Abstract
A plethora of modified nucleotides extends the chemical and conformational space for natural occurring RNAs. tRNAs constitute the class of RNAs with the highest modification rate. The extensive modification modulates their overall stability, the fidelity and efficiency of translation. However, the impact of nucleotide modifications on the local structural dynamics is not well characterized. Here we show that the incorporation of the modified nucleotides in tRNAfMet from Escherichia coli leads to an increase in the local conformational dynamics, ultimately resulting in the stabilization of the overall tertiary structure. Through analysis of the local dynamics by NMR spectroscopic methods we find that, although the overall thermal stability of the tRNA is higher for the modified molecule, the conformational fluctuations on the local level are increased in comparison to an unmodified tRNA. In consequence, the melting of individual base pairs in the unmodified tRNA is determined by high entropic penalties compared to the modified. Further, we find that the modifications lead to a stabilization of long-range interactions harmonizing the stability of the tRNA’s secondary and tertiary structure. Our results demonstrate that the increase in chemical space through introduction of modifications enables the population of otherwise inaccessible conformational substates.
INTRODUCTION
tRNAs show the highest modification rates of all known RNAs. Among the over 170 modified nucleotides known so far at least 123 occur in tRNAs (1–5). Remarkably, in Escherichia coli one percent of the genome codes for proteins involved in post-transcriptional nucleotide modification. This highlights the functional impact of the increased chemical diversity on tRNA biology (6–13).
However, irrespective of the number and kind of modifications and of the vast different primary structures (14), all tRNAs adopt the same secondary (cloverleaf) and highly folded tertiary structure (L-shape). This iconic three dimensional structure was first shown for yeast tRNAPhe (15). It is virtually identical for all tRNAs across the biological kingdom. As common for all tRNAs, its secondary cloverleaf structure is divided into five parts, the acceptor stem (Acc. stem), the dihydrouridine arm (D-arm), the anticodon stem loop (ACSL), a variable loop and the TΨC-arm. The typical L-shaped tertiary structure is mainly formed by a plethora of interactions between the D-arm and TΨC-arm.
Modifications within tRNA are classified by their functional role (16). Modifications in the anticodon stem loop (ACSL) modulate the cognate codon-anticodon interaction and therefore have a regulatory function during decoding (17,18). In contrast, modifications in the core of the tRNA influence the tertiary fold and thus are mainly found at the interface between D- and TΨC-arm. They maintain the balance between flexibility and stability within the L-shaped fold (19). The structural impact of modifications depends on their type and position within the tRNA. It ranges from influencing the hydrophobic character of a base, its stacking ability and base pairing properties up to the net charge of the nucleotide (reviewed in (20)).
Among all tRNAs, tRNAfMet possesses not only a unique function but also at least three unique structural features. In bacteria, it initiates translation by decoding the start codon AUG of the mRNA. tRNAfMet directly binds to the P-site of the ribosome, whereas elongator-tRNAs first bind to the ribosomal A-site and then translocate to the P-site. The ability of initiator tRNAfMet to bind directly to the P-site is attributed to the formylated methionine. Structurally, tRNAfMet contains three uniquely conserved GC base pairs (21,22) in the ACSL (G29-C41, G30-C40 and G31-C39). A C1-A72 mismatch functions as recognition site for the formylation (23). The base pair A11-U24 represents a purine-pyrimidine pair, whereas all other tRNA contain a pyrimidine-purine base pair at this position (23). In addition, the structure contains the wobble-like Cm32:A38 base pair (24). The atomic details of the structure were solved by X-ray crystallography (25) and the most recent structure exhibits a resolution of 3.1 Å (24). Besides the Cm32 modification in the ACSL, four core modifications were found: 4-thiouridine 8 (s4U8), dihydrouridine 20 (D20), ribothymidine 54 (T54) and pseudouridine 55 (Ψ55). All the core modifications except dihydrouridine are generally assumed to thermally stabilize the tRNA structure due to the higher structural rigidity of the nucleosides introduced by these modifications (20,26).
A number of studies have focused on the role of singular modifications regarding structure or function (19,27–29). For example, it could be shown that T54 has a high impact on the thermal stability of elongator tRNAMet (29). Thiolation of U8 changes base pairing properties, functions as UV protective in tRNAs and stabilizes tRNA structure (20,30,31). The thio-modification at position U8 is also highly conserved in bacteria.
Exhibiting a non-aromatic and non-planar base, dihydrouridine cannot contribute to stacking interactions in helices and thus is supposed to increase the flexibility of the D-arm (19,27). Pseudouridine (Ψ) being one of the most abundant base modification, has, due to its C–C sugar bond, an additional imino group involved in hydrogen bonding, e.g. with localized water molecules (27,32). Moreover, it can base pair with all four canonical bases. The Ψ modification can be found over the whole tRNA primary structure, but Ψ55 is highly conserved. However, little is known about the impact of the entirety of modifications on the local and global dynamics of a tRNA molecule (33). It has been hypothesized that the modifications lead to a stabilization of tRNA structure by simultaneously maintaining its flexibility needed for their proper biological function. Comparative structural analyses revealed that the overall structures of tRNAs are identical (34,35), although modifications can increase the surface area of tRNA up to 20% (26). Interestingly, unmodified tRNAs can adopt several structural conformations (34,36–42), highlighting the fact that in several cases the modifications are needed to restrict the conformational space. Along this line, it could be shown that in vitro in absence or at low concentrations (<2 mM) of Mg2+, the tertiary fold of non-modified are less stable than modified tRNAs (35,43). Further, some results hint at a variation in the local stabilities that influence the structural behavior of the tRNAs at saturating (44) (>5 mM) concentrations of Mg2+. Recently, the structural comparison between the crystal structure of E. coli tRNAPhe and the yeast analogue revealed a different angle between the acceptor and anticodon stems as well as a different arrangement of a triplet base pair (42). On the other hand, experiments investigating differences between non-modified and modified E. coli tRNAVal showed that modifications do not alter the global structure (34). It is important to note, that the concentration of Mg2+ critically influences the globular arrangement of the tRNA and under non-saturating concentrations globular structural differences might occur between modified and non-modified tRNAs (45). Investigation of local base pair dynamics were performed by deuterium exchange rates at 5°C showing a difference in dynamics between non-modified and modified tRNA. However, detailed investigations about local base pair dynamics are still missing.
Here, we investigate the impact of modifications on the structural dynamics of E. coli tRNAfMet at single nucleotide resolution by nuclear magnetic resonance (NMR) spectroscopy. We performed a comprehensive analysis between modified and non-modified tRNAs on different time scales to evaluate the local and global dynamics. We further investigated the global stability and structural plasticity of both tRNAs in comparison. As reported before, we find that the introduction of modifications does not alter the global structure of tRNA by means of nucleotide interactions responsible for the formation of secondary and tertiary interactions (34–44). However, our data clearly shows that the introduction of nucleotide modifications preserves or even enhances local dynamics. This affects also long-range interactions of nucleobases that get stabilized over a wide temperature range. Although the modifications are silent with respect to the overall structure, in consequence of these two effects the thermodynamic stability of tRNAfMet’s secondary and tertiary structure gets harmonized. Through this harmonization the two seemingly opposing properties of having a structural stable but at the same time flexible tRNA molecule are enabled. Our results lead to new insights for understanding how the modifications interfere with and modulate local dynamics and are correlated to the global stability of tRNAs.
MATERIALS AND METHODS
Preparation of unmodified tRNAfMet
The DNA template of 77 nt tRNAfMet from E. coli is flanked by two restriction sites and a hammerhead ribozyme: (transcribed sequence underlined) GAATTC (EcoRI) – TAATACGACTCACTATAG (T7 promotor) – GGACCCCGCGCTGATGAGTCCGTGAGGACGAAAGACCGTCTTCGGACGGTCTC (hammerhead ribozyme) – CGCGGGGUGGAGCAGCCUGGUAGCUCGUCGGGCUCAUAACCCGAAGAUCGUCGGUUCAAAUCCGGCCCCCGCAACCA (tRNAfMet) – TATG (NdeI).
Plasmid DNA was purchased from Genscript (New Jersey, USA). DNA was amplified by polymerase chain reaction (PCR) using T7 forward primer (5′-TAATACGACTCACTATAGG-3′) and tRNA reverse primer (5′-TGGTTGCGGGGGCC-3′). Primers were purchased from Eurofins MWG Operon (Ebersberg, Germany).
13C–15N-labeled 77 nt tRNAfMet was synthesized by in vitro transcription with T7 RNA polymerase from PCR product as described in literature (46,47). The 13C–15N-labeled (rATP, rCTP, rGTP, rUTP) nucleotides for transcription were purchased from Silantes (Munich, Germany). The construct was purified by preparative polyacrylamide gel electrophoresis according to standard protocols (48). The construct was folded in water for 5 min at 95°C and immediately diluted 10-fold with ice-cold water. The RNA was buffer-exchanged into NMR buffer (25 mM potassium phosphate, 200 mM KCl, 5 mM MgCl2, pH 6.2).
Preparation of modified tRNAfMet
The plasmid DNA template (pBStRNAfmetY2) (49) was kindly provided by Emanuelle Schmitt. The gene of interest is flanked by a lpp promoter, a 5′-flanking sequence, and a rrnC terminator sequence (50–52). The plasmid was transformed into competent JM101tr cells.
For expression of native tRNAfMet, pre-culture was grown in 5 ml LB medium at 37°C for 4 h. Afterwards, the culture was centrifuged at 4000 g for 5 min and the supernatant was discarded. The pellet was resuspended in 100 ml M9 medium, which contained uniformly labeled 13C-glucose and 15N-ammonium chloride as the sole carbon and nitrogen supplies. Cell cultures were grown at 37°C overnight. On the next day, the culture was transferred to 2 l of uniformly labeled 13C,15N-M9 medium for further expression. The culture was incubated at 37°C for 24 h, the culture was harvested, and the supernatant was discarded.
The cell pellet obtained from the expression was resuspended in 10 ml of buffer T1 (1 mM Tris–HCl, pH 7.4, 10 mM MgCl2). The suspension was mixed with one equivalent of T1-saturated phenol. Thereafter, the mixture was vortexed for 3 min and then slowly shaken for 1 h. After incubation, the preparation was centrifuged at 10 000 g and RT for 1 h to separate the phenol and the water phase. The water phase was carefully recovered, whereas the phenol phase was mixed again with 2 mL of buffer T1 for counter-extraction. The total soluble phase, which contains total nucleic acids from bacteria, was mixed with 1 volume equivalent chloroform to remove remaining phenol. Again, phase separation was achieved by centrifugation at 10 000 g and 25°C for 20 min. After recovering of the soluble phase, the preparation was mixed with 5 M NaCl solution to reach a final concentration of 0.5 M and then mixed with 2.5 volume equivalents of ice-cold ethanol. Before centrifuging the mixture at 10 000 g and 4°C for 1 h, the mixture was vortexed for 10 s. The supernatant was discarded, and the pellet was resuspended in 5 ml of 1 M sodium chloride. The preparation was again centrifuged at 10 000 g and 4°C for 1 h. Ice-cold ethanol (2.5 volume equivalents) was added to the supernatant and centrifuged again at 10 000 g and 4°C for 1 h. The supernatant was discarded, and the pellet was resuspended in 2 ml of 1.8 M Tris–HCl buffer (pH 8.0). Afterwards, the preparation was incubated at 37°C for 2 h to insure a full deacylation of all tRNA molecules. The preparation, which contains total tRNA extract without any aminoacylation, was mixed with 0.2 ml 5 M NaCl and 2.5 volume equivalents ice-cold ethanol for precipitation. After centrifuging, the preparation at 10 000 g and 4°C for 1 h, the pellet was resuspended in 4 ml of T2 buffer (20 mM Tris–HCl, pH 7.5, 0,1 mM EDTA, 0.2 M NaCl) for further purification by anion exchange chromatography.
