Abstract
The opportunistic pathogen Acinetobacter baumannii possesses stress tolerance strategies against host innate immunity and antibiotic killing. However, how the host-pathogen-antibiotic interaction affects the overall molecular regulation of bacterial pathogenesis and host response remains unexplored. Here, we simultaneously investigate proteomic changes in A. baumannii and macrophages following infection in the absence or presence of the polymyxins. We discover that macrophages and polymyxins exhibit complementary effects to disarm several stress tolerance and survival strategies in A. baumannii, including oxidative stress resistance, copper tolerance, bacterial iron acquisition and stringent response regulation systems. Using the spoT mutant strains, we demonstrate that bacterial cells with defects in stringent response exhibit enhanced susceptibility to polymyxin killing and reduced survival in infected mice, compared to the wild-type strain. Together, our findings highlight that better understanding of host-pathogen-antibiotic interplay is critical for optimization of antibiotic use in patients and the discovery of new antimicrobial strategy to tackle multidrug-resistant bacterial infections.
Author summary
Bacterial response towards antibiotics is sensitive to the surrounding environment; however, how the host immune microenvironment affects the response of opportunistic pathogen Acinetobacter baumannii towards polymyxins remains largely unexplored. In this study, we established a host-pathogen-antibiotic tripartite in vitro model to examine the complex tripartite molecular interplay from both bacteria and macrophages perspectives, mimicking the physiological infection-treatment condition. This is the first comprehensive proteomics dataset for A. baumannii interacting with host cells, with an extraordinary coverage (i.e., 50%) of A. baumannii proteome even in infection conditions. For the first time, we report that macrophages and polymyxins utilize complementary mechanisms to disarm several A. baumannii stress tolerance strategies to persist in the macrophages. Our host-pathogen-antibiotic mechanistic study and the discovery of potential druggable targets (e.g., bacterial stringent stress response regulator SpoT) represent a significant advance, paving way to antibiotic optimization and drug discovery to tackle multidrug-resistant bacterial infections.
Introduction
Antimicrobial resistance (AMR) has been recognized as a major global threat with severe health and economic consequences [1,2]. One of the most important emerging pathogens is multidrug-resistant (MDR) Acinetobacter baumannii which causes severe pneumonia, bacteremia and other infections, especially in immunocompromised patients [3]. Indeed, carbapenem-resistant A. baumannii has been highlighted by the World Health Organization (WHO) as one of the three top-priority pathogens (Priority 1: Critical) urgently requiring the discovery and development of novel antibiotics [4]. Unfortunately, the increasing emergence and spread of antibiotic resistance has thus far outcompeted the discovery of novel antibiotics [5,6]. Notwithstanding the recently approved siderophore cephalosporin cefiderocol to treat extensively drug-resistant Gram-negative infections [7], the ‘old’ polymyxins (i.e., polymyxin B and colistin) are often the only therapeutic option for otherwise untreatable bacterial infections, including those caused by A. baumannii [8,9]. Polymyxins are membrane-targeting cyclic lipopeptide antibiotics that are active against many of the MDR Gram-negative bacterial pathogens responsible for nosocomial infections, such as A. baumannii [10–15]. However, polymyxin resistance has been increasingly reported since their re-introduction to the clinic in the early 2000s, with sub-optimal use as a major contributor [16,17]. Therefore, optimization of the dosage regimens of polymyxins is urgently required to maximize the antibacterial efficacy, reduce the emergence of resistance and preserve their clinical utility.
Current antibiotic pharmacokinetics/pharmacodynamics (PK/PD) are significantly limited by a lack of understanding of the complex interplay between the host, pathogen and drug [9,18]. Most studies emphasize either pathogen-drug, host-pathogen or host-drug bipartite interactions at any one time, neglecting the effect of a dynamic host environment which includes modulation of antibiotic activity against bacteria by the innate immune system. As A. baumannii is an opportunistic pathogen, better understanding of its interaction with host innate immune cells is critical for the optimization of antibiotic treatment strategies [19,20]. In this regard, macrophages are a critical component of early host responses to invading bacteria, capable of phagocytosing a low number of bacteria prior to the subsequent recruitment of neutrophils [21]. In vivo ablation of alveolar macrophages has also been shown to exacerbate bacterial burdens in mice infected by A. baumannii [21,22]. In A. baumannii, several virulence and immune evasion factors have been identified which assist the pathogen to evade neutrophil chemotaxis, including the cytotoxic outer membrane protein A (OmpA), immunogenic lipopolysaccharide, phagocytosis-preventing capsular polysaccharide, and the phenylacetic acid catabolic pathway [23–26]. The specific bacterial machineries that facilitate A. baumannii persistence in macrophages remain to be elucidated. Understanding the correlative global protein expression profiles of both the host and pathogen is critical for the identification of key biochemical pathways and protein targets underlying the host-pathogen interaction [27]. To the best of our knowledge, no study has yet examined the molecular interactions between macrophages and interacting (adherent and intracellular) A. baumannii in the presence of antibiotics, limiting our understanding on how the unique macrophage microenvironment affects the bacterial response. Technical challenges exist particularly in sufficient coverage of bacterial proteins for proteomic analysis upon interaction with host cells [27].
Here we employed proteomics to elucidate the complex interplay among MDR A. baumannii, human macrophages and polymyxins. Using differentiated THP-1 human macrophages (THP-1-dMs) and the model MDR isolate A. baumannii AB5075, we simultaneously investigated proteome changes of both the pathogen and host in the absence or presence of the polymyxins. Interacting bacterial fraction was focused in this study and sufficient numbers of bacterial cells were obtained for proteomic analysis by a two-step differential centrifugation method. With 1,954 bacterial proteins and 4,320 mammalian proteins identified, we discovered the unique proteome signatures that underlie macrophage immune-inflammatory responses following infection, and A. baumannii tolerance strategies in the presence of macrophage and polymyxin. Finally, an A. baumannii transposon mutant library was employed to demonstrate that impairment of bacterial redox stress resistance, iron acquisition and stringent response regulators significantly enhanced bacterial killing by polymyxin. Our findings provide critical mechanistic information for optimizing polymyxin use in patients.
Results
Proteomic profiling of A. baumannii
We examined whether macrophages (THP-1-dMs) alter molecular response of A. baumannii AB5075 towards 30 mg/L polymyxin B (PMB) treatment or vice versa, with a focus on the interacting bacterial fraction (S1 Fig). A total of 1,954 proteins were identified across all bacteria-associated samples that covered approximately 50% of the AB5075 proteome. To the best of our knowledge, this is the first comprehensive proteomics dataset for A. baumannii interacting with host cells. The principal component analysis (PCA) revealed that the expression data of bacteria treated with polymyxin B only (A. baumannii [AB] + PMB) exhibited the most distinct separation from untreated bacterial controls in the first component, followed by the polymyxin B-treated infection group (AB + THP-1-dMs + PMB) and THP-1-dMs infection alone group (AB + THP-1-dMs) (S2A Fig). Compared to the untreated bacterial controls, AB + THP-1-dMs + PMB resulted in 607 differentially expressed proteins (DEPs) followed by AB + PMB (549 DEPs) and AB + THP-1-dMs (202 DEPs), indicating that perturbations in the AB5075 proteome were predominantly driven by polymyxin treatment (S2B Fig).
THP-1-dMs induced modulation of redox and iron metabolism in A. baumannii
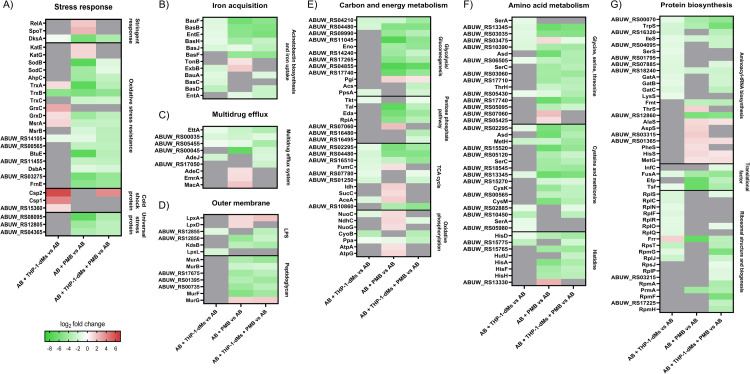
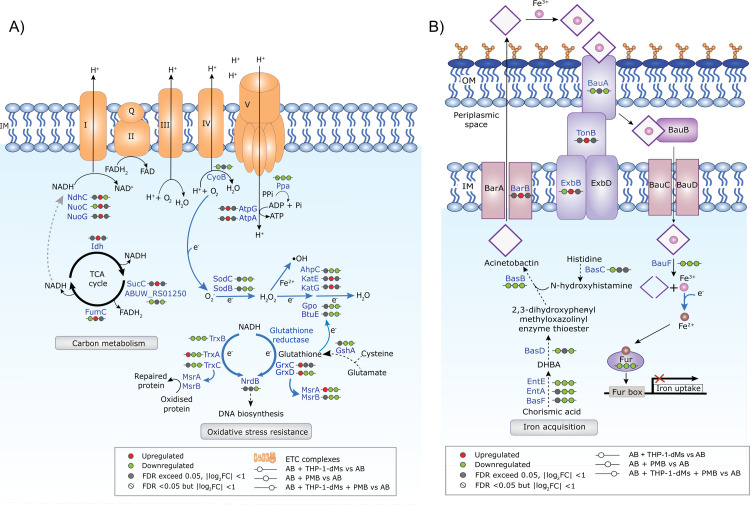
In response to THP-1-dMs, interacting bacteria uniquely upregulated several cold shock proteins namely Csp2 (ABUW_RS12225), Csp1 (ABUW_RS13055) and putative cold shock protein (ABUW_RS15360) with respective log2FC values of 6.12, 4.12 and 1.23, compared to the untreated controls (Fig 1A). Interacting bacteria also upregulated oxidative stress resistance-associated thioredoxin TrxA, glutaredoxins GrxC and GrxD, peptide-methionine (S)-S-oxide reductase MsrA, and an oxidative damage protection protein (ABUW_RS17615) (Fig 1A and S1 Table). THP-1-dMs infection alone also led to downregulation in bacterial iron acquisition systems. Several acinetobactin biosynthetic enzymes were significantly downregulated in the AB + THP-1-dMs group, including 2,3-dihydroxybenzoate-AMP ligase EntE, non-ribosomal peptide synthetase BasD, BasC and BasB, TonB-dependent siderophore receptors BauA, TonB-dependent receptor (ABUW_RS14495), FhuE (ABUW_RS08070), MotA/TolQ/ExbB proton channel family protein (ExbB, ABUW_RS16650), and acinetobactin utilization protein BauF (Fig 1B). Fig 2A and 2B show the schematics of oxidative stress resistance and iron acquisition pathways, respectively, with up- and down-regulated DEPs.
Fig 1. Polymyxins and THP-1-dMs cause unique proteomic changes in A. baumannii.
Heatmaps showing protein expression changes of AB5075 in A) stress response; B) metal acquisition; C) multidrug efflux; D) outer membrane; E) carbon and energy metabolism; F) amino acid metabolism; and G) protein biosynthesis in respective comparison groups. The colour code shows log2FC values of differentially expressed proteins with FDR <0.05 compared to that of untreated controls. Grey indicates protein expression exceeded an FDR of 0.05. Data are presented as mean of three biological replicates per experimental group. AB, wild-type A. baumannii AB5075; PMB, polymyxin B; THP-1-dMs, macrophage-like THP-1 cells.
