Summary
Wheat stem (or black) rust, caused by Puccinia graminis f. sp. tritici (Pgt), has been historically among the most devastating global fungal diseases of wheat. The recent occurrence and spread of new virulent races such as Ug99 have prompted global efforts to identify and isolate more effective stem rust resistance (Sr) genes. Here, we report the map‐based cloning of the Ug99‐effective SrTm5 gene from diploid wheat Triticum monococcum accession PI 306540 that encodes a typical coiled‐coil nucleotide‐binding leucine‐rich repeat protein. This gene, designated as Sr22b, is a new allele of Sr22 with a rare insertion of a large (13.8‐kb) retrotransposon into its second intron. Biolistic transformation of an ~112‐kb circular bacterial artificial chromosome plasmid carrying Sr22b into the susceptible wheat variety Fielder was sufficient to confer resistance to stem rust. In a survey of 168 wheat genotypes, Sr22b was present only in cultivated T. monococcum subsp. monococcum accessions but absent in all tested tetraploid and hexaploid wheat lines. We developed a diagnostic molecular marker for Sr22b and successfully introgressed a T. monococcum chromosome segment containing this gene into hexaploid wheat to accelerate its deployment and pyramiding with other Sr genes in wheat breeding programmes. Sr22b can be a valuable component of gene pyramids or transgenic cassettes combining different resistance genes to control this devastating disease.
Keywords: Sr22b, stem rust, resistance gene, CC‐NBS‐LRR, introgression, wheat
Introduction
Wheat is an important cereal crop that contributes a substantial proportion of the calories and proteins consumed by humankind. Reducing yield losses inflicted by pathogens can contribute to grain yield improvements that are required to feed a growing world population. Puccinia graminis f. sp. tritici (Pgt), the causal agent of wheat stem rust (or black rust), has historically been a devastating fungal disease of tetraploid and hexaploid wheat. In the past, this pathogen was effectively controlled by growing resistant wheat varieties and eradicating alternate host (Berberis vulgaris) plants around cereal fields (Roelfs, 1982, 1985; Singh et al., 2015).
After the year 1998, this disease became a major concern again after the emergence and spread of the Pgt race TTKSK (Ug99) and its variants (henceforth the Ug99 race group), which were virulent on the majority of resistance genes deployed worldwide, including resistance genes Sr31 and Sr38 (Pretorius et al., 2000; Singh et al., 2006, 2011). In recent years, additional highly virulent Pgt races unrelated to Ug99, such as TRTTF, TKTTF and TTRTF (Olivera et al., 2012, 2015; Patpour et al., 2017; Tesfaye et al., 2020), have been detected in outbreaks in Africa (Olivera et al., 2015), Asia (Shamanin et al., 2016, 2018) and Europe (Bhattacharya, 2017; Olivera et al., 2017). Due to the threat of these new virulent Pgt races, there is an urgent need to identify and isolate new effective Sr genes to diversify the sources of resistance in wheat breeding programmes.
Over 60 stem rust resistance genes (Sr1–Sr61) have been assigned official designations (Chen et al., 2020; Zhang et al., 2020), among which a large proportion were introgressed from wild wheat relatives (Singh et al., 2015). The diploid wheat species Triticum monococcum (einkorn, genome Am), comprising of the domesticated T. monococcum ssp. monococcum and the wild T. monococcum ssp. aegilopoides, is closely related to T. urartu (genome Au), the donor of the A genome in polyploid wheat (Dvorak and McGuire, 1988). T. monococcum harbours several valuable rust resistance genes, including the leaf rust resistance genes LrTM16 (Sodkiewicz and Strzembicka, 2008) and Lr63 (Kolmer and Anderson, 2010); the stripe rust resistance loci QYrtm.pau‐2A and QYrtb.pau‐5A (Chhuneja et al., 2008) and Yr34 (Chen et al., 2021); and the stem rust resistance genes Sr21 (Chen et al., 2015; The, 1973), Sr22 (Gerechter‐Amitai et al., 1971), Sr35 (McIntosh et al., 1984), SrTm4 (Briggs et al., 2015) and Sr60 and SrTm5 (Chen et al., 2018a).
Triticum monococcum chromosomes can recombine with the A‐genome chromosomes of polyploid wheat, particularly in the presence of the ph1b mutation (Dubcovsky and Luo, 1995). This feature has fuelled interest of scientists and breeders in the identification and isolation of stem rust resistance genes from this species and its transfer to commercial wheat cultivars. Among the six stem rust resistance genes derived from T. monococcum, four officially named ones (Sr21, Sr22, Sr35 and Sr60) have been successfully cloned and transferred into hexaploid wheat so far (Chen et al., 2020; Chen et al., 2018b; Saintenac et al., 2013; Steuernagel et al., 2016). The first three are Ug99‐resistance genes encoding typical coiled‐coil nucleotide‐binding leucine‐rich repeat (CC‐NBS‐LRR) proteins (Chen et al., 2018b; Saintenac et al., 2013; Steuernagel et al., 2016), whereas Sr60 encodes a different type of protein with two putative kinase domains (Chen et al., 2020).
Cultivated T. monococcum accession PI 306540 was identified as having a unique resistance response to five Pgt isolates (Rouse and Jin, 2011a, b), which was subsequently associated with the presence of stem rust resistance genes SrTm4, Sr21, Sr60 and SrTm5 (Briggs et al., 2015; Chen et al., 2018a, b). SrTm5 was previously mapped to the same region as Sr22 on the long arm of chromosome 7Am, and showed good levels of resistance (IT = ; to ;1) to several Pgt races, including TTKSK, TTKST and MCCFC (Chen et al., 2018a). Based on its mapped location and its different resistance profiles from Sr22, it was hypothesized that SrTm5 could be a novel allele of Sr22 or a tightly linked gene (Chen et al., 2018a).
In this study, we describe the map‐based cloning of the stem rust resistance gene SrTm5, and confirm that it is a new allele of the cloned gene Sr22. SrTm5 was roughly 96% identical to the reported Sr22 proteins and showed a characteristic insertion of 13.8‐kb retrotransposon in its second intron. We successfully introgressed a T. monococcum chromosome segment carrying SrTm5 into hexaploid wheat and developed a diagnostic molecular marker to accelerate its deployment in wheat breeding programmes.
Results
Assessment of stem rust responses
At the seedling stage, the SrTm5 monogenic line TmR54‐3 exhibited high levels of resistance (Its = ; to ;1) to Pgt races 34PKUSC, 34MTGSM and TTKSK, but was susceptible (ITs = 3+) to the other three races BCCBC, 21C3CTTTM and RTJRM. By contrast, its sister line TmS57‐57 without SrTm5 displayed susceptible infection types (ITs = 3+) to all the tested races (Figure S1a and Table S1). When inoculated with race 34PKUSC, selected F5 families from the SrTm5 segregating mapping population showed infection types that ranged from ‘;’ to ‘1’ in resistant plants, and from ‘3’ to ‘4’ in susceptible plants (Figure S1b).
To quantify the infected leaf area, we measured the percentage of the leaf area covered with Pgt pustules on six independent infected leaves of TmR54‐3 and TmS57‐57 using the software ASSESS version 2. For SrTm5‐avirulent races 34PKUSC, 34MTGSM and TTKSK, the average percentage was significantly lower (P < 0.001) in plants carrying SrTm5 than in those without the gene (Figure S1).
Map‐based cloning of SrTm5
The initial mapping of SrTm5 suggested that this gene was either a novel allele of Sr22 (TraesCS7A02G499600) or a tightly linked gene (Chen et al., 2018a). Since Sr22 is located on the long arm of chromosome 7A at 689.9 Mb (Chinese Spring RefSeq v1.0; The International Wheat Genome Sequencing Consortium, 2018), we developed Cleaved Amplified Polymorphic Sequence (CAPS) markers pkw4974 (690.9 Mb) and pkw5009 (688.2 Mb) (Table 1) flanking the Sr22 locus. Subsequently, we used these two markers to screen a population of 1132 plants (2264 gametes) from the cross PI 306540 × PI 272557, and we found 51 plants carrying recombination events within this region (2.7 Mb or 2.3 cM). Evaluations of progeny of these plants with race 34PKUSC confirmed that SrTm5 was located within this region. Using nine new markers spanning the 2.7 Mb (Table 1), we further delimited the SrTm5 candidate region to a 0.08‐cM interval (140.4 kb, CS RefSeq v1.0 coordinates) flanked by CAPS markers pkw4995 and pkw4999 (Figure 1b).
