In this study, Zheng et al. investigated how BEN factors, a recently recognized DNA binding module, identify their targets in humans. They characterize several mammalian BEN domain (BD) factors, including from two NACC family BTB-BEN proteins and from BEND3, which has four BDs, and provide structural insights into sequence-specific DNA binding by mammalian BEN proteins.
Keywords: DNA binding activity, gene regulation, protein–DNA structure, transcription factor
Abstract
The BEN domain is a recently recognized DNA binding module that is present in diverse metazoans and certain viruses. Several BEN domain factors are known as transcriptional repressors, but, overall, relatively little is known of how BEN factors identify their targets in humans. In particular, X-ray structures of BEN domain:DNA complexes are only known for Drosophila factors bearing a single BEN domain, which lack direct vertebrate orthologs. Here, we characterize several mammalian BEN domain (BD) factors, including from two NACC family BTB-BEN proteins and from BEND3, which has four BDs. In vitro selection data revealed sequence-specific binding activities of isolated BEN domains from all of these factors. We conducted detailed functional, genomic, and structural studies of BEND3. We show that BD4 is a major determinant for in vivo association and repression of endogenous BEND3 targets. We obtained a high-resolution structure of BEND3-BD4 bound to its preferred binding site, which reveals how BEND3 identifies cognate DNA targets and shows differences with one of its non-DNA-binding BEN domains (BD1). Finally, comparison with our previous invertebrate BEN structures, along with additional structural predictions using AlphaFold2 and RoseTTAFold, reveal distinct strategies for target DNA recognition by different types of BEN domain proteins. Together, these studies expand the DNA recognition activities of BEN factors and provide structural insights into sequence-specific DNA binding by mammalian BEN proteins.
Sequence-specific transcription factors (TFs) are responsible for orchestrating all aspects of development and physiology. Together with general chromatin and epigenetic factors, the specific DNA binding properties of TFs allow them to identify appropriate cohorts of target genes for transcriptional regulation, either by themselves or as combinations of TFs (Iwafuchi-Doi and Zaret 2016; Reiter et al. 2017). The executive-level activities of TFs to instruct or repress cell fates have long been recognized (Davis et al. 1987), but reached a new apex with the finding that specific TF combinations could induce the pluripotent state (Takahashi and Yamanaka 2006; Takahashi et al. 2007). This discovery elicited an ongoing chase for other TF combinations that are sufficient to instruct a diverse and ever-growing catalog of cell identities (Feng et al. 2008; Szabo et al. 2010; Vierbuchen et al. 2010; Huang et al. 2011; Yamamizu et al. 2013).
Such efforts rely on functional resources for systematic manipulation of TFs (Lambert et al. 2018; Ng et al. 2021). With comprehensive efforts to annotate transcription factors in most species, it is increasingly rare to identify novel families, especially in well-studied species. However, we and others recently added to the repertoire of metazoan TFs with our studies of BEN domain proteins. “BEN” was previously a domain of unknown function (DUF1172) within diverse proteins. As several of these have nuclear and/or chromatin functions, it was renamed for the exemplar proteins BANP, E5R, and NACC1 (Abhiman et al. 2008). For example, we found that BEN proteins Drosophila Insensitive (Duan et al. 2011) and mammalian BEND6 (Dai et al. 2013a) are nuclear inhibitors of Notch signaling and regulate neural development. Nevertheless, the specific function of BEN domains was unknown until we surprisingly found that BEN domains from multiple proteins bind specific DNA sequences and direct them to their cognate sites genome-wide (Dai et al. 2013b, 2015; Ueberschär et al. 2019). The Schedl laboratory (Aoki et al. 2012; Fedotova et al. 2018) independently reported that Drosophila BEN factors directly recognize sequence motifs within Fab-7, which separates two enhancers within the Bithorax complex, and are functional components of this chromatin boundary element. We established broader roles for BEN factors as repressors and insulators (Dai et al. 2013b; Ueberschär et al. 2019).
To date, structural analyses of DNA binding by BEN proteins have mostly focused on Drosophila. Although many mammalian BEN proteins are known to influence transcription and/or interact with chromatin factors (Kaul et al. 2003; Korutla et al. 2007, 2009; Sathyan et al. 2011; Dai et al. 2013a; Xuan et al. 2013; Saksouk et al. 2014; Khan et al. 2015), there are few insights into their direct abilities to recognize target genes specifically via cognate sites in the genome. Mammalian BTB-BEN domain proteins RBB/NACC2 (Xuan et al. 2013) and NACC1 (Nakayama et al. 2020) were shown to interact with specific DNA sequences via BEN domains, but their relationship to endogenous targets was only addressed selectively. Very recently, the mammalian BEN domain factor BANP was shown to bind CGCG core motifs and to be methylation-sensitive (Grand et al. 2021). In addition, while the fly BEN proteins and mammalian factors above harbor single BEN domains, mammals encode proteins with multiple BEN domains (Abhiman et al. 2008), which have been less studied. Of these, BEND3 is an appealing candidate as a representative multi-BEN factor bearing four tandem BEN domains. We previously characterized BEND3 as a heterochromatin-associated repressor (Sathyan et al. 2011; Khan et al. 2015), and the Déjardin laboratory (Saksouk et al. 2014) further analyzed its role in switching from constitutive to facultative heterochromatin.
Importantly, despite many hints that mammalian BEN domains access DNA, little is known about their underlying structural features that permit sequence-specific recognition. In this study, we demonstrate direct sequence-specific binding activities of mammalian BEN proteins NACC1, NACC2, and BEND3. We reveal overlapping site preferences of the NACC-BEN domains, as well as distinct modes of target interaction. With BEND3, we found that its fourth BEN domain (BD4) plays a key role in the direct recognition and repression of target genes. ChIP-seq studies confirmed that BD4 target sites are a major determinant of the endogenous genomic occupancy of BEND3. We solved the structure of BD4 in complex with its preferred DNA binding site, which reveals how BEND3 identifies its cognate target sites in vivo and why the BD1 domain of BEND3 does not associate with DNA. In addition, by combining our previous structural studies of Drosophila BEN domains with recent improvements in structural predictions, we classified two general strategies for how BEN domains mediate sequence-specific DNA recognition. These data provide a new foundation to interpret and dissect the molecular functions of mammalian BEN proteins.
Results
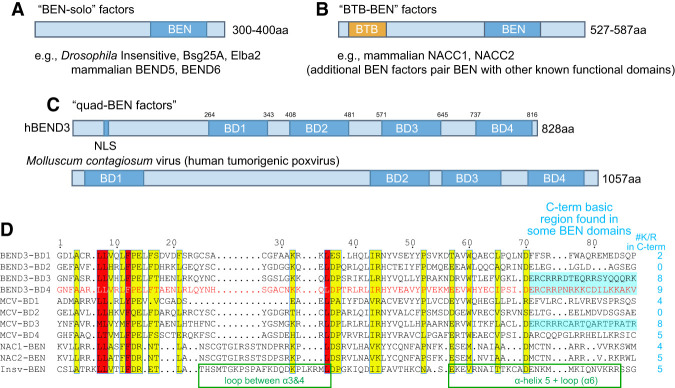
Diverse domain layouts of mammalian BEN proteins
We previously analyzed proteins whose only feature was one BEN domain (BEN-solo factors) (Fig. 1A). However, BEN factors have other layouts. Several proteins fuse a BEN domain to a distinct known functional domain, such as mammalian NACC1 and NACC2 (also known as BEND8 and BEND9, respectively), which are homologous factors that have an N-terminal BTB (Broad complex, Tramtrack, and Bric-a-brac)/POZ (poxvirus and zinc finger) domain and a C-terminal BEN domain (Fig. 1B). Other BEN proteins contain other domains, such as C2H2 zinc fingers or MCAFN, SCLM1, or RNaseT2 domains (Abhiman et al. 2008). Another class of proteins contains multiple (two, three, or four) BEN domains. For example, conserved mammalian BEND3 proteins are quadruple BEN (quad-BEN) domain factors (Fig. 1C,D). Although the functional rationale of four BEN domains is unknown, Molluscum contagiosum virus (MCV), a DNA poxvirus with a large (190-kb) genome (Senkevich et al. 1996), encodes an analogous quad-BEN protein named MC036R (Fig. 1C). The BEN domains of BEND3 and MC036R are more related to each other than to other BEN factors (Abhiman et al. 2008). In particular, some of the BEN domains in these quad-BEN factors bear C-terminal regions with large numbers of basic residues (Fig. 1D). The comparable region of Drosophila BEN-solo factors (e.g., Insv) is involved in binding nucleic acid (Dai et al. 2013b); however, other BEN domains have few or no basic residues in their C-terminal regions (Fig. 1D). These observations hint at potential functional diversity of BEN domains.
Figure 1.
Representative subclasses of DNA-binding BEN factors. (A) The most well-characterized BEN factors include Drosophila and mammalian proteins that encode a single BEN domain and no other recognizable functional domains (“BEN-solo” factors). (B) Some BEN factors are fused to another known domain, such as the BTB domain; diverse BEN proteins bear other domains (Abhiman et al. 2008). (C) The conserved vertebrate protein BEND3 and a poxvirus protein (MCV) contain four BEN domains (“quad-BEN” factors). (D) Alignment of the four BEN domains of human BEND3 and MCV-M036R, human NACC1 and NACC2 BEN domains, and the Drosophila Insv-BEN domain, whose structure we solved (Dai et al. 2013a). Core motifs that define the BEN domain are evident, but these mammalian/viral BEN domains contain characteristic residues. Note that MCV-BD4 is most similar to BEND3-BD4.
To investigate DNA binding by mammalian BEN proteins, we used protein-binding microarrays (PBMs) and high-throughput systematic evolution of ligands by exponential enrichment (HT-SELEX) to assay BEN domains from NACC proteins and different BEN domains of BEND3. In the PBM technique, all possible 8-mers are represented multiple times within an array so that specific binding of a candidate recombinant protein can be inferred from de novo motif analysis of the bound oligos (Weirauch et al. 2013; Narasimhan et al. 2015). To increase confidence in enriched motifs, we used two arrays (ME and HK) bearing nonoverlapping oligonucleotide sets. With HT-SELEX, we sequenced and then analyzed the bound populations of molecules selected from a randomized oligo pool after all three selection cycles (Nitta et al. 2015). The data were analyzed with AutoSeed, a de novo motif discovery program that detects local maxima of most enriched gapped k-mers, and motifs were constructed semimanually with parameters shown on headers of the motifs in Supplemental Table S2. These independent methods yielded mutually supportive data.
