Abstract
Two rare 2-phenoxychromone derivatives, 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2), were isolated from the areal parts of Artemisia commutata and A. glauca, respectively, for the first time. Being rare in nature, the inhibition potentialities of 1 and 2 against SARS-CoV-2 was investigated using multistage in silico techniques. At first, molecular similarity and fingerprint studies were conducted for 1 and 2 against co-crystallized ligands of eight different COVID-19 enzymes. The carried-out studies indicated the similarity of 1 and 2 with TTT, the co-crystallized ligand of COVID-19 Papain-Like Protease (PLP), (PDB ID: 3E9S). Therefore, molecular docking studies of 1 and 2 against the PLP were carried out and revealed correct binding inside the active site exhibiting binding energies of −18.86 and −18.37 Kcal/mol, respectively. Further, in silico ADMET in addition to toxicity evaluation of 1 and 2 against seven models indicated the general safety and the likeness of 1 and 2 to be drugs. Lastly, to authenticate the binding and to investigate the thermodynamic characters, molecular dynamics (MD) simulation studies were conducted on 1 and PLP.
Keywords: Artemisia commutate, 2-phenoxychromones, SARS-CoV-2, COVID-19 Papain-Like Protease, molecular docking, molecular fingerprints, ADMET, MD simulations
1. Introduction
Natural products have been utilized in different folklores to treat many illnesses since ancient times [1,2,3]. The research in the field of natural product chemistry identified various bioactive metabolites that are engaged with the bioactivities such as diterpenes [4], phenolics [5], sesquiterpenes [6,7], steroids [8], flavonoids [9,10], alkaloids [11], saponins [12,13,14], isochromenes [15], benzofuran glucosides [16], and α-pyrones [17,18].
The chemical composition as well as the biological activities of plants that belong to the genus Artemisia L. (family Asteraceae) drew the attention of scientists to explore and investigate. For example, anhydroaustricin, epiashantin, and matricarin are biologically active compounds that were isolated from A. albida [19], A. sieversiana [20], and A. austriaca [21], respectively. Besides, cirsineol is an active lactone that was reported in A. umbrosa [22]. Also, the presence of sesquiterpene lactones and flavonoids were reported in A. tschernieviana [23], A. albida [24], and A. santolinifolia [25], respectively. On the other hand, the bioactivities and phyto analysis of essential oils were studied for A. kasakorum [26], A. lercheana and A. sieversiana [27], A. umbrosa [28], A. gurganica [29], A. proceriformis [30], A. terrae-albae [31], A. keiskeana [32], A. littoricola, A. mandshurica [33], A. commutata [34], in addition to five plants belonging to the Artemisia species [35].
A. commutata [36] and A. glauca [37] are perennial herbs that are cultivated in northern and eastern Kazakhstan [38]. The lack of scientific records about the phytochemistry and the biological effects of these two species encouraged us to examine their phytochemical properties aiming at the isolation of promising secondary metabolites.
The WHO declared on December 28, 2021, that the confirmed cases of COVID-19 infections on a global base are 280,119,931. Sadly, 5,403,662 of them are dead [39]. In regard to these bad numbers, massive work is demanded from scientists all over the world to find a cure.
The field of computer-aided/based (in silico or computational) drug design and discovery is a fast-growing approach that has shown several successes and advancements during the last decade [40]. The structural informatics explosion allowed the identification of the exact chemical and physical properties of molecules. Consequently, the ability to compare different structures and predict a specific biological activity became much easier [41]. Also, the huge advancement in the field of proteomic enabled scientists to expect the biding and hence the activity of a molecule and target enzyme [42]. Our teamwork reported the utilization of the in silico methods to suggest a treatment that could be effective against COVID-19 in many reports [43,44,45,46,47].
We here in this manuscript, report the isolation and structure elucidation of two rare 2-phenoxychromone derivatives, 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2) from the areal parts of A. commutata and A. glauca, respectively, for the first time. Because of the lack of research on the isolated compounds and the severe need to find a cure against COVID-19, the potential inhibitory effects of 1 and 2 against COVID-19 were investigated depending on a multiphase in silico screening method. The potentialities of 1 and 2 against COVID-19 Papain-Like Protease were confirmed according to molecular similarity, structure fingerprint, molecular docking, and molecular dynamics simulations studies. Additionally, ADMET and toxicity properties of 1 and 2 were studied to predict their likeness to be used as drugs.
2. Results
2.1. Isolation of Compounds
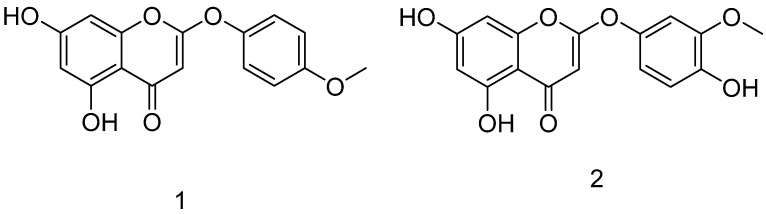
The areal parts of A. commutata, (940 g) and A. glauca (1.02 kg) were collected from the western part of the Altai Mountains. The dried plants were extracted with chloroform. The chloroform extract of A. commutata was subjected to chromatographic purifications steps to afford the amorphous powder of 1. Compound 1 was identified to be 6-demethoxy-4`-O-capillarsine (Figure 1). Identification of 1 was done depending on 1H and 13C NMR (Table S1, in the Supplementary Materials), comparing the published data. Compound 1 was previously isolated from the leaves of Mimosa tenuiflora [48], A. capillaris [49], and A. rupestris [50]. The chloroform extract of A. glauca was subjected to chromatographic purification steps to give the amorphous powder of 2. Compound 2 was identified to be 5,7,4′-trihydroxy, 3′-methoxy-2-phenoxychromone (Figure 1). Compound 2 was identified depending on 1H and 13C NMR (Table S2, in the Supplementary Materials) compared to the published data. Compound 2 was previously isolated from Mimosa tenuiflora [48].
Figure 1.
The chemical structures 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2).
2.2. Molecular Similarity Studies
To understand the principle of molecular similarity, we have to understand that the bioactivity of any compound is a result of some well-known interactions with a specific protein target. These protein–ligand interactions depend on specific physical and chemical interactions such as hydrogen bonding and hydrophobic interactions. Consequently, the similarity in the chemical structures of two compounds will give a similarity in the number and position of hydrogen bond donors, acceptors, and hydrophobic centers in addition to the steric configuration. This similarity could cause a noticeable similarity in the bioactivity too [51].
