Keywords: intestinal epithelium, marker, microRNA, Paneth, tuft
Abstract
MicroRNA-mediated regulation is critical for the proper development and function of the small intestinal (SI) epithelium. However, it is not known which microRNAs are expressed in each of the cell types of the SI epithelium. To bridge this important knowledge gap, we performed comprehensive microRNA profiling in all major cell types of the mouse SI epithelium. We used flow cytometry and fluorescence-activated cell sorting with multiple reporter mouse models to isolate intestinal stem cells, enterocytes, goblet cells, Paneth cells, enteroendocrine cells, tuft cells, and secretory progenitors. We then subjected these cell populations to small RNA-sequencing. The resulting atlas revealed highly enriched microRNA markers for almost every major cell type (https://sethupathy-lab.shinyapps.io/SI_miRNA/). Several of these lineage-enriched microRNAs (LEMs) were observed to be embedded in annotated host genes. We used chromatin-run-on sequencing to determine which of these LEMs are likely cotranscribed with their host genes. We then performed single-cell RNA-sequencing to define the cell type specificity of the host genes and embedded LEMs. We observed that the two most enriched microRNAs in secretory progenitors are miR-1224 and miR-672, the latter of which we found is deleted in hominin species. Finally, using several in vivo models, we established that miR-152 is a Paneth cell-specific microRNA.
NEW & NOTEWORTHY In this study, first, microRNA atlas (and searchable web server) across all major small intestinal epithelial cell types is presented. We have demonstrated microRNAs that uniquely mark several lineages, including enteroendocrine and tuft. Identification of a key marker of mouse secretory progenitor cells, miR-672, which we show is deleted in humans. We have used several in vivo models to establish miR-152 as a specific marker of Paneth cells, which are highly understudied in terms of microRNAs.
INTRODUCTION
The epithelium of the mammalian small intestine is a continuous cell monolayer that functions as a critical interface between the contents of the gut lumen and the host. It is composed of five major postmitotic, fully differentiated cell types: one absorptive (enterocytes) and four secretory (goblet cells, Paneth cells, tuft cells, and enteroendocrine cells), which are continually produced by a population of actively cycling stem cells located at the base of the intestinal crypts. Together they perform several vital physiological functions including nutrient absorption, energy homeostasis, innate immunity, and tissue regeneration. Disruption of these functions can lead to debilitating health conditions including, but not limited to, malabsorptive disorders, diabetes, inflammatory bowel disease, and intestinal cancer. Numerous protein factors have been identified as specific markers and critical regulators of intestinal epithelial cell (IEC) types. In addition, noncoding RNAs have emerged as important regulators of the structure and function of the small intestinal epithelium. Strikingly, McKenna et al. (1) showed that eliminating intestinal epithelial expression of the RNase III enzyme Dicer, which mediates canonical microRNA (miRNA) biogenesis, leads to disorganized small intestinal architecture, as well as defective cell type allocation, and compromised barrier function.
Following this work, we used the Sox9-EGFP reporter mouse model to show that cell fractions enriched for enterocytes and enteroendocrine cells are associated with miR-194 and miR-7, respectively, and that the loss of gut microbes leads to a significant change in the small intestinal epithelial miRNA landscape (2). In a follow-up study, we demonstrated that miR-7 is enriched in enteroendocrine progenitor cells, its expression level is highly sensitive to obesogenic high-fat diet, and it regulates both epithelial growth and enteroendocrine cell abundance in enteroids (3). Despite these advances, the miRNA profiles of most IEC types, including canonical stem, secretory progenitor, tuft, Paneth, and goblet cells, remain undefined. Therefore, it is not known whether any intestinal epithelial lineages express specific miRNA markers.
To bridge this important knowledge gap, we performed the first comprehensive profiling analysis of miRNAs across all major epithelial cell types of the mouse small intestine. We first used flow cytometry and fluorescence activated cell sorting with multiple reporter mouse models to isolate all major cell types. We then employed a multiomic strategy, comprising small RNA-seq, chromatin run-on sequencing (ChRO-seq), and single-cell RNA-sequencing (scRNA-seq) to identify high-confidence microRNA markers of each lineage. We also leveraged several additional in vivo mouse models to provide corroborative evidence that miR-1224 and miR-672 are highly enriched in secretory progenitor cells that give rise to both enteroendocrine and tuft cells and that miR-152 is a Paneth cell-specific miRNA. The comprehensive miRNA atlas is provided at the following website: https://sethupathy-lab.shinyapps.io/SI_miRNA/.
EXPERIMENTAL PROCEDURES
Mouse Models
The following mice were utilized: 1- to 3-mo-old male and female wild-type C57BL/6, 6- and 12-mo-old male wild-type albino C57BL6/2J (B62J), 7-mo-old female Sox9-EGFP (4), 6-mo-old male and female Lgr5-EGFP (5), 2- to 3-mo-old male and female Defa6-Cre; CAG-tdTomato (6), 6- to 7-mo-old male and female Prox1-GFP, 2 mo old male Pou2f3−/−; Smart13 (7), 2- to 6-mo-old male and female Defa6-DTR (8, 9), and 2- to 3-mo-old male and female Mist1-Cre; CAG-tdTomato mice (10). All animal procedures were performed with the approval and authorization of the Institutional Animal Care and Use Committee at each participating institution. Mice were used in these experiments due to their tractability to genetic manipulation and the availability of a wide array of appropriate experimental reagents. Mice were housed in well-ventilated cages under 12-h light/dark cycles with free access to water and standard chow in addition to tubing for environmental enrichment. During experimentation, the mice were monitored at regular intervals to determine their well-being, and at the time of tissue collection the mice were anesthetized by CO2 inhalation and euthanized by means of cervical dislocation. To induce tdTomato expression in Mist1-Cre; CAG-tdTomato mice, the mice were injected intraperitoneally with 1 mg of tamoxifen 3 days before tissue collection.
Cell Sorting
Three distinct mouse-based cell marker systems were used to sort intestinal stem cells (ISCs) (Sox9-EGFP; Lgr5-EGFP; and Cd24). Mouse intestinal epithelial cells from the jejunum of Sox9-EGFP and Lgr5-EGFP mice were dissociated and prepared for fluorescence-activated cell sorting (FACS) as described previously (11), while jejunal crypts were isolated for Cd24-Low cell sorts. For Sox9-EGFP and Lgr5-EGFP: CD31-APC (BioLegend, San Diego, CA, Cat. No. 102416), CD45-APC (BioLegend, Cat. No. 1032124), Annexin-V-APC (Life Technologies, Carlsbad, CA, Cat. No. A35110), and Sytox-Blue (Life Technologies, Cat. No. S34857) staining were used to exclude endothelial cells, immune cells, apoptotic cells, and nonviable cells, respectively. The gating parameters of FACS sorting were described previously (11). For Cd24-Low cell sorts, UEA-FITC (Vector Laboratories, Burlingame, CA, Cat. No. FL-1061), CD45-FITC (BioLegend, San Diego, CA, Cat. No. 553080), and propidium iodide (BioLegend, Cat. No. 421301) staining were used to exclude goblet/Paneth, endothelial, and nonviable cells, respectively. In addition, for these sorts CD24-Pac Blue (BioLegend, Cat. No. 101819) and EpCAM-PECγ7 (BioLegend, Cat. No. 118215) staining were used to positively select for CD24+ epithelial cells. The Sox9 and Lgr5 sorts were performed using a Mo-Flo XDP cell sorter (Beckman-Coulter, Fullerton, CA) at the University of North Carolina Flow Cytometry Core Facility. Sorting of Cd24-Low cells was conducted at North Carolina State University, College of Veterinary Medicine using a Mo-Flo XDP cell sorter (Beckman-Coulter, Fullerton, CA).
The Prox1 sorts (3) were performed using BD FACS Aria Fusion Fluorescence Activated Cell Sorter (BD Biosciences, San Jose, CA) at Cornell University Flow Cytometry Core Facility at the Biotechnology Resource Center. The cells from jejunal crypts were sorted directly into cold DMEM or lysis buffer. Goblet cells (12–15) and tuft cells (16) from the entire small intestine were sorted as Live, CD45−EpCAM+CD24−CD166−UEA-I+ and Live, CD45−EpCAM+CD24+SiglecF+, respectively, using a BD Biosciences FACsAria Fusion Cell Sorter at Cornell University College of Veterinary Medicine or a BD Biosciences Aria II Cell Sorter at the University of Washington. Staining of these cells was achieved by use of the following antibodies and reagents: Aqua Live/Dead fixable viability stain (Thermo Fisher Scientific), anti-CD24 Per CP Cy5.5 (Thermo Fisher Scientific, clone M1/69), anti-CD45 eFluor (Thermo Fisher Scientific, clone 30-F11), anti-CD166 Fitc (Thermo Fisher Scientific, eBio ALC 48), anti-EpCAM PE (Thermo Fisher Scientific, clone G8.8), anti-SiglecF (BD Biosciences, E50-2440), and biotinylated Ulex Europaeus Agglutinin I (UEA-I) (Vector Laboratories, Burlingame, CA). The cells were sorted into cold sort buffer and spun down, and the cell pellets were immediately snap frozen. Defa6-expressing Paneth cells and Mist1-expressing Paneth cells from jejunal crypts were sorted as Live, CD45−EpCAM+tdTomato+ cells and Live, CD45−CD24+EpCAM+tdTomato+ cells, respectively, at North Carolina State University, College of Veterinary Medicine using a Mo-Flo XDP cell sorter (Beckman-Coulter, Fullerton, CA) and antibodies described previously for the Cd24-Low sorts. The cells were sorted into cold lysis buffer and snap frozen.
