Summary
Genome editing technologies operate by inducing site-specific DNA perturbations that are resolved by cellular DNA repair pathways. Products of genome editors include DNA breaks generated by CRISPR-associated nucleases, base modifications induced by base editors, DNA flaps created by prime editors and integration intermediates formed by site-specific recombinases and transposases associated to CRISPR systems. Here, we discuss the cellular processes that repair CRISPR-generated DNA lesions and describe strategies to obtain desirable genomic changes through modulation of DNA repair pathways. Advances in our understanding of the DNA repair circuitry, in conjunction with the rapid development of innovative genome editing technologies, promise to greatly enhance our ability to improve food production, combat environmental pollution, develop cell-based therapies and cure genetic and infectious diseases.
Introduction
Genome stability is constantly threatened by DNA lesions induced by endogenous and environmental DNA damaging agents (Ciccia and Elledge, 2010). Living organisms have developed a network of cellular pathways, collectively referred to as the DNA damage response (DDR), that maintains genome stability in response to DNA damage. Highly conserved cellular DNA repair processes include double-strand break (DSB) repair, single-strand break (SSB) repair, mismatch repair (MMR), and base and nucleotide excision repair (BER and NER, respectively). Each of these pathways is specialized in the detection and resolution of distinct types of DNA lesions. DNA repair pathways are extensively interconnected, as highlighted by recent genetic investigation of the DDR network in human cells (Olivieri et al., 2020).
Genome editing tools, such as CRISPR-based technologies (Doudna, 2020; Knott and Doudna, 2018; Liu et al., 2022), generate targeted DNA lesions that are resolved through a complex interplay of DNA repair pathways (Chen et al., 2021b; Hussmann et al., 2021; Koblan et al., 2021a). Site-specific DNA nucleases introduce DSBs, which can undergo error-prone or error-free repair, resulting in DNA mutagenesis or the insertion of desired changes, respectively, at a locus of interest (Figure 1) (Jasin and Haber, 2016). Inactivated DNA nucleases fused to or co-expressed with DNA modifying enzymes generate a variety of DNA lesions and structures, including SSBs, modified bases, abasic sites, mismatched nucleotides, DNA flaps and integration intermediates, whose resolution can lead to desired base substitutions and deletions or insertions of interest (Figure 1) (Anzalone et al., 2020). These DSB-free genome editors mitigate the cellular risks associated with the induction of genomic DSBs by site-specific DNA nucleases. Based on their distinct features, genome editors display different strengths and weaknesses, as highlighted in (Anzalone et al., 2020; Hayward and Ciccia, 2021; Newby and Liu, 2021). In this review, we discuss genome editing technologies with an emphasis on the distinct types of DNA lesions induced by genome editors. In addition, we highlight the main players of the DNA repair pathways implicated in the resolution of those DNA lesions and discuss applications and limitations of genome editing technologies.
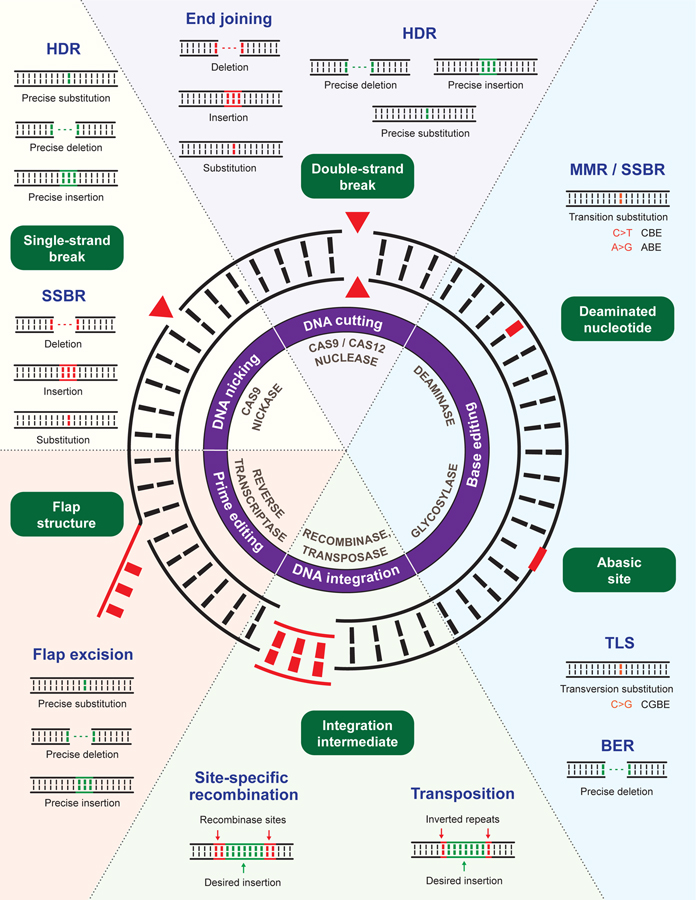
Figure 1. Site-specific DNA perturbations induced by CRISPR-based genome editing technologies and DNA repair processes that resolve them.
Schematic of the five major CRISPR-based genome editing technologies and six site-specific DNA lesions/structures generated by them. Enzymatic activities of genome editors generating the depicted DNA lesions/structures, DNA repair processes involved in their resolution, and outcomes of the repair events are also illustrated. BER (base excision repair), HDR (homology-directed repair), MMR (mismatch repair), SSBR (single-strand break repair) and TLS (translesion synthesis).
DSB-based Genome Editing
The induction of site-specific DSBs to modify the genome is the cornerstone of genome editing technologies (Jasin and Haber, 2016). Indeed, the discovery that genome editing is stimulated by DSB formation induced by meganucleases (e.g., I-SceI) spearheaded the search and development of programmable nucleases (Jasin and Haber, 2016; Rouet et al., 1994). The generation of site-specific genome editing tools was initially enabled by combining the non-specific FokI nuclease domain to sequence-specific DNA binding proteins, such as zinc-finger binding domains and TAL effectors (Bibikova et al., 2003; Boch et al., 2009; Kim and Kim, 2014; Moscou and Bogdanove, 2009; Urnov et al., 2005). More recently, the emergence of RNA-guided genome editing systems that employ CRISPR-Cas proteins has accelerated the use of DSB-based genome editing technologies.
DSB Induction by Programmable Nucleases Derived from CRISPR-Cas Systems
CRISPR–Cas is a prokaryotic adaptive defense system that targets and eliminates invading nucleic acids to circumvent predation by viruses and other mobile genetic elements (Koonin et al., 2017). Specificity for DNA/RNA targets is achieved through small non-coding RNAs, known as guide RNAs (gRNAs), which constitute the cellular memory of past infections (Gasiunas et al., 2012; Jinek et al., 2012; Schneider, 2020; Shivram et al., 2021). In the case of a secondary encounter with the same infectious agent, gRNAs form base pairs with the intruder’s genome, leading to nucleic acid cleavage by Cas proteins, followed by its elimination (Garneau et al., 2010; Nussenzweig and Marraffini, 2020).
CRISPR systems are classified into two classes based on the architecture of the effector complexes (Makarova et al., 2020). In Class 1 CRISPR systems, which are the most abundant in prokaryotes, DNA interference is mediated by a multi-protein complex, known as CRISPR-associated complex for antiviral defense (Cascade), through its Cas3 helicase/nuclease subunit (Brouns et al., 2008). The Cascade complex has been harnessed for genome engineering applications to induce site-specific deletions, introduce new genomic sequences, and regulate gene transcription (Cameron et al., 2019; Chen et al., 2020; Csorgo et al., 2020; Pickar-Oliver and Gersbach, 2019). Class 2 CRISPR systems are composed of single-subunit effector nucleases with distinct, and highly diverse, functional properties (Makarova et al., 2020; Stella et al., 2017). For example, different Class 2 effectors display nuclease activities directed against either ssDNA (Harrington et al., 2018), dsDNA (Gasiunas et al., 2012; Jinek et al., 2012; Karvelis et al., 2020; Liu et al., 2019; Pausch et al., 2020) or RNA (Abudayyeh et al., 2017; Abudayyeh et al., 2016; Cox et al., 2017). Within this class, the type II and V systems are based on the RNA-guided DNA endonuclease families of Cas9 and Cas12. Cas9 employs two distinct nuclease domains to achieve DSB formation, an HNH domain that cleaves the gRNA-targeted strand and a RuvC domain that cleaves the non-targeted DNA strand. Inactivation of either of the nuclease domains generates a nickase Cas9 (nCas9) mutant (Gasiunas et al., 2012; Jinek et al., 2012), while inactivation of both nuclease domains generates a catalytically-dead Cas9 (dCas9) (Qi et al., 2013). Cas9 nickases and catalytically-dead variants constitute the building blocks of DSB-free genome editors. The Cas12 family is characterized by multiple variants with unique features (Makarova et al., 2020; Yan et al., 2019). Cas12a is the type V nuclease most widely utilized for genome editing applications (Zetsche et al., 2015). Additional Cas12 variants include Cas12b (Strecker et al., 2019a; Teng et al., 2018), Cas12d (also known as CasY) (Burstein et al., 2017), Cas12e (also known as CasX) (Burstein et al., 2017; Liu et al., 2019), Cas12f (also known as Cas14) (Harrington et al., 2018), Cas12j (also known as CasΦ) (Pausch et al., 2020), Cas12h, Cas12i and Cas12c (Yan et al., 2019). A subset of Cas12 enzymes (e.g., Cas12a, Cas12i) process their own CRISPR gRNA array without accessory factors (Fonfara et al., 2016; Liao and Beisel, 2021; Yan et al., 2019), thus enabling multiplexed genome editing and gene regulation (Campa et al., 2019; Zetsche et al., 2017). Certain Cas12 nucleases (e.g., Cas14, CasY, CasX, CasΦ) are characterized by a small size, which could facilitate in vivo genome editing applications using viral vectors with limited packaging size (Kim et al., 2021a; Wang et al., 2020a; Xu et al., 2021b). Recently, by reconstructing the evolution of the Cas9 and Cas12 endonucleases, a family of highly abundant transposon-encoded RNA-guided systems, called OMEGA, was discovered in both prokaryotes and eukaryotic cells (Altae-Tran et al., 2021; Karvelis et al., 2021). Although these widespread programmable nucleases, including IscB, IsrB and TnpB, hold great promise for genome targeting and editing, their activities and biological relevance are still not fully characterized.
Cellular Mechanisms that Repair DSBs Introduced by Programmable Nucleases
DSBs introduced by DNA nucleases are primarily repaired by end joining mechanisms, which are often error-prone, or homology-directed repair (HDR), which is typically error-free (Figure 2) (Scully et al., 2019). End joining involves the direct ligation of the two DSB ends, with or without end processing. End processing can lead to the addition or removal of nucleotides at the DSB ends, resulting in insertion/deletion (indel) mutations. End joining can occur through non-homologous end joining (NHEJ) or microhomology-mediated end joining (MMEJ) (Chang et al., 2017). Distinct from end joining, HDR generally utilizes homologous sequences located on sister chromatids or, less commonly, homologous chromosomes to repair DSBs (Chen et al., 2018b). The HDR machinery can also repair DSBs using exogenous DNA donor templates, enabling the insertion of a wide array of desired modifications at defined genomic loci for precision genome editing applications.
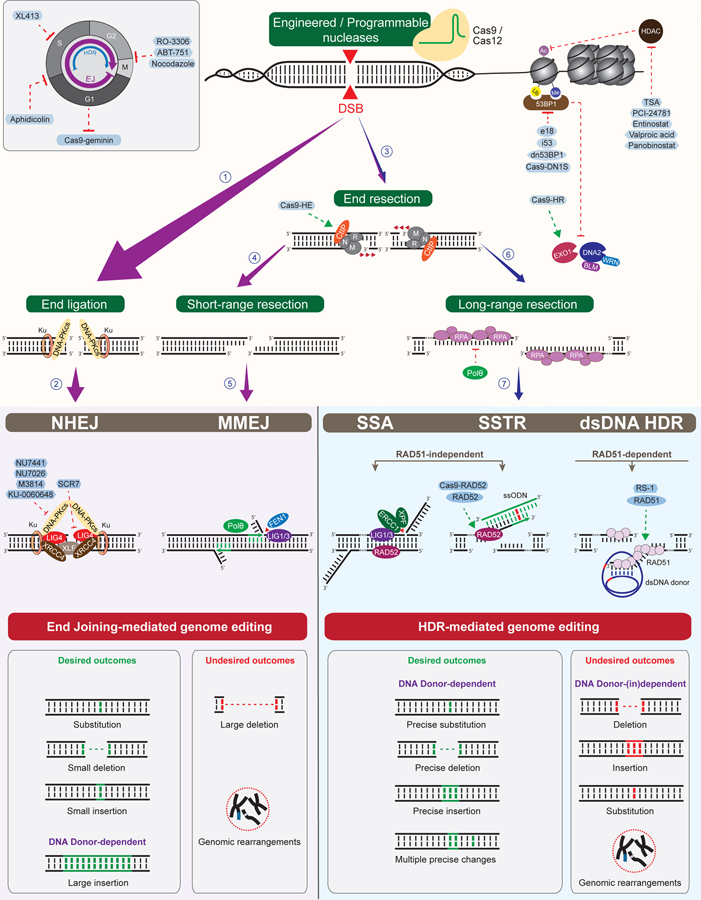
Figure 2. Repair of DSBs induced by site-specific DNA nucleases and strategies to stimulate precision genome editing.
DNA double-strand breaks generated by engineered or programmable nucleases can either be repaired by end joining (left) or HDR mechanisms (right). While end joining can occur throughout the cell cycle, HDR is confined to the S and G2 phases of the cell cycle when a sister chromatid is available for recombination (top left). The choice between end joining and HDR is regulated by DNA end resection. DSB ends that require minimal or no end processing can be ligated by NHEJ through the activities of Ku, DNAPKcs, XLF, XRCC4 and LIG4 (1–2). Short-range end resection catalyzed by MRE11-RAD50-NBS1 (MRN) in complex with CtIP can expose regions of microhomology, which can undergo annealing and end joining by MMEJ (3–5). MMEJ is mediated by Polθ, which extends the annealed ends, followed by the removal of 5’ flaps by FEN1 and DNA ligation by LIG1/3 (5). End joining-mediated genome editing creates substitutions and small indel mutations, and can also generate large insertions in the presence of linearized dsDNA donors (bottom left, in green). End joining events can also result in the formation of large deletions and chromosomal rearrangements (bottom left, in red). Long-range resection is catalyzed by EXO1 or by DNA2 in complex with BLM or WRN and is inhibited by 53BP1 bound to histones with H4K20me2 (me) and H2AK15ub (ub) marks (top right). Long-range resection results in the generation of 3’ ssDNA tails that initiate HDR events (6–7). Annealing of homologous sequences by RAD52, followed by excision of 3’ flaps by XPF-ERCC1 and DNA ligation by LIG1/3 promotes the joining of DSB ends by SSA. SSA causes deletions and potential genomic rearrangements (bottom right, in red). In the presence of ssODNs or dsDNA donors, SSTR or dsDNA donor-dependent HDR (dsDNA HDR) promote the generation of precise DNA substitutions, deletions, insertions and complex mutations (bottom right, in green). While SSTR is mediated by RAD52-dependent annealing of 3’ ssDNA tails to DNA donors, followed by templated DNA synthesis, dsDNA HDR is catalyzed by the recombinase RAD51, which promotes the invasion of 3’ ssDNA tails into homologous sequences of DNA donors to initiate DNA synthesis. Precision genome editing can be stimulated by end joining inhibitors or activators of end resection and HDR. Enhanced HDR can also be obtained by alteration of the cell cycle (top left) and chromatin structure (top right), modulation of the activity of DNA repair factors, expression of engineered DNA repair variants, fusion of DNA repair proteins to Cas nucleases, and modification of DNA donor molecules. HDR modulators are highlighted in blue (see also Table 1). While largely error-free, SSTR and dsDNA HDR can also cause deletions, insertions, point mutations and genomic rearrangements when conducted inaccurately (bottom right, in red). These events can occur in a manner dependent or independent from DNA donors.
End Joining-mediated Genome Editing
Non-Homologous End Joining
NHEJ, the predominant DSB repair pathway in mammalian systems, takes place with rapid kinetics throughout the cell cycle (Figure 2, steps 1–2) (Hustedt and Durocher, 2016; Mao et al., 2008). After a DSB has formed, the broken ends are quickly bound and protected by the Ku70–Ku80 (Ku) heterodimer (Chang et al., 2017; Mimitou and Symington, 2010). Ku then recruits and activates the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), whose kinase activity is essential for NHEJ (Frit et al., 2019). Ku also recruits the NHEJ factors XRCC4, XLF and DNA ligase IV (LIG4) (Zhao et al., 2019). NHEJ factors initially form a long-range synaptic complex that tethers the two DSB ends (Chen et al., 2021c; Graham et al., 2016; Stinson and Loparo, 2021). DNA-PKcs autophosphorylation leads to its dissociation from DNA and the transition to a short-range synaptic complex in which the DSB ends are closely aligned to enable end processing and subsequent ligation by LIG4 (Chen et al., 2021c; Graham et al., 2016; Stinson et al., 2020). The short-range synaptic complex ensures that the DNA ends undergo ligation as soon as they become compatible, thereby restricting DNA end processing and minimizing mutagenesis (Stinson et al., 2020).
Microhomology-mediated End Joining
MMEJ was originally discovered in NHEJ-deficient cells as a backup pathway for DSB repair (Sfeir and Symington, 2015). Similar to NHEJ, MMEJ joins the broken DSB ends without the need for homologous templates (Figure 2, steps 3–5). However, unlike NHEJ, MMEJ requires initial DSB end resection and functions independently of Ku and LIG4 (Sfeir and Symington, 2015). Moreover, MMEJ operates on short homologous sequences (“microhomologies”), ranging between ~5 and 25 bp in mammalian cells, that lie close to both DSB ends (McVey and Lee, 2008). MMEJ is initiated by 5’ to 3’ resection of the DSB ends by the MRE11–RAD50–NBS1 (MRN) nuclease complex together with its stimulatory factor CtIP (Anand et al., 2016; Reginato and Cejka, 2020; Truong et al., 2013). MRN-CtIP-mediated resection of the DSB ends has two important consequences. First, it promotes the removal of Ku and DNA-PKcs, thereby inhibiting NHEJ-mediated repair (Chanut et al., 2016; Deshpande et al., 2020; Reginato et al., 2017). Second, by exposing potential ssDNA microhomologies, it allows the annealing of the broken ends, followed by end trimming and extension by the DNA polymerase Polθ (Brambati et al., 2020; Ramsden et al., 2021; Zahn et al., 2021). The FEN1 endonuclease can then remove the resulting 5′ flaps and DNA ligase I or III ligates the remaining nicks to complete DSB repair (Brambati et al., 2020; Ramsden et al., 2021). Recent studies have also implicated RAD17 and the associated RAD9-HUS1-RAD1 (9–1-1) complex in MMEJ (Hussmann et al., 2021), albeit the role of these factors remains to be elucidated. Although MMEJ and NHEJ protect cells against the severe threat posed by DSBs, when they operate inaccurately they can cause genomic rearrangements, such as chromosomal translocations, intra- and inter-chromosomal deletions and insertions, and end-to-end chromosomal fusions (Dahiya et al., 2021; Ramsden and Nussenzweig, 2021).
DNA Repair Outcomes Induced by End Joining during Genome Editing
During genome editing, reconstitution of the target sequence by error-free NHEJ-mediated repair (i.e., in the absence of end processing) of Cas9-induced DSBs results in repeated Cas9 cleavage cycles until mutagenic NHEJ events block gRNA target recognition (Brinkman et al., 2018). Therefore, Cas9-mediated genome editing ultimately enriches for mutagenic outcomes induced by templated or non-templated nucleotide insertions and deletions (Figure 2, bottom left) (Allen et al., 2019; Chakrabarti et al., 2019; Chen et al., 2019; Leenay et al., 2019; Shen et al., 2018; Shou et al., 2018; van Overbeek et al., 2016). In line with these observations, the DNA polymerase Polλ was recently shown to promote mutagenic fill-in of Cas9-mediated DSBs, and this activity is inhibited by NHEJ factors (Hussmann et al., 2021). In the presence of microhomologies flanking Cas9-induced DSBs, MMEJ-mediated DSB repair can result in the formation of Polθ-dependent deletions (Taheri-Ghahfarokhi et al., 2018). The frequency of MMEJ-induced deletions correlates positively with GC base content, microhomology length and proximity to the DSB site (Allen et al., 2019; Chakrabarti et al., 2019; Shen et al., 2018). While microhomologies with mismatches can also generate deletions, the presence of mismatches reduces the frequency of these events (Allen et al., 2019).
Recent studies have shown that the mutational profiles generated by end joining events at Cas9-induced DSBs are largely reproducible and depend on the sequence context of the Cas9-targeted site (Allen et al., 2019; Chakrabarti et al., 2019; Chen et al., 2019; Leenay et al., 2019; Shen et al., 2018; Shou et al., 2018; van Overbeek et al., 2016). The majority of the reproducible mutations at Cas9-induced DSBs correspond to NHEJ-dependent single base insertions and small deletions and MMEJ-mediated deletions (Allen et al., 2019; Chakrabarti et al., 2019; Chen et al., 2019; Shen et al., 2018; van Overbeek et al., 2016). Single base insertions are suppressed by the kinase activity of ATM (Bermudez-Cabrera et al., 2021). Together, the above observations led to the development of machine learning models (i.e., inDelphi, FORECasT, SPROUT, Lindel) that can predict the main mutational signatures resulting from end joining-mediated repair of Cas9-induced DSBs (Allen et al., 2019; Chen et al., 2019; Leenay et al., 2019; Shen et al., 2018).
Applications of End Joining-mediated Genome Editing
End joining-mediated DSB repair is employed in numerous genome editing applications (Pickar-Oliver and Gersbach, 2019). Given its mutagenic nature, end joining is often harnessed to disrupt the functionality of coding and non-coding elements. End joining-mediated gene disruption has been used to study the function of genes in cellular and animal models, including previously genetically intractable systems, such as monkey embryos (Niu et al., 2014), or newly established genetic models, like the killifish (Harel et al., 2016). Additionally, high-throughput screening technologies with gRNA libraries that induce loss-of-function perturbations have been used to annotate functional genetic elements and discover new cancer dependencies (Shalem et al., 2015). Interestingly, the generation of multiple site-specific DSBs has also enabled the modeling of chromosomal deletions and translocations frequently associated with human cancer (Brunet and Jasin, 2018). In addition, both NHEJ and MMEJ have been exploited to insert new sequences into the genome (Figure 2, bottom left) (Nakade et al., 2014; Schmid-Burgk et al., 2016; Suzuki and Izpisua Belmonte, 2018; Suzuki et al., 2016). End joining-based approaches enable the efficient insertion of DNA payloads in biological systems not proficient in HDR-mediated DSB repair, such as non-dividing cells (e.g., neuronal cells) and certain human organoid models (Artegiani et al., 2020; Nami et al., 2018; Suzuki et al., 2016).
End joining-mediated gene disruption has been successfully employed to revert the effects induced by pathogenic mutations in pre-clinical or clinical settings. This approach has shown promise for the treatment of sickle cell disease and β-thalassemia, with multiple ex vivo strategies developed during clinical trials to inactivate BCL11A in hematopoietic stem and progenitor cells (Ferrari et al., 2021; Frangoul et al., 2021; Mullard, 2020). Clinical studies have also shown the potential of end joining-dependent approaches for the in vivo disruption of transthyretin in patients with transthyretin amyloidosis (Gillmore et al., 2021). Moreover, recent studies have highlighted the feasibility of end joining-mediated strategies for disrupting T cell receptors and the PD-1 immune checkpoint regulator in T cells of patients with refractory cancer (Ellis et al., 2021; Stadtmauer et al., 2020). In addition, strategies that rely on the joining of two paired Cas9-induced DSBs have been utilized to excise mutant exons of the dystrophin gene and restore its open reading frame in cells from patients with Duchenne muscular dystrophy (DMD) or DMD animal models (Amoasii et al., 2018; Amoasii et al., 2017; Choi and Koo, 2021; Min et al., 2019). Similar dual DSB-based strategies developed for excising an aberrant splicing donor site in the CEP290 gene are currently being used in clinical trials for in vivo correction of the CEP290 splicing defects that underlie Leber congenital amaurosis (Maeder et al., 2019; Quinn et al., 2020). Related approaches have also been employed to delete expanded trinucleotide repeats in cell lines derived from patients with Huntington’s disease (HD), fragile X syndrome or myotonic dystrophy type 1 (DM1) and in animal models of HD, DM1 or Friedrich’s ataxia (Monteys et al., 2017; Mosbach et al., 2019; Ouellet et al., 2017; Provenzano et al., 2017; Shin et al., 2016; van Agtmaal et al., 2017; Xie et al., 2016). Finally, approaches that take advantage of the predictable mutation outcome of MMEJ-mediated DSB repair have been deployed to correct pathogenic microduplications in cells from patients with limb-girdle muscular dystrophy and Hermansky–Pudlak syndrome (Iyer et al., 2019; Shen et al., 2018). In this regard, the use of machine learning-based algorithms that predict the mutational outcome of end joining events (Allen et al., 2019; Chen et al., 2019; Leenay et al., 2019; Shen et al., 2018) may further facilitate clinical applications of end joining-mediated genome editing.
Donor-dependent Homology-directed Repair
HDR promotes the repair of DSBs using endogenous or exogenous homologous DNA templates (Gallagher and Haber, 2018; Jasin and Haber, 2016). Although HDR is a largely error-free repair mechanism, if not properly regulated, it can cause genomic rearrangements and thus result in genomic instability (Al-Zain and Symington, 2021). HDR requires extensive 5’ to 3’ resection of DNA ends, which is initiated by the MRN-CtIP complex and then extended by the exonuclease EXO1 or by the helicase-nuclease DNA2 in complex with the BLM or WRN helicases (Cejka and Symington, 2021). End resection results in the generation of 3’ ssDNA tails that promote the search of homologous sequences to be used as templates for repair (Figure 2, steps 6–7). In genome editing, different forms of exogenous donor templates are used to repair site-specific DSBs, including double-strand DNA (dsDNA) donors, short or long single-strand oligodeoxynucleotides (ssODN) or chromatinized templates (Cruz-Becerra and Kadonaga, 2020; Li et al., 2019; Yeh et al., 2019). The repair of DSBs using dsDNA and ssODN donors occurs through RAD51-dependent or -independent mechanisms, respectively, as discussed below.
RAD51-dependent HDR
The RAD51 recombinase mediates homology search and strand exchange between broken DNA strands and dsDNA repair templates (Figure 2, step 7). Following RAD51-mediated strand invasion and formation of a displacement loop (D-loop) structure, the invading strand is thought to be elongated by the DNA polymerase δ using the homologous sequence as a template (McVey et al., 2016). DNA synthesis can then be followed by the dissociation of the invading DNA strand and its reannealing to the second DSB end through a process known as synthesis-dependent strand annealing (SDSA) (McVey et al., 2016). Alternatively, if the second DSB end is captured by the D-loop structure, double Holliday junctions can form, which then undergo either dissolution by the BLM-TOPO III complex or resolution by Holliday junction resolvases (Matos and West, 2014). Although it remains unclear how the choice between SDSA and double Holliday junction formation is regulated, both pathways have been implicated in repairing DSBs using dsDNA donors (Kan et al., 2014). In the absence of second end capture, the repair of nuclease-mediated DSBs can occur through break-induced replication (BIR), a process that entails extensive DNA synthesis carried out at migrating D-loop structures (Kramara et al., 2018; Llorente et al., 2008). BIR has been shown to cause DNA mutagenesis and chromosomal rearrangements (Malkova and Haber, 2012). Interestingly, recent studies have identified mechanisms that promote second end capture and D-loop dissociation to restrict mutagenic BIR (Pham et al., 2021).
RAD51-independent HDR
RAD51-independent HDR pathways include single-strand annealing (SSA) and single-stranded template repair (SSTR) (Figure 2, step 7) (Gallagher and Haber, 2018). SSA shares similarities with MMEJ in its ability to promote the joining of DSB ends with partial or complete sequence homology. Unlike MMEJ, however, SSA utilizes long flanking sequences of homology, which can span several hundred nucleotides in mammalian cells (Bhargava et al., 2016). SSA is mediated by the HDR protein RAD52, which binds the resected ssDNA ends and anneals regions of homology, resulting in the deletion of intervening sequences (Bhargava et al., 2016).