Table 1.
Primers used in the present study
| Marker | ID in CS RefSeq v1.1 | Primer sequence 5'–3' (Forward) | Primer sequence 5'–3' (Reverse) | Size (bp) | Enzyme | Function |
|---|---|---|---|---|---|---|
| pkw4974 | TraesCS7A02G497400 | GCACTCCAGGTGTCGCTCAG | ACCATTTCTCGCCGCTGTTC | 619 | HaeIII | Fine mapping |
| pkw4982 | TraesCS7A02G498200 | GTATGTGAAATAGAAAATGGGCAAC | CATAAGATTGCTGCCAAAGAACT | 944 | MfeI | Fine mapping |
| pkw4984 | TraesCS7A02G498400 | CCATTTGCTCCCACGAACA | CCCCATCAAGCCACTCTAT | 607 | MboII | Fine mapping |
| Pkw4990 | TraesCS7A02G499000 | TGAAAGGGAAGGTGAAGGA | AGGTGGAGGTTAAGGCGAG | 970 | BsaJI | Fine mapping |
| pkw4995 | TraesCS7A02G499500 | CTCAGAACACGGCTTCAACA | GATCACATGGACCTTCATCG | 900 | SspI | Fine mapping |
| Tm5F3R4 | TraesCS7A02G499600 | TGGAGAAAGTGGACAAGAT | GCTGCTCTATCTTCGGTTG | 971 | PvuII | Fine mapping |
| TM5TF3R3 | TraesCS7A02G499600 | GGATTTAGGGTTTCGGGGA | CCAACTACCACCACGGACG | 1137 | – | Fine mapping |
| pkw4997 | TraesCS7A02G499700 | TATGCCCAAAAGGAGTAGG | TACATCCTGTAGGACAAAACTG | 709 | AccI | Fine mapping |
| pkw4999 | TraesCS7A02G499900 | TGTCTACTGCATGAAGTTCAACC | AGCGGTCTCATTGACGGAA | 799 | AatII | Fine mapping |
| pkw5001 | TraesCS7A02G500100 | CGGTGTAGCATACCATTTCG | TTTCTTGTAGAGCGGGAGC | 1448 | – | Fine mapping |
| pkw5003 | TraesCS7A02G500300 | CTGTTGCTCAACGCCCATCTC | GATCACGTCGGGCATGAACTTATA | 675 | SmaI | Fine mapping |
| pkw5009 | TraesCS7A02G500900 | TCTTGCTGTTGCTTGGCTGTC | TGTCCCGCCTGTTGTTCCT | 1205 | SphI | Fine mapping |
| TM5TF2R2 | TraesCS7A02G499600 | GCACTGAGACTCCTCGGTGATGT | CACTCATATTACCCCCTTCCTTACC | 673 | – | MAS |
| A120F6R6 | TraesCS7A02G499600 | AAGAACTTGCTGCCGGACAT | AATCTTGTACCTTGAAAATCTGTCG | 108 | – | Expression analysis |
| HL‐F61R60 | TraesCS7A02G499600 | GTTGCAGAGTTTTCGGGTTTACC | GGCTTTCCGATGAAGTCATAGAA | 109 | – | Expression analysis |
| 4997QF2R2 | TraesCS7A02G499700 | CCAAAAGGAGTAGGAGTACA | ACGCATCATATCAAAGAAAC | 260 | – | Semi‐quantitative PCR |
| 4998QF5R5 | TraesCS7A02G499800 | CATTCTAAAGGTGTGATGGATTA | ATTGGCCTTTCTGAGGTTGG | 272 | ‐ | Semi‐quantitative PCR |
| TM5AF6R8 | TraesCS7A02G499600 | CTAGACAATTACATCAAGGTATA | GGGTATCAATCCAATCATCTCAATA | 1688 | Sequencing | |
| TM5AF4R4 | TraesCS7A02G499600 | GGTGTCCTCTCTCTGTAAACTGG | ATCTATTTGCTCGTCTCGTAACATA | 649 | Sequencing | |
| cfa2049 | – | TAATTTGATTGGGTCGGAGC | CGTGTCGATGGTCTCCTTG | – | Introgression | |
| wmc405 | – | GTGCGGAAAGAGACGAGGTT | TATGTCCACGTTGGCAGAGG | – | Introgression | |
| cfd68 | – | TTTGCAGCATCACACGTTTT | AAAATTGTATCCCCCGTGGT | – | Introgression | |
| gwm260 | – | GCCCCCTTGCACAAATC | CGCAGCTACAGGAGGCC | – | Introgression | |
| barc108 | – | GCGGGTCGTTTCCTGGAAATTCATCTAA | GCGAAATGATTGGCGTTACACCTGTTG | – | Introgression | |
| barc121 | – | ACTGATCAGCAATGTCAACTGAA | CCGGTGTCTTTCCTAACGCTATG | – | Introgression | |
| wmc790 | – | AATTAAGATAGACCGTCCATATCATCCA | CGACAACGTACGCGCC | – | Introgression |
Figure 1.
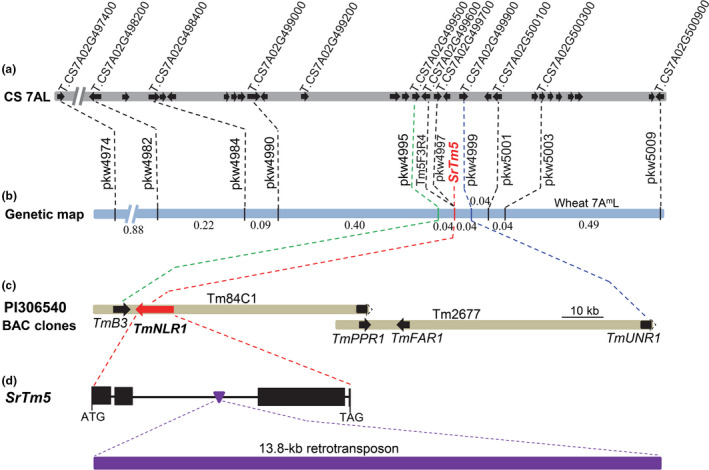
Map‐based cloning of SrTm5. (a) Collinear region on chromosome arm 7AL of Chinese Spring (RefSeq v1.1). Arrows represent genes. (b) High‐density genetic map of SrTm5 using 2264 segregating gametes. (c) Predicted genes in the SrTm5 candidate region constructed with two overlapping BACs from the resistant parent PI 306540. Dotted lines in arrows indicate deleted partial gene coding regions in BACs. (d) Gene structure of SrTm5 in PI 306540. Black rectangles indicate exons and black lines indicate introns; the purple inverted triangle in the second intron indicates the insertion of a retrotransposon.
Only three complete genes (TraesCS7A02G499600, TraesCS7A02G499700 and TraesCS7A02G499800) were annotated in the Chinese Spring reference genome within this region (Figure 1a). To determine if additional genes were present in the orthologous region in T. monococcum, we screened the bacterial artificial chromosome (BAC) library of resistant parent PI 306540 using the two flanking markers (pkw4995 and pkw4999) and two markers completely linked to SrTm5 (Tm5F3R4 and pkw4997). We obtained two overlapping BAC clones designated hereafter as Tm84C1 and Tm2677. Sequencing and annotation of these two selected BACs (Figure 1c; GenBank accession MZ327628) showed no additional genes in the SrTm5 candidate region in PI 306540 (146.5 kb) relative to Chinese Spring.
We designated the T. monococcum orthologues of Chinese Spring genes TraesCS7A02G499600, TraesCS7A02G499700 and TraesCS7A02G499800 as TmNLR1, TmPPR1 and TmFAR1 respectively. TmPPR1 encodes a protein containing pentatricopeptide repeat domains, whereas TmFAR1 encodes a far1‐related sequence 5‐like protein. We were not able to detect transcripts of these two genes in the leaves of SrTm5‐resistant T. monococcum plants infected with Pgt (Figure S2), suggesting that they are unlikely candidate genes for SrTm5.
