Abstract
Rationale
Current guidelines do not sufficiently capture the heterogeneous nature of asthma; a more detailed molecular classification is needed. Metabolomics represents a novel and compelling approach to derive asthma endotypes (i.e., subtypes defined by functional and/or pathobiological mechanisms).
Objectives
To validate metabolomic-driven endotypes of asthma and explore their underlying biology.
Methods
In the Genetics of Asthma in Costa Rica Study (GACRS), untargeted metabolomic profiling, similarity network fusion, and spectral clustering was used to identify metabo-endotypes of asthma, and differences in asthma-relevant phenotypes across these metabo-endotypes were explored. The metabo-endotypes were recapitulated in the Childhood Asthma Management Program (CAMP), and clinical differences were determined. Metabolomic drivers of metabo-endotype membership were investigated by meta-analyzing findings from GACRS and CAMP.
Measurements and Main Results
Five metabo-endotypes were identified in GACRS with significant differences in asthma-relevant phenotypes, including prebronchodilator (p-ANOVA = 8.3 × 10−5) and postbronchodilator (p-ANOVA = 1.8 × 10−5) FEV1/FVC. These differences were validated in the recapitulated metabo-endotypes in CAMP. Cholesterol esters, trigylcerides, and fatty acids were among the most important drivers of metabo-endotype membership. The findings suggest dysregulation of pulmonary surfactant homeostasis may play a role in asthma severity.
Conclusions
Clinically meaningful endotypes may be derived and validated using metabolomic data. Interrogating the drivers of these metabo-endotypes has the potential to help understand their pathophysiology.
Keywords: asthma, metabolomics, endotyping, metabo-endotypes
At a Glance Commentary
Scientific Knowledge on the Subject
Asthma is an extremely heterogeneous condition; however, current therapeutic approaches do not take this heterogeneity into account.
What This Study Adds to the Field
We propose five validated asthma metabo-endotypes that differ in clinically important lung function phenotypes. These metabo-endotypes provide an improved understanding of asthma heterogeneity and a step toward precision medicine for this common condition.
Asthma affects 26 million children and adults in the United States and remains a leading cause of morbidity (1). Asthma is characterized by variable reversible airflow obstruction, nonspecific airway hyperresponsiveness, and airway inflammation; however, there is substantial heterogeneity in its etiology, pathology, and manifestation (2). Current guidelines for defining asthma, which categorize cases from mild to severe, do not sufficiently capture this heterogeneity, leading to suboptimal management strategies in certain subgroups (3). A more detailed molecular classification is needed.
It is hypothesized there are multiple asthma endotypes (i.e., subtypes defined by functional or pathobiological mechanisms) that confer clinically meaningful differences in patient outcomes (4). Treatments and management strategies based on these underlying pathobiological mechanisms, rather than a “one-size-fits-all” approach, may be more effective in terms of improved outcomes and optimized use of healthcare resources. The relative contribution of genetics and environment to the formation of these mechanistically driven endotypes is likely to vary between endotypes. Metabolomics reflects genetics, environmental factors, and their interactions (5) and as the -ome closest to phenotype provides insight into the physiological state of an individual. As such, it represents a novel and compelling approach to identify asthma endotypes.
Interrogating high-dimensional omic datasets to infer biological meaning can be challenging. Clustering methods have proven powerful in the identification of molecular subtypes of asthma that differ by atopic status, eosinophil count, and cytokine concentrations (4, 6–10), but to date, none have taken an untargeted approach leveraging the global metabolome. In this study, we aim to derive clinically meaningful “metabo-endotypes” of asthma and to validate these findings in an independent population.
Some of the results of this study have been previously reported in the form of an abstract (11), and some of the results of this study have been previously reported in the form of a preprint (https://doi.org/10.21203/rs.3.rs-358819/v1). Metabolomic data were generated as part of the NHLBI Trans-Omics for Precision Medicine Initiative (TOPMed). These data will be released to the scientific community in their entirety via NIH-designated repositories according to the TOPMed data release timeline. Full details can be found at https://www.nhlbiwgs.org/topmed-data-access-scientific-community. All statistical analyses were conducted using freely available packages in R version 4.0.0; all such packages are stated and referenced in the Methods and eMethods in the online supplement.
Methods
Study Populations
The study populations have previously been described. The Genetics of Asthma in Costa Rica Study (GACRS) (12) recruited 1,165 children with asthma aged 6–14 years (physician’s diagnosis and ⩾2 respiratory symptoms or asthma attacks in the prior year). At enrollment, all children completed a protocol including questionnaires, blood collection, and spirometry conducted with a Survey Tach Spirometer (Warren E. Collins) in accordance with American Thoracic Society recommendations (12). Written parental and participating child consent and/or assent was obtained. The study was approved by the Mass General Brigham Human Research Committee at Brigham and Women’s Hospital, protocol #2000-P-001130/55, and the Hospital Nacional de Niños. All children who had available plasma samples with sufficient volume were selected for this current study.
The Childhood Asthma Management Program (CAMP) (13) (Clinicaltrials.gov: NCT00000575) is a completed randomized clinical trial of inhaled treatments for mild to moderate asthma (symptoms for >6 months in the year prior to interview and provocative concentration of methacholine causing a 20% drop in FEV1 [PC20] < 12.5 mg/ml) in children aged 5–12 at baseline. All children completed a similar protocol to GACRS. The study was approved by the institutional review board of Mass General Brigham Healthcare (protocol #1999-P-001549/29), by all participating clinical centers and the Data Coordinating Center. Child assent and parental written consent was obtained. Participants who had available plasma samples with sufficient volume from the end of the trial visit (5 to 6 years after baseline) were selected for this current study (see the online supplement).
Metabolomic Profiling
Metabolomic profiling was conducted using four complementary liquid chromatography tandem mass spectrometry (LC-MS) platforms as part of the TOPMed initiative (14). Three nontargeted LC-MS methods using high resolution, accurate mass (HRAM) profiling measured 1) polar and nonpolar lipids (C8-pos); 2) free fatty acids, bile acids, and metabolites of intermediate polarity (C18-neg); and 3) polar metabolites including amino acids, acylcarnitines, and amines (HILIC-pos). An additional targeted LC-MS profiling method measured intermediary metabolites including purines and pyrimidines, and acyl CoAs (Amide-neg) (15, 16). Datasets were restricted to those metabolites that were present in both GACRS and CAMP, resulting in a total of 589 metabolites for analysis (C8-pos n = 205; C18-neg n = 88; HILIC-pos n = 195; and Amide-neg n = 101). Of these, 398 of 589 (67.6%) were confirmed at the Metabolite Standards Initiative level 1 with authentic standards. Full details are in the online supplement (Tables E1 and E2). We generated metabolite residuals for each individual regressing on sex, body mass index (BMI), and age to account for the potential influence of these factors on the metabolome and to ensure they did not drive subgroup formation. All analyses were conducted on the residuals.
Statistical Methods
Derivation of “metabo-endotypes” in GACRS
We grouped 1,151 subjects from the GACRS based on their metabolite residuals into distinct metabolomic-driven endotypes, using Similarity Network Fusion (SNF) (R package SNFtool version 2.2) (17) and spectral clustering. SNF is a patient-centered method for integrating “omic” data through the construction and integration of patient networks (17, 18) (Figure E1) in which patient similarity networks are constructed from nodes (asthma cases) connected by edges (similarity in metabolomic profile between cases). The similarity between any two given asthma cases is based on the Euclidean distance between those two cases with a scaled exponential similarity kernel to determine the weight of the edge (17). For this kernel, two parameters must be set: k, the number of neighbors, and α, the decay rate of the exponential (17). In this study, we set the parameters k = 288 and α = 0.8. k was computed based on the recommended algorithm n/c, in which n is number of participants and c is the expected number of clusters, and α was set based on the recommended setting in Wang and colleagues, 2014 (17). We hypothesized c = 4 asthma endotypes based on previously published work (7, 19–21).
We considered each of the four metabolite platforms as separate “omics”, built a network for each platform, and then fused the platforms in the network fusion step of SNF, a nonlinear method based on message-passing theory (22). To compute the fused matrix, for each platform-specific patient similarity network, the k-nearest neighbor approach is used to measure the local affinity by creating a kernel matrix where the similarity of nonneighbors is set to zero. The kernel matrices for each network are then used to iteratively update each normalized patient similarity matrix until it converges to one final patient similarity network (17). During the fusion procedure, patient-to-patient connections are accentuated if they occurred in multiple platforms and dropped if the connections were weak or only supported by a single platform (17, 18).
