Abstract
An ongoing pandemic of newly emerged SARS-CoV-2 has puzzled many scientists and health care policymakers around the globe. The appearance of the virus was accompanied by several distinct antigenic changes, specifically spike protein which is a key element for host cell entry of virus and major target of currently developing vaccines. Some of these mutations enable the virus to attach to receptors more firmly and easily. Moreover, a growing number of trials are demonstrating higher transmissibility and, in some of them, potentially more serious forms of illness related to novel variants. Some of these lineages, especially the Beta variant of concern, were reported to diminish the neutralizing activity of monoclonal and polyclonal antibodies present in both convalescent and vaccine sera. This could imply that these independently emerged variants could make antiviral strategies prone to serious threats. The rapid changes in the mutational profile of new clades, especially escape mutations, suggest the convergent evolution of the virus due to immune pressure. Nevertheless, great international efforts have been dedicated to producing efficacious vaccines with cutting-edge technologies. Despite the partial decrease in vaccines efficacy against worrisome clades, most current vaccines are still effective at preventing mild to severe forms of disease and hospital admission or death due to coronavirus disease 2019 (COVID-19). Here, we summarize existing evidence about newly emerged variants of SARS-CoV-2 and, notably, how well vaccines work against targeting new variants and modifications of highly flexible mRNA vaccines that might be required in the future.
Keywords: Escape mutations, Neutralizing activity, SARS-CoV-2, COVID-19, Spike protein, Vaccines efficacy, Variants of concern
Background
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has become the most concerning issue in the world, causing rapid growth in the number of cases. Coronaviruses belong to the family called Coronaviridae, responsible for a variety of acute and chronic diseases in humans and other animals [1]. Among this family, several coronaviruses can infect humans, and four of them—HKU-1, NL63, OC43, and 229E—cause common colds and mild respiratory infections in patients with healthy immune responses [2]. There are three strains capable of initiating outbreaks. SARS-CoV and MERS-CoV are causative agents of severe acute respiratory syndrome outbreaks with manifestations of atypical pneumonia in China and the Middle East, dating back to 2002 and 2013, respectively [3–5]. A novel coronavirus with a gene sequence 96% similar to a bat coronavirus has emerged since December 2019, with the presentation of pneumonia linked to the seafood market in Wuhan, China. The infectious agent is called 2019-novel coronavirus (2019-nCoV) or severe acute respiratory coronavirus 2, SARS-CoV-2 [6, 7]. Until the end of 2021, over 280 million cases and more than 5.4 million fatalities related to COVID-19 were reported, according to World Health Organization, WHO [8].
Since the emergence of SARS-CoV-2 in December 2019 [6], many molecular variants have been found related to the spike (S) glycoprotein, which enables the virus to enter the host cell more effectively. D614G was the first notable variation found in April 2020 and dominated the wild type of virus found in Wuhan, China [9]. This shift stabilized S protein and made it more transmissible than the wild type. Following the D614G shift, there have been several distinct variants, including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529) variants raising concerns about more fatal and rapid-spreading variants [10–13]. Data are revealing that some of these escape mutations, especially E484K, have emerged as converging evolution, and there might be similar independent arising mutations [14]. To cope with hazards of novel viruses and worrisome variants emerging, international collaborations pointed the efforts toward producing efficacious vaccines and therapeutics. Although many other mutations occurred in the path of viral evolution, we will highlight variants with more important antigenic modifications and their respective locations they are involved. Furthermore, we will discuss evidence of immune evasion by variants of concern and currently available vaccines' potential to prevent disease caused by SARS-CoV-2. This review aims to classify different novel variants of SARS-CoV-2 based on their antigenic properties, especially variants of concern that may hinder efforts to tackle ongoing pandemic, exploring evidence of escaping from the immune system and assessing current conditions vaccine efficacy against these emerging lineages.
