Abstract
The nuclear factor-κB (NF-κB) signaling pathway is one of the most well-studied pathways related to inflammation, and its involvement in aging has attracted considerable attention. As aging is a complex phenomenon and is the result of a multi-step process, the involvement of the NF-κB pathway in aging remains unclear. To elucidate the role of NF-κB in the regulation of aging, different systems biology approaches have been employed. A multi-omics data-driven approach can be used to interpret and clarify unknown mechanisms but cannot generate mechanistic regulatory structures alone. In contrast, combining this approach with a mathematical modeling approach can identify the mechanistics of the phenomena of interest. The development of single-cell technologies has also helped clarify the heterogeneity of the NF-κB response and underlying mechanisms. Here, we review advances in the understanding of the regulation of aging by NF-κB by focusing on omics approaches, single-cell analysis, and mathematical modeling of the NF-κB network.
Keywords: aging, mathematical model, nuclear factor kappa B, systems biology, transcription factors
Introduction
The transcription factor nuclear factor-κB (NF-κB) is responsible for regulating genes associated with inflammation [1]. NF-κB is linked not only to diseases such as cancer but also to the aging process [2]. Aging is an unavoidable time-dependent decline in physiological organ function. By disrupting the homeostasis of health, aging rapidly increases the risk of death from cancer, diabetes, or heart disease [3,4].
As we age, our bodies accumulate senescent cells [5,6]. Notably, the activation of NF-κB signaling promotes cell senescence [7]; NF-κB activation has been known to shorten the lifespan of fruit flies [8] and mice [9,10]. Furthermore, an increase in NF-κB DNA-binding activity has been reported in dermal fibroblasts and renal tissues derived from elderly individuals [11,12]. Therefore, a comprehensive understanding and control of the NF-κB system may shed light on how to prevent age-related diseases and prolong lifespan.
NF-κB is the primary regulator of senescence-associated secretory phenotype (SASP), which consists of inflammatory cytokines (interleukin [IL]-6 and IL-8), proteases (matrix metalloproteinases), chemokines (monocyte chemoattractant proteins and macrophage inflammatory proteins), and growth factors (granulocyte–macrophage colony-stimulating factor and transforming growth factor-β), and has a significant function in aging [13,14]. SASP is responsible for maintaining tissue homeostasis by removing unwanted senescent cells and triggers an inflammatory response that recruits immune cells, such as granulocytes, macrophages, natural killer (NK) cells, and T cells [15]. Thus, if not properly regulated, this system can cause pathological inflammation.
With age, our bodies become more susceptible to harmful inflammatory conditions, such as cytokine storms (a state of excessive cytokine production) [16]. Senescent cells have higher basal levels of various inflammatory cytokines and chemokines [17]. More importantly, in response to IL-1β, lipopolysaccharide (LPS), and tumor necrosis factor (TNF)-α stimulation, the induction of various inflammatory mediators is enhanced in senescent cells compared to that in non-senescent cells, and LPS stimulation causes significantly higher levels of NF-κB nuclear translation [17]. For example, elderly individuals infected with the coronavirus disease 2019 (COVID-19) are likely to have very high rates of adverse health effects and mortality due to cytokine storms [16].
Aging and senescence are caused by various developmental signals and different types of stresses and are considered to be the results of a multistep process [18,19]. Although various induced senescent cell models [20] and aging animal models [2] have been reported to elucidate the complex system, still lacking is an overall picture that recapitulates the cellular transmission and spatial interactions of aging to understand and overcome the unfavorable effects of aging. The utilization of multi-omics methods, such as transcriptomic, epigenetic, and proteomic approaches, has contributed to the elucidation of complex mechanisms. However, these methods cannot determine how age-related diseases can be prevented because they alone cannot generate mechanistic regulatory structures [21]. By contrast, investigating aging and senescence by integrating data-driven time-course omics with mathematical modeling approaches will expand our understanding of how the whole system of aging is regulated and predict the individual outcomes of the system against genetic and environmental factors. Thus, here, ‘systems biology' can be termed as a set of approaches to uncover complex mechanistic structures through analysis of comprehensive time-course omics data and mathematical models to identify, dissect, and manipulate the molecular regulatory network of aging.
In this review, we summarize how NF-κB regulates aging systems and how multi-omics and mathematical modeling approaches have contributed to the elucidation of transcriptional regulation of NF-κB and aging. We also discuss the dynamic properties of the NF-κB system and the significance of oscillation and non-oscillation dynamics in the regulation of downstream target genes.
NF-κB pathway
Since its discovery as a nuclear factor that binds to DNA elements in the intronic enhancer of the kappa light chain gene of B cells, NF-κB has been investigated for its function as a transcription factor [22]. The NF-κB network consists of seven family members: p105/p50 (NFKB1), p100/p52 (NFKB2), p65 (RELA), RelB (RELB), and c-Rel (REL); these form homodimers or heterodimers and acquire the ability to bind to DNA differently. Among these proteins, only p65 (RELA), RelB, and c-Rel contain the carboxy-terminal transactivation domain (TAD) that activates the transcription of NF-κB target genes [23,24]. In most cells, the p50/p65 heterodimer is a major NF-κB transcription factor. Homo- or heterodimers of p50 and p52 cannot promote transcription due to the absence of TAD but instead bind to the κB site sequence and act as transcriptional repressors [25].
The NF-κB transcription factor constitutes a homo- or heterodimeric protein that binds to a 5'-GGGRNNNYCC-3’ (where G, C, R, Y, and N are guanine, cytosine, purine, pyrimidine, and any nucleotide base, respectively) consensus κB site sequence consisting of 10 nucleotides [26]. The DNA-binding motifs of most transcription factors, including NF-κB, have been identified by the Encyclopedia of DNA Elements (ENCODE) Project Consortium using both chromatin immunoprecipitation (ChIP) and next-generation sequencing [27–29]. To date, several different kB sites have been identified despite being limited to the p50/p65 heterodimer [30].
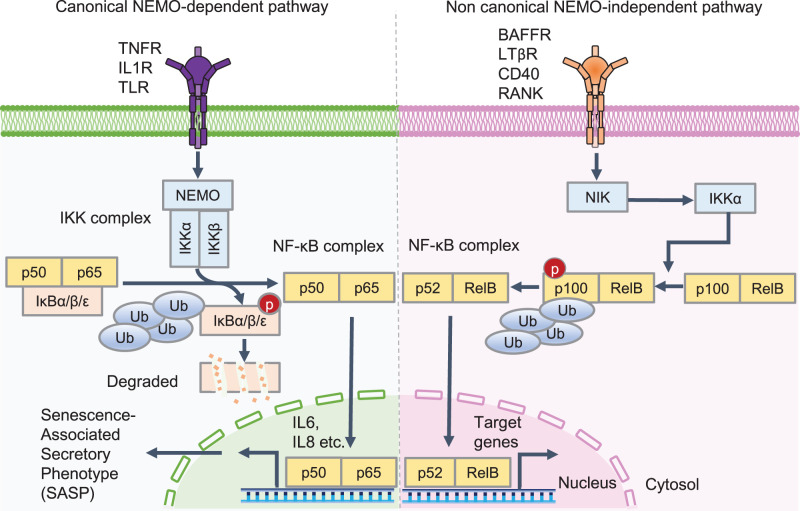
The NF-κB network is explained by two major pathways: the canonical NF-κB essential modulator (NEMO)-dependent pathway and the non-canonical NEMO-independent pathway (Figure 1) [31,32]. The canonical pathway is activated by stimuli such as TNF-α, IL-1β, and LPS, which leads to the phosphorylation of the inhibitor of κB (IκB) kinase (IKK) complex; the latter is composed of IKKα, IKKβ, and NEMO. The activation of the IKK complex phosphorylates IκB proteins (IκBα, IκBβ, and IκBε), leading to proteasome-mediated proteolysis of IκB proteins and allowing the NF-κB complex (p50/p65) to enter the nucleus [33].
Figure 1. Overview of the canonical and non-canonical NF-κB pathway.
In the canonical nuclear factor-κB (NF-κB) essential modulator (NEMO)-dependent pathway, the inhibitor of κB (IκB) kinase (IKK) complex is activated by activated tumor necrosis factor receptor (TNFR), interleukin (IL)-1 receptor (IL1R), and Toll-like receptor (TLR)s. IKK complex activation induces proteasome-mediated proteolysis of IκB proteins and allows the NF-κB complex (p50/p65) to accumulate in the nucleus. p50/p65 dimers bind to DNA and regulate the transcription of senescence-associated secretory phenotype genes, such as IL-6 and IL-8. In the non-canonical NEMO-independent pathway, NF-κB-inducing kinase (NIK) phosphorylates IKKα and leads to the phosphorylation of p100. This process induces subsequent ubiquitination and partial degradation of p100 by the proteasome to form the NF-κB complex (p52/RelB). p52/RelB dimers enter the nucleus and regulate downstream target genes.
The non-canonical pathway is NEMO-independent but NF-κB-inducing kinase (NIK)- and IKKα-dependent [34,35]. It is activated through receptors such as B-cell activating factor receptor [36,37], lymphotoxin beta receptor [38], cluster of differentiation (CD) 40 [39], and receptor activator of NF-κB [40]. Signal transduction by receptors activates the NIK/IKKα complex, which phosphorylates p100 and is processed into p52, which allows RelB to form the NF-κB complex (p52/RelB) that moves into the nucleus [34,35]. This translocated complex then binds to DNA and induces its target gene expression, including inflammatory cytokines and modulators of the NF-κB pathway.
NF-κB and cellular senescence
Accumulating senescent cells is one of the major causes of aging-related disorders [4–6]. It has been reported that senescent cells increase with age in many mice and human tissues, such as the adipose tissue, liver, kidney, and skeletal muscle [41]. Cellular senescence was first reported in 1961 by Leonard Hayflick using human fibroblast cells to relay the limitations of mitotic capacity with replicative stress (RS), known as the Hayflick limitation [42]. Senescent cells are stable, terminal-growth-arrest cells that can be characterized by flat, large-shaped cell morphology, telomere shortening, promotion of cyclin-dependent kinase inhibitor p16 [43] or p21 [44], and increased senescence-associated β-galactosidase (SA-β-gal) activity. SA-β-gal was one of the first published biomarkers of aging, and helped to demonstrate the accumulation of cells with aging characteristics in various mammalian aging-related diseases and in aging tissues [44]. The removal of p16-expressing senescent cells suppresses aging and extends the lifespan of mice, indicating that cellular senescence is an important factor in aging research [43,45].
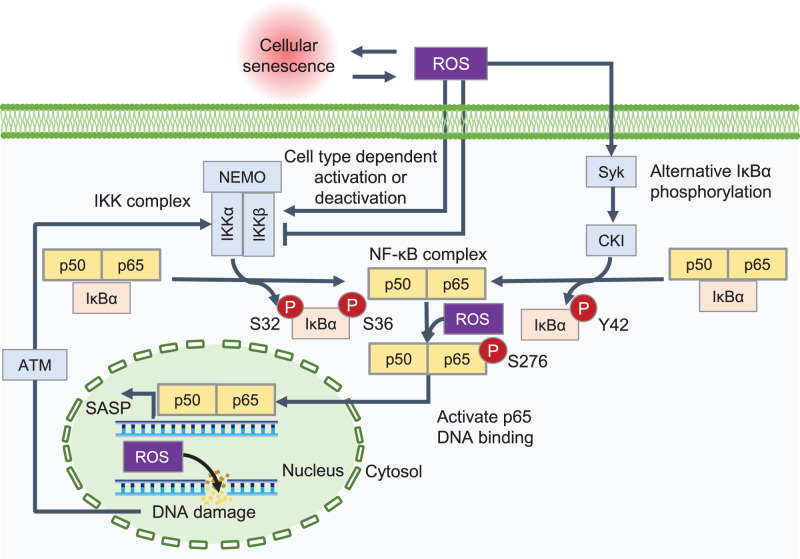
Reactive oxygen species (ROS), such as hydrogen peroxide (H2O2), and DNA damage accumulate with cellular senescence and crosstalk with NF-κB signaling (Figure 2) [46,47]. ROS induce DNA damage, which results in IKK activation via activation of ataxia-telangiectasia mutated kinase [48]. Typical IκBα is phosphorylated on S32 and S36 by the IKK complex upon NF-κB activation, whereas H2O2 phosphorylates IκBα on Y42, a process mediated by the spleen tyrosine kinase–casein kinase II pathway [49–51]. H2O2 directly activates [52–54] or inactivates [55–57] NF-κB via the IKK complex depending on the cell type and influences DNA binding through the phosphorylation of p65 at S276 [58].
Figure 2. Activation of NF-κB in cellular senescence via ROS.
Cellular senescence leads to the accumulation of reactive oxygen species (ROS), which can induce senescence. ROS interact with nuclear factor-κB (NF-κB) at various locations during signal transduction and induce DNA damage. DNA damage induces activation of NF-κB via ataxia-telangiectasia mutated (ATM) kinase activating the inhibitor of κB (IκB) kinase (IKK) complex. ROS are thought to directly affect downstream targets by activating and deactivating the IKK complex, which occurs in a cell type-specific manner. Compared with canonical IKK activation, ROS can alternatively activate the NF-κB complex through the spleen tyrosine kinase (Syk)–casein kinase II (CKII) pathway by phosphorylation of IκBα at Y42. Phosphorylation of p65 at S276 by ROS activates the DNA binding of p65, resulting in greater NF-κB activation (adapted from [47]).
In human skin keratinocytes, the overexpression of c-Rel, a subunit of NF-κB, induces premature senescence [59]. The inhibition of H2O2 in c-Rel-overexpressing keratinocytes decreases the expression of SA-β-gal, suggesting that NF-κB is involved in cellular senescence. In addition, the overexpression of p50/p65 in keratinocytes induces cell growth arrest along with p21 activation [60]. Inflammatory SASPs (such as IL-6 and IL-8, among others) are regulated by NF-κB and not only are characterized as senescence markers but also form autocrine loops and induce growth arrest [61,62]. Microarray analysis of senescent human fibroblasts has shown that the expression of SASP genes is up-regulated and that the inhibition of NF-κB leads to cells escaping cell-cycle arrest [7].