We applied spectral clustering (23) to both platform-specific networks and the fused network to identify metabolomic-driven clusters within each platform. Spectral clustering is a method derived from graph theory to cluster patients using a similarity network as the input. This method first calculates the relevant eigenvectors of a Laplacian matrix of the similarity network, and then cluster the patients on those eigenvectors using k-means clustering (23) We used two approaches, the rotational cost approach (17) and the eigengap approach (24), to identify the optimal number of clusters for each platform. For the eigengap approach, the optimal number of clusters is determined by the largest eigengap (i.e., difference between consecutive eigenvectors) (24). For the rotational cost approach, the optimal number of clusters is calculated based on minimizing the value of a cost-function over all possible rotations that best aligns the eigenvectors with the canonical coordinate system (25).
Exploration clinical characteristics of metabo-endotypes
We examined whether the omic-derived alterations in biological pathway between the clusters (metabo-endotypes) resulted in measurable clinical or epidemiological differences using one-way ANOVA for continuous variables and chi-square tests for categorical variables.
Validation of metabo-endotypes
In CAMP, we used the label propagation classifier approach as a graph-based semisupervised machine learning method to predict the metabo-endotype (as defined in GACRS) (17). The label propagation algorithm is an iterative process where labeled nodes in a network (i.e., GACRS patients in a combined CAMP and GACRS patient similarity matrix) propagate their label to its nearest neighbors to assign a label to unlabeled nodes (i.e., CAMP patient) (26). To use this approach, we first constructed a similarity matrix using SNF combining the GACRS and CAMP populations using their metabolite residual data, then applied the classifier to assign each of the CAMP validation subjects to one of the GACRS-defined metabo-endotypes. In this way, an individual in metabo-endotype 1 in CAMP had a similar metabolomic profile to an individual in metabo-endotype 1 in GACRS, and individuals in metabo-endotype 2 in CAMP were metabolomically similar to those in GACRS metabo-endotype 2, and so on. We then assessed the clinical and phenotypic characteristics of CAMP subjects within these metabo-endotypes as before.
We considered our metabo-endotypes as validated if the clinical characteristics that differentiate the asthma metabo-endotypes generated in GACRS also differentiated the asthma metabo-endotypes in CAMP.
Identification of metabolomic drivers of meta-endotypes
We used independent logistic regression models and a one-endotype-versus-the-rest approach to identify the metabolites that contributed the most to the formation of each metabo-endotype. To generate a single effect estimate for the contribution of each metabolite to the generation of the metabo-endotypes, we meta-analyzed the GACRS and CAMP results using a random effects model and the R package meta (version 4.18–0). We restricted to those metabolites that were in a concordant direction of effect in the two cohorts, had a P value less than 0.05 in both cohorts, and applied a Bonferroni correction to the meta-analyzed P value assuming 589 metabolites. To explore the biology of these significant metabolites, we employed ChemRICH (analysis of chemical similarity enrichment) (27). ChemRICH is a metabolite enrichment approach that automatically detects and labels nonoverlapping sets of metabolites based on their structural and chemical similarity, rather than their biological annotations, then uses Kolmogorov-Smirnoff statistics to test the significance of differential regulation among the defined metabolite sets between two conditions of interest. For each of the one-endotype-versus-the-rest analyses, we entered the effect estimates and P values for the 485 metabolites that could be assigned Simplified Molecular Input Line Entry System (SMILES) IDs and therefore could be included in the ChemRICH online tool to identify the metabolite sets of interest.
All analyses were conducted in R version 4.0.0, with the exception of ChemRICH, which was performed using the web interface at http://chemrich.fiehnlab.ucdavis.edu.
Results
Study Population
In GACRS, 1,151 subjects had plasma samples available for metabolomic profiling, and in CAMP, 911 subjects had suitable plasma samples extracted at the end of trial (Table 1). In the original CAMP trial, no significant difference in lung function outcomes between the study arms was found (13). The GACRS discovery population was slightly younger (mean age, 9.22 yr [SD, 1.88 yr], vs. 12.94 yr [SD, 2.14 yr]) in CAMP and had a lower BMI (mean, 18.28 kg/m2 [SD, 3.77 kg/m2] vs. mean, 21.42 kg/m2 [SD, 4.70 kg/m2]). Furthermore, all GACRS subjects were Hispanic, whereas CAMP subjects were primarily White (69.2%) with only approximately 10% Hispanic subjects.
Table 1.
Characteristics of the Discovery and Validation Populations
| Discovery GACRS (n = 1,151) | Validation CAMP (n = 911) | |
|---|---|---|
| Age, yr, mean (SD) | 9.22 (1.88) | 12.94 (2.14) |
| Sex, male, n (%) | 682 (59.3%) | 549 (60.3%) |
| Sex, female, n (%) | 469 (40.7%) | 362 (39.7%) |
| Height, cm, mean (SD) | 132.66 (11.85) | 155.89 (13.35) |
| Weight, kg, mean (SD) | 33.02 (11.49) | 53.23 (17.32) |
| BMI, kg/m2, mean (SD) | 18.28 (3.77) | 21.42 (4.70) |
| Race | ||
| White, n (%) | — | 630 (69.2%) |
| Black, n (%) | — | 117 (12.8%) |
| Hispanic, n (%)* | 1,151 (100%) | 84 (9.2%) |
| Other, n (%) | — | 80 (8.8%) |
| Treatment arm† | ||
| Budesonide, n (%) | — | 270 (29.6%) |
| Nedocromil, n (%) | — | 269 (29.5%) |
| Placebo, n (%) | — | 372 (40.8%) |
Definition of abbreviations: BMI = body mass index; CAMP = Childhood Asthma Management Program; GACRS = Genetics of Asthma in Costa Rica Study.
GACRS represents a unique population isolate where participants were selected on the basis of having six or more great-grandparents born within the central valley of Costa Rica.
The CAMP population was from a completed clinical trial.
GACRS Metabo-Endotypes
The optimal numbers of clusters of GACRS asthmatics were determined to be two, two, two, and three based on metabolite residuals from the C8-pos, C18-neg, HILIC-pos, and Amide-neg platforms, respectively. We applied SNF to fuse the networks from the four platforms, with convergence after 10 iterations, and performed spectral clustering. We determined five was the optimal number of clusters, containing 213, 270, 222, 232, and 214 asthma cases, respectively, and we designated these as the asthma metabo-endotypes (Figure E2). We compared the ways in which individuals clustered between each platform with how they clustered to form metabo-endotypes based on the fused networks (Figure E3). Individuals in metabo-endotype 1 and 2 tended to cluster within these groupings in all platforms, whereas overall the fused metabo-endotype clusterings were most similar to those of the Amide-neg platform (adjusted Rand index = 0.297) (Table E3).
There was no difference between the clusters in terms of sex, age, BMI, vitamin D concentration, or current smoking status (P > 0.5) (Table 2). However, there was a significant difference across the endotypes in measures of lung function: prebronchodilator FEV1/FVC ratio (P = 8.25 × 10−5) for which metabo-endotype 2 had the lowest ratio (mean = 83.1%; range = 50.6–98.8%) and metabo-endotype 3 the highest (mean = 86.5%; range = 64.2–99.9%) and postbronchodilator FEV1/FVC ratio (P = 1.82 × 10−5). Again, metabo-endotype 2 (mean = 85.9%; range = 52.2–100%) was the lowest and metabo-endotype 3 (mean = 89.1%, range = 69.0–100%) the highest. The same pattern was observed when considering percent predicted FEV1/FVC ratio (prebronchodilator P = 4.46 × 10−5; postbronchodilator P = 1.00 × 10−5) (Figure 1). Consequently, metabo-endotype 2 and metabo-endotype 3 were considered to be of greatest interest, representing poor and good lung function, respectively.
Table 2.