Fig 2. A. baumannii differentially remodels oxidative stress resistance and iron homeostasis following different treatments.
Schematic diagram illustrating protein networks in AB5075 associated with A) carbon metabolism and oxidative stress resistance, and B) acinetobactin biosynthesis and iron acquisition. Red and green indicate upregulated and downregulated differentially expressed proteins, respectively. Data are presented as mean of three biological replicates per experimental group. IM, inner membrane; OM, outer membrane; ETC, electron transport chain; Gpo, glutathione peroxidase (ABUW_RS11455); GshA, glutamate—cysteine ligase (ABUW_RS00565); NrdB, ribonucleotide-diphosphate reductase subunit beta (ABUW_RS15470); DHBA, 2,3-dihydroxybenzoic acid; EntA, SDR family oxidoreductase (ABUW_RS10080).
Polymyxin B perturbed bacterial redox, energy, iron homeostasis and stringent response in A. baumannii
The bacterial proteome signature corresponding to antibiotic treatment alone (i.e., the AB + PMB group) includes a unique upregulation of catalases KatE and KatG, and severe downregulation of superoxide dismutases SodB (ABUW_RS05940) and SodC (ABUW_RS01670), with log2FC values of 1.46, 1.37, -7.32 and -4.08, respectively (Figs 1A and 2A). Notably, two thioredoxins (TrxA and TrxC), thioredoxin-disulfide reductase (TrxB), GrxD, and glutathione peroxidases BtuE (ABUW_RS18150) were substantially downregulated (log2FC values of -7.20, -2.23, -5.73, -4.31, and -7.21, respectively) (Figs 1A and 2A). Furthermore, polymyxin B treatment alone downregulated AB5075 protein quality control systems, including the DsbC family protein (ABUW_RS03275), DsbA family oxidoreductase (FrnE, ABUW_RS17415), thiol:disulfide interchange protein DsbA/DsbL (DsbA, ABUW_RS18725), MsrA and peptide-methionine (R)-S-oxide reductase MsrB (Fig 1A).
Interestingly, we observed a unique upregulation in tricarboxylic acid (TCA) cycle and oxidative phosphorylation proteins with polymyxin B treatment alone. These upregulated proteins included NADP-dependent isocitrate dehydrogenase Idh (ABUW_RS05265), ADP-forming succinate-CoA ligase beta subunit SucC, class II fumarate hydratase FumC, isocitrate lyase AceA (ABUW_RS14030), type I NADH dehydrogenase subunits NuoC, NdhC, and NuoG (involved in shuttling electrons from NADH to quinones in the electron transport chain [ETC]), F1Fo ATP synthase subunits AtpA (ABUW_RS18175) and AtpG (Figs 1E and 2A). In addition, three proteins responsible for energy-mediated uptake of iron complexes, acinetobactin export ABC transporter permease/ATP-binding subunit BarB, energy transducer TonB (ABUW_RS16655) and ExbB, were all mildly upregulated (log2FC values of 1.53, 1.01 and 1.80, respectively; Figs 1B and 2B). Conversely, there was a dramatic downregulation in acinetobactin biosynthetic enzymes EntE, BasB, BasF, and utilization protein BauF (log2FC values of -5.11, -5.19, -4.61 and -6.13, respectively; Figs 1B and 2B).
Stringent response regulators are critical in bacterial physiological activities promoting survival during stress [28], and several were significantly perturbed in response to polymyxin B alone, such as guanosine tetra- or penta-phosphate (p)ppGpp synthetase/hydrolase and RNA polymerase-binding protein DksA. There was a unique upregulation in bifunctional (p)ppGpp synthetase/guanosine-3’,5’-bis(diphosphate) 3’-pyrophosphohydrolase SpoT (ABUW_RS01520; log2FC, 2.46) and (p)ppGpp synthetase RelA (ABUW_RS16040; log2FC, 1.91) (Fig 1A); while, intriguingly, a dramatic downregulation of DksA (log2FC, -6.09) (Fig 1A).
Tripartite condition perturbed energy, stringent response, redox and metal homeostasis in A. baumannii
To determine whether THP-1-dMs affected the adaptive responses of interacting bacteria towards subsequent polymyxin B treatment, we compared the bacterial proteome signatures from the tripartite condition (i.e., AB + THP-1-dMs + PMB) with those of the bipartite condition (i.e., AB + PMB). Despite a high level of similarity in DEPs between the two groups, polymyxin-induced upregulation in energy production and ATP-dependent systems observed in the AB + PMB group was abrogated in the AB + THP-1-dMs + PMB group. For example, upregulation in SucC, NuoG, AtpA and AtpG observed in response to polymyxin B alone was reduced to a level approximating that of the untreated control in the AB + THP-1-dMs + PMB group (Fig 1E). Notably, the expression of NdhC changed from a log2FC of 1.35 when exposed only to polymyxin B to a log2 FC of -1.32 in the tripartite condition (Fig 1E). Energy-dependent systems that were uniquely upregulated only in response to polymyxin B were also abrogated in the presence of THP-1-dMs. These included multidrug efflux pump membrane proteins (MacA, AdeC, and EmrA/EmrK family multidrug efflux transporter periplasmic adaptor subunit [EmrA, ABUW_RS14655]), ATP-binding protein MlaF (ABUW_RS01880), methionine ABC transporter ATP-binding protein MetN (ABUW_RS07465), lipoprotein-releasing ABC transporter ATP-binding protein LolD, ATP-dependent Clp protease ATP-binding subunit ClpX, energy transducer TonB, and acinetobactin export associated BarB (Fig 1B, 1C and S1–S3 Tables).
Interacting bacteria isolated from the AB + THP-1-dMs + PMB group exhibited insignificant differences in KatE, KatG, GrxC, Csp1 and putative cold shock protein (ABUW_RS15360) compared to the untreated control (Fig 1A). However, unique proteome remodeling in the iron acquisition systems of interacting bacteria was also observed in this group, as shown by the tripartite-specific upregulation in TonB-dependent siderophore receptor (ABUW_RS18465) and downregulation in siderophore achromobactin biosynthesis protein AcsC (ABUW_RS10625) (S3 Table). In the interacting bacteria, the tripartite condition also severely downregulated the copper resistance system multicopper oxidase (ABUW_RS16135; log2FC, -3.00) and abrogated infection-specific upregulation in the heavy-metal associated domain-containing protein CopZ (ABUW_RS13140) observed in the AB + THP-1-dMs group (S1–S3 Tables). Interestingly, nickel and cobalt homeostasis associated RcnB family protein (ABUW_RS02965) was upregulated (log2FC, 3.16) solely in the tripartite condition (S3 Table). Of note, upregulation in SpoT and RelA observed in the AB + PMB group was both reduced to a level approximating that of the untreated control in the AB + THP-1-dMs + PMB group (Fig 1A).
Proteomic profiling of THP-1-dMs
We further examined the proteomics of THP-1-dMs samples to better understand the effects of A. baumannii infection, polymyxin B treatment, and their combination from the host (macrophages) perspective (S1 Fig). In total, approximately 4,320 mammalian proteins were identified across all samples involving THP-1-dMs. The PCA score plot revealed that the most distinct separation was between the AB + THP-1-dMs group and the untreated THP-1-dMs control (S2C Fig). Expression data of THP-1-dMs in the AB + THP-1-dMs + PMB group was closely clustered with those of the AB + THP-1-dMs group; whereas the data of the THP-1-dMs + PMB group were closely clustered with the untreated THP-1-dMs control (S2C Fig). Compared with the untreated control group, there were 98, 94 and 2 DEPs with the AB + THP-1-dMs, AB + THP-1-dMs + PMB, and THP-1-dMs + PMB groups, respectively (S2D Fig). Collectively, our data show that the THP-1-dMs proteome was predominantly modulated by AB5075 infection, and 1-h exposure to 30 mg/L polymyxin B had little effect on THP-1-dMs whether alone or in the presence of AB5075 infection. Pathway analysis revealed that infected THP-1-dMs from the AB + THP-1-dMs group were enriched in platelet activation and signaling, high density lipoprotein (HDL) remodeling, heme homeostasis and apoptosis (S3 Fig).
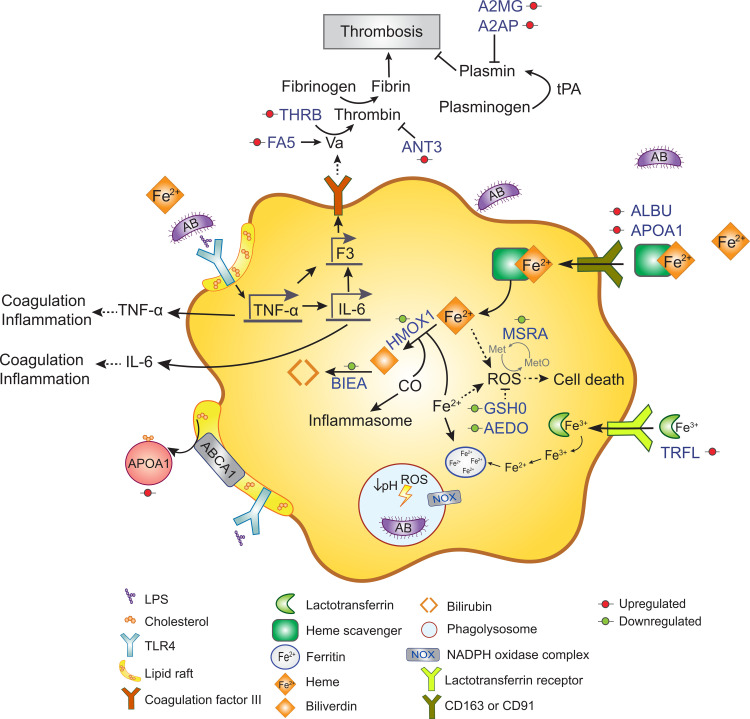
Compared to the untreated control, THP-1-dMs in the AB + THP-1-dMs group at 4 h post infection exhibited a significant upregulation of iron-binding proteins, including lactotransferrin (TRFL; log2FC, 2.01), heme scavenging associated proteins (e.g., serum albumin [ALBU; log2FC, 2.01], hemoglobin subunit alpha [HBA1; log2FC, 1.13), and apolipoprotein A-I [APOA1; log2FC, 1.74]) (Fig 3). However, downregulation was observed in heme catabolic enzymes, such as heme oxygenase 1 (HMOX1; log2FC, -1.64) and biliverdin reductase A (BIEA; log2FC, -2.68) (Fig 3). Notably, host cellular antioxidants were depleted in AB5075-infected THP-1-dMs with downregulation in the modifier subunit of glutamate-cysteine ligase GSH0 (log2FC, -1.51) and 2-aminoethanethiol dioxygenase AEDO (log2FC, -2.73) (Fig 3). Intriguingly, our findings reveal AB5075-induced upregulation of clotting cascade enzymes in infected THP-1-dMs. This was shown by the increased expression of coagulation factor V (FA5; log2FC, 3.37), prothrombin (THRB; log2FC, 1.27), fibrinolysis inhibitor alpha-2-antiplasmin (A2AP; log2FC, 1.45), and alpha-2-macroglobulin (A2MG; log2FC, 1.35) in the AB + THP-1-dMs group (Fig 3). Of note, anti-coagulant antithrombin-III (ANT3) was also upregulated in infected THP-1-dMs (log2FC, 1.52; Fig 3).
