Abstract
Background
Complete deletion of both the short arm of chromosome 1 (1p) and the long arm of chromosome 19 (19q), known as 1p/19q codeletion, is a mutation that can occur in gliomas. It occurs in a type of glioma known as oligodendroglioma and its higher grade counterpart known as anaplastic oligodendroglioma. Detection of 1p/19q codeletion in gliomas is important because, together with another mutation in an enzyme known as isocitrate dehydrogenase, it is needed to make the diagnosis of an oligodendroglioma. Presence of 1p/19q codeletion also informs patient prognosis and prediction of the best drug treatment. The main two tests in use are fluorescent in situ hybridisation (FISH) and polymerase chain reaction (PCR)‐based loss of heterozygosity (LOH) assays (also known as PCR‐based short tandem repeat or microsatellite analysis). Many other tests are available. None of the tests is perfect, although PCR‐based LOH is expected to have very high sensitivity.
Objectives
To estimate the sensitivity and specificity and cost‐effectiveness of different deoxyribonucleic acid (DNA)‐based techniques for determining 1p/19q codeletion status in glioma.
Search methods
We searched MEDLINE, Embase and BIOSIS up to July 2019. There were no restrictions based on language or date of publication. We sought economic evaluation studies from the results of this search and using the National Health Service Economic Evaluation Database.
Selection criteria
We included cross‐sectional studies in adults with glioma or any subtype of glioma, presenting raw data or cross‐tabulations of two or more DNA‐based tests for 1p/19q codeletion. We also sought economic evaluations of these tests.
Data collection and analysis
We followed procedures outlined in the Cochrane Handbook for Diagnostic Test Accuracy Reviews. Two review authors independently screened titles/abstracts/full texts, performed data extraction, and undertook applicability and risk of bias assessments using QUADAS‐2. Meta‐analyses used the hierarchical summary ROC model to estimate and compare test accuracy. We used FISH and PCR‐based LOH as alternate reference standards to examine how tests compared with those in common use, and conducted a latent class analysis comparing FISH and PCR‐based LOH. We constructed an economic model to evaluate cost‐effectiveness.
Main results
We included 53 studies examining: PCR‐based LOH, FISH, single nucleotide polymorphism (SNP) array, next‐generation sequencing (NGS), comparative genomic hybridisation (CGH), array comparative genomic hybridisation (aCGH), multiplex‐ligation‐dependent probe amplification (MLPA), real‐time PCR, chromogenic in situ hybridisation (CISH), mass spectrometry (MS), restriction fragment length polymorphism (RFLP) analysis, G‐banding, methylation array and NanoString. Risk of bias was low for only one study; most gave us concerns about how patients were selected or about missing data. We had applicability concerns about many of the studies because only patients with specific subtypes of glioma were included. 1520 participants contributed to analyses using FISH as the reference, 1304 participants to analyses involving PCR‐based LOH as the reference and 262 participants to analyses of comparisons between methods from studies not including FISH or PCR‐based LOH.
Most evidence was available for comparison of FISH with PCR‐based LOH (15 studies, 915 participants): PCR‐based LOH detected 94% of FISH‐determined codeletions (95% credible interval (CrI) 83% to 98%) and FISH detected 91% of codeletions determined by PCR‐based LOH (CrI 78% to 97%). Of tumours determined not to have a deletion by FISH, 94% (CrI 87% to 98%) had a deletion detected by PCR‐based LOH, and of those determined not to have a deletion by PCR‐based LOH, 96% (CrI 90% to 99%) had a deletion detected by FISH. The latent class analysis suggested that PCR‐based LOH may be slightly more accurate than FISH. Most other techniques appeared to have high sensitivity (i.e. produced few false‐negative results) for detection of 1p/19q codeletion when either FISH or PCR‐based LOH was considered as the reference standard, although there was limited evidence. There was some indication of differences in specificity (false‐positive rate) with some techniques. Both NGS and SNP array had high specificity when considered against FISH as the reference standard (NGS: 6 studies, 243 participants; SNP: 6 studies, 111 participants), although we rated certainty in the evidence as low or very low. NGS and SNP array also had high specificity when PCR‐based LOH was considered the reference standard, although with much more uncertainty as these results were based on fewer studies (just one study with 49 participants for NGS and two studies with 33 participants for SNP array).
G‐banding had low sensitivity and specificity when PCR‐based LOH was the reference standard. Although MS had very high sensitivity and specificity when both FISH and PCR‐based LOH were considered the reference standard, these results were based on only one study with a small number of participants. Real‐time PCR also showed high specificity with FISH as a reference standard, although there were only two studies including 40 participants.
We found no relevant economic evaluations. Our economic model using FISH as the reference standard suggested that the resource‐optimising test depends on which measure of diagnostic accuracy is most important. With FISH as the reference standard, MLPA is likely to be cost‐effective if society was willing to pay GBP 1000 or less for a true positive detected. However, as the value placed on a true positive increased, CISH was most cost‐effective. Findings differed when the outcome measure changed to either true negative detected or correct diagnosis. When PCR‐based LOH was used as the reference standard, MLPA was likely to be cost‐effective for all measures of diagnostic accuracy at lower threshold values for willingness to pay. However, as the threshold values increased, none of the tests were clearly more likely to be considered cost‐effective.
Authors' conclusions
In our review, most techniques (except G‐banding) appeared to have good sensitivity (few false negatives) for detection of 1p/19q codeletions in glioma against both FISH and PCR‐based LOH as a reference standard. However, we judged the certainty of the evidence low or very low for all the tests. There are possible differences in specificity, with both NGS and SNP array having high specificity (fewer false positives) for 1p/19q codeletion when considered against FISH as the reference standard. The economic analysis should be interpreted with caution due to the small number of studies.
Keywords: Humans; Brain Neoplasms; Brain Neoplasms/genetics; Chromosomes, Human, Pair 1; Chromosomes, Human, Pair 1/genetics; Cost-Benefit Analysis; Cross-Sectional Studies; Diagnostic Tests, Routine; DNA; Glioma; Glioma/diagnosis; Glioma/genetics; Oligodendroglioma; State Medicine
Plain language summary
Comparing different methods of determining whether gliomas are missing arms 1p and 19q of the chromosomes
Why is improving the detection of 1p/19q codeletion in glioma important?
Gliomas are a type of brain tumour (cancer). There are different types of glioma, with different changes in their genetic material. One of the possible genetic changes is the loss of parts of two of our 23 chromosomes. When both a specific part of chromosome 1 and a specific part of chromosome 19 are missing, it is known as '1p/19q codeletion'. 1p/19q codeletion is used to diagnose a glioma known as an oligodendroglioma. Presence of 1p/19q codeletion can also tell us how long a patient with a glioma may survive and which is the best medicine to treat that patient.
What is the aim of this review?
We wanted to find out which is the most accurate and cost‐effective way to identify 1p/19q codeletion in gliomas.
What is studied in the review?
The review examined and compared all methods to detect 1p/19q codeletion that are based on the deoxyribonucleic acid (DNA, which contains the information for an organism to develop, survive and reproduce) of the tumour. These include tests known as FISH and CISH, which are performed directly on tumour tissue and a number of other tests that are based on DNA extracted from the tumour tissue including: PCR‐based LOH, real‐time PCR, MLPA, SNP array, CGH array and NGS. None of these tests is perfect, so there is no 'gold standard' against which to compare them. The two most commonly used tests (FISH and PCR‐based LOH) were used as the best available reference tests against which to examine the others.
What are the main results of the review?
We found 53 studies. Most tests were good at identifying instances of 1p/19q codeletion (meaning they were tests with good 'sensitivity') that had been identified by either of the two common tests. However, there were some differences in how well the tests were able to rule out 1p/19q codeletion when it did not seem to be present (the 'specificity' of the test). NGS and SNP arrays were better at this (i.e. having fewer 'false‐positives' results) when considered against FISH as the reference test. The cost per correct diagnosis was lowest for MLPA, although this was not a firm finding because the amount of evidence was small.
How reliable are results of the studies in this review?
Our certainty in the evidence was low or very low, because there were few studies for most of the tests and there were limitations to almost all the studies. Similarly, the economic analysis must be interpreted with caution due to the relatively small number of studies.
To whom do the results of this review apply?
The ways in which the tests were performed were thought to be representative of how they would be performed in practice. However, many of the studies included people with specific types of gliomas, so the results might not be representative of all people with gliomas.
What are the implications of this review?
The limited evidence suggests that currently used techniques show good sensitivity for detection of 1p/19q codeletion. NGS and SNP arrays may have higher specificity when FISH is the reference standard, but this comes at greater cost per test.
How up‐to‐date is this review?
The latest search for studies took place in August 2019.
Summary of findings
Summary of findings 1. Accuracy of tests for 1p/19q codeletion in people with glioma: assuming FISH is the reference standard.
|
Review question: what is the best method to detect 1p/19q codeletion in gliomas? Patients/population: adults with glioma Role: 1p/19q status is used for diagnosis, to inform treatment decisions and to give information on prognosis (survival) Index tests: any test Threshold for index tests: any threshold Reference standards: FISH Studies: cross‐sectional studies Setting: any setting; gliomas are typically diagnosed by a neuropathologist | ||||||
| Test | Number of participants (studies) | Accuracy | Overall prevalence (95% CrI) | Interpretation: assuming 31 people out of 100 with glioma will have a FISH‐detected 1p/19q codeletion and 69 people without the codeletion. | Certainty of the evidence (GRADE) | |
| Sensitivity (95% CrI) | Specificity (95% CrI) | |||||
| CISH | 38 (1) | 1.00 (0.84 to 1.00) | 0.92 (0.33 to 1.00) | 0.31 | 31 people will be given the correct positive result and 0 people will be given a false‐negative result. 63 people will be given a correct negative result and 6 people will be given a false‐positive result. |
Low‐certainty evidence: downgraded due to high imprecision. |
| PCR‐based LOH | 915 (15) | 0.94 (0.83 to 0.98) | 0.94 (0.87 to 0.98) | 0.31 | 29 people will be given the correct positive result and 2 people will be given a false‐negative result. 65 people will be given a correct negative result and 4 people will be given a false‐positive result. |
Low‐certainty evidence: downgraded due to risk of bias and indirectness. |
| Real‐time PCR | 40 (2) | 0.81 (0.20 to 0.99) | 1.00 (0.95 to 1.00) | 0.31 | 25 people will be given the correct positive result and 6 people will be given a false‐negative result. 69 people will be given a correct negative result and 0 people will be given a false‐positive result. |
Very low‐certainty evidence: downgraded due to high risk of bias, high imprecision and indirectness. |
| MLPA | 33 (2) | 0.96 (0.44 to 1.00) | 0.68 (0.20 to 0.95) | 0.31 | 30 people will be given the correct positive result and 1 person will be given a false‐negative result. 47 people will be given a correct negative result and 22 people will be given a false‐positive result. |
Very low‐certainty evidence: downgraded due to risk of bias, high imprecision and indirectness. |
| CGH | 75 (4) | 0.95 (0.59 to 1.00) | 0.99 (0.90 to 1.00) | 0.31 | 29 people will be given the correct positive result and 2 people will be given a false‐negative result. 68 people will be given a correct negative result and 1 person will be given a false‐positive result. |
Low‐certainty evidence: downgraded due to risk of bias and imprecision. |
| aCGH | 39 (3) | 1.00 (0.89 to 1.00) | 0.91 (0.55 to 0.99) | 0.31 | 31 people will be given the correct positive result and 0 people will be given a false‐negative result. 63 people will be given a correct negative result and 6 people will be given a false‐positive result. |
Very low‐certainty evidence: downgraded due to risk of bias, imprecision and indirectness. |
| SNP array | 111 (6) | 0.90 (0.57 to 0.99) | 0.97 (0.84 to 1.00) | 0.31 | 28 people will be given the correct positive result and 3 people will be given a false‐negative result. 67 people will be given a correct negative result and 2 people will be given a false‐positive result. |
Very low‐certainty evidence: downgraded due to risk of bias, imprecision and indirectness. |
| NGS | 243 (6) | 0.94 (0.75 to 0.99) | 1.00 (0.99 to 1.00) | 0.31 | 29 people will be given the correct positive result and 2 people will be given a false‐negative result. 69 people will be given a correct negative result and 0 people will be given a false‐positive result. |
Low‐certainty evidence: downgraded due to risk of bias and indirectness. |
| MS | 10 (1) | 1.00 (0.60 to 1.00) | 1.00 (0.70 to 1.00) | 0.31 | 31 people will be given the correct positive result and 0 people will be given a false‐negative result. 69 people will be given a correct negative result and 0 people will be given a false‐positive result. |
Very low certainty evidence: downgraded due to high risk of bias and imprecision. |
| NanoString | 16 (1) | 0.85 (0.11 to 1.00) | 0.80 (0.10 to 1.00) | 0.31 | 26 people will be given the correct positive result and 5 people will be given a false‐negative result. 55 people will be given a correct negative result and 14 people will be given a false‐positive result. |
Very low certainty evidence: downgraded due to high risk of bias and high imprecision. |
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CrI: credible interval; CISH: chromogenic in situ hybridisation; FISH: fluorescent in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS: mass spectrometry; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism.
Summary of findings 2. Accuracy of tests for 1p/19q codeletion in people with glioma: assuming PCR‐based LOH is the reference standard.
|
Review question: what is the best method to detect 1p/19q codeletion in gliomas? Patients/population: adults with glioma Role: 1p/19q status is used for diagnosis, to inform treatment decisions and to give information on prognosis (survival) Index tests: any test Threshold for index tests: any threshold Reference standards: PCR‐based LOH Studies: cross‐sectional studies Setting: any setting; gliomas are typically diagnosed by a neuropathologist | ||||||
| Test | Number of participants (studies) | Accuracy | Overall prevalence (95% CrI) | Interpretation: assuming 31 people out of 100 with glioma will have a PCR‐detected 1p/19q codeletion and 69 people without the codeletion | Certainty of the evidence (GRADE) | |
| Sensitivity (95% CrI) | Specificity (95% CrI) | |||||
| FISH | 915 (15) | 0.91 (0.78 to 0.97) | 0.96 (0.90 to 0.99) | 0.31 | 28 people will be given the correct positive result and 3 people will be given a false‐negative result. 66 people will be given a correct negative result and 3 people will be given a false‐positive result. |
Low‐certainty evidence: downgraded due to risk of bias and indirectness. |
| Real‐time PCR | 10 (1) | 1.00 (0.77 to 1.00) | NA | 0.31 | 31 people will be given the correct positive result and 0 people will be given a false‐negative result. Results are not provided for those without the codeletion. | Very low‐certainty evidence: downgraded due to risk of bias, imprecision and indirectness. |
| MLPA | 18 (1) | 1.00 (0.74 to 1.00) | 1.00 (0.83 to 1.00) | 0.31 | 31 people will be given the correct positive result and 0 people will be given a false‐negative result. 69 people will be given a correct negative result and 0 people will be given a false‐positive result. |
Very low‐certainty evidence: downgraded due to high risk of bias, imprecision and indirectness. |
| CGH | 151 (6) | 0.94 (0.74 to 0.99) | 0.98 (0.91 to 1.00) | 0.31 | 29 people will be given the correct positive result and 2 people will be given a false‐negative result. 68 people will be given a correct negative result and 1 people will be given a false‐positive result. |
Low‐certainty evidence: downgraded due to risk of bias and indirectness. |
| aCGH | 57 (4) | 1.00 (0.97 to 1.00) | 0.96 (0.75 to 1.00) | 0.31 | 31 people will be given the correct positive result and 0 people will be given a false‐negative result. 66 people will be given a correct negative result and 3 people will be given a false‐positive result. |
Low‐certainty evidence: downgraded due to high risk of bias. |
| SNP array | 33 (2) | 0.97 (0.50 to 1.00) | 1.00 (0.92 to 1.00) | 0.31 | 30 people will be given the correct positive result and 1 person will be given a false‐negative result. 69 people will be given a correct negative result and 0 people will be given a false‐positive result. |
Very low‐certainty evidence: downgraded due to risk of bias and high imprecision. |
| NGS | 49 (1) | 1.00 (0.86 to 1.00) | 0.98 (0.64 to 1.00) | 0.31 | 31 people will be given the correct positive result and 0 people will be given a false‐negative result. 68 people will be given a correct negative result and 1 people will be given a false‐positive result. |
Very low‐certainty evidence: downgraded due to risk of bias, imprecision and indirectness. |
| MS | 50 (1) | 1.00 (0.85 to 1.00) | 1.00 (0.94 to 1.00) | 0.31 | 31 people will be given the correct positive result and 0 people will be given a false‐negative result. 69 people will be given a correct negative result and 0 people will be given a false‐positive result. |
Very low‐certainty evidence: downgraded due to risk of bias and imprecision. |
| G‐banding | 21 (1) | 0.00 (0.00 to 0.20) | 1.00 (0.78 to 1.00) | 0.31 | 0 people will be given the correct positive result and 31 people will be given a false‐negative result. 69 people will be given a correct negative result and 0 people will be given a false‐positive result. |
Very low‐certainty evidence: downgraded due to high risk of bias, high imprecision and indirectness. |
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CrI: credible interval; CISH: chromogenic in situ hybridisation; FISH: fluorescent in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS: mass spectrometry; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism.
Background
Gliomas are a group of brain tumours arising within the central nervous system. Different types of gliomas can show different changes in genetic information. Some of these genetic changes can serve as diagnostic, prognostic and predictive biomarkers. Diagnostic biomarkers help to establish which specific type of glioma is present. Prognostic biomarkers give information about the likely clinical outcome or prognosis for a patient with glioma, and predictive biomarkers indicate the likelihood of response to a particular treatment. One of the possible genetic changes that may be present is the loss of parts of chromosome 1 and chromosome 19, known as codeletion of chromosomal arms 1p and 19q. 1p/19q codeletion is most commonly found in a type of glioma called an oligodendroglioma and is a diagnostic biomarker for this glioma. In addition, 1p/19q codeletion acts as a prognostic and predictive biomarker for glioma because it informs patient prognosis and treatment strategy.
In this review, we aimed to determine the most accurate way of testing whether a glioma has codeletion of chromosomal arms 1p and 19q. There are costs to patients, their families, health services and society in general associated with glioma. One review of studies found that the estimated cost of clinical care for a patient with glioma ranged between USD 4755 and USD 42,907 (reported costs were all converted into 2013 US dollars using an exchange rate based on purchasing power parities) (Messali 2014). These studies were carried out before particular chemotherapies became the standard of care for different types of glioma, which would also increase the treatment costs. In addition to an integrated full review of economic evaluations, this review features an economic decision model as a further level of evidence synthesis. The use of an economic decision model allows consideration of the resource implications of tests for diagnosis of codeletion of chromosomal arms 1p and 19q. We used this approach because we anticipated that we would identify limited economic evidence for inclusion in the review.
Target condition being diagnosed
Gliomas are thought to arise from stem or progenitor cells in the central nervous system and they share some features with glial cells. Glial cells have several functions including supporting and insulating neurons. Age‐adjusted incidence rates for all gliomas (ICD‐O‐3 morphology codes 9380–9480) range from 4.67 to 5.73 per 100,000 persons, with varied survival rates (Ostrom 2014). One review of population‐based studies found that the lowest grade glioma, called pilocytic astrocytoma (World Health Organization (WHO) grade I), has the highest five‐year relative survival rate at 57.3% to 97.3%; while the highest grade glioma, glioblastoma (WHO grade IV), has the poorest survival with only 0.1% to 8.9% of people surviving five years after diagnosis (Ostrom 2014).
Loss of a chromosome arm can be complete (where the whole chromosome arm is lost) or partial (where only part of the chromosomal arm is lost). Complete deletion of both the short arm of chromosome 1 (1p) and the long arm of chromosome 19 (19q) (1p/19q codeletion) is a mutation that can occur in gliomas. The codeletion is thought to be an early event in the development of cancer (Pinkham 2015), that is due to an unbalanced whole‐arm translocation between chromosomes 1 and 19 with the loss of the resulting hybrid chromosome (Griffin 2006; Jenkins 2006). As described below, the 1p/19q codeletion is a diagnostic, prognostic and predictive biomarker in glioma. We are not interested in partial loss of 1p or 19q (or both), as these partial deletions do not share the diagnostic, prognostic and predictive abilities of the complete 1p/19q codeletion.
According to the WHO, the diagnosis of oligodendroglioma (a type of glioma) and anaplastic (high‐grade) oligodendroglioma requires the demonstration of both an isocitrate dehydrogenase (IDH) gene family mutation and 1p/19q codeletion (Louis 2016).
One systematic review and meta‐analysis of the prognostic value of chromosomal 1p/19q codeletion in low‐grade (WHO grade II) and high‐grade/anaplastic (WHO grade III) tumours found a summary hazard ratio (HR) for mortality of 0.28 (95% confidence interval (CI) 0.13 to 0.62; 9 studies) favouring 1p/19q codeletion after adjusting for age, extent of resection, IDH‐1 mutation and type of therapy (Hu 2016). Another systematic review and meta‐analysis that evaluated the association between codeletion (versus no codeletion) of 1p/19q and overall survival among people with different grades and types of gliomas found that 1p/19q codeletion was associated with increased overall survival (HR 0.43, 95% CI 0.35 to 0.53; 14 studies) (Zhao 2014). There were similar results in both low‐grade tumours (HR 0.45, 95% CI 0.30 to 0.68; 5 studies) and high‐grade gliomas (HR 0.41, 95% CI 0.31 to 0.53; 6 studies). This is akin to the results also seen for astrocytic tumours (HR 0.52, 95% CI 0.36 to 0.75; 3 studies) and oligodendroglial tumours (HR 0.41, 95% CI 0.30 to 0.56; 9 studies) (Zhao 2014). This review also observed no evidence of a difference in the HR for overall survival between studies using two different techniques (polymerase chain reaction (PCR)‐based loss of heterozygosity (LOH) and fluorescence in situ hybridisation (FISH)) to assess the status of chromosomal arms 1p and 19q (Zhao 2014).
1p/19q codeletion predicts response to chemotherapy in anaplastic oligodendrogliomas. The European Organisation for Research and Treatment of Cancer (EORTC) study 26951 was a phase III trial comparing radiotherapy (RT) with RT plus adjuvant chemotherapy with procarbazine, lomustine and vincristine (PCV) in people with newly diagnosed anaplastic oligodendroglioma (van den Bent 2013). An exploratory analysis of long‐term follow‐up found a trend towards increased survival for people with 1p/19q codeletion from adjuvant PCV. In people with 1p/19q codeletion, fewer than half died during follow‐up in the RT plus PCV group (and therefore median overall survival was not reached) versus a median survival of 112 months in the RT group (HR 0.56, 95% CI 0.31 to 1.03) (van den Bent 2013). In people with non‐codeleted 1p/19q, the median overall survival was 25 months in the RT plus PCV group versus 21 months in the RT group (HR 0.83, 95% CI 0.62 to 1.10) (van den Bent 2013). Similarly, long‐term follow‐up of the Radiation Therapy Oncology Group (RTOG) study 9402, which also compared PCV plus RT with RT alone in people with pure and mixed anaplastic oligodendrogliomas, found that the median survival of those with codeleted tumours treated with PCV plus RT was twice that of people receiving RT (14.7 years with PCV plus RT versus 7.3 years with RT; HR 0.59, 95% CI 0.37 to 0.95; P = 0.03) (Cairncross 2013). For people with non‐codeleted tumours, there was no evidence of a difference in median survival by treatment arm (2.6 years with PCV plus RT versus 2.7 years with RT; HR 0.85, 95% CI 0.58 to 1.23; P = 0.39) (Cairncross 2013).
1p/19q codeletion 1p can be absolute (i.e. loss in the presence of the normal number of other chromosomes), or relative if it occurs in the presence of polysomy (when cells contain at least one more copy of a chromosome than normal) or polyploidy (when cells contain more than two sets of chromosomes). Several studies have suggested that people with relative 1p/19q codeletions (deletions in the presence of polysomy or polyploidy) have a worse prognosis (progression‐free survival or overall survival) than people with absolute 1p/19q codeletions, with some studies suggesting that prognosis in people with relative codeletions may be similar to that of people with no codeletion (Chamberlain 2015; Jiang 2014; Ren 2013; Snuderl 2009). In all these studies, classification of polysomy occurred when more than 30% of nuclei had more than two 1q and 19p signals, as assessed by FISH. Although there are limitations to these studies, for example treatment was not standardised, these findings suggest that diagnosing absolute deletions is more important. In this review, our interest was primarily in detection of absolute deletions. We were also interested in diagnosing situations where one copy of 1p/19q had been lost and the other copy duplicated (also termed copy‐neutral LOH). Combinations of chromosomal deletions in oligodendrogliomas and the corresponding signals in FISH are presented in a schematic representation in Figure 1.
1.

Combinations of chromosomal deletions in oligodendrogliomas and the corresponding signals in fluorescent in situ hybridisation (FISH) in a schematic representation.
In all parts of the figure, chromosome 1 and chromosome 19 are presented in separate frames to visualise the combination of FISH signals. The 1p probes and the 19q probes are red, and the reference probes (1q and 19p) are green. The approximate labelling sites are indicated in the chromosomal schematics. An unrelated chromosome (2) is also shown. Below each frame a schematic representation of the nuclear hybridisation signals as they appear on FISH images. (A): normal cell with diploid set of chromosomes. There are two red signals each, for chromosomal arms 1p and 19q, as well as two green signals each for chromosomal arms 1q and 19p. (B): the most common constellation in oligodendrogliomas with absolute 1p/19q codeletion in a diploid set of chromosomes. Loss of one red signal in chromosome 1p and in 19q and two green signals for each 1q and 19p. (C): relative codeletion with example of polysomy of chromosome 19 and chromosome 2. (D): 1p/19q codeletion in tetraploid cells, resulting in two red and four green signals for both, 1p and 19q tests. (E): complex deletion patterns can be found in a small proportion of oligodendrogliomas, often associated with anaplastic histological types. In this example, there are diploid cells (left, 30%), triploid cells (centre, 30%) and tetraploid cells (right, 40%).
Index test(s)
This review assessed the sensitivity and specificity of any deoxyribonucleic acid (DNA)‐based techniques that can be used on tumour tissue to directly evaluate 1p/19q codeletion status. These include the following.
Fluorescent in situ hybridisation (FISH).
Chromogenic in situ hybridisation (CISH).
PCR‐based LOH assays (also known as PCR‐based – short tandem repeat or microsatellite analysis).
Restriction fragment length polymorphism (RFLP) analysis.
Comparative quantitative PCR (a form of real‐time PCR).
Multiplex‐ligation‐dependent probe amplification (MLPA).
Comparative genomic hybridisation (CGH).
Array comparative genomic hybridisation (aCGH).
Single nucleotide polymorphism (SNP) arrays.
Methylation arrays.
Next‐generation sequencing (NGS).
These techniques are briefly described in Table 3. There is no perfect (100% sensitive, 100% specific) 'gold standard' test for 1p/19q codeletion status: each of the above tests could theoretically produce false‐positive or false‐negative (or both) results, as described in Table 4.
1. Techniques that can be used to detect 1p/19q codeletion.
| Technique | Brief description |
| FISH | FISH testing uses fluorescently labelled probes that are designed to hybridise to specific chromosomal locations. It can be performed on FFPE, and on fresh or frozen tissue. In this technique tissue architecture is preserved. To test for chromosome 1p/19q codeletion, chromosomes 1 and 19 are analysed on separate slides. FISH probes corresponding to regions of 1p or 19q labelled using 1 colour, and control probes on 1q or 19p labelled in another colour (as 1q and 19p seem to remain unaffected) are used. Many commercially available probes hybridise to loci at 1p36 and 19q13, although the FISH probes used at different centres may not target exactly the same loci (Pinkham 2015). Normal nuclei show a diploid signal ratio of 2/2 (2 signals from 1p or 19q and 2 signals from 1q or 19p). Absolute deletions will theoretically result in 1 signal from 1p or 19q in the presence of 2 signals from the control loci. There is no consensus on cut‐offs to diagnose codeletion. This is demonstrated by the fact that the EORTC study 26951 and the RTOG study 9402 used slightly different criteria (Pinkham 2015). Some laboratories define cut‐offs based on the percentage of cells with deleted and imbalanced signals, some define cut‐offs based on ratios calculated by dividing the total number of test probes by the total number of control probes, and some combine percentage and ratio cut‐offs. |
| CISH | This is a very similar technique to FISH, but instead of using fluorescent labelling, the probes are labelled with a marker such as biotin, digoxigenin or dinitrophenyl, and then this marker is detected using antibodies or streptavidin (that binds biotin) that is conjugated to enzymes such as horseradish peroxidase or alkaline phosphatase. The presence of the probe can then be visualised in the presence of a substrate that undergoes a colour change in the presence of the enzyme. The advantages of CISH is that it does not require a fluorescence microscope and staining is permanent. |
| PCR‐based LOH assays | This technique analyses polymorphic microsatellites that are dispersed throughout the genome. Different alleles have different numbers of repeats. PCR amplification of regions containing polymorphic microsatellites can therefore result in different length PCR products. If an individual is heterozygous (has 2 different alleles) for a microsatellite, PCR of this region will result in 2 different length products. If heterozygosity is lost, only 1 length product will be obtained. An individual must be heterozygous for a microsatellite for it to be informative, and DNA from normal tissue is required to determine this. LOH can be determined by comparing the ratio of PCR products of different lengths obtained from normal and tumour tissue. Primers that amplify regions containing microsatellites on 1p and 19q can be used to determine whether 1p and 19q are codeleted. However, there is no consensus on location or number of microsatellites analysed. |
| RFLP analysis | LOH can also be detected using RFLP analysis. In RFLP, restriction enzymes that recognise specific sequences are used to cut DNA, resulting in fragments of specific sizes. Different alleles may contain cut sites, or the DNA fragment that the restriction enzyme produces after digestion may be expected to differ due to different numbers of repeats in different alleles. Therefore, in a similar manner to PCR, LOH can be detected through loss of fragments of a specific size from informative loci (where an individual is heterozygous in normal tissue). |
| Comparative quantitative PCR | Comparative quantitative PCR compares the amount of PCR product obtained from 1p/19q with PCR product obtained from other chromosomal regions. If a deletion is present, less PCR product will be obtained. This technique has the advantages that heterozygosity at loci is not required, neither is a sample of normal tissue. |
| MLPA | MLPA uses probes designed to hybridise to specific regions of the genome that have been split into 2. Each probe 'half' also contains sequences corresponding to universal forward and reverse binding sites for PCR primers, and 1 'half' contains a region of varying length to help identify the probe later. The primers are hybridised to denatured sample DNA (e.g. from a tumour). The next step is ligation. Only probe halves that are hybridised to adjacent sequences on the sample DNA will be ligated together. PCR, using primers corresponding to the universal binding sides contained in the probes, is used to amplify the probes. Only those probe halves that were ligated together will be amplified to any extent, as it is only these products that contain the binding sites for both the forward and reverse PCR primers. The PCR products can then be separated by length, and quantified. The results are then normalised internally (by comparing reference probes with target probes), and then compared with reference samples. Heterozygous deletions can be identified as a probe ratio of 0.5 will be observed, and heterozygous duplications from a probe ratio of 1.5. Usually, probe ratios < 0.7 or > 1.3 are regarded as indicative of a heterozygous deletion (copy number change from 2 to 1 allele) or duplication (copy number change from 2 to 3 alleles), respectively (Eijk‐Van Os 2011). |
| CGH | In CGH, differentially labelled genomes from the tumour (the test genome) and normal tissue (the control genome, which does not need to be from the same person) are simultaneously hybridised to normal metaphase chromosomes. Changes in copy number, caused for example by loss or gain of regions, will alter the ratio of the 2 genomes. If 2 different fluorochromes are used to mark the genomes (or detect the labels), changes in copy number can be revealed from the relative intensities of fluorochromes used to detect the 2 genomes. CGH detects DNA sequence copy number changes relative to the mean copy number in the entire tumour sample. However, signals can be normalised using the sex chromosomes, which may help if a tumour is known to be normal for these chromosomes. |
| aCGH | aCGH follow the same principles as CGH, but instead of the 2 genomes being competitively hybridised to metaphase chromosomes, they are hybridised to a microarray. The theoretical resolution of aCGH is greater than that of traditional CGH. |
| SNP arrays | An SNP array is a type of DNA microarray. SNP arrays allow both copy number status and genotype to be determined, allowing detection of both losses and copy‐neutral LOH. SNPs are variations at a single position in a DNA sequence. Since individuals usually inherit 1 copy of each SNP position from each parent, the individual's genotype at a SNP site is typically either AA, AB or BB. To detect abnormalities using SNP arrays, sample DNA is fragmented, labelled and hybridised to an array containing immobilised allele‐specific oligonucleotide probes (1 probe for each allele). The signal intensity associated with each probe is then measured. Copy number changes can be detected from the intensity of signal. By comparing the result for each SNP with those from normal tissue, or by using a hidden Markov model, LOH can be detected. In the rare case of 2:2 tetraploidy, it is possible that SNP arrays will not be able to distinguish absolute from relative deletions. |
| Methylation arrays | Genome‐wide DNA methylation array data can also be used to detect 1p/19q status, as reported in Capper 2018b. In methylation arrays, specific regions of the genome that may be modified by methylation are investigated. The array has 2 probes for each region, 1 for the methylated and 1 for the unmethylated. To detect copy number variations, the signal from both probes (the methylated and unmethylated) for a specific region are added together and compared with a reference genome. |
| NGS | NGS refers to post‐Sanger sequencing technologies including sequencing‐by‐synthesis, sequencing‐by‐ligation and ion semiconductor sequencing. While traditional Sanger sequencing sequences a single DNA sequence, NGS is capable of sequencing multiple sequences simultaneously. Techniques have been developed to detect LOH and copy number variations using NGS. Deletions can be detected by relative perturbations in the read depth. LOH can be detected when the ratio of alleles at a heterozygous SNP site is perturbed. |
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; DNA: deoxyribonucleic acid; EORTC: European Organisation for Research and Treatment of Cancer; FFPE: formalin‐fixed, paraffin‐embedded tissue; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; NGS: next‐generation sequencing; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; RTOG: Radiation Therapy Oncology Group; SNP: single nucleotide polymorphism.
2. Theoretical ways in which false‐positive and false‐negative results could be obtained from the various techniques.
| Technique | Potential ways false‐positive results could be obtained | Potential ways false‐negative results could be obtained |
| FISH | Focal deletions at regions that the target probes hybridise could lead to false‐positive results as these cannot be distinguished from whole arm deletions (as only 1 probe per chromosome arm is normally used). | False‐negative results could be obtained if there has been a loss of heterozygosity without copy number reduction. |
| Depending on the way that deletions are diagnosed (i.e. the cut‐off used and whether it depends on the ratio of test probes to control probes), aberrations that lead to disproportionate gain in control probe loci (i.e. 1q and 19p) could lead to false‐positive results. | False‐negative results could be obtained if non‐neoplastic nuclei are assessed. | |
| The way that the tumour tissue is sectioned to prepare it for FISH could lead to 'truncation artefact'. Nuclei may be transected, which may lead to them containing incomplete genetic material. False‐positive results may be obtained from normal tissue in the presence of excessive truncation artefact. | Excessive truncation artefact in neoplastic tissue could lead to false‐negative results. | |
| CISH | As for FISH. | As for FISH. |
| PCR‐based LOH assays | PCR cannot distinguish between relative and absolute deletions, so people with relative deletions will be given false‐positive results. | If tumour samples are heavily contaminated with normal tissue, PCR products for both alleles will be obtained in a ratio that would give a false‐negative result. |
| Depending on primer spacing and the number of informative loci, the technique may detect focal rather than whole arm deletions. | ||
| Imbalanced polysomy, e.g. gain of 1 copy of chromosome 1 and 19, may result in allelic imbalance and be interpreted as loss of heterozygosity. | ||
| RFLP analysis | Cannot distinguish between relative and absolute deletions, so people with relative deletions will be given false‐positive results. | If tumour samples are heavily contaminated with normal tissue, digestion products for both alleles will be obtained in a ratio that would give a false‐negative result. |
| Depending on the regions analysed, it is possible that this technique may detect focal rather than whole arm deletions. | ||
| Imbalanced polysomy, e.g. gain of 1 copy of chromosome 1 and 19, may result in allelic imbalance and be interpreted as loss of heterozygosity. | ||
| Comparative quantitative PCR | PCR cannot distinguish between absolute deletion and relative deletions in the presence of polyploidy (i.e. those deletions that would give a 2:4 ratio/equivalent with FISH). | If tumour samples are heavily contaminated with normal tissue the amount of PCR product obtained would result in a false‐negative result. |
| Polysomy which causes the PCR product from control regions to increase could result in false‐positive results. | False‐negative results could be obtained if there has been an LOH without copy number reduction. | |
| Aneuploidy which causes the PCR product from control regions to decrease could result in false‐negative results. | ||
| MLPA | Cannot distinguish between absolute deletion and relative deletions in the presence of polyploidy (i.e. those deletions that would give a 2:4 ratio/equivalent with FISH). | If tumour samples are heavily contaminated with normal tissue, a false‐negative result may arise. |
| SNPs at primer binding sites, as single mismatches at ligation sites can inhibit ligation. | False‐negative results could be obtained if there has been an LOH without copy number reduction. | |
| CGH | Cannot distinguish between absolute deletion and relative deletions in the presence of polyploidy (i.e. those deletions that would give a 2:4 ratio/equivalent with FISH). | If tumour samples are heavily contaminated with normal tissue, a false‐negative result may arise. |
| False‐negative results could be obtained if there has been an LOH without copy number reduction. | ||
| aCGH | As for CGH. | As for CGH. |
| SNP arrays | Cannot distinguish between absolute deletion and relative deletions in the presence of polyploidy arising from whole genome duplication after the codeletion event (i.e. those deletions that would give a 2:4 ratio/equivalent with FISH). | If tumour samples are heavily contaminated with normal tissue, a false‐negative result may arise. |
| Methylation arrays | Cannot distinguish between absolute deletion and relative deletions in the presence of polyploidy arising from whole genome duplication after the codeletion event (i.e. those deletions that would give a 2:4 ratio/equivalent with FISH). | If tumour samples are heavily contaminated with normal tissue, a false‐negative result may arise. |
| False‐negative results could be obtained if there has been an LOH without copy number reduction. | ||
| NGS | Cannot distinguish between absolute deletion and relative deletions in the presence of polyploidy arising from whole genome duplication after the codeletion event (i.e. those deletions that would give a 2:4 ratio/equivalent with FISH). | If tumour samples are heavily contaminated with normal tissue, a false‐negative result may arise. |
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; FFPE: formalin‐fixed, paraffin‐embedded tissue; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; NGS: next‐generation sequencing; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; SNP: single nucleotide polymorphism.
Clinical pathway
Prior test(s)
Before testing for 1p/19q codeletion status, tumours undergo histological assessment. 1p/19q status is determined in tumours with histological appearances of gliomas, typically with morphological appearances suggestive of oligodendroglioma and usually after an initial set of histological special stains (immunohistochemistry) assessing the status of the most common IDH mutation (R132H) and of ATRX expression. Within IDH‐mutation tumours, 1p/19q codeletion tumours have different prognosis and different treatments, so 1p/19q deletions are only relevant for diagnosis of oligodendroglioma if there is IDH mutation (although there are very rare exceptions to this).
Role of index test(s)
As described previously, the codeletion has diagnostic, prognostic and predictive abilities in glioma. The results of testing for 1p/19q status are used for diagnosis, to inform treatment decisions and to give information on prognosis (survival). It is usual for testing to be done once, using one technique. Patients may be misdiagnosed, are likely to receive suboptimal treatments (although there is no good evidence on what the effect of this will be), and receive inaccurate prognostic information if given false‐positive or false‐negative results for 1p/19q status.
Alternative test(s)
All DNA‐based techniques that are used to determine 1p/19q status in tumour tissue were eligible.
Rationale
European guidelines recommend that 1p/19q status is evaluated to support a diagnosis of oligodendroglioma and for prognosis, and that treatment decisions are based on the 1p/19q status (Stupp 2014; Weller 2017). WHO guidelines required the demonstration of both an IDH mutation and 1p/19q codeletion for the diagnosis of oligodendroglioma and anaplastic oligodendroglioma (Louis 2016). Current guidance from the National Institute for Health and Care Excellence (NICE) recommend testing 1p/19q codeletion to identify oligodendrogliomas, and the adjuvant chemotherapeutic recommended after surgery for people with grade III glioma varies according to 1p/19q status (NICE 2018).
cIMPACT‐NOW 2 (the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy) guidance has suggested that in the setting of a diffuse astrocytic‐appearing WHO grade II or III glioma that has IDH mutation as well as loss of ATRX nuclear expression or strong, diffuse p53 immunopositivity, a diagnosis of diffuse astrocytoma, IDH‐mutant or anaplastic astrocytoma, IDH‐mutant can be rendered in the absence of 1p/19q testing (Louis 2018). However, for the diagnosis of oligodendroglioma and anaplastic oligodendroglioma assessment of both 1p/19q codeletion and IDH mutation is still required.
There are several different methods for determining 1p/19q status and no clear consensus regarding the optimal method. The two most common methods for routine diagnostic use are FISH‐ and PCR‐based LOH assays (Woehrer 2015). In the 2017 UK Cytogenomic External Quality Assessment Service (CEQAS) report, of the 35 enrolled laboratories, 25 laboratories used FISH, one laboratory used MLPA, four laboratories used arrays and one laboratory used quantitative PCR.
This review should go some way to answering the question "Do molecular subtyping techniques improve treatment selection, prediction and prognostication in people with brain and spinal cord tumours?", one of the top 10 topics identified by the James Lind Alliance Neuro‐Oncology Priority Setting Partnership (James Lind Alliance). The National Cancer Research Institute Brain Clinical Studies Group has identified this as an area for future research.
The final element of the review was to consider the costs and cost‐effectiveness of alternative methods of assessing 1p/19q status. Each method of 1p/19q assessment incurs costs such as staff costs, laboratory costs and clinic costs.
Objectives
To estimate the sensitivity and specificity and cost‐effectiveness of different deoxyribonucleic acid (DNA)‐based techniques for determining 1p/19q codeletion status in glioma.
Secondary objectives
If sufficient studies are identified, we aimed to break down each technique by relevant features, for example the region analysed/probes used and the cut‐off used to classify 1p/19q status.
We further aimed to critically appraise and summarise current evidence on the resource use, costs and cost‐effectiveness of techniques for determining 1p/19q status in gliomas (conduct a full integrated review of economic evidence) and assess the cost‐effectiveness of the different approaches of determining 1p/19q status using a decision model.
Methods
Criteria for considering studies for this review
Types of studies
Types of studies for diagnostic test accuracy review
Cross‐sectional studies that used two or more DNA‐based tests to assess 1p/19q status in tumour tissue from the same set of people.
To be included, studies needed to present either raw data or classified results for patients for at least two DNA‐based tests. Studies that reported only on concordance of test results were excluded. Studies with data for just one person were excluded.
Types of studies for the full integrated review of economic evidence
We sought economic evaluations (cost‐effectiveness analyses, cost‐utility analyses and cost‐benefit analyses) conducted alongside any study designs or as part of a modelling exercise.
Participants
Adults aged 18 years or over with glioma or any subtype of glioma, which would typically be diagnosed by a neuropathologist.
Studies of people recruited because they were all determined by a specific technique to be 1p/19q codeleted (or all 1p/19q non‐codeleted) were excluded.
Index tests
Any DNA‐based technique that is used to determine 1p/19q status in tumour tissue.
Studies that assessed 1p/19q status by immunohistochemistry were excluded.
Studies that assessed 1p/19q status from blood samples or by imaging (i.e. magnetic resonance imagining, computed tomography, positron emission tomography) were excluded.
Target conditions
Absolute 1p/19q codeletion (1p/19q codeletion in the absence of polysomy).
Reference standards
As described in Table 4, each test can potentially generate false‐positive and false‐negative results. As such, there is no true 'gold standard' reference test. However, in order to estimate the sensitivity and specificity of each test, we considered two alternative reference standards. These were selected as the two tests that are most commonly used so are most familiar to people considering using alternative tests.
Using FISH as the reference standard, which can be interpreted as assuming that FISH has 100% sensitivity and 100% specificity.
Using PCR‐based LOH assays as the reference standard, which can be interpreted as assuming that PCR‐based LOH assays have 100% sensitivity and 100% specificity.
No reference standard: using latent class methodology, it is theoretically possible to estimate the sensitivity and specificity of a number of tests without making the strong assumption that any one test is 100% sensitive and 100% specific, although other strong assumptions are required. Further details are provided in the 'Statistical analysis and data synthesis for the diagnostic test accuracy review' section.
Use of FISH or PCR‐based LOH assays was not an eligibility criterion: all studies that used two or more tests to assess 1p/19q status in tumour tissue from the same set of people were included in the review.
Search methods for identification of studies
Electronic searches
Electronic searches for the diagnostic test accuracy review
We searched MEDLINE Ovid (from 1946 to 31 July 2019), Embase Ovid (from 1974 to 2019 week 30) and BIOSIS Citation Index (from 1969 to 1 August 2019). The search strategies are given in Appendix 1.
We also searched for studies available in PubMed that were not available in MEDLINE using the syntax 'pubmednotmedline[sb]' (all years).
There were no restrictions based on language or date of publication.
Electronic searches for the full integrated review of economic evidence
We screened the search in MEDLINE and Embase in Appendix 1 for suitable economic evaluation studies at the same time as screening for study inclusion in the diagnostic test accuracy (DTA) review. In addition, we searched the NHS Economic Evaluation Database (EED) up to the end of March 2015, when the last records were added to that database. This search used key individual clinical terms from the main search strategy (Appendix 1), both alone and in combination, to identify suitable economic evaluations.
Searching other resources
Searching other resources for the diagnostic test accuracy review
We searched Open Grey (www.opengrey.eu/, all available years to 1 August 2019) using the free‐text terms from our MEDLINE search ((("chromosome 1" OR 1p) AND ("chromosome 19" OR 19q)) OR (1p19q OR "1p/19q" OR (1p* NEAR/3 19q*))) AND (glioma* OR astrocytoma* OR astroblastoma* OR ependymoma* OR subependymoma* OR oligodendroglioma* OR oligoastrocytoma* OR pleomorphic xanthoastrocytoma* OR glioblastoma* OR GBM* OR ganglioglioma* OR gliosarcoma* OR gangliocytoma* OR ((glial* OR glioneuronal* OR brain*) AND (tumor* OR tumour* OR cancer* OR neoplasm*))).
We searched for relevant material in dissertations and theses using ProQuest Dissertations & Theses Global (search.proquest.com/pqdtglobal/dissertations/), using the same strategy as for Open Grey but limiting to all fields except full text (all available years to 1 August 2019). We also searched the Networked Digital Library of Theses and Dissertations (search.ndltd.org/index.php) (all available years to 1 August 2019).
The Society of Neuro‐Oncology (SNO) and its partner associations the EANO and The Japan Society of Neuro‐Oncology hold meetings where relevant research may be presented. We searched for abstracts from these meetings and other relevant conferences via the Web of Science Conference Proceedings Citation Index (CPCI‐S) (from 1990 to 1 August 2019). We translated the BIOSIS search for CPCI‐S as both databases are hosted on Web of Science.
We also searched for any ongoing studies via the WHO International Clinical Trials Registry Platform (ICTRP) (all available years to 1 August 2019). The search strategy is given in Appendix 1.
We examined the reference lists of included studies to identify any additional studies.
We examined results of searches of these other resources for both the DTA and economic components of the review.
Data collection and analysis
Selection of studies
Selection of diagnostic test accuracy studies
Two review authors independently screened titles and abstracts using EPPI‐Reviewer 4 or EPPI‐Reviewer Web (Beta). In case of disagreement, a third review author independently screened the title and abstract and decided on potential relevance. Full‐text articles were then independently screened in duplicate. Disagreements were resolved by consensus, with discussion with a third review author if necessary.
Selection of economic studies
Studies that met the inclusion criteria for DTAs were screened by one review author to assess if any of the clinically relevant studies could possibly meet the economic inclusion criteria. Had any potentially relevant studies been identified they would have been screened by two review authors.
Data extraction and management
Data extraction and management of diagnostic test accuracy studies
Two review authors independently performed data extraction onto a data extraction form split between Excel and EPPI‐Reviewer 4/EPPI‐Reviewer Web (Beta). Disagreements were resolved by consensus, with discussion with a third review author if necessary. We extracted data on the following items.
Study characteristics
Author.
Year.
Country.
Whether the study compared two or compared multiple techniques for determining 1p/19q status.
Population characteristics
Number of participants.
Population source and setting.
Inclusion/exclusion criteria.
Tumour subtype and grade.
Prior testing.
Age.
Gender.
Karnofsky performance status.
First diagnosis or recurrent disease.
Prevalence of 1p/19 codeletion.
Index tests (per test performed)
Technique.
Tumour sample type (i.e. formalin‐fixed, paraffin‐embedded (FFPE) or frozen tissue).
Region(s) analysed.
Cut‐off/threshold used to determine 1p/19q status.
Method of determining threshold and whether it was prespecified.
Raw test result data
We extracted the raw data from each individual study as a contingency table of cross‐classified test results. For studies comparing two tests, this was a 2 × 2 table. Regardless of whether the study treated one of the tests as a reference standard, overall in the review we did not label results as true positives but rather 'positive on both tests'; not true negatives but rather 'negative on both tests'; not false positives but rather 'positive on test A and negative on test B'; and not false negatives but rather 'negative on test A and positive on test B'. In situations where more than two tests were compared the data formed tables of higher dimensions. For example, if three tests were compared then the table formed was 2 × 2 × 2, that is, eight cells of cross‐classified results.
We extracted the researchers' classifications of test results (i.e. we did not attempt to reclassify test results even if raw data were available if the researchers had made classifications). If raw data were presented and classifications had not been made, but thresholds for classification were reported, we used these to classify test results. In situations where classifications were not made, raw data were presented, and the threshold to be used to interpret the raw data were not specified, we applied a threshold ourselves and explain our choices of threshold in the results.
Data extraction and management of economic evaluation studies
We adapted a data extraction form for economic investigations based on the format and guidelines used to produce structured abstracts of economic evaluations for inclusion in the NHS EED to the specific requirements of this review. The following data were to be collected from the economic studies.
Type of evaluations.
Sources of effectiveness data.
Cost data.
Sources of cost data.
Sources of outcome valuations.
Analytical approach.
Outcome valuations (e.g. utility values).
Sources of outcome valuations.
Cost‐effectiveness data (e.g. incremental cost‐effectiveness ratios (ICER)).
Analytic approach.
Assessment of methodological quality
Assessment of methodological quality in included diagnostic test accuracy studies
Two review authors independently assessed the applicability and risk of bias of included studies using the QUADAS‐2 tool (Whiting 2011). We resolved disagreements by consensus, with discussion with a third review author if necessary. We tailored the tool to our review, and the tailored form of the tool, along with how we judged risk of bias and applicability in each study is described in Appendix 2. We illustrated assessments using the robvis tool (McGuinness 2020).
Assessment of methodological quality in included economic studies
If any relevant economic evaluations were identified then these were to be assessed for bias in two stages. The first stage was to involve assessing the risk of bias from the sources of the DTA effectiveness data. Summary effect sizes from systematic reviews used as data inputs in model‐based economic evaluations were to be assessed using the ROBIS tool (Whiting 2016). The second stage was to assess the overall methodological quality of the economic component of the evaluation. Evaluations carried out alongside studies were to be assessed with reference to items included in the CHEERS (Consolidated Health Economic Evaluation Reporting Standards) checklist for reporting (Husereau 2013) and model‐based economic evaluations were to be assessed using the NICE methodology checklist (NICE 2014).
Statistical analysis and data synthesis
Statistical analysis and data synthesis for the diagnostic test accuracy review
No diagnostic test is free of errors (Bossuyt 2021); each of the tests can potentially generate false‐positive and false‐negative results. FISH and PCR‐based LOH are the most commonly used tests, so are most familiar to users of the tests. Furthermore, PCR‐based LOH is expected to have very high sensitivity (Table 3). We performed three analyses as follows.
Using FISH as the reference standard.
Using PCR‐based LOH assay as the reference standard.
Latent class analysis comparing FISH with PCR‐based LOH.
For each analysis following the first two strategies, the raw cross‐classified test result data from all studies that included the respective reference standard were first relabelled as 'true positive', 'false negative', 'true negative' and 'false negative' (2 × 2 table), based on the reference standard test result. If a study compared more than one test with the reference standard, multiple 2 × 2 tables were derived.
For analysis with each of the respective reference standards, we performed bivariate meta‐analyses of the sensitivity and false‐positive rate (1 – specificity) of each index test, assuming binomial likelihood for the number of 'true positive' and 'true negative' test results (Chu 2006; Reitsma 2005). This approach allows for heterogeneity in sensitivity and specificity across studies and for between‐study correlation in these measures. In our main analyses, we assumed that this between‐study correlation and the standard deviation (heterogeneity) parameters were shared (i.e. identical) across tests. This was because there were small numbers of studies for many of the tests, such that there were few or no data to inform estimation of test‐specific correlation and heterogeneity parameters. This unified modelling approach allowed tests to be included in the analysis even if they were only evaluated in a single study: between‐study heterogeneity and correlation are allowed for by 'borrowing' these parameters from the data on other tests. The model did not account for within‐study correlations arising from a study evaluating two or more tests against the same reference standard.
The bivariate meta‐analysis model can be used to produce summary operating points (summary sensitivities and specificities) with 95% confidence or credible regions. Drawing on the equivalence of the bivariate model and the hierarchical summary receiver operating characteristic (HSROC) model (Rutter 2001) in the absence of covariates, the bivariate model can also be used to produce summary receiver operating characteristic curves (Arends 2008; Harbord 2007). We displayed summary operating points for each test with 95% credible intervals (CrIs). By default, we also displayed a 95% credible region (ellipse) and HSROC curve. Plotting of credible ellipses relies on an assumption of approximate bivariate normality of the summary estimates on the logit scale. Where this assumption was clearly violated due to skew, we omitted the summary ellipse and plotted only 95% CrIs. More specifically, ellipses were omitted for tests with summary sensitivity or specificity greater than 99%, which we found to correspond to large skew on the logit scale. HSROC curves are omitted from plots where there was no variability in one of the two accuracy dimensions (sensitivity or specificity) across studies. Prediction ellipses were not plotted.
Because neither FISH nor PCR‐based LOH assays are likely to be true 'gold standards', we applied latent class meta‐analysis methods to the data for FISH and PCR‐based LOH (Chu 2009; Dendukuri 2012; Walter 1999). These methods provide estimates of sensitivity and specificity based on a probabilistic definition of disease state, rather than requiring classification of test results as true positives, false negatives, true negatives and false positives. We assumed multinomial likelihoods for the 2 × 2 table of cross‐classified (FISH × PCR‐based LOH assay) test results from each study. The four probability parameters are defined as functions of the study‐level sensitivity and specificity of each of the two tests and the (unknown) prevalence of 1p/19q codeletion status among people with glioma in the study. We assumed bivariate normal distributions for logit‐transformed sensitivity and specificity of each of the two tests across studies.
In latent class analyses, it is important to allow for the possibility that tests are positively correlated within disease states, usually referred to as 'conditional dependence' (Vacek 1985). In addition to 'conditional independence' models, we fitted models that allowed for conditional dependencies through the inclusion of within‐study covariance terms (Chu 2009; Dendukuri 2012). These covariance parameters are naturally bound in magnitude by functions of test sensitivity and specificity (Chu 2009; Dendukuri 2012).
The advantage of latent class methods is that they do not make the unrealistic assumption that one of the tests is a gold standard. However, in order to relax this assumption, it is often necessary to make other assumptions. This is to avoid problems with parameter identifiability (Jones 2010), which are introduced by recognising that study‐level prevalence and the sensitivity and specificity of the 'reference standard' are all in fact unknown. To reduce the number of parameters that need to be estimated, we assumed again that between‐study heterogeneity and between‐study correlation parameters are shared across tests. We additionally performed an analysis in which we assumed that PCR‐based LOH had a sensitivity of at least 95%. PCR‐based LOH ought to have a sensitivity close to 100% (no false‐negative results) in research contexts. False‐negative results on this test can only be obtained if there is excessive contamination of tumour samples with normal tissue. In a research context, we would expect great care to be taken to minimise the risk of contamination with normal tissue. An informative prior distribution (a uniform (0.95, 1.00) prior on sensitivity) was, therefore, used to constrain the sensitivity of this test to be at least 95%.
We had planned to extend the latent class analysis approach to the complex structure of our data (for multiple studies involving different selections of test and different numbers of tests), which would involve novel methodological development of the statistical models. Prioritisation of work in response to the COVID‐19 pandemic prevented this development work from happening.
Meta‐analysis models were fitted in the Bayesian statistical software WinBUGS (Lunn 2000). For models comparing tests against PCR‐based LOH or FISH as a reference standard, vague normal prior distributions were assumed for the mean sensitivity and mean false‐positive rate of each test on the logit scale, with a mean of 0 and variance of 100. In latent class models, these were replaced with uniform(0,1) priors on the probability scale, following observed poor mixing of Markov Chain Monte Carlo (MCMC) chains and bimodal posterior distributions with the initial prior distributions.
Standard deviations of logit(sensitivity) and of logit(false‐positive rate) across studies were given uniform(0,2) prior distributions. Between‐study correlation parameters were given uniform(0,1) priors.
In latent class analyses, each study‐specific prevalence parameter was assigned a uniform(0,1) prior. Within‐study covariance parameters, representing conditional dependencies between tests, were assumed to be non‐negative and were assigned uniform priors across the range zero to their theoretical maximums (Chu 2009; Dendukuri 2012).
In addition to summary operating points, we estimated differences in (summary) sensitivity and in specificity between index tests, which we present with 95% CrIs (Takwoingi 2013).
Investigations of heterogeneity
Investigations of heterogeneity planned for the diagnostic test accuracy review
Where sufficient number of studies assessed the same index test, we planned to investigate the impact of the following index test characteristics and population characteristics.
Tumour sample type (i.e. FFPE or frozen tissues).
Region(s) analysed.
Cut‐off/threshold used to determine 1p/19q status.
Study prevalence of 1p/19q codeletion.
Tumour subtype and grade.
We did not perform these investigations of heterogeneity due to small numbers of studies for specific tests.
Sensitivity analyses
Sensitivity analyses planned for the diagnostic test accuracy review
For tests evaluated in four or more studies against the same 'reference standard' (FISH or PCR‐based LOH), we performed a sensitivity analysis in which accuracy data were meta‐analysed separately for each test (i.e. with test‐specific between‐study heterogeneity and correlation parameters). Prior distributions for these analyses were the same as in the main analysis.
If sufficient data were available, we planned to perform sensitivity analyses by restricting analyses to studies judged not to be at high risk of bias or low applicability.
Assessment of reporting bias
Assessment of reporting bias in the diagnostic test accuracy review
Because of uncertainty about the determinants of publication bias for diagnostic accuracy studies and the inadequacy of tests for detecting funnel plot asymmetry (Deeks 2005), we did not perform tests aimed at detecting publication bias.
Summary of findings for the diagnostic test accuracy review
We presented the summary diagnostic accuracy results for key tests in a summary of findings table, selecting for inclusion the tests that are relevant to current clinical practice (PCR‐based LOH, FISH, aCGH, SNP array, NGS, MLPA and real‐time PCR). We assessed confidence in each result following the GRADE approach (Guyatt 2008; Schünemann 2008). We rated overall certainty in the evidence for each test as 'high', 'moderate', 'low' or 'very low' considering risk of bias, imprecision, inconsistency, indirectness and publication bias, all of which may lead to downgrading the certainty of the evidence (see Appendix 3).
An issue when using GRADE to rate the certainty of the evidence is that test accuracy is considered a surrogate for outcomes that are important to patients and can only provide indirect evidence of impact on patient‐important outcomes (Schünemann 2008). As we described in the Background section, the codeletion has diagnostic, prognostic and predictive abilities in glioma, and all the tests described have the same risk of adverse events associated with the test as they all require some biopsied tumour material. Therefore, we assumed that testing using the most accurate test will improve patient‐important outcomes. We used the indirectness domain to downgrade the certainty of the evidence if studies had low applicability to our review question using QUADAS‐2. We also considered publication bias, but note that there is uncertainty about the determinants of publication bias for diagnostic accuracy studies and tests for detecting funnel plot asymmetry are inadequate (Deeks 2005).
Full integrated review of economic evidence and economic model
Economic evidence
Characteristics and results of included economic evaluations were to be summarised using additional tables, supplemented by a narrative summary to compare and evaluate methods used and principal results between studies. This includes the currency and price year of costs, incremental cost and ICERs. If it were not possible to express costs in this way, then we planned to express these results as the most recent international dollars value using implicit price deflators for gross domestic product (GDP) and GDP Purchasing Power Parities. Where possible, unit cost data were also to be combined and summarised (Shemilt 2019). This review was to be conducted according to current guidance on the use of economics methods in the preparation and maintenance of Cochrane Reviews (Shemilt 2019).
Economic model
We built a decision tree using TreeAge software (TreeAge 2021) to estimate the expected cost of: 1. a true positive diagnosis, 2. a true negative diagnosis and 3. a correct diagnosis for each of the diagnostic tests (Appendix 4). The decision tree was based on the estimated diagnostic accuracies of the testing strategies calculated in the meta‐analysis. In terms of cost, the economic model included only those costs associated with carrying out the test. The model adopted a health service perspective and had a very short time horizon covering the diagnostic process only. Thus, the model did not include subsequent patient costs related to further treatment and did not include health outcomes beyond diagnosis.
Estimation of model parameters
Intervention costs were derived from both expert opinion from within the Newcastle upon Tyne Hospitals NHS Foundation Trust based on internal costings and existing literature. This information was provided by G Cuthbert, Consultant Clinical Scientist, Newcastle Genetics Laboratory (21 September 2020). Following advice from clinical expertise in the review team, FISH and CISH costs and real‐time PCR and PCR‐based costs were grouped due the similarity of the resources involved. This source provided costs for FISH and CISH, real‐time PCR and PCR‐based LOH, MLPA and SNP array. The overall costs included the staff, consumables, equipment and overheads (heat, power, light, etc.) associated with preparing the sample, running the analysis and feeding back findings. The costs for NGS and aCGH were derived from existing literature. All costs are reported in 2020 pounds sterling, and shown in Appendix 4 (Table A4.1). Where necessary, costs were converted into 2020 pounds sterling using the EPPI‐Centre Cost Converter (CCEMG 2019).
No cost for the G‐banding, karyotyping, mass spectrometry (MS) and NanoString techniques and CGH were identified. Content experts advised that these tests were not routinely performed in the health system (the UK NHS) that provided the data, and were likely to be used only in research settings. Consequently, these tests are not included in the model.
The model was designed to generate the expected costs per true‐positive diagnosis, per true‐negative diagnosis and per correct diagnosis. Given the sensitivities and specificities of the different diagnostic tests, and the prevalence rates of glioma in the various studies, we calculated the diagnostic accuracy classifications for the various testing strategies as:
true positive (TP) rate = prevalence × sensitivity;
true negative (TN) rate = (1 – prevalence) × specificity;
correct diagnosis (CD) rate = (prevalence × sensitivity) + ((1 – prevalence) × specificity).
We estimated the prevalence as the proportion of condition‐positive individuals across all the studies included in the meta‐analysis. This was limited to studies that had low/unclear risk of bias. The true‐positive rate can also be thought of as the number of true‐positive diagnoses divided by the total number of people in the study. The true‐negative rate can be thought of as the true‐negative diagnoses divided by the total number people in the study. The correct diagnosis rate is the sum of the true‐positive rate and true‐negative rate.
Base‐case analysis
Once the true‐positive, true‐negative and correct diagnosis rates were calculated, the diagnostic tests were compared in terms of both their cost and their diagnostic accuracy. Diagnostic tests that were dominated and extendedly dominated were first removed from the analysis, and the remaining strategies were then compared in terms of their ICERs. A dominated strategy is a strategy with both higher costs and worse outcomes than the next less costly strategy. In an incremental analysis, an extendedly dominated strategy is a strategy that has an ICER that is higher than the ICER of the next, more effective but more costly, alternative strategy. The ICER shows the estimated additional cost that would be needed for an additional unit of benefit from a strategy. Ranking the diagnostic strategies by cost and comparing the incremental costs and yields between increasingly costly diagnostic strategies allowed for the calculation of incremental costs per additional true positive, true negative and correct diagnosis, and the identification of dominated and extendedly dominated options.
Sensitivity analysis
We carried out a probabilistic sensitivity analysis (PSA) to assess the statistical precision surrounding estimates of cost‐effectiveness. Unlike a deterministic sensitivity analysis, a PSA allows uncertainty surrounding the estimates used in the model to be examined simultaneously. In a PSA, uncertain parameters are characterised using appropriate probability distributions around the point estimate for that parameter rather than a single value. Using a Monte Carlo simulation, a set of parameter values is then drawn by randomly sampling from the distribution and cost, and cost‐effectiveness is estimated. This process is repeated multiple times so that uncertainty around the model outputs can be presented. In our analysis, we repeated the Monte Carlo simulation process 10,000 times to assess the robustness of the conclusions. We characterised the prevalence and measures of sensitivity and specificity as beta distributions. As there was only one point estimate for the cost of each of the tests, we characterised the cost parameters as triangular distributions, with the minimum and maximum specified as 25% below and 25% above the likeliest value. The specific distributions used for each of the parameters are shown in Appendix 5. We presented results for a range of values (GBP 0 to GBP 10,000) for a decision‐maker's willingness to pay for a unit of outcome.
Results
Results of the search
Searches of MEDLINE, Embase, BIOSIS, PubMed, Web of Science CPCI‐S, ICTRP, OpenGrey, Proquest Dissertations & Theses and NDLTD identified 5427 records, and one record was identified through other sources. After removal of 2418 duplicate records, we screen the titles and abstracts of 3010 records. We selected 238 records to assess at full text. Fifty‐three studies (in 78 publications) met the inclusion criteria for test accuracy studies and were included in the review (Figure 2). We excluded 86 full‐text records (Characteristics of excluded studies table). Six studies are awaiting classification (Characteristics of studies awaiting classification table), and five studies are ongoing (Characteristics of ongoing studies table).
2.
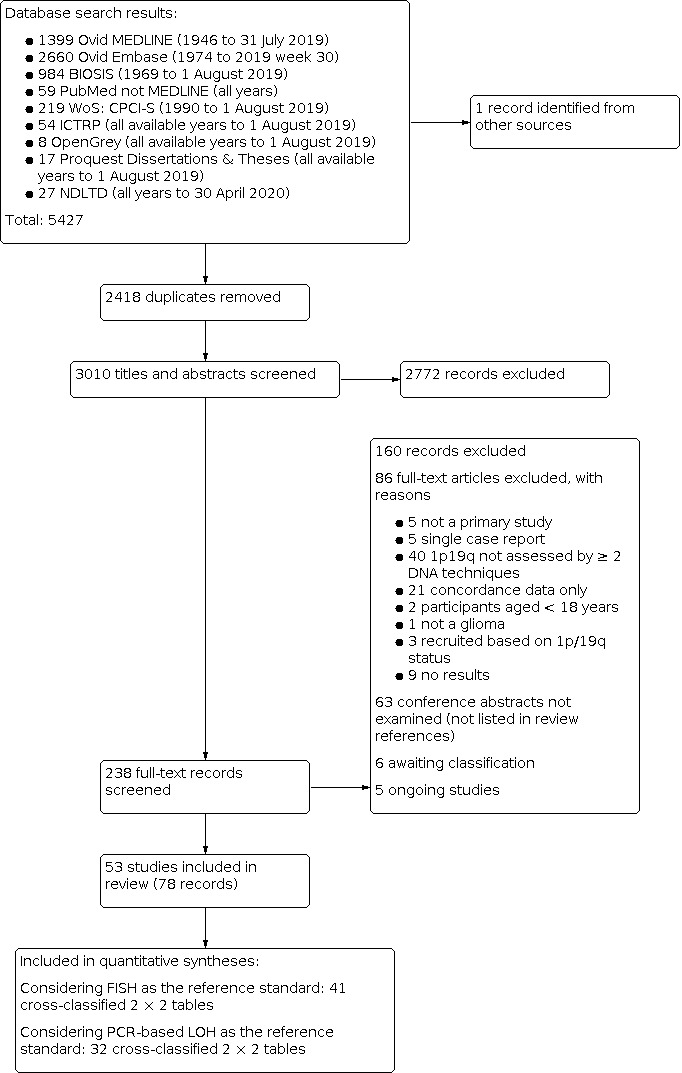
PRISMA flow chart.
None of the included studies also met the economic inclusion criteria. There were no studies identified from the NHS EED database and no economic evaluations from grey literature.
We classified the tests used into test categories based on the authors' description of the tests (see Table 3). We also grouped several PCR‐based techniques that used some form of real‐time PCR‐based technology: comparative quantitative PCR (investigated in Chaturbedi 2012), quantitative microsatellite PCR (investigated in Nigro 2001), and semi‐quantitative PCR (investigated in Ariza 2010). We included studies that used three additional techniques: MS (investigated in Pesenti 2017), NanoString (Armanious 2017), and G‐banding/karyotyping (Dahlback 2009; Dahlback 2011; Ransom 1992a; Ransom 1992b; Schrock 1994).
We initially tried to distinguish PCR‐based LOH performed with comparison to normal DNA (extracted from normal tissue or blood from the same patient) from PCR‐based LOH performed without this normal DNA sample. One of the studies we included compared assays with and without comparison to normal DNA (Hatanpaa 2003a; Hatanpaa 2003b). However, we found that several studies initially classified as 'PCR‐based LOH performed with comparison to normal DNA' stated that, in the absence of normal DNA from the same patient, PCR was performed without the normal DNA sample (e.g. Horbinski 2012 and Clark 2013). Therefore, we combined these two categories.
We included 39 studies that performed two categories of test on the same set of participants (Ariza 2010; Armanious 2017; Bigner 1999; Bouvier 2004; Broholm 2008; Byeon 2014; Chaturbedi 2012; Cieply 2004; Clark 2013; Cowell 2004; D'Haene 2019; Dahlback 2009; Dubbink 2016; Gadji 2009; Ghasimi 2016; Harada 2011; Hinrichs 2016; Horbinski 2012; Jeuken 2006; Jha 2011; Kato 2019; Kolhe 2016; Lass 2013; Lhotska 2015; Na 2019; Natte 2005; Nigro 2001; Park 2019; Paxton 2015; Ransom 1992a; Ransom 1992b; Scheie 2006; Schrock 1994; Sim 2018a (glioblastoma cohort); Sim 2018b (oligodendroglial cohort); Thakur 2012; Thomas 2017; Tsiatis 2010; Wiestler 2014), seven studies performed three categories of test on the same participants (Burger 2001; Dahlback 2011; Hatanpaa 2003a (assay development and non‐blinded validation cohort); Hatanpaa 2003b (blinded validation cohort); Mohapatra 2006; Pesenti 2017 (note that in this study four tests were investigated but a maximum of three were applied to the same participants); Smith 1999), and one study performed four test categories on the same participants (Blesa 2009). In addition, several studies performed multiple variants of tests on the same participants (Belaud‐Rotureau 2006; Duval 2014; Duval 2015; Hatanpaa 2003a (assay development and non‐blinded validation cohort); Hatanpaa 2003b (blinded validation cohort 3); Horbinski 2012; Senetta 2013; Srebotnik‐Kirbis 2016; Uchida 2019).
All possible test comparisons from the included studies are shown in Table 5. The studies can be visualised as a network of test comparisons (Figure 3). From Table 4 and Figure 3, it is clear that there is the most information for a comparison between FISH and PCR.
3. All possible 2 × 2 test comparisons from the included studies.
| Test categories | aCGH | CGH | CISH | FISH | G‐banding | Methylation array | MLPA | MS | NanoString | NGS | PCR‐based LOH | RFLP | Real‐time PCR | SNP array |
| aCGH | — | — | — | Blesa 2009; Byeon 2014; Mohapatra 2006; Pesenti 2017 | — | — | Blesa 2009 | Pesenti 2017 | — | — | Blesa 2009; Cowell 2004; Mohapatra 2006 | — | — | — |
| CGH | — | — | — | Burger 2001; Hatanpaa 2003a (assay development and non‐blinded validation cohort); Hatanpaa 2003b (blinded validation cohort); Smith 1999 | Dahlback 2009; Dahlback 2011; Schrock 1994 | — | Jeuken 2006 | — | — | — | Bigner 1999; Burger 2001; Dahlback 2011; Hatanpaa 2003a (assay development and non‐blinded validation cohort); Hatanpaa 2003b (blinded validation cohort); Smith 1999 | — | — | — |
| CISH | — | — | — | Lass 2013 | — | — | — | — | — | — | — | — | — | — |
| FISH | — | — | — | Belaud‐Rotureau 2006a; Duval 2014b; Duval 2015c; Horbinski 2012d; Senetta 2013e; Srebotnik‐Kirbis 2016f; Uchida 2019g | — | — | Blesa 2009; Natte 2005 | Pesenti 2017 | Armanious 2017 | D'Haene 2019; Kato 2019; Na 2019; Park 2019; Sim 2018a (glioblastoma cohort)h; Sim 2018b (oligodendroglial cohort); Thomas 2017 | Blesa 2009; Bouvier 2004; Broholm 2008; Burger 2001; Cieply 2004; Clark 2013; Gadji 2009; Hatanpaa 2003a (assay development and non‐blinded validation cohort); Hatanpaa 2003b (blinded validation cohort); Horbinski 2012; Jha 2011; Mohapatra 2006; Pesenti 2017; Scheie 2006; Smith 1999 | — | Chaturbedi 2012; Nigro 2001 | Ghasimi 2016; Hinrichs 2016; Kolhe 2016; Lhotska 2015; Paxton 2015; Thakur 2012 |
| G‐banding | — | — | — | — | — | — | — | — | — | — | Dahlback 2011 | Ransom 1992a; Ransom 1992b | — | — |
| Methylation array | — | — | — | — | — | — | Wiestler 2014 | — | — | — | — | — | — | — |
| MLPA | — | — | — | — | — | — | — | — | — | — | Blesa 2009 | — | — | — |
| MS | — | — | — | — | — | — | — | — | — | — | Pesenti 2017 | — | — | — |
| NanoString | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| NGS | — | — | — | — | — | — | — | — | — | — | Dubbink 2016 | — | — | — |
| PCR‐based LOH | — | — | — | — | — | — | — | — | — | — | Hatanpaa 2003a (assay development and non‐blinded validation cohorti); Hatanpaa 2003b (blinded validation cohorti) | — | Ariza 2010 | Harada 2011; Tsiatis 2010 |
| RFLP | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Real time PCR | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| SNP array | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; NGS: next‐generation sequencing; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; SNP: single nucleotide polymorphism.
aBelaud‐Rotureau 2006 performed FISH with manual analysis with the 1p36.3 (D1Z2)/1q12 (D1Z1) and 19q13.3/19pter probe set, manual analysis with the 1p36/1q25 and 19q13/19p13 Abbott Vysis probe set, and automatic analysis (Metafer 4, Metasystems, Althlussheim, Germany) with the 1p36/1q25 and 19q13/19p13 Abbott Vysis probe set. bDuval 2014 performed FISH and immunoFISH (FISH with immunohistochemistry against Ki67 (MIB‐1)), and used two different cut‐offs for both – a "combination" cut‐off (which was based on the number of cells showing a deletion) and a "ratio" cut‐off (based on the ratio of signals for 1p to 1q and 19q and 19p). cDuval 2015 performed FISH with automated analysis (Metafer 4 software (Metasystem) using the "1p19q tile‐sampling classifier") and FISH with manual analysis. dHorbinski 2012 performed FISH with two different cut‐offs in addition to PCR‐based LOH (target‐ploidy control ratio < 0.87, with ≥ 20% of nuclei showing deletion and target‐ploidy control ratio < 0.75, with ≥ 20% of nuclei showing deletion). eSenetta 2013 performed FISH with two different cut‐offs (cut‐off ratios 1p ≤ 0.8 and 19q ≤ 0.8 and cut‐off ratios 1p ≤ 0.7 and 19q ≤ 0.8). fSrebotnik‐Kirbis 2016 FISH variants on fresh tissue cytospins and on FFPE sections. gUchida 2019 performed FISH with two different criteria for judging whether a deletion was present (signals of 1p or 19q < signals of 1q or 19p or single signal of 1q or 19q and two signals of 1q or 19p; in both cases the cut‐off value was set at 20%). hIn Sim 2018a and Sim 2018b, glioblastoma cohort FISH was compared to NGS or aCGH (or both). We categorised NGS or aCGH (or both) as NGS for the purposes of this table. iHatanpaa 2003a and Hatanpaa 2003b performed PCR‐based LOH with or without comparison to normal DNA (in addition to CGH and FISH). This study developed a cut‐off for PCR‐based LOH without comparison to normal DNA in one set of participants (Hatanpaa 2003a assay development and non‐blinded validation cohort) and validated it in another set of participants (Hatanpaa 2003b blinded validation cohort).
3.
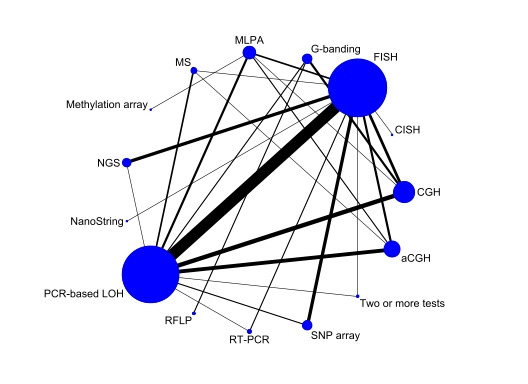
Network plot of the included studies. The size of the circles represents the number of test results for a test category. The thickness of the lines is proportional to the number of studies making the comparison. Note that the FISH and PCR circles include comparison within test categories.
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS; mass spectrometry; NGS: next‐generation sequencing; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; RT: real‐time; SNP: single nucleotide polymorphism.
Details of the 53 included studies, including country and population source and setting, inclusion and exclusion criteria, and population characteristics (including age, gender, Karnofsky performance status) and test categories analysed can be found in the Characteristics of included studies table. In most cases, glioma was diagnosed using histopathology.
Raw data from the included studies
Raw data for the included studies extracted as contingency tables of cross‐classified test results by tumour subtype (where possible, and as described by the study authors) can be found in Appendix 6. Separate tables are presented for comparisons of four test categories/variants (2 × 2 × 2 × 2 table; Appendix 6, Table A6.1), three test categories/variants (2 × 2 × 2 table; Appendix 6, Table A6.2) and two test categories/variants (2 × 2 table; Appendix 6, Table A6.3). Studies that compared more than two test categories/variants may be found in multiple tables if only a subset of the tests were performed on a proportion of the participants. For example, if a study compared four tests but a subset of participants only had three tests, the study appears in both the 'three test' and the 'four test' tables. However, each study participant is only included in a single table. Therefore, if interest is in a comparison of two particular tests, it may be necessary to sum across or within (or both) tables, to obtain the relevant 2 × 2 table. Details of the tests used (tumour sample type, region(s) analysed, cut‐off used) are also presented in these tables. Details of specific decisions we made during data extraction are provided in Appendix 7 and Appendix 8.
Regions analysed
Where the regions on 1p analysed by the different tests used in the different studies were reported in sufficient detail, we mapped the regions to the regions on 1p. These are shown diagrammatically, where available, in Figure 4 (studies comparing four tests and comparing three tests), Figure 5 (studies comparing two tests) and Figure 6 (comparative listing for FISH and PCR‐based LOH). Regions analysed on chromosome 19 are listed in the Characteristics of included studies table. It is to be expected that the more regions are analysed by a technique, the more reliably a codeletion of whole chromosomal arms will be detected.
4.
Graphical representation of regions analysed in studies comparing four tests (panel A) and studies comparing three tests (panel B), as listed in Appendix 6 (Tables A6.1 and A6.2). The top of the figure indicates a graphical representation of chromosome 1 (adapted from the GRCh38/hg38 assembly). The figure legend indicates the different methods, with different colour codes for FISH, depending on the origin or manufacturer of the probes. In each section, the first author of the study is represented on top, and the techniques on the left of the table. The graphical representation indicates the position of the probe or primer on the chromosome.
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS; mass spectrometry; NGS: next‐generation sequencing; PCR: polymerase chain reaction; RT‐PCR: real‐time polymerase chain reaction; SNP: single nucleotide polymorphism.
5.

