Abstract
Preprotein translocation across the outer and inner envelope membranes of chloroplasts is an energy-dependent process requiring ATP hydrolysis. Several precursor proteins analyzed so far have been found to be imported into isolated chloroplasts equally well in the dark in the presence of ATP as in the light where ATP is supplied by photophosphorylation in the chloroplasts themselves. We demonstrate here that precursors of two maize (Zea mays L. cv Golden Cross Bantam) ferredoxin isoproteins, pFdI and pFdIII, show distinct characteristics of import into maize chloroplasts. pFdI, a photosynthetic ferredoxin precursor, was efficiently imported into the stroma of isolated maize chloroplasts both in the light and in the dark. In contrast pFdIII, a non-photosynthetic ferredoxin precursor, was mostly mis-sorted to the intermembrane space of chloroplastic envelopes as an unprocessed precursor form in the light but was efficiently imported into the stroma and processed to its mature form in the dark. The mis-sorted pFdIII, which accumulated in the intermembrane space in the light, could not undergo subsequent import into the stroma in the dark, even in the presence of ATP. However, when the mis-sorted pFdIII was recovered and used for a separate import reaction, pFdIII was capable of import into the chloroplasts in the dark. pFNRII, a ferredoxin-NADP+ reductase isoprotein precursor, showed import characteristics similar to those of pFdIII. Moreover, pFdIII exhibited similar import characteristics with chloroplasts isolated from wheat (Pennisetum americanum) and pea (Pisum sativum cv Alaska). These findings suggest that the translocation of precursor proteins across the envelope membranes of chloroplasts may involve substrate-dependent light-regulated mechanisms.
Most chloroplast proteins are nuclear-encoded and synthesized as precursor proteins in the cytosol before import into the organelle. The precursor proteins generally contain an amino-terminal extension called the transit peptide, which is required for translocation across the envelope membranes of the chloroplast (Schnell, 1995; May and Soll, 1999). Proteolytic processing of the transit peptide occurs in the stroma by a stromal metalloprotease. Translocation of precursor proteins across the outer and inner envelope membranes have been shown to be mediated by two import apparatus called the Toc (translocon at the outer envelope) and Tic (translocon at the inner envelope) complexes (Schnell et al., 1990; Schnell et al., 1991; Schnell et al., 1994; Nielsen et al., 1997; Caliebe and Soll, 1999; Chen and Schnell, 1999; Keegstra and Froehlich, 1999; Keegstra and Cline, 1999). The Toc complex consists of at least four proteins, Toc34, Toc75, Toc160, and newly identified Toc64 (Kessler et al., 1994; Ma et al., 1996; Bolter et al., 1998; Bauer et al., 2000; Chen et al., 2000; Hirohashi and Nakai, 2000; Sohrt and Soll, 2000). Toc34 and Toc160 are GTP-binding proteins that interact directly with the precursor proteins during early recognition stage at the outer membrane (Seedorf et al., 1995; Kouranov and Schnell, 1997; Jarvis et al., 1998; Sveshnikova et al., 2000). Toc64 contains three tetratricopeptide repeats and shows relatively low but significant degree of sequence similarity to amidase and phosphatase (Sohrt and Soll, 2000). Toc64 also functions early in preprotein recognition at the surface of the chloroplast as a docking protein for a guidance complex consisting of a precursor protein and a cytosolic chaperone such as Hsp70 and/or 14-3-3 proteins (May and Soll, 2000). Toc75 may function as a protein conductance channel at the outer envelope membrane (Tranel and Keegstra, 1996). Several candidates for the components of the Tic complex have been identified. Tic20 and Tic22 may form a core protein translocation apparatus (Kouranov and Schnell, 1997; Kouranov et al., 1998), whereas Tic110 may function as a stromal docking site for the molecular chaperones such as ClpC and Cpn60 (Lubeck et al., 1996; Nielsen et al., 1997; Jackson et al., 1998). However, precise roles of these putative Tic components on precursor protein translocation across the inner envelope membrane remain unclear.
Successive translocation of precursor proteins across the outer and the inner envelope membranes is an energy-dependent process requiring ATP and/or GTP hydrolysis (Pain and Blobel, 1987; Theg et al., 1989; Kouranov and Schnell, 1997; Young et al., 1999). GTP-binding and subsequent GTP-hydrolysis by Toc34 (and/or Toc160) has been proposed to regulate the initial stages of the translocation across the outer envelope (Kouranov and Schnell, 1997; Young et al., 1999). Low concentrations of ATP (<100 μm) are required at the surface or within the intermembrane space, possibly so the molecular chaperones can bind and stabilize the precursor proteins across the outer envelope (Olsen and Keegstra, 1992). Higher concentrations of ATP (>1 mm) are required to complete translocation of the precursor proteins across the outer and inner envelope membranes. This ATP requirement has been thought to be due to the presence of stromal ATPase, which provides the driving force for the directional protein translocation across the envelope membranes (Pain and Blobel, 1987). The most likely candidates for such stromal ATPase are molecular chaperones like ClpC, Cpn60, or Hsp70 (Kessler and Blobel, 1996; Nielsen et al., 1997). Proteolytic processing of the translocated precursor proteins by the stromal metalloprotease does not require ATP hydrolysis.
In in vitro import experiments, the complete translocation of precursor proteins into the stroma of isolated chloroplasts can be observed both in the dark and in the light (Pain and Blobel, 1987; Schindler et al., 1987; Cline et al., 1989; Theg et al., 1989). ATP is an absolute requirement with exogenous addition of ATP necessary under dark conditions while in the light, ATP is synthesized by the organelle via photosynthetic reactions. Various precursor proteins have been used in such experiments, and results have suggested that there are essentially no mechanistic differences between import reactions in the light or in the dark once sufficient ATP is present (Theg et al., 1989; Pilon et al., 1992; Pilon et al., 1995). To clarify this, we have compared protein import under both light and dark conditions of a variety of precursor proteins, which exhibit different expression patterns in the plant. Our results suggest that particular precursor proteins demonstrate distinct import characteristics in the light or in the dark and that these observations are common to chloroplasts isolated from various sources.
