Abstract
Subgroup J Avian leukosis virus (ALV-J) is an important pathogen of poultry tumor diseases. Since its discovery, it has caused significant economic losses to the poultry industry. Thus, the rapid detection of molecular level with strong specificity is particularly important whether poultry are infected with ALV-J. In this study, we designed primers and probe for real-time fluorescent reverse-transcription recombinase-aided amplification assay (RT-RAA) based on the ALV-J gp85 sequence. We had established a real-time fluorescent RT-RAA method and confirmed this system by verifying the specificity and sensitivity of the primers and probe. In addition, repeatability tests and clinical sample regression tests were used for preliminary evaluation of this detection method. The sensitivity of established method was about 101 copies/μL, and the repeatability of the CV of the CT value is 4%, indicating repeatability is good. Moreover, there was no cross-reactivity with NDV, IBV, IBDV, H9N2, MDV, and REV, and other avian leukosis virus subgroups, such as subgroups A, B, C, D, K and E. Importantly, the real-time fluorescent RT-RAA completed the test within 30 min at a constant temperature of 41°C. Forty-two clinical samples with known background were tested, and the test results were coincided with 100%. Overall, these results suggested that the real-time fluorescent RT-RAA developed in this study had strong specificity, high sensitivity, and good feasibility. The method is simple, easy, and portable, that is suitable for clinical and laboratory diagnosis, and provides technical support for the prevention and control of ALV-J.
Key words: subgroup J Avian leukosis virus, reverse-transcription recombinase-aided amplification assay, gp85, constant temperature detection
INTRODUCTION
Avian leukemia (AL) caused by the avian leukosis virus (ALV), belongs to the Alpharetrovirus genus of the family Retroviridae (Crittenden, 1981; Bai et al., 1995). AL is a type of neoplastic disease, mainly caused by the proliferation of hematopoietic cells such as lymphocytic AL. Depending on the transmission mode, ALV can be classified as endogenous and exogenous virus (Crittenden, 1981). According to the host range, viral envelope interference, and cross-neutralization mode of ALV, exogenous ALVs can be further divided into 11 different subgroups, including subgroup A, B, C, D, J, and K from chicken (Payne, 1998; Venugopal, 1999). ALV-J has the strongest pathogenicity and transmission ability in ALV (Li et al., 2017). ALV-J was first detected in broiler flocks in China in 1999 (Spencer et al., 2000). Subsequently, there were cases of ALV-J infection and tumors in commercial laying hens and local chickens in 2004 (Xu et al., 2004; Sun et al., 2007). Moreover, the outbreak of ALV-J caused serious economic losses to the poultry industry, leading to bankruptcy of some hen farms after 2008 (Lai et al., 2011; Mao et al., 2020). Currently, there is no effective vaccine for AL. China's prevention and treatment of AL is mainly based on an eradication plan (Dai et al., 2020) to achieve the effect of purification. Fortunately, the outbreak of AL in commercial broilers and laying hens has decreased since 2013 (Wang et al., 2018). Previous study analyzed the molecular characteristics and epidemic trend of ALV-J in Chinese native chickens from 2013 to 2018, showing that ALV-J gp85 and U3 of native chicken isolates had formed a unique local chicken branch, and gene deletion in the 3′UTR region was a unique molecular feature of ALV-J in China (Ma et al., 2020). It had been reported that the outbreak of ALV-J in white-feathered broilers in parts of the Jiangxi province of China had occurred again since the end of 2018 (Li et al., 2021). According to their results, they have shown that the new mutant ALV-J was being bred in local chickens.
Detection is a particularly important part of the AL eradication plan. The current methods for detecting ALV mainly include virus isolation, ELISA, conventional PCR, real-time fluorescent quantitative PCR (qPCR), immunofluorescence test (IFA), and Loop-mediated isothermal amplification (LAMP). We all known the virus isolation is the gold standard for detected ALV. However, culturing CEF or DF-1 cells requires more than 7 d to obtain final accurate results, and other methods such as ELISA (Yun et al., 2013) need to assist, but cannot distinguish subgroups (Garc et al., 2003). Conventional PCR, qPCR, and IFA can be used to distinguish subgroups but require expensive and complex equipment such as quantitative PCR fluorescent machines and fluorescent microscopes, which were suitable for laboratory diagnosis (Smith et al., 1998; Bagust et al., 2004; Li et al., 2012; Qin et al., 2013; Gao et al., 2014; Chen et al., 2018). LAMP detection had applications in clinical, but it required 63°C reactions in a water bath or incubator, which was limited in the field (Zhang et al., 2010). However, the reverse-transcription recombinase aided amplification assay (RT-RAA) is a constant temperature nucleic acid amplification technology. The RAA and recombinase polymerase amplification assay (RPA) have similar amplification principles. The difference between the two is the source of recombinase. The RPA recombinase is derived from Phage T4, and the RAA recombinase is derived from bacteria or fungus. In recent years, RAA technology can be applied to human diseases, pets, aquatic, poultry infectious diseases, genetic diseases, etc. For example, Xue and Wang both reported applications on the COVID-19 (Wang et al., 2020; Xue et al., 2020). Fan reported on the clinical testing of African swine fever virus (Fan et al., 2020). The RAA principle is that the recombinase and primers formed a complex of recombinase and primers at a constant temperature of 35 to 42°C and invades the DNA double-stranded nucleic acid template. At the invading site, the recombinase will open the double-stranded DNA, while the single-stranded binding protein maintains the double-stranded template in an open-stranded state. Next, the DNA polymerase binds to the 3′end of the primer and begins to synthesize a new strand. The subsequent amplified product can be used as a template again, so that it was repeated continuously to complete the amplification of the target gene (Guo et al., 2020; Wang et al., 2020). Overall, the RT-RAA method has some advantages, like high sensitivity, strong specificity, short detection time, simple operation, no expensive instruments. If combined with fluorescent probes, real-time detection and rapid on-site detection can be achieved.
