Abstract
The assembly of light-harvesting chlorophyll-binding proteins (LHCPs) is coordinated with chlorophyll biosynthesis during chloroplast development. The ATP-independent chaperone known as chloroplast signal recognition particle 43 (cpSRP43) mediates post-translational LHCP targeting to the thylakoid membrane, and also participates in tetrapyrrole biosynthesis (TBS). How these distinct actions of cpSRP43 are controlled has remained unclear. Here, we demonstrate that cpSRP43 effectively protects several TBS proteins from heat-induced aggregation and enhances their stability during leaf greening and heat shock. While the substrate-binding domain (SBD) of cpSRP43 is sufficient for chaperoning LHCPs, stabilization of TBS clients requires the chromodomain 2 of the protein. Strikingly, cpSRP54 – which activates cpSRP43’s LHCP-targeted function – inhibits the chaperone activity of cpSRP43 towards TBS proteins. High temperature weakens the interaction of cpSRP54 with cpSRP43, thus freeing cpSRP43 to interact with and protect the integrity of TBS proteins. Our data indicate that the temperature-sensitivity of the cpSRP43-cpSRP54 complex enables cpSRP43 to serve as an autonomous chaperone for thermoprotection of TBS proteins.
Introduction
Chlorophyll (Chl) enables oxygenic photoautotrophs to absorb solar energy, transfer excitation energy, and initiate charge separation in the light reactions of photosynthesis1. At the thylakoid membrane, Chl is bound to various Chl-binding proteins of photosystems I and II, and their light-harvesting antenna complexes (LHCs)2,3. The availability of newly synthesized Chl is critical for the stability, proper folding, and membrane integration of Chl-binding proteins, including the light-harvesting Chl-binding proteins (LHCPs)4,5. However, how plants coordinate the biogenesis of LHC apoproteins with de novo synthesis of Chl to meet the dynamic demand for Chl during LHC assembly under changing environmental conditions remains unclear6,7.
Chl is synthesized via the magnesium (Mg) branch of tetrapyrrole biosynthesis (TBS), which also produces three other essential end-products – heme, siroheme and phytochromobilin – in plants8,9. Glutamyl-tRNA reductase (GluTR) is the rate-limiting enzyme in TBS, and together with glutamate 1-semialdehyde aminotransferase it catalyzes the synthesis of 5-aminolevulinic acid (ALA). Eight ALA molecules are assembled into protoporphyrin IX (Proto), which is subsequently distributed between the magnesium (Mg) and iron branches of TBS for Chl and heme production, respectively. Mg chelatase (MgCh), which is composed of the catalytic CHLH and the two AAA+ subunits CHLD and CHLI, catalyzes the insertion of Mg2+ into Proto to produce Mg-protoporphyrin IX (MgP)10,11, which is further converted into MgP monomethylester (MgPMME) and protochlorophyllide (Pchlide). GENOMES UNCOUPLED 4 (GUN4) binds to Proto and MgP, as well as CHLH, to stimulate MgCh activity12–14. Although the essential enzymatic steps in TBS are well documented15, it is still not entirely known how land plants and green algae adjust ALA synthesis and channel metabolic intermediates to ensure the adequate formation of end products and avoid the accumulation of phototoxic free tetrapyrrole metabolites.
Increased temperature owing to climate change poses a severe threat to plant viability16. Heat stress impairs the activity and stability of proteins and alters the fluidity of phospholipid membranes, and can cause irreversible damage and cell death17,18. Plants have evolved sophisticated mechanisms that enhance thermotolerance, which is highly related to the multifaceted regulation of Chl metabolism19,20. For example, the RNA-binding protein FCA is required for the thermal induction of PROTCHLOROPHYLLIDE OXIDOREDUCTASE (POR) transcription and thus contributes to the maintenance of POR upon exposure to rising temperature21. However, little is known about how plants post-translationally fine-tune Chl biosynthesis in response to heat stress.
The chloroplast signal recognition particle (cpSRP) pathway mediates post-translational LHCP transport within chloroplasts22,23. LHCPs imported into the stroma of chloroplasts are kept soluble by forming a ‘transit complex’ with the cpSRP, which itself consists of the subunits cpSRP43 and cpSRP5424,25. cpSRP43 is an ATP-independent chaperone that is responsible for protecting LHCPs from aggregation26, while cpSRP54 serves to allosterically activates its LHCP-targeted chaperone activity27,28. cpSRP54 also mediates interaction of the cpSRP complex with the SRP receptor cpFtsY to deliver LHCPs to the translocase Albino 3 (Alb3) at the thylakoid membrane29,30. Alb3 then triggers the release of LHCP from cpSRP43 and mediates the integration of LHCP into the thylakoid membrane31, where assembly of Chls into the LHCs takes place.
Apart from its role in the delivery of LHCPs to the thylakoid membrane, cpSRP43 is also implicated in TBS. It enhances the stability of GluTR to support efficient ALA synthesis in Arabidopsis32, which is compatible with a role of cpSRP43 in the coordination of LHCP biogenesis and Chl biosynthesis. However, the molecular mechanism that controls the action of cpSRP43 in these two processes, and the physiological significance of cpSRP43’s chaperone function under adverse environmental conditions, remain unclear. In this study, we report that cpSRP43 efficiently protects GluTR and two vital components of MgCh, CHLH and its positive regulator GUN4, from heat-induced aggregation, contributing to enhanced stability of these proteins during leaf greening and heat shock stress. Unexpectedly, the action of cpSRP43 on these TBS proteins is inhibited by cpSRP54, indicating that its LHCP- and TBS-directed functions are mutually exclusive. Our data suggest that a heat-sensitive interaction between cpSRP43 and cpSRP54 regulates the ability of cpSRP43 to switch between these two roles: as a dedicated chaperone for LHCP translocation and as an autonomous chaperone to protect mature TBS proteins from heat stress.
Results
cpSRP43 interacts with and stabilizes multiple TBS proteins
To identify novel TBS clients for cpSRP43, the metabolic flows through this pathway were re-examined in seedlings of (i) the Arabidopsis cpsrp43 knockout mutant (designated chaos)33, (ii) a cpSRP43-OX line constitutively expressing CaMV 35S::cpSRP43 in the chaos background, and (iii) a wild-type control (Landsberg erecta, Ler-0) (Fig. 1a and 1b). The chaos mutant showed growth retardation and pale-green leaves. The weak pigmentation of chaos correlated with lower Chl levels, diminished ALA synthesis rate, and lower steady-state levels of Chl precursors including MgP, MgPMME, Pchlide, and chlorophyllide (Chlide) (Fig. 1c–1e). In contrast, the heme content of chaos was WT-like (Fig. 1f). The defect in Chl biosynthesis was completely rescued by constitutive overexpression of cpSRP43 (Fig. 1a–1e). These results point to a specific requirement for cpSRP43 to maintain efficient Chl synthesis.
Fig. 1. cpSRP43 is required for the stabilization of GluTR, CHLH and GUN4.
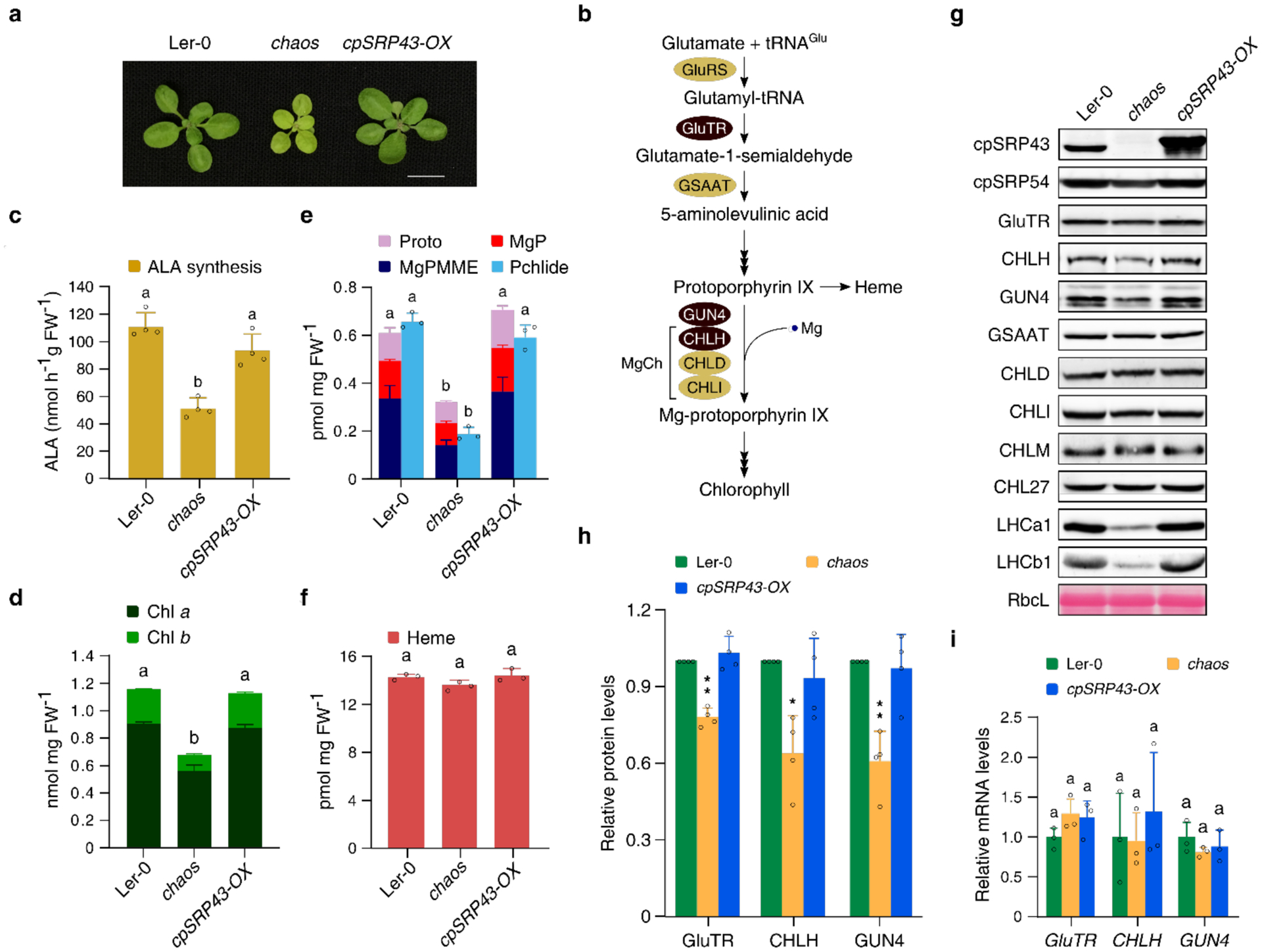
a, A representative image of 18-day-old chaos, cpSRP43-OX and wild-type (Ler-0) plants grown under normal conditions (18 h light/6 h dark, 100 μmol photons m−2 s−1). Scale bar: 1 cm. b, Scheme of the TBS pathway in plants. The key enzymes regulated by cpSRP43 are highlighted by reddish brown shadow. GluRS, glutamyl-tRNA synthetase; GluTR, glutamyl-tRNA reductase; GSAAT, glutamate 1-semialdehyde aminotransferase; MgCh, magnesium chelatase; GUN4, genomes uncoupled 4; CHLD and CHLI, two AAA+ subunits of MgCh. c-f, ALA synthesis rate (c) and levels of Chl (d), the indicated TBS metabolic intermediates (e), and heme (f) in 18-day-old Ler-0, chaos and cpSRP43-OX grown under the same conditions as in a; MgP, Mg-protoporphyrin IX; MgPMME, MgP monomethylester; Pchlide, protochlorophyllide. FW, fresh weight. g, Steady-state levels of TBS proteins in 18-day-old Ler-0, chaos and cpSRP43-OX grown as in a were quantified by immunoblotting using the indicated antibodies. The Ponceau S-stained large subunit of RuBisCO (RbcL) is shown as a loading control. h, Semiquantitative analysis with Image J software (NIH) of the immunoblots in g from four biological replicates. The relative amounts of GluTR, CHLH and GUN4 in chaos and cpSRP43-OX were normalized to the levels in Ler-0. The data are plotted as means ± s.d. (n = 4). The statistical analyses were performed using two-tailed Student’s t-tests. The asterisks indicate significant differences compared to protein levels in Ler-0: * P < 0.05, **P < 0.01. i, Relative mRNA levels of GluTR, CHLH and GUN4 in 18-day-old Ler-0, chaos and cpSRP43-OX grown as in a. Gene expression level was calculated relative to that in Ler-0, using SAND as the reference gene. All values in this Figure are plotted as means ± s.d. (n = 3–4). The small open circles represent the individual data points. Letters above histograms indicate significant differences, as determined by one-way ANOVA with Tukey test (P < 0.05).
