Abstract
This overview of the molecular pathology of lung cancer includes a review of the most salient molecular alterations of the genome, transcriptome, and the epigenome. The insights provided by the growing use of next-generation sequencing (NGS) in lung cancer will be discussed, and interrelated concepts such as intertumor heterogeneity, intratumor heterogeneity, tumor mutational burden, and the advent of liquid biopsy will be explored. Moreover, this work describes how the evolving field of molecular pathology refines the understanding of different histologic phenotypes of non-small-cell lung cancer (NSCLC) and the underlying biology of small-cell lung cancer. This review will provide an appreciation for how ongoing scientific findings and technologic advances in molecular pathology are crucial for development of biomarkers, therapeutic agents, clinical trials, and ultimately improved patient care.
The goal of this review is to provide a focused review of the most clinically relevant aspects of thoracic molecular pathology. Historically, genomic alterations (GAs) that were considered drivers in lung cancer were interrogated in panels using an à la carte approach with techniques such as Sanger sequencing, pyrosequencing, immunohistochemistry (IHC), and fluorescence in situ hybridization (FISH), which limited reportable GAs to select genes (Yu et al. 2009; Kim et al. 2013a; Sholl et al. 2013). Mutations in epidermal growth factor receptor (EGFR) and translocations in the anaplastic lymphoma kinase (ALK) gene and the ROS1 gene were the first three alterations recognized as clinically actionable drivers, and comprised the majority of the initial clinical genetic testing in lung cancer (Gaughan and Costa 2011).
As the use of next-generation sequencing (NGS) became more cost-effective and widely used, it revealed additional actionable GAs. Evidence has accumulated to prove that NGS is an accurate technology with performance that is concordant or better than conventional methods (Hinrichs et al. 2015; Ali et al. 2016). Advances in information about actionable GAs from comprehensive genomic profiling (CGP) has propelled precision medicine forward with the aim to provide evidence-based interpretation of GAs for targeted therapy (Coco et al. 2015; Suh et al. 2016).
The increase in the number of genes and GAs covered by NGS has expanded the variety of GAs identified as actionable in lung cancer. A shift toward NGS-based testing in advanced lung cancer has decreased the number of lung cancer cases where a driver is not identified. Increasingly, case reports describe patients with novel drivers identified by NGS that would not have been identified if they were evaluated solely by hotspot-based panels (Capelletti et al. 2014).
Historically, lung cancer was divided categorically into non-small-cell lung cancer (NSCLC) (∼85%) and small-cell lung cancer (SCLC) (∼15%) by pathologist review of hematoxylin and eosin (H&E) stained slides. NSCLC is a heterogeneous category of entities that includes lung adenocarcinoma (LUAD) as the most common histologic cancer type (∼50%), squamous cell carcinoma of the lung (SqCCL) as the second most common type (∼30%), large-cell lung carcinoma (LCLC) (∼10%), and combinations of histologic phenotypes such as adenosquamous carcinoma (ASC), and rare histologic entities, such as atypical carcinoid (AC) tumor, bronchial gland carcinoma, and sarcomatoid carcinoma, collectively accounting for the remaining types of NSCLC (∼10%) (Pikor et al. 2013; Chan and Hughes 2015; Stevic and Milenkovic 2016; Chirieac and Kobzik 2017; Li and Lu 2018; Melosky 2018; Weissferdt 2018).
In clinical practice, lung cancer patients typically undergo an initial biopsy to determine whether there are histomorphologic features on H&E that are able to diagnostically distinguish between NSCLC or SCLC. Performance of IHC typically will assist in determining the subtypes of NSCLC. IHC that favors NSCLC that is LUAD is typically positive for CK7 and TTF-1, and negative for p63, CK5/6, and p40. IHC that favors NSCLC that is SqCCL is typically positive for p63, CK5/6, and p40 and negative for TTF-1 and CK7. The art of establishing a diagnosis may be challenging as many pathologists are aware that, in practice, some tumors might not have “read the book” regarding conventional IHC staining patterns. In such cases, other special stains such as mucin can assist (e.g., mucin is typically positive for LUAD, and negative for SqCCL). ASCs are a rarely encountered tumor that is biphasic, meaning there is a component that is LUAD and a component that is SqCCL. These components by definition have at least 10% each of malignant squamous and glandular components. Neuroendocrine IHC markers (CD56, chromogranin, and synaptophysin) are typically positive for neuroendocrine carcinomas such as SCLC and LCLC. In NSCLC, the tumor proportion score (TPS) for programmed death ligand-1 (PD-L1) as determined by IHC (e.g., PD-L1 IHC 22C3 pharmDx) assists in determining eligibility for immunotherapy. While tissue is submitted for NGS, rapid testing for mutations in EGFR, KRAS, and BRAF are selectively tested via a rapid method such as pyrosequencing, while rearrangements in ALK and ROS1 can be rapid identified via FISH. When NGS results are made available, these results can corroborate the results of rapid testing, and potentially provide other GAs that may be actionable targets.
The Centers for Medicare & Medicaid Services (CMS) regulates all laboratory testing via the Clinical Laboratory Improvement Amendments of 1988 (CLIA) certification process. Commercial laboratories and leading cancer centers that develop their molecular panels for obtaining analyses to guide patient care must have laboratories that are CLIA-certified. For instance, the PD-L1 IHC 22C3 pharmDx assay was approved by the FDA for identifying NSCLC patients eligible (TPS ≥ 1%) for treatment with pembrolizumab, an assay available at NeoGenomics Laboratories, which is CLIA-certified.
Particularly in lung biopsies, there is a balance between the need to acquire adequate tissue for H&E, IHC, rapid testing, and/or NGS, which are weighed against mitigating the risk of adverse events such as iatrogenic pneumothorax. A discussion between the clinician and patient must occur to communicate the risk balanced with the diagnostic benefit gained from a biopsy to determine that the patient is clearly providing an informed consent to undergo a procedure with the goal to retrieve tissue for diagnostic purposes.
Given the sheer volume, breadth, and rapid evolution of current literature describing the molecular pathology of lung cancer to date, the intention of this review is to distill only the most salient points to complement the other reviews in this collection. One general caveat about our understanding of lung molecular pathology is that clear molecular distinctions between different subtypes of lung cancer is complicated by some studies that molecularly characterize NSCLC without distinguishing between histologic types.
Herein, we describe the genomic, transcriptomic, epigenomic, and proteomic differences between different histologic types of lung cancer with relevance to research and therapeutic strategies.
ONCOGENOTYPES
Oncogenotypes are molecular alterations that include genomic mutations, alterations that cause dysregulation in mRNA expression, dysregulation in expression of miRs, and epigenomic changes.
Mutations
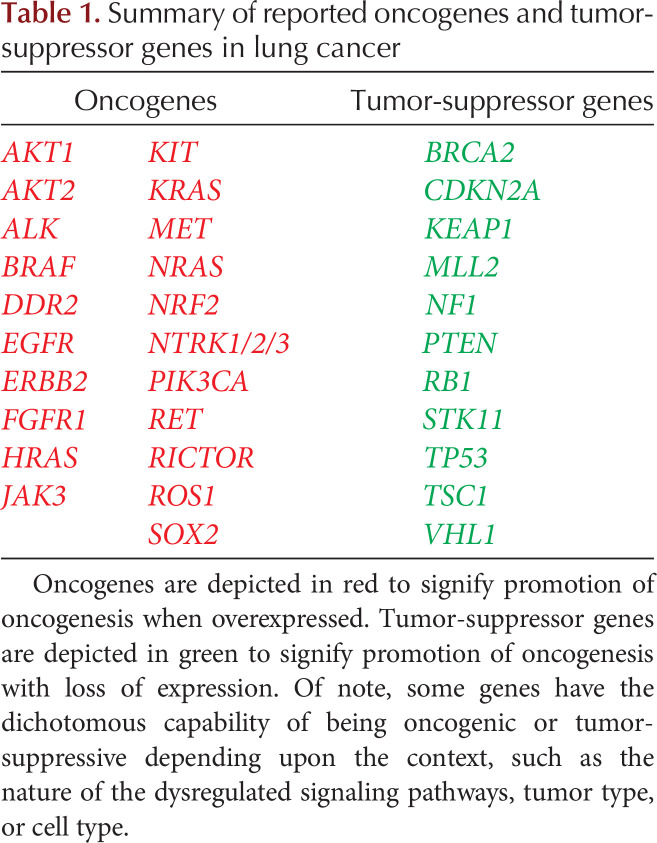
As NGS-based DNA sequencing has become increasingly available, the knowledge about different types of mutations that are potentially actionable beyond the genetic biomarkers with associated FDA-approved targeted therapies has expanded (Chan and Hughes 2015; Tafe et al. 2016). An oncogene is a gene in which activating mutations confer gain-of-function to a gene, which ultimately promotes oncogenesis. Conversely, a tumor-suppressor gene (TSG) is a gene in which inactivating mutations confer loss-of-function to a gene, which ultimately promotes oncogenesis. Significant oncogenes and TSGs in lung cancer are summarized in Table 1.
Table 1.
Summary of reported oncogenes and tumor-suppressor genes in lung cancer
Molecular characteristics of lung cancer are critical for patient care and are included in the National Comprehensive Cancer Network (NCCN) guidelines (National Comprehensive Cancer Network 2021). They are also fundamental for epidemiological studies, such as genome-wide association studies (GWAS) (Chang et al. 2015; Schabath et al. 2016; Inamura 2017; Bossé and Amos 2018; de Groot et al. 2018; Li et al. 2019b; Tam et al. 2019; Tang et al. 2019). The advancement of the use of clinically relevant GAs (CRGAs) for patient care is the consequence of evidence-based studies and clinical trials.
Efforts to characterize the diversity of GAs recurrently observed in lung cancer are motivated by the clinical need to determine the likelihood of response to specific therapies based on specific lung cancer genotypes (Shim et al. 2017). An advantage of NGS testing is that it can identify GAs, such as NTRK fusions that are rare in lung cancer, but have proven benefits in other tumor types (Tafe et al. 2016).
The interpretation of NGS-based test results to determine the most appropriate course of therapy often requires a review of the most recent literature and guidelines. Many current clinical trials for patients with lung cancer require knowledge about mutated or wild-type genes for eligibility.
As an overall class, tyrosine kinase inhibitors (TKIs) have been effective as targeted therapies for a variety of genotypes of NSCLC. Different TKIs have activity specifically targeted for mutations in EGFR, ERBB2 (HER2), BRAF, and rearrangements in ALK, RET, and ROS1. The most common genes with clinically relevant GAs are summarized below.
EGFR Family
In LUAD, the most clinically relevant and prevalent GAs are in exons 19 and 21 of the EGFR gene. The most prevalent and well characterized of these are EGFR exon 19 deletions and the L858R mutation in exon 21 (Sholl et al. 2009; Yu et al. 2009). These GAs are predictive of response to targeted therapy with EGFR-targeted TKIs, such as osimertinib, gefitinib, erlotinib, and afatinib, to abrogate the constitutive TKI-activating effect conferred by these EGFR mutations. Osimertinib was historically provided once patients developed EGFR T790M as a resistance mechanism to the other EGFR inhibitors; however, more recently, the phase III FLAURA study established osimertinib as first-line standard of care (SoC) for EGFR-mutated NSCLC in patients with locally advanced or metastatic NSCLC (Nan et al. 2017; Soria et al. 2018). In the FLAURA trial, frontline use of osimertinib was associated with better outcome than other EGFR TKIs (erlotinib or gefitinib). Frontline use of osimertinib results in a different portfolio of resistance mutations, such as EGFR C797S and MET amplification, which will be discussed in more depth later in the context of therapy-induced tumor heterogeneity (Sequist et al. 2011).
Other drivers in the EGFR family include ERBB2 (HER2) alterations, predominantly exon 20 insertions, but also point mutations, and amplifications, and VEGFR amplifications. ERBB2 (HER2) up-regulation is typically associated with low PD-L1 expression; whereas VEGFR up-regulation correlates positively with PD-L1 expression, although this has been mainly characterized in other cancer types (Xue et al. 2017). Monocertinib and amivantamab are FDA approved for NSCLC with ERBB2 exon 20 insertions. Monoclonal antibodies that target VEGFR include bevacizumab and ramucirumab (Remon et al. 2019).
ALK Rearrangements
In 3% to 7% of NSCLC, carcinogenesis is driven by a translocation that causes the fusion of the echinoderm microtubule-associated protein-like 4 (EML4) gene promoter region, with the ALK gene tyrosine kinase domain known as the EML4-ALK fusion oncogene (Shroff et al. 2018). EML4-ALK fusions are more prevalent in younger (≤35 yr) patients with LUAD (16.9%) and in patients with a never-smoker history (Chen et al. 2019). Current FDA-approved TKIs that target ALK fusions include crizotinib, ceritinib, alectinib, brigatinib, and lorlatinib; while entrectinib has been under investigation (Awad and Shaw 2014). Recent NCCN guidelines for NSCLC (Version 6.2021) clarified brigatinib (category 1) as a preferred first-line therapy option for patients with ALK rearrangement-positive metastatic NSCLC (National Comprehensive Cancer Network 2021). Moreover, brigatinib and lorlatinib are currently FDA approved for ALK fusions.
KRAS Alterations
Mutations in the small GTPase Kirsten Rat Sarcoma (KRAS) are identified in ∼25% of NSCLCs (Sholl et al. 2015). The overall importance of developing KRAS inhibitors is highlighted in a pan-cancer analysis reporting general RAS mutations (KRAS, NRAS, and HRAS) as a driver in ∼25% of all cancer types. KRAS gene mutations are the most common type of RAS gene mutation, with a prevalence of 85% out of all RAS-mutated cancer types (Hobbs et al. 2016; Li et al. 2018b). Although development of successful KRAS-targeted therapies has been challenging, recent studies demonstrate evidence of clinical activity with the KRAS G12C inhibitor AMG 510 targeted against LUADs harboring the KRAS G12C mutation (Cox et al. 2014; Chan and Hughes 2015; Canon et al. 2019; Fakih et al. 2019; Moore et al. 2020). In May 2021, the FDA approved sotorasib (AMG 510) (Amgen), which is a first-in-class KRAS-G12C inhibitor for NSCLC, with an ongoing phase 3 study (NCT04303780) for the comparison of AMG 510 and docetaxel KRAS-G12C-mutated NSCLC subjects. Further exploration includes combinations that pair sotorasib with PD1/PDL1 blockers, MEK inhibitors, and SHP2 inhibitors (Mullard 2021).
KRAS mutation testing has clinical utility in other ways. For example, based on the BATTLE-2 trial, the presence of a KRAS mutation predicts lack of response to EGFR inhibition. The BATTLE-2 trial stratified NSCLCs based on KRAS mutational status to determine its effect on clinical responsiveness to erlotinib and reported that patients with KRAS-mutated tumors experienced a statistically longer progression-free survival (PFS) if not treated with the EGFR TKI erlotinib, although a significant impact on overall survival (OS) was not observed (Papadimitrakopoulou et al. 2016). In addition, as KRAS mutations tend to be a mutually exclusive driver in LUAD prior to first-line therapy and can often be identified quickly by a rapid tissue test or liquid biopsy, it can lead to more rapid initiation of therapy or clinical trial enrollment.
ROS1 Rearrangements
The c-ROS oncogene 1 (ROS1) is a receptor tyrosine kinase gene that acts as a driver in 1% to 2% of NSCLC, typically in never-smokers or light smokers. FDA-approved TKIs for ROS1-rearranged NSCLC include crizotinib and entrectinib (Remon et al. 2019). ROS1-rearranged NSCLC is associated with PD-L1 expression, whereas EGFR-mutated NSCLC is negatively associated with PD-L1 expression (Lee et al. 2019).
MET Alterations
The mesenchymal-to-epithelial transition (MET) factor transmembrane tyrosine kinase receptor gene is a proto-oncogene that binds hepatocyte growth factor (HGF). Activating MET GAs predominantly consist of MET gene amplification or MET gene exon 14 (METex14) splice-site mutations. NSCLCs with METex14 mutations often have concomitant MDM2 and cyclin-dependent kinase 4 (CDK4) amplifications (Frampton et al. 2015; Paik et al. 2015b; Awad et al. 2016; Schrock et al. 2016).
MET GAs are identified as primary changes and as resistance mechanisms to targeted therapy, such as resistance to EGFR inhibitor therapy in EGFR-mutated LUADs and NSCLCs harboring MET GAs often respond to MET inhibition (Paik et al. 2015a). For example, MET inhibition of NSCLCs with concurrent EGFR and MET abnormalities may respond to dual blockade of EGFR and MET (Pérez-Ramírez et al. 2015a; Shi et al. 2016b). Activation of ERBB3 signaling via MET amplification may mediate resistance to EGFR TKIs (Engelman et al. 2007). Cigarette smoke is a factor that enhances c-MET signaling in lung cancer and consequently may limit responsiveness to EGFR TKI therapy (Tu et al. 2018).
BRAF Alterations
A BRAF inhibitor, vemurafenib, can be used if dabrafenib and trametinib combination therapy is unable to be tolerated in lung cancers harboring BRAF V600E mutations. BRAF mutations and BRAF fusions, such as between AGK and BRAF, may arise as resistance mechanisms to therapy with EGFR or ALK inhibitors (Vojnic et al. 2019; Boyle et al. 2020). The potential for BRAF-targeted therapy after a BRAF GA is identified as the resistance mechanism is under investigation (Vojnic et al. 2019). Trametinib, a MEK inhibitor that has been studied in combination with the BRAF inhibitor, dabrafenib, is a combination therapy that has shown activity against NSCLC, and is FDA approved for BRAF-mutated NSCLC (Solassol et al. 2019).
RET Rearrangements
RET rearrangements have been reported in 1.2% of NSCLCs, predominantly LUADs. RET rearrangements have been found to be mutually exclusive with both EGFR mutations and ALK rearrangements. RET rearrangements have been associated with younger age, nonsmoking history, papillary morphology, and mucin production (Tsuta et al. 2014). Success with treating RET-rearranged lung cancer with targeted therapies may depend on use of highly selective RET inhibitors (Bronte et al. 2019). Selpercatinib and pralsetinib are FDA approved for NSCLCs that harbor RET fusions.
NTRK Fusions
Neurotrophic receptor tyrosine kinase (NTRK) fusions in the NTRK1, NTRK2, and NTRK3 genes are identified in <1% of NSCLCs (Farago et al. 2018). Identification of NTRK fusions in lung cancer has been limited by availability of assays that accurately and comprehensively detect these fusions. However, given the potent clinical activity and FDA approval of TRK inhibitors in NTRK-mutated solid tumors, identification of such fusions with multiplexed NGS-based fusion assays is encouraged (Amatu et al. 2016; Cocco et al. 2018; Garinet et al. 2018; Kummar and Lassen 2018; Yan and Zhang 2018; Huang and Feng 2019). Recent NCCN guidelines for NSCLC (Version 6.2021) clarified that NTRK1/2/3 gene fusions are established predictive molecular biomarkers (National Comprehensive Cancer Network 2021). Larotrectinib and entrectinib are FDA approved for NTRK fusion-positive cancers.
PI3K/AKT1/PTEN /mTOR Pathway
Up-regulation of the PI3K/AKT1/PTEN pathway is important in that it can be a mechanism of resistance to TKIs. In the event of this up-regulation, PI3K, AKT, mTOR, and NF-κB are potential targets for effective inhibition (Pérez-Ramírez et al. 2015b).
Beyond the Genome
Beyond the characterization of the underlying GAs enriched in different lung cancer subtypes, it is also important to understand the dysregulation in cell signaling attributed to these GAs (Zhang et al. 2019d). Dysregulation in cell signaling with oncogenic consequences may also result from transcriptomic or epigenomic alterations (Duruisseaux and Esteller 2018; Kim and Kim 2018; Teixeira et al. 2019).
mRNA Expression
RNA sequencing, also referred to as “RNA-seq” or gene expression profiling (GEP), is a method for evaluating the sequences and/or expression levels of the mRNA transcriptomes of biological specimens. In some scenarios, RNA sequencing provides more accurate identification of GAs that are difficult to comprehensively detect by DNA sequencing, such as splice variants, gene fusions, or dysregulated signaling pathways that may not have a clearly defined underlying driver (Dhanasekaran et al. 2014; Fernandez-Cuesta et al. 2015; Li et al. 2017b). Moreover, comparison of GEP results in a significant number of different histologic tumor types, which provides insight into which genes are typically dysregulated in a given tumor type (Lim et al. 2018b; Cai et al. 2019a).
Single-cell RNA-sequencing (scRNA-seq) is a recent approach that has been shown to provide insight into the lung tumor microenvironment (TME). With scRNA-seq, it is possible to characterize different stromal cell and immune cell subtypes (Lambrechts et al. 2018; Ma et al. 2019; Tian et al. 2019). scRNA-seq can also provide insight into the heterogeneity of tumor subpopulations, or identify the presence or absence of therapy-responsive or therapy-resistant subpopulations (Kim et al. 2015; Levitin et al. 2018; Damiani et al. 2019).
Comparison of the RNA sequence of lung tumor with paired nontumor lung tissue facilitates studies of transcriptomic dysregulation, such as changes in PD-L1, PD-L2, and VEGFR expression in NSCLC tumors. Furthermore, recurrent patterns of dysregulation specifically observed in tumor tissue have prognostic implications in NSCLC (Bang et al. 2017; Uhlen et al. 2017).
GEP is also a useful adjunct to determine the anatomic site of origin in cancers of unknown primary (CUP) origin, which traditionally relies heavily on IHC to deduce the most probable anatomic site of origin (Lin and Liu 2014; Kandalaft and Gown 2016; Selves et al. 2018). Oftentimes, selection of treatment options is reliant on confirming the anatomic site of origin (Greco and Pavlidis 2009; Tomuleasa et al. 2017). GEP can reveal distinct differential patterns of RNA expression in different cancer types and subtypes oftentimes with use of less tumor tissue than the extensive IHC panels used in challenging CUP cases (Greco et al. 2010, 2012; Hainsworth et al. 2013; Handorf et al. 2013; Varadhachary 2013; Bentley et al. 2014; Hainsworth and Greco 2014; Kerr et al. 2014; Wei et al. 2014; Greco et al. 2015; Hoadley et al. 2018; Lyu and Haque 2018). Although traditionally IHC has been the main method for diagnosis and biomarker evaluation in CUP, recent data demonstrate a commensurate level of performance compared to evaluation by DNA-seq and/or RNA-seq (Pavlidis et al. 2015; Conway et al. 2019).
MicroRNAs (miRs)
Knowledge is growing about the importance and diversity of the regulatory roles that noncoding RNAs (ncRNAs) play in normal cellular function, as well as the consequences of dysregulation of ncRNAs for the development of lung cancer (Guan et al. 2012; Siegler et al. 2012; Ludwig et al. 2016; Wang et al. 2016; Dhawan et al. 2018; Li et al. 2018a; Liu et al. 2018a,b; Sonea et al. 2018; Deng et al. 2019; Li and Fu 2019; Ling and Yuen 2019; Tutar and Tutar 2019). miRs are a significant portion of ncRNAs. miRs range from ∼21 to 25 nucleotides in length, and inhibit target mRNAs through sequestration and subsequent degradation via the RNA-induced silencing complex mRNA, a process effectively referred to as RNA interference (Chipman and Pasquinelli 2019). miRs can promote or suppress oncogenesis and metastasis in the context of transcriptomic dysregulation (Griffiths-Jones et al. 2006; Chan and Wang 2015; Coebergh van den Braak et al. 2018; Kim et al. 2018a; Lamichhane et al. 2018; Wu et al. 2019a).
Some miRs have oncogenic behavior, such as oncomiRs, which inhibit TSGs when up-regulated. Conversely, other miRs are oncogenic when down-regulated, such as tumor-suppressive miRs (TS-miRs or anti-oncomiRs) (Liu et al. 2019). A significant number of miRs are capable of functioning as either tumor-suppressive or oncomiRs depending on the context of the dysregulated tumor type (Svoronos et al. 2016). Given the nonstringent range of an average of ∼100–200 targets that typically bind to a given miR, it is not uncommon for a miR that predominantly functions as an oncomiR in most contexts, to occasionally serve as a TS-miR in rare instances. Conversely, a miR that predominantly functions as a TS-miR in most contexts could occasionally function as an oncomiR (Svoronos et al. 2016). However, some miRs are known to consistently function as oncomiRs, such as miR-21, and others, such as miR-126 and miR-153, consistently function as TS-miRs (Hamada et al. 2012; Chen et al. 2015b; Bica-Pop et al. 2018; Huang et al. 2018b; Liu et al. 2019).
miR expression profiling is an approach that uses differentially expressed miRs to offer additional diagnostic and/or prognostic information, particular in cases where traditional pathologic data is limited or noncontributory. For instance, miR expression profiling with a panel of 92 differentially expressed miRs can help identify the site of origin in cases of CUP, which is valuable in that many treatment decisions and clinical trials are based on tumor type/site of origin (Rosenfeld et al. 2008; Barker et al. 2009; Rosenwald et al. 2010; Varadhachary et al. 2011; Meiri et al. 2012; Pentheroudakis et al. 2013). Moreover, miR-17, miR-190b, and miR-375 can accurately differentiate between SCLC and NSCLC. Other differentially expressed miRs have been specifically attributed to epithelial-to-mesenchymal transition (EMT) and metastatic potential (Wu et al. 2019b). Progress has been made in characterizing differentially expressed miRs in lung cancer to serve as the basis for development of miR profiles with potential for diagnostic, prognostic, or predictive clinical utility (Hu et al. 2010; Guan et al. 2012; Gallach et al. 2017; Yerukala Sathipati and Ho 2017).
miRs also are being considered in the development of therapeutic strategies for lung cancer. In general, a therapeutic approach to mitigate miR dysregulation either targets an up-regulated oncomiR by using antisense oligonucleotides; or alternatively, the down-regulation of a TS-miR may be mitigated by administering a synthetic miR that mimics the deficient TS-miR (Lu et al. 2018; Weidle et al. 2019). Notable miRs that are consistently dysregulated in lung cancer include oncomiRs miR-192 and miR-662 and TS-miR miR-374a (Võsa et al. 2011; Kuo et al. 2013; Chen et al. 2015a; Shu et al. 2016; Filipska et al. 2018). Significant miRs in lung cancer are summarized in Table 2.
Table 2.