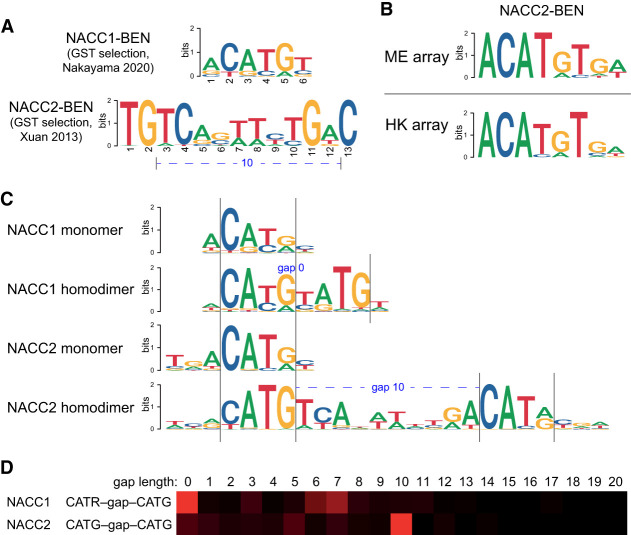
Modes of sequence-specific DNA binding by NACC family BEN domain proteins
The BEN domains of NACC2 (Xuan et al. 2013) and NACC1 (Nakayama et al. 2020) were reported to harbor specific target preferences from limited sequencing of probes recovered from cyclic amplification and selection of target (CASTing) assays. These works seem to indicate that NACC1 and NACC2 prefer largely distinct target sites, although some limited similarity can be discerned (Fig. 2A). Our PBM experiments with NACC2-BEN revealed similar motifs enriched from both arrays, which include the palindrome ACATGT (Fig. 2B; Supplemental Fig. S1). Notably, this matches well to the recently described NACC1-BEN target site, even though this result was obtained by sequencing only 18 target clones (Nakayama et al. 2020). Thus, the BEN domains of NACC1 and NACC2 may harbor more similar DNA binding activities than suspected.
Figure 2.
Modes of DNA recognition by BEN domains of mammalian NACC proteins. (A) Prior studies suggested distinct, sequence-specific, binding activities of the BEN domains from NACC1 (Nakayama et al. 2020) and NACC2 (Xuan et al. 2013). (B) Preferred binding sites for mammalian BEN domains, determined from independent protein-binding microarrays (PBMs, ME, and HK designs). PBM analyses of NACC2-BEN reveal a similar motif dissimilar to a prior study, but nearly identical to that determined for NACC1-BEN. (C,D) HT-SELEX assays reveal distinct types of preferred binding sites that reconcile past and current studies. (C) The NACC1-BEN domain selects a palindromic site that is similar to the NACC2-BEN PBM site, but also enriches sequences bearing dual palindromes that imply an allosteric binding mechanism. (D) These sites exhibit only mild preference for distinct spacing. The NACC2-BEN domain selects a simple palindrome similar to that of NACC1-BEN (C), but also selects dual sites with a highly preferred spacing (D). These data allow us to show that the previously reported NACC2-BEN site (A) overlaps the roughly double palindromic sequence, but lacks most of the flanking adjacent sequences that define the individual sites.
To study this further, we subjected both BEN domains of NACC1 and NACC2 to HT-SELEX. These assays demonstrate that BEN domains of NACC1 and NACC2 bind to very similar, partially palindromic, monomeric sites with an ACATGY core consensus sequence. As is often observed in HT-SELEX, the earlier selection cycles showed higher incidence of the monomeric sites, as they are more prominent in the initial pool. However, the later cycles enabled longer motifs to be recovered and revealed cooperatively binding dimeric palindrome motifs for both NACC1 and NACC2. However, these exhibited distinct preferences for different spacing and orientation configurations. NACC1 binds mostly as a monomer or to a dimeric composite (overlapping) site (Fig. 2C); the monomeric sites appear to be overlapping, but this may reflect protein binding into the major groove on the opposite side of the DNA strand, as is commonly observed when TF complexes bind cooperatively to composite sites (Jolma et al. 2015). There is also broad enrichment to other spacings with loose spacing preference (Fig. 2D). On the other hand, NACC2 enriched for a dimeric palindromic site, the spacing of which was consistent with the motif made from the previous site (Xuan et al. 2013), although the HT-SELEX site model is distinct (Fig. 2C). The stringent spacing preference between the individual palindromic sites of NACC2 (10 nt between CATG cores) (Fig. 2D) likely means that cooperativity is not mediated only through oscillatory DNA allostery (Kim et al. 2013), but may involve protein–protein interaction between BEN domain monomers.
We note that our previous crystal structure of the Drosophila Bsg25A BEN–target DNA complex yielded two homodimeric BEN domain assemblies bound to two end-to-end stacked duplexes of its palindromic target sequence (Dai et al. 2015). Although we did not find a notable population of such dual homodimeric sites in Bsg25A ChIP-seq data (Ueberschär et al. 2019), these findings may suggest that certain classes of BEN domain proteins could engage higher-order complexes.
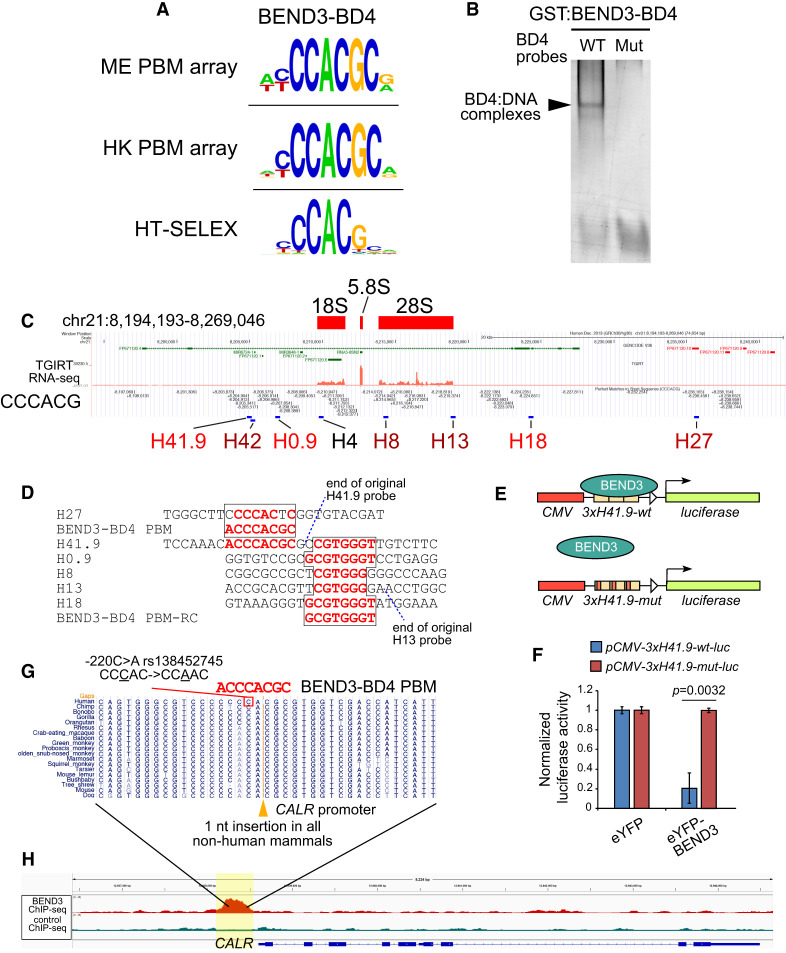
The BD4 domain of BEND3 harbors specific DNA binding activity
We and others reported that BEND3 can associate with DNA (Khan et al. 2015; Aghajanirefah et al. 2016), but DNA binding by individual BEN domains of a multi-BEN factor has not been tested previously. We focused on the first and fourth BEN domains (designated BD1 and BD4) of BEND3, since BD1 was shown to mediate protein–protein interactions with PICH (Pitchai et al. 2017), while BD4 was implicated in its localization to heterochromatin (Sathyan et al. 2011). Interestingly, while BD1 did not meet thresholds for motif enrichment, BD4 reproducibly selected nearly identical motifs (YCCACGC) in independent microarray panels (Fig. 3A; Supplemental Fig. S1). We conducted additional HT-SELEX assays with BEND3-BD4 and found that the selected sequences clearly enrich for a YCCACG motif that was nearly identical to the PBM motifs (Fig. 3A). Finally, we used gel shift assays to show that BD4 efficiently bound to this motif, and that point mutations in the target sequence abrogated BD4–DNA complexes (Fig. 3B).
Figure 3.
The BEND3-BD4 binding site explains direct binding at rDNA and CALR. (A) We subjected the BEND3-BD1 and BEND3-BD4 domains to PBM assays, with only BD4 enriching for a candidate binding site that was consistent between both array designs. We also subjected BEND3-BD4 to HT-SELEX assays, which retrieved a similar apparently monomeric site, as well as a longer consensus that includes a palindrome. (B) Gel shift assay using recombinant GST fusion protein confirmed that BEND3-BD4 binds directly and specifically to a probe bearing a CCCACGC core. (C) UCSC genome browser view at a representative human rDNA repeat. Total RNA-seq highlights rRNA transcripts, and sequence matches to the core BEND3-BD4 motif are marked. rDNA regions previously assayed for BEND3 association are labeled, of which the promoter-proximal H41.9 region was bound most strongly by BEND3 (Khan et al. 2015). (D) Alignment of BD4 sequence matches in BEND3 target regions in rRNA. Note that H41.9 contains two adjacent BD4 sites. (E) H41.9 wild-type and mutant transcriptional reporters. (F) eYFP-BEND3 represses H41.9-wt but not H41.9-mut reporters. Data are mean ± SD; n = 3 experiments; two-tailed t-test applied. (G) The conserved basal promoter of the CALR gene contains a human-specific nucleotide divergence, which coincides with a SNP in multiple schizophrenia and bipolar cases (Aghajanirefah et al. 2016). (H) The human-specific CALR promoter sequence that is affected in psychiatric disease resides within a strong BEND3 ChIP-seq peak.
Overall, these in vitro binding and selection assays with NACC proteins and BEND3 show that mammalian BEN domains have modular activities to recognize DNA.