Graphical representation of regions analysed in studies comparing two tests, as listed in Appendix 6 (Table A6.3). For legend to symbols, see Figure 4. Studies that are already represented in the three‐ or four‐test comparisons are omitted (see footnotes). Studies comparing FISH with FISH used different parameters, see Table A6.3 for details.
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; FISH: fluorescence in situ hybridisation; MLPA: multiplex‐ligation‐dependent probe amplification; MS; mass spectrometry; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism.
6.
Comparative listing of all studies using FISH (panel A) and all studies using PCR‐based LOH (panel B). Studies appear in alphabetical order of first author. For legend to symbols, see Figure 4.
FISH: fluorescence in situ hybridisation; PCR: polymerase chain reaction.
Methodological quality of included studies
Risk of bias
QUADAS‐2 risk of bias assessments for studies that performed two or more categories of test are shown in Figure 7, and assessments for studies that performed two or more FISH variants are shown in Appendix 9.
7.
Risk of bias and applicability concerns summary: review authors' judgements about each domain for each included study
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS; mass spectrometry; NGS: next‐generation sequencing; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; SNP: single nucleotide polymorphism. Note: Horbinski 2012 used two FISH variants. The judgements for variant 1 are shown (cut‐off: target‐ploidy control ratio was less than 0.87, with at least 20% of nuclei showing deletion). Hatanpaa 2003a and Hatanpaa 2003b used two PCR‐based LOH variants. The judgements shown are for PCR compared with normal DNA.
Only one study (comparing variants of FISH) was rated at low risk of bias across all domains (Senetta 2013). How patients were selected for inclusion into the study was unclear in most studies. We judged that a consecutive or random series of patients were selected for inclusion for only a minority of studies. It was clear that some studies included a non‐random sample of patients, or used a case‐control design, and we rated these studies at high risk of bias.
Hatanpaa 2003 selected participants based on them having concordant results on at least two tests investigated in Smith 1999 (majority of participants) or Burger 2001 (one participant). Because of this, we assessed risk of bias due to patient selection to be high in this study. The participants included in Hatanpaa 2003a and Hatanpaa 2003b were removed from the data extraction from Smith 1999 and Burger 2001. However, if the results for the tests investigated in Smith 1999 and Burger 2001 are considered for all three studies together, the risk of bias should be lower, as participants were not selected on the basis of concordant results into Smith 1999 and Burger 2001.
We rated many of the index test methods at unclear or high risk of bias. This was because it was frequently not reported whether the test results were interpreted without the knowledge of the other tests being compared or because the threshold used to classify results was not reported, or both.
Several studies presented raw data but did not classify the results of the test and the authors did not report a cut‐off. In these cases, we classified the results using cut‐off points we regarded as clinically realistic (see Appendix 2). In such instances, we judged that the lack of prespecification of the cut‐off points and the lack of blinding to other test results did not cause bias as our choice of cut‐off point was not influenced by the data. However, it should be noted that even in these studies the study authors had frequently made some judgements that we relied upon to determine 1p/19q status, for example whether there was LOH at a particular locus.
We generally assumed that cut‐off to determine 1p/19q status for interpretation had been prespecified, provided these thresholds were stated in the methods section of the publication. In some cases, it was clear that the cut‐off was not prespecified or that the results of one test were interpreted with full knowledge of the results of at least one other test that was performed. In this situation, we judged the index test domain at high risk of bias.
Many of the studies were rated at high risk of bias on the domain relating to flow and timing of participants. This was always because of missing data. We had predefined a cut‐off of 5% of the proportion of the enrolled population being excluded for a study to be rated at high risk of bias, and many studies had missing data for at least 5% of participants. Some studies that compared more than two tests may have been rated at high risk of bias because of missing data for one or more tests, but the results for some comparisons had no missing data and were at low risk of bias. For example, in Pesenti 2017, all participants had two of the tests investigated (MS and PCR‐based LOH). However, this study was rated at high risk of bias because only a small proportion of participants had results for either aCGH or FISH. In Belaud‐Rotureau 2006, all participants had results for the comparison of manual versus automatic analysis using the Abbott Vysis probe set. However, fewer than half of the included participants had results for manual analysis using the 1p36.3 (D1Z2)/1q12 (D1Z1) and 19q13.3/19pter probes.
Dubbink 2016 compared NGS with PCR‐based LOH and was assessed at low risk of bias on the flow and timing domain. However, participants in Dubbink 2016 were from a randomised trial (EORTC 26951). In this trial, 1p and 19q status was determined by FISH in participants with sufficient tissue. These previously obtained FISH results were not reported for the participants included in Dubbink 2016. Similarly, Wiestler 2014 compared methylation array with MLPA in the biomarker cohort of the NOA‐04 trial. In this trial, MLPA was used to detect 1p/19q codeletion, and PCR‐based LOH was also used in participants with leukocyte DNA available. There was no comparison of the results of PCR‐based LOH with MLPA or methylation array (or both).
Applicability
QUADAS‐2 applicability assessments for studies that performed two or more categories of test are shown in Figure 7, and assessments for studies that performed two or more FISH variants are shown in Appendix 9.
We had concerns over the applicability of the included participants for many studies. This is because many of the studies only included participants with specific subtypes of glioma. The results of these studies may not be applicable to all gliomas. We only had low concerns over the applicability of the patient population if the study had included at least patients with both astrocytomas and oligodendrogliomas of several grades, or if the study stated that all gliomas were eligible for inclusion. We had high concerns over the applicability of the included participants in Ransom 1992a and Ransom 1992b. However, these two studies could be considered one larger study: they appeared to have been one study that was then subdivided by subtype of glioma. If they were combined, we would have had low concerns over applicability.
We had low concerns regarding the applicability of the index tests in most studies. We felt that it would be rare that the index tests were conducted or interpreted in a manner that differed from our review question, and, therefore, we had low concerns even in situations where there was minimal detail regarding the index test (e.g. in conference abstracts). We had low concerns regarding the applicability of G‐banding/karyotyping in studies that performed this test, as results were given for the entire genome, although it should be noted that this technique was rarely used with the expressed purpose of determining 1p/19q status in the studies identified. We had concerns over the index test in a couple of studies. In Sim 2018a (glioblastoma cohort), we had concerns over the aCGH or NGS test (or both). The use of one test or another test (or both) was not applicable to our review question. In Thomas 2017, although we could extract 1p/19q NGS results, there was no attempt to use NGS to determine 1p/19q status in the paper, so it did not appear to represent how the test would be used for this purpose in practice. In Duval 2015, the automated FISH analysis was performed on archival slides that had been stored at −20 °C. The results of the automated analysis were compared with results of the initial manual analysis that had probably been performed when the slides were 'fresh'. Discordances in this study were attributed to degradation of the FISH signals during storage, and this storage is non‐standard.
Findings
Results of test accuracy
We report results for analyses of each test using FISH as a reference standard followed by results for each test using PCR‐based LOH as a reference standard. We then present results of the latent class analysis comparing FISH with PCR‐based LOH and provide a comparison of results based on the two reference standards. Finally, we present the limited data available from studies that did not include FISH or PCR‐based LOH, and that compared different variants of FISH and PCR‐based LOH.
Using fluorescent in situ hybridisation as the reference standard
From the included studies that performed FISH and at least one other test that was not a FISH variant, we created 41 cross‐classified 2 × 2 tables (from 33 studies, 1520 participants) in which FISH was treated as the reference standard. FISH has been compared directly with 10 different test categories: CISH (one comparison), PCR‐based LOH (15 comparisons), real‐time PCR (two comparisons), MLPA (two comparisons), CGH (four comparisons), aCGH (three comparisons), SNP array (six comparisons), NGS (six comparisons), MS (one comparison) and NanoString (one comparison). Of these 33 studies contributing to the simultaneous analysis, 26 studies compared FISH with one other test category, six studies compared FISH with two other test categories and one study compared FISH with three other test categories.
The main results from the bivariate meta‐analysis model are presented in Figure 8 (forest plots) and Figure 9 (summary receiver operating characteristic plots for tests that had been examined in sufficient studies to draw them), and are summarised in Table 1.
8.
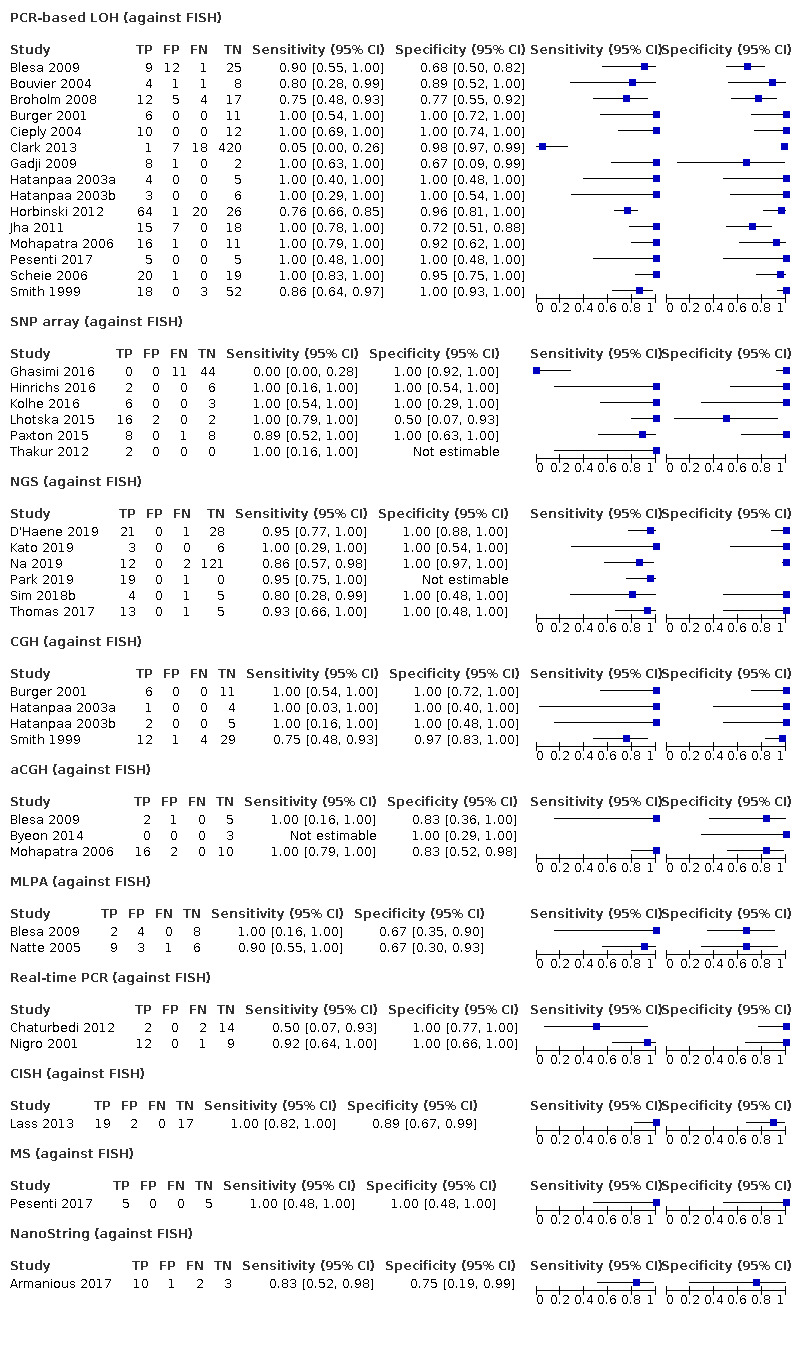
Forest plot of tests: 1 PCR‐based LOH (against FISH), 2 SNP array (against FISH), 3 NGS (against FISH), 4 CGH (against FISH), 5 aCGH (against FISH), 6 MLPA (against FISH), 7 real‐time PCR (against FISH), 8 CISH (against FISH), 9 MS (against FISH), 10 NanoString (against FISH).
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CI: confidence interval; CISH: chromogenic in situ hybridisation; FISH: fluorescence in situ hybridisation; FN: false negative; FP: false positive; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS; mass spectrometry; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism; TN: true negative; TP: true positive.
9.

Receiver operating characteristic plots obtained using FISH as the reference standard (panel A) and PCR‐based LOH as the reference standard (panel B) for tests with four or more studies and variation in both sensitivity and specificity. Summary estimates of sensitivity and specificity with 95% credible regions are included along with a hierarchical summary receiver operating characteristic (HSROC) line.
CGH: comparative genomic hybridisation; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism.
CISH: results using FISH as reference standard. Only one study (with 38 participants) contributed data on CISH against FISH (Data table 1). The estimated sensitivity was 1.00 (95% CrI 0.84 to 1.00) and specificity was 0.92 (95% CrI 0.33 to 1.00). Our GRADE assessment was of low certainty due to high imprecision.
1. Test.

CISH (against FISH)
PCR‐based LOH: results using FISH as reference standard. 15 studies (915 participants) provided data on PCR‐based LOH against FISH (Data table 2), making this the comparison with the strongest evidence base. Central estimates were high with of sensitivity of 0.94 (95% CrI 0.83 to 0.98) and specificity of 0.94 (95% CrI 0.87 to 0.98), indicating high concordance with FISH results. Our GRADE assessment was of low certainty due to the risk of bias in the individual study results and indirectness. Because we had more than four studies, we included PCR‐based LOH in the sensitivity analysis with test‐specific between‐study heterogeneity and correlation parameters. The results were identical within the level of precision presented (see Table 6).
2. Test.
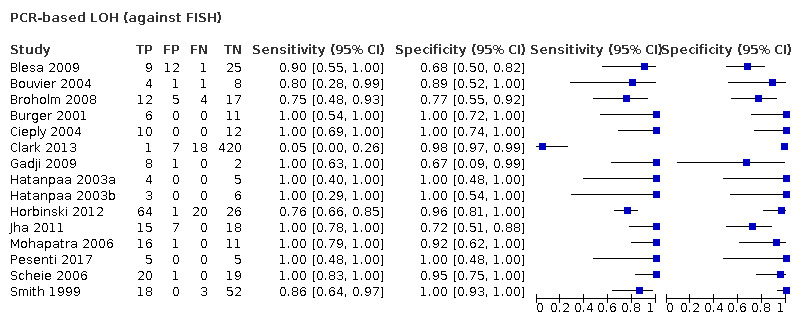
PCR‐based LOH (against FISH)
4. Results of the main analysis (simultaneous analysis of all tests against FISH), with results of the sensitivity analysis (separate analyses for each test with four or more studies) using FISH as the reference standard.
| Test | Number of studies | Main analysis | Sensitivity analysis | ||
| Sensitivity (95% CrI) | Specificity (95% CrI) | Sensitivity (95% CrI) | Specificity (95% CrI) | ||
| CISH | 1 | 1.00 (0.84 to 1.00) | 0.92 (0.33 to 1.00) | Insufficient studies | |
| PCR‐based LOH | 15 | 0.94 (0.83 to 0.98) | 0.94 (0.87 to 0.98) | 0.94 (0.83 to 0.98) | 0.94 (0.87 to 0.98) |
| Real‐time PCR | 2 | 0.81 (0.20 to 0.99) | 1.00 (0.95 to 1.00) | Insufficient studies | |
| MLPA | 2 | 0.96 (0.44 to 1.00) | 0.68 (0.20 to 0.95) | Insufficient studies | |
| CGH | 4 | 0.95 (0.59 to 1.00) | 0.99 (0.90 to 1.00) | 0.90 (0.64 to 0.99) | 0.99 (0.91 to 1.00) |
| aCGH | 3 | 1.00 (0.89 to 1.00) | 0.91 (0.55 to 0.99) | Insufficient studies | |
| SNP array | 6 | 0.90 (0.57 to 0.99) | 0.97 (0.84 to 1.00) | 0.90 (0.57 to 0.99) | 0.97 (0.84 to 1.00) |
| NGS | 6 | 0.94 (0.75 to 0.99) | 1.00 (0.99 to 1.00) | 0.93 (0.83 to 0.98) | 1.00 (0.99 to 1.00) |
| MS | 1 | 1.00 (0.60 to 1.00) | 1.00 (0.70 to 1.00) | Insufficient studies | |
| NanoString | 1 | 0.85 (0.11 to 1.00) | 0.80 (0.10 to 1.00) | Insufficient studies | |
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; CrI: credible interval; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS; mass spectrometry; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism.
RFLP: results using FISH as reference standard. No studies compared RFLP with FISH.
Real‐time PCR: results using FISH as reference standard. Two comparisons (40 participants) provided data on real‐time PCR against FISH (Data table 3). Sensitivity was estimated to be 0.81 (95% CrI 0.20 to 0.99) and specificity to be 1.00 (95% CrI 0.95 to 1.00). Our GRADE assessment was of very low certainty, with low precision for sensitivity as well as risk of bias and indirectness.
3. Test.

Real‐time PCR (against FISH)
MLPA: results using FISH as reference standard. Two comparisons (33 participants) provided data on MLPA against FISH (Data table 4). Central estimates of sensitivity was 0.96 (95% CrI 0.44 to 1.00) and specificity was 0.68 (95% CrI 0.20 to 0.95), the latter result arising from seven cases (out of 21) in which MLPA identified a deletion when FISH did not. Our GRADE assessment was of very low certainty.
4. Test.

MLPA (against FISH)
CGH: results using FISH as reference standard. Four comparisons (75 participants) provided data on CGH against FISH (Data table 5). Central estimates of sensitivity was 0.95 (95% CrI 0.59 to 1.00) and specificity was 0.99 (95% CrI 0.90 to 1.00), providing some evidence of high specificity in relation to FISH. Our GRADE assessment was of low certainty. The sensitivity analysis gave a similar result, although with a slightly different result for sensitivity, perhaps not surprising given that sensitivity was estimated imprecisely in both analyses (see Table 6).
5. Test.
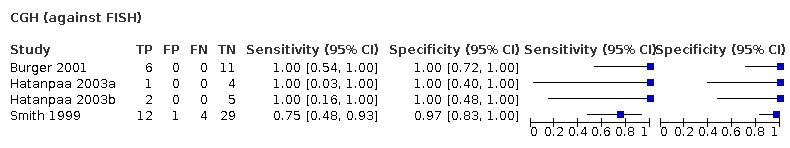
CGH (against FISH)
aCGH: results using FISH as reference standard. Three comparisons (39 participants) provided data on aCGH against FISH (Data table 6). Sensitivity was estimated to be high at 1.00 (95% CrI 0.89 to 1.00) although specificity was imprecisely estimated at 0.91 (95% CrI 0.55 to 0.99). Our GRADE assessment was of very low certainty.
6. Test.
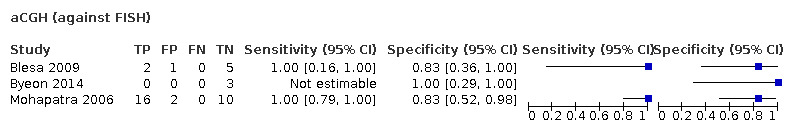
aCGH (against FISH)
SNP arrays: results using FISH as reference standard. Six comparisons (111 participants) provided data on SNP arrays against FISH (Data table 7). Central estimates of sensitivity was 0.90 (95% CrI 0.57 to 0.99) and specificity was 0.97 (95% CrI 0.84 to 1.00). Our GRADE assessment was of very low certainty. The sensitivity analysis gave a very similar result (see Table 6).
7. Test.
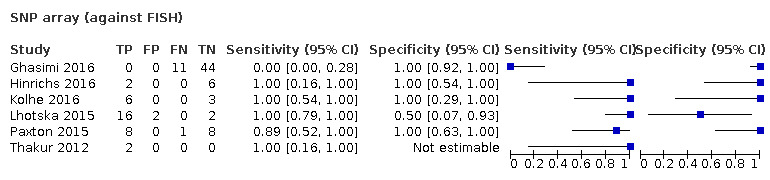
SNP array (against FISH)
Methylation arrays: results using FISH as reference standard. No studies compared methylation arrays with FISH.
NGS: results using FISH as reference standard. Six comparisons (243 participants) provided data on NGS against FISH (Data table 8). Central estimates of sensitivity was 0.94 (95% CrI 0.75 to 0.99) and specificity was 1.00 (95% CrI 0.99 to 1.00). Our GRADE assessment was of low certainty. Results of the sensitivity analysis were very similar (Table 6).
8. Test.
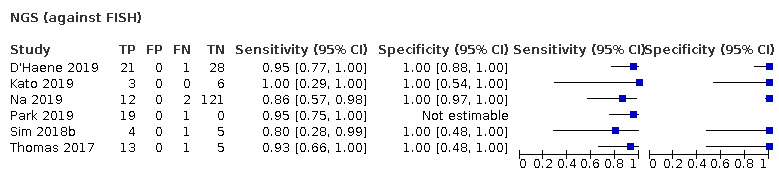
NGS (against FISH)
MS: results using FISH as reference standard. Very little information was available for MS against FISH, with just one comparison involving 10 participants (Data table 9), leaving us with very low certainty.
9. Test.

MS (against FISH)
NanoString: results using FISH as reference standard. Very little information was available for NanoString against FISH, with just one comparison involving 16 participants (Data table 10), leaving us with very low certainty.
10. Test.