Jejunal Crypt Isolation
Harvested small intestine was measured and divided into three equal segments. The middle region was considered jejunum. Subsequent to luminal flushing with ice-cold phosphate-buffered saline (PBS), the tissue was longitudinally cut and subjected to incubation in 3 mM EDTA in ice-cold PBS with 1% (vol/vol) primocin for 15 min at 4°C. The mucosa of the intestinal pieces was gently scrapped of mucus, shaken in ice-cold PBS with 1% (vol/vol) primocin for 2 min, and incubated in fresh 3 mM EDTA in ice cold PBS with 1% (vol/vol) primocin for 40 min at 4°C. After 2–6 min of gentle manual shaking in ice-cold PBS with 1% (vol/vol) primocin, the intestinal pieces were inspected microscopically (×50 magnification) for detached intestinal crypts, diluted 1:2 with ice-cold PBS with 1% (vol/vol) primocin, filtered with a 70-μm cell strainer, and collected by pelleting with centrifugation at 110 g for 10 min at 4°C. For RNA extraction, collected pellets were resuspended in 200 μL of lysis buffer (Buffer RL, Norgen Biotek), vortexed for 10 s, and stored at −80°C.
RNA Extraction and Real-Time qPCR
Total RNA was isolated using the Total or Single-cell RNA Purification kit (Norgen Biotek, Thorold, ON, Canada). High Capacity RNA to cDNA kit (Life Technologies, Grand Island, NY) was used for reverse transcription of RNA. TaqMan microRNA Reverse Transcription kit (Life Technologies) was used for reverse transcription of miRNA. Both miRNA and gene expression qPCR were performed using TaqMan assays (Life Technologies) with either TaqMan Universal PCR Master Mix (miRNA qPCR) or TaqMan Gene Expression Master Mix (mRNA qPCR) per the manufacturer’s protocol on a Bio-Rad CFX96 Touch Real Time PCR Detection System (Bio-Rad Laboratories, Richmond, CA). Reactions were performed in duplicate or triplicate using either U6 (miRNA qPCR) (Life Technologies, Assay ID: 001973), Rps9 (mouse mRNA qPCR) (Life Technologies, Assay ID: Mm00850060_s1), or Rps9 (canine mRNA qPCR, Assay ID: Cf02694829_u1) as the normalizer. miRNA expression was assayed with the use of the following probes: dme-miR-7 (Life Technologies, Assay ID: 000268), hsa-miR-30b (Life Technologies, Assay ID: 000602), mmu-miR-31 (Life Technologies, Assay ID: 000185), hsa-miR-152 (Life Technologies, Assay ID: 000475), hsa-miR-194 (Life Technologies, Assay ID: 000493), and hsa-miR-375 (Life Technologies, Assay ID: 000564). Gene expression was assayed with the use of the following probes: Chga (Life Technologies, Assay ID: Mm00514341_m1), Dclk1 (Life Technologies, Assay ID: Mm00444950_m1), Defa20 (Life Technologies, Assay ID: Mm00842045_g1), Hopx (Life Technologies, Assay ID: Mm00558630_m1), Lgr5 (Life Technologies, Assay ID: Mm00438890_m1), Lyz (Life Technologies, Assay ID: Cf02642933_m1), Lyz1 (Life Technologies, Assay ID: Mm00657323_m1), and Sis (Life Technologies, Assay ID: Mm01210305_m1).
Small RNA Library Preparation and Sequencing
The small RNA sequencing of cells from the various cell sorts was conducted at Genome Sequencing Facility of Greehey Children’s Cancer Research Institute at University of Texas Health Science Center at San Antonio. Libraries were prepared using the TriLink CleanTag Small RNA Ligation kit (TriLink Biotechnologies, San Diego, CA). Seven to eight libraries were sequenced per lane with single-end 50× on the HiSeq2500 platform. All cell sorts were sequenced individually with the exception of the Lgr5-EGFP sorts in which case two pools of samples, one including sorts from two mice and the other including sorts from three mice, were sequenced.
scRNA-Seq Library Preparation and Sequencing
Mouse jejunal crypts from 12-mo-old male WT B62J mice were isolated as previously described. Isolated crypts were resuspended in an ice-cold solution of PBS with 0.04% (wt/vol) bovine serum albumin and pelleted at 1,000 g at 4°C for 5 min. The crypts were subsequently digested with 0.3 U/mL dispase I in Hanks’ balanced salt solution (HBSS) at 37°C for 12 min with gentle agitation. After stopping dispase I activity with an addition of fetal bovine serum to a final concentration of 10% (vol/vol), the single-cell suspension was filtered, pelleted at 500 g at 4°C for 5 min, washed with cold HBSS, filtered again, and then resuspended in an ice-cold solution of PBS with 0.04% (wt/vol) bovine serum albumin. Before submission, single-cell suspensions were triturated by pipetting and evaluated for total viable cell number by using trypan blue staining with a TC20 automated cell counter (Bio-Rad Laboratories, Richmond, CA). Single-cell RNA sequencing of these samples was performed at the Cornell University Biotechnology Resource Center. Libraries were prepared using the 10× Genomics Chromium preparation kit (10× Genomics, Pleasanton, CA).
ChRO-Seq Library Preparation
ChRO-seq was performed as previously described (17, 18) on jejunal crypts isolated from 6- to 7-mo-old adult male Prox1-GFP mice on chow (LabDiet, St. Louis, MO, Laboratory Rodent Diet 500I) (n = 2) or high-fat diet (Research Diets, New Brunswick, NJ, D12492) (n = 2) for 18–20 wk. To isolate chromatin from mouse crypts, samples were pelleted and resuspended in 1× NUN buffer [20 mM HEPES, 7.5 mM MgCl2, 0.2 mM EDTA, 0.3 M NaCl, 1 M urea, 1% (vol/vol) NP-40, 1 mM DTT, 50 U/mL SUPERase In RNase Inhibitor (Thermo Fisher Scientific, AM2694), 1× Protease Inhibitor Cocktail (Roche, 11873580001)]. The 1× NUN buffer containing samples were incubated in an Eppendorf Thermomixer (Eppendorf, Hamburg, Germany) at 12°C and shaking at 2,000 rpm for 30 min before centrifugation at 12,500 g for 30 min at 4°C. Samples were then washed with wash buffer [1 mL 50 mM Tris·HCl pH 7.5, 40 U/mL SUPERase inhibitor] and stored in chromatin storage buffer (50 mM Tris·HCl pH 8.0, 25% (vol/vol) glycerol, 5 mM magnesium acetate, 0.1 mM EDTA, 5 mM DTT, and 40 U/mL SUPERase In RNase Inhibitor). Bioruptor (Diagenode, Denville, NJ) was used to solubilize the chromatin into the storage buffer.
To perform run-on reaction with the solubilized chromatin, samples were mixed with an equal volume of two times run-on reaction mix [10 mM Tris·HCl pH 8.0, 5 mM MgCl2, 1 mM DTT, 300 mM KCl, 400 μM ATP, 0.8 μM CTP, 400 μM GTP, 400 μM UTP, 40 μM Biotin-11-CTP (Perkin Elmer, Waltham, MA, NEL542001EA), 100 ng of yeast tRNA (VWR, Radnor, PA, 80054-306), 0.8 U/μL SUPERase In RNase Inhibitor, 1% (wt/vol) Sarkosyl]. The samples were then incubated in an Eppendorf Thermomixer at 37°C for 5 min (700 rpm). The reaction was stopped by adding TRIzol LS (Life Technologies, 10296-010) and pelleted with GlycoBlue (Ambion, AM9515) to visualize the pellet of nascent RNA. The RNA pellet was resuspended in diethylpyrocarbonate (DEPC)-treated water and heat denatured at 65°C for 40 s. To fragment RNA molecules, the reaction of base hydrolysis was performed by incubating RNA with 0.2 N NaOH on ice for 4 min. Base hydrolysis was stopped by adding 1 M Tris·HCl pH 6.8. The RNA was purified by binding to streptavidin beads (NEB, Ipswich, MA, S1421S) followed by TRIzol extraction.
To construct ChRO-seq libraries, the RNAs were processed through the following procedures: 1) 3′ adapter ligation with T4 RNA Ligase 1 (NEB, M0204); 2) Streptavidin bead binding followed by TRIzol extraction; 3) 5′ de-capping with RNA 5′ pyrophosphohydrolase (RppH, NEB, M0356); 4) 5′ end phosphorylation using T4 polynucleotide kinase (NEB, M0201); 5) 5′ adapter ligation with T4 RNA Ligase 1 (NEB, M0204); the 5′ adaptor contained a 6-nucleotide unique molecular identifier (UMI) to allow for bioinformatic detection and elimination of PCR duplicates in data processing steps; 6) Streptavidin bead binding followed by TRIzol extraction; 7) cDNA synthesis by reverse transcription using SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, 18090010); and 8) library amplification by PCR using the Q5 High-Fidelity DNA Polymerase (NEB, M0491). Libraries were sequenced (5′ single end; single-end 75×) using the NextSeq500 high-throughput sequencing system (Illumina, San Diego, CA) at the Cornell University Biotechnology Resource Center.
miR-672 Genomic Analysis
The 100-bp RefSeq mRNA sequence for mouse miR-672 (NR_030430) was used to query the genomes of all available mammalian species, including Gorilla gorilla, Macaca mullatta, Sapajus apella, Mus musculus, Cavia porcellus, Oryctolagus cuniculus, Felis catus, Canis lupus, Equus caballus, Sus scrofa, Ovis aries, Bos taurus, Myotis lucifugus, Balaenoptera musculus, Loxodonta africana, and Dasypus novemcinctus, with BLASTN to acquire the genomic sequences of the genes for miR-672 in the corresponding species. To characterize hominin genomic loci that harbor the miR-672 gene in non-hominin primates, the genomic sequence in G. gorilla that includes the miR-672 gene and 1,450 bp upstream and downstream of this gene was used to query the genomes of Pan paniscus, Pan troglodytes, and Homo sapiens as well as Macaca mullatta. The homologous genomic sequences obtained from hominin species were then compared with G. gorilla by pairwise BLAST analysis. Multiple DNA sequence alignment for the genomic sequences of the miR-672 gene that we acquired for mammalian placental species was performed with CLUSTAL W software that is included in the MEGA software suite.
