Abstract
Parkinson’s disease (PD) is a disabling neurodegenerative disorder in which multiple cell types, including dopaminergic and cholinergic neurons, are affected. The mechanisms of neurodegeneration in PD are not fully understood, limiting the development of therapies directed at disease-relevant molecular targets. C. elegans is a genetically tractable model system that can be used to disentangle disease mechanisms in complex diseases such as PD. Such mechanisms can be studied combining high-throughput molecular profiling technologies such as transcriptomics and metabolomics. However, the integrative analysis of multi-omics data in order to unravel disease mechanisms is a challenging task without advanced bioinformatics training. Galaxy, a widely-used resource for enabling bioinformatics analysis by the broad scientific community, has poor representation of multi-omics integration pipelines. We present the integrative analysis of gene expression and metabolite levels of a C. elegans PD model using GAIT-GM, a new Galaxy tool for multi-omics data analysis. Using GAIT-GM, we discovered an association between branched-chain amino acid metabolism and cholinergic neurons in the C. elegans PD model. An independent follow-up experiment uncovered cholinergic neurodegeneration in the C. elegans model that is consistent with cholinergic cell loss observed in PD. GAIT-GM is an easy to use Galaxy-based tool for generating novel testable hypotheses of disease mechanisms involving gene-metabolite relationships.
Subject terms: Alzheimer's disease, Transcriptomics, Data integration, Genetics of the nervous system, Metabolomics
Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disorder that is characterized by motor symptoms including resting tremor, muscle rigidity, and slowness of movement1, as well as non-motor symptoms including sleep disturbances and autonomic dysfunction2. Several populations of neurons degenerate in PD, including dopaminergic neurons in the substantia nigra, and cholinergic neurons in the dorsal motor nucleus of the vagus and the nucleus basalis of Meynert3. Current treatment options mitigate symptoms of the disease but do not offer a cure, emphasizing the urgent need for greater understanding of disease mechanisms to guide the design of new potential therapeutics.
C. elegans is a small nematode worm with an exceptionally high degree of genetic tractability that can be used to uncover biological mechanisms in a tissue and cell type-specific manner. C. elegans possess orthologs for 60–80% of human genes, and the worm nervous system utilizes many of the same neurotransmitters as in mammals, including dopamine, acetylcholine, γ-amino butyric acid (GABA), and glutamate4. In our recently developed C. elegans model of PD, RNAi-mediated reduction of the branched-chain amino acid (BCAA) transferase bcat-1 recapitulated several features of PD, including age-dependent and progressive motor dysfunction characterized by spasm-like ‘curling’ behavior. bcat-1 reduction also promoted the loss of dopaminergic cell bodies and degeneration of dopaminergic neurites, with a concomitant decline in dopaminergic neuron function5,6. Transcriptomics and metabolomics profiling were used to study underlying molecular mechanisms, and analysis of each of these datasets separately pointed to an important role of mitochondria in driving disease6,7.
Integration of gene expression and metabolomics data requires advanced bioinformatics skills that are not always within the reach of the biologists who develop and test disease models. However, the experimental scientists are in an advantageous position to interpret the molecular data due to their profound knowledge of the biology of the system and of the significance of analysis results. Therefore, user-friendly bioinformatics tools that provide experimentalists with access to advanced omics data integration analysis methods are needed. Galaxy is a widely used open-source community development platform with an easy to understand GUI interface and a ‘mix and match’ pipeline building philosophy that makes it attractive to users with limited bioinformatic background. Galaxy has become one of the most successful resources for reproducible omics data analysis and already contains a plethora of tools to analyze gene expression, and metabolomics8. However, tools for the integrative analysis of transcriptomics and metabolomics data are not readily available in Galaxy.
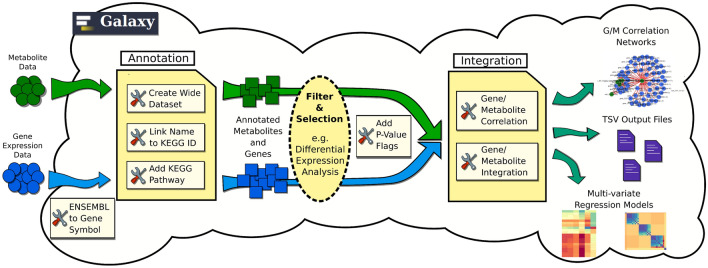
GAIT-GM is a new Galaxy tool developed to address these limitations. GAIT-GM leverages the plethora of tools available in Galaxy for the separate analysis of transcriptomics and metabolomics data to create an integration module that combines significant gene expression and metabolite annotations. This allows for studies of both data-driven and biologically oriented relationships among genes and metabolites, while seamlessly managing the data formatting issues that are endemic in trying to integrate these data types. GAIT-GM integration is based on the joint mapping of genes and metabolites to the KEGG database and in the application of dimension reduction9 and clustering10 techniques to combine gene expression and metabolite level signals. In addition to this pathway-based integration, GAIT-GM includes unbiased approaches that directly explore links between metabolites and gene expression by modeling metabolite changes as a function of transcriptional regulation (Fig. 1). We have applied these tools to the bcat-1(RNAi) C. elegans transcriptomics and metabolomics datasets6 in order to gain deeper insights into bcat-1-associated PD phenotypes. Using GAIT-GM, a previously underappreciated relationship between BCAA metabolism and cholinergic signaling were identified. This relationship was independently tested with new experimental data. These independent follow-up experiments with C. elegans confirmed that bcat-1 reduction causes cholinergic neurodegeneration, paralleling the loss of cholinergic neurons in PD.