Differences in Characteristics between the Five GACRS Asthma Metabo-Endotypes
| GACRS Variable | Endotype 1 |
Endotype 2 |
Endotype 3 |
Endotype 4 |
Endotype 5 |
P Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 213 | n = 270 | n = 222 | n = 232 | n = 214 | ||||||||
| Demographic characteristics | Sex, male, n (%) | 125 (58.7%) | 160 (59.3%) | 136 (61.3%) | 133 (57.3%) | 128 (59.8%) | 0.941 | |||||
| Age, mean (SD) | 9.21 (1.82) | 9.23 (1.83) | 9.27 (1.84) | 9.14 (2.02) | 9.28 (1.87) | 0.932 | ||||||
| BMI, mean (SD) | 18.3 (3.84) | 18.4 (3.66) | 18.4 (3.83) | 18.3 (3.81) | 17.9 (3.73) | 0.577 | ||||||
| Serum vitamin D, ng/ml, mean (%) | 37.4 (11.6) | 37.6 (10.8) | 35.2 (9.0) | 37.5 (12.6) | 38.0 (14.3) | 0.870 | ||||||
| Current smoking exposure, yes, n (%) | 53 (24.9%) | 68 (25.2%) | 53 (23.9%) | 58 (25.0%) | 52 (24.3%) | 0.995 | ||||||
| Lung function | Prebronchodilator FEV1, L, mean (SD) | 1.76 (0.48) | 1.78 (0.46) | 1.78 (0.48) | 1.79 (0.56) | 1.77 (0.53) | 0.980 | |||||
| Prebronchodilator FVC, L, mean (SD) | 2.09 (0.57) | 2.16 (0.56) | 2.08 (0.58) | 2.13 (0.64) | 2.11 (0.63) | 0.588 | ||||||
| Prebronchodilator FEV1/FVC, mean (SD) | 84.34 (8.60) | 83.14 (6.64) | 86.16 (7.03) | 83.81 (8.52) | 84.43 (8.36) | 8.25 × 10−5* | ||||||
| Postbronchodilator FEV1, L, mean (SD) | 1.85 (0.52) | 1.87 (0.48) | 1.86 (0.48) | 1.88 (0.57) | 1.85 (0.53) | 0.942 | ||||||
| Postbronchodilator FVC, L, mean (SD) | 2.13 (0.59) | 2.19 (0.56) | 2.1 (0.56) | 2.17 (0.64) | 2.14 (0.63) | 0.595 | ||||||
| Postbronchodilator FEV1/FVC, mean (SD) | 87.10 (7.55) | 85.87 (6.06) | 89.10 (5.82) | 87.15 (7.22) | 86.97 (7.45) | 1.82 × 10−5* | ||||||
| % predicted prebronchodilator FEV1/FVC, mean (SD) | 94.80 (9.55) | 93.00 (7.50) | 96.90 (7.78) | 94.20 (9.52) | 95.00 (9.22) | 4.46 × 10−5* | ||||||
| % predicted postbronchodilator FEV1/FVC, mean (SD) | 98.00 (8.46) | 96.60 (6.81) | 100.0 (6.37) | 97.90 (8.04) | 97.80 (8.24) | 1.00 × 10−5* | ||||||
| Indices of asthma severity | Use of oral steroids in previous year, yes, n (%) | 96 (45.1%) | 156 (57.8%) | 118 (53.2%) | 126 (54.3%) | 93 (43.5%) | 0.007* | |||||
| Use of inhaled steroids in previous year, yes, n (%) | 138 (64.8%) | 91 (33.7%) | 139 (62.6%) | 123 (53.0%) | 97 (45.3%) | 4.97 × 10−13* | ||||||
| Use of short-acting β2-agonists in previous year, yes, n (%) | 25 (11.7%) | 82 (30.4%) | 17 (7.7%) | 45 (19.4%) | 47 (22.0%) | 3.25 × 10−10* | ||||||
| Any asthma medication in previous year, yes, n (%) | 209 (98.1%) | 256 (94.8%) | 217 (97.7%) | 229 (98.7%) | 204 (95.3%) | 0.046* | ||||||
| Ever hospitalized for asthma, yes, n (%) | 80 (37.6%) | 117 (43.3%) | 95 (42.8%) | 106 (45.7%) | 90 (42.1%) | 0.522 | ||||||
| Ever visited ER for asthma, yes, n (%) | 208 (97.7%) | 259 (95.9%) | 214 (96.4%) | 224 (96.6%) | 206 (96.3%) | 0.886 | ||||||
| Allergic phenotypes and eosinophil concentrations | Log10 blood eosinophils, cells/μl, mean (SD) | 2.56 (0.41) | 2.67 (0.35) | 2.55 (0.46) | 2.58 (0.41) | 2.60 (0.43) | 0.009* | |||||
| Eosinophilic asthma (untransformed count > 300 cells/μl), yes, n (%) | 127 (59.6%) | 202 (74.8%) | 142 (64.0%) | 136 (58.6%) | 143 (66.8%) | 0.006 | ||||||
| Log10 IgE, kU/L, mean (SD) | 2.52 (0.70) | 2.48 (0.69) | 2.52 (0.65) | 2.45 (0.69) | 2.57 (0.61) | 0.344 | ||||||
| Number of positive skin prick tests, mean (SD) | 3.12 (1.77) | 3.02 (1.91) | 2.94 (1.89) | 3.14 (1.83) | 3.02 (1.77) | 0.784 | ||||||
| Prevalent hay fever, yes, n (%) | 79 (37.1%) | 74 (27.4%) | 80 (36.0%) | 66 (28.4%) | 67 (31.3%) | 0.076 | ||||||
| Prevalent atopic dermatitis, yes, n (%) | 10 (4.7%) | 12 (4.4%) | 7 (3.2%) | 8 (3.4%) | 17 (7.9%) | 0.142 | ||||||
Definition of abbreviations: BMI = body mass index; ER = emergency room; GACRS = Genetics of Asthma in Costa Rica Study.
P value is based on the ANOVA test for continuous variables and the chi-square test for categorical variables.
Significant at the 95% confidence level.
Figure 1.
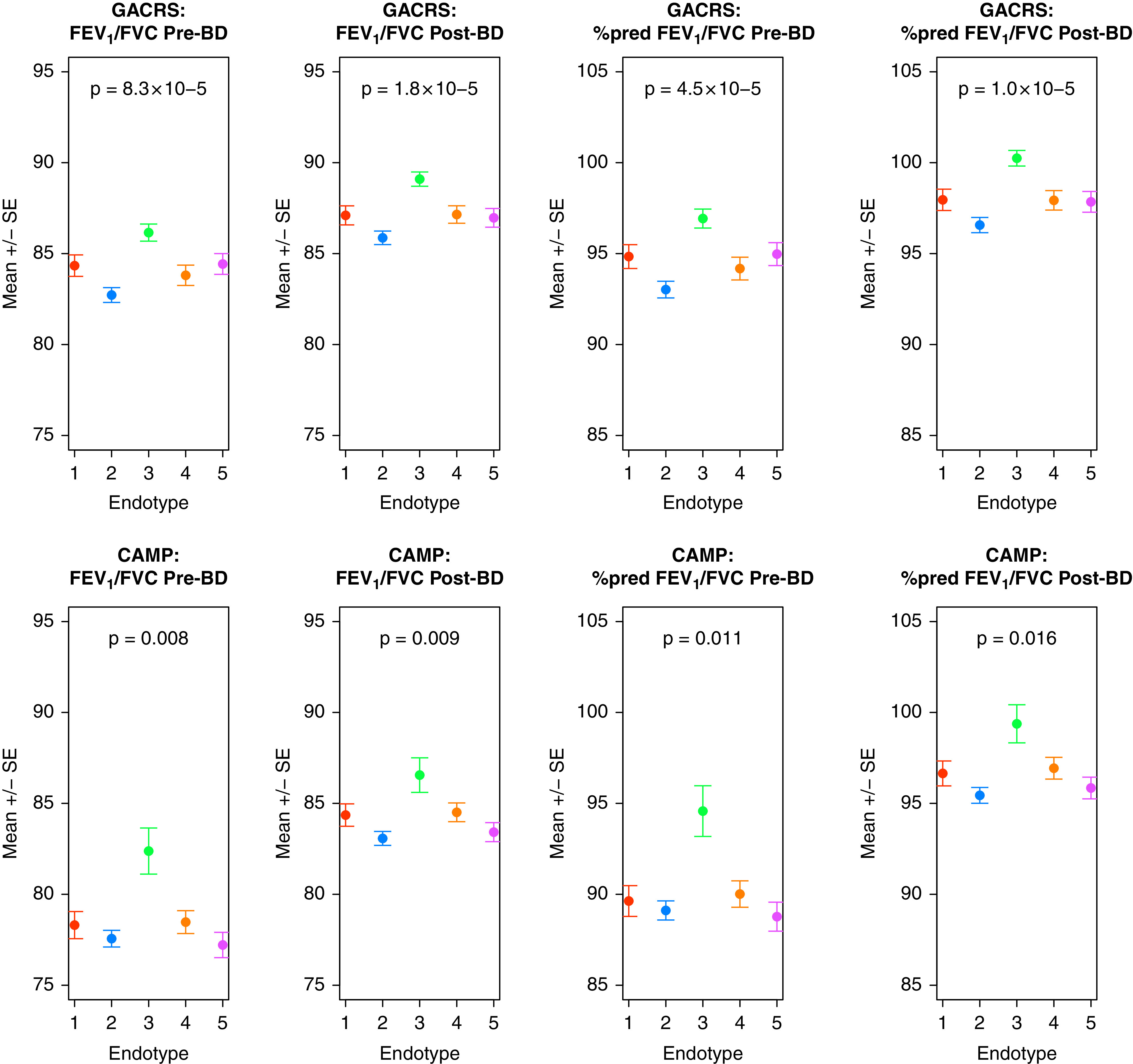
FEV1/FVC ratio before and after bronchodilator (BD) demonstrating significant difference across metabo-endotypes in both the Genetics of Asthma in Costa Rica Study (GACRS) and the Childhood Asthma Management Program (CAMP). Mean and standard errors for the specified metric in each metabo-endotype are shown. P values are derived from a one-way ANOVA test comparing the continuous variables across the five metabo-endotypes.