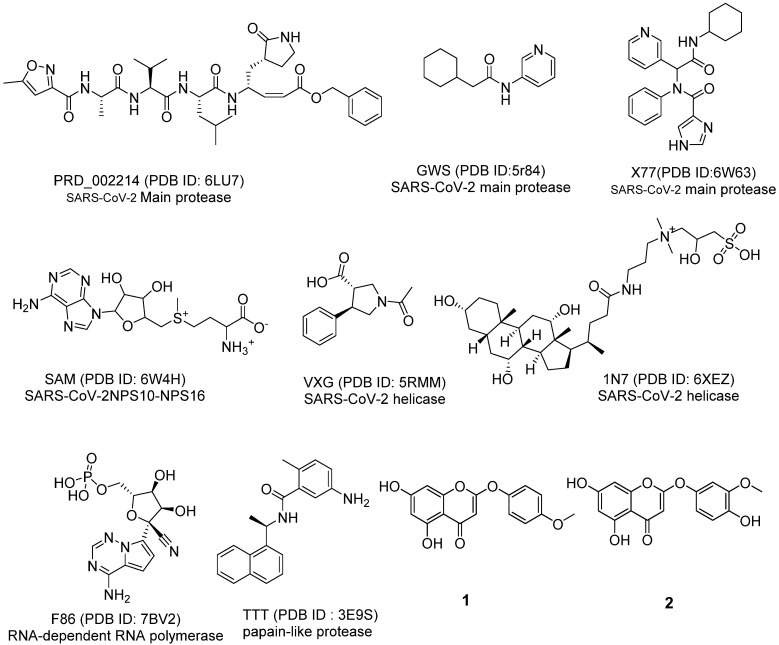
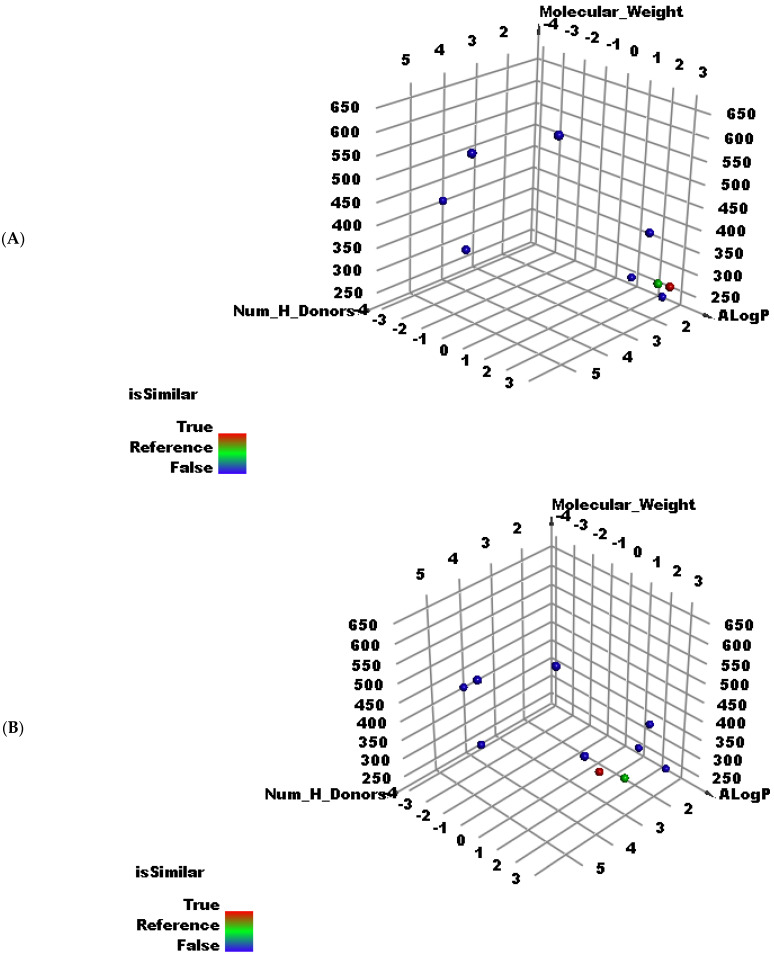
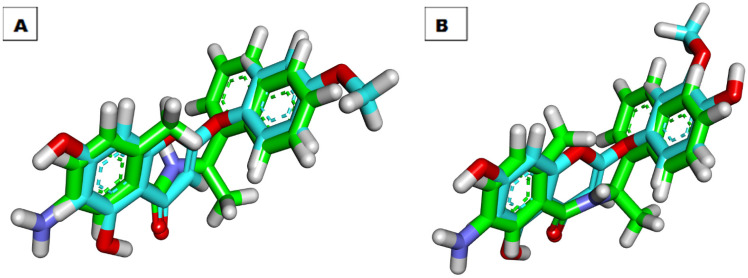
To evaluate the expected anti-COVID-19 potentialities of compounds 1 and 2, the chemical structures of them were compared with the chemical structures of eight different co-crystallized ligands of eight crucial COVID-19 proteins (Figure 2) in a structural similarity experiment. Utilizing Discovery Studio software, the following molecular properties were measured to 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2) as well as the eight ligands; partition coefficient (ALog p) [52], molecular weight (M. W) [53], H- bond donors (HBD) [54] and acceptors (HBA) [55], rotatable bonds [56], aromatic rings [57], heterocyclic rings [58], along with the molecular fractional polar surface area (MFPSA) [59]. The results revealed the great similarity between compounds 1 and 2 and the co-crystallized ligands (TTT) of SARS-CoV-PLP (PDB ID: 3E9S) (Table 1, Figure 3A,B).
Figure 2.
The co-crystallized ligands of coronavirus proteins and compounds 1 and 2.
Table 1.
Structural properties of compounds 1, 2 and TTT.
| Compound | ALog p | M. W | HBA | HBD | Rotatable Bonds | Rings | Aromatic Rings | MFPSA | Minimum Distance |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3.129 | 300.263 | 6 | 2 | 3 | 3 | 2 | 0.302 | 0 |
| 2 | 2.887 | 316.262 | 7 | 3 | 3 | 3 | 2 | 0.359 | 0 |
| TTT | 3.647 | 304.386 | 2 | 2 | 3 | 3 | 3 | 0.171 | 0.723 |
Figure 3.
The results of similarity analysis of compound 1 (A), compound 2 (B).
2.3. Fingerprints Studies
The molecular fingerprint is an in silico method that analyzes the similarities of two molecules or more. Molecular fingerprint explores the property profiles of a certain compound, the technique computes these properties in the forms of bits vectors examining the existence, absence, and frequencies it in the reference and target compounds.
The fingerprint approach examined the existence or absence of the these parameters: charge [60], hybridization [61], H-bond acceptor and donor [62], negative and positive ionizables [63], halogen [64], aromatic [65], and ALogP [66]. The experiment was achieved depending on Discovery Studio. The results verified the similarity of 6-demethoxy-4`-O-capillarsine (1), tenuflorin C (2), and TTT (Table 2).
Table 2.
Fingerprint similarity between compounds 1, 2 and TTT.
| Compound | Similarity | SA | SB | SC |
|---|---|---|---|---|
| 1 | 1 | 299 | 0 | 0 |
| 2 | 1 | 289 | 0 | 0 |
| TTT | 0.579 | 276 | 178 | 23 |
SA: The number bits present in both the isolated 2-phenoxychromone derivative and TTT. SB: The number of bits in the isolated 2-phenoxychromone derivative but not TTT. SC: The number of bits in TTT but not the isolated 2-phenoxychromone derivative.
2.4. Pharmacophoric Features and Flexible Alignment Studies
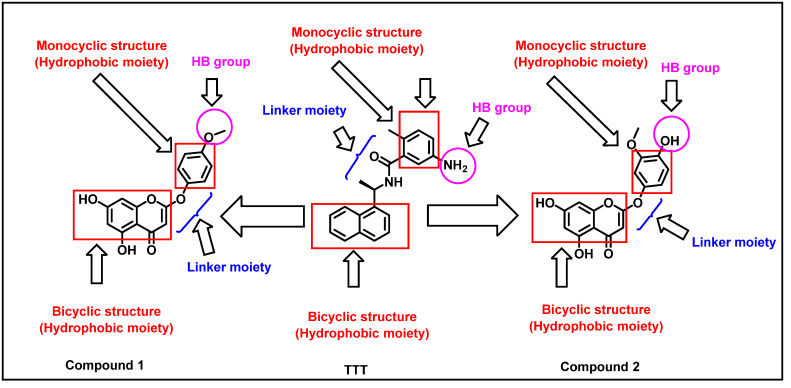
The reported main pharmacophoric features of SARS-CoV-2 Papain-Like protease inhibitors (PLPIs) are two hydrophobic systems separated be a linker [67]. As shown in Figure 4, the co-crystallized ligand (TTT) has naphthyl and phenyl groups as hydrophobic centers. The two hydrophobic groups are separated by hydrophilic linker (amide group). In addition, the terminal phenyl group is substituted by amino group as a hydrogen bonding group.
Figure 4.
Compounds 1 and 2 have the same pharmacophoric features of the co-crystallized ligand of SARS CoV-2 PLP(TTT).
Similarly, compounds 1 and 2 have coumarin moiety as the first hydrophobic center in addition to phenyl group as the second hydrophobic moiety. In each compound, the two hydrophobic centers were separated by oxygen atom as a hydrophilic linker. This linker facilitates these compounds to have the same configuration of TTT. Additionally, the two phenyl rings were substituted by hydrophilic groups as which can form hydrogen bonds with the target receptors as TTT. From medicinal chemistry point of view, this investigation revealed that there is a great similarity between the two tested molecules and the co-crystallized ligand (TTT).
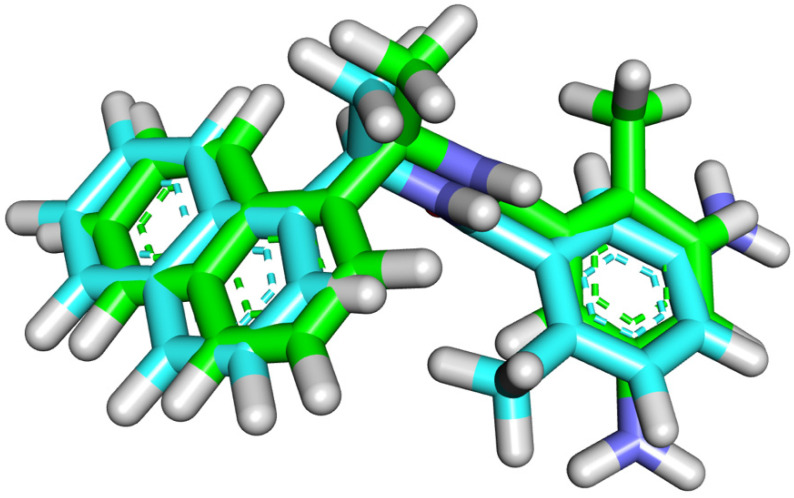
3D- Flexible alignment of the compounds 1 and 2 with TTT was presented in Figure 5. From the figure, it is possible to observe that, in general, the structure of compounds 1 and 2 have a good overlap with the reference molecule (TTT). In addition, the two tested compounds showed the same spatial orientation of TTT due to the presence of the flexible linker at the center of these compounds.