NanoString (against FISH)
G‐banding: results using FISH as reference standard. No studies compared G‐banding with FISH.
Overview across tests: using FISH as reference standard. Differences between tests in sensitivity and specificity are shown in Table 7. There are suggestions of differences between tests such that specificity of NGS may be higher than for PCR‐based LOH and SNP array, and that real‐time PCR may have higher specificity than PCR‐based LOH, aCGH and MLPA.
5. Differences in sensitivity (upper right triangle) and specificity (bottom left triangle) from analyses using FISH as the reference standard. Values are test in column minus test in row (95% credible interval). Positive values favour the test defining the column, negative values favour the test defining the row.
| Test | PCR‐based LOH | SNP array | NGS | CGH | aCGH | MLPA | Real‐time PCR | CISH | MS | NanoString |
| PCR‐based LOH | — | −0.03 (−0.37 to 0.1) | 0 (−0.19 to 0.12) | 0.01 (−0.35 to 0.13) | 0.05 (−0.05 to 0.16) | 0.02 (−0.5 to 0.14) | −0.12 (−0.74 to 0.09) | 0.05 (−0.1 to 0.16) | 0.05 (−0.34 to 0.16) | −0.08 (−0.82 to 0.11) |
| SNP array | −0.03 (−0.11 to 0.1) | — | 0.04 (−0.17 to 0.37) | 0.03 (−0.31 to 0.37) | 0.09 (−0.03 to 0.42) | 0.04 (−0.46 to 0.38) | −0.08 (−0.7 to 0.29) | 0.09 (−0.07 to 0.42) | 0.08 (−0.29 to 0.42) | −0.04 (−0.79 to 0.32) |
| NGS | −0.06 (−0.13 to −0.02) | −0.03 (−0.16 to 0) | — | 0 (−0.35 to 0.2) | 0.05 (−0.05 to 0.25) | 0.01 (−0.5 to 0.21) | −0.12 (−0.74 to 0.14) | 0.05 (−0.1 to 0.24) | 0.04 (−0.33 to 0.24) | −0.08 (−0.82 to 0.16) |
| CGH | −0.05 (−0.12 to 0.04) | −0.01 (−0.14 to 0.07) | 0.01 (0 to 0.1) | — | 0.05 (−0.06 to 0.4) | 0.01 (−0.49 to 0.35) | −0.11 (−0.74 to 0.26) | 0.05 (−0.1 to 0.4) | 0.04 (−0.32 to 0.4) | −0.07 (−0.82 to 0.29) |
| aCGH | 0.03 (−0.08 to 0.4) | 0.06 (−0.09 to 0.42) | 0.09 (0.01 to 0.45) | 0.08 (−0.03 to 0.44) | — | −0.03 (−0.56 to 0.06) | −0.18 (−0.8 to 0) | 0 (−0.16 to 0.1) | 0 (−0.39 to 0.1) | −0.14 (−0.88 to 0.02) |
| MLPA | 0.25 (−0.02 to 0.75) | 0.28 (0 to 0.77) | 0.32 (0.05 to 0.8) | 0.3 (0.03 to 0.79) | 0.2 (−0.22 to 0.71) | — | −0.11 (−0.74 to 0.39) | 0.03 (−0.11 to 0.55) | 0.03 (−0.32 to 0.55) | −0.08 (−0.83 to 0.41) |
| Real‐time PCR | −0.06 (−0.13 to 0) | −0.02 (−0.16 to 0.03) | 0 (−0.01 to 0.05) | −0.01 (−0.09 to 0.04) | −0.09 (−0.45 to 0) | −0.31 (−0.8 to −0.04) | — | 0.18 (−0.03 to 0.8) | 0.17 (−0.2 to 0.79) | 0.02 (−0.73 to 0.68) |
| CISH | 0.02 (−0.09 to 0.62) | 0.05 (−0.1 to 0.64) | 0.08 (0 to 0.67) | 0.07 (−0.05 to 0.66) | −0.01 (−0.36 to 0.57) | −0.19 (−0.72 to 0.4) | 0.08 (−0.01 to 0.67) | — | 0 (−0.39 to 0.15) | −0.14 (−0.88 to 0.05) |
| MS | −0.05 (−0.13 to 0.24) | −0.02 (−0.15 to 0.26) | 0 (−0.01 to 0.3) | −0.01 (−0.09 to 0.28) | −0.08 (−0.44 to 0.19) | −0.3 (−0.79 to 0.04) | 0 (−0.04 to 0.29) | −0.07 (−0.66 to 0.19) | — | −0.13 (−0.87 to 0.23) |
| NanoString | 0.14 (−0.08 to 0.84) | 0.16 (−0.07 to 0.87) | 0.2 (0 to 0.9) | 0.18 (−0.03 to 0.88) | 0.09 (−0.31 to 0.81) | −0.09 (−0.67 to 0.66) | 0.2 (0 to 0.9) | 0.08 (−0.5 to 0.81) | 0.18 (−0.13 to 0.89) | — |
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS: mass spectrometry; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism.
Technical details
The main results above arise from a single, simultaneous analysis in which the same amount of heterogeneity was assumed for all tests, so a small number of studies for a specific test does not adversely affect our ability to estimate heterogeneity in the random‐effects model. Estimated between‐study standard deviation and correlation parameters from both the main analysis and the sensitivity analysis are shown in Appendix 10 (Table A10.1). Several of these estimates are likely heavily driven by the prior distributions, due to the limited data available. The estimated heterogeneity standard deviation in the main analysis was 1.84 (95% CrI 1.42 to 1.99) on the logit(sensitivity) scale and 1.36 (95% CrI 0.78 to 1.94) on the logit(1 – specificity) scale).
Using polymerase chain reaction‐based loss of heterozygosity as the reference standard
From the included studies that performed PCR‐based LOH and at least one other test that was not a PCR‐based LOH variant, we created 32 cross‐classified 2 × 2 tables (from 22 studies, 1304 participants) in which PCR‐based LOH was treated as the reference standard. PCR‐based LOH has been compared directly with nine different test categories: FISH (15 comparisons, the same as those summarised in 'Using FISH as the reference standard'), real‐time PCR (one comparison), MLPA (one comparison), CGH (six comparisons), aCGH (four comparisons), SNP array (two comparisons), NGS (one comparison), G‐banding (one comparison), and MS (one comparison). Of these 22 studies contributing to the simultaneous analysis, 14 studies compared PCR‐based LOH with one other test category, six studies compared PCR‐based LOH with two other test categories, and two studies compared PCR‐based LOH with three other test categories.
The main results from the bivariate meta‐analysis model are presented in Figure 20 (forest plots) and Figure 9 (summary receiver operating characteristic plots for tests that had been examined in sufficient studies to draw them), and are summarised in Table 2).
10.
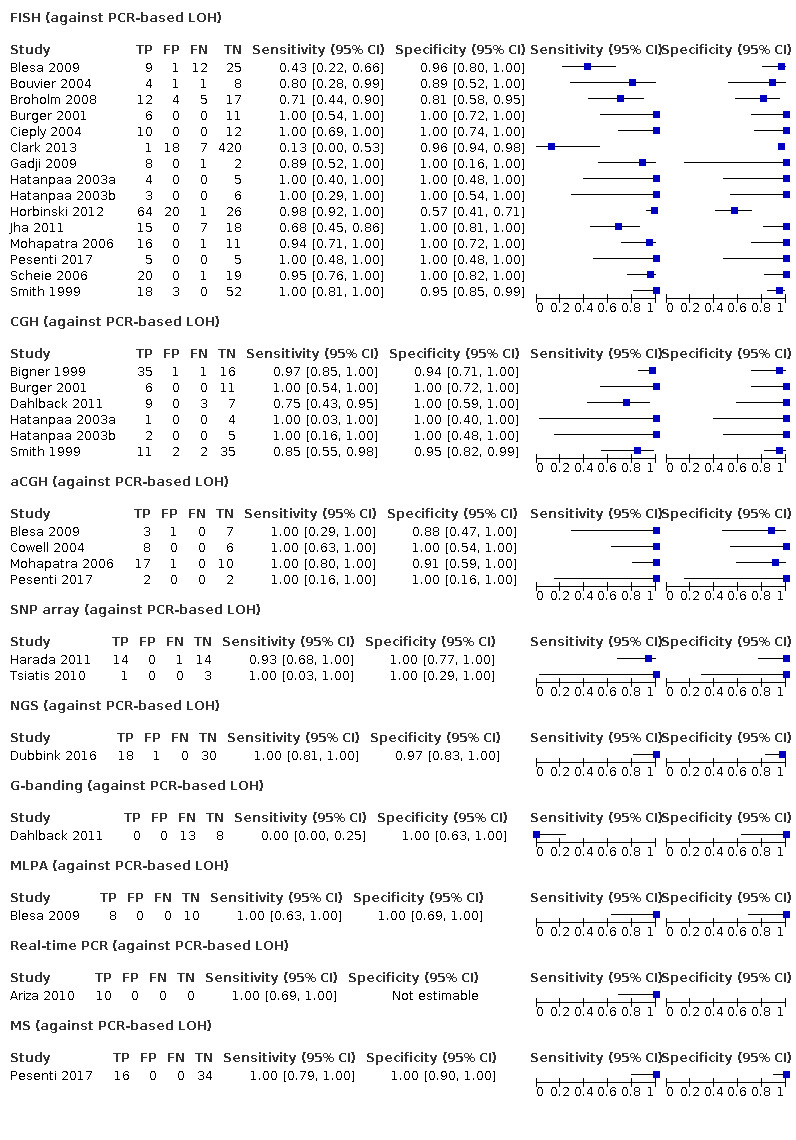
Forest plot of tests: 11 FISH (against PCR‐based LOH), 12 CGH (against PCR‐based LOH), 13 aCGH (against PCR‐based LOH), 14 SNP array (against PCR‐based LOH), 15 NGS (against PCR‐based LOH), 16 G‐banding (against PCR‐based LOH), 17 MLPA (against PCR‐based LOH), 18 real‐time PCR (against PCR‐based LOH), 19 MS (against PCR‐based LOH).
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; FISH: fluorescence in situ hybridisation; FN: false negative; FP: false positive; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS; mass spectrometry; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism; TN: true negative; TP: true positive.
FISH: results using PCR‐based LOH as reference standard. Fifteen comparisons (915 participants) provided data on FISH against PCR‐based LOH (Data table 11). The central estimate of sensitivity was 0.91 (95% CrI 0.78 to 0.97), which is slightly lower than the sensitivity of PCR‐based LOH at detecting deletions determined by FISH (0.94, based on the same data). The central estimate of specificity was 0.96 (95% CrI 0.90 to 0.99), slightly higher than the converse (0.94, again based on the same data). A sensitivity analysis with test‐specific between‐study heterogeneity and correlation parameters gave the same results (Table 8). Our GRADE assessment was of low certainty due to risk of bias and indirectness.
11. Test.
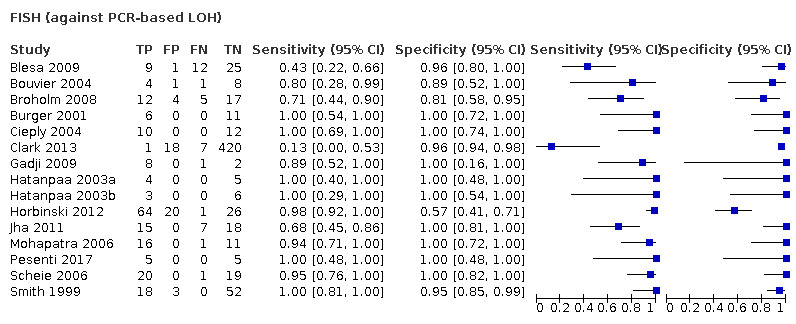
FISH (against PCR‐based LOH)
6. Results of the main analysis (simultaneous analysis of all tests against PCR‐based LOH), with results of the sensitivity analysis (separate analyses for each test with four or more studies) using PCR‐based LOH as the reference standard.
| Test | Number of studies | Main analysis | Sensitivity analysis | ||
| Sensitivity (95% CrI) | Specificity (95% CrI) | Sensitivity (95% CrI) | Specificity (95% CrI) | ||
| FISH | 15 | 0.91 (0.78 to 0.97) | 0.96 (0.90 to 0.99) | 0.91 (0.78 to 0.97) | 0.96 (0.90 to 0.99) |
| Real‐time PCR | 1 | 1.00 (0.77 to 1.00) | No data | Insufficient studies | |
| MLPA | 1 | 1.00 (0.74 to 1.00) | 1.00 (0.83 to 1.00) | Insufficient studies | |
| CGH | 6 | 0.94 (0.74 to 0.99) | 0.98 (0.91 to 1.00) | 0.93 (0.78 to 0.99) | 0.97 (0.91 to 1.00) |
| aCGH | 4 | 1.00 (0.97 to 1.00) | 0.96 (0.75 to 1.00) | 1.00 (0.97 to 1.00) | 0.95 (0.78 to 1.00) |
| SNP array | 2 | 0.97 (0.50 to 1.00) | 1.00 (0.92 to 1.00) | Insufficient studies | |
| NGS | 1 | 1.00 (0.86 to 1.00) | 0.98 (0.64 to 1.00) | Insufficient studies | |
| MS | 1 | 1.00 (0.85 to 1.00) | 1.00 (0.94 to 1.00) | Insufficient studies | |
| G‐banding | 1 | 0.00 (0.00 to 0.20) | 1.00 (0.78 to 1.00) | Insufficient studies | |
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CrI: credible interval; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS: mass spectrometry; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism.
CISH: results using PCR‐based LOH as reference standard. No studies compared CISH with PCR‐based LOH.
RFLP: results using PCR‐based LOH as reference standard. No studies compared RFLP with PCR‐based LOH.
Real‐time PCR: results using PCR‐based LOH as reference standard. One comparison (10 participants) provided data on real‐time PCR against PCR‐based LOH (Data table 12). The two techniques were completely concordant for the 10 participants, with all participants having positive results by both tests. Sensitivity was estimated to be 1.00 (95% CrI 0.77 to 1.00). Specificity was not estimable (as there were no negative cases by either test). Our GRADE assessment was of very low certainty, due to risk of bias, the small number of participants tested and indirectness.
12. Test.

Real‐time PCR (against PCR‐based LOH)
MLPA: results using PCR‐based LOH as reference standard. One comparison (18 participants) provided data on MLPA against PCR‐based LOH (Data table 13). Results were completely concordant for the 18 participants. Central estimates of sensitivity was 1.00 (95% CrI 0.74 to 1.00) and specificity was 1.00 (95% CrI 0.83 to 1.00). Our GRADE assessment was of very low certainty, due to high risk of bias, the small number of participants tested and indirectness.
13. Test.

MLPA (against PCR‐based LOH)
CGH: results using PCR‐based LOH as reference standard. Six comparisons (151 participants) provided data on CGH against PCR‐based LOH (Data table 14). Central estimates of sensitivity was 0.94 (95% CrI 0.74 to 0.99) and specificity was 0.98 (95% CrI 0.91 to 1.00), providing some evidence of high specificity in relation to PCR‐based LOH. Our GRADE assessment was of low certainty due to risk of bias and indirectness. The sensitivity analysis gave very similar results (see Table 8).
14. Test.
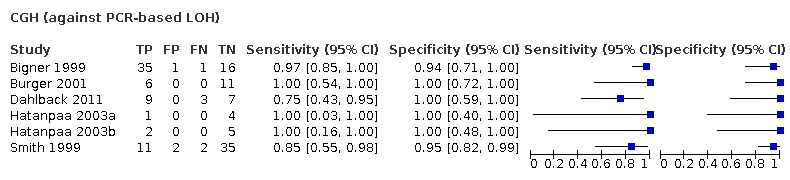
CGH (against PCR‐based LOH)
aCGH: results using PCR‐based LOH as reference standard. Four comparisons (57 participants) provided data on aCGH against PCR‐based LOH (Data table 15). Sensitivity (1.00, 95% CrI 0.97 to 1.00) and specificity (0.96, 95% CrI 0.75 to 1.00) were estimated to be high. Our GRADE assessment was of low certainty due to high risk of bias. The sensitivity analysis gave very similar results (see Table 8).
15. Test.
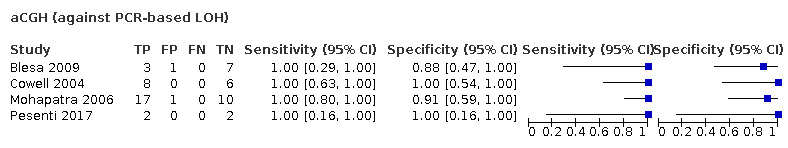
aCGH (against PCR‐based LOH)
SNP arrays: results using PCR‐based LOH as reference standard. Two comparisons (33 participants) provided data on SNP arrays against PCR‐based LOH (Data table 16). Sensitivity was estimated at 0.97 (95% CrI 0.50 to 1.00) and specificity at 1.00 (95% CrI 0.92 to 1.00). Our GRADE assessment was of very low certainty due to risk of bias and high imprecision in the estimate of sensitivity.
16. Test.
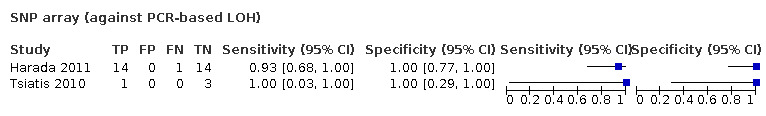
SNP array (against PCR‐based LOH)
Methylation arrays: results using PCR‐based LOH as reference standard. No studies compared methylation arrays with PCR‐based LOH.
NGS: results using PCR‐based LOH as reference standard. One comparison (49 participants) provided data on NGS against PCR‐based LOH (Data table 17). Sensitivity was estimated at 1.00 (95% CrI 0.86 to 1.00) and specificity at 0.98 (95% CrI 0.64 to 1.00). Our GRADE assessment was of very low certainty due to risk of bias, imprecision (of the estimate of specificity) and indirectness.
17. Test.

NGS (against PCR‐based LOH)
MS: results using PCR‐based LOH as reference standard. One comparison (50 participants) provided data on MS against PCR‐based LOH (Data table 18). Sensitivity was estimated at 1.00 (95% CrI 0.85 to 1.00) and specificity at 1.00 (95% CrI 0.94 to 1.00). Our GRADE assessment was of very low certainty due to risk of bias and the small number of participants tested.
18. Test.

MS (against PCR‐based LOH)
NanoString: results using PCR‐based LOH as reference standard. No studies compared NanoString with PCR‐based LOH.
G‐banding: results using PCR‐based LOH as reference standard. One comparison (21 participants) provided data on G‐banding against PCR‐based LOH (Data table 19). Sensitivity was estimated at 0.00 (95% CrI 0.00 to 0.20) and specificity at 1.00 (95% CrI 0.78 to 1.00). The poor estimate of sensitivity for G‐banding/karyotyping is based on a single study in which none of 13 PCR‐detected 1p/19q codeletions were identified. Our GRADE assessment was of very low certainty due to high risk of bias, high imprecision and indirectness.
19. Test.

G‐banding (against PCR‐based LOH)
Overview across tests: PCR‐based LOH as reference standard. Differences between tests in sensitivity and specificity are shown in Table 9. There is a suggestion of greater sensitivity of aCGH than CGH. Other than differences relating to the poor estimated sensitivity for G‐banding/karyotyping, there were no other apparent differences between tests in either sensitivity or specificity.
7. Differences in sensitivity (upper right triangle) and specificity (bottom left triangle) from analyses using PCR‐based LOH as the reference standard. Values are test in column minus test in row (95% credible interval). Positive values favour the test defining the column, negative values favour the test defining the row.
| Test | FISH | CGH | aCGH | SNP array | NGS | G‐banding | MLPA | Real‐time PCR | MS |
| FISH | — | 0.03 (−0.17 to 0.17) | 0.09 (0.03 to 0.22) | 0.05 (−0.41 to 0.19) | 0.09 (−0.05 to 0.21) | −0.9 (−0.97 to −0.68) | 0.08 (−0.17 to 0.21) | 0.08 (−0.14 to 0.21) | 0.09 (−0.06 to 0.21) |
| CGH | −0.02 (−0.08 to 0.05) | — | 0.05 (0 to 0.25) | 0.02 (−0.44 to 0.22) | 0.05 (−0.08 to 0.25) | −0.93 (−0.99 to −0.65) | 0.05 (−0.2 to 0.25) | 0.05 (−0.16 to 0.25) | 0.05 (−0.09 to 0.25) |
| aCGH | 0 (−0.07 to 0.2) | 0.02 (−0.06 to 0.22) | — | −0.03 (−0.5 to 0.01) | 0 (−0.14 to 0.03) | −1 (−1 to −0.79) | 0 (−0.26 to 0.03) | 0 (−0.23 to 0.03) | 0 (−0.15 to 0.03) |
| SNP array | −0.04 (−0.1 to 0.04) | −0.02 (−0.09 to 0.06) | −0.04 (−0.24 to 0.04) | — | 0.03 (−0.09 to 0.49) | −0.95 (−1 to −0.43) | 0.03 (−0.2 to 0.49) | 0.03 (−0.17 to 0.49) | 0.03 (−0.1 to 0.49) |
| NGS | −0.02 (−0.08 to 0.32) | 0 (−0.07 to 0.34) | −0.02 (−0.21 to 0.31) | 0.02 (−0.05 to 0.36) | — | −1 (−1 to −0.69) | 0 (−0.25 to 0.13) | 0 (−0.22 to 0.13) | 0 (−0.14 to 0.13) |
| G‐banding | −0.04 (−0.1 to 0.17) | −0.01 (−0.09 to 0.19) | −0.04 (−0.24 to 0.16) | 0 (−0.07 to 0.21) | −0.02 (−0.35 to 0.18) | — | 1 (0.6 to 1) | 1 (0.63 to 1) | 1 (0.69 to 1) |
| MLPA | −0.04 (−0.1 to 0.12) | −0.02 (−0.09 to 0.14) | −0.04 (−0.24 to 0.12) | 0 (−0.07 to 0.16) | −0.02 (−0.35 to 0.13) | 0 (−0.21 to 0.15) | — | 0 (−0.21 to 0.25) | 0 (−0.14 to 0.25) |
| Real‐time PCR | 0.45 (−0.08 to 0.98) | 0.47 (−0.06 to 1) | 0.43 (−0.17 to 0.99) | 0.49 (−0.03 to 1) | 0.42 (−0.22 to 1) | 0.47 (−0.09 to 1) | 0.48 (−0.06 to 1) | — | 0 (−0.14 to 0.22) |
| MS | −0.04 (−0.1 to 0.02) | −0.02 (−0.09 to 0.04) | −0.04 (−0.24 to 0.02) | 0 (−0.08 to 0.05) | −0.02 (−0.36 to 0.03) | 0 (−0.21 to 0.05) | 0 (−0.16 to 0.05) | −0.49 (−1 to 0.02) | — |
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS: mass spectrometry; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism.
Technical details
The main results above arose from a single, simultaneous analysis in which the same amount of heterogeneity was assumed for all tests (see Appendix 10, Table A10.2). The estimated heterogeneity standard deviation in the main analysis was 1.66 (95% CrI 1.09 to 1.98) on the logit(sensitivity) scale and 1.27 (95% CrI 0.69 to 1.93) on the logit(1 – specificity) scale. Several of the estimates presented above are likely to be heavily driven by the prior distributions, due to the limited data available. Note that the prior distributions forced correlation parameters to be non‐negative and standard deviation parameters to be no greater than 2.
Latent class analysis
A total of 915 participants from 15 studies provided data on both FISH and PCR‐based LOH. We present results from three latent class models applied to these data in Table 10. Point estimates of sensitivity were sensitive to the assumptions we made, although CrIs had a large degree of overlap. Our preferred model from a theoretical perspective had been the model allowing for conditional dependencies between FISH and PCR‐based LOH test results. However, we observed no improvement in model fit (as measured by residual deviance) between this model and the conditional independence model despite including many more parameters, so we focus on results from the conditional independence model. The results suggest that PCR‐based LOH may be slightly more accurate than FISH, consistent with our a priori expectation that PCR‐based LOH has near‐perfect sensitivity. Sensitivity of PCR‐based LOH was estimated as 0.97 (95% CrI 0.90 to 1.00) and sensitivity of FISH as 0.95 (95% CrI 0.83 to 1.00). Specificity of PCR‐based LOH was estimated as 0.98 (95% CrI 0.91 to 1.00) and specificity of FISH as 0.97 (95% CrI 0.93 to 0.99). As explained in the methods section, we were unable to include the other tests in the latent class analysis.
8. Results from latent class analyses of FISH and PCR‐based LOH.
| Analysis | Sensitivity of FISH (95% CrI) | Specificity of FISH (95% CrI) | Sensitivity of PCR‐based LOH (95% CrI) | Specificity of PCR‐based LOH (95% CrI) |
| Assuming conditional independence | 0.95 (0.83 to 1.00) |
0.97 (0.93 to 0.99) |
0.97 (0.90 to 1.00) |
0.98 (0.91 to 1.00) |
| Allowing for conditional dependencies | 0.90 (0.74 to 0.99) |
0.96 (0.91 to 0.99) |
0.94 (0.80 to 0.99) |
0.98 (0.89 to 1.00) |
| With an informative prior distribution forcing sensitivity of PCR‐based LOH to be ≥ 95% | 0.92 (0.78 to 0.99) |
0.96 (0.90 to 0.99) |
0.97 (0.95 to 1.00) |
0.98 (0.88 to 1.00) |
These results are derived from 910 participants from 16 studies for whom test results were available for both FISH and PCR‐based LOH.
CrI: credible interval; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; PCR: polymerase chain reaction.
Comparison of results obtained when using fluorescent in situ hybridisation or polymerase chain reaction‐based loss of heterozygosity as the reference standard
A comparison of the results obtained using FISH as the reference standard and using PCR‐based LOH as the reference standard (for test categories that were included in both analyses) is shown in Table 11. Note that different studies contributed to the two analyses for each test category and care is required not to over interpret the results. Apart from a general observation that methods are generally in good agreement with both FISH and PCR‐based LOH, there is little to discern between the two sets of analyses. There is a suggestion that MLPA may label more cases as codeletions when FISH does not than when PCR‐based LOH does not (specificity 0.68, 95% CI 0.20 to 0.95 when FISH is the reference versus 1.00, 95% CI 0.83 to 1.00 when PCR‐based LOH is the reference) and that real‐time PCR may miss more cases that FISH detects as codeletions than PCR‐based LOH detects as codeletions (sensitivity 0.81, 95% CI 0.20 to 0.99 when FISH is the reference versus 1.00, 95% CI 0.77 to 1.00 when PCR‐based LOH is the reference).
9. Comparison of results obtained when using FISH or PCR‐based LOH as the reference standard.
| Test | Reference standard | Number of studies | Number people with disease | Number people without disease | Total people in meta‐analysis | Sensitivity (95% CrI) | Specificity (95% CrI) |
| Real‐time PCR | FISH | 2 | 17 | 23 | 40 | 0.81 (0.20 to 0.99) | 1.00 (0.95 to 1.00) |
| PCR‐based LOH | 1 | 10 | 0 | 10 | 1.00 (0.77 to 1.00) | N/A | |
| MLPA | FISH | 2 | 12 | 21 | 33 | 0.96 (0.44 to 1.00) | 0.68 (0.20 to 0.95) |
| PCR‐based LOH | 1 | 8 | 10 | 18 | 1.00 (0.74 to 1.00) | 1.00 (0.83 to 1.00) | |
| CGH | FISH | 4 | 25 | 50 | 75 | 0.95 (0.59 to 1.00) | 0.99 (0.90 to 1.00) |
| PCR‐based LOH | 6 | 70 | 81 | 151 | 0.94 (0.74 to 0.99) | 0.98 (0.91 to 1.00) | |
| aCGH | FISH | 3 | 18 | 21 | 39 | 1.00 (0.89 to 1.00) | 0.91 (0.55 to 0.99) |
| PCR‐based LOH | 4 | 30 | 27 | 57 | 1.00 (0.97 to 1.00) | 0.96 (0.75 to 1.00) | |
| SNP array | FISH | 6 | 46 | 65 | 111 | 0.90 (0.57 to 0.99) | 0.97 (0.84 to 1.00) |
| PCR‐based LOH | 2 | 16 | 17 | 33 | 0.97 (0.50 to 1.00) | 1.00 (0.92 to 1.00) | |
| NGS | FISH | 6 | 78 | 165 | 243 | 0.94 (0.75 to 0.99) | 1.00 (0.99 to 1.00) |
| PCR‐based LOH | 1 | 18 | 31 | 49 | 1.00 (0.86 to 1.00) | 0.98 (0.64 to 1.00) | |
| MS | FISH | 1 | 5 | 5 | 10 | 1.00 (0.60 to 1.00) | 1.00 (0.70 to 1.00) |
| PCR‐based LOH | 1 | 16 | 34 | 50 | 1.00 (0.85 to 1.00) | 1.00 (0.94 to 1.00) |
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CrI: credible interval; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS; mass spectrometry; N/A: not applicable; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism.
Results for other comparison of tests
Six studies (262 participants) did not include FISH or PCR‐based LOH. One study (71 participants) compared CGH with MLPA (Jeuken 2006; Data table 20), finding them to be highly concordant. One study (99 participants) compared methylation array with MLPA (Wiestler 2014; Data table 21), observing seven cases in which MLPA identified a deletion but methylation array did not (specificity 0.85, 95% CrI 0.71 to 0.94). Two studies (65 participants) compared G‐banding with CGH (Dahlback 2009; Schrock 1994); we present the results of this comparison, including also a result from a third study that also included PCR‐based LOH (Dahlback 2011; included in the analyses above) in Data table 22 (total 75 participants). G‐banding found no deletion detections using CGH. Finally, two comparisons (27 participants) were made between G‐banding and RFLP (Ransom 1992a; Ransom 1992b; Data table 23). Again G‐banding failed to identify any instances in which RFLP detected a deletion.
20. Test.

CGH (against MLPA)
21. Test.