Summary of microRNAs (miRs) in lung cancer
| miRs | Lung adenocarcinoma | Squamous cell cancer of the lung | Non-small-cell lung cancer | Atypical carcinoid | Neuroendocrine carcinoma |
|---|---|---|---|---|---|
| OncomiRs | miR-155 miR-16 miR-19a miR-20a miR-26a miR- 31 miR-93 miR-99a miR-100 miR-106b miR-135b miR-200b miR-200c (Molina-Pinelo et al. 2014) |
miR-192 (Filipska et al. 2018) miR-662 (Filipska et al. 2018) miR-21 PTEN JAG1 and WNT1 (Gao et al. 2011) miR-205 |
miR-138 Sox4 (Bai et al. 2019) miR-21 PTEN JAG1 and WNT1 (Zhang et al. 2010; Bica-Pop et al. 2018) miR-96 GPC3 (Fei et al. 2018) miR-224 TNFAIP1 and SMAD4 (Cui et al. 2015) |
miR-100 mTOR miR-34a SIRT1 and TP53 (Demes et al. 2016) |
miR-21 JAG1 and WNT1 (Demes et al. 2016) |
| TS-miRs | miR-374a TGFA (Shu et al. 2016) miR33a-5p (Kuo et al. 2013) |
miR-374a TGFA (Võsa et al. 2011) miR-345-5p (Chen et al. 2015a) miR-143 EGFR (Zhang et al. 2016) miR-134 EGFR (Qin et al. 2016) miR-182 Cortactin (Li et al. 2018d) miR-4317 FGF9 and CCND2 (He et al. 2018) miR-181a-5p HMGB2 (Li et al. 2018c) miR-183 MTA1 (Yang et al. 2018) miR-200c (Ceppi et al. 2010) miR-153 AKT (Chen et al. 2015b; Yuan et al. 2015) |
MicroRNAs (miRs) with select representative complementary targets in lung cancer. MiRs are short RNA sequences that can promote or suppress oncogenesis, and metastasis in the context of transcriptomic dysregulation. OncomiRs target tumor-suppressor genes, whereas tumor-suppressor miRs (TS-miRs) target oncogenes. Although this table represents miRs and examples of select relevant targets, miRs often have multiple targets.
Epigenomic Changes
Epigenomic or epigenetic changes are generally defined as changes in gene expression that are not the result of an underlying alteration in the genomic sequence, but rather an external modification. The prefix “epi” signifies that the changes observed are not in the genome proper; rather they are changes made “over” the genome.
Modifications that result from epigenomic changes include regulation by transcriptional deactivation mediated by miRs, methylation “silencing” of DNA by DNA methyltransferases (DNMTs), or transcriptional deactivation by chromatin condensation via histone deacetylation by histone deacetylases (HDACs). Conversely, transcriptional activation is regulated by chromatin loosening via histone acetylation by histone acetyltransferases (HATs) (Langevin et al. 2015).
Therapeutic strategies to alter epigenomic features of DNA are under investigation (Jakopovic et al. 2013; Duruisseaux and Esteller 2018). For instance, the role of HDAC inhibitors (HDACis) and DNMT inhibitors (DNMTis) to effectively restore the expression of epigenetically silenced TSGs is actively under investigation in lung cancer in preclinical models and clinical trials (Schiffmann et al. 2016; Zheng et al. 2016; Chen et al. 2018b; Damaskos et al. 2018; Jia et al. 2018; Yamada et al. 2018). HDACis in combination with immune checkpoint inhibition (ICI) and other targeted therapies have demonstrated clinical benefit (Beg and Gray 2016; Zheng et al. 2016; Terranova-Barberio et al. 2017; Gandhi et al. 2018; Suraweera et al. 2018; Banik et al. 2019). HDACis cause therapeutic benefit in lung cancer due to transcriptional activation in cancer cells and components of the TME, such as T cells (Zheng et al. 2016; Gandhi et al. 2018).
Single-cell chromatin immunoprecipitation followed by sequencing (scChIP-seq) is a technique that is useful in identifying areas of epigenomic dysregulation (Grosselin et al. 2019). Studies are underway to analyze sputum with droplet digital PCR (ddPCR) to develop a methylation-specific ddPCR profile for identification of early-stage lung cancer (Leng et al. 2017b; Su et al. 2018).
Studies are increasingly offering an integrated view of the genomic, transcriptomic, epigenomic, and proteomic alterations that occur in lung cancer (Lazar et al. 2013; Chooback et al. 2017; Stewart et al. 2019; Teixeira et al. 2019). Comprehensive integration of multi-omics data has revealed important findings, such as distinct mutation profiles and GEP findings observed in heavy smokers compared with nonsmokers (Li et al. 2015; Cardona et al. 2019). Multi-omics data also enables an evidence-based approach to identify site of origin in tumors with unknown site of origin by clinical history and histopathology (Hoadley et al. 2018).
Another noteworthy study used whole-genome sequencing (WGS) to reveal recurrent noncoding alterations in LUADs that did not have alterations in the RTK/RAS/RAF pathway genes such as EGFR, BRAF, ALK, RET, ROS1, and KRAS. The LUADs in this study had a high frequency of TP53 mutations and high mutation burden, as well as focal deletions targeting the promoter or transcription start site of STK11 and KEAP1 (n = 3), along with promoter mutations associated with the increased expression of ILF2 (Carrot-Zhang et al. 2021).
A large-scale study from the Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium of the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA) performed WGS with integrative analyses of 2658 whole-cancer genomes and their matching normal tissues across 38 tumor types (Campbell et al. 2020).
INTERTUMOR HETEROGENEITY AND DEVELOPMENT OF ACQUIRED VULNERABILITIES
Recurrently observed types of dysregulation by different tumor types serve as an underlying biologic explanation for different clinical behaviors of specific tumor types. Pathways that are uniquely up-regulated or down-regulated are potentially clinically relevant when considering clinical response to a given therapy. For instance, SqCCL and SCLC cell lines with up-regulated squalene epoxidase (SQLE) have been shown to be particularly responsive to squalene epoxidase inhibition (SQLEi) (Cirmena et al. 2018; Mahoney et al. 2019).
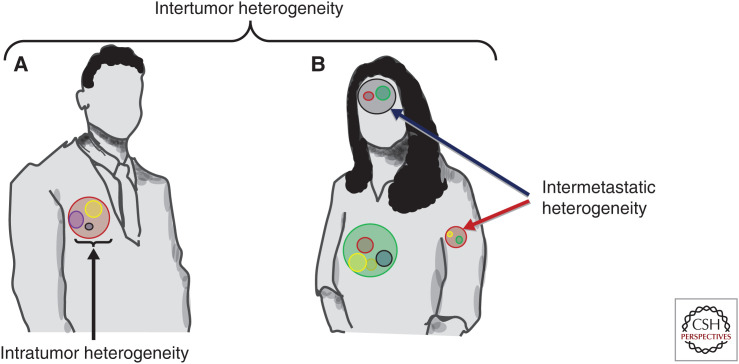
Intertumor heterogeneity is defined as differences in tumors with the same histologic type in separate patients. Intertumor heterogeneity accounts for the observation that the same tumor type may behave differently in different patients (Fig. 1 highlights different types of heterogeneity). Differences in behavior may include distinguishing metrics such as traditional pathologic features when comparing morphologic appearance; potential for invasion and/or metastases; TME, which includes the immune TME (ITME); proliferative index; and growth patterns (Sun et al. 2014; Donnem et al. 2015; Busch et al. 2016; Soo et al. 2018; DeNardo and Ruffell 2019; Shaul and Fridlender 2019; Togashi et al. 2019). Determination of critical differences in intertumor heterogeneity is critical to understanding why different patients with the same histologic tumor type have different responses when all given the same therapy. These critical differences in intertumor heterogeneity motivate the need to further explore the variety of molecular subtypes that comprise tumor types (The Cancer Genome Atlas Research Network 2012a; Müller et al. 2016).
Figure 1.
Intertumor heterogeneity, intratumor heterogeneity (ITH) and intermetastatic heterogeneity. Patient A and Patient B both share the diagnosis of lung adenocarcinoma (LUAD). Intertumor heterogeneity is defined as the difference between the molecular alterations in the primary LUAD from Patient A and Patient B. ITH is defined as the molecular alterations between subpopulations within the same tumor as depicted in Patient A as represented by the yellow, purple, and gray spheres within the tumor. Intermetastatic heterogeneity is defined as the difference in the molecular alterations that comprise the subpopulations when comparing metastatic sites as depicted in Patient B with metastases in her brain (dark blue arrow) and in her left humerus (red arrow).
Other distinguishing features between tumors may be differences in mutational status, gene expression, microsatellite instability (MSI) status, extent of chromosomal instability (CIN), viral status, and/or epigenomic differences, such as methylation status (Perez-Moreno et al. 2012; Clinical Lung Cancer Genome Project (CLCGP); Network Genomic Medicine (NGM) 2013; Pikor et al. 2013; Stinchcombe 2013; Hashida and Daibata 2014; Fang et al. 2015; Robinson et al. 2016; Guo et al. 2017; Inamura 2017; Zhang et al. 2017; Kim et al. 2018b; Testa et al. 2018; Cai et al. 2019b; Hu et al. 2019a; Rudin et al. 2019b; Cirenajwis et al. 2020). Protein expression studies via IHC have been performed to provide a more comprehensive context of the proteomic outcome of GAs (Lee et al. 2019). Protein expression that is up-regulated and down-regulated in lung cancer has also been characterized by mass spectrometry (MS) (Carvalho et al. 2017; Cheung and Juan 2017).
Overall, studies of intertumor heterogeneity focus on key biomarkers that offer insight for clinically meaningful outcomes, such as response to chemotherapy or ICI responsiveness. Moreover, there has been an effort to provide clarity about the extent to which lung cancer can be meaningfully further subdivided into molecular subtypes. For instance, molecular subtypes of LUAD have been proposed based upon established mutations, such as in EGFR and TP53, in addition to differentially expressed genes, such as these five differentially expressed genes with prognostic significance (RTKN2, ADAM6, SPINK1, COL3A1, and COL1A2) (West et al. 2012; Chen et al. 2017; Hu et al. 2019a). The actionable GAs specifically enriched in a given subtype may collectively confer responsiveness to a targeted agent or specific therapeutic approach (Kim et al. 2013b; Minna et al. 2014; Derks et al. 2018; Huang et al. 2018a; Barazas et al. 2019; Rudin et al. 2019a).
Subtypes that have GAs that inactivate TSGs are unable to be approached via direct targeting the inactivated TSG. However, inactivation of a TSG may confer a vulnerability that is exploitable by using an approach using a targeted agent that is especially responsive for a TSG-inactivated subtype, a concept called synthetic lethality. For instance, cases of NSCLC with SMARCA4 loss are specifically sensitive to CDK4/6 inhibitors. Synthetic lethality is also an approach that may be considered for inhibiting oncogenes that are difficult to inhibit with traditional therapeutic strategies, such as KRAS. Consequently, agents that are able to exploit the synthetic lethality imparted by KRAS-mutated NSCLC are currently under investigation (Shimomura et al. 2019).
Non-Small-Cell Lung Cancer
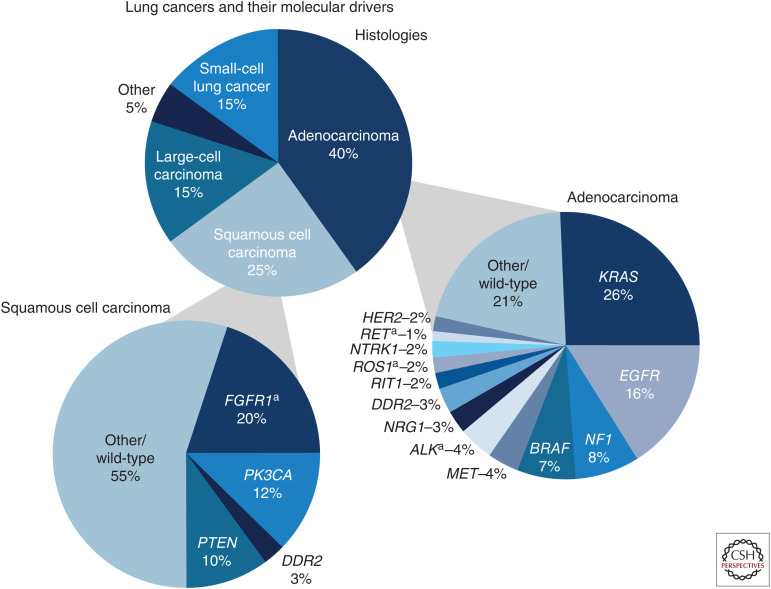
The major histologic types of NSCLC as described in the 2015 WHO classification are summarized below (Kim et al. 2013b; Travis et al. 2015; Inamura 2017). Figure 2 highlights the drivers in lung adenocarcinoma and squamous cell carcinoma of the lung.
Figure 2.
Comparison of drivers in lung adenocarcinoma and squamous cell carcinoma of the lung (SqCCL). The top pie chart shows the categorization of lung cancer into subtypes. The bottom right pie chart shows the most common drivers reported in lung adenocarcinoma. The bottom left pie chart shows the most common drivers reported in squamous cell carcinoma of the lung. (a) Includes both gene amplications and mutations. For additional information, see Rosell et al. (2016). (Figure is freely available from the American Cancer Society website.)
Lung Adenocarcinoma
LUAD is the most common histologic cancer type of NSCLC, and is typically comprised of a combination of several major morphologic subtypes, which include solid, papillary, micropapillary and lepidic growth patterns (Solis et al. 2012). Molecular characterization of LUADs focuses on actionable drivers that predict potential response to targeted therapy, such as LUADs with EGFR, BRAF, and ERBB2 mutations, and ALK, ROS1, RET, and NTRK fusions, and METex14/amplification; while mutations that have proven to be more challenging to target, such as those in KRAS, TP53, and PIK3CA, are also characterized in a significant subset of LUAD studies (Jordan et al. 2017). In untreated LUAD, KRAS mutations, EGFR mutations, and ALK rearrangements tend to be mutually exclusive (Dong et al. 2016).
A TCGA comprehensive study using whole-exome sequencing (WES) of 230 LUADs reported common mutations in KRAS (33%), EGFR (14%), BRAF (10%), PIK3CA (7%), MET (7%), and RIT1 (2%). Mutated TSGs included TP53 (46%), STK11 (17%), KEAP1 (17%), NF1 (11%), RB1 (4%), and CDKN2A (4%). Mutations in chromatin-modifying genes included SETD2 (9%), ARID1A (7%), and SMARCA4 (6%) and mutations in the RNA splicing genes included RBM10 (8%) and U2AF1 (3%). Recurrent mutations in MGA (8%) were primarily loss-of-function (frameshift and nonsense) and were mutually exclusive with focal MYC amplification (The Cancer Genome Atlas Research Network 2014). Other studies have reported AXL and GAS6 coexpression to have potential prognostic implications in LUAD (Seike et al. 2017).
The major histologic patterns of LUAD subtypes with characterized GAs are summarized below.
Mucinous Lung Adenocarcinoma
A study of mucinous LUADs reported a higher prevalence of KRAS mutations (62%) in mucinous LUADs than in general LUADs (46%), reducing options for targeted therapy (Dong et al. 2016). Nonmucinous LUADs had a lower prevalence of KRAS mutations (13%) and a higher prevalence of EGFR mutations. NRG1 fusions that activate the ERBB2/ERBB3 signaling pathway were also enriched in mucinous LUADs (Garinet et al. 2018).
Mucinous and Nonmucinous Micropapillary Lung Adenocarcinoma Subtypes
Mucinous micropapillary LUADs have a higher prevalence of ERBB2 mutations, ALK rearrangements, and a lower prevalence of EGFR mutations when compared to LUADs with a nonmucinous micropapillary component (Choi et al. 2013; Kamata et al. 2016). LUADs with a nonmucinous micropapillary component are enriched with EGFR, KRAS, and BRAF mutations (Ninomiya et al. 2009; Travis et al. 2011; Cai et al. 2016).
Papillary, Lepidic, Acinar, and Hobnail Lung Adenocarcinoma Subtypes
LUADs with papillary, lepidic, acinar, or hobnail morphological components are enriched with EGFR mutations (Shim et al. 2011; Travis et al. 2011; Cai et al. 2016; Dong et al. 2016).
Solid Lung Adenocarcinoma Subtype
KRAS mutations are frequently found in tumors with a solid growth pattern (Rekhtman et al. 2013; Dong et al. 2016). ALK rearrangements are enriched in LUADs with solid or cribriform patterns (Dong et al. 2016).
Signet Ring Cell Lung Adenocarcinoma Subtype
ALK rearrangements are enriched in LUADs with a signet ring cell component (Dong et al. 2016).
Squamous Cell Carcinoma of the Lung
SqCCL is the second most common histologic cancer type (∼30%) of NSCLC and enriched with mutations in FGFR1, NRF2, AKT1, and DDR2 with comparison to LUAD (Pikor et al. 2013).
A genomic study by TCGA of 178 SqCCLs by CGP determined that the most common mutations were in the TP53 (81%) MLL2 (20%), PIK3CA (16%), and PTEN (15%) genes, and other phosphoinositide 3-kinase (PI3K) pathway genes. Inactivating mutations in the CDKN2A gene that encode the p16INK4A and p14ARF proteins were identified in 72% of SqCCL. Squamous differentiation gene mutations were reported in 44% of SqCCL cases, with 8% identified in the NOTCH1 gene. Other genes with prevalent mutations were KEAP1 (12%), NFE2L2 (19%), and HLA-A (3%) (The Cancer Genome Atlas Research Network 2012b).
TP53 mutations and PIK3CA, FGFR1, and SOX2 amplifications are more common in SqCCLs compared with LUADs; whereas EGFR mutations are much less common in SqCCLs (8% in nonsmokers with SqCCL and 2.1% of smokers with SqCCL compared with LUADs (Heist et al. 2012; Perez-Moreno et al. 2012; Rooney et al. 2013). Rare targetable GAs in SqCCL, such as fusions in RET and ALK, have been reported, but it is difficult to know whether or not the SqCCL cases with these rare changes have an undetected adenocarcinoma component to them since tumors can be morphologically heterogeneous and the often small samples used for diagnosis may not fully represent the morphological subtypes within the tumor (Kenmotsu et al. 2014; Hirsch et al. 2018; Zhang et al. 2019c). The MS4A1 gene has been found to be significantly up-regulated in SqCCLs that arise due to exposure to asbestos (Wright et al. 2012).
Large-Cell Lung Carcinoma
A genomic study that evaluated 78 samples of LCLC reported the most frequent mutations in TP53 (71%) and RB1 (26%). Mutations in the PI3K/AKT/mTOR pathway were identified in 15% of LCLCs with specific mutations in the PIK3CA (3%), PTEN (4%), AKT2 (4%), RICTOR (5%), and MTOR (1%) genes. Activating mutations were detected in the KRAS (6%), FGFR1 (5%), KIT (4%), ERBB2 (4%), HRAS (1%), and EGFR (1%) genes (Miyoshi et al. 2017). A study of neuroendocrine carcinomas reported that FGFR2 mutations were exclusively found in LCLC and not in SCLC (Vollbrecht et al. 2015). A study of 17 LCLCs reported 24% with KRAS mutations (Naidoo et al. 2016). A study that stratified LCLC based upon mutational status of RB1 concluded that the RB1 wild-type LCLCs had a more favorable response with a platinum-gemcitabine chemotherapy combination when compared with a platinum-etoposide combination (Derks et al. 2018).
A recent study positively correlated LCLC with PD-L1 expression compared with other histologic types of NSCLC, which supports investigation of effectiveness of ICI in LCLC (Chan et al. 2019a).
Typical and Atypical Carcinoid
Typical carcinoid (TC) and atypical carcinoid (AC) of the lung are characterized by GAs that inactivate genes affecting histone methylation, such as SWI/SNF complex subunit mutations (ARID1A, SMARC1, SMARCA2, SMRCA4). Additional alterations include mutations of CBX6, EZH2, EIF1AX, and E3 ubiquitin ligase genes (Pelosi et al. 2017).
Adenosquamous Carcinoma
ASC of the lung likely has a monoclonal origin for the SqCCL and LUAD components because microdissection of these components in two studies of 53 total ASCs demonstrated a highly convergent mutation rate between components (Vassella et al. 2015; Shi et al. 2016c). Mutations typically associated with LUADs and SqCCLs are identified in ASCs, including a first report of a novel TPD52L1-ROS1 fusion variant in ASC (Zhu et al. 2016).
Pulmonary Sarcomatoid Carcinoma
Activating GAs in MET are particularly prevalent in pulmonary sarcomatoid carcinomas, namely, METex14 splice-site mutations (22%) (Liu et al. 2016).
TP53 mutations have been reported in up to 74% of pulmonary sarcomatoid carcinomas, and KRAS mutations in 34%. Other less common mutations that are consistently reported are in the EGFR, BRAF, ERBB2 (HER2), RET, ALK, AKT1, JAK3, NRAS, and PIK3CA genes. A high percentage of pulmonary sarcomatoid cancers have high PD-L1 expression (from 53% to 69%), warranting further investigation regarding the potential benefit of ICI in this cancer subtype (Karim et al. 2018; Weissferdt 2018).
Bronchial Mucoepidermoid Carcinoma
Bronchial mucoepidermoid carcinoma is extremely rare. It is characterized by a specific translocation, t(11;19)(q21;p13), which results in an MECT1-MAML2 fusion. This same fusion is also a characteristic of salivary mucoepidermoid carcinoma (Fois et al. 2017; Szymanski et al. 2018).
Small-Cell Lung Cancer
In a study of 110 SCLCs, nearly all tumors had concomitant inactivation of the TP53 and RB1 genes resulting from complex genomic rearrangements. In two SCLCs, chromothripsis, which is defined as a massive genomic rearrangement during a single catastrophic event, was identified as an oncogenic cause with overexpression of cyclin D1 leading to effective inactivation of Rb1 (George et al. 2015).
A study of 98 undifferentiated SCLCs reported the most common mutations in the TP53 (86%), RB1 (54%) and MLL2 (17%), RICTOR (10%), KIT (7%), PIK3CA (6%), EGFR (5%), PTEN (5%), KRAS (5%), MCL1 (4%), FGFR1 (4%), BRCA2, (4%), TSC1 (3%), NF1 (3%), EPHA3 (3%), and CCND1 genes (Ross et al. 2014). Mutations in JAK3, NRAS, RB1, and VHL1 were exclusively found in SCLC and not in LCLC (Vollbrecht et al. 2015). A study of SCLCs in Chinese patients demonstrated fewer mutations in Wnt and Notch signaling pathways compared to Western studies, emphasizing the importance of diverse patient cohorts for the genomic understanding of lung cancer (Hu et al. 2019b). Molecular subtypes within SCLC have been proposed based upon the differential expression of ASCL1, NeuroD1, YAP1, and POU2F3 (Tsai et al. 2017; Chang et al. 2018; Ujhazy and Lindwasser 2018; Rudin et al. 2019a).
Malignant Pleural Mesothelioma
Malignant pleural mesothelioma (MPM) is characterized by frequent inactivation of TSGs, including enrichment of inactivating mutations in the cyclin-dependent kinase inhibitor 2A/alternative reading frame (CDKN2A/ARF) gene. An association between CDKN2A copy number loss in MPM tumor cells, stromal p16 immunoreactivity, and high pulmonary asbestos fiber count was reported with the suggestion that CDKN2A alterations in MPMs might indicate a history of asbestos exposure (Sekido 2013; Kettunen et al. 2019). The NF2/Hippo and BRCA-associated protein 1 (BAP1) pathways are also frequently mutated in MPM (Arzt et al. 2014; Miyanaga et al. 2015; Joseph et al. 2017; Felley-Bosco 2018). Overall, MPM is enriched in mutations in pathways involving TP53/DNA repair, the cell cycle, MAPK, and PI3K/AKT (Hylebos et al. 2016). In cases of MPM that are challenging to diagnose due to complex morphology and nonconclusive IHC results, molecular testing may provide additional diagnostic clues.
In a TCGA study of 74 MPMs, epithelioid MPMs had strong expression of the VISTA immune-checkpoint gene. WES revealed a group of MPMs with TP53 and SETDB1 mutations as well as concomitant loss of heterozygosity (LOH) that affected more than 80% of the genome (Hmeljak et al. 2018). In another TCGA study of 42 malignant mesotheliomas (including pleural [55%], peritoneal [26%], and pericardial [5%]), the most common aberrations were in the BAP1 (47.6%), NF2 (38.1%), CDKN2A/B (35.7%), and TP53 (16.7%) genes. BAP1 aberrations consisted of mutations (50%), loss (25%), rearrangements (5%), and multiple alterations (20%). NF2 aberrations consisted of mutations (81.3%), loss (12.5%), and multiple alterations (6.3%) (Kato et al. 2016). Current strategies under investigation for the treatment of MPM include poly-ADP ribose polymerase (PARP), CDK, and mTOR inhibitors as well as ICI and epigenetic modulators (Tolani et al. 2018).
Loss-of-function mutations in BAP1 and DNA repair genes predict improved OS following platinum chemotherapy (Hassan et al. 2019). A six-miR signature including miR-21-5p, miR-23a-3p, miR-30e-5p, miR-221-3p, miR-222-3p, and miR-31-5p can also predict improved OS in MPM (Ahmadzada et al. 2018).
INTRATUMOR HETEROGENEITY AND CLONALITY
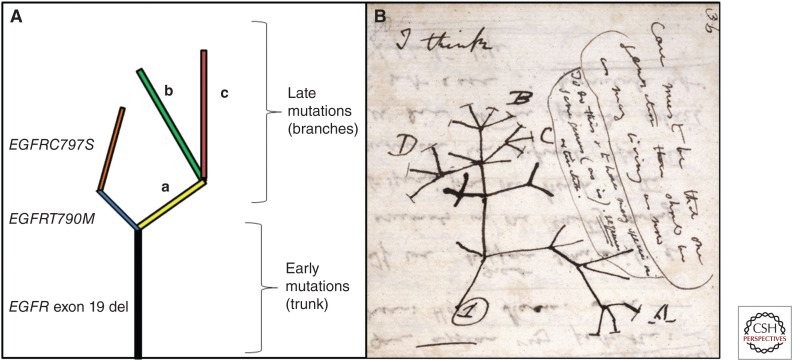
GAs shared by the majority or all regions of a tumor are called trunk mutations, or clonal mutations. In contrast, branch mutations are unique to a subclonal population within a tumor. Clonal mutations tend to reflect early events in tumor evolution, in contrast to subclonal mutations that tend to occur later (see Fig. 3). Clonal evolution is characterized as GAs becoming more “branched” as the tumor evolves from early precursor lesions, such as adenocarcinoma in situ (AIS), to progressively more advanced lesions, such as minimally invasive adenocarcinoma (MIA), then invasive LUAD (Vinayanuwattikun et al. 2016). One study reported that mutations in the TP53, KEAP1, STK11, and EGFR genes tend to be clonal as oposed to subclonal with the conclusion that these drivers have an earlier role in progression (Fig. 1; Shi et al. 2016a).
Figure 3.
