p were found, but only mutants with single-amino-acid substitutions were investigated further. The CDC42I46M and CDC42S71P mutant alleles were isolated from haploids that showed defective invasive growth (Table 3, Fig. 1A). Five CDC42 mutants causing a nonfilamentous phenotype (CDC42N26I, CDC42R68S, CDC42E100G, CDC42S158T, and CDC42L160P) as well as four alleles of CDC42 conferring hyperfilamentation (CDC42A30T, CDC42D65N, CDC42N92D, and CDC42E95K) were identified in diploid strains on low-ammonium medium (Table 3, Fig. 1B). Importantly, none of the isolated CDC42 mutant genes caused significant defects in the growth rate (Table 3), demonstrating that they did not affect functions essential for cell division. Moreover, strains harboring the mutant alleles did not display a detectable temperature-sensitive growth phenotype when grown at 37°C.
TABLE 3.
Regulation of growth and development by different CDC42 alleles
| Developmental phenotypea | Mutation | Growth rateb (h−1) |
|---|---|---|
| Haploid invasive growth | ||
| + | None | 0.34 |
| − | I46M | 0.34 |
| − | S71P | 0.33 |
| Diploid pseudohyphal growth | ||
| + | None | 0.37 |
| − | N26I | 0.39 |
| ++ | A30T | 0.32 |
| ++ | D65N | 0.38 |
| +/− | R68S | 0.39 |
| ++ | N92D | 0.36 |
| ++ | E95K | 0.38 |
| − | E100G | 0.36 |
| − | S158T | 0.33 |
| − | L160P | 0.38 |
Haploid invasive growth was determined by expression of CDC42 alleles in a haploid cdc42Δ mutant strain and growth on solid SC−Leu medium for 4 days before performing a wash assay. +, wild-type invasiveness; −, complete loss of invasive growth. Pseudohyphal growth was determined by expression of CDC42 alleles in a diploid cdc42Δ/cdc42Δ mutant strain and growth on solid SLAD+Ura medium for 5 days. Symbols indicate the degree of filamentation: +, wild type; ++, enhanced; +/−, reduced; −, undetectable.
Growth rate was determined in liquid SC−Leu medium.
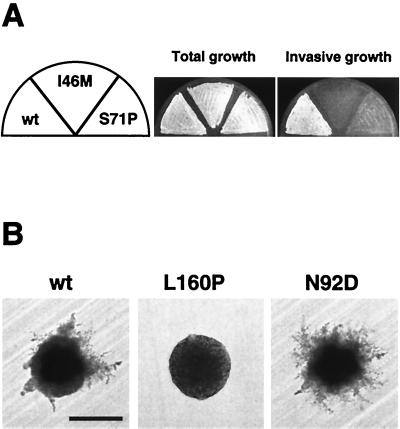
FIG. 1.
Cdc42p mutants causing specific defects in yeast development. (A) Haploid invasive growth of strains expressing wild-type CDC42 (wt), CDC42I46M (I46M), or CDC42S71P (S71P). Plasmids carrying the different CDC42 alleles were introduced into the haploid cdc42Δ strain RH2197 (MATα) by plasmid shuffling. Resulting strains were patched on SC−Leu medium for 4 days. After incubation, the plate was photographed before (Total growth) and after (Invasive growth) cells were washed off the agar surface. (B) Pseudohyphal development of diploid strains expressing wild-type CDC42 (wt), CDC42L160P (L160P), or CDC42N92D (N92D). Plasmids carrying the different CDC42 alleles were introduced into the diploid cdc42Δ/cdc42Δ strain RH2199. Resulting strains were streaked to obtain single colonies on nitrogen starvation medium (SLAD+Ura). Plates were incubated at 30°C, and representative colonies were photographed after 3 days of growth. Bar, 100 μm.
These results demonstrate that specific single-amino-acid exchanges within Cdc42p are sufficient to separate the function of this Rho-type GTPase required for cell division from its regulatory function in cellular development.
Cdc42p regulates haploid invasive growth by mechanisms both dependent on and independent of FLO11 expression.
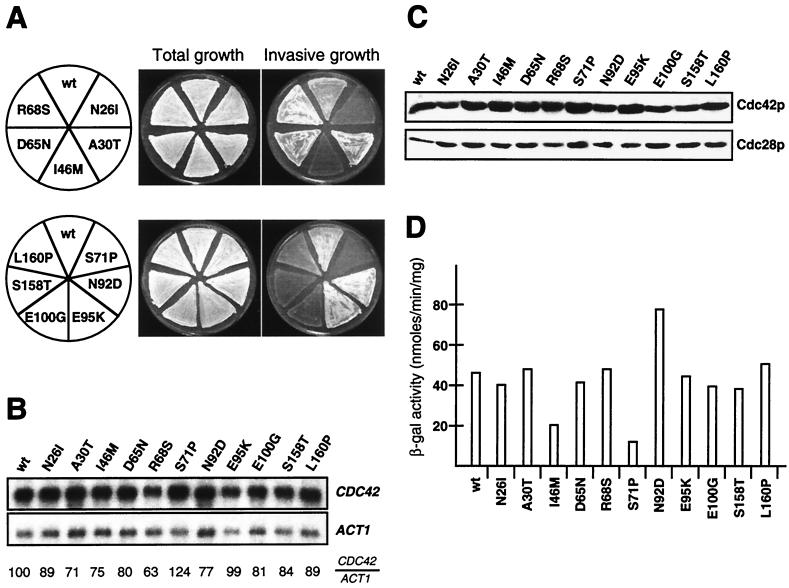
Expression of dominant activated forms of CDC42, CDC42G12V or CDC42Q61L, not only induces pseudohyphal growth in diploids (41) but also suppresses defective invasive growth caused by loss of RAS2 in haploid cells (40). This suggests that the functions of CDC42 in regulating haploid invasive growth and diploid pseudohyphal development overlap. Therefore, we tested the effects of all 11 isolated CDC42 alleles on invasive growth when expressed in a haploid background. CDC42 mutant genes were introduced into a haploid MATa cdc42Δ strain by plasmid shuffling, and invasive growth was assayed on medium rich in nitrogen (Fig. 2A). We found that with the exception of R68S, all amino acid exchanges in Cdc42p causing reduced pseudohyphal development in diploid cells also lead to a suppression of invasive growth in haploids. All amino acid substitutions conferring hyperfilamentation in diploids supported invasive growth in haploid cells. CDC42 mRNA and intracellular Cdc42 protein levels were measured in all strains to exclude that the invasive growth phenotypes found were due to altered expression or stability of Cdc42p mutant proteins (Fig. 2B and C). No significant differences were found between the wild type and any of the mutant forms of CDC42, demonstrating that developmental phenotypes are caused by altered function and not by intracellular amounts of the different Cdc42p mutant proteins.
FIG. 2.
