Abstract
The control of mRNA stability plays a central role in regulating gene expression patterns. Recent studies have revealed that codon composition in the open reading frame determines mRNA stability in multiple organisms. Based on genome‐wide correlation approaches, this previously unrecognized role for the genetic code is attributable to the kinetics of the codon‐decoding process by the ribosome. However, complementary experimental analyses are required to clarify the codon effects on mRNA stability and the related cotranslational mRNA decay pathways, for example, those triggered by aberrant ribosome stalling. In the current study, we performed a set of reporter‐based analyses to define codon‐mediated mRNA decay and ribosome stall‐dependent mRNA decay in zebrafish embryos. Our analysis showed that the effect of codons on mRNA stability stems from the decoding process, independent of the ribosome quality control factor Znf598 and stalling‐dependent mRNA decay. We propose that codon‐mediated mRNA decay is rather triggered by transiently slowed ribosomes engaging in a productive translation cycle in zebrafish embryos.
Keywords: codons, mRNA degradation, ribosome, tRNA, zebrafish
Subject Categories: RNA Biology, Translation & Protein Quality
Parallel analysis of codon effects (PACE) system differentiates codon‐mediated mRNA degradation from ribosome stall/Znf598‐dependent decay in zebrafish embryos.
Introduction
During the translation of mRNA into protein, a sequence of codons is decoded by tRNAs on the ribosome. While the translation elongation process is strictly controlled, the speed of ribosome movement passing through the codons is not uniform. For example, each codon is decoded with variable efficiency due to the difference in tRNA availability (Varenne et al, 1984; Hussmann et al, 2015; Weinberg et al, 2016). Particular pairs of amino acids and codons inhibit peptide bond formation and/or decoding (Doerfel et al, 2013; Ude et al, 2013; Gamble et al, 2016; Schuller et al, 2017). The nascent polypeptide may interact with the ribosome exit tunnel and slow the ribosome (Ito & Chiba, 2013; Wilson et al, 2016). As a result, ribosomes traverse along the open reading frame (ORF) with distinct kinetics unique to each coding sequence (Choi et al, 2018).
Nonuniform movement of the ribosome is not always harmful and is instead often beneficial to the cell (Stein & Frydman, 2019). In bacteria, programmed ribosome stalling by specific nascent peptides acts as a regulatory switch to control protein expression (Ito & Chiba, 2013; Wilson et al, 2016). Ribosome slowdown provides additional time for nascent chain folding and complex formation (Pechmann & Frydman, 2013; Pechmann et al, 2014; Buhr et al, 2016; Weinberg et al, 2016; Stein et al, 2019). The 5´ end of the ORF tends to contain a stretch of uncommon codons that is decoded inefficiently, optimizing the ribosome traffic immediately downstream of the initiation codon (Tuller et al, 2010; Weinberg et al, 2016). Hence, cells harness a nonuniform translation elongation process to control protein output.
In addition to proteome quality, ribosome movement impacts the stability of mRNAs. Transcriptome‐wide analysis of mRNA half‐life in budding yeast revealed a correlation between codon frequency and mRNA stability: some codons are enriched in stable mRNAs, whereas others are enriched in unstable mRNAs (Presnyak et al, 2015). Similarly, the stability of thousands of maternal mRNAs in zebrafish embryos is attributable to their codon composition (Bazzini et al, 2016; Mishima & Tomari, 2016). A correlation between codons and mRNA stability has been reported in multiple organisms based on analyses of endogenous mRNAs and ORFenome reporter mRNAs, indicating a conserved role for codons in determining mRNA stability (hereafter called codon‐mediated decay) (Bazzini et al, 2016; Boël et al, 2016; Harigaya & Parker, 2016; Hia et al, 2019; Narula et al, 2019; Wu et al, 2019; Forrest et al, 2020).
Because codon‐mediated decay is dependent on translation, the ribosome bridges the gap between codons and mRNA stability (Presnyak et al, 2015; Bazzini et al, 2016; Mishima & Tomari, 2016; Narula et al, 2019; Wu et al, 2019; Forrest et al, 2020). Indeed, codon effects on mRNA stability negatively correlate with tRNA availability and ribosome density relative to A‐site codons in yeast (Hanson et al, 2018). These correlations led to a model wherein codon optimality determines mRNA stability: codons that are slowly decoded due to low tRNA availability confer a mRNA‐destabilizing effect, whereas codons with high tRNA availability are decoded smoothly and confer a mRNA‐stabilizing effect (Presnyak et al, 2015). A subsequent study in yeast revealed that the recruitment of Not5 to the E site of the slowed ribosome and ubiquitination of ribosomal protein eS7 by Not4 are both crucial for codon‐mediated decay (Buschauer et al, 2020). As Not5 and Not4 are components of the Ccr4‐Not deadenylase complex, these mechanisms explain how slow ribosomes promote mRNA deadenylation and decay.
Codon‐mediated decay accompanies mRNA deadenylation not only in yeast but also in zebrafish and humans (Presnyak et al, 2015; Bazzini et al, 2016; Mishima & Tomari, 2016; Webster et al, 2018; Wu et al, 2019). Furthermore, recent analyses in human cells revealed that the codon stability coefficient (CSC), ribosome density relative to the A site, and tRNA availability are correlated with one another (Narula et al, 2019; Wu et al, 2019; Forrest et al, 2020). Although tRNA availability is a complex parameter influenced by multiple factors, such as tRNA levels, tRNA aminoacylation levels, and amino acid levels, these studies suggest conservation of the molecular mechanism underlying codon‐mediated decay. However, genome‐wide analyses and ORFeome rely on a mere correlation between mRNA stability and codon composition on a genome‐wide scale and therefore lack a direct and experimental measurement of each single codon effect on mRNA stability (Presnyak et al, 2015; Bazzini et al, 2016; Hanson et al, 2018; Narula et al, 2019; Wu et al, 2019; Forrest et al, 2020).
Moreover, previous works may include effects from related but distinct mRNA decay pathways that also act via ORFs. One such example is a translation quality control pathway that monitors elongating ribosomes. Slowed ribosomes often induce collisions with trailing ribosomes and form specific structures called disomes and trisomes, a subset of which is recognized by the E3 ubiquitin ligase Hel2 in yeast. Hel2 ubiquitinates specific ribosomal proteins in the 40S subunit to initiate ribosome‐associated quality control (RQC) and endonucleolytic cleavage of mRNAs via no‐go decay (NGD) (Joazeiro, 2019; Inada, 2020). RQC is conserved in mammals, in which the Hel2 ortholog Znf598 acts as a collision sensor (Garzia et al, 2017; Juszkiewicz & Hegde, 2017; Sundaramoorthy et al, 2017; Juszkiewicz et al, 2018). In contrast, the occurrence of NGD is controversial in vertebrates: the established RQC substrate, consecutive Lys AAA codons, does not promote apparent mRNA decay in vertebrates (Juszkiewicz & Hegde, 2017), whereas recruitment of the Znf598‐4EHP‐GIGYF1/2 complex on paused ribosomes promotes mRNA decay (Weber et al, 2020). Hence, it remains unclear whether the codon effects captured by genome‐wide analyses in vertebrates are entirely attributable to codon‐mediated decay since they may include the effect of NGD‐related pathways. A simplified experimental approach that recapitulates the essence of codon‐mediated decay should complement genome‐wide analysis and better define the ribosome status that triggers cotranslational mRNA decay pathways.
Herein, we analyzed codon‐mediated decay in zebrafish embryos using an artificial yet simplified reporter system. Our approach allowed us to capture every single codon effect on mRNA stability in a defined context. Using this system, we evaluated the correlations between codon effects, ribosome occupancy, and tRNA levels. We also characterized the mRNA decay pathway induced by ribosome stalls in zebrafish and analyzed its relationship to codon‐mediated decay. Finally, we compared the effect of ribosome slowdown at destabilizing codons on protein production to that of other ribosome‐stalling events. Our data provide a better definition of codon‐mediated decay in zebrafish as a molecular event that occurs during slow but productive translation elongation cycles.
Results
Development of a codon‐tag reporter system that recapitulates codon‐mediated decay in zebrafish embryos
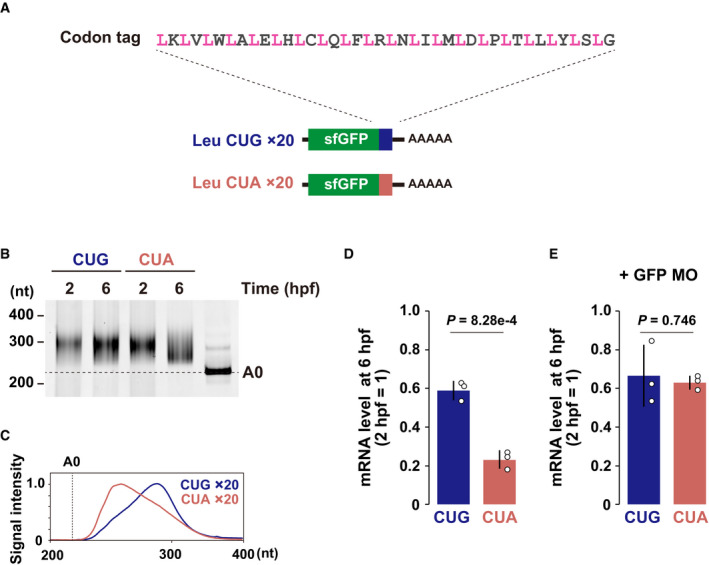
We designed an artificial codon‐tag sequence to analyze the codon effects on mRNA stability in a defined sequence context (Fig 1A. The rationale behind the design of the codon‐tag sequence is described in the Materials and Methods). This sequence contains a codon to be tested (e.g., a codon for leucine [Leu]) 20 times, each separated by a codon encoding one of the 20 amino acids. The sequence was inserted at the 3´ end of the superfolder green fluorescent protein (sfGFP) ORF since nonoptimal codons close to the 3´ end efficiently promote codon‐mediated deadenylation (Mishima & Tomari, 2016). We then compared the CUG and CUA Leu codon tags, whose differential effects on deadenylation were previously reported in zebrafish embryos (Mishima & Tomari, 2016). In vitro synthesized, capped, and polyadenylated reporter mRNAs were injected into 1‐cell stage zebrafish embryos and analyzed two hours post fertilization (hpf) (before the maternal‐to‐zygotic transition [MZT]) and 6 hpf (after the MZT). Analysis of the poly(A) tail lengths by polyadenylation test (PAT) assays revealed that the CUA codon tag promoted deadenylation compared to the CUG codon tag (Fig 1B and C). Consistent with the poly(A) tail status, quantitative reverse transcription–PCR (qRT–PCR) showed that the CUA codon‐tag reporter was less stable than the CUG codon‐tag reporter (Fig 1D). The observed difference in mRNA stability was a cotranslational effect because inhibition of translation initiation using an antisense morpholino oligonucleotide (MO) specific to the GFP ORF abolished the destabilizing effect of the CUA codon (Fig 1E). Hence, our codon‐tag reporters recapitulated the translation‐dependent effect of codons on mRNA deadenylation and degradation in zebrafish embryos.
Figure 1. Development of a codon‐tag reporter system.
-
AA scheme of the codon‐tag reporter system. A codon to be tested (pink, in this case leucine) and 1 of the 20 sense codons (gray) are alternately repeated 20 times. This codon‐tag sequence was inserted at the end of the sfGFP ORF.
-
BPoly(A) tail analysis of codon‐tag reporter mRNAs at 2 and 6 hpf. The developmental stages are shown above. The lane labeled A0 shows the 3´ UTR fragment without a poly(A) tail.
-
CQuantification of the PAT assay at 6 hpf in (B).
-
D, EqRT–PCR analysis of codon‐tag reporter mRNA levels at 6 hpf relative to 2 hpf in the absence (D) or the presence (E) of translation‐blocking GFP MO. Bar charts show the average of three independent biological replicates. Error bars show standard deviation (SD). Individual data points are shown as dots. P‐values were calculated using two‐sided Student’s t‐test.
