Abstract
Advances in technology are only beginning to reveal the complex interactions between hosts and their resident microbiota that have co-evolved over centuries. In this review, we present compelling evidence that implicates the host-associated microbiome in the generation of 11β-hydroxyandrostenedione, leading to the formation of potent 11-oxy-androgens. Microbial steroid-17,20-desmolase cleaves the side-chain of glucocorticoids (GC), including cortisol (and its derivatives of cortisone, 5α-dihydrocortisol, and also (allo)- 3α, 5α-tetrahydrocortisol), but not 3α−5β-tetrahydrocortisol) and drugs (prednisone and dexamethasone). In addition to side-chain cleavage, we discuss the gut microbiome’s robust potential to transform a myriad of steroids, mirroring much of the host’s metabolism. We also explore the overlooked role of intestinal steroidogenesis and efflux pumps as a potential route for GC transport into the gut. Lastly, we propose several health implications from microbial steroid-17,20-desmolase function, including aberrant mineralocorticoid, GC, and androgen receptor signaling in colonocytes, immune cells, and prostate cells, which may exacerbate disease states.
Keywords: glucocorticoid; cortisol; microbiome; steroid-17,20-desmolase; 11-oxy-androgen; hydroxysteroid dehydrogenase; sterolbiome; GALF
Graphical abstract
1. Introduction
11β-hydroxyandrostenedione (11β-OHAD) was for decades largely ignored in the study of vertebrate androgens, with the exception of teleost fishes. However, 11β-OHAD is now recognized to play an important role as a precursor to potent 11-oxy-androgens that strongly activate nuclear androgen receptors (AR). While it is now considered that the 11β-hydroxylation of androstenedione by CYP11B1 is the major route of 11β-OHAD in the adrenal gland, the enzyme(s) responsible for the peripheral side-chain cleavage of 11β-OHAD is less clear. Recent in vitro and in vivo work cast doubt on the role of the enzyme CYP17A1 in generating 11β-OHAD through side-chain cleavage of cortisol, leaving two possibilities: (1) an unknown, peripheral host enzyme and/or (2) gut bacterial metabolism of cortisol and derivatives (e.g. 5α-dihydrocortisol, (allo)-3α,5α-tetrahydrocortisol).1,2 In this review, we evaluate the compelling evidence that the host-associated microbiome is implicated in the production of 11β-OHAD, leading to formation of 11-oxy-androgens. Indeed, in recent years much progress has been made developing the molecular biology, enzymology, and structural biology of a microbial pathway known as steroid-17,20-desmolase that catalyzes the side-chain cleavage of cortisol and derivatives such as 5α-dihydrocortisol, (allo)-3α,5α-tetrahydrocortisol). Gaps in our knowledge of the intestinal metabolism of cortisol by gut microbes are indicated that promise to shed light on the potential importance of microbial metabolism of cortisol to 11-oxy-androgens.
1.1. Microbiome as a virtual endocrine organ
The human body harbors approximately the same number of microbial cells, collectively termed the microbiota, as human cells.3 The microbiota inhabit a number of ecosystems comprising the human body, primarily the gut, urinary tract, genitals, and skin, and play a vital role in maintaining human health.4,5 The complex gut microbiome varies significantly cross-sectionally between both healthy and diseased individuals6–8 and longitudinally throughout the lifespan of the individual.5,9 Host genetics, diet, geographical location, lifestyle, mode of delivery (birth), and pharmaceutical interventions impact the composition and function of the microbiota.10–13 Much of the research conducted in the microbiome field has focused on understanding the associations between the gut microbiota and health outcomes involving changes in host metabolism, immunity, behavior, and many other systems.14–16 Elucidating the ‘gut-brain axis’ is currently of major interest, the crosstalk between the brain, central and enteric nervous system, and gut that is partially facilitated by the gut microbiome.17–20
In 1993, Lyte and Ernst first defined the field of microbial endocrinology.21 While the field is still in its infancy, we are beginning to elucidate the bidirectional host-microbe and microbe-microbe interactions involving hormones. The two major groups of hormones involved in mediating host-microbe interactions are neurohormones and steroid hormones. The microbiota is not only capable of producing and secreting hormones, but also modulating host production of endogenous hormones.21 The interplay between the gut microbiome and host endocrinology can be evidenced by the ability of Lactobacillus helveticus and Bifidobacterium longum to reduce cortisol levels and anxiety-like behavior in rats and humans.22 Additionally, host steroid hormones, including bile acids, can change the composition and function of the gut microbiome. For example, the opportunistic gut pathogen, Clostridioides difficile, is inhibited by the host-derived bile acid chenodeoxycholic acid23, yet requires host taurocholic acid for spore germination and growth.24 Conversely, the gut microbiome can reciprocally impact steroid hormones which, in turn, influence host physiology. For instance, 21-deoxycortisol is a gut microbial metabolite of cortisol that acts as a potent inhibitor of 11β-hydroxysteroid dehydrogenase 2 (11β-HSD2), an enzyme responsible for controlling levels of active glucocorticoid (GC), cortisol, in the body.25,26 Gut microbiota are also capable of converting cortisol to the androgen precursor, 11β-OHAD27, which is the primary focus of this review.
The host-microbe interactions that define this virtual endocrine organ are still poorly understood, yet recent research is beginning to focus on potential therapeutic targets. It is therefore important to work out microbial biochemical pathways involved in the synthesis or metabolism of signaling molecules that represent communication between commensal prokaryotes and host eukaryotes in order to determine their role in physiology and pathophysiology.
1.2. Human cortisol metabolism
Corticosteroids are 21-carbon steroid hormones that are synthesized in the adrenal cortex of vertebrates from cholesterol. The two main classes of corticosteroids, GCs and mineralocorticoids, play an important role in regulating many physiological functions in the host, including inflammation, stress response, immune response, macronutrient metabolism, electrolyte balance, and water retention. The two main GC in humans are cortisol and corticosterone, which structurally differ by the presence or absence of a hydroxyl group at the C17 position, respectively. In humans, the concentration of circulating cortisol is approximately 10–20 fold higher than corticosterone. The local concentration of active GC is tightly controlled in many tissues by the regulated action of 11β-hydroxysteroid dehydrogenase isoforms 1 and 2 (11β-HSD1/2). Cortisol and corticosterone can be interconverted to their respective inactive forms when the C11 hydroxyl group is oxidized, forming cortisone and 11-dehydrocorticosterone.28
Stimulation of the central nervous system through stress activates hypothalamic-pituitary-adrenal axis (HPA) signaling. This initiates the secretion of corticotropin releasing hormone (CRH) from the hypothalamus that consequently signals adrenocorticotropic hormone (ACTH) release from the pituitary gland, stimulating GC synthesis in the adrenal glands. Because of low cortisol stores in adrenocortical cells, cortisol is primarily synthesized de novo upon activation of the HPA axis. Approximately 10–15 mg of cortisol is secreted daily from the adrenal glands in humans.29 The biosynthetic precursor for cortisol is cholesterol, which comes from: (1) uptake from circulating low-density lipoprotein or high-density lipoprotein, (2) mobilization of stored cholesteryl esters in lipid droplets through hormone sensitive lipase, and (3) endogenous synthesis from acetyl-CoA. Once synthesized, cortisol is released into systemic circulation where it acts on peripheral tissues by binding and signaling through nuclear glucocorticoid receptor (GR), and modulating gene expression involved with inflammation, homeostasis, growth, and energy production.28 Cortisol also affects physiology through non-genomic mechanisms.30 The majority of cortisol in circulation is protein bound to cortisol binding globulin (CBG) and approximately only 5% of circulating cortisol is free and biologically active. Protein bound cortisol has a longer half-life of between 70 and 120 minutes compared to the half-life of free cortisol ranging to a few minutes.28 Once arrived at the target tissue, cortisol and other hydrophobic steroid hormones diffuse across cytoplasmic membranes, but may be effluxed out of the cell by active transporters to regulate the intracellular concentrations of steroids.31
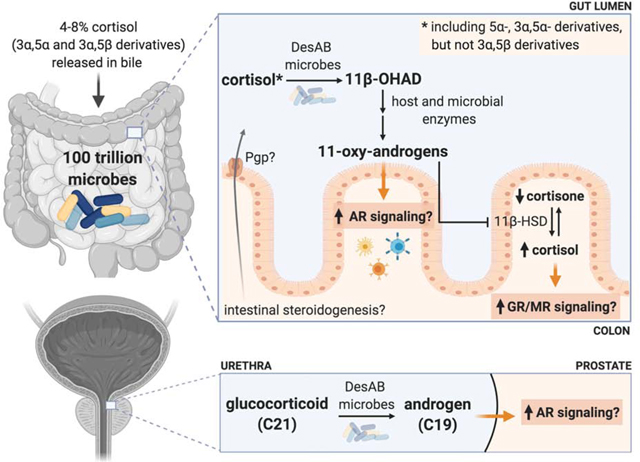
The major site of host peripheral cortisol metabolism occurs in the liver, where cortisol can be reduced, oxidized, hydroxylated, and conjugated. The solubility of these cortisol metabolites is increased by either sulfation or glucuronidation to facilitate their main route of excretion through urine. Approximately 4–8% of cortisol and its derivatives in humans are released in bile, entering into the ileum upon ingestion of a meal and excreted through feces.28 Radiometabolism studies in a variety of species have revealed significant sex and species differences regarding excretion route of GCs.32
Psychological stress measurement in diverse vertebrates, particularly in conservation studies, is accomplished in a “feedback free” manner through the measurement of fecal derivatives of cortisol and side-chain cleavage products of cortisol, including 11β-OHAD and its ring-A reduced and 17-hydroxy derivatives.32 Early studies showed that intravenous administration of C14 labeled cortisol to human patients resulted in both urinary and fecal excretion of C14 labeled metabolites.33 While numerous authors have remarked that gut microbial metabolism of cortisol in humans can be disregarded due to the relatively minor enterohepatic circulation of cortisol (relative to corticosterone), much evidence has emerged since the 1950s that support microbial cortisol metabolism, and potential alternative routes for GC entry into the GI tract (discussed in Section 2).
1.3. Discovery of microbial desmolase activity
One of the earliest clinical observations that suggested microbial side-chain cleavage, or steroid-17,20-desmolase activity, was obtained after administration of rectal hydrocortisone (cortisol) as a treatment for ulcerative colitis. Rectal infusion led to a 100-fold spike in urinary excretion of C19 17-ketosteroids.34 Interestingly, this effect was suppressed when the hydrocortisone treatment was given in conjunction with oral neomycin, a broad spectrum antibiotic that accumulates in the GI tract, suggesting that the gut bacteria are responsible for the biotransformation.35 Direct evidence of microbial side-chain cleavage of cortisol by fecal microbiota came a decade later when cortisol-spiked samples of human and rat feces yielded notable conversion of various C19 metabolites.36 Additional work from the 1970s and early 1980s confirmed that desmolase activity in normal human feces was of bacterial origin, but the microbe(s) responsible for this action had not been identified.2,37
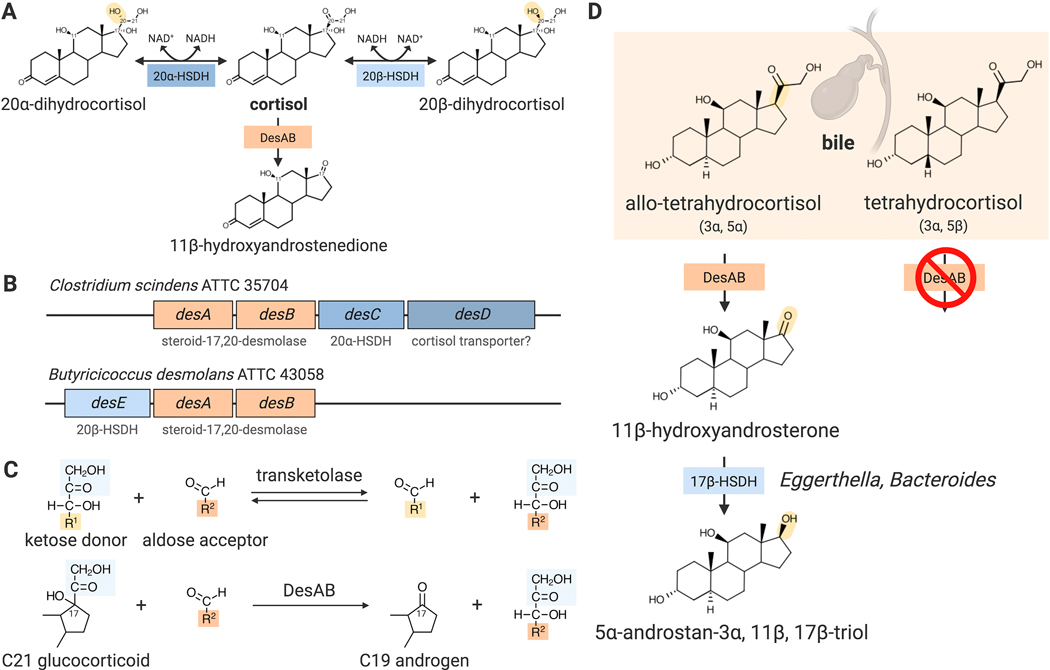
In 1984, the first human fecal bacterium with demonstrated steroid-17,20-desmolase activity was isolated.38 Originally referred to as “Clostridium strain 19,” the isolate was provisionally named Clostridium scindens (type strain American Type Culture Collection (ATCC) 35704) which means “to cut” owing to steroid-17,20-desmolase activity 39. C. scindens is a gram-positive, spore-forming, motile, obligate anaerobe that was shown to metabolize both cortisol and bile acids. When investigating steroid-17,20-desmolase activity in C. scindens, it was determined that this microbe expressed NADH-dependent 20α-hydroxysteroid dehydrogenase (HSDH) activity.40 Importantly, the expression of steroid-17,20-desmolase and NAD(H)-dependent corticosteroid 20α-HSDH activities were tightly regulated by the presence of cortisol and cortisone (but not corticosterone), indicating an evolved specificity of microbial transcriptional control for host steroids.
