Abstract
Background and Purpose
Stroke is the leading cause of death and long-term disability worldwide. Previous genome-wide association studies identified 51 loci associated with stroke (mostly ischemic) and its subtypes among predominantly European populations. Using whole-genome sequencing (WGS) in ancestrally-diverse populations from the Trans-Omics for Precision Medicine (TOPMed) Program, we aimed to identify novel variants, especially low-frequency or ancestry-specific variants, associated with all stroke (AS), ischemic stroke (IS) and its subtypes [large artery (LAS), cardioembolic (CES), and small vessel (SVS)], and hemorrhagic stroke (HS) and its subtypes [intracerebral (ICH) and subarachnoid (SAH)].
Methods
WGS data were available for 6,833 stroke cases and 27,116 controls, including 22,315 European, 7,877 African American, 2,616 Hispanic/Latino, 850 Asian, 54 Native American and 237 other ancestry participants. In TOPMed, we performed single variant association analysis examining 40 million common variants and aggregated association analysis focusing on rare variants. We also combined TOPMed European populations with over 28,000 additional European participants from the UK BioBank (UKBB) genome-wide array data through meta-analysis.
Results
In the single variant association analysis in TOPMed, we identified one novel locus 13q33 for LAS at whole genome-wide significance (P<5.00E-9) and four novel loci at genome-wide significance (P<5.00E-8), all of which need conformation in independent studies. Lead variants in all five loci are low-frequency but are more common in non-European populations. An aggregation of synonymous rare variants within the gene C6orf26 demonstrated suggestive evidence of association for HS (P<3.11E-6). By meta-analyzing European ancestry samples in TOPMed and UKBB, we replicated several previously reported stroke loci including PITX2, HDAC9, ZFHX3, and LRCH1.
Conclusions
We represent the first association analysis for stroke and its subtypes using WGS data from ancestrally-diverse populations. While our findings suggest the potential benefits of combining WGS data with populations of diverse genetic backgrounds to identify possible low-frequency or ancestry-specific variants, they also highlight the need to increase genome coverage and sample sizes.
Keywords: Single nucleotide polymorphism genetics, stroke ischemic, stroke hemorrhagic
Introduction
Stroke is the second leading cause of premature mortality and a leading cause of long-term disability worldwide1, 2. The pathogenesis of stroke is heterogeneous and multifactorial. Ischemic stroke (IS), which accounts for 87% of all stroke cases, shows an estimated heritability of approximately 38% and substantial variation across its three subtypes [cardioembolic stroke (CES), 33%; large artery stroke (LAS), 40%; and small vessel stroke (SVS), 16%]3, 4. Hemorrhagic stroke (HS) is less common, with heritability estimated at over 40% for its two subtypes [intracerebral hemorrhage (ICH), 44%; and subarachnoid hemorrhage (SAH), 41%]5, 6.
Previous genome-wide association studies (GWAS) and Exomechip analysis have identified 51 loci associated with stroke types and subtypes, 32 of which were reported at genome-wide significance in the largest trans-ethnic meta-analysis of stroke consisting of more than half a million participants from the MEGASTROKE Consortium7. Most of these published studies focused on all stroke (AS), IS, and its subtypes. The majority of these studies identified loci associated with common variants [minor allele frequency (MAF) >1%] and, in aggregate, explain a limited proportion of the phenotypic variation (0.6%−1.8%)7. In addition, the participants included in these association studies were predominantly of European ancestry3, 7–20. Previous epidemiological studies have demonstrated an excess incidence of stroke cases in African American (AA) and Hispanic ancestry populations compared to European ancestry populations in the United States21–23. These observations reinforce the importance of exploring all stroke types and subtypes in ancestrally-diverse populations.
In the current analysis, we performed the first whole genome sequencing (WGS) analysis for multiple stroke subtypes in an ancestrally-diverse population from the Trans-Omics for Precision Medicine (TOPMed) program, aiming to uncover additional novel loci, especially those driven by low-frequency variants and variants more common in non-European populations. We also attempted to refine previously reported loci using our WGS data with more comprehensive characterizations of the genome.
Methods
Study overview and stroke adjudication
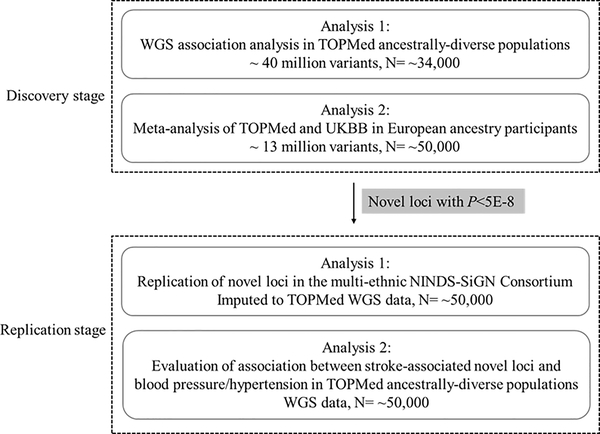
TOPMed data are available on dbGAP (ARIC, phs001211; BioMe: phs001644; CHS, phs001368; FHS, phs000974; JHS, phs000964; MESA, phs001416; WHI, phs001237). In the discovery stage, we performed two GWAS analyses, one focused on TOPMed multi-ethnic samples with denser coverage of the genome using WGS data and the other combining TOPMed and UK BioBank (UKBB) European ancestry samples to increase statistical power while focusing on relatively common variants (Fig. 1). In TOPMed, we included 6,833 incident stroke cases (5,616 IS cases and 1,080 HS cases) and 27,116 controls in our association analyses from the freeze6 data. These participants were from six cohort studies and one biobank: the Atherosclerosis Risk in Communities Study (ARIC) 24–26, the Cardiovascular Health Study (CHS) 27, 28, the Framingham Heart Study (FHS) 29–34, the Jackson Heart Study (JHS) 35–40, the Multi-Ethnic Study of Atherosclerosis (MESA) 41, 42, the Women’s Health Initiative (WHI) 43, and the BioMe™ Biobank (BioMe) (Supplemental Table I). In addition, 4,474 IS cases, 959 ICH cases, 1,194 SAH cases, and up to 24,000 controls of European ancestry from the UKBB were selected for analysis. These participants in the discovery stage represented six ancestral groups based on self-reported ancestry, namely Europeans (n=22,315), AA (n=7,877), Hispanics (n=2,616), Asians (n=850), Native Americans (n=54), and others (primarily South Asian, mixed heritage, and other racial/ethnic groups, n=237, Table 1). All studies were approved by local Institutional Review Boards and written informed consent was obtained from each participant.