The BEND3-BD4 motif underlies known examples of BEND3 genomic occupancy
As mentioned, DNA association by BEND3 has been suggested (Saksouk et al. 2014; Khan et al. 2015), but neither direct nor sequence-specific DNA binding by BEN domains of BEND3 has been studied. Thus, we asked whether our in vitro site selection assays provided insights into the binding of BEND3 with any specific genomic locations. Two targets have been reported; namely, the association of BEND3 with ribosomal (rDNA) loci (Khan et al. 2015) and with the calreticulin (CALR) gene (Aghajanirefah et al. 2016).
Clusters of rDNA loci are found on several chromosomes (McStay and Grummt 2008), and our previous work indicated that BEND3 associates with rDNA and represses accumulation of rRNA transcripts (Khan et al. 2015). Although BEND3 was shown to associate with specific rDNA genomic regions in gel shift assays, it was not known what sequences mediate this association. Strikingly, the rDNA region of strongest BEND3 association (H41.9) contains a perfect match to the BD4 PBM/SELEX motif (Fig. 3C), with another perfect BD4 PBM match directly upstream (2 nt away) of the assayed H41.9 region (Fig. 3D). It is expected that the fragments assayed using ChIP-qPCR would include both neighboring sites. Thus, the presence of dual high-affinity BEND3-BD4 sites may underlie the recruitment of BEND3 to this rDNA region.
To test this notion, we assayed a CMV>luciferase reporter bearing a multimer of the H41.9 region (slightly expanded to include the paired BD4 motifs) and a companion reporter with point mutations in each of the BD4 motifs (Fig. 3E). When normalized to their respective expression in the presence of eYFP control, eYFP-BEND3 strongly repressed the wild-type 3xH41.9wt-luc reporter, but not the mutant version (Fig. 3F). As controls, we tested other mammalian BEN factors (BEND5 and BEND6), and these do not regulate 3xH41.9wt-luc or 3xH41.9mut-luc (Supplemental Fig. S2A,B). Moreover, deletion of the BD4 domain abrogated the ability of BEND3 to repress this reporter (Supplemental Fig. S2C,D). Thus, BEND3 can directly and specifically repress a target gene via BD4 motif matches. We further note that our prior survey showed that BEND3 associates with additional genomic regions within rDNA loci (Khan et al. 2015). Notably, most of these directly contain strong nucleotide matches to the BEND3-BD4 motif (H0.9 and H18) or reside very close to such matches and were likely to overlap with assayed ChIP fragments (H42, H8 and H13) (Fig. 3C). Thus, the regulation of rDNA by BEND3 may involve multiple locations of association.
The association of BEND3 with the CALR locus was recognized via a complex path. Originally, mutations within the CALR promoter were found to be associated with human psychiatric disorders (Farokhashtiani et al. 2011). The −220C>A allele (rs138452745) was of special interest, as it is located in a region of human-specific divergence within an otherwise well-conserved mammalian sequence. Moreover, the −220C>A mutation is associated with increased CALR expression, implying that it may affect a repressor binding site (Esmaeilzadeh-Gharehdaghi et al. 2011). Interestingly, proteomic analysis of probes that compare the wild-type and mutant human alleles showed that the normal sequence is preferentially bound by a single factor, BEND3 (Aghajanirefah et al. 2016). However, the precise relationship of BEND3 to binding these alleles was not studied further. Strikingly, the wild-type BEND3 −220C site resides within a perfect 7/7-nt match to the BD4 PBM motif (Fig. 3G). Thus, BEND3 is likely the direct CALR repressor that is affected in these psychiatric disorders.
Overall, it is striking that both of the known BEND3 genomic targets are directly explained by the presence of perfect matches to the preferred binding sites determined from in vitro assays. Moreover, taken together with previous CALR studies (Aghajanirefah et al. 2016), recruitment of BEND3 by BD4 sites may suffice for target repression, at least at certain loci.
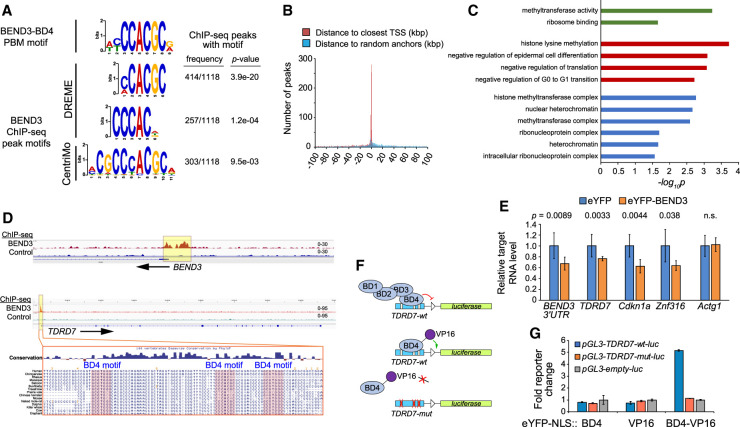
Genome-wide enrichment of the BD4 motif in BEND3 ChIP-seq data
To broaden these findings, we analyzed ChIP-seq data from NTERA2 cells, which express substantial BEND3 (Kurniawan et al. 2022). Inspection of rDNA loci revealed broad binding across the entire genic and intergenic regions, but this was also evident in control data (Supplemental Fig. S3). However, ChIP-seq analysis of rDNA loci is problematic due to their highly repetitive nature, as is the case for all multicopy loci. On the other hand, we observed a robust BEND3-specific ChIP-seq peak precisely and specifically on the CALR region bearing the BD4 motif, but nowhere else across the gene (Fig. 3H). Thus, BEND3 ChIP-seq data were specifically related to the BD4 motif at this known target.
Upon systematic analysis of BEND3 ChIP-seq data, we were pleased to find that aggregate BEND3 peaks were strongly enriched for BD4-like motifs. In particular, de novo motif finding using DREME (Bailey 2011) and CentriMo (Bailey and Machanick 2012) showed that top hits, in terms of both significance and frequency, closely resembled the BD4 PBM site (Fig. 4A). Thus, genomic association of BEND3 correlates well with the presence of this motif from in vitro studies of isolated BD4. Although previously reported as a heterochromatin factor, we note that BEND3 ChIP-seq peaks with BD4 motifs were preferentially located near transcription start sites (Fig. 4B; Supplemental Fig. S3). The associated genes were enriched for numerous gene ontology (GO) terms involved in histone methylation, heterochromatin, regulation of cell cycle, and inhibition of differentiation (Fig. 4C). Consistent with these observations, loss of BEND3 from the NTERA2 stem-like model promotes their differentiation (Kurniawan et al. 2022), and we also observed that BEND3 is predominantly expressed in pluripotent cell types (Supplemental Fig. S4).
Figure 4.
BEND3 associates directly with BD4 motifs genome-wide. (A) The top motifs recovered from BEND3 ChIP-seq peaks from NTERA2 cells closely resemble the BEND3-BD4 motif identified from our in vitro binding assays. P-values of motif enrichment were generated using Fisher's exact test. (B) BEND3 peaks bearing BD4 sites are strongly localized near core promoters. (C) Representative gene ontology (GO) terms associated with BEND3 targets bearing BD4 sites. (D) IGV views of BEND3 and control ChIP-seq data at BEND3 and TDRD7. BEND3 ChIP-seq peaks in both genes are associated with multiple BD4 sites, as illustrated for TDRD7. (E) Transcript levels of BEND3-BD4 target genes are repressed upon ectopic expression of eYFP-BEND3. (F) Schematic of TDRD7-wt and mutant transcriptional reporters, and rationale to infer direct recruitment of BD4 to its cognate sites in TDRD7 using a BD4-VP16 activator fusion. (G) Luciferase reporter assays show that BD4 and VP16 domains are neutral, but BD4-VP16 specifically activates the TDRD7-wt reporter. Data in E and G are mean ± SD; n = 3 experiments; two-tailed t-test applied.
Since BEND3 was previously characterized as a repressor (Sathyan et al. 2011; Saksouk et al. 2014; Khan et al. 2015), we tested the response of newly identified BEND3 targets with BD4 motifs. Some BEND3 ChIP peaks had single sites, but others had multiple sites. For example, BEND3 exhibits prominent binding near its own alternative promoters, which contain seven matches to the core BD4 motif within a <2-kb interval, while a strong BEND3 ChIP-seq peak at the TDRD7 promoter encompasses three conserved BD4 motifs within only ∼70 bp (Fig. 4D). We checked the steady-state levels of these and other targets following overexpression of eYFP-BEND3, compared with control eYFP transfection. To distinguish endogenous BEND3, we assayed a 3′ UTR amplicon that is not included in the eYFP-BEND3 vector. While Atcg1 did not change, we observed that BEND3, TDRD7, Cdkn1a, and Znf316 transcripts were all reduced in response to ectopic BEND3 (Fig. 4E). These data broaden the impact of BEND3 in transcriptional repression, and also indicate that BEND3 is autoregulatory, as is a theme for many other regulatory factors.
Given notable and functional binding of BEND3 at TDRD7 (Fig. 4D,E), we used this for additional, stringent tests of the notion that the BEND3-BD4 domain and its cognate site were directly responsible for target regulation. To do so, we fused the BD4 domain alone to VP16. In principle, while full-length BEND3 is a repressor, BD4-VP16 might be a synthetic activator of targets bearing BEND3-BD4 sites (Fig. 4F). We tested its properties on a TDRD7-wt-luc reporter or a variant reporter with point mutations of BD4 sites. Indeed, while all controls were inert, BD4-VP16 activated TDRD7-wt-luc, but did not affect TDRD7-mut-luc (Fig. 4G). Thus, BD4 motifs are specific, conserved sequences that recruit the quad-BEN domain factor BEND3 to regulatory targets. We note this differs from a recent study of NACC1, whose BEN domain was concluded as insufficient to interact with chromatin in cells (based on the mobility behavior of GFP fusion proteins) (Nakayama et al. 2020).
Structural basis for sequence-specific DNA binding by the BEND3-BD4 domain
These data motivated us to generate atomic insights into DNA recognition by BEND3. Although the structure of BEND3-BD1 in complex with PICH was determined (Pitchai et al. 2017), there are limited structural studies for DNA binding by mammalian BEN factors (Nakayama et al. 2020).