RESULTS
Ferredoxin Isoprotein Precursors
Plant-type ferredoxin, an electron-transfer protein, is a small (11 kD), soluble, acidic protein distributed in various plant plastids. In maize (Zea mays L. cv Golden Cross Bantam), at least six ferredoxin isoproteins (FdI–FdVI) have been found and among them; FdI and FdIII are the major ferredoxin isoproteins in leaves and in roots, respectively (Kimata and Hase, 1989; Hase et al., 1991; Suzuki et al., 1991). In leaves, expression of FdI was found to be largely induced by light, whereas small amounts of FdIII are constitutively expressed. Thus FdI has been regarded as a photosynthetic ferredoxin and FdIII as a non-photosynthetic ferredoxin. Both are nuclear-encoded and synthesized in the cytosol as a larger precursor protein with an amino-terminal transit peptide. Sequence identity between the transit peptides of the two precursors is quite low (25%), whereas the mature domains show relatively high sequence identity (67%).
In Vitro Protein Import Assay in the Light Versus in the Dark
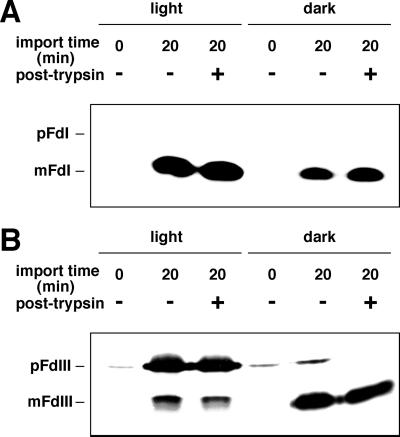
In vitro import reactions were carried out under illumination or in the dark using in vitro synthesized ferredoxin isoprotein precursors (pFdI and pFdIII) and freshly isolated maize chloroplasts. Under dark conditions, 5 mm Mg-ATP was added as an energy source. As shown in Figure 1A, pFdI was efficiently imported into chloroplasts both in the light and in the dark and processed to the mature protein (mFdI). The import efficiency in the dark did seem to be slightly lower than that in the light. Non-photosynthetic pFdIII was also efficiently imported in the dark as shown in Figure 1B. After the import reaction in the dark, mFdIII was completely resistant to trypsin treatment, whereas the pFdIII, which was recovered with the chloroplasts, was completely digested. This indicated that this pFdIII was bound to the surface of the chloroplasts. However, in the light, conversion to trypsin-resistant mature FdIII (mFdIII) was inefficient and accumulation of trypsin-resistant pFdIII was observed.
Figure 1.
In vitro protein import assay of pFdI and pFdIII in the light versus in the dark. Intact chloroplasts were incubated with in vitro-synthesized pFdI (A) or pFdIII (B) in a standard import reaction either in the light or in the dark as described in “Materials and Methods.” Reactions, which were incubated in the dark, had 5 mm Mg-ATP added to them prior to incubation. The chloroplasts were reisolated and protease accessibility after import reaction was assayed by the incubation of reisolated chloroplasts with 60 μg/mL trypsin on ice for 20 min. Proteins were separated by centrifugation and analyzed by SDS-PAGE followed by fluorography.
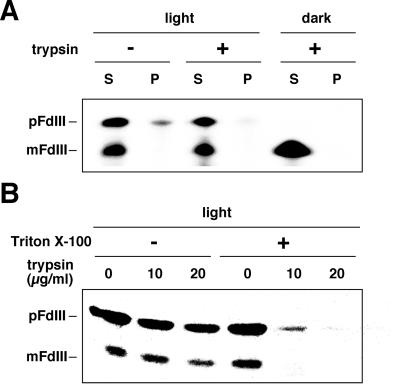
As shown in Figure 2A, the trypsin-resistant pFdIII, which accumulated in the light was recovered in the soluble fraction after osmotic rupture of chloroplasts. When the membrane integrity of the chloroplasts was destroyed by the addition of 0.2% (w/v) Triton X-100, the accumulated pFdIII could now be digested by low concentrations of trypsin (Fig. 2B). These results indicate that pFdIII accumulated in the interior soluble compartment of chloroplasts (intermembrane space or stroma) during import under light conditions.
Figure 2.
pFdIII accumulated in the light was present in a soluble compartment of chloroplasts. A, pFdIII was incubated with chloroplasts as described in Figure 1. After the incubation, chloroplasts were recovered by centrifugation and treated with or without trypsin. The chloroplasts were reisolated and resuspended in a hypotonic buffer. Soluble (S) and membrane (P) fractions were separated by centrifugation and analyzed by SDS-PAGE followed by fluorography. Note that all trypsin-resistant pFdIII proteins were recovered in the soluble fraction of chloroplasts. B, After import of pFdIII either in the light or in the dark, trypsin treatment (0, 10, or 20 μg/mL of trypsin, on ice for 5 min) was performed as indicated either in the presence or absence of 0.2% (w/v) Triton X-100.
To exclude the possibility that this result was due to an insufficient ATP supply by the photosynthetic reactions, we repeated the import experiment under light conditions in the presence of 5 mm Mg-ATP and obtained essentially similar results (data not shown). In the following experiments, unless otherwise stated, 5 mm Mg-ATP was included in the import assays carried out in the light.
Localization of pFdIII to the Intermembrane Space of Chloroplast Envelopes in the Light
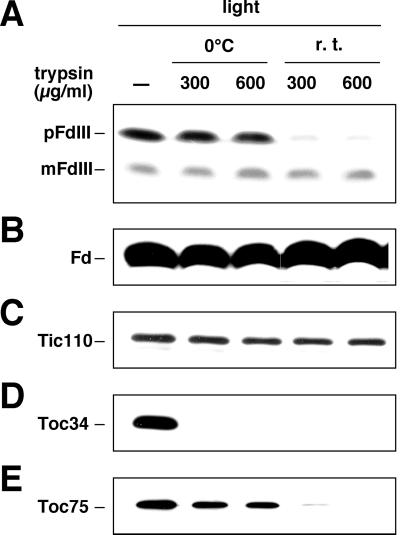
Certain proteases, most notably trypsin, are able to destroy the membrane integrity of the outer envelope of chloroplasts and thereby degrade proteins within the intermembrane space while leaving the stromal proteins undigested (Jackson et al., 1998; Kouranov et al., 1998). This selective proteolysis technique was used to analyze the precise location of the pFdIII accumulated in the light. When chloroplasts were incubated on ice for 10 min with 5- or 10-fold higher concentrations of trypsin as used in the experiments described in Figure 1, the accumulated pFdIII was not degraded (Fig. 3A). However, when the trypsin digestion was carried out at room temperature for 10 min, conditions under which trypsin has been shown to destroy the membrane integrity of the outer envelope but not that of the inner envelope, the accumulated pFdIII was largely degraded (Fig. 3A). The small amount of mature FdIII, which was thought to be localized to the stroma, remained undigested (Fig. 3A).