Figure 1.
Galaxy pipeline construction enabled by GAIT-GM. Metabolomics and gene expression data are mapped to KEGG pathways. Relevant features are identified using existing statistical analysis approaches. Changes in metabolite levels are modeled as a result of changes in gene expression. Pipelines for data-driven and biology-informed integration are easily created.
Results
GAIT-GM Annotation Tool effectively maps genes and metabolites to pathways
Annotation using GAIT-GM text mining (see Supplementary Methods) resulted in 110/110 metabolites (100%) mapping to KEGG compound IDs and 3096/3146 genes (98%) mapping to KEGG IDs. While some gene and metabolite KEGG IDs did not have associated pathways, 79 metabolites were mapped to a total of 75 unique pathways, and 1187 genes mapped to a total of 115 unique pathways (Fig. S1). Pathways where both metabolites and genes were found included valine, leucine and isoleucine degradation; alanine, aspartate and glutamate metabolism; glycolysis / gluconeogenesis; and citrate cycle (TCA cycle), which recapitulates the pathway identification obtained at the separate analysis of these data6.
GAIT-GM Integrative Tools characterize the gene-metabolite co-variation network
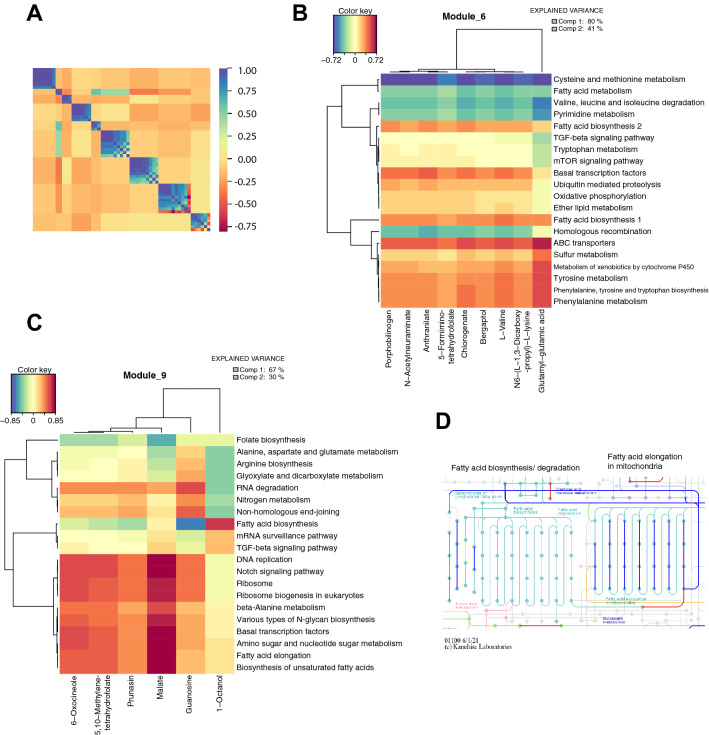
We first hypothesized that metabolic changes in bcat-1(RNAi) worms may relate to global transcriptional patterns. To explore this idea, we used MMC to cluster metabolites into modules and summarized pathway transcriptional activity by obtaining metagene profiles using PANA9 (see “Methods”). MMC identified nine metabolite modules (Fig. 2A). Several of these modules consisted of metabolites related to BCAA metabolism, including L-valine (Module 6), succinyl-CoA (Module 3), and thiamin diphosphate (Module 1). sPLS analysis of Module 6 revealed a negative correlation between L-valine and the valine, leucine and isoleucine degradation pathway (Fig. 2B), which is consistent with the known increase of BCAAs in the bcat-1 background6,11. Also consistent with previous findings was the positive correlation between L-valine and the oxidative phosphorylation pathway, since bcat-1 knockdown was reported to increase mitochondrial respiration6. In addition, L-valine was positively correlated with the phenylalanine, tyrosine and tryptophan biosynthesis pathway and negatively correlated with homologous recombination, entirely consistent with previously identified upregulation and downregulation of these pathways, respectively, in bcat-1(RNAi) worms6. In summary, integrative results involving L-valine recapitulated previous observations and provided confidence on the power of the GAIT-GM approach to recover meaningful biological insights.
Figure 2.