Figure 5.
(A) Flexible alignment of compounds 1 (turquoise) TTT (green). (B) Flexible alignment of compounds 2 (turquoise) with TTT (green).
2.5. Docking Studies
To examine the obtained results from structural similarity and fingerprint studies, molecular docking experiments were achieved for 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2) against PLP (PDB ID: 3E9S) using TTT, the co-crystallized ligand, as a reference. We considered the binding free energies (∆G) besides the binding modes as the bases of evaluation.
Firstly, a validation process has proceeded via the re-docking of TTT against PLP. The obtained RMSD value between the two poses was 2.1 ᵒA confirming the validity of the docking method (Figure 6).
Figure 6.
Superimposition of the co-crystallized molecule (Turquoise) and the docking pose (dark green) of the same molecule.
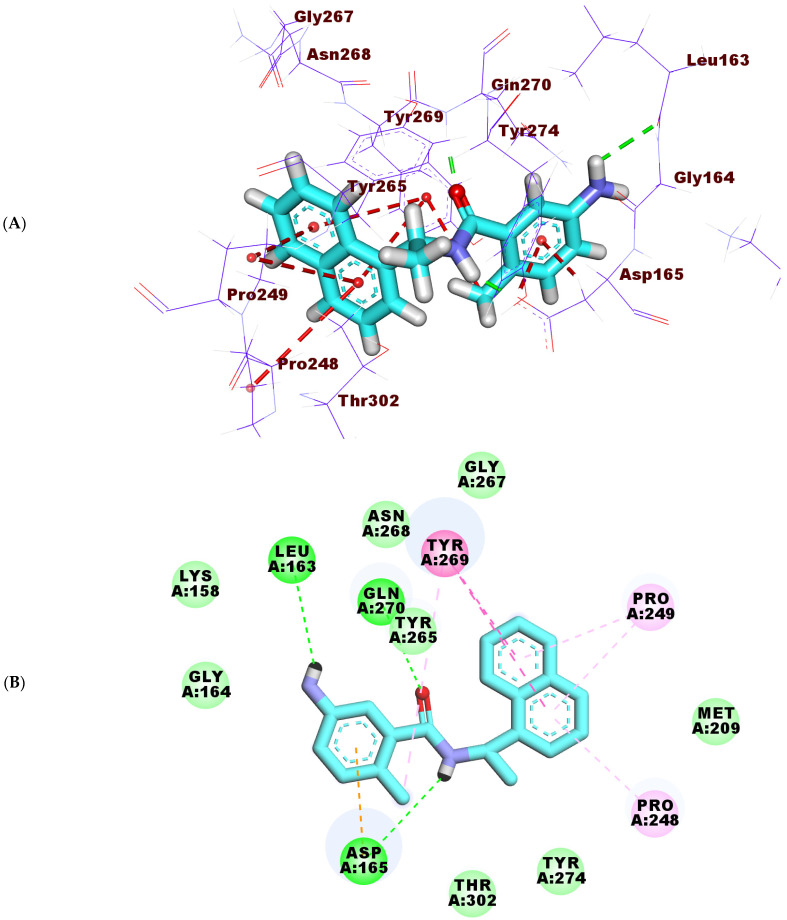
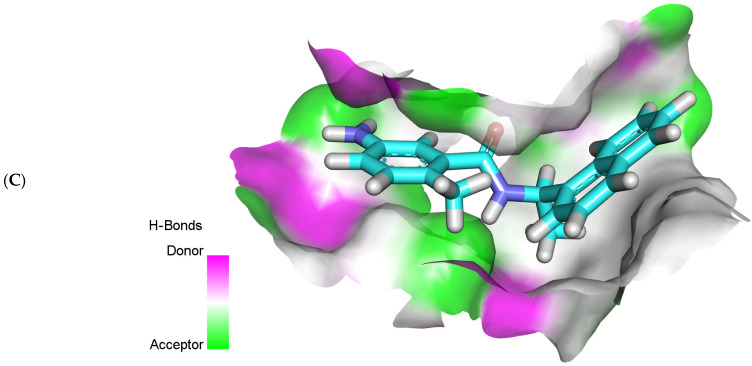
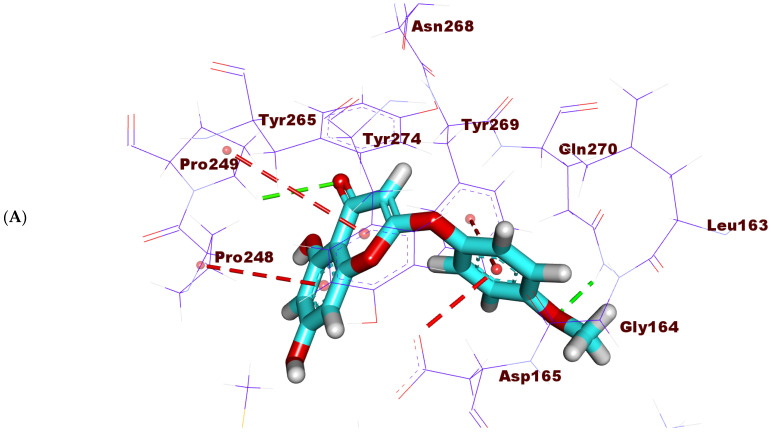
The binding mode of TTT showed a free energy of −20.32 kcal/mol. The p-toluidine part occupied the first pocket of PLP forming one H- bond with the amino acid, Leu163. The same moiety was engaged hydrophobically in two interactions with the amino acids Asp165 and Tyr269. In addition, the amide linker formed two H- bonds with the amino acids Asp165 and Gln270. The naphthalene moiety was buried in the second pocket making five hydrophobic interactions with Pro249, Pro248, and Tyr269 (Figure 7A–C).
Figure 7.
(A) 3D, (B) 2D and (C) Surface mapping of TTT inside the active site of PLP.
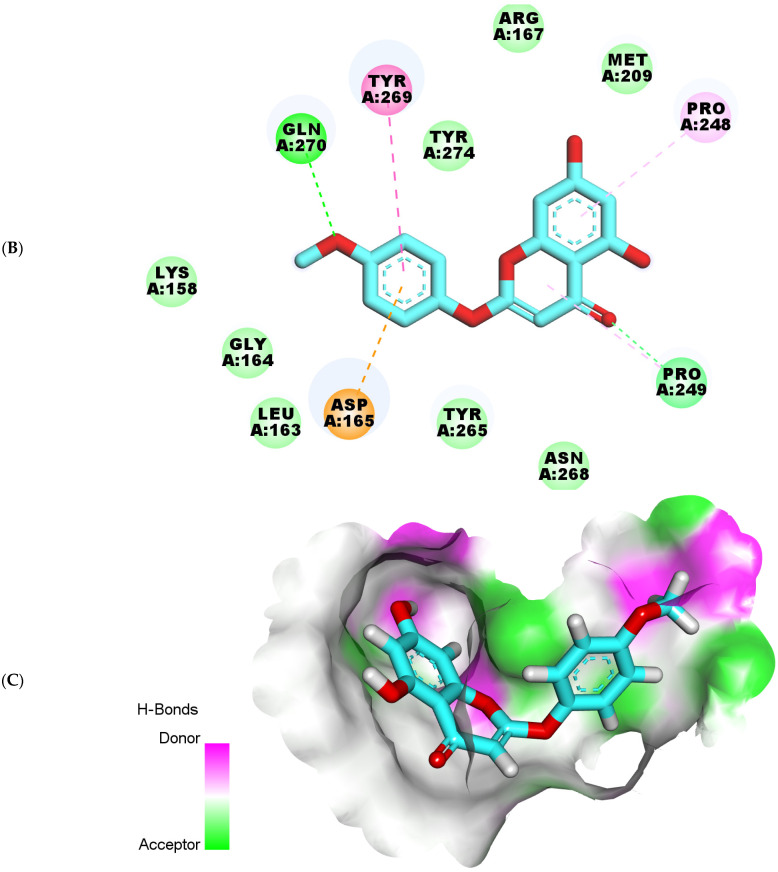
The binding mode of 6-demethoxy-4`-O-capillarsine (1) exhibited a binding free energy of -18.86 kcal/mol. The anisole moiety was directed into the first pocket of PLP forming one H- bond with the amino acid, Gln270, in addition to two hydrophobic interactions with the amino acids, Tyr269 and Asp165. The 5,7-dihydroxy-4H-chromen-4-one part occupied the second pocket of PLP forming a H- bond with the amino acid Pro249 besides two hydrophobic bonds with the amino acids Pro248 and Pro249 (Figure 8A–C).