Fig 3. A. baumannii induces the activation of coagulation cascade in infected THP-1-dMs.
Schematic diagram illustrating protein networks involved with the coagulation cascade and iron-heme homeostasis in THP-1-dMs infected by AB5075. Red and green circles indicate upregulated and downregulated differentially expressed proteins, respectively. Data are presented as mean of three biological replicates per experimental group.
In the THP-1-dMs + PMB group, THP-1-dMs downregulated transcriptional regulator SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (SMCE1; log2FC, -2.08) and upregulated intracellular trafficking associated TBC1 domain family member 15 (TBC15; log2FC, 1.83). When the AB + THP-1-dMs + PMB and AB + THP-1-dMs groups were compared, downregulation was revealed in glycolytic enzyme ATP-dependent 6-phosphofructokinase, muscle type (PFKAM; log2FC, -4.23), while upregulation in phospholipid phosphatase 6 (PLPP6; log2FC, 2.56).
Impaired bacterial oxidative stress resistance, iron homeostasis and stringent response regulation enhanced polymyxin B killing and reduced bacterial virulence in vivo
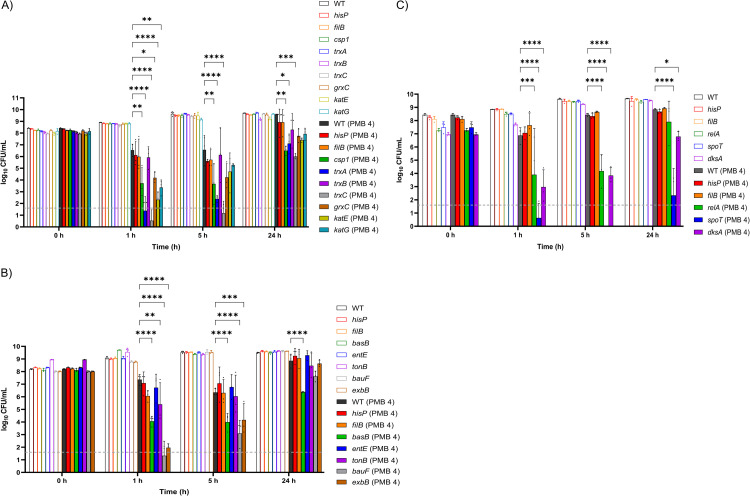
Next, we investigated the role of identified candidate targets in affecting bacterial killing by polymyxin B. As the wild-type A. baumannii AB5075 exhibited rapid tolerance towards polymyxins [29], 4 mg/L polymyxin B (i.e., 16× minimum inhibitory concentration [MIC]) was employed to compare the in vitro killing kinetics of the wild-type and mutants. Consistent with our hypothesis generated from the proteomics results above, enhanced bacterial killing by polymyxin B was observed with the selected AB5075 transposon mutants in redox stress resistance, namely those with disrupted trxC, trxA, katE, katG and csp1 (Fig 4A). At 1 and 5 h post polymyxin B treatment (4 mg/L), trxC and trxA mutants both showed 4- to 6 log10 CFU/mL additional killing compared to that of the wild-type, with >3 log10 CFU/mL additional killing at 24 h (Fig 4A). Enhanced polymyxin B killing was also observed with the katG and katE mutants at 1 h (at least 3 and 4 log10 CFU/mL), the csp1 mutant at 1, 5 and 24 h (at least ~3 log10 CFU/mL), and the grxC mutant at 1 and 5 h (>2 log10 CFU/mL) (Fig 4A). Polymyxin B time-kill kinetics of AB5075 mutants with T26 transposon-mutated pseudogenes hisP (ABUW_RS06545) or filB (ABUW_RS15750) mirrored those of the wild-type strain (Fig 4).
Fig 4. Impaired stress response machineries in A. baumannii enhances polymyxin killing.
Time-kill profiles of AB5075 transposon mutants with disrupted A) redox stress resistance genes; B) iron acquisition genes; C) stringent response regulatory genes following 4 mg/L polymyxin B (PMB) treatment. Data shown as mean ± SD of three biological replicates. Two-way ANOVA with Tukey’s HSD post-hoc test, * p <0.05, ** p <0.01, *** p <0.001, **** p <0.0001. Dashed line indicates the limit of detection.
AB5075 transposon mutants with disrupted iron acquisition genes were more susceptible to the initial killing by polymyxin B, compared to that of the wild-type strain. Much better polymyxin B killing was evident against bauF and exbB mutants, such as at 1 h with additional killing of 6 and 5 log10 CFU/mL, respectively (Fig 4B). With the basB mutant, >2 log10 CFU/mL killing was observed at 1 and 5 h, and 1.95 log10 CFU/mL killing with the tonB mutant at 1 h (Fig 4B).
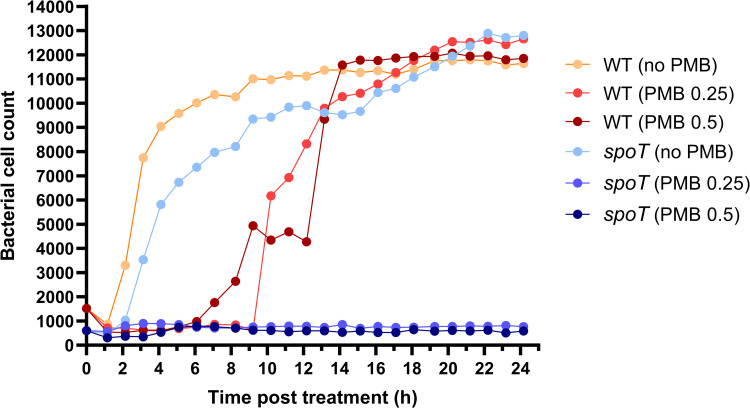
Notably, with polymyxin B treatment alone, inactivation in the bifunctional (p)ppGpp synthetase/hydrolase spoT resulted in the greatest enhancement of killing (6–8 log10 CFU/mL) in the tested mutants across all timepoints (Fig 4C). This dramatically enhanced killing matched the observations of our time-lapse imaging study which showed low cell counts of the spoT mutant across 24 h when treated with polymyxin B at 0.25 and 0.5 mg/L (Fig 5). In contrast, at both polymyxin B concentrations growth of the wild-type AB5075 was suppressed only over the first 7–10 h followed by rapid regrowth that approximated the control values by ~14 h (Fig 5). Interestingly, our imaging data revealed that, compared to the wild-type AB5075, spoT mutants exhibited a relatively smaller cell size at 0 h. AB5075 wild-type showed a mean (± SD) cell area of 3.35 ± 0.92 μm2 at 0 h; whereas spoT mutants exhibited a much lower mean (± SD) cell area of 2.58 ± 1.13 μm2 (Welch’s t-test, p <0.0001). At 24 h, the mean (± SD) cell area of spoT mutants treated with 0.5 mg/L polymyxin B did not increase (2.98 ± 1.43 μm2); while the mean (± SD) cell area of untreated spoT mutants increased to 4.91 ± 3.42 μm2, comparable to those of wild-type cells regardless of polymyxin B treatment (4.31 ± 2.56 μm2, 4.56 ± 2.81 μm2 and 4.58 ± 2.85 μm2 for untreated, 0.25 mg/L and 0.5 mg/L polymyxin B treated wild-type, respectively).
Fig 5. Time-lapse live-cell imaging reveals different growth kinetics of spoT mutant following polymyxin treatment.
ImageJ was employed for imaging analysis and cell count of AB5075 wild-type (WT) and spoT mutant following polymyxin B (PMB) treatment at 0.25 or 0.5 mg/L, and no treatment (i.e., controls).
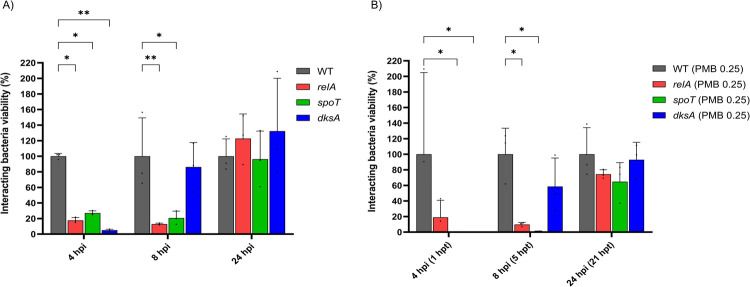
Inactivation of stringent response regulator dksA also led to enhanced polymyxin B killing, with ~4 log10 CFU/mL additional killing of the dksA mutants at 1 and 5 h and 2 log10 CFU/mL additional killing at 24 h (Fig 4C). Additionally, relA mutants exhibited reduced tolerance to initial polymyxin B exposure with additional killing of 2.97 and 4 log10 CFU/mL at 1 and 5 h, respectively (Fig 4C). Subsequently, we compared the antibacterial activity of polymyxin B against the three stringent response-associated mutants spoT, dksA, and relA to that of the wild-type in a macrophage infection microenvironment. Herein, 0.25 mg/L polymyxin B (i.e., 1×MIC) was employed to prevent overkill of interacting bacterial cells and allow comparison of the antibiotic killing dynamics between the interacting wild-type and mutants. Interestingly, during early THP-1-dMs infection in the absence of polymyxin B, all three mutants had significantly fewer interacting bacteria at 4 h post infection, compared to that of the wild-type (Fig 6A). At 4 h (i.e., following 1 h of polymyxin B treatment), the antibiotic treated wild-type group exhibited 2.7 log10 CFU/mL interacting bacteria; however, at this time no interacting bacteria were detected with the dksA mutant while the spoT mutant was reduced by 99.5% compared to that of the wild-type (Fig 6B). At 8 h (i.e., 5 h post polymyxin B treatment), the viability of the interacting spoT and relA mutants was significantly reduced (by >98% and 90%, respectively), compared to that of the wild-type (Fig 6B).
Fig 6. Impaired stringent response reduces interacting bacterial growth in the presence of THP-1-dMs.
Interacting bacterial growth of AB5075 wild-type (WT) and transposon mutants with disrupted stringent response regulatory genes following MOI 1,000 infection of THP-1-dMs in the A) absence and B) presence of 0.25 mg/L polymyxin B (PMB) treatment. Data shown as mean ± SD of three biological replicates compared to that of the wild-type. Two-way ANOVA with Tukey’s HSD post-hoc test, * p <0.05, ** p <0.01.