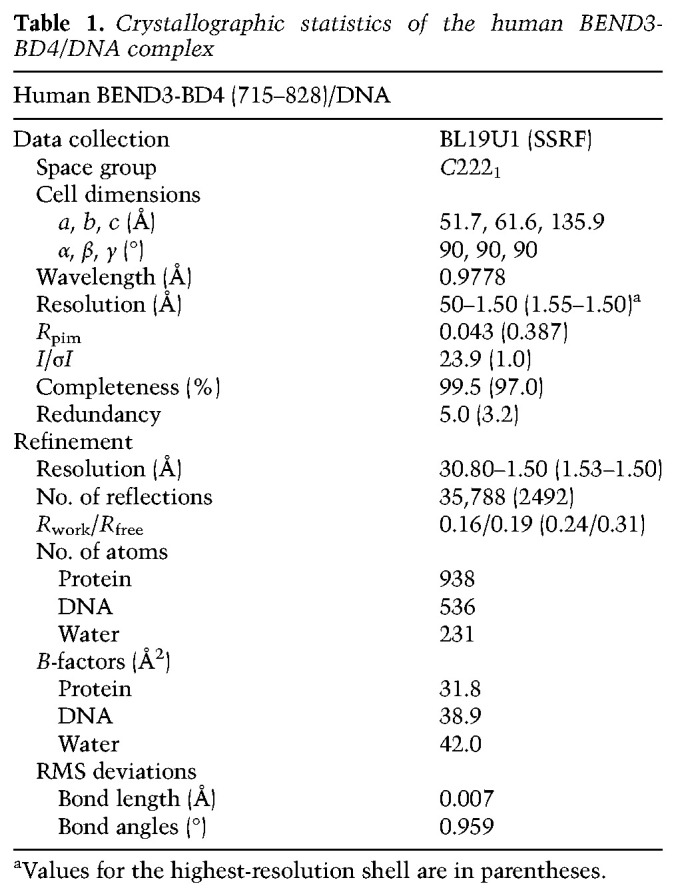
We assayed a range of conditions to cocrystallize the BD4 domain with different DNA targets that shared the consensus core-binding sequence. We eventually obtained high-quality crystals that diffracted to 1.5 Å resolution using BEND3-BD4 protein spanning residues 715–828 in complex with a DNA duplex (5′-GGACCCACGCAGC-3′/3′-CTGGGTGCGTCGG-5′), forming a 12-bp complementary duplex with 5′ G overhangs (Table 1). Each asymmetric unit contained one complex in which one BEND3-BD4 bound an individual DNA duplex target. Two symmetry-related BD4/DNA complexes adopted an end-to-end stacking pattern along the DNA duplex in the tertiary structure of the complex (Supplemental Fig. S5). We also investigated the interaction between proteins in the tertiary structure and found only one hydrogen bond formed between Gln731 and Arg778′ from two symmetry-related molecules (Mol A and Mol A′sym) and one stacking interaction formed between Arg746 and Arg746′′ from another two symmetry-related molecules (Mol A and Mol A′′sym) (Supplemental Fig. S5).
Table 1.
Crystallographic statistics of the human BEND3-BD4/DNA complex
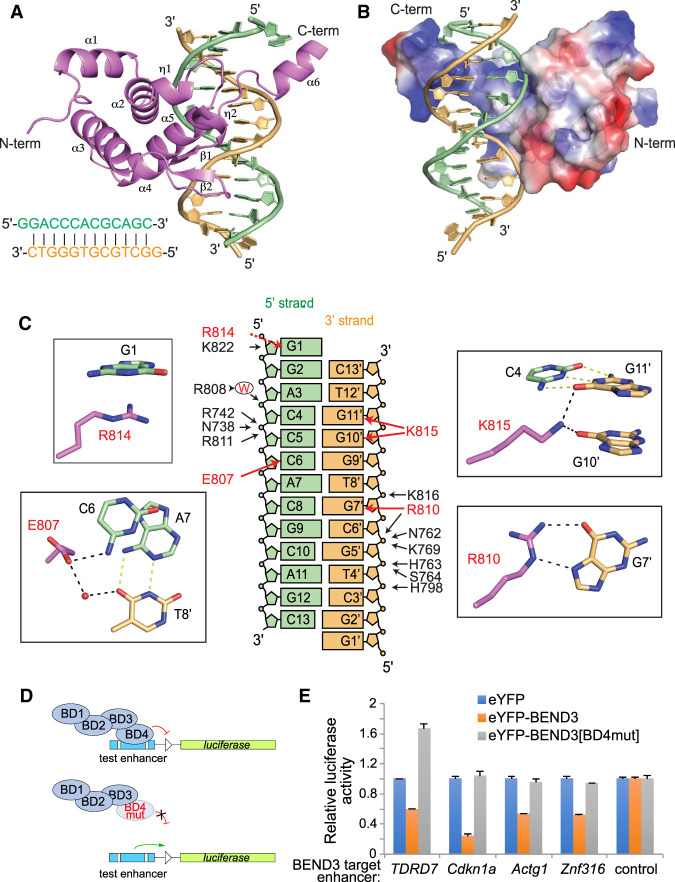
We depict the overall structure of the BD4/DNA complex in cartoon representation (Fig. 5A) and in electrostatic surface representation (Fig. 5B). The folding topology of the BEND3-BD4 domain is composed of six α helices, two short helical turns (η1 and η2), and two short β strands (Fig. 5A). The BD4 fold generates a basic positively charged binding channel along the major groove encompassing one-half of the DNA helix (Fig. 5B). We provide composite omit electron density maps of the DNA target bound to BEND3-BD4 protein and of specific DNA-interacting residues of BEND3-BD4 in Supplemental Figure S6.
Figure 5.
Tertiary structure of the BEND3-BD4/DNA complex. (A) Tertiary structure of the BEND3-BD4 domain in complex with the DNA duplex shown in cartoon representation, with the BD4 domain in violet and the two strands of DNA complex in orange and pale green, respectively. The sequence of the 13-mer DNA duplex is shown at the bottom left. (B) Electrostatic surface representation of the BD4 domain in complex with DNA. (C) Summary of base contacts (red arrows) and sugar–phosphate interactions (black arrows) in the complex. The red capital W represents water. The details of the base interactions are shown in the black rectangle. G1 is stacking on R814. The side chain of E807 is hydrogen-bonded to the 4-NH2 of C6 in the 5′ strand and forms water-mediated interaction with T8′ in the 3′ strand. Both O6 of G10′ and G11′ form interactions with the side chain of K815. The Hoogsteen edge of G7′ forms hydrogen bonds with the side chain of R810. (D) Schematic for testing transcriptional regulation by wild-type and mutant BEND3 proteins. As guided by the structure, we mutated three BD4 residues involved in sequence-specific base contacts (E807V, R810L, and R814L). (E) Assays of transcriptional reporters of four BEND3 targets show that they are repressed by eYFP-BEND3, but not by eYFP-BEND3[BD4mut].
We identified a large number of intermolecular contacts formed between the DNA duplex and protein in the complex, which include base interactions as well as extensive nonbase interactions. Helix α5 and the loop between α5 and α6 of the BEND3-BD4 domain are located in the major groove of the DNA duplex, while the long loop between η2 and α3 is stretched close to the minor groove of the DNA duplex (Fig. 5A). We have summarized the intermolecular contacts between the BEND3-BD4 protein (shown in violet) and the two strands of the 13-mer DNA duplex (the 5′ strand and 3′ strand are shown in green and orange, respectively, in Fig. 5C).
Intermolecular contacts between the BEND3-BD4 domain and the 5′ strand (colored green) are subdivided into base interactions (red arrows) and sugar–phosphate interactions (black arrows) in the left panels in Figure 5C. At one end of the duplex, the G1 overhang base is involved in a stacking interaction with Arg814 located toward the C terminus of BD4 (shown with a red dotted arrow in Fig. 5C). In addition, G1 is further stabilized by interaction between the side chain of Lys822 and the ribose of G1 (Fig. 5C; Supplemental Fig. S7). Moreover, E807 also interacts with O4 of T8′ via a water molecule (Fig. 5C). Residues Asn738, Arg742 from α2, and Arg811 located in the loop region between α5 and α6 form hydrogen bonds with the phosphate of C5 in the 5′ DNA strand (Fig. 5C; Supplemental Fig. S7). The base of C5 also forms van der Waals contacts with the side chain of R811. Finally, we found that a water molecule mediates the interaction between Arg808 and C4 (Fig. 5C; Supplemental Fig. S7).
Intermolecular contacts between the BEND3-BD4 domain and the 3′ DNA strand (colored orange) are also subdivided into base-specific interactions (red arrows) and sugar–phosphate interactions (black arrows) in the right panels of Figure 5C. Toward one end of the 3′ strand, the side chain of Lys815 (located in the loop region between α5 and α6) forms two hydrogen bond interactions with the bases of G10′ and G11′, and van der Waals contacts with the base of C4 (which pairs with G11′) (Fig. 5C; Supplemental Fig. S7).
Beside these base-specific interactions, the main chain of Cys817 forms one hydrogen bond with the phosphate of G9′, and the side chain of Cys817 forms van der Waals contacts with the sugar of G9′ (Supplemental Fig. S7). The side chain of Lys816 (located in α6) forms one hydrogen bond with the phosphate of T8′, while Pro812 forms a partial stacking interaction with the base of T8 (Fig. 5C; Supplemental Fig. S7). The main chain of Arg810 forms one indirect interaction with the phosphate of G7′ through one water molecule, and the side chain of Arg810 forms hydrogen bonding interaction with the Hoogsteen edge of G7′ (Fig. 5C; Supplemental Fig. S7). Furthermore, the phosphates of C6′–G5′–T4′ were recognized by extensive hydrogen bond interaction and van der Waals contacts with BD4. Arg810, Asn762, and Lys769 participate in the non-base-specific interaction with the phosphate of C6′ (Fig. 5C; Supplemental Fig. S7). Several other residues interact with the phosphate of G5′, in which His763 and Ser764 form hydrogen bond interactions, while the nearby Asp806, Asn762, Ala766, and Cys767 form van der Waals contacts. His798 contacts the phosphate of T4′ by hydrogen bonding (Fig. 5C; Supplemental Fig. S7).
The residues of BD4 involved in base-specific interaction with two strands of the DNA duplex (including Glu807, Arg810, Arg814, and Lys815) are all clustered in the C-terminal 807–815 segment of the BD4 protein (Fig. 5C). The electrostatic surface representation of BD4 indicated a basic positively charged binding channel formed by the C terminus of BD4, which facilitates the interaction with the negatively charged DNA (Fig. 5B). The residues of BEND3-BD4 involved in the sugar–phosphate nonbase interaction are distributed from α2 to α6, including the loop regions. These interactions shaped the conformation of the loop region of BD4, as well as stabilized the overall structure of BD4, which facilitate the specific binding of BEND3-BD4 to the DNA binding site. Taken together, the base-specific interactions combined with sugar–phosphate interactions revealed the specific binding pattern of the BEND3-BD4 domain with its 5′-ACCCACGCAG-3′/3′-TGGGTGCGTC-5′ binding site.
