SUMMARY
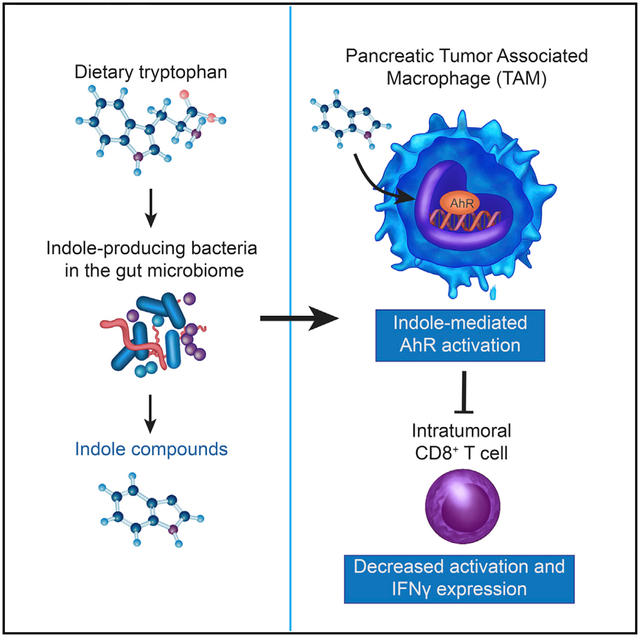
The aryl hydrocarbon receptor (AhR) is a sensor of products of tryptophan metabolism and a potent modulator of immunity. Here, we examined the impact of AhR in tumor-associated macrophage (TAM) function in pancreatic ductal adenocarcinoma (PDAC). TAMs exhibited high AhR activity and Ahr-deficient macrophages developed an inflammatory phenotype. Deletion of Ahr in myeloid cells or pharmacologic inhibition of AhR reduced PDAC growth, improved efficacy of immune checkpoint blockade, and increased intra-tumoral frequencies of IFNγ+CD8+ T cells. Macrophage tryptophan metabolism was not required for this effect. Rather, macrophage AhR activity was dependent on Lactobacillus metabolization of dietary tryptophan to indoles. Removal of dietary tryptophan reduced TAM AhR activity and promoted intra-tumoral accumulation of TNFα+IFNγ+CD8+ T cells; provision of dietary indoles blocked this effect. In patients with PDAC, high AHR expression associated with rapid disease progression and mortality, as well as with an immune-suppressive TAM phenotype, suggesting conservation of this regulatory axis in human disease.
Graphical abstract
In brief
AhR directs macrophage polarization. Hezaveh et al. identified a key role for AhR in tumor macrophage function in pancreatic cancer, with AhR suppressing inflammatory T cell infiltration and promoting growth. AhR was activated by gut microbiome-produced tryptophan metabolites and human disease showed association of macrophage AHR expression and worse outcomes.
INTRODUCTION
Despite significant improvements in cancer therapies, outcomes for patients afflicted with pancreatic ductal adenocarcinoma (PDAC) remain grim as PDAC exhibits resistance to therapeutic approaches, including checkpoint blockade. Interestingly, microbiota appear to impact outcomes of PDAC. In long-term PDAC survivors, increased microbiome diversity correlates with survivorship and immunologic features of the tumor microenvironment (TME). Furthermore, fecal microbiota transplant (FMT) from long-term survivors reduced tumor size in mouse models, whereas FMT from short-term survivors (STS) resulted in larger tumor sizes and increases in CD4+ FOXP3+ T cells (Riquelme et al., 2019). Thus, the microbiota can promote or inhibit tumor progression in PDAC, impacting the TME cellular composition.
An immunologically important class of bacterial metabolites is generated by metabolization of the amino acid tryptophan (Trp) to indole and related compounds (herein, collectively referred to as indoles) (Roager and Licht, 2018). Indoles are essential for mucosal barrier integrity and suppression of inflammation by activation of the transcriptional regulator AhR (Monteleone et al., 2011). Upon binding of indoles in the cytoplasm, AhR translocates to the nucleus. Ahr activation exerts potent effects on T cells, dendritic cells, and macrophages. Macrophages are a major component of the immune infiltrate in PDAC, providing stromal support for tumor growth (DeNardo and Ruffell, 2019) and resistance to chemotherapy (Halbrook et al., 2019). AhR activation can drive macrophages to acquire an immune-suppressive phenotype. AhR induces the immune-suppressive cytokine interleukin (IL)-10 (Shinde et al., 2018) and drives the expression of transforming growth factor (TGF)-α, TGF-β, and arginase (Arg1) (Franchini et al., 2019; Rothhammer et al., 2018). Importantly, AhR expression is elevated in myeloid-lineage cells relative to other cell types (Goudot et al., 2017; Shinde et al., 2018), which is suggestive of increased sensitivity to AhR ligands.
Based on these findings, we asked whether the microbiota could drive immune suppression in the PDAC TME by inducing AhR activity in tumor-associated macrophages (TAMs). To test this, we deleted Ahr in PDAC TAMs or inhibited AhR activity pharmacologically. Loss of AhR function caused a reduction in tumor size, and Ahr-deficient TAMs acquired a proinflammatory phenotype with increased intra-tumoral IFNγ+ CD8+ T cells. Microbial production of indole compounds was the key driver of AhR activity. In human PDAC samples, AHR expression was associated with rapid disease progression and mortality, as well as with an immune-suppressive TAM phenotype. Thus, microbial metabolites activate AhR in macrophages, driving macrophage polarization and PDAC outcomes.
RESULTS
Deletion of AhR drives inflammatory polarization of TAMs and CD8+ T cell infiltration in the PDAC TME
To understand the role of macrophages in PDAC, we used an orthotopic model of pancreatic cancer utilizing the mT4 organoid line cloned from a primary tumor isolated from Trp53+/LSL-R172H Kras+/LSL-G12D Pdx1-Cre (KPC) mice (Boj et al., 2015). First, we performed RNA-seq comparing transcriptomes of intra-tumoral versus tissue-resident macrophages. Compared with tissue-resident macrophages, PDAC TAMs exhibited increased expression of 880 genes, including the pro-tumor genes Arg1, Nos2, and Cd274 (the gene encoding PD-L1) (Figures 1A and S1A). Ingenuity pathway gene set enrichment analysis (iGSEA) predicted increased AhR activity in TAMs relative to the healthy tissue-resident macrophages (Figure S1B) and there was an increased expression of the AhR-responsive gene Cyp1b1 in TAMs compared with the resident macrophages (Figures 1A and 1B). To test the prediction that macrophage AhR was required for suppression of immunity in PDAC, we deleted AhR from Mφ by crossing Lyz2cre/+ × AhRfl/fl mice (Lyz2cre/+Ahrfl/fl) (Shinde et al., 2018). Since the Lyz2-CRE promoter can show activity in several myeloid cell types, we first tested the relative activity of CRE in TAMs by crossing Lyz2cre/+ × Cg-Gt(ROSA) 26Sortm9(CAG-tdTomato)Hze mice (Madisen et al., 2010), examining tdTomato expression in TAMs and DCs. In the tumors, 98% of the CD11b+F4/80+ TAMs expressed tdTomato (Figure S1C), contrasting with intra-tumoral CD11c+ DCs where 20% were tdTomato+ (Figure S1C), showing Lyz2cre/+ is an appropriate model to study macrophage AhR function in PDAC.
Figure 1. Deletion of AhR in macrophages drives inflammatory polarization of TAMs and CD8+ T cells in the PDAC TME.
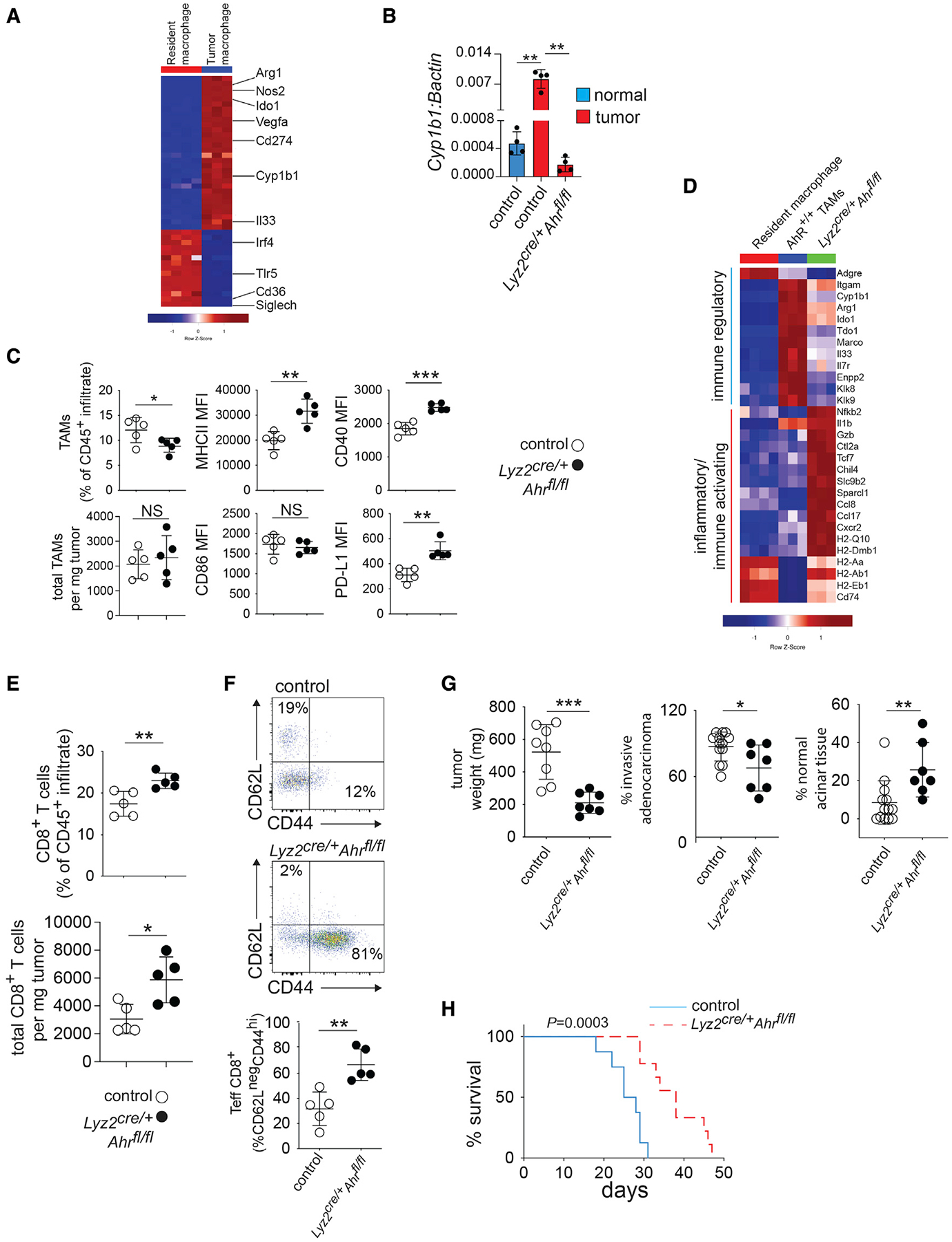
(A) CD45+CD11b+F4/80+ macrophages were enriched by flow cytometry sorting from normal pancreas (i.e., resident macrophages) or PDAC tumors 14 days after tumor implantation. Heatmap shows hierarchical clustering depicting differentially expressed RNA transcripts (>2.0 logFC, FDR p < 0.01). Each column represents an individual mouse.
(B) Macrophages were sorted from normal pancreas or tumors from mice of the indicated genotype, and mRNA expression of Cyp1b1 relative to βactin was determined by qRT-PCR.
(C) Expression of surface markers in TAMs from tumor-bearing mice of the indicated genotype were determined by flow cytometry. MFI, mean fluorescence intensity.
(D) Heatmaps showing differential expression (FDR, <0.01; logFC, >1) of selected inflammatory or immune-regulatory markers in FACS-sorted TAMs and healthy tissue macrophages as described in (A and B).
(E) Intra-tumoral CD3+CD8+ T cell numbers as a percent of the CD45+ immune infiltrate were determined by flow cytometry in d14 tumors in Lyz2cre/+Ahrfl/fl versus control tumor-bearing mice.
(F) Percent of CD44- and CD62L-positive intra-tumoral CD8+ T cells from samples described in (E) was determined by flow cytometry.
(G) Day-14 tumor weight and pathology in mice of the indicated genotype.
(H) Survival curves for tumor-bearing mice of the indicated genotype. n = 8 mice per group. (see also Figure S1). * p <.05, ** p <.01, *** p <.001.
Importantly, whereas WT TAMs exhibited a 10-fold increase in expression of Cyp1b1 compared with control macrohages, Lyz2cre/+Ahrfl/fl TAMs had an abrogation of relative Cyp1b1, demonstrating the dependence on AhR for Cyp1b1 expression (Figure 1B). AhR deletion reduced the percentage of TAMs in the CD45+ infiltrate but did not reduce the absolute numbers of TAMs (Figure 1C). However, AhR deletion increased the expression of MHCII, CD40, and PD-L1, indicating increased TAM activation (Figure 1C). Ahr deletion altered the expression of 416 genes (Figures S1D and S1E). TAMs from Lyz2cre/+Ahrfl/fl tumors showed a decreased immune-regulatory transcriptional signature as compared with TAMs from control tumors, characterized by reductions in the expression of Arg1 and Ido1 (Figure 1D). In agreement with protein expression data (Figure 1C), TAMs from Lyz2cre/+Ahrfl/fl tumors were more proinflammatory with increased Il1b, Gzb, and MHCII genes expression (Figure 1D). TAMs from PDAC tumors can proliferate in situ and contribute to fibrosis-expressing genes for extracellular matrix (ECM) proteins (Zhu et al., 2017). When the TAMs lacked AhR, function genes involved in cell-cycle progression were decreased and the TAMs showed increased expression of pro-apoptotic genes, as well as caspases, suggesting reduced proliferation and self-renewal capability (Figure S1F). Likewise, the TAM ECM transcriptional signature was decreased in the absence of AhR (Figure S1G). Thus, the data suggest that AhR is a key driver of the TAM pro-tumor phenotype, and when AhR function is abrogated, TAM phenotypic polarization is shifted to a proinflammatory state.