Fig. 1.
Study design.
Table 1.
Age, sex and ancestry distributions according to stroke types and subtypes among participants in TOPMed and UKBB
| Stroke | Age (years)1 | Women (%) | N | N by ancestry |
|||||
|---|---|---|---|---|---|---|---|---|---|
| European | AA | Hispanic/Latino | Asian | Native American | Other | ||||
|
| |||||||||
| TOPMed | |||||||||
| AS 2 | 75.1±8.8 | 87.4 | 6,833 | 5,294 | 1,022 | 360 | 116 | 20 | 21 |
| IS 2 | 75.1±8.7 | 87.1 | 5,616 | 4,307 | 884 | 316 | 80 | 12 | 17 |
| CES | 78.9±7.3 | 92.5 | 1,459 | 1,276 | 122 | 39 | 17 | 5 | - |
| LAS | 75.5±7.2 | 91.8 | 352 | 296 | 33 | 19 | 4 | - | - |
| SVS | 74.7±7.8 | 93.1 | 898 | 692 | 132 | 43 | 29 | 2 | - |
| HS 2 | 74.5±8.8 | 90.7 | 1,080 | 862 | 127 | 43 | 36 | 8 | 4 |
| ICH | 75.1±8.0 | 94.3 | 716 | 592 | 68 | 19 | 31 | 6 | - |
| SAH | 70.5±8.3 | 96.6 | 208 | 167 | 26 | 9 | 5 | 1 | - |
| Control | 74.6±11.3 | 64.5 | 27,116 | 17,021 | 6,855 | 2,256 | 734 | 34 | 216 |
| UKBB | |||||||||
| IS | 61.4±6.6 | 35.3 | 4,474 | 4,474 | - | - | - | - | - |
| ICH | 60.9±6.8 | 43.0 | 959 | 959 | - | - | - | - | - |
| SAH | 58.1±7.2 | 60.7 | 1,194 | 1,194 | - | - | - | - | - |
| Control for IS | 56.6±8.1 | 33.3 | 24,000 | 24,000 | - | - | - | - | - |
| Control for ICH | 56.6±8.1 | 33.3 | 4,800 | 4,800 | - | - | - | - | - |
| Control for SAH | 56.6±8.2 | 33.3 | 5,970 | 5,970 | - | - | - | - | - |
AS, all stroke; IS, ischemic stroke; CES, cardioembolic stroke; LAS, large artery stroke; SVS, small vessel stroke; HS, hemorrhagic stroke; ICH, intracerebral hemorrhage; SAH, subarachnoid hemorrhage; AA, African American.
Age of stroke cases indicated age at incident stroke and age of controls indicated age at the last follow-up.
Some AS, IS and HS cases were unclassified.
All stroke cases were adjudicated by physicians in each participating study in the six cohort studies. Baseline stroke cases were excluded from the analysis. In BioMe, the identification of stroke cases was based on the 10th revision of the International Statistical Classification of Diseases and Related Health Problems (ICD). IS cases in CHS, MESA, and WHI studies were further divided into CES, LAS, and SVS according to the Trial of Org 10172 in Acute Stroke Treatment (TOAST) criteria44. HS cases were further divided into ICH and SAH. Details are provided in the Supplemental Methods.
Whole-genome sequencing in TOPMed
A total of 106,809 samples (freeze6) underwent ~30× WGS using DNA extracted from blood samples at designated sequencing centers. Harmonization, joint calling and quality control (QC) procedures are described on the TOPMed website (https://www.nhlbiwgs.org/topmed-whole-genome-sequencing-methods-freeze-6).
Genotyping and imputation in UKBB
Genotyping of 500,000 UKBB participants was performed using either the Affymetrix UK BiLEVE Axiom array or the Affymetrix UK Biobank Axiom® array. QC procedures were performed at both the variants and the sample level, and detailed information is provided on the UKBB website (http://www.ukbiobank.ac.uk/wp-content/uploads/2014/04/UKBiobank_genotyping_QC_documentation-web-1.pdf). Imputation was performed based on reference panels from the Haplotype Reference Consortium (HRC), UK10K, and the 1000 Genome Phase 3 using MACH (http://csg.sph.umich.edu/abecasis/MACH/index.html). Genetic variants with MAF>0.1% and imputation quality score R2>0.3 were included in the association analysis.
Single variant association analysis
Approximately 40 million genetic variants with minor allele count (MAC)>20 were included in the single variant association analysis in TOPMed at the discovery stage. We first tested the association of each variant with stroke types and subtypes treated as dichotomous outcomes using a logistic model adjusted for age, sex, ancestry, study, the first 10 principal components (PCs), and accounting for relatedness using a genetic relationship matrix (GRM) by pooling all TOPMed studies together. Due to the extremely unbalanced case/control ratios, the Scalable and Accurate Implementation of Generalized mixed model (SAIGE) software45 was used to conduct the association analysis. In addition to the ancestry-combined analyses, ancestry-specific analyses were performed in European and AA ancestry populations. All single variant association analyses were performed on the University of Michigan ENCORE server (https://encore.sph.umich.edu). For the UKBB, association analyses of over 16 million genetic variants with IS and the two subtypes of HS (ICH and SAH) were performed after adjustment for age, sex, and the first 10 PCs using PLINK (http://zzz.bwh.harvard.edu/plink/plink2.shtml) and SAIGE (only for IS where we identified one significant locus using PLINK). We combined summary statistics from TOPMed and UKBB in European ancestry populations using fixed-effect inverse-variance-weighted meta-analysis implemented in METAL (https://genome.sph.umich.edu/wiki/METAL_Documentation, Supplemental Table II and III). Approximately 13 million variants available in both TOPMed and UKBB were included in the meta-analysis. Novel genetic variants associated with stroke outcomes were defined as those that showed P<5E-9 (whole genome-wide significance) 46 and P<5E-8 (genome-wide significance) and were located more than 1Mb of any reported loci. There have been reports about inflated odds ratios (OR) using the SAIGE algorithm47 for very rare alleles. As an alternative analysis algorithm, we further implemented the Firth algorithm for association testing, which uses a penalized likelihood approach to estimate coefficients48, in all unrelated TOPMed samples (removing cousins and closer relatives).