The proviral DNA of the ALV genome is arranged as 5′LTR-leader-gag-pol-env-3′LTR, with a total length of 7,200 bp to 7,800 bp (Wang et al., 2018). According to the low homology rate between the sequence of ALV-J gp85 and other exogenous subgroups (Bova et al., 1988; Bai et al, 1995), primers and probes can be designed on the sequence of gp85 to distinguish the subgroups of ALV (Venugopal, 1999). In this study, we designed the RT-RAA primers and probes according to the ALV-J gp85 gene sequence, and established the real-time fluorescent RT-RAA isothermal amplification detection method for ALV-J.
MATERIALS AND METHODS
Virus and Clinical Plasma Samples
Viruses subgroup A Avian Leukemia virus (ALV-A, DQ365814), subgroup J Avian Leukemia virus (ALV-J, MT175600), subgroup K Avian Leukemia virus (ALV-K, KP686142), Newcastle disease virus (NDV, JF950510), Infectious bronchitis virus (IBV, KR605489), Infections bursal disease virus (IBDV, AF416621), and H9N2 subgroup avian influenza virus (H9N2, MN064851) were stored in our laboratory. Marek's Disease Virus (MDV, L37202) vaccine was purchased from Harbin Pharmaceutical Group Bio-vaccine Co., Ltd., Harbin, China. Avian reticuloendothelial hyperplasia (REV) provided by Wen’s Foodstuff Group Co, Ltd., Yunfu, China. The gp85 gene fragment of subgroup B Avian Leukemia virus (ALV-B, HM446005), subgroup C Avian Leukemia virus (ALV-B, MT783248), subgroup D Avian Leukemia virus (ALV-D, D10653), and subgroup E Avian Leukemia virus (ALV-E, AY013303) were synthesized by the Sangon Biotech Co., Ltd. (Shanghai, China).
A total of 42 clinical serum samples with known clinical background were collected from Guangdong Aihealth Biotechnology Co., Ltd., Qingyuan, China, of which 13 were positive samples and 29 were negative samples. The nucleic acids of 42 samples were extracted according to the recommended steps of the Viral DNA/RNA isolation AxyPrep Body Fluid Viral DNA/RNA Miniprep Kit (China Guangzhou Suyan Biotechnology Co., Ltd.) and stored at −80°C.
Reagents and Instruments
Viral DNA/RNA isolation AxyPrep Body Fluid Viral DNA/RNA Miniprep Kit was purchased from Guangzhou Suyan Biotechnology Co., Ltd. The RT-RAA Freeze-dried powder was purchased from Hangzhou ZC Bio-Sci & Tech Co., Ltd., Hangzhou, China and real-time fluorescent quantitative PCR instrument (Model: CFX96, Bio-Rad Laboratories, Shanghai, China).
Design of Primers and Probes
Referred to the gp85 gene sequence of ALV-J published on GenBank (GenBank accession number: Z46390, MT175600, MN066150, MN066151, MN066152, MK940585, MN066140, JQ935966, JX848322, KC149971, JF932004, KU170199, DQ115805, JF932004 and JQ935966), we used DNAStar and DNAMAN (v9.0) to compare multiple sequences. According to the gene alignment and analysis of the conserved regions, the real-time fluorescent RT-RAA detection primers and probe (Table 1) were designed. The amplified fragment was 148 bp. The primers and probe were synthesized by the Sangon Biotech Co., Ltd.
Table 1.
Sequences of primers and probe for real-time RT-RAA assays for ALV-J.
| Primer | Sequence (5’-3’) | Location |
|---|---|---|
| JF-Primer | CAACAAGCAAGAAAGACCCGGAGAAGACAC | 5363-5392 |
| JR-Primer | CACACGTTTCCTGGTTGTTGCAATAGATG | 5482-5510 |
| JP-Probe | TTCTTTCAAATGATACTTGTGTGCGTGGTTAT/i6FAMdT/ /THF/ /iBHQ1dT/TTCCGTTGTCCCAGG[C3-spacer] | 5419-5468 |
Abbreviations: ALV-J, J Avian leukosis virus; RT-RAA, reverse-transcription recombinase-aided amplification assay.