Figure 3.
pFdIII accumulated in the intermembrane space of chloroplasts in the light. After import of pFdIII in the light, trypsin treatment was carried out either on ice or at room temperature (r.t.) for 10 min as indicated. Proteins were separated by SDS-PAGE and then analyzed by fluorography (A) or by immunoblotting using antisera raised against ferredoxin (B), Tic110 (C), Toc34 (D), or Toc75 (E).
Proteins of known localizations were analyzed using the same system. A stromal ferredoxin and Tic110, an inner envelope membrane protein that is oriented toward the stroma, were resistant to trypsin digestion at room temperature (Fig. 3, B and C). Toc34, an outer envelope membrane protein with a large domain exposed to the cytosol, was easily degraded by trypsin on ice (Fig. 3D). Toc75, an integral outer envelope membrane protein, is known to be degraded when trypsin is able to penetrate the outer envelope membrane (Jackson et al., 1998; Kouranov et al., 1998). Toc75 was digested by trypsin at room temperature but not on ice (Fig. 3E). Taken together, pFdIII accumulated under light conditions in the intermembrane space between the outer and inner envelope membranes of chloroplasts as a soluble protein.
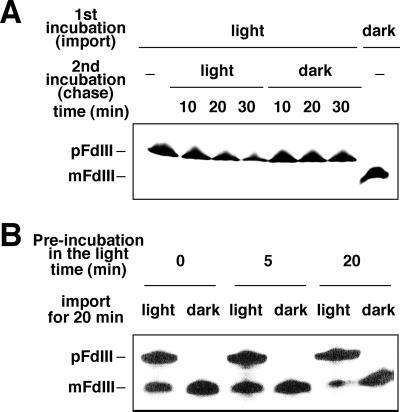
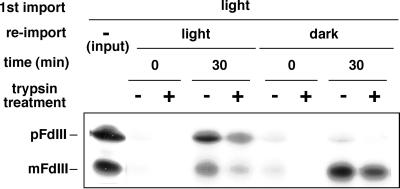
After Incubation in the Light, Accumulated pFdIII Did Not Undergo Subsequent Import into the Stroma in the Dark
Next, we analyzed whether pFdIII induced to accumulate in the intermembrane space by light would be able to be transported to the stroma by subsequent incubation of the chloroplasts in the dark. As shown in Figure 4A, the accumulated pFdIII was not processed to its mature form during a subsequent 30-min incubation in the dark, even in the presence of excess ATP, indicating that the pFdIII accumulated in the intermembrane space in the light was no longer translocated across the inner envelope membrane of chloroplast. It is noteworthy that, in the light, the accumulated pFdIII was not processed to its mature form but rather degraded significantly. This suggests that the pFdIII accumulated in the intermembrane space is unstable under light conditions.
Figure 4.
Accumulated pFdIII did not undergo subsequent import into the stroma in the dark. A, Chase experiment of accumulated pFdIII. After import of pFdIII in the light (first incubation), chloroplasts were recovered and further incubated in the dark (second incubation) and analyzed as described in Figure 1. B, Chloroplasts were preincubated in the import buffer in the light for the indicated time. Then in vitro-synthesized pFdIII was added to each reaction mixture. The mixture was further incubated for 20 min in the light or in the dark and analyzed as above.
It was possible that exposure of the chloroplasts to light might result in damage that affected the chloroplast's ability to import pFdIII. To exclude this possibility, chloroplasts were preincubated in the presence of light and then tested for the ability to import pFdIII in the dark. As shown in Figure 4B, the preilluminated chloroplasts were not impaired in their ability to import pFdIII under dark conditions.
pFdIII Recovered after Light-Induced Accumulation Is a Monomer and Can Be Re-Imported into Chloroplasts under Dark Conditions
To analyze the pFdIII induced by light to accumulate in the intermembrane space, we performed three independent assays on protein recovered from this system.
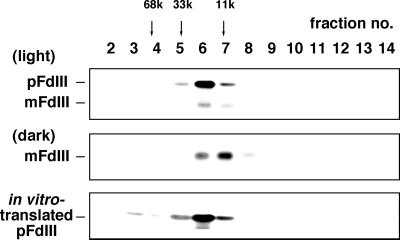
First, the protein was analyzed by gel filtration (Fig. 5). The elution profile of the recovered protein was equivalent to that of in vitro translated pFdIII, and both of these eluted slightly before the mature FdIII, which is produced in chloroplasts incubated in the dark. The elution profiles of the recovered protein and the in vitro translated protein were in good agreement with that expected from the monomeric size of the protein, and we have concluded that the pFdIII accumulated in the intermembrane space existed as the free monomeric form.
Figure 5.
Gel filtration chromatography of pFdIII accumulated in the light. After import of pFdIII in the light or in the dark, chloroplasts were recovered and their soluble fractions purified as described in Figure 2. Gel filtration analysis was performed with a Superose 6 column (Pharmacia Biotech, Piscataway, NJ) that had been equilibrated with 20 mm HEPES-KOH (pH 8.0) and 150 mm KCl using a SMART system (Pharmacia Biotech). Fractions obtained by gel filtration were analyzed as described in Figure 2. As a control, in vitro-translated pFdIII was also chromatographed and analyzed.