To validate the BD4 structure, we conducted reporter assays using experimentally validated BEND3 targets. As noted, Tdrd7, Cdkn1a, Actg1, and Znf316 all have prominent BEND3 ChIP-seq peaks bearing BD4 motifs (Supplemental Fig. S3) and can be repressed by ectopic BEND3 (Fig. 4E). We cloned regions including these BEND3 ChIP-seq peaks into luciferase reporters and tested their response to ectopic eYFP, eYFP-BEND3, or a variant with neutralizing substitutions at three charged base-contacting residues (E807V, R810L, and R814L) in BD4 (i.e., BEND3[BD4mut]) (Fig. 5D). Our tests revealed that all four target enhancers conferred BEND3-mediated repression that was fully abrogated upon mutation of three BD4 residues (Fig. 5E), validating their importance for target recognition.
Comparison of the BEND3-BD4/DNA complex with other solved BEN domain complexes
With our knowledge of a functional BEND3-BD4/DNA binding complex, we sought insights into how variant BEN domains mediated nucleic acid interactions or are prevented from doing so.
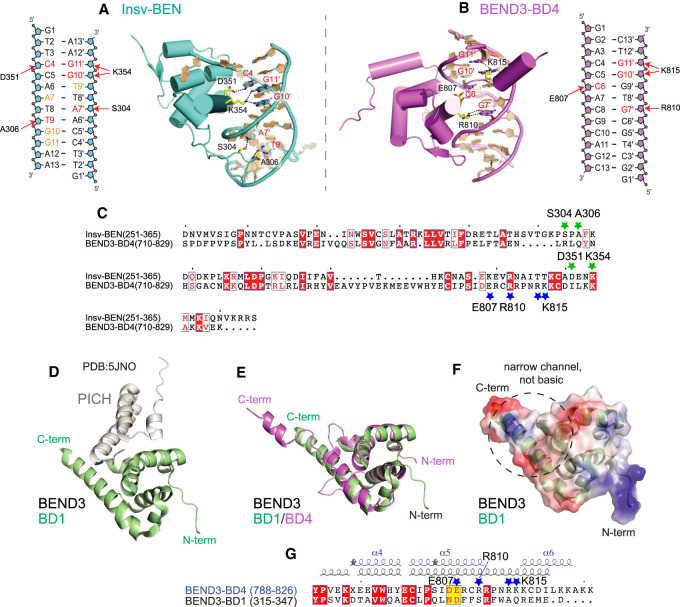
The BEND3-BD4 and Insv-BEN domains recognize distinct DNA motifs. To understand the mechanistic basis underlying this diversity, we compared the structures of BEND3-BD4–DNA and Insv-BEN–DNA complexes (Fig. 6A–C; Supplemental Fig. S8). Both BEND3-BD4 and Insv-BEN proteins are composed mainly of helices and two short β sheets (Supplemental Fig. S8). Unlike Insv-BEN that interacts with a palindromic DNA binding site (TTCCAATTGGAA) with a binding ratio of 2:1 (Supplemental Fig. S8C), BEND3-BD4 selectively binds a core sequence motif (5′-ACCCACGCAG-3′/3′-TGGGTGCGTC-5′) with a binding ratio of 1:1 (Fig. 6B), implying differences in recognition of target DNA between these two proteins. To simplify analysis, we chose only one selected Insv-BEN protein/DNA complex and the BEND3-BD4/DNA complex for comparison (Fig. 6A,B).
Figure 6.
Comparison of BEN domains that mediate DNA or protein interactions. (A–C) Comparison of base-specific interactions between the BEND3-BD4/DNA complex and the Insv-BEN/DNA complex (PDB: 4IX7). (A) Schematic and structural details of base-specific intermolecular hydrogen bonding contacts in the Insv-BEN/DNA complex. (B) Diagram and structural details of base-specific intermolecular hydrogen bonding contacts between BEND3-BD4 and the DNA target (involved residues are colored red). (C) Sequence alignment of Insv-BEN and BEND3-BD4. Conserved residues are colored white in the red boxes. The green and blue stars label positions of residues involved in base-specific interaction by the Insv-BEN and BEND3-BD4 domains, respectively. (D–G) Comparison of the BEND3-BD4/DNA complex with the BEND3-BD1/PICH-TPR complex (PDB: 5JNO) shows that their DNA-binding and protein-binding interaction surfaces are mostly distinct. (D) Cartoon view of the BEND3-BD1/PICH-TPR complex, with the BD1 domain in green and PICH in gray. (E) Superposition of the BD1 (green) and BD4 (violet) domains of BEND3. (F) Electrostatic surface representation of the BD1 domain of BEND3. BD1 lacks features that enable DNA binding by BD4 (Fig. 5B); namely, an open channel lined with basic residues that makes multiple contacts with DNA. (G) Sequence and secondary structure alignments of the C-terminal regions of the BD1 and BD4 domains of BEND3. Conserved residues are colored white in the red boxes, and the adoption of α helix, short helix turns (η), and β strand is labeled above. The dark-blue stars mark positions of residues involved in base-specific interaction in the BD4 domain.
Both Insv-BEN and BEND3-BD4 proteins form a positively charged DNA binding channel on their surface (Supplemental Fig. S8B,D). As shown in Figure 6, A and B, the Insv-BEN protein not only forms base-specific interactions with the DNA duplex in the major groove (C4, G11′, and G10′), but also forms base-specific interaction in the minor groove (A7′ and T9). However, as shown in Figure 6B, BEND3-BD4 protein only forms base-specific interactions with the DNA duplex in the major groove (C6, G7′, G10′, and G11′). The residues Glu807, Arg810, and Lys815 of BEND3-BD4 protein involved in DNA motif recognition (Fig. 6B) are all located in the C terminus of the BEN domain (shown with blue stars in Fig. 6C). The residues Ser304, Ala306, Asp351, and Lys354 of Insv-BEN protein involved in DNA interaction are located not only in the C terminus of the BEN domain, but also in the middle (shown with green stars in Fig. 6C). Most of the base-specific interacting residues in the BEND3-BD4 domain and Insv-BEN are distinct (Fig. 6C). This appears to account for the diverse DNA-binding properties of these two BEN domains.
The structure of the BEND3-BD1 domain in complex with PICH (BD1-NTPR; PDB: 5JNO) was reported (Pitchai et al. 2017). The cartoon representation of the structure of the latter complex is shown in Figure 6D, in which BD1 is shown in green and PICH is shown in gray. The tertiary structural alignment of the BD1 and BD4 folds show that their helical segments align well (Fig. 6E). The electrostatic surface representation of BD1 is shown in Figure 6F. Unlike BD4, whose basic C terminus is involved in DNA recognition (Fig. 5B), there is no obvious positively charged channel located in the C terminus of BD1 (Fig. 6F). Moreover, sequence and secondary structure alignments of the BD1 and BD4 C termini show that the four charged residues in BD4 that participate in specific recognition of its DNA motif (Glu807, Arg810, Arg814, and Lys815) (labeled by stars in Fig. 6G) are not conserved in BD1. Thus, BD1 and BD4 have structural distinctions that are associated with different functional properties, despite their overall similar folds.
Overall, Drosophila BEN domains and mammalian BEND3-BD4 have partially distinct surfaces with which they recognize DNA; Insv/Bsg25A/Elba2, NACC1/NACC2, and BEND3 all bind quite different target sequences; and BEND3-BD1 does not seem to bind DNA specifically. These findings may suggest that the BEN domain is a scaffold that has been flexibly deployed for both DNA recognition and protein recognition.
BEN domains interact with DNA through distinct strategies
The NACC1-BEN domain shows higher homology with Insv-BEN than with BEND3-BD4 (Fig. 1D). In addition, the NACC1-BEN structure determined by NMR spectroscopy suggested that NACC1-BEN is more similar to Insv-BEN in protein folding and DNA interaction pattern. They both contain five α helices and bind DNA through α5 and the loop between α3 and α4 (Nakayama et al. 2020). In contrast, BEND3-BD4 contains six α helices and interacts with DNA by the “α5–loop–α6” region in the C-terminal region (Fig. 5).
To broaden our findings that NACC1-BEN, Insv-BEN, and BEND3-BD4 use different strategies to bind DNA, we investigated predicted structures of other BEN domains. Recent monumental advances in protein structural prediction provide valuable tools to explore 3D structure with unprecedented efficacy (Baek et al. 2021; Tunyasuvunakool et al. 2021). As predicted structures for several proteomes are available from EBI (https://www.alphafold.ebi.ac.uk), we downloaded relevant Drosophila and human protein models and isolated their BEN domains. To benchmark these, we compared the predicted structures of Insv-BEN and BEND3-BD4 with our determinations from X-ray crystallography. We found that they overlap well, with minor differences in the disposition of BD4 α6 (Supplemental Fig. S9). Therefore, even though AlphaFold2 is unaware of the cognate DNA ligands, its predictions of BEN domain structures have utility for further exploration.
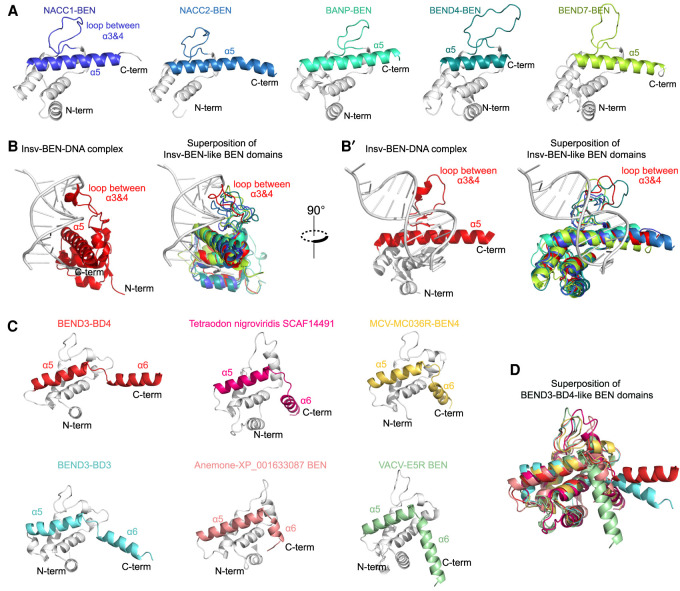
We found that many human BEN domains (including from NACC1, NACC2, BANP, BEND4, and BEND7) bore clear similarities in their overall predicted tertiary structures (Fig. 7A). These BEN domains all contain an α5 helix and middle loop between α3/α4, and were relatively well superimposed onto our solved Insv-BEN:DNA complex (Fig. 7B,B′). This strongly suggests a shared strategy for DNA recognition by BEN domains, as originally characterized for Drosophila Insv and Bsg25A (Dai et al. 2013b, 2015). We collectively term these as “type I” BEN domains, and distinguish them from non-DNA-binding BEN domains (“type 0”; e.g., BEND3-BD1).
