Abstract
Background.
Pathologist and computational assessments have been used to evaluate immunohistochemistry (IHC) in epidemiologic studies. We compared Definiens Tissue Studio® to pathologist scores for 17 markers measured in breast tumor tissue microarrays (TMAs) [AR, CD20, CD4, CD8, CD163, EPRS, ER, FASN, H3K27, IGF1R, IR, Ki67, phospho-mTOR, PR, PTEN, RXR, and VDR].
Methods.
5 914 Nurses’ Health Study participants, diagnosed 1976–2006 (NHS) and 1989–2006 (NHS-II), were included. IHC was conducted by the Dana-Farber/Harvard Cancer Center Specialized Histopathology Laboratory. The percent of cells staining positive was assessed by breast pathologists. Definiens output was used to calculate a weighted average of percent of cells staining positive across TMA cores for each marker. Correlations between pathologist and computational scores were evaluated with Spearman correlation coefficients. Receiver-operator characteristic curves were constructed, using pathologist scores as comparison.
Results.
Spearman correlations between pathologist and Definiens assessments ranged from weak (RXR, rho=−0.05; CD163, rho=0.10) to strong (Ki67, rho=0.79; pmTOR, rho=0.77). The area under the curve was >0.70 for all markers except RXR.
Conclusion.
Our data indicate that computational assessments exhibit variable correlations with interpretations made by an expert pathologist, depending on the marker evaluated. This study provides evidence supporting the use of computational platforms for IHC evaluation in large-scale epidemiologic studies, with the caveat that pilot studies are necessary to investigate agreement with expert assessments. In sum, computational platforms may provide greater efficiency and facilitate high-throughput epidemiologic analyses.
Keywords: breast cancer, epidemiology, immunohistochemistry, computational pathology
1. Introduction
Immunohistochemistry (IHC) is frequently used to measure expression of putative biomarkers in tumor tissue microarrays (TMAs) constructed from formalin-fixed paraffin-embedded (FFPE) tissue samples. An expert pathologist’s interpretation of IHC assays has generally been considered the gold standard in large-scale epidemiologic investigations. Often these manually assigned scores are semi-quantitative, incorporating both intensity and extent of immunoreactivity [1]. This approach is limited, however, in that it is expensive, time-consuming, reliant on subjective scoring parameters, and potentially prone to bias [2].
An alternative strategy is to utilize computational platforms which, in theory, provide a more objective assessment of the extent and/or intensity of immunoreactivity [3]. Previous studies have generally demonstrated good agreement between pathologist- and computationally-generated scores for estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor (HER2) [4–8]. However, whether this agreement extends to other biomarkers, including those exhibiting cytoplasmic staining and reactivity in stromal cells, is not well understood. Understanding the extent to which automated platforms concur with pathologist assessments can help in determining the appropriate analytic method to evaluate immunoreactivity, particularly in analyses using archival FFPE tissues.
In this study, we evaluated the performance of the semi-automated Definiens TissueStudio® platform compared to pathologist assessment of 17 markers measured in breast tumors collected nationally over several decades [AR (androgen receptor), CD20, CD4, CD8, and CD163, EPRS (glutamyl-prolyl-tRNA synthetase), ER (estrogen receptor), FASN (fatty acid synthase), H3K27 (histone 3 lysine 27 trimethylation), IGF1R (insulin-like growth factor 1 receptor), IR (insulin receptor), Ki67 (marker of proliferation Ki-67), phospo-mTOR (phosphorylated mammalian target of rapamycin), PR (progesterone receptor), PTEN (phosphatase and tensin homolog), RXR (retinoic acid receptor), and VDR (vitamin D receptor)].
2. Materials and Methods
2.1. Study population.
The TMAs included in this study contain tissue from 5 914 participants of the Nurses’ Health Study (NHS) and Nurses’ Health Study II (NHS-II) who were diagnosed with in situ or invasive breast carcinomas between 1976–2006 (NHS) and 1989–2006 (NHS-II). Eligibility criteria and procurement of tumor tissue have previously been described in detail [9]. Briefly, tumor tissue was requested from pathology departments at hospitals treating NHS/NHS-II participants who have no history of cancer other than non-melanoma skin cancer prior to their breast cancer diagnosis. For each participant, three 0.6 mm tumor tissue cores were selected as representative of the tumor and placed into TMAs. This study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. Informed consent was obtained from all NHS and NHS-II participants.
2.2. Selection of tissue markers and immunohistochemistry.
We selected those markers for which IHC had previously been completed and for which both pathologist and automated scores were available. We sought to utilize the large amount of IHC data collected in the NHS/NHS-II cohorts and therefore we included both clinically useful (e.g., ER, PR) and investigational markers in our analysis. HER2 was not included as automated data was not available.
For ER, PR, and AR, both pathologist and automated scores were generated for the majority of cases. For the remaining markers, a single TMA was manually scored for comparison to the Definiens data. For IR, H3K27, RXR, VDR, and Ki67, the manually scored TMA contained 159 cases. For CD163, CD20, CD8, CD4, EPRS, FASN, IGF1R, pmTOR, and PTEN, the manually scored TMA contained 132 cases. The markers evaluated in this study are expressed by tumor cells, stromal cells, and/or inflammatory cells, and exhibit either nuclear or cytoplasmic localization (see Table 1 for the cell type and localization evaluated for each marker). Staining of most markers was evaluated in tumor cells only, although for CD4, CD8, and CD20, staining of stromal lymphocytes was examined. Additionally, EPRS and CD163 were assessed separately in tumor and stromal cells. We excluded 182 cases that were missing pathologist or Definiens data for all of the 17 markers. Missing data could be attributed to core loss during sectioning or staining, staining artifacts that obscure the evaluation of a core, tissue folding within a core, or lack of tumor cells in a core following sectioning. For ER, PR, and AR, comparisons between pathologist evaluation and computational assessment were made on this final dataset containing 4 673 cases from NHS and 1 241 cases from NHS-II. For all other markers, comparisons were made for 159 cases (IR, H3K27, RXR, VDR, Ki67) or 132 cases (CD163, CD20, CD8, CD4, EPRS, FASN, IGF1R, pmTOR, PTEN), because only one TMA was evaluated by the pathologist for those markers.
Table 1.
Antibody, cell type, localization, and pathologist evaluation for each biomarker.
| Protein | Antibody | Dilution | Cell type | Localization | Evaluation |
|---|---|---|---|---|---|
| AR | Dako (clone AR441) | 1:200 | Tumor | Nucleus | % of cells expressing the proteins Negative Low positive = 1 – 10% Positive > 10% |
| ER | Dako (clone 1D5) | 1:200 | Tumor | Nucleus | |
| H3K27 | Millipore 07–449 (rabbit polyclonal) | 1:800 | Tumor | Nucleus | |
| PR | Dako (clone PgR636) | 1:50 | Tumor | Nucleus | |
| VDR | Novus NBP1–19478 (rabbit polyclonal) | 1:500 | Tumor | Nucleus | |
| Ki67 | Vector Labs VP-RM04 (clone SP6) | 1:250 | Tumor | Nucleus | % of positive cells High proliferation > 14% Low proliferation ≤ 14% |
| CD4 | Dako 7310 (clone 4B12) | 1:80 | Stromal lymphocytes | Cytoplasm | % of stromal region containing lymphocytes expressing the marker Low < 10% Moderate = 10 – 50% High > 50% |
| CD8 | Dako 7103 (clone C8/144B) | 1:100 | Stromal lymphocytes | Cytoplasm | |
| CD20 | Dako 0755 (clone L26) | 1:500 | Stromal lymphocytes | Cytoplasm | |
| CD163 | Vector Labs VP-C374 (clone 10D6) | 1:250 | Tumor Stromal macrophages | Cytoplasm Cytoplasm | % of cells expressing the proteins Negative Low positive = 1 – 10% Positive > 10% |
| EPRS | Sigma HPA026490 (rabbit polyclonal) | 1:100 | Tumor Stroma | Cytoplasm Cytoplasm | |
| FASN | ENZO ADI-905–069 (rabbit polyclonal) | 1:300 | Tumor | Cytoplasm | |
| IGF1R | Santa Cruz 713 (clone C-20) | 1:100 | Tumor | Cytoplasm | |
| IR | Millipore 05–1104 (clone CT-3) | 1:50 | Tumor | Cytoplasm | |
| pmTOR | Cell Signaling 2976 (clone 49F9) | 1:50 | Tumor | Cytoplasm | |
| PTEN | Cell Signaling 9559 (clone 138G6) | 1:50 | Tumor | Cytoplasm | |
| RXR | Santa Cruz sc-553 (clone D-20) | 1:300 | Tumor | Cytoplasm |
IHC assays were performed on 5-μm sections using standard protocols and commercially sourced antibodies (Table 1). 3,3’-diaminobenzidine was used as the chromogen and all slides were counterstained with hematoxylin. Appropriate positive and negative controls were included in all IHC experiments. IHC protocols were carried out using a Dako Autostainer (Dako Corporation, Carpinteria, CA, USA) at the Dana-Farber/Harvard Cancer Center Specialized Histopathology Laboratory, Boston, MA, USA. ER, PR, AR, H3K27, RXR, and PTEN were stained in multiple batches; all other markers were stained in a single batch. Batch was included as a covariate in downstream analyses. TMAs were digitized at 40x magnification using the Panoramic SCAN 150 whole slide scanner (3DHISTECH Ltd., Budapest, Hungary).