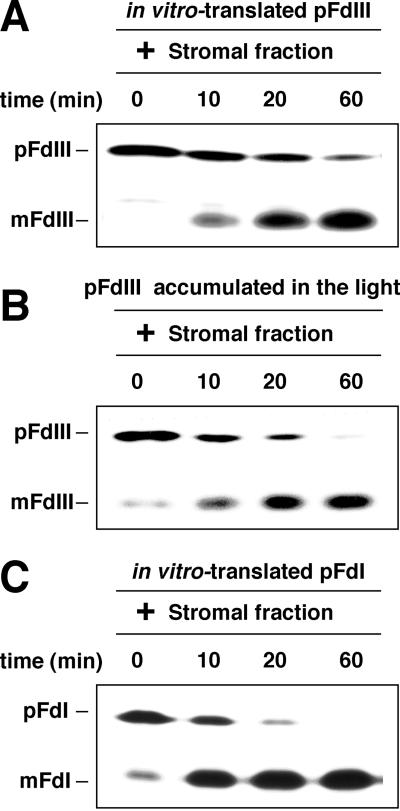
Next, we assessed the sensitivity of the protein to the metalloproteinase present in the stromal fraction of maize chloroplasts. This enzyme can convert pFdIII to the mature form of the protein. As shown in Figure 6, the accumulated pFdIII was efficiently processed to its mature form upon incubation with the 10-fold excess amount of concentrated stromal fraction as was in vitro-translated pFdIII, although the rate of processing of the pFdIII was somewhat slower than that of the pFdI.
Figure 6.
pFdIII accumulated in the light was able to be processed by stromal processing protease. The soluble fraction of the chloroplast, which contained the pFdIII accumulated under light conditions, was incubated with a concentrated stromal fraction (10-fold excess amounts as compared with that had been contained in the chloroplasts used in the initial import reaction) that had been prepared from maize chloroplasts as indicated B. As control experiments, in vitro-synthesized pFdIII (A) and pFdI (C) were also separately incubated with the concentrated stromal fraction.
Finally, we repeated the import experiments with the recovered pFdIII and fresh chloroplasts. The recovered pFdIII could be efficiently imported into chloroplasts and processed to its mature form in the dark (Fig. 7). Under light conditions, once again protease-resistant pFdIII accumulated in the intermembrane space. In all these analyses, pFdIII, which accumulated in the intermembrane space, was indistinguishable from in vitro-translated pFdIII.
Figure 7.
pFdIII recovered after light-induced accumulation in the intermembrane space was able to be reimported into chloroplastic stroma in the dark. The soluble fraction of the chloroplasts, which contained this pFdIII, was incubated with fresh chloroplasts either in the light or in the dark and then analyzed as described in Figure 1.
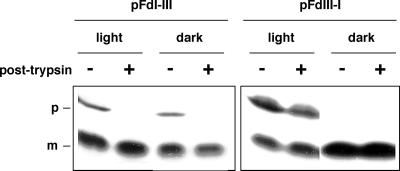
The Presequence But Not the Mature Moiety of pFdIII Was Responsible for Accumulation of Unprocessed Form of Precursor in the Intermembrane Space in the Light
As mentioned above, pFdIII and pFdI show relatively high sequence identity to each other in the mature domains, but little sequence similarity between their presequences. To clarify whether the presequence or the mature moiety of pFdIII was responsible for its interesting import characteristics, we constructed two chimeric genes. Each encoded a fusion protein consisting of the presequence of one Fd and the mature sequence of the other. As shown in Figure 8, light-induced accumulation of precursor proteins in the intermembrane space clearly depended on the presequence of pFdIII.
Figure 8.
The presequence of pFdIII was responsible for accumulation of the unprocessed form of precursor in the intermembrane space in the light. Two chimeric proteins were translated in vitro: One consists of the presequence of pFdI and mature domain of FdIII (pFdI-III) and the other consists of the presequence of pFdIII and mature domain of FdI (pFdIII-I). These chimeric proteins and authentic pFdI and pFdIII were used independently in in vitro import assays with chloroplasts in the light or in the dark. After import, chloroplasts were recovered and analyzed as described in Figure 1.
Light-Induced Mis-Sorting of Precursor Proteins to the Intermembrane Space Also Occurred with Other Precursor Proteins and in Chloroplasts Prepared from Pea or Wheat Plants
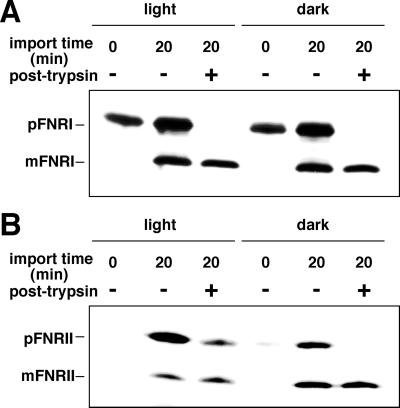
It was necessary to confirm that this phenomenon was not specific to pFdIII and maize chloroplasts. In vitro import experiments with maize pFNRI and pFNRII (precursors of ferredoxin-NADP+ reductase isoproteins) and maize chloroplasts demonstrated that a protease-resistant unprocessed form of pFNRII accumulated under light conditions, whereas pFNRI was imported efficiently both in light and dark conditions (Fig. 9).
Figure 9.
Accumulation of precursor proteins in the intermembrane space in the light was observed in the case of pFNRII. In vitro import assays of pFNRI (A) and pFNRII (B) with maize chloroplasts were carried out as described in Figure 1.