All novel loci that showed genome-wide significance (P<5E-8) were carried forward to the replication stage (Fig. 1). Replication was performed using data from the National Institute of Neurological Disorders and Stroke (NINDS) Stroke Genetics Network (NINDS-SiGN, 16,851 cases and 32,473 controls, IS, CES, LAS, and SVS were available for testing)17. WHI was included in both SiGN and TOPMed, and meta-analysis combining all studies in SiGN was performed after excluding samples from WHI. Since higher blood pressure is a risk factor for developing stroke, we also sought to determine whether any of our novel stroke loci were associated with blood pressure or hypertension in the TOPMed Blood Pressure Working Group. Details are provided in the Supplemental Methods.
To dissect association signals at previously established stroke loci, we performed stepwise conditional analysis within each known stroke locus that harbored at least two variants showing P<1E-5 with any stroke type or subtype in TOPMed and UKBB using individual level data.
We followed the STREGA (Strengthening the reporting of genetic association studies) reporting guideline, and a flow diagram is presented in Fig. 1.
Aggregated rare variant association analysis
In the aggregated association analysis, rare variants with MAF<1% were combined using various gene-based aggregation units that were based on high confidence loss of function (hcLoF), missense, protein-altering indels, and synonymous variants (defined based on GENCODE). Associations of aggregated units with AS, IS and HS were tested with adjustment for age, sex, ancestry, study, the first 10 PCs, and GRM using a logistic mixed model implemented in the GENESIS package49. We used two sampling approaches to overcome the unbalanced case/control ratios. The main approach was to include participants from WHI only, as this study contributed 70.5% of stroke cases and had a relatively balanced case/control ratio (Supplemental Table IV). The other exploratory approach was to randomly match case and control participants on a 1:3 ratio based on study, ancestry, and sex (Supplemental Table IV). Both burden test and SNP-set Sequence Kernel Association Test (SKAT) were performed for each gene region harboring more than one variant and a total MAC>20 (Supplemental Table V). Gene-wide and suggestively significant regions were defined as those with P<(0.05/(number of tested regions×four types of aggregation units×two types of association testing methods)) and P<0.05/(number of tested regions), respectively.
Functional annotation of the novel loci
Bioinformatic follow-up was performed for each novel locus using a comprehensive functional annotation database constructed with the whole genome sequence annotator (WGSA50, including GTEx51, DANN52 and Eigen-PC53 scores) and a custom UCSC analysis data hub visualizing enhancer and repressor activities, DNase I hypersensitive sites (DHS) and transcribed regions in selected tissues (Supplemental Methods).
Results
Single variant association analysis
In the discovery stage, we performed two GWAS analyses, one focused on multi-ethnic samples in TOPMed with denser coverage of the genome using WGS data and the other combining European samples in TOPMed and UKBB to increase statistical power while focusing on relatively common variants. Among 6,833 stroke cases and 27,116 controls in TOPMed, 65.7% are of European ancestry and 69.1% are females (Table 1). Genomic inflation factors ranged from 0.967 to 1.103, indicating limited evidence of population stratification in the association analyses (Supplemental Table II and III).
In the ancestry-combined association analysis in TOPMed, we identified one novel locus at whole genome-wide significance (13q33-rs181401679 for LAS, P=3.67E-9) and three additional novel loci showing genome-wide significant associations (7q22-rs141857337 for HS, RAP1GAP2-rs60380775 and AUTS2-rs150022429 for IS, P<5E-8) (Table 2, Supplemental Fig. I). The MAFs of the lead variants ranged from 0.1% (13q33-rs181401679) to 2.2% (RAP2GAP2-rs60380775) in all TOPMed samples. They are either monomorphic (7q22-rs141857337 and AUST2-rs150022429) or extremely rare (13q33-rs181401679 and RAP2GAP2-rs60380775, MAF=0.1%) in European populations. Ancestry-specific analysis in European and AA ancestry populations indicated that the identified significant associations were mainly driven by signals in AA populations at three loci (7q22, AUTS2, and RAP1GAP2, Supplemental Table VI). At the 13q33 locus associated with LAS in the combined analysis, no ancestry-specific results could be produced due to a MAC that was below our cut-off (MAC<20). However, we observed considerably higher MAF in Hispanic ancestry cases (MAF=10.5%, Supplemental Table VII), suggesting that the identified association might be driven by the Hispanic subgroup. In the AA-specific analysis, we identified one additional novel locus at genome-wide significance, TEX13C-rs145400922 on chromosome X for association with CES (P=2.40E-8, Table 2). Among these five novel loci, two of them (RAP1GAP2 and TEX13C) harbored multiple variants with P<5E-8, while the top variant at each of the other three loci (7q22, AUTS2, and 13q33) was the only one showing P<5E-8. All of them harbored multiple variants with P<1E-5, ranging from two to 21. Associations of the five novel loci with each stroke type and subtype are presented in Supplemental Table VIII. The OR values that were observed for the novel loci using the SAIGE algorithm, especially for the two extremely rare variants at 7q22 and 13q33, were larger than those observed using the Firth algorithm in unrelated TOPMed samples (Table 2). The P values using both algorithms were similar, but associations at AUTS2 and 13q33 were not genome-wide significant after Firth correction (P>5E-8, Table 2; recall the Firth correction analysis excluded related individuals, and included 5,564 out of 5,616 cases and 21,756 out of 27,116 controls for IS, and similar reductions for other types and subtypes).
Table 2.