Next, we sought to understand how AhR-mediated alterations in TAM polarization impact T cells in the TME. We found that the frequency and number of CD8+ T cells were increased within the TME of Lyz2cre/+Ahrfl/fl mice (Figure 1E). Moreover, intra-tumoral CD8+ T cells from Lyz2cre/+Ahrfl/fl mice exhibited a change from a naive (CD62L+CD44lo) to an effector/memory (CD62LnegCD44hi) phenotype (Figure 1F), suggesting increased T cell activation. Moreover, tumor weight and the extent of invasive adenocarcinoma were decreased in Lyz2cre/+Ahrfl/fl mice (Figure 1G), leading to improved survival compared with littermate controls (Figure 1H). Further, we observed decreased tumor burden in Lyz2cre/+Ahrfl/fl mice which bear tumors from several other KPC-derived tumor cell lines (Figure S1H), indicating that lack of AhR in TAMs reduces tumor burden across multiple PDAC tumor models.
Pharmacologic inhibition of AhR promotes inflammation in the TME and improves responses to therapy
To test whether pharmacologic inhibition of AhR could impact the immune infiltrate phenotype and tumor growth, we treated mice with the AhR inhibitor CH223191 (Shinde et al., 2018). TAMs from tumor-bearing mice receiving CH223191 had increased expression of MHCII, CD40, and PD-L1, increases in IFNγ and granzyme B-expressing CD8+ T cells, and enhanced tumor control (Figures 2A–2C). Since CH223191 inhibits AhR activity in all cell types, we tested the contribution of macrophage AhR function to these results by repeating AhR inhibition experiments using Lyz2cre/+Ahrfl/fl mice. On day 14, tumors had a 50% reduction in weight when the mice received CH223191 in the littermate control groups. In contrast, the addition of CH223191 did not have any additional effect on tumor control in Lyz2cre/+Ahrfl/fl mice (Figure 2D), indicating that the effect of the drug is through targeting of AhR in macrophages. AhR has been reported to directly affect tumor cell growth (Murray et al., 2014); therefore, we tested the impact of AhR deletion in PDAC cells on tumor growth. Deletion of AhR in tumor cells by CRISPR (Figure 2E) nullified PDAC cell responses to AhR agonists in vitro (Figure 2F); however, there was no effect on tumor growth in vivo compared with mock PDAC tumors (Figure 2G). These data suggest that macrophage-specific AhR function is the main contributor to AhR-dependent tumor growth.
Figure 2. Pharmacologic inhibition of AhR promotes inflammation in the TME and improves responses to immune therapy.

(A) CD45+CD11b+F4/80+ TAMs were analyzed by flow cytometry in d14 tumors from B6 mice +/− treatment with the AhR inhibitor CH223191 as described in STAR Methods. MFI, mean fluorescence intensity.
(B) Tumor infiltrating CD3+CD8+ T cells were analyzed from mice described in (A) by flow cytometry. GZMB-granzyme B.
(C) Tumor weight of d14 tumors from mice described in (A).
(D) D14 tumor weight from mice of the indicated genotype +/− CH223191 treatment as described in (A).
(E) Representative Western blot showing loss of AhR expression in mT4 cell clone transfected with control versus AhR guide RNA.
(F) Cyp1a1 quantification by qRT-PCR of control and mT4 cultures treated with the AhR agonists ITE and FICZ.
(G) B6 mice were orthotopically implanted with the indicated mT4 clones +/− CH223191 beginning 4 days after implantation. Weights are shown for d14 tumors.
(H) Survival curves of mice treated with AhR antagonist, anti-PD-L1 blockade, or a combination of the two versus controls. n = 10 mice per group.
(I) Survival curves of iKPC mice treated with AhR antagonist versus controls 12 weeks post-tamoxifen treatment. n = 14 mice per group. * p <.05, ** p <.01, ***, NS-not significant.
Since PD-L1 was increased in TAMs from both Lyz2cre/+Ahrfl/fl (Figure 1C) and CH223191-treated mice (Figure 2B), we tested for a potential effect of dual AhR and PD-L1 blockade. Control mice (isotype IgG-treated) had a median survival of 25 days (Figure 2H). Administration of either αPD-L1 IgG or AhR inhibitor alone increased survival; however, combining αPD-L1 IgG and AhR inhibition had the strongest effect by extending the survival 2-fold compared with the control treatment and by increasing the survival up to 50% higher than either therapy alone (Figure 2H). We then tested the effect of AhR inhibition on autochthonous PDAC tumors using a tamoxifen inducible KPC (iKPC) model of PDAC (Maddipati and Stanger, 2015). Median survival for the control group was 28 days post-initiation of treatment (Figure 2I). In contrast, mice treated with AhR inhibitor had improved survival (Figure 2I), suggesting AhR inhibition can improve outcomes for both in situ pancreatic adenocarcinoma, as well as orthotopic tumors. Hence, the data show that AhR is a central driver of TAM function, and suppression of inflammatory T cell maturation, and its inhibition improves responses to immune checkpoint blockade.
Macrophage AhR activity shapes the transcriptional landscape in PDAC
To understand the impact of macrophage AhR function on the TME, we conducted CyTOF and single-cell RNA sequence (scRNA-seq) analysis of day-14 tumors (Halaby et al., 2019; Levine et al., 2015). We identified 32 clusters of intra-tumoral immune cells by CyTOF, with the majority being myeloid or granulocytic (Figures 3A and S2A). There were 3 macrophage clusters identified by the expression of F4/80 (clusters 12, 13, and 22). Macrophage cluster 22 expressed the immune-regulatory polarization markers CD206 and PD-L1, as well as the inflammatory monocyte marker Ly6C, suggesting monocytic origin (Figures 3B and S2A). In contrast, macrophage clusters 12 and 13 were Ly6Cneg, with macrophage cluster 12 expressing PD-L1, MHCII, and Ly6G compared with cluster 13, which had the lowest staining of all three markers (Figures 3B and S2A). Furthermore, we identified 2 populations of CD11c+ dendritic cells (DCs, clusters 14 and 16) differentiated by the expression of PD-L1 (in cluster 14) and the integrin CD103 (in cluster 16) (Figure S2A). In addition, DC cluster 14 had expression of several markers found in the macrophage clusters, including CD11b, CD44, and Nos2 (iNOS). CD8+ T cells appeared as 5 related clusters (clusters 11, 15, 17, 18, and 28) differentiated by the expression of T bet, a key transcription factor driving inflammatory Th1 T cell lineage commitment (Szabo et al., 2000) and CX3CR1, a marker of inflammatory intra-tumoral T cells (Yamauchi et al., 2020) (Figure S2A). Deletion of TAM Ahr did not impact cell clustering or the relative composition of the immune infiltrate (Figures 2B and S2C); however, loss of Ahr reduced iNOS, CCL4, and TNFα detectable by CyTOF in all macrophage clusters (clusters 12, 13, and 22), suggesting alteration in functional activity (Figure 3C).
Figure 3. Macrophage AhR activity shapes the immune transcriptional landscape in PDAC.
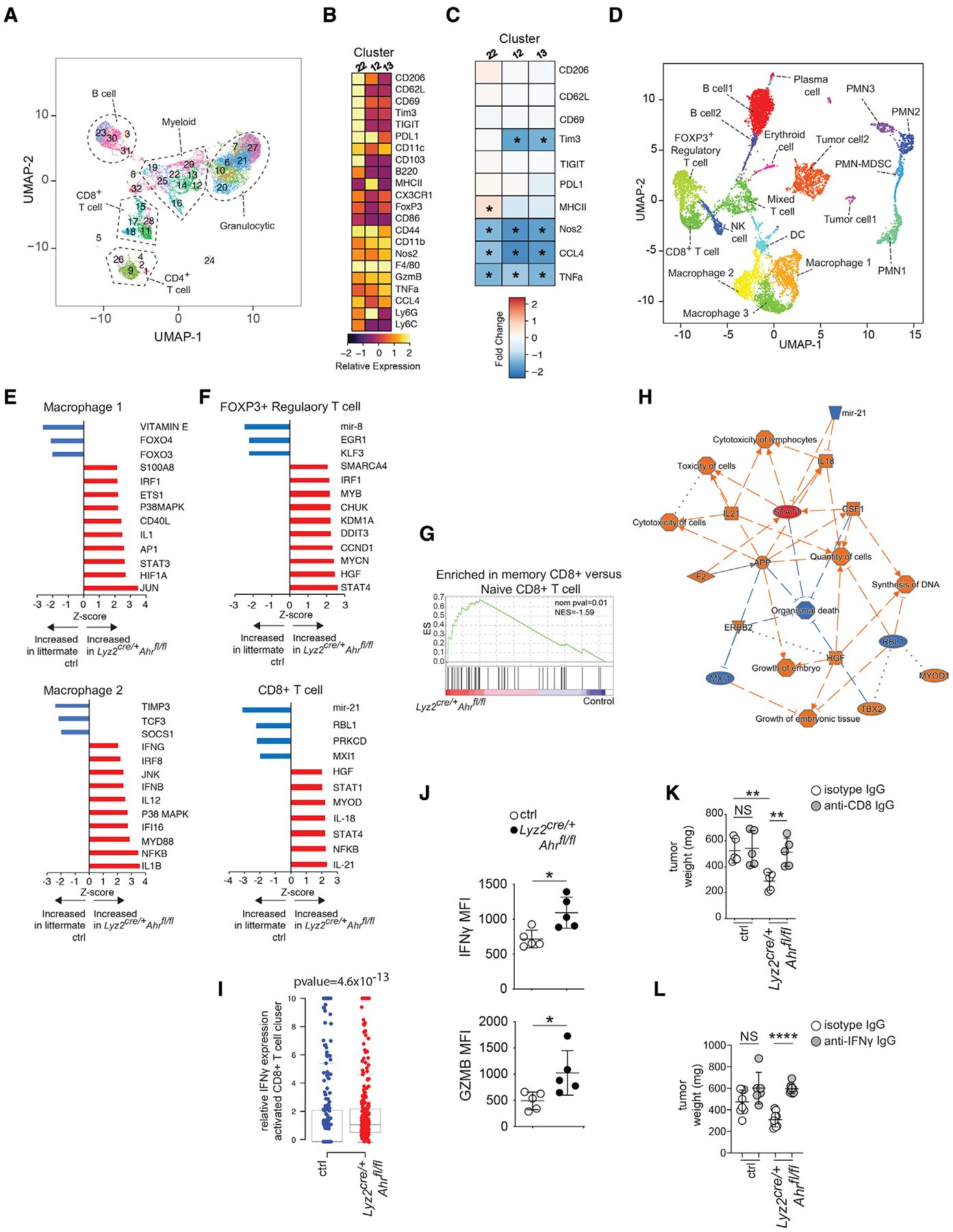
(A) UMAP and phenograph analysis of the CD45+ infiltrate in d14 PDAC tumors showing the major immune subpopulations identified by CyTOF analysis. Data from 4 (control) or 5 (Lyz2cre/+Ahrfl/fl) biological replicates were concatenated in the plots.
(B) Heatmap depicting relative expression of the indicated markers in d14 tumors of Lyz2cre/+Ahrfl/fl-tumor-bearing mice versus controls for the 3 F4/80+ macrophage clusters (Cluster 12, 13, and 22).
(C) Heatmap showing relative expression of indicated markers in each TAM cluster in Lyz2cre/+Ahrfl/f versus control determined by CyTOF. Red squares indicate increased expression compared with control baseline, whereas blue squares indicate decreased expression relative to control baseline.
(D) UMAP showing scRNA-seq data of pooled whole day-14 tumor samples for both Lyz2cre/+Ahrfl/fl and control tumors. n = 3 mice per group. PMN- poly morphonuclear leukocyte.
(E) iGSEA analysis of the macrophage clusters 1 and 2 for enrichment of genes associated with the indicated pathways.
(F) iGSEA analysis of the clusters FOXP3+ regulatory T cell and CD8+ T cell for enrichment of genes associated with the indicated pathways.
(G) GSEA plot of CD8+ T cell cluster for enrichment of genes associated with memory CD8+ T cell differentiation was done with MSigDB (C7) ES = enrichment score.
(H) Summary network analysis of scRNA-seq data for the activated CD8+ T cell cluster showing the interactions between upstream regulators, downstream genes, and physiological functions of activated CD8+ T cells. Red symbols/lines indicate activation, whereas blue ones indicate inhibition.
(I) Relative IFNγ single-cell gene expression in the activated CD8+ T cells cluster.
(J) Mean fluorescence intensity of IFNγ (left) and granzyme B (right) in Lyz2cre/+Ahrfl/fl and control CD8+ T cells from day-14 tumors as determined by flow cytometry.
(K and L) Effect of CD8+ T cell depletion (K) or IFNγ blockade (L) on PDAC tumor weight in Lyz2cre/+Ahrfl/fl and control tumor-bearing mice. Tumors were collected at day 14. (see also Figure S2). * p <.05, ** p <.01, **** p <.0001, NS- not significant.
To understand the transcriptomic changes in the TME caused by loss of macrophage Ahr, we performed scRNA-seq analysis of 10,000 cells from Lyz2cre/+Ahrfl/fl and control tumors. We identified 18 clusters of cells in the tumor, including multiple clusters of macrophages, T cells, and granulocytes (Figures 3D and S2D). Similar to the CyTOF results, we identified 3 clusters of macrophages (Figure 3D). Macrophage cluster 1 expressed Arg1 and Tgm2 (a gene involved in macrophage remodeling of ECM in tumors) (Afik et al., 2016), whereas macrophage clusters 2 and 3 expressed Klf4 encoding the transcription factor Krüppel-like factor 4, a driver of regulatory macrophage function in the TME (Liao et al., 2011) (Figure S2E). Further distinguishing between macrophage cluster 2 and 3, cluster 2 expressed the immune-suppressive macrophage marker Chil3/Ym1 (Zhu et al., 2020), whereas there was an enrichment in cluster 3 of mRNA for Il1b and Ptgs2 (the gene encoding prostaglandin-endoperoxide synthase 2) (Figure S2E). Thus, cumulatively, the data show that the macrophage populations in the PDAC TME exhibit a transcriptional profile with characteristics of pro-tumor, alternative polarization states.
Macrophage clusters 1 and 2 showed a significant transcriptional change in Lyz2cre/+Ahrfl/fl tumors. iGSEA analysis showed that in Lyz2cre/+Ahrfl/fl tumors, macrophage cluster 1 was enriched for expression of target genes for several interferon-induced transcription factors (Jun/Ap1, STAT3, and IRF1), signal transduction networks, and inflammatory mediators, with reduction of genes driven by the immune-regulatory transcription factors Foxo3 (Dejean et al., 2009) and Foxo4 (Zhu et al., 2015) (Figure 3E). iGESA analysis of macrophage cluster 2 showed an even stronger enrichment of interferon response pathways in Lyz2-cre/+Ahrfl/fl tumors, including IFNγ, IRF8, IFI16, IFNβ, and the MYD88 pathway (Figure 3E). Surprisingly, iGSEA analysis of cluster 3 showed that loss of AhR had relatively little impact with no enrichment of genes associated with inflammation or interferon responses (Figure S2F), suggesting a differential effect of AhR deletion on TAM populations in the PDAC TME.
