Summary
Ectopic deposition of visceral adipose tissue (VAT) in abdomen is usually accompanied with systematic chaos of energy metabolism, a higher risk of cardiovascular diseases and type II diabetes. Here, we identified a previously unexplored gene Scd5 as a master regulator of fat distribution, which alone plays a significant role in determining the VAT accumulation. Firstly, zebrafish scd5 had the highest homology with human SCD5 compared to other SCDs in mouse and rat. We then observed that scd5-homozygous mutant zebrafish displayed a puffy, short and rounded apple-shaped figure. Whole-mount micro-CT scan showed that excessive VAT deposition and short spine are responsible for the abnormal body ratio. And the supplementation of ω3-polyunsaturated fatty acid (ω3-PUFA) in dietary significantly decreased VAT accumulation in scd5−/− zebrafish. Lastly, transcriptional analyses revealed that the Wnt, PPAR, C/EBP, and fat synthesis signaling pathways are significantly affected in the VAT of scd5−/− mutant and restored by ω3-PUFA.
Subject areas: Genetics, Molecular genetics, Molecular biology
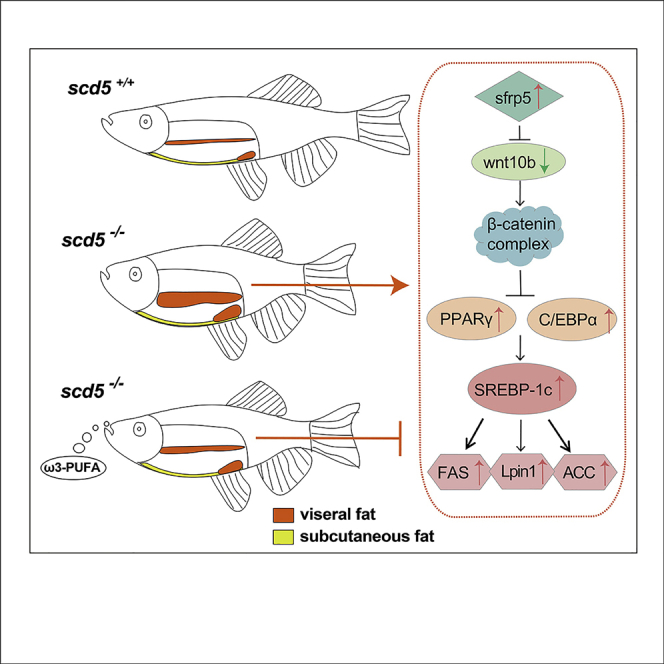
Graphical abstract
Highlights
-
•
Zebrafish scd5 is a better match of homolog to human SCD5
-
•
scd5 deficiency induced significant VAT depositions in zebrafish
-
•
Supplementation of ω3-PUFA significantly reduced the VAT deposition in scd5 mutants
Genetics; Molecular genetics; Molecular biology
Introduction
Obesity is a global epidemic and a primary risky factor for multiple metabolic diseases, and it also harms body shape fitness and self-confidence in physical appearance (González-Muniesa et al., 2017; Shungin et al., 2015). The overall mass and distribution of body fat are two primary aspects for analyzing obesity phenotypes, and both of which are of clinical and aesthetic medical interests (Elagizi et al., 2018; Rask-Andersen et al., 2019; Schleinitz et al., 2014; Tchernof and Després, 2013). Ectopic distribution of body fat, especially the “centralized adiposity”, is typically associated with the unfavorable “apple-shaped stature”. It is represented by excess visceral fat deposition in abdomen, with significantly higher level of serum lipids and chronic inflammatory factors, hence lead to higher risks of dyslipidemia, cardiovascular disease, and type 2 diabetes (T2D), even compared with the similar level of “total adiposity” (Canoy et al., 2007; Lotta et al., 2018; Oliveira et al., 2011; Wang et al., 2005).
Fat accumulation and distribution are regulated by both genetic and environmental factors, and shows significant heritability (González-Muniesa et al., 2017; Li and Lu, 2019). In genetics, obesity is commonly regarded as a complex polygenic phenotype, and a few master regulators were identified to be able to exert significant effects on fatty content and distribution (Cohen et al., 2014; Fathzadeh et al., 2020; Minchin et al., 2015). Currently, genome-wide association analysis (GWAS) have uncovered more than 300 single-nucleotide polymorphisms (SNPs) which are associated with BMI, WHR, or other adiposity traits (Goodarzi 2018; Shungin et al., 2015; Yılmaz and Gezmen Karadağ, 2020). Some of the locus is at or near the classic obesity-relevant genes, such as LEP, FTO, MC4R etc. However, the majority of the genes harboring or near those predicted locus are not investigated for their functions yet (Pigeyre et al., 2016). Among these, SCD5 is an interesting candidate of fat distribution regulators.
SCD5 is one of two stearoyl-CoA desaturases (SCDs) family members in human. SCDs are evolutionarily conserved endoplasmic reticulum (ER) enzymes in vertebrates which catalyzes the D9-cis desaturation of palmitoyl- and stearoyl-CoA, and converts them into palmitoleoyl- and oleoyl-CoA, respectively (Igal and Sinner, 2020; Paton and Ntambi, 2009). SCDs are critical intersections in lipid metabolism. SCD5, only conserved in several model animals, is rarely studied. All functional studies of SCD5 conducted so far were carried out on in vitro cultured cell models (Angelucci et al., 2018; Burhans et al., 2015; Lu et al., 2020). The other SCDs family member, SCD, has been extensively investigated for its roles in metabolism and several disease progression, including cancer in diverse mammalian organisms and cultured cells (Y. Li et al., 2017; Sampath et al., 2007).
Rodents are traditionally utilized to investigate obesity, and currently, zebrafish begins to rise as a novel model system. Zebrafish have highly similarity in lipid metabolism with mammals, and the small size of zebrafish facilitates the whole-body investigation and fast-track screening (Den Broeder et al., 2015; Salmerón 2018). Previously, we and other groups employed zebrafish to identify several fat distribution regulators of obesity, hepatic steatosis, and fat distribution, and showed that zebrafish are a feasible and convenient tools for genetic modification, phenotype observation, and drug screening (Fei et al., 2017, 2020; Loh et al., 2020; McMenamin et al., 2013; Minchin et al., 2015; Yao et al., 2018).
Here, we analyzed the homology of SCDs in zebrafish, mice, rat, and human, and zebrafish scd had the highest homology with human SCD5. Tissue expression profile of scd in zebrafish also had high similarity with SCD5 in human. Therefore, we generated the homozygous mutant zebrafish of scd (to prevent confusion with human SCD, referred to as scd5 in this study), which displayed significantly ectopic visceral fat accumulation at adult stages. Spines and body length of which are significantly shorter, BMI and the zWHtR are significantly increased. Further study indicates that mutation of scd5 inhibited the canonical Wnt-β-catenin signal and induced PPAR, C/EBP, and fat synthesis signaling pathways to promote visceral fat accumulation; the VAT accumulation also promotes fatty acid oxidative metabolism and gluconeogenesis. Dietary intervention of ω3-PUFA supplement on scd5−/− could significantly decrease visceral fat accumulation and restore these key genes.
Results
SCD5 is a candidate of fat distribution regulator
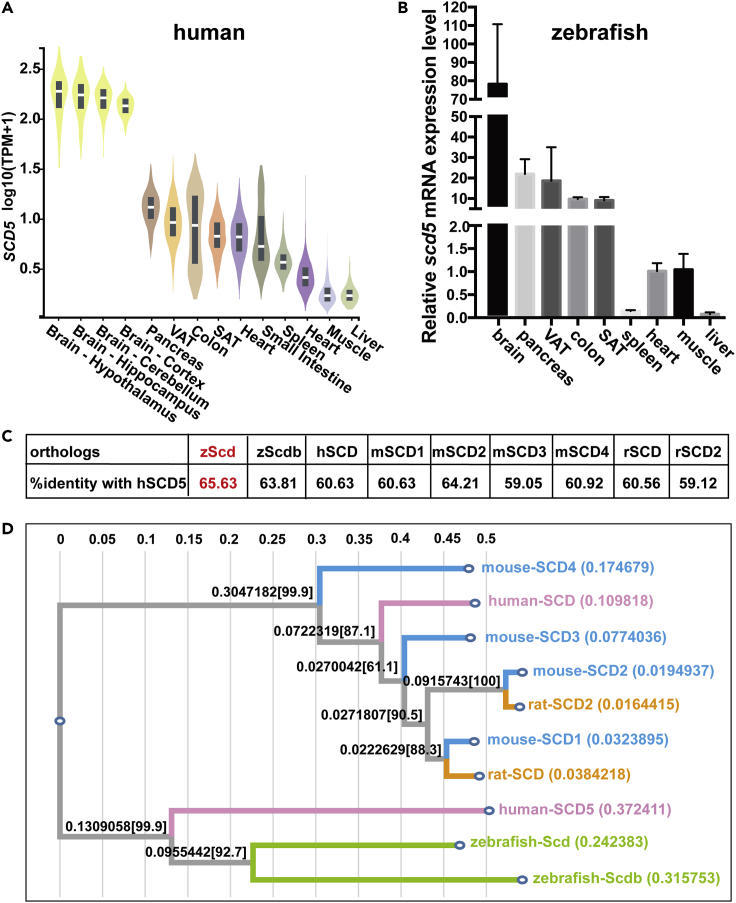
The waist-to-hip ratio adjusted for body mass index (WHRadjBMI) is an important indicator to describe centralized obesity. According to a meta-analysis conducted on 700,000 individuals of European ancestry (Pulit et al., 2019), three SNPs (rs10007189, rs17006523, and rs7682394) were significantly associated with WHRadjBMI (Table 1). The positions of the three SNPs are close to SCD5 (chr4: 82,629,539–82,798,796) at the chromosome, and eQTL data from GTEx demonstrating that the three SNPs affect the expression of SCD5 in brain and visceral adipose, respectively (GTEx Consortium, 2013) (Table 2). Given that SCD5 is predominantly expressed in brain, visceral fat, and pancreas (Figure 1A), this suggests that SCD5 plays an important role on fat distribution, and it is necessary to conduct functional studies on it.
