Abstract
BACKGROUND
Autophagy is an intracellular catabolic process of degrading and recycling proteins and organelles to modulate various physiological and pathological events, including cell differentiation and development. Emerging data indicate that autophagy is closely associated with male reproduction, especially the biosynthetic and catabolic processes of sperm. Throughout the fate of sperm, a series of highly specialized cellular events occur, involving pre-testicular, testicular and post-testicular events. Nonetheless, the most fundamental question of whether autophagy plays a protective or harmful role in male reproduction, especially in sperm, remains unclear.
OBJECTIVE AND RATIONALE
We summarize the functional roles of autophagy in the pre-testicular (hypothalamic–pituitary–testis (HPG) axis), testicular (spermatocytogenesis, spermatidogenesis, spermiogenesis, spermiation) and post-testicular (sperm maturation and fertilization) processes according to the timeline of sperm fate. Additionally, critical mechanisms of the action and clinical impacts of autophagy on sperm are identified, laying the foundation for the treatment of male infertility.
SEARCH METHODS
In this narrative review, the PubMed database was used to search peer-reviewed publications for summarizing the functional roles of autophagy in the fate of sperm using the following terms: ‘autophagy’, ‘sperm’, ‘hypothalamic–pituitary–testis axis’, ‘spermatogenesis’, ‘spermatocytogenesis’, ‘spermatidogenesis’, ‘spermiogenesis’, ‘spermiation’, ‘sperm maturation’, ‘fertilization’, ‘capacitation’ and ‘acrosome’ in combination with autophagy-related proteins. We also performed a bibliographic search for the clinical impact of the autophagy process using the keywords of autophagy inhibitors such as ‘bafilomycin A1’, ‘chloroquine’, ‘hydroxychloroquine’, ‘3-Methyl Adenine (3-MA)’, ‘lucanthone’, ‘wortmannin’ and autophagy activators such as ‘rapamycin’, ‘perifosine’, ‘metformin’ in combination with ‘disease’, ‘treatment’, ‘therapy’, ‘male infertility’ and equivalent terms. In addition, reference lists of primary and review articles were reviewed for additional relevant publications. All relevant publications until August 2021 were critically evaluated and discussed on the basis of relevance, quality and timelines.
OUTCOMES
(i) In pre-testicular processes, autophagy-related genes are involved in the regulation of the HPG axis; and (ii) in testicular processes, mTORC1, the main gate to autophagy, is crucial for spermatogonia stem cell (SCCs) proliferation, differentiation, meiotic progression, inactivation of sex chromosomes and spermiogenesis. During spermatidogenesis, autophagy maintains haploid round spermatid chromatoid body homeostasis for differentiation. During spermiogenesis, autophagy participates in acrosome biogenesis, flagella assembly, head shaping and the removal of cytoplasm from elongating spermatid. After spermatogenesis, through PDLIM1, autophagy orchestrates apical ectoplasmic specialization and basal ectoplasmic specialization to handle cytoskeleton assembly, governing spermatid movement and release during spermiation. In post-testicular processes, there is no direct evidence that autophagy participates in the process of capacitation. However, autophagy modulates the acrosome reaction, paternal mitochondria elimination and clearance of membranous organelles during fertilization.
WIDER IMPLICATIONS
Deciphering the roles of autophagy in the entire fate of sperm will provide valuable insights into therapies for diseases, especially male infertility.
Keywords: autophagy / hypothalamic-pituitary-testis axis / spermatogenesis / acrosome biogenesis / acrosome reaction / paternal mitochondria elimination / ectoplasmic specialisation / sperm maturation / erectile dysfunction / fertilisation
Introduction
Autophagy
Autophagy is a ‘self-eating’ catabolic process of degrading cytoplasmic materials in lysosomes, which plays a fundamental role in various physiological or pathological processes (Mizushima and Levine, 2020). Our knowledge of autophagy is dramatically expanding day by day. The 60-year developmental history of autophagy (Fig. 1a) reflects the progress of science and leads to a new era in our understanding of autophagy in human health. Known as a double-edged sword, autophagy can serve as protective mechanism by eliminating damaged organelles and providing energy for cellular renovation, yet, autophagy may also contribute to cell damage (Shintani and Klionsky, 2004).
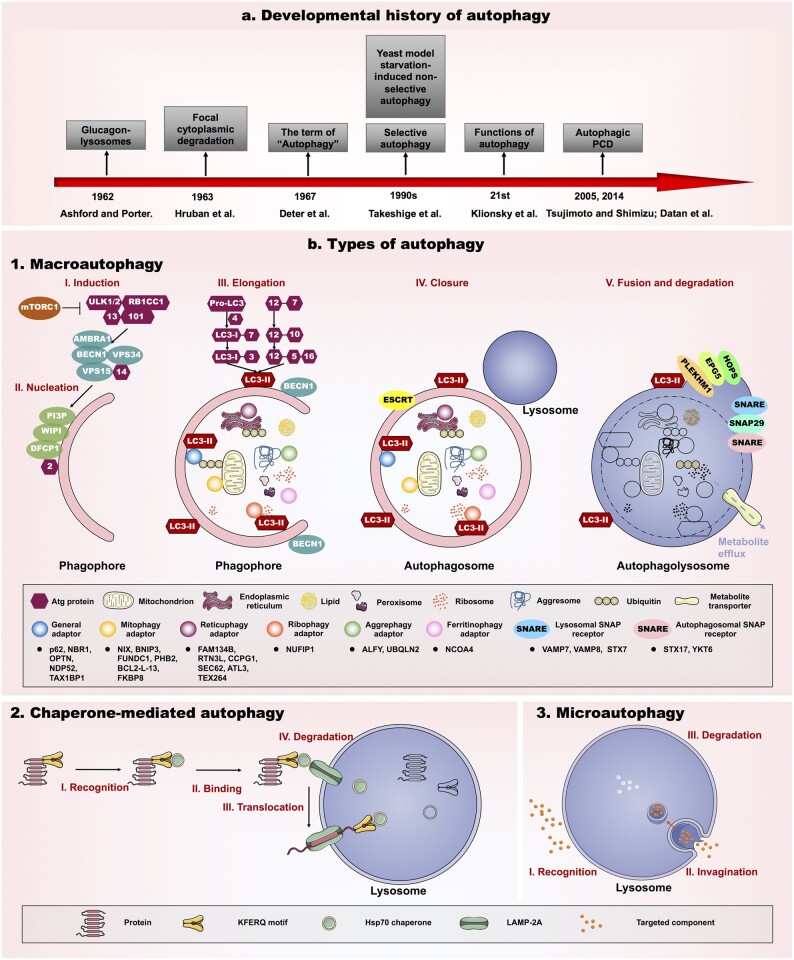
Figure 1.
The developmental history and types of autophagy. (a) Developmental history of autophagy. In 1962, autophagy originated from an observation that increased lysosomes migrated toward organelles in response to the addition of glucagon in rat liver cells. In 1963, a detailed ultrastructure of ‘focal cytoplasmic degradation’ was described; the term ‘autophagy’ was proposed as a part of lysosomal function. In the 1990s, autophagy started to be investigated extensively due to starvation-induced non-selective autophagy, followed by selective autophagy. In the early 21st century, autophagy was demonstrated as one of the repair mechanisms as damaged organelles, membranes, and proteins were degraded for generating energy and building new proteins and membranes through autophagy-mediated cellular metabolism. In 2005 and 2014, autophagic cell death was identified as a novel way of programmed cell death (PCD). (b) Types of autophagy. 1. Macroautophagy is a conserved dynamic process, which consists of induction (phagophore formation), nucleation, elongation, closure (autophagosome formation), fusion (autophagosome and lysosome into autophagolysosome) and degradation, thus degrading organelles and proteins for the synthesis of new macromolecules or as a source of energy. Involved signaling pathways are as follows: I. Induction: mTORC1 inhibits ULK complex, which comprises ULK1/2 (also named ATG1), RB1CC1 (also named ATG17), ATG13 and ATG101, leading to the translocation of the complex to the phagophore and initiating the autophagy. II. Nucleation: ULK complex activates PI3K complex (BECN1-VPS34-VPS15-ATG14) by the phosphorylation of AMBRA1 and BECN1, generating phosphatidylinositol 3-phosphate (PI3P) on phagophore membrane. PI3P recruits WIPI, DFCP1 and ATG2 to promote autophagosome formation. III. Elongation: it is regulated by two conjugated systems of LC3-II (also named ATG8) and ATG12–ATG5–ATG16 complex. LC3 precursor is hydrolyzed by ATG4 to form LC3-I, which interacts with ATG7 and ATG3, forming LC3-II (also known as LC3-PE). IV. Closure: LC3-II and ESCRT regulate the closure of phagophore, thus facilitating the autophagosome formation. V. Fusion and degradation: after closure, the mature autophagosome and a lysosome fuse into an autophagolososome, which degrades the dispensable organelles and proteins. Autophagosomal SNAP receptors (STX17, YKT6) interact with SNAP29, which binding to lysosomal SNAP receptors (VAMP7, VAMP8 and STX7), together with HOPS complex, EPG5 and PLEKHM1 promoting the fusion. Due to the difference of destructive targets, macroautophagy is divided into non-selective autophagy targeting bulk cytosol and selective autophagy targeting specific cargos, such as mitochondrion, endoplasmic reticulum, lipid, peroxisome, ribosome, aggresome and ferritin. The specific cargo directly recognizes LC3 or indirectly recognizes LC3 through ubiquitin with specific cargo adapters, such as general adaptors (p62, NBR1, OPTN, NDP52, TAX1BP1), mitophagy adapters (NIX, BNIP3, FUNDC1, PHB2, BCL2-L-13, FKBP8), reticuphagy adapters (FAM134B, RTN3L, CCPG1, SEC62, ATL3, TEX264), ribophagy adapter (NUFIP1), aggrephagy adaptors (ALFY, UBQLN2), and ferritinophagy adaptor (NCOA4). 2. In chaperone-mediated autophagy (CMA), proteins carrying KFERQ motif are recognized by the Hsp70 chaperone, which interacts with lysosome membrane protein LAMP-2A, leading the translocation of the bound protein into the lysosome and degradation. 3. Microautophagy is characterized by the direct engulfment of cytoplasmic material into the lysosome through invagination and pinching off.
Autophagy-related genes (Atgs) and enzymes are identified in the three types of autophagy: macroautophagy, chaperone-mediated autophagy (CMA) and microautophagy (Fig. 1b). Firstly, macroautophagy, as the major form of autophagy, is a conserved dynamic process, which consists of induction (phagophore formation), nucleation, elongation, closure (autophagosome formation), fusion (autophagosome and lysosome into autophagolysosome) and degradation, thus degrading organelles and proteins for the synthesis of new macromolecules or as a source of energy. Due to the difference of destructive targets, macroautophagy (hereafter called autophagy) is divided into non-selective autophagy (targeting bulk cytosol) and selective autophagy (targeting specific organelles), such as mitophagy, lipophagy, pexophagy, ribophagy, reticulophagy, aggrephagy and ferritinophagy. The involved signaling pathways and key molecules are shown in Fig. 1b. Secondly, in CMA, proteins carrying the KFERQ motif are recognized by the Hsp70 chaperone, which interacts with lysosome membrane protein LAMP-2A, leading the translocation of the bound protein into the lysosome (Boya et al., 2013). Thirdly, microautophagy is characterized by the direct engulfment of cytoplasmic material into the lysosome through invagination and pinching off. To sustain homeostasis, autophagy, together with the ubiquitin-proteasome system (UPS), constitutes the major cellular quality control systems for the degradation and disposal of organelles (Pohl and Dikic, 2019). Depending on these emerging regulatory processes and functions, autophagy regulates cell growth, survival and cell death (Dai et al., 2020).
The fate of sperm
As shown in Table I, the knockdown/knockout (KD/KO) of Atgs in mammalian testis has shown the role of specific molecules in the regulation of distinct testicular cells. Sperm is the male reproductive cell or gamete that fuses with an oocyte to form a fertilized oocyte that develops into an individual, so sperm is the key to male fertility (Kimble and Page, 2007). The fate of sperm refers to an intricate process from the origin of sperm to its disappearance. Sperm is produced by spermatogenesis in the male reproductive gland, the testis, whose functions are controlled by the hypothalamic–pituitary–gonadal (HPG) axis. Once completing spermatogenesis in the testis, sperm move into the epididymis to mature into spermatozoa. Subsequently, mature sperm-containing semen is ejaculated into the female reproductive tract and for fertilization of the oocyte. Overall, the fate of sperm may be categorized as pre-testicular, testicular, post-testicular according to the timeline (Krausz, 2011).
Table I.
Autophagy-related genes KD/KO in the mammalian testis/testicular cells.
| Autophagy- related gene | Autophagy- related process | KD/KO model | Fertility | Phenotype | Functions in testis | Reference(s) |
|---|---|---|---|---|---|---|
| Macroautophagy | ||||||
| Atg5 | Phagophore formation | cKO in germ cells | Subfertile | Induce sperm counts and motility reduction, misshapen sperm heads and tails, abnormal mitochondria and acrosome distribution | Elongating spermatid development, sperm individualization during spermiogenesis | Huang et al. (2021) |
| cKO in Sertoli cells | Infertile | Disrupt cytoskeleton structures and ectoplasmic specialization assembly | Ectoplasmic specialization assembly | Liu et al. (2016) | ||
| cKO in Leydig cells | Subfertile | Suppress testosterone synthesis, affect sexual behavior | Testosterone synthesis | Gao et al. (2018) | ||
| Atg7 | Phagophore, autophagosome formation | cKO in germ cells | Subfertile | Inhibit spermatozoa flagella biogenesis and cytoplasm removal | Spermatozoa flagella biogenesis and cytoplasm removal during spermiogenesis | Shang et al. (2016) |
| cKO in germ cells | Infertile | A defect in acrosome biogenesis | Acrosome biogenesis | Wang et al. (2014) | ||
| cKO in Sertoli cells | Subfertile | Disrupt cytoskeleton structures and ectoplasmic specialization assembly | Ectoplasmic specialization assembly | Liu et al. (2016) | ||
| cKO in Leydig cells | Subfertile | Suppress testosterone synthesis, affect sexual behavior | Testosterone synthesis | Gao et al. (2018) | ||
| KD in rat primary Leydig cells | – | Suppress testosterone biosynthesis | Testosterone biosynthesis | Ma et al. (2018) | ||
| KD in rat primary Sertoli cells | – | Promote androgen-binding protein expression | Autophagic clearance of androgen-binding protein | Ma et al. (2015) | ||
| Beclin 1 | Phagophore formation | KD in TM3 mouse Leydig cells | – | Decrease testosterone production | Steroidogenesis | Li et al. (2011) |
| Tfeb | Lysosomal biogenesis | KD in GC-1 mouse spg cells | – | Not affect spermatogonial differentiation, but significantly reduce cell migration in GC-1 cells | Spermatogonial cell migration | Liu et al. (2018) |
| Atg9, Atg12, Atg14, Atg16L, LC3, Dram1, Lamp1, Lamp2, p62 | Phagophore, autophagosome, autolysosome formation | No KD/KO in testis | ||||
| Chaperon-mediated autophagy | ||||||
| Ppp1cc | HSC70 substrate | KO mice | Infertile | Impair spermiogenesis, meiosis, induce polyploid spermatids | Spermiogenesis | Varmuza et al. (1999) |
| KO mice | Infertile | Disruptions in spermatogenesis that begin during prepubertal testicular development, and continue into adulthood, often resulting in loss of germ cells to the point of Sertoli cell-only syndrome. | Chromatin condensation and acrosome development | Forgione et al. (2010) | ||
| cKO in germ cells | Infertile | Induce sperm counts reduction, misshapen sperm | Spermiogenesis | Sinha et al. (2013) | ||
| Lamp2a | Chaperon-mediated autophagy | No KD/KO in testis | ||||
| Mitophagy | ||||||
| Atg32, Nix | Mitophagy | No KD/KO in testis | ||||
| mTORC1-autophagy pathway | ||||||
| Mtor | mTORC1/mTORC2 component | KD in rat primary Sertoli cells | – | Reduce androgen- binding protein expression | Autophagic clearance of androgen binding protein | Ma et al. (2015) |
| cKO in Sertoli cells | Infertile | Induce testicular atrophy, loss of Sertoli cell polarity, germ cell premature release/apoptosis, loss of pachytene spermatocytes and spermatids, sperm abnormalities | Sertoli cell polarity, germ cell development through the pachytene spermatocyte stage | Boyer et al. (2016) | ||
| cKO in germ cells | Infertile | Result in smaller testis and no sperm, impair spermatogonial proliferation | Spermatogonial proliferation and differentiation | Serra et al. (2017) | ||
| cKO in germ cells | Subfertile | Induce age-dependent perturbation of testicular development, diminished spermatogonial pool and germ cell population | Spermatogonial proliferation and differentiation | Cao et al. (2020) | ||
| Raptor | mTORC1 component | cKO in germ cells | Infertile | Block spermatogonia proliferated and differentiation, result in Sertoli cell-only testes by adulthood | SSCs pool maintenance | Serra et al. (2019) |
| cKO in germ cells | Infertile | Induce smaller testes and infertility | Meiotic arrest and sex chromosomes silence | Xiong et al. (2017) | ||
| cKO in germ cells | Infertile | Impair spermatogenesis and induce progressive loss of spermatogonia | SSCs proliferation | Wang et al. (2017) | ||
| cKO in Sertoli cells | Infertile | Cause severe tubular degeneration in the neonatal testis, azoospermia in adult mice with disruption of cytoskeletal organization | Sertoli cell cytoskeletal organization and polarity | Xiong et al. (2018) | ||
| Rictor | mTORC2 component | cKO in germ cells | Sterile | Impair spermatogonial differentiation potential, cell–cell junctions, BTB dynamics, and spermiogenesis | Spermatogonial differentiation and intercellular adhesion | Bai et al. (2018) |
| Akt1/2 | Upstream of mTOR | KD in rat primary Sertoli cells | – | Disrupt Sertoli cell tight junction barrier | BTB function | Mok et al. (2015) |
| Lama2 | Upstream of mTORC1 | KD in rat primary Sertoli cells | – | Perturb F-actin and MTs organization in Sertoli cells | Sertoli cell BTB dynamics | Gao et al. (2017) |
| Tsc1 | mTORC1 inhibitor | cKO in germ cells | Subfertile | Induce testicular developmental defects, partial spermatogenic arrest, excessive germ cell loss, sperm count reduction and subfertility; mTORC1 activation promotes spermatogonial differentiation at the expense of germline maintenance | Spermatogonial differentiation | Wang et al. (2016) |
| Bif1, Uvrag, Ambra1 | Downstream of mTORC1 | No KD/KO in testis | ||||
| ULK1 | ULK complex | KD in goat primary Sertoli cells | – | Decrease cell viability and expressions of goat Sertoli cell marker genes (ABP, AMH, FASL and GATA4) | Sertoli cell function, viability | Pang et al. (2019) |
| ULK2 | ULK complex | KD in swine Sertoli cells | – | Inhibit swine Sertoli cell autophagy | Sertoli cell function | Ran et al. (2018) |
| Atg13, Atg101, Fip200 | ULK complex | No KD/KO in testis | ||||
| Vps34, Vps15, Atg14L | VPS34 complex, autophagosome formation | No KD/KO in testis | ||||
ABP, sex hormone binding globulin; AMH, anti-Mullerian hormone; BTB, blood-testis barrier; cKO, conditional knockout; FASL, Fas ligand; GATA4, GATA binding protein 4; GC-1 cells, mouse spermatogonial cell lines; KD, knockdown; KO, knockout; MT, microtubule; mTOR, mammalian target of rapamycin; SCC, spermatogonia stem cell; TM3 cells, mouse Leydig cell lines; ULK1, Unc-51 like autophagy activating kinase.
In pre-testicular processes, the HPG axis is the neuroendocrine network that regulates sexual development and reproduction (Navarro and Tena-Sempere, 2011). In the hypothalamus, kisspeptin (a neuropeptide), following neurokinin B and dynorphin, signals directly to GnRH neurons to orchestrate pulsatile GnRH release (Skorupskaite et al., 2014). GnRH binds to a membrane receptor in the pituitary, which secretes LH and follicle-stimulating hormone (FSH) (Kaprara and Huhtaniemi, 2018). After release from the pituitary, LH and FSH interact with the LH/FSH receptor in testicular cells, respectively, to initiate and maintain spermatogenesis.