There were 3 T cell clusters identified in the TME, including a FOXP3+ T reg cell cluster, a mixed population of CD4+ and CD8+ T cells, and a population of Tc1-like CD8+ T cells (St Paul and Ohashi, 2020) with expression of Ifng (Figure S2D). The CD8+ T cell cluster also showed expression of the exhaustion marker Lag3 (Figure S2D); however, expression of other T cell exhaustion markers (e.g., PD1, TIM3, and CTLA4) were low, suggesting that the population was not functionally exhausted per se. Transcriptomic analysis of the FOXP3+ T reg cell cluster showed that loss of Ahr in macrophages caused enrichment for a number of pathways associated with plasticity and loss of T reg cell suppressive function, including STAT4 (Cuadrado et al., 2018) and IRF1 (Fragale et al., 2008) (Figure 3F). In addition, there was enrichment of several pathways involved in T reg cell homeostasis in the FOXP3+ T reg cell cluster in Lyz2cre/+Ahrfl/fl tumors, including CHUK/IKKA and MYB (Chen et al., 2015; Dias et al., 2017), as well as the stress response/apoptosis pathway driven by DDIT3/CHOP (Figure 3F). Together, these results suggest that the loss of Mφ AhR may promote a switch to an inflammatory phenotype in FOXP3+ T reg cells with increased cellular stress and population turnover.
We also observed inflammatory transcriptional changes in the CD8+ T cell cluster in Lyz2cre/+Ahrfl/fl tumors compared with controls. There was enrichment of IFN responsive pathways (e.g., STAT1, STAT4, NFKB, and IL-12) in the CD8+ T cell cluster (Figure 3F). This showed that the intra-tumoral CD8+ T cells were responding to the increased inflammatory milieu in the TME of Lyz2cre/+Ahrfl/fl tumors with heightened functional maturation. Supporting this prediction, GSEA analysis of the differentially expressed genes from the CD8+ T cell clusters identified enrichment of upregulated genes characteristic of memory CD8+ T cells (Figure 3G), suggesting a potentially key role for this population for the reduced tumor burden seen in macrophage Ahr-deficient tumors.
Network analysis of the transcriptional signatures impacted in the CD8+ T cell cluster from Lyz2cre/+Ahrfl/fl tumors identified a key role for STAT1 in the observed global transcriptional pathway alterations (Figure 3H). Since STAT1 is a driver of IFNγ signaling, we predicted that increased IFNγ may be a significant contributor to inflammation and CD8+ T cell activation in the intra-tumoral milieu of Lyz2cre/+Ahrfl/fl tumors. In agreement with this, the CD8+ T cell cluster showed increased Ifng expression when macrophage AhR was deleted compared with controls (Figure 3I). This was confirmed at the protein level by flow cytometry where we observed increased IFN-γ in CD8+ T cells from Lyz2cre/+Ahrfl/fl tumors compared with control tumors (Figure 3J). Because IFNγ is a driver of anti-tumor immunity (Braümuller et al., 2013) (Shankaran et al., 2001), we tested the impact of IFNγ on tumor growth in PDAC. Depletion of CD8+ T cells abrogated the effect of macrophage Ahr deletion on tumor size, showing that CD8+ T cells are needed for control of tumor growth (Figure 3K). We found that antibody blockade of IFNγ did not impact tumor size in controls (Figure 3L); however, IFNγ blockade in Lyz2cre/+Ahrfl/fl tumor-bearing mice rescued tumor growth to control levels, showing that increased IFNγ production by CD8+ T cells is an important driver of the reduced tumor burden observed in Lyz2cre/+Ahrfl/fl tumors (Figure 3L).
Indole-producing microbiota drive suppression in the TME promoting tumor growth
Next, we investigated sources of ligands that may drive AhR activity in the TME. Because AhR is a receptor for ligands generated by mammalian and bacterial metabolization of the amino acid Trp (Shinde and McGaha, 2018), we focused on these pathways. Trp is metabolized to the AhR ligand kynurenine by the enzyme indoleamine 2,3 dioxygenase 1 (IDO1) (Shinde and McGaha, 2018); however, neither the inhibition of IDO1 by treatment of tumor-bearing mice with 1-methyl tryptophan (1MT) (Ravishankar et al., 2015) nor the deletion of Ido1 impacted tumor growth (Figures 4A and S3A) or AhR activity in TAMs (Figure S3B). In contrast, administration of broad-spectrum antibiotics reduced tumor size (Figure 4A). Bacteria belonging to the genus Lactobacillus are gut commensals with an ability to produce indoles from Trp metabolization (Roager and Licht, 2018). Lactobacilli, with a sensitivity to the antibiotic ampicillin (Amp) and resistance to vancomycin (Vanc), are important drivers of AhR activity in myeloid cells in inflammatory disease (Rothhammer et al., 2016). We hypothesized that removal of Lactobacilli by administration of Amp would phenocopy observations in Lyz2cre/+Ahrfl/fl mice. In agreement with this prediction, Amp treatment, but not Vanc, reduced tumor weight. Moreover, the TAMs in Amp-treated mice showed increased expression of MHCII and PD-L1 (Figure 4C) and reduced expression of AhR-responsive genes (Cyp1a1, Cyp1b1, and Cyp1a2) (Figure S3C), indicating that Amp exposure reduces TAM AhR transcriptional activity. Furthermore, Amp treatment increased infiltration of activated CD8+ T cells (Figure 4D). Importantly, intra-tumoral IFNγ+TNFα+CD8+ T cells increased 5-fold in Amp-treated mice compared with Vanc-treated groups (Figure 4E). AhR function was required for this effect as Amp treatment had no effect on tumor size in Lyz2cre/+Ahrfl/fl mice (Figure 4F). These data suggest that Amp-sensitive microbes may promote tumor growth and immune suppression in PDAC.
Figure 4. Indole-producing microbiota drive immune suppression in the TME.
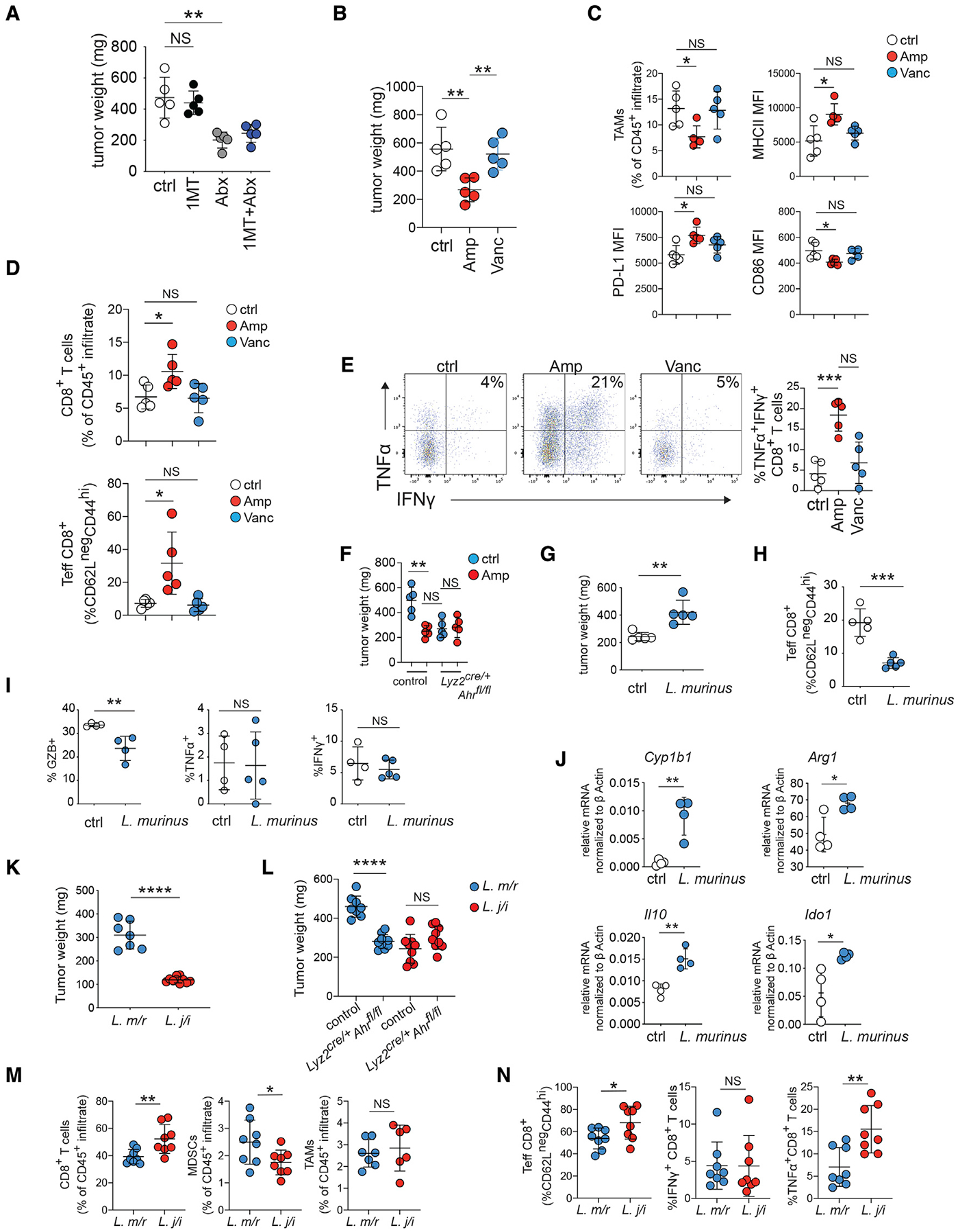
(A) B6 pancreatic tumor-bearing mice were treated with D1MT or broad-spectrum antibiotics. Tumor weight was determined 14 days after implantation.
(B) Tumor-bearing B6 mice were placed on drinking water containing the indicated antibiotic as described in STAR Methods. Tumor weight was determined 14 days after implantation.
(C) CD45+CD11b+F4/80+ TAMs from the tumors in (B) were analyzed for the indicated markers by flow cytometry.
(D) CD3+CD8+ T cells from the tumors in (B) were analyzed for the indicated markers by flow cytometry.
(E) Percentage of tumor infiltrating CD3+CD8+ T cells, expressing IFNγ and TNFα from the tumors in (B) was determined by flow cytometry. Plots to the left are representative pseudocolor dot plots from each group gated on CD3+CD8+ T cells.
(F) Tumor-bearing B6 mice of the indicated genotype were placed on drinking water containing ampicillin (Amp) as described in (B) and tumor weight was determined 14 days after implantation.
(G) Day-14 tumor weight in germ-free mice with a L. murinus microbiome compared with controls.
(H and I) Total effector CD8+ T cells (CD62LloCD44hi) as a percentage of the CD45+ infiltrate (H) and the percentage of CD3+CD8+ T cells expressing granzyme B (GZB), TNFα, and IFNγ (I) was determined by flow cytometry in day-14 tumors in inoculated versus control germ-free tumor-bearing mice.
(J) CD45+CD11b+F4/80+ TAMs were sorted from day-14 tumors from germ-free B6 mice +/−, an L. murinus microbiome. The mRNA indicated were measured by q-rtPCR and normalized against Bactin as described in STAR Methods.
(K and L) Day-14 tumor weight from B6 mice (J) or Lyz2cre/+Ahrfl/fl mice or littermate controls (L) with a microbiome containing L. murinus+L. reuteri (L. m/r) or L. johnsonii+L. intestinalis (L. j/i).
(M) Percent of intra-tumoral CD8+ T cells, CD11b+GR1+MHCIIlo MDSCs, and CD11b+F4/80+ TAMs in the CD45+ infiltrate of tumors from (J) was determined by flow cytometry.
(N) Percentage of Teff, IFNγ+, and TFNα+ CD8+ T cells from tumors described in (J) was determined by flow cytometry. (see also Figure S3). * p <.05, ** p <.01, *** p <.001, **** p <.0001, NS- not significant.
When we analyzed the gut microbiome of antibiotic-treated, tumor-bearing mice by 16S ribosomal sequencing, we found that that Amp treatment increased fecal microbial alpha-diversity and operational taxonomic units compared with controls (Figure S3D), while reducing the relative abundance of Lactobacillus (Figure S3E). Lactobacillus made up 10% of the total bacteria taxa detected in control microbiome, dropping to 3.7% in Amp-treated mice (Figure S4A). This contrasted with Vanc treatment, which reduced overall microbial diversity but increased the relative Lactobacillus abundance (Figures S3E and S4A). Analysis of the Lactobacillus species present in the fecal microbiome indicated that the most abundant species was Lactobacillus murinus, which constituted approximately 50% of the total Lactobacillus detected overall in controls and the large majority of Lactobacillus detected in Vanc-treated mice (Figure S4A). L. murinus can reduce inflammation in the central nervous system via indole production (Wilck et al., 2017), suggesting a potential link between antibiotic treatment, the modulation of L. murinus in the fecal microbiome, and intra-tumoral immune modulation
Next, we compared 4 Lactobacillus species cultured from the gastrointestinal tract (GIT) of C57BL/6J mice for the ability to produce indoles by mass spectrometry analysis. Reports indicate that L. johnsonii possesses limited ability to produce the AhR ligand indole-3-aldehyde (IAld), whereas L. reuteri and L. murinus can both produce anti-inflammatory indoles (Wilck et al., 2017; Zelante et al., 2013). In agreement with this, we found that neither L. johnsonni nor L. intestinalis produced IAld or another AhR ligand indole lactic acid (ILA) (Figures S4B and S4C). In contrast, ILA and IAld were produced by both L. reuteri and L. murinus cultures, with L. murinus showing higher relative production of both the metabolites compared with L. reuteri (Figures S4B and S4C). An examination of indoles and amino acids or their derivatives present in the Lactobacillus cultures showed that L. murinus and L. reuteri cultures had similar profiles that were compositionally distinct from L. intestinalis and L. johnsonii (Figures S4C and S4D). Since L. murinus can suppress inflammation via production of indoles (Wilck et al., 2017), we then tested whether this microbe could impact tumor growth and inflammation in PDAC. For this, we gavaged germ-free mice with L. murinus prior to implantation of PDAC tumors, examining growth and immune characteristics. Transplanted L. murinus was stable, and we could detect it in the feces 30 days after gavage (Figure S4E). Importantly, mice gavaged with L. murinus had increased fecal ILA (Figure S4F). Tumors in mice with L. murinus were larger than tumors in control germ-free mice (Figure 4G), suggesting L. murinus promoted PDAC tumor growth. Compared with the control group, mice with L. murinus microbiota had decreased numbers of activated and granzyme B-expressing CD8+ T cells, although there was no change in TNFα and IFNγ expression (Figures 4H and 4I). TAMs sorted from 14 day tumors showed that the presence of a L. murinus microbiome increased the expression of Cyp1b1 compared with controls, demonstrating that L. murinus microbiota correlate with increased AhR activity (Figure 4J). Moreover, the TAMs exhibited increases in A expression of the pro-tumor genes Arg1, Ido1, and Il10 (Figure 4J), suggesting that L. murinus could drive TAMs to acquire an immunosuppressive program.