The metabo-endotypes differed in the use of oral (P = 0.007) and inhaled (P = 4.97 × 10−13) corticosteroids and the use of β2-agonists (P = 3.25 × 10−10). Asthma cases in metabo-endotype 2 were the most likely to have taken oral steroids (57.8%) or β2-agonists (30.4%) in the previous year, but the least likely to have taken inhaled steroids (33.7%). Metabo-endotype 3 had the lowest number who reported the use of β2-agonists in the previous year (Figure E4).
Finally, there was evidence of a significant difference in eosinophil concentrations across the metabo-endotypes. Metabo-endotype 2 had the highest concentrations of blood eosinophils (log10 eosinophil count = 2.67 cells/μl; range = 1.0–3.41 cells/μl) and the highest percentage of individuals with eosinophilic asthma (74.8%) defined as untransformed eosinophil count > 300 cells/μl (28) (Figure E5).
Validating Metabo-Endotypes in CAMP
The recapitulated metabo-endotypes contained 99, 375, 45, 207, and 185 CAMP cases in metabo-endotypes 1, 2, 3, 4, and 5, respectively. Individuals within the same number metabo-endotype across the two cohorts can be considered metabolomically equivalent; therefore, we sought to determine if they also displayed the same clinical differences.
The significant difference across metabo-endotypes for FEV1/FVC ratio before and after bronchodilator validated in CAMP with an almost identical pattern, with individuals in metabo-endotype3 demonstrating the best lung function (Table 3 and Figure 1). Given that before sample collection, CAMP subjects had been randomized to differing treatment regimens, we could not directly compare medication use, and no significant differences were observed (Figure E6).
Table 3.
Differences in Characteristics between the Five CAMP Asthma Metabo-Endotypes
| CAMP Variable | Endotype 1 |
Endotype 2 |
Endotype 3 |
Endotype 4 |
Endotype 5 |
P Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 99 | n = 375 | n = 45 | n = 207 | n = 185 | ||||||||
| Demographic characteristics | Sex, male, n (%) | 58 | (58.6%) | 229 | (61.1%) | 32 | (71.1%) | 125 | (60.4%) | 105 | (56.8%) | 0.496 |
| Age, mean (SD) | 13.06 | (2.38) | 12.91 | (2.10) | 12.32 | (1.61) | 12.95 | (2.19) | 13.08 | (2.12) | 0.288 | |
| BMI, mean (SD) | 21.18 | (4.60) | 21.43 | (4.66) | 20.74 | (4.93) | 21.31 | (4.19) | 21.84 | (5.31) | 0.598 | |
| Serum vitamin D, ng/ml, mean (%) | 33.94 | (14.61) | 31.55 | (14.15) | 30.49 | (13.59) | 28.3 | (14.31) | 26.75 | (11.86) | 4.83 × 10−5* | |
| Current smoking exposure, yes, n (%) | 9 | (9.1%) | 55 | (14.7%) | 6 | (13.3%) | 26 | (12.6%) | 32 | (17.3%) | 0.399 | |
| Race, White, n (%) | 72 | (72.7%) | 262 | (69.9%) | 32 | (71.1%) | 140 | (67.6%) | 124 | (67.0%) | 0.644 | |
| Lung phenotypes | Prebronchodilator FEV1, L, mean (SD) | 2.55 | (0.70) | 2.54 | (0.74) | 2.51 | (0.65) | 2.62 | (0.80) | 2.54 | (0.71) | 0.771 |
| Prebronchodilator FVC, L, mean (SD) | 3.29 | (0.99) | 3.29 | (0.94) | 3.08 | (0.85) | 3.36 | (1.05) | 3.30 | (0.91) | 0.518 | |
| Prebronchodilator FEV1/FVC, mean (SD) | 78.31 | (7.39) | 77.56 | (8.89) | 82.38 | (8.51) | 78.47 | (9.03) | 77.21 | (9.46) | 0.008* | |
| Postbronchodilator FEV1, L, mean (SD) | 2.78 | (0.78) | 2.76 | (0.77) | 2.68 | (0.67) | 2.85 | (0.85) | 2.79 | (0.75) | 0.614 | |
| Postbronchodilator FVC, L, mean (SD) | 3.32 | (1.00) | 3.34 | (0.94) | 3.12 | (0.83) | 3.4 | (1.07) | 3.36 | (0.92) | 0.506 | |
| Postbronchodilator FEV1/FVC, mean (SD) | 84.36 | (6.11) | 83.07 | (7.42) | 86.56 | (6.83) | 84.51 | (7.38) | 83.42 | (7.05) | 0.009* | |
| % predicted prebronchodilator FEV1/FVC, mean (SD) | 89.6 | (8.38) | 89.1 | (10.2) | 94.6 | (9.34) | 90.0 | (10.4) | 88.8 | (10.8) | 0.011* | |
| % predicted postbronchodilator FEV1/FVC, mean (SD) | 96.6 | (6.79) | 95.4 | (8.43) | 99.4 | (7.03) | 96.9 | (8.59) | 95.8 | (8.05) | 0.016* | |
| Indices of asthma severity | Use of corticosteroid since last visit, yes, n (%) | 21 | (21.2%) | 68 | (18.1%) | 8 | (17.8%) | 34 | (16.4%) | 29 | (15.7%) | 0.787 |
| Use of albuterol since last visit, yes, n (%) | 80 | (80.8%) | 313 | (83.5%) | 33 | (73.3%) | 170 | (82.1%) | 151 | (81.6%) | 0.530 | |
| Ever hospitalized for asthma, yes, n (%) | 10 | (10.1%) | 36 | (9.6%) | 2 | (4.4%) | 25 | (12.1%) | 19 | (10.3%) | 0.659 | |
| Ever visited ER for asthma, yes, n (%) | 33 | (33.3%) | 129 | (34.4%) | 10 | (22.2%) | 77 | (37.2%) | 54 | (29.2%) | 0.238 | |
| Allergic phenotypes and eosinophil concentrations | Log10 blood eosinophils (cells/μl), mean (SD) | 2.47 | (0.40) | 2.43 | (0.48) | 2.45 | (0.36) | 2.40 | (0.48) | 2.31 | (0.58) | 0.050* |
| Eosinophilic asthma (untransformed count > 300 cells/μl), yes, n (%) | 54 | (54.5%) | 196 | (52.3%) | 24 | (53.3%) | 104 | (50.2%) | 79 | (42.7%) | 0.343 | |
| Log10 IgE, kU/L, mean (SD) | 2.77 | (0.62) | 2.64 | (0.62) | 2.50 | (0.70) | 2.64 | (0.68) | 2.56 | (0.69) | 0.086 | |
| Number of positive skin prick tests, mean (SD) | 6.09 | (3.69) | 6.14 | (4.42) | 4.70 | (3.53) | 6.20 | (4.37) | 5.99 | (4.25) | 0.312 | |
| Prevalent hay fever, yes, n (%) | 47 | (47.5%) | 165 | (44.0%) | 29 | (64.4%) | 88 | (42.5%) | 93 | (50.3%) | 0.061 | |
| Prevalent atopic dermatitis, yes, n (%) | 23 | (23.2%) | 98 | (26.1%) | 15 | (33.3%) | 59 | (28.5%) | 60 | (32.4%) | 0.371 | |
Definition of abbreviations: BMI = body mass index; CAMP = Childhood Asthma Management Program; ER = emergency room.
P value is based on the ANOVA test for continuous variables and the chi-square test for categorical variables.
Significant at the 95% confidence level.
The differences in log10 eosinophil count and hay fever prevalence seen in GACRS were borderline significant across the metabo-endotypes in CAMP (P = 0.050 and P = 0.061, respectively). However, the patterns differed (Figure E5). In addition, there was evidence that vitamin D concentrations differed significantly across the metabo-endotypes in CAMP (4.83 × 10−5).