Integration analysis highlights gene-metabolite pathway relationships. (A) Modulated Modularity Clustering (MMC) analysis in which a smooth correlation ordering is performed in order to differentiate blocks of metabolites with the same behavior. Metabolites were grouped into 9 different modules. Blue is positively correlated and red is negatively correlated. (B,C) Heatmaps28 of Modules 6 (B) and 9 (C) after a sparse PLS (sPLS) analysis was performed with the Gene/Metabolite Integration Tool28. Metagenes were estimated from the PANA approach9. (D) Representations of C. elegans fatty acid metabolic pathways in KEGG31. Genes significantly downregulated in bcat-1(RNAi) worms are blue, and genes significantly upregulated in bcat-1(RNAi) worms are red.
Module 9 included the TCA cycle metabolite, malate (Fig. 2C), which is thought to be decreased upon bcat-1 knockdown along with other TCA cycle substrates6. In Module 9, malate was highly positively correlated with DNA replication and Notch signaling (Fig. 2C), two pathways known to be downregulated in bcat-1(RNAi) worms6, thus maintaining the expected directional relationship. Interestingly, both Modules 6 and 9 involved several pathways related to fatty acid metabolism, including fatty acid biosynthesis, fatty acid elongation, and biosynthesis of unsaturated fatty acids (Fig. 2B,C). Closer examination of the fatty acid metabolic pathways revealed that the associated genes are largely downregulated, particularly those functioning in fatty acid elongation in mitochondria (Fig. 2D). Given that mitochondrial respiration is increased in bcat-1(RNAi) worms6, it possible that fatty acid elongation is being downregulated in favor of fatty acid breakdown and utilization as a substrate for TCA cycle activity. Fatty acid metabolism was not previously highlighted in single omics analyses of the bcat-1(RNAi) worms, indicating that GAIT-GM integrated analysis may be used to reveal new potential relationships within networks of co-regulated genes and metabolites.
GAIT-GM data-driven analysis points to potential cholinergic neuron defects
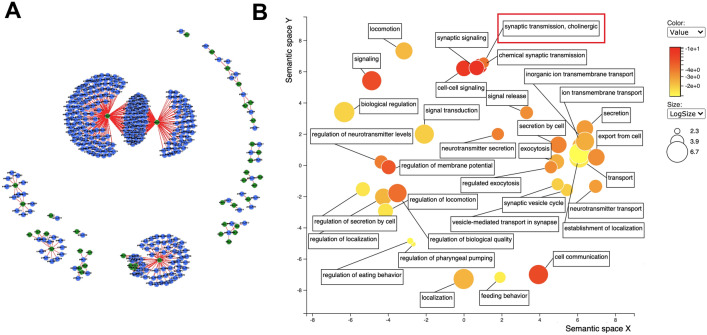
To explore possible gene-metabolite relationships not captured by the pathway analysis, we performed an unbiased discovery analysis using the Pearson correlation between individual differentially expressed metabolites and genes (Fig. 3). Two prominent metabolite hubs with 226 and 60 highly correlated genes, respectively, were identified (Fig. 3A). In the larger cluster of genes, which are positively correlated with the metabolites retinoate and amylopectin, several functional terms related to neurotransmission were significantly overrepresented. These include chemical synaptic transmission, neurotransmitter secretion, and synaptic vesicle cycle (Fig. 3B). The known role of retinoic acid in neuronal development and maintenance12 offers a possible explanation for the high number of neuronal genes correlated with this metabolite.
Figure 3.
Correlation analysis between individual genes and metabolites points to potential cholinergic defects. (A) Output from the Gene/Metabolite Correlation Tool showing the 500 strongest correlations between individual genes and metabolites. Metabolites are represented in green, while genes are represented in blue. Red lines represent positive correlations and light blue lines signify negative correlations. (B) Gene Ontology analysis visualized by REVIGO for the largest cluster from (A) shows functional enrichment for neurotransmission, and in particular cholinergic synaptic transmission (red box). Gene lists were analyzed using gProfiler29 and visualized using REVIGO30.
The second largest cluster was positively correlated with arachidonyltrifluoromethane, a derivative of arachidonic acid, which also plays a role in neuronal function13. However, in contrast to the first cluster, genes in the second largest cluster were functionally enriched for processes related to reproduction, including germ cell development, germ-line stem cell division, and cellular process involved in reproduction in multicellular organism (Fig. S2). These findings suggest the interesting possibility that bcat-1 disruption may affect reproduction in addition to neuronal health. Thus, GAIT-GM analyses offer several avenues by which to further investigate bcat-1 regulation of biological and disease processes.