However, these results did not rule out a general role for microbiota in the suppression of immunity in the TME rather than a specific effect of L. murinus. Thus, we utilized a microbiota transplant approach, directly comparing tumor growth in mice with a microbiome containing indole-producing bacteria (i.e., L. murinus and L. reuteri, hereafter referred to as L. m/r) to mice with a microbiome containing Lactobacillus that do not robustly produce indoles (i.e., L. johnsonii and L. intestinalis, hereafter referred to as L. j/I). Similar to germ-free mice, the L. m/r group had increased fecal ILA compared with the L. j/i group (Figure S4G). Strikingly, there was a large difference in tumor weight between the groups at day 14 (Figure 4K), suggesting that the presence of indole-producing bacteria in the GIT contributes to tumor growth. To test the prediction that the increased tumor burden in the L. m/r group was due to indole-induced TAM AhR activity, we repeated the experiment using Lyz2cre/+Ahrfl/fl mice. We found that Lyz2cre/+Ahrfl/fl mice with an L. m/r microbiome had reduced tumor weight compared with L. m/r littermate controls (Figure 4L). In contrast, the loss of TAM AhR function had no impact on tumor size in L. j/I mice (Figure 4L), suggesting that the increased tumor weight in L. m/r mice is due to TAM AhR activity. The difference in tumor weight was accompanied with alteration of the TME. There was an increase in intra-tumoral CD8+ T cells and a reduction in myeloid derived suppressor cells (MDSCs) in the tumors from the L. j/I group compared with tumors from the L. m/r group (Figure 4M). The percentages of effector CD8+ T cells were significantly increased in the tumors of L. j/i mice compared with tumors from the L. m/r group, which paralleled an increase in TNFα+-CD8+ T cells (Figure 4N). Combined with the germ-free experiments, the data show that indole-producing bacteria increase AhR transcriptional responses and promote an immunosuppressive TME in PDAC.
Trp and the indoles IAA and ILA in the diet promote immune suppression and pancreatic tumor growth
Since Trp metabolization is a key mechanism of AhR ligand generation (Shinde and McGaha, 2018), we tested whether the removal of dietary Trp would phenocopy effects of Ahr deletion in macrophages. Removal of Trp from the diet was well tolerated over the course of the experiment; however, at day 10, tumors were 2-fold smaller in the dietary Trpneg group compared with controls (Figure 5A). We then reasoned that if the reduction of PDAC burden in mice in a Trpneg diet was due to reduced indole production by the microbiome, we should be able to rescue tumor growth by provision of dietary indoles. To test this, we gavaged dietary Trpneg mice with indoles (indole-3-acetic acid [IAA], ILA, or IAld) after tumor implantation. In agreement with our hypothesis, both IAA and ILA supplementation abrogated the effect of a Trpneg diet, resulting in tumor weight that was comparable with that of the dietary Trp+ controls (Figure 5B). Further examination of the TME by flow cytometry showed that removal of Trp from the diet did not impact the percentage of MDSCs (Figure 5C); however, lack of dietary Trp increased the number of TNFα+IFNγ+ and proliferating CD8+ T cells, an effect that was reversed by dietary IAA or ILA (Figure 5D). In the dietary Trpneg group, TAMs decreased as a percentage of the immune infiltrate and showed decreased expression of MHCII compared with controls, suggesting a less activated phenotype (Figure 5E). Supplementation of the diet with IAA or ILA rescued this effect increasing the percentage of TAMs and MHCII expression to levels comparable with that of the Trp+ diet controls (Figure 5E). Moreover, in the absence of dietary Trp, TAMs had decreased expression of Il10 and Arg1 and Cyp1b1 (Figure 5F). Importantly, IAA supplementation in dietary Trpneg mice increased Il10, Arg1, Cyp1a1, and Cyp1a2 expression, suggesting that IAA activates AhR and drives an immunosuppressive TAM phenotype (Figure 5F). Ultimately, the data suggest that dietary Trp is a key driver of immune phenotype in the PDAC TME by serving as a source of indoles via microbiome metabolization.
Figure 5. Dietary Trp and indoles promote immune suppression and PDAC growth.
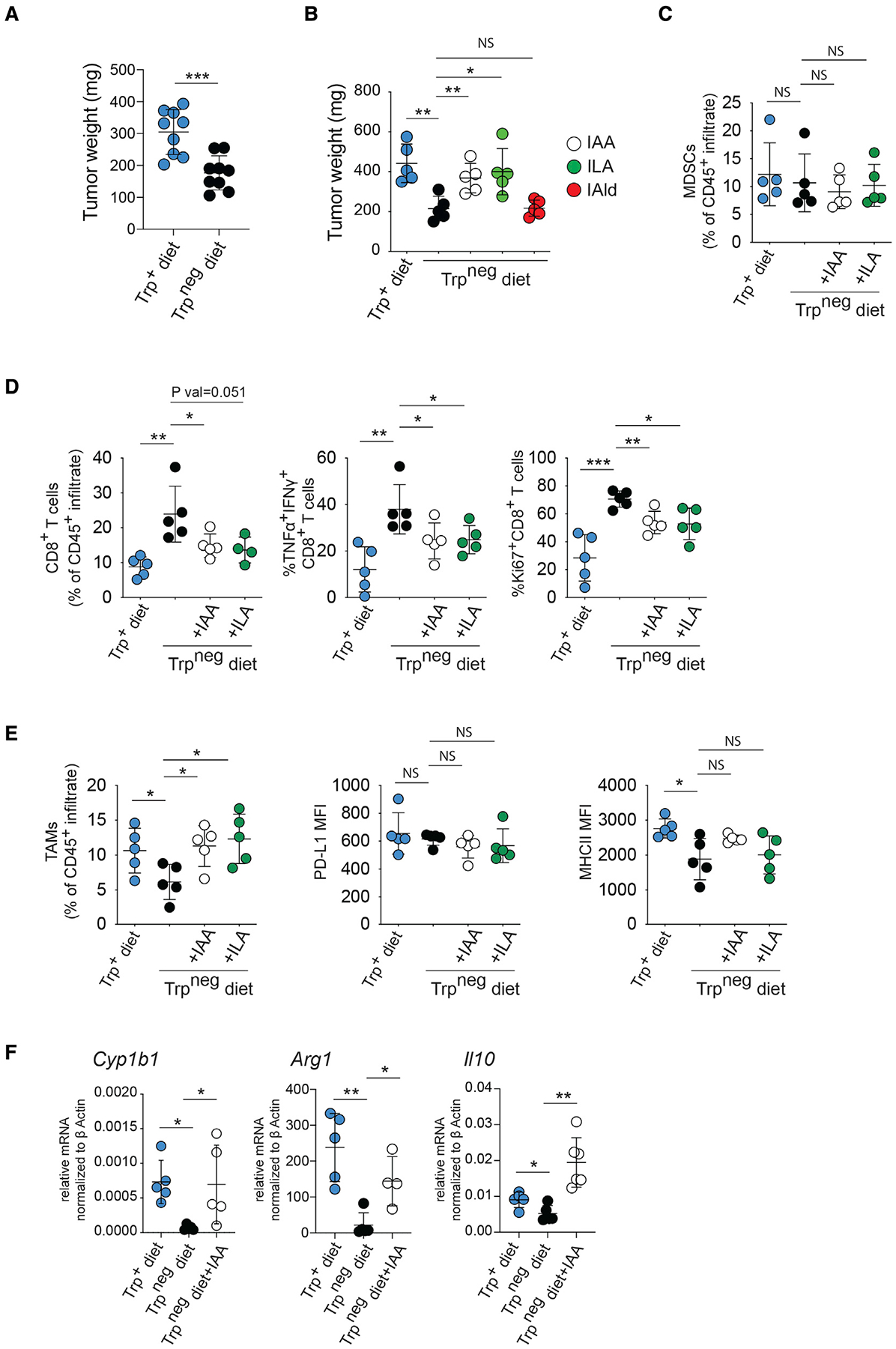
(A) Day-14 PDAC tumor weight in mice on control or Trp-free diet.
(B) Day-14 tumor size in B6 mice on chow +/− Trp with some groups receiving daily gavage with the indicated indole.
(C–E) Day-14 tumors were collected from B6 mice on Trp+/− diet with or without daily IAA or ILA gavage, and flow cytometry analysis of the intra-tumoral in filtrates was performed for the markers indicated. MDCSs were defined as CD11b+GR1+MHCIIlo, and TAMs were defined as CD11b+F4/80+; the T cells were CD3+CD8+ cells. MFI, mean fluorescence intensity.
(F) CD45+CD11b+F4/80+ TAMs were sorted from day-14 tumors from mice treated with IAA as in (C). The mRNA indicated were then measured by qrt-PCR and normalized against βactin as described in STAR Methods. (see also Figure S4). * p <.05, ** p <.01, *** p <.001, NS- not significant.
AHR expression and activity correlates with patient outcomes in human pancreatic ductal adenocarcinoma
Pan-cancer analysis showed that PDAC exhibits higher AHR compared with most other cancer types (Figure 6A). We then examined correlation of AHR expression with survival. In PDAC patients, AHR expression below the median value was associated with improved overall survival (OS) compared with patients above the median (Figures 6B, panel 1 and S5A), and when we grouped patients into quartiles based on relative AHR, we observed the best OS in the lowest AHR quartile (Q1), whereas the other expression quartiles did not show differential OS. This suggested a benefit for patients with low AHR expression. Although the difference in OS among the quartiles was not significant (Figure 6B, panel 2), hazard ratios suggested that AHR expression was a risk factor for death (Figure S5B). Thus, we surmised that a threshold of AHR expression may compromise survival in PDAC. In agreement with this, Q1 showed an improvement in OS when compared with all other PDAC patients (Figure 6B, panel 3). Thus, overall, the relationship of AHR to OS suggests that low AHR expression may be beneficial for survival, but even moderate expression may negatively impact outcomes.
Figure 6. AHR expression and activity correlates with patient outcomes in human PDAC.
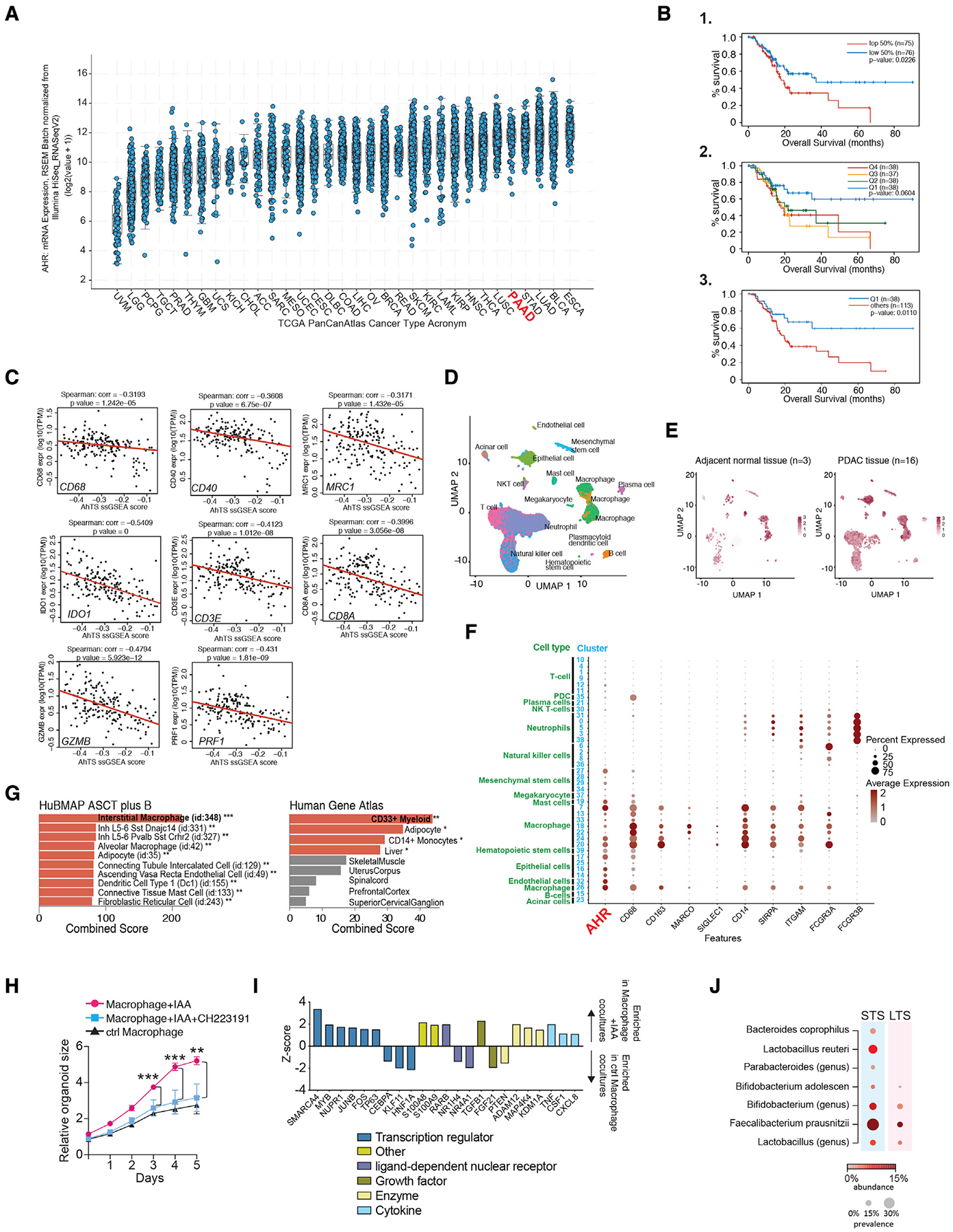
(A) Pan-cancer TCGA analysis showing relative AHR expression across 32 cancer types.
(B) Overall survival of the PAAD (PDAC) TCGA patient dataset grouped based on relative AHR expression. Patients were grouped based on 1-median AHR expression. 2-quartiles of AHR expression. Q1 < Q2 < Q3 < Q4. 3- Q1 survival compared with all other quartiles combined.
(C) ssGSEA analysis was performed by examining correlation between the AhR transcriptional signature and the indicated genes in the PAAD-TCGA dataset. Red line is the quartile regression as described in STAR Methods.
(D) UMAP showing scRNA-seq data of 46,244 cells from human PDAC tumor samples and 8,542 cells from adjacent unaffected tissue.