NF-κB maintains cellular senescence by promoting DNA repair and genome stability. Wang et al. [63] compared the rate of immortalization of primary fibroblasts from RelA/p65 (−/−) mouse fibroblasts and RelA/p65 (+/+) cells and found that RelA/p65 (−/−) cells undergo immortalization at a faster rate owing to early DNA mutation and gene deletion caused by genome instability. In a study of NF-κB-dependent effects and mechanisms underlying ROS generation in normal human lung fibroblasts, ROS were reported to cause cell cycle arrest and lead to premature senescence [64]. Interestingly, a previous report showed that the inhibition of NF-κB signaling up-regulates p53-p21 expression and promotes premature senescence [64]. These reports indicate that NF-κB signaling may not only function as a mechanism to promote aging but also have completely opposite functions, suggesting the possibility of differences in functions among cell types and regulatory mechanisms that have not yet been elucidated.
NF-κB and aging
The role of the NF-κB pathway has attracted increasing attention in the field of aging research [65,66]. Persistent NF-κB activation in chronic inflammation leads to the promotion of aging via the shortening of telomeres [67] and increased expression of γ-H2AX protein [68], which are both widely known aging markers [4,69].
Progressive decline in immune response, including a reduction in proliferative effector responses and a decrease in T cell signaling, has been observed in aging [70]. T cells have several subtypes, including CD8+ T cells (killer T cells), CD4+ T cells (helper T cells), and regulatory CD4+ CD25+ T cells [71]. Human CD8+ T cells are further classified as naïve, central memory (T(CM)), and effector memory cells (T(EM) and T(EMRA)) [72,73]. A Naïve CD8+ T cells and T(CM) CD8+ T cells in the elderly have been found to be more sensitive to apoptosis than those in young individuals [70]. In contrast, T(EM) and T(EMRA) CD8+ T cells are less sensitive, and there is no significant difference in their levels between younger and older individuals [70]. TNF-α production is known to accelerate aging in humans [74–77]. However, the expression of TNF receptors (TNFRI and TNFRII) in CD8+ T cells has been reported to be similar between young and aged subjects [70]. Thus, heterogeneity in the age and cell type response in CD8+ T cells can be attributed to the difference in TNFR downstream NF-κB signal transduction. Because T cells play a central role in cellular immunity, the mechanistic association between NF-κB and aging in T cells will provide a deeper understanding of age-related immune responses.
In terms of the immune response in aging, interesting findings from an animal model of aging, the naked mole-rat (NMR, Heterocephalus glaber), have been reported. NMRs are relatively long-lived rodents for their size, and the risk of mortality does not appear to increase with age [78]. Compared with those in mice, macrophages from NMRs have been reported to activate Toll-like receptors in response to LPS stimulation, initiate NF-κB, and produce large amounts of cytokines to enhance immune responses [79]. Hilton et al. [80] collected single-cell RNA sequencing (scRNA-seq) data from spleen derived from NMRs and mice and found NMRs lack canonical NK cells. In addition, LPS was intraperitoneally administered to NMRs and mice, and their spleens were harvested for bulk RNA-seq and scRNA-seq [80]. Subsequently, gene set enrichment analysis (GSEA) revealed that LPS induces the NF-κB inflammatory pathway in both organisms and that NMRs have a subset of LPS-responsive cells that are not present in mice. While no direct reports indicate that NMR aging is linked to NF-κB, elucidation of NF-κB system regulation in NMR may lead to a deeper understanding of aging.
Studies on human fibroblasts from elderly people [11] and animal organs, such as the liver, kidney, and brain, have revealed that the nuclear concentration of NF-κB increases with age. In aging livers, a significant age-related increase in NF-κB binding activity, along with increased levels of nuclear p52 and p65 proteins in the rat liver, has been reported [81]. Interestingly, the expression of NF-κB mRNA (NFKB1, NFKB2, RELA, and REL) and its inhibitor transcripts (NFKBIA and NFKBIB) have not shown statistically significant age-related changes [81]. This result indicates that post-translational modifications occur during liver aging and may affect nuclear localization and binding activity during aging. In rat kidney, decreased IκBα levels and increased nuclear p65 protein levels indicate that NF-κB activity is enhanced by a reduction in IκBα levels [82]. This increase in NF-κB activity is accompanied by an increase in the mRNA and protein levels of cyclooxygenase 2, from which ROS is derived in the body. The roles of IKK-β and NF-κB in mouse brain aging have also been reported. Zhang et al. [10] studied the hypothalamus and cortical tissues of young (3–4-month-old), middle-aged (11–13-month-old), and old (22–24-month-old) C57BL/6 mice and found that p-RelA expression increases in the hypothalamus and cortical tissues as mice age and that preventing the activation of IKK-β and NF-κB in these tissues can prolong their lifespan.
The relationship between NF-κB activation and aging has been demonstrated using aging animal models. Senescence-accelerated mice (SAM), which have been used as an animal model for pathological aging, show increased levels of oxidative stress-related dysfunction and inflammation [83]. SAM prone 8 (SAMP8), a strain of SAM, is short-lived with accelerated aging and exhibits age-related pathologies, such as learning and memory deficits, similar to those in human aging [84]. Pro-inflammatory, pro-apoptotic, and pro-oxidative states are markedly increased in the lungs of aged SAMP8 compared with those of SAM resistant 1 [85]. Forman et al. [85] showed that, in SAMP8 hearts, the nuclear concentration of NF-κB (p50, p52, and p65) is increased, while the levels of cytoplasmic IκBα and IκBβ are reduced.
Experiments on transgenic Nfkb1−/− mice have also contributed to uncovering the relationship between NF-κB and aging [2]. Nfkb1−/− mice lack both the p105 precursor and p50 subunits and demonstrate persistent induction of p65/p65 homodimer formation [86]. The phenotype of this transgenic mouse shows accelerated apparent senescence, shortened lifespan, telomere damage, and a chronic inflammatory state, which leads to the production of SASP. A cellular level analysis of Nfkb1−/− mice showed decreases in proliferation speed and apoptosis and increases in SA-β-gal positive cells, expression of cyclin-dependent kinase inhibitors p16 and p21, and γ-H2AX accumulation [87]. These data indicate that the loss of Nfkb1 leads to premature senescence in mammals.
Multi-omics regulation of NF-κB and aging
What occurs within our bodies as we age? To answer this question, multi-omics analysis has been vigorously performed to understand transcriptomic, epigenetic, and proteomic regulation [88]. Attempts to collect omics data on aging phenotypes have produced several databases, such as Human Aging Genomic Resources (HAGR) [89], SeneQuest [90], and the Aging Atlas [91], which help us to integrate data and provide a bird's eye view of the biological pathways involved in the aging process. HAGR (http://genomics.senescence.info/) is a collection of databases on human and model organism aging that can be searched for age-related genes, drugs, and genetic variants [89]. SeneQuest (https://senequest.net/) is a database of genes related to senescence and is maintained by the International Cell Senescence Association [90]. The Aging Atlas (https://ngdc.cncb.ac.cn/aging/index) is a multi-omics database for aging biology that enables researchers to find information from genomics, epigenomics, transcriptomics, proteomics, metabolomics, and pharmacogenomics data at either the bulk or single-cell level [91]. These databases were developed with graphical user interface-based platforms and enable biologists to readily access omics data related to aging and senescence.
Hereafter, we discuss omics research investigating the relationship between NF-κB and aging (Table 1).
Table 1. Omics studies of NF-κB and aging.
| Omics | Methodology | Sample | Result | Reference |
|---|---|---|---|---|
| Transcriptome regulation | Microarray | Naïve (CD44low) and memory (CD44high) CD4+ T cells derived from young (2–3-month-old) and aged (28-month-old) mice | Upstream analysis of differentially expressed genes during aging was performed, and NF-κB was identified as a potential age regulator | [92] |
| Transcriptome regulation | RNA-seq | pHBEC senescence induced by RS and CSE | Under both RS and CSE stress, gene set enrichment analysis indicated dysfunction in the regulation of ROS, proteasome degradation, and NF-κB signaling | [93] |
| Transcriptome regulation | miRNA sequencing | Peripheral arterial and venous blood of young (8-week-old) and aged (22-month-old) rats | miR-136-3p and miR-503-3p are differentially expressed with aging and are regulated by NF-κB and SIRT1 | [95] |
| Transcriptome regulation | scRNA-seq | Lung, heart, and artery tissues derived from young (4–6-year-old) and old (18–21-year-old) cynomolgus monkeys | ACE2 expression increases with age in alveolar epithelial barrier, cardiomyocytes, and vascular endothelial cells; IL-7 accumulates in aged cardiopulmonary tissues and induces ACE2 expression in human vascular endothelial cells in an NF-κB-dependent manner | [98] |
| Transcriptome regulation | Microarray | Sun-unexposed skin tissue of healthy males aged 19 to 86 years | Metabolic activity and cellular damage associated with NF-κB pathways increase in the middle aged (30–45 years old) | [99] |
| Transcriptome regulation | scRNA-seq | Human upper eyelid skin samples collected from young (18–28-year-old), middle-aged (35–48-year-old), and aged (70–76-year-old) groups | NF-κB signaling pathway is up-regulated with aging in several cell types, including epidermal basal cells, mitotic cells, granular cells, and spinous cells | [101] |
| Transcriptome and epigenetic regulation | RNA-seq ChIP-seq (H3K27ac) | Brain tissues derived from young (<60-year-old) and old (>60-year-old) humans and young (3-month-old) and aged (18-month-old) mice | The expression levels of regulators of the NF-κB pathway, TNFRSF1A, NFKBIA, and TMED4, increase in the aged brain, and NF-κB and BCL3 binding annotations are concentrated in up-regulated genes | [106] |
| Transcriptome and epigenetic regulation | Microarray ChIP-seq | ChIP-seq data from ENCODE [27–29] and the list of kidney age-regulated genes from Rodwell et al. [12]. Additionally, TNF-, IFNγ-, and IL-6-treated human renal proximal tubular epithelial cells | Both transcriptomic and epigenetic analyses showed that the expression levels of NF-κB, STAT1, and STAT3 increase with renal aging | [108] |
| Transcriptome and epigenetic regulation | RNA-seq ChIP-seq (H3K4me3 and H3K27ac) | Heart, liver, cerebellum, and olfactory bulb derived along with primary cultures of neural stem cells from young (3-month-old), middle-aged (12-month-old), and aged (29-month-old) mice | Both transcriptomic and epigenetic analyses of heart, liver, and cerebellum and functional enrichment analysis showed TNF-α signaling via NF-κB is up-regulated as age increases | [109] |
| Protein regulation | Nano LC-MS/MS | Senescent human diploid IMR-90 fibroblasts induced by etoposide or infection with oncogenic H-RasV12 | In senescent IMR-90 cells, the NF-κB p65 subunit was found to be one of the most significantly enriched transcriptional regulators bound to chromatin | [113] |
| Protein regulation | Nano LC–MS/MS | Marmoset senescent TPC induced by RS and TPC from young (2–3-year-old) and aged (10–15-year-old) marmoset monkeys | In both the RS in vitro model and aged cell in vivo model, NF-κB signaling is altered by aging | [114] |
| Protein regulation | Nano LC–MS/MS | Tear samples from health humans (18–83 years old) | Upstream analysis of 17 tear fluid-derived proteins, which were correlated with donor age, showed that the NF-κB complex acts as a transcriptional regulator | [115] |
Abbreviations: NF-κB, nuclear factor-kappa B; pHBEC, primary human bronchial epithelial cell; RS, replicative stress; CSE, cigarette smoke extract; ROS, reactive oxygen species; SIRT1, sirtuin-1; scRNA-seq, single-cell RNA sequencing; ACE2, angiotensin-converting enzyme 2; ENCODE, Encyclopedia of DNA Elements; TNF, tumor necrosis factor; IFN, interferon; IL, interleukin; STAT, signal transducer and activator of transcription; Nano LC–MS/MS, nanoscale liquid chromatography coupled to tandem mass spectrometry; TPC, testicular peritubular cells.
Omics 1. Transcriptome regulation of NF-κB and aging
Transcriptomic changes during the aging of various cells and tissue samples indicate a relationship between NF-κB and aging. To explore the cause of the decline in immune response, transcriptomic profiles of naïve (CD44low) and memory (CD44high) CD4+ T cells, which are impaired by the aging process, were derived from young and aged mice and examined by Taylor et al. [92]. To determine probable upstream regulators, cis-regulatory analysis of differentially expressed genes during aging was performed, and NF-κB was identified as a potential age regulator [92].
Transcriptome changes in senescent cells induced by RS and cigarette smoke exposure also indicate that NF-κB is related to aging [93]. Cigarette smoke exposure and RS are well-known senescence inducers [94]. Voic et al. [93] performed RNA-seq in passaged cigarette smoke extract (CSE)-exposed and non-CSE-exposed senescent primary human bronchial epithelial cells and showed that GSEA indicated dysfunction in the regulation of ROS, proteasome degradation, and NF-κB signaling under either RS or CSE. miRNA analysis has also been conducted to explore the influence of aging. Transcriptome analysis of miRNA from the peripheral blood of young and aged rats showed that miR-136-3p and miR-503-3p are differentially expressed with aging and are regulated by NF-κB and SIRT1 [95].
Aging has been reported to increase the risk of severe COVID-19 [96,97]. Ma et al. [98] created a single-cell transcriptomic atlas of the cardiopulmonary system of aged cynomolgus monkeys and reported the involvement of NF-κB in age-related susceptibility to severe acute respiratory syndrome coronavirus disease 2 (SARS-CoV-2). The authors reported that the expression of angiotensin-converting enzyme 2 (ACE2), a receptor for SARS-CoV-2, increases with age in the alveolar epithelial barrier, cardiomyocytes, and vascular endothelial cells. They also found that aged cardiopulmonary tissues accumulate IL-7 and induce ACE2 expression in human vascular endothelial cells in an NF-κB-dependent manner.
Interestingly, the aging of the skin is thought to differ slightly from that of other tissues because it is caused by both internal factors (such as senescent cells) and external factors (such as ultraviolet light [UV]). Haustead et al. [99] focused on skin aging caused by internal factors by analyzing the transcriptome of sun-unexposed skin tissue from healthy males aged 19 to 86 years. In middle-aged subjects (30–45 years old), the authors found gene enrichment in response to DNA damage stimuli and positive regulation of the NF-κB cascade via IKK. Regarding the relationship between UV light and NF-κB, O'Dea et al. [100] reported that UV light does not induce but amplifies NF-κB activity. Using mathematical modeling, the authors showed that UV-induced activation of NF-κB occurs not only through the IKK-mediated degradation pathway of IκBs but also through the IKK-independent pathway. Single-cell transcriptomic analysis has also been used to reveal the involvement of NF-κB in human skin aging. Zou et al. [101] collected scRNA-seq data from the eyelid skin of healthy individuals of various age groups and identified 11 canonical cell types and 6 basal cell types in skin tissues. Their analysis showed that the NF-κB signaling pathway was up-regulated with aging in several cell types, including epidermal basal cells, mitotic cells, granular cells, and spinous cells.