Anion exchange chromatography was performed to purify the native tRNAfMet construct. The corresponding protocol was modified (49). For this, a HiPrep Q Sepharose Fast-Flow column (GE Healthcare) was connected to an Äkta® purifier system (GE Healthcare) and RNA of various concentration was injected. The flow rate was set to 5 ml/min. Unbound RNA fragments were eluted with one column volume (CV, 53 ml) at a sodium chloride concentration of 0.48 M. A gradient was applied from 0.48 to 0.52 M sodium chloride. The change of salt concentration per hour was 0.06 M/h. An isocratic flow step at 0.52 M was then applied for either 5 CV, 6 CV or 8 CV. A second gradient (0.06 M/h) was applied from 0.516 to 0.544 M sodium chloride followed by a further, but steeper gradient (2 M/h) from 0.544 to 1 M to elute all remaining RNA. Eluate was collected and analyzed with denaturing PAGE.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
150 ng of total RNA per sample was digested to nucleosides using 0.6 U nuclease P1 from P. citrinum (Sigma-Aldrich), 0.2 U snake venom phosphodiesterase from C. adamanteus (Worthington), 0.2 U bovine intestine phosphatase (Sigma-Aldrich), 10 U benzonase (Sigma-Aldrich), 200 ng Pentostatin (Sigma-Aldrich) and optional 500 ng tetrahydrouridine (Merck-Millipore) in 5 mM Tris (pH 8) and 1 mM magnesium chloride for two hours at 37°C. 30 ng of digested RNA was mixed with 25 ng of internal standard (13C stable isotope-labeled nucleosides from E. coli) and subjected to LC–MS analysis. Nucleoside levels were measured using an Agilent 1260 Infinity system in combination with an Agilent 6470 Triple Quadrupole mass spectrometer equipped with an electrospray ion source (ESI)). The solvents consisted of 5 mM ammonium acetate buffer (pH 5.3, adjusted with acetic acid; solvent A) and LC–MS grade acetonitrile (solvent B; Honeywell). A C18 reverse HPLC column (Synergi™ 4 μM particle size, 80 Å pore size, 250 × 2.0 mm; Phenomenex) was used at a temperature of 35°C and a constant flow rate of 0.35 ml/min was applied. The compounds were eluted with a linear gradient of 0–8% solvent B over 10 min, followed by 8–40% solvent B over 10 min. Initial conditions were regenerated with 100% solvent A for 10 min. The four main nucleosides were detected photometrically at 254 nm via a diode array detector (DAD). The following ESI parameters were used: gas temperature 300°C, gas flow 7 l/min, nebulizer pressure 60 psi, sheath gas temperature 400°C, sheath gas flow 12 l/min, capillary voltage 3000 V, nozzle voltage 0 V. The MS was operated in the positive ion mode using Agilent MassHunter software in the dynamic MRM (multiple reaction monitoring) mode. For absolute quantification, internal and external calibration was applied as described in Thüring et al. (53). The total amount of modified nucleosides was normalized to the amount of injected RNA molecules.
Circular dichroism (CD)
For the determination of the melting behavior of the tRNAfMet, CD melting profiles were measured. One sample from each construct was therefore prepared with an absorption at 260 nm of ∼1–2 AU.
All CD spectra and melting curves were acquired on a JASCO J-810 spectropolarimeter with constant influx of nitrogen to prevent formation of ozone. The spectra were acquired in a quartz cuvette with 1 mm path length with a sample volume of 200 μl. Melting curves were acquired at constant wavelength (265 nm), where largest change in molar ellipticity upon heat denaturation was expected, typically using a temperature ramp rate of 0.5°C/min.
The melting curves were evaluated according to the method of Marky and Breslauer (54). In short, two baselines representing the maximal and minimal value of CD ellipticity were determined. The percentage of folded RNA was then calculated by normalizing the measured CD ellipticity as described. Percentage of folded RNA was then non-linear regressed against the temperature with the following equation:
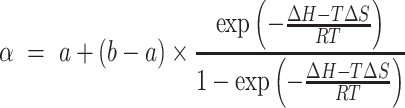 |
NMR spectroscopy
All experiments were performed at 25°C in NMR buffer (25 mM potassium phosphate, 200 mM KCl, 5 mM MgCl2, pH 6.2) and 5–10% D2O. All spectra were referenced to DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid). Nitrogen-15 and Carbon-13 chemical shifts were indirectly referenced using the ratio of the gyromagnetic ratios of proton to 15N (0.101329118) and 13C (0.251449530), respectively (55). NMR experiments were performed on 700, 800, 900 or 950 MHz NMR spectrometer equipped with a 5 mm, z-axis gradient 1H {13C, 15N} TCI cryogenic probe. NMR Experiments were analyzed using Bruker Biospin software TopSpin 3.5. Assignments were performed using the software Sparky 3.114 (56).
2D-[1H,15N]-BEST-TROSY and 2D-[1H,15N]-HSQC experiments
BEST-TROSY (Band-selective excitation short-transient – transverse relaxation-optimized spectroscopy) and HSQC (Heteronuclear single quantum coherence) experiments were performed to observe 15N–1H correlations of uniformly 13C,15N-isotope labeled tRNAfMet (57–59). Moreover, signal of imino protons can only be detected for nucleotides in stable base pairs, which protects the imino proton from fast solvent exchange (46). However, the ‘imino’ proton of the 5,6-dihydrouridine (D) can be observed even if the nucleoside is not part of a stable base pair due to its higher pKa (19). For BEST-TROSY experiments, the pulse program with the modification proposed by Brutscher and coworkers was used (58,59). For each experiment, the used parameters are listed in the respective figure caption.
2D-[1H,15N]-HNN-COSY and 3D-[1H,15N,15N]-HNN-COSY experiments
The HNN-COSY experiment correlates a NH group with a hydrogen-bonded N-atom by a hydrogen bond scalar coupling (HBC) (60). 2D-[1H,15N]-HNN-COSY and 3D-[1H,15N,15N]-HNN-COSY experiments were acquired with 45 ms and 30 ms 15N–15N transfer delay, respectively.
2D-[1H,1H]-NOESY and 3D-[H1H,15N,1H]-SOFAST-HMQC-NOESY experiments
2D-[1H,1H]-NOESY and 3D-[1H,15N,1H]-SOFAST-HMQC-NOESY experiments were acquired using a pulse program with jump-return water suppression (2D-NOESY) or Watergate water suppression (3D-NOESY) and a carrier frequency switch between the direct and indirect dimension. For the direct dimension, the proton carrier frequency was set to the water resonance frequency (4.7 ppm), whereas it was switched to either 8.85 ppm or 7.29 ppm in the indirect dimension. The nitrogen carrier frequency was set to 153.25 ppm. All spectra were recorded with a spectral width of 25 ppm in the direct dimension, 16 ppm in the indirect proton dimension and to either 19.2 ppm or 22.5 ppm in the indirect nitrogen dimension. The mixing time for the through-space coherence transfer was set to 150 ms.
Sofast-1H,31P HMQC experiments
For the HP-correlation across hydrogen bond a sofast-HMQC experiment (61) was conducted on a Bruker AVIII 700MHz spectrometer equipped with a cryogenic-QCI HCNP probe. The transfer timer was set to 16.6ms, and pulses were applied at frequencies of 12.5, 0 and 135 ppm for 1H, 31P and 15N, respectively. Selective proton pulses were applied with a bandwidth of 4.8 ppm. Hard 31P pulses were applied with a bandwidth of 5.81 kHz and 15N decoupling with a bandwidth of 1 kHz. The recycling delay was set to 0.5 s.
Determination of 15N relaxation parameters
15N relaxation parameters were measured as described (62,63) and were determined with the program DynamicsCenter, version 2.6.2 (Bruker Biospin). A delay of either 1.5 s at a magnetic field of 600 MHZ or 2.8 s at a magnetic field of 800 MHz was used as relaxation delay. For determination of the 15N spin-lattice relaxation rate R1 up to 20 mixing times between 20 ms and 2.8 s were applied. In case of the 15N spin-spin relaxation rate, multiples of the 16.96 ms CPMG length of up to 12 were applied. For an accurate error estimation in all measurements, two mixing times were repeated.
In short, the three parameters comprising of the spin-lattice relaxation rate R1, the spin-spin relaxation rate R2, and the steady-state heteronuclear NOE (hetNOE) were measured by a series of 2D-[1H,15N]-HSQC experiments as described in the following.
Determination of 15N-spin-lattice relaxation rates (R1) were conducted by measuring a series of inversion-recovery experiments with varying recovery delays  as a pseudo-3D-[1H,15N]-HSQC-
as a pseudo-3D-[1H,15N]-HSQC-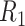 experiment. Fitting of the time-modulated signal intensities with the following equation yields the 15N-spin-lattice relaxation rate:
experiment. Fitting of the time-modulated signal intensities with the following equation yields the 15N-spin-lattice relaxation rate:
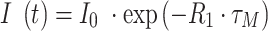 |
The 15N-spin-spin relaxation rate (R2) describes the decay of the transverse magnetization by loss of phase coherence, which is mainly driven by dipolar interaction with adjacent spins. The obtained signal intensity is plotted against the recovery delays and fitted with a mono-exponential function resulting in the 15N-R2 value:
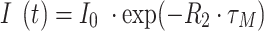 |
The 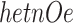 is a cross-relaxation phenomenon(64) and thus can be used to calculate the 1H-15N cross relaxation (63). The
is a cross-relaxation phenomenon(64) and thus can be used to calculate the 1H-15N cross relaxation (63). The 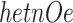 of each imino group was determined by either an interleaved 2D-[1H,15N]-HSQC-
of each imino group was determined by either an interleaved 2D-[1H,15N]-HSQC-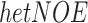 at 600 MHz or a pseudo-3D-[1H,15N]-HSQC-
at 600 MHz or a pseudo-3D-[1H,15N]-HSQC-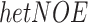 at a field strength of 800 MHz. Both experiments measure one set of data with prior proton saturation and another set without proton saturation. The
at a field strength of 800 MHz. Both experiments measure one set of data with prior proton saturation and another set without proton saturation. The 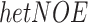 value is extracted from both sets by calculating the quotient of saturated and unsaturated signal intensity:
value is extracted from both sets by calculating the quotient of saturated and unsaturated signal intensity:
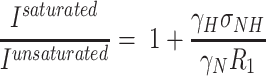 |
Model-free analysis
The determined relaxation parameters were used as input for model-free analysis (65,66) with the program relax, version (4.1.3) (67,68) using a virtual machine provided by NMRBox (69). Analysis was performed at two temperatures (298 and 313 K) with the imino bond length and imino chemical shift anisotropy determined by Grishaev et al. (70). Parameters of the motion models as described in the relax manual were fit to the data. Thereby, the diffusion seed paradigm was used as described at 298 K (71). The resulting diffusion tensor was fixed for the Model-free analysis at 313 K, the global rotational correlation time and the 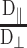 ratio was calculated as described by Fushman (72). The Model-free selection was performed using the Akaike's Information Criterion (AIC) (73).
ratio was calculated as described by Fushman (72). The Model-free selection was performed using the Akaike's Information Criterion (AIC) (73).