Source data are available online for this figure.
Parallel analysis of codon effects on mRNA stability
Having validated the codon‐tag reporter system using the two Leu codons, we constructed a library of codon‐tag reporters to analyze multiple codon effects in parallel (hereafter called parallel analysis of codon effects; PACE). We prepared a mixture of codon‐tag reporter mRNAs by plasmid cloning and in vitro transcription for this approach. We then injected a mixture of reporter mRNAs into zebrafish embryos and performed RNA sequencing (RNA‐Seq) at two different time points (2 hpf and 6 hpf) to quantify the reads mapped on each tag sequence (Fig 2A). Relative codon‐tag levels 6 hpf compared to 2 hpf were calculated as the effect of each codon on mRNA stability. Although four codon tags were not included due to technical limitations in library preparation and/or sequencing (UCU, UCA, GGG, and a stop codon UAG), this approach allowed us to compare the effect of 58 sense codons on mRNA stability in a defined sequence context with reasonable reproducibility (Figs 2B and EV1A).
Figure 2. Parallel analysis of codon effects (PACE) in zebrafish embryos.
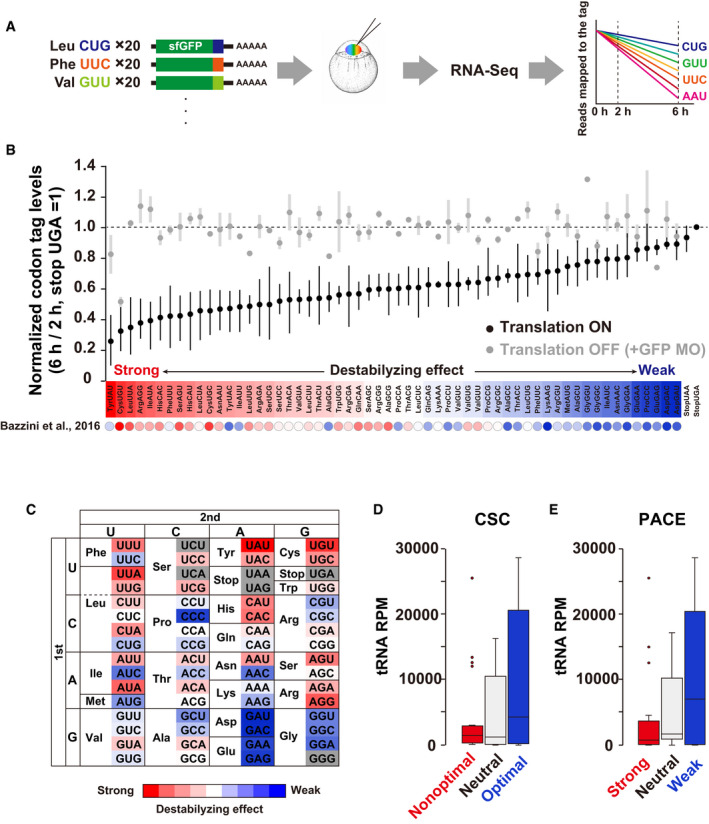
- Scheme of parallel analysis of codon effects (PACE). Codon‐tag reporter mRNAs were pooled as a library and co‐injected into fertilized zebrafish eggs. Embryos were collected at 2 and 6 hpf and subjected to RNA‐Seq analysis. Reads mapped on each codon‐tag region were quantified to calculate each codon’s effect on mRNA stability.
- Results of PACE in zebrafish embryos. Black circles show the relative stability of reporter mRNAs under normal translation (averages of three biological replicates). The stability of a codon‐tag reporter with a UGA stop codon was set to one. Gray circles show the relative stability of reporter mRNAs under the condition in which translation initiation was inhibited by GFP MO (averages of two biological replicates). Error bars represent max and minimum data points. The relative effect of each codon on mRNA stability is shown as a color gradient from red (destabilizing) to blue (stabilizing). CSCs in Bazzini et al, 2016 are also indicated below as circles with the same color gradient.
- PACE results shown in (B) are presented as a codon table.
- Box plot of tRNA amounts for nonoptimal (red, 20 codons), neutral (light gray, 18 codons), and optimal (blue, 20 codons) codons based on CSCs (Bazzini et al, 2016).
- Box plot of tRNA levels for codons with strong (red, 20 codons), neutral (light gray, 18 codons), and weak (blue, 20 codons) destabilizing effects based on PACE.
Data information: In (D) and (E), the box represents the interquartile range (IQR), with the median indicated as a black horizontal line in the box. The whiskers represent the variation within 1.5 IQR outside the upper and lower quartiles. Outliers are shown as dots. RPM, reads per million mapped reads.
Source data are available online for this figure.
Figure EV1. Validation of PACE.
-
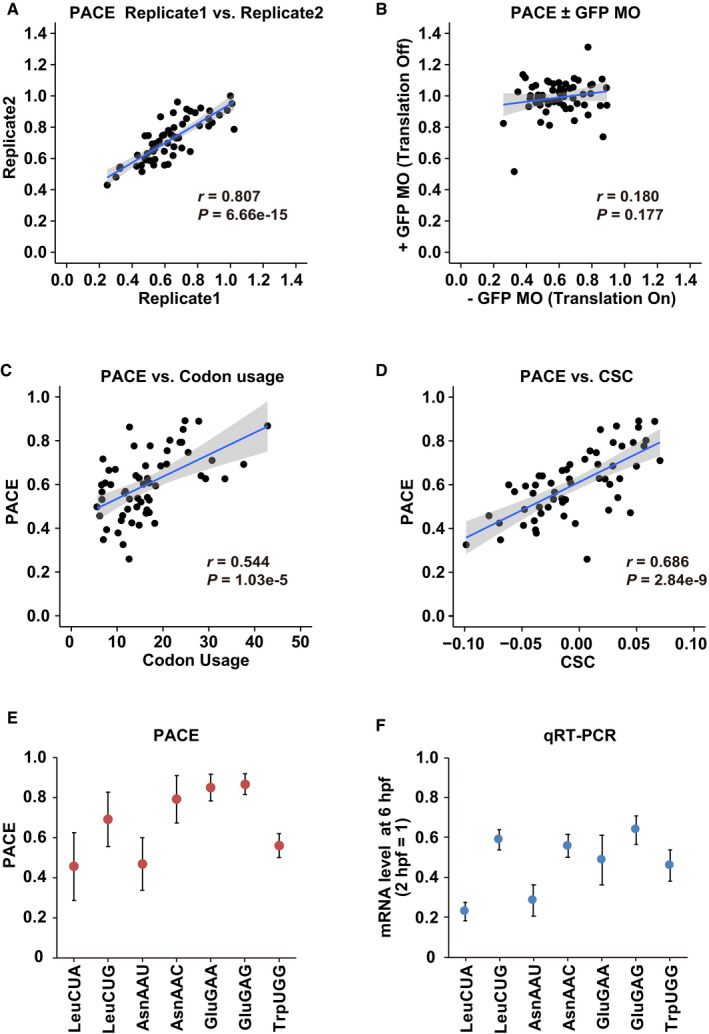
AA scatter plot showing the reproducibility of the codon effects measured by PACE in two replicates.
-
BA scatter plot comparing the codon effects measured by PACE in the absence (x‐axis) and in the presence of GFP MO (y‐axis).
-
CA scatter plot comparing codon usage in the zebrafish genome (per thousand codons, x‐axis) and the codon effects measured by PACE (y‐axis).
-
DA scatter plot comparing CSCs (Bazzini et al, 2016, x‐axis) and the codon effects measured by PACE (y‐axis).
-
E, FComparison of the codon effects measured by PACE (E) with the results of qRT–PCR (F). mRNA levels at 6 hpf relative to 2 hpf are shown. The experiments were repeated three times. Error bars show SD.
Data information: In (A–D), the regression line is shown in blue and the 95% confidence interval is shown in gray. r represents Pearson's correlation. The significance was calculated using Student’s t‐test.
Source data are available online for this figure.
Several lines of evidence indicated that PACE was a valid approach for the analysis of codon‐mediated decay. First, PACE detected codon‐specific effects on mRNA stability. Consistent with the pilot experiment using Leu CUG and CUA reporter mRNAs, the effects were variable between synonymous codons; for example, among three synonymous codons for isoleucine (Ile), AUU and AUA exhibited a more substantial destabilizing effect than AUC (Fig 2B and C). A similar difference was observed in most codon boxes, although some two‐codon boxes (histidine [His], aspartic acid [Asp], and glutamic acid [Glu]) and the glycine (Gly) codon box displayed limited variability. Second, the variable effects were dependent on translation. Inhibition of translation initiation by GFP MO stabilized almost all codon‐tag reporters to a similar level, reducing the difference between codons (Figs 2B and EV1B). Third, PACE exhibited good correlations with codon usage in the zebrafish genome (Pearson correlation coefficient r = 0.544; Fig EV1C) and CSC (r = 0.686, Fig EV1D), both of which have been used to infer codon effects on mRNA stability in previous genome‐wide studies (Bazzini et al, 2016; Mishima & Tomari, 2016). Specifically, codons classified as optimal or nonoptimal based on CSC were clearly separated in PACE (Fig 2B). We also confirmed the accuracy of PACE by qRT–PCR analysis of individual reporter mRNAs (Fig EV1E and F).
Using CSC and tRNA sequencing, Bazzini et al showed that nonoptimal codons tend to have fewer corresponding tRNAs than optimal codons in zebrafish (Bazzini et al, 2016) (Fig 2D). This trend was also true with the codon effects measured using PACE. When we divided codons into three groups based on their destabilizing effects in PACE (strong, neutral, and weak), we observed that codons with strong destabilizing effects tended to have fewer corresponding tRNAs (Fig 2E). Overall, we concluded that PACE successfully captured cotranslational, codon‐driven effects on mRNA stability in zebrafish embryos.
Codon effects correlate with ribosome elongation speed and tRNA levels
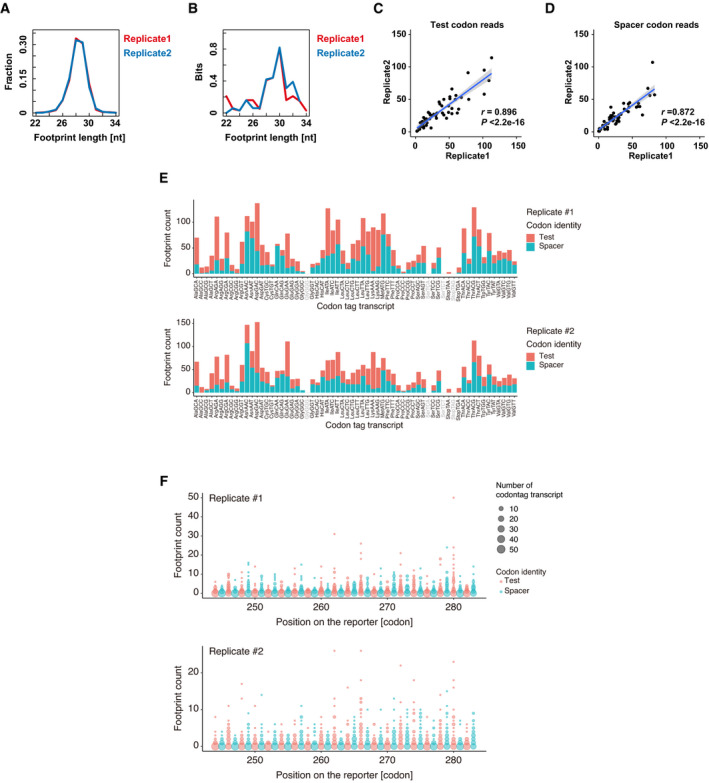
Next, we examined whether the codon effects measured by PACE correlated with the ribosome elongation process. To this end, we performed ribosome footprint profiling with PACE reporters in zebrafish embryos (Ingolia et al, 2009, 2011; Han et al, 2020). We collected embryos injected with the PACE library at 4 hpf, a middle time point between 2 hpf and 6 hpf, to capture ribosome movement during the process of codon‐mediated decay. Embryos were snap‐frozen and treated with cycloheximide after cell lysis to avoid aberrant effects of the compound on ribosome occupancy at codons (Hussmann et al, 2015; Weinberg et al, 2016). The obtained reads fell into 28–30‐nt‐length fragments, a hallmark of ribosome footprints (Fig EV2A). Assessment of Kullback–Leibler divergence ensured the presence of robust three‐nucleotide periodicity in the 28–30‐nt fragments (Fig EV2B). To analyze ribosome occupancy on codon tags, we counted the footprints with the A‐site position placed on the given test codon and then normalized that by the footprints with the A site at the spacer codons (Fig 3A). This calculation allowed us to estimate the density of ribosomes waiting for decoding test codons without a complex normalization procedure. The numbers of footprints were well correlated between the two replicates (Fig EV2C–E). The distribution of footprints along codon tags was not uniform, and while not all codon positions yielded footprints, this tendency was also well correlated in replicates (Fig EV2F). We thus combined reads from two replicates to calculate ribosome density.