Novel loci identified in the whole-genome single variant association analyses
| Variant | Chr:pos (Build 38) | Gene/Region | CA/NCA | CAF (EA/AA/HA,%)1 | Outcome | N (case/control) | OR | 95%CI | P |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| TOPMed ancestry-combined analysis using SAIGE 2 | |||||||||
| rs141857337 | 7:362834 | 7q22 | A/G | 0/0.7/0.3 | HS | 1,080/27,116 | 455.40 | 50.99, 4067.00 | 4.26E-8 |
| rs150022429 | 7:69400857 | AUTS2 | C/T | 0/1.1/0.2 | IS | 5,616/27,116 | 5.28 | 2.91, 9.58 | 4.56E-8 |
| rs181401679 | 13:104244508 | 13q33 | G/C | 0.05/0.2/0.4 | LAS | 352/11,274 | 1.52E8 | 2.91E5, 7.96E10 | 3.67E-9 |
| rs60380775 | 17:2969421 | RAP1GAP2 | T/C | 0.06/8.4/2.0 | IS | 5,616/27,116 | 1.74 | 1.44, 2.12 | 1.51E-8 |
| TOPMed ancestry-combined analysis using the Firth algorithm 3 | |||||||||
| rs141857337 | 7:362834 | 7q22 | A/G | 0/0.7/0.3 | HS | 1,072/21,756 | 12.94 | 5.75, 26.98 | 4.90E-8 |
| rs150022429 | 7:69400857 | AUTS2 | C/T | 0/1.1/0.2 | IS | 5,564/21,756 | 3.39 | 2.20, 5.13 | 1.31E-7 |
| rs181401679 | 13:104244508 | 13q33 | G/C | 0.05/0.2/0.4 | LAS | 349/10,721 | 34.01 | 11.33, 92.40 | 9.68E-8 |
| rs60380775 | 17:2969421 | RAP1GAP2 | T/C | 0.05/8.5/2.1 | IS | 5,564/21,756 | 1.69 | 1.41, 2.01 | 1.48E-8 |
| TOPMed AA-specific analysis using SAIGE 2 | |||||||||
| rs145400922 | X:125619640 | TEX13C | G/A | 0.02/3.6/0.6 | CES | 122/2,090 | 11.02 | 4.74, 25.63 | 2.40E-8 |
| TOPMed AA-specific analysis using the Firth algorithm 3 | |||||||||
| rs145400922 | X:125619640 | TEX13C | G/A | 0.03/3.7/0.6 | CES | 122/1,994 | 1.37 | 4.74, 25.63 | 2.90E-8 |
CA, coded allele; NCA, non-coded allele; CAF, coded allele frequency; EA, European ancestry; AA, African American; HA, Hispanic; HS, hemorrhagic stroke; IS, ischemic stroke; LAS, large artery stroke; CES, cardioembolic stroke.
The CAF of each lead variant in each ancestral population was calculated using the associated cases and controls.
Analysis using SAIGE included all TOPMed samples.
Analysis using the Firth algorithm included unrelated TOPMed samples.
In the European-specific meta-analysis of TOPMed and UKBB for IS, ICH, and SAH, we did not identify additional novel loci. A previously reported locus PITX2 reached whole genome-wide significance for association with IS in the meta-analysis using the SAIGE results (rs1906611, P=4.68E-9, Supplemental Table IX). None of the five novel loci we discovered in TOPMed were available in UKBB due to their extremely low MAFs or monomorphism in Europeans.
We sought replication of the five novel loci in Table 2 in the multi-ethnic SiGN Consortium. Although the coverage of variants in SiGN was improved through imputation using the TOPMed WGS data as the reference panel, it remains difficult to capture genetic variants with MAF<0.5%. As a result, only two loci (AUTS2-rs150022429 and RAP1GAP2-rs60380775) were available in SiGN and the other three loci were not available due to their low MAFs. However, neither of the two loci showed evidence of association with IS in SiGN (P>0.05, Supplemental Table X). The AA samples in SiGN (1,323 IS cases and 2,383 controls) provided 80% power to detect OR>1.75 for AUTS2-rs150022429 and OR>1.27 for RAP1GAP2-rs60380775. In the analysis of our five novel loci with blood pressure phenotypes in TOPMed, all loci were available for testing except for TEX13C on chromosome X. None of the four novel loci showed evidence of association with SBP, DBP or hypertension (P>0.05, Supplemental Table XI).
Assessment of previously reported stroke loci
Full association results of the 51 previously reported stroke loci (72 unique variants) in UKBB, TOPMed ancestry-combined analysis, and meta-analysis of TOPMed and UKBB are presented in Supplemental Table XI. In TOPMed alone, four of the 51 known loci were not available (polymorphic only in East Asians) and three achieved at least nominal statistical significance [P<1.06E-3 (0.05/47)] for the same stroke phenotype using the reported variants (PITX2 and ZFHX3 for CES, and HDAC9 for LAS, Supplemental Table XII). Seven regions harbored more than two variants with P<1E-5 (ALDH1A2 and PITX2 for CES, PMF1 for HS, TBX3, CYP4F12, and SLC6A11 for IS, SH3PXD2A-OBFC1 for AS), but no additional signals were identified at these loci in the stepwise conditional analysis (Supplemental Table XIII). In UKBB, seven of the 51 reported stroke loci were not available (monomorphism or extremely low MAFs in Europeans) and four loci (PITX2, LRCH1, HDAC9, and ZFHX3) with IS were nominally significant at P<1.14E-3 using the reported variants (0.05/44). Four regions harbored more than two variants with P<1E-5 (PITX2, HDAC9, and HABP2 for IS, and ITPK1 for ICH), but no additional variants showed P<1E-5 in the 1Mb regions (Data not shown).
Aggregated rare variant association analysis
In the aggregated association analysis, we focused on rare genetic variants and three stroke types (AS, IS and HS). No gene region reached gene-wide significance, but one gene region aggregated using synonymous rare variants, C6orf26 showed suggestive significance [P<3.12E-6 (0.05/16,051 regions)] for HS in WHI (Supplemental Table XIV). Similar P values were observed in the burden and the SKAT tests (P=1.33E-6 and 4.59E-7, respectively, Supplemental Table XIV). In the randomly selected samples, only 4 rare variants with a total MAC of 10 were included in the analysis due to smaller sample size compared to using all WHI samples (Supplemental Table IV), which is below the MAC cutoff we used and were excluded from the analysis. Associations of this region using other aggregation units with stroke outcomes were not significant (Supplemental Table XIV). This region is located about 400kb away from a previously reported common variant SLC22A7-rs16896398 for association with AS7. Among the nine rare variants included in this region, rs61747887 showed the highest MAF of 0.9%, and is more frequent in European compared to AA populations (MAF=1.5% and 0.5%, respectively). It showed nominal association with HS in the ancestry-combined and European-specific analysis (P=9.94E-5 and 5.23E-5, respectively) while no evidence of association was observed in AA-specific analysis (P=0.80). The rare variant rs61747887 we observed is not in LD with the reported common variant SLC22A7-rs16896398 (r2<0.1), which showed no evidence of association with AS or HS in TOPMed ancestry-combined analysis (P=0.44 and 0.48, respectively). In GTEx, rs61747887 is associated with gene expression levels of CUL7 (P=1.5E-5) and RP1–20C7.6 (P=4.4E-5) in brain tissue. In addition, seven previously reported loci harbored nearby genes (±500kb) that showed evidence of association with at least one of the three stroke types [P<9.80E-4 (0.05/51 known loci), Supplemental Table XV].