Regulation of haploid invasive growth and FLO11 expression by CDC42 alleles. (A) Haploid invasive growth of strains expressing wild-type CDC42 (wt) and CDC42 alleles encoding proteins with single-amino-acid substitutions as indicated. Plasmids carrying the different CDC42 alleles were introduced into the haploid cdc42Δ strain RH2442 (MATa), and the resulting strains were patched on SC−Leu medium for 4 days. After incubation, plates were photographed before (Total growth) and after (Invasive growth) cells were washed off the agar surface. (B) Autoradiogram showing steady-state mRNA levels of CDC42 alleles in the strains described for panel A. ACT1 gene expression was used as an internal standard. Relative expression levels of CDC42 alleles (CDC42/ACT1) are shown and were obtained using a Phosphor-Imaging scanner. Numbers represent mean values of three independent measurements and were obtained by normalizing CDC42 transcript levels to ACT1 levels and to wild-type CDC42 (wt). Standard deviation was below 20%. (C) Expression levels of Cdc42 proteins were determined in extracts from strains described for panel A by Western blot analysis using a polyclonal anti-Cdc42p antibody. As an internal control, expression levels of Cdc28p were measured in the same extracts using a polyclonal anti-Cdc28p antibody (lower panel). (D) FLO11-lacZ expression levels. β-Galactosidase (β-gal) activity was measured in strains carrying FLO11-lacZ on a plasmid (B3782). Bars depict means of three independent measurements, with standard deviations not exceeding 15%.
Invasive growth development in haploid cells is regulated by at least two signaling pathways, the Kss1p MAPK cascade and the cAMP protein kinase A pathway. Both pathways regulate expression of FLO11, encoding a cell surface flocculin required for invasive growth (32, 54), and FLO11 expression is well correlated with the invasiveness of yeast cells. Therefore, we determined expression of the FLO11-lacZ reporter gene (54) in all haploid strains containing the isolated CDC42 mutant genes (Fig. 2D). We found that FLO11 expression was clearly reduced in strains carrying either the CDC42I46M or the CDC42S71P mutant allele. The I46M mutation caused a reduction to 45% compared to a wild-type control, whereas expression of FLO11 was reduced to 26% of the wild-type level in strains expressing the S71P mutant protein. An increase in FLO11 expression to 175% was found in strains expressing the N92D mutant protein. No significant changes in FLO11 expression levels were detected in strains carrying any of the other CDC42 mutant alleles, although most of them caused clearly detectable changes in invasive growth behavior. These measurements indicate that in haploid cells, Cdc42p might regulate invasive growth by different mechanisms: one that affects expression of FLO11 by acting via the Kss1p MAPK cascade, and a second one that must operate independently of changes in FLO11 expression levels.
We further tested whether mating of MATa haploid cells is affected by the amino acid exchanges in Cdc42p that we found affected haploid invasive growth. However, none of the mutations significantly influenced pheromone-induced growth arrest, induction of FUS1-lacZ expression, formation of mating projections, or formation of diploid cells (data not shown). This is in agreement with earlier studies showing that Cdc42p does not appear to directly regulate pheromone-mediated gene expression (27, 44, 47).
Cdc42p regulates pseudohyphal development by affecting PH cell morphogenesis and invasive growth.
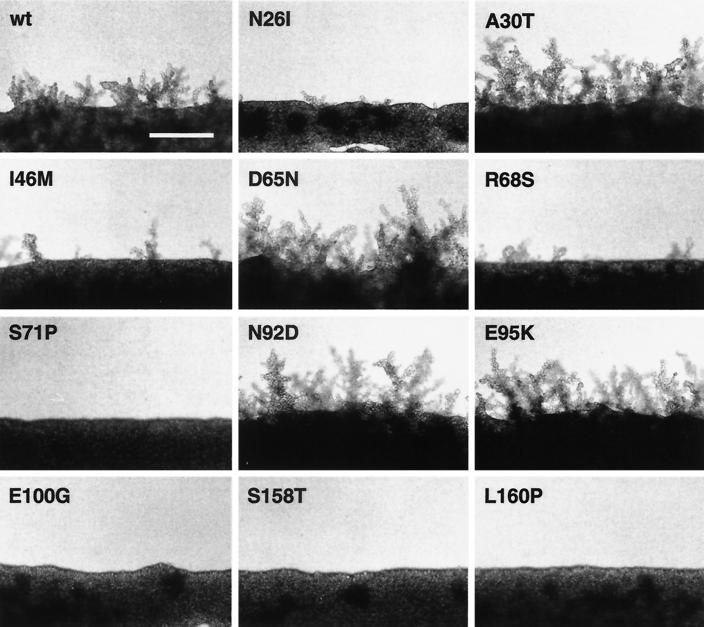
Pseudohyphal development is a composite of several cellular changes, including alterations in cell morphogenesis and invasive growth behavior. We tested the effects of all 11 CDC42 mutant alleles on diploid pseudohyphal growth in more detail. Mutant alleles were introduced into a diploid cdc42Δ/cdc42Δ strain by plasmid shuffling, and diploid pseudohyphal development was assayed on nitrogen starvation medium (Fig. 3). We found that both alleles of CDC42 that were isolated from haploids as suppressing invasive growth, CDC42I46M and CDC42S71P, also led to reduced pseudohyphal development in diploids. We further characterized each of the diploid CDC42 mutant strains with respect to changes in cell shape upon nitrogen starvation and the ability to invade agar as described earlier (38). The shape of cells was determined by defining three morphological groups: long PH cells, oval YF cells, and round YF cells (Table 4). Substrate invasion was measured by determining the ratio of invasive to noninvasive cells after growth on nitrogen starvation medium. Characterization of all CDC42 mutant alleles by these criteria defined three different classes.
FIG. 3.
Regulation of pseudohyphal development by CDC42 alleles. Shown is the growth on nitrogen starvation medium of diploid strains expressing wild-type CDC42 (wt) or mutant CDC42 alleles leading to single-amino-acid substitutions as indicated. Plasmids carrying the different CDC42 alleles were introduced into the diploid cdc42Δ/cdc42Δ strain RH2199 by plasmid shuffling. Pseudohyphal development of resulting strains was photographed after 4 days of growth on SLAD+Ura medium. Bar, 100 μm.
TABLE 4.