2.3. Pathologist review.
Immunoreactivity was manually assessed by expert breast pathologists (L.C.C. or G.M.B.) in the Department of Pathology at Beth Israel Deaconess Medical Center. For CD4, CD8, and CD20, each core was scored according to the percentage of the stromal region containing lymphocytes expressing the marker, using the categories of low (<10%), moderate (10–50%), and high (>50%). Ki67 was scored continuously as the percentage of positive cells. For the remaining markers, each core was scored according to the percentage of cells expressing the protein, using the categories of negative, low positive (1–10%), and positive (>10%). Cases were classified as negative or low if all evaluable cores were scored negative or low, low positive or moderate if at least one core was scored low positive or moderate (but none scored positive or high), and positive or high if at least one core was scored positive or high. We secondarily defined cases using the average of pathologist’s scores across a participant’s cores.
2.4. Semi-automated scoring.
Semi-automated scores were generated using the Definiens TissueStudio® (Definiens AG, Munich, Germany) computational pathology system. This software requires the user (Y.J.H. and M.E.P.) to first create an algorithm for each stain. To create these algorithms, 12 random cores were chosen on one TMA for each stain, per the manufacturer’s instructions. Regions of interest were annotated and used to train Definiens to recognize nuclei, tumor and stromal regions, and to optimize the stain detection threshold. The stain-specific algorithms are then applied to the raw TMA images to quantify staining, without further manual annotation of individual tissue cores. The representative training TMAs were included in the study set.
For each marker, the number of cells or nuclei staining positive were summed across a participant’s cores and divided by the sum of the total number of cells/nuclei present across each participant’s cores. This quantity was then multiplied by 100 to generate a weighted average percent positive for each participant. We excluded participants with <100 cells/nuclei across evaluable cores.
Weighted average percent positive = (# positive cells, core 1 + # positive cells, core 2, + # positive cells, core 3) / (total # cells, core 1 + total # cells, core 2 + total # cells, core 3) * 100
2.5. Statistical analysis.
For each marker, we calculated Spearman’s rank-order correlation coefficients between the continuous percent positive obtained from Definiens and both case-level pathologist definitions (ordinal and average pathologist assessments). In addition, we calculated Spearman’s correlation coefficients between the Definiens percent positive and pathologist score for each core. For the Definiens data, we calculated intraclass correlation coefficients across cores. For the pathologist data, Fleiss’ kappa coefficients were calculated for all markers except Ki67, for which the intraclass correlation coefficient was computed. Raw percent agreement and Cohen’s kappa coefficients were calculated to evaluate agreement between the pathologist and Definiens assessments at the core and case level, by categorizing the Definiens percent positive using the same cutpoints as the pathologist interpretation.
To assess the sensitivity and specificity of the Definiens-derived percent positive, we generated receiver-operator characteristic (ROC) curves using the pathologist assessment for each marker as the gold standard. Pathologist assessments were collapsed into positive and negative cases, where positive included low positive and positive, or moderate and high, according to the marker analyzed.
3. Results
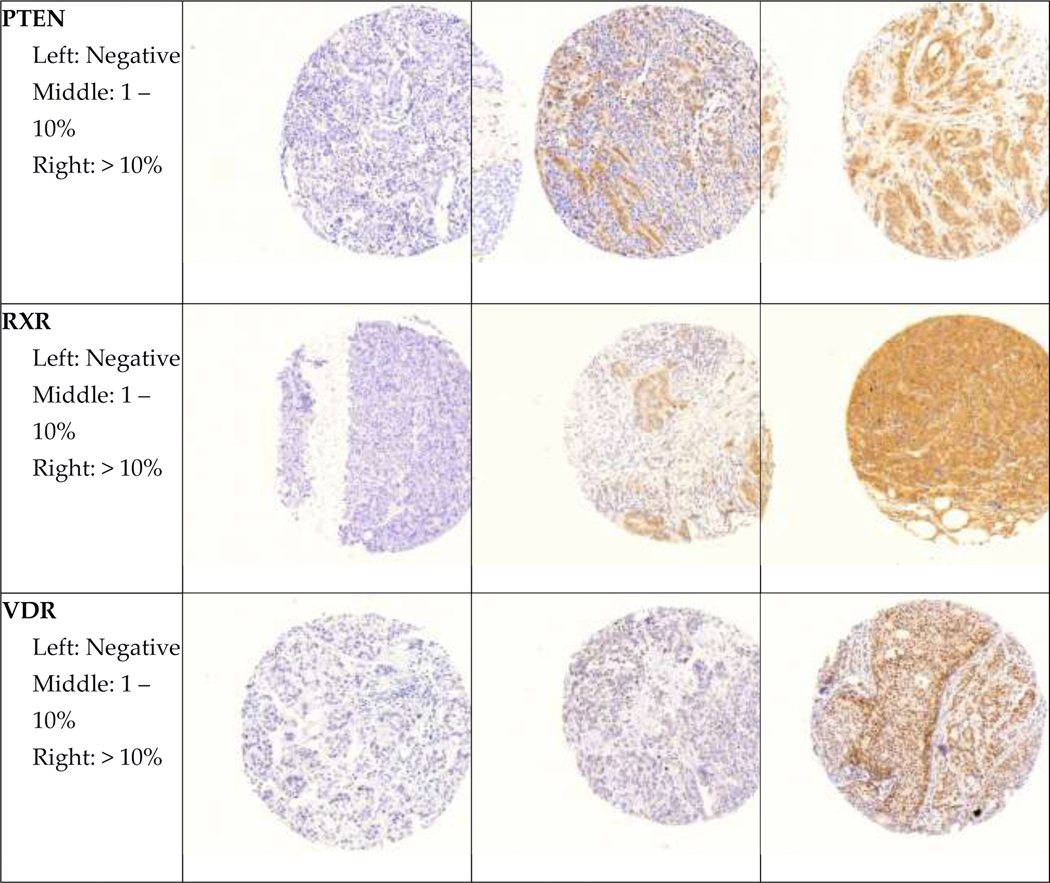
Representative images of all immunohistochemical markers by pathologist-assigned category are shown in Figure 1. Mean age at diagnosis and selected tumor characteristics for participants in both cohorts are shown in Table 2. In both cohorts, most participants were diagnosed with stage I and II, node negative tumors. Roughly half of participants were diagnosed with moderately differentiated tumors.
Figure 1. Representative images of immunohistochemical stains.