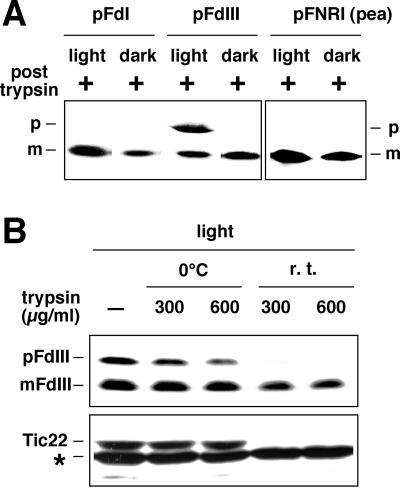
Chloroplasts isolated from other plants also demonstrated the mis-sorting of pFdIII in in vitro import assays. pFdIII accumulated in its unprocessed form under light conditions in both pea (Pisum sativum cv Alaska) (Fig. 10A) and wheat (Pennisetum americanum) chloroplasts (data not shown). Such accumulation of protease-resistant precursor under the light import conditions was not observed in the case of pFdI and also in the case of pea pFNRI, which has been widely used for in vitro import assay. To demonstrate again the localization of the accumulated pFdIII in the intermembrane space of pea chloroplasts, we compared the trypsin accessibility of the accumulated pFdIII with that of pea Tic22 protein, which has already been proven to reside in the intermembrane space (Kouranov et al., 1998). As shown in Figure 10B, the endogenenous Tic22 and the accumulated pFdIII were equally degraded by the high concentration of trypsin in the condition that trypsin was capable of permeating the outer but not the inner envelope membrane. These results suggest that mis-sorting of certain precursor proteins to the intermembrane space in the light is not restricted to the pFdIII and maize chloroplasts and might be observed in a range of precursor proteins and chloroplasts derived from a variety of plant sources.
Figure 10.
Light-induced mis-sorting of precursor proteins to the intermembrane space also occurred in chloroplasts prepared from pea. A, Maize pFdI and pFdIII and pea pFNRI were used independently in in vitro import assays with chloroplasts in the light or in the dark for 20 min. After import, chloroplasts were recovered, and protease-resistant proteins were analyzed as described in Figure 1. B, After import of pFdIII in the light into the pea chloroplasts, trypsin treatment was carried out either on ice or at room temperature (r.t.) for 10 min as indicated. Proteins were separated by SDS-PAGE and then analyzed by fluorography for FdIII (upper) or by immunoblotting using antisera raised against pea Tic22 (lower). Note that an asterisk corresponds to a position of an unknown thylakoid protein, which was cross-reacted with the anti-pea Tic22 serum.
DISCUSSION
We have demonstrated that maize FdIII, the non-photosynthetic ferredoxin, exhibited distinct import patterns in the chloroplast under light and dark conditions. In the presence of light, the precursor form accumulated in the intermembrane space of the chloroplast envelope membranes, whereas in the dark, the protein was processed correctly. In contrast, maize photosynthetic FdI was imported equally well by isolated chloroplasts in both the light and the dark, as has been reported for leaf-specific ferredoxins from other plants (Theg et al., 1989; Pilon et al., 1992; Pilon et al., 1995; Rensink et al., 2000). Once pFdIII had been mis-sorted in the isolated chloroplast, subsequent incubation in the dark in the presence of ATP did not correct the localization of the protein. This suggested that the mis-sorted product had come to a dead end. However the accumulated pFdIII, which was present as a soluble monomer, could be easily processed to the mature form when exposed to the stromal processing protease. Also, once recovered from the intermembrane space, the pFdIII could still be imported correctly into intact chloroplasts. These results suggest that neither modification nor interaction with other protein factor(s) to form a translocation-incompetent structure occurred during the translocation of pFdIII across the outer membrane in the light. Mis-sorting of precursor proteins to the intermembrane space in the light occurred in chloroplasts prepared from various sources including maize, wheat, and pea. Furthermore, since mis-sorting was not restricted to pFdIII and was also observed in the case of pFNRII, this phenomenon might be common for a subset of precursor proteins in plants. We propose that the translocation of precursor proteins across the envelope membranes of chloroplasts may involve substrate-dependent light-regulated mechanisms.
pFdIII is constitutively expressed in leaves regardless of light exposure, and the redox potential of FdIII (−345 mV) is significantly higher than that of photosynthetic FdI (−423 mV; Kimata and Hase, 1989; Akashi et al., 1999). Accumulation of pFdIII in the intermembrane space has not been observed in vivo in maize leaves grown in light conditions. It is possible that if pFdIII were mis-sorted to the intermembrane space in vivo that it would be rapidly degraded as shown in Figure 4. The exact physiological consequences of the mis-sorting of pFdIII has not been demonstrated in this paper. The most likely scenario is that in the presence of light, excess FdIII in the chloroplast is unfavorable as the higher redox potential of FdIII would affect electron transport in the photosynthetic plastids (Onda et al., 2000; Yonekura-Sakakibara et al., 2000). pFdIII import might be regulated such that immediately upon exposure to light, it is excluded from the stroma of chloroplasts. Even immediately after exposure to light, as shown in Figure 4B, normal import of pFdIII into stroma under dark conditions was observed suggesting that this tightly regulated process may involve light-dependent signal transduction in the chloroplasts. Light-dependent signal transduction in plants regulates gene expression in both the nucleus and chloroplasts (Li et al., 1993; Deng, 1994; Fankhauser and Chory, 1999). However, it is an intriguing hypothesis that chloroplasts also regulate protein import so as to rapidly establish the desired protein composition in the organelle in response to light. Reversible GTP binding and/or phosphorylation of Toc34, a preprotein receptor, may be involved in such light-dependent regulation of protein import (Sveshnikova et al., 2000). We would also emphasize that the presequence of pFdIII carried sufficient information for light-dependent mis-sorting, suggesting that protein sorting may be regulated at the stage where the presequence is being recognized by any factor(s) during its import process.
Many precursor proteins, which have so far been shown to be imported into isolated chloroplasts equally well in the light as in the dark are photosynthetic proteins (Pain and Blobel, 1987; Schindler et al., 1987; Cline et al., 1989; Theg et al., 1989). Therefore, if the import of other non-photosynthetic plastid proteins is tested in the presence of light, it might be expected that other examples of proteins with differences in light- versus dark-induced import processes will be found. Wan et al. (1996) reported that some precursors can be imported equally efficiently into two types of plastids, chloroplasts and leucoplasts, that others are imported preferentially into one type of plastid versus the other, and that the ability of plastids to import different proteins correlated with the in vivo steady state levels of these proteins. Thus, different types of plastids might contain distinct import apparatus whose preferences to precursors are somewhat different but overlapped. It is interesting that several differentially expressed isoforms of Toc components have been identified (Jarvis et al., 1998; Bauer et al., 2000; Hirohashi and Nakai, 2000; S. Kikuchi, T. Hirohashi, and M. Nakai, unpublished data). This suggests that these isoforms are built up into distinct Toc complexes each of which support the import of a distinct subset of precursor proteins (Muckel and Soll, 1996). Reinbothe et al. (1995a, 1995b, 2000) recently reported that import into chloroplasts of pPORA, a precursor of the NADPH:protochlorophyllide oxidoreductase A, uses a second substrate-dependent site. Therefore it is possible that differential binding of pFdIII to distinct Toc complexes might be responsible for the import characteristics described here. Tic22, a peripheral component of the protein translocation apparatus of the chloroplastic inner envelope, was shown to be localized to the intermembrane space by a novel pathway that is distinct from the general import pathway (Kouranov et al., 1999). In the presence of light, pFdIII might enter such an import pathway for the subset of precursor proteins whose final destination is the intermembrane space. Competition experiments with different precursor proteins and inhibition experiments with various antibodies against Toc components will help to identify the exact import pathway of pFdIII used under light conditions (Cline et al., 1993).