Table 1.
Detailed information of three locus associated with WHRadjBMI
| CHR | |||||||
|---|---|---|---|---|---|---|---|
| POS | SNP | Tested Allele | Other Allele | Freq_Tested | Beta | P | N |
| 4: 84078182 | rs10007189 | A | G | 0.0423 | 0.0131 | 0.005472 | 626896 |
| 4: 84074997 | rs17006523 | T | C | 0.0392 | 0.0123 | 0.01157 | 615055 |
| 4: 83645606 | rs7682394 | C | A | 0.5536 | 0.0036 | 0.04949 | 626725 |
Table 2.
eQTL data of three locus associated with SCD5 expression
| Gene Symbol | SNP | Tested Allele | Other Allele | P | NES | Tissue |
|---|---|---|---|---|---|---|
| SCD5 | rs10007189 | A | G | 0.0000054 | -0.75 | Brain - Hypothalamus |
| SCD5 | rs17006523 | T | C | 0.0000054 | -0.75 | Brain - Hypothalamus |
| SCD5 | rs7682394 | C | A | 0.0000019 | -0.16 | Adipose - Visceral (Omentum) |
Figure 1.
Zebrafish Scd is a better match of homolog to human SCD5
(A) Tissue expression profile of SCD5 in human from GTEx.
(B) Tissue expression profile of Scd in zebrafish.
(C) Amino acid similarity between hSCD5 and Scd homologs in several model animals.
(D) Phylogenetic tree of SCDs based on amino acid sequences alignment. Zebrafish scd displayed highest homolog with human SCD5. The data are represented as mean ± SD
Zebrafish scd is highly homologous to human SCD5
Owing to a series of evolutionary events, the number of homologs of SCDs varies among species. Human have two SCD family members, SCD and SCD5. One of the commonly used model animals, the mouse, has four Scd family members, Scd1–Scd4, which have different tissue specificity (Cohen et al., 2002; Paton and Ntambi, 2009). Rat has two orthologs, Scd and Scd2. Zebrafish also has two orthologs, scd and scdb (Castro et al., 2011). Comparison of the amino acid sequences of these homologs with human SCD5 revealed that zebrafish Scd has the highest sequence identity, 65.63% (Figure 1C). And the orthologs' C-termini are more conserved compared to their N-termini (Figure S1). Protein evolution analysis of these homologs revealed higher homology between human SCD5 and zebrafish scds, especially scd, than that between mouse and rat scds (Figure 1D).
Further, we analyzed the expression levels of scd in major organs and tissues of zebrafish. Results showed that the relative expression of scd was at highest in zebrafish brain and significantly higher in pancreas and VAT than in colon, SAT, spleen, muscle, heart, and liver. The expression pattern was highly similar to that of SCD5 in human (Figures 1A and 1B). Therefore, zebrafish scd is a better match of homolog to human SCD5.
scd5-deficient zebrafish showed an apple-shaped body with visceral fat deposits and short spines
To investigate the function and molecular mechanism of scd5, we generated scd5 knockout zebrafish to carry out a series of analyses. As we previously reported, the scd5 mutant zebrafish with a 10bp deletion in the third exon were generated via CRISPR/Cas9 technique and the translation was early terminated at exon 4 (Figures 2A and S2) (Yao et al., 2018). The heterozygous mutant zebrafish displayed no apparent appearance changes in entire life, while homozygous mutant exhibited significant differences in body shape and body size after about 1mpf (Figure 2B).
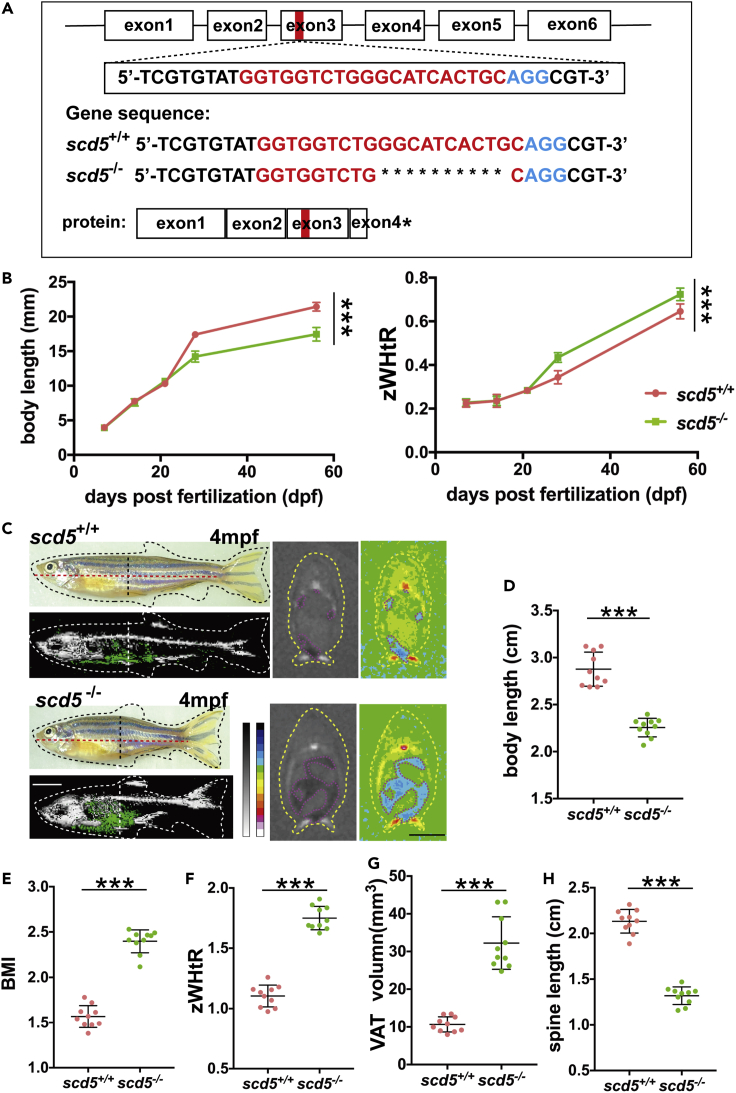
Figure 2.
scd5-deficient zebrafish showed an apple-shaped body with visceral fat deposits and short spines
(A) Diagram of target site on exon three of zebrafish scd5 genomic DNA, representative sequencing results of scd5+/+ and scd5−/− at six dpf larvae, red letter indicates N20 sequence, blue letter indicates NGG.
(B) scd5−/− zebrafish exhibited differences in body length and body shape at around 30 dpf compared to scd5+/+ (n = 20).
(C) Phenotype, the 3D reconstruction and section view of micro-CT scans on 4mpf scd5−/− zebrafish and their wild-type siblings. Black dotted lines indicate sectional position, red dotted line indicates body length, and yellow dotted curve indicates the outline of zebrafish. Blue area in section view indicates visceral adipose tissue. White scale bar = 5mm, Black scale bar = 2mm. D-H. 4mpf scd5−/− had significant decrease in body
(D) and spine (H) length, significant increases in BMI
(E), zWHtR (F) and VAT (G) (n = 10).
The data are represented as mean ± SD ∗∗∗p < 0.001 represents statistical significance compared with wild-type controls (unpaired t test).
Considering the known gender differences in body shape between male and female zebrafish, we used leaner and relatively uniformed male zebrafish for morphological observation and quantitative analyses (Löhr and Hammerschmidt 2011). Under pair-feeding conditions, four mpf scd5 homozygous knockout (scd5−/−) male adult zebrafish had puffy abdomen and significantly shorter body length than wild type (n = 10) (Figures 2C and 2D). BMI was calculated with weight and body length, and scd5−/− males displayed significantly increased BMI (Figure 2E). To better characterize the phenotype of body shape and fat distribution in zebrafish, we defined a zebrafish version of waist-height ratio to describe the degree of apple-shape: zWHtR (zebrafish version of WHtR, body circumference at the pelvic fin position-body length ratio), and scd5−/− zebrafish showed significantly increased zWHtR (Figure 2F). These results indicate that scd5−/− had an apple-shaped obese figure.