A few years later, Bokkenheuser identified two additional isolates with steroid-17,20-desmolase activity, Eubacterium desmolans (now known as Butyricicoccus desmolans ATCC 43058) and Clostridium cadaveris AGR2141, from cat feces and New York sewage, respectively.41 Because cortisol undergoes enterohepatic circulation in cats, they hypothesized that this animal would naturally harbor steroid-17,20-desmolase-encoding microbes in the gut. B. desmolans is a gram-positive, non-spore forming, and chemoorganotrophic obligate anaerobe. In addition to steroid-17,20-desmolase activity, B. desmolans and C. cadaveris reduce 20-keto steroids by 20β-HSDH, distinct from the 20α-HSDH activity of C. scindens (Figure 1A-B).41
Fig. 1. Cortisol metabolism by the bacterial steroid desmolase pathway.
(A) Cortisol is side-chain cleaved by DesAB and/or reversibly reduced/oxidized by NAD(H)-dependent 20α- and 20β-hydroxysteroid dehydrogenases (HSDHs), encoded by desC and desE, respectively. (B) Genomic structure of steroid-17,20-desmolase operon in gut microbes, C. scindens ATCC 35704 and B. desmolans ATCC 43058. (C) Metabolism of glucocorticoids by DesAB parallels the biochemistry of sugar transketolation. (D) Released in bile, allo-tetrahydrocortisol, but not tetrahydrocortisol, is a substrate for DesAB, forming 11β-hydroxyandrosterone, which can be further metabolized by other gut microbes belonging to the genus Eggerthella and Bacteroides by 17β-HSDH activity.
1.4. Human 11β-hydroxyandrostenedione metabolism
The relevance of bacterial steroid-17,20-desmolase is in the importance of the formation of its end-product, 11β-OHAD, to host physiology. 11β-OHAD is a C19 steroid endogenously synthesized in the mammalian adrenal. Even though 11β-OHAD is a major adrenal product that was first discovered in 1955, it has been largely overlooked until recently because its biological significance eluded researchers for many decades. Recent prolific work from the Swart and Storbeck laboratories has shown that 11β-OHAD is a weak androgen that when metabolized in steroid-responsive peripheral tissues, yields potent androgens implicated in androgen-dependent diseases, such as castration resistant prostate cancer, congenital adrenal hyperplasia, and polycystic ovary syndrome (PCOS).42–47 Through the synergistic actions of host enzymes 5α-reductase (SRD5A), 11β-HSD2, and 17β-hydroxysteroid dehydrogenase (17β-HSD), 11β-OHAD can be converted to the potent 11-oxy-androgens, 11-keto-testosterone (11-KT) and 11-keto-dihydrotestosterone (11-KDHT), whose androgenic activity is on par with dihydrotestosterone (DHT).48
It was first postulated by Dorfman in the 1950s that C21 GCs gave rise to C19 androgens through a side-chain cleavage, or lyase, reaction in the adrenal.49 However, with significant advances of analytical tools, much of the work over the next few decades led to the conclusion that 11β-OHAD was formed through 11β-hydroxylation (by CYP11B1) of androstenedione, a C19 precursor molecule of the androgen pathway. It was believed that human cytochrome P450 17A1 (CYP17A1, 17α-hydroxylase, 17,20-lyase) is responsible for side-chain cleavage of cortisol because the enzyme was shown to cleave the C17-C20 carbon-carbon bond of other adrenal steroids, particularly the conversion of 17-hydroxypregnenolone and 17-hydroxyprogesterone to dehydroepiandrosterone (DHEA) and androstenedione, respectively.50 Shackleton et al. studied patients who were deficient in CYP17A1, and thus deficient in cortisol, 11β-OHAD, and androstenedione production. Intriguingly, oral administration of cortisol to this group resulted in urinary excretion of 11β-OHAD metabolites.1 It was later confirmed that cortisol was not a substrate for the enzyme CYP17A1 expressed in H295R cells, an adrenal cell model.47 Taken together, these results suggest that unknown peripheral enzyme(s), expressed either by the host, or the microbiota, are responsible for the conversion of cortisol to 11β-OHAD.
2. Gut bacteria metabolize cortisol
2.1. The Steroid-17,20-Desmolase (DesAB) Pathway
The observation that anaerobic gut microbes were capable of side-chain cleavage of cortisol (but not corticosterone) became a focus of research in the Bokkenheuser and Hylemon laboratories in the 1980s. A key feature of steroid-17,20-desmolase and pyridine nucleotide-dependent 20α-HSDH activities in C. scindens ATCC 35704 is induction by the addition of cortisol to the growth medium.51 Additional inducers included substrates for DesAB and 20α-HSDH (DesC) such as cortisone and 11-desoxycortisol, as well as the non-substrate 17α-hydroxyprogesterone.52 However, deoxycorticosterone, progesterone, 4-pregnen-17α,20β-diol-3-one, and 4-pregnen-11β,17α,20β,21-tetrol-3-one were not inducers of steroid-17,20-desmolase and 20α-HSDH activities.52 Both enzymatic functions were lost following dialysis in the presence of EDTA, indicating the importance of metal cations. Indeed, addition of divalent metal cations along with NAD+ partially restored steroid-17,20-desmolase and 20α-HSDH activities.52 Molecular oxygen is not required for steroid-17,20-desmolase activity in extracts of C. scindens ATCC 35704, which is a strictly anaerobic bacterium.52 The substrate-specificity for NAD(H)-dependent 20α-HSDH from C. scindens ATCC 35704 differs markedly from most vertebrate 20α-HSDHs which are involved in progesterone metabolism.27,53
The steroid-17,20-desmolase and pyridine nucleotide-dependent 20β-HSDH activities were also reported for Clostridium cadaveris AGR2141 and Butyricicoccus desmolans ATCC 43058 (formerly Eubacterium desmolans) which were reported to be synthesized “constitutively” in culture. This was concluded after observations of undiminished synthesis after weekly transfers of the organism for 6 months, although, in fact, the regulation of this activity was not determined.41 Substrate specificity for side-chain cleavage by B. desmolans was similar to that observed in cell extracts of C. scindens with the exception of the inability to metabolize 20α-dihydrocortisol derivatives, and the ability to side-chain cleave 20β-dihydrocortisol derivatives.41 Research on bacterial steroid-17,20-desmolase stagnated for the next three decades. Then in 2007, a draft genome of C. scindens ATCC 35704 was submitted to NCBI as a reference genome for the Human Microbiome Project. Shortly thereafter, interest in steroid-17,20-desmolase was rekindled in the Hylemon lab.
We knew that the biochemical reaction catalyzed by bacterial desmolase would be unlike that of the vertebrate host as well as aerobic soil fungi and bacteria54, which encode cytochrome P450 monooxygenase requiring NADPH and molecular oxygen. Instead, we predicted that it would be an alternate form of carbon-carbon lyase, but had no clear candidates after examining the genomic annotations. We decided on an unbiased genome-wide transcriptomic approach (RNA-Seq), taking advantage of the cortisol-inducibility of steroid-17,20-desmolase. RNA-Seq analysis resulted in identification of a gene cluster comprising three structural genes and a transport protein that was highly co-expressed after cortisol-induction encoding what appeared to be involved in sugar metabolism. The first two genes encode putative N-terminal and C-terminal transketolase predicted to be involved in sugar metabolism in the pentose-phosphate pathway. There was a clear biochemical analogy between sugar transketolation and steroid-17,20-desmolase leading us to pursue this gene cluster further (Figure 1C). The initial confirmation that this gene cluster was functionally involved in cortisol metabolism came from expressing the zinc-dependent medium chain dehydrogenase, DesC, in E. coli and characterizing the purified recombinant enzyme. The recombinant 40kDa (subunit) 20α-HSDH exhibited NADH-dependent activity with cortisol, but not 20α- or 20β-dihydrocortisol, and NAD+-dependent activity with 20α-dihydrocortisol. The 20α-HSDH is specific for those substrates with 17α-hydroxy and 21-hydroxyl groups.27
More recently, we purified recombinant DesAB (rDesAB) and confirmed that the predicted desA and desB genes from C. scindens encoded steroid-17,20-desmolase activity. We developed a novel enzyme-linked continuous spectrophotometric assay to quantify steroid-17,20-desmolase activity, linking conversion of 11β-OHAD to 11β-hydroxy-testosterone (11β-OHT) by recombinant 17β-HSDH from Cochliobolus lunatus, allowing quantification of conversion of NADPH to NADP+ at 340 nm.55 The Km and kcat kinetic constants with cortisol as the substrate were determined to be 4.96 ± 0.57 μM and 0.87 ± 0.076 min−1, respectively. Substrate specificity analysis using rDesAB revealed a requirement of 17,21-dihydroxy-20-ketosteroids for desmolase activity.55 Importantly, we observed that while tetrahydrocortisol (3α-,5β-) is not a substrate for DesAB.55 This is in contrast to the original characterization of C. scindens ATCC 35704 which report side-chain cleavage of tetrahydrocortisol.40 We determined that allo-tetrahydrocortisol (3α-, 5α-) is side-chain cleaved by both whole cells of C. scindens ATCC 35704 as well as rDesAB.56 The secretion of cortisol in bile is in the form of conjugated 3α-,5β- and 3α-,5α-derivatives. The ability of C. scindens ATCC 35704 to side-chain cleave allo-tetrahydrocortisol to 11β-hydroxyandrosterone is physiologically significant as 200–400 micrograms is secreted in bile each day57 (Figure 1D). Since both cortisol and allo-tetrahydrocortisol have planar trans-A/B rings, while the cis-A/B rings of tetrahydrocortisol are non-planar, these features appear to be important for substrate-binding.