Figure EV2. Validation of ribosome footprint analysis.
-
ADistribution of the ribosome footprint lengths derived from codon‐tag sequences. The results of the two replicates are plotted separately in red and blue.
-
BEvaluation of the codon periodicity by calculating the relative entropy of information content (Kullback–Leibler divergence) of each reading frame using ribosome footprints aligned to the codon‐tag reporter sequences. In this analysis, higher bits indicate the presence of periodicity in a particular reading frame. The results of the two replicates are plotted separately in red and blue.
-
C, DScatter plots showing the reproducibility of ribosome footprint analysis using PACE reporters. Two replicates for test codon reads (C) and spacer codon reads (D) are shown. The regression line is shown in blue and the 95% confidence interval is shown in gray. r, Pearson's correlation. Significance was calculated by Student’s t‐test.
-
ENumbers of ribosome footprints with a test codon (pink) or a spacer codon (turquoise) in the corresponding A‐site position are shown for each codon‐tag transcript. The results of the two replicates are plotted separately.
-
FDistributions of the ribosome footprints along the codon‐tag sequences. The size of the circles indicates the number of codon‐tag transcripts that generated footprints at each codon‐tag position. The y‐axis indicates the number of counted footprints. The results of the two replicates are plotted separately.
Source data are available online for this figure.
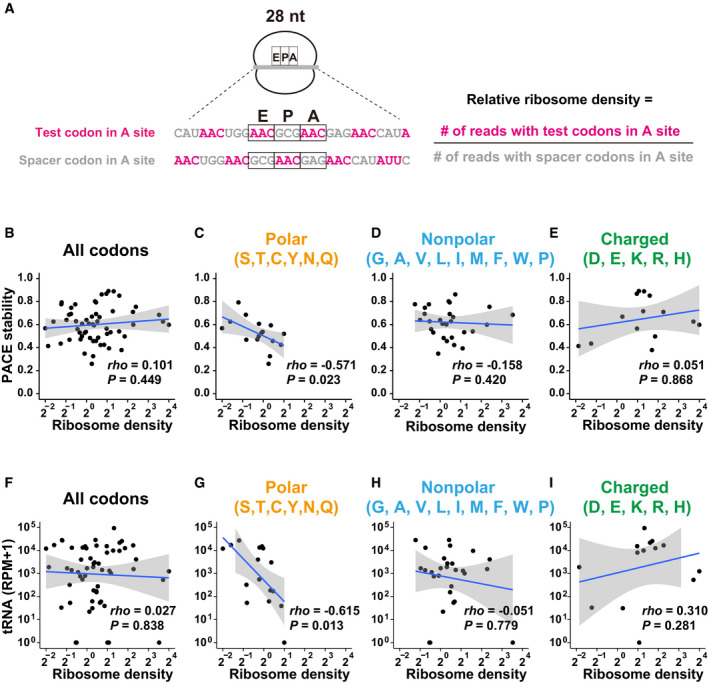
Figure 3. Analysis of the ribosome decoding speed at codon‐tag sequences.
-
AScheme of ribosome footprint analysis using PACE reporter mRNAs. An example of a 28‐nt footprint is shown. The relative ribosome density of each codon at the A site was calculated using the formula shown on the right.
-
B–EScatter plots showing a correlation between relative ribosome density (28–30‐nt footprints; x‐axis) and the codon effects measured by PACE (y‐axis). Each dot represents a single codon. Graphs for all codons (B), codons for polar amino acids (C), nonpolar amino acids (D), and charged amino acids (E) are shown.
-
F–IScatter plots showing a correlation between relative ribosome density (28–30‐nt footprints; x‐axis) and the tRNA levels (y‐axis, Bazzini et al, 2016). Each dot represents a single codon. Graphs for all codons (F), codons for polar amino acids (G), nonpolar amino acids (H), and charged amino acids (I) are shown. RPM, reads per million mapped reads.
Data information: In (B–I), the regression line is shown in blue and the 95% confidence interval is shown in gray. P‐values were calculated using Student’s t‐test. rho, Spearman's correlation.
Source data are available online for this figure.
We first analyzed a correlation between the codon effects measured using PACE and the ribosome density for all codons tested, but no clear correlation was observed (Fig 3B). Because translation of a codon tag produces a nascent peptide enriched with a particular amino acid locally, the nascent peptide within the ribosome exit tunnel may impact the speed of ribosome traverse. Therefore, we divided the codon tags into three groups based on the characteristics of the encoded amino acid: polar, nonpolar, and charged. This classification revealed that codon tags encoding polar amino acids exhibited a negative correlation between the codon effects and the ribosome density: the higher the ribosome density on a given codon, the stronger the destabilization effect (Fig 3C). On the other hand, this correlation was not observed with codons for nonpolar or charged amino acids (Fig 3D and E). We compared the ribosome density of tested codons to the corresponding tRNA levels to further analyze the relationship between the ribosome density and the decoding process. Consistent with the analysis of ribosome density and the PACE results, a negative correlation between ribosome density and tRNA levels was observed with codons for polar amino acids (Fig 3F–I). Although there was a technical limitation (see discussion), our PACE approach confirmed that codon‐mediated decay is connected to the slower codon‐decoding process at the A site by tRNA, at least for polar amino acid codons.
Alteration of codon effects in response to changes in tRNA availability
Next, we modulated tRNA availability in zebrafish embryos to experimentally validate the connection between codon effects and the decoding process. The bacterial enzyme AnsB, which hydrolyzes asparagine (Asn) to aspartic acid, reduces aminoacylated tRNAAsn and increases the duration of the ribosome’s occupancy on Asn codons in human cells (Loayza‐Puch et al, 2016). Overexpression of AnsB in zebrafish embryos by mRNA injection caused morphological defects at the shield stage (Fig 4A). Analysis of charged tRNA levels in these embryos by OXOPAP (Gaston et al, 2008) followed by qRT–PCR revealed that aminoacylated tRNAAsn GUU was specifically reduced in the presence of AnsB (Fig 4B). We then examined whether the stable Asn AAC codon‐tag reporter mRNA was subject to codon‐mediated decay in the presence of AnsB. We found that the AAC codon‐tag reporter mRNA was less stable in AnsB‐expressing embryos than in control embryos (Fig 4C). In contrast, the stability of the unrelated codon‐tag reporter mRNA for Leu CUG was not affected by AnsB. The PAT assay revealed that deadenylation of the AAC codon‐tag reporter mRNA was promoted in AnsB‐expressing embryos (Fig 4D and E), whereas the poly(A) tail status of the CUG codon‐tag reporter mRNA was not affected (Fig 4F and G). Hence, experimental modulation of a particular tRNA availability (tRNAAsn GUU) alters the effect of the corresponding codon (AAC, which is decoded by tRNAAsn GUU).
Figure 4. Modulation of charged tRNA levels in zebrafish embryos.
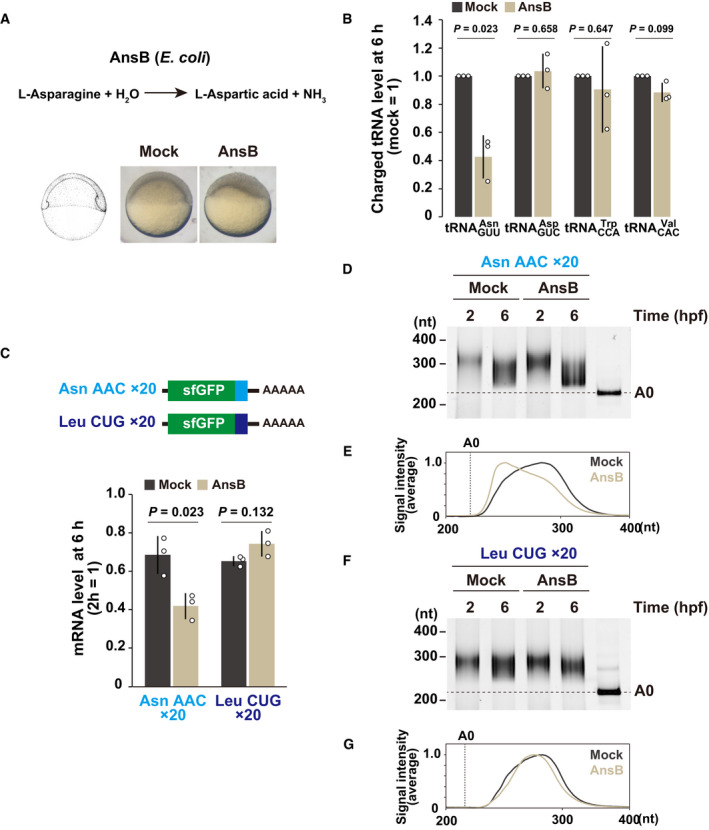
- AnsB causes asparagine deprivation by converting asparagine to aspartic acid. Mock‐injected and AnsB‐overexpressing zebrafish embryos at 6 hpf are shown.
- Charged tRNA levels in mock‐injected and AnsB‐overexpressing embryos at 6 hpf. Total and charged tRNAs were measured by qRT–PCR coupled to the OXOPAP assay. The relative charged tRNA level in mock‐injected embryos were set to one.
- qRT–PCR analysis of Asn AAC and Leu CUG codon‐tag reporter mRNAs at 6 hpf relative to 2 hpf in mock‐injected and AnsB‐overexpressing embryos.
- Poly(A) tail analysis of Asn AAC codon‐tag reporter mRNA at 2 and 6 hpf. The developmental stages are shown above as hpf. The lane labeled A0 shows the 3´ UTR fragment without a poly(A) tail.
- Quantification of the PAT assay at 6 hpf in (D).
- Poly(A) tail analysis of Leu CUG codon‐tag reporter mRNA at 2 and 6 hpf.
- Quantification of the PAT assay at 6 hpf in (F).
Data information: In (B) and (C), bar charts show the average of three independent biological replicates. Error bars show SD. Individual data points are shown as dots. P‐values were calculated using two‐sided Student’s t‐test.
Source data are available online for this figure.
To further explore the effect of asparagine deprivation on codon‐mediated decay, we performed PACE in the presence of AnsB (Fig EV3A). Consistent with the qRT–PCR analysis, AnsB changed Asn AAC to a destabilizing codon in PACE. Asn AAU, initially classified as a destabilizing codon, also gained a destabilizing effect to a lesser extent. As a result, Asn AAC and AAU were ranked as the 1st and 6th codons relatively destabilized in the presence of AnsB, respectively (Fig EV3B). We also noted that a few other codons such as arginine codons obtained a destabilizing effect with AnsB. These changes might be a consequence of eIF2α phosphorylation and the integrated stress response (Pakos‐Zebrucka et al, 2016) (Fig EV3C), as genes involved in amino acid metabolism and transport, including the arginase gene arg2, were upregulated in AnsB‐expressing embryos (Fig EV3D). Overall, we concluded that asparagine deprivation conferred destabilizing effects to Asn codons. These analyses support the idea that tRNA availability is a major determinant of codon‐mediated deadenylation and mRNA degradation in zebrafish embryos.