Generation of Plasmid Standard
The plasmid standard was constructed based on the RT-TAA primer designed fragment of the ALV-J gp85 gene sequence (the gp85 reference strain SCAU1903, GenBank accession number MT175600.1). The primer sequence was quoted from this paper which published by Wang in 2018 (Table 2). A 545 bp fragment of the gp85 gene was cloned into the PMD19-T vector to quantify DNA copy number. After the plasmid successfully constructing, the plasmid concentration measured by the ultra-micro nucleic acid analyzer was about 20.229 ng/µL, and the copy number of the plasmid was about 5.70 × 109 copies/µL calculated by the formula.
Table 2.
PCR primers for plasmid construction.
| Primer | Sequence (5’-3’) | Gen Localization | Source |
|---|---|---|---|
| ALV-F | GGATGAGGTGACTAAGAAAG | 5258-5277 | (Wang et al., 2018) |
| ALV-R | CGAACCAAAGGTAACACACG | 5783-5802 | (Wang et al., 2018) |
RT-RAA Primers Verification
In order to verify the fragment amplified by RT-RAA primers, the template was the nucleic acid of ALV-J SCAU1903 strain, PCR reaction system: JF forward (10 µM) 1.0 µL, JR reverse (10 µM) 1.0 µL, ddH2O 6.0 µL, 2 × Es Taq MasterMix 10.0 µL, proviral DNA 2 µL. The reaction procedure was: 95°C 3 min, 95°C 30 s, 58°C 30 s, 72°C 30 s, 35 cycles, 72°C extension for 2 min. Finally, the obtained product was analyzed by 1% agarose gel electrophoresis.
Establishment and Optimization of RT-RAA Reaction System
Preparation of the real-time RT-RAA reaction system: Each test sample corresponds to a RT-RAA Freeze-dried powder tube (single-stranded DNA-binding protein, DNA polymerase, recombinase, MLV reverse transcriptase, ATP, dNTP mix). The reaction components in each RT-RAA reaction tube and the volume were shown in Table 3. After preparing the reaction system as described above, we flicked the bottom of the tube to expel the bubbles and centrifuged at low speed for 10 s. The amplification temperature of the real-time RT-RAA reaction system was selected to 41°C, so the amplification protocol of the system was 41°C 60s, 41°C 30s, 40 cycles, and the fluorescence signal was collected. After the reaction, the positive control had a typical amplification curve, and the peak time was ≤19.5 min (Ct value ≤39); and the negative control has no amplification curve appeared, or the peak time was >19.5 min (Ct value >39), indicating valid results.
Table 3.
Real-time RT-RAA reaction system preparation table.
| RT-RAA reaction system | Usage |
|---|---|
| RT-RAA Freeze-dried powder | 1 tube |
| A Buffer | 25 μL |
| B Buffer | 2.5 μL |
| JF-Primer (10 µM) | 2 μL |
| JR-Primer (10 µM) | 2 μL |
| JP-Probe (10 µM) | 0.6 μL |
| H2O | 12.9 μL |
| RNA | 5 μL |
| Total | 50 μL |
Sensitivity Analysis of ALV-J Real-time RT-RAA Assay
Diluting the prepared plasmid in a 10-fold ratio. We used 5 concentration gradients as templates, which were 1 × 105, 1 × 104, 1 × 103, 1 × 102 and 1 × 101 copies/µL. And then 5 µL plasmid in different diluted concentrations as the template that performed the real-time RT-RAA amplification according to the optimized reaction conditions described above.
Specificity Analysis of ALV-J Real-Time RT-RAA Assay
Using ALV-J nucleic acid as a positive control and ddH2O as a negative control. Five uL template of ALV-A, ALV-B, ALV-C, ALV-D, ALV-K, ALV-E, NDV, IBV, IBDV, H9N2, MDV, and REV was respectively taken to detect the specificity and performed the real-time RT-RAA amplification according to the above optimized reaction conditions RT-RAA amplification.
Repeatability Analysis of ALV-J Real-Time RT-RAA Assay
In order to verify the reproducibility of the method, the optimized reaction system was used for 3 times repeated detections with ALV-J nucleic acid, and ddH2O was used as a negative control.
Evaluation of the RT-RAA Assay Using Avian Clinical Samples
Real-time fluorescent RT-RAA detection was performed on the nucleic acids of the above 42 clinical samples, and the results were compared with known results. Among them, 13 were positive samples and 29 were negative samples.