SSTR promotes the repair of DSBs using ssODN templates (Gallagher and Haber, 2018; Storici et al., 2006). Given its higher efficiency and fidelity relative to HDR processes that use dsDNA templates and the facile access to synthesized and chemically modified ssODNs, SSTR is the most frequently utilized HDR mechanism for genome editing (Richardson et al., 2016; Yeh et al., 2019). SSTR shares similarities with multiple HDR pathways. Like SDSA, SSTR is thought to be stimulated by DSB resection (Canny et al., 2018; Gallagher and Haber, 2018; Nambiar et al., 2019), and both HDR processes involve unidirectional conversion tracts due to strand pairing and extension of one of the DSB ends (Gallagher and Haber, 2018; Kan et al., 2017; Paix et al., 2017). However, unlike SDSA, SSTR functions independently of RAD51 (Bothmer et al., 2017; Richardson et al., 2018). Instead, SSTR requires RAD52, similar to SSA, and reminiscent of RNA-templated HDR (Gallagher et al., 2020; Keskin et al., 2014; Mazina et al., 2017; Storici et al., 2006). In addition to being used as templates for HDR events, ssODNs can also directly integrate into homologous genomic sites at Cas9-induced breaks through mechanisms that remain to be fully established (Kan et al., 2017). Besides RAD52, the Fanconi anemia pathway has also been proposed to be involved in SSTR and dsDNA donor-dependent HDR through unresolved mechanisms (Richardson et al., 2018; Wienert et al., 2020). Altogether, several important aspects of the molecular mechanisms underlying SSTR remain to be elucidated, including the involvement of DSB resection machineries and the processes that mediate strand invasion, D-loop extension, and second end capture.
Applications of HDR-mediated Genome Editing
The ability of HDR to install any desired nucleotide change into the genome makes it an attractive pathway for disease modeling. HDR-based approaches also hold great promise for clinical management of inherited and acquired human diseases (Doudna, 2020). For example, a pig model of Huntington’s disease (HD) was recently developed by replacing exon 1 of the pig HTT gene with the corresponding exon of the human HTT gene containing a 150-CAG repeat, enabling the recapitulation in mammalian models of the selective neurodegeneration induced by CAG triplet expansion in HD patients (Yan et al., 2018). The clinical potential of CRISPR-mediated HDR is also highlighted by the enhanced antitumor activity exhibited by T cells with a chimeric antigen receptor inserted by HDR at the T cell receptor locus (Eyquem et al., 2017). Pooled knock-in HDR-based strategies to insert large DNA cargos into the T cell receptor locus of primary human T cells have enabled the identification of gene constructs that improve T cell-mediated antitumor activity (Roth et al., 2020). Furthermore, saturation mutagenesis experiments that used HDR to install all possible substitution mutations into genomic regions of interest in haploid cells have allowed the interrogation of clinically actionable genes at a single-base level (Findlay et al., 2014; Findlay et al., 2018). Despite the success of these studies, the low frequency of HDR events relative to concomitant mutations induced at the targeted loci by error-prone end joining-mediated repair has limited the widespread adoption of high-throughput knock-in screens using HDR. HDR-based approaches have also been employed for endogenous gene tagging with genetic markers, enabling the detection of the subcellular localization of proteins, isolation of native protein complexes, and temporal regulation of protein function (Chen et al., 2018a; Cho et al., 2021; Dalvai et al., 2015; Leonetti et al., 2016; Miyaoka et al., 2014; Natsume et al., 2016). Finally, HDR-mediated strategies have been used to promote gene drive, a process that stimulates non-Mendelian inheritance of genetic elements, to potentially eradicate harmful species causing vector-borne diseases (Champer et al., 2016).
DDR Modulating Strategies to Promote HDR-based Precision Genome Editing
One of the major determinants of DSB repair pathway choice is DNA end resection. As discussed above, DNA end resection promotes DSB repair by MMEJ and HDR, while generating DSB ends not suitable for NHEJ-mediated repair (Symington and Gautier, 2011). In mammalian cells, end resection operates with slower kinetics than NHEJ. It is regulated by the cell cycle and chromatin environment, ensuring that HDR occurs in the S and G2 phases when an appropriate repair template (i.e., a sister chromatid) is present (Figure 2, top left) (Hustedt and Durocher, 2016). Ultimately, DSB repair pathway choice dictates genome editing outcomes, and competition between alternative DSB repair pathways can generate mosaicism among edited alleles. Consequently, strategies to regulate DSB repair pathway choice by either stimulating or counteracting specific DSB repair pathways enable more efficient and precise genome editing.
Given the fast-acting nature of NHEJ, inhibiting this pathway is an attractive strategy for precision genome editing (Figure 2, step 2). In this regard, efforts to inhibit the core NHEJ factors have produced mixed results in human cells. For example, pharmacological inhibition of Ku70/80 marginally stimulates HDR (Table 1) (Riesenberg and Maricic, 2018). LIG4 inhibition by the small-molecule SCR7 or viral proteins that induce its proteasomal degradation are reported to increase HDR in certain human and mouse cells, although this effect was more limited in other cellular contexts (Chu et al., 2015; Greco et al., 2016; Gutschner et al., 2016; Hu et al., 2018b; Maruyama et al., 2015; Pinder et al., 2015; Robert et al., 2015; Song et al., 2016; Srivastava et al., 2012; Yang et al., 2016; Zhang et al., 2017). Such variability in effect might be partially due to the usage of different chemical derivatives of SCR7 (Yeh et al., 2019). Similarly, small-molecule inhibition of DNA-PKcs with NU7441, NU7026 and M3814 has also been reported to increase HDR (Riesenberg et al., 2019; Robert et al., 2015; Suzuki et al., 2016). Dual inactivation of both MMEJ and NHEJ also holds the promise of synergistically enhancing HDR. For example, combinatorial inactivation of NHEJ and MMEJ proteins, such as Ku70/80 and Polθ, prevents off-target integration of dsDNA donors (Zelensky et al., 2017). Given that inactivation of end joining is associated with exacerbation of sensitivity to various genotoxic agents and increased carcinogenesis (Davis and Chen, 2013; Ramsden et al., 2021), the impact of these HDR-modulating treatments on genomic integrity and cell viability needs to be carefully investigated.
Table 1. Strategies utilized to improve CRISPR-based HDR at Cas9-induced DSBs.
The mechanisms of action for each strategy is indicated, along with the type of treatment, the delivery method for Cas9, the DNA donor type, the cell lines utilized, and the effect observed on DSB repair. References of the studies that employ the indicated strategies are also listed. AAV (adeno-associated virus), AAV6 (adeno-associated virus type 6), AdV (adenovirus), Ad4 (adenovirus type 4), Ad5 (adenovirus type 5), AECs (alveolar epithelial cells), CRISPRa/i (CRISPR activation and interference), DBCO (dibenzylcyclooctyne), hESCs (human embryonic stem cells), hHSCs (human hematopoietic stem cells), hHSPCs (human hematopoietic stem and progenitor cells), hiPSCs (human induced pluripotent stem cells), hPSCs (human pluripotent stem cells), HUVECs (human umbilical vein endothelial cells), IDLV (integrase defective lentiviral vector), LCL (lymphoblastoid B cell line, EBV-transformed), MEFs (mouse embryonic fibroblasts), mESCs (mouse embryonic stem cells), miPSCs (mouse induced pluripotent stem cells), PBMCs (peripheral blood mononuclear cells), PCV (porcine circovirus 2 Rep protein), RNP (ribonucleoprotein), scAAV (self-complementary adeno-associated virus), ssAAV (single-stranded adeno-associated virus).
| Strategy | Proposed mechanism of action | Treatment | Treatment format | Delivery of Cas9 | Donor type | Cell line | Effect | References |
|---|---|---|---|---|---|---|---|---|
| Optimizing the donor DNA | Improved annealing of the DNA donor to the asymmetrically released 3’ end of the non-target strand following Cas9 cleavage | Asymmetric ssODN | Oligonucleotide | RNP | ssODN | • HEK293 | ↑ HDR | (Richardson et al., 2016) |
| Generation of DNA donor ends that facilitate HDR | dsDNA donor with 3’ overhangs | Plasmid, oligonucleotide | RNP | dsDNA | • HEK293 • mESCs • Mouse zygotes |
↑ HDR | (Hirotsune et al., 2020; Liang et al., 2017) | |
| Chromatin-mediated stimulation of HDR | Chromatinized dsDNA donor | Plasmid | Plasmid | dsDNA | • MCF10A • HeLa |
↑ HDR | (Cruz-Becerra and Kadonaga, 2020) | |
| Inhibition of DNA donor degradation | ssODN with phosphorothioate-modified ends | Oligonucleotide | Plasmid, mRNA | ssODN | • U2OS • RPE1 • Rat C6 • Rat, mouse and zebrafish embryos |
↑ HDR | (Renaud et al., 2016) | |
| Inhibition of DNA donor multimerization | dsDNA donor with biotin-modified 5’ ends | DNA amplicon | mRNA | dsDNA | • Medaka zygotes | ↓ Illegitimate DNA donor integration | (Gutierrez-Triana et al., 2018) | |
| Enhanced DNA donor uptake | dsDNA or ssDNA/dsDNA hybrid donor with Cas9 target sequences | Oligonucleotide + RNP | RNP | dsDNA, ssDNA/dsDNA | • T cells • B cells • NK cells • hHSPCs |
↑ HDR | (Nguyen et al., 2020; Shy et al., 2021) | |
| gRNA-mediated localization of the DNA donor to the Cas9-induced DSB | gRNA-ssODN covalently linked fusion | Oligonucleotide + RNP | RNP | ssODN | • HEK293 | ↑ HDR | (Lee et al., 2017) | |
| gRNA-S1m aptamer-streptavidin/biotin-ssODN complex | Oligonucleotide + RNP | RNP | ssODN | • hPSCs • HEK293 |
↑ HDR | (Carlson-Stevermer et al., 2017) | ||
| Cas9-mediated localization of the DNA donor to the DSB | Cas9-avidin/biotin-ssODN complex | Oligonucleotide + RNP / plasmid / mRNA | RNP, plasmid, mRNA | ssODN | • HEK293 • Mouse zygotes |
↑ HDR | (Ma et al., 2017b) | |
| ssODN covalently tethered to a Cas9-PCV fusion | Oligonucleotide + RNP | RNP | ssODN | • HEK293 • U2OS |
↑ HDR | (Aird et al., 2018) | ||
| ssODN covalently tethered to a Cas9-SNAP tag fusion | Oligonucleotide + RNP | RNP | ssODN | • HEK293 • K562 • mESCs |
↑ HDR | (Savic et al., 2018) | ||
| DBCO- ssODN covalently tethered to an azido-modified Cas9 | Oligonucleotide + RNP | RNP | ssODN | • HEK293 • Mouse zygotes |
↑ HDR | (Ling et al., 2020) | ||
| Controlling the cell cycle | G1/S phase synchronization: DNA polymerase α inhibition | Aphidicolin | Small molecule | RNP | ssODN | • HEK293 • Neonatal dermal fibroblasts |
↑ HDR | (Lin et al., 2014) |
| S phase synchronization: CDC7 inhibition | XL413 | Small molecule | RNP | ssODN, dsDNA | • HEK293 • HeLa • U-251 • K562 • hiPSCs • Primary HSPCs • Primary T cells |
↑ HDR ↓ NHEJ |
(Wienert et al., 2020) | |
| G2/M phase synchronization: inhibition of microtubule polymerization | Nocodazole | Small molecule | RNP | ssODN, dsDNA | • HEK293 | ↑ HDR | (Lin et al., 2014) | |
| ABT-751 | Small molecule | Plasmid | dsDNA | • hESCs • hiPSCs |
↑ HDR | (Yang et al., 2016) | ||
| G2/M phase synchronization: CDK1 inhibition | RO-3306 | Small molecule | mRNA | AAV6 | • hHSPCs | ↑ HDR ↓ End joining |
(Lomova et al., 2019) | |
| Restricting Cas9 function during the cell cycle | Limiting Cas9 activity to the S/G2 phase | Cas9-geminin fusion | Plasmid | Plasmid | dsDNA | • HEK293 | ↑ HDR | (Gutschner et al., 2016) |
| mRNA | mRNA | AAV6 | • hHSPCs | ↑ HDR ↓ NHEJ |
(Lomova et al., 2019) | |||
| Plasmid | Plasmid | dsDNA | • HEK293 | ↑ HDR | (Charpentier et al., 2018) | |||
| Cas9 injection in the S phase | mRNA, RNP | mRNA, RNP | dsDNA | • Mouse zygotes | ↑ HDR | (Abe et al., 2020) | ||
| Cas9 injection in the G2 phase | mRNA | mRNA | dsDNA | • Mouse embryos | ↑ HDR | (Gu et al., 2018) | ||
| Promoting open chromatin | Histone deacetylase (HDAC) inhibition, acetylation-mediated regulation of DSB repair factors | Trichostatin A (TSA), PCI-24781 | Small molecule | Plasmid, RNP | dsDNA, ssODN | • Pig fetal fibroblasts • Pig embryos |
↑ HDR, MMEJ | (Li et al., 2020b) |
| HDAC inhibition | TSA | Small molecule | Cas9-expressing cell line | None | • HepG2 | ↑ End joining | (Chakrabarti et al., 2019) | |
| Entinostat, Panobinostat | Small molecule | Plasmid, AdV | dsDNA | • HEK293 • HeLa • HT29 |
↑ HDR, End joining | (Liu et al., 2020a) | ||
| Valproic acid | Small molecule | Plasmid | dsDNA | • hESCs • hiPSCs |
↑ HDR | (Takayama et al., 2017) | ||
| Inhibiting NHEJ | Ku depletion | shRNA, siRNA | Plasmid, siRNA | Plasmid | dsDNA | • HEK293 | ↑ HDR ↓ NHEJ |
(Chu et al., 2015; Robert et al., 2015) |
| LIG4 inhibition | shRNA, siRNA | Plasmid, siRNA | Plasmid | dsDNA | • HEK293 | ↑ HDR ↓ NHEJ |
(Chu et al., 2015; Robert et al., 2015) | |
| SCR7 | Small molecule | Cas9-expressing cell lines, mRNA | dsDNA, ssODN | • A549 • MelJuSo • mDC2.4 • Mouse zygotes |
↑ HDR ↓ NHEJ |
(Maruyama et al., 2015) | ||
| Plasmid | dsDNA | • HEK293 | ↑ HDR ↓ NHEJ |
(Chu et al., 2015; Pinder et al., 2015; Robert et al., 2015) | ||||
| Plasmid, Cas9-expressing cell line | dsDNA, ssODN | • HEK293 • hiPSCs |
No HDR stimulation | (Gutschner et al., 2016; Riesenberg and Maricic, 2018; Zhang et al., 2017) | ||||
| Expression of the Ad4/Ad5 proteins E1B55K and E4orf6 | Plasmid | Plasmid | dsDNA | • HEK293 • Mouse Burkitt lymphoma cells |
↑ HDR ↓ NHEJ |
(Chu et al., 2015; Robert et al., 2015) | ||
| DNA-PKcs inhibition | siRNA | siRNA | Plasmid | dsDNA | • HEK293 | ↑ HDR ↓ NHEJ |
(Robert et al., 2015) | |
| NU7441 | Small molecule | Plasmid | dsDNA, ssODN | • HEK293 • MEFs |
↑ HDR ↓ NHEJ |
(Robert et al., 2015) | ||
| Plasmid | dsDNA | • hiPSC | ↑ HDR | (Zhang et al., 2017) | ||||
| RNP | None | • HEK293 | ↑ MMEJ ↓ NHEJ |
(van Overbeek et al., 2016) | ||||
| Plasmid | dsDNA | • U2OS | ↑ HDR | (Canny et al., 2018) | ||||
| NU7026 | Small molecule | Plasmid | dsDNA | • HEK293 | ↑ HDR ↓ NHEJ |
(Suzuki et al., 2016) | ||
| KU-0060648 (also inhibits PI3K) | Small molecule | Plasmid | dsDNA, ssODN | • HEK293 • MEFs |
↑ HDR ↓ NHEJ |
(Robert et al., 2015) | ||
| M3814 | Small molecule | RNP | ssODN | • K562 • hiPSCs |
↑ HDR ↓ End joining |
(Riesenberg et al., 2019) | ||
| Promoting end resection | Inhibition of 53BP1 | i53 (inhibitor of 53BP1) | Plasmid, AAV | Plasmid, RNP | ssODN, dsDNA | • U2OS • HEK293 • K562 • MCF10A • MEFs |
↑ HDR | (Canny et al., 2018) |
| e18 (enhanced RAD18 variant) | Plasmid, mRNA | Plasmid, mRNA | ssODN, dsDNA | • HEK293 • HeLa • U2OS • hESCs |
↑ HDR, MMEJ ↓ NHEJ |
(Nambiar et al., 2019) | ||
| Cas9-DN1S (dominant negative 53BP1 mutant) fusion | Plasmid, RNP | Plasmid, RNP | dsDNA, scAAV, ssAAV | • HEK293 • K562 • LCL • Jurkat • Patient-derived B lymphocytes |
↑ HDR ↓ NHEJ |
(Jayavaradhan et al., 2019) | ||
| Engagement of CtIP | Cas9-HE (minimal N-terminal CtIP fragment) fusion | Plasmid, mRNA | Plasmid, mRNA | dsDNA | • HEK293 • HCT116 • RG37 • hiPSCs • Rat oocytes |
↑ HDR, MMEJ ↓ NHEJ |
(Charpentier et al., 2018) | |
| Engagement of EXO1 | Cas9-HR (EXO1 fragment) fusion | Plasmid | Plasmid | dsDNA | • A549 • H1299 • K562 |
↑ HDR | (Hackley, 2021) | |
| Inhibiting MMEJ | POLQ inactivation | POLQ knockout | Cas9-mediated gene knockout | Plasmid | None | • miPSCs | ↓ MMEJ | (Mateos-Gomez et al., 2015) |
| dsDNA | • MEFs • mESCs |
↑ HDR | (Mateos-Gomez et al., 2017) | |||||
| Promoting strand annealing | RAD52 stimulation | RAD52 overexpression | Plasmid | Plasmid | ssODN | • hiPSCs • HEK293 |
↑ HDR | (Paulsen et al., 2017) |
| Cas9-Rad52 (yeast homolog) fusion | Plasmid | Plasmid | dsDNA, ssODN | • HEK293 • PK15 |
↑ HDR | (Shao et al., 2017) | ||
| Promoting DNA recombination | RAD51 stimulation | RS-1 | Small molecule | Plasmid | dsDNA | • HEK293 • U2OS |
↑ HDR | (Pinder et al., 2015) |
| RAD51 overexpression | mRNA | mRNA | dsDNA | • Rabbit embryos | ↑ HDR | (Song et al., 2016) | ||
| RAD51 localization | Expression of miCas9 (fusion of BRCA2 exon 27 to SpCas9) | Plasmid, RNP | Plasmid, RNP | ssODN, dsDNA | • Human fibroblasts • AECs • hiPSCs • Ad293 • hHSCs • Jurkat |
↑ HDR ↓ End joining |
(Ma et al., 2020) | |
| BRCA1 modulation | Expression of the BRCA1 variant BRCA1M1775R | Plasmid | Plasmid | dsDNA | • HEK293 | ↑ HDR | (Pinder et al., 2015) | |
| Enhancing DSB repair | Stimulation of DSB repair events | Cas9-POLD3 fusion | Plasmid, mRNA, RNP | Plasmid, mRNA, RNP | ssODN | • HEK293 • RPE1 • BJ • PBMCs • hESCs |
↑ HDR, NHEJ | (Reint et al., 2021) |
| Combining multiple HDR regulators | Stimulation of G1 to S phase transition and G/M phase synchronization | Cyclin D1 overexpression + nocodazole treatment | Plasmid + small molecule | Plasmid | dsDNA | • hiPSCs | ↑ HDR | (Zhang et al., 2017) |
| G2/M phase synchronization followed by G1/S phase synchronization | Nocodazole + aphidicolin treatment | Small molecule | RNP | ssODN | • hESCs | ↑ HDR | (Lin et al., 2014) | |
| G2/M phase synchronization + degradation of Cas9 in the G1 phase | RO-3306 + Cas9-geminin fusion | Small molecule + mRNA | mRNA | AAV6 | • hHSPCs | ↑ HDR ↓ End joining |
(Lomova et al., 2019) | |
| Confinement of Cas9 activity to the G2 phase + localization of the DNA donor to the Cas9-induced DSB | Injection of a Cas9-streptavidin fusion and a biotin-dsDNA donor in the G2 phase | mRNA + DNA amplicon | mRNA | dsDNA | • Mouse embryos | ↑ HDR | (Gu et al., 2018) | |
| Promotion of strand annealing and end resection | Expression of RAD52 and dominant-negative 53BP1 (dn53BP1) | Plasmid | Plasmid | ssODN | • hiPSCs • HEK293 |
↑ HDR | (Paulsen et al., 2017) | |
| Inhibition of MMEJ and NHEJ | Disruption of POLQ and Ku | Cas9-mediated gene knockout | Plasmid | dsDNA | • mESCs | ↓ Illegitimate DNA donor integration | (Zelensky et al., 2017) | |
| Modulation of multiple DSB repair proteins | CRISPY mix (NU7026 + TSA + MLN4924 + NSC 15520) | Small molecule | nCas9-expressing cell line, plasmid | ssODN | • hiPSCs • hESCs |
↑ HDR ↓ End joining |
(Riesenberg and Maricic, 2018) | |
| End resection initiation and extension | eRAD18 (e18)-Cas9-CtIP fusion | Plasmid | Plasmid | dsDNA | • HEK293 | ↑ HDR | (Richardson et al., 2020) | |
| Cell cycle modulation + inhibition of NHEJ | XLF413 + cold shock + NU7441 + SCR7 | Small molecule + temperature change | RNP | ssODN | • hiPSCs | ↑ HDR ↓ End joining |
(Maurissen and Woltjen, 2020) | |
| Promotion of end resection + inhibition of NHEJ | CRISPRa/i-mediated activation of CDK1 and repression of Ku80 | Plasmid, lentivirus | Cas9-expressing cell lines | dsDNA | • HEK293 • HeLa |
↑ HDR | (Ye et al., 2018) | |
| Stimulation of RAD51-mediated strand invasion + chromatin opening at target site | RAD51 overexpression + valproic acid treatment | Plasmid + small molecule | Plasmid | dsDNA | • hESCs • hiPSCs |
↑ HDR | (Takayama et al., 2017) | |
| Limiting cell toxicity associated with DSB formation and/or donor DNA delivery | Transient inhibition of p53 | Expression of a dominant-negative p53 truncated form (GSE56) | mRNA | RNP | AAV6 | • hHSPCs | ↑ HDR | (Schiroli et al., 2019) |
| Transient inhibition of p53 + cell cycle modulation | Expression of the Ad5 protein E4orf6/7 and the dominant-negative p53 mutant protein GSE56 | mRNA | RNP | AAV6, IDLV | • hHSPCs | ↑ HDR, End joining | (Ferrari et al., 2020) | |
| Transient inhibition of p53 | Overexpression of MDM2 (p53 antagonist) | Plasmid | RNP | ssODN | • RPE1 | ↑ HDR | (Haapaniemi et al., 2018) | |
| Inhibition of apoptotic cell death | Overexpression of BCL-XL | Plasmid, lentivirus | Plasmid | dsDNA, ssODN | • hiPSCs • mESCs |
↑ HDR, NHEJ | (Li et al., 2018c) | |
| Of Unknown Mechanism | Unknown for genome editing; β3-adrenergic receptor activation | L755507 | Small molecule | Plasmid | dsDNA, ssODN | • mESCs • HeLa • K562 • hiPSCs • Neural stem cells • HUVECs • CRL-2097 |
↑ HDR ↓ End joining |
(Yu et al., 2015) |
| Unknown for genome editing; ER-Golgi transport inhibition | Brefeldin A | Small molecule | Plasmid | dsDNA | • mESCs | ↑ HDR | ||
| Unknown; potential contribution of cell cycle-dependent and -independent effects | Cold shock (32°C for 1–2 days) | Temperature change | mRNA | ssODN | • HEK293 • hiPSCs |
↑ HDR | (Guo et al., 2018) | |
| Unknown | SHROOM1 depletion / deletion | siRNA, Cas9-mediated gene knockout | Plasmid, mRNA | dsDNA, ssODN | • HEK293T • HCT116 • Hepa1-6 • Mouse embryos |
↑ HDR | (Zhao et al., 2020) | |
| Unknown | Resveratrol | Small molecule | Plasmid | dsDNA | • Porcine fetal fibroblasts | ↑ HDR | (Li et al., 2017) |
Because of the central role of DSB resection in the choice between end joining and HDR, stimulation of DSB resection has shown great promise in promoting HDR. TP53-binding protein 1 (53BP1), a key inhibitor of end resection, is recruited to chromatin through the binding of its Tudor and UDR domains to histone H4 dimethylated at Lys20 (H4K20me2) and histone H2A monoubiquitylated at Lys15 (H2AK15ub), respectively (Figure 2, top right) (Botuyan et al., 2006; Fradet-Turcotte et al., 2013). Multiple strategies have been developed to stimulate HDR by inhibiting 53BP1 function (Table 1). Expression of an engineered ubiquitin variant that binds the Tudor domain of 53BP1 (i53, inhibitor of 53BP1) has been shown to prevent 53BP1 binding to chromatin and stimulate HDR (Canny et al., 2018). Similarly, a dominant-negative 53BP1 (dn53BP1) and an engineered variant of RAD18 (e18) stimulate HDR by competing with 53BP1 for binding H2AK15ub through their ubiquitin binding domains, thereby occluding 53BP1 from DSBs (Nambiar et al., 2019; Paulsen et al., 2017). Fusing Cas9 to dn53BP1 or e18 (Jayavaradhan et al., 2019; Richardson et al., 2020) also stimulates HDR with potentially improved specificity, given the targeted inhibition of 53BP1 function at Cas9-induced DSBs. Notably, some of these 53BP1 targeting strategies stimulate HDR without altering off-target editing by Cas9 or compromising cellular viability (Jayavaradhan et al., 2019; Nambiar et al., 2019; Paulsen et al., 2017). Additional studies have shown that combinatorial inactivation of 53BP1 along with upregulation of factors downstream of end resection enhances HDR rates. For example, co-expression of dn53BP1 and RAD52 additively enhances HDR using ssODN in human cells (Paulsen et al., 2017). This combination promotes end resection by blocking 53BP1 with dn53BP1, while stimulating SSTR by overexpressing RAD52 (Paulsen et al., 2017; Richardson et al., 2018; Shao et al., 2017). Likewise, fusions of the ubiquitin-binding domains of RAD18 or RNF169 to BRCA1, a key HDR factor required for RAD51 loading, have been shown to promote HDR likely by blocking 53BP1, while promoting homologous recombination (Bashir et al., 2020). Finally, the possibility of activating HDR in the G1 phase has been demonstrated by restoring the BRCA1-PALB2-BRCA2 interaction (required for RAD51 loading) and activating CtIP-mediated end resection in G1 by expressing a phospho-mimetic CtIP in a 53BP1 knock out cell line (Orthwein et al., 2015). It remains to be investigated if this approach can promote HDR with sufficient efficiency and without cellular toxicity to enable editing of non-dividing cells for clinical applications. Moreover, since loss of 53BP1 is associated with hyper-resection and mutagenic repair by SSA (Ochs et al., 2016), the consequences of transiently inhibiting 53BP1 on genomic integrity need to be carefully evaluated.
In addition to 53BP1 regulators, proteins that stimulate end resection (e.g., CtIP and EXO1), DNA recombination factors (e.g., RAD52), and DNA polymerase subunits (e.g., POLD3) have been shown to promote HDR when fused to Cas9 (Table 1) (Charpentier et al., 2018; Hackley, 2021; Ma et al., 2020; Reint et al., 2021; Shao et al., 2017). Such Cas9 fusions can reduce the pleiotropic consequences of DDR manipulation caused by overexpression of DNA repair factors. Interestingly, phage-encoded single-strand DNA annealing proteins improve HDR 1,000-fold in bacteria through their interactions with host single-strand DNA binding proteins (Filsinger et al., 2021). However, it remains to be investigated whether the expression of recombination-promoting viral protein could similarly stimulate HDR at Cas9-induced breaks in mammalian cells. Cell cycle modulation has also been shown to stimulate HDR by ensuring Cas9-mediated DSB formation at the HDR-permissive S and G2 phases of the cell cycle (Table 1) (Abe et al., 2020; Gu et al., 2018; Gutschner et al., 2016; Lin et al., 2014; Lomova et al., 2019; Wienert et al., 2020). Given that end joining pathways also repair DSBs in the S and G2 phases, inhibition of end joining has been shown to synergistically enhance HDR rates in combination with cell cycle modulation (Maurissen and Woltjen, 2020).