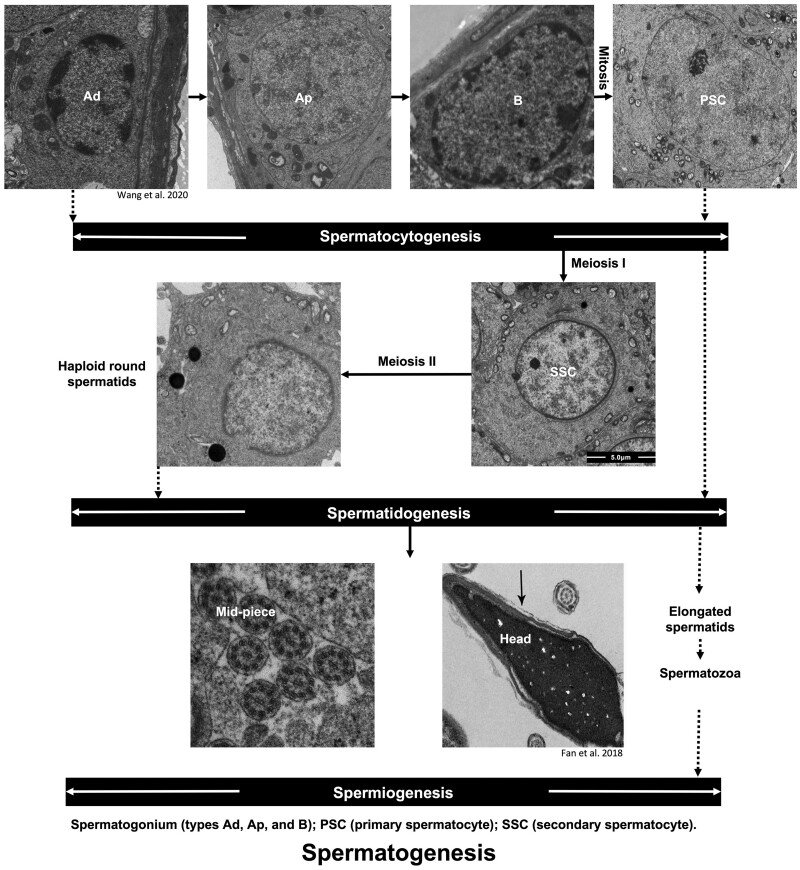
In testicular processes, the testis is responsible for producing spermatozoa through spermatogenesis. Testicular deficiency often leads to spermatogenic failure caused by conditions other than obstruction or HPG dysfunction. The entire process of spermatogenesis can be divided into three stages (Fig. 2), as follows: (i) during spermatocytogenesis, spermatogonia undergo mitosis to develop into primary spermatocytes (PSC); (ii) during spermatidogenesis, the PSC undergo meiosis I to form secondary spermatocytes, which divide into haploid round spermatids via meiosis II; and (iii) during spermiogenesis, round spermatids are differentiated into the elongated spermatids and then spermatozoa (Cheng and Mruk, 2012; Staub and Johnson, 2018). The last step in the testis is spermiation. Elongated spermatids are then released from the Sertoli cells into the seminiferous tubule lumen.
Figure 2.
The process of spermatogenesis. The entire process of spermatogenesis can be divided into three stages as follows: Spermatocytogenesis: spermatogonia undergo mitosis to develop into primary spermatocytes; Spermatidogenesis: the primary spermatocytes undergo meiosis I to form secondary spermatocytes, which divide into haploid round spermatids via meiosis II; and spermiogenesis: round spermatids are differentiated into the elongated spermatids and then spermatozoa.
In post-testicular processes, after spermatogenesis and spermiation, spermatozoa are transported from the testis to the epididymis for obtaining progressive motility and fertility (Sullivan and Mieusset, 2016). Subsequently, mature sperm are ejaculated into the female reproductive tract to undergo capacitation and activation (Jin and Yang, 2017). Eventually, the fusion of the sperm and the oocyte into a zygote, namely fertilization, represents the disappearance of sperm and appearance of a new individual, which indicates the final step for the fate of sperm.
Recently, increasing evidence has shown that autophagy functions in multiple mammalian organs or systems, including the embryo (Yoshii et al., 2016), placenta (Agrawal et al., 2015), liver (Komatsu et al., 2005), brain (Kaushik et al., 2011), heart (Nakai et al., 2007; Lu et al., 2016), skeletal muscle (Masiero et al., 2009), intestine (Cadwell et al., 2008), pancreas (Ebato et al., 2008), kidney (Hartleben et al., 2010), and the male (Gao et al., 2018) and female reproductive systems (Gawriluk et al., 2014) (Table II). Atgs have been implicated in numerous cellular events in the fate of sperm, such as GnRH secretion, LH, FSH and testosterone production, ectoplasmic specialization (ES) assembly, acrosome biogenesis and paternal mitochondria elimination (PME), shedding light on new therapeutic targets in the autophagy pathway for male subfertility or infertility (Nian et al., 2019). To date, there has been no effective and timely summary on the role of autophagy in the entire fate of sperm. Given that autophagy is known as a double-edged sword, the most fundamental question of whether autophagy plays a protective or a harmful role in the regulation of sperm remains unclear. In this narrative review, we will summarize the role of autophagy in the entire fate of sperm, including pre-testicular (HPG axis), testicular (spermatogenesis and spermiation) and post-testicular (sperm maturation and fertilization), and evaluate the clinical impact of autophagy on the sperm and male fertility. Our review will provide a better understanding of autophagy in the fate of sperm, laying the foundation for the further study of sperm, which can help us identify more therapeutic targets for male subfertility or infertility.
Table II.
The functions of autophagy in mammalian systems.
| Organs | Functions | Autophagy-related gene or protein changes | Selected references |
|---|---|---|---|
| The male reproductive system | Testosterone biosynthesis | Atg5/7↓ | Gao et al. (2018) and Ma et al. (2018) |
| Acrosome biogenesis | Atg7, Sirt1, Tbc1d20↓ | Liu et al. (2017), Sidjanin et al. (2016) and Wang et al. (2014) | |
| Spermiogenesis | Atg7↓ | Shang et al. (2016) | |
| Ectoplasmic specialization | Atg5/7↓ | Liu et al. (2016) | |
| Androgen binding protein metabolism | Atg7, mTOR↓ | Ma et al. (2015) | |
| The female reproductive system | Preventing excessive loss of oocytes in the neonatal ovaries | Beclin 1, Atg7↓ | Gawriluk et al. (2011) and Song et al. (2015) |
| Promoting progesterone synthesis | Beclin 1↓ | Gawriluk et al. (2014) | |
| Follicle atresia | LC3-II/LC3-I↑ | Choi et al. (2011) and Serke et al. (2009) | |
| Embryogenesis | Preimplantation development/protein synthesis | Atg5, Beclin 1↓ | Tsukamoto et al. (2008) and Yue et al. (2003) |
| Nervous system development | Ambra1↓ | Fimia et al. (2007) | |
| Preventing neonatal lethality/survival during neonatal starvation | Atg3/5/7/8/9a/16L1↓ | Komatsu et al. (2005), Kuma et al. (2004), Saitoh et al. (2008, 2009), Sou et al. (2008) and Yoshii et al. (2016) | |
| Placenta | Preventing placental infection and preterm labor | LC3-II/LC3-I, Atg4/7/16L1↑ and p62↓ | Agrawal et al. (2015) and Cao et al. (2016) |
| Liver | Constitutive turnover of cytoplasmic components | Atg7↓ | Komatsu et al. (2005) |
| Brain | Regulating food intake and energy balance | Atg7↓ | Kaushik et al. (2011) |
| Metabolic regulation/central control of feeding, energy and body weight balance | Atg5/7, LC3-II/LC3-I↓ | Meng and Cai (2011) | |
| Prevent neurodegenerative disease | Atg5/7, LC3-II/LC3-I, Atg12↓ | Hara et al. (2006) and Komatsu et al. (2006) | |
| Axonal homeostasis | Atg5/7↓ | Komatsu et al. (2007) and Nishiyama et al. (2007) | |
| Atg9, ULK1, Beclin 1, LC3-II↓ | Ko et al. (2020) | ||
| Heart | Maintaining cardiomyocyte size and global cardiac structure and function/adaption to hemodynamic stress | Atg5/7, LC3-II/LC3-I↓ and p62↑ | Nakai et al. (2007) |
| Angiogenesis | Beclin 1, Atg5, LC3-II/LC3-I ↓ and p62↑ | Lu et al. (2016) | |
| Skeletal Muscle | Preserving muscle mass and to maintain myofiber integrity | Atg7↓ | Masiero et al. (2009) |
| Intestine | Maintaining Paneth cells function | Atg5/16L1↓ | Cadwell et al. (2008) |
| Pancreas | Maintaining pancreatic β-cell volume and function | Atg7↓ | Ebato et al. (2008) and Jung et al. (2008) |
| Adaption to high-fat diet | Atg7↓ | Ebato et al. (2008) | |
| Kidney | Maintaining podocyte integrity | Atg5↓ | Hartleben et al. (2010) |
| Maintaining proximal tubule cell homeostasis and protecting against ischemic injury | Atg5↓ | Kimura et al. (2011) |
mTOR, mammalian target of rapamycin.
Pre-testicular processes
Role of autophagy in HPG axis
Atg5 deficient mice show neuronal dysfunction and hypogonadism (Yoshii et al., 2016), implying the key role of autophagy in HPG axis. Warburg Micro syndrome (WARBM) is a rare autosomal recessive genetic disease characterized by defective neurodevelopmental and ophthalmological phenotypes, such as microcephaly, microcornea, optic atrophy, lower limb spasticity and hypogonadotropic hypogonadism (Nian et al., 2019). RAB18, a member of small G protein, has been demonstrated to be a causative gene of WARBM in human. Rab18−/− mice neurons exhibit abnormal lysosomal transport and autophagosome marker LC3-II expression, suggesting the involvement of aberrant autophagy activities. Furthermore, RAB18 protein colocalizes with the lysosomal regulator RAB7 protein, which is upregulated in Rab18-deficient neurons, indicating a compensatory effect of Rab7 (Nian et al., 2019).
Rab3GAP2, another causative gene of WARBM, is reported to be a guanosine nucleotide exchange factor for activating Rab18. Rab3GAP2 mutation in Drosophila leads to the disorganization of autophagosomals/late endosomal compartments and lysosomal transport, finally perturbing autolysosome morphology. Meanwhile, Rab3GAP2, RAB18 and ATG6/BECN1 (subunits of Vps34 complexes) are co-located on autophagosomal and autolysosomal membranes. Consequently, the Rab3GAP-RAB18 module regulates autolysosomal maturation via its interaction with the Vps34 complexes. Taken together, Rab3GAP-RAB18-VPS34-mediated autolysosomal maturation may contribute to the development of WARBM, affecting gonadal function (Takats et al., 2021).
In addition to Rab3GAP and RAB18, mutations in GnRHR are also associated with hypogonadotropic hypogonadism. Houck et al. (2014) show that a missense mutation E90K in GnRHR induces misfolded proteins due to molecular chaperones Hsp70 and Hsp40 JB12. Interaction between the Hsp40 and Vps34 (autophagy initiation) complex permits the selective degradation of membrane proteins via an autophagy pathway.
Microcystin-leucine arginine (MC-LR), a kind of toxin produced by cyanobacterial, is reported to induce GnRH neurons apoptosis, resulting in a reduction of serum testosterone and spermatogenesis disruption in mice. An autophagy inhibitor (3-MA, PI3K inhibitor) aggravates MC-LR-induced apoptotic cell death in GT1-7 mouse hypothalamic GnRH neuronal cell line, implying a potential protective role of PI3K from apoptosis of GnRH neurons upon MC-LR exposure (Jin et al., 2021).
GnRH and kisspeptin/neurokinin B/dynorphin neurons in the hypothalamus regulate the age-related physiological decline in energy metabolism, hormone regulation and reproduction. Mammalian target of rapamycin (mTOR), autophagy and SIRT1 have been recognized as critical factors or pathways in hypothalamus-mediated aging progression (Kim and Choe, 2019).
ZNF216 is an identified causative gene for Gordon Holmes syndrome, characterized by ataxia, dementia and hypogonadotropic hypogonadism in humans. KD of RNF216 leads to the augmentation of BECN1 and migration defects in the GN11 immature GnRH neuronal cell line, which can be rescued by the autophagy inhibitors chloroquine (CQ) and 3-MA. Meanwhile, rapamycin (an autophagy activator) can suppress the GN11 cell migration. Therefore, RNF216 regulates GnRH neuron migration via inhibiting BECN1-mediated autophagy, suggesting a potential contribution of autophagy to hypogonadotropic hypogonadism (Li et al., 2019).
KiSS1 plays a key role in the activation of the HPG axis in regulating the onset of puberty and reproductive function. KiSS1-derived peptides, i.e. kisspeptins, signal through the G-protein coupled receptor GPR54 (Corno and Perego, 2019). In breast cancer cells, KiSS1 has been reported to down-regulate two Atgs (ATG5 and ATG7) and inhibit conversion of LC3-I to LC3-II, whereas KiSS1 KD leads to the down-regulation of p62 and up-regulation of Beclin 1. Therefore, KiSS1 inhibition is associated with promotion of autophagy (Kaverina et al., 2017).
Kisspeptin preserves mitochondrial function by inducing mitophagy and autophagy in the hippocampus of aging rat brains and in a human neuronal cell line via a series of signaling pathways, including Ca2+/CaM-dependent protein kinase β (CaMKKβ), AMP-activated protein kinase (AMPK) and Unc-51 like autophagy activating kinase (ULK1) (Mattam et al., 2021).
Environmental endocrine disruptors can disturb HPG axis by autophagy. Cadmium (Cd) decreases the serum concentrations of GnRH, FSH, LH and testosterone. Naringenin (Nar) suppresses MDA and H2O2 production and protects the testis from Cd-induced autophagy by downregulating P62 and LC3-II expression. Therefore, Nar protects the testis from Cd-induced toxicity (Wang et al., 2021). Paternal exposure to arsenic results in oxidative stress, autophagy and mitochondrial impairment in the HPG axis of pubertal male offspring. Specifically, autophagic cell death-related genes and proteins, such as Atg3, Atg5, Beclin 1, p62, Atg12, PI3K and mTOR, are disturbed in the HPG tissues of the pubertal male mice offspring (F1-generation) (Ommati et al., 2019). Similarly, in the mature male offspring, arsenic induces autophagic alterations and mitochondrial impairments in the HPG-sperm axis through AMPK/tuberous sclerosis complex (TSC) (tuberous sclerosis complex)/mTOR and LC3-related pathways (Ommati et al., 2020). Zearalenone, a non-steroidal estrogen mycotoxin, can induce mitochondrial dysfunctions through overproduction of reactive oxygen species (ROS) and aberrant autophagy pathways, finally disturbing the synthesis and secretion of mammalian sex steroid hormones (Zheng et al., 2019).
Autosomal dominant familial neurohypophyseal diabetes insipidus (adFNDI) is a progressive and inherited neurodegenerative disorder. The mutation of vasopressin (VP) from posterior pituitary nerve terminals can cause adFNDI, which is rescued by the inhibition of autophagy. Consequently, autophagy is a prosurvival mechanism in cells expressing an adFNDI mutant VP transgene (Castino et al., 2005a). Subsequently, Castino et al. (2005b) demonstrated that autophagy-mediated cell death is a two-hit process: the first hit of autophagy by cellular stress degrades the misfolded mutant protein, thus autophagy is pro-survival; whereas, a second insult triggers an autophagy-dependent apoptosis.
Testicular processes
Spermatogenesis is a dynamic process that allows the development of a diploid spermatogonium (SG) into haploid elongated spermatids in the seminiferous tubules (Wang and Proud, 2011). In this process, a well-defined progression of mitosis, meiosis and morphological transformations occurs in spermatogonia, spermatocytes and spermatids (Griswold, 2016). Normal spermatogenesis requires a balance of degradation and energy supply to maintain cellular metabolic homeostasis. Interestingly, autophagy is a unique catabolic pathway that participates in diverse physiological processes, especially cell residual bodies disposal, structural reconstruction, growth and development (Dikic and Elazar, 2018). Increasing studies have revealed that autophagy is involved in multifarious environmental toxicant-induced injury of testicular cells, including Sertoli cells, Leydig cells, spermotogonium and primary and second spermatocytes (Table III). Deciphering the role of autophagy in spermatogenesis will provide insights into the treatment of male infertility. Herein, we will clarify the role of autophagy in spermatogenesis (spermatocytogenesis, spermatidogenesis and spermiogenesis) (Roosen-Runge, 1962; Hess and Renato de Franca, 2008) and spermiation.
Table III.
Autophagy in environmental toxicants-induced mammalian testicular cell injury.
| Toxin (environmental source) | Affected testicular cell | Autophagy- related gene or protein changes | Resulted in testicular pathology | In human/animal/ cell model | Reference |
|---|---|---|---|---|---|
| DEHP | Leydig cells | LC3-II, Atg5, Beclin 1↑ | Decrease serum testosterone, induce oxidative stress and cell apoptosis | Kunming mice | Sun et al. (2018) |
| GC-1 cells | LC3-II, Beclin 1, Atg5, LC3-II/ LC3-I↑ | Induce oxidative stress, increase autophagic vacuoles number | Mouse GC-1 spg cell line | Gan et al. (2020) | |
| Bisphenol A and nonylphenol | Leydig cells and sperm | Beclin 1, Atg5/12, LC3↑ | Induce spermatogenic epithelium atrophy, germ cell loss, changes of hormones in serum and oxidative stress | Prepubertal Sprague Dawley rats | Su et al. (2018) |
| Copper | GC-1 cells | Atg3, Atg5, p62, LC3-II/LC3-I, Beclin 1, Atg5, p62↑ | Alter cell viability and morphology, induce oxidative stress-mediated mitochondrial dysfunction | Mouse GC-1 spg cell line | Kang et al. (2019) |
| Aflatoxin B1 | Leydig cells and sperm |
LC3, Beclin 1, Atg5, p62↑ p-mTOR/mTOR↓ |
Reduce serum testosterone level, impair sperm, induce the atrophic seminiferous tubules, vacuole-like changes of spermatogenic epithelium, and oxidative stress | Kunming mice | Huang et al. (2019) |
| Leydig cells | LC3, Beclin 1, p62↑ | lower serum T, LH and FSH levels, reduce Leydig cell number | Sprague Dawley rats | Chen et al. (2019b) | |
| Cisplatin | Leydig cells | LC3-II, Atg5↑ | Inhibit cell proliferation and vitality | MLTC-1 cell line | Yang et al. (2018) |
| TOCP | Leydig TM3 cells | LC3-II/LC3-I, Atg5, Beclin 1 ↑ | Inhibit cell viability and testosterone output, increase autophagic vacuoles and oxidative stress | Mouse Leydig TM3 cell line | Liu et al. (2016) |
| SSCs | LC3-II/LC3-I, Atg5, Beclin 1 ↑ | Inhibit viability and proliferation of rat SSCs | Primary SSCs of rats | Liu et al. (2015) | |
| Fluoride | Sertoli cells |
Beclin 1, p62↑ LC3, Atg5↓ |
No significant morphological alterations | Primary Sertoli cells of mice | Feng et al. (2019) |
| Leydig cells | LC3, Beclin 1, Atg5↑ | Increase autophagosomes number | Kunming mice | Zhang et al. (2017) | |
| 4-Nonylphenol | Sertoli cells |
Beclin 1, Atg3/5/7/12↑ LC3-II/LC3-I↑ |
Stimulate the formation of autophagosomes | Sprague Dawley rats | Duan et al. (2017) |
| SCOTP | SSCs | LC3-II/LC3-I, Atg5, Beclin 1↑ | Decrease cell viability and increase autophagic vacuoles | Primary SSCs of rats | Xu et al. (2016) |
| Arsenic | Leydig tumor cells | LC3, Atg7, Beclin 1, Vps34↑ | Impair lysosomes function and induce accumulation of autophagosomes | MLTC-1 | Liang et al. (2020) |
| Zinc oxide nanoparticles | Leydig cells | LC3-II, Atg5, Beclin 1↑ | Disrupt seminiferous epithelium, decrease sperm density and serum testosterone levels | Kunming mice and TM3 cells | Shen et al. (2019) |
| Methyl mercury | Sperm |
LC3-II, Beclin 1↑ p-mTOR/mTOR↓ |
Reduce sperm count and motility, impair the seminiferous tubule, induce apoptosis and oxidative stress | Sprague Dawley rats | Chen et al. (2019a) |
| Nicotine | Leydig cells | Beclin 1, LC3B↑ | Induce serum testosterone reduction and more autophagosomes | C57BL/6 J mice and TM3 cells | Zhao et al. (2018) |
| Cadmium | SSCs | Beclin 1, LC3B↑ | Increase testicular organ coefficients, seminiferous tubular atrophy, SSCs falling off the inner lining, reduce germ cell layers of disorderly arrangements | Wistar rats | Wang et al. (2017) |
| Spermotogonium, Sertoli/ Leydig cells, primary/second spermatocytes, GC-1/GC-2/TM3/TM4 cells |
Beclin 1, LC3-II/LC3-I↑ p-mTOR/mTOR (in vivo)↓ |
Reduce sperm count and motility, impair the seminiferous tubule, induce autophagy, apoptosis and oxidative stress | Sprague Dawley rats, GC-1/GC-2/TM3/TM4 cells | Wang et al. (2020) | |
| Acrolein | Leydig cells | Beclin 1↑ | Suppress proliferation and viability of Leydig cells, decrease testosterone, stimulate autophagy | Primary Leydig cells of mice | Gu et al. (2017) |
DEHP, di-2-ethyl hexyl phthalate; GC-1 cells, mouse spermatogonial cell lines; GC-2 cells, mouse spermatocyte cell lines; MLTC-1, mouse Leydig tumor cell lines; mTOR, mammalian target of rapamycin; SCOTP, saligenin cyclic-O-tolyl phosphate; SSCs, spermatogonial stem cells; TM3 cells, mouse Leydig cell lines; TM4 cells, mouse Sertoli cell lines; TOCP, tri-ortho-cresyl phosphate.