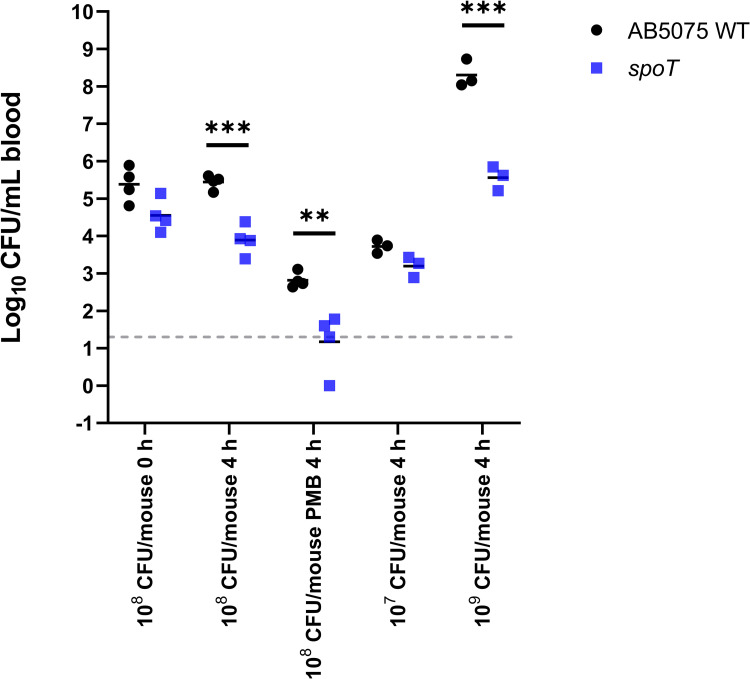
To validate our in vitro findings, we employed spoT mutants as an example to examine the role of bacterial stringent response in the survivability in an immunocompetent mouse bacteremia model. At 6 h after bacterial inoculation (in the absence of polymyxin B), significantly reduced bacterial load in blood was observed in mice infected by the spoT mutant at both 108 and 109 CFU/mouse inoculum, compared to that of AB5075 wild-type infection control (Fig 7). Using the same mouse bacteremia model with intravenous polymyxin B (4 mg/kg, the maximum dose in mice via intravenous administration), the average bacterial count of spoT mutant in blood at 4 h reduced to below the limit of detection of 1.30 log10 CFU/mL, showing at least >1.65 log10 CFU/mL reduction compared to that of the wild-type AB5075 (Fig 7).
Fig 7. spoT mutant exhibits reduced virulence in vivo.
Bacterial load of AB5075 wild-type (WT) and spoT mutant in blood in a mouse bacteremia model with and without polymyxin B (PMB) treatment (4 mg/kg, intravenous). At least three biological replicates per experimental group were examined. Multiple unpaired t-tests, ** p <0.01, *** p <0.001. Dashed line indicates the limit of detection.
Discussion
Our study is the first to examine the complex tripartite interplay among A. baumannii, macrophages and an antibiotic using an in vitro host-pathogen-drug model. Using correlative proteomics, we discovered several potential pathogen-directed or host-directed candidate proteins and pathways for future therapeutic targeting. Correlative proteomic profiling of infected macrophages and their interacting bacteria allowed an integrative analysis of the interaction between innate immune cells and pathogen during the infection. A high multiplicity of infection (MOI; 1,000) was employed to achieve a sufficient number of interacting bacterial cells for proteomic analysis. Considering the rapid killing by polymyxins [30], 4 h post infection and 1 h post treatment was examined. The polymyxin concentration of 30 mg/L used in this proteomic study is achievable in lung epithelial lining fluid in patients following aerosolized polymyxins [31]. We identified diverse responses and tolerance strategies of A. baumannii towards polymyxin B, macrophages, and the combination. It was revealed that macrophages and polymyxin B exhibited complementary effects to disarm several tolerance and survival strategies of A. baumannii, most notably oxidative stress resistance, copper tolerance, bacterial iron acquisition and stringent response regulation systems. Importantly, we have identified several novel pathogen-directed targets from the redox stress response system, iron acquisition and stringent response regulation that warrant further investigations for developing potential therapeutic strategies to enhance polymyxin efficacy against MDR A. baumannii.
A major finding here is the association of polymyxin activity with (p)ppGpp-mediated stringent response in AB5075. As nutritional stress (e.g., starvation of amino acids, carbon, fatty acids, phosphate and iron) can trigger (p)ppGpp biosynthesis [28,32–34], it is very likely that the upregulation of bifunctional (p)ppGpp synthetase/guanosine-3’,5’-bis(diphosphate) 3’-pyrophosphohydrolases SpoT and (p)ppGpp synthetase RelA in A. baumannii after polymyxin treatment (Fig 1A) was triggered by polymyxin-induced nutritional stress. This is supported by the downregulation of proteins involved in amino acid biosynthesis, glycolysis, and iron uptake systems (Fig 1B, 1E and 1F), as well as the previously reported transcriptional perturbations of fatty acids biosynthesis in A. baumannii following polymyxin treatment [35]. Alarmone (p)ppGpp promotes bacterial survival in adverse growth conditions via global reprogramming of bacterial cellular and metabolic activities, during which (p)ppGpp binds to the RNA polymerase to regulate gene expression [36]. The outcome of this process is reduced transcription of ribosomal and transfer RNAs necessary for translation in order to balance protein biosynthesis with nutrient availability [36–38]. In the present study, we observed the downregulation of translational machinery such as elongation factors FusA, EfP, and Tsf (ABUW_RS06040) in polymyxin-treated A. baumannii, representative of stringent response activation (Fig 1G). The crucial role of (p)ppGpp-mediated stringent response in governing oxidative stress resistance, heat shock response, antibiotic tolerance and virulence has been recently reported in F. tularensis, B. subtilis and P. aeruginosa [39–41]. Several recent studies on relA-deleted A. baumannii unveiled a global regulatory role of relA in bacterial adherence, biofilm formation, and Ade multidrug efflux family; as well as a reduced virulence in a Galleria mellonella model and neutropenic murine pneumonia model [42–44]. Here, we firstly identified the key role of (p)ppGpp-mediated stringent response in the host-pathogen-drug interplay using proteomics, and subsequently demonstrated that the transposon mutants relA (ABUW_RS16040) and especially spoT (ABUW_RS01520) in AB5075 were much more susceptible to polymyxin killing (Fig 4C). Our findings highlight the importance of the (p)ppGpp-mediated stringent response in polymyxin tolerance in A. baumannii. The prolonged (up to 24 h) suppression of bacterial replication of polymyxin-treated spoT mutants observed in our time-lapse imaging study (Fig 5) further highlights the crucial role of spoT mediated nutritional regulation and downstream signaling in polymyxin tolerance.
Besides (p)ppGpp synthetase/hydrolase, our study also highlights the critical role of another stringent response regulator DksA in mediating A. baumannii tolerance towards polymyxins. DksA exerts a synergistic regulatory role in the (p)ppGpp-mediated stringent response in Salmonella enterica [45]. In nutritionally-starved Salmonella, DksA plays a key role in regulating the NAD(P)H/NAD(P)+ redox balance that fuels downstream antioxidant systems, including the thioredoxin superfamily, through fine-tuning discrete steps in central carbon metabolism [46]. In line with this notion, we discovered dramatic downregulation of DksA protein abundance in polymyxin-treated A. baumannii along with the downregulation of NAD(P)H-dependent antioxidant systems, including thioredoxin and glutaredoxin, and protein repair associated Dsb systems (Figs 1A and 2A). Notably, our transposon mutant study further confirmed that singly-gene disruption of relA, spoT or dksA significantly decreased the growth of interacting A. baumannii during early macrophage infection. Consistent with our tripartite proteomics data showing complementary action between macrophages and polymyxins in disarming the upregulation of (p)ppGpp synthetase/hydrolase SpoT (Fig 1A), a low concentration of polymyxin B (0.25 mg/L) in the THP-1-dMs infection system is sufficient to significantly reduce the survival of interacting spoT mutants compared to the wild-type strain (Fig 6B). Importantly, our in vivo study further validated that disruption of spoT reduced A. baumannii survivability and polymyxin treatment cleared most of the spoT mutants in blood of the infected mice (Fig 7). Collectively, these results show a potential therapeutic benefit of targeting the A. baumannii stringent response regulation (not limited to relA) to reduce bacterial virulence and enhance polymyxin killing, including macrophage-associated interacting bacteria.
Furthermore, our study also discovered several proteins of oxidative stress resistance and metal homeostasis systems that underlie A. baumannii pathogenesis and polymyxin tolerance. During infection, A. baumannii can induce reactive oxygen species (ROS) production in macrophages [47] and phagocytes such as macrophages employ respiratory burst (i.e., a rapid increase in the production of ROS during phagocytosis) to kill bacteria [48]. Our results here highlight the distinct oxidative stress resistance machineries in A. baumannii in response to macrophages and polymyxins. Interacting A. baumannii may utilize cold shock proteins and proteins of the thioredoxin superfamily to facilitate their tolerance against redox stress and lysosomal pH encountered in macrophages (Fig 1A). This finding is new as the use of cold shock proteins to resist redox stress has not been reported in A. baumannii. In the literature, cold shock protein CspA in Brucella melitensis plays an important role in resistance to acidic and hydrogen peroxide stresses, allowing bacterial replication in J774.A1 murine macrophages [49]. The thioredoxin superfamily includes both the thioredoxin and glutathione/glutaredoxin systems, and serves as a key antioxidant system through regulation of protein dithiol/disulfide balance [50,51]. It is also involved in DNA and protein repair by transferring reducing equivalents to ribonucleotide reductase, Msr and Dsb systems, as well as regulating the activity of redox-sensitive transcription factors [52]. Our finding of the upregulation of TrxA in interacting A. baumannii with macrophages is in agreement with the known roles of TrxA in facilitating bacterial intracellular replication in epithelial cells or macrophage-like cells, resistance to hydrogen peroxide, and in vivo virulence of several bacterial species, including A. baumannii [53–55]. Moreover, we demonstrated here a critical link between A. baumannii oxidative stress resistance systems and polymyxin antibacterial efficacy. In A. baumannii polymyxins induce upregulation of H2O2-hydrolyzing catalases and their killing is potentially mediated by a hydroxyl radical death pathway [56,57]. Enhanced bacterial killing of A. baumannii has been reported with a combination of colistin and pro-oxidant curcumin that synergistically increased ROS production [58]. The results of our transposon mutant time-kill kinetics highlight the crucial role of catalases (KatE, KatG), thioredoxin (TrxA, TrxC) and glutaredoxin (GrxC) in A. baumannii to resist polymyxins (Fig 4A). Notably, the increased expression of catalases, thioredoxin superfamily, and cold shock proteins in interacting A. baumannii was abrogated in the tripartite condition (Fig 1A), indicating complementary antibacterial activity between macrophages and polymyxins. Together, our findings highlight the potential of targeting such oxidative stress resistance systems as to enhance polymyxin activity.
Maintenance of metal homeostasis is crucial for both the host and pathogen [59]. Metals serve as cofactors in diverse biochemical processes, but are toxic at high concentrations [59,60]. During bacterial infection, the host innate immune system could harness copper toxicity as an antimicrobial strategy [61]. The mechanisms underlying copper toxicity include rapid inactivation of iron-sulfur dehydratase family [62]; incorrect periplasmic disulfide bond formation [63]; and Fenton and Haber-Weiss reaction-mediated hydroxyl radical generation [64]. Attenuated virulence of A. baumannii mutants with disrupted copper resistance genes in Galleria mellonella and mouse pneumonia infection models highlight the importance of copper tolerance in bacterial virulence within host [65,66]. Our proteomics results indicate that interacting A. baumannii cells likely utilized glutaredoxins (GrxC, GrxD) and a putative copper chaperone CopZ (ABUW_RS13140) as a copper tolerance strategy to survive within macrophages. Glutaredoxin minimizes the incorrect formation of disulfide bonds in cellular proteins resulting from copper-induced oxidation of sulfhydryl groups [67,68]. In Escherichia coli, inactivation of the disulfide bond isomerase dsbC increases the sensitivity to copper toxicity through impairing bacterial ability to resolve copper-catalyzed non-native disulfides [63]. Interestingly, our proteomics results indicate that polymyxin treatment likely sensitizes interacting bacteria towards host-induced copper toxicity by preventing the upregulation of putative copper chaperone CopZ, and by inducing downregulation of the bacterial Dsb system (S1–S3 Tables and Fig 1A). Downregulation of multicopper oxidase (ABUW_RS16135) was only observed in the tripartite condition (S3 Table). ABUW_RS16135 encodes for a CopA homolog (87.4% identity with CopA [AMQ95338.1] in A. baumannii), and shares similarity with periplasmic copper oxidase PcoA (BCZ13020.1) in A. baumannii (83.6% identity) which detoxifies oxidizing Cu1+ to less damaging Cu2+ ions [69,70]. Such tripartite-specific downregulation in CopA (ABUW_RS16135) suggests a novel cooperative action of macrophages and polymyxins in perturbing A. baumannii defense against toxic cuprous ions. Overall, our findings indicate a complementary antibacterial mechanism between macrophages and polymyxins against A. baumannii by both impairing bacterial copper detoxification.