RESULTS
RT-RAA Primer Verification
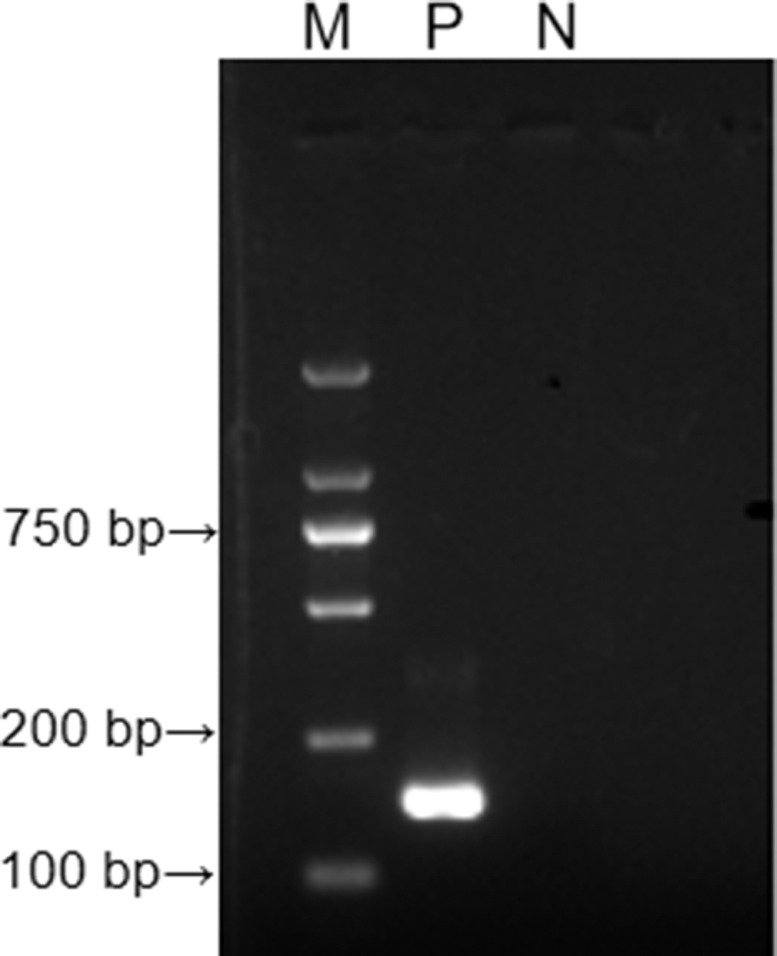
With ALV-J SCAU1903 nucleic acid as a positive control and ddH2O as a negative control, PCR amplification was performed on the designed RT-RAA primer pair. The result was shown in Figure 1. The target amplified fragment was consistent with the expected result, with a size of 148 bp and no secondary structure in primers. It shows that the designed RT-RAA primer can specifically detect ALV-J.
Figure 1.
The results of agarose electrophoresis of PCR amplified products with ALV-J RT-RAA primers. M: D 2000 Marker; P: positive control, that is, ALV-J nucleic acid; N: negative control, that is, ddH2O. Abbreviations: ALV-J, J Avian leukosis virus; RT-RAA, reverse-transcription recombinase-aided amplification assay.
Establishment and Optimization of RT-RAA Reaction System
In order to determine the most suitable reaction temperature for the primers and probe, the temperature gradient was explored between 39°C and 42°C. The results showed that at 41°C, the primers and probe listed in Table 1 can reach their fluorescence intensity in the shortest time. Therefore, it was determined that the optimal reaction temperature of the RT-RAA method established in this study was 41°C. The optimal reaction conditions for RT-RAA were: JF-Primer (10 µM) 2.0 µL, JR-Primer (10 µM) 2.0 µL, JP-Probe (10 µM) 0.6 µL, A Buffer 40.9 µL, B Buffer 2.5 µL, RNA 5 µL, and total 50 µL system.
Sensitivity Analysis of ALV-J Real-Time RT-RAA Assay
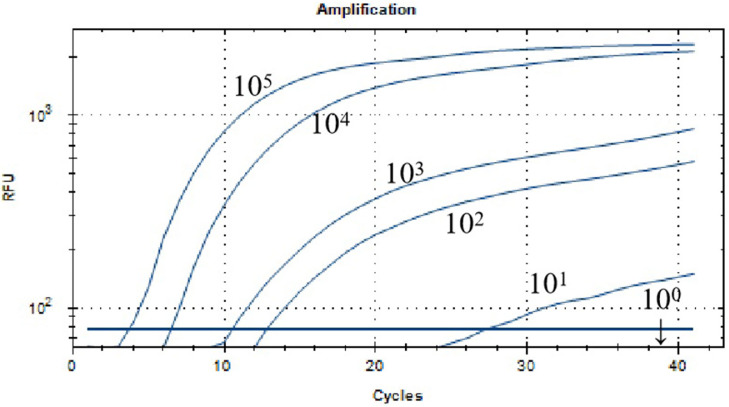
The result of sensitivity analysis of ALV-J real-time RT-RAA assay was shown in Figure 2. When the plasmid concentration was 1 × 101 copies/µL, there was still fluorescence signal that was, the lowest detection line of this method was 1 × 101 copies/µL.
Figure 2.
Sensitivity analysis of the real-time RT-RAA assay for ALV-J. The dilution range of ALV-J plasmid was 105 to 101 copies/μL, and the minimum detection line was 101 copies/μL. Abbreviations: ALV-J, J Avian leukosis virus; RT-RAA, reverse-transcription recombinase-aided amplification assay.