Emerging studies have also highlighted the role of the chromatin context in regulating DSB repair pathway choice. Indeed, Cas9 cleavage efficiency has been shown to correlate positively with chromatin accessibility or transcription of the target sequences (Chen et al., 2017b; Chen et al., 2016; Daer et al., 2017; Horlbeck et al., 2016; Jensen et al., 2017; Schep et al., 2021). It has also been observed that NHEJ-mediated repair of Cas9-induced DSBs is more efficient in euchromatin, while MMEJ and SSTR are more active in certain heterochromatin contexts (Schep et al., 2021). Thus, besides modulating the activity of Cas9, the chromatin context can influence DSB repair pathway balance at Cas9-induced breaks (Chakrabarti et al., 2019; Schep et al., 2021). For example, inhibition of the H3K27 methyltransferase EZH2 enhances NHEJ-mediated DSB repair at H3K27me3-marked heterochromatin domains at the expense of MMEJ (Schep et al., 2021), while chromatinization of dsDNA donors has been reported to stimulate HDR events (Cruz-Becerra and Kadonaga, 2020). Future approaches that manipulate the chromatin context may provide new strategies to obtain specific DNA repair outcomes.
Undesirable DNA Repair Outcomes Associated with Cas9-induced DSBs
DSBs are dangerous DNA lesions that can impair cell survival or lead to mutagenesis of the genome (Burgio and Teboul, 2020; Scully et al., 2019). The consequences of DSB formation include the generation of indels, gross chromosomal rearrangements and p53 activation. The nature of these undesirable outcomes associated with Cas9-induced DSBs are discussed below.
Mutational Consequences of DSB Induction and Repair
Illegitimate and Undesirable DNA Donor Recombination
While HDR is primarily an error-free repair pathway, several lines of evidence suggest that exogenous donors can promote unintended mutagenesis at DSBs (Figure 2, bottom right). For example, dsDNA donors are prone to multiple head-to-tail integrations or concatenation at on- and off-target loci, which is often accompanied by indel formation, suggesting the involvement of NHEJ and MMEJ in these events (Paulis et al., 2015; Roberts et al., 2017; Skryabin et al., 2020; Zelensky et al., 2017). Additionally, inaccurate recombination events occurring using ssODNs have also been described (Boel et al., 2018; Gallagher et al., 2020; Paix et al., 2017; Rivera-Torres et al., 2017). These errors are attributed to mutagenic DNA synthesis during SSTR (Boel et al., 2018; Gallagher et al., 2020; Paix et al., 2017). Furthermore, it has also been shown that cellular cytidine deaminases can trigger the deamination of ssODN bases, resulting in base substitutions in genomic DNA (Lei et al., 2018).
Gross chromosomal rearrangements and large deletions
Gross chromosomal rearrangements (GCRs) include chromosomal translocations, inversions, duplications, deletions, and other complex lesions associated with human cancers (Dahiya et al., 2021). Mounting evidence points toward the induction of complex chromosomal lesions by CRISPR-Cas nucleases (Figure 2, bottom) (Liu et al., 2021a). It was recently shown that DSBs introduced by Cas9 cause the formation of micronuclei and chromosome bridges, which subsequently result in catastrophic genomic rearrangements, such as chromothripsis (Leibowitz et al., 2021). The generation of micronuclei and chromosome bridges has been attributed to the presence of unrepaired Cas9-induced DSBs during cell division (Leibowitz et al., 2021). The formation of chromosomal rearrangements induced by chromothripsis can result from NHEJ, MMEJ and HDR events (Hastings et al., 2009; Ly and Cleveland, 2017; Piazza et al., 2017; Rausch et al., 2012; Stephens et al., 2011). The generation of complex rearrangements is favored by the presence of multiple DSBs induced by site-specific nucleases at on- and off-target sites (Brunet and Jasin, 2018; Frock et al., 2015; Maddalo et al., 2014; Pristyazhnyuk et al., 2019).
Large genomic deletions (>250 bp) at single Cas9-induced DSBs have also been reported in mouse and human cell lines (Figure 2, bottom left) (Adikusuma et al., 2018; Adikusuma et al., 2017; Alanis-Lobato et al., 2021; Cullot et al., 2019; Kosicki et al., 2018; Owens et al., 2019; Shin et al., 2017; Weisheit et al., 2020; Zuo et al., 2017). End resection and MMEJ factors have been implicated in the generation of large deletions (Kosicki et al., 2020; Owens et al., 2019). However, the size distributions of large deletions cannot be predicted by the location of microhomologies alone, thereby implicating multiple pathways in their generation. Genome editing in mouse and human embryos results in a high frequency of large deletions and complex rearrangements causing loss of heterozygosity (Adikusuma et al., 2018; Alanis-Lobato et al., 2021; Egli et al., 2018; Papathanasiou et al., 2021; Zuccaro et al., 2020). The occurrence of these rearrangements has challenged recent observations of inter-homolog homologous recombination (IH-HR) following Cas9 cleavage in human embryos (Ma et al., 2018; Ma et al., 2017a), a process that is thought to be stimulated by RAD51 (Wilde et al., 2021). Indeed, targeting the POU5F1 gene in human embryos caused a high frequency of large deletions that spanned 4 to 20 kb (Alanis-Lobato et al., 2021), while a single Cas9-induced DSB at a pericentromeric locus in human embryos was found to yield segmental and whole chromosome losses (Zuccaro et al., 2020). These studies highlight the risks of DSB-based genome editing in human embryos (Adikusuma et al., 2018; Alanis-Lobato et al., 2021; Egli et al., 2018; Zuccaro et al., 2020).
p53 Activation and Induction of Apoptosis
DSBs activate the tumor suppressor p53, which preserves genome stability by triggering cell cycle arrest, cellular senescence and apoptosis (Bieging et al., 2014). In genome editing applications, DSBs induced by Cas9 have been shown to cause p53-dependent cell cycle arrest in non-transformed human cells (Haapaniemi et al., 2018; Ihry et al., 2018). Surprisingly, mild, but sustained, p53 activation was also observed following expression of Cas9 itself without gRNA (Enache et al., 2020). These studies suggested that genome editing experiments may result in the selection of cell clones with a dysfunctional p53-dependent tumor suppressor response (Enache et al., 2020; Yla-Herttuala, 2018), raising safety concerns for the use of edited cells in ex vivo therapeutic applications. Additional studies have shown that Cas9-induced p53 activation results in reduced proliferation and functional impairment of hematopoietic stem and progenitor cells, which could be partially overcome by transient p53 inhibition (Ferrari et al., 2020; Schiroli et al., 2019). In addition to p53 mutations, KRAS mutations have also been suggested to confer a selective advantage to cells undergoing Cas9-mediated DSB formation (Sinha et al., 2021). The toxicity induced by DSB formation is particularly apparent when high copy number loci or repetitive sequences are targeted (Aguirre et al., 2016; Castanon et al., 2020; Kuscu et al., 2017; Shen and Ideker, 2017). Interestingly, the cellular toxicity associated with targeting repeat sequences in the human genome has been exploited as a kill-switch for Cas9-expressing cells (Castanon et al., 2020).
Future Challenges of DSB-based Genome Editing
As previously discussed, DSB-based genome editing is influenced by the sequence context of the target locus. While the main mutagenic outcome of Cas9-induced DSBs can be predicted (Allen et al., 2019; Chakrabarti et al., 2019; Chen et al., 2019; Leenay et al., 2019; Shen et al., 2018; Shou et al., 2018; van Overbeek et al., 2016), the precise mixture of indel outcomes of end joining pathways remains largely unpredictable. Recent high-throughput technologies to study the effects of genetic perturbations on DNA repair products (i.e., Repair-seq) have shown the high complexity of the repair events occurring at Cas9-induced DSBs and defined the end-processing enzymes responsible for the generation of specific DSB repair products (Hussmann et al., 2021). Future studies to define the influence of chromatin contexts and genetic backgrounds on the identity of the DSB repair products generated by DNA repair enzymes should provide a more comprehensive view of DSB repair events, opening potential avenues to modulate DSB repair for obtaining specific outcomes of interest.
DSB-based genome editing is also affected by the cellular and tissue context. However, there is currently limited understanding of the cell- and tissue-specific regulation of DSB repair events, limiting the implementation of CRISPR technologies for clinical applications (Ferreira da Silva et al., 2021a). In non-dividing cells (e.g., neurons), HDR is non-functional and installation of specific insertions into the genome has only been achieved through integration of DNA donors by end joining pathways (Suzuki and Izpisua Belmonte, 2018; Suzuki et al., 2016). However, due to the high frequency and indiscriminate nature of NHEJ, as well as the low fidelity of MMEJ, off-target integration of dsDNA donors used in such approaches remains a concern. Future work that elucidates the DSB repair processes occurring in terminally differentiated cells is needed to enhance the accuracy of end joining-mediated precision editing.
In proliferating cells, HDR-mediated genome editing is the most precise pathway for DSB-based genome editing, as previously discussed. However, its relatively low frequency remains a major barrier for DSB-based precision genome engineering. DDR modulation is therefore a key strategy for stimulating DSB-based precision genome editing, particularly in contexts (e.g., in vivo editing) that do not allow for the enrichment of cells with desirable edits. As previously discussed, several DSB repair modulating strategies have been designed to stimulate HDR based precision genome editing (Table 1). A comparative approach to evaluate the impact of these strategies on on-target or off-target editing is required to identify the most effective strategy in locus- or cell type-specific contexts. Additionally, future efforts are needed to develop approaches for the delivery of DNA repair modulators for precision genome editing applications in vivo.
SSB-based Genome Editing
SSB-based genome editing approaches rely on the use of CRISPR-Cas nickase mutant proteins. nCas9 proteins carry mutations in either the RuvC (e.g., D10A) or HNH (e.g., H840A, N863A) nuclease domains (Nishimasu et al., 2014). nCas9-based approaches have been utilized to generate single or paired nicks into the genome for end joining-mediated gene disruption or HDR-dependent precision genome editing (Davis and Maizels, 2014; Mali et al., 2013; Ran et al., 2013), as discussed below.
Repair of CRISPR-induced Single Nicks
Single nicks can stimulate HDR-mediated precision genome editing, although with lower efficiency than DSBs (Davis and Maizels, 2014). Distinct from DSBs, nicks are asymmetric lesions that disrupt only one DNA strand. Multiple cellular repair mechanisms can stimulate nick-induced HDR using dsDNA donors or ssODNs (Figure 3, steps 1–7 and 10–13) (Maizels and Davis, 2018; Vriend and Krawczyk, 2017). Repair of nicks by HDR is more efficient on the coding strand relative to the non-coding strand (Davis and Maizels, 2014). Furthermore, nicks are repaired by ssODN-mediated HDR using strand-specific mechanisms (Davis and Maizels, 2014). ssODNs complementary to the nicked strand hybridize to the 3′ end of the nicked target, which then initiates DNA synthesis using the annealed donor as a template through SSTR-like mechanisms (Davis and Maizels, 2014, 2016; Maizels and Davis, 2018) (Figure 3, steps 2–9). On the other hand, ssODNs complementary to the intact DNA strand have been proposed to be directly incorporated into the genome (Davis and Maizels, 2014, 2016; Kan et al., 2017) (Figure 3, steps 10–15).
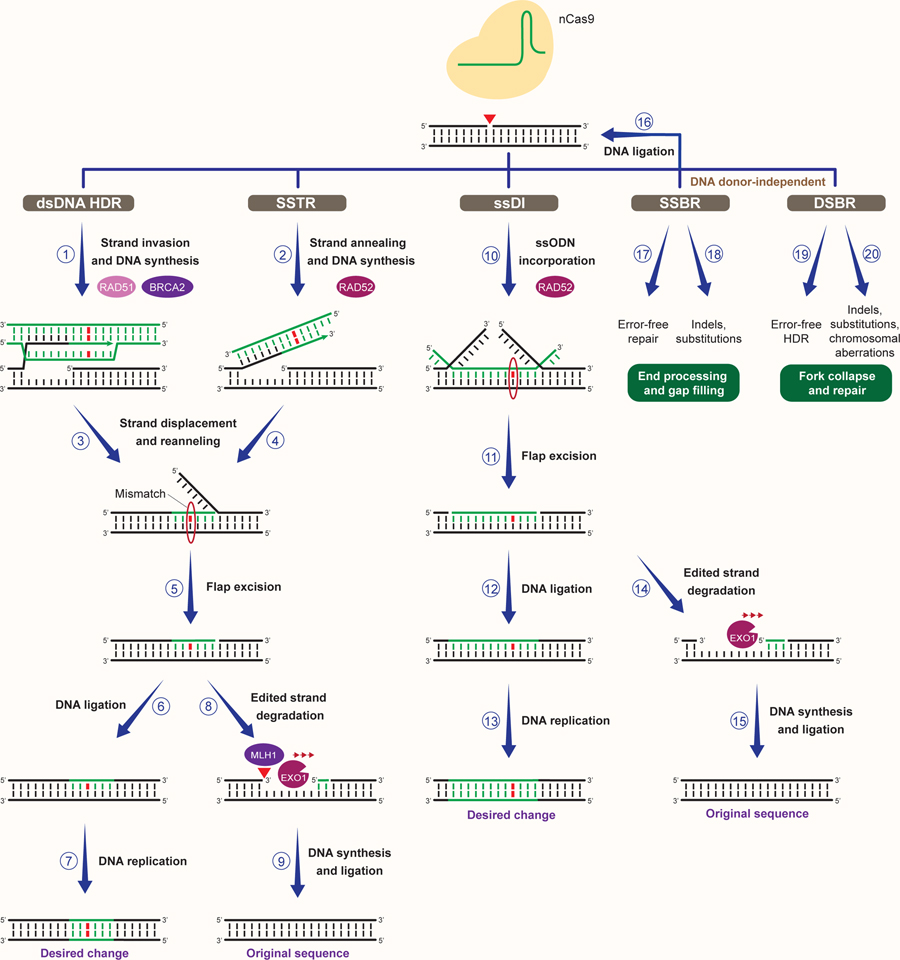
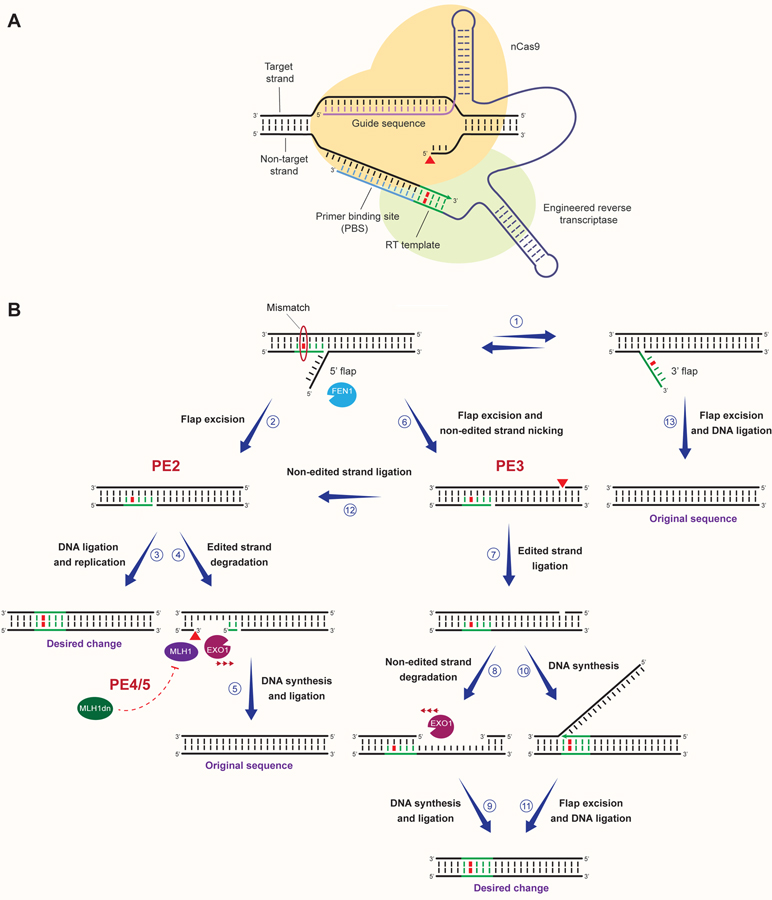
Figure 3. Repair of CRISPR-induced SSBs for precision genome editing.
Multiple RAD51-dependent and -independent cellular mechanisms act at SSBs generated by nCas9 to promote the installment of genomic changes. In the presence of dsDNA donors, dsDNA donor-dependent HDR (dsDNA HDR) can lead to BRCA2- and RAD51-mediated invasion of the 3’ end of the nicked genomic DNA strand into the donor DNA, followed by templated DNA synthesis (1). ssODNs complementary to the nicked genomic DNA strand can instead serve as a template for DNA synthesis following RAD52-dependent ssODN annealing to the nicked strand through a SSTR-like process (2). Displacement of the genomic DNA strand from dsDNA or ssDNA donors and its reannealing to the parental strand can lead to the generation of a 5’ flap and the formation of heteroduplex DNA containing a mismatch between the edited and the parental sequence (3–4). Flap excision by 5’ flap endonucleases, followed by DNA ligation and DNA replication can then lead to the incorporation of the desired change (5–7). Annealing of ssODNs complementary to the non-nicked DNA strand through single-strand DNA incorporation (ssDI) can result in the formation 5’ and 3’ flap structures and the generation of heteroduplex DNA with a mismatch (10). Incorporation of the desired change can then occur upon flap excision by 3’ and 5’ endonucleases, DNA ligation and DNA replication (10–13). Recognition of mismatches between the edited and parental strand by the MMR machinery can cause EXO1-dependent degradation of the edited strand, restoring the original DNA sequence (8–9 and 14–15). Religation of the nCas9-induced nick can also result in the restoration of the original DNA sequence (16). Alternatively, DNA end processing and gap filling by SSB repair (SSBR) can lead to error-free repair or formation of indels and substitutions (17–18). Unrepaired SSBs can be converted into DSBs during DNA replication, resulting in the collapse of replication forks and their subsequent repair by either error-free HDR or error-prone DSB repair (DSBR), which can cause indels, substitutions and chromosomal aberrations (19–20).
The repair of nCas9-induced nicks is controlled by DNA end resection factors and canonical HDR proteins in a manner dependent on the orientation of the nick and the use of dsDNA or ssODN donors (Maizels and Davis, 2018). While dsDNA donor-mediated HDR at nicks requires the RPA complex, BRCA2 and RAD51 (Davis and Maizels, 2014; Vriend and Krawczyk, 2017), ssODN-mediated genome editing at single nicks is inhibited by RAD51 and can occur independently from resection (Bothmer et al., 2017; Davis and Maizels, 2014; Maizels and Davis, 2018; Vriend and Krawczyk, 2017). Interestingly, the fusion of RAD51 mutants to nCas9 results in increased efficiency of ssODN-mediated HDR, while overexpression of wild-type RAD51 decreases ssODN-mediated HDR at nCas9-induced SSBs (Rees et al., 2019). ssODN-mediated HDR occurs with greater efficiency at nicks generated by Cas9 D10A than at Cas9 N863A-induced SSBs, possibly due to different accessibility of the DNA strand cleaved by the two nickase mutants (Bothmer et al., 2017). To further improve the efficiency and accuracy of Cas9 D10A-mediated genome editing, high-fidelity Cas9 D10A variants that display greater specificity and reduced off-targets have been derived (Wang et al., 2021b). Given the potential for accurate genome editing events promoted by Cas9 D10A, more fundamental studies dissecting the cellular mechanisms involved in the repair of nCas9-induced SSBs are needed.
Repair of Paired Nicks Induced by Cas9 Nickases
Paired nCas9 proteins targeting opposite DNA strands with PAM recognition sites facing outward with respect to each other (PAM-out orientation) have been shown to generate DSBs (Bothmer et al., 2017; Ran et al., 2013). Dual nicking by Cas9 D10A and N863A can generate DSBs with 5’ or 3’ overhangs, respectively (Bothmer et al., 2017). Cas9 D10A-induced DSBs with 5’ overhangs are repaired by HDR more efficiently relative to DSBs with 3’ overhangs induced by Cas9 N863A (Bothmer et al., 2017). Given that HDR requires the generation of 3’ ssDNA tails, DSBs with 5’ overhangs necessitate nucleolytic processing to initiate HDR. HDR events at Cas9 D10A-induced DSBs require RAD51 and BRCA2 when a dsDNA template is utilized for repair, while they occur through SSTR-like, RAD51- and BRCA2-independent processes when using ssODNs (Bothmer et al., 2017). DSBs with 3’ overhangs generated by Cas9 N863A have, instead, been shown to undergo preferentially MMEJ-dependent repair, resulting in the generation of insertions (Bothmer et al., 2017). nCas9-induced DSBs can also be repaired through NHEJ (Ran et al., 2013). Distinct from paired nCas9 systems with PAM-out orientation, nCas9 strategies that employ gRNAs with PAM sites facing inward with respect to each other (PAM-in orientation) do not efficiently generate DSBs possibly due to inadequate DNA strand separation, resulting in a low frequency of editing events (Bothmer et al., 2017; Mali et al., 2013; Ran et al., 2013).
Alternative strategies using paired nickases that generate nicks on the same DNA strand of the target site or on both the target site and a dsDNA donor molecule have been developed to promote HDR without the formation of DSBs (Chen et al., 2017a; Davis and Maizels, 2014; Goncalves et al., 2012; Hyodo et al., 2020; Nakajima et al., 2018; Wang et al., 2021b). These nCas9-based strategies have been shown to display greater efficiency than single nick-dependent approaches (Chen et al., 2017a; Davis and Maizels, 2014; Goncalves et al., 2012; Hyodo et al., 2020; Nakajima et al., 2018), and in some instances they were reported to be as efficient as DSB-based strategies (Hyodo et al., 2020).
Applications and Current Limitations of SSB-based Genome Editing Approaches
Paired nCas9 that generate DSBs have been utilized for end joining-mediated gene disruption or gene insertion in human cell lines and mouse zygotes, and for HDR-dependent editing of human cell lines, including human embryonic stem cells (Bothmer et al., 2017; Mali et al., 2013; Ran et al., 2013). These approaches suffer from the same limitations discussed above for DSB-inducing nucleases. However, they display higher specificity and lower indel formation relative to wild-type Cas9-based approaches (Bothmer et al., 2017; Ran et al., 2013).
DSB-independent single and dual nick-based approaches have been utilized for generating gene mutations by HDR in human cell lines, including human embryonic stem cells and iPSCs (Bothmer et al., 2017; Chen et al., 2017a; Davis and Maizels, 2014; Goncalves et al., 2012; Hyodo et al., 2020; Li and Margolis, 2021; Nakajima et al., 2018; Rees et al., 2019; Wang et al., 2021b). nCas9-based approaches have also been employed in human cell lines to induce the contraction of trinucleotide repeats associated to neurological diseases (Cinesi et al., 2016). DSB-independent nick-based strategies do not lead to the activation of p53-dependent responses associated with DSB-inducing nucleases (Hyodo et al., 2020). While nicks are significantly less toxic lesions than DSBs, the introduction of hundreds of nicks into the genome can reduce cellular viability (Smith et al., 2020). Thus, the impact of off-target nicking by nCas9 needs to be further explored, and such studies could benefit from recently developed assays that efficiently capture SSBs in genomes (Cao et al., 2020; Cao et al., 2019; Elacqua et al., 2021; Sriramachandran et al., 2020). In addition, processing of nCas9-induced SSBs can lead to indels and nucleotide substitutions (Figure 3, step 18), which are suppressed by RAD51 and BRCA2 and depend on DNA2-mediated gap extension and Polθ-dependent mutagenic DNA synthesis (Bothmer et al., 2017; Davis and Maizels, 2014; Zhang et al., 2021). In some instances, nicks introduced by genome editing machineries can also be converted into DSBs, causing mutagenic events. For example, when replication forks encounter a nick, they can convert it into a one-ended DSB (Kuzminov, 2001), resulting in replication fork collapse (Figure 3, step 20). Alternatively, endogenous cytidine deaminases can cause DSB formation by generating additional nicks on the opposite strand of the nCas9-induced SSB (Lei et al., 2018). However, single nicks introduced by Cas9 nickases lead to limited formation of indels relative to DSBs generated by wild-type Cas9 or paired nCas9 (Bothmer et al., 2017; Davis and Maizels, 2014; Rees et al., 2019; Zhang et al., 2021), suggesting a reduced contribution of error-prone mechanisms to the repair of nCas9-induced single nicks. Given their overall higher fidelity, SSB-based HDR strategies may prove to be more attractive than DSB-based HDR approaches for precision genome editing applications that require greater accuracy (e.g., clinical applications).
DNA Deamination-mediated Genome Editing
Direct modification of nucleotides in DNA sequences or RNA transcripts can be achieved through base editing (Rees and Liu, 2018). Here, we focus on DNA base editing, which utilizes two main types of editing machineries: adenine base editors (ABE) and cytosine base editors (CBE) (Molla and Yang, 2019; Porto et al., 2020). We refer readers interested in RNA base editing to recent reviews covering this topic (Pickar-Oliver and Gersbach, 2019; Rees and Liu, 2018).
Development and Architecture of Base Editors
Base editors are programmable molecular machines that deaminate base(s) at targeted loci within a window of high activity (Anzalone et al., 2020; Huang et al., 2021b), without introducing DSBs and with limited activation of DDR signaling or induction of apoptosis (Kuscu et al., 2017; Wang et al., 2020c). To achieve potent site-specific and targeted base deamination, base editors harbor multiple modules with distinct functions required for a) localization to desired genomic regions, b) chemical modification of targeted DNA base(s), and c) inhibition of key DNA repair processes (only for CBE). Base editors use the programmable genome search and targeting ability of CRISPR systems to locate and engage the desired targeted base(s). Similar to other CRISPR systems, genomic targeting of base editors requires the formation of a three-stranded RNA-DNA structure (R-loop) between the gRNA and the genomic sequence complementary to the gRNA (Figure 4) (Lapinaite et al., 2020). Base editors also contain natural or engineered ssDNA-specific deaminases that modify bases within ssDNA exposed by the R-loop structure, thereby exhibiting activity on a restricted window of accessible genomic bases. In particular, CBE consists of a cytidine deaminase (e.g., APOBEC1/3, AID or CDA) fused to a dCas9 or a nCas9 with an active HNH nuclease domain (Figure 4) (Hess et al., 2016; Komor et al., 2016; Ma et al., 2016; Nishida et al., 2016). Cytidine deaminases play critical roles in promoting mutagenesis during innate and adaptive immunity (Conticello et al., 2007), and are also responsible for a significant fraction of the genomic mutations observed in human cancers (Buisson et al., 2019; Burns et al., 2013; Nik-Zainal et al., 2012; Roberts and Gordenin, 2014). CBEs divert the mutagenic activity of ssDNA-specific cytidine deaminases toward genomic bases of interest within the R-loop. Since natural enzymes that deaminate adenosine in ssDNA are not currently known, ABEs were derived as a result of protein engineering and directed evolution experiments. In particular, Gaudelli et al. evolved the prokaryotic tRNA-specific adenosine deaminase TadA (Wolf et al., 2002) to acquire a deoxyadenosine deaminase activity that can operate on ssDNA (Figure 4) (Gaudelli et al., 2017). Of note, CBEs, but not ABEs, harbor an additional module that functions as a uracil DNA glycosylase inhibitor (UGI). UGI is a DNA mimic peptide isolated from PBS2, a unique bacteriophage that has a genome rich in uracil instead of thymine residues (Takahashi and Marmur, 1963). UGI potently inhibits UNG, the major DNA glycosylase involved in the removal of uracil from DNA (Wang and Mosbaugh, 1989). During base editing, UNG inhibition allows uracil intermediates generated by CBE to persist in genomic DNA, enabling the subsequent engagement of DNA repair pathways that lead to base substitution, as discussed below.