Figure EV3. PACE under asparagine deprivation conditions.
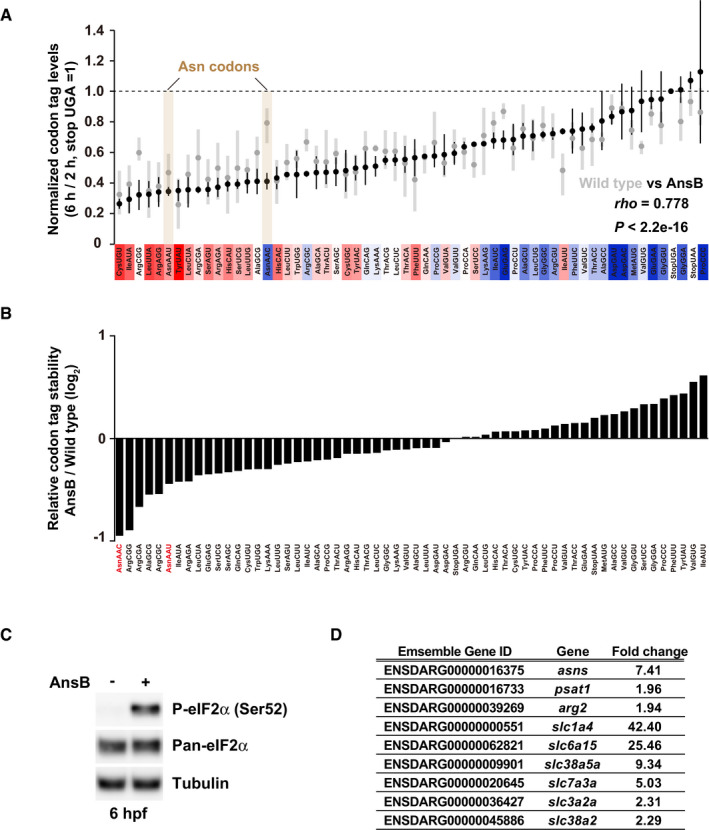
- Results of PACE in AnsB‐expressing zebrafish embryos. Black circles show the relative stability of the reporter mRNAs (averages of two biological replicates) in AnsB‐expressing embryos. Gray circles show the relative stability of reporter mRNAs in wild‐type embryos presented in Fig 2B. The stability of a codon‐tag reporter with a UGA stop codon is set to one. Error bars represent maximum and minimum data points. The relative effect of each codon on mRNA stability in wild‐type embryos measured in Fig 2B is shown as a color gradation with red (destabilizing) to blue (stabilizing). rho, Spearman's correlation. Significance was calculated by Student’s t‐test.
- Relative change in codon‐tag reporter stability between wild‐type and AnsB‐expressing embryos. PACE results in AnsB‐expressing embryos were divided by PACE results in wild‐type embryos and are shown as bar charts. Asn codons are indicated in red.
- Western blotting to detect phospho‐eIF2α (Ser51) and pan‐eIF2α at 6 hpf in the absence or in the presence of AnsB. Tubulin was detected as a loading control.
- A list of genes related to amino acid metabolism or transport and upregulated in AnsB‐expressing embryos compared to wild‐type embryos at 6 hpf. Fold changes assessed by RNA‐Seq and ensemble gene ID are shown.
Source data are available online for this figure.
Codon‐mediated decay occurs independently of Znf598
In theory, the slowed ribosome at the decoding step may drive other types of cotranslational mRNA decay such as NGD. Thus, we tested whether the codon effects observed in our PACE experiments represented codon‐mediated decay or a mixed output with NGD.
As NGD has not been previously reported in zebrafish, we first had to investigate whether the ribosome stalls induce NGD in zebrafish embryos. For this purpose, we focused on the ribosome‐stalling sequence from human cytomegalovirus (hCMV) upstream ORF2 (uORF2), whose peptide arrests the ribosome on the stop codon together with eRF1 (Matheisl et al, 2015). Here, we hypothesized that hCMV uORF2 causes ribosome stalls strong enough to induce NGD in zebrafish embryos. When we inserted codon‐optimized hCMV uORF2 at the 3´ end of the sfGFP ORF, protein output was significantly reduced in zebrafish embryos (Fig 5A and B). As reported, mutating the diproline motif at the end of uORF2, critical amino acids for ribosome stalling, partially restored the protein output (Matheisl et al, 2015). We then measured the stability of these reporter mRNAs and found that hCMV uORF2 reduced mRNA stability in a diproline motif‐dependent manner (Fig 5C). Notably, the PAT assay detected no significant deadenylation of the reporter mRNAs (Fig 5D and E). These results suggested that hCMV uORF2 induces mRNA decay in a deadenylation‐independent manner, distinct from codon‐mediated decay.
Figure 5. The effect of ribosome stalling on mRNA stability.
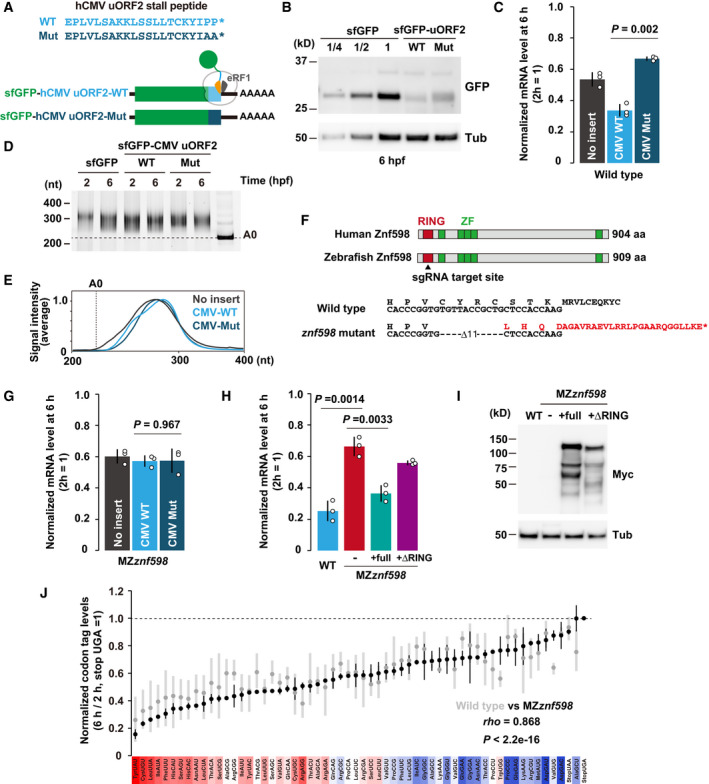
- A scheme of the hCMV uORF2 reporter mRNAs. Wild‐type hCMV uORF2 (light blue) or its mutant (dark blue) was inserted at the end of the sfGFP ORF. Asterisks indicate stop codons.
- Western blotting to detect the sfGFP levels at 6 hpf. Tubulin (Tub) was detected as a loading control.
- qRT–PCR analysis of hCMV uORF2 reporter mRNA levels in wild‐type embryos at 6 hpf relative to 2 hpf.
- Poly(A) tail analysis of hCMV uORF2 reporter mRNAs at 2 and 6 hpf. The developmental stages are shown above as hpf. The lane labeled A0 shows the 3´ UTR fragment without a poly(A) tail.
- Quantification of the PAT assay at 6 hpf in (D).
- Scheme of human and zebrafish Znf598 proteins. The sgRNA target site is indicated by an arrowhead. Genome sequences of wild‐type and znf598 mutant zebrafish are shown below. The predicted amino acid sequence of mutated Znf598 is shown in red. The asterisk indicates the premature stop codon.
- qRT–PCR analysis of hCMV uORF2 reporter mRNA levels in MZznf598 embryos at 6 hpf relative to 2 hpf.
- qRT–PCR analysis of hCMV uORF2 reporter mRNA levels in wild‐type embryos, MZznf598 embryos and MZznf598 embryos injected with a mRNA encoding Myc‐tagged full‐length (full) or RING domain‐deleted (ΔRING) Znf598. mRNA levels at 6 hpf relative to 2 hpf are shown.
- Western blotting to detect the Myc‐tagged Znf598 proteins at 6 hpf. Tubulin (Tub) was detected as a loading control.
- Results of PACE in MZznf598 zebrafish embryos. Black circles show the relative stability of the reporter mRNAs (averages of two biological replicates) in MZznf598 embryos. Gray circles show the relative stability of reporter mRNAs in wild‐type embryos shown in Fig 2B. The stability of a codon‐tag reporter with a UGA stop codon was set to one. Error bars represent maximum and minimum data points. The relative effect of each codon on mRNA stability in wild‐type embryos measured in Fig 2B is shown as a color gradient from red (destabilizing) to blue (stabilizing). rho, Spearman's correlation. Significance was calculated by Student’s t‐test.
Data information: In (C), (G), and (H), bar charts show the average of three independent biological replicates. Error bars show SD. Individual data points are shown as dots. P‐values were calculated using two‐sided Student’s t‐test.
Source data are available online for this figure.
RQC‐coupled NGD in yeast requires the E3 ubiquitin ligase Hel2 and subsequent ubiquitination of the ribosomal protein uS10 (Ikeuchi et al, 2019). To characterize mRNA decay induced by hCMV uORF2, we generated a zebrafish mutant of Znf598, a vertebrate homolog of Hel2, using CRISPR–Cas9. We obtained a strain containing an 11‐bp deletion in exon 1 of the znf598 locus, which was predicted to induce a frameshift and produce a protein product truncated within the RING finger domain (Fig 5F). Embryos lacking both maternal and zygotic Znf598 (MZznf598) were obtained by crossing homozygous znf598 mutants. znf598 mRNA was significantly reduced in MZznf598 (Fig EV4A), likely reflecting the premature termination of translation followed by nonsense‐mediated mRNA decay (Maquat, 2004). The detailed phenotype of our znf598 mutant will be described elsewhere. Analysis of the reporter mRNA stability revealed that hCMV uORF2 did not induce mRNA decay in MZznf598 embryos (Fig 5G). Rescue experiments revealed that the mRNA decay activity of hCMV uORF2 was restored by full‐length Znf598 but not by a deletion construct lacking the RING domain (Fig 5H and I). These results indicate that specific ribosome stalls induced mRNA decay in a Znf598‐dependent manner in zebrafish embryos.
Figure EV4. Validation of MZznf598 mutant.
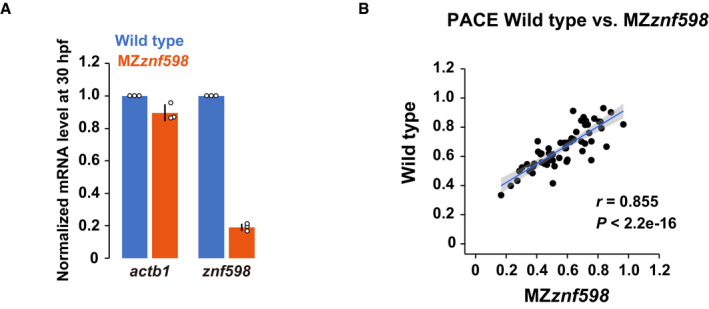
- qRT–PCR analysis of actb1 and znf598 mRNA levels in wild‐type and MZznf598 embryos at 24 hpf. The results are normalized to 18S rRNA levels. The mRNA levels in wild type were set to one. The experiments were repeated three times. Error bars show SD. Individual data points are shown as dots.
- A scatter plot comparing the codon effects measured by PACE in MZznf598 (x‐axis) and wild‐type (y‐axis) embryos. The regression line is shown in blue, and the 95% confidence interval is shown in gray. r, Pearson's correlation. Significance was calculated by Student’s t‐test.