Omics 2. Epigenetic regulation of NF-κB and aging
When histones are subjected to epigenetic modifications, such as methylation and acetylation, chromatin changes its shape to form either euchromatin, where transcription factors can likely bind to DNA and transcription is actively carried out, or heterochromatin, where DNA aggregates and transcription factors cannot easily bind, which can be monitored by ChIP-seq [102,103]. Epigenetic changes have been shown to be a feature of aging [69]. For example, disturbance of histone H3 trimethylation at lysine 4 or lysine 27 (H3K4me3 or H3K27me3, respectively) has been reported to affect the lifespan of Caenorhabditis elegans [104,105]. In the brains of aged humans and mice, a general loss of H3K27ac has been observed [106]. In addition, deletion of SIRT6, a deacetylase of H3K9ac, results in excessive activation of NF-κB signaling and accelerated aging [107]. Here, we introduce epigenetic studies that elucidate the relationship between NF-κB and aging.
Brown et al. compared transcription factor changes during kidney aging using data from the ChIP-seq dataset from the ENCODE Consortium [27–29] and genome-wide maps of transcription factor occupancy obtained from ChIP-seq data of human cells [108]. The authors identified associations between ENCODE data and kidney age-related genes derived from transcriptional profiles [12] and found NF-κB, signal transducer and activator of transcription (STAT)1, and STAT3 to be promising candidates whose activities increase with age in the epithelial compartment of the renal cortex [108]. They also showed that DNA variants common to RELA and NFKB1 are associated with renal function and chronic kidney disease in genetic association studies, showing that genetic variants in NF-κB contribute to the phenotype of renal aging.
Chronic inflammation aging profiling using RNA-seq and H3K27ac ChIP-seq of human and mouse brain samples was performed by Cheng et al. [106]. The expression levels of regulators of the NF-κB pathway — TNFRSF1A, NFKBIA, and TMED4 — were increased in the aged brain; NF-κB and BCL3 binding annotations were enriched in up-regulated genes, whereas the expression of GATA-3, which is related to the regulation of neuronal functions, was down-regulated [106].
Systematic studies of transcriptomic and epigenomic changes in several tissues and species during aging have been reported. Benayoun et al. [109] generated ChIP-seq (H3K4me3 and H3K27ac) and RNA-seq datasets from tissues of young, middle-aged, and old C57BL/6N male mice. Transcriptomic, epigenomic analyses of the heart, liver, and cerebellum and functional enrichment analysis showed that TNF-α signaling via NF-κB increases as age increases [109]. The authors also used public human [110], rat [111], and African turquoise killifish (Nothobranchius furzeri) [112] transcriptome data to determine whether the effects of aging are conserved in multiple vertebrates; they found that functional enrichment showed TNF-α signaling via NF-κB is commonly regulated by aging.
Omics 3. Protein regulation of NF-κB and aging
While genetic and epigenetic regulation is important, it is also essential to understand the function of proteins to identify changes in physiological phenomena. Here, we introduce the relationship between NF-κB and aging at the proteomic level. Chien et al. [113] performed a proteomic analysis of proteins bound to chromatin in senescent IMR-90 cells and found that the NF-κB p65 subunit is one of the most significantly enriched transcriptional regulators. Stöckl et al. [114] reported proteome and secretome analyses of RS induced in testicular peritubular cells derived from marmoset monkeys and showed impaired protein secretion, altered NF-κB signaling, and reduced contractility.
The effect of aging on tear fluid samples and its association with NF-κB was reported by Nättinen et al. [115], who performed proteomics analysis using healthy human tear samples collected from 115 subjects. In their study, 17 tear proteins related to inflammation, immune response, and cell death were found to correlate with donor age. Upstream analysis of these proteins indicated that the NF-κB complex acts as a transcription regulator.
In vitro and in vivo multilayer omics analyses have shown that NF-κB signaling is highly related to senescence and aging. Data-driven omics analyses in aging have revealed that the perturbation of aging activates the promoter and enhancer regions of NF-κB target genes, promotes binding of NF-κB to the κB site, and increases transcription of NF-κB target genes.
Mathematical modeling of NF-κB and aging
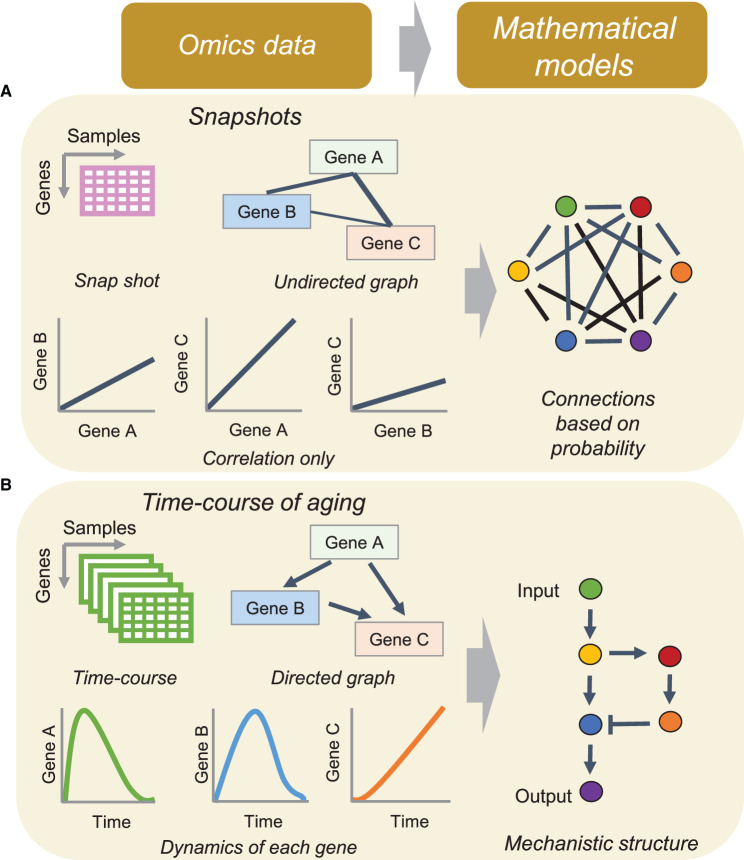
Data-driven omics analysis provides valuable information regarding the relationship between NF-κB and aging. Snapshot omics studies do not provide information on the underlying mechanistic regulatory structures; only the correlations between genes based on their expressions can be obtained [21]. From these static datasets, we can construct only mathematical models (e.g. regression models [116]) based on the probability of correlation between genes (Figure 3A). By contrast, time-course omics data on aging, along with knowledge of biological aging, show a directed graph between the gene regulatory network (GRN) and the possible activity dynamics of each gene. By integrating these data with mathematical models and performing numerical simulations, we can predict the mechanistic structure of biological systems and even manipulate them (Figure 3B). Thus, integration between time-course omics data and mathematical models can provide a mechanistic structure to understand the entire system of aging.
Figure 3. Differences between time-course and snapshot omics analysis.
(A) Time-course aging omics showing a directed graph between the gene regulatory network and the dynamics of each gene. By integrating these data with mathematical models, mechanisms of biological functions can be obtained. (B) Snapshot omics studies do not provide information on the underlying regulatory mechanisms but provide only information regarding the correlations between genes. For these datasets, we can only construct mathematical models based on the probability of correlation among genes.
Dynamic behaviors obtained from a mathematical simulation can also provide important insights into biological functions in silico, elucidating sensitivity and robustness of regulatory mechanisms and filling the gap between experimentally observable data and theoretical regulatory principles [117,118]. Biological systems can be represented using several networks, such as the ordinary differential equation (ODE) model, Boolean model, and Bayesian network [119–121].
ODE models are most commonly used to quantitatively understand biological systems [122,123]. The ODE model is said to be ordinary because it contains only one independent variable, which is basically time. It assumes that species are present in well-mixed compartments and that concentrations can be considered to be continuous [119], which generally makes the ODE model unsuitable for representing processes such as diffusion, spatial heterogeneity, and stochasticity [124]. Each variable represents the concentration of one component (e.g. gene), and how it changes over time depends on the initial value of each variable, the concentrations of other variables, and fixed kinetic parameters. Hence, if we attempt to represent the biological system with the ODE model, we need to estimate the kinetic parameters that correspond to the time-series data, which increases the computational cost by expanding the model.
The Boolean network is a type of dynamic model that is based on quantitative logical rules [120,121,125]. Based on the interaction between genes, regulation (e.g. transcriptional regulation) can be termed as active or inactive. In other words, this can be stated as Boolean logical values: OFF (‘0') or ON (‘1'). The regulation network between nodes, for example, genes, phosphorylation, or ubiquitination, can be connected with ‘logic gates' found in digital electronic circuits (e.g. AND, OR, and NOT gates) [120,121]. Each logic gate has a set of logical rules that connect the input to the output. Time in the Boolean network takes integer values, and nodes are either ‘OFF' or ‘ON' depending on the activation or repression, respectively, of each factor, which allows us to simulate dynamic behavior.
A Bayesian network reconstructs a gene regulatory expression network between nodes to statistically evaluate different connections in a network with conditional probability [126,127]. One characteristic of the Bayesian network is that it simplifies high-dimensional data into a simple graph. In a Bayesian network graph, the nodes represent the variables (e.g. gene), and the edges represent the links between genes. Displaying probability distributions as a directed graph makes graphical analysis possible. Calculating these probabilities according to the occurrence path makes it possible to quantify the probability of the occurrence of causal relations with complex pathways.
Mathematical models using the Boolean network have been proposed to elucidate the regulatory network between NF-κB and aging [128–130]. A Boolean network model-based GRN of the SASP activated by DNA damage was proposed by Meyer et al. [128]. They simulated a model of pathway interactions between p53/p16-induced cell cycle arrest, NF-κB-regulated SASP, and IL-1/IL-6-driven inflammatory activity based on knowledge of biological aging. From their work, NEMO was identified as a target for mechanistic inhibition of IL-6 and IL-8 and was experimentally validated using NEMO knockout murine dermal fibroblasts.
In addition to the knowledge-based mathematical model, Schwab et al. integrated data-driven omics analysis and the Boolean model to clarify hidden knowledge under NF-κB signaling and aging [129,130]. The authors used a binarized time-series gene expression dataset of muscle samples [131] from young and elderly healthy male humans to construct a Boolean network and found behavioral changes in NF-κB signaling during aging [129].
A combination of sc-RNA-seq of human hematopoietic stem cells (HSCs) derived from young (19–40-year-old) and aged (61–70-year-old) human individuals [132] and a Boolean network was used to capture the heterogeneity of aging in NF-κB signaling-regulated genes [130]. With aging, the number of HSCs increases, but their activity becomes highly heterogeneous and impaired, which is known to be related to NF-κB signaling-induced inflammation [133]. Using pseudo time-series of single-cell data in dormant and active HSCs, Schwab et al. [130] reconstructed specific regulatory networks for each individual and performed attractor analysis to show that single-cell dynamics of NF-κB target genes capture heterogeneity in mechanisms and regulatory networks of aging HSCs. This study shows that the integration of omics analysis and mathematical models is a powerful tool to elucidate hidden mechanisms between NF-κB and aging. Moreover, it might be possible to elucidate transcriptional heterogeneity in other tissues using scRNA-seq aging data with time-course omics and mathematical integration methods [134].
Notably, aging is a physiological phenomenon that progresses over several decades. By contrast, cellular senescence can be induced by a variety of stimuli in a rather short time period [20]. In other words, time-series information on signal transduction is important for phenomena that change rapidly, such as cellular senescence, and a model that allows a structural understanding of signal pathways, such as the ODE model, is considered suitable. Thus, the selection of an appropriate model for each dataset in aging and senescence should be considered before building the model.
Dynamic properties of NF-κB regulation
Based on scRNA-seq analysis of aging cells and tissues, such as T-cells [135], aorta and coronary arteries [136], pancreas [137], microglia cells [138], lung [139], skin [101,140], and muscle [141], transcriptional heterogeneity has been reported to increase with aging [134]. Understanding the heterogeneity in aging will make it possible to identify new markers that distinguish between actual and biological age and provide optimal treatment options for individuals [142]. Moreover, single-cell dynamics of NF-κB target genes capture heterogeneity in the mechanisms and regulatory networks of aging HSCs [130]. Dysregulation of nuclear localization of NF-κB leads to various pathological conditions [143]; therefore, controlling the localization of NF-κB might shed light on the heterogeneity in aging, and research on the regulation of NF-κB dynamics is needed. Thus, in this chapter, we first introduce what has been elucidated in the regulatory systems of NF-κB dynamics, then examine how NF-κB dynamics change the response to different types of input stimuli from mathematical research findings.
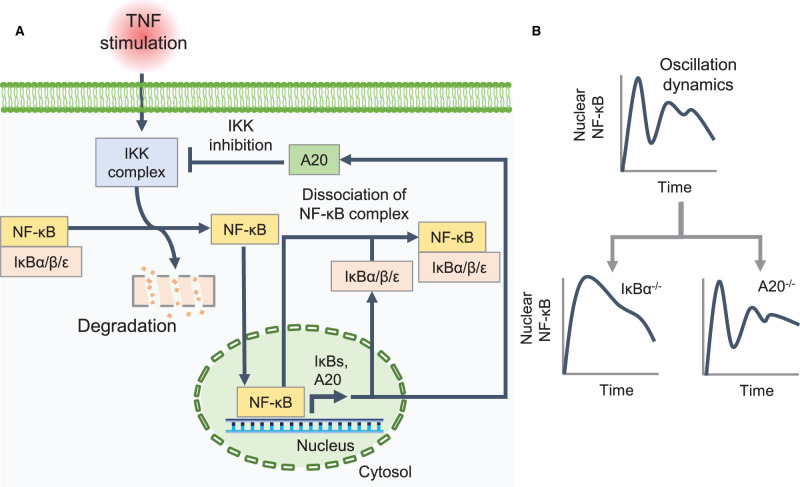
Along with the activation of inflammation-related genes, NF-κB has multiple negative feedback mechanisms and constitutes a complex regulatory mechanism. These negative feedback loops shape the characteristic oscillation dynamics of NF-κB nuclear localization (Figure 4). IκBs (NFKBIA [IκBα], IKBKB [IκBβ], and IKBKE [IκBε]) and TNFAIP3 (A20) are target genes of NF-κB; when NF-κB is activated, IκBs are released from the DNA binding domain of NF-κB, and A20 suppresses IKK activation [144–146]. These two negative feedback processes have distinct functions in the regulatory system of NF-κB dynamics. IκBα protein is synthesized upon stimulation of the NF-κB pathway and rapidly transferred to the nucleus; it strips NF-κB bound to DNA and transfers it to the cytoplasm [145]. Therefore, knockout of IKBA abolishes NF-κB oscillation, which induces sustained activation of NF-κB target genes [147,148]. IκBβ, which has high homology with IκBα, is reported to be unable to provide the same adequate negative feedback as IκBα [149]. In contrast to IκBα, IκBε has been reported to show delayed negative feedback due to transcriptional delay [150]. Using the ODE model, Longo et al. [151] showed that this delayed negative feedback by IκBε controls dampened oscillation of NF-κB. Interestingly, A20 mainly shapes the late NF-κB response rather than the initial response, and thus, knockout of TNFAIP3 does not directly affect NF-κB localization [147].
