Abstract
We characterized the photobiology of light-activated chloroplast transcription and transcript abundance in mature primary leaves by using the following two systems: transplastomic promoter-reporter gene fusions in tobacco (Nicotiana tabacum), and phytochrome (phyA, phyB, and hy2) and cryptochrome (cry1) mutants of Arabidopsis. In both dicots, blue light and UV-A radiation were the major signals that activated total chloroplast and psbA, rbcL, and 16S rrn transcription. In contrast, transcription activities in plants exposed to red and far-red light were 30% to 85% less than in blue light/UV-A, depending on the gene and plant species. Total chloroplast, psbA, and 16S rrn transcription were 60% to 80% less in the Arabidopsis phyA mutant exposed to blue light/UV-A relative to wild type, thus definitively linking phyA signaling to these photoresponses. To our knowledge, the major role of phyA in mediating the blue light/UV-A photoresponses is a new function for phyA in chloroplast biogenesis at this stage of leaf development. Although rbcL expression in plants exposed to UV-A was 50% less in the phyA mutant relative to wild type, blue light-induced rbcL expression was not significantly affected in the phyA, phyB, and cry1 mutants. However, rbcL expression in blue light was 60% less in the phytochrome chromophore mutant, hy2, relative to wild type, indicating that another phytochrome species (phyC, D, or E) was involved in blue light-induced rbcL transcription. Therefore, at least two different phytochromes, as well as phytochrome-independent photosensory pathways, mediated blue light/UV-A-induced transcription of chloroplast genes in mature leaves.
During chloroplast development, light quality and quantity coordinate nuclear and chloroplast gene expression needed for the proper assembly and function of the multisubunit photosynthetic enzymes and pigment-protein complexes (Mayfield et al., 1995; Goldschmidt-Clermont, 1998; Leon et al., 1998). The effects of light on chloroplast biogenesis are mediated by the co-action of several photoreceptors. They include the phytochromes (Quail, 1994; Pratt, 1995; Smith, 1995; Chory, 1997), the protochlorophyllide holochrome (Thompson and White 1991; Barnes et al., 1996), and the blue light/UV-A photoreceptors such as the flavoprotein, cryptochrome (Cashmore et al., 1999), and photosystem-generated redox potentials (Danon and Mayfield, 1994; Escoubas et al., 1995; Alfonso et al., 2000). The photoreceptors initiate signaling pathways that interact with each other and with pathways initiated by developmental and plastid-derived signals (Fuglevand et al., 1996; Casal and Mazzella, 1998; Lopez-Juez et al., 1998; Neff and Chory, 1998) to modulate gene expression at transcriptional and post-transcriptional levels (Thompson and White 1991; Quail, 1994; Mayfield et al., 1995).
A great deal of information has been obtained from genetic, biochemical, and molecular studies on the photoreceptors and pathways involved in light-regulated nuclear gene expression (Bowler et al., 1994; Quail, 1994; Smith, 1995; Barnes et al., 1996; Chamovitz and Deng, 1996; Chory, 1997; Zhong et al., 1997; Cashmore et al., 1999). However, much less is known about the individual photoreceptor species and photosensory pathways that modulate gene expression in chloroplasts. In seedlings, red and far-red light, mediated by phytochrome, are primary signals that control the levels of light-induced chloroplast mRNA (Link, 1982; Thompson et al., 1983; Zhu et al., 1985) and RNA polymerase activities (Bottomley, 1970; Dubell and Mullet, 1995; Christopher, 1996). Although phyA and the downstream nuclear protein, DET1, have been shown to be involved (Pepper et al., 1994; Dubell and Mullet, 1995; Christopher, 1996; Christopher and Hoffer, 1998), the effects of mutants for individual phytochrome species on chloroplast transcription have not been examined directly.
Many of the studies of phytochrome involvement in chloroplast gene expression have used seedlings as experimental material and have focused on de-etiolation (Link, 1982; Thompson et al., 1983; Zhu et al., 1985; Bowler et al., 1994; Christopher and Mullet, 1994; Dubell and Mullet, 1995). In contrast, we know very little about the photobiology of light-activated transcription of chloroplast genes in green primary leaves. This is important because several light-regulatory mechanisms essential for photosynthetic efficiency and adaptation occur in mature leaves (Melis, 1991; Aro et al., 1993). In addition, the action spectrum for various light-regulated processes (Fluhr et al., 1986; Cosgrove, 1994; Mohr, 1994) and the types of phytochromes that predominate in tissues (Chory et al., 1989; Quail, 1994) change during leaf development. The types of RNA polymerases predominating in plastids also change during leaf development, from a nuclear-encoded T7 phage-type in immature plastids to a plastid-encoded Escherichia coli-like RNA polymerase (PEP) in mature chloroplasts (Igloi and Kössel, 1992; Iratni et al., 1994; Allison et al., 1996; Hajdukiewicz et al., 1997). The PEP initiates transcription from ς70-type promoters upstream from many chloroplast genes such as rbcL, psbA, and the 16S rrn operon, which encode the large subunit of Rubisco, the D1 protein of photosystem II, and the ribosomal RNAs, respectively. Light modulates the association of the PEP with promoters via sigma factors and protein phosphorylation (Baginski et al., 1997; Isono et al., 1997; Kim et al., 1998; Kanamaru et al., 1999; Tan and Troxler, 1999)
Photoreceptor mutants and chimeric promoter-reporter gene fusions in transplastomic chloroplasts potentially represent useful systems with which to begin defining the photobiology of chloroplast gene expression in mature leaves. Therefore, in this study we determined the effects of spectral quality on chloroplast transcription and mRNA accumulation in the following two systems: transplastomic tobacco (Nicotiana tabacum) lines containing the uidA reporter gene driven by the rbcL and 16S rrn promoters; and wild-type and several photoreceptor mutants (phyA, phyB, hy2, and cyrptochrome1 [cry1]) of Arabidopsis. The mutants phyA and phyB are defective in the major phytochrome species, A and B, respectively (Quail, 1994), whereas hy2 is defective in chromophore biosynthesis, making it severely deficient in all phytochrome activities (Smith, 1995). The cry1 mutation is impaired in a high-fluence blue light photoreceptor, cry1 (Cashmore et al., 1999). Mature phyA and cry1 plants grown in white light resemble the wild-type phenotype (Whitelam et al., 1993; Ahmad et al., 1998), whereas phyB and hy2 are yellow-green. We provide direct evidence that blue light and UV-A, but not red or far-red light, were primary signals for activating chloroplast transcription in mature leaves and that phyA mediated the psbA and 16S rrn, but not the rbcL, photoresponses. Another phytochrome species, as well as a distinct blue photosensory pathway, also influenced chloroplast transcription.