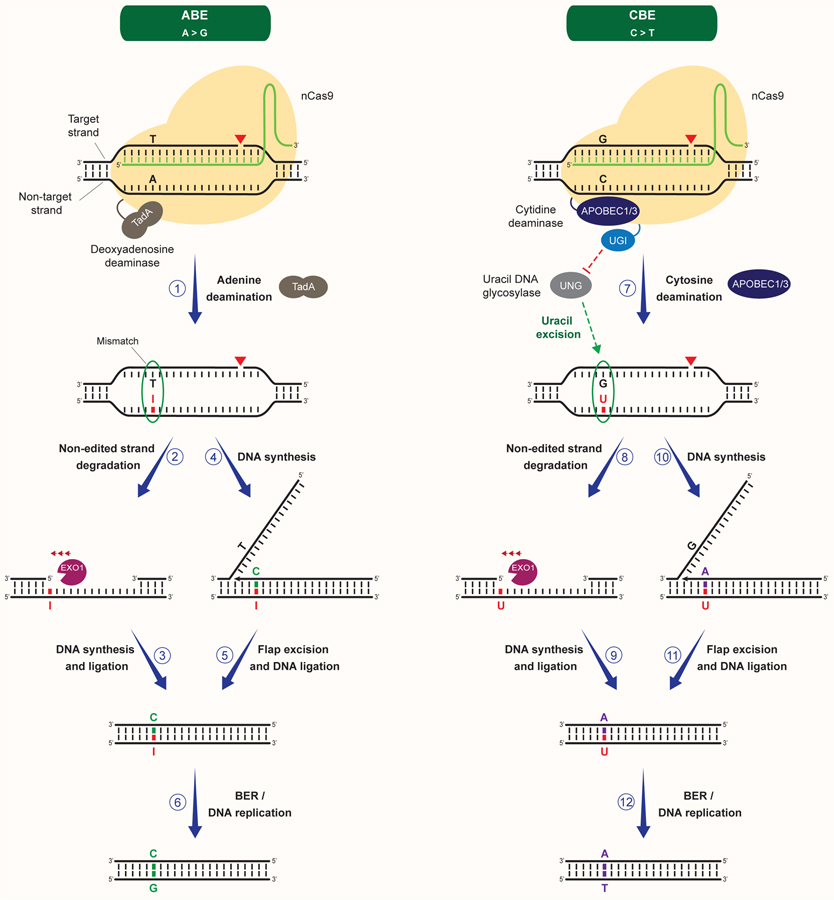
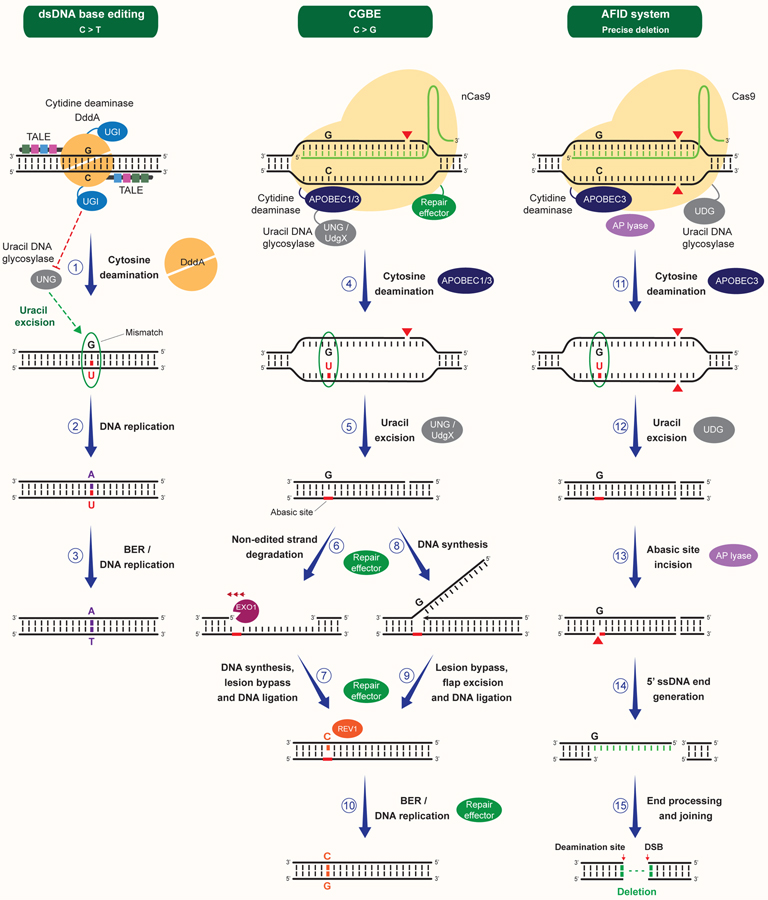
Figure 4. Repair of site-specific base lesions generated by canonical base editors.
Generation of site-specific base transitions by the canonical base editors ABE and CBE. Traditional ABE is constituted of a fusion of nCas9 to the TadA deoxyadenosine deaminase. ABE promotes the deamination of adenines located within the ssDNA of the R-loop generated upon gRNA pairing to the targeted DNA strand (1). Adenine deamination leads to the formation of inosine and the generation of a I:T mismatch, which can be recognized by the MMR machinery (1). nCas9-dependent nicking of the non-edited strand can promote its degradation by EXO1 and its subsequent resynthesis using the edited strand as a template, resulting in the incorporation of a cytosine opposite to inosine (2–3). Alternatively, replacement of the T with a C can occur upon DNA synthesis initiated by the 3’ end of the nicked non-edited strand, followed by strand displacement, excision of the resulting 5’ flap and DNA ligation (4–5). These events can occur in a MMR-dependent manner or result from long-patch repair of the nCas9-induced SSB. T to C substitution can also occur following religation of the nicked strand and DNA replication (not shown). Subsequent DNA replication or BER can lead to the replacement of the I with a G and the generation of a A:T->G:C transition (6). Distinct from ABE, CBE is traditionally constituted of a fusion of nCas9 to the cytidine deaminase APOBEC1/3, which catalyzes the deamination of cytosines into uracils within the ssDNA of the gRNA-containing R-loop (7). CBE inhibits uracil excision by UNG through a UGI peptide fused to nCas9, resulting in the persistence of a G:U mismatch, which can be recognized by MMR proteins (7). Similar to ABE, CBE then catalyzes the nicking of the non-edited strand, favoring its degradation or its displacement and excision, followed by the replacement of the G with an A upon DNA synthesis (7–11). G to A substitution can also occur upon DNA replication (not shown). Subsequent DNA replication or BER can lead to the replacement of the U with a T and the generation of a C:G->T:A transition (12).
Mechanisms of Base Editing-dependent Lesion Repair
Base deamination can result from exposure to environmental DNA damaging agents, occur spontaneously upon DNA decay, or be induced by the action of cellular deaminases and base editors. Hydrolytic deamination of the exocyclic amine of adenine or cytosine yields the non-canonical inosine or uracil bases, respectively (Alseth et al., 2014; Nabel et al., 2012). During DNA replication, inosine is read by DNA polymerases as guanine, generating I:T to I:C substitutions, while uracil is read as thymine, causing U:G to U:A changes.
Modified bases are normally detected, excised and replaced by the BER pathway (Beard et al., 2019; Caldecott, 2020). BER is initiated by specialized DNA glycosylases, which catalyze the cleavage of the glycosidic bond between the nitrogenous base and the deoxyribose sugar of specific damaged nucleotides, thereby generating an abasic site (also known as an apurinic/apyrimidinic site or AP site) (Krokan and Bjoras, 2013). Abasic sites are among the most prevalent DNA lesions in the genome, and it is estimated that ~10,000 abasic sites are formed in every cell of the human body each day (Barnes and Lindahl, 2004). If left unrepaired, abasic sites threaten genomic integrity, as they can interfere with DNA replication and transcription (Thompson and Cortez, 2020). In mammalian cells, uracil DNA glycosylases, such as UNG and SMUG1, prevent genomic instability by removing uracils. UNG, the main uracil DNA glycosylase, has two isoforms, UNG1 and UNG2, that remove uracil from mitochondrial and nuclear DNA, respectively (Krokan and Bjoras, 2013). Distinct from uracil, inosine is thought to be excised from DNA by the AAG glycosylase (Alseth et al., 2014). Abasic sites generated by DNA glycosylases are subsequently processed by AP site DNA lyases (AP lyases), such as APEX1 or APEX2, with help from other BER factors, such as PARP1 and XRCC1 (Beard et al., 2019). XRCC1 acts as scaffold for BER factors and regulates PARP1, preventing BER misregulation and genomic instability (Demin et al., 2021; Lindahl, 2000). APEX1 is the main AP lyase that initiates abasic site repair by incising the phosphate backbone on the 5’ side of the abasic site to create a SSB (Beard et al., 2019). De novo DNA synthesis by DNA polymerases then fills the remaining gap using the undamaged strand as a template. Two main SSB repair pathways that differ in the length of DNA synthesis promote the filling of gaps resulting from the processing of DNA base or backbone lesions: short-patch repair promotes the synthesis of a single nucleotide by the DNA polymerase β, while long-patch repair leads to the synthesis of 2–12 nucleotides catalyzed by the DNA polymerases β, δ or ε (Caldecott, 2014). During long-patch repair, a 5ʹ ssDNA flap structure is generated by strand displacement DNA synthesis and then subsequently resolved by the FEN1 endonuclease (Prasad et al., 2001). Finally, DNA ligase I or III seals the remaining nick in the phosphate backbone, restoring the integrity of the double helix (Cappelli et al., 1997; Prasad et al., 1996).
Excision of inosine and uracil by the BER pathway reestablishes the original sequence and, therefore, counteracts the activity of base editors. Consequently, efficient base editing requires inhibiting BER to stabilize the modified base(s). As discussed above, in the case of cytosine deamination by CBE, this effect is achieved by inhibiting UNG using the UGI peptide (Figure 4). Interestingly, the fusion of one or multiple UGI peptides to CBEs and the ectopic expression of UGI enhance base conversion efficiency by blocking BER initiation (Komor et al., 2016; Komor et al., 2017; Wang et al., 2017). In ABE experiments, strategies to inhibit inosine excision are not required, given the limited rate of inosine removal by cellular glycosylases. Indeed, it was shown that cells lacking AAG or expressing fusions of ABE to inactive forms of human AAG or E. coli EndoV to block cellular AAG from accessing inosine do not improve ABE-mediated editing efficiency (Gaudelli et al., 2017). These results suggest that AAG is inefficient at excising inosine from genomic DNA, possibly because inosine is not frequently found in genomic DNA, given the absence of cellular deoxyadenosine deaminases. Consequently, ABE-mediated deoxyadenosine deamination leads to limited engagement of BER, thus yielding base editing products of high purity.
The U:G or I:T mismatches generated by base editors can be recognized and resolved by MMR (Figure 4, steps 1–3 and 7–9), a pathway that corrects errors introduced by DNA polymerases during DNA replication (Sanders et al., 2021). MMR removes mismatches by catalyzing the degradation of the newly synthesized DNA strand containing the incorrect base and restoring the correct sequence using the parental DNA strand as a template (Jiricny, 2013). To repair mismatches, MMR must distinguish the correct from the incorrect DNA strand. In eukaryotes, strand discrimination is achieved through the recognition of SSBs and gaps occurring on the newly synthesized DNA strand during DNA replication (Cortez, 2019; Hsieh and Zhang, 2017; Kunkel and Erie, 2015; Schanz et al., 2009). SSBs and gaps on the 5’ side of the mismatch provide entry points for EXO1-mediated DNA degradation, resulting in the processing of the newly synthesized DNA strand and the removal of the incorrect DNA base (Constantin et al., 2005; Goellner et al., 2015; Zhang et al., 2005). Gap filling by the replicative DNA polymerases δ or ε, followed by ligation by DNA ligase I, then restores DNA integrity. To promote the removal of the non-edited base of the mismatch by MMR, CBE and ABE fusions containing nCas9 introduce a nick on the 5’ side of the mismatch on the non-edited strand, thus favoring its degradation by EXO1 (Figure 4, steps 2 and 8) (Gaudelli et al., 2017; Gu et al., 2021; Komor et al., 2016). DNA synthesis can then result in the incorporation of an A opposite to U or a C opposite to I (Figure 4, steps 3 and 9). This process can also occur through a round of DNA replication or following DNA synthesis initiated by the 3’ end of the nick, accompanied by the displacement of the non-edited strand and the excision of the resulting 5’ flap (Gu et al., 2021) (Figure 4, steps 4–5 and 10–11). Nick-induced DNA synthesis can take place upon recognition of the mismatch by the MMR machinery or result from long-patch repair of the nCas9-induced SSB. Subsequent DNA replication or BER would then replace the U and I with T and G, respectively, leading to C:G->T:A and A:T->G:C base transitions (Figure 4, steps 6 and 12) (Gaudelli et al., 2017; Komor et al., 2016; Nishida et al., 2016).
Although base editing can also yield byproducts, such as indels and transversion mutations, these outcomes are generated with relatively low frequency at the targeted locus. Indels originate from end joining-mediated repair of DSBs (Koblan et al., 2021a), likely resulting from nCas9-mediated nicking of the non-edited strand and APEX1-dependent nicking of the edited strand at abasic sites generated upon removal of the edited bases by DNA glycosylases. Additionally, DNA synthesis across abasic sites by error-prone translesion synthesis (TLS) polymerases can lead to the insertion of incorrect bases, thereby generating transversion mutations (Koblan et al., 2021a). Indeed, by modulating the formation of BER intermediates during base editing, non-canonical base editors that generate predictable transversion mutations and small deletions have recently been developed.
Generation of Transversion Mutations and Precise Deletions using Non-Canonical Base Editors
As discussed above, canonical base editors generate transition mutations, with transversion mutations occurring occasionally as byproducts. Although the generation of transversion mutations by base editors can be predicted from machine learning models (Arbab et al., 2020; Marquart et al., 2021), it remains infrequent and context-specific. Importantly, genetic deletion of UNG or addition of multiple UGI peptides to base editors leads to a reduced formation of transversion mutations, suggesting that these events might result from the accumulation of abasic sites due to enhanced removal of uracil (Komor et al., 2016; Komor et al., 2017; Kurt et al., 2021).
In mammalian cells, abasic sites can be shielded from DNA processing enzymes through crosslinking to HMCES (Mohni et al., 2019). Alternatively, abasic sites can be bypassed using HDR or TLS (Thompson and Cortez, 2020). In particular, the TLS polymerase REV1, which exhibits an intrinsic dCMP transferase activity, incorporates cytosines across abasic sites, thereby leading to C:G->G:C transversion mutations following cytosine deamination and uracil removal (Chan et al., 2013; Kim et al., 2011; Lin et al., 1999; Nelson et al., 1996). Accordingly, loss of REV1 was reported to decrease the formation of C:G->G:C transversions during AID-mediated somatic hypermutation in mice (Jansen et al., 2006). Therefore, stimulation of REV1-mediated DNA synthesis across abasic sites could potentially increase the frequency of C:G->G:C transversions. These findings suggest that stabilization of abasic sites generated from uracil excision may favor error-prone DNA repair events and enhance the generation of transversion mutations by base editors. Based on these observations, novel base editors that exploit the repair of abasic sites have been developed to generate C:G->G:C transversions (Figure 5, steps 4–10). These non-canonical base editors, named CGBEs for “C-to-G base editors”, consist of a modified CBE in which the UGI has been removed or replaced by a uracil DNA glycosylase, such as UNG (Kurt et al., 2021; Zhao et al., 2021b) or UdgX (Koblan et al., 2021a). In certain CGBEs, nCas9 has also been fused to DNA repair effectors, such as the BER factors XRCC1, DNA ligase III and DNA polymerase β, RBMX, EXO1, or POLD2 (Chen et al., 2021a; Koblan et al., 2021a). The observation that ABE catalyzes the deamination of cytosine in a restricted sequence context (TC motifs) in human cells (Kim et al., 2019) has also led to the derivation of an ABE variant with increased C:G to G:C editing at TC sequences (Jeong et al., 2021). Therefore, a set of CGBEs displaying complementary features based on sequence context preference, editing purity and efficiency are currently available for genome editing studies.
Figure 5. Repair of site-specific base lesions generated by non-canonical base editors.
Modification of dsDNA and generation of transversion mutations or predictable deletions by non-canonical base editors (DddA, CGBE and AFID system). dsDNA-dependent base editing in the mitochondrial genome can be obtained using a split DddA deaminase fused to TALE arrays, which provide site-specific DNA binding. Reconstituted DddA deaminates cytosines located at the TALE DNA binding site, generating uracil and inhibiting its excision through a UGI peptide fused to DddA (1). The resulting G:U mismatch can then be resolved through DNA replication and BER events, leading to the generation of a C:G->T:A transition (2–3). CGBE enzymes are derived from CBE, whereby the UGI peptide has been removed or replaced with the uracil DNA glycosylase UNG or UdgX. Similar to CBE, CGBE promotes site-specific cytosine deamination and nicks the non-edited strand (4). However, in the case of CGBE, the uracil is then excised by uracil DNA glycosylases, resulting in the formation of an abasic site (5). The non-edited strand can then be degraded by EXO1 and undergo resynthesis by the replicative DNA polymerases (6–7). Upon encountering the abasic site, the replicative DNA polymerases are replaced by translesion DNA polymerases, such as REV1, which can insert a cytosine opposite to the abasic site (7). Insertion of a C opposite to the abasic site can also occur upon DNA synthesis initiated by the 3’ end of the nicked non-edited strand, followed by the bypass of the abasic site by translesion DNA polymerases and the excision of the 5’ flap resulting from strand displacement DNA synthesis (8–9). DNA replication or abasic site repair by AP lyases and other BER enzymes can then lead to the replacement of the abasic site with a G, resulting in a C:G->G:C transversion (10). The generation of transversion mutations is stimulated by DNA repair effectors fused to CGBE (6–10). AFID systems are based on a fusion of Cas9 to APOBEC3 and a bacterial uracil DNA glycosylase. Similar to CGBE, AFID systems promote cytosine deamination into uracil, which is then excised by the uracil DNA glycosylase, generating an abasic site (11–12). However, distinct from CGBE, AFID systems introduce a DSB near the PAM sequence and express an AP lyase, which introduces a nick at the abasic site, resulting in the generation of a DNA end with a 5’ ssDNA tail (13–14). Degradation of the 5’ ssDNA tail and DSB repair by NHEJ results in predictable deletions spanning from the deaminated cytosine to the site of the Cas9-induced DSB (15).
Recently developed non-canonical base editing approaches also include the APOBEC-Cas9 fusion-induced deletion (AFID) system, which introduces and processes uracil bases to generate small predictable targeted deletions (Wang et al., 2020b). The AFID system combines a Cas9 nuclease, a cytidine deaminase, a uracil DNA glycosylase, and an AP lyase in a single construct (Figure 5, steps 11–15). The target base(s) is deaminated into uracil(s) by the cytidine deaminase. The uracil DNA glycosylase then generates an abasic site, which is subsequently nicked by the AP lyase. Additionally, Cas9 cuts the two strands on the 3’ side of the nick, thus exposing the ssDNA region in between the nick and the DSB. These steps generate two non-compatible DNA ends (blunt end and ssDNA-containing end), requiring nucleolytic degradation of the ssDNA region before subsequent blunt ligation of the two ends by NHEJ. Therefore, AFID systems generate small deletions spanning from the deaminated C to the Cas9 cleavage site (Wang et al., 2020b). Improvements in the prediction of cytosine deaminase activity based on the sequence context, combined with the use of Cas9 PAM variants appropriate for the locus of interest, should enable a more precise generation of desired small deletions, which would be particularly useful for generating in-frame protein deletion mutants. Additionally, the accuracy of the deletions generated by AFID could be further improved by modulating the activity of the DNA repair factors fused to Cas9. Future work will be required to optimize the architecture of non-canonical base editors to maximize the generation of C:G->G:C transversions and small deletions and enhance product purity in different cellular systems.
Base Editing on dsDNA
Canonical base editors are unable to edit bases in the context of dsDNA due to the ssDNA-specific activity of cytidine and adenosine deaminases. Furthermore, since RNA molecules cannot enter mitochondria, the mitochondrial genome is not a substrate for conventional CBE and ABE editors (Gammage et al., 2018). However, recent studies have shown that DddA, a toxin secreted by bacteria to induce replication stress and genomic instability in neighboring bacteria, displays cytidine deaminase activity on dsDNA (de Moraes et al., 2021). To determine whether DddA could be used for base editing on dsDNA, its toxic deaminase domain was split into two non-toxic halves and fused to two TALE arrays, which enables the in situ reconstitution of a functional cytidine deaminase domain upon DNA binding (Mok et al., 2020). The resulting DddA-derived cytosine base editor (DdCBE) was shown to efficiently convert C:G to T:A in nuclear dsDNA of mammalian cells, and similar editing events were observed in mitochondrial dsDNA following the fusion of DdCBE to a mitochondrial-targeting signal (Mok et al., 2020) (Figure 5, steps 1–3). Although lacking DSB repair, mitochondria possess functional BER and MMR pathways that enable the repair of base lesions generated by base editing (Fu et al., 2020). Accordingly, fusion of one or two UGI peptides to DdCBE was shown to enhance its editing efficiency and decrease base editing byproducts, probably through inhibition of UNG1. Besides its use for editing the mitochondrial genome in mammalian cells, DdCBE has also been successfully exploited to modify mitochondria in mice and zebrafish embryos (Lee et al., 2021; Sabharwal et al., 2021). The use of DdCBE has enabled the direct manipulation of mitochondrial DNA (mtDNA) in living systems with single-base resolution without requiring the formation of R-loop structures necessary for the activity of ssDNA-specific deaminases and in the absence of mitochondrial DSBs, which cause mtDNA degradation and potent induction of innate immune signaling events (Tigano et al., 2021). The development of DdCBE is a prime example of how mining bacterial genomes can lead to the discovery of unexpected enzymatic activities that can be harnessed to create new genome editing tools. Further advancements in the generation of base editing tools for mtDNA will provide novel opportunities to correct pathogenic mtDNA variants that cause mitochondrial disorders (Murphy and Hartley, 2018) and will enable a better understanding of mtDNA repair mechanisms.
Applications of Canonical and Non-Canonical Base Editors
Base editors have been successfully applied to introduce targeted genomic modifications in many cellular and animal models (Molla and Yang, 2019). Furthermore, base editing has allowed investigators to model and correct genetic variants in both dividing and non-dividing cells (Billon et al., 2020; Gaudelli et al., 2017; Komor et al., 2016; Newby and Liu, 2021; Yeh et al., 2018), inactivate genes through the insertion of premature STOP codons (Billon et al., 2017; Kuscu et al., 2017) or splice site mutations (Gapinske et al., 2018; Kluesner et al., 2021; Yuan et al., 2018), disrupt multiple loci without the formation of undesired chromosomal translocations and mosaicism (Kim et al., 2017a; Webber et al., 2019), and study coding and non-coding nucleotide variants at scale (Cheng et al., 2021; Cuella-Martin et al., 2021; Després et al., 2020; Hanna et al., 2021; Hess et al., 2016; Huang et al., 2021a; Jun et al., 2020; Kweon et al., 2020; Li et al., 2018a; Ma et al., 2016; Sangree et al., 2021; Xu et al., 2021a). With all three modes of currently available base editors, CBE (C:G->T:A), ABE (A:T->G:C), and CGBE (C:G->G:C), a complete set of mutation types can be theoretically obtained by multistep editing (Zhao et al., 2021b). For example, the target C of C:G can be initially edited into U by CBE (U:G), and U:G can in turn be converted into T:A by cellular DNA replication and repair mechanisms. Alternatively, C:G can be edited into G:C by CGBE, followed by CBE-mediated conversion of the nascent G:C pair into A:T. This strategy could then convert a targeted C into T (CBE), G (CGBE), or A (CGBE + CBE). Similarly, A can be edited into G (ABE), C (ABE + CGBE), or T (ABE + CGBE + CBE). However, multi-step editing is difficult to achieve at high efficiency and specificity due to the limitations of base editing discussed below. Given its ability to precisely induce base transition mutations at single-nucleotide resolution, base editing has been employed to generate new cellular and animal models of cancer and various genetic syndromes (Annunziato et al., 2020; Rosello et al., 2021; Ryu et al., 2018; Zafra et al., 2018). Especially promising is the prospect of using base editing to revert disease-causing mutations (Doudna, 2020; Porto et al., 2020). For example, base editing has already been applied to rescue animal models of Hutchinson-Gilford progeria syndrome, sickle cell disease, tyrosinemia, Duchenne muscular dystrophy, phenylketonuria, and deafness, and to reduce cholesterol levels in mice and primates (Koblan et al., 2021b; Levy et al., 2020; Musunuru et al., 2021; Newby et al., 2021; Rossidis et al., 2018; Rothgangl et al., 2021; Ryu et al., 2018; Song et al., 2020; Villiger et al., 2018; Yeh et al., 2020). These remarkable pre-clinical results bode well for future applications of base editing in the treatment of many human diseases.
Current Limitations of Base Editing Approaches
Despite its high efficiency, base editing is restricted to a window of high deaminase activity (~ 5–8 bp for Cas9-based CBE and ABE) located at a specific position from the PAM sequence. Furthermore, the editing outcome and efficiency of base editing can be affected by the sequence context of the targeted locus. The presence of multiple editable bases within the activity window — and occasionally outside of the window — can result in undesired bystander mutations. Although prediction methods for the outcome and efficiency of base editing guides have been developed based on studies in HEK293 and mouse ES cells (Arbab et al., 2020; Marquart et al., 2021; Wang et al., 2021c), these methods require further refinement through studies in other cellular systems.
To overcome the above limitations, base editors have been extensively engineered for improving their targeting scope, efficiency and specificity (Anzalone et al., 2020; Arbab et al., 2020; Fu et al., 2021; Gaudelli et al., 2020; Gehrke et al., 2018; Grunewald et al., 2020; Hu et al., 2018a; Jeong et al., 2021; Kim et al, 2017b; Kleinstiver et al., 2019; Koblan et al., 2018; Li et al., 2018b; Richter et al., 2020; Sakata et al., 2020; Walton et al, 2020; Wang et al., 2021a; Wang et al., 2018; Zafra et al., 2018; Zhang et al., 2020a; Zhang et al., 2020b). These studies have led to the development of a large set of base editors with distinct PAM requirements and editing windows, higher on-target editing efficiency and decreased generation of off-target events. In addition, to generate concomitant A:T->G:C and C:G->T:A changes, base editors with both ABE and CBE activities have been developed (Grunewald et al., 2020; Li et al., 2020a; Sakata et al., 2020; Zhang et al., 2020b). However, there are currently no tools available to simultaneously introduce combinatorial transition and transversion mutations. It is expected that future developments will provide a greater array of base editing tools with multiple distinct features, allowing for even more precise genomic changes, including combinations of selected mutations, in the locus and cell type of interest. At the same time, given that the SSBs and abasic sites generated during base editing can potentially interfere with DNA replication and transcription, additional studies of the impact of base editors on genomic stability in the cell type of interest would be especially important.
Error-prone DNA Synthesis-based Genome Editing
DNA synthesis promoted at loci of interest enables the rewriting of genomic DNA sequences for precision genome editing. As discussed above, HDR-mediated genome editing strategies entail the copying of a synthetic donor template by cellular DNA polymerases at sites of DSB or SSB formation. Additionally, certain non-canonical base editors exploit error-prone TLS polymerases to insert cytosines across abasic sites and generate transversion mutations. Recent studies have successfully employed a nCas9 fusion to PolI3M, an E. coli DNA polymerase PolI mutant engineered for low-fidelity, to initiate DNA synthesis from nCas9-induced nicks in the genomes of both bacteria and yeast (Halperin et al., 2018; Tou et al., 2020). Although this system, known as EvolvR, is efficient in generating a spectrum of transition and transversion mutations, the identity of the mutations introduced is not predictable. Furthermore, it remains to be established whether EvolvR, or a similar approach that relies on nCas9-fusions to other error-prone DNA polymerases, can be successfully applied to mammalian systems.