The steady-state levels of TBS proteins in chaos were compared with those in cpSRP43-OX and Ler-0 (Fig. 1g and 1h). In agreement with our previous observations, amounts of GluTR were reduced by approximately 25% in chaos, while levels of MgP methyltransferase (CHLM), and MgPMME oxidative cyclase (CHL27) were unaltered32. Similar to GluTR, the steady-state levels of CHLH and GUN4 were significantly decreased in chaos compared with Ler-0, and were restored to wild-type levels in cpSRP43-OX. Quantitative real-time PCR (qRT-PCR) analyses showed that transcript levels of CHLH and GUN4 were comparable in all three lines (Fig. 1i), indicating that the reduced contents of CHLH and GUN4 contents in chaos were not due to transcriptional regulation. Finally, lack of cpSRP5424,25 led to slightly increased steady-state levels of GluTR and CHLH and a wild-type-like level of GUN4, despite the retarded growth and pale-green leaf phenotype observed in the cpsrp54 knockout mutant ffc34 (Supplementary Fig. 1). These results indicate that cpSRP43, but not cpSRP54, is required for the stability of TBS proteins.
To determine whether cpSRP43’s role in stabilizing CHLH and GUN4 depends on a direct protein-protein interaction, we carried out bimolecular fluorescence complementation (BiFC) assays. Both CHLH and GUN4 directly interacted with cpSRP43, whereas a negative control (CHLM) did not (Fig. 2a and Supplementary Fig. 2). In vitro pull-down assays verified these interactions. Both CHLH and GUN4 were detected in the eluted fraction following incubation with GST-cpSRP43, but not with GST alone (Fig. 2b and 2c). These data, along with equivalent experiments previously carried out with cpSRP43 and GluTR32, demonstrate that cpSRP43 physically interacts with GluTR, CHLH, and GUN4. Nevertheless, neither cpSRP43 nor GST affected MgCh activity, while GUN4 enhanced MgCh activity by approximately 12-fold (Supplementary Fig. 3), excluding a role for cpSRP43 in the control of MgCh’s enzymatic activity. Together, these results suggest a post-translational role for cpSRP43 in stabilizing several TBS proteins involved in chlorophyll biosynthesis via direct interactions.
Fig. 2. Physical interactions of cpSRP43 with CHLH and GUN4.
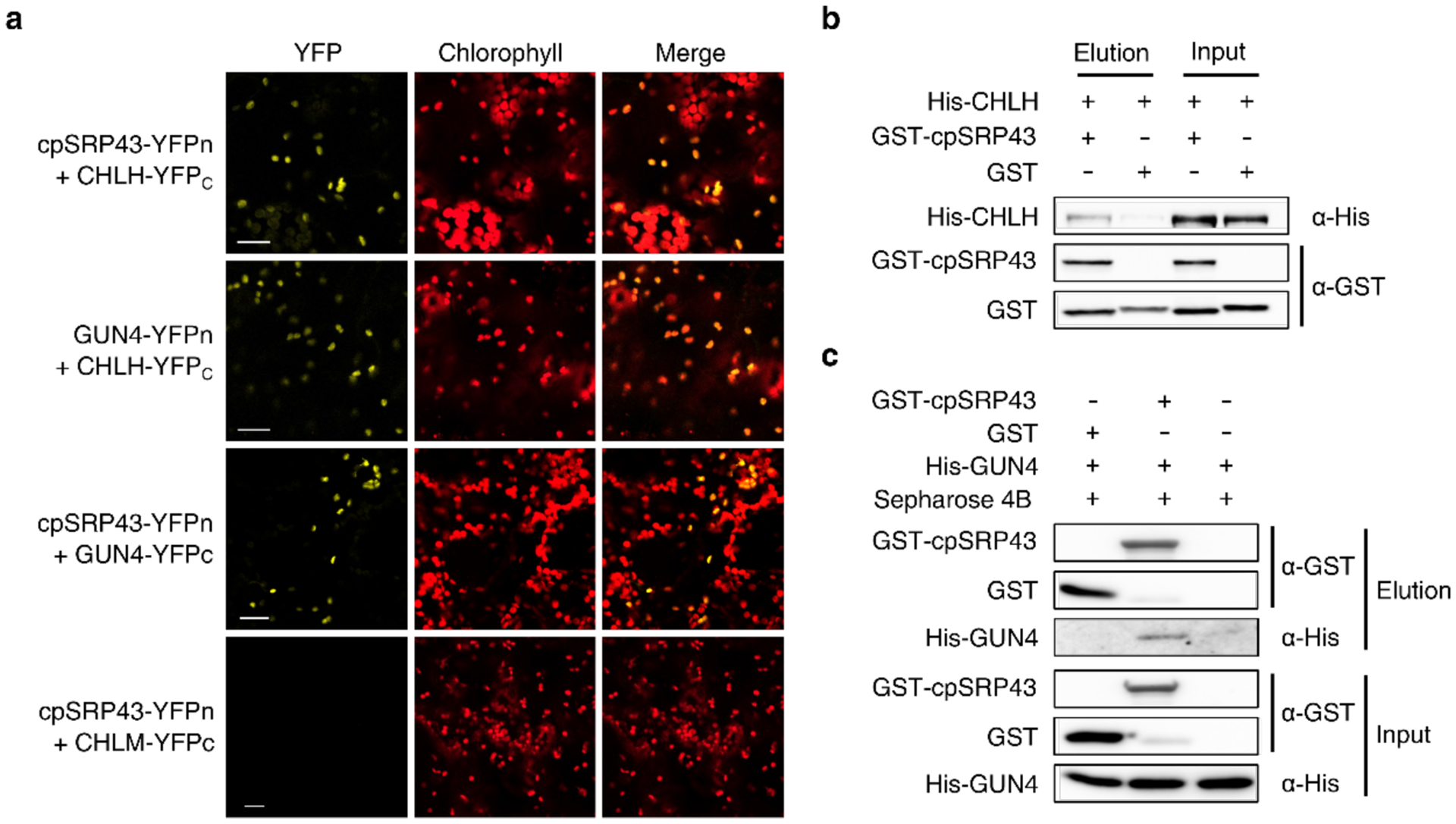
a, Bimolecular Fluorescence Complementation (BiFC) demonstrates that cpSRP43 interacts with CHLH and GUN4. Co-expression of cpSRP43-YFPn with CHLM-YFPc served as the negative control, and that of GUN4-YFPn with CHLH-YFPc served as the positive control Scale bars, 20 μm. Expression of the fused cpSRP43, CHLH, GUN4 and CHLM in N. benthamiana was proven by immunoblot analysis shown in Supplementary Fig. 2. b and c, CHLH and GUN4 bound to cpSRP43 were detected by in vitro GST pull-down assays. Recombinant purified GST-cpSRP43 (5 μM) was used as bait and incubated with His-tagged CHLH or GUN4 (10 μM). The proteins bound to GST-cpSRP43 or GST were eluted with buffer containing 10 mM glutathione. Input and elution fractions were analyzed by immunoblot using the indicated antibodies.
cpSRP43 protects mature TBS proteins from heat-induced aggregation
To explore the potential role of cpSRP43 as a molecular chaperone for TBS proteins, we considered whether it might assist the folding of these clients upon entry into the chloroplast, as it does for LHCPs. To mimic the chaperone activity of cpSRP43 on unfolded clients in vitro, we used a light-scattering assay to monitor the aggregation of urea-denatured clients upon rapid dilution of the denaturant. cpSRP43 displayed no or at most weak protection of denatured GluTR, CHLH and GUN4 at all concentrations tested (Fig. 3a–3c, red lines), suggesting that it is insufficient for the de novo folding of newly imported TBS proteins.
Fig. 3. cpSRP43 effectively chaperones TBS proteins.