Sensitivity Analyses
To confirm that the significant differences in lung function metrics across the metabo-endotypes were not confounded by inhaled corticosteroid (ICS) use, we ran analysis of covariance models for the four significant lung metrics: prebronchodilator FEV1/FVC, postbronchodilator FEV1/FVC, percent predicted prebronchodilator FEV1/FVC, and percent predicted postbronchodilator FEV1/FVC, adjusting for ICS use in the previous year in GACRS and corticosteroid use since the last visit in CAMP. The between–metabo-endotype differences retained significance in both GACRS and CAMP for all four metrics with this additional adjustment (Table E4). These results indicate that differences in lung metrics across metabo-endotypes are not being driven by steroid usage in those with poorer lung function.
Key Metabolite Metabo-Endotype Drivers
In a meta-analysis of GACRS and CAMP, 147, 256, 161, 332, and 269 metabolites were significantly associated with membership of metabo-endotypes 1, 2, 3, 4, and 5, respectively, after Bonferroni correction and restriction to those metabolites with concordant directions of effect and a P value less than 0.05 in each individual cohort (Tables E5–E9). There was some crossover in the metabolites associated with each metabo-endotype (Table E10 and Figure E7), although the direction of effect often differed. For example, 9,10-diHOME, which has been shown to correlate with lung function (29), was lower among the individuals in metabo-endotype 3 who had the best lung function (β = −0.400; P = 2.23 × 10−15) relative to all other metabo-endotypes, but at higher concentrations among those in metabo-endotype 2 who had the worst (β = 0.595; P = 1.78 × 10−28). Similarly, two polyunsaturated fatty acids (PUFAs), linoleic acid (β = 0.595; P = 1.78 × 10−28) and arachidonic acid (β = −0.564; P = 1.04 × 10−5), which are also thought to play key roles in lung function (29, 30), were lower in metabo-endotype 2 relative to all other metabo-endotypes. We also identified a number of metabolites unique to each metabo-endotype (Table E11).
ChemRICH analysis was used to determine overall metabolomic pathway enrichment or depletion for each metabo-endotype (Figure 2, Figures E8–E10, and Table E12). Overall, lipids, and in particular triglycerides and phospholipid concentrations, were among the greatest drivers of membership. Metabo-endotype3, which had the lowest degree of lung obstruction, was characterized by depletion of hydroxy and unsaturated fatty acids (false discovery rate [FDR] = 4.6 × 10−19 for both sets), carnitines (FDR = 1.4 × 10−9), and cholesterol esters (FDR = 1.7 × 10−9) and by enrichment of triglycerides (FDR = 4.6 × 10−19) among others (Figure E3 and Table E12). In contrast, metabo-endotype2, which demonstrated a high degree of lung obstruction, was characterized by depletion of triglycerides (FDR = 2.6 × 10−19 for both saturated and unsaturated triglycerides), unsaturated phosphatidylcholines and lysophosphatidylcholines (FDR = 2.6 × 10−19 for both), and unsaturated fatty acids (FDR = 2.3 × 10−13), among others (Figure 2 and Table E12).
Figure 2.
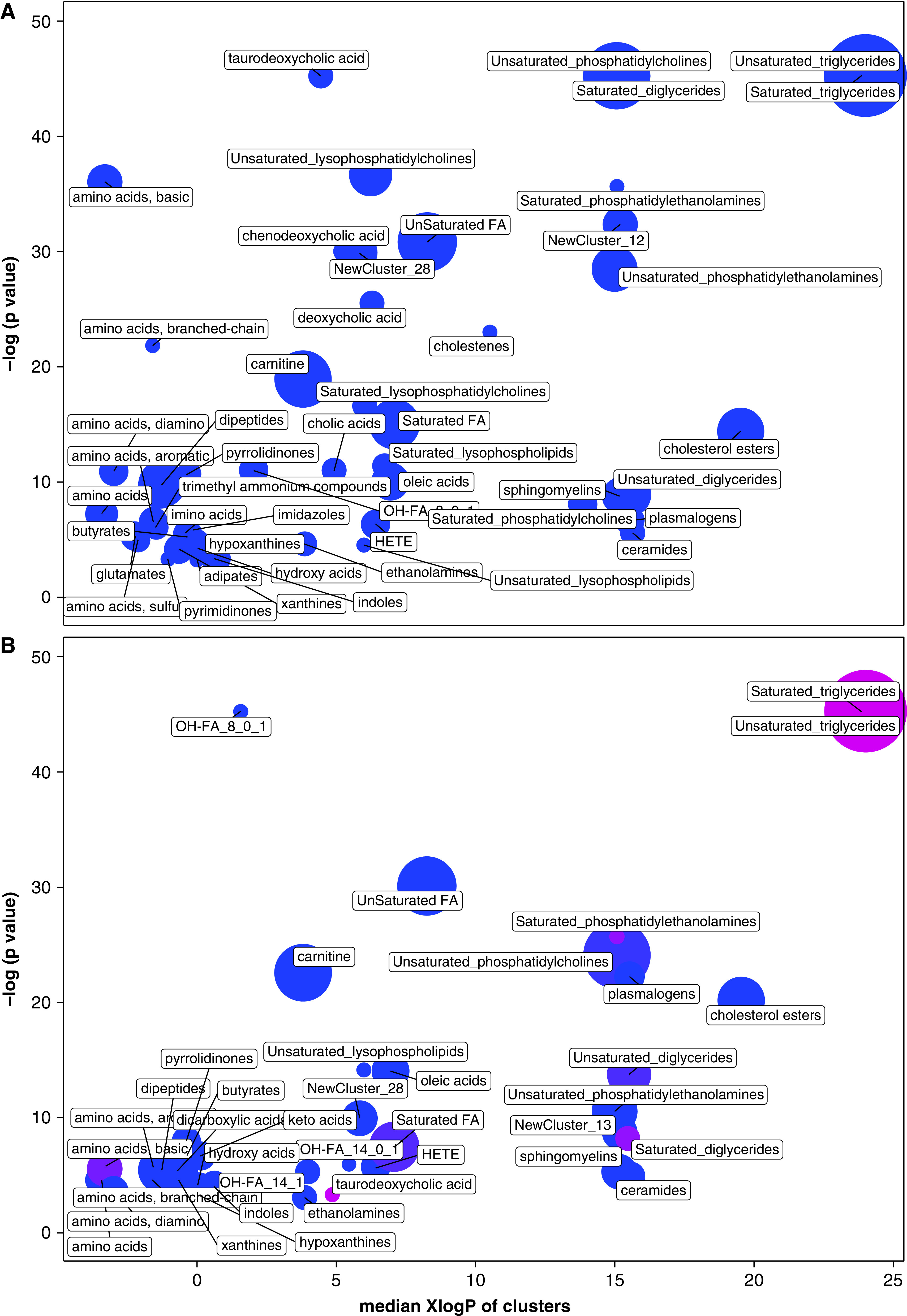
Chemical similarity enrichment analysis of (A) metabo-endotype 2 membership versus membership in any other metabo-endotype and (B) metabo-endotype 2 membership versus membership in any other metabo-endotype. Figures include all enriched metabolite sets based on false discovery rate < 0.05. Each circle represents a set; circle sizes represent the total number of metabolites in each set. The color corresponds to the proportion of increased (red) or decreased (blue) compounds. Purple circles have both increased and decreased metabolites. The y-axis shows the significance of the enrichment for a given metabolite set. The x-axis shows the median XlogP of clusters, which is a measure of the average lipophilicity of the set, based on the octanol/water partition of the component metabolites. FA = fatty acids; HETE = hydroxyeicosatetraenoic acids; OH-FA = hydroxy-fatty acids.
Discussion
In this study, we identified and validated five asthma metabo-endotypes with differing lung function and clinical characteristics driven by distinct metabolomic pathways.
Although several studies have attempted to identify asthma endotypes (31, 32), these have been somewhat limited and have tended to use one of two general approaches: a priori definitions of a phenotype based on characteristics of subjects, or pathobiologic differences in sputum or bronchoscopy specimens (4, 33–36). The resulting endotypes have often demonstrated high overlap in important clinical features, rendering them challenging for clinical use. More importantly, they provide little information on underlying mechanisms. Among the few studies using omic data to derive clusters (31, 32, 37–39), sample sizes have been small; however, results support the existence of multiple heterogeneous asthma subtypes with differing molecular profiles and pathophysiological pathways. Although several studies have incorporated metabolomics into their exploration of endotypes (6, 8, 9, 40, 41), to date none have leveraged unsupervised clustering of the global blood metabolome to subphenotype individuals with asthma.