Functional annotation of the novel loci
At each of the five significant or suggestive novel loci listed in Table 2, the lead variant and its LD proxies (r2≥0.4) were examined using both the functional annotation database constructed from WGSA (Supplemental Table XVI) and the customized UCSC genome browser (Supplemental Fig. II). At the 7q22 locus, the lead variant rs141857337 and its two LD proxies all showed Eigen-PC score>0 (functional, Supplemental Table XVI) and overlapped with enhancer, repressor, and DHS in brain and ventricle tissues (Supplemental Fig. II A). At the RAP1GAP2 locus, an LD proxy rs115318048 that is in moderate LD with the lead variant (r2=0.48) overlapped with enhancer activity in all selected tissues (Supplemental Fig. II D). At the TEX13C locus, the lead variant rs145400922 showed DANN scores>0.9 (deleterious, Supplemental Table XVI).
Discussion
We present the first WGS association analysis for stroke and subtypes in ancestrally-diverse populations. We identified five possible novel loci harboring low-frequency lead variants in the single variant association analysis and one suggestively gene-wide significant gene in the aggregate association analysis indicating independent signals from rare variants at an established region. We were unable to replicate two of the single variant association signals that were available using independent data from the SiGN Consortium. While our findings suggest the potential benefits of combining WGS data with populations of diverse genetic backgrounds to identify possible low-frequency or ancestry-specific variants associated with stroke, they also highlight some of the accompanying challenges including the requirement for very large numbers of stroke cases for discovery especially in the face of, phenotypic complexity compounded by the current paucity of appropriately-powered replication samples.
In our discovery analysis in TOPMed, approximately 40 million genetic variants with MAC>20 were examined in the single variant association analysis, which is five times the number of variants examined in the MEGASTROKE Consortium after imputation using the 1000 Genome reference panel7. The substantial improvement in the coverage of the genome using WGS coupled with ancestrally-diverse populations facilitated the identification of ancestry-specific low-frequency variants associated with stroke especially among non-European populations. Among the five novel loci we identified in the single variant association analysis in TOPMed, two of them harbored lead variants that are monomorphic in European ancestry populations (7q22 and AUTS2) and three of the lead variants show low MAFs in Europeans (MAF<0.1% for 13q33 and RAP1GAP2, and MAF=0.2% for TEX13C). Previous GWAS analyses focusing on European ancestry populations and relatively common variants (MAF≥1%) would not be able to capture these low-frequency and ancestry-specific variants. Of note, RAP1GAP2 encodes a GTPase-activating protein that activates the small guanine-nucleotide-binding protein Rap1 in platelets and interacts with synaptotagmin-like protein 1 and Rab27 and regulates secretion of dense granules from platelets at sites of endothelial damage. This gene has been reported for suggestive association with sudden cardiac arrest54 and genome-wide significant association with white blood cell indices55, 56, but none of the reported variants are in LD with the lead variant we identified for association with IS in TOPMed (r2<0.02). In addition to the novel findings in the single variant association analysis, examination of rare variants (MAF<1% and total MAC>20) derived from WGS through aggregated analysis in TOPMed highlighted a rare variant, rs61747887, at an established region, SLC22A7, which is not in LD with the reported common variant rs16896398 (r2<0.1)7. Gene expression data from GTEx helped to prioritize CUL7 at this locus, whose encoded protein is a component of an E3 ubiquitin-protein ligase complex. Previous studies in mouse models have demonstrated its important role in vascular morphogenesis57 and improved cardiac function after myocardial infarction58.
Unlike published GWAS analyses focusing on limited numbers of stroke types or subtypes, a major strength of our study is our ability to perform a comprehensive analysis for AS, including the two stroke types (IS and HS), and the five subtypes (CES, LAS, SVS, ICH, and SAH). Previous studies have identified shared genetic loci across different subtypes (SH2B3 was associated with both LAS and SVS and ABO was associated with both LAS and CES) as well as subtype-specific loci (EDNRA, LINC01492, TSPAN2, and HDAC9 were associated with LAS only and PITX2 and NKX2–5 were associated with CES only)7, 10, 11, 17, 18. These findings indicate both shared biological pathways and risk factors across stroke subtypes and subtype-specific mechanisms. We observed similar results at PITX2 and HDAC9 in TOPMed. At the PITX2 locus, all reported variants exhibiting more significant associations for CES compared to IS (smallest P=9.01E-7 and 0.011 for CES and IS, respectively, Supplemental Table XII). At HDAC9 locus, reported variants showed nominal association with LAS but not IS (smallest P=4.24E-4 and 0.151 for LAS and IS, respectively, Supplemental Table XII). In addition, the ZFHX3 locus has been primarily reported for association with CES, but we also observed nominal association with SVS in TOPMed (smallest P=5.10E-4 and 3.98E-3 for CES and SVS, respectively, Supplemental Table XII). In the evaluation of the five novel loci across stroke types and subtypes in TOPMed, the 13q33 locus significantly associated with LAS (P=3.67E-9) was nominally associated with SAH (P=0.022) and the TEX13C suggestively associated with CES (P=2.54E-8) was nominally associated with SVS (P=0.028, Supplemental Table VIII), suggesting these loci may impact pathways important across multiple stroke subtypes.
Our study has several limitations. First, only two out of the five novel loci are present in the SiGN replication dataset and can be attempted for replication. The fact that these novel loci are relatively rare made it difficult to find proper replication datasets for the other three loci. Second, the sample sizes in TOPMed remained limited compared to published GWAS, with the largest meta-analysis incorporating over 67,000 stroke cases and 454,000 controls7. This situation likely contributed to the relatively small numbers of reported loci that were confirmed in our analysis and the failure to identify independent signals at reported loci. Moreover, over 70% of our sample involves Europeans, making it challenging to definitively identify heterogeneity of the associated loci across diverse ancestral groups. Third, some of the cases were not grouped into subtypes, especially in HS where more than half of the cases did not have subtype classification. This missingness further limited statistical power to identify novel findings for these subtypes.
In conclusion, we performed the first association analysis for stroke types and subtypes using WGS data in ancestrally-diverse populations. Through single variant and aggregate association analyses, we identified five novel loci that harbored low-frequency variants and showed ancestry-specificity and confirmed one reported gene region at genome-wide significance. These findings require replication in additional well powered sample set when available. Our findings indicate that dense coverage of the genome, large sample sizes, increased representation of ancestrally-diverse participants, and detailed classification of stroke cases are essential to the identification of novel findings and better characterization of stroke-associated loci.
Supplementary Material
Acknowledgements
We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
ARIC: The authors thank the staff and participants of the ARIC study for their important contributions.
FHS: We acknowledge the dedication of the FHS study participants without whom this research would not be possible.
JHS: The authors wish to thank the staffs and participants of the JHS.
WHI: The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf.