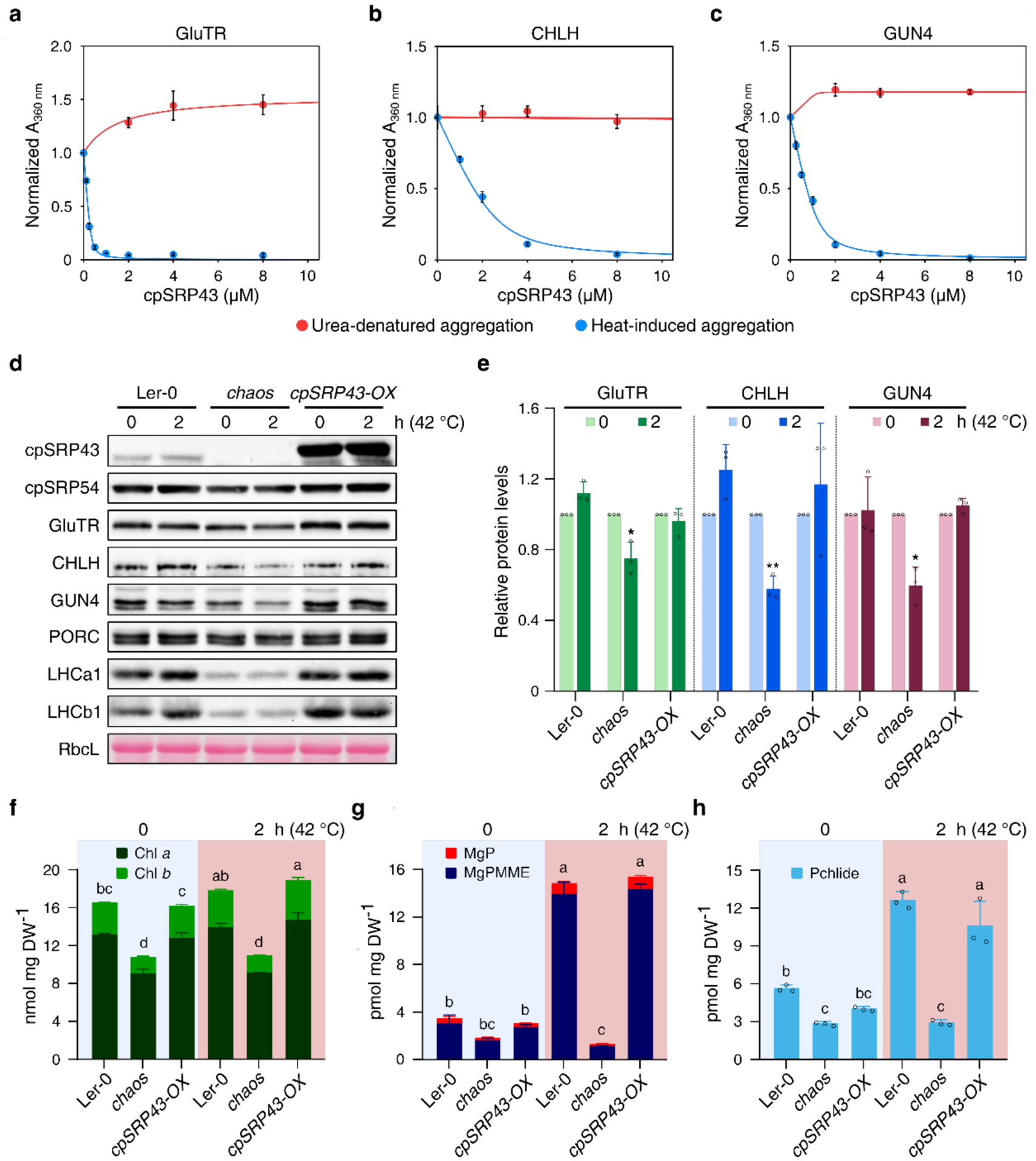
a-c, In vitro light-scattering assays were used to measure the ability of cpSRP43 to suppress the aggregation of GluTR (a), CHLH (b) and GUN4 (c) subjected to either heat-induced stress (blue) or chemical denaturation (red). Turbidity values at 360 nm were normalized to the value obtained in the absence of cpSRP43 under each condition and plotted as means ± s.e. (n = 3). The data were fitted to Eq. 1 in the Methods and yielded Ksol values of 0.02 ± 0.01 μM, 0.3 ± 0.06 μM, and 0.15 ± 0.05 μM, respectively, for the protection of GluTR, CHLH, and GUN4 from heat-induced aggregation. Ksol values for urea-denatured aggregation were not determined, as no protection was detected under such conditions. Clients were tested at the following concentrations: 2.7 μM (GluTR), 1 μM (CHLH) and 5 μM (GUN4). d, Steady-state levels of indicated TBS proteins and LHCPs in Ler-0, chaos, cpSRP43-OX seedlings before and after 2 h of heat treatment at 42 °C were quantified by immunoblotting using the indicated antibodies. The Ponceau S-stained RbcL is shown as a loading control. e, Semiquantitative analysis with Image J software (NIH) of the immunoblots from (d). The relative amounts of GluTR (green), CHLH (blue) and GUN4 (reddish brown) in Ler-0, chaos and cpSRP43-OX were normalized to their levels before heat treatment. The small open circles represent the individual data points. The data are plotted as means ± s.d. (n = 3). The small open circles represent the individual data points. The statistical analyses were performed using two-tailed Student’s t-tests. The asterisks indicate significant differences compared to protein levels prior to heat treatment: * P < 0.05, **P < 0.01. f-h, Levels of Chl (f), MgP and MgPMME (g), and Pchlide (h) in Ler-0, chaos and cpSRP43-OX prior to or after heat treatment were detected by HPLC. DW, dry weight. The data are plotted as means ± s.d. (n = 3). The small open circles represent the individual values. Letters above histograms indicate significant differences as determined by two-way ANOVA with Tukey test (P < 0.05).
We next tested whether cpSRP43 can protect folded TBS proteins from stress-induced misfolding and aggregation by carrying out light-scattering assays on soluble, folded TBS proteins that were briefly exposed to elevated temperature. After incubation of GluTR, CHLH and GUN4 at 44 °C for 5 min, we observed significant turbidity in the samples, indicating heat-induced misfolding and aggregation. Addition of cpSRP43 resulted in dose-dependent mitigation of aggregation (Fig. 3a–3c, blue lines), with apparent solubilization constants (Ksol) of 0.15 μM, 0.3 μM and 0.02 μM for GUN4, CHLH and GluTR, respectively. These values are comparable to those observed during cpSRP43-mediated protection of urea-denatured LHCPs (Ksol ~0.1 μM)26,27, suggesting that cpSRP43 effectively protects these TBS proteins from heat-induced aggregation.
In light of the apparent in vitro prevention of heat-induced aggregation of TBS proteins by cpSRP43, the role of cpSRP43 during heat shock stress was explored in planta. We monitored and compared the steady-state levels of TBS proteins in wild-type, chaos, and cpSRP43-OX seedlings exposed to short-term heat shock. Intriguingly, heat treatment at 42 °C for 2 hours (h) reduced the levels of GluTR, CHLH and GUN4 in chaos by approximately 20%–40%, whereas the levels of these proteins were not diminished by heat treatment in wild type and cpSRP43-OX (Fig. 3d and 3e). Notably, although the Chl levels in all lines analyzed showed no change after a 2-h heat treatment at 42 °C (Fig. 3f), we observed greatly increased levels of the analyzed Chl precursors, including MgP, MgPMME and Pchlide, in both wild type and cpSRP43-OX, but not in chaos seedlings (Fig. 3g and 3h). qRT-PCR and immunoblotting analyses indicate that neither cpSRP transcripts nor levels of cpSRP43 and cpSRP54 were altered after a 2-h heat treatment at 42 °C (Fig. 3d and Supplementary Fig. 4), excluding the possibility that increased cpSRP43 expression or degradation of cpSRP54 is involved in thermotolerance. These results suggest that cpSRP43 plays a key role in preserving the metabolic flow through the Chl biosynthesis pathway by protecting TBS proteins against denaturation during heat shock.
Both the substrate-binding domain (SBD) and chromodomain 2 (CD2) of cpSRP43 are required for thermostability of TBS proteins
cpSRP43 is a modular protein comprising multiple structural and functionally distinct domains. The N-terminal chromodomain (CD1), four ankyrin-repeat motifs (Ank1-Ank4), and a 20-amino acid bridging helix (BH) together form the SBD, which is both necessary and sufficient to protect LHCP from aggregation27 (Fig. 4a). Two C-terminal chromodomains, CD2 and CD3, mediate interactions with cpSRP54 and Alb3, respectively35–37. Previous biophysical work showed that the SBD, CD2, or CD3 domains fold independently, and deletion of one does not lead to unfolding of the other domains27,28,40. We examined the structural requirements for cpSRP43’s chaperone activity towards TBS proteins. Deletion of the SBD essentially abolished cpSRP43-mediated protection of GluTR, CHLH and GUN4 against heat-induced aggregation (Fig. 4b–4e). Unexpectedly, although cpSRP43-SBD exhibits slightly higher chaperone activity on LHCP than full-length, mature cpSRP43 (cpSRP43-FL) does27,28, its chaperone activity towards GluTR, CHLH and GUN4 was more than 20-fold lower than that of cpSRP43-FL (Fig. 4b–4e). In contrast, fusion of either CD2 or CD3 to the SBD was sufficient to restore effective protection of GluTR, CHLH and GUN4 under heat stress, with Ksol values comparable to those observed with cpSRP43-FL (Fig. 4b–4e). Thus, effective chaperoning of TBS proteins by cpSRP43 requires not only the SBD, but also at least one additional C-terminal chromodomain.
Fig. 4. The chaperone activity of cpSRP43 towards TBS proteins requires both its SBD and at least one of the two C-terminal CDs.
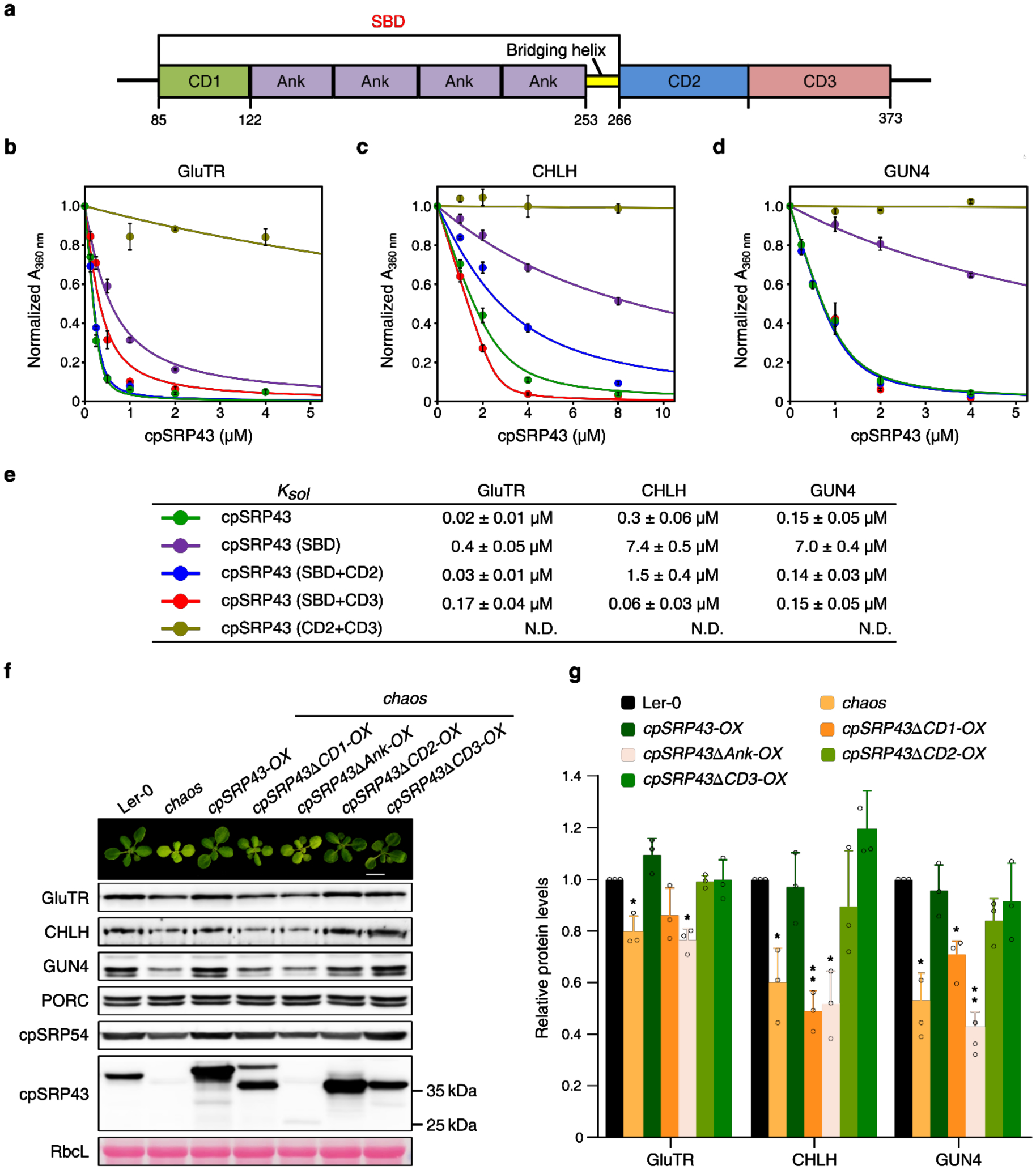
a, Schematic overview of cpSRP43 domains.CD, chromodomain; Ank, ankyrin repeats; BH, bridging helix; SBD, substrate-binding domain. b-e, Heat-induced aggregation of GluTR (b), CHLH (c) and GUN4 (d) was monitored in the presence of wild-type or truncated cpSRP43. Turbidity at 360 nm was normalized to the value in the absence of the cpSRP43 variant under each condition, and is plotted as the mean ± s.e. (n = 3), and fitted to Eq. 1 in the Methods to obtain the Ksol values in (e). N.D., not detected. f, A representative image (the top panel) and steady-state levels of indicated cpSRP43, cpSRP54 and TBS proteins (lower panels) in 18-day-old Ler-0, chaos, and the indicated chaos complementation lines under normal conditions (18 h light/ 6 h dark, 100 mmol photon m−2 s−1). Ponceau S-stained RbcL is shown as a loading control. g, Semiquantitative analysis with Image J software (NIH) of the immunoblots in (h) from three biological replicates. The relative amounts of GluTR, CHLH and GUN4 in chaos and various chaos complementation lines were normalized to the levels in Ler-0. Data are plotted as means ± s.d. (n = 3), and the small open circles represent the individual values. The statistical analyses were performed using two-tailed Student’s t-tests. The asterisks indicate significant differences compared to protein levels in Ler-0: * P < 0.05, ** P < 0.01.