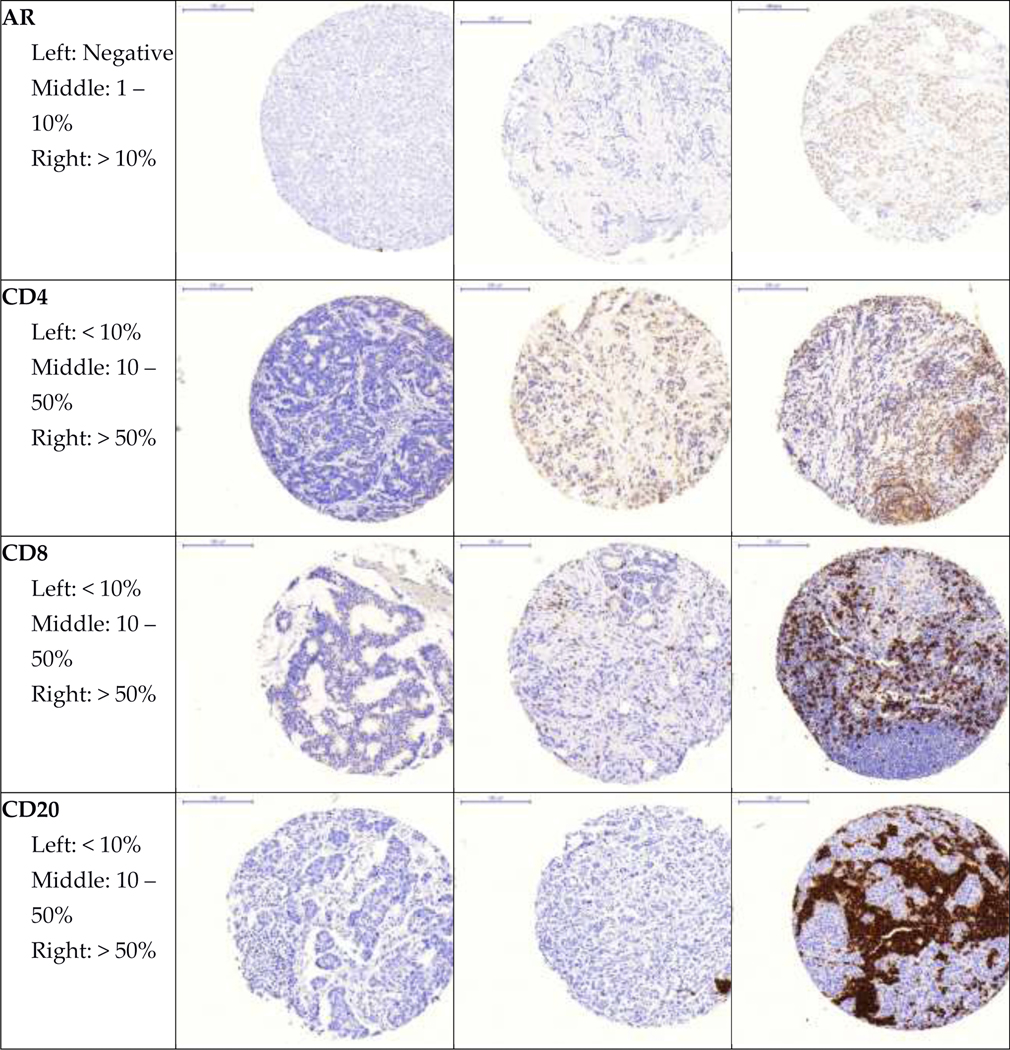
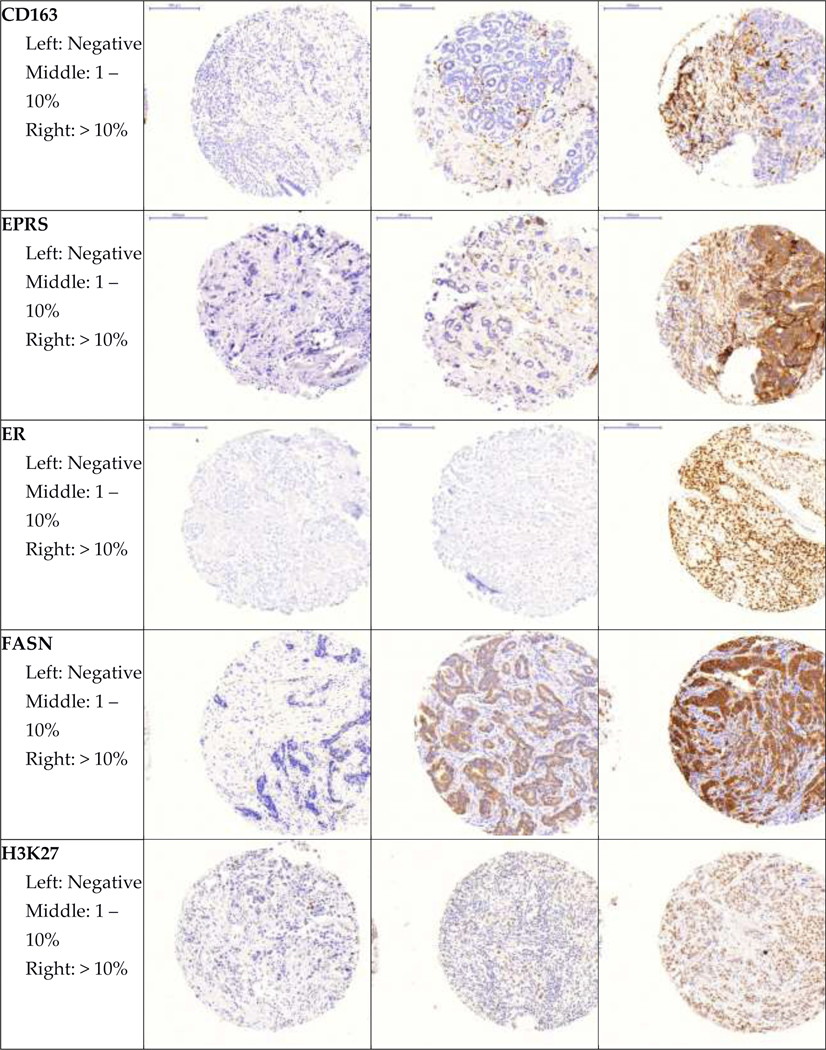
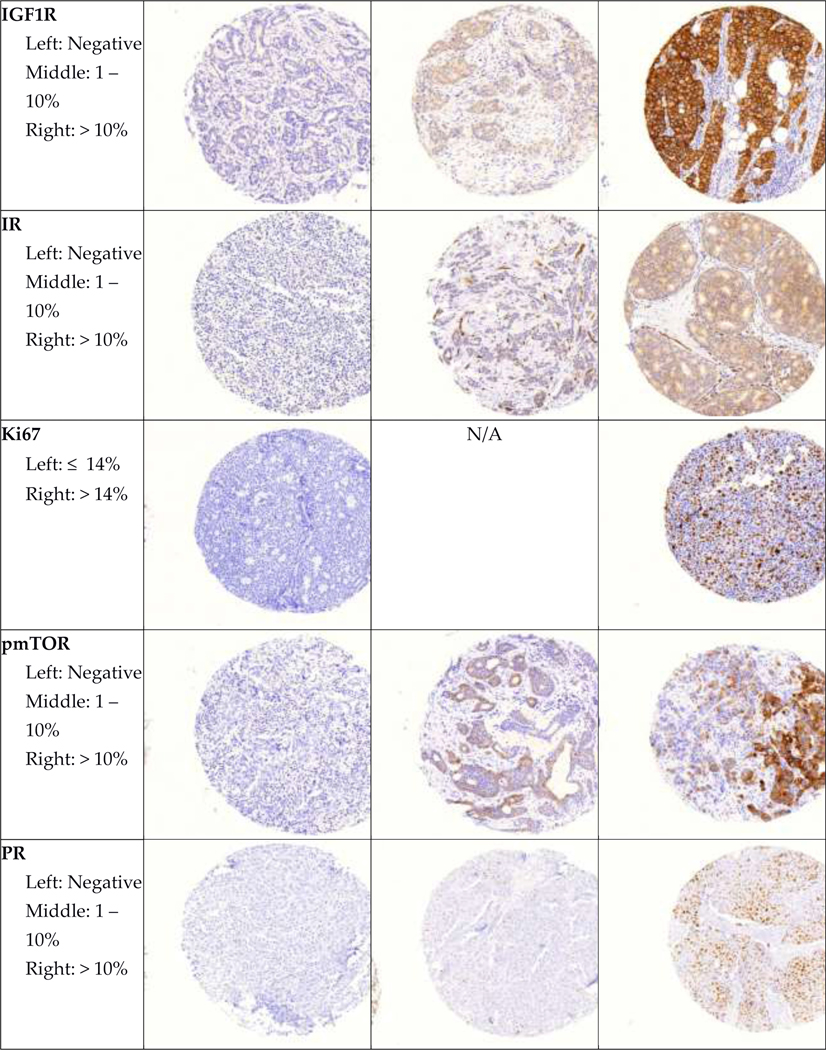
Representative images of each marker by pathologist-assigned category are shown.