Figure 7.
Distinct strategies for BEN domains to recognize DNA (A) AlphaFold2 predicts that several mammalian BEN domains (type I) have overall similar structures, including a substantial loop between α helices 3 and 4 and a long α helix 5. (B,B′) Both of these features are shared with DNA-binding contact regions in the Insv-BEN tertiary structure (PDB: 4IX7, in red), indicating that these BEN domains (type I) likely use similar strategies to interact with DNA. (C) The BEND3-BD4 domain contains a short loop in the C terminus that produces an α helix 6 (α6) and provides a channel to load DNA. RoseTTAFold predicts that several other BEN domains (type II) share this short “helix (α5)–loop–helix (α6)” structure, implying a shared strategy for DNA recognition. (D) Superposition of BEND3-BD4 and other predicted type II BEN domains predicted by RoseTTAFold.
BEND3-BD4 appeared distinct among human BEN domains in bearing a C-terminal loop and α6 helix, but in fact the BD3 domain of BEND3 also adopts a similar fold with a central basic channel formed by its C-terminal helix α5–loop–helix α6 region, and a small loop between α3/α4 (Fig. 7C). We then considered BEN domains encoded by other metazoans as well as viruses. Because many of these are not included in the current EBI AlphaFold database, we used RoseTTAFold (https://robetta.bakerlab.org) to predict their structures. From this, we recovered additional metazoan BEN domains (e.g., from fugu SCAF14491 and from sea anemone XP_001633087) and even viral BEN domains (e.g., from MCV-M036R and from VACV-E5R) with overall similarity to BEND3-BD4 (Fig. 7D). All of these superimpose fairly well onto BEND3-BD4, excepting the angle of the very C-terminal α6 of these BEN domains, whose disposition is slightly different in these predictions due to the preceding loop. We collectively refer to this distinct subset of BEN DNA-binding motifs as “type II” BEN domains.
Discussion
BEN domain proteins as an emerging class of transcription factors
These studies substantially expand our understanding of DNA binding by mammalian BEN factors. Despite growing appreciation of the impacts of BEN factors on chromatin organization and gene expression, there is comparatively little knowledge of sequence-specific recognition by mammalian BEN domains and how this relates to their genomic occupancy, as there is in Drosophila (Dai et al. 2013b, 2015; Ueberschär et al. 2019). Such knowledge is critical to interpret direct regulatory interactions. In this study, we clarify the site preference of NACC2, which is different from previously reported (Xuan et al. 2013) but instead quite similar to a motif recently ascribed to NACC1 (Nakayama et al. 2020). We also provide evidence that the BD4 domain of BEND3 binds DNA specifically and is a major driver for its endogenous genomic recruitment. More importantly, we found that different BEN domains use different strategies to interact with DNA. We propose two subclasses of DNA-binding BEN domains (Insv-BEN, Bsg25A-BEN, and NACC-BEN as representatives of type I, and BEND3-BD4 as a prototype of type II); other BEND domains may be platforms for protein–protein interaction (type 0; e.g., BEND3-BD1). Together with recently available structural prediction software, these efforts will facilitate functional interrogation of other BEN proteins in the future.
These data also serve as foundation to dissect the mechanisms and biology of BEND3. Several previous studies link BEND3 to gene repression and/or heterochromatin dynamics (Sathyan et al. 2011; Saksouk et al. 2014; Khan et al. 2015; Aghajanirefah et al. 2016), but the lack of knowledge of direct DNA binding activity limits the interpretation of these studies. Strikingly, we located optimal BD4 binding sites at both the previously described targets (rDNA and CALR) and show that BEND3 can repress many other newly identified targets via BD4 sites. These genes are typical euchromatic genes, but BEND3 was previously suggested as a specific factor for switching heterochromatic state. While general chromatin factors are usually studied with respect to heterochromatin dynamics, often in self-perpetuating feedback loops (Laugesen et al. 2019), it has also been proposed that sequence-specific factors could drive locus-specific heterochromatin (Bulut-Karslioglu et al. 2012). BEND3 may be positioned to play such a role, and may be thematically related to functions of Drosophila BEN proteins recently studied with respect to chromatin boundaries and insulators (Fedotova et al. 2018, 2019; Ueberschär et al. 2019). Finally, our analyses suggest that a viral quad-BEN domain protein (MCV-M036R) likely uses strategies similar to those of BEND3 to associate with DNA. As viral genomes often exploit homologs of cellular factors to hijack or rewire host gene expression during their life cycles (Liu et al. 2020), these findings are likely of relevance to gene regulation by Molluscum contagiosum virus.
Intersection of BEN factors with other transcriptional regulatory strategies
The GC-rich binding site of BEND3-BD4 overlaps that of certain other transcription factors, notably of Wilms’ tumor gene 1 (WT1) (Rauscher et al. 1990) and multiple members of the early growth response 1–4 (EGR1–4) family (Christy and Nathans 1989; Nardelli et al. 1991), all of which are multi-zinc-finger proteins whose individual DNA binding domains interact with distinct portions of the binding site. The BEND3-BD4 site is particularly similar to EGR family proteins, which can directly recognize CGCCCACGC motifs (Jolma et al. 2013), and inclusion of EGR sites among the catalog of known DNA binding motifs (http://jaspar.genereg.net) underlies its recovery in CentriMo analysis. However, it is clear from this work that BEND3-BD4 independently recognizes a similar site via a convergent—and structurally distinct—strategy. EGR family proteins and WT1 have distinct transcriptional regulatory effects with, broadly speaking, EGR proteins involved in transcriptional activation and WT1 as a transcriptional repressor (Go et al. 2019). Since a portion of EGR binding sites match perfectly to optimal BEND3-BD4 sites, and such a motif is sufficient to recruit BEND3 in cells, there may be competing regulatory effects of BEND3 and EGR factors at individual target sites. This hypothesis remains to be tested.
There is recent interest in phase-separated liquid droplets in heterochromatin organization (Larson et al. 2017; Strom et al. 2017; Sanulli et al. 2019; Wang et al. 2019). This made us wonder whether highly clustered BEND3 might confer distinct regulatory effects. We have previously shown that BEND3 accumulates in cytologically visible domains at repeats (Sathyan et al. 2011; Khan et al. 2015). If BEND3 is recruited to a high local valency of sites, potentially within tandem repeats that are characteristic of certain heterochromatic loci, it is plausible that this could trigger condensate formation. Potentially consistent with this is our inference that NACC-BEN domains may be involved in higher-order complexes, as suggested by a preferred spacing in compound binding site configurations in SELEX experiments.
Finally, Déjardin and colleagues (Saksouk et al. 2014) further suggested that BEND3 switches heterochromatic state, in which it is repelled by 5mC in constitutive heterochromatin but binds unmethylated DNA and recruits PRC2 to maintain facultative heterochromatin. However, further mechanistic evaluation was not possible due to a lack of data on direct binding sites of BEND3. Provocatively, BD4 motifs contain CG (Fig. 3A); thus, BEND3 may itself be a methylation-sensitive heterochromatin factor. While this work was in preparation, Schübeler and colleagues (Grand et al. 2021) reported that another mammalian BEN protein (BANP) binds to CG cores and is a methylation-sensitive transcriptional activator. Thus, there may be a larger theme of how mammalian gene regulatory networks incorporate BEN domain proteins as modular factors that can be influenced by DNA modification as well as drive chromatin dynamics and structure.
Materials and methods
Expression of recombinant proteins
We used PCR to amplify BEN domains of NACC2 and BEND3 with flanking AscI and Sbf1 restriction sites, and cloned them into the corresponding sites within the expression construct pTH6838, a T7 promoter-driven GST expression vector. For gel shift experiments, the coding region corresponding to human BEND3 amino acids 546–828 was cloned into a pGEX-5x plasmid downstream from a GST tag and transformed into the E.coli BL21 strain. Expression of fusion proteins was induced with 0.1 mM IPTG for 24 h at 18°C. E.coli were harvested at 4°C and then lysed with 100 μg/mL lysozyme (Sigma-Aldrich) in NETN buffer (100 mM NaCl, 20 mM Tris, 0.5 mM EDTA at pH 8.0) for 20 min on ice. The expressed protein was purified with EZview Red glutathione affinity gel (Sigma-Aldrich). Cloning primers are listed in Supplemental Table S2.
Protein-binding microarray (PBM) analysis
PBM assays were performed following the procedure described previously (Lam et al. 2011; Weirauch et al. 2013). Each GST-tagged protein sample was expressed with PURExpress in vitro protein synthesis kit (New England Biolabs), and the binding specificity was analyzed in duplicates on two different double-stranded DNA microarray designs (HK and ME) with different probe sequences. Calculation of 8-mer Z-scores and E-scores was performed as described (Berger et al. 2006). PBM data were generated with motifs derived using Top10AlignZ (Weirauch et al. 2014). The PBM data are provided as Supplemental Data Sets S1–S4.
High-throughput (HT-SELEX) analysis
HT-SELEX was modified from our previous approach (Jolma et al. 2013) to use glutathione-coated magnetic beads (Sigma-Aldrich G0924-1ML) in the step where the protein–DNA complexes are separated from unbound DNA. Otherwise, the assay similarly used IVT-produced proteins as PBM, and the selection reactions were carried out in a buffer of 140 mM KCl, 5 mM NaCl, 1 mM K2HPO4, 2 mM MgSO4, 100 µM EGTA, 1 mM ZnSO4, and 20 mM HEPES-HCl (pH 7). After selections, the ligands were amplified by PCR and then subjected to Illumina sequencing. Data analysis was performed as described (Nitta et al. 2015), where automatic detection of a sequence pattern defining local maxima was followed by semimanual generation of seeds that were then used to construct multinomial-1 or multinomial-2 position frequency matrices for the TF target specificity. The raw HT-SELEX data are available in the NCBI Short Read Archive (SRA) and European Nucleotide Archive (ENA) under accession PRJEB49150.