Budding patterns, morphology, and invasive growth behavior of diploid cdc42 mutants
| Genotype | Budding patterna (% of cells)
|
Cell shapeb (% of cells)
|
Invasive growthc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Long PH
|
Oval YF
|
Round YF
|
||||||||
| Bipolar | Random | Unipolar | +N | −N | +N | −N | +N | −N | ||
| CDC42 | 72 | 8 | 20 | 4 | 29 | 54 | 57 | 42 | 14 | + |
| CDC42N26I | 75 | 6 | 19 | 1 | 3 | 48 | 61 | 51 | 36 | − |
| CDC42A30T | 70 | 5 | 25 | 6 | 55 | 46 | 39 | 48 | 6 | + |
| CDC42I46M | 74 | 5 | 21 | 1 | 6 | 49 | 49 | 50 | 45 | − |
| CDC42D65N | 51 | 8 | 41 | 8 | 40 | 66 | 55 | 26 | 5 | + |
| CDC42R68S | 67 | 2 | 31 | 12 | 33 | 59 | 55 | 29 | 12 | + |
| CDC42S71P | 81 | 5 | 14 | 0 | 2 | 37 | 35 | 63 | 63 | − |
| CDC42N92D | 57 | 8 | 35 | 20 | 61 | 59 | 33 | 21 | 6 | + |
| CDC42E95K | 60 | 10 | 30 | 11 | 45 | 73 | 40 | 16 | 15 | + |
| CDC42E100G | 69 | 6 | 25 | 1 | 6 | 43 | 55 | 56 | 39 | − |
| CDC42S158T | 70 | 6 | 24 | 0 | 2 | 44 | 34 | 56 | 64 | − |
| CDC42L160P | 73 | 7 | 20 | 0 | 1 | 51 | 37 | 49 | 62 | − |
| bni1/bni1 | 13 | 77 | 10 | 0 | 0 | 2 | 6 | 98 | 94 | + |
Budding patterns of strains were determined after growth to logarithmic phase in nitrogen-rich medium.
Cell shape patterns were determined after growth in nitrogen-rich medium (+N) or on nitrogen starvation plates (−N).
Invasive growth was determined after growth on nitrogen starvation plates.
The first class includes six alleles of CDC42, CDC42N26I, CDC42I46M, CDC42S71P, CDC42E100G, CDC42S158T, and CDC42L160P, which are impaired for pseudohyphal development (Fig. 3). These mutants are unable both to produce regular amounts of long PH cells and to invade the agar in response to nitrogen starvation (Table 4). A second class comprises four CDC42 alleles, CDC42A30T, CDC42D65N, CDC42N92D, and CDC42E95K, which cause a hyperfilamentous growth phenotype (Fig. 3). The most prominent phenotype of mutants in this class is that they produce significantly more cells of the long PH type. A single CDC42 allele, CDC42R68S, defines a third class of mutations. Although expression of this allele impairs filament formation of diploids, PH cell morphogenesis is normal and cells invade the agar in a manner almost indistinguishable from that of cells expressing wild-type CDC42 (Fig. 3, Table 4). This result correlates with our finding that haploids expressing the CDC42R68S allele display normal invasive growth behavior (Fig. 2A). Thus, regulation of cellular functions other than PH cell morphogenesis or invasiveness appears to be impaired in the mutants carrying the R68S mutant protein that causes a nonfilamentous phenotype.
Mutations in Cdc42p blocking PH cell morphogenesis in response to nitrogen starvation do not severely affect morphology or polarity of the YF.
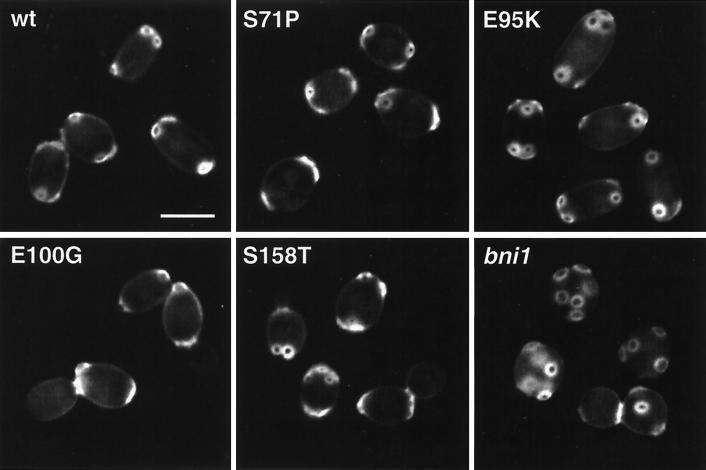
Several mutations in actin or in proteins associated with the actin cytoskeleton have been found to affect pseudohyphal development, including Bni1p, Tpm1p, Srv2p, and certain variants of Act1p (5, 38). However, these mutations not only suppress PH cell morphogenesis in response to nitrogen starvation, but additionally cause characteristic morphological and polarity defects in the yeast form when grown in nutrient-rich medium. Typical phenotypes include a random budding pattern, partially depolarized actin, and consequently a very high proportion of round YF cells. Therefore, we characterized all CDC42 mutations with respect to their effects on bud site selection, actin distribution, and cell morphology when strains were grown in nutrient-rich medium. Bud site selection patterns were determined by staining bud scars of exponentially growing YF cells with Calcofluor (Fig. 4) and dividing them into three groups: bipolar, random, and unipolar (Table 4). Actin was visualized by staining YF cells with rhodamine-phalloidin (Fig. 5). Cell shape patterns of exponentially growing cells in nitrogen-rich medium were determined as described above and directly compared to the patterns obtained by growth under nitrogen starvation conditions (Table 4).
FIG. 4.
Chitin localization. Shown are representative YF cells of diploid strains expressing wild-type CDC42 (wt) or CDC42 mutant alleles (S71P, E95K, E100G, and S158T) or carrying a bni1 mutation (RH2488). Strains were grown to the logarithmic growth phase in nutrient-rich medium, fixed, and stained with Calcofluor before fluorescence imaging. Bar, 5 μm.
FIG. 5.