2.2. Metabolism of cortisol and 11β-hydroxyandrostenedione by the gut microbiome
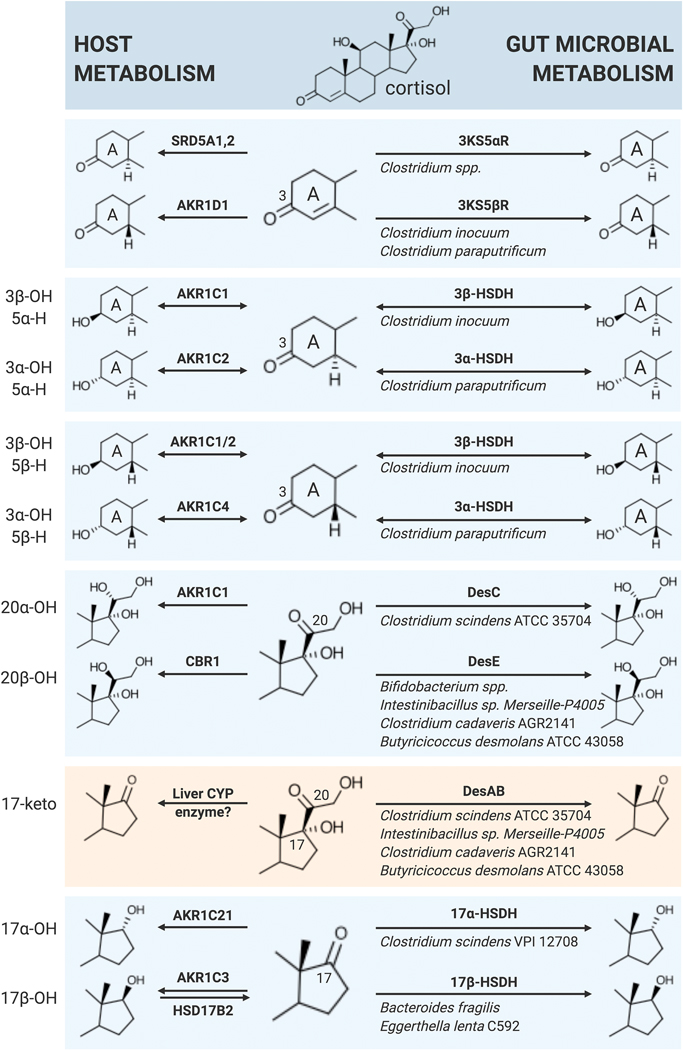
Side-chain cleavage of cortisol by bacterial DesAB is one pathway in the complex vertebrate gut sterolbiome.58,59 However, radiometric studies of cortisol incubated with rat and human feces identified the formation of numerous metabolites (Table 1). Under anaerobic conditions, bacteria utilize numerous substrates, including GC and other steroids as electron acceptors.2 Ring-A metabolism includes conversion of cortisol by gut bacteria to dihydrocortisol (5β-H) and tetrahydrocortisol (3α-hydroxy,5β-H) in addition to 3β-hydroxy/5β-H, 3α-hydroxy/5α-H, and 3β-hydroxy/5α-H derivatives. These observations indicate that gut microbiota encode 3α-HSDHs and 3β-HSDHs that recognize cortisol. Indeed, NAD(P)(H)-dependent 3α-HSDHs in the short chain dehydrogenase (SDR) family have been reported that recognize bile acids, although corticosteroids have not been tested as possible substrates.60,61 3β-HSDHs were recently reported to be involved in bile acid60,61 and cholesterol62 metabolism. Bacterial HSDHs of a given stereo- and regio-specificity are often encoded by more than one species, and may represent convergent evolution from more than one protein family. For instance, gut bacterial 3β-HSDHs have been reported in both the SDR and aldo/keto reductase families.60,61 Clostridium paraputrificum and Clostridium innocuum were shown to possess 3α-HSDH and 3β-HSDH, respectively, involved in steroid metabolism.63 The genes encoding these enzymes in C. paraputrificum and C. innocuum have yet to be identified. The expression of bacterial enzymes active against Ring-A functional groups indicate that both cortisol and (allo)tetrahydrocortisol produced by the host may be modified in the GI tract.
Table 1.
Metabolites generated from radiolabeled cortisol spiked into fecal samples.a
| Steroid | Trivial name |
|---|---|
| 5β-pregnan-3α, 11 β,17,21-tetrol-20-one | tetrahydrocortisol |
| 4-pregnen-17, 21-diol-3,20-dione | 21-deoxycortisol |
| 5β-pregnan-3α, 17α,21-triol-20-one | tetrahydro-21-deoxycortisol |
| 5α-androstane-3α,11 β-diol-17-one | 11α-hydroxyetiocholanolone |
| 5β-androstane-3α,11β-diol-17-one | 11 β-hydroxyetiocholanolone |
Data from Cerone-McLernon et al. 19812.
The results of Cerone-McLernon et al (1981) also indicate that gut bacteria encode steroid 5β- and 5α-reductases. Indeed, exogenous cortisol was reported to be converted to tetrahydrocortisol and allo-tetrahydrocortisol in fecal suspensions.2 Work performed decades ago established that Clostridium paraputrificum and Clostridium innocuum possess 3-oxo-4–5β-reductase activity against progesterone and testosterone.63 Previous studies with cell extracts and purified pyridine-nucleotide and flavin-dependent bile acid-4–5β-reductases encoded by baiCD and baiH genes in C. scindens indicate that gut microbial metabolism of C4=C5 may be reversible.64,65 Thus, the ratio of host secreted biliary 5α- and 5β-reduced GC may be altered by oxidation and epimerization reactions by concerted action of gut microbes encoding steroid 5α- and 5β-reductases. Further studies are needed to determine this, but it is well known that there is a radial redox gradient from the mucosal surface to the center of the lumen, suggesting a stronger tendency for oxidation (formation of 3-oxo-Δ4) near the microaerophilic mucosa, and reduction (3α-hydroxy, 3β-hydroxy and 5α-H, 5β-H) in the anaerobic lumen. The combination of microbial side-chain cleavage of allo-tetrahydrocortisol, and the possibility of gut microbiota altering the ratio of host biliary secreted ring-A reduced derivatives in vivo will be an important aspect of cortisol metabolism to determine.
Following side-chain cleavage of cortisol, bacterial ring-A active enzymes are capable of converting 11β-OHAD to a diversity of reduced products(Table 1).2 The human microbiome is also capable of reducing the 17-ketone to androgens (17βhydroxy) and epi-derivatives (17α-hydroxy). Indeed, Bacteroides fragilis was previously shown to express pyridine nucleotide-dependent 17β-HSDH.66 17β-HSDH activity has also been reported in Eggerthella lenta strain C592.67 The genes encoding microbial enzymes involved in metabolism of the Δ4 and 17-keto have yet to be identified and characterized. The human intestinal epithelium also expresses 17β-HSD isoforms 2 and 4, which catalyze the oxidative conversion of biologically active estradiol and testosterone (T) to inactive estrone and androstenedione (A4).68 Thus, the luminal steroid metabolome is likely an equilibrium between the mucosal oxidation to inactive 17-keto, and the gut microbial reduction to 17β-hydroxy as a direct means of removing excess reducing equivalents from the cell, and perhaps indirect benefit through inter-kingdom signaling.
Gut bacteria are also capable of forming epi-testosterone derivatives through expression of 17α-HSDH activity, which has been reported in C. scindens VPI 1270869, a strain that lacks steroid-17,20-desmolase activity, although screening of likely candidate genes was unable to identify the gene encoding this enzyme.67 C. scindens ATCC 35704 which harbors the desABCD genes involved in steroid-17,20-desmolase, lacks detectable 17-HSDH activities.27 Originally, C. scindens was named based on the expression of steroid-17,20-desmolase activity.40 However, we found that this activity is in fact rare among C. scindens isolates.27 Functional variation between strains of bacterial species is an important concept in gastrointestinal microbiology. This is particularly critical to understand given the advent of 16S rDNA sequencing of complex, host-associated communities. Even if 16S rDNA sequence can establish a particular species is present, it is not clear that the particular strain(s) in the sample harbors genes encoding functions of interest such as steroid-17,20-desmolase or 17α-HSDH. Instead, the gene itself must be detected by metagenomic sequencing of bacterial genomes in the sample, or by quantitative PCR. Thus, inter-individual and life-long inter-personal variation in the gut microbial metagenomic gene content involved in steroid biotransformations by intestinal bacteria (genotype) determine the extent to which microbes can contribute to the steroid metabolome (phenotype).
These observations indicate that the microbiome may be capable of altering the structure of host corticosteroids and 11β-OHAD through vitamin B1-dependent, pyridine-nucleotide, as well as flavin-dependent enzymes. Host generated tetrahydrocortisol in the liver that is secreted into the GI tract in bile might be converted to cortisol by bacteria such as C. paraputrificum under microaerophilic conditions close to the mucosa, and reduced to allo-tetrahydrocortisol by organisms such as C. innocuum followed by side-chain cleavage, resulting in 11β-hydroxyandrosterone. The metabolic potential of each individual’s microbiome is determined at least in part by the ‘sterolbiome’ present as well as the relative expression and activity of sterolbiome enzymes (Figure 2). Much work is needed to understand the interaction between sterolbiome enzymes expressed by diverse facultative and strictly anaerobic members of the normal microbiota. So far, the steroid-17,20-desmolase pathway is the best understood and characterized relating to cortisol metabolism.
Fig. 2. Summary of similarities between host (left) and gut microbial (right) steroid metabolism.
3KS5αR, 3-ketosteroid-5α-reductase; 3KS5βR, 3-ketosteroid-5β-reductase; 3α-HSDH, 3α-hydroxysteroid dehydrogenase; 3β-HSDH, 3β-hydroxysteroid dehydrogenase; 17α-HSDH, 17α-hydroxysteroid dehydrogenase; 17β-HSDH, 17β-hydroxysteroid dehydrogenase; AKR1C1, aldo-keto reductase 1C1; AKR1C2, aldo-keto reductase 1C2; AKR1C3, aldo-keto reductase 1C3; AKR1C4, aldo-keto reductase 1C4; AKR1D1, aldo-keto reductase 1D1; AKR1C21, aldo-keto reductase 1C21; CBR1, carbonyl reductase 1; DesAB, steroid-17,20-desmolase; DesC, 20α-hydroxysteroid dehydrogenase; DesE, 20β-hydroxysteroid dehydrogenase; HSD17B2, 17β-hydroxysteroid dehydrogenase; SRD5A1,2, steroid-5α-reductase 1,2.
3. Structural biology of cortisol metabolizing enzymes
3.1. Steroid-17,20-desmolase (DesAB)
The desA and desB genes from C. scindens ATCC 35704 encode 296 and 327 amino acid proteins that were previously annotated as N-terminal and C-terminal subunits of transketolase, respectively. However, desA and desB deduced amino acid sequences share only 34% identity with other sugar transketolases in the BLAST database.27 Pfam searches predict that DesA has a transketolase thiamine diphosphate binding domain (PF00456) and DesB has a transketolase C-terminal domain (PF02780) and transketolase pyrimidine binding domain (PF02779). rDesAB from C. scindens has a native molecular mass of approximately 142 kDa, suggesting that it is a heterotetramer composed of rDesA and rDesB subunits that are 32.7 kDa and 38.4 kDa, respectively.55 CLUSTAL-Omega alignment shows that DesAB from C. scindens, B. desmolans, C. cadaveris, P. lymphophilum, and Arcanobacterium urinimassiliense share conserved substrate-binding, metal chelating, and active-site binding residues with transketolases from humans and Lactobacillus salivarius.55 These residues include: metal binding residues (DesA: D153, E155), TPP binding residues (DesA: H74, G154, N183, K253; DesB: F95), as well as active site residues (DesA: H34, H268; DesB: E70) and substrate binding residues (DesB: H121, H132, R178).55 Steroid-17,20-desmolase is the first example of a steroid transketolase. It will be interesting to determine if experimentally verified sugar transketolases are capable of side-chain cleavage of steroids. Comparison of substrate-binding sites and active-site amino acid and cofactor geometry are expected to reveal the mechanism of steroid-17,20-desmolase and suggest how sugar transketolase might be evolutionarily co-opted to shift from sugar to steroid metabolism.
Currently, DesAB has yet to be crystallized. Crystallization of DesAB will allow us to begin computational screens for small molecule inhibitors to block enzymatic activity. However, further work is needed to determine if DesAB is a potential therapeutic target for decreasing androgen production in vivo.
3.2. 20α/β-hydroxysteroid dehydrogenases (DesC, DesE)
Hydroxysteroid dehydrogenases (HSDHs) catalyze the pyridine nucleotide-dependent reversible oxidation and reduction of steroid hormones.70 HSDHs fall within three different large and diverse protein superfamilies: aldo-keto reductase (AKR), medium-chain dehydrogenase/reductase (MCDR), or short-chain dehydrogenase/reductase.27,70 Although AKR HSDHs have been primarily found in mammals71, we recently reported 3β-HSDH in the AKR family from Eggerthella CAG:298.60 HSDHs within the SDR and MCDR superfamilies have also been discovered within the gut microbiome.27,61,72,73
DesE, or NADH-dependent cortisol 20β-HSDH, is a member of the SDR superfamily.73,74 The SDR superfamily is one of the largest, with members across all three domains of life and diverse substrate-specificities.75 SDR enzymes are non-metalloenzymes with a typical length of 250 amino acid residues70 that recognize substrates ranging from sugars to steroids.76 Due to their dependence on pyridine nucleotides to catalyze oxidation and reduction, they are characterized by Rossmann fold domains for cofactor binding.77 The Rossmann fold domain consists of 6–7 β-strands surrounded by 3–4 α-helices on each side.77,78 The N-terminal region of SDR proteins typically binds the cofactor and contains a Gly-rich sequence while the C-terminal region binds substrates.76 The majority have a conserved Tyr in the catalytic site with nearby Ser and Lys residues.78 Additionally, the folding pattern of SDR members is more highly conserved than their underlying amino acid sequences78, causing extreme difficulty in prediction of their substrate-specificities by homology search.