These conclusions are supported by measurements of the binding of cpSRP43 variants for TBS clients using microscale thermophoresis (MST). Either the SBD or CD2+CD3 displayed greatly weakened binding to GluTR and GUN4 (Supplementary Fig. 5), whereas the binding of SBD-CD2 and SBD-CD3 to the TBS proteins were comparable to that of cpSRP43-FL (Supplementary Fig. 5). Together, these results show that, in addition to the SBD of cpSRP43, at least one of the C-terminal CDs is required for stable interactions with and protection against heat-induced aggregation of these TBS proteins.
To verify these conclusions in vivo, we monitored TBS protein levels in leaf extracts from wild type, chaos, and a set of chaos complementation lines constitutively expressing wild-type or truncated cpSRP43 variants32. Immunoblot analyses revealed lower steady-state levels of GluTR, CHLH and GUN4 in the cpSRP43ΔCD1-OX and cpSRP43ΔAnk-OX transgenic lines (Fig. 4f and 4g), indicating an essential role for the SBD in stabilizing these clients. Accordingly, expression of cpSRP43ΔCD1 and cpSRP43ΔAnk failed to rescue the chaos phenotypes, including pale-green leaves and reduced levels of Chl and Chl precursors (Supplementary Fig. 6). In contrast, expression of either SBD-CD3 (cpSRP43ΔCD2) or SBD-CD2 (cpSRP43ΔCD3) re-established wild-type leaf pigmentation in the corresponding chaos complementation lines (Fig. 4f and 4g and Supplementary Fig. 6). These data reinforce the important role of both the SBD and at least one of the C-terminal chromodomains in maintaining the stability of TBS clients under normal growth conditions.
To further assess the binding of cpSRP43 variants to TBS proteins in vivo, we carried out co-immunoprecipitation assays using leaf extracts from seedlings expressing FLAG-tagged cpSRP43-OX, cpSRP43ΔCD2-OX, and cpSRP43ΔCD3-OX. Intriguingly, cpSRP43ΔCD3 immunoprecipitated GluTR as efficiently as did wild-type cpSRP43 (Fig. 5a). In contrast, cpSRP43ΔCD2 did not pull down GluTR, suggesting while CD3 may substitute CD2 for interaction with GluTR in vitro that CD2 itself is essential and irreplaceable for interaction with TBS clients in vivo. While the interaction of cpSRP43 with CHLH and GUN4 can be detected by BiFC and in vitro pull-down assays (Fig. 2), cpSRP43 did not immunoprecipitate CHLH and GUN4 (Supplementary Fig. X), likely due to the presence of detergent during lysis that could disrupte these interactions.
Fig. 5. The CD2 domain of cpSRP43 is indispensable for the stability of TBS proteins in planta.
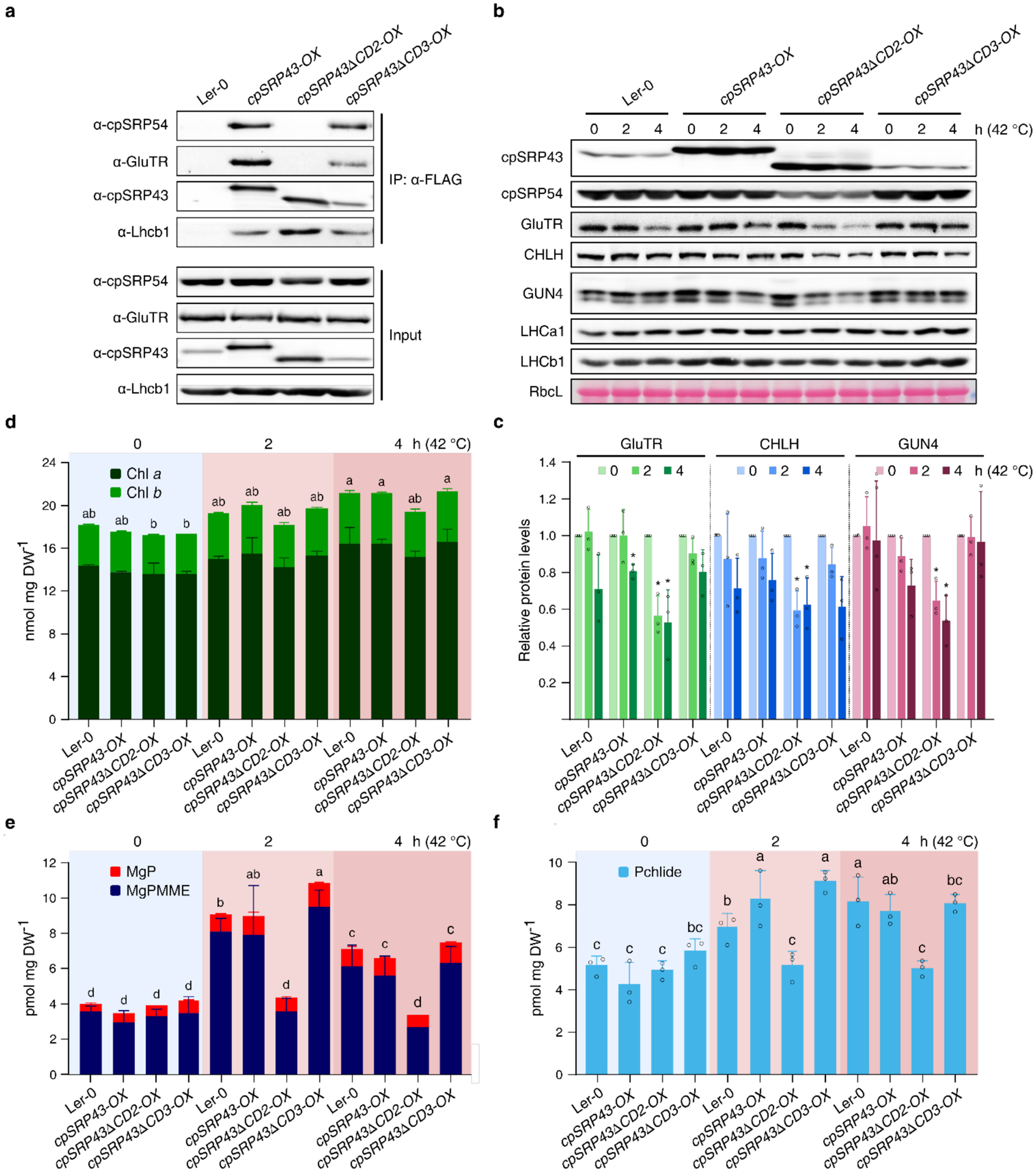
a, Co-immunoprecipitation assay demonstrates the abolished binding affinities of cpSRP43 for GluTR and cpSRP54 in the absence of CD2 domain in vivo. Total chloroplast extracts from Arabidopsis transgenic lines overexpressing cpSRP43-FLAG (cpSRP43-OX), cpSRP43 truncated CD2-FLAG (cpSRP43ΔCD2-OX), cpSRP43 truncated CD3-FLAG (cpSRP43ΔCD3-OX) and wild type (Ler-0, used as negative control) were incubated with the anti-FLAG affinity gels. The cpSRP43 interaction partners were identified by immunoblotting using the indicated antibodies. b, Steady-state levels of cpSRP43, cpSRP54, TBS proteins and LHCPs in 18-day-old Ler-0, cpSRP43-OX, cpSRP43-ΔCD2- OX and cpSRP43-ΔCD3-OX prior to (0 h at 42°C) or after 2–4 h heat treatment (2–4 h at 42°C) were detected by immunoblotting using the indicated antibodies. The Ponceau S-stained RbcL is shown as loading control. c, Semiquantitative analysis with Image J software (NIH) of the immunoblots in b. The relative amounts of GluTR (green), CHLH (blue) and GUN4 (reddish brown) were normalized to their levels prior to exposure to elevated temperature (0 h at 42°C). The data are plotted as means ± s.d. (n = 3). Small open circles represent the individual values. The statistical analysis was performed by using two-tailed Student’s t-tests. The asterisks indicate significant differences relative to each ecotype prior to heat treatment (0 h at 42 °C): * P < 0.05, ** P < 0.01. d-f, Levels of Chl (d), MgP and MgPMME (e) and Pchlide (f) in Ler-0, chaos and cpSRP43-OX prior to or after 2–4 h heat treatment were determined by HPLC. DW, dry weight. The data are plotted as means ± s.d. (n = 3). The small open circles represent the individual values. Letters above histograms indicate significant differences as determined by two-way ANOVA with Tukey test (P < 0.05).
Finally, we explored the contributions of CD2 and CD3 to the thermoprotection of TBS proteins in vivo. After heat treatment at 42 °C for 2–4 h, the steady-state levels of GluTR, CHLH and GUN4 decreased in cpSRP43ΔCD2-OX seedlings compared with wild-type, cpSRP43-OX and cpSRP43ΔCD3-OX seedlings (Fig. 5b and 5c). During heat treatment, levels of Chl precursors in cpSRP43ΔCD2-OX, including MgP, MgPMME and Pchlide, were significantly lower than that in the control seedlings (Fig. 5d–5f). Taken together, these results demonstrate that both SBD and CD2 play key roles in the stabilization of TBS clients under short-term heat shock conditions.
cpSRP54 inhibits the chaperone activity of cpSRP43 towards TBS proteins
The chaperone activity of cpSRP43 on LHCPs is stimulated by cpSRP54-dependent allosteric regulation27,28. The RRKRp10 segment of the cpSRP54M domain (designated cpSRP54M peptide) binds to CD2 and triggers a conformational change in cpSRP43-SBD, thereby enhancing its chaperone activity towards LHCP by 6-fold and restoring the activity of multiple chaperone-defective mutants (Supplementary Fig. 7)27,28. Surprisingly, the cpSRP54M peptide substantially diminished the efficacy of cpSRP43 in preventing heat-induced aggregation of GluTR, CHLH and GUN4 in vitro (Fig. 6a–6c). The complete cpSRP54M domain and full-length mature cpSRP54 had the same inhibitory effect on chaperone activity as the cpSRP54M peptide (Supplementary Fig. 8). The increase in turbidity seen in the presence of cpSRP43 and cpSRP54 was not due to heat-induced aggregation of cpSRP43, cpSRP54M peptide or the cpSRP54M domain, as heat treatment did not generate detectable aggregation in the absence of client protein (Supplementary Fig. 8, gray lines). While aggregation was detected when cpSRP43 was incubated with full-length mature cpSRP54 during heat stress, turbidity increased substantially in the presence of GluTR or GUN4, highlighting the contribution of the clients to the aggregation (Supplementary Fig. 8, gray vs. red lines). These results demonstrate that the interaction with cpSRP54 inhibits the chaperone activity of cpSRP43 towards TBS proteins during heat stress, in contrast to the role of cpSRP54 as an enhancer of cpSRP43’s chaperone activity on LHCPs.
Fig. 6. cpSRP54 inhibits the chaperone function of cpSRP43 for TBS proteins.