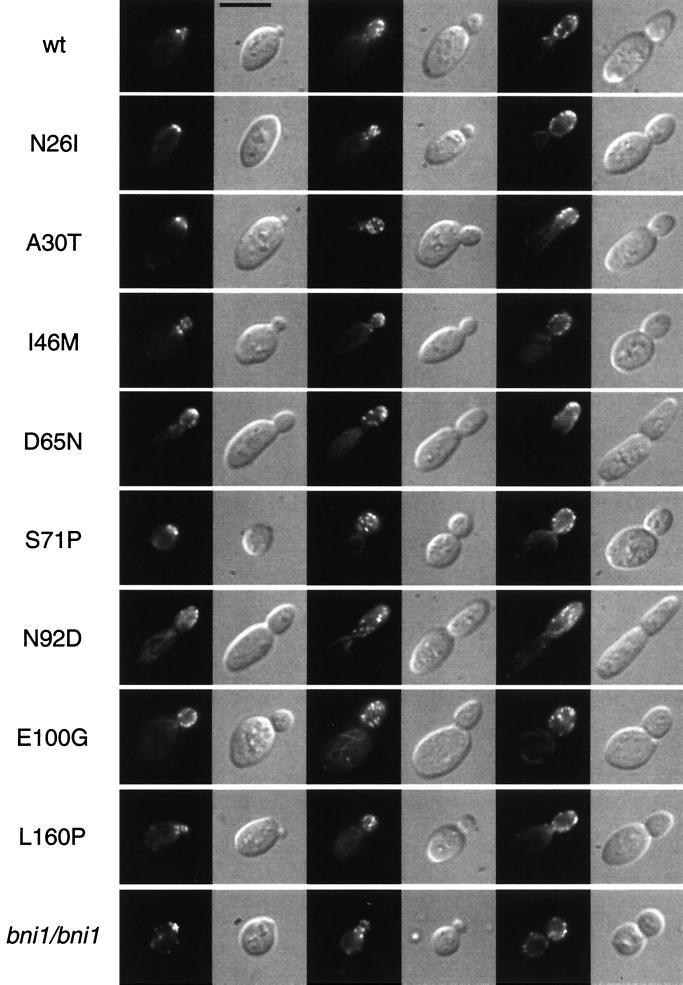
Actin localization. Shown are representative YF cells at different stages of the cell cycle of diploid strains expressing wild-type CDC42 (wt) or CDC42 mutant alleles (N26I, A30T, I46M, D65N, S71P, N92D, E100G, or L160P) or carrying a bni1 mutation (RH2488). Strains were grown to the logarithmic growth phase in nutrient-rich medium, fixed, and stained with the actin fluorescent stain rhodamine-phalloidin before observation using either fluorescence microscopy or DIC microscopy. Bar, 5 μm.
This analysis revealed that none of the Cdc42p mutations suppressing pseudohyphal development and invasive growth (N26I, I46M, S71P, E100G, S158T, or L160P) significantly affected the morphology or polarity of cells grown in the yeast form. None of these mutations caused a random budding pattern as found for a bni1 mutant strain but led to regular bipolar budding (Fig. 4, Table 4). Actin distribution in the N26I, I46M, E100G, S158T, and L160P mutants was indistinguishable from that in the CDC42 wild-type strain (Fig. 5). A higher percentage of mother cells containing actin patches at early stages of the cell division cycle were found for the S71P mutant. However, this phenotype was clearly less pronounced compared to a bni1 mutant strain (Fig. 5). The cell morphology patterns of the N26I, I46M, E100G, S158T, or L160P mutants were normal, with 43 to 51% oval YF cells (Table 4, Fig. 4). The S71P mutation had a slight but distinct effect on the yeast form morphology, with 37% of cells being oval YF cells. In contrast, only 2% of bni1 mutant cells were oval and 98% were round. Thus, with respect to cell polarity, distribution of actin patches, and yeast form morphology, the N26I, I46M, E100G, S158T, and L160P mutants do not resemble any of the bni1, tpm1, or certain act1 mutant strains. However, these CDC42 mutants have a specific block in their morphological response to nitrogen starvation conditions and are unable to grow invasively (Table 4). In contrast, bni1 and tpm1 mutations do not affect agar invasion (Table 4) (38).
Cdc42p mutations causing hyperfilamentous growth (A30T, D65N, N92D, and E95K) did not significantly alter the bud site selection patterns of the yeast form (Table 4, Fig. 4). Alterations in cell morphology patterns could be detected in the D65N, N92D, and E95K mutants, where higher proportions of elongated cells were found when strains were grown in nutrient-rich medium (Table 4). No changes in the yeast form morphology pattern was found for the A30T mutation, although this mutant produces a much higher amount of long PH cells under nitrogen starvation conditions (Table 4). The actin staining patterns of the A30T, D65N, N92D, and E95K mutants were not significantly different from those of a CDC42 wild-type strain (Fig. 5). In summary, the D65N, N92D, and E95K mutants exhibit hyperpolarized growth under both nutrient-rich and starvation conditions, partly explaining their hyperfilamentous growth phenotype. In contrast, the A30T mutant is hyperinducible for morphology changes specifically in response to nitrogen starvation.
Localization of Ste20p to the bud tip by Cdc42p is not sufficient for PH cell morphogenesis.
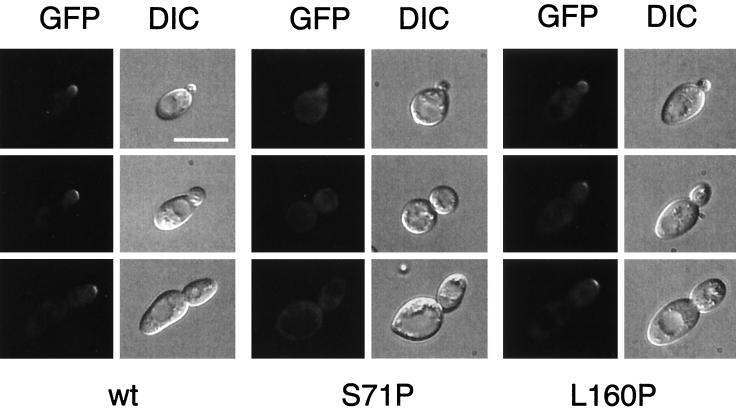
Several lines of evidence suggest that the Ste20p kinase is necessary for Cdc42p-dependent induction of pseudohyphal development and that interactions between Cdc42p and the CRIB domain of Ste20p are necessary for this induction (27, 40, 41, 47). Moreover, Ste20p is localized to the sites of polarized growth in emerging buds, while the CRIB-deleted Ste20p shows a general cytoplasmic staining (47). These results led to the view that Cdc42p is necessary for proper localization of Ste20p to the sites of polarized growth and that this localization of Ste20p during cell growth might be important for the development of PH cells. Therefore, we determined the localization of a GFP-Ste20p fusion protein in strains expressing CDC42 mutant alleles causing specific defects in pseudohyphal development. Diploid N26I, I46M, S71P, E100G, S158T, or L160P mutant strains were transformed with a plasmid carrying a GFP-STE20 fusion gene under the control of the endogenous STE20 promoter. Expression of this STE20prom-GFP-STE20 fusion gene is sufficient to rescue the filamentous growth defect of an ste20Δ/ste20Δ mutant strain (data not shown). The resulting strains were analyzed for localization of the GFP-Ste20p protein by fluorescence microscopy (Fig. 6). We predicted that if localization of Ste20p to the tip of the emerging bud was necessary for PH cell morphogenesis, all CDC42 mutant alleles causing a nonfilamentous growth phenotype should display general cytoplasmic staining. However, we found that only the S71P mutation in Cdc42p causes general cytoplasmic staining of cells by GFP-Ste20p (Fig. 6). In all other strains expressing either the N26I, I46M, E100G, S158T, or L160P mutants, GFP-Ste20p was found to be localized to the tip of emerging cells (for L160P, see Fig. 6), although these strains are unable to form pseudohyphae. Thus, localization of Ste20p to the bud tip of emerging daughter cells alone is not sufficient to induce PH cell morphogenesis.
FIG. 6.