Measurement of selective water inversion recovery experiments
The solvent exchange rates of imino protons were measured as previously described (74–76). The resulting pseudo-2D-[1H,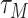 ]-jump-return experiments were measured at six different temperatures from 10°C to 55°C. In total, 18 recovery delays were applied in a range from 1 ms to 3200 ms. The buffer contained 25 mM KPi (pH 6.2), 200 mM KCl and 5 mM MgCl2. The experiment was recorded with 40 scans and 18 delays for τM. Carrier frequencies were set to 4.697 ppm (direct dimension) and 153.5 ppm (15N). The spectral width was set to 25 ppm. Radiofrequency field strength of the proton hard pulse were set to 13.47 kHz. The hard pulse for 15N decoupling was set to 6.41 kHz. The relaxation delay was set to 5 s and the acquisition time was set to 51.2 ms. Garp4 was used as a decoupling scheme during acquisition. Analysis of the imino exchange rate was applied according to the proposed theory (74–76).
]-jump-return experiments were measured at six different temperatures from 10°C to 55°C. In total, 18 recovery delays were applied in a range from 1 ms to 3200 ms. The buffer contained 25 mM KPi (pH 6.2), 200 mM KCl and 5 mM MgCl2. The experiment was recorded with 40 scans and 18 delays for τM. Carrier frequencies were set to 4.697 ppm (direct dimension) and 153.5 ppm (15N). The spectral width was set to 25 ppm. Radiofrequency field strength of the proton hard pulse were set to 13.47 kHz. The hard pulse for 15N decoupling was set to 6.41 kHz. The relaxation delay was set to 5 s and the acquisition time was set to 51.2 ms. Garp4 was used as a decoupling scheme during acquisition. Analysis of the imino exchange rate was applied according to the proposed theory (74–76). 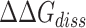 values were calculated by subtracting the individual
values were calculated by subtracting the individual 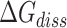 values of the non-modified tRNA from the modified tRNA values.
values of the non-modified tRNA from the modified tRNA values.
RESULTS
Modified tRNAfMet was obtained by overexpression in 13C–15N-enriched minimal medium followed by a non-denaturing purification mainly through ion-exchange chromatography. High amounts (1.6 mg/l) of pure labeled tRNAfMet suitable for NMR experiments were achieved (Supplementary Figure S1).
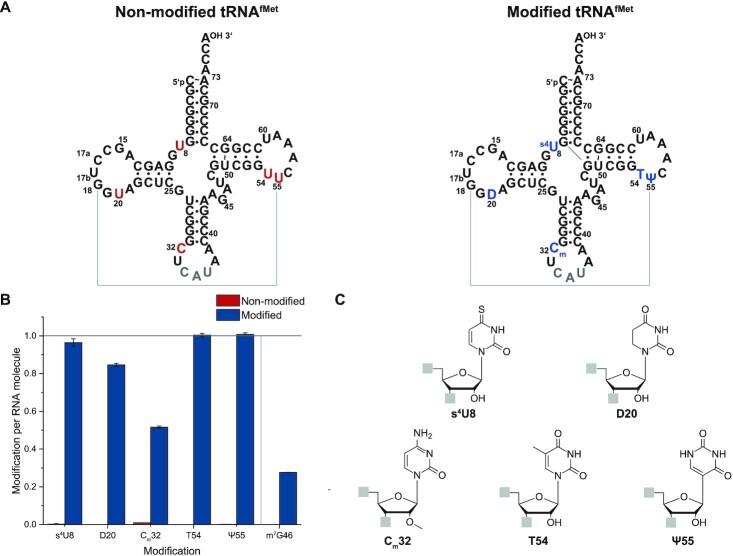
The degree of modification was evaluated by liquid chromatography-tandem mass spectrometry (LC-MS/MS). All known modifications present in E. coli’s tRNAfMet were detected. For the four modified nucleotides within the core of tRNAfMet, degrees of modification between 85 ± 1% for D20 and 101 ± 1% for Ψ55 are found (Figure 1 and Supplementary Figure S2).
Figure 1.
Quantification of modified nucleosides levels in the non-modified (red) and modified (blue) tRNAfMet constructs. (A) Secondary structure of non-modified and modified tRNAfMet. (B) LC–MS based quantification of modified nucleosides levels in tRNAfMet. Both tRNAfMet constructs were digested to nucleosides and analyzed via LC-MS. The amount of each modification was normalized to the amount of injected RNA molecules. The occurrence of one modified nucleoside per RNA molecule is highlighted with a black line. The modified nucleosides are s4U8 (thiouridine 8), D20 (5,6-dihydrouridine 20), Cm32 (2′O-methylcytidine 32), T54 (ribothymidine 54), Ψ55 (pseudouridine 55), and m7G46 (7-methylguanosine 46). The measured relative abundance for m7G46 is based on the endogenous isoacceptor of tRNAfMet in E. coli. In E. coli K strains, the initiator tRNAfMet is encoded by four genes that differ by a single nucleotide at position 46 resulting in two tRNAfMet species (79,80): metZ, metW and metV encode tRNAfMet1 with m7G46 and metY encode tRNAfMet2 with A46. Therefore, the cellular tRNAfMet pool consists of 75% tRNAG46fMet1, and 25% of tRNAA46fMet2 due to the presence of three genes encoding for m7G46 and only one gene encoding for A46 (80,81). In contrast, the results here show only a relative abundance of 27.7% and thus, the second isoacceptor reflecting the tRNAA46fMet2 of interest is successfully overexpressed. (C) Chemical structure of analyzed modified nucleosides.
The methoxy modification of Cm32 shows only a relative abundance of 52% ± 1% and thus representing a hypomodification of Cm32 in our sample. In general, it was shown that the degree of modification varies with growth and stress conditions, e.g. growth temperature or starvation (77,78). However, it was recently shown that the Cm32 modification of initiator tRNAfMet in E. coli is not affected by different growth conditions and temperatures (77).
The measured relative abundance of 27.7% ± 0.1% for m7G46 is based on the endogenous isoacceptor of tRNAfMet in E. coli. In E. coli K strains, the initiator tRNAfMet is encoded by four genes resulting in two tRNAfMet species (79): metZ, metW and metV encode tRNAfMet1 and metY encode tRNAfMet2. The tRNAs differ by a single nucleotide at position 46, where tRNAfMet1 contains an m7G46 and tRNAfMet2 an A46 (80). Therefore, the cellular tRNAfMet pool consists of 75% tRNAG46fMet, and 25% of tRNAA46fMet due to the presence of three genes encoding for m7G46 and only one gene encoding for A46 (80,81). In contrast, the results here show only a relative abundance of 27.7% and thus, the second isoacceptor reflecting the tRNAfMet of interest is successfully overexpressed.
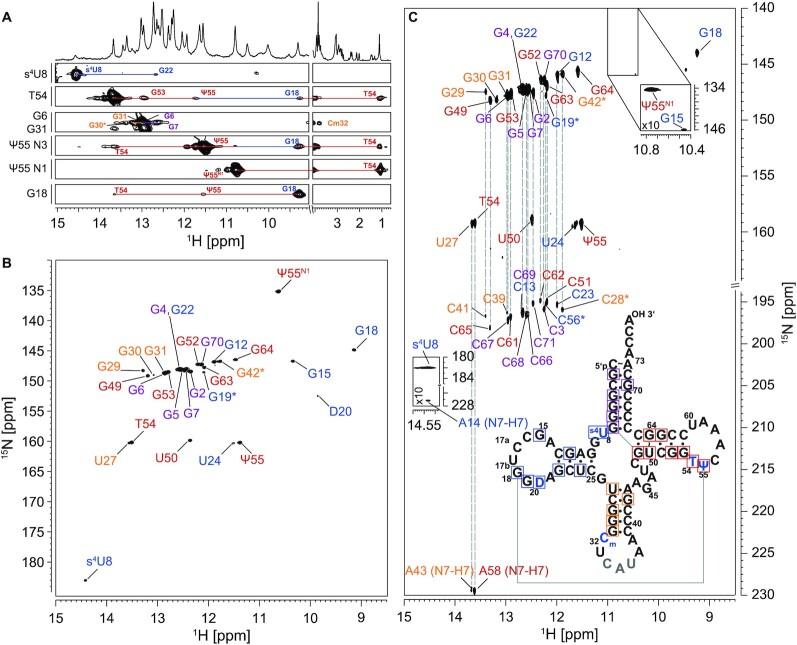
To enable the investigation of tRNA dynamics at nucleotide resolution, we used solution-state NMR spectroscopy to map the base pairing interactions. Therefore, we assigned all imino proton resonances of nucleobases involved in stable hydrogen bonds for both modified and non-modified tRNAfMet (Supplementary Table S1 and Supplementary Table S2). So far, only a subset of signals in the modified tRNA were assigned (82) and to our knowledge there is no non-modified tRNAfMet NMR assignment published yet. Here, we assigned all resonances for the modified tRNA corresponding to 96% of all base-paired residues (Figure 2, Supplementary Figure S3, Supplementary Table S3). The TΨC-arm and the acceptor stem were assigned completely and unambiguously. For the non-modified tRNA three resonances cannot unambiguously be assigned (Figure 3, Supplementary Figure S4).
Figure 2.
Selected [1H,1H]-strips of a 3D-[1H,15N,1H]-SOFAST-HMQC-NOESY (A), 2D-[1H,15N]-BEST-TROSY (B) and 2D-[1H,15N]-BEST-TROSY-HNN-COSY experiment (C) of modified tRNAfMet at 25°C. Secondary structure of the modified tRNAfMet is shown in (C). Unambiguously assigned imino peaks are highlighted in either purple (acceptor stem), blue (D-arm), yellow (ACSL), or red (TΨC-arm). Experimental details are provided in an extended figure caption in SI.
Figure 3.
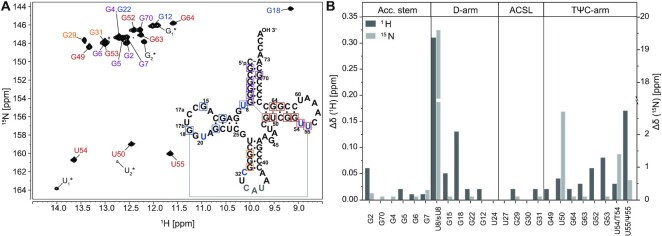
2D-[1H,15N]-BEST-TROSY experiment of unmodified tRNAfMet with secondary structure (A) and analysis of chemical shift differences between modified and non-modified tRNA (B). (A) The BEST-TROSY spectrum of the non-modified tRNAfMet was acquired at 25°C. The secondary structure of the non-modified tRNAfMet is shown. Unambiguously assigned imino peaks are highlighted in either purple (acceptor stem), blue (D-arm), orange (ACSL) or red (TΨC-arm). (B) Chemical shift difference between the imino resonances of the non-modified and modified tRNAfMet construct was calculated as absolute values of the difference between corresponding imino signals (Supplementary Table S1 and Supplementary Table S2). Experimental details are provided in an extended figure caption in SI.
As tRNAfMet contains the highest content of GC base pairs among all bacterial tRNAs (83), the chemical shift dispersion is reduced leading to resonance overlap in the guanine imino group region. The non-modified tRNA only harbors the four canonical nucleotides and does not contain any modification. This renders the assignment of the non-modified tRNA more challenging than of its modified counterpart. However, the strong NOE cross peak caused by the G64:U50 wobble base pair can be used as a starting point resulting in the complete assignment of the TΨC-arm. Furthermore, the acceptor stem can then be assigned starting from the tertiary interaction G7:G49. By overlaying the spectra of the modified construct, the two guanosine resonances of the D-arm and the three guanosine resonances of the ACSL could be assigned due to an identical pattern. This results in assignment of 72% of base-paired imino groups in the non-modified tRNA (Supplementary Table S3).