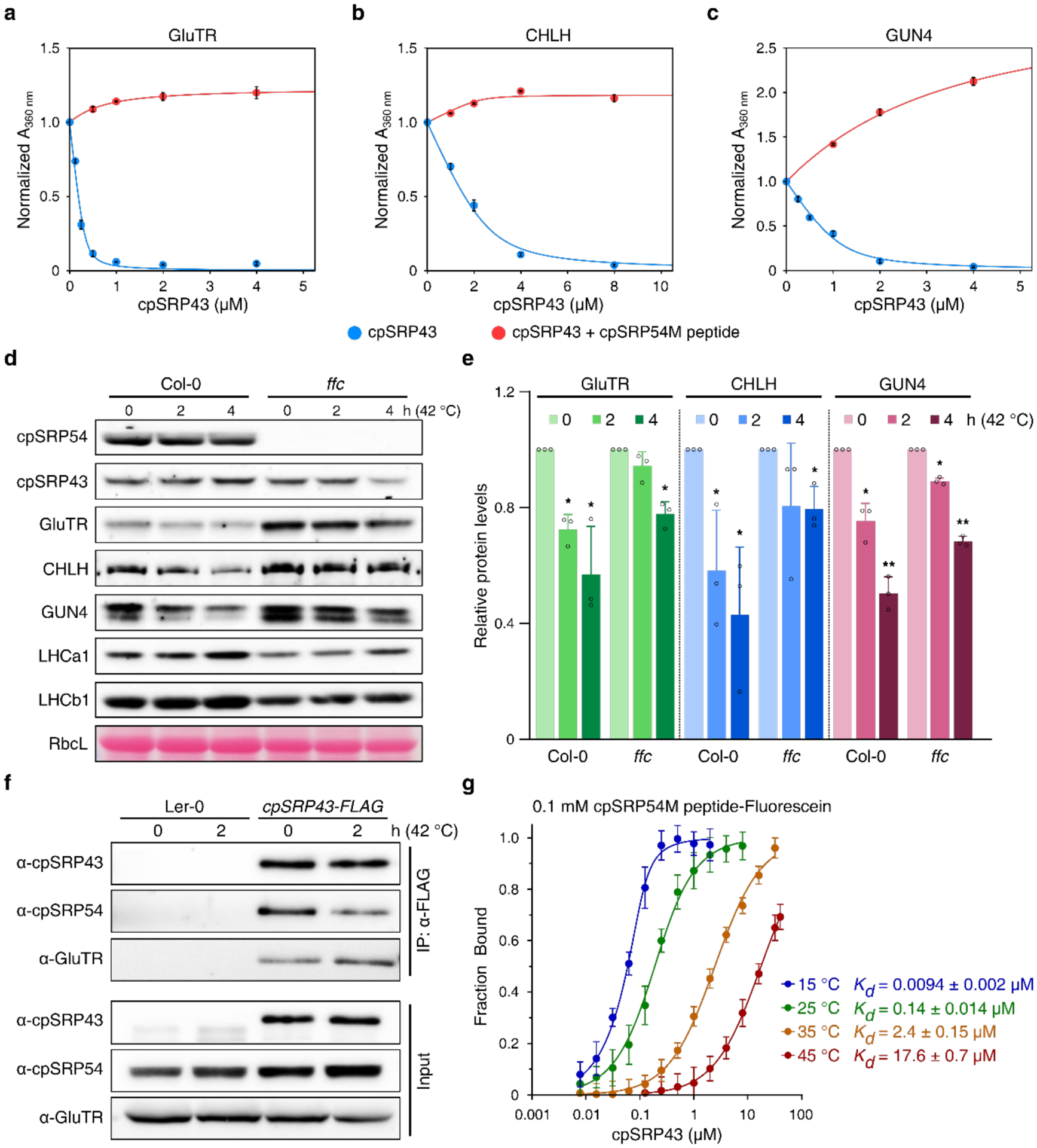
a-c, Heat-induced aggregation of GluTR (a), CHLH (b) and GUN4 (c) was monitored in the presence of apo-cpSRP43 (blue) or cpSRP43 in combination with 50 μM cpSRP54M peptide (red). The turbidity at 360 nm was normalized to the value obtained in the absence of cpSRP43 under each condition, plotted as means ± s.e. (n = 3), and fitted to Eq. 1 in the Methods. The data for apo cpSRP43 are the same as those depicted in Fig. 3a and are shown here for comparison purposes. Ksol values for cpSRP43 in the presence of cpSRP54M peptide were not obtained, as no protection was detected. d, Steady-state levels of cpSRP43, cpSRP54, indicated TBS proteins and LHCPs in the 18-day-old wild-type (Col-0) and the cpsrp54-deficient mutant (ffc) prior to (0 h at 42 °C) or after 2–4 h of heat treatment (2–4 h at 42 °C) were quantified by immunoblotting using the indicated antibodies. The Ponceau S-stained RbcL is shown as loading control. e, Semiquantitative analysis with Image J software (NIH) of the immunoblots in d. The relative amounts of GluTR (green), CHLH (blue) and GUN4 (reddish brown) in Col-0 and ffc were normalized to their levels prior to exposure to elevated temperature (42 °C 0 h). The data are plotted as means ± s.d. (n = 3). The small open circles represent the individual values. The statistical analysis was performed by using two-tailed Student’s t-tests. The asterisks indicate significant differences relative to protein levels in each ecotype prior to heat treatment (0 h at 42 °C): * P < 0.05, ** P < 0.01. f, Co-immunoprecipitation assay to determine the changes in the interaction of cpSRP43 with cpSRP54 or with GluTR prior to or after heat treatment. Total chloroplast extracts from wild type (Ler-0, used as negative control) and transgenic Arabidopsis lines overexpressing cpSRP43-FLAG were incubated with anti-FLAG affinity beads. cpSRP43 interaction partners were recovered by centrifugation and detected by immunoblotting using the indicated antibodies. g, Equilibrium titrations to determine the binding affinity of the cpSRP54M peptide for cpSRP43 measured at 15, 25, 35 and 45 °C. Binding affinities were detected based on fluorescence anisotropy change of fluorescein-labelled cpSRP54M peptide. The lines are fits of the data to Eq. 2 in the Methods, which yield the indicated Kd values, plotted as means ± s.e. (n = 3).
To verify these results in planta, the steady-state levels of TBS proteins during heat shock in wild type and cpsrp54-knockout ffc seedlings were compared with those grown under standard conditions. Immunoblot analyses showed enhanced stability of TBS proteins, including GluTR, CHLH and GUN4, in the ffc mutant relative to wild type after a 2- or 4-h heat treatment at 42 °C (Fig. 6d and 6e). Consistently, the levels of Chl precursors, including MgP, MgPMME and Pchlide, during heat treatment were more stable in ffc than that in wild type (Supplementary Fig. 9). These results reveal a negative effect of cpSRP54 on the stability of TBS proteins subjected to short-term heat stress.
High temperature causes cpSRP43 to dissociate from cpSRP54 and bind TBS proteins
Given the contrasting effects of cpSRP54 on cpSRP43’s chaperone functions for LHCPs and TBS proteins, we asked whether cpSRP54-cpSRP43 complex formation is modulated in response to heat stress. We carried out co-immunoprecipitation assays to measure the binding of cpSRP43 to TBS proteins and to cpSRP54 before and after heat treatment, using seedlings of wild-type and transgenic plants overexpressing FLAG-tagged cpSRP43. After a 2-h heat treatment at 42 °C, less cpSRP54 was bound to cpSRP43, and a corresponding increase in the amount of GluTR immunoprecipitated with cpSRP43 was observed (Fig. 6f). To monitor temperature-dependent changes in the binding affinity of cpSRP54 for cpSRP43, we carried out fluorescence anisotropy measurements using a fluorescein-labeled cpSRP54M peptide. Remarkably, the interaction of cpSRP54M peptide with cpSRP43 was hypersensitive to temperature: the equilibrium dissociation constant (Kd) for their binding increased by an order of magnitude for every 10 °C rise in temperature (Fig. 6g). Taken together, our results suggest that heat stress triggers the release of cpSRP54 from cpSRP43, which increases the pool of apo cpSRP43 that binds with and protects TBS proteins against heat stress.
Discussion
In oxygenic photosynthetic organisms, Chl biosynthesis must be tightly regulated, as free Chl and most of its precursors are phototoxic38. Many auxiliary factors, including chaperones, scaffold proteins, proteases, thioredoxins and porphyrin-binding proteins, are required for efficient Chl synthesis in the light and immediate shutdown of Chl production in the dark39. Our work here identifies a novel post-translational mechanism, mediated by the molecular chaperone cpSRP43, which stabilizes TBS proteins and maintains Chl biosynthesis pathway during leaf greening and heat shock, and reveals how this chaperone can switch between promoting the biogenesis of LHC proteins and safeguarding Chl biosynthesis.
Together with our previous work32, this study shows that a chaperone function of cpSRP43 is involved in stabilizing three vital TBS proteins – GluTR, CHLH and GUN4 – under normal growth and heat-stress conditions (Figs. 1 and 3). We previously reported that cpSRP43 reduces the tendency of an N-terminal fragment of GluTR to aggregate following urea-induced denaturation, although >20 μM cpSRP43 is required for complete protection32. However, cpSRP43 had weak or no impact on full-length mature GUN4, CHLH or GluTR after urea denaturation, whereas it was highly effective in shielding these proteins from the disruptive consequences of heat stress (Fig. 3a). These observations suggest that, in contrast to its canonical role in protecting unfolded LHCPs during their biogenesis, this novel chaperone activity of cpSRP43 acts primarily on mature, folded TBS proteins to prevent misfolding and aggregation during chloroplast development and heat shock.
The protective role of cpSRP43 on TBS proteins has markedly different molecular requirements compared to its chaperone function for LHCPs. While its SBD is necessary and sufficient for the latter27,28, the protection of TBS proteins requires an additional C-terminal chromodomain (Fig. 4 and Supplementary Fig. 5). Although CD3 can functionally substitute for CD2 in vitro (Fig. 4b–4e), in vivo studies point to an essential role for both the SBD and CD2 in the stabilization of TBS proteins subjected to heat stress (Fig. 5). While the reason for the strict requirement for CD2 in the chaperoning of TBS proteins in vivo remains to be determined, the mechanism by which cpSRP43 interact with LHCP is distinct from that with TBS proteins.
The participation of cpSRP43 in both LHCP biogenesis and Chl synthesis also raises the question of how these distinct functions are regulated. Our work here points to a key role for cpSRP54 in orchestrating this coordination. cpSRP54 stimulates cpSRP43’s chaperone activity on LHCPs by stabilizing the cpSRP43-SBD domain in a closed, tightly folded conformation27,28. Surprisingly, our data show that cpSRP54 has the opposite effect on cpSRP43’s engagement with TBS clients (Fig. 6). Thus, the steady-state levels of TBS proteins are significantly higher in cpsrp54 (ffc) plants than in the wild type, which supports the idea that free cpSRP43, rather than the cpSRP43-cpSRP54 complex, serves as the active chaperone for TBS proteins under heat stress. Given that CD2 forms the binding site for the cpSRP54M domain40 and is required for cpSRP43 to provide thermoprotection of TBS proteins (Fig. 5), we speculate that TBS protein protection requires additional client contacts with CD2 that are antagonized by cpSRP54. Binding of cpSRP54 to cpSRP43 could occlude the TBS binding sites on CD2, or force cpSRP43 to adopt a conformation that is incompatible with TBS interaction. Although the mechanistic details behind the cpSRP54-induced inhibition of cpSRP43’s chaperone activity towards TBS proteins remain to be understood, these results provide compelling evidence for the notion that cpSRP54 acts as a switch that can direct cpSRP43 to either LHCP delivery or to the protection of TBS clients.