RESULTS
We initially examined the effects of spectral quality on chloroplast transcription and mRNA accumulation in two transplastomic lines of tobacco. Each line differed in the gene-specific promoters driving transcription of the uidA reporter gene. The two promoters were derived from the 16S rrn operon and the light-activated rbcL gene, respectively. The non-consensus type plastid promoter transcribed by the nuclear-encoded T7 phage-type was not present in the 16S rrn promoter-uidA transgene (Zoubenko et al., 1994). A third transplastomic line, which had a promoterless uidA transgene, served as a negative control, as did wild-type tobacco.
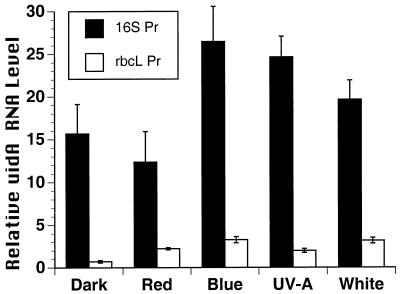
In Figure 1, a uidA gene-specific probe was used to detect uidA mRNA levels in transgenic plants exposed to red, blue, or white light or UV-A radiation, and the uidA RNA levels were quantitated. Steady-state uidA mRNA levels driven by the 16S rrn promoter were higher in plants exposed to blue light, white light, or UV-A, relative to red light and dark controls (Fig. 1). The highest mRNA levels were observed in the blue light and UV-A treatments. For the 16S rrn promoter, similar uidA mRNA levels occurred in plants exposed to red light or darkness (Fig. 1). UidA mRNA levels driven by the rbcL promoter were slightly higher in plants exposed to blue and white light relative to red light, UV-A, or darkness (Fig. 1). About 2-fold higher levels of uidA mRNA were detected in the line with the rbcL promoter exposed to red light relative to dark controls. Overall, uidA mRNA levels were 80% to 90% lower when driven by the rbcL promoter relative to the 16S rrn promoter (Fig. 1), which is in agreement with differences in the expression of these genes from other species and experiments (Rapp et al., 1992; Dubell and Mullet, 1995; Shiina et al., 1998). This experiment suggested that the respective promoters driving uidA transcription controlled differences in mRNA accumulation in response to light quality, although RNA stability effects could not be ruled out.
Figure 1.
Analysis of the effects of spectral quality on transcription of chloroplast promoters (rbcL and 16S rrn) fused to the uidA gene in transplastomic tobacco plants. The transplastomic lines contained the 16S rrn promoter-uidA gene (16S Pr), rbcL promoter-uidA gene (rbcL Pr), or the promoterless uidA gene. Dark-adapted plants were exposed to 12 h of red or blue light at 18 ± 2 μmol m−2 s−1, white light at 100 ± 10 μmol m−2 s−1, or UV-A radiation at 18 ± 2 μmol m−2 s−1. Equal amounts of total cell RNA (12 μg) from each treatment were analyzed. Radioactivity for the bands corresponding to uidA mRNA was quantitated for each treatment from three separate duplicated experiments to estimate the relative mRNA level. Values from the promoterless control were subtracted from the values for plants containing the promoter-bearing constructs. Means ± sd are shown.
It is possible that sequences in the untranslated leader, which is fused to the uidA coding sequence in the rbcL construct, could influence chloroplast mRNA accumulation, via mRNA stability, in response to light (Shiina et al., 1998). Thus, in Figure 2 we used the lysed chloroplast run-on transcription assay to determine transcription rates directly in chloroplasts from the transgenic lines exposed to darkness, red, and blue light. These treatments were chosen because they represented the major extremes in uidA mRNA accumulation (Fig. 1).
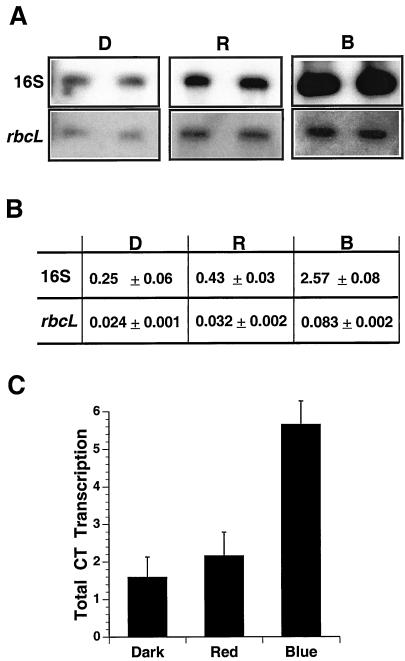
Figure 2.
Analysis of total and promoter-specific chloroplast transcription rates in response to spectral quality in chloroplasts from mature leaves of tobacco. A, The lysed run-on transcription assay was performed on equal amounts of purified chloroplasts from dark-adapted transplastomic tobacco plants containing the 16S rrn promoter-uidA gene (16S rrn) or the rbcL promoter-uidA gene (rbcL) exposed to 12 h of additional darkness (D), or equal fluences of red (R) or blue (B) light (18 ± 2 μmol m−2 s−1). Transcripts produced in the lysed run-on assay were hybridized to non-radioactive uidA gene-specific and vector-specific DNAs attached to nylon membranes. To visualize bands, rbcL-labeled panels were exposed to x-ray film 5-fold longer than for the 16S panels. B, Radioactivity hybridized to gene-specific DNAs was quantitated, after subtracting background hybridization to the vector, for each treatment from two replicates of two independent experiments. Transcription activity (means ± sd) is expressed as fmol [32P]UMP incorporated (1 × 108 chloroplasts)−1 (10 min)−1. C, Total chloroplast transcription rates were measured in wild type treated as in A and B. Rates are expressed as pmol [32P]UMP incorporated (1 × 108 chloroplasts)−1 (10 min)−1. The data represent the means ± sd of two independent experiments with two replicates.
Promoter-specific and total chloroplast transcription were measured in mature tobacco leaves (Fig. 2). Transcription activities from the 16S rrn and rbcL promoters were over 5- and 2-fold higher, respectively, in plants exposed to blue light relative to red light or darkness (Fig. 2, A and B). Overall, transcription from the rbcL promoter was less than one-tenth of transcription from the 16S rrn promoter (Fig. 2B), which resembled differences in mRNA levels (Fig. 1). Transcription from both promoters was moderately higher in plants exposed to red light compared with darkness. Total chloroplast transcription in plants exposed to blue light was over 2-fold higher than red light and darkness (Fig. 2C). In summary, blue light significantly stimulated total and promoter-specific transcription in tobacco chloroplasts (Fig. 2), and this stimulation was correlated with an increase in steady-state mRNA levels (Fig. 1).