Sources of Funding
WGS for the TOPMed program was supported by the National Heart, Lung and Blood Institute (NHLBI). See below for study-specific acknowledgments and omics support information. Centralized read mapping and genotype calling, along with variant quality metrics and filtering, were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Phenotype harmonization, data management, sample-identity QC, and general study coordination were provided by the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I).
ARIC: The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HHSN268201700005I).
BioMe: Funding support for the BioMe study was provided through the National Human Genome Research Institute (NIH U01HG007417).
CHS: This research was supported by contracts HHSN268201200036C, HHSN268200800007C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and grants U01HL080295 and U01HL130114 from the National Heart, Lung, and Blood Institute (NHLBI), with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided by R01AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
FHS: This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The Framingham Heart Study (FHS) acknowledges the support of contracts NO1-HC-25195 and HHSN268201500001I from the National Heart, Lung and Blood Institute, contract 75N92019D00031 and grant supplement R01 HL092577-06S1 for this research. This work was also supported in part by grant U01DK078616.
JHS: The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), the Mississippi State Department of Health (HHSN268201800015I/HHSN26800001) and the University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I and HHSN268201800012I) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute for Minority Health and Health Disparities (NIMHD). The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
MESA: MESA and the MESA SHARe projects are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, and UL1-TR-001420. Also supported by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
WHI: The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C.
NINDS-SiGN: This research has been conducted using the National Institute of Neurological Disorders and Stroke (NINDS)-SiGN resource (NINDS U01NS06920).
UKBB: This research has been conducted using the UK Biobank Resource (access number: 19746).
For each participating study in TOPMed, the phs numbers, omics center names, and omics center support are summarized in the table below.
L.M.R. was funded by NHLBI grant T32 HL129982. C.D.A. was supported in this work through NINDS R01 NS103924. P.L.A. was funded by NHLBI grant 5R01HL136574-04.
| TOPMed Accession # | TOPMed Project | Parent Study Short Name | Omics Center | Omics Support | Omics Type |
|---|---|---|---|---|---|
|
| |||||
| phs001211 | AFGen | ARIC AFGen | Broad Genomics | 3R01HL092577-06S1 | WGS |
| phs001211 | VTE | ARIC | Baylor | 3U54HG003273-12S2 / HHSN268201500015C | WGS |
| phs001644 | BioMe | BioMe | Baylor | HHSN268201600033I | WGS |
| phs001644 | BioMe | BioMe | MGI | HHSN268201600037I | WGS |
| phs001368 | CHS | CHS | Baylor | HHSN268201600033I | WGS |
| phs001368 | VTE | CHS VTE | Baylor | 3U54HG003273-12S2 / HHSN268201500015C | WGS |
| phs000974 | AFGen | FHS AFGen | Broad Genomics | 3R01HL092577-06S1 | WGS |
| phs000974 | FHS | FHS | Broad Genomics | 3U54HG003067-12S2 | WGS |
| phs000964 | JHS | JHS | NWGC | HHSN268201100037C | WGS |
| phs001416 | AA_CAC | MESA AA_CAC | Broad Genomics | HHSN268201500014C | WGS |
| phs001416 | MESA | MESA | Broad Genomics | 3U54HG003067-13S1 | WGS |
| phs001237 | WHI | WHI | Broad Genomics | HHSN268201500014C | WGS |
Disclosures
Dr. Moscati reports salary support from Regeneron Pharmaceuticals outside the submitted work. Dr Anderson reports grants from National Institutes of Health of the United States, grants from American Heart Association, grants from Massachusetts General Hospital, grants from Bayer AG, and personal fees from ApoPharma, Inc. outside the submitted work. Dr. Rosand reports grants from National Institutes of Health during the conduct of the study; personal fees from Boehringer Ingelheim outside the submitted work. Dr. Xu reports grants from NIH during the conduct of the study; grants from AHA outside the submitted work; in addition, Dr. Xu has a patent to U.S. patent application: methods for diagnosing ischemia. pending and with royalties paid. Dr. Do reports grants from AstraZeneca, grants and non-financial support from Goldfinch, personal fees from Variant Bio, and scientific co-founder, scientific consultant and equity holder from Pensieve Health outside the submitted work. Dr. Psaty reports serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. Dr. Silver reports personal fees from Medicolegal malpractice review, personal fees from Women’s Health Initiative, personal fees from Best Doctors Inc., personal fees from Ebix, personal fees from Medlink, and personal fees from Medscape outside the submitted work. Dr. Liu reports grants from National Institute of Health 1R01DK125403 during the conduct of the study.
Non-standard Abbreviations and Acronyms
- AA
African American
- ARIC
Atherosclerosis Risk in Communities Study
- AS
all stroke
- BioMe
BioMe™ Biobank
- CES
cardioembolic stroke
- CHS
Cardiovascular Health Study
- DBP
diastolic blood pressure
- DHS
DNase I hypersensitive sites
- FHS
Framingham Heart Study
- GWAS
genome-wide association studies
- GRM
genetic relationship matrix
- hcLoF
high confidence loss of function
- HS
hemorrhagic stroke
- ICD
International Statistical Classification of Diseases and Related Health Problems
- ICH
intracerebral hemorrhage
- IS
ischemic stroke
- JHS
Jackson Heart Study
- LAS
Large artery stroke
- LD
linkage disequilibrium
- MAC
minor allele count
- MAF
minor allele frequency
- MESA
Multi-Ethnic Study of Atherosclerosis
- NINDS
National Institute of Neurological Disorders and Stroke
- OR
odds ratio
- PC
principal component
- QC
quality control
- SAH
subarachnoid hemorrhage
- SAIGE
Scalable and Accurate Implementation of Generalized mixed model
- SBP
systolic blood pressure
- SiGN
Stroke Genetics Network
- SKAT
SNP-set Sequence Kernel Association Test
- STREGA
Strengthening the reporting of genetic association studies
- SVS
small vessel stroke
- TOAST
Trial of Org 10172 in Acute Stroke Treatment
- TOPMed
Trans-Omics for Precision Medicine
- UKBB
UK Biobank
- WGS
whole genome sequencing
- WGSA
whole genome sequence annotator
- WHI
Women’s Health Initiative
Footnotes
References