Plants normally experience elevated temperatures in the early afternoon, especially during summertime. Plants have evolved strategies to cope with this daily heat shock, maintain cell integrity and preserve the integrity of crucial cellular processes41. Our results provide evidence that cpSRP43 helps to maintain the stability of TBS clients during short-term heat treatment, thereby maintaining the metabolic flow during Chl biosynthesis during peak sunlight hours (Figs. 3 and 5). The inhibitory effect of cpSRP54 on the chaperone activity of cpSRP43 towards TBS proteins further raises questions as to how this chaperone activity is switched on during heat stress. Remarkably, co-immunoprecipitation assays indicated reduced interactions of cpSRP43 with cpSRP54 and increased interaction with TBS proteins upon heat treatment. Quantitative measurements in vitro further showed that the cpSRP54-cpSRP43 interaction is highly sensitive to increases in temperature, weakening ~10-fold with every 10 °C rise (Fig. 6g). These results support a model in which higher temperature drives dissociation of the complex, thereby freeing cpSRP43 for the stabilization of TBS proteins (Fig. 7).
Fig. 7. Model for dual participation of cpSRP43 in Chl synthesis and LHCP transport in plants.
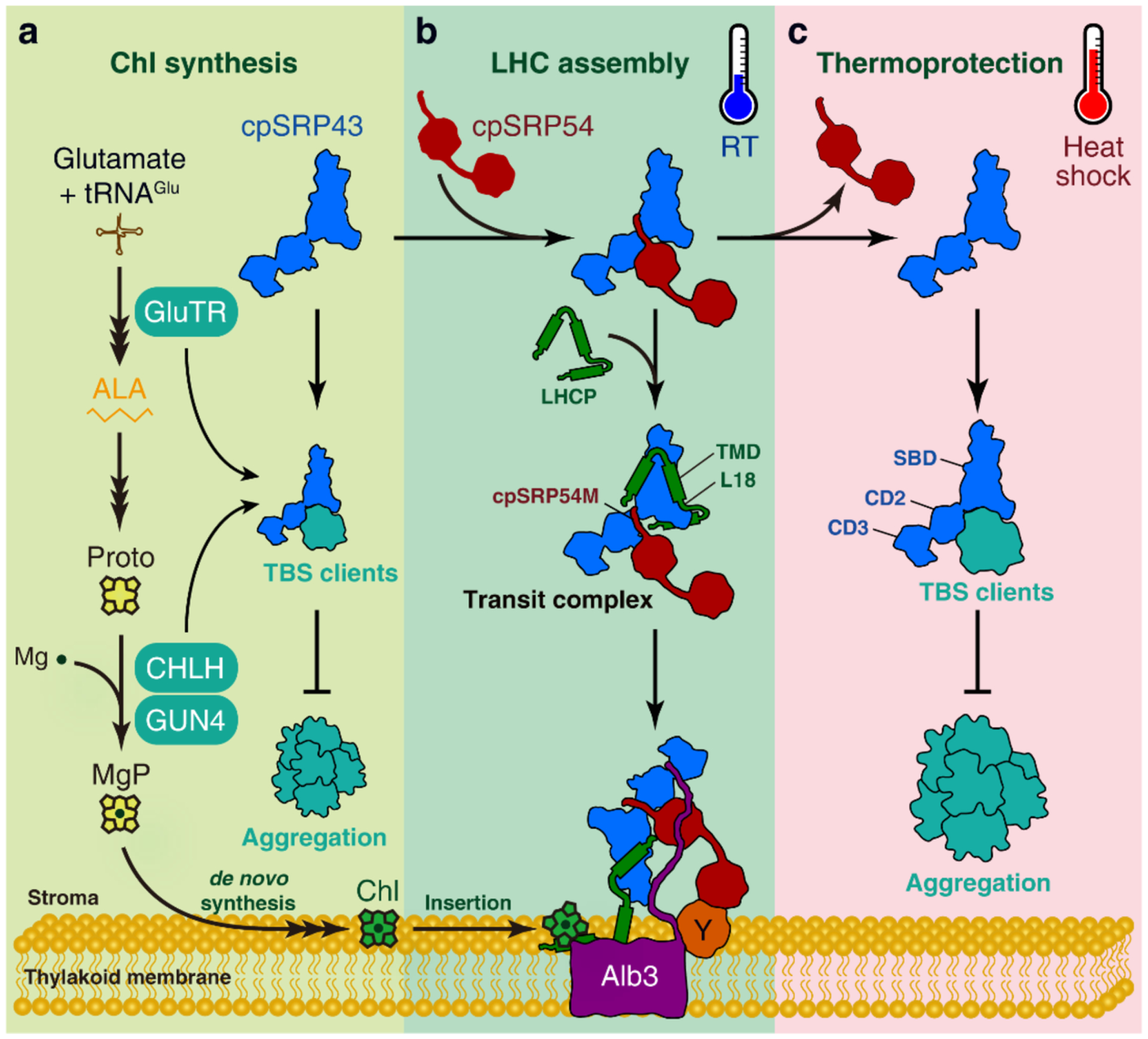
cpSRP43 harbours two distinct chaperone activities: as a general chaperone for protection of TBS proteins (a, c), or as a dedicated chaperone to mediate the protection and delivery of LHCPs (b). The switch between these two activities is regulated by cpSRP54 binding. Under normal conditions, a minor portion of apo cpSRP43 optimizes Chl biosynthesis by protecting crucial TBS proteins, including GluTR, CHLH and GUN4, against aggregation (a). Meanwhile, a majority of cpSRP43 was bound to cpSRP54 with high affinity (b). cpSRP54 optimizes the conformation of the cpSRP43-SBD domain for interaction with and protection of LHCP, while inhibiting the chaperone activity of cpSRP43 towards TBS proteins and thus limits the access of cpSRP43 to TBS proteins. At the thylakoid membrane, cpSRP54 binds to cpFtsY (Y), and the stromal domain of Alb3 interacts with cpSRP43 to trigger the release of LHCP and mediate insertion into the thylakoid membrane. The participation of cpSRP43 in both Chl synthesis and LHCP transport ensures that plants produce sufficient amounts of Chl for LHC assembly during leaf greening. In contrast, when plants experience short-term heat shock stress (c), the affinity of cpSRP54 for cpSRP43 is greatly weakened, increasing the available pool of apo cpSRP43 that can use both its SBD and CD2 to effectively protect TBS proteins from heat-induced aggregation. ALA, 5-aminolevulinic acid. L18, L18 motif of LHCP interacting with cpSRP43. RT, room temperature. TMD, transmembrane domain.
In summary, this study shows that, in addition to its role as an LHCP chaperone in conjunction with cpSRP54, the chloroplast chaperone cpSRP43 has an autonomous function as a heat-inducible protectant of the TBS proteins GluTR, CHLH and GUN4 in response to rising temperatures. Under normal growth conditions (Fig. 7a and 7b), the majority of cpSRP43 acts together with cpSRP54 to transport newly imported and unfolded mature LHCPs to the Alb3 translocase for insertion into the thylakoid membrane, while a smaller pool of free cpSRP43 interacts with GluTR, CHLH and GUN4 to protect these clients from aggregation. During heat shock (Fig. 7C) when TBS proteins are increasingly susceptible to aggregation, cpSRP54 dissociates from cpSRP43, generating a larger pool of free cpSRP43 that can be rapidly re-purposed as a stress-responsive chaperone to interact with and protect TBS clients from heat-induced misfolding and aggregation. The cpSRP54-cpSRP43 interaction serves as a thermostat that enables cpSRP43 to toggle between these distinct chaperone functions, and thus cater to different proteostatic demands in response to changing environmental conditions.
Methods
Plant materials and growth conditions
The Arabidopsis thaliana chaos (cpsrp43 null mutant) and ffc (cpsrp54 knockout mutant, stock CS850421 from ABRC) lines used here, and the various T3 populations of chaos complementation lines constitutively expressing wild-type or truncated FLAG-tagged cpSRP43 have been described previously32. The Arabidopsis ecotypes Columbia-0 (Col-0, for ffc) and Landsberg-0 (Ler-0, for chaos and its complementation lines) were used as wild-type controls. After a 2-day stratification in darkness at 4 °C, Arabidopsis seeds were sown in soil, and germinating young seedlings were grown under standard long-day conditions (18 h light/6 h dark, 100 μmol photons m−2 s−1, 22 °C, and 70% relative humidity). Heat stress was applied by transferring 18-day-old seedlings from standard long-day conditions to constant light (100 μmol photons m−2 s−1) and a temperature of 42 °C for the indicated time. 14- to 18-day-old Arabidopsis seedlings were harvested for analyses of pigment levels, gene expression and protein amounts. For the BiFC assay, 3-week-old tobacco (Nicotiana benthamiana) plants grown under long-day conditions were used to transiently express proteins of interest.
Analysis of TBS intermediates and end-products
Rosette leaves (40–60 μg) were harvested and weighed to determine the fresh weight (FW) of plants grown under standard conditions. In contrast, rosette leaves from plants exposed to heat stress treatment were harvested, frozen, ground in liquid nitrogen, and lyophilized to determine the dry weight (DW) of leaf materials. TBS intermediates and end-products were extracted from frozen or lyophilized leaf powders in 300–600 μL of ice-cold pigment-extraction buffer (acetone: 0.2 M NH4OH, 9:1, v/v) at −20 °C for at least 1 h. After centrifugation (14,000 g, 20 min, and 4 °C), the supernatant was subjected to high-performance liquid chromatography (HPLC). After removal of the supernatant, non-covalently bound heme was extracted from the pellet using AHD buffer (acetone: hydrochloric acid: dimethylsulfoxide, 10:0.5:2, v/v/v), and centrifuged at 14,000 g for 20 min at room temperature. HPLC analyses were conducted using the Agilent 1100 or 1290 HPLC system equipped with a diode array and fluorescence detectors (Agilent Technologies), essentially as described previously42.
Measurement of ALA synthesis capacity
To determine ALA synthesis rates in seedlings of the various lines of interest, whole rosette leaves were excised from 18-day-old plants, weighed to determine FW, and incubated in 5 mL of 50 mM Tris-HCl (pH 7.2) containing 40 mM levulinic acid (Sigma-Aldrich) for 3 h under normal conditions. Leaf materials were then frozen and homogenized in liquid nitrogen before resuspension in 0.5 mL of 20 mM potassium phosphate buffer (pH 7.2). After centrifugation at 12,000 g for 5 min at 4 °C, 0.4 mL of the homogenate was mixed with 0.1 mL of ethyl acetoacetate (Sigma-Aldrich) and boiled for 10 min. The chilled samples were then mixed with 0.5 mL of Ehrlich’s reagent [373 mL acetic acid, 90 mL of 70% (v/v) perchloric acid, 1.55 g of HgCl2, 9.10 g of 4-dimethylamino benzaldehyde, and 500 mL of double-distilled water] and centrifuged at 12,000 g for 5 min at 4 °C. The absorption of the ALA pyrrole was measured at 525, 553 and 600 nm. The ALA content was calculated using a standard dilution curve of authentic ALA solutions (Sigma-Aldrich) and normalized to the incubation time and FW of leaf material used.
RNA extraction and qRT-PCR
Total RNA was extracted from Arabidopsis leaves frozen in liquid nitrogen using the citric-acid extraction method43. Aliquots (2 μg) of DNase-treated RNA were reverse transcribed into cDNA with oligo-dT18 using Revert Aid reverse transcriptase (Thermo Fisher Scientific). qPCR was carried out in the CFX96-C1000 96-well plate thermocycler (Bio-Rad) by using 2×qPCR Mastermix (Bimake). SAND (At2g28390) was routinely used as the reference gene. Calculation of relative gene expression was done with the Biorad CFX-manager software (1.6) using the 2−ΔΔCt method. Primers for qRT-PCR are listed in Supplementary Table 1.