(E) UMAPs of scRNA-seq from indicated samples showing relative AHR expression.
(F) Normalized expression and per cluster percentage expression of the genes indicated for each cell cluster for the scRNA-sequencing analysis described in (D).
(G) GSEA of AHR+25 most similar genes for the indicated gene sets. Bars show relative p value, orange bars = p value <0.05, gray bars were not significant.
(H) 5-day growth curve of human PDAC PDO cultured with PBMC-derived macrophages treated as indicated prior to initiation of the co-culture. Each data point represents the mean value for triplicate samples +/− the standard deviation.
(I) Z score for iGSEA analysis of differential gene set enrichment of PDAC PDO/macrophage co-culture at day 5. Colored bars correspond to selected classifiers.
(J) The relative prevalence and abundance of indole-producing bacterial taxa in the tumor microbiome for short-term surviving (STS) versus long-term surviving (LTS) PDAC patients. Results are sorted by odds ratio of bacterial presence in STS versus LTS in descending order. (see also Figure S5). ** p <.01, *** p <.001.
A follow-on prediction from these results is that AhR activity may impact survival by altering immunity in the TME. To test this, we applied a human AhR transcriptional signature (AhTS) (Goudot et al., 2017), examining correlation with T cell and macrophage gene expression in the PDAC TCGA cohort. We observed moderate negative correlation of the AhTS with expression of the macrophage markers CD68, CD206 (MRC1), and CD40, suggesting macrophage density was negatively impacted by AhR activity (Figure 6C). Genes associated with CD8+ T cells and effector function also showed a negative correlation with the AhTS (Figure 6C). Thus, the data show that AhR gene signatures correlate with a paucity of T cells transcriptional signatures in the TME and a cold tumor overall, whereas reduced AhTSs predicts increased cytolytic CD8+ T cell presence and inflammation.
We then examined AHR expression in a single-cell dataset from treatment naive PDAC patients and non-malignant tumor adjacent pancreas tissue (Steele et al., 2020). AHR had the highest and broadest expression in epithelial and macrophage clusters (Figures 6D–6F). In macrophage clusters, AHR expression showed a high degree of similarity with CD163 (a gene associated with alternative polarization) (Ambarus et al., 2012) and SIRPA (a key inhibitor of phagocytosis of self-cells) (Figure 6F), suggesting that AHR expression is associated with regulatory TAM populations in the PDAC TME. We then compared the similarity of AHR expression across all cell clusters, identifying the top 25 genes most similar to AHR in relative level and distribution of cellular expression (Figure S5C). Using this dataset, we performed gene set enrichment analysis to test for general cellular expression patterns. When we probed the HuBMAP ASCT plus B dataset, the highest cellular enrichment indicated was interstitial macrophages (Figure 6G). Similarly, for the Human Gene Atlas, the gene set enriched for CD33+ myeloid cells and CD14+ monocytes (Figure 6G). Thus, AHR is highly expressed in subsets of alternatively activated TAMs in the PDAC TME and is associated with gene set enrichment in myeloid cells across tissue types.
In addition to immune-suppressive function, macrophages can provide direct stromal support to PDAC tumors. Since AhR can induce expression of growth factors, including VEGF and TGFα, we postulated that AhR activation in macrophage may drive a phenotype that could directly support tumor growth. To test this, we co-cultured PBMC-derived macrophage (Halaby et al., 2019) exposed to IAA with PDAC patient-derived organoids (PDO). In the presence of control PBMC-derived macrophages, organoids doubled in size over 5 days (Figure 6D). Co-culture of the PDO with IAA-exposed macrophages increased PDO growth compared with control cultures (Figure 6D). Importantly, addition of the AhR antagonist CH223191 to the macrophages prior to co-culture abrogated the increased growth effect, showing that IAA stimulation improved PDO growth due to the activation of macrophage AhR (Figure 6D). RNA-seq analysis showed that co-culture with IAA-exposed macrophages altered the expression of 235 genes in the PDO (Table S1). iGSEA enrichment analysis of the differentially expressed genes in the PDO co-cultured with control macrophages versus IAA-treated macrophages showed that indole-treated macrophage cultures increased enrichment of pathways associated with PDAC growth and metastasis and reduced enrichment of genes associated with negative regulation of PDAC proliferative potential (e.g., the CEBPA pathway) (Lourenço and Coffer, 2017) (Figure 6D). Altogether, the data show that indoles induce macrophages to acquire a PDAC-supporting phenotype by an AhR-driven mechanism.
Riquelme et al. described a set of PDAC that survived more than 5 years post-resection (long-term survivors [LTS]), identifying local TME and fecal microbial diversity and enrichment for taxa in the LTS patients correlating with this effect (Riquelme et al., 2019). Importantly, FMT experiments suggested that, although microbiota from LTS patients reduced tumor PDAC burden in mice, FMTs from patients who survived less than 5 years post-surgery (matched STS) had a detrimental effect on tumor burden (Riquelme et al., 2019). This result implied that the microbiota from STS contain taxa that could actively contribute to PDAC progression worsening disease. A number of bacterial taxa from the Bacteroides, Bifidobacterium, Clostridium, and Lactobacillus genera are prominent producers of indoles (Cervantes-Barragan et al., 2017; Dodd et al., 2017; Elsden et al., 1976; Smith and Macfarlane, 1996), and we hypothesized that the worse outcomes in STS may correlate with increased abundance and/or prevalence of indole-producing bacterial taxa in the local tumor microbiome. Thus, we examined the Riquleme et al. intra-tumoral microbiome dataset to test the prevalence and relative abundance of a set of 20 indole-producing (i.e., IAA, ILA, and IAld) bacterial taxa (Table S2), examining their prevalence within the samples of the cohorts and the average relative abundance between STS and LTS patient groups (Figure 6F). We found an increased prevalence of L. reuteri in STS over LTS, with 5/21 STS patients displaying a relative abundance of L. reuteri above 2%, whereas none of the LTS patients displayed any detectable relative abundance of L. reuteri upon filtering. Similar increases were seen for other indole-producing bacteria or genera of known indole-producing bacteria, such as Bacteroides coprophilus, Faecalibacterium prausnitzi, and the Lactobacillus and Bifidobacterum genera (Figure 6F). Thus, the data suggest that enrichment of indole-producing bacteria in the local TME correlates with poor response to resection and OS in PDAC.
DISCUSSION
In some cancers, the gut microbiota influence therapeutic responses to checkpoint inhibitor therapy (Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2018). The local tumor microbiota in PDAC influences survival independent of immune therapy (Riquelme et al., 2019). However, mechanisms directly linking either the local tumor or the gut microbiome to cancer progression are not well understood. We examined the connection between AhR and microbial indole production extensively, characterizing the immune TME and directly testing the ability of indole-producing bacteria and indoles to activate AhR and promote immune suppression and tumor growth. We found that macrophage AhR function promotes the expression of Arg1 and Il10 in TAMs and inhibits IFNγ expression in CD8+ T cells. The observation that AHR expression correlates with poor outcomes and reduced immune signatures in human PDAC and indole-exposed human Mφ support PDAC organoid growth in an AhR-dependent mechanism suggests analogous function in human disease.
AhR is an important sensor for bacterial indoles produced via utilization of Trp as an energy source (Shinde and McGaha, 2018). In this report, we demonstrated that loss of the gut microbiome, or removal of dietary Trp phenocopied macrophage Ahr deletion; importantly, the effect could be rescued by provision of indoles. A recent report suggested that the microbiome can promote tumor growth by TLR-dependent stimulation of innate inflammation locally with enrichment of Bifidobacterium (Pushalkar et al., 2018). Bifidobacterium species were not a significant constituent of the microbiome in our mice; however, the Bifidobacterium genus was enriched in the microbiome of the STS in our human PDAC microbiome analysis, albeit to a much lesser extent when compared with other taxa. Nevertheless, Bifidobacterium spp. are known producers of ILA (Aragozzini et al., 1979; Russell et al., 2013), indicating that the association with oncogenesis in animal models and poor outcomes in human PDAC may be linked to the capacity to produce indoles. This prediction is strengthened by our results, directly comparing the tumorigenic potential of Lactobacillus species that were differentiated by the ability to produce indoles. The fact that indole-producing Lactobacilli drastically increase tumor size, with an increase in MDSC numbers and a reduction in activated CD8+ T cells and TNFα production, suggests that indole production by microbiota is an important force influencing immunity in the TME. Recently, a role for TAM AhR function was also identified in glioblastoma multiforme (GBM), driving CD39 expression and impairing T cell responses (Takenaka et al., 2019). This study, along with the data from our paper, strongly supports the prediction that macrophage AhR is a central driver of TAM function responding to multiple inputs to drive an immune-suppressive phenotype in the TME.
Therapeutic targeting of the microbiome has been a sought-after goal for cancer treatment. As such, the approaches had generally fallen into two categories: (1) FMT or (2) specific gut enrichment of bacterial consortia associated with therapeutic response. Indeed, a recent report has shown that a single FMT from melanoma patients responsive to αPD-1 therapy to αPD-1 refractive recipients promoted the accumulation of CD8+ T cells and acquired responsiveness to αPD1 therapy (Davar et al., 2021). However, this approach may be limited in utility because of the lack of knowledge regarding specific taxa driving positive therapeutic responses and variability of key taxa engraftment. Our results suggest that targeting immunologic responses to the microbiome could have meaningful impact on therapy, augmenting microbiome manipulation approaches or potentially bypassing the need to manipulate the microbiome by precise inhibition of the response to microbial metabolites.
In conclusion, we found that macrophage AhR has a strong impact on tumor growth and the TME in pancreatic cancer. The link we identified between AhR, immune function, indoles, and microbiome constituents may provide useful prognostics for predicting patient outcomes.
Limitations of the study
Our study leaves open several questions. The chief among them is why the microbiome appears to serve a dominant role in AhR activation in our study, whereas well-known, Trp metabolizing mammalian enzymatic pathways appear to play a negligible role in the modulation of AhR function. In contrast to our findings, others have reported PDAC tumor cells can produce L-Kyn via an IDO- or TDO-dependent mechanism driving AhR activity (Wang et al., 2020). This dichotomy with our current study is likely reflective of the diverse and variable role of the microbiome versus IDO in shaping the TME in a heterogeneous disease, such as PDAC. Thus, it will be important in future experiments to examine conditions that predicate IDO versus microbiome contribution to AhR function in the TME. Second, in contrast to other reports, we could not detect intra-tumoral bacteria by PCR or 16S sequencing. The reasons for the dichotomy are not clear; however, our use of defined, cultured mouse feces-isolated bacteria may impact dissemination from the gut to the tumor. Third, although we show that pharmacologic inhibition of AhR improves responses to therapy, there are limitations to the application of AhR blockade in cancer due to the variable extent and composition of the tumor immune infiltrate and microbiome taxa. Finally, there is a difference in AhR-ligand-binding affinities between mouse and human (Flaveny and Perdew, 2009; Hubbard et al., 2015) that must be considered when applying data derived from mouse models to clinical disease. Nevertheless, a first-in-human clinical trial targeting AhR has been initiated in patients with advanced solid lung, colorectal, and urothelial tumors (clinicalTrials.gov identifier: NTC04069026), indicating potential applicability of AhR inhibition in multiple cancer types. Ultimately, these trials will provide a definitive test as to whether AhR is a legitimate target for cancer therapy.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Tracy L. McGaha (tmcgaha@uhnresearch.ca).
Materials availability
This study did not generate new unique reagents.