There were significant differences in asthma-relevant phenotypic characteristics across our metabo-endotypes. Metabo-endotypes 2 and 3 were of greatest interest, demonstrating the highest and lowest degree of airflow obstruction, respectively, as well as differences in the usage of oral corticosteroids and β2-agonists. We recapitulated these metabo-endotypes in an independent population and observed almost identical differences in FEV1/FVC. Although we did not observe the same differences in medication usage in CAMP that were seen in GACRS, we hypothesize that this is because CAMP blood was collected at the end of a clinical trial, which would have dictated use of steroids in the previous year, as well as differences in the prescribing and therapeutic approaches applied by the respective health systems of the Costa Rican–based discovery cohort, and U.S.-based validation cohort. In contrast, FEV1/FVC, which was significantly different between metabo-endotypes in both populations, provides a more objective measure. Additional adjustment for steroid usage did not nullify the significant differences in lung obstruction metrics across the metabo-endotypes, suggesting steroids are not the driving their formation.
There were several other differences of interest between the discovery metabo-endotypes. In GACRS, metabo-endotype 2, which had the greatest degree of lung obstruction, displayed the highest blood eosinophil counts and proportion of eosinophilic asthmatics. This was not observed in CAMP, which may be explained by the underlying differences in immune phenotypes between the two populations, with CAMP displaying significantly higher log(IgE) concentrations (mean [SD], 2.63 kU/L [0.65 kU/L] vs. 2.50 kU/L [0.67 kU/L] in GACRS, P = 1.57 × 10−5), prevalence of hay fever (53.2% vs. 31.8%; P < 2.2 × 10−16) and eczema (28.0% vs. 4.7%; P < 2.2 × 10−16), and mean number of total skin prick tests (6.05 [2.38] vs. 3.05 [1.84]; P < 2.2 × 10−16) across the total population.
Individuals in metabo-endotype 3 had the best lung function and were also found to have the lowest concentrations of fatty acids, carnitines, and cholesterol esters, all of which have previously been associated with asthma and lung function (42–44). There is growing evidence that cholesterol plays a particularly important role in pulmonary physiology, serving as a primary source of antioxidant vitamin D and a promotor of surfactant production for alveolar epithelial type II cells (43). Pulmonary surfactant (45) lines the inner surface of the lung and works to lower surface tension and prevent alveolar collapse as well as playing a role in innate immune defense (46). Consequently, dysregulation of surfactant homeostasis has been implicated in pulmonary diseases and reduced lung function in both adults and children. However, excessive amounts of cholesterol have been shown to impair surfactant production, which may explain why concentrations were lower among patients with asthma in metabo-endotype 3 relative to the other metabo-endotypes (43).
Pulmonary surfactant also has multiple integrated and highly regulated lipid metabolite components, including phospholipids (phosphatidylcholines, phosphatidylglycerols, and phosphatidylethanolamines), triglycerides, cholesterols, fatty acids, and sphingomyelins (47), which were found to be among the greatest drivers of metabo-endotype membership and therefore lung function. There was evidence of an enrichment of both saturated and unsaturated triglycerides in metabo-endotype 3, whereas there was a depletion of these metabolite sets in metabo-endotype 2. Interestingly, although some studies suggest that trigylceride concentrations are higher in asthma cases than controls, others report no association (48), and to date no studies have compared concentrations of triglycerides within asthma cases. Further work is therefore needed to disentangle the role of triglycerides in asthmatic lung function. Metabo-endotype 2 was also associated with decreased concentrations of both n3 and n6 long-chain PUFAs. PUFAs have been shown to play a role in pulmonary function and disease through their role in the maintenance of the proinflammatory–proresolvin pathways (30, 49) but again are also important in the regulation of pulmonary surfactant homeostasis.
A potential difference in blood eosinophil count between asthma metabo-endotypes is in agreement with existing subphenotyping studies of asthmatics. Clustering of nine clinical variables identified and validated in the Airways Disease Endotyping for Personalized Therapeutics (ADEPT) and Unbiased Biomarkers for the Prediction of Respiratory Disease Outcomes (U-BIOPRED) cohorts identified four groups with distinct clinical and biomarker profiles, one of which had a “moderate, hyperresponsive, eosinophilic” phenotype, with moderate asthma control, mild airflow obstruction, and predominant Type-2 inflammation (21). Similarly, three separate studies of exhaled breath samples showed that models based on volatile organic compounds (VOCs) could classify asthma as eosinophilic or neutrophilic with high accuracy (8, 40, 50). Eosinophil count also forms a component of the commonly cited T-helper type 2 (Th2)-high and Th2-low asthma endotypes (51). However, such subgroups have not yet demonstrated clear clinical utility (31). More promisingly, endotypes based on gene expression profiles in participants from U-BIOPRED were shown to differ in their responses to oral corticosteroids (39), whereas another study on exhaled breath demonstrated that concentrations of VOCs could help to predict steroid responsiveness (41).
To be of use, it is crucial that omic-driven endotypes have the ability to inform therapeutic and management approaches. Metabo-endotypes are uniquely positioned to offer twofold translational potential: first, the assignment of individuals to subgroups that can receive treatment targeted to their particular disorder, and second the identification of the therapeutic targets within those subgroups underlying that treatment. For example, supplementation with specific metabolites that can help to restore the pulmonary surfactant homeostasis imbalance in metabo-endotype2. Furthermore, the measurement of the metabolome is relatively inexpensive compared with other omics. Yet, to date, no omic-driven endotypes have been translated into clinical practice, and additional work is required before the metabo-endotypes reported in this manuscript could be employed in the management of asthma. This should include validation of these metabo-endotypes in larger and more diverse populations, assessment of their stability over time, and targeted quantification of metabolites that may have biomarker or therapeutic target potential. However, if successful, these metabo-endotypes could have an important impact on a globally significant disorder.
There were several limitations to these analyses. All participants were under 18 years old at blood collection. Additional work in older populations is required to determine the generalizability of the findings to adult-onset asthma, although we note that early-onset asthma may represent the larger public health burden owing to its higher prevalence (52). Cluster analysis is a descriptive method, and groups can be defined even when there is no underlying structure in the data; however, we addressed this using two separate methods to define the optimal number of cluster and by assessing the clinical characteristics of the clusters in two different populations. In the determination of the patient similarity network from which the metabo-endotypes were derived, all four metabolomic platforms were weighted equally as per the SNF methodology. However, this does not take into account differences in the number and breadth of metabolites measured in each platform. It should be noted these clusters are based on a single time point. The cross-sectional nature therefore makes it challenging to determine whether the observed differences in lung function are the cause of the metabolomic differences or an effect of these differences. Similarly, although we determined these differences were not confounded by steroid use, we cannot rule out additional unmeasured confounding. Repeated sampling is necessary to assess the temporal stability of these metabo-endotypes, and this could also help to address issues of cause and effect. Encouragingly, previous clustering studies in asthmatic populations have demonstrated good longitudinal cluster stability (21).
There is both a genetic and environmental component to the metabolomic profiles from which the endotypes are derived. This environmental component encompasses early life exposures. Further work in cohorts with more extensive longitudinal data is required to disentangle the influence of such exposures. Metabo-endotypes were derived via metabolomic profiling of blood. The utility of blood for asthma studies is supported by the literature (53) and has the benefits of being readily accessible, vital for the development of clinically translatable biomarkers. However, future studies should address the replicability of these metabo-endotypes in different biosamples, particularly those closest to the lung such as sputum. There were also underlying differences in the two populations in terms of age, BMI, and, most strikingly, race. These differences may explain why we did not see greater replication of between–metabo-endotype clinical characteristics in the two populations. For example, there is evidence that prevalence of immune phenotypes can vary by race, as a product of both underlying genetic susceptibility and exposure profile (54), which may be masking between–metabo-endotype differences. However, we do note that percent predicted FEV1/FVC, which accounts for race in its calculation, did replicate between the Hispanic GACRS population and the multiethnic CAMP population.
There are also several strengths to this study. It is the largest endotyping study of asthma to date, the first to employ a unique design using a bottom-up approach from molecular signatures to clinical endotypes, unlike the majority of studies that have clustered based on phenotype, potentially missing mechanistic information. The patient similarity networks were generated using state-of-the-art metabolomic profiling platforms, providing broad coverage and highly reproducible data. We leveraged machine learning approaches to derive endotypes, and most crucially, we were able to validate our findings in an independent population with comparable metabolomic, phenotypic, and clinical data, despite underlying differences between the cohorts.