Methylation array (against MLPA)
22. Test.
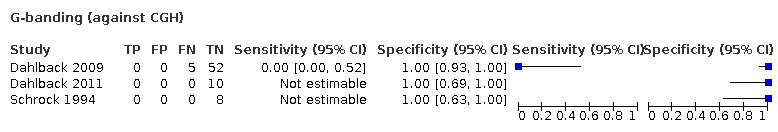
G‐banding (against CGH)
23. Test.
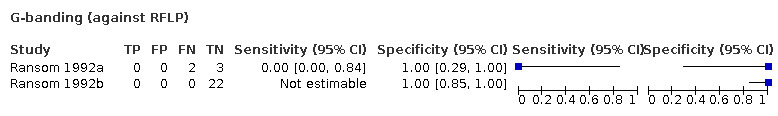
G‐banding (against RFLP)
Seven studies compared different versions of FISH. Belaud‐Rotureau 2006 performed FISH with manual analysis with the 1p36.3 (D1Z2)/1q12 (D1Z1) and 19q13.3/19pter probe set, manual analysis with the 1p36/1q25 and 19q13/19p13 Abbott Vysis probe set, and automatic analysis (Metafer 4, Metasystems, Althlussheim, Germany) with the 1p36/1q25 and 19q13/19p13 Abbott Vysis probe set. Ten participants were tested with all three FISH variants, and 13 with two FISH variants. For all participants, the results obtained were concordant. Duval 2014 performed FISH and immunoFISH (FISH with immunohistochemistry against Ki67 (MIB‐1)), and used two different cut‐offs for both: a "combination" cut‐off (which was based on the number of cells showing a deletion) and a "ratio" cut‐off (based on the ratio of signals for 1p to 1q and 19q and 19p). Twenty participants were positive on all four FISH variants, and 14 negative on all four variants, but one obtained a negative result with ImmunoFISH using the "combination" cut‐off despite obtaining positive results with the other variants; and one was positive on FISH (both cut‐offs) but negative on ImmunoFISH (both cut‐offs). Duval 2015 performed FISH with automated analysis (Metafer 4 software (Metasystem) using the "1p19q tile‐sampling classifier") and FISH with manual analysis. There were discordant results for four of the 29 participants, Senetta 2013 performed FISH with two different cut‐offs (cut‐off ratios 1p of 0.8 or less and 19q of 0.8 or less and cut‐off ratios 1p of 0.7 or less and 19q of 0.8 or less). There were discordant results for 16 of the 143 participants. Srebotnik‐Kirbis 2016 performed FISH on fresh tissue cytospins and on FFPE sections. Results were concordant for all 12 participants. Uchida 2019 performed FISH with two different criteria for judging whether a deletion was present (signals of 1p or 19q less than signals of 1q or 19p or single signal of 1p or 19q and two signals of 1q or 19p; in both cases the cut‐off value was set at 20%). Results were discordant for five of the 141 participants. Horbinski 2012 performed FISH with two different cut‐offs (target‐ploidy control ratio less than 0.87, with at least 20% of nuclei showing deletion and target‐ploidy control ratio less than 0.75, with at least 20% of nuclei showing deletion), in addition to PCR‐based LOH. The FISH results using the two different cut‐offs were discordant for eight of the 111 participants. Raw results for these studies are presented in Appendix 6; we were unable to include the results of these studies in our statistical analyses.
There were two comparisons (in one publication) that compared different version of PCR‐based LOH. The study performed PCR‐based LOH with or without comparison to normal DNA (in addition to CGH and FISH). This study developed a cut‐off for PCR‐based LOH without comparison to normal DNA in one set of participants (Hatanpaa 2003a) and validated it in another set of participants (Hatanpaa 2003b). The results for both variants of PCR‐based LOH were concordant. Raw results for these studies are presented in Appendix 6; we were unable to include the results of these studies in our statistical analyses.
Results of model‐based economic evaluation
The base‐case economic analysis results along with results of the probabilistic sensitivity analyses are shown in Table 12 (for FISH as reference standard) and Table 13 (for PCR‐based LOH as reference standard). These were based on the overall prevalence (the proportion of condition positive participants in the population tested) of 0.31 among all the studies included in the meta‐analysis, the costs in Appendix 4 and the diagnostic accuracy results in Figure 8 and Figure 20. Many of the results derive from estimates of accuracy that are based on small numbers of studies, so caution is required in the interpretation of the point estimates of cost‐effectiveness from the deterministic analysis.
10. Costs and diagnostic accuracy of diagnostic tests to evaluate 1p/19q status codeletion (FISH as reference standard).
| Inputs | Deterministic analysis | Probabilistic sensitivity analysis | ||||||||
| Incremental cost per TP detected | ||||||||||
| Diagnostic test | Cost (GBP) | Effect (TP rate) | Incremental cost (GBP) | Incremental effect | ICER (GBP) | Prob of being CE at WTP of GBP 0 per TP | Prob of being CE at WTP of GBP 500 per TP | Prob of being CE at WTP of GBP 1000 per TP | Prob of being CE at WTP of GBP 5000 per TP | Prob of being CE at WTP of GBP 10,000 per TP |
| MLPA | 73 | 0.27 | — | — | — | 100% | 100% | 95% | 46% | 26% |
| PCR‐based LOH | 142 | 0.25 | — | — | Dominated | 0% | 0% | 0% | 0% | 0% |
| Real‐time PCR | 142 | 0.24 | — | — | Dominated | 0% | 0% | 2% | 2% | 1% |
| CISH | 186 | 0.30 | 113 | 0.03 | 3827 | 0% | 0% | 3% | 39% | 48% |
| aCGH | 233 | 0.29 | — | — | Dominated | 0% | 0% | 0% | 13% | 25% |
| SNP array | 257 | 0.23 | — | — | Dominated | 0% | 0% | 0% | 0% | 0% |
| NGS | 571 | 0.28 | — | — | Dominated | 0% | 0% | 0% | 0% | 0% |
| Incremental cost per TN detected | ||||||||||
| Diagnostic test | Cost (GBP) | Effect (TN rate) | Incremental cost (GBP) | Incremental effect | ICER (GBP) | Prob of being CE at WTP of GBP 0 per TN | Prob of being CE at WTP of GBP 500 per TN | Prob of being CE at WTP of GBP 1000 per TN | Prob of being CE at WTP of GBP 5000 per TN | Prob of being CE at WTP of GBP 10,000 per TN |
| MLPA | 73 | 0.45 | — | — | — | 100% | 16% | 1% | 0% | 0% |
| PCR‐based LOH | 142 | 0.65 | — | — | Dominated | 0% | 30% | 28% | 13% | 7% |
| Real‐time PCR | 142 | 0.66 | 69 | 0.22 | 326 | 0% | 54% | 71% | 73% | 67% |
| CISH | 186 | 0.59 | — | — | Dominated | 0% | 0% | 0% | 2% | 2% |
| aCGH | 233 | 0.66 | — | — | Dominated | 0% | 0% | 0% | 9% | 15% |
| SNP array | 257 | 0.66 | — | — | Dominated | 0% | 0% | 0% | 3% | 5% |
| NGS | 571 | 0.69 | 498 | 0.24 | 2111 | 0% | 0% | 0% | 0% | 4% |
| Incremental cost per CD | ||||||||||
| Diagnostic test | Cost (GBP) | Effect (CD rate) | Incremental cost (GBP) | Incremental effect | ICER (GBP) | Prob of being CE at WTP of GBP 0 per CD | Prob of being CE at WTP of GBP 500 per CD | Prob of being CE at WTP of GBP 1000 per CD | Prob of being CE at WTP of GBP 5000 per CD | Prob of being CE at WTP of GBP 10,000 per CD |
| MLPA | 73 | 0.72 | — | — | — | 100% | 23% | 4% | 0% | 0% |
| PCR‐based LOH | 142 | 0.90 | — | — | Dominated | 0% | 27% | 24% | 2% | 0% |
| Real‐time PCR | 142 | 0.91 | 69 | 0.19 | 362 | 0% | 47% | 54% | 27% | 18% |
| CISH | 186 | 0.89 | — | — | Dominated | 0% | 3% | 10% | 12% | 9% |
| aCGH | 233 | 0.95 | 160 | 0.24 | 673 | 0% | 0% | 8 | 58% | 60% |
| SNP array | 257 | 0.89 | — | — | Dominated | 0% | 0% | 0% | 0% | 0% |
| NGS | 571 | 0.97 | 498 | 0.25 | 1968 | 0% | 0% | 0% | 1% | 13% |
Tests ordered by cost. Diagnostic accuracy figures rounded to 3 decimal places. ICERs based upon exact values for incremental outcomes and costs. aCGH: array comparative genomic hybridisation; CD: correct diagnosis; CE: cost‐effective; CISH: chromogenic in situ hybridisation; FISH: fluorescence in situ hybridisation; ICER: incremental cost‐effectiveness ratio; LOH: loss of heterozygosity; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism; TN: true negative; TP: true positive; WTP: willingness to pay.
11. Costs and diagnostic accuracy of diagnostic tests to evaluate 1p/19q status codeletion (PCR‐based LOH as reference standard).
| Inputs | Deterministic analysis | Probabilistic sensitivity analysis | ||||||||
| Incremental cost per true positive detected | ||||||||||
| Diagnostic test | Cost (GBP) | Effect (true positive rate) | Incremental cost (GBP) | Incremental effect | ICER (GBP) | Prob of being CE at WTP of GBP 0 per TP | Prob of being CE at WTP of GBP 500 per TP | Prob of being CE at WTP of GBP 1000 per TP | Prob of being CE at WTP of GBP 5000 per TP | Prob of being CE at WTP of GBP 10,000 per TP |
| MLPA | 73 | 0.28 | — | — | — | 100% | 99% | 94% | 61% | 45% |
| Real‐time PCR | 142 | 0.28 | — | — | Dominated | 0% | 1% | 6% | 27% | 29% |
| FISH | 186 | 0.26 | — | — | Dominated | 0% | 0% | 0% | 0% | 0% |
| aCGH | 233 | 0.30 | 160 | 0.02 | 7507 | 0% | 0% | 0% | 12% | 24% |
| SNP array | 257 | 0.28 | — | — | Dominated | 0% | 0% | 0% | 0% | 2% |
| NGS | 571 | 0.29 | — | — | Dominated | 0% | 0% | 0% | 0% | 0% |
| Incremental cost per true negative detected | ||||||||||
| Diagnostic test | Cost (GBP) | Effect (true negative rate) | Incremental cost (GBP) | Incremental effect | ICER (GBP) | Prob of being CE at WTP of GBP 0 per TN | Prob of being CE at WTP of GBP 500 per TN | Prob of being CE at WTP of GBP 1000 per TN | Prob of being CE at WTP of GBP 5000 per TN | Prob of being CE at WTP of GBP 10,000 per TN |
| MLPA | 73 | 0.63 | — | — | — | 100% | 99% | 94% | 61% | 49% |
| FISH | 186 | 0.64 | — | — | Extendedly dominated | 0% | 1% | 6% | 10% | 9% |
| aCGH | 233 | 0.62 | — | — | Dominated | 0% | 0% | 0% | 5% | 6% |
| SNP array | 257 | 0.65 | 184 | 0.02 | 8686 | 0% | 0% | 0% | 24% | 35% |
| NGS | 571 | 0.65 | — | — | Dominated | 0% | 0% | 0% | 0% | 1% |
| Incremental cost per correct diagnosis | ||||||||||
| Diagnostic test | Cost (GBP) | Effect (correct diagnosis rate) | Incremental cost (GBP) | Incremental effect | ICER (GBP) | Prob of being CE at WTP of GBP 0 per CD | Prob of being CE at WTP of GBP 500 per CD | Prob of being CE at WTP of GBP 1000 per CD | Prob of being CE at WTP of GBP 5000 per CD | Prob of being CE at WTP of GBP 10,000 per CD |
| MLPA | 73 | 0.91 | — | — | — | 100% | 99% | 93% | 55% | 42% |
| FISH | 186 | 0.90 | — | — | Dominated | 0% | 1% | 4% | 3% | 2% |
| aCGH | 233 | 0.92 | — | — | Extendedly dominated | 0% | 0% | 2% | 18% | 20% |
| SNP array | 257 | 0.93 | 0.02 | 184 | 10,372 | 0% | 0% | 1% | 24% | 29% |
| NGS | 571 | 0.94 | 0.03 | 498 | 15,971 | 0% | 0% | 0% | 0% | 7% |
Tests ordered by cost. Diagnostic accuracy figures rounded to 3 decimal places. True Negative and Correct Diagnosis rates could be calculated for real‐time PCR because a specificity value could not be calculated for this test. aCGH: array comparative genomic hybridisation; CD: correct diagnosis; CE: cost‐effective; FISH: fluorescence in situ hybridisation; ICER: incremental cost‐effectiveness ratio; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism; TN: true negative; TP: true positive; WTP: willingness to pay.
When FISH was used as the reference standard (Table 12), and if a decision were to be made on cost alone, then MLPA had almost 100% chance of being considered the least costly. With regard to the incremental cost per true positive detected, MLPA had 95% or higher chance of being considered cost‐effective given a willingness to pay (WTP) up to GBP 1000 per true positive. As WTP per true positive increases, the probability of other tests being considered cost‐effective increases. However, of the seven tests compared, none had a probability of being cost‐effective above 50% when WTP per true positive detected was GBP 10,000.
When considering true negatives, the real‐time PCR test had the highest probability (54% to 67%) of being cost‐effective at a WTP per additional true negative detected of between GBP 500 and GBP 10,000. None of the other tests had a probability of being cost‐effective above 30% when WTP per additional true negative detected was between GBP 500 and GBP 10,000.
For a correct diagnosis, real‐time PCR had the highest probability of being cost‐effective at a WTP of GBP 500 and GBP 1000, and the aCGH had the highest probability of being cost‐effective at a WTP of GBP 5000 and GBP 10,000. However, for none of the seven tests compared over the ranges of WTP between GBP 500 and GBP 10,000 was the probability of test being cost‐effective over 60%.
When PCR‐LOH was used as the reference standard, the results indicated that MLPA had a 100% probability of being considered the least costly of the five tests compared (Table 13). MLPA also had the highest probability of being cost‐effective in terms of true positives, true negative and correct diagnoses at a WTP up to GBP 10,000. However, at GBP 5000 and GBP 10,000, no test had a probability of being cost‐effective above 55%.
Discussion
Summary of main results
Test accuracy
We found limited evidence about most of the available techniques for detecting 1p/19q codeletions. Most techniques, with the exception of G‐banding, provided point estimates indicating very good sensitivity (i.e. produced few false‐negative results) for detection of 1p/19q codeletion when either FISH or PCR‐based LOH were considered as the reference standard. There was some evidence for differences in specificity (false‐positive rate) with some techniques. However, we caution against ranking the tests included in this review based on point estimates of sensitivity and specificity. Our latent class analysis was not conclusive, but suggested that PCR‐based LOH may be more accurate than FISH. This concords with our a prior assumption that PCR‐based LOH has very high sensitivity.
G‐banding had low sensitivity and specificity when PCR‐based LOH was the reference standard, suggesting that G‐banding may not be a suitable test for 1p/19q analysis. G‐banding is not in current routine clinical use for 1p/19q analysis and most of the studies investigating this technique were older, with the last study investigating this technique published in 2011. Although MS had very high sensitivity and specificity when both FISH and PCR‐based LOH were considered the reference standard, these results should be treated with caution because they were based on only one study, which had a small number of participants. MS is not in current clinical use for 1p/19q analysis but further research may be indicated in this area.
Both NGS and SNP array had high specificity when considered against FISH. For both of these, there were six studies including a good number of participants (243 for NGS and 111 for SNP array). NGS and SNP array also had high specificity when PCR‐based LOH was considered the reference standard, although the CrIs were much wider as these results were based on fewer studies (just one study with 49 participants for NGS and two studies with 33 participants for SNP array). Real‐time PCR also showed high specificity with FISH as a reference standard, although there were only two studies including 40 participants in this analysis. It seems unsurprising that NGS and SNP array had high specificity given that these two techniques are capable of looking at the whole chromosome arm.
A further technology that has gained importance in neuropathology diagnostics is methylation array, combined with algorithmic tumour classification (Capper 2018a; Jaunmuktane 2019; Pickles 2020). The readout of these methylation arrays also returns a copy number assay that has the added benefit of directly demonstrating chromosomal aberrations, including 1p/19q codeletion (Capper 2018b). The estimated costs in clinical practice have been calculated in one of the UK studies (Jaunmuktane 2019).
Complete hemizygous losses of 1p are tightly associated with 19q loss and oligodendroglial phenotype, whereas partial 1p deletions alone are mainly observed in astrocytic tumours, including the IDH‐wildtype glioblastoma, and are not associated with 19q loss. As these tumours represent biologically distinct entities, they are associated with a different (in this case poorer) prognosis (Vogazianou 2010). Therefore, false positives in detection of 1p loss caused by partial 1p deletions are of key clinical importance in 1p/19q analysis because they may result in incorrect diagnosis and treatment. See also illustrations of regions analysed on 1p in the different studies in Figure 4; Figure 6; Figure 7; Figure 8; Figure 9; Figure 20; and Figure 34.
11.
Economic model
CISH: chromogenic in situ hybridisation; MLPA: multiplex‐ligation‐dependent probe amplification; RT‐PCR: real‐time polymerase chain reaction.
The results for test accuracy are in concordance with Zhao 2014, who found that there was no difference in the HR for overall survival between studies using two different techniques (PCR‐based LOH and FISH) to assess the status of chromosomal arms 1p and 19q.
Cost‐effectiveness
We identified no economic evaluations relevant to this study question. There is thus a paucity of evidence about the optimum testing strategy for 1p/19q codeletion in the management of glioma.
Results from the economic model extend the DTA results, highlighting which tests appear on average to have lower accuracy (or – for extendedly dominated tests – not sufficiently better test accuracy) and at higher cost. The analysis incorporates the imprecision surrounding diagnostic performance and test costs. Which test appeared most likely to be cost‐effective depended on which measure of diagnostic performance was considered and the value placed on a unit change in that measure.
Taking FISH as the reference standard, MLPA was the most likely to be cost‐effective if society were willing to pay GBP 1000 per true‐positive case detected. At willingness to pay thresholds higher than this, no test was clearly superior. However, if the outcome were true‐negative cases detected then providing society were willing to pay over GBP 500, real‐time PCR was most likely to be cost‐effective.
When PCR‐based LOH was the reference standard, MLPA was the most likely to be considered cost‐effective if society were willing to pay GBP 5000 for either a true‐positive case detected, true‐negative case detected or a correct diagnosis. However, at threshold values higher than this, no test was clearly more likely to be considered cost‐effective.
Overall, even when accounting for the imprecision in estimates, the results showed that cost‐effectiveness is sensitive to the choice of the reference standard and the decision maker's willingness to pay for additional benefit.
Strengths and weaknesses of the review
To our knowledge this is the first systematic review of the DTA of different techniques for assessing 1p/19q codeletion in glioma. We undertook a thorough search, applied systematic methods and assessed results for risk of bias using the QUADAS‐2 tool.
The review has some limitations, however. None of the available tests is perfect, and we undertook analyses assuming either FISH or PCR‐based LOH as reference standards. In these analyses, none of the investigated tests was superior to the reference standard assumed. Related to this, we were unable to include the results of studies that did not investigate either FISH or PCR‐based LOH in the statistical synthesis.
For most techniques, there was a relatively small number of studies available, most of which had few participants. This meant that some studies produced empty cells in the 2 × 2 comparisons created. Our search was undertaken in July 2019, and further studies may have been reported since then. Furthermore, most studied had risks of bias: only one study was at low risk of bias across all domains of QUADAS‐2. Where reported, we used authors' classifications and thresholds, and it is possible that some of the thresholds used did not fully exploit the potential of the investigated technique.
We were unable to distinguish between absolute and relative deletions in most studies. Even when studies used techniques that can, in theory, distinguish between absolute and relative deletions, few did so. Furthermore, loss of 1p and 19q in combination with 1q or 19p (or both) was considered by some studies to count as 1p/19q codeletion, and in others not. When we had to interpret the results of techniques, we did so by looking for the presence/absence of 1p and 19q without consideration of 1q and 19p.
On a technical note, we did not allow for within‐study correlations when performing the statistical synthesis. In addition, we did not extract some of the information contained within studies that did not address our objectives. For example, some studies had correlated their test results with prognosis, which provides information about which test results were more likely to be correct in the case of discordant test results.
The structure of the economic model was relatively simple, and we did not allow for the possibility of using multiple tests or sequences of tests in diagnosis. Furthermore, since it is based on the results of the meta‐analysis, the economic model has all the same strengths and limitations as that analysis. In particular, cost‐effectiveness results are based on a small number of studies, each typically with a small number of participants. For example, the MLPA technique had the highest probability of being cost‐effective when the threshold value was GBP 1000 or less when PCR‐based LOH was used as the reference standard. This is primarily due to lower costs. However, the sensitivity and specificity values were based on a single study with a sample size of 18. The imprecision associated with this is shown when the threshold was higher (e.g. GBP 10,000). At these higher thresholds, no test was clearly cost‐effective.
Another weakness was that we obtained costs from a single hospital provider, supplemented by expert opinion. These are best thought of as illustrative and it is for readers to judge how applicable these costs are to their own setting. Alternative costs could in principle be derived from a sample of hospitals, and by costing each aspect of resource use needed to provide a test, using micro‐costing techniques. However, such costs may vary substantially between hospitals and between countries. Although we accounted for the uncertainty in the costs in the PSA by including the parameters as triangular distributions, other distributions (e.g. gamma) would be preferable if better data were available.
We also considered only the costs of the tests themselves and measures of diagnostic accuracy. We did not include the impact on health and subsequent costs of management (both to health services and the patient and their families) which may occur following the use of the diagnostic test. Specifically, the consequences of false positives, false negatives and correct diagnoses may have different resource implications. The inclusion of longer‐term costs and consequences of the diagnosis would be useful for future evaluations of tests for 1p/19q codeletion in people with glioma.
Applicability of findings to the review question
The majority of studies recruited participants with particular types of glioma, rather than a cross‐section of all gliomas, meaning that our results may not be applicable to all glioma subtypes. In general, we judged that tests were conducted as we would expect them to be performed in practice, although some tests are probably outdated (G‐banding/karyotyping) and others have not yet been put into practice and are probably more experimental in nature (e.g. NanoString and MS).
Authors' conclusions
Implications for practice.
Current guidelines recommend that 1p/19q codeletion should be evaluated to support a diagnosis of oligodendroglioma and to predict the chemosensitivity and prognosis of these patients (NICE 2018; Stupp 2014); however, there is no consensus as to the best approach. Our review judged the certainty of evidence for all tests to be low or very low, making it difficult to make recommendations for practice. We found little evidence to support the use of G‐banding/karyotyping for 1p/19q analysis. However, all other techniques appeared to have high sensitivity when compared against fluorescent in situ hybridisation (FISH) or polymerase chain reaction (PCR)‐based loss of heterozygosity (LOH) (which itself is thought to be highly sensitive) as a reference standard. Among currently considered techniques, next‐generation sequencing (NGS) and single nucleotide polymorphism (SNP) array had the strongest evidence of high specificity against FISH and PCR‐based LOH, and these two techniques also have the advantage of being able to detect other abnormalities simultaneously.
The various tests differed in costs, but which test would make the best use of resources depends on which measure of diagnostic accuracy is most important. Taking FISH as a reference standard and focusing on the ability to make a correct diagnosis, all the tests except multiplex‐ligation‐dependent probe amplification (MLPA) and chromogenic in situ hybridisation (CISH) were inefficient. MLPA was less costly and was less able to make a true diagnosis than CISH, based on very limited data, but CISH was estimated to cost an additional GBP 9032 per additional true‐positive case diagnosed. When PCR‐based LOH was used as the reference standard, MLPA was the dominant strategy on average. It is for the decision‐maker to judge whether the benefits of a test with potentially better diagnostic performance are worth any extra cost.
Implications for research.
For the comparison of most techniques with FISH or PCR‐based LOH, we identified a relatively small numbers of studies, most of which had few participants. We were unable to reach conclusions about several techniques with promising results due to a sparsity of data. Further research on promising tests is warranted. For example, mass spectroscopy had high sensitivity and specificity when both FISH and PCR‐based LOH were the reference standard, but our results should be treated with caution because they are based on only one study, which had a small number of participants.
Because none of the available tests is perfect, our results are limited by our assumptions about which can be taken as a reference standard. A future analysis of the data would recognise that all tests are imperfect, for example by assuming a latent class model. We plan such an analysis for an update of this review.
Our economic model addresses an evidence gap on the efficiency of genetic testing in the management of people with glioma. However, further evidence is required for a full evaluation of the cost‐effectiveness of the different tests. Once more evidence on the diagnostic performance of the techniques is available, a more detailed modelling study will be able to determine the most cost‐effective diagnostic testing strategy for this patient group. This analysis should more fully incorporate uncertainties using techniques such as probabilistic sensitivity analysis. A more complete evaluation would also seek to explore how test results are used and the implications of this on patient management and health.
History
Protocol first published: Issue 8, 2019
Acknowledgements
We thank Robin Grant (Co‐ordinating Editor) and Gail Quinn (Managing Editor) from the Cochrane Gynaecological, Neuro‐oncology and Orphan Cancer Group (GNOC) for editorial guidance. We also thank Joanne Platt (Information Specialist, GNOC) for aiding with the early development of the search strategy. We are grateful to Luke McGuinness for creating customised QUADAS‐2 plots and Hung‐Yuan Cheng for creating the network plot. We also thank Hung‐Yuan Cheng and Tony Ades for reading and commenting on a draft of the protocol. We thank the Cochrane DTA team and all of our external peer reviewers, including Helen Bulbeck, Mike Jenkinson and Dwayne Boyers.
This project was supported by the National Institute for Health Research (NIHR), via Cochrane Programme Grant funding (16/114/18) to the Cochrane Gynaecological, Neuro‐oncology and Orphan Cancers Group. AM and JPTH were supported in part by Cancer Research UK (grant numbers C18281/A19169 and C18281/A29019). LS was supported by an NIHR Systematic Review Fellowship (RM‐SR‐2017‐09‐028). JPTH is an NIHR Senior Investigator (NF‐SI‐0617‐10145) and is supported by NIHR Bristol Biomedical Research Centre at University Hospitals Bristol and Weston NHS Foundation Trust and the University of Bristol. The views and opinions expressed herein are those of the review authors and do not necessarily reflect those of the Systematic Reviews Programme, NIHR, National Health Service (NHS), the Department of Health and Social Care or Cancer Research UK.
Appendices
Appendix 1. Database search strategies
In this review, we aimed to include all tests for 1p/19q codeletion that have been studied comparatively, and consequently did not have a predefined list of eligible index tests. Therefore, decided to focus the search strategy based on the population (people with glioma) and the target condition (codeletion of chromosomes 1 and 19).
Ovid MEDLINE(R) and Epub Ahead of Print, In‐Process & Other Non‐Indexed Citations and Daily <1946 onwards>
1. exp glioma/
2. (glioma* or astrocytoma* or astroblastoma* or ependymoma* or subependymoma* or oligodendroglioma* or oligoastrocytoma* or pleomorphic xanthoastrocytoma* or glioblastoma* or GBM* or ganglioglioma* or gliosarcoma* or gangliocytoma* or ((glial* or glioneuronal* or brain*) and (tumor* or tumour* or cancer* or neoplasm*))).mp.
3. 1 or 2
4. Chromosomes, Human, Pair 1/ or (chromosome 1 or 1p).mp.
5. Chromosomes, Human, Pair 19/ or (chromosome 19 or 19q).mp.
6. (1p?19q* or "1p/19q" or (1p* adj3 19q*)).mp.
7. 4 and 5
8. 6 or 7
9. 3 and 8
Ovid Embase <1974 onwards>
1. exp glioma/
2. (glioma* or astrocytoma* or astroblastoma* or ependymoma* or subependymoma* or oligodendroglioma* or oligoastrocytoma* or pleomorphic xanthoastrocytoma* or glioblastoma* or GBM* or ganglioglioma* or gliosarcoma* or gangliocytoma* or ((glial* or glioneuronal* or brain*) and (tumor* or tumour* or cancer* or neoplasm*))).mp.
3. 1 or 2
4. Chromosome 1/ or chromosome 1p/ or (chromosome 1 or 1p).mp.
5. chromosome 19/ or chromosome 19q/ or (chromosome 19 or 19q).mp.
6. 4 and 5
7. (1p?19q* or "1p/19q" or (1p* adj3 19q*)).mp.
8. 6 or 7
9. 3 and 8
BIOSIS Citation Index <1969 onwards>
#1 TS=(glioma* or astrocytoma* or astroblastoma* or ependymoma* or subependymoma* or oligodendroglioma* or oligoastrocytoma* or pleomorphic xanthoastrocytoma* or glioblastoma* or GBM* or ganglioglioma* or gliosarcoma* or gangliocytoma* or ((glial* or glioneuronal* or brain*) and (tumor* or tumour* or cancer* or neoplasm*)))
#2 TS=(1p*19q* OR “1p/19q”)
#3 TS=(("chromosome 1" OR 1p) AND ("chromosome 19" OR 19q))
#4 #2 or #3
#5 #1 and #4
WHO International Clinical Trials Registry Platform (ICTRP)
Results from each of the following search lines were downloaded and deduplicated in EndNote.
Search 1: 1p* and 19q*
Search 2: 1p19q or 1p/19q
Search 3: glioma* and diagnostic test or astrocytoma* and diagnostic test or astroblastoma* and diagnostic test or ependymoma* and diagnostic test or subependymoma* and diagnostic test or oligodendroglioma* and diagnostic test or oligoastrocytoma* and diagnostic test or pleomorphic xanthoastrocytoma* and diagnostic test or glioblastoma* and diagnostic test or GBM* and diagnostic test or ganglioglioma* and diagnostic test or gliosarcoma* and diagnostic test or gangliocytoma* and diagnostic test or glial tumor* and diagnostic test or glial tumour* and diagnostic test or glial cancer* and diagnostic test or glial neoplasm* and diagnostic test or glioneuronal tumor* and diagnostic test or glioneuronal tumour* and diagnostic test or glioneuronal cancer* and diagnostic test or glioneuronal neoplasm* and diagnostic test or brain tumor* and diagnostic test or brain tumour* and diagnostic test or brain cancer* and diagnostic test or brain neoplasm* and diagnostic test
Search 4: glioma* and diagnostic assessment or astrocytoma* and diagnostic assessment or astroblastoma* and diagnostic assessment or ependymoma* and diagnostic assessment or subependymoma* and diagnostic assessment or oligodendroglioma* and diagnostic assessment or oligoastrocytoma* and diagnostic assessment or pleomorphic xanthoastrocytoma* and diagnostic assessment or glioblastoma* and diagnostic assessment or GBM* and diagnostic assessment or ganglioglioma* and diagnostic assessment or gliosarcoma* and diagnostic assessment or gangliocytoma* and diagnostic assessment or glial tumor* and diagnostic assessment or glial tumour* and diagnostic assessment or glial cancer* and diagnostic assessment or glial neoplasm* and diagnostic assessment or glioneuronal tumor* and diagnostic assessment or glioneuronal tumour* and diagnostic assessment or glioneuronal cancer* and diagnostic assessment or glioneuronal neoplasm* and diagnostic assessment or brain tumor* and diagnostic assessment or brain tumour* and diagnostic assessment or brain cancer* and diagnostic assessment or brain neoplasm* and diagnostic assessment or brainstem tumor* and diagnostic assessment or brainstem tumour* and diagnostic assessment or brainstem cancer* and diagnostic assessment or brainstem neoplasm* and diagnostic assessment
Search 5: glioma* and DTA or astrocytoma* and DTA or astroblastoma* and DTA or ependymoma* and DTA or subependymoma* and DTA or oligodendroglioma* and DTA or oligoastrocytoma* and DTA or pleomorphic xanthoastrocytoma* and DTA or glioblastoma* and DTA or GBM* and DTA or ganglioglioma* and DTA or gliosarcoma* and DTA or gangliocytoma* and DTA or glial tumor* and DTA or glial tumour* and DTA or glial cancer* and DTA or glial neoplasm* and DTA or glioneuronal tumor* and DTA or glioneuronal tumour* and DTA or glioneuronal cancer* and DTA or glioneuronal neoplasm* and DTA or brain tumor* and DTA or brain tumour* and DTA or brain cancer* and DTA or brain neoplasm* and DTA or brainstem tumor* and DTA or brainstem tumour* and DTA or brainstem cancer* and DTA or brainstem neoplasm* and DTA
Appendix 2. Review‐specific tailoring of QUADAS‐2
Domain 1: patient selection
Risk of bias
Was a consecutive or random sample of patients enrolled?
Yes: if a consecutive sample or a random sample of eligible participants was included in the study.
No: if a non‐consecutive sample or a non‐random sample of eligible participants was included in the study.
Unclear: if it was not clear whether a consecutive sample or a random sample of eligible participants was included in the study.
Was a case‐control (or 'two‐gate') design avoided?
Yes: if the study had a single set of inclusion criteria.
No: if the study had more than one set of inclusion criteria.
Unclear: if the inclusion criteria for the study are not clear.
Did the study avoid inappropriate exclusions?
Yes: if all patients with glioma were included.
No: if a subset of patients with glioma were excluded due to subclassification/severity of glioma.
Unclear: if the inclusion criteria for the study were not clear.
Overall: could the selection of patients have introduced bias?
We took highest concern from any individual signalling question as our overall judgement (i.e. risk of bias was classified as low if the response to all three questions was 'yes'; high if the response to any question was 'no'; and unclear if the response to any question was 'unclear' and the criteria for high risk of bias were not fulfilled).
Applicability
Were there concerns that the included patients do not match the review question?
High: if the study population included patients who would not have undergone testing in real practice, for example healthy controls.
Low: if the study included only a clinically relevant population that would have undergone testing in real practice.
Unclear: if the inclusion criteria for the study are not clear.
Domain 2: index test
Risk of bias
Were the index test results interpreted without knowledge of the results of the other tests being compared?
Yes: if the index test was objective or if subjective was interpreted without the knowledge of the results of other tests for 1p/19q codeletion. The first test to be interpreted was judged to be interpreted without knowledge of the results of the other tests even if it was not explicitly reported that it was interpreted 'blind' or without the knowledge of other test results.
No: if test was subjective and interpreted with the knowledge of the results of other tests for 1p/19q codeletion.
Unclear: if the test was subjective and it was unclear whether it was interpreted with the knowledge of other tests for 1p/19q codeletion.
If a threshold was used, was it prespecified?
Yes: if the definition of what was considered to be a positive test result was defined before testing was performed (we judged that if a threshold was reported in the methods section that it was prespecified).
No: if the definition of what was considered to be a positive test result was defined after testing was performed and based on the results.
Unclear: if it was unclear whether the definition of what was considered to be a positive test result was defined before testing was performed or if the threshold used was not reported.
Overall: could the conduct or interpretation of the index test have introduced bias?
We took highest concern from any individual signalling question as our overall judgement.
If the threshold was not prespecified and patients were not classified, and if we applied our own classification, we judged this domain as low risk of bias because we were not trying to maximise concordance between tests.
Applicability: were there concerns that the index test, its conduct or its interpretation differ from the review question?
High: if there were concerns that the index test, its conduct or its interpretation differed from the review question.
Low: if there were no concerns that the index test, its conduct or its interpretation differed from the review question.
Unclear: if the description of the index test was inadequate.
Domain 3: reference standard
We envisaged that many studies would have compared two or more tests without necessarily designating a reference standard.
In addition, as we planned a latent class analysis, which allows for an imperfect reference standard, the risk of bias signalling question regarding whether the reference standard was likely to correctly classify the target condition was omitted.
For similar reasons, we decided that the applicability question was not relevant.
We completed domain 2 for each test that was compared.
Domain 4: flow and timing
We modified some of the wording of the signalling questions to reflect the fact that studies may not have designated a reference standard.
Risk of bias
Was there an appropriate interval between the tests being compared?
We envisaged that most tests would be performed on biopsied material.
Yes: if all tests were performed on biopsied tumour material collected on one occasion.
No: if tests were performed on tumour material collected at different time points.
Unclear: if it was unclear whether the tests were performed on the same material.
Were all patients included in the analysis?
Yes: if all participants were included in the analysis, or if participants were excluded because they did not meet inclusion criteria or if withdrawals were less than 5% of the enrolled population (arbitrarily selected cut‐off).
No: if any participants were excluded from the analysis because of uninterpretable results, because of inability to undergo index test or reference standard or for unclear reasons.
Overall: could the patient flow have introduced bias?
We took the highest concern from any individual signalling question as our overall judgement.
Appendix 3. Domains to be considered when judging the strength of the body of evidence
Domains to be considered when judging the strength of the body of evidence, based on GRADE.
| Domain | Explanation |
| Risk of bias | Based on results of risk of bias assessments. Certainty in the evidence base was downgraded if most of the evidence was from studies not judged to be at low risk of bias. |
| Imprecision | Certainty in the evidence base was downgraded if the estimate of the effect size from a meta‐analysis was not precise. We downgraded by 2 levels if (i) the upper 95% confidence limit for either sensitivity or specificity was more than 2 times the lower limit or (ii) the total sample size was 50 or less; and by 1 level if an upper limit was more than 1.4 times the lower limit. |
| Inconsistency | Certainty in the evidence base was downgraded if there was unexplained heterogeneity or variability in results across studies. |
| Indirectness | Based on QUADAS‐2 assessments of applicability. Certainty in the evidence base was downgraded if most of the evidence was from studies judged to have low applicability to the review question. |
| Publication bias | Certainty in the evidence base was downgraded if we uncovered evidence of publication bias. |
Appendix 4. Economic model and estimated costs of diagnostic tests
Figure 34 provides the basic model structure for the model‐based economic analysis. The model illustrates a choice between three alternatives: the multiplex‐ligation‐dependent probe (MLPA), real‐time polymerase chain reaction (PCR) and chromogenic in situ hybridisation (CISH) tests only. In the analysis, all available tests for each reference standard were included in the model. The blue square is a decision node, which represents a point of choice between the different tests. The green circles are chance nodes, which represent chance events characterised by probabilities (the chance nodes to the left indicate the chance of positive and negative test results, the green chance nodes to the right indicate true disease status). The red triangles are terminal nodes, which represent the final outcomes in terms of diagnosis from the alternative decision tree pathways.
Table A4.1 provides the estimated costs of diagnostic tests.
Table A4.1. Estimated costs of diagnostic tests
| Cost Item | Unit cost (GBP) | Source |
| FISH | 185.95 | The Newcastle upon Tyne Hospitals NHS Foundation Trust (2020) |
| CISH | 185.95 | The Newcastle upon Tyne Hospitals NHS Foundation Trust (2020) |
| PCR‐based LOH | 142.21 | The Newcastle upon Tyne Hospitals NHS Foundation Trust (2020) |
| Real‐time PCR | 142.21 | The Newcastle upon Tyne Hospitals NHS Foundation Trust (2020) |
| MLPA | 73.08 | The Newcastle upon Tyne Hospitals NHS Foundation Trust (2020) |
| aCGH | 233.47 | Sagoo GS, Mohammed S, Barton G, Norbury G, Ahn JW, Ogilvie CM, et al. Cost effectiveness of using array‐CGH for diagnosing learning disability. Applied Health Economic and Health Policy 2015;13:421‐32. |
| SNP array | 256.80 | The Newcastle upon Tyne Hospitals NHS Foundation Trust (2020) |
| NGS | 570.87 | Marino P, Touzani R, Perrier L, Rouleau E, Kossi DS, Zhaomin Z, et al. Cost of cancer diagnosis using next‐generation sequencing targeted gene panels in routine practice: a nationwide French study. European Journal of Human Genetics 2018;26:314‐23. |
| aCGH: array comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; FISH: fluorescent in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism. | ||
Appendix 5. Distributions used in the probabilistic sensitivity analyses
Abbreviations used: aCGH: array comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; FISH: fluorescence in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS; mass spectrometry; N/A: not applicable; NGS: next‐generation sequencing; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; SD: standard deviation; SNP: single nucleotide polymorphism.
Table A5.1. Analysis using PCR‐based LOH as reference standard
| Parameter description | Probability distribution |
| Prevalence | Beta (mean = 0.31, SD = 0.13) |
| Sensitivity of MLPA | Beta (α = 9, β = 1) |
| Specificity of MLPA | Beta (α = 11, β = 1) |
| Cost of MLPA | Triangular (minimum 55.35, likeliest 73.08, maximum 93.35) |
| Sensitivity of real‐time PCR | Beta (α = 11, β = 1) |
| Specificity of real‐time PCR | N/A |
| Cost of real‐time PCR | Triangular (minimum 106.66, likeliest 142.21, maximum 177.76) |
| Sensitivity of FISH | Beta (α = 196, β = 37) |
| Specificity of FISH | Beta (α = 638, β = 48) |
| Cost of FISH | Triangular (minimum 139.46, likeliest 185.95, maximum 232.44) |
| Sensitivity of aCGH | Beta (α = 31, β = 1) |
| Specificity of aCGH | Beta (α = 26, β = 3) |
| Cost of aCGH | Triangular (minimum 175.10, likeliest 233.47, maximum 291.84) |
| Sensitivity of SNP array | Beta (α = 16, β = 2) |
| Specificity of SNP array | Beta (α = 18, β = 1) |
| Cost of SNP array | Triangular (minimum 192.60, likeliest 256.80, maximum 321.00) |
| Sensitivity of NGS | Beta (α = 19, β = 1) |
| Specificity of NGS | Beta (α = 31, β = 2) |
| Cost of NGS | Triangular (minimum 428.15, likeliest 570.87, maximum 713.59) |
Table A5.2. Analysis using FISH as reference standard
| Parameter description | Probability distribution |
| Prevalence | Beta (mean = 0.31, SD = 0.13) |
| Sensitivity of MLPA | Beta (α = 12, β = 2) |
| Specificity of MLPA | Beta (α = 15, β = 8) |
| Cost of MLPA | Triangular (minimum 55.35, likeliest 73.08, maximum 93.35) |
| Sensitivity of real‐time PCR | Beta (α = 15, β = 4) |
| Specificity of real‐time PCR | Beta (α = 24, β = 1) |
| Cost of real‐time PCR | Triangular (minimum 106.66, likeliest 142.21, maximum 177.76) |
| Sensitivity of PCR‐based LOH | Beta (α = 196, β = 48) |
| Specificity of PCR‐based LOH | Beta (α = 633, β = 42) |
| Cost of PCR‐based LOH | Triangular (minimum 106.66, likeliest 142.21, maximum 177.76) |
| Sensitivity of CISH | Beta (α = 20, β = 1) |
| Specificity of CISH | Beta (α = 18, β = 3) |
| Cost of CISH | Triangular (minimum 139.46, likeliest 185.95, maximum 232.44) |
| Sensitivity of aCGH | Beta (α = 19, β = 1) |
| Specificity of aCGH | Beta (α = 19, β = 4) |
| Cost of aCGH | Triangular (minimum 175.10, likeliest 233.47, maximum 291.84) |
| Sensitivity of SNP array | Beta (α = 35, β = 13) |
| Specificity of SNP array | Beta (α = 64, β = 3) |
| Cost of SNP array | Triangular (minimum 192.60, likeliest 256.80, maximum 321.00) |
| Sensitivity of NGS | Beta (α = 73, β = 7) |
| Specificity of NGS | Beta (α = 166, β = 1) |
| Cost of NGS | Triangular (minimum 428.15, likeliest 570.87, maximum 713.59) |
Appendix 6. Raw data from included studies
Table A6.1. Raw results for comparisons of four test categories or variants within categories
| Study | Tests included | Tumour type | Tumour grade | ++++ | +++‐ | ++‐+ | +‐++ | ‐+++ | ++‐‐ | +‐‐+ | ‐‐++ | ‐++‐ | ‐+‐+ | +‐+‐ | +‐‐‐ | ‐+‐‐ | ‐‐+‐ | ‐‐‐+ | ‐‐‐‐ | Total with test results | Notes |
| Blesa 2009 | A: aCGH B: FISH C: MLPA D: PCR |
Anaplastic oligodendroglioma | III | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | Some differences between Table 2 in text and the Supplementary Table. Since the Supplementary Table had more information, we extracted from this. Relative deletions on FISH were classified as a negative result as they were not absolute deletions. |
| Oligodendroglioma | II | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |||
| Anaplastic oligoastrocytoma | III | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | |||
| Duval 2014 | A: FISH (combination cut‐off based on number of cells showing a deletion) B: FISH (ratio cut‐off based on the ratio of signals for 1p to 1q and 19q and 19p) C: FISH (immunoFISH with combination cut‐off) D: FISH (immunoFISH with ratio cut‐off) |
Oligodendroglioma | II | 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 11 | FISH assessments were made independently by 2 observers. In 1 case, a participant was classified as having the codeletion by 1 observer but not the other when using the ratio method to interpret the results of ImmunoFISH. Raw data were available from the 2 observers, and we averaged the raw data and applied the reported cut‐off to come to a consensus classification. |
| Anaplastic oligodendroglioma | III | 13 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 22 | |||
| Glioblastoma with oligodendroglioma component | IV | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | |||
| Hatanpaa 2003a (assay development and non‐blinded validation cohort) | A: CGH B: FISH C: PCR (comparison to normal DNA) D: PCR (microsatellite) |
Astrocytoma | II | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | Participants from Smith 1999. We removed D1S534 from the regions analysed by PCR with comparison to normal DNA as this was not used to assess 1p/19q status. We used the histological diagnosis from this paper (rather than from Smith 1999). |
| Astrocytoma | III | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |||
| Oligodendroglioma | II | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | |||
| Hatanpaa 2003b (blinded validation cohort) | A: CGH B: FISH C: PCR (comparison to normal DNA) D: PCR (microsatellite) |
Astrocytoma | II | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Participants from Smith 1999. We removed D1S534 from the regions analysed by PCR with comparison to normal DNA as this was not used to assess 1p/19q status. We used the histological diagnosis from this paper (rather than from Smith 1999). Note: 1 of the ++++ tumours (T246, oligodendroglioma) (quote) "was found to have partial LOH on 1p, a finding confirmed by comparison with the allelic pattern derived from normal tissue dissected from this case (Table 2). In this tumor, heterozygosity was only preserved at one locus of eight assessed on 1p and 19q. In light of previous studies (Bello et al, 2000; Bigner et al, 1999; Smith et al, 1999), this tumor may represent a rare aberration from the usual extent of LOH on 1p in oligodendrogliomas. Although the clinical significance of this finding of partial LOH is not known, the tumor is probably best classified as having a high likelihood of clinically relevant LOH on 1p, considering that heterozygosity was preserved at only one locus and that the tumor was histologically an oligodendroglioma". |
| Mixed | II | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | |||
| Oligodendroglioma | II | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | |||
| Hatanpaa 2003b (blinded validation cohort) | A: CGH B: FISH C: PCR (comparison to normal DNA) D: PCR (microsatellite) |
Mixed | III | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Participant from Burger 2001. We have used the histological diagnosis from this paper (rather than from Burger 2001). |
++++: number of people positive on all tests (meaning that 1p19q codeletion found with all tests); +++‐: positive on test A, positive on test B, positive on test C, negative on test D; ++‐+: positive on test A, positive on test B, negative on test C, positive on test D; +‐++: positive on test A, negative on test B, positive on test C, positive on test D; ‐+++: negative on test A; positive on test B; positive on test C; positive on test D; ++‐‐: positive on test A; positive on test B; negative on test C, negative on test D; +‐‐+: positive on test A, negative on test B; negative on test C, positive on test D; ‐‐++: negative on test A, negative on test B, positive on test C, positive on test D; ‐++‐: negative on test A, positive on test B, positive on test C, negative on test D; ‐+‐+: Negative on test A, positive on test B, negative on test C, positive on test D; +‐+‐: Positive on test A, negative on test B, positive on test C, negative on test D; +‐‐‐: positive on test A, negative on test B, negative on test C, negative on test D; ‐+‐‐: negative on test A, positive on test B, negative on test C, negative on test D; ‐‐+‐: negative on test A, negative on test B, positive on test C, negative on test D; ‐‐‐+: negative on test A, negative on test B, negative on test C, positive on test D; ‐‐‐‐: negative on all tests.
Table A6.2. Raw results for comparisons of three test categories or variants within categories
| Study | Tests included | Tumour type | Grade | +++ | ++‐ | +‐+ | ‐++ | +‐‐ | ‐‐+ | ‐+‐ | ‐‐‐ | Total with test results | Notes |
| Belaud‐Rotureau 2006 | A: FISH (1p36.3 (D1Z2)/1q12 (D1Z1) and 19q13.3/19pter probes and manual analysis) B: FISH (1p36/1q25 and 19q13/19p13 Abbott Vysis probe set and manual analysis) C: FISH (1p36/1q25 and 19q13/19p13 Abbott Vysis probe set and automated analysis) |
Oligodendroglioma | III | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | — |
| Oligoastrocytoma | II | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | |||
| Oligoastrocytoma | III | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 7 | |||
| Blesa 2009 (aCGH vs FISH vs PCR) | A: aCGH B: FISH C: PCR |
Anaplastic oligoastrocytoma | III | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Some differences between Table 2 in text and the Supplementary Table. Since the Supplementary Table had more information, we extracted from this. Relative deletions on FISH were classified as a negative result as they were not absolute deletions. |
| Oligodendroglioma | II | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |||
| Blesa 2009 (FISH vs MLPA vs PCR) | A: FISH B: MLPA C: PCR |
Anaplastic oligodendroglioma | III | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | Some differences between Table 2 in text and the Supplementary Table. Since the Supplementary Table had more information, we extracted from this. Relative deletions on FISH were classified as a negative result as they were not absolute deletions. |
| Oligodendroglioma | II | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 6 | |||
| Anaplastic oligoastrocytoma | III | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |||
| Blesa 2009 (aCGH vs MLPA vs PCR) | A: aCGH B: MLPA C: PCR |
Oligodendroglioma | II | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | Some differences between Table 2 in text and the Supplementary Table. Since the Supplementary Table had more information, we extracted from this. Relative deletions on FISH were classified as a negative result as they were not absolute deletions. |
| Burger 2001 | A: CGH B: FISH C: PCR |
Oligodendroglioma | II | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | We used the "review" diagnosis (still done without knowledge of the results of the tests). Overall results for PCR‐based LOH for 1p and 19q not given. We assumed that if results were homozygous or indeterminant at all loci examined that codeletion was present. We excluded the second sample for participant T276. We removed participant T272 because they were included in Hatanpaa 2003b. |
| Malignant oligodendroglioma | III | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |||
| Astrocytoma | II | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | |||
| Astrocytoma | III | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 6 | |||
| Malignant mixed glioma | III | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |||
| Glioblastoma | IV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | |||
| Dahlback 2011 | A: CGH B: G‐banding C: PCR |
Oligoastrocytoma | II | 0 | 0 | 4 | 0 | 0 | 3 | 0 | 6 | 13 | We excluded people with failure results. We could not make the numbers that we derived from Table 1 correspond to what was reported in the text. |
| Oligodendroglioma | II | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 1 | 5 | |||
| Hatanpaa 2003a (assay development and non‐blinded validation cohort) (FISH vs PCR with comparison to normal DNA vs PCR without comparison to normal DNA) | A: FISH B: PCR (comparison to normal DNA) C: PCR (microsatellite) |
Astrocytoma | II | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Participants from Smith 1999. We removed D1S534 from the regions analysed by PCR with comparison to normal DNA as this was not used to assess 1p/19q status. We used the histological diagnosis from this paper (rather than from Smith 1999). |
| Oligodendroglioma | II | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |||
| Oligodendroglioma | III | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |||
| Hatanpaa 2003b (blinded validation cohort) (FISH vs PCR with comparison to normal DNA vs PCR without comparison to normal DNA) | A: FISH B: PCR (comparison to normal DNA) C: PCR (microsatellite) |
Astrocytoma | IV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Participants from Smith 1999. We removed D1S534 from the regions analysed by PCR with comparison to normal DNA as this was not used to assess 1p/19q status. We used the histological diagnosis from this paper (rather than from Smith 1999). |
| Oligodendroglioma | II | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |||
| Horbinski 2012 | A: FISH (cut‐off < 0.87) B: FISH (cut‐off < 0.75) C: PCR |
Oligodendroglioma | II, III | 61 | 15 | 3 | 0 | 5 | 1 | 0 | 26 | 111 | Discrepancy between text (and what they calculated in Table 2) and Table 1 – we used text and Table 2. |
| Mohapatra 2006a | A: aCGH B: FISH C: PCR |
"Oligodendroglial tumours" | II, III | 16 | 0 | 1 | 0 | 1 | 0 | 0 | 10 | 28 | 32 tumours included in the study, had results for 28 and results not broken down by tumour subtype, apart for the discordant cases. |
| Pesenti 2017 (aCGH vs MS vs PCR) | A: aCGH B: MS C: PCR |
Oligodendroglioma | II | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | — |
| Anaplastic oligodendroglioma | III | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |||
| Glioblastoma | IV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | |||
| Pesenti 2017 (FISH vs MS vs PCR) | A: FISH B: MS C: PCR |
Oligodendroglioma | II | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | — |
| Anaplastic oligodendroglioma | III | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |||
| Astrocytoma | II | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |||
| Glioblastoma | IV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 4 | |||
| Smith 1999 | A: CGH B: FISH C: PCR |
Astrocytoma | III | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | Quote: "LOH, FISH, and CGH were performed as previously described (Cliby et al., 1993; Ritland et al., 1995; Qian et al., 1995; Mohapatra et al., 1995, 1998; Piper et al., 1995)". Overall results for 1p and 19q for PCR not given. We assumed that if results were confirmed allelic loss/presumed allelic loss/homozygous at all loci on 1p except the most centromeric (D1S534‐ removed from list of loci examined) and all loci on 19q examined that codeletion was present. Overall results for FISH also not reported. We defined codeletion as hemizygous deletion of 1p36, 1q13.1‐q13.2 and 19q13.3. |
| Astrocytoma | IV | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 17 | 18 | |||
| Mixed | II | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |||
| Mixed | III | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | |||
| Mixed | IV | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 3 | |||
| Oligodendroglioma | II | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 9 | |||
| Oligodendroglioma | III | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 6 | |||
| Oligodendroglioma | IV | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
+++: number of people (or tumours (a: studies where we consider the unit of analysis was a tumour)) positive on all tests (meaning that 1p19q codeletion found with all tests); ++‐: positive on test A, positive on test B, negative on test C; +‐+: positive on test A, negative on test B, positive on test C; ‐++: negative on test A, positive on test B, positive on test C; +‐‐: positive on test A, negative on test B, negative on test C; ‐‐+: negative on test A, negative on test B, positive on test C; ‐+‐: negative on test A, positive on test B, negative on test C; ‐‐‐: negative on all tests.
Table A6.3. Raw results for comparisons of two test categories or variants within categories
| Study | Tests included | Tumour subtype | Tumour grade | ++ | +‐ | ‐+ | ‐‐ | Total with test results | Notes |
| Ariza 2010a | A: PCR B: real‐time PCR |