Scott and Theg (1996) have studied protein transport across the envelope membranes of chloroplasts and have characterized a new chloroplast import intermediate. They interfered with the chloroplast protein transport through the use of a chemical poison, HgCl2, or by simply halting the in vitro import reaction by placing it on ice. The intermediate identified had completely traversed the outer envelope membrane but had not yet reached the stroma. It was present in the protease-protected precursor form and reached at least the surface of the inner envelope. It could be chased into the stroma, suggesting that the intermediate was a productive member of the import pathway. From these results, they concluded that two distinct protein translocation machineries are present in both envelope membranes and that they are able to operate independently of one another. In the present study, the pFdIII accumulated in the intermembrane space was unable to undergo subsequent import into the stroma in the dark (Fig. 4A) but was able to be reimported into the stroma of the fresh chloroplasts (Fig. 7). This might indicate that distinct import-competent structures are required for pFdIII to be translocated across the outer or inner envelope membrane. Our present data alternatively might suggest that there are no accessible entrance sites on the surface of the inner envelope membrane for precursor proteins that have been inappropriately liberated into the intermembrane space from the outer membrane. In this case, Tic might be more rigidly and mechanically connected to Toc machinery than has been thought (Schnell and Blobel, 1993; Akita et al., 1997). This is in contrast to two protein translocons at the mitochondrial outer and inner membranes, Tom and Tim, the function of which is not dependent on physical connection (Neupert, 1997; Pfanner and Meijer, 1997). Mitoplasts whose outer membrane is disrupted by osmotic shock while the inner membrane remains intact are able to import various matrix-targeted precursor proteins directly via Tim. This suggests that Tim functions independently of Tom. It is interesting that Tim23, a key component of Tim, was recently reported to link the inner and outer mitochondrial membranes (Donzeau et al., 2000). Tethering the inner membrane translocase to the outer membrane has been proposed to facilitate the transfer of precursor proteins from the TOM complex to the TIM complex and increase the efficiency of protein import. Thus Tom and Tim might also be mechanically coupled to each other.
We are currently investigating how pFdIII is mislocalized to the intermembrane space of chloroplasts in the light and also how light affects the protein sorting mechanisms of chloroplasts.
MATERIALS AND METHODS
Plant Materials and Intact Chloroplast Isolation
Maize (Zea mays L. cv Golden Cross Bantam) and wheat (Pennisetum americanum) seedlings were grown on vermuculite at 26°C to 30°C in a light regime (14-h-light/10-h-dark cycles) for 7 to 11 d. Pea (Pisum sativum cv Alaska) seedlings were grown as described previously (Endo et al., 1994). Intact chloroplasts were isolated from leaves as described previously (Nakai et al., 1994). After the final centrifugation at 3,000 rpm for 4 min, chloroplasts were washed once with the import buffer (50 mm HEPES-KOH, pH 7.5, 330 mm sorbitol) and resuspended in the same buffer at a final concentration of 0.8 to 1.0 mg/mL of chlorophyll. The chlorophyll concentration was determined according to the published procedure.
In Vitro Transcription and Translation of Precursor Proteins
Plasmids used for in vitro transcription/translation of ferredoxin isoprotein precursors (pFdI and pFdIII) and ferredoxin-NADP+ reductase isoprotein precursors (pFNRI and pFNRII) were described previously (Suzuki et al., 1991; Onda et al., 2000). The mRNAs for various precursor proteins were synthesized using an in vitro transcription system (Ribomax, Promega, Madison, WI) with SP6 RNA polymerase. The resulting mRNAs were translated in a wheat germ extract (Promega) at 30°C for 90 min in the presence of l-[4,5-3H]Leu, and the translation mixtures were kept at −80°C until use.
In Vitro Import Experiment
In vitro chloroplast import was carried out in the import buffer (50 mm HEPES-KOH, pH 7.5, 330 mm sorbitol) containing 1.25 mm Leu, 0.5 to 0.8 mg chlorophyll chloroplasts, and the translation mixtures. The volume of the standard import mixture was 200 μL. Each import mixture was incubated in a glass test tube with gentle shaking at 30°C under light illumination for the desired time. Import reactions in the dark were carried out using the same import mixture supplemented with 5 mm Mg-ATP. After the incubation, the mixtures were immediately chilled on ice and washed with 1 mL of ice-cold import buffer. Chloroplasts were reisolated by centrifugation at 3,000 rpm for 1 min.
Protease accessibility after import reaction was analyzed by the incubation of reisolated chloroplasts with 60 μg/mL trypsin on ice for 20 min. After the incubation, trypsin was inactivated by the addition of 5-fold molar excess of soybean trypsin inhibitor.
Chloroplasts were lysed in the SDS-polyacrylamide gel electrophoresis sample buffer, followed by heating to 95°C for 5 min. The solubilized proteins were separated on a 12.5% (w/v) SDS-polyacrylamide gel. The gels were then treated with EN3HANCE (New England Nuclear, Boston) and subjected to fluorography.
General Methods
Published methods were used for SDS-polyacrylamide gel electrophoresis, western-blotting, and protein assays. Fluorograms were quantified with an image scanner and the National Institutes of Health image software.
