Abstract
Coregulators for nuclear receptors (NR) are factors that either enhance or repress their transcriptional activity. Both coactivators and corepressors have been shown to use similar but functionally distinct NR interacting determinants containing the core motifs LxxLL and ΦxxΦΦ, respectively. These interactions occur through a hydrophobic cleft located on the surface of the ligand-binding domain (LBD) of the NR and are regulated by ligand-dependent activation function 2 (AF-2). In an effort to identify novel coregulators that function independently of AF-2, we used the LBD of the orphan receptor RVR (which lacks AF-2) as bait in a yeast two-hybrid screen. This strategy led to the cloning of a nuclear protein referred to as CIA (coactivator independent of AF-2 function) that possesses both repressor and activator functions. Strikingly, we observed that CIA not only interacts with RVR and Rev-ErbAα in a ligand-independent manner but can also form complexes with estrogen receptor alpha (ERα) and ERβ in vitro and enhances ERα transcriptional activity in the presence of estradiol (E2). CIA-ERα interactions were found to be independent of AF-2 and enhanced by the antiestrogens EM-652 and ICI 182,780 but not by 4-hydroxytamoxifen and raloxifene. We further demonstrate that CIA-ERα interactions require the presence within CIA of a novel bifunctional NR recognition determinant containing overlapping LxxLL and ΦxxΦΦ motifs. The identification and functional characterization of CIA suggest that hormone binding can create a functional coactivator interaction interface in the absence of AF-2.
Nuclear receptors belong to a superfamily of transcription factors that modulate hormone-regulated physiological pathways involved in reproduction, development, growth, and metabolism (38). Members of the nuclear receptor superfamily have been shown to possess the dual ability to activate and repress the expression of target genes through the recruitment of coactivators and corepressors (reviewed in references 21 and 41). These regulatory proteins associate mostly in a ligand-dependent manner with the ligand-binding domain (LBD) of the receptor. A short helical motif that is located at the C-terminal end of the LBD and is referred to as activation function 2 (AF-2) has been shown to play a central role in coregulator-receptor interaction, as its integrity is essential for ligand-dependent coactivator binding (12, 18, 31, 50) whereas its deletion favors corepressor binding (52, 66). Comparative analysis of the crystal structures of several unliganded and liganded nuclear receptors has revealed that the AF-2 helix appears to take a distinct configuration in the presence of ligand, suggesting that ligand binding modified the conformation of the LBD and promotes the recruitment of coactivators through the formation of a novel interacting surface (reviewed in reference 42).
A large number of coactivators have been characterized to date. These proteins generally possess multiple functional domains which cooperate to maximize receptor activity through diverse mechanisms. These include making direct contacts with chromatin remodeling complexes, the basal transcription machinery, and the p300/CBP cointegrators, as well as exercising their own enzymatic activities, such as ubiquitin ligase, ATPase, protease, kinase, and histone acetyltransferase (reviewed in references 21 and 41). On the other hand, the precise function of several coactivators remains to be elucidated. A more limited number of potential nuclear receptor corepressors have also been identified (8, 10, 26, 51, 65). Of these, N-CoR and SMRT, which are related, have been shown to encode large proteins containing autonomous repressor domains involved in both adapter-dependent and -independent recruitment of histone deacetylases (24, 28, 32, 43).
The ligand-dependent interaction between coactivators and nuclear receptors has been shown to be mediated by a helical motif consisting of the sequence LxxLL (where L is leucine and x is any amino acid) (23, 35), referred to as the NR box (14, 16) or the LXD motif (40). The specificity of interaction between a given coactivator and various nuclear receptors may depend on the number, the appropriate spacing, and especially the sequences surrounding each NR box (14, 40). These motifs interact with a region on the surface of the nuclear receptor's LBD that forms a hydrophobic cleft (18, 37). It consists of a surface shaped by helices 3, 5, 6, and 12 (AF-2) of the LBD which makes direct contacts with the LxxLL helical motifs present in coactivators. This interacting determinant becomes functional when the cognate ligand binds the LBD, an event that repositions the AF-2 helix and results in the formation of the complete interaction surface. Recently, it has been shown that nuclear receptors interact with the corepressors N-CoR and SMRT via a similar mechanism (27, 44, 47). Each corepressor interaction domain possesses a core helical motif with the consensus sequence ΦxxΦΦ, where Φ is a hydrophobic residue. It has also been proposed that the length of these helices present in corepressors is extended in comparison with that of the coactivator LxxLL helices, therefore providing a molecular basis for the discrimination by the liganded receptors between these two classes of regulatory proteins (47). The corepressor helical motifs apparently recognize either the same or an overlapping site that participates in coactivator NR box binding to nuclear receptors. This observation suggests that competition between corepressors and coactivators for a common binding site on nuclear receptors is an important component of hormone signaling.
The steroid hormone 17β-estradiol (E2) is essential for the normal development and maintenance of reproductive functions and also plays important roles in neural, bone, and cardiovascular physiology (13). In particular, this hormone regulates the growth and development of the mammary gland and can stimulate the proliferation of breast cancer cells (15). Consequently, considerable efforts have been made to develop potent antiestrogens that could act as therapeutic agents for the treatment and possible prevention of breast cancer (reviewed in reference 30). However, while antiestrogens such as 4-hydroxytamoxifen (OHT) behave as antagonists in the breast, these molecules often induce estrogen-like effects in the uterus and other peripheral tissues. These compounds were shown to display agonistic properties in both cell type- and promoter-specific manners in transient-transfection assays using cloned receptors (5, 39, 46, 64). Synthetic ligands that display such diversity in their action are currently referred to as selective estrogen receptor (ER) modulators (SERMs). While the molecular basis of tissue- and gene-specific effects of SERMs is not completely understood, structural analysis of diethylstilbestrol-, OHT-, and raloxifene (RAL)-bound ERα revealed that each compound induces a specific helix 12 conformation (7, 53). In particular, the bulkier side chains present in OHT and RAL prevent the AF-2 helix from folding normally and reposition it to the site of coactivator interaction, thus prohibiting coactivator binding (7, 53). These findings suggest that SERM-induced changes in the conformation of the LBD may indeed result in the formation of novel surfaces that could promote interactions with specific sets of coactivators and corepressors.
Among members of the superfamily of nuclear receptors exists a class of receptors whose discovery preceded the identification of their cognate ligands and which are therefore referred to as orphan receptors. The identification of orphan receptors and the further characterization of both their modes of action and physiological roles, as well as the identification of their ligands, have greatly contributed to our understanding of the molecular mechanisms underlying nuclear receptor signaling and unveiled several novel hormone response systems (reviewed in references 19 and 33). Although AF-2-mediated coactivator interaction plays an important role in nuclear receptor transactivation, this mechanism may not be unique, as some orphan members of the nuclear receptor superfamily lack AF-2 (reviewed in reference 19). In an effort to identify novel potential coactivators that function independently of AF-2, we performed a yeast two-hybrid screen using the LBD of RVR (NR1D2), an orphan nuclear receptor lacking AF-2 (49). Here we report the identification of a novel coactivator for nuclear receptors referred to as CIA. In addition to its interaction with RVR, CIA displayed specific ligand-inducible interaction and coactivator activity with ERα (NR3A1) and ERβ (NR3A2). Delineation of the molecular determinants of CIA-ERα binding revealed that AF-2 is also dispensable for ligand-dependent interaction. We further demonstrate that CIA-ERα interaction requires the presence within CIA of a novel nuclear receptor recognition determinant containing overlapping ΦxxΦΦ and LxxLL motifs. Finally, CIA-ERα interactions were found to be differentially regulated by SERMs.