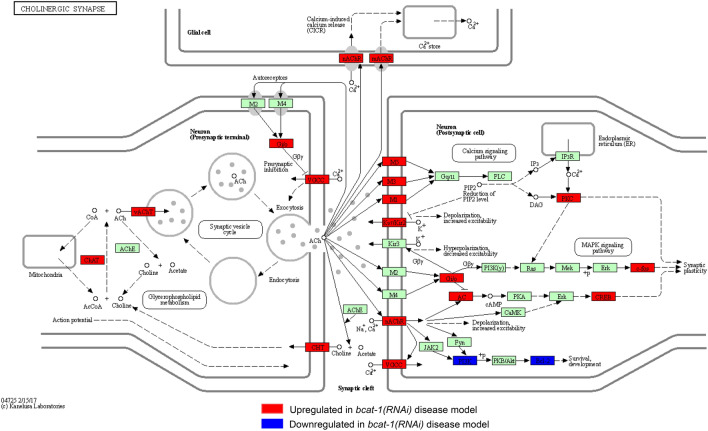
Here were focus on the impact of bcat-1 on the regulation of neuronal function and its role in neurodegenerative disease. We identified a significant functional enrichment related to cholinergic synaptic transmission (Fig. 3B) in GO analysis of the larger cluster. Further examination of cholinergic synapse components revealed that the associated genes are largely upregulated in bcat-1(RNAi) worms, including the vesicular acetylcholine transporter VACHT/unc-17, and the choline acetyltransferase CHAT/cha-1 (Fig. 4). These findings suggested to us that bcat-1(RNAi) worms may have abnormal cholinergic signaling and/or damage to cholinergic neurons which may contribute to PD-like disease.
Figure 4.
KEGG pathway for the human cholinergic synapse31 with highlighted genes representing those that are affected in the C. elegans bcat-1(RNAi) PD model. Gene orthologs significantly upregulated in bcat-1(RNAi) worms are red, gene orthologs significantly downregulated in bcat-1(RNAi) worms are blue, and all other genes with KEGG annotations are green.
Follow-up confirmation of cholinergic degeneration in the C. elegans PD model
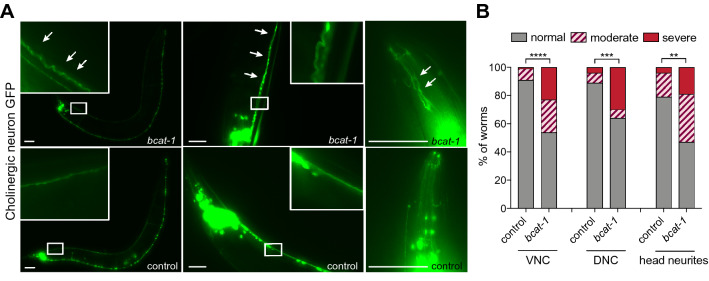
To investigate the possibility of cholinergic defects, we crossed transgenic worms expressing GFP in cholinergic neurons with a neuronal RNAi-sensitive strain14, and fed these animals either bcat-1 or control (empty vector) RNAi starting at the onset of adulthood. On day 8 of adulthood, when C. elegans are considered aged, we imaged cholinergic neurons expressing GFP and quantified neurodegeneration by assessing neurite morphology. Remarkably, RNAi-mediated reduction of bcat-1 resulted in significant degeneration of cholinergic neurites in the head, ventral nerve cord, and dorsal nerve cord (Fig. 5). Specifically, cholinergic neurites in bcat-1(RNAi) worms had more severe wavy appearances and defasciculation compared with control RNAi-treated animals (Fig. 5). These findings indicate that BCAA metabolism plays a crucial role in the health of cholinergic neurons and illustrate the power of GAIT-GM to integrate metabolite and gene expression data to reveal novel mechanisms of disease.
Figure 5.
bcat-1 RNAi-treated worms exhibit cholinergic neurodegeneration. (A,B) Cholinergic neuron projections in the head, ventral nerve cord (VNC), and dorsal nerve cord (DNC) of RNAi-sensitive worms (unc119p::sid-1;unc-17p::GFP) were scored for the degree of degenerated appearance on day 8 following bcat-1 or control RNAi treatment during adulthood. In (A), representative images of DNC (left), VNC (middle), and head neurites (right) are shown, with arrows indicating abnormal neurite regions. Scale bars, 50 μm. Quantification in (B). Chi-square test in, n ≥ 60 worms per group. **p < 0.01; ***p < 0.001, ****p < 0.0001.
Discussion
Parkinson’s disease (PD) is increasingly recognized as a multi-system disorder involving multiple brain regions and neurotransmitter systems, yet the mechanisms causing neurodegeneration of various neuronal subtypes is largely unknown. The separate analysis of gene expression and metabolomics data in a C. elegans model of PD resulted in valuable new discoveries, in particular that bcat-1 knockdown in worms alters levels of TCA cycle components and increases mitochondrial respiration. Reducing mitochondrial activity with low-dose treatment of sodium azide mitigates PD-like motor dysfunction and dopaminergic neurodegeneration in bcat-1(RNAi) worms6, underscoring the power of these technologies.