To directly observe the fat distribution and skeletal conditions in scd5 knockout zebrafish, we performed whole-mount noninvasive micro-CT to investigate abdominal adipose tissue and skeletal structure. Reconstruction of the 3D model showed increased abdominal visceral adipose tissue (VAT) in the scd5 deficiency model. Statistics of VAT volume showed significant increase, while SAT volume showed no change (Figures 2C, 2G and S3). In addition, spines of scd5−/− zebrafish were significantly short (Figure 2H), while measurements of the skull and tailfin showed no significant differences. These results indicate that the increase of abdomen circumference in scd5−/− was caused by ectopic visceral fat accumulation, while the shortened body length was attributed to the shorten spine.
ω3-PUFA significantly rescued the ectopic adipose deposition of scd5 mutant
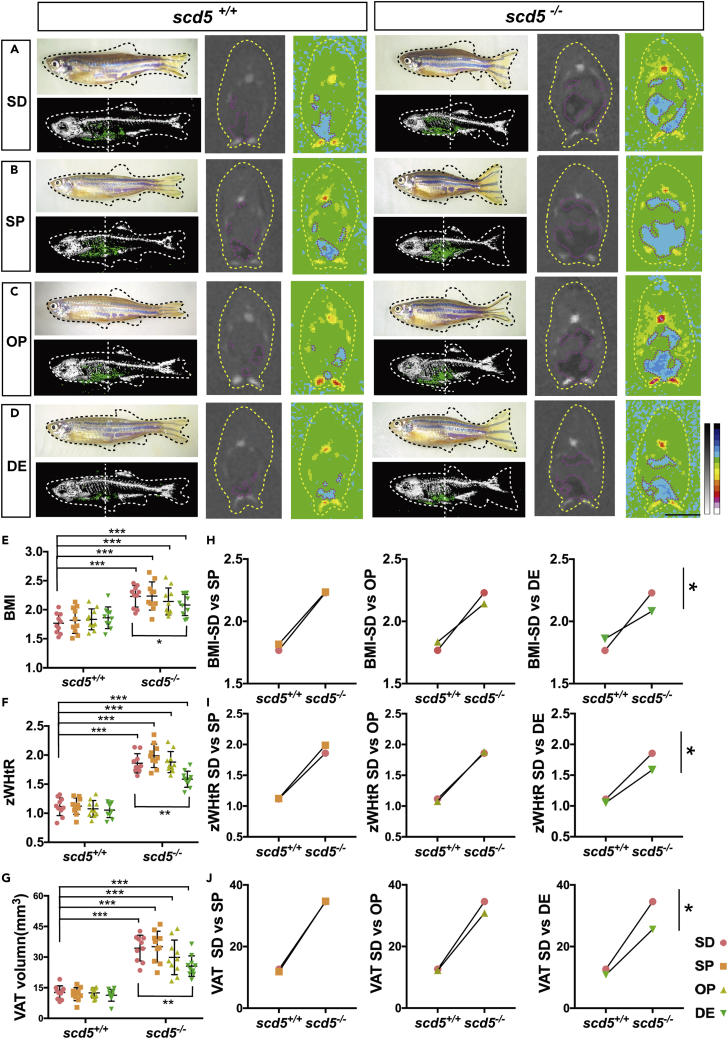
To investigate whether the apple-shaped obese figure caused by scd5 deficiency can be reversed, we performed a dietary intervention experiment. Four groups of 1-month-old male scd5+/+ and scd5−/− zebrafish were fed separately for three months (n = 10 in each group). The control group were fed on the standard diet (SD) and the experimental group was supplemented with different types of fatty acids in addition to SD Three combinations of fatty acids with different degrees of unsaturation were used in experimental group, saturated fatty acids (SFA) stearic acid and palmitic acid (SP), mono unsaturated fatty acids (MUFA) oleic acid and palmitoleic acid (OP), and ω3-PUFA DHA and EPA (DE) (see Table S2 for details of diet composition). After the treatment, micro-CT was performed to investigate fat distribution and skeletal condition (Figures 3A–3D). To compare the effects of the four different dietary, we measured body length, spine length, and VAT, calculated BMI and zWHtR, and performed two-way ANOVA analysis (Figures 3E–3G).
Figure 3.
ω3-PUFA significantly rescued the ectopic adipose deposition of scd5 mutant
(A–D) Appearance view, the 3D reconstruction, and section view of micro-CT scans on four different dietary supplementations fed scd5+/+ and scd5−/− zebrafish. White dotted vertical line indicates sectional position, blue area indicates adipose tissues. Scale bar = 2mm.
(E–G) Data statistics on BMI, zWHtR, and VAT volumes of standard diet (SD), SD plus SFA (SP), SD plus MUFA (OP), and SD plus ω3-PUFA (DE) groups. Data are represented as mean ± SD two-way ANOVA results of every groups compared to scd5+/+ -SD were shown, and the simple effects within the same genotypes were also shown. H-J, two-way ANOVA results of three diets vs control for scd5+/+ and scd5−/−. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 represents statistical significance of interaction between genotype and dietary. n = 10 for each group.
Firstly, we compared every groups to the scd5+/+ control diet (scd5+/+SD); the results showed that the scd5+/+ experimental groups (scd5+/+SP∖OP∖DE) displayed no significantly differences for all parameters. Regardless of the dietary formulation, scd5−/− had significantly higher BMI, zWHtR, and VAT values than scd5+/+SD, shorter body length and spine length, and unaffected SAT (Figures 3E–3G and S4A–S4C), which is consistent with former result.
Although the degree of obesity was heavier in scd5−/− than in scd5+/+, the degree of VAT deposition was different in dietary-intervened scd5−/− zebrafish. Considering the biochemical function of scd5, we hypothesized that MUFA would have stronger rescue effect. But in fact, surprisingly, it was ω3-PUFA. Two-way ANOVA showed significant interaction between genotype and dietary in zWHtR and VAT volumes, and multiple comparisons on scd5+/+ vs scd5−/− for each diet suggesting that ω3-PUFA supplementation reduced BMI, zWHtR, and VAT volumes (Figures 3H–3J and S4D–S4F). Another multiple comparison on simple effects within same genotype samples showed the same results; scd5−/−DE group showed decreased BMI, zWHtR, and VAT compared to the scd5−/−SD, while the scd5+/+ showed no differences between four dietaries (Figures 3E–3G). Given that the body length of scd5−/− was not significantly different in three FA treatment groups, ω3-PUFA mainly reduced the VAT accumulation, and the FA addition had no rescue effect on shortened spine due to scd5 deficiency.
Though the two-way ANOVA analysis showed no significance compared to SD control group, SP group showed a consistent increasing trend in the BMI, zWHtR, and VAT, while the OP group showed a decreasing trend in the three parameters (Table 3).
Table 3.
Percentage increase of scd5−/− over means of scd5+/+
| SD | SP | OP | DE | |
|---|---|---|---|---|
| BMI | 26.2% | 27.0% | 16.79% | 11.8% |
| zWHtR | 27% | 33.4% | 24.4% | 20.2% |
| VAT | 171.5% | 196.4% | 139.5% | 126.4% |
Four DEG sets were screened from RNA-seq analysis
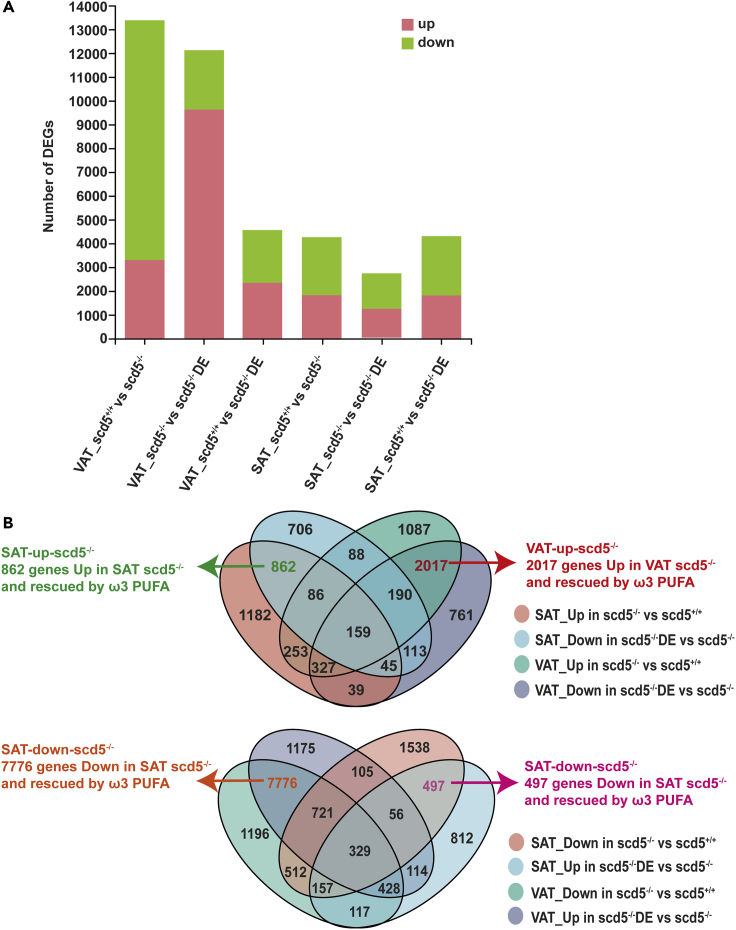
To explore the underlying molecular mechanisms beneath the phenotype change, we performed RNA-seq analysis. Differentially expressed genes (DEGs) were detected in VAT sample and SAT sample from three strains (scd5+/+, scd5−/−, and scd5−/− with dietary supplement of ω3-PUFA, referred to as scd5−/−DE) (Figure 4A) (Table S1. Detailed information of DEGs. Reference to Figure 4A).