Table 2.
Characteristics of participants in the Nurses’ Health Study cohorts with breast tumor tissue.
| Characteristic | NHS (N = 4,673) | NHS II (N = 1,241) | |
|---|---|---|---|
|
| |||
| Age at diagnosis, mean (SD) | 61.3 (9.0) | 46.5 (5.2) | |
| Gradea, n (%) | Well-differentiated | 938 (23.6) | 152 (15.4) |
| Moderately differentiated | 2144 (53.9) | 540 (54.7) | |
| Poorly/undifferentiated | 894 (22.5) | 296 (30.0) | |
| Missing | 697 | 253 | |
| Tumor size, n (%) | ≤1.0 cm | 1230 (29.9) | 237 (23.9) |
| 1.1 – 2.0 cm | 1611 (39.1) | 412 (41.6) | |
| 2.1 – 4.0 cm | 932 (22.6) | 272 (27.5) | |
| >4.0 cm | 346 (8.4) | 69 (7.0) | |
| Missing or carcinoma in situ | 554 | 251 | |
| Node status, n (%) | Node negative | 3444 (73.7) | 884 (71.8) |
| 1 – 3 nodes positive | 741 (15.9) | 225 (18.3) | |
| 4 – 9 nodes positive | 256 (5.5) | 103 (8.4) | |
| 10+ nodes positive | 157 (3.4) | 17 (1.4) | |
| Metastatic at diagnosis | 75 (1.6) | 3 (0.2) | |
| Missing | 0 | 9 | |
| Stage, n (%) | In situ | 523 (11.7) | 247 (20.3) |
| 1 | 2168 (48.3) | 483 (39.7) | |
| 2 | 1237 (27.6) | 345 (28.4) | |
| 3 | 483 (10.8) | 138 (11.4) | |
| 4 | 75 (1.7) | 3 (0.3) | |
| Missing | 187 | 25 | |
Grade was obtained from centralized review of tumor tissue or pathology reports if centralized review was unavailable.
Percentages exclude the number of missing for each variable. Percentages may not add up to 100 due to rounding.
Spearman correlations between pathologist and Definiens assessments are shown in Table 3, and ranged from weak (RXR, rho=−0.05; CD163 in stroma, rho=0.10) to strong (Ki67, rho=0.79; pmTOR, rho=0.77). Correlations were ≥0.55 for 10 of the 17 markers analyzed (PR, IR, Ki67, CD163 (tumor), CD20, CD8, EPRS (tumor), IGF1R, pmTOR, and PTEN). There were no appreciable differences in the strength of the correlations when we used the average case definition (average of pathologist’s scores across cores) versus the categorical case definition (cases classified as negative/low positive/positive or low/moderate/high). We observed modest correlations for both ER (rho=0.43, categorical; rho=0.46, average) and PR (rho=0.66, categorical; rho=0.70, average). In general, we found that the mean Definiens percent positive was highest among cases classified by the pathologist as positive, and lowest among those classified as negative (Table 3). One notable exception is RXR, for which the mean percent positive was 93%, independent of the pathologist’s classification. Agreement across cores was generally good: for pathologist scores, Fleiss’ kappa ranged from 0.64 (EPRS in stroma) to 0.83 (H3K27), while for Definiens percent positive, intraclass correlation coefficients ranged from 0.48 (CD20) to 0.87 (AR).
Table 3.
Comparison between Definiens percent positive and pathologist assessment of immunohistochemical markers.
| Marker | Pathologist score | N | Definiens % positive, mean (SD) | Rhoa, Categorical case definition | Rhob, Average case definition | Kappac, Pathologist (SE) | ICCd, Definiens (95% CI) |
|---|---|---|---|---|---|---|---|
|
| |||||||
| ER | Positive | 3866 | 49.8 (19.3) | 0.43 | 0.46 | 0.73 (0.005) | 0.82 (0.81–0.82) |
| Low Positive | 285 | 40.0 (23.9) | |||||
| Negative | 874 | 21.0 (20.7) | |||||
|
| |||||||
| PR | Positive | 2588 | 59.2 (29.3) | 0.66 | 0.70 | 0.70 (0.005) | 0.63 (0.62–0.65) |
| Low Positive | 313 | 15.4 (22.8) | |||||
| Negative | 1086 | 11.0 (19.0) | |||||
|
| |||||||
| AR | Positive | 3077 | 46.0 (22.6) | 0.37 | 0.42 | 0.66 (0.005) | 0.87 (0.86–0.88) |
| Low Positive | 172 | 26.4 (23.3) | |||||
| Negative | 890 | 24.9 (21.6) | |||||
|
| |||||||
| IR | Positive | 79 | 67.5 (17.4) | 0.56 | 0.58 | 0.79c | 0.71 (0.70–0.73) |
| Low Positive | 3 | 34.6 (34.8) | |||||
| Negative | 41 | 41.3 (19.6) | |||||
|
| |||||||
| H3K27 | Positive | 116 | 45.7 (11.8) | 0.26 | 0.31 | 0.83c | 0.83 (0.81–0.84) |
| Low Positive | 3 | 32.4 (18.2) | |||||
| Negative | 13 | 33.1 (17.0) | |||||
|
| |||||||
| VDR | Positive | 104 | 56.1 (15.1) | 0.52 | 0.52 | 0.77c | 0.78 (0.76–0.79) |
| Low Positive | 4 | 23.7 (21.6) | |||||
| Negative | 11 | 13.1 (12.5) | |||||
|
| |||||||
| RXR | Positive | 72 | 93.2 (6.3) | −0.05 | −0.01 | 0.80c | 0.62 (0.60–0.65) |
| Low Positive | 3 | 93.1 (5.3) | |||||
| Negative | 15 | 93.5 (5.6) | |||||
|
| |||||||
| Ki67 | High proliferation | 16 | 17.5 (23.2) | 0.79 | 0.79 | 0.83 (0.72–0.90)c | 0.80 (0.78–0.81) |
| Low proliferation | 13 | 2.5 (2.6) | |||||
|
| |||||||
| CD163, tumor | Positive | 17 | 82.9 (14.6) | 0.57 | 0.56 | 0.66c | 0.68 (0.65–0.70) |
| Low Positive | 72 | 48.4 (24.9) | |||||
| Negative | 27 | 27.6 (21.2) | |||||
|
| |||||||
| CD163, stroma | Positive | 65 | 12.7 (4.6) | 0.10 | 0.04 | 0.76c | 0.52 (0.48–0.56) |
| Low Positive | 57 | 11.9 (5.6) | |||||
| Negative | 0 | -- | |||||
|
| |||||||
| CD20 | High | 2 | 61.6 (10.3) | 0.64 | 0.64 | 0.80c | 0.48 (0.44–0.52) |
| Moderate | 20 | 13.8 (10.2) | |||||
| Low | 90 | 1.1 (1.8) | |||||
|
| |||||||
| CD8 | High | 7 | 39.3 (12.9) | 0.72 | 0.75 | 0.71c | 0.69 (0.67–0.72) |
| Moderate | 43 | 15.6 (11.7) | |||||
| Low | 68 | 4.3 (4.1) | |||||
|
| |||||||
| CD4 | High | 11 | 15.7 (20.7) | 0.52 | 0.54 | 0.73c | 0.68 (0.65–0.70) |
| Moderate | 31 | 6.3 (4.1) | |||||
| Low | 78 | 3.3 (3.8) | |||||
|
| |||||||
| EPRS, tumor | Positive | 5 | 80.2 (11.4) | 0.63 | 0.68 | 0.79c | 0.72 (0.70–0.75) |
| Low Positive | 64 | 58.2 (21.3) | |||||
| Negative | 47 | 29.9 (15.4) | |||||
|
| |||||||
| EPRS, stroma | Positive | 24 | 9.8 (6.6) | 0.45 | 0.49 | 0.64c | 0.60 (0.57–0.64) |
| Low Positive | 78 | 4.1 (3.6) | |||||
| Negative | 11 | 2.0 (1.8) | |||||
|
| |||||||
| FASN | Positive | 51 | 87.8 (8.0) | 0.49 | 0.51 | 0.73c | 0.72 (0.70–0.75) |
| Low Positive | 64 | 73.1 (20.3) | |||||