Figure 4. Negative feedback mechanism in NF-κB dynamics.
(A) Upon inhibitor of κB (IκB) kinase (IKK) complex activation by tumor necrosis factor (TNF) stimulation, nuclear factor-κB (NF-κB) is released by IκBs (IκBα, IκBβ, and IκBε) and enters the nucleus. Along with the activation of inflammation-related genes, NF-κB induces IκBs and A20. When NF-κB is activated, IκBs are released from the NF-κB DNA-binding domain, and A20 suppresses IKK activation. IκBα, which rapidly transfers to the nucleus upon synthesis, is the primary component of this negative feedback and has the function of stripping NF-κB bound to DNA and transferring it to the cytoplasm. Due to this negative feedback system, NF-κB moves in and out of the nucleus, resulting in an oscillating behavior. (B) IKBA knockout abolishes NF-κB oscillations. By contrast, A20 mainly shapes the late NF-κB response rather than the initial response, and thus, knockout of TNFAIP3 does not directly affect NF-κB localization (adapted from [147]).
Many mathematical models include a negative feedback mechanism by IκBs or A20 in NF-κB signaling to identify the mechanism of NF-κB oscillations or their encoding or decoding properties. The difference between oscillating and non-oscillating NF-κB dynamics was investigated by Mothes et al. [152] using the ODE model. They found that NF-κB can exhibit heterogeneity of dynamics from the same stimulus, which is caused by a change in intracellular parameters in the pathway. They also indicated that the concentration of NF-κB and the transcriptional rate constant of IκBα are important parameters that change the dynamics and fold change of NF-κB. Benary and Wolf extended the ODE model developed by Lipniacki et al. [153] to investigate the impact of the regulation of beta-transduction repeat containing protein (β-TrCP)-mediated IκB degradation [154]. NF-κB is activated when IκBs are degraded, and this process is mediated by β-TrCP. In this study, they found that enhancing the β-TrCP-mediated degradation of IκB increases the steady-state concentration of nuclear NF-κB.
A20 is also an important feedback system, along with IκBs. Mothes et al. [155] used a modular modeling approach to determine the impact of different A20 feedback implementations on the dynamics of NF-κB. The authors used three different models (Lipniacki et al. [153], Ashall et al. [146], and Murakawa et al. [156]), which had different A20 implementations against IKK inhibition, and combined them with a common IκBα–A20 negative feedback core module. The authors also analyzed the effects of a wide range of changes in A20 feedback strength, IκBα feedback strength, and TNF-α stimulus strength on NF-κB dynamics. The results showed that A20 feedback strength and TNF-α stimulation strength had different effects on the initial and long-term NF-κB concentrations depending on the models analyzed. Based on experimental validation using TNF-α-stimulated HeLa cells, Murakawa et al. [156] best described the dynamic features of NF-κB. This indicates that regulation described in their ODE model [156], in which both TNF-α-dependent and -independent regulation increases the amount of active IKK and its inhibition by A20, is imprinted in TNF-α-stimulated HeLa cell systems. Using their modular modeling approach, we can elucidate the mechanism by which A20 negative feedback inhibits IKK depending on the cell type.
Positive feedback regulates the TNF-α pathway. A positive feedback mechanism within the upstream kinase signaling complex induces the switch-like activation of NF-κB [157]. TNF-α itself is also transcriptionally regulated by NF-κB, which is reported to function as a feedforward-like rather than positive feedback regulation because several additional regulations, such as splicing control, mRNA degradation, pro-TNF-α activation, and secretion, are needed to fully activate TNF-α [144,158].
Recent reports indicate that the oscillation dynamics of NF-κB nuclear localization are functionally important for biological systems [148,159]. Adelaja et al. [159] reported that different stimuli, such as TNF-α, Pam3-Cys-SK4 (P3CSK), polyinosinic:polycytidylic acid (Poly(I:C)), LPS, and CpG, induce the activation of NF-κB with different dynamics. Using the features of different stimuli and machine learning, they identified six dynamic features encoding NF-κB dynamics: activation speed, peak amplitude, oscillatory dynamics, total activity, duration, and ratio of early to late activity. They further utilized the ODE model to visualize the underlying mechanism and indicated four key characteristics: ligand half-life, receptor translocation and replenishment rates, dose response of adaptor proteins, and deactivation kinetics of adaptors. This study illustrated that cytokine stimuli (TNF-α) and bacterial pathogenic stimuli (P3CSK, Poly(I:C), LPS, and CpG) are recognized by cells as differences in dynamics and induce different biological functions.
Cheng et al. [148] identified NF-κB dynamics as important properties regulating epigenetic changes induced by different input stimuli in macrophage activation. In particular, they showed that non-oscillatory NF-κB liberates chromatin by disrupting nucleosome–histone and DNA interactions. This finding not only indicates that NF-κB oscillation influences chromatin accessibility and regulates gene expression but also presents important evidence that NF-κB dynamics have significant implications for biological systems.
scRNA-seq has been used to reveal the heterogeneity of NF-κB dynamics. Lane et al. [160] revealed the relationship between live-cell imaging of nuclear NF-κB dynamics and scRNA-seq gene expression induced by LPS stimulation using the same single cell. By comparing clustered cells based on NF-κB dynamics observed by microfluidic single-cell imaging and scRNA-seq, the authors found that strong and long-lasting nuclear p65 signaling correlated with increased expression of pro-inflammatory cytokines. Since pro-inflammatory cytokines increase with aging, sustained nuclear p65 activation might be a promising target for the treatment of aging.
As shown in this chapter, the nuclear NF-κB localization pattern is regulated by multiple feedback mechanisms, and NF-κB dynamics are meaningful for the regulation of downstream target genes. Mathematical models, in particular the ODE models, have played important roles in elucidating NF-κB dynamics. How nuclear NF-κB dynamics are involved in aging and what the results of NF-κB dynamics disturbance would be in terms of aging remain unclear; however, these areas seem to be promising targets for preventing age-related diseases.
Next, we will look closer at NF-κB mathematical models for a deeper theoretical understanding of NF-κB dynamics and consider the possibility of regulating aging.
Single-cell analysis and mathematical modeling of the NF-κB network
The unique properties of oscillation dynamics of the NF-κB signaling pathway have been studied and reviewed using mathematical models [161–165]. As previously mentioned, the dynamic properties of NF-κB regulation during aging are not yet fully understood. Thus, utilization of NF-κB mathematical models in aging phenomena, such as decline in immune response, may provide clues to understanding the mechanistic structure of NF-κB dynamics. Here, through a mathematical model of NF-κB, we explore how the heterogeneity of NF-κB dynamics is regulated at the single-cell level and aim to gain insights into the relationship between NF-κB dynamics and aging.
The control of heterogeneity in NF-κB dynamics has been studied along with the development of technologies that allow live imaging observation of single cells; for instance, microfluidic devices allow constant imaging under various conditions specified by the researcher.
When NF-κB activity is measured at the bulk population level using methods such as western blot (WB) analysis, the behavior of single cells is masked in the underlying system [166]. For example, an analog response to a gradual increase in activation and an all-or-nothing digital response may produce similar WBs even though the number of activated cells and the amount of activity per cell may be different [166]. Thus, single-cell level observation is a powerful tool to elucidate NF-κB heterogeneity.
Cells regulate inflammation, caused by cell damage or microbial invasion, by detecting changes in the surrounding environment through NF-κB signaling. However, in the actual cellular environment, the concentration of NF-κB ligands around the cells may fluctuate randomly even when no immune or inflammatory response occurs. Therefore, the NF-κB pathway must distinguish between this external ‘noise' and significant increases in ligands associated with the immune response. To overcome this problem, a system regulated as ‘ON’ or ‘OFF,' called the switch-like system, is activated only when a sufficiently strong signal exceeding the internal threshold is applied in NF-κB signal transduction [167–169]. This increases the robustness of cellular decision-making in noisy environments, resulting in cell-to-cell heterogeneity in transcriptional response [170,171].
To clarify the heterogeneity of NF-κB activation among single cells, Kellogg et al. [167] and Tay et al. [169] focused on the encoding and decoding of the duration and concentration of ligand of the input information by the NF-κB system. Experimental observation of single-cell NF-κB activation by LPS [167] or TNF-α [169] treatment suggested that NF-κB activation at the microscopic level is similar to a switch, with the strength of the induced signal altering the stochastic all-or-nothing response in each cell. Using a hybrid stochastic–deterministic mathematical model, Kellogg et al. [167] found that the integral value (calculated by multiplying ‘concentration' by ‘duration') of stimulation determines the activation of NF-κB in single cells and that the sustained weak stimulation results in heterogeneous activation and timing delay, which is transmitted to gene expression. By contrast, transient strong stimulation at the same site results in rapid and uniform dynamics [167]. These results indicate that it is possible to control the phenotype of a cell via the strength of the input and to control the activation probability of a single cell. They also indicate that a switch-like NF-κB system response to bacterial pathogenic stimuli results in a heterogeneous reaction in the cell population and protects against chronic activation by external ‘noise' in the environment. With aging, SASP and chronic inflammation (inflammaging) occur; the ‘concentration' and ‘duration' of NF-κB perturbation may be said to increase with aging, causing an ‘ON' state in the switch-like NF-κB system.
To understand the sources of internal ‘noise' under intercellular heterogenic responses in the NF-κB system, Bass et al. [172] combined single-transcript measurements with mathematical models to study transcriptional noise in NF-κB-regulated genes. A main cause of gene expression noise in a single cell is the flow of promoters between transcriptionally active and inactive states, a process known to induce ‘transcriptional bursting' [173]. By analyzing the changes in gene expression noise and transcriptional bursts at endogenous NF-κB target promoters before and after TNF-α stimulation, they found that TNF-α primarily activates transcription by increasing the burst size while maintaining burst frequency at gene promoters with relatively high basal values of histone 3-acetylation, which marks the open chromatin status [172]. The authors simulated the transcriptional bursting behavior using a two-promoter state model in which a promoter switches the ‘OFF' state to the ‘ON' state [172]. In this model, the transcription process is described by two main features: burst size, defined as the average number of mRNAs produced per burst (gene activation event), and burst frequency, defined as the frequency at which bursts occur [174]. Using this model, Bass et al. [172] showed that positive feedback of TNF-α amplifies the noise of gene expression by transcription through burst size, forming a subset of cells that express high levels of TNF-α protein. The effect of ‘transcriptional bursting' in aging has not been reported, but since the levels of TNF-α are known to increase with aging [74–77], this internal ‘noise' might provide clues to elucidate the heterogeneity in aging between cells and individuals.
To understand whether cells respond to absolute cytokine levels or to the rate of change in the dynamic concentration of cytokines around the cells, Son et al. [175] studied how the NF-κB pathway responds to time-varying immune inputs, such as increases, decreases, and fluctuations in cytokine signaling. From experimental single-cell observations, they found that NF-κB activity responds to differences in absolute cytokine concentrations, not the cytokine concentrations themselves. Sensitivity analysis using their ODE modeling suggested that A20 and IκBα negative feedback mechanisms accurately detect the dynamics of extracellular cytokines. This work indicates that changes in environmental cytokine concentrations are important for signal processing and that expression of some NF-κB target genes (TNFAIP3 and CCL2) correspond closely to the TNF-α dose at a given time during ramping.
The impact on the rate of change in the localization of NF-κB has also been reported [176]. Nuclear NF-κB levels vary considerably among cells, even in unstimulated cells. To elucidate the mechanism underlying this heterogeneity, Lee et al. [176] confirmed the localization of p65 (RELA) upon TNF-α stimulation in single cells, indicating that the rate of change rather than the absolute amount present in the nucleus is important for the transcriptional activity of NF-κB. Using mathematical modeling, Goentoro et al. [177] found that an incoherent feedforward loop (I1-FFL) (Figure 5) initiated by competition for κB motif binding provides the pre-ligand state required for fold-change detection [176]. In the I1-FFL model, NF-κB and its competitors competitively bind to the κB motif and transcribe the target genes downstream. In the absence of I1-FFL regulation, the transcriptional dynamics of IL-8, TNFAIP3, and NFKBIA could not be reproduced when TNF-α stimulation was applied to experimentally obtained dynamics, whereas the I1-FFL model reproduced the experimental dynamics [176]. The I1-FFL model involves transcription and translation of the competitive factor, and its inhibition results in a structure that causes a time delay [176]. These findings indicate that the transcriptional activity of NF-κB is controlled by two contradictory controls, that is, both activation and repression of the same gene target.
Figure 5. Schematic diagrams of simple and I1-FFL models.
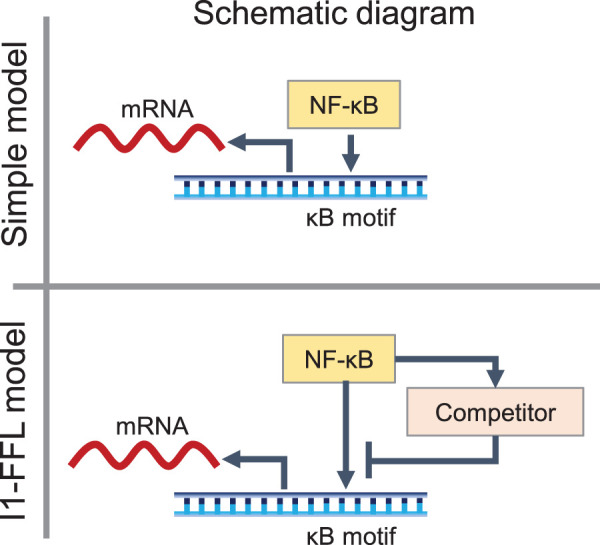
In a simple model, nuclear factor kappa B (NF-κB) binds to the κB motif, which results in the transcription of downstream target genes. In the incoherent feedforward loop (I1-FFL model), NF-κB and its competitors competitively bind to the κB motif and initiate transcription of downstream target genes.