Tumor evolution and consequential intratumor heterogeneity in lung cancer. (A) Early clonal mutations that are shared between high numbers of the tumor subpopulations are depicted in the trunk. These early mutations are often described as founder mutations, which are commonly found in genes such as EGFR, MET, KRAS, BRAF, and TP53. Later subclonal mutations are depicted in the branches, which are limited to specific tumor subpopulation(s). Later mutations are often described as private mutations when only reported in a unique tumor subpopulation. Later mutations are commonly found in genes such as PIK3CA, NF1, and EP300 and can occur as resistance mutations. In this figure, the early truncal EGFR exon 19 deletion is clonal. The EGFR T790M and EGFR C797S are later subclonal resistance mutations, which can be identified after treatment with early-generation EGFR inhibitors. For simplicity, mutations in other branches are referred to as a, b, and c. (B) A sketch by Charles Darwin similarly depicting evolutionary relationships with a trunk and branches. (Panel B, which is in the public domain in the UK, was reprinted from Darwin 1837–1838.)
In lung cancer, intratumor heterogeneity (ITH) has been quantified using the ITH index (ITHi) (Zhang et al. 2019f). Factors associated with high ITH include higher numbers of mutations, both clonal and subclonal, and copy number alterations. High ITH is associated with exposure to exogenous mutagens (e.g., cigarette smoker). Studies of pre-neoplastic nontumor tissue with mutagen exposure have shown areas of clonal expansion with higher numbers of mutations compared to nonexposed tissue (Martincorena et al. 2015, 2018; Martincorena 2019; Yizhak et al. 2019). The U.S. Cancer Moonshot initiative has recommended the development of a pre-cancer atlas. This initiative and others are supporting efforts to better understand the evolution of pre-neoplastic “nontumor” tissue, to clones/subclones, to early precursor tumor tissue, to late tumor tissue with a goal to help define sentinel genomic events and enable earlier detection of cancer (Spira et al. 2017; Janiszewska and Polyak 2018; Lippman et al. 2018; Srivastava et al. 2018a,b).
Enrichment of specific GAs in subpopulations that comprise ITH may be informative in anticipating tumor behavior. For instance, in LUADs that were post-therapy with EGFR TKIs, identification of biallelic inactivation of the TP53 and RB1 genes predicted transformation into SCLC, which is an established resistance mechanism to EGFR TKIs (Fig. 4; Sequist et al. 2011; Lee and Um 2017).
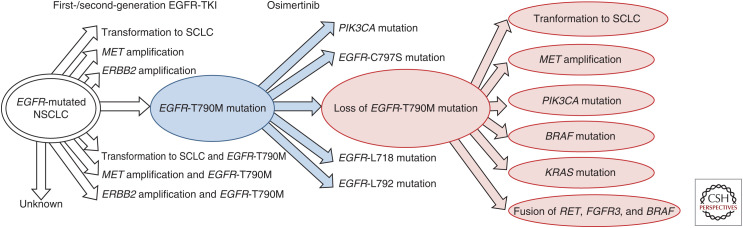
Figure 4.
Mechanisms of resistance after therapy with epidermal growth factor receptor (EGFR) inhibitors. This overview shows the most commonly reported resistance mechanisms (white arrows) that confer resistance to treatment with first-/second-generation EGFR tyrosine kinase inhibitors in EGFR-mutated non-small-cell lung cancer (NSCLC). In lung tumors harboring an EGFR T790M mutation, subsequent treatment with osimertinib imparts a selective advantage to subpopulations with resistance to osimertinib (blue arrows), which includes EGFR wild-type NSCLC with loss of EGFR T790 with other concomitant alterations (red connectors) (Remon et al. 2019).
Prior to the FDA approval of osimertinib as a first-line treatment of EGFR-mutated NSCLC, treatment of NSCLC with first- or second-generation EGFR TKIs (first-/second-generation EGFR TKIs) frequently became ineffectual due to resistance to treatment via a mutation at EGFR T790M. Consequently, EGFR T790M-mutated NSCLCs were treated with osimertinib, a third-generation EGFR inhibitor that binds irreversibly to C797; however, a concomitant cis-oriented mutation at C797S could then occur and confer resistance to osimertinib (Mitsudomi and Yatabe 2010; Niederst et al. 2015; Yu et al. 2015). A therapeutic strategy to minimize the heterogeneity of populations with EGFR resistance mutations at T790M and C797S with agents that are synthetic lethal is actively under investigation (Vyse et al. 2017).
Although mutation at EGFR T790M is the most common mechanism attributed to development of resistance to first-/second-generation EGFR TKIs, other resistance mechanisms, such as MET amplification and histologic transformation to SCLC, can occur. Transformation to SCLC is characterized by enrichment with mutations in retinoblastoma 1 gene (RB1), tumor protein p53 gene (TP53), and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α gene (PIK3CA) with retention of the original EGFR mutation (Yu et al. 2013; Zhang and Yuan 2016; Lee et al. 2017; Liu 2018; Remon et al. 2019).
The AURA3 trial reported resistance mechanisms to osimertinib, such as loss of EGFR T790M in ∼50% of patients, acquired EGFR mutations in 21%, MET amplification in 19%, ERBB2 (HER2) amplification in 5%, PIK3CA mutation and/or amplification in 5%, oncogenic fusions (RET, NTRK, and FGFR) in 4%, cell-cycle mutations in 4%, and BRAF V600E mutations in 3% (Papadimitrakopoulou et al. 2018).
Any changes in the evolution of the populations of clones/subclones that reflect ITH are of great importance when considering therapeutic agents, particularly if a clonal/subclonal population has a gene mutation that confers resistance to therapy (Amirouchene-Angelozzi et al. 2017; Sheng et al. 2018; Testa et al. 2018; Williams et al. 2018). “Game theory” is a conceptual approach to understanding tumor evolution. “Game theory” strategies can enable studies of the dynamic changes in subpopulations contributing to a tumor's ITH and the impact of specific treatments and their timing in an effort to determine treatment conditions that are beneficial to patients. Variations in treatment include duration of treatment, breaks in treatment, the sequence of agents given in a treatment regimen, or use of collateral agents. The dynamic changes to consider include how the cancer cells and the TME change following treatment (Kaznatcheev et al. 2019).
The identification of resistance mutations that arise as a consequence of therapies emphasize the importance of serial biopsies, as the mutation(s) that account for the majority of tumor cells that drive the progression of cancer may behave fluidly, as opposed to static tumor populations that remain fixed over time. Frustratingly, this presents a challenge to clinicians and pathologists alike to seek to determine the “moving target” of the driver mutation that would be most amenable to therapy.
The TRAcking non-small cell lung Cancer Evolution through therapy (Rx) (TRACERx) study aims to characterize recurrent patterns of evolution in ITH that are observed in multiple patients with NSCLC to determine or “track” populations with actionable drivers or resistant populations (Jamal-Hanjani et al. 2017; Negrao et al. 2018). Ongoing studies are actively investigating agents that demonstrate a response in EGFR inhibitor-resistant tumor cell populations, such as poly(ADP-ribose) polymerase 1 (PARP-1) inhibitors (Marcar et al. 2019).
Intermetastatic heterogeneity is defined as the entirety of heterogeneity of clones/subclones within all of the metastatic lesions that arise from an original primary lung cancer (Caswell and Swanton 2017). Intrametastatic heterogeneity is defined as the heterogeneity within a single metastasis (Vignot et al. 2013; Roper et al. 2019; Zhang et al. 2019f). The determination of whether there are therapy-resistant clones/subclones within metastatic sites is a clinically relevant consideration when making management decisions to continue a given therapy or initiate a new therapeutic option (Sosa Iglesias et al. 2018; Zhang et al. 2018a).
In summary, the evolution of the overall heterogeneity of the primary lung tumor and metastatic sites ideally should be used to inform the type(s) of therapy, and sequence of therapy administration in lung cancer (Pakkala and Ramalingam 2018).
TUMOR MUTATIONAL BURDEN (TMB)
High TMB is under investigation as a biomarker for patients with lung cancer to predict clinical benefit from ICI (Rizvi et al. 2015; Goodman et al. 2017; Gandara et al. 2018; Greillier et al. 2018; Hellmann et al. 2018; Hendriks et al. 2018; Maleki Vareki 2018; Chan et al. 2019b; Ready et al. 2019; Samstein et al. 2019). TMB is defined as the number of somatic nonsynonymous single-nucleotide variants (SNVs), insertions, and deletions per megabase covered by a genomic panel. Currently, the PD-L1 TPS, which is defined as the percentage of viable tumor cells with at least partial membrane staining for PD-L1, is the only biomarker with FDA-approved indications to predict response to ICI in lung cancer. However, assessment of PD-L1 expression in the collected specimen is difficult to standardize and does not always comprehensively reflect the PD-L1 expression in the entire lung cancer due to possible complicating factors such as heterogeneity of PD-L1 expression and small biopsies with a paucity of available tumor cells for evaluation (McLaughlin et al. 2016; Terra et al. 2017; Khunger et al. 2018; Velcheti et al. 2018; O'Malley et al. 2019). Accurate assessment is further complicated by interassay and interobserver variability. Even with high quantity and quality tissue for PD-L1 assessment, PD-L1 assessments do not clearly predict which patients will or will not respond to ICI. Consequently, alternative biomarkers for ICI responsiveness, such as TMB, are desirable. Regardless of biomarker, the benefit imparted by ICI needs to be considered in context of the risk of immune-related adverse events (irAEs) (Puzanov et al. 2017; Martins et al. 2019; Ricciuti et al. 2019).
When TMB is evaluated based upon a given targeted sequencing panel, the TMB reported may vary from other targeted sequencing panels given factors such as differences in the areas of genomic coverage (Büttner et al. 2019). An approach that addresses this challenge is for targeted sequencing panels to correlate the extent to which a targeted TMB is commensurate with the TMB determined by WES (Meléndez et al. 2018; Butler et al. 2019). Efforts for standardization and harmonization of TMB to minimize variability across NGS assays are ongoing (Chang et al. 2019; Stenzinger et al. 2019).
A comparison of single-region and multiregional tissue tumor mutational burden (tTMB) with blood TMB (bTMB) in a series of LUAD, SqCCL, and lymphoepithelioma-like carcinoma (LELC) concluded that single-region tTMB correlated more closely with multiregional tTMB than bTMB (Zhang et al. 2019e). Another study proposed that the quality of the mutations that comprise TMB should be considered to provide context rather than solely focusing on the TMB number. This study found that relatively more widespread clonal mutations had greater potential in eliciting T-cell immunoreactivity and predicting sensitivity to ICI than relatively more private subclonal mutations (McGranahan et al. 2016). The extent to which mutational status, such as MET-activating and STK11-inactivating mutations, contribute to tumor immunotolerance is another ongoing area of investigation (Saigi et al. 2019).
High TMB (TMB-H) may result from GAs that cause an underlying deficiency in DNA damage response (DDR) pathways, such as tumors with homologous recombination deficiency (HRD), deficient DNA polymerase ε or DNA polymerase δ1 (POLE or POLD1) proofreading, alterations of the base excision repair (BER) pathway, deficient mismatch repair (dMMR), and deficient ERCC1-mediated repair (Warth et al. 2016; Takamochi et al. 2017; Bever and Le 2018; Heeke et al. 2018; Knijnenburg et al. 2018; Penault-Llorca and Radosevic-Robin 2018; Song et al. 2018b; Chabanon et al. 2019; Chae et al. 2019b; Park and Pursell 2019; Diossy et al. 2021).
Increased TMB has also been associated with carcinogen exposure in tobacco smokers that develop lung cancer (Alexandrov et al. 2016; Davis et al. 2017; Nagahashi et al. 2018; Norum and Nieder 2018; Phillips 2018; Song et al. 2018a; Blons et al. 2019; Cardona et al. 2019; Chae et al. 2019b). Of note, TMB-H is associated with global DNA hypomethylation, increased CIN, and GAs in the TP53 and APOBEC genes. TMB-H is not often observed in lung cancers with EGFR GAs (Cai et al. 2018; Nagahashi et al. 2018; Phillips 2018; Zhong et al. 2018).
Increased TMB is not the only measure of overall tumor antigenicity. Viral-related antigenicity is another cause of up-regulation of host immune response and immunogenicity. Viral-related antigenicity is associated with responsiveness to ICI, even if the corresponding TMB is relatively low.
Whether the underlying cause of immunogenicity is mutant protein antigenicity or viral, there are specific histologic findings that correspond with a strong immunogenic response to the tumor, such as tumor-infiltrating lymphocytes (TILs), peritumoral stromal lymphocytes, and/or tumor-adjacent lymph node–like structures termed tertiary lymphoid structures (TLSs) (Al-Shibli et al. 2008; Donnem et al. 2015; Schalper et al. 2015; Anichini et al. 2018; Soo et al. 2018; Blons et al. 2019).
A digital pathology-based method to quantify tumor immunogenic response was developed by the Society for Immunotherapy of Cancer (SITC), which was initially defined as an immunoscore predictive biomarker in colon cancer (Pagès et al. 2018). Ongoing studies are investigating the potential of higher immunoscores to predict response to ICI in patients with multiple other tumor types (Kirilovsky et al. 2016; Gnjatic et al. 2017; Zeng et al. 2018), including NSCLC (Donnem et al. 2015, 2016; Schalper et al. 2015; Paulsen et al. 2017; Brahmer et al. 2018; Soo et al. 2018).
CELL-FREE DNA (cfDNA) AND CIRCULATING TUMOR CELLS (CTCs)
Circulating tumor DNA (ctDNA) derived from circulating cell-free DNA (cfDNA) and circulating tumor cells (CTCs) are detectable in most cases of advanced stage lung cancer and evidence supports their clinical utility in guiding therapy (Duréndez-Sáez et al. 2017; Razavi et al. 2017; Lim et al. 2018a; Zhang et al. 2018c; Cheng et al. 2019; Zhao et al. 2019). Liquid biopsies with assessment of mutations in ctDNA are a particularly helpful approach for identifying mutations in tumors that are not amenable to biopsy (Calabuig-Fariñas et al. 2016; Foy et al. 2017). Quantification of CTCs in lung cancer may have prognostic value with correlation between post-therapy decreases in CTCs and therapeutic response. With advanced technologies, CTCs can be isolated for sequencing, tested with other methods such as IHC, or even be used to create patient-derived xenografts (PDX) (Hofman et al. 2011a,b, 2019; Tan et al. 2016; Carter et al. 2017; Drapkin et al. 2018).
In a clinical context, ctDNA results can be complementary to solid tumor biposies, since both methods have advantages and disadvantages; however, the cost for testing with multiple methods may be too cost-prohibitive for widespread simultaneous testing with debatable improved clinical benefit (Aggarwal et al. 2018). An advantage of ctDNA over solid tissue testing is the ability to detect clonal and subclonal mutations arising from the primary and all metastatic sites if ctDNA is released from all tumor sites into the circulating blood. This advantage provides value for detecting mutations in tumors with heterogeneity among metastatic sites and/or within the primary site. Traditional biopsies of solid tumor are limited to mutational analysis of only the sampled tumor due to the impractical costs and risks of performing multiple biopsies in patients with multiple metastases (Santarpia et al. 2018a; Stanta and Bonin 2018). After initiation of targeted therapy, ctDNA testing of blood samples can be performed to evaluate for resistance mutations, reducing the risks and costs of repeat biopsies (Zill et al. 2018).
Detection of TMB and PD-L1 with ctDNA and CTCs, respectively, is being explored with comparison to tumor tissue biopsy TMB and PD-L1 results and evaluation as potential biomarkers for response to ICI in NSCLC (Chae et al. 2019a). Significant PD-L1 heterogeneity was observed in the CTCs (Koh et al. 2019).
Liquid biopsies can also be a reliable method for detecting circulating miRs, which has led to the development of miR profiles to detect circulating differentially expressed miRs in lung cancer (Hu et al. 2010; Deng et al. 2014; Huang et al. 2014; Wu et al. 2015; Leng et al. 2017a; Yang et al. 2017; Zaporozhchenko et al. 2018; Calabrese et al. 2019). Methylation patterns of ctDNA have potential to differentiate between cancer types and tumor tissue site origins (Moss et al. 2018).
The chronologic appearance of GAs throughout the evolution of lung cancer has been characterized in solid tumor tissue, and the evaluation of ctDNA data from serial liquid biopsies has helped to enrich the understanding of the evolution of different tumor subpopulations (Sequist et al. 2011; Vinayanuwattikun et al. 2016; Jamal-Hanjani et al. 2017; Ma et al. 2017; Klein et al. 2018). Detection of GAs by ctDNA analysis that are observed relatively early in tumor evolution, such as KRAS mutations in pancreatic ductal adenocarcinoma (PDAC), in combination with blood testing for CA19-9 is a promising strategy to enable screening and earlier diagnosis of PDAC, a cancer that is typically not detected until presentation of symptoms at an advanced stage (Cohen et al. 2017). Given that NSCLC is also not usually diagnosed until symptoms present at an advanced stage, efforts to enable earlier diagnosis have likewise been invested in studies using ctDNA as an adjunct to other specific biomarkers such as miRs (exosomal and non-exosomal), DNA methylation patterns, protein markers, metabolites and CTCs to enable early-stage diagnosis of NSCLC (Deng et al. 2014; Liang et al. 2018b; Oxnard et al. 2018; Santarpia et al. 2018b; Calabrese et al. 2019).
Serial liquid biopsies offer the benefit of determining any changes in the intermetastatic heterogeneity during the post-therapy evolution of the populations of resistant clones/subclones (e.g., EGFR TKI resistance) (Imamura et al. 2016; Oxnard et al. 2016; Lim et al. 2018a). An approach using serial liquid biopsies to track the decrease of relevant mutant allele fractions (MAFs) in ctDNA is under investigation as a surrogate metric to determine the presence/extent of a therapeutic response after chemotherapy, radiotherapy, or targeted therapy (Raja et al. 2018; Gao et al. 2019; Phallen et al. 2019). Clearance of detectable ctDNA mutations occurs rapidly after effective therapy with the MAFs representing a snapshot of the immediate rate of tumor cell death at the time of liquid biopsy (Gao et al. 2019). Postoperative increases in ctDNA and/or CTCs may serve as predictive surrogate metrics for early tumor recurrence (Liang et al. 2018a; Moding et al. 2018).
The fragmentation profile of the overall ctDNA is a feature that may have potential utility in distinguishing cancer patients from healthy patients. A machine learning-based study that evaluated the fragmentation lengths from WGS of cfDNA demonstrated shorter ctDNA fragmentation in lung cancer patients than cfDNA in healthy patients (Cristiano et al. 2019).
CLINICAL TRANSLATIONS
Recent advances in molecular pathology and associated technologies outpace their translation to clinical care. Triage is needed to focus clinical translation efforts on technologies most likely to truly benefit patients. Recent guidelines have focused on recommending a translational approach informed by the benefits of using targeted TKIs (Kalemkerian et al. 2018; Lindeman et al. 2018; Planchard et al. 2018; Zhang et al. 2019a). ICI for standard of care is typically most beneficial when no drivers are identified in the cancer to indicate targeted therapy (Herbst et al. 2016; Zago et al. 2016; Carbone et al. 2017; Bironzo and Di Maio 2018; Bylicki et al. 2018; Hirsch et al. 2018; Osmani et al. 2018; Shroff et al. 2018; Dong et al. 2019; Forde et al. 2019; West et al. 2019). For instance, the KEYNOTE 024 and KEYNOTE 042 trials support the first-line use pembrolizumab in advanced NSCLC with a level of PD-L1 expression of at least 50%, only in the absence of EGFR and ALK alterations (Mok et al. 2019; Reck et al. 2019).
A therapeutic approach in lung cancer that is gaining momentum is the use of additive or synergistic therapy ICI combinations with other complementary treatments such as chemotherapy, radiotherapy or targeted agents, such as EGFR TKIs, VEGFR TKIs, or HDACis, as well as the use of dual agents for ICI such as concomitant PD-1/PD-L1 and cytotoxic T-lymphocyte antigen 4 (CTLA-4) inhibition (Le et al. 2017; Fukumura et al. 2018; Qiao et al. 2018a,b; Drapkin and Farago 2019; Jiang et al. 2019; Pavan et al. 2019; Rocco et al. 2019). Studies in other cancer types have supported potential clinical benefit with ICI combination strategies (Bastos and Zaharcu 2016; Chen et al. 2018a; Tang et al. 2018; Burrack et al. 2019; Merhi et al. 2019; Rini et al. 2019; Sicklick et al. 2019; Taberna et al. 2019; von der Grün et al. 2019).
Moreover, pathologic analyses of major pathological response in NSCLC after neoadjuvant treatment is actively under investigation, which have corroborated the prognostic significance of major pathological response (MPR), which has been determined by assessment of the percentage of residual tumor cells (65% for LUAD and 10% for non-LUAD) to assign a prognostic score to NSCLC cases that are post-neoadjuvant therapy. Assessment of a tumor regression grade (TRG) to quantify the percentage of residual tumor cells has been used in the setting of other tumor types, and clinical utility of TRG in NSCLC will likely assist in determination of prognosis.
Incorporation of NGS testing in a way that is complementary to other methods, such as PCR, FISH, and IHC, may optimally benefit treatment decisions (Dorward et al. 2017; Sholl 2017; Osmani et al. 2018; Dong et al. 2019). The NCCN provides guidance about molecular testing in their guidelines, which are derived from an evidence-based approach based on publications and professional consensus of expert panels (National Comprehensive Cancer Network 2021).
Despite efforts by the NCCN to provide guidelines and recommendations, universal incorporation of these recommendations has not been adopted across all clinical practice patterns (MacLean et al. 2016; Gutierrez et al. 2017; Gierman et al. 2019). The National Lung Cancer Round Table (NLCRT) has enacted task groups, such as the Triage for Appropriate Treatment (TAT) group, to advocate for patients in the interest of guideline-adherent diagnosis and treatment. This effort includes gathering of data to better understand barriers to guideline-recommended care (Kim et al. 2019).
The NCCN guidelines acknowledge the reality of clinical practice in resource-limited situations and have developed general guidelines for best patient care in such situations. The NCCN guidelines also recognize the need for balance between simpler but rapid testing with more comprehensive (but often slower and more expensive) testing, such as NGS, to identify GAs that may be less common but are nonetheless actionable with targeted therapy or may qualify patients for clinical trials or off-label therapies (Jordan et al. 2017). Another important consideration about molecular testing in lung cancer addressed by the NCCN guidelines is the need to triage the limited amount of tumor tissue often obtained with biopsies and fine needle aspirations for the most clinically needed diagnostic tests (Bubendorf et al. 2017; Brainard and Farver 2019).
Furthermore, studies such as the Interrogating Cancer Etiology Using Proactive Genetic Testing (INTERCEPT) study determined that pathogenic germline variants contributed to cancer in 13.3% of the 3000 patients in the study (Mandelker and Zhang 2018; Lincoln et al. 2020; Fiala et al. 2021; Samadder et al. 2021). The importance of studies such as the INTERCEPT study helped to inform a joint consensus recommendation to establish standards and guidelines for the interpretation and reporting of sequence variants in cancer as published by the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. This study emphasized that NGS reporting should include germline variants with known evidence of clinical impact (Li et al. 2017a).
ARTIFICIAL INTELLIGENCE, DEEP LEARNING, AND FUTURE DIRECTIONS
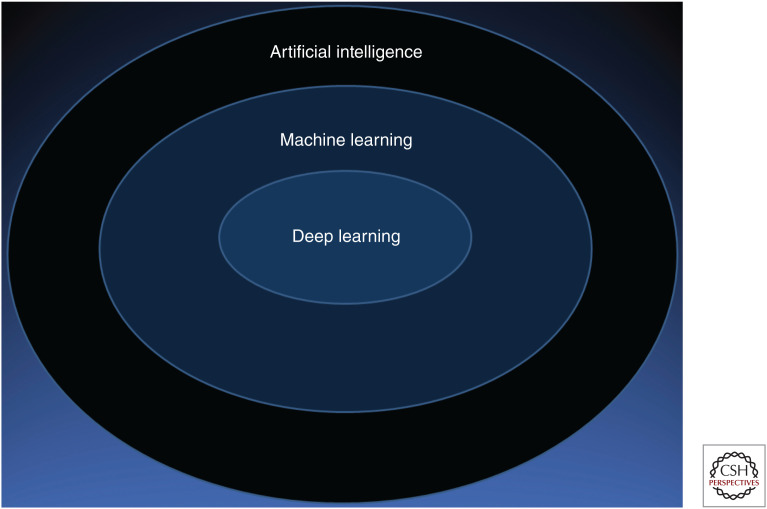
Development of research tools to confirm, challenge, and expand our understanding of lung cancer is critical (Washetine et al. 2018). Artificial intelligence (AI), with a deep learning (DL)-based approach, is a potentially useful tool to facilitate interpretation of the massive amounts of data that are being produced by research and clinical genetic testing, data that ordinarily requires a labor-intensive effort for humans to interpret (Yoon et al. 2018; Zhou et al. 2018; AlQuraishi 2019; Amin et al. 2019; Clark et al. 2019; Kuleshov et al. 2019; Li et al. 2019a; Shah et al. 2019; Thormann et al. 2019). Studies involving DL had initially focused on digital pathology applications (Cruz-Roa et al. 2013; Wang et al. 2014, 2018b; Xie et al. 2015; Djuric et al. 2017; Nalisnik et al. 2017; Saha et al. 2017; Sharma et al. 2017; Xu et al. 2017; Ehteshami Bejnordi et al. 2018; Khosravi et al. 2018; Liu et al. 2018c; Rakhlin et al. 2018; Saltz et al. 2018; Venkat et al. 2018; Yan et al. 2018; Zhang et al. 2018b; Hu and Yu 2019; Pell et al. 2019; Shapcott et al. 2019). DL via convolutional neural networks (CNNs) has set a precedent for its diagnostic predictive utility of histomorphologic and radiologic features, commensurate with the performance of pathologists and radiologists in some contexts (Fig. 5; Cruz-Roa et al. 2013, 2015; Yu et al. 2016; Teramoto et al. 2017; Alsubaie et al. 2018; Borkowski et al. 2018; Coudray et al. 2018; Hosny et al. 2018; Wang et al. 2018a; Ardila et al. 2019; Gertych et al. 2019; Wei et al. 2019; Zhang et al. 2019b,g,h).
Figure 5.
Artificial intelligence, machine learning, and deep learning. Artificial intelligence is a broadly defined technique that uses logic to mimic human intelligence. Machine learning is a subset of artificial intelligence that uses algorithms to improve effectiveness of performance. Deep learning is a subset of machine learning that uses neural networks to process identified data to train optimal performance with respect to a specific task (e.g., recognition of histologic features in a particular tumor type).