Oligodendrogliomas | NR | 10 | 0 | 0 | 0 | 10 | 69 astrocytomas also studied but information that we could extract not reported. |
| Armanious 2017a | A: FISH B: NanoString | Oligodendroglioma, glioblastoma and oligoastrocytoma | NR | 10 | 2 | 1 | 3 | 16 | — |
| Belaud‐Rotureau 2006 (manual vs automatic analysis with the 1p36/1q25 and 19q13/19p13 Abbott Vysis probe set) | A: FISH (1p36/1q25 and 19q13/19p13 Abbott Vysis probe set and manual analysis) B: FISH (1p36/1q25 and 19q13/19p13 Abbott Vysis probe set and automated analysis) |
Astrocytoma | II | 0 | 0 | 0 | 6 | 6 | — |
| Oligodendroglioma | III | 1 | 0 | 0 | 0 | 1 | — | ||
| Oligoastrocytoma | II | 1 | 0 | 0 | 5 | 6 | — | ||
| Bigner 1999 | A: CGH B: PCR |
Oligodendroglioma | II | 15 | 0 | 1 | 5 | 21 | By CGH, some participants had (quote) "Copy number changes of partial chromosome arms". It was unclear what the authors meant by this – whether the chromosome arm was only partial and then there was copy number loss or whether this should be considered partial (i.e. not complete) arm loss. However, from the text it was clear that the authors considered that when this had occurred on 1p/19q that this was considered a codeletion. We excluded recurrent tumours and tumours from participants who were aged < 18 years. |
| Oligoastrocytoma | II | 1 | 0 | 0 | 0 | 1 | |||
| Anaplastic oligodendroglioma | III | 16 | 1 | 0 | 3 | 20 | |||
| Anaplastic oligoastrocytoma | III | 2 | 0 | 0 | 4 | 6 | |||
| Glioblastoma | IV | 1 | 0 | 0 | 3 | 4 | |||
| Astrocytoma | II | 0 | 0 | 0 | 1 | 1 | |||
| Blesa 2009 (aCGH vs PCR) | A: aCGH B: PCR |
Anaplastic oligoastrocytoma | III | 0 | 0 | 0 | 1 | 1 | Some differences between Table 2 in text and the Supplementary Table. Since the Supplementary Table had more information, we extracted from this. |
| Blesa 2009 (FISH vs PCR) | A: FISH B: PCR |
Oligodendroglioma | II | 2 | 0 | 1 | 4 | 7 | Some differences between Table 2 in text and the Supplementary Table. Since the Supplementary Table had more information, we extracted from this. Relative deletions on FISH were classified as a negative result as they were not absolute deletions. |
| Oligoastrocytoma | II | 1 | 0 | 0 | 7 | 8 | |||
| Anaplastic oligodendroglioma | III | 3 | 0 | 6 | 0 | 9 | |||
| Anaplastic oligoastrocytoma | III | 0 | 0 | 1 | 5 | 6 | |||
| Blesa 2009 (MLPA vs PCR) | A: MLPA B: PCR |
Oligodendroglioma | II | 1 | 0 | 0 | 1 | 2 | Some differences between Table 2 in text and the Supplementary Table. Since the Supplementary Table had more information, we extracted from this. |
| Bouvier 2004 | A: FISH B: PCR |
Glioblastoma | IV | 0 | 1 | 0 | 2 | 3 | 2 oligodendroglioma grade II participants and 2 oligodendroglioma grade III participants who were classified as having the codeletion by PCR (concordant with FISH) were then described as having (quote) "a partial deletion on 19q for the 19q13.32 and not for the 19q13.12". |
| Oligodendroglioma | II | 2 | 0 | 1 | 1 | 4 | |||
| Oligodendroglioma | III | 2 | 0 | 0 | 1 | 3 | |||
| Oligoastrocytoma | II | 0 | 0 | 0 | 3 | 3 | |||
| Oligoastrocytoma | III | 0 | 0 | 0 | 1 | 1 | |||
| Broholm 2008 | A: FISH B: PCR |
Oligodendroglioma | II | 3 | 0 | 1 | 0 | 4 | — |
| Anaplastic oligodendroglioma | III | 4 | 0 | 0 | 1 | 5 | |||
| Oligoastrocytoma | II | 2 | 0 | 1 | 2 | 5 | |||
| Anaplastic oligoastrocytoma | III | 2 | 0 | 1 | 2 | 5 | |||
| Astrocytoma | II | 1 | 0 | 0 | 4 | 5 | |||
| Anaplastic astrocytoma | III | 0 | 2 | 1 | 1 | 4 | |||
| Glioblastoma | IV | 0 | 2 | 1 | 7 | 10 | |||
| Byeon 2014 | A: aCGH B: FISH |
Rhabdoid glioblastoma | NR | 0 | 0 | 0 | 3 | 3 | — |
| Cieply 2004 | A: FISH B: PCR |
Gliomas (oligodendroglioma, mixed tumours, astrocytoma) | NR | 10 | 0 | 0 | 12 | 22 | 2 cases (not included) had borderline results for deletion by FISH, and indeterminate results by PCR. |
| Chaturbedi 2012a | A: FISH B: real‐time PCR |
Anaplastic oligodendroglioma | III | 0 | 2 | 0 | 5 | 7 | Comparative quantitative PCR cut‐offs were determined post hoc. Eventually they did use all the prespecified marker or reference genes. We could not recreate their numbers for concordance for 1p and 19q. |
| Mixed oligoastrocytoma | II | 0 | 0 | 0 | 2 | 2 | |||
| Oligodendroglioma | II | 2 | 0 | 0 | 7 | 9 | |||
| Clark 2013 | A: FISH B: PCR |
Glioblastoma | IV | 1 | 18 | 7 | 420 | 446 | Another cut‐off for FISH mentioned in discussion. |
| Cowell 2004 | A: aCGH B: PCR |
Low‐grade oligodendroglioma | NR | 4 | 0 | 0 | 2 | 6 | Unclear if all the PCR regions were used to inform the PCR result. |
| Anaplastic oligodendroglioma | NR | 3 | 0 | 0 | 2 | 5 | |||
| Mixed oligoastrocytoma | NR | 1 | 0 | 0 | 2 | 3 | |||
| D'Haene 2019a | A: FISH B: NGS |
Gliomas | I–IV | 21 | 1 | 0 | 28 | 50 | Results likely to include > 1 sample from the same patient: quote: "A retrospective collection of samples, which consisted of 52 glioma samples from 47 patients". (Presume there is an error and it should read 53 glioma samples.) Excluded 3 with non‐informative results on the NGS panel. Note: these results have required a lot of interpretation. Distinguishing glioma from non‐glioma samples was difficult, and is based on the text in section 2.3 (quote) "concordant positive results were obtained for 21 of the 22 glioma samples (95.4% sensitivity) … Among the 31 gliomas that did not show a 1p/19q codeletion by FISH, 28 showed neither patterns of 1p/19q loss of heterozygosity (LOH) by NGS, as defined by our criteria (three were non‐informative)". |
| Dahlback 2009 | A: CGH B: G‐banding |
Glioblastoma | IV | 0 | 3 | 0 | 40 | 43 | — |
| Glioblastoma – multifocal | IV | 0 | 0 | 0 | 1 | 1 | |||
| Glioblastoma with granular cell component | IV | 0 | 0 | 0 | 1 | 1 | |||
| Glioblastoma with oligodendroglial component | IV | 0 | 1 | 0 | 4 | 5 | |||
| Giant cell glioblastoma | IV | 0 | 0 | 0 | 1 | 1 | |||
| Gliosarcoma | IV | 0 | 1 | 0 | 5 | 6 | |||
| Dahlback 2011 (CGH vs G‐banding) | A: CGH B: G‐banding |
Fibrillary astrocytoma | II | 0 | 0 | 0 | 8 | 8 | Excluded 5 participants with fibrillary astrocytoma without high resolution‐CGH results and 1 participant with a failed result. 1 participant with fibrillary astrocytoma had loss of 1p36 and monosomy 19. In the text, they implied that this does not count as 1p19q codeletion. Quote: "None of the astrocytic tumors displayed the complete 1p/19q codeletion (i.e., loss of both arms 1p and 19q). However, one fibrillary astrocytoma showed partial loss of 1p, one showed partial loss of 19q, and one tumor sample showed loss of 1p36 and monosomy 19". |
| Gemistocytic astrocytoma | II | 0 | 0 | 0 | 2 | 2 | |||
| Dahlback 2011 (CGH vs PCR) | A: CGH B: PCR |
Oligodendroglioma | II | 1 | 0 | 0 | 0 | 1 | — |
| Dahlback 2011 (G‐banding vs PCR) | A: G‐banding B: PCR |
Oligoastrocytoma | II | 0 | 0 | 0 | 1 | 1 | — |
| Oligodendroglioma | II | 0 | 0 | 2 | 0 | 2 | |||
| Dubbink 2016 | A: NGS B: PCR |
Anaplastic oligodendrogliomas and anaplastic oligoastrocytomas | NR | 18 | 1 | 0 | 30 | 49 | — |
| Duval 2015 | A: FISH (automated analysis) B: FISH (manual analysis) |
Pilocytic astrocytoma | I | 0 | 0 | 0 | 1 | 1 | Quote: "In this control series, two cases were incomplete for 19q (broken slides) and 5 cases were non interpretable for 1p because of a total lack of telomeric fluorescent signal ("R" signal)". Automated method on archival slides that had been stored at −20 °C. We assumed from the discussion that the cut‐off used the combination + ratio method. |
| Astrocytoma | III | 0 | 0 | 0 | 1 | 1 | |||
| Oligodendroglioma | II | 4 | 0 | 0 | 0 | 4 | |||
| Oligodendroglioma | III | 0 | 0 | 1 | 0 | 1 | |||
| Oligoastrocytoma | II | 1 | 0 | 0 | 3 | 4 | |||
| Oligoastrocytoma | III | 3 | 1 | 2 | 5 | 11 | |||
| Oligoastrocytoma | IV | 0 | 0 | 0 | 2 | 2 | |||
| Glioblastoma with oligodendroglial component | NR | 0 | 0 | 0 | 4 | 4 | |||
| Dysembryoplastic neuroepithelial tumour | I | 0 | 0 | 0 | 1 | 1 | |||
| Gadji 2009 | A: FISH B: PCR |
Anaplastic oligoastrocytoma | NR | 1 | 0 | 0 | 1 | 2 | We extracted the WHO 2007 classification of tumours. |
| Anaplastic oligodendroglioma | NR | 6 | 0 | 1 | 0 | 7 | |||
| Oligodendroglioma | NR | 1 | 0 | 0 | 0 | 1 | |||
| Gliosarcoma | NR | 0 | 0 | 0 | 1 | 1 | |||
| Ghasimi 2016 | A: FISH B: SNP array |
Mixed (grade II–IV gliomas) | II–IV | 0 | 11 | 0 | 44 | 55 | Potential error in paper: is the 1q control probe for FISH 1q25? Results have required intense interpretation. Text stated that 55 people had results from both techniques. Then stated that FISH detected 14 samples with codeletion. From Supplementary Table 3 it appeared that only 11 of these had data from both techniques. We assumed that the phrase (quote) "none was detected by SNP array data" to mean that none was found to have a codeletion by SNP array (not just the 14 with FISH results). Some participants aged < 18 years. |
| Harada 2011 | A: PCR B: SNP array |
Oligodendroglioma | NR | 9 | 0 | 0 | 0 | 9 | Have classified the papers' "partial deletions" with no LOH (as not full loss). Within the cohort some participants aged < 18 years (age range 14–82 years). The participant with pineal parenchymal tumour was excluded (not a glioma). |
| Anaplastic oligodendroglioma | NR | 4 | 1 | 0 | 0 | 5 | |||
| Oligodendroglioma + oligosarcoma | NR | 1 | 0 | 0 | 0 | 1 | |||
| Fibrillary astrocytoma | NR | 0 | 0 | 0 | 3 | 3 | |||
| Astrocytoma | NR | 0 | 0 | 0 | 2 | 2 | |||
| Anaplastic astrocytoma | NR | 0 | 0 | 0 | 7 | 7 | |||
| Glioblastoma | NR | 0 | 0 | 0 | 2 | 2 | |||
| Hatanpaa 2003a (assay development and non‐blinded validation cohort) (PCR vs ≥ 2 tests) | A: PCR B: ≥ 2 of CGH, FISH and PCR |
Astrocytoma | II | 0 | 0 | 0 | 1 | 1 | This was a participant we could not match to those included in Smith 1999 or Burger 2001. |
| Hinrichs 2016 | A: FISH B: SNP array |
Glioblastoma with an oligodendroglial component (GBM‐O) | IV | 2 | 0 | 0 | 6 | 8 | Figures based on our interpretation of SNP array results in figure 3 of the paper. |
| Jeuken 2006a | A: CGH B: MLPA |
Oligodendroglioma | II | 8 | 0 | 0 | 0 | 8 | Overall results for 1p and 19q not given using MLPA, and threshold to do so not reported. We assumed that if all probes were lost, or the majority were lost and those that were not lost were flanked on both sides by probes that were lost, that loss had occurred (stated in paper: "ratios of adjacent probes should be taken into consideration for the assessment of the presence of gains or losses". We ignored the results for the most centromeric 1p probe (NOTCH2). CGH: references 3 papers. Reference 25 (Jeuken et al. Journal of Neuropathology & Experimental Neurology 1999;58:606‐12): "Detection thresholds for losses and gains of chromosomal regions (19, 20) were set at 0.8 and 1.2 respectively. Aberrations with a ratio of 0.6 or 1.4 were called clear copy number changes and a ratio larger than 1.6 were called high‐copy number gains. CGH only detects copy number changes of chromosomal regions larger than 2Mb (20, 36, 37)". Reference 26 (Jeuken et al. Journal of Pathology 2001;194:81‐7) References the reference above. "Analysis was performed using QUIPS CGH software (Applied Imaging, UK) and the standard thresholds for gains (1.2) and losses (0.8) were used. Aberrations with a ratio less than 0.6 or more than 1.4 were called clear copy number changes, whereas a ratio larger than 1.6 was called a high copy number gain". Reference 27 (Jeuken et al. Journal of Neurosurgery 2002, 96:559‐64) – not located. |
| Anaplastic oligodendroglioma | III | 12 | 0 | 0 | 2 | 14 | |||
| Oligoastrocytoma | II | 2 | 0 | 0 | 4 | 6 | |||
| Anaplastic oligoastrocytoma | III | 3 | 0 | 0 | 12 | 15 | |||
| Pilocytic astrocytoma | I | 0 | 0 | 0 | 1 | 1 | |||
| Astrocytoma | II | 0 | 0 | 1 | 4 | 5 | |||
| Anaplastic astrocytoma | III | 0 | 0 | 0 | 1 | 1 | |||
| Ependymoma | II | 0 | 0 | 0 | 3 | 3 | |||
| Anaplastic ependymoma | III | 0 | 0 | 0 | 1 | 1 | |||
| Glioblastoma | IV | 0 | 0 | 0 | 17 | 17 | |||
| Jha 2011 | A: FISH B: PCR |
Oligodendroglioma | II | 10 | 0 | 3 | 3 | 16 | Although cut‐off for number of loci that needed to have LOH not explicitly reported, it was reported how many cases had combined loss of 1p and 19q by PCR and from that it seems that LOH of 1 marker on each of the chromosome arms was sufficient to count as codeletion. |
| Anaplastic oligodendroglioma | III | 5 | 0 | 4 | 5 | 14 | |||
| Glioblastoma | IV | 0 | 0 | 0 | 10 | 10 | |||
| Kato 2019a | A: FISH B: NGS |
Glioma | II–IV | 3 | 0 | 0 | 6 | 9 | We assumed that (quote) "Our sequence pipeline and also FISH identified 1p19q codeletion only in these 3 cases" means that neither test found any other codeletions. |
| Kolhe 2016 | A: FISH B: SNP array |
Glioblastoma | IV | 1 | 0 | 0 | 1 | 2 | Table very small in the conference abstract. We assumed '+' meant codeleted. We used histological diagnoses. |
| Infiltrating mixed glioma, oligoastrocytoma | NR | 1 | 0 | 0 | 0 | 1 | |||
| Anaplastic oligoastrocytoma | III | 0 | 0 | 0 | 1 | 1 | |||
| Anaplastic astrocytoma | III | 1 | 0 | 0 | 1 | 2 | |||
| Anaplastic oligodendroglioma | III | 2 | 0 | 0 | 0 | 2 | |||
| Malignant mixed oligoastrocytoma | IV | 1 | 0 | 0 | 0 | 1 | |||
| Lass 2013 | A: CISH B: FISH |
Astrocytoma | II | 0 | 0 | 0 | 4 | 4 | Astrocytoma: could not include participant 24 (ID 56240) as no 19q CISH result (however, as 1p retained, and no 1p19q codeletion on FISH, this participant could be classified as negative on both tests). Excluded participant 25 as aged < 18 years. Anaplastic astrocytoma: CISH result correct – FISH was repeated and tumour was codeleted. Oligodendroglioma: CISH result correct – confirmed by microsatellite PCR and FISH was repeated and tumour was codeleted. Anaplastic oligodendroglioma: we excluded participant 16 (ID56604) because no results on initial FISH, and on investigation it seemed the results depended on where the tumour sample was from. |
| Anaplastic astrocytoma | III | 0 | 1 | 0 | 9 | 10 | |||
| GBM | IV | 0 | 0 | 0 | 1 | 1 | |||
| Oligodendroglioma | II | 8 | 1 | 0 | 1 | 10 | |||
| Anaplastic oligodendroglioma | III | 5 | 0 | 0 | 0 | 5 | |||
| Oligoastrocytoma | II | 5 | 0 | 0 | 1 | 6 | |||
| Anaplastic oligoastrocytoma | III | 1 | 0 | 0 | 0 | 1 | |||
| Pilocytic astrocytoma | I | 0 | 0 | 0 | 1 | 1 | |||
| Lhotska 2015 | A: FISH B: SNP array |
Oligodendroglioma and oligoastrocytoma | II | 16 | 0 | 2 | 2 | 20 | Cut‐off to interpret SNP array results not reported, and participants not classified. We set the criteria for codeletion as 1 copy of (or homozygous for) 1p36.33p11.2 or 1p31.1p12 or 1p31.3p31.1 AND 1 copy of/homozygous for 19q12q13.43 or 19q13.2q13.43 or 19q13.32q13.43. |
| Na 2019 | A: FISH B: NGS |
Astrocytoma and anaplastic astrocytoma | II/III | 0 | 0 | 0 | 23 | 23 | The study also looked at CCNE1 on 19q. But this did not seem to have been considered when calculating concordance. Quote: "The copy number loss of 1p/19q genes detected in NGS was compared with FISH and the results were concordant in all cases of ODs [oligodendroglioma]. Given that about 20% of ODs are related to incomplete 1p/19q‐codeletion [15], some mismatches between the copy number loss of CCNE1 gene (NGS) and 19q deletion (FISH) can be explained by the distant genomic loci of CCNE1 and the FISH‐probe target region". Some participants aged < 18 years. |
| Oligodendroglioma and anaplastic oligodendroglioma | II/III | 12 | 0 | 0 | 4 | 16 | |||
| Glioblastoma | IV | 0 | 1 | 0 | 85 | 86 | |||
| Diffuse midline glioma | IV | 0 | 1 | 0 | 9 | 10 | |||
| Natte 2005 | A: FISH B: MLPA |
Oligodendroglioma | NR | 0 | 1 | 0 | 0 | 1 | — |
| Anaplastic oligodendroglioma | NR | 9 | 0 | 3 | 3 | 15 | |||
| Oligoastrocytoma | NR | 0 | 0 | 0 | 2 | 2 | |||
| Anaplastic oligoastrocytoma | NR | 0 | 0 | 0 | 1 | 1 | |||
| Nigro 2001a | A: FISH B: Real‐time PCR |
Oligodendroglioma | NR | 3 | 0 | 0 | 4 | 7 | Figure 5 in the paper implies that some samples were tetraploid. It is not clear how this was detected (i.e. by which technique). In addition, they show an image of the FISH results for 1 case (8758) which they describe as tetraploid. This showed a 1 red: 2 green dot pattern so it was unclear how this case could be tetraploid. Therefore, we assumed that none of the tumours actually were tetraploid. |
| Anaplastic oligodendroglioma | NR | 5 | 1 | 0 | 4 | 10 | |||
| Oligoastrocytoma | NR | 4 | 0 | 0 | 1 | 5 | |||
| Park 2019 | A: FISH B: NGS |
Oligodendroglioma | NR | 10 | 0 | 0 | 0 | 10 | — |
| Anaplastic oligodendroglioma | NR | 9 | 1 | 0 | 0 | 10 | |||
| Paxton 2015 | A: FISH B: SNP array |
Oligodendroglioma | NR | 8 | 1 | 0 | 0 | 9 | Selected because positive on FISH assay. Quote: "One case (1p19q‐03), with diploid copy number along 1p and 19q arms, was tetraploid across the remainder of the genome (Fig. 1a). Genotyping data from the array indicate that 1p and 19q were co deleted prior to a doubling of the genome, so that although diploid in number, the 1p and 19q arms are still deleted in the context of the entire genome". This is a relative rather than an absolute deletion. |
| Glioblastoma | NR | 0 | 0 | 0 | 8 | 8 | Selected because negative on FISH assay | ||
| Pesenti 2017 | A: MS B: PCR |
Astrocytoma | II | 0 | 0 | 0 | 4 | 4 | — |
| Anaplastic astrocytoma | III | 0 | 0 | 0 | 3 | 3 | |||
| Oligodendroglioma | II | 8 | 0 | 0 | 0 | 8 | |||
| Anaplastic oligodendroglioma | III | 1 | 0 | 0 | 0 | 1 | |||
| Glioblastoma | IV | 0 | 0 | 0 | 20 | 20 | |||
| Ransom 1992a | A: G‐banding B: RFLP |
Juvenile pilocytic astrocytoma | II | 0 | 0 | 0 | 1 | 1 | We removed participants with no growth (G‐banding/karyotyping), and those who we thought had no results for RFLP because 1p/19q not listed in the informative arms retained, lost or gained; we also removed participants aged < 18 years. Details of tests taken from Ransom 1992b. |
| Oligodendroglioma | II | 0 | 0 | 1 | 1 | 2 | |||
| Oligodendroglioma | III | 0 | 0 | 1 | 0 | 1 | |||
| Ependymoma | I | 0 | 0 | 0 | 1 | 1 | |||
| Ransom 1992b | A: G‐banding B: RFLP |
Astrocytoma | III | 0 | 0 | 0 | 1 | 1 | We removed participants with no growth (G‐banding/karyotyping), and those who we thought had no results for RFLP because 1p/19q not listed in the informative arms retained, lost or gained. |
| Astrocytoma | IV | 0 | 0 | 0 | 18 | 18 | |||
| Mixed oligoastrocytoma | III | 0 | 0 | 0 | 2 | 2 | |||
| Mixed oligoastrocytoma | IV | 0 | 0 | 0 | 1 | 1 | |||
| Scheie 2006 | A: FISH B: PCR |
Oligodendroglioma | II | 7 | 0 | 0 | 4 | 11 | Another cut‐off for FISH mentioned. |
| Oligodendroglioma | III | 4 | 0 | 0 | 1 | 5 | |||
| Oligoastrocytoma | II | 8 | 0 | 0 | 6 | 14 | |||
| Oligoastrocytoma | III | 1 | 0 | 1 | 8 | 10 | |||
| Schrock 1994 | A: CGH B: G‐banding |
Anaplastic astrocytoma | III | 0 | 0 | 0 | 2 | 2 | Participants aged < 18 years excluded. |
| Glioblastoma | IV | 0 | 0 | 0 | 6 | 6 | |||
| Senetta 2013b | A: FISH (cut‐off ratios 1p ≤ 0.8 and 19q ≤ 0.8) B: FISH (cut‐off ratios 1p ≤ 0.7 and 19q ≤ 0.8) |
Oligodendroglioma | II | 25 | 6 | 0 | 17 | 48 | — |
| Oligodendroglioma | III | 17 | 5 | 0 | 19 | 41 | |||
| Oligoastrocytoma | II | 5 | 1 | 0 | 12 | 18 | |||
| Oligoastrocytoma | III | 2 | 1 | 0 | 17 | 20 | |||
| Glioblastoma with an oligodendroglial component (GBM‐O) | IV | 3 | 3 | 0 | 10 | 16 | |||
| Sim 2018a (glioblastoma cohort) | A: FISH B: NGS or aCGH (or both) |
Glioblastoma | NR | 0 | 2 | 0 | 73 | 75 | Excluded the 5 recurrent samples from 4 participants. These were negative on both tests. Quote: "Primary and recurrent tumors from three patients revealed intact 1p or 19q by either technique. Three tumor samples (two recurrent and one primary tumor) from one patient showed partial deletion of 1p36 by aCGH and/or WES [whole exome sequencing], but FISH revealed no deletion of 1p or 19q in the primary or secondary recurrent tumors and no 1p deletion in the first recurrence". |
| Sim 2018b (oligodendroglial cohorta) | A: FISH B: NGS |
Anaplastic oligodendroglioma | NR | 4 | 1 | 0 | 1 | 6 | We used the original diagnoses. |
| Anaplastic oligoastrocytoma | NR | 0 | 0 | 0 | 3 | 3 | |||
| Oligoastrocytoma | NR | 0 | 0 | 0 | 1 | 1 | |||
| Smith 1999 (CGH vs FISH) | A: CGH B: FISH |
Astrocytoma | IV | 0 | 0 | 0 | 1 | 1 | Quote: "LOH, FISH, and CGH were performed as previously described (Cliby et al., 1993; Ritland et al., 1995; Qian et al., 1995; Mohapatra et al., 1995, 1998; Piper et al., 1995)". Overall results for FISH not reported. We defined codeletion as hemizygous deletion of 1p36, 1q13.1‐q13.2 and 19q13.3. |
| Smith 1999 (CGH vs PCR) | A: CGH B: PCR |
Astrocytoma | III | 0 | 0 | 0 | 1 | 1 | Quote: "LOH, FISH, and CGH were performed as previously described (Cliby et al., 1993; Ritland et al., 1995; Qian et al., 1995; Mohapatra et al., 1995, 1998; Piper et al., 1995)". Overall results for 1p and 19q for PCR not given. We assumed that if results were confirmed allelic loss/presumed allelic loss/homozygous at all loci on 1p except the most centromeric (D1S534 – removed from list of loci examined) and all loci on 19q examined that codeletion was present. |
| Astrocytoma | IV | 0 | 0 | 0 | 4 | 4 | |||
| Smith 1999 (FISH vs PCR) | A: FISH B: PCR |
Astrocytoma | II | 0 | 0 | 0 | 1 | 1 | Quote: "LOH, FISH, and CGH were performed as previously described (Cliby et al., 1993; Ritland et al., 1995; Qian et al., 1995; Mohapatra et al., 1995, 1998; Piper et al., 1995)". Overall results for 1p and 19q for PCR not given. We assumed that if results were confirmed allelic loss/presumed allelic loss/homozygous at all loci on 1p except the most centromeric (D1S534 – removed from list of loci examined) and all loci on 19q examined that codeletion was present. Overall results for FISH also not reported. We defined codeletion as hemizygous deletion of 1p36, 1q13.1‐q13.2 and 19q13.3. |
| Astrocytoma | III | 0 | 0 | 0 | 1 | 1 | |||
| Astrocytoma | IV | 1 | 0 | 0 | 15 | 16 | |||
| Mixed | II | 0 | 0 | 0 | 1 | 1 | |||
| Mixed | III | 0 | 0 | 0 | 3 | 3 | |||
| Oligodendroglioma | II | 3 | 0 | 0 | 2 | 5 | |||
| Oligodendroglioma | III | 1 | 0 | 0 | 0 | 1 | |||
| Srebotnik‐Kirbis 2016 | A: FISH (cytospins) B: FISH (FFPE) |
Oligodendroglioma | II | 5 | 0 | 0 | 0 | 5 | Excluded participants with uninterpretable test results. Participants with 1p/19q imbalance were counted as not having the deletion (quote: "Imbalance was defined as a relative loss of target signals in comparison with controls, with target signals >1 (ex. 2/3, 2/4, 3/4, etc.)". Also tested a group of 19 non‐oligodendroglial tumours. Could not extract results for these as 7/19 were not gliomas and results just for the participants with glioma were not presented. |
| Anaplastic oligodendroglioma | III | 1 | 0 | 0 | 0 | 1 | |||
| Anaplastic oligoastrocytoma | III | 1 | 0 | 0 | 5 | 6 | |||
| Thakur 2012a | A: FISH B: SNP array |
Oligodendroglioma | NR | 2 | 0 | 0 | 0 | 2 | FISH was presumably also on FFPE samples. |
| Thomas 2017 | A: FISH B: NGS |
Anaplastic oligodendroglioma, anaplastic oligoastrocytoma | NR | 13 | 1 | 0 | 5 | 19 | We assumed only 1 participant with a false‐positive result and all other results concordant. Quote: "A total of 19 patients had available tissue with adequate DNA quality and quantity for gene sequencing analysis, including 14 patients with confirmed 1p/19q codeletion and 5 with 1p/19q intact … One patient thought to have 1p/19q codeletion on FISH had a glioblastoma‐like signature with PTEN, CDKN2B, CDKN2AP16INK4A, and CDKN2AP14ARF with no evidence of 1p/19q loss or IDH1 or 2 mutation on gene sequencing, suggesting a false‐positive 1p/19q codeletion on FISH". |
| Tsiatis 2010 | A: PCR B: SNP array |
Anaplastic mixed oligoastrocytoma | NR | 0 | 0 | 0 | 1 | 1 | We excluded case n5 as aged < 18 years. |
| Anaplastic oligodendroglioma | NR | 1 | 0 | 0 | 0 | 1 | |||
| Astrocytoma | NR | 0 | 0 | 0 | 1 | 1 | |||
| Low‐grade glioneural tumour | NR | 0 | 0 | 0 | 1 | 1 | |||
| Uchida 2019 | A: FISH (deletion criterion of 1p or 19q signals < signals of 1q or 19p) B: FISH (deletion criterion of single signal of 1p or 19q and 2 signals of 1q or 19p) |
Glioblastoma | IV | 1 | 5 | 0 | 135 | 141 | — |
| Wiestler 2014 | A: Methylation array B: MLPA |
Anaplastic astrocytoma, Anaplastic oligoastrocytoma, Anaplastic oligodendroglioma | III | 39 | 1 | 7 | 52 | 99 | Reference 12 in this paper: Wick et al. (2009) "Detection of chromosome arms 1p and 19q deletions was performed by a multiplex ligation‐dependent probe assay (Salsa MLPA, P088 lots 0305 and 0706, MRC Holland, Amsterdam, the Netherlands).16 Chromosomal regions were scored as under‐ or overrepresented if two or more loci on 1p or 19q adjacent to each other exhibited a gene dosage ratio less than 70% or more than 130% relative to the reference value. In the 59 patients from whom leukocyte DNA was available, we additionally performed microsatellite‐based loss of heterozygosity analyses for allelic losses on 1p and 19q. At least five microsatellite loci on each arm were analyzed.17,18". No comparison of the results from these 2 techniques. MLPA methods from this reference. Quote: "In the few discordant 1p/19q cases (1p/19q codeleted based on MLPA, 1p/19q intact as per HM450), the clinical course and pathological characteristics tended to accord to the HM450 data … This may be explained by the rather low threshold chosen in the initial MLPA assessment where two adjacent gene loci with a gene dosage ratio of less than 70% were considered as evidence of chromosome arm deletion". |
++: number of people (or tumours (astudies where we consider the unit of analysis was a tumour)) positive on both tests (meaning that 1p19q codeletion found with both tests); +‐: positive on test A, negative on test B; ‐+: negative on test A, positive on Test B; ‐‐: negative on both tests; aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CISH: chromogenic in situ hybridisation; FISH: fluorescent in situ hybridisation; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; NGS: next‐generation sequencing; NR: not reported; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; SNP: single nucleotide polymorphism. bSenetta 2013 had data on other FISH criteria, and comparisons could be made for subsets of participants. These are shown in Appendix 8.
Appendix 7. Data extraction details
We attempted to include participants only once, that is, we extracted one result per person, as 1p/19q codeletion status is thought to be stable. Where it was not clear if results were per person, we extracted data once per tumour. This meant that where possible if it was clear that the same participant contributed multiple samples, we extracted results for only one sample and excluded results for the other samples. It was generally the case that if a study included multiple samples from the same participant, then primary and recurrent tumours were included, and, in this scenario, we used the result for the primary tumour. For example, we excluded recurrent cases from Bigner 1999 and Sim 2018a Glioblastoma cohort, and the second tumour sample from the same participant in Burger 2001. However, in some cases it was not possible to extract just one result per participant despite it being clear that some participants must have contributed more than one sample, because sufficient individual participant data were not presented (e.g. D'Haene 2019).
In some cases the same participants were included in multiple studies. In this situation, we tried to include participants only once in the analyses. For example, Hatanpaa 2003a and Hatanpaa 2003b studied the performance of multiplex microsatellite polymerase chain reaction (PCR) and capillary electrophoresis interpreted without comparison to normal DNA (from normal tissue or blood) in tumour samples that had produced concordant results after being tested with at least of two of the following methods: comparative genomic hybridisation (CGH), fluorescent in situ hybridisation (FISH) and PCR‐based microsatellite analysis with comparison to normal deoxyribonucleic acid (DNA) from the same participant. They report using tumour samples from Johns Hopkins Hospital that had been investigated in Smith 1999, and one sample from Burger 2001. We attempted to match the tumour samples up using case numbers, although this required some interpretation as it appeared that histological diagnoses may have altered between publications. However, we were unable to match one participant (T117 in Hatanpaa 2003a; Hatanpaa 2003b) to any of the included participants in Smith 1999. We excluded the matched participants from data sets for Smith 1999 and Burger 2001. During this analysis, the possibility of further overlap in participants between these publications arose as two of the participants in Burger 2001 had the same case numbers as those in Hatanpaa 2003b that were linked to participants in Smith 1999. All three studies used samples from Johns Hopkins Hospital. However, as Burger 2001 made no mention of using samples that had already been tested in Smith 1999, we decided that we could not conclude that these were the same participants.
Where possible, we excluded results for participants aged less than 18 years. However, again in some cases it was clear that some participants were aged less than 18 years (e.g. because the lower bound of the age range was less than 18 years) but it was not possible to exclude results for participants aged less than 18 years as individual participant data with all relevant characteristics were not presented (e.g. Ghasimi 2016; Harada 2011; Na 2019).
We excluded participants without some form of glioma. For example, we excluded one participant with pineal parenchymal tumour from Harada 2011.
If 1p/19q codeletion status was not reported, we examined the status of 1p and 19q, in combination with any reported polysomy to classify participants as having 1p/19q codeletion. However, we did not examine the status of 1q or 19p. This means that some of the participants/tumours that were assessed as having 1p/19q codeletion may have additionally lost 1q or 19p. There were some studies that judged 1p/19q codeletion not to be present if 1q or 19p was lost; if this was the case, we extracted the authors' classifications.
There were several instances where we had to classify the results of at least one test in a study. For example, Smith 1999 compared PCR‐based LOH assay with FISH and CGH. Although not explicitly reported in this publication, CGH presumably looked genome wide; there were a number of PCR markers examined along 1p (D1S468, D1S1612, D1S1597, D1S199, D1S1665, D1S1728, D1S1588, D1S1675, D1S187, D1S534) and 19q (D19S213, D19S569, D19S422, D19S219, DM, D19S112, D19S412, D19S596, HRC, D19S589, D19S218); and target FISH probes were used that hybridise to 1p36 and to both 19q13.2‐q13.2 (AKT2) and 19q13.3. Cut‐offs were not prespecified or explicitly reported, but the paper defined "minimal deletion regions" on 1p36 (D1S468‐D1S1612) and 19q13.3 (D19S412‐D19S596). It was unclear if these were to be taken as the criteria for judging deletion of 1p or 19q, and classification of participants based on the results of FISH and PCR‐based LOH was not reported in the paper (CGH results were classified), although it must have occurred as the paper reported concordances and correlations among PCR‐based LOH, FISH and CGH by chromosome arm. Instead, individual participant data for each PCR marker and FISH probe were presented. We assumed that the aim of this paper was to identify the localisation of common deletion regions on 1p and 19q in gliomas, and, given the individual participant data presented, we decided to classify the FISH and PCR results ourselves. 1p was considered lost by PCR if all markers on 1p, with the exception of D1S534, showed either confirmed allelic loss, presumed allelic loss, were homozygous or were indeterminant; and by FISH if there was a hemizygous deletion in the 1p36 probe. 19q was considered lost by PCR if all markers on 19q showed either confirmed allelic loss, presumed allelic loss, were homozygous or were indeterminant; and by FISH if there was a hemizygous deletion in both the 19q13.2‐q13.2 (AKT2) and 19q13.3 probe. 1p/19q codeletion was present if both 1p and 19q were lost.
Burger 2001 also did not classify results for PCR‐based LOH, although it did state that some results strongly suggested loss of a chromosome arm. We classified the results with the threshold that all informative markers needed to be homozygous.
Lhotska 2015 did not specify a threshold for determining whether the 1p/19q codeletion was present by single nucleotide polymorphism (SNP) array, and did not classify. We set the criteria for codeletion as one copy of (or homozygous for, suggestive of copy neutral LOH) 1p36.33p11.2 or 1p31.1p12 or 1p31.3p31.1 AND one copy of/homozygous for 19q12q13.43 or 19q13.2q13.43 or 19q13.32q13.43.
Jeuken 2006 prespecified cut‐off for particular MLPA probes (probe ratio 0.8 or less) but not for deletion of a chromosome arm. Again, we classified the results. We assumed that if all probes were lost, or the majority were lost and those that were not lost were flanked by probes that were lost, that loss had occurred, ignoring the results for the most centromeric 1p probe (NOTCH2).
We also interpreted the results for the SNP array in Hinrichs 2016.
There were other situations where we had to make judgements regarding 1p/19q status based on reported cytogenetic abnormalities or the presence, loss or gain of chromosomal regions (e.g. Ransom 1992a; Ransom 1992b; Schrock 1994).
In other papers, even though classifications had been made, we had to interpret the results. For example, in Bigner 1999, some participants had the CGH result (quote) "copy number changes of partial chromosome arms". We derived from the text that the authors considered that when this had occurred on 1p/19q that a codeletion was present. In Dahlback 2009 and Dahlback 2011, the criteria for 1p/19q codeletion based on the results of CGH were not reported (high‐resolution CGH (HR‐CGH), was used in these studies). There were some classifications made in the text, and raw results were also presented. We attempted to identify participants with classifications of 1p/19q codeletion to confirm that they had results from other tests, but we had difficulty identifying which participants would have been classified as having the codeletion. The data extraction for Dahlback 2009 was mainly based on the following text: "Combined 1p/19q loss was not observed by G‐banding analysis. The HR‐CGH data revealed 1p/19q loss in only five tumours (3 GB, 1 GS, and 1 GB‐OD)". In Dahlback 2011, it was reported that "HR‐CGH could be performed in 18 of the 22 tumours where LOH‐PCR results were available. Both methods found the complete 1p/19q codeletion in nine samples. The PCR‐based method detected a codeletion in a sample where HR‐CGH showed loss of 19q, but only partial loss of 1p. Two tumours that were normal by LOH‐PCR showed partial loss of 19q by HR‐CGH and one displayed partial loss of 1p where no aberrations were found by the former method". Numbers with positive results for HR‐CGH and PCR by histological diagnosis were also reported. However, not all the numbers tallied. PCR results were categorised in a table. We used these, and the raw HR‐CGH results and the text to try and interpret the CGH results. In Duval 2014, FISH assessments were made independently by two observers. In one case, a participant was classified as having the codeletion by one observer but not the other when using the ratio method to interpret the results of ImmunoFISH (FISH with immunohistochemistry against Ki67 (MIB‐1)). Raw data were available from the two observers, and we averaged the raw data and applied the reported cut‐off to come to a consensus classification. The results in Ghasimi 2016 required careful interpretation. In the text it stated that 55 people had results from both techniques, and that FISH detected 14 people with codeletion. From their Supplementary Table 3 it appeared that only 11 of these people had results also from SNP array. We assumed that the phrase "none was detected by SNP array" to mean that no one had the codeletion by SNP array (and not those with positive FISH results). Thomas 2017 made no direct comparison between the results obtained by FISH and next‐generation sequencing (NGS). A discrepant result is described in detail, we have, therefore, assumed that all other cases were concordant. We attempted to extract results for just gliomas from D'Haene 2019. The results we extracted were based on the text "concordant positive results were obtained for 21 of the 22 glioma samples (95.4% sensitivity)." and "Among the 31 gliomas that did not show a 1p/19q codeletion by FISH, 28 showed neither patterns of 1p/19q loss of heterozygosity (LOH) by NGS, as defined by our criteria (three were non‐informative)". Kato 2019 was reported as a conference abstract only. We assumed that the sentence "our sequence pipeline and also FISH identified 1p19q codeletion only in these 3 cases" meant that neither test found any other codeletions.
In Blesa 2009, there were some differences between Table 2 in their text and their Supplementary Table. Since their Supplementary Table had more information, we extracted from this. In Horbinski 2012, there were discrepancies between text (and what they calculated in their Table 2) and Table 1; we used the text and their Table 2.
In some cases, results were classified for 1p and 19q separately. We only extracted participants who had results for both 1p and 19q from at least two tests, and classified all other participants as having missing results. In some cases, results would have been deducible: for example if 1p or 19q was retained then the participant could not have the codeletion. However, the result is not deducible in the inverse situation (i.e. if 1p or 19q is deleted but a result is not available for the other chromosome arm).
As described in the introduction, copy neutral loss of heterozygosity was classified as a loss.
Also as described in the introduction, we were interested in diagnosing absolute 1p/19q codeletions (rather than relative codeletions). Some studies classified participants/tumours as have relative codeletions or as having imbalance. We grouped these participants/tumours with participants/tumours without a codeletion. In addition, we made any exception to our rule of extracting the researchers' classifications of test results if it was clear that some of the participants or tumours classified as having a 1p/19q codeletion had a relative codeletion that was detectable by the technique used.
Cut‐offs/thresholds used
The cut‐offs/thresholds used to classify 1p/19q status were often not described or incompletely reported.
As stated previously, where available, we extracted the researchers' classifications of test results (i.e. we did not attempt to reclassify test results even if raw data were available if the researchers had made classifications). In some studies, it appeared that the cut‐offs/thresholds used to classify codeletion would classify both partial deletions of 1p/19q and full arm deletions of 1p/19q as codeletions or that the cut‐offs/thresholds used did not fully exploit the potential of a particular test, or both.
In studies investigating PCR‐based LOH, it was sometimes clear that although participants were classified as having the codeletion, it was more likely that they had partial deletions. For example, in Bouvier 2004, codeletion was investigated using three microsatellite markers on 1p and three on 19q (1p36.23 (D1S1612), 1p34.2 (D1S447), 1p13.3 (D1S252) and on 19q13.32 (D19S412 and D19S219), 19p13.12 (D19S226)). Criteria for LOH were prespecified ("LOH was scored when signal intensity was <0.5 or >2 in a tumor sample") but it was unclear how many of the microsatellite markers needed to show LOH to be classified as a codeletion. Four participants classified by the authors as having the codeletion were then described as having "partial deletion on 19q for the 19q13.32 and not for the 19q13.12". Clark 2013 stated that "to be considered codeleted the majority of informative microsatellite loci on both 1p and 19q had to show LOH". Eight glioblastomas tested positive for the codeletion by PCR. However, in the discussion, they stated that "in the 8 cases in this subset that met our initial LOH codeletion criteria, usually only a single 1p or 19q microsatellite remained intact. For there to be true whole‐arm codeletion, all microsatellites should be lost on 1p and 19q. Because none of the tumours in this cohort showed complete loss of microsatellites on both arms". In Gadji 2009, it was unclear how many markers needed to show LOH, and some participants who were classified as having 1p/19q codeletion appeared to be heterozygous at numerous markers. In the discussion they stated "additionally, across our population the 1p‐/19q‐ codeletion was located in different regions of chromosome arms 1 and 19, from comprising the whole chromosomal arm deletion to partial terminal chromosomal arm or interstitial chromosomal deletion" implying that not all participants classified as having the codeletion had it. In Jha 2011, although the cut‐off for number of loci that needed to have LOH was not explicitly reported, it seemed that LOH of one marker on each of the chromosome arms was sufficient to count as codeletion.
Other tests also had issues. For example, 1p loss by real‐time PCR in Nigro 2001 was defined by copy number less than 1.58 in two or more sequential loci (rather than along the whole arm). In Wiestler 2014, the criteria for arm deletion on MLPA was initially two adjacent loci with a gene dosage ratio less than 70% (again, rather than along the whole arm). In Ghasimi 2016, although the SNP array used could look along the whole of 1p and 19q, they chose to only look at the results for regions corresponding to the location of the FISH probes, meaning that the full potential of the SNP array was not exploited.
'Perfect' tests
We extracted results without regard to whether any of the tests was assumed to be a perfect test.
However, several of the included studies predefined the reference standard test and calculated sensitivity and specificity for the other test based on this. For example, Scheie 2006 designated PCR‐based LOH as the reference standard when comparing FISH and PCR‐based LOH and Nigro 2001 designated FISH as the reference standard for the comparison of real‐time PCR with FISH.
Some studies repeated analyses or tested discordant cases using a third technique. For example, Lass 2013 repeated FISH analysis or performed PCR‐based LOH (or both) for cases with discordant FISH and CISH results.
Other studies looked at the clinical course/prognosis of participants with discordant results to determine which test was more likely to be correct, for example Wiestler 2014 and Senetta 2013. We did not systematically extract this information as it was outside the scope of this review.
Appendix 8. Extra comparisons in Senetta 2013
Senetta 2013 assessed 1p/19q status using different fluorescent in situ hybridisation (FISH) criteria:
two different ratio cut‐offs (a cut‐off ratio of 0.8 or less was used to define 1p and 19q allelic losses; in addition for 1p a more stringent ratio cut‐off of 0.7 or less was applied);
percentage of neoplastic nuclei carrying 1p and 19q deletions of 50% or greater; and
combination of the ratio cut‐offs and percentage of neoplastic nuclei carrying 1p deletions of 50% or greater.
They also considered polysomy (30% or greater of nuclei carrying three or more control signals for both arms).
We extracted into our main outcomes table the comparison between the two different ratio cut‐offs (FISH ratio cut‐offs of 0.8 or less for 1p and 19q; and 0.7 or less for 1p and 0.8 or less for 19q) as this comparison included all participants.
We could not compare the results of the second set of criteria (percentage of neoplastic nuclei carrying 1p and 19q deletions of 50% or less) with any of the other sets of criteria.
For the third set of criteria, we could make some comparisons with the ratio cut‐off alone for the 132 of the 143 cases with data on the percentage of deleted nuclei, as shown below.
| Cut‐off A | Cut‐off B | ++ | +‐ | ‐+ | ‐‐ |
| Ratio cut‐off 1p ≤ 0.8 and 19q ≤ 0.8 | Ratio cut‐off 1p ≤ 0.8 and 19q ≤ 0.8 plus ≥ 50% of 1p deleted nuclei | 47 | 16 | 0 | 69 |
| Ratio cut‐off 1p ≤ 0.7 and 19q ≤ 0.8 | Ratio cut‐off 1p ≤ 0.7 and 19q ≤ 0.8 plus ≥ 50% of 1p deleted nuclei | 42 | 5 | 0 | 85 |
++: positive using both cut‐offs (meaning that 1p19q codeletion found with both cut‐offs); +‐: positive with cut‐off A, negative with cut‐off B; ‐+: negative on cut‐off A, positive on cut‐off B; ‐‐: negative with both cut‐offs.
If we considered the relative deletions (deletions in the presence of 30% or greater of nuclei carrying three or more control signals for both arms) as being negative we could calculate the following numbers (assuming that we had data for 132 cases):
| Cut‐off A | Cut‐off B | ++ | +‐ | ‐+ | ‐‐ |
| Ratio cut‐off 1p ≤ 0.8 and 19q ≤ 0.8 | Ratio cut‐off 1p ≤ 0.8 and 19q ≤ 0.8 plus < 30% nuclei carrying 3 or more control signals for both arms | 58 | 5 | 0 | 69 |
| Ratio cut‐off 1p ≤ 0.7 and 19q ≤ 0.8 | Ratio cut‐off 1p ≤ 0.7 and 19q ≤ 0.8 plus < 30% nuclei carrying 3 or more control signals for both arms | 42 | 5 | 0 | 85 |
++: positive using both cut‐offs (meaning that 1p19q codeletion found with both cut‐offs); +‐: positive with cut‐off A, negative with cut‐off B; ‐+: negative on cut‐off A, positive on cut‐off B; ‐‐: negative with both cut‐offs.
Appendix 9. QUADAS‐2 risk of bias assessments for studies that performed two or more FISH variants
QUADAS‐2 risk of bias and applicability assessments for studies that assessed at least two fluorescence in situ hybridisation (FISH) variants are shown in Figure 35.
12.
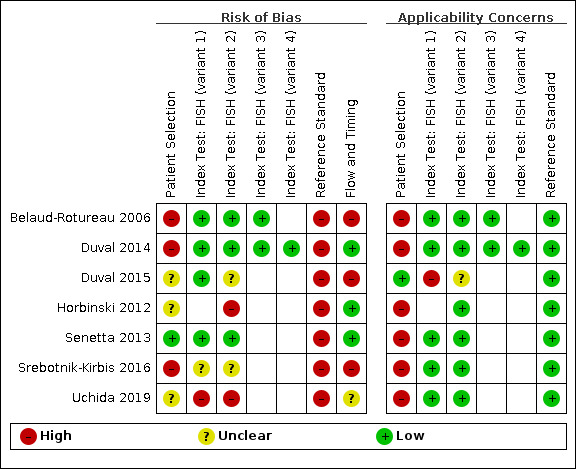
Risk of bias and applicability concerns summary for studies that assessed at least two FISH variants: review authors' judgements about each domain for each included study. Note: the judgements for FISH variant 1 for Horbinski 2012 are shown in Figure 7.
FISH: fluorescence in situ hybridisation.
The tests were as follows.
Belaud‐Rotureau 2006: FISH variant 1 was FISH with the 1p36.3 (D1Z2)/1q12 (D1Z1) and 19q13.3/19pter probes and manual analysis, FISH variant 2 was FISH with the 1p36/1q25 and 19q13/19p13 Abbott Vysis probe set and manual analysis and FISH variant 3 was FISH with 1p36/1q25 and 19q13/19p13 Abbott Vysis probe set and automated analysis.
Duval 2014: FISH variant 1 was a combination cut‐off (based on number of cells showing a deletion), FISH variant 2 was a ratio cut‐off (based on the ratio of signals for 1p to 1q and 19q and 19p), FISH variant 3 was immunoFISH with a combination cut‐off, FISH variant 4 was immunoFISH with a ratio cut‐off.
Duval 2015: FISH variant 1 was automated FISH analysis, FISH variant 2 was manual FISH analysis.
Horbinski 2012: FISH variant 1 was a cut‐off of a target‐ploidy control ratio of less than 0.87, FISH variant 2 was a cut‐off of a target‐ploidy control ratio of less than 0.75; in both cases at least 20% of nuclei needed to show deletion. This study is also in Figure 7.
Senetta 2013: FISH variant 1 was FISH with cut‐off ratios 1p of 0.8 or less and 19q of 0.8 or less, FISH variant 2 was cut‐off ratios 1p of 0.7 or less and 19q of 0.8 or less.
Srebotnik‐Kirbis 2016: FISH variant 1 was FISH performed on cytospins, FISH variant 2 was FISH performed on FFPE sections.
Uchida 2019: FISH variant 1 was deletion criterion of 1p or 19q signals < signals of 1q or 19p, FISH variant 2 was deletion criterion of single signal of 1p or 19q and two signals of 1q or 19p; in both cases the cut‐off value was set at 20%.
Appendix 10. Standard deviations and between‐study correlations
Table A10.1. Standard deviations and between‐study correlation from analyses using FISH as the reference standard
| Standard deviation of logit(sensitivity) (95% CrI) |
Standard deviation of logit(1 – specificity) (95% CrI) |
Between‐study correlation (95% CrI) | ||
| Main analysis (shared parameters) | 1.84 (1.42 to 1.99) | 1.36 (0.78 to 1.94) | 0.36 (0.03 to 0.78) | |
| Sensitivity analysis | PCR‐based LOH | 1.80 (1.27 to 1.99) | 1.39 (0.79 to 1.95) | 0.29 (0.02 to 0.73) |
| SNP array | 1.80 (1.22 to 1.99) | 1.55 (0.46 to 1.98) | 0.66 (0.07 to 0.98) | |
| NGS | 0.54 (0.03 to 1.78) | 0.96 (0.05 to 1.95) | 0.50 (0.03 to 0.98) | |
| CGH | 1.11 (0.07 to 1.96) | 0.83 (0.05 to 1.93) | 0.45 (0.02 to 0.97) | |
CGH: comparative genomic hybridisation; CrI: credible interval; LOH: loss of heterozygosity; NGS: next‐generation sequencing; PCR: polymerase chain reaction; SNP: single nucleotide polymorphism.
Table A10.2. Standard deviations and between‐study correlation from analyses using PCR as the reference standard
| Standard deviation of logit(sensitivity) (95% CrI) | Standard deviation of logit(1 – specificity) (95% CrI) | Between‐study correlation (95% CrI) | ||
| Main analysis (shared parameters) | 1.66 (1.09 to 1.98) | 1.27 (0.69 to 1.93) | 0.23 (0.01 to 0.66) | |
| Sensitivity analysis | FISH | 1.71 (1.14 to 1.99) | 1.43 (0.78 to 1.97) | 0.22 (0.01 to 0.65) |
| CGH | 1.01 (0.08 to 1.94) | 0.73 (0.03 to 1.91) | 0.49 (0.02 to 0.97) | |
| aCGH | 0.97 (0.04 to 1.95) | 0.81 (0.04 to 1.92) | 0.50 (0.03 to 0.98) | |
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CrI: credible interval; FISH: fluorescence in situ hybridisation.
Data
Presented below are all the data for all of the tests entered into the review.
Tests. Data tables by test.
| Test | No. of studies | No. of participants |
|---|---|---|
| 1 CISH (against FISH) | 1 | 38 |
| 2 PCR‐based LOH (against FISH) | 15 | 915 |
| 3 Real‐time PCR (against FISH) | 2 | 40 |
| 4 MLPA (against FISH) | 2 | 33 |
| 5 CGH (against FISH) | 4 | 75 |
| 6 aCGH (against FISH) | 3 | 39 |
| 7 SNP array (against FISH) | 6 | 111 |
| 8 NGS (against FISH) | 6 | 243 |
| 9 MS (against FISH) | 1 | 10 |
| 10 NanoString (against FISH) | 1 | 16 |
| 11 FISH (against PCR‐based LOH) | 15 | 915 |
| 12 Real‐time PCR (against PCR‐based LOH) | 1 | 10 |
| 13 MLPA (against PCR‐based LOH) | 1 | 18 |
| 14 CGH (against PCR‐based LOH) | 6 | 151 |
| 15 aCGH (against PCR‐based LOH) | 4 | 57 |
| 16 SNP array (against PCR‐based LOH) | 2 | 33 |
| 17 NGS (against PCR‐based LOH) | 1 | 49 |
| 18 MS (against PCR‐based LOH) | 1 | 50 |
| 19 G‐banding (against PCR‐based LOH) | 1 | 21 |
| 20 CGH (against MLPA) | 1 | 71 |
| 21 Methylation array (against MLPA) | 1 | 99 |
| 22 G‐banding (against CGH) | 3 | 75 |
| 23 G‐banding (against RFLP) | 2 | 27 |
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Ariza 2010.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Not explicitly reported. Included a series of 69 astrocytomas and 10 oligodendrogliomas (although data could not be extracted for the 69 astrocytomas). Prior testing Not explicitly reported but presumably histopathological diagnosis, as a series of 69 astrocytomas and 10 oligodendrogliomas was included. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 10 Country: Spain Population source and setting: NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: PCR‐based LOH and real‐time PCR PCR‐based LOH Tumour sample type: NR Region(s) analysed: NR Cut‐off: NR Real‐time PCR Tumour sample type: NR Region(s) analysed: 1p (SPAG17, ATG4), 19q (DPY19L3, RPS9), 1q (PYGO2 and GREM2), 19p (COPE, FUT3) Cut‐off: NR Additional details: semiquantitative real‐time PCR, quote: "Semiquantitative real‐time PCR of telomeric and centromeric sequences on 1 p (SPAG17, ATG4), 19q (DPY19L3, RPS9), 1q (PYGO2 and GREM2), and 19p (COPE, FUT3) … the deltadeltaCt method was used for relative quantification of PCR products". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | Conference abstract | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Armanious 2017.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: FFPE specimens with histological diagnosis by a neuropathologist and confirmed FISH results. Prior testing Histopathological diagnosis and FISH |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 16 Country: Canada Population source and setting: NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and NanoString FISH Tumour sample type: NR Region(s) analysed: NR Cut‐off: NR NanoString Tumour sample type: FFPE Region(s) analysed: NR Cut‐off: NR Additional details: quote: "Nanostring nCounter CNV assay … Samples were run on the nCounter CNV assay and analyzed by the nSolver software". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | Conference abstract | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (NanoString) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Belaud‐Rotureau 2006.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: grade II or III glioma, sufficient frozen material for FISH analysis and histological control of frozen sections performed during the imprint procedure. Prior testing Examination of haematoxylin and eosin‐stained paraffin sections. Glial‐fibrillary acid‐protein and MIB‐1 immunostaining. Classified and graded according to WHO 2000. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 23 Country: France Population source and setting: Neurosurgery Department of the University Hospital Centre of Bordeaux, France. July 1995 to September 2002 Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: unclear. Previously untreated glioma |
||
| Index tests |
3 tests: FISH (variant 1), FISH (variant 2) and FISH (variant 3) FISH (variant 1) Tumour sample type: touch imprints of frozen tumours Region(s) analysed: 1p36.3 (D1Z2)/1q12 (D1Z1), 19q13.3/19pter Cut‐off: quote: "For each chromosome probe mix, hybridization signals of control and test probes were counted per nucleus, which was classified as follows: (1) In case of deletion, the ratio of control and test probes was 2/1, partially in conjunction with 4/2, 3/1, 4/1 ratios. (2) An imbalance hybridization pattern was associated to a disproportion of the ratio of control and test probes signals (3/2, 4/3, 5/3, etc.). Such pattern does not prove an LOH, which should be further determined by ancillary techniques. (3) A normal pattern (no deletion, no imbalance) was associated to an equal ratio of control and test probes signals (2/2, 4/4). At least 200 tumour cell nuclei were assessed. The cut‐off values, i.e. the percentage of deleted patterns required to assess a deletion for each 1p36 or 19q13 probes were determined to be the mean + 3 SD of the percentage of deleted nuclei on control tissues (reactive lymphadenitis, n=5) [45]. A tumour was classified as deleted if the percentage (%) of deleted nuclei exceeded the cut‐off value of the probe set. In the other cases, it was classified as (1) normal if the percentage of deleted plus imbalanced nuclei was less than the cutoff or (2) imbalanced if the percentage of imbalanced nuclei or the sum of imbalanced plus deleted nuclei was greater than or equal to the cut‐off". Comment: cut‐off determined to be 10%. Additional details: manual analysis with 1p36.3 (D1Z2)/1q12 (D1Z1) and 19q13.3/19pter probe set FISH (variant 2) Tumour sample type: touch imprints of frozen tumours Region(s) analysed: 1p36/1q25, 19q13/19p13 (Abbott Vysis) Cut‐off: quote: "For each chromosome probe mix, hybridization signals of control and test probes were counted per nucleus, which was classified as follows: (1) In case of deletion, the ratio of control and test probes was 2/1, partially in conjunction with 4/2, 3/1, 4/1 ratios. (2) An imbalance hybridization pattern was associated to a disproportion of the ratio of control and test probes signals (3/2, 4/3, 5/3, etc.). Such pattern does not prove an LOH, which should be further determined by ancillary techniques. (3) A normal pattern (no deletion, no imbalance) was associated to an equal ratio of control and test probes signals (2/2, 4/4). At least 200 tumour cell nuclei were assessed. The cut‐off values, i.e. the percentage of deleted patterns required to assess a deletion for each 1p36 or 19q13 probes were determined to be the mean + 3 SD of the percentage of deleted nuclei on control tissues (reactive lymphadenitis, n=5) [45]. A tumour was classified as deleted if the percentage (%) of deleted nuclei exceeded the cut‐off value of the probe set. In the other cases, it was classified as (1) normal if the percentage of deleted plus imbalanced nuclei was less than the cut‐off or (2) imbalanced if the percentage of imbalanced nuclei or the sum of imbalanced plus deleted nuclei was greater than or equal to the cut‐off". Comment: cut‐off determined to be 6%. Additional details: manual analysis with the 1p36/1q25 and 19q13/19p13 Abbott Vysis probe set. FISH (variant 3) Tumour sample type: touch imprints of frozen tumours Region(s) analysed: 1p36/1q25, 19q13/19p13 (Abbott Vysis) Cut‐off: quote: "For each chromosome probe mix, hybridization signals of control and test probes were counted per nucleus, which was classified as follows: (1) In case of deletion, the ratio of control and test probes was 2/1, partially in conjunction with 4/2, 3/1, 4/1 ratios. (2) An imbalance hybridization pattern was associated to a disproportion of the ratio of control and test probes signals (3/2, 4/3, 5/3, etc.). Such pattern does not prove an LOH, which should be further determined by ancillary techniques. (3) A normal pattern (no deletion, no imbalance) was associated to an equal ratio of control and test probes signals (2/2, 4/4). At least 200 tumour cell nuclei were assessed. The cut‐off values, i.e. the percentage of deleted patterns required to assess a deletion for each 1p36 or 19q13 probes were determined to be the mean + 3 SD of the percentage of deleted nuclei on control tissues (reactive lymphadenitis, n=5) [45]. A tumour was classified as deleted if the percentage (%) of deleted nuclei exceeded the cut‐off value of the probe set. In the other cases, it was classified as (1) normal if the percentage of deleted plus imbalanced nuclei was less than the cut‐off or (2) imbalanced if the percentage of imbalanced nuclei or the sum of imbalanced plus deleted nuclei was greater than or equal to the cut‐off". Comment: cut‐off determined to be 6%. Additional details: automatic analysis (Metafer 4, Metasystems, Althlussheim, Germany) with the 1p36/1q25 and 19q13/19p13 Abbott Vysis probe se |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | All tests were performed on frozen tissue samples. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Bigner 1999.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: tumours composed partially or completely of neoplastic oligodendroglia (only cases on which there was agreement between 2 or 3 of the 3 observers for histological classification were included); frozen tissue and normal peripheral lymphocytes available. Prior testing Histopathological classification |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 53 Country: USA Population source and setting: NR Age: mean: 39.9 years, standard deviation: 11.8 years Gender: 64.2% male Karnofsky performance status: NR First diagnosis/recurrent disease: unclear. We excluded the recurrences for people whose primary tumour was also included in the study. Some additional participants had had prior surgery or RT, unclear if the tumours studied were recurrences. |
||
| Index tests |