Source data are available online for this figure.
This observation led us to test the impact of Znf598 on codon effects measured using PACE (Fig 5J). Overall, we found that most codon effects were maintained in MZznf598 compared to wild type, with a high correspondence (correlation coefficients 0.855 [Pearson] and 0.868 [Spearman]) (Figs 5J and EV4B). However, we noted that a few codons exhibited altered effects in MZznf598. For example, Tryptophan (Trp) UGG codon tag reporter mRNA was more stable in MZznf598 than in the wild type. Since ribosome stalls formed by consecutive UGG codons are subject to RQC in yeast (Dimitrova et al, 2009; Mizuno et al, 2021), the Trp UGG codon tag might induce mRNA decay in part via the NGD‐like pathway in zebrafish embryos. We draw two conclusions from these results: (i) Znf598‐dependent NGD‐like activity exists in zebrafish embryos, and (ii) most, if not all, of the codon effects measured using PACE are distinct from Znf598‐dependent mRNA decay.
Codon‐mediated decay occurs during productive translation
To further characterize the nature of the ribosome slowdown that leads to codon‐mediated decay, we performed a tandem ORF reporter assay that could quantitatively detect ribosome stalling in mammalian cells (Juszkiewicz & Hegde, 2017; Sundaramoorthy et al, 2017). Since cell sorting is difficult due to the large size of cells in early embryos, we utilized luciferases instead of fluorescent proteins to monitor protein outputs. Renilla luciferase (Rluc) and firefly luciferase (Fluc) were separated by two porcine teschovirus‐1 2A (P2A) sequences that cause skipping of peptide bond formation (Kim et al, 2011), and a sequence to be tested for the efficiency of translation elongation was inserted between the two P2A sequences (Fig 6A). If the inserted sequence inhibited the elongation process of the ribosome, the Fluc activity relative to the Rluc activity would be reduced in this experimental setup. In vitro synthesized mRNA was injected into 1‐cell stage zebrafish embryos, and luciferase activities were measured 3 hpf to quantify protein levels before the completion of codon‐mediated decay.
Figure 6. Analysis of the translation elongation rate in zebrafish embryos.
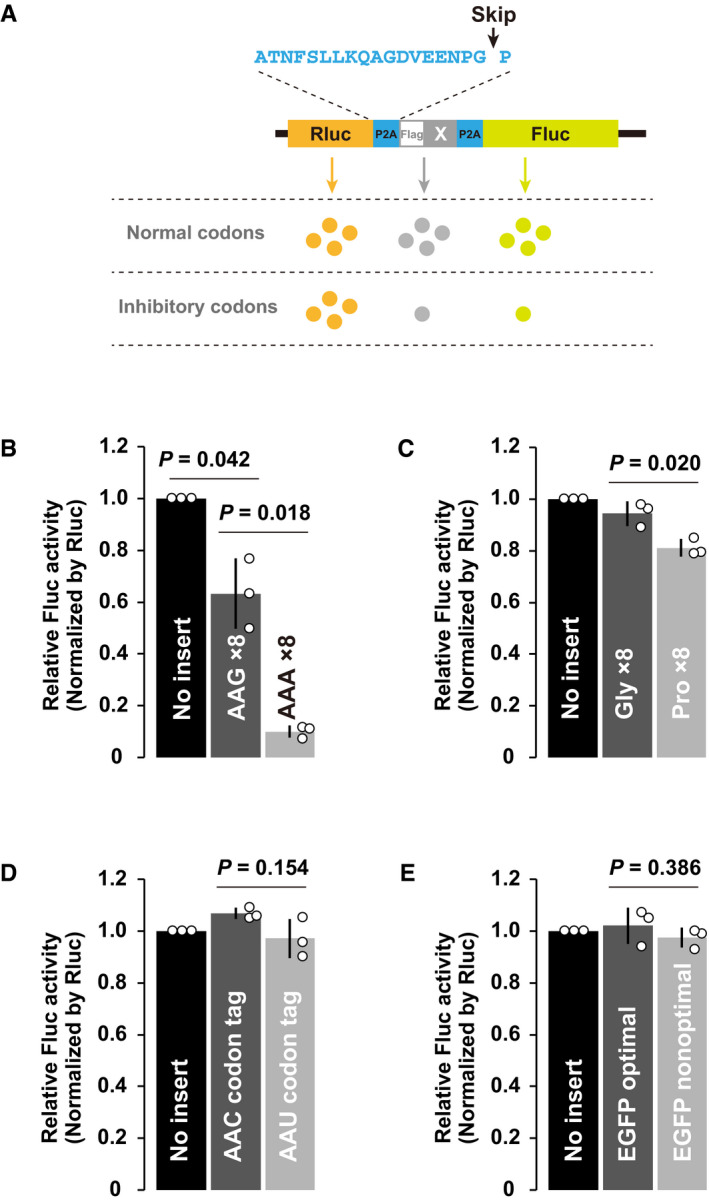
- A schematic of the tandem ORF assay. The Rluc ORF (orange) and Fluc ORF (light green) are separated by two P2A translation skipping sequences (light blue). A sequence to be tested (shown as X in gray) is inserted between the two P2A sequences.
- Results of the tandem ORF assay with Lys AAG×8 and Lys AAA×8 sequences.
- Results of the tandem ORF assay with Gly×8 and Pro×8 sequences.
- Results of the tandem ORF assay with Asn AAC and Asn AAU codon tags.
- Results of the tandem ORF assay using EGFP ORFs with different codon optimalities.
Data information: In (B–E), normalized Fluc activity with no insert was set to one. All experiments were repeated three times, and the average Fluc activity is shown as bar charts. Error bars show SD. Individual data points are shown as dots. P‐values were calculated using two‐sided Student’s t‐test.
Source data are available online for this figure.
To verify the sensitivity of the assay, we first analyzed the effect of consecutive Lys AAA codons that are known to induce ribosome stalling, collision, and RQC in other organisms (Ito‐Harashima et al, 2007; Juszkiewicz & Hegde, 2017; Sundaramoorthy et al, 2017; Tesina et al, 2020). Insertion of eight consecutive AAG codons caused an ~40% reduction in Fluc activity, likely reflecting the inhibitory effect of the positively charged nascent chain per se. Insertion of eight consecutive AAA codons further reduced Fluc activity to ~10%, resulting in a sixfold decrease compared to AAG codons (Fig 6B). AAA codons caused detectable levels of frameshifting (~ 14% in total), as previously reported (Juszkiewicz & Hegde, 2017), but frameshifting alone was not sufficient to explain the overall reduction in Fluc activity (Fig EV5A). An endogenous stall‐prone sequence derived from ZCRB1 (Arthur et al, 2015; Han et al, 2020) caused a significant yet milder reduction in Fluc activity (~35%) in a Lys AAA codon‐dependent manner accompanied by frameshifting (~ 20% in total) (Fig EV5B–D). We also tested the effect of tandem proline codons, which inhibit peptide bond formation, as an example of ribosome slowdown rescued by eIF5A (Schuller et al, 2017). Insertion of eight consecutive proline codons caused an ~20% reduction in Fluc activity (Fig 6C). Hence, our assay was sensitive enough to detect a variety of aberrant ribosome slowdown events in zebrafish embryos.
Figure EV5. Additional analysis of the translation elongation rate in zebrafish embryos.
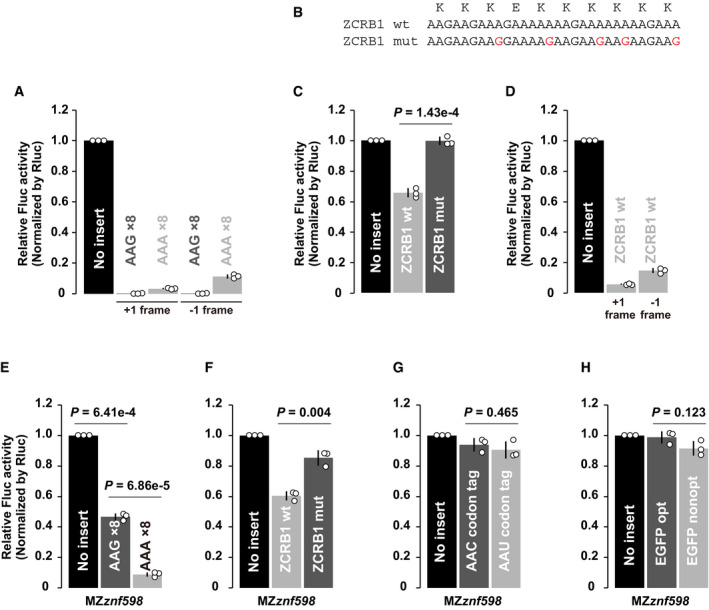
- Results of the tandem ORF assay to detect frameshifting with Lys AAG×8 or Lys AAA×8 sequences. One or two guanine nucleotides (G or GG) were inserted after Lys AAG×8 or Lys AAA×8 sequences to detect protein products from +1 or −1 frame.
- Wild‐type (wt) or mutated (mut) stalling sequences of human ZCRB1. Mutated nucleotides were indicated in red. The amino acid sequence is shown above.
- Results of the tandem ORF assay with wild‐type (wt) or mutated (mut) stalling sequences of ZCRB1.
- Results of the tandem ORF assay to detect frameshifting with a ZCRB1‐stalling sequence. One or two guanine nucleotides (G or GG) were inserted after the ZCRB1 sequence to detect protein products from +1 or −1 frame.
- Results of the tandem ORF assay with Lys AAG×8 or Lys AAA×8 sequences in MZznf598 embryos.
- Results of the tandem ORF assay with wild‐type (wt) or mutated (mut) stalling sequences of ZCRB1 in MZznf598 embryos.
- Results of the tandem ORF assay with Asn AAC and Asn AAU codon tags in MZznf598 embryos.
- Results of the tandem ORF assay using EGFP ORFs with different codon optimalities in MZznf598 embryos.
Data information: In all experiments, normalized Fluc activity with no insert was set to one. All experiments were repeated three times, and the average Fluc activity is shown as bar charts. Error bars show SD. Individual data points are shown as dots. P‐values were calculated using two‐sided Student’s t‐test.
Source data are available online for this figure.
We then analyzed the effect of two synonymous AAC and AAU Asn codon tags, which exerted opposite effects on mRNA stability in PACE (Fig 2B and C). We found that Asn codon tags did not significantly alter the Fluc/Rluc ratios (Fig 6D). This result suggests that ribosomes are not significantly retarded on the destabilizing codons. To confirm this observation in other codons, we compared two enhanced GFP (EGFP) ORFs encoded by either optimal or nonoptimal codons throughout the ORF (Mishima & Tomari, 2016). The inserted nonoptimal EGFP ORF did not significantly alter the Fluc/Rluc ratio (Fig 6E). Hence, the translation of >200 nonoptimal codons was still more efficient than that of a short, stall‐prone polyA stretch and polyproline sequence. Similar results were observed in MZznf598 embryos (Fig EV5E–H). These results demonstrate that the ribosome slowdown required for inducing codon‐mediated decay is weaker than the ribosome stall events that elicit specific rescue systems.
Discussion
In this study, we developed a reporter‐based assay called PACE to analyze codon‐mediated decay in zebrafish embryos. PACE was sensitive enough to detect translation‐dependent, codon‐specific effects on mRNA stability in zebrafish embryos (Fig 2). PACE relies on relatively simple bioinformatics analysis and is potentially applicable to other organisms capable of undergoing mRNA injection or transfection. Optimization of the codon‐tag sequence and inclusion of the four codon tags missed during its construction or RNA‐Seq will broaden the utility of PACE. Recapitulation of the events that occur during mRNA maturation, such as binding RNA‐regulatory proteins in the nucleus and introducing chemical modifications, should also be considered in future studies.