Paneth Cell Depletion
PC-DTR (Defa6-DTR) mice were used as previously described (8, 9). In brief, PC-DTR mice were generated on C57BL6/J backgrounds by transgenic insertion of a HA-tagged human diphtheria toxin receptor into the Cryptdin-2 (Defa6) promoter on the surface of Paneth cells in the University of Iowa Transgenic Mouse Core. Paneth cells were depleted via an intraperitoneal injection with either 40 ng/g body weight diphtheria toxin (2 μg/μL solution) in PBS.
Mouse Enteroid Culture
Jejunal crypts were isolated from 6-mo-old male WT B62J mice as previously described. The isolated crypts (day 0) were grown into Reduced Growth Factor Matrigel (Corning, Corning, NY, Cat. No. 356231). Advanced DMEM/F12 (Gibco, Gaithersburg, MD, Cat. No. 12634-028) supplemented with GlutaMAX (Gibco, Cat. No. 35050-061), Pen/Strep (Gibco, Cat. No. 15140), HEPES (Gibco, Cat. No. 15630-080), N2 supplement (Gibco, Cat. No. 17502-048), 50 ng/mL EGF (R&D Systems, Minneapolis, MN, Cat. No. 2028-EG), 100 μg/mL Noggin (PeproTech, Rocky Hill, NJ, Cat. No. 250-38), 250 ng/μL murine R-spondin (R&D Systems, Cat. No. 3474-RS-050), and 10 mM Y-27632 (Sigma-Aldrich, St. Louis, MO, Cat. No. Y0503) was added. At day 3, enteroid cultures were replenished with fresh media and growth factors except for Y-27632. Enteroids at day 5 were harvested for RNA isolation.
Canine Enteroids
Canine enteroids were cultured and maintained according to previously published methods (19). In brief, jejunal crypts containing adult ISCs were isolated using cold EDTA chelation. Isolated crypts were grown in Matrigel [Corning Matrigel Growth Factor Reduced (GFR) Basement Membrane Matrix] for the first 2 days in the presence of a complete medium with ISC growth factors (CMGF+) (19) containing 10 μM rho-associated kinase inhibitor (ROCKi), Y-27632 (StemGent), and 2.5 μM glycogen synthase kinase 3β (GSK3β) inhibitor CHIR99021 (StemGent). After 2 days of culture, canine enteroids were then grown in CMGF+ media without Rock and GSK3 inhibitor supplements to allow stem cell differentiation, and the medium was changed every 2 days. After complete cell differentiation on day 7, enteroids were harvested for RNA isolation.
Bioinformatics Analysis
Small RNA-sequencing reads were aligned to the mouse genome (mm9) and quantified using miRquant 2.0 as previously described by Kanke et al. (20), with the exception that raw miRNA counts were further analyzed using DESeq2 (1.30) to obtain normalized counts and determine significance. miRNA annotation was performed using miRbase (r18 for mouse). Single-cell RNA-sequencing was performed using 10× genomics cellranger software (v3.0.1) and aligned to the mouse genome (mm10, v1.2.0 provided by 10× genomics) to get a cell count matrix. Single-cell RNA-seq data were subsequently analyzed using Seurat (v3.2.1). To be included in the analysis, genes were required to be present in at least three cells and cells had to have greater than 750 genes with less than 15% of the cell reads aligning to mitochondrial genes. The two mouse samples were combined using the Seurat integration anchor methodology. Clustering was accomplished using Seurat’s FindClusters function with a resolution of 0.5. Cell clustering was visualized with a UMAP. Clusters were assigned cell types using markers reported by Haber et al. (21), and unassigned or immune-like clusters were discarded, resulting in 2,614 wild-type cells.
For ChRO-seq analysis, we used an established pipeline (22) to process sequencing data. Briefly, read quality was assessed using FastQC. PCR deduplication was performed by collapsing UMIs followed by UMI trimming using PRINSEQ lite 0.20.2 (23). Adapters were trimmed using cutadapt 1.16 with a maximum 10% error rate, minimum 2-bp overlap, and minimum 20 quality score. Processed reads with a minimum length of 15 bp were mapped to the mm9 genome modified with the addition of a single copy of the human Pol I ribosomal RNA complete repeating unit (GenBank: BK000964.3) with Burrows-Wheeler Aligner (BWA). The location of the RNA polymerase active site was represented by a single base that denotes the 5′ end of the nascent RNA, which corresponds to the position on the 3′ end of each sequenced read. Supplemental Table S2 (all Supplemental figures and tables are available at https://github.com/Sethupathy-Lab/2021_AJP_Shanahan_et_al_supplemental) provides the mapping statistics of the ChRO-seq experiments. Data were converted to bigwig format using bedtools and UCSC bedGraphToBigWig (24) for visualization and identification of transcriptional regulatory elements (TREs). Bigwig files from identical conditions were merged and normalized to a total signal of 1 × 106 before visualization. Genomic loci snapshots were generated using Gviz. The location of the RNA polymerase active site was represented by a single base that denotes the 3′ end (ChRO-seq) or 5′ end (leChRO-seq) of the nascent RNA, which corresponds to the position on the 5′ or 3′ end of each sequenced read, respectively.
Statistics
In most figure panels, quantitative data are reported as an average of biological replicates ± standard error of the mean. In all analyses, statistical differences were assessed by two-tailed Student’s t test with threshold P value <0.05, unless otherwise specifically noted.
Patient and Public Involvement
Patients or the public were not involved in the design, or conduct, or reporting, or dissemination plans of our research.
RESULTS
Each Major Intestinal Epithelial Cell Type Exhibits a Unique MicroRNA Profile
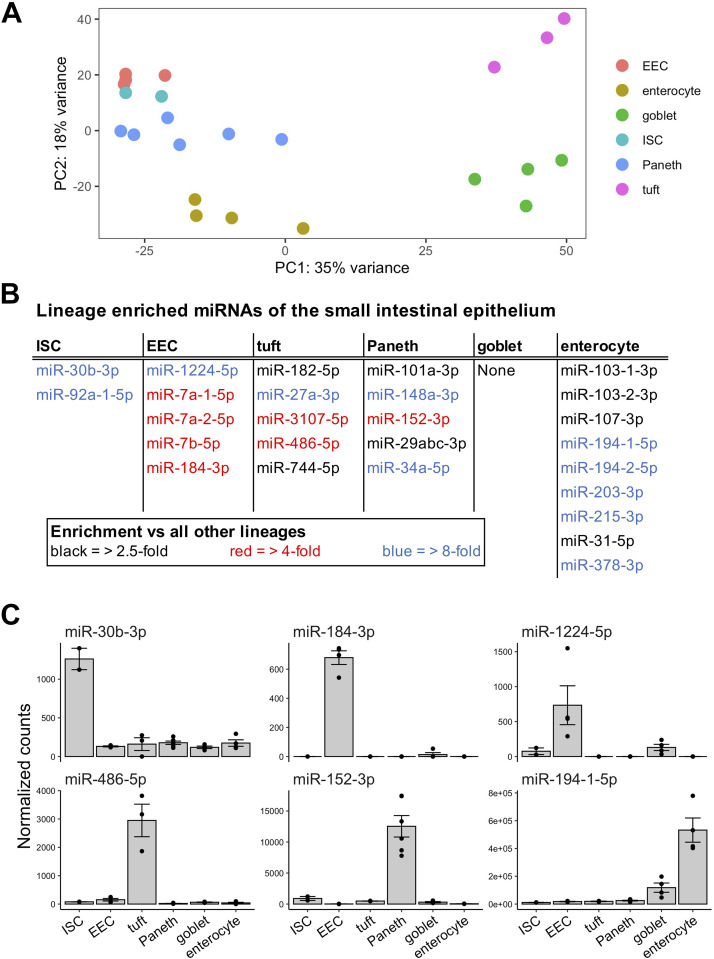
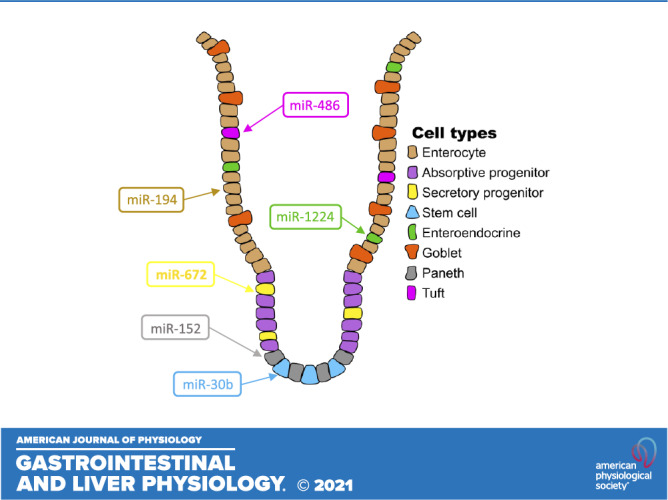
Each major small intestinal epithelial cell type can be sorted by taking advantage of the expression of specific protein markers (2, 4, 12, 25–29). We isolated cell populations enriched for intestinal stem cells (ISCs) (Supplemental Fig. S1, A and B) (2), enteroendocrine cells (EECs) (2), tuft cells (Supplemental Fig. S2A), Paneth cells (Supplemental Fig. S2B), goblet cells (Supplemental Fig. S2C), and enterocytes (2) by performing fluorescence-activated cell sorting (FACS) or flow cytometry on small intestinal epithelia from various reporter mice or C57BL/6 mice, respectively (Table 1). We validated each of the cell fractions through real-time RT-qPCR analysis of a panel of well-established cell type specific markers: Lgr5 for ISCs (Supplemental Fig. S3A), Dclk1 for tuft cells (Supplemental Fig. S3B), Lyz1 and Defa20 for Paneth cells (Supplemental Fig. S3C), Muc2 for goblet cells (Supplemental Fig. S3D), Sis for enterocytes (Supplemental Fig. S3E), and Chga for EECs (30–36). Small RNA-seq analysis (Experimental Procedures, Supplemental Table S1) of these cell fractions showed that each intestinal epithelial cell type exhibits a clearly distinguishing miRNA expression pattern (Fig. 1A) and revealed at least two miRNAs that are greater than fourfold enriched (adjusted P < 0.05) in each cell type (with the exception of goblet cells) relative to all other IEC types (Fig. 1, B and C). Moreover, we found that one or more miRNAs are greater than eightfold enriched (adjusted P < 0.05, average normalized counts > 500) in each of EECs, tuft cells, and Paneth cells relative to each of the other lineages. Particularly notable and heretofore unreported lineage-enriched miRNAs (LEMs) include miR-1224-5p for EECs, miR-486-5p for tuft cells, and miR-152-3p for Paneth cells (Fig. 1C).