Role of autophagy in spermatocytogenesis
Spermatocytogenesis is the first stage of spermatogenesis. In this process, a diploid spermatogonial stem cell (SSC) undergoes mitosis to renew the stem cell pool or differentiate into two diploid PSC (Staub and Johnson, 2018). In non-primates, spermatogonia are composed of A-single (As), A-paired (Apr) and A-aligned (Aal), and Type B spermatogonia (Lie et al., 2009). In primates, this compartment consists of three subtypes (Fig. 2): (i) Type A dark (Ad), namely SSCs, renew themselves or generate differentiating Type A pale (Ap) spermatogonia; (ii) Type A pale (Ap) undergo mitosis to acquire Type B spermatogonia; and (iii) Type B spermatogonia produce PSC (Clermont, 1966; Di Persio et al., 2017). In terms of the causal relationship of Ap and Ad spermatogonia, other seemingly contradictory views are also proposed, such as that Ap spermatogonia are the SSCs, or that both of Ap and Ad cells are the SSCs simultaneously (Fouquet and Dadoune, 1986; Hermann et al., 2009). Regardless of the debate on Ap and Ad, what can be confirmed is that Type B spermatogonia will differentiate into PSC via mitosis. In this process, autophagy delicately exerts its bilateral effects.
Mammalian target of rapamycin
mTOR, a serine/threonine kinase, is a major regulator of cell growth, survival, metabolism and immunity (Saxton and Sabatini, 2017). As a core component, mTOR forms two distinct signaling complexes, mTOR Complex 1 (mTORC1) and mTORC2 by binding specific proteins Raptor and Rictor, respectively (Kim et al., 2002; Sarbassov et al., 2004; Jacinto et al., 2006). Differences in Raptor and Rictor sensitivity to rapamycin determine the differences in mTORC1 and mTORC2 function (Jacinto et al., 2004; Sarbassov et al., 2006). mTORC2 can phosphorylate Akt at Ser473 (Sarbassov et al., 2005) and regulate cell survival, metabolism and cytoskeletal organization (Cybulski and Hall, 2009) via AGC family kinases (PKA, PKG and PKC) (Cybulski et al., 2009). mTORC1 mainly regulates cell growth, proliferation, apoptosis, energy metabolism and autophagy (Kim et al., 2002; Yang et al., 2013). Noticeably, mTORC1 is the main gateway to autophagy (Rabanal-Ruiz et al., 2017) by modulating the localization of transcription factor EB, a major transcriptional regulator of lysosomal and autophagy genes (Settembre et al., 2013; Kim and Guan, 2015). Activation of mTORC1 by various nutrients and growth factors leads to the inhibition of autophagy through the phosphorylation of multiple autophagy-related proteins, such as ULK1, ATG13, AMBRA1 and ATG14L, which normally promote autophagy initiation and autophagosome nucleation (Kim and Guan, 2015).
Increasing studies have shown that mTOR regulates sperm quality in human (Silva et al., 2015, 2019). Diverse environmental toxicants, such as cadmium, di-2-ethyl hexyl phthalate, fine particle matters, nonylphenol and silica nanoparticles, induce testicular injury and regulate autophagy via mTOR signaling in mice and rats (Huang et al., 2016; Li et al., 2017; Ren et al., 2019, 2020; Zhang et al., 2020b). In mammals, sperm count, motility and morphology present a significant positive correlation with the phosphorylated levels of p70S6 kinase (Silva et al., 2015; Xu et al., 2016). Rapamycin inhibits spermatogenesis by suppressing mTOR-p70S6 kinase to alter the autophagy status in male rats (Liu et al., 2017b). Herein, this section will review the role of mTORC1 in spermatogenesis to indicate the regulation of sperm fate by autophagy.
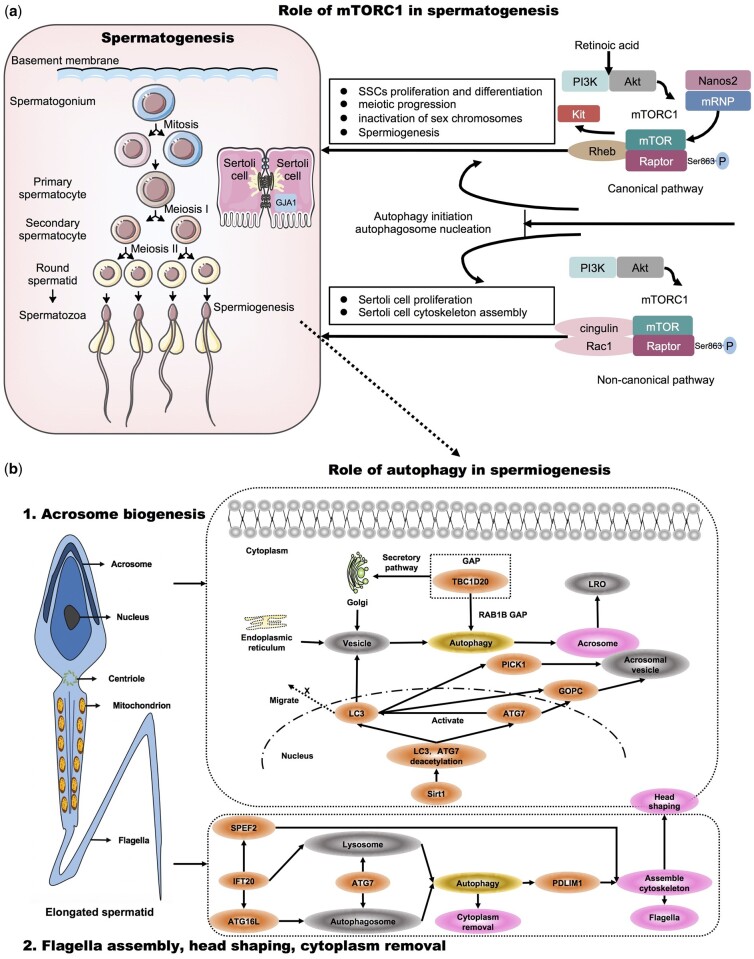
mTORC1, as a central modulator in stem cell homeostasis (Yilmaz et al., 2006; Chen et al., 2008; Gan and DePinho, 2009), is critical for maintenance of the pool of SSCs (Serra et al., 2019). Imbalances of SCC self-renewal and differentiation before meiosis can cause spermatogenesis disruption, even male infertility (Busada et al., 2015b). Although germ cell conditional knockout (cKO) mice for mTORC1-specific component Raptor were viable and healthy, spermatogonial proliferation was reduced in the neonatal testis, and blocked in the juvenile and adult testis, suggesting that mTORC1 is autonomously required for SCC proliferation and differentiation (Serra et al., 2017, 2019). Further, high phosphorylation of Raptor by raptor overexpression induced rapid growth of cultured SSCs, indicating that proliferation of SSCs requires phosphorylation of the mTORC1 component Raptor at Ser863 (Wang et al., 2017b). A recent study shows that conditional ablation of Raptor causes infertility due to meiotic arrest and impaired inactivation of sex chromosomes in the male germline (Xiong et al., 2017). Male germline cKO of Rheb, a critical component for mTORC1 activation, leads to defects of meiotic progression and spermiogenesis (Baker et al., 2014). Collectively, mTORC1 is crucial for SCCs proliferation and differentiation meiotic progression, silencing of sex chromosomes and spermiogenesis (Fig. 3).
Figure 3.
Role of autophagy in spermatogenesis. (a) Role of mTORC1 in spermatogenesis. mTORC1, the main gate to autophagy, is crucial for spermatogonia stem cell (SCCs) proliferation, differentiation, meiotic progression, inactivation of sex chromosomes and spermiogenesis. Firstly, proliferation of SSCs requires phosphorylation of the mTORC1 component Raptor at Ser863. Secondly, conditional ablation of Raptor causes infertility due to meiotic arrest and impaired inactivation of sex chromosomes in the male germline. Thirdly, Rheb, a critical component for mTORC1 activation, is required for meiotic progression and spermiogenesis. Consistently, retinoic acid (RA), as a requisite driver of spermatogonia differentiation and entrance into meiosis, regulates the PI3K/AKT/mTOR pathway to induce Kit translation during spermatogonial differentiation. In addition, Nanos2, an RNA-binding protein, interacts with mRNP to repress mTORC1 signaling by trapping mTOR, providing a post-transcriptional buffering system for SSCs homeostasis. Apart from germ cells, mTOR also plays a pivotal role in Sertoli cells. The regulation of Sertoli cell proliferation by FSH requires the PI3K/AKT/mTORC1 pathway. Independent of the canonical Rheb/mTORC1 pathway, Raptor dominates Sertoli cell cytoskeletal organization and polarity by affecting cingulin expression and Rac1 activity. Consistently, mTOR regulates gap junction alpha-1 (GJA1) distribution in Sertoli cells and is essential for progression through the pachytene spermatocyte stage. Finally, mTORC1 regulates spermatogenesis by inhibiting autophagy initiation and autophagosome nucleation. (b) Role of autophagy in spermiogenesis. In terms of the role of autophagy in spermiogenesis, autophagy-related genes participate in acrosome biogenesis, flagella assembly, head shaping and the removal of cytoplasm from elongating spermatids. 1. Acrosome biogenesis. TBC1D20 facilitates autophagy flux by its RAB1B GAP function, and regulates the formation of acrosome, which is a LRO. Sirt1 regulates spermiogenesis by stimulating autophagy. I. The depletion of Sirt1 disrupts LC3 and ATG7 deacetylation, provoking the redistribution of LC3 from the nucleus to the cytoplasm. II. Golgi-derived vesicles fail to recruit LC3. And III. nucleus-associated acrosomal vesicles are unable to recruit GOPC and PICK1. ATG7 not only partially targets GOPC to control acrosome biogenesis, but also motivates LC3, which initiates autophagy. 2. Flagella assembly, head shaping, cytoplasm removal. ATG7 is required for spermatozoa flagella biogenesis and cytoplasm removal during spermiogenesis. IFT20, as a Golgi transport protein, contributes to the formation of autophagosome by delivering ATG16L, and lysosome biogenesis by regulating the post-Golgi transport of acid hydrolases. SPEF2 is elementary for microtubule-mediated transport in axonemal CP assembly and sperm head shaping. IFT20 interacts with SPEF2 to regulate the flagella. The autophagy-lysosome pathway regulates spermatid differentiation by degrading PDLIM1 to facilitate cytoskeleton organization. LRO, lysosome-related organelle; GOPC, Golgi-associated PDZ- and coiled-coil motif-containing protein; PDLIM1, PDZ and LIM domain 1; SPEF2, sperm flagellar 2.
When it comes to the upstream and downstream signaling of mTORC1 in the spermatogenesis, Zhou et al. (2015) firstly confirm that Nanos2, an RNA-binding protein, interacts with messenger ribonucleoprotein (mRNP) to repress mTORC1 signaling by trapping mTOR, providing a post-transcriptional buffering system for SSCs homeostasis. Meanwhile, GILZ is indispensable for the modulation of mTORC1 in SSCs (La et al., 2018). Lin28a promotes SSCs proliferation through regulating mTOR and PI3K/AKT in dairy goats (Ma et al., 2016). Retinoic acid (RA), as a requisite driver of spermatogonial differentiation and meiosis, regulates the PI3K/AKT/mTOR pathway to induce Kit translation during spermatogonial differentiation in mice (Busada et al., 2015a).
Apart from the role in germ cells, mTOR also plays a pivotal role in Sertoli cells. The regulation of Sertoli cell proliferation by FSH requires the PI3K/AKT/mTORC1 pathway (Riera et al., 2012). Loss of the mTORC1 component Raptor in Sertoli cells leads to severe tubular degeneration in the neonatal testis, azoospermia and cytoskeletal organization disruption in adult mice (Xiong et al., 2018). Independent of the canonical Rheb/mTORC1 pathway, Raptor dominates Sertoli cell cytoskeletal organization and polarity by affecting cingulin expression and Rac1 activity (Xiong et al., 2018). Sertoli cell cKO mice for Mtor exhibit similar phenotypes, but a further study demonstrates that mTOR regulates gap junction alpha-1 (GJA1) distribution in Sertoli cells and is essential for progression through the pachytene spermatocyte stage (Boyer et al., 2016). Given that the mTORC2 component Rictor also controls spermatogonial differentiation and intercellular adhesion (Bai et al., 2018), the functions of mTORC1 and mTORC2 overlap partially. mTORC1 and mTORC2 may synergistically orchestrate blood–testis barrier (BTB) dynamics by intercellular adhesion and cytoskeleton. To check this assumption, much work is needed to investigate whether mTOR complexes exert their effects on the F-actin via drebrin E, paladin, formins, filamins, Eps8, the Arp2/3 complex or others (Mok et al., 2013).
Our previous research demonstrated that the PI3K inhibitor 3-MA rescues apoptosis by partially aggravating the reduction of the autophagy flux in cadmium-treated mouse spermatogonia, while in cadmium-treated mouse spermatocyte cells, 3-MA rescued apoptosis by inhibiting autophagy (Wang et al., 2020). The results imply that autophagy exerts different effects on spermatogonial cells and spermatocyte cells in response to external stimuli. Actually, in PSC, autophagy acts as a protective mechanism for mitochondrial abnormalities induced by Trl gene mutations (Dorogova et al., 2014). In spermatogonia, except from pro-survival function, autophagy implements a pro-death function.
Mancilla et al. (2015) demonstrated that deleting glutathione (GSH)-induced autophagy promotes cell survival by antagonizing apoptosis via the AMPK-independent pathway in the mouse spermatogonia cell line, GC-1 cells. Similarly, autophagy plays a cytoprotective role in nutrient-deprived GC-1 cells by the ANKRD49 and NF-κB pathways (Wang et al., 2015). Instead, Tri-ortho-cresyl phosphate (TOCP)-mediated autophagy contributes to cell death. TOCP, as widespread plasticizers and flame retardants, have been known to induce testicular toxicity since 1987 (Somkuti et al., 1987). Recently, TOCP was further reported to decrease the viability of spermatogonia and motivate autophagy, which was confirmed by increases in LC3-II/LC3-I, ATG5 and BECN1 in mice and rats (Chen et al., 2012; Liu et al., 2015). Intriguingly, the cell cycle and apoptosis present no noticeable changes (Liu et al., 2015). According to the three updated criteria for identifying autophagic cell death as proposed by Shen and Codogno (2011), the first criterion is that apoptosis is not involved in the issue. Therefore, it is reasonable to suspect that autophagy contributes to autophagic cell death, which is constitutes a pro-death mechanism in TOCP-induced spermatogonia injury. Nonetheless, in terms of this study, more experimental evidence, such as the activation of an autophagy flux and rescue by an autophagy inhibitor, is required for verifying these speculations.
Role of autophagy in spermatidogenesis
During spermatidogenesis, each diploid PSC develops into four haploid round spermatids through meiosis, which occupies an absolute central position in the process. One PSC undergoes one round of DNA replication and cell division (Meiosis I) to produce two haploid secondary spermatocytes, which subsequently proceed through the second cell division (Meiosis II) to produce four equal haploid round spermatids (Wang et al., 2017a). Meanwhile, a series of distinctive cellular events occur, including programmed DNA double-strand break formation, homologous recombination, crossover formation and resolution (Sung and Klein, 2006).
The study on the role of autophagy in spermatidogenesis originates in 1986. Chemes (1986) proved that Sertoli cells phagocytized and digested meiotic spermatocyte residual bodies by autophagy in a testosterone-independent manner. Recent research showed that autophagy and apoptosis are synchronously provoked in the heat-treated mouse spermatocyte cell line, GC-2 cells. Dramatically, Atg7-mediated downregulation of autophagy reduced apoptosis in heat-induced GC-2 cells, indicating that autophagy and apoptosis act as partners to promote cell death (Zhang et al., 2012).
Apart from being pro-death, autophagy seems to undertake a dual mission in regulating chromatoid bodies (CBs) during spermatidogenesis. CBs are a typical cytoplasmic features of haploid round sperm cells, consisting of RNA and RNA-binding proteins, and are unique ribonucleoprotein (RNP) granules (Meikar et al., 2011). Despite appearing in late pachytene spermatocytes, the CB-like granules are immediately condensed into a single granule after meiosis and maintain their character throughout the differentiation of round spermatids (Kotaja and Sassone-Corsi, 2007). Due to the accumulation of plentiful PIWI-interacting RNA (piRNA) in the CBs of round spermatids, the CB is deemed to be responsible for piRNA-targeted RNA regulation (Gomes Fernandes et al., 2018). Remarkably, piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis (Goh et al., 2015). Therefore, it is rational to suspect that CBs govern meiotic transcripts during spermatidogenesis.
Interestingly, Da Ros et al. (2017) discovered that the LC3B-interacting protein FYCO1, a novel CB component, initiates the intracellular transport of autophagic vesicles, which regulates the integrity of RNP granules in haploid male germ cells. Furthermore, both the agonists and antagonists of autophagy aggravate the cellular defects (fragmented CB) in the Fyco1 cKO germ cells, manifesting that autophagy devotes itself to two distinct events: the clearance of CB materials and the maintenance of CB homeostasis, synchronously (Da Ros et al., 2017). Consistent with the perception, autophagy can protect genomic stability by degrading retrotransposon RNA (Guo et al., 2014), and PIWI proteins/piRNA in the CB are the targets of degradation after autophagy activation (Siomi et al., 2011; Da Ros et al., 2017). Taken together, autophagy plays three roles during spermatidogenesis: (i) pro-death in spermatocyte, (ii) clearance of CB materials in round spermatids and (iii) maintenance of genomic stability in CBs by degrading PIWI proteins/piRNA in round spermatids.
The regulation of spermatidogenesis by autophagy was confirmed in Ppp1cc and Raptor KO mice, respectively. Ppp1cc2, as a substrate of the CMA regulator Hsc70 (Bonam et al., 2019; Itoh et al., 2019), was required for chromatin condensation and acrosome development (Forgione et al., 2010). Ppp1cc KO mice exhibited meiosis arrest during spermatidogenesis and spermiogenesis, presenting polyploid spermatids (Varmuza et al., 1999). Similarly, conditional ablation of mTORC1 component Raptor in the male germline caused meiotic arrest (Xiong et al., 2017). Therefore, the cross-talk between meiosis and autophagy-related proteins needs further exploration during spermatidogenesis.
Role of autophagy in spermiogenesis
Spermiogenesis refers to the final stage of spermatogenesis when round spermatids become elongated and then develop into spermatozoa through drastic morphological changes. Spermiogenesis involves the reshaping of the nucleus, rearrangement of mitochondria and development of flagellum and acrosome (Tanaka and Baba, 2005). These orchestrated physiological processes require a cellular homeostasis between degradation and recycling of cytoplasmic components. Recently, growing evidence indicates that autophagy, as a unique catabolic pathway (Ktistakis and Tooze, 2016), is involved in spermiogenesis. Autophagy-related proteins (LC3, ATG5, ATG16, BECN1, p62, mTOR, AMPKα 1/2 and PINK1) and their upstream regulators are functionally active in human spermatozoa, suggesting that autophagy may regulate sperm motility (Aparicio et al., 2016). Specifically, LC3 and ATG7 are increased dramatically from the round to elongated spermatids (Yang et al., 2017). Besides, autophagosomes in spermatozoa originate from the bilayer separation membrane of the chrysanthemum flower center, which is converted from endoplasmic reticulum (Yang et al., 2017), implying the regulation of spermiogenesis by autophagy.