The stimulatory effects of blue light on tobacco chloroplast transcription and RNA accumulation raise the question of whether this response was specific to tobacco or whether it existed in other plant species. The types of photoreceptors involved in mediating the activation of chloroplast transcription by blue light in mature leaves are also not known. Therefore, to answer these questions we measured total (Fig. 3) and gene-specific (Fig. 4) chloroplast transcription in mature dark-adapted wild-type Arabidopsis and phyA mutant plants exposed to five spectral regimes. The phyA mutant was chosen because previous research with far-red light suggested that phyA was involved in chloroplast transcription in etiolated pea (Dubell and Mullet, 1995) and barley (Christopher, 1996) seedlings.
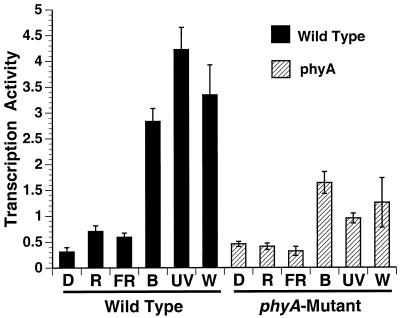
Figure 3.
Analysis of the influence of phyA and light quality on total chloroplast transcription rates in mature leaves of Arabidopsis. Plants were dark-adapted for 36 h and were then exposed to 12 h of additional darkness (D), or 18 ± 2 μmol m−2 s−1 of red light (R), far-red light (fR), blue light (B), or UV-A radiation (UV) or 100 ± 10 μmol m−2 s−1 white light (W) as described in “Materials and Methods.” Lysed run-on transcription activities (means ± sd) are expressed as pmol [32P]UMP incorporated (1 × 108 chloroplasts)−1 (10 min)−1. The data represent four independent experiments with two replicates each.
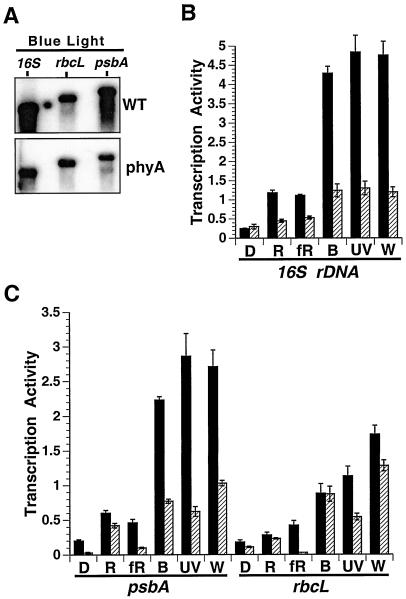
Figure 4.
Analysis of the influence of phyA and light quality on gene-specific chloroplast transcription rates in mature leaves of wild-type (WT) and the phyA mutant of Arabidopsis. A, Transcription of the rbcL, psbA, and 16S genes were measured in purified chloroplasts using the lysed run-on assay as described in “Materials and Methods.” A representative blot from the blue light treatment is shown. B, Quantitation of 16S rrn transcription in wild-type (black bars) and the phyA mutant (hatched bars). Light treatments are as described in Figure 3. C, Quantitations of rbcL and psbA transcription are shown. Transcription activities (means ± sd) for all genes are expressed as fmol [32P]UMP incorporated (1 × 108 chloroplasts)−1 (10 min)−1. The data represent four independent experiments with two replicates each.
Total chloroplast transcription was highest in dark-adapted wild-type plants exposed to blue light, white light, or UV-A radiation (Fig. 3). Red and far-red light had minimal stimulatory effects on transcription relative to the dark controls (Fig. 3). The level of chloroplast transcription in plants exposed to blue light, white light, and UV-A radiation was lower by 40%, 56%, and 72%, respectively, in the phyA mutant relative to wild-type plants. Hence, blue light and UV-A significantly induced total chloroplast transcription in mature leaves of wild-type Arabidopsis plants as in tobacco and this response was attenuated in the phyA mutant.
In Figure 4 we determined the effects of spectral quality on the transcription of two light-responsive chloroplast genes, psbA and rbcL, as well as the 16S rrn in wild-type and phyA mutant plants. The overall values of gene-specific transcription rates descended in the following order: 16S rrn > psbA > rbcL. In general, transcription of each gene was higher in wild-type plants exposed to all light regimes relative to dark controls (Fig. 4). However, the degree of transcriptional activation depended on the specific gene and light treatment. PsbA, rbcL, and 16S rrn transcription were stimulated 12-, 8-, and 18-fold, respectively, in wild-type plants exposed to white light relative to dark controls (Fig. 4). Transcription of these genes increased markedly, from 11- to 16-fold, in wild-type plants exposed to blue light or UV-A radiation relative to dark controls (Fig. 4). However, psbA, rbcL, and 16S rrn transcription were 30% to 80% lower in wild-type plants exposed to red or far-red light relative to blue light, white light, and UV-A radiation. The significant activation of chloroplast gene transcription in response to blue light and UV-A (Fig. 4) resembled the response of total chloroplast transcription to these light regimes (Fig. 3). In general, rbcL transcription rates in Arabidopsis chloroplasts were 10- to 20-fold higher than in tobacco chloroplasts (Figs. 2 and 4). This could be due to differences between the two species, leaf age, or other factors.
16S rrn, psbA, and rbcL transcription were 20% to 80% less, depending on the light treatment, in the phyA mutant compared with wild-type plants (Fig. 4), especially psbA and 16S rrn transcription in blue light, white light, and UV-A. In contrast, rbcL transcription in the phyA mutant exposed to blue or red light was similar to the wild-type controls (Fig. 4C). These results indicate that phyA is involved in blue light-, UV-A-, red light-, and far-red light-induced 16S rrn and psbA transcription and UV-A-induced rbcL transcription. However, phyA is not involved in mediating blue light- and red light-induced rbcL transcription. In addition, rbcL, psbA, and 16S rrn transcription were higher in the phyA mutant exposed to blue light relative to red light and darkness, suggesting that another photoreceptor besides phyA was also involved in light-activated chloroplast transcription.