ACKNOWLEDGMENTS
We thank Tomohiro Matsumura and Hitoshi Sakakibara for various maize cDNAs and Yuichi Fujita for advice in the initial stages of this work. We also thank Singo Kikuchi and Hiroo Oguchi for valuable discussions.
Footnotes
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan.
LITERATURE CITED
- Akashi T, Matsumura T, Ideguchi T, Iwakiri K, Kawakatsu T, Taniguchi I, Hase T. Comparison of the electrostatic binding sites on the surface of ferredoxin for two ferredoxin-dependent enzymes, ferredoxin-NADP+ reductase and sulfite reductase. J Biol Chem. 1999;274:29399–29405. doi: 10.1074/jbc.274.41.29399. [DOI] [PubMed] [Google Scholar]
- Akita M, Nielsen E, Keegstra K. Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J Cell Biol. 1997;10:983–994. doi: 10.1083/jcb.136.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;403:203–207. doi: 10.1038/35003214. [DOI] [PubMed] [Google Scholar]
- Bolter B, May T, Soll J. A protein import receptor in pea chloroplasts, Toc86, is only a proteolytic fragment of a larger polypeptide. FEBS Lett. 1998;441:59–62. doi: 10.1016/s0014-5793(98)01525-7. [DOI] [PubMed] [Google Scholar]
- Caliebe A, Soll J. News in chloroplast protein import. Plant Mol Biol. 1999;39:641–645. doi: 10.1023/a:1006170321840. [DOI] [PubMed] [Google Scholar]
- Chen K, Chen X, Schnell DJ. Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts. Plant Physiol. 2000;122:813–822. doi: 10.1104/pp.122.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schnell DJ. Protein import into chloroplasts. Trends Cell Biol. 1999;9:222–227. doi: 10.1016/s0962-8924(99)01554-8. [DOI] [PubMed] [Google Scholar]
- Cline K, Fulsom DR, Viitanen PV. An imported thylakoid protein accumulated in the stroma when insertion into thylakoids is inhibited. J Biol Chem. 1989;264:14225–14232. [PubMed] [Google Scholar]
- Cline K, Henry R, Li C, Yuan J. Multiple pathways for protein transport into or across the thylakoid membrane. EMBO J. 1993;12:4105–4114. doi: 10.1002/j.1460-2075.1993.tb06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW. Fresh view of light signal transduction in plants. Cell. 1994;76:423–426. doi: 10.1016/0092-8674(94)90107-4. [DOI] [PubMed] [Google Scholar]
- Donzeau M, Kaldi K, Adam A, Paschen S, Wanner G, Guiard B, Bauer MF, Neupert W, Brunner M. Tim23 links the inner and outer mitochondrial membranes. Cell. 2000;101:401–412. doi: 10.1016/s0092-8674(00)80850-8. [DOI] [PubMed] [Google Scholar]
- Endo T, Kawakami M, Goto A, America T, Weisbeek P, Nakai M. Chloroplast protein import chloroplast envelopes and thylakoids have different abilities to unfold proteins. Eur J Biochem. 1994;225:403–409. doi: 10.1111/j.1432-1033.1994.00403.x. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light receptor kinases in plants. Curr Biol. 1999;9:R123–R126. doi: 10.1016/s0960-9822(99)80078-5. [DOI] [PubMed] [Google Scholar]
- Hase T, Kimata Y, Yonekura K, Matsumura T, Sakakibara H. Molecular cloning and differential expression of the maize ferredoxin gene family. Plant Physiol. 1991;96:77–83. doi: 10.1104/pp.96.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirohashi T, Nakai M. Molecular cloning and characterization of maize Toc34, a regulatory component of the protein import machinery of chloroplast. Biochim Biophys Acta. 2000;1491:309–314. doi: 10.1016/s0167-4781(00)00043-9. [DOI] [PubMed] [Google Scholar]
- Jackson DT, Froehlich JE, Keegstra K. The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J Biol Chem. 1998;273:16583–16588. doi: 10.1074/jbc.273.26.16583. [DOI] [PubMed] [Google Scholar]
- Jarvis P, Chen LJ, Li H, Peto CA, Fankhauser C, Chory J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- Keegstra K, Cline K. Protein import and routing systems of chloroplasts. Plant Cell. 1999;11:557–570. doi: 10.1105/tpc.11.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Froehlich JE. Protein import into chloroplasts. Curr Opin Plant Biol. 1999;2:471–476. doi: 10.1016/s1369-5266(99)00021-7. [DOI] [PubMed] [Google Scholar]
- Kessler F, Blobel G. Interaction of the protein import and folding machineries of the chloroplast. Proc Natl Acad Sci USA. 1996;93:7684–7689. doi: 10.1073/pnas.93.15.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F, Blobel G, Patel HA, Schnell DJ. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- Kimata Y, Hase T. Localization of ferredoxin isoproteins in mesophyll and bundle sheath cells in maize leaf. Plant Physiol. 1989;89:1193–1197. doi: 10.1104/pp.89.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol. 1998;143:991–1002. doi: 10.1083/jcb.143.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A, Schnell DJ. Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J Cell Biol. 1997;139:1677–1685. doi: 10.1083/jcb.139.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A, Wang H, Schnell DJ. Tic22 is targeted to the intermembrane space of chloroplasts by a novel pathway. J Biol Chem. 1999;274:25181–25186. doi: 10.1074/jbc.274.35.25181. [DOI] [PubMed] [Google Scholar]
- Li HM, Washburm T, Chory J. Regulation of gene expression by light. Curr Opin Cell Biol. 1993;5:455–460. doi: 10.1016/0955-0674(93)90011-e. [DOI] [PubMed] [Google Scholar]
- Lubeck J, Soll J, Akita M, Nielsen E, Keegstra K. Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO J. 1996;15:4230–4238. [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Kouranov A, LaSala SE, Schnell DJ. Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J Cell Biol. 1996;134:315–327. doi: 10.1083/jcb.134.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T, Soll J. Chloroplast precursor protein translocon. FEBS Lett. 1999;452:52–56. doi: 10.1016/s0014-5793(99)00527-x. [DOI] [PubMed] [Google Scholar]
- May T, Soll J. 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell. 2000;12:53–64. doi: 10.1105/tpc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckel E, Soll J. A protein import receptor of chloroplasts is inserted into the outer envelope membrane by a novel pathway. J Biol Chem. 1996;271:23846–23852. doi: 10.1074/jbc.271.39.23846. [DOI] [PubMed] [Google Scholar]