The acrosome is also a unique lysosome-related membranous organelle (MO) in the anterior part of the sperm nucleus (Berruti and Paiardi, 2011). The acrosome carries hydrolytic enzymes to facilitate sperm penetrating the zona pellucida (Jin et al., 2011; Ozturk et al., 2017). For the first time, Hartree proposed the correlation between the acrosome and the lysosome in 1975 (Hartree, 1975). Controversially, Martínez-Menárguez et al. argued in 1996 that acrosomes were independent of endosomes or lysosomes due to the absence of late endosomal marker cation-dependent and non-dependent mannose 6-phosphate receptor in acrosomal vesicles and preantral vesicles (Martinez-Menarguez et al., 1996). Ten years later, both Berruti et al. (2010) and Hu et al. (2007) demonstrated that the acrosome was indeed a lysosome-related organelle (LRO). Furthermore, acrosome biogenesis, as a hinge of spermiogenesis (Kang-Decker et al., 2001), was linked closely with autophagy.
Generally, acrosome biogenesis is divided into four major phases: Golgi, cap, acrosome and maturation phases (Khawar et al., 2019). The first phase corresponds to the perspective that acrosome is derived from Golgi (Berruti and Paiardi, 2011). Research verifies that Golgi can be regulated by TBC1D20 (Haas et al., 2007), and the disruption of TBC1D20 results in testicular abnormalities in mice (Park et al., 2014), as well as in humans (Liegel et al., 2013). Thus, TBC1D20 may mediate the testicular function via regulating Golgi. TBC1D20, as a member of GAPs, interacts with GTP and subsequently, the ‘active’ GTP-bound RAB-GTPase is returned to the ‘inactive’ GDP-bound state (Frasa et al., 2012). Strikingly, Sidjanin et al. (2016) showed that TBC1D20 facilitates autophagy flux by its RAB1B GAP function, and regulates the formation of acrosome in mice, suggesting that TBC1D20 may regulate acrosome biogenesis via autophagy.
In parallel, Sirt1 regulates acrosome biogenesis by modulating autophagy flux during spermiogenesis in mice. Liu et al. found that the depletion of Sirt1 undermined spermiogenesis by stimulating autophagy in mice spermatids, including three successional cellular events: (i) the depletion of Sirt1 disrupted LC3 and ATG7 deacetylation, leading to the redistribution of LC3 from the nucleus to the cytoplasm; (ii) Golgi-derived vesicles failed to recruit LC3; and (iii) nucleus-associated acrosomal vesicles were unable to recruit Golgi-associated PDZ- and coiled-coil motif-containing protein (GOPC) and PICK1 (Liu et al., 2017a). In addition, for this comprehensive research, further studies proved that ATG7 not only partially targeted GOPC to control acrosome biogenesis (Wang et al., 2014), but is also essential for activation of LC3, which initiated autophagy (Tanida et al., 2012). Even, ATG7 was required for spermatozoa flagella biogenesis and cytoplasm removal during mice spermiogenesis (Shang et al., 2016). In terms of the mechanism, research elucidated that the autophagy-lysosome pathway regulates spermatid differentiation by degrading negative cytoskeleton regulator PDZ and LIM domain 1 (PDLIM1) to facilitate cytoskeleton organization (Shang et al., 2016). Coincident with this assertion, in moss, ATG5- and ATG7-mediated autophagy promote the flagellated motile sperm differentiation and cytoplasmic elimination (Sanchez-Vera et al., 2017). Therefore, whether in mice or moss, autophagy is indispensable for spermatid differentiation, especially in acrosome biogenesis and flagella biogenesis.
When it comes to flagella biogenesis, it is important to mention intraflagellar transport 20 (IFT20). During mouse spermiogenesis, IFT20, as a Golgi transport protein, interacts with the sperm flagellar 2 (SPEF2) to regulate the development of the sperm flagella (Lehti et al., 2017). SPEF2 is critical for microtubule-mediated transport in axonemal CP assembly and sperm head reshaping in mouse (Lehti et al., 2017). Consistently, in humans, homozygous mutations in SPEF2 lead to multiple morphological abnormalities of the sperm flagella and male infertility (Liu et al., 2020). These solid data support that IFT20 and SPEF2 are required for sperm tail formation and head reshaping during spermiogenesis. Although LC3 and ubiquitin are well-balanced in the testis of the Ift20 mutant mice (Zhang et al., 2016), it is not enough to deny the correlation between IFT20 and autophagy because of the scarcity of autophagy flow evaluation in this limited study. Remarkably, IFT20 contributes to autophagosome formation by delivering ATG16L (Pampliega et al., 2013) and to lysosome biogenesis by regulating the post-Golgi transport of acid hydrolases (Finetti et al., 2020).
Hence, autophagy participates in acrosome biogenesis, flagella assembly, shaping of the head and the removal of elongating spermatid cytoplasm during spermiogenesis (Fig. 3).
Role of autophagy in spermiation
Spermiation is the release of elongated spermatids from Sertoli cells into the seminiferous tubule lumen prior to the epididymis (Cheng and Mruk, 2010). This process is guided by a testis-specific, actin-based anchoring junction, apical ES (aES) in the Sertoli cell-spermatid interface (Mruk et al., 2008). Apart for spermatids movement and release, aES also contributes to shaping the spermatid head (Toyama et al., 2003; Mruk and Cheng, 2004). To achieve this goal, aES needs to undergo tightly and timely managed restructuring (Berruti and Paiardi, 2014).
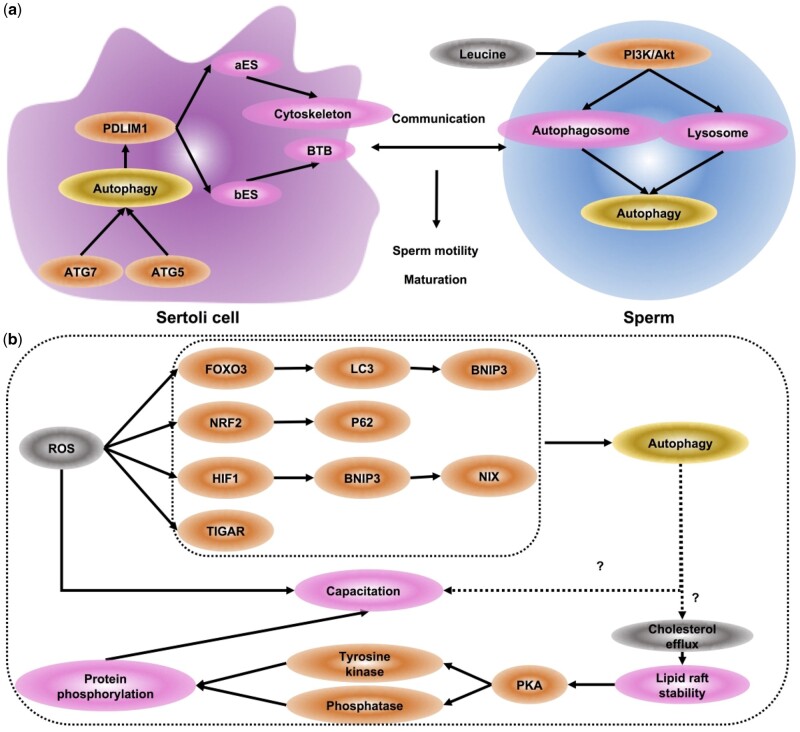
Currently, autophagy has been demonstrated to be required for ES assembly. ES encompasses two parts: (i) aES, at the Sertoli cell-spermatids interface and (ii) basal ES (bES), at the Sertoli–Sertoli cell interface (Cheng and Mruk, 2010, 2012). Sertoli cell-specific KO of Atg5 or Atg7 results in spermatozoa with malformed heads and low motility, affecting the fertility of male mice (Liu et al., 2016). Meanwhile, defective autophagy in Sertoli cells perturbs the degradation of PDLIM1, which is vital for cytoskeleton assembly (Liu et al., 2016). Consequently, through PDLIM1, autophagy not only affects bES to regulate BTB, but also mediates aES to govern spermatids movement and release during spermiation (Fig. 4a).
Figure 4.
Role of autophagy in sperm maturation. (a) Sertoli cells. Cell-specific knockout of Atg5 or Atg7 results in spermatozoa with malformed heads and low motility. Through PDLIM1, autophagy not only affects bES to regulate BTB, but also mediates aES to handle cytoskeleton, governing spermatids movement and release during spermiation. In sperm: autophagy can affect sperm motility by inhibiting the fusion of autophagosome and lysosomes through the PI3K/Akt-dependent pathway in leucine-treated zebrafish. (b) Capacitation. ROS regulates autophagy by transcriptional and post-transcriptional regulation, including ROS-FOXO3-LC3/BNIP3-autophagy, ROS-NRF2-P62-autophagy, ROS-HIF1-BNIP3/NIX-autophagy and ROS-TIGAR-autophagy. Cholesterol efflux alters lipid raft stability during capacitation. PKA can stimulate protein phosphorylation and capacitation or acrosomal exocytosis by activating tyrosine kinase and/or inhibiting protein phosphatase. Nonetheless, so far, there is no direct evidence that autophagy participates in the process of capacitation. It may be a good potential target for exploring the molecular mechanisms in capacitation disruption-induced male infertility. PDLIM1, PDZ and LIM domain 1; aES, apical ectoplasmic specialization; bES, basal ectoplasmic specialization; BTB, blood–testis barrier.
Post-testicular processes
Role of autophagy in sperm maturation
For fertilization, elongated spermatids have to undergo maturation. Sperm maturation contains three highly orchestrated processes: (i) obtaining motility and fertility in the epididymis, (ii) capacitation in the female reproductive tract and (iii) activation upon the approach of ovary and sperm.
Firstly, sperm flows into the rete testis in Sertoli cell-secreted fluid. With the action of their flagella and smooth muscle contraction, sperm then enter the caput epididymis. In the epididymis, sperm maturation involves both changing flagellum beat and acquiring the characteristics necessary for effective contact with the oocyte (Sullivan and Mieusset, 2016). The sperm cannot attain fertilization ability until they reach the proximal end of the epididymis. The motility and fertility capacities of sperm are completed in the cauda epididymis.
Secondly, capacitation, as the penultimate step of mammalian sperm maturation, occurs in the female reproductive tract. When semen is ejaculated into a female vagina, the sperm leaves the semen and enters the uterine cavity and fallopian tube via the cervical canal. Then female genital tract secrets alpha and beta-amylase, which degrades the acrosome surface glycoprotein. Meanwhile, the sperm launches a series of orchestrated biochemical reactions, including phosphorylation, alkalinization and hyperpolarization, dependent on CFTR and PKA, preparing for penetrating out layer of the oocyte (Puga Molina et al., 2017). Additionally, the increase in Ca2+ influx allows the sperm tail greater motility (Ren et al., 2001). Finally, capacitation renders sperms competent to fertilize an oocyte.
Notably, a dramatic change of ROS occurs in sperm during capacitation (Fig. 4b). Despite the detrimental effect of high concentrations of ROS, low concentrations of ROS are beneficial for sperm fertilization. It is pivotal to balance the homeostasis between ROS and autophagy. ROS regulates autophagy by transcriptional and post-transcriptional regulation, including ROS-FOXO3-LC3/BNIP3-autophagy, ROS-NRF2-P62-autophagy, ROS-HIF1-BNIP3/NIX-autophagy and ROS-TIGAR-autophagy (Li et al., 2015). In turn, autophagy can regulate ROS levels through CMA, the mitotic pathway and the P62 delivery pathway (Li et al., 2015). Also, autophagy can affect sperm motility by inhibiting the fusion of autophagosomes and lysosomes through the PI3K/AKT-dependent pathway in leucine-treated zebrafish (Zhang et al., 2017).
Given that cholesterol efflux alters lipid raft stability and distribution during capacitation of boar spermatozoa (Shadan et al., 2004), cholesterol efflux may have close relationship with capacitation. Recent studies have shown a strong link between the PKA pathway and sperm capacitation. PKA can stimulate protein phosphorylation and capacitation or acrosomal exocytosis by activating tyrosine kinase and/or inhibiting protein phosphatase (Puga Molina et al., 2017).
Nonetheless, so far, there is no direct evidence that autophagy participates in the process of capacitation (Fig. 4b). It may be a good potential target for exploring the molecular mechanism in capacitation dysfunction-induced male infertility.
Role of autophagy in fertilization
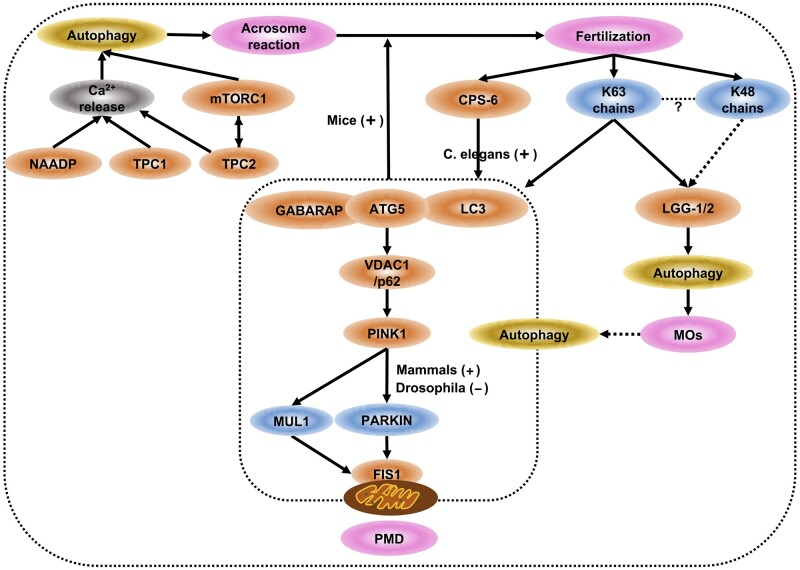
Fertilization is a fundamental process in reproduction (Ohto et al., 2016). In this process, the oocyte and sperm achieve mutual recognition, and fuse into a zygote, which then develops into a new individual, allowing for the continuity of a species (Okabe, 2013).
Classically, the acrosome reaction is one of the most critical steps in fertilization. This may be due to the fact that acrosome reaction allows the sperm to release acrosomal enzymes, dissolve the radiation corona and penetrate the zona pellucida (Tosti and Menezo, 2016). Recent research shows that NAADP and the two-pore channel (TPC) protein 1 participate in the acrosome reaction by Ca2+ release in mammalian spermatozoa (Arndt et al., 2014). The regulation of TPCs by autophagy is involved in multiple cells, such as neural cells (Pereira et al., 2017), cardiomyocytes (Garcia-Rua et al., 2016) and cancer cells (Sun and Yue, 2018). TPC2 mediates mTORC1 (Chang et al., 2020), which is the main gateway to autophagy (Rabanal-Ruiz et al., 2017). In turn, mTORC1 controls lysosomal Ca2+ release by TPC2 (Ogunbayo et al., 2018). Despite the interaction of TPC2 and mTORC1, there is no direct evidence that autophagy regulates the acrosome reaction through TPCs. This is waiting to be clarified.
During fertilization, most animals only inherit the mitochondrial genome from the maternal parent (Ankel-Simons and Cummins, 1996). The paternal mitochondria are eliminated in the embryo, although the sperm provides DNA, centrioles, some cytoplasm and organelles for the offspring (Kaneda et al., 1995). The underlying mechanism of PME during fertilization remains unclear.
Fertilization-triggered autophagy can degrade paternal mitochondria (Sato and Sato, 2011; Zhou et al., 2011), and post-fertilization autophagy of sperm organelles prevents paternal mitochondrial DNA (mtDNA) transmission in Caenorhabditis elegans (Al Rawi et al., 2011). Zhou et al. (2016) found that CPS-6 migrated from the parent mitochondrial intermembrane to the matrix after fertilization for the degradation of mtDNA. Loss of cps-6 disrupts autophagy, PME and embryogenesis, revealing that CPS-6 regulates PME by interacting with maternal autophagy and proteasome machinery upon fertilization in C. elegans (Zhou et al., 2016). Tsukamoto et al. discovered that autophagy-deficient sperm (sperm-specific KO ATG5) and oocytes without ATG5 in mice could undergo regular fertilization and fuse into a zygote, but failed to develop into four-cell and eight-cell stages. In contrast wild-type sperm and autophagy-deficient oocytes were available to develop normally into embryos, suggesting that fertilization-triggered autophagy was essential for the development of mammalian early embryos (Tsukamoto et al., 2008).
In mice embryos, this autophagic degradation process is dependent on the E3 ubiquitin ligases PARKIN and MUL1, mitochondrial outer membrane protein FIS1, autophagy adaptor p62 and PINK1 kinase (Rojansky et al., 2016). Owing to mitochondrial depolarization stabilizing PINK1, both PINK1 and PARKIN are recruited to damaged mitochondria, and latent PARKIN is activated for mitophagy in Parkinson’s disease (Kawajiri et al., 2010; Matsuda et al., 2010). PINK1/PARKIN-mediate mitophagy through voltage-dependent anion channel 1 and p62/SQSTM1 (Geisler et al., 2010). Although in Drosophila, autophagy and endocytosis regulate PME by a PARKIN-independent pathway (Politi et al., 2014), PINK1/PARKIN-mediated mitophagy may be essential for PME in mammals.
Another study demonstrated that fertilization induces ubiquitination and recruitment of LGG-1/LGG-2 (the two LC3 homologs in worms) around the flagellum mid-piece; after fertilization, paternal mitochondria and MOs are engulfed in autophagosomes and degraded during the first zygote divisions (Al Rawi et al., 2012). Furthermore, the depletion of lgg-1 led to the death of 95% animals at or before the L1 larval stage, implying that disruption of PME may undermine embryogenesis, although there were other zygotic defects due to the loss of LGG-1 (Levine and Elazar, 2011; Sato and Sato, 2011). Djeddi et al. (2015) verified that sperm-inherited organelle clearance in C. elegans relied on LC3-dependent autophagosomes. The above results suggest that both proteasomal and autophagic degradation participate in PME and MOs clearance upon fertilization in C. elegans.
In parallel, Hajjar et al. confirmed that sperm-derived mitochondria and MOs, which cluster together, are simultaneously decreased during fertilization in C. elegans. K48-linked ubiquitin chains (K48 chains), which degrade proteasomes, arise on MOs and vanish very quickly; meanwhile, K63-linked ubiquitin chains (K63 chains), which recruit autophagosomal markers (LGG-1/2) to MOs, emerge on MOs early and remain throughout the first several cell divisions (Hajjar et al., 2014). These findings suggest that K63 chain-mediated autophagy modulates PME and MOs clearance, and K48 chains only participate in MOs clearance. However, the interaction mechanism between K63 chains and K48 chains in PME or MOs clearance remains an enigma.
Song et al. (2016) showed that sperm mitophagy in pig and rhesus monkey relied on p62-dependent autophagy, valosin-containing protein-mediated dislocation and recruitment of ubiquitin to the 26S proteasome. They concluded that autophagy and the UPS contributed to sperm mitophagy after mammalian fertilization (Song et al., 2016).
Interestingly, Al Rawi et al. (2011) and Sato and Sato (2011) claim that ubiquitin was only related to MOs but not paternal mitochondria, despite the fact that autophagosomes indeed degrade both MOs and paternal mitochondria. In view of this, Levine and Elazar put forward two potential roles of paternal MOs by suggested that (i) MOs provided membranes for autophagosome formation and (ii) MOs launched autophagy in the process of PME (Levine and Elazar, 2011).
However, this theory was challenged by Luo et al. (2013). They proposed that autophagy was not involved in PME after fertilization by using two transgenic mice, separately labeling mitochondria and LC3, although some autophagy-related proteins, such as p62/SQSTM1 and LC3 indeed localized near the sperm mitochondria before the two-cell stage. Additionally, most motile sperm eliminated their mtDNA before fertilization (Luo and Sun, 2013). Thus, in mice, maternal inheritance of mtDNA may be a passive process as a result of pre-fertilization sperm mtDNA elimination and uneven mitochondrial distribution in embryos (Luo and Sun, 2013). Whether this is the case awaits further research.
To sum up, autophagy may regulate the acrosome reaction, PME and MOs clearance during fertilization (Fig. 5).
Figure 5.