Historically, correlation of histomorphologic features with underlying molecular alterations has been limited by the amount of work required to complete such studies (Shim et al. 2011; Song et al. 2013; Hu et al. 2014). As DL-based studies using CNNs has advanced, the potential to provide an enhanced understanding of the relationship between histomorphologic features and molecular alterations has expanded (Coudray et al. 2018; Couture et al. 2018; Mobadersany et al. 2018; Saltz et al. 2018; Cheerla and Gevaert 2019; Kather et al. 2019; Montalto and Edwards 2019). This approach may provide unique insights into how to best detect DL-predicted drivers, and which signaling pathways are activated as a method to predict response to targeted therapy (Way et al. 2018; Chiu et al. 2019; Han et al. 2019; Luo et al. 2019). Moreover, DL may provide insight into the accurate identification of molecular subtypes and recurrent mutational signatures (Huang et al. 2018c; Yu et al. 2020). DL has also demonstrated promise in classifying tumor types based on RNA-seq analysis (Lyu and Haque 2018). CancerSEEK is an innovative approach that uses a DL-based method to determine tumor-type specific mutations in liquid biopsies to determine the anatomic site of origin in CUP cases (Cohen et al. 2018). Availability of high-quality genomic data via reliable databases or data repositories will be crucial for future DL-based studies (Avsec et al. 2019).
With appropriate supervision and quality control, the ongoing technical advances in AI have potential to enable and refine the interpretation of clinical data. Advancement in these tools to perform meaningful analyses of large datasets may enhance our ability to practice precision medicine (Boersma et al. 2019).
CONCLUDING REMARKS
The art of translating advances in science and technology into improvement in patient care involves leveraging available tools and knowledge. Although this review represents only a snapshot in our understanding of the molecular pathology of lung cancer, it also offers a contextual view of the ongoing advances in lung cancer to improve patient care and the development of more accurate biomarkers to inform therapeutic decisions.
COMPETING INTEREST STATEMENT
The authors declare no relevant conflicts of interest.
Footnotes
Editors: Christine M. Lovly, David P. Carbone, and John D. Minna
Additional Perspectives on Lung Cancer: Disease Biology and Its Potential for Clinical Translation available at www.perspectivesinmedicine.org
REFERENCES
- Aggarwal C, Thompson JC, Black TA, Katz SI, Fan R, Yee SS, Chien AL, Evans TL, Bauml JM, Alley EW, et al. 2019. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol 5: 173–180. 10.1001/jamaoncol.2018.4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadzada T, Reid G, Kao S. 2018. Biomarkers in malignant pleural mesothelioma: current status and future directions. J Thorac Dis 10: S1003–S1007. 10.21037/jtd.2018.04.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal F, Totoki Y, Fujimoto A, Nakagawa H, Shibata T, et al. 2016. Mutational signatures associated with tobacco smoking in human cancer. Science 354: 618–622. 10.1126/science.aag0299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SM, Hensing T, Schrock AB, Allen J, Sanford E, Gowen K, Kulkarni A, He J, Suh JH, Lipson D, et al. 2016. Comprehensive genomic profiling identifies a subset of crizotinib-responsive ALK-rearranged non-small cell lung cancer not detected by fluorescence in situ hybridization. Oncologist 21: 762–770. 10.1634/theoncologist.2015-0497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlQuraishi M. 2019. End-to-end differentiable learning of protein structure. Cell Syst 8: 292–301.e3. 10.1016/j.cels.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shibli KI, Donnem T, Al-Saad S, Persson M, Bremnes RM, Busund LT. 2008. Prognostic effect of epithelial and stromal lymphocyte infiltration in non-small cell lung cancer. Clin Cancer Res 14: 5220–5227. 10.1158/1078-0432.CCR-08-0133 [DOI] [PubMed] [Google Scholar]
- Alsubaie N, Shaban M, Snead D, Khurram A, Rajpoot N. 2018. A multi-resolution deep learning framework for lung adenocarcinoma growth pattern classification. In Medical image understanding and analysis (ed. Nixon M, Mahmoodi S, Zwiggelaar R), pp. 3–11. Springer, Berlin. [Google Scholar]
- Amatu A, Sartore-Bianchi A, Siena S. 2016. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open 1: e000023. 10.1136/esmoopen-2015-000023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin N, McGrath A, Chen YPP. 2019. Evaluation of deep learning in non-coding RNA classification. Nat Mach Intell 1: 246–256. 10.1038/s42256-019-0051-2 [DOI] [Google Scholar]
- Amirouchene-Angelozzi N, Swanton C, Bardelli A. 2017. Tumor evolution as a therapeutic target. Cancer Discov 7: 805–817. 10.1158/2159-8290.CD-17-0343 [DOI] [PubMed] [Google Scholar]
- Anichini A, Tassi E, Grazia G, Mortarini R. 2018. The non-small cell lung cancer immune landscape: emerging complexity, prognostic relevance and prospective significance in the context of immunotherapy. Cancer Immunol Immunother 67: 1011–1022. 10.1007/s00262-018-2147-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, et al. 2019. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 25: 954–961. 10.1038/s41591-019-0447-x [DOI] [PubMed] [Google Scholar]
- Arzt L, Quehenberger F, Halbwedl I, Mairinger T, Popper HH. 2014. BAP1 protein is a progression factor in malignant pleural mesothelioma. Pathol Oncol Res 20: 145–151. 10.1007/s12253-013-9677-2 [DOI] [PubMed] [Google Scholar]
- Avsec Ž, Kreuzhuber R, Israeli J, Xu N, Cheng J, Shrikumar A, Banerjee A, Kim DS, Beier T, Urban L, et al. 2019. The Kipoi repository accelerates community exchange and reuse of predictive models for genomics. Nat Biotechnol 37: 592–600. 10.1038/s41587-019-0140-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad MM, Shaw AT. 2014. ALK inhibitors in non-small cell lung cancer: crizotinib and beyond. Clin Adv Hematol Oncol 12: 429–439. [PMC free article] [PubMed] [Google Scholar]
- Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P, Heng JC, Dahlberg SE, Jänne PA, Verma S, et al. 2016. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J Clin Oncol 34: 721–730. 10.1200/JCO.2015.63.4600 [DOI] [PubMed] [Google Scholar]
- Bai Y, Zhang G, Chu H, Li P, Li J. 2019. The positive feedback loop of lncRNA DANCR/miR-138/Sox4 facilitates malignancy in non-small cell lung cancer. Am J Cancer Res 9: 270–284. [PMC free article] [PubMed] [Google Scholar]
- Bang MS, Kang K, Lee JJ, Lee YJ, Choi JE, Ban JY, Oh CH. 2017. Transcriptome analysis of non-small cell lung cancer and genetically matched adjacent normal tissues identifies novel prognostic marker genes. Genes Genomics 39: 277–284. 10.1007/s13258-016-0492-5 [DOI] [Google Scholar]
- Banik D, Moufarrij S, Villagra A. 2019. Immunoepigenetics combination therapies: an overview of the role of HDACs in cancer immunotherapy. Int J Mol Sci 20: 2241. 10.3390/ijms20092241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazas M, Gasparini A, Huang Y, Küçükosmanoğlu A, Annunziato S, Bouwman P, Sol W, Kersbergen A, Proost N, de Korte-Grimmerink R, et al. 2019. Radiosensitivity is an acquired vulnerability of PARPi-resistant BRCA1-deficient tumors. Cancer Res 79: 452–460. 10.1158/0008-5472.CAN-18-2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker EV, Cervigne NK, Reis PP, Goswami RS, Xu W, Weinreb I, Irish JC, Kamel-Reid S. 2009. microRNA evaluation of unknown primary lesions in the head and neck. Mol Cancer 8: 127. 10.1186/1476-4598-8-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos B, Zaharcu A. 2016. Synergistic combination of PD-1 and EGFR antibodies: a proposed phase 1 clinical trial using nivolumab and cetuximab. Int J Radiat Oncol Biol Phys 94: 929. 10.1016/j.ijrobp.2015.12.238 [DOI] [Google Scholar]
- Beg AA, Gray JE. 2016. HDAC inhibitors with PD-1 blockade: a promising strategy for treatment of multiple cancer types? Epigenomics 8: 1015–1017. 10.2217/epi-2016-0066 [DOI] [PubMed] [Google Scholar]
- Bentley TG, Schroeder BE, Schnabel CA, Erlander MG, Hsiao WC, Ortendahl JD, Broder MS. 2014. Cost effectiveness of a 92-gene assay for the diagnosis of metastatic cancer. J Med Econ 17: 527–537. 10.3111/13696998.2014.909817 [DOI] [PubMed] [Google Scholar]
- Bever KM, Le DT. 2018. DNA repair defects and implications for immunotherapy. J Clin Invest 128: 4236–4242. 10.1172/JCI122010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bica-Pop C, Cojocneanu-Petric R, Magdo L, Raduly L, Gulei D, Berindan-Neagoe I. 2018. Overview upon miR-21 in lung cancer: focus on NSCLC. Cell Mol Life Sci 75: 3539–3551. 10.1007/s00018-018-2877-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bironzo P, Di Maio M. 2018. A review of guidelines for lung cancer. J Thorac Dis 10: S1556–S1563. 10.21037/jtd.2018.03.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blons H, Garinet S, Laurent-Puig P, Oudart JB. 2019. Molecular markers and prediction of response to immunotherapy in non-small cell lung cancer, an update. J Thorac Dis 11: S25–S36. 10.21037/jtd.2018.12.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma S, Khuperkar D, Verhagen BMP, Sonneveld S, Grimm JB, Lavis LD, Tanenbaum ME. 2019. Multi-color single-molecule imaging uncovers extensive heterogeneity in mRNA decoding. Cell 178: 458–472.e19. 10.1016/j.cell.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkowski AA, Wilson CP, Borkowski SA, Deland LA, Mastorides SM. 2018. Using Apple machine learning algorithms to detect and subclassify non-small cell lung cancer. arXiv 1808.08230v2
- Bossé Y, Amos CI. 2018. A decade of GWAS results in lung cancer. Cancer Epidemiol Biomarkers Prev 27: 363–379. 10.1158/1055-9965.EPI-16-0794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle TA, Quinn GP, Schabath MB, Muñoz-Antonia T, Saller JJ, Duarte LF, Hair LS, Teer JK, Chiang DY, Leary R, et al. 2020. A community-based lung cancer rapid tissue donation protocol provides high-quality drug-resistant specimens for proteogenomic analyses. Cancer Med 9: 225–237. 10.1002/cam4.2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer JR, Govindan R, Anders RA, Antonia SJ, Sagorsky S, Davies MJ, Dubinett SM, Ferris A, Gandhi L, Garon EB, et al. 2018. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of non-small cell lung cancer (NSCLC). J Immunother Cancer 6: 75. 10.1186/s40425-018-0382-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brainard J, Farver C. 2019. The diagnosis of non-small cell lung cancer in the molecular era. Mod Pathol 32: 16–26. 10.1038/s41379-018-0156-x [DOI] [PubMed] [Google Scholar]
- Bronte G, Ulivi P, Verlicchi A, Cravero P, Delmonte A, Crino L. 2019. Targeting RET-rearranged non-small-cell lung cancer: future prospects. Lung Cancer (Auckl) 10: 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubendorf L, Lantuejoul S, de Langen AJ, Thunnissen E. 2017. Nonsmall cell lung carcinoma: diagnostic difficulties in small biopsies and cytological specimens: number 2 in the series “pathology for the clinician” edited by Peter Dorfmuller and Alberto Cavazza. Eur Respir Rev 26: 170007. 10.1183/16000617.0007-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack AL, Spartz EJ, Raynor JF, Wang I, Olson M, Stromnes IM. 2019. Combination PD-1 and PD-L1 blockade promotes durable neoantigen-specific T cell-mediated immunity in pancreatic ductal adenocarcinoma. Cell Rep 28: 2140–2155.e6. 10.1016/j.celrep.2019.07.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch SE, Hanke ML, Kargl J, Metz HE, MacPherson D, Houghton AM. 2016. Lung cancer subtypes generate unique immune responses. J Immunol 197: 4493–4503. 10.4049/jimmunol.1600576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M, Konigshofer Y, Clement O, Liu L, Zhao C, Arcila ME, Zehir A, Kohle R, Magliocco AM, Gligorich K, et al. 2019. Tumor mutational burden reference materials for assay standardization. J Clin Oncol 37: e14746–e14746. 10.1200/JCO.2019.37.15_suppl.e14746 [DOI] [Google Scholar]
- Büttner R, Longshore JW, López-Ríos F, Merkelbach-Bruse S, Normanno N, Rouleau E, Penault-Llorca F. 2019. Implementing TMB measurement in clinical practice: considerations on assay requirements. ESMO Open 4: e000442. 10.1136/esmoopen-2018-000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylicki O, Paleiron N, Rousseau-Bussac G, Chouaid C. 2018. New PDL1 inhibitors for non-small cell lung cancer: focus on pembrolizumab. Onco Targets Ther 11: 4051–4064. 10.2147/OTT.S154606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai YR, Dong YJ, Wu HB, Liu ZC, Zhou LJ, Su D, Chen XJ, Zhang L, Zhao YL. 2016. Micropapillary: a component more likely to harbour heterogeneous EGFR mutations in lung adenocarcinomas. Sci Rep 6: 23755. 10.1038/srep23755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Bai H, Wang Z, Wang S, Duan J, Gao S, He J, Wang J. 2018. MA06.07 genetic and epigenetic alterations are associated with tumor mutation burden in non-small cell lung cancer. J Thorac Oncol 13: S377. 10.1016/j.jtho.2018.08.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Lin S, Girard L, Zhou Y, Yang L, Ci B, Zhou Q, Luo D, Yao B, Tang H, et al. 2019a. LCE: an open web portal to explore gene expression and clinical associations in lung cancer. Oncogene 38: 2551–2564. 10.1038/s41388-018-0588-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai MC, Chen M, Ma P, Wu J, Lu H, Zhang S, Liu J, Zhao X, Zhuang G, Yu Z, et al. 2019b. Clinicopathological, microenvironmental and genetic determinants of molecular subtypes in KEAP1/NRF2-mutant lung cancer. Int J Cancer 144: 788–801. 10.1002/ijc.31975 [DOI] [PubMed] [Google Scholar]
- Calabrese F, Lunardi F, Pezzuto F, Fortarezza F, Vuljan SE, Marquette C, Hofman P. 2019. Are there new biomarkers in tissue and liquid biopsies for the early detection of non-small cell lung cancer? J Clin Med 8: 414. 10.3390/jcm8030414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabuig-Fariñas S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. 2016. Circulating tumor cells versus circulating tumor DNA in lung cancer—which one will win? Transl Lung Cancer Res 5: 466–482. 10.21037/tlcr.2016.10.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Getz G, Korbel JO, Stuart JM, Jennings JL, Stein LD, Perry MD, Nahal-Bose HK, Ouellette BFF, Li CH, et al. 2020. Pan-cancer analysis of whole genomes. Nature 578: 82–93. 10.1038/s41586-020-1969-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, Gaida K, Holt T, Knutson CG, Koppada N, et al. 2019. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575: 217–223. 10.1038/s41586-019-1694-1 [DOI] [PubMed] [Google Scholar]
- Capelletti M, Dodge ME, Ercan D, Hammerman PS, Park SI, Kim J, Sasaki H, Jablons DM, Lipson D, Young L, et al. 2014. Identification of recurrent FGFR3-TACC3 fusion oncogenes from lung adenocarcinoma. Clin Cancer Res 20: 6551–6558. 10.1158/1078-0432.CCR-14-1337 [DOI] [PubMed] [Google Scholar]
- Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, Felip E, van den Heuvel MM, Ciuleanu TE, Badin F, et al. 2017. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med 376: 2415–2426. 10.1056/NEJMoa1613493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona AF, Rojas L, Zatarain-Barrón ZL, Ruiz-Patiño A, Ricaurte L, Corrales L, Martín C, Freitas H, Cordeiro de Lima VC, Rodriguez J, et al. 2019. Multigene mutation profiling and clinical characteristics of small-cell lung cancer in never-smokers vs. heavy smokers (Geno1.3-CLICaP). Front Oncol 9: 254. 10.3389/fonc.2019.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrot-Zhang J, Yao X, Devarakonda S, Deshpande A, Damrauer JS, Silva TC, Wong CK, Choi HY, Felau I, Robertson AG, et al. 2021. Whole-genome characterization of lung adenocarcinomas lacking alterations in the RTK/RAS/RAF pathway. Cell Rep 34: 108707. 10.1016/j.celrep.2021.108707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L, Rothwell DG, Mesquita B, Smowton C, Leong HS, Fernandez-Gutierrez F, Li Y, Burt DJ, Antonello J, Morrow CJ, et al. 2017. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med 23: 114–119. 10.1038/nm.4239 [DOI] [PubMed] [Google Scholar]
- Carvalho AS, Cuco CM, Lavareda C, Miguel F, Ventura M, Almeida S, Pinto P, de Abreu TT, Rodrigues LV, Seixas S, et al. 2017. Bronchoalveolar lavage proteomics in patients with suspected lung cancer. Sci Rep 7: 42190. 10.1038/srep42190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell DR, Swanton C. 2017. The role of tumour heterogeneity and clonal cooperativity in metastasis, immune evasion and clinical outcome. BMC Med 15: 133. 10.1186/s12916-017-0900-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceppi P, Mudduluru G, Kumarswamy R, Rapa I, Scagliotti GV, Papotti M, Allgayer H. 2010. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res 8: 1207–1216. 10.1158/1541-7786.MCR-10-0052 [DOI] [PubMed] [Google Scholar]
- Chabanon RM, Muirhead G, Krastev DB, Adam J, Morel D, Garrido M, Lamb A, Henon C, Dorvault N, Rouanne M, et al. 2019. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J Clin Invest 129: 1211–1228. 10.1172/JCI123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae YK, Davis AA, Agte S, Pan A, Simon NI, Iams WT, Cruz MR, Tamragouri K, Rhee K, Mohindra N, et al. 2019a. Clinical implications of circulating tumor DNA tumor mutational burden (ctDNA TMB) in non-small cell lung cancer. Oncologist 24: 820–828. 10.1634/theoncologist.2018-0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae YK, Davis AA, Raparia K, Agte S, Pan A, Mohindra N, Villaflor V, Giles F. 2019b. Association of tumor mutational burden with DNA repair mutations and response to anti-PD-1/PD-L1 therapy in non-small-cell lung cancer. Clin Lung Cancer 20: 88–96.e6. 10.1016/j.cllc.2018.09.008 [DOI] [PubMed] [Google Scholar]
- Chan BA, Hughes BG. 2015. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res 4: 36–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SH, Wang LH. 2015. Regulation of cancer metastasis by microRNAs. J Biomedl Sci 22: 9. 10.1186/s12929-015-0113-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AW, Chau SL, Tong JH, Chow C, Kwan JSH, Chung LY, Lung RW, Tong CY, Tin EK, Law PP, et al. 2019a. The landscape of actionable molecular alterations in immunomarker-defined large-cell carcinoma of the lung. J Thorac Oncol 14: 1213–1222. 10.1016/j.jtho.2019.03.021 [DOI] [PubMed] [Google Scholar]
- Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, Peters S. 2019b. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 30: 44–56. 10.1093/annonc/mdy495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JS, Chen LT, Shan YS, Lin SF, Hsiao SY, Tsai CR, Yu SJ, Tsai HJ. 2015. Comprehensive analysis of the incidence and survival patterns of lung cancer by histologies, including rare subtypes, in the era of molecular medicine and targeted therapy: a nation-wide cancer registry-based study from Taiwan. Medicine (Baltimore) 94: e969. 10.1097/MD.0000000000000969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MT, Penson A, Desai NB, Socci ND, Shen R, Seshan VE, Kundra R, Abeshouse A, Viale A, Cha EK, et al. 2018. Small-cell carcinomas of the bladder and lung are characterized by a convergent but distinct pathogenesis. Clin Cancer Res 24: 1965–1973. 10.1158/1078-0432.CCR-17-2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Sasson A, Srinivasan S, Golhar R, Greenawalt DM, Geese WJ, Green G, Zerba K, Kirov S, Szustakowski J. 2019. Bioinformatic methods and bridging of assay results for reliable tumor mutational burden assessment in non-small-cell lung cancer. Mol Diagn Ther 23: 507–520. 10.1007/s40291-019-00408-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerla A, Gevaert O. 2019. Deep learning with multimodal representation for pancancer prognosis prediction. Bioinformatics 35: i446–i454. 10.1093/bioinformatics/btz342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li X, Chen X. 2015a. Prognostic significance of tissue miR-345 downregulation in non-small cell lung cancer. Int J Clin Exp Med 8: 20971–20976. [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Zhang EN, Zhong ZK, Jiang MZ, Yang XF, Zhou DM, Wang XW. 2015b. MicroRNA-153 expression and prognosis in non-small cell lung cancer. Int J Clin Exp Pathol 8: 8671–8675. [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhang Y, Parra E, Rodriguez J, Behrens C, Akbani R, Lu Y, Kurie JM, Gibbons DL, Mills GB, et al. 2017. Multiplatform-based molecular subtypes of non-small-cell lung cancer. Oncogene 36: 1384–1393. 10.1038/onc.2016.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Ali N, Boasberg P, Ho AS. 2018a. Clinical remission of cutaneous squamous cell carcinoma of the auricle with cetuximab and nivolumab. J Clin Med 7: 10. 10.3390/jcm7010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu X, Li Y, Quan C, Zheng L, Huang K. 2018b. Lung cancer therapy targeting histone methylation: opportunities and challenges. Comput Struct Biotechnol J 16: 211–223. 10.1016/j.csbj.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Teng X, Zhang J, Huang K, Shen Q, Cao H, Luo H, Yuan Y, Teng X. 2019. Molecular features of lung adenocarcinoma in young patients. BMC Cancer 19: 777. 10.1186/s12885-019-5978-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YH, Chen YC, Lin E, Brien R, Jung S, Chen YT, Lee W, Hao Z, Sahoo S, Kang HM, et al. 2019. Hydro-seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells. Nat Commun 10: 2163. 10.1038/s41467-019-10122-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CHY, Juan HF. 2017. Quantitative proteomics in lung cancer. J Biomed Sci 24: 37. 10.1186/s12929-017-0343-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman LB, Pasquinelli AE. 2019. miRNA targeting: growing beyond the seed. Trends Genet 35: 215–222. 10.1016/j.tig.2018.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirieac LR, Kobzik L. 2017. Pathology and molecular pathology of lung cancer. In Pathology and epidemiology of cancer (ed. Loda M, et al. ), pp. 367–390. Springer, Berlin. [Google Scholar]
- Chiu YC, Chen HIH, Zhang T, Zhang S, Gorthi A, Wang LJ, Huang Y, Chen Y. 2019. Predicting drug response of tumors from integrated genomic profiles by deep neural networks. BMC Med Genomics 12: 18. 10.1186/s12920-018-0460-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi IH, Lee B, Han J, Yi CA, Choi YS, Ahn JS. 2013. Micropapillary mucinous adenocarcinoma of the lung: a brief case report. Korean J Pathol 47: 603–605. 10.4132/KoreanJPathol.2013.47.6.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooback N, Ho C, Shen Y, Tsang ES, Zhao Y, Mungall AJ, Moore R, Lim HJ, Renouf DJ, Gelmon KA, et al. 2017. Whole genome and transcriptome sequencing of lung cancer: options for personalized cancer treatment. J Clin Oncol 35: e20567–e20567. 10.1200/JCO.2017.35.15_suppl.e20567 [DOI] [Google Scholar]
- Cirenajwis H, Lauss M, Planck M, Vallon-Christersson J, Staaf J. 2020. Performance of gene expression-based single sample predictors for assessment of clinicopathological subgroups and molecular subtypes in cancers: a case comparison study in non-small cell lung cancer. Brief Bioinformatics 21: 729–740. 10.1093/bib/bbz008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirmena G, Franceschelli P, Isnaldi E, Ferrando L, De Mariano M, Ballestrero A, Zoppoli G. 2018. Squalene epoxidase as a promising metabolic target in cancer treatment. Cancer Lett 425: 13–20. 10.1016/j.canlet.2018.03.034 [DOI] [PubMed] [Google Scholar]
- Clark MM, Hildreth A, Batalov S, Ding Y, Chowdhury S, Watkins K, Ellsworth K, Camp B, Kint CI, Yacoubian C, et al. 2019. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci Transl Med 11: eaat6177. 10.1126/scitranslmed.aat6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Lung Cancer Genome Project (CLCGP); Network Genomic Medicine (NGM). 2013. A genomics-based classification of human lung tumors. Sci Transl Med 5: 209ra153–209ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco E, Scaltriti M, Drilon A. 2018. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 15: 731–747. 10.1038/s41571-018-0113-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coco S, Truini A, Vanni I, Dal Bello MG, Alama A, Rijavec E, Genova C, Barletta G, Sini C, Burrafato G, et al. 2015. Next generation sequencing in non-small cell lung cancer: new avenues toward the personalized medicine. Curr Drug Targets 16: 47–59. 10.2174/1389450116666141210094640 [DOI] [PubMed] [Google Scholar]
- Coebergh van den Braak RRJ, Sieuwerts AM, Lalmahomed ZS, Smid M, Wilting SM, Bril SI, Xiang S, van der Vlugt-Daane M, de Weerd V, van Galen A, et al. 2018. Confirmation of a metastasis-specific microRNA signature in primary colon cancer. Sci Rep 8: 5242. 10.1038/s41598-018-22532-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, Schmidt CM, Yip-Schneider MT, Allen PJ, Schattner M, et al. 2017. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci 114: 10202–10207. 10.1073/pnas.1704961114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, et al. 2018. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359: 926–930. 10.1126/science.aar3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway AM, Mitchell C, Kilgour E, Brady G, Dive C, Cook N. 2019. Molecular characterisation and liquid biomarkers in carcinoma of unknown primary (CUP): taking the “U” out of “CUP.” Br J Cancer 120: 141–153. 10.1038/s41416-018-0332-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N, Tsirigos A. 2018. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med 24: 1559–1567. 10.1038/s41591-018-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture HD, Williams LA, Geradts J, Nyante SJ, Butler EN, Marron JS, Perou CM, Troester MA, Niethammer M. 2018. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer 4: 30. 10.1038/s41523-018-0079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. 2014. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov 13: 828–851. 10.1038/nrd4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, Jensen SØ, Medina JE, Hruban C, White JR, et al. 2019. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570: 385–389. 10.1038/s41586-019-1272-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Roa AA, Arevalo Ovalle JE, Madabhushi A, Gonzalez Osorio FA. 2013. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. Med Image Comput Comput Assist Interv 16: 403–410. [DOI] [PubMed] [Google Scholar]