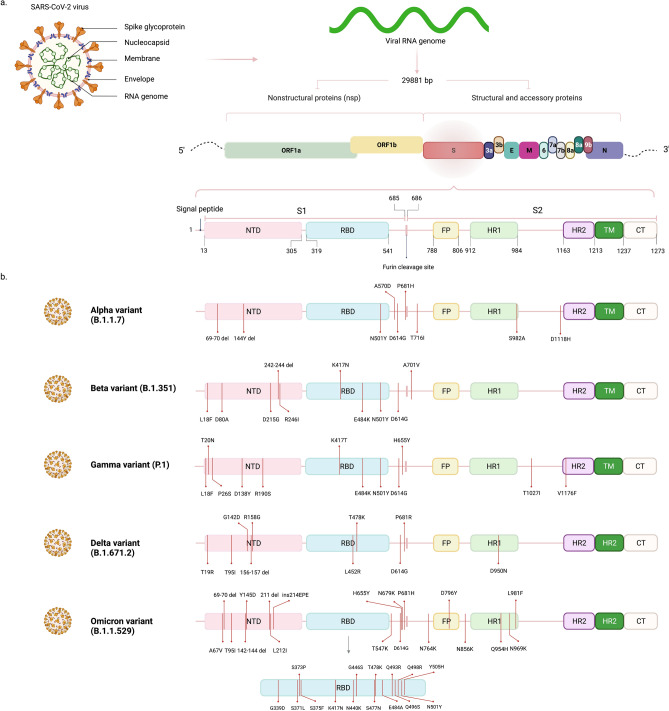
SARS-CoV-2 is an enveloped positive-sense single-stranded RNA virus with a genome of around 30 kb [6]. The viral genome comprises several distinct regions responsible for encoding virus components. These areas include multiple open reading frames (ORF), spike surface glycoprotein (S), membrane (M), nucleocapsid protein (N), and envelope (E) [15].
The majority of genome sequence pertains to ORFs specifically two overlapping ORF1a and ORF1b. These 2 ORFs encode for polypeptides cleaved by viral proteases (Mpro and PLpro) into 16 nonstructural proteins [16]. Envelope, nucleocapsid, and membrane are structural proteins essential for the assembly and morphological characteristics of the virus. Nucleocapsid acts as a package for viral RNA and ribonucleoprotein (RNP) complex [17]. In SARS-CoV-2, N protein is necessary for viral RNA replication and also raised hopes for its potential to be the target of new drugs due to its high immunogenicity [18]. Envelope of coronaviruses promotes pathogenicity by making cation channels, budding of the virus, and mediating releasing from host cell’s Endoplasmic reticulum-Golgi intermediate compartment (ERGIC) [19]. For assembling virus particles, membrane protein is the key to interacting with other structural proteins [20]. Spike glycoprotein has a pivotal role for SARS-CoV-2 entry to cells and transmissibility. Thus, we cover mutations and structural changes in S protein with corresponding extended effects on escaping from the immune system or antiviral therapies and their potential for reinfection.
Mutations are the inevitable byproducts of viral replications [21], and there are several mechanisms to trim these errors. Coronaviruses encode enzymes responsible for RNA proofreading and capping, including 3′-5′ exoribonuclease (ExoN), an endoribonuclease (NendoU), and capping enzymes in the nonstructural proteins (NSPs) region of the genome. [22]. Despite these inherent abilities, vaccination may hamper the diversity of escape mutations [23], whereas convalescent plasma therapy of immunocompetent individuals with chronic infection may be attributed to the emergence of new mutants with resistance to the neutralizing antibodies [24].
Classification of variants
It is crucial to understand the specific definitions and criteria for the variants classification system currently used for SARS-CoV-2, which includes variants of concern (VOC), variants of interest (VOI), and variants under monitoring (VUM), before delineating their features. Using classification methods for distinct clades, leading public health organizations can address current and unforeseen issues provoked by these variants. Since May 31, 2021, the WHO has labeled each variant with the Greek Alphabet letters to avoid stigmatizing and be more convenient to use [25].
According to the WHO and Centers for Disease Control and Prevention (CDC), the VOC is designated as a variant responsible for (1) increased transmissibility or impacting the COVID-19 epidemiology or (2) more severe disease manifestations or higher virulence or 3) Causing drop in the effectiveness of preventive, diagnostic, or therapeutic tools [26, 27]. This classification is dynamic based on current evidence of posing a threat to public health. As of September 27, 2021, there are four VOCs considered by WHO: (1) Alpha variant (B.1.1.7), (2) Beta variant (B.1.351), (3) Gamma variant (P.1), and (4) Delta variant (B.1.617.2) [26].