2 tests: CGH and PCR‐based LOH CGH Tumour sample type: frozen Region(s) analysed: genome wide Cut‐off: ratio of 0.85 indicated a loss. PCR Tumour sample type: frozen Region(s) analysed: FGR, MYCL1, AMY2B (1p); D19S217, D19S112, D19S412, STD, D19S596, D19S180, D19S254, D19S218 (19q) Cut‐off: a reduction in intensity > 50% in the tumour lane compared to the corresponding blood lane was scored as LOH. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | Both tests were performed on frozen tumour material. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Blesa 2009.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Not explicitly reported but only oligodendrogliomas and oligoastrocytomas included. Prior testing Histopathological diagnosis according to WHO 2007 classification. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 52 Country: Spain Population source and setting: Virgen de la Salud Hospital (Toledo, Spain), Clinic Hospital (Barcelona, Spain), and Xeral‐Cies Hospital (Vigo, Spain). Time period NR Age: mean: 45.8 years, standard deviation: 12.5 years Gender: 55.8% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
4 tests: aCGH, FISH, MLPA and PCR aCGH Tumour sample type: fresh‐frozen Region(s) analysed: whole genome Cut‐off: quote: "Genomic imbalances were determined on the basis of the log2 of the Cy3/Cy5 ratios of the average of 3 clone replicates, and regions were considered to have a gain or loss of DNA if at least 2 consecutive clones exceeded the +/‐0.25 threshold". Additional details: used a (quote) "whole human genome CGH array developed in collaboration with Dr. Klaas Kok at the University Medical Centre Groningen (Department of Genetics, Groningen, The Netherlands)". FISH Tumour sample type: FFPE Region(s) analysed: 1p36/1q25, 19p13/19q13 (Vysis Inc., Downers Grove, Illinois, USA) Cut‐off: quote: "Tumors were considered as having loss when an unbalanced 1p/1q (1 vs 2 signals) or 19p/19q (2 vs 1 signal) was identified in more than 25% of tumor cells. In addition, loss with ploidy was noted when an unbalanced pattern 1p/1q of 2 versus 4 signals or an unbalanced 19p/19q of 4 versus 2 signals was detected". MLPA Tumour sample type: unclear, either FFPE or frozen Region(s) analysed: NR. Salsa kit P088, MRC‐Holland, Amsterdam, the Netherlands Cut‐off: losses scored when ≥ 3 test probes exhibited a ratio < 0.7. Additional details: Salsa kit P088, MRC‐Holland, Amsterdam, the Netherlands PCR Tumour sample type: fresh‐frozen or FFPE Region(s) analysed: 1p: D1S199, D1S186, D1S162, D1S312, D1S226; 19q: D19S918, D19S112, D19S206 Cut‐off: LOH pattern, when the shorter allele presented a relatively high peak and the longer allele a low peak (never > 12% of the height of the shorter allele), or when the intensity of the shorter allele was less than that of the longer allele. When all markers were homozygous, LOH was scored as positive. Additional details: PCR‐based LOH (microsatellite) without the need for comparison to normal DNA. References Hatanpaa 2003a. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Bouvier 2004.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. The study included 35 patients operated on or biopsied at the Department of Neurosurgery (la Timone hospital, Marseille) between June and December 2001. This included (quote) "one grade III astrocytoma, 14 glioblastomas, 10 oligodendrogliomas (five grades II and III) and four mixed oligoastrocytomas (three grade II and one grade III). One oligodendroglioma grade II was reclassified as gliomatosis after neuroimaging review. Five cortectomies for epilepsy were used as controls". Prior testing Histopathological diagnosis according to the WHO classification (version not specified). |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 14 Country: France Population source and setting: Department of Neurosurgery, la Timone hospital, Marseille, France. June to December 2001 Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and PCR FISH Tumour sample type: frozen smear Region(s) analysed: 1p36.33 vs 1q12; 19q13.3 vs 19p13.2 Cut‐off: NR PCR Tumour sample type: frozen Region(s) analysed: 1p36.23 (D1S1612), 1p34.2 (D1S447), 1p13.3 (D1S252) and on 19q13.32 (D19S412 and D19S219), 19p13.12 (D19S226). Cut‐off: LOH was scored when signal intensity was < 0.5 or > 2 in a tumour sample. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Broholm 2008.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. Quote: "46 patients were included in the study. The material consisted of 10 oligodendrogliomas (5 WHO Grade II (OII) and 5 Grade III (AOIII)), 10 mixed oligoastrocytomas (5 WHO Grade II (OAII) and 5 Grade III (AOAIII)), 10 astrocytomas (5 WHO Grade II (AII) and 5 Grade III (AAIII)) and 11 glioblastomas, WHO Grade IV (GBMIV)". Prior testing Histopathological diagnosis according to WHO 2000 classification. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 38 Country: Denmark Population source and setting: FFPE tissue from the Laboratory of Neuropathology, Rigshospitalet, Copenhagen, Denmark. Time period NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and PCR FISH Tumour sample type: FFPE Region(s) analysed: 1p36.33 and 19q13 (Vysis #40218 and #38967, Vysis, Des Plaines, Illinois, USA) Cut‐off: quote: "The FISH‐sum in % was calculated (amount of cells with only one or none fluorescence signal for the investigated probe in relation to the total cell count). The conclusion – loss (Pos.) or no loss (N) was noted. For 1p loss the FISH‐sum in % had to be higher than the calculated cut‐off level at 43.24% and for 19q more than 55.12%". PCR Tumour sample type: FFPE Region(s) analysed: D1S164 (1p34.4), D1S496 (1p34.4), D1S199 (1p36.1), D1S468 (1p36.3), D1S2736 (1p36.3); D19S867 (19q13.3), D19S888 (19q13.4), D19S572 (19q13.4), D19S210 (19q13.4) Cut‐off: allelic ratio < 0.5 or > 1.65 Additional details: PCR‐based LOH (microsatellite) without the need for comparison to normal DNA. References Hatanpaa 2003a. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Burger 2001.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. The study included 18 cases of classic and borderline examples of infiltrating gliomas of oligodendroglioma, mixed gliomas (oligoastrocytomas), fibrillary astrocytomas of varying grade, and difficult‐to‐classify intermediate lesions (note, 1 glioma was included in Hatanpaa 2003b and is extracted as part of that study). Prior testing Initial histopathological diagnosis, followed by review and diagnosis according to WHO 2000 classification (independent of the results of CGH, FISH and LOH microsatellite analysis). |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 17 Country: USA Population source and setting: NR Age: mean: 39.5 years, standard deviation: 12.7 years Gender: 41.2% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
3 tests: CGH, FISH and PCR CGH Tumour sample type: FFPE (apart from 1 case, where fresh tissue was available) Region(s) analysed: genome wide Cut‐off: scored as loss if the relative loss < 0.8 FISH Tumour sample type: FFPE (apart from 1 case, where fresh tissue was available) Region(s) analysed: 1p36, 1q24, 19p13.1, 19q13.1‐q13.2, 19q13.3 Cut‐off: NR PCR Tumour sample type: FFPE (apart from 1 case, where fresh tissue was available) Region(s) analysed: 1p: D1S226, D1S312, D1S162, D1S186, D1S199, D1S243; 19p: D19S206, D19S412, D19S112, D19S197, D19S400, D19S422, D19S570 Cut‐off: NR. Overall results for 1p and 19q not given. We assumed that if results were homozygous or indeterminant at all loci examined that codeletion was present. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | No | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Byeon 2014.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: treated for rhabdoid glioblastoma Prior testing Histopathological diagnosis. Electron microscopy |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 3 Country: Republic of Korea Population source and setting: Seoul National University Hospital, Yonsei Severance Hospital, and Soon Cheon Yang University Bucheon Hospital, Republic of Korea. 2004–2011 Agea: mean: 35.6 years, standard deviation: NR; range: 20–45 years Gendera: 20.0% male Karnofsky performance status: NR aFor whole population: 5 participants included in the study, only 3 were tested with aCGH and FISH. |
||
| Index tests |
2 tests: aCGH and FISH aCGH Tumour sample type: FFPE Region(s) analysed: genome wide Cut‐off: NR Additional details: used a (quote) "MacArray Karyo (Macrogen, Seoul, South Korea), which consisted of 4365 human bacterial artificial chromosome clones". FISH Tumour sample type: NR Region(s) analysed: Vysis probes (Abbott Laboratories, Abbott Park, Illinois, USA). 1p36 and 19q13 (from DOI: 10.1593/tlo.12328, reference 12 in the paper). Cut‐off: < 0.8 (from doi.org/10.1111/j.1440‐1789.2006.00735.x, which is referenced by reference 12 in the paper). |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Chaturbedi 2012.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. Quote: "We included DNA samples from 44 OTs, 9 paired blood lymphocytes collected from patients during surgical resection of their gliomas, and 14 glioblastoma multiformes (GBMs) in this study. Breaking down of 44 OT samples included in analysis are 21 WHO grade II oligodendroglioma (OG), 8 WHO Grade II mixed oligo‐astrocytoma (OA), 15 either WHO grade III anaplastic oligodendroglioma (AO) or WHO grade III anaplastic mixed oligo‐astrocytoma (AOA)". Prior testing Histopathological diagnosis |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 18 Country: USA Population source and setting: University of California, Irvine, USA and University of Arkansas for Medical Sciences, USA. Time period NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and real‐time PCR FISH Tumour sample type: FFPE Region(s) analysed: 1p36/1q24, 19q13/19p13 Cut‐off: quote: "A normal ratio is considered 1.0 and any ratio <0.80 is considered deletion of the region of interest" Real‐time PCR Tumour sample type: frozen Region(s) analysed: 1p: E2F2 (1p36) and NOTCH2 (1p13‐11) (also looked at CAMTA1 (1p36.31‐p36.23), but then excluded). 19q: PLAUR (19q13). Reference genes ERC2 (3p14.3), SPAG16 (2q34) and SPOCK1 (5q31). However: (quote) "A ratio of 1:1 between selected marker and reference genes in autosomal chromosomes is expected in normal cells while changes in this ratio in tumor DNA would suggest CNV, either deletion or amplification, in the studied gene of interest. Considering the inherent genome instability of cancer cells, we analyzed the stability of three reference genes in tumor samples and found amplification of SPAG16 in some OT. To mitigate this, we took the average of two ratios of ERC2 and SPOCK1 for most tumors. For other samples, the two reference gene ratios showing the most concordance were used to take a mean and SD. With consideration of 10%–20% variation inherited with real‐time PCR, the mean values of the marker and reference ratio was taken for determination of deletion (<0.8) or amplification (>1.2), Shown in Table 1, there was a gain at the 1p marker gene CAMTA1 (1p36.31‐23) in both GBM and OT, which were not found in other two 1p marker genes E2F2 (1p36) and NOTCH2 (1p13‐p11). Thus average of these two 1p marker genes ratio to reference gene were taken to determine 1p deletion status (value <0.80 is considered 1p deleted)". Cut‐off: marker/reference < 0.8 per gene. Mean of E2F2 (1p36) and NOTCH2 (1p13‐p11) marker genes ratio to reference gene were taken to determine 1p deletion status (value < 0.80 is considered 1p deleted). Additional details: comparative quantitative PCR |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on samples obtained at the same time. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| If a threshold was used, was it pre‐specified? | No | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Cieply 2004.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. 24 cases of gliomas (11 oligodendrogliomas, 5 mixed tumours, 8 astrocytomas) and 10 cases of non‐neoplastic tissue were analysed. Prior testing Presumably histopathological diagnosis, but not explicitly reported. No other tests reported. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 22 Country: USA Population source and setting: NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and PCR FISH Tumour sample type: FFPE Region(s) analysed: NR. Quote: "For FISH, the Vysis dual‐colour probe sets were utilized with a standard approach". Cut‐off: NR PCR Tumour sample type: FFPE Region(s) analysed: 1p34‐36, 19q13. Quote: "The PCR assay used up to 10 sets of primers for short tandem repeats that localize to 1p34‐36 and 19q13". Cut‐off: NR |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | Conference abstract | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Clark 2013.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: glioblastoma cases in the Hillman Cancer registry. Exclusion criteria: cases of recurrent or treated (or both) glioma. Prior testing Histopathological diagnosis (WHO 2007 classification). |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 446 Country: USA Population source and setting: Hillman Cancer Registry, University of Pittsburgh, USA. 2002–2010 Agea: median: 63 years, interquartile range: NR; range: 18–89 years Gendera: 58.5% male Karnofsky performance status: NR First diagnosis/recurrent disease: 100% first diagnosis. Cases of recurrent glioma were excluded. aFor whole population: there were 532 cases in the complete glioblastoma cohort, 491 had upfront 1p/19q testing, 446 had results for both tests. |
||
| Index tests |
2 tests: FISH and PCR FISH Tumour sample type: FFPE Region(s) analysed: 1p36/1q25, 19q13/19p36 (Abbott Molecular, Des Plaines, Illinois, USA). Cut‐off: quote: "Codeletion was counted if the 1p36/1q25 and 19q13/19p13 ratios were both below 0.87 and at least 20% of tumour nuclei showed relative deletion". PCR Tumour sample type: FFPE Region(s) analysed: D1S1172, D1S226, D1S162, D1S1161, D1S199, D1S407, D1S171, D19S112, D19S206, D19S559 (from 2007 onwards, comprising 75% of the total cohort) Cut‐off: quote: "To be considered codeleted the majority of informative microsatellite loci on both 1p and 19q had to show LOH". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. |
||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Cowell 2004.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. However, (quote) "To assess the usefulness of CGHa for the identification of LOH events in low‐grade brain tumors, we used DNA from a series of 3 different tumor subtypes whose LOH status had previously been determined using microsatellite markers (23). The tumors had been grouped according to histopathological diagnosis and divided into low‐grade oligodendrogliomas (LGO), anaplastic oligodendrogliomas (AO), and mixed oligoastrocytomas (MOA). From the original series of tumors classified using microsatellite markers, we analyzed representative samples that consisted of 6 LGOs, 5 AOs, and 3 MOAs. In each of these groups were examples of tumors where there was clear presence or absence of LOH for at least one of the microsatellite markers on each of the 1p and 19q chromosome arms". Prior testing Presumably histopathological diagnosis, although not explicitly reported. To be included in this study LOH status had to have previously been determined using microsatellite markers (PCR‐based LOH). |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 14 Country: USA Population source and setting: Cleveland Clinic Foundation Department of Neurosurgery. Time period NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: aCGH and PCR aCGH Tumour sample type: frozen Region(s) analysed: genome wide. Quote: "A genome‐wide resource of ~6,000 FISH‐mapped, gene/marker content‐verified, sequenced BAC clones (22) from the RPCI‐11 human BAC library are represented as immobilized DNA targets on glass slides for array‐based CGH analysis, as previously described (20). Each clone is spotted in triplicate at 280 µm intervals (see http://genomics.roswellpark.org for a complete list of clones)". Cut‐off: quote: "In general, background variation was considered to extend between ratios of 1.2 and 0.8 for diploid tumors on the linear scale and hybridization ratios outside of these values were considered losses or gains of genetic material". PCR Tumour sample type: frozen Region(s) analysed: 1p36 (D1S552, D1S1612, D1S468), 1p31 (D1S551, D1S430), 19q13.4 (D19S254, D19S572) Cut‐off: NR |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
D'Haene 2019.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: quote: "samples were selected according to the availability of NGS results obtained by the Ion AmpliSeq CHP (v2, Thermo Fisher Scientific, Carlsbad, CA, United States) and the availability of the 1p/19q codeletion status, as determined by FISH". Exclusion criteria: NGS results were excluded if they were low quality. Prior testing Presumably histopathological diagnosis (WHO 2016 classification for glioma samples), although not explicitly reported. Had to have NGS and FISH results. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 50 Country: Belgium Population source and setting: Department of Pathology, Erasme Hospital, Brussels, Belgium. Time period NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and NGS FISH Tumour sample type: FFPE Region(s) analysed: 1p36/1q25 and 19p13/19q13 (Vysis Inc., Downers Grove, Illinois, USA) Cut‐off: quote: "The codeletion was considered to be positive in cases where the ratio of red (1p or 19q) versus green (1q or 19p) signals were lower than 0.8 after scoring at least 50 nuclei". NGS Tumour sample type: FFPE Region(s) analysed: 55 SNPs along the full length of chromosomes 1 and 19 (30 on chromosome 1 and 25 on chromosome 19). Quote: "These SNPs were selected based on the high polymorphic value of their minor allele frequency, as reported on the National Center for Biotechnology Information (NCBI) dbSNP database, as well as from previous reports [24,28]. The average distance between SNPs is 5.2 Mb in 1p, 8.0 Mb in 1q, 1.9 Mb in 19p, and 2.1 Mb in 19q". SNPs: 1p: rs7663, rs169957, rs309481, rs159525, rs157208, rs6425953, rs7315, rs7903, rs504816, rs7374, rs87061, rs11811946, rs5680, rs191142, rs54396, rs106075, rs1132, rs8888, rs6604120, rs8128; 1q: rs2275073, rs347303, rs4575136, rs898114, rs1342566, rs12744553, rs2802849, rs1770214, rs6692892, rs16848862, rs2275073, rs347303, rs4575136, rs898114, rs1342566; 19p: rs13345388, rs7256720, rs36115836, rs164020, rs57167556, rs2114724, rs7246440, rs10419689, rs8107776, rs4808732; 19q: rs7283, rs2542297, rs33841, rs12852, rs1291, rs17628, rs166539, rs3817, rs10113, rs8355, rs11573, rs193040, rs3814, rs10217, rs10448. Cut‐off: quote: "To detect 1p/19q LOH, we firstly applied a quality criterion based on the SNP coverage. The test was considered optimal, suboptimal, or non‐informative, according to the number of SNPs that were covered by fewer than 250 reads (Table 3). Secondly, the allelic frequencies (AF) for each SNP (with more than 250×) were annotated. Homozygous SNPs with the same nucleotide as that of the reference genome will have an AF of approximately 100%, while homozygous SNPs with a nucleotide that differs from the reference genome will have an AF of approximately 0%. Heterozygous SNPs will have an AF of approximately 50%. However, because NGS provides a semi‐quantitative measure based on the number of reads [19], we established the following confidence intervals: 90–100% or 0–10% for homozygous markers, and 40–60% for heterozygous markers. These confidence intervals were defined based on the analysis of 12 nontumor samples. Imbalances of 1p and 19q markers due to LOH were scored when their AFs were outside the established ranges for homozygosity or heterozygosity (i.e., 10–40% or 60–90%) (Figure 1). We defined the criterion for the 1p/19q codeletion as the absence of any heterozygous markers in these chromosomal arms, together with the presence of at least one heterozygous marker in the opposite arm. No codeletion was scored if at least one heterozygous marker was present in 1p or 19q. A whole chromosome arm LOH is observed when there are no heterozygous markers present in either arm". Additional details: custom‐designed glioma NGS‐targeted panel. Sequenced using an Ion Torrent Personal Genome Machine (Thermo Fisher). |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Dahlback 2009.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: primary GBM (including GBM, multifocal GBM, gliosarcoma, giant cell GBM, GBM with granular cell component, GBM with oligodendroglial component). Aged ≥ 16 years at time of surgery. Prior testing Histopathological diagnosis according to the WHO 2007 classification. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 57 Country: Norway Population source and setting: Department of Neurosurgery, Rikshospitalet, Oslo, Norway. January 2005 to January 2008 Age: mean: 61.7 years, standard deviation: 9.4 years Gender: 57.9% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: CGH and G‐banding CGH Tumour sample type: fresh‐frozen Region(s) analysed: genome wide Cut‐off: quote: "Aberrations were scored whenever the case profile and the reference profile did not overlap with a significance level of 99%". Additional details: high‐resolution CGH. Quote: "CGH was performed on DNA from these samples according to the manufacturer's protocol and analyzed according to Kallioniemi et al., (1992) with the modifications described by Kraggerud et al., (2000) and Teixeira et al. (2004). Final evaluations of the CGH results used dynamic standard reference intervals (D‐SRI) as described by Ribeiro et al. (2006)". G‐banding Tumour sample type: fresh Region(s) analysed: genome wide Cut‐off: N/A Additional details: quote: "Chromosome preparations were G‐banded using Wright stain and karyotyped according to the ISCN (2005) guidelines". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Dahlback 2011.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: Grade II glioma; aged ≥ 16 years at time of surgery Prior testing Histopathological diagnosis according to the WHO 2007 classification. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 32 Country: Norway Population source and setting: The Department of Neurosurgery, Oslo University Hospital, Rikshospitalet, Oslo, Norway. January 2005 to October 2008 Age: mean: 38.4 years, standard deviation: 8.7 years Gender: 65.6% male Karnofsky performance status: NR First diagnosis/recurrent disease: 78.1% primary (25/32 participants with results on ≥ 2 tests), 21.9% recurrent (7/32 participants) |
||
| Index tests |
3 tests: CGH, G‐banding and PCR CGH Tumour sample type: fresh‐frozen Region(s) analysed: genome wide Cut‐off: from Dahlback 2009: quote "Aberrations were scored whenever the case profile and the reference profile did not overlap with a significance level of 99%". Additional details: quote: "HR‐CGH [high‐resolution CGH] was performed according to the manufacturer's protocol and analyzed as previously described (Dahlback et al., 2009)". Dahlback 2009: quote: "CGH was performed on DNA from these samples according to the manufacturer's protocol and analyzed according to Kallioniemi et al., (1992) with the modifications described by Kraggerud et al., (2000) and Teixeira et al. (2004). Final evaluations of the CGH results used dynamic standard reference intervals (D‐SRI) as described by Ribeiro et al. (2006)". G‐banding Tumour sample type: fresh Region(s) analysed: whole genome Cut‐off: not applicable Additional details: quote: "Chromosome preparations were G‐banded using Wright stain and karyotyped according to the ISCN (2009)". PCR Tumour sample type: fresh‐frozen Region(s) analysed: at least 4 of 6 microsatellite markers on 1p35‐36 (D1S2660, D1S507, D1S199, D1S2734, D1S1676, D1S247) and 19q13 (D19S918, D19S219, D19S112, D19S412, D19S596, D19S206) used. Cut‐off: As described in Scheie D, Cvancarova M, Mork S, Skullerud K, Andresen PA, Benestad I, Helseth E, Meling T, Beiske K. 2008. Can morphology predict 1p/19q loss in oligodendroglial tumors? Histopathology 53:578‐87: results were defined as LOH‐positive when the peak areas of fluorescent intensity curves, corresponding to PCR products from individual primer sets, showed a relative reduction of ≥ 40% when the products from tumour DNA were compared with those from normal DNA. Unclear how many markers had to display LOH. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Dubbink 2016.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria for EORTC 26951: quote: "Patients were eligible for this study if they had been diagnosed by the local pathologist with an anaplastic oligodendroglioma or anaplastic mixed oligoastrocytoma with at least 25% oligodendroglial elements; had at least three of five anaplastic characteristics (high cellularity, mitosis, nuclear abnormalities, endothelial proliferation, and necrosis); were between 16 and 70 years old; had an Eastern Cooperative Oncology Group (ECOG) performance status (PS) of 0 to 2; had provided written informed consent; had not undergone prior chemotherapy or RT to the skull; had no diseases interfering with follow‐up; and had adequate hematologic, renal, and hepatic function (WBC [white blood cell] count ≥ 3.0 × 109/L, platelets ≥ 100 × 109/L, serum creatinine < 120 μmol/L, and serum bilirubin < 25 μmol/L)". How participants were selected for this study was NR. Prior testing Histopathological diagnosis. For participants with sufficient tissue to assess 1p and 19q status, this was performed by FISH in EORTC 26951. FISH results for the participants included in this study were NR. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 49 Country: Austria, Belgium, Finland, France, Germany, Hungary, Italy, the Netherlands, Sweden, UK Population source and setting: quote "Forty‐nine glioma tissues were collected between 1997 and 2003 during the European Organization of Research and Treatment of Cancer study 26951 on adjuvant procarbazine, lomustine, and vincristine chemotherapy of anaplastic oligodendrogliomas and anaplastic oligoastrocytomas". Agea*: median: 48.6 years in the RT plus PCV arm; 49.8 years in the RT arm, interquartile range: NR; range: 18.6–68.7 years in the RT plus PCV arm; 19.2–68.7 years in the RT only arm. Gendera: 57.6% male Karnofsky performance status: NR First diagnosis/recurrent disease: unclear. Patients with "newly diagnosed" anaplastic oligodendrogliomas or anaplastic oligoastrocytomas recruited into the trial. However, some had had previous resections for lower‐grade tumours. aFor whole population: results are for the 368 patients included in EORTC 26951, only 49 samples included in this study. |
||
| Index tests |
2 tests: NGS and PCR‐based LOH NGS Tumour sample type: FFPE Region(s) analysed: chromosome 1p: SNP rs7663, position 16112795; SNP rs169957, position 19683301; SNP rs169885, position 21628545; SNP rs742358, position 22459170; SNP rs309481, position 23210600; SNP rs189882, position 24868045; SNP rs9259, position 25168124; SNP rs7491, position 25895238; SNP rs159525, position 26213991; SNP rs7504, position 27238150; SNP rs6564, position 28212975; SNP rs157208, position 29245406; SNP rs6425953, position 36168038; SNP rs7686, position 38268918; SNP rs7315, position 40306898; SNP rs7903, position 45976472; SNP rs504816, position 53307957; SNP rs7374, position 55316322; SNP rs87061, position 60594980; SNP rs11811946, position 65952428; SNP rs5680, position 71477315; SNP rs191142, position 76990862; SNP rs12754569, position 85462971; SNP rs54396, position 88776278; SNP rs106075, position 91604522, SNP rs1132, position 95394352; SNP rs8888, position 101338324; SNP rs6604120, position 109289487; SNP rs8128, position 115110683; Chromosome 19q: SNP rs7283, position 30106659, SNP rs2542297, position 31883906; SNP rs33841, position 34011248; SNP rs12852, position 35615179; SNP rs1291, position 38229378; SNP rs17628, position 39926509; SNP rs166539, position 40931717; SNP rs3817, position 44090195; SNP rs10113, position 47112648; SNP rs8355, position 48833800; SNP rs6521, position 49519873; SNP rs11573, position 51359497; SNP rs193040, position 53073605; SNP rs3814, position 53611187; SNP rs10217, position 56030428; SNP rs10448, position 59093239 Cut‐off: quote: "A SNP was considered to be imbalanced or relatively lost when the variant B‐allele frequency of a heterozygous SNP was either higher than 55% or lower than 45%. All variant frequencies between 45% and 55% were considered not to be aberrant. Similarly, cut‐off lines were indicated at 5% and 95%, if not otherwise stated … Typical oligodendroglial co‐deletion of 1p and 19q was defined as equivalent of all informative SNP on both chromosomal arms". Additional details: quote: "A custom primer panel was designed that includes SNPs on chromosomes 1p and 19q using the Ion AmpliSeq Designer 2.0 (ThermoFisher Scientific Inc.).23 Highly polymorphic SNPs on both chromosomes were selected via the NCBI SNP database (http://www.ncbi.nlm.nih.gov/SNP, last accessed September 17, 2013) with a global minor allele frequency of at least 45% to obtain a high number of informative SNPs in each assay. The mean SNP density for chromosomes 1p and 19q was set arbitrarily to approximately 1SNP per 3.5Mb and 1 SNP per 2 Mb, respectively, yielding a total of 29 SNPs on chromosome 1p and 16 SNPs on chromosome 19q that covered the entire chromosomal arms (Figure 1A). Selected SNPs and their chromosomal localization (SNP database 138) are shown in Table 2. Next‐generation targeted sequencing was performed by semiconductor sequencing with the Ion Torrent Personal Genome Machine". PCR Tumour sample type: FFPE Region(s) analysed: D1S199 (locus: 1p36.13), D1S513 (1p35.2), D1S197 (1p32.3), D1S2806 (1p31.3), D1S495 (1p21.1), D19S875 (19q12), D19S198 (19q13.2), D19S412 (19q13.32), D19S606 (19q13.32), D19S572 (19q13.42) Cut‐off: quote: "Allelic losses were assessed based on the analysis of multiple informative markers, as described elsewhere.10 Typical oligodendroglial co‐deletion of 1p and 19q was defined as equivalent imbalance of all informative SNPs on both chromosomal arms. If not all informative markers were lost, a chromosome was considered partially lost". Additional details: PCR without the need for comparison to normal DNA: reference Hatanpaa 2003a, and state no normal tissue available. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Duval 2014.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: oligodendrogliomas with FFPE tissue with previously established 1p/19q status by FISH. Prior testing Histopathological diagnosis according to WHO 2007 classification. Previously established 1p/19q status by FISH. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 36 Country: Canada Population source and setting: NR Age: median: 55 years, interquartile range: NR; range: 26–82 years Gender: 38.9% male Karnofsky performance status: NR First diagnosis/recurrent disease: 16.7% recurrent tumours |
||
| Index tests |
4 tests: FISH (variant 1), FISH (variant 2), FISH (variant 3) and FISH (variant 4) FISH (variant 1) Tumour sample type: FFPE Region(s) analysed: 1p36/19q13 Dual‐Color Probe kit (Abbott Molecular, Abbott Park, Illinois, USA) Cut‐off: combination cut‐off: quote: "The cut‐off of nuclei that had to show deletion was calculated on a series of 10 non‐neoplastic brain tissue samples (from epilepsy surgery cases and autopsy brains). This cut‐off was calculated using mean +3 SD and was set at 50% for both 1p and 19q. Cases above the cutoff were considered deleted and those under the cut‐off were considered normal or imbalanced according to the literature guidelines [43,44]". FISH (variant 2) Tumour sample type: FFPE Region(s) analysed: 1p36/19q13 Dual‐Color Probe kit (Abbott Molecular, Abbott Park, Illinois, USA) Cut‐off: ratio cut‐off: ratio ≤ 0.8 was considered to indicate a deletion; quote: "For each case the signal ratio of red signals to green signals per cell was also established. A ratio ≤0.8 was considered to indicate a deletion whereas a ratio between 0.8 and 1.1 was considered to indicate a normal status on the chromosomal arm. A ratio over 1.1 was considered to indicate polysomy and was classified in the imbalanced status subgroup [2,30]". FISH (variant 3) Tumour sample type: FFPE Region(s) analysed: 1p36/19q13 Dual‐Color Probe kit (Abbott Molecular, Abbott Park, Illinois, USA) Cut‐off: combination cut‐off: quote: "cut‐off at the median value of our tumor series which corresponds to a value of 65% for both 1p and 19q". Additional details: ImmunoFISH. IHC performed against Ki67 (MIB‐1). FISH (variant 4) Tumour sample type: FFPE Region(s) analysed: 1p36/19q13 Dual‐Color Probe kit (Abbott Molecular, Abbott Park, Illinois, USA) Cut‐off: ratio cut‐off: ratio ≤ 0.8 was considered to indicate a deletion; quote: "For the ratio method, established values were the same as for the FISH". Additional details: ImmunoFISH. IHC performed against Ki67 (MIB‐1). |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Duval 2015.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR Prior testing Not explicitly reported but presumably histopathological diagnosis |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 29 Country: France Population source and setting: Centre Hospitalier Universitaire de Rennes (France). 2010–2015 Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH (variant 1) and FISH (variant 2) FISH (variant 1) Tumour sample type: FFPE Region(s) analysed: 1p36/1q25 and 19q13/19p13 (Dual Color Probe kit, Abbott Molecular, Abbott Park, Illinois, USA). Cut‐off: combination + ratio method. Cases deleted in 1 method and normal in the other were considered deleted. Combination method: deletion status combinations (for 1p and 19q, control/test): 2/0, 2/1, 3/0, 3/1, 4/1, 4/2, 5/2, 6/2, 6/3. Normal status combinations 2/2, 1/2. (Imbalance status combinations 1/3, 1/4, 2/3, 2/4, 2/5, 3/3, 3/4, 4/3, 4/4, 4/5, 5/4, 5/5, 5/6.) The cut‐off value for the number of nuclei that had to show deletion was 55% for both 1p and 19q for deletion status (and 20% for imbalance status). Ratio method: the signal ratio of test:control probes ≤ 0.8 was considered to indicate a deletion. Additional details: automated analysis using the Metafer 4 software (Metasystem) using the "1p19q tile‐sampling classifier". FISH (variant 2) Tumour sample type: FFPE Region(s) analysed: 1p36/1q25 and 19q13/19p13 (Dual Color Probe kit, Abbott Molecular, Abbott Park, Illinois, USA). Cut‐off: combination + ratio method. Cases deleted in 1 method and normal in the other were considered deleted. Combination method: deletion status combinations (for 1p and 19q, control/test): 2/0, 2/1, 3/0, 3/1, 4/1, 4/2, 5/2, 6/2, 6/3. Normal status combinations 2/2, 1/2. (Imbalance status combinations 1/3, 1/4, 2/3, 2/4, 2/5, 3/3, 3/4, 4/3, 4/4, 4/5, 5/4, 5/5, 5/6.) The cut‐off value for the number of nuclei that had to show deletion was 55% for both 1p and 19q for deletion status (and 20% for imbalance status). Ratio method: the signal ratio of test:control probes ≤ 0.8 was considered to indicate a deletion. Additional details: manual analysis |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Gadji 2009.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: initially diagnosed oligodendroglial or oligoastrocytomal brain tumour Prior testing Histopathological diagnosis: tumours were reviewed and classified according to the WHO 2007 classification. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 11 Country: Canada Population source and setting: Sherbrooke University hospital, Canada. 1993–2007 Age: mean: 38.8 years, standard deviation: 8.3 years Gender*: 35.1% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR *For whole population: 37 participants were included, but only 11 had both tests. |
||
| Index tests |
2 tests: FISH and PCR FISH Tumour sample type: FFPE or fresh (touch‐preparation smear) Region(s) analysed: 1p36/1q25, 19p13/19q13 (Vysis LSI; Abbott Molecular, Des Plaines, Illinois, USA) Cut‐off: quote: "A case was considered deleted with 1p, 19q, or both when the scored nuclei displayed an imbalance between green and red signals". PCR Tumour sample type: NR Region(s) analysed: chromosome 1 (8 test markers on the short arm and 2 control markers on the long arm): D1S2795 (1p36.31), D1S2666 (1p36.23), D1S244 (1p36.22), D1S2676 (1p34.3), D1S2729 (1p34.3), D1S2722 (1p34.2), D1S508 (1p36.31~p36.21), D1S2734 (1p35.36), D1S252 (1q21), D1S2346 (1q22); chromosome 19 (9 test markers on the long arm and 1 control marker on the short arm): D19S412 (19q13.32), D19S559 (19q13.32), D19S200 (19q13.2), D19S397 (19q13.14), D19S422 (19q13.13), D19S425 (19q13.12), D19S416 (19q13.11), D19S112 (19q13.3), D19S556 (19p13.13). All markers were obtained from IDT (Coralville, Iowa, USA). Cut‐off: quote: "An absence or a significant reduction in intensity of >50% in the tumor lane compared with the corresponding blood and saliva lanes was scored as LOH". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Ghasimi 2016.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: availability of FFPE tissue. No other criteria were reported. This study included 33 grade II–III gliomas and 58 glioblastomas. Prior testing Presumably histopathological diagnosis, although not explicitly reported. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 55 Country: Sweden Population source and setting: Umeå University Hospital, Sweden. Time period NR Agea: median: 58 years, interquartile range: NR; range: 15–80 years Gendera: 59.3% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR aFor whole population: the data are for the 59 participants who had SNP array data, of whom 55 also had FISH data. |
||
| Index tests |
2 tests: FISH and SNP array FISH Tumour sample type: FFPE Region(s) analysed: 1p36/1q13, 19p13/19q13 (Vysis, Illinois, USA) Cut‐off: 1p36/1q25 ratios < 0.88 and 19q13/19p13 ratios < 0.74 in > 12% of the cells were considered as deleted. SNP array Tumour sample type: FFPE Region(s) analysed: regions corresponding to the location of the FISH probes Cut‐off: quote: "For comparison between FISH and ASCAT, we extracted the median total copy number from the ASCAT profiles for the genomic regions corresponding to the FISH probes. These copy number data were subsequently used to mimic the sample classification based on FISH data, by calculating the same ratios and using the same cutoff values that had been used for classification by FISH". Additional details: "Illumina HumanOmni1‐Quad BeadChips. The ASCAT algorithm [26] (version 2.0) was used to calculate somatic whole‐genome allele‐specific copy number profiles (ASCAT‐profiles), as well as estimates of tumor cell content and tumor cell ploidy". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Harada 2011.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: samples submitted for routine clinical analysis of 1p/19q loss. Prior testing Not explicitly reported, but presumably histopathological diagnosis. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 29 Country: USA Population source and setting: Johns Hopkins Medical Institutions. 2010 Agea: mean: 42.7 years (48.3 years in participants with oligodendrogliomas, 37.7 years in participants with non‐oligodendroglioma tumours, standard deviation: 15.3 yearsb; range: 14–82 years Gendera: 53.3% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR aIncluded 1 participant with pineal parenchymal tumour of intermediate differentiation, who was excluded. bThe standard error of the mean was 4.47 years in participants with oligodendrogliomas and 3.35 years in participants with non‐oligodendroglioma tumours. The standard deviation was calculated from these figures. |
||
| Index tests |
2 tests: PCR and SNP array PCR Tumour sample type: FFPE Region(s) analysed: chromosome 1: D1S199, D1S186, D1S162, D1S312, D1S226; chromosome 19: D19S918, D19S112, D19S206 Cut‐off: NR Additional details: Multiplex PCR (as described in Hatanpaa 2003a and Hatanpaa 2003b). Fluorescent labelled PCR products were detected by capillary electrophoresis with use of the ABI 3130 Genetic Analyzer and GeneMapper software version 4 (Applied Biosystems, Carlsbad, California, USA). SNP array Tumour sample type: FFPE Region(s) analysed: genome wide Cut‐off: NR Additional details: quote: "Illumina Infinium II SNP array with 300K markers (HumanCytoSNP‐12, Illumina Inc., San Diego, CA)". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Hatanpaa 2003a.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: tested for 1p/19q codeletion by ≥ 2 of the following 3 reference methods with concordant results: CGH, FISH and PCR‐based microsatellite analysis with comparison to normal DNA from the same participant. Prior testing Tested for 1p/19q codeletion by ≥ 2 of the following 3 reference methods with concordant results: CGH, FISH and PCR‐based microsatellite analysis with comparison to normal DNA from the same participant. Appeared that histopathological grading was redone from the original studies. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 10 Country: USA Population source and setting: FFPE glioma specimens with concordant results in Smith 1999 or Burger 2001 (note: we could not match 1 tumour, T117, to tumours described in either study) Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR in this publication. Smith 1999 included both primary and recurrent tumour specimens. |
||
| Index tests |
4 tests: CGH, FISH, PCR (with comparison to normal DNA) and PCR (without comparison to normal DNA) CGH Tumour sample type: NR Region(s) analysed: NR Cut‐off: Smith 1999 references Mohapatra G, Kim DH, Feuerstein BG. Detection of multiple gains and losses of genetic material in ten glioma cell lines by comparative genomic hybridization. Genes, Chromosomes & Cancer 1995;13;86‐93. In this publication: quote: "Definition of CGH ratio thresholds to define ratios that were indicative of changes in DNA copy number, we performed 21 CGH experiments using normal control DNA. We calculated average ratio changes and standard deviations by using the software program cghprofstats. new (Piper et al., 1994). The average ratio for all 21 hybridizations was 0.99 (range 0.9‐1.1). The average standard deviation was 0.04 (range 0.02‐0.06). Taking these findings into consideration, we chose upper and lower ratio thresholds of 1.2 and 0.8, respectively. Any change in ratio in excess of these thresholds was interpreted as indicative of DNA copy number changes only if found in both forward and reverse experiments. Amplifications were defined both by a ratio >2.0 and by visual inspection". Piper J, Rutovitz D, Sudar D, Kallioniemi A, Kallioniemi O, Waldman FM, et al. Computer image analysis of comparative genomic hybridization. Cytometry 1995;19:10‐26 also cited. FISH Tumour sample type: NR Region(s) analysed: from Smith 1999: 1p36, 1q24, 19p13.1, 19q13.1‐q13.2, 19q13.3 Cut‐off: for Smith 1999 we defined codeletion defined as hemizygous deletion of 1p36, 1q13.1‐q13.2 and 19q13.3. What defined hemizygous deletion NR. Also cited Qian J, Bostwick DG, Takahashi S, Borell TJ, Herath JF, Lieber MM et al. Chromosomal anomalies in prostatic intraepithelial neoplasia and carcinoma detected by fluorescence in situ hybridization. Cancer Research 1995;55:5408‐14. In this paper (quote) "abnormal autosomal loss required ≥55% nuclei with zero or one signal". Unclear if this threshold was used. PCR (with comparison to normal DNA) (referred to as PCR‐based LOH below. Note: the risk of bias and applicability judgements for this PCR variant appear inFigure 7) Tumour sample type: NR Region(s) analysed: 1p: D1S468, D1S1612, D1S1597, D1S199, D1S1665, D1S1728, D1S1588, D1S1675, D1S187; 19q: D19S213, D19S569, D19S422. D19S219, SM, S19S112, S19S412, D19S596, HRC, D19S589, D19S218 Cut‐off: For Smith 1999 we defined codeletion as all markers showing confirmed allelic loss, presumed allelic loss, were homozygous or were indeterminant. Additional details: PCR with comparison to normal DNA PCR (without comparison to normal DNA) Tumour sample type: FFPE Region(s) analysed: chromosome 1: D1S162, D1S226, D1S199, D1S186, D1S312; chromosome 19: D19S112, D19S918, D19S206. Cut‐off: presence of 1 allele or LOH pattern A or B at all loci ("LOH pattern A, consisted of a shorter allele (the allele measuring fewer nucleotides in length) with a relatively high peak and a longer allele with a diminutive peak (Fig. 6). The height of the longer allele was never more than 12% of the height of the shorter allele … LOH pattern B, the intensity of the shorter allele was less than that of the longer allele"). Additional details: PCR‐based LOH (microsatellite) without the need for comparison to normal DNA. Quote: "Multiplex PCR amplification of microsatellite loci followed by high‐resolution PCR product sizing by capillary electrophoresis on formalin‐fixed, paraffin‐embedded tissue". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| If a threshold was used, was it pre‐specified? | No | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | No | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Hatanpaa 2003b.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: tested for 1p/19q codeletion by ≥ 2 of the following 3 reference methods with concordant results: CGH, FISH and PCR‐based microsatellite analysis with comparison to normal DNA from the same participant. Prior testing Tested for 1p/19q codeletion by ≥ 2 of the following 3 reference methods with concordant results: CGH, FISH and PCR‐based microsatellite analysis with comparison to normal DNA from the same participant. Appeared that histopathological grading was redone from the original studies. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 9 Country: USA Population source and setting: FFPE glioma specimens with concordant results in Smith 1999 or Burger 2001 (note: we could not match 1 tumour, T117, to tumours described in either study) Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR in this publication. Smith 1999 included both primary and recurrent tumour specimens. |
||
| Index tests |
4 tests: CGH, FISH, PCR (with comparison to normal DNA) and PCR (without comparison to normal DNA) CGH Tumour sample type: NR Region(s) analysed: NR Cut‐off: Smith 1999 references Mohapatra G, Kim DH, Feuerstein BG. Detection of multiple gains and losses of genetic material in ten glioma cell lines by comparative genomic hybridization. Genes, Chromosomes & Cancer 1995;13;86‐93. In this publication: "Definition of CGH ratio thresholds to define ratios that were indicative of changes in DNA copy number, we performed 21 CGH experiments using normal control DNA. We calculated average ratio changes and standard deviations by using the software program cghprofstats.new (Piper et al., 1994). The average ratio for all 21 hybridizations was 0.99 (range 0.9‐1.1). The average standard deviation was 0.04 (range 0.02‐0.06). Taking these findings into consideration, we chose upper and lower ratio thresholds of 1.2 and 0.8, respectively. Any change in ratio in excess of these thresholds was interpreted as indicative of DNA copy number changes only if found in both forward and reverse experiments. Amplifications were defined both by a ratio >2.0 and by visual inspection". Piper J, Rutovitz D, Sudar D, Kallioniemi A, Kallioniemi O, Waldman FM, et al. Computer image analysis of comparative genomic hybridization. Cytometry 1995;19:10‐26 also cited. FISH Tumour sample type: NR Region(s) analysed: from Smith 1999: 1p36, 1q24, 19p13.1, 19q13.1‐q13.2, 19q13.3 Cut‐off: for Smith 1999 we defined codeletion defined as hemizygous deletion of 1p36, 1q13.1‐q13.2 and 19q13.3. What defined hemizygous deletion NR. Also cited Qian J, Bostwick DG, Takahashi S, Borell TJ, Herath JF, Lieber MM et al. Chromosomal anomalies in prostatic intraepithelial neoplasia and carcinoma detected by fluorescence in situ hybridization. Cancer Research 1995;55:5408‐14. In this paper (quote) "abnormal autosomal loss required ≥55% nuclei with zero or one signal". Unclear if this threshold was used. PCR (with comparison to normal DNA) (referred to as PCR‐based LOH below. Note: the risk of bias and applicability judgements for this PCR variant appear inFigure 7) Tumour sample type: NR Region(s) analysed: 1p: D1S468, D1S1612, D1S1597, D1S199, D1S1665, D1S1728, D1S1588, D1S1675, D1S187; 19q: D19S213, D19S569, D19S422. D19S219, SM, S19S112, S19S412, D19S596, HRC, D19S589, D19S218 Cut‐off: for Smith 1999 we defined codeletion as all markers showing confirmed allelic loss, presumed allelic loss, were homozygous or were indeterminant. Additional details: PCR with comparison to normal DNA PCR (without comparison to normal DNA) Tumour sample type: FFPE Region(s) analysed: chromosome 1: D1S162, D1S226, D1S199, D1S186, D1S312; chromosome 19: D19S112, D19S918, D19S206. Cut‐off: presence of 1 allele or LOH pattern A or B at all loci ("LOH pattern A, consisted of a shorter allele (the allele measuring fewer nucleotides in length) with a relatively high peak and a longer allele with a diminutive peak (Fig. 6). The height of the longer allele was never more than 12% of the height of the shorter allele … LOH pattern B, the intensity of the shorter allele was less than that of the longer allele"). Additional details: PCR‐based LOH (microsatellite) without the need for comparison to normal DNA. Quote: "Multiplex PCR amplification of microsatellite loci followed by high‐resolution PCR product sizing by capillary electrophoresis on formalin‐fixed, paraffin‐embedded tissue". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Hinrichs 2016.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: GBM‐O. Cases were selected from the 28 diagnosed cases (quote) "based on availability of tissue and diversity of clinical diagnostic markers". Prior testing Histopathological diagnosis using WHO 2007 classification. FISH for EGFR and 1p/19q. Immunohistochemistry for IDH1. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 8 Country: USA Population source and setting: Emory University Hospitals, USA. 2007–2011 Age: mean: 55.0 years, standard deviation: 17.3 years Gender: 75% male Karnofsky performance status: NR First diagnosis/recurrent disease: 87.5% (7/8) primary tumours, 12.5% (1/8) secondary tumours |
||
| Index tests |
2 tests: FISH and SNP array FISH Tumour sample type: FFPE Region(s) analysed: 1p36/1q25, 19p13/19q13 (Vysis LSI probe sets, Abbott Molecular). From Appin CL, Gao J, Chisolm C, Torian M, Alexis D, Vincentelli C, et al. Glioblastoma with oligodendroglioma component (GBM‐O): molecular genetic and clinical characteristics. Brain Pathology 2013;23:454‐61 (reference 2 in this study). Cut‐off: quote: "1p and 19q deletions were considered present if ≥10% of cells contained the respective deletions". From Appin CL, Gao J, Chisolm C, Torian M, Alexis D, Vincentelli C, et al. Glioblastoma with oligodendroglioma component (GBM‐O): molecular genetic and clinical characteristics. Brain Pathology 2013;23:454‐61 (reference 2 in this study). SNP array Tumour sample type: FFPE Region(s) analysed: genome wide Cut‐off: segmented log2 ratio of −0.135 for losses and −0.45 for homozygous deletions Additional details: quote: "Illumina HumanCytoSNP‐12v2.1‐FFPE SNP arrays … Data were processed and analyzed with BioDiscovery Nexus software (Hawthorne, CA) using SNPRank segmentation". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | |||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Horbinski 2012.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: oligodendrogliomas. Exclusion criteria: recurrent or treated (or both) gliomas; children aged < 18 years Prior testing Histopathological diagnosis according to WHO criteria at the time of initial biopsy. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 111 Country: USA Population source and setting: University of Pittsburgh. 2002–2010 Age: median: Grade II oligodendroglioma: 42 years. Grade III oligodendroglioma: 49 years. Interquartile range: NR; range: 19–80 years (Grade II oligodendroglioma 19–79 years; Grade III oligodendroglioma 25–80 years) Gender: 56.8% male Karnofsky performance status: NR First diagnosis/recurrent disease: first diagnosis (cases of recurrent glioma were excluded) |
||
| Index tests |
3 tests: FISH (variant 1), FISH (variant 2) and PCR FISH (variant 1) (referred to as FISH below. Note: the risk of bias and applicability judgements for this FISH variant appear inFigure 7) Tumour sample type: FFPE Region(s) analysed: 1p36/1q25 19q13 /19p13 (Abbott Molecular, Des Plaines, Illinois, USA) Cut‐off: target‐ploidy control ratio was < 0.87, with ≥ 20% of nuclei showing deletion FISH (variant 2) Tumour sample type: FFPE Region(s) analysed: 1p36/1q25 19q13 /19p13 (Abbott Molecular, Des Plaines, Illinois) Cut‐off: target‐ploidy control ratio was < 0.75, with ≥ 20% of nuclei showing deletion PCR Tumour sample type: FFPE Region(s) analysed: chromosome 1: D1S1172, D1S226, D1S162, D1S1161, D1S199, D1S407, D1S171; chromosome 19: D19S112, D19S206 Cut‐off: at least half of all informative microsatellite loci on both 1p and 19q had to show LOH to be designated as having 1p/19q codeletion. Quote: "When available, patient‐matched germline DNA from a peripheral blood sample was used as a control. When normal tissue was not available, peak height ratios falling outside 2 SDs beyond the mean of previously validated normal values for each polymorphic allele paring were assessed as showing LOH". Additional details: from Horbinski C, Hamilton RL, Nikiforov Y, Pollack IF. Association of molecular alterations, including BRAF, with biology and outcome in pilocytic astrocytomas. Acta Neuropathologica 2010;119:641‐49) "Polymerase chain reaction was performed, and the products were analyzed using capillary gel electrophoresis on GeneMapper ABI 3730 (Applied Biosystems, Foster City, CA)". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| If a threshold was used, was it pre‐specified? | No | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Jeuken 2006.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. Quote: "Eighty‐eight specimens obtained from glioma patients treated in the Department of Neurosurgery of the Radboud University Nijmegen Medical Centre, The Netherlands, were selected". Prior testing Histopathological diagnosis (WHO 2000 classification). 79/88 participants were previously analysed by conventional CGH. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 71 Country: the Netherlands Population source and setting: Department of Neurosurgery of the Radboud University Nijmegen Medical Center, the Netherlands. Time period NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: CGH and MLPA CGH Tumour sample type: snap‐frozen Region(s) analysed: genome wide Cut‐off: 0.8 for losses and 1.2 for gains MLPA Tumour sample type: snap frozen or FFPE Region(s) analysed: 1p: TNFRSF4, GB1, SKII, TP72, PARK7, EPHA8, RUNX3, PTAFR, STK22C, MYCL1, FAF1, PPAP2B, CYP2J2, LPHN2, SOYS, NARS, NOTCH2; 19q: CCNE1, PDCD5, UPK1A, TGFB1, ZNF342, PPP1R15A, BAX, BC‐2 (kit P088; MRC‐Holland, Amsterdam, the Netherlands) Cut‐off: ratio ≤ 0.8 per probe. Overall results for 1p and 19q not given. We assumed that if all probes were lost, or the majority were lost and those that were not lost were flanked by probes that were that lost that loss had occurred (stated in paper: "ratios of adjacent probes should be taken into consideration for the assessment of the presence of gains or losses"). We ignored the results for the most centromeric 1p probe (NOTCH2). |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | We presumed that both tests were performed on samples obtained at the same time. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (CGH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Jha 2011.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. 40 gliomas including 16 oligodendrogliomas grade‐II (O‐II), 14 oligodendrogliomas grade III (AO‐III) and 10 GBMs were selected for this study. Prior testing Histopathological diagnosis according to the WHO 2007 classification. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 40 Country: India Population source and setting: Neurosurgery Department of All India Institute of Medical Sciences, New Delhi, India. Time period NR Age: mean: 37.3 years, standard deviation: 10.8 years Gender: 80% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and PCR FISH Tumour sample type: FFPE Region(s) analysed: 1p36/1q25, 19q13/1p36 (using a locus‐specific probe for 1p36 and 19q13) (Vysis, Downers Grove, Illinois, USA). Cut‐off: an interpretation of deletion or imbalance was made if > 20% of the nuclei showed test to reference ratio of 1/2 or 0/2. PCR Tumour sample type: fresh‐frozen Region(s) analysed: 1p: D1S1184 (1P31.1), D1S1592 (1P36.13), D1S548 (1P36.23), D1S1608 (1P36.32); 19q: D19S431 (19q12), D19S718 (19q13.2), D19S559 (19q13.32), D19S601 (19q13.41) Cut‐off: a complete loss of band or reduction in intensity of > 50% in the tumour lane in comparison with the corresponding blood lane was scored as LOH. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Kato 2019.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. 9 "grade II–IV gliomas" seem to have been studied. Prior testing Presumably tumour grading, but this was not reported explicitly. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 9 Country: Japan Population source and setting: NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and NGS FISH Tumour sample type: NR Region(s) analysed: NR Cut‐off: NR NGS Tumour sample type: FFPE or PAXgene‐fixed paraffin‐embedded Region(s) analysed: NR Cut‐off: NR Additional details: MiSeq (Illumina) processed by Genome Jack (Mitsubishi Space Software Inc.) |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | Conference abstract | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Kolhe 2016.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. Quote: "Adult brain tumors". Prior testing Histopathological diagnosis |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 9 Country: USA Population source and setting: NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and SNP array FISH Tumour sample type: NR Region(s) analysed: NR Cut‐off: NR SNP array Tumour sample type: FFPE Region(s) analysed: genome wide Cut‐off: NR Additional details: OncoScan assay, Affymetrix, Inc. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | Conference abstract | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Lass 2013.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Not explicitly reported. Quote: "Formalin fixed paraffin‐embedded (FFPE) tissue of 42 consecutive brain tumor biopsies with previously established 1p/19q status by FISH was available for a comparative analysis of CISH … FISH analysis of 1p/19q was initiated in all cases during diagnostic work‐up and based on morphological features resembling oligodendroglioma". Prior testing Histopathological diagnosis according to WHO 2007 classification. All tumours had previously established 1p/19q status by FISH. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 38 Country: Germany Population source and setting: NR Age: mean: 43.2 years, standard deviation: 14.8 years Gender: 52.6% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: CISH and FISH CISH Tumour sample type: FFPE Region(s) analysed: 1p36/1q25, 19q13/19p13 Cut‐off: 50% of cells had to show deletion FISH Tumour sample type: FFPE Region(s) analysed: 1p36/1q25, 19q13/19p13 (ZytoVision) Cut‐off: 50% of cells had to show deletion |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Lhotska 2015.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: low grade (WHO grade II) oligodendroglioma and oligoastrocytoma Prior testing Not explicitly reported but presumably histopathological diagnosis |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 20 Country: Czech Republic Population source and setting: Department of Neurosurgery, Central Military Hospital and 1st Faculty of Medicine, Charles University, Prague, Czech Republic and the Department of Neurosurgery, Regional Hospital, Liberec, Czech Republic. 2005–2014 Agea: 56.5% aged ≤ 50 years (median age 34.5 years); 43.5% aged > 50 years (median age 57 years) Genderb: 55.6% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR aFor whole population: the study included 23 participants with oligodendroglioma or oligoastrocytoma, results for both tests available for 20. bFor whole population: this result included participants with astrocytoma (1p/19q status not investigated). 23 participants with oligodendroglioma or oligoastrocytoma. Results for both tests available for 20. |