- Cruz-Roa A, Arévalo J, Judkins A, Madabhushi A, González F. 2015. A method for medulloblastoma tumor differentiation based on convolutional neural networks and transfer learning. In Proceedings of the 11th International Symposium on Medical Information Processing and Analysis, 968103, Volume 9681. [Google Scholar]
- Cui R, Meng W, Sun HL, Kim T, Ye Z, Fassan M, Jeon YJ, Li B, Vicentini C, Peng Y, et al. 2015. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc Natl Acad Sci 112: E4288–E4297. 10.1073/pnas.1502068112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaskos C, Tomos I, Garmpis N, Karakatsani A, Dimitroulis D, Garmpi A, Spartalis E, Kampolis CF, Tsagkari E, Loukeri AA, et al. 2018. Histone deacetylase inhibitors as a novel targeted therapy against non-small cell lung cancer: where are we now and what should we expect? Anticancer Res 38: 37–43. [DOI] [PubMed] [Google Scholar]
- Damiani C, Maspero D, Di Filippo M, Colombo R, Pescini D, Graudenzi A, Westerhoff HV, Alberghina L, Vanoni M, Mauri G. 2019. Integration of single-cell RNA-seq data into population models to characterize cancer metabolism. PLoS Comput Biol 15: e1006733. 10.1371/journal.pcbi.1006733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin CR. 1837–1838. Notebook B [Transmutation of species (1837–1838)], CUL-DAR121 (transcribed by Kees Rookmaaker). Darwin Online, http://darwin-online.org.uk
- Davis AA, Chae YK, Agte S, Pan A, Mohindra NA, Villaflor VM, Giles FJ. 2017. Association of tumor mutational burden with smoking and mutation status in non-small cell lung cancer (NSCLC). J Clin Oncol 35: 24–24. 10.1200/JCO.2017.35.7_suppl.24 [DOI] [PubMed] [Google Scholar]
- de Groot PM, Wu CC, Carter BW, Munden RF. 2018. The epidemiology of lung cancer. Transl Lung Cancer Res 7: 220–233. 10.21037/tlcr.2018.05.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demes M, Aszyk C, Bartsch H, Schirren J, Fisseler-Eckhoff A. 2016. Differential miRNA-expression as an adjunctive diagnostic tool in neuroendocrine tumors of the lung. Cancers (Basel) 8: 38. 10.3390/cancers8040038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo DG, Ruffell B. 2019. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 19: 369–382. 10.1038/s41577-019-0127-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Chen H, Liu H, Ai J, Li Y, Pirooznia M, Borgia JA, Liptay MJ, Fidler MJ, Bonomi P. 2014. Circulating microRNA profiling for early detection of non-small cell lung cancer. J Clin Oncol 32: e22051–e22051. 10.1200/jco.2014.32.15_suppl.e22051 [DOI] [Google Scholar]
- Deng Q, Hu H, Yu X, Liu S, Wang L, Chen W, Zhang C, Zeng Z, Cao Y, Xu-Monette ZY, et al. 2019. Tissue-specific microRNA expression alters cancer susceptibility conferred by a TP53 noncoding variant. Nat Commun 10: 5061. 10.1038/s41467-019-13002-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derks JL, Leblay N, Thunnissen E, van Suylen RJ, den Bakker M, Groen HJM, Smit EF, Damhuis R, van den Broek EC, Charbrier A, et al. 2018. Molecular subtypes of pulmonary large-cell neuroendocrine carcinoma predict chemotherapy treatment outcome. Clin Cancer Res 24: 33–42. 10.1158/1078-0432.CCR-17-1921 [DOI] [PubMed] [Google Scholar]
- Dhanasekaran SM, Balbin OA, Chen G, Nadal E, Kalyana-Sundaram S, Pan J, Veeneman B, Cao X, Malik R, Vats P, et al. 2014. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nat Commun 5: 5893. 10.1038/ncomms6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan A, Scott JG, Harris AL, Buffa FM. 2018. Pan-cancer characterisation of microRNA across cancer hallmarks reveals microRNA-mediated downregulation of tumour suppressors. Nat Commun 9: 5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diossy M, Sztupinszki Z, Borcsok J, Krzystanek M, Tisza V, Spisak S, Rusz O, Timar J, Csabai I, Fillinger J, et al. 2021. A subset of lung cancer cases shows robust signs of homologous recombination deficiency associated genomic mutational signatures. NPJ Precis Oncol 5: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuric U, Zadeh G, Aldape K, Diamandis P. 2017. Precision histology: how deep learning is poised to revitalize histomorphology for personalized cancer care. NPJ Precis Oncol 1: 22. 10.1038/s41698-017-0022-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YJ, Cai YR, Zhou LJ, Su D, Mu J, Chen XJ, Zhang LI. 2016. Association between the histological subtype of lung adenocarcinoma, EGFR/KRAS mutation status and the ALK rearrangement according to the novel IASLC/ATS/ERS classification. Oncol Lett 11: 2552–2558. 10.3892/ol.2016.4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Li B, Lin D, Zhou Q, Huang D. 2019. Advances in targeted therapy and immunotherapy for non-small cell lung cancer based on accurate molecular typing. Front Pharmacol 10: 230. 10.3389/fphar.2019.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnem T, Hald SM, Paulsen EE, Richardsen E, Al-Saad S, Kilvaer TK, Brustugun OT, Helland A, Lund-Iversen M, Poehl M, et al. 2015. Stromal CD8+ T-cell density—a promising supplement to TNM staging in non–small cell lung cancer. Clin Cancer Res 21: 2635–2643. 10.1158/1078-0432.CCR-14-1905 [DOI] [PubMed] [Google Scholar]
- Donnem T, Kilvaer TK, Andersen S, Richardsen E, Paulsen EE, Hald SM, Al-Saad S, Brustugun OT, Helland A, Lund-Iversen M, et al. 2016. Strategies for clinical implementation of TNM-immunoscore in resected nonsmall-cell lung cancer. Ann Oncol 27: 225–232. 10.1093/annonc/mdv560 [DOI] [PubMed] [Google Scholar]
- Dorward DA, Walsh K, Oniscu A, Wallace WA. 2017. Molecular pathology of non-small cell lung cancer. Diagnostic Histopathology 23: 450–457. 10.1016/j.mpdhp.2017.08.002 [DOI] [Google Scholar]
- Drapkin BJ, Farago AF. 2019. Unexpected synergy reveals new therapeutic strategy in SCLC. Trends Pharmacol Sci 40: 295–297. 10.1016/j.tips.2019.03.005 [DOI] [PubMed] [Google Scholar]
- Drapkin BJ, George J, Christensen CL, Mino-Kenudson M, Dries R, Sundaresan T, Phat S, Myers DT, Zhong J, Igo P, et al. 2018. Genomic and functional fidelity of small cell lung cancer patient-derived xenografts. Cancer Discov 8: 600–615. 10.1158/2159-8290.CD-17-0935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duréndez-Sáez E, Azkárate A, Meri M, Calabuig-Fariñas S, Aguilar-Gallardo C, Blasco A, Jantus-Lewintre E, Camps C. 2017. New insights in non-small-cell lung cancer: circulating tumor cells and cell-free DNA. J Thorac Dis 9: S1332–s1345. 10.21037/jtd.2017.06.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duruisseaux M, Esteller M. 2018. Lung cancer epigenetics: from knowledge to applications. Semin Cancer Biol 51: 116–128. 10.1016/j.semcancer.2017.09.005 [DOI] [PubMed] [Google Scholar]
- Ehteshami Bejnordi B, Mullooly M, Pfeiffer RM, Fan S, Vacek PM, Weaver DL, Herschorn S, Brinton LA, van Ginneken B, Karssemeijer N, et al. 2018. Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod Pathol 31: 1502–1512. 10.1038/s41379-018-0073-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. 2007. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316: 1039–1043. 10.1126/science.1141478 [DOI] [PubMed] [Google Scholar]
- Fakih M, O'Neil B, Price TJ, Falchook GS, Desai J, Kuo J, Govindan R, Rasmussen E, Morrow PKH, Ngang J, et al. 2019. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors. J Clin Oncol 37: 3003–3003. 10.1200/JCO.2019.37.15_suppl.3003 [DOI] [Google Scholar]
- Fang W, Hong S, Chen N, He X, Zhan J, Qin T, Zhou T, Hu Z, Ma Y, Zhao Y, et al. 2015. PD-L1 is remarkably over-expressed in EBV-associated pulmonary lymphoepithelioma-like carcinoma and related to poor disease-free survival. Oncotarget 6: 33019–33032. 10.18632/oncotarget.5028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farago AF, Taylor MS, Doebele RC, Zhu VW, Kummar S, Spira AI, Boyle TA, Haura EB, Arcila ME, Benayed R, et al. 2018. Clinicopathologic features of non-small-cell lung cancer harboring an NTRK gene fusion. JCO Precis Oncol 2018: PO.18.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei X, Zhang J, Zhao Y, Sun M, Zhao H, Li S. 2018. miR-96 promotes invasion and metastasis by targeting GPC3 in non-small cell lung cancer cells. Oncol Lett 15: 9081–9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felley-Bosco E. 2018. Special issue on mechanisms of mesothelioma heterogeneity: highlights and open questions. Int J Mol Sci 19: 3560. 10.3390/ijms19113560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cuesta L, Sun R, Menon R, George J, Lorenz S, Meza-Zepeda LA, Peifer M, Plenker D, Heuckmann JM, Leenders F, et al. 2015. Identification of novel fusion genes in lung cancer using breakpoint assembly of transcriptome sequencing data. Genome Biol 16: 7. 10.1186/s13059-014-0558-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala EM, Jayakumaran G, Mauguen A, Kennedy JA, Bouvier N, Kemel Y, Fleischut MH, Maio A, Salo-Mullen EE, Sheehan M, et al. 2021. Prospective pan-cancer germline testing using MSK-IMPACT informs clinical translation in 751 patients with pediatric solid tumors. Nat Cancer 2: 357–365. 10.1038/s43018-021-00172-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipska M, Skrzypski M, Czetyrbok K, Stokowy T, Stasiłojć G, Supernat A, Jassem J, Żaczek AJ, Bigda J. 2018. MiR-192 and miR-662 enhance chemoresistance and invasiveness of squamous cell lung carcinoma. Lung Cancer 118: 111–118. 10.1016/j.lungcan.2018.02.002 [DOI] [PubMed] [Google Scholar]
- Fois AG, Diana G, Arcadu A, Marras V, Crivelli P, Putzu C, Ginesu GC, Canu S, Pirina P. 2017. Bronchial mucoepidermoid carcinoma: a case report. Int J Surg Case Rep 31: 159–162. 10.1016/j.ijscr.2017.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde PM, Scherpereel A, Tsao AS. 2019. Use of immune checkpoint inhibitors in mesothelioma. Curr Treat Options Oncol 20: 18. 10.1007/s11864-019-0613-x [DOI] [PubMed] [Google Scholar]
- Foy V, Fernandez-Gutierrez F, Faivre-Finn C, Dive C, Blackhall F. 2017. The clinical utility of circulating tumour cells in patients with small cell lung cancer. Transl Lung Cancer Res 6: 409–417. 10.21037/tlcr.2017.07.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, Akimov M, Bufill J, Lee C, Jentz D, et al. 2015. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 5: 850–859. 10.1158/2159-8290.CD-15-0285 [DOI] [PubMed] [Google Scholar]
- Fukumura D, Kloepper J, Amoozgar Z, Duda DG, Jain RK. 2018. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol 15: 325–340. 10.1038/nrclinonc.2018.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach S, Jantus-Lewintre E, Calabuig-Farinas S, Montaner D, Alonso S, Sirera R, Blasco A, Uso M, Guijarro R, Martorell M, et al. 2017. MicroRNA profiling associated with non-small cell lung cancer: next generation sequencing detection, experimental validation, and prognostic value. Oncotarget 8: 56143–56157. 10.18632/oncotarget.18603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, Rittmeyer A, Fehrenbacher L, Otto G, Malboeuf C, et al. 2018. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 24: 1441–1448. 10.1038/s41591-018-0134-3 [DOI] [PubMed] [Google Scholar]
- Gandhi L, Janne PA, Opyrchal M, Ramalingam SS, Rybkin II, Hafez N, Raez LE, Gabrilovich D, Wang F, Ordentlich P, et al. 2018. Efficacy and safety of entinostat (ENT) and pembrolizumab (PEMBRO) in patients with non-small cell lung cancer (NSCLC) previously treated with anti-PD-(L)1 therapy. J Clin Oncol 36: 9036–9036. 10.1200/JCO.2018.36.15_suppl.9036 [DOI] [Google Scholar]
- Gao W, Shen H, Liu L, Xu J, Xu J, Shu Y. 2011. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol 137: 557–566. 10.1007/s00432-010-0918-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Shang Y, Wang X, Ma Y, Yang F, Wang J, Chen K, Zhang Y. 2019. Application of circulating tumor DNA for dynamic monitoring of advanced non-small cell lung cancer treatment response: an open-label, multicenter, prospective, observational study protocol. Thorac Cancer 10: 1310–1315. 10.1111/1759-7714.13031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinet S, Laurent-Puig P, Blons H, Oudart JB. 2018. Current and future molecular testing in NSCLC, what can we expect from new sequencing technologies? J Clin Med 7: 144. 10.3390/jcm7060144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughan EM, Costa DB. 2011. Genotype-driven therapies for non-small cell lung cancer: focus on EGFR, KRAS and ALK gene abnormalities. Ther Adv Med Oncol 3: 113–125. 10.1177/1758834010397569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G, et al. 2015. Comprehensive genomic profiles of small cell lung cancer. Nature 524: 47–53. 10.1038/nature14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertych A, Swiderska-Chadaj Z, Ma Z, Ing N, Markiewicz T, Cierniak S, Salemi H, Guzman S, Walts AE, Knudsen BS. 2019. Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci Rep 9: 1483. 10.1038/s41598-018-37638-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierman HJ, Goldfarb S, Labrador M, Weipert CM, Getty B, Skrzypczak SM, Catasus C, Carbral S, Singaraju M, Singleton N, et al. 2019. Genomic testing and treatment landscape in patients with advanced non-small cell lung cancer (aNSCLC) using real-world data from community oncology practices. J Clin Oncol 37: 1585–1585. 10.1200/JCO.2019.37.15_suppl.1585 [DOI] [Google Scholar]
- Gnjatic S, Bronte V, Brunet LR, Butler MO, Disis ML, Galon J, Hakansson LG, Hanks BA, Karanikas V, Khleif SN, et al. 2017. Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer 5: 44. 10.1186/s40425-017-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. 2017. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Therapeut 16: 2598–2608. 10.1158/1535-7163.MCT-17-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco FA, Pavlidis N. 2009. Treatment for patients with unknown primary carcinoma and unfavorable prognostic factors. Semin Oncol 36: 65–74. 10.1053/j.seminoncol.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Greco FA, Spigel DR, Yardley DA, Erlander MG, Ma XJ, Hainsworth JD. 2010. Molecular profiling in unknown primary cancer: accuracy of tissue of origin prediction. Oncologist 15: 500–506. 10.1634/theoncologist.2009-0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco FA, Oien K, Erlander M, Osborne R, Varadhachary G, Bridgewater J, Cohen D, Wasan H. 2012. Cancer of unknown primary: progress in the search for improved and rapid diagnosis leading toward superior patient outcomes. Ann Oncol 23: 298–304. 10.1093/annonc/mdr306 [DOI] [PubMed] [Google Scholar]
- Greco FA, Lennington WJ, Spigel DR, Hainsworth JD. 2015. Poorly differentiated neoplasms of unknown primary site: diagnostic usefulness of a molecular cancer classifier assay. Mol Diagn Ther 19: 91–97. 10.1007/s40291-015-0133-8 [DOI] [PubMed] [Google Scholar]
- Greillier L, Tomasini P, Barlesi F. 2018. The clinical utility of tumor mutational burden in non-small cell lung cancer. Transl Lung Cancer Res 7: 639–646. 10.21037/tlcr.2018.10.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144. 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F, Dahmani A, Lameiras S, Reyal F, Frenoy O, et al. 2019. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet 51: 1060–1066. 10.1038/s41588-019-0424-9 [DOI] [PubMed] [Google Scholar]
- Guan P, Yin Z, Li X, Wu W, Zhou B. 2012. Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J Exp Clin Cancer Res 31: 54. 10.1186/1756-9966-31-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Liu S, Zhang S, Chen Q, Zhang M, Quan P, Sun X. 2017. Human papillomavirus infection as a prognostic marker for lung adenocarcinoma: a systematic review and meta-analysis. Oncotarget 8: 34507–34515. 10.18632/oncotarget.15671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez ME, Choi K, Lanman RB, Licitra EJ, Skrzypczak SM, Pe Benito R, Wu T, Arunajadai S, Kaur S, Harper H, et al. 2017. Genomic profiling of advanced non-small cell lung cancer in community settings: gaps and opportunities. Clin Lung Cancer 18: 651–659. 10.1016/j.cllc.2017.04.004 [DOI] [PubMed] [Google Scholar]
- Hainsworth JD, Greco FA. 2014. Gene expression profiling in patients with carcinoma of unknown primary site: from translational research to standard of care. Virchows Arch 464: 393–402. 10.1007/s00428-014-1545-2 [DOI] [PubMed] [Google Scholar]
- Hainsworth JD, Rubin MS, Spigel DR, Boccia RV, Raby S, Quinn R, Greco FA. 2013. Molecular gene expression profiling to predict the tissue of origin and direct site-specific therapy in patients with carcinoma of unknown primary site: a prospective trial of the Sarah Cannon research institute. J Clin Oncol 31: 217–223. 10.1200/JCO.2012.43.3755 [DOI] [PubMed] [Google Scholar]
- Hamada S, Satoh K, Fujibuchi W, Hirota M, Kanno A, Unno J, Masamune A, Kikuta K, Kume K, Shimosegawa T. 2012. MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res 10: 3–10. 10.1158/1541-7786.MCR-11-0272 [DOI] [PubMed] [Google Scholar]
- Han Y, Yang J, Qian X, Cheng WC, Liu SH, Hua X, Zhou L, Yang Y, Wu Q, Liu P, et al. 2019. DriverML: a machine learning algorithm for identifying driver genes in cancer sequencing studies. Nucleic Acids Res 47: e45–e45. 10.1093/nar/gkz096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handorf CR, Kulkarni A, Grenert JP, Weiss LM, Rogers WM, Kim OS, Monzon FA, Halks-Miller M, Anderson GG, Walker MG, et al. 2013. A multicenter study directly comparing the diagnostic accuracy of gene expression profiling and immunohistochemistry for primary site identification in metastatic tumors. Am J Surg Pathol 37: 1067–1075. 10.1097/PAS.0b013e31828309c4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida Y, Daibata M. 2014. Considerations on the link between Merkel cell polyomavirus and lung cancer. Lung Cancer Manag 3: 297–299. 10.2217/lmt.14.20 [DOI] [Google Scholar]
- Hassan R, Morrow B, Thomas A, Walsh T, Lee MK, Gulsuner S, Gadiraju M, Panou V, Gao S, Mian I, et al. 2019. Inherited predisposition to malignant mesothelioma and overall survival following platinum chemotherapy. Proc Natl Acad Sci 116: 9008–9013. 10.1073/pnas.1821510116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Chen SY, Yang Z, Zhang J, Wang W, Liu MY, Niu Y, Wei XM, Li HM, Hu WN, et al. 2018. miR-4317 suppresses non-small cell lung cancer (NSCLC) by targeting fibroblast growth factor 9 (FGF9) and cyclin D2 (CCND2). J Exp Clin Cancer Res CR 37: 230. 10.1186/s13046-018-0882-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeke AL, Pishvaian MJ, Lynce F, Xiu J, Brody JR, Chen WJ, Baker TM, Marshall JL, Isaacs C. 2018. Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis Oncol 2018: PO.17.00286. 10.1200/PO.17.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heist RS, Sequist LV, Engelman JA. 2012. Genetic changes in squamous cell lung cancer: a review. J Thorac Oncol 7: 924–933. 10.1097/JTO.0b013e31824cc334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers S, Salman P, et al. 2018. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 378: 2093–2104. 10.1056/NEJMoa1801946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks LE, Rouleau E, Besse B. 2018. Clinical utility of tumor mutational burden in patients with non-small cell lung cancer treated with immunotherapy. Transl Lung Cancer Res 7: 647–660. 10.21037/tlcr.2018.09.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, et al. 2016. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387: 1540–1550. 10.1016/S0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- Hinrichs JW, van Blokland WT, Moons MJ, Radersma RD, Radersma-van Loon JH, de Voijs CM, Rappel SB, Koudijs MJ, Besselink NJ, Willems SM, et al. 2015. Comparison of next-generation sequencing and mutation-specific platforms in clinical practice. Am J Clin Pathol 143: 573–578. 10.1309/AJCP40XETVYAMJPY [DOI] [PubMed] [Google Scholar]
- Hirsch FR, Kerr KM, Bunn PA, Kim ES, Obasaju C, Pérol M, Bonomi P, Bradley JD, Gandara D, Jett JR, et al. 2018. Molecular and immune biomarker testing in squamous-cell lung cancer: effect of current and future therapies and technologies. Clin Lung Cancer 19: 331–339. 10.1016/j.cllc.2018.03.014 [DOI] [PubMed] [Google Scholar]
- Hmeljak J, Sanchez-Vega F, Hoadley KA, Shih J, Stewart C, Heiman D, Tarpey P, Danilova L, Drill E, Gibb EA, et al. 2018. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov 8: 1548–1565. 10.1158/2159-8290.CD-18-0804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et al. 2018. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173: 291–304.e6. 10.1016/j.cell.2018.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs GA, Der CJ, Rossman KL. 2016. RAS isoforms and mutations in cancer at a glance. J Cell Sci 129: 1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman V, Bonnetaud C, Ilie MI, Vielh P, Vignaud JM, Fléjou JF, Lantuejoul S, Piaton E, Mourad N, Butori C, et al. 2011a. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res 17: 827–835. 10.1158/1078-0432.CCR-10-0445 [DOI] [PubMed] [Google Scholar]
- Hofman V, Ilie MI, Long E, Selva E, Bonnetaud C, Molina T, Vénissac N, Mouroux J, Vielh P, Hofman P. 2011b. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay and the isolation by size of epithelial tumor cell method. Int J Cancer 129: 1651–1660. 10.1002/ijc.25819 [DOI] [PubMed] [Google Scholar]
- Hofman V, Heeke S, Marquette CH, Ilie M, Hofman P. 2019. Circulating tumor cell detection in lung cancer: but to what end? Cancers (Basel) 11: 262. 10.3390/cancers11020262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, Bussink J, Gillies RJ, Mak RH, Aerts H. 2018. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med 15: e1002711. 10.1371/journal.pmed.1002711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Yu Z. 2019. Diagnosis of mesothelioma with deep learning. Oncol Lett 17: 1483–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, et al. 2010. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol 28: 1721–1726. 10.1200/JCO.2009.24.9342 [DOI] [PubMed] [Google Scholar]
- Hu H, Pan Y, Li Y, Wang L, Wang R, Zhang Y, Li H, Ye T, Zhang Y, Luo X, et al. 2014. Oncogenic mutations are associated with histological subtypes but do not have an independent prognostic value in lung adenocarcinoma. Onco Targets Ther 7: 1423–1437. 10.2147/OTT.S58900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Zhou Y, Wang Q, Yang Z, Shi Y, Chi Q. 2019a. Gene expression classification of lung adenocarcinoma into molecular subtypes. IEEE/ACM Trans Comput Biol Bioinform 17: 1187–1197. [DOI] [PubMed] [Google Scholar]
- Hu J, Wang Y, Zhang Y, Yu Y, Chen H, Liu K, Yao M, Wang K, Gu W, Shou T. 2019b. Comprehensive genomic profiling of small cell lung cancer in Chinese patients and the implications for therapeutic potential. Cancer Med 8: 4338–4347. 10.1002/cam4.2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FW, Feng FY. 2019. A tumor-agnostic NTRK (TRK) inhibitor. Cell 177: 8. 10.1016/j.cell.2019.02.049 [DOI] [PubMed] [Google Scholar]
- Huang J, Wu J, Li Y, Li X, Yang T, Yang Q, Jiang Y. 2014. Deregulation of serum microRNA expression is associated with cigarette smoking and lung cancer. Biomed Res Int 2014: 364316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Ni M, Chalishazar MD, Huffman KE, Kim J, Cai L, Shi X, Cai F, Zacharias LG, Ireland AS, et al. 2018a. Inosine monophosphate dehydrogenase dependence in a subset of small cell lung cancers. Cell Metab 28: 369–382.e5. 10.1016/j.cmet.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Xia J, Wang L, Wang X, Ma X, Deng Q, Lu Y, Kumar M, Zhou Z, Li L, et al. 2018b. miR-153 suppresses IDO1 expression and enhances CAR T cell immunotherapy. J Hematol Oncol 11: 58. 10.1186/s13045-018-0600-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PJ, Chiu LY, Lee CC, Yeh YM, Huang KY, Chiu CH, Tang P. 2018c. mSignatureDB: a database for deciphering mutational signatures in human cancers. Nucleic Acids Res 46: D964–D970. 10.1093/nar/gkx1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylebos M, Van Camp G, van Meerbeeck JP, Op de Beeck K. 2016. The genetic landscape of malignant pleural mesothelioma: results from massively parallel sequencing. J Thorac Oncol 11: 1615–1626. 10.1016/j.jtho.2016.05.020 [DOI] [PubMed] [Google Scholar]
- Imamura F, Uchida J, Kukita Y, Kumagai T, Nishino K, Inoue T, Kimura M, Oba S, Kato K. 2016. Monitoring of treatment responses and clonal evolution of tumor cells by circulating tumor DNA of heterogeneous mutant EGFR genes in lung cancer. Lung Cancer 94: 68–73. 10.1016/j.lungcan.2016.01.023 [DOI] [PubMed] [Google Scholar]
- Inamura K. 2017. Lung cancer: understanding its molecular pathology and the 2015 WHO classification. Front Oncol 7: 193. 10.3389/fonc.2017.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakopovic M, Thomas A, Balasubramaniam S, Schrump D, Giaccone G, Bates SE. 2013. Targeting the epigenome in lung cancer: expanding approaches to epigenetic therapy. Front Oncol 3: 261. 10.3389/fonc.2013.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, et al. 2017. Tracking the evolution of non-small-cell lung cancer. N Engl J Med 376: 2109–2121. 10.1056/NEJMoa1616288 [DOI] [PubMed] [Google Scholar]
- Janiszewska M, Polyak K. 2018. A confetti trail of tumour evolution. Nat Cell Biol 20: 639–641. 10.1038/s41556-018-0110-7 [DOI] [PubMed] [Google Scholar]