An integrative analysis of the transcriptomics and metabolomics data has the potential to reveal deeper insights into the interplay between gene expression and metabolic changes associated with PD. Existing methods for the integration of metabolomics and gene expression data are often based on multivariate dimension reduction techniques that inform on the covariation structures between transcript and metabolite levels15,16. These methods rarely incorporate existing knowledge, which hampers the interpretation of analysis results. Pathway databases such as KEGG, that include both metabolites and genes, can be used to map features to a common biological process and to guide integrative analysis and interpretation. However, mapping to pathways such as in KEGG is limited by the sparsity of metabolites captured in particular metabolic pathways. Conversely, many detected metabolites that cannot be mapped to the pathway data are eventually left aside in the integrative analysis. Moreover, despite the clear evidence of the importance of using metabolite identifiers17, names are still prevalent, impeding the use of databases that rely on identifiers. Finally, existing pathway tools have limited or no choices for the preprocessing of gene expression and metabolomics data, thus fragmenting the analysis pipeline.
To meet these challenges, we developed GAIT-GM, a Galaxy-based tool for the integrative analysis of multi-omics data. Galaxy is a platform widely used by experimentalists who analyze their own omics data because it allows flexible configuration of analysis pipelines to accommodate a wide range of experimental designs and analysis needs. However, few tools for integrative analysis of transcriptomics and metabolomics datasets exist in Galaxy. The GAIT-GM tool fills a gap in multi-omics data analysis and enables both hypothesis-driven and unbiased hypothesis-generating integrated data analysis in a well-supported open source environment. The tool provides a user-friendly, modular analytical framework that enables the construction of reproducible pipelines and transparent sharing of analytical approaches and results. Additionally, there are two important yet unmet needs in the analysis of metabolomics data specifically developed here: the efficient mapping of compound names to KEGG, and the analysis of the heterogeneity and co-regulation with gene expression to improve functional characterization.
Here, unbiased analysis of the association between metabolites and gene expression in bcat-1(RNAi) versus control C. elegans using GAIT-GM led to the discovery of cholinergic neurons as a vulnerable cell type to bcat-1 disruption. We observed severe degeneration and defasciculation of cholinergic axons/neurites in aged bcat-1(RNAi) worms. Consistent with this, several populations of cholinergic cells degenerate in PD, including in the dorsal motor nucleus of the vagus and the nucleus basalis of Meynert3. In particular, the loss of cholinergic neurons in the pedunculopontine nucleus (PPN) is implicated in gait abnormalities and akinesia in PD18, and lesions of the PPN in monkeys produce parkinsonian symptoms19,20. To our knowledge the present study is the first to link bcat-1 with cholinergic neurodegeneration. C. elegans unc-17 and cha-1 cholinergic synaptic mutants show highly similar motor dysfunction phenotypes to bcat-1(RNAi) worms21, further supporting a connection between BCAA metabolism and cholinergic neuron health. Follow-up studies may wish to dissect which BCAAs are particularly important for cholinergic signaling and/or viability using BCAA and metabolite supplementation, as well as RNAi knockdown of corresponding enzymes.
Our results herein suggest that BCAA metabolic defects may contribute to the vulnerability of multiple cell types in PD, providing a potential unifying mechanism of PD neurodegeneration. We previously found that bcat-1 reduction promotes dopaminergic neurodegeneration5,6, and we now show cholinergic degeneration in this PD model, as well. Genome-wide association studies have pointed to other BCAA pathway genes, methylcrotonoylCoA carboxylase 1 (MCCC1) and branched-chain ketoacid dehydrogenase kinase (BCKDK), as being associated with PD22, further implicating BCAA metabolism in PD pathogenesis. Moreover, metabolomics studies of PD patient urine and cerebrospinal fluid have shown elevated levels of BCAAs23 and altered TCA cycle metabolites24, respectively, consistent with our findings in the bcat-1(RNAi) C. elegans model6.
As demonstrated here, the use of GAIT-GM to uncover previously unknown relationships between genes, metabolites, and functional processes can generate novel testable hypotheses for continued investigation and discovery. Interestingly, we also observed an association between bcat-1 and reproduction pathways; since bcat-1 is known to regulate longevity in C. elegans11 and C. elegans lifespan regulators are functionally related to human longevity GWAS genes5, these findings may suggest another avenue of study that could lend insights into the common trade-off in evolution between reproduction and lifespan. Taken together, our present analyses provide greater understanding of the complex relationship between BCAA metabolism and neuronal health, and our new GAIT-GM tool can be widely applied to biological questions requiring transcriptomic and metabolomic data integration.
Methods
All method details for the generation of transcriptomics and metabolomics datasets were previously published6.
Transcriptomics
TU3311 uIs60 (unc-119p::sid-1, unc119p::yfp) worms were fed either bcat-1 or control RNAi starting at the L4 stage. On day 5 of adulthood, neurons were isolated and RNA-sequenced25 for five independent collections from each feeding group. Using Galaxy, reads were mapped to the C. elegans genome (WS245) using RNA STAR, mapped reads that overlap with gene features were counted using htseq-count (mode = union), and differential gene expression analysis was performed using DESeq2. Raw sequencing reads are available at National Center for Biotechnology Information BioProject PRJNA599166. A total of 3,146 differentially-expressed genes (false discovery rate < 0.05) were selected for the integrated analysis.
