Abstract
Wsc1 and Mid2 are highly O-glycosylated cell surface proteins that reside in the plasma membrane of Saccharomyces cerevisiae. They have been proposed to function as mechanosensors of cell wall stress induced by wall remodeling during vegetative growth and pheromone-induced morphogenesis. These proteins are required for activation of the cell wall integrity signaling pathway that consists of the small G-protein Rho1, protein kinase C (Pkc1), and a mitogen-activated protein kinase cascade. We show here by two-hybrid experiments that the C-terminal cytoplasmic domains of Wsc1 and Mid2 interact with Rom2, a guanine nucleotide exchange factor (GEF) for Rho1. At least with regard to Wsc1, this interaction is mediated by the Rom2 N-terminal domain. This domain is distinct from the Rho1-interacting domain, suggesting that the GEF can interact simultaneously with a sensor and with Rho1. We also demonstrate that extracts from wsc1 and mid2 mutants are deficient in the ability to catalyze GTP loading of Rho1 in vitro, providing evidence that the function of the sensor-Rom2 interaction is to stimulate nucleotide exchange toward this G-protein. In a related line of investigation, we identified the PMT2 gene in a genetic screen for mutations that confer an additive cell lysis defect with a wsc1 null allele. Pmt2 is a member of a six-protein family in yeast that catalyzes the first step in O mannosylation of target proteins. We demonstrate that Mid2 is not mannosylated in a pmt2 mutant and that this modification is important for signaling by Mid2.
The cell wall of the budding yeast Saccharomyces cerevisiae is required to maintain cell shape and integrity (3, 23). The cell must remodel this rigid structure during vegetative growth and during pheromone-induced morphogenesis. Wall remodeling is monitored and regulated by the cell integrity signaling pathway controlled by the Rho1 GTPase. Two essential functions have been identified for Rho1. First, it serves as an integral regulatory subunit of the 1,3-β-glucan synthase (GS) complex that stimulates GS activity in a GTP-dependent manner (7, 36). A pair of closely related genes, FKS1 and FKS2, encode alternative catalytic subunits of the GS complex (6, 13, 32, 41) that are the presumed targets of Rho1 activity.
A second essential function of Rho1 is to bind and activate protein kinase C (20, 33), which is encoded by PKC1 (29, 47). Loss of PKC1 function, or of any of the components of the mitogen-activated protein kinase (MAPK) cascade under its control (28), results in a cell lysis defect that is attributable to a deficiency in cell wall construction (26, 27, 36). The MAPK cascade is a linear pathway comprising a MEKK (BCK1 4, 25), a pair of redundant MEKs (MKK1/2 14), and a MAPK (MPK1 24). One of the consequences of signaling through the MAPK cascade is the activation of the serum response factor-like transcription factor Rlm1 (48). Signaling through Rlm1 regulates the expression of at least 25 genes, most of which have been implicated in cell wall biogenesis (18).
Cell wall integrity signaling is induced in response to several environmental stimuli. First, signaling is activated persistently in response to growth at elevated temperatures (e.g., 37 to 39°C 19), consistent with the finding that null mutants in many of the pathway components display cell lysis defects only when cultivated at high temperature. Second, hypo-osmotic shock induces a rapid, but transient activation of signaling (5, 19). Third, treatment with mating pheromone stimulates signaling at a time that is coincident with the onset of morphogenesis (1a, 8). Indeed, mutants defective in cell integrity signaling undergo cell lysis during pheromone-induced morphogenesis (8). Finally, agents that cause cell wall stress, such as the chitin antagonist calcofluor white, also activate signaling (21).
The mechanism by which information regarding the state of the cell wall is transmitted to the intracellular signaling apparatus remains an open question. However, several regulators of Rho1 activity have been identified. Rom1 and Rom2 comprise a redundant pair of guanine nucleotide exchange factors (GEFs) for Rho1 (35). Additionally, Bem2 and Sac7 are GTPase-activating proteins (GAPs) for Rho1 (22, 37, 43). Two major cell surface sensors for the activation of cell integrity signaling have also been described. One of these, known variously as Wsc1 (46), Hcs77 (9), and Slg1 (16), is dedicated to signaling wall stress during vegetative growth (40). The primary function of the other, called Mid2, is to signal wall stress that results from pheromone-induced morphogenesis (40). Null mutations in WSC1 and MID2, in combination, result in a severe cell lysis defect, indicating that the functions of these genes are partially redundant for vegetative growth (21, 40).
Both Wsc1 and Mid2 are transmembrane proteins that reside in the plasma membrane (21, 30, 40, 46). Their overall structures are similar in that they possess small cytoplasmic domains, each has a single transmembrane region, and their extracellular domains are rich in Ser/Thr residues. These Ser/Thr-rich regions are highly O mannosylated (21, 30, 40), probably resulting in extension and stiffening of the polypeptide. Therefore, the extracellular domains of Wsc1 and Mid2 have been proposed to act as rigid probes of the extracellular matrix (40). Despite the broad similarity between the proteins, their cytoplasmic domains do not appear to be structurally related. Here we show that the cytoplasmic domains of both Wsc1 and Mid2 interact with the Rom2 GEF and that this interaction stimulates GTP loading of Rho1. We also provide evidence for the importance of O mannosylation of Mid2 for signaling, a modification that we demonstrate is specifically catalyzed by the PMT2-encoded protein mannosyl transferase.
MATERIALS AND METHODS
Strains, growth conditions, and transformations.
The S. cerevisiae strains used in this study are listed in Table 1. Yeast cultures were grown in YEPD (1% Bacto yeast extract, 2% Bacto Peptone, 2% glucose). Synthetic minimal (SD) medium (42) supplemented with the appropriate nutrients was used to select for plasmid maintenance and gene replacement. Yeast transformations were carried out by the lithium acetate method (15). Escherichia coli DH5α was used to propagate all plasmids. E. coli cells were cultured in Luria broth medium (1% Bacto Tryptone, 0.5% Bacto yeast extract, 1% NaCl) and transformed by standard methods.
TABLE 1.
S. cerevisiae strains used
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| 1783 | MATaa | I. Herskowitz |
| 1784 | MATαa | I. Herskowitz |
| 1788 | MATa/MATαa | I. Herskowitz |
| DL251 | MATa/MATα bck1Δ::URA3/bck1Δ::URA3a | 25 |
| DL376 | MATa pkc1Δ::LEU2a | 26 |
| DL1985 | MATa wsc1Δ::LEU2a | 40 |
| DL1986 | MATα wsc1Δ::LEU2a | 40 |
| DL1987 | MATa/MATα wsc1Δ::LEU2/wsc1Δ::LEU2a | 40 |
| DL2069 | MATa/MATα rom2Δ::URA3/rom2Δ::URA3a | This study |
| DL2276 | SFY526b | Clontech Laboratories |
| DL2278 | MATa mid2Δ::URA3a | 40 |
| DL2282 | MATa mid2Δ::URA3 wsc1Δ::LEU2a | 40 |
| DL2393 | MATa cdc28-13 (D13au)c | 40 |
| DL2394 | MATa/MATα mid2Δ::URA3/mid2Δ::URA3a | 40 |
| DL2407 | MATad | W. Tanner |
| DL2408 | MATa pmt2Δ::LEU2d | W. Tanner |
| DL2435 | MATa cdc28-13 mid2Δ::URA3c | 40 |
| DL2440 | MATa cdc28-13 pmt2Δ::LEU2c | This study |
| DL2468 | MATa pmt2Δ::LEU2a | This study |
| DL2550 | MATa wsc1Δ::LEU2 pmt2-1a | This study |
EG123 background: leu2-3, 112trp1-1 ura3-52 his4 can1.
MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3, 112 Canr gal4-542 gal80-538 URA3::GAL1-lacZ.
D13au background: ade1 his2 leu2-3, 112 trp1-1a ura3ΔNS.
SEY6211 background: leu2-3, 112 ura3-52 his3-Δ200 ade2-101 trp1-Δ901 suc2-Δ9.
Synthetic cell lysis screen and isolation of PMT2.
S. cerevisiae strain DL1985 (wsc1Δ) was grown in YEPD to an optical density at 600 nm of 0.6, washed, and resuspended in 0.9% KCl to a density of 3 × 107 cells/ml. The cells were then irradiated for 30 s with a 254-nm UV lamp (0.5 J m−2 s−1), which resulted in an approximately 90% loss in plating efficiency. The irradiated cells were harvested, resuspended in YEPD with 10% sorbitol, and grown overnight in the dark at 30°C. They were then plated onto YEPD with 10% sorbitol plates (approximately 100 colonies/plate) and placed at room temperature for 2 to 3 days. The colonies were then replica plated onto YEPD without sorbitol and incubated at 34°C for 2 days. Colonies that exhibited a cell lysis phenotype at the higher temperature, as assessed by microscopic examination, were chosen for further analysis. Nine candidates were transformed with WSC1 on centromeric vector pRS316. Synthetic lysis candidates were selected as those that were viable on YEPD at 34°C when WSC1 was expressed from centromeric plasmid pRS316 (44).
To identify the gene responsible for the wsc1Δ-additive defect of one candidate (DL2550), this strain was transformed with a library of genomic yeast DNA in centromeric plasmid, pRS314 (25). Transformants were grown at room temperature and replicated onto YEPD at 34°C. Plasmids were rescued from colonies arising at the nonpermissive temperature. One clone possessed a 3.5-kb insert that contained one complete open reading frame (ORF) (PMT2) and 699 bp representing the 3′ end of another (LTE2). Deletion analysis of this clone revealed that the PMT2 gene was sufficient for suppression.
Wild-type PMT2 and pmt2-1 were amplified by PCR from DL1985 and DL2550 genomic DNA, respectively, with 378 bp of 5′ flanking sequence. The PCR products were inserted blunt ended into the SmaI site of centromeric vector pRS314 (44). The complete DNA sequences of the inserts were determined. Sequence analysis was performed by the Johns Hopkins University Biosynthesis and Sequencing Facility with oligonucleotides synthesized by the Biochemistry Core Facility. PCR was performed with Pfu polymerase (Stratagene). Both PMT2 and pmt2-1 were transformed into DL2550 to assess complementation.
Genomic deletions of PMT2.
For construction of the pmt2Δ allele, plasmid pBDis (pmt2Δ::LEU2 in pUC19) (provided by W. Tanner) was digested with Apa1 and Spe1, and the resultant 2.4-kb fragment was purified and used to transform 1783 and DL2393 to leucine prototrophy. Deletion of PMT2 in Leu+ transformants was confirmed by PCR in both strains.
Construction of mid2 C-terminal and S/T deletion mutants.
The mid2 C-terminal deletion mutant was constructed by PCR amplification of the first 768 bp of the MID2 ORF with 632 bp of 5′ noncoding sequence. This fragment was inserted blunt ended into the Sma1 site of pRS315[GFPuv] (42) to generate a fusion protein with green fluorescent protein (GFP) at the C terminus. The S/T mutant was constructed by the PCR overlap extension method (11). Two separate PCRs were used to amplify the MID2 ORF 5′ and 3′ of the S/T-rich region (amino acids [aa] 31 to 172). To amplify the first 90 bp of the MID2 ORF with 632 bp 5′ to the translation start site, a left junction primer was used with a primer that lies 5′ to the coding sequence. The coding sequence 3′ of the S/T-encoded region was amplified through the final coding base by using a right junction primer that contained a 15-bp complementary flanking region to the left junction primer and a primer that lies at the 3′ end of the coding sequence. This generated overlapping 5′ and 3′ regions of MID2 without the S/T-encoded region. The products of these reactions were combined in a third reaction to generate the mid2 amino acids 31 to 172 (aa 31–172)Δ allele, which was then inserted blunt ended into the Sma1 site of pRS315[GFPuv]. Both deletion alleles were confirmed by DNA sequence analysis.
Two-hybrid plasmids and assays.
The sequences encoding the C-terminal tails of WSC1 (aa 298 to 378) and MID2 (aa 252 to 376) were PCR amplified. The amplified fragments were cloned in frame into two-hybrid vectors pGAD424 and pGBT9 (Clontech Laboratories, Inc.) via the BamHI-PstI sites. The ROM2 coding region was amplified and cloned in-frame into the two-hybrid vectors via the BamHI-SmaI sites. The RHO1 wild-type, RHO1G22A, and RHO1Q68L coding regions were PCR amplified without their terminal CAAX box motifs from vectors carrying the respective alleles (provided by Y. Takai) and cloned into pGAD424 and pGBT9 via the BamHI-PstI sites. The various ROM2 domain fusions were constructed by PCR amplification of the respective regions with a genomic ROM2 clone (gift of Mike Hall), and the fragments were cloned via the BamHI-PstI sites into pGAD424, except for the ROM2 (aa 1 to 660) fusion, which was cloned via BamHI-SmaI. All fusions were confirmed by DNA sequence analysis. Primers are available upon request.
Yeast strain SFY526 (Clontech Laboratories, Inc.) was cotransformed with pGAD424 expressing a Gal4-activation domain (ADGal4) fusion protein and pGBT9 expressing a Gal4 DNA-binding domain (DBDGal4) fusion protein. The resultant transformants were patched onto nitrocellulose filters and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (1 mg/ml) for β-galactosidase activity. The filters were developed at 30°C for 6 to 8 h.
Pheromone-induced killing.
To test for sensitivity to killing by α-factor, MATa strains were grown in synthetic minimal medium containing a limiting amount of calcium (100 μM CaCl2 12) for 24 h, subcultured in the same medium, and grown to an A600 of 0.5 to 1.0 (5 × 106 to 1 × 107). Cells were then treated with α-factor (8 μg/ml; Sigma), and viability was measured over time by plating onto YEPD.
Immunodetection of Mid2HA and Wsc1HA.