Figure 8.
(A) 3D, (B) 2D and (C) Surface mapping of 6-demethoxy-4`-O-capillarsine (1) inside the active site of PLP.
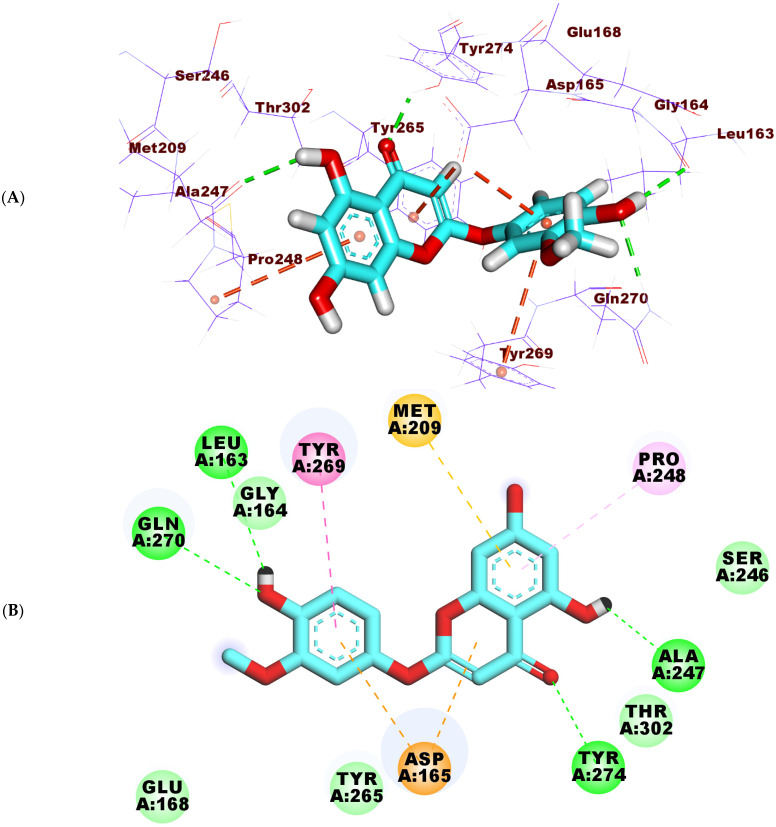
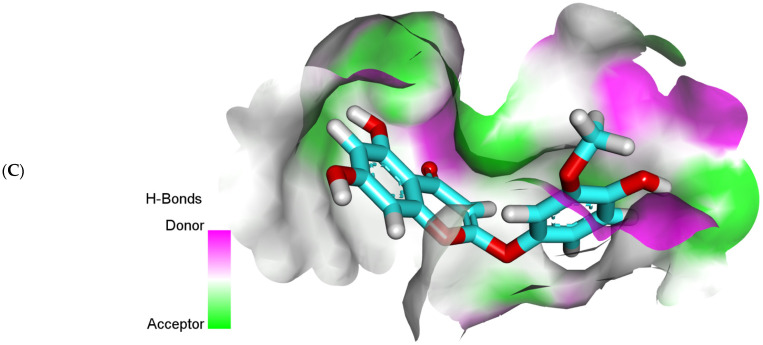
Tenuflorin C (2) bonded to PLP exhibiting a binding energy of −18.37 kcal/mol. The 2-methoxyphenol part was directed into the first pocket of PLP with two H-bonds with the amino acids Gln270 and Leu163. Additionally, the same moiety was engaged hydrophobically in two interactions with the amino acids Tyr269 and Asp165. Further, the 5,7-dihydroxy-4H-chromen-4-one part occupied the second pocket of PLP forming two H- bonds with Tyr274 and Ala247. Finally, it was incorporated hydrophobically in three bonds with the amino acids Pro248, Asp165 and Met209 (Figure 9A–C).
Figure 9.
(A) 3D, (B) 2D, and (C) surface mapping of tenuflorin C (2) inside the active site of PLP.
2.6. In Silico ADMET Study
There is always a need to examine the ADMET properties of new molecules especially in the stage of designing to decrease the possibility of late-stage attrition. Also, the identification of ADMET properties provides good information regarding the amount and frequency of dosing as well as the toxicity [68].
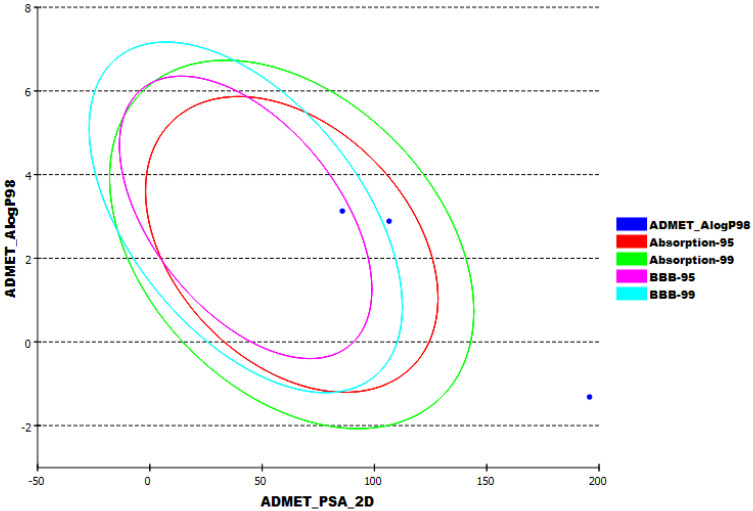
The in silico ADMET parameters were determined for 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2) using Discovery Studio software and utilizing Remdesivir as a reference. The results were illustrated in Figure 10 and Table 3.
Figure 10.
The expected ADMET study.
Table 3.
Binding free energies (∆G in Kcal/mol) of the 6-demethoxy-4`-O-capillarsine (1), tenuflorin C (2) and TTT against PLP.
| Compound | ∆G [Kcal/mol] |
|---|---|
| 1 | −18.86 |
| 2 | −18.37 |
| TTT | −20.32 |
The results revealed that 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2) have low and very low chance to penetrate BBB, respectively. Further, 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2) exhibited good aqueous solubility and intestinal absorption. Finally, 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2) were predicted to be a CYP2D6 non-inhibitor and to be able to bind plasma protein by a ratio of more than 90%.
2.7. In Silico Toxicity Studies
The predicted toxicity levels of 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2) were investigated in silico using Discovery Studio software against seven toxicity models. Remdesivir was used as a reference. As shown in Table 4, 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2) were predicted to be non-carcinogenic and have high TD50 values of 144.939 and 113.277 mg/kg/day, respectively. The values were more than 12-fold that of remdesivir (9.2458 mg/kg/day). Further, 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2) showed high rat maximum tolerated dose (MTD) values of 0.289735 and 0.44318 g/kg, respectively, compared to remdesivir (0.234965 g/kg). The rat oral LD50 values for 1 and 2 were 0.363122 and 0.549081 g/kg, respectively, and regarding the rat chronic LOAEL model, their values were 0.0221904 and 0.0362493, respectively. Finally, 6-demethoxy-4`-O-capillarsine (1) and tenuflorin C (2) were predicted to have mild ocular irritancy and to have no irritancy against the skin model.
Table 4.
Toxicity properties of compounds.
| Compound | FDA Rodent Carcinogenicity | Carcinogenicity TD50 a | Rat MTD b | Rat Oral LD50 b | LOAEL b | Ocular Irritancy c |
Skin Irritancy c |
|---|---|---|---|---|---|---|---|
| 1 | Not a carcinogen |
144.939 | 0.289735 | 0.363122 | 0.0221904 | + | - |
| 2 | Not a carcinogen |
113.277 | 0.44318 | 0.549081 | 0.0362493 | + | - |
| remdesivir | Not a carcinogen |
9.2458 | 0.234965 | 0.308859 | 0.0037911 | + | + |
a Unit: mg·kg−1·day−1., b Unit: g·kg−1., c - = nonirritant, + = mild.
2.8. Molecular Dynamics (MD) Simulation Studies
The main advantage of molecular dynamics (MD) simulation is its ability to recognize the flexibility of the protein–ligand complex. This advantage enables the MD studies to estimate accurately the thermodynamics and kinetics that take place during the drug–enzyme binding [69].