Conclusions
Asthma represents a spectrum of disorders with heterogeneous etiologies and clinical presentations; yet its clinical definition has remained unchanged for more than 50 years (31). A significant proportion of patients with asthma do not respond to the “one-size-fits-all” management approach, and it is these patients who are responsible for the majority of the asthma-related economic burden (55). This study, which is by far the largest to leverage metabolomics for asthma endotyping and the first to use an unsupervised metabolome-wide clustering approach, proposes five novel validated asthma metabo-endotypes with differing asthma relevant characteristics. These metabo-endotypes provide strong candidates for more precise asthma management strategies while informing on underlying mechanisms, paving the way for more personalized approaches to asthma management.
Acknowledgments
Acknowledgment
The authors thank the studies and participants who provided biological samples and data for TOPMed. They also thank the participants who provided biological samples and data for GACRS and CAMP, and all staff involved in these studies. They thank the NHLBI TOPMed Initiative for its support. For a full list of TOPMed collaborators, please see https://www.nhlbiwgs.org/topmed-banner-authorship.
Footnotes
The Trans-Omics in Precision Medicine (TOPMed) program was supported by the NHLBI. Metabolomic profiling for "NHLBI TOPMed: Childhood Asthma Management Program (CAMP)" (phs001726) and “NHLBI TOPMed: The Genetic Epidemiology of Asthma in Costa Rica (GACRS)” (phs000988) was performed at the Broad Institute and at Beth Israel (HHSN268201600034I). Core support, including phenotype harmonization, data management, sample-identity quality control, and general program coordination, was provided by the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). CAMP and GACRS were also supported by P01 HL132825 from the NHLBI. Additional support was provided by K01HL146980 (R.S.K.); R01HL141826 (M.H. and J.L.-S.); NHLBI R01HL123915 (J.L.-S.); NHLBI R01HL155742 (M.M. and J.L.-S.); NIH R01HL135142, NHLBI R01 HL137927, and NHLBI R01 HL147148 (M.H.C.); NHLBI R01 HL139634 (M.J.M.); NHLBI K01HL153941 (S.H.C.); HLF 20180290 and HLF 20200693 from the Swedish Heart Lung Foundation (C.E.W.); 2016-02798 from the Swedish Research Council (C.E.W.); and K08 HL136928, U01 HL089856, R01 HL147148, and R01 HL135142 from the NIH (B.D.H.). The funding bodies played no role in the design or conduct of the study, the collection, management, analysis or interpretation of the data, the preparation, review or approval of the manuscript nor the decision to submit the manuscript for publication.
Author Contributions: Conceptualization: R.S.K., J.L.-S., and S.T.W. Methodology: R.S.K., K.M.M., B.D.H., and S.H.C. Validation: M.H. Formal analysis: R.S.K. and K.M.M. Investigation: R.S.K. and J.L.-S. Resources: R.S.K, J.L.-S., S.T.W., C.B.C., R.G., M.H.C., and J.C.C. Data curation: R.S.K, K.M.M., and M.H. Writing, original draft preparation: R.S.K. and J.L.-S. Writing, review and editing: K.M.M., M.H., B.D.H., C.B.C., R.G., M.H.C., C.E.W., M.J.M., S.H.C., J.C.C., and S.T.W. Supervision: J.L.-S., S.T.W., M.H.C., and C.E.W. Funding acquisition: R.S.K, J.L.-S., S.T.W., and J.C.C. R.S.K. has full access to all the data in the study and takes responsibility for the integrity of the data analysis.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202105-1268OC on November 12, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Nunes C, Pereira AM, Morais-Almeida M. Asthma costs and social impact. Asthma Res Pract . 2017;3:1. doi: 10.1186/s40733-016-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med . 2006;355:2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 3. Boulet LP, FitzGerald JM, Reddel HK. The revised 2014 GINA strategy report: opportunities for change. Curr Opin Pulm Med . 2015;21:1–7. doi: 10.1097/MCP.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 4. Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. National Heart, Lung, and Blood Institute’s Severe Asthma Research Program Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med . 2010;181:315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fiehn O. Metabolomics: the link between genotypes and phenotypes. Plant Mol Biol . 2002;48:155–171. [PubMed] [Google Scholar]
- 6. Reinke SN, Gallart-Ayala H, Gómez C, Checa A, Fauland A, Naz S, et al. Metabolomics analysis identifies different metabotypes of asthma severity. Eur Respir J . 2017;49:160740. doi: 10.1183/13993003.01740-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lefaudeux D, De Meulder B, Loza MJ, Peffer N, Rowe A, Baribaud F, et al. U-BIOPRED Study Group U-BIOPRED clinical adult asthma clusters linked to a subset of sputum omics. J Allergy Clin Immunol . 2017;139:1797–1807. doi: 10.1016/j.jaci.2016.08.048. [DOI] [PubMed] [Google Scholar]
- 8. Sinha A, Desiraju K, Aggarwal K, Kutum R, Roy S, Lodha R, et al. Exhaled breath condensate metabolome clusters for endotype discovery in asthma. J Transl Med . 2017;15:262. doi: 10.1186/s12967-017-1365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Comhair SA, McDunn J, Bennett C, Fettig J, Erzurum SC, Kalhan SC. Metabolomic endotype of asthma. J Immunol . 2015;195:643–650. doi: 10.4049/jimmunol.1500736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Howrylak JA, Moll M, Weiss ST, Raby BA, Wu W, Xing EP. Gene expression profiling of asthma phenotypes demonstrates molecular signatures of atopy and asthma control. J Allergy Clin Immunol . 2016;137:1390–1397.e6. doi: 10.1016/j.jaci.2015.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kelly RS, Mendez KM, Huang M, Hobbs BD, Clish CB, Gerszten R, et al. Metabo-endotypes of asthma reveal clinically important differences in lung function [abstract] Am J Respir Crit Care Med . 2021;203:A4327. doi: 10.1164/rccm.202105-1268OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kelly RS, Virkud Y, Giorgio R, Celedón JC, Weiss ST, Lasky-Su J. Metabolomic profiling of lung function in Costa-Rican children with asthma. Biochim Biophys Acta Mol Basis Dis . 2017;1863:1590–1595. doi: 10.1016/j.bbadis.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Szefler S, Weiss S, Tonascia J, Adkinson NF, Bender B, Cherniack R, et al. Childhood Asthma Management Program Research Group Long-term effects of budesonide or nedocromil in children with asthma. N Engl J Med . 2000;343:1054–1063. doi: 10.1056/NEJM200010123431501. [DOI] [PubMed] [Google Scholar]
- 14.Taliun DHD, Kessler MD, Carlson J, Szpiech ZA, Torres R, Gagliano Taliun SA, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program Nature 2021;590:290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. IBDMDB Investigators Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature . 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mazzilli KM, McClain KM, Lipworth L, Playdon MC, Sampson JN, Clish CB, et al. Identification of 102 correlations between serum metabolites and habitual diet in a metabolomics study of the prostate, lung, colorectal, and ovarian cancer trial. J Nutr . 2020;150:694–703. doi: 10.1093/jn/nxz300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang B, Mezlini AM, Demir F, Fiume M, Tu Z, Brudno M, et al. Similarity network fusion for aggregating data types on a genomic scale. Nat Methods . 2014;11:333–337. doi: 10.1038/nmeth.2810. [DOI] [PubMed] [Google Scholar]
- 18. Li CX, Wheelock CE, Sköld CM, Wheelock AM. Integration of multi-omics datasets enables molecular classification of COPD. Eur Respir J . 2018;51:1701930. doi: 10.1183/13993003.01930-2017. [DOI] [PubMed] [Google Scholar]