- 1.GBD 2015 mortality and causes of death collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specifi c mortality for 249 causes of death, 1980–2015: A systematic analysis for the global burden of disease study 2015 (vol 388, pg 1459, 2016). Lancet. 2016;388:1459–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feigin VL, Norrving B, Mensah GA. Global burden of stroke. Circ Res. 2017;120:439–448. [DOI] [PubMed] [Google Scholar]
- 3.Holliday EG, Maguire JM, Evans TJ, Koblar SA, Jannes J, Sturm JW, Hankey GJ, Baker R, Golledge J, Parsons MW, et al. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nature Genetics. 2012;44:1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevan S, Traylor M, Adih-Samii P, Malik R, Paul NLM, Jackson C, Farrall M, Rothwell PM, Sudlow C, Dichgans M, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43:3161–3167. [DOI] [PubMed] [Google Scholar]
- 5.Devan WJ, Falcone GJ, Anderson CD, Jagiella JM, Schmidt H, Hansen BM, Jimenez-Conde J, Giralt-Steinhauer E, Cuadrado-Godia E, Soriano C, et al. Heritability estimates identify a substantial genetic contribution to risk and outcome of intracerebral hemorrhage. Stroke. 2013;44:1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korja M, Silventoinen K, McCarron P, Zdravkovic S, Skytthe A, Haapanen A, Faire U, Pedersen NL, Christensen K, Koskenvuo M, et al. Genetic epidemiology of spontaneous subarachnoid hemorrhage nordic twin study. Stroke. 2010;41:2458–2462. [DOI] [PubMed] [Google Scholar]
- 7.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese A, Lann SW, Gretarsdottir S, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nature Genetics. 2018;50:524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, Debette S, Lumley T, Folsom AR, van den Herik EG, et al. Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njølstad I, Nyrnes A, et al. A sequence variant in zfhx3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, Fornage M, Ikram MA, Malik R, Bevan S, et al. Genetic risk factors for ischaemic stroke and its subtypes (the metastroke collaboration): A meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Stroke Genetics C, Wellcome Trust Case Control C, Bellenguez C, Bevan S, Gschwendtner A, Spencer CC, Burgess AI, Pirinen M, Jackson CA, Traylor M, et al. Genome-wide association study identifies a variant in hdac9 associated with large vessel ischemic stroke. Nat Genet. 2012;44:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams FMK, Carter AM, Hysi PG, Surdulescu G, Hodgkiss D, Soranzo N, Traylor M, Bevan S, Dichgans M, Rothwell PMW, et al. Ischemic stroke is associated with the abo locus: The euroclot study. Ann Neurol. 2014;75:166–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD, Anderson CD, Brouwers HB, Valant V, Battey TWK, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. American Journal of Human Genetics. 2014;94:511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilarski LL, Achterberg S, Devan WJ, Traylor M, Malik R, Lindgren A, Pare G, Sharma P, Slowik A, Thijs V, et al. Meta-analysis in more than 17,900 cases of ischemic stroke reveals a novel association at 12q24.12. Neurology. 2014;83:678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Traylor M, Makela KM, Kilarski LL, Holliday EG, Devan WJ, Nalls MA, Wiggins KL, Zhao W, Cheng Y, Achterberg S, et al. A novel mmp12 locus is associated with large artery atherosclerotic stroke using a genome-wide age-at-onset informed approach. Plos Genetics. 2014;10:e1004469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carty CL, Keene KL, Cheng YC, Meschia JF, Chen WM, Nalls M, Bis JC, Kittner SJ, Rich SS, Tajuddin S,et al. Meta-analysis of genome-wide association studies identifies genetic risk factors for stroke in african americans. Stroke. 2015;46:2063–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NINDS Stroke Genetics Network (SiGN); International Stroke Genetics Consortium (ISGC). Loci associated with ischaemic stroke and its subtypes (sign): A genome-wide association study. Lancet Neurology. 2016;15:174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik R, Traylor M, Pulit SL, Bevan S, Hopewell JC, Holliday EG, Zhao W, Abrantes P, Amouyel P, Attia JR, et al. Low-frequency and common genetic variation in ischemic stroke the metastroke collaboration. Neurology. 2016;86:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Traylor M, Malik R, Nalls MA, Cotlarciuc I, Radmanesh F, Thorleifsson G, Hanscombe KB, Langefeld C, Saleheen D, Rost NS, et al. Genetic variation at 16q24.2 is associated with small vessel stroke. Ann Neurol. 2017;81:383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada Y, Sakuma J, Takeuchi I, Yasukochi Y, Kato K, Oguri M, Fujimaki T, Horibe H, Muramatsu M, Sawabe M, et al. Identification of six polymorphisms as novel susceptibility loci for ischemic or hemorrhagic stroke by exome-wide association studies. Int J Mol Med. 2017;39:1477–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howard VJ, Kleindorfer DO, Judd SE, McClure LA, Safford MM, Rhodes JD, Cushman M, Moy CS, Soliman EZ, Kissela BM, et al. Disparities in stroke incidence contributing to disparities in stroke mortality. Ann Neurol. 2011;69:619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kissela B, Schneider A, Kleindorfer D, Khoury J, Miller R, Alwell K, Woo D, Szaflarski J, Gebel J, Moomaw C, et al. Stroke in a biracial population - the excess burden of stroke among blacks. Stroke. 2004;35:426–431. [DOI] [PubMed] [Google Scholar]
- 23.Gardener H, Sacco RL, Rundek T, Battistella V, Cheung YK, Elkind MSV. Race and ethnic disparities in stroke incidence in the northern manhattan study. Stroke. 2020;51:1064–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The atherosclerosis risk in communities (aric) study: Design and objectives. The aric investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 25.Rosamond WD, Folsom AR, Chambless LE, Wang CH, McGovern PG, Howard G, Copper LS, Shahar E. Stroke incidence and survival among middle-aged adults: 9-year follow-up of the atherosclerosis risk in communities (aric) cohort. Stroke. 1999;30:736–743. [DOI] [PubMed] [Google Scholar]
- 26.The national survey of stroke. National institute of neurological and communicative disorders and stroke. Stroke. 1981;12:I1–91. [PubMed] [Google Scholar]
- 27.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, et al. The cardiovascular health study: Design and rationale. Ann Epidemiol. 1991;1:263–276. [DOI] [PubMed] [Google Scholar]
- 28.Longstreth WT, Bernick C, Fitzpatrick A, Cushman M, Knepper L, Lima J, Furberg CD. Frequency and predictors of stroke death in 5,888 participants in the cardiovascular health study. Neurology. 2001;56:368–375. [DOI] [PubMed] [Google Scholar]