MD simulations can explore the dynamic structural information of the protein–ligand complex and provide plentiful information on the energetic changes that resulted from the interactions between protein and ligand. Such information could be very useful to understand the structure–function relationship of the protein–ligand complex interactions [70]. In order to authenticate the binding and to investigate the thermodynamic characters of the isolated compounds, molecular dynamics (MD) simulation studies were conducted to 6-demethoxy-4`-O-capillarsine, 1, as it showed a bitter binding energy, against PLP.
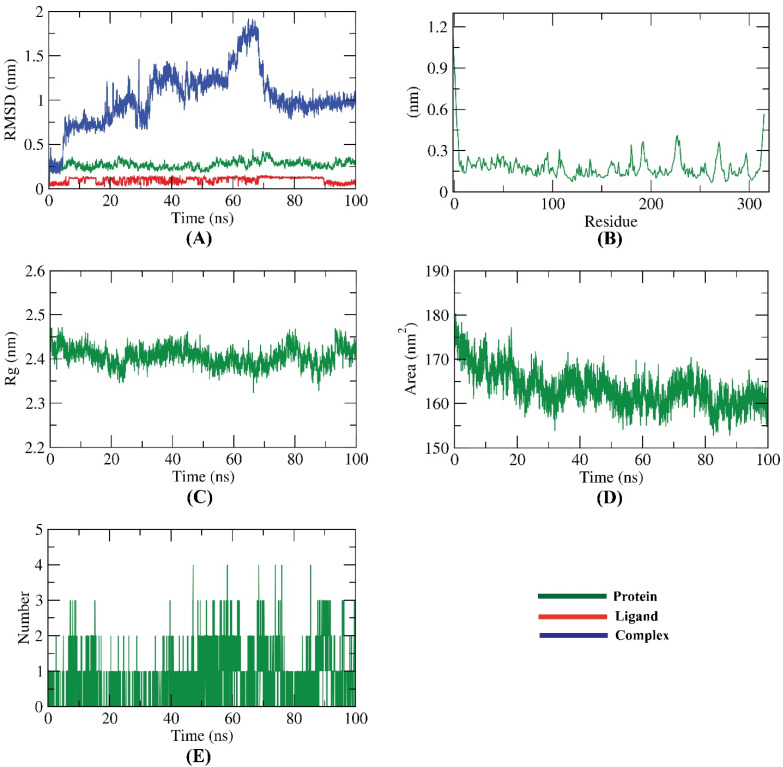
The atomical dynamic movements and conformational variations of backbone atoms of the 6-demethoxy-4`-O-capillarsine -PLP complex were calculated by RMSD to detect their stability upon ligand bonded and apo states. It was observed that the protein and ligand exhibit lower RMSD with no major fluctuations indicating their greater stability. The 6-demethoxy-4`-O-capillarsine -PLP complex was slightly fluctuating till 70 ns~ and stabled later (Figure 11A). The flexibility of PLP was evaluated in terms of RMSF (Figure 11B) to get more information about the protein regions that had been fluctuating during the simulation. It can be understood that the binding of 6-demethoxy-4`-O-capillarsine does not make PLP very flexible. The compactness of the 6-demethoxy-4`-O-capillarsine -PLP complex was evaluated through the examination of the radius of gyration (Rg) (Figure 11C). Decreasing fluctuation during the simulation study is an indication of the higher compactness of the system. The Rg of the 6-demethoxy-4`-O-capillarsine -PLP complex was found to be similar throughout the simulation period. Interaction between 6-demethoxy-4`-O-capillarsine -PLP complexes and solvents was evaluated through the calculation of solvent accessible surface area (SASA) over the simulation period. So, SASA of the 6-demethoxy-4`-O-capillarsine -PLP complex was calculated to analyze the extent of the conformational changes that occurred during the interaction. Interestingly, PLP featured a reduction of the surface area showing a relatively lower SASA value than the starting period (Figure 11D). Hydrogen bonding in the 6-demethoxy-4`-O-capillarsine -PLP complex is essential to stabilize the structure. It was noticed that the highest number of conformations of the PLP formed up to three H- bonds with the 6-demethoxy-4`-O-capillarsine (Figure 11E).
Figure 11.
Molecular dynamics simulations results; (A) RMSD values of 6-demethoxy-4`-O-capillarsine, PLP and 6-demethoxy-4`-O-capillarsine -PLP complex during MD runs. (B) RMSF for PLP in the MD run. (C) Radius of gyration of PLP in the MD run. (D) SASA of PLP in the MD run. (E) H- bonding between 6-demethoxy-4`-O-capillarsine -PLP complex in the MD run.
3. Experimental
3.1. Isolation and Structure Elucidation of Compounds
A total of 0.94 kg of Artemisia commutata and 1.04 kg of Artemisia glauca areal parts were extracted with chloroform. Successive chromatographic methods led to the isolation of compounds (1 and 2). Detailed description in the method part in Supplementary Materials.
3.2. Molecular Similarity
Molecular Similarity of 6-demethoxy-4`-O-capillarsine (1), tenuflorin C (2) against the co-crystallized ligands of SARS-CoV-2 was carried out calculated using Discovery Studio 4.0 (See the method part in Supplementary Materials).
3.3. Fingerprint Study
A fingerprint study of 6-demethoxy-4`-O-capillarsine (1), tenuflorin C (2) against the eight co-crystallized ligands of SARS-CoV-2 was carried out calculated using Discovery Studio 4.0. (See Supplementary Materials)
3.4. DFT
The DFT parameters were calculated for 6-demethoxy-4`-O-capillarsine (1), tenuflorin C (2) using Discovery Studio software [71]. (See the method part in Supplementary Materials).
3.5. Docking Studies
The docking investigation was carried out for 6-demethoxy-4`-O-capillarsine (1), tenuflorin C (2) using MOE2014 software and Discovery Studio 4.0 [72,73,74] (See the method part in Supplementary Materials).
3.6. ADMET
ADMET descriptors of 6-demethoxy-4`-O-capillarsine (1), tenuflorin C (2) were determined using Discovery Studio 4.0. [75,76] (See the method part in Supplementary Materials).
3.7. Toxicity Studies
Seven toxicity parameters of 6-demethoxy-4`-O-capillarsine (1), tenuflorin C (2) were calculated using Discovery Studio 4.0 [77,78,79] (See the method part in Supplementary Materials).
3.8. Molecular Dynamics Simulations
The system was prepared using the web-based CHARMM-GUI [80,81,82] interface utilizing CHARMM36 force field [83] and NAMD 2.13 [84] package. The TIP3P explicit solvation model was used (See the method part in Supplementary Materials).
Supplementary Materials
The following are available online.