Thus far, enzymes encoded by two desE genes have been characterized, one from Butyricicoccus desmolans ATCC 43058 and another from Bifidobacterium adolescentis strain L2–32.73,74 We recently solved the 2.0Å apo- and 2.2Å holoenzyme structures of NADH-dependent 20β-HSDH from B. adolescentis (PDB ID: 6M9U, 6OW4) (Figure 3). A bifunctional 3α,20β-HSDH from Streptomyces exfoliatus (previously reported as S. hydrogenans) (PDB ID: 2HSD)79 has also been crystallized, although it shares little amino acid sequence identity (~30%) with DesE. The holoenzyme structure reveals that B. adolescentis DesE binds NADH within its Rossmann fold domain composed of 6 α-helices around a collection of 7β-strands. This protein has a catalytic tetrad consisting of Asn-Ser-Tyr-Lys. Its catalytic mechanism is predicted to begin with a proton transfer from Ser to the C-20 carbonyl group of cortisol. Then, a hydride transfer likely occurs from NADH to the C-20 carbonyl leading to a proton relay from NADH to Ser, Tyr to 2’OH ribose, and bulk solvent back to the catalytic Tyr. This protein also contains a large, flexible N-terminal region that, upon mutagenesis and a combination of gel-filtration chromatography and thermal circular dichroism spectroscopy, was revealed to play a critical role in protein stability.73
Fig. 3. Structural Biology of DesC and DesE.
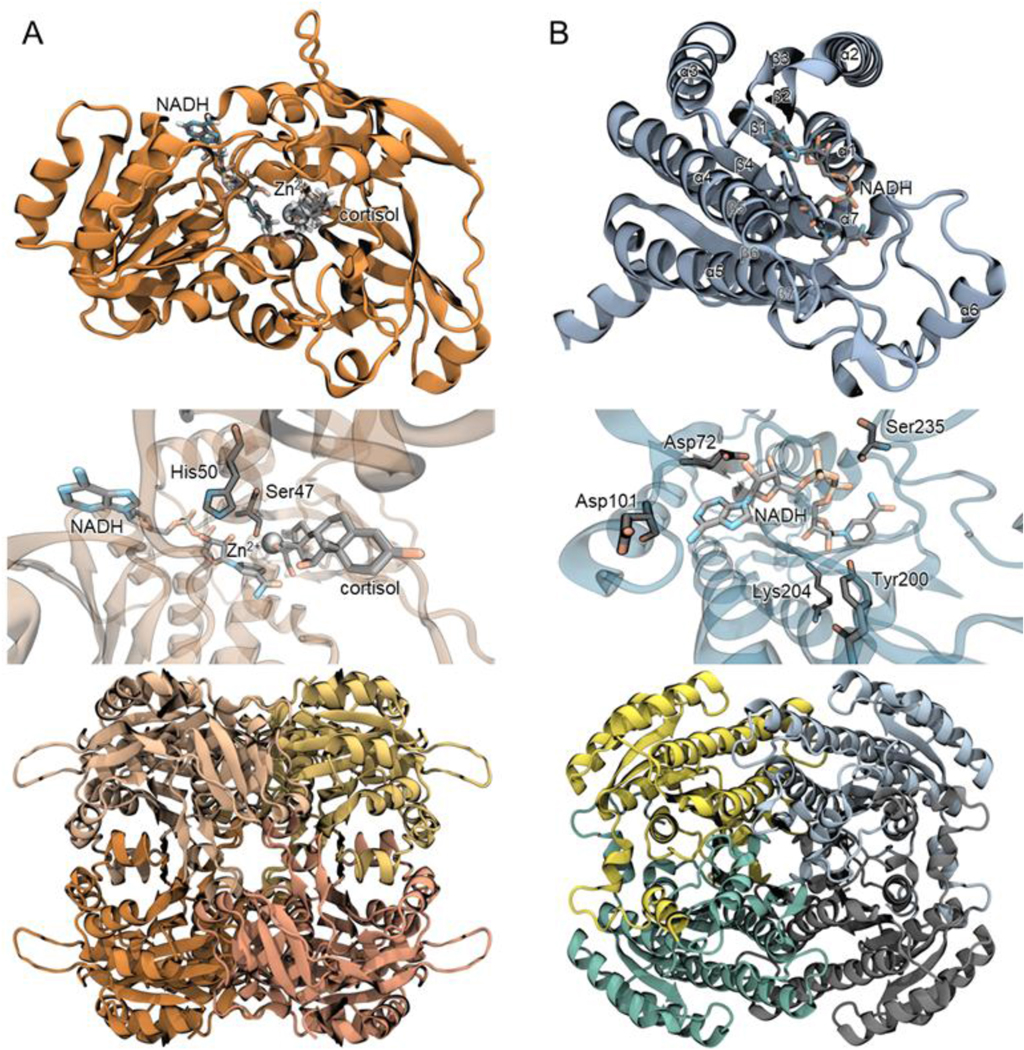
(A, top) DesC (PDB accession: 4OH1) monomer depicted with NADH and cortisol docking simulated by quantum mechanical molecular modeling. (Middle) DesC binding pocket showing predicted residues involved in a proton relay to replenish catalytic residue protonation. (Bottom) DesC homotetramer. (B, top) DesE (PDB accessions: 6M9U, 6OW4) monomer displaying a Rossmann fold with 7 central β-strands and surrounding α-helices. (Middle) DesE binding pocket showing NADH-binding residues Asp72, Asp101, Tyr200, Lys204, Ser235. (Bottom) DesE homotetramer.
The microbial steroid-17,20-desmolase pathway, encoded by the desABCD operon in C. scindens ATCC 35704, includes a 20α-HSDH (DesC) that is a member of the MCDR superfamily.72 The MCDR superfamily is continually growing, and now is close in complexity to the SDR superfamily.78 MCDR members differ from SDRs in that they are metal-dependent, but both contain Rossmann fold domain for NAD(P)(H) binding.80
Zinc-dependent C. scindens DesC apo-form has been crystallized at 2.0 Å resolution (PDB ID: 4OH1)72 (Figure 3). DesC is tetrameric with its NAD(P)(H)-binding Rossmann fold located near the C-terminus. Through a combination of hybrid quantum mechanics and classical molecular mechanics simulations, this enzyme mechanism is predicted to proceed through an ordered bi-bi reaction in which NAD(H) binds to initiate the reaction and is the last to leave. The most likely mechanism was a concerted proton transfer from Ser47 along with a hydride from NADH to cortisol, forming 20α-dihydrocortisol. Ser and His residues are proposed to be involved in a proton relay to replenish catalytic residue protonation states.72
Structural biology studies of host HSDHs have led to the development of many pharmaceutical inhibitors aiming to mitigate various steroid driven diseases.81 Due to both the regiospecific (17- vs. 20-HSDH) and stereospecific (20α- vs. 20β-HSDH) nature of HSDHs and their distinct actions in steroid metabolism, inhibitors must be identified for specific isoforms. Crystal structures of HSDHs aid in discerning how inhibitors bind in detail. Additionally, these structures can be leveraged for large in silico screens to find new inhibitors.81 For example, this approach has been applied to 11β-HSD type 1 regarding therapy for metabolic syndrome and obesity82, as well as 17-βHSD type 1 for breast cancer.83 Due to the growing potential for microbial HSDHs in steroid metabolism and signaling, along with other microbial enzymes such as steroid17,20-desmolase, the same strategy of utilizing crystallography in inhibitor discovery may be applied to the gut microbiome for disease therapy.
4. Implications for human health
4.1. Intestinal Steroidogenesis
The assumption that intestinal cortisol could only derive from bile led Bokkenheuser, who had originally isolated desmolase-expressing gut bacteria, to conclude that cortisol was not the physiological substrate for steroid-17,20-desmolase because it does not undergo enterohepatic circulation.40 From a microbiological perspective, it is difficult to explain the evolution of a tightly regulated pathway, the expression of which is transcriptionally controlled by cortisol, if cortisol and its derivatives are not present in the lumen of the intestines.
While the adrenal gland is the major focus for the study of GC synthesis, there are extra-adrenal mucosal epithelial surfaces where steroidogenesis takes place. To be sure, despite their small size compared to other GC producing tissues, the adrenal gland is responsible for the bulk of serum GC levels as judged by adrenalectomized rodents84. Extra-adrenal synthesis of GC is instead thought to be important locally84,85, and GCs have been shown to be produced in thymus86, lung87, skin88, and the gastrointestinal tract.89,90 Yet, ex vivo studies of GC production rate indicate that extra-adrenal tissues may be capable of generating significant amounts of GC due to their size relative to the adrenal.
In particular, the adult human gastrointestinal tract is 300 m2 epithelium separating 100 trillion microbes from access to the bloodstream and vital organs. Not surprisingly, complex mechanisms have evolved to maintain intestinal homeostasis: developing immune tolerance of commensal microbes and protection against pathogens. Central in this struggle are intestinal epithelial cells (IECs), which physically separate microbes and immune cells. Evidence in the 1990’s began to accumulate that the intestines, and IECs in the crypts, are sites of steroidogenesis aimed at regulating immune responses at the intestinal barrier. It was demonstrated that both the duodenum and colon expressed P450C17 mRNA and were capable of converting [4-14C] pregnenolone to dehydroepiandrosterone (DHEA) ex vivo, indicating the expression of CYP17 in the intestine.91–93 The expression of CYP11A1, CYP17 and CYP11B1 as well as cortisol biosynthesis was observed in human colorectal cell lines (Caco-2 & HT-29) as well as primary human intestinal cell lines and colorectal tumor tissue.92,94
While the regulation of adrenal cortisol biosynthesis is well understood95, the regulation of intestinal steroidogenesis is less well characterized.92 Steroidogenic Factor 1 (SF-1) is a key nuclear receptor regulating steroid biosynthesis genes in the adrenal, but is not expressed in the intestine.96,97 Indeed, intestinal synthesis of GC does not respond to ACTH.98 Instead, liver receptor homolog 1 (LRH-1) appears to govern steroidogenesis in the gut, and is highly expressed in intestinal crypts.99 siRNA in human CRC cell lines shows that down-regulation of LRH-1 significantly reduces both the expression of steroidogenic enzymes and the synthesis of cortisol.92,94 Tissue-specific deletion of LRH-1 in IECs was shown to disrupt GC synthesis, resulting in greater susceptibility of conditional knockout mice to colitis.100 While inflammation appears to be a key trigger for inducing intestinal GC synthesis, additional mechanisms regulating intestinal steroidogenesis remain poorly characterized.101 Of relevance to bacterial metabolism of cortisol, IEC steroidogenesis provides another route of GC efflux into the GI tract distinct from biliary excretion. Importantly, transmembrane efflux pumps that export cortisol have been studied and shown to be expressed in the apical membrane of IEC.102
It was previously believed that GC transport across cell membranes proceeds by simple diffusion, but this notion was disproved starting in the 1970s with the discovery of a transmembrane efflux protein named P-glycoprotein (Pgp), also known as multidrug resistant protein 1 (MDR1).103,104 Pgp is encoded by the mdr1 gene and is expressed in liver, kidneys, intestines, brain, adrenal, and placenta. This ATP hydrolysis-dependent transporter has broad substrate specificity, including drugs, lipids, xenobiotics, peptides, bilirubin, cardiac glycosides, in addition to the steroids and GC that will be discussed in more detail here. Pgp plays a critical role in mediating efflux and protecting cells from reaching toxic concentrations of endogenous and exogenous substrates.105
In vitro and in vivo studies have demonstrated that Pgp limits the intracellular accumulation of specific GC. LLC-PK, a pig kidney epithelium-derived cell line, stably expressing human Pgp (L-MDR1 isoform), shows preferential efflux based on GC structure: 6α-methylprednisolone > prednisolone > betamethasone > dexamethasone/prednisone > cortisol.106 A nonpolar substituent at the C6 position and a hydroxyl at the C11 position increased Pgp efflux, whereas bulky substituents at C16 decreased Pgp affinity.106 Rats showed differential absorption of GC in the small intestine following the same pattern of Pgp substrate specificity. After administering the rats with 0.1 mM of steroid using an in situ loop technique, the concentration of steroid was monitored throughout the small intestine for 30 minutes. In the ileum, 33% of 6α-methylprednisolone, 45% of prednisolone, 63% of dexamethasone, and 69% of cortisol disappeared, suggesting the remaining steroid detected in the lumen was effluxed by Pgp.107 This contrasts with other steroids examined, such as progesterone, estradiol, and testosterone that had 100% clearance from the ileum107, potentially due to Pgp lacking or having decreased specificity for these substrates.31 Further, in vitro transepithelial flux of cortisol in the basolateral to apical direction was measured approximately 3–4 fold greater than the reverse direction.106 More studies are required to determine if cortisol synthesized in IECs is exported to the lumen of the gut through this mechanism, and if this is an important source of cortisol for conversion to 11β-OHAD by gut bacteria such as C. scindens ATCC 35704.