Role of autophagy in fertilization. Autophagy regulates the acrosome reaction, PME and MOs clearance during fertilization. (a) Acrosome reaction. NAADP and the two-pore channel (TPC) protein 1 participate in the acrosome reaction by Ca2+ release in mammalian spermatozoa. TPC2 mediates mTORC1, which is the main gateway to autophagy; in turn, mTORC1 controls lysosomal Ca2+ release by TPC2. Autophagy may operate the acrosome reaction through TPCs. (b) PME and MOs clearance. Both autophagy and the ubiquitin-proteasome system contribute to PME and MOs after fertilization. In C. elegans, CPS-6 regulates PME, and LGG-1/LGG-2 (LC3 homologs) operate PME and MOs clearance by interacting with maternal autophagy and proteasome machinery upon fertilization; K63 chains-mediated autophagy may modulate PME and MOs clearance, and K48 chains may participate in MOs clearance. However, the crosstalk of K63 chains and K48 chains remains an enigma. In mice, fertilization-triggered autophagy regulates the development of mammalian early embryos by ATG5; this autophagic degradation process is dependent on the E3 ubiquitin ligases PARKIN and MUL1, mitochondrial outer membrane protein FIS1, autophagy adaptor p62 and PINK1 kinase. Ubiquitin is only related to MOs but not PME, despite the fact that autophagosomes degrade both of MOs and paternal mitochondria. Two potential roles of paternal MOs are that MOs provide membranes for autophagosome formation or MOs launch autophagy through the process of PME. Autophagy is not involved in PME after fertilization: in mice, maternal inheritance of mtDNA may be a passive process as a result of pre-fertilization sperm mtDNA elimination and uneven mitochondrial distribution in embryos. The actual situation awaits further research. PME, paternal mitochondrial elimination; MOs, membranous organelles; TPC, the two-pore channel; mtDNA, mitochondria DNA.
Clinical impact of autophagy on the fate of sperm
Dysfunctional autophagy contributes to many diseases, which determine the clinical impacts of autophagy. Currently, autophagy activators (such as rapamycin, perifosine, metformin) and autophagy inhibitors (such as bafilomycin A1, CQ, lucanthone, wortmannin) have been investigated for clinical and translational medicine (Table IV) and present good prospects in clinical application.
Table IV.
The clinical prospects of autophagy activators and inhibitors.
| Drugs | Diseases | Functions | Possible mechanism | In human/animal/cell model | Reference |
|---|---|---|---|---|---|
| Autophagy activators | |||||
| Rapamycin | Breast cancer | Inhibits proliferation of the endoplasmic reticulum-positive MCF-7 cell line | Rapidly stimulate mTOR non-specifically after medium replacement | MCF-7 breast cancer cell line | Chang et al. (2007) |
| Transplanted tumors | Inhibits tumor growth at any stage of development | – | – | Eng et al. (1984) | |
| Pancreatic cancer | Regulates cell growth and cyclin D1 expression | Constitutively actives FRAP-p70s6K pathway and inhibits cyclin D1 expression | MiaPaCa-2 and Panc-1 human pancreatic cancer cells and a pancreatic cancer tissue sample | Grewe et al. (1999) | |
| Vascular disease | Reduces vascular inflammation | Suppresses macrophage proliferation | Mice | Boada et al. (2020) | |
| Type 2 diabetes | Improves insulin resistance and hepatic steatosis | Enhances autophagy by the inhibition of mTOR pathway | T2DM rats | Zhou and Ye (2018) | |
| Myotrophic lateral sclerosis | Reduces neuronal loss and TDP43 inclusions; expands regulatory T lymphocytes with slow progression in ALS patients | Activates autophagy | Four human NB cell lines (AS, NGP, BE2, and KCNR); mice carrying xenograft NB tumors | Mandrioli et al. (2018) | |
| Facial angiofibromas | Appears effective and safe for treatment of TSC-related facial angiofibromas | – | TB3 cells | Koenig et al. (2018) | |
| Improves ovarian function and reproductive longevity | – | – | Hine (2017) | ||
| Perifosine | Neuroblastoma | increases apoptosis and inhibits neuroblastoma tumor cell growth in vitro and in vivo | Decreases AKT phosphorylation | Four human NB cell lines (AS, NGP, BE2, and KCNR); mice carrying xenograft NB tumors | Li et al. (2010) |
| Neuroblastoma | Attenuates brain-derived neurotrophic factor/TrkB-induced chemoresistance | Inhibits AKT | TB3 cells | Li et al. (2011) | |
| Metformin | Glioblastoma | Inhibits growth of human glioblastoma cells and enhances therapeutic response | Activates AMPK, Redd1 and inhibits mTOR pathway | Four human glioblastoma cell lines, U87 (ATCC HTB-14), LN18 (ATCC CRL-2610), U251 and SF767 | Sesen et al. (2015) |
| Metformin and Rapamycin | Prostate tumors | Inhibits progression of prostatic intraepithelial neoplasia lesions to adenocarcinomas in the ventral prostate | Down-regulates mTORC1 signaling | HiMyc mice | Saha et al. (2015) |
| Metformin | Reproductive health, gynecological cancer | Inhibits progression of prostatic intraepithelial neoplasia lesions to adenocarcinomas in the ventral prostate | Upstream activation of AMPK, resulting in inhibition of the mTOR pathway. | HiMyc mice | Saha et al. (2015) |
| Autophagy inhibitors | |||||
| Bafilomycin A1 | Pediatric B-cell acute lymphoblastic leukemia | Inhibits and kills pediatric B-cell acute lymphoblastic leukemia cells | Targets both autophagy and apoptosis by disassociating the Beclin 1–Vps34 complex | – | Yuan et al. (2015) |
| Microcephaly | Inhibits ZIKV entry and prevents the spread of the infection by interfering with viral maturation | Inhibits V-ATPase | – | Sabino et al. (2019) | |
| Tongue squamous cell carcinoma | Increases the sensitivity of tongue squamous cell carcinoma cells to cisplatin | Inhibition of the lysosomal uptake of platinum ions but not autophagy | – | Chu et al. (2018) | |
| Chloroquine | Breast cancer | Enhances the efficacy of tumor cell killing by combination with chemotherapeutic drugs and radiation | – | – | Maycotte et al. (2012) |
| Colon cancer | Enhances the chemotherapeutic activity of 5-fluorouracil in a colon cancer cell line via cell cycle alteration | Anti-cancer effect of 5-FU via cell cycle inhibition | Human colon cancer DLD-1 cells | Choi et al. (2012) | |
| 3-MA or chloroquine | Glioblastomas | Improves the efficacy of curcumin/temozolomide combination therapy | Increases apoptosis | C6, U251MG and U87MG cell lines; primary astrocytes | Zanotto-Filho et al. (2015) |
| Malignant gliomas | Enhances temozolomide cytotoxicity | Blocks autophagy and triggers endoplasmic reticulum stress, increasing the chemosensitivity of glioma cells to temozolomide | Subcutaneously implanted U87MG tumors from mice | Golden et al. (2014) | |
| Glioma | Potentiates temozolomide cytotoxicity | Inhibits mitochondrial autophagy | Tumor cells derived from a glioblastoma patient and human U87-MG glioblastoma cells | Hori et al. (2015) | |
| Hydroxy chloroquine | Advanced solid tumors and melanoma | Augments cell death in preclinical models | Blocks autophagy | – | Rangwala et al. (2014a,b) |
| Lucanthone | Breast cancer | Induces apoptosis via cathepsin D accumulation and enhances vorinostat-mediated cell death in breast cancer models. | Induces lysosomal membrane permeabilization | p53(+/+) and p53(−/−) HCT116 cells | Carew et al. (2011) |
| Wortmannin | Ovarian cancer | Enhances cisplatin-induced apoptosis | Activates PI3K/Akt signaling pathway | A2780 ovarian adenocarcinoma cell line and A2780cis | Zhao et al. (2014) |
3-MA, 3-methylade nine; AKT, protein kinase B; ALS, amyotrophic lateral sclerosis; AMPK, AMP-activated protein kinase; MCF-7 cells, human breast cancer cell lines; mTOR, mammalian target of rapamycin; NB tumor, neuroblastoma; T2DM rat, type 2 diabetes mellitus rat; TrkB, neurotrophic receptor tyrosine kinase 2; TSC, tuberous sclerosis complex; ZIKV, Zika virus.
It has been clearly shown that autophagy could be associated with infertility, in particular for patients with cryptorchidism (Yefimova et al., 2019). Cryptorchidism, i.e. undescended testis, is recognized as one of the strongest risk factors for infertility in adulthood (Gurney et al., 2017). Autophagy is increased in cryptorchid testis resulting in abnormal spermatozoa, thus autophagy maybe a potential target to improve sperm quality in cryptorchid men (Yefimova et al., 2019).
During pre-testicular processes, the dysregulation of HPG axis can lead to male subfertility of infertility. GnRH decreases cell proliferation by inhibiting the AKT/ERK1/2 pathway in pancreatic cancer (Suo et al., 2019). GnRH-II antagonist trptorelix-1 (Trp-1) induces autophagosome formation, AKT phosphorylation reduction and c-Jun NH (2) terminal kinase phosphorylation elevation in prostate cancer cells, indicating that GnRH-II antagonists-mediated autophagic degradation may contribute to the treatment of prostate cancer (Kim et al., 2009). Therefore, GnRH-mediated autophagy pathway may hold promise in the treatment of pancreatic or prostate cancer.
Pituitary tumors can disturb the HPG axis, influencing pre-testicular regulation. Dopamine agonists bromocriptine and cabergoline (CAB) have been utilized for the therapies of pituitary prolactinomas and other neuroendocrine tumors. Low-dose CAB is able to induce autophagy but fails to suppress cell growth. However, combination of chloroquine (CQ, autophagy inhibitor) with CAB facilitates the accumulation of p62/caspase-8/LC3-II, indicating blockage of autophagic cycles and involvement of apoptosis. Collectively, combined use of CAB and CQ may increase clinical effectiveness for the treatment of pituitary adenomas (PAs), making it an attractive option in pituitary tumor-induced male infertility (Lin et al., 2017).
Conversely, DEP domain-containing mechanistic target of rapamycin (mTOR)-interacting protein (DEPTOR), as a key regulator of mTOR, inhibits mTOR Complex 1 (mTORC1) and 2 (mTORC2) by decreasing E3 ubiquitin ligase βTrCP1. DEPTOR enhanced autophagic cell death to confer cells sensitivity to CAB in PA (Yao et al., 2019). In line with this, dopamine receptor D5 (DRD5) activation can inhibit tumor growth by autophagic cell death with mTOR pathway (Leng et al., 2017).
In pituitary prolactinoma, FOXP1-induced lncRNA CLRN1-AS1 represses cell proliferation, promotes apoptosis and inhibits autophagy by inactivation of Wnt/β-catenin signaling, thus acting as a tumor suppressor (Wang et al., 2019).
Low-dose (1.25 μM) tetrandrine (Tet) induces autophagy by down-regulation of MAPK/STAT3 in PA; higher-dose (5.0 μM) Tet induces caspase-dependent apoptosis. Meanwhile, autophagy inhibitors enhance Tet-induced caspase activity and apoptotic cell death, indicating that autophagy inhibition enhances the anti-PA effect of Tet (Lyu et al., 2021).
Schaaf–Yang syndrome (SHFYNG), is a neurodevelopmental disorder with neonatal hypotonia, feeding difficulties, hypogonadism, intellectual disability and sleep apnea, caused by pathogenic variants in the paternal copy of MAGEL2. The Magel2-null mouse model and fibroblast cell lines from individuals with SHFYNG exhibit the up-regulation of mTOR and the down-regulation of autophagy, which can be rescued by the mTOR inhibitor rapamycin. mTOR may be a potential target for the treatments of SHFYNG-induced hypogonadism (Crutcher et al., 2019).
Male late-onset hypogonadism (LOH), one of the most common diseases, affects male fertility. Testosterone replacement therapy is the main clinical treatment but has obvious side effects. It has been reported that methylpyrimidine-fused tricyclic diterpene analogs 29 could stimulate autophagy by the AMPK/mTOR pathway and furthermore increase testosterone levels and improve sperm qualities with few side effects in androgen deficiency-aging male rats. Therefore, methylpyrimidine-fused tricyclic diterpene analogs may be used as a new type of potential anti-LOH agent by regulating autophagy (Bai et al., 2019).
Non-obstructive azoospermia (NOA), from pre-testicular and testicular factors, is identified in ∼10% of infertile males. Over-expression of hsa_circ_0000116 is presented in patients with NOA. Furthermore, hsa_circ_0000116 may affect male fertility through a hsa_circ_0000116-miR-449-autophagy-related competing endogenous RNA network, suggesting its predictive value in testicular sperm retrieval (Lv et al., 2020).
Cordycepin (COR), an active constituent of the nutrient powerhouse Cordyceps militaris Linn., ameliorates age-related testicular dysfunction in rats, including sperm motility, progressiveness and average path/straight line velocity. Meanwhile, the expression of spermatogenesis-related protein, SIRT1, and autophagy-related mTORC1 are ameliorated by COR in aged rats, implying the potential therapeutic value of COR in male sexual dysfunction by regulating mTORC1 (KopalLi et al., 2019).
Aflatoxin B1 (AFB1), a potential endocrine disrupter, reduces serum testosterone (T), LH and FSH levels, and the expression of testosterone biosynthesis-related genes. AFB1 induces Leydig cells apoptosis by suppressing AMPK/mTOR-mediated autophagy flux pathway (Chen et al., 2019). KD of Beclin 1 decreases expression of StAR (a testosterone biosynthesis marker) and testosterone production in Leydig cells. Rapamycin, an autophagy activator, enhances steroidogenesis in primary Leydig cells from aged, but not young, rats (Li et al., 2011). Therefore, autophagic deficiency is related to steroidogenic declines in aged rat Leydig cells.
Excessive activation of autophagy is linked with testicular cell injuries induced by environmental endocrine disruptors. Co-exposure to fluoride and arsenic disrupts the intestinal flora balance and induces testicular autophagy by altering autophagic flux, increasing Beclin1 and LC3-II expression, and decreasing p62 expression in offspring rats (Liu et al., 2021). Perfluoroundecanoic acid (PFUnA), one of long-chain perfluoroalkyl carboxylic acids, downregulates Lhb and Fshb expression in the pituitary and serum FSH/LH/testosterone levels. Meantime, PFUnA reduces Leydig cell number and induces autophagy by suppressing the phosphorylation of mTOR, AKT1/2 and ERK1/2 in the testis of pubertal male rats (Yan et al., 2021).
Apart from the clinical impacts on pre-testicular and testicular diseases, autophagy pathway-related activators and inhibitors may be potential therapies for post-testicular diseases, especially in erectile dysfunction (ED). ED is a problem getting or keeping an erection for satisfactory sexual performance (Najari and Kashanian, 2016). ED can affect libido (sexual interest), orgasm or ejaculation, leading to accumulation of sperm in the epididymis, with a decline of sperm motility.
ED is common in patients with diabetes mellitus. Increasing evidence shows that diabetes mellitus-induced ED (DMED) involves autophagy. Rapamycin, an mTOR inhibitor, can lead to inhibition of AMPK/mTOR and PI3K/AKT/mTOR pathways by mTORC1 (raptor)/p70S6K but not the mTORC2-related pathway. Lin et al. (2018) found that rapamycin significantly ameliorated erectile function and nitric oxide/cGMP pathway repression in rats with DMED by inhibiting mTORC1 (raptor)/p70S6K to promote autophagy and inactivate apoptosis. In parallel, Ding et al. (2020) reported that Simvastatin alleviated DMED in rats by enhancing AMPK pathway-induced autophagy.
Similarly, glucagon-like peptide-1 (GLP-1) analog liraglutide is reported to improve erectile function by promoting autophagy with different pathways in rats with DMED. Liraglutide reduces oxidative stress and downregulates Ras homolog gene family (RhoA) and Rho-associated protein kinase (ROCK) 2. Therefore, liraglutide exerts protective effects on ED by regulating oxidative stress, autophagy and the RhoA/ROCK pathway, providing preclinical evidence for a potential treatment for DMED (Yuan et al., 2020).
Furthermore, transplantation of human urine-derived stem cells (USCs) ameliorates erectile function and cavernosal endothelial function by promoting autophagy of corpus cavernosal endothelial cells (CCECs) in rats with DMED. In vitro, advanced glycation end products (AGEs) were used to mimic the diabetic situation. AGEs-treated CCECs exhibited fewer LC3 puncta formation and lower LC3-II, BECN1 and PCNA expression but increased p62 and cleaved-caspase3 expression, which could be rescued by USCs. More importantly, the repaired erectile function could be abolished by the autophagy inhibitor 3-MA, demonstrating the future clinical perspectives of autophagy-related molecules in ED (Zhang et al., 2019).
miR-301a-3p-enriched exosome significantly recovers erectile function in rats and corpus cavernous smooth muscle cells (CCSMCs) by stimulating autophagy and inhibiting apoptosis. Interestingly, the overexpression of PTEN or TLR4 reverses the therapeutic effects of miR-301a-3p in CCSMCs, indicating that miR-301a-3p modulates PTEN/HIF-1α/TLR4 signaling, autophagy and apoptosis for the treatment of ED. However, the cross-talk of PTEN/HIF-1α/TLR4 signaling and autophagy remains to be further investigated in ED (Liang et al., 2021).
In contrast, icariside II (ICAII) and metformin (MET) can improve erectile function and down-regulate AGEs and receptor of AGEs (RAGE), but ICAII and MET up-regulate the PI3K/AKT/mTOR signaling to reduce the excessive mitochondrial autophagy of corpus cavernosum smooth muscle cells (CCSMCs) (Zhang et al., 2020a). These data suggest that both autophagy defects and autophagy overshoot can lead to ED. Autophagy may be a critical therapeutic target for DMED.
Conclusions, outstanding questions and future perspectives
Autophagy is a critical lysosomal pathway that maintains cell function and survival through the degradation of cellular components such as organelles and proteins (Czaja et al., 2013). In specific mammalian organ or system, autophagy plays different functions (Table II). Herein, we first present a comprehensive overview on the regulation of autophagy on the entire fate of the sperm, including spermatogenesis (consisting of spermatocytogenesis, spermatidogenesis and spermiogenesis), sperm maturation and fertilization. We also emphasize, in different sections, the specific issues that deserve attention by researchers in future studies.
During spermatocytogenesis, autophagy acts as a protective mechanism for mitochondrial abnormalities induced by Trl gene mutations in PSC; meanwhile, autophagy exerts its bilateral effects on spermatogonium: pro-death and pro-survival under stress. During spermatidogenesis, autophagy and apoptosis acts as partners to promote spermatocyte death; while in haploid round spermatids, autophagy devotes itself to two distinct events: the clearance of CB material and the maintenance of CB homeostasis synchronously. During spermiogenesis, autophagy participates in acrosome biogenesis, flagella assembly and head shaping, and the removal of cytoplasm from elongating spermatids.
After spermatogenesis, through PDLIM1, autophagy not only affects bES to regulate BTB, but also mediates aES to handle cytoskeleton, governing spermatid movement and release during spermiation. A dramatic change of ROS occurs in sperm during capacitation. Proverbially, ROS regulates autophagy by transcriptional and post-transcriptional regulation, including ROS-FOXO3-LC3/BNIP3-autophagy, ROS-NRF2-P62-autophagy, ROS-HIF1-BNIP3/NIX-autophagy and ROS-TIGAR-autophagy (Li et al., 2015). Nonetheless, so far, there is no direct evidence that autophagy participates in the process of capacitation. It may be a good potential target for exploring the molecular mechanisms in capacitation disruption-induced male infertility.
During fertilization, the most striking cellular events are acrosome reaction and PME. Autophagy may be involved in the acrosome reaction through TPCs. When it comes to PME, three popular mechanisms are involved: (i) both autophagy and UPS contribute to PME and MOs after fertilization; (ii) MOs provide membranes for autophagosome formation and launch autophagy in the process of PME; and (iii) autophagy is absent in PME after fertilization in mice. Actually, autophagy may regulate PME before fertilization in mammals.
Definitely, we really appreciate researchers who have contributed significantly to the study of Atgs KD/KO in mammalian testis (Table I). They have opened the prelude to the regulation of autophagy on sperm fate, and even verified the specific mechanism of certain autophagy molecules on sperm regulation. Early innovative explorations of eminent investigators helped us to prepare this article for reproductive biologists interested in understanding the intriguing regulation of autophagy on the fate of sperm.
Future studies are needed to answer the following questions:
Whether and how do mTORC1 and mTORC2 synergistically orchestrate BTB dynamics by intercellular adhesion and cytoskeleton?
How does autophagy exert its bilateral effects, pro-death and pro-survival, in environmental toxicant-induced spermatogonial cell injury?
What is the cross-talk between meiosis and autophagy-related proteins during spermatidogenesis?
Whether and how does autophagy participate in capacitation?
Despite the interaction of TPC2 and mTORC1, whether is there direct evidence that autophagy operates the acrosome reaction through TPCs?
What is the interaction mechanism between K63 chains and K48 chains in PME or MOs clearance?
How and when is sperm mtDNA eliminated in mammals, and is there direct evidence that autophagy participates in PME or MOs clearance?