Reverse transcription-based Genome Editing
Mechanism of Prime Editing
Reverse transcriptases synthesize complementary DNA strands from an RNA template, transferring genetic information from RNA to DNA molecules (Martin-Alonso et al., 2021). Recent studies have exploited the activity of a CRISPR-based engineered reverse transcriptase paired with a multifunctional prime editing gRNA (pegRNA) to introduce sequences of interest into the genome without DSB intermediates or exogenous DNA templates (Anzalone et al., 2019). This technology, known as prime editing, leverages the possibility to extend the 3’ end of gRNAs with sequences of interest and exploits the ability of nicked DNA strands to prime DNA synthesis by reverse transcriptases when annealed to RNA templates of interest. Prime editors are composed of a modified Moloney murine leukemia virus (M-MLV) reverse transcriptase conjugated to nCas9 (Figure 6A). The pegRNA carries a reverse transcription template that encodes the desired sequence to be incorporated in the genome, a primer binding site (PBS) to prime the reverse transcriptase, and a regular gRNA sequence for nCas9-mediated genomic targeting. The nick introduced by nCas9 (HNH mutant) on the non-target strand enables pairing of the generated 3’ ssDNA end with its complementary sequence located within the PBS. This DNA:RNA heteroduplex primer initiates RNA-templated DNA synthesis by the reverse transcriptase, resulting in the extension of the non-target DNA strand. Strand extension produces a 3’ flap structure containing the desired modifications. The edited flap competes for base pairing with the cognate target strand containing the original sequence, generating an equilibrium between a 3’ flap containing the edited sequence and a 5’ flap lacking the edited sequence (Figure 6B, step 1). The observation that prime editing can be applied successfully in both yeast and mammalian cells implies that the machinery that resolves prime editing intermediates is conserved among eukaryotes (Anzalone et al., 2019). Indeed, DNA flaps are structures frequently processed during DNA replication and repair. In particular, 5’ flap structures can be removed by the FEN1, EXO1, and DNA2 nucleases, while 3’ flaps are generally cleaved by endonucleases of the XPF family (Balakrishnan and Bambara, 2013; Ciccia et al., 2008; Sertic et al., 2020; Zheng et al., 2020). Given the equilibrium between 5’ and 3’ flaps in prime editing intermediates, editing outcomes vary depending on which flap is excised, such that 3’ edited flap cleavage restores the original genomic sequence, while 5’ flap excision leads to incorporation of the edited sequence (Figure 6B, steps 2 and 13). Consistently, knockdown of the 5’ flap endonuclease FEN1 reduces the frequency of intended edits in prime editing experiments (Chen et al., 2021b). Furthermore, given the presence of a nick on the edited strand, the MMR machinery is targeted to that DNA strand to promote EXO1-mediated degradation of the mismatch generated by prime editing and restore the original sequence (Figure 6B, steps 4–5). Thus, loss of MMR proteins or EXO1, or inhibition of the MMR pathway stimulates the efficiency of prime editing (Chen et al., 2021b; Ferreira da Silva et al., 2021b). Installing silent mutations in the vicinity of the edit of interest can also increase prime editing efficiency by weakening the recognition of the edited mismatch by the MMR machinery (Chen et al., 2021b). Moreover, prime editing efficiency can be stimulated by introducing a nick on the non-edited strand with a second gRNA, which can favor EXO1-mediated degradation of the non-edited strand or promote its displacement and excision, accompanied by DNA resynthesis using the edited strand as a template (Figure 6B, steps 6–11). Alternatively, enhanced installment of precise insertions or deletions can also be achieved by using paired pegRNAs that generate complementary DNA sequences on both DNA strands, thus limiting the formation of DNA mismatches (Figure 7A–B) (Anzalone et al., 2021; Choi et al., 2021b; Jiang et al., 2021; Lin et al., 2021; Zhuang et al., 2021).
Figure 6. Repair of DNA lesions generated by prime editing.
A) Modules that constitute prime editors. Prime editors consist of the fusion of an engineered reverse transcriptase (RT) to nCas9. The reverse transcriptase utilizes an RNA:DNA heteroduplex formed upon the annealing of the nicked non-target DNA strand to a primer binding site (PBS) in the pegRNA. The 3’ end of the nicked DNA strand is then extended by the reverse transcriptase using the pegRNA sequence containing the desired edits as template (RT template). B) Processing of DNA intermediates generated by prime editors. Prime editor-mediated reverse transcription leads to the generation of a 3’ flap containing the edit, in equilibrium with a 5’ flap that does not contain the edit (1). Formation of the 5’ flap is accompanied by the generation of heteroduplex DNA containing a mismatch between the edited strand and the parental strand (1). In PE2 prime editing systems, excision of the 5’ flap by FEN1 can be followed by resolution of the mismatch upon ligation of the nicked strand and DNA replication, leading to the incorporation of the desired change (2–3). Recognition of the mismatch by the MMR machinery can lead to MLH1- and EXO1-dependent degradation of the nicked edited strand, restoring the original sequence (4–5). In PE3 prime editing systems, introduction of a nick on the 5’ side of the mismatch using a second gRNA can engage the MMR machinery on the non-edited strand and promote its degradation by EXO1, resulting in the incorporation of the desired change following DNA synthesis (6–9). Incorporation of the edits of interest in PE3 systems can also occur upon DNA synthesis initiated by the 3’ end of the nicked non-edited strand, followed by strand displacement and excision of the resulting 5’ flap (10–11). Engagement of the MMR machinery on the nicked edited strand can lead to the restoration of the original sequence also in PE3 systems (12, 4–5). To suppress MMR-mediated restoration of the original sequence, the PE4 and PE5 prime editing systems combine respectively PE2- and PE3-based approaches with the transient expression of a dominant negative MLH1 protein (MLH1dn) (4). Excision of the 3’ flap containing the edited sequence results in MMR-independent restoration of the original sequence (13).
Figure 7. Processing of DNA intermediates induced by paired prime editors to generate precise substitutions, insertions and/or deletions.
A) Design of paired prime editing strategies. A pair of prime editors is targeted to opposite DNA strands to generate two 3’ flaps with complementary sequences and desired modifications. B) Mechanisms for the generation of precise substitutions, insertions and/or deletions using paired prime editors. Substitutions (left) can be obtained using paired prime editors that generate two complementary 3’ flaps containing the desired edit(s) (green) (1). Annealing of the two edited 3’ flaps and the two corresponding 5’ flaps containing the parental DNA sequence, followed by excision of the annealed 5’ flaps and ligation of the nicked strands, can then lead to the incorporation of the desired substitution(s) (2–3). Precise insertions and deletions (middle) can be induced using paired prime editors that generate two 3’ flaps that contain complementary (purple) and non-complementary (lighter and darker purple) sequences of the desired insert adjacent to the genomic sequence to be replaced (4). Annealing of the two edited 3’ flaps and the two parental 5’ flaps, followed by excision of the annealed 5’ flaps, filling of ssDNA gaps within the insert sequence and DNA ligation, can lead to the replacement of the genomic sequence of interest with the desired insert (5–6). Deletions (right) can be obtained using paired prime editors that generate 3’ flaps (blue and red) complementary to genomic sequences upstream and downstream of the generated flaps (blue and red) (7). Annealing of the 3’ flaps with their complementary genomic sequences, followed by excision of the corresponding annealed 5’ flaps and ligation of the nicked strands, can result in the deletion of the genomic sequence located between the two 3’ flaps (8–9). Paired prime editors can be utilized for the insertion of serine recombinase sites (e.g., attP or attB sites), enabling site-specific integration of gene-size fragments upon transfection of donor gene constructs and expression of serine recombinases (e.g., Bxb1) (10). Site-specific integration can also be obtained using a prime editor fused to Bxb1 (not shown).
Prime Editing Applications
Prime editing is a highly versatile technology with multiple advantages over other genome editing approaches. Distinct from HDR, prime editing does not rely on DSB formation or synthetic exogenous DNA donor molecules to replace targeted genomic sequences. In addition, prime editors can edit the genomic region containing the PAM, allowing for the introduction of silent mutations to block potential multi-turnover nCas9 cleavage, a current limitation of base editors. Prime editing also enables the incorporation of a greater variety of desirable genomic changes relative to base editing. Although still in its early stages, highly versatile approaches and applications for prime editing have already been developed. Indeed, prime editing has been exploited to efficiently generate transition and transversion mutations, small insertions and deletions, and combinations of mutations in cancer and primary cells (Aida et al., 2020; Anzalone et al., 2019; Benamozig et al., 2021; Habib et al., 2021; Kweon et al., 2021; Liu et al., 2020b; Petri et al., 2021), patient-derived organoids (Schene et al., 2020), zebrafish (Petri et al., 2021), flies (Bosch et al., 2021), bacteria (Tong et al., 2021) and plants (Lin et al., 2020). Small genomic insertions can be introduced at higher efficiency, although with increased indel formation, using prime editors that generate DSBs and engage end joining pathways (Peterka et al., 2021). Moreover, prime editing has been utilized for saturation mutagenesis studies, allowing the interrogation of single genomic bases without introducing DSBs or editing byproducts (Erwood et al., 2021). Prime editing has also been used to develop genome-based systems that enable multiplex recording of cell lineage histories, transcriptional activities and signaling events (Chen et al., 2021d; Choi et al., 2021a). Importantly, recent studies have demonstrated that prime editing can be used to correct pathogenic mutations in adult mice (Böck et al., 2021; Jang et al., 2021; Liu et al., 2021b), suggesting a promising future for this technology in therapeutic applications.
Limitations and Current Improvements of Prime Editing
The current main limitations of prime editing are its lower editing efficiency and more complex gRNA design relative to other precision genome editing technologies, such as base editing. However, recent reports in human cells and plants have shown that high-efficiency pegRNAs can be designed based on Cas9 activity prediction models and sequence contexts, highlighting, for example, that editing efficiency is influenced by the length of the PBS sequence and the reverse transcriptase template, and by the GC base content and secondary structure of the insert sequence (Kim et al., 2021b; Koeppel et al., 2021; Lin et al., 2021). It is expected that prime editing will follow the same path of other genome editing technologies, such as base editing, leading to the design of more efficient prime editors. Such editors could be achieved, for example, by improving the processivity and fidelity of the reverse transcriptase, engineering the 3’ extension of the gRNA, developing machine learning algorithms to predict pegRNA efficiency in various cellular systems, and fusing, co-expressing or inhibiting DNA repair factors to modulate prime editing outcome. Indeed, recent studies have already shown that prime editing efficiency can be enhanced by engineering pegRNAs and prime editors and modulating cellular DNA repair. For instance, adding hairpin structures to the 3’ end of the pegRNA, which contains the reverse transcriptase template sequence and the PBS, protects pegRNAs from degradation by cellular exonucleases and enhances prime editing efficiency (Chai et al., 2021; Liu et al., 2021c; Nelson et al., 2021). Prime editing events can also be enhanced by optimizing the codon sequence of the reverse transcriptase (Chen et al., 2021b), by increasing the localization of prime editors into the nucleus using multiple nuclear localization signal sequences (Chen et al., 2021b; Liu et al., 2021b), by introducing mutations in Cas9 that improve its nickase activity (Chen et al., 2021b), and by expressing a reverse transcriptase fusion that is recruited to prime editing sites through a MS2 RNA aptamer fused to the pegRNA (Chai et al., 2021). Furthermore, prime editing efficiency can be stimulated by co-expressing a dominant negative version of the MMR protein MLH1 to disrupt MMR-mediated degradation of the edited DNA bases (Chen et al., 2021b), or by fusing the DNA binding domain of RAD51 to prime editors to stabilize the pairing of the pegRNA to the nicked target DNA strand (Song et al., 2021). Prime editor fusions to small peptides derived from DNA repair proteins have also been shown to display greater editing efficiency, although their mechanisms of action still remain to be elucidated (Velimirovic et al., 2021). Recent studies have also reported improvement of prime editing efficiency through marker-free co-selection of edits simultaneously introduced in a gene of interest and ATP1A1, an essential gene encoding for the main subunit of the sodium/potassium pump (Levesque et al, 2021).
Prime editing strategies relying on dual nicking events (e.g., PE3, paired prime editors) can lead to the formation of DSBs (Figures 6B and 7B), resulting in increased frequency of indel byproducts relative to single nick-based prime editing approaches (e.g., PE2) (Anzalone et al., 2019; Chen et al., 2021b; Lin et al., 2021). However, the frequency of indel byproducts, which is significantly lower than that obtained using Cas9 nucleases (Anzalone et al., 2021; Anzalone et al., 2019), can be reduced by inhibiting MMR (Chen et al., 2021b). Prime editing is highly specific, showing a low frequency of pegRNA-dependent off-target edits and no observable pegRNA-independent off-targets in plants and human cells (Gao et al., 2021a; Jin et al., 2021). Whole-genome sequencing of plants has also revealed that prime editors do not change retrotransposon copy number or telomere structure (Jin et al., 2021). While prime editing leads to minimal perturbations of the cellular transcriptome (Anzalone et al., 2019), the M-MLV reverse transcriptase utilized for prime editing has been shown to promote the readthrough of STOP codons through its interaction with the translation pre-termination complex (Tang et al., 2016; Zheng et al., 2021), thereby potentially affecting protein synthesis. However, this activity can be disrupted by deleting the RNase H domain of the M-MLV reverse transcriptase, without resulting in a reduction of prime editing efficiency (Zheng et al., 2021). Given their smaller size, prime editors lacking the RNase H domain of the reverse transcriptase have been employed for in vivo editing following delivery by adeno-associated virus (AAV) vectors (Böck et al., 2021; Gao et al., 2021b).
Retron-based Genome Editing
Retrons are bacterial genetic elements composed of a reverse transcriptase and a non-coding RNA that generate chimeric RNA-DNA molecules to protect cells against phages (Millman et al., 2020). CRISPR-dependent retron systems encode a gRNA fused to a retron RNA, which is used as a template by a reverse transcriptase to produce a multi-copy ssDNA product tethered to the retron RNA (Simon et al., 2019). CRISPR/retron systems have been employed to generate ssDNA donors in cellulo to serve as templates for HDR-mediated introduction of targeted substitutions in the genomes of bacteria, yeast and human cells (Lim et al., 2020; Sharon et al., 2018; Zhao et al., 2021a). In their current format, however, these systems rely on the generation of DSBs and provide limited HDR stimulation in human cells (Zhao et al., 2021a). DSB- and CRISPR-independent retron systems that rely on the recombineering of ssDNA products into the bacterial genome have recently been developed for high-throughput generation of nucleotide variants and other genome engineering applications (Farzadfard et al., 2021; Schubert et al., 2021). ssDNA-based recombineering approaches have also enabled the generation of mutant yeast strains at scale through RAD51-independent annealing of ssDNA oligos to the lagging strand of replication forks (Barbieri et al., 2017). However, whether CRISPR-independent retron systems can be employed to engineer eukaryotic cells remains to be determined.
Integration-based Genome Editing
CRISPR-associated Recombinase Systems
Insertion of large DNA sequences is critical for many biomedical applications, including gene therapy (High and Roncarolo, 2019). Site-specific insertion of genetic payloads can be achieved by HDR or NHEJ approaches. However, as discussed above, these methods suffer from the introduction of DSB intermediates and the generation of editing byproducts.
Site-specific recombinases are widely used as genetic tools for their ability to enable robust and stable DNA manipulations (Merrick et al., 2018; Olorunniji et al., 2016). To enhance their specificity, recombinases have been fused to genome editing effectors. The fusion of serine recombinase domains to programmable TALE and zinc-finger effectors and to dCas9 has resulted in site-directed insertion or deletion of desired DNA sequences at exogenous recombinase recognition sites or at endogenous genomic pseudo-sites recognized by recombinases, albeit with limited efficiency (Chaikind et al., 2016; Gersbach et al., 2011; Gordley et al., 2009; Mercer et al., 2012). Recent studies led to the development of improved CRISPR-based recombinase systems that employ prime editing to insert recognition sequences of site-specific recombinases (e.g., attB or attP sites) using single or paired pegRNAs, enabling targeted integration of gene-size payloads into the genome by serine recombinases (e.g., Bxb1) ectopically expressed or fused to prime editors (Anzalone et al., 2021; Ioannidi et al., 2021) (Figure 7B, step 10).
CRISPR-associated Transposases
Transposases are self-contained enzymatic machineries that insert or excise DNA segments in the genome (Rebollo et al., 2012). Transposases act by recognizing and excising cognate donor sites located at the ends of the transposable element (Muñoz-López and García-Pérez, 2010). The transposable element is then integrated to new non-homologous insertion sites. Transposition requires multiple DNA repair activities. For instance, loss of host NHEJ repair factors affect piggyBat excision and repair of the broken donor backbone in yeast (She et al., 2015). Transposition results in the duplication of the transposon end sequences originating from the repair of ssDNA gaps generated during the integration of the transposon into the genome (Hickman and Dyda, 2016).
Recent computational analysis of bacterial genomes revealed the association of bacterial Tn7-like transposons with nuclease-defective CRISPR systems (Peters et al., 2017), suggesting the potential existence of RNA-guided transposition in bacteria (Faure et al., 2019). Subsequent studies successfully reconstituted minimal RNA-guided transposition using type I and type V CRISPR systems, resulting in site-specific insertion of large DNA fragments in bacteria (Klompe et al., 2019; Strecker et al., 2019b). These studies led to the characterization of INsert Transposable Elements by Guide RNA-Assisted TargEting (INTEGRATE) and CRISPR-ASsociated Transposase (CAST) systems for site-specific transposition of genetic cargos into the bacterial genome. While INTEGRATE is based on the CRISPR–Cas type I-F system, which lacks the Cas3 nuclease–helicase and is associated with the transposition protein TniQ, CAST employs a Tn7-like transposon associated with the CRISPR–Cas type V-K system, which contains a catalytically inactive Cas12k nuclease (Klompe et al., 2019; Strecker et al., 2019b). Comparative studies have shown that the INTEGRATE system is more efficient and displays higher purity of products relative to CAST (Rubin et al., 2021; Vo et al., 2021).
Alternative approaches led to the development of an engineered synthetic RNA-guided transposition system, called Cas-Transposon (Cas-Tn), by fusing the Himar1 transposase to dCas9 to induce site-specific transposition of DNA cargos into plasmids in bacteria (Chen and Wang, 2019). However, it remains to be determined whether the Cas-Tn system allows integration of cargos into the bacterial and eukaryotic genomes. More recent studies have reported the development of a transposition-based system that relies on the fusion of an engineered piggyBac transposase domain to Cas9 (Pallarès-Masmitjà et al., 2021). This system enables site-specific integration of multi-kilobase piggyBac transposon constructs into the mammalian genome (Pallarès-Masmitjà et al., 2021).
Applications and Current Limitations of CRISPR-associated Integration-based Systems
CRISPR-associated DNA integration systems generally operate without formation of DSBs, and do not require the presence of DNA sequences homologous to the targeted genomic sequence in DNA donor molecules (Anzalone et al., 2021; Ioannidi et al., 2021; Klompe et al., 2019; Strecker et al., 2019b). While CRISPR-associated recombinase systems require prior insertion of exogenous sequences into the genome, CRISPR-based transposition enables integration of large DNA cargos at any genomic sequence of interest. However, CRISPR-based transposition is not scarless, resulting in the incorporation of transposon end sequences into the genome and the duplication of targeted genomic sites (Klompe et al., 2019; Strecker et al., 2019b). Further studies will be required to minimize the amount of undesired genetic changes induced by CRISPR-associated transposition. Improved CRISPR-associated transposon systems will surely emerge upon further characterization of their structural features and molecular mechanisms (Halpin-Healy et al., 2020; Park et al., 2021; Petassi et al., 2020; Querques et al., 2021; Saito et al., 2021; Shen et al., 2021; Vo et al., 2021; Xiao et al., 2021). RNA-guided transposases have been exploited for high-throughput gene disruption studies in bacteria (Chen et al., 2021e) and for locus- and species-specific genetic manipulation of bacteria in complex microbial communities (Rubin et al., 2021; Vo et al., 2021). This approach has the potential to revolutionize the manipulation of bacterial communities in human health-related environments, such as the gut microbiome, and symbiotic agricultural microbial communities. Additional work is, however, necessary to achieve programmable transposition using these approaches in mammalian cells. PiggyBac transposase-based CRISPR systems have been shown to promote site-specific integration of transgenes in mammalian cell lines and in the mouse liver (Pallarès-Masmitjà et al., 2021). However, these systems result in indel formation due their reliance on Cas9-induced DSBs, which stimulate the insertional activity of the piggyBac transposase (Pallarès-Masmitjà et al., 2021).
Prime editing-dependent recombinase systems have been shown to enable efficient and site-specific integration of large DNA cargos into the genome of mammalian cells (Anzalone et al., 2021; Ioannidi et al., 2021). These approaches have been employed for inserting fluorescent markers and antibiotic resistance gene cassettes, labeling proteins with fluorescent tags, producing and secreting therapeutically relevant enzymes, and correcting pathogenic genomic inversions (Anzalone et al., 2021; Ioannidi et al., 2021). The recent discovery of a multitude of microbial serine recombinases with diverse target sites (Durrant et al., 2021) should provide enhanced efficiency, flexibility and multiplexing capabilities to recombinase-based systems. Altogether, technologies relying on CRISPR-associated DNA integration hold great promise for clinical applications requiring the integration of fully functional therapeutic genes, the replacement of gene exons or the insertion of engineered constructs, such as chimeric antigen receptors, in safe harbor loci, while minimizing the safety concerns associated with the random insertion of viral constructs as in current gene therapy strategies.
Conclusions and Future Perspectives
Programmable technologies enabling the introduction of site-specific DSBs have sparked a revolution in genome editing. In particular, the repurposing of natural CRISPR systems into powerful genome editing technologies and the development of new tools for genome engineering are transforming biology and medicine (Doudna, 2020; Kulkarni et al., 2021; Pickar-Oliver and Gersbach, 2019) and offering encouraging perspectives for the treatment of debilitating human diseases (Frangoul et al., 2021; Gillmore et al., 2021; Lu et al., 2020; Newby and Liu, 2021; Porteus, 2019; Stadtmauer et al., 2020; Xu et al., 2019). Moreover, DSB-free technologies, such as CRISPR-dependent base editing, prime editing and DNA integration, have allowed unprecedented interrogation of genomes with high precision at single base resolution and enabled highly efficient insertion of DNA constructs at sites of interest. These technologies hold great promise for the correction of disease-causing mutations and the development of mutation-specific or gene replacement therapies. The next generation of precision genome editing tools are likely to result from fundamental discoveries obtained by studying the wide diversity of prokaryotic organisms and the broad variety of anti-phage immune mechanisms (Hampton et al., 2020; Hug et al., 2016). For instance, recent studies have discovered several novel bacterial defense systems with unique properties and activities that might bolster the development of future genome engineering technologies (Altae-Tran et al., 2021; Doron et al., 2018; Gao et al., 2020; Karvelis et al., 2021; Rousset et al., 2021). These tools will help us define the fundamentals of organismal life, combat environmental pollution, produce improved food, and understand and treat genetic and infectious human diseases.
Acknowledgments
We thank Richard Baer, Alex Chavez, David Liu, Lorraine Symington and members of the Billon and Ciccia laboratories for insightful discussions and comments on the manuscript. We apologize to our colleagues whose work could not be cited due to space constraints. This work was supported by the generous start-up package from the University of Calgary, Cumming School of Medicine, the Arnie Charbonneau Cancer Institute and the Robson DNA Science Centre to P.B and NIH grants R01CA197774, R01CA227450 and P01CA174653 to A.C.