TmNLR1 is an orthologue of the cloned stem rust resistance gene Sr22 (TraesCS7A02G499600) (Steuernagel et al., 2016) and therefore an excellent candidate gene for SrTm5. In PI 306540, the TmNLR1 gene spans 19715 bp from start to stop codons, including the insertion of a 13.8‐kb gypsy‐like retrotransposon in the second intron (Figure 1d). Comparing the TmNLR1 genomic region with the full‐length complementary DNA (cDNA) of TmNLR1, we determined that this gene contains four exons. The 2817 bp coding sequence encodes a typical CC‐NBS‐LRR protein containing 938 amino acids that were 95.7%–96.7% identical to six reported Sr22‐resistant protein haplotypes (Figure S3).
Three lines of evidence support TmNLR1 as the best candidate for SrTm5. First, TmNLR1 is the only candidate gene that is expressed in infected leaves of the resistant parent. Second, the TmNLR1 allele from PI 306540 shares the diagnostic amino acids present in known Sr22‐resistant alleles, whereas PI 272557 shares the diagnostic amino acids for the susceptible alleles (V381L, S605F/Y and G655D, BLOSUM62 scores = 1, −2 and −1, Table S2). Finally, sequencing of TmNLR1 in T. monococcum accession PI 277131‐2, which was previously postulated to possess SrTm5 (Rouse and Jin, 2011a), confirmed the presence of a gene 100% identical to TmNLR1. Based on these results, we selected TmNLR1 for further functional characterizations.
Validation of TmNLR1 by transgenic complementation
To test if TmNLR1 was sufficient to confer resistance to Pgt, we transformed the Ug99‐susceptible wheat variety Fielder with the PI 306540 circular BAC plasmid Tm84C1, which includes two complete genes TmB3 and TmNLR1, and about 70% of the coding sequence of TmPPR1 (Figure 1c). Gene TmB3 is orthologous to Chinese Spring gene TraesCS7A02G499500 and encodes a B3 domain‐containing protein likely to be involved in plant growth and development (Peng and Weselake, 2013; Waltner et al., 2005). Among them, only TmNLR1 was expressed in infected leaves and co‐segregated with the disease phenotypes.
We obtained eight independent T0 transgenic plants, for which we confirmed the presence of the TmNLR1 transgene using markers Tm5F3R4, TM5TF2R2 and TM5TF3R3 (Table 1). We genotyped more than 20 T1 plants from each transgenic family, and all except one showed significant segregation distortion from the 3 : 1 (transgenic/non‐transgenic) segregation expected from a single copy of transgene, with an excess of non‐transgenic plants (Table S3). We also genotyped T2 plants derived from one single positive T1 plant per event. Families T2‐Tm505‐15, T2‐Tm514‐2, T2‐Tm517‐1, T2‐Tm548‐3, T2‐Tm554‐2 and T2‐Tm558‐7 were fixed for the transgene (all plants are positive). Families T2‐Tm515‐6 and T2‐Tm547‐3 displayed a distorted segregation ratio from the expected 3+ : 1‐ with an excess of non‐transgenic plants close to a 1 : 1 segregation (Table S3). Taken together, these results suggest some segregation distortion against the transgene.
Transcript levels of TmNLR1 in all transgenic T1 families were significantly higher than in the susceptible control Fielder (P < 0.01), but only five of them (T1‐Tm514, T1‐Tm515, T1‐Tm517, T1‐Tm548 and T1‐Tm554) were expressed at similar levels as in the introgression of the T. monococcum chromosome segment including SrTm5 into Fielder (positive control, see later) (Figure S4).
Roughly 25 T2 plants from each transgenic event and the untransformed control Fielder were challenged with Pgt race TTKSK (isolate 04KEN156/04). All plants from T2 transgenic families T2Tm514‐2 and T2Tm517‐1 fixed for the transgene showed high levels of resistance (Figure 2a), whereas resistance in Tm515‐6 T2 plants perfectly co‐segregated with the presence of the transgene (Figure S5). Measures of the percentage of leaf area covered by Pgt pustules was significantly lower (P < 0.0001) in the resistant transgenic plants of these three families (ranging from 1.3% to 9.2%) than in the non‐transgenic Fielder control (ranging from 10.3% to 24.6%) (Figure 2b). The progeny of the other five transgenic families displayed susceptible reactions similar to Fielder in all plants suggesting that the resistance gene was broken or damaged during the bombardment insertion. These transgenic families were discarded for further analysis.
Figure 2.
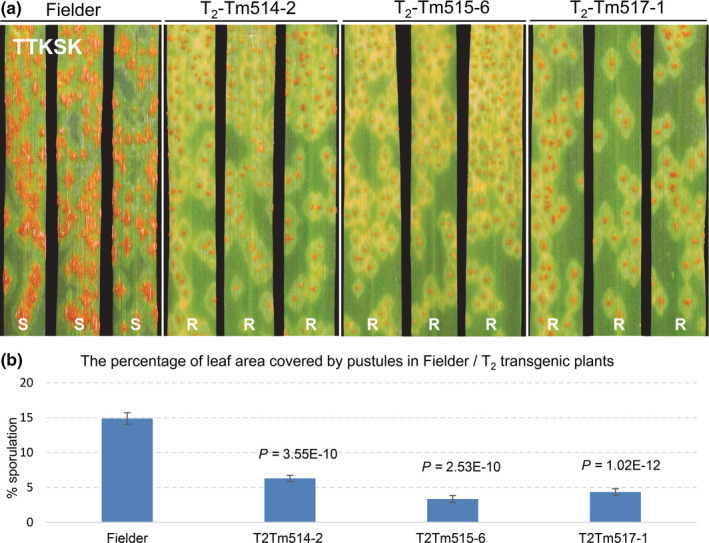
Gene TmNLR1 confers resistance when transferred into the susceptible wheat variety Fielder. (a) Reactions to Pgt race TTKSK (isolate 04KEN156/04) in Fielder control and three transgenic families T2Tm514‐2, T2Tm515‐6 and T2Tm517‐1. S, susceptible; R, resistant. (b) The average percentage of the leaf area covered by Pgt pustules was measured using the software ASSESS v.2. More than 20 independent T2 plants were evaluated. Error bars are standard errors of the mean.
To test if the transgenic plants had the same resistance profile as the natural SrTm5 gene in monogenic line TmR54‐3, we inoculated transgenic family T2Tm514‐2 (homozygous for the transgene) with another two Pgt races RTJRM and 21C3CTTTM, which are virulent on SrTm5 in T. monococcum. Plants from T2Tm514‐2 showed susceptible reactions similar to Fielder when challenged with SrTm5‐virulent races RTJRM and 21C3CTTTM (Figure S6) but were resistant when challenged with TTKSK (Figure 2), suggesting similar race specificity between the transgene and natural SrTm5 in T. monococcum.
Taken together, the map‐based cloning and transgenic complementation results demonstrate that SrTm5 is an allele of the cloned gene Sr22. Based on its different resistance profiles (Table S4), we designated the R1 (Schomburgk/PI 660256) and R4 (PI 190945) haplotypes as allele Sr22a, and SrTm5 as allele Sr22b. This nomenclature has been approved by the Catalogue of Gene Symbols for wheat.
Effect of Pgt inoculation on transcript levels of Sr22b
We analysed Sr22b transcript levels relative to ACTIN in the monogenic line TmR54‐3 by qRT‐PCR. We found no significant transcriptional differences between plants inoculated with Sr22b‐avirulent Pgt race 34PKUSC and mock inoculated with water at 1, 3 and 6 days post inoculation (dpi) (Figure 3), suggesting that Sr22b is not induced by the presence of the Pgt pathogen. We also compared the transcript levels of Sr22a in T. monococcum accession PI 190945 and Sr22b in T. monococcum line TmR54‐3 before inoculation and found no significant differences between them (Figure S7).
Figure 3.