Table 1.
Sorting strategies for all major intestinal epithelial cell types
| Small Intestinal Cell Type | Mouse Model | FACS Gating Scheme |
|---|---|---|
| Intestinal stem cells | Sox9-EGFP | Low |
| Lgr5-EGFP | High | |
| C57BL/6 | Live, CD45−EpCAM+UEA-I−CD24low | |
| Enterocytes | Sox9-EGFP | Negative |
| Goblet cells | C57BL/6 | Live, CD45−EpCAM+UEA-I+CD24−CD166− |
| Paneth cells | Defa6-Cre; CAG-tdTomato | High |
| Enteroendocrine cells | Sox9-EGFP | High |
| Tuft cells | C57BL/6 | Live, CD45−EpCAM+SiglecF+CD24+ |
| Secretory progenitor cells | Prox1-GFP | High |
List of small intestinal epithelial cell types and the sorting methods used to isolate each type.
Figure 1.
MicroRNAs display robust lineage-specific expression patterns in the intestinal epithelium. A: principal component analysis of variance stabilized transformed (VST) counts from sorted cell populations shows clustering of samples by small intestine epithelial cell type. Sorted cell populations represented include enteroendocrine cell (EEC, n = 3 mice), enterocyte (n = 4 mice), goblet (n = 4 mice), intestinal stem cell (ISC, n = 2 pools of mice), Paneth (n = 6 mice), and tuft (n = 3 mice). B: microRNA enrichment in each of the six main small intestinal cell types (excluding intermediate progenitor populations). Base microRNA enrichment is defined as an average expression > 500, adjusted P value < 0.05, and fold change > 2.5 versus each other cell type (black). Stringent microRNA enrichment requires fold change > 4 versus each other cell type (blue). Lineage specificity requires an even more stringent requirement of fold change > 8 versus each other cell type (red). C: normalized expression of a subset of lineage-enriched microRNAs across different intestinal epithelial cell types. Error bars represent standard error.
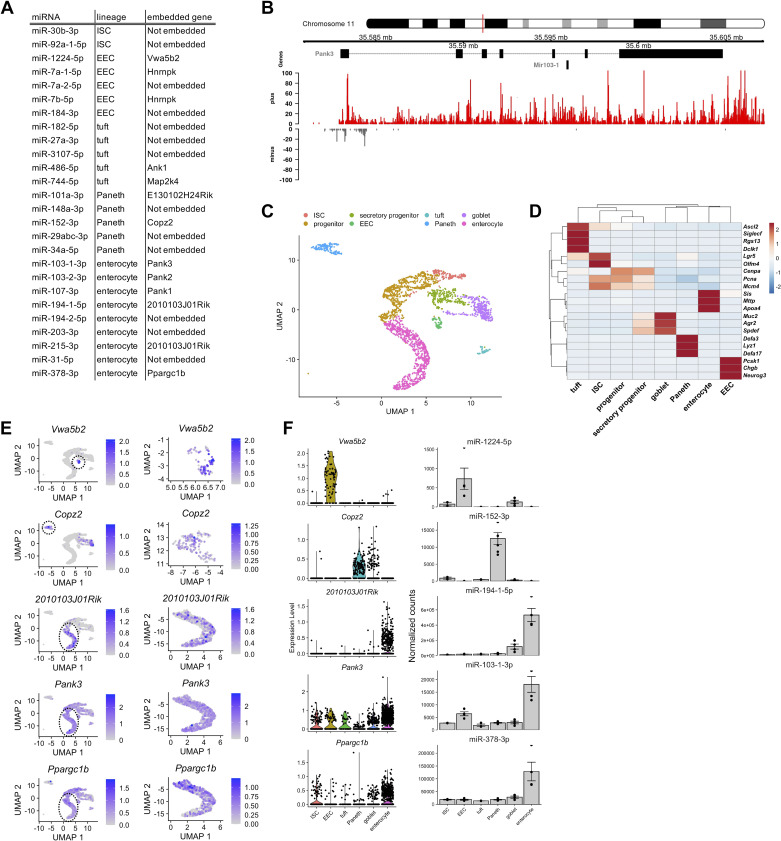
Single-Cell Analyses Reveal That Five Different MicroRNAs Exhibit the Same Cell Type Enrichment as the Host Genes in Which They Are Embedded
MiRNAs embedded within host genes often display expression patterns that are similar to the host genes (37). We first determined that 13 of the 26 LEMs (Fig. 1B) are embedded within 11 previously annotated genes (Fig. 2A). We then performed chromatin run-on sequencing (ChRO-seq) (Experimental Procedures, Supplemental Table S2) of jejunal crypts from adult male Prox1-GFP mice, which facilitated the genome-wide identification of active regulatory elements. This analysis revealed that for each of the 13 LEMs, the nearest upstream active promoter is the same as the promoter for the gene in which it is embedded (Fig. 2B, Supplemental Fig. S4A), suggesting that these 13 miRNAs are likely co-transcribed with their host genes. However, the expression patterns of co-transcribed genes and miRNAs can still differ due to extensive post-transcriptional regulation during miRNA biogenesis (38).
Figure 2.
Single-cell analysis and chromatin run-on sequencing reveals expression patterns of genes that host lineage-enriched microRNAs. A: table indicating lineage-enriched microRNAs (first column), the lineage in which the microRNA is enriched (second column), and the host gene (third column). MicroRNAs not located within a host gene are listed as “Not embedded” in the third column. B: UCSC browser image showing the miR-152 locus embedded within the Copz2 gene (top). The site of miR-152 transcriptional initiation was identified using signal from chromatin run-on sequencing (ChRO-seq) on crypts from 6- to 7-mo-old male Prox1-GFP mice (n = 2 mice). Normalized ChRO-seq signal on the plus strand is shown in red and on the minus strand is shown in gray. C: uniform manifold projection and approximation (UMAP) projection of single-cell RNA-seq data from intestinal epithelial crypts from 12-mo-old male B62J mice (n = 2 mice, n = 2,614 cells). All major intestinal epithelial cell types were detected and annotated based on known markers. D: expression of known cell type marker genes across all of the clusters. Expression is scaled by row. E: expression pattern and level of the same genes shown in (E) visualized on the full UMAP projection (left) and the specific cell type cluster in which its embedded microRNA is enriched (right). Dotted line circles in the full UMAP projection (left) indicate the cluster being enlarged in the right. F: expression of host gene (left; log normalized expression) and embedded microRNA (right; DESeq2 normalized expression) for Vwa5b2/miR-1224, Copz2/miR-152, 2010103J01Rik/miR-194, Pank3/miR-103, and Ppargc1b/miR-378. Error bars represent standard error.
To resolve this further, we next sought to define the expression patterns of the 11 host genes of interest at high resolution by performing single-cell RNA-seq (scRNA-seq) of jejunal crypts from adult male B62J mice (Experimental Procedures, Supplemental Table S3). We were able to identify several distinct clusters of cell populations and unambiguously assign each of them to an established jejunal epithelial cell type (Fig. 2C) based on the expression of distinguishing marker genes (Fig. 2D) (21). Of the 11 host gene/miRNA pairs, 6 exhibited discordant expression patterns (Supplemental Fig. S4B), suggesting that although these pairs are co-transcribed, their mature expression levels are uncoupled likely due to differential regulation at the post-transcriptional level, which has been documented previously (38). Notably, five host genes exhibited the same cell-type enrichment as their hosted miRNAs (Vwa5b2/miR-1224-5p, Copz2/miR-152-3p, 2010103J01Rik/miR-194-1-5p/miR-215-3p, Pank3/miR-103-1-3p, and Ppargc1b/miR-378-3p) (Fig. 2, E and F), indicating that the regulation of these pairs is tightly coupled.
We next mined published human small intestinal scRNA-seq datasets to evaluate the expression patterns of four of the five host genes of interest for which a human ortholog is annotated (VWA5B2, COPZ2, PANK3, and PPARGC1B). We found that each of these genes, except COPZ2, exhibited the same cell type enrichment observed in our studies of the mouse small intestine (Supplemental Fig. S4C). COPZ2 was not robustly detected in the single-cell data, so future investigations of additional adult human datasets as they become available are merited. Taken together, these results strongly suggest that the hosted miRNAs, miR-152-3p, miR-1224-5p, miR-103-1-3p, and miR-378-3p, are likely enriched in Paneth cells, EECs, and enterocytes, respectively, in the small bowel epithelium (Fig. 1B).