MATERIALS AND METHODS
CIA isolation and interaction assay in yeast.
Saccharomyces cerevisiae Y190 [MATa gal4 gal180 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL→lacZ LYS2::GAL(UAS)→HIS3 cyh] (a gift from Stephen Elledge) containing bait plasmid pAS1-RVR (amino acids [aa] 286 to 509) was transformed with a human fetal kidney library (19 to 23 weeks) (MATCHMAKER Library; Clontech, Palo Alto, Calif.) and plated on SD medium lacking tryptophan, leucine, and histidine and containing 50 mM 3-aminotriazole (17). His+ colonies having β-galactosidase activity, as determined by a filter lift assay, were further characterized via standard techniques (3). The library plasmids were recovered by isolating total yeast DNA, electroporated into Escherichia coli HB101, and isolated on a minimal medium lacking leucine and containing ampicillin. For the interaction assays, CIA was retransformed into Y190 and mated to strain Y187 (MATα gal4 gal80 his3 trp-901 ade2-101 ura2-3,112 met URA3::GAL→lacZ; a gift from Stephen Elledge) containing various baits (SNF1, lamin, CDK1, p53, hERRα [NR3B1], rERRβ [NR3B2], hRevErbAα [NR1D1], mRVR, hGR [NR3C1], and hRARα [NR1B1]). The baits were assayed for transactivation using pGAD-CIA as described elsewhere (36).
Plasmid constructs and reagents.
To construct the yeast two-hybrid bait, pCMXmRVR (described in reference 49) was digested with BstXI, end filled with Klenow, and further digested with BamHI. The 1.2-kb fragment containing the LBD was subcloned into pAS1 (17) digested with NcoI, end filled with Klenow, and digested with BamHI. pCMXhCIA was constructed by PCR using forward oligonucleotide (introducing a consensus start site [underlined]) 5′-ACGGAATTCGTACCATGGCGCCTTTGTCCTACGGC-3′ and reverse oligonucleotide 5′-GCGCGAATTCTCAGTAATGCCTCTGGTA-3′. The PCR product was digested with EcoRI and cloned into pCMX. Expression plasmid pGFPhCIA was constructed as follows. pCMXhCIA was digested with EcoRI, and a fragment containing the CIA open reading frame (ORF) was cloned into pEGFP-C2 (Clontech) digested with EcoRI. pGSTmRVR was constructed by cutting pCMXmRVR with NcoI and BamHI, end filling with Klenow, and cloning the insert into SmaI-cut vector pGEX-2T (Pharmacia Biotech). All of the glutathione S-transferase (GST)-RVR LBD deletions were constructed as follows. A PCR using specific 5′ and 3′ oligonucleotides was performed, and the products were digested with BamHI and MfeI. The fragments were cloned between the BamHI and EcoRI sites of the pGEX-2T vector. pGSThCIA was constructed as follows. pCMXhCIA was digested with BamHI and Asp718I and end filled with Klenow, and the 1,030-bp fragment was cloned into the SmaI site of pGEX-3X (Pharmacia Biotech, Piscataway, N.J.). The various Gal4 DNA-binding domain (DBD)-CIA fusion proteins were generated as follows. Fragments of CIA were PCR amplified using VENT DNA polymerase (NEB, Beverly, Mass.), with specific primers that included new unique restriction sites at both the 5′ and 3′ ends (SacI and EcoRI, respectively). PCR fragments were then digested with SacI/EcoRI and cloned into the corresponding sites of pCMX-Gal4(DBD). The pCMX, pCMXhERα, TKLuc, 2C-vERE-TKLuc, pS2-Luc, and pS2(ΔERE)-Luc reporters were all previously described (58), as well as the mouse mammary tumor virus (MMTV)-Luc construct (25). 3C-TREPal-TKLuc was constructed by cloning three copies of a TREpal (61) consensus oligonucleotide between the HindIII and BamHI sites of TKLuc. All of the hERα and hCIA mutants were constructed by PCR site-directed mutagenesis using Pfu polymerase (Stratagene, La Jolla, Calif.), and the smallest possible fragment containing the mutation(s) was sequenced, cut out, and reinserted into the template plasmid to eliminate the risk of undesired mutations. The integrity of each construct described here was confirmed by DNA sequencing. SRC-1 was a gift of Joe Torchia, University of Western Ontario, London, Ontario, Canada. E2, all-trans retinoic acid, 3-iodothyroxine, progesterone, and dexamethasone were all obtained from Sigma Chemical Co., St. Louis, Mo. 1,25-Dihydroxyvitamin D was a generous gift of Alain Moreau, Institut de Recherches Cliniques de Montréal, Montréal, Québec, Canada. EM-652 and ICI 182,780 were synthesized in the medicinal chemistry division of the Laboratory of Molecular Endocrinology, CHUL Research Center, Québec, Québec, Canada. OHT was kindly provided by D. Salin-Drouin, Besins-Iscovesco, Paris, France.
Protein expression.
The various bait protein constructs were transformed in either E. coli DH5α (GSTmRVR LBD) or BL21DE3pLysE (GSThCIA), and protein expression was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 3 h (0.05 mM IPTG in the case of GSThCIA [LXXAA]). Bacterial extracts were prepared via sonication (DH5α) or freeze-thaw cycles (BL21DE3pLysE). The extracts were aliquoted in an ethanol-dry ice bath and stored at −80°C.
Cell culture and transfection.
COS-1 and HeLa cells were obtained from the American Type Culture Collection. Cells were routinely cultured in Dulbecco's minimal essential medium (DMEM) containing penicillin (25 U/ml), streptomycin (25 U/ml), and 10% fetal calf serum (FCS) at 37°C with 5% CO2. Twenty-four hours prior to transfection, the cells were split and seeded into 12-well dishes. At this stage, the medium was changed for phenol red-free DMEM supplemented with antibiotics and 10% charcoal-dextran-treated FCS. Cells were transfected using the calcium phosphate-DNA coprecipitation method (20). Typically, 0.5 μg of reporter plasmid, 0.2 μg of the internal control (pCMVβGal), 50 ng of receptor expression vector, 100 ng of coactivator expression vector, and carrier (Bluescript pKS II, to a total of 1 μg per well) were added to the cells. After 12 to 14 h, cells were washed twice with phosphate-buffered saline and treated with either 10−8 M E2 or carrier (ethanol) for 24 h in phenol red-free DMEM supplemented with 10% stripped FCS. Cells were then washed and harvested in potassium phosphate lysis buffer containing 1% Triton X-100. Luciferase and β-galactosidase assays were performed as previously described (56). All of the transfection results presented here are averages of at least two independent experiments performed in triplicate. Green fluorescent protein (GFP) and CIAhGFP intracellular localization experiments were conducted as follows. At 24 h prior to transfection, HeLa cells were seeded into a six-well dish with DMEM supplemented with 10% FCS. GFP or CIA-GFP was transiently transfected at 2 μg per well using calcium phosphate-DNA coprecipitation. After transfection, cells were washed twice with phosphate-buffered saline and incubated in fresh medium for several hours. Pictures were taken using a charge-coupled device camera mounted on a Zeiss Axiovert-135 microscope. Image capture and analysis were performed via Northern Eclipse software (EMPIX).
Northern blotting.