Subcellular localization of GFP-Ste20p in CDC42 pseudohyphal mutants. Shown are cells of diploid strains that express wild-type CDC42 (wt), CDC42S71P (S71P), or CDC42L160P (L160P) and harbor plasmid pME1760 encoding GFP-Ste20p under the control of the STE20 promoter. Living cells at different stages of the cell cycle were chosen for photography according to their bud size and were viewed by either fluorescence microscopy (GFP) or DIC microscopy. Identical results were obtained with centromere-based plasmid pME1759, although with markedly decreased fluorescence signals due to lower expression of GFP-Ste20p. Bar, 10 μm.
Cdc42p developmental mutations alter interaction patterns with several downstream effectors.
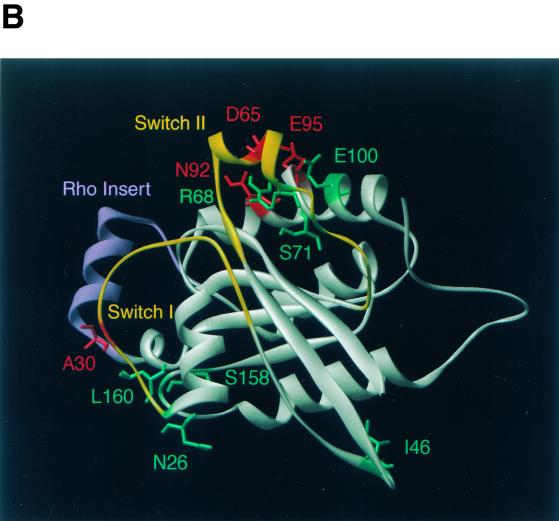
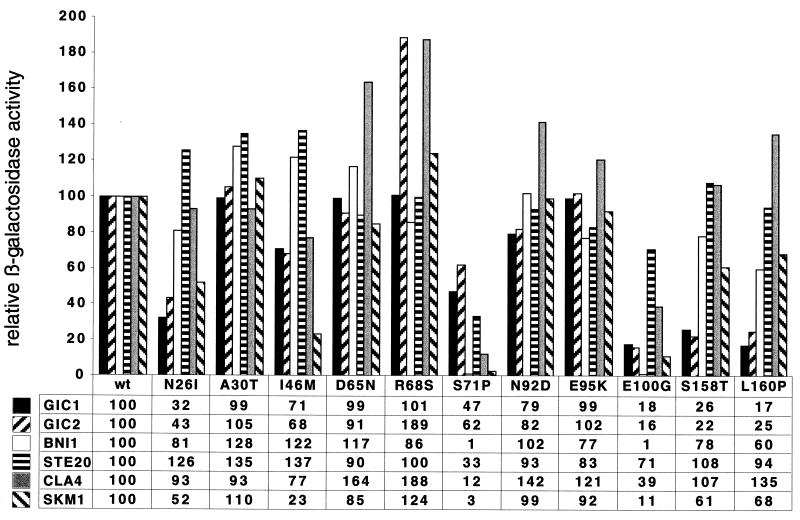
The specific developmental defects caused by the novel mutations in CDC42 obtained here suggested that these Cdc42p mutants do not display general biochemical defects but might have altered interaction patterns with downstream effectors. Indeed, analysis of the mutations in a three-dimensional structure model of S. cerevisiae Cdc42p revealed that most mutations were located on the surface of Cdc42p, predicting altered interactions with other proteins (Fig. 7). To test this prediction, we performed a two-hybrid analysis between all Cdc42p mutants and the Gic1p, Gic2p, Bni1p, Ste20p, Cla4p, and Skm1p effector proteins. The C188S mutation was additionally introduced into all mutants to prevent localization of the proteins to the plasma membrane. The interaction of mutant proteins with the diverse effectors was then measured and compared to that of Cdc42C188Sp as the control (Fig. 8).
FIG. 7.
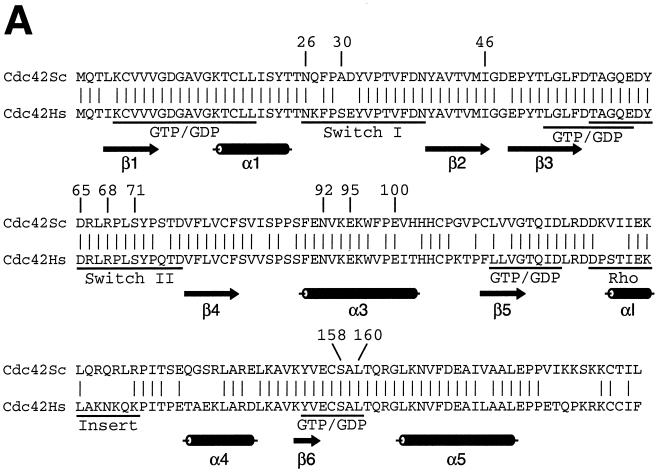
Cdc42p developmental mutations and structural model of Cdc42p. (A) Sequence alignment of Cdc42p from S. cerevisiae (Cdc42Sc) and from human (Cdc42Hs). Vertical lines indicate identical residues. Known GTP-binding/hydrolysis domains (GTP/GDP), switch I and switch II domains, and the Rho insert domain are underlined. Numbers indicate residues that were identified by mutations in the yeast Cdc42p sequence in this study. (B) Three-dimensional structure model of S. cerevisiae Cdc42p was obtained by homology modeling of the primary structure of S. cerevisiae Cdc42p using the Swiss-Model service (19) and is based on the X-ray crystal structure of Cdc42Hs (43, 53). Amino acid residues identified in this study are indicated in different colors based on the phenotypes caused by their exchange. Substitutions of green residues were found to suppress pseudohyphal or invasive growth, and exchanges of red residues enhanced pseudohyphal development. Switch I and switch II domains are colored yellow, and the Rho insert domain is shown in purple.
FIG. 8.
Two-hybrid protein interactions between Cdc42 proteins and effector proteins Gic1p, Gic2p, Bni1p, Ste20p, Cla4p, and Skm1p. All Cdc42p proteins measured (wild-type [wt] or mutant proteins N26I, A30T, I46M, D65N, R68S, S71P, N92D, E95K, E100G, S158T, and L160P) carry the C188S mutation to prevent plasma membrane localization. Bars and values show relative β-galactosidase activities normalized to the activities obtained for wild-type Cdc42p (wt) and the different effectors, with values set to 100 (for absolute units, see Materials and Methods). The value shown for each interaction represents the average of at least four independent β-galactosidase assays, each done in triplicate.