Previous studies have estimated codon effects by calculating the correlation between codon frequency and mRNA stability for thousands of endogenous mRNAs (Presnyak et al, 2015; Bazzini et al, 2016; Harigaya & Parker, 2016; Mishima & Tomari, 2016; Forrest et al, 2020). These analyses took advantage of averaging codon effects in multiple different positions on the transcriptome. However, the correlation tended to be modest in part due to the presence of factors that affect mRNA stability independent of codons (e.g., RNA‐binding proteins and microRNAs) (Medina‐Muñoz et al, 2021). The ORFome approach minimizes this disadvantage by fixing the 5´ and 3´ untranslated region (UTR) sequences, yet it also depends on a correlation study, and its target is limited to endogenous mRNA sequences that are under evolutionary constraints (Bazzini et al, 2016; Narula et al, 2019; Wu et al, 2019). In contrast, PACE places each codon in a single artificial context to directly evaluate that codon’s effect on mRNA stability. As the findings of PACE in this study were largely consistent with those of CSCs, our study provides additional, experimental validation of previously reported codon effects on mRNA stability in zebrafish (Figs 2B–E, and EV1D). Furthermore, we showed several lines of evidence indicating that codon effects stem from the decoding process in zebrafish. First, codons with destabilizing effects had lower corresponding tRNA levels than those with stabilizing effects (Fig 2E). Second, the occupancy of the ribosome with an A‐site codon for a polar amino acid was negatively correlated with that codon’s effect on mRNA stability (Fig 3C) and the corresponding tRNA level (Fig 3G). Third, experimental reduction of Asn tRNA availability conferred destabilizing effects to Asn codons (Figs 4 and EV3). Together, our study supports a view wherein a slow ribosome waiting for decoding by tRNAs triggers mRNA degradation in eukaryotes.
One limitation of our analysis was that a correlation between the codon effect and the decoding process was not observed with codons for nonpolar and charged codons (Fig 3D and E). Although the exact reason for this is unknown, we noted that ribosome occupancy and tRNA levels were poorly correlated with these codons (Fig 3H and I). As either local or net hydrophobicity and the charge of the nascent peptides affects ribosome movement and functionality (Lu & Deutsch, 2008; Charneski & Hurst, 2013; Chadani et al, 2016, 2017; Weinberg et al, 2016; Dao Duc & Song, 2018), one possibility is that ribosome occupancy on these codon tags predominantly reflects the effect of nascent peptides; therefore, precise measurement of the ribosome waiting for decoding is hampered. Future studies are necessary to confirm whether the decoding process of codons for nonpolar and charged amino acids also serves as a determinant of mRNA stability in zebrafish.
Recent studies in yeast have provided mechanistic insights into codon‐mediated decay and NGD, establishing them as distinct cotranslational mRNA decay pathways (Webster et al, 2018; D’Orazio et al, 2019; Ikeuchi et al, 2019; Buschauer et al, 2020). However, their definition is less clear in other organisms due to the lack of a connection between cis‐elements and trans‐acting factors. We showed that ribosome stalling by hCMV uORF2 caused mRNA decay in a deadenylation‐independent and Znf598‐dependent manner, providing the first example of NGD in zebrafish embryos (Fig 5A–I). Irrespective of that, we found that the codon effects were largely maintained even in the absence of Znf598 (Fig 5J). Hence, cellular mechanisms clearly distinguish ribosome slowdown during the decoding process from ribosome stalls by a problematic RNA or peptide and promote specific downstream mRNA decay pathways in zebrafish.
How does the cell distinguish ribosomes for codon‐mediated decay and those for NGD? Although previous yeast studies have provided insights, it is unclear whether the underlying molecular mechanisms for recognizing slow ribosomes and stalled ribosomes are evolutionally conserved in higher eukaryotes. While Not5 is not conserved in vertebrates, Not3 shares similarity with Not5 in the N‐terminal domain required for E‐site binding. It is, therefore, possible that the yeast Not3 ortholog CNOT3 anchors the CCR4‐NOT complex to the slow ribosome via the E site in vertebrates (Collart & Panasenko, 2017; Buschauer et al, 2020). Ubiquitination site(s) on the ribosome and downstream NGD effectors are also conserved in vertebrates (Garzia et al, 2017; Juszkiewicz & Hegde, 2017; Matsuo et al, 2017; Sundaramoorthy et al, 2017; Juszkiewicz et al, 2018, 2020; D’Orazio et al, 2019; Ikeuchi et al, 2019; Weber et al, 2020). Future studies will investigate the conservation and biological function of codon‐mediated decay and NGD. As exemplified by our analysis using the znf598 mutant, PACE will be a valuable tool for comparing codon effects in multiple mutant backgrounds in such studies.
Although the ribosome slows down locally on a destabilizing codon, our tandem ORF assay revealed that such slowdown had a limited impact on the overall translation elongation rate (Fig 6). This assay was sensitive enough to detect the inhibitory effects of stall‐prone consecutive AAA codons and polyproline codons. Therefore, we propose that ribosome slowdown during codon‐mediated decay does not reach a nonproductive status and maintains a productive translation cycle. This model is reasonable given that codon‐mediated decay regulates the stability of thousands of endogenous protein‐coding mRNAs for essential metabolic genes and maternal genes (Presnyak et al, 2015; Bazzini et al, 2016; Mishima & Tomari, 2016; Forrest et al, 2020). One remaining question in codon‐mediated decay is how such transient slowness of the ribosome leads to robust deadenylation and mRNA degradation. In the case of RQC/NGD, a prolonged and robust ribosome stall causes a collision with the following ribosome, which becomes a substrate for Hel2/Znf598 (Juszkiewicz et al, 2018; Ikeuchi et al, 2019; Matsuo et al, 2020). While the collision itself is estimated to occur when the elongation rate decreases several fold (~6 codons/second to ~1 codon/s) (Juszkiewicz et al, 2018), live imaging of RQC revealed that a much longer stall is required for RQC to occur (~0.12 codons/s) (Goldman et al, 2021). Our study indicated that ribosome slowdown required for triggering codon‐mediated decay does not reach such a prolonged ribosome stall (Fig 6). Among the two deadenylases in the Ccr4‐Not complex, Caf1 is involved in deadenylation related to codon optimality in yeast (Webster et al, 2018). As Caf1 is a processive deadenylase, only a transient interaction between the Ccr4–Not complex and a slow ribosome, possibly mediated by E‐site recognition, may be sufficient to initiate continuous deadenylation (Viswanathan et al, 2004). Alternatively, ubiquitination of eS7 by Not4 acts as an additional scaffold on the ribosome to stably recruit the Ccr4–Not complex after transient slowdown (Buschauer et al, 2020). Kinetics analysis of codon‐mediated decay using a defined reporter mRNA is required to understand its detailed mechanism of action.
Materials and Methods
Zebrafish
The zebrafish AB strain was raised and maintained at 28.5°C under standard laboratory conditions according to the Animal Experiment Protocol (2018‐46) at Kyoto Sangyo University. Fertilized eggs were obtained by natural breeding. Embryos were developed in system water at 28.5°C. Bright field images were acquired using a SteREO Lumar V12 microscope and an AxioCam MRc camera with ZEN software (Zeiss, Jena, Germany) using spline mode.
Microinjection
To synthesize mRNA for microinjection, RNA was transcribed from a linearized plasmid DNA template using the SP6‐Scribe Standard RNA IVT Kit (CELLSCRIPT) and purified using the RNeasy Mini Kit (QIAGEN). The m7G cap structure was added to the purified RNA using the ScriptCap m7G Capping System (CELLSCRIPT). Capped mRNA was purified using the RNeasy Mini Kit. For sfGFP reporter mRNAs, the poly(A) tail was added using the A‐Plus Poly(A) Polymerase Tailing Kit (CELLSCRIPT), and the mRNA was then purified using the RNeasy Mini Kit. Purified mRNAs were diluted in water to the following concentrations: sfGFP reporter mRNAs and PACE reporter library: 50 ng/μl; Myc‐Znf598 mRNAs: 100 ng/μl; AnsB: 50 ng/μl; tandem ORF reporter mRNAs: 50 ng/μl. GFP MO was injected as previously described (Mishima & Tomari, 2016). Microinjection was performed using an IM300 Microinjector (NARISHIGE). Approximately 1,000 pl of solution was injected per embryo within 15 min after fertilization.
Generation of a znf598 mutant strain using CRISPR–Cas9
sgRNA targeting exon 1 of zebrafish znf598 (ENSDARG00000014945) was designed using CRISPRscan (Moreno‐Mateos et al, 2015). The DNA template for sgRNA synthesis was prepared by PCR using gene‐specific primer and sgRNA tail primer. sgRNAs were transcribed in vitro using T7 RNA Polymerase (TaKaRa) and purified with ProbeQuant G‐50 microcolumns (Cytiva). m7G‐capped nCas9 mRNA was prepared with the SP6‐Scribe Standard RNA IVT Kit and ScriptCap m7G Capping System (CELLSCRIPT) using pCS2+nCas9n as a template (Jao et al, 2013). sgRNA and nCas9 mRNA were mixed at 50 ng/μl each and injected into fertilized eggs. Embryos were raised to adulthood, and the fishes were crossed with AB fish. F1 embryos were screened by PCR analysis using MultiNA (SHIMADZU). DNA sequences were determined by cloning PCR fragments and Sanger sequencing. We identified a strain with an 11‐bp deletion in exon 1, as shown in Fig 5F. Siblings of F1 fishes were raised to adulthood, and the fish harboring a heterozygous mutation were identified by fin‐clipping and PCR genotyping. The F1 fish were crossed with AB to dilute any nonspecific mutations, and the F2 fish were used to obtain homozygous mutants. Primer sequences are shown in Appendix Table S1
Plasmid construction
To construct reporter plasmids, pCS2+ (Rupp et al, 1994) was modified by PCR to generate pCS2+neo using primers y683fw, y683rv, y684fw, and y684rv. The suv39h1a 3´ UTR was excised from the previously described EGFP reporter plasmid (Mishima & Tomari, 2016) and cloned into pCS2+neo by XhoI and XbaI. The amino acid sequence of the sfGFP ORF was converted to nucleotide sequences comprising the most frequent synonymous codons in E. coli using EMBOSS Backtranseq (http://www.ebi.ac.uk/Tools/st/emboss_backtranseq/). The first 25 nucleotides of the sfGFP ORF were identical to the EGFP ORF used in a previous study to maintain the target site for GFP MO (Mishima & Tomari, 2016). EcoRI and EcoRV sites were inserted before the stop codon. The sfGFP DNA fragment was synthesized by GeneArt Strings DNA Fragments service (Life Technologies) and cloned into the pCS2+neo‐suv39h1a 3´ UTR using NcoI and XhoI. DNA oligonucleotides encoding hCMV uORF2 were annealed and cloned into the pCS2+neo‐sfGFP‐suv39h1a 3´ UTR using EcoRI and EcoRV.
To construct the tandem ORF reporter plasmid pLuc, Rluc‐P2A‐3×FLAG and P2A‐Fluc fragments were amplified from psiCHECK2‐2A‐3xFLAG‐SBP‐2A (Han et al, 2020) using primer pairs KI019×KI020 and KI021×KI022, respectively. The amplified DNA fragments contained a homologous sequence with EcoRI and XhoI sites, either downstream of Rluc‐P2A‐3×FLAG or upstream of P2A‐Fluc. Both fragments were assembled and cloned into pCS2+ between BamHI and XbaI sites using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs). DNA fragments encoding test sequence were prepared by annealing complementary oligonucleotides as follows: y921 for AAG×8; y922 for AAA×8; y933 for AAG×8 +1 frame; y 934 for AAG×8 −1 frame; y935 for AAA×8 +1 frame; y936 for AAA×8 −1 frame; y937 for AAU codon tag; y938 for AAC codon tag; y942 for Pro×8; y943 for ZCRB1 wt; y961 for ZCRB1 wt +1 frame; y962 for ZCRB1 wt −1 frame; y963 for Gly×8; y964 for ZCRB1 mut. Optimal and nonoptimal EGFP that was previously described (Mishima & Tomari, 2016) were amplified by PCR using y939 and y940 (optimal GFP) or y939 and y941 (nonoptimal GFP). Each fragment was cloned into pLuc at EcoRI and XhoI sites using DNA Ligation Kit (TaKaRa).