Protein extraction and western-blot analysis
Whole 14- to 18-day-old rosette seedlings were harvested from 3 to 6 individual plants of each genotype. When plants grown under normal conditions, freshly harvested leaves were ground in liquid nitrogen and total leaf proteins were extracted from the powder using 2× Laemmli buffer [100 mM Tris-HCl pH 6.8, 4% (w/v) SDS, 20% (v/v) glycerol, 200 mM DTT, and 0.01% Bromophenol Blue] and incubated at 95 °C for 10 min. Protein concentration was determined and is expressed relative to leaf fresh weight. For heat-stress experiments, rosette leaves were harvested before or after heat treatment and ground in liquid nitrogen. Total leaf protein was extracted from frozen plant material into PEB buffer [2% (w/v) SDS, 56 mM NaCO3, 12% (w/v) sucrose, and 2 mM EDTA] by heating the samples for 20 min at 70 °C. Protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). All samples in PEB buffer were diluted to the same protein concentration, supplemented with 56 mM DTT, and incubated at 70 °C for 5 min. Aliquots (15 μg) of protein were subjected to SDS-PAGE, transferred to nitrocellulose membranes (GE Healthcare), and probed with specific antibodies. Antibodies against GluTR (1:1000), GSAAT (1:2000), GUN4 (1:2000), PORC (1:1000) and CHLM (1:500) were generated in our laboratory; those for CHL27 (AS06122, 1:1000), LHCa1 (AS01005, 1:2500) and LHCb1 (AS09522, 1:2500) were purchased from Agrisera (Vännäs). Antibodies against cpSRP43 (1:2500) and cpSRP54 (1:2500) were kindly donated by Prof. Danja Schünemann (Ruhr University, Bochum, Germany), those directed against CHLH (1:1000) and CHLI (1:5000) were kindly provided by Prof. Da-Peng Zhang (Tsinghua University, China) and Prof. Meizhong Luo (Huazhong Agricultural University, China), respectively. Immunoblotting signals visualized by Clarity™ Western ECL (Bio-Rad) were detected with a CCD camera (Intas Biopharmaceuticals).
BiFC assay
cDNA fragments encoding full-length cpSRP43 or GUN4 were cloned into the pSPYNE vector (fusing them to the N-terminal half of YFP, nYFP), whereas the full-length coding sequences of CHLH, GUN4 and CHLM were cloned into pSPYCE (fusing them to the C-terminal half of YFP, cYFP). The BiFC constructs were transiently transformed into the abaxial epidermal cells of Nicotiana benthamiana leaves using Agrobacterium tumefaciens strain GV2260. After 3 days in darkness, the infiltrated leaf areas were analyzed for YFP fluorescence with an LSM 800 confocal microscope (Carl Zeiss, Jena, Germany). YFP signals were detected at Ex/Em 514/530–555 nm, while Chl fluorescence was visualized at Ex/Em 514/600–700 nm.
Expression and purification of recombinant proteins
cDNA fragments encoding wild-type or truncated cpSRP43 proteins were cloned into the pET-28a expression vector using the primers listed in Table S1. Expression and purification of His-cpSRP43, His-GluTR, His-GUN4, His-tagged MgCh subunits from Oryza sativa (OsCHLH, OsCHLD, and OsCHLI), GST and GST-cpSRP43 were reported previously32,42. Expression and purification of truncated cpSRP43 protein was similar with that for wild-type cpSRP43. The cpSRP54M peptide (QKAPPGTARRKRKAC) was synthesized by Eton Bioscience (99% purity).
Mature cpSRP54 and the cpSRP54M domain were expressed from pMAL plasmids, yielding fusions linked to N-terminal maltose-binding protein (MBP). BL21 (DE3) cells transformed with heterologous expression vectors were added (at a volumetric ratio of 1:200) to an overnight culture in LB medium and grown at 37 °C. Cells were induced with 0.5 mM IPTG at an optical density (OD600) of 0.6 and grown at 37 °C for a further 4 h. Cells were harvested by centrifugation (5,000 g for 15 min at 4 °C), resuspended in lysis buffer (20 mM HEPES pH7.5 and 300 mM KCl), and lysed in a French press. The clarified cell lysate was then incubated with Ni-NTA agarose pre-equilibrated in lysis buffer for 2 h. Columns were washed with 20 column volumes (CV) of wash buffer (20 mM HEPES pH 7.5, 1 M NaCl, 20 mM imidazole), eluted with elution buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 200 mM imidazole), and dialyzed overnight into MonoS buffer (50 mM Tris, 150 mM NaCl, 2 mM DTT, 1 mM EDTA) containing 1% thrombin to cleave off the MBP. Clients were further purified on a MonoS column using a 0 to 1M NaCl gradient in MonoS buffer. Purified cpSRP54 and cpSRP54 M-domain proteins were concentrated and stored in LSA buffer (50 mM HEPES, 200 mM NaCl, pH 7.5) supplemented with 20% (v/v) glycerol at −80 °C.
In vitro MgCh assay
The MgCh assay was performed as described previously44 and overall 150 μL mixture containing 2.5 μM CHLH, 1 μM CHLD, 1 μM CHLI, 2.5 μM GUN4 in assay buffer (50 mM Tricine-KOH, pH 8.0, 15 mM MgCl2, 2 mM DTT, 1 mM ATP and 10 μM Proto). His-tagged cpSRP43, cpSRP43 (CD2+CD3), CHLH, GUN4, and Proto in MgCh assay buffer (overall 100 μL) were pre-incubated in darkness for 30 min on ice, while CHLD and CHLI were separately pre-incubated in 50 μL of MgCh assay buffer. Then the two mixtures were warmed for 5 min at 30°C and combined in the 96-well plate to start the assay. MgP formation was continuously monitored by spectrofluorometry using a 96-well plate reader mounted on a Hitachi F7000 fluorescence photometer. Excitation and emission wavelengths were set to 416 nm and 595 nm, respectively, with slit widths of 5 nm (ex) and 10 nm (em). The delay time for each scan was 5 min and the whole san time was indicated in the figure legend. Absolute amounts of MgP formed were calculated from a standard curve. A 30-μL sample of the reaction solution was boiled in Laemmli buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 200 mM DTT, and 0.01% Bromophenol Blue) and applied to a 12% SDS-PAGE to determine the protein levels at the end of the reaction.
Isolation of intact chloroplasts
Four-week-old Arabidopsis seedlings were harvested and homogenized in HB buffer [0.45 M sorbitol, 20 mM Tricine-KOH pH 8.4, 10 mM EDTA, 10 mM NaHCO3, 0.1% (w/v) bovine serum albumin]. The homogenate was then filtrated through two layers of Miracloth (Calbiochem). The resulting suspension was centrifuged at 500 g for 8 min at 4 °C, and the pellet gently resuspended in RB buffer (0.3 M sorbitol, 20 mM Tricine-KOH, pH 8.0, 5 mM MgCl2 and 2.5 mM EDTA). The suspension was applied to a two-step Percoll gradient [40% (v/v) and 80% (v/v) in RB buffer]. After centrifugation, intact chloroplasts were collected from the interface between the Percoll solutions, and washed twice with RB buffer. The pellet was resuspended in lysis Buffer (20 mM Tricine-KOH, 2.5 mM EDTA, 5 mM MgCl2, and the cOmplete protease inhibitor cocktail). The supernatant was either used directly or frozen in liquid nitrogen for further applications.
In vitro pull-down assay
In vitro GST pull-down analysis was performed as described previously32 with the following modifications. Purified recombinant GST and GST-cpSRP43 were used as baits, and incubated with purified His-GUN4 or His-CHLH at 4 °C overnight in binding buffer [25 mM Tris-HCl, pH 7.8, 150 mM NaCl, 5 mM MgCl2, 10% (v/v) glycerol, and the cOmplete protease inhibitor cocktail]. A 10 μL aliquot of an equilibrated 50% (v/v) slurry of Glutathione 4B Agarose (GE Healthcare) was added to each tube and incubated for 1 h at 80 rpm at 4 °C. The agarose was collected by centrifugation (2,800 g, 30 sec, and 4 °C), and washed three to four times with binding buffer. The proteins bound to GST-cpSRP43 or GST were eluted with binding buffer containing 10 mM reduced glutathione, denatured in Laemmli buffer, and finally subjected to SDS-PAGE and immunoblot analyses.
Co-immunoprecipitation analysis
Intact chloroplasts (100 μg of Chl) isolated from wild-type (Ler-0) or transgenic plants expressing cpSRP43-FLAG, cpSRP43 (SBD+CD2)-FLAG or cpSRP43 (SBD+CD3)-FLAG were solubilized in binding buffer (25 mM Tris-HCl pH 7.8, 150 mM NaCl, 5 mM MgCl2, 10% (v/v) glycerol, and cOmplete protease inhibitor cocktail). The Chl concentration of extracted chloroplasts were adjusted to 1–1.2 μg μL−1 Chl. Thylakoids then were solubilized with 1% (w/v) n-dodecyl-β-D-maltoside (β-DM) in binding buffer for 10 min on ice. After centrifugation (14,000 g, 10 min, 4 °C), the supernatant was incubated at room temperature for 2 h with 10 μL of anti-FLAG affinity gel (Biotool). When analyzed the protein binding affinity after heat treatment, the incubation was performed at 4 °C for 1 h. Then, the beads were pelleted by centrifugation (2,800 g, 30 sec, and 4 °C) and washed four times to remove non-specifically bound proteins. Finally, cpSRP43, cpSRP43 (SBD+CD2), or cpSRP43 (SBD+CD3)-FLAG and their interaction partners were eluted from the beads with Laemmli buffer, and subjected to SDS-PAGE and immunoblot analyses.
MST assay
All MST reagents and consumables were purchased from NanoTemper Technologies (Munich, Germany). His-GUN4 was labeled using the Protein Labeling Kit RED-NHS. The labeling reaction was performed according to the manufacturer’s instructions in the supplied labeling buffer applying a concentration of 20 μM protein (molar dye: protein ratio ≈ 3:1) at 4 °C for 30 min. Unreacted dye was removed with the supplied dye removal column equilibrated with PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.5). The degree of labeling was determined using UV/VIS spectrophotometry at 650 and 280 nm and was typically 0.75. The labeled His-GUN4 was adjusted to 40 nM with MST buffer (50 mM Tris-HCl pH 7.8, 150 mM NaCl, 10 mM MgCl2) supplemented with 0.05 % Tween 20. The ligand protein cpSRP43 and its truncated proteins were dissolved in PBS buffer and a series of 16 1:1 dilutions was prepared using the same buffer, producing ligand concentrations ranging from 6.1 nM to 200 μM. For the measurement, each ligand dilution was mixed with one volume of labeled His-GUN4, which led to a final concentration of His-GUN4 of 20 nM and final ligand concentrations ranging from 3.05 nM to 100 μM. After 10 min incubation followed by centrifugation at 14,000 g for 5 min at 4 °C, the samples were loaded into Premium Monolith NT.115 Capillaries. MST was measured using a Monolith NT.115 instrument (NanoTemper Technologies) at a constant temperature of 30 °C. Instrument parameters were adjusted to 60% LED power and medium MST power. Data of three independently pipetted measurements were analyzed (MO.Affinity Analysis software version 2.1.3, NanoTemper Technologies) using the signal from an MST-on time of 2.5 s. The Kd is estimated by fitting the equation:
where f(c) is the fraction bound at a given ligand concentration c; Unbound is the Fnorm signal of the target alone; Bound is the Fnorm signal of the complex; Kd is the dissociation constant or binding affinity; and ctarget is the final concentration of target in the assay.