| Negative | 4 | 32.1 (33.6) | |||||
|
| |||||||
| IGF1R | Positive | 5 | 84.8 (8.6) | 0.72 | 0.78 | 0.78c | 0.77 (0.75–0.79) |
| Low Positive | 79 | 55.5 (22.4) | |||||
| Negative | 37 | 15.2 (15.9) | |||||
|
| |||||||
| pmTOR | Positive | 28 | 65.8 (19.4) | 0.77 | 0.85 | 0.66c | 0.76 (0.74–0.78) |
| Low Positive | 63 | 31.9 (22.7) | |||||
| Negative | 27 | 3.3 (4.4) | |||||
|
| |||||||
| PTEN | Positive | 9 | 81.3 (13.6) | 0.71 | 0.78 | 0.79c | 0.77 (0.75–0.78) |
| Low Positive | 73 | 34.8 (25.4) | |||||
| Negative | 35 | 6.4 (7.8) | |||||
For cases with both pathologist and Definiens data, Spearman correlations were calculated between ordinal pathologist-assigned scores and continuous percent positive obtained from Definiens (with the exception of Ki67, where cores were manually scored continuously). For pathologist assessments, low positive was defined as 1–10% positive cells, while positive was defined as >10% positive cells. For pathologist assessment of CD20, CD8, and CD4, low was defined as <10%, moderate as 10–50%, and high as >50%. For Ki67, low expression/proliferation was defined as ≤14% positive cells.
For cases with both pathologist and Definiens data, Spearman correlations were calculated between average pathologist-assigned scores and continuous percent positive obtained from Definiens. Average pathologist-assigned scores were obtained by averaging the ordinal scores for each participant’s cores.
Fleiss’ kappa was used to assess agreement in pathologist-assigned scores across each participant’s tumor cores (maximum of 3) using all available pathologist data. Due to small sample sizes, the standard error could not be calculated for all markers other than ER, PR, and AR. For Ki67, intraclass correlation coefficients and 95% confidence intervals were computed.
Intraclass correlation coefficients and 95% confidence intervals were computed across each participant’s tumor cores (maximum of 3) using all available Definiens data. CI=confidence interval; ICC=intraclass correlation coefficient; SD=standard deviation; SE=standard error
Raw percent agreement between pathologist’s scores and dichotomized Definiens percent positive ranged from moderate (Ki67, 58.6%) to perfect (CD163, 100%). Kappa statistics were more modest, ranging from no agreement (IGF1R, kappa=0.07, 95% CI −0.02, 0.17) to moderate (CD20, kappa=0.71, 95% CI 0.54, 0.89) (Table 4). Kappa values ≥0.35 were observed for 6 of the 17 markers (ER, PR, CD20, CD8, FASN, pmTOR); kappa could not be computed for 6 markers (IR, H3K27, RXR, CD163 (tumor and stroma), EPRS (tumor)) because all tumors were classified as positive using the Definiens percent positive data at the cutpoints employed by the pathologists to assign positivity. We observed similar results when we categorized the continuous Definiens percent positive for each core, finding that that agreement between pathologist and Definiens assessments ranged from no agreement to moderate (Supplementary Table 1). At the core level, correlations were similar in magnitude across cores for all markers and the strength of the correlations was similar to that observed at the case-level.
Table 4.
Agreement between dichotomized Definiens percent positive and pathologist-assigned scores.
| Marker | Pathologist score | Definiens positive, N (%) | Definiens negative, N (%) | Percent agreement (%) | Cohen’s kappa (95% CI) |
|---|---|---|---|---|---|
|
| |||||
| ER | Low positive/Positive | 4106 (86.7) | 45 (15.7) | 86.5 | 0.36 (0.33–0.40) |
| Negative | 632 (13.3) | 242 (84.3) | |||
|
| |||||
| PR | Low positive/Positive | 2838 (79.5) | 63 (15.1) | 80.1 | 0.38 (0.35–0.41) |
| Negative | 732 (20.5) | 354 (84.9) | |||
|
| |||||
| AR | Low positive/Positive | 3168 (80.1) | 81 (43.8) | 79.1 | 0.13 (0.10–0.16) |
| Negative | 786 (19.9) | 104 (56.2) | |||
|
| |||||
| IR | Low positive/Positive | 82 (66.7) | 0 | 66.7 | N/A |
| Negative | 41 (33.3) | 0 | |||
|
| |||||
| H3K27 | Low positive/Positive | 119 (90.2) | 0 | 90.2 | N/A |
| Negative | 13 (9.9) | 0 | |||
|
| |||||
| VDR | Low positive/Positive | 108 (91.5) | 0 | 91.6 | 0.15 (−0.11–0.42) |
| Negative | 10 (8.5) | 1 (100) | |||
|
| |||||
| RXR | Low positive/Positive | 75 (83.3) | 0 | 83.3 | N/A |
| Negative | 15 (16.7) | 0 | |||
|
| |||||
| Ki67 | High proliferation | 4 (100) | 12 (48.0) | 58.6 | 0.23 (0.02–0.44) |
| Low proliferation | 0 | 13 (52.0) | |||
|
| |||||
| CD163, tumor | Low positive/Positive | 89 (76.7) | 0 | 76.7 | N/A |
| Negative | 27 (23.3) | 0 | |||
|
| |||||
| CD163, stroma | Low positive/Positive | 122 (100) | 0 | 100 | N/A |
| Negative | 0 | 0 | |||
|
| |||||
| CD20 | Moderate/High | 14 (93.3) | 8 (8.3) | 92.0 | 0.71 (0.54–0.89) |
| Low | 1 (6.7) | 89 (91.8) | |||
|
| |||||
| CD8 | Moderate/High | 34 (85.0) | 16 (20.5) | 81.4 | 0.61 (0.46–0.75) |
| Low | 6 (15.0) | 62 (79.5) | |||
|
| |||||
| CD4 | Moderate/High | 11 (61.1) | 31 (30.4) | 68.3 | 0.20 (0.03–0.36) |
| Low | 7 (38.9) | 71 (69.6) | |||
|
| |||||
| EPRS, tumor | Low positive/Positive | 69 (59.5) | 0 | 59.5 | N/A |
| Negative | 47 (40.5) | 0 | |||
|
| |||||
| EPRS, stroma | Low positive/Positive | 90 (92.8) | 12 (75.0) | 83.2 | 0.20 (−0.04–0.45) |
| Negative | 7 (7.2) | 4 (25.0) | |||
|
| |||||
| FASN | Low positive/Positive | 115 (97.5) | 0 | 97.5 | 0.39 (−0.15–0.93) |
| Negative | 3 (2.5) | 1 (100) | |||
|
| |||||
| IGF1R | Low positive/Positive | 84 (70.6) | 0 | 71.1 | 0.07 (−0.02–0.17) |
| Negative | 35 (29.4) | 2 (100) | |||
|
| |||||
| pmTOR | Low positive/Positive | 91 (87.5) | 0 | 89.0 | 0.62 (0.45–0.80) |
Percent agreement and kappa statistics were calculated by dichotomizing the Definiens percent positive, using the following cutpoints for positivity: ≥10% (CD20, CD8, and CD4); >14% (Ki67); or ≥1% (all other markers).