4.2. Bacterial steroid-17,20-desmolase in the urinary tract
Urine represents the major route of GC excretion in the human body.108 Recent interest in 11-oxo androgen precursors44,109 and the search for urinary tract microbiome correlates to prostate cancer risk110 may find a convergence at steroid-17,20-desmolase. Metagenomic sequencing of surgically removed prostate tissue revealed a high abundance (~60% relative abundance) of Propionibacteriaceae.111 Within this bacterial family are organisms such as Propionibacterium acnes and Propionimicrobium lymphophilum both of which are recognized as pro-inflammatory pathobionts which have been isolated from prostate tissue after radical prostatectomy.112–114 A recent metagenomic study of urine from control patients (n=65) vs. men with prostate cancer (n=65) revealed a significant correlation with abundance of Propionimicrobium lymphophilum, although a potential role of this microbial taxa in prostate cancer was not suggested.110
To determine the key gut microbial players encoding desAB, we performed extensive phylogenetic analysis of >40,000 proteins in NCBI and Uniprot similar to DesAB from C. scindens ATCC 35704.56 To our surprise, the desAB genes encoding steroid-17,20-desmolase are also detected in the genomes of urinary tract bacterial isolates.56 Functional network-based clustering revealing that two taxa inhabiting the urinary tract, P. lymphophilum and Arcanobacterium urinimassiliense, encode the desABE gene cluster and are thus genetically capable of forming 11-oxy-androgens.56 We then showed that pure cultures of P. lymphophilum ACS-093-V-SCH5 convert cortisol as well as cortisone, allo-tetrahydrocortisol (but not tetrahydrocortisol), prednisone, prednisolone, dexamethasone and 9-fluorocortisol to C19 side-chain cleavage products.56 Interestingly, rDesAB shows approximately two-fold greater relative activity against exogenous substrates, prednisone (100%) and prednisolone (71.47%), relative to cortisone (37.80%) and cortisol (34.57%).56 Desmolase activity was also observed in vivo with mice mono-associated with C. scindens and orally administered dexamethasone. The C19, side-chain cleaved product of dexamethasone was detected in the cecum and serum, suggesting that this metabolism may not only affect the gut locally, but also systemically.115
Urinary cortisol concentrations range from 130 nM to 276 nM in healthy individuals, far more than the ~0.1 to 1.0 nM level of androgen necessary to activate AR.116 A urinary metagenome harboring desAB genes may put some men at risk for prostate cancer through the potential to continuously produce 11-oxy-androgens in the urinary tract. Indeed, we measured proliferation of LNCaP cells, an androgen-sensitive human prostate adenocarcinoma cell line, in the presence of steroid-17,20-desmolase products relative to dihydrotestosterone (DHT), and found that 1,4-androstadiene-3,11,17-trione, resulting from side-chain cleavage of prednisone, caused significant proliferation relative to both DHT and 11-keto-androstenedione (11-KA4).56 Additional work will be necessary to determine whether this proliferation is due to AR-activation, and determine the extent of metabolism for each of these androgens in culture. However, this is important because replacement GC (like dexamethasone or prednisone) are co-prescribed with abiraterone in the treatment of castration-resistant prostate cancer through inhibiting adrenal androgen biosynthesis.117 Importantly, 11-oxy-androgen precursors, such as 11β-OHAD have been shown recently to be the preferred substrates for aldo-keto reductase 1C3 (AKR1C3): 11-KA4 ≥ 11-keto-5α-androstanedione (11-K-5α-A4) > 11-keto-adrenosterone (11-KAST) > A4 > androsterone (AST) > 5α-dione. AKR1C3 is an enzyme with 17β-reductase activity in prostate cancer tissue.43 Further work will be needed to determine a link, if any, between bacterial steroid-17,20-desmolase activity in the urinary tract and prostate cancer risk and progression.
Interestingly, recent work identified changes in the gut microbiome following androgen receptor-axis target therapies.118 Akkermansia muciniphila was recently shown to be enriched in rectal swab samples in men taking abiraterone for castration-resistant prostate cancer, relative to control patients not taking the drug.118 A subsequent study showed that A. muciniphila is enriched through the cleavage and utilization of acetate from abiraterone-acetate as an energy source.118 In addition, a suggested mechanism for the efficacy of abiraterone acetate is the increased microbial production of vitamin K2 in prostate cancer patients through interactions with A. muciniphila.119 These results indicate the need for additional studies to determine the role of microbial metabolism of drugs involved in prostate cancer treatment on disease progression, and whether particular microbial species or genes may be predictive of treatment success.
4.3. Microbial metabolites of cortisol and host 11β-HSD function
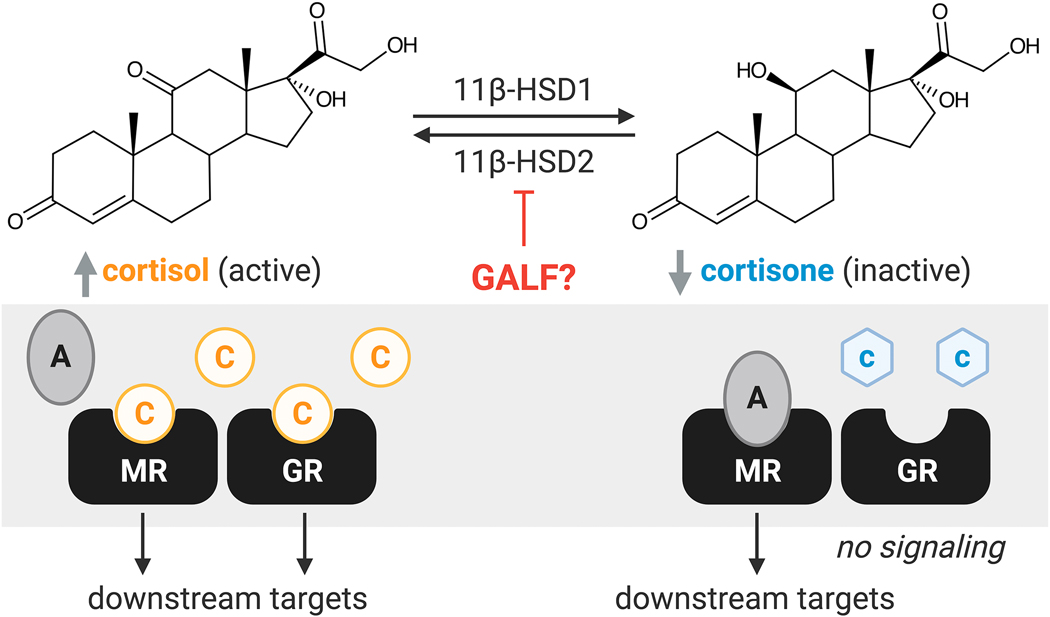
The enzyme 11β-HSD2 is viewed as a “guardian” of the mineralocorticoid receptor (MR).120–122 11β-HSD2 converts intracellular cortisol (active GC) to cortisone (inactive GC) (Figure 4). This is important because cortisol, but not cortisone, can promiscuously signal through both GR and MR (Figure 4). Indeed, cortisol binds to MR with the same affinity as aldosterone, the principal mineralocorticoid in humans, but is at concentrations 100–1000 fold higher in circulation. Genetic defect in 11β-HSD2 results in a severe form of hypertension known as apparent mineralocorticoid excess (AME), symptoms of which can be reversibly observed by pharmacological inhibition of 11β-HSD2 (e.g. glycyrrhetinic acid (GA) from licorice).121,123 The two isoforms, 11β-HSD1 and 11β-HSD2, are expressed in various tissues throughout the body and act as reductase and dehydrogenase, respectively. 11β-HSD2 is expressed in the distal colon, distal renal tubules, sweat and salivary glands, placenta, and vascular endothelial wall, and primarily oxidizes cortisol to cortisone. 11β-HSD1 is expressed in the liver, adipose tissue, and brain, and mainly reduces cortisone to cortisol, amplifying local GC action.28 Inducible levels of 11β-HSD have been detected in immune cells, including macrophages and peripheral blood mononuclear cells, indicating that 11β-HSD function may be critical for maintaining local, active GC hormone levels to regulate inflammation.124 11β-HSD1 function in the colon also influences host bile acid function and microbial composition in a diet dependent mechanism. 11β-HSD1 deficient mice (Hsd11b1Del1/Del1), characterized by their impaired post-prandial bile acid release and altered bile acid composition, were fed a Western diet (41% fat with 0.2% cholesterol) for two weeks, resulting in an increased relative abundance of Bacteroidaceae.125
Fig. 4. Potential effect of glycyrrhetinic acid-like factors (GALFs) on 11β-hydroxysteroid dehydrogenase 1 and 2 (11β-HSD1/2).
11β-HSD1/2 play a critical role in regulating the local concentration of active glucocorticoid, cortisol (C), by conversion to inactive glucocorticoid, cortisone (c). Presence of GALFs may inhibit 11β-HSD leading to aberrant glucocorticoid receptor (GR) and mineralocorticoid receptor (MR) signaling as cortisol competes with aldosterone (A) for signaling through MR.
The glycyrrhetinic acid-like factors (GALF) hypothesis postulates that host derived and bacterial GC metabolites inhibit 11β-HSD2 in a similar mechanism to GA found in licorice, resulting in hypertension in some individuals.126 The association between GA and AME has been established for decades.127 High licorice consumption results in inhibition of 11β-HSD2 in the kidneys leading to enhanced sodium retention, edema, and hypertension, which is caused by activation of MR by excess cortisol.128 Endogenous compounds, such as 11β-hydroxy-testosterone (11β-OHT) and 3α,5α-TH-11β-OHT, have also been identified to potently inhibit 11β-HSD2 and 11β-HSD1 dehydrogenase activity with IC50 values of 0.35–4.5 μM and 5.0–9.0 μM, respectively.26,129,130 Infusion of these compounds into rats led to increased blood pressure.123 Notably, these C19 GALFs can also be generated microbially from the DesAB product, 11β-OHAD, which is described in greater detail in other reviews.126,131 Similar observations were made with some progesterone derivatives, which increased potency to inhibit 11β-HSD2 and 11β-HSD1 dehydrogenase activity, that are produced through 21-dehydroxylation by members of the gut microbiome. It is suspected that the generation of GALFs from corticosterone resulted in the steroid hypertension observed in the Honour rat model, which could be reversed by oral antibiotic treatment.132–134 The gut microbiome has been shown to be causally involved in hypertension, and several mechanisms are likely to be involved.126,131 Questions remain about the importance of gut microbial metabolism in the generation of potential steroid GALFs, particularly whether the concentrations of metabolites are sufficient to affect blood pressure through inhibition of renal 11β-HSD2. Individuals with polymorphisms in HSD11B2, a subset of essential hypertensive patients, that reduce functional activity may be especially susceptible to effects from endogenous steroid GALFs.
GALFs produced by microbial side-chain cleavage of cortisol and 3α-, 5α- and 17β-reduction may also function to inhibit 11β-HSD2 locally in colonocytes or other MR responsive tissues. 11β-HSD2 and COX-2 mRNA increase coordinately in adenoma tissue relative to healthy surrounding tissue.135 Treatment of APC+/min mice with GA was shown to significantly reduce adenoma size and number, and reduce COX-2 expression.136 A mechanism was proposed by studying mouse adenocarcinoma cells (CT26) transfected with human 11β-HSD2 cDNA and treated with corticosterone.137 GA treatment of CT26 cells significantly enhanced COX-2 inhibition. Gene knock-down of 11β-HSD2 also reduced adenocarcinoma growth, which was reversed by addition of GR antagonist RU486.137 GC are potent COX-2 inhibitors, and COX-2 a target for prevention of colorectal cancer (CRC).138 By inhibiting 11β-HSD2 oxidation to cortisone, GC concentration of cortisol remains high intracellularly, inhibiting COX-2 and dampening local inflammation. Side-chain cleavage of allo-tetrahydrocortisol, and 17β-reduction, yielding 11β-hydroxyandrosterone by the microbiota could conceivably act in a manner similar to GA. Further studies are needed to determine the role of bacterial steroid-17,20-desmolase on 11β-HSD2 function and the role of endogenous GALFs in prevention of CRC.