Gel shift assay
The wild-type and mutant BD4 domain probes used were CTAAACACCCACGCGCCGTGGGTTGTCTT and CTAAACCTTTCTACGCTAAAACCTGTCTT, respectively. Complementary oligonucleotides were annealed using a thermocycler. For the gel shift assay, 70 ng of each probe and 1 mM GST fusion protein were mixed and incubated with 10 mM Tris (pH 7.4), 100 mM NaCl, and 1 mM MgCl2 for 30 min. The reactions were then loaded with 6% sucrose and resolved on a 6% nondenaturing polyacrylamide gel (Thermo Fisher Scientific) at room temperature using TBE running buffer. The gel was stained with ethidium bromide for 15 min and visualized with UV.
Dual-luciferase reporter assay
To generate reporter constructs for rDNA H41.9 enhancer, 3× wild-type or mutant H41.9 5′ region was prepared with recursive PCR and cloned into the HindIII site between the CMV promoter and firefly luciferase CDS of a pcDNA-luciferase plasmid. To generate reporter constructs for BEND3 binding sites, BEND3 ChIP-seq peaks on Tdrd7, Actg1, Znf316, and Cdkn1a gene loci were amplified and cloned into a pGL3-promoter vector (Promega) between EcoRI and XhoI sites. The primers are listed in Supplemental Table S1. To generate the BD4 domain point mutation construct, nucleotides corresponding to E807, R810, and R814 were replaced with V, L, and L, respectively, by point mutation cloning. To test BD4 mediated transactivating activity, the coding region corresponding to human BEND3 amino acids 646∼828 was tagged with a SV40 nuclear localization signal. The VP16 transactivating domain was then cloned downstream from BD4.
Transient transfections were performed using Lipofectamine 3000 (Thermo Fisher Scientific) or Hieff Trans liposomal transfection reagent (Yeasen) according to the manufacturer's instructions. For each well of a 96-well plate, 35 ng of pGL3 reporter, 80 ng of BEND3 construct (wild type, mutated, or empty), and 5 ng of Renilla plasmid were transfected for 24–48 h before being lysed for measuring luciferase activity using the dual-luciferase reporter assay kit (Promega or Vazyme). The relative fold change in reporter activity of the construct with the BD4 motif relative to control was calculated from three biological replicates.
ChIP-seq data analysis
BEND3 and IgG control ChIP-seq libraries are described in Kurniawan et al. (2022). The raw reads were aligned to the hg38 genome using Bowtie2 without removing duplicates, which identified 61,506,063 reads for BEND3 ChIP-seq and 66,692,850 reads for the control set. BEND3 binding peaks were called using the MACS2 algorithm (version 2.1.1.20160309) with default parameters (Feng et al. 2012). The resulting 3719 BEND3 binding peaks were further filtered with the ENCODE blacklist. For de novo motif discovery or motif mapping, 500 bp flanking the top 1000 summits were sent for analysis using MEME suite (Bailey et al. 2015). LiftOver was used for reference genome conversion. SAMtools and Bedtools were used for file processing and format conversion. IGV software was used to visualize the peaks and motifs in the genome.
Protein expression and purification
The corresponding DNA sequence of the human BEND3-BD4 domain spanning from 715 to 828 residues was cloned into a modified pRSFDuet-1 vector with a ubiquitin-like protease (ULP1) cleavage site existing between a preceding 6xHis-SUMO tag and the target protein. Proteins were overexpressed in E.coli B834 (DE3) in M9 medium containing selenomethionine. The fusion proteins were purified by a Ni-NTA affinity column (GE Healthcare), and the 6xHis-SUMO tag was cleaved by ULP1 along with dialysis in buffer containing 20 mM Tris (pH 8.0), 500 mM NaCl, and 5 mM β-mercaptoethanol. After dialysis, the proteins were reloaded onto a second Ni-NTA affinity column (GE Healthcare) to remove the cleaved 6xHis-SUMO tag. The proteins were further purified by chromatography using a HiTrap Heparin SP column (GE Healthcare), followed by gel filtration on a HiLoad Superdex 75 16/60 column (GE Healthcare). The final protein sample for crystallization was concentrated to ∼8 mg/mL in buffer containing 20 mM HEPES (pH 6.5), 150 mM NaCl, 5 mM DTT, and 20 mM MgCl2.
BEND3-BD4–DNA complex preparation and crystallization
To generate the BEND3-BD4–DNA complex, a 13-mer DNA duplex (5′-GGACCCACGCAGC-3′/3′-CTGGGTGCGTCGG-5′) containing a central binding site was incubated with human BEND3-BD4 (715–828) in a 1:1.5 molar ratio for 1 h on ice. The 13-mer DNA duplex (purchased from Sangon Biotech) was purified by ethanol precipitation, annealed for 5 min at 95°C, and cooled for 30 min on ice before incubation with human BEND3-BD4 (715–828) protein. Crystals of the BEND3-BD4–DNA complex were obtained under 0.1 M BIS-TRIS (pH 6.5) and 25% PEG3350 conditions. The crystals were directly flash-frozen in liquid nitrogen for data collection.
Structure determination
All X-ray diffraction data for the BEND3-BD4–DNA complex were collected at 100 K on beamline BL19U1 at the Shanghai Synchrotron Radiation Facility (SSRF) and processed with HKL2000 (HKL Research). The space group used was C2221. We solved the tertiary structure of the BEND3-BD4–DNA complex by molecular replacement using the Insv-BEN(251-365)–DNA complex (PDB: 4IX7) as a search model. Then, the model was manually built and adjusted using the program Coot (Emsley et al. 2010), followed by refinement of the structure with PHENIX (Adams et al. 2002). The Rwork and Rfree of the final model were 0.16 and 0.19, respectively. There was one molecule of the BEND3-BD4–DNA complex in the asymmetric unit (ASU). Each DNA duplex molecule bound to one protein molecule. The crystallographic statistics of X-ray data collection and refinement are listed in Table 1.
Data access
The protein binding microarray (PBM) data and statistics are provided as Supplemental Data Sets S1–S4. The raw HT-SELEX data are available in the NCBI Short Read Archive (SRA) and European Nucleotide Archive (ENA) under accession PRJEB49150. The coordinates of the BEND3-BD4:DNA complex were deposited in the RSCB Protein Data Bank (PDB) under accession 7W27. The GEO accession number for BEND3 ChIP-seq data is GSE151235.
Supplementary Material
Acknowledgments
We thank Nan Pang for helping to clone BEN domain constructs for protein expression. Work in A.R.'s group was supported by the National Key Research and Development Project of China (2021YFC2300300) and the National Natural Science Foundation of China (32022039, 31870810, 91940302, and 91640104). Work in S.G.P.’s group was supported by the National Science Foundation (1243372 and 1818286) and National Institutes of Health (NIH; GM125196). Work in Y.Y.’s group was supported by the National Natural Science Foundation of China (32170550), State Key Laboratory Special Fund 2060204, the Special Research Fund for Central Universities, Peking Union Medical College (2020-RC310-003 and 2020-RC310-011), and CAMS (Chinese Academy of Medical Sciences) Innovation Fund for Medical Sciences (CIFMS) 2021-I2M-1-019. Work in E.C.L.’s group was supported by the NIH (R01-NS083833 and R01-NS074037) and Memorial Sloan Kettering Core Grant P30-CA008748.
Author contributions: L. Zheng, L. Zhang, and A.R. determined the structure of the BEND3-BD4 domain. J.L., L.N., and Y.Y. performed the functional and cellular analyses of BEND3. J.L. and Y.Y. performed BEN domain structural comparisons using Alphafold2 and RoseTTA. Q.D., A.W.H.Y., A.J., and T.R.H. performed binding specificity determination analyses of GST-BEN domain proteins. M.K. and S.G.P. contributed the BEND3 ChIP-seq data. A.R., T.R.H., D.J.P., Y.Y., and E.C.L. supervised the project and helped interpret data. A.R., Y.Y., and E.C.L. wrote the manuscript with input from all coauthors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348993.121.
Competing interest statement
The authors declare no competing interests.