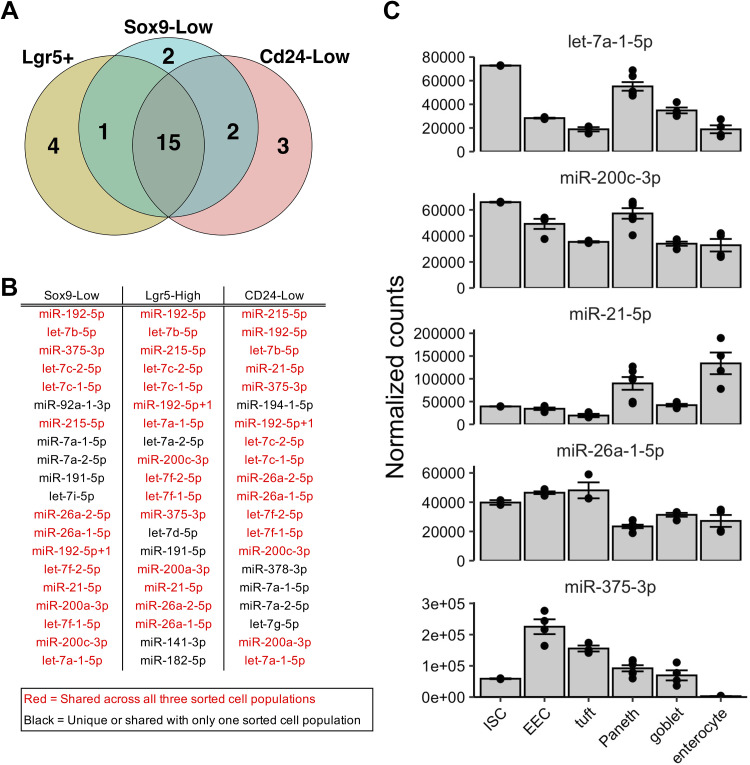
None of the MicroRNAs Most Highly Expressed in ISCs Are Significantly Enriched in ISCs
ISCs are critical for the maturation of all IEC types. To further explore the miRNA profile of ISCs, we performed small RNA-seq on two additional sorted intestinal epithelial cell populations that have previously been shown to be enriched for stem cell features: Sox9-Low cells (4) and Cd24-Low cells (25). Bioinformatic analysis of the small RNA-seq data revealed that 15 miRNAs are shared among the top 20 most highly expressed miRNAs in Lgr5+ cells and each of the two additional ISC-enriched cell populations (Fig. 3, A and B). Notably, however, none of these 15 miRNAs are significantly enriched in ISCs relative to each of the other major differentiated cell types analyzed in this study (Fig. 3C). For example, although miR-21-5p and miR-375-3p are among the 15 most highly expressed miRNAs in ISCs, they are even more highly expressed in most other differentiated cell types (Fig. 3C). Also, while let-7a-1-5p and miR-200c-3p are indeed the most highly expressed in ISCs, other cell types (such as Paneth cells) exhibit highly comparable levels (Fig. 3C).
Figure 3.
The microRNAs most highly expressed in intestinal stem cells (ISCs) are not enriched in ISCs. A: Venn diagram of the top 20 expressed microRNAs across three different sorted subpopulations enriched for ISCs: Lgr5+ (n = 2 pools of mice), Sox9-Low (n = 4 mice), or Cd24-Low (n = 4 mice). B: list of the top 20 expressed microRNAs across all three ISC-enriched sorted cell populations, arranged in order of expression. Red indicates those microRNAs that are identified among the top 20 in all three ISC-enriched sorted cell populations. C: normalized expression of a subset of highly expressed microRNAs in ISC across major small intestinal epithelial cell types. Error bars represent standard error.
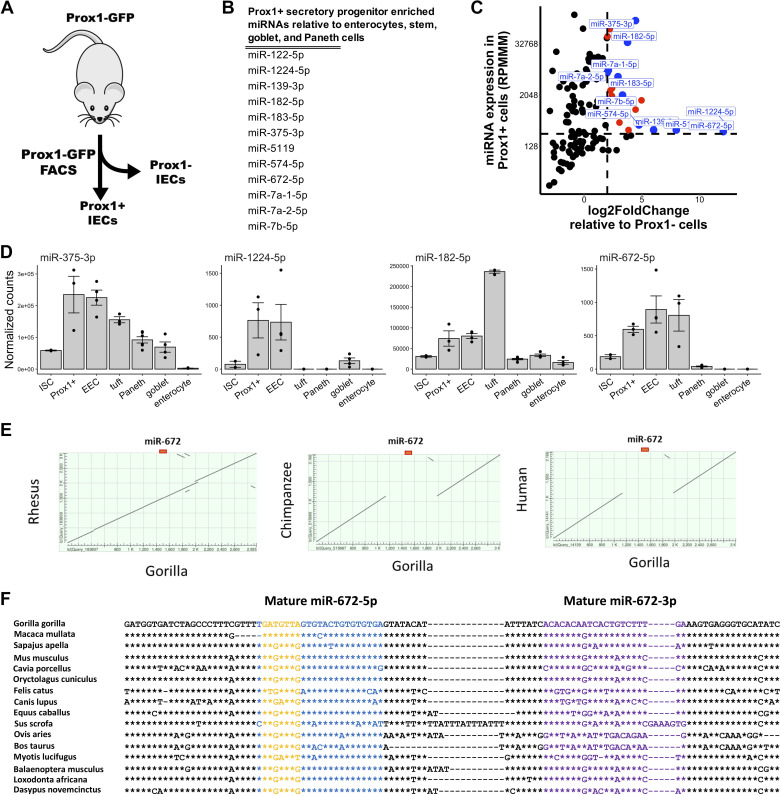
miR-1224 and miR-672 Are Secretory Progenitor Cell Enriched MicroRNAs
We additionally sought to determine whether secretory progenitor cells exhibit enrichment for any miRNAs. Progenitor cells that give rise to both tuft cells and EECs are specifically marked by Prox1 (28). Using Prox1-GFP reporter mice, we isolated Prox1+ and Prox1− cells by FACS (Fig. 4A) (Supplemental Fig. S5A). RT-qPCR analysis showed that, relative to Prox1− cells, Prox1+ cells are enriched for expression of the EEC marker Chga and the tuft cell marker Dclk1, as well as the postulated reserve ISC marker Hopx (Supplemental Fig. S5B) (39, 40). Small RNA-seq identified 12 miRNAs that are enriched in Prox1+ cells relative to ISCs, goblet cells, Paneth cells, and enterocytes (Fig. 4B). Further analysis showed that miR-1224 and miR-672 are the most enriched in Prox1+ cells relative to Prox1− cells (Fig. 4, C and D), whereas miR-375 and miR-182 are the most highly expressed in Prox1+ cells (Fig. 4, C and D).
Figure 4.
miR-1224 and miR-672 are Prox1+ secretory progenitor cell enriched microRNAs. A: schematic of experimental plan for analysis of Prox1+ small intestinal secretory progenitor cells. Prox1+ and Prox1− cells were isolated from the jejunum of Prox1-GFP mice and subjected to small RNA-seq analysis. B: list of microRNAs enriched in jejunal Prox1+ secretory progenitor cells relative to stem cells, enterocytes, goblet cells, and Paneth cells (enteroendocrine and tuft cells not included since they are derived from Prox1+ secretory progenitors). MicroRNA enrichment is defined as an average expression > 500 and fold change > 1.5 versus each listed cell type. C: scatter plot of expression levels (reads per million mapped to microRNAs, RPMMM; y-axis) of microRNAs in isolated Prox1+ cells and log2-fold change (x-axis) of microRNA expression in Prox1+ cells relative to Prox1− cells. Prox1+ cell enriched microRNAs are depicted in orange and those with adjusted P value < 0.05 are depicted in blue. D: normalized expression of miR-375-3p, miR-1224-5p, miR-182-5p, and miR-672-5p across cell fractions enriched for ISCs, Prox1+ secretory progenitor cells, enteroendocrine cells (EECs), tuft cells, Paneth cells, goblet cells, and enterocytes. E: dot plot of the 3-kb locus centered on the miR-672 gene in the rhesus, chimpanzee, and human genomes relative to the gorilla genome. F: multiple DNA sequence alignment of the miR-672 gene in various placental mammalian species. The mature -5p, mature -3p, and seed sequences are depicted in blue, purple, and yellow, respectively. Position identities relative to gorilla are denoted with asterisks and gaps are denoted with hyphens. Error bars represent standard error.