To determine whether fold-change detection in localization of NF-κB is universally implemented in NF-κB target genes, Wong et al. [176] used an I1-FFL model and single-cell live imaging to show that even genes with low levels of TNF-induced transcription induced by NF-κB are responsive to the fold-change in nuclear NF-κB [178]. Using live cell imaging, the authors tracked nuclear NF-κB after TNF-α stimulation and quantified the number of transcripts by RNA fluorescence in situ hybridization in the same cells. They found that the presence of TNF-α induces transcripts of low and high abundance target genes correlated with similar intensity of nuclear NF-κB fold change [178]. The I1-FFL model implements fold-change detection from competition for κB motif binding and shows that fold-change detection can be reproduced across the range of experimentally measured transcripts [178]. This study suggests that cells use the same mechanistic model for low and high levels of transcription genes and distinguish them by adopting different kinetic parameters. Thus, multiple biological mechanisms regulate transcriptional output while maintaining the robustness of NF-κB fold-change detection.
The studies summarized in this chapter show that NF-κB regulation at the single-cell level has been elucidated using mathematical models. A switch-like system and fold-change detection lead to robustness in NF-κB regulation. Linking these systems with aging will further elucidate the mechanistic structure of NF-κB signal transduction regulation in aging as a promising target for research.
Discussion
In recent years, drug discovery and development based on systems biology have attracted increasing interest in the scientific and industrial fields [179,180]. Human disease models and multi-layered omics data can improve the success rate of drug development and predict the safety and efficacy of drugs in patients [181]. Systems biology is a powerful tool for deepening understanding of diseases, such as neurodegenerative diseases [182] and cancer [183]. Drugs related to NF-κB signaling have also been identified using systems biology [184].Pabon et al. [184] reported a network-centric drug discovery approach to predict drugs that inhibit NF-κB signaling and elucidate the network response. By integrating transcriptome analysis, machine learning, structural analysis, and live cell imaging, the authors showed that the maturation of multiprotein complexes required for IKK activation can be inhibited, thus effectively inhibiting NF-κB signaling. Oppelt et al. [185] used a mathematical model to identify factors — IKK and free cytoplasmic IκBα—that contribute to increased hepatotoxicity caused by the combination of TNF-α and the anti-inflammatory drug diclofenac. They not only proposed a new mathematical model of NF-ĸB but also used it to assess the safety of drugs.
Although cell fate is determined by the crosstalk of complex signaling networks, the priority in drug efficacy studies to date has been to confirm the efficacy of a single target. Because the phenotypic system of aging is a product of various signaling networks, a systems biology approach can allow us to view the entire system from a bird's eye view and extract its characteristics. This approach is expected to be applied not only to drug discovery but also to research on general commercial products for aging.
As introduced in this review, the application of mathematical models can elucidate complexes hidden under signal pathways. In the future, combining mathematical models of different signaling pathways for a certain disease or symptom will be important to comprehensively confirm the effects, which will lead to better understanding and treatment of the target disease. Methods are also being developed to accelerate the integration and utilization of the crosstalk of multiple models [121,186–189]. Software tools, such as MATLAB, Copasi [190], Berkeley Madonna [191], and CellDesigner [192,193], are used as modeling tools. For logical modeling (e.g. Boolean networks), tools such as BoolNet [194], MaBoSS [195], GINsim [196], and PyBoolNet [197] are widely used and are summarized by the CoLoMoTo consortium (http://www.colomoto.org/). Our group developed a Python-based ODE modeling and analysis tool for signaling systems called BioMASS [198], which allows multiple sets of parameters to be optimized simultaneously, generating multiple candidate parameters that describe the desired signaling dynamics. Using these tools allows us to integrate two or more separately developed models and perform parameter estimation to fit the experimental data of individual researchers.
Conclusion
In addition to the classical roles of NF-κB, such as inflammation and response during immunity, the role of the NF-κB pathway should not be overlooked in terms of aging. In this review, we outlined the relationship between aging and NF-κB signaling, the contribution of multi-omics analysis to aging research, and the mathematical models indicating the relationship between NF-κB and aging. We summarized the dynamic properties of the NF-κB system and showed that NF-κB dynamics are meaningful for the regulation of downstream target genes. The integration of mathematical models and observation of cell dynamics, such as immunoprecipitation, live single-cell observation, and scRNA-seq analysis, is a powerful tool for deepening understanding of biological phenomena. A combination of both omics analysis and mathematical models will enable us to understand the whole system of aging; these research results are expected to be widely implemented in society in the near future.
Acknowledgements
The authors wish to acknowledge Dr. Sho Tabata for fruitful discussions on the manuscript. M.O. was supported by the JST-CREST Grant No. JPMJCR21N3, JST-Mirai Program No. JPMJMI19G7, and Moonshot R&D Grant No. JPMJMS2021.
Abbreviations
- ACE2
angiotensin-converting enzyme 2
- CD
cluster of differentiation
- ChIP
chromatin immunoprecipitation
- CKII
casein kinase II
- COVID-19
coronavirus disease 2019
- CSE
cigarette smoke extract
- GRN
gene regulatory network
- GSEA
gene set enrichment analysis
- H2O2
hydrogen peroxide
- H3K27ac
H3 acetylation at lysine 27
- H3K4me3
histone H3 trimethylation at lysine 4
- HAGR
Human Aging Genomic Resources
- HSCs
hematopoietic stem cells
- I1-FFL
incoherent feedforward loop
- IKK
inhibitor of κB kinase
- IL
interleukin
- IκB
inhibitor of κB
- LPS
lipopolysaccharide
- NEMO
NF-κB essential modulator
- NF-κB
nuclear factor-κB
- NIK
NF-κB inducing kinase
- NK
natural killer
- NMR
naked mole-rat
- ODE
ordinary differential equation
- P3CSK
Pam3-Cys-SK4
- pHBEC
primary human bronchial epithelial cell
- Poly(I:C)
polyinosinic:polycytidylic acid
- ROS
reactive oxygen species
- RS
replicative stress
- SAM
senescence-accelerated mouse
- SAMP8
senescence-accelerated mouse prone 8
- SARS-CoV-2
severe acute respiratory syndrome coronavirus disease 2
- SASP
senescence-associated secretory phenotype
- SA-β-gal
senescence-associated beta-galactosidase
- scRNA-seq
single-cell RNA sequencing
- STAT
signal transducer and activator of transcription
- TAD
transactivation domain
- TNF
tumor necrosis factor
- UV
ultraviolet
- WB
western blot
- β-TrCP
beta-transduction repeat containing protein
- β-TrCP
beta-transduction repeat containing protein
Data Availability
Data availability statements do not apply to this document as no new data were created or analyzed as part of this study.
Competing Interests
M.H. was an employee of Rohto Pharmaceutical Co., Ltd. at the time of contributing to the manuscript. This work was partially performed under the condition of an invention agreement between Osaka University and Rohto Pharmaceutical.
References
- 1.Pahl, H.L. (1999) Activators and target genes of Rel/NF-kB transcription factors. Oncogene 18, 6853–6866 10.1038/sj.onc.1203239 [DOI] [PubMed] [Google Scholar]
- 2.García-García, V.A., Alameda, J.P., Page, A. and Casanova, M.L. (2021) Role of nf-κb in ageing and age-related diseases: lessons from genetically modified mouse models. Cells 10, 1–29 10.3390/cells10081906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaeberlein, M. (2013) Longevity and aging. F1000Prime Rep. 5, 1–8 10.12703/P5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Childs, B.G., Gluscevic, M., Baker, D.J., Laberge, R.M., Marquess, D., Dananberg, J.et al. (2017) Senescent cells: an emerging target for diseases of ageing. Nat. Rev. Drug Discov. 16, 718–735 10.1038/nrd.2017.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamakoshi, K., Takahashi, A., Hirota, F., Nakayama, R., Ishimaru, N., Kubo, Y.et al. (2009) Real-time in vivo imaging of p16Ink4a reveals cross talk with p53. J. Cell Biol. 186, 393–407 10.1083/jcb.200904105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker, D.J., Perez-Terzic, C., Jin, F., Pitel, K., Niederländer, N.J., Jeganathan, K.et al. (2008) Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat. Cell Biol. 10, 825–836 10.1038/ncb1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rovillain, E., Mansfield, L., Caetano, C., Alvarez-Fernandez, M., Caballero, O.L., Medema, R.H.et al. (2011) Activation of nuclear factor-kappa B signalling promotes cellular senescence. Oncogene 30, 2356–2366 10.1038/onc.2010.611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moskalev, A. and Shaposhnikov, M. (2011) Pharmacological inhibition of NF-κB prolongs lifespan of Drosophila melanogaster. Aging (Albany, NY) 3, 391–394 10.18632/aging.100314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasegawa, Y., Saito, T., Ogihara, T., Ishigaki, Y., Yamada, T., Imai, J.et al. (2012) Blockade of the nuclear factor-κB pathway in the endothelium prevents insulin resistance and prolongs life spans. Circulation 125, 1122–1133 10.1161/CIRCULATIONAHA.111.054346 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, G., Li, J., Purkayastha, S., Tang, Y., Zhang, H., Yin, Y.et al. (2013) Hypothalamic programming of systemic ageing involving IKK-β, NF-κB and GnRH. Nature 497, 211–216 10.1038/nature12143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kriete, A., Mayo, K.L., Yalamanchili, N., Beggs, W., Bender, P., Kari, C.et al. (2008) Cell autonomous expression of inflammatory genes in biologically aged fibroblasts associated with elevated NF-kappaB activity. Immun. Ageing 5, 1–8 10.1186/1742-4933-5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodwell, G.E.J., Sonu, R., Zahn, J.M., Lund, J., Wilhelmy, J., Wang, L.et al. (2004) A transcriptional profile of aging in the human kidney. PLoS Biol. 2, e427 10.1371/journal.pbio.0020427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salminen, A., Kauppinen, A. and Kaarniranta, K. (2012) Emerging role of NF-κB signaling in the induction of senescence-associated secretory phenotype (SASP). Cell. Signal. 24, 835–845 10.1016/j.cellsig.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 14.Lopes-Paciencia, S., Saint-Germain, E., Rowell, M.C., Ruiz, A.F., Kalegari, P. and Ferbeyre, G. (2019) The senescence-associated secretory phenotype and its regulation. Cytokine 117, 15–22 10.1016/j.cyto.2019.01.013 [DOI] [PubMed] [Google Scholar]
- 15.Prata, L.G.P.L., Ovsyannikova, I.G., Tchkonia, T. and Kirkland, J.L. (2018) Senescent cell clearance by the immune system: emerging therapeutic opportunities. Semin. Immunol. 40, 101275 10.1016/j.smim.2019.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nidadavolu, L.S. and Walston, J.D. (2021) Underlying vulnerabilities to the cytokine storm and adverse COVID-19 outcomes in the aging immune system. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 76, E13–E18 10.1093/gerona/glaa209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budamagunta, V., Manohar-Sindhu, S., Yang, Y., He, Y., Traktuev, D.O., Foster, T.C.et al. (2021) Senescence-associated hyper-activation to inflammatory stimuli in vitro. Aging (Albany, NY) 13, 19088–19107 10.18632/aging.203396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Deursen, J.M. (2014) The role of senescent cells in ageing. Nature 509, 439–446 10.1038/nature13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumari, R. and Jat, P. (2021) Mechanisms of cellular senescence: cell cycle arrest and senescence associated secretory phenotype. Front. Cell Dev. Biol. 9, 1–24 10.3389/fcell.2021.64559320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrova, N.V., Velichko, A.K., Razin, S.V. and Kantidze, O.L. (2016) Small molecule compounds that induce cellular senescence. Aging Cell 15, 999–1017 10.1111/acel.12518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke, M.A. and Fisher, J. (2020) Executable cancer models: successes and challenges. Nat. Rev. Cancer 20, 343–354 10.1038/s41568-020-0258-x [DOI] [PubMed] [Google Scholar]
- 22.Sen, R. and Baltimore, D. (1986) Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 46, 705–716 10.1016/0092-8674(86)90346-6 [DOI] [PubMed] [Google Scholar]
- 23.O'Dea, E. and Hoffmann, A. (2010) The regulatory logic of the NF-kappaB signaling system. Cold Spring Harb. Perspect. Biol. 2, 1–13 10.1101/cshperspect.a000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oeckinghaus, A. and Ghosh, S. (2009) The nuclear factor-kB (NF-kB) family of transcription factors. Cold Spring Harb. Perspect. Biol. 1, 1–15 10.1101/cshperspect.a000034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong, H., May, M.J., Jimi, E. and Ghosh, S. (2002) The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell 9, 625–636 10.1016/S1097-2765(02)00477-X [DOI] [PubMed] [Google Scholar]