Figure 4.
Transcriptome analysis of VAT of scd5−/− and scd5−/− DE
(A) Histogram of number of DEGs obtained from the comparative analysis between the six samples. Pink square indicates upregulated, green square indicates downregulated (n = 2).
(B) Venn analysis of DEGs. Each elliptical box represents different gene groups. Four genes sets were obtained by intersection. VAT-up-scd5−/− (2017 genes which were upregulated in VAT sample of scd5−/− compared to scd5+/+, restored in VAT sample of scd5−/− DE, and not significantly different in SAT samples of all three strains) , VAT-down-scd5−/− (7,776 genes which were downregulated in the VAT of scd5−/− compared to scd5+/+, restored in the VAT of scd5−/− DE, and have no significantly difference in the SAT of scd5−/− and scd5−/− DE), SAT-up-scd5−/− (862 genes which were upregulated in the SAT of scd5−/− compared to scd5+/+ and restored in the SAT of scd5−/− DE, and have no significantly difference in the VAT of all three strains), and SAT-down-scd5−/− (497 genes which were downregulated in the SAT of scd5−/− compared to scd5+/+, restored in the SAT of scd5−/− DE, and have no significantly difference in the VAT of scd5−/− and scd5−/− DE).
Results showed comparative analysis of VAT in scd5+/+ vs scd5−/− and scd5−/− vs scd5−/−DE both yielded more than 12,000 DEGs, and the former analysis detected a large proportion of downregulated genes in scd5−/−, while the latter analysis detected a larger proportion of upregulated genes in scd5−/−DE versus scd5−/−. The total number of DEGs for scd5−/−DE compared to scd5+/+ was reduced to less than 5,000, and the proportion of up-regulated genes to downregulated genes was equal (Figure 4A). DEGs of three comparative analysis of SAT sample also exhibit quantitative differences, although not as large as for VAT. Therefore, we cross-referenced up- and downregulated genes under different analyses to screen for meaningful sets of genes.
Venn analysis is shown in Figure 4B; four important gene sets were screened. We focused on those genes whose expression was significantly altered in VAT and SAT of scd5-deficient zebrafish and restored to normal levels by ω3-PUFA, named VAT-up-scd5−/−, VAT-down-scd5−/−, SAT-up-scd5−/−, and SAT-down-scd5−/−.
Clustering analysis indicates fat anabolism-associated pathways are important effector pathway of scd5 knockout
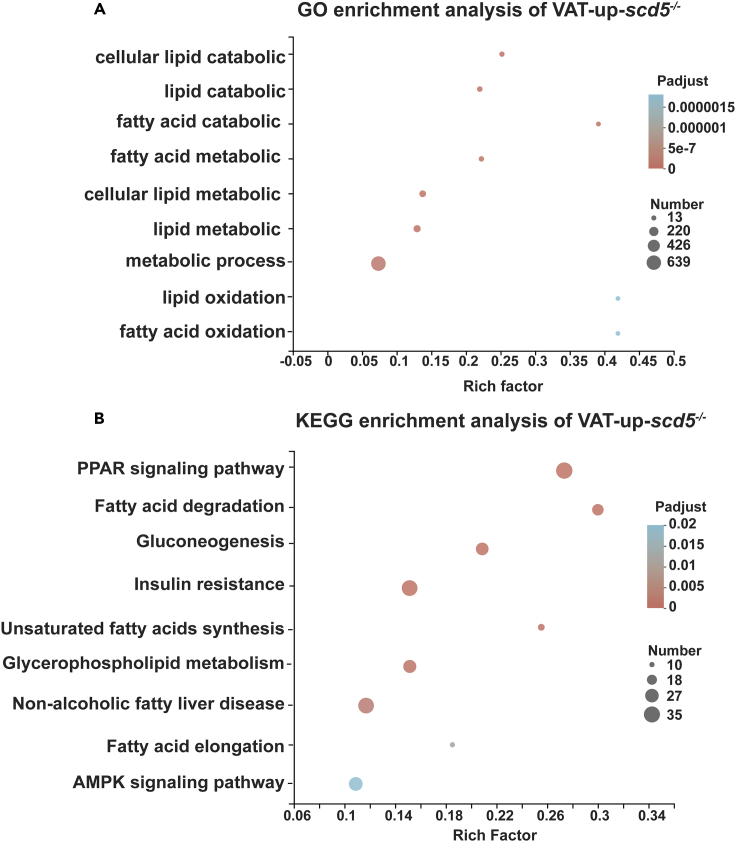
Clustering analysis of DEGs was conducted in the four gene sets. Gene Ontology (GO) clustering analysis regarding the biological processes (BP) was performed. Figure 5A shows significant GO terms on the VAT-up-scd5−/− gene sets, mainly clustered in lipid catabolism and metabolism-related processes. VAT-down-scd5−/− genes were mainly associated with RNA transcription and translation processes, SAT-up-scd5−/− clustering analysis showed no significant results, and SAT-down-scd5−/− clustering analysis was associated with chemotaxis and immune response (Figure S2).
Figure 5.
Significantly enriched pathways in VAT-up-scd5−/− gene sets
(A) GO enrichment analysis showed genes enriched in lipid anabolism-associated pathways.
(B) KEGG enrichment pathways include PPAR, C/EBP, fat synthesis, and fatty acid degradation/elongation pathways.
The latter three gene-sets-related pathways were indirectly correlated with VAT accumulation phenotype. We performed KEGG pathway enrichment analysis of the VAT-up-scd5−/− gene sets to screen for affected signaling pathways (Figure 5B). Results showed that the PPAR pathway, which plays an important role in fat differentiation (Janani and Ranjitha Kumari, 2015; Tang and Lane 2012; Y.-X. Wang 2010), was the most significant. Besides, fatty acid degradation, gluconeogenesis, insulin resistance, and non-alcoholic fatty liver disease (NAFLD) were also significantly affected in the scd5−/− mutant (Figure 5B).
The Wnt, PPAR, C/EBP, fat synthesis and fatty acid oxidation, and gluconeogenesis signaling pathways are significantly affected in the scd5−/− mutant and restored by ω3-PUFA
Analysis of the KEGG pathway yielded distinct differentially enriched pathways, including cellular signaling pathways such as the PPAR pathway and disease-related pathways such as insulin sensitivity, NAFLD, etc. In-depth analysis of these pathways will further reveal the cellular signaling networks acted upon by scd5.