The criteria required to be fit as VOI are (1) Genome changes associated with the viral transmissibility, diseases severity, resistance to neutralizing antibodies, diagnostic or treatment approaches, and (2) evidence of causing a high number of infection or outbreak clusters in countries with increasing relative prevalence or identified as an emerging threat to global public health [26, 27]. Currently, two variants fall under the category of VOI by WHO as of September 27, 2021: (1) Lambda variant (C.37) and (2) Mu variant (B.1621) [26]. For VOCs and VOIs, CDC classification may differ from those of WHO due to different COVID-19 situations in other regions [27].
Besides VOC and VOI, the term variants under monitoring (VUM) is applied to the emerged strains with the speculation of initiating public health threats; however, current evidence is not sufficient around the epidemiological risk of new genetic changes until the mounting data reveal the possible dangerous characteristics. Hence, this category is highly dynamic that even former VOCs or VOIs may fall under this classification for a specific period [26].
The first notable mutation that appeared in April 2020 in Wuhan was the D614G mutation. It was identified by carrying a single amino acid substitution in spike protein at residue 614 from an aspartic acid (D) to glycine (G) (D614G). This G-form is now the globally prevalent form and is potentially more transmissible [28]. The S protein of SARS-CoV-2 consists of two subunits S1, S2. S1 subunit, which harbors the RBD, interacts with ACE2 (angiotensin-converting enzyme 2) of the host cell. S2 subunit, on the other hand, acts as a fusion machine by forming a six-helical bundle via heptad repeat domains 1 and 2 (HR1 and HR2) [29].
Current evolving ideas around better fitness acquired by D614G substitution could be categorized into claims about cleavage efficiency, more open conformation, higher density, and stabilization of the S protein conferred by this mutation [30].
The crucial step before the membrane fusion of the S2 subunit is the cleavage of the S protein at the furin cleavage site at the S1/S2 junction. This process is mediated by furin protease and with the aid of a cellular enzyme called transmembrane protease serine 2 (TMPRSS2) [31]. In addition to the surface protease of TMPRSS2, another crucial activator of the spike protein is cathepsin l (CTSL) [32]. This lysosomal protease has been shown to mediate the cleavage of the spike protein of SARS-CoV and MERS-CoV [33, 34]. Cathepsin L acts in the endosomal pathway of viral entry and discloses the fusion peptide of the S2 subunit, following the cleavage of the spike protein into two subunits [35]. This enzymatic activity leads to the release of viral genome content into the host cell by viral membrane fusion, while infection with SARS-CoV-2 is also reported to enhance CTSL gene expression [36]; thus, targeting this pathway might confer new insights into the treatment of COVID-19 even though viral entry might not be blocked entirely [32].
Some studies, including both in silico and in vitro, have analyzed the cleavage modifications after D614G mutation and revealed that it might improve the furin cleavage capability and also its binding to the S protein [37–39], while one study reported lower cleavage potency after the acquisition of D614G mutation [40].
Regarding conformational alterations subsequent to D614G mutation, one comprehensive study demonstrated that this substitution turned interactions between the spike protomers more symmetrical and exhibited more open conformation for RBD of the spike (75:25 ratio of open states to closed ones). These findings may imply that more exposed RBDs are accessible for antibody epitopes; thus, the mutation may sensitize the virus to neutralizing antibodies [38, 41, 42].
Studies on the spike density changes related to this mutation revealed that using virus-like particles (VLPs) harboring D614G mutation, the total amount of virion spike protein and S1:S2 ratio was substantially higher compared to the original type (D614) [30, 41].
It was previously thought that with changing one letter in the RNA sequence for the D614G variant, the S protein has a more open conformation and better ACE2 affinity [9]. However, newer studies suggest that it does not contribute to an increase in cell binding affinity or being resistant to neutralizing antibodies [42]. The D614G substitution made the virion produce more S protein and less shedding of the S1 subunit. Additionally, the considerable decrease in S1 subunit shedding is shown to link with viral entry enhancement [30, 41]. This may partly explain why the chance of infection might have increased over time [41].