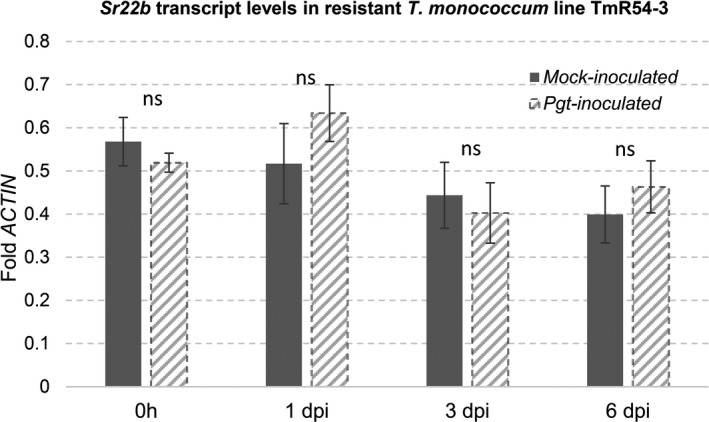
Transcript levels of Sr22b in mock‐inoculated and Pgt‐inoculated T. monococcum plants. Leaves were collected from Sr22b monogenic line TmR54‐3 at four time points: 0 h, 1 dpi, 3 dpi and 6 dpi. Plants were grown in growth chambers at 22 °C day/20 °C night with 16 h light/8 h dark. Transcript levels were expressed as fold‐ACTIN (n = 4). ns = not significant; Error bars are standard errors of the mean.
Sr22b is present only in T. monococcum
The dominant marker TM5TF2R2 was designed based on the special polymorphism (the insertion of repetitive sequence in the second intron) that differentiates Sr22b from the cloned Sr22‐resistant haplotypes and all susceptible alleles. The forward primer was designed in the second intron and the reverse primer in the inserted retrotransposon. Amplification with PCR marker TM5TF2R2 at an annealing temperature of 60 °C generates an amplicon of 673 bp only when the gene Sr22b is present (Figure S8). Using this marker, we evaluated a collection of 165 wheat accessions, including 89 accessions of T. monococcum, 23 of T. turgidum and 53 of T. aestivum. PCR products were present only in 13 (14.6%) of the Triticum monococcum accessions but were absent in all tetraploid and hexaploid wheat lines tested in this study (Table S5). These observations were consistent with Sanger sequencing results using two pairs of primers TM5AF6R8 and TM5AF4R4 (Table 1), which were designed to amplify the LRR region of Sr22. The 13 T. monococcum accessions with the retrotransposon insertion, all carry the Sr22b haplotype in the LRR coding region, whereas all the other accessions have different haplotypes in the coding region and lack the retrotransposon insertion.
We then used the TM5TF2R2 marker to explore the presence of Sr22b in T. monococcum accessions PI 355538, PI 362610 and PI 377668 from the Balkans (Table S6), which were previously postulated to carry an unknown Pgt resistance gene different from Sr21 based on their different resistance reactions to races BCCBC and MCCFC (Chen et al., 2018b). We found that these three lines have Sr22b, which can explain their resistance to Pgt race MCCFC but susceptibility to BCCBC. This was confirmed by phenotyping 48 plants with race 34PKUSC in three F2 populations derived from crosses between PI 355538, PI 362610 and PI 377668 and the susceptible accession PI 272557. Genotyping with marker TM5TF2R2 showed that all plants in which the 673‐bp fragment was amplified were resistant, whereas all plants without PCR products were susceptible. Moreover, we sequenced the coding regions of Sr22 from PI 355538, PI 362610 and PI 377668, and found that they were all 100% identical to Sr22b in PI 306540. These results confirmed that the resistance to MCCFC and 34PKUSC in these accessions was conferred by Sr22b.
Introgression of Sr22b into hexaploid wheat background
Figure 4a describes the crosses involved in the generation of the Sr22b introgression into hexaploid wheat. The diagnostic marker TM5TF2R2 and the closely linked CAPS marker pkw4974 (Table 1) were used for monitoring the presence of T. monococcum chromatin during backcrosses and for the final selection of BC3F2 plants homozygous for Sr22b. We confirmed the absence of stem rust resistance genes Sr13, Sr60, Sr21 and SrTm4 from the parental lines using diagnostic or closely linked markers (Briggs et al., 2015; Chen et al., 2020; Chen et al., 2018b; Zhang et al., 2017).
Figure 4.
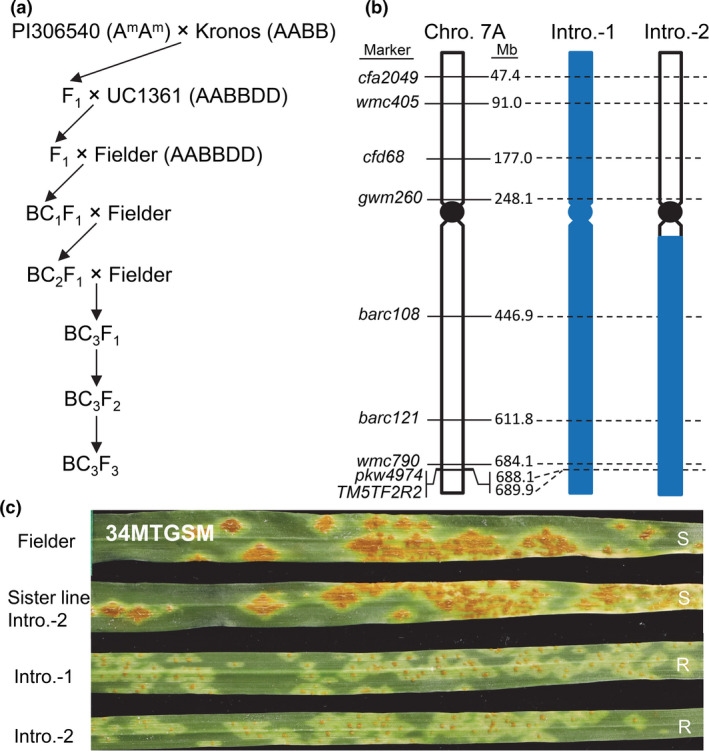
Introgression of Sr22b into common wheat background. (a) The procedure for the production of Sr22b introgression lines. Markers TM5TF2R2 and pkw4974 (digested with HaeIII; Table 1) were used for confirming the presence of T. monococcum chromatin. (b) Markers on chromosome 7A were used to determinate the length of the introgression segments. The physical locations of polymorphic markers were based on the Chinese Spring reference genome Refseq v1.0. Blue rectangles indicate T. monococcum chromatin. (c) Infection types from Fielder control, introgression lines Intro.‐1 and Intro.‐2 and its sister line (named ‘Sister line Intro.‐2’) lacking Sr22b. BC3F3 plants were challenged with Pgt race 34MTGSM. S, susceptible; R, resistant.
To determine the size of the 7Am chromosome region introgressed into hexaploid wheat, we first screened lines PI 306540, Kronos and Fielder for polymorphisms using 23 SSR markers distributed along chromosome 7A. We obtained seven polymorphic markers (Table 1) and determined their physical locations in the Chinese Spring reference genome (Refseq v1.0; Figure 4b). We genotyped 13 BC3F1 plants with markers TM5TF2R2 and pkw4974, and detected five plants with the 7AmL introgression. BC3F1 plants 1, 3, 4 and 5 carried the 7AmL alleles for all the tested markers extending from 47.4 Mb to 689.9 Mb suggesting that they are disomic 7Am (7A) substitution lines (Intro. 1 henceforth). The T. monococcum segment in plant number 2 extended from 446.9 Mb (barc108) to 689.9 Mb (TM5TF2R2), indicating a translocation of part of the long arm (referred hereafter as Intro.2, Figure S9). All these plants exhibited good levels of fertility when self‐pollinated.
Homozygous BC3F3 plants from these introgression lines challenged with Chinese Pgt race 34MTGSM showed good levels of resistance, whereas the recurrent parent Fielder and its sister line lacking Sr22b were completely susceptible (Figure 4c). Small amounts of BC3F3 seeds from the introgression lines are available by request from the senior authors. After the seed is increased, it will be deposited in the National Small Grain Collection in the United States and in the Chinese Crop Germplasm Resources Information System (CGRIS) in China.
Discussion
In this study, we confirmed that SrTm5 is a new allele of Sr22, officially designated as Sr22b. The stem rust resistance gene Sr22 was previously identified to encode a coiled‐coil nucleotide‐binding leucine‐rich repeat protein, which confers broad‐spectrum resistance to commercially important Pgt races, including the Ug99 race group (Steuernagel et al., 2016). Sr22b and Sr22a both confer strong levels of resistance to Pgt races TTKSK (Ug99), TTKST, MCCFC, 34MTGSM and 34PKUSC, but differ in that Sr22b is susceptible to races BCCBC, 21C3CTTTM, RTJRM, QFCSC, TRTTF and TTTTF and Sr22a is not (Table S4). These results suggest that the Sr22a allele (R1 and R4 haplotypes) confers a broader resistance to tested Pgt races than Sr22b (Table S4). We currently do not know whether the other four Sr22‐resistant haplotypes (R2, R3, R5 and R6, Figure S3) have different resistance profiles because monogenic lines are not available for these haplotypes.