||
| Index tests |
2 tests: FISH and SNP array FISH Tumour sample type: fresh Region(s) analysed: 1p36/1q25, 19q13/19p13 (Vysis probes; Abbott Molecular, Des Plaines, Illinois, USA) Cut‐off: 5% for deletion SNP array Tumour sample type: fresh Region(s) analysed: genome wide Cut‐off: NR and participants not classified. We set the criteria for codeletion as 1 copy of (or homozygous for) 1p36.33p11.2 or 1p31.1p12 or 1p31.3p31.1 AND 1 copy of (or homozygous for) 19q12q13.43 or 19q13.2q13.43 or 19q13.32q13.43. Additional details: human CytoSNP‐12 BeadChip (Illumina, San Diego, California) |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | All tests performed with tumour tissues taken during routine neurosurgical procedures and peripheral blood taken after the procedures. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| If a threshold was used, was it pre‐specified? | No | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | No | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Mohapatra 2006.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Not explicitly reported, but all glioma samples had oligodendroglial features on histopathological examination. Prior testing Not explicitly reported, but presumably histopathological diagnosis. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 28 Country: USA Population source and setting: Massachusetts General Hospital. 1999–2004 Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
3 tests: aCGH, FISH and PCR aCGH Tumour sample type: FFPE Region(s) analysed: not explicitly reported but 100 BACs (bacterial artificial chromosome) over chromosome 1, 50 BACs over chromosome 19 Cut‐off: quote: "Segments were considered to represent true losses or gains according to whether their associated absolute mean log2 ratio levels were greater than (2 × σ/√n) With σ estimated to be 0.58, the empirical estimate of the SD of the standardized segment means for 1q, and n equal to the number of clones in the given segment. For example, a 19q segment that contains 26 clones was considered to represent loss if its estimated mean level was less than −0.23. A 1p segment that contains 57 clones was considered to represent loss if its estimated mean level was less than −0.16. A 1p segment that contains 30 clones was considered to represent loss if its estimated mean level was less than −0.22". Additional details: a BAC array was constructed containing 200 targets that represented chromosomes 1, 7, 19, and X. FISH Tumour sample type: FFPE Region(s) analysed: 1p36.2/1q21, 19q13.3/19p13.3 Cut‐off: quote: "Relative copy numbers for 1p/1q and 19q/19p were counted, and a ratio of 0.7 or less for 1p:1q and/or 19q:19p was considered a loss". PCR Tumour sample type: FFPE Region(s) analysed: 1p: D1S508, D1S199, D1S2734; 19q: D19S219, D19S112, D19S412. Cut‐off: NR |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | All tests were performed on the same FFPE tissue. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Unclear | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Na 2019.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: FFPE specimens of primary diffuse glioma Prior testing Histopathological diagnosis according to WHO 2016 classification and the update series of the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy‐Not Official WHO (cIMPACT‐NOW). Quote: "Ancillary tests used in initial diagnosis included IDH1 (R132H), p53, and ATRX immunostaining and fluorescence in situ hybridization (FISH)‐based detection of 1p/19q‐codeletion. Select cases were immunostained by BRAF (VE1) or by paired set of H3.3K27M and H3K27me3 (midline location). After diagnosis, most representative FFPE specimens were tested for TERT mutation, MGMT methylation, and analyzed by NGS". |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 135 Country: Republic of Korea Population source and setting: Severance Hospital, Republic of Korea. March 2017 to May 2018 Agea: mean: 51.0 years, standard deviation: 16.3 years Gender: 59.3% male Karnofsky performance status: NR First diagnosis/recurrent disease: unclear. Described as "primary" diffuse gliomas. aThere are 5 participants aged < 18 years in this analysis. We have not excluded them as we are unable to link their individual patient data to the 1p/19q result for all of the tumours. |
||
| Index tests |
2 tests: FISH and NGS FISH Tumour sample type: FFPE Region(s) analysed: 1p36/1q25, 19q13/19p13 (Vysis, Abbott Molecular, Illinois, USA) Cut‐off: deletion was defined as signal ratios of > 50% for region of interest to control probe. NGS Tumour sample type: FFPE Region(s) analysed: 1p: NRAS, MYCL1; 19q: ERCC1, ERCC2, AKT2 Cut‐off: quote: "The genes with lower than 0.7‐fold change relative to average levels were considered to exhibit significant copy number loss". Additional details: Illumina TruSight Tumor 170 (TST‐170) panel |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that both tests were performed on the same sample for each case. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Natte 2005.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. 19 oligodendroglial tumours from 19 participants were analysed. Prior testing Histopathological diagnosis according to WHO classification. Quote: "From all tumors FISH data were available". |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 19 Country: the Netherlands Population source and setting: NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and MLPA FISH Tumour sample type: FFPE Region(s) analysed: 1p36(D1S32)/1cen(PUC 1.77) and 19q13(BAC127F23)/19p13(BAC2310A1) Cut‐off: 0.8 MLPA Tumour sample type: FFPE Region(s) analysed: 1p36.33, 1p36, 1p36.3,1p34.3‐1p32.1, 1p13.2, 1p22‐21, 1p13.3, 19q13.3, 19q13.3, 19q13, 19q13.43. Quote: "The MLPA kit was assembled by MRC‐Holland (Amsterdam, The Netherlands). Details of MLPA and probes can be found at http://www.mlpa.com". Cut‐off: the principal decision rule for a deletion was that for 1p ≥ 4 probes (of 7?) and for 19q ≥ 2 focus probes (of 4?) had normalised peak heights ≥ 0.25 below the median normalised peak height of the reference probes. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Nigro 2001.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: oligodendrogliomas and oligoastrocytomas Prior testing Histopathological diagnosis |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 22 Country: USA Population source and setting: University of California, San Francisco. Time period NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and real‐time PCR FISH Tumour sample type: FFPE Region(s) analysed: 1p36/1q24, 19p13/19q13.3 Cut‐off: ratio of target to control probes of ≤ 0.85 scored as a loss Real‐time PCR Tumour sample type: FFPE Region(s) analysed: D1S468, D1S214, D1S2736, D1S2783, D1S514, D19S408, D19S596, D19S867, D19S418, D19S926. Not all primers run for all samples. Cut‐off: copy numbers < 1.58 in ≥ 2 sequential loci (requirement for loss at ≥ 2 sequential loci at least for 1p). Additional details: real‐time quantitative PCR, quantitative microsatellite analysis. Utilises a probe designed to bind to CA repeats. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Park 2019.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: oligodendroglial tumours (oligodendroglioma or anaplastic oligodendroglioma) with 1p/19q FISH results available Prior testing Presumably histopathological diagnosis, although not explicitly stated. FISH |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 20 Country: Republic of Korea Population source and setting: Asan Medical Center, Seoul, Korea. January 2015 to December 2016 Age: mean: 51.7 years, standard deviation: 11.6 years Gender: 45% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and NGS FISH Tumour sample type: FFPE Region(s) analysed: 1p36 and 19q13 (Vysis, Downers Grove, Illinois, USA) Cut‐off: combined target‐to‐control signal ratio < 0.75 or cut‐off of a nucleus with a 1 or 0 target signal > 50%. NGS Tumour sample type: FFPE Region(s) analysed: whole genome. 1p and 19q specific: chr1:1‐ 125000000 and chr19:26500001‐59128983 Cut‐off: quote: "The targeted NGS panel did not cover the whole chromosome; therefore, chromosome 1p deletion and 19q deletion were defined as complete segmental loss covered by NGS panel within chr1:1‐ 125000000 and chr19:26500001‐59128983, respectively, based on the hg19 human reference genome. Partial segmental loss within regions was classified as negative". CNV plots obtained using the log 2 ratio were reviewed manually by 2 pathologists. Additional details: "MiSeq (Illumina, Inc., San Diego, CA, USA) with OncoPanel AMCv3 (OP‐AMCv3, developed in‐house by Asan‐CCGD) to include the exons of 199 genes (575,147 bp) and partial introns from 8 genes often rearranged in cancer (209,397 bp) to detect fusion genes and additional small (10,534 bp) specific single nucleotide polymorphism loci for CNV analysis. Overall, the panel covered 823,971 bp". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Paxton 2015.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: cases were selected retrospectively from confirmed glioma diagnoses based on positive FISH results for either 1p/19q codeletions (9) or EGFR amplification (8). Prior testing FISH (to determine 1p/19q status). Presumably histopathological diagnosis, although not explicitly reported. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 17 Country: USA Population source and setting: NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and SNP array FISH Tumour sample type: NR Region(s) analysed: NR Cut‐off: NR SNP array Tumour sample type: FFPE Region(s) analysed: whole genome Cut‐off: NR Additional details: OncoScan array |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that both tests were performed on the same archival sample for each participant. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Pesenti 2017.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: diffuse glioma; availability of tumour and peripheral blood specimens Prior testing Presumably histopathological diagnosis |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 50 Country: Italy Population source and setting: Fondazione IRCCS Ca' Granda, Ospedale Maggiore Policlinico di Milano, Italy. December 2013 to November 2016 Agea: median: 53 years, interquartile range: NR; range: 21–81 years Gender: 56.0% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR aFor whole population: all 50 participants had 1 tests (MS and PCR‐based LOH), only a subset had aCGH or FISH. |
||
| Index tests |
4 tests: aCGH, FISH, MS and PCR‐based LOH aCGH Tumour sample type: FFPE Region(s) analysed: genome wide Cut‐off: quote: "The aberration filter was set to detect a minimum of five consecutive probes/region, and the minimum absolute average log ratio (MAALR) was ± 0.25. A second analysis was run with a MAALR of ± 0.15 (again with a minimum number of five probes/region), to detect low level mosaicism". Additional details: SurePrint G3 Human CGH 4 × 180K, Agilent Technologies, Santa Clara, California, USA MS Tumour sample type: FFPE Region(s) analysed: rs3737577 (1p21.2), rs59317557 (1p21.2), rs2038366 (1p21.2), rs859104 (1p21.3), rs17378384 (1p31.3), rs2455638 (1p32.1), rs550663 (1p33), rs586057 (1p34.3), rs624971 (1p34.3), rs16866144 (1p35.2), rs11247639 (1p36.11), rs2473287 (1p36.12), rs7512426 (1p36.21), rs809972 (1p36.22), rs4908744 (1p36.23), rs6426368 (1p36.32), rs28503746 (19q13.2), rs67421541 (19q13.2), rs12611404 (19q13.2), rs6070 (19q13.33), rs1674139 (19q13.33), rs1807277 (19q13.33), rs186585 (19q13.41), rs28702875 (19q13.41), rs11666952 (19q13.42), rs36629 (19q13.42) and rs437229 (19q13.43) Cut‐off: the following equation was used to quantitatively define the LOH status: (N2/N1)/((N2/N1) + (T2/T1)) where N1 and N2 were the frequencies of Allele 1 and Allele 2 found in peripheral blood lymphocyte DNA and T1 and T2 were those of the corresponding alleles in tumour DNA. LOH was defined as detected with the value obtained using this formula was < 0.3 or > 0.7. LOH/NO LOH status was defined by the presence of ≥ 2 informative SNPs per chromosome arm with concordant results, 1 of which was located in a centromeric region and the other at a telomeric locus. Additional details: MassARRAY iPLEX platform (Agena Bioscience, San Diego, California, USA), based on MALDI‐TOF MS. PCR performed as a first step. PCR Tumour sample type: FFPE Region(s) analysed: D1S1592 (1p36.13), D1S548 (1p36.23), D1S2694 (1p36.23), D1S2666 (1p36.23), D1S1612 (1p36.23), D1S468 (1p36.32), D19S412 (19q13.32), D19S596 (19q13.33) and D19S206 (19q13.41) Cut‐off: the peak height derived from each allele amplified from both tumor and corresponding normal DNA was compared. The formula (T1/T2)/(N1/N2) was applied, where T1 and T2 were the peak heights of the alleles detected in tumor DNA, and N1 and N2 were the peak heights produced from peripheral blood lymphocyte DNA. LOH was considered present when the result of the calculation was < 0.50. For values > 1.00, the ratio was converted to 1/[(T1/T2)/(N1/N2)] and, again, LOH was considered present if the resulting value was < 0.50. Additional details: analysed by capillary gel electrophoresis using Gene Mapper software on an ABI 3130XL system FISH Tumour sample type: FFPE Region(s) analysed: p36/1q25 and 19q13/19p13 (ZytoVision, Bremerhaven, Germany) Cut‐off: quote: "Interpretation of FISH images was performed accordingly to Ambros et al, 2001 [37]: normal pattern was defined by the presence of an equal number of control/green and target/red signals (i.e. control/target ratio: 2/2, 3/3, 4/4, etc), deletion pattern was characterized by the presence of at least two control/green signals but only one or zero target/red signals (i.e. control/target ratio: 2/1, 2/0, 3/1, etc); finally imbalance pattern was identified by the presence of more than 1 target/red signal (i.e. control/target ratio: 3/2, 4/2, 4/3, etc). A sample was considered positive for 1p/19q codeletion when more than 50% of nuclei per chromosome arm displayed a typical deletion pattern". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Ransom 1992a.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: participants with oligodendroglioma, pilocytic astrocytoma, or ependymoma Prior testing Tumours were classified morphologically according to the WHO 1993 classification and were graded by the St Anne/Mayo method (Daumas‐Duport 1988). |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 5 Country: USA Population source and setting: location NR. May 1988 to June 1990 Age: mean: 45.0 years, standard deviation: 17.0 years Gender: 80% male Karnofsky performance status: NR First diagnosis/recurrent disease: 80% (4/5) first diagnosis, 20% (1/5) recurrent disease |
||
| Index tests |
2 tests: G‐banding and RFLP G‐banding Tumour sample type: NR Region(s) analysed: genome wide Cut‐off: N/A Additional details: from Ransom 1992b: "cytogenetically analyzed using previously described methods (Jenkins et al., 1989)". RFLP Tumour sample type: frozen Region(s) analysed: 1p: DIZ2 (1p36.3), AMY (1p21), NGFB (1p22.1); 19q D19S8 (19q13.2), S19S7 (19cen‐q12) Cut‐off: NR Additional details: from Ransom 1992b: "Paired blood and tumor DNA specimens were digested with various restriction enzymes and electrophoresed on agarose gels. Southern blotting was performed, and nylon membranes were hybridized under high stringency to a series of probes detecting RFLPs on all human chromosomes (Feinberg and Vogelstein, 1984; Southern, 1975). The resulting autoradiographs were then examined for signal intensity. Quantitative densitometry was applied to autoradiographs in cases were subjective interpretation was not immediately obvious. A normal range for relative tumor/leukocyte DNA allele intensity was established using the 3'HVR probe, which detects multiple alleles on chromosome 16 and is frequently heterozygous. The probe 3'HVR was chosen because of its high PIC score and because chromosome 16 is rarely lost in gliomas (James et al., 1988; this report). Quantitative results were then objectively classified into the categories of loss, no loss, or indeterminate, as determined by comparison to normal range values". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Ransom 1992b.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: diffuse astrocytomas of gemistocytic, fibrillary and protoplasmic type; astroblastomas; mixed oligoastrocytomas. Exclusion criteria: pure oligodendrogliomas, pilocytic astrocytomas, subependymal giant cell astrocytomas and ependymomas. Prior testing Tumours were morphologically classified using the WHO 1993 classification and were graded by the St Anne/Mayo method (Daumas‐Duport 1988). |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 22 Country: USA Population source and setting: source and setting NR. Collected between May 1988 and June 1990 Age: mean: 53.5 years, standard deviation: 14.3 years Gender: 63.6% male Karnofsky performance status: NR First diagnosis/recurrent disease: first diagnosis 86.4% (19/22); 13.6% recurrent (3/22) |
||
| Index tests |
2 tests: G‐banding and RFLP G‐banding Tumour sample type: NR Region(s) analysed: genome wide Cut‐off: NA Additional details: quote: "cytogenetically analyzed using previously described methods (Jenkins et al., 1989)". RFLP Tumour sample type: frozen Region(s) analysed: 1p: DIZ2 (1p36.3), AMY (1p21), NGFB (1p22.1); 19q D19S8 (19q13.2) and S19S7 (19cen‐q12) Cut‐off: NR Additional details: quote: "Paired blood and tumor DNA specimens were digested with various restriction enzymes and electrophoresed on agarose gels. Southern blotting was performed, and nylon membranes were hybridized under high stringency to a series of probes detecting RFLPs on all human chromosomes (Feinberg and Vogelstein, 1984; Southern, 1975). The resulting autoradiographs were then examined for signal intensity. Quantitative densitometry was applied to autoradiographs in cases were subjective interpretation was not immediately obvious. A normal range for relative tumor/leukocyte DNA allele intensity was established using the 3'HVR probe, which detects multiple alleles on chromosome 16 and is frequently heterozygous. The probe 3'HVR was chosen because of its high PIC score and because chromosome 16 is rarely lost in gliomas (James et al., 1988; this report). Quantitative results were then objectively classified into the categories of loss, no loss, or indeterminate, as determined by comparison to normal range values". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Scheie 2006.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: people with a supratentorial oligodendroglial tumour Prior testing Histopathological diagnosis according to WHO 2000 classification. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 40 Country: Norway Population source and setting: Rikshospitalet‐Radiumhospitalet Medical Center (Oslo, Norway). 2000–2004. Age: mean: 43.1 years, standard deviation: NR; range: 19–66 years Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: 95% first diagnosis (38/40); 5% recurrent disease (2/40) |
||
| Index tests |
2 tests: FISH and PCR FISH Tumour sample type: fresh or fresh‐frozen Region(s) analysed: 1p36.3 (D1Z2)/D1Z hybridising to the pericentric region (Q‐biogene, Heidelberg, Germany), 19q (D19S238E)/telomeric region on chromosome 19p (Vysis Inc, Downers Grove, Illinois, USA). Cut‐off: a tumour was defined as FISH positive when FISH‐sum (proportion of cells with FISH‐LOH and FISH‐imbalance) exceeded the mean plus 3 standard deviations value in control specimens from non‐neoplastic brain tissue. The presence of 0 or 1 1p36‐ or 19q‐ signal was reported as FISH‐LOH. Losses with any disproportion (signal ratios 3/2, 4/3, 4/2, 5/3, etc. were defined as FISH‐imbalance. Cut‐offs: 27.7% for 1p and 33.2% for 19q Additional details: FISH on touch preparations PCR Tumour sample type: fresh‐frozen or FFPE Region(s) analysed: ≥ 4 of the following for chromosome 1 and 19: chromosome 1: D1S2660, D1S507, D1S199, D1S2734, D1S1676, D1S247; chromosome 19: D19S918, D19S219, D19S112, D19S412, D19S596, D19S206 Cut‐off: results were defined as LOH‐positive when the peak areas of fluorescent intensity curves, corresponding to PCR products from individual primer sets, showed a relative reduction of ≥ 40% when the products from tumour DNA were compared with those from normal DNA. Unclear how many markers had to display LOH. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that both tests were performed on the same sample for each participant. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Schrock 1994.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR. Quote: "Nine human malignant gliomas" studied. Prior testing Histopathological diagnosis according to WHO 1993 classification. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 8 Country: Germany Population source and setting: NR Age: mean: 57.5 years, standard deviation: 12.8 years Gender: 62.5% male Karnofsky performance status: NR First diagnosis/recurrent disease: 87.5% (7/8) primary tumours, 12.5% (1/8) recurrent tumour |
||
| Index tests |
2 tests: CGH and G‐banding CGH Tumour sample type: frozen Region(s) analysed: genome wide Cut‐off: quote: "For each case evaluation of chromosomal imbalances and amplification sites in gliomas was performed both by visual inspection and calculation of fluorescence ratio profiles. For visual inspection digitized FITC and TRITC images of 10 reference metaphase spreads and the corresponding ratio images were analyzed.29 A five color lookup table was established according to the results of CGH with test DNAs from cell populations with specific monosomies and trisomies (S. du Manoir et al, manuscript in preparation). Chromosomes were identified using DAPI banding patterns. Photographs were taken from the screen with Agfa RS 50 color slide film. For fluorescence ratio profiles computer programs were developed on the basis of TCL‐Image (TNO Institute of Applied Physics, Delft, The Netherlands) running on a Macintosh Quadra 950. After determination of the chromosomal axis, individual FITC/TRITC profiles were calculated for each chromosome. Mean ratio profiles were determined from 10 metaphases. The central line in the profiles (Figures 2 and 5) represents the most frequently measured fluorescence ratio for each reference metaphase spread. The left and right vertical lines define threshold values for underrepresentation and overrepresentation of chromosome material (S. du Manoir et al, manuscript in preparation)". G‐banding Tumour sample type: fresh Region(s) analysed: genome wide Cut‐off: not applicable |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Senetta 2013.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Brain tumours with an oligodendroglial component with diagnostic agreement between 2 observers. Recurrences were excluded. Prior testing Histopathological diagnosis using the WHO 2007 classification. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 143 Country: Italy Population source and setting: Department of Medical Sciences, University of Turin. January 2004 to March 2012 Age: mean: 51.5 years, standard deviation: NR; range: 22–81 years Gender: 56.6% male Karnofsky performance status: NR First diagnosis/recurrent disease: 100% first diagnosis |
||
| Index tests |
2 tests: FISH (variant 1) and FISH (variant 2) FISH (variant 1) Tumour sample type: FFPE Region(s) analysed: 1p36/1q25 and 19q13/19p13 (Vysis, Abbott Molecular Europe, Wiesbaden, Germany) Cut‐off: ratios 1p ≤ 0.8 and 19q ≤ 0.8 FISH (variant 2) Tumour sample type: FFPE Region(s) analysed: 1p36/1q25 and 19q13/19p13 (Vysis, Abbott Molecular Europe, Wiesbaden, Germany) Cut‐off: ratios 1p ≤ 0.7 and 19q ≤ 0.8 |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | The same FISH preparations were used, and 2 different thresholds were applied. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Sim 2018a.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: glioblastoma Prior testing Histopathological diagnosis (WHO 2007 classification) |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 75 Country: Republic of Korea Population source and setting: Samsung Medical Center, Seoul, Korea. 2011–2014 Age: mean: 52.8 years, standard deviation: NR; range: 21–76 years Gender: 53.3% male Karnofsky performance status: NR First diagnosis/recurrent disease: not clear. We excluded recurrent samples from participants who also contributed samples from their primary tumour. |
||
| Index tests |
2 tests: FISH and NGS or aCGH (or both) FISH Tumour sample type: FFPE Region(s) analysed: 1p36 and 19q13 (Vysis, Downers Grove, Illinois, USA) Cut‐off: 1p deletion as a combined target‐to‐control signal ratio < 0.75 or cut‐off of a nucleus with a 1 or 0 target signal > 50%. 19q deletion as a combined target‐to‐control signal ratio < 0.8 and a nucleus cut‐off with a 1 or 0 target signal > 30% NGS or aCGH (or both) Tumour sample type: fresh frozen Region(s) analysed: genome wide Cut‐off: whole arm losses Additional details: aCGH (Afilent SurePrint G3 Human CGH 4x180k array) or whole exome sequencing (Illumina TruSeq Exome capture kit or the Agilent SureSelect kit and either the Illumina HiSeq 2000 or HiSeq 2500) (or both) |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Unclear | ||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Sim 2018b.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion: oligodendroglial tumour Prior testing Presumably histopathological diagnosis, although not explicitly reported |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 10 Country: Republic of Korea Population source and setting: NR Age: mean: 45.7 years, standard deviation: 13.5 years Gender: 60% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and NGS FISH Tumour sample type: FFPE Region(s) analysed: 1p36 and 19q13 (Vysis, Downers Grove, Illinois, USA) Cut‐off: 1p deletion as a combined target‐to‐control signal ratio < 0.75 or cut‐off of a nucleus with a 1 or 0 target signal > 50%. 19q deletion as a combined target‐to‐control signal ratio < 0.8 and a nucleus cut‐off with a 1 or 0 target signal > 30% NGS Tumour sample type: fresh frozen Region(s) analysed: genome wide Cut‐off: whole arm losses Additional details: whole exome sequencing (Illumina TruSeq Exome capture kit or the Agilent SureSelect kit and either the Illumina HiSeq 2000 or HiSeq 2500) |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that both tests were performed on tumour material harvested at the same time point. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Could the patient flow have introduced bias? | Low risk | ||
Smith 1999.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: diffuse gliomas Prior testing Histopathological diagnosis according to WHO 1993 classification. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 79 Country: USA Population source and setting: Mayo Clinic in Rochester, MN, Johns Hopkins Hospital in Baltimore, Maryland, USA, and the University of California at San Francisco, USA. Time period NR. Cases subsequently included in Hatanpaa 2003a and Hatanpaa 2003b were removed. Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: 72.2% (57/79) primary glioma specimens; 27.8% (22/79) recurrent glioma specimens |
||
| Index tests |
3 tests: CGH, FISH and PCR CGH Tumour sample type: NR Region(s) analysed: NR Cut‐off: reference Mohapatra G, Kim DH, Feuerstein BG. Detection of multiple gains and losses of genetic material in ten glioma cell lines by comparative genomic hybridization. Genes, Chromosomes & Cancer 1995;13;86‐93. In this publication: "Definition of CGH ratio thresholds to define ratios that were indicative of changes in DNA copy number, we performed 21 CGH experiments using normal control DNA. We calculated average ratio changes and standard deviations by using the software program cghprofstats.new (Piper et al., 1994). The average ratio for all 21 hybridizations was 0.99 (range 0.9‐1.1). The average standard deviation was 0.04 (range 0.02‐0.06). Taking these findings into consideration, we chose upper and lower ratio thresholds of 1.2 and 0.8, respectively. Any change in ratio in excess of these thresholds was interpreted as indicative of DNA copy number changes only if found in both forward and reverse experiments. Amplifications were defined both by a ratio >2.0 and by visual inspection". Piper J, Rutovitz D, Sudar D, Kallioniemi A, Kallioniemi O, Waldman FM, et al. Computer image analysis of comparative genomic hybridization. Cytometry 1995;19:10‐26 also cited. FISH Tumour sample type: NR Region(s) analysed: 1p36, 1q24, 19p13.1, 19q13.1‐q13.2 and 19q13.3. Cut‐off: we defined codeletion as hemizygous deletion of 1p36, 1q13.1‐q13.2 and 19q13.3. What defined hemizygous deletion NR. Also cited Qian J, Bostwick DG, Takahashi S, Borell TJ, Herath JF, Lieber MM et al. Chromosomal anomalies in prostatic intraepithelial neoplasia and carcinoma detected by fluorescence in situ hybridization. Cancer Research 1995;55:5408‐14. In this paper (quote) "abnormal autosomal loss required ≥55% nuclei with zero or one signal". Unclear if this threshold was used. PCR Tumour sample type: NR Region(s) analysed: 1p: D1S468, D1S1612, D1S1597, D1S199, D1S1665, D1S1728, D1S1588, D1S1675, D1S187; 19q: D19S213, D19S569, D19S422. D19S219, SM, S19S112, S19S412, D19S596, HRC, D19S589, D19S218 Cut‐off: we defined codeletion as all markers showing confirmed allelic loss, presumed allelic loss, were homozygous or were indeterminant. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH or PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Srebotnik‐Kirbis 2016.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria No further details reported. Included an oligodendroglial tumour and non‐oligodendroglial tumour cohort. We have only extracted data for the oligodendroglial tumour cohort. Prior testing Histopathological diagnosis, according to the WHO 2007 classification. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 12 Country: Slovenia Population source and setting: tissue samples from non‐consecutive patients who underwent surgical resection or biopsy to the Institute of Pathology, Faculty of Medicine, University of Ljubljana. December 2011 to November 2015. Age: mean: 47.8 years, standard deviation: 10.5 years Gender: 50% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH (variant 1) and FISH (variant 2) FISH (variant 1) Tumour sample type: fresh tissue cytospins Region(s) analysed: 1p36/1q25 and 19q13/19p13 (Vysis paired probes, Abbott Laboratories, Abbott Park, Illinois, USA) Cut‐off: quote: "Deletion was defined as a nucleus showing none or one target signal, and 2 or more control signals (ex. 1/2, 0/2, 1/3 etc.) … A tumour sample was considered positive for 1p or 19q deletion when it displayed a percentage of nuclei with deletion above the cut‐off value for that probe, specifically … 30% for 1p and 19% for 19q on cytospins". FISH (variant 2) Tumour sample type: FFPE tissue section Region(s) analysed: 1p36/1q25 and 19q13/19p13 (Vysis paired probes, Abbott Laboratories, Abbott Park, Illinois, USA) Cut‐off: quote: "Deletion was defined as a nucleus showing none or one target signal, and 2 or more control signals (ex. 1/2, 0/2, 1/3 etc.) … A tumour sample was considered positive for 1p or 19q deletion when it displayed a percentage of nuclei with deletion above the cut‐off value for that probe, specifically, 43% for 1p and 33% for 19q in tissue sections" and "the cut‐off value of 50 %, which is often reported in the literature, was also included in the analysis of FISH results for FFPE sections". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | High risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Thakur 2012.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR Prior testing NR |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 2 Country: USA Population source and setting: NR Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: FISH and SNP array FISH Tumour sample type: NR Region(s) analysed: NR Cut‐off: NR SNP array Tumour sample type: FFPE Region(s) analysed: genome wide Cut‐off: NR Additional details: quote: "Affymetrix GeneChip human mapping 250L Nsp I array". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | Conference abstract | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Thomas 2017.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Nested in a phase II study. Inclusion criteria: diagnosis of anaplastic oligodendroglioma or anaplastic oligoastrocytoma according to the WHO 2000 classification (mixed tumours should have a minimum of 25% oligodendroglial elements); aged ≥18 years; Karnofsky performance status ≥ 60; adequate organ and bone marrow function including a granulocyte count ≥ 1.5 × 109/L, platelet count of ≥ 100 × 109/L, aspartate aminotransferase ≤ 2× UNL, serum creatinine ≤1.5 × UNL and bilirubin ≤ 1.5 × UNL. Exclusion criteria: systemic or non‐contiguous leptomeningeal metastases; prior cranial RT or systemic chemotherapy; other concurrent malignancy with the exception of cervical carcinoma in situ or basal cell carcinoma of the skin; serious illness that would interfere with the prescribed treatment; pregnancy or lactation; refusal to use effective contraception. Prior testing Histopathological diagnosis according to the WHO 2000. Quote: "Within 2 weeks of starting treatment, all patients were evaluated with a complete history, physical and neurological examination, contrast enhanced MRI [magnetic resonance imaging], a biochemistry panel, and complete blood count and underwent screening for hepatitis B and C and HIV". |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 19 Country: USA Population source and setting: phase II study Agea: median: 44 years, interquartile range: NR; range: 30–66 years Gender: 65.9% male Karnofsky performance status: median 90, range 70–100 First diagnosis/recurrent disease: unclear. Quote: "Patients with newly diagnosed AO [anaplastic oligodendroglioma] or AOA [anaplastic oligoastrocytoma] were eligible to participate in this prospective multicenter phase II study". aFor whole population: 41 people in the phase II study, only 19 had available tissue with adequate DNA quality and quantity for NGS. |
||
| Index tests |
2 tests: FISH and NGS FISH Tumour sample type: NR Region(s) analysed: NR Cut‐off: NR NGS Tumour sample type: NR Region(s) analysed: genome wide Cut‐off: NR Additional details: MSK‐IMPACT: a hybridisation capture‐based sequencing assay utilising an Illumina HiSeq 2500 platform. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. FISH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| Could the selection of patients have introduced bias? | Low risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | No | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | High | ||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
Tsiatis 2010.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria NR Prior testing NR, but presumably histopathological diagnosis. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 4 Country: USA Population source and setting: NR Age: mean: 43.3 years, standard deviation: 8.2 years Gender: 25% male Karnofsky performance status: NR First diagnosis/recurrent disease: NR |
||
| Index tests |
2 tests: PCR and SNP array PCR Tumour sample type: FFPE Region(s) analysed: D1S199, D1S186, D1S162, D1S312, D1S226, D19S918, D19S112, D19S206 Cut‐off: NR Additional details: quote: "Samples were run on an ABI 3100 following multiplex PCR amplification of 5 STRs on chromosome 1p (D1S199, D1S186, D1S162, D1S312, D1S226) and 3 STRs on chromosome 19q (D19S918, D19S112, D19S206)". SNP array Tumour sample type: FFPE Region(s) analysed: genome wide Cut‐off: NR Additional details: quote: "Array analysis was performed using the Affymetrix genome‐wide human SNP array 6.0 platform (906,600 SNPs) according to protocol. Data were analyzed with Partek Genomics Suite". |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. PCR‐based LOH used as reference standard in some of our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | Conference abstract | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | Low concern | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Uchida 2019.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria: primary glioblastoma. Tested by FISH Prior testing Presumably histopathological diagnosis, although this was not explicitly reported. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 141 Country: Japan Population source and setting: The Department of Neurosurgery, University of Kagoshima. 2009–2016 Age: NR Gender: NR Karnofsky performance status: NR First diagnosis/recurrent disease: unclear. Primary GBM |
||
| Index tests |
2 tests: FISH (variant 1) and FISH (variant 2) FISH (variant 1) Tumour sample type: NR Region(s) analysed: 1p36/1q25, 19p13/19q13 (Vysis LSI DNA probes) Cut‐off: 20%. Criteria for judging whether a deletion was present: signals of 1p or 19q < signals of 1q or 19p. FISH (variant 2) Tumour sample type: NR Region(s) analysed: 1p36/1q25, 19p13/19q13 (Vysis LSI DNA probes) Cut‐off: 20%. Criteria for judging whether a deletion was present: single signal of 1p or 19q and 2 signals of 1q or 19p. |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | This was 1 test analysed with 2 different cut‐offs. | ||
| Comparative | |||
| Notes | Conference abstract | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| If a threshold was used, was it pre‐specified? | No | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| If a threshold was used, was it pre‐specified? | No | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | High risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | Unclear | ||
| Could the patient flow have introduced bias? | Unclear risk | ||
Wiestler 2014.
| Study characteristics | |||
| Patient Sampling |
Inclusion/exclusion criteria Inclusion criteria for NOA‐04: adults with centrally confirmed diagnosis of a WHO grade III anaplastic glioma, Karnofsky performance score ≥ 70, no prior systemic chemotherapy or RT to the brain, and adequate bone marrow reserve, liver and renal functions, and stable or decreasing corticosteroid dose within 14 days before random assignment. Prior testing Histopathological diagnosis according to WHO 1993 and WHO 2000 classifications. |
||
| Patient characteristics and setting |
Number of participants/tumours with results for 1p/19q status by ≥ 2 DNA‐based tests: 99 Country: Germany Population source and setting: people enrolled in the NOA‐04 trial from 39 sites in Germany. Time period NR Agea: median: 42 years in the RT arm and 41.5 years in the procarbazine, lomustine and vincristine/temozolomide arm, interquartile range: NR; range: 23–74 years Genderb: 57.7% male Karnofsky performance status**: median 90, range 70–100 First diagnosis/recurrent disease: NR, although presumably first diagnosis. aFor whole population: these data were for the biomarker cohort (the 115 participants with sufficient amount and quantity of tumour DNA). Methylation array and MLPA data was only available for 99 participants. bFor whole population: these data were for the modified intention‐to‐treat population of NOA‐04. This included 274 participants, we had data on both tests for 99 participants. |
||
| Index tests |
2 tests: methylation array and MLPA Methylation array Tumour sample type: FFPE Region(s) analysed: genome wide Cut‐off: quote: "Copy number aberrations were detected from the HM450 data as described.20,22 Copy number plots were manually analysed for 1p/19q codeletion". Additional details: HM450 BeadChip (Illumina, San Diego, California, USA) MLPA Tumour sample type: FFPE Region(s) analysed: used Salsa MLPA P088, MRC Holland, Amsterdam, the Netherlands Cut‐off: quote: "Chromosomal regions were scored as under‐ or overrepresented if two or more loci on 1p or 19q adjacent to each other exhibited a gene dosage ratio less than 70% or more than 130% relative to the reference value". Additional details: Salsa MLPA, P088 lots 0305 and 0706, MRC Holland, Amsterdam, the Netherlands |
||
| Target condition and reference standard(s) | Target condition was absolute 1p/19q deletion. No tests used as reference standard in our analyses. | ||
| Flow and timing | We presumed that all tests were performed on biopsied tumour material collected on 1 occasion. | ||
| Comparative | |||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Could the selection of patients have introduced bias? | Unclear risk | ||
| Are there concerns that the included patients and setting do not match the review question? | High | ||
| DOMAIN 2: Index Test (NanoString) | |||
| DOMAIN 2: Index Test (aCGH) | |||
| DOMAIN 2: Index Test (NGS) | |||
| DOMAIN 2: Index Test (G‐banding) | |||
| DOMAIN 2: Index Test (FISH (variant 4)) | |||
| DOMAIN 2: Index Test (SNP array) | |||
| DOMAIN 2: Index Test (PCR (with comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (PCR (without comparison to normal DNA)) | |||
| DOMAIN 2: Index Test (CISH) | |||
| DOMAIN 2: Index Test (MS) | |||
| DOMAIN 2: Index Test (RFLP) | |||
| DOMAIN 2: Index Test (PCR‐based LOH) | |||
| DOMAIN 2: Index Test (NGS or aCGH (or both)) | |||
| DOMAIN 2: Index Test (Methylation array) | |||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Unclear | ||
| Could the conduct or interpretation of the index test have introduced bias? | Unclear risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (FISH) | |||
| DOMAIN 2: Index Test (FISH (variant 1)) | |||
| DOMAIN 2: Index Test (FISH (variant 2)) | |||
| DOMAIN 2: Index Test (FISH (variant 3)) | |||
| DOMAIN 2: Index Test (Real‐time PCR) | |||
| DOMAIN 2: Index Test (MLPA) | |||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Were the index test results interpreted without knowledge of the results of the other tests being compared? | Yes | ||
| Could the conduct or interpretation of the index test have introduced bias? | Low risk | ||
| Are there concerns that the index test, its conduct, or interpretation differ from the review question? | Low concern | ||
| DOMAIN 2: Index Test (CGH) | |||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | No | ||
| Could the reference standard, its conduct, or its interpretation have introduced bias? | High risk | ||
| Are there concerns that the target condition as defined by the reference standard does not match the question? | Low concern | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| Could the patient flow have introduced bias? | High risk | ||
aCGH: array comparative genomic hybridisation; CGH: comparative genomic hybridisation; CI: confidence interval; CISH: chromogenic in situ hybridisation; DNA: deoxyribonucleic acid; EORTC: European Organisation for Research and Treatment of Cancer; FFPE: formalin‐fixed, paraffin‐embedded; FISH: fluorescent in situ hybridisation; IDH: isocitrate dehydrogenase; LOH: loss of heterozygosity; MLPA: multiplex‐ligation‐dependent probe amplification; MS: mass spectrometry; NGS: next‐generation sequencing; NR: not reported; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism; RT: radiotherapy; SNP: single nucleotide polymorphism; WHO: World Health Organization.
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Afyounian 2017 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Alentorn 2014 | Concordance only data. |
| Aoki 2015a | 1p19q not assessed by ≥ 2 DNA techniques. |
| Aoki 2015b | 1p19q not assessed by ≥ 2 DNA techniques. |
| Assem 2009 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Assem 2012 | No results. |
| Bady 2013 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Ballester 2017 | Single case report. |
| Becker 2017 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Bienkowski 2018 | No results. |
| Boudreau 2004 | Not a primary study. |
| Brat 2015 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Buckley 2011 | Concordance only data. |
| Burgenske 2017 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Bystricka 2011 | Concordance only data. |
| Carrato 2006 | Concordance only data. |
| Carrato 2010 | Recruited based on 1p/19q status. |
| Carter 2016 | Concordance only data. |
| Castilla 2003 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Chernova 2003 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Cieply 2005 | Concordance only data. |
| Durand 2010 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Eckel‐Passow 2017 | Concordance only data. |
| Fontaine 2007 | Concordance only data. |
| Franco‐Hernandez 2009a | Concordance only data. |
| Franco‐Hernandez 2009b | 1p19q not assessed by ≥ 2 DNA techniques. |
| French 2005 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Garber 2016 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Hartmann 2005 | Not a primary study. |
| Hashimoto 2002 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Hench 2018 | Concordance only data. |
| Horbinski 2008 | Single case report. |
| Horbinski 2011 | Concordance only data. |
| Ida 2018 | Participants aged < 18 years. |
| Idbaih 2008 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Joo 2013 | Single case report. |
| Juratli 2012 | No results. |
| Kamoun 2015 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Kashofer 2018 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Kim 2016 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Kitange 2004 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Kitange 2005 | No results. |
| Klink 2010 | No results. |
| Klink 2011 | No results |
| Kouwenhoven 2009 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Kuo 2009 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Kuo 2013 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Kwon 2019 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Lautenschlaeger 2013 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Levine 2018 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Liu 2014 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Magnani 2003 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Martinez 2005 | Single case report. |
| Marucci 2012 | 1p19q not assessed by ≥ 2 DNA techniques. |
| McDonald 2005 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Mohapatra 2011 | No results. |
| Molinari 2010 | Concordance only data. |
| Mrachek 2018 | Recruited based on 1p/19q status. |
| Mur 2013 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Myung 2011 | Not a glioma. |
| Narasimhaiah 2010 | Concordance only data. |
| Neill 2015 | Single case report. |
| Nielsen 2007 | Not a primary study. |
| Parizi‐Robinson 2004 | Concordance only data. |
| Payne 2008 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Pekmezci 2016 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Pietsch 2015 | Participants aged < 18 years. |
| Pina‐Oviedo 2012 | Concordance only data. |
| Pinkham 2015 | Not a primary study. |
| Pinto 2008 | No results. |
| Ramkissoon 2015 | No results. |
| Rolston 2002 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Roy 2012 | Concordance only data. |
| Satomi 2018 | Concordance only data. |
| Satomi 2019 | Concordance only data. |
| Scheinin 2014 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Schiavo 2009 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Serrano 2015 | Concordance only data. |
| Tauziede‐Espariat 2018 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Walker 2000 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Woehrer 2015 | Not a primary study. |
| Xiu 2015 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Yokogami 2018 | Concordance only data. |
| Yoshimoto 2002 | 1p19q not assessed by ≥ 2 DNA techniques. |
| Zacher 2017 | Recruited based on 1p/19q status. |
| Zheng 2019 | Concordance only data. |
DNA: deoxyribonucleic acid.
Characteristics of studies awaiting classification [ordered by study ID]
Ducray 2011.
| Patient Sampling | |
| Patient characteristics and setting | |
| Index tests | |
| Target condition and reference standard(s) | |
| Flow and timing | |
| Comparative | |
| Notes | No full‐text |
Hazra 2006.
| Patient Sampling | |
| Patient characteristics and setting | |
| Index tests | |
| Target condition and reference standard(s) | |
| Flow and timing | |
| Comparative | |
| Notes | No full‐text |
McDonald 2003.
| Patient Sampling | |
| Patient characteristics and setting | |
| Index tests | |
| Target condition and reference standard(s) | |
| Flow and timing | |
| Comparative | |
| Notes | No full‐text |
Meunier 2005.
| Patient Sampling | |
| Patient characteristics and setting | |
| Index tests | |
| Target condition and reference standard(s) | |
| Flow and timing | |
| Comparative | |
| Notes | No full‐text |
Monnot 2007.
| Patient Sampling | |
| Patient characteristics and setting | |
| Index tests | |
| Target condition and reference standard(s) | |
| Flow and timing | |
| Comparative | |
| Notes | No full‐text |
Sebastian 2003.
| Patient Sampling | |
| Patient characteristics and setting | |
| Index tests | |
| Target condition and reference standard(s) | |
| Flow and timing | |
| Comparative | |
| Notes | No full‐text |
Characteristics of ongoing studies [ordered by study ID]
ACTRN12618000006246.
| Study name | Access to innovative molecular diagnostic PROfiling for paediatric brain tumours (application of innovative molecular profiling techniques to improve diagnosis of paediatric central nervous system tumours and develop an accredited Australasian molecular profiling service) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
JPRN‐UMIN000003196.
| Study name | Genetic analysis of prognosis‐related factors in gliomas: methylation of MGMT, LOH of 1p/19q, and mutation of IDH1/2 |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT00031538.
| Study name | Genetic analysis of brain tumors (a prospective national study to molecularly and genetically characterize human gliomas: the Glioma Molecular Diagnostic Initiative) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT01004887.
| Study name | Study of tissue and blood samples from patients with high‐grade glioma (diagnostic and prognostic markers in high‐grade glioma) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
NCT03336931.
| Study name | Precision medicine for children with cancer (a multicenter prospective study of the feasibility and clinical value of a diagnostic service for identifying therapeutic targets and recommending personalised treatment for children and adolescents with high‐risk cancer) |
| Target condition and reference standard(s) | |
| Index and comparator tests | |
| Starting date | |
| Contact information | |
| Notes |
Differences between protocol and review
We made the following changes from the protocol (McAleenan 2019).
Criteria for considering studies for this review
We excluded studies with data for just one person.
We had planned to contact authors of studies where only concordance data were reported rather than contingency tables of cross‐classified results, and of studies where it was clear that at least two tests were applied but results were not reported. However, due to resource constraints and a larger than anticipated number of included studies, we were unable to do this.
We added a clarification that studies were only eligible if participants had not been recruited based on their 1p/19q status (i.e. if all participants were 1p/19q positive or if all participants were 1p/19q negative on one test).
Selection of studies
We had planned to retrieve full texts of all titles and abstracts that had been deemed relevant by at least one review author. However, we changed this due to the very large number of papers this would have produced, and used a third review author to arbitrate in cases of disagreement.
Data extraction and management
In situations where classifications were not made, raw data were presented, and the threshold to be used to interpret the raw data were not specified, we had planned to contact study authors to enquire regarding the threshold to be used, and only to apply our own threshold based on our own expertise if we did not receive a response. However, we applied our own thresholds to interpret the data in all cases.
Assessment of methodological quality
We made some refinements to our tailoring of QUADAS‐2.
Statistical analysis and data synthesis
We had planned to investigate whether the index characteristics such as tumour sample type (i.e. FFPE or frozen tissues), region(s) analysed and cut‐off/threshold used to determine 1p/19q status, and population characteristics such as prevalence of 1p/19q codeletion and tumour subtype and grade contributed to heterogeneity. We had planned to perform sensitivity analyses restricting to direct comparative studies and by restricting analyses to studies judged not to be at high risk of bias or low applicability. Due limited amounts of data and resource constraints, we did not complete these analyses.
Economic model
As well as reporting the incremental cost per additional true‐positive diagnosis (as stated in the protocol), we also reported the incremental cost per additional true‐negative diagnosis and incremental cost per correct diagnosis.
Contributions of authors
SD undertook the searches. AM, LS, SD, ESL and CF performed title/abstract screening. AM, LS, ESL and JPTH performed full‐text screening. AM, LS, CK, ESL, AP, JPTH and KMK performed data extraction. AM, LS, CK, JPTH and KMK undertook QUADAS‐2 assessments. HEJ performed statistical analyses. AK and TR performed the economic analyses. JPTH performed GRADE assessments. AMM managed the project. JPTH, LV and HEJ provided methodological expertise. KMK, SB, CF and CW provided content expertise. AM, HEJ, AK, TR, SB, LV, JPTH and KMK drafted the manuscript. JPTH, SD and CK entered the review into Review Manager 5. All review authors commented on the manuscript.
Sources of support
Internal sources
University of Bristol, UK
External sources
-
National Institute for Health Research, UK
This work is supported by a National Institute for Health Research Systematic Reviews Cochrane Programme Grant 16/144/18
-
Cancer Research UK, UK
AM and JPTH are supported in part by Cancer Research UK (grant number C18281/A19169)
The views and opinions expressed are those of the review authors and do not necessarily reflect those of the NIHR, its Systematic Reviews Programme, the National Health Service (NHS), the Department of Health, or Cancer Research UK.
Declarations of interest
AM: none HEJ: none AK: none TR: none LS: none SD: none CK: none ESL: none CLF: none AP: none CW: none SJ: none SB: none LV: none JPTH: none KMK: none
New
References
References to studies included in this review
Ariza 2010 {published data only}
- Ariza A, Carrato C, Lopez MD, Domingo-Sabat M, Lopez MT, Beyer K.Distinction between complete and partial 1P/19Q losses in gliomas: a novel user-friendly approach. Laboratory Investigation 2010;90(Supp 1):375A. [Google Scholar]
Armanious 2017 {published data only}
- Armanious H, Izevbaye I.Nanostring copy number variation assay is very sensitive in identifying EGFR amplification but is less sensitive in identifying deletions in 1p/19q and PTEN compared to FISH in brain tumors samples. Laboratory Investigation 2017;97(Suppl 1):525A. [Google Scholar]
Belaud‐Rotureau 2006 {published data only}
- Belaud-Rotureau MA, Meunier N, Eimer S, Turmo M, Dubus P, Vital A, et al.Automatic assessment of 1P36-19Q13 status in gliomas by interphase FISH assay. Virchows Archiv 2005;447(2):218. [Google Scholar]
- Belaud-Rotureau MA, Meunier N, Eimer S, Vital A, Loiseau H, Merlio JP.Automatized assessment of 1p36-19q13 status in gliomas by interphase FISH assay on touch imprints of frozen tumours. Acta Neuropathologica 2006;111(3):255-63. [DOI] [PubMed] [Google Scholar]
Bigner 1999 {published data only}
- Bigner SH, Matthews MR, Rasheed BK, Wiltshire RN, Friedman HS, Friedman AH, et al.Molecular genetic aspects of oligodendrogliomas including analysis by comparative genomic hybridization. American Journal of Pathology 1999;155(2):375-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon RE, Herndon IJ, West B, Reardon D, Wiltshire R, Rasheed BK, et al.Survival analysis of presumptive prognostic markers among oligodendrogliomas. Cancer 2005;104(8):1693-9. [DOI] [PubMed] [Google Scholar]
Blesa 2009 {published data only}
- Blesa D, Mollejo M, Ruano Y, Lope AR, Fiano C, Ribalta T, et al.Novel genomic alterations and mechanisms associated with tumor progression in oligodendroglioma and mixed oligoastrocytoma. Journal of Neuropathology & Experimental Neurology 2009;68(3):274-85. [DOI] [PubMed] [Google Scholar]
Bouvier 2004 {published data only}
- Bouvier C, Roll P, Quilichini B, Metellus P, Calisti A, Gilles S, et al.Deletions of chromosomes 1p and 19q are detectable on frozen smears of gliomas by FISH: usefulness for stereotactic biopsies. Journal of Neuro-Oncology 2004;68(2):141-9. [DOI] [PubMed] [Google Scholar]
Broholm 2008 {published data only}
- Broholm H, Born PW, Guterbaum D, Dyrbye H, Laursen H.Detecting chromosomal alterations at 1p and 19q by FISH and DNA fragment analysis – a comparative study in human gliomas. Clinical Neuropathology 2008;27(6):378-87. [DOI] [PubMed] [Google Scholar]
Burger 2001 {published data only}
- Burger PC, Minn AY, Smith JS, Borell TJ, Jedlicka AE, Huntley BK, et al.Losses of chromosomal arms 1p and 19q in the diagnosis of oligodendroglioma. A study of paraffin-embedded sections. Modern Pathology 2001;14(9):842-53. [DOI] [PubMed] [Google Scholar]
Byeon 2014 {published data only}
- Byeon SJ, Cho HJ, Baek HW, Park CK, Choi SH, Kim SH, et al.Rhabdoid glioblastoma is distinguishable from classical glioblastoma by cytogenetics and molecular genetics. Human Pathology 2014;45(3):611-20. [DOI] [PubMed] [Google Scholar]
Chaturbedi 2012 {published data only}
- Chaturbedi A, Yu L, Linskey ME, Zhou YH.Detection of 1p19q deletion by real-time comparative quantitative PCR. Biomarker Insights 2012;7:9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturbedi A, Yu L, Zhou YH.Detection of 1P/19Q deletion in gliomas by real-time comparative quantitative PCR. Neuro-Oncology 2011;3(iii):80. [Google Scholar]