This complex analysis of 72 different combinations revealed specific changes in the effector interaction patterns for the six Cdc42p mutants causing defects in pseudohyphal development and invasive growth (N26I, I46M, S71P, E100G, S158T, and L160P). Four distinct interaction patterns were found. For N26I, S158T, and L160P, binding to Gic1p was clearly reduced (between 17 and 32% of the control), and interaction with Gic2p was reduced to 22 to 43%. In addition, the interaction of these mutants with Skm1p and Bni1p was partially reduced (between 52 and 81% of the control). However, no alterations in binding to Ste20p and Cla4p were found. The I46M mutant protein was specifically reduced in its interaction with Skm1p, with a binding efficiency of only 23% compared to the control. No strong alterations were detected for the binding of I46M to the other effectors. The most dramatic changes for effector binding were measured for the S71P and E100G mutant proteins. Whereas interaction of S71P with Gic1p and Gic2p was only partially altered, binding to Bni1p, Ste20p, Cla4p, and Skm1p was strongly reduced. Yet another pattern was found for the E100G mutant protein. Binding of E100G to Gic1p, Gic2p, Bni1p, and Skm1p was strongly reduced, whereas interaction with Ste20p and Cla4p was only partially affected. In case of the nonfilamentous R68S mutant, no reduction in effector binding was measured, but a somewhat higher affinity to Gic2p and Cla4p was found. No significant alterations in effector binding were detectable for any of the hyperfilamentous A30T, D65N, N92D, or E95K variants.
In summary, the effects on pseudohyphal development and invasive growth of the nonfilamentous N26I, I46M, S71P, E100G, S158T, and L160P Cdc42p variants can be correlated with alterations in the binding to distinct subsets of downstream effectors. This suggests that several of the Cdc42p mutants isolated here are specific effector mutants.
DISCUSSION
Evidence for specific Cdc42p signaling mutations.
Here, we have isolated novel Cdc42p mutant proteins that have lost the ability to confer PH cell morphogenesis and invasive growth in response to nitrogen starvation. A crucial finding of our study is that many of these mutants do not affect polarity or cell morphogenesis during yeast form proliferation, as measured by morphology indices, actin staining, and bud scar distribution. The developmental defects caused by the N26I, I46M, E100G, S158T, and L160P mutations are very specific and are clearly different from previously identified mutations in Cdc42p that cause either lethality (e.g., G12V, K16A, T35A, D38A, Y40K, Q61L, and D118A) or temperature sensitivity (e.g., K5A, Y32K, V36T, V44A, D76A, and W97R) and that lead to significant changes in actin distribution or cell morphology and polarity (10, 24, 25, 37, 49, 65). Mutations in Cdc42p identified here also differ from mutations in actin-associated proteins such as Bni1p or Tpm1p. Diploid bni1 and tpm1 mutants display altered cell morphology already under yeast form growth conditions (and consequently are suppressed for PH morphogenesis), but unlike the N26I, I46M, E100G, S158T, and L160P mutants, have no significant invasive growth defects (38). Phenotypically, the CDC42 mutants isolated here more strongly resemble mutants with alterations in the known signaling pathways that control pseudohyphal growth. For instance, mutations in components of the pseudohyphal and invasive growth Kss1p-MAPK cascade (e.g., Ste20p, Ste11p, Ste7p, Ste12p, and Tec1p) as well as mutations in Ras2p, Gpa2p, or Tpk2p specifically suppress PH morphogenesis and invasive growth. Much like the N26I, I46M, E100G, S158T, and L160P mutations isolated here, Kss1p-MAPK and cAMP signaling mutations do not affect cell polarity or morphology during yeast form proliferation. Expression of FLO11, a gene reporting activities of both the Kss1p-MAPK and the cAMP pathways, is also reduced in some of these Cdc42p mutants, whereas activated expression of FUS1 and formation of mating projections in response to pheromone are not affected. Thus, several of the Cdc42p mutations uncovered in this study appear to be specific signaling mutations rather than general morphological mutations because the mutants display transcriptional and morphological defects only in response to certain nutritional signals.
We also found several mutations in Cdc42p that cause alterations in cell morphology independently of the growth conditions. These include the hyperfilamentous D65N, N92D, and E95K and to some degree the nonfilamentous S71P mutation. However, these mutations also differ from other morphological mutations of Cdc42p, as they do not cause lethality or temperature sensitivity. Because these mutations also affect invasive growth, they might cause both general morphological alterations and signaling defects.
Novel Cdc42p effector mutations point to Gic1p, Gic2p, and Skm1p as development-specific Cdc42p effectors.
A surprising outcome of our study is the finding that several of the Cdc42p mutations that suppress pseudohyphal and invasive growth confer specific defects in binding to Gic1p and Gic2p (S158T and L160P) or Skm1p (I46M). These effectors of Cdc42p have not yet been described as being involved in regulation of pseudohyphal and invasive growth. Gic1p and Gic2p are required for cell polarization but are dispensable for MAPK signal transduction and thus have been suggested to link Cdc42p to dynamic rearrangements of the actin cytoskeleton (4, 6). Interestingly, Gic1p and Gic2p act in a pathway for signaling from Cdc42p to the actin cytoskeleton that operates in parallel with a pathway that includes Bni1p, Msb3p, and Msb4p (3). Because the Gic1p/Gic2p pathway and the Bni1p/Msb3p/Msb4p pathways are largely redundant in function, it has been proposed that each of the pathways may be optimized for distinct growth conditions. One interpretation of our results is that the Gic1p/Gic2p pathway might be optimized for cell polarization in response to the nutritional signals that induce pseudohyphal and invasive growth. Whether interaction of Cdc42p with the Bni1p/Msb3p/Msb4p pathway is also essential for pseudohyphal development cannot be concluded from our study, because the two mutations blocking interaction with Bni1p (S71P and E100G) also diminish binding to other effectors. However, these mutants allow the conclusion that binding of Cdc42p to Bni1p is not essential for all functions of this effector. Both the S71P and E100G mutations completely abolish Bni1p binding, yet these mutants do not display the polarity or morphology defects caused by a bni1 mutation. This finding is not unexpected, because Bni1p interacts with several other proteins, such as Rho1p, Rho3p, Rho4p, Pfy1p, and Spa2p (13, 16, 22). Yet S71P and E100G are novel mutations as defining residues of Cdc42p that are essential for binding to Bni1p.
Our results indicate that interaction between Cdc42p and Gic1p/Gic2p is required for pseudohyphal development. However, this function of Cdc42p alone cannot be sufficient for a full developmental response. This interpretation can be made based on the finding that the I46M mutant displays only minimal alterations in Gic1p/Gic2p binding but has a much more pronounced defect in binding to Skm1p. This points towards Skm1p as a further effector of Cdc42p that affects pseudohyphal and invasive growth.
Our study did not uncover mutations in Cdc42p that exclusively block binding to Ste20p. The only mutant showing significantly reduced interaction with Ste20p is S71P. This finding correlates with the fact that S71P is the only mutant found here that affects both localization of GFP-Ste20p to the bud tip and expression of the Kss1p-MAPK target gene FLO11. However, although reduced binding of Cdc42p to Ste20p causes some of the phenotypes that would be expected based upon previous studies defining Ste20p as an important pseudohyphal effector, interpretation of these data is complicated by the fact that S71P also affects interaction of Cdc42p with other effectors.