- Jia D, Augert A, Kim DW, Eastwood E, Wu N, Ibrahim AH, Kim KB, Dunn CT, Pillai PS, Gazdar AF, et al. 2018. Crebbp loss drives small cell lung cancer and increases sensitivity to HDAC inhibition. Cancer Discov 8: 1422–1437. 10.1158/2159-8290.CD-18-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Guo F, Liu X, Li X, Qin Q, Shu P, Li Y, Wang Y. 2019. Continuous targeted kinase inhibitors treatment induces upregulation of PD-L1 in resistant NSCLC. Sci Rep 9: 3705. 10.1038/s41598-018-38068-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, Chang MT, Ni A, Kundra R, Jonsson P, et al. 2017. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov 7: 596–609. 10.1158/2159-8290.CD-16-1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph NM, Chen YY, Nasr A, Yeh I, Talevich E, Onodera C, Bastian BC, Rabban JT, Garg K, Zaloudek C, et al. 2017. Genomic profiling of malignant peritoneal mesothelioma reveals recurrent alterations in epigenetic regulatory genes BAP1, SETD2, and DDX3X. Mod Pathol 30: 246–254. 10.1038/modpathol.2016.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalemkerian GP, Narula N, Kennedy EB, Biermann WA, Donington J, Leighl NB, Lew M, Pantelas J, Ramalingam SS, Reck M, et al. 2018. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J Clin Oncol 36: 911–919. 10.1200/JCO.2017.76.7293 [DOI] [PubMed] [Google Scholar]
- Kamata T, Yoshida A, Shiraishi K, Furuta K, Kosuge T, Watanabe S, Asamura H, Tsuta K. 2016. Mucinous micropapillary pattern in lung adenocarcinomas: a unique histology with genetic correlates. Histopathology 68: 356–366. 10.1111/his.12763 [DOI] [PubMed] [Google Scholar]
- Kandalaft PL, Gown AM. 2016. Practical applications in immunohistochemistry: carcinomas of unknown primary site. Arch Pathol Lab Med 140: 508–523. 10.5858/arpa.2015-0173-CP [DOI] [PubMed] [Google Scholar]
- Karim NA, Schuster J, Eldessouki I, Gaber O, Namad T, Wang J, Xie C, Morris JC. 2018. Pulmonary sarcomatoid carcinoma: University of Cincinnati experience. Oncotarget 9: 4102–4108. 10.18632/oncotarget.23468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, et al. 2019. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 25: 1054–1056. 10.1038/s41591-019-0462-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Tomson BN, Buys TP, Elkin SK, Carter JL, Kurzrock R. 2016. Genomic landscape of malignant mesotheliomas. Mol Cancer Ther 15: 2498–2507. 10.1158/1535-7163.MCT-16-0229 [DOI] [PubMed] [Google Scholar]
- Kaznatcheev A, Peacock J, Basanta D, Marusyk A, Scott JG. 2019. Fibroblasts and alectinib switch the evolutionary games played by non-small cell lung cancer. Nat Ecol Evol 3: 450–456. 10.1038/s41559-018-0768-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenmotsu H, Serizawa M, Koh Y, Isaka M, Takahashi T, Taira T, Ono A, Maniwa T, Takahashi S, Mori K, et al. 2014. Prospective genetic profiling of squamous cell lung cancer and adenosquamous carcinoma in Japanese patients by multitarget assays. BMC Cancer 14: 786. 10.1186/1471-2407-14-786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr SE, Schnabel CA, Sullivan PS, Zhang Y, Huang VJ, Erlander MG, Brachtel EF, Dry SM. 2014. A 92-gene cancer classifier predicts the site of origin for neuroendocrine tumors. Mod Pathol 27: 44–54. 10.1038/modpathol.2013.105 [DOI] [PubMed] [Google Scholar]
- Kettunen E, Savukoski S, Salmenkivi K, Böhling T, Vanhala E, Kuosma E, Anttila S, Wolff H. 2019. CDKN2A copy number and p16 expression in malignant pleural mesothelioma in relation to asbestos exposure. BMC Cancer 19: 507. 10.1186/s12885-019-5652-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi P, Kazemi E, Imielinski M, Elemento O, Hajirasouliha I. 2018. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine 27: 317–328. 10.1016/j.ebiom.2017.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khunger M, Bordeaux J, Dakappagari N, Vaupel C, Khunger A, Hu B, Schalper KA, Rimm DL, Velcheti V. 2018. Tumor PD-L1 heterogeneity in non-small cell lung cancer: does biopsy size and volume matter? J Clin Oncol 36: 12058–12058. 10.1200/JCO.2018.36.15_suppl.12058 [DOI] [Google Scholar]
- Kim D, Kim DH. 2018. Epigenome-based precision medicine in lung cancer. Methods Mol Biol 1856: 57–85. 10.1007/978-1-4939-8751-1_4 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Oh SY, Kim WS, Kim SJ, Yoo GH, Kim WD, Lee KY. 2013a. Clinical investigation of EGFR mutation detection by pyrosequencing in lung cancer patients. Oncol Lett 5: 271–276. 10.3892/ol.2012.950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Hyun S, Mendiratta S, Kim J, Pecot Chad V, Larsen Jill E, Zubovych I, Seo Bo Y, Kim J, Eskiocak B, Chung H, et al. 2013b. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell 155: 552-566. 10.1016/j.cell.2013.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KT, Lee HW, Lee HO, Kim SC, Seo YJ, Chung W, Eum HH, Nam DH, Kim J, Joo KM, et al. 2015. Single-cell mRNA sequencing identifies subclonal heterogeneity in anti-cancer drug responses of lung adenocarcinoma cells. Genome Biol 16: 127. 10.1186/s13059-015-0692-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yao F, Xiao Z, Sun Y, Ma L. 2018a. MicroRNAs and metastasis: small RNAs play big roles. Cancer Metastasis Rev 37: 5–15. 10.1007/s10555-017-9712-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Pierce CM, Robinson LA. 2018b. Impact of viral presence in tumor on gene expression in non-small cell lung cancer. BMC Cancer 18: 843. 10.1186/s12885-018-4748-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ES, Roy UB, Ersek JL, King J, Smith RA, Martin N, Martins R, Moore A, Silvestri GA, Jett J. 2019. Updates regarding biomarker testing for non-small cell lung cancer: considerations from the national lung cancer roundtable. J Thorac Oncol 14: 338–342. 10.1016/j.jtho.2019.01.002 [DOI] [PubMed] [Google Scholar]
- Kirilovsky A, Marliot F, El Sissy C, Haicheur N, Galon J, Pagès F. 2016. Rational bases for the use of the immunoscore in routine clinical settings as a prognostic and predictive biomarker in cancer patients. Int Immunol 28: 373–382. 10.1093/intimm/dxw021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EA, Hubbell E, Maddala T, Aravanis A, Beausang JF, Filippova D, Gross S, Jamshidi A, Kurtzman K, Shen L, et al. 2018. Development of a comprehensive cell-free DNA (cfDNA) assay for early detection of multiple tumor types: the Circulating Cell-free Genome Atlas (CCGA) study. J Clin Oncol 36: 12021–12021. 10.1200/JCO.2018.36.15_suppl.12021 [DOI] [Google Scholar]
- Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, Fan H, Shen H, Way GP, Greene CS, et al. 2018. Genomic and molecular landscape of dna damage repair deficiency across the Cancer Genome Atlas. Cell Rep 23: 239–254.e6. 10.1016/j.celrep.2018.03.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh Y, Yagi S, Akamatsu H, Kanai K, Hayata A, Tokudome N, Akamatsu K, Higuchi M, Kanbara H, Nakanishi M, et al. 2019. Heterogeneous expression of programmed death receptor-ligand 1 on circulating tumor cells in patients with lung cancer. Clin Lung Cancer 20: 270–277.e1. 10.1016/j.cllc.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Kuleshov V, Ding J, Vo C, Hancock B, Ratner A, Li Y, Ré C, Batzoglou S, Snyder M. 2019. A machine-compiled database of genome-wide association studies. Nat Commun 10: 3341. 10.1038/s41467-019-11026-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummar S, Lassen UN. 2018. TRK inhibition: a new tumor-agnostic treatment strategy. Target Oncol 13: 545–556. 10.1007/s11523-018-0590-1 [DOI] [PubMed] [Google Scholar]
- Kuo PL, Liao SH, Hung JY, Huang MS, Hsu YL. 2013. MicroRNA-33a functions as a bone metastasis suppressor in lung cancer by targeting parathyroid hormone related protein. Biochim Biophys Acta 1830: 3756–3766. 10.1016/j.bbagen.2013.02.022 [DOI] [PubMed] [Google Scholar]
- Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, Bassez A, Decaluwé H, Pircher A, Van den Eynde K, et al. 2018. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med 24: 1277–1289. 10.1038/s41591-018-0096-5 [DOI] [PubMed] [Google Scholar]
- Lamichhane S, Thachil T, De Ieso P, Gee H, Andrew Moss S, Milic N. 2018. Prognostic role of microRNAs in human non-small-cell lung cancer: a systematic review and meta-analysis. Dis Markers 2018: 8309015. 10.1155/2018/8309015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin SM, Kratzke RA, Kelsey KT. 2015. Epigenetics of lung cancer. Transl Res 165: 74–90. 10.1016/j.trsl.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar V, Suo C, Orear C, van den Oord J, Balogh Z, Guegan J, Job B, Meurice G, Ripoche H, Calza S, et al. 2013. Integrated molecular portrait of non-small cell lung cancers. BMC Med Genomics 6: 53. 10.1186/1755-8794-6-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le AD, Alzghari SK, Jean GW, La-Beck NM. 2017. Update on targeted therapies for advanced non-small cell lung cancer: nivolumab in context. Ther Clin Risk Manag 13: 223–236. 10.2147/TCRM.S104343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Um SW. 2017. Intratumoral heterogeneity and biomarkers for transformation into small cell lung carcinomas from lung adenocarcinomas. J Thorac Dis 9: 4248–4250. 10.21037/jtd.2017.10.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Lee J, Kim S, Kim S, Youk J, Park S, An Y, Keam B, Kim DW, Heo DS, et al. 2017. Clonal history and genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J Clin Oncol 35: 3065–3074. 10.1200/JCO.2016.71.9096 [DOI] [PubMed] [Google Scholar]
- Lee J, Park CK, Yoon HK, Sa YJ, Woo IS, Kim HR, Kim SY, Kim TJ. 2019. PD-L1 expression in ROS1-rearranged non-small cell lung cancer: a study using simultaneous genotypic screening of EGFR, ALK, and ROS1. Thorac Cancer 10: 103–110. 10.1111/1759-7714.12917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Q, Lin Y, Jiang F, Lee CJ, Zhan M, Fang H, Wang Y, Jiang F. 2017a. A plasma miRNA signature for lung cancer early detection. Oncotarget 8: 111902–111911. 10.18632/oncotarget.22950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng S, Wu G, Klinge DM, Thomas CL, Casas E, Picchi MA, Stidley CA, Lee SJ, Aisner S, Siegfried JM, et al. 2017b. Gene methylation biomarkers in sputum as a classifier for lung cancer risk. Oncotarget 8: 63978–63985. 10.18632/oncotarget.19255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin HM, Yuan J, Sims PA. 2018. Single-cell transcriptomic analysis of tumor heterogeneity. Trends Cancer 4: 264–268. 10.1016/j.trecan.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Fu XD. 2019. Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat Rev Genet 20: 503–519. 10.1038/s41576-019-0135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lu H. 2018. Adenosquamous carcinoma of the lung. OncoTargets Ther 11: 4829–4835. 10.2147/OTT.S164574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xiao X, Ji X, Liu B, Amos CI. 2015. RNA-seq analysis of lung adenocarcinomas reveals different gene expression profiles between smoking and nonsmoking patients. Tumour Biol 36: 8993–9003. 10.1007/s13277-015-3576-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ, Younes A, et al. 2017a. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 19: 4–23. 10.1016/j.jmoldx.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhao K, Tian H. 2017b. Integrated analysis of differential expression and alternative splicing of non-small cell lung cancer based on RNA sequencing. Oncol Lett 14: 1519–1525. 10.3892/ol.2017.6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qin F, Hu F, Xu H, Sun G, Han G, Wang T, Guo M. 2018a. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci 8: 2. 10.1186/s13578-018-0202-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Balmain A, Counter CM. 2018b. A model for RAS mutation patterns in cancers: finding the sweet spot. Nat Rev Cancer 18: 767–777. 10.1038/s41568-018-0076-6 [DOI] [PubMed] [Google Scholar]
- Li S, Yang J, Xia Y, Fan Q, Yang KP. 2018c. Long noncoding RNA NEAT1 promotes proliferation and invasion via targeting miR-181a-5p in non-small cell lung cancer. Oncol Res 26: 289–296. 10.3727/096504017X15009404458675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang H, Gong H, Yuan Y, Li Y, Wang C, Li W, Zhang Z, Liu M, Liu H, et al. 2018d. miR-182 suppresses invadopodia formation and metastasis in non-small cell lung cancer by targeting cortactin gene. J Exp Clin Cancer Res 37: 141. 10.1186/s13046-018-0824-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Chapuy B, Varelas X, Sebastiani P, Monti S. 2019a. Identification of candidate cancer drivers by integrative Epi-DNA and gene expression (iEDGE) data analysis. Sci Rep 9: 16904. 10.1038/s41598-019-52886-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xiao X, Bosse Y, Gorlova O, Gorlov I, Han Y, Byun J, Leighl N, Johansen JS, Barnett M, et al. 2019b. Genetic interaction analysis among oncogenesis-related genes revealed novel genes and networks in lung cancer development. Oncotarget 10: 1760–1774. 10.18632/oncotarget.26678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Huang J, Wang B, Liu Z, He J, Liang W. 2018a. The role of liquid biopsy in predicting post-operative recurrence of non-small cell lung cancer. J Thorac Dis 10: S838–S845. 10.21037/jtd.2018.04.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Zhao Y, Huang W, Liang H, Zeng H, He J. 2018b. Liquid biopsy for early stage lung cancer. J Thorac Dis 10: S876–S881. 10.21037/jtd.2018.04.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M, Kim CJ, Sunkara V, Kim MH, Cho YK. 2018a. Liquid biopsy in lung cancer: clinical applications of circulating biomarkers (CTCs and ctDNA). Micromachines (Basel) 9: 100. 10.3390/mi9030100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SB, Tan SJ, Lim WT, Lim CT. 2018b. A merged lung cancer transcriptome dataset for clinical predictive modeling. Sci Data 5: 180136. 10.1038/sdata.2018.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Liu H. 2014. Immunohistochemistry in undifferentiated neoplasm/tumor of uncertain origin. Arch Pathol Lab Med 138: 1583–1610. 10.5858/arpa.2014-0061-RA [DOI] [PubMed] [Google Scholar]
- Lincoln SE, Nussbaum RL, Kurian AW, Nielsen SM, Das K, Michalski S, Yang S, Ngo N, Blanco A, Esplin ED. 2020. Yield and utility of germline testing following tumor sequencing in patients with cancer. JAMA Netw Open 3: e2019452. 10.1001/jamanetworkopen.2020.19452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, Colasacco C, Dacic S, Hirsch FR, Kerr K, et al. 2018. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 142: 321–346. 10.5858/arpa.2017-0388-CP [DOI] [PubMed] [Google Scholar]
- Ling YH, Yuen KWY. 2019. Point centromere activity requires an optimal level of centromeric noncoding RNA. Proc Natl Acad Sci 116: 6270–6279. 10.1073/pnas.1821384116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman SM, Abate-Shen C, Colbert Maresso KL, Colditz GA, Dannenberg AJ, Davidson NE, Disis ML, DuBois RN, Szabo E, Giuliano AR, et al. 2018. AACR white paper: shaping the future of cancer prevention—a roadmap for advancing science and public health. Cancer Prev Res (Phila) 11: 735–778. 10.1158/1940-6207.CAPR-18-0421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. 2018. Small cell lung cancer transformation from EGFR-mutated lung adenocarcinoma: a case report and literatures review. Cancer Biol Ther 19: 445–449. 10.1080/15384047.2018.1435222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jia Y, Stoopler MB, Shen Y, Cheng H, Chen J, Mansukhani M, Koul S, Halmos B, Borczuk AC. 2016. Next-generation sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable MET gene mutations. J Clin Oncol 34: 794–802. 10.1200/JCO.2015.62.0674 [DOI] [PubMed] [Google Scholar]
- Liu H, Lei C, He Q, Pan Z, Xiao D, Tao Y. 2018a. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol Cancer 17: 64. 10.1186/s12943-018-0765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang H, Li Y, Wang R, Li Y, Zhang H, Ren D, Liu H, Kang C, Chen J. 2018b. HOTAIR, a long noncoding RNA, is a marker of abnormal cell cycle regulation in lung cancer. Cancer Sci 109: 2717–2733. 10.1111/cas.13745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kohlberger T, Norouzi M, Dahl GE, Smith JL, Mohtashamian A, Olson N, Peng LH, Hipp JD, Stumpe MC. 2018c. Artificial intelligence–based breast cancer nodal metastasis detection: insights into the black box for pathologists. Arch Pathol Lab Med 143: 859–868. 10.5858/arpa.2018-0147-OA [DOI] [PubMed] [Google Scholar]
- Liu B, Shyr Y, Cai J, Liu Q. 2019. Interplay between miRNAs and host genes and their role in cancer. Brief Funct Genomics 18: 255–266. 10.1093/bfgp/elz002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhan Y, Feng J, Luo J, Fan S. 2018. MicroRNAs associated with therapy of non-small cell lung cancer. Int J Biol Sci 14: 390–397. 10.7150/ijbs.22243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stahler C, Meese E, et al. 2016. Distribution of miRNA expression across human tissues. Nucleic Acids Res 44: 3865–3877. 10.1093/nar/gkw116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P, Ding Y, Lei X, Wu FX. 2019. Deepdriver: predicting cancer driver genes based on somatic mutations using deep convolutional neural networks. Front Genet 10: 13. 10.3389/fgene.2019.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu B, Haque A. 2018. Deep learning based tumor type classification using gene expression data. In Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, pp. 89–96. Association for Computing Machinery, Washington, DC. [Google Scholar]
- Ma P, Fu Y, Cai MC, Yan Y, Jing Y, Zhang S, Chen M, Wu J, Shen Y, Zhu L, et al. 2017. Simultaneous evolutionary expansion and constraint of genomic heterogeneity in multifocal lung cancer. Nat Commun 8: 823. 10.1038/s41467-017-00963-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma KY, Schonnesen AA, Brock A, Van Den Berg C, Eckhardt SG, Liu Z, Jiang N. 2019. Single-cell RNA sequencing of lung adenocarcinoma reveals heterogeneity of immune response-related genes. JCI Insight 4: e121387. 10.1172/jci.insight.121387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean E, Louder A, Saverno K, Smith G, Mardekian J, Brunis C, Ward M, Sweetman R, Pasquale M. 2016. Molecular testing patterns in metastatic non-small cell lung cancer. Am J Manag Care 22: e60–e67. [PubMed] [Google Scholar]
- Mahoney CE, Pirman D, Chubukov V, Sleger T, Hayes S, Fan ZP, Allen EL, Chen Y, Huang L, Liu M, et al. 2019. A chemical biology screen identifies a vulnerability of neuroendocrine cancer cells to SQLE inhibition. Nat Commun 10: 96. 10.1038/s41467-018-07959-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki Vareki S. 2018. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer 6: 157. 10.1186/s40425-018-0479-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelker D, Zhang L. 2018. The emerging significance of secondary germline testing in cancer genomics. J Pathol 244: 610–615. 10.1002/path.5031 [DOI] [PubMed] [Google Scholar]
- Marcar L, Bardhan K, Gheorghiu L, Dinkelborg P, Pfäffle H, Liu Q, Wang M, Piotrowska Z, Sequist LV, Borgmann K, et al. 2019. Acquired resistance of EGFR-mutated lung cancer to tyrosine kinase inhibitor treatment promotes PARP inhibitor sensitivity. Cell Rep 27: 3422–3432.e3424. 10.1016/j.celrep.2019.05.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I. 2019. Somatic mutation and clonal expansions in human tissues. Genome Med 11: 35. 10.1186/s13073-019-0648-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, et al. 2015. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348: 880–886. 10.1126/science.aaa6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, Cagan A, Murai K, Mahbubani K, Stratton MR, et al. 2018. Somatic mutant clones colonize the human esophagus with age. Science 362: 911–917. 10.1126/science.aau3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins F, Sofiya L, Sykiotis GP, Lamine F, Maillard M, Fraga M, Shabafrouz K, Ribi C, Cairoli A, Guex-Crosier Y, et al. 2019. Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat Rev Clin Oncol 16: 563–580. 10.1038/s41571-019-0218-0 [DOI] [PubMed] [Google Scholar]
- McGranahan N, Furness AJS, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, et al. 2016. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351: 1463–1469. 10.1126/science.aaf1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J, Han G, Schalper KA, Carvajal-Hausdorf D, Pelekanou V, Rehman J, Velcheti V, Herbst R, LoRusso P, Rimm DL. 2016. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol 2: 46–54. 10.1001/jamaoncol.2015.3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri E, Mueller WC, Rosenwald S, Zepeniuk M, Klinke E, Edmonston TB, Werner M, Lass U, Barshack I, Feinmesser M, et al. 2012. A second-generation microRNA-based assay for diagnosing tumor tissue origin. Oncologist 17: 801–812. 10.1634/theoncologist.2011-0466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meléndez B, Van Campenhout C, Rorive S, Remmelink M, Salmon I, D'Haene N. 2018. Methods of measurement for tumor mutational burden in tumor tissue. Transl Lung Cancer Res 7: 661–667. 10.21037/tlcr.2018.08.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melosky B. 2018. Advanced typical and atypical carcinoid tumours of the lung: management recommendations. Curr Oncol 25: S86–S93. 10.3747/co.25.3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhi M, Raza A, Inchakalody V, Zar AR, Uddin S, Dermime S. 2019. Immunotherapeutic strategies in patients with advanced head and neck squamous cell carcinoma. Ann Transl Med 7: S22. 10.21037/atm.2019.01.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minna JD, Gazdar A, Augustyn A, Britt R, Carstens R, Dospoy P, Gao B, Girard L, Hight S, Huffman K, et al. 2014. Abstract IA21: developing a new functional classification of lung cancer based on tumor acquired vulnerabilities. Clin Cancer Res 20: IA21. 10.1158/1078-0432.14AACRIASLC-IA21 [DOI] [Google Scholar]
- Mitsudomi T, Yatabe Y. 2010. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J 277: 301–308. 10.1111/j.1742-4658.2009.07448.x [DOI] [PubMed] [Google Scholar]
- Miyanaga A, Masuda M, Tsuta K, Kawasaki K, Nakamura Y, Sakuma T, Asamura H, Gemma A, Yamada T. 2015. Hippo pathway gene mutations in malignant mesothelioma: revealed by RNA and targeted exon sequencing. J Thorac Oncol 10: 844–851. 10.1097/JTO.0000000000000493 [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Umemura S, Matsumura Y, Mimaki S, Tada S, Makinoshima H, Ishii G, Udagawa H, Matsumoto S, Yoh K, et al. 2017. Genomic profiling of large-cell neuroendocrine carcinoma of the lung. Clin Cancer Res 23: 757–765. 10.1158/1078-0432.CCR-16-0355 [DOI] [PubMed] [Google Scholar]
- Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velázquez Vega JE, Brat DJ, Cooper LAD. 2018. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci 115: E2970–E2979. 10.1073/pnas.1717139115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moding EJ, Diehn M, Wakelee HA. 2018. Circulating tumor DNA testing in advanced non-small cell lung cancer. Lung Cancer 119: 42–47. 10.1016/j.lungcan.2018.02.019 [DOI] [PubMed] [Google Scholar]
- Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, Castro G Jr, Srimuninnimit V, Laktionov KK, Bondarenko I, et al. 2019. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 393: 1819–1830. 10.1016/S0140-6736(18)32409-7 [DOI] [PubMed] [Google Scholar]
- Molina-Pinelo S, Pastor MD, Suarez R, Romero-Romero B, González De la Peña M, Salinas A, García-Carbonero R, De Miguel MJ, Rodríguez-Panadero F, Carnero A, et al. 2014. MicroRNA clusters: dysregulation in lung adenocarcinoma and COPD. Eur Respir J 43: 1740–1749. 10.1183/09031936.00091513 [DOI] [PubMed] [Google Scholar]
- Montalto MC, Edwards R. 2019. And they said it couldn't be done: predicting known driver mutations from H&E slides. J Pathol Inform 10: 17. 10.4103/jpi.jpi_91_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AR, Rosenberg SC, McCormick F, Malek S. 2020. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov 19: 533–552. 10.1038/s41573-020-0068-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, Samet Y, Maoz M, Druid H, Arner P, et al. 2018. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun 9: 5068. 10.1038/s41467-018-07466-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. 2021. FDA approves first-in-class KRAS inhibitor. Nat Rev Drug Discov 20: 496. 10.1038/d41573-021-00098-4 [DOI] [PubMed] [Google Scholar]
- Müller MF, Ibrahim AE, Arends MJ. 2016. Molecular pathological classification of colorectal cancer. Virchows Arch 469: 125–134. 10.1007/s00428-016-1956-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahashi M, Sato S, Yuza K, Shimada Y, Ichikawa H, Watanabe S, Takada K, Okamoto T, Okuda S, Lyle S, et al. 2018. Common driver mutations and smoking history affect tumor mutation burden in lung adenocarcinoma. J Surg Res 230: 181–185. 10.1016/j.jss.2018.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo J, Santos-Zabala ML, Iyriboz T, Woo KM, Sima CS, Fiore JJ, Kris MG, Riely GJ, Lito P, Iqbal A, et al. 2016. Large cell neuroendocrine carcinoma of the lung: clinico-pathologic features, treatment, and outcomes. Clin Lung Cancer 17: e121–e129. 10.1016/j.cllc.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalisnik M, Amgad M, Lee S, Halani SH, Velazquez Vega JE, Brat DJ, Gutman DA, Cooper LAD. 2017. Interactive phenotyping of large-scale histology imaging data with HistomicsML. Sci Rep 7: 14588. 10.1038/s41598-017-15092-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Xie C, Yu X, Liu J. 2017. EGFR TKI as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer. Oncotarget 8: 75712–75726. 10.18632/oncotarget.20095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Comprehensive Cancer Network. 2021. Non-Small Cell Lung Cancer (Version 6.2021). Retrieved from https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf [Google Scholar]
- Negrao MV, Quek K, Zhang J, Sepesi B. 2018. TRACERx: tracking tumor evolution to impact the course of lung cancer. J Thorac Cardiovasc Surg 155: 1199–1202. 10.1016/j.jtcvs.2017.10.134 [DOI] [PubMed] [Google Scholar]
- Niederst MJ, Hu H, Mulvey HE, Lockerman EL, Garcia AR, Piotrowska Z, Sequist LV, Engelman JA. 2015. The allelic context of the C797S mutation acquired upon treatment with third-generation EGFR inhibitors impacts sensitivity to subsequent treatment strategies. Clin Cancer Res 21: 3924–3933. 10.1158/1078-0432.CCR-15-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya H, Hiramatsu M, Inamura K, Nomura K, Okui M, Miyoshi T, Okumura S, Satoh Y, Nakagawa K, Nishio M, et al. 2009. Correlation between morphology and EGFR mutations in lung adenocarcinomas significance of the micropapillary pattern and the hobnail cell type. Lung Cancer 63: 235–240. 10.1016/j.lungcan.2008.04.017 [DOI] [PubMed] [Google Scholar]