The different Pgt resistance profiles of Sr22a and Sr22b were associated with more than 30 polymorphisms, located mostly within the LRR region (Figure S3). The LRR domain of plant NLR genes is known to play a major role in pathogen recognition specificity, and diversifying selection drives higher levels of sequence variation (Dodds et al., 2006; Jiang et al., 2007; Krasileva and Dahlbeck, 2010). The different resistance profiles of Sr22a and Sr22b provide a useful tool to study the recognition mechanisms between Sr22 and the corresponding Avr proteins.
Insertions of large retrotransposons into functional genes is not a rare phenomenon in wheat, and can result in loss of function if inserted in the coding region. Insertions in introns may or may not have functional effects in the expression of the gene. For example, the gene Zfp69 is disrupted by a inserted retrotransposon in its intron, which generates a truncated mRNA (Scherneck et al., 2009) and insertion of retrotransposons into the intron of Maize waxy gene caused alternative splicing (Varagona and Purugganan, 1992). Unlike these genes, the large retrotransposon insertion in the intron of Sr22b did not affect its expression levels or function (Figure S7). We used this distinctive retrotransposon insertion in Sr22b to develop a diagnostic marker for this allele.
The complete coding region, UTRs and the inserted retrotransposon of Sr22b were too large to clone into a binary vector for Agrobacterium‐mediated transformation, so we performed biolistic transformation using the circular BAC plasmid Tm84C1, which carries the 103.4‐kb genomic fragment of PI 306540 and the 8.1‐kb vector backbone sequence. Transformation with DNA fragments or circular plasmids larger than 100 kb has been previously reported in several plant species, such as tobacco (Wang et al., 2015), potato (Ercolano et al., 2004) and rice (Wang et al., 2015), but we are not aware of similar examples in wheat. Very large genes transformed by bombardment can be broken and disrupted (Liu et al., 2019; Makarevitch and Svitashev, 2003; Svitashev et al., 2002), which can explain the five confirmed transformation events that were susceptible to Pgt.
Fortunately, three independent events showed strong levels of resistance after infection with Pgt race TTKSK, indicating that the whole Sr22b gene was integrated into the plant genome in these three transgenic lines. We observed a significant segregation distortion against the transgene both in T1 and T2 families (Table S3), but the distortion was not that strong, and we were able to recover plants homozygous for the different transformation events that showed stable resistance to Pgt.
Sr22b was successfully introgressed into the common wheat variety Fielder, where it conferred good levels of resistance to Pgt (Figure 4). However, the sizes of the T. monococcum introgression are quite large, including the whole 7Am chromosome or most of the long arm of chromosome 7Am (Figure S9). More work will be needed to reduce the length of the introgressed T. monococcum chromosome segment to minimize potential linkage drag. Fortunately, recombination between the A and Am chromosomes can be restored to normal levels through using the ph1b mutation (Dubcovsky et al., 1995). The diagnostic marker for Sr22b and the flanking SSR markers (Figure S9; Table 1) will be useful tools to develop shorter T. monococcum introgression lines carrying Sr22b.
Sr22b is only present in few cultivated T. monococcum accessions but absent in all tested polyploid wheats, indicating that it has the potential to improve Ug99 resistance in a wide range of modern wheat cultivars. However, since Sr22b is susceptible to several Pgt races, it would be necessary to combine with other resistance genes to provide a broader virulence spectrum. Sr genes that are susceptible to race TTKSK but effective to other Pgt races could be considered as candidates for combination with Sr22b. Examples of these complementary genes include Sr60 (Chen et al., 2020), Sr8155B1 (Nirmala et al., 2017), Sr_TRTTF (Hiebert et al., 2017) and Sr9e (Olivera et al., 2012).
The cloning of SrTm5 demonstrated that it is a new allele of Sr22 and brings close to completion the characterization of all previously mapped stem rust resistance genes in T. monococcum (Sr21, Sr22, Sr35 and Sr60). The only mapped gene that has not been cloned yet is the recessive resistance gene SrTm4 (Briggs et al., 2015). This information expands our understanding of the role of different stem rust resistance gene combinations in the adaptation of diploid wheat to this damaging rust pathogen and provides an entry point to understand the recognition specificity of different Sr22 alleles to different Pgt races and effectors. From a practical point of view, the identification of Sr22b, its transfer to hexaploid wheat and the reliable diagnostic marker developed in this study provide a useful tool to diversify the Sr genes deployed in modern wheat breeding programmes.
Methods
T. monococcum materials and mapping populations
As a source of SrTm5, we used T. monococcum subsp. monococcum accession PI 306540, which was collected in Romania and that was previously shown to express the high levels of resistance to different Pgt races (Rouse and Jin, 2011a). PI 306540 was crossed with T. monococcum cultivated accession PI 272557, which does not carry any known Sr genes (Rouse and Jin, 2011b). Since PI 306540 carries multiple Sr genes, we selected F5 families segregating only for SrTm5 from the cross PI 306540 × PI 272557 (Chen et al., 2018a). A total of 2264 segregating gametes were used to construct a high‐density genetic map of SrTm5. From this population, we selected the monogenic F5 line TmR54‐3 homozygous for SrTm5 (without any of the other resistance genes) and the sister control line TmS57‐57 carrying no stem rust resistance gene.
A collection of 92 accessions of T. monococcum, 23 of T. turgidum and 53 of T. aestivum obtained from the US Department of Agriculture National Small Grains Collection (USDA‐NSGC, https://npgsweb.ars‐grin.gov/gringlobal/search) or the Chinese Crop Germplasm Resources Information System (CGRIS, http://www.cgris.net/cgris_english.html) were used to test the presence/absence of SrTm5.
Stem rust evaluation
Previously, infection types of SrTm5 to multiple Pgt races were reported, including TTKSK (isolate 04KEN156/04), TTKST (06KEN19v3), MCCFC (59KS19), QTHJC (75ND717C), QFCSC (06ND76C), SCCSC (09ID73‐2), TTTTF (01MN84A‐1‐2), TRTTF (06YEM34‐1) and TKTTF (13ETH18‐1 and 13GER15‐1) (Chen et al., 2018a). In this study, stem rust seedling tests were carried out in three institutions: Peking University Institute of Advanced Agricultural Sciences, Weifang, China; USDA‐ARS Cereal Disease Laboratory, Minnesota, USA; and University of California, Davis, USA. Selected sister lines TmR54‐3 and TmS57‐57 were re‐evaluated with race TTKSK (04KEN156/04). To expand the resistance profile of SrTm5, we also evaluated these lines with North American race BCCBC (09CA115‐2) and Chinese races 34MTGSM (20GSA1), 21C3CTTTM (20GH13), RTJRM (mutant strain, 20IAS11) and 34PKUSC (19IAS08) (Li et al., 2016, 2018; Zhao et al., 2015). The origin and virulence/avirulence profiles of these Pgt races are presented in supplemental Table S1. Procedures for inoculation and scoring infection types (ITs) were as previously reported (Rouse et al., 2011; Stakman and Stewart, 1962).
For plants carrying critical recombination events in the high‐density map, we preformed progeny tests including at least 25 progenies. These plants were inoculated with Chinese Pgt race 34PKUSC, and the percentage of the leaf area covered with pustules was estimated using the software ASSESS version 2.0 (American Phytopathological Society, St Paul, MN, USA) as reported previously (Lamari, 2008).