Full‐length and ΔRING Znf598 ORF fragments were amplified by RT‐PCR using primers y624 and y775 and cloned into pCS2+MT using XhoI and XbaI. AnsB was amplified by PCR from the genomic DNA of E. coli DH5α using primers y603 and cloned into pCS2+SBP‐HA (Mishima & Tomari, 2016) using EcoRI and XhoI.
Primer sequences are shown in Appendix Table S1.
PACE library
The codon‐tag sequence was designed with the following considerations. We decided on 20 for the number of test codons based on our previous study, in which we substituted 21 Leu CUG codons in the codon‐optimized EGFP to Leu CUA and observed a change in the deadenylation rate (Mishima & Tomari, 2016). We included spacer codons: (i) to avoid generating highly consecutive nucleotide and amino acid sequences, (ii) to stably maintain the sequences during plasmid construction, and (iii) to obtain unique reads in RNA‐Seq and ribosome footprint profiling. We referred codon usage of the zebrafish genome and avoided uncommon codons in the spacer codons. The order of the spacer codons was adjusted to average the local hydrophobicity.
Oligonucleotides encoding codon‐tag sequences for all codons were synthesized as 64 pairs of single‐strand DNA fragments. Complementary oligonucleotides were annealed and cloned into the pCS2+neo‐sfGFP‐suv39h1a 3´ UTR using EcoRI and EcoRV. The sequence of the cloned plasmids was confirmed by Sanger sequencing. Three codon tags for UCU, UCA, and UAG could not be cloned for unknown reasons. The cloned plasmids were mixed at an equimolar concentration and linearized by NotI. Capped and polyadenylated mRNAs were prepared using the mixed templates and purified as described above. The homogenous length of the PACE library mRNAs was confirmed by electrophoresis. The codon‐tag sequences are listed in Appendix Table S2.
RNA sequencing
Total RNA was extracted from 40–50 embryos injected with PACE library mRNAs using TRI Reagent (Molecular Research Center) and purified using the RNeasy Mini Kit (QIAGEN). After RNA integrity was confirmed using an Agilent RNA 6000 Nano Chip (Agilent Technologies, USA), total RNA samples were treated with Ribo‐Zero (Illumina) and used for RNA‐seq library preparation using an Illumina TruSeq Stranded Total RNA Library Prep Kit (Illumina, USA) according to the manufacturer’s instructions with multiplexing. The pooled libraries were sequenced on HiSeq4000 or NextSeq500 (Illumina) sequencing platforms with single‐end sequencing of 65 (HiSeq4000) or 76 bp (NextSeq500) lengths.
Ribosome footprint profiling
Ribosome footprint profiling using zebrafish embryos was performed as previously described (Han et al, 2020; Mito et al, 2020). Briefly, 50–60 zebrafish embryos injected with PACE reporter mRNAs were snap‐frozen at the sphere stage and lysed with lysis buffer containing cycloheximide at 100 μg/ml (Sigma–Aldrich). An S‐400 HR gel filtration spin column (Cytiva) was used to isolate the ribosomes. RNA fragments ranging from 26 to 34 nt were selected for monosome profiling. PCR‐amplified DNA libraries were gel‐excised and sequenced with the HiSeq4000 platform (Illumina).
Data analysis
RNA‐seq reads for PACE analysis were analyzed using Galaxy on public servers (Goecks et al, 2010). Single‐ended reads matched to the codon‐tag sequences were counted by Sailfish (Patro et al, 2014) with default parameters using the sequences listed in Appendix Table S2 as references. To calculate the stability of the codon‐tag reporter mRNAs, the read numbers at 6 hpf were divided by the read numbers at 2 hpf. The stability of each codon‐tag reporter mRNA was normalized to the value of a UGA codon tag reporter mRNA. The results from two or three biological replicates were averaged and used for the analysis.
Sequences from the ribosome profiling data were aligned against ncRNA sequences (GRCz11/danRer11 noncoding RNA gene dataset) and mitochondrial gene sequences using Bowtie2 (aligned sequences were removed). The remaining sequences were aligned to the codon‐tag reporter sequences or the zebrafish genome (GRCz11/danRer11) using TopHat. Specific A‐site assignment was determined by sequences aligned to the zebrafish genome, calibrated from footprints mapped to the beginning of the CDSes. The raw footprint read count at the test codon or spacer codon (as described in Fig 3A) was counted for further calculations.
qRT–PCR
qRT–PCR analysis of mRNAs was performed as previously described (Mishima & Tomari, 2016). Briefly, total RNA was prepared using TRI Reagent (Molecular Research Center), and cDNA was synthesized using the PrimeScript RT reagent kit with gDNA eraser (TaKaRa). A random hexamer was used for cDNA synthesis. qRT–PCR was performed using SYBR Premix Ex TaqII (Tli RNaseH Plus) and the Thermal Cycler Dice Real Time System (TaKaRa). The results were analyzed based on standard curves from a serial dilution of cDNA. The data were normalized using 18S rRNA as a reference, and the mRNA level at 6 hpf was normalized to the data at 2 hpf. The primer sequences are listed in Appendix Table S1.
Measurement of charged tRNA levels
To measure total tRNA and the charged tRNAs, we modified the OXOPAP assay (Gaston et al, 2008) and combined it with qRT–PCR analysis. Total RNA was purified using TRI reagent and dissolved in storage buffer (50 mM NaOAc pH 5.2 and 1 mM EDTA). For charged tRNA, 1 μg of the total RNA was incubated in oxidization buffer (50 mM NaIO4 and 100 mM NaOAc pH 5.2) supplemented with RNasin (Promega) for 30 min at 25°C to oxidize the nonacylated tRNA 3´ end. For total tRNA, 1 μg of the total RNA was incubated in control buffer (50 mM NaCl and 100 mM NaOAc pH 5.2) supplemented with RNasin for 30 min at 25°C. Reactions were quenched with 100 mM glucose, incubated for 5 min at 25°C, and purified with G‐25 columns (Cytiva). RNA samples were deacylated by adding 3.4 μl of 1 M Tris‐HCl pH 9.0 and incubated for 30 min at 37°C. RNA samples were purified by ethanol precipitation and dissolved in 6.5 μl of RNase‐free water. Four microliters of the RNA sample was denatured at 65°C for 5 min and polyadenylated by the Poly(A) Polymerase Tailing Kit (CELLSCRIPT) in a 10‐μl reaction in the presence of 2 U ePAP at 37°C for 60 min. A total of 2.5 μl of the polyadenylated sample was used for cDNA synthesis by the PrimeScript RT reagent kit with gDNA eraser (TaKaRa), with the y625 primer in Appendix Table S1. Reverse transcription was performed at 50°C for 30 min.
qRT–PCR was performed as described above using the tRNA‐specific forward primers shown in Appendix Table S1. Specific amplification was confirmed by dissociation curve analysis followed by gel electrophoresis and sequencing. Charged tRNA levels relative to total tRNA levels were calculated using the ΔΔCt method (ΔCt of total tRNA sample – ΔCt of charged tRNA sample). The charged tRNA levels of the AnsB overexpression experiment were normalized to those of the mock injection experiment.
PAT assay
The PAT assay was performed as previously described (Mishima & Tomari, 2016). Briefly, 150 ng of total RNA was incubated with 75 U of yeast poly(A) polymerase (PAP) (Affymetrix) in the presence of GTP/ITP mix at 37°C for 60 min. cDNA was synthesized at 44°C for 15 min using the PrimeScript RT reagent kit with gDNA eraser (TaKaRa) and a y300 PAT universal C10 primer. PAT‐PCR was performed using a 3´ UTR‐specific forward primer and a y300 PAT universal C10 primer with GoTaq Green Master Mix (Promega). The primer sequences are provided in Appendix Table S1. The PCR products were separated by 6% PAGE in 0.5×TBE. Gels were stained with GelRed (Biotium) and the signals were detected using LAS4000 (Cytiva) or Amersham Imager 680 (Cytiva). Signals were quantified using the “Plot profiles” function of ImageJ software (http://imagej.nih.gov/ij/).
Western blotting
Proteins were detected using anti‐Myc (MBL Life science My3 mouse monoclonal, 1:4,000), anti‐GFP (MBL Life science No. 598 rabbit polyclonal, 1:3,000), anti‐Phospho eIF2α (Ser51)(Cell Signaling Technology #3398 rabbit monoclonal, 1:1,000), anti‐eIF2α (Cell Signaling Technology #9722 rabbit polyclonal, 1:4,000), and anti‐α‐tubulin‐pAb HRP‐DirecT (MBL Life science PM054‐7 rabbit polyclonal 1: 10,000) antibodies. The signals were developed using Luminata Forte (Millipore) or ImmunoStar LD (FUJIFILM WaKo Chemicals) and detected using Amersham Imager 680 (Cytiva).
Luciferase assay
The luciferase assay was performed essentially as previously described (Mishima et al, 2012). Briefly, 5–10 embryos were collected 3 hpf and lysed in passive lysis buffer (Promega). 10 μl of lysate containing one embryo was used for the assay. The luciferase activities were measured using the Dual‐Glo Luciferase Assay System and a GloMax 20/20 luminometer (Promega). The intensity of Fluc activity was normalized to the intensity of Rluc activity, and the normalized Fluc activity was further normalized to the control experiment (no insert).
Author contributions
Yuichiro Mishima: Conceptualization; Formal analysis; Supervision; Funding acquisition; Investigation; Methodology; Writing – original draft; Project administration. Peixun Han: Formal analysis; Validation; Visualization. Kota Ishibashi: Validation; Investigation; Methodology. Seisuke Kimura: Resources; Formal analysis; Funding acquisition; Methodology; Writing – review and editing. Shintaro Iwasaki: Resources; Formal analysis; Funding acquisition; Validation; Investigation; Methodology; Writing – review and editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
YM designed the project. YM and KI conceived the experiments. PH and SI analyzed the ribosome profiling data. SK provided the RNA‐Seq data. YM and SI wrote the manuscript. All the authors discussed the results and approved the manuscript for submission.
Disclosure statement and competing interests
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
This work was supported by the Japan Society for the Promotion of Science (JSPS) (JP18H02370), the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (JP17H05593, JP17H05662), the Japan Agency for Medical Research and Development (AMED) (AMED PRIME, JP21gm6310017), and the Inamori Foundation to YM. SI was supported by JSPS (JP17H04998 and JP19K22406), MEXT (JP20H05784), AMED (AMED‐CREST, JP21gm1410001), RIKEN (Pioneering Project “Biology of Intracellular Environments” and Aging Project), and the Takeda Science Foundation. SK was supported by JSPS (JP21H02513) and MEXT‐Supported Program for the Strategic Research Foundation at Private Universities (S1511023). PH was an International Program Associate of RIKEN.
We thank Yukihide Tomari and Kaori Kiyokawa for their support during the initial phase of this project and Ariel Bazzini for sharing zebrafish CSC and tRNA sequencing data and engaging in discussions. We thank our laboratory members for discussions and their critical comments on the project, Kimi Wakabayashi for fish maintenance, Kaori Kaminoyama and Tomoaki Sakamoto for RNA‐Seq, and Mari Mito for technical assistance. We thank the Shirahide Laboratory at the Institute for Quantitative Biology for performing the RNA‐Seq. DNA libraries for ribosome profiling were sequenced by the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, which is supported by the National Institutes for Health (NIH) Instrumentation Grant (S10 OD018174). Computational analysis was supported by the supercomputer HOKUSAI Sailing Ship in RIKEN.