- Norum J, Nieder C. 2018. Tobacco smoking and cessation and PD-L1 inhibitors in non-small cell lung cancer (NSCLC): a review of the literature. ESMO Open 3: e000406. 10.1136/esmoopen-2018-000406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley DP, Yang Y, Boisot S, Sudarsanam S, Wang JF, Chizhevsky V, Zhao G, Arain S, Weiss LM. 2019. Immunohistochemical detection of PD-L1 among diverse human neoplasms in a reference laboratory: observations based upon 62,896 cases. Mod Pathol 32: 929–942. 10.1038/s41379-019-0210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani L, Askin F, Gabrielson E, Li QK. 2018. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol 52: 103–109. 10.1016/j.semcancer.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxnard GR, Thress KS, Alden RS, Lawrance R, Paweletz CP, Cantarini M, Yang JCH, Barrett JC, Jänne PA. 2016. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol 34: 3375–3382. 10.1200/JCO.2016.66.7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxnard GR, Maddala T, Hubbell E, Aravanis A, Zhang N, Venn O, Valouev A, Shen L, Patel S, Jamshidi A, et al. 2018. Genome-wide sequencing for early stage lung cancer detection from plasma cell-free DNA (cfDNA): the Circulating Cancer Genome Atlas (CCGA) study. J Clin Oncol 36: LBA8501. 10.1200/JCO.2018.36.18_suppl.LBA8501 [DOI] [Google Scholar]
- Pagès F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, Lugli A, Zlobec I, Rau TT, Berger MD, et al. 2018. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 391: 2128–2139. 10.1016/S0140-6736(18)30789-X [DOI] [PubMed] [Google Scholar]
- Paik PK, Drilon A, Fan PD, Yu H, Rekhtman N, Ginsberg MS, Borsu L, Schultz N, Berger MF, Rudin CM, et al. 2015a. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 5: 842–849. 10.1158/2159-8290.CD-14-1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik PK, Drilon A, Yu H, Rekhtman N, Ginsberg MS, Borsu L, Schultz N, Berger MF, Rudin CM, Ladanyi M. 2015b. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 5: 842–849. 10.1158/2159-8290.CD-14-1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakkala S, Ramalingam SS. 2018. Personalized therapy for lung cancer: striking a moving target. JCI Insight 3: e120858. 10.1172/jci.insight.120858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitrakopoulou V, Lee JJ, Wistuba II, Tsao AS, Fossella FV, Kalhor N, Gupta S, Byers LA, Izzo JG, Gettinger SN, et al. 2016. The BATTLE-2 study: a biomarker-integrated targeted therapy study in previously treated patients with advanced non-small-cell lung cancer. J Clin Oncol 34: 3638–3647. 10.1200/JCO.2015.66.0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitrakopoulou V, Wu YL, Han JY, Ahn MJ, Ramalingam SS, John T, Okamoto I, Yang JCH, Bulusu K, Laus G, et al. 2018. LBA51Analysis of resistance mechanisms to osimertinib in patients with EGFR T790M advanced NSCLC from the AURA3 study. Ann Oncol 29: viii741. 10.1093/annonc/mdy424.064 [DOI] [Google Scholar]
- Park VS, Pursell ZF. 2019. POLE proofreading defects: contributions to mutagenesis and cancer. DNA Repair (Amst) 76: 50–59. 10.1016/j.dnarep.2019.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen EE, Kilvaer TK, Khanehkenari MR, Al-Saad S, Hald SM, Andersen S, Richardsen E, Ness N, Busund LT, Bremnes RM, et al. 2017. Assessing PDL-1 and PD-1 in non-small cell lung cancer: a novel immunoscore approach. Clin Lung Cancer 18: 220–233.e228. 10.1016/j.cllc.2016.09.009 [DOI] [PubMed] [Google Scholar]
- Pavan A, Attili I, Pasello G, Guarneri V, Conte PF, Bonanno L. 2019. Immunotherapy in small-cell lung cancer: from molecular promises to clinical challenges. J Immunother Cancer 7: 205. 10.1186/s40425-019-0690-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis N, Khaled H, Gaafar R. 2015. A mini review on cancer of unknown primary site: a clinical puzzle for the oncologists. J Adv Res 6: 375–382. 10.1016/j.jare.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pell R, Oien K, Robinson M, Pitman H, Rajpoot N, Rittscher J, Snead D, Verrill C. 2019. The use of digital pathology and image analysis in clinical trials. J Pathol Clin Res 5: 81–90. 10.1002/cjp2.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi G, Sonzogni A, Harari S, Albini A, Bresaola E, Marchio C, Massa F, Righi L, Gatti G, Papanikolaou N, et al. 2017. Classification of pulmonary neuroendocrine tumors: new insights. Transl Lung Cancer Res 6: 513–529. 10.21037/tlcr.2017.09.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penault-Llorca F, Radosevic-Robin N. 2018. Tumor mutational burden in non-small cell lung cancer—the pathologist's point of view. Transl Lung Cancer Res 7: 716–721. 10.21037/tlcr.2018.09.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentheroudakis G, Pavlidis N, Fountzilas G, Krikelis D, Goussia A, Stoyianni A, Sanden M, St Cyr B, Yerushalmi N, Benjamin H, et al. 2013. Novel microRNA-based assay demonstrates 92% agreement with diagnosis based on clinicopathologic and management data in a cohort of patients with carcinoma of unknown primary. Mol Cancer 12: 57. 10.1186/1476-4598-12-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno P, Brambilla E, Thomas R, Soria JC. 2012. Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin Cancer Res 18: 2443–2451. 10.1158/1078-0432.CCR-11-2370 [DOI] [PubMed] [Google Scholar]
- Pérez-Ramírez C, Cañadas-Garre M, Jiménez-Varo E, Faus-Dáder MJ, Calleja-Hernández MA. 2015a. MET: a new promising biomarker in non-small-cell lung carcinoma. Pharmacogenomics 16: 631–647. 10.2217/pgs.15.11 [DOI] [PubMed] [Google Scholar]
- Pérez-Ramírez C, Cañadas-Garre M, Molina MA, Faus-Dáder MJ, Calleja-Hernández MA. 2015b. PTEN and PI3K/AKT in non-small-cell lung cancer. Pharmacogenomics 16: 1843–1862. 10.2217/pgs.15.122 [DOI] [PubMed] [Google Scholar]
- Phallen J, Leal A, Woodward BD, Forde PM, Naidoo J, Marrone KA, Brahmer JR, Fiksel J, Medina JE, Cristiano S, et al. 2019. Early noninvasive detection of response to targeted therapy in non-small cell lung cancer. Cancer Res 79: 1204–1213. 10.1158/0008-5472.CAN-18-1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DH. 2018. Mutational spectra and mutational signatures: insights into cancer aetiology and mechanisms of DNA damage and repair. DNA Repair (Amst) 71: 6–11. 10.1016/j.dnarep.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikor L, Ramnarine VR, Lam S, Lam W. 2013. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer 82: 179–189. 10.1016/j.lungcan.2013.07.025 [DOI] [PubMed] [Google Scholar]
- Planchard D, Popat S, Kerr K, Novello S, Smit EF, Faivre-Finn C, Mok TS, Reck M, Van Schil PE, Hellmann MD, et al. 2018. Metastatic non-small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 29: iv192–iv237. 10.1093/annonc/mdy275 [DOI] [PubMed] [Google Scholar]
- Puzanov I, Diab A, Abdallah K, Bingham CO, Brogdon C, Dadu R, Hamad L, Kim S, Lacouture ME, LeBoeuf NR, et al. 2017. Managing toxicities associated with immune checkpoint inhibitors: consensus recommendations from the Society for Immunotherapy of Cancer (SITC) Toxicity Management Working Group. J Immunother Cancer 5: 95. 10.1186/s40425-017-0300-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao M, Jiang T, Ren S, Zhou C. 2018a. Combination strategies on the basis of immune checkpoint inhibitors in non-small-cell lung cancer: where do we stand? Clin Lung Cancer 19: 1–11. 10.1016/j.cllc.2017.06.005 [DOI] [PubMed] [Google Scholar]
- Qiao M, Jiang T, Zhou C. 2018b. Shining light on advanced NSCLC in 2017: combining immune checkpoint inhibitors. J Thorac Dis 10: S1534–S1546. 10.21037/jtd.2018.04.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Wei F, Zhang J, Wang X, Li B. 2016. miR-134 inhibits non-small cell lung cancer growth by targeting the epidermal growth factor receptor. J Cell Mol Med 20: 1974–1983. 10.1111/jcmm.12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja R, Kuziora M, Brohawn PZ, Higgs BW, Gupta A, Dennis PA, Ranade K. 2018. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin Cancer Res 24: 6212–6222. 10.1158/1078-0432.CCR-18-0386 [DOI] [PubMed] [Google Scholar]
- Rakhlin A, Shvets A, Iglovikov V, Kalinin AA. 2018. Deep convolutional neural networks for breast cancer histology image analysis. In Image analysis and recognition (ed. Campilho A, et al. ), pp. 737–744. Springer, Berlin. [Google Scholar]
- Razavi P, Li BT, Abida W, Aravanis A, Jung B, Shen R, Hou C, De Bruijn I, Gnerre S, Lim RS, et al. 2017. Performance of a high-intensity 508-gene circulating-tumor DNA (ctDNA) assay in patients with metastatic breast, lung, and prostate cancer. J Clin Oncol 35: LBA11516. 10.1200/JCO.2017.35.18_suppl.LBA11516 [DOI] [Google Scholar]
- Ready N, Hellmann MD, Awad MM, Otterson GA, Gutierrez M, Gainor JF, Borghaei H, Jolivet J, Horn L, Mates M, et al. 2019. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol 37: 992–1000. 10.1200/JCO.18.01042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, Gottfried M, Peled N, Tafreshi A, Cuffe S, et al. 2019. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol 37: 537–546. 10.1200/JCO.18.00149 [DOI] [PubMed] [Google Scholar]
- Rekhtman N, Ang DC, Riely GJ, Ladanyi M, Moreira AL. 2013. KRAS mutations are associated with solid growth pattern and tumor-infiltrating leukocytes in lung adenocarcinoma. Mod Pathol 26: 1307–1319. 10.1038/modpathol.2013.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remon J, Ahn MJ, Girard N, Johnson M, Kim DW, Lopes G, Pillai RN, Solomon B, Villacampa G, Zhou Q. 2019. Advanced-stage non-small cell lung cancer: advances in thoracic oncology 2018. J Thorac Oncol 14: 1134–1155. 10.1016/j.jtho.2019.03.022 [DOI] [PubMed] [Google Scholar]
- Ricciuti B, Genova C, De Giglio A, Bassanelli M, Dal Bello MG, Metro G, Brambilla M, Baglivo S, Grossi F, Chiari R. 2019. Impact of immune-related adverse events on survival in patients with advanced non-small cell lung cancer treated with nivolumab: long-term outcomes from a multi-institutional analysis. J Cancer Res Clin Oncol 145: 479–485. 10.1007/s00432-018-2805-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rini BI, Plimack ER, Stus V, Gafanov R, Hawkins R, Nosov D, Pouliot F, Alekseev B, Soulières D, Melichar B, et al. 2019. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 380: 1116–1127. 10.1056/NEJMoa1816714 [DOI] [PubMed] [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. 2015. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348: 124–128. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LA, Jaing CJ, Pierce Campbell C, Magliocco A, Xiong Y, Magliocco G, Thissen JB, Antonia S. 2016. Molecular evidence of viral DNA in non-small cell lung cancer and non-neoplastic lung. Br J Cancer 115: 497–504. 10.1038/bjc.2016.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco D, Della Gravara L, Battiloro C, Gridelli C. 2019. The role of combination chemo-immunotherapy in advanced non-small cell lung cancer. Expert Rev Anticancer Ther 19: 561–568. 10.1080/14737140.2019.1631800 [DOI] [PubMed] [Google Scholar]
- Rooney M, Devarakonda S, Govindan R. 2013. Genomics of squamous cell lung cancer. Oncologist 18: 707–716. 10.1634/theoncologist.2013-0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper N, Gao S, Maity TK, Banday AR, Zhang X, Venugopalan A, Cultraro CM, Patidar R, Sindiri S, Brown AL, et al. 2019. APOBEC mutagenesis and copy-number alterations are drivers of proteogenomic tumor evolution and heterogeneity in metastatic thoracic tumors. Cell Rep 26: 2651–2666.e6. 10.1016/j.celrep.2019.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell R, Karachaliou N. 2016. Large-scale screening for somatic mutations in lung cancer. Lancet 387: 1354–1356. 10.1016/S0140-6736(15)01125-3 [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, et al. 2008. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol 26: 462–469. 10.1038/nbt1392 [DOI] [PubMed] [Google Scholar]
- Rosenwald S, Gilad S, Benjamin S, Lebanony D, Dromi N, Faerman A, Benjamin H, Tamir R, Ezagouri M, Goren E, et al. 2010. Validation of a microRNA-based qRT-PCR test for accurate identification of tumor tissue origin. Mod Pathol 23: 814–823. 10.1038/modpathol.2010.57 [DOI] [PubMed] [Google Scholar]
- Ross JS, Wang K, Elkadi OR, Tarasen A, Foulke L, Sheehan CE, Otto GA, Palmer G, Yelensky R, Lipson D, et al. 2014. Next-generation sequencing reveals frequent consistent genomic alterations in small cell undifferentiated lung cancer. J Clin Pathol 67: 772–776. 10.1136/jclinpath-2014-202447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, Heymach JV, Johnson JE, Lehman JM, MacPherson D, et al. 2019a. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 19: 289–297. 10.1038/s41568-019-0133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, Heymach JV, Johnson JE, Lehman JM, MacPherson D, et al. 2019b. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 19: 289–297. 10.1038/s41568-019-0133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha M, Chakraborty C, Arun I, Ahmed R, Chatterjee S. 2017. An advanced deep learning approach for Ki-67 stained hotspot detection and proliferation rate scoring for prognostic evaluation of breast cancer. Sci Rep 7: 3213. 10.1038/s41598-017-03405-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saigi M, Alburquerque-Bejar JJ, Sanchez-Cespedes M. 2019. Determinants of immunological evasion and immunocheckpoint inhibition response in non-small cell lung cancer: the genetic front. Oncogene 38: 5921–5932. 10.1038/s41388-019-0855-x [DOI] [PubMed] [Google Scholar]
- Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, Samaras D, Shroyer KR, Zhao T, Batiste R, et al. 2018. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep 23: 181–193.e7. 10.1016/j.celrep.2018.03.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadder NJ, Riegert-Johnson D, Boardman L, Rhodes D, Wick M, Okuno S, Kunze KL, Golafshar M, Uson PLS Jr, Mountjoy L, et al. 2021. Comparison of universal genetic testing vs guideline-directed targeted testing for patients with hereditary cancer syndrome. JAMA Oncol 7: 230–237. 10.1001/jamaoncol.2020.6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein RM, Lee C-H, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, et al. 2019. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 51: 202–206. 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia M, Funel N, Ali A, Giovannetti E. 2018a. Liquid biopsies to optimize therapeutic efficacy in unresponsive lung cancer patients. Exp Opin Drug Metab Toxicol 14: 761–763. 10.1080/17425255.2018.1491965 [DOI] [PubMed] [Google Scholar]
- Santarpia M, Liguori A, D'Aveni A, Karachaliou N, Gonzalez-Cao M, Daffinà MG, Lazzari C, Altavilla G, Rosell R. 2018b. Liquid biopsy for lung cancer early detection. J Thorac Dis 10: S882–S897. 10.21037/jtd.2018.03.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schabath MB, Cress D, Muñoz-Antonia T. 2016. Racial and ethnic differences in the epidemiology and genomics of lung cancer. Cancer Control 23: 338–346. 10.1177/107327481602300405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalper KA, Brown J, Carvajal-Hausdorf D, McLaughlin J, Velcheti V, Syrigos KN, Herbst RS, Rimm DL. 2015. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst 107: dju4335. 10.1093/jnci/dju435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffmann I, Greve G, Jung M, Lübbert M. 2016. Epigenetic therapy approaches in non-small cell lung cancer: update and perspectives. Epigenetics 11: 858–870. 10.1080/15592294.2016.1237345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrock AB, Frampton GM, Suh J, Chalmers ZR, Rosenzweig M, Erlich RL, Halmos B, Goldman J, Forde P, Leuenberger K, et al. 2016. Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J Thorac Oncol 11: 1493–1502. 10.1016/j.jtho.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Seike M, Kim CH, Zou F, Noro R, Chiba M, Ishikawa A, Kappaunugi S, Kubota K, Gemma A. 2017. AXL and GAS6 co-expression in lung adenocarcinoma as a prognostic classifier. Oncol Rep 37: 3261–3269. 10.3892/or.2017.5594 [DOI] [PubMed] [Google Scholar]
- Sekido Y. 2013. Molecular pathogenesis of malignant mesothelioma. Carcinogenesis 34: 1413–1419. 10.1093/carcin/bgt166 [DOI] [PubMed] [Google Scholar]
- Selves J, Long-Mira E, Mathieu MC, Rochaix P, Ilié M. 2018. Immunohistochemistry for diagnosis of metastatic carcinomas of unknown primary site. Cancers (Basel) 10: 108. 10.3390/cancers10040108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. 2011. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 3: 75ra26. 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Kendall F, Khozin S, Goosen R, Hu J, Laramie J, Ringel M, Schork N. 2019. Artificial intelligence and machine learning in clinical development: a translational perspective. NPJ Digit Med 2: 69. 10.1038/s41746-019-0148-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapcott M, Hewitt KJ, Rajpoot N. 2019. Deep learning with sampling in colon cancer histology. Front Bioeng Biotechnol 7: 52. 10.3389/fbioe.2019.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P. 2017. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph 61: 2–13. 10.1016/j.compmedimag.2017.06.001 [DOI] [PubMed] [Google Scholar]
- Shaul ME, Fridlender ZG. 2019. Tumour-associated neutrophils in patients with cancer. Nat Rev Clin Oncol 16: 601–620. 10.1038/s41571-019-0222-4 [DOI] [PubMed] [Google Scholar]
- Sheng S, Margarida Bernardo M, Dzinic SH, Chen K, Heath EI, Sakr WA. 2018. Tackling tumor heterogeneity and phenotypic plasticity in cancer precision medicine: our experience and a literature review. Cancer Metastasis Rev 37: 655–663. 10.1007/s10555-018-9767-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Hua X, Zhu B, Ravichandran S, Wang M, Nguyen C, Brodie SA, Palleschi A, Alloisio M, Pariscenti G, et al. 2016a. Somatic genomics and clinical features of lung adenocarcinoma: a retrospective study. PLoS Med 13: e1002162. 10.1371/journal.pmed.1002162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Oh YT, Zhang G, Yao W, Yue P, Li Y, Kanteti R, Riehm J, Salgia R, Owonikoko TK, et al. 2016b. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett 380: 494–504. 10.1016/j.canlet.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Shi X, Wu H, Lu J, Duan H, Liu X, Liang Z. 2016c. Screening for major driver oncogene alterations in adenosquamous lung carcinoma using PCR coupled with next-generation and Sanger sequencing methods. Sci Rep 6: 22297. 10.1038/srep22297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim HS, Lee DH, Park EJ, Kim SH. 2011. Histopathologic characteristics of lung adenocarcinomas with epidermal growth factor receptor mutations in the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society lung adenocarcinoma classification. Arch Pathol Lab Med 135: 1329–1334. 10.5858/arpa.2010-0493-OA [DOI] [PubMed] [Google Scholar]
- Shim HS, Choi YL, Kim L, Chang S, Kim WS, Roh MS, Kim TJ, Ha SY, Chung JH, Jang SJ, et al. 2017. Molecular testing of lung cancers. J Pathol Transl Med 51: 242–254. 10.4132/jptm.2017.04.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Yamamoto Y, Ochiya T. 2019. Synthetic lethality in lung cancer-from the perspective of cancer genomics. Medicines (Basel) 6: 38. 10.3390/medicines6010038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl L. 2017. Molecular diagnostics of lung cancer in the clinic. Transl Lung Cancer Res 6: 560–569. 10.21037/tlcr.2017.08.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl LM, Yeap BY, Iafrate AJ, Holmes-Tisch AJ, Chou YP, Wu MT, Goan YG, Su L, Benedettini E, Yu J, et al. 2009. Lung adenocarcinoma with EGFR amplification has distinct clinicopathologic and molecular features in never-smokers. Cancer Res 69: 8341–8348. 10.1158/0008-5472.CAN-09-2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl LM, Weremowicz S, Gray SW, Wong KK, Chirieac LR, Lindeman NI, Hornick JL. 2013. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. J Thorac Oncol 8: 322–328. 10.1097/JTO.0b013e31827db604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl LM, Aisner DL, Varella-Garcia M, Berry LD, Dias-Santagata D, Wistuba II, Chen H, Fujimoto J, Kugler K, Franklin WA, et al. 2015. Multi-institutional oncogenic driver mutation analysis in lung adenocarcinoma: the Lung Cancer Mutation Consortium Experience. J Thorac Oncol 10: 768–777. 10.1097/JTO.0000000000000516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff GS, de Groot PM, Papadimitrakopoulou VA, Truong MT, Carter BW. 2018. Targeted therapy and immunotherapy in the treatment of non-small cell lung cancer. Radiol Clin North Am 56: 485–495. 10.1016/j.rcl.2018.01.012 [DOI] [PubMed] [Google Scholar]
- Shu XO, Liu Y, Cai Q, Wu H. 2016. MiR-374a suppresses lung adenocarcinoma cell proliferation and invasion by targeting TGFA gene expression. Carcinogenesis 37: 567–575. 10.1093/carcin/bgw038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicklick JK, Kato S, Okamura R, Schwaederle M, Hahn ME, Williams CB, De P, Krie A, Piccioni DE, Miller VA, et al. 2019. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med 25: 744–750. 10.1038/s41591-019-0407-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegler J, Rizzo A, Tauler J, Moreno L, Garcia JGN. 2012. A myosin light chain kinase pseudogene (MYLKP) contributes to lung cancer proliferation and tumor growth. Am J Respir Crit Care Med 185: A6370. [Google Scholar]
- Solassol J, Vendrell JA, Senal R, Audran P, Leenhardt F, Quantin X. 2019. Challenging BRAF/EGFR co-inhibition in NSCLC using sequential liquid biopsies. Lung Cancer 133: 45–47. 10.1016/j.lungcan.2019.05.003 [DOI] [PubMed] [Google Scholar]
- Solis LM, Behrens C, Raso MG, Lin HY, Kadara H, Yuan P, Galindo H, Tang X, Lee JJ, Kalhor N, et al. 2012. Histologic patterns and molecular characteristics of lung adenocarcinoma associated with clinical outcome. Cancer 118: 2889–2899. 10.1002/cncr.26584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonea L, Buse M, Gulei D, Onaciu A, Simon I, Braicu C, Berindan-Neagoe I. 2018. Decoding the emerging patterns exhibited in non-coding RNAs characteristic of lung cancer with regard to their clinical significance. Curr Genomics 19: 258–278. 10.2174/1389202918666171005100124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Zhu H, Guo Z, Wu W, Sun W, Zhang Y. 2013. Correlation of EGFR mutation and predominant histologic subtype according to the new lung adenocarcinoma classification in Chinese patients. Medical Oncology 30: 645. 10.1007/s12032-013-0645-1 [DOI] [PubMed] [Google Scholar]
- Song K, Bi JH, Qiu ZW, Felizardo R, Girard L, Minna JD, Gazdar AF. 2018a. A quantitative method for assessing smoke associated molecular damage in lung cancers. Transl Lung Cancer Res 7: 439–449. 10.21037/tlcr.2018.07.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Cheng G, Xu C, Wang W, Shao Y, Zhang Y. 2018b. Clinicopathological characteristics of POLE mutation in patients with non-small-cell lung cancer. Lung Cancer 118: 57–61. 10.1016/j.lungcan.2018.02.004 [DOI] [PubMed] [Google Scholar]
- Soo RA, Chen Z, Yan Teng RS, Tan HL, Iacopetta B, Tai BC, Soong R. 2018. Prognostic significance of immune cells in non-small cell lung cancer: meta-analysis. Oncotarget 9: 24801–24820. 10.18632/oncotarget.24835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, et al. 2018. Osimertinib in untreated EGFR-mutated advanced non–small-cell lung cancer. N Engl J Med 378: 113–125. 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- Sosa Iglesias V, Giuranno L, Dubois LJ, Theys J, Vooijs M. 2018. Drug resistance in non-small cell lung cancer: a potential for NOTCH targeting? Front Oncol 8: 267. 10.3389/fonc.2018.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spira A, Yurgelun MB, Alexandrov L, Rao A, Bejar R, Polyak K, Giannakis M, Shilatifard A, Finn OJ, Dhodapkar M, et al. 2017. Precancer atlas to drive precision prevention trials. Cancer Res 77: 1510–1541. 10.1158/0008-5472.CAN-16-2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Ghosh S, Kagan J, Mazurchuk R. 2018a. The precancer atlas (PCA). Trends Cancer 4: 513–514. 10.1016/j.trecan.2018.06.003 [DOI] [PubMed] [Google Scholar]
- Srivastava S, Ghosh S, Kagan J, Mazurchuk R, Boja E, Chuaqui R, Chavarria-Johnson E, Davidsen T, Eary J, Haim T, et al. 2018b. The making of a precancer atlas: promises, challenges, and opportunities. Trends Cancer 4: 523–536. 10.1016/j.trecan.2018.06.007 [DOI] [PubMed] [Google Scholar]
- Stanta G, Bonin S. 2018. Overview on clinical relevance of intra-tumor heterogeneity. Front Med (Lausanne) 5: 85. 10.3389/fmed.2018.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzinger A, Allen JD, Maas J, Stewart MD, Merino DM, Wempe MM, Dietel M. 2019. Tumor mutational burden standardization initiatives: recommendations for consistent tumor mutational burden assessment in clinical samples to guide immunotherapy treatment decisions. Genes Chromosomes Cancer 58: 578–588. 10.1002/gcc.22733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevic R, Milenkovic B. 2016. Tracheobronchial tumors. J Thorac Dis 8: 3401–3413. 10.21037/jtd.2016.11.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PA, Welsh EA, Slebos RJC, Fang B, Izumi V, Chambers M, Zhang G, Cen L, Pettersson F, Zhang Y, et al. 2019. Proteogenomic landscape of squamous cell lung cancer. Nat Commun 10: 3578. 10.1038/s41467-019-11452-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe TE. 2013. Molecular subtypes of non-small-cell lung cancer. In Personalized management of lung cancer, pp. 6–20. Future Medicine, London. [Google Scholar]
- Su Y, Fang HB, Jiang F. 2018. An epigenetic classifier for early stage lung cancer. Clin Epigenetics 10: 68. 10.1186/s13148-018-0502-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh JH, Johnson A, Albacker L, Wang K, Chmielecki J, Frampton G, Gay L, Elvin JA, Vergilio JA, Ali S, et al. 2016. Comprehensive genomic profiling facilitates implementation of the national comprehensive cancer network guidelines for lung cancer biomarker testing and identifies patients who may benefit from enrollment in mechanism-driven clinical trials. Oncologist 21: 684–691. 10.1634/theoncologist.2016-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YH, Lin SW, Hsieh CC, Yeh YC, Tu CC, Chen KJ. 2014. Treatment outcomes of patients with different subtypes of large cell carcinoma of the lung. Ann Thorac Surg 98: 1013–1019. 10.1016/j.athoracsur.2014.05.012 [DOI] [PubMed] [Google Scholar]