- Nakai M, Goto A, Nohara T, Sugita D, Endo T. Identification of the SecA protein homolog in pea chloroplasts and its possible involvement in thylakoidal protein transport. J Biol Chem. 1994;269:31338–31341. [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen LJ, Keegstra K. The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J Biol Chem. 1992;267:433–439. [PubMed] [Google Scholar]
- Onda Y, Matsumura T, Kimata-Ariga Y, Sakakibara H, Sugiyama T, Hase T. Differential interaction of maize root Ferredoxin:NADP+ oxidoreductase with photosynthetic and non-photosynthetic ferredoxin isoproteins. Plant Physiol. 2000;123:1037–1046. doi: 10.1104/pp.123.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain D, Blobel G. Protein import into chloroplasts requires a chloroplast ATPase. Proc Natl Acad Sci USA. 1987;84:3288–3292. doi: 10.1073/pnas.84.10.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, Meijer M. The Tom and Tim machine. Curr Biol. 1997;7:100–103. doi: 10.1016/s0960-9822(06)00048-0. [DOI] [PubMed] [Google Scholar]
- Pilon M, de Kruijff B, Weisbeek PJ. New insights into the import mechanism of the ferredoxin precursor into chloroplasts. J Biol Chem. 1992;267:2548–2556. [PubMed] [Google Scholar]
- Pilon M, Wienk H, Sips W, de Swaaf M, Talboom I, van't Hof R, de Korte-Kool G, Demel R, Weisbeek P, de Kruijff B. Functional domains of the ferredoxin transit sequence involved in chloroplast import. J Biol Chem. 1995;270:3882–3893. doi: 10.1074/jbc.270.8.3882. [DOI] [PubMed] [Google Scholar]
- Reinbothe S, Mache R, Reinbothe C. A second, substrate-dependent site of protein import into chloroplasts. Proc Natl Acad Sci USA. 2000;97:9795–9800. doi: 10.1073/pnas.160242597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Runge S, Apel K. Enzymatic product formation impairs both the chloroplast receptor-binding function as well as translocation competence of the NADPH: protochlorophyllide oxidoreductase, a nuclear-encoded plastid precursor protein. J Cell Biol. 1995a;129:299–308. doi: 10.1083/jcb.129.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Runge S, Reinbothe C, van Cleve B, Apel K. Substrate-dependent transport of the NADPH:protochlorophyllide oxidoreductase into isolated plastids. Plant Cell. 1995b;7:161–172. doi: 10.1105/tpc.7.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink WA, Schnell DJ, Weisbeek PJ. The transit sequence of ferredoxin contains different domains for translocation across the outer and inner membrane of the chloroplast envelope. J Biol Chem. 2000;275:10265–10271. doi: 10.1074/jbc.275.14.10265. [DOI] [PubMed] [Google Scholar]
- Schindler C, Hracky R, Soll J. Protein transport in chloroplasts: ATP is prerequisite. Z Naturforsch. 1987;42c:103–108. [Google Scholar]
- Schnell DJ. Shedding light on the chloroplast protein import machinery. Cell. 1995;83:521–524. doi: 10.1016/0092-8674(95)90090-x. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Blobel G. Identification of intermediates in the pathway of protein import into chloroplasts and their localization to envelope contact sites. J Cell Biol. 1993;120:103–115. doi: 10.1083/jcb.120.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell DJ, Blobel G, Pain D. The chloroplast import receptor is an integral membrane protein of chloroplast envelope contact sites. J Cell Biol. 1990;111:1825–1838. doi: 10.1083/jcb.111.5.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell DJ, Blobel G, Pain D. Signal peptide analogs derived from two chloroplast precursors interact with the signal recognition system of the chloroplast envelope. J Biol Chem. 1991;266:3335–3342. [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G. Isolation of components of the chloroplast protein import machinery. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- Scott SV, Theg SM. A new chloroplast protein import intermediate reveals distinct translocation machineries in the two envelope membranes: energetics and mechanistic implications. J Cell Biol. 1996;132:63–75. doi: 10.1083/jcb.132.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf M, Waegemann K, Soll J. A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 1995;7:401–411. doi: 10.1046/j.1365-313x.1995.7030401.x. [DOI] [PubMed] [Google Scholar]
- Sohrt K, Soll J. Toc64, a new component of the protein translocon of chloroplasts. J Cell Biol. 2000;148:1213–1221. doi: 10.1083/jcb.148.6.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Izumihara K, Hase T. Plastid import of iron-sulfur cluster assembly of photosynthetic and non-photosynthetic ferredoxin isoproteins in maize. Plant Physiol. 1991;97:375–380. doi: 10.1104/pp.97.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveshnikova N, Soll J, Schleiff E. Toc34 is a preprotein receptor regulated by GTP and phosphorylation. Proc Natl Acad Sci USA. 2000;97:4873–4978. doi: 10.1073/pnas.080491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theg SM, Bauerle C, Olsen LJ, Selman BR, Keegstra K. Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J Biol Chem. 1989;264:6730–6736. [PubMed] [Google Scholar]
- Tranel PJ, Keegstra K. A novel, bipartite transit peptide targets OEP75 to the outer membrane of the chloroplastic envelope. Plant Cell. 1996;8:2093–2104. doi: 10.1105/tpc.8.11.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Blakeley SD, Dennis DT, Ko K. Transit peptides play a major role in the preferential import of proteins into leucoplasts and chloroplasts. J Biol Chem. 1996;271:31227–31233. doi: 10.1074/jbc.271.49.31227. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Onda Y, Ashikari T, Tanaka Y, Kusumi T, Hase T. Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs of maize. Plant Physiol. 2000;122:887–894. doi: 10.1104/pp.122.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ME, Keegstra K, Froehlich JE. GTP promotes the formation of early-import intermediates but is not required during the translocation step of protein import into chloroplasts. Plant Physiol. 1999;121:237–244. doi: 10.1104/pp.121.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]