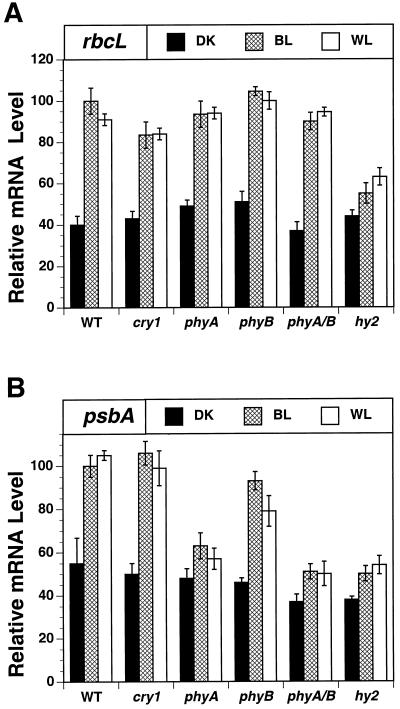
We focused next on identifying photoreceptors involved in blue light-induced rbcL transcription. We analyzed rbcL expression in five mutants, cry1, phyA, phyB, hy2, and the phyA/phyB double mutant (Fig. 5A). The degree of rbcL mRNA accumulation was 10% lower in the cry1 mutant relative to wild type in blue and white light (Fig. 5A). This experiment indicates that cry1 is not the major photoreceptor mediating light-induced accumulation of rbcL mRNA. No effect of the phyA and phyB mutants and the double mutant phyA/phyB were observed on light-induced rbcL expression (Fig. 5). However, blue light-induced rbcL mRNA accumulation was 60% lower in the hy2 mutant relative to wild-type plants. The results indicate another phytochrome species (phyC, D, or E) was involved in blue light-induced rbcL expression.
Figure 5.
Analysis of the influence of blue light, cry1, and phytochrome on rbcL and psbA expression in Arabidopsis chloroplasts from mature leaves. Relative rbcL (A) and psbA (B) mRNA levels were determined by quantitating radioactive bands on RNA gel blots that hybridized with gene-specific probes. The total cell RNAs were isolated from dark-adapted wild-type (W.T.) and cry1, phyA, phyB, phyA/phyB, and hy2 mutant plants exposed to 15 h of additional darkness (DK), or 18 ± 2 μmol m−2 s−1 blue (BL) or white 100 ± 10 μmol m−2 s−1 (WL) light. RNA levels in wild-type plants exposed to blue light were set at 100. The values for the other treatments are expressed relative to 100.
PsbA transcription was 6- to 10-fold higher in the phyA mutant exposed to blue light relative to dark controls (Fig. 4). Thus, the phyA mutant did not abolish the stimulatory effect of blue light on psbA transcription. This raised the question whether any other photoreceptors besides phyA were involved in blue light-induced psbA expression. Therefore, we analyzed psbA expression in the same series of mutants (Fig. 5B). The accumulation of psbA mRNA was over 40% lower in the phyA, digenic phyA/phyB, and hy2 mutants exposed to blue or white light relative to wild-type controls, whereas psbA mRNA levels were 10% to 20% lower in the phyB mutant. No additive effect of phyB with phyA was observed. In the hy2 mutant a moderate 10% to 15% increase in psbA mRNA levels occurred in blue and white light compared with dark controls.
DISCUSSION
Blue Light/UV-A Activate Chloroplast Transcription in Mature Leaves of Two Dicots
In this study we employed two complementary systems, transplastomic promoter-reporter gene fusions in tobacco and Arabidopsis photoreceptor mutants, as a means to begin elucidating the photosensory pathways that regulate chloroplast gene expression in mature leaves. Previous research on the role of phytochrome in chloroplast gene expression depended on varying light quality and fluence treatments of wild-type plants, particularly, seedlings (Bottomley, 1970; Link, 1982; Thompson et al., 1983; Zhu et al., 1985; Dubell and Mullet, 1995; Christopher, 1996). To our knowledge no studies have used the two systems described here to examine the photobiology of chloroplast transcription and to measure chloroplast transcription directly in a phytochrome mutant. Although the Arabidopsis system allowed the analysis of the effects of phytochrome and cryptochrome mutants on light-activated chloroplast transcription and mRNA accumulation, the routine transformation of Arabidopsis chloroplasts is not yet possible. Therefore, the tobacco system enabled us to analyze, in parallel, the photobiology of light-activated promoter-reporter gene fusions in transgenic chloroplasts. We provide direct evidence that blue light and UV-A are major signals responsible for the activation of chloroplast transcription in the two dicots, whereas red and far-red light had much smaller effects.
Although we measured transcription and RNA accumulation in response to light, RNA stability also contributes to chloroplast mRNA abundance (Mayfield et al., 1995; Shiina et al., 1998). In the tobacco plants exposed to red light, uidA mRNA levels driven by the rbcL promoter increased more than the corresponding transcription rate (Figs. 1 and 2). Red light may have differentially enhanced the stability of the rbcL-5′-uidA RNA. In Arabidopsis, rbcL and psbA mRNA accumulation (Fig. 5) was correlated with transcription rates (Fig. 4), but did not increase proportionally relative to dark controls because mRNA levels remained high in the dark due to enhanced stability of these RNAs (Kim et al., 1993). Also, we cannot rule out an influence of RNA stability in the hy2 background. The approach used here can be combined with chloroplast transcription inhibitors such as tagetitoxin to assess the roles of RNA stability in the photobiology of chloroplast gene expression.
Detection of phyA-Dependent and -Independent Photosensory Pathways
In Arabidopsis, phyA signaling was definitively linked to blue light/UV-A-induced chloroplast gene transcription. The 60% to 80% decrease in blue light-induced psbA and 16S rrn transcription in the phyA mutant is an indication that phyA is playing a major role in these photoresponses. The phytochromes absorb blue light in vitro and in vivo (Butler et al., 1964; Mohr, 1994) and phyA has been proposed to play at least a minor role in blue light responses for some time, independent of and dependent on cryptochrome (Reed, 1999; Lin, 2000) However, phyA has been primarily associated with mediating the high-irradiance responses to far-red light, particularly during germination, seedling de-etiolation and establishment, chloroplast development, and the response to very low fluence light (Chory et al., 1989; Bowler et al., 1994; Quail 1994; Dubell and Mullet 1995; Smith, 1995; Barnes et al., 1996; Furuya and Schäfer, 1996; Neff and Chory, 1998). The finding that phyA is playing a major role in mediating the response of chloroplast transcription to blue light/UV-A in mature leaves is, to our knowledge, a new function for this photoreceptor at this stage of chloroplast biogenesis. Additional examples of phyA mediating blue light responses are cotyledon expansion and hypocotyl inhibition (Whitelam et al., 1993; Neff and Chory, 1998), seed germination (Shinomura et al., 1996), and Lhcb gene induction (Hamazato et al., 1997).
There is precedence for a role of phyA in mature green tissue. Although phyA levels and gene expression drop precipitously in illuminated plants (Quail, 1994; Smith, 1995; Reed, 1999), its' levels can increase during dark-adaptation (Hunt and Pratt, 1980; Smith, 1995). In addition, a small amount of phyA may be adequate to regulate processes in mature tissue as less than 5% of the total phytochrome levels are required for phyA interactions in vitro (Ahmad et al., 1998). PhyA may modulate chloroplast transcription by sensing circadian periods (Zhong et al., 1997), which also affect chloroplast transcription (Krupinska, 1992; Nakahira et al., 1998).