What is the function of Atgs with no KD/KO model in the testis (Table I)?
Answers to these questions, will be beneficial to interpret the roles of autophagy in the fate of sperm, but may also be helpful to diagnose and treat cases of relevant male infertility.
Data availability
The data underlying this article are available in the article.
Acknowledgements
The authors thank the editors and reviewers for their significant contributions during the revision period.
Authors’ roles
M.W. and Y.Z.Z. designed the study and edited the final text. M.W. and P.S. collected the data from publications, developed the database and wrote the manuscript. M.W. prepared the figures. M.W. and L.Z. prepared the tables. M.W., Y.Z.Z., L.M. and M.Z. contributed to the manuscript revision and critical discussion.
Funding
This study was supported by Ministry of Science and Technology of the People’s Republic of China (National Key R&D Program of China, No. 2020YFA0803900) and Wuhan University (Wuhan University Medical Development Plan, No. TFJC2018001).
Conflict of interest
None declared.
References
- Agrawal V, Jaiswal MK, Mallers T, Katara GK, Gilman-Sachs A, Beaman KD, Hirsch E.. Altered autophagic flux enhances inflammatory responses during inflammation-induced preterm labor. Sci Rep 2015;5:9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V.. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 2011;334:1144–1147. [DOI] [PubMed] [Google Scholar]
- Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V.. Allophagy: a macroautophagic process degrading spermatozoid-inherited organelles. Autophagy 2012;8:421–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankel-Simons F, Cummins JM.. Misconceptions about mitochondria and mammalian fertilization: implications for theories on human evolution. Proc Natl Acad Sci U S A 1996;93:13859–13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio IM, Espino J, Bejarano I, Gallardo-Soler A, Campo ML, Salido GM, Pariente JA, Pena FJ, Tapia JA.. Autophagy-related proteins are functionally active in human spermatozoa and may be involved in the regulation of cell survival and motility. Sci Rep 2016;6:33647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt L, Castonguay J, Arlt E, Meyer D, Hassan S, Borth H, Zierler S, Wennemuth G, Breit A, Biel M. et al. NAADP and the two-pore channel protein 1 participate in the acrosome reaction in mammalian spermatozoa. Mol Biol Cell 2014;25:948–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Xie J, Xing Y, Wang LT, Xie J, Yang F, Liu T, Liu M, Tang J, Yi Z. et al. Synthesis and biological evaluation of methylpyrimidine-fused tricyclic diterpene analogs as novel oral anti-late-onset hypogonadism agents. Eur J Med Chem 2019;176:21–40. [DOI] [PubMed] [Google Scholar]
- Bai S, Cheng L, Zhang Y, Zhu C, Zhu Z, Zhu R, Cheng CY, Ye L, Zheng K.. A germline-specific role for the mTORC2 component Rictor in maintaining spermatogonial differentiation and intercellular adhesion in mouse testis. Mol Hum Reprod 2018;24:244–259. [DOI] [PubMed] [Google Scholar]
- Baker MD, Ezzati M, Aloisio GM, Tarnawa ED, Cuevas I, Nakada Y, Castrillon DH.. The small GTPase Rheb is required for spermatogenesis but not oogenesis. Reproduction 2014;147:615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruti G, Paiardi C.. Acrosome biogenesis: revisiting old questions to yield new insights. Spermatogenesis 2011;1:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruti G, Paiardi C.. The dynamic of the apical ectoplasmic specialization between spermatids and Sertoli cells: the case of the small GTPase Rap1. Biomed Res Int 2014;2014:635979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruti G, Ripolone M, Ceriani M.. USP8, a regulator of endosomal sorting, is involved in mouse acrosome biogenesis through interaction with the spermatid ESCRT-0 complex and microtubules. Biol Reprod 2010;82:930–939. [DOI] [PubMed] [Google Scholar]
- Boada C, Zinger A, Tsao C, Zhao P, Martinez JO, Hartman K, Naoi T, Sukhoveshin R, Sushnitha M, Molinaro R. et al. Rapamycin-loaded biomimetic nanoparticles reverse vascular inflammation. Circ Res 2020;126:25–37. [DOI] [PubMed] [Google Scholar]
- Bonam SR, Ruff M, Muller S.. HSPA8/HSC70 in immune disorders: a molecular rheostat that adjusts chaperone-mediated autophagy substrates. Cells 2019;8:849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Reggiori F, Codogno P.. Emerging regulation and functions of autophagy. Nat Cell Biol 2013;15:713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer A, Girard M, Thimmanahalli DS, Levasseur A, Celeste C, Paquet M, Duggavathi R, Boerboom D.. mTOR regulates gap junction Alpha-1 protein trafficking in sertoli cells and is required for the maintenance of spermatogenesis in mice. Biol Reprod 2016;95:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busada JT, Chappell VA, Niedenberger BA, Kaye EP, Keiper BD, Hogarth CA, Geyer CB.. Retinoic acid regulates Kit translation during spermatogonial differentiation in the mouse. Dev Biol 2015a;397:140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busada JT, Niedenberger BA, Velte EK, Keiper BD, Geyer CB.. Mammalian target of rapamycin complex 1 (mTORC1) Is required for mouse spermatogonial differentiation in vivo. Dev Biol 2015b;407:90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008;456:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew JS, Espitia CM, Esquivel JA II, Mahalingam D, Kelly KR, Reddy G, Giles FJ, St N.. Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis. J Biol Chem 2011;286:6602–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Macones C, Mysorekar IU.. ATG16L1 governs placental infection risk and preterm birth in mice and women. JCI Insight 2016;1:e86654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Lin ZB, Tong MH, Zhang YL, Li YP, Zhou YC.. Mechanistic target of rapamycin kinase (Mtor) is required for spermatogonial proliferation and differentiation in mice. Asian J Androl 2020;22:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castino R, Davies J, Beaucourt S, Isidoro C, Murphy D.. Autophagy is a prosurvival mechanism in cells expressing an autosomal dominant familial neurohypophyseal diabetes insipidus mutant vasopressin transgene. FASEB J 2005a;19:1021–1023. [DOI] [PubMed] [Google Scholar]
- Castino R, Isidoro C, Murphy D.. Autophagy-dependent cell survival and cell death in an autosomal dominant familial neurohypophyseal diabetes insipidus in vitro model. FASEB J 2005b;19:1024–1026. [DOI] [PubMed] [Google Scholar]
- Chang FS, Wang Y, Dmitriev P, Gross J, Galione A, Pears C.. A two-pore channel protein required for regulating mTORC1 activity on starvation. BMC Biol 2020;18:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SB, Miron P, Miron A, Iglehart JD.. Rapamycin inhibits proliferation of estrogen-receptor-positive breast cancer cells. J Surg Res 2007;138:37–44. [DOI] [PubMed] [Google Scholar]
- Chemes H. The phagocytic function of Sertoli cells: a morphological, biochemical, and endocrinological study of lysosomes and acid phosphatase localization in the rat testis. Endocrinology 1986;119:1673–1681. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Liu Y, Zheng P.. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med 2008;205:2397–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JX, Xu LL, Mei JH, Yu XB, Kuang HB, Liu HY, Wu YJ, Wang JL.. Involvement of neuropathy target esterase in tri-ortho-cresyl phosphate-induced testicular spermatogenesis failure and growth inhibition of spermatogonial stem cells in mice. Toxicol Lett 2012;211:54–61. [DOI] [PubMed] [Google Scholar]
- Chen N, Lin M, Liu N, Wang S, Xiao X.. Methylmercury-induced testis damage is associated with activation of oxidative stress and germ cell autophagy. J Inorg Biochem 2019a;190:67–74. [DOI] [PubMed] [Google Scholar]
- Chen X, Li C, Chen Y, Ni C, Chen X, Zhang L, Xu X, Chen M, Ma X, Zhan H. et al. Aflatoxin B1 impairs Leydig cells through inhibiting AMPK/mTOR-mediated autophagy flux pathway. Chemosphere 2019;233:261–272. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Mruk DD.. A local autocrine axis in the testes that regulates spermatogenesis. Nat Rev Endocrinol 2010;6:380–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Mruk DD.. The blood-testis barrier and its implications for male contraception. Pharmacol Rev 2012;64:16–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Jo M, Lee E, Choi D.. Induction of apoptotic cell death via accumulation of autophagosomes in rat granulosa cells. Fertil Steril 2011;95:1482–1486. [DOI] [PubMed] [Google Scholar]
- Choi JH, Yoon JS, Won YW, Park BB, Lee YY.. Chloroquine enhances the chemotherapeutic activity of 5-fluorouracil in a colon cancer cell line via cell cycle alteration. APMIS 2012;120:597–604. [DOI] [PubMed] [Google Scholar]
- Chu HY, Wang W, Chen X, Jiang YE, Cheng R, Qi X, Zhong ZM, Zeng MS, Zhu XF, Sun CZ.. Bafilomycin A1 increases the sensitivity of tongue squamous cell carcinoma cells to cisplatin by inhibiting the lysosomal uptake of platinum ions but not autophagy. Cancer Lett 2018;423:105–112. [DOI] [PubMed] [Google Scholar]
- Clermont Y. Renewal of spermatogonia in man. Am J Anat 1966;118:509–524. [DOI] [PubMed] [Google Scholar]
- Corno C, Perego P.. KiSS1 in regulation of metastasis and response to antitumor drugs. Drug Resist Updat 2019;42:12–21. [DOI] [PubMed] [Google Scholar]
- Crutcher E, Pal R, Naini F, Zhang P, Laugsch M, Kim J, Bajic A, Schaaf CP.. mTOR and autophagy pathways are dysregulated in murine and human models of Schaaf-Yang syndrome. Sci Rep 2019;9:15935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski N, Hall MN.. TOR complex 2: a signaling pathway of its own. Trends Biochem Sci 2009;34:620–627. [DOI] [PubMed] [Google Scholar]
- Cybulski N, Polak P, Auwerx J, Ruegg MA, Hall MN.. mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc Natl Acad Sci U S A 2009;106:9902–9907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja MJ, Ding WX, Donohue TM Jr, Friedman SL, Kim JS, Komatsu M, Lemasters JJ, Lemoine A, Lin JD, Ou JH. et al. Functions of autophagy in normal and diseased liver. Autophagy 2013;9:1131–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Ros M, Lehtiniemi T, Olotu O, Fischer D, Zhang FP, Vihinen H, Jokitalo E, Sironen A, Toppari J, Kotaja N.. FYCO1 and autophagy control the integrity of the haploid male germ cell-specific RNP granules. Autophagy 2017;13:302–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai E, Han L, Liu J, Xie Y, Kroemer G, Klionsky DJ, Zeh HJ, Kang R, Wang J, Tang D.. Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy 2020;16:2069–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Persio S, Saracino R, Fera S, Muciaccia B, Esposito V, Boitani C, Berloco BP, Nudo F, Spadetta G, Stefanini M. et al. Spermatogonial kinetics in humans. Development 2017;144:3430–3439. [DOI] [PubMed] [Google Scholar]
- Dikic I, Elazar Z.. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 2018;19:349–364. [DOI] [PubMed] [Google Scholar]
- Ding F, Shan C, Li H, Zhang Y, Guo C, Zhou Z, Zheng J, Shen W, Dai Q, Ouyang Q. et al. Simvastatin alleviated diabetes mellitus-induced erectile dysfunction in rats by enhancing AMPK pathway-induced autophagy. Andrologia 2020;8:780–792. [DOI] [PubMed] [Google Scholar]
- Djeddi A, Al Rawi S, Deuve JL, Perrois C, Liu YY, Russeau M, Sachse M, Galy V.. Sperm-inherited organelle clearance in C. elegans relies on LC3-dependent autophagosome targeting to the pericentrosomal area. Development 2015;142:1705–1716. [DOI] [PubMed] [Google Scholar]
- Dorogova NV, Fedorova EV, Bolobolova EU, Ogienko AA, Baricheva EM.. GAGA protein is essential for male germ cell development in Drosophila. Genesis 2014;52:738–751. [DOI] [PubMed] [Google Scholar]
- Duan P, Hu C, Quan C, Yu T, Huang W, Chen W, Tang S, Shi Y, Martin FL, Yang K.. 4-Nonylphenol induces autophagy and attenuates mTOR-p70S6K/4EBP1 signaling by modulating AMPK activation in Sertoli cells. Toxicol Lett 2017;267:21–31. [DOI] [PubMed] [Google Scholar]
- Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E. et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab 2008;8:325–332. [DOI] [PubMed] [Google Scholar]
- Eng CP, Sehgal SN, Vezina C.. Activity of rapamycin (AY-22,989) against transplanted tumors. J Antibiot (Tokyo) 1984;37:1231–1237. [DOI] [PubMed] [Google Scholar]
- Feng Z, Liang C, Manthari RK, Wang C, Zhang J.. Effects of fluoride on autophagy in mouse Sertoli cells. Biol Trace Elem Res 2019;187:499–505. [DOI] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P. et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007;447:1121–1125. [DOI] [PubMed] [Google Scholar]
- Finetti F, Cassioli C, Cianfanelli V, Onnis A, Paccagnini E, Kabanova A, Baldari CT.. The intraflagellar transport protein IFT20 controls lysosome biogenesis by regulating the post-Golgi transport of acid hydrolases. Cell Death Differ 2020;27:310–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgione N, Vogl AW, Varmuza S.. Loss of protein phosphatase 1c{gamma} (PPP1CC) leads to impaired spermatogenesis associated with defects in chromatin condensation and acrosome development: an ultrastructural analysis. Reproduction 2010;139:1021–1029. [DOI] [PubMed] [Google Scholar]
- Fouquet JP, Dadoune JP.. Renewal of spermatogonia in the monkey (Macaca fascicularis). Biol Reprod 1986;35:199–207. [DOI] [PubMed] [Google Scholar]
- Frasa MA, Koessmeier KT, Ahmadian MR, Braga VM.. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev Mol Cell Biol 2012;13:67–73. [DOI] [PubMed] [Google Scholar]
- Gan B, DePinho RA.. mTORC1 signaling governs hematopoietic stem cell quiescence. Cell Cycle 2009;8:1003–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Yang D, Yang S, Wang J, Wei J, Chen J.. Di-2-ethylhexyl phthalate (DEHP) induces apoptosis and autophagy of mouse GC-1 spg cells. Environ Toxicol 2020;35:292–299. [DOI] [PubMed] [Google Scholar]
- Gao F, Li G, Liu C, Gao H, Wang H, Liu W, Chen M, Shang Y, Wang L, Shi J. et al. Autophagy regulates testosterone synthesis by facilitating cholesterol uptake in Leydig cells. J Cell Biol 2018;217:2103–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Chen H, Lui WY, Lee WM, Cheng CY.. Basement membrane laminin alpha2 regulation of BTB dynamics via its effects on F-Actin and microtubule cytoskeletons is mediated through mTORC1 signaling. Endocrinology 2017;158:963–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rua V, Feijoo-Bandin S, Rodriguez-Penas D, Mosquera-Leal A, Abu-Assi E, Beiras A, Maria Seoane L, Lear P, Parrington J, Portoles M. et al. Endolysosomal two-pore channels regulate autophagy in cardiomyocytes. J Physiol 2016;594:3061–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawriluk TR, Hale AN, Flaws JA, Dillon CP, Green DR, Rucker EB III. Autophagy is a cell survival program for female germ cells in the murine ovary. Reproduction 2011;141:759–765. [DOI] [PubMed] [Google Scholar]
- Gawriluk TR, Ko C, Hong X, Christenson LK, Rucker EB.. Beclin-1 deficiency in the murine ovary results in the reduction of progesterone production to promote preterm labor. Proc Natl Acad Sci U S A 2014;111:E4194–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W.. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 2010;12:119–131. [DOI] [PubMed] [Google Scholar]
- Goh WS, Falciatori I, Tam OH, Burgess R, Meikar O, Kotaja N, Hammell M, Hannon GJ.. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev 2015;29:1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden EB, Cho HY, Jahanian A, Hofman FM, Louie SG, Schonthal AH, Chen TC.. Chloroquine enhances temozolomide cytotoxicity in malignant gliomas by blocking autophagy. Neurosurg Focus 2014;37:E12. [DOI] [PubMed] [Google Scholar]
- Gomes Fernandes M, He N, Wang F, Van Iperen L, Eguizabal C, Matorras R, Roelen BAJ, Chuva De Sousa Lopes SM.. Human-specific subcellular compartmentalization of P-element induced wimpy testis-like (PIWIL) granules during germ cell development and spermatogenesis. Hum Reprod 2018;33:258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe M, Gansauge F, Schmid RM, Adler G, Seufferlein T.. Regulation of cell growth and cyclin D1 expression by the constitutively active FRAP-p70s6K pathway in human pancreatic cancer cells. Cancer Res 1999;59:3581–3587. [PubMed] [Google Scholar]
- Griswold MD. Spermatogenesis: the commitment to meiosis. Physiol Rev 2016;96:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YP, Yang XM, Luo P, Li YQ, Tao YX, Duan ZH, Xiao W, Zhang DY, Liu HZ.. Inhibition of acrolein-induced autophagy and apoptosis by a glycosaminoglycan from Sepia esculenta ink in mouse Leydig cells. Carbohydr Polym 2017;163:270–279. [DOI] [PubMed] [Google Scholar]
- Guo H, Chitiprolu M, Gagnon D, Meng L, Perez-Iratxeta C, Lagace D, Gibbings D.. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun 2014;5:5276. [DOI] [PubMed] [Google Scholar]
- Gurney JK, McGlynn KA, Stanley J, Merriman T, Signal V, Shaw C, Edwards R, Richiardi L, Hutson J, Sarfati D.. Risk factors for cryptorchidism. Nat Rev Urol 2017;14:534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AK, Yoshimura S, Stephens DJ, Preisinger C, Fuchs E, Barr FA.. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci 2007;120:2997–3010. [DOI] [PubMed] [Google Scholar]
- Hajjar C, Sampuda KM, Boyd L.. Dual roles for ubiquitination in the processing of sperm organelles after fertilization. BMC Dev Biol 2014;14:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006;441:885–889. [DOI] [PubMed] [Google Scholar]
- Hartleben B, Godel M, Meyer-Schwesinger C, Liu S, Ulrich T, Kobler S, Wiech T, Grahammer F, Arnold SJ, Lindenmeyer MT. et al. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest 2010;120:1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartree EF. The acrosome-lysosome relationship. J Reprod Fertil 1975;44:125–126. [DOI] [PubMed] [Google Scholar]
- Hermann BP, Sukhwani M, Simorangkir DR, Chu T, Plant TM, Orwig KE.. Molecular dissection of the male germ cell lineage identifies putative spermatogonial stem cells in rhesus macaques. Hum Reprod 2009;24:1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess RA, Renato de Franca L.. Spermatogenesis and cycle of the seminiferous epithelium. Adv Exp Med Biol 2008;636:1–15. [DOI] [PubMed] [Google Scholar]
- Hine C. Rapamycin keeps the reproductive clock ticking. Sci Transl Med 2017;9:eaan4296. [DOI] [PubMed] [Google Scholar]
- Hori YS, Hosoda R, Akiyama Y, Sebori R, Wanibuchi M, Mikami T, Sugino T, Suzuki K, Maruyama M, Tsukamoto M, Mikuni N. et al. Chloroquine potentiates temozolomide cytotoxicity by inhibiting mitochondrial autophagy in glioma cells. J Neurooncol 2015;122:11–20. [DOI] [PubMed] [Google Scholar]
- Houck SA, Ren HY, Madden VJ, Bonner JN, Conlin MP, Janovick JA, Conn PM, Cyr DM.. Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol Cell 2014;54:166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu ZZ, Valencia JC, Huang H, Chi A, Shabanowitz J, Hearing VJ, Appella E, Wu C.. Comparative bioinformatics analyses and profiling of lysosome-related organelle proteomes. Int J Mass Spectrom 2007;259:147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Liu Y, Zhang S, Yap YT, Li W, Zhang D, Gardner A, Zhang L, Song S, Hess RA. et al. Autophagy core protein ATG5 is required for elongating spermatid development, sperm individualization and normal fertility in male mice. Autophagy 2021;17:1753–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Cao Z, Zhang J, Ji Q, Li Y.. Aflatoxin B1 promotes autophagy associated with oxidative stress-related PI3K/AKT/mTOR signaling pathway in mice testis. Environ Pollut 2019;255:113317. [DOI] [PubMed] [Google Scholar]
- Huang W, Quan C, Duan P, Tang S, Chen W, Yang K.. Nonylphenol induced apoptosis and autophagy involving the Akt/mTOR pathway in prepubertal Sprague-Dawley male rats in vivo and in vitro. Toxicology 2016;373:41–53. [DOI] [PubMed] [Google Scholar]