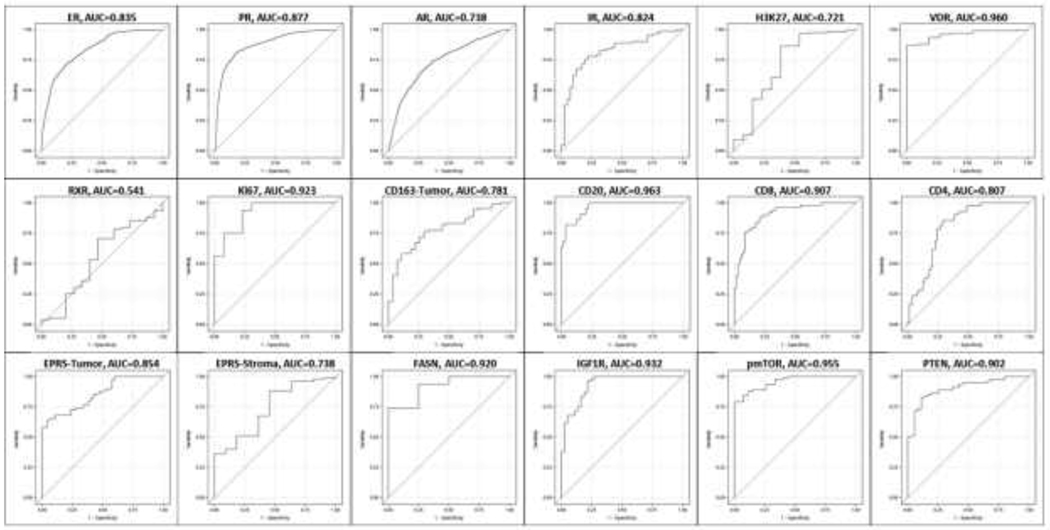
The overlap between pathologist classifications and Definiens percent positive varied by marker (Figure 2). For some markers (ER, AR, IR, H3K27, RXR, CD163 in stroma), there was substantial overlap of Definiens percent positive across pathologist-assigned categories. For others (Ki67, CD8, CD20, FASN, IGF1R, pmTOR, PTEN), there was very little to no overlap. To evaluate the predictive ability of the Definiens percent positive, using the binary pathologist assessment as the gold standard, we constructed ROC curves for each marker (Figure 3). The best performing markers were VDR, Ki67, CD20, CD8, FASN, IGF1R, pmTOR, and PTEN (area under the curve (AUC) >0.9), while the worst performing was RXR (AUC=0.541). For the remaining markers, the AUC was >0.7.
Figure 2. Definiens percent positive by pathologist-assigned category.
The distribution of Definiens percent positive by pathologist-assigned category. The overlap between between pathologist-assigned category and Definiens percent positive varied by marker, with some exhibiting substantial overlap (ER, AR, IR, H3K27, RXR, CD163 in stroma) and others exhibiting little or no overlap (Ki67, CD8, CD20, FASN, IGF1R, pmTOR, PTEN). ER, PR, AR, IR, H3K27, VDR, RXR, CD163-tumor, EPRS, FASN, IGF1R, pmTOR, PTEN: Left panel, negative; middle panel, low positive (1–10%); right panel, positive (>10%). CD163-stroma: Left panel, low positive (1–10%); right panel, positive (>10%). No cases were manually scored as negative. Ki67: Left panel, low proliferation (≤14%); right panel, high proliferation (>14%). CD20, CD8, CD4: Left panel, low (<10%); middle panel, moderate (10–50%); right panel, high (>50%).
Figure 3. Receiver-operator characteristic curves.
The binary pathologist assessment (any positive versus negative, high/moderate versus low, or >14% (the cutpoint for Ki67) was used as the gold standard. No ROC curve was generated for CD163 in stroma because no cases were assigned negative by the pathologist. The best performing markers were VDR, Ki67, CD20, CD8, FASN, IGF1R, pmTOR, and PTEN (area under the curve (AUC) > 0.9), while the worst performing was RXR (AUC = 0.541). For the remaining markers, the AUC was above 0.7.
The correlation matrix for all markers, assessed by the pathologist (top) or Definiens (bottom) is shown in Supplementary Table 2; in general, the markers we included were weakly correlated, with stronger correlations observed between ER and PR (rho=0.66), EPRS in tumor and stroma (rho=0.64), and CD20, CD8, and CD4 (rho=0.46–0.59).
4. Discussion
We observed moderate to strong correlations between Definiens and pathologist assessments of immunoreactivity for most of the markers we analyzed. Kappa statistics between pathologist’s scores and categorized Definiens percent positive ranged from no agreement to moderate agreement, but for some markers, all tumors were categorized as positive using the Definiens data, indicating that cutpoints for computationally derived data may require marker-specific optimization.
For the clinically used breast tumor biomarkers, ER, PR, HER2, and Ki67, several studies have examined correlations between automated systems and pathologist interpretations of immunoreactivity. Camp et al. identified strong correlations between pathologist evaluations of ER status and AQUA algorithms in 340 lymph node positive breast tumors (r=0.884), and showed that the automated analysis had slightly better reproducibility (automated, r=0.824; pathologist, r=0.732) [4]. Other groups have found similarly strong correlations for ER and PR using automated algorithms designed with MatLab (ER, rho=0.74; PR, rho=0.62) [5], Ariol (Leica Biosystems) (ER, rho=0.89; PR, rho=0.88) [10], and QuPath (Queen’s University Belfast) (ER, rho=0.892; PR, rho=0.887) [11]. Strong agreement between pathologist and automated evaluations of ER and PR obtained with QCA, Ariol, TMALab II (Aperio), and TMAx (Beecher Instruments) software (ER, kappa 0.75–0.91; PR, kappa 0.65–0.91) [6,7,10,12] has also been demonstrated.
Kappa statistics to compare agreement between the HER2 HercepTest score and analysis using the Ariol, TMALabII, and TMAx systems ranged from 0.53 (TMALabII) to 0.72 (Ariol/TMAx) [10,12]. For Ki67, moderate to strong associations have been observed across multiple computational platforms (ImageJ, kappa=0.57; Ariol, kappa=0.64; Slidepath Tissue IA, kappa=0.70; Ventana Virtuoso, intraclass correlation coefficient=0.974; QuPath, rho=0.729) [11,13–16].
In our data, we observed modest correlations for ER (rho=0.43) and stronger correlations for PR (rho=0.66) and Ki67 (rho=0.79), which is broadly in line with the studies previously discussed. When we dichotomized the continuous Definiens data using the same cutpoints as the pathologist’s interpretations, we found generally weaker agreement with the pathologist’s score (ER, kappa=0.36; PR, kappa=0.38; Ki67, kappa=0.23), which differs from the previously discussed studies that observed generally stronger agreement. One potential reason may be due to differences in the level of automation of the software used. Definiens, for example, requires user input to train the software to automatically segregate tumor and stromal regions. Following application of the training algorithm, no further input from the user is required to define regions of interest (e.g., tumor cells) for analysis. Other systems require manual annotation of regions of interest for all tumors, which could avoid potentially problematic situations such as tissue folding or nonspecific staining.