Metabolomics
CF512 fem-1(hc17); fer-15(b26) worms were fed either bcat-1 or control RNAi starting at the L4 stage. On day 5 of adulthood, 6 sets of independent replicates from each feeding group were collected for metabolomics6. Raw data are available at Dryad (https://doi.org/10.5061/dryad.5mkkwh72q). Metabolites were annotated through mummichog (level 3 confidence26), and those that were significantly different (p < 0.05) between the two groups were further examined. If a metabolic feature had multiple putative annotations, the annotation with the least difference between the theoretical and observed mass was retained, and if multiple features were assigned the same putative annotation, the putative annotation with the least difference between the theoretical and observed mass was retained. Adducts and isomers were manually curated for a final list of 110 significantly increased/decreased metabolites selected for the integrated analysis.
GAIT-GM tool
GAIT-GM (Galaxy Annotation and Integration Tools for Genes and Metabolites) consists of two major analysis modules (Fig. 1, see Supplementary Methods for details) that jointly identify, classify, group and link gene expression and metabolomics data. The annotation tool maps gene and metabolites to the KEGG database to enable subsequent pathway-based integration. This novel tool implements a text mining approach to parse metabolite names and match them to KEGG compound IDs. Ambiguous metabolites such as lipids of the same type but different formula’s (i.e. sphingomyelins) are assigned to the same KEGG ID and considered a metabolite class. Gene names are maximally mapped to KEGG through a combination of databases cross-referencing as described27. The integration tool provides different options for the clustering and summarization of each omic data modality, as well as methods for integrative analysis (see integration workflow examples in Fig. S3 and Fig. S4). Metabolites might be grouped by KEGG pathway ID, by metabolite class (i.e. all sphingomyelins), or by abundance patterns (e.g. using the Modulated Modularity Clustering (MMC) tool10. Similarly, genes can be grouped by pathway and pathway metagenes representing the expression trend in the pathway are computed using an implementation of the PANA algorithm9. Using correlation tool, bipartite networks are created to explore the relationship between metabolite and gene expression levels in an unbiased-way and without prior knowledge (Fig. S4). Alternatively, metabolite-gene associations can be studied by multivariate regression methods (Sparse Partial Least Squares or sPLS28) where any grouping of explanatory (typically pathway-specific genes or metagenes) and response (i.e. metabolite classes of MMC clusters) variables might be combined. Further information on the GAIT-GM use is provided in the Supplementary User Guide, Supplementary Table 1, and Supplementary Methods. The tool has been implemented as conda and pypi packages and available at the galaxy toolshed repository (see “Data availability” section).
Integrative analysis of C. elegans transcriptomics and metabolomics data
Five biological samples of each experimental condition and omics type were used for integrative analysis. Significant genes and metabolites were uploaded to GAIT-GM and annotated to KEGG pathways using the annotation tool. We used GAIT-GM to analyze the pair-wise correlation between individual metabolites and genes and obtained the bipartite network containing the top 500 metabolite-gene pairs with the largest absolute correlation values. Additionally, we analyzed the relationship between metabolite changes and pathway gene expression changes using the multivariate regression method available in the GAIT-GM. For this, we used KEGG pathway metagenes as explanatory variables and MMC-derived clusters for metabolites as response variables.
Gene ontology
Gene lists were analyzed using gProfiler29 with the following settings: C. elegans organism, only annotated genes, g:SCS threshold, user threshold 0.05, ENTREZGENE_ACC. REVIGO30 was used to cluster and plot GO terms with a q value < 0.05.
Validation experiments
The following C. elegans strain was used for validation experiments: CQ491 vsIs48 [unc-17p::gfp]; vIs69 [pCFJ90 (myo-2p::mCherry + unc-119p::sid-1)]. This strain was generated by crossing LX929 vsIs48 [unc-17p::gfp] with LC108 uIs69 [pCFJ90 (myo-2p::mCherry + unc-119p::sid-1)]14, both of which were obtained from the C. elegans Genetics Center. Worms were maintained at 20 °C on standard high growth medium (HG) plates seeded with OP50 E. coli. Following synchronization by bleaching, worms were transferred at day 1 of adulthood to standard nematode growth medium (NGM) plates supplemented with carbenicillin and isopropyl β-D-1-thiogalactopyranoside (IPTG), seeded with HT115 RNAi E. coli (either bcat-1 or L4440 empty vector control). RNAi plates were pre-induced with 0.1 M IPTG at 1 h before transfer, and worms were transferred onto fresh RNAi plates every 2–3 days. On day 8, animals were mounted on 2% agarose pads in M9 and sodium azide for cholinergic neuron imaging. Images were captured on a Nikon Eclipse Ti inverted microscope and processed using Nikon NIS elements software. Neurite morphology was scored as normal, moderately degenerated, or severely degenerated at the anterior portion of the ventral and dorsal nerve cords, as well as the neurites projecting from cholinergic neurons in the head ganglia. At least 15 worms were imaged per condition in each replicate.