Transformants of yeast strains DL2407 and DL2408 expressing either Mid2-hemagglutinin (HA) (Mid2HA) or Wsc1-HA (Wsc1HA) from YEp352 (40) were grown in YEPD to an A600 of approximately 1.0. Cells were harvested from 50 ml of culture medium, washed with water, and resuspended in 400 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 50 mM KF, 1% Triton X-100) containing protease inhibitors (20 μg of leupeptin, 20 μg of benzamidine, 10 μg of pepstatin, and 40 μg of aprotinin per ml, plus 1 mM phenylmethylsulfonyl fluoride). Cells were broken by vigorous vortexing for 5 min at 4°C in the presence of an equivalent volume of glass beads (0.3 mm in diameter). The beads and cell debris were cleared by centrifugation for 15 min at 13,000 × g. The supernatant was removed and stored in 33% glycerol. The protein concentration was determined by the bicinchoninic acid protein assay reagent (Pierce). Crude extract (50 μg) was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on an 8% polyacrylamide gel followed by immunoblotting as described previously (19).
Activation of Mpk1HA by pheromone treatment.
Yeast strains DL2436, DL2437, and DL2440 expressing Mpk1-HA (Mpk1HA) from pRS424 (19) were tested for α-factor-induced activation of Mpk1HA as described previously (1a, 40). Quantitation of incorporation of 32P into myelin basic protein by Mpk1HA was measured with a Fuji phosphorimager.
GEF assays.
The GDP-GTP exchange activity of various strains towards Rho1 was assayed by measuring the binding of [35S]GTPγS to HA-Rho1 (Rho1HA) as described by Schmidt et al. (43) with the following modifications. Strains were grown in either YEPD or YEPD supplemented with 10% sorbitol (where indicated) at 30°C for extract preparation. Whole-cell extracts from various strains were prepared by resuspending cells in extraction buffer (20 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 2.5 mM EDTA, 1 mM dithiothreitol, protease inhibitors), followed by lysis by vortexing for 4 min with glass beads. The cell debris was cleared by centrifugation at 13,000 × g for 5 min. Whole-cell extracts from cells expressing Rho1HA from pRS424 were prepared by resuspending cells in Rho1 extraction buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 0.1 mM EDTA, 0.1% NP-40, 10% glycerol, and protease inhibitors), followed by lysis with glass beads and centrifugation, as described above. Rho1HA was immunoprecipitated from 200 μg of whole-cell extract as described previously (43). The immune complex was resuspended in 50 μl of extraction buffer and incubated with 1 μM [35S]GTPγS–0.75 mM l-α-dimyristoyl phosphatidylcholine in the presence of 30 μg of whole-cell extract from various yeast strains at 25°C. The reaction was stopped by adding 1 ml of ice-cold stop buffer (20 mM Tris-HCl [pH 7.5], 25 mM MgCl2, 100 mM NaCl) at various time points between 1 and 5 min. The mixture was filtered through Whatman GF/C filters by using a vacuum manifold and washed three times with cold stop buffer. The [35S]GTPγS trapped on the filters was measured by liquid scintillation. The GEF activity on Rho1HA was calculated by subtracting the radioactive counts from a control reaction in which immunoprecipitated Rho1HA was mock treated with extraction buffer only. The GEF activity towards Rho1HA in the various mutant extracts was expressed as a percentage of the activity in the wild-type extract by using time points that were within the linear range for nucleotide exchange.
RESULTS
The cytoplasmic tails of Mid2 and Wsc1 interact with Rom2.
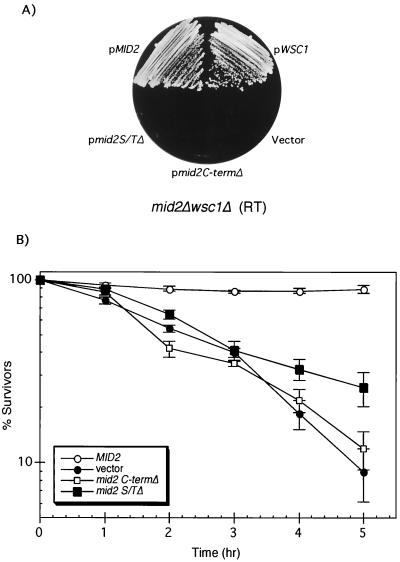
Mid2 and Wsc1 (also known as Hcs77) have been proposed to act as sensors of cell wall stress (9, 21, 40, 46). One criterion for classification of a transmembrane protein as a sensor is the importance of both the extracellular and cytoplasmic domains for function. This has been demonstrated for Wsc1 (30), but not for Mid2. Therefore, we constructed a pair of mid2 mutants with deletions of either the N-terminal Ser/Thr-rich extracellular domain (aa 31 to 172) or the C-terminal cytoplasmic tail (aa 257 to 376). Both forms were fused at their C termini to GFP, cloned into a centromeric plasmid, and expressed under the control of the MID2 promoter to assess function. Although both mutant forms localized normally to the cell periphery (not shown), neither was capable of suppressing the cell lysis defect of a mid2Δ wsc1Δ strain, in contrast to full-length Mid2::GFP (Fig. 1A). Moreover, these mid2 alleles were deficient in complementing the pheromone-induced death of a mid2Δ mutant (Fig. 1B), confirming the requirement of both the cytoplasmic and extracellular domains for Mid2 function.
FIG. 1.
The S/T-rich and C-terminal (C-term) domains of Mid2 are essential for its function. (A) Transformants of a mid2Δ wsc1Δ mutant (DL2282) expressing centromeric plasmids pRS314[MID2::GFP], pRS314 [mid2S/TΔ::GFP], pRS314[mid2CΔ:GFP], pRS314[WSC1], and pRS314-GFP (vector) were streaked onto YEPD and incubated at room temperature (RT) for 3 days. (B) A mid2Δ strain (DL2278) was transformed with pRS314[MID2], pRS314[mid2S/TΔ], pRS314[mid2CΔ], or pRS314 (vector) and grown in SD with limiting calcium (100 μM CaCl2) at 30°C to an A600 of approximately 0.6. α-Factor (8 μg/ml) was added to the cultures, and samples were tested for viability at the indicated times by plating onto YEPD. Plates were scored after 2 days at room temperature. The results shown are the mean and standard deviation from three independent experiments.
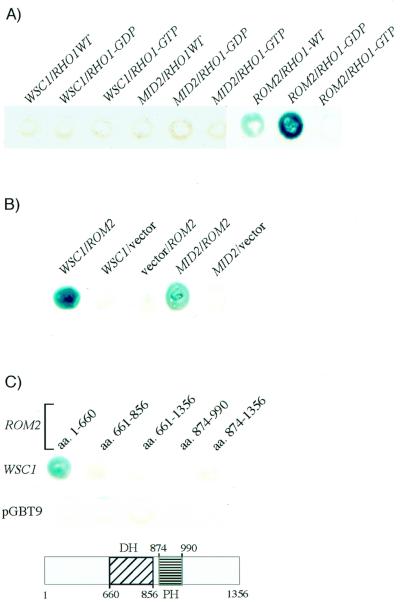
If Mid2 and Wsc1 act as sensors of cell wall stress, they should signal to one or more of the intracellular components of the cell wall integrity signaling pathway. The two most likely candidates for interaction with the cell surface sensors are the Rho1 GTPase and its GEFs, Rom1 and Rom2, because they reside at the plasma membrane (31, 38). Therefore, we tested Mid2 and Wsc1 for interaction with Rho1 and Rom2 by two-hybrid analysis. The sequences encoding the cytoplasmic domains of Wsc1 (aa 298 to 378) and Mid2 (aa 252 to 376) were fused to the Gal4 activation domain (ADGal4) of a two-hybrid vector (pGAD424; Clontech). Three forms of Rho1 (35) were fused to the DBDGal4 of two-hybrid vector pGBT9 (Clontech): wild type, GDP bound (rho1-G22A), and GTP bound (RHO1-Q68L). Figure 2A shows that neither Wsc1 nor Mid2 could interact with any form of Rho1. As expected, ADGal4-Rom2 interacted strongly with the GDP-bound form of Rho1, less well with wild-type Rho1, and not at all with the GTP-bound form.