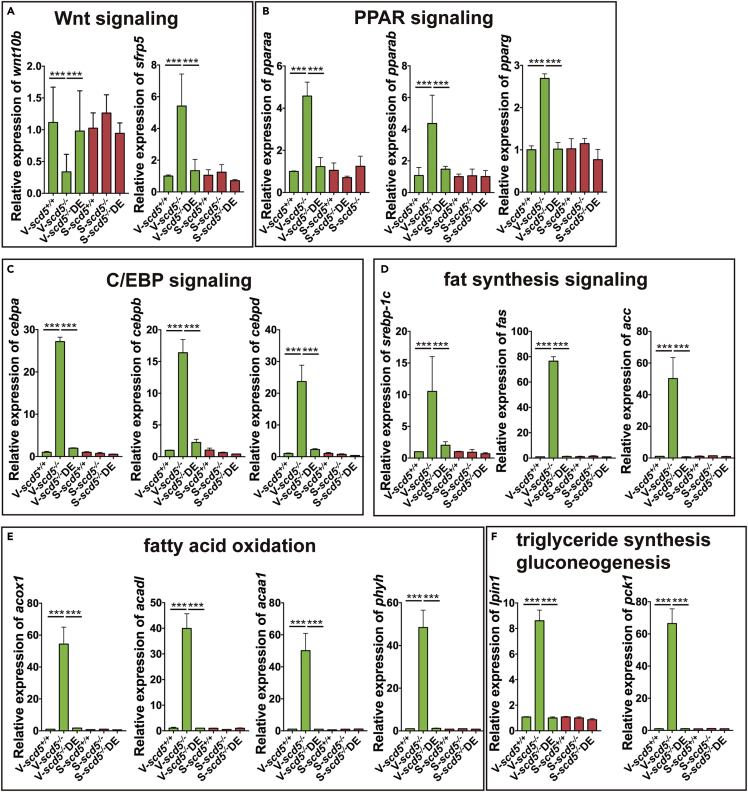
For instance, the insulin resistance pathway and its key node gene srebp-1c, fas, and acc which are closely related to adipogenesis and fatty acid biosynthesis were affected (Kim et al., 1998; Payne et al., 2009). In the NAFLD pathway, srebp-1c, PPARs, and C/EBPs were significantly affected. Our previous work suggested a potential relationship between the Wnt signaling pathway and SCD (Yao et al., 2018). SCD5 shares the same biochemical functions as SCD, which gives us a hint that SCD5 might also be associated with Wnt signaling pathways. Results showed that canonical Wnt signal directly affected the upstream of PPARγ (Cristancho and Lazar 2011; Tang and Lane 2012). All these affected genes were restored in VAT sample of ω3-PUFA fed zebrafish.
We performed qPCR assays for key nodes in these signaling pathways. Results showed that the expression of wnt10b, which regulates Wnt signaling, was suppressed in VAT sample of scd5−/−, and its inhibitor sfrp5 was upregulated, as were PPAR, C/EBP signaling, and fatty acid synthesis-related genes srebp-1c, fas, acc, etc (Figures 6A–6D). The expression of these genes was restored in scd5−/−DE. All these genes were unaffected in SAT sample.
Figure 6.
Relative expression of key nodes from main enriched pathways
(A–F). Main enriched pathways include Wnt, PPAR, C/EBP, fat synthesis, fatty acid oxidation, triglyceride synthesis, and gluconeogenesis signaling pathways. n = 4. The data are represented as mean ± SD ∗∗∗p < 0.001 represents statistical significance compared with wild-type controls (one-way ANOVA).
The GO-enriched lipolytic and metabolic pathway effector genes were largely overlapped, and we analyzed expression levels of several key node genes using qPCR. These genes include fatty acid beta-oxidation pathway-related acox1, acadl, and acaa1, alpha-oxidation of 3-methyl branched fatty acids pathway-related phyh, and gluconeogenesis regulates associated pck1, and lpin1, which encodes enzyme that catalyzes the penultimate step in triglyceride synthesis including the dephosphorylation of phosphatidic acid to yield diacylglycerol ((Fang et al., 2021; He et al., 2020; Schofield and McDonough 2007; Wang et al., 2021; Xu et al., 2020). Results showed that all these genes were significantly upregulated in VAT of scd5−/−, and restored by ω3-PUFA (Figures 6E–6F).
These results indicate that scd5 knockout leads to suppression of canonical Wnt signaling, activation of PPAR, C/EBP, fat synthesis, fatty acid oxidation, triglyceride synthesis, and gluconeogenesis in VAT, and that ω3-PUFA supplementation rescued the abnormalities at the genetic level.
Discussion
Characterization of Scd5 as a novel master regulator of VAT accumulation and fat distribution
Data from the GIANT and UK Biobank WHRadjBMI meta-analysis and the GTEx database suggest that human Scd5 is a strong candidate for fat distribution regulator (Tables 1 and 2). Interestingly, Scd5 is evolutionally conserved in humans, zebrafish, chimpanzees, and cattle except the most popular rodent model mouse and rat (Castro et al., 2011) (Figure 1). In addition, expression analysis in various major tissues also showed that the expression pattern of zebrafish scd is similar to that of human Scd5. Therefore, in this study, to prevent confusion, we refer to zebrafish scd as scd5. This research conducted the first functional study of scd5 using zebrafish and it turns out that scd5 monogenic mutation resulted in a very pronounced VAT accumulation effect and shorter spine length, while SAT volume was unaffected (Figure S3).
Validation of scd5 as another master regulator of fat accumulation and distribution added a new piece of jigsaw to comprehensive understanding of the mechanism. Also, identification of such master regulators will benefit both the accurate diagnosis and intervention of obesity-relevant diseases.
The regulatory mechanism and functional differences between SCD5 and SCD in fatty tissue accumulation and distribution
Humans have two SCDs, SCD5 and SCD, both of which play rate-limiting enzymes in the biosynthesis of MUFA, only the mechanism and location of expression are very different.
In scd5−/− zebrafish VAT, the ligand wnt10b of the canonical Wnt pathway was significantly downregulated compared to wild type, whereas the repressor protein sfrp5 was upregulated (Figure 6A). Inhibition of the wnt10b-mediated canonical pathway was clinically observed to be associated with the obesity phenotype (Christodoulides et al., 2006; R. Wang et al., 2014) because the wnt10b-mediated pathway inhibits the expression of PPARγ and C/EBPα, keeping adipocyte progenitors in an undifferentiated state (Ross et al., 2000). Once the lipogenesis process begins, the PPARγ and C/EBPα promote the expression of srebp-1c as well as downstream fas, acc, and lipin1 (Hu et al., 2012; Kim et al., 1998; Lu et al., 2018). This regulatory process is represented in figure abstract in a concise flowchart to show how these genes are affected and interacted. Therefore, scd5 deficiency may promote differentiation of visceral adipose progenitor cells and adipose tissue accumulation by inhibiting the canonical Wnt pathway, and thus affecting the expression of PPAR, C/EBP, and fat synthesis pathway in VAT. In addition, fatty acid oxidation and gluconeogenesis pathway also induced in VAT sample of scd5−/−, indicated enhanced fat synthesis/metabolism flux.
SCD is widely expressed almost everywhere, while SCD5 is mainly expressed in visceral fat, brain, and pancreas (Igal and Sinner, 2020; Paton and Ntambi, 2009; Wang et al., 2005) (Figure 1A). Previous studies identified two SNPs associated with SCD5 expression in hypothalamus, which is the main center to integrate appetite signals (Yengo et al., 2018) (Table 2). AMPK and mTOR signaling pathway were two complementary mechanisms of central energy-sensing regulation (Hardie et al. 2012). In hypothalamus, activation of AMPK promotes neuropeptide Y (NPY) and leads to increased food intake, while it inhibits the anorexigenic neuropeptide proopiomelanocortin (POMC), which suppresses food intake under satiety conditions. In addition, mTOR, which is inhibited by AMPK activation, reduces feeding behavior in response to leptin (Huynh et al., 2016). We analyzed relative expression level of these important signals; results showed the prkaa1 and npy were significantly activated, while the pomca and mtor were significantly inhibited (Figure S7). This indicates that AMPK signaling pathway in hypothalamus might be the potential candidates of scd5 effect.
Mouse Scd1 were homologous to human Scd. Quite the opposite of scd5−/− zebrafish, Scd1 knockout mice exhibited a lipid-reducing, weight-losing phenotype (Ntambi et al., 2002) downregulated adipogenesis-related genes serbp-1c, fas, and acc may be directly responsible (Paton and Ntambi, 2009). Scd1 functioned in Wnt-YAP pathway activation loop, different from scd5 in zebrafish (Kikuchi and Tsukamoto 2020; Torres et al. 2019). SCD-associated SNP analysis was also consistent with animal experiments. A cohort study showed decreased BMI and waist circumference and improved insulin sensitivity in homozygous for rare alleles of four SNP loci of SCD (Warensjö et al., 2007).
Supplementation of ω3-PUFA could significantly intervene apple-shaped figure; refined and personalized dietary need to be further investigated
Based on our research and clinical data, scd5 is certainly a potentially interesting therapeutic target, and dietary supplementation with a combination of both types of ω3-PUFA, EPA and DHA could substantially reduce VAT accumulation. For non-fatal but long-lasting health effects of apple-shaped body, dietary supplementation seems to be more appropriate than drastic measures such as gene therapy.
However, because SCD has the same biochemical function as SCD5, with opposite effects in fat accumulation and distribution, and its expression is widely distributed, it may be a hindrance to precise ω3-PUFA supplementation and intervention in apple-shaped body. In the dietary intervention experiment, supplementation with MUFA, the direct product of SCDs, had no significant rescue effect, probably because dietary intake of MUFA was distributed throughout the body, restoring the pathway of widely expressed SCD. Only a small fraction of MUFA acting on visceral fat, brain, and pancreas restored the pathway of scd5 action. In contrast, PUFA tends to act more in the brain (Lauritzen et al., 2016; Sun et al., 2018); thus, dietary intake of ω3-PUFA is more effective in rescuing scd5 mutations. Nevertheless, dietary supplementation of ω3-PUFA can also compensate for both SCD and SCD5, making it difficult to distinguish the desaturation function of both SCDs in the same compartment. In future, tissue-specific supplementation of ω3-PUFA may serve as a practical intervention for patients with apple-shaped stature due to SCD5 expression deficiency or decline.