Few studies have compared assessment methods for markers beyond ER, PR, HER2, and Ki67 in breast cancer. Bolton et al. compared pathologist scoring of ERβ (intensity and percent staining) and aromatase (intensity only) to the percent staining and intensity values obtained with Ariol, TMALab II, and TMAx in 440 invasive breast tumors. Agreement with pathologist scores was excellent across automated systems for ERβ (kappa 0.80–0.86), but less strong for aromatase (TMALabII/TMAx, kappa 0.65–0.67; Ariol, kappa=0.41) [12]. Howat et al. compared pathologist scoring of epidermal growth factor receptor (EGFR) and cytokeratins 5/6 (CK5/6) to that obtained using the Ariol system in 8 267 breast cancers collected through the Breast Cancer Association Consortium. Both markers were manually scored continuously based on the percent of positive cells as weak, intermediate, and strong staining intensities. Moderate agreement was observed between pathologist and automated assessments for both markers (EGFR, kappa=0.44; CK5/6, kappa=0.49) [10].
Comparisons specifically using Definiens software have been performed in prostate and esophageal cancers, but to our knowledge, not in breast cancer. Braun et al. compared pathologist scoring of ERG, SLC45A3, and TMPRSS2 in 630 prostate cancer cases to automated assessments obtained from Definiens TissueStudio, finding strong correlations between the two methods (ERG, rho=0.94; SLC45A3, rho=0.92; TMPRSS2, rho=0.90) [17]. In 153 esophageal adenocarcinomas, Feuchtinger et al. compared pathologist scoring of EGFR, pEGFR, β-catenin, E-cadherin, and HER2 to scores obtained from Definiens Developer XD2 software, and observed strong correlations for all markers (EGFR, r=0.79; pEGFR, r=0.89; β-catenin, r=0.71; E-cadherin, r=0.68; HER2, r=0.74) [18].
Apart from ER, PR, and Ki67, the markers we investigated have not been previously examined in this capacity. However, our results are broadly in line with previous studies[10,12,17,18], in that we observed moderate to strong correlations for many of the markers we analyzed, including those with nuclear and cytoplasmic staining patterns in tumor, stromal, and inflammatory cells.
We also calculated the AUC for each marker, using the binary positive/negative pathologist score as the gold standard. In general, the Definiens-derived percent positive was able to discriminate cases as well as the pathologist score for most markers, with AUC >0.8. Previous studies have indicated near-perfect discrimination for ER, PR, and HER2 across the Aperio, Ariol, TMAx, and QuPath systems (ER, AUC 0.96–1.00; PR, AUC 0.93–0.99; HER2, AUC 0.93–0.97) and very strong discrimination for Ki67 using the QuPath system (AUC 0.88) [10–12]. Performance using the nuclear algorithm developed in MatLab was lower, however (ER, AUC=0.85; PR, AUC=0.74) [5].
In our group, Definiens algorithm development is iterative, and algorithms may be refined several times before final release of data for analysis. Recently, we have begun incorporating pathologist data into algorithm development to improve the quality of the data. However, the biomarker data used in this study have been collected over many years and for the markers we evaluated, pathologist data were not used in algorithm development. Our results therefore speak to the need for fine-tuning of algorithms to better match pathologist evaluations. More specific machine learning approaches, in conjunction with pathologist input, are likely required to reduce the chance of misclassification in epidemiologic studies.
Strengths of our study include the use of tissues from a population-based, prospective cohort study and the evaluation of multiple antibodies exhibiting different staining patterns. Use of data collected within a population-based cohort allowed us to leverage real-world data collected across different pathology departments, with varying tissue processing protocols, making our results more generalizable to a wider group of researchers. In our study, we compared a semi-automated computational platform to pathologist assessments. Up to 12 cores per stain were used for algorithm training. Given the variability inherent in tumor tissue, this represents a potential limitation of the software as algorithms may have misclassified regions due to insufficient training data. Because the Definiens platform does not require manual annotation of regions by a trained reviewer following the initial development of each stain’s algorithm, it is also possible that benign tissue, non-tumor cells, or inappropriate regions (e.g., folded tissue, spurious staining) could have been included in the quantitative percent positive. This potential misclassification is a limitation and could have negatively affected the correlation and agreement with the pathologist’s assessment. Pathologist scores were used as the gold standard in this analysis. Because the two study pathologists evaluated different markers, we were unable to compare scores between pathologists, and thus could not assess measurement error in the pathologist reports. Another potential limitation is the small sample size for markers other than ER, PR, and AR, which may affect the reliability of the results.
5. Conclusion
In our study, we compared pathologist to computational assessments across a wide variety of immunohistochemical markers, including those with nuclear and cytoplasmic staining and those staining tumor and stromal cell types. Our data indicate that, for some markers, the Definiens semi-automated digital image analysis system can provide results comparable to those obtained by an expert pathologist. The strength of the correlation varied widely across markers, however. Pilot studies to examine the agreement between pathologist and computational assessments are therefore critical, as agreement may be dependent on the marker type, cellular compartment, or other parameters. Such pilot studies may allow for fine-tuning of algorithms, which may be necessary to improve agreement with pathologist’s evaluations prior to wider study implementation. Computational image analysis is a promising approach for the evaluation of immunohistochemical data in large-scale epidemiologic research. Computational evaluations for IHC can provide greater efficiency for epidemiologic studies, allowing increases in the scope of assessments that may not be feasible using manual review.
Supplementary Material
Highlights.
Computational methods to assess immunohistochemistry are often used in epidemiology
We compared Definiens output to pathologist scores for 17 markers in breast cancer
Correlations ranged from weak (RXR, rho = −0.05) to strong (Ki67, rho = 0.79)
Area under the curve > 0.70 was observed for all markers except RXR
Pilot studies are key; computational methods can aid high-throughput analyses
Acknowledgements
The authors gratefully acknowledge Andrew H. Beck and Benjamin Glass of the Beth Israel Deaconess Medical Center Department of Pathology for their technical guidance with the Definiens platform. We thank Dana-Farber/Harvard Cancer Center in Boston, MA for the use of the Specialized Histopathology Core, which provided histology and immunohistochemistry service. Dana-Farber/Harvard Cancer Center is supported in part by an NCI Cancer Center Support Grant # NIH 5 P30 CA06516. We would also like to thank the participants and staff of the Nurses’ Health Study and Nurses’ Health Study-II for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Financial support
This work was supported by the National Institutes of Health/National Cancer Institute (UM1 CA186107, U01 CA176726, and P01 CA87969); M.R.R. was supported by an NCI training grant (T32CA009001). The funding agencies had no role in the study design or interpretation of the data.