FIG. 2.
Wsc1 and Mid2 interact with Rom2, a GEF for Rho1. (A) Yeast strain SFY526 expressing pGBT9[WSC1], pGBT9[MID2], or pGBT9[ROM2] was transformed with either pGAD424[RHO1] (wild type [WT]), pGAD424[rho1-Q68L] (GDP bound), or pGAD424[rho1-G22A] (GTP bound). The resultant transformants were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside for β-galactosidase activity. (B) SFY526 transformants expressing pGBT9[WSC1], pGBT9[MID2], or pGBT9 (vector) were transformed with either pGAD424[ROM2] or pGAD424 (vector). The resultant transformants were tested for β-galactosidase activity. (C) Segments of the ROM2 ORF, corresponding to recognized domains, were cloned into pGAD424. SFY526 expressing pGBT9[WSC1] or pGBT9 was transformed with the various domain fusions of ROM2 and tested for β-galactosidase activity. A schematic of the Rom2 protein delineating its various domains is shown. PH, pleckstrin homology domain; DH, Dbl homology domain.
We next tested the Wsc1 and Mid2 cytoplasmic domain fusions for interaction with DBDGal4-Rom2. We chose Rom2 rather than Rom1 for these experiments because ROM2 appears to be functionally more important than ROM1 (35). Figure 2B shows that both of these putative sensors interact with Rom2, the Wsc1-Rom2 interaction being stronger than that of Mid2-Rom2. Rom2 is a large protein with several functional domains. To determine which domain of Rom2 interacts with Wsc1, several additional fusions were constructed and tested for interaction (Fig. 2C). Among these, only a fusion of the N-terminal 660 aa displayed interaction with Wsc1. Unfortunately, the interaction between the cytoplasmic domain of Mid2 and Rom2 was too weak to dissect.
Wsc1 and Mid2 regulate guanine nucleotide exchange toward Rho1.
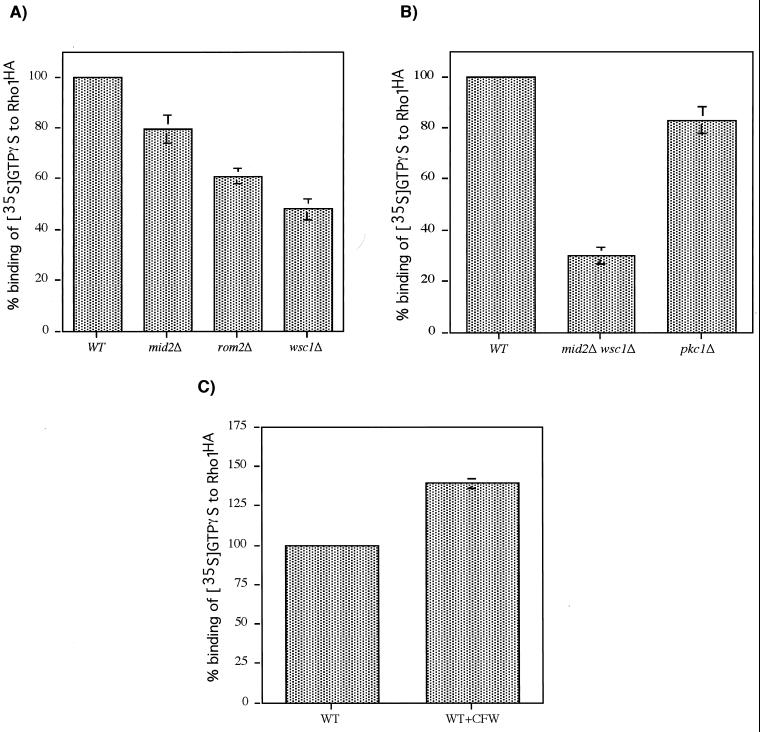
To determine the function of the interaction of the cell surface sensors with Rom2, we examined the effect of their absence on Rom1 or -2-catalyzed guanine nucleotide exchange activity toward Rho1. The rate at which crude extracts from various mutants catalyzed loading of [35S]GTPγS onto immunoprecipitated Rho1-HA was measured. Figure 3A shows that extract from a mid2Δ mutant was only slightly impaired for exchange activity, whereas a wsc1Δ mutant retained approximately 50% of wild-type activity. These results are consistent with the relatively minor role that Mid2 plays in vegetative growth. A rom2Δ mutant also retained approximately 60% exchange activity, presumably reflecting Rom1 activity. A mid2Δ wsc1Δ mutant, which was cultivated in the presence of 1 M sorbitol for osmotic support, retained approximately 30% of wild-type activity (Fig. 3B). As a control, we tested extract from a pkc1Δ strain, which also requires osmotic support. Pkc1 is critical for cell wall integrity signaling, but acts downstream of Rho1. As expected, the pkc1Δ extract displayed exchange activity that was only slightly diminished compared to that of the wild type (Fig. 3B). Therefore, the role of Mid2 and Wsc1 in cell wall integrity signaling is to activate Rom1 or -2-catalyzed guanine nucleotide exchange toward Rho1.
FIG. 3.
Wsc1 and Mid2 regulate guanine nucleotide exchange activity toward Rho1HA. (A) The ability of crude extracts from wild-type (WT [1788]), mid2Δ (DL2394), rom2Δ (DL2069), and wsc1Δ (DL1987) strains to catalyze loading of [35S]GTPγS onto immunoprecipitated Rho1HA was measured with a filter binding assay (see Materials and Methods). Rho1HA was immunoprecipitated from wild-type (1788) cells overexpressing pRS424[RHO1HA] by using the 12CA5 antibody. The rate of [35S]GTPγS loading onto Rho1HA by the mutant extracts was expressed as a percentage of the rate catalyzed by the wild-type extract. All strains were grown in YEPD. (B) The ability of crude extracts from wild type (WT; 1783), mid2Δwsc1Δ (DL2282), and pkc1Δ (DL376) strains to catalyze loading of [35S]GTPγS onto immunoprecipitated Rho1HA was measured. All strains were grown in YEPD supplemented with 10% sorbitol for osmotic support. (C) The effect of in vivo-generated cell wall stress on in vitro-catalyzed guanine nucleotide exchange on immunoprecipitated Rho1HA was measured. Wall stress was induced in wild-type (1783) cells grown in YEPD at 23°C by treatment for 90 min with 40 μg of calcofluor white per ml.