Not only the assignment of resonances in the presence of modifications is highly facilitated, but in turn also the mapping of tertiary interactions within the RNA. By NMR spectroscopy it is possible to characterize the interaction network around the modified nucleotides s4U8, Cm32, T54 and Ψ55 and monitor the unstacked residue D20.
In general, comparison of the chemical shifts for the nucleotides in the two tRNA species enables to decipher regions where the introduction of modified nucleotides leads to structural changes. Not unsurprisingly, residues in the proximity of the modifications show the strongest changes in chemical shifts as these residues experience a different chemical environment. However, the most eminent changes are observable for those in the D- and TΨC-arm (Figure 3B).
Thiouridine (s4U8), that exhibits prominent chemical shifts of its imino group (δ (1H): 14.5 ppm/δ (15N): 183 ppm), resides in a base triplet formed with A14 and A21 (s4U8:A14:A21). The Watson–Crick Hoogsteen interaction between U8-N3H3 and A14-N7 can be directly mapped via the HNN-COSY experiment through its 2hJN3N7 coupling (Figure 2C). The interaction between A21 and A14 is only eminent in the respective NOESY through-space connectivity. However, the formation of this interaction can be further corroborated from the close spatial proximity of residue G22 to U8 as eminent from their imino-to-imino cross peak (Figure 2A, Supplementary Figure S3C).
The neighboring residues T54 and Ψ55 are involved in many different non-Watson–Crick like base pairing interactions that are essential to establish the tertiary interaction between the D- and the TΨC-arm. The intra-loop interaction between T54 and A58 is mediated through formation of hydrogen bonds between the T54 Watson-Crick side and the A58 Hoogsteen edge, again detectable with the HNN-COSY experiment (84) (Figure 2C). Further, the NOESY connectivity of T54’s methyl group resonating at 1 ppm (Figure 2A) to G18, G53 and Ψ55 confirms the close proximity of these nucleotides to each other as determined by crystallography and highlights the long-range interaction between nucleotides from the D- and TΨC-arm. Further, the long range interaction established through the Levitt base pair (85) between G15 and C48 can be detected in the spectra of the modified tRNA (Figure 2).
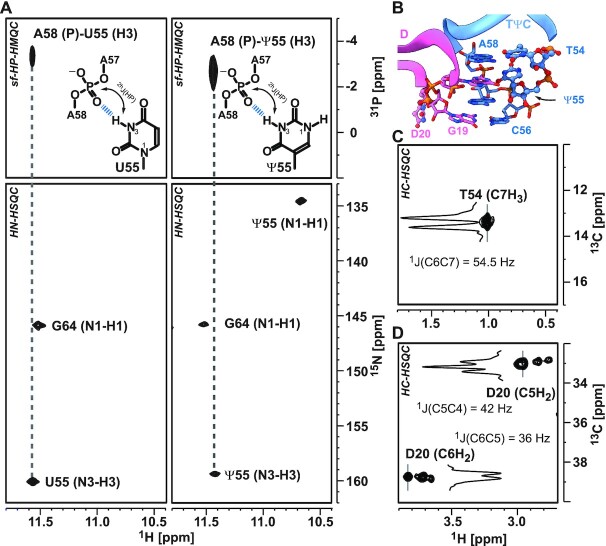
Within the latter loop a sharp turn in the sugar-phosphate backbone is formed that is stabilized through a hydrogen bond between the N3H3 group of Ψ55 and the OP group of A58. This interaction can be monitored via a long-range HP-correlation experiment (86,87) (Figure 4). Interestingly, the very same interaction between residues U55 and A58 can also be observed in the non-modified tRNA. Furthermore, no rotation of Ψ55’s nucleobase around the C1′–C5 torsion to facilitate a hydrogen bond between N1H1 and the phosphate backbone is observed.
Figure 4.
NMR spectroscopic signatures of modified residues. (A) Characterization of N3H3-phosphate group hydrogen bonds through sofast-1H,31P HMQC experiments. The H3 atom of residues U55 (left panel) and Ψ55 (right panel) with the phosphate group of residues A58 is correlated and related to the according HN cross peaks of the corresponding 1H15N HSQC, which are given in the panels below. (B) Placement of modified residues within the core region, which are establishing the interaction between D- (pink) and TΨC-loop (blue) as mapped by crystal structure (PDB: 3CW6). (C) The C7H3 cross peak of T54 detected in 1H,13C HSQC and (D) the methylene-group cross peaks of D20 C5H2 and C6H2 in 1H,13C HSQC. The inserts in (C) and (D) reflect the traces along the 13C-dimension to highlight the multiplet structure and reveal the respective 1JCC couplings. Experimental details are provided in an extended figure caption in SI.
To investigate the base pair stability, we measured proton exchange rates of the imino protons with the surrounding water. Solvent-exchange rates report on the kinetics of the replacement of imino protons with water protons. This exchange process can only occur in open states of base pairs. Thus, higher exchange rates are indicative for less stable base pairing interactions leading to faster exchange kinetics. The reduced solvent-exchange rates of imino groups in dihydrouridines compared to uridines allows the detection of the unstacked and unpaired nucleotide D20 through its HN-correlation (Figure 2B). Besides this, the ethylene group of D20 can easily be assigned in HC-correlation spectra due to the absence of other RNA resonances within this spectral region (Figure 4). Due to its unstacked nature, the line widths of the ethylene group are rather narrow, and the exhibited multiplicity of the peaks are utilized to unambiguously assign the resonances. Interestingly, for each of the CH2 groups three distinct peaks with different intensities can be observed. This spectral feature reports on a conformational exchange between at least three distinct structural states for the residue D20.
The 2′-O methyl group of residue Cm32 is also clearly detectable resonating at δ (1H) 3.76 ppm/δ (13C) 60.2 ppm in the HC-correlation spectra (Supplementary Figure S3D). It can be used as starting point for the assignment of the ACSL arm. So, the three conserved base pairs G31:C39, G30:C40 and G29:C41 are assigned. Although our assignment is in agreement with the conformational model presented in the crystal structure of the tRNAfMet, we do not detect the predicted A38(+)Cm32 base pair, even at low pH values (pH 5.5) (24).
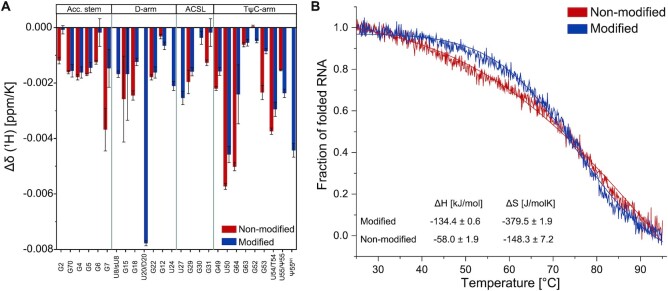
With the assigned NMR spectroscopic fingerprints of the modified and non-modified tRNA at hand we can now follow their structural changes with temperatures (Figure 5A, Supplementary Figure S5–S6). Temperature coefficients report on the extend of changes of chemical shifts with changes in temperatures. As the chemical shift is sensitive to the surrounding local structures, larger temperature coefficients can be attributed to larger temperature induced changes in local structure. Regions susceptible to temperature variations are affected to a larger degree and also seem to be more extended in the non-modified than the modified tRNA. This is also reflected in the CD-spectroscopic melting curve where the determined fraction of unfolded RNA is plotted against the temperature (Figure 5B). For the modified tRNAfMet, ΔH and ΔS values of −134.4 ± 0.6 kJ/mol and −379.5 ± 1.9 J/mol K were determined, respectively; in comparison, −58.0 ± 1.9 kJ/mol and −148.3 ± 7.2 J/mol K were determined for the non-modified tRNAfMet. Here, a significant decrease in helicity — as a measure for the foldedness of the tRNA — is observed already at ambient temperatures (>30°C). This behavior indicates an earlier melting of tertiary interactions in the non-modified compared to the modified tRNA, whereas — under the chosen conditions (200 mM K+ and 5 mM Mg2+, pH 6.2) — the individual secondary structures are of comparable stability.
Figure 5.
Temperature coefficients of the proton chemical shifts (A) and CD melting profiles of the non-modified (blue) and modified (grey) tRNAfMet. (A) Temperature coefficients of the proton chemical shifts for the non-modified tRNA ((1H), blue bars) and for the modified tRNAfMet construct ((1H), red bars). The temperature coefficients were determined through a linear fit of the measured chemical shifts at temperatures ranging from 5 to 45°C (Supplementary Figure S4–S5). (B) CD melting profiles were measured and analyzed as described in the method section. The resulting fitting parameters are shown.
Local stability of tRNA
In order to analyze the local dynamics of tRNAfMet, we measured 15N spin relaxation data and solvent-exchange rates (Supplementary Tables S4–S7, S13–S14 and Supplementary Figures S7–S10).
Spin relaxation data and solvent-exchange rates are sensitive to dynamics on different motional timescales (74,88,89). Spin relaxation data of nuclei such as 13C and 15N contain information about the dynamics in the range of ps to ns through modulation of the underlying spin interactions (88). On the other hand, the solvent exchange rates represent the exchange between solvent protons and imino protons based on base pair opening and base flipping on a ms timescale (74,89).
The relaxation data were collected for the modified and non-modified tRNA at two magnetic fields (600 MHz and 800 MHz) and for one field at two different temperatures (25°C and 40°C) (Supplementary Figures S7–S8). The relaxation data are characterized by the longitudinal R1 rate, the transversal R2 rate and the hetNOE as an indication for cross-relaxation between 1H and 15N. Unsurprisingly, the R1 values for both tRNAs increase with higher magnetic field from 0.4 rad/s to 0.6 rad/s, where the non-modified tRNA exhibit a larger increase than the modified tRNA. The transversal R2 rate is determined to around 20 rad/s, which is constant at both magnetic fields for the modified tRNA, but increases up to 30 rad/s for the non-modified construct. This trend is also reflected in the R2/R1 ratio, which is constant between 30 and 60 for the modified tRNA at both magnetic fields, but increases from 30 at 600 MHz up to 90 at 800 MHz in the case of the non-modified tRNA. In contrast, the hetNOE shows values between 0.6 and 0.9, which is expected for a sizeable RNA. Furthermore, it shows no large deviation between both magnetic fields and tRNAs. At an elevated temperature of 40°C, the temperature-induced decrease of the hetNOE and R2/R1 ratio is observed showing the temperature-induced flexibility of the tRNAs. However, no larger deviation was observed between both tRNAs.
In order to compare the three different datasets in more detail we performed a model-free analysis (65,66) with the program RELAX (Supplementary Table S8-S12) (67,68). As expected for a structured RNA, the 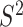 order parameters range between 0.75 and 0.98 for both tRNAs. In general, the
order parameters range between 0.75 and 0.98 for both tRNAs. In general, the 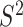 order parameter is a normalized indication for the flexibility, where
order parameter is a normalized indication for the flexibility, where 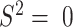 corresponds to a highly flexible and a
corresponds to a highly flexible and a 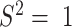 is equal to a highly rigid bond vector. For both tRNAs similar global rotational correlation times which indirect reflects the molecular shape of either 17.4 ns (non-modified) or 20.4 ns (modified) and an isotropic diffusion value of the axially symmetric tensor on the order of 107 s–1 were determined. The determined values are expected for a large RNA molecule with a molecular mass of 26 kDa (89–91) and reflect an overall identical shape of both molecules (Table 1).
is equal to a highly rigid bond vector. For both tRNAs similar global rotational correlation times which indirect reflects the molecular shape of either 17.4 ns (non-modified) or 20.4 ns (modified) and an isotropic diffusion value of the axially symmetric tensor on the order of 107 s–1 were determined. The determined values are expected for a large RNA molecule with a molecular mass of 26 kDa (89–91) and reflect an overall identical shape of both molecules (Table 1).