One of the most intriguing of the Prox1+ cell-enriched miRNAs is miR-672-5p, because it is not present in the human genome. In mice, this miRNA is embedded in a large intron of the gene neurite extension and migration factor (Nexmif). Data mining of NCBI-deposited genomes allowed for the identification of the miR-672 gene in 120 placental mammals in addition to five species present in the GenBank database. Interestingly, in hominin species (Bonobo, Chimpanzee, and Human), an ∼1 kb genomic sequence of the Nexmif intron that harbors the miR-672 gene is deleted (Fig. 4E). Although the mature sequences of miR-672-5p appear to be broadly conserved across placental mammals, the nucleotides at two positions in the seed region differ for Old World monkey and Ape compared with other mammals (Fig. 4F).
miR-486 is a Tuft Cell-Enriched MicroRNA
We next sought to validate miR-486 as a tuft cell-enriched miRNA. To do this, we isolated jejunal crypts from a mouse model in which the transcription factor Pou2f3, which is required for tuft cell differentiation, has been knocked out (Supplemental Fig. S6A) (41, 42). RT-qPCR analysis of IEC marker expression showed that, as expected, jejunal crypts from these Pou2f3−/− mice exhibit greatly reduced (∼12-fold) expression of the tuft cell marker Dclk1 relative to wild-type (Supplemental Fig. S6B), which verifies the depletion of tuft cells. We also found that miR-486 expression is remarkably downregulated (∼3-fold) in these tuft cell-depleted crypts relative to WT (Supplemental Fig. S6C), which was not observed for five other miRNAs tested (Supplemental Fig. S6D).
miR-152 is a Paneth Cell Enriched MicroRNA
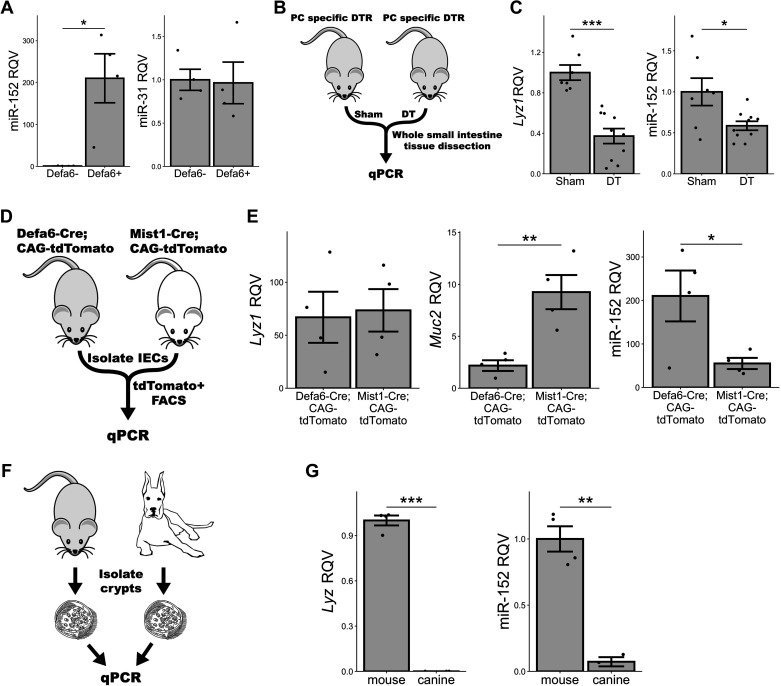
Similar to what we observed in the small RNA-seq analysis, RT-qPCR analysis of sorted Defa6+ cells showed that miR-152 is enriched in Paneth cells, whereas other miRNAs, such as miR-31, are not (Fig. 5A). To validate that Paneth cells are enriched for miR-152-3p, we leveraged three different in vivo models in which Paneth cell identity is affected or Paneth cells are depleted. In the first model, Defa6-DTR, Paneth cells are depleted by diphtheria toxin (DT) injection into mice that are modified to express diphtheria toxin receptor (DTR) in a Paneth-cell specific manner (Fig. 5B) (43). As expected, we found that the abundance of the classic Paneth cell marker, Lyz1, is significantly reduced in whole intestinal tissue upon Paneth cell depletion (Fig. 5C). We also detected subtle effects on the expression levels of Muc2 and Dclk1, but not nearly to the levels observed for Lyz1 (Supplemental Fig. S7A and Fig. 5C), indicating that DT had the intended effect. Importantly, we observed a significant reduction of miR-152 expression (Fig. 5C), consistent with our finding that miR-152 is a Paneth cell-enriched miRNA.
Figure 5.
miR-152 is a Paneth cell-enriched microRNA. A: RT-qPCR data for miR-152 and miR-31 in sorted Defa6 expressing tdTomato+ cells from Defa6-Cre; CAG-tdTomato mice (Defa6+) (n = 4 mice) relative to Defa6 nonexpressing tdTomato- cells (Defa6−) (n = 4 mice). B: schematic of experimental plan for analyzing Paneth cells under two different conditions: normal and Paneth-diminished. Defa6-DTR mice were either sham treated or administered diphtheria toxin (DT) and then small intestinal whole tissue was dissected for RT-qPCR analysis. C: RT-qPCR data for Lyz1 and miR-152 in DT-treated Defa6-DTR mice (n = 7 mice) relative to sham treatment (n = 9 mice). D: schematic of experimental plan for analyzing Paneth cells from Defa6-Cre; CAG-tdTomato and Mist1-Cre; CAG-tdTomato mice. tdTomato+ cells were sorted and subjected to RT-qPCR analysis. E: RT-qPCR data for Lyz1, Muc2, and miR-152 in sorted Mist1-Cre; CAG-tdTomato mice (n = 4 mice) relative to those derived from Defa6-Cre; CAG-tdTomato mice (n = 4 mice). F: schematic of experimental plan for analyzing mouse and canine jejunal enteroids. Mouse (B62J) and canine jejunal crypts were isolated and used to culture enteroids which were subsequently subjected to RT-qPCR analysis. G: RT-qPCR data for canine Lyz, mouse Lyz1, and miR-152 in canine jejunal enteroids (n = 3 replicates) relative to mouse jejunal enteroids (n = 4 replicates). *P < 0.05, **P < 0.01, ***P < 0.005 by two-tailed Student’s t test. RQV, relative quantitative value.
For the second model, we used a Mist1 disrupted mouse (10). In intestinal crypts, Mist1 is specifically expressed by Paneth cells. Its expression promotes proper Paneth cell differentiation, whereas the loss of Mist1 leads to abnormal Paneth cells that also exhibit goblet cell features (10). We reasoned that if miR-152 is a Paneth cell-specific marker, its expression may be reduced in abnormal Paneth cells of Mist1 disrupted mice relative to canonical Paneth cells of Mist1 wild-type mice. To test this, we sorted cells from both Defa6-Cre; CAG-tdTomato and Mist1-Cre; CAG-tdTomato mice (Supplemental Fig. S7B) and assessed gene and miRNA expression levels by RT-qPCR analysis (Fig. 5D). Although Lyz1 is unchanged between Defa6-Cre; CAG-tdTomato and Mist1-Cre; CAG-tdTomato Paneth cells, we noted that Muc2, normally a marker for goblet cells, is increased in expression in Mist1-Cre; CAG-tdTomato Paneth cells relative to Defa6-Cre; CAG-tdTomato Paneth cells, thus signifying the loss of a pure Paneth cell character and the gain of a hybrid Paneth-goblet cell intermediate cell type (Fig. 5E and Supplemental Fig. S7C) (10). Moreover, we find that the rise of this intermediate cell type in place of Paneth cells in the Mist1-Cre; CAG-tdTomato mouse is accompanied by a significant reduction in miR-152 expression (Fig. 5E). Since Copz2, the host gene for the miR-152 gene, is a direct target of Mist1, reduced miR-152 expression may be the direct result of the loss of Mist1 (44–46).
Finally, for the third model, we selected a species, canine, that is devoid of histologically discernible canonical Paneth cells (47, 48). Specifically, we isolated jejunal crypts from both B62J mice and canines, established enteroids ex vivo, and performed RT-qPCR (Fig. 5F). We demonstrated that not only are canine enteroids lacking in lysozyme expression, as expected, but also that they exhibit greatly reduced expression of miR-152 relative to mouse jejunal enteroids (Fig. 5G). Taken together, these three models add substantial support to the notion that miR-152 is a miRNA marker of canonical, fully differentiated Paneth cells.
DISCUSSION
In this study, we have presented the first comprehensive profiling of miRNAs across all major cell types of the mouse small intestinal epithelium. Although we have previously shown that miR-7 is enriched in mouse small intestinal epithelial enteroendocrine progenitor cells, the identity of miRNAs enriched in Paneth cells, tuft cells, goblet cells, and ISCs has been unknown (3). Treveil et al. (49) used a mouse enteroid system to enrich for either goblet or Paneth cells to define miRNAs associated with either cell type. However, in such an explant system, sample preparations are still highly heterogenous, and the absence of in vivo luminal and basolateral influences may alter miRNA expression. To bridge the knowledge gap, we used flow cytometry and FACS-isolated populations of enriched cell types from reporter mice for small RNA-seq analysis. With the assistance of additional genome-scale technologies, scRNA-seq and ChRO-seq, we provided the most comprehensive and high-resolution picture to date of miRNA expression in the small intestine. We found that all major cell types exhibit unique miRNA profiles. Moreover, by leveraging several different experimental in vivo systems, we validated that miR-152 is a specific miRNA marker of Paneth cells. We also find that miR-486 is enriched in small intestinal tuft cells. Recent work suggests that overexpression of another miRNA, miR-195, can suppress the expression of a canonical tuft cell marker, Dclk1 (50). We detected endogenous miR-195 only at exceedingly low levels in the murine intestinal epithelium; however, it is important to note that our analysis is restricted to the jejunum and future investigations should extend to other regions of the small intestine.
There are three important caveats concerning the interpretation of the data in this study. First, we have only determined the LEMs of cell types located within the small intestinal epithelial layer. We have made no comparison with cells outside of the small intestinal epithelium. For instance, although we report that miR-486 is highly enriched in small intestinal tuft cells relative to other IECs, other studies have reported that miR-486 is also highly expressed in other cell types such as skeletal muscle cells and myeloid cells (51–53). Likewise, intestinal secretory progenitor cell-enriched miR-375 is known to be highly expressed also in pancreatic islet beta cells (54). Second, it is important to emphasize that while the FACS-isolated fractions used for our analysis are indeed highly enriched for certain cell types, they are not 100% pure, homogenous cell populations. Finally, we have not made any determination of miRNA expression in subtypes of specific cell types. For example, it has long been known that there are several different subtypes of enteroendocrine cells (55). Previous single-cell transcriptomic analyses suggest that there are also molecular subtypes of tuft cells, Paneth cells, ISCs, and enterocytes (21, 56, 57). Single-cell analysis of miRNA expression patterns would be ideal to achieve this level of resolution; however, while this technique is emerging onto the scene, it is not yet in widespread use. Once optimized, this approach will be powerful for achieving even higher resolution on miRNA profiles.