BAC library screening and sequencing
A non‐gridded BAC library from PI 306540 with roughly 5× genome equivalents was available at the Wheat Molecular Genetics Laboratory, University of California, Davis (Chen et al., 2020). A PCR screening was performed using increasingly diluted library samples following the manufacturer’s instruction (Amplicon Express Inc., Pullman, WA). Screening of the BAC library with PCR markers pkw4995, Tm5F3R4, pkw4997 and pkw4999 yielded two positive BAC clones Tm84C1 and Tm2677. High quality BAC DNAs were extracted using Qiagen Large‐Construct Kits (Qiagen, Hilden, Germany) and sequenced with Wideseq at Purdue University (https://purdue.ilabsolutions.com/landing/808). Repetitive elements were identified and annotated using the Cereal Repeat Sequences Database (https://wheat.pw.usda.gov/ITMI/Repeats/blastrepeats3.html). Candidate genes were annotated using the published reference genomes (The International Wheat Genome Sequencing Consortium, 2018; Walkowiak et al., 2020), and confirmed using the BLASTN/BLASTX searches available at National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/). Expression profiles were determined with the Wheat Expression Browser (expVIP, http://www.wheat‐expression.com/).
Wheat transformation
Bacterial artificial chromosome clone Tm84C1 containing 103 429 bp of T. monococcum PI 3065040 genomic sequence (GenBank accession MZ327628) was cloned into vector pCC1BAC (8128 bp). The cloned T. monococcum region carries complete genes TmB3 and TmNLR1 and a partial sequence of gene TmPPR1 (missing 30% of the distal coding region). Biolistic transformation was performed using a PDS1000/He particle bombardment system (Bio‐Rad, Hercules, CA, USA). The cloned BAC Tm84C1 was co‐transformed with plasmid pAHC20, which carries bialaphos (BAR) selectable marker gene. BAC DNAs were mixed in a 1 : 1 (1 : 1 for BAC DNA and pAHC20) molar ratio prior to bombardment. Transformation was performed using the Ug99‐susceptible spring wheat variety Fielder by biolistic bombardment as described previously (Zhang et al., 2015).
Positive transgenic plants were identified using dominant or codominant PCR markers Tm5F3R4, TM5TF2R2 and TM5TF3R3 (Table 1). Expression levels of TmNLR1 in transgenic plants were assessed by quantitative real‐time PCR (qRT‐PCR) with primer pairs HL‐F61R60. About 25 T2 transgenic seeds from each transgenic event were germinated and tested for their responses to Pgt race TTKSK (Ug99).
qRT‐PCR analysis
Plants from SrTm5 monogenic line TmR54‐3 were mock inoculated or Pgt inoculated in two independent chambers under the same environmental condition: 22 °C day/20 °C night and 16 h light/8 h dark. Total RNAs were extracted from leaves of different plants collected immediately after inoculation (0 h) and 1, 3 and 6 days post inoculation (dpi) using Spectrum Plant Total RNA Kit (Sigma‐Aldrich, Saint Louis, MO, USA). First‐strand cDNA was synthesized using the Applied Biosystems™ High‐Capacity cDNA Reverse Transcription Kits. qRT‐PCR reactions were performed on a QuantStudio™ 5 real‐time PCR system (Thermo Fisher Scientific, Inc., Waltham, MA, USA) using Fast SYBR GREEN reagents. PCR primers A120F6R6 (Table 1, 97% efficiency) were used to evaluate the effect of Pgt inoculation on SrTm5. Transcript levels were determined in four biological replicates and expressed as fold‐ACTIN levels as described previously (Pearce and Vanzetti, 2013).
Introgression of T. monococcum segments carrying SrTm5 into hexaploid wheat
The diploid wheat accession PI 306540 (AmAm) was used for transferring T. monococcum gene SrTm5 to hexaploid wheat variety Fielder using T. durum wheat variety Kronos (AABB) as bridging species (The, 1973). The F1 triploid plants from the cross of PI 306540 × Kronos were crossed with hexaploid wheat variety Clear White (UC1361), and the resulting F1 plants were backcrossed three times to the recurrent spring common wheat line Fielder. PCR markers TM5TF2R2 and pkw4974 (Table 1) were used to validate the presence of the introgressed T. monococcum segments during backcrossing. Five BC3F1 plants carrying alien chromosome segments were self‐pollinated and characterized with 23 simple sequence repeat (SSR) markers across chromosome 7A to analyse the length of introgressed T. monococcum segments. Subsequently, we selected BC3F2 plants homozygous for the introgressed T. monococcum segment to generate seeds. The resulting progeny were inoculated with Pgt race 34MTGSM.
Conflicts of interest
The authors declare that they have no conflict of interests.
Author contributions
JL and MNR performed most of the experimental work; YW and CG designed the transgenic experiments. BL performed the biolistic transformation and obtained T1 seeds. LeiH contributed qRT‐PCR and filled the gaps of BAC sequence; HnaL contributed primers development; TL performed part of the phenotyping experiments; WZ created the mapping population and contributed sequence analyses. SC analysed the data and wrote the first version of the manuscript. YW, SC and JD proposed and supervised the project, obtained the funding and generated the final version of the paper. All authors revised the manuscript and provided suggestions.
Supporting information
Figure S1 Reactions to six Pgt races 34PKUSC, 34MTGSM, TTKSK, BCCBC, 21C3CTTTM and RTJRM.
Figure S2 Semi‐quantitative PCR products from markers 4997QF2R2 (260 bp, TraesCS7A02G499700), 4998QF5R5 (272 bp, TraesCS7A02G499800) and ACTINF1R1 (ACTIN).
Figure S3 SrTm5 protein sequence analysis. Multiple sequence alignment between SrTm5 and reported Sr22‐resistant and susceptible protein sequences (Steuernagel et al. 2016).
Figure S4 Transcript levels of TmNLR1 in transgenic T1 families (three positive plants per event, n = 3).
Figure S5 Reactions to Pgt race TTKSK (Ug99) in transgenic family T2Tm515‐6.
Figure S6 Transgenic family T2Tm514‐2 homozygous for the transgene were inoculated with two SrTm5‐virulent Pgt races RTJRM and 21C3CTTTM.
Figure S7 Transcript levels and infection types of Sr22a and Sr22b in T. monococcum background.
Figure S8 PCR products from the Sr22b diagnostic marker TM5TF2R2.
Figure S9 Markers across chromosome 7A were used to analyse the length of introgressed T. monococcum segments.
Table S1 Avirulence/virulence formulae of Pgt races, and their responses to SrTm5.
Table S2 Comparison of SrTm5 protein with polymorphisms that discriminate perfectly between Sr22‐susceptible and ‐resistant haplotypes from Steuernagel et al. (2016).
Table S3 Segregation ratios in T1 and T2 transgenic families detected using PCR markers Tm5F3R4, TM5TF2R2 and TM5TF3R3 (Table 1).
Table S4 Resistance profiles of Sr22b (=SrTm5) and Sr22a (haplotypes R1 and R4) to multiple Pgt races.
Table S5 A collection of 92 accessions of T. monococcum, 23 of T. turgidum and 53 of T. aestivum was used to test the presence of Sr22b.
Table S6 Geographic distribution of T. monococcum accessions, and their reactions against Pgt races TTKSK, MCCFC and 34PKUSC.
Acknowledgements
We thank Dr. Jianhui Wu of Northwest Agriculture & Forestry University, Shanxi, China, for providing the Pgt race 34PKUSC. Work at JD laboratory was supported by the Howard Hughes Medical Institute and by the Agriculture and Food Research Initiative Competitive Grant 2017‐67007‐25939 (WheatCAP) from the USDA National Institute of Food and Agriculture (NIFA). Work at SC laboratory was supported by the Provincial Technology Innovation Program of Shandong. Work at YW laboratory was supported by the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2020000003). Work at the USDA‐ARS was supported by the USDA‐ARS National Plant Disease Recovery System.
Luo, J. , Rouse, M. N. , Hua, L. , Li, H. , Li, B. , Li, T. , Zhang, W. , Gao, C. , Wang, Y. , Dubcovsky, J. and Chen, S. (2022) Identification and characterization of Sr22b, a new allele of the wheat stem rust resistance gene Sr22 effective against the Ug99 race group. Plant Biotechnol. J., 10.1111/pbi.13737
Contributor Information
Yanpeng Wang, Email: yanpengwang@genetics.ac.cn.
Jorge Dubcovsky, Email: jdubcovsky@ucdavis.edu.
Shisheng Chen, Email: shisheng.chen@pku-iaas.edu.cn.