Altogether, the effector mutants isolated here are very helpful for dissecting the distinct functions of Cdc42p, although we cannot exclude that some of the functions that are impaired in these mutants are due to reduced binding or activation of further effectors of Cdc42p. This might also explain the fact that none of the hyperfilamentous A30T, D65N, N92D, or E95K variants display altered binding patterns to the effectors tested here. However, the novel mutants found in this study point towards Gic1p/Gic2p and Skm1p as effectors that, apart from Ste20p, might be important for morphological responses to nutritional signals. Whether each of these effectors acts independently or whether and how functions of Ste20p, Gic1p/Gic2p, and Skm1p are connected to control pseudohyphal development remains to be investigated.
New insights into Cdc42p structure and effector binding.
A growing compendium of mutations in CDC42 together with both the solution and the crystal structures of human Cdc42Hs have defined several functional domains within Cdc42p (14, 24, 43, 50, 53). Four domains have been implicated in the binding and hydrolysis of GTP, and three regions—switch I, switch II, and Rho insert—are involved in protein-protein interactions with downstream effector proteins (Fig. 7). With the exception of Rho insert, all of these domains are highly conserved between human Cdc42Hs and S. cerevisiae Cdc42p (Fig. 7A). Further important information on Cdc42p structure and effector binding domains comes from studies using specific effector mutants that are selectively impaired for binding to certain effectors but not to others. For instance, the V44A mutation selectively blocks binding of Cdc42p to Gic1p, Gic2p, and Cla4p but not to Bni1p, Skm1p, or Ste20p (49). Our study has identified several residues of Cdc42p previously unknown to be required for selective effector binding. Although specificity is not absolutely clear-cut in all mutants, significant differences can be observed. Residues S158 and L160 show selectivity for interaction with Gic1p and Gic2p. These two residues map to a conserved region of Cdc42p that has been implicated in GTP binding and hydrolysis. However, the S158T and L160P mutations are not likely to inhibit the GTPase activity of the protein, because neither binding to Bni1p, Ste20p, or Cla4p nor functions essential for cell division are affected. A more likely explanation is that residues S158 and L160 are involved in effector binding. N26 is another residue that is required for binding to Gic1p/Gic2p (and to a certain extent to Skm1p) but not for interaction with Bni1p, Ste20p, or Cla4p. N26 maps to the end of switch I, a region that, together with switch II, is among the regions of Cdc42p that display the most significant flexibility in nuclear magnetic resonance measurement of the protein (33). The three-dimensional structure model of Cdc42p shows that N26, S158, and L160 are in close proximity on the surface of Cdc42p (Fig. 7), defining this region of the protein as important for binding of Gic1p/Gic2p. Another residue important for discrimination between distinct effectors is I46, due to the selective binding pattern of the I46M mutant that is impaired for binding to Skm1p. I46 is located in the loop between switch I and switch II, a region that also includes residue V44, which is required for selective binding to Gic1p/Gic2p and Cla4p (49). Our study has further identified S71, located within the switch II region, and E100, residing within helix α3, as residues that are crucial for binding to diverse effector proteins. These residues do not appear to be selective for a single effector. However, they still define a part of the protein that appears to be highly important for binding to Bni1p. The effects of the S71P and E100G mutations on binding of Cdc42p to Gic1p/Gic2p (S71P) or Ste20p and Cla4p (E100G) are by far less pronounced than effects measured on interaction with Bni1p (with 1% of binding capacity left). Because the two residues are very close to each other on the Cdc42p surface (Fig. 7), this region might be an important binding site for Bni1p.
Taken together, our results help to improve our understanding of the functions and functional domains of the essential GTPase Cdc42p in S. cerevisiae. Given the high degree of conservation of Cdc42p throughout the eukaryotic kingdom, mutations identified here might help to dissect the distinct functions of Cdc42p in other organisms.
ACKNOWLEDGMENTS
We thank Maria Meyer for excellent technical assistance during the course of this work. We are grateful to Charles Boone, Roger Brent, J. Hegemann, D. Johnson, D. Lew, M. Peter, and S. Rupp for generously providing plasmids and strains. We thank Sven Krappmann and Naimeh Taheri for helpful comments on the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Volkswagenstiftung.
REFERENCES
- 1.Adams A E, Pringle J R. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 1991;194:729–731. doi: 10.1016/0076-6879(91)94054-g. [DOI] [PubMed] [Google Scholar]
- 2.Benton B K, Tinkelenberg A, Gonzalez I, Cross F R. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol. 1997;17:5067–5076. doi: 10.1128/mcb.17.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi E, Chiavetta J B, Chen H, Chen G C, Chan C S, Pringle J R. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol Biol Cell. 2000;11:773–793. doi: 10.1091/mbc.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown J L, Jaquenoud M, Gulli M P, Chant J, Peter M. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 1997;11:2972–2982. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cali B M, Doyle T C, Botstein D, Fink G R. Multiple functions for actin during filamentous growth of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1873–1889. doi: 10.1091/mbc.9.7.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G C, Kim Y J, Chan C S. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 1997;11:2958–2971. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G C, Zheng L, Chan C S. The LIM domain-containing Dbm1 GTPase-activating protein is required for normal cellular morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1376–1390. doi: 10.1128/mcb.16.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross F R, Tinkelenberg A H. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 9.Cvrckova F, De Virgilio C, Manser E, Pringle J R, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 10.Davis C R, Richman T J, Deliduka S B, Blaisdell J O, Collins C C, Johnson D I. Analysis of the mechanisms of action of the Saccharomyces cerevisiae dominant lethal cdc42G12V and dominant negative cdc42D118A mutations. J Biol Chem. 1998;273:849–858. doi: 10.1074/jbc.273.2.849. [DOI] [PubMed] [Google Scholar]
- 11.Eby J J, Holly S P, van Drogen F, Grishin A V, Peter M, Drubin D G, Blumer K J. Actin cytoskeleton organization regulated by the PAK family of protein kinases. Curr Biol. 1998;8:967–970. doi: 10.1016/s0960-9822(98)00398-4. [DOI] [PubMed] [Google Scholar]
- 12.Epp J A, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7:921–929. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- 13.Evangelista M, Blundell K, Longtine M S, Chow C J, Adames N, Pringle J R, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 14.Feltham J L, Dotsch V, Raza S, Manor D, Cerione R A, Sutcliffe M J, Wagner G, Oswald R E. Definition of the switch surface in the solution structure of Cdc42Hs. Biochemistry. 1997;36:8755–8766. doi: 10.1021/bi970694x. [DOI] [PubMed] [Google Scholar]