One critical substitution from C to U at site 14408 in RNA-dependent RNA polymerase gene (RdRp P323L) showed an association with higher infectivity in the D614G variant [9, 43]. These modifications in the genome result in a fitter variant with more negligible founder effect or genetic drift. Clinical findings are suggestive that D614G mutation is responsible for higher viral load in the upper respiratory tract but not disease severity [44]. Following the D614G change, several SARS-CoV-2 variants have emerged, threatening health care resources and spreading more efficiently.
Alpha variant (B.1.1.7)
In September 2020, a highly transmissible variant of SARS-CoV-2, B.1.1.7 also referred to as VOC 202,012/01, 20I/501Y.Y1, and Alpha variant by WHO on May 31, 2021 [26] was reported in the United Kingdom [10]. The high number of mutations associated with B.1.1.7, including 14 amino acid altering non-synonymous mutations and 3 deletions, was unprecedented. Most of the mutations occurred to the S protein, including 69-70 del, Y144 del, N501Y, A570D, P681H, T716I, S982A, and D1118H (Fig. 1) [45]. N501Y took place in motif 501 of RBD, while asparagine (N) residue was substituted with tyrosine (Y). N501 forms part of the binding loop in the contact region of ACE2, forming a hydrogen bond with Y41 in ACE2. Tyrosine residue at this position makes interaction with intramolecular salt bridges on ACE2, thus stabilizing the spike-ACE2 complex [46, 47]. Following the small structural changes, N501Y gained the advantage of higher affinity, but it can still be neutralized by antibodies [48, 49]. In P681H, the change occurred to the furin cleavage site between the S1 and S2 subunits. These areas are crucial for attaching and fusion to host cells and viral replication. The amount of viral replication and its respective pathogenesis was weakened based on animal models and human cells for analyzing the impact of cleavage site deletion on viral replication [50]. The cleavage site between S1 and S2 is essential for the virus to infect the cell as it separates two subunits and enables S1 and S2 to function properly. Therefore, P681H mutation could have a potential impact on virus infectivity. This mutation has also been observed several times in the past [51]. Deletion 69–70 is another change in a sequence that has occurred multiple times previously, especially in Denmark's mink cluster V variant. It is created by deletion of 6 bases coding for histidine and valine at positions 69 and 70 in the N-terminal of viral S protein [52]. Currently, del 69–70 and N501Y are the two mutations causing significant concerns among scientists since clinical data del 69–70 was shown to minimize vulnerability to neutralization against polyclonal antibodies present in convalescent sera [53]. Also, N501Y was described to augment-binding affinity to ACE2 [54]. There is evidence that N501Y causes up to 56% higher transmissibility in individuals [55]. Based on an analysis of community cases affected by the B.1.1.7 variant in England, it is estimated that this lineage is 61% more lethal than wild strain and could explain more severe diseases and health hazards associated with this particular lineage [56].
Fig. 1.
a SARS-CoV-2 structure and RNA genome encoding for different structural and nonstructural protein constituents. b Spike mutational profile for variants of concern (VOCs). Note that all VOCs contain D614G substitution and, N501Y is present in all of them except the Delta variant. Substitutions in the Beta and Gamma variants are highly similar, especially escape mutations of E484K and K417N/T with the evidence of immune evasion. The Delta variant with L452R mutation inside the RBD region shares similar features to the Beta and Gamma variant in the context of increased transmissibility and immune escape. The Omicron variant is the latest VOC as of December 2021 and harbors many mutations at the spike; with 15 mutations in RBD, Omicron is a highly contagious variant with threatening immune evasion capabilities
Beta variant (B.1.351)
This variant has appeared first in South Africa and was detected in isolates as early as October 2020 and since then has raised concerns for health care and policymakers around the world [11]. This variant was also labeled the Beta variant by WHO on May 31, 2021 [26]. B.1.351 is known for its extensive number of mutations in different parts of the virus ranging from ORFs to envelop protein. This variant emerged independently from the Alpha variant (B