Total RNA was collected from different tissues of 17.5-day postcoitus mouse embryos and processed with TRIZOL reagent (Life Technologies, Gaithersburg, Md.). Poly(A)+ RNA was prepared using a QuickPrep Micro mRNA purification kit (Pharmacia Biotech). The samples were separated by electrophoresis in a 1% agarose–0.4% formaldehyde–1× MOPS (morpholinepropanesulfonic acid) gel and transferred onto a nylon membrane (Hybond N; Amersham Life Sciences, Amersham, United Kingdom) in 20× SSC buffer (1× SSC is 0.15 M NaCl and 0.015 M Na3citrate · 2H2O at pH 7.0). The RNA was UV cross-linked to the membrane and then prehybridized at 42°C for 1 h in a buffer containing 50% formamide, 5× SSPE (1× SSPE is 0.15 M NaCl, 0.01 M NaH2PO4 · H2O, and 1 mM EDTA at pH 7.4), 5× Denhardt's solution, 1% glycine, and 100 mg of denatured salmon sperm DNA ml−1. After prehybridization, the membranes were hybridized overnight at 42°C in a solution of 50% formamide, 5× SSPE, 1× Denhardt's solution, 0.3% sodium dodecyl sulfate (SDS), 10% (wt/vol) dextran sulfate, 100 mg of denatured salmon sperm DNA ml−1, and 6 × 105 cpm of 32P-labeled probe ml−1. Membranes were then washed as follows: 2× SSC–1% SDS at 55°C for 20 min, followed by 0.2× SSC–0.1% SDS for 20 min at 55°C and for another 20 min at 65°C. They were then autoradiographed at −80°C on X-Omat film (Eastman Kodak Company, Rochester, N.Y.).
GST pull-down assays.
Fusion proteins were incubated with glutathione-Sepharose beads (Pharmacia Biotech) for 20 min at 4°C. The beads were then centrifuged, washed four times with GST-binding buffer containing 20 mM HEPES (pH 7.9), 150 mM KCl, 5 mM MgCl2, 0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 0.1 mM phenylmethylsulfonyl fluoride, and 1 μM leupeptin. Beads were then resuspended in 150 μl of GST-binding buffer with bovine serum albumin (BSA) at 20 μg ml−1, the appropriate hormone (or carrier), and 5 μl of in vitro-translated protein using TNT rabbit reticulocyte lysate (Promega, Madison, Wis.). The reaction mixtures were incubated for 1 h 30 min at 4°C with mild agitation. The complexes were then centrifuged and washed in GST-binding buffer, twice with BSA at 20 μg ml−1 and twice without BSA. Samples were then resuspended in 2× SDS sample buffer and boiled for 5 min prior to separation by standard SDS–10% polyacrylamide gel electrophoresis. Gels were then fixed, treated with the fluorographic reagent Amplify (Amersham Life Sciences), dried, and autoradiographed at −80°C on X-Omat film. To ensure the presence of equal amounts of bait proteins, purified extracts from the various baits were previously separated by SDS-PAGE, stained with Coomassie blue, and compared. Equivalent amounts of bait proteins were then used for each pull-down experiment.
Nucleotide sequence accession number.
The GenBank accession number of the CIA clone sequence reported here is AF230533.
RESULTS
Identification of CIA.
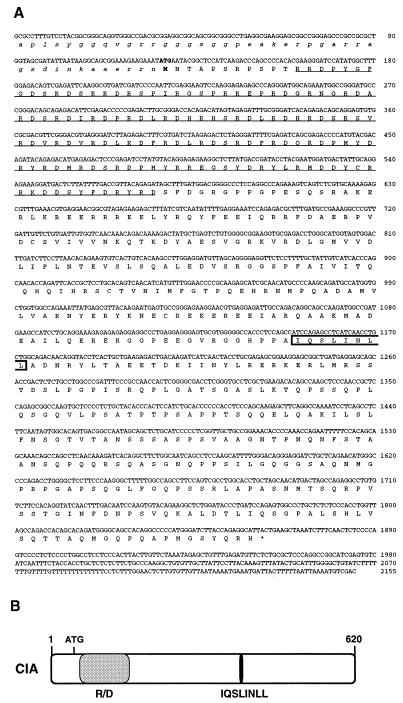
In an effort to identify novel coregulatory proteins that would act independently of AF-2, we used the LBD of RVR, an orphan nuclear receptor with no AF-2 (49), as bait in a yeast two-hybrid screen. A standard procedure was followed (11), and several cDNA clones were obtained from a human fetal kidney library. One of the clones whose interaction with RVR was potent and specific in the standard yeast liquid β-galactosidase assay (data not shown) and whose ORF encoded a novel protein was chosen for further studies. Sequencing of this 2,155-bp clone revealed a long ORF (620 amino acids) starting at the 5′ end of the DNA fragment (Fig. 1A). Since extensive screening using pairs of exonic primers (Genome Systems, St. Louis, Mo.) and rapid amplification of cDNA ends did not generate a clone with a longer 5′ end, we assume that the encoded protein starts at the first methionine residue (position 43). Sequence comparison studies failed to reveal significant homology with known proteins. However, as shown in Fig. 1A and B, two identifiable features are present in the ORF: (i) overlapping LxxLL and ΦxxΦΦ helices (boxed in Fig. 1A, discussed below) recently shown to mediate nuclear receptor-coactivator and -corepressor interactions, respectively, and (ii) an amino-terminal arginine- and aspartic acid-rich region (RD-rich region, underlined in Fig. 1A). A similar but shorter RD-rich domain is also found in the coactivator TRAP220/DRIP205 (48, 60). On the basis of its interaction with an orphan receptor lacking AF-2 and other characteristics that will be discussed below, we named this clone CIA (coactivator independent of AF-2).
FIG. 1.
CIA clone sequence and features. (A) The sequence obtained from human fetal kidney cDNA is 2,155 bp long and contains a 620-aa ORF. The arginine- and aspartic acid-rich region is underlined, and the overlapping LxxLL and ΦxxΦΦ nuclear receptor interaction motifs are boxed. The first putative methionine is in bold, and the amino acid residues preceding it are noncapitalized. Nucleotides are numbered on the right. (B) Schematic representation of CIA. The arginine- and aspartic acid-rich region (R/D) of the protein is represented by the gray box, and the location of the overlapping ΦxxΦΦ and LxxLL motifs is indicated by the black box.
Tissue expression and cellular localization of CIA.
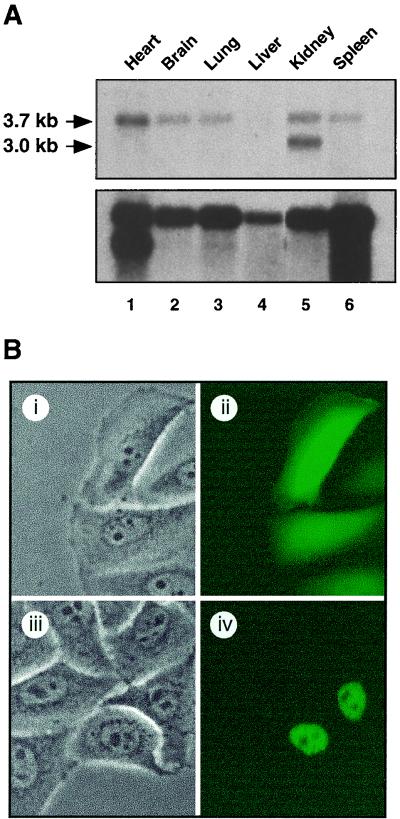
In order to determine if CIA expression exhibits tissue specificity, we performed Northern blot assays using poly(A)+ mRNA extracted from various tissues of a fetal (embryonic day 17.5) mouse. As shown in Fig. 2A (top), we observed expression of mouse CIA mRNA in all of the tissues tested. The level of expression was very low in embryonic liver but high in the heart and kidneys. Note the presence of two transcripts in the embryonic kidney (lane 5). The difference between the two transcripts is about 700 nucleotides and is due to the use of an alternative polyadenylation signal, which can be found on expressed sequence tag clones (data not shown).