- 26.Mulero, M.C., Wang, V.Y.F., Huxford, T. and Ghosh, G. (2019) Genome reading by the NF-κB transcription factors. Nucleic Acids Res. 47, 9967–9989 10.1093/nar/gkz739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerstein, M.B., Kundaje, A., Hariharan, M., Landt, S.G., Yan, K.K., Cheng, C.et al. (2012) Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 10.1038/nature11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, J., Zhuang, J., Iyer, S., Lin, X.Y., Whitfield, T.W., Greven, M.C.et al. (2012) Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22, 1798–1812 10.1101/gr.139105.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunham, I., Kundaje, A., Aldred, S.F., Collins, P.J., Davis, C.A., Doyle, F., et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, F.E. and Ghosh, G. (1999) Regulation of DNA binding by Rel/NF-κB transcription factors: structural views. Oncogene 18, 6845–6852 10.1038/sj.onc.1203224 [DOI] [PubMed] [Google Scholar]
- 31.Hayden, M.S. and Ghosh, S. (2012) NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 26, 203–234 10.1101/gad.183434.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, Q., Lenardo, M.J. and Baltimore, D. (2017) 30 years of NF-κB: a blossoming of relevance to human pathobiology. Cell 168, 37–57 10.1016/j.cell.2016.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Courtois, G. and Fauvarque, M.O. (2018) The many roles of ubiquitin in NF-κB signaling. Biomedicines 6, 1–47 10.3390/biomedicines6020043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun, S.C. (2017) The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 17, 545–558 10.1038/nri.2017.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cildir, G., Low, K.C. and Tergaonkar, V. (2016) Noncanonical NF-κB signaling in health and disease. Trends Mol. Med. 22, 414–429 10.1016/j.molmed.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 36.Claudio, E., Brown, K., Park, S., Wang, H. and Siebenlist, U. (2002) BAFF-induced NEMO-independent processing of NF-κB2 in maturing B cells. Nat. Immunol. 3, 958–965 10.1038/ni842 [DOI] [PubMed] [Google Scholar]
- 37.Kayagaki, N., Yan, M., Seshasayee, D., Wang, H., Lee, W., French, D.M.et al. (2002) BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-κB2. Immunity 17, 515–524 10.1016/S1074-7613(02)00425-9 [DOI] [PubMed] [Google Scholar]
- 38.Dejardin, E., Droin, N.M., Delhase, M., Haas, E., Cao, Y., Makris, C.et al. (2002) The lymphotoxin-receptor induces different patterns of gene expression via two NF-B pathways. Immunity 17, 525–535 10.1016/S1074-7613(02)00423-5 [DOI] [PubMed] [Google Scholar]
- 39.Coope, H.J., Atkinson, P.G.P., Huhse, B., Belich, M., Janzen, J., Holman, M.J.et al. (2002) CD40 regulates the processing of NF-κB2 p100 to p52. EMBO J. 21, 5375–5385 10.1093/emboj/cdf542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novack, D.V., Yin, L., Hagen-Stapleton, A., Schreiber, R.D., Goeddel, D.V., Ross, F.P.et al. (2003) The IκB function of NF-κB2 p100 controls stimulated osteoclastogenesis. J. Exp. Med. 198, 771–781 10.1084/jem.20030116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krishnamurthy, J., Torrice, C., Ramsey, M.R., Kovalev, G.I., Al-Regaiey, K., Su, L.et al. (2004) Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 114, 1299–1307 10.1172/JCI22475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayflick, L. and Moorhead, P.S. (1961) The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621 10.1016/0014-4827(61)90192-6 [DOI] [PubMed] [Google Scholar]
- 43.Baker, D.J., Childs, B.G., Durik, M., Wijers, M.E., Sieben, C.J., Zhong, J.et al. (2016) Naturally occurring p16 Ink4a-positive cells shorten healthy lifespan. Nature 530, 184–189 10.1038/nature16932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan, R.G., Ives, S.J., Lesniewski, L.A., Cawthon, R.M., Andtbacka, R.H.I., Dirk Noyes, R.et al. (2013) Age-related telomere uncapping is associated with cellular senescence and inflammation independent of telomere shortening in human arteries. Am. J. Physiol. Heart Circ. Physiol. 305, H251–H258 10.1152/ajpheart.00197.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker, D.J., Wijshake, T., Tchkonia, T., Lebrasseur, N.K., Childs, B.G., Van De Sluis, B.et al. (2011) Clearance of p16 Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 10.1038/nature10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blaser, H., Dostert, C., Mak, T.W. and Brenner, D. (2016) TNF and ROS crosstalk in inflammation. Trends Cell Biol. 26, 249–261 10.1016/j.tcb.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 47.Morgan, M.J. and Liu, Z.G. (2011) Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 21, 103–115 10.1038/cr.2010.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janssens, S. and Tschopp, J. (2006) Signals from within: the DNA-damage-induced NF-κB response. Cell Death Differ. 13, 773–784 10.1038/sj.cdd.4401843 [DOI] [PubMed] [Google Scholar]
- 49.Schieven, G.L., Kirihara, J.M., Myers, D.E., Ledbetter, J.A. and Uckun, F.M. (1993) Reactive oxygen intermediates activate NF-κB in a tyrosine kinase- dependent mechanism and in combination with vanadate activate the p56(lck) and p59(fyn) tyrosine kinases in human lymphocytes. Blood 82, 1212–1220 10.1182/blood.v82.4.1212.1212 [DOI] [PubMed] [Google Scholar]
- 50.Takada, Y., Mukhopadhyay, A., Kundu, G.C., Mahabeleshwar, G.H., Singh, S. and Aggarwal, B.B. (2003) Hydrogen peroxide activates NF-κB through tyrosine phosphorylation of iκBα and serine phosphorylation of p65. Evidence for the involvement of IκBα kinase and Syk protein-tyrosine kinase. J. Biol. Chem. 278, 24233–24241 10.1074/jbc.M212389200 [DOI] [PubMed] [Google Scholar]
- 51.Schoonbroodt, S., Ferreira, V., Best-Belpomme, M., Boelaert, J.R., Legrand-Poels, S., Korner, M.et al. (2000) Crucial role of the amino-terminal tyrosine residue 42 and the carboxyl-terminal PEST domain of iκBα in NF-κB activation by an oxidative stress. J. Immunol. 164, 4292–4300 10.4049/jimmunol.164.8.4292 [DOI] [PubMed] [Google Scholar]
- 52.Herscovitch, M., Comb, W., Ennis, T., Coleman, K., Yong, S., Armstead, B.et al. (2008) Intermolecular disulfide bond formation in the NEMO dimer requires Cys54 and Cys347. Biochem. Biophys. Res. Commun. 367, 103–108 10.1016/j.bbrc.2007.12.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaspers, I., Zhang, W., Fraser, A., Samet, J.M. and Reed, W. (2001) Hydrogen peroxide has opposing effects on IKK activity and IκBα breakdown in airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 24, 769–777 10.1165/ajrcmb.24.6.4344 [DOI] [PubMed] [Google Scholar]
- 54.Kamata, H., Manabe, T., Sin ichi, O., Kamata, K. and Hirata, H. (2002) Hydrogen peroxide activates IκB kinases through phosphorylation of serine residues in the activation loops. FEBS Lett. 519, 231–237 10.1016/S0014-5793(02)02712-6 [DOI] [PubMed] [Google Scholar]
- 55.Reynaert, N.L., van der Vliet, A., Guala, A.S., McGovern, T., Hristova, M., Pantano, C.et al. (2006) Dynamic redox control of NF-κB through glutaredoxin-regulated S-glutathionylation of inhibitory κB kinase β. Proc. Natl Acad. Sci. U.S.A. 103, 13086–13091 10.1073/pnas.0603290103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byun, M.-S., Jeon, K.-I., Choi, J.-W., Shim, J.-Y. and Jue, D.-M. (2002) Dual effect of oxidative stress on NF-κB activation in HeLa cells. Exp. Mol. Med. 34, 332–339 10.1038/emm.2002.47 [DOI] [PubMed] [Google Scholar]
- 57.Korn, S.H., Wouters, E.F.M., Vos, N. and Janssen-Heininger, Y.M.W. (2001) Cytokine-induced activation of nuclear factor-γB is inhibited by hydrogen peroxide through oxidative inactivation of IκB kinase. J. Biol. Chem. 276, 35693–35700 10.1074/jbc.M104321200 [DOI] [PubMed] [Google Scholar]
- 58.Nowak, D.E., Tian, B., Jamaluddin, M., Boldogh, I., Vergara, L.A., Choudhary, S.et al. (2008) Rela Ser 276 phosphorylation is required for activation of a subset of NF-κB-dependent genes by recruiting cyclin-dependent kinase 9/cyclin T1 complexes. Mol. Cell. Biol. 28, 3623–3638 10.1128/mcb.01152-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernard, D., Gosselin, K., Monte, D., Vercamer, C., Bouali, F., Pourtier, A.et al. (2004) Involvement of Rel/nuclear factor-κB transcription factors in keratinocyte senescence. Cancer Res. 64, 472–481 10.1158/0008-5472.CAN-03-0005 [DOI] [PubMed] [Google Scholar]
- 60.Seitz, C.S., Deng, H., Hinata, K., Lin, Q. and Khavari, P.A. (2000) Nuclear factor κB subunits induce epithelial cell growth arrest. Cancer Res. 60, 4085–4092 [PubMed] [Google Scholar]
- 61.Acosta, J.C., O'Loghlen, A., Banito, A., Guijarro, M.V., Augert, A., Raguz, S.et al. (2008) Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133, 1006–1018 10.1016/j.cell.2008.03.038 [DOI] [PubMed] [Google Scholar]
- 62.Kuilman, T., Michaloglou, C., Vredeveld, L.C.W., Douma, S., van Doorn, R., Desmet, C.J.et al. (2008) Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133, 1019–1031 10.1016/j.cell.2008.03.039 [DOI] [PubMed] [Google Scholar]
- 63.Wang, J., Jacob, N.K., Ladner, K.J., Beg, A., Perko, J.D., Tanner, S.M.et al. (2009) Rela/p65 functions to maintain cellular senescence by regulating genomic stability and DNA repair. EMBO Rep. 10, 1272–1278 10.1038/embor.2009.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sfikas, A., Batsi, C., Tselikou, E., Vartholomatos, G., Monokrousos, N., Pappas, P.et al. (2012) The canonical NF-κB pathway differentially protects normal and human tumor cells from ROS-induced DNA damage. Cell. Signal. 24, 2007–2023 10.1016/j.cellsig.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao, J., Li, X., McGowan, S., Niedernhofer, L.J. and Robbins, P.D. (2015) NF-κB activation with aging: characterization and therapeutic inhibition. In NF-kappa B: Methods and Protocols (May, M.J., ed.), pp. 543–557, Springer New York, New York, NY: [DOI] [PubMed] [Google Scholar]
- 66.Osorio, F.G., Soria-Valles, C., Santiago-Fernández, O., Freije, J.M.P. and López-Otín, C. (2016) Chapter Four - NF-κB signaling as a driver of ageing. In International Review of Cell and Molecular Biology (Jeon, K.W., and Galluzzi, L., eds), pp. 133–174, Academic Press, Cambridge, MA. [DOI] [PubMed] [Google Scholar]
- 67.Tichy, E.D., Ma, N., Sidibe, D., Loro, E., Kocan, J., Chen, D.Z.et al. (2021) Persistent NF-κB activation in muscle stem cells induces proliferation-independent telomere shortening. Cell Rep. 35, 109098 10.1016/j.celrep.2021.109098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee, W.P., Hou, M.C., Lan, K.H., Li, C.P., Chao, Y., Lin, H.C.et al. (2016) Helicobacter pylori-induced chronic inflammation causes telomere shortening of gastric mucosa by promoting PARP-1-mediated non-homologous end joining of DNA. Arch. Biochem. Biophys. 606, 90–98 10.1016/j.abb.2016.07.014 [DOI] [PubMed] [Google Scholar]
- 69.López-Otín, C., Blasco, M.A., Partridge, L., Serrano, M. and Kroemer, G. (2013) The hallmarks of aging. Cell 153, 1194–1217 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gupta, S., Su, H., Agrawal, S. and Gollapudi, S. (2018) Molecular changes associated with increased TNF-α-induced apoptotis in naïve (TN) and central memory (TCM) CD8+ T cells in aged humans. Immun. Ageing 15, 1–10 10.1186/s12979-017-0109-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar, B.V., Connors, T.J. and Farber, D.L. (2018) Human T cell development, localization, and function throughout life. Immunity 48, 202–213 10.1016/j.immuni.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, N. and Bevan, M.J. (2011) CD8+ t cells: foot soldiers of the immune system. Immunity 35, 161–168 10.1016/j.immuni.2011.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mousset, C.M., Hobo, W., Woestenenk, R., Preijers, F., Dolstra, H. and van der Waart, A.B. (2019) Comprehensive phenotyping of T cells using flow cytometry. Cytom. Part A 95, 647–654 10.1002/cyto.a.23724 [DOI] [PubMed] [Google Scholar]
- 74.Fagiolo, U., Cossarizza, A., Scala, E., Fanales–Belasio, E., Ortolani, C., Cozzi, E.et al. (1993) Increased cytokine production in mononuclear cells of healthy elderly people. Eur. J. Immunol. 23, 2375–2378 10.1002/eji.1830230950 [DOI] [PubMed] [Google Scholar]
- 75.Bruunsgaard, H., Andersen-Ranberg, K., Hjelmborg, J.V.B., Pedersen, B.K. and Jeune, B. (2003) Elevated levels of tumor necrosis factor alpha and mortality in centenarians. Am. J. Med. 115, 278–283 10.1016/S0002-9343(03)00329-2 [DOI] [PubMed] [Google Scholar]
- 76.Trzonkowski, P., Myśliwska, J., Godlewska, B., Szmit, E., Łukaszuk, K., Wiȩckiewicz, J.et al. (2004) Immune consequences of the spontaneous pro-inflammatory status in depressed elderly patients. Brain Behav. Immun. 18, 135–148 10.1016/S0889-1591(03)00111-9 [DOI] [PubMed] [Google Scholar]
- 77.Penninx, B.W.J.H., Kritchevsky, S.B., Newman, A.B., Nicklas, B.J., Simonsick, E.M., Rubin, S.et al. (2004) Inflammatory markers and incident mobility limitation in the elderly. J. Am. Geriatr. Soc. 52, 1105–1113 10.1111/j.1532-5415.2004.52308.x [DOI] [PubMed] [Google Scholar]
- 78.Ruby, J.G., Smith, M. and Buffenstein, R. (2018) Naked mole-rat mortality rates defy Gompertzian laws by not increasing with age. Elife 7, 1–18 10.7554/eLife.31157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng, J., Yuan, Z., Yang, W., Xu, C., Cong, W., Lin, L.et al. (2017) Comparative study of macrophages in naked mole rats and ICR mice. Oncotarget 8, 96924–96934 10.18632/oncotarget.19661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hilton, H.G., Rubinstein, N.D., Janki, P., Ireland, A.T., Bernstein, N., Fong, N.L.et al. (2019) Single-cell transcriptomics of the naked molerat reveals unexpected features of mammalian immunity. PLoS Biol 17, e3000528 10.1371/journal.pbio.3000528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Helenius, M.A., Kyrylenko, S., Vehviläinen, P. and Salminen, A. (2001) Characterization of aging-associated up-regulation of constitutive nuclear factor-kappa B binding activity. Antioxid. Redox Signal. 3, 147–156 10.1089/152308601750100669 [DOI] [PubMed] [Google Scholar]
- 82.Kim, H.-J., Kim, K.-W., Yu, B.-P. and Chung, H.-Y. (2000) The effect of age on cyclooxygenase-2 gene expression. Free Radic. Biol. Med. 28, 683–692 10.1016/S0891-5849(99)00274-9 [DOI] [PubMed] [Google Scholar]
- 83.Hosokawa, M. (2002) A higher oxidative status accelerates senescence and aggravates age-dependent disorders in SAMP strains of mice. Mech. Ageing Dev. 123, 1553–1561 10.1016/S0047-6374(02)00091-X [DOI] [PubMed] [Google Scholar]