A currently unexplored area is whether bacterial steroid-17,20-desmolase activity is found in members of the skin microbiome.59 Cortisol levels in sweat are similar to concentrations in saliva.139 There is evidence that pregnanolone is side-chain cleaved by axillary bacteria.140 The side-chain cleavage of cortisol may be relevant to eccrine sweat glands which have been shown to have co-localization of MR and isoforms of 11β-HSD.141,142 The generation of 11β-HSD inhibitors from microbial metabolism of cortisol may be expected to affect GC and salt concentrations in sweat, thus affecting skin microbial community structure.59 Future studies may consider the role of skin microbial metabolism as it relates to 11β-HSD function and MR agonism.
4.4. Steroids, the gut microbiota, and intestinal immunity
Sex differences have been observed in autoimmune diseases. The expression of AR has been confirmed in innate immune cell lineages including neutrophils, macrophages, mast cells, as well as adaptive immune cells, such as B and T cells.143 Gonadectomy alters the gut microbiome in both males and females.144 Non-obese, diabetic mouse models demonstrated that exposure to androgens or transplant of gut microbiota from male mice conferred protection to type 1 diabetes in female mice.145 Currently, very little is understood about the mechanisms involved in the regulation of the gut microbiome by sex steroids, and vice versa.144 It is known that estrogen as well as androgen levels in serum, feces, and urine, are altered by bacterial β-glucuronidase enzymes.146 However, mechanistic studies examining alterations to the steroid nucleus and side chain by the gut microbiome have yet to be undertaken (Figure 2).
Roughly 5–15% of reproductive-age women suffer from polycystic ovary syndrome (PCOS).147 PCOS is associated with increased circulating androgens and higher neutrophil counts, which can be reduced to normal after treatment with anti-androgen therapeutics, such as flutamide.148 Dysregulation of cortisol and elevated adrenal androgens such as DHEA(S), as well as 11β-OHAD, are associated with PCOS.149 Recent studies have established altered gut microbial communities in patients with PCOS relative to controls.144 Fecal transplant from women with PCOS vs. control into germ-free mice revealed a mechanistic association between the conjugated bile acid glycodeoxycholic acid, intestinal IL-22 secretion, and improvement of PCOS symptoms.150 Intriguingly, metagenomic sequencing of stool microbiome for functional gene content reported bile acid metabolism as second to a large shift in genes predicted to be involved in steroid hormone biosynthesis.150 Taken together with the adrenocortical hyper-reactivity to ACTH characterized by PCOS, and the observation that alleviation of symptoms proceeded through bile acid-intestinal immune signaling, these data indicate that the effects of microbial cortisol and 11-oxy-androgen metabolism on mucosal immunity in PCOS should be further pursued.
Clostridium scindens emerges as a particularly interesting microbial species, which is composed of strains that collectively generate secondary bile acids that have been shown to regulate serotonin production by enterochromaffin cells151 and potentially modulate regulatory T cells in the gut mucosa through production of allo-secondary bile acids that bind retinoic acid receptor-related orphan receptor γ (RORγT).152 Yet, only some strains of C. scindens (e.g. ATCC 35704, SO76, SO77) have been shown to side-chain cleave cortisol to 11β-OHAD.27 C. scindens is a low abundance member of the microbiota (103–105 colony forming units per gram wet weight stool) and only one of a few species capable of forming hydrophobic bile acids65,153 are also capable of steroid- 17,20-desmolase activity.56 Thus, this species may have a disproportionate effect on host physiology through steroid signaling via host nuclear receptors and G-protein coupled receptors.
5. Conclusions
The human microbiome is recognized as a ‘virtual endocrine organ’ that encodes steroid metabolizing enzymes paralleling many host steroid metabolizing capabilities. Specific gut and urinary bacteria expressing DesAB can side-chain cleave GC, forming C19 androgens that can be further metabolized by both host tissue and the microbiome to form potent 11-oxy-androgens. There are additional microbial enzymes involved in the metabolism of C3, C4-C5, C17 that contribute to the 11-oxy-androgen metabolome, but remain to be discovered. The extent to which gut microbial steroid metabolism impacts host health is still unknown, but further mechanistic work is warranted as DesAB may be linked to many disease states such as hypertension, prostate cancer, colorectal cancer, and polycystic ovary syndrome. Clinical studies with these patient populations will reveal if associations exist between DesAB microbes and disease status. Our discovery and characterization of the steroid-17,20-desmolase pathway provides a molecular basis to quantify and correlate microbial genes and host metabolites with disease.
Many key questions still remain that can be elucidated using gnotobiotic animal models and genetically modified microbes paired with multi-omics analyses. Some of these questions include: (1) if DesAB affects the gut-brain axis and (2) whether C. scindens is a keystone species that regulates immune function through biotransformations of steroids and bile acids, influencing immune tolerance (3) whether there are sex-differences relating to steroid-17,20-desmolase, HSDHs, in both the GI tract and reproductive tracts of males and females (4) whether steroid-17,20-desmolase activity in the gut and reproductive tract affect the immune and reproductive systems in early development. Another main research focus is to determine the detailed “rules” governing microbial steroid metabolism, such as substrate-specificity and kinetics of pure enzymes and whole cells in both in vitro and in vivo systems of varying microbiome complexity and redox potential. These studies will allow us to work towards rationally manipulating the microbiome to generate desired steroid metabolites to potentially mitigate disease and improve human health. Taken together, through the metabolism of steroids, the human microbiome should be considered as an extension of the host endocrine system.
Highlights.
The human microbiome encodes steroid metabolizing enzymes, paralleling host enzymes.
Gut and urinary bacteria side-chain cleave glucocorticoids, resulting in androgens.
The extent to which gut microbial steroid metabolism impacts host health is still unknown.
Acknowledgements
We gratefully acknowledge Rafael C. Bernardi, Department of Physics at Auburn University, for support in rendering DesC and DesE crystal structure figures. This work was supported by grants from the National Institutes of Health (R01GM134423 and R03AI147127). The graphical abstract and figures 1, 2, and 4 were created using Biorender.com.
Abbreviations:
- 11β-HSD1/2
11β-hydroxysteroid dehydrogenase isoforms 1 and 2
- 11β-OHAD
11β-hydroxyandrostenedione
- 11-K-5α-A4
11-keto-5α-androstanedione
- 11-KA4
11-keto-androstenedione
- 11-KAST
11-keto-adrenosterone
- 11-KT
11-keto-testosterone
- 11-KDHT
11-keto-dihydrotestosterone
- 17β-HSD
17β-hydroxysteroid dehydrogenase
- A4
androstenedione
- ACTH
adrenocorticotropic hormone
- AKR
aldo-keto reductase
- AKR1C3
aldo-keto reductase 1C3
- AME
apparent mineralocorticoid excess
- AR
androgen receptor
- AST
androsterone
- CBG
cortisol binding globulin
- CRC
colorectal cancer
- CRH
corticotropin releasing hormone
- DHEA
dehydroepiandrosterone
- DHT
dihydrotestosterone
- DesAB
steroid-17,20-desmolase
- DesC
20α-hydroxysteroid dehydrogenase
- DesE
20β-hydroxysteroid dehydrogenase
- GA
glycyrrhetinic acid
- GALF
glycyrrhetinic acid-like factors
- GC
glucocorticoid
- GR
glucocorticoid receptor
- HPA
hypothalamic-pituitary-adrenal axis
- HSDH
hydroxysteroid dehydrogenase
- IEC
intestinal epithelial cells
- LRH-1
liver receptor homolog 1
- MCDR
medium-chain dehydrogenase/reductase
- MDR1
multidrug resistant protein 1
- MR
mineralocorticoid receptor
- PCOS
polycystic ovary syndrome
- Pgp
P-glycoprotein
- rDesAB
recombinant DesAB
- SDR
short chain dehydrogenase
- SF-1
steroidogenic factor 1
- SRD5A
5α-reductase
- T
testosterone
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shackleton CHL, Neres MS, Hughes BA, Stewart PM & Kater CE 844 17-Hydroxylase/C17,20-lyase (CYP17) is not the enzyme responsible for side-chain cleavage of cortisol and its metabolites. Steroids 73, 652–656 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Shackleton CHL, Neres MS, Hughes BA, Stewart PM & Kater CE 17-Hydroxylase/C17,20-lyase (CYP17) is not the enzyme responsible for side-chain cleavage of cortisol and its metabolites. Steroids 73, 652–656 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Sender R, Fuchs S. & Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 14, e1002533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shreiner AB, Kao JY & Young VB The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 31, 69–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young VB The role of the microbiome in human health and disease: an introduction for clinicians. BMJ 356, j831 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Lin L. & Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 18, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagpal R. et al. Gut microbiome and aging: Physiological and mechanistic insights. Nutr. Heal. aging 4, 267–285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koren O. et al. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput. Biol. 9, e1002863 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David LA et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yatsunenko T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasolli E. et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 176, 649–662.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy M, Blacher E. & Elinav E. Microbiome, metabolites and host immunity. Curr. Opin. Microbiol. 35, 8–15 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Rooks MG & Garrett WS Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fung TC, Olson CA & Hsiao EY Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 20, 145–155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carabotti M, Scirocco A, Maselli MA & Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 28, 203–209 (2015). [PMC free article] [PubMed] [Google Scholar]
- 18.Bercik P, Collins SM & Verdu EF Microbes and the gut-brain axis. Neurogastroenterol. Motil. Off. J. Eur. Gastrointest. Motil. Soc. 24, 405–413 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Cryan JF et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 99, 1877–2013887 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Sampson TR & Mazmanian SK Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 17, 565–576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roshchina VV New Trends and Perspectives in the Evolution of Neurotransmitters in Microbial, Plant, and Animal Cells. Adv. Exp. Med. Biol. 874, 25–77 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Messaoudi M. et al. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br. J. Nutr. 105, 755–764 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Sorg JA & Sonenshein AL Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J. Bacteriol. 191, 1115–1117 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winston JA & Theriot CM Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 41, 44–50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bokkenheuser VD, Winter J, Dehazya P. & Kelly WG Isolation and characterization of human fecal bacteria capable of 21-dehydroxylating corticoids. Appl. Environ. Microbiol. 34, 571–575 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Latif SA, Sheff MF, Ribeiro CE & Morris DJ Selective inhibition of sheep kidney 11 beta-hydroxysteroid dehydrogenase isoform 2 activity by 5 alpha-reduced (but not 5 beta) derivatives of adrenocorticosteroids. Steroids 62, 230–237 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Ridlon JM. et al. Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J. Lipid Res 54, 2437–49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goulding N. & Flower R. Glucocorticoids. (Springer; Basel AG, 2001). [Google Scholar]
- 29.Esteban NV et al. Daily cortisol production rate in man determined by stable isotope dilution/mass spectrometry. J. Clin. Endocrinol. Metab. 72, 39–45 (1991). [DOI] [PubMed] [Google Scholar]