Author Contributions
Conceptualization, Y.M.S., R.N.S. and A.M.M.; Funding acquisition, A.A.A.; Investigation, Y.M.S., R.A.J., R.N.S., C.A., M.I., S.T., W.D., A.M.M. and I.H.E.; Software, E.B.E., I.H.E. and A.M.M.; Writing—review & editing, A.A.A., E.B.E., I.H.E. and A.M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Princess Nourah Bint Abdulrahman University Researchers Support Project number (PNURSP2022R116), Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia. This research was funded by the Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan (Grant OR11465530 «Creation and replenishment of the collection of industrially valuable microorganisms, study and preservation of their biological diversity for the needs of biotechnology, medicine and agriculture» Grant AP13067578 «Comprehensive phytochemical study of a number of endemic plants of Kazakhstan» and Grant AP08051842 «Study of the composition and biological activity of the essential oils of plants of Central and Southeast Asia»). This research was funded by Princess Nourah Bint Abdulrahman University Researchers Support Project number (PNURSP2022R116), Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Metwaly A.M., Ghoneim M.M., Eissa I.H., Elsehemy I.A., Mostafa A.E., Hegazy M.M., Afifi W.M., Dou D. Traditional ancient Egyptian medicine: A review. Saudi J. Biol. Sci. 2021;28:5823–5832. doi: 10.1016/j.sjbs.2021.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han X., Yang Y., Metwaly A.M., Xue Y., Shi Y., Dou D. The Chinese herbal formulae (Yitangkang) exerts an antidiabetic effect through the regulation of substance metabolism and energy metabolism in type 2 diabetic rats. J. Ethnopharmacol. 2019;239:111942. doi: 10.1016/j.jep.2019.111942. [DOI] [PubMed] [Google Scholar]
- 3.Chu X., Sun B., Huang Q., Peng S., Zhou Y., Zhang Y. Quantitative knowledge presentation models of traditional Chinese medicine (TCM): A review. Artif. Intell. Med. 2020;103:101810. doi: 10.1016/j.artmed.2020.101810. [DOI] [PubMed] [Google Scholar]
- 4.Zhanzhaxina A., Suleimen Y., Metwaly A.M., Eissa I.H., Elkaeed E.B., Suleimen R., Ishmuratova M., Akatan K., Luyten W. In Vitro and In Silico Cytotoxic and Antibacterial Activities of a Diterpene from Cousinia alata Schrenk. J. Chem. 2021;2021:5542455. doi: 10.1155/2021/5542455. [DOI] [Google Scholar]
- 5.Hegazy M.M., Metwaly A.M., Mostafa A.E., Radwan M.M., Mehany A.B.M., Ahmed E., Enany S., Magdeldin S., Afifi W.M., ElSohly M.A. Biological and chemical evaluation of some African plants belonging to Kalanchoe species: Antitrypanosomal, cytotoxic, antitopoisomerase I activities and chemical profiling using ultra-performance liquid chromatography/quadrupole-time-of-flight mass spectrometer. Pharmacogn. Mag. 2021;17:6. [Google Scholar]
- 6.Imieje V.O., Zaki A.A., Metwaly A.M., Eissa I.H., Elkaeed E.B., Ali Z., Khan I.A., Falodun A. Antileishmanial Derivatives of Humulene from Asteriscus hierochunticus with in silico Tubulin Inhibition Potential. Rec. Nat. Prod. 2021;16:150. doi: 10.25135/rnp.253.21.01.1945. [DOI] [Google Scholar]
- 7.Jalmakhanbetova R., Elkaeed E.B., Eissa I.H., Metwaly A.M., Suleimen Y.M. Synthesis and Molecular Docking of Some Grossgemin Amino Derivatives as Tubulin Inhibitors Targeting Colchicine Binding Site. J. Chem. 2021;2021:5586515. doi: 10.1155/2021/5586515. [DOI] [Google Scholar]
- 8.Suleimen Y.M., Metwaly A.M., Mostafa A.E., Elkaeed E.B., Liu H.-W., Basnet B.B., Suleimen R.N., Ishmuratova M.Y., Turdybekov K.M., Van Hecke K. Isolation, Crystal Structure, and In Silico Aromatase Inhibition Activity of Ergosta-5, 22-dien-3β-ol from the Fungus Gyromitra esculenta. J. Chem. 2021;2021:5529786. doi: 10.1155/2021/5529786. [DOI] [Google Scholar]
- 9.Ghoneim M.M., Afifi W.M., Ibrahim M., Elagawany M., Khayat M.T., Aboutaleb M.H., Metwaly A.M. Biological evaluation and molecular docking study of metabolites from Salvadora Persica L. Growing in Egypt. Pharmacogn. Mag. 2019;15:232. [Google Scholar]
- 10.Liu L., Luo S., Yu M., Metwaly A.M., Ran X., Ma C., Dou D., Cai D. Chemical Constituents of Tagetes patula and Their Neuroprotecting Action. Nat. Prod. Commun. 2020;15 doi: 10.1177/1934578X20974507. [DOI] [Google Scholar]
- 11.Metwaly A.M., Ghoneim M.M., Musa A. Two new antileishmanial diketopiperazine alkaloids from the endophytic fungus Trichosporum sp. Derpharmachemica. 2015;7:322–327. [Google Scholar]
- 12.Yassin A.M., El-Deeb N.M., Metwaly A.M., El Fawal G.F., Radwan M.M., Hafez E.E. Induction of apoptosis in human cancer cells through extrinsic and intrinsic pathways by Balanites aegyptiaca furostanol saponins and saponin-coated silvernanoparticles. Appl. Biochem. Biotechnol. 2017;182:1675–1693. doi: 10.1007/s12010-017-2426-3. [DOI] [PubMed] [Google Scholar]
- 13.Sharaf M.H., El-Sherbiny G.M., Moghannem S.A., Abdelmonem M., Elsehemy I.A., Metwaly A.M., Kalaba M.H. New combination approaches to combat methicillin-resistant Staphylococcus aureus (MRSA) Sci. Rep. 2021;11:4240. doi: 10.1038/s41598-021-82550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metwaly A.M., Lianlian Z., Luqi H., Deqiang D. Black ginseng and its saponins: Preparation, phytochemistry and pharmacological effects. Molecules. 2019;24:1856. doi: 10.3390/molecules24101856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metwaly A.M., Kadry H.A., Atef A., Mohammad A.-E.I., Ma G., Cutler S.J., Ross S.A. Nigrosphaerin A a new isochromene derivative from the endophytic fungus Nigrospora sphaerica. Phytochem. Lett. 2014;7:1–5. doi: 10.1016/j.phytol.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y.-M., Ran X.-K., Riaz M., Yu M., Cai Q., Dou D.-Q., Metwaly A.M., Kang T.-G., Cai D.-C. Chemical constituents of stems and leaves of Tagetespatula L. and its fingerprint. Molecules. 2019;24:3911. doi: 10.3390/molecules24213911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metwaly A.M., Fronczek F.R., Ma G., Kadry H.A., Atef A., Mohammad A.-E.I., Cutler S.J., Ross S.A. Antileukemic α-pyrone derivatives from the endophytic fungus Alternaria phragmospora. Tetrahedron Lett. 2014;55:3478–3481. doi: 10.1016/j.tetlet.2014.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metwaly A.M., Wanas A.S., Radwan M.M., Ross S.A., ElSohly M.A. New α-Pyrone derivatives from the endophytic fungus Embellisia sp. Med. Chem. Res. 2017;26:1796–1800. doi: 10.1007/s00044-017-1889-7. [DOI] [Google Scholar]
- 19.Suleimenov E., Raldugin V., Adekenov S. Anhydroaustricin from Artemisia albida. Chem. Nat. Compd. 2008;44:541–542. doi: 10.1007/s10600-008-9118-8. [DOI] [Google Scholar]
- 20.Suleimenov E., Smagulova F., Seidakhmetova R., Aksartov R., Raldugin V., Adekenov S. 4-Epiashantin from Artemisia sieversiana. Chem. Nat. Compd. 2007;43:232–233. doi: 10.1007/s10600-007-0090-5. [DOI] [Google Scholar]
- 21.Kikhanova Z.S., Iskakova Z.B., Dzhalmakhanbetova R., Seilkhanov T., Ross S., Suleimen E. Constituents of Artemisia austriaca and their biological activity. Chem. Nat. Compd. 2013;49:967–968. doi: 10.1007/s10600-013-0796-5. [DOI] [Google Scholar]
- 22.Tashenov E., Dzhalmakhanbetova R., Smagulova F., Dudkin R., Gorovoi P., Suleiman E., Ross S. Cirsilineol and cubreuva lactone from Artemisia umbrosa and their biological activity. Chem. Nat. Compd. 2013;49:97–98. doi: 10.1007/s10600-013-0517-0. [DOI] [Google Scholar]
- 23.Sisengalieva G., Suleimen E., Ishmuratova M.Y., Iskakova Z.B., Van Hecke K. Constituents of Artemisia tschernieviana and their biological activity. Chem. Nat. Compd. 2015;51:544–547. doi: 10.1007/s10600-015-1337-1. [DOI] [Google Scholar]
- 24.Suleimenov E., Smagulova F., Morozova O., Raldugin V., Bagryanskaya I.Y., Gatilov Y.V., Yamovoi V., Adekenov S. Sesquiterpene lactones and flavonoids from Artemisia albida. Chem. Nat. Compd. 2005;41:689–691. doi: 10.1007/s10600-006-0013-x. [DOI] [Google Scholar]