The EMBO Journal (2022) 41: e109256.
Data availability
The datasets produced in this study are available in the following databases:
RNA‐Seq data: Gene Expression Omnibus GSE173179 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173179) and GSE185915 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185915).
Ribosome footprint data: Gene Expression Omnibus GSE173604 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173604).
References
- Arthur LL, Pavlovic‐Djuranovic S, Koutmou KS, Green R, Szczesny P, Djuranovic S (2015) Translational control by lysine‐encoding A‐rich sequences. Sci Adv 1: e1500154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, del Viso F, Moreno‐Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, Giraldez AJ (2016) Codon identity regulates mRNA stability and translation efficiency during the maternal‐to‐zygotic transition. EMBO J 35: 2087–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boël G, Letso R, Neely H, Price WN, Wong K‐H, Su M, Luff JD, Valecha M, Everett JK, Acton TB et al (2016) Codon influence on protein expression in E. coli correlates with mRNA levels. Nature 529: 358–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr F, Jha S, Thommen M, Mittelstaet J, Kutz F, Schwalbe H, Rodnina MV, Komar AA (2016) Synonymous codons direct cotranslational folding toward different protein conformations. Mol Cell 61: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschauer R, Matsuo Y, Sugiyama T, Chen Y‐H, Alhusaini N, Sweet T, Ikeuchi K, Cheng J, Matsuki Y, Nobuta R et al (2020) The Ccr4‐Not complex monitors the translating ribosome for codon optimality. Science 368: eaay6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadani Y, Niwa T, Chiba S, Taguchi H, Ito K (2016) Integrated in vivo and in vitro nascent chain profiling reveals widespread translational pausing. Proc Natl Acad Sci USA 113: E829–E838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadani Y, Niwa T, Izumi T, Sugata N, Nagao A, Suzuki T, Chiba S, Ito K, Taguchi H (2017) Intrinsic ribosome destabilization underlies translation and provides an organism with a strategy of environmental sensing. Mol Cell 68: 528–539 [DOI] [PubMed] [Google Scholar]
- Charneski CA, Hurst LD (2013) Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol 11: e1001508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Grosely R, Prabhakar A, Lapointe CP, Wang J, Puglisi JD (2018) How messenger RNA and nascent chain sequences regulate translation elongation. Annu Rev Biochem 87: 421–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Panasenko OO (2017) The Ccr4‐Not complex: architecture and structural insights. Sub‐Cell Biochem 83: 349–379 [DOI] [PubMed] [Google Scholar]
- D’Orazio KN, Wu CC‐C, Sinha N, Loll‐Krippleber R, Brown GW, Green R (2019) The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay. eLife 8: 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Duc K, Song YS (2018) The impact of ribosomal interference, codon usage, and exit tunnel interactions on translation elongation rate variation. PLoS Genet 14: e1007166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova LN, Kuroha K, Tatematsu T, Inada T (2009) Nascent peptide‐dependent translation arrest leads to Not4p‐mediated protein degradation by the proteasome. J Biol Chem 284: 10343–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV (2013) EF‐P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339: 85–88 [DOI] [PubMed] [Google Scholar]
- Forrest ME, Pinkard O, Martin S, Sweet TJ, Hanson G, Coller J (2020) Codon and amino acid content are associated with mRNA stability in mammalian cells. PLoS One 15: e0228730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble CE, Brule CE, Dean KM, Fields S, Grayhack EJ (2016) Adjacent codons act in concert to modulate translation efficiency in yeast. Cell 166: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T et al (2017) The E3 ubiquitin ligase and RNA‐binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat Commun 8: 16056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston KW, Rubio MAT, Alfonzo JD (2008) OXOPAP assay: for selective amplification of aminoacylated tRNAs from total cellular fractions. Methods 44: 170–175 [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Afgan E, Ananda G, Baker D, Blankenberg D, Chakrabarty R, Coraor N, Goecks J et al (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman DH, Livingston NM, Movsik J, Wu B, Green R (2021) Live‐cell imaging reveals kinetic determinants of quality control triggered by ribosome stalling. Mol Cell 81: 1830–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Shichino Y, Schneider‐Poetsch T, Mito M, Hashimoto S, Udagawa T, Kohno K, Yoshida M, Mishima Y, Inada T et al (2020) Genome‐wide survey of ribosome collision. Cell Rep 31: 107610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson G, Alhusaini N, Morris N, Sweet T, Coller J (2018) Translation elongation and mRNA stability are coupled through the ribosomal A‐site. RNA 24: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Parker R (2016) Analysis of the association between codon optimality and mRNA stability in Schizosaccharomyces pombe . BMC Genom 17: 895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hia F, Yang SF, Shichino Y, Yoshinaga M, Murakawa Y, Vandenbon A, Fukao A, Fujiwara T, Landthaler M, Natsume T et al (2019) Codon bias confers stability to human mRNAs. EMBO Rep 20: e48220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH (2015) Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet 11: e1005732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, Beckmann R, Inada T (2019) Collided ribosomes form a unique structural interface to induce Hel2‐driven quality control pathways. EMBO J 38: e100276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T (2020) Quality controls induced by aberrant translation. Nucleic Acids Res 48: 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS (2009) Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Chiba S (2013) Arrest peptides: Cis ‐acting modulators of translation. Annu Rev Biochem 82: 171–202 [DOI] [PubMed] [Google Scholar]
- Ito‐Harashima S, Kuroha K, Tatematsu T, Inada T (2007) Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev 21: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L‐E, Wente SR, Chen W (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA 110: 13904–13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CAP (2019) Mechanisms and functions of ribosome‐associated protein quality control. Nat Rev Mol Cell Biol 20: 368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Hegde RS (2017) Initiation of quality control during poly(A) translation requires site‐specific ribosome ubiquitination. Mol Cell 65: 743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, Hegde RS (2018) ZNF598 is a quality control sensor of collided ribosomes. Mol Cell 72: 469–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Slodkowicz G, Lin Z, Freire‐Pritchett P, Peak‐Chew S‐Y, Hegde RS (2020) Ribosome collisions trigger cis‐acting feedback inhibition of translation initiation. eLife 9: 1–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee S‐R, Li L‐H, Park H‐J, Park J‐H, Lee KY, Kim M‐K, Shin BA, Choi S‐Y (2011) High cleavage efficiency of a 2A peptide derived from porcine teschovirus‐1 in human cell lines, Zebrafish and Mice. PLoS One 6: e18556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza‐Puch F, Rooijers K, Buil LCM, Zijlstra J, F. Oude Vrielink J, Lopes R, Ugalde AP, van Breugel P, Hofland I, Wesseling J et al (2016) Tumour‐specific proline vulnerability uncovered by differential ribosome codon reading. Nature 530: 490–494 [DOI] [PubMed] [Google Scholar]
- Lu J, Deutsch C (2008) Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol 384: 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE (2004) Nonsense‐mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Matheisl S, Berninghausen O, Becker T, Beckmann R (2015) Structure of a human translation termination complex. Nucleic Acids Res 43: 8615–8626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K et al (2017) Ubiquitination of stalled ribosome triggers ribosome‐associated quality control. Nat Commun 8: 159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Tesina P, Nakajima S, Mizuno M, Endo A, Buschauer R, Cheng J, Shounai O, Ikeuchi K, Saeki Y et al (2020) RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1. Nat Struct Mol Biol 27: 323–332 [DOI] [PubMed] [Google Scholar]
- Medina‐Muñoz SG, Kushawah G, Castellano LA, Diez M, DeVore ML, Salazar MJB, Bazzini AA (2021) Crosstalk between codon optimality and cis‐regulatory elements dictates mRNA stability. Genome Biol 22: 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Fukao A, Kishimoto T, Sakamoto H, Fujiwara T, Inoue K (2012) Translational inhibition by deadenylation‐independent mechanisms is central to microRNA‐mediated silencing in zebrafish. Proc Natl Acad Sci USA 109: 1104–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Tomari Y (2016) Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Mol Cell 61: 874–885 [DOI] [PubMed] [Google Scholar]
- Mito M, Mishima Y, Iwasaki S (2020) Protocol for disome profiling to survey ribosome collision in humans and zebrafish. STAR Protocols 1: 100168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno M, Ebine S, Shounai O, Nakajima S, Tomomatsu S, Ikeuchi K, Matsuo Y, Inada T (2021) The nascent polypeptide in the 60S subunit determines the Rqc2‐dependency of ribosomal quality control. Nucleic Acids Res 49: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno‐Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ (2015) CRISPRscan: designing highly efficient sgRNAs for CRISPR‐Cas9 targeting in vivo . Nat Methods 12: 982–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narula A, Ellis J, Taliaferro JM, Rissland OS (2019) Coding regions affect mRNA stability in human cells. RNA 25: 1751–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos‐Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM (2016) The integrated stress response. EMBO Rep 17: 1374–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Mount SM, Kingsford C (2014) Sailfish enables alignment‐free isoform quantification from RNA‐seq reads using lightweight algorithms. Nat Biotechnol 32: 462–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Frydman J (2013) Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol 20: 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Chartron JW, Frydman J (2014) Local slowdown of translation by nonoptimal codons promotes nascent‐chain recognition by SRP in vivo . Nat Struct Mol Biol 21: 1100–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen Y‐H, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker K, Graveley B et al (2015) Codon optimality is a major determinant of mRNA stability. Cell 160: 1111–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp RA, Snider L, Weintraub H (1994) Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev 8: 1311–1323 [DOI] [PubMed] [Google Scholar]
- Schuller AP, Wu CCC, Dever TE, Buskirk AR, Green R (2017) eIF5A functions globally in translation elongation and termination. Mol Cell 66: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein KC, Frydman J (2019) The stop‐and‐go traffic regulating protein biogenesis: how translation kinetics controls proteostasis. J Biol Chem 294: 2076–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein KC, Kriel A, Frydman J (2019) Nascent polypeptide domain topology and elongation rate direct the cotranslational hierarchy of Hsp70 and TRiC/CCT. Mol Cell 75: 1117–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ (2017) ZNF598 and RACK1 regulate mammalian ribosome‐associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol Cell 65: 751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesina P, Lessen LN, Buschauer R, Cheng J, Wu CC, Berninghausen O, Buskirk AR, Becker T, Beckmann R, Green R (2020) Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts. EMBO J 39: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y (2010) An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141: 344–354 [DOI] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K (2013) Translation elongation factor EF‐P alleviates ribosome stalling at polyproline stretches. Science 339: 82–85 [DOI] [PubMed] [Google Scholar]
- Varenne S, Buc J, Lloubes R, Lazdunski C (1984) Translation is a non‐uniform process. J Mol Biol 180: 549–576 [DOI] [PubMed] [Google Scholar]
- Viswanathan P, Ohn T, Chiang YC, Chen J, Denis CL (2004) Mouse CAF1 can function as a processive deadenylase/3′‐5′ ‐exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J Biol Chem 279: 23988–23995 [DOI] [PubMed] [Google Scholar]
- Weber R, Chung M‐Y, Keskeny C, Zinnall U, Landthaler M, Valkov E, Izaurralde E, Igreja C (2020) 4EHP and GIGYF1/2 mediate translation‐coupled messenger RNA decay. Cell Rep 33: 108262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MW, Chen YH, Stowell JAW, Alhusaini N, Sweet T, Graveley BR, Coller J, Passmore LA (2018) mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4‐not nucleases. Mol Cell 70: 1089–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, Bartel DP (2016) Improved ribosome‐footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep 14: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Arenz S, Beckmann R (2016) Translation regulation via nascent polypeptide‐mediated ribosome stalling. Curr Opin Struct Biol 37: 123–133 [DOI] [PubMed] [Google Scholar]
- Wu Q, Medina SG, Kushawah G, DeVore ML, Castellano LA, Hand JM, Wright M, Bazzini AA (2019) Translation affects mRNA stability in a codon‐dependent manner in human cells. eLife 8: 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]