We next examined the effect of cell wall stress generated in vivo on Rom-catalyzed nucleotide exchange activity in vitro. Wall stress was induced by a 90-min treatment with the chitin antagonist calcofluor white (at 40 μg/ml). Figure 3C shows that this treatment, which strongly activates signaling to Mpk1 (data not shown), increased guanine nucleotide exchange activity towards Rho1 by approximately 40% as compared with unstressed cells growing at 23°C. This increase is not the result of increased expression of Rom1 or Rom2 (not shown) and presumably reflects a change in the sensor-exchange factor complex that is preserved in extracts.
Isolation of mutants that display additive cell lysis defects with wsc1Δ.
To identify additional components of the cell wall integrity signaling pathway that may be important for signaling from the cell surface, we isolated mutants that display a cell lysis defect in combination with a wsc1Δ mutation. A haploid wsc1Δ strain (DL1985), which only displays a cell lysis defect when cultivated at 39°C (not shown), was mutagenized with UV light (see Materials and Methods). Mutagenized cells were cultivated in the presence of 10% sorbitol for osmotic support and replicate plated at 34°C in the absence of sorbitol to identify mutants for which this temperature was restrictive for growth. Microscopic examination of candidates revealed those that underwent cell lysis at the restrictive temperature. Cell lysis mutants identified from this screen were transformed with the centromeric plasmid pRS316[WSC1] to identify those whose defects were dependent on wsc1Δ. Five cell lysis mutants whose defects are additive with wsc1Δ were identified from this screen. One of these (DL2550) is discussed below.
The PMT2-encoded protein mannosyl transferase contributes to cell wall integrity signaling by modification of Mid2.
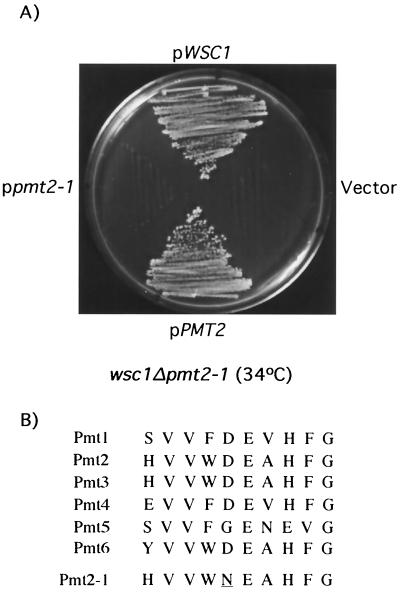
A backcross of DL2550 to wsc1Δ (DL1986) was first conducted to confirm that the wsc1Δ-additive cell lysis defect of this mutant strain segregated as a single mutation (not shown). To isolate the gene responsible for the wsc1Δ-additive defect, this mutant was transformed with a genomic yeast library in the centromeric vector pRS314. Plasmids that suppressed the cell lysis defect of DL2550 at 34°C were rescued and subjected to DNA sequence and deletion analysis. Among seven suppressing plasmids isolated, three harbored WSC1, two contained MID2, and two carried the PMT2 gene, a member of a six-gene family that encode protein mannosyl transferases (45). We reported previously that MID2 expressed from a centromeric plasmid is capable of suppressing the cell lysis defect of a wsc1Δ mutant (40). Although additional copies of either MID2 or PMT2 suppressed the growth defect of DL2550 at 34°C, only PMT2 was capable of suppression at 37°C (not shown). Because the parental wsc1Δ strain is unimpaired for growth at 37°C, we concluded that DL2550 did not carry a debilitating mutation in MID2. Therefore, PMT2 was isolated by PCR from DL2550 and from its isogenic wild-type strain (EG123) and cloned into the centromeric vector pRS316. The PMT2 gene from wild-type cells complemented the cell lysis defect of DL2550 (Fig. 4A). In contrast, the PMT2 allele isolated from DL2550 failed to complement this mutant. Therefore, we concluded that a mutation in PMT2 (designated pmt2-1) was responsible for the defect in DL2550. DNA sequence analysis of pmt2-1 revealed that it contains a single base change within the coding sequence that converts Asp92 to Asn. This change is in a region that is highly conserved among the yeast Pmt isoforms (Fig. 4B). A double wsc1Δ pmt2Δ mutant was constructed and found to be viable only in the presence of osmotic support (i.e., 10% sorbitol [data not shown]), indicating that pmt2-1 retains partial function. In contrast, a mid2Δ pmt2Δ mutant was viable, even at 37°C (data not shown). This result suggests that Pmt2 functions in parallel with Wsc1, but on the same pathway as Mid2.
FIG. 4.
wsc1Δ and pmt2-1 mutations exhibit an additive cell lysis phenotype. (A) Yeast strain DL2550 (wsc1Δ pmt2-1) was transformed with centromeric plasmids pRS314[WSC1], pRS314[PMT2], pRS314[pmt2-1], and pRS314 (vector). Transformants were streaked onto YEPD plates for 2 days at 34°C. (B) Amino acid sequence alignment of the six yeast Pmt isoforms through the region surrounding the pmt2-1 mutation, which results in replacement of Asp92 with Asn (underlined).
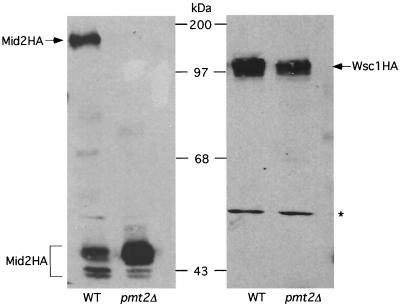
The Pmt enzymes are responsible for the first step in O-linked protein glycosylation in yeast. This involves attachment of a mannose to the side-chain hydroxyl group of seryl and threonyl residues in target proteins (45). There appears to be some functional overlap among the various Pmt isoforms, because loss of function of any one PMT gene does not result in apparent phenotypic defects, whereas some combinations of pmt mutations are deleterious (45). We demonstrated previously that both Wsc1 and Mid2 are O mannosylated on their extracellular domains (40). Additionally, Ketela et al. (21) showed that a pmt1Δ pmt2Δ mutant was defective for modification of Mid2, but did not report results obtained with single pmt mutants. Therefore, we examined the possibility that Pmt2 function is required in a wsc1Δ mutant because this enzyme is solely responsible for modification of Mid2. We expressed a fully functional, C-terminally epitope-tagged form of Mid2 (Mid2HA 40) in a pmt2Δ mutant and its isogenic wild-type strain. Figure 5 (left panel) shows that Mid2HA isolated from pmt2Δ cells migrates on SDS-PAGE with an apparent molecular mass of approximately 45 kDa, very close to its predicted mass. In contrast, the fully modified form of this protein, isolated from wild-type cells, migrates with a much larger apparent molecular mass (160 to 180 kDa 40) (Fig. 5). This form of Mid2HA was not detected in pmt2Δ cells, indicating that Pmt2 is the only protein mannosyl transferase isoform capable of modifying Mid2. Consistent with this interpretation, pmt1, -3, or -4 deletion mutants modified Mid2HA normally (not shown). In contrast to these results, a similarly epitope-tagged form of Wsc1 (Wsc1HA 40) was fully modified in a pmt2Δ mutant (Fig. 5, right panel).
FIG. 5.