Specificity Analysis of ALV-J Real-Time RT-RAA Assay
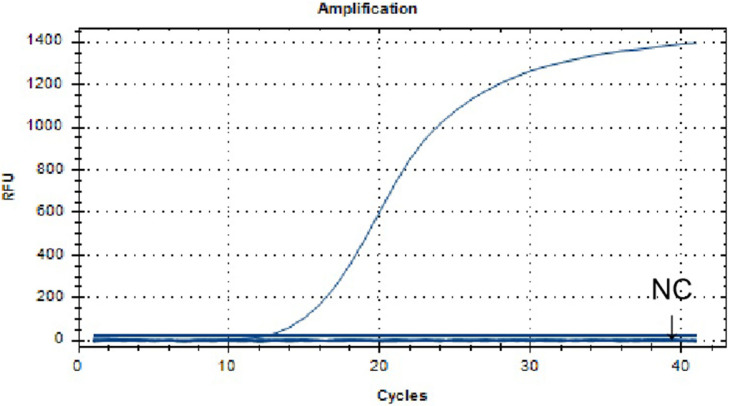
The results of specificity analysis of ALV-J real-time RT-RAA assay were shown in Figure 3. The positive control ALV-J nucleic acid had a fluorescent signal, the negative control ddH2O and the nucleic acid of other viruses had no fluorescent signal. It shows that the established real-time RT-RAA method can distinguish ALV-A, ALV-B, ALV-C, ALV-D, ALV-K, ALV-E, NDV, IBV, IBDV, H9N2, MDV, and REV, and specifically detect ALV-J.
Figure 3.
Specificity analysis of the real-time RT-RAA assay for ALV-J. Detection signals were recorded by real-time fluorescence RT-RAA with ALV-J (SCAU1903), while no signal detected from the thirteen samples including other subgroups ALVs, IBV, H9N2, REV, MDV, NDV, IBDV and negative controls. Abbreviations: ALV-J, J Avian leukosis virus; IBV, infectious bronchitis virus; IBDV, infections bursal disease virus; MDV, Marek's disease virus; NDV, Newcastle disease virus; REV, Avian reticuloendothelial hyperplasia; RT-RAA, reverse-transcription recombinase-aided amplification assay.
Repeatability Analysis of ALV-J Real-Time RT-RAA Assay
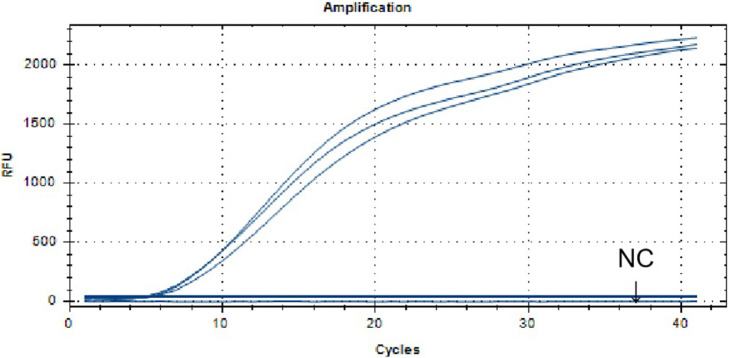
The results of repeatability analysis of ALV-J real-time RT-RAA assay were shown in Figure 4. The results showed that the amplification reaction peak time was the same, the curve shape was similar, the negative control has no amplification, the CV of the CT value was 4%, indicating good repeatability.
Figure 4.
Repeatability analysis of the real-time RT-RAA assay for ALV-J. The ALV-J nucleic acid was positive, the detection signal curve of 3 times repeated experiments was basically the same and the CV of the CT value was 4%. Abbreviations: ALV-J, J Avian leukosis virus; RT-RAA, reverse-transcription recombinase-aided amplification assay.
Evaluation of the RT-RAA Assay Using Avian Clinical Samples
The results of evaluation of the RT-RAA assay using avian clinical samples were shown in Table 4. The result of real-time fluorescent RT-RAA test result was 100% coincident with the negative and positive results of 42 samples with known background, of which the positive rate was 31% and the negative rate was 69%. The test results reveal that the real-time fluorescent RT-RAA has good consistency.
Table 4.
The results of Clinical test by the real-time RT-RAA assay for ALV-J.
| Sample | Known | RT-RAA | Coincidence rate(%) |
|---|---|---|---|
| Positive | 13 | 13 | 100 |
| Negative | 29 | 29 | 100 |
| Total | 42 | 42 | / |
Abbreviations: ALV-J, J Avian leukosis virus; RT-RAA, reverse-transcription recombinase-aided amplification assay.
DISCUSSION
Since the discovery of ALV, it has brought huge economic losses to the poultry industry around the world. Since China implemented the eradication plan, the outbreak of AL has decreased. However, according to the research in 2018, some provinces in China almost simultaneously broke out of myelocytoma disease caused by mutant ALV-J (Li et al., 2019; Zhou et al., 2019). Studies had shown that ALV-J can be detected and isolated from commercial live vaccines, which can inhibit the immune effect of the vaccine (Wang et al., 2018; Wang et al., 2021). Because of the high level of vertical and horizontal transmission of ALV-J, effective vaccines and drugs were useless (Venugopal, 1999), it was important to improve the eradication efficiency and speed up the eradication process. As the detection in the eradication plan is particularly important, therefore, it is urgent to establish a detection method that is simple to operate, fast, and low in cost.