FIG. 2.
Expression and intracellular localization of CIA. (A, top) Northern blot of poly(A)+ mRNA from 17.5-day postcoitus mouse embryos, probed with the human CIA cDNA clone showing expression of a 3.7-kb mRNA in the heart, brain, lung, kidneys, and spleen. Note the presence of a second 3-kb mRNA in the kidneys. (A, bottom) A β-actin probe was used as control for mRNA integrity. Equal loading was assessed via ethidium bromide staining (not shown). (B) CIA is localized to the nucleus. Phase-contrast (i and iii) and fluorescence (ii and iv) pictures of HeLa cells transiently transfected with pGFP (i and ii) and pGFPhCIA (iii and iv) expression vectors.
To assess the intracellular localization of CIA, transient-transfection experiments were performed with HeLa cells using a fusion of GFP with the entire ORF of CIA. GFP alone was distributed evenly throughout the cell (Fig. 2B, part ii), whereas the CIA-GFP fusion protein was restricted to the nucleus (Fig. 2B, part iv). Similar results were obtained when the experiments were performed with COS-1 cells (data not shown). Therefore, the CIA-GFP fusion protein is strictly nuclear when transfected into mammalian cells.
In vitro interaction of CIA with nuclear receptors.
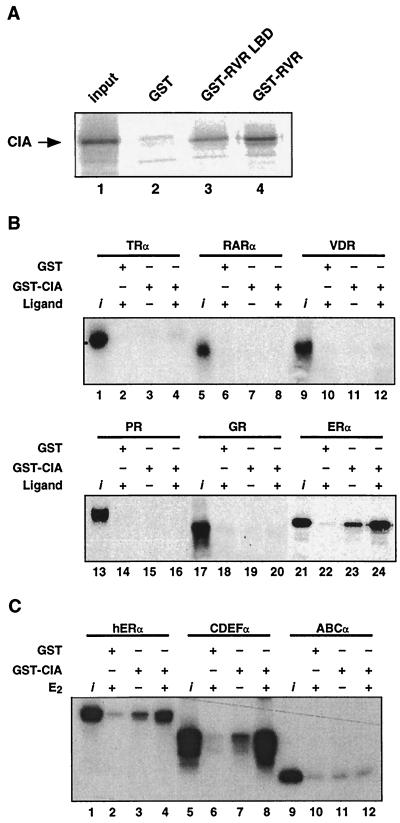
To confirm the interaction of CIA with RVR previously observed in yeast, in vitro GST pull-down experiments were performed. As expected from the screening interaction, in vitro-translated, [35S]methionine-labeled CIA interacted with both the GST-LBD-RVR and GST-RVR fusion proteins (Fig. 3A). To test whether the observed interaction was specific to RVR, a GST fusion protein containing aa 55 to 395 of CIA, sufficient to bind RVR (data not shown), and including the putative nuclear receptor interacting motifs was used to perform pull-down experiments in combination with in vitro-translated, [35S]methionine-labeled nuclear receptors (Fig. 3B). The GST-CIA fusion protein did not interact with T3Rα, RARα, or VD3R in the presence or absence of the cognate ligand. The fusion protein also did not interact with the steroid receptors GR and PR. No interaction was observed when the GST-CIA fusion protein was tested with other nuclear receptors not shown here: the mineralocorticoid receptor, RXRα, T3Rβ, and the more closely related orphan nuclear receptor RORα (NR1F1). Strikingly, the GST-CIA fusion protein interacted with ERα (Fig. 3B, bottom) and ERβ (data not shown), and these interactions were greatly enhanced in the presence of E2. Therefore, CIA shows in vitro specificity of interaction for two distinct classes of nuclear receptors: the classic steroid receptors ERα and ERβ and the orphan nuclear receptors RVR and Rev-erbAα (data not shown).
FIG. 3.
CIA in vitro interaction shows specificity for RVR and ERα. (A) [35S]methionine-labeled CIA interacts in vitro with the GST-RVR LBD (lane 3) and full-length GST-RVR (lane 4) but not with the GST control protein (lane 2). The input represents 10% of the labeled CIA used in the assay (lane 1). (B) [35S]methionine-labeled ERα specifically interacts in both ligand-independent and -dependent fashions with GST-CIA. The assay was performed in the presence of either 10−7 M E2 (lanes 22 and 24) or the vehicle (ethanol, lane 23). None of the other nuclear receptors tested, including TRα, RARα, VD3R, PR, and GR, demonstrated any interaction with GST-CIA (lanes 1 to 20) either in the absence or in the presence of the appropriate ligands (T3Rα, 3-iodothyronine at 10−6 M; RARα, all-trans retinoic acid at 10−6 M; VD3R, 1,25-dihydroxyvitamin D3 at 10−7 M; PR, progesterone at 10−7 M; GR, dexametasone at 10−7 M). (C) Interaction between CIA and ERα requires the C-terminal portion of ERα. Full-length [35S]methionine-labeled ERα proteins and the same protein with the amino-terminal domain truncated (CDEFα) or the carboxy-terminal domain truncated (ABCα) were incubated with GST-CIA. Pull-down assays were performed as described in Materials and Methods.
To further probe the CIA interaction with ERα, we constructed N-terminal and C-terminal truncated versions of ERα and tested them for the ability to interact with CIA in vitro (Fig. 3C). Ligand-dependent interaction of CIA with the LBD-containing C-terminal construct of ERα (CDEFα) appeared to be more potent than with the wild-type receptor (compare lanes 7 and 8 to lanes 3 and 4). Conversely, the N-terminal construct of ERα (ABCα) did not interact with CIA. These results show that CIA interaction with ERα occurs through the C-terminal extremity of the receptor, in a manner similar to binding to RVR.
CIA is a specific transcriptional coactivator of ERα.
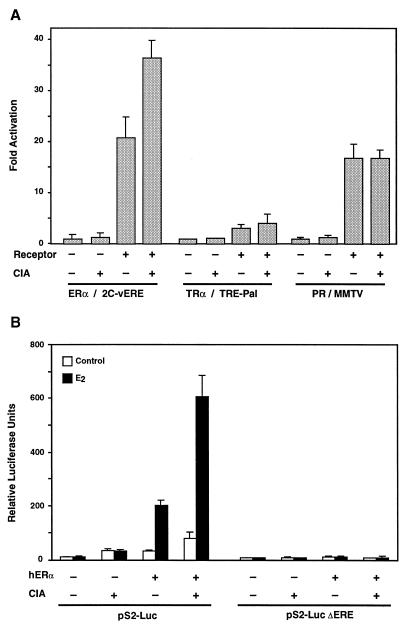
To test the possibility that CIA acts as a coactivator for ERα, transient-transfection experiments were performed with COS-1 cells (Fig. 4). When cotransfected with various nuclear receptors in the presence of their ligands, CIA was only able to potentiate ERα transcriptional activity on a reporter gene driven by the thymidine kinase (TK) promoter linked to two copies of the consensus vitellogenin estrogen response element (vERE) by approximately twofold. Conversely, CIA was unable to influence either T3Rα or PR transcriptional activity on the TREpTK and MMTV promoters, respectively (Fig. 4A). We then tested the ability of CIA to potentiate ERα transcriptional activity on a natural promoter. The pS2 promoter has been shown to contain a classical inverted palindromic ERE that is essential for ER binding and responsiveness to E2 (6). As shown in Fig. 4B, the addition of CIA to ERα leads to a sharp increase in transcriptional activity in the presence of E2. Figure 4B also shows that the induction of the pS2 promoter by the ERα-CIA combination is totally dependent on the presence of an intact ERE. Taken together, these results confirm the specificity of interaction of CIA for ERα observed previously in GST pull-down assays and clearly indicate that CIA is a potent transcriptional coactivator of ERα.