Table 1.
Description of the diffusion tensor obtained by the model-free analysis of the 15N relaxation parameters. 15N spin relaxation data of non-modified and modified tRNAfMet were analyzed with the program RELAX (67,68) resulting in the diffusion tensor and the model-free parameters (Supplementary Table S8-S12) of both RNAs. The parameters and the explanation of the diffusion tensor are given in the first two columns, the determined value at the given temperature in the following columns
| Unmodified tRNAfMet | Modified tRNAfMet | ||||
|---|---|---|---|---|---|
| 25°C | 40°C | 25°C | 40°C | ||
| Rotational correlation time | τm [ns] | 17.4 | 9.05 | 20.4 | 13.0 |
| Isotropic part of the diffusion tensor | D iso [106 rad/s] | 9.56 | 18.4 | 8.19 | 12.8 |
| Anisotropic part of the diffusion tensor | D a [106 rad/s] | 5.94 | 11.2 | −6.04 | −8.22 |
| Ratio between parallel to orthogonal part of the diffusion tensor | D ratio | 1.78 | 1.76 | 0.41 | 0.47 |
| Spherical angles describing the orientation of the main axis in the diffusion tensor | Theta [rad] | 0.59 | 0.59 | 1.90 | 1.90 |
| Phi [rad] | 1.57 | 1.57 | 2.95 | 2.95 | |
Out of the ten models (Supplementary Table S8), the best models of the model-free analysis describing the 15N relaxation parameter are listed in Supplementary Table S9-S12. The models are characterized with an increasing complexity starting by the simplest model M0, and ends in the most complex model M8 which is described by five parameters. Hereby, models M0, M1 and M9 assume very fast local dynamics (τf < 10 ps) with either no further parameter, the S2 order parameter or an additional Rex contribution on the ms timescale. Further complexity of the models is introduced with a local correlation time τe of the group of interest representing the timescale of the motion. The models M5 to M8 describe a more complex behavior of the imino group. Here, the order parameter as well as the local correlation time are divided into a fast (τf < 100 ps) and a slow (τs > 100 ps) motion assuming that the group of interest is involved both in a very fast and a slower dynamic motion.
Thereby, the 15N spins of the imino group in the non-modified tRNA show a strong tendency to motion models which involve the exchange parameter Rex at 25°C. At 40°C, the selected models of the non-modified tRNA comprises only simpler ones such as M1 or M2 suggesting a potential melting of the exchange states. On the contrary, models comprising no exchange were selected for almost all 15N spins of the modified tRNA (models M2, M4–7) suggesting one stable conformational state with slow global dynamics and a few motional hotspots.
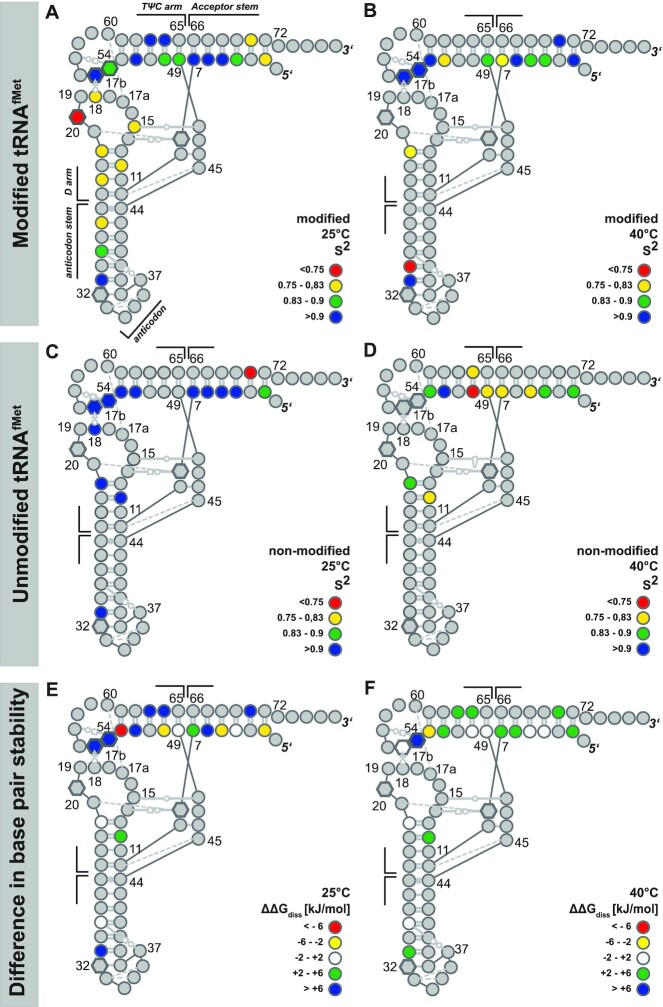
For the native, modified tRNA we find that at 25°C, judging from the order parameters, the local dynamics in the anticodon domain (anticodon-stem and D-arm) are generally higher compared to those in the TΨC and acceptor stem (Figure 6A). This indicates that the local dynamic features are similar for the pairs of coaxial stacked helices in the tRNAfMet.
Figure 6.
Comparison of local dynamics and base pair stabilities for modified and unmodified tRNAfMet. Nucleotide-resolved representation of the L-shaped structure that is color coded to represent the S2 order parameter for (A) native tRNA at 25°C and (B) at 40°C; (C) unmodified tRNA at 25°C and (D) at 40°C. The base interactions are annotated according to the nomenclature of Leontis and Westhof (112), modified residues are represented by hexagons instead of circles. The differences in the stabilities of corresponding base pairs (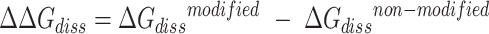 ) are given for (E) 25°C and (F) 40°C. Positive values shown in blue and green mean that modified tRNA is more stable than unmodified tRNA. In contrast, negative
) are given for (E) 25°C and (F) 40°C. Positive values shown in blue and green mean that modified tRNA is more stable than unmodified tRNA. In contrast, negative 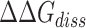 values shown in yellow and red show that unmodified tRNA is more stable than modified tRNA. White highlighted nucleotides represent only a minor difference in
values shown in yellow and red show that unmodified tRNA is more stable than modified tRNA. White highlighted nucleotides represent only a minor difference in 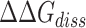 between both tRNAs. All data is summarized in Supplementary Tables S9-S12 and S15-S16.
between both tRNAs. All data is summarized in Supplementary Tables S9-S12 and S15-S16.
In the acceptor stem only residues close to the CCA-end exhibit increased local dynamics. Residue D20 is the most dynamic on the fast time-scale, presumably because it is not involved in stacking interactions with any other residue. Increasing the temperature to 40°C results in a general increase in local dynamics of the modified tRNA (Figure 6B). This is especially manifested for residues residing in the helical stacking region of TΨC and acceptor stem and further for those in the anticodon stem close to the anti-codon loop. However, at the elbow region of the native tRNA where the D- and TΨC-arm interact, the local dynamics are not increased (T54, Ψ55).
The non-modified tRNA exhibits at 25°C rather uniform low dynamics with S2 > 0.9 (Figure 6C). At this temperature, only residues close to the CCA-end in the acceptor stem exhibit increased local fluctuation on the fast timescale, as has been already observed for the modified tRNA. This behavior changes at elevated temperatures (Figure 6D). For nearly all residues in the non-modified tRNA, a decrease in the order parameter and hence an increase in local dynamics is observable at 40°C. Similar to the modified tRNA, the largest increase is manifested at the helix junction. However, for the elbow region a different behavior is observed, in contrast to the native tRNA, the transcribed tRNA does not exhibit a relative decrease of dynamics in the elbow region upon increased temperature.
Comparing the dynamics of tertiary interactions represented by the nucleotides D20, G18, T54 and Ψ55 to secondary interactions (e.g. the acceptor stem), only minor differences between these interactions can be found in the S2 order parameter and the local correlation time τe for both tRNAs at 25°C. At a temperature of 40°C, a difference between secondary and tertiary interactions is observable. In case of the modified tRNA, this difference is represented by the timescale of the local dynamics, which is on the ns timescale for the secondary interactions and on the ps timescale for tertiary interactions. In contrast, the tertiary interactions were not detectable for the non-modified tRNA due to melting of the tertiary structure.
The comparison of the exchange rate Rex being a measure for conformational exchange on the slow ms timescale indicates a more stable conformation for the modified tRNAfMet (Supplementary Table S9-S12). We report that at a temperature of 25°C only five imino groups in the modified tRNA exhibit a conformational exchange according to the model-free results with largest contributions in the D-arm possibly due to the impact of D20, whereas 12 imino groups evenly distributed over the sequence feature conformational exchange in the non-modified tRNA. However, increasing the temperature to 40°C results in a completely different situation. In the modified tRNA, conformational exchange is now found for imino groups in the TΨC-arm rather than in the D-arm, albeit with similar Rex rates. Interestingly, the non-modified tRNA exhibits no Rex rates at all at this temperature. This suggests that the conformational exchange being observable at room temperature for the non-modified tRNA ceases and only one conformation is found at elevated temperatures. In the case of the modified tRNA, the shift of conformational exchange on the ms timescale from D-arm to TΨC-arm could be explained with the beginning of melting of tertiary interactions and thus a destabilized conformation at the elbow region. Notably, this trend follows the CD-spectroscopic melting curves showing an initial decrease of folded RNA fraction for the non-modified tRNA (Figure 5B, blue line).
Similar to the relaxation data, the solvent-exchange rates of imino protons report on the stability and hence local base-pairing dynamics (74). From the temperature profile of solvent exchange rates, the thermodynamic parameters of base pair opening can be deduced. The observed base-pair stabilities range at 25°C between 20.9 and 47.4 kJ/mol for the native tRNA and between 15.0 and 32 kJ/mol for the non-modified tRNA. In the range between 20 and 40°C, the native tRNA exhibits very stable base pairs and those are mostly more stable than the corresponding base pairs within the non-modified tRNA; this is indicated by the difference of the free dissociation enthalpy 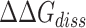 between the modified and non-modified tRNA (Figure 6E and F). Thereby, a positive value of
between the modified and non-modified tRNA (Figure 6E and F). Thereby, a positive value of 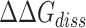 corresponds to a more stable base pairing in the modified tRNA. It is interesting to note that the modified tRNA has a high enthalpic contribution to base-pair stability whereas the non-modified tRNA exhibits comparably low base-pair dissociation enthalpy but mostly positive dissociation entropy (Figure 5B, Figure 6E and F, Supplementary Tables S15 and S16). Notably, temperature increases are less impactful for the native than for the unmodified tRNA. At elevated temperatures, the gross of base pairs is more stable in the modified compared to the unmodified tRNA; especially the base- pairing stabilities in the elbow region and at the helix junctions exhibit a higher relative stability.
corresponds to a more stable base pairing in the modified tRNA. It is interesting to note that the modified tRNA has a high enthalpic contribution to base-pair stability whereas the non-modified tRNA exhibits comparably low base-pair dissociation enthalpy but mostly positive dissociation entropy (Figure 5B, Figure 6E and F, Supplementary Tables S15 and S16). Notably, temperature increases are less impactful for the native than for the unmodified tRNA. At elevated temperatures, the gross of base pairs is more stable in the modified compared to the unmodified tRNA; especially the base- pairing stabilities in the elbow region and at the helix junctions exhibit a higher relative stability.