References
- Abhiman S, Iyer LM, Aravind L. 2008. BEN: a novel domain in chromatin factors and DNA viral proteins. Bioinformatics 24: 458–461. 10.1093/bioinformatics/btn007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954. 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- Aghajanirefah A, Nguyen LN, Ohadi M. 2016. BEND3 is involved in the human-specific repression of calreticulin: implication for the evolution of higher brain functions in human. Gene 576: 577–580. 10.1016/j.gene.2015.10.040 [DOI] [PubMed] [Google Scholar]
- Aoki T, Sarkeshik A, Yates J, Schedl P. 2012. Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. Elife 1: e00171. 10.7554/eLife.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, et al. 2021. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373: 871–876. 10.1126/science.abj8754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659. 10.1093/bioinformatics/btr261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Machanick P. 2012. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40: e128. 10.1093/nar/gks433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, Noble WS. 2015. The MEME suite. Nucleic Acids Res 43: W39–W49. 10.1093/nar/gkv416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW 3rd, Bulyk ML. 2006. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol 24: 1429–1435. 10.1038/nbt1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, Perrera V, Scaranaro M, de la Rosa-Velazquez IA, van de Nobelen S, Shukeir N, Popow J, Gerle B, Opravil S, Pagani M, et al. 2012. A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol 19: 1023–1030. 10.1038/nsmb.2382 [DOI] [PubMed] [Google Scholar]
- Christy B, Nathans D. 1989. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci 86: 8737–8741. 10.1073/pnas.86.22.8737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Andreu-Agullo C, Insolera R, Wong LC, Shi SH, Lai EC. 2013a. BEND6 is a nuclear antagonist of Notch signaling during self-renewal of neural stem cells. Development 140: 1892–1902. 10.1242/dev.087502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Ren AM, Westholm JO, Serganov AA, Patel DJ, Lai EC. 2013b. The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev 27: 602–614. 10.1101/gad.213314.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Ren A, Westholm JO, Duan H, Patel DJ, Lai EC. 2015. Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family. Genes Dev 29: 48–62. 10.1101/gad.252122.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000. 10.1016/0092-8674(87)90585-X [DOI] [PubMed] [Google Scholar]
- Duan H, Dai Q, Kavaler J, Bejarano F, Medranda G, Nègre N, Lai EC. 2011. Insensitive is a corepressor for Suppressor of Hairless and regulates Notch signalling during neural development. EMBO J 30: 3120–3133. 10.1038/emboj.2011.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeilzadeh-Gharehdaghi E, Banan M, Farashi S, Mirabzadeh A, Farokhashtiani T, Hosseinkhani S, Heidari A, Najmabadi H, Ohadi M. 2011. Support for down-tuning of the calreticulin gene in the process of human evolution. Prog Neuropsychopharmacol Biol Psychiatry 35: 1770–1773. 10.1016/j.pnpbp.2011.06.009 [DOI] [PubMed] [Google Scholar]
- Farokhashtiani T, Mirabzadeh A, Olad Nabi M, Magham ZG, Khorshid HR, Najmabadi H, Ohadi M. 2011. Reversion of the human calreticulin gene promoter to the ancestral type as a result of a novel psychosis-associated mutation. Prog Neuropsychopharmacol Biol Psychiatry 35: 541–544. 10.1016/j.pnpbp.2010.12.012 [DOI] [PubMed] [Google Scholar]
- Fedotova A, Aoki T, Rossier M, Mishra RK, Clendinen C, Kyrchanova O, Wolle D, Bonchuk A, Maeda RK, Mutero A, et al. 2018. The BEN domain protein insensitive binds to the Fab-7 chromatin boundary to establish proper segmental identity in Drosophila. Genetics 210: 573–585. 10.1534/genetics.118.301259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedotova A, Clendinen C, Bonchuk A, Mogila V, Aoki T, Georgiev P, Schedl P. 2019. Functional dissection of the developmentally restricted BEN domain chromatin boundary factor insensitive. Epigenetics Chromatin 12: 2. 10.1186/s13072-018-0249-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. 2008. PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci 105: 6057–6062. 10.1073/pnas.0711961105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740. 10.1038/nprot.2012.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go CK, Gross S, Hooper R, Soboloff J. 2019. EGR-mediated control of STIM expression and function. Cell Calcium 77: 58–67. 10.1016/j.ceca.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand RS, Burger L, Grawe C, Michael AK, Isbel L, Hess D, Hoerner L, Iesmantavicius V, Durdu S, Pregnolato M, et al. 2021. BANP opens chromatin and activates CpG-island-regulated genes. Nature 596: 133–137. 10.1038/s41586-021-03689-8 [DOI] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. 2011. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475: 386–389. 10.1038/nature10116 [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. 2016. Cell fate control by pioneer transcription factors. Development 143: 1833–1837. 10.1242/dev.133900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. 2013. DNA-binding specificities of human transcription factors. Cell 152: 327–339. 10.1016/j.cell.2012.12.009 [DOI] [PubMed] [Google Scholar]
- Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. 2015. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527: 384–388. 10.1038/nature15518 [DOI] [PubMed] [Google Scholar]
- Kaul R, Mukherjee S, Ahmed F, Bhat MK, Chhipa R, Galande S, Chattopadhyay S. 2003. Direct interaction with and activation of p53 by SMAR1 retards cell-cycle progression at G2/M phase and delays tumor growth in mice. Int J Cancer 103: 606–615. 10.1002/ijc.10881 [DOI] [PubMed] [Google Scholar]
- Khan A, Giri S, Wang Y, Chakraborty A, Ghosh AK, Anantharaman A, Aggarwal V, Sathyan KM, Ha T, Prasanth KV, et al. 2015. BEND3 represses rDNA transcription by stabilizing a NoRC component via USP21 deubiquitinase. Proc Natl Acad Sci 112: 8338–8343. 10.1073/pnas.1424705112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Brostromer E, Xing D, Jin J, Chong S, Ge H, Wang S, Gu C, Yang L, Gao YQ, et al. 2013. Probing allostery through DNA. Science 339: 816–819. 10.1126/science.1229223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korutla L, Degnan R, Wang P, Mackler SA. 2007. NAC1, a cocaine-regulated POZ/BTB protein interacts with CoREST. J Neurochem 101: 611–618. 10.1111/j.1471-4159.2006.04387.x [DOI] [PubMed] [Google Scholar]
- Korutla L, Wang P, Jackson TG, Mackler SA. 2009. NAC1, a POZ/BTB protein that functions as a corepressor. Neurochem Int 54: 245–252. 10.1016/j.neuint.2008.12.008 [DOI] [PubMed] [Google Scholar]
- Kurniawan F, Chetlangia N, Kamran M, Redon CE, Pongor L, Sun Q, Lin Y-C, Mohan V, Shaqildi O, Asoudegi D, et al. 2022. BEND3 safeguards pluripotency by repressing differentiation-associated genes. PNAS (in press). [DOI] [PMC free article] [PubMed]
- Lam KN, van Bakel H, Cote AG, van der Ven A, Hughes TR. 2011. Sequence specificity is obtained from the majority of modular C2H2 zinc-finger arrays. Nucleic Acids Res 39: 4680–4690. 10.1093/nar/gkq1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT. 2018. The human transcription factors. Cell 172: 650–665. 10.1016/j.cell.2018.01.029 [DOI] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547: 236–240. 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A, Højfeldt JW, Helin K. 2019. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol Cell 74: 8–18. 10.1016/j.molcel.2019.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hong T, Parameswaran S, Ernst K, Marazzi I, Weirauch MT, Fuxman Bass JI. 2020. Human virus transcriptional regulators. Cell 182: 24–37. 10.1016/j.cell.2020.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B, Grummt I. 2008. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol 24: 131–157. 10.1146/annurev.cellbio.24.110707.175259 [DOI] [PubMed] [Google Scholar]
- Nakayama N, Sakashita G, Nagata T, Kobayashi N, Yoshida H, Park SY, Nariai Y, Kato H, Obayashi E, Nakayama K, et al. 2020. Nucleus accumbens-associated protein 1 binds DNA directly through the BEN domain in a sequence-specific manner. Biomedicines 8: 608. 10.3390/biomedicines8120608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan K, Lambert SA, Yang AW, Riddell J, Mnaimneh S, Zheng H, Albu M, Najafabadi HS, Reece-Hoyes JS, Fuxman Bass JI, et al. 2015. Mapping and analysis of Caenorhabditis elegans transcription factor sequence specificities. Elife 4: e06967. 10.7554/eLife.06967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardelli J, Gibson TJ, Vesque C, Charnay P. 1991. Base sequence discrimination by zinc-finger DNA-binding domains. Nature 349: 175–178. 10.1038/349175a0 [DOI] [PubMed] [Google Scholar]
- Ng AHM, Khoshakhlagh P, Rojo Arias JE, Pasquini G, Wang K, Swiersy A, Shipman SL, Appleton E, Kiaee K, Kohman RE, et al. 2021. A comprehensive library of human transcription factors for cell fate engineering. Nat Biotechnol 39: 510–519. 10.1038/s41587-020-0742-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta KR, Jolma A, Yin Y, Morgunova E, Kivioja T, Akhtar J, Hens K, Toivonen J, Deplancke B, Furlong EE, et al. 2015. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. Elife 4: e04837. 10.7554/eLife.04837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchai GP, Kaulich M, Bizard AH, Mesa P, Yao Q, Sarlos K, Streicher WW, Nigg EA, Montoya G, Hickson ID. 2017. A novel TPR–BEN domain interaction mediates PICH–BEND3 association. Nucleic Acids Res 45: 11413–11424. 10.1093/nar/gkx792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher FJ 3rd, Morris JF, Tournay OE, Cook DM, Curran T. 1990. Binding of the Wilms’ tumor locus zinc finger protein to the EGR-1 consensus sequence. Science 250: 1259–1262. 10.1126/science.2244209 [DOI] [PubMed] [Google Scholar]
- Reiter F, Wienerroither S, Stark A. 2017. Combinatorial function of transcription factors and cofactors. Curr Opin Genet Dev 43: 73–81. 10.1016/j.gde.2016.12.007 [DOI] [PubMed] [Google Scholar]
- Saksouk N, Barth TK, Ziegler-Birling C, Olova N, Nowak A, Rey E, Mateos-Langerak J, Urbach S, Reik W, Torres-Padilla ME, et al. 2014. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol Cell 56: 580–594. 10.1016/j.molcel.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD, Narlikar GJ. 2019. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575: 390–394. 10.1038/s41586-019-1669-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyan KM, Shen Z, Tripathi V, Prasanth KV, Prasanth SG. 2011. A BEN-domain-containing protein associates with heterochromatin and represses transcription. J Cell Sci 124: 3149–3163. 10.1242/jcs.086603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich TG, Bugert JJ, Sisler JR, Koonin EV, Darai G, Moss B. 1996. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science 273: 813–816. 10.1126/science.273.5276.813 [DOI] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547: 241–245. 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo E, Rampalli S, Risueño RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. 2010. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468: 521–526. 10.1038/nature09591 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Zidek A, Bridgland A, Cowie A, Meyer C, Laydon A, et al. 2021. Highly accurate protein structure prediction for the human proteome. Nature 596: 590–596. 10.1038/s41586-021-03828-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueberschär M, Wang H, Zhang C, Kondo S, Aoki T, Schedl P, Lai EC, Wen J, Dai Q. 2019. BEN-solo factors partition active chromatin to ensure proper gene activation in Drosophila. Nat Commun 10: 5700. 10.1038/s41467-019-13558-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. 2010. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463: 1035–1041. 10.1038/nature08797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gao Y, Zheng X, Liu C, Dong S, Li R, Zhang G, Wei Y, Qu H, Li Y, et al. 2019. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol Cell 76: 646–659.e6. 10.1016/j.molcel.2019.08.019 [DOI] [PubMed] [Google Scholar]
- Weirauch MT, Cote A, Norel R, Annala M, Zhao Y, Riley TR, Saez-Rodriguez J, Cokelaer T, Vedenko A, Talukder S, et al. 2013. Evaluation of methods for modeling transcription factor sequence specificity. Nat Biotechnol 31: 126–134. 10.1038/nbt.2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. 10.1016/j.cell.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan C, Wang Q, Han X, Duan Y, Li L, Shi L, Wang Y, Shan L, Yao Z, Shang Y. 2013. RBB, a novel transcription repressor, represses the transcription of HDM2 oncogene. Oncogene 32: 3711–3721. 10.1038/onc.2012.386 [DOI] [PubMed] [Google Scholar]
- Yamamizu K, Piao Y, Sharov AA, Zsiros V, Yu H, Nakazawa K, Schlessinger D, Ko MS. 2013. Identification of transcription factors for lineage-specific ESC differentiation. Stem Cell Reports 1: 545–559. 10.1016/j.stemcr.2013.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.