Supplementary Information
Acknowledgements
We acknowledge the support of HiPerGator the University of Florida High performance Computing platform. In particular, the UFL Galaxy instance. We thank the C. elegans Genetics Center for strains (P40 OD010440), and the Confocal Imaging Facility at Princeton University.
Author contributions
D.E.M., A.M.M., R.K., C.T.M., V.K., G.W.M., A.C., and L.M.M. designed the research; F.H., O.M. and A.M.M. coded, implemented and tested the GAIT_GM tool; A.M.M. wrote GAIT-GM documentation. L.M.M. and A.C. supervised GAIT-GM design and implementation. D.E.M., A.M.M., R.K., V.K. performed research and analyzed the data; D.E.M., A.C., and L.M.M. wrote the main manuscript. A.M.M. wrote the supplementary methods and User Guide.
Funding
This work has been supported by the National Institute of Health SECIM grant U24 DK097209 (LMM) and R03 CA222444 (AC, LMM), U2C ES030163 (GWM) and R01 ES023839 (GWM), Pioneer Award DP1 GM119167 (CTM) and the Glenn Foundation for Medical Research CNV1001899 (CTM), Ruth L. Kirschstein National Research Service Award F32 AG062036 (DEM), and start-up fund from the Medical College of Georgia at Augusta University (DEM).
Data availability
GAIT-GM scripts, test data, Galaxy xmls are available at https://github.com/secimTools/gait-gm, as a PyPi repository (https://pypi.org/project/gait-gm/ ), and as a bioconda package (https://anaconda.org/bioconda/gait-gm ). A detailed Galaxy User Guide providing step-by-step instructions for running each tool in Galaxy is included as Supplementary Material. All tools are deposited in the Galaxy ToolShed for download and installation (https://toolshed.g2.bx.psu.edu/view/malex/gait_gm/ec9ee8edb84d ). r-mixomincs version 6.3.228, python version 3.7 and R version 4.1.1 and SECOMTools version 21.6.3. Raw RNAseq reads are available at National Center for Biotechnology Information BioProject PRJNA599166 and raw metabolomics data are available at Dryad (https://doi.org/10.5061/dryad.5mkkwh72q).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lauren M. McIntyre, Email: mcintyre@ufl.edu
Ana Conesa, Email: ana.conesa@csic.es.
Danielle E. Mor, Email: dmor@augusta.edu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-07238-9.
References
- 1.Hoehn MM, Yahr MD. Parkinsonism: onset, progression and mortality. Neurology. 1967;17:427–442. doi: 10.1212/WNL.17.5.427. [DOI] [PubMed] [Google Scholar]
- 2.Lim SY, Fox SH, Lang AE. Overview of the extranigral aspects of Parkinson disease. Arch. Neurol. 2009;66:167–172. doi: 10.1001/archneurol.2008.561. [DOI] [PubMed] [Google Scholar]
- 3.Forno LS. Neuropathology of Parkinson's disease. J. Neuropathol. Exp. Neurol. 1996;55:259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Corsi AK, Wightman B, Chalfie M. A transparent window into biology: A primer on Caenorhabditis elegans. WormBook. 2015 doi: 10.1895/wormbook.1.177.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao V, Kaletsky R, Keyes W, Mor DE, Wong AK, Sohrabi S, Murphy CT, Troyanskaya OG. An integrative tissue-network approach to identify and test human disease genes. Nat. Biotechnol. 2018 doi: 10.1038/nbt.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mor DE, Sohrabi S, Kaletsky R, Keyes W, Tartici A, Kalia V, Miller GW, Murphy CT. Metformin rescues Parkinson's disease phenotypes caused by hyperactive mitochondria. Proc. Natl. Acad. Sci. USA. 2020;117:26438–26447. doi: 10.1073/pnas.2009838117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mor DE, Murphy CT. Mitochondrial hyperactivity as a potential therapeutic target in Parkinson's disease. Transl. Med. Aging. 2020;4:117–120. doi: 10.1016/j.tma.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirpich AS, Ibarra M, Moskalenko O, Fear JM, Gerken J, Mi X, Ashrafi A, Morse AM, McIntyre LM. SECIMTools: A suite of metabolomics data analysis tools. BMC Bioinform. 2018;19:151. doi: 10.1186/s12859-018-2134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponzoni I, Nueda M, Tarazona S, Götz S, Montaner D, Dussaut J, Dopazo J, Conesa A. Pathway network inference from gene expression data. BMC Syst. Biol. 2014;8(Suppl 2):S7. doi: 10.1186/1752-0509-8-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone EA, Ayroles JF. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genet. 2009;5:e100079. doi: 10.1371/journal.pgen.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mansfeld J, Urban N, Priebe S, et al. Branched-chain amino acid catabolism is a conserved regulator of physiological ageing. Nat. Commun. 2015;6:10043. doi: 10.1038/ncomms10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat. Rev. Neurosci. 2007;8:755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- 13.Bosetti F. Arachidonic acid metabolism in brain physiology and pathology: Lessons from genetically altered mouse models. J. Neurochem. 2007;102:577–586. doi: 10.1111/j.1471-4159.2007.04558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat. Methods. 2010;7:554–559. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavill R, Jennen D, Kleinjans J, Briedé JJ. Transcriptomic and metabolomic data integration. Brief. Bioinform. 2016;17:891–901. doi: 10.1093/bib/bbv090. [DOI] [PubMed] [Google Scholar]