Notwithstanding these interpretative caveats and technical limitations, the results of this study have uncovered some heretofore unknown features of the miRNA expression profile of the mouse small intestinal epithelium. In particular, we highlight the finding that miR-152 is a Paneth cell-specific miRNA. This may be especially technically useful because current Paneth cell markers are all limited to secreted protein factors, which may be confounding if they are depleted in the in vivo source, such as in the event of a hypersecretion episode that outruns replenishment by de novo protein production. Furthermore, the fact that certain IECs are enriched for specific miRNAs does immediately provoke questions about the functional significance of these miRNAs. For example, the enrichment of miR-1224 in EECs may indicate that miR-1224 regulates the production and secretion of metabolic hormones in response to changes in nutrient intake. Alternatively, miR-1224 may serve to maintain the differentiated state of mature EECs. Possibly the loss of miR-1224 in EECs allows for their dedifferentiation to ISCs, since it is known that enteroendocrine cells, as well as IEC types, have this latent potency which is revealed under certain environmental challenges (28). Yet another possibility is that miR-1224 serves as a regulatory check, or block, on further specification of the EEC lineage. This possibility has pathophysiological implications as well. Specifically, given that EECs are critical for energy homeostasis, it is plausible that dysfunction of one or more of the EEC lineage miRNAs identified in this study could lead to EEC defects, which in turn would predispose to conditions such as diabetes. The investigation of these possibilities is outside of the scope of this study, but future work in this area is most definitely warranted.
Perhaps one of the more intriguing LEMs that could be investigated functionally is miR-672, which we found to be highly expressed in both the EEC and tuft lineages. Interestingly, the divergence of its seed sequence in Old World monkeys and Apes from other placental mammals, and even more interestingly, the deletion of the miRNA altogether in hominins, suggests an intriguing possibility that its loss may contribute to unique features of hominins. The host gene of miR-672, neurite extension and migration factor (Nexmif), is thought to control neurite outgrowth. What, if any, role miR-672 plays in this process, as well as possibly in EEC and tuft biology, remains to be determined.
SUPPLEMENTAL DATA
Supplemental Figs. S1–S7 and Supplemental Tables S1–S3: https://github.com/Sethupathy-Lab/2021_AJP_Shanahan_et_al_supplemental.
GRANTS
Financial support was provided by the ADA Pathway to Stop Diabetes Award, 1-16-ACE-47 (awarded to P. Sethupathy).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
M.T.S. and P.S. conceived and designed research; M.T.S., O.O.O., Y-H.H., A.P.S., B.C.E.P., M.B., B.S., J.E.C., H.G., D.K.S., R.C., S.D., J.v.M., and P.S. performed experiments; M.T.S., M.K., Y.-H.H., K.K.-L., and P.S. analyzed data; M.T.S., M.K., O.O.O., Y-H.H., N.A.K., J.P.M., K.A., S.J.M., C.M.D., E.D.T.W., and P.S. interpreted results of experiments; M.T.S., M.K., K.K-L., and P.S. prepared figures; M.T.S., M.K., and P.S. drafted manuscript; M.T.S., M.K., O.O.O., Y-H.H., K.K-L., A.P.S., B.C.E.P., K.A., S.J.M., J.v.M., C.M.D., E.D.T.W., and P.S. edited and revised manuscript; M.T.S., M.K., O.O.O., Y-H.H., K.K-L., A.P.S., B.C.E.P., M.B., B.S., J.E.C., H.G., R.C., N.A.K., J.P.M., K.A., S.J.M., S.D., J.v.M., C.M.D., E.D.T.W., and P.S. approved final version of manuscript.
REFERENCES
- 1.McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, Kaestner KH. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology 139: 1654–1664.e1, 2010. doi: 10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peck BC, Mah AT, Pitman WA, Ding S, Lund PK, Sethupathy P. Functional transcriptomics in diverse intestinal epithelial cell types reveals robust microRNA sensitivity in intestinal stem cells to microbial status. J Biol Chem 292: 2586–2600, 2017. doi: 10.1074/jbc.M116.770099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh AP, Hung YH, Shanahan MT, Kanke M, Bonfini A, Dame MK, Biraud M, Peck BCE, Oyesola OO, Freund JM, Cubitt RL, Curry EG, Gonzalez LM, Bewick GA, Tait-Wojno ED, Kurpios NA, Ding S, Spence JR, Dekaney CM, Buchon N, Sethupathy P. Enteroendocrine progenitor cell-enriched miR-7 regulates intestinal epithelial proliferation in an Xiap-dependent manner. Cell Mol Gastroenterol Hepatol 9: 447–464, 2020. doi: 10.1016/j.jcmgh.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol 296: G1108–G1118, 2009. doi: 10.1152/ajpgi.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265, 2009. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 6.Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, Glickman JN, Tschurtschenthaler M, Hartwig J, Hosomi S, Flak MB, Cusick JL, Kohno K, Iwawaki T, Billmann-Born S, Raine T, Bharti R, Lucius R, Kweon MN, Marciniak SJ, Choi A, Hagen SJ, Schreiber S, Rosenstiel P, Kaser A, Blumberg RS. Paneth cells as a site of origin for intestinal inflammation. Nature 503: 272–276, 2013. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, Miller CN, Pollack JL, Nagana Gowda GA, Fontana MF, Erle DJ, Anderson MS, Locksley RM, Raftery D, von Moltke J. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity 49: 33–41.e7, 2018. doi: 10.1016/j.immuni.2018.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lueschow SR, Stumphy J, Gong H, Kern SL, Elgin TG, Underwood MA, Kalanetra KM, Mills DA, Wong MH, Meyerholz DK, Good M, McElroy SJ. Loss of murine Paneth cell function alters the immature intestinal microbiome and mimics changes seen in neonatal necrotizing enterocolitis. PLoS One 13: e0204967, 2018. doi: 10.1371/journal.pone.0204967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger JN, Gong H, Good M, McElroy SJ. Dithizone-induced Paneth cell disruption significantly decreases intestinal perfusion in the murine small intestine. J Pediatr Surg 54: 2402–2407, 2019. doi: 10.1016/j.jpedsurg.2019.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dekaney CM, King S, Sheahan B, Cortes JE. Mist1 expression is required for Paneth cell maturation. Cell Mol Gastroenterol Hepatol 8: 549–560, 2019. doi: 10.1016/j.jcmgh.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mah AT, Van Landeghem L, Gavin HE, Magness ST, Lund PK. Impact of diet-induced obesity on intestinal stem cells: hyperproliferation but impaired intrinsic function that requires insulin/IGF1. Endocrinology 155: 3302–3314, 2014. doi: 10.1210/en.2014-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong VW, Stange DE, Page ME, Buczacki S, Wabik A, Itami S, van de Wetering M, Poulsom R, Wright NA, Trotter MW, Watt FM, Winton DJ, Clevers H, Jensen KB. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol 14: 401–408, 2012. doi: 10.1038/ncb2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol 8: 198–210, 2015. doi: 10.1038/mi.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nefzger CM, Jarde T, Rossello FJ, Horvay K, Knaupp AS, Powell DR, Chen J, Abud HE, Polo JM. A versatile strategy for isolating a highly enriched population of intestinal stem cells. Stem Cell Rep 6: 321–329, 2016. doi: 10.1016/j.stemcr.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith NR, Davies PS, Levin TG, Gallagher AC, Keene DR, Sengupta SK, Wieghard N, El Rassi E, Wong MH. Cell adhesion molecule CD166/ALCAM functions within the crypt to orchestrate murine intestinal stem cell homeostasis. Cell Mol Gastroenterol Hepatol 3: 389–409, 2017. doi: 10.1016/j.jcmgh.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGinty JW, Ting HA, Billipp TE, Nadjsombati MS, Khan DM, Barrett NA, Liang HE, Matsumoto I, von Moltke J. Tuft-cell-derived leukotrienes drive rapid anti-helminth immunity in the small intestine but are dispensable for anti-protist immunity. Immunity 52: 528–541.e7, 2020. doi: 10.1016/j.immuni.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung YH, Huang S, Dame MK, Yu Q, Yu QC, Zeng YA, Camp JG, Spence JR, Sethupathy P. Chromatin regulatory dynamics of early human small intestinal development using a directed differentiation model. Nucleic Acids Res 49: 726–744, 2021. doi: 10.1093/nar/gkaa1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinh TA, Sritharan R, Smith FD, Francisco AB, Ma RK, Bunaciu RP, Kanke M, Danko CG, Massa AP, Scott JD, Sethupathy P. Hotspots of aberrant enhancer activity in fibrolamellar carcinoma reveal candidate oncogenic pathways and therapeutic vulnerabilities. Cell Rep 31: 107509, 2020. doi: 10.1016/j.celrep.2020.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandra L, Borcherding DC, Kingsbury D, Atherly T, Ambrosini YM, Bourgois-Mochel A, Yuan W, Kimber M, Qi Y, Wang Q, Wannemuehler M, Ellinwood NM, Snella E, Martin M, Skala M, Meyerholz D, Estes M, Fernandez-Zapico ME, Jergens AE, Mochel JP, Allenspach K. Derivation of adult canine intestinal organoids for translational research in gastroenterology. BMC Biol 17: 33, 2019. doi: 10.1186/s12915-019-0652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanke M, Baran-Gale J, Villanueva J, Sethupathy P. miRquant 2.0: an expanded tool for accurate annotation and quantification of microRNAs and their isomiRs from small RNA-sequencing data. J Integr Bioinform 13: 307, 2016. doi: 10.2390/biecoll-jib-2016-307. [DOI] [PubMed] [Google Scholar]