References
- Bhattacharya, S. (2017) Deadly new wheat disease threatens Europe's crops. Nature, 542, 145–146. [DOI] [PubMed] [Google Scholar]
- Briggs, J. , Chen, S. , Zhang, W. , Nelson, S. , Dubcovsky, J. and Rouse, M.N. (2015) Mapping of SrTm4, a recessive stem rust resistance gene from diploid wheat effective to Ug99. Phytopathology, 105, 1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Guo, Y. , Briggs, J. , Dubach, F. , Chao, S. , Zhang, W. , Rouse, M.N. et al. (2018a) Mapping and characterization of wheat stem rust resistance genes SrTm5 and Sr60 from Triticum monococcum . Theor. Appl. Genet. 131, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Hegarty, J. , Shen, T. , Hua, L. , Li, H. , Luo, J. , Li, H. et al. (2021) Stripe rust resistance gene Yr34 (synonym Yr48) is located within a distal translocation of Triticum monococcum chromosome 5AmL into common wheat. Theor. Appl. Genet. 134, 2197–2211. 10.1007/s00122-021-03816-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Rouse, M.N. , Zhang, W. , Jin, Y. , Akhunov, E. , Wei, Y. and Dubcovsky, J. (2015) Fine mapping and characterization of Sr21, a temperature‐sensitive diploid wheat resistance gene effective against the Puccinia graminis f. sp. tritici Ug99 race group. Theor. Appl. Genet. 128, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Rouse, M.N. , Zhang, W. , Zhang, X. , Guo, Y. , Briggs, J. and Dubcovsky, J. (2020) Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. New Phytol. 225, 948–959. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Zhang, W. , Bolus, S. , Rouse, M.N. and Dubcovsky, J. (2018b) Identification and characterization of wheat stem rust resistance gene Sr21 effective against the Ug99 race group at high temperature. PLoS Genet. 14, e1007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhuneja, P. , Kaur, S. , Garg, T. , Ghai, M. , Kaur, S. , Prashar, M. , Bains, N.S. et al. (2008) Mapping of adult plant stripe rust resistance genes in diploid A genome wheat species and their transfer to bread wheat. Theor. Appl. Genet. 116, 313–324. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.M. , Teh, T. , Wang, C.I. , Ayliffe, M.A. , Kobe, B. et al. (2006) Direct protein interaction underlies gene‐for‐gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl Acad. Sci. USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky, J. , Luo, M. and Dvorak, J. (1995) Differentiation between homoeologous chromosomes 1A of wheat and 1Am of Triticum monococcum and its recognition by the wheat Ph1 locus. Proc. Natl Acad. Sci. USA, 92, 6645–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak, J. , McGuire, P.E. and Cassidy, B. (1988) Apparent sources of the A genomes of wheats inferred from the polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome, 30, 680–689. [Google Scholar]
- Ercolano, M.R. , Ballvora, A. , Paal, J. , Steinbiss, H.‐H. , Salamini, F. and Gebhardt, C. (2004) Functional complementation analysis in potato via biolistic transformation with BAC large DNA fragments. Mol. Breeding, 13, 15–22. [Google Scholar]
- Gerechter‐Amitai, Z. , Wahl, I. , Vardi, A. and Zohary, D. (1971) Transfer of stem rust seedling resistance from wild diploid einkorn to tetraploid durum wheat by means of a triploid hybrid bridge. Euphytica, 20, 281–285. [Google Scholar]
- Hiebert, C.W. , Rouse, M.N. , Nirmala, J. and Fetch, T. (2017) Genetic mapping of stem rust resistance to Puccinia graminis f. sp. tritici race TRTTF in the Canadian wheat cultivar Harvest. Phytopathology, 107, 192–197. [DOI] [PubMed] [Google Scholar]
- Jiang, H. , Wang, C. , Ping, L. , Tian, D. and Yang, S. (2007) Pattern of LRR nucleotide variation in plant resistance genes. Plant Sci. 173, 253–261. [Google Scholar]
- Kolmer, J.A. , Anderson, J.A. and Flor, J.M. (2010) Chromosome location, linkage with simple sequence repeat markers, and leaf rust resistance conditioned by gene Lr63 in wheat. Crop Sci. 50, 2392–2395. [Google Scholar]
- Krasileva, K.V. , Dahlbeck, D. and Staskawicz, B.J. (2010) Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine‐rich repeat domain with the cognate oomycete effector. Plant Cell, 22, 2444–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamari, L. (2008) ASSESS 2.0: image analysis software for plant disease quantification. St Paul, MN: American Phytopathological Society. [Google Scholar]
- Li, T.Y. , Ma, Y.C. , Wu, X.X. , Chen, S. , Xu, X.F. , Wang, H. , Cao, Y.Y. et al. (2018) Race and virulence characterization of Puccinia graminis f. sp. tritici in China. PLoS ONE, 13, e0197579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Wu, X. , Xu, X. , Wang, W. and Cao, Y. (2016) Postulation of seedling stem rust resistance genes of Yunnan wheat cultivars in China. Plant Protect. Sci. 52, 242–249. [Google Scholar]
- Liu, J. , Nannas, N.J. , Fu, F.‐F. , Shi, J. , Aspinwall, B. , Parrott, W.A. and Dawe, R.K. (2019) Genome‐scale sequence disruption following biolistic transformation in rice and maize. Plant Cell, 31, 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch, I. , Svitashev, S. and Somers, D. (2003) Complete sequence analysis of transgene loci from plants transformed via microprojectile bombardment. Plant Mol. Biol. 52, 421–432. [DOI] [PubMed] [Google Scholar]
- McIntosh, R. , Dyck, P. , Cusick, J. and Milne, D. (1984) Cytogenetical studies in wheat XIII. Sr35, a third gene from Triticum monococcum for resistance to Puccinia graminis tritici . Z. Pflanzenzucht, 92, 1–14. [Google Scholar]
- Nirmala, J. , Saini, J. , Newcomb, M. , Olivera, P. , Gale, S. , Klindworth, D. , Elias, E. et al. (2017) Discovery of a novel stem rust resistance allele in durum wheat that exhibits differential reactions to Ug99 isolates. G3‐Genes Genom. Genet. 7, 3481–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera, P. , Jin, Y. , Rouse, M. , Badebo, A. , Fetch, T. Jr , Singh, R. and Yahyaoui, A. (2012) Races of Puccinia graminis f. sp. tritici with combined virulence to Sr13 and Sr9e in a field stem rust screening nursery in Ethiopia. Plant Dis. 96, 623–628. [DOI] [PubMed] [Google Scholar]
- Olivera, P. , Newcomb, M. , Szabo, L.J. , Rouse, M. , Johnson, J. , Gale, S. , Luster, D.G. et al. (2015) Phenotypic and genotypic characterization of race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in southern Ethiopia in 2013–14. Phytopathology, 105, 917–928. [DOI] [PubMed] [Google Scholar]
- Olivera, P. , Newcomb, M. , Flath, K. , Sommerfeldt‐Impe, N. , Szabo, L. , Carter, M. , Luster, D. et al. (2017) Characterization of Puccinia graminis f. sp. tritici isolates derived from an unusual wheat stem rust outbreak in Germany in 2013. Plant Pathol. 66, 1258–1266. [Google Scholar]
- Patpour, M. , Hovmoller, M. , Hansen, J. , Justesen, A. , Thach, T. , Rodriguez‐Algaba, J. , Hodson, D. et al. (2017) Epidemics of yellow rust and stem rust in Southern Italy 2016–2017. In: BGRI 2018 Technical Workshop.