- 84.Takeda, T. (2009) Senescence-accelerated mouse (SAM) with special references to neurodegeneration models, SAMP8 and SAMP10 mice. Neurochem. Res. 34, 639–659 10.1007/s11064-009-9922-y [DOI] [PubMed] [Google Scholar]
- 85.Forman, K., Vara, E., García, C., Kireev, R., Cuesta, S., Acuña-Castroviejo, D.et al. (2016) Influence of aging and growth hormone on different members of the NFkB family and IkB expression in the heart from a murine model of senescence-accelerated aging. Exp. Gerontol. 73, 114–120 10.1016/j.exger.2015.11.005 [DOI] [PubMed] [Google Scholar]
- 86.Jurk, D., Wilson, C., Passos, J.F., Oakley, F., Correia-Melo, C., Greaves, L.et al. (2014) Chronic inflammation induces telomere dysfunction and accelerates ageing in mice. Nat. Commun. 2, 4172 10.1038/ncomms5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bernal, G.M., Wahlstrom, J.S., Crawley, C.D., Cahill, K.E., Pytel, P., Liang, H.et al. (2014) Loss of Nfkb1 leads to early onset aging. Aging (Albany, NY) 6, 931–942 10.18632/aging.100702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dato, S., Crocco, P., Rambaldi Migliore, N. and Lescai, F. (2021) Omics in a digital world: the role of bioinformatics in providing new insights into human aging. Front. Genet. 12, 1–17 10.3389/fgene.2021.689824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tacutu, R., Thornton, D., Johnson, E., Budovsky, A., Barardo, D., Craig, T.et al. (2018) Human ageing genomic resources: new and updated databases. Nucleic Acids Res. 46, D1083–D1090 10.1093/nar/gkx1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gorgoulis, V., Adams, P.D., Alimonti, A., Bennett, D.C., Bischof, O., Bishop, C.et al. (2019) Cellular senescence: defining a path forward. Cell 179, 813–827 10.1016/j.cell.2019.10.005 [DOI] [PubMed] [Google Scholar]
- 91.Liu, G.H., Bao, Y., Qu, J., Zhang, W., Zhang, T., Kang, W.et al. (2021) Aging atlas: a multi-omics database for aging biology. Nucleic Acids Res. 49, D825–D830 10.1093/nar/gkaa894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taylor, J., Reynolds, L., Hou, L., Lohman, K., Cui, W., Kritchevsky, S.et al. (2017) Transcriptomic profiles of aging in naïve and memory CD4+ cells from mice. Immun. Ageing 14, 1–14 10.1186/s12979-017-0092-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Voic, H., Li, X., Jang, J.H., Zou, C., Sundd, P., Alder, J.et al. (2019) RNA sequencing identifies common pathways between cigarette smoke exposure and replicative senescence in human airway epithelia. BMC Genomics 20, 1–12 10.1186/s12864-018-5409-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nyunoya, T., Monick, M.M., Klingelhutz, A., Yarovinsky, T.O., Cagley, J.R. and Hunninghake, G.W. (2006) Cigarette smoke induces cellular senescence. Am. J. Respir. Cell Mol. Biol. 35, 681–688 10.1165/rcmb.2006-0169OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang, H., Zhou, Y., Yin, Z., Chen, L., Jin, L., Cui, Q.et al. (2020) Transcriptome analysis of common and diverged circulating miRNAs between arterial and venous during aging. Aging (Albany, NY) 12, 12987–13004 10.18632/aging.103385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang, J., Litvinova, M., Liang, Y., Wang, Y., Wang, W., Zhao, S.et al. (2020) Changes in contact patterns shape the dynamics of the COVID-19 outbreak in China. Science 368, 1481–1486 10.1126/science.abb8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Onder, G., Rezza, G. and Brusaferro, S. (2020) Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA - J. Am. Med. Assoc. 323, 1775–1776 10.1001/jama.2020.4683 [DOI] [PubMed] [Google Scholar]
- 98.Ma, S., Sun, S., Li, J., Fan, Y., Qu, J., Sun, L.et al. (2021) Single-cell transcriptomic atlas of primate cardiopulmonary aging. Cell Res. 31, 415–432 10.1038/s41422-020-00412-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haustead, D.J., Stevenson, A., Saxena, V., Marriage, F., Firth, M., Silla, R., et al. (2016) Transcriptome analysis of human ageing in male skin shows mid-life period of variability and central role of NF-κB. Sci. Rep. 6, 26846 10.1038/srep26846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O'Dea, E.L., Kearns, J.D. and Hoffmann, A. (2008) UV as an amplifier rather than inducer of NF-κB activity. Mol. Cell 30, 632–641 10.1016/j.molcel.2008.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zou, Z., Long, X., Zhao, Q., Zheng, Y., Song, M., Ma, S.et al. (2021) A single-cell transcriptomic atlas of human skin aging. Dev. Cell 56, 383–397.e8 10.1016/j.devcel.2020.11.002 [DOI] [PubMed] [Google Scholar]
- 102.Furey, T.S. (2012) ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 13, 840–852 10.1038/nrg3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park, P.J. (2009) ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 10, 669–680 10.1038/nrg2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Greer, E.L., Maures, T.J., Hauswirth, A.G., Green, E.M., Leeman, D.S., Maro, G.S.et al. (2010) Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466, 383–387 10.1038/nature09195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maures, T.J., Greer, E.L., Hauswirth, A.G. and Brunet, A. (2011) The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell 10, 980–990 10.1111/j.1474-9726.2011.00738.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheng, H., Xuan, H., Green, C.D., Han, Y., Sun, N., Shen, H.et al. (2018) Repression of human and mouse brain inflammaging transcriptome by broad gene-body histone hyperacetylation. Proc. Natl Acad. Sci. U.S.A. 115, 7611–7616 10.1073/pnas.1800656115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kawahara, T.L.A., Michishita, E., Adler, A.S., Damian, M., Berber, E., Lin, M.et al. (2009) SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell 136, 62–74 10.1016/j.cell.2008.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.O'Brown, Z.K., Van Nostrand, E.L., Higgins, J.P. and Kim, S.K. (2015) The inflammatory transcription factors NFκB, STAT1 and STAT3 drive age-associated transcriptional changes in the human kidney. PLoS Genet. 11, e1005734 10.1371/journal.pgen.1005734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Benayoun, B.A., Pollina, E.A., Singh, P.P., Mahmoudi, S., Harel, I., Casey, K.M.et al. (2019) Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 29, 697–709 10.1101/gr.240093.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ardlie, K.G., DeLuca, D.S., Segrè, A.V., Sullivan, T.J., Young, T.R., Gelfand, E.T.et al. (2015) The genotype-tissue expression (GTEx) pilot analysis: multiissue gene regulation in humans. Science 348, 648–660 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu, Y., Fuscoe, J.C., Zhao, C., Guo, C., Jia, M., Qing, T.et al. (2014) A rat RNA-seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 5, 3230 10.1038/ncomms4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Baumgart, M., Groth, M., Priebe, S., Savino, A., Testa, G., Dix, A.et al. (2014) RNA-seq of the aging brain in the short-lived fish N. furzeri - conserved pathways and novel genes associated with neurogenesis. Aging Cell 13, 965–974 10.1111/acel.12257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chien, Y., Scuoppo, C., Wang, X., Fang, X., Balgley, B., Bolden, J.E.et al. (2011) Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 25, 2125–2136 10.1101/gad.17276711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stöckl, J.B., Schmid, N., Flenkenthaler, F., Drummer, C., Behr, R., Mayerhofer, A.et al. (2020) Proteomic insights into senescence of testicular peritubular cells from a nonhuman primate model. Cells 9, 1–16 10.3390/cells9112498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nättinen, J., Jylhä, A., Aapola, U., Mäkinen, P., Beuerman, R., Pietilä, J.et al. (2019) Age-associated changes in human tear proteome. Clin. Proteomics 16, 1–11 10.1186/s12014-019-9233-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eberly, L.E. (2007) Correlation and Simple Linear Regression. In Topics in Biostatistics (Ambrosius, W.T., ed.), pp. 143–164, Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 117.Westont, A.D. and Hood, L. (2004) Systems biology, proteomics, and the future of health care: toward predictive, preventative, and personalized medicine. J. Proteome Res. 3, 179–196 10.1021/pr0499693 [DOI] [PubMed] [Google Scholar]
- 118.Gunawardena, J. (2014) Models in biology: ‘accurate descriptions of our pathetic thinking’. BMC Biol. 12, 29 10.1186/1741-7007-12-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kestler, H.A., Wawra, C., Kracher, B. and Kühl, M. (2008) Network modeling of signal transduction: establishing the global view. BioEssays 30, 1110–1125 10.1002/bies.20834 [DOI] [PubMed] [Google Scholar]
- 120.Kholodenko, B., Yaffe, M.B. and Kolch, W. (2012) Computational approaches for analyzing information flow in biological networks. Sci. Signal. 5, 1–15 10.1126/scisignal.2002961 [DOI] [PubMed] [Google Scholar]
- 121.Schwab, J.D., Kühlwein, S.D., Ikonomi, N., Kühl, M. and Kestler, H.A. (2020) Concepts in Boolean network modeling: what do they all mean? Comput. Struct. Biotechnol. J. 18, 571–582 10.1016/j.csbj.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang, B. and Chen, Y. (2020) Overview of gene regulatory network interference based on differential equation models. Curr. Protein Pept. Sci. 21, 1054–1059 10.2174/1389203721666200213103350 [DOI] [PubMed] [Google Scholar]
- 123.Lebl, J. (2013) Ordinary differential equations. In Computational Toxicology: Volume II (Reisfeld, B. and Mayeno, A.N., eds), pp. 475–498, Humana Press, Totowa, NJ [Google Scholar]
- 124.Chen, W.W., Niepel, M. and Sorger, P.K. (2010) Classic and contemporary approaches to modeling biochemical reactions. Genes Dev. 24, 1861–1875 10.1101/gad.1945410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang, R.S., Saadatpour, A. and Albert, R. (2012) Boolean modeling in systems biology: an overview of methodology and applications. Phys. Biol. 9, 055001 10.1088/1478-3975/9/5/055001 [DOI] [PubMed] [Google Scholar]
- 126.Seixas, F.L., Zadrozny, B., Laks, J., Conci, A. and Muchaluat Saade, D.C. (2014) A Bayesian network decision model for supporting the diagnosis of dementia, Alzheimer's disease and mild cognitive impairment. Comput. Biol. Med. 51, 140–158 10.1016/j.compbiomed.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 127.Pe'er, D. (2005) Bayesian network analysis of signaling networks: a primer. Sci. STKE 2005, 1–13 10.1126/stke.2812005pl4 [DOI] [PubMed] [Google Scholar]
- 128.Meyer, P., Maity, P., Burkovski, A., Schwab, J., Müssel, C., Singh, K., et al. (2017) A model of the onset of the senescence associated secretory phenotype after DNA damage induced senescence. PLoS Comput. Biol. 13, 1–30 10.1371/journal.pcbi.1005741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schwab, J.D., Siegle, L., Kühlwein, S.D., Kühl, M. and Kestler, H.A. (2017) Stability of signaling pathways during aging—a Boolean network approach. Biology (Basel) 6, 1–12 10.3390/biology6040046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schwab, J.D., Ikonomi, N., Werle, S.D., Weidner, F.M., Geiger, H. and Kestler, H.A. (2021) Reconstructing Boolean network ensembles from single-cell data for unraveling dynamics in the aging of human hematopoietic stem cells. Comput. Struct. Biotechnol. J. 19, 5321–5332 10.1016/j.csbj.2021.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Welle, S., Brooks, A.I., Delehanty, J.M., Needler, N. and Thornton, C.A. (2003) Gene expression profile of aging in human muscle. Physiol. Genomics 14, 149–159 10.1152/physiolgenomics.00049.2003 [DOI] [PubMed] [Google Scholar]
- 132.Ratliff, M.L., Garton, J., James, J.A. and Webb, C.F. (2020) ARID3a expression in human hematopoietic stem cells is associated with distinct gene patterns in aged individuals. Immun. Ageing 17, 1–15 10.1186/s12979-020-00198-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.De Haan, G. and Lazare, S.S. (2018) Aging of hematopoietic stem cells. Blood 131, 479–487 10.1182/blood-2017-06-746412 [DOI] [PubMed] [Google Scholar]
- 134.Uyar, B., Palmer, D., Kowald, A., Murua Escobar, H., Barrantes, I., Möller, S.et al. (2020) Single-cell analyses of aging, inflammation and senescence. Ageing Res. Rev. 64, 101156 10.1016/j.arr.2020.101156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Martinez-Jimenez, C.P., Eling, N., Chen, H., Vallejos, C.A., Kolodziejczyk, A.A., Connor, F.et al. (2017) Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science 355, 1433–1436 10.1126/science.aah4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang, W., Zhang, S., Yan, P., Ren, J., Song, M., Li, J.et al. (2020) A single-cell transcriptomic landscape of primate arterial aging. Nat. Commun. 11, 1–13 10.1038/s41467-020-15997-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Enge, M., Arda, H.E., Mignardi, M., Beausang, J., Bottino, R., Kim, S.K.et al. (2017) Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171, 321–330.e14 10.1016/j.cell.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hammond, T.R., Dufort, C., Dissing-Olesen, L., Giera, S., Young, A., Wysoker, A.et al. (2019) Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6 10.1016/j.immuni.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Angelidis, I., Simon, L.M., Fernandez, I.E.. Strunz, M., Mayr, C.H., Greiffo, F.R.et al. (2019) An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 10, 1–17 10.1038/s41467-019-08831-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Solé-Boldo, L., Raddatz, G., Schütz, S., Mallm, J.P., Rippe, K., Lonsdorf, A.S.et al. (2020) Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun. Biol. 3, 1–12 10.1038/s42003-020-0922-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hernando-Herraez, I., Evano, B., Stubbs, T., Commere, P.H., Jan Bonder, M., Clark, S., et al. (2019) Ageing affects DNA methylation drift and transcriptional cell-to-cell variability in mouse muscle stem cells. Nat. Commun. 10, 1–11 10.1038/s41467-019-12293-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ferrucci, L., Gonzalez-Freire, M., Fabbri, E., Simonsick, E., Tanaka, T., Moore, Z.et al. (2020) Measuring biological aging in humans: a quest. Aging Cell 19, 1–21 10.1111/acel.13080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang, M., McKay, D., Pollard, J.W. and Lewis, C.E. (2018) Diverse functions of macrophages in different tumor microenvironments. Cancer Res. 78, 5492–5503 10.1158/0008-5472.CAN-18-1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mitchell, S., Vargas, J. and Hoffmann, A. (2016) Signaling via the NFκB system. Wiley Interdiscip. Rev. Syst. Biol. Med. 8, 227–241 10.1002/wsbm.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hoffmann, A., Levchenko, A., Scott, M.L. and Baltimore, D. (2002) The IκB-NF-κB signaling module: temporal control and selective gene activation. Science 298, 1241–1245 10.1126/science.1071914 [DOI] [PubMed] [Google Scholar]