It has been well-documented that cry1, cry2, and NPH1 are three of the known blue light photoreceptors in Arabidopsis (Christie et al., 1998; Cashmore et al., 1999). However, cry1 did not significantly influence blue light-activated psbA and rbcL expression (Fig. 5) and, previously, blue light-induced psbD expression (Christopher and Hoffer, 1998). NPH1 and the photolabile cry2 operate at much lower fluences (Christie et al., 1998; Guo et al., 1999) than the high fluences used here. High, but not low fluence, light activates chloroplast transcription in dicots and monocots (Gamble and Mullet, 1989; Christopher and Mullet, 1994; Dubell and Mullet, 1995; Christopher, 1996). We did not test the cry2 and nph1 mutants. However, with the recent finding that phyA can phosphorylate cry1 and cry2 (Ahmad et al., 1998), it would be valuable to test the cry1-cry2 double, phyA-cry1-cry2 triple, and phyC, D, and E, mutants.
PhyA was not involved in blue light-induced rbcL transcription (Fig. 4C). PhyA involvement also varies for blue light-induced nuclear gene expression (Oelmüller and Kendrick, 1991). Our results with the hy2 and cry1 mutants (Fig. 5A) suggest that another phytochrome species such as phyC, D, or E was involved in rbcL expression, with a minor modulatory role for cry1. The results with psbA expression (Fig. 5B) indicate that there were phyA- and phyB-dependent and phytochrome-independent modes of blue light-induced psbA expression. Therefore, light-induced chloroplast transcription involves multiple phytochromes and blue light/UV-A signaling, as does seedling development (Quail, 1994; Smith, 1995; Chory, 1997; Casal and Mazzella, 1998).
The lack of a role for phyA in blue light-activated rbcL transcription raises questions about the mechanism regulating the photobiology and gene selectivity of the transcription apparatus. The light-responsiveness of rbcL, psbA, and 16S rrn transcription depends on the PEP (Allison et al., 1996), which predominates in mature leaves. Multiple photosensory pathways can interact by regulating distinct transcription factors that modulate the PEP. Several nuclear-encoded sigma factors have been identified in chloroplasts (Isono et al., 1997; Kanamaru et al., 1999; Tan and Troxler, 1999) and some are light-induced. Therefore, it seems reasonable to hypothesize that blue light acting via phyA would regulate at least one sigma factor, whereas a phyA-independent sigma factor may control rbcL transcription.
Developmental Change in Action Spectrum for Chloroplast Transcription
In etiolated monocot and dicot seedlings phyA is the predominant phytochrome species (Quail, 1994; Smith, 1995) and it has been extensively shown that red and far-red light are the primary signals stimulating chloroplast transcription and mRNA accumulation (Bottomley, 1970; Link, 1982; Thompson et al., 1983; Zhu et al., 1985). In etiolated pea the activation of chloroplast transcription is a high-irradiance response to far-red light mediated by phyA (Dubell and Mullet, 1995). The only exception is the psbD gene, which is selectively activated by high-fluence blue light from a blue light-responsive promoter in etiolated seedlings (Christopher and Mullet, 1994; Chen et al., 1995). The major stimulatory effects of red and far-red light on chloroplast transcription in dicot seedlings are related to the stimulation of chloroplast and leaf development (Chory et al., 1989; Dubell and Mullet, 1995). In contrast, we observed that continuous far-red and red light stimulated chloroplast transcription in mature leaves to a much smaller degree than did blue light/UV-A. We postulate that the action spectrum for activating chloroplast transcription changed during plant development. Similar developmental changes in action spectrum have been reported for rbcS and chs gene expression (Fluhr et al., 1986; Mohr, 1994), hypocotyl development (Cosgrove, 1994), and the accumulation of chloroplast glyceraldehyde-3-phosphate dehydrogenase (Mohr, 1994). PhyA signaling in mature, relative to etiolated, tissue may be different because of changes in phyA substrate specificities, interactions with other pathways, or the optical properties of the tissue.
Evolution of Phytochrome-Mediated Blue Light/UV-A-Activated Chloroplast Transcription
From an evolutionary standpoint the emphasis on blue light responsive transcription in chloroplasts is consistent with the blue light-activation of photosynthesis genes (psbA and psbD) in the cyanobacterium Synechococcus (Tsinoremas et al., 1994). In a similar manner, high-fluence blue light rather than red light activates transcription of the barley chloroplast psbA promoter when heterologously expressed in Synechococcus (Tsinoremas et al., 1999). Cyanobacteria are considered to be ancestors to the plant chloroplasts. However, no cryptochrome genes were found in a completely sequenced cyanobacterial genome, whereas several novel variants of phytochromes were identified, one of which influenced gene expression for the light harvesting apparatus (Kaneko et al., 1996; Kehoe and Grossman, 1996; Lamparter et al., 1997; Yeh et al., 1997). This suggests that photosensory pathways linking phytochrome and photosynthesis gene expression were established prior to the endosymbiotic events that gave rise to plant chloroplasts. As ancestral genes for phytochrome (Quail, 1994) and chloroplast proteins such as Lhcb were transferred to the nucleus, the photoregulation of genes remaining in the chloroplast co-evolved with the phytochromes to coordinate chloroplast and nuclear gene expression for the stoichiometric production of photosystem subunits. Because transcription of genes encoding photosynthesis functions is lower in the phyA mutant exposed to white light, this raises the question of whether the ability of the photosynthetic apparatus to adapt to high light is impaired in the mutant. Measurement of photosynthetic activity in wild-type and phyA mutant plants under increasing light intensity would answer this question.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type seeds from the Columbia and Landsberg erecta ecotypes (Arabidopsis) were purchased from Lehle Seeds (Round Rock, TX). Seeds of the cry1, phyA, phyB, and hy2 mutants were obtained from the Arabidopsis Biological Resource Center (Ohio State University). Seeds were planted in flats containing water-saturated Jiffy potting mix (Bentonville, AR) and chilled (5°C) overnight (14 h). Plants were exposed to a photoperiod of 12 h of darkness and 12 h of white light (fluorescent, 100 ± 10 μmol m−2 s−1) at 22°C to 26°C for 28 d. To obtain dark-adapted seedlings, the 28-d-old plants were placed in complete darkness for 36 h. Following dark-adaptation, plants were maintained in the dark or were exposed to the light sources (described below) for 12 and 15 h (indicated in Figure Legends) before harvesting. Tissue was harvested by quick freezing in liquid nitrogen or was used fresh for chloroplast isolation. All manipulations of dark-grown plants were performed in complete darkness or under a dim green safelight as previously described (Christopher, 1996).