Footnotes
Declaration of interests
The authors have filed a patent application related to the development of a new method for the detection of precision genome editing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe T, Inoue KI, Furuta Y, and Kiyonari H (2020). Pronuclear Microinjection during S-Phase Increases the Efficiency of CRISPR-Cas9-Assisted Knockin of Large DNA Donors in Mouse Zygotes. Cell reports 31, 107653. [DOI] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. (2017). RNA targeting with CRISPR-Cas13. Nature 550, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, Hughes J, Pomerantz RT, and Thomas PQ (2018). Large deletions induced by Cas9 cleavage. Nature 560, E8–E9. [DOI] [PubMed] [Google Scholar]
- Adikusuma F, Williams N, Grutzner F, Hughes J, and Thomas P (2017). Targeted Deletion of an Entire Chromosome Using CRISPR/Cas9. Mol Ther 25, 1736–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, et al. (2016). Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov 6, 914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida T, Wilde JJ, Yang L, Hou Y, Li M, Xu D, Lin J, Qi P, Lu Z, and Feng G (2020). Prime editing primarily induces undesired outcomes in mice. bioRxiv, 2020.2008.2006.239723. [Google Scholar]
- Aird EJ, Lovendahl KN, St Martin A, Harris RS, and Gordon WR (2018). Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun Biol 1, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zain AM, and Symington LS (2021). The dark side of homology-directed repair. DNA Repair (Amst) 106, 103181. [DOI] [PubMed] [Google Scholar]
- Alanis-Lobato G, Zohren J, McCarthy A, Fogarty NME, Kubikova N, Hardman E, Greco M, Wells D, Turner JMA, and Niakan KK (2021). Frequent loss of heterozygosity in CRISPR-Cas9-edited early human embryos. Proc Natl Acad Sci U S A 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen F, Crepaldi L, Alsinet C, Strong AJ, Kleshchevnikov V, De Angeli P, Palenikova P, Khodak A, Kiselev V, Kosicki M, et al. (2019). Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat Biotechnol. 37, 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseth I, Dalhus B, and Bjoras M (2014). Inosine in DNA and RNA. Curr Opin Genet Dev 26, 116–123. [DOI] [PubMed] [Google Scholar]
- Altae-Tran H, Kannan S, Demircioglu FE, Oshiro R, Nety SP, McKay LJ, Dlakic M, Inskeep WP, Makarova KS, Macrae RK, et al. (2021). The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoasii L, Hildyard JCW, Li H, Sanchez-Ortiz E, Mireault A, Caballero D, Harron R, Stathopoulou TR, Massey C, Shelton JM, et al. (2018). Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362, 86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoasii L, Long C, Li H, Mireault AA, Shelton JM, Sanchez-Ortiz E, McAnally JR, Bhattacharyya S, Schmidt F, Grimm D, et al. (2017). Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Science translational medicine 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R, Ranjha L, Cannavo E, and Cejka P (2016). Phosphorylated CtIP Functions as a Co-factor of the MRE11-RAD50-NBS1 Endonuclease in DNA End Resection. Mol Cell 64, 940–950. [DOI] [PubMed] [Google Scholar]
- Annunziato S, Lutz C, Henneman L, Bhin J, Wong K, Siteur B, van Gerwen B, de Korte-Grimmerink R, Zafra MP, Schatoff EM, et al. (2020). In situ CRISPR-Cas9 base editing for the development of genetically engineered mouse models of breast cancer. EMBO J 39, e102169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone AV, Gao XD, Podracky CJ, Nelson AT, Koblan LW, Raguram A, Levy JM, Mercer JAM, and Liu DR (2021). Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nature biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone AV, Koblan LW, and Liu DR (2020). Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nature biotechnology 38, 824–844. [DOI] [PubMed] [Google Scholar]
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbab M, Shen MW, Mok B, Wilson C, Matuszek Z, Cassa CA, and Liu DR (2020). Determinants of Base Editing Outcomes from Target Library Analysis and Machine Learning. Cell 182, 463–480 e430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X, Joore I, Chuva de Sousa Lopes S, van Zon J, Tans S, and Clevers H (2020). Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat Cell Biol 22, 321–331. [DOI] [PubMed] [Google Scholar]
- Balakrishnan L, and Bambara RA (2013). Flap Endonuclease 1. Annual Review of Biochemistry 82, 119–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri EM, Muir P, Akhuetie-Oni BO, Yellman CM, and Isaacs FJ (2017). Precise Editing at DNA Replication Forks Enables Multiplex Genome Engineering in Eukaryotes. Cell 171, 1453–1467 e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DE, and Lindahl T (2004). Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 38, 445–476. [DOI] [PubMed] [Google Scholar]
- Bashir S, Dang T, Rossius J, Wolf J, and Kuhn R (2020). Enhancement of CRISPR-Cas9 induced precise gene editing by targeting histone H2A-K15 ubiquitination. BMC Biotechnol 20, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard WA, Horton JK, Prasad R, and Wilson SH (2019). Eukaryotic Base Excision Repair: New Approaches Shine Light on Mechanism. Annu Rev Biochem 88, 137–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benamozig O, Baudrier L, and Billon P (2021). Chapter Twelve - A detection method for the capture of genomic signatures: From disease diagnosis to genome editing. In Methods in Enzymology, Eichman BF, ed. (Academic Press; ), pp. 251–282. [DOI] [PubMed] [Google Scholar]
- Bermudez-Cabrera HC, Culbertson S, Barkal S, Holmes B, Shen MW, Zhang S, Gifford DK, and Sherwood RI (2021). Small molecule inhibition of ATM kinase increases CRISPR-Cas9 1-bp insertion frequency. Nature communications 12, 5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava R, Onyango DO, and Stark JM (2016). Regulation of Single-Strand Annealing and its Role in Genome Maintenance. Trends Genet 32, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, and Carroll D (2003). Enhancing gene targeting with designed zinc finger nucleases. Science 300, 764. [DOI] [PubMed] [Google Scholar]
- Bieging KT, Mello SS, and Attardi LD (2014). Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer 14, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon P, Bryant EE, Joseph SA, Nambiar TS, Hayward SB, Rothstein R, and Ciccia A (2017). CRISPR-Mediated Base Editing Enables Efficient Disruption of Eukaryotic Genes through Induction of STOP Codons. Mol Cell 67, 1068–1079 e1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon P, Nambiar TS, Hayward SB, Zafra MP, Schatoff EM, Oshima K, Dunbar A, Breinig M, Park YC, Ryu HS, et al. (2020). Detection of Marker-Free Precision Genome Editing and Genetic Variation through the Capture of Genomic Signatures. Cell reports 30, 3280–3295 e3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, and Bonas U (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Böck D, Rothgangl T, Villiger L, Schmidheini L, Mathis N, Ioannidi E, Kreutzer S, Kontarakis Z, Rimann N, Grisch-Chan HM, et al. (2021). Treatment of a metabolic liver disease by in vivo prime editing in mice. bioRxiv, 2021.2008.2017.456632. [Google Scholar]
- Boel A, De Saffel H, Steyaert W, Callewaert B, De Paepe A, Coucke PJ, and Willaert A (2018). CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Disease Models & Mechanisms 11, dmm035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch JA, Birchak G, and Perrimon N (2021). Precise genome engineering in Drosophila using prime editing. Proc Natl Acad Sci U S A 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Phadke T, Barrera LA, Margulies CM, Lee CS, Buquicchio F, Moss S, Abdulkerim HS, Selleck W, Jayaram H, et al. (2017). Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nature communications 8, 13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, and Mer G (2006). Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambati A, Barry RM, and Sfeir A (2020). DNA polymerase theta (Poltheta) - an error-prone polymerase necessary for genome stability. Curr Opin Genet Dev 60, 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, de Haas M, Holland HA, Akhtar W, and van Steensel B (2018). Kinetics and Fidelity of the Repair of Cas9-Induced Double-Strand DNA Breaks. Mol Cell 70, 801–813 e806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, and van der Oost J (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet E, and Jasin M (2018). Induction of Chromosomal Translocations with CRISPR-Cas9 and Other Nucleases: Understanding the Repair Mechanisms That Give Rise to Translocations. Adv Exp Med Biol 1044, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson R, Langenbucher A, Bowen D, Kwan EE, Benes CH, Zou L, and Lawrence MS (2019). Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgio G, and Teboul L (2020). Anticipating and Identifying Collateral Damage in Genome Editing. Trends in Genetics 36, 905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Temiz NA, and Harris RS (2013). Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 45, 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, and Banfield JF (2017). New CRISPR-Cas systems from uncultivated microbes. Nature 542, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW (2014). DNA single-strand break repair. Experimental Cell Research 329, 2–8. [DOI] [PubMed] [Google Scholar]
- Caldecott KW (2020). Mammalian DNA base excision repair: Dancing in the moonlight. DNA Repair (Amst) 93, 102921. [DOI] [PubMed] [Google Scholar]
- Cameron P, Coons MM, Klompe SE, Lied AM, Smith SC, Vidal B, Donohoue PD, Rotstein T, Kohrs BW, Nyer DB, et al. (2019). Harnessing type I CRISPR–Cas systems for genome engineering in human cells. Nature biotechnology 37, 1471–1477. [DOI] [PubMed] [Google Scholar]
- Campa CC, Weisbach NR, Santinha AJ, Incarnato D, and Platt RJ (2019). Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat Methods 16, 887–893. [DOI] [PubMed] [Google Scholar]
- Canny MD, Moatti N, Wan LCK, Fradet-Turcotte A, Krasner D, Mateos-Gomez PA, Zimmermann M, Orthwein A, Juang YC, Zhang W, et al. (2018). Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat Biotechnol 36, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Wu X, Zhou J, Wu H, Liu L, Zhang Q, DeMott MS, Gu C, Wang L, You D, et al. (2020). Nick-seq for single-nucleotide resolution genomic maps of DNA modifications and damage. Nucleic Acids Res 48, 6715–6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Salazar-Garcia L, Gao F, Wahlestedt T, Wu CL, Han X, Cai Y, Xu D, Wang F, Tang L, et al. (2019). Novel approach reveals genomic landscapes of single-strand DNA breaks with nucleotide resolution in human cells. Nat Commun 10, 5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, and Frosina G (1997). Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J Biol Chem 272, 23970–23975. [DOI] [PubMed] [Google Scholar]
- Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, and Saha K (2017). Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat Commun 8, 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon O, Smith CJ, Khoshakhlagh P, Ferreira R, Güell M, Said K, Yildiz R, Dysart M, Wang S, Thompson D, et al. (2020). CRISPR-mediated biocontainment. bioRxiv, 2020.2002.2003.922146. [Google Scholar]
- Cejka P, and Symington LS (2021). DNA End Resection: Mechanism and Control. Annual Review of Genetics 55, 285–307. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang Y, Wang J, Qiao D, Zhang Y, Xin C, Zhou Y, Wang X-C, and Chen Q-J (2021). MS2 RNA aptamer enhances prime editing in rice. bioRxiv, 2021.2010.2020.465209. [Google Scholar]
- Chaikind B, Bessen JL, Thompson DB, Hu JH, and Liu DR (2016). A programmable Cas9-serine recombinase fusion protein that operates on DNA sequences in mammalian cells. Nucleic Acids Res 44, 9758–9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti AM, Henser-Brownhill T, Monserrat J, Poetsch AR, Luscombe NM, and Scaffidi P (2019). Target-Specific Precision of CRISPR-Mediated Genome Editing. Molecular Cell 73, 699–713.e696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champer J, Buchman A, and Akbari OS (2016). Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet 17, 146–159. [DOI] [PubMed] [Google Scholar]
- Chan K, Resnick MA, and Gordenin DA (2013). The choice of nucleotide inserted opposite abasic sites formed within chromosomal DNA reveals the polymerase activities participating in translesion DNA synthesis. DNA Repair (Amst) 12, 878–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HHY, Pannunzio NR, Adachi N, and Lieber MR (2017). Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nature Reviews Molecular Cell Biology 18, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanut P, Britton S, Coates J, Jackson SP, and Calsou P (2016). Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nature communications 7, 12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Khedher AHY, Menoret S, Brion A, Lamribet K, Dardillac E, Boix C, Perrouault L, Tesson L, Geny S, et al. (2018). CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat Commun 9, 1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Zou W, Xu H, Liang Y, and Huang B (2018a). Efficient labeling and imaging of protein-coding genes in living cells using CRISPR-Tag. Nat Commun 9, 5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Feng W, Lim PX, Kass EM, and Jasin M (2018b). Homology-Directed Repair and the Role of BRCA1, BRCA2, and Related Proteins in Genome Integrity and Cancer. Annu Rev Cancer Biol 2, 313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Park JE, Paa P, Rajakumar PD, Prekop HT, Chew YT, Manivannan SN, and Chew WL (2021a). Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat Commun 12, 1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PJ, Hussmann JA, Yan J, Knipping F, Ravisankar P, Chen P-F, Chen C, Nelson JW, Newby GA, Sahin M, et al. (2021b). Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652.e5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lee L, Naila T, Fishbain S, Wang A, Tomkinson AE, Lees-Miller SP, and He Y (2021c). Structural basis of long-range to short-range synaptic transition in NHEJ. Nature 593, 294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SP, and Wang HH (2019). An Engineered Cas-Transposon System for Programmable and Site-Directed DNA Transpositions. CRISPR J 2, 376–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Choi J, Nathans JF, Agarwal V, Martin B, Nichols E, Leith A, Lee C, and Shendure J (2021d). Multiplex genomic recording of enhancer and signal transduction activity in mammalian cells. bioRxiv, 2021.2011.2005.467434. [Google Scholar]
- Chen W, McKenna A, Schreiber J, Haeussler M, Yin Y, Agarwal V, Noble WS, and Shendure J (2019). Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Research 47, 7989–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Ren ZH, Tang N, Chai G, Zhang H, Zhang Y, Ma J, Wu Z, Shen X, Huang X, et al. (2021e). Targeted genetic screening in bacteria with a Cas12k-guided transposase. Cell reports 36, 109635. [DOI] [PubMed] [Google Scholar]
- Chen X, Janssen JM, Liu J, Maggio I, t Jong AEJ, Mikkers HMM, and Goncalves M (2017a). In trans paired nicking triggers seamless genome editing without double-stranded DNA cutting. Nat Commun 8, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu J, Janssen JM, and Goncalves M (2017b). The Chromatin Structure Differentially Impacts High-Specificity CRISPR-Cas9 Nuclease Strategies. Mol Ther Nucleic Acids 8, 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Rinsma M, Janssen JM, Liu J, Maggio I, and Goncalves MA (2016). Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res 44, 6482–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu J, Zhi S, Zheng Q, Ma W, Huang J, Liu Y, Liu D, Liang P, and Songyang Z (2020). Repurposing type I-F CRISPR-Cas system as a transcriptional activation tool in human cells. Nat Commun 11, 3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Li Y, Qi Q, Xu P, Feng R, Palmer L, Chen J, Wu R, Yee T, Zhang J, et al. (2021). Single-nucleotide-level mapping of DNA regulatory elements that control fetal hemoglobin expression. Nature Genetics 53, 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NH, Cheveralls KC, Brunner A-D, Kim K, Michaelis AC, Raghavan P, Kobayashi H, Savy L, Li JY, Canaj H, et al. (2021). OpenCell: proteome-scale endogenous tagging enables the cartography of human cellular organization. bioRxiv, 2021.2003.2029.437450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, and Koo T (2021). CRISPR technologies for the treatment of Duchenne muscular dystrophy. Mol Ther. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Chen W, Minkina A, Chardon FM, Suiter CC, Regalado SG, Domcke S, Hamazaki N, Lee C, Martin B, et al. (2021a). A temporally resolved, multiplex molecular recorder based on sequential genome editing. bioRxiv, 2021.2011.2005.467388. [Google Scholar]
- Choi J, Chen W, Suiter CC, Lee C, Chardon FM, Yang W, Leith A, Daza RM, Martin B, and Shendure J (2021b). Precise genomic deletions using paired prime editing. Nature biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, and Kuhn R (2015). Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nature biotechnology 33, 543–548. [DOI] [PubMed] [Google Scholar]
- Ciccia A, and Elledge SJ (2010). The DNA damage response: making it safe to play with knives. Mol Cell 40, 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, and West SC (2008). Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem 77, 259–287. [DOI] [PubMed] [Google Scholar]
- Cinesi C, Aeschbach L, Yang B, and Dion V (2016). Contracting CAG/CTG repeats using the CRISPR-Cas9 nickase. Nature communications 7, 13272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin N, Dzantiev L, Kadyrov FA, and Modrich P (2005). Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J Biol Chem 280, 39752–39761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Yang Z, and Neuberger MS (2007). DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol 94, 37–73. [DOI] [PubMed] [Google Scholar]
- Cortez D (2019). Replication-Coupled DNA Repair. Mol Cell 74, 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, and Zhang F (2017). RNA editing with CRISPR-Cas13. Science 358, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Becerra G, and Kadonaga JT (2020). Enhancement of homology-directed repair with chromatin donor templates in cells. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorgo B, Leon LM, Chau-Ly IJ, Vasquez-Rifo A, Berry JD, Mahendra C, Crawford ED, Lewis JD, and Bondy-Denomy J (2020). A compact Cascade-Cas3 system for targeted genome engineering. Nat Methods 17, 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuella-Martin R, Hayward SB, Fan X, Chen X, Huang J-W, Taglialatela A, Leuzzi G, Zhao J, Rabadan R, Lu C, et al. (2021). Functional interrogation of DNA damage response variants with base editing screens. Cell 184, 1081–1097.e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullot G, Boutin J, Toutain J, Prat F, Pennamen P, Rooryck C, Teichmann M, Rousseau E, Lamrissi-Garcia I, Guyonnet-Duperat V, et al. (2019). CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun 10, 1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daer RM, Cutts JP, Brafman DA, and Haynes KA (2017). The Impact of Chromatin Dynamics on Cas9-Mediated Genome Editing in Human Cells. ACS Synth Biol 6, 428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya R, Hu Q, and Ly P (2021). Mechanistic origins of diverse genome rearrangements in cancer. Seminars in Cell & Developmental Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalvai M, Loehr J, Jacquet K, Huard CC, Roques C, Herst P, Cote J, and Doyon Y (2015). A Scalable Genome-Editing-Based Approach for Mapping Multiprotein Complexes in Human Cells. Cell Rep 13, 621–633. [DOI] [PubMed] [Google Scholar]
- Davis AJ, and Chen DJ (2013). DNA double strand break repair via non-homologous end-joining. Transl Cancer Res 2, 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, and Maizels N (2014). Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc Natl Acad Sci U S A 111, E924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, and Maizels N (2016). Two Distinct Pathways Support Gene Correction by Single-Stranded Donors at DNA Nicks. Cell reports 17, 1872–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moraes MH, Hsu F, Huang D, Bosch DE, Zeng J, Radey MC, Simon N, Ledvina HE, Frick JP, Wiggins PA, et al. (2021). An interbacterial DNA deaminase toxin directly mutagenizes surviving target populations. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demin AA, Hirota K, Tsuda M, Adamowicz M, Hailstone R, Brazina J, Gittens W, Kalasova I, Shao Z, Zha S, et al. (2021). XRCC1 prevents toxic PARP1 trapping during DNA base excision repair. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Myler LR, Soniat MM, Makharashvili N, Lee L, Lees-Miller SP, Finkelstein IJ, and Paull TT (2020). DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. Sci Adv 6, eaay0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després PC, Dubé AK, Seki M, Yachie N, and Landry CR (2020). Perturbing proteomes at single residue resolution using base editing. Nature communications 11, 1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, and Sorek R (2018). Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA (2020). The promise and challenge of therapeutic genome editing. Nature 578, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant MG, Fanton A, Tycko J, Hinks M, Chandrasekaran S, Perry NT, Schaepe J, Du PP, Bintu L, Bassik MC, et al. (2021). Large-scale discovery of recombinases for integrating DNA into the human genome. bioRxiv, 2021.2011.2005.467528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D, Zuccaro MV, Kosicki M, Church GM, Bradley A, and Jasin M (2018). Inter-homologue repair in fertilized human eggs? Nature 560, E5–E7. [DOI] [PubMed] [Google Scholar]
- Elacqua JJ, Ranu N, DiIorio SE, and Blainey PC (2021). DENT-seq for genome-wide strand-specific identification of DNA single-strand break sites with single-nucleotide resolution. Genome Res 31, 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis GI, Sheppard NC, and Riley JL (2021). Genetic engineering of T cells for immunotherapy. Nat Rev Genet 22, 427–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enache OM, Rendo V, Abdusamad M, Lam D, Davison D, Pal S, Currimjee N, Hess J, Pantel S, Nag A, et al. (2020). Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat Genet 52, 662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwood S, Bily TMI, Lequyer J, Yan J, Gulati N, Brewer RA, Zhou L, Pelletier L, Ivakine EA, and Cohn RD (2021). Saturation variant interpretation using CRISPR prime editing. bioRxiv, 2021.2005.2011.443710. [DOI] [PubMed] [Google Scholar]
- Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gonen M, and Sadelain M (2017). Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, Gharaei N, Citorik RJ, and Lu TK (2021). Efficient retroelement-mediated DNA writing in bacteria. Cell Systems 12, 860–872.e865. [DOI] [PubMed] [Google Scholar]
- Faure G, Shmakov SA, Yan WX, Cheng DR, Scott DA, Peters JE, Makarova KS, and Koonin EV (2019). CRISPR-Cas in mobile genetic elements: counter-defence and beyond. Nat Rev Microbiol 17, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari G, Thrasher AJ, and Aiuti A (2021). Gene therapy using haematopoietic stem and progenitor cells. Nat Rev Genet 22, 216–234. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Jacob A, Beretta S, Unali G, Albano L, Vavassori V, Cittaro D, Lazarevic D, Brombin C, Cugnata F, et al. (2020). Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nature biotechnology 38, 1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira da Silva J, Meyenberg M, and Loizou JI (2021a). Tissue specificity of DNA repair: the CRISPR compass. Trends in Genetics. [DOI] [PubMed] [Google Scholar]
- Ferreira da Silva J, Oliveira G, Arasa-Verge E, Moretton A, Thimelthaler G, Jiricny J, and Loizou JI (2021b). Prime Editing Efficiency and Fidelity are Enhanced in the Absence of Mismatch Repair. bioRxiv, 2021.2009.2030.462548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filsinger GT, Wannier TM, Pedersen FB, Lutz ID, Zhang J, Stork DA, Debnath A, Gozzi K, Kuchwara H, Volf V, et al. (2021). Characterizing the portability of phage-encoded homologous recombination proteins. Nat Chem Biol 17, 394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GM, Boyle EA, Hause RJ, Klein JC, and Shendure J (2014). Saturation editing of genomic regions by multiplex homology-directed repair. Nature 513, 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, Janizek JD, Huang X, Starita LM, and Shendure J (2018). Accurate classification of BRCA1 variants with saturation genome editing. Nature 562, 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovič M, Le Rhun A, and Charpentier E (2016). The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521. [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, et al. (2013). 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangoul H, Altshuler D, Cappellini MD, Chen YS, Domm J, Eustace BK, Foell J, de la Fuente J, Grupp S, Handgretinger R, et al. (2021). CRISPR-Cas9 Gene Editing for Sickle Cell Disease and beta-Thalassemia. N Engl J Med 384, 252–260. [DOI] [PubMed] [Google Scholar]
- Frit P, Ropars V, Modesti M, Charbonnier JB, and Calsou P (2019). Plugged into the Ku-DNA hub: The NHEJ network. Prog Biophys Mol Biol 147, 62–76. [DOI] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, and Alt FW (2015). Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nature biotechnology 33, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Li Q, Liu X, Tu T, Lv X, Yin X, Lv J, Song Z, Qu J, Zhang J, et al. (2021). Human cell based directed evolution of adenine base editors with improved efficiency. Nature communications 12, 5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Tigano M, and Sfeir A (2020). Safeguarding mitochondrial genomes in higher eukaryotes. Nat Struct Mol Biol 27, 687–695. [DOI] [PubMed] [Google Scholar]
- Gallagher DN, and Haber JE (2018). Repair of a Site-Specific DNA Cleavage: Old-School Lessons for Cas9-Mediated Gene Editing. ACS Chem Biol 13, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher DN, Pham N, Tsai AM, Janto AN, Choi J, Ira G, and Haber JE (2020). A Rad51-independent pathway promotes single-strand template repair in gene editing. PLOS Genetics 16, e1008689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammage PA, Moraes CT, and Minczuk M (2018). Mitochondrial Genome Engineering: The Revolution May Not Be CRISPR-Ized. Trends Genet 34, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Altae-Tran H, Bohning F, Makarova KS, Segel M, Schmid-Burgk JL, Koob J, Wolf YI, Koonin EV, and Zhang F (2020). Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369, 1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Fu Z-C, Li X, Wang Y, Wei J, Li G, Wang L, Wu J, Xue W, Huang X, et al. (2021a). No observable guide-RNA-independent off-target mutation induced by prime editor. bioRxiv, 2021.2004.2009.439109. [Google Scholar]
- Gao Z, Haldrup J, Ravendran S, Mikkelsen NS, Mikkelsen JG, and Bak RO (2021b). A truncated reverse transcriptase enhances prime editing by split AAV vectors. bioRxiv, 2021.2011.2005.467423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapinske M, Luu A, Winter J, Woods WS, Kostan KA, Shiva N, Song JS, and Perez-Pinera P (2018). CRISPR-SKIP: programmable gene splicing with single base editors. Genome Biol 19, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, and Moineau S (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, and Siksnys V (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109, E2579–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, and Liu DR (2017). Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Lam DK, Rees HA, Sola-Esteves NM, Barrera LA, Born DA, Edwards A, Gehrke JM, Lee SJ, Liquori AJ, et al. (2020). Directed evolution of adenine base editors with increased activity and therapeutic application. Nat Biotechnol 38, 892–900. [DOI] [PubMed] [Google Scholar]
- Gehrke JM, Cervantes O, Clement MK, Wu Y, Zeng J, Bauer DE, Pinello L, and Joung JK (2018). An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat Biotechnol 36, 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersbach CA, Gaj T, Gordley RM, Mercer AC, and Barbas CF 3rd (2011). Targeted plasmid integration into the human genome by an engineered zinc-finger recombinase. Nucleic Acids Res 39, 7868–7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmore JD, Gane E, Taubel J, Kao J, Fontana M, Maitland ML, Seitzer J, O’Connell D, Walsh KR, Wood K, et al. (2021). CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N Engl J Med 385, 493–502. [DOI] [PubMed] [Google Scholar]
- Goellner EM, Putnam CD, and Kolodner RD (2015). Exonuclease 1-dependent and independent mismatch repair. DNA Repair (Amst) 32, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves MA, van Nierop GP, Holkers M, and de Vries AA (2012). Concerted nicking of donor and chromosomal acceptor DNA promotes homology-directed gene targeting in human cells. Nucleic Acids Res 40, 3443–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordley RM, Gersbach CA, and Barbas CF 3rd (2009). Synthesis of programmable integrases. Proc Natl Acad Sci U S A 106, 5053–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TG, Walter JC, and Loparo JJ (2016). Two-Stage Synapsis of DNA Ends during Non-homologous End Joining. Mol Cell 61, 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco GE, Matsumoto Y, Brooks RC, Lu Z, Lieber MR, and Tomkinson AE (2016). SCR7 is neither a selective nor a potent inhibitor of human DNA ligase IV. DNA Repair (Amst) 43, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald J, Zhou R, Lareau CA, Garcia SP, Iyer S, Miller BR, Langner LM, Hsu JY, Aryee MJ, and Joung JK (2020). A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nature biotechnology 38, 861–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Posfai E, and Rossant J (2018). Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat Biotechnol 36, 632–637. [DOI] [PubMed] [Google Scholar]
- Gu S, Bodai Z, Cowan QT, and Komor AC (2021). Base editors: Expanding the types of DNA damage products harnessed for genome editing. Gene and Genome Editing 1, 100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Mintier G, Ma-Edmonds M, Storton D, Wang X, Xiao X, Kienzle B, Zhao D, and Feder JN (2018). ‘Cold shock’ increases the frequency of homology directed repair gene editing in induced pluripotent stem cells. Sci Rep 8, 2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Triana JA, Tavhelidse T, Thumberger T, Thomas I, Wittbrodt B, Kellner T, Anlas K, Tsingos E, and Wittbrodt J (2018). Efficient single-copy HDR by 5’ modified long dsDNA donors. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Haemmerle M, Genovese G, Draetta GF, and Chin L (2016). Post-translational Regulation of Cas9 during G1 Enhances Homology-Directed Repair. Cell Rep 14, 1555–1566. [DOI] [PubMed] [Google Scholar]
- Haapaniemi E, Botla S, Persson J, Schmierer B, and Taipale J (2018). CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 24, 927–930. [DOI] [PubMed] [Google Scholar]
- Habib O, Habib G, Hwang G-H, and Bae S (2021). Comprehensive analysis of prime editing outcomes in human embryonic stem cells. bioRxiv, 2021.2004.2012.439533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackley CR (2021). A Novel Set of Cas9 Fusion Proteins to Stimulate Homologous Recombination: Cas9-HRs. CRISPR J 4, 253–263. [DOI] [PubMed] [Google Scholar]
- Halperin SO, Tou CJ, Wong EB, Modavi C, Schaffer DV, and Dueber JE (2018). CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 560, 248–252. [DOI] [PubMed] [Google Scholar]
- Halpin-Healy TS, Klompe SE, Sternberg SH, and Fernandez IS (2020). Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system. Nature 577, 271–274. [DOI] [PubMed] [Google Scholar]
- Hampton HG, Watson BNJ, and Fineran PC (2020). The arms race between bacteria and their phage foes. Nature 577, 327–336. [DOI] [PubMed] [Google Scholar]
- Hanna RE, Hegde M, Fagre CR, DeWeirdt PC, Sangree AK, Szegletes Z, Griffith A, Feeley MN, Sanson KR, Baidi Y, et al. (2021). Massively parallel assessment of human variants with base editor screens. Cell 184, 1064–1080.e1020. [DOI] [PubMed] [Google Scholar]
- Harel I, Valenzano DR, and Brunet A (2016). Efficient genome engineering approaches for the short-lived African turquoise killifish. Nat Protoc 11, 2010–2028. [DOI] [PubMed] [Google Scholar]
- Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, Cofsky JC, Kyrpides NC, Banfield JF, and Doudna JA (2018). Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, and Lupski JR (2009). A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5, e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward SB, and Ciccia A (2021). Towards a CRISPeR understanding of homologous recombination with high-throughput functional genomics. Current Opinion in Genetics & Development 71, 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess GT, Frésard L, Han K, Lee CH, Li A, Cimprich KA, Montgomery SB, and Bassik MC (2016). Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nature methods 13, 1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman AB, and Dyda F (2016). DNA Transposition at Work. Chem Rev 116, 12758–12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High KA, and Roncarolo MG (2019). Gene Therapy. New England Journal of Medicine 381, 455–464. [DOI] [PubMed] [Google Scholar]
- Hirotsune S, Kiyonari H, Jin M, Kumamoto K, Yoshida K, Shinohara M, Watanabe H, Wynshaw-Boris A, and Matsuzaki F (2020). Enhanced homologous recombination by the modulation of targeting vector ends. Sci Rep 10, 2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck MA, Witkowsky LB, Guglielmi B, Replogle JM, Gilbert LA, Villalta JE, Torigoe SE, Tjian R, and Weissman JS (2016). Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P, and Zhang Y (2017). The Devil is in the details for DNA mismatch repair. Proc Natl Acad Sci U S A 114, 3552–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, et al. (2018a). Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Shi Z, Guo X, Jiang B, Wang G, Luo D, Chen Y, and Zhu YS (2018b). Ligase IV inhibitor SCR7 enhances gene editing directed by CRISPR-Cas9 and ssODN in human cancer cells. Cell Biosci 8, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Li G, Wu J, Liang J, and Wang X (2021a). Identification of pathogenic variants in cancer genes using base editing screens with editing efficiency correction. Genome Biology 22, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TP, Newby GA, and Liu DR (2021b). Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat Protoc 16, 1089–1128. [DOI] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, et al. (2016). A new view of the tree of life. Nat Microbiol 1, 16048. [DOI] [PubMed] [Google Scholar]
- Hussmann JA, Ling J, Ravisankar P, Yan J, Cirincione A, Xu A, Simpson D, Yang D, Bothmer A, Cotta-Ramusino C, et al. (2021). Mapping the genetic landscape of DNA double-strand break repair. Cell 184, 5653–5669.e5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustedt N, and Durocher D (2016). The control of DNA repair by the cell cycle. Nat Cell Biol 19, 1–9. [DOI] [PubMed] [Google Scholar]
- Hyodo T, Rahman ML, Karnan S, Ito T, Toyoda A, Ota A, Wahiduzzaman M, Tsuzuki S, Okada Y, Hosokawa Y, et al. (2020). Tandem Paired Nicking Promotes Precise Genome Editing with Scarce Interference by p53. Cell Rep 30, 1195–1207 e1197. [DOI] [PubMed] [Google Scholar]
- Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, Kommineni S, Chen J, Sondey M, Ye C, et al. (2018). p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med 24, 939–946. [DOI] [PubMed] [Google Scholar]
- Ioannidi EI, Yarnall MTN, Schmitt-Ulms C, Krajeski RN, Lim J, Villiger L, Zhou W, Jiang K, Roberts N, Zhang L, et al. (2021). Drag-and-drop genome insertion without DNA cleavage with CRISPR-directed integrases. bioRxiv, 2021.2011.2001.466786. [Google Scholar]
- Iyer S, Suresh S, Guo D, Daman K, Chen JCJ, Liu P, Zieger M, Luk K, Roscoe BP, Mueller C, et al. (2019). Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 568, 561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Jo DH, Cho CS, Shin JH, Seo JH, Yu G, Gopalappa R, Kim D, Cho S-R, Kim JH, et al. (2021). Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nature Biomedical Engineering. [DOI] [PubMed] [Google Scholar]
- Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, and de Wind N (2006). Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med 203, 319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, and Haber JE (2016). The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair (Amst) 44, 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayavaradhan R, Pillis DM, Goodman M, Zhang F, Zhang Y, Andreassen PR, and Malik P (2019). CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat Commun 10, 2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KT, Floe L, Petersen TS, Huang J, Xu F, Bolund L, Luo Y, and Lin L (2017). Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett 591, 1892–1901. [DOI] [PubMed] [Google Scholar]
- Jeong YK, Lee S, Hwang GH, Hong SA, Park SE, Kim JS, Woo JS, and Bae S (2021). Adenine base editor engineering reduces editing of bystander cytosines. Nat Biotechnol. [DOI] [PubMed] [Google Scholar]
- Jiang T, Zhang X-O, Weng Z, and Xue W (2021). Deletion and replacement of long genomic sequences using prime editing. Nature biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Lin Q, Luo Y, Zhu Z, Liu G, Li Y, Chen K, Qiu JL, and Gao C (2021). Genome-wide specificity of prime editors in plants. Nat Biotechnol. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J (2013). Postreplicative mismatch repair. Cold Spring Harb Perspect Biol 5, a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Lim H, Chun H, Lee JH, and Bang D (2020). Single-cell analysis of a mutant library generated using CRISPR-guided deaminase in human melanoma cells. Communications Biology 3, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Y, Ruis B, Lin S, and Hendrickson EA (2014). The mechanism of gene targeting in human somatic cells. PLoS Genet 10, e1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Y, Ruis B, Takasugi T, and Hendrickson EA (2017). Mechanisms of precise genome editing using oligonucleotide donors. Genome Res 27, 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis T, Bigelyte G, Young JK, Hou Z, Zedaveinyte R, Budre K, Paulraj S, Djukanovic V, Gasior S, Silanskas A, et al. (2020). PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res 48, 5016–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis T, Druteika G, Bigelyte G, Budre K, Zedaveinyte R, Silanskas A, Kazlauskas D, Venclovas Č, and Siksnys V (2021). Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, and Storici F (2014). Transcript-RNA-templated DNA recombination and repair. Nature 515, 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Lee JM, Moon SB, Chin HJ, Park S, Lim Y, Kim D, Koo T, Ko JH, and Kim YS (2021a). Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nature biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, and Kim J-S (2014). A guide to genome engineering with programmable nucleases. Nature Reviews Genetics 15, 321–334. [DOI] [PubMed] [Google Scholar]
- Kim HK, Yu G, Park J, Min S, Lee S, Yoon S, and Kim HH (2021b). Predicting the efficiency of prime editing guide RNAs in human cells. Nat Biotechnol 39, 198–206. [DOI] [PubMed] [Google Scholar]
- Kim HS, Jeong YK, Hur JK, Kim JS, and Bae S (2019). Adenine base editors catalyze cytosine conversions in human cells. Nat Biotechnol 37, 1145–1148. [DOI] [PubMed] [Google Scholar]
- Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, Chung E, Kim S, and Kim JS (2017a). Highly efficient RNA-guided base editing in mouse embryos. Nature biotechnology 35, 435–437. [DOI] [PubMed] [Google Scholar]
- Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR (2017b). Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol 35, 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Mudrak SV, and Jinks-Robertson S (2011). The dCMP transferase activity of yeast Rev1 is biologically relevant during the bypass of endogenously generated AP sites. DNA Repair (Amst) 10, 1262–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfo I, et al. (2019). Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol 37, 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klompe SE, Vo PLH, Halpin-Healy TS, and Sternberg SH (2019). Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature 571, 219–225. [DOI] [PubMed] [Google Scholar]
- Kluesner MG, Lahr WS, Lonetree CL, Smeester BA, Qiu X, Slipek NJ, Claudio Vazquez PN, Pitzen SP, Pomeroy EJ, Vignes MJ, et al. (2021). CRISPR-Cas9 cytidine and adenosine base editing of splice-sites mediates highly-efficient disruption of proteins in primary and immortalized cells. Nat Commun 12, 2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott GJ, and Doudna JA (2018). CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblan LW, Arbab M, Shen MW, Hussmann JA, Anzalone AV, Doman JL, Newby GA, Yang D, Mok B, Replogle JM, et al. (2021a). Efficient C•G-to-G•C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nature biotechnology 39, 1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, Maianti JP, Raguram A, and Liu DR (2018). Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nature Biotechnology 36, 843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblan LW, Erdos MR, Wilson C, Cabral WA, Levy JM, Xiong ZM, Tavarez UL, Davison LM, Gete YG, Mao X, et al. (2021b). In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature 589, 608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppel J, Peets EM, Weller J, Pallaseni A, Liberante F, and Parts L (2021). Predicting efficiency of writing short sequences into the genome using prime editing. bioRxiv, 2021.2011.2010.468024. [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, and Liu DR (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW, Kim YB, Badran AH, and Liu DR (2017). Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 3, eaao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, and Zhang F (2017). Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M, Allen F, and Bradley A (2020). Cas9-induced large deletions and small indels are controlled in a convergent fashion. bioRxiv, 2020.2008.2005.216739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, and Bradley A (2018). Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36, 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramara J, Osia B, and Malkova A (2018). Break-Induced Replication: The Where, The Why, and The How. Trends Genet 34, 518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan HE, and Bjoras M (2013). Base excision repair. Cold Spring Harb Perspect Biol 5, a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni JA, Witzigmann D, Thomson SB, Chen S, Leavitt BR, Cullis PR, and van der Meel R (2021). The current landscape of nucleic acid therapeutics. Nat Nanotechnol 16, 630–643. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, and Erie DA (2015). Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu Rev Genet 49, 291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt IC, Zhou R, Iyer S, Garcia SP, Miller BR, Langner LM, Grunewald J, and Joung JK (2021). CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol 39, 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Parlak M, Tufan T, Yang J, Szlachta K, Wei X, Mammadov R, and Adli M (2017). CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nature methods 14, 710–712. [DOI] [PubMed] [Google Scholar]
- Kuzminov A (2001). Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci U S A 98, 8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon J, Jang A-H, Shin HR, See J-E, Lee W, Lee JW, Chang S, Kim K, and Kim Y (2020). A CRISPR-based base-editing screen for the functional assessment of BRCA1 variants. Oncogene 39, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon J, Yoon JK, Jang AH, Shin HR, See JE, Jang G, Kim JI, and Kim Y (2021). Engineered prime editors with PAM flexibility. Mol Ther 29, 2001–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinaite A, Knott GJ, Palumbo CM, Lin-Shiao E, Richter MF, Zhao KT, Beal PA, Liu DR, and Doudna JA (2020). DNA capture by a CRISPR-Cas9-guided adenine base editor. Science 369, 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Lee S, Baek G, Kim A, Kang BC, Seo H, and Kim JS (2021). Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. Nat Commun 12, 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Mackley VA, Rao A, Chong AT, Dewitt MA, Corn JE, and Murthy N (2017). Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenay RT, Aghazadeh A, Hiatt J, Tse D, Roth TL, Apathy R, Shifrut E, Hultquist JF, Krogan N, Wu Z, et al. (2019). Large dataset enables prediction of repair after CRISPR-Cas9 editing in primary T cells. Nature biotechnology 37, 1034–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Chen H, Xue W, Yang B, Hu B, Wei J, Wang L, Cui Y, Li W, Wang J, et al. (2018). APOBEC3 induces mutations during repair of CRISPR-Cas9-generated DNA breaks. Nat Struct Mol Biol 25, 45–52. [DOI] [PubMed] [Google Scholar]
- Leibowitz ML, Papathanasiou S, Doerfler PA, Blaine LJ, Sun L, Yao Y, Zhang C-Z, Weiss MJ, and Pellman D (2021). Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nature Genetics 53, 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti MD, Sekine S, Kamiyama D, Weissman JS, and Huang B (2016). A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc Natl Acad Sci U S A 113, E3501–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque S, Mayorga D, Fiset J-P, Goupil C, Duringer A, Loiselle A, Bouchard E, Agudelo D, Doyon Y (2021). Marker-free coselection for successive rounds of prime editing in human cells. bioRxiv. doi: 10.1101/2021.11.02.464583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JM, Yeh WH, Pendse N, Davis JR, Hennessey E, Butcher R, Koblan LW, Comander J, Liu Q, and Liu DR (2020). Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat Biomed Eng 4, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang R, Meng X, Chen S, Zong Y, Lu C, Qiu JL, Chen YH, Li J, and Gao C (2020a). Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat Biotechnol 38, 875–882. [DOI] [PubMed] [Google Scholar]
- Li G, Zhang X, Wang H, Liu D, Li Z, Wu Z, and Yang H (2020b). Increasing CRISPR/Cas9-mediated homology-directed DNA repair by histone deacetylase inhibitors. Int J Biochem Cell Biol 125, 105790. [DOI] [PubMed] [Google Scholar]
- Li G, Zhang X, Zhong C, Mo J, Quan R, Yang J, Liu D, Li Z, Yang H, and Wu Z (2017). Small molecules enhance CRISPR/Cas9-mediated homology-directed genome editing in primary cells. Sci Rep 7, 8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Beckman KA, Pessino V, Huang B, Weissman JS, and Leonetti MD (2019). Design and specificity of long ssDNA donors for CRISPR-based knock-in. bioRxiv, 178905. [Google Scholar]
- Li PP, and Margolis RL (2021). Use of single guided Cas9 nickase to facilitate precise and efficient genome editing in human iPSCs. Scientific reports 11, 9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li Y, Yang S, Huang S, Yan M, Ding Y, Tang W, Lou X, Yin Q, Sun Z, et al. (2018a). CRISPR-Cas9-mediated base-editing screening in mice identifies DND1 amino acids that are critical for primordial germ cell development. Nat Cell Biol 20, 1315–1325. [DOI] [PubMed] [Google Scholar]
- Li X, Wang Y, Liu Y, Yang B, Wang X, Wei J, Lu Z, Zhang Y, Wu J, Huang X, et al. (2018b). Base editing with a Cpf1-cytidine deaminase fusion. Nature biotechnology 36, 324–327. [DOI] [PubMed] [Google Scholar]
- Li XL, Li GH, Fu J, Fu YW, Zhang L, Chen W, Arakaki C, Zhang JP, Wen W, Zhao M, et al. (2018c). Highly efficient genome editing via CRISPR-Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res 46, 10195–10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Potter J, Kumar S, Ravinder N, and Chesnut JD (2017). Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J Biotechnol 241, 136–146. [DOI] [PubMed] [Google Scholar]
- Liao C, and Beisel CL (2021). The tracrRNA in CRISPR Biology and Technologies. Annual Review of Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H, Jun S, Park M, Lim J, Jeong J, Lee JH, and Bang D (2020). Multiplex Generation, Tracking, and Functional Screening of Substitution Mutants Using a CRISPR/Retron System. ACS Synth Biol 9, 1003–1009. [DOI] [PubMed] [Google Scholar]
- Lin Q, Jin S, Zong Y, Yu H, Zhu Z, Liu G, Kou L, Wang Y, Qiu JL, Li J, et al. (2021). High-efficiency prime editing with optimized, paired pegRNAs in plants. Nature biotechnology 39, 923–927. [DOI] [PubMed] [Google Scholar]
- Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, Wang Y, Anzalone AV, Raguram A, Doman JL, et al. (2020). Prime genome editing in rice and wheat. Nat Biotechnol 38, 582–585. [DOI] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, and Doudna JA (2014). Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 3, e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Xin H, Zhang Y, Wu X, Yuan F, and Wang Z (1999). The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res 27, 4468–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T (2000). Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat Res 462, 129–135. [DOI] [PubMed] [Google Scholar]
- Ling X, Xie B, Gao X, Chang L, Zheng W, Chen H, Huang Y, Tan L, Li M, and Liu T (2020). Improving the efficiency of precise genome editing with site-specific Cas9-oligonucleotide conjugates. Sci Adv 6, eaaz0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Chen S, Rose A, Chen D, Cao F, Zwinderman M, Kiemel D, Aissi M, Dekker FJ, and Haisma HJ (2020a). Inhibition of histone deacetylase 1 (HDAC1) and HDAC2 enhances CRISPR/Cas9 genome editing. Nucleic Acids Res 48, 517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Lin Q, Jin S, and Gao C (2022). The CRISPR-Cas toolbox and gene editing technologies. Mol Cell. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Orlova N, Oakes BL, Ma E, Spinner HB, Baney KLM, Chuck J, Tan D, Knott GJ, Harrington LB, et al. (2019). CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 566, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang W, Xin C, Yin J, Shang Y, Ai C, Li J, Meng F-L, and Hu J (2021a). Global detection of DNA repair outcomes induced by CRISPR–Cas9. Nucleic Acids Research 49, 8732–8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Liang S-Q, Zheng C, Mintzer E, Zhao YG, Ponnienselvan K, Mir A, Sontheimer EJ, Gao G, Flotte TR, et al. (2021b). Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nature communications 12, 2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li X, He S, Huang S, Li C, Chen Y, Liu Z, Huang X, and Wang X (2020b). Efficient generation of mouse models with the prime editing system. Cell Discov 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yang G, Huang S, Li X, Wang X, Li G, Chi T, Chen Y, Huang X, and Wang X (2021c). Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Research 31, 1134–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Smith CE, and Symington LS (2008). Break-induced replication: what is it and what is it for? Cell Cycle 7, 859–864. [DOI] [PubMed] [Google Scholar]
- Lomova A, Clark DN, Campo-Fernandez B, Flores-Bjurstrom C, Kaufman ML, Fitz-Gibbon S, Wang X, Miyahira EY, Brown D, DeWitt MA, et al. (2019). Improving Gene Editing Outcomes in Human Hematopoietic Stem and Progenitor Cells by Temporal Control of DNA Repair. Stem Cells 37, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Xue J, Deng T, Zhou X, Yu K, Deng L, Huang M, Yi X, Liang M, Wang Y, et al. (2020). Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med 26, 732–740. [DOI] [PubMed] [Google Scholar]
- Ly P, and Cleveland DW (2017). Rebuilding Chromosomes After Catastrophe: Emerging Mechanisms of Chromothripsis. Trends Cell Biol 27, 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Marti-Gutierrez N, Park S-W, Wu J, Hayama T, Darby H, Van Dyken C, Li Y, Koski A, Liang D, et al. (2018). Ma et al. reply. Nature 560, E10–E23. [DOI] [PubMed] [Google Scholar]
- Ma H, Marti-Gutierrez N, Park S-W, Wu J, Lee Y, Suzuki K, Koski A, Ji D, Hayama T, Ahmed R, et al. (2017a). Correction of a pathogenic gene mutation in human embryos. Nature 548, 413–419. [DOI] [PubMed] [Google Scholar]
- Ma L, Ruan J, Song J, Wen L, Yang D, Zhao J, Xia X, Chen YE, Zhang J, and Xu J (2020). MiCas9 increases large size gene knock-in rates and reduces undesirable ontarget and off-target indel edits. Nature communications 11, 6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Zhuang F, Hu X, Wang B, Wen XZ, Ji JF, and Xi JJ (2017b). Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res 27, 578–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang J, Yin W, Zhang Z, Song Y, and Chang X (2016). Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods 13, 1029–1035. [DOI] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, et al. (2014). In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 516, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS, Bumcrot D, Chao H, Ciulla DM, DaSilva JA, et al. (2019). Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med 25, 229–233. [DOI] [PubMed] [Google Scholar]
- Maizels N, and Davis L (2018). Initiation of homologous recombination at DNA nicks. Nucleic Acids Res 46, 6962–6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, et al. (2020). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, and Church GM (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology 31, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, and Haber JE (2012). Mutations arising during repair of chromosome breaks. Annu Rev Genet 46, 455–473. [DOI] [PubMed] [Google Scholar]
- Mao Z, Bozzella M, Seluanov A, and Gorbunova V (2008). Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 7, 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquart KF, Allam A, Janjuha S, Sintsova A, Villiger L, Frey N, Krauthammer M, and Schwank G (2021). Predicting base editing outcomes with an attention-based deep learning algorithm trained on high-throughput target library screens. Nature communications 12, 5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Alonso S, Frutos-Beltran E, and Menendez-Arias L (2021). Reverse Transcriptase: From Transcriptomics to Genome Editing. Trends Biotechnol 39, 194–210. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, and Ploegh HL (2015). Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nature biotechnology 33, 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, and Sfeir A (2015). Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez PA, Kent T, Deng SK, McDevitt S, Kashkina E, Hoang TM, Pomerantz RT, and Sfeir A (2017). The helicase domain of Poltheta counteracts RPA to promote alt-NHEJ. Nat Struct Mol Biol 24, 1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, and West SC (2014). Holliday junction resolution: regulation in space and time. DNA Repair (Amst) 19, 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurissen TL, and Woltjen K (2020). Synergistic gene editing in human iPS cells via cell cycle and DNA repair modulation. Nat Commun 11, 2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina OM, Keskin H, Hanamshet K, Storici F, and Mazin AV (2017). Rad52 Inverse Strand Exchange Drives RNA-Templated DNA Double-Strand Break Repair. Mol Cell 67, 19–29 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Khodaverdian VY, Meyer D, Cerqueira PG, and Heyer WD (2016). Eukaryotic DNA Polymerases in Homologous Recombination. Annu Rev Genet 50, 393–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, and Lee SE (2008). MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet 24, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AC, Gaj T, Fuller RP, and Barbas CF 3rd (2012). Chimeric TALE recombinases with programmable DNA sequence specificity. Nucleic Acids Res 40, 11163–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick CA, Zhao J, and Rosser SJ (2018). Serine Integrases: Advancing Synthetic Biology. ACS Synth Biol 7, 299–310. [DOI] [PubMed] [Google Scholar]
- Millman A, Bernheim A, Stokar-Avihail A, Fedorenko T, Voichek M, Leavitt A, Oppenheimer-Shaanan Y, and Sorek R (2020). Bacterial Retrons Function In Anti-Phage Defense. Cell 183, 1551–1561 e1512. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, and Symington LS (2010). Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 29, 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min YL, Li H, Rodriguez-Caycedo C, Mireault AA, Huang J, Shelton JM, McAnally JR, Amoasii L, Mammen PPA, Bassel-Duby R, et al. (2019). CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv 5, eaav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Chan AH, Judge LM, Yoo J, Huang M, Nguyen TD, Lizarraga PP, So PL, and Conklin BR (2014). Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat Methods 11, 291–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohni KN, Wessel SR, Zhao R, Wojciechowski AC, Luzwick JW, Layden H, Eichman BF, Thompson PS, Mehta KPM, and Cortez D (2019). HMCES Maintains Genome Integrity by Shielding Abasic Sites in Single-Strand DNA. Cell 176, 144–153 e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok BY, de Moraes MH, Zeng J, Bosch DE, Kotrys AV, Raguram A, Hsu F, Radey MC, Peterson SB, Mootha VK, et al. (2020). A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla KA, and Yang Y (2019). CRISPR/Cas-Mediated Base Editing: Technical Considerations and Practical Applications. Trends Biotechnol 37, 1121–1142. [DOI] [PubMed] [Google Scholar]
- Monteys AM, Ebanks SA, Keiser MS, and Davidson BL (2017). CRISPR/Cas9 Editing of the Mutant Huntingtin Allele In Vitro and In Vivo. Mol Ther 25, 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbach V, Poggi L, and Richard G-F (2019). Trinucleotide repeat instability during double-strand break repair: from mechanisms to gene therapy. Current Genetics 65, 17–28. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, and Bogdanove AJ (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501. [DOI] [PubMed] [Google Scholar]
- Mullard A (2020). Gene-editing pipeline takes off. Nat Rev Drug Discov 19, 367–372. [DOI] [PubMed] [Google Scholar]
- Muñoz-López M, and García-Pérez JL (2010). DNA transposons: nature and applications in genomics. Curr Genomics 11, 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP, and Hartley RC (2018). Mitochondria as a therapeutic target for common pathologies. Nat Rev Drug Discov 17, 865–886. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Chadwick AC, Mizoguchi T, Garcia SP, DeNizio JE, Reiss CW, Wang K, Iyer S, Dutta C, Clendaniel V, et al. (2021). In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 593, 429–434. [DOI] [PubMed] [Google Scholar]
- Nabel CS, Manning SA, and Kohli RM (2012). The curious chemical biology of cytosine: deamination, methylation, and oxidation as modulators of genomic potential. ACS Chem Biol 7, 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakade S, Tsubota T, Sakane Y, Kume S, Sakamoto N, Obara M, Daimon T, Sezutsu H, Yamamoto T, Sakuma T, et al. (2014). Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nature communications 5, 5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Zhou Y, Tomita A, Hirade Y, Gurumurthy CB, and Nakada S (2018). Precise and efficient nucleotide substitution near genomic nick via noncanonical homology-directed repair. Genome Res 28, 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar TS, Billon P, Diedenhofen G, Hayward SB, Taglialatela A, Cai K, Huang JW, Leuzzi G, Cuella-Martin R, Palacios A, et al. (2019). Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nat Commun 10, 3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nami F, Basiri M, Satarian L, Curtiss C, Baharvand H, and Verfaillie C (2018). Strategies for In Vivo Genome Editing in Nondividing Cells. Trends Biotechnol 36, 770–786. [DOI] [PubMed] [Google Scholar]
- Natsume T, Kiyomitsu T, Saga Y, and Kanemaki MT (2016). Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep 15, 210–218. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, and Hinkle DC (1996). Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382, 729–731. [DOI] [PubMed] [Google Scholar]
- Nelson JW, Randolph PB, Shen SP, Everette KA, Chen PJ, Anzalone AV, An M, Newby GA, Chen JC, Hsu A, et al. (2021). Engineered pegRNAs improve prime editing efficiency. Nature biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby GA, and Liu DR (2021). In vivo somatic cell base editing and prime editing. Molecular Therapy 29, 3107–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby GA, Yen JS, Woodard KJ, Mayuranathan T, Lazzarotto CR, Li Y, Sheppard-Tillman H, Porter SN, Yao Y, Mayberry K, et al. (2021). Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature 595, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DN, Roth TL, Li PJ, Chen PA, Apathy R, Mamedov MR, Vo LT, Tobin VR, Goodman D, Shifrut E, et al. (2020). Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nature biotechnology 38, 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. (2012). Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, et al. (2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, and Nureki O (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. (2014). Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843. [DOI] [PubMed] [Google Scholar]
- Nussenzweig PM, and Marraffini LA (2020). Molecular Mechanisms of CRISPR-Cas Immunity in Bacteria. Annu Rev Genet 54, 93–120. [DOI] [PubMed] [Google Scholar]
- Ochs F, Somyajit K, Altmeyer M, Rask MB, Lukas J, and Lukas C (2016). 53BP1 fosters fidelity of homology-directed DNA repair. Nat Struct Mol Biol 23, 714–721. [DOI] [PubMed] [Google Scholar]
- Olivieri M, Cho T, Álvarez-Quilón A, Li K, Schellenberg MJ, Zimmermann M, Hustedt N, Rossi SE, Adam S, Melo H, et al. (2020). A Genetic Map of the Response to DNA Damage in Human Cells. Cell 182, 481–496.e421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olorunniji Femi J., Rosser Susan J., and Stark WM (2016). Site-specific recombinases: molecular machines for the Genetic Revolution. Biochemical Journal 473, 673–684. [DOI] [PubMed] [Google Scholar]
- Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, et al. (2015). A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ouellet DL, Cherif K, Rousseau J, and Tremblay JP (2017). Deletion of the GAA repeats from the human frataxin gene using the CRISPR-Cas9 system in YG8R-derived cells and mouse models of Friedreich ataxia. Gene Therapy 24, 265–274. [DOI] [PubMed] [Google Scholar]
- Owens DDG, Caulder A, Frontera V, Harman JR, Allan AJ, Bucakci A, Greder L, Codner GF, Hublitz P, McHugh PJ, et al. (2019). Microhomologies are prevalent at Cas9-induced larger deletions. Nucleic Acids Res 47, 7402–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Goldman DH, Kulaga H, Grzelak MJ, Rasoloson D, Paidemarry S, Green R, Reed RR, and Seydoux G (2017). Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc Natl Acad Sci U S A 114, E10745–E10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallarès-Masmitjà M, Ivančić D, Mir-Pedrol J, Jaraba-Wallace J, Tagliani T, Oliva B, Rahmeh A, Sánchez-Mejías A, and Güell M (2021). Find and cut-and-transfer (FiCAT) mammalian genome engineering. Nature communications 12, 7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papathanasiou S, Markoulaki S, Blaine LJ, Leibowitz ML, Zhang C-Z, Jaenisch R, and Pellman D (2021). Whole chromosome loss and genomic instability in mouse embryos after CRISPR-Cas9 genome editing. Nature communications 12, 5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-U, Tsai AW-L, Mehrotra E, Petassi MT, Hsieh S-C, Ke A, Peters JE, and Kellogg EH (2021). Structural basis for target site selection in RNA-guided DNA transposition systems. Science 373, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulis M, Castelli A, Lizier M, Susani L, Lucchini F, Villa A, and Vezzoni P (2015). A pre-screening FISH-based method to detect CRISPR/Cas9 off-targets in mouse embryonic stem cells. Sci Rep 5, 12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen BS, Mandal PK, Frock RL, Boyraz B, Yadav R, Upadhyayula S, Gutierrez-Martinez P, Ebina W, Fasth A, Kirchhausen T, et al. (2017). Ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR-Cas9 genome editing. Nat Biomed Eng 1, 878–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, Knott GJ, Jacobsen SE, Banfield JF, and Doudna JA (2020). CRISPR-CasPhi from huge phages is a hypercompact genome editor. Science 369, 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petassi MT, Hsieh SC, and Peters JE (2020). Guide RNA Categorization Enables Target Site Choice in Tn7-CRISPR-Cas Transposons. Cell 183, 1757–1771 e1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterka M, Akrap N, Li S, Wimberger S, Hsieh P-P, Degtev D, Bestas B, Barr J, van de Plassche S, Mendoza-Garcia P, et al. (2021). Harnessing DSB repair to promote efficient homology-dependent and -independent prime editing. bioRxiv, 2021.2008.2010.455572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE, Makarova KS, Shmakov S, and Koonin EV (2017). Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc Natl Acad Sci U S A 114, E7358–E7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri K, Zhang W, Ma J, Schmidts A, Lee H, Horng JE, Kim DY, Kurt IC, Clement K, Hsu JY, et al. (2021). CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nature biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham N, Yan Z, Yu Y, Faria Afreen M, Malkova A, Haber JE, and Ira G (2021). Mechanisms restraining break-induced replication at two-ended DNA double-strand breaks. EMBO J 40, e104847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Wright WD, and Heyer WD (2017). Multi-invasions Are Recombination Byproducts that Induce Chromosomal Rearrangements. Cell 170, 760–773 e715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickar-Oliver A, and Gersbach CA (2019). The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol 20, 490–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder J, Salsman J, and Dellaire G (2015). Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res 43, 9379–9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH (2019). A New Class of Medicines through DNA Editing. N Engl J Med 380, 947–959. [DOI] [PubMed] [Google Scholar]