FIG. 4.
CIA is a potent and specific ERα transcriptional activator. (A) A luciferase reporter gene linked with various hormone-responsive promoters (2C-vERE-TKLuc, 3C-TREpal-TKLuc, and MMTV-Luc) was transfected into COS-1 cells along with expression vectors for CIA, ERα, TRα, and PR in the combinations shown. Results are expressed as fold induction over the reporter alone in the presence of the appropriate ligand (ERα, 10−8 M E2; TRα, 10−8 M 3-iodothyronine; PR, 10−8 M progesterone). (B) Ligand-dependent activation of ERα on the pS2-Luc reporter gene is enhanced by cotransfection of CIA. No effect of CIA is observed on the pS2 promoter when its ERE site is mutated (pS2-Luc ΔERE).
Activation and repression function of CIA.
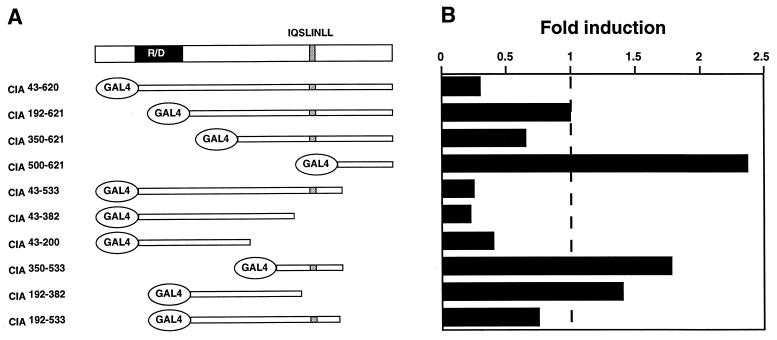
Coactivators can possess an autonomous activation function and/or recruit other coregulatory factors. Using fragments of CIA fused to the Gal4 DBD (Fig. 5A), we tested whether such an autonomous activation function is present within CIA. Unexpectedly, CIA43-620, which contains the whole coding region downstream of the putative ATG fused to the Gal4 DBD, displayed repressor activity (Fig. 5B). Given the coactivator function previously observed in cotransfection with ERα (Fig. 4), this result suggests that CIA may act as both a coactivator and a corepressor, depending on how CIA is brought into the proximity of the transcriptional unit. Further deletion analysis revealed a complex structure-function relationship within CIA. Results shown in Fig. 5 suggest that the region surrounding the R/D domain possesses repressor activity while the carboxy-terminal region of CIA, on its own, has transcriptional activation potential.
FIG. 5.
CIA contains both activation and repression functions. (A) Schematic representation of CIA and Gal4 DBD fusion proteins. (B) Activity of Gal4 DBD-CIA fusion constructs on a three-copy Gal4 UAS-TK luciferase reporter gene. Results are expressed as fold induction over the reporter in the presence of the Gal4 DBD expression vector.
CIA contains a bifunctional interacting determinant (BID) for nuclear receptor recognition.
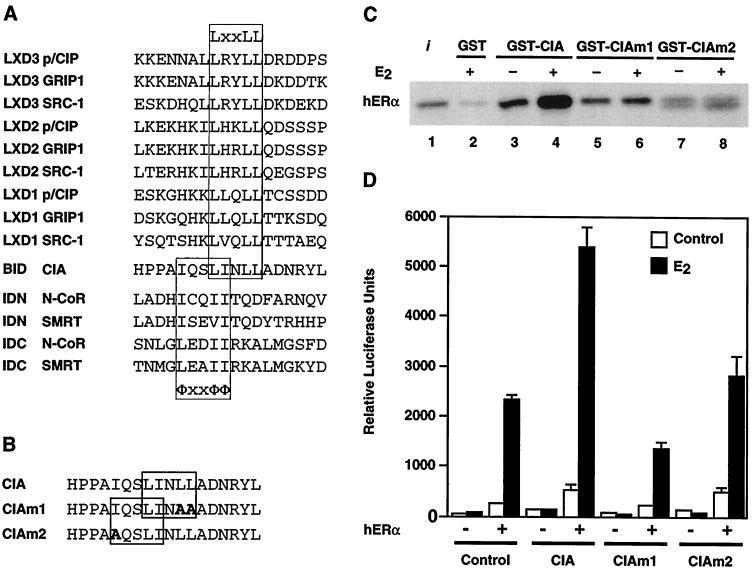
Comparison of the amino acid sequence of the CIA region (aa 53 to 395) that is required to bind to RVR and ERα with a series of previously characterized coactivator- and corepressor-nuclear receptor recognition sequences revealed the presence of overlapping corepressor (ΦxxΦΦ) and coactivator (LxxLL) binding motifs in CIA (Fig. 6A). This unique characteristic of CIA may partially explain our observation that CIA interacts with ERα in both ligand-independent and -dependent manners and with RVR which is devoid of an AF-2 helix. To test this hypothesis, we introduced point mutations into both the LxxLL and ΦxxΦΦ helices and assayed for E2-dependent and -independent CIA interaction with ERα (Fig. 6B). As shown in Fig. 6C, mutation of two conserved leucine residues within the coactivator LxxLL motif (CIAm1) abolished E2-inducible CIA-ERα interaction. However, basal ligand-independent CIA-ERα interaction was only slightly affected by these mutations. On the other hand, mutant CIAm2, which contains a mutation of the first isoleucine residue of the corepressor ΦxxΦΦ motif, displayed almost no interaction. We next tested the ability of each mutant to potentiate ERα transcriptional activity in transient-transfection assays (Fig. 6D). In agreement with the results obtained with the in vitro interaction assay, both mutants were unable to enhance ERα transactivation. Taken together, these results demonstrate that both overlapping LxxLL and ΦxxΦΦ core motifs are required for full functionality of the CIA-nuclear receptor interaction determinant.
FIG. 6.
Both ΦxxΦΦ and LxxLL core motifs participate in the CIA-ERα interaction. (A) Sequence alignment of p160 family member coactivator LxxLL and SMRT/N-CoR corepressor ΦxxΦΦ core motifs with the CIA BID. (B) Sequence of the CIA BID. Bold highlights indicate residues that have been mutated in CIAm1 (boxed LxxLL motif) and CIAm2 (boxed ΦxxΦΦ motif). (C) Ligand-dependent interaction between GST-CIA and 35S-labeled ERα (lanes 3 to 6) is disrupted by mutation of the LxxLL motif in CIAm1. Basal and ligand-independent interactions between GST-CIA and 35S-labeled ERα (lanes 7 and 8) are disrupted by mutation of the ΦxxΦΦ motif in CIAm2. (D) Coactivator function of CIA mutants assayed in vivo. Transfections were performed with COS-1 cells using the pS2-Luc reporter and pCMX-based expression vectors as described in Materials and Methods. Transfected cells were treated with either 10−8 M E2 or the carrier (ethanol) for 24 h postinfection.