- 30.Groeneweg FL, Karst H, de Kloet ER & Joëls M. Rapid non-genomic effects of corticosteroids and their role in the central stress response. J. Endocrinol. 209, 153–167 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Ueda K. et al. Human P-glycoprotein transports cortisol, aldosterone, and dexamethasone, but not progesterone. J. Biol. Chem. 267, 24248–24252 (1992). [PubMed] [Google Scholar]
- 32.Palme R, Rettenbacher S, Touma C, El-Bahr SM & Möstl E. Stress hormones in mammals and birds: comparative aspects regarding metabolism, excretion, and noninvasive measurement in fecal samples. Ann. N. Y. Acad. Sci. 1040, 162–171 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Bradlow HL, Frazell EL, Gallagher TF & Hellman L. Tracer studies of the absorption and fate of steroid hormones in man. J. Clin. Invest. 35, 1033–1044 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nabarro J, Moxham A, Walker G. & Slater JD Rectal hydrocortisone. Br. Med. J. 2, 272–274 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wade AP, Slater JD, Kellie AE & Holliday ME Urinary excretion of 17ketosteroids following rectal infusion of cortisol. J. Clin. Endocrinol. Metab. 19, 444–453 (1959). [DOI] [PubMed] [Google Scholar]
- 36.Gustafsson JA. & Sjövall J. Steroids in germfree and conventional rats. 5. Identification of C19 steroids in faeces from germfree rats. Eur. J. Biochem. 6, 227–235 (1968). [DOI] [PubMed] [Google Scholar]
- 37.Eriksson H. & Gustafsson JA Excretion of steroid hormones in adults. Steroids in faeces from adults. Eur. J. Biochem. 18, 146–150 (1971). [DOI] [PubMed] [Google Scholar]
- 38.Bokkenheuser VD, Morris GN, Ritchie AE, Holdeman LV & Winter J. Biosynthesis of androgen from cortisol by a species of Clostridium recovered from human fecal flora. J. Infect. Dis. 149, 489–494 (1984). [DOI] [PubMed] [Google Scholar]
- 39.Morris GN, Winter J, Cato EP, Ritchie AE & Bokkenheuser V. Clostridium scindens sp. nov., a Human Intestinal Bacterium with Desmolytic Activity on Corticoids. Int. J. Syst. Evol. Microbiol. 35, 478–481 (1985). [Google Scholar]
- 40.Winter J. et al. Mode of action of steroid desmolase and reductases synthesized by Clostridium ‘scindens’ (formerly Clostridium strain 19). J. Lipid Res. 25, 1124–1131 (1984). [PubMed] [Google Scholar]
- 41.Bokkenheuser VD, Winter J, Morris GN & Locascio S. Steroid desmolase synthesis by Eubacterium desmolans and Clostridium cadavaris. Appl. Environ. Microbiol. 52, 1153–1156 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnard M, Mostaghel EA, Auchus RJ & Storbeck K-H The role of adrenal derived androgens in castration resistant prostate cancer. J. Steroid Biochem. Mol. Biol. 197, 105506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnard M. et al. 11-Oxygenated androgen precursors are the preferred substrates for aldo-keto reductase 1C3 (AKR1C3): Implications for castration resistant prostate cancer. J. Steroid Biochem. Mol. Biol. 183, 192–201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pretorius E. et al. 11-Ketotestosterone and 11-Ketodihydrotestosterone in Castration Resistant Prostate Cancer: Potent Androgens Which Can No Longer Be Ignored. PLoS One 11, e0159867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swart AC & Storbeck K-H 11β-Hydroxyandrostenedione: Downstream metabolism by 11βHSD, 17βHSD and SRD5A produces novel substrates in familiar pathways. Mol. Cell. Endocrinol. 408, 114–123 (2015). [DOI] [PubMed] [Google Scholar]
- 46.du Toit T. et al. Profiling adrenal 11β-hydroxyandrostenedione metabolites in prostate cancer cells, tissue and plasma: UPC(2)-MS/MS quantification of 11βhydroxytestosterone, 11keto-testosterone and 11keto-dihydrotestosterone. J. Steroid Biochem. Mol. Biol 166, 54–67 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Bloem LM, Storbeck K-H, Schloms L. & Swart AC 11β-hydroxyandrostenedione returns to the steroid arena: biosynthesis, metabolism and function. Molecules 18, 13228–13244 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pretorius E, Arlt W. & Storbeck K-H A new dawn for androgens: Novel lessons from 11-oxygenated C19 steroids. Mol. Cell. Endocrinol. 441, 76–85 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Rubin BL, Dorfman RI & Dorfman A. Adrenocorticotropic hormone (ACTH) in urine. J. Clin. Endocrinol. Metab. 14, 154–160 (1954). [DOI] [PubMed] [Google Scholar]
- 50.Yoshimoto FK & Auchus RJ The diverse chemistry of cytochrome P450 17A1 (P450c17, CYP17A1). J. Steroid Biochem. Mol. Biol. 151, 52–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krafft AE & Hylemon PB Purification and characterization of a novel form of 20 alpha-hydroxysteroid dehydrogenase from Clostridium scindens. J. Bacteriol. 171, 2925–2932 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krafft AE, Winter J, Bokkenheuser VD & Hylemon PB Cofactor requirements of steroid-17–20-desmolase and 20 alpha-hydroxysteroid dehydrogenase activities in cell extracts of Clostridium scindens. J. Steroid Biochem. 28, 49–54 (1987). [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Dufort I, Rheault P. & Luu-The V. Characterization of a human 20alpha-hydroxysteroid dehydrogenase. J. Mol. Endocrinol. 25, 221–228 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Sih CJ & Rahim AM Mechanisms Of Steroid Oxidation By Microorganisms. III. Enzymatic Mechanism Of Ring A Aromatization. J. Pharm. Sci. 52, 1075–1080 (1963). [DOI] [PubMed] [Google Scholar]
- 55.Devendran S, Mythen SM & Ridlon JM The desA and desB genes from Clostridium scindens ATCC 35704 encode steroid-17,20-desmolase. J. Lipid Res. 59, 1005–1014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ly LK et al. Bacterial steroid-17,20-desmolase is a taxonomically rare enzymatic pathway that converts prednisone to 1,4-androstanediene-3,11,17trione, a metabolite that causes proliferation of prostate cancer cells. J. Steroid Biochem. Mol. Biol. 199, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peterson RE & Pierce CE The metabolism of corticosterone in man. J. Clin. Invest. 39, 741–757 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ridlon JM & Bajaj JS The human gut sterolbiome: Bile acid-microbiome endocrine aspects and therapeutics. Acta Pharmaceutica Sinica B 5, 99–105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ridlon JM Conceptualizing the Vertebrate Sterolbiome. Appl. Environ. Microbiol. 86, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mythen SM, Devendran S, Méndez-García C, Cann I. & Ridlon JM Targeted Synthesis and Characterization of a Gene Cluster Encoding NAD(P)H-Dependent 3α-, 3β-, and 12α-Hydroxysteroid Dehydrogenases from Eggerthella CAG:298, a Gut Metagenomic Sequence. Appl. Environ. Microbiol 84, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devlin AS & Fischbach MA A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 11, 685–690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kenny DJ et al. Cholesterol Metabolism by Uncultured Human Gut Bacteria Influences Host Cholesterol Level. Cell Host Microbe 28, 245–257.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stokes NA & Hylemon PB Characterization of delta 4–3-ketosteroid-5 beta-reductase and 3 beta-hydroxysteroid dehydrogenase in cell extracts of Clostridium innocuum. Biochim. Biophys. Acta 836, 255–261 (1985). [DOI] [PubMed] [Google Scholar]
- 64.Kang D-J, Ridlon JM, Moore DR 2nd, Barnes S. & Hylemon PB. Clostridium scindens baiCD and baiH genes encode stereo-specific 7alpha/7beta-hydroxy-3-oxo-delta4-cholenoic acid oxidoreductases. Biochim. Biophys. Acta 1781, 16–25 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ridlon JM, Kang D-J & Hylemon PB Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–59 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Winter J, O’Rourke-Locascio S, Bokkenheuser VD, Mosbach EH & Cohen BI Reduction of 17-keto steroids by anaerobic microorganisms isolated from human fecal flora. Biochim. Biophys. Acta 795, 208–211 (1984). [DOI] [PubMed] [Google Scholar]
- 67.Harris SC Discovery and characterization of bile acid and steroid metabolism pathways in gut-associated microbes. (Virginia Commonwealth University, 2017). [Google Scholar]
- 68.English MA, Stewart PM & Hewison M. Estrogen metabolism and malignancy: analysis of the expression and function of 17beta-hydroxysteroid dehydrogenases in colonic cancer. Mol. Cell. Endocrinol. 171, 53–60 (2001). [DOI] [PubMed] [Google Scholar]
- 69.de Prada P, Setchell KD & Hylemon PB Purification and characterization of a novel 17 alpha-hydroxysteroid dehydrogenase from an intestinal Eubacterium sp. VPI 12708. J. Lipid Res 35, 922–929 (1994). [PubMed] [Google Scholar]
- 70.Penning TM Molecular endocrinology of hydroxysteroid dehydrogenases. Endocr. Rev. 18, 281–305 (1997). [DOI] [PubMed] [Google Scholar]
- 71.Penning TM, Wangtrakuldee P. & Auchus RJ Structural and Functional Biology of Aldo-Keto Reductase Steroid-Transforming Enzymes. Endocr. Rev. 40, 447–475 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bernardi RC et al. Bacteria on steroids: the enzymatic mechanism of an NADH-dependent dehydrogenase that regulates the conversion of cortisol to androgen in the gut microbiome. bioRxiv 2020.06.12.149468 (2020). doi: 10.1101/2020.06.12.149468 [DOI] [Google Scholar]
- 73.Doden HL et al. Structural and biochemical characterization of 20β-hydroxysteroid dehydrogenase from Bifidobacterium adolescentis strain L2–32. J. Biol. Chem. 294, 12040–12053 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Devendran S, Méndez-García C. & Ridlon JM Identification and characterization of a 20β-HSDH from the anaerobic gut bacterium Butyricicoccus desmolans ATCC 43058. J. Lipid Res. 58, 916–925 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Filling C. et al. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277, 25677–25684 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Kallberg Y, Oppermann U, Jörnvall H. & Persson B. Short-chain dehydrogenases/reductases (SDRs). Eur. J. Biochem. 269, 4409–4417 (2002). [DOI] [PubMed] [Google Scholar]
- 77.Rossmann MG, Moras D. & Olsen KW Chemical and biological evolution of nucleotide-binding protein. Nature 250, 194–199 (1974). [DOI] [PubMed] [Google Scholar]
- 78.Kavanagh KL, Jörnvall H, Persson B. & Oppermann U. Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65, 3895–3906 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ghosh D, Wawrzak Z, Weeks CM, Duax WL & Erman M. The refined three-dimensional structure of 3 alpha,20 beta-hydroxysteroid dehydrogenase and possible roles of the residues conserved in short-chain dehydrogenases. Structure 2, 629–640 (1994). [DOI] [PubMed] [Google Scholar]
- 80.Persson B, Hedlund J. & Jörnvall H. Medium- and short-chain dehydrogenase/reductase gene and protein families : the MDR superfamily. Cell. Mol. Life Sci. 65, 3879–3894 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Penning TM Human hydroxysteroid dehydrogenases and pre-receptor regulation: insights into inhibitor design and evaluation. J. Steroid Biochem. Mol. Biol. 125, 46–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang H. et al. Structure-based virtual screening for identification of novel 11beta-HSD1 inhibitors. Eur. J. Med. Chem. 44, 1167–1171 (2009). [DOI] [PubMed] [Google Scholar]
- 83.Schuster D. et al. Discovery of nonsteroidal 17beta-hydroxysteroid dehydrogenase 1 inhibitors by pharmacophore-based screening of virtual compound libraries. J. Med. Chem. 51, 4188–4199 (2008). [DOI] [PubMed] [Google Scholar]
- 84.Kostadinova F, Schwaderer J, Sebeo V. & Brunner T. Why does the gut synthesize glucocorticoids? Ann. Med. 46, 490–497 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Talabér G, Jondal M. & Okret S. Extra-adrenal glucocorticoid synthesis: immune regulation and aspects on local organ homeostasis. Mol. Cell. Endocrinol. 380, 89–98 (2013). [DOI] [PubMed] [Google Scholar]
- 86.Talaber G, Jondal M. & Okret S. Local glucocorticoid production in the thymus. Steroids 103, 58–63 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Provost PR, Boucher E. & Tremblay Y. Glucocorticoid metabolism in the developing lung: adrenal-like synthesis pathway. J. Steroid Biochem. Mol. Biol. 138, 72–80 (2013). [DOI] [PubMed] [Google Scholar]
- 88.Slominsk A. et al. Steroidogenesis in the skin: implications for local immune functions. J. Steroid Biochem. Mol. Biol 137, 107–123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Phan TS, Merk VM & Brunner T. Extra-adrenal glucocorticoid synthesis at epithelial barriers. Genes Immun. 20, 627–640 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Ahmed A, Schmidt C. & Brunner T. Extra-Adrenal Glucocorticoid Synthesis in the Intestinal Mucosa: Between Immune Homeostasis and Immune Escape. Front. Immunol. 10, 1438 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dalla Valle L, Belvedere P, Simontacchi C. & Colombo L. Extraglandular hormonal steroidogenesis in aged rats. J. Steroid Biochem. Mol. Biol. 43, 1095–1098 (1992). [DOI] [PubMed] [Google Scholar]