According to research results, the ALV-J gp85 gene and other exogenous ALV subgroups only show about 40% homology (Bai et al., 1995; Smith et al., 1998). Therefore, we designed the RT-RAA primers and probe with ALV-J gp85 gene. Through verified primers and probes experiments, there was no crossover with other subgroups (ALV-A, ALV-B, ALV-C, ALV-D, ALV-E, ALV-K) and other viruses (IBV, H9N2, REV, MDV, NDV, and IBDV), indicating that the established RT-RAA method can distinguish ALV-J from other subgroups and other viruses, and can specifically detect ALV-J. Compared with ELISA, the real-time RT-RAA can overcome the interference caused by the integration of endogenous viral nucleic acid into the host body and the false positive phenomenon.
The real-time RT-RAA had a minimum detection line of 101 copies/uL, which was 100 times that of the conventional PCR (Gao et al., 2014), similar to the results of the QPCR (Dai et al., 2015). However, the traditional PCR and qPCR were suitable for laboratory testing, but large-scale testing in the clinical field was limited. The sensitivity of the LAMP (Zhang et al., 2010) is the same, but if no paraffin oil is used, the LAMP method may have the possibility of cross-contamination.
The real-time RT-AA was compared with existing detection methods in detection time. The analysis time of traditional RT-PCR and RT-qPCR (excluding nucleic acid extraction) is more than 2 h and requires expensive equipment with high temperature and variable temperature. The detection time of ELISA is generally 2 h 30 min, and the detection samples are limited. For example, because IDEXX products are interfered by the integration of endogenous viruses in serum and egg into the host genome. It takes about a week for IFA and pathogen separation test results, expensive equipment and high requirements for test personnel. The time of LAMP detection is about 45 min and does not require expensive equipment. Nevertheless, LAMP once opened the lid and it was easily to form aerosol pollution. However, the detection time of RT-RAA is 5 to 30 min, which greatly saves the detection time and does not require heavy and expensive equipment and professionals.
In this study, we detected the clinical samples with known background to evaluate the established RT-RAA method. Among the 42 samples detected, 13 positive samples had a positive rate of 31%, and 29 negative samples had a negative rate of 69%, which was consistent with the known background results. The ALV-J nucleic acid was repeated 3 times, and the CV of the CT value was 4%, indicating that the method established in this test has good sensitivity and repeatability.
Overall, the real-time RT-RAA we established can complete the detection within 30 min at a constant temperature of 41°C, which has high sensitivity, good repeatability, and strong specificity. It needs only simple equipment to complete the detection quickly. Trial and clinical sample testing have shown that the RT-RAA detection method is practical in laboratory and clinical diagnosis. In addition, this real-time RT-RAA is helpful for epidemiological surveillance and disease detection that may develop into a clinical detection program for ALV-J in the poultry industry.
ACKNOWLEDGMENTS
This work was supported by the Key Research and Development Program of Guangdong Province (2020B020222001), the National Natural Science Foundation of China (grants nos., 31902252, 31972659, 31802185), the Natural Science Foundation of Guangdong Province (grant no. 2019A1515012006 and S2013030013313), the Construction of Modern Agricultural Science and Technology Innovation Alliance in Guangdong Province (2020KJ128), the Special Project of National Modern Agricultural Industrial Technology System (CARS-41), the National Modern Agricultural Industry Science and Technology Innovation Center in Guangzhou (2018kczx01), the National Key R&D Program of China (2017YFD0502001), the Creation of a Triple Chimeric Vaccine (rIBV-ND-H9), using Avian Infectious Bronchitis Attenuated D90 as a Vector (2017KZDM008) and the Guangzhou Science and Technology Bureau (GZKTP201930).
DISCLOSURES
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled.