- Porto EM, Komor AC, Slaymaker IM, and Yeo GW (2020). Base editing: advances and therapeutic opportunities. Nat Rev Drug Discov 19, 839–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, and Wilson SH (2001). DNA polymerase beta -mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J Biol Chem 276, 32411–32414. [DOI] [PubMed] [Google Scholar]
- Prasad R, Singhal RK, Srivastava DK, Molina JT, Tomkinson AE, and Wilson SH (1996). Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J Biol Chem 271, 16000–16007. [DOI] [PubMed] [Google Scholar]
- Pristyazhnyuk IE, Minina J, Korablev A, Serova I, Fishman V, Gridina M, Rozhdestvensky TS, Gubar L, Skryabin BV, and Serov OL (2019). Time origin and structural analysis of the induced CRISPR/cas9 megabase-sized deletions and duplications involving the Cntn6 gene in mice. Sci Rep 9, 14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano C, Cappella M, Valaperta R, Cardani R, Meola G, Martelli F, Cardinali B, and Falcone G (2017). CRISPR/Cas9-Mediated Deletion of CTG Expansions Recovers Normal Phenotype in Myogenic Cells Derived from Myotonic Dystrophy 1 Patients. Mol Ther Nucleic Acids 9, 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, and Lim WA (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querques I, Schmitz M, Oberli S, Chanez C, and Jinek M (2021). Target site selection and remodelling by type V CRISPR-transposon systems. Nature 599, 497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J, Musa A, Kantor A, McClements ME, Cehajic-Kapetanovic J, MacLaren RE, and Xue K (2020). Genome-Editing Strategies for Treating Human Retinal Degenerations. Human Gene Therapy 32, 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden DA, Carvajal-Garcia J, and Gupta GP (2021). Mechanism, cellular functions and cancer roles of polymerase-theta-mediated DNA end joining. Nature Reviews Molecular Cell Biology. [DOI] [PubMed] [Google Scholar]
- Ramsden DA, and Nussenzweig A (2021). Mechanisms driving chromosomal translocations: lost in time and space. Oncogene 40, 4263–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. (2012). Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148, 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, and Mager DL (2012). Transposable Elements: An Abundant and Natural Source of Regulatory Sequences for Host Genes. Annual Review of Genetics 46, 21–42. [DOI] [PubMed] [Google Scholar]
- Rees HA, and Liu DR (2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19, 770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HA, Yeh WH, and Liu DR (2019). Development of hRad51-Cas9 nickase fusions that mediate HDR without double-stranded breaks. Nat Commun 10, 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginato G, Cannavo E, and Cejka P (2017). Physiological protein blocks direct the Mre11-Rad50-Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev 31, 2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginato G, and Cejka P (2020). The MRE11 complex: A versatile toolkit for the repair of broken DNA. DNA Repair (Amst) 91–92, 102869. [DOI] [PubMed] [Google Scholar]
- Reint G, Li Z, Labun K, Keskitalo S, Soppa I, Mamia K, Tolo E, Szymanska M, Meza-Zepeda LA, Lorenz S, et al. (2021). Rapid genome editing by CRISPR-Cas9-POLD3 fusion. eLife 10, e75415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud JB, Boix C, Charpentier M, De Cian A, Cochennec J, Duvernois-Berthet E, Perrouault L, Tesson L, Edouard J, Thinard R, et al. (2016). Improved Genome Editing Efficiency and Flexibility Using Modified Oligonucleotides with TALEN and CRISPR-Cas9 Nucleases. Cell Rep 14, 2263–2272. [DOI] [PubMed] [Google Scholar]
- Richardson CD, Kazane KR, Feng SJ, Zelin E, Bray NL, Schafer AJ, Floor SN, and Corn JE (2018). CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat Genet 50, 1132–1139. [DOI] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, and Corn JE (2016). Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34, 339–344. [DOI] [PubMed] [Google Scholar]
- Richardson RR, Steyert M, Inen J, Khim S, Romanowski AJ, Altas B, and Poulopoulos A (2020). Cas9 fusions for precision in vivo editing. bioRxiv, 2020.2007.2015.199620. [Google Scholar]
- Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW, Wilson C, Koblan LW, Zeng J, Bauer DE, et al. (2020). Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol 38, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenberg S, Chintalapati M, Macak D, Kanis P, Maricic T, and Paabo S (2019). Simultaneous precise editing of multiple genes in human cells. Nucleic Acids Res 47, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenberg S, and Maricic T (2018). Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat Commun 9, 2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Torres N, Banas K, Bialk P, Bloh KM, and Kmiec EB (2017). Insertional Mutagenesis by CRISPR/Cas9 Ribonucleoprotein Gene Editing in Cells Targeted for Point Mutation Repair Directed by Short Single-Stranded DNA Oligonucleotides. PLoS One 12, e0169350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Barbeau M, Ethier S, Dostie J, and Pelletier J (2015). Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med 7, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B, Haupt A, Tucker A, Grancharova T, Arakaki J, Fuqua MA, Nelson A, Hookway C, Ludmann SA, Mueller IA, et al. (2017). Systematic gene tagging using CRISPR/Cas9 in human stem cells to illuminate cell organization. Mol Biol Cell 28, 2854–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, and Gordenin DA (2014). Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer 14, 786–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosello M, Vougny J, Czarny F, Mione MC, Concordet JP, Albadri S, and Del Bene F (2021). Precise base editing for the in vivo study of developmental signaling and human pathologies in zebrafish. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossidis AC, Stratigis JD, Chadwick AC, Hartman HA, Ahn NJ, Li H, Singh K, Coons BE, Li L, Lv W, et al. (2018). In utero CRISPR-mediated therapeutic editing of metabolic genes. Nat Med 24, 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Li PJ, Blaeschke F, Nies JF, Apathy R, Mowery C, Yu R, Nguyen MLT, Lee Y, Truong A, et al. (2020). Pooled Knockin Targeting for Genome Engineering of Cellular Immunotherapies. Cell 181, 728–744 e721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothgangl T, Dennis MK, Lin PJC, Oka R, Witzigmann D, Villiger L, Qi W, Hruzova M, Kissling L, Lenggenhager D, et al. (2021). In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nat Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, and Jasin M (1994). Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14, 8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F, Dowding J, Bernheim A, Rocha EPC, and Bikard D (2021). Prophage-encoded hotspots of bacterial immune systems. bioRxiv, 2021.2001.2021.427644. [Google Scholar]
- Rubin BE, Diamond S, Cress BF, Crits-Christoph A, Lou YC, Borges AL, Shivram H, He C, Xu M, Zhou Z, et al. (2021). Species- and site-specific genome editing in complex bacterial communities. Nature Microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, Kim HS, Kim DE, Lee H, Chung E, et al. (2018). Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nature biotechnology 36, 536–539. [DOI] [PubMed] [Google Scholar]
- Sabharwal A, Kar B, Restrepo-Castillo S, Holmberg SR, Mathew ND, Kendall BL, Cotter RP, WareJoncas Z, Seiler C, Nakamaru-Ogiso E, et al. (2021). The FusX TALE Base Editor (FusXTBE) for Rapid Mitochondrial DNA Programming of Human Cells In Vitro and Zebrafish Disease Models In Vivo. The CRISPR Journal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Ladha A, Strecker J, Faure G, Neumann E, Altae-Tran H, Macrae RK, and Zhang F (2021). Dual modes of CRISPR-associated transposon homing. Cell 184, 2441–2453 e2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata RC, Ishiguro S, Mori H, Tanaka M, Tatsuno K, Ueda H, Yamamoto S, Seki M, Masuyama N, Nishida K, et al. (2020). Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat Biotechnol 38, 865–869. [DOI] [PubMed] [Google Scholar]
- Sanders MA, Vöhringer H, Forster VJ, Moore L, Campbell BB, Hooks Y, Edwards M, Bianchi V, Coorens THH, Butler TM, et al. (2021). Life without mismatch repair. bioRxiv, 2021.2004.2014.437578. [Google Scholar]
- Sangree AK, Griffith AL, Szegletes ZM, Roy P, DeWeirdt PC, Hegde M, McGee AV, Hanna RE, and Doench JG (2021). Benchmarking of SpCas9 variants enables deeper base editor screens of BRCA1 and BCL2. bioRxiv, 2021.2008.2018.456848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic N, Ringnalda FC, Lindsay H, Berk C, Bargsten K, Li Y, Neri D, Robinson MD, Ciaudo C, Hall J, et al. (2018). Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanz S, Castor D, Fischer F, and Jiricny J (2009). Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc Natl Acad Sci U S A 106, 5593–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schene IF, Joore IP, Oka R, Mokry M, van Vugt AHM, van Boxtel R, van der Doef HPJ, van der Laan LJW, Verstegen MMA, van Hasselt PM, et al. (2020). Prime editing for functional repair in patient-derived disease models. Nat Commun 11, 5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep R, Brinkman EK, Leemans C, Vergara X, van der Weide RH, Morris B, van Schaik T, Manzo SG, Peric-Hupkes D, van den Berg J, et al. (2021). Impact of chromatin context on Cas9-induced DNA double-strand break repair pathway balance. Mol Cell 81, 2216–2230 e2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiroli G, Conti A, Ferrari S, Della Volpe L, Jacob A, Albano L, Beretta S, Calabria A, Vavassori V, Gasparini P, et al. (2019). Precise Gene Editing Preserves Hematopoietic Stem Cell Function following Transient p53-Mediated DNA Damage Response. Cell Stem Cell 24, 551–565 e558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Honing K, Ebert TS, and Hornung V (2016). CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism. Nature communications 7, 12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A (2020). A short history of guide RNAs: The intricate path that led to the discovery of a basic biological concept. EMBO Rep 21, e51918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert MG, Goodman DB, Wannier TM, Kaur D, Farzadfard F, Lu TK, Shipman SL, and Church GM (2021). High-throughput functional variant screens via in vivo production of single-stranded DNA. Proceedings of the National Academy of Sciences 118, e2018181118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Panday A, Elango R, and Willis NA (2019). DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol 20, 698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sertic S, Quadri R, Lazzaro F, and Muzi-Falconi M (2020). EXO1: A tightly regulated nuclease. DNA Repair 93, 102929. [DOI] [PubMed] [Google Scholar]
- Sfeir A, and Symington LS (2015). Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci 40, 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, and Zhang F (2015). High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 16, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Ren C, Liu Z, Bai Y, Chen Z, Wei Z, Wang X, Zhang Z, and Xu K (2017). Enhancing CRISPR/Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52. Int J Biochem Cell Biol 92, 43–52. [DOI] [PubMed] [Google Scholar]
- Sharon E, Chen SA, Khosla NM, Smith JD, Pritchard JK, and Fraser HB (2018). Functional Genetic Variants Revealed by Massively Parallel Precise Genome Editing. Cell 175, 544–557 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She W, Cambouris CB, and Craig NL (2015). Different DNA repair pathways are required following excision and integration of the DNA cut & paste transposon piggyBat in Saccharomyces cerevisiae. bioRxiv, 015289. [Google Scholar]
- Shen JP, and Ideker T (2017). Correcting CRISPR for copy number. Nat Genet 49, 1674–1675. [DOI] [PubMed] [Google Scholar]
- Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O, Cassa CA, Liu DR, Gifford DK, and Sherwood RI (2018). Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Gomez-Blanco J, Petassi MT, Peters JE, Ortega J, and Guarné A (2021). Structural basis for DNA targeting by the Tn7 transposon. bioRxiv, 2021.2005.2024.445525. [DOI] [PubMed] [Google Scholar]
- Shin HY, Wang C, Lee HK, Yoo KH, Zeng X, Kuhns T, Yang CM, Mohr T, Liu C, and Hennighausen L (2017). CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat Commun 8, 15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JW, Kim KH, Chao MJ, Atwal RS, Gillis T, MacDonald ME, Gusella JF, and Lee JM (2016). Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet 25, 4566–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivram H, Cress BF, Knott GJ, and Doudna JA (2021). Controlling and enhancing CRISPR systems. Nat Chem Biol 17, 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou J, Li J, Liu Y, and Wu Q (2018). Precise and Predictable CRISPR Chromosomal Rearrangements Reveal Principles of Cas9-Mediated Nucleotide Insertion. Mol Cell 71, 498–509 e494. [DOI] [PubMed] [Google Scholar]
- Shy BR, Vykunta V, Ha A, Roth TL, Talbot A, Nguyen DN, Chen YY, Blaeschke F, Vedova S, Mamedov MR, et al. (2021). Hybrid ssDNA repair templates enable high yield genome engineering in primary cells for disease modeling and cell therapy manufacturing. bioRxiv, 2021.2009.2002.458799. [Google Scholar]
- Simon AJ, Ellington AD, and Finkelstein IJ (2019). Retrons and their applications in genome engineering. Nucleic Acids Research 47, 11007–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Barbosa K, Cheng K, Leiserson MDM, Jain P, Deshpande A, Wilson DM, Ryan BM, Luo J, Ronai Ze.A., et al. (2021). A systematic genome-wide mapping of oncogenic mutation selection during CRISPR-Cas9 genome editing. Nature communications 12, 6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin BV, Kummerfeld DM, Gubar L, Seeger B, Kaiser H, Stegemann A, Roth J, Meuth SG, Pavenstadt H, Sherwood J, et al. (2020). Pervasive head-to-tail insertions of DNA templates mask desired CRISPR-Cas9-mediated genome editing events. Sci Adv 6, eaax2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Castanon O, Said K, Volf V, Khoshakhlagh P, Hornick A, Ferreira R, Wu CT, Guell M, Garg S, et al. (2020). Enabling large-scale genome editing at repetitive elements by reducing DNA nicking. Nucleic Acids Res 48, 5183–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CQ, Jiang T, Richter M, Rhym LH, Koblan LW, Zafra MP, Schatoff EM, Doman JL, Cao Y, Dow LE, et al. (2020). Adenine base editing in an adult mouse model of tyrosinaemia. Nat Biomed Eng 4, 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Yang D, Xu J, Zhu T, Chen YE, and Zhang J (2016). RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun 7, 10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Lim JM, Min S, Oh J-S, Kim DY, Woo J-S, Nishimasu H, Cho S-R, Yoon S, and Kim HH (2021). Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain. Nature communications 12, 5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriramachandran AM, Petrosino G, Mendez-Lago M, Schafer AJ, Batista-Nascimento LS, Zilio N, and Ulrich HD (2020). Genome-wide Nucleotide-Resolution Mapping of DNA Replication Patterns, Single-Strand Breaks, and Lesions by GLOE-Seq. Mol Cell 78, 975–985 e977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Nambiar M, Sharma S, Karki SS, Goldsmith G, Hegde M, Kumar S, Pandey M, Singh RK, Ray P, et al. (2012). An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 151, 1474–1487. [DOI] [PubMed] [Google Scholar]
- Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E, Mangan PA, Kulikovskaya I, Gupta M, Chen F, et al. (2020). CRISPR-engineered T cells in patients with refractory cancer. Science 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S, Alcon P, and Montoya G (2017). Class 2 CRISPR-Cas RNA-guided endonucleases: Swiss Army knives of genome editing. Nat Struct Mol Biol 24, 882–892. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. (2011). Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson BM, and Loparo JJ (2021). Repair of DNA Double-Strand Breaks by the Nonhomologous End Joining Pathway. Annual Review of Biochemistry 90, 137–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson BM, Moreno AT, Walter JC, and Loparo JJ (2020). A Mechanism to Minimize Errors during Non-homologous End Joining. Mol Cell 77, 1080–1091 e1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Snipe JR, Chan GK, Gordenin DA, and Resnick MA (2006). Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol Cell Biol 26, 7645–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker J, Jones S, Koopal B, Schmid-Burgk J, Zetsche B, Gao L, Makarova KS, Koonin EV, and Zhang F (2019a). Engineering of CRISPR-Cas12b for human genome editing. Nat Commun 10, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker J, Ladha A, Gardner Z, Schmid-Burgk JL, Makarova KS, Koonin EV, and Zhang F (2019b). RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, and Izpisua Belmonte JC (2018). In vivo genome editing via the HITI method as a tool for gene therapy. J Hum Genet 63, 157–164. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, et al. (2016). In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, and Gautier J (2011). Double-strand break end resection and repair pathway choice. Annu Rev Genet 45, 247–271. [DOI] [PubMed] [Google Scholar]
- Taheri-Ghahfarokhi A, Taylor BJM, Nitsch R, Lundin A, Cavallo AL, Madeyski-Bengtson K, Karlsson F, Clausen M, Hicks R, Mayr LM, et al. (2018). Decoding non-random mutational signatures at Cas9 targeted sites. Nucleic Acids Res 46, 8417–8434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi I, and Marmur J (1963). Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis. Nature 197, 794–795. [DOI] [PubMed] [Google Scholar]
- Takayama K, Igai K, Hagihara Y, Hashimoto R, Hanawa M, Sakuma T, Tachibana M, Sakurai F, Yamamoto T, and Mizuguchi H (2017). Highly efficient biallelic genome editing of human ES/iPS cells using a CRISPR/Cas9 or TALEN system. Nucleic Acids Res 45, 5198–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Zhu Y, Baker SL, Bowler MW, Chen BJ, Chen C, Hogg JR, Goff SP, and Song H (2016). Structural basis of suppression of host translation termination by Moloney Murine Leukemia Virus. Nature communications 7, 12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Cui T, Feng G, Guo L, Xu K, Gao Q, Li T, Li J, Zhou Q, and Li W (2018). Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov 4, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PS, and Cortez D (2020). New insights into abasic site repair and tolerance. DNA Repair (Amst) 90, 102866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigano M, Vargas DC, Tremblay-Belzile S, Fu Y, and Sfeir A (2021). Nuclear sensing of breaks in mitochondrial DNA enhances immune surveillance. Nature 591, 477–481. [DOI] [PubMed] [Google Scholar]
- Tong Y, Jørgensen TS, Whitford CM, Weber T, and Lee SY (2021). A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing. Nature communications 12, 5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tou CJ, Schaffer DV, and Dueber JE (2020). Targeted Diversification in the S. cerevisiae Genome with CRISPR-Guided DNA Polymerase I. ACS Synth Biol 9, 1911–1916. [DOI] [PubMed] [Google Scholar]
- Truong LN, Li Y, Shi LZ, Hwang PY, He J, Wang H, Razavian N, Berns MW, and Wu X (2013). Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci U S A 110, 7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, and Holmes MC (2005). Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651. [DOI] [PubMed] [Google Scholar]
- van Agtmaal EL, Andre LM, Willemse M, Cumming SA, van Kessel IDG, van den Broek W, Gourdon G, Furling D, Mouly V, Monckton DG, et al. (2017). CRISPR/Cas9-Induced (CTGCAG)n Repeat Instability in the Myotonic Dystrophy Type 1 Locus: Implications for Therapeutic Genome Editing. Mol Ther 25, 24–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek M, Capurso D, Carter MM, Thompson MS, Frias E, Russ C, Reece-Hoyes JS, Nye C, Gradia S, Vidal B, et al. (2016). DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Mol Cell 63, 633–646. [DOI] [PubMed] [Google Scholar]
- Velimirovic M, Zanetti LC, Shen MW, Fife JD, Lin L, Cha M, Akinci E, Barnum D, Yu T, and Sherwood RI (2021). Peptide fusion improves prime editing efficiency. bioRxiv, 2021.2009.2022.461415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villiger L, Grisch-Chan HM, Lindsay H, Ringnalda F, Pogliano CB, Allegri G, Fingerhut R, Haberle J, Matos J, Robinson MD, et al. (2018). Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med 24, 1519–1525. [DOI] [PubMed] [Google Scholar]
- Vo PLH, Ronda C, Klompe SE, Chen EE, Acree C, Wang HH, and Sternberg SH (2021). CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat Biotechnol 39, 480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend LE, and Krawczyk PM (2017). Nick-initiated homologous recombination: Protecting the genome, one strand at a time. DNA Repair (Amst) 50, 1–13. [DOI] [PubMed] [Google Scholar]
- Walton RT, Christie KA, Whittaker MN, Kleinstiver BP (2020). Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang F, and Gao G (2020a). CRISPR-Based Therapeutic Genome Editing: Strategies and In Vivo Delivery by AAV Vectors. Cell 181, 136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xue W, Yan L, Li X, Wei J, Chen M, Wu J, Yang B, Yang L, and Chen J (2017). Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell Res 27, 1289–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xue W, Zhang H, Gao R, Qiu H, Wei J, Zhou L, Lei YN, Wu X, Li X, et al. (2021a). Eliminating base-editor-induced genome-wide and transcriptome-wide off-target mutations. Nat Cell Biol 23, 552–563. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu J, Janssen JM, Le Bouteiller M, Frock RL, and Goncalves M (2021b). Precise and broad scope genome editing based on high-specificity Cas9 nickases. Nucleic Acids Res 49, 1173–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Yang J, Zhong Z, Vanegas JA, Gao X, and Kolomeisky AB (2021c). A general theoretical framework to design base editors with reduced bystander effects. Nature communications 12, 6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zong Y, Lin Q, Zhang H, Chai Z, Zhang D, Chen K, Qiu JL, and Gao C (2020b). Precise, predictable multi-nucleotide deletions in rice and wheat using APOBEC-Cas9. Nat Biotechnol 38, 1460–1465. [DOI] [PubMed] [Google Scholar]
- Wang X, Ding C, Yu W, Wang Y, He S, Yang B, Xiong YC, Wei J, Li J, Liang J, et al. (2020c). Cas12a Base Editors Induce Efficient and Specific Editing with Low DNA Damage Response. Cell Rep 31, 107723. [DOI] [PubMed] [Google Scholar]
- Wang X, Li J, Wang Y, Yang B, Wei J, Wu J, Wang R, Huang X, Chen J, and Yang L (2018). Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat Biotechnol 36, 946–949. [DOI] [PubMed] [Google Scholar]
- Wang Z, and Mosbaugh DW (1989). Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J Biol Chem 264, 1163–1171. [PubMed] [Google Scholar]
- Webber BR, Lonetree CL, Kluesner MG, Johnson MJ, Pomeroy EJ, Diers MD, Lahr WS, Draper GM, Slipek NJ, Smeester BA, et al. (2019). Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat Commun 10, 5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisheit I, Kroeger JA, Malik R, Klimmt J, Crusius D, Dannert A, Dichgans M, and Paquet D (2020). Detection of Deleterious On-Target Effects after HDR-Mediated CRISPR Editing. Cell Rep 31, 107689. [DOI] [PubMed] [Google Scholar]
- Wienert B, Nguyen DN, Guenther A, Feng SJ, Locke MN, Wyman SK, Shin J, Kazane KR, Gregory GL, Carter MAM, et al. (2020). Timed inhibition of CDC7 increases CRISPR-Cas9 mediated templated repair. Nature communications 11, 2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde JJ, Aida T, Del Rosario RCH, Kaiser T, Qi P, Wienisch M, Zhang Q, Colvin S, and Feng G (2021). Efficient embryonic homozygous gene conversion via RAD51-enhanced interhomolog repair. Cell 184, 3267–3280 e3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J, Gerber AP, and Keller W (2002). tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J 21, 3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Wang S, Han R, Li Z, Gabel C, Mukherjee IA, and Chang L (2021). Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition. Molecular Cell 81, 4457–4466.e4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie N, Gong H, Suhl JA, Chopra P, Wang T, and Warren ST (2016). Reactivation of FMR1 by CRISPR/Cas9-Mediated Deletion of the Expanded CGG-Repeat of the Fragile X Chromosome. PLOS ONE 11, e0165499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Wang J, Liu Y, Xie L, Su B, Mou D, Wang L, Liu T, Wang X, Zhang B, et al. (2019). CRISPR-Edited Stem Cells in a Patient with HIV and Acute Lymphocytic Leukemia. N Engl J Med 381, 1240–1247. [DOI] [PubMed] [Google Scholar]
- Xu P, Liu Z, Liu Y, Ma H, Xu Y, Bao Y, Zhu S, Cao Z, Wu Z, Zhou Z, et al. (2021a). Genome-wide interrogation of gene functions through base editor screens empowered by barcoded sgRNAs. Nat Biotechnol. [DOI] [PubMed] [Google Scholar]
- Xu X, Chemparathy A, Zeng L, Kempton HR, Shang S, Nakamura M, and Qi LS (2021b). Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol Cell. [DOI] [PubMed] [Google Scholar]
- Yan S, Tu Z, Liu Z, Fan N, Yang H, Yang S, Yang W, Zhao Y, Ouyang Z, Lai C, et al. (2018). A Huntingtin Knockin Pig Model Recapitulates Features of Selective Neurodegeneration in Huntington’s Disease. Cell 173, 989–1002 e1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Hunnewell P, Alfonse LE, Carte JM, Keston-Smith E, Sothiselvam S, Garrity AJ, Chong S, Makarova KS, Koonin EV, et al. (2019). Functionally diverse type V CRISPR-Cas systems. Science 363, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Scavuzzo MA, Chmielowiec J, Sharp R, Bajic A, and Borowiak M (2016). Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci Rep 6, 21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Wang C, Hong L, Sun N, Chen D, Chen S, and Han F (2018). Programmable DNA repair with CRISPRa/i enhanced homology-directed repair efficiency with a single Cas9. Cell Discov 4, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CD, Richardson CD, and Corn JE (2019). Advances in genome editing through control of DNA repair pathways. Nature Cell Biology 21, 1468–1478. [DOI] [PubMed] [Google Scholar]
- Yeh WH, Chiang H, Rees HA, Edge ASB, and Liu DR (2018). In vivo base editing of post-mitotic sensory cells. Nature communications 9, 2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh WH, Shubina-Oleinik O, Levy JM, Pan B, Newby GA, Wornow M, Burt R, Chen JC, Holt JR, and Liu DR (2020). In vivo base editing restores sensory transduction and transiently improves auditory function in a mouse model of recessive deafness. Science translational medicine 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yla-Herttuala S (2018). CRISPR/Cas9 and p53: An Odd Couple Requiring Relationship Management. Mol Ther 26, 2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, Liu H, La Russa M, Xie M, Ding S, et al. (2015). Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16, 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ma Y, Huang T, Chen Y, Peng Y, Li B, Li J, Zhang Y, Song B, Sun X, et al. (2018). Genetic Modulation of RNA Splicing with a CRISPR-Guided Cytidine Deaminase. Mol Cell 72, 380–394 e387. [DOI] [PubMed] [Google Scholar]
- Zafra MP, Schatoff EM, Katti A, Foronda M, Breinig M, Schweitzer AY, Simon A, Han T, Goswami S, Montgomery E, et al. (2018). Optimized base editors enable efficient editing in cells, organoids and mice. Nature biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn KE, Jensen RB, Wood RD, and Doublie S (2021). Human DNA polymerase theta harbors DNA end-trimming activity critical for DNA repair. Mol Cell 81, 1534–1547 e1534. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zelensky AN, Schimmel J, Kool H, Kanaar R, and Tijsterman M (2017). Inactivation of Pol theta and C-NHEJ eliminates off-target integration of exogenous DNA. Nature communications 8, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, et al. (2017). Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nature biotechnology 35, 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JP, Li XL, Li GH, Chen W, Arakaki C, Botimer GD, Baylink D, Zhang L, Wen W, Fu YW, et al. (2017). Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol 18, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen L, Zhu B, Wang L, Chen C, Hong M, Huang Y, Li H, Han H, Cai B, et al. (2020a). Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat Cell Biol 22, 740–750. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhu B, Chen L, Xie L, Yu W, Wang Y, Li L, Yin S, Yang L, Hu H, et al. (2020b). Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nature biotechnology 38, 856–860. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Davis L, and Maizels N (2021). Pathways and signatures of mutagenesis at targeted DNA nicks. PLoS Genet 17, e1009329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, and Li GM (2005). Reconstitution of 5’-directed human mismatch repair in a purified system. Cell 122, 693–705. [DOI] [PubMed] [Google Scholar]
- Zhao B, Chen S-AA, Lee J, and Fraser HB (2021a). Bacterial retrons enable precise gene editing in human cells. bioRxiv, 2021.2003.2029.437260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Watanabe G, Morten MJ, Reid DA, Rothenberg E, and Lieber MR (2019). The essential elements for the noncovalent association of two DNA ends during NHEJ synapsis. Nat Commun 10, 3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Li J, Li S, Xin X, Hu M, Price MA, Rosser SJ, Bi C, and Zhang X (2021b). Glycosylase base editors enable C-to-A and C-to-G base changes. Nature biotechnology 39, 35–40. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhang H, Xiong T, Wang J, Yang D, Zhu D, Li J, Yang Y, Sun C, Zhao Y, et al. (2020). Suppression of SHROOM1 Improves In Vitro and In Vivo Gene Integration by Promoting Homology-Directed Repair. Int J Mol Sci 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Liang S-Q, Liu B, Liu P, Kwan S-Y, Wolfe SA, and Xue W (2021). Development of a flexible split prime editor using truncated reverse transcriptase. bioRxiv, 2021.2008.2026.457801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Meng Y, Campbell JL, and Shen B (2020). Multiple roles of DNA2 nuclease/helicase in DNA metabolism, genome stability and human diseases. Nucleic Acids Research 48, 16–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Liu J, Wu H, Zhu Q, Yan Y, Meng H, Chen PR, and Yi C (2021). Increasing the efficiency and precision of prime editing with guide RNA pairs. Nature chemical biology. [DOI] [PubMed] [Google Scholar]
- Zuccaro MV, Xu J, Mitchell C, Marin D, Zimmerman R, Rana B, Weinstein E, King RT, Palmerola KL, Smith ME, et al. (2020). Allele-Specific Chromosome Removal after Cas9 Cleavage in Human Embryos. Cell 183, 1650–1664 e1615. [DOI] [PubMed] [Google Scholar]
- Zuo E, Huo X, Yao X, Hu X, Sun Y, Yin J, He B, Wang X, Shi L, Ping J, et al. (2017). CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol 18, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]