- Suraweera A, O'Byrne KJ, Richard DJ. 2018. Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front Oncol 8: 92. 10.3389/fonc.2018.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoronos AA, Engelman DM, Slack FJ. 2016. Oncomir or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res 76: 3666–3670. 10.1158/0008-5472.CAN-16-0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski LJ, Molas-Torreblanca K, Bawab R, Kim E, Don D, Mascarenhas L, Stanley P, Zhou S, Shillingford N. 2018. Bronchial mucoepidermoid carcinoma with the classic MAML2 gene rearrangement in a 2-year-old boy. Pediatr Dev Pathol 21: 480–485. 10.1177/1093526617707855 [DOI] [PubMed] [Google Scholar]
- Taberna M, Oliva M, Mesía R. 2019. Cetuximab-containing combinations in locally advanced and recurrent or metastatic head and neck squamous cell carcinoma. Front Oncol 9: 383. 10.3389/fonc.2019.00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafe LJ, Pierce KJ, Peterson JD, de Abreu F, Memoli VA, Black CC, Pettus JR, Marotti JD, Gutmann EJ, Liu X, et al. 2016. Clinical genotyping of non-small cell lung cancers using targeted next-generation sequencing: utility of identifying rare and co-mutations in oncogenic driver genes. Neoplasia 18: 577–583. 10.1016/j.neo.2016.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamochi K, Takahashi F, Suehara Y, Sato E, Kohsaka S, Hayashi T, Kitano S, Uneno T, Kojima S, Takeuchi K, et al. 2017. DNA mismatch repair deficiency in surgically resected lung adenocarcinoma: microsatellite instability analysis using the Promega panel. Lung Cancer 110: 26–31. 10.1016/j.lungcan.2017.05.016 [DOI] [PubMed] [Google Scholar]
- Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. 2019. Benefits and limitations of genome-wide association studies. Nat Rev Genet 20: 467–484. 10.1038/s41576-019-0127-1 [DOI] [PubMed] [Google Scholar]
- Tan CL, Lim TH, Lim T, Tan DS, Chua YW, Ang MK, Pang B, Lim CT, Takano A, Lim AS, et al. 2016. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget 7: 23251–23262. 10.18632/oncotarget.8136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Yu JX, Hubbard-Lucey VM, Neftelinov ST, Hodge JP, Lin Y. 2018. The clinical trial landscape for PD1/PDL1 immune checkpoint inhibitors. Nat Rev Drug Discov 17: 854–855. 10.1038/nrd.2018.210 [DOI] [PubMed] [Google Scholar]
- Tang W, Lei Y, Su J, Zhang C, Fu R, Kang J, Yan H, Yang X, Tu H, Wu Y, et al. 2019. TNM stages inversely correlate with the age at diagnosis in ALK-positive lung cancer. Transl Lung Cancer Res 8: 144–154. 10.21037/tlcr.2019.03.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira VH, Pipinikas CP, Pennycuick A, Lee-Six H, Chandrasekharan D, Beane J, Morris TJ, Karpathakis A, Feber A, Breeze CE, et al. 2019. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat Med 25: 517–525. 10.1038/s41591-018-0323-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto A, Tsukamoto T, Kiriyama Y, Fujita H. 2017. Automated classification of lung cancer types from cytological images using deep convolutional neural networks. Biomed Res Int 2017: 4067832. 10.1155/2017/4067832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra S, Mansfield AS, Dong H, Peikert T, Roden AC. 2017. Temporal and spatial heterogeneity of programmed cell death 1-ligand 1 expression in malignant mesothelioma. Oncoimmunology 6: e1356146. 10.1080/2162402X.2017.1356146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova-Barberio M, Thomas S, Ali N, Pawlowska N, Park J, Krings G, Rosenblum MD, Budillon A, Munster PN. 2017. HDAC inhibition potentiates immunotherapy in triple negative breast cancer. Oncotarget 8: 114156–114172. 10.18632/oncotarget.23169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa U, Castelli G, Pelosi E. 2018. Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers (Basel) 10: 248. 10.3390/cancers10080248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2012a. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2012b. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489: 519–525. 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2014. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511: 543–550. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann A, Halachev M, McLaren W, Moore DJ, Svinti V, Campbell A, Kerr SM, Tischkowitz M, Hunt SE, Dunlop MG, et al. 2019. Flexible and scalable diagnostic filtering of genomic variants using G2P with Ensembl VEP. Nat Commun 10: 2373. 10.1038/s41467-019-10016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Dong X, Freytag S, Lê Cao KA, Su S, JalalAbadi A, Amann-Zalcenstein D, Weber TS, Seidi A, Jabbari JS, et al. 2019. Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments. Nat Methods 16: 479–487. 10.1038/s41592-019-0425-8 [DOI] [PubMed] [Google Scholar]
- Togashi Y, Shitara K, Nishikawa H. 2019. Regulatory T cells in cancer immunosuppression—implications for anticancer therapy. Nat Rev Clin Oncol 16: 356–371. 10.1038/s41571-019-0175-7 [DOI] [PubMed] [Google Scholar]
- Tolani B, Acevedo LA, Hoang NT, He B. 2018. Heterogeneous contributing factors in MPM disease development and progression: biological advances and clinical implications. Int J Mol Sci 19: 238. 10.3390/ijms19010238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomuleasa C, Zaharie F, Muresan MS, Pop L, Fekete Z, Dima D, Frinc I, Trifa A, Berce C, Jurj A, et al. 2017. How to diagnose and treat a cancer of unknown primary site. J Gastrointestin Liver Dis 26: 69–79. 10.15403/jgld.2014.1121.261.haz [DOI] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, et al. 2011. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol 6: 244–285. 10.1097/JTO.0b013e318206a221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, et al. 2015. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 10: 1243–1260. 10.1097/JTO.0000000000000630 [DOI] [PubMed] [Google Scholar]
- Tsai HK, Lehrer J, Alshalalfa M, Erho N, Davicioni E, Lotan TL. 2017. Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma. BMC Cancer 17: 759. 10.1186/s12885-017-3729-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuta K, Kohno T, Yoshida A, Shimada Y, Asamura H, Furuta K, Kushima R. 2014. RET-rearranged non-small-cell lung carcinoma: a clinicopathological and molecular analysis. Br J Cancer 110: 1571–1578. 10.1038/bjc.2014.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu CY, Cheng FJ, Chen CM, Wang SL, Hsiao YC, Chen CH, Hsia TC, He YH, Wang BW, Hsieh IS, et al. 2018. Cigarette smoke enhances oncogene addiction to c-MET and desensitizes EGFR-expressing non-small cell lung cancer to EGFR TKIs. Mol Oncol 12: 705–723. 10.1002/1878-0261.12193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutar E, Tutar Y. 2019. Non-coding RNAs in lung cancer. J Thorac Dis 11: S245–S248. 10.21037/jtd.2019.01.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. 2017. A pathology atlas of the human cancer transcriptome. Science 357: eaan2507. 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]
- Ujhazy P, Lindwasser OW. 2018. Small cell lung cancer: updates and new concepts. Transl Lung Cancer Res 7: 1–3. 10.21037/tlcr.2018.02.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadhachary G. 2013. New strategies for carcinoma of unknown primary: the role of tissue-of-origin molecular profiling. Clin Cancer Res 19: 4027–4033. 10.1158/1078-0432.CCR-12-3030 [DOI] [PubMed] [Google Scholar]
- Varadhachary GR, Spector Y, Abbruzzese JL, Rosenwald S, Wang H, Aharonov R, Carlson HR, Cohen D, Karanth S, Macinskas J, et al. 2011. Prospective gene signature study using microRNA to identify the tissue of origin in patients with carcinoma of unknown primary. Clin Cancer Res 17: 4063–4070. 10.1158/1078-0432.CCR-10-2599 [DOI] [PubMed] [Google Scholar]
- Vassella E, Langsch S, Dettmer MS, Schlup C, Neuenschwander M, Frattini M, Gugger M, Schäfer SC. 2015. Molecular profiling of lung adenosquamous carcinoma: hybrid or genuine type? Oncotarget 6: 23905–23916. 10.18632/oncotarget.4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velcheti V, Patwardhan PD, Liu FX, Chen X, Cao X, Burke T. 2018. Real-world PD-L1 testing and distribution of PD-L1 tumor expression by immunohistochemistry assay type among patients with metastatic non-small cell lung cancer in the United States. PLoS ONE 13: e0206370. 10.1371/journal.pone.0206370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat A, Mohlman J, Leventhal S, Gyulassy A, Pascucci V, Salama M. 2018. Application of a convolutional neural network to distinguish Burkitt lymphoma from diffuse large B-cell lymphoma. Am J Clin Pathol 150: S119. 10.1093/ajcp/aqy099.286 [DOI] [PubMed] [Google Scholar]
- Vignot S, Frampton GM, Soria JC, Yelensky R, Commo F, Brambilla C, Palmer G, Moro-Sibilot D, Ross JS, Cronin MT, et al. 2013. Next-generation sequencing reveals high concordance of recurrent somatic alterations between primary tumor and metastases from patients with non-small-cell lung cancer. J Clin Oncol 31: 2167–2172. 10.1200/JCO.2012.47.7737 [DOI] [PubMed] [Google Scholar]
- Vinayanuwattikun C, Le Calvez-Kelm F, Abedi-Ardekani B, Zaridze D, Mukeria A, Voegele C, Vallée M, Purnomosari D, Forey N, Durand G, et al. 2016. Elucidating genomic characteristics of lung cancer progression from in situ to invasive adenocarcinoma. Sci Rep 6: 31628. 10.1038/srep31628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojnic M, Kubota D, Kurzatkowski C, Offin M, Suzawa K, Benayed R, Schoenfeld AJ, Plodkowski AJ, Poirier JT, Rudin CM, et al. 2019. Acquired BRAF rearrangements induce secondary resistance to EGFR therapy in EGFR-mutated lung cancers. J Thorac Oncol 14: 802–815. 10.1016/j.jtho.2018.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht C, Werner R, Walter RF, Christoph DC, Heukamp LC, Peifer M, Hirsch B, Burbat L, Mairinger T, Schmid KW, et al. 2015. Mutational analysis of pulmonary tumours with neuroendocrine features using targeted massive parallel sequencing: a comparison of a neglected tumour group. Br J Cancer 113: 1704–1711. 10.1038/bjc.2015.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Grün J, Rodel F, Brandts C, Fokas E, Guckenberger M, Rödel C, Balermpas P. 2019. Targeted therapies and immune-checkpoint inhibition in head and neck squamous cell carcinoma: where do we stand today and where to go? Cancers (Basel) 11: 472. 10.3390/cancers11040472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Võsa U, Vooder T, Kolde R, Fischer K, Välk K, Tõnisson N, Roosipuu R, Vilo J, Metspalu A, Annilo T. 2011. Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes, Chromosomes Cancer 50: 812–822. 10.1002/gcc.20902 [DOI] [PubMed] [Google Scholar]
- Vyse S, Howitt A, Huang PH. 2017. Exploiting synthetic lethality and network biology to overcome EGFR inhibitor resistance in lung cancer. J Mol Biol 429: 1767–1786. 10.1016/j.jmb.2017.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cruz-Roa A, Basavanhally A, Gilmore H, Shih N, Feldman M, Tomaszewski J, González F, Madabhushi A. 2014. Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J Med Imaging (Bellingham) 1: 034003. 10.1117/1.JMI.1.3.034003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Ma X, Zhu C, Guo L, Li Q, Liu M, Zhang J. 2016. The prognostic value of long non coding RNAs in non small cell lung cancer: a meta-analysis. Oncotarget 7: 81292–81304. 10.18632/oncotarget.13223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chen A, Yang L, Cai L, Xie Y, Fujimoto J, Gazdar A, Xiao G. 2018a. Comprehensive analysis of lung cancer pathology images to discover tumor shape and boundary features that predict survival outcome. Sci Rep 8: 10393. 10.1038/s41598-018-27707-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Dong N, Dai W, Rosario SD, Xing EP. 2018b. Classification of breast cancer histopathological images using convolutional neural networks with hierarchical loss and global pooling. In Image analysis and recognition (ed. Campilho A, Karray F, ter Haar Romeny B), pp. 745–753. Springer, Berlin. [Google Scholar]
- Warth A, Körner S, Penzel R, Muley T, Dienemann H, Schirmacher P, von Knebel-Doeberitz M, Weichert W, Kloor M. 2016. Microsatellite instability in pulmonary adenocarcinomas: a comprehensive study of 480 cases. Virchows Arch 468: 313–319. 10.1007/s00428-015-1892-7 [DOI] [PubMed] [Google Scholar]
- Washetine K, Heeke S, Bonnetaud C, Kara-Borni M, Ilie M, Lassalle S, Butori C, Long-Mira E, Marquette CH, Cohen C, et al. 2018. Establishing a dedicated lung cancer biobank at the University Center Hospital of Nice (France). Why and how? Cancers (Basel) 10: 220. 10.3390/cancers10070220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way GP, Sanchez-Vega F, La K, Armenia J, Chatila WK, Luna A, Sander C, Cherniack AD, Mina M, Ciriello G, et al. 2018. Machine learning detects Pan-cancer Ras pathway activation in the Cancer Genome Atlas. Cell Rep 23: 172–180.e3. 10.1016/j.celrep.2018.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei IH, Shi Y, Jiang H, Kumar-Sinha C, Chinnaiyan AM. 2014. RNA-seq accurately identifies cancer biomarker signatures to distinguish tissue of origin. Neoplasia 16: 918–927. 10.1016/j.neo.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JW, Tafe LJ, Linnik YA, Vaickus LJ, Tomita N, Hassanpour S. 2019. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci Rep 9: 3358. 10.1038/s41598-019-40041-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidle UH, Birzele F, Nopora A. 2019. MicroRNAs as potential targets for therapeutic intervention with metastasis of non-small cell lung cancer. Cancer Genomics Proteomics 16: 99–119. 10.21873/cgp.20116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissferdt A. 2018. Pulmonary sarcomatoid carcinomas: a review. Adv Anat Pathol 25: 304–313. 10.1097/PAP.0000000000000202 [DOI] [PubMed] [Google Scholar]
- West L, Vidwans SJ, Campbell NP, Shrager J, Simon GR, Bueno R, Dennis PA, Otterson GA, Salgia R. 2012. A novel classification of lung cancer into molecular subtypes. PLoS ONE 7: e31906. 10.1371/journal.pone.0031906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West H, McCleod M, Hussein M, Morabito A, Rittmeyer A, Conter HJ, Kopp HG, Daniel D, McCune S, Mekhail T, et al. 2019. Atezolizumab in combination with carboplatin plus nab-paclitaxel chemotherapy compared with chemotherapy alone as first-line treatment for metastatic non-squamous non-small-cell lung cancer (IMpower 130): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol 20: 924–937. 10.1016/S1470-2045(19)30167-6 [DOI] [PubMed] [Google Scholar]
- Williams MJ, Werner B, Heide T, Curtis C, Barnes CP, Sottoriva A, Graham TA. 2018. Quantification of subclonal selection in cancer from bulk sequencing data. Nat Genet 50: 895–903. 10.1038/s41588-018-0128-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C, Francis S, Tan M, Martins M, Winterford C, Davidson M, Duhig EE, Clarke B, Hayward NK, Yang IA, et al. 2012. MS4A1 dysregulation in asbestos-related lung squamous cell carcinoma is due to CD20 stromal lymphocyte expression. PLoS ONE 7: e34943. 10.1371/journal.pone.0034943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Li L, Li S. 2015. Circulating microRNA-21 as a biomarker for the detection of various carcinomas: an updated meta-analysis based on 36 studies. Tumour Biol 36: 1973–1981. 10.1007/s13277-014-2803-2 [DOI] [PubMed] [Google Scholar]
- Wu KL, Tsai YM, Lien CT, Kuo PL, Hung AJ. 2019a. The roles of microRNA in lung cancer. Int J Mol Sci 20: 1611. 10.3390/ijms20071611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SG, Chang TH, Liu YN, Shih JY. 2019b. MicroRNA in lung cancer metastasis. Cancers (Basel) 11: 265. 10.3390/cancers11020265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Kong X, Xing F, Liu F, Su H, Yang L. 2015. Deep voting: a robust approach toward nucleus localization in microscopy images. Med Image Comput Comput Assist Interv 9351: 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Jia Z, Wang LB, Ai Y, Zhang F, Lai M, Chang EI. 2017. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinformatics 18: 281. 10.1186/s12859-017-1685-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Hu M, Li P, Ma J, Xie L, Teng F, Zhu Y, Fan B, Mu D, Yu J. 2017. Relationship between expression of PD-L1 and tumor angiogenesis, proliferation, and invasion in glioma. Oncotarget 8: 49702–49712. 10.18632/oncotarget.17922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Amann JM, Tanimoto A, Taniguchi H, Shukuya T, Timmers C, Yano S, Shilo K, Carbone DP. 2018. Histone deacetylase inhibition enhances the antitumor activity of a MEK inhibitor in lung cancer cells harboring RAS mutations. Mol Cancer Ther 17: 17–25. 10.1158/1535-7163.MCT-17-0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Zhang W. 2018. Precision medicine becomes reality-tumor type-agnostic therapy. Cancer Commun (Lond) 38: 6. 10.1186/s40880-018-0274-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Wang X, Lu L, Summers RM. 2018. Deeplesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J Med Imaging (Bellingham) 5: 036501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hu Z, Zhou Y, Zhao G, Lei Y, Li G, Chen S, Chen K, Shen Z, Chen X, et al. 2017. The clinical use of circulating microRNAs as non-invasive diagnostic biomarkers for lung cancers. Oncotarget 8: 90197–90214. 10.18632/oncotarget.21644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CL, Zheng XL, Ye K, Ge H, Sun YN, Lu YF, Fan QX. 2018. MicroRNA-183 acts as a tumor suppressor in human non-small cell lung cancer by down-regulating MTA1. Cell Physiol Biochem 46: 93–106. 10.1159/000488412 [DOI] [PubMed] [Google Scholar]
- Yerukala Sathipati S, Ho SY. 2017. Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Sci Rep 7: 7507. 10.1038/s41598-017-07739-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhak K, Aguet F, Kim J, Hess JM, Kübler K, Grimsby J, Frazer R, Zhang H, Haradhvala NJ, Rosebrock D, et al. 2019. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364: eaaw0726. 10.1126/science.aaw0726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HJ, Robinson S, Blair Christian J, Qiu JX, Tourassi GD. 2018. Filter pruning of Convolutional Neural Networks for text classification: a case study of cancer pathology report comprehension. In IEEE EMBS International Conference on Biomedical & Health Informatics, BHI 2018, pp. 345–348. Las Vegas, NV, March 4–7. [Google Scholar]
- Yu J, Kane S, Wu J, Benedettini E, Li D, Reeves C, Innocenti G, Wetzel R, Crosby K, Becker A, et al. 2009. Mutation-specific antibodies for the detection of EGFR mutations in non-small-cell lung cancer. Clin Cancer Res 15: 3023–3028. 10.1158/1078-0432.CCR-08-2739 [DOI] [PubMed] [Google Scholar]
- Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M, Riely GJ. 2013. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 19: 2240–2247. 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HA, Tian SK, Drilon AE, Borsu L, Riely GJ, Arcila ME, Ladanyi M. 2015. Acquired resistance of EGFR-mutant lung cancer to a T790M-specific EGFR inhibitor: emergence of a third mutation (C797S) in the EGFR tyrosine kinase domain. JAMA Oncol 1: 982–984. 10.1001/jamaoncol.2015.1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KH, Zhang C, Berry GJ, Altman RB, Ré C, Rubin DL, Snyder M. 2016. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 7: 12474. 10.1038/ncomms12474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KH, Wang F, Berry GJ, Ré C, Altman RB, Snyder M, Kohane IS. 2020. Classifying non-small cell lung cancer types and transcriptomic subtypes using convolutional neural networks. J Am Med Inform Assoc 27: 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Du W, Wang Y, Xu C, Wang J, Zhang Y, Wang H, Ju J, Zhao L, Wang Z, et al. 2015. Suppression of AKT expression by miR-153 produced anti-tumor activity in lung cancer. Int J Cancer 136: 1333–1340. 10.1002/ijc.29103 [DOI] [PubMed] [Google Scholar]
- Zago G, Muller M, van den Heuvel M, Baas P. 2016. New targeted treatments for non-small-cell lung cancer—role of nivolumab. Biologics 10: 103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaporozhchenko IA, Morozkin ES, Ponomaryova AA, Rykova EY, Cherdyntseva NV, Zheravin AA, Pashkovskaya OA, Pokushalov EA, Vlassov VV, Laktionov PP. 2018. Profiling of 179 miRNA expression in blood plasma of lung cancer patients and cancer-free individuals. Sci Rep 8: 6348. 10.1038/s41598-018-24769-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D, Yu Y, Zhou R, Liao W. 2018. A novel immunoscore signature to predict survival in patients with gastric cancer: implications for immunotherapy. J Clin Oncol 36: 84–84. 10.1200/JCO.2018.36.5_suppl.84 [DOI] [Google Scholar]
- Zhang K, Yuan Q. 2016. Current mechanism of acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitors and updated therapy strategies in human nonsmall cell lung cancer. J Cancer Res Ther 12: C131–C137. 10.4103/0973-1482.200613 [DOI] [PubMed] [Google Scholar]
- Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. 2010. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clinica Chimica Acta 411: 846–852. 10.1016/j.cca.2010.02.074 [DOI] [PubMed] [Google Scholar]
- Zhang HB, Sun LC, Ling L, Cong LH, Lian R. 2016. miR-143 suppresses the proliferation of NSCLC cells by inhibiting the epidermal growth factor receptor. Exp Ther Med 12: 1795–1802. 10.3892/etm.2016.3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang DC, Shi L, Zhu B, Min Z, Jin J. 2017. Genome analyses identify the genetic modification of lung cancer subtypes. Semin Cancer Biol 42: 20–30. 10.1016/j.semcancer.2016.11.005 [DOI] [PubMed] [Google Scholar]
- Zhang J, Späth SS, Marjani SL, Zhang W, Pan X. 2018a. Characterization of cancer genomic heterogeneity by next-generation sequencing advances precision medicine in cancer treatment. Prec Clin Med 1: 29–48. 10.1093/pcmedi/pby007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu L, Summers RM, Kebebew E, Yao J. 2018b. Convolutional invasion and expansion networks for tumor growth prediction. IEEE Trans Med Imaging 37: 638–648. 10.1109/TMI.2017.2774044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zheng H, Zhan Y, Long M, Liu S, Lu J, Zang H, Fan S. 2018c. Detection and application of circulating tumor cell and circulating tumor DNA in the non-small cell lung cancer. Am J Cancer Res 8: 2377–2386. [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Leighl NB, Wu YL, Zhong WZ. 2019a. Emerging therapies for non-small cell lung cancer. J Hematol Oncol 12: 45. 10.1186/s13045-019-0731-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Sun X, Dang K, Li K, Guo XW, Chang J, Yu ZQ, Huang FY, Wu YS, Liang Z, et al. 2019b. Toward an expert level of lung cancer detection and classification using a deep convolutional neural network. Oncologist 24: 1159–1165. 10.1634/theoncologist.2018-0908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wang H, Xie Q, Cao H, Wu F, Di Wu DB, Wan Y. 2019c. Identification of potential diagnostic and therapeutic target genes for lung squamous cell carcinoma. Oncol Lett 18: 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Ma Y, Hu X, Zheng Y, Chen X. 2019d. Targeting PRMT5/Akt signalling axis prevents human lung cancer cell growth. J Cell Mol Med 23: 1333–1342. 10.1111/jcmm.14036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chang L, Yang Y, Fang W, Guan Y, Wu A, Hong S, Zhou H, Chen G, Chen X, et al. 2019e. The correlations of tumor mutational burden among single-region tissue, multi-region tissues and blood in non-small cell lung cancer. J Immunother Cancer 7: 98. 10.1186/s40425-019-0581-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chang L, Yang Y, Fang W, Guan Y, Wu A, Hong S, Zhou H, Chen G, Chen X, et al. 2019f. Intratumor heterogeneity comparison among different subtypes of non-small-cell lung cancer through multi-region tissue and matched ctDNA sequencing. Mol Cancer 18: 7. 10.1186/s12943-019-0939-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen P, McGough M, Xing F, Wang C, Bui M, Xie Y, Sapkota M, Cui L, Dhillon J, et al. 2019g. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat Mach Intell 1: 236–245. 10.1038/s42256-019-0052-1 [DOI] [Google Scholar]
- Zhao W, Liu Y, Jenkins BD, Cheng R, Harris BN, Zhang W, Xie J, Murrow JR, Hodgson J, Egan M, et al. 2019. Tumor antigen-independent and cell size variation-inclusive enrichment of viable circulating tumor cells. Lab Chip 19: 1860–1876. 10.1039/C9LC00210C [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zhao W, Yan C, Watson CC, Massengill M, Xie M, Massengill C, Noyes DR, Martinez GV, Afzal R, et al. 2016. HDAC inhibitors enhance T-cell chemokine expression and augment response to PD-1 immunotherapy in lung adenocarcinoma. Clin Cancer Res 22: 4119–4132. 10.1158/1078-0432.CCR-15-2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Li H, Cheng Y. 2018. P107 the study of relationship between tumor burden (TMB) and molecular typing of non-small cell lung cancer (NSCLC). J Thorac Oncol 13: S1087. 10.1016/j.jtho.2018.10.133 [DOI] [Google Scholar]
- Zhou J, Theesfeld CL, Yao K, Chen KM, Wong AK, Troyanskaya OG. 2018. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat Genet 50: 1171–1179. 10.1038/s41588-018-0160-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu VW, Upadhyay D, Schrock AB, Gowen K, Ali SM, Ou SH. 2016. TPD52L1-ROS1, a new ROS1 fusion variant in lung adenosquamous cell carcinoma identified by comprehensive genomic profiling. Lung Cancer 97: 48–50. 10.1016/j.lungcan.2016.04.013 [DOI] [PubMed] [Google Scholar]
- Zill OA, Banks KC, Fairclough SR, Mortimer SA, Vowles JV, Mokhtari R, Gandara DR, Mack PC, Odegaard JI, Nagy RJ, et al. 2018. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin Cancer Res 24: 3528–3538. 10.1158/1078-0432.CCR-17-3837 [DOI] [PubMed] [Google Scholar]