Cieply 2004 {published data only}
- Cieply K, Couce MS, Hunt J.Molecular assessment of malignant gliomas: a combined approach using FISH and PCR for determining 1p and 19q status. Journal of Molecular Diagnostics 2004;6(4):428. [Google Scholar]
Clark 2013 {published data only}
- Clark K, Nikiforova M, Hamilton R, Horbinski C.Histology trumps apparent 1P/19Q codeletion in glioblastomas. Journal of Neuropathology and Experimental Neurology 2012;71(6):548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KH, Villano JL, Nikiforova MN, Hamilton RL, Horbinski C.1p/19q testing has no significance in the workup of glioblastomas. Neuropathology and Applied Neurobiology 2013;39(6):706-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Cowell 2004 {published data only}
- Cowell JK, Barnett GH, Nowak NJ.Characterization of the 1p/19q chromosomal loss in oligodendrogliomas using comparative genomic hybridization arrays (CGHa). Journal of Neuropathology & Experimental Neurology 2004;63(2):151-8. [DOI] [PubMed] [Google Scholar]
D'Haene 2019 {published data only}
- D'Haene N, Melendez B, Blanchard O, De Neve N, Lebrun L, Campenhout C, et al.Design and validation of a gene-targeted, next-generation sequencing panel for routine diagnosis in gliomas. Cancers 2019;11(6):04. [DOI] [PMC free article] [PubMed] [Google Scholar]
Dahlback 2009 {published data only}
- Dahlback HS, Brandal P, Meling TR, Gorunova L, Scheie D, Heim S.Genomic aberrations in 80 cases of primary glioblastoma multiforme: pathogenetic heterogeneity and putative cytogenetic pathways. Genes Chromosomes and Cancer 2009;48(10):908-24. [DOI] [PubMed] [Google Scholar]
Dahlback 2011 {published data only}
- Dahlback HS, Gorunova L, Brandal P, Scheie D, Helseth E, Meling TR, et al.Genomic aberrations in diffuse low-grade gliomas. Genes, Chromosomes & Cancer 2011;50(6):409-20. [DOI] [PubMed] [Google Scholar]
Dubbink 2016 {published data only}
- Dubbink E, Atmodimedjo PN, Marion RM, Kros JM, den Bent MJ, Dinjens WN.Sensitive and specific detection of 1p/19q codeletion in gliomas by next generation sequencing. European Journal of Cancer 2014;6:53. [Google Scholar]
- Dubbink H, Atmodimedjo P, Marion R, Kros JM, den Bent MJ, Dinjens WN.Diagnostic application of targeted next-generation sequencing for detection of chromosomal losses and allelic imbalances: sensitive and specific diagnostic testing of 1p/19q co-deletion in gliomas. Journal of Molecular Diagnostics 2015;17(6):817. [Google Scholar]
- Dubbink HJ, Atmodimedjo PN, Marion R, Krol NM, Riegman PH, Kros JM, et al.Diagnostic detection of allelic losses and imbalances by next-generation sequencing: 1p/19q co-deletion analysis of gliomas. Journal of Molecular Diagnostics 2016;18(5):775-86. [DOI] [PubMed] [Google Scholar]
- Dubbink HJ, Atmodimedjo PN, Marion R, Riegman PH, Kros JM, den Bent MJ, et al.Diagnostic detection of allelic losses and imbalances by next-generation sequencing: 1p/19q co-deletion analysis of gliomas. Journal of Molecular Diagnostics 2016;18(6):1009. [DOI] [PubMed] [Google Scholar]
- Dubbink HJ.Correction: diagnostic detection of allelic losses and imbalances by next-generation sequencing: 1p/19q co-deletion analysis of gliomas [the Journal of Molecular Diagnostics (2016) 18(5) (775-786) (S1525157816300903) (10.1016/j.jmoldx.2016.06.002]. Journal of Molecular Diagnostics 2016;18(6):933. [DOI] [PubMed] [Google Scholar]
Duval 2014 {published data only}
- Duval C, Tayrac M, Sanschagrin F, Michaud K, Gould PV, Saikali S.ImmunoFISH is a reliable technique for the assessment of 1p and 19q status in oligodendrogliomas. PLoS One 2014;9(6):e100342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould P, Duval C, Tayrac M, Sanschagrin F, Michaud K, Saikali S.ImmunoFISH is a reliable technique for the assessment of 1p and 19q status in oligodendrogliomas. Journal of Neuropathology and Experimental Neurology 2014;73(6):632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Duval 2015 {published data only}
- Duval C, Tayrac M, Michaud K, Cabillic F, Paquet C, Gould PV, et al.Automated analysis of 1p/19q status by FISH in oligodendroglial tumors: rationale and proposal of an algorithm. PLoS One 2015;10(7):e0132125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould PV, Duval C, Tayrac M, Sanschagrin F, Michaud K, Saikali S.Automated analysis of 1p/19q status by FISH in oligodendroglial tumours. Canadian Journal of Neurological Sciences 2015;42(Suppl 2):S3. [Google Scholar]
- Saikali S, Tayrac M, Sanschagrin F, Michaud K, Gould PV.Comparison between manual and automated assessment of 1p-19q status in gliomas by FISH assay on paraffin embedded tissue: a need for standardization. Canadian Journal of Neurological Sciences 2013;40(1):119. [Google Scholar]
Gadji 2009 {published data only}
- Drouin R, Gadji M, Tsanaclis AM, Fortin D.Genetic characterization of oligodendrogliomas. Chromosome Research 2007;15(Suppl 1):221-2. [Google Scholar]
- Gadji M, Fortin D, Tsanaclis AM, Drouin R.Is the 1p/19q deletion a diagnostic marker of oligodendrogliomas? Cancer Genetics & Cytogenetics 2009;194(1):12-22. [DOI] [PubMed] [Google Scholar]
- Gadji M.Identification and impacts of genetic anomalies in the genesis, clinical development and treatment of gliomas [PhD thesis] [Identification et impacts des anomalies génétiques dans la genèse, l'évolution clinique et le traitement des gliomes]. ProQuest Dissertations & Theses Global 2010;NR83337:285. [Google Scholar]
Ghasimi 2016 {published data only}
- Ghasimi S, Wibom C, Dahlin AM, Brannstrom T, Golovleva I, Andersson U, et al.Genetic risk variants in the CDKN2A/B, RTEL1 and EGFR genes are associated with somatic biomarkers in glioma. Journal of Neuro-Oncology 2016;127(3):483-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Harada 2011 {published data only}
- Harada S, Henderson LB, Eshleman JR, Gocke CD, Burger P, Griffin CA, et al.Genomic changes in gliomas by single nucleotide polymorphism (SNP) array in formalin-fixed paraffin-embedded (FFPE) tissue. Laboratory Investigation 2011;1:381A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S, Henderson LB, Eshleman JR, Gocke CD, Burger P, Griffin CA, et al.Genomic changes in gliomas detected using single nucleotide polymorphism array in formalin-fixed, paraffin-embedded tissue: superior results compared with microsatellite analysis. Journal of Molecular Diagnostics 2011;13(5):541-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Hatanpaa 2003a {published data only}
- Hatanpaa KJ, Burger PC, Eshleman JR, Murphy KM, Berg KD.Molecular diagnosis of oligodendroglioma in paraffin sections. Laboratory Investigation 2003;83(3):419-28. [DOI] [PubMed] [Google Scholar]
Hatanpaa 2003b {published data only}
- Hatanpaa KJ, Burger PC, Eshleman JR, Murphy KM, Berg KD.Molecular diagnosis of oligodendroglioma in paraffin sections. Laboratory Investigation 2003;83(3):419-28. [DOI] [PubMed] [Google Scholar]
Hinrichs 2016 {published data only}
- Hinrichs BH, Newman S, Appin CL, Dunn W, Cooper L, Pauly R, et al.Farewell to GBM-O: genomic and transcriptomic profiling of glioblastoma with oligodendroglioma component reveals distinct molecular subgroups. Acta Neuropathologica Communications 2016;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Horbinski 2012 {published data only}
- Horbinski C, Nikiforova MN, Hobbs J, Bortoluzzi S, Cieply K, Dacic S, et al.The importance of 10q status in an outcomes-based comparison between 1p/19q fluorescence in situ hybridization and polymerase chain reaction-based microsatellite loss of heterozygosity analysis of oligodendrogliomas. Journal of Neuropathology & Experimental Neurology 2012;71(1):73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jeuken 2006 {published data only}
- Jeuken J, Comelissen S, Boots-Sprenger S, Gijsen S, Wesseling P.Multiplex ligation-dependent probe amplification: a diagnostic tool for simultaneous identification of different genetic markers in glial tumors. Journal of Molecular Diagnostics 2006;8(4):433-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeuken JW, Boots-Sprenger SH, Wesseling P, Natte R, Eijk R, Eilers P, et al.Re: multiplex ligation dependent probe amplification for the detection of 1P and 19Q loss in oligodendroglial tumors (multiple letters). Brain Pathology 2005;15(4):364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jha 2011 {published data only}
- Jha P, Sarkar C, Pathak P, Sharma MC, Kale SS, Gupta D, et al.Detection of allelic status of 1p and 19q by microsatellite-based PCR versus FISH: limitations and advantages in application to patient management. Diagnostic Molecular Pathology 2011;20(1):40-7. [DOI] [PubMed] [Google Scholar]
Kato 2019 {published data only}
- Kato Y, Yanagita E, Wang L, Aimono E, Tanaka S, Nishihara H.Detection of 1p19q codeletion by targeted sequencing for glioma genotyping [abstract]. Brain Pathology 2019;29(Suppl 1):118. [Google Scholar]
- Nishihara H, Tanishima S, Yuzawa S, Wang L, Yamaguchi S, Kobayashi H, et al.Genotyping of glioma including 1P19Q codeletion by targeted sequencing. Neuro-Oncology 2016;18(Suppl 6):vi118. [Google Scholar]
- Nishihara H.Genotyping of glioma including 1p19q codeletion by targeted sequencing [SST5-2]. Cancer Science 2018;109(Suppl 1):813. [Google Scholar]
Kolhe 2016 {published data only}
- Kolhe R, Chaubey A, DuPont BR, Lee WS, Mondal AK, Rojiani A.Utility of whole genome single nucleotide polymorphism microarray (SNPM) and targeted somatic mutations analysis in the evaluation of adult brain tumors. Laboratory Investigation 2016;1:432A. [Google Scholar]
Lass 2013 {published data only}
- Lass U, Hartmann C, Capper D, Herold-Mende C, Deimling A, Meiboom M, et al.Chromogenic in situ hybridization is a reliable alternative to fluorescence in situ hybridization for diagnostic testing of 1p and 19q loss in paraffin-embedded gliomas. Brain Pathology 2013;23(3):311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Lhotska 2015 {published data only}
- Lhotska H, Zemanova Z, Cechova H, Ransdorfova S, Lizcova L, Kramar F, et al.Genetic and epigenetic characterization of low-grade gliomas reveals frequent methylation of the MLH3 gene. Genes, Chromosomes & Cancer 2015;54(11):655-67. [DOI] [PubMed] [Google Scholar]
- Lhotska H, Zemanova Z, Kramar F, Lizcova L, Svobodova K, Ransdorfova S, et al.Molecular cytogenetic analysis of chromosomal aberrations in cells of low grade gliomas and its contribution for tumour classification [Molekularne cytogeneticka analyza chromozomovych aberaci v bunkach nizkostupnovych gliomu a jeji prinos pro klasifikaci nadoru]. Klinicka Onkologie 2014;27(3):183-91. [DOI] [PubMed] [Google Scholar]
- Lhotska H.Unbalanced changes in cancer cells genome and its role in cancer pathogenesis [PhD thesis] [Nebalancované zm?ny v genomu nádorových bun?k a jejich úloha v patogenezi onemocn?ní]. Prague (Czech Republic): Univerzita Karlova, 2017. [Google Scholar]
Mohapatra 2006 {published data only}
- Mohapatra G, Betensky RA, Miller ER, Carey B, Gaumont LD, Engler DA, et al.Glioma test array for use with formalin-fixed, paraffin-embedded tissue: array comparative genomic hybridization correlates with loss of heterozygosity and fluorescence in situ hybridization. 97th Annual Meeting of the American Association for Cancer Research; 2006 April 1-5; Washington (DC). [DOI] [PMC free article] [PubMed]
- Mohapatra G, Betensky RA, Miller ER, Carey B, Gaumont LD, Engler DA, et al.Glioma test array for use with formalin-fixed, paraffin-embedded tissue: array comparative genomic hybridization correlates with loss of heterozygosity and fluorescence in situ hybridization. Journal of Molecular Diagnostics 2006;8(2):268-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Na 2019 {published data only}
- Na K, Kim HS, Shim HS, Chang JH, Kang SG, Kim SH.Targeted next-generation sequencing panel (TruSight Tumor 170) in diffuse glioma: a single institutional experience of 135 cases. Journal of Neuro-Oncology 2019;142(3):445-54. [DOI] [PubMed] [Google Scholar]
Natte 2005 {published data only}
- Natte R, Eijk R, Eilers P, Cleton-Jansen AM, Oosting J, Kouwenhove M, et al.Multiplex ligation-dependent probe amplification for the detection of 1p and 19q chromosomal loss in oligodendroglial tumors. Brain Pathology 2005;15(3):192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Nigro 2001 {published data only}
- Nigro JM, Takahashi MA, Ginzinger DG, Law M, Passe S, Jenkins RB, et al.Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real-time quantitative polymerase chain reaction assay. American Journal of Pathology 2001;158(4):1253-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Park 2019 {published data only}
- Park H, Chun SM, Shim J, Oh JH, Cho EJ, Hwang HS, et al.Detection of chromosome structural variation by targeted next-generation sequencing and a deep learning application. Scientific Reports 2019;9(1):3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Paxton 2015 {published data only}
- Paxton CN, Rowe LR, South ST.Observations of the genomic landscape beyond 1p19q deletions and EGFR amplification in Glioma. Cytogenetic and Genome Research 2014;142(3):247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxton CN, Rowe LR, South ST.Observations of the genomic landscape beyond 1p19q deletions and EGFR amplification in glioma. Molecular Cytogenetics 2015;8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Pesenti 2017 {published data only}
- Pesenti C, Paganini L, Fontana L, Veniani E, Runza L, Ferrero S, et al.Mass spectrometry-based assay for the molecular diagnosis of glioma: concomitant detection of chromosome 1p/19q codeletion, and IDH1, IDH2, and TERT mutation status. Oncotarget 2017;8(34):57134-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Ransom 1992a {published data only}
- Ransom DT, Ritland SR, Kimmel DW, Moertel CA, Dahl RJ, Scheithauer BW, et al.Cytogenetic and loss of heterozygosity studies in ependymomas, pilocytic astrocytomas, and oligodendrogliomas. Genes Chromosomes and Cancer 1992;5(4):348-56. [DOI] [PubMed] [Google Scholar]
Ransom 1992b {published data only}
- Ransom DT, Ritland SR, Moertel CA, Dahl RJ, O'Fallon JR, Scheithauer BW, et al.Correlation of cytogenetic analysis and loss of heterozygosity studies in human diffuse astrocytomas and mixed oligo-astrocytomas. Genes Chromosomes and Cancer 1992;5(4):357-74. [DOI] [PubMed] [Google Scholar]
Scheie 2006 {published data only}
- Scheie D, Andresen PA, Cvancarova M, Bo AS, Helseth E, Skullerud K, et al.Fluorescence in situ hybridization (FISH) on touch preparations: a reliable method for detecting loss of heterozygosity at 1p and 19q in oligodendroglial tumors. American Journal of Surgical Pathology 2006;30(7):828-37. [DOI] [PubMed] [Google Scholar]
- Scheie D.1p/19q loss in oligodendroglial tumors-methodological, histological, radiological and clinical aspects. Clinical Neuropathology 2012;31(6):464. [Google Scholar]
- Scheie D.1p/19q-loss in Oligodendroglial Tumours. Methodological, Histological, Radiological and Clinical Aspects [PhD thesis]. Oslo (Norway): University of Oslo, 2012. [ISBN:9788282641883] [Google Scholar]
Schrock 1994 {published data only}
- Schrock E, Thiel G, Lozanova T, du Manoir S, Meffert MC, Jauch A, et al.Comparative genomic hybridization of human malignant gliomas reveals multiple amplification sites and nonrandom chromosomal gains and losses. American Journal of Pathology 1994;144(6):1203-18. [PMC free article] [PubMed] [Google Scholar]
Senetta 2013 {published data only}
- Senetta R, Verdun di Cantogno L, Chiusa L, Castellano I, Gugliotta P, Sapino A, et al.A "weighted" fluorescence in situ hybridization strengthens the favorable prognostic value of 1p/19q codeletion in pure and mixed oligodendroglial tumors. Journal of Neuropathology & Experimental Neurology 2013;72(5):432-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sim 2018a {published data only}
- Sim J, Nam DH, Kim Y, Lee IH, Choi JW, Sa JK, et al.Comparison of 1p and 19q status of glioblastoma by whole exome sequencing, array-comparative genomic hybridization, and fluorescence in situ hybridization. Medical Oncology 2018;35(5):60. [DOI] [PubMed] [Google Scholar]
- Suh YL, Sim J.Comparison of 1p19q status by FISH and whole exome sequencing and/or array-CGH in glioblastoma. Clinical Neuropathology 2016;35(4):228. [Google Scholar]
Sim 2018b {published data only}
- Sim J, Nam DH, Kim Y, Lee IH, Choi JW, Sa JK, et al.Comparison of 1p and 19q status of glioblastoma by whole exome sequencing, array-comparative genomic hybridization, and fluorescence in situ hybridization. Medical Oncology 2018;35(5):60. [DOI] [PubMed] [Google Scholar]
- Suh YL, Sim J.Comparison of 1p19q status by FISH and whole exome sequencing and/or array-CGH in glioblastoma. Clinical Neuropathology 2016;35(4):228. [Google Scholar]
Smith 1999 {published data only}
- Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, Mohapatra G, et al.Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene 1999;18(28):4144-52. [DOI] [PubMed] [Google Scholar]
Srebotnik‐Kirbis 2016 {published data only}
- Limbaeck-Stokin C, Srebotnik-Kirbis I, Zupan A, Glavac D, Popovic M.Brush cytology for FISH based detection of 1p/19q loss in oligodendroglial tumors. Clinical Neuropathology 2012;31(4):256. [Google Scholar]
- Srebotnik-Kirbis I, Limback-Stokin C.Application of brush cytology for FISH-based detection of 1p/19q codeletion in oligodendroglial tumors. Journal of Neuro-Oncology 2016;129(3):415-22. [DOI] [PubMed] [Google Scholar]
Thakur 2012 {published data only}
- Thakur S, Alvarez K, Weindel M, Li MM, Monzon FA.Validation of affymetrix 250K Nsp arrays for clinical use in the evaluation of genomic copy number alteration and loss of heterozygosity in cancer. Journal of Molecular Diagnostics 2012;14(6):707-8. [Google Scholar]
Thomas 2017 {published data only}
- Thomas AA, Abrey LE, Terziev R, Raizer J, Martinez NL, Forsyth P, et al.Multicenter phase II study of temozolomide and myeloablative chemotherapy with autologous stem cell transplant for newly diagnosed anaplastic oligodendroglioma. Neuro-Oncology 2017;19(10):1380-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Tsiatis 2010 {published data only}
- Tsiatis AC, Hafez MJ, Jedlicka AE, Maitra A, Murphy KM, Eshleman JR.Use of SNP arrays to assess loss of heterozygosity in gliomas. 101st Annual Meeting of American Association for Cancer Research; 2010 April 17-21; Washington (DC). [DOI: 10.1158/1538-7445.AM10-1781] [DOI]
Uchida 2019 {published data only}
- Uchida H, Yonezawa H, Hirano H, Yoshimoto K.Usefulness and pitfalls of 1p/ 19q-codeletion analysis by FISH method in glioblastoma. Brain Pathology 2019;29(Suppl 1):120. [Google Scholar]
Wiestler 2014 {published data only}
- Wiestler B, Capper D, Hovestadt V, Sill M, Jones D, Hartmann C, et al.Assessing CpG island methylator phenotype, 1p/19q codeletion, and MGMT promoter methylation from epigenome-wide data in the biomarker cohort of the NOA-04 trial. Neuro-Oncology 2014;16(12):1630-8. [DOI] [PMC free article] [PubMed]
- Wiestler B, Capper D, Hovestadt V, Sill M, Jones D, Hartmann C, et al.Determining the glioma CpG island methylator phenotype, 1p/19q codeletion, and MGMT promoter methylation from epigenome-wide methylation data in the biomarker cohort of the NOA-04 trial. Journal of Clinical Oncology 2014;32:15. [DOI: 10.1200/jco.2014.32.15_suppl.2017] [DOI] [PMC free article] [PubMed]
References to studies excluded from this review
Afyounian 2017 {published data only}
- Afyounian E, Annala M, Nykter M.Segmentum: a tool for copy number analysis of cancer genomes. BMC Bioinformatics 2017;18(1):215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Alentorn 2014 {published data only}
- Alentorn A, Thuijl HF, Marie Y, Alshehhi H, Carpentier C, Boisselier B, et al.Clinical value of chromosome arms 19q and 11p losses in low-grade gliomas. Neuro-Oncology 2014;16(3):400-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Aoki 2015a {published data only}
- Aoki K, Suzuki H, Nakamura H, Motomura K, Mizoguchi M, Abe T, et al.Prognostic model of lower grade gliomas. Journal of Clinical Oncology 2015;33(15 Suppl 1):2038. [Google Scholar]
Aoki 2015b {published data only}
- Aoki K, Suzuki H, Nakamura H, Motomura K, Ohka F, Mizoguchi M, et al.Prognostic significance of genetic alterations from comprehensive analysis in lower-grade gliomas. Neuro-Oncology 2015;5:v91. [Google Scholar]
Assem 2009 {published data only}
- Assem M, Sibenaller Z, Marner J, Bair T, Ryken TC.Genomic profiling as a tool to improve diagnosis and treatment of malignant gliomas. Proceedings of the American Association for Cancer Research Annual Meeting 2009;50:341. [Google Scholar]
Assem 2012 {published data only}
- Assem M, Sibenaller Z, Agarwal S, Al-Keilani MS, Alqudah MA, Ryken TC.Enhancing diagnosis, prognosis, and therapeutic outcome prediction of gliomas using genomics. Omics a Journal of Integrative Biology 2012;16(3):113-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Bady 2013 {published data only}
- Bady P, Kurscheid S, Delorenzi M, Hegi ME.The genetic and epigenetic context of MGMT methylation in glioma may impact the predictive and prognostic value. Neuro-Oncology 2013;3:iii141-2. [Google Scholar]
Ballester 2017 {published data only}
- Ballester LY, Huse JT, Tang G, Fuller GN.Molecular classification of adult diffuse gliomas: conflicting IDH1/IDH2, ATRX, and 1p/19q results. Human Pathology 2017;69:15-22. [DOI] [PubMed] [Google Scholar]
Becker 2017 {published data only}
- Becker AP, Bell EH, Fleming J, McElroy JP, Fabian D, Beyer S, et al.Comprehensive assessment of ATRX mutation, protein expression, and alternative lengthening of telomeres (ALT) phenotype in grade II and III gliomas. Journal of Clinical Oncology 2017;35(15 Suppl 1):2064. [Google Scholar]
Bienkowski 2018 {published data only}
- Bienkowski M, Wohrer A, Moser P, Kitzwogerer M, Ricken G, Strobel T, et al.Molecular diagnostic testing of diffuse gliomas in the real-life setting: a practical approach. Clinical Neuropathology 2018;37(4):166-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Boudreau 2004 {published data only}
- Boudreau CR, Liau LM.Molecular characterization of brain tumors. Clinical Neurosurgery 2004;51:81-90. [PubMed] [Google Scholar]
Brat 2015 {published data only}
- Brat DJ, Fehrenbach A, Finocchiaro G, Verhaak RG, Friedman W, Al-Dape KD, et al.Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. New England Journal of Medicine 2015;372(26):2481-98. [DOI] [PMC free article] [PubMed]
Buckley 2011 {published data only}
- Buckley PG, Alcock L, Heffernan J, Woods J, Brett F, Stallings RL, et al.Loss of chromosome 1p/19q in oligodendroglial tumors: refinement of chromosomal critical regions and evaluation of internexin immunostaining as a surrogate marker. Journal of Neuropathology & Experimental Neurology 2011;70(3):177-82. [DOI] [PubMed] [Google Scholar]
Burgenske 2017 {published data only}
- Burgenske D, Eckel-Passow J, Decker P, Kosel M, Youland R, Remonde D, et al.Clinical and molecular analyses of long term survivors of glioblastoma. Neuro-Oncology 2017;19(Suppl 6):vi98. [Google Scholar]
Bystricka 2011 {published data only}
- Bystricka D, Zemanova Z, Kramar F, Lizcova L, Ransdorfova S, Dostalova-Merkerova M, et al.Genomic changes in 37 patients with low-grade diffuse gliomas. Chromosome Research 2011;1:S146-7. [Google Scholar]
Carrato 2006 {published data only}
- Carrato C, Beyer K, Zamora L, Granada I, Roussos I, Balana C, et al.1p/19q loss in gliomas: microsatellite amplification in comparison with FISH. Laboratory Investigation 2006;86(Suppl 1):284A. [Google Scholar]
Carrato 2010 {published data only}
- Carrato C, Beyer K, Md L, Domingo-Sabat M, Lopez MT, Ariza A.A rapid, user-friendly approach for distinguishing between partial and complete 1p/19q losses in gliomas. Brain Pathology 2010;1:60. [Google Scholar]
Carter 2016 {published data only}
- Carter JH, McNulty S, Cottrell C, Heusel J.Targeted NGS in molecular subtyping of lower-grade diffuse gliomas: application of the WHO's 2016 revised criteria for CNS tumors. Journal of Molecular Diagnostics 2016;18(6):1009. [DOI] [PubMed] [Google Scholar]
Castilla 2003 {published data only}
- Castilla EA, Prayson RA, Hartke M, Staugaitis SM, Barnett GH, Tubbs RR.Discordance in allelic losses on chromosomes 1p and 19q in gliomas. Modern Pathology 2003;16(1):289A. [Google Scholar]
Chernova 2003 {published data only}
- Chernova OB, Barnett GH, Cowell JK.Rapid detection of allelic losses in brain tumours using microsatellite repeat markers and high-performance liquid chromatography. British Journal of Cancer 2003;88(12):1889-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Cieply 2005 {published data only}
- Cieply KM, Sherer C, Hunt J, Hamilton RL.Assessment of 1p and 19q status in pediatric oligodendrogliomas by FISH. Journal of Molecular Diagnostics 2005;7(5):688. [Google Scholar]
Durand 2010 {published data only}
- Durand KS, Guillaudeau A, Weinbreck N, De Armas R, Robert S, Chaunavel A, et al.1p19q LOH patterns and expression of p53 and Olig2 in gliomas: relation with histological types and prognosis. Modern Pathology 2010;23(4):619-28. [DOI] [PubMed] [Google Scholar]
Eckel‐Passow 2017 {published data only}
- Eckel-Passow J, Decker P, Hughes E, Kollmeyer T, Kosel M, Burgenske D, et al.Clinical sensitivity and specificity of illumina methylation array for classifying adult gliomas into who groups. Neuro-Oncology 2017;19(Suppl 6):vi181. [Google Scholar]
Fontaine 2007 {published data only}
- Fontaine D, Monnot S, Vandenbos F, Paquis P, Michiels JF, Bannwarth S, et al.DNA extraction by FTATM technology: application for rapid detection of 1p/19q deletions in gliomas. Neuropathology and Applied Neurobiology 2007;33(3):360-3. [DOI] [PubMed] [Google Scholar]
Franco‐Hernandez 2009a {published data only}
- Franco-Hernandez C, Martinez-Glez V, Campos JM, Isla A, Vaquero J, Gutiérrez M, et al.Allelic status of 1p and 19q in oligodendrogliomas and glioblastomas: multiplex ligation-dependent probe amplification versus loss of heterozygosity. Cancer Genetics & Cytogenetics 2009;190(2):93-6. [DOI] [PubMed] [Google Scholar]
Franco‐Hernandez 2009b {published data only}
- Franco-Hernandez C, Martinez-Glez V, Torres-Martin M, Campos JM, Isla A, Vaquero J, et al.Identification of genetic alterations by multiple ligation-dependent probe amplification (MLPA) analysis in oligodendrogliomas. Neurocirugia (Asturias, Spain) 2009;20(2):117-23. [PubMed] [Google Scholar]
French 2005 {published data only}
- French PJ, Swagemakers SM, Nagel JH, Kouwenhoven MC, Brouwer E, van der Spek, et al.Gene expression profiles associated with treatment response in oligodendrogliomas. Cancer Research 2005;65(24):11335-44. [DOI] [PubMed] [Google Scholar]
Garber 2016 {published data only}
- Garber ST, Hashimoto Y, Weathers SP, Xiu J, Gatalica Z, Verhaak RG, et al.Immune checkpoint blockade as a potential therapeutic target: surveying CNS malignancies. Neuro-Oncology 2016;18(10):1357-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Hartmann 2005 {published data only}
- Hartmann C, Mueller W, Lass U, Kamel-Reid S, Deimling A.Molecular genetic analysis of oligodendroglial tumors. Journal of Neuropathology & Experimental Neurology 2005;64(1):10-4. [DOI] [PubMed] [Google Scholar]
Hashimoto 2002 {published data only}
- Hashimoto N, Murakami M, Sasajima H, Takahashi Y, Mineura K.Diagnostic molecular genetics and its application to clinical decision in brain tumors. In: Watanabe K, editors(s). Developments in Neuroscience, Proceedings. 1247 edition. Amsterdam (the Netherlands): Elsevier Science Bv, 2002:221-9. [Google Scholar]
Hench 2018 {published data only}
- Hench J, Bihl M, Bratic Hench I, Hoffmann P, Tolnay M, Bösch Al Jadooa N, et al.Satisfying your neuro-oncologist: a fast approach to routine molecular glioma diagnostics. Neuro-Oncology 2018;20(12):1682-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Horbinski 2008 {published data only}
- Horbinski C, Dacic S, Cieply K, Brat DJ, McClendon RE, Chu CT.Chordoid glioma: molecular characterization of four cases. Laboratory Investigation 2008;88(Suppl 1):321A. [Google Scholar]
Horbinski 2011 {published data only}
- Horbinski C, Nikiforova M, Hobbs J, Cieply K, Hamilton R.Prospective comparison of FISH versus PCR-based microsatellite LOH in the evaluation of gliomas for 1p/19q codeletion. Journal of Neuropathology and Experimental Neurology 2011;70(6):526. [Google Scholar]
Ida 2018 {published data only}
- Ida C, Zepeda-Mendoza C, Praska C, Balcom J, Swanson K, Barr Fritcher E, et al.Recurrent unusual patterns in clinical molecular profiling of adult diffuse gliomas. Journal of Neuropathology and Experimental Neurology 2018;77(6):482.
Idbaih 2008 {published data only}
- Idbaih A, Kouwenhoven M, Jeuken J, Carpentier C, Gorlia T, Kros JM, et al.Chromosome 1p loss evaluation in anaplastic oligodendrogliomas. Neuropathology 2008;28(4):440-3. [DOI] [PubMed] [Google Scholar]
Joo 2013 {published data only}
- Joo M, Park SH, Chang SH, Kim H, Choi CY, Lee CH, et al.Cytogenetic and molecular genetic study on granular cell glioblastoma: a case report. Human Pathology 2013;44(2):282-8. [DOI] [PubMed] [Google Scholar]
Juratli 2012 {published data only}
- Juratli TA, Kirsch M, Geiger K, Klink B, Leipnitz E, Pinzer T, et al.The prognostic value of IDH mutations and MGMT promoter status in secondary high-grade gliomas. Journal of Neuro-Oncology 2012;110(3):325-33. [DOI] [PubMed] [Google Scholar]
Kamoun 2015 {published data only}
- Kamoun A, Ducray F, Dehais C, Idbaih A, Elarouci N, Letouze E, et al.Integrated genomic analysis of oligodendroglial tumors identifies distinct molecular subgroups within 1p/19q co-deleted oligodendrogliomas. Neuro-Oncology 2015;5:v95. [Google Scholar]
Kashofer 2018 {published data only}
- Kashofer K, Halbwedl I, Winter E, Hoefler G, Asslaber M.Expanding on WHO guideline compliant molecular testing of central nervous system tumours by low density whole genome sequencing. Virchows Archiv 2018;473(Suppl 1):s28. [Google Scholar]
Kim 2016 {published data only}
- Kim L, Xiu J, Judy K, Evans J, Andrews D, Farrell C.Results of molecular profiling for recurrent malignant gliomas reveal significant changes in biomarkers compared to mostly treatment naive tumors that could impact treatment decision. Neuro-Oncology 2016;18(Suppl 6):vi117-8. [Google Scholar]
Kitange 2004 {published data only}
- Kitange GJ, Misra A, Law M, Passe S, Kollymeyer T, Mauer M, et al.Analysis of genetic dosage anomalies using comparative genomic hybridization array (CGHa) in human oligodendrogliomas. Proceedings of the American Association for Cancer Research Annual Meeting 2004;45:386. [Google Scholar]
Kitange 2005 {published data only}
- Kitange G, Misra A, Law M, Passe S, Kollmeyer TM, Maurer M, et al.Chromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas. Genes, Chromosomes & Cancer 2005;42(1):68-77. [DOI] [PubMed] [Google Scholar]
Klink 2010 {published data only}
- Klink B, Schlingelhof B, Klink M, Stout-Weider K, Patt S, Schrock E.Glioblastomas with oligodendroglial component – common origin of the different histological parts and genetic subclassification. Analytical Cellular Pathology 2010;33(1):37-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Klink 2011 {published data only}
- Klink B, Schlingelhof B, Klink M, Stout-Weider K, Patt S, Schrock E.Glioblastomas with oligodendroglial component-common origin of the different histological parts and genetic subclassification. Cellular Oncology 2011;34(3):261-75. [DOI] [PubMed] [Google Scholar]
Kouwenhoven 2009 {published data only}
- Kouwenhoven MC, Gorlia T, Kros JM, Ibdaih A, Brandes AA, Bromberg JE, et al.Molecular analysis of anaplastic oligodendroglial tumors in a prospective randomized study: a report from EORTC study 26951. Neuro-Oncology 2009;11(6):737-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kuo 2009 {published data only}
- Kuo LT, Kuo KT, Lee MJ, Wei CC, Scaravilli F, Tsai JC, et al.Correlation among pathology, genetic and epigenetic profiles, and clinical outcome in oligodendroglial tumors. International Journal of Cancer 2009;124(12):2872-9. [DOI] [PubMed] [Google Scholar]
Kuo 2013 {published data only}
- Kuo LT, Tsai SY, Chang CC, Kuo KT, Huang AP, Tsai JC, et al.Genetic and epigenetic alterations in primary-progressive paired oligodendroglial tumors. PLoS One 2013;8(6):e67139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Kwon 2019 {published data only}
- Kwon MJ, Suh YL, Cho H, Kang SY.Molecular subgrouping of gliomatosis cerebri according to the 2016 World Health Organization classification of tumors of the central nervous system. Brain Pathology 2019;29(Suppl 1):43-4. [Google Scholar]
Lautenschlaeger 2013 {published data only}
- Lautenschlaeger T, Juratli TA, McElroy J, Meng W, Huebner A, Geiger KD, et al.Genome-wide CNV analysis identifies loci putatively associated with delayed time to first recurrence in IDH-mutant low-grade astrocytomas. Neuro-Oncology 2013;3:iii145. [Google Scholar]
Levine 2018 {published data only}
- Levine AB, Yip S.Implementation of the 2016 who classification for infiltrating glioma: the VGH experience. Laboratory Investigation 2018;98(Suppl 1):660. [Google Scholar]
Liu 2014 {published data only}
- Liu EZ, Abreu FB, Hickey WF, Ronan LK, Fadul CE, Otis L, et al.Development of a 1p19q LOH assay for glioma patients using capillary electrophoresis. Journal of Molecular Diagnostics 2014;16(6):789. [Google Scholar]
Magnani 2003 {published data only}
- Magnani I, Moroni RF, Beghini A, Larizza L.Multiple alterations of chromosome arms 1p and 19q detected by FISH analysis correlate with LOH heterogeneity in gliomas. Annales de Genetique 2003;46(2-3):165. [Google Scholar]
Martinez 2005 {published data only}
- Martinez R, Schackert HK, Kirsch M, Paulus W, Joos S, Schackert G.Comparative genetic analysis of metachronous anaplastic oligoastrocytomas with extended recurrence-free interval. Journal of Neuro-Oncology 2005;72(2):95-102. [DOI] [PubMed] [Google Scholar]
Marucci 2012 {published data only}
- Marucci G, Di Oto, Farnedi A, Panzacchi R, Ligorio C, Foschini MP.Nogo-A: a useful marker for the diagnosis of oligodendroglioma and for identifying 1p19q codeletion. Human Pathology 2012;43(3):374-80. [DOI] [PubMed] [Google Scholar]
McDonald 2005 {published data only}
- McDonald JM, See SJ, Tremont IW, Colman H, Gilbert MR, Groves M, et al.The prognostic impact of histology and 1p/19q status in anaplastic oligodendroglial tumors. Cancer 2005;104(7):1468-77. [DOI] [PubMed] [Google Scholar]
Mohapatra 2011 {published data only}
- Mohapatra G, Engler DA, Starbuck KD, Kim JC, Bernay DC, Scangas GA, et al.Genome-wide comparison of paired fresh frozen and formalin-fixed paraffin-embedded gliomas by custom BAC and oligonucleotide array comparative genomic hybridization: facilitating analysis of archival gliomas. Acta Neuropathologica 2011;121(4):529-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Molinari 2010 {published data only}
- Molinari C, Iorio P, Medri L, Ballardini M, Guiducci G, Cremonini AM, et al.Chromosome 1p and 19q evaluation in low-grade oligodendrogliomas: a descriptive study. International Journal of Molecular Medicine 2010;25(1):145-51. [PubMed] [Google Scholar]
Mrachek 2018 {published data only}
- Mrachek EK, Batchala P, Patel S, Fadul C, Schiff D, Williams E, et al.Neuroradiology concordance with neuropathology in predicting 1p/19q-codeletion in IDH-mutant lower-grade gliomas. Journal of Neuropathology and Experimental Neurology 2018;77(6):483. [Google Scholar]
Mur 2013 {published data only}
- Mur P, Mollejo M, Ruano Y, Lope AR, Fiano C, García JF, et al.Codeletion of 1p and 19q determines distinct gene methylation and expression profiles in IDH-mutated oligodendroglial tumors. Acta Neuropathologica 2013;126(2):277-89. [DOI] [PubMed] [Google Scholar]
Myung 2011 {published data only}
- Myung JK, Byeon SJ, Kim B, Suh J, Kim SK, Park CK, et al.Papillary glioneuronal tumors: a review of clinicopathologic and molecular genetic studies. American Journal of Surgical Pathology 2011;35(12):1794-805. [DOI] [PubMed] [Google Scholar]
Narasimhaiah 2010 {published data only}
- Narasimhaiah DA, Godfraind C.Fluorescence in situ hybridization (FISH) for 1p/19q in oligodendrogliomas – the need for consensus criteria and uniform guidelines. Brain Pathology 2010;1:55. [Google Scholar]
Neill 2015 {published data only}
- Neill S, Hill K, Adewumi D, Holder C, Rossi M, Schniederjan M.Disseminated oligodendroglioma-like leptomeningeal neoplasm in a 31 year-old patient. Journal of Neuropathology and Experimental Neurology 2015;74(6):635. [Google Scholar]
Nielsen 2007 {published data only}
- Nielsen KV, Rasmussen BB, Balslev E, Poulsen TS, Schonau A, Ejlertsen B.Loss of heterozygosity on chromosomes 1 and 19 in primary brain tumors. Ugeskrift for Laeger 2007;169(2):147. [PubMed] [Google Scholar]
Parizi‐Robinson 2004 {published data only}
- Parizi-Robinson M, Hatanpaa K, Yegappan M, Siegelman MH.Molecular diagnosis of oligodendrogliomas using capillary electrophoresis to detect loss of heterozygosity on chromosomes 1p and 19q. Journal of Molecular Diagnostics 2004;6(4):428. [Google Scholar]
Payne 2008 {published data only}
- Payne C, Parkinson J, Cook R, Wheeler H, Robinson B, McDonald K.Oligodendrogliomas with LOH 1P/19Q: identifying genes associated with therapeutic sensitivity. Neuro-Oncology 2008;10(5):802. [Google Scholar]
Pekmezci 2016 {published data only}
- Pekmezci M, Walsh KM, Molinaro A, Decker PA, Hansen HM, Sicotto H, et al.Molecularly defined adult gliomas with known ATRX status: does TERT have an additional prognostic role. Laboratory Investigation 2016;1:435A. [Google Scholar]
Pietsch 2015 {published data only}
- Pietsch T, Gessi M, Muhlen A, Dörner E, Last A, Sundar P.Implementation of next generation copy number and mutational analysis in routine neuropathology: molecular inversion profiling is a helpful tool in differential diagnostics and prognostification of pediatric brain tumors. Neuro-Oncology 2015;3:iii30-iii.
Pina‐Oviedo 2012 {published data only}
- Pina-Oviedo S, Alvarez K, Powell SZ, Chang CC, Monzon FA.Utility of whole genome amplification (WGA) to enable virtual karyotyping with SNP arrays in paraffin-embedded brain tumor biopsies with limited tissue. Laboratory Investigation 2012;1:434A-5A. [Google Scholar]
Pinkham 2015 {published data only}
- Pinkham MB, Telford N, Whitfield GA, Colaco RJ, O'Neill F, McBain CA.FISHing tips: what every clinician should know about 1p19q analysis in gliomas using fluorescence in situ hybridisation. Clinical Oncology (Royal College of Radiologists) 2015;27(8):445-53. [DOI] [PubMed] [Google Scholar]
Pinto 2008 {published data only}
- Pinto LW, Araujo MB, Vettore AL, Wernersbach L, Leite AC, Chimelli LM, et al.Glioblastomas: correlation between oligodendroglial components, genetic abnormalities, and prognosis. Virchows Archiv 2008;452(5):481-90. [DOI] [PubMed] [Google Scholar]
Ramkissoon 2015 {published data only}
- Ramkissoon SH, Bi WL, Schumacher SE, Ramkissoon LA, Haidar S, Knoff D, et al.Clinical implementation of integrated whole-genome copy number and mutation profiling for glioblastoma. Neuro-Oncology 2015;17(10):1344-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rolston 2002 {published data only}
- Rolston R, Dacic S, Kondziolka D, Sasatomi E, Swalsky PA, Hamilton R, et al.Detection of 1p/19q deletion by microdissection genotyping identifies high grade gliomas patients with long survival. Laboratory Investigation 2002;82(1):302A. [Google Scholar]
Roy 2012 {published data only}
- Roy S, Hagenkord JM, Durso MB, Kash SF, Hamilton RL, Nikiforova MN.Comparison of SNP array karyotyping, FISH and LOH analysis for diagnosis of gliomas. Journal of Molecular Diagnostics 2012, 2012;14(6):698. [Google Scholar]
Satomi 2018 {published data only}
- Satomi K, Yamasaki K, Yoshida A, Wakai S, Matsushita Y, Narita Y, et al.Development of a novel fish probe for detection of 1p/19q codeletion in routine glioma diagnosis. Journal of Pathology 2018;246(Suppl 1):S17. [Google Scholar]
Satomi 2019 {published data only}
- Satomi K, Yamasaki K, Yoshida A, Wakai S, Matsushita Y, Narita Y, et al.Development of a novel FISH probe for detection of 1p/19q codeletion in routine glioma diagnosis. Brain Pathology 2019;29(Suppl 1):124-5. [Google Scholar]
Scheinin 2014 {published data only}
- Scheinin I, Thuijl HF, Sie D, Essen HF, Eijk PP, Rustenburg F, et al.A novel approach to copy number assessment by whole genome sequencing reveals extensive spatial heterogeneity in diffuse low-grade glioma. Cancer Research 2014;74(19 Suppl 1):3426. [Google Scholar]
Schiavo 2009 {published data only}
- Schiavo N, Brunelli M, Martignoni G, Menestrina F, Gobbo S, Eccher A, et al.Patterns of 1p/19q chromosomal abnormalities among glial neoplasms. Virchows Archiv 2009;1:S400. [Google Scholar]
Serrano 2015 {published data only}
- Serrano J, Forrester L, Kannan K, Faustin A, Thomas C, Capper D, et al.Improving molecular diagnostics with 450K methylation array in clinical neuropathology. Journal of Molecular Diagnostics 2015;17(6):815-6. [Google Scholar]
Tauziede‐Espariat 2018 {published data only}
- Tauziede-Espariat A, Saffroy R, Pages M, Pallud J, Legrand L, Besnard A, et al.Cerebellar high-grade gliomas do not present the same molecular alterations as supratentorial high-grade gliomas and may show histone H3 gene mutations. Clinical Neuropathology 2018;37(5):209-16. [DOI] [PubMed] [Google Scholar]
Walker 2000 {published data only}
- Walker C, Joyce KA, Machell Y, Thompson-Hehir J, Sibson DR, Broome J, et al.Analysis of genetic alterations in microdissected gliomas. Brain Pathology 2000;10(4):576. [Google Scholar]
Woehrer 2015 {published data only}
- Woehrer A, Hainfellner JA.Molecular diagnostics: techniques and recommendations for 1p/19q assessment. CNS Oncology 2015;4(5):295-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Xiu 2015 {published data only}
- Xiu J, Spetzler D, Bender R, Ghazalpour A, Gatalica Z, Reddy SK, et al.Tumor profiling on 1245 gliomas and paired tumor study on 19 high grade gliomas. Journal of Clinical Oncology 2015;33(15 Suppl 1):2058. [Google Scholar]
Yokogami 2018 {published data only}
- Yokogami K, Yamasaki K, Matsumoto F, Yamashita S, Saito K, Tacheva A, et al.Impact of PCR-based molecular analysis in daily diagnosis for the patient with gliomas. Brain Tumor Pathology 2018;35(3):141-7. [DOI] [PubMed] [Google Scholar]
Yoshimoto 2002 {published data only}
- Yoshimoto K, Iwaki T, Inamura T, Fukui M, Tahira T, Hayashi K.Multiplexed analysis of post-PCR fluorescence-labeled microsatellite alleles and statistical evaluation of their imbalance in brain tumors. Japanese Journal of Cancer Research 2002;93(3):284-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Zacher 2017 {published data only}
- Zacher A, Kaulich K, Stepanow S, Wolter M, Koehrer K, Felsberg J, et al.Molecular diagnostics of gliomas using next generation sequencing of a glioma-tailored gene panel. Brain Pathology 2017;27(2):146-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Zheng 2019 {published data only}
- Zheng S, Alfaro-Munoz K, Wei W, Wang X, Wang F, Eterovic AK, et al.Prospective clinical sequencing of adult glioma. Molecular Cancer Therapeutics 2019;18(5):991-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
References to studies awaiting assessment
Ducray 2011 {published data only}
- Ducray F.Prognostic and predictive biomarkers of gliomas in adults. Pratique Neurologique - FMC 2011;2(2):64-70. [Google Scholar]
Hazra 2006 {published data only}
- Hazra, Asmita.Prevalence of Alterations at Microsatellite Loci on 1p and 19q in Primary Human Gliomas (thesis). Ann Arbor (MI), 2006. [Google Scholar]
McDonald 2003 {published data only}
- McDonald M, Aldape K, Taylor E, Huang J, Hess K, Sawaya R, et al.Genetic and epigenetic alterations of oligodendrogliomas revealed by gene mapping and pathway transcriptome profiling. Proceedings of the American Association for Cancer Research Annual Meeting 2003;44:1216. [Google Scholar]
Meunier 2005 {published data only}
- Meunier N.Fluorescence in situ (FISH) assay for the detection of 1p36 and 19q13 deletions in gliomas (technical improvements for a retrospective study of 25 patients). hdl.handle.net/10068/770157 (accessed prior to 28 January 2022).
Monnot 2007 {published data only}
- Monnot S.1p / 19q deletions, in gliomas (study of 46 Nice patients and implementation of new detection methods) [Délétions 1p/19q, dans les gliomes (étude de 46 patients niçois et mise en place de nouvelles méthodes de détection)]. hdl.handle.net/10068/803964 (accessed prior to 28 January 2022).
Sebastian 2003 {published data only}
- Sebastian S, Lister D, Rasheed BK, McLendon RE, Friedman HS, Bigner DD, et al.Validation of a semi-automated loss of heterozygosity assay for chromosomes 1 and 19 in human gliomas using commercially available fluorescent-labeled human MapPairs(R) and capillary electrophoresis. Proceedings of the American Association for Cancer Research Annual Meeting 2003;44:702. [Google Scholar]
References to ongoing studies
ACTRN12618000006246 {unpublished data only}
- ACTRN12618000006246.Access to innovative molecular diagnostic PROfiling for paediatric brain tumours [Application of innovative molecular profiling techniques to improve diagnosis of paediatric central nervous system tumours and develop an accredited Australasian molecular profiling service]. anzctr.org.au/ACTRN12618000006246 (first received 10 January 2018). [AIM-BRAIN PROject]
JPRN‐UMIN000003196 {unpublished data only}
- JPRN-UMIN000003196.Genetic analysis of prognosis-related factors in gliomas: methylation of MGMT, LOH of 1p/19q, and mutation of IDH1/2. upload.umin.ac.jp/cgi-open-bin/ctr_e/ctr_view.cgi?recptno=R000003871 (first received 22 February 2010).
NCT00031538 {unpublished data only}
- NCT00031538.Genetic analysis of brain tumors [A prospective national study to molecularly and genetically characterize human gliomas: the Glioma Molecular Diagnostic Initiative]. clinicaltrials.gov/ct2/show/NCT00031538 (first received 7 March 2002).
NCT01004887 {published data only}
- NCT01004887.Study of tissue and blood samples from patients with high-grade glioma [Diagnostic and prognostic markers in high-grade glioma]. clinicaltrials.gov/ct2/show/NCT01004887 (first received 30 October 2009).
NCT03336931 {published data only}
- NCT03336931.Precision medicine for children with cancer [A multicenter prospective study of the feasibility and clinical value of a diagnostic service for identifying therapeutic targets and recommending personalised treatment for children and adolescents with high-risk cancer]. clinicaltrials.gov/ct2/show/NCT03336931 (first received 8 November 2017).
Additional references
Arends 2008
- Arends LR, Hamza TH, Houwelingen JC, Heijenbrok-Kal MH, Hunink MG, Stijnen T.Bivariate random effects meta-analysis of ROC curves. Medical Decision Making 2008;28(5):621-38. [DOI] [PubMed] [Google Scholar]
Bossuyt 2021
- Bossuyt PM.Chapter 4: Understanding the design of test accuracy studies. Draft version (21 May 2021) for inclusion. In: Deeks JJ, Bossuyt PM, Leeflang MM, Takwoingi Y, editor(s). Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 2. London: Cochrane. Available from training.cochrane.org/handbook-diagnostic-test-accuracy/PDF/v2.
Cairncross 2013
- Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J, et al.Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. Journal of Clinical Oncology 2013;31(3):337-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Capper 2018a
- Capper D, Jones DT, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al.DNA methylation-based classification of central nervous system tumours. Nature 2018;555(7697):469-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Capper 2018b
- Capper D, Stichel D, Sahm F, Jones DT, Schrimpf D, Sill M, et al.Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathologica 2018;136(2):181-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
CCEMG 2019
- CCEMG – EPPI-Centre.CCEMG – EPPI-Centre Cost Converter (v.1.6, 29 April 2019). eppi.ioe.ac.uk/costconversion/ (accessed prior to 28 January 2022).
Chamberlain 2015
- Chamberlain MC, Born D.Prognostic significance of relative 1p/19q codeletion in oligodendroglial tumors. Journal of Neuro-Oncology 2015;125(2):249-51. [DOI] [PubMed] [Google Scholar]
Chu 2006
- Chu H, Cole SR.Bivariate meta-analysis of sensitivity and specificity with sparse data: a generalized linear mixed model approach. Journal of Clinical Epidemiology 2006;59(12):1331-2; author reply 1332-3. [DOI] [PubMed] [Google Scholar]
Chu 2009
- Chu HT, Chen SN, Louis TA.Random effects models in a meta-analysis of the accuracy of two diagnostic tests without a gold standard. Journal of the American Statistical Association 2009;104(486):512-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Daumas‐Duport 1988
- Daumas-Duport C, Scheithauer B, O'Fallon J, Kelly P.Grading of astrocytomas: a simple and reproducible method. Cancer 1988;62(10):2152-65. [DOI] [PubMed] [Google Scholar]
Deeks 2005
- Deeks JJ, Macaskill P, Irwig L.The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. Journal of Clinical Epidemiology 2005;58(9):882-93. [DOI] [PubMed] [Google Scholar]
Dendukuri 2012
- Dendukuri N, Schiller I, Joseph L, Pai M.Bayesian meta-analysis of the accuracy of a test for tuberculous pleuritis in the absence of a gold standard reference. Biometrics 2012;68(4):1285-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Eijk‐Van Os 2011
- Eijk-Van Os PG, Schouten JP.Multiplex ligation-dependent probe amplification (MLPA(R)) for the detection of copy number variation in genomic sequences. Methods in Molecular Biology 2011;688:97-126. [DOI] [PubMed] [Google Scholar]
Griffin 2006
- Griffin CA, Burger P, Morsberger L, Yonescu R, Swierczynski S, Weingart JD, et al.Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. Journal of Neuropathology and Experimental Neurology 2006;65(10):988-94. [DOI] [PubMed] [Google Scholar]
Guyatt 2008
- Guyatt GH, Oxman AD, Vist GE, Kunz R, Falck-Ytter Y, Alonso-Coello P, et al.GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ 2008;336(7650):924-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Harbord 2007
- Harbord RM, Deeks JJ, Egger M, Whiting P, Sterne JA.A unification of models for meta-analysis of diagnostic accuracy studies. Biostatistics 2007;8(2):239-51. [DOI] [PubMed] [Google Scholar]
Hu 2016
- Hu N, Richards R, Jensen R.Role of chromosomal 1p/19q co-deletion on the prognosis of oligodendrogliomas: a systematic review and meta-analysis. Interdisciplinary Neurosurgery: Advanced Techniques and Case Management 2016;5:58-63. [Google Scholar]
Husereau 2013
- Husereau D, Drummond M, Petrou S, Carswell C, Moher D, Greenberg D, et al.Consolidated Health Economic Evaluation Reporting Standards (CHEERS) statement. BMJ 2013;346:f1049. [DOI] [PubMed] [Google Scholar]
James Lind Alliance
- James Lind Alliance Priority Setting Partnerships.Neuro-oncology top 10. www.jla.nihr.ac.uk/priority-setting-partnerships/neuro-oncology/top-10-priorities/ (accessed 8 October 2018).
Jaunmuktane 2019
- Jaunmuktane Z, Capper D, Jones DT, Schrimpf D, Sill M, Dutt M, et al.Methylation array profiling of adult brain tumours: diagnostic outcomes in a large, single centre. Acta Neuropathologica Communications 2019;7(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jenkins 2006
- Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, et al.A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Research 2006;66(20):9852-61. [DOI] [PubMed] [Google Scholar]
Jiang 2014
- Jiang H, Ren X, Zhang Z, Zeng W, Wang J, Lin S.Polysomy of chromosomes 1 and 19: an underestimated prognostic factor in oligodendroglial tumors. Journal of Neuro-Oncology 2014;120(1):131-8. [DOI] [PubMed] [Google Scholar]
Jones 2010
- Jones G, Johnson WO, Hanson TE, Christensen R.Identifiability of models for multiple diagnostic testing in the absence of a gold standard. Biometrics 2010;66(3):855-63. [DOI] [PubMed] [Google Scholar]
Louis 2016
- Louis DN, Perry A, Reifenberger G, Deimling A, Figarella-Branger D, Cavenee WK, et al.The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathologica 2016;131(6):803-20. [DOI] [PubMed] [Google Scholar]
Louis 2018
- Louis DN, Giannini C, Capper D, Paulus W, Figarella-Branger D, Lopes MB, et al.cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathology 2018;135:639-42. [DOI] [PubMed] [Google Scholar]
Lunn 2000
- Lunn DJ, Thomas A, Best N, Spiegelhalter D.WinBUGS – a Bayesian modelling framework: concepts, structure, and extensibility. Statistics and Computing 2000;10(4):325-37. [Google Scholar]
McGuinness 2020
- McGuinness LA, Higgins JP.Risk-Of-Bias VISualization (robvis): an R package and Shiny web app for visualizing risk-of-bias assessments. Research Synthesis Methods 2021;12(1):55-61. [DOI] [PubMed] [Google Scholar]
Messali 2014
- Messali A, Villacorta R, Hay JW.A review of the economic burden of glioblastoma and the cost effectiveness of pharmacologic treatments. PharmacoEconomics 2014;32:1201-12. [DOI] [PubMed] [Google Scholar]
NICE 2014
- National Institute for Health and Care Excellence.Appendix H: appraisal checklists, evidence tables, GRADE and economic profiles. In: Developing NICE Guidelines: the Manual. London (UK): National Institute for Health and Care Excellence, 2014:1-43. [Google Scholar]
NICE 2018
- National Institute for Health and Care Excellence.Brain tumours (primary) and brain metastases in adults. www.nice.org.uk/guidance/ng99 (accessed 8 October 2018).
Ostrom 2014
- Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, et al.The epidemiology of glioma in adults: a state of the science review. Neuro-Oncology 2014;16(7):896-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Pickles 2020
- Pickles JC, Fairchild AR, Stone TJ, Brownlee L, Merve A, Yasin SA, et al.DNA methylation-based profiling for paediatric CNS tumour diagnosis and treatment: a population-based study. Lancet Child & Adolescent Health 2020;4(2):121-30. [DOI] [PubMed] [Google Scholar]
Reitsma 2005
- Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH.Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. Journal of Clinical Epidemiology 2005;58(10):982-90. [DOI] [PubMed] [Google Scholar]
Ren 2013
- Ren X, Jiang H, Cui X, Cui Y, Ma J, Jiang Z, et al.Co-polysomy of chromosome 1q and 19p predicts worse prognosis in 1p/19q codeleted oligodendroglial tumors: FISH analysis of 148 consecutive cases. Neuro-Oncology 2013;15(9):1244-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rutter 2001
- Rutter CM, Gatsonis CA.A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Statistics in Medicine 2001;20(19):2865-84. [DOI] [PubMed] [Google Scholar]
Schünemann 2008
- Schünemann HJ, Oxman AD, Brozek J, Glasziou P, Jaeschke R, Vist GE, et al.Grading quality of evidence and strength of recommendations for diagnostic tests and strategies. BMJ 2008;336(7653):1106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Shemilt 2019
- Shemilt I, Aluko P, Graybill E, Craig D, Henderson C, Drummond M, et al, on behalf of the Campbell and Cochrane Economics Methods Group.Chapter 20: Economic evidence. In: Higgins JP, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, et al, editors(s). Cochrane Handbook for Systematic Reviews of Interventions. 2nd edition. Chichester (UK): John Wiley & Sons, 2019:507-25. [Google Scholar]
Snuderl 2009
- Snuderl M, Eichler AF, Ligon KL, Vu QU, Silver M, Betensky RA, et al.Polysomy for chromosomes 1 and 19 predicts earlier recurrence in anaplastic oligodendrogliomas with concurrent 1p/19q loss. Clinical Cancer Research 2009;15(20):6430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Stupp 2014
- Stupp R, Brada M, den Bent MJ, Tonn JC, Pentheroudakis G, ESMO Guidelines Working Group.High-grade glioma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Annals of Oncology 2014;25:93-101. [DOI] [PubMed] [Google Scholar]
Takwoingi 2013
- Takwoingi Y, Leeflang MM, Deeks JJ.Empirical evidence of the importance of comparative studies of diagnostic test accuracy. Annals of Internal Medicine 2013;158(7):544-54. [DOI] [PubMed] [Google Scholar]
TreeAge 2021 [Computer program]
- TreeAge Pro 2021, R1.Williamstown (MA): TreeAge Software, 2021. Available at www.treeage.com.
Vacek 1985
- Vacek PM.The effect of conditional dependence on the evaluation of diagnostic tests. Biometrics 1985;41(4):959-68. [PubMed] [Google Scholar]
van den Bent 2013
- den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kouwenhoven MC, Delattre JY, et al.Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. Journal of Clinical Oncology 2013;31(3):344-50. [DOI] [PubMed] [Google Scholar]
Vogazianou 2010
- Vogazianou AP, Chan R, Bäcklund LM, Pearson DM, Liu L, Langford CF, et al.Distinct patterns of 1p and 19q alterations identify subtypes of human gliomas that have different prognoses. Neuro-Oncology 2010;12(7):664-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Walter 1999
- Walter SD, Irwig L, Glasziou PP.Meta-analysis of diagnostic tests with imperfect reference standards. Journal of Clinical Epidemiology 1999;52(10):943-51. [DOI] [PubMed] [Google Scholar]
Weller 2017
- Weller M, den Bent M, Tonn JC, Stupp R, Preusser M, Cohen-Jonathan-Moyal E, et al.European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncology 2017;18(6):e315-29. [DOI] [PubMed] [Google Scholar]
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al.QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529-36. [DOI] [PubMed] [Google Scholar]
Whiting 2016
- Whiting P, Savovic J, Higgins JP, Caldwell DM, Reeves BC, Shea B, et al.ROBIS: a new tool to assess risk of bias in systematic reviews was developed. Journal of Clinical Epidemiology 2016;69:225-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
WHO 1993
- Kleihues P, Burger PC, Scheithauer BW.Histological Typing of Tumours of the Central Nervous System. Berlin (Germany): Springer-Verlag, 1993. [Google Scholar]
WHO 2000
- Kleihues P, Cavenee WK, eds.World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Nervous System. Lyon (France): International Agency for Research on Cancer (IARC), 2000. [Google Scholar]
WHO 2007
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, eds.WHO Classification of Tumours of the Central Nervous System. 4th edition. Lyon (France): International Agency for Research on Cancer (IARC), 2007. [Google Scholar]
WHO 2016
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK.WHO Classification of Tumours of the Central Nervous System. Revised 4th edition. Lyon (France): International Agency for Research on Cancer (IARC), 2016. [Google Scholar]
Zhao 2014
- Zhao J, Ma W, Zhao H.Loss of heterozygosity 1p/19q and survival in glioma: a meta-analysis. Neuro-Oncology 2014;16(1):103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
References to other published versions of this review
McAleenan 2019
- McAleenan A, Jones HE, Kernohan A, Faulkner CL, Palmer A, Dawson S, et al.Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma. Cochrane Database of Systematic Reviews 2019, Issue 8. Art. No: CD013387. [DOI: 10.1002/14651858.CD013387] [DOI] [PMC free article] [PubMed] [Google Scholar]