Limitations of the study
Our article identifies scd5 as a novel regulator of visceral fat deposition, which has therapeutic and cosmetic value. Several immediate questions arise from our study. For example, scd5 deficiency in zebrafish leads to a short spine, suggesting that scd5 also plays an important role in spine development. Furthermore, considering that scd5 is a fatty acid desaturase, analysis of saturated/unsaturated fatty acid fluxes within the VAT following scd5 deficiency will provide insights into the mechanism of it.
Ethics approval and consent to participate
All procedures are approved by the institutional animal care committee of Children's Hospital of Fudan University, China.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Plasmid: pCas9 | Addgene | Cat:42876 |
| Chemicals, peptides, and recombinant proteins | ||
| Oleic acid | TCI | CAS: 112-80-1 |
| palmitoleic acid | TCI | CAS: 373-49-9 |
| Stearic acid | TCI | CAS: 57-11-4 |
| Palmitic acid | TCI | CAS: 57-10-3 |
| DHA | TCI | CAS: 6217-54-5 |
| Trizol | Ambion | Cat:15596-026 |
| EPA | TCI | CAS: 10417-94-4 |
| Critical commercial assays | ||
| PrimeScript™ RT Reagent Kit | TaKaRa | Cat:RR047A |
| Deposited data | ||
| SRA RNA-seq data | This paper | PRJNA770361 |
| Experimental models: Organisms/strains | ||
| AB strain | Our lab | N/A |
| Scd5-KO | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism | Prism | https://www.graphpad.com/scientific-software/prism |
| Amira | Thermo Fisher Scientific | https://www.thermofisher.cn/cn/zh/home/industrial/electron-microscopy/electron-microscopy-instruments-workflow-solutions/3d-visualization-analysis-software/amira-life-sciences-biomedical/amira-cell-biology.html |
| CLUSTALW | Kyoto University Bioinformatics Center | https://www.genome.jp/tools-bin/clustalw |
| Adobe Illustrator | Adobe | https://www.adobe.com/cn/products/illustrator.html |
| Image J | Schneider et al. (2012) | https://imagej.nih.gov/ij/ |
| Oligonucleotides | ||
| Primers for qPCR, see Table S3 | This paper | N/A |
Resource availability
Lead contact
Further information and requests about this study should be directed to and will be fulfilled by the lead contact, Qiang Li (liq@fudan.edu.cn).
Material availability
This study did not generate new unique reagents. All requests for resources and reagents should be directed to the lead contact and will be made available on request after completion of a Materials Transfer Agreement.
Experiment model and subject details
Animals
3-month-old male AB strain and mutant Zebrafish were maintained under standard conditions at 28.5°C in a 14 h/10 h light/dark cycle with approval by Animal Care Committee of Children's hospital of Fudan University.
Method details
Multiple alignment and phylogenetic trees analysis
Multiple alignments of SCDs homologs and phylogenetic trees analysis were performed using the CLUSTALW (https://www.genome.jp/tools-bin/clustalw).
CDS sequence accession number were as followed. human SCD5: NM_024906, human SCD: NM_005063, zebrafish scd: NM_198815, zebrafish scdb: NM_001020705, mouse Scd1: AF509567.1, mouse Scd2: NM_009128.2, mouse Scd3: NM_024450.2, mouse Scd4: NM_183216.3, rat Scd: NM_139192.2, rat Scd2: NM_031841.2.
Generation of scd5-/- strain
Generation of scd5 heterozygous mutant have been described previously(Yao et al. 2018), heterozygous mutants were incrossed to obtain homozygous mutants. Healthy, uniformly sized zebrafish were selected for the follow-up experimental program.
Micro-CT
Zebrafish were fixed overnight in 8% PFA at 4°C and embedded in 1% agarose; The samples were then scanned using the QuantumGX microCT Imaging System (PerkinElmer) with X-ray parameters set at 70 kV, 114 μA, 36 mm field diameter, and 72 μm resolution. Three-dimensional images were reconstructed using Analyze software (AnalyzeDirect Inc., Overland Park, USA), signals were processed in the grayscale range of 1,880-2,050 for adipose tissue and 2,600-9,000 for bone. Volume of VAT and SAT were analyzed with Amira software (Thermo Fisher, USA)
Dietary, feeding and measurement
FA supplemented diets were based on control diet (Larval AP100, Zeigler Bros, Inc. PA, USA), which formulated to meet the nutritional requirements of zebrafish. 70g FA/kg body weight were supplemented to control diet to make experimental diets, nutritional profile of each group sees Table S1. scd5+/+ and scd5-/- were randomly selected to different group. Zebrafish were fed 6% of wild type's mean body weight control diet, and weighed every week to adjust feed quantity. 1g fat produce 9 kcal of calories, 70g FA supplement increase 630 kcal caloric intake per kg body weight. 1g control diet produce 4.04 kcal of calories, to ensure that the caloric intake of zebrafish in each group was consistent, an additional 155.9 g control diet was added to the control group per kg body weight. At the end of the trial, zebrafish were anaesthetized to measure weight and body length, spine length and body circumference at pelvic fin position were measured after micro-CT with image J (Schneider et al. 2012). The VAT content was estimated by the following method. Figure 2A shows longitudinal section of each fish at the pelvic fin position. With this slice as the center, four more longitudinal sections were taken at 0.5 mm intervals anteriorly and posteriorly, for a total of nine sections covering the most abundant area of visceral fat in zebrafish. The area of VAT in each section was measured using Photoshop, and the summarized value was used as an indicator of the visceral fat content in zebrafish and for comparison and analysis between experimental groups.
RNA isolation, library construction, and high-throughput sequencing
Zebrafish were euthanized on ice. VAT and SAT sample were isolated by dissection using forceps, adipose tissues from 3 fish were pooled for one sample, and the total RNA was extracted using TRIzol® Reagent according the manufacturer’s instructions (Invitrogen) and genomic DNA was removed using DNase I (TaKara). Then RNA quality was determined by 2100 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). Only high-quality RNA sample (OD260/280=1.8∼2.2, OD260/230≥2.0, RIN≥6.5, 28S:18S≥1.0, >10μg) was used to construct sequencing library.
RNA-seq transcriptome librariy was prepared following TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA) using 5μg of total RNA. Shortly, messenger RNA was isolated according to polyA selection method by oligo(dT) beads and then fragmented by fragmentation buffer firstly. Secondly double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, CA) with random hexamer primers (Illumina). Then the synthesized cDNA was subjected to end-repair, phosphorylation and ‘A’ base addition according to Illumina’s library construction protocol. Libraries were size selected for cDNA target fragments of 200–300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by TBS380, paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq 4000 (2 × 150bp read length).
Bioinformatics analysis of RNA-Seq data
To identify DEGs between two different samples, the expression level of each transcript was calculated according to the fragments per kilobase of exon per million mapped reads (FRKM) method. RSEM (http://deweylab.biostat.wisc.edu/rsem/) was used to quantify gene abundances. R statistical package software EdgeR (Empirical analysis of Digital Gene Expression in R, http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html) was utilized for differential expression analysis, the screening threshold is: |log2FC| >=1 & padjust <0.05. In addition, functional-enrichment analysis including GO and KEGG were performed to identify which DEGs were significantly enriched in GO terms and metabolic pathways at Bonferroni-corrected P-value ≤0.05 compared with the whole-transcriptome background. GO functional enrichment and KEGG pathway analysis were carried out by Goatools and KOBAS.
Real-time quantitative polymerase chain reaction (RT-qPCR) analysis
RT-qPCR was performed in a CFX96 Real-Time System (BIO-RAD, USA). β-actin was employed as the internal standard. Real-time qPCR was performed using Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. The sequences of the primers are shown in Table S3. For each sample, the test and control reactions were run in triplicate. After the reaction, we used the threshold cycle (Ct) values of 2-ΔΔCT to calculate the relative expression in different samples by qRT software provided for the CFX96 Real-Time System.
Quantification and statistical analysis
Statistical analyses were performed using the Student’s t-test or one-way analysis of variance (ANOVA) with the Bonferroni–Dunn multiple comparison test, depending on the number of comparisons, using GraphPad Prism version 7 (GraphPad Software Inc., San Diego, CA, USA). A p-value less than 0.05 denoted a statistically significant difference.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant number: 81771632, 81271509) to Qiang Li, Natural Science Foundation of Shanghai (21ZR1410100) to Qiang Li, and National Key Research and Development Program of China (2020YFA0803202) to Xu Wang, National Natural Science Foundation of China (81402582, 82172884) to Xu Wang, Natural Science Foundation of Shanghai (18ZR1404500) to Xu Wang.