Seeds of wild-type and transplastomic tobacco (Nicotiana tabacum var. Ottowa) lines pLAA24A, pLAA25A (Zoubenko et al., 1994), and pWW11 (Shiina et al., 1998) were used. The chloroplast genome of line pLAA25A contains the promoterless uidA gene, whereas lines pLAA24A and pWW11 contained the uidA gene under the control of the tobacco chloroplast promoters for the 16S rRNA and rbcL genes, respectively. Two copies of each transgene, one in each copy of the inverted repeat, are present in the plastid genomes. Tobacco plants were grown on Sunshine Mix No. 4 (SunGro Horticulture, Bellevue, WA) under a photoperiod of 10 h of darkness and 14 h of light at 24°C to 27°C for 42 d. Plants were dark-adapted for 36 h prior to exposing them to 12 h red, blue, UV-A, and white light treatments (described below). They were then harvested for RNA or chloroplast isolation.
Light Sources and Conditions
Fluences for red, far-red, blue, and white light were measured using a quantum photometer (LI-COR, LI-1000, Lincoln, NE) and a radiometer (model 65, YSI-Kettering). Fluences of UV-A radiation (W m−2) were measured using a UVX radiometer with a UVX-36 sensor (UVP, Inc., Upland, CA). Fluences of visible light and UV-A radiation were normalized using the radiometer and corrected for detection efficiency. Red light (18 ± 2 μmol m−2 s−1) of 650 to 670 nm band width with a peak at 658 nm was obtained by passing white light (tungsten halogen 300 W, EXR 54392, Sylvania, Danvers, MA) through a red interference filter (CBS-R, Carolina Biological Supply, Burlington, NC) and filtered through clear heat-absorbing glass. Far-red light (18 ± 2 μmol m−2 s−1) greater than 700 nm was obtained by passing incandescent light (60 W, GE) through a far-red cut-off filter (CBS FR 750, Carolina Biological Supply). Red and far-red light sources were cooled with a fan. Blue light (18 ± 2 μmol m−2 s−1) from 410 to 480 nm with a peaks at 440 and 420 nm was obtained by the following two means depending on the number of plants treated: (a) For plants in small pots, white light (tungsten halogen 300 W, EXR 54392, Sylvania) was passed through a narrow band width blue interference filter (B-1, Edmund Scientific, Barrington, NJ); and (b) for large flats used in chloroplast isolation, light from 100% actinic blue fluorescent light bulbs (40 W T12, 420 nm peak, Coralife, Carson CA) was used. UV-A light (75–85 W m−2, radiometer-determined equivalent to 18 ± 2 μmol m−2 s−1) from 330 to 405 nm with a peak at 365 nm (90% of total emission contained in 330–395 nm) was obtained by using a black light blue bulb (F15T8 BLB-15 W, Sylvania) housed in a lamp (Spectronics, Westbury, NY). White light (100 ± 10 μmol m−2 s−1) was obtained from cool-white bulbs (F40CW-RS-WM 34 W, GE W-Miser).
Chloroplast Isolation and Transcription Assays
Arabidopsis and tobacco chloroplasts were isolated from leaves as described (Hoffer and Christopher, 1997). Intact chloroplasts were counted in a hemacytometer using a phase contrast microscope. Chloroplast transcription activity was assayed using [α-32P]UTP and 1 × 108 of purified chloroplasts at a final concentration of 9.1 × 108 chloroplasts mL−1. Total chloroplast transcription activities from four experiments of duplicate samples (Arabidopsis) or two experiments with duplicate samples (tobacco) were expressed as pmol [32P]UMP incorporated (1 × 108 chloroplasts)−1 (10 min)−1. Radiolabeled run-on transcripts were hybridized as described (Christopher, 1996) to non-radiolabeled gene-specific, single-stranded antisense RNAs (1 pmol each psbA, rbcL, 16S rRNA, or pSK vector; Rapp et al., 1992), or to 0.5 pmol (160 ng) of a 485-bp uidA gene-specific DNA fragment. Gene-specific DNAs and RNAs were separated by agarose gel electrophoresis and transferred to nylon membranes. The levels of radioactivity hybridized to the membranes were determined by liquid scintillation counting of excised bands and by using an AMBIS 4000 Image Acquisition and Analysis System (AMBIS, Inc., San Diego). Values of counts hybridized to the pSK vector (Stratagene Cloning Systems, San Diego) were subtracted from the gene-specific values. Gene-specific transcription activities were expressed as fmol [32P]UMP incorporated (1 × 108 chloroplasts)−1 (10 min)−1 (kb)−1.
RNA and DNA Gel-Blot Hybridization and Analysis
Total cell RNA was isolated from frozen leaf tissue (Arabidopsis and tobacco) by extraction with acid phenol (pH 4.5) and was quantitated spectrophotometrically as described (Hoffer and Christopher, 1997). RNA was separated by electrophoresis on 1.5% (w/v) formaldehyde-1.2% (w/v) agarose gels (16 mm MOPS [3-(N-morpholino)-propanesulfonic acid], 4 mm NaOAc, and 1 mm EDTA, pH 7.0). RNA gel blots (Genescreen) containing equal amounts of total cell RNA (12 μg) per lane were hybridized with radiolabeled gene-specific probes as previously described (Christopher, 1996).
Heterologous antisense RNA probes were derived from linearized recombinant plasmids with DNA inserts specific for the barley chloroplast genes, 16S rRNA, rbcL, and psbA (Rapp et al., 1992). The RNA probes were synthesized and radiolabeled with [α-32P]UTP (>800 Ci/mm, ICN Pharmaceuticals) using T3 and T7 RNA polymerases. A 485-bp PCR product internal to the uidA gene (Jefferson et al., 1987) was made using primers 5′-TGCGGTCACTCATTACGGCA and 5′-AGTATCTCTATTGGAAGTGG. The PCR contained 50 ng of plasmid DNA template (pGUS1 for uidA), 2.5 units of Taq DNA polymerase, 50 mm KCl, 10 mm Tris-HCl (pH 9.0), 1% (w/v) Triton X-100, and 0.2 mm for each of the dNTPs. The PCR consisted of 40 cycles of 1 min at 94°C, 1 min at 45°C, and 1.5 min at 72°C. The resulting PCR product was gel-purified, diluted, and used to make DNA blots to hybridize with radiolabeled RNAs generated in the lysed plastid run-on transcription assays and to make a radiolabeled uidA probe. The uidA-specific PCR product was labeled with [α-32P]dATP (>800 Ci/mm, ICN Pharmaceuticals) by the method of Schowalter and Sommer (1989).
ACKNOWLEDGMENT
The authors would like to thank Dr. Lori Allison for generously providing seeds of the transplastomic tobacco lines (pLAA24A, pLAA25A, and pWW11).
Footnotes
This work was supported by the U.S. Department of Energy Biosciences Program (grant no. DE–FG03–97ER20273 to D.A.C.). College of Tropical Agriculture and Human Resources Journal Series 4521.