Data code and availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication key resources table under NCBI GEO accession ID GSE171603.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD8a | BioXCell | Cat# BE0117, RRID: AB_10950145 |
| Anti-mouse INFg Ab | BioXCell | Cat# BE0055, RRID: AB_1107694 |
| IgG1 isotype control | BioXCell | Cat# BE0083, RRID: AB_ 1107784 |
| IgG2a isotype control | BioXCell | Cat# BE0090, RRID: AB_1107780 |
| CD45 (mouse), clone 30-F11 | Fluidigm | cat# 3089005B |
| Ly6C (mouse), clone HK1.4 | Biolegend | cat# 128001 |
| GzmB (mouse), clone QA 16a02 | Biolegend | cat# 372202 |
| CD44 (mouse), clone IM7 | Biolegend | cat# 103051 |
| CD69 (mouse), clone H1.2F3 | Fluidigm | cat# 3143004B |
| CTLA4 (mouse), clone UC104B9 | Biolegend | cat# 106302 |
| F4/80 (mouse), clone BM8 | Fluidigm | cat# 3146008B |
| CD11b (mouse), clone M1/70 | Fluidigm | cat# 3148003B |
| Nos2 (mouse), clone W16030C | Biolegend | cat# 696802 |
| Ly6G (mouse), clone 1A8 | Biolegend | cat# 127637 |
| CD25 (mouse), clone 3C7 | Biolegend | cat# 101913 |
| CD3e (mouse), clone 145-2C11 | Fluidigm | cat# 3152004B |
| CD8a (mouse), clone 536.7 | Fluidigm | cat# 3153012B |
| CD103 (mouse), clone Ber-ACT8 | Novus Bio | cat# NBP1-97564 |
| PDL1 (mouse), clone 29E.2A3 | Biolegend | cat# 329719 |
| Tim3 (mouse), clone F38-2E2 | Biolegend | cat# 345019 |
| CD62L (mouse), clone MEL-14 | Fluidigm | cat# 3160008B |
| TBET (mouse), clone 4B10 | Biolegend | cat# 644825 |
| FOXP3 (mouse), clone MF-14 | Biolegend | cat# 126401 |
| CD86 (mouse), clone IT2.2 | Biolegend | cat# 305435 |
| CX3CR1 (mouse), clone SSA011F11 | Fluidigm | cat# 3164023B |
| INFg (mouse), clone XMG1.2 | Fluidigm | cat# 3165003B |
| B220 (mouse), clone RA36B2 | Biolegend | cat# 103249 |
| CCL4 (mouse), clone W15194A | Biolegend | cat# 625504 |
| TNFa (mouse), clone mp6xt22 | Fluidigm | cat# 3162002B |
| CD206 (mouse), clone C068C2 | Fluidigm | cat# 3169021B |
| CD4 (mouse), clone RM45 | Fluidigm | cat #3172003B |
| CD11c (mouse), clone N418 | Biolegend | cat# 117301 |
| PD1 (mouse), clone RMP1-30 | Biolegend | cat# 109113 |
| TIGIT (mouse), clone 4D4/m1 | Biolegend | cat# 156102 |
| MHCII (mouse), clone m5/114.15.2 | Fluidigm | cat# 3209006B |
| Bacterial and Virus Strains | ||
| Lactobacillus murinus strain NM26_J9 | This Paper | NCBI Genome Assembly GCA_004793535.1 |
| Lactobacillus reuteri strain NM11_1-41 | This Paper | NCBI Genome Assembly GCA_004793875.1 |
| Lactobacillus johnsonii strain NM60_B2-8 | This Paper | NCBI Genome Assembly GCA_004793575.1 |
| Lactobacillus intestinalis strain NM61_E11 | This paper | NCBI Genome Assembly GCA_004793775.1 |
| Biological Samples | ||
| Healthy human peripheral blood mononuclear cells (PBMC) | The Princess Margaret Cancer Centre | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 3-Indoleacetic acid (IAA) | Sigma Aldrich | Cat# I3750, CAS: 87-51-4 |
| Indole-3-Lactic Acid (ILA) | Sigma Aldrich | Cat# I5508, CAS: 832-97-3 |
| ITE | Tocris | Cat# 1803, CAS: 448906-42-1 |
| FICZ | Millipore Sigma | Cat# SML1489, CAS: 172922-91-7 |
| CH223191 | Cayman Chemicals | Cat# 16154, CAS: 301326-22-7 |
| Streptomycin sulfate | Millipore Sigma | Cat# S9137, CAS: 3810-74-0 |
| Clindamycin hydrochloride | Millipore Sigma | Cat# C5269, CAS: 21462-39-5 |
| Vancomycin hydrochloride | Millipore Sigma | Cat# V2002, CAS: 1404-93-9 |
| Indole-3-Carboxyaldehyde (IAId) | Millipore Sigma | Cat# 129445, CAS: 487-89-8 |
| Ampicillin | Millipore Sigma | Millipore Sigma |
| Metabolomics Amino Acid Mix Standard | Cambridge Isotopes Laboratories Inc | Cat# MSK-A2-1.2 |
| peptone-tryptone media | Sigma-Aldrich | cat# BP1421 |
| Man-Rogosa-Sharpe (MRS) agar | Sigma-Aldrich | cat# 69964 |
| tamoxifen | Sigma-Aldrich | cat# T5648 |
| corn oil | Sigma Aldrich | cat# C8267 |
| Maxpar Staining Buffer | Fluidigm | cat # 201068 |
| TruStain Fc blocking buffer | Biolegend | cat# 101320 |
| Cell_Id cisplatin | Fluidigm | cat# 201064 |
| Cell-ID multiplex Barcoding kit | Fluidigm | cat # 201060 |
| Foxp3 transcription factor staining kit | eBioscience | cat# 00-5523-00 |
| EQ beads | Fluidigm | cat# 201078 |
| Critical Commercial Assays | ||
| SMART-Seq v4 Ultra Low Input 20 536 RNA Kit for Sequencing | Takara | cat# R400752 |
| NexteraXT DNA Library Preparation | Illumina | cat# FC-131-1096 |
| NexteraXT Index Kit V1 or V2 Set A | 10X Genomics | cat# PN-1000121 & PN-1000127 |
| Platinum Green Hot Start PCR 2x Master Mix | Invitrogen | cat# 13001013 |
| Monarch PCR & DNA Cleanup Kit | New England Biolabs | cat # T1030S |
| Deposited Data | ||
| Raw RNA sequencing data | This paper | NCBI GEO accession ID GSE171603 |
| Experimental Models: Cell Lines | ||
| mT4 PDAC cell | Dr. David Tuveson (CSHL) | Cell, 160(1-2), 324-338. https://doi.org/10.1016/j.cell.2014.12.021 |
| mT3 PDAC cell line | Dr. David Tuveson (CSHL) | Cell, 160(1-2), 324-338. https://doi.org/10.1016/j.cell.2014.12.021 |
| mT5 PDAC cell line | Dr. David Tuveson (CSHL) | Cell, 160(1-2), 324-338. https://doi.org/10.1016/j.cell.2014.12.021 |
| Patient derived organoid model | The University Health Network Living Biobank, ON Canada | PPTO.46 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Jackson Laboratory | JAX:000664 |
| Mouse: B6.Ahrfl/fl | Jackson Laboratory | JAX:006203 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | Jackson Laboratory | JAX:007909 |
| Mouse: Krastm4Tyj Trp53tm1Brn Tg(Pdx1-cre/Esr1*)#Dam/J | Jackson Laboratory | Jax: 032429 |
| Mouse: B6.Lyz2-CRE | Jackson Laboratory | Jax: 004781 |
| Oligonucleotides | ||
| AhR CRISPR guide RNA (CTCCACTATCCAAGATTACC) | GenScript | CrRNA 3 |
| Mouse: Cyp1a1 F: 5’-CAAT5’-CCCTTCTCAAATGTCCTGTAGTG-3’ R: 5’- CCCTTCTCAAATGTCCTGTAGTG-3’, |
ThermoFisher Scientific | N/A |
| Mouse: Cyp1b1 F: 5’-CCACCAGCCTTAGTGCAGAC-3’ R: 5’-GGCCAGGACGGAGAAGAGT-3’ |
ThermoFisher Scientific | N/A |
| Mouse: Actb F: 5’-AAGAGCTATGAGCTGCCTGA-3’ R: 5’-TACGGATGTCAACGTCACAC-3’ |
ThermoFisher Scientific | N/A |
| NM26 (L. Murinus) F: 5’- CCACATGCTAGTGAGCGTATC-3’ R: 5’- GTCCAGTTTCTTCTCGCTTCT-3’ |
ThermoFisher Scientific | N/A |
| NM11 (L. Reuteri) F: 5’- GGACGCTTAGACCGCAATGTA-3’ R: 5’- TCTCAACACCCGCCTTAATC-3’ |
ThermoFisher Scientific | N/A |
| Mouse: Arg-1 F: 5’-CTCCAAGCCAAAGTCCTTAGAG-3’ R: 5’-GGAGCTGTTCATTAGGGACATCA-3’ |
ThermoFisher Scientific | N/A |
| Mouse: IL-10 F: 5’- ATTTTAATAAGCTCCAAGACCAAGGT-3’ R: 5’-CTGCAGGTGTTTTAGCTTTTCATTT-3’ |
ThermoFisher Scientific | N/A |
| mOUSE: IDO-1 F: 5’- GAGGATGCGTGACTTTGTGGA-3’ R: 5’-TCCCAGACCCCCTCATACAG-3’ |
ThermoFisher Scientific | N/A |
| Recombinant DNA | ||
| pSpCas9(BB)-2A-GFP | GenScript | Cat# PX458 |
| Software and Algorithms | ||
| FlowJo v10 | BD Biosciences | https://www.flowjo.com/ |
| Cytobank | Beckmann Coulter | https://www.mybeckman.ca/flow-cytometry/software/cytobank-premium |
| infercnv v1.6.0 | infercnv v1.6.0 inferCNV of the Trinity CTAT Project. https://github.com/broadinstitute/inferCNV |
https://github.com/broadinstitute/infercnv |
| Seurat v4.0.3 | Satija et al., 2015 | https://satijalab.org/seurat/ |
| Harmony v0.1.0 | Korsunsky et al., 2019 | https://github.com/immunogenomics/harmony |
| R v4.0.3 | R Core Team, 2021 | R Core Team, 2021 https://www.r-project.org/ |
| Prism 7 | GraphPad | graphpad.com/scientific-software/prism/ |
| Other | ||
| Trp negative chow diet | Envigo | cat# TD.01084 |
| Amino acid chow diet | Envigo | cat# TD.08126 |
| X-tremeGENE 9 DNA Transfection Reagent | Millipore-Sigma | cat# XTG9-RO |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C57BL6/J (B6), B6.Ido1−/−, B6.Ahrfl/fl, B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J, tamoxifen-inducible (i)KPC mice (Krastm4Tyj Trp53tm1Brn Tg(Pdx1-cre/Esr1*)#Dam/J), and B6.Lyz2CRE+/− mice were obtained from Jackson Laboratories and maintained under specific pathogen-free conditions in the Princess Margaret Cancer Centre animal facility. Germ free B6 mice were obtained from and maintained at the University of Toronto germ free core facility. All mice were cared for in accordance with the Canadian Institutional Animal Care and Use Committee guidelines. Female mice 10–12 weeks of age were used for all experiments. Protocols were approved by the Princess Margaret Cancer Centre Animal Care Committee.
METHODS DETAIL
Tumor implantation and tumor induction
Mice were anesthetized with 2% isoflurane in oxygen. A lateral incision was made on the abdominal wall of each mouse and tumors were implanted orthotopically in the pancreas with 10×103 cells of the KPC primary pancreatic adenocarcinoma organoid cell line mT3, mT4, or mT5 (a gift from Dr. David Tuveson) (Boj et al., 2015) resuspended in 80μl of Matrigel (VWR scientific, Cat. # 354234) diluted in PBS at a 1:4 ratio Matrigel/PBS and injected into the tail of the pancreas. Tumor growth was monitored by palpation, and mice were sacrificed 2 weeks after the tumor implantation. At the endpoint, tumors, large intestine fecal and blood specimens were harvested and processed for further analysis.
For induction of tumors in iKPC mice, 8-week-old mice were injected intraperitoneally with tamoxifen dissolved in corn oil for 5 consecutive days at 75mg/kg body weight in a volume of 200μl. 12 weeks after cessation of tamoxifen treatment mice received AhR inhibitor CH22319 intraperitoneally three times a week at a dose of 100 μg/mouse in a 200μl total injection volume.
Isolation of bacteria from mouse intestine
L. reuteri, L. intestinalis, L. johnsonii, and L. murinus strains were isolated from the small intestine of B6 mice through anaerobic culturing on MRS media. Isolated colonies were selected for 16S PCR amplification (Platinum™ Green Hot Start PCR 2x Master Mix) using the universal 16S primers, 8F (8F: AGA GTT TGA TCC TGG CTC AG) and 1492R (1492R: GGT TAC CTT GTT ACG ACT T) (Weisburg et al., 1991) followed by a clean-up step (Monarch® PCR & DNA Cleanup Kit (5 μg), Sanger sequencing of the 16S PCR product using the 1492R primer was performed at The Centre for Applied Genomics, Toronto, ON. Bacterial species were identified by alignment of the 16S sequence using BLASTn against the NCBI database with an alignment of at least 99.5%.
Bacterial culture for metabolomics analysis
Bacteria from glycerol stock were plated on MRS agar plates and incubated anaerobically at 37°C for at least 3 days, after which visible colonies were selected and cultured anaerobically in MRS broth at 37°C. Cultures were incubated at 37 °C for 72hrs. two culture sets per condition were prepared (second set is for checking CFU/OD). Bacteria cultures were then collected and resuspended in 1 mL of test medium (MRS or peptone-tryptone (PT) media +/− tryptophan) and incubated overnight at 37°C anaerobically. Bacteria cultures were centrifuged at 4000xg for 10 minutes and supernatants were collected for metabolomics analysis.
Antibiotic treatment to remove microbiota
Mice were treated 1 g/l ampicillin, 1 g/l metronidazole, 1 g/l neomycin, and 0.5 g/l vancomycin in their drinking water, which was replaced daily for the course of the experiment. Tumor implantations occurred 3d after mice were placed on antibiotic containing water. When indicated mice were placed on drinking water containing only ampicillin or vancomycin at the concentrations indicated above.
Microbiota transplantation in SPF mice
Female B6 were treated for two weeks with an antibiotic solution (ATBx) containing streptomycin (5 mg/ml), and clindamycin (0.1 mg/ml) added to sterile drinking water. Solutions and bottles were changed 3 times a week. After two weeks ATBx treatment was stopped, and the mice were gavaged with pooled L. murinus and L. reuteri or L. johnsonii and L. intestinalis cultures. Each gavage contained approximately 108 CFU per 200uL (corresponding to an OD 5). Mice received bacterial culture 3 times a week by oral gavage using animal feeding needles before undergoing orthotopic tumor implantation and once/week after the tumor implantation until experimental end points.
Germ free microbiota transplantation
Germ free mice were received single oral gavage of 200ul of bacterial culture (L. murinus) 4 weeks before undergoing orthotopic tumor implantation. Control mice received 200ul of sterile phosphate buffered saline.
Bulk RNA-sequencing and analysis
RNA samples were quantified by qubit (Life Technologies) and an Agilent Bioananlyzer assessed the RNA quality. All samples had RIN above 8. SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech #634894) was used per manufacturer’s instructions for amplification of RNA and subsequent cDNA synthesis. AMPure XP Bead (Agencourt AMPure beads XP PCR, Beckman Coulter A63881) purification was done manually for the first amplification set. A bead ratio of 1x was used (50μl of AMPure XP beads to 50μl cDNA PCR product with 1μl of 10x lysis buffer added, as per Clontech instructions), and purified cDNA was eluted in 17μl elution buffer provided by Clontech. All samples were quantitated using a Bioanalyzer 2100 Instrument (Agilent Genomics). All samples proceeded through NexteraXT DNA Library Preparation (Illumina FC-131–1096) using NexteraXT Index Kit V1 or V2 Set A (FC-131–1002 or FC-131–2001) following the manufacturer’s instructions. An aliquot of all samples was first normalized to 150pg/μl with Nuclease-Free Water (Ambion), then the normalized sample aliquot was used as input material into the NexteraXT DNA Library Prep. AMPure XP bead purification was done using 0.9x bead ratio to sample volume, and all samples were eluted in 22μl of Resuspension Buffer (Illumina). As with the Amplification sets, manual bead purification was done for the first Library set. All samples were run on Agilent Bioanalyzer 2100 using High Sensitivity DNA chips. A portion of this library pool was sent to an outside vendor for sequencing on an Illumina NextSeq HighOutput single read. An average of 400M reads were obtained per pool, with an average of 40M reads/sample across the entire data set.
Raw fastq sequencing reads were aligned against the respective reference genome sequence (GRCm38/mm10 or GRCh37/hg19) using the STAR aligned v2.5.0c (Dobin et al., 2013), discarding all non-uniquely aligned reads. For read counting per annotated gene, we have utilized the STAR function “–quantMode GeneCounts”, counting reads matching exons of the Ensembl V75 Genes annotation. Further processing was performed with the R Bioconductor package edgeR v.3.14.0 (Robinson et al., 2010) using non-stranded reads. Reads were normalized for intra- and inter-sample variances using the functions “calcNormFactors” and “estimateTagwiseDisp”, resulting in counts-per-million (CPM) for each gene. Differential gene expression analysis was performed with the functions “glmQLFit” and “glmQLFTest”, reporting p-value, false-discovery rate (FDR) and log2 fold-changes between any possible pair-wise comparison and gene.
scRNA-sequencing and analysis
Tumors from three mice per group were digested, pooled, and stained with DAPI. Live cells were FACS-sorted into buffer (PBS + 2% FBS), washed 2 times with PBS + 0.04% BSA, and then 10×103 cells were mixed with 10X Genomics Chromium single-cell RNA master mix, followed by loading onto a 10X Chromium chip according to the manufacturer’s protocol to obtain single-cell cDNA. Libraries were subsequently prepared and sequenced using a NovaSeq sequencer (Illumina).