DISCUSSION
The modifications in tRNAs are numerous and modulate the recognition and binding of their cellular interaction partners like aminoacyl-tRNA synthetases (aaRS), mRNAs and ribosomes (92). Although modifications in tRNAs are highly abundant, they are not equally distributed over their conserved structure but occur clustered within the structural core and the anticodon stem loop (ACSL) (93). Within the full-length initiator tRNAfMet from E. coli produced by overexpression in minimal medium the core modifications (s4U8, D20, T54, Ψ55) are incorporated almost to completion (85–101% ± 1%) whereas the modification in the ACSL is only found in a subset of the molecules. In the presence of 5 mM Mg2+-ions the modified tRNA exhibits a higher structural stability as its non-modified counterpart, whose tertiary interactions melt earlier as evidenced by CD-melting experiments. This is also represented in the larger (more negative) Gibbs free energy for the modified tRNA in the ambient temperature range. Based on crystallographic structures and solution state NMR experiments, it is expected that modified tRNA and non-modified tRNA have a similar global structure (36,37,40,43,94,95). Through assignment of the imino groups of both forms of tRNAfMet as well as the carbon resonances of the introduced modifications we could confirm the predicted secondary and tertiary interactions, with exception of the protonated base pair in the anticodon stem-loop. This could be an indication that in the crystal structure of the tRNAfMet a conformation of the ACSL is frozen out which is under ambient conditions only populated to a small extent (<10%), rendering it invisible in standard NMR experiments employed here (96).
Due to the different chemical environments, the chemical shifts of the imino groups vary in regions where nucleobases are modified. Moreover, very large differences can be found for all core modifications whereas the ACSL modification Cm32 has a smaller influence on the chemical shifts of the nearby imino groups. From earlier theoretical and experimental works it is deduced that modifications restrict the conformational flexibility of the tRNA (10,97–100). However, the dynamic behavior of nucleotides in tRNAfMet is quite different. At 25°C the non-modified tRNA seems to be less dynamic than the modified counterpart as the former exhibits on average slightly higher order parameters. This more rigid-like behavior on the fast ps- to ns-timescale is complemented by exchange reactions between conformational substates found in 12 residues of the non-modified tRNA. This conformational exchange, however, has to occur on the slower μs- to ms-timescale. Furthermore, similarly in the modified tRNA, five residues show conformational exchange, and for the resonance of D20 three distinct peaks are observed in the [1H,13C]-HSQC spectrum, also indicating exchange between three different conformational substates. Upon temperature increase to 40°C, there are no significant changes in the order parameters and thus also no changes in the local dynamics of the nucleotides in the modified tRNA. In contrast, for the non-modified tRNA, the order parameters drop significantly upon temperature increase and thus the dynamics tremendously increase. This is supported by the CD-spectroscopic melting profiles, which show for the modified tRNA almost no decrease until a temperature of about 50°C; thus it has a more temperature-stable tertiary structure. Both findings agree with the dynamics occurring on a slower time scale as investigated via the solvent-exchange rates. Here, the base pairs of the modified tRNAfMet are typically around 6 kJ/mol more stable than in the non-modified tRNA at temperatures between 25°C and 40°C. Above 50°C, no difference in (base-pair) stability due to simultaneous melting of secondary structure elements can be determined. Similarly, for tRNAVal it was found by HD-exchange rates (34) that the local base pair stabilities of selected nucleotides are higher. In summary, these findings lead to two main conclusions: (i) Although the modifications for the tRNAfMet are clustered in the core region, a remote effect of stabilization throughout all base-pairs of the whole molecule is observed. This is in line with earlier findings that showed that modifications affect correlated motions within tRNAs (101), a mechanism that could mediate the overall stabilizing effect of nucleotide modifications throughout the molecule. (ii) From the differential temperature profile of order parameters and the higher degree of fast dynamics at 25°C, the stabilization of the modified tRNA has to be attributed to entropic effects. In other words, increased fast local conformational dynamics lead to a stabilization of the tRNA’s tertiary structure. Such local structural fluctuations were also observed in molecular dynamic simulations (34,103), however they are not manifested as differences in the tertiary structures of the various tRNAfMet solved so far (Supplementary Figure S11). Importantly, the local dynamics do not negatively impact the tertiary structure of the tRNA as the global structure remains intact. The introduction of modifications can therefore regarded to be essential for a harmonization of the stabilities of secondary and tertiary structure elements in tRNAfMet.
In this respect, an interesting example is the modification of uridine to dihydrouridine in the D-loop that is highly conserved in all three domains of life. Consistent with reported results (19), our analysis finds D20 as the most flexible nucleotide in the modified tRNA. At 25°C it exhibits a low 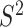 order parameter of 0.52 and additionally can be observed to exist in at least three different conformational substates which may only exchange on the slower timescale. Such low order parameters are typically found for nucleotides which are highly flexible, such as looped nucleotides pointing towards the solvent surface, e.g. the second nucleotide in YNMG loops (91). Interestingly, the D20 nucleotide is located close to those modified nucleotides which establish the tertiary interactions responsible for the formation and stabilization of the elbow region of the L-shaped structure (T54 and Ψ55) (104). Our analysis shows that pseudouridylation introduces stability to the tertiary structure as we cannot detect a signal for U55 in the non-modified tRNA at higher temperatures (>45°C). In line with reported results for tRNA folds, the rigidity of Ψ55 and nearby nucleotides is maintained with an
order parameter of 0.52 and additionally can be observed to exist in at least three different conformational substates which may only exchange on the slower timescale. Such low order parameters are typically found for nucleotides which are highly flexible, such as looped nucleotides pointing towards the solvent surface, e.g. the second nucleotide in YNMG loops (91). Interestingly, the D20 nucleotide is located close to those modified nucleotides which establish the tertiary interactions responsible for the formation and stabilization of the elbow region of the L-shaped structure (T54 and Ψ55) (104). Our analysis shows that pseudouridylation introduces stability to the tertiary structure as we cannot detect a signal for U55 in the non-modified tRNA at higher temperatures (>45°C). In line with reported results for tRNA folds, the rigidity of Ψ55 and nearby nucleotides is maintained with an 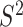 order parameter of 0.90 and 0.93 at 25 and 40°C, respectively. To date, the impact of Ψ is highly discussed (33). In general, it is shown that Ψ stabilizes tRNA folds but in other RNA the stabilizing effect is not shown with clear evidence. For the tRNAfMet here, we assume that the interplay of flexibility of D20 and the stabilization of Ψ55 is needed to stabilize the very important elbow region of the L-shape, while Ψ acts through enthalpic and D through an entropic mechanism. Besides the increased thermodynamic stability that might be of biological importance during encounters of environmental stresses, the enhanced local dynamics as evidenced for D20 and other residues will play a role during functional interactions. For example, during translation initiation the tRNAfMet must retain its overall shape while allowing a certain degree of flexibility during correct accommodation at the right position within the 30S subunit and towards the start codon at the mRNA (21,105,106). A delicate interplay of rigidity and flexibility is necessary. While at the same time rigidity of the anticodon stem is required for translocation during translation, structural fluctuations are required for building the correct codon:anticodon (102). Functional evidence that the nucleotide modifications are causal for the balance of local fluctuations and overall stability is provided by ribosome binding studies where the
order parameter of 0.90 and 0.93 at 25 and 40°C, respectively. To date, the impact of Ψ is highly discussed (33). In general, it is shown that Ψ stabilizes tRNA folds but in other RNA the stabilizing effect is not shown with clear evidence. For the tRNAfMet here, we assume that the interplay of flexibility of D20 and the stabilization of Ψ55 is needed to stabilize the very important elbow region of the L-shape, while Ψ acts through enthalpic and D through an entropic mechanism. Besides the increased thermodynamic stability that might be of biological importance during encounters of environmental stresses, the enhanced local dynamics as evidenced for D20 and other residues will play a role during functional interactions. For example, during translation initiation the tRNAfMet must retain its overall shape while allowing a certain degree of flexibility during correct accommodation at the right position within the 30S subunit and towards the start codon at the mRNA (21,105,106). A delicate interplay of rigidity and flexibility is necessary. While at the same time rigidity of the anticodon stem is required for translocation during translation, structural fluctuations are required for building the correct codon:anticodon (102). Functional evidence that the nucleotide modifications are causal for the balance of local fluctuations and overall stability is provided by ribosome binding studies where the 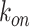 values are identical, but the
values are identical, but the 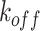 values for unmodified tRNAs are largely increased compared to those containing modifications (107,108). Besides ribosome binding, the modifications’ impact on local flexibility can also play a crucial role in the accuracy and fidelity of translation (108,109).
values for unmodified tRNAs are largely increased compared to those containing modifications (107,108). Besides ribosome binding, the modifications’ impact on local flexibility can also play a crucial role in the accuracy and fidelity of translation (108,109).
So far, it was shown that modified residues can either increase the rigidity and thermal stability of the structure of tRNA or have exactly the opposite effect (10,110). Until a similar comparative mapping of local structural dynamics is undertaken for more full length tRNAs, it remains to be seen if the findings provided here for tRNAfMet are generally applicable to all tRNAs. It will be especially insightful to understand how the local dynamics interfere, modulate and direct the incorporation of the modified nucleotides themselves (111).
DATA AVAILABILITY
Chemical shifts and relaxation data are deposited at BMRB under accession code 51144.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Emmanuelle Schmitt for providing E. coli JM101tr cells and the tRNA plasmid for overexpression in cells. We thank Harald Schwalbe for continuous support and discussion on this project. We also thank Jens Wöhnert and Martin Hengesbach for help and insightful discussions. We further thank Annika Kotter, Frank Löhr and Christian Richter for excellent technical assistance.
This study made use of NMRbox: National Center for Biomolecular NMR Data Processing and Analysis, a Biomedical Technology Research Resource (BTRR), which is supported by NIH grant P41GM111135 (NIGMS). Molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
Contributor Information
Thomas Biedenbänder, Institute for Organic Chemistry and Chemical Biology, Center for Biomolecular Magnetic Resonance (BMRZ), Johann Wolfgang Goethe-Universität, Frankfurt am Main 60438, Germany; Institute of Chemistry and Department Life, Light & Matter, University of Rostock, Rostock 18059, Germany.
Vanessa de Jesus, Institute for Organic Chemistry and Chemical Biology, Center for Biomolecular Magnetic Resonance (BMRZ), Johann Wolfgang Goethe-Universität, Frankfurt am Main 60438, Germany.
Martina Schmidt-Dengler, Institut für pharmazeutische und biomedizinische Wissenschaften (IPBW), Johannes Gutenberg-Universität, Mainz 55128, Germany.
Mark Helm, Institut für pharmazeutische und biomedizinische Wissenschaften (IPBW), Johannes Gutenberg-Universität, Mainz 55128, Germany.
Björn Corzilius, Institute of Chemistry and Department Life, Light & Matter, University of Rostock, Rostock 18059, Germany.