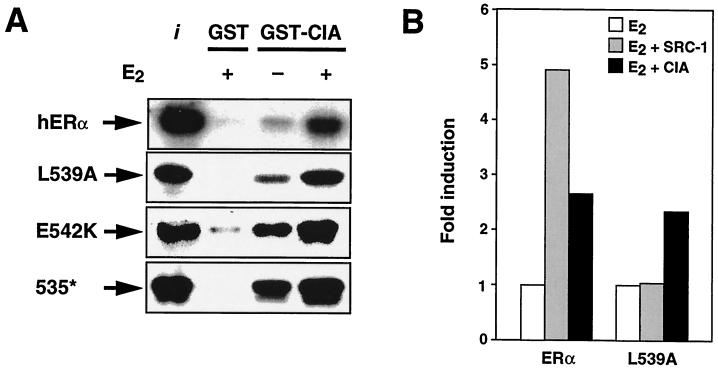
Given the observation that the ΦxxΦΦ core motif present in CIA is required for interaction, we next tested the possibility that the ERα AF-2 helix is dispensable for CIA interaction. To verify this hypothesis, three ERα AF-2 mutants were tested for the ability to interact with CIA using GST pull-down assays. As shown in Fig. 7A, mutant ERαL539A, which has been previously shown to be unable to interact in a ligand-dependent fashion with the SRC-1 coactivator (59, 63), displays both ligand-independent and -dependent binding to CIA. Similarly, mutant ERαE542K, in which the glutamic acid residue involved in forming the charge clamp required for AF-2 function is replaced by a lysine residue, displays stronger ligand-dependent interaction while still being inducible by E2 (Fig. 7A). Since interaction of distinct NR boxes has been shown to depend on specific residues within the different AF-2 motifs (9), we further tested CIA interaction with an ERα mutant lacking the entire AF-2 helical motif. As shown in Fig. 7A (bottom), mutant ERα535*, which lacks AF-2, exhibits both stronger ligand-independent and -dependent interactions with CIA, as expected if CIA utilizes the corepressor-like ΦxxΦΦ core motif in its interaction with ERα. Finally, to confirm these results in vivo, transient transfections were performed using the ERαL539A AF-2 null mutant (Fig. 7B). When ERαL539A was cotransfected with SRC-1, ligand-dependent transactivation could not be restored. In contrast, when the ERα AF-2 mutant was cotransfected with CIA, enhancement of ligand-dependent transactivation activity could still be observed.
FIG. 7.
ERα AF-2 domain is dispensable for CIA interaction. (A) GST pulldown assays showing the interaction between GST-CIA and both the wild-type and mutant AF-2 domains of ERα. L539A and E542K are point mutations within the AF-2 domain, while 535* is deletion of all of AF-2 through insertion of a stop codon at position 535. (B) In contrast to SRC-1, CIA can still enhance ligand-dependent activation of an ERα–AF-2 null mutant. Transfections were performed with COS-1 cells using the 2C-vERE-TKLuc reporter and pCMX-based expression vectors as described in Materials and Methods. Transfected cells were treated with either 10−8 M E2 or the carrier (ethanol) for 24 h posttransfection.
CIA interaction with hERα is differentially regulated by SERMs.
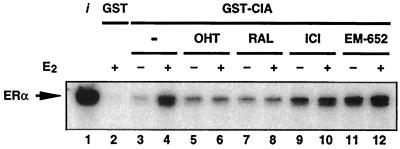
The finding that CIA appears to possess a bifunctional nuclear receptor recognition motif that may allow it to recognize nuclear receptors in both corepressor-receptive (ligand- and AF-2-independent) and coactivator-receptive (ligand-dependent) modes combined with the observation that CIA is expressed at high levels in human breast cancer cells (data not shown) led us to test the abilities of different SERMs to modulate CIA-ERα interactions using pull-down experiments. As shown in Fig. 8, addition of either OHT or RAL had no effect on basal CIA-ERα interactions (lanes 5 and 7) but completely abrogated the E2-induced interactions (lanes 6 and 8). Remarkably, the addition of ICI 182,780 and EM-652, SERMs that function as pure antiestrogens in both the breast and the uterus, led to an increase in CIA-ERα interaction (lanes 9 and 11) and are inefficient at blocking the E2-induced interaction (lanes 10 and 12). These results show for the first time the abilities of distinct classes of SERMs to differentially regulate the interaction of a specific coactivator with ERα LBD.
FIG. 8.
CIA interaction with ERα is differentially regulated by SERMs. Ligand-dependent enhancement of the CIA-ERα interaction (lanes 3 and 4) is inhibited by the partial antagonists OHT and RAL (compare lanes 6 and 8 with lane 4) but is not significantly altered by the pure antiestrogens ICI 182,780 and EM-652 (compare lanes 10 and 12 with lane 4).
DISCUSSION
In this report, we describe the identification and functional characterization of a novel nuclear receptor coregulator referred to as CIA. CIA is distinct from other known coregulators in that it possesses a BID for the nuclear receptor recognition consisting of overlapping ΦxxΦΦ and LxxLL core motifs. This bifunctional motif allows CIA to bind specifically to the orphan nuclear receptors Rev-erbA and RVR that lack an AF-2 domain and to ERα in a ligand-dependent but AF-2-independent manner. These observations indicate that hormone binding can create a functional coregulator interaction surface in the absence of the AF-2 helix and suggests an evolutionary path through which the LBD could have progressively adapted to both corepressor and coactivator binding in a ligand-regulated manner.
Initial analysis of the CIA sequence revealed little information about its putative role and mechanism of action. As shown in Fig. 1B, the only identifiable motifs are an RD-rich region and overlapping ΦxxΦΦ and LxxLL core motifs. The cluster of RD represents a highly hydrophilic region of alternating positively and negatively charged residues which are most likely exposed at the surface of the protein. The coactivator TRAP220/DRIP205 also contains such an RD-rich region, which is only 14 amino acids long and was suggested to be involved in either oligomerization of the protein or DNA binding (48, 60). It is possible that it plays a similar role in CIA, although the RD-rich domain is considerably longer. On the other hand, the ΦxxΦΦ and LxxLL core motifs have been clearly shown to mediate ligand-independent and -dependent interactions between nuclear receptors and corepressors and coactivators, respectively. Indeed, recent studies have shown that a key feature of hormone signaling consists of a ligand-dependent coregulator exchange in which the AF-2 domain and both the ΦxxΦΦ and LxxLL core motifs play a predominant role (reviewed in reference 21). In this model, the presence of the AF-2 helix is inhibitory to corepressor binding but is critical for coactivator interaction. Strikingly, CIA appears to interact in a ligand-dependent fashion with nuclear receptors through a related but distinct mechanism. First, the sequence of the CIA BID can be simultaneously aligned with both the LxxLL motifs present in many coactivators and the ΦxxΦΦ motifs present in the corepressors N-CoR and SMRT (Fig. 6A). Mutational analysis showed that the carboxy end of the extended helix is required for ligand-dependent interaction with ERα but not for basal contact with CIA. However, the amino end of the helix is important for both basal and ligand-enhanced CIA interactions with ERα. These results suggest that while being bifunctional, the CIA BID helix functions as an integrated unit. Second, ligand-enhanced ERα-CIA interaction does not require intact AF-2 since CIA still interacts with the ERαL539A, ERαE542K, and ERα535* mutants, suggesting that the ligand-induced charged-clamp model does not apply to the CIA BID helix. Interestingly, dissection of the LxxLL core motif using combinatorial peptide libraries has shown that a certain class of synthetic LxxLL-containing peptides, referred to as class III LxxLL motifs, interacts strongly with AF-2 ERα mutants (9). Class III LxxLL motifs are preceded by a serine or threonine and an isoleucine or leucine and can be found in the RIP140 coactivator and the orphan nuclear receptors SHP and DAX-1. Therefore, the class III motifs may play a role in the interaction of these proteins with ERs. However, the sequence of the CIA BID does not match the consensus for class III LxxLL motifs as defined in that study (9). Third, the CIA BID helix is not analogous to the extended helices present in N-CoR which constitute high-affinity interaction determinants with the unliganded RARs and T3Rs (47). In particular, the spacing of the hydrophobic residues is clearly distinct between CIA BID and the N-CoR extended helices (Fig. 6A). Thus, while the molecular mechanism of ERα-CIA contact is more similar to that of corepressor interaction with nuclear receptors, it is evidently distinct from it, since ligand binding promotes association instead of dissociation of the coregulator. Taken together, these observations demonstrate that the CIA BID represents a novel class of nuclear receptor recognition determinants.