REFERENCES
- Bova C.A., Olsen J.C., Swanstrom R. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J. Virol. 1988;62:75–83. doi: 10.1128/jvi.62.1.75-83.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagust T.J., Fenton S.P., Reddy M.R. Detection of subgroup J avian leukosis virus infection in Australian meat-type chickens. Vet. J. 2004;82:701–706. doi: 10.1111/j.1751-0813.2004.tb12163.x. [DOI] [PubMed] [Google Scholar]
- Bai J., Howes K., Payne L.N., Skinner M.A. Sequence of host-range determinants in the env gene of a full-length, infectious proviral clone of exogenous avian leukosis virus HPRS-103 confirms that it represents a new subgroup (designated J) J. Gen. Virol. 1995;76:181–187. doi: 10.1099/0022-1317-76-1-181. [DOI] [PubMed] [Google Scholar]
- Chen P., Zhao Z.J., Chen Y.Y., Zhang J., Yun L.F., Zheng X.C., Liao M., Cao W.S. Corrigendum to "Development and application of a SYBR green real-time PCR for detection of the emerging avian leukosis subgroup K". Poult. Sci. 2018;97:3763. doi: 10.3382/ps/pey384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden L.B. Exogenous and endogenous leukosis virus genes–a review. Avian Pathol. 1981;10:101–112. doi: 10.1080/03079458108418464. [DOI] [PubMed] [Google Scholar]
- Dai M., Feng M., Liu D., Cao W., Liao M. Development and application of SYBR Green I real-time PCR assay for the separate detection of subgroup J Avian leukosis virus and multiplex detection of avian leukosis virus subgroups A and B. Virol. J. 2015;12:52. doi: 10.1186/s12985-015-0291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M., Li S., Shi K., Liao J., Sun H., Liao M. Systematic identification of host immune key factors influencing viral infection in PBL of ALV-J infected SPF chicken. Viruses. 2020;12:114. doi: 10.3390/v12010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Li L., Zhao Y., Liu Y., Liu C., Wang Q., Dong Y., Wang S., Chi T., Song F., Sun C., Wang Y., Ha D., Zhao Y., Bao J., Wu X., Wang Z. Clinical validation of two recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of African swine fever virus. Front. Microbiol. 2020;11:1696. doi: 10.3389/fmicb.2020.01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Yun B.L., Wang Q., Jiang L.L., Zhu H.B., Gao Y.N., Qin L.T., Wang Y.Q., Qi X.L., Gao H.L., Wang X.M., Gao Y.L. Development and application of a multiplex PCR method for rapid differential detection of subgroup A, B, and J avian leukosis viruses. J. Clin. Microbiol. 2014;52:37–44. doi: 10.1128/JCM.02200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M., El-Attrache J., Riblet S.M., Lunge V.R., Fonseca A.S., Villegas P., Ikuta N. Development and application of reverse transcriptase nested polymerase chain reaction test for the detection of exogenous avian leukosis virus. Avian Dis. 2003;47:41–53. doi: 10.1637/0005-2086(2003)047[0041:DAAORT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Guo L., Sun X.H., Wang X.G., Liang C., Jiang H.P., Gao Q.Q., Dai M.Y., Qu B., Fang S., Mao Y.H., Chen Y.C., Feng G.H., Gu Q., Wang R.R., Zhou Q., Li W. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020;6:34. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H., Zhang H., Ning Z., Chen R., Zhang W., Qing A., Xin C., Yu K., Cao W., Liao M. Isolation and characterization of emerging subgroup J avian leukosis virus associated with hemangioma in egg-type chickens. Vet. Microbiol. 2011;151:275–283. doi: 10.1016/j.vetmic.2011.03.037. [DOI] [PubMed] [Google Scholar]
- Li H., Tan M., Zhang F., Ji H., Zeng Y., Yang Q., Tan J., Huang J., Su Q., Huang Y., Kang Z. Diversity of Avian leukosis virus subgroup J in local chickens, Jiangxi, China. Sci. Rep. 2021;11:4797. doi: 10.1038/s41598-021-84189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang P., Lin L., Shi M., Gu Z., Huang T., Mo M.L., Wei T., Zhang H., Wei P. The emergence of the infection of subgroup J avian leucosis virus escalated the tumour incidence in commercial yellow chickens in Southern China in recent years. Transbound Emerg. Dis. 2019;66:312–316. doi: 10.1111/tbed.13023. [DOI] [PubMed] [Google Scholar]
- Li X., Li D.Q., Zhao P., Cui Z.Z. The ALV-A/B specific antibodies correlation between ELISA and IFA detection in chicken serum. Chin. J. Virol. 2012;28:615–620. [PubMed] [Google Scholar]
- Li Y., Cui S., Li W.H., Wang Y.X., Cui Z.Z., Zhao P., Chang S. Vertical transmission of avian leukosis virus subgroup J (ALV-J) from hens infected through artificial insemination with ALV-J infected semen. Bmc Vet. Res. 2017;13:204. doi: 10.1186/s12917-017-1122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M.G., Yu M.M., Chang F.F., Xing L.X., Bao Y.L., Wang S.Y., Farooque M., Li X.Y., Liu P., Chen Y.T., Qi X.L., Pan Q., Gao L., Li K., Liu C.J., Zhang Y.P., Cui H.Y., Wang X.M., Sun Y.M., Gao Y.L. Molecular characterization of avian leukosis virus subgroup J in Chinese local chickens between 2013 and 2018. Poult. Sci. 2020;99:5286–5296. doi: 10.1016/j.psj.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.Q., Su Q., Li J.P., Jiang T.Z., Wang Y.X. Avian leukosis virus contamination in live vaccines: a retrospective investigation in China. Vet. Microbiol. 2020;246 doi: 10.1016/j.vetmic.2020.108712. [DOI] [PubMed] [Google Scholar]