Boris Fürtig, Institute for Organic Chemistry and Chemical Biology, Center for Biomolecular Magnetic Resonance (BMRZ), Johann Wolfgang Goethe-Universität, Frankfurt am Main 60438, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Funding Agency (DFG) in Collaborative Research Center 902: Molecular principles of RNA-based regulation; B.F. is supported by the DFG in graduate school CLIC [GRK 1986]; work at BMRZ was supported by the state of Hesse (HMWK); V.D.J. was supported by Boehringer Ingelheim Fonds; T.B. has been supported by the Joachim Herz Foundation through an Add-on Fellowship for Interdisciplinary Life Science; B.C. acknowledges funding through DFG [CO 802/2-2, CO 802/4-1]. Funding for open access charge: University Frankfurt.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dunin-Horkawicz S., Czerwoniec A., Gajda M.J., Feder M., Grosjean H., Bujnicki J.M.. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006; 34:D145–D149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Czerwoniec A., Dunin-Horkawicz S., Purta E., Kaminska K.H., Kasprzak J.M., Bujnicki J.M., Grosjean H., Rother K.. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009; 37:D118–D121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Machnicka M.A., Milanowska K., Oglou O.O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M.et al.. MODOMICS: a database of RNA modification pathways - 2013 update. Nucleic Acids Res. 2013; 41:D262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boccaletto P., MacHnicka M.A., Purta E., Pitkowski P., Baginski B., Wirecki T.K., De Crécy-Lagard V., Ross R., Limbach P.A., Kotter A.et al.. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021; 22:375–392. [DOI] [PubMed] [Google Scholar]
- 6. Phizicky E.M., Hopper A.K.. tRNA biology charges to the front. Genes Dev. 2010; 24:1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raina M., Ibba M.. tRNAs as regulators of biological processes. Front. Genet. 2014; 5:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barraud P., Tisné C.. To be or not to be modified: miscellaneous aspects influencing nucleotide modifications in tRNAs. IUBMB Life. 2019; 71:1126–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006; 34:721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Motorin Y., Helm M.. tRNA stabilization by modified nucleotides. Biochemistry. 2010; 49:4934–4944. [DOI] [PubMed] [Google Scholar]
- 11. Motorin Y., Helm M.. RNA nucleotide methylation. Wiley Interdiscipl. Rev.: RNA. 2011; 2:611–631. [DOI] [PubMed] [Google Scholar]
- 12. El Yacoubi B., Bailly M., De Crécy-Lagard V.. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012; 46:69–95. [DOI] [PubMed] [Google Scholar]
- 13. Ling J., Söll D. Transfer RNA function and evolution. RNA Biology. 2018; 15:423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dube S.K., Marcker K.A., Clark B.F.C., Cory S.. The nucleotide sequence of N-formyl-methionyl-transfer RNA. Products of complete digestion with ribonuclease T1 and pancreatic ribonuclease and derivation of their sequences. Eur. J. Biochem. 1969; 8:244–255. [DOI] [PubMed] [Google Scholar]
- 15. Kim S.H., Quigley G.J., Suddath F.L., Mcpherson A., Sneden D., Kim J.J., Weinzierl J., Rich A.. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973; 179:285–288. [DOI] [PubMed] [Google Scholar]
- 16. Machnicka M.A., Olchowik A., Grosjean H., Bujnicki J.M.. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014; 11:1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Agris P.F., Vendeix F.A.P., Graham W.D.. tRNA’s Wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007; 366:1–13. [DOI] [PubMed] [Google Scholar]
- 18. Grosjean H., de Crécy-Lagard V., Marck C.. Deciphering synonymous codons in the three domains of life: Co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010; 584:252–264. [DOI] [PubMed] [Google Scholar]
- 19. Dyubankova N., Sochacka E., Kraszewska K., Nawrot B., Herdewijn P., Lescrinier E.. Contribution of dihydrouridine in folding of the D-arm in tRNA. Org. Biomol. Chem. 2015; 13:4960–4966. [DOI] [PubMed] [Google Scholar]
- 20. Lorenz C., Lünse C.E., Mörl M.. tRNA modifications: impact on structure and thermal adaptation. Biomolecules. 2017; 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mandal N., Mangroo D., Dalluge J.J., McCloskey J.A., RajBhandary U.L.. Role of the three consecutive G:C base pairs conserved in the anticodon stem of initiator tRNAs in initiation of protein synthesis in Escherichia coli. RNA. 1996; 2:473–482. [PMC free article] [PubMed] [Google Scholar]
- 22. Shepherd J., Ibba M.. Bacterial transfer RNAs. FEMS Microbiol. Rev. 2015; 39:280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shetty S., Bhattacharyya S., Varshney U.. Is the cellular initiation of translation an exclusive property of the initiator tRNAs. RNA Biol. 2015; 12:675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barraud P., Schmitt E., Mechulam Y., Dardel F., Tisné C.. A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis. Nucleic Acids Res. 2008; 36:4894–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woo N.H., Roe B.A., Rich A.. Three-dimensional structure of escherichia coli initiator tRNAfmet. Nature. 1980; 286:346–351. [DOI] [PubMed] [Google Scholar]
- 26. Bjork G.R., Ericson J.U., Gustafsson C.E.D., Hagervall T.G., Jonsson Y.H., Wikstrom P.M.. Transfer RNA modification. Annu. Rev. Biochem. 1987; 56:263–285. [DOI] [PubMed] [Google Scholar]
- 27. Newby M.I., Greenbaum N.L.. Investigation of overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl Acad. Sci. U.S.A. 2002; 99:12697–12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dalluge J.J., Hashizume T., Sopchik A.E., McCloskey J.A., Davis D.R.. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res. 1996; 24:1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Davanloo P., Sprinzl M., Watanabe K., Albani M., Kersten H.. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res. 1979; 6:1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kramer G.F., Baker J.C., Ames B.N.. Near-UV stress in salmonella typhimurium: 4-Thiouridine in tRNA, ppGpp, and apppgpp as components of an adaptive response. J. Bacteriol. 1988; 170:2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Čavužić M., Liu Y.. Biosynthesis of sulfur-containing tRNA modifications: a comparison of bacterial, archaeal, and eukaryotic pathways. Biomolecules. 2017; 7:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pascal A., Westhof E.. Effects of pseudouridylation on tRNA hydration and dynamics: A theoretical approach. 1998; 10.1128/9781555818296.ch6. [DOI]
- 33. Westhof E. Pseudouridines or how to draw on weak energy differences. Biochem. Biophys. Res. Commun. 2019; 520:702–704. [DOI] [PubMed] [Google Scholar]
- 34. Vermeulen A., McCallum S.A., Pardi A.. Comparison of the global structure and dynamics of native and unmodified tRNAval. Biochemistry. 2005; 44:6024–6033. [DOI] [PubMed] [Google Scholar]
- 35. Serebrov V., Vassilenko K., Kholod N., Gross H.J., Kisselev L.. Mg 2+ binding and structural stability of mature and in vitro synthesized unmodified escherichia coli tRNA phe. Nucleic Acids Res. 1998; 26:2723–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sampson J.R., Uhlenbeck O.C.. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. U.S.A. 1988; 85:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perret V., Garcia A., Puglisi J., Grosjean H., Ebel J.P., Florentz C., Giegé R.. Conformation in solution of yeast tRNAAsp transcripts deprived of modified nucleotides. Biochimie. 1990; 72:735–743. [DOI] [PubMed] [Google Scholar]
- 38. Behlen L.S., Sampson J.R., DiRenzo A.B., Uhlenbeck O.C.. Lead-catalyzed cleavage of yeast tRNAPhe mutants. Biochemistry. 1990; 29:2515–2523. [DOI] [PubMed] [Google Scholar]
- 39. Behlen L.S., Sampson J.R., Uhlenbeck O.C.. An ultraviolet light-induced crosslink in yeast tRNA(Phe). Nucleic Acids Res. 1992; 20:4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Derrick W.B., Horowitz J.. Probing structural differences between native and in vitro transcribed escherichia coli valine transfer RNA: evidence for stable base modification-dependent conformers. Nucleic Acids Res. 1993; 21:4948–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maglott E.J., Deo S.S., Przykorska A., Glick G.D.. Conformational transitions of an unmodified tRNA: implications for RNA folding. Biochemistry. 1998; 37:16349–16359. [DOI] [PubMed] [Google Scholar]
- 42. Byrne R.T., Konevega A.L., Rodnina M. V, Antson A.A. The crystal structure of unmodified tRNA Phe from Escherichia coli. Nucleic Acids Res. 2010; 38:4154–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hall K.B., Sampson J.R., Uhlenbeck O.C., Redfield A.G.. Structure of an unmodified tRNA molecule. Biochemistry. 1989; 28:5794–5801. [DOI] [PubMed] [Google Scholar]
- 44. Schauss J., Kundu A., Fingerhut B.P., Elsaesser T.. Magnesium contact ions stabilize the tertiary structure of transfer RNA: electrostatics mapped by two-dimensional infrared spectra and theoretical simulations. J. Phys. Chem. B. 2021; 125:740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Friederich M.W., Gast F.U., Vacano E., Hagerman P.J.. Determination of the angle between the anticodon and aminoacyl acceptor stems of yeast phenylalanyl tRNA in solution. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:4803–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fürtig B., Richter C., Wöhnert J., Schwalbe H.. NMR spectroscopy of RNA. ChemBioChem. 2003; 4:936–962. [DOI] [PubMed] [Google Scholar]
- 47. Guilleres J., Lopez P.J., Proux F., Launay H., Dreyfus M.. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc. Natl Acad. Sci. U.S.A. 2005; 102:5958–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bains J.K., Blechar J., de Jesus V., Meiser N., Zetzsche H., Fürtig B., Schwalbe H., Hengesbach M.. Combined smFRET and NMR analysis of riboswitch structural dynamics. Methods. 2019; 153:22–34. [DOI] [PubMed] [Google Scholar]
- 49. Mechulam Y., Guillon L., Yatime L., Blanquet S., Schmitt E.. Protection-Based assays to measure Aminoacyl-tRNA binding to translation initiation factors. Methods in Enzymology. 2007; 430:Academic Press Inc; 265–281. [DOI] [PubMed] [Google Scholar]
- 50. Ponchon L., Catala M., Seijo B., El Khouri M., Dardel F., Nonin-Lecomte S., Tisné C.. Co-expression of RNA-protein complexes in escherichia coli and applications to RNA biology. Nucleic Acids Res. 2013; 41:e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Young R.A. Transcription termination in the escherichia coli ribosomal RNA operon rrnC. J. Biol. Chem. 1979; 254:12725–12731. [PubMed] [Google Scholar]
- 52. Meinnel T., Blanquet S.. Maturation of pre-tRNA(fMet) by Escherichia coli RNase p is specified by a guanosine of the 5′-flanking sequence. J. Biol. Chem. 1995; 270:15908–15914. [DOI] [PubMed] [Google Scholar]
- 53. Thüring K., Schmid K., Keller P., Helm M.. Analysis of RNA modifications by liquid chromatography–tandem mass spectrometry. Methods. 2016; 107:48–56. [DOI] [PubMed] [Google Scholar]
- 54. Marky L.A., Breslauer K.J.. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987; 26:1601–1620. [DOI] [PubMed] [Google Scholar]
- 55. Wishart D.S., Bigam C.G., Yao J., Abildgaard F., Dyson H.J., Oldfield E., Markley J.L., Sykes B.D.. 1H, 13C and 15N chemical shift referencing in biomolecular nMR. J. Biomol. NMR. 1995; 6:135–140. [DOI] [PubMed] [Google Scholar]
- 56. Lee W., Tonelli M., Markley J.L.. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015; 31:1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bodenhausen G., Ruben D.J.. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980; 69:185–189. [Google Scholar]