- 29.Dawber TR, Kannel WB. The framingham study. An epidemiological approach to coronary heart disease. Circulation. 1966;34:553–555. [DOI] [PubMed] [Google Scholar]
- 30.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The framingham offspring study. Design and preliminary data. Prev Med. 1975;4:518–525. [DOI] [PubMed] [Google Scholar]
- 31.Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, D’Agostino RB Sr, Fox CS, Larson MG, Murabito JM, et al. The third generation cohort of the national heart, lung, and blood institute’s framingham heart study: Design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–1335. [DOI] [PubMed] [Google Scholar]
- 32.Carandang R, Seshadri S, Beiser A, Kelly-Hayes M, Kase CS, Kannel WB, Wolf PA. Trends in incidence, lifetime risk, severity, and 30-day mortality of stroke over the past 50 years. JAMA. 2006;296:2939–2946. [DOI] [PubMed] [Google Scholar]
- 33.Seshadri S, Beiser A, Kelly-Hayes M, Kase CS, Au R, Kannel WB, Wolf PA. The lifetime risk of stroke: Estimates from the framingham study. Stroke. 2006;37:345–350. [DOI] [PubMed] [Google Scholar]
- 34.Wolf PA, Kannel WB, Dawber TR. Prospective investigations: The framingham study and the epidemiology of stroke. Adv Neurol. 1978;19:107–120. [PubMed] [Google Scholar]
- 35.Taylor HA Jr. The jackson heart study: An overview. Ethn Dis. 2005;15:S6–1-3. [PubMed] [Google Scholar]
- 36.Carpenter MA, Crow R, Steffes M, Rock W, Heilbraun J, Evans G, Skelton T, Jensen R, Sarpong D. Laboratory, reading center, and coordinating center data management methods in the jackson heart study. Am J Med Sci. 2004;328:131–144. [DOI] [PubMed] [Google Scholar]
- 37.Fuqua SR, Wyatt SB, Andrew ME, Sarpong DF, Henderson FR, Cunningham MF, Taylor HA Jr. Recruiting african-american research participation in the jackson heart study: Methods, response rates, and sample description. Ethn Dis. 2005;15:S6–18-29. [PubMed] [Google Scholar]
- 38.Payne TJ, Wyatt SB, Mosley TH, Dubbert PM, Guiterrez-Mohammed ML, Calvin RL, Taylor HA Jr, Williams DR. Sociocultural methods in the jackson heart study: Conceptual and descriptive overview. Ethn Dis. 2005;15:S6–38-48. [PubMed] [Google Scholar]
- 39.Wyatt SB, Akylbekova EL, Wofford MR, Coady SA, Walker ER, Andrew ME, Keahey WJ, Taylor HA, Jones DW. Prevalence, awareness, treatment, and control of hypertension in the jackson heart study. Hypertension. 2008;51:650–656. [DOI] [PubMed] [Google Scholar]
- 40.Keku E, Rosamond W, Taylor HA Jr., Garrison R, Wyatt SB, Richard M, Jenkins B, Reeves L, Sarpong D. Cardiovascular disease event classification in the jackson heart study: Methods and procedures. Ethn Dis. 2005;15:S6–62-70. [PubMed] [Google Scholar]
- 41.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR Jr, Kronmal R, Liu K, et al. Multi-ethnic study of atherosclerosis: Objectives and design. Am J Epidemiol. 2002;156:871–881. [DOI] [PubMed] [Google Scholar]
- 42.Kawasaki R, Xie J, Cheung N, Lamoureux E, Klein R, Klein BE, Cotch MF, Sharrett AR, Shea S, Wong TY, et al. Retinal microvascular signs and risk of stroke: The multi-ethnic study of atherosclerosis (mesa). Stroke. 2012;43:3245–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Design of the women’s health initiative clinical trial and observational study. The women’s health initiative study group. Control Clin Trials. 1998;19:61–109. [DOI] [PubMed] [Google Scholar]
- 44.Adams HP Jr., Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, Marsh EE 3rd. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. Toast. Trial of org 10172 in acute stroke treatment. Stroke. 1993;24:35–41. [DOI] [PubMed] [Google Scholar]
- 45.Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, LeFaive J, VandeHaar P, Gagliano SA, Gifford A, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50:1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin DY. A simple and accurate method to determine genomewide significance for association tests in sequencing studies. Genet Epidemiol. 2019;43:365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mbatchou J, Barnard L, Backman J, Marcketta A, Kosmicki JA, Ziyatdinov A, Benner C, O’Dushlaine C, Barber M, Boutkov B, et al. Computationally efficient whole genome regression for quantitative and binary traits. bioRxiv 2020.06.19.162354. [DOI] [PubMed] [Google Scholar]
- 48.Firth D Bias Reduction of Maximum Likelihood Estimates. Biometrika. 1993; 80:27–38. [Google Scholar]
- 49.Gogarten SM, Sofer T, Chen H, Yu CY, Brody JA, Thornton TA, Rice KM, Conomos MP. Genetic association testing using the genesis r/bioconductor package. Bioinformatics. 2019;35:5346–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X, White S, Peng B, Johnson AD, Brody JA, Li AH, Huang Z, Carroll A, Wei P, Gibbs R, et al. Wgsa: An annotation pipeline for human genome sequencing studies. J Med Genet. 2016;53:111–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gamazon ER, Segre AV, van de Bunt M, Wen XQ, Xi HS, Hormozdiari F, Ongen H, Konkashbaev A, Derks EM, Aguet F, et al. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nature Genetics. 2018;50:956–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quang D, Chen YF, Xie XH. Dann: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ionita-Laza I, McCallum K, Xu B, Buxbaum JD. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nature Genetics. 2016;48:214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aouizerat BE, Vittinghoff E, Musone SL, Pawlikowska L, Kwok PY, Olgin JE, Tseng ZH. Gwas for discovery and replication of genetic loci associated with sudden cardiac arrest in patients with coronary artery disease. BMC Cardiovasc Disord. 2011;11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, Mead D, Bouman H, Riveros-Mckay F, Kostadima MA, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, Schoech A, Pasaniuc B, Price AL. Leveraging polygenic functional enrichment to improve gwas power. Am J Hum Genet. 2019;104:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arai T, Kasper JS, Skaar JR, Ali SH, Takahashi C, DeCaprio JA. Targeted disruption of p185/cul7 gene results in abnormal vascular morphogenesis. Proc Natl Acad Sci U S A. 2003;100:9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hassink RJ, Nakajima H, Nakajima HO, Doevendans PA, Field LJ. Expression of a transgene encoding mutant p193/cul7 preserves cardiac function and limits infarct expansion after myocardial infarction. Heart. 2009;95:1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.