Mid2 is a specific substrate of the Pmt2 protein O-d-mannosyl transferase. Extracts from wild-type (WT [1783]) or pmt2Δ (DL2468) cells expressing either Mid2HA (left panel) or Wsc1HA (right panel) from high-copy plasmid YEp352 were subjected to SDS-PAGE analysis followed by immunoblot analysis with the 12CA5 antibody. The asterisk denotes a protein that cross-reacts with this antibody. Molecular mass markers are indicated in kilodaltons.
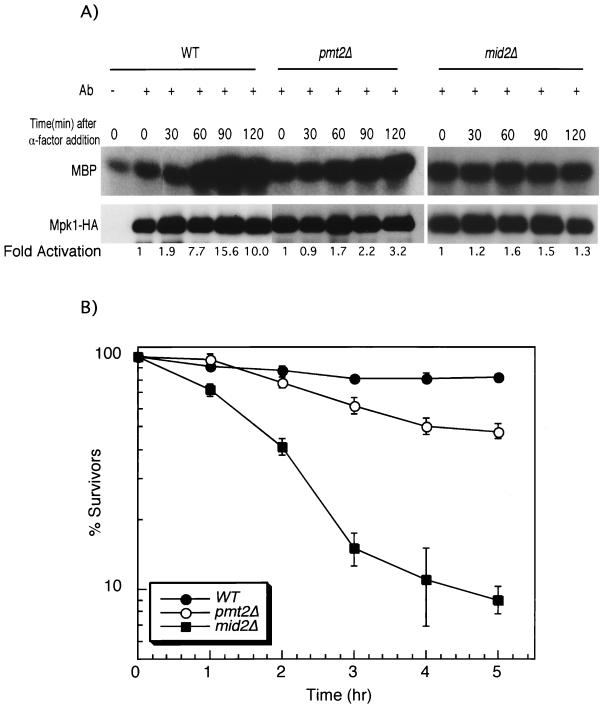
Mid2 is required for activation of the Mpk1 MAPK in response to mating pheromone treatment (21, 40). To determine if Pmt2-catalyzed modification of Mid2 is important for signaling by this sensor, we measured pheromone-induced activation of Mpk1HA in a pmt2Δ strain. Indeed, Mpk1HA activation was compromised in the pmt2Δ mutant (Fig. 6A). However, in contrast to a mid2Δ mutant, the pmt2Δ mutant retained some ability to activate Mpk1HA. A GFP-tagged form of Mid2 (40) localized normally to the cell surface in a pmt2Δ mutant (not shown), indicating that the deficiency in signaling observed in this mutant is not attributable to mislocalization of Mid2.
FIG. 6.
(A) PMT2 is important for pheromone-induced activation of Mpk1. Cultures of cdc28-13 (wild type [WT]; DL2393), cdc28-13 mid2Δ (DL2435), and cdc28-13 pmt2Δ (DL2440) strains expressing Mpk1HA from pRS425 were synchronized by arrest in G1 at 37°C for 90 min, followed by treatment with 50 nM α-factor. Mpk1HA was immunoprecipitated (with the 12CA5 antibody) from extracts of cultures taken at the indicated time points. Protein kinase assays were conducted with myelin basic protein (MBP) as the substrate. The lower panel represents an immunoblot of Mpk1HA immunoprecipitates. Mpk1 is not activated in this strain background by temperature upshift (1a). (B) PMT2 is important for survival of α-factor treatment. Wild-type (1783), pmt2Δ (DL2468), and mid2Δ (DL2278) strains were grown in SD medium with limiting calcium (100 μM CaCl2) at 30°C to an A600 of approximately 0.6. α-Factor (8 μg/ml) was added to the cultures, and samples were tested for viability at the indicated times by plating onto YEPD. Plates were scored after 2 days at room temperature. The results shown are the mean and standard deviation from three independent experiments.
Mid2 function is required for survival of cells treated with mating pheromone (12). Therefore, as a further test of the importance of O mannosylation for Mid2 function, we examined a pmt2Δ mutant for its ability to survive treatment with α-factor. Figure 6B shows that a MATa pmt2Δ strain displayed some sensitivity to pheromone, but not as much as an isogenic mid2Δ mutant, consistent with the observed retention of some signaling capability in the pmt2Δ mutant. We conclude from these experiments that O mannosylation of the extracellular domain of Mid2 is important for signaling by this molecule in response to cell wall stress generated either during vegetative growth or by pheromone-induced morphogenesis.
DISCUSSION
The Wsc1 and Mid2 cell surface sensors interact with and regulate the Rom2 GEF for the Rho1 GTPase.
Wsc1 and Mid2 act in parallel to stimulate cell wall integrity signaling. In the absence of one of these cell surface proteins, the other is essential. Although their C-terminal intracellular domains are not similar at the primary sequence level, their genetic interactions suggested that these sensors share common intracellular targets (40). In this study, we demonstrated by two-hybrid analysis that the intracellular domains of both Wsc1 and Mid2 interact with Rom2, a GEF for Rho1. Additionally, at least Wsc1 interacts specifically with the N-terminal domain of Rom2 (aa 1 to 660), which is not conserved among other known guanine nucleotide exchange proteins. The conserved DBL-homologous domain (aa 661 to 856) interacts with Rho1 and possesses the nucleotide exchange activity of this protein (35). Therefore, it seems likely that Rom2 (and probably Rom1) can interact simultaneously with a sensor and with Rho1.
The results of in vitro guanine nucleotide exchange experiments indicate that the function of the interaction between the sensors and Rom2 is to stimulate nucleotide exchange on Rho1. Specifically, we found that extract from a wsc1Δ mid2Δ mutant was severely impaired in its ability to catalyze loading of GTPγS onto Rho1, retaining only about 30% of the wild-type level of activity. The residual activity may be due to the presence of the minor sensors Wsc2 and Wsc3 in these extracts.
There are other examples in which regulation of a small G-protein is mediated by its GEF. The best studied of these is the mammalian Ras GEF known as mSos1 (39), which is recruited to the plasma membrane by an adapter protein (Grb2) in response to receptor activation. In this way, it is thought that mSos1 is targeted to its effector Ras, which resides at the membrane. The neuronal Ras-GEF p140 Ras-GRF is activated in response to elevated calcium levels through its N-terminal domain (1). This involves association with membranes and as-yet-unidentified cellular components that are required for calcium-induced activation. In yeast, activation of the Rho-type GTPase Cdc42 in response to pheromone treatment is mediated by the Cdc24 GEF (49). This stimulation requires an interaction between Cdc24 and the βγ subunit of the trimeric G-protein regulated by the pheromone receptors. Finally, the N-terminal domain of the yeast Ras-GEF Cdc25 may promote homodimerization as a means of regulating its activity (2). Our finding that Rom2 activation requires Wsc1 or Mid2 suggests that this GEF may be regulated by direct interaction with a transmembrane receptor. Alternatively, an unknown adapter protein may mediate this interaction.
O mannosylation of the extracellular domain of Mid2 is catalyzed specifically by Pmt2 and is important for signaling.