- 25.Suleimen E., Dzhalmakhanbetova R., Ishmuratova M.Y. Flavonoids from Artemisia santolinifolia. Chem. Nat. Compd. 2014;50:918–919. doi: 10.1007/s10600-014-1117-3. [DOI] [Google Scholar]
- 26.Suleimenov E., Ozek T., Demirci F., Demirci B., Baser K., Adekenov S. Component composition and antimicrobial activity of essential oil from Artemisia kasakorum. Chem. Nat. Compd. 2008;44:263–265. doi: 10.1007/s10600-008-9034-y. [DOI] [Google Scholar]
- 27.Suleimenov E., Ozek T., Demirci F., Demirci B., Baser K., Adekenov S. Component composition of essential oils of Artemisia lercheana and A. sieversiana of the flora of Kazakhstan. Antimicrobial activity of A. sieversiana essential oil. Chem. Nat. Compd. 2009;45:120–123. doi: 10.1007/s10600-009-9232-2. [DOI] [Google Scholar]
- 28.Suleimen E.M., Dudkin R.V., Gorovoi P.G., Wang M., Khan I., Ross S.A. Composition and Bioactivity of Artemisia umbrosa Essential Oil. Chem. Nat. Compd. 2014;50:545–546. doi: 10.1007/s10600-014-1011-z. [DOI] [Google Scholar]
- 29.Suleimen E.M., Ibataev Z.A., Iskakova Z.B., Ishmuratova M.Y. Constituent Composition and Biological Activity of Essential Oil from Artemisia gurganica. Chem. Nat. Compd. 2015;51:1184–1185. doi: 10.1007/s10600-015-1526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sampietro D.A., Lizarraga E.F., Ibatayev Z.A., Omarova A.B., Suleimen Y.M., Catalán C.A.N. Chemical composition and antimicrobial activity of essential oils from Acantholippia deserticola, Artemisia proceriformis, Achillea micrantha and Libanotis buchtormensis against phytopathogenic bacteria and fungi. Nat. Prod. Res. 2016;30:1950–1955. doi: 10.1080/14786419.2015.1091453. [DOI] [PubMed] [Google Scholar]
- 31.Suleimen E.M., Ibataev Z.A., Iskakova Z.B., Ishmuratova M.Y., Ross S.A., Martins C.H.G. Constituent Composition and Biological Activity of Essential Oil from Artemisia terrae-albae. Chem. Nat. Compd. 2016;52:173–175. doi: 10.1007/s10600-016-1584-9. [DOI] [Google Scholar]
- 32.Suleimen E.M., Sisengalieva G.G., Adilkhanova A.A., Dudkin R.V., Gorovoi P.G., Iskakova Z.B. Composition and Biological Activity of Essential Oil from Artemisia keiskeana. Chem. Nat. Compd. 2019;55:154–156. doi: 10.1007/s10600-019-02641-7. [DOI] [Google Scholar]
- 33.Suleimen E.M., Dudkin R.V., Gorovoi P.G., Wang M., Khan I., Ross S.A. Constituent Compositions of Essential Oils from Artemisia littoricola and A. mandshurica. Chem. Nat. Compd. 2015;51:790–792. doi: 10.1007/s10600-015-1414-5. [DOI] [Google Scholar]
- 34.Shatar S., Altantsetseg S., Darijimaa S. The essential oil composition of six Artemisia species from Mongolia. J. Essent. Oil Bear. Plants. 1999;2:56–67. [Google Scholar]
- 35.Ozek G., Suleimen Y., Tabanca N., Doudkin R., Gorovoy P.G., Goger F., Wedge D.E., Ali A., Khan I.A., Baser K.H. Chemical Diversity and Biological Activity of the Volatiles of Five Artemisia Species from Far East Russia. Agricultural Research Service University, Natural Products Utilization; Oxford, MS, USA: 2014. [Google Scholar]
- 36.Vasilyeva A.P.G., Goloskokov V.P., Zaitseva L.G. Flora of Kazakhstan. Alma-Ata. 1966;9:106–107. [Google Scholar]
- 37.Plantarium Artemisia glauca, Taxon Details. [(accessed on 23 November 2021)]. Available online: https://www.plantarium.ru/page/view/item/3891.html.
- 38.Vasilyeva A.P.G., Goloskokov V.P., Zaitseva L.G. Flora of Kazakhstan, Science. V N. V. Pavlova, Alma-Ata; 1966. [Google Scholar]
- 39.WHO WHO Coronavirus (COVID-19) Dashboard. [(accessed on 25 December 2021)]. Available online: https://covid19.who.int/
- 40.Bharatam P.V. Computer-Aided Drug Design. Springer; Singapore: 2021. pp. 137–210. [DOI] [Google Scholar]
- 41.Sabe V.T., Ntombela T., Jhamba L.A., Maguire G.E., Govender T., Naicker T., Kruger H.G. Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: A review. Eur. J. Med. Chem. 2021;224:113705. doi: 10.1016/j.ejmech.2021.113705. [DOI] [PubMed] [Google Scholar]
- 42.Sliwoski G., Kothiwale S., Meiler J., Lowe E.W. Computational methods in drug discovery. Pharmacol. Rev. 2014;66:334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jalmakhanbetova R.I., Suleimen Y.M., Oyama M., Elkaeed E.B., Eissa I., Suleimen R.N., Metwaly A.M., Ishmuratova M.Y. Isolation and In Silico Anti-COVID-19 Main Protease (Mpro) Activities of Flavonoids and a Sesquiterpene Lactone from Artemisia sublessingiana. J. Chem. 2021;2021:5547013. doi: 10.1155/2021/5547013. [DOI] [Google Scholar]
- 44.Al-Karmalawy A.A., Dahab M.A., Metwaly A.M., Elhady S.S., Elkaeed E.B., Eissa I.H., Darwish K.M. Molecular Docking and Dynamics Simulation Revealed the Potential Inhibitory Activity of ACEIs Against SARS-CoV-2 Targeting the hACE2 Receptor. Front. Chem. 2021;9:661230. doi: 10.3389/fchem.2021.661230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alesawy M.S., Abdallah A.E., Taghour M.S., Elkaeed E.B., H Eissa I., Metwaly A.M. In Silico Studies of Some Isoflavonoids as Potential Candidates against COVID-19 Targeting Human ACE2 (hACE2) and Viral Main Protease (Mpro) Molecules. 2021;26:2806. doi: 10.3390/molecules26092806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Demerdash A., Metwaly A.M., Hassan A., El-Aziz A., Mohamed T., Elkaeed E.B., Eissa I.H., Arafa R.K., Stockand J.D. Comprehensive virtual screening of the antiviral potentialities of marine polycyclic guanidine alkaloids against SARS-CoV-2 (COVID-19) Biomolecules. 2021;11:460. doi: 10.3390/biom11030460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eissa I.H., Khalifa M.M., Elkaeed E.B., Hafez E.E., Alsfouk A.A., Metwaly A.M. In Silico Exploration of Potential Natural Inhibitors against SARS-CoV-2 nsp10. Molecules. 2021;26:6151. doi: 10.3390/molecules26206151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.León L., Maldonado E., Cruz A., Ortega A. Tenuiflorins A–C: New 2-Phenoxychromones from the Leaves of Mimosa tenuiflora. Planta Med. 2004;70:536–539. doi: 10.1055/s-2004-827154. [DOI] [PubMed] [Google Scholar]
- 49.Hashidoko Y., Tahara S., Mizutani J. 2-Phenoxychromones and a structurally related flavone from leaves ofRosa rugosa. Phytochemistry. 1991;30:3837–3838. doi: 10.1016/0031-9422(91)80131-J. [DOI] [Google Scholar]
- 50.Aisa H.A., Zhao Y., He C. A 2-phenoxychromone from Artemisia rupestris. Chem. Nat. Compd. 2006;42:16–18. doi: 10.1007/s10600-006-0026-5. [DOI] [Google Scholar]
- 51.Nasser M., Salim N., Hamza H., Saeed F., Rabiu I. Improved deep learning based method for molecular similarity searching using stack of deep belief networks. Molecules. 2021;26:128. doi: 10.3390/molecules26010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turchi M., Cai Q., Lian G. An evaluation of in-silico methods for predicting solute partition in multiphase complex fluids–A case study of octanol/water partition coefficient. Chem. Eng. Sci. 2019;197:150–158. doi: 10.1016/j.ces.2018.12.003. [DOI] [Google Scholar]
- 53.Sullivan K.M., Enoch S.J., Ezendam J., Sewald K., Roggen E.L., Cochrane S. An adverse outcome pathway for sensitization of the respiratory tract by low-molecular-weight chemicals: Building evidence to support the utility of in vitro and in silico methods in a regulatory context. Appl. In Vitro Toxicol. 2017;3:213–226. doi: 10.1089/aivt.2017.0010. [DOI] [Google Scholar]
- 54.Altamash T., Amhamed A., Aparicio S., Atilhan M. Effect of hydrogen bond donors and acceptors on CO2 absorption by deep eutectic solvents. Processes. 2020;8:1533. doi: 10.3390/pr8121533. [DOI] [Google Scholar]