- 92.Bouguen G, Dubuquoy L, Desreumaux P, Brunner T. & Bertin B. Intestinal steroidogenesis. Steroids 103, 64–71 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Dalla Valle L. et al. Occurrence of cytochrome P450c17 mRNA and dehydroepiandrosterone biosynthesis in the rat gastrointestinal tract. Mol. Cell. Endocrinol. 111, 83–92 (1995). [DOI] [PubMed] [Google Scholar]
- 94.Sidler D. et al. Colon cancer cells produce immunoregulatory glucocorticoids. Oncogene 30, 2411–2419 (2011). [DOI] [PubMed] [Google Scholar]
- 95.Gjerstad JK, Lightman SL & Spiga F. Role of glucocorticoid negative feedback in the regulation of HPA axis pulsatility. Stress 21, 403–416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ramayya MS. et al. Steroidogenic factor 1 messenger ribonucleic acid expression in steroidogenic and nonsteroidogenic human tissues: Northern blot and in situ hybridization studies. J. Clin. Endocrinol. Metab 82, 1799–1806 (1997). [DOI] [PubMed] [Google Scholar]
- 97.Mueller M. et al. Differential regulation of glucocorticoid synthesis in murine intestinal epithelial versus adrenocortical cell lines. Endocrinology 148, 1445–1453 (2007). [DOI] [PubMed] [Google Scholar]
- 98.Atanasov AG et al. Cell cycle-dependent regulation of extra-adrenal glucocorticoid synthesis in murine intestinal epithelial cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 22, 4117–4125 (2008). [DOI] [PubMed] [Google Scholar]
- 99.Mueller M. et al. The nuclear receptor LRH-1 critically regulates extra-adrenal glucocorticoid synthesis in the intestine. J. Exp. Med. 203, 2057–2062 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Seitz C. et al. The orphan nuclear receptor LRH-1/NR5a2 critically regulates T cell functions. Sci. Adv. 5, eaav9732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Noti M, Corazza N, Mueller C, Berger B. & Brunner T. TNF suppresses acute intestinal inflammation by inducing local glucocorticoid synthesis. J. Exp. Med. 207, 1057–1066 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hunter J, Jepson MA, Tsuruo T, Simmons NL & Hirst BH Functional expression of P-glycoprotein in apical membranes of human intestinal Caco-2 cells. Kinetics of vinblastine secretion and interaction with modulators. J. Biol. Chem. 268, 14991–14997 (1993). [PubMed] [Google Scholar]
- 103.Juliano RL & Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim. Biophys. Acta 455, 152–162 (1976). [DOI] [PubMed] [Google Scholar]
- 104.van Kalken CK et al. Cortisol is transported by the multidrug resistance gene product P-glycoprotein. Br. J. Cancer 67, 284–289 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Leschziner GD, Andrew T, Pirmohamed M. & Johnson MR ABCB1 genotype and PGP expression, function and therapeutic drug response: a critical review and recommendations for future research. Pharmacogenomics J. 7, 154–179 (2007). [DOI] [PubMed] [Google Scholar]
- 106.Yates CR et al. Structural determinants of P-glycoprotein-mediated transport of glucocorticoids. Pharm. Res. 20, 1794–1803 (2003). [DOI] [PubMed] [Google Scholar]
- 107.Nakayama A, Eguchi O, Hatakeyama M, Saitoh H. & Takada M. Different absorption behaviors among steroid hormones due to possible interaction with P-glycoprotein in the rat small intestine. Biol. Pharm. Bull. 22, 535–538 (1999). [DOI] [PubMed] [Google Scholar]
- 108.Schiffer L. et al. Human steroid biosynthesis, metabolism and excretion are differentially reflected by serum and urine steroid metabolomes: A comprehensive review. J. Steroid Biochem. Mol. Biol. 194, 105439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.du Toit T. & Swart AC The 11β-hydroxyandrostenedione pathway and C11-oxy C(21) backdoor pathway are active in benign prostatic hyperplasia yielding 11keto-testosterone and 11keto-progesterone. J. Steroid Biochem. Mol. Biol. 196, 105497 (2020). [DOI] [PubMed] [Google Scholar]
- 110.Shrestha E. et al. Profiling the Urinary Microbiome in Men with Positive versus Negative Biopsies for Prostate Cancer. J. Urol. 199, 161–171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cavarretta I. et al. The Microbiome of the Prostate Tumor Microenvironment. Eur. Urol. 72, 625–631 (2017). [DOI] [PubMed] [Google Scholar]
- 112.Davidsson S. et al. Frequency and typing of Propionibacterium acnes in prostate tissue obtained from men with and without prostate cancer. Infect. Agent. Cancer 11, 26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leheste JR et al. P. acnes-Driven Disease Pathology: Current Knowledge and Future Directions. Front. Cell. Infect. Microbiol. 7, 81 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Williams GD Two Cases of Urinary Tract Infection Caused by Propionimicrobium lymphophilum. J. Clin. Microbiol. 53, 3077–3080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R. & Goodman AL Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.de Launoit Y, Veilleux R, Dufour M, Simard J. & Labrie F. Characteristics of the biphasic action of androgens and of the potent antiproliferative effects of the new pure antiestrogen EM-139 on cell cycle kinetic parameters in LNCaP human prostatic cancer cells. Cancer Res. 51, 5165–5170 (1991). [PubMed] [Google Scholar]
- 117.Rice MA, Malhotra SV & Stoyanova T. Second-Generation Antiandrogens: From Discovery to Standard of Care in Castration Resistant Prostate Cancer. Front. Oncol. 9, 801 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sfanos KS et al. Compositional differences in gastrointestinal microbiota in prostate cancer patients treated with androgen axis-targeted therapies. Prostate Cancer Prostatic Dis. 21, 539–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Daisley BA. et al. Abiraterone acetate preferentially enriches for the gut commensal Akkermansia muciniphila in castrate-resistant prostate cancer patients. Nat. Commun 11, 4822 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Edwards CR et al. Localisation of 11 beta-hydroxysteroid dehydrogenase-tissue specific protector of the mineralocorticoid receptor. Lancet (London, England) 2, 986–989 (1988). [DOI] [PubMed] [Google Scholar]
- 121.Funder JW, Pearce PT, Smith R. & Smith AI Mineralocorticoid action: target tissue specificity is enzyme, not receptor, mediated. Science 242, 583–585 (1988). [DOI] [PubMed] [Google Scholar]
- 122.Chapman K, Holmes M. & Seckl J. 11β-hydroxysteroid dehydrogenases: intracellular gate-keepers of tissue glucocorticoid action. Physiol. Rev. 93, 1139–1206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Funder JW Apparent mineralocorticoid excess. J. Steroid Biochem. Mol. Biol. 165, 151–153 (2017). [DOI] [PubMed] [Google Scholar]
- 124.Thieringer R. et al. 11 Beta-hydroxysteroid dehydrogenase type 1 is induced in human monocytes upon differentiation to macrophages. J. Immunol. 167, 30–35 (2001). [DOI] [PubMed] [Google Scholar]
- 125.Johnson JS et al. 11β-hydroxysteroid dehydrogenase-1 deficiency alters the gut microbiome response to Western diet. J. Endocrinol. 232, 273–283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Morris DJ & Ridlon JM Glucocorticoids and gut bacteria: ‘The GALF Hypothesis’ in the metagenomic era. Steroids 125, 1–13 (2017). [DOI] [PubMed] [Google Scholar]
- 127.Stewart PM et al. Mineralocorticoid activity of liquorice: 11-beta-hydroxysteroid dehydrogenase deficiency comes of age. Lancet (London, England) 2, 821–824 (1987). [DOI] [PubMed] [Google Scholar]
- 128.Borst JGG et al. [Excretion of water and electrolytes during a 24-hour period and under influence of licorice extract]. Acta Clin. Belg. 5, 405–409 (1950). [PubMed] [Google Scholar]
- 129.Latif SA, Pardo HA, Hardy MP & Morris DJ Endogenous selective inhibitors of 11beta-hydroxysteroid dehydrogenase isoforms 1 and 2 of adrenal origin. Mol. Cell. Endocrinol. 243, 43–50 (2005). [DOI] [PubMed] [Google Scholar]
- 130.Morris DJ, Latif SA, Hardy MP & Brem AS Endogenous inhibitors (GALFs) of 11beta-hydroxysteroid dehydrogenase isoforms 1 and 2: derivatives of adrenally produced corticosterone and cortisol. J. Steroid Biochem. Mol. Biol 104, 161–168 (2007). [DOI] [PubMed] [Google Scholar]
- 131.Morris DJ & Brem AS Role of gut metabolism of adrenal corticosteroids and hypertension: clues gut-cleansing antibiotics give us. Physiol. Genomics 51, 83–89 (2019). [DOI] [PubMed] [Google Scholar]
- 132.Honour J. The possible involvement of intestinal bacteria in steroidal hypertension. Endocrinology 110, 285–287 (1982). [DOI] [PubMed] [Google Scholar]
- 133.Honour JW, Borriello SP, Ganten U. & Honour P. Antibiotics attenuate experimental hypertension in rats. J. Endocrinol. 105, 347–350 (1985). [DOI] [PubMed] [Google Scholar]
- 134.Honour JW Historical perspective: gut dysbiosis and hypertension. Physiological genomics 47, 443–446 (2015). [DOI] [PubMed] [Google Scholar]
- 135.Yang S, Jiang L. & Zhang M-Z 11β-Hydroxysteroid Dehydrogenase Type II is a Potential Target for Prevention of Colorectal Tumorigenesis. J. oncobiomarkers 1, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jiang L. et al. Epithelial-specific deletion of 11β-HSD2 hinders Apcmin/+ mouse tumorigenesis. Mol. Cancer Res. 11, 1040–1050 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang M-Z et al. Inhibition of 11beta-hydroxysteroid dehydrogenase type II selectively blocks the tumor COX-2 pathway and suppresses colon carcinogenesis in mice and humans. J. Clin. Invest 119, 876–885 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Marnett LJ & DuBois RN COX-2: a target for colon cancer prevention. Annu. Rev. Pharmacol. Toxicol. 42, 55–80 (2002). [DOI] [PubMed] [Google Scholar]
- 139.Jia M, Chew WM, Feinstein Y, Skeath P. & Sternberg EM Quantification of cortisol in human eccrine sweat by liquid chromatography - tandem mass spectrometry. Analyst 141, 2053–2060 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gower DB & Ruparelia BA Olfaction in humans with special reference to odorous 16-androstenes: their occurrence, perception and possible social, psychological and sexual impact. J. Endocrinol. 137, 167–187 (1993). [DOI] [PubMed] [Google Scholar]
- 141.Kenouch S. et al. Human skin as target for aldosterone: coexpression of mineralocorticoid receptors and 11 beta-hydroxysteroid dehydrogenase. J. Clin. Endocrinol. Metab. 79, 1334–1341 (1994). [DOI] [PubMed] [Google Scholar]
- 142.Hirasawa G. et al. Colocalization of 11 beta-hydroxysteroid dehydrogenase type II and mineralocorticoid receptor in human epithelia. J. Clin. Endocrinol. Metab. 82, 3859–3863 (1997). [DOI] [PubMed] [Google Scholar]
- 143.Trigunaite A, Dimo J. & Jørgensen TN Suppressive effects of androgens on the immune system. Cell. Immunol. 294, 87–94 (2015). [DOI] [PubMed] [Google Scholar]
- 144.Thackray VG Sex, Microbes, and Polycystic Ovary Syndrome. Trends Endocrinol. Metab. 30, 54–65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Markle JGM et al. Sex differences in the gut microbiome drive hormonedependent regulation of autoimmunity. Science 339, 1084–1088 (2013). [DOI] [PubMed] [Google Scholar]
- 146.Colldén H. et al. The gut microbiota is a major regulator of androgen metabolism in intestinal contents. Am. J. Physiol. Endocrinol. Metab 317, E1182–E1192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Turcu AF, Nanba AT & Auchus RJ The Rise, Fall, and Resurrection of 11-Oxygenated Androgens in Human Physiology and Disease. Horm. Res. Paediatr. 89, 284–291 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.La J-J. et al. Androgen receptor influences on body defense system via modulation of innate and adaptive immune systems: lessons from conditional AR knockout mice. Am. J. Pathol 181, 1504–1512 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Turcu AF, Rege J, Auchus RJ & Rainey WE 11-Oxygenated androgens in health and disease. Nat. Rev. Endocrinol. 16, 284–296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Qi X. et al. Gut microbiota-bile acid-interleukin-22 axis orchestrates polycystic ovary syndrome. Nat. Med. 25, 1225–1233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yano JM et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hang S. et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature 576, 143–148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ridlon JM, Harris SC, Bhowmik S, Kang D-J & Hylemon PB Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 1251 7, 22–39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]