Structural studies and the use of a combinatorial phage display approach to dissect determinants of coactivator-nuclear receptor interactions have further demonstrated that the ability of coregulators and synthetic peptides to interact with ER is regulated by the characteristics of the bound ligand (7, 9, 45, 53). This property of coregulator-nuclear receptor interactions could potentially be used to classify natural compounds and drugs according to their abilities to promote interactions between nuclear receptors and selected recognition determinants. Here we have shown that the interaction of CIA with ERα is differentially regulated by SERMs. Both EM-652 and ICI 182,780, which have been characterized as pure antiestrogens in reproductive tissues (34, 57, 62), effectively promote CIA binding to ERα (Fig. 8). On the other hand, RAL and OHT clearly antagonize CIA-ERα interaction. These results indicate that EM-652 and ICI 182,780 influence coregulator binding to ERα in a manner that is different from that of RAL and OHT, thereby placing these two sets of compounds in truly distinct mechanistic classes.
Transient-transfection experiments clearly support a role for CIA as a ligand-dependent coactivator of ERs (Fig. 4). CIA is also expressed in breast cancer cell lines (data not shown), indicating that it may have a role in modulating the response of ERs to estrogens and antiestrogens in hormone-dependent cancers. Paradoxically, the pure antiestrogens EM-652 and ICI 182,780, while promoting CIA-ERα interaction in vitro, do not enhance ERα transcriptional activity when cotransfected with CIA in COS cells (data not shown). Similarly, transfection of CIA did not reverse the constitutive repressor activity of RVR and Rev-erbA. These results suggest that the interactions between ERα and CIA in the presence of EM-652 or between CIA and RVR or Rev-erbA in the absence of a known ligand may not lead to the formation of transcriptionally competent complexes. Structure-function analyses of CIA have revealed that CIA possesses autonomous repression domains (Fig. 5). Taken together, these results indicate that CIA may not only contain a bifunctional nuclear receptor interaction determinant but could also play both positive and negative roles in regulating nuclear receptor functions. This hypothesis is in agreement with the recent characterization of mouse Zac1, a promoter- and cell line-dependent dual transcriptional coactivator and corepressor of nuclear receptors (29). Further studies will not only define more precisely the functional and physiological significance of CIA-ER and CIA–Rev-erbA interactions, but the isolation of CIA could prove to be an important tool for the identification of ligands for members of the Rev-erbA family of orphan nuclear receptors.
Based on the results presented here, we conclude that CIA represents a novel class of ligand-regulated coactivators that mediates their action independently of AF-2 integrity. This study and the many recent studies cited above have clearly demonstrated that small variations on a common theme, the ΦxxΦΦ and LxxLL motifs, can generate tremendous diversity in the mechanisms and specificity of ligand-regulated coregulator-nuclear receptor interactions. Thus, identification and characterization of novel coactivators such as CIA are important to promote our understanding of nuclear receptor transcriptional regulation. Better comprehension of this regulation will help in the development of a global scheme of nuclear receptor action in development, physiology, and disease states and will ultimately be used to design fitting therapies.
ACKNOWLEDGMENTS
Financial support was provided by the Canadian Institute for Health Research (CIHR). A. N. Moraitis is the recipient of a training grant from the Fonds de la Recherche en Santé du Québec. V. Giguère holds a CIHR senior scientist career award.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X-Y, Sauter G, Kallioniemi O-P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 3.Bartel P, Chien C T, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. BioTechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 4.Bautista S, Valles H, Walker R L, Anzick S, Zeillinger R, Meltzer P, Theillet C. In breast cancer, amplification of the steroid receptor coactivator gene AIB1 is correlated with estrogen and progesterone receptor positivity. Clin Cancer Res. 1998;4:2925–2929. [PubMed] [Google Scholar]
- 5.Berry M, Metzger D, Chambon P. Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J. 1990;9:2811–2818. doi: 10.1002/j.1460-2075.1990.tb07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry M, Nunez A-M, Chambon P. Estrogen-responsive element of the human pS2 gene is an imperfectly palindromic sequence. Proc Natl Acad Sci USA. 1989;86:1218–1222. doi: 10.1073/pnas.86.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engström L, Greene G L, Gustafsson J-Å, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 8.Burris T P, Nawaz Z, Tsai M J, O'Malley B W. A nuclear hormone receptor-associated protein that inhibits transactivation by the thyroid hormone and retinoic acid receptors. Proc Natl Acad Sci USA. 1995;92:9525–9529. doi: 10.1073/pnas.92.21.9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang C-Y, Norris J D, Grøn H, Paige L A, Hamilton P T, Kenan D J, Fowlkes D, McDonnell D P. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors α and β. Mol Cell Biol. 1999;19:8226–8239. doi: 10.1128/mcb.19.12.8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 11.Chien C T, Bartel P L, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collingwood T N, Rajanayagam O, Adams M, Wagner R, Cavailles V, Kalkhoven E, Matthews C, Nystrom E, Stenlof K, Lindstedt G, Tisell L, Fletterick R J, Parker M G, Chatterjee V K. A natural transactivation mutation in the thyroid hormone β receptor: impaired interaction with putative transcriptional mediators. Proc Natl Acad Sci USA. 1997;94:248–253. doi: 10.1073/pnas.94.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couse J F, Korach K S. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- 14.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson N E, Lippman M E. The role of estrogens in growth regulation of breast cancer. Crit Rev Oncol. 1989;1:89–111. [PubMed] [Google Scholar]
- 16.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 17.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 18.Feng W, Ribeiro R C J, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 19.Giguère V. Orphan nuclear receptors: from gene to function. Endocr Rev. 1999;20:689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- 20.Giguère V, Shago M, Zirngibl R, Tate P, Rossant J, Varmuza S. Identification of a novel isoform of the retinoic acid receptor γ expressed in the mouse embryo. Mol Cell Biol. 1990;10:2335–2340. doi: 10.1128/mcb.10.5.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 22.Guan X Y, Xu J, Anzick S L, Zhang H, Trent J M, Meltzer P S. Hybrid selection of transcribed sequences from microdissected DNA: isolation of genes within amplified region at 20q11–13.2 in breast cancer. Cancer Res. 1996;56:3446–3450. [PubMed] [Google Scholar]