- 16.Meng C, Zeleznik OA, Thallinger GG, Kuster B, Gholami AM, Culhane AC. Dimension reduction techniques for the integrative analysis of multi-omics data. Brief. Bioinform. 2016;17:628–641. doi: 10.1093/bib/bbv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kind T, Scholz M, Fiehn O. How large is the metabolome? A critical analysis of data exchange practices in chemistry. PLoS ONE. 2009;4:e5440. doi: 10.1371/journal.pone.0005440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pahapill PA, Lozano AM. The pedunculopontine nucleus and Parkinson's disease. Brain. 2000;123:1767–1783. doi: 10.1093/brain/123.9.1767. [DOI] [PubMed] [Google Scholar]
- 19.Kojima J, Yamaji Y, Matsumura M, Nambu A, Inase M, Tokuno H, Takada M, Imai H. Excitotoxic lesions of the pedunculopontine tegmental nucleus produce contralateral hemiparkinsonism in the monkey. Neurosci. Lett. 1997;226:111–114. doi: 10.1016/S0304-3940(97)00254-1. [DOI] [PubMed] [Google Scholar]
- 20.Aziz TZ, Davies L, Stein J, France S. The role of descending basal ganglia connections to the brain stem in Parkinsonian Akinesia. Br. J. Neurosurg. 1998;12:245–249. doi: 10.1080/02688699845078. [DOI] [PubMed] [Google Scholar]
- 21.Sohrabi S, Mor DE, Kaletsky R, Keyes W, Murphy CT. High-throughput behavioral screen in C. elegans reveals Parkinson's disease drug candidates. Commun. Biol. 2021;4:203. doi: 10.1038/s42003-021-01731-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nalls MA, Pankratz N, Lill CM, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luan H, Liu LF, Tang Z, Zhang M, Chua KK, Song JX, Mok VC, Li M, Cai Z. Comprehensive urinary metabolomic profiling and identification of potential noninvasive marker for idiopathic Parkinson's disease. Sci. Rep. 2015;5:13888. doi: 10.1038/srep13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willkommen D, Lucio M, Moritz F, Forcisi S, Kanawati B, Smirnov KS, Schroeter M, Sigaroudi A, Schmitt-Kopplin P, Michalke B. Metabolomic investigations in cerebrospinal fluid of Parkinson's disease. PLoS ONE. 2018;13:e0208752. doi: 10.1371/journal.pone.0208752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaletsky R, Lakhina V, Arey R, Williams A, Landis J, Ashraf J, Murphy CT. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature. 2016;529:92–96. doi: 10.1038/nature16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schymanski EL, Jeon J, Gulde R, Fenner K, Ruff M, Singer HP, Hollender J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014;48:2097–2098. doi: 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- 27.Hernández-de-Diego R, Tarazona S, Martínez-Mira C, Balzano-Nogueira L, Furió-Tarí P, Pappas GJ, Jr, Conesa A. PaintOmics 3: A web resource for the pathway analysis and visualization of multi-omics data. Nucleic Acids Res. 2018;46:W503–W509. doi: 10.1093/nar/gky466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohart F, Gautier B, Singh A, Lê Cao KA. mixOmics: An R package for 'omics feature selection and multiple data integration. PLoS Comput. Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GAIT-GM scripts, test data, Galaxy xmls are available at https://github.com/secimTools/gait-gm, as a PyPi repository (https://pypi.org/project/gait-gm/ ), and as a bioconda package (https://anaconda.org/bioconda/gait-gm ). A detailed Galaxy User Guide providing step-by-step instructions for running each tool in Galaxy is included as Supplementary Material. All tools are deposited in the Galaxy ToolShed for download and installation (https://toolshed.g2.bx.psu.edu/view/malex/gait_gm/ec9ee8edb84d ). r-mixomincs version 6.3.228, python version 3.7 and R version 4.1.1 and SECOMTools version 21.6.3. Raw RNAseq reads are available at National Center for Biotechnology Information BioProject PRJNA599166 and raw metabolomics data are available at Dryad (https://doi.org/10.5061/dryad.5mkkwh72q).