- Payne L.N. HPRS-103: a retro virus strikes back. The emergence of subgroup J avian leukosis virus. Avian Pathol. 1998;27(Supp. l)):S36–S45. [Google Scholar]
- Qin L.T., Gao Y.L., Ni W., Sun M.Y., Wang Y.Q., Yin C.H., Qi X.L., Gao H.L., Wang X.M. Development and application of real-time PCR for detection of subgroup J avian leukosis virus. J. Clin. Microbiol. 2013;51:149–154. doi: 10.1128/JCM.02030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.M., Brown S.R., Howes K., McLeod S., Arshad S.S., Barron G.S., Venugopal K., McKay J.C., Payne L.N. Development and application of polymerase chain reaction (PCR) tests for the detection of subgroup J avian leukosis virus. Virus Res. 1998;54:87–98. doi: 10.1016/s0168-1702(98)00022-7. [DOI] [PubMed] [Google Scholar]
- Spencer J.L., Chan M., Nadin-Davis S. Relationship between egg size and subgroup J avian leukosis virus in eggs from broiler breeders. Avian Pathol. 2000;29:617–622. doi: 10.1080/03079450020016878. [DOI] [PubMed] [Google Scholar]
- Sun S.H., Cui Z.Z. Epidemiological and pathological studies of subgroup J avian leukosis virus infections in Chinese local “yellow” chickens. Avian Pathol. 2007;36:221–226. doi: 10.1080/03079450701332345. [DOI] [PubMed] [Google Scholar]
- Venugopal K. Avian leukosis virus subgroup J: a rapidly evolving group of oncogenic retroviruses. Res. Vet. Sci. 1999;67:113–119. doi: 10.1053/rvsc.1998.0283. [DOI] [PubMed] [Google Scholar]
- Wang J., Cai K., He X., Shen X., Wang J., Liu J., Xu J., Qiu F., Lei W., Cui L., Ge Y., Wu T., Zhang Y., Yan H., Chen Y., Yu J., Ma X., Shi H., Zhang R., Li X., Gao Y., Niu P., Tan W., Wu G., Jiang Y., Xu W., Ma X. Multiple-centre clinical evaluation of an ultrafast single-tube assay for SARS-CoV-2 RNA. Clin. Microbiol. Infect. 2020;26:1076–1081. doi: 10.1016/j.cmi.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Lin L., Li H., Shi M., Gu Z., Wei P. Full-length genome sequence analysis of an avian leukosis virus subgroup J (ALV-J) as contaminant in live poultry vaccine: the commercial live vaccines might be a potential route for ALV-J transmission. Transbound Emerg. Dis. 2018;65:1103–1106. doi: 10.1111/tbed.12841. [DOI] [PubMed] [Google Scholar]
- Wang P.K., Li M., Li H.J., Bi Y.Y., Lin L.L., Shi M.Y., Huang T., Mo M.L., Wei T.C., Wei P. ALV-J-contaminated commercial live vaccines induced pathogenicity in three-yellow chickens: one of the transmission routes of ALV-J to commercial chickens. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.K., Lin L.L., Li H.J., Yang Y.L., Huang T., Wei P. Diversity and evolution analysis of glycoprotein GP85 from avian leukosis virus subgroup J isolates from chickens of different genetic backgrounds during 1989-2016: coexistence of five extremely different clusters. Arch. Virol. 2018;163:377–389. doi: 10.1007/s00705-017-3601-0. [DOI] [PubMed] [Google Scholar]
- Wang S., Huang B., Ma X., Liu P., Wang Y., Zhang X., Zhu L., Fan Q., Sun Y., Wang K. Reverse-transcription recombinase-aided amplification assay for H7 subtype avian influenza virus. Transbound Emerg. Dis. 2020;67:877–883. doi: 10.1111/tbed.13411. [DOI] [PubMed] [Google Scholar]
- Xu B., Dong W., Yu C., He Z., Lv Y., Sun Y., Feng X., Li N., Lee L.F., Li M. Occurrence of avian leukosis virus subgroup J in commercial layer flocks in China. Avian Pathol. 2004;33:13–17. doi: 10.1080/03079450310001636237a. [DOI] [PubMed] [Google Scholar]
- Xue G., Li S., Zhang W., Du B., Cui J., Yan C., Huang L., Chen L., Zhao L., Sun Y., Li N., Zhao H., Feng Y., Wang Z., Liu S., Zhang Q., Xie X., Liu D., Yao H., Yuan J. Reverse-transcription recombinase-aided amplification assay for rapid detection of the 2019 novel coronavirus (SARS-CoV-2) Anal. Chem. 2020;92:9699–9705. doi: 10.1021/acs.analchem.0c01032. [DOI] [PubMed] [Google Scholar]
- Yun B.L., Li D.L., Zhu H.B., Liu W., Qin L.T., Liu Z.S., Wu G., Wang Y.Q., Qi X.L., Gao H.L., Wang X.M., Gao Y.L. Development of an antigen-capture ELISA for the detection of avian leukosis virus p27 antigen. J. Virol. Methods. 2013;187:278–283. doi: 10.1016/j.jviromet.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Zhang X., Liao M., Jiao P., Luo K., Zhang H., Ren T., Zhang G., Xu C., Xin C., Cao W. Development of a loop-mediated isothermal amplification assay for rapid detection of subgroup J avian leukosis virus. J. Clin. Microbiol. 2010;48:2116–2121. doi: 10.1128/JCM.02530-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D.F., Xue J.W., Zhang Y., Wang G.H., Feng Y.S., Hu L.P., Shang Y.L., Cheng Z.Q. Outbreak of myelocytomatosis caused by mutational avian leukosis virus subgroup J in China, 2018. Transbound Emerg. Dis. 2019;66:622–626. doi: 10.1111/tbed.13096. [DOI] [PubMed] [Google Scholar]