LITERATURE CITED
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998;1:939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- Alfonso M, Perewoska I, Kirilovsky D. Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803: involvement of the cytochrome b6/f complex. Plant Physiol. 2000;122:505–515. doi: 10.1104/pp.122.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison LA, Simon LD, Maliga P. Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 1996;15:2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Aro E-M, Virgin I, Andersson B. Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143:113–134. doi: 10.1016/0005-2728(93)90134-2. [DOI] [PubMed] [Google Scholar]
- Baginski S, Tiller K, Link G. Transcription factor phosphorylation by a protein kinase associated with chloroplast RNA polymerase from mustard (Sinapis alba) Plant Mol Biol. 1997;34:181–189. doi: 10.1023/a:1005802909902. [DOI] [PubMed] [Google Scholar]
- Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua N-H. Far-red light blocks greening of Arabidopsis seedlings via a phytochrome-A-mediated change in plastid development. Plant Cell. 1996;8:601–615. doi: 10.1105/tpc.8.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley W. Deoxyribonucleic acid-dependent ribonucleic acid polymerase activity of nuclei and plastids from etiolated peas and their response to red and far-red light in vivo. Plant Physiol. 1970;45:608–611. doi: 10.1104/pp.45.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua N-H. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Butler WL, Hendricks SB, Siegelman HW. Action spectra of phytochrome in vitro. Photochem Photobiol. 1964;3:521–528. [Google Scholar]
- Casal JJ, Mazzella MA. Conditional synergism between cryptochrome 1 and phytochrome b is shown by the analysis of phyA, phyB, and hy4 simple, double and triple mutants in Arabidopsis. Plant Physiol. 1998;118:19–25. doi: 10.1104/pp.118.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- Chamovitz DA, Deng X-W. Light signaling in plants. Crit Rev Plant Sci. 1996;15:455–478. [Google Scholar]
- Chen SCG, Wu SP, Lo PK, Mon DP, Chen LFO. Regulation of plastid photosynthetic psbK-I-D-C gene expression by light in rice plants. Physiol Plant. 1995;93:617–623. [Google Scholar]
- Chory J. Light modulation of vegetative development. Plant Cell. 1997;9:1225–1234. doi: 10.1105/tpc.9.7.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saginich R, Pratt L, Ausubel F. Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell. 1989;1:867–880. doi: 10.1105/tpc.1.9.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- Christopher DA. Leaf development and phytochrome modulate the activation of psbD-psbC transcription by high-fluence blue light in barley chloroplasts. Photosyn Res. 1996;47:239–251. doi: 10.1007/BF02184285. [DOI] [PubMed] [Google Scholar]
- Christopher DA, Hoffer PH. DET1 represses a chloroplast blue light-responsive promoter in a developmental and tissue-specific manner in Arabidopsis thaliana. Plant J. 1998;14:1–11. doi: 10.1046/j.1365-313x.1998.00078.x. [DOI] [PubMed] [Google Scholar]
- Christopher DA, Mullet JE. Separate photosensory pathways coregulates blue light/ultraviolet-A-activated psbD-psbC transcription and light-induced D2 and CP43 degradation in barley (Hordeum vulgare) chloroplasts. Plant Physiol. 1994;104:1119–1129. doi: 10.1104/pp.104.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. Photomodulation of growth. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in Plants. Boston: Kluwer Academic Publishers; 1994. pp. 631–638. [Google Scholar]
- Danon A, Mayfield SP. Light-regulated translation of chloroplast messenger RNAs through redox potential. Science. 1994;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- Dubell AN, Mullet JE. Differential transcription of pea chloroplast genes during light-induced leaf development: continuous far-red light activates chloroplast transcription. Plant Physiol. 1995;109:105–112. doi: 10.1104/pp.109.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubas JM, Lomas M, LaRoche J, Falkowski PG. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr R, Moses P, Morelli G, Coruzzi G, Chua N-H. Expression dynamics of the pea rbcS multigene family and organ distribution of the transcripts. EMBO J. 1986;5:2063–2071. doi: 10.1002/j.1460-2075.1986.tb04467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglevand G, Jackson JA, Jenkins GI. UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell. 1996;8:2347–2357. doi: 10.1105/tpc.8.12.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya M, Schäfer Photoperception and signaling of induction reactions by different phytochromes. Trends Plant Sci. 1996;1:301–307. [Google Scholar]
- Gamble PE, Mullet JE. Blue light regulates the accumulation of two psbD-psbC transcripts in barley chloroplasts. EMBO J. 1989;8:2785–2794. doi: 10.1002/j.1460-2075.1989.tb08424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Coordination of nuclear and chloroplast gene expression in plant cells. Int Rev Cyt. 1998;177:115–180. doi: 10.1016/s0074-7696(08)62232-9. [DOI] [PubMed] [Google Scholar]
- Guo H, Duong H, Ma N, Lin C. The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent transcriptional mechanism. Plant J. 1999;19:279–287. doi: 10.1046/j.1365-313x.1999.00525.x. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz PTJ, Allison LA, Maliga P. The two RNA polymerases encoded by the nuclear and plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997;16:4041–4048. doi: 10.1093/emboj/16.13.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazato F, Shinomura T, Hanazawa H, Chory J, Furuya M. Fluence and wavelength requirements for Arabidopsis Cab gene induction by different phytochromes. Plant Physiol. 1997;115:1533–1540. doi: 10.1104/pp.115.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffer PH, Christopher DA. Structure and blue light-responsive transcription of a chloroplast psbD promoter from Arabidopsis thaliana, Plant Physiol. 1997;115:213–222. doi: 10.1104/pp.115.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt RE, Pratt LH. Radioimmunoassay of phytochrome content in green, light-grown oats. Plant Cell Environ. 1980;3:91–95. [Google Scholar]
- Igloi GL, Kössel H. The transcriptional apparatus of chloroplasts. Crit Rev Plant Sci. 1992;10:525–558. [Google Scholar]
- Iratni RL, Andreeva A, Mache R, Lerbs-Mache S. Regulation of rDNA transcription in chloroplasts: promoter exclusion by constitutive repression. Genes Dev. 1994;8:2928–2938. doi: 10.1101/gad.8.23.2928. [DOI] [PubMed] [Google Scholar]