- Itoh K, Kondoh G, Miyachi H, Sugai M, Kaneko Y, Kitano S, Watanabe H, Maeda R, Imura A, Liu Y. et al. Dephosphorylation of protamine 2 at serine 56 is crucial for murine sperm maturation in vivo. Sci Signal 2019;12:eaao7232. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B.. SIN1/MIP1 maintains Rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 2006;127:125–137. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN.. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 2004;6:1122–1128. [DOI] [PubMed] [Google Scholar]
- Jin H, Hou J, Meng X, Ma T, Wang B, Liu Z, Sha X, Ding J, Han X.. Microcystin-leucine arginine induced the apoptosis of GnRH neurons by activating the endoplasmic reticulum stress resulting in a decrease of serum testosterone level in mice. Ecotoxicol Environ Saf 2021;208:111748. [DOI] [PubMed] [Google Scholar]
- Jin M, Fujiwara E, Kakiuchi Y, Okabe M, Satouh Y, Baba SA, Chiba K, Hirohashi N.. Most fertilizing mouse spermatozoa begin their acrosome reaction before contact with the zona pellucida during in vitro fertilization. Proc Natl Acad Sci U S A 2011;108:4892–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SK, Yang WX.. Factors and pathways involved in capacitation: how are they regulated? Oncotarget 2017;8:3600–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW. et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab 2008;8:318–324. [DOI] [PubMed] [Google Scholar]
- Kaneda H, Hayashi J, Takahama S, Taya C, Lindahl KF, Yonekawa H.. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc Natl Acad Sci U S A 1995;92:4542–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Z, Qiao N, Liu G, Chen H, Tang Z, Li Y.. Copper-induced apoptosis and autophagy through oxidative stress-mediated mitochondrial dysfunction in male germ cells. Toxicol In Vitro 2019;61:104639. [DOI] [PubMed] [Google Scholar]
- Kang-Decker N, Mantchev GT, Juneja SC, McNiven MA, van Deursen JM.. Lack of acrosome formation in Hrb-deficient mice. Science 2001;294:1531–1533. [DOI] [PubMed] [Google Scholar]
- Kaprara A, Huhtaniemi IT.. The hypothalamus-pituitary-gonad axis: tales of mice and men. Metabolism 2018;86:3–17. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, Cuervo AM, Singh R.. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab 2011;14:173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina N, Borovjagin AV, Kadagidze Z, Baryshnikov A, Baryshnikova M, Malin D, Ghosh D, Shah N, Welch DR, Gabikian P. et al. Astrocytes promote progression of breast cancer metastases to the brain via a KISS1-mediated autophagy. Autophagy 2017;13:1905–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, Eguchi H, Hattori N.. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett 2010;584:1073–1079. [DOI] [PubMed] [Google Scholar]
- Khawar MB, Gao H, Li W.. Mechanism of acrosome biogenesis in mammals. Front Cell Dev Biol 2019;7:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM.. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002;110:163–175. [DOI] [PubMed] [Google Scholar]
- Kim DK, Yang JS, Maiti K, Hwang JI, Kim K, Seen D, Ahn Y, Lee C, Kang BC, Kwon HB, Cheon J. et al. A gonadotropin-releasing hormone-II antagonist induces autophagy of prostate cancer cells. Cancer Res 2009;69:923–931. [DOI] [PubMed] [Google Scholar]
- Kim K, Choe HK.. Role of hypothalamus in aging and its underlying cellular mechanisms. Mech Ageing Dev 2019;177:74–79. [DOI] [PubMed] [Google Scholar]
- Kim YC, Guan KL.. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest 2015;125:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Page DC.. The mysteries of sexual identity. The germ cell’s perspective. Science 2007;316:400–401. [DOI] [PubMed] [Google Scholar]
- Kimura T, Takabatake Y, Takahashi A, Kaimori JY, Matsui I, Namba T, Kitamura H, Niimura F, Matsusaka T, Soga T. et al. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol 2011;22:902–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SH, Apple EC, Liu Z, Chen L.. Age-dependent autophagy induction after injury promotes axon regeneration by limiting NOTCH. Autophagy 2020;16:2052–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig MK, Bell CS, Hebert AA, Roberson J, Samuels JA, Slopis JM, Tate P, Northrup H; TREATMENT Trial Collaborators. Efficacy and safety of topical rapamycin in patients with facial angiofibromas secondary to tuberous sclerosis complex: the TREATMENT randomized clinical trial. JAMA Dermatol 2018;154:773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E. et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006;441:880–884. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005;169:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL Jr, Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z.. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A 2007;104:14489–14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopalli SR, Cha KM, Lee SH, Hwang SY, Lee YJ, Koppula S, Kim SK.. Cordycepin, an active constituent of nutrient powerhouse and potential medicinal mushroom Cordyceps militaris Linn., ameliorates age-related testicular dysfunction in rats. Nutrients 2019;11:906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Sassone-Corsi P.. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol 2007;8:85–90. [DOI] [PubMed] [Google Scholar]
- Krausz C. Male infertility: pathogenesis and clinical diagnosis. Best Pract Res Clin Endocrinol Metab 2011;25:271–285. [DOI] [PubMed] [Google Scholar]
- Ktistakis NT, Tooze SA.. Digesting the expanding mechanisms of autophagy. Trends Cell Biol 2016;26:624–635. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N.. The role of autophagy during the early neonatal starvation period. Nature 2004;432:1032–1036. [DOI] [PubMed] [Google Scholar]
- La HM, Chan AL, Legrand JMD, Rossello FJ, Gangemi CG, Papa A, Cheng Q, Morand EF, Hobbs RM.. GILZ-dependent modulation of mTORC1 regulates spermatogonial maintenance. Development 2018;145:dev165324. [DOI] [PubMed] [Google Scholar]
- Lehti MS, Zhang FP, Kotaja N, Sironen A.. SPEF2 functions in microtubule-mediated transport in elongating spermatids to ensure proper male germ cell differentiation. Development 2017;144:2683–2693. [DOI] [PubMed] [Google Scholar]
- Leng ZG, Lin SJ, Wu ZR, Guo YH, Cai L, Shang HB, Tang H, Xue YJ, Lou MQ, Zhao W. et al. Activation of DRD5 (dopamine receptor D5) inhibits tumor growth by autophagic cell death. Autophagy 2017;13:1404–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Elazar Z.. Development. Inheriting maternal mtDNA. Science 2011;334:1069–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Li D, Liu H, Cao B-B, Jiang F, Chen D-N, Li J-D.. RNF216 regulates the migration of immortalized GnRH neurons by suppressing Beclin1-mediated autophagy. Front Endocrinol (Lausanne) 2019;10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Tan J, Miao Y, Lei P, Zhang Q.. ROS and autophagy: interactions and molecular regulatory mechanisms. Cell Mol Neurobiol 2015;35:615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Luo X, Zhu Y, Zhao L, Li L, Peng Q, Ma M, Gao Y.. ATM signals to AMPK to promote autophagy and positively regulate DNA damage in response to cadmium-induced ROS in mouse spermatocytes. Environ Pollut 2017;231:1560–1568. [DOI] [PubMed] [Google Scholar]
- Li WR, Chen L, Chang ZJ, Xin H, Liu T, Zhang YQ, Li GY, Zhou F, Gong YQ, Gao ZZ. et al. Autophagic deficiency is related to steroidogenic decline in aged rat Leydig cells. Asian J Androl 2011;13:881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Oh DY, Nakamura K, Thiele CJ.. Perifosine-induced inhibition of Akt attenuates brain-derived neurotrophic factor/TrkB-induced chemoresistance in neuroblastoma in vivo. Cancer 2011;117:5412–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tan F, Liewehr DJ, Steinberg SM, Thiele CJ.. In vitro and in vivo inhibition of neuroblastoma tumor cell growth by AKT inhibitor perifosine. J Natl Cancer Inst 2010;102:758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng Z, Manthari RK, Wang C, Han Y, Fu W, Wang J, Zhang J.. Arsenic induces dysfunctional autophagy via dual regulation of mTOR pathway and Beclin1-Vps34/PI3K complex in MLTC-1 cells. J Hazard Mater 2020;391:122227. [DOI] [PubMed] [Google Scholar]
- Liang L, Zheng D, Lu C, Xi Q, Bao H, Li W, Gu Y, Mao Y, Xu B, Gu X.. Exosomes derived from miR-301a-3p-overexpressing adipose-derived mesenchymal stem cells reverse hypoxia-induced erectile dysfunction in rat models. Stem Cell Res Ther 2021;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie PP, Cheng CY, Mruk DD.. Coordinating cellular events during spermatogenesis: a biochemical model. Trends Biochem Sci 2009;34:366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liegel RP, Handley MT, Ronchetti A, Brown S, Langemeyer L, Linford A, Chang B, Morris-Rosendahl DJ, Carpanini S, Posmyk R. et al. Loss-of-function mutations in TBC1D20 cause cataracts and male infertility in blind sterile mice and Warburg micro syndrome in humans. Am J Hum Genet 2013;93:1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wang T, Ruan Y, Liu K, Li H, Wang S, Li M, Liu J.. Rapamycin supplementation may ameliorate erectile function in rats with streptozotocin-induced type 1 diabetes by inducing autophagy and inhibiting apoptosis, endothelial dysfunction, and corporal fibrosis. J Sex Med 2018;15:1246–1259. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Wu ZR, Cao L, Zhang Y, Leng ZG, Guo YH, Shang HB, Zhao WG, Zhang X, Wu ZB.. Pituitary tumor suppression by combination of cabergoline and chloroquine. J Clin Endocrinol Metab 2017;102:3692–3703. [DOI] [PubMed] [Google Scholar]
- Liu C, Lv M, He X, Zhu Y, Amiri-Yekta A, Li W, Wu H, Kherraf ZE, Liu W, Zhang J. et al. Homozygous mutations in SPEF2 induce multiple morphological abnormalities of the sperm flagella and male infertility. J Med Genet 2020;57:31–37. [DOI] [PubMed] [Google Scholar]
- Liu C, Song Z, Wang L, Yu H, Liu W, Shang Y, Xu Z, Zhao H, Gao F, Wen J. et al. Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 2017a;144:441–451. [DOI] [PubMed] [Google Scholar]
- Liu C, Wang H, Shang Y, Liu W, Song Z, Zhao H, Wang L, Jia P, Gao F, Xu Z. et al. Autophagy is required for ectoplasmic specialization assembly in Sertoli cells. Autophagy 2016;12:814–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ML, Wang JL, Wei J, Xu LL, Yu M, Liu XM, Ruan WL, Chen JX.. Tri-ortho-cresyl phosphate induces autophagy of rat spermatogonial stem cells. Reproduction 2015;149:163–170. [DOI] [PubMed] [Google Scholar]
- Liu P, Li R, Tian X, Zhao Y, Li M, Wang M, Ying X, Yuan J, Xie J, Yan X. et al. Co-exposure to fluoride and arsenic disrupts intestinal flora balance and induces testicular autophagy in offspring rats. Ecotoxicol Environ Saf 2021;222:112506. [DOI] [PubMed] [Google Scholar]
- Liu S, Huang L, Geng Y, He J, Chen X, Xu H, Li R, Wang Y, Ding Y, Liu X.. Rapamycin inhibits spermatogenesis by changing the autophagy status through suppressing mechanistic target of rapamycin-p70S6 kinase in male rats. Mol Med Rep 2017b;16:4029–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Xu L, Shen J, Wang J, Ruan W, Yu M, Chen J.. Involvement of oxidative stress in tri-ortho-cresyl phosphate-induced autophagy of mouse Leydig TM3 cells in vitro. Reprod Biol Endocrinol 2016;14:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hu Y, Wang L, Xu C.. Expression of transcriptional factor EB (TFEB) in differentiating spermatogonia potentially promotes cell migration in mouse seminiferous epithelium. Reprod Biol Endocrinol 2018;16:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Yao Y, Hu Z, Hu C, Song Q, Ye J, Xu C, Wang AZ, Chen Q, Wang QK.. Angiogenic factor AGGF1 activates autophagy with an essential role in therapeutic angiogenesis for heart disease. PLoS Biol 2016;14:e1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo SM, Ge ZJ, Wang ZW, Jiang ZZ, Wang ZB, Ouyang YC, Hou Y, Schatten H, Sun QY.. Unique insights into maternal mitochondrial inheritance in mice. Proc Natl Acad Sci U S A 2013;110:13038–13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo SM, Sun QY.. Autophagy is not involved in the degradation of sperm mitochondria after fertilization in mice. Autophagy 2013;9:2156–2157. [DOI] [PubMed] [Google Scholar]
- Lv MQ, Zhou L, Ge P, Li YX, Zhang J, Zhou DX.. Over-expression of hsa_circ_0000116 in patients with non-obstructive azoospermia and its predictive value in testicular sperm retrieval. Andrology-Us 2020;8:1834–1843. [DOI] [PubMed] [Google Scholar]
- Lyu L, Hu Y, Yin S, Wang L, Ye F, Wang M, Zhou Y, Ma W, Chen C, Jiang Y. et al. Autophagy inhibition enhances anti-pituitary adenoma effect of tetrandrine. Phytother Res 2021;35:4007–4021. [DOI] [PubMed] [Google Scholar]
- Ma F, Zhou Z, Li N, Zheng L, Wu C, Niu B, Tang F, He X, Li G, Hua J.. Lin28a promotes self-renewal and proliferation of dairy goat spermatogonial stem cells (SSCs) through regulation of mTOR and PI3K/AKT. Sci Rep 2016;6:38805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Yang HZ, Xu LM, Huang YR, Dai HL, Kang XN.. Testosterone regulates the autophagic clearance of androgen binding protein in rat Sertoli cells. Sci Rep 2015;5:8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhou Y, Zhu YC, Wang SQ, Ping P, Chen XF.. Lipophagy contributes to testosterone biosynthesis in male rat Leydig cells. Endocrinology 2018;159:1119–1129. [DOI] [PubMed] [Google Scholar]
- Mandrioli J, D'Amico R, Zucchi E, Gessani A, Fini N, Fasano A, Caponnetto C, Chio A, Dalla Bella E, Lunetta C. et al. ; RAP-ALS Investigators Group. Rapamycin treatment for amyotrophic lateral sclerosis: Protocol for a phase II randomized, double-blind, placebo-controlled, multicenter, clinical trial (RAP-ALS trial). Medicine (Baltimore) 2018;97:e11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M.. Autophagy is required to maintain muscle mass. Cell Metab 2009;10:507–515. [DOI] [PubMed] [Google Scholar]
- Mancilla H, Maldonado R, Cereceda K, Villarroel-Espindola F, Montes de Oca M, Angulo C, Castro MA, Slebe JC, Vera JC, Lavandero S. et al. Glutathione depletion induces spermatogonial cell autophagy. J Cell Biochem 2015;116:2283–2292. [DOI] [PubMed] [Google Scholar]
- Martinez-Menarguez JA, Geuze HJ, Ballesta J.. Evidence for a nonlysosomal origin of the acrosome. J Histochem Cytochem 1996;44:313–320. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F. et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 2010;189:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattam U, Talari NK, Paripati AK, Krishnamoorthy T, Sepuri NBV.. Kisspeptin preserves mitochondrial function by inducing mitophagy and autophagy in aging rat brain hippocampus and human neuronal cell line. Biochim Biophys Acta Mol Cell Res 2021;1868:118852. [DOI] [PubMed] [Google Scholar]
- Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A.. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy 2012;8:200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikar O, Da Ros M, Korhonen H, Kotaja N.. Chromatoid body and small RNAs in male germ cells. Reproduction 2011;142:195–209. [DOI] [PubMed] [Google Scholar]
- Meng Q, Cai D.. Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IkappaB kinase beta (IKKbeta)/NF-kappaB pathway. J Biol Chem 2011;286:32324–32332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B.. Autophagy in human diseases. N Engl J Med 2020;383:1564–1576. [DOI] [PubMed] [Google Scholar]
- Mok KW, Chen H, Lee WM, Cheng CY.. rpS6 regulates blood-testis barrier dynamics through Arp3-mediated actin microfilament organization in rat Sertoli cells. An in vitro study. Endocrinology 2015;156:1900–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok KW, Mruk DD, Cheng CY.. Regulation of blood-testis barrier (BTB) dynamics during spermatogenesis via the “Yin” and “Yang” effects of mammalian target of rapamycin complex 1 (mTORC1) and mTORC2. Int Rev Cell Mol Biol 2013;301:291–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk DD, Cheng CY.. Cell-cell interactions at the ectoplasmic specialization in the testis. Trends Endocrinol Metab 2004;15:439–447. [DOI] [PubMed] [Google Scholar]
- Mruk DD, Silvestrini B, Cheng CY.. Anchoring junctions as drug targets: role in contraceptive development. Pharmacol Rev 2008;60:146–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najari BB, Kashanian JA.. Erectile dysfunction. JAMA 2016;316:1838. [DOI] [PubMed] [Google Scholar]
- Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M. et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med 2007;13:619–624. [DOI] [PubMed] [Google Scholar]
- Navarro VM, Tena-Sempere M.. Neuroendocrine control by kisspeptins: role in metabolic regulation of fertility. Nat Rev Endocrinol 2011;8:40–53. [DOI] [PubMed] [Google Scholar]
- Nian FS, Li LL, Cheng CY, Wu PC, Lin YT, Tang CY, Ren BS, Tai CY, Fann MJ, Kao LS. et al. Rab18 collaborates with Rab7 to modulate lysosomal and autophagy activities in the nervous system: an overlapping mechanism for Warburg Micro Syndrome and Charcot-Marie-Tooth Neuropathy Type 2B. Mol Neurobiol 2019;56:6095–6105. [DOI] [PubMed] [Google Scholar]
- Nishiyama J, Miura E, Mizushima N, Watanabe M, Yuzaki M.. Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy 2007;3:591–596. [DOI] [PubMed] [Google Scholar]
- Ogunbayo OA, Duan J, Xiong J, Wang Q, Feng X, Ma J, Zhu MX, Evans AM.. mTORC1 controls lysosomal Ca(2+) release through the two-pore channel TPC2. Sci Signal 2018;11:eaao5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto U, Ishida H, Krayukhina E, Uchiyama S, Inoue N, Shimizu T.. Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization. Nature 2016;534:566–569. [DOI] [PubMed] [Google Scholar]
- Okabe M. The cell biology of mammalian fertilization. Development 2013;140:4471–4479. [DOI] [PubMed] [Google Scholar]
- Ommati MM, Heidari R, Manthari RK, Tikka Chiranjeevi S, Niu R, Sun Z, Sabouri S, Zamiri MJ, Zaker L, Yuan J. et al. Paternal exposure to arsenic resulted in oxidative stress, autophagy, and mitochondrial impairments in the HPG axis of pubertal male offspring. Chemosphere 2019;236:124325. [DOI] [PubMed] [Google Scholar]
- Ommati MM, Manthari RK, Tikka C, Niu R, Sun Z, Sabouri S, Zamiri MJ, Ahmadi HN, Ghaffari H, Heidari R, Wang J.. Arsenic-induced autophagic alterations and mitochondrial impairments in HPG-S axis of mature male mice offspring (F1-generation): a persistent toxicity study. Toxicol Lett 2020;326:83–98. [DOI] [PubMed] [Google Scholar]
- Ozturk N, Steger K, Schagdarsurengin U.. The impact of autophagy in spermiogenesis. Asian J Androl 2017;19:617–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM.. Functional interaction between autophagy and ciliogenesis. Nature 2013;502:194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J, Han L, Liu Z, Zheng J, Zhao J, Deng K, Wang F, Zhang Y.. ULK1 affects cell viability of goat Sertoli cell by modulating both autophagy and apoptosis. In Vitro Cell Dev Biol Anim 2019;55:604–613. [DOI] [PubMed] [Google Scholar]
- Park AK, , LiegelRP, , RonchettiA, , EbertAD, , GeurtsA, , Sidjanin DJ.. Targeted disruption of Tbc1d20 with zinc-finger nucleases causes cataracts and testicular abnormalities in mice. BMC Genet 2014;15:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira GJ, Antonioli M, Hirata H, Ureshino RP, Nascimento AR, Bincoletto C, Vescovo T, Piacentini M, Fimia GM, Smaili SS.. Glutamate induces autophagy via the two-pore channels in neural cells. Oncotarget 2017;8:12730–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl C, Dikic I.. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019;366:818–822. [DOI] [PubMed] [Google Scholar]
- Politi Y, Gal L, Kalifa Y, Ravid L, Elazar Z, Arama E.. Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Dev Cell 2014;29:305–320. [DOI] [PubMed] [Google Scholar]