Author contributions
Qi Zhang: Conceptualization, Investigation, Writing—original draft. Shaoyang Sun: Methodology, Visualization. Yinglan Zhang: Methodology, Investigation. Xu Wang: Conceptualization, Writing—original draft, Supervision, Writing—review & editing. Qiang Li: Conceptualization, Supervision, Writing—review & editing.
Declaration of interests
Authors declare that they have no competing interests.
Published: March 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103916.
Contributor Information
Xu Wang, Email: wangxu2013@fudan.edu.cn.
Qiang Li, Email: liq@fudan.edu.cn.
Supplemental information
Data and code availability
Sequencing data reported in this paper has been deposited to SRA (PRJNA770361) and is publically available.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Reference
- Angelucci C., D'Alessio A., Iacopino F., Proietti G., Di Leone A., Masetti R., Sica G. Pivotal role of human stearoyl-CoA desaturases (SCD1 and 5) in breast cancer progression: oleic acid-based effect of SCD1 on cell migration and a novel pro-cell survival role for SCD5. Oncotarget. 2018;9:24364–24380. doi: 10.18632/oncotarget.25273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Broeder M.J., Marjo J., Kopylova V.A., Kamminga L.M., Legler J. Zebrafish as a model to study the role of peroxisome proliferating-activated receptors in adipogenesis and obesity. PPAR Res. 2015;2015:358029. doi: 10.1155/2015/358029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans M.S., Flowers M.T., Harrington K.R., Bond L.M., Guo C.A., Anderson R.M., Ntambi J.M. Hepatic oleate regulates adipose tissue lipogenesis and fatty acid oxidation. J. lipid Res. 2015;56:304–318. doi: 10.1194/jlr.M054429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canoy D., Boekholdt S.M., Wareham N., Luben R., Welch A., Bingham S., Buchan I., Day N., Khaw K.T. Body fat distribution and risk of coronary heart disease in men and women in the European prospective investigation into cancer and nutrition in norfolk cohort: a population-based prospective study. Circulation. 2007;116:2933–2943. doi: 10.1161/CIRCULATIONAHA.106.673756. [DOI] [PubMed] [Google Scholar]
- Castro L.F., Wilson J.M., Gonçalves O., Galante-Oliveira S., Rocha E., Cunha I. The evolutionary history of the stearoyl-CoA desaturase gene family in vertebrates. BMC Evol. Biol. 2011;11:132. doi: 10.1186/1471-2148-11-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulides C., Scarda A., Granzotto M., Milan G., Dalla Nora E., Keogh J., De Pergola G., Stirling H., Pannacciulli N., Sethi J.K., et al. WNT10B mutations in human obesity. Diabetologia. 2006;49:678–684. doi: 10.1007/s00125-006-0144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P., Miyazaki M., Socci N.D., Hagge-Greenberg A., Liedtke W., Soukas A.A., Sharma R., Hudgins L.C., Ntambi J.M., Friedman J.M. Role for stearoyl-CoA desaturase-1 in leptin-mediated weight loss. Science (New York, N.Y.) 2002;297:240–243. doi: 10.1126/science.1071527. [DOI] [PubMed] [Google Scholar]
- Cohen P., Levy J.D., Zhang Y., Frontini A., Kolodin D.P., Svensson K.J., Lo J.C., Zeng X., Ye L., Khandekar M.J. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho A.G., Lazar M.A. Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cel. Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagizi A., Kachur S., Lavie C.J., Carbone S., Pandey A., Ortega F.B., Milani R.V. An overview and update on obesity and the obesity paradox in cardiovascular diseases. Prog. Cardiovasc. Dis.diseases. 2018;61:142–150. doi: 10.1016/j.pcad.2018.07.003. [DOI] [PubMed] [Google Scholar]
- Fang X., Du Z., Duan C., Zhan S., Wang T., Zhu M., Shi J., Meng J., Zhang X., Yang M., et al. Beinaglutide shows significantly beneficial effects in diabetes/obesity-induced nonalcoholic steatohepatitis in ob/ob mouse model. Life Sci. 2021;270:118966. doi: 10.1016/j.lfs.2020.118966. [DOI] [PubMed] [Google Scholar]
- Fathzadeh M., Li J., Rao A., Cook N., Chennamsetty I., Seldin M., Zhou X., Sangwung P., Gloudemans M.J., Keller M., et al. FAM13A affects body fat distribution and adipocyte function. Nat. Commun. 2020;11:1465. doi: 10.1038/s41467-020-15291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei F., Sun S., Li Q., Pei Z., Wang L., Zhang R., Luo F., Yu M., Wang X. Combinatorial Normalization of Liver-Derived Cytokine Pathways Alleviates Hepatic Tumor-Associated Cachexia in Zebrafish. Cancer research. 2020;81:873–884. doi: 10.1158/0008-5472.CAN-20-2818. [DOI] [PubMed] [Google Scholar]
- Fei F., Sun .Y., Yao Y.-X., Wang X. Generation and phenotype analysis of zebrafish mutations of obesity-related genes lepr and mc4r. Acta Physiol. Sin. 2017;69:61–69. [PubMed] [Google Scholar]
- González-Muniesa P., Mártinez-González M.A., Hu F.B., Després J.P., Matsuzawa Y., Loos R.J.F., Moreno L.A., Bray G.A., Martinez J.A. Obesity. Nat. Rev. Dis. primers. 2017;3:17034. doi: 10.1038/nrdp.2017.34. [DOI] [PubMed] [Google Scholar]
- Goodarzi M.O. Genetics of obesity: what genetic association studies have taught us about the biology of obesity and its complications. Lancet. Diabetes Endocrinology. 2018;6:223–236. doi: 10.1016/S2213-8587(17)30200-0. [DOI] [PubMed] [Google Scholar]
- Hardie D.G., Ross F.A., Hawley S.A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cel. Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A., Chen X., Tan M., Chen Y., Lu D., Zhang X., Dean J.M., Razani B., Lodhi I.J. Acetyl-CoA derived from hepatic peroxisomal β-oxidation inhibits autophagy and promotes steatosis via MTORC1 activation. Mol. Cell. 2020;79:30–42.e4. doi: 10.1016/j.molcel.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Wang F., Li X., Rogers C.Q., Liang X., Finck B.N., Mitra M.S., Zhang R., Mitchell D.A., You M. Regulation of hepatic lipin-1 by ethanol: role of AMP-activated protein kinase/sterol regulatory element-binding protein 1 signaling in mice. Hepatol. (Baltimore, Md.) 2012;55:437–446. doi: 10.1002/hep.24708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh M.K., Kinyua A.W., Yang D.J., Kim K.W. Hypothalamic AMPK as a regulator of energy homeostasis. Neural plasticity. 2016;2016:2754078. doi: 10.1155/2016/2754078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igal R.A., Sinner D.I. Stearoyl-CoA desaturase 5 (SCD5), a Δ-9 fatty acyl desaturase in search of a function. Biochim. Biophys. Acta Mol. Cel. Biol. Lipids. 2020;1866:158840. doi: 10.1016/j.bbalip.2020.158840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janani C., Ranjitha Kumari B.D. PPAR gamma gene--a review. Diabetes Metab. Syndr. 2015;9:46–50. doi: 10.1016/j.dsx.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Tsukamoto H. Stearoyl-CoA desaturase and tumorigenesis. Chemico-biological interactions. 2020;316:108917. doi: 10.1016/j.cbi.2019.108917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.B., Wright H.M., Wright M., Spiegelman B.M. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc. Natl. Acad. Sci. U S A. 1998;95:4333–4337. doi: 10.1073/pnas.95.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen L., Brambilla P., Mazzocchi A., Harsløf L.B., Ciappolino V., Agostoni C. DHA effects in brain development and function. Nutrients. 2016;8:6. doi: 10.3390/nu8010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Lu Q. Gene-environment interactions on body fat distribution. Int. J. Mol. Sci. 2019;20:3690. doi: 10.3390/ijms20153690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Rong Y., Bao L., Nie B., Ren G., Zheng C., Amin R., Arnold R.D., Jeganathan R.B., Huggins K.W. Suppression of adipocyte differentiation and lipid accumulation by stearidonic acid (SDA) in 3T3-L1 cells. Lipids Health Dis. 2017;16:181. doi: 10.1186/s12944-017-0574-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh N.Y., Minchin J.E.N., Pinnick K.E., Verma M., Todorčević M., Denton N., Moustafa J.E., Kemp J.P., Gregson C.L., Evans D.M., et al. RSPO3 impacts body fat distribution and regulates adipose cell biology in vitro. Nat. Commun. 2020;11:2797. doi: 10.1038/s41467-020-16592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löhr H., Hammerschmidt M. Zebrafish in endocrine systems: recent advances and implications for human disease. Annu. Rev. Physiol. 2011;73:183–211. doi: 10.1146/annurev-physiol-012110-142320. [DOI] [PubMed] [Google Scholar]