- 15.Fujiwara T, Tanaka K, Inoue E, Kikyo M, Takai Y. Bni1p regulates microtubule-dependent nuclear migration through the actin cytoskeleton in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8016–8027. doi: 10.1128/mcb.19.12.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 18.Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 19.Guex N, Peitsch M C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 20.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press; 1991. [PubMed] [Google Scholar]
- 21.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a Human G1 and S Phase Protein Phosphatase That Associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 22.Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen R-P, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional Myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 24.Johnson D I. Cdc42: an essential rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozminski K G, Chen A J, Rodal A A, Drubin D G. Functions and functional domains of the GTPase Cdc42p. Mol Biol Cell. 2000;11:339–354. doi: 10.1091/mbc.11.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kübler E, Mösch H-U, Rupp S, Lisanti M P. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- 27.Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall J E, Thomas D Y. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li R, Debreceni B, Jia B, Gao Y, Tigyi G, Zheng Y. Localization of the PAK1-, WASP-, and IQGAP1-specifying regions of Cdc42. J Biol Chem. 1999;274:29648–29654. doi: 10.1074/jbc.274.42.29648. [DOI] [PubMed] [Google Scholar]
- 30.Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 32.Lo W S, Dranginis A M. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh A P, Guo W, Nicholson L K, Oswald R E. Backbone dynamics of inactive, active, and effector-bound Cdc42Hs from measurements of 15N relaxation parameters at multiple field strengths. Biochemistry. 1999;38:12547–12557. doi: 10.1021/bi9913707. [DOI] [PubMed] [Google Scholar]
- 34.Lorenz M C, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackay D J, Hall A. Rho GTPases. J Biol Chem. 1998;273:20685–20688. doi: 10.1074/jbc.273.33.20685. [DOI] [PubMed] [Google Scholar]
- 36.Martin H, Mendoza A, Rodriguez-Pachon J M, Molina M, Nombela C. Characterization of SKM1, a Saccharomyces cerevisiae gene encoding a novel Ste20/PAK-like protein kinase. Mol Microbiol. 1997;23:431–444. doi: 10.1046/j.1365-2958.1997.d01-1870.x. [DOI] [PubMed] [Google Scholar]
- 37.Miller P J, Johnson D I. Characterization of the Saccharomyces cerevisiae cdc42–1ts allele and new temperature-conditional-lethal cdc42 alleles. Yeast. 1997;13:561–572. doi: 10.1002/(SICI)1097-0061(199705)13:6<561::AID-YEA114>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 38.Mösch H-U, Fink G R. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mösch H-U, Graf R, Braus G H. Sequence-specific initiator elements focus initiation of transcription to distinct sites in the yeast TRP4 promoter. EMBO J. 1992;11:4583–4590. doi: 10.1002/j.1460-2075.1992.tb05560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mösch H-U, Kübler E, Krappmann S, Fink G R, Braus G H. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:1325–1335. doi: 10.1091/mbc.10.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mösch H-U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naqvi S N, Zahn R, Mitchell D A, Stevenson B J, Munn A L. The WASP homologue Las17p functions with the WIP homologue End5p/verprolin and is essential for endocytosis in yeast. Curr Biol. 1998;8:959–962. doi: 10.1016/s0960-9822(98)70396-3. [DOI] [PubMed] [Google Scholar]
- 43.Nassar N, Hoffman G R, Manor D, Clardy J C, Cerione R A. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat Struct Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- 44.Oehlen L J, Cross F R. The role of Cdc42 in signal transduction and mating of the budding yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:8556–8559. doi: 10.1074/jbc.273.15.8556. [DOI] [PubMed] [Google Scholar]
- 45.Osman M A, Cerione R A. Iqg1p, a yeast homologue of the mammalian IQGAPs, mediates Cdc42p effects on the actin cytoskeleton. J Cell Biol. 1998;142:443–455. doi: 10.1083/jcb.142.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Owen D, Mott H R, Laue E D, Lowe P N. Residues in Cdc42 that specify binding to individual CRIB effector proteins. Biochemistry. 2000;39:1243–1250. doi: 10.1021/bi991567z. [DOI] [PubMed] [Google Scholar]
- 47.Peter M, Neiman A M, Park H O, van Lohuizen H, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 48.Pringle J R. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- 49.Richman T J, Sawyer M M, Johnson D I. The Cdc42p GTPase is involved in a G2/M morphogenetic checkpoint regulating the apical-isotropic switch and nuclear division in yeast. J Biol Chem. 1999;274:16861–16870. doi: 10.1074/jbc.274.24.16861. [DOI] [PubMed] [Google Scholar]
- 50.Rittinger K, Walker P A, Eccleston J F, Nurmahomed K, Owen D, Laue E, Gamblin S J, Smerdon S J. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- 51.Roberts R L, Fink G R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 52.Roberts R L, Mösch H-U, Fink G R. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- 53.Rudolph M G, Wittinghofer A, Vetter I R. Nucleotide binding to the G12V-mutant of Cdc42 investigated by X-ray diffraction and fluorescence spectroscopy: two different nucleotide states in one crystal. Protein Sci. 1999;8:778–787. doi: 10.1110/ps.8.4.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rupp S, Summers E, Lo H J, Madhani H, Fink G R. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shannon K B, Li R. The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast. Mol Biol Cell. 1999;10:283–296. doi: 10.1091/mbc.10.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon M N, De Virgilio C, Souza B, Pringle J R, Abo A, Reed S I. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature. 1995;376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- 58.Sloat B F, Adams A, Pringle J R. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1981;89:395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stevenson B J, Ferguson B, De Virgilio C, Bi E, Pringle J R, Ammerer G, Sprague G F., Jr Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes Dev. 1995;9:2949–2963. doi: 10.1101/gad.9.23.2949. [DOI] [PubMed] [Google Scholar]
- 60.Umikawa M, Tanaka K, Kamei T, Shimizu K, Imamura H, Sasaki T, Takai Y. Interaction of Rho1p target Bni1p with F-actin-binding elongation factor 1alpha: implication in Rho1p-regulated reorganization of the actin cytoskeleton in Saccharomyces cerevisiae. Oncogene. 1998;16:2011–2016. doi: 10.1038/sj.onc.1201724. [DOI] [PubMed] [Google Scholar]
- 61.Ward M P, Gimeno C J, Fink G R, Garrett S. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol Cell Biol. 1995;15:6854–6863. doi: 10.1128/mcb.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zahner J E, Harkins H A, Pringle J R. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Z, Leung T, Manser E, Lim L. Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol Cell Biol. 1995;15:5246–5257. doi: 10.1128/mcb.15.10.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng Y, Cerione R, Bender A. Control of the yeast bud-site assembly GTPase Cdc42: catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994;269:2369–2372. [PubMed] [Google Scholar]
- 65.Ziman M, O'Brien J M, Ouellette L A, Church W R, Johnson D I. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol Cell Biol. 1991;11:3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]