- 55.Wan Y., Tian Y., Wang W., Gu S., Ju X., Liu G. In silico studies of diarylpyridine derivatives as novel HIV-1 NNRTIs using docking-based 3D-QSAR, molecular dynamics, and pharmacophore modeling approaches. RSC Adv. 2018;8:40529–40543. doi: 10.1039/C8RA06475J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Escamilla-Gutiérrez A., Ribas-Aparicio R.M., Córdova-Espinoza M.G., Castelán-Vega J.A. In silico strategies for modeling RNA aptamers and predicting binding sites of their molecular targets. Nucleosides Nucleotides Nucleic Acids. 2021;40:798–807. doi: 10.1080/15257770.2021.1951754. [DOI] [PubMed] [Google Scholar]
- 57.Kaushik A.C., Kumar A., Bharadwaj S., Chaudhary R., Sahi S. Bioinformatics Techniques for Drug Discovery. Springer; Singapore: 2018. Ligand-Based Approach for In-silico Drug Designing; pp. 11–19. [Google Scholar]
- 58.Rarey M., Dixon J.S. Feature trees: A new molecular similarity measure based on tree matching. J. Comput.-Aided Mol. Des. 1998;12:471–490. doi: 10.1023/A:1008068904628. [DOI] [PubMed] [Google Scholar]
- 59.Zhang H., Ren J.-X., Ma J.-X., Ding L. Development of an in silico prediction model for chemical-induced urinary tract toxicity by using naïve Bayes classifier. Mol. Divers. 2019;23:381–392. doi: 10.1007/s11030-018-9882-8. [DOI] [PubMed] [Google Scholar]
- 60.Ieritano C., Campbell J.L., Hopkins W.S. Predicting differential ion mobility behaviour in silico using machine learning. Analyst. 2021;146:4737–4743. doi: 10.1039/D1AN00557J. [DOI] [PubMed] [Google Scholar]
- 61.Taha M., Ismail N.H., Ali M., Rashid U., Imran S., Uddin N., Khan K.M. Molecular hybridization conceded exceptionally potent quinolinyl-oxadiazole hybrids through phenyl linked thiosemicarbazide antileishmanial scaffolds: In silico validation and SAR studies. Bioorganic Chem. 2017;71:192–200. doi: 10.1016/j.bioorg.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 62.Chu H., He Q.-X., Wang J., Hu Y., Wang Y.-Q., Lin Z.-H. In silico design of novel benzohydroxamate-based compounds as inhibitors of histone deacetylase 6 based on 3D-QSAR, molecular docking, and molecular dynamics simulations. New J. Chem. 2020;44:21201–21210. doi: 10.1039/D0NJ04704J. [DOI] [Google Scholar]
- 63.Opo F.A.D.M., Rahman M.M., Ahammad F., Ahmed I., Bhuiyan M.A., Asiri A.M. Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Sci. Rep. 2021;11:4049. doi: 10.1038/s41598-021-83626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heidrich J., Sperl L.E., Boeckler F.M. Embracing the diversity of halogen bonding motifs in fragment-based drug discovery—construction of a diversity-optimized halogen-enriched fragment library. Front. Chem. 2019;7:9. doi: 10.3389/fchem.2019.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szatylowicz H., Stasyuk O.A., Krygowski T.M. Calculating the aromaticity of heterocycles. Adv. Heterocycl. Chem. 2016;120:301–327. [Google Scholar]
- 66.Andreeva E., Raevsky O. Lipophilicity of organic compounds calculated using structural similarity and molecular physicochemical descriptors. Pharm. Chem. J. 2009;43:258. doi: 10.1007/s11094-009-0280-5. [DOI] [Google Scholar]
- 67.Alesawy M.S., Elkaeed E.B., Alsfouk A.A., Metwaly A.M., Eissa I.J.M. In Silico Screening of Semi-Synthesized Compounds as Potential Inhibitors for SARS-CoV-2 Papain-Like Protease: Pharmacophoric Features, Molecular Docking, ADMET, Toxicity and DFT Studies. Molecules. 2021;26:6593. doi: 10.3390/molecules26216593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Norinder U., Bergström C.A. Prediction of ADMET properties. ChemMedChem Chem. Enabling Drug Discov. 2006;1:920–937. doi: 10.1002/cmdc.200600155. [DOI] [PubMed] [Google Scholar]
- 69.Hansson T., Oostenbrink C., van Gunsteren W. Molecular dynamics simulations. Curr. Opin. Struct. Biol. 2002;12:190–196. doi: 10.1016/S0959-440X(02)00308-1. [DOI] [PubMed] [Google Scholar]
- 70.Liu X., Shi D., Zhou S., Liu H., Liu H., Yao X. Molecular dynamics simulations and novel drug discovery. Expert Opin. Drug Discov. 2018;13:23–37. doi: 10.1080/17460441.2018.1403419. [DOI] [PubMed] [Google Scholar]
- 71.Parmar D.R., Soni J.Y., Guduru R., Rayani R.H., Kusurkar R.V., Vala A.G., Talukdar S.N., Eissa I.H., Metwaly A.M., Khalil A. Discovery of new anticancer thiourea-azetidine hybrids: Design, synthesis, in vitro antiproliferative, SAR, in silico molecular docking against VEGFR-2, ADMET, toxicity, and DFT studies. Bioorganic Chem. 2021;115:105206. doi: 10.1016/j.bioorg.2021.105206. [DOI] [PubMed] [Google Scholar]
- 72.Amer H.H., Alotaibi S.H., Trawneh A.H., Metwaly A.M., Eissa I.H. Anticancer activity, spectroscopic and molecular docking of some new synthesized sugar hydrazones, Arylidene and α-Aminophosphonate derivatives. Arab. J. Chem. 2021;14:103348. doi: 10.1016/j.arabjc.2021.103348. [DOI] [Google Scholar]
- 73.El-Adl K., Sakr H.M., Yousef R.G., Mehany A.B., Metwaly A.M., Elhendawy M.A., Radwan M.M., ElSohly M.A., Abulkhair H.S., Eissa I.H. Discovery of new quinoxaline-2 (1H)-one-based anticancer agents targeting VEGFR-2 as inhibitors: Design, synthesis, and anti-proliferative evaluation. Bioorganic Chem. 2021;114:105105. doi: 10.1016/j.bioorg.2021.105105. [DOI] [PubMed] [Google Scholar]
- 74.Eissa I.H., Ibrahim M.K., Metwaly A.M., Belal A., Mehany A.B., Abdelhady A.A., Elhendawy M.A., Radwan M.M., ElSohly M.A., Mahdy H.A. Design, molecular docking, in vitro, and in vivo studies of new quinazolin-4 (3H)-ones as VEGFR-2 inhibitors with potential activity against hepatocellular carcinoma. Bioorganic Chem. 2021;107:104532. doi: 10.1016/j.bioorg.2020.104532. [DOI] [PubMed] [Google Scholar]
- 75.Yousef R.G., Sakr H.M., Eissa I.H., Mehany A.B., Metwaly A.M., Elhendawy M.A., Radwan M.M., ElSohly M.A., Abulkhair H.S., El-Adl K. New quinoxaline-2 (1 H)-ones as potential VEGFR-2 inhibitors: Design, synthesis, molecular docking, ADMET profile and anti-proliferative evaluations. New J. Chem. 2021;45:16949–16964. doi: 10.1039/D1NJ02509K. [DOI] [PubMed] [Google Scholar]
- 76.Eissa I.H., El-Helby A.-G.A., Mahdy H.A., Khalifa M.M., Elnagar H.A., Mehany A.B., Metwaly A.M., Elhendawy M.A., Radwan M.M., ElSohly M.A. Discovery of new quinazolin-4 (3H)-ones as VEGFR-2 inhibitors: Design, synthesis, and anti-proliferative evaluation. Bioorganic Chem. 2020;105:104380. doi: 10.1016/j.bioorg.2020.104380. [DOI] [PubMed] [Google Scholar]
- 77.El-Adl K., El-Helby A.-G.A., Ayyad R.R., Mahdy H.A., Khalifa M.M., Elnagar H.A., Mehany A.B., Metwaly A.M., Elhendawy M.A., Radwan M.M. Design, synthesis, and anti-proliferative evaluation of new quinazolin-4 (3H)-ones as potential VEGFR-2 inhibitors. Bioorganic Med. Chem. 2021;29:115872. doi: 10.1016/j.bmc.2020.115872. [DOI] [PubMed] [Google Scholar]
- 78.El-Helby A.-G.A., Sakr H., Ayyad R.R., Mahdy H.A., Khalifa M.M., Belal A., Rashed M., El-Sharkawy A., Metwaly A.M., Elhendawy M.A. Design, synthesis, molecular modeling, in vivo studies and anticancer activity evaluation of new phthalazine derivatives as potential DNA intercalators and topoisomerase II inhibitors. Bioorganic Chem. 2020;103:104233. doi: 10.1016/j.bioorg.2020.104233. [DOI] [PubMed] [Google Scholar]
- 79.Eissa I.H., Metwaly A.M., Belal A., Mehany A.B., Ayyad R.R., El-Adl K., Mahdy H.A., Taghour M.S., El-Gamal K.M., El-Sawah M.E. Discovery and antiproliferative evaluation of new quinoxalines as potential DNA intercalators and topoisomerase II inhibitors. Arch. Pharm. 2019;352:1900123. doi: 10.1002/ardp.201900123. [DOI] [PubMed] [Google Scholar]
- 80.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 81.Brooks B.R., Brooks C.L., III, Mackerell A.D., Jr., Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y., et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Best R.B., Zhu X., Shim J., Lopes P.E., Mittal J., Feig M., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article.