In addition, mammalian hosts can activate nutritional immunity through restricting the availability of nutrient metals to invading bacteria [71]. Our proteomics results indicate that infected macrophages induced host iron sequestration via increased cellular uptake of lactotransferrin and heme scavengers from the environment (Fig 3), corroborating with the observed upregulation of lactotransferrin and heme scavenging associated haptoglobin in an A. baumannii lung infection proteomic study in mice [72]. Attenuation of bacterial siderophore biosynthesis, supplementation of iron competitors (i.e., gallium) and iron scavenger (i.e., host-derived transferrin) can reduce bacterial survival and virulence (e.g., A. baumannii) in vitro and in vivo [73–75]. Additionally, our host proteomics results revealed that A. baumannii-infected macrophages downregulated the host heme catabolism, which possibly activated innate immune responses [76]. Depletion of heme catabolic enzymes HMOX1 and BIEA in infected macrophages leads to intracellular accumulation of pro-oxidative and pro-inflammatory free labile heme [77,78]. Free heme may act as damage-associated molecular patterns (DAMPs), amplifying the innate immune response through induction of toll-like receptor (TLR)-mediated production of pro-inflammatory tumor necrosis factor TNF-α, and NADPH oxidase (NOX)-dependent ROS production in phagocytes [76,79]. In line with this, the recent proteomic study of A. baumannii lung infection in mice indicated the activation of NOX signaling as an important host defense strategy; whereas the role of heme in this process warrants further investigation [72,80]. On the other hand, inefficient neutralization of excessive free heme leads to heme toxicity and sepsis [81]. In addition, an overwhelming pro-inflammatory response during Acinetobacter infection may activate the coagulation cascade and lead to thrombosis and disseminated intravascular coagulation [82]. Our proteomics findings indicate that better understanding of A. baumannii-induced coagulation may assist with the treatment of A. baumannii infection.
In the present study, polymyxin-treated A. baumannii cells modulated their ferric iron acquisition systems by downregulating acinetobactin biosynthetic enzymes at 1 h post treatment (Figs 1B and 2B), indicating bacterial attempt to reduce the iron-catalyzed Fenton reaction and ROS production during early polymyxin exposure. Similarly, downregulation in siderophore-mediated iron acquisition in interacting bacterial cells (Figs 1B and 2B) and extracellular bacterial cells [83] following macrophage infection indicates a common stress tolerance strategy in A. baumannii through regulating the siderophore activities. Iron is an essential enzymatic cofactor in a wide variety of fundamental metabolic pathways, including antioxidant systems (e.g., iron-containing superoxide dismutases and catalases), central carbon metabolism (e.g., iron-sulfur domain-containing aconitase, succinate dehydrogenase, and NADH dehydrogenase), and DNA biosynthesis and repair [84–86]. Therefore, suppression of iron uptake can cause iron starvation and subsequent activation of bacterial iron acquisition system as a feedback mechanism. In agreement with this, we observed that polymyxin-treated A. baumannii upregulated their TonB protein and MotA/TolQ/ExbB proton channel for active uptake of ferric iron complexes, likely to cope with iron starvation (Figs 1B and 2B). Further, our AB5075 transposon mutant screening demonstrated, for the first time, the importance of iron acquisition proteins BasB, EntE, MotA/TolQ/ExbB proton channel (ExbB) and iron assimilation protein BauF in mediating polymyxin tolerance in AB5075. Indeed, disruption of bacterial heavy metal homeostasis by ionophore 2-(dimethylamino) methyl-5,7-dichloro-8-hydroxyquinoline (PBT2) dramatically sensitizes polymyxin-resistant Gram-negative (e.g., A. baumannii) to polymyxin killing [87]. Taken together, our findings highlight the therapeutic potential of targeting heavy metal homeostasis in A. baumannii (e.g., iron acquisition) to enhance polymyxin killing.
Conclusions
With the emergence of polymyxin-resistant A. baumannii threatening the utility of this important last-line class of antibiotics, innovative strategies to preserve their clinical efficacy (e.g., development of novel adjuvant therapies) are urgently needed. Our proteomics findings in the A. baumannii-macrophage-polymyxin interactions discovered that key bacterial stress responses and tolerance strategies towards polymyxins and/or macrophages may serve as pathogen-directed therapeutic target candidates. In particular, the stringent response regulator spoT is exemplified as how a single gene/protein modulation can affect A. baumannii tolerance towards polymyxins in vitro and in mice. Our findings highlight the significant potential of deciphering the complex tripartite interactions between host, pathogen and drug in optimizing antibiotic use and expediting antibiotic discovery against MDR pathogens.
Materials and methods
Bacterial strains, media and antibiotic
Acinetobacter baumannii AB5075 (wild-type [WT]) is a recently characterized MDR isolate collected from a wound of a patient at the Walter Reed Army Medical Center [88]. The A. baumannii AB5075-UW transposon mutant library with targeted gene disruption via single T26 transposon insertion was purchased from the University of Washington [89]. The AB5075 transposon mutants used in this study are shown in S4 Table and were validated using colony polymerase chain reaction. Logarithmic-phase cultures of AB5075 WT were prepared from -80°C frozen stock, plated onto nutrient agar (NA), and incubated aerobically at 37°C for 16 to 18 h. A single bacterial colony was then inoculated into cation-adjusted Mueller-Hinton broth (CaMHB) and incubated overnight at 37°C with shaking (200 rpm) for 16–18 h. Overnight cultures were diluted 100-fold in pre-warmed CaMHB and further grown at 37°C to reach an optical density (OD) of 0.5 at 600 nm. AB5075 transposon mutants were similarly prepared using Luria-Bertani (LB) agar supplemented with 10 mg/L tetracycline hydrochloride (Catalog: T7660-5G; Sigma). Polymyxin B sulphate (PMB; Catalog: 86–40302) was obtained from Betapharma (Shanghai, China). Sterile stock solutions were prepared using Milli-Q water (Millipore, USA) filtered through a 0.22-μm syringe filter (Sartorius, Germany).
Mammalian cell line
THP-1 (ATCC TIB-202), a human leukemia monocytic cell line, was maintained in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 25 mM of 4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid (HEPES) buffer solution and 10% fetal bovine serum (FBS; Lot 15703; Bovogen, Victoria, Australia) and incubated at 37°C in a humidified atmosphere containing 5% CO2. THP-1 cells were differentiated into macrophage-like THP-1 cells (THP-1-dMs) via 48 h of treatment with 25 nM of phorbol 12-myristate 13-acetate (PMA; Santa Cruz Biotechnology; Texas, USA), after which the media was removed and replaced with fresh pre-warmed RPMI 1640 media supplemented with 25 mM HEPES and 10% FBS. Differentiated cells were allowed to rest for at least 24 h in PMA-free media prior to experiments.
THP-1-dMs infection and polymyxin treatment
To obtain sufficient interacting bacterial proteins for mass spectrometric analysis, AB5075 infection of THP-1-dMs was performed in two T175 flasks (Corning; New York, USA) at a multiplicity of infection (MOI) of 1,000; the content of each flask was subsequently pooled to obtain a single sample. For host cell proteomics samples, infection was performed in 6-well plates (Corning; Jiangsu Province, China) and proceeded for 4 h in RPMI 1640 media supplemented with 25 mM HEPES and 10% heat-inactivated FBS at 37°C (5% CO2). For antibiotic-treated samples, polymyxin B 30 mg/L was added at 3 h post infection meaning cells were exposed to polymyxin B for 1 h. At 4 h, cell monolayers were washed twice with ice cold Dulbecco’s phosphate-buffered saline (DPBS, 1×; Gibco, Paisley, UK) to remove non-interacting extracellular bacteria. To isolate interacting bacteria from infected THP-1-dMs, cells were firstly lysed in 1% Triton X-100 for 1 h on ice. A two-step differential centrifugation strategy was then performed to separate interacting bacterial cells from host cell debris [90]. Firstly, the sample was centrifuged at 600 × g and 4°C for 5 min to pellet host cell debris. Second, the supernatant containing interacting bacteria released from lysed host cells was centrifuged at 3,220 × g for 20 min (at 4°C) to pellet bacteria. The resulting bacterial pellet was thrice washed with ice cold DPBS (1×) to minimize contamination from both residual host proteins and Triton X-100. The final bacterial pellets were stored at -20°C until further analysis. The respective controls of THP-1-dMs and AB5075 were treated in the same manner. Three biological replicates were included for each experimental condition.
LC-MS/MS sample preparation and analysis
To extract proteins from infected THP-1-dMs, cell monolayers were washed twice with ice cold DPBS (1×) to remove residual media prior to the treatment with lysis buffer 1% sodium deoxycholate (SDC; Sigma, D6750-100G) in 100 mM HEPES (pH 8.5). For bacterial protein extraction, the previously isolated bacterial pellet was resuspended with 1% SDC in 100 mM HEPES (pH 8.5). SDC-treated samples were heated at 95°C for 10 min prior to probe sonication at output strength 3 and 50% duty cycle. Sample protein was then quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific; Illinois, USA). The total protein was normalized to 150 μg followed by protein denaturation and alkylation with 10 mM bond-breaker tris-(2-carboxyethyl) phosphine (TCEP) and 40 mM chloroacetamide (CAA). Samples were then subjected to overnight trypsin digestion at 37°C. SDC was subsequently removed by 1% formic acid precipitation and the sample peptides purified using the Eppendorf PerfectPure C18 ZipTip protocol. Acetonitrile in eluted samples was removed by a vacuum concentrator. Peptide samples were then reconstituted in 0.1% formic acid and sonicated in a water bath prior to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Mass spectrometric analysis were performed by the Monash Proteomics & Metabolomics Facility. Briefly, a Dionex UltiMate 3000 RSLCnano system was employed with an Acclaim PepMap RSLC analytical column (75 μm × 50 cm, nanoViper, C18, 2 μm, 100Å; Thermo Scientific) and an Acclaim PepMap 100 trap column (100 μm × 2 cm, nanoViper, C18, 5 μm, 100Å, Thermo Scientific). Tryptic peptides of bacterial cells were analyzed on an Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Fisher Scientific) operated in data-dependent acquisition (DDA). For AB5075, raw files were analyzed with MaxQuant v1.6.5.0 and statistical analysis of the label-free quantification (LFQ) intensities was performed in Perseus v1.6.2.3. The AB5075 proteome was annotated using the NCBI Assembly Database (https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/963/815/GCF_000963815.1_ASM96381v1/). For THP-1-dMs, the tryptic peptides were analyzed on a QExactive Plus Mass Spectrometer (Thermo Scientific) operated in data-independent acquisition (DIA) mode. The raw files of THP-1-dMs data were analyzed with Spectronaut v13 Laika (Biognosys) to obtain relative LFQ values using in-house standard parameters. The expression dataset of each experimental groups was visualized with a principal component analysis (PCA) score plot generated using MetaboAnalyst and Venn diagram generated using InteractiVenn [91,92]. Differences in protein expression levels were evaluated through comparison of the mean LFQ intensities among all experimental groups and expressed as the log2 fold change (log2 FC). Significance was determined using a two-sided, two-sample t-test with a false discovery rate (FDR) adjusted p-value. Differentially expressed proteins (DEPs) with a log2FC >1 or <-1 and FDR <0.05 were considered statistically significant. Statistically significant bacterial DEPs were subjected to pathway analysis according to Clusters of Orthologous Groups (COGs) and predicted by eggNOG [93] and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using R. Statistical significance of enrichment analysis was examined using Fisher’s Exact Test (FDR <0.2). THP-1-dMs DEPs were functionally analyzed on KEGG and Reactome pathways using WebGestalt with the pathway enrichment significance set at FDR <0.05 using the Benjamini-Hochberg method [94,95].