- 19. Fitzpatrick AM, Teague WG, Meyers DA, Peters SP, Li X, Li H, National Institutes of Health/National Heart, Lung, and Blood Institute Severe Asthma Research Program Heterogeneity of severe asthma in childhood: confirmation by cluster analysis of children in the National Institutes of Health/National Heart, Lung, and Blood Institute Severe Asthma Research Program. J Allergy Clin Immunol . 2011;127:382–389.e1-13. doi: 10.1016/j.jaci.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chang TS, Lemanske RF, Jr, Mauger DT, Fitzpatrick AM, Sorkness CA, Szefler SJ, et al. Childhood Asthma Research and Education (CARE) Network Investigators Childhood asthma clusters and response to therapy in clinical trials. J Allergy Clin Immunol . 2014;133:363–369. doi: 10.1016/j.jaci.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Loza MJ, Djukanovic R, Chung KF, Horowitz D, Ma K, Branigan P, et al. ADEPT (Airways Disease Endotyping for Personalized Therapeutics) and U-BIOPRED (Unbiased Biomarkers for the Prediction of Respiratory Disease Outcome Consortium) investigators Validated and longitudinally stable asthma phenotypes based on cluster analysis of the ADEPT study. Respir Res . 2016;17:165. doi: 10.1186/s12931-016-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearl J. Probabilistic reasoning in intelligent systems: networks of plausible inference. San Francisco, CA: Elsevier Inc; 1988. [Google Scholar]
- 23.Ng AY, Jordan MI, Weiss Y. In: Advances in neural information processing systems. Leen T, Dietterrich T, Tresp V, editors. Cambridge, MA: MIT Press; 2002. On spectral clustering: analysis and an algorithm; pp. 849–856. [Google Scholar]
- 24. von Luxburg U. A tutorial on spectral clustering. Stat Comput . 2007;17:395–416. [Google Scholar]
- 25.Zelnik-Manor L, Perona P. In: Advances in neural information processing systems 17. Saul LK, Weiss Y, Bottou L, editors. Cambridge, MA: MIT Press; 2005. Self-tuning spectral clustering; pp. 1601–1608. [Google Scholar]
- 26.Zhu X, Ghahramani Z.2002. http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.13.8280
- 27. Barupal DK, Fiehn O. Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci Rep . 2017;7:14567. doi: 10.1038/s41598-017-15231-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bakakos A, Loukides S, Bakakos P. Severe eosinophilic asthma. J Clin Med . 2019;8:1375. doi: 10.3390/jcm8091375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balgoma D, Yang M, Sjödin M, Snowden S, Karimi R, Levänen B, et al. Linoleic acid-derived lipid mediators increase in a female-dominated subphenotype of COPD. Eur Respir J . 2016;47:1645–1656. doi: 10.1183/13993003.01080-2015. [DOI] [PubMed] [Google Scholar]
- 30. Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature . 2014;510:92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Svenningsen S, Nair P. Asthma endotypes and an overview of targeted therapy for asthma. Front Med (Lausanne) . 2017;4:158. doi: 10.3389/fmed.2017.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tyler SR, Bunyavanich S. Leveraging -omics for asthma endotyping. J Allergy Clin Immunol . 2019;144:13–23. doi: 10.1016/j.jaci.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Canonica GW, Ferrando M, Baiardini I, Puggioni F, Racca F, Passalacqua G, et al. Asthma: personalized and precision medicine. Curr Opin Allergy Clin Immunol . 2018;18:51–58. doi: 10.1097/ACI.0000000000000416. [DOI] [PubMed] [Google Scholar]
- 34. Wangberg H, Woessner K. Choice of biologics in asthma endotypes. Curr Opin Allergy Clin Immunol . 2021;21:79–85. doi: 10.1097/ACI.0000000000000708. [DOI] [PubMed] [Google Scholar]
- 35. Nadif R, Febrissy M, Andrianjafimasy MV, Le Moual N, Gormand F, Just J, et al. Endotypes identified by cluster analysis in asthmatics and non-asthmatics and their clinical characteristics at follow-up: the case-control EGEA study. BMJ Open Respir Res . 2020;7:e000632. doi: 10.1136/bmjresp-2020-000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Conrad LA, Cabana MD, Rastogi D. Defining pediatric asthma: phenotypes to endotypes and beyond. Pediatr Res . 2021;90:45–51. doi: 10.1038/s41390-020-01231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Howrylak JA, Fuhlbrigge AL, Strunk RC, Zeiger RS, Weiss ST, Raby BA, Childhood Asthma Management Program Research Group Classification of childhood asthma phenotypes and long-term clinical responses to inhaled anti-inflammatory medications. J Allergy Clin Immunol . 2014;133:1289–1300. doi: 10.1016/j.jaci.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peters MC, Mekonnen ZK, Yuan S, Bhakta NR, Woodruff PG, Fahy JV. Measures of gene expression in sputum cells can identify TH2-high and TH2-low subtypes of asthma. J Allergy Clin Immunol . 2014;133:388–394. doi: 10.1016/j.jaci.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bigler J, Boedigheimer M, Schofield JPR, Skipp PJ, Corfield J, Rowe A, et al. U-BIOPRED Study Group ‖ A severe asthma disease signature from gene expression profiling of peripheral blood from U-BIOPRED cohorts. Am J Respir Crit Care Med . 2017;195:1311–1320. doi: 10.1164/rccm.201604-0866OC. [DOI] [PubMed] [Google Scholar]
- 40. Ibrahim B, Basanta M, Cadden P, Singh D, Douce D, Woodcock A, et al. Non-invasive phenotyping using exhaled volatile organic compounds in asthma. Thorax . 2011;66:804–809. doi: 10.1136/thx.2010.156695. [DOI] [PubMed] [Google Scholar]
- 41. van der Schee MP, Palmay R, Cowan JO, Taylor DR. Predicting steroid responsiveness in patients with asthma using exhaled breath profiling. Clin Exp Allergy . 2013;43:1217–1225. doi: 10.1111/cea.12147. [DOI] [PubMed] [Google Scholar]
- 42. Rago D, Rasmussen MA, Lee-Sarwar KA, Weiss ST, Lasky-Su J, Stokholm J, et al. Fish-oil supplementation in pregnancy, child metabolomics and asthma risk. EBioMedicine . 2019;46:399–410. doi: 10.1016/j.ebiom.2019.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gowdy KM, Fessler MB. Emerging roles for cholesterol and lipoproteins in lung disease. Pulm Pharmacol Ther . 2013;26:430–437. doi: 10.1016/j.pupt.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wendell SG, Baffi C, Holguin F. Fatty acids, inflammation, and asthma. J Allergy Clin Immunol . 2014;133:1255–1264. doi: 10.1016/j.jaci.2013.12.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Palombo JD, Lydon EE, Chen PL, Bistrian BR, Forse RA. Fatty acid composition of lung, macrophage and surfactant phospholipids after short-term enteral feeding with n-3 lipids. Lipids . 1994;29:643–649. doi: 10.1007/BF02536099. [DOI] [PubMed] [Google Scholar]
- 46. Choi Y, Jang J, Park HS. Pulmonary surfactants: a new therapeutic target in asthma. Curr Allergy Asthma Rep . 2020;20:70. doi: 10.1007/s11882-020-00968-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parra E, Pérez-Gil J. Composition, structure and mechanical properties define performance of pulmonary surfactant membranes and films. Chem Phys Lipids . 2015;185:153–175. doi: 10.1016/j.chemphyslip.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 48. Su X, Ren Y, Li M, Zhao X, Kong L, Kang J. Association between lipid profile and the prevalence of asthma: a meta-analysis and systemic review. Curr Med Res Opin . 2018;34:423–433. doi: 10.1080/03007995.2017.1384371. [DOI] [PubMed] [Google Scholar]
- 49. Han S, Mallampalli RK. The role of surfactant in lung disease and host defense against pulmonary infections. Ann Am Thorac Soc . 2015;12:765–774. doi: 10.1513/AnnalsATS.201411-507FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brinkman P, Wagener AH, Hekking PP, Bansal AT, Maitland-van der Zee AH, Wang Y, U-BIOPRED Study Group Identification and prospective stability of electronic nose (eNose)-derived inflammatory phenotypes in patients with severe asthma. J Allergy Clin Immunol . 2019;143:1811–1820.e7. doi: 10.1016/j.jaci.2018.10.058. [DOI] [PubMed] [Google Scholar]
- 51. Wenzel SE, Schwartz LB, Langmack EL, Halliday JL, Trudeau JB, Gibbs RL, et al. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am J Respir Crit Care Med . 1999;160:1001–1008. doi: 10.1164/ajrccm.160.3.9812110. [DOI] [PubMed] [Google Scholar]
- 52. Dharmage SC, Perret JL, Custovic A. Epidemiology of asthma in children and adults. Front Pediatr . 2019;7:246. doi: 10.3389/fped.2019.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tiotiu A. Biomarkers in asthma: state of the art. Asthma Res Pract . 2018;4:10. doi: 10.1186/s40733-018-0047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Litonjua AA, Celedón JC, Hausmann J, Nikolov M, Sredl D, Ryan L, et al. Variation in total and specific IgE: effects of ethnicity and socioeconomic status. J Allergy Clin Immunol . 2005;115:751–757. doi: 10.1016/j.jaci.2004.12.1138. [DOI] [PubMed] [Google Scholar]
- 55. Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J . 2014;43:343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]