- Pearce, S. , Vanzetti, L.S. and Dubcovsky, J. (2013) Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION 1 . Plant Physiol. 163, 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, F.Y. and Weselake, R.J. (2013) Genome‐wide identification and analysis of the B3 superfamily of transcription factors in Brassicaceae and major crop plants. Theor. Appl. Genet. 126, 1305–1319. [DOI] [PubMed] [Google Scholar]
- Pretorius, Z.A. , Singh, R.P. , Wagoire, W.W. and Payne, T.S. (2000) Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 84, 203. [DOI] [PubMed] [Google Scholar]
- Roelfs, A.P. (1982) Effects of Barberry eradication. Plant Dis. 66, 177. [DOI] [PubMed] [Google Scholar]
- Roelfs, A. (1985) Wheat and rye stem rust. In Roelfs, A.P. & Bushnell, W. R. (Eds.), Diseases, Distribution, Epidemiology, and Control, pp. 3–37. Orlando FL: Academic Press. 10.1016/B978-0-12-148402-6.50009-2 [DOI] [Google Scholar]
- Rouse, M. and Jin, Y. (2011a) Genetics of resistance to race TTKSK of Puccinia graminis f. sp. tritici in Triticum monococcum . Phytopathology, 101, 1418–1423. [DOI] [PubMed] [Google Scholar]
- Rouse, M. and Jin, Y. (2011b) Stem rust resistance in A ‐ genome diploid relatives of wheat. Plant Dis. 95, 941–944. [DOI] [PubMed] [Google Scholar]
- Rouse, M. , Wanyera, R. , Njau, P. and Jin, Y. (2011) Sources of resistance to stem rust race Ug99 in spring wheat germplasm. Plant Dis. 95, 762–766. [DOI] [PubMed] [Google Scholar]
- Saintenac, C. , Zhang, W. , Salcedo, A. , Rousse, M. , Trick, H. , Akhunov, E. and Dubcovsky, J. (2013) Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science, 341, 783–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherneck, S. , Nestler, M. , Vogel, H. , Blüher, M. , Block, M.‐D. , Diaz, M.B. , Herzig, S. et al. (2009) Positional cloning of zinc finger domain transcription factor Zfp69, a candidate gene for obesity‐associated diabetes contributed by mouse locus Nidd/SJL . PLoS Genet. 5, e1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamanin, V. , Salina, E. , Wanyera, R. , Zelenskiy, Y. , Olivera, P. and Morgounov, A. (2016) Genetic diversity of spring wheat from Kazakhstan and Russia for resistance to stem rust Ug99. Euphytica, 212, 287–296. [Google Scholar]
- Shamanin, V. , Salina, E. , Zelenskiv, Y. , Kokhmetova, A. , Patpour, M. , Holmoller, M. , Olivera, P. et al. (2018) Large scale wheat stem rust outbreaks in Western Siberia/Northern Kazakhstan in 2015–2017. BGRI 2018 Technical Workshop. https://www.globalrust.org/content/large‐926scale‐wheat‐stem‐rust‐outbreaks‐western‐siberia‐northern‐kazakhstan‐2015‐2017.
- Singh, R.P. , Hodson, D.P. , Jin, Y. , Huerta‐Espino, J. , Kinyua, M.G. , Wanyera, R. , Njau, P. et al. (2006) Current status, likely migration and strategies to mitigate the threat to wheat production from race Ug99 (TTKS) of stem rust pathogen. CAB Rev: Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 1, 1–13. [Google Scholar]
- Singh, R.P. , Hodson, D.P. , Huerta‐Espino, J. , Jin, Y. , Bhavani, S. , Njau, P. , Herrera‐Foessel, S. et al. (2011) The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 49, 465–481. [DOI] [PubMed] [Google Scholar]
- Singh, R.P. , Hodson, D.P. , Jin, Y. , Lagudah, E.S. , Ayliffe, M.A. , Bhavani, S. , Rouse, M.N. et al. (2015) Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology, 105, 872–884. [DOI] [PubMed] [Google Scholar]
- Sodkiewicz, W. , Strzembicka, A. and Apolinarska, B. (2008) Chromosomal location in triticale of leaf rust resistance genes introduced from Triticum monococcum . Plant Breed. 127, 364–367. [Google Scholar]
- Stakman, E.C. , Stewart, D.M. and Loegering, W.Q. (1962) Identification of physiologic races of Puccinia graminis var. tritici . United States Department of Agriculture Research Service E‐617. Washington DC.
- Steuernagel, B. , Periyannan, S.K. , Hernandez‐Pinzon, I. , Witek, K. , Rouse, M.N. , Yu, G. , Hatta, A. et al. (2016) Rapid cloning of disease‐resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652–655. [DOI] [PubMed] [Google Scholar]
- Svitashev, S.K. , Pawlowski, W.P. , Makarevitch, I. , Plank, D.W. and Somers, D.A. (2002) Complex transgene locus structures implicate multiple mechanisms for plant transgene rearrangement. Plant J. 32, 433–445. [DOI] [PubMed] [Google Scholar]
- Tesfaye, T. , Chala, A. , Shikur, E. , Hodson, D. and Szabo, L.J. (2020) First report of TTRTF race of wheat stem rust, Puccinia graminis f. sp. tritici, in Ethiopia. Plant Dis. 104, 293. [Google Scholar]
- The International Wheat Genome Sequencing Consortium . (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science, 361, eaar7191. [DOI] [PubMed] [Google Scholar]
- The, T. (1973) Chromosome location of genes conditioning stem rust resistance transferred from diploid to hexaploid wheat. Nat. New Biol. 241, 256. [DOI] [PubMed] [Google Scholar]
- Varagona, M.J. , Purugganan, M. and Wessler, S.R. (1992) Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell, 4, 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkowiak, S. , Gao, L. , Monat, C. , Haberer, G. , Kassa, M.T. , Brinton, J. , Ramirez‐Gonzalez, R.H. et al. (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature, 588, 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltner, J.K. , Peterson, F.C. , Lytle, B.L. and Volkman, B.F. (2005) Structure of the B3 domain from Arabidopsis thaliana protein At1g16640. Protein Sci. 14, 2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Zeng, H. , Zhou, X.U. , Huang, F. , Peng, W. , Liu, L. , Xiong, W. et al. (2015) Transformation of rice with large maize genomic DNA fragments containing high content repetitive sequences. Plant Cell Rep. 34, 1049–1061. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Zhang, P. , Dodds, P. and Lagudah, E. (2020) How Target‐sequence Enrichment and Sequencing (TEnSeq) pipelines have catalysed resistance gene cloning in the wheat‐rust pathosystem. Front. Plant Sci. 11, 678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Liu, J. , Zhang, Y. , Yang, Z. and Gao, C. (2015) Biolistic genetic transformation of a wide range of Chinese elite wheat (Triticum aestivum L.) varieties. J. Genet. Genom. 42, 39–42. [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Chen, S. , Abate, Z. , Nirmala, J. , Rouse, M.N. and Dubcovsky, J. (2017) Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group. Proc. Natl Acad. Sci. USA, 114, E9483–9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Zhao, S. , Chen, X. , Wang, Z. , Wang, L. , Yao, J. , Chen, W. et al. (2015) Determination of the role of Berberis spp. in wheat stem rust in China. Plant Dis. 99, 1113–1117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Reactions to six Pgt races 34PKUSC, 34MTGSM, TTKSK, BCCBC, 21C3CTTTM and RTJRM.
Figure S2 Semi‐quantitative PCR products from markers 4997QF2R2 (260 bp, TraesCS7A02G499700), 4998QF5R5 (272 bp, TraesCS7A02G499800) and ACTINF1R1 (ACTIN).
Figure S3 SrTm5 protein sequence analysis. Multiple sequence alignment between SrTm5 and reported Sr22‐resistant and susceptible protein sequences (Steuernagel et al. 2016).
Figure S4 Transcript levels of TmNLR1 in transgenic T1 families (three positive plants per event, n = 3).
Figure S5 Reactions to Pgt race TTKSK (Ug99) in transgenic family T2Tm515‐6.
Figure S6 Transgenic family T2Tm514‐2 homozygous for the transgene were inoculated with two SrTm5‐virulent Pgt races RTJRM and 21C3CTTTM.
Figure S7 Transcript levels and infection types of Sr22a and Sr22b in T. monococcum background.
Figure S8 PCR products from the Sr22b diagnostic marker TM5TF2R2.
Figure S9 Markers across chromosome 7A were used to analyse the length of introgressed T. monococcum segments.
Table S1 Avirulence/virulence formulae of Pgt races, and their responses to SrTm5.
Table S2 Comparison of SrTm5 protein with polymorphisms that discriminate perfectly between Sr22‐susceptible and ‐resistant haplotypes from Steuernagel et al. (2016).
Table S3 Segregation ratios in T1 and T2 transgenic families detected using PCR markers Tm5F3R4, TM5TF2R2 and TM5TF3R3 (Table 1).
Table S4 Resistance profiles of Sr22b (=SrTm5) and Sr22a (haplotypes R1 and R4) to multiple Pgt races.
Table S5 A collection of 92 accessions of T. monococcum, 23 of T. turgidum and 53 of T. aestivum was used to test the presence of Sr22b.
Table S6 Geographic distribution of T. monococcum accessions, and their reactions against Pgt races TTKSK, MCCFC and 34PKUSC.