Functional investigation of AB5075 mutants
The time-kill kinetics of polymyxin B against AB5075 wild-type and its transposon mutants were examined. Logarithmic-phase broth cultures (in CaMHB) of respective bacterial isolates were centrifuged at 3,220 × g for 20 min to pellet bacteria. Pellets were subsequently resuspended in pre-warmed RPMI 1640 media supplemented with 25 mM HEPES and 10% heat-inactivated FBS to yield an inoculum of ~108 CFU/mL, followed by polymyxin B treatment at 4 mg/L. Treated cultures were incubated at 37°C with shaking at 200 rpm. At baseline (0 h) and 1, 5, and 24 h post polymyxin B treatment, viable bacteria were quantified by manually plating 25 μL of appropriately diluted bacterial suspension onto nutrient agar followed by incubation at 37°C for 16–18 h prior to colony counting.
To examine the interacting growth of AB5075 WT and candidate transposon mutants following THP-1-dMs infection and polymyxin B treatment, THP-1-dMs were firstly infected at MOI 1,000 on 24-well plates and incubated at 37°C (5% CO2). At 3 h post infection, non-interacting extracellular bacteria were discarded and the cell monolayer was washed twice with pre-warmed DPBS (1×) before being replaced with pre-warmed cell culture media containing 0.25 mg/L polymyxin B. The experimental culture was further incubated at 37°C (5% CO2). At 1, 5, and 21 h post polymyxin B treatment, cell monolayers were washed twice with ice cold DPBS (1×) to remove non-interacting extracellular bacteria and THP-1-dMs were lysed in 1% Triton X-100 for 1 h on ice. The lysates containing the released interacting bacteria from infected THP-1-dMs were centrifuged at 10,000 × g for 10 min to pellet bacterial cells and remove the residual Triton X-100. The bacterial pellet was resuspended in 0.9% NaCl solution and 25 μL of bacterial suspension was spot-plated on NA following appropriate serial dilution, and then incubated at 37°C for 16–18 h prior to colony counting.
Time-lapse live-cell imaging
Logarithmic-phase cultures of AB5075 wild-type and spoT mutant were firstly pelleted at 3,220 × g for 20 min and resuspended in RPMI 1640 media supplemented with HEPES and 10% heat-inactivated FBS. To ensure sufficient and non-overcrowding of bacterial cells throughout the 24-h continuous imaging, bacterial cell density was adjusted accordingly by examination on EVE Cell Counting Slides using a Leica DMi8 Inverted Microscope (Leica Microsystems) at 20× magnification. Images were taken at baseline (0 h) using untreated controls. Additionally, 40 μL of the adjusted bacterial cultures were added to respective treatment wells of μ-Slide VI (IBIDI) pre-loaded with 60 μL of media containing no polymyxin B (0 mg/L) or polymyxin B at 0.25 mg/L (1× minimum inhibitory concentration [MIC]) or 0.5 mg/L (2×MIC). The treatment slide was incubated on the microscope stage at 37°C. Using 20× magnification, twelve spots per treatment group were marked and traced, and images at all spots were collected every 10 min for 24 h. The time-lapse videos for each treatment group are attached in S1–S6 Videos. For analysis of bacterial cell size changes over time, images of the same spot collected every 1 h for 24 h were examined using ImageJ [96].
Mouse bacteremia model
An immunocompetent mouse bacteremia model was employed to examine the survivability of spoT mutants and AB5075 wild-type, and polymyxin B efficacy in vivo (Monash University, Clayton, Victoria, Australia). Female Swiss mice (8-week-old, weight of 24–32 g) were housed with food and water available ad libitum and were used in the immunocompetent mouse bacteremia model (Monash University, Clayton, Victoria, Australia). Mice were randomly assigned to different experimental groups. Early logarithmic-phase cultures (100 μL) of AB5075 wild-type or spoT transposon mutants were intravenously administered into each female Swiss mouse to establish bloodstream infection (bacterial inoculum of 1×107, 108 or 109 CFU/mouse). At 2 h after inoculation, polymyxin B was administered intravenously at 4 mg/kg to the treatment group. Mice were euthanized at 0 h and 4 h post polymyxin B treatment using 100% carbon dioxide inhalation, and blood was collected using cardiac puncture. Mouse blood was serially diluted with 0.9% saline and spirally plated on nutrient agar plates to determine bacterial load in blood (CFU/mL). At least three independent biological replicates per experimental group were examined in this study. All animal experiments were approved by the Monash University Animal Ethics Committee and performed in compliance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
Quantification and statistical analysis
Bacterial viable count in the in vitro time-kill assay and THP-1-dMs infection study were quantified using spot assay on nutrient agar plate (25 μL per spot; 40 CFU/mL as the limit of detection) and statistically analyzed using two-way ANOVA followed by Tukey’s HSD post-hoc test on GraphPad Prism 9.1.0. In the mouse bacteremia study, bacterial load in blood was quantified by spirally spreading on nutrient agar plates (50 μL; 20 CFU/mL as the limit of detection). Multiple unpaired t-tests were performed in GraphPad Prism 9.1.0 to analyze the in vivo bacterial count, with p-value <0.05 defined as statistically significant. The sample size used in each experiment is specified in the methodology sections and figure legends.
Supporting information
(TIF)
A) PCA score plot of A. baumannii proteomic profiles showing the relatedness of the dataset within and across the experimental groups of untreated AB5075 (i.e., controls; AB), polymyxin B treated AB5075 (AB + PMB), interacting AB5075 from infected THP-1-dMs (AB + THP-1-dMs), and interacting AB5075 from polymyxin B treated THP-1-dMs infection (AB + THP-1-dMs + PMB). B) Venn diagram showing common and unique sets of differentially expressed proteins of A. baumannii (log2FC >1 or <-1, FDR <0.05) between different comparison groups. C) PCA score plot of THP-1-dMs proteomic profiles showing the relatedness of the dataset within and across the experimental groups of untreated THP-1-dMs (i.e., controls; THP-1-dMs), AB5075-infected THP-1-dMs (AB + THP-1-dMs), polymyxin B treated THP-1-dMs (THP-1-dMs + PMB), and polymyxin B treated and AB5075-infected THP-1-dMs (AB + THP-1-dMs + PMB). D) Venn diagram showing common and unique sets of differentially expressed proteins of THP-1-dMs (log2FC >1 or <-1, FDR <0.05) between different comparison groups. Three biological replicates were employed in each experimental group.
(TIF)
Volcano plot generated using WebGestalt showing the enriched KEGG and Reactome pathways (Benjamini-Hochberg FDR <0.05) in THP-1-dMs at 4 h post infection with AB5075 in the absence of polymyxin B.
(TIF)
DEPs, differentially expressed proteins; AB, wild-type Acinetobacter baumannii AB5075; PMB, polymyxin B; THP-1-dMs, macrophage-like THP-1 cells.
(XLSX)
DEPs, differentially expressed proteins; AB, wild-type Acinetobacter baumannii AB5075; PMB, polymyxin B; THP-1-dMs, macrophage-like THP-1 cells.
(XLSX)
DEPs, differentially expressed proteins; AB, wild-type Acinetobacter baumannii AB5075; PMB, polymyxin B; THP-1-dMs, macrophage-like THP-1 cells.
(XLSX)
(XLSX)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
Acknowledgments
We thank the technical assistance provided by Monash Proteomics & Metabolomics Facility (MPMF), which operates BPA-enabled (Bioplatforms Australia) / NCRIS-enabled (National Collaborative Research Infrastructure Strategy) infrastructure. We thank Dr. Jiping Wang and Mr. Ke (Kenny) Chen for their assistance in planning and performing experiments on mice; and Dr. Phillip Bergen for proofreading the article.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the manuscript and its Supporting Information files. The mass spectrometry proteomics data has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD028502.
Funding Statement
This research was funded by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health, R01 AI132681 and AI146160 (T.Q.Z. and J.L.; https://grants.nih.gov/grants/funding/r01.htm). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. Z.Y.K. was supported by Monash Graduate Scholarship (https://www.monash.edu/study/fees-scholarships/scholarships/find-a-scholarship/monash-graduate-scholarship-mgs). J.L. is an Australian National Health and Medical Research Council (NHMRC) Principal Research Fellow (APP1157909) [https://www.nhmrc.gov.au/funding/manage-your-funding/personnel-and-salary-support-packages] and T.N. is an Australian Research Council Future Fellow (FT170100313) [https://www.arc.gov.au/grants/discovery-program/future-fellowships]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tagliabue A, Rappuoli R. Changing priorities in vaccinology: antibiotic resistance moving to the top. Front Immunol. 2018;9:1068. doi: 10.3389/fimmu.2018.01068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'neill J. Antimicrobial resistance. 2014 [September 14, 2021]. Available from: https://amr-review.org/.
- 3.Centres for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. Atlanta, GA: US Department of Health and Human Services, CDC; 2019. [Google Scholar]
- 4.World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics 2017 [September 14, 2021]. Available from: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1.
- 5.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther. 2015;40(4):277. [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. Antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline 2019 [September 14, 2021]. Available from: https://www.who.int/publications/i/item/9789240000193.