- Isono K, Shimizu M, Yoshimoto, Niwa Y, Satoh K, Yokota A, Kobayashi H. Leaf-specifically expressed genes for polypeptides destined for chloroplasts with domains of ς70 factors of bacterial RNA polymerases in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:14948–14953. doi: 10.1073/pnas.94.26.14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3301–3307. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru K, Fujiwara M, Seki M, Katagiri T, Nakamura M, Mochizuki N, Nagatani A, Shinozaki K, Tanaka K, Takahashi H. Plastidic RNA Polymerase ς factors in Arabidopsis. Plant Cell Physiol. 1999;40:832–842. doi: 10.1093/oxfordjournals.pcp.a029612. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803: II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:185–209. doi: 10.1093/dnares/3.3.185. [DOI] [PubMed] [Google Scholar]
- Kehoe DM, Grossman AR. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science. 1996;273:1409–1411. doi: 10.1126/science.273.5280.1409. [DOI] [PubMed] [Google Scholar]
- Kim MK, Christopher DA, Mullet JE. Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol Biol. 1993;22:447–463. doi: 10.1007/BF00015975. [DOI] [PubMed] [Google Scholar]
- Kim MK, Christopher DA, Mullet JE. ADP-dependent phosphorylation regulates association of a DNA-binding complex with the barley chloroplast psbD blue light-responsive promoter. Plant Physiol. 1998;119:663–670. doi: 10.1104/pp.119.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupinska K. Transcriptional control of plastid gene expression during development of barley primary foliage leaves under a daily light-dark regime. Planta. 1992;186:294–303. doi: 10.1007/BF00196259. [DOI] [PubMed] [Google Scholar]
- Lamparter T, Mittmann F, Gärtner W, Börner T, Hartmann E, Hughs J. Characterization of recombinant phytochrome from the cyanobacterium Synechocystis. Proc Natl Acad Sci USA. 1997;94:11792–11797. doi: 10.1073/pnas.94.22.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon P, Arroyo A, Mackenzie S. Nuclear control of plastid and mitochondrial development in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:453–480. doi: 10.1146/annurev.arplant.49.1.453. [DOI] [PubMed] [Google Scholar]
- Lin C. Plant blue-light receptors. Trend Plant Sci. 2000;5:337–342. doi: 10.1016/s1360-1385(00)01687-3. [DOI] [PubMed] [Google Scholar]
- Link G. Phytochrome control of plastid mRNA in mustard (Sinapis alba L.) Planta. 1982;154:81–86. doi: 10.1007/BF00385501. [DOI] [PubMed] [Google Scholar]
- Lopez-Juez E, Jarvis RP, Takeuchi A, Page AM, Chory J. New Arabidopsis cue mutants suggest a close connection between plastid- and phytochrome-regulation of nuclear gene expression. Plant Physiol. 1998;118:803–815. doi: 10.1104/pp.118.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Yohn CB, Cohen A, Danon A. Regulation of chloroplast gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:147–166. [Google Scholar]
- Melis A. Dynamics of photosynthetic membrane composition and function. Biochim Biophys Acta. 1991;1058:87–106. [Google Scholar]
- Mohr H. Coaction between pigment systems. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in Plants. Boston: Kluwer Academic Publishers; 1994. pp. 353–376. [Google Scholar]
- Nakahira Y, Baba K, Yoneda A, Shiina T, Toyoshima Y. Circadian-regulated transcription of the psbD light-responsive promoter in wheat chloroplasts. Plant Physiol. 1998;118:1079–1088. doi: 10.1104/pp.118.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Chory J. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998;118:27–36. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmüller R, Kendrick RE. Blue light is required for survival of the tomato phytochrome-deficient aurea mutant and the expression of four nuclear genes coding for plastidic proteins. Plant Mol Biol. 1991;16:293–299. doi: 10.1007/BF00020560. [DOI] [PubMed] [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J. DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell. 1994;78:109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- Pratt LH. Phytochromes: differential properties, expression patterns and molecular evolution. Photochem Photobiol. 1995;61:10–21. [Google Scholar]
- Quail PH. Phytochrome genes and their expression. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in Plants. Boston: Kluwer Academic Publishers; 1994. pp. 71–104. [Google Scholar]
- Rapp JC, Baumgartner BJ, Mullet JE. Quantitative analysis of transcription and RNA levels of 15 barley chloroplast genes: transcription rates and mRNA levels vary over 300-fold, predicted mRNA stabilities vary 30-fold. J Biol Chem. 1992;267:21404–21411. [PubMed] [Google Scholar]
- Reed JW. Phytochromes are Pr-ipatetic kinases. Curr Opin Plant Biol. 1999;2:393–397. doi: 10.1016/s1369-5266(99)00011-4. [DOI] [PubMed] [Google Scholar]
- Schowalter DB, Sommer SS. The generation of radiolabeled DNA and RNA probes with polymerase chain reaction. Anal Biochem. 1989;177:90–94. doi: 10.1016/0003-2697(89)90019-5. [DOI] [PubMed] [Google Scholar]
- Shiina T, Allison LA, Maliga P. rbcL transcript levels in tobacco plastids are independent of light: reduced dark transcription rate is compensated by increased mRNA stability. Plant Cell. 1998;10:1713–1722. doi: 10.1105/tpc.10.10.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. Physiological and ecological function within the phytochrome family. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:289–315. [Google Scholar]
- Tan S, Troxler RF. Characterization of two chloroplast RNA polymerase ς-factors from Zea mays: photoregulation and differential expression. Proc Natl Acad Sci USA. 1999;96:5316–5321. doi: 10.1073/pnas.96.9.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WF, Everett M, Polans NO, Jorgansen RA, Palmer JD. Phytochrome control of RNA levels in developing pea and mung-bean leaves. Planta. 1983;158:487–500. doi: 10.1007/BF00397240. [DOI] [PubMed] [Google Scholar]
- Thompson WF, White MJ. Physiological and molecular studies of light-regulated nuclear genes in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:423–466. [Google Scholar]
- Tsinoremas NF, Kawakami AK, Christopher DA. High-fluence blue light stimulates transcription from a higher plant chloroplast psbA promoter expressed in cyanobacteria, Synechococcus sp. strain PCC7942. Plant Cell Physiol. 1999;40:448–452. doi: 10.1093/oxfordjournals.pcp.a029562. [DOI] [PubMed] [Google Scholar]
- Tsinoremas NF, Schaefer M, Golden SS. Blue and red light reversibly control psbA expression in the cyanobacterium Synechococcus sp strain PCC 7942. J Biol Chem. 1994;269:16143–16147. [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-C, Wu S-H, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- Zhong HH, Resnick AS, Straume M, McClung CR. Effects of synergistic signaling by phytochrome A and cryptochrome on circadian clock-regulated catalase expression. Plant Cell. 1997;9:947–955. doi: 10.1105/tpc.9.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YS, Kung SD, Bogorad L. Phytochrome control of levels of mRNA complementary to plastid and nuclear genes of maize. Plant Physiol. 1985;7:371–376. doi: 10.1104/pp.79.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoubenko OV, Allison LA, Svab Z, Maliga P. Efficient targeting of foreign genes into the tobacco plastid genome. Nucleic Acids Res. 1994;22:3819–3824. doi: 10.1093/nar/22.19.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]