We identified a recessive point mutation in the PMT2 gene (pmt2-1) in a genetic screen for defect additivity with a wsc1Δ mutation. PMT2 is a member of a six-gene family that encodes protein mannosyl transferases (45). These enzymes catalyze the addition of the first of several mannosyl residues to the side-chain hydroxyl groups of seryl and threonyl residues in target proteins. The Pmts reside in the endoplasmic reticulum and possess seven membrane-spanning domains (45). The Asp92 residue of Pmt2 that was mutated in pmt2-1 resides in a loop between membrane domains 1 and 2, which is located on the lumenal face of the endoplasmic reticulum, which appears to be important for dimer formation (8a).
We and others have shown that Wsc1 and Mid2 are O mannosylated (21, 30, 40). Although loss of PMT2 function alone results in no apparent phenotypic defect (45), our finding that a pmt2 mutation exacerbates the cell lysis defect of a wsc1Δ mutation suggested that it might be critical for Mid2 modification. Analysis of the Mid2 protein in a pmt2Δ strain revealed that the protein mannosyl transferase encoded by PMT2 was indeed specifically required for Mid2 modification. Normal modification of Mid2 was observed in other pmtΔ mutants. In contrast, Wsc1 was modified normally in a pmt2Δ mutant. In fact, single deletion mutants in PMT1-6 all modified Wsc1 normally (B. Philip, unpublished data), suggesting redundancy in function with regard to this target.
Pmt1 and Pmt2 have been isolated together in complex and are thought to act as a heterodimer (45). However, our results indicate that Pmt2 can function normally in the absence of Pmt1, at least for modification of Mid2. Members of the Pmt family may form homodimers as well as heterodimers as a mechanism to enhance combinatorial substrate selectivity. Mid2 is the first example of a protein that is modified specifically by Pmt2 (45).
Three lines of evidence establish the importance of O mannosylation in Mid2 function. First, the additive cell lysis defect of a pmt2 mutation with a wsc1Δ mutation (but not with a mid2Δ mutation) suggested that unmodified Mid2 is impaired for signaling during vegetative growth. Second, a direct measure of Mid2 function is its ability to signal to the Mpk1 MAPK in response to pheromone-induced morphogenesis (40). We found that this signaling was deficient, but not completely defective in a pmt2Δ mutant. Third, Mid2 function is required for survival of cells treated with mating pheromone (12). We found that a pmt2Δ strain was sensitive to pheromone-induced death; however, it was not as sensitive as a mid2Δ strain. Thus, unmodified Mid2 was deficient for function by all known criteria.
The extracellular domains of both Wsc1 and Mid2 are very rich in seryl and threonyl residues. It is these domains that are O mannosylated (30, 40). In yeast, this modification consists of several (four or five) mannosyl residues in α-1,2 and α-1,3 linkages (45). When many such modifications are present in a stretch of seryl/threonyl residues, as appears to be the case for both Wsc1 and Mid2, they induce the polypeptide to adopt a stiff and extended conformation (17). We have proposed previously (40) that these cell surface proteins may function as molecular probes that span the periplasmic space to interact directly with the cell wall. The importance of O mannosylation for Mid2 function is consistent with this model.
Protein O mannosylation has been implicated previously in the maintenance of yeast cell wall integrity, but the nature of its involvement has been unclear. Many integral cell wall proteins are O mannosylated, and pmt2Δ pmt3Δ and pmt2Δ pmt4Δ mutants display osmotic-remedial cell lysis defects (45). Additionally, certain triple pmtΔ mutants (i.e., pmt1, -2, and -4Δ and pmt2, -3, and -4Δ) are inviable, but not rescued by osmolytes, suggesting severe cell wall defects. Our demonstration of the importance of O mannosylation for Mid2 function indicates a critical role for this modification in the transmission of cell wall stress signals and explains, at least in part, the involvement of the Pmts in the maintenance of cell wall integrity.
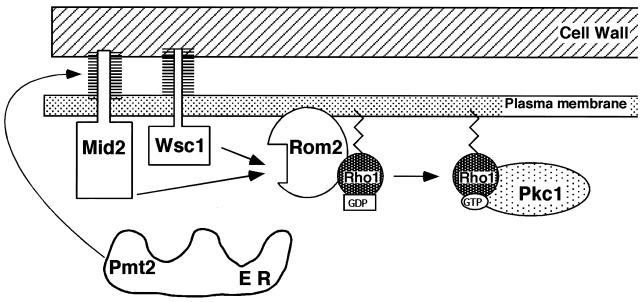
Figure 7 incorporates the observations made in this study into a model for Mid2 and Wsc1 function. When these sensors are activated by cell wall perturbations, perhaps by direct contact with the cell wall, they stimulate Rom2 (and presumably Rom1) activity towards Rho1. This promotes exchange of GDP for GTP, thereby activating Rho1 for signal transmission through Pkc1. The mechanism by which Mid2 and Wsc1 transmit wall perturbation signals to Rom1 and -2 remains obscure.
FIG. 7.
A model for the function of Wsc1 and Mid2 in the transmission of cell wall stress signals to Rho1. ER, endoplasmic reticulum.
ACKNOWLEDGMENTS
We thank Widmar Tanner for PMT plasmids, mutants, and valuable discussion; Yoshimi Takai for Rho1 mutant alleles; Mike Hall for a ROM2 plasmid; and Mathu Rajavel for MID2 reagents.
This work was supported by grants from the NIH (GM48533) and the American Cancer Society (Faculty Research Award 446) to D.E.L.