- 7.Oliva A, Ceccarelli G, De Angelis M, Sacco F, Miele MC, Mastroianni CM, et al. Cefiderocol for compassionate use in the treatment of complicated infections caused by extensively and pan-resistant Acinetobacter baumannii. J Glob Antimicrob Resist. 2020;23:292–6. doi: 10.1016/j.jgar.2020.09.019 [DOI] [PubMed] [Google Scholar]
- 8.Yang Q, Pogue JM, Li Z, Nation RL, Kaye KS, Li J. Agents of last resort: an update on polymyxin resistance. Infect Dis Clin North Am. 2020;34(4):723–50. doi: 10.1016/j.idc.2020.08.003 [DOI] [PubMed] [Google Scholar]
- 9.Nang SC, Azad MA, Velkov T, Zhou QT, Li J. Rescuing the last-line polymyxins: achievements and challenges. Pharmacol Rev. 2021;73(2):679–728. doi: 10.1124/pharmrev.120.000020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Centre for Disease Prevention and Control. Rapid risk assessment. Carbapenem-resistant Acinetobacter baumannii in healthcare settings. Stockholm: ECDC; 2016. [Google Scholar]
- 11.Mirnejad R, Heidary M, Bahramian A, Goudarzi M, Pournajaf A. Evaluation of polymyxin B susceptibility profile and detection of drug resistance genes among Acinetobacter baumannii clinical isolates in Tehran, Iran during 2015–2016. Mediterr J Hematol Infect Dis. 2018;10(1):e2018044. doi: 10.4084/MJHID.2018.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahamat A, Bertrand X, Moreau B, Hommel D, Couppie P, Simonnet C, et al. Clinical epidemiology and resistance mechanisms of carbapenem-resistant Acinetobacter baumannii, French Guiana, 2008–2014. Int J Antimicrob Agents. 2016;48(1):51–5. doi: 10.1016/j.ijantimicag.2016.03.006 [DOI] [PubMed] [Google Scholar]
- 13.Strateva T, Sirakov I, Stoeva T, Stratev A, Dimov S, Savov E, et al. Carbapenem-resistant Acinetobacter baumannii: current status of the problem in four Bulgarian university hospitals (2014–2016). J Glob Antimicrob Resist. 2019;16:266–73. doi: 10.1016/j.jgar.2018.10.027 [DOI] [PubMed] [Google Scholar]
- 14.Gales AC, Seifert H, Gur D, Castanheira M, Jones RN, Sader HS. Antimicrobial susceptibility of Acinetobacter calcoaceticus–Acinetobacter baumannii complex and Stenotrophomonas maltophilia clinical isolates: results from the SENTRY antimicrobial surveillance program (1997–2016). Open Forum Infect Dis. 2019;6(Supplement_1):S34–S46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shortridge D, Gales AC, Streit JM, Huband MD, Tsakris A, Jones RN. Geographic and temporal patterns of antimicrobial resistance in Pseudomonas aeruginosa over 20 years from the SENTRY antimicrobial surveillance program, 1997–2016. Open Forum Infect Dis. 2019;6(Supplement_1):S63–S8. doi: 10.1093/ofid/ofy343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lima WG, Brito JCM, Cardoso BG, Cardoso VN, de Paiva MC, de Lima ME, et al. Rate of polymyxin resistance among Acinetobacter baumannii recovered from hospitalized patients: a systematic review and meta-analysis. Eur J Clin Microbiol Infect Dis. 2020;39(8):1427–38. doi: 10.1007/s10096-020-03876-x [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Cao Y, Yi L, Liu J-H, Yang Q. Emergent polymyxin resistance: end of an era? Open Forum Infect Dis. 2019;6(10):ofz368. doi: 10.1093/ofid/ofz368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorsted A, Nielsen EI, Friberg LE. Pharmacodynamics of immune response biomarkers of interest for evaluation of treatment effects in bacterial infections. Int J Antimicrob Agents. 2020;56(3):106059. doi: 10.1016/j.ijantimicag.2020.106059 [DOI] [PubMed] [Google Scholar]
- 19.García-Patiño MG, García-Contreras R, Licona-Limón P. The immune response against Acinetobacter baumannii, an emerging pathogen in nosocomial infections. Front Immunol. 2017;8:441. doi: 10.3389/fimmu.2017.00441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W. Host innate immune responses to Acinetobacter baumannii infection. Front Cell Infect Microbiol. 2020;10(486):486. doi: 10.3389/fcimb.2020.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu H, KuoLee R, Harris G, Van Rooijen N, Patel GB, Chen W. Role of macrophages in early host resistance to respiratory Acinetobacter baumannii infection. PLoS One. 2012;7(6):e40019. doi: 10.1371/journal.pone.0040019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruhn KW, Pantapalangkoor P, Nielsen T, Tan B, Junus J, Hujer KM, et al. Host fate is rapidly determined by innate effector-microbial interactions during Acinetobacter baumannii bacteremia. J Infect Dis. 2015;211(8):1296–305. doi: 10.1093/infdis/jiu593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skerniškytė J, Karazijaitė E, Lučiūnaitė A, Sužiedėlienė E. OmpA protein-deficient Acinetobacter baumannii outer membrane vesicles trigger reduced inflammatory response. Pathogens. 2021;10(4):407. doi: 10.3390/pathogens10040407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erridge C, Moncayo-Nieto OL, Morgan R, Young M, Poxton IR. Acinetobacter baumannii lipopolysaccharides are potent stimulators of human monocyte activation via Toll-like receptor 4 signalling. J Med Microbiol. 2007;56(2):165–71. doi: 10.1099/jmm.0.46823-0 [DOI] [PubMed] [Google Scholar]
- 25.Cress BF, Englaender JA, He W, Kasper D, Linhardt RJ, Koffas MA. Masquerading microbial pathogens: capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol Rev. 2014;38(4):660–97. doi: 10.1111/1574-6976.12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhuiyan MS, Ellett F, Murray GL, Kostoulias X, Cerqueira GM, Schulze KE, et al. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc Natl Acad Sci USA. 2016;113(34):9599–604. doi: 10.1073/pnas.1523116113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Hu M, Yu K, Zeng X, Liu X. Mass spectrometry-based proteomic approaches to study pathogenic bacteria-host interactions. Protein Cell. 2015;6(4):265–74. doi: 10.1007/s13238-015-0136-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p) ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13(5):298–309. doi: 10.1038/nrmicro3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y, Lu J, Han ML, Jiang X, Azad MA, Patil NA, et al. Polymyxins bind to the cell surface of unculturable Acinetobacter baumannii and cause unique dependent resistance. Adv Sci. 2020;7(15):2000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owen RJ, Li J, Nation RL, Spelman D. In vitro pharmacodynamics of colistin against Acinetobacter baumannii clinical isolates. J Antimicrob Chemother. 2007;59(3):473–7. doi: 10.1093/jac/dkl512 [DOI] [PubMed] [Google Scholar]
- 31.Boisson M, Jacobs M, Grégoire N, Gobin P, Marchand S, Couet W, et al. Comparison of intrapulmonary and systemic pharmacokinetics of colistin methanesulfonate (CMS) and colistin after aerosol delivery and intravenous administration of CMS in critically ill patients. Antimicrob Agents Chemother. 2014;58(12):7331–9. doi: 10.1128/AAC.03510-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3 ‘, 5 ‘-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266(9):5980–90. [PubMed] [Google Scholar]
- 33.Vinella D, Albrecht C, Cashel M, d’Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol. 2005;56(4):958–70. doi: 10.1111/j.1365-2958.2005.04601.x [DOI] [PubMed] [Google Scholar]
- 34.Spira B, Silberstein N, Yagil E. Guanosine 3’, 5’-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol. 1995;177(14):4053–8. doi: 10.1128/jb.177.14.4053-4058.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, Aye SM, Ahmed MU, Han M-L, Li C, Song J, et al. Pan-transcriptomic analysis identified common differentially expressed genes of Acinetobacter baumannii in response to polymyxin treatments. Mol Omics. 2020;16(4):327–38. doi: 10.1039/d0mo00015a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Artsimovitch I, Patlan V, Sekine S-i, Vassylyeva MN, Hosaka T, Ochi K, et al. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117(3):299–310. doi: 10.1016/s0092-8674(04)00401-5 [DOI] [PubMed] [Google Scholar]
- 37.Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–70. doi: 10.1146/annurev.genet.38.072902.091347 [DOI] [PubMed] [Google Scholar]
- 38.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68(5):1128–48. doi: 10.1111/j.1365-2958.2008.06229.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma Z, King K, Alqahtani M, Worden M, Muthuraman P, Cioffi CL, et al. Stringent response governs the oxidative stress resistance and virulence of Francisella tularensis. PLoS One. 2019;14(10):e0224094. doi: 10.1371/journal.pone.0224094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martins D, McKay G, Sampathkumar G, Khakimova M, English AM, Nguyen D. Superoxide dismutase activity confers (p) ppGpp-mediated antibiotic tolerance to stationary-phase Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2018;115(39):9797–802. doi: 10.1073/pnas.1804525115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schäfer H, Beckert B, Frese CK, Steinchen W, Nuss AM, Beckstette M, et al. The alarmones (p) ppGpp are part of the heat shock response of Bacillus subtilis. PLoS Genet. 2020;16(3):e1008275. doi: 10.1371/journal.pgen.1008275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung H-W, Kim K, Islam MM, Lee JC, Shin M. Role of ppGpp-regulated efflux genes in Acinetobacter baumannii. J Antimicrob Chemother. 2020;75(5):1130–4. doi: 10.1093/jac/dkaa014 [DOI] [PubMed] [Google Scholar]
- 43.Kim K, Islam M, Jung H-w, Lim D, Kim K, Lee S-G, et al. ppGpp signaling plays a critical role in virulence of Acinetobacter baumannii. Virulence. 2021;12(1):2122–32. doi: 10.1080/21505594.2021.1961660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pérez-Varela M, Tierney AR, Kim J-S, Vázquez-Torres A, Rather P. Characterization of RelA in Acinetobacter baumannii. J Bacteriol. 2020;202(12):e00045–20. doi: 10.1128/JB.00045-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J-S, Liu L, Fitzsimmons LF, Wang Y, Crawford MA, Mastrogiovanni M, et al. DksA–DnaJ redox interactions provide a signal for the activation of bacterial RNA polymerase. Proc Natl Acad Sci USA. 2018;115(50):E11780–E9. doi: 10.1073/pnas.1813572115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henard CA, Bourret TJ, Song M, Vázquez-Torres A. Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J Biol Chem. 2010;285(47):36785–93. doi: 10.1074/jbc.M110.160960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato Y, Unno Y, Miyazaki C, Ubagai T, Ono Y. Multidrug-resistant Acinetobacter baumannii resists reactive oxygen species and survives in macrophages. Sci Rep. 2019;9(1):1–12. doi: 10.1038/s41598-018-37186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cross AR, Segal AW. The NADPH oxidase of professional phagocytes—prototype of the NOX electron transport chain systems. Biochim Biophys Acta Bioenerg. 2004;1657(1):1–22. doi: 10.1016/j.bbabio.2004.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Wang S, Wu Q. Cold shock protein A plays an important role in the stress adaptation and virulence of Brucella melitensis. FEMS Microbiol Lett. 2014;354(1):27–36. doi: 10.1111/1574-6968.12430 [DOI] [PubMed] [Google Scholar]
- 50.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036 [DOI] [PubMed] [Google Scholar]
- 51.Sun J, Hang Y, Han Y, Zhang X, Gan L, Cai C, et al. Deletion of glutaredoxin promotes oxidative tolerance and intracellular infection in Listeria monocytogenes. Virulence. 2019;10(1):910–24. doi: 10.1080/21505594.2019.1685640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ezraty B, Gennaris A, Barras F, Collet J-F. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol. 2017;15(7):385–96. doi: 10.1038/nrmicro.2017.26 [DOI] [PubMed] [Google Scholar]