Light-scattering assay
Light-scattering assays of urea-denatured proteins were performed as described previously27,45. GUN4, CHLH and GluTR were purified as soluble proteins and each was then denatured by buffer exchange into 8 M urea, 10 mM Tris, 100 mM Na2HPO4, pH 8.0 using P6 DG resin (Bio-Rad). Reactions were initiated by adding 97 μL of the indicated concentrations of cpSRP43 in LSA buffer to 3 μL of the urea-denatured client protein to yield the final client concentration (2.7 μM GluTR, 5 μM GUN4 or 1 μM CHLH), and monitored by recording the optical density at 360 nm for 5–10 minutes.
For heat-induced aggregation, purified client proteins in LSA buffer were centrifuged for 30 min at 100,000 rpm (TLA100, Beckman Coulter) and mixed with an equal volume of cpSRP43 at the indicated concentrations in the same buffer at 4 °C. The chaperone-client mixtures were incubated in a water bath at 44 °C for 5 min, while controls were kept at 4 °C, and then placed on ice for 5 min. The optical density at 360 nm was recorded, normalized to that of the client without cpSRP43, and plotted as a function of cpSRP43 concentration. Where possible, data that showed concentration-dependent protection by cpSRP43, were fit to:
| Eq. 1 |
where [cpSRP43] is the total cpSRP43 concentration, [client] the concentration of client (5 μM GUN4, 1 μM CHLH, or 2.7 μM GluTR), and Ksol is the apparent solubilization constant (i.e. the degree to which cpSRP43 protects against client aggregation. In cases where data that could not be fitted in this way (i.e. where no protection was detectable), a smoothing spline was plotted, but no Ksol was calculated (Ksol = N.D.).
Fluorescence anisotropy
Experiments were carried out as previously described27,32. Binding of cpSRP43 to the cpSRP54M peptide (QKQKAPPGTARRKRKAC) was detected by changes in the fluorescence anisotropy of fluorescein covalently attached to the C-terminal cysteine. Anisotropy measurements were performed in 50 mM HEPES (pH 7.5), 200 mM NaCl at 15, 25, 35, and 45 °C on a FluoroLog 3–22 (Yobin Yvon), using 100 nM fluorescein-labeled cpSRP54 M peptide and the indicated concentrations of cpSRP43. The samples were excited at 500 nm, and fluorescence anisotropy recorded at 527 nm. To obtain the equilibrium dissociation constant for the cpSRP43-cpSRP54M complex, the data were fit to:
| Eq. 2 |
in which Aobs is the observed anisotropy value, A0 is the anisotropy value of 54M-fluorescein alone, ΔA is the change in anisotropy at saturating concentrations of cpSRP43, and Kd is the equilibrium dissociation constant for the interaction between cpSRP43 and 54M-fluorescein.
Supplementary Material
Acknowledgements
We thank Danja Schünemann (Ruhr-Universität Bochum) for providing us the chaos and ffc mutants, the GST-cpSRP43 expression vector, and antibodies against cpSRP43 and cpSRP54, and Paul Hardy for critical reading of the manuscript. This work was supported by the Chinese Scholarship Council to S.J., Grant R35 GM136321 and DOE.DE-SC0020661 to A.S. and S.S., the Deutsche Forschungsgemeinschaft to B.G (FOR2092, GR 936/18-1, and SFB TRR175, subproject C04) and to P.W. (WA 4599/2-1).
Footnotes
Competing Interests
The authors declare no competing interests.
Data availability
All data are available from the corresponding authors on request.
References
- 1.Mochizuki N et al. The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci 15, 488–498, doi: 10.1016/j.tplants.2010.05.012 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Jarvis P & Lopez-Juez E Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14, 787–802, doi: 10.1038/nrm3702 [pii] (2013). [DOI] [PubMed] [Google Scholar]
- 3.Allen JF, de Paula WB, Puthiyaveetil S & Nield J A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci 16, 645–655, doi: 10.1016/j.tplants.2011.10.004 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Plumley GF & Schmidt GW Light-Harvesting Chlorophyll a/b Complexes: Interdependent Pigment Synthesis and Protein Assembly. The Plant cell 7, 689–704, doi: 10.1105/tpc.7.6.689 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dall’Osto L, Bressan M & Bassi R Biogenesis of light harvesting proteins. Biochim Biophys Acta 1847, 861–871, doi: 10.1016/j.bbabio.2015.02.009S0005-2728(15)00034-1 [pii] (2015). [DOI] [PubMed] [Google Scholar]
- 6.Wang P & Grimm B Connecting Chlorophyll Metabolism with Accumulation of the Photosynthetic Apparatus. Trends Plant Sci, doi: 10.1016/j.tplants.2020.12.005 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Wang P & Grimm B Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynthesis research, doi: 10.1007/s11120-015-0154-5 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Brzezowski P, Richter AS & Grimm B Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim Biophys Acta 1847, 968–985, doi: 10.1016/j.bbabio.2015.05.007S0005-2728(15)00088-2 [pii] (2015). [DOI] [PubMed] [Google Scholar]
- 9.Tanaka R & Tanaka A Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58, 321–346, doi: 10.1146/annurev.arplant.57.032905.105448 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Masuda T Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth Res 96, 121–143, doi: 10.1007/s11120-008-9291-4 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Adams NBP et al. The active site of magnesium chelatase. Nature plants 6, 1491–1502, doi: 10.1038/s41477-020-00806-9 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Larkin RM, Alonso JM, Ecker JR & Chory J GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906, doi: 10.1126/science.1079978 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Peter E & Grimm B GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Molecular plant 2, 1198–1210, doi: 10.1093/mp/ssp072 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Richter AS et al. Phosphorylation of GENOMES UNCOUPLED 4 Alters Stimulation of Mg Chelatase Activity in Angiosperms. Plant Physiol 172, 1578–1595, doi: 10.1104/pp.16.01036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka R, Kobayashi K & Masuda T Tetrapyrrole Metabolism in Arabidopsis thaliana. Arabidopsis Book 9, e0145, doi: 10.1199/tab.0145 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu JK Abiotic Stress Signaling and Responses in Plants. Cell 167, 313–324, doi: 10.1016/j.cell.2016.08.029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen B, Retzlaff M, Roos T & Frydman J Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol 3, a004374, doi: 10.1101/cshperspect.a004374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sangwan V, Orvar BL, Beyerly J, Hirt H & Dhindsa RS Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J 31, 629–638, doi: 10.1046/j.1365-313x.2002.01384.x (2002). [DOI] [PubMed] [Google Scholar]
- 19.Wang QL, Chen JH, He NY & Guo FQ Metabolic Reprogramming in Chloroplasts under Heat Stress in Plants. Int J Mol Sci 19, doi: 10.3390/ijms19030849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu S, Ding Y & Zhu C Sensitivity and Responses of Chloroplasts to Heat Stress in Plants. Front Plant Sci 11, 375, doi: 10.3389/fpls.2020.00375 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha JH, Lee HJ, Jung JH & Park CM Thermo-Induced Maintenance of Photo-oxidoreductases Underlies Plant Autotrophic Development. Dev Cell 41, 170–179 e174, doi: 10.1016/j.devcel.2017.03.005 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Ziehe D, Dünschede B & Schünemann D Molecular mechanism of SRP-dependent light-harvesting protein transport to the thylakoid membrane in plants. Photosynthesis research 138, 303–313, doi: 10.1007/s11120-018-0544-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akopian D, Shen K, Zhang X & Shan SO Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem 82, 693–721, doi: 10.1146/annurev-biochem-072711-164732 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schünemann D et al. A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc Natl Acad Sci U S A 95, 10312–10316, doi: 10.1073/pnas.95.17.10312 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Henry R, Yuan J, Cline K & Hoffman NE A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proceedings of the National Academy of Sciences of the United States of America 92, 3789–3793, doi: 10.1073/pnas.92.9.3789 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaru-Ampornpan P et al. ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat Struct Mol Biol 17, 696–702, doi: 10.1038/nsmb.1836 [pii] (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang FC et al. Conformational dynamics of a membrane protein chaperone enables spatially regulated substrate capture and release. Proc Natl Acad Sci U S A 113, E1615–1624, doi: 10.1073/pnas.1524777113 [pii] (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegel A et al. A Disorder-to-Order Transition Activates an ATP-Independent Membrane Protein Chaperone. J Mol Biol 432, 166708, doi: 10.1016/j.jmb.2020.11.007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaru-Ampornpan P, Chandrasekar S & Shan SO Efficient interaction between two GTPases allows the chloroplast SRP pathway to bypass the requirement for an SRP RNA. Mol Biol Cell 18, 2636–2645, doi: 10.1091/mbc.e07-01-0037 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandrasekar S, Sweredoski MJ, Sohn CH, Hess S & Shan SO Co-evolution of Two GTPases Enables Efficient Protein Targeting in an RNA-less Chloroplast Signal Recognition Particle Pathway. J Biol Chem 292, 386–396, doi: 10.1074/jbc.M116.752931 [pii] (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore M, Harrison MS, Peterson EC & Henry R Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. The Journal of biological chemistry 275, 1529–1532, doi: 10.1074/jbc.275.3.1529 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Wang P et al. Chloroplast SRP43 acts as a chaperone for glutamyl-tRNA reductase, the rate-limiting enzyme in tetrapyrrole biosynthesis. Proc Natl Acad Sci U S A 115, E3588–E3596, doi: 10.1073/pnas.1719645115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klimyuk VI et al. A chromodomain protein encoded by the arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell 11, 87–99 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amin P et al. Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant physiology 121, 61–70, doi: 10.1104/pp.121.1.61 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonas-Straube E, Hutin C, Hoffman NE & Schunemann D Functional analysis of the protein-interacting domains of chloroplast SRP43. J Biol Chem 276, 24654–24660, doi: 10.1074/jbc.M100153200 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Dunschede B, Bals T, Funke S & Schunemann D Interaction studies between the chloroplast signal recognition particle subunit cpSRP43 and the full-length translocase Alb3 reveal a membrane-embedded binding region in Alb3 protein. J Biol Chem 286, 35187–35195, doi: 10.1074/jbc.M111.250746 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falk S & Sinning I The C terminus of Alb3 interacts with the chromodomains 2 and 3 of cpSRP43. J Biol Chem 285, le25–26; author reply le26–28, doi: 10.1074/jbc.L110.160093 (2010b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Apel K & Hirt H Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55, 373–399, doi: 10.1146/annurev.arplant.55.031903.141701 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Herbst J, Hey D & Grimm B Posttranslational control of tetrapyrrole biosynthesis: Interacting proteins, chaperones, auxiliary factors. Advances in botanical research 91, 163–194 (2019). [Google Scholar]
- 40.Holdermann I et al. Chromodomains read the arginine code of post-translational targeting. Nat Struct Mol Biol 19, 260–263, doi: 10.1038/nsmb.2196 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Kotak S et al. Complexity of the heat stress response in plants. Current opinion in plant biology 10, 310–316, doi: 10.1016/j.pbi.2007.04.011 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Wang P, Richter AS, Kleeberg JRW, Geimer S & Grimm B Post-translational coordination of chlorophyll biosynthesis and breakdown by BCMs maintains chlorophyll homeostasis during leaf development. Nat Commun 11, 1254, doi: 10.1038/s41467-020-14992-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onate-Sanchez L & Vicente-Carbajosa J DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1, 93, doi: 10.1186/1756-0500-1-93 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou S, Sawicki A, Willows RD & Luo M C-terminal residues of oryza sativa GUN4 are required for the activation of the ChlH subunit of magnesium chelatase in chlorophyll synthesis. FEBS Lett 586, 205–210, doi: 10.1016/j.febslet.2011.12.026 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Jaru-Ampornpan P et al. Mechanism of an ATP-independent protein disaggregase: II. distinct molecular interactions drive multiple steps during aggregate disassembly. J Biol Chem 288, 13431–13445, doi: 10.1074/jbc.M113.462861 [pii] (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding authors on request.