- Puga Molina LC, Pinto NA, Torres Rodriguez P, Romarowski A, Vicens Sanchez A, Visconti PE, Darszon A, Trevino CL, Buffone MG.. Essential role of CFTR in PKA-dependent phosphorylation, alkalinization, and hyperpolarization during human sperm capacitation. J Cell Physiol 2017;232:1404–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabanal-Ruiz Y, Otten EG, Korolchuk VI.. mTORC1 as the main gateway to autophagy. Essays Biochem 2017;61:565–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, Schuchter LM, Torigian DA, Panosian JT, Troxel AB. et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy 2014a;10:1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, Tan KS, Heitjan DF, Rodgers G, Gallagher M. et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy 2014b;10:1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran M, Li Z, Cao R, Weng B, Peng F, He C, Chen B.. miR-26a suppresses autophagy in swine Sertoli cells by targeting ULK2. Reprod Domest Anim 2018;53:864–871. [DOI] [PubMed] [Google Scholar]
- Ren D, Navarro B, Perez G, Jackson AC, Hsu S, Shi Q, Tilly JL, Clapham DE.. A sperm ion channel required for sperm motility and male fertility. Nature 2001;413:603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Huang J, Wei J, Zang Y, Zhao Y, Wu S, Zhao X, Zhou X, Sun Z, Lu H.. Maternal exposure to fine particle matters cause autophagy via UPR-mediated PI3K-mTOR pathway in testicular tissue of adult male mice in offspring. Ecotoxicol Environ Saf 2020;189:109943. [DOI] [PubMed] [Google Scholar]
- Ren L, Liu J, Zhang J, Wang J, Wei J, Li Y, Guo C, Sun Z, Zhou X.. Silica nanoparticles induce spermatocyte cell autophagy through microRNA-494 targeting AKT in GC-2spd cells. Environ Pollut 2019;255:113172. [DOI] [PubMed] [Google Scholar]
- Riera MF, Regueira M, Galardo MN, Pellizzari EH, Meroni SB, Cigorraga SB.. Signal transduction pathways in FSH regulation of rat Sertoli cell proliferation. Am J Physiol Endocrinol Metab 2012;302:E914–923. [DOI] [PubMed] [Google Scholar]
- Rojansky R, Cha MY, Chan DC.. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. Elife 2016;5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosen-Runge EC. The process of spermatogenesis in mammals. Biol Rev Camb Philos Soc 1962;37:343–377. [DOI] [PubMed] [Google Scholar]
- Sabino C, Basic M, Bender D, Elgner F, Himmelsbach K, Hildt E.. Bafilomycin A1 and U18666A efficiently impair ZIKV infection. Viruses 2019;11:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Blando J, Tremmel L, DiGiovanni J.. Effect of metformin, rapamycin, and their combination on growth and progression of prostate tumors in HiMyc mice. Cancer Prev Res (Phila) 2015;8:597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T. et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A 2009;106:20842–20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008;456:264–268. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vera V, Kenchappa CS, Landberg K, Bressendorff S, Schwarzbach S, Martin T, Mundy J, Petersen M, Thelander M, Sundberg E.. Autophagy is required for gamete differentiation in the moss Physcomitrella patens. Autophagy 2017;13:1939–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM.. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 2004;14:1296–1302. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM.. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 2006;22:159–168. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM.. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005;307:1098–1101. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K.. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 2011;334:1141–1144. [DOI] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM.. mTOR signaling in growth, metabolism, and disease. Cell 2017;168:960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serke H, Vilser C, Nowicki M, Hmeidan FA, Blumenauer V, Hummitzsch K, Losche A, Spanel-Borowski K.. Granulosa cell subtypes respond by autophagy or cell death to oxLDL-dependent activation of the oxidized lipoprotein receptor 1 and toll-like 4 receptor. Autophagy 2009;5:991–1003. [DOI] [PubMed] [Google Scholar]
- Serra N, Velte EK, Niedenberger BA, Kirsanov O, Geyer CB.. The mTORC1 component RPTOR is required for maintenance of the foundational spermatogonial stem cell pool in micedagger. Biol Reprod 2019;100:429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra ND, Velte EK, Niedenberger BA, Kirsanov O, Geyer CB.. Cell-autonomous requirement for mammalian target of rapamycin (Mtor) in spermatogonial proliferation and differentiation in the mousedagger. Biol Reprod 2017;96:816–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesen J, Dahan P, Scotland SJ, Saland E, Dang VT, Lemarie A, Tyler BM, Brem H, Toulas C, Cohen-Jonathan Moyal E. et al. Metformin inhibits growth of human glioblastoma cells and enhances therapeutic response. PLoS One 2015;10:e0123721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, Ballabio A.. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 2013;14:283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadan S, James PS, Howes EA, Jones R.. Cholesterol efflux alters lipid raft stability and distribution during capacitation of boar spermatozoa. Biol Reprod 2004;71:253–265. [DOI] [PubMed] [Google Scholar]
- Shang Y, Wang H, Jia P, Zhao H, Liu C, Liu W, Song Z, Xu Z, Yang L, Wang Y. et al. Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy 2016;12:1575–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Codogno P.. Autophagic cell death: Loch Ness monster or endangered species? Autophagy 2011;7:457–465. [DOI] [PubMed] [Google Scholar]
- Shen J, Yang D, Zhou X, Wang Y, Tang S, Yin H, Wang J, Chen R, Chen J.. Role of autophagy in zinc oxide nanoparticles-induced apoptosis of mouse LEYDIG cells. Int J Mol Sci 2019;20:4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ.. Autophagy in health and disease: a double-edged sword. Science 2004;306:990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidjanin DJ, Park AK, Ronchetti A, Martins J, Jackson WT.. TBC1D20 mediates autophagy as a key regulator of autophagosome maturation. Autophagy 2016;12:1759–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JV, Cabral M, Correia BR, Carvalho P, Sousa M, Oliveira PF, Fardilha M.. mTOR signaling pathway regulates sperm quality in older men. Cells 2019;8:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JV, Freitas MJ, Correia BR, Korrodi-Gregorio L, Patricio A, Pelech S, Fardilha M.. Profiling signaling proteins in human spermatozoa: biomarker identification for sperm quality evaluation. Fertil Steril 2015;104:845–856.e8. [DOI] [PubMed] [Google Scholar]
- Sinha N, Puri P, Nairn AC, Vijayaraghavan S.. Selective ablation of Ppp1cc gene in testicular germ cells causes oligo-teratozoospermia and infertility in mice. Biol Reprod 2013;89:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA.. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 2011;12:246–258. [DOI] [PubMed] [Google Scholar]
- Skorupskaite K, George JT, Anderson RA.. The kisspeptin-GnRH pathway in human reproductive health and disease. Hum Reprod Update 2014;20:485–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somkuti SG, Lapadula DM, Chapin RE, Lamb JC, Abou-Donia MB.. Testicular toxicity following oral administration of tri-o-cresyl phosphate (TOCP) in roosters. Toxicol Lett 1987;37:279–290. [DOI] [PubMed] [Google Scholar]
- Song WH, Yi YJ, Sutovsky M, Meyers S, Sutovsky P.. Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc Natl Acad Sci U S A 2016;113:E5261–E5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song ZH, Yu HY, Wang P, Mao GK, Liu WX, Li MN, Wang HN, Shang YL, Liu C, Xu ZL, Sun QY. et al. Germ cell-specific Atg7 knockout results in primary ovarian insufficiency in female mice. Cell Death Dis 2015;6:e1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y. et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 2008;19:4762–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub C, Johnson L.. Review: spermatogenesis in the bull. Animal 2018;12:s27–s35. [DOI] [PubMed] [Google Scholar]
- Su Y, Quan C, Li X, Shi Y, Duan P, Yang K.. Mutual promotion of apoptosis and autophagy in prepubertal rat testes induced by joint exposure of bisphenol A and nonylphenol. Environ Pollut 2018;243:693–702. [DOI] [PubMed] [Google Scholar]
- Sullivan R, Mieusset R.. The human epididymis: its function in sperm maturation. Hum Reprod Update 2016;22:574–587. [DOI] [PubMed] [Google Scholar]
- Sun W, Yue J.. TPC2 mediates autophagy progression and extracellular vesicle secretion in cancer cells. Exp Cell Res 2018;370:478–489. [DOI] [PubMed] [Google Scholar]
- Sun Y, Shen J, Zeng L, Yang D, Shao S, Wang J, Wei J, Xiong J, Chen J.. Role of autophagy in di-2-ethylhexyl phthalate (DEHP)-induced apoptosis in mouse Leydig cells. Environ Pollut 2018;243:563–572. [DOI] [PubMed] [Google Scholar]
- Sung P, Klein H.. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol 2006;7:739–750. [DOI] [PubMed] [Google Scholar]
- Suo L, Chang X, Xu N, Ji H.. The anti-proliferative activity of GnRH through downregulation of the Akt/ERK pathways in pancreatic cancer. Front Endocrinol (Lausanne) 2019;10:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats S, Levay L, Boda A, Toth S, Simon-Vecsei Z, Rubics A, Varga A, Lippai M, Lorincz P, Glatz G. et al. The Warburg Micro Syndrome-associated Rab3GAP-Rab18 module promotes autolysosome maturation through the Vps34 Complex I. FEBS J 2021;288:190–211. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Baba T.. Gene expression in spermiogenesis. Cell Mol Life Sci 2005;62:344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Yamasaki M, Komatsu M, Ueno T.. The FAP motif within human ATG7, an autophagy-related E1-like enzyme, is essential for the E2-substrate reaction of LC3 lipidation. Autophagy 2012;8:88–97. [DOI] [PubMed] [Google Scholar]
- Tosti E, Menezo Y.. Gamete activation: basic knowledge and clinical applications. Hum Reprod Update 2016;22:420–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama Y, Maekawa M, Yuasa S.. Ectoplasmic specializations in the Sertoli cell: new vistas based on genetic defects and testicular toxicology. Anat Sci Int 2003;78:1–16. [DOI] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N.. Autophagy is essential for preimplantation development of mouse embryos. Science 2008;321:117–120. [DOI] [PubMed] [Google Scholar]
- Varmuza S, Jurisicova A, Okano K, Hudson J, Boekelheide K, Shipp EB.. Spermiogenesis is impaired in mice bearing a targeted mutation in the protein phosphatase 1cgamma gene. Dev Biol 1999;205:98–110. [DOI] [PubMed] [Google Scholar]
- Wang C, Tan C, Wen Y, Zhang D, Li G, Chang L, Su J, Wang X.. FOXP1-induced lncRNA CLRN1-AS1 acts as a tumor suppressor in pituitary prolactinoma by repressing the autophagy via inactivating Wnt/beta-catenin signaling pathway. Cell Death Dis 2019;10:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang Z, Xiong Z, Dai H, Zou Z, Jia C, Bai X, Chen Z.. mTORC1 activation promotes spermatogonial differentiation and causes subfertility in mice. Biol Reprod 2016;95:97. [DOI] [PubMed] [Google Scholar]
- Wang H, Wan H, Li X, Liu W, Chen Q, Wang Y, Yang L, Tang H, Zhang X, Duan E. et al. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res 2014;24:852–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HL, Fan SS, Pang M, Liu YH, Guo M, Liang JB, Zhang JL, Yu BF, Guo R, Xie J. et al. The Ankyrin repeat domain 49 (ANKRD49) augments autophagy of serum-starved GC-1 cells through the NF-kappaB pathway. PLoS One 2015;10:e0128551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhu H, Lin S, Wang K, Wang H, Liu Z.. Protective effect of naringenin against cadmium-induced testicular toxicity in male SD rats. J Inorg Biochem 2021;214:111310. [DOI] [PubMed] [Google Scholar]
- Wang L, Xu Z, Khawar MB, Liu C, Li W.. The histone codes for meiosis. Reproduction 2017a;154:R65–R79. [DOI] [PubMed] [Google Scholar]
- Wang M, Guo Y, Wang M, Zhou T, Xue Y, Du G, Wei X, Wang J, Qi L, Zhang H. et al. The Glial cell-derived neurotrophic factor (GDNF)-responsive phosphoprotein landscape identifies raptor phosphorylation required for spermatogonial progenitor cell proliferation. Mol Cell Proteomics 2017b;16:982–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang XF, Li YM, Chen N, Fan Y, Huang WK, Hu SF, Rao M, Zhang YZ, Su P.. Cross-talk between autophagy and apoptosis regulates testicular injury/recovery induced by cadmium via PI3K with mTOR-independent pathway. Cell Death Dis 2020;11:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Proud CG.. mTORC1 signaling: what we still don't know. J Mol Cell Biol 2011;3:206–220. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Yan J, Yin F, Li L, Qin YG, Meng CY, Lu RF, Guo L.. Role of autophagy in cadmium-induced testicular injury. Hum Exp Toxicol 2017;36:1039–1048. [DOI] [PubMed] [Google Scholar]
- Xiong M, Zhu Z, Tian S, Zhu R, Bai S, Fu K, Davis JG, Sun Z, Baur JA, Zheng K. et al. Conditional ablation of Raptor in the male germline causes infertility due to meiotic arrest and impaired inactivation of sex chromosomes. FASEB J 2017;31:3934–3949. [DOI] [PubMed] [Google Scholar]
- Xiong Z, Wang C, Wang Z, Dai H, Song Q, Zou Z, Xiao B, Zhao AZ, Bai X, Chen Z.. Raptor directs Sertoli cell cytoskeletal organization and polarity in the mouse testis. Biol Reprod 2018;99:1289–1302. [DOI] [PubMed] [Google Scholar]
- Xu H, Shen L, Chen X, Ding Y, He J, Zhu J, Wang Y, Liu X.. mTOR/P70S6K promotes spermatogonia proliferation and spermatogenesis in Sprague Dawley rats. Reprod Biomed Online 2016;32:207–217. [DOI] [PubMed] [Google Scholar]
- Xu LL, Liu ML, Wang JL, Yu M, Chen JX.. Saligenin cyclic-o-tolyl phosphate (SCOTP) induces autophagy of rat spermatogonial stem cells. Reprod Toxicol 2016;60:62–68. [DOI] [PubMed] [Google Scholar]
- Yan H, Li C, Zou C, Xin X, Li X, Li H, Li Y, Li Z, Wang Y, Chen H. et al. Perfluoroundecanoic acid inhibits Leydig cell development in pubertal male rats via inducing oxidative stress and autophagy. Toxicol Appl Pharmacol 2021;415:115440. [DOI] [PubMed] [Google Scholar]
- Yang F, Wei Y, Liao B, Wei G, Qin H, Pang X, Wang J.. Lycium barbarum polysaccharide prevents cisplatin-induced MLTC-1 cell apoptosis and autophagy via regulating endoplasmic reticulum stress pathway. Drug Des Devel Ther 2018;12:3211–3219. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP.. mTOR kinase structure, mechanism and regulation. Nature 2013;497:217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Ahmed N, Wang L, Chen H, Waqas Y, Liu T, Haseeb A, Bangulzai N, Huang Y, Chen Q.. In vivo autophagy and biogenesis of autophagosomes within male haploid cells during spermiogenesis. Oncotarget 2017;8:56791–56801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Tang H, Zhang Y, Zhang QF, Liu XY, Liu YT, Gu WT, Zheng YZ, Shang HB, Wang Y, Huang JY. et al. DEPTOR inhibits cell proliferation and confers sensitivity to dopamine agonist in pituitary adenoma. Cancer Lett 2019;459:135–144. [DOI] [PubMed] [Google Scholar]
- Yefimova MG, Buschiazzo A, Burel A, Lavault MT, Pimentel C, Jouve G, Jaillard S, Jegou B, Bourmeyster N, Ravel C.. Autophagy is increased in cryptorchid testis resulting in abnormal spermatozoa. Asian J Androl 2019;21:570–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ.. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 2006;441:475–482. [DOI] [PubMed] [Google Scholar]
- Yoshii SR, Kuma A, Akashi T, Hara T, Yamamoto A, Kurikawa Y, Itakura E, Tsukamoto S, Shitara H, Eishi Y. et al. Systemic analysis of Atg5-null mice rescued from neonatal lethality by transgenic ATG5 expression in neurons. Dev Cell 2016;39:116–130. [DOI] [PubMed] [Google Scholar]
- Yuan N, Song L, Zhang S, Lin W, Cao Y, Xu F, Fang Y, Wang Z, Zhang H, Li X. et al. Bafilomycin A1 targets both autophagy and apoptosis pathways in pediatric B-cell acute lymphoblastic leukemia. Haematologica 2015;100:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Ma D, Gao X, Wang J, Li R, Liu Z, Wang T, Wang S, Liu J, Liu X.. Liraglutide ameliorates erectile dysfunction via regulating oxidative stress, the RhoA/ROCK pathway and autophagy in diabetes mellitus. Front Pharmacol 2020;11:1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N.. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 2003;100:15077–15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotto-Filho A, Braganhol E, Klafke K, Figueiro F, Terra SR, Paludo FJ, Morrone M, Bristot IJ, Battastini AM, Forcelini CM. et al. Autophagy inhibition improves the efficacy of curcumin/temozolomide combination therapy in glioblastomas. Cancer Lett 2015;358:220–231. [DOI] [PubMed] [Google Scholar]
- Zhang C, Luo D, Li T, Yang Q, Xie Y, Chen H, Lv L, Yao J, Deng C, Liang X. et al. Transplantation of human urine-derived stem cells ameliorates erectile function and cavernosal endothelial function by promoting autophagy of corpus cavernosal endothelial cells in diabetic erectile dysfunction rats. Stem Cells Int 2019;2019:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li S, Li S, Zhang S, Wang Y, Jin S, Zhao C, Yang W, Liu Y, Fang D. et al. Effect of icariside II and metformin on penile erectile function, glucose metabolism, reaction oxygen species, superoxide dismutase, and mitochondrial autophagy in type 2 diabetic rats with erectile dysfunction. Transl Androl Urol 2020a;9:355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yao Y, Pan J, Guo X, Han X, Zhou J, Meng X.. Maternal exposure to Di-(2-ethylhexyl) phthalate (DEHP) activates the PI3K/Akt/mTOR signaling pathway in F1 and F2 generation adult mouse testis. Exp Cell Res 2020b;394:112151. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang X, Liu Y, Su Z, Dawar FU, Dan H, He Y, Gui JF, Mei J.. Leucine mediates autophagosome-lysosome fusion and improves sperm motility by activating the PI3K/Akt pathway. Oncotarget 2017;8:111807–111818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhu Y, Shi Y, Han Y, Liang C, Feng Z, Zheng H, Eng M, Wang J.. Fluoride-induced autophagy via the regulation of phosphorylation of mammalian targets of rapamycin in mice Leydig cells. J Agric Food Chem 2017;65:8966–8976. [DOI] [PubMed] [Google Scholar]
- Zhang M, Jiang M, Bi Y, Zhu H, Zhou Z, Sha J.. Autophagy and apoptosis act as partners to induce germ cell death after heat stress in mice. PLoS One 2012;7:e41412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li W, Zhang Y, Zhang L, Teves ME, Liu H, Strauss JF III, Pazour GJ, Foster JA, Hess RA, Zhang Z.. Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. Mol Biol Cell 2016;27:3705–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J-X, Liu H, Lv J, Yang X-J.. Wortmannin enhances cisplatin-induced apoptosis in human ovarian cancer cells in vitro. Eur Rev Med Pharmacol Sci 2014;18:2428–2434. [PubMed] [Google Scholar]
- Zhao X, Xu W, Wu J, Zhang D, Abou-Shakra A, Di L, Wang Z, Wang L, Yang F, Qiao Z.. Nicotine induced autophagy of Leydig cells rather than apoptosis is the major reason of the decrease of serum testosterone. Int J Biochem Cell Biol 2018;100:30–41. [DOI] [PubMed] [Google Scholar]
- Zheng W, Feng N, Wang Y, Noll L, Xu S, Liu X, Lu N, Zou H, Gu J, Yuan Y. et al. Effects of zearalenone and its derivatives on the synthesis and secretion of mammalian sex steroid hormones: a review. Food Chem Toxicol 2019;126:262–276. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li H, Li H, Nakagawa A, Lin JL, Lee ES, Harry BL, Skeen-Gaar RR, Suehiro Y, William D. et al. Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Science 2016;353:394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li H, Xue D.. Elimination of paternal mitochondria through the lysosomal degradation pathway in C. elegans. Cell Res 2011;21:1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ye S.. Rapamycin improves insulin resistance and hepatic steatosis in type 2 diabetes rats through activation of autophagy. Cell Biol Int 2018;42:1282–1291. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Shirakawa T, Ohbo K, Sada A, Wu Q, Hasegawa K, Saba R, Saga Y.. RNA binding protein Nanos2 organizes post-transcriptional buffering system to retain primitive state of mouse spermatogonial stem cells. Dev Cell 2015;34:96–107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article are available in the article.