REFERENCES
- 1.Buchsbaum R, Telliez J-B, Goonesekera S, Feig L A. The N-terminal pleckstrin, coiled-coil, and IQ domains of the exchange factor Ras-GRF act cooperatively to facilitate activation by calcium. Mol Cell Biol. 1996;16:4888–4896. doi: 10.1128/mcb.16.9.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Buehrer B M, Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen R E, Michaeli T, Van Aelst L, Ballester R. A role for the noncatalytic N terminus in the function of Cdc25, a Saccharomyces cerevisiae Ras-guanine nucleotide exchange factor. Genetics. 2000;154:1473–1484. doi: 10.1093/genetics/154.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cid V J, Durán A, del Rey F, Snyder M P, Nombela C, Sánchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costigan C, Gehrung S, Snyder M. A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol. 1992;12:1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davenport K R, Sohaskey M, Kamada Y, Levin D E. A second osmosensing signal transduction pathway in yeast. J Biol Chem. 1995;270:30157–30161. doi: 10.1074/jbc.270.50.30157. [DOI] [PubMed] [Google Scholar]
- 6.Douglas C M, Foor F, Marrinan J A, Morin N, Nielsen J B, Dahl A M, Mazur P, Baginsky W, Li W, El-Sherbeini M, Clemas J A, Mandala S M, Frommer B R, Kurtz M B. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-D-glucan synthase. Proc Natl Acad Sci USA. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drgonova J, Drgon T, Tanaka K, Kollar R, Chen G-C, Ford R A, Chan C S M, Takai Y, Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- 8.Errede B, Cade R M, Yashar B M, Kamada Y, Levin D E, Irie K, Matsomoto K. Dynamics and organization of MAP kinase signal pathways. Mol Reprod Dev. 1995;42:477–485. doi: 10.1002/mrd.1080420416. [DOI] [PubMed] [Google Scholar]
- 8a.Girrbach V, Zeller T, Priesmeier M, Strahl-Bolsinger S. Structure-function analysis of the dolichyl phosphate-mannose: protein O-mannosyltransferase ScPmt1p. J Biol Chem. 2000;275:19288–19296. doi: 10.1074/jbc.M001771200. [DOI] [PubMed] [Google Scholar]
- 9.Gray J V, Ogas J P, Kamada Y, Stone M, Levin D E, Herskowitz I. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 1997;16:4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill J E, Muers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 11.Ho S-N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 12.Iida H, Nakamura H, Ono T, Okumura M S, Anraku Y. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and mating. Mol Cell Biol. 1994;14:8259–8271. doi: 10.1128/mcb.14.12.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue S B, Takewaki N, Takasuka T, Mio T, Adachi M, Fujii Y, Miyamoto C, Arisawa M, Furuichi Y, Watanabe T. Characterization and gene cloning of 1,3-β-D-glucan synthase from Saccharomyces cerevisiae. Eur J Biochem. 1995;231:845–854. doi: 10.1111/j.1432-1033.1995.tb20770.x. [DOI] [PubMed] [Google Scholar]
- 14.Irie K, Takase M, Lee K S, Levin D E, Araki H, Matsumoto K, Oshima Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol Cell Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacoby J J, Nilius S M, Heinisch J J. A screen for upstream components of the yeast protein kinase C signal transduction pathway identifies the product of the SLG1 gene. Mol Gen Genet. 1998;258:148–155. doi: 10.1007/s004380050717. [DOI] [PubMed] [Google Scholar]
- 17.Jentoft N. Why are proteins O-glycosylated? Trends Biochem Sci. 1990;15:291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 18.Jung U S, Levin D E. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signaling pathway. Mol Microbiol. 1999;34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- 19.Kamada Y, Jung U S, Piotrowski J, Levin D E. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 20.Kamada Y, Qadota H, Python C P, Anraku Y, Ohya Y, Levin D E. Activation of yeast protein kinase C by Rho1 GTPase. J Biol Chem. 1996;271:9193–9195. doi: 10.1074/jbc.271.16.9193. [DOI] [PubMed] [Google Scholar]
- 21.Ketela T, Green R, Bussey H. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J Bacteriol. 1999;181:3330–3340. doi: 10.1128/jb.181.11.3330-3340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y J, Francisco L, Chen G C, Marcotte E, Chan C S M. Control of cellular morphogenesis by the Ipl2/Bem2 GTP-activating protein: possible role of protein phosphorylation. J Cell Biol. 1994;127:1381–1394. doi: 10.1083/jcb.127.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klis F M. Review: cell wall assembly in yeast. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 24.Lee K S, Irie K, Gotoh Y, Watanabe Y, Araki H, Nishida E, Matsumoto K, Levin D E. A yeast mitogen-activated protein kinase homolog (Mpk1) mediates signalling by protein kinase C. Mol Cell Biol. 1993;13:3067–3075. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K S, Levin D E. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol Cell Biol. 1992;12:172–182. doi: 10.1128/mcb.12.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin D E, Bartlett-Heubusch E. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin D E, Bowers B, Chen C, Kamada Y, Watanabe M. Dissecting the protein kinase C/MAP kinase signaling pathway of Saccharomyces cerevisiae. Cell Mol Biol Res. 1994;40:229–239. [PubMed] [Google Scholar]
- 28.Levin D E, Errede B. The proliferation of MAP kinase signaling pathways in yeast. Curr Opin Cell Biol. 1995;7:197–202. doi: 10.1016/0955-0674(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 29.Levin D E, Fields F O, Kunisawa R, Bishop J M, Thorner J. A candidate protein kinase C gene PKC1, is required for the S. cerevisiae cell cycle. Cell. 1990;62:213–224. doi: 10.1016/0092-8674(90)90360-q. [DOI] [PubMed] [Google Scholar]
- 30.Lodder A L, Lee T K, Ballester R. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics. 1999;152:1487–1499. doi: 10.1093/genetics/152.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manning B D, Padmanabha R, Snyder M. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1829–1844. doi: 10.1091/mbc.8.10.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazur P, Morin N, Baginsky W, El-Sherbeini M, Clemas J A, Nielsen J B, Foor F. Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol Cell Biol. 1995;15:5671–5681. doi: 10.1128/mcb.15.10.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono T, Suzuki T, Anraku Y, Iida H. The MID2 gene encodes a putative integral membrane protein with a Ca(2+)-binding domain and shows mating pheromone-stimulated expression in Saccharomyces cerevisiae. Gene. 1994;151:203–208. doi: 10.1016/0378-1119(94)90657-2. [DOI] [PubMed] [Google Scholar]
- 35.Ozaki K, Tanaka K, Imamura H, Hihara T, Kamayema T, Nonaka H, Hirano H, Matsuura Y, Takai Y. Rom1p and Rom2p are small GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP-binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:2196–2207. [PMC free article] [PubMed] [Google Scholar]
- 36.Paravicini G, Cooper M, Friedli L, Smith D J, Carpentier J-L, Klig L S, Payton M A. The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol Cell Biol. 1992;12:4896–4905. doi: 10.1128/mcb.12.11.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. Interactions between the bud emergence proteins Bem1 and Bem2 and the Rho-type GTPases in yeast. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qadota H, Python C P, Inoue S B, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin D E, Ohya Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 39.Quilliam L A, Khosravi-Far R, Huff S Y, Der C J. Guanine nucleotide exchange factors: activators of the Ras superfamily of proteins. Bioessays. 1995;17:395–404. doi: 10.1002/bies.950170507. [DOI] [PubMed] [Google Scholar]
- 40.Rajavel M, Philip B, Buehrer B M, Errede B, Levin D E. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3969–3976. doi: 10.1128/mcb.19.6.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ram A F J, Brekelmans S S C, Oehlen L J W M, Klis F M. Identification of two cell cycle regulated genes affecting the β-1,3-glucan content of cell wall in Saccharomyces cerevisiae. FEBS Lett. 1995;358:165–170. doi: 10.1016/0014-5793(94)01418-z. [DOI] [PubMed] [Google Scholar]
- 42.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 43.Schmidt A, Bickle M, Beck T, Hall M. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- 44.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strahl-Bolsinger S, Gentzsch M, Tanner W. Protein O-mannosylation. Biochim Biophys Acta. 1999;1426:297–307. doi: 10.1016/s0304-4165(98)00131-7. [DOI] [PubMed] [Google Scholar]
- 46.Verna J, Lodder A, Lee K, Vagts A, Ballester R. A family of genes required for the maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe M, Chen C-Y, Levin D E. Saccharomyces cerevisiae PKC1 encodes a protein kinase C (PKC) homolog with a substrate specificity similar to that of mammalian PKC. J Biol Chem. 1994;269:16829–16836. [PubMed] [Google Scholar]
- 48.Watanabe Y, Takaesu G, Hagiwara M, Irie K, Matsumoto K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:2615–2623. doi: 10.1128/mcb.17.5.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Z-S, Leung T, Manser E, Lim L. Pheromone signaling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol Cell Biol. 1995;15:5246–5257. doi: 10.1128/mcb.15.10.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]