Single cell raw data was demultiplexed and converted to FASTQ format with Illumina bcl2fastq. We then processed the fastq data with the Cell Ranger Single-Cell Software Suite Version 1.2 (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger) with default parameters according to the 10x genomics guidelines, to align reads against the mouse reference genome (GRCm38/mm10) and to further assign all reads to genes and individual cells based on the barcode information. We filtered all duplicated reads and those not uniquely mapped. Next, Seurat v2.2.1 (Butler et al., 2018) we used to process counts data. We excluded cells with less than 200 detected genes or > 5% mitochondrial transcripts. Expression data was normalized with the NormalizeData function, using the “LogNormalize” approach, and further scaled by sequencing depth. Clustering of cells based on gene expression was performed on all cells from both conditions with default parameters (function FindClusters), as well as a cluster-specific marker analysis (FindAllMarkers function), using only genes that were expressed at least in 25% of cells of each particular cluster. Different cell types were assigned to clusters based on known surface markers uniquely expressed in the particular cluster and found as a significant marker (FDR < 0.05). For each defined cluster/cell type, we have performed individual differential gene expression analysis between WT and Lyz2cre/+Ahrfl/fl cells using the FindMarkers function for genes expressed in at least 25% of cells of each particular cluster. For visualization, violin plots of gene expressions were generated based on normalized gene expression data.
Pan cancer TCGA analysis
Gene expression levels were accessed from 10,071 samples from 32 studies curated by cBioPortal.org (cite Gao et al. PMID 23550210) as “TCGA PanCancer Atlas Studies”. Samples were grouped by TCGA PanCancer Atlas Cancer Type Acronym and mRNA expression values displayed as RSEM Batch normalized from Illumina HiSeq RNASeqV2. Cancer types were sorted by expression median. Data points were coloured by the presence of specific mutation types as curated by cBioPortal. The full bookmarked query is available as: https://www.cbioportal.org/results/plots?cancer_study_list=laml_tcga_pan_can_atlas_2018%2Cacc_tcga_pan_can_atlas_2018%2Cblca_tcga_pan_can_atlas_2018%2Clgg_tcga_pan_can_atlas_2018%2Cbrca_tcga_pan_can_atlas_2018%2Ccesc_tcga_pan_can_atlas_2018%2Cchol_tcga_pan_can_atlas_2018%2Ccoadread_tcga_pan_can_atlas_2018%2Cdlbc_tcga_pan_can_atlas_2018%2Cesca_tcga_pan_can_atlas_2018%2Cgbm_tcga_pan_can_atlas_2018%2Chnsc_tcga_pan_can_atlas_2018%2Ckich_tcga_pan_can_atlas_2018%2Ckirc_tcga_pan_can_atlas_2018%2Ckirp_tcga_pan_can_atlas_2018%2Clihc_tcga_pan_can_atlas_2018%2Cluad_tcga_pan_can_atlas_2018%2Clusc_tcga_pan_can_atlas_2018%2Cmeso_tcga_pan_can_atlas_2018%2Cov_tcga_pan_can_atlas_2018%2Cpaad_tcga_pan_can_atlas_2018%2Cpcpg_tcga_pan_can_atlas_2018%2Cprad_tcga_pan_can_atlas_2018%2Csarc_tcga_pan_can_atlas_2018%2Cskcm_tcga_pan_can_atlas_2018%2Cstad_tcga_pan_can_atlas_2018%2Ctgct_tcga_pan_can_atlas_2018%2Cthym_tcga_pan_can_atlas_2018%2Cthca_tcga_pan_can_atlas_2018%2Cucs_tcga_pan_can_atlas_2018%2Cucec_tcga_pan_can_atlas_2018%2Cuvm_tcga_pan_can_atlas_2018&Z_SCORE_THRESHOLD=2.0&RPPA_SCORE_THRESHOLD=2.0&data_priority=0&profileFilter=0&case_set_id=all&gene_list=AHR&geneset_list=%20&tab_index=tab_visualize&Action=Submit&plots_horz_selection=%7B%22dataType%22%3A%22clinical_attribute%22%2C%22selectedDataSourceOption%22%3A%22CANCER_TYPE_ACRONYM%22%7D&plots_vert_selection=%7B%22selectedGeneOption%22%3A196%2C%22selectedDataSourceOption%22%3A%22rna_seq_v2_mrna%22%2C%22logScale%22%3A%22true%22%7D&plots_coloring_selection=%7B%22colorByCopyNumber%22%3A%22false%22%7D
Analysis of human PDAC scRNA sequencing
To examine transcriptomics in human PDAC we analyzed a previously published scRNA sequencing dataset using a similar methodology (Steele et al., 2020). Briefly, cells with high mitochondrial reads were filtered out (>10%), as well as cells that were outliers due to oversequencing (total_counts=4836; > 95th percentile) and cells with too few genes (n_genes=581; < 5th percentile). Using the filtered cells, we combined all cells from the PDAC tissue and controls using Harmony (Korsunsky et al., 2019) to create sample and group non-specific clusters that were annotated using the SCSA algorithm (Cao et al., 2020). We then manually resolved any ambiguous annotations using the original publication groupings and known cell-type markers as a reference. UMAP visualization of the was done using the Seurat RunUMAP command with parameters set to 30 dimensions, 10 nearest neighbors, 2 components, 400 epochs, and minimum distance of 0.1. We compared AHR expression to our own marker gene set of interest and all genes expressed across all cells. Expression similarity was done on a per-cluster basis, using the clusters inferred from Harmony. For both the mean gene expression and percent-expressed per cluster, we calculated the euclidean distances between AhR and all other genes (n=32,738). We then combined the distance metric using the mean value of both these distances and selected the top 25 for further analysis.
PDAC organoid culture
To visualize the effect of indole acetic acid (IAA)-treated macrophages on organoid growth, human monocyte-derived macrophages (MDMs) were grown in the presence of hM-CSF for 5 days. They were then treated with 250 μM IAA overnight or left untreated (control). Macrophages were lifted off the plates using PBS+ 2% FBS+ 2mM EDTA. They were then labeled with DiD for 5 minutes at 37°C and washed twice in complete medium. GFP-labeled PDAC organoid (Pancreatic Organoid, Tumor, PMLB PPTO.46 from the University Health Network Living Biobank, Toronto, Canada) domes were grown for 4 days in matrigel. The PDAC organoids were then freed from the matrigel domes by gently pipetting up and down. Ice cold advanced DMEM/F12 (AD/F12) was added to the organoids and they were centrifuged down. Cell rinse solution was then added to the pellet to depolymerize matrigel. PDAC organoids and macrophages were then co-cultured in the 24-GLAnCE system (D’Arcangelo et al., 2020) for visualization over a period of 5 days.
For sorting of organoid cells from macrophage/organoid co-cultures, IAA-treated or untreated unlabeled macrophages were co-cultured in matrigel domes with GFP-ve PDAC organoid for 4 days. Domes were then disrupted by pipetting up and down and addition of TRyple enzyme. Single cell suspension containing macrophages and tumor cells were then stained with anti-CD45 antibody and Zombie violet viability dye. Cells were then sorted directly into RNA lysis buffer using the MoFlo Astrios EQ cell sorter (Beckmann Coulter).
Metabolomic Sample Preparation
Briefly, 20 μl of bacterial cultures supernatants or mouse serum were mixed with 80 μl of ice-cold methanol spiked with 5 μl of a 1:1 dilution of an amino acid standard in methanol. The mixture was then incubated at −20°C for 1 hr and centrifuged for 10 minutes at maximum speed in a refrigerated countertop centrifuge. The supernatant was then collected and ran on a mass spectrometer at Dalhousie University Biological Mass Spectrometry Facility. Data analysis was performed using the Skyline software platform
Time-of-Flight mass cytometry (CyTOF)
CD45+ cells were enriched from single cell suspensions from orthotopic PDAC tumors using the positive selection kit and biotin labeled anti-CD45.2 antibody StemCell. Enriched cells were resuspended in Maxpar Staining Buffer, blocked with TruStain Fc blocking buffer for 10 minutes and then resuspended in antibody staining cocktail for 30 Cells were washed twice and incubated in 1 μM Cell_Id cisplatin for 5 minutes. Cisplatin was then quenched by addition of PBS + 5% FBS. Cells were then washed, fixed and permeabilized then barcoded using the Cell-ID multiplex Barcoding kit. Pooled cells were then stained with intracellular antibodies using the permeabilization buffer from Foxp3 transcription factor staining kit. They were then washed twice with perm. Buffer and resuspended in 1.6% paraformaldehyde in PBS containing 0.3% saponin and 125 nM Iridium (Fluidigm, Cat. # 201192A). They were stored in this solution at 4°C until the day of acquisition when they were centrifuged down, washed with Maxpar buffer and resuspended in Maxpar water + EQ beads solution, and analyzed on a Helios Mass Cytometer (Fluidigm) at the Sickkids CYTOF facility.
Flow Cytometry
PDAC tumors were digested using 100 U/ml collagenase IV and 50 u/ml DNase I in complete RPMI medium at 37C. Single cell PDAC preparations were stained with antibody cocktail at a concentration of 1:300 in FACS buffer (PBS + 2% FBS) for 45 minutes at 4°C in the presence of a fixable viability dye. Cells were then washed twice with FACS buffer, fixed in 4% paraformaldehyde for 10 minutes, washed again with FACS buffer and assessed for fluorescence on a BD Fortessa flow cytometer. For staining of intracellular cytokines cells were first treated for 4 hrs with PMA (5 ng/ml) and ionomycin (500ng/ml) in the presence of Brefeldin A. Surface staining was performed as described above, followed by fixation, permeabilization and staining with intracellular antibodies using the from Foxp3 transcription factor staining kit according to manufacturer’s recommendations.
For sorting experiments, mouse tumor cells were stained with anti-CD45, anti-CD11b anti-F4/80 (TAMs) on a MoFlo (Beckman Coulter) cell sorter. For flow cytometric analysis at least 105 events were collected on the LSR Fortessa flow cytometer (BD Biosciences).
INFγ blocking or αCD8 T cell delpetion
Orthotopic PDAC surgery was performed on AhRfl/fl and AhRfl/fl LMC mice. At day 3 post-surgery the mice were injected intraperitoneally with 200 μg InVivoMab α-mouse INFγ IgG1 antibody or IgG1 isotype control every 3 days for up to 2 weeks.
In one set of experiments to deplete CD8+ T cells, 3 days post-surgery mice received 200μg of αmouse CD8 IgG2a or IgG2A isotype control in an initial injection and 100μg every third day thereafter for up to 2 weeks.
Tryptophan-deficient diet and indole gavage
One day before PDAC orthotopic surgery, regular mouse chow was replaced by an amino acid control diet or a tryptophan-deficient diet. The diet was maintained for 14 days until the experimental endpoint.
In one set of experiments mice were gavaged with 40μg/kg of IAA, IAld (both from Cayman Chemicals) or ILA (Sigma Aldrich) dissolved in 200μl of water every day beginning 4 days after orthotopic tumor injection.
Metabolomics
Targeted metabolomic profiling of cell culture supernatant was performed using liquid chromatography tandem mass spectrometry as previously described (de Vries et al., 2020). Briefly, 20 μl of cell culture supernatant was mixed with 80 μl of cold methanol and 5ul of internal standard (Isotopically labeled amino acids, 1.25mM, PN MSK-A2–1.2, Cambridge Isotope Laboratories), incubated for 30 minutes at −20 C, centrifuged at 13, 000xg for 5 min at 4 C. Samples were further diluted 10-fold prior to analysis. Quality control (QC) samples were prepared by pooling 10 μl of each sample. All samples including QC’s were separated using a Cortecs T3 2.1 × 50 mm, 2.7 μm column (Waters Inc.) using (A) water with 0.1% formic acid and (B) acetonitrile with 0.1% formic acid solvent system coupled to a Sciex Qtrap 5500 triple quadrupole linear ion trap tandem mass spectrometer. The data acquisition included 151 transitions. Data were captured using Analyst, version 1.6.2 software (Sciex); peak integration was performed using Skyline, version 4.1 (Pino et al., 2020). An in-house R script (R-4.0.2) was used for data processing. Metabolites with low peak heights (<5000) and peaks with variable retention times (>30% CV) were excluded from further analysis. Metabolite peak heights were normalized using the mean of nearest QC samples. Statistical analysis and heatmaps were generated with MetaboAnalyst (Xia and Wishart, 2016).
16S rRNA gene sequencing
The V4 hypervariable region of the 16S rRNA gene was amplified using uniquely barcoded forward and reverse sequencing primers to allow for multiplexing. Amplification reactions were performed using 12.5 μL of KAPA2G Robust HotStart ReadyMix, 1.5 μL of 10 μM forward and reverse primers, 7.5 uL of sterile water and 2 μL of DNA. The V4 region was amplified by cycling the reaction at 95°C for 3 minutes, 18x or 24x cycles of 95°C for 15 seconds, 50°C for 15 seconds and 72°C for 15 seconds, followed by a 5-minute 72°C extension. All amplification reactions were done in triplicate, pooled together, and checked on a 1% agarose TBE gel. Pooled trip-licates were quantified using PicoGreen and combined by even concentrations. The library was then purified using Ampure XP beads and loaded on to the Illumina MiSeq for sequencing, according to manufacturer instructions (Illumina, San Diego, CA). Sequencing was performed using the V2 (150bp × 2) chemistry. A single-species (Pseudomonas aeruginosa DNA), a mock community (Zymo Microbial Standard: https://www.zymoresearch.de/zymobiomics-community-standard) and a template-free negative control were included in the sequencing run.
Analysis of the bacterial microbiome
The last base was removed from all sequences using cutadapt v.1.18. Sequences were assembled and quality trimmed using –fastq_mergepairs with a –fastq_trunctail set at 5, a –fastq_minqual set at 3, and minimum and maximum assemble lengths set at 243 and 263 (+/− 10 from the mean) base pairs. Assembled sequences were quality filtered using –fastq_filter with a –fastq_maxee set at 1.0. The trimmed data was then processed following the UNOISE pipeline. Sequences were first de-replicated and sorted to remove singletons, then denoised and chimeras were removed using the unoise3 command. Assembled sequences were mapped back to the chimera-free denoised sequences at 97% identity OTUs. Taxonomy assignment was executed using SINTAX, available through USEARCH, and the UNOISE compatible Ribosomal Database Project (RDP) database version 16, with a minimum confidence cutoff of 0.8 (Wang et al., 2007). OTU sequences were aligned using align_seqs.py v.1.9.1 through QIIME1 (Caporaso et al., 2010). Sequences that did not align were removed from the dataset and a phylogenetic tree of the filtered aligned sequence data was made using FastTree (Price et al., 2009). The OTU table, including taxonomic assignments, was imported into MicrobiomeAnalyst (Chong et al., 2020; Dhariwal et al., 2017) for comparisons of alpha diversity and to generate bar plots of taxon relative abundances. Data were filtered to remove OTUs with low counts or variance using default parameters. Data were scaled using Total sum scaling (TSS) to remove variability between samples based on sequencing depth.