- 53.May HC, Yu J-J, Zhang H, Wang Y, Cap AP, Chambers JP, et al. Thioredoxin-A is a virulence factor and mediator of the type IV pilus system in Acinetobacter baumannii. PLoS One. 2019;14(7):e0218505. doi: 10.1371/journal.pone.0218505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bjur E, Eriksson-Ygberg S, Aslund F, Rhen M. Thioredoxin 1 promotes intracellular replication and virulence of Salmonella enterica serovar Typhimurium. Infect Immun. 2006;74(9):5140–51. doi: 10.1128/IAI.00449-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng C, Dong Z, Han X, Wang H, Jiang L, Sun J, et al. Thioredoxin A is essential for motility and contributes to host infection of Listeria monocytogenes via redox interactions. Front Cell Infect Microbiol. 2017;7:287. doi: 10.3389/fcimb.2017.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henry R, Crane B, Powell D, Deveson Lucas D, Li Z, Aranda J, et al. The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J Antimicrob Chemother. 2015;70(5):1303–13. doi: 10.1093/jac/dku536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sampson TR, Liu X, Schroeder MR, Kraft CS, Burd EM, Weiss DS. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob Agents Chemother. 2012;56(11):5642–9. doi: 10.1128/AAC.00756-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaur A, Sharma P, Capalash N. Curcumin alleviates persistence of Acinetobacter baumannii against colistin. Sci Rep. 2018;8(1):1–11. doi: 10.1038/s41598-017-17765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Becker KW, Skaar EP. Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol Rev. 2014;38(6):1235–49. doi: 10.1111/1574-6976.12087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chandrangsu P, Rensing C, Helmann JD. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol. 2017;15(6):338–50. doi: 10.1038/nrmicro.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Djoko KY, Cheryl-lynn YO, Walker MJ, McEwan AG. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem. 2015;290(31):18954–61. doi: 10.1074/jbc.R115.647099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci USA. 2009;106(20):8344–9. doi: 10.1073/pnas.0812808106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hiniker A, Collet J-F, Bardwell JC. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J Biol Chem. 2005;280(40):33785–91. doi: 10.1074/jbc.M505742200 [DOI] [PubMed] [Google Scholar]
- 64.Ladomersky E, Petris MJ. Copper tolerance and virulence in bacteria. Metallomics. 2015;7(6):957–64. doi: 10.1039/c4mt00327f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alquethamy SF, Khorvash M, Pederick VG, Whittall JJ, Paton JC, Paulsen IT, et al. The role of the CopA copper efflux system in Acinetobacter baumannii virulence. Int J Mol Sci. 2019;20(3):575. doi: 10.3390/ijms20030575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams CL, Neu HM, Alamneh YA, Reddinger RM, Jacobs AC, Singh S, et al. Characterization of Acinetobacter baumannii copper resistance reveals a role in virulence. Front Microbiol. 2020;11:16. doi: 10.3389/fmicb.2020.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Almárcegui RJ, Navarro CA, Paradela A, Albar JP, von Bernath D, Jerez CA. New copper resistance determinants in the extremophile Acidithiobacillus ferrooxidans: a quantitative proteomic analysis. J Proteome Res. 2014;13(2):946–60. doi: 10.1021/pr4009833 [DOI] [PubMed] [Google Scholar]
- 68.De Benedetto ML, Capo CR, Ferri A, Valle C, Polimanti R, Carrì MT, et al. Glutaredoxin 1 is a major player in copper metabolism in neuroblastoma cells. Biochim Biophys Acta Gen Subj. 2014;1840(1):255–61. doi: 10.1016/j.bbagen.2013.09.008 [DOI] [PubMed] [Google Scholar]
- 69.Bazzi W, Abou Fayad AG, Nasser A, Haraoui L-P, Dewachi O, Abou-Sitta G, et al. Heavy metal toxicity in armed conflicts potentiates AMR in A. baumannii by selecting for antibiotic and heavy metal co-resistance mechanisms. Front Microbiol. 2020;11:68. doi: 10.3389/fmicb.2020.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Djoko KY, Xiao Z, Wedd AG. Copper resistance in E. coli: the multicopper oxidase PcoA catalyzes oxidation of copper (I) in CuICuII-PcoC. ChemBioChem. 2008;9(10):1579–82. doi: 10.1002/cbic.200800100 [DOI] [PubMed] [Google Scholar]
- 71.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen–host interface. Nat Rev Microbiol. 2012;10(8):525–37. doi: 10.1038/nrmicro2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X, Liu X, Horvatovich P, Hu Y, Zhang J. Proteomics landscape of host-pathogen interaction in Acinetobacter baumannii infected mouse lung. Front Genet. 2021;12. doi: 10.3389/fgene.2021.563516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Léséleuc L, Harris G, KuoLee R, Xu HH, Chen W. Serum resistance, gallium nitrate tolerance and extrapulmonary dissemination are linked to heme consumption in a bacteremic strain of Acinetobacter baumannii. Int J Med Microbiol. 2014;304(3–4):360–9. doi: 10.1016/j.ijmm.2013.12.002 [DOI] [PubMed] [Google Scholar]
- 74.Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, Actis LA. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun. 2012;80(3):1015–24. doi: 10.1128/IAI.06279-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin L, Pantapalangkoor P, Tan B, Bruhn KW, Ho T, Nielsen T, et al. Transferrin iron starvation therapy for lethal bacterial and fungal infections. J Infect Dis. 2014;210(2):254–64. doi: 10.1093/infdis/jiu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Figueiredo RT, Fernandez PL, Mourao-Sa DS, Porto BN, Dutra FF, Alves LS, et al. Characterization of heme as activator of Toll-like receptor 4. J Biol Chem. 2007;282(28):20221–9. doi: 10.1074/jbc.M610737200 [DOI] [PubMed] [Google Scholar]
- 77.Duvigneau JC, Esterbauer H, Kozlov AV. Role of heme oxygenase as a modulator of heme-mediated pathways. Antioxidants. 2019;8(10):475. doi: 10.3390/antiox8100475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ryter SW. Significance of heme and heme degradation in the pathogenesis of acute lung and inflammatory disorders. Int J Mol Sci. 2021;22(11):5509. doi: 10.3390/ijms22115509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Graça-Souza AV, Arruda MAB, de Freitas MS, Barja-Fidalgo C, Oliveira PL. Neutrophil activation by heme: implications for inflammatory processes. Blood. 2002;99(11):4160–5. doi: 10.1182/blood.v99.11.4160 [DOI] [PubMed] [Google Scholar]
- 80.Qiu H, KuoLee R, Harris G, Chen W. Role of NADPH phagocyte oxidase in host defense against acute respiratory Acinetobacter baumannii infection in mice. Infect Immun. 2009;77(3):1015–21. doi: 10.1128/IAI.01029-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Larsen R, Gozzelino R, Jeney V, Tokaji L, Bozza FA, Japiassú AM, et al. A central role for free heme in the pathogenesis of severe sepsis. Sci Transl Med. 2010;2(51):51ra71. doi: 10.1126/scitranslmed.3001118 [DOI] [PubMed] [Google Scholar]
- 82.Baldeo C, Isache C, Baldeo C, Bajwa A. A case of disseminated intravascular coagulation secondary to Acinetobacter lwoffii and Acinetobacter baumannii bacteremia. IDCases. 2015;2(3):70–1. doi: 10.1016/j.idcr.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Méndez JA, Mateos J, Beceiro A, Lopez M, Tomás M, Poza M, et al. Quantitative proteomic analysis of host—pathogen interactions: a study of Acinetobacter baumannii responses to host airways. BMC Genom. 2015;16(1):1–21. doi: 10.1186/s12864-015-1608-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharma A, Sharma D, Verma SK. In silico study of iron, zinc and copper binding proteins of Pseudomonas syringae pv. lapsa: emphasis on secreted metalloproteins. Front Microbiol. 2018;9:1838. doi: 10.3389/fmicb.2018.01838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pecsi I, Hards K, Ekanayaka N, Berney M, Hartman T, Jacobs WR Jr, et al. Essentiality of succinate dehydrogenase in Mycobacterium smegmatis and its role in the generation of the membrane potential under hypoxia. mBio. 2014;5(4):e01093–14. doi: 10.1128/mBio.01093-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puig S, Ramos-Alonso L, Romero AM, Martínez-Pastor MT. The elemental role of iron in DNA synthesis and repair. Metallomics. 2017;9(11):1483–500. doi: 10.1039/c7mt00116a [DOI] [PubMed] [Google Scholar]
- 87.De Oliveira DM, Bohlmann L, Conroy T, Jen FE-C, Everest-Dass A, Hansford KA, et al. Repurposing a neurodegenerative disease drug to treat Gram-negative antibiotic-resistant bacterial sepsis. Sci Transl Med. 2020;12(570):570. doi: 10.1126/scitranslmed.abb3791 [DOI] [PubMed] [Google Scholar]
- 88.Zurawski DV, Thompson MG, McQueary CN, Matalka MN, Sahl JW, Craft DW, et al. Genome sequences of four divergent multidrug-resistant Acinetobacter baumannii strains isolated from patients with sepsis or osteomyelitis. J Bacteriol. 2012;194(6):1619–20. doi: 10.1128/JB.06749-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, et al. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J Bacteriol. 2015;197(12):2027–35. doi: 10.1128/JB.00131-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu Y, Zhang Q, Hu M, Yu K, Fu J, Zhou F, et al. Proteomic analyses of intracellular Salmonella enterica serovar Typhimurium reveal extensive bacterial adaptations to infected host epithelial cells. Infect Immun. 2015;83(7):2897–906. doi: 10.1128/IAI.02882-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49(W1):W388–W96. doi: 10.1093/nar/gkab382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015;16(1):1–7. doi: 10.1186/s12859-015-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47(D1):D309–D14. doi: 10.1093/nar/gky1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. doi: 10.1093/nar/gkz401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rivals I, Personnaz L, Taing L, Potier M-C. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics. 2007;23(4):401–7. doi: 10.1093/bioinformatics/btl633 [DOI] [PubMed] [Google Scholar]
- 96.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. doi: 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
A) PCA score plot of A. baumannii proteomic profiles showing the relatedness of the dataset within and across the experimental groups of untreated AB5075 (i.e., controls; AB), polymyxin B treated AB5075 (AB + PMB), interacting AB5075 from infected THP-1-dMs (AB + THP-1-dMs), and interacting AB5075 from polymyxin B treated THP-1-dMs infection (AB + THP-1-dMs + PMB). B) Venn diagram showing common and unique sets of differentially expressed proteins of A. baumannii (log2FC >1 or <-1, FDR <0.05) between different comparison groups. C) PCA score plot of THP-1-dMs proteomic profiles showing the relatedness of the dataset within and across the experimental groups of untreated THP-1-dMs (i.e., controls; THP-1-dMs), AB5075-infected THP-1-dMs (AB + THP-1-dMs), polymyxin B treated THP-1-dMs (THP-1-dMs + PMB), and polymyxin B treated and AB5075-infected THP-1-dMs (AB + THP-1-dMs + PMB). D) Venn diagram showing common and unique sets of differentially expressed proteins of THP-1-dMs (log2FC >1 or <-1, FDR <0.05) between different comparison groups. Three biological replicates were employed in each experimental group.
(TIF)
Volcano plot generated using WebGestalt showing the enriched KEGG and Reactome pathways (Benjamini-Hochberg FDR <0.05) in THP-1-dMs at 4 h post infection with AB5075 in the absence of polymyxin B.
(TIF)
DEPs, differentially expressed proteins; AB, wild-type Acinetobacter baumannii AB5075; PMB, polymyxin B; THP-1-dMs, macrophage-like THP-1 cells.
(XLSX)
DEPs, differentially expressed proteins; AB, wild-type Acinetobacter baumannii AB5075; PMB, polymyxin B; THP-1-dMs, macrophage-like THP-1 cells.
(XLSX)
DEPs, differentially expressed proteins; AB, wild-type Acinetobacter baumannii AB5075; PMB, polymyxin B; THP-1-dMs, macrophage-like THP-1 cells.
(XLSX)
(XLSX)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
20× magnification; video speed at 7 frames per second (fps).
(AVI)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the manuscript and its Supporting Information files. The mass spectrometry proteomics data has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD028502.