- 58. Favier A., Brutscher B.. Recovering lost magnetization: polarization enhancement in biomolecular NMR. J. Biomol. NMR. 2011; 49:9–15. [DOI] [PubMed] [Google Scholar]
- 59. Solyom Z., Schwarten M., Geist L., Konrat R., Willbold D., Brutscher B.. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J. Biomol. NMR. 2013; 55:311–321. [DOI] [PubMed] [Google Scholar]
- 60. Dingley A.J., Nisius L., Cordier F., Grzesiek S.. Direct detection of N−H⋯N hydrogen bonds in biomolecules by NMR spectroscopy. Nat. Protoc. 2008; 3:242–248. [DOI] [PubMed] [Google Scholar]
- 61. Schanda P., Brutscher B.. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 2005; 127:8014–8015. [DOI] [PubMed] [Google Scholar]
- 62. Shajani Z., Varani G.. NMR studies of dynamics in RNA and DNA by 13C relaxation. Biopolymers. 2007; 86:348–359. [DOI] [PubMed] [Google Scholar]
- 63. Ferrage F. Protein dynamics by 15n nuclear magnetic relaxation. Methods Mol. Biol. 2012; 831:141–163. [DOI] [PubMed] [Google Scholar]
- 64. Abragam A. The Principles Of Nuclear Magnetism. 1961; Oxford: Clarendon Press. [Google Scholar]
- 65. Lipari G., Szabo A.. Model-Free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982; 104:4546–4559. [Google Scholar]
- 66. Lipari G., Szabo A.. Model-Free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 1982; 104:4559–4570. [Google Scholar]
- 67. Auvergne E.J., Gooley P.R.. Optimisation of NMR dynamic models. I. Minimisation algorithms and their performance within the model-free and brownian rotational diffusion spaces. J. Biomol. NMR. 2008; 40:107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Auvergne E.J., Gooley P.R.. Optimisation of NMR dynamic models II. A new methodology for the dual optimisation of the model-free parameters and the brownian rotational diffusion tensor. J. Biomol. NMR. 2008; 40:121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Maciejewski M.W., Schuyler A.D., Gryk M.R., Moraru I.I., Romero P.R., Ulrich E.L., Eghbalnia H.R., Livny M., Delaglio F., Hoch J.C.. NMRbox: a resource for biomolecular NMR computation. Biophys. J. 2017; 112:1529–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grishaev A., Yao L., Ying J., Pardi A., Bax A.. Chemical shift anisotropy of imino 15N nuclei in watson-crick base pairs from magic angle spinning liquid crystal NMR and nuclear spin relaxation. J. Am. Chem. Soc. 2009; 131:9490–9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. D’Auvergne E.J., Gooley P.R.. Set theory formulation of the model-free problem and the diffusion seeded model-free paradigm. Mol. Biosyst. 2007; 3:483–494. [DOI] [PubMed] [Google Scholar]
- 72. Fushman D. Determination of Protein Dynamics Using 15 N Relaxation Measurements. 2002; John Wiley & Sons, Ltd; 283–308. [Google Scholar]
- 73. d’Auvergne E.J., Gooley P.R.. The use of model selection in the model-free analysis of protein dynamics. J. Biomol. NMR. 2003; 25:25–39. [DOI] [PubMed] [Google Scholar]
- 74. Rinnenthal J., Klinkert B., Narberhaus F., Schwalbe H.. Direct observation of the temperature-induced melting process of the salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res. 2010; 38:3834–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Steinert H.S., Rinnenthal J., Schwalbe H.. Individual basepair stability of DNA and RNA studied by NMR-detected solvent exchange. Biophys. J. 2012; 102:2564–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wagner D., Rinnenthal J., Narberhaus F., Schwalbe H.. Mechanistic insights into temperature-dependent regulation of the simple cyanobacterial hsp17 RNA thermometer at base-pair resolution. Nucleic Acids Res. 2015; 43:5572–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Galvanin A., Vogt L.-M., Grober A., Freund I., Ayadi L., Bourguignon-Igel V., Bessler L., Jacob D., Eigenbrod T., Marchand V.et al.. Bacterial tRNA 2′-O-methylation is dynamically regulated under stress conditions and modulates innate immune response. Nucleic Acids Res. 2020; 48:12833–12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Han L., Phizicky E.M.. A rationale for tRNA modification circuits in the anticodon loop. RNA. 2018; 24:1277–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Berlyn M.K.B. Linkage map of escherichia coli K-12, edition 10: the traditional map. Microbiol. Mol. Biol. Rev. 1998; 62:1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mandal N., RajBhandary U.L.. Escherichia coli b lacks one of the two initiator tRNA species present in e. coli K-12. J. Bacteriol. 1992; 174:7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kapoor S., Das G., Varshney U.. Crucial contribution of the multiple copies of the initiator tRNA genes in the fidelity of tRNA fMet selection on the ribosomal P-site in Escherichia coli. Nucleic Acids Res. 2011; 39:202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wallis N.G., Dardel F., Blanquet S.. Heteronuclear NMR studies of the interactions of 15N-Labeled methionine-specific transfer RNAs with Methionyl-tRNA transformylase. Biochemistry. 1995; 34:7668–7677. [DOI] [PubMed] [Google Scholar]
- 83. Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J.. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009; 37:D159–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wöhnert J., Dingley A.J., Stoldt M., Görlach M., Grzesiek S., Brown L.R.. Direct identification of NH⋯N hydrogen bonds in non-canonical base pairs of RNA by NMR spectroscopy. Nucleic Acids Res. 1999; 27:3104–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Levitt M., Dhu A.. Detailed molecular model for transfer ribonucleic acid AMINO-ACID ARM. 1969; [DOI] [PubMed]
- 86. Duchardt-Ferner E., Ferner J., Wöhnert J.. Rapid identification of noncanonical RNA structure elements by direct detection of OH⋯O = p, NH⋯O = p, and NH 2⋯O = p hydrogen bonds in solution NMR spectroscopy. Angew. Chem. - Int. Ed. 2011; 50:7927–7930. [DOI] [PubMed] [Google Scholar]
- 87. Wolter A.C., Weickhmann A.K., Nasiri A.H., Hantke K., Ohlenschläger O., Wunderlich C.H., Kreutz C., Duchardt-Ferner E., Wöhnert J.. A stably protonated adenine nucleotide with a highly shifted pKa value stabilizes the tertiary structure of a GTP-Binding RNA aptamer. Angew. Chem. Int. Ed. 2017; 56:401–404. [DOI] [PubMed] [Google Scholar]
- 88. Palmer A.G. NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 2004; 104:3623–3640. [DOI] [PubMed] [Google Scholar]
- 89. Marušič M., Schlagnitweit J., Petzold K.. RNA dynamics by NMR spectroscopy. Chem Bio Chem. 2019; 20:2685–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Akke M., Fiala R., Jiang F., Patel D., Palmer A.G.. Base dynamics in a UUCG tetraloop RNA hairpin characterized by 15N spin relaxation: correlations with structure and stability. RNA. 1997; 3:702–709. [PMC free article] [PubMed] [Google Scholar]
- 91. Ferner J., Villa A., Duchardt E., Widjajakusuma E., Wöhnert J., Stock G., Schwalbe H.. NMR and MD studies of the temperature-dependent dynamics of RNA YNMG-tetraloops. Nucleic Acids Res. 2008; 36:1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Alexander R.W., Eargle J., Luthey-Schulten Z.. Experimental and computational determination of tRNA dynamics. FEBS Lett. 2010; 584:376–386. [DOI] [PubMed] [Google Scholar]
- 93. Grosjean H., Edqvist J., Stråby K.B., Giegé R.. Enzymatic formation of modified nucleosides in tRNA: dependence on tRNA architecture. J. Mol. Biol. 1996; 255:67–85. [DOI] [PubMed] [Google Scholar]
- 94. Chu W.C., Horowitz J.. 19F NMR of 5-fluorouracil-substituted transfer RNA transcribed in vitro: resonance assignment of fluorouracil-guanine base pairs. Nucleic Acids Res. 1989; 17:7241–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Harrington K.M., Nazarenko I.A., Uhlenbeck O.C., Dix D.B., Thompson R.C.. In vitro analysis of translational rate and accuracy with an unmodified tRNA. Biochemistry. 1993; 32:7617–7622. [DOI] [PubMed] [Google Scholar]
- 96. Kimsey I.J., Petzold K., Sathyamoorthy B., Stein Z.W., Al-Hashimi H.M.. Visualizing transient Watson–Crick-like mispairs in DNA and RNA duplexes. Nature. 2015; 519:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Auffinger P., Louise-May S., Westhof E.. Molecular dynamics simulations of solvated yeast tRNAAsp. Biophys. J. 1999; 76:50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cabello-Villegas J., Winkler M.E., Nikonowicz E.P.. Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem-loops of escherichia coli tRNAphe. J. Mol. Biol. 2002; 319:1015–1034. [DOI] [PubMed] [Google Scholar]
- 99. Stuart J.W., Koshlap K.M., Guenther R., Agris P.F.. Naturally-occurring modification restricts the anticodon domain conformational space of tRNAPhe. J. Mol. Biol. 2003; 334:901–918. [DOI] [PubMed] [Google Scholar]
- 100. Durant P.C., Bajji A.C., Sundaram M., Kumar R.K., Davis D.R.. Structural effects of hypermodified nucleosides in the escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s 2U, mcm5U, mcm5s2U, mnm 5s2U, t6A, and ms2t6A. Biochemistry. 2005; 44:8078–8089. [DOI] [PubMed] [Google Scholar]
- 101. Zhang X., Walker R.C., Phizicky E.M., Mathews D.H.. Influence of sequence and covalent modifications on yeast tRNA dynamics. J. Chem. Theory Comput. 2014; 10:3473–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wohlgemuth I., Pohl C., Mittelstaet J., Konevega A.L., Rodnina M. V.. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos. Trans. R. Soc. B: Biol. Sci. 2011; 366:2979–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Godwin R.C., Macnamara L.M., Alexander R.W., Salsbury F.R.. Structure and dynamics of tRNA met containing core substitutions. 2018; 10.1021/acsomega.8b00280. [DOI] [PMC free article] [PubMed]
- 104. Quigley G.J., Wang A.H.J., Seeman N.C., Suddath F.L., Rich A., Sussman J.L., Kim S.H.. Hydrogen bonding in yeast phenylalanine transfer RNA. Proc. Natl Acad. Sci. U.S.A. 1975; 72:4866–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lancaster L., Noller H.F.. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005; 20:623–632. [DOI] [PubMed] [Google Scholar]
- 106. Shetty S., Shah R.A., Chembazhi U. V., Sah S., Varshney U.. Two highly conserved features of bacterial initiator tRNAs license them to pass through distinct checkpoints in translation initiation. Nucleic Acids Res. 2017; 45:2040–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Fahlman R.P., Dale T., Uhlenbeck O.C.. Uniform binding of aminoacylated transfer RNAs to the ribosomal a and p sites. Mol. Cell. 2004; 16:799–805. [DOI] [PubMed] [Google Scholar]
- 108. Yarian C., Townsend H., Czestkowski W., Sochacka E., Malkiewicz A.J., Guenther R., Miskiewicz A., Agris P.F.. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 2002; 277:16391–16395. [DOI] [PubMed] [Google Scholar]
- 109. Urbonavičius J., Qian Q., Durand J.M.B., Hagervall T.G., Björk G.R.. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001; 20:4863–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kawai G., Yamamoto Y., Kamimura T., Masegi T., Sekine M., Hata T., Iimori T., Watanabe T., Miyazawa T., Yokoyama S.. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 2002; 31:1040–1046. [DOI] [PubMed] [Google Scholar]
- 111. Barraud P., Gato A., Heiss M., Catala M., Kellner S., Tisné C.. Time-resolved NMR monitoring of tRNA maturation. Nat. Commun. 2019; 10:3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Leontis N.B., Westhof E.. Geometric nomenclature and classification of RNA base pairs. RNA. 2001; 7:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Chemical shifts and relaxation data are deposited at BMRB under accession code 51144.