- 23.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 24.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 25.Hollenberg S M, Evans R M. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell. 1988;55:899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- 26.Horlein A J, Naar A M, Heinzel T, Torchia J, Glass B, Kurokawa R, Ryan A, Kamel Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 27.Hu X, Lazar M A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 28.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 29.Huang S M, Stallcup M R. Mouse Zac1, a transcriptional coactivator and repressor for nuclear receptors. Mol Cell Biol. 2000;20:1855–1867. doi: 10.1128/mcb.20.5.1855-1867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan V C, Morrow M. Tamoxifen, raloxifene, and the prevention of breast cancer. Endocr Rev. 1999;20:253–278. doi: 10.1210/edrv.20.3.0368. [DOI] [PubMed] [Google Scholar]
- 31.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Glass B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 32.Kao H Y, Downes M, Ordentlich P, Evans R M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 33.Kliewer S A, Lehmann J M, Willson T M. Orphan nuclear receptors: shifting endocrinology into reverse. Science. 1999;284:757–760. doi: 10.1126/science.284.5415.757. [DOI] [PubMed] [Google Scholar]
- 34.Labrie F, Labrie C, Belanger A, Simard J, Gauthier S, Luu-The V, Merand Y, Giguere V, Candas B, Luo S, Martel C, Singh S M, Fournier M, Coquet A, Richard V, Charbonneau R, Charpenet G, Tremblay A, Tremblay G, Cusan L, Veilleux R. EM-652 (SCH 57068), a third generation SERM acting as pure antiestrogen in the mammary gland and endometrium. J Steroid Biochem Mol Biol. 1999;69:51–84. doi: 10.1016/s0960-0760(99)00065-5. [DOI] [PubMed] [Google Scholar]
- 35.Le Douarin B, Nielsen A L, Garnier J-M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1α and TIF1β in the epigenic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J W, Ryan F, Swaffield J C, Johnston S A, Moore D D. Interaction of thyroid-hormone receptor with a conserved transcriptional mediator. Nature. 1995;374:91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- 37.Mak H Y, Hoare S, Henttu P M, Parker M G. Molecular determinants of the estrogen receptor-coactivator interface. Mol Cell Biol. 1999;19:3895–3903. doi: 10.1128/mcb.19.5.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McInerney E M, Katzenellenbogen B S. Different regions in activation function-1 of the human estrogen receptor required for antiestrogen- and estradiol-dependent transcription activation. J Biol Chem. 1996;271:24172–24178. doi: 10.1074/jbc.271.39.24172. [DOI] [PubMed] [Google Scholar]
- 40.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenna N J, Lanz R B, O'Malley B W. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 42.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 43.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3a, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 44.Nagy L, Kao H Y, Love J D, Li C, Banayo E, Gooch J T, Krishna V, Chatterjee K, Evans R M, Schwabe J W. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norris J D, Paige L A, Christensen D J, Chang C Y, Huacani M R, Fan D, Hamilton P T, Fowlkes D M, McDonnell D P. Peptide antagonists of the human estrogen receptor. Science. 1999;285:744–746. doi: 10.1126/science.285.5428.744. [DOI] [PubMed] [Google Scholar]
- 46.Paech K, Webb P, Kuiper G G J M, Nilsson S, Gustafsson J-Å, Kushner P J, Scanlan T S. Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 47.Perissi V, Staszewski L M, McInerney E M, Kurokawa R, Krones A, Rose D W, Lambert M H, Milburn M V, Glass C K, Rosenfeld M G. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 49.Retnakaran R, Flock G, Giguère V. Identification of RVR, a novel orphan nuclear receptor that acts as a negative transcriptional regulator. Mol Endocrinol. 1994;8:1234–1244. doi: 10.1210/mend.8.9.7838156. [DOI] [PubMed] [Google Scholar]
- 50.Saatcioglu F, Lopez G, West B L, Zandi E, Feng W, Lu H, Esmaili A, Apriletti J W, Kushner P J, Baxter J D, Karin M. Mutations in the conserved C-terminal sequence in thyroid hormone receptor dissociate hormone-dependent activation from interference with AP-1 activity. Mol Cell Biol. 1997;17:4687–4695. doi: 10.1128/mcb.17.8.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sande S, Privalsky M L. Identification of Tracs (T-3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 52.Schulman I G, Juguilon H, Evans R M. Activation and repression by nuclear hormone receptors: hormone modulates an equilibrium between active and repressive states. Mol Cell Biol. 1996;16:3807–3813. doi: 10.1128/mcb.16.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 54.Tanner M M, Tirkkonen M, Kallioniemi A, Collins C, Stokke T, Karhu R, Kowbel D, Shadravan F, Hintz M, Kuo W L, et al. Increased copy number at 20q13 in breast cancer: defining the critical region and exclusion of candidate genes. Cancer Res. 1994;54:4257–4260. [PubMed] [Google Scholar]
- 55.Tanner M M, Tirkkonen M, Kallioniemi A, Isola J, Kuukasjarvi T, Collins C, Kowbel D, Guan X Y, Trent J, Gray J W, Meltzer P, Kallioniemi O P. Independent amplification and frequent co-amplification of three nonsyntenic regions on the long arm of chromosome 20 in human breast cancer. Cancer Res. 1996;56:3441–3445. [PubMed] [Google Scholar]
- 56.Tini M, Otulakowski G, Breitman M L, Tsui L-T, Giguère V. An everted repeat mediates retinoic acid induction of the γF-crystallin gene: evidence of a direct role for retinoids in lens development. Genes Dev. 1993;7:295–307. doi: 10.1101/gad.7.2.295. [DOI] [PubMed] [Google Scholar]
- 57.Tremblay A, Tremblay G B, Labrie C, Labrie F, Giguère V. EM-800, a novel antiestrogen, acts as a pure antagonist of the transcriptional functions of estrogen receptors α and β. Endocrinology. 1998;139:111–118. doi: 10.1210/endo.139.1.5702. [DOI] [PubMed] [Google Scholar]
- 58.Tremblay G B, Tremblay A, Copeland N G, Gilbert D J, Jenkins N A, Labrie F, Giguère V. Cloning, chromosomal localization and functional analysis of the murine estrogen receptor β. Mol Endocrinol. 1997;11:353–365. doi: 10.1210/mend.11.3.9902. [DOI] [PubMed] [Google Scholar]
- 59.Tremblay G B, Tremblay A, Labrie F, Giguère V. Ligand-independent activation of the estrogen receptors α and β by mutations of a conserved tyrosine can be abolished by antiestrogens. Cancer Res. 1998;58:877–881. [PubMed] [Google Scholar]
- 60.Treuter E, Johansson L, Thomsen J S, Wärnmark A, Leers J, Pelto-Huikko M, Sjöberg M, Wright A P, Spyrou G, Gustafsson J Å. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J Biol Chem. 1999;274:6667–6677. doi: 10.1074/jbc.274.10.6667. [DOI] [PubMed] [Google Scholar]
- 61.Umesono K, Giguère V, Glass C K, Rosenfeld M G, Evans R M. Retinoic acid and thyroid hormone induce gene expression through a common responsive element. Nature. 1988;336:262–265. doi: 10.1038/336262a0. [DOI] [PubMed] [Google Scholar]
- 62.Wakeling A E, Bowler J. ICI 182,780, a new antioestrogen with clinical potential. J Steroid Biochem Mol Biol. 1992;43:173–177. doi: 10.1016/0960-0760(92)90204-v. [DOI] [PubMed] [Google Scholar]
- 63.White R, Sjöberg M, Kalkhoven E, Parker M G. Ligand-independent activation of the estrogen receptor by mutation of a conserved tyrosine. EMBO J. 1997;16:1427–1435. doi: 10.1093/emboj/16.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang N N, Venugopalan M, Hardikar S, Glasebrook A. Identification of an estrogen response element activated by metabolites of 17β-estradiol and raloxifene. Science. 1996;273:1222–1225. doi: 10.1126/science.273.5279.1222. [DOI] [PubMed] [Google Scholar]
- 65.Zamir I, Dawson J, Lavinsky R M, Glass C K, Rosenfeld M G, Lazar M A. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci USA. 1997;94:14400–14405. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Hu X, Lazar M A. A novel role for helix 12 of retinoid X receptor in regulating repression. Mol Cell Biol. 1999;19:6448–6457. doi: 10.1128/mcb.19.9.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]