- 21.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, Tirosh I, Beyaz S, Dionne D, Zhang M, Raychowdhury R, Garrett WS, Rozenblatt-Rosen O, Shi HN, Yilmaz O, Xavier RJ, Regev A. A single-cell survey of the small intestinal epithelium. Nature 551: 333–339, 2017. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu T, Wang Z, Chou SP, Danko CG. Discovering transcriptional regulatory elements from run-on and sequencing data using the web-based dREG gateway. Curr Protoc Bioinformatics 66: e70, 2019. doi: 10.1002/cpbi.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864, 2011. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26: 2204–2207, 2010. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Furstenberg RJ, Gulati AS, Baxi A, Doherty JM, Stappenbeck TS, Gracz AD, Magness ST, Henning SJ. Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am J Physiol Gastrointest Liver Physiol 300: G409–G417, 2011. doi: 10.1152/ajpgi.00453.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, Sangiorgi E, Capecchi MR, Kuo CJ. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA 109: 466–471, 2012. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Moltke J, Ji M, Liang HE, Locksley RM. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529: 221–225, 2016. doi: 10.1038/nature16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, et al. Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell 21: 78–90.e6, 2017. doi: 10.1016/j.stem.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheahan BJ, Freeman AN, Keeley TM, Samuelson LC, Roper J, Hasapis S, Lee CL, Dekaney CM. Epithelial regeneration after doxorubicin arises primarily from early progeny of active intestinal stem cells. Cell Mol Gastroenterol Hepatol 12: 119–140, 2021. doi: 10.1016/j.jcmgh.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007, 2007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 31.Gerbe F, Brulin B, Makrini L, Legraverend C, Jay P. DCAMKL-1 expression identifies Tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology 137: 2179–2180, 2009. doi: 10.1053/j.gastro.2009.06.072. [DOI] [PubMed] [Google Scholar]
- 32.Heitz PU, Wegmann W. Identification of neoplastic Paneth cells in an adenocarcinoma of the stomach using lysozyme as a marker, and electron microscopy. Virchows Arch A Pathol Anat Histol 386: 107–116, 1980. doi: 10.1007/BF00432648. [DOI] [PubMed] [Google Scholar]
- 33.Eisenhauer PB, Harwig SS, Lehrer RI. Cryptdins: antimicrobial defensins of the murine small intestine. Infect Immun 60: 3556–3565, 1992. doi: 10.1128/iai.60.9.3556-3565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss AA, Babyatsky MW, Ogata S, Chen A, Itzkowitz SH. Expression of MUC2 and MUC3 mRNA in human normal, malignant, and inflammatory intestinal tissues. J Histochem Cytochem 44: 1161–1166, 1996. doi: 10.1177/44.10.8813081. [DOI] [PubMed] [Google Scholar]
- 35.Slabý J, Lojda Z, Kraml J, Kolínská J. Immunohistochemical localization of intestinal glycosidases. Acta Univ Carol Med Monogr 77: 105–111, 1977. [PubMed] [Google Scholar]
- 36.Cetin Y, Muller-Koppel L, Aunis D, Bader MF, Grube D. Chromogranin A (CgA) in the gastro-entero-pancreatic (GEP) endocrine system. II. CgA in mammalian entero-endocrine cells. Histochemistry 92: 265–275, 1989. doi: 10.1007/BF00500540. [DOI] [PubMed] [Google Scholar]
- 37.Wang D, Lu M, Miao J, Li T, Wang E, Cui Q. Cepred: predicting the co-expression patterns of the human intronic microRNAs with their host genes. PLoS One 4: e4421, 2009. doi: 10.1371/journal.pone.0004421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steiman-Shimony A, Shtrikman O, Margalit H. Assessing the functional association of intronic miRNAs with their host genes. RNA 24: 991–1004, 2018. doi: 10.1261/rna.064386.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez LM, Williamson I, Piedrahita JA, Blikslager AT, Magness ST. Cell lineage identification and stem cell culture in a porcine model for the study of intestinal epithelial regeneration. PLoS One 8: e66465, 2013. doi: 10.1371/journal.pone.0066465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li N, Yousefi M, Nakauka-Ddamba A, Jain R, Tobias J, Epstein JA, Jensen ST, Lengner CJ. Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Rep 3: 876–891, 2014. doi: 10.1016/j.stemcr.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, Bruschi M, Harcus Y, Zimmermann VS, Taylor N, Maizels RM, Jay P. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529: 226–230, 2016. doi: 10.1038/nature16527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita J, Ohmoto M, Yamaguchi T, Matsumoto I, Hirota J. Skn-1a/Pou2f3 functions as a master regulator to generate Trpm5-expressing chemosensory cells in mice. PLoS One 12: e0189340, 2017. doi: 10.1371/journal.pone.0189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang C, Sherman MP, Prince LS, Bader D, Weitkamp JH, Slaughter JC, McElroy SJ. Paneth cell ablation in the presence of Klebsiella pneumoniae induces necrotizing enterocolitis (NEC)-like injury in the small intestine of immature mice. Dis Model Mech 5: 522–532, 2012. doi: 10.1242/dmm.009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian X, Jin RU, Bredemeyer AJ, Oates EJ, Błazewska KM, McKenna CE, Mills JC. RAB26 and RAB3D are direct transcriptional targets of MIST1 that regulate exocrine granule maturation. Mol Cell Biol 30: 1269–1284, 2010. doi: 10.1128/MCB.01328-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Direnzo D, Hess DA, Damsz B, Hallett JE, Marshall B, Goswami C, Liu Y, Deering T, Macdonald RJ, Konieczny SF. Induced Mist1 expression promotes remodeling of mouse pancreatic acinar cells. Gastroenterology 143: 469–480, 2012. doi: 10.1053/j.gastro.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin RU, Mills JC. RAB26 coordinates lysosome traffic and mitochondrial localization. J Cell Sci 127: 1018–1032, 2014. doi: 10.1242/jcs.138776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghoos Y, Vantrappen G. The cytochemical localization of lysozyme in Paneth cell granules. Histochem J 3: 175–178, 1971. doi: 10.1007/BF01002560. [DOI] [PubMed] [Google Scholar]
- 48.Sheahan DG, Jervis HR. Comparative histochemistry of gastrointestinal mucosubstances. Am J Anat 146: 103–131, 1976. doi: 10.1002/aja.1001460202. [DOI] [PubMed] [Google Scholar]
- 49.Treveil A, Sudhakar P, Matthews ZJ, Wrzesiński T, Jones EJ, Brooks J, Ölbei M, Hautefort I, Hall LJ, Carding SR, Mayer U, Powell PP, Wileman T, Di Palma F, Haerty W, Korcsmáros T. Regulatory network analysis of Paneth cell and goblet cell enriched gut organoids using transcriptomics approaches. Mol Omics 16: 39–58, 2020. doi: 10.1039/c9mo00130a. [DOI] [PubMed] [Google Scholar]
- 50.Kwon MS, Chung HK, Xiao L, Yu TX, Wang SR, Piao JJ, Rao JN, Gorospe M, Wang JY. MicroRNA-195 regulates Tuft cell function in the intestinal epithelium by altering translation of DCLK1. Am J Physiol Cell Physiol 320: C1042–C1054, 2021. doi: 10.1152/ajpcell.00597.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beg F, Wang R, Saeed Z, Devaraj S, Masoor K, Nakshatri H. Inflammation-associated microRNA changes in circulating exosomes of heart failure patients. BMC Res Notes 10: 751, 2017. doi: 10.1186/s13104-017-3090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horak M, Novak J, Bienertova-Vasku J. Muscle-specific microRNAs in skeletal muscle development. Dev Biol 410: 1–13, 2016. doi: 10.1016/j.ydbio.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 53.Wang L-S, Li L, Li L, Chu S, Shiang K-D, Li M, Sun H-Y, Xu J, Xiao F-J, Sun G, Rossi JJ, Ho YWei, Bhatia R. MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood 125: 1302–1313, 2015. doi: 10.1182/blood-2014-06-581926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230, 2004. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 55.Beumer J, Puschhof J, Bauzá-Martinez J, Martínez-Silgado A, Elmentaite R, James KR, et al. High-resolution mRNA and secretome atlas of human enteroendocrine cells. Cell 182: 1062–1064, 2020. doi: 10.1016/j.cell.2020.08.005. [DOI] [PubMed] [Google Scholar]
- 56.Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, Ashenberg O, Su C-W, Smillie C, Shekhar K, Chen Z, Wu C, Ordovas-Montanes J, Alvarez D, Herbst RH, Zhang M, Tirosh I, Dionne D, Nguyen LT, Xifaras ME, Shalek AK, von Andrian UH, Graham DB, Rozenblatt-Rosen O, Shi HN, Kuchroo V, Yilmaz OH, Regev A, Xavier RJ. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell 175: 1307–1320.e22, 2018. doi: 10.1016/j.cell.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moor AE, Harnik Y, Ben-Moshe S, Massasa EE, Rozenberg M, Eilam R, Bahar Halpern K, Itzkovitz S. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell 175: 1156–1167.e15, 2018. doi: 10.1016/j.cell.2018.08.063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figs. S1–S7 and Supplemental Tables S1–S3: https://github.com/Sethupathy-Lab/2021_AJP_Shanahan_et_al_supplemental.