Abbreviations:
- AR
androgen receptor
- CD
cluster of differentiation
- EPRS
glutamyl-prolyl-tRNA synthetase
- ER
estrogen receptor
- FASN
fatty acid synthase
- FFPE
formalin-fixed, paraffin-embedded
- H3K27
histone 3 lysine 27 trimethylation
- IGF1R
insulin-like growth factor 1 receptor
- IHC
immunohistochemistry
- IR
insulin receptor
- Ki67
marker of proliferation Ki-67
- mTOR
mammalian target of rapamycin
- NHS
Nurses’ Health Study
- PR
progesterone receptor
- PTEN
phosphatase and tensin homolog
- ROC
receiver operator characteristic
- RXR
retinoic acid receptor
- TMA
tissue microarray
- VDR
vitamin D receptor
Footnotes
Declarations of interest: none
Data Sharing Statement
Data sharing continues to be a critical component of the Nurses’ Health Study (NHS) and Nurses’ Health Study II (NHS-II). The NHS/NHS-II uses a data enclave approach to data sharing, developed with input from and approval of the External Advisory Committee and the NCI. Our study website (https://www.nurseshealthstudy.org/researchers) is accessible to the public and includes a description of each cohort, links to all questionnaires, descriptions of available biospecimens, and guidelines for external users. ‘
Conflict of interest
The authors declare they have no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Walker RA, Quantification of immunohistochemistry--issues concerning methods, utility and semiquantitative assessment I., Histopathology. 49 (2006) 406–10. 10.1111/j.1365-2559.2006.02514.x. [DOI] [PubMed] [Google Scholar]
- [2].Aeffner F, Wilson K, Martin NT, Black JC, Hendriks CLL, Bolon B, Rudmann DG, Gianani R, Koegler SR, Krueger J, Young GD, The Gold Standard Paradox in Digital Image Analysis: Manual Versus Automated Scoring as Ground Truth, Arch. Pathol. Lab. Med 141 (2017) 1267–1275. 10.5858/arpa.2016-0386-RA. [DOI] [PubMed] [Google Scholar]
- [3].Taylor CR, Levenson RM, Quantification of immunohistochemistry--issues concerning methods, utility and semiquantitative assessment II., Histopathology. 49 (2006) 411–24. 10.1111/j.1365-2559.2006.02513.x. [DOI] [PubMed] [Google Scholar]
- [4].Camp RL, Chung GG, Rimm DL, Automated subcellular localization and quantification of protein expression in tissue microarrays, Nat. Med 8 (2002) 1323–1328. 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- [5].Rexhepaj E, Brennan DJ, Holloway P, Kay EW, McCann AH, Landberg G, Duffy MJ, Jirstrom K, Gallagher WM, Novel image analysis approach for quantifying expression of nuclear proteins assessed by immunohistochemistry: application to measurement of oestrogen and progesterone receptor levels in breast cancer., Breast Cancer Res. 10 (2008) R89. 10.1186/bcr2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Turbin DA, Leung S, Cheang MCU, Kennecke HA, Montgomery KD, McKinney S, Treaba DO, Boyd N, Goldstein LC, Badve S, Gown AM, van de Rijn M, Nielsen TO, Gilks CB, Huntsman DG, Automated quantitative analysis of estrogen receptor expression in breast carcinoma does not differ from expert pathologist scoring: a tissue microarray study of 3,484 cases, Breast Cancer Res. Treat 110 (2008) 417–426. 10.1007/s10549-007-9736-z. [DOI] [PubMed] [Google Scholar]
- [7].Diaz LK, Sahin A, Sneige N, Interobserver agreement for estrogen receptor immunohistochemical analysis in breast cancer: a comparison of manual and computer-assisted scoring methods., Ann. Diagn. Pathol 8 (2004) 23–7. http://www.ncbi.nlm.nih.gov/pubmed/15129906 (accessed January 25, 2019). [DOI] [PubMed] [Google Scholar]
- [8].Lehr H-A, Jacobs TW, Yaziji H, Schnitt SJ, Gown AM, Quantitative Evaluation of HER-2/ neu Status in Breast Cancer by Fluorescence In Situ Hybridization and by Immunohistochemistry With Image Analysis, Am. J. Clin. Pathol 115 (2001) 814–822. 10.1309/AJ84-50AK-1X1B-1Q4C. [DOI] [PubMed] [Google Scholar]
- [9].Colditz GA, Hankinson SE, The Nurses’ Health Study: lifestyle and health among women, Nat. Rev. Cancer 5 (2005) 388–396. 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- [10].Howat WJ, Blows FM, Provenzano E, Brook MN, Morris L, Gazinska P, Johnson N, McDuffus L-A, Miller J, Sawyer EJ, Pinder S, van Deurzen CHM, Jones L, Sironen R, Visscher D, Caldas C, Daley F, Coulson P, Broeks A, Sanders J, Wesseling J, Nevanlinna H, Fagerholm R, Blomqvist C, Heikkilä P, Ali HR, Dawson S-J, Figueroa J, Lissowska J, Brinton L, Mannermaa A, Kataja V, Kosma V-M, Cox A, Brock IW, Cross SS, Reed MW, Couch FJ, Olson JE, Devillee P, Mesker WE, Seyaneve CM, Hollestelle A, Benitez J, Perez JIA, Menéndez P, Bolla MK, Easton DF, Schmidt MK, Pharoah PD, Sherman ME, García-Closas M, Performance of automated scoring of ER, PR, HER2, CK5/6 and EGFR in breast cancer tissue microarrays in the Breast Cancer Association Consortium, J. Pathol. Clin. Res 1 (2015) 18–32. 10.1002/cjp2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bankhead P, Fernández JA, McArt DG, Boyle DP, Li G, Loughrey MB, Irwin GW, Harkin DP, James JA, McQuaid S, Salto-Tellez M, Hamilton PW, Integrated tumor identification and automated scoring minimizes pathologist involvement and provides new insights to key biomarkers in breast cancer, Lab. Investig 98 (2018) 15–26. 10.1038/labinvest.2017.131. [DOI] [PubMed] [Google Scholar]
- [12].Bolton KL, Garcia-Closas M, Pfeiffer RM, Duggan MA, Howat WJ, Hewitt SM, Yang XR, Cornelison R, Anzick SL, Meltzer P, Davis S, Lenz P, Figueroa JD, Pharoah PDP, Sherman ME, Assessment of Automated Image Analysis of Breast Cancer Tissue Microarrays for Epidemiologic Studies, Cancer Epidemiol. Biomarkers Prev 19 (2010) 992–999. 10.1158/1055-9965.EPI-09-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Abubakar M, Howat WJ, Daley F, Zabaglo L, McDuffus L-A, Blows F, Coulson P, Raza Ali H, Benitez J, Milne R, Brenner H, Stegmaier C, Mannermaa A, Chang-Claude J, Rudolph A, Sinn P, Couch FJ, Tollenaar RAEM, Devilee P, Figueroa J, Sherman ME, Lissowska J, Hewitt S, Eccles D, Hooning MJ, Hollestelle A, WM Martens J, HM van Deurzen C, kConFab Investigators MK Bolla Q. Wang M. Jones M. Schoemaker A. Broeks FE van Leeuwen L. Van’t Veer AJ Swerdlow N. Orr M. Dowsett D. Easton MK Schmidt PD Pharoah M. Garcia-Closas, High-throughput automated scoring of Ki67 in breast cancer tissue microarrays from the Breast Cancer Association Consortium, J. Pathol. Clin. Res 2 (2016) 138–153. 10.1002/cjp2.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Konsti J, Lundin M, Joensuu H, Lehtimäki T, Sihto H, Holli K, Turpeenniemi-Hujanen T, Kataja V, Sailas L, Isola J, Lundin J, Development and evaluation of a virtual microscopy application for automated assessment of Ki-67 expression in breast cancer., BMC Clin. Pathol 11 (2011) 3. 10.1186/1472-6890-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mohammed ZMA, McMillan DC, Elsberger B, Going JJ, Orange C, Mallon E, Doughty JC, Edwards J, Comparison of visual and automated assessment of Ki-67 proliferative activity and their impact on outcome in primary operable invasive ductal breast cancer., Br. J. Cancer 106 (2012) 383–8. 10.1038/bjc.2011.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhong F, Bi R, Yu B, Yang F, Yang W, Shui R, A Comparison of Visual Assessment and Automated Digital Image Analysis of Ki67 Labeling Index in Breast Cancer., PLoS One. 11 (2016) e0150505. 10.1371/journal.pone.0150505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Braun M, Kirsten R, Rupp NJ, Moch H, Fend F, Wernert N, Kristiansen G, Perner S, Quantification of protein expression in cells and cellular subcompartments on immunohistochemical sections using a computer supported image analysis system., Histol. Histopathol 28 (2013) 605–10. http://www.ncbi.nlm.nih.gov/pubmed/23361561 (accessed May 19, 2016). [DOI] [PubMed] [Google Scholar]
- [18].Feuchtinger A, Stiehler T, Jütting U, Marjanovic G, Luber B, Langer R, Walch A, Image analysis of immunohistochemistry is superior to visual scoring as shown for patient outcome of esophageal adenocarcinoma., Histochem. Cell Biol 143 (2015) 1–9. 10.1007/s00418-014-1258-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.