- 146.Ashall, L., Horton, C.A., Nelson, D.E., Paszek, P., Harper, C.V., Sillitoe, K.et al. (2009) Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science 324, 242–246 10.1126/science.1164860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Werner, S.L., Kearns, J.D., Zadorozhnaya, V., Lynch, C., O'Dea, E., Boldin, M.P.et al. (2008) Encoding NF-κB temporal control in response to TNF: distinct roles for the negative regulators iκBα and A20. Genes Dev. 22, 2093–2101 10.1101/gad.1680708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cheng, Q.J., Ohta, S., Sheu, K.M., Spreafico, R., Adelaja, A., Taylor, B.et al. (2021) NF-κB dynamics determine the stimulus specificity of epigenomic reprogramming in macrophages. Science 372, 1349–1353 10.1126/science.abc0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fagerlund, R., Behar, M., Fortmann, K.T., Lin, Y.E., Vargas, J.D. and Hoffmann, A. (2015) Anatomy of a negative feedback loop: the case of IκB. J. R. Soc. Interface 12, 20150262 10.1098/rsif.2015.0262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kearns, J.D., Basak, S., Werner, S.L., Huang, C.S. and Hoffmann, A. (2006) Iκbε provides negative feedback to control NF-κB oscillations, signaling dynamics, and inflammatory gene expression. J. Cell Biol. 173, 659–664 10.1083/jcb.200510155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Longo, D.M., Selimkhanov, J., Kearns, J.D., Hasty, J., Hoffmann, A. and Tsimring, L.S. (2013) Dual delayed feedback provides sensitivity and robustness to the NF-κB signaling module. PLoS Comput. Biol. 9, e1003112 10.1371/journal.pcbi.1003112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mothes, J., Busse, D., Kofahl, B. and Wolf, J. (2015) Sources of dynamic variability in NF-κB signal transduction: a mechanistic model. BioEssays 37, 452–462 10.1002/bies.201400113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lipniacki, T., Paszek, P., Brasier, A.R., Luxon, B. and Kimmel, M. (2004) Mathematical model of NF- κB regulatory module. J. Theor. Biol. 228, 195–215 10.1016/j.jtbi.2004.01.001 [DOI] [PubMed] [Google Scholar]
- 154.Benary, U. and Wolf, J. (2019) Controlling nuclear NF-κB dynamics by β-TrCP- insights from a computational model. Biomedicines 7, 8–13 10.3390/biomedicines7020040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mothes, J., Ipenberg, I., Arslan, S.Ç, Benary, U., Scheidereit, C. and Wolf, J. (2020) A quantitative modular modeling approach reveals the effects of different A20 feedback implementations for the NF-kB signaling dynamics. Front. Physiol. 11, 896 10.3389/fphys.2020.00896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Murakawa, Y., Hinz, M., Mothes, J., Schuetz, A., Uhl, M., Wyler, E., et al. (2015) RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-κ B pathway. Nat. Commun. 6, 7367 10.1038/ncomms8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shinohara, H., Behar, M., Inoue, K., Hiroshima, M., Yasuda, T., Nagashima, T.et al. (2014) Positive feedback within a kinase signaling complex functions as a switch mechanism for NF-κB activation. Science 344, 760–764 10.1126/science.1250020 [DOI] [PubMed] [Google Scholar]
- 158.Caldwell, A.B., Birnbaum, H.A., Cheng, Z., Hoffmann, A. and Vargas, J.D. (2014) Network dynamics determine the autocrine and paracrine signaling functions of TNF. Genes Dev. 28, 2120–2133 10.1101/gad.244749.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Adelaja, A., Taylor, B., Sheu, K.M., Liu, Y., Luecke, S. and Hoffmann, A. (2021) Six distinct NFκB signaling codons convey discrete information to distinguish stimuli and enable appropriate macrophage responses. Immunity 54, 916–930.e7 10.1016/j.immuni.2021.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lane, K., Van Valen, D., DeFelice, M.M., Macklin, D.N., Kudo, T., Jaimovich, A.et al. (2017) Measuring signaling and RNA-seq in the same cell links gene expression to dynamic patterns of NF-κB activation. Cell Syst. 4, 458–469.e5 10.1016/j.cels.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mitchell, S., Tsui, R. and Hoffmann, A. (2015) Studying NF-κB Signaling with Mathematical Models. In NF-kappa B: Methods and Protocols (May, M.J., ed.), pp. 647–661, Springer New York, New York, NY [DOI] [PubMed] [Google Scholar]
- 162.Basak, S., Behar, M. and Hoffmann, A. (2012) Lessons from mathematically modeling the NF-κB pathway. Immunol. Rev. 246, 221–238 10.1111/j.1600-065X.2011.01092.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cheong, R., Hoffmann, A. and Levchenko, A. (2008) Understanding NF-κB signaling via mathematical modeling. Mol. Syst. Biol. 4, 192 10.1038/msb.2008.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lipniacki, T. and Kimmel, M. (2007) Deterministic and stochastic models of NFκB pathway. Cardiovasc. Toxicol. 7, 215–234 10.1007/s12012-007-9003-x [DOI] [PubMed] [Google Scholar]
- 165.Williams, R.A., Timmis, J. and Qwarnstrom, E.E. (2014) Computational models of the NF-κB signalling pathway. Computation 2, 131–158 10.3390/computation2040131 [DOI] [Google Scholar]
- 166.Jeknić, S., Kudo, T. and Covert, M.W. (2019) Techniques for studying decoding of single cell dynamics. Front. Immunol. 10, 1–12 10.3389/fimmu.2019.00755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kellogg, R.A., Tian, C., Lipniacki, T., Quake, S.R. and Tay, S. (2015) Digital signaling decouples activation probability and population heterogeneity. Elife 4, e08931 10.7554/eLife.08931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shah, N.A. and Sarkar, C.A. (2011) Robust network topologies for generating switch-like cellular responses. PLoS Comput. Biol. 7, e1002085 10.1371/journal.pcbi.1002085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tay, S., Hughey, J.J., Lee, T.K., Lipniacki, T., Quake, S.R. and Covert, M.W. (2010) Single-cell NF-B dynamics reveal digital activation and analogue information processing. Nature 466, 267–271 10.1038/nature09145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Michida, H., Imoto, H., Shinohara, H., Yumoto, N., Seki, M., Umeda, M.et al. (2020) The number of transcription factors at an enhancer determines switch-like gene expression. Cell Rep. 31, 107724 10.1016/j.celrep.2020.107724 [DOI] [PubMed] [Google Scholar]
- 171.Wibisana, J.N., Inaba, T., Shinohara, H., Yumoto, N., Hayashi, T., Umeda, M.et al. (2021) Enhanced transcriptional heterogeneity mediated by NF-κB super-enhancers. bioRxiv 10.1101/2021.07.13.452147, Dec. 06, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bass, V.L., Wong, V.C., Bullock, M.E., Gaudet, S. and Miller-Jensen, K. (2021) TNF stimulation primarily modulates transcriptional burst size of NF-κB-regulated genes. Mol. Syst. Biol. 17, 1–20 10.15252/msb.202010127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Klindziuk, A. and Kolomeisky, A.B. (2021) Understanding the molecular mechanisms of transcriptional bursting. Phys. Chem. Chem. Phys. 23, 21399–21406 10.1039/D1CP03665C [DOI] [PubMed] [Google Scholar]
- 174.Nicolas, D., Zoller, B., Suter, D.M. and Naef, F. (2018) Modulation of transcriptional burst frequency by histone acetylation. Proc. Natl Acad. Sci. U.S.A. 115, 7153–7158 10.1073/pnas.1722330115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Son, M., Wang, A.G., Tu, H.L., Metzig, M.O., Patel, P., Husain, K.et al. (2021) NF-κB responds to absolute differences in cytokine concentrations. Sci. Signal. 14, 1–13 10.1126/scisignal.aaz4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lee, R.E.C., Walker, S.R., Savery, K., Frank, D.A. and Gaudet, S. (2014) Fold change of nuclear NF-κB determines TNF-induced transcription in single cells. Mol. Cell 53, 867–879 10.1016/j.molcel.2014.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Goentoro, L., Shoval, O., Kirschner, M.W. and Alon, U. (2009) The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol. Cell 36, 894–899 10.1016/j.molcel.2009.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wong, V.C., Mathew, S., Ramji, R., Gaudet, S. and Miller-Jensen, K. (2019) Fold-change detection of NF-κB at target genes with different transcript outputs. Biophys. J. 116, 709–724 10.1016/j.bpj.2019.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Behl, T., Kaur, I., Sehgal, A., Singh, S., Bhatia, S., Al-Harrasi, A.et al. (2021) Bioinformatics accelerates the major tetrad: a real boost for the pharmaceutical industry. Int. J. Mol. Sci. 22, 1–28 10.3390/ijms22126184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mohammadi, E., Benfeitas, R., Turkez, H., Boren, J., Nielsen, J., Uhlen, M.et al. (2020) Applications of genome-wide screening and systems biology approaches in drug repositioning. Cancers 12, 1–24 10.3390/cancers12092694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nassar, S.F., Raddassi, K. and Wu, T. (2021) Single-cell multiomics analysis for drug discovery. Metabolites 11, 729 10.3390/metabo11110729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Parikshak, N.N., Gandal, M.J. and Geschwind, D.H. (2015) Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet. 16, 441–458 10.1038/nrg3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pathmanathan, S., Grozavu, I., Lyakisheva, A. and Stagljar, I. (2021) Drugging the undruggable proteins in cancer: a systems biology approach. Curr. Opin. Chem. Biol. S1367–5931, 1–9 10.1016/j.cbpa.2021.07.004 [DOI] [PubMed] [Google Scholar]
- 184.Pabon, N.A., Zhang, Q., Cruz, J.A., Schipper, D.L., Camacho, C.J. and Lee, R.E.C. (2019) A network-centric approach to drugging TNF-induced NF-κB signaling. Nat. Commun. 10, 1–9 10.1038/s41467-019-08802-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Oppelt, A., Kaschek, D., Huppelschoten, S., Sison-Young, R., Zhang, F., Buck-Wiese, M.et al. (2018) Model-based identification of TNFα-induced IKKβ-mediated and iκBα-mediated regulation of NFκB signal transduction as a tool to quantify the impact of drug-induced liver injury compounds. NPJ Syst. Biol. Appl. 4, 23 10.1038/s41540-018-0058-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Olivier, B.G., Swat, M.J. and Moné, M.J. (2016) Modeling and simulation tools: from systems biology to systems medicine. In Systems Medicine (Schmitz, U. and Wolkenhauer, O., eds), pp. 441–463, Springer New York, New York, NY: [DOI] [PubMed] [Google Scholar]
- 187.Lubbock, A.L.R. and Lopez, C.F. (2021) Programmatic modeling for biological systems. Curr. Opin. Syst. Biol. 27, 100343 10.1016/j.coisb.2021.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gyori, B.M., Bachman, J.A., Subramanian, K., Muhlich, J.L., Galescu, L. and Sorger, P.K. (2017) From word models to executable models of signaling networks using automated assembly. Mol. Syst. Biol. 13, 954 10.15252/msb.20177651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ghosh, S., Matsuoka, Y., Asai, Y., Hsin, K.Y. and Kitano, H. (2011) Software for systems biology: from tools to integrated platforms. Nat. Rev. Genet. 12, 821–832 10.1038/nrg3096 [DOI] [PubMed] [Google Scholar]
- 190.Hoops, S., Gauges, R., Lee, C., Pahle, J., Simus, N., Singhal, M.et al. (2006) COPASI - A COmplex PAthway SImulator. Bioinformatics 22, 3067–3074 10.1093/bioinformatics/btl485 [DOI] [PubMed] [Google Scholar]
- 191.Macey, R.I. and Oster, G. (2001) Berkeley Madonna: Modeling and Analysis of Dynamic Systems, University of California, Berkeley, CA [Google Scholar]
- 192.Matsuoka, Y., Funahashi, A., Ghosh, S. and Kitano, H. (2014) Modeling and simulation using cellDesigner. Methods Mol. Biol. 1164, 121–145 10.1007/978-1-4939-0805-9_11 [DOI] [PubMed] [Google Scholar]
- 193.Funahashi, A., Matsuoka, Y., Jouraku, A., Morohashi, M., Kikuchi, N. and Kitano, H. (2008) Celldesigner 3.5: a versatile modeling tool for biochemical networks. Proc. IEEE 96, 1254–1265 10.1109/JPROC.2008.925458 [DOI] [Google Scholar]
- 194.Müssel, C., Hopfensitz, M. and Kestler, H.A. (2010) BoolNet-an R package for generation, reconstruction and analysis of Boolean networks. Bioinformatics 26, 1378–1380 10.1093/bioinformatics/btq124 [DOI] [PubMed] [Google Scholar]
- 195.Stoll, G., Viara, E., Barillot, E. and Calzone, L. (2012) Continuous time boolean modeling for biological signaling: application of Gillespie algorithm. BMC Syst. Biol. 6, 1–18 10.1186/1752-0509-6-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Naldi, A., Berenguier, D., Fauré, A., Lopez, F., Thieffry, D. and Chaouiya, C. (2009) Logical modelling of regulatory networks with GINsim 2.3. BioSystems 97, 134–139 10.1016/j.biosystems.2009.04.008 [DOI] [PubMed] [Google Scholar]
- 197.Klarner, H., Streck, A. and Siebert, H. (2017) Pyboolnet: a Python package for the generation, analysis and visualization of boolean networks. Bioinformatics 33, 770–772 10.1093/bioinformatics/btw682 [DOI] [PubMed] [Google Scholar]
- 198.Imoto, H., Zhang, S. and Okada, M. (2020) A computational framework for prediction and analysis of cancer signaling dynamics from rna sequencing data—application to the erbb receptor signaling pathway. Cancers 12, 1–13 10.3390/cancers12102878 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data availability statements do not apply to this document as no new data were created or analyzed as part of this study.