QUANTIFICATION AND STATISTICAL ANALYSIS
Number of mice and experiments, and statistical tests are reported in each figure legend. Analyses were performed using GraphPad Prism 8 software. Statistical significance was calculated using t-test (unpaired) or Wilcoxon rank-sum test or one-way ANOVA or with Kaplan-Meier analysis followed by the log-rank test. Error bars represent standard deviation and p values <.05 were considered statistically significant (* p <.05, ** p <.01, *** p <.001, **** p <.0001, NS- not significant).
Supplementary Material
Highlights.
AhR controls tumor-associated macrophage function in pancreatic cancer
AhR is activated in tumor macrophages by microbiome-produced tryptophan metabolites
Inhibition of AhR function improves T cell function, inhibiting tumor growth
AhR expression is enriched in human tumor macrophages and correlates with survival
ACKNOWLEDGMENTS
We thank Kate Banks, Karen Parisien, and Amy Cao from the University of Toronto Germ Free Mouse Core for assistance in establishing the germ free orthotopic PDAC mouse model. In addition, we thank Dr. Kieran Manion for assistance with graphical abstract images. We also thank Dr. Hal Berman for pathologic assessment of mouse PDAC tumors. This work was supported by NIH grants R01AR067763, R01CA190449, R01CA255670, the Terry Fox Research Institute New Frontiers Program, Medicine by Design-Canada First Research Excellence Fund, the John R. Evans Leaders Fund, and project grants from the Canadian Institutes of Health Research (T.L.M.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2022.01.006.
DECLARATION OF INTERESTS
The authors have no conflicting interests to declare.
REFERENCES
- Afik R, Zigmond E, Vugman M, Klepfish M, Shimshoni E, Pasmanik-Chor M, Shenoy A, Bassat E, Halpern Z, Geiger T, et al. (2016). Tumor macrophages are pivotal constructors of tumor collagenous matrix. J. Exp. Med 213, 2315–2331. 10.1084/jem.20151193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambarus CA, Krausz S, van Eijk M, Hamann J, Radstake TR, Reedquist KA, Tak PP, and Baeten DL (2012). Systematic validation of specific phenotypic markers for in vitro polarized human macrophages. J. Immunol. Methods 375, 196–206. 10.1016/j.jim.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Aragozzini F, Ferrari A, Pacini N, and Gualandris R (1979). Indole-3-lactic acid as a tryptophan metabolite produced by Bifidobacterium spp. Appl. Environ. Microbiol 38, 544–546. 10.1128/AEM.38.3.544-546.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boj SF, Hwang C-I, Baker LA, Chio IIC, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, et al. (2015). Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338. 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braümuller H, Wieder T, Brenner E, Aßmann S, Hahn M, Alkhaled M, Schilbach K, Essmann F, Kneilling M, Griessinger C, et al. (2013). T-helper-1-cell cytokines drive cancer into senescence. Nature 494, 361–365. 10.1038/nature11824. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Wang X, and Peng G (2020). SCSA: a cell type annotation tool for single-cell RNA-seq data. Front. Genet 11, 490. 10.3389/fgene.2020.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, et al. (2017). Lactobacillus reuteri induces gut intraepithelial CD4+CD8alphaalpha+T cells. Science 357, 806–810. 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Willette-Brown J, Wu X, Hu Y, Howard OM, Hu Y, and Oppenheim JJ (2015). IKKalpha is required for the homeostasis of regulatory T cells and for the expansion of both regulatory and effector CD4 T cells. FASEB J 29, 443–454. 10.1096/fj.14-259564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Liu P, Zhou G, and Xia J (2020). Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc 15, 799–821. 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- Cuadrado E, van den Biggelaar M, de Kivit S, Chen Y-Y, Slot M, Doubal I, Meijer A, van Lier RAW, Borst J, and Amsen D (2018). Proteomic analyses of human regulatory T cells reveal adaptations in signaling pathways that protect cellular identity. Immunity 48, 1046–1059.e6. 10.1016/j.immuni.2018.04.008. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo E, Wu NC, Chen T, Shahaj A, Cadavid JL, Huang L, Ailles L, and McGuigan AP (2020). Gels for live analysis of compartmentalized environments (GLAnCE): a tissue model to probe tumour phenotypes at tumour-stroma interfaces. Biomaterials 228, 119572. 10.1016/j.biomaterials.2019.119572. [DOI] [PubMed] [Google Scholar]
- Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin J-M, Morrison RM, Deblasio RN, Menna C, Ding Q, Pagliano O, et al. (2021). Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 371, 595–602. 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, de Vries S, Curtis BA, Zhou H, Penny S, Feussner K, Pinto DM, Steinert M, Cohen AM, von Schwartzenberg K, and Archibald JM (2020). Heat stress response in the closest algal relatives of land plants reveals conserved stress signaling circuits. Plant J 103, 1025–1048. 10.1111/tpj.14782. [DOI] [PubMed] [Google Scholar]
- Dejean AS, Beisner DR, Ch’en IL, Kerdiles YM, Babour A, Arden KC, Castrillon DH, DePinho RA, and Hedrick SM (2009). Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat. Immunol 10, 504–513. 10.1038/ni.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo DG, and Ruffell B (2019). Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol 19, 369–382. 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhariwal A, Chong J, Habib S, King IL, Agellon LB, and Xia J (2017). MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 45, W180–W188. 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S, D’Amico A, Cretney E, Liao Y, Tellier J, Bruggeman C, Almeida FF, Leahy J, Belz GT, Smyth GK, et al. (2017). Effector regulatory T cell differentiation and immune homeostasis depend on the transcription factor Myb. Immunity 46, 78–91. 10.1016/j.immuni.2016.12.017. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, and Sonnenburg JL (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652. 10.1038/nature24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsden SR, Hilton MG, and Waller JM (1976). The end products of the metabolism of aromatic amino acids by Clostridia. Arch. Microbiol 107, 283–288. 10.1007/BF00425340. [DOI] [PubMed] [Google Scholar]
- Flaveny CA, and Perdew GH (2009). Transgenic humanized AHR mouse reveals differences between human and mouse AHR ligand selectivity. Mol. Cell. Pharmacol 1, 119–123. 10.4255/mcpharmacol.09.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragale A, Gabriele L, Stellacci E, Borghi P, Perrotti E, Ilari R, Lanciotti A, Remoli AL, Venditti M, Belardelli F, and Battistini A (2008). IFN regulatory factor-1 negatively regulates CD4+ CD25+ regulatory T cell differentiation by repressing Foxp3 expression. J. Immunol 181, 1673–1682. 10.4049/jimmunol.181.3.1673. [DOI] [PubMed] [Google Scholar]
- Franchini AM, Myers JR, Jin G-B, Shepherd DM, and Lawrence BP (2019). Genome-wide transcriptional analysis reveals novel AhR targets that regulate dendritic cell function during influenza A virus infection. Immunohorizons 3, 219–235. 10.4049/immunohorizons.1900004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudot C, Coillard A, Villani A-C, Gueguen P, Cros A, Sarkizova S, Tang-Huau T-L, Bohec M, Baulande S, Hacohen N, et al. (2017). Aryl hydrocarbon receptor Controls Monocyte Differentiation into dendritic Cells versus Macrophages. Immunity 47, 582–596.e6. 10.1016/j.immuni.2017.08.016. [DOI] [PubMed] [Google Scholar]
- Halaby MJ, Hezaveh K, Lamorte S, Ciudad MT, Kloetgen A, MacLeod BL, Guo M, Chakravarthy A, Medina TDS, Ugel S, et al. (2019). GCN2 drives macrophage and MDSC function and immunosuppression in the tumor microenvironment. Sci. Immunol 4. 10.1126/sciimmunol.aax8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbrook CJ, Pontious C, Kovalenko I, Lapienyte L, Dreyer S, Lee H-J, Thurston G, Zhang Y, Lazarus J, Sajjakulnukit P, et al. (2019). Macrophage-released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell Metab 29, 1390–1399.e6. 10.1016/j.cmet.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TD, Murray IA, Bisson WH, Lahoti TS, Gowda K, Amin SG, Patterson AD, and Perdew GH (2015). Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci. Rep 5, 12689. 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, and Raychaudhuri S (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir E-a.D., Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197. 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, et al. (2011). Kruppel-like factor 4 regulates macrophage polarization. J. Clin. Invest 121, 2736–2749. 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço AR, and Coffer PJ (2017). A tumor suppressor role for C/EBPalpha in solid tumors: more than fat and blood. Oncogene 36, 5221–5230. 10.1038/onc.2017.151. [DOI] [PubMed] [Google Scholar]
- Maddipati R, and Stanger BZ (2015). Pancreatic cancer metastases harbor evidence of polyclonality. Cancer Discov 5, 1086–1097. 10.1158/2159-8290.CD-15-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, and Gajewski TF (2018). The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108. 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone I, Rizzo A, Sarra M, Sica G, Sileri P, Biancone L, MacDonald TT, Pallone F, and Monteleone G (2011). Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 141, 237–248. 248.e1. 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Murray IA, Patterson AD, and Perdew GH (2014). Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat. Rev. Cancer 14, 801–814. 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino LK, Searle BC, Bollinger JG, Nunn B, MacLean B, and MacCoss MJ (2020). The Skyline ecosystem: informatics for quantitative mass spectrometry proteomics. Mass Spectrom. Rev 39, 229–244. 10.1002/mas.21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, and Arkin AP (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol 26, 1641–1650. 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, et al. (2018). The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov 8, 403–416. 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravishankar B, Liu H, Shinde R, Chaudhary K, Xiao W, Bradley J, Koritzinsky M, Madaio MP, and McGaha TL (2015). The amino acid sensor GCN2 inhibits inflammatory responses to apoptotic cells promoting tolerance and suppressing systemic autoimmunity. Proc. Natl. Acad. Sci. USA 112, 10774–10779. 10.1073/pnas.1504276112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, San Lucas A, et al. (2019). Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 178, 795–806.e12. 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roager HM, and Licht TR (2018). Microbial tryptophan catabolites in health and disease. Nat. Commun 9, 3294. 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a bio-conductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V, Borucki DM, Tjon EC, Takenaka MC, Chao C-C, Ardura-Fabregat A, de Lima KA, Gutiérrez-Vázquez C, Hewson P, Staszewski O, et al. (2018). Microglial control of astrocytes in response to microbial metabolites. Nature 557, 724–728. 10.1038/s41586-018-0119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, Chao CC, Patel B, Yan R, Blain M, et al. (2016). Type I inter-ferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med 22, 586–597. 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, Anderson SE, and Flint HJ (2013). Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res 57, 523–535. 10.1002/mnfr.201200594. [DOI] [PubMed] [Google Scholar]
- Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, and Schreiber RD (2001). IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 410, 1107–1111. 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- Shinde R, Hezaveh K, Halaby MJ, Kloetgen A, Chakravarthy A, da Silva Medina T, Deol R, Manion KP, Baglaenko Y, Eldh M, et al. (2018). Apoptotic cell-induced AhR activity is required for immunological tolerance and suppression of systemic lupus erythematosus in mice and humans. Nat. Immunol 19, 571–582. 10.1038/s41590-018-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde R, and McGaha TL (2018). The aryl hydrocarbon receptor: connecting immunity to the microenvironment. Trends Immunol 39, 1005–1020. 10.1016/j.it.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EA, and Macfarlane GT (1996). Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J. Appl. Bacteriol 81, 288–302. 10.1111/j.1365-2672.1996.tb04331.x. [DOI] [PubMed] [Google Scholar]
- St Paul M, and Ohashi PS (2020). The roles of CD8+ T cell subsets in anti-tumor immunity. Trends Cell Biol 30, 695–704. 10.1016/j.tcb.2020.06.003. [DOI] [PubMed] [Google Scholar]
- Steele NG, Carpenter ES, Kemp SB, Sirihorachai V, The S, Delrosario L, Lazarus J, Amir E-AD, Gunchick V, Espinoza C, et al. (2020). Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nat. Cancer 1, 1097–1112. 10.1038/s43018-020-00121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, and Glimcher LH (2000). A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669. 10.1016/S0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Takenaka MC, Gabriely G, Rothhammer V, Mascanfroni ID, Wheeler MA, Chao C-C, Gutiérrez-Vázquez C, Kenison J, Tjon EC, Barroso A, et al. (2019). Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat. Neurosci 22, 729–740. 10.1038/s41593-019-0370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tang W, Yang S, He P, Wang J, Gaedcke J, Ströbel P, Azizian A, Ried T, Gaida MM, et al. (2020). NOd/RUNX3/kynurenine metabolic signaling enhances disease aggressiveness in pancreatic cancer. Int. J. Cancer 146, 3160–3169. 10.1002/ijc.32733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, and Cole JR (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol 73, 5261–5267. 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, and Lane DJ (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol 173, 697–703. 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mähler A, Balogh A, Markó L, et al. (2017). Salt-responsive gut commensal modulates TH17 axis and disease. Nature 551, 585–589. 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, and Wishart DS (2016). Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics 55, 14.10.1–14.10.91. 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Hoki T, Oba T, Saito H, Attwood K, Sabel MS, Chang AE, Odunsi K, and Ito F (2020). CX3CR1-CD8+ T cells are critical in anti-tumor efficacy but functionally suppressed in the tumor microenvironment. JCI Insight 5. 10.1172/jci.insight.133920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. (2013). Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385. 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zhu M, Goetsch SC, Wang Z, Luo R, Hill JA, Schneider J, Morris SM Jr., and Liu Z-P (2015). FoxO4 promotes early inflammatory response upon myocardial infarction via endothelial Arg1. Circ. Res 117, 967–977. 10.1161/CIRCRESAHA.115.306919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Lönnblom E, Förster M, Johannesson M, Tao P, Meng L, Lu S, and Holmdahl R (2020). Natural polymorphism of Ym1 regulates pneumonitis through alternative activation of macrophages. Sci. Adv 6. 10.1126/sciadv.aba9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Herndon JM, Sojka DK, Kim K-W, Knolhoff BL, Zuo C, Cullinan DR, Luo J, Bearden AR, Lavine KJ, et al. (2017). Tissue-resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity 47, 597. 10.1016/j.immuni.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