- Lotta L.A., Wittemans L.B.L., Zuber V., Stewart I.D., Sharp S.J., Luan J., Day F.R., Li C., Bowker N., Cai L., et al. Association of genetic variants related to gluteofemoral vs abdominal fat distribution with type 2 diabetes, coronary disease, and cardiovascular risk factors. JAMA. 2018;320:2553–2563. doi: 10.1001/jama.2018.19329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Cao Y., Xiao J., Song M., Ho C.T. Molecular mechanisms of the anti-obesity effect of bioactive ingredients in common spices: a review. Food Funct. 2018;9:4569–4581. doi: 10.1039/c8fo01349g. [DOI] [PubMed] [Google Scholar]
- Lu X., Zhang Y., Chen L., Wang Q., Zeng Z., Dong C., Qi Y., Liu Y. Whole exome sequencing identifies SCD5 as a novel causative gene for autosomal dominant nonsyndromic deafness. Eur. J. Med. Genet. 2020;63:103855. doi: 10.1016/j.ejmg.2020.103855. [DOI] [PubMed] [Google Scholar]
- McMenamin S.K., Minchin J.E., Gordon T.N., Rawls J.F., Parichy D.M. Dwarfism and increased adiposity in the Gh1 mutant zebrafish vizzini. Endocrinology. 2013;154:1476–1487. doi: 10.1210/en.2012-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin J.E., Dahlman I., Harvey C.J., Mejhert N., Singh M.K., Epstein J.A., Arner P., Torres-Vázquez J., Rawls J.F. Plexin D1 determines body fat distribution by regulating the type V collagen microenvironment in visceral adipose tissue. Proc. Natl. Acad. Sci. U S A. 2015;112:4363–4368. doi: 10.1073/pnas.1416412112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntambi J.M., Miyazaki M., Stoehr J.P., Lan H., Kendziorski C.M., Yandell B.S., Song Y., Cohen P., Friedman J.M., Attie A.D. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc. Natl. Acad. Sci. U S A. 2002;99:11482–11486. doi: 10.1073/pnas.132384699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A., Lopes C., Severo M., Rodríguez-Artalejo F., Barros H. Body fat distribution and C-reactive protein--a principal component analysis. Nutr. Metab. Cardiovasc. Dis. 2011;21:347–354. doi: 10.1016/j.numecd.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Paton C.M., Ntambi J.M. Biochemical and physiological function of stearoyl-CoA desaturase. Am. J. Physiol. Endocrinol. Metab. 2009;297:E28–E37. doi: 10.1152/ajpendo.90897.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne V.A., Au W.S., Lowe C.E., Rahman S.M., Friedman J.E., O'Rahilly S., Rochford J.J. C/EBP transcription factors regulate SREBP1c gene expression during adipogenesis. Biochem. J. 2009;425:215–223. doi: 10.1042/BJ20091112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigeyre M., Fereshteh T.Y., Kaur Y., Meyre D. Recent progress in genetics, epigenetics and metagenomics unveils the pathophysiology of human obesity. Clin. Sci. 2016;130:943–986. doi: 10.1042/CS20160136. [DOI] [PubMed] [Google Scholar]
- Pulit S.L., Stoneman C., Morris A.P., Wood A.R., Glastonbury C.A., Tyrrell J., Yengo L., Ferreira T., Marouli E., Ji Y., et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019;28:166–174. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask-Andersen M., Torgny K., Weronica E.E., Johansson Å. Genome-wide association study of body fat distribution identifies adiposity loci and sex-specific genetic effects. Nat. Commun. 2019;10:339. doi: 10.1038/s41467-018-08000-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S.E., Hemati N., Longo K.A., Bennett C.N., Lucas P.C., Erickson R.L., MacDougald O.A. Inhibition of adipogenesis by wnt signaling. Science (New York, N.Y.) 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- Salmerón C. Adipogenesis in fish. J. Exp. Biol. 2018;221:jeb161588. doi: 10.1242/jeb.161588. [DOI] [PubMed] [Google Scholar]
- Sampath H., Miyazaki M., Dobrzyn A., Ntambi J.M. Stearoyl-CoA desaturase-1 mediates the pro-lipogenic effects of dietary saturated fat. J. Biol. Chem. 2007;282:2483–2493. doi: 10.1074/jbc.M610158200. [DOI] [PubMed] [Google Scholar]
- Schleinitz D., Böttcher Y., Blüher M., Kovacs P. The genetics of fat distribution. Diabetologia. 2014;57:1276–1286. doi: 10.1007/s00125-014-3214-z. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Wayne S.R., Eliceiri K.W. NIH image to ImageJ: 25 Years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield C.J., McDonough M.A. Structural and mechanistic studies on the peroxisomal oxygenase phytanoyl-CoA 2-hydroxylase (PhyH) Biochem. Soc. Trans. 2007;35:870–875. doi: 10.1042/BST0350870. [DOI] [PubMed] [Google Scholar]
- Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E., et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G.Y., Simonyi A., Fritsche K.L., Chuang D.Y., Hannink M., Gu Z., Greenlief C.M., Yao J.K., Lee J.C., Beversdorf D.Q. Docosahexaenoic acid (DHA): an essential nutrient and a nutraceutical for brain health and diseases. Prostaglandins, Leukot. Essent. fatty Acids. 2018;136:3–13. doi: 10.1016/j.plefa.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q.Q., Lane M.D. Adipogenesis: from stem cell to adipocyte. Annu. Rev. Biochem. 2012;81:715–736. doi: 10.1146/annurev-biochem-052110-115718. [DOI] [PubMed] [Google Scholar]
- Tchernof A., Després J.P. Pathophysiology of human visceral obesity: an update. Physiol. Rev. 2013;93:359–404. doi: 10.1152/physrev.00033.2011. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres V.I., Godoy J.A., Inestrosa N.C. Modulating wnt signaling at the root: porcupine and wnt acylation. Pharmacol. Ther. 2019;198:34–45. doi: 10.1016/j.pharmthera.2019.02.009. [DOI] [PubMed] [Google Scholar]
- Wang J., Yu L., Schmidt R.E., Su C., Huang X., Gould K., Cao G. Characterization of HSCD5, a novel human stearoyl-CoA desaturase unique to primates. Biochem. Biophys. Res. Commun. 2005;332:735–742. doi: 10.1016/j.bbrc.2005.05.013. http://www.sciencedirect.com/science/article/pii/S0006291X05009617 [DOI] [PubMed] [Google Scholar]
- Wang R., Hong J., Liu R., Chen M., Xu M., Gu W., Zhang Y., Ma Q., Wang F., Shi J., et al. SFRP5 acts as a mature adipocyte marker but not as a regulator in adipogenesis. J. Mol. Endocrinol. 2014;53:405–415. doi: 10.1530/JME-14-0037. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li X., Cao Y., Xiao C., Liu Y., Jin H., Cao Y. Effect of the ACAA1 gene on preadipocyte differentiation in sheep. Front. Genet. 2021;12:649140. doi: 10.3389/fgene.2021.649140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.X. PPARs: diverse regulators in energy metabolism and metabolic diseases. Cell Res. 2010;20:124–137. doi: 10.1038/cr.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Rimm E.B., Stampfer M.J., Willett W.C., Hu F.B. Comparison of abdominal adiposity and overall obesity in predicting risk of type 2 diabetes among men. Am. J. Clin. Nutr. 2005;81:555–563. doi: 10.1093/ajcn/81.3.555. [DOI] [PubMed] [Google Scholar]
- Warensjö E., Ingelsson E., Lundmark P., Lannfelt L., Syvänen A.C., Vessby B., Risérus U. Polymorphisms in the SCD1 gene: associations with body fat distribution and insulin sensitivity. Obes. (Silver Spring Md.) 2007;15:1732–1740. doi: 10.1038/oby.2007.206. [DOI] [PubMed] [Google Scholar]
- Xu D., Wang Z., Xia Y., Shao F., Xia W., Wei Y., Li X., Qian X., Lee J.H., Du L., et al. The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature. 2020;580:530–535. doi: 10.1038/s41586-020-2183-2. [DOI] [PubMed] [Google Scholar]
- Yao Y., Sun S., Wang J., Fei F., Dong Z., Ke A.W., He R., Wang L., Zhang L., Ji M.B., et al. Canonical wnt signaling remodels lipid metabolism in zebrafish hepatocytes following ras oncogenic insult. Cancer Res. 2018;78:5548–5560. doi: 10.1158/0008-5472.CAN-17-3964. [DOI] [PubMed] [Google Scholar]
- Yengo L., Sidorenko J., Kemper K.E., Zheng Z., Wood A.R., Weedon M.N., Frayling T.M., Hirschhorn J., Yang J., Visscher P.M. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yılmaz B., Gezmen Karadağ M. The current review of adolescent obesity: the role of genetic factors. J. Pediatr. Endocrinol. Metab. JPEM. 2020;34:151–162. doi: 10.1515/jpem-2020-0480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data reported in this paper has been deposited to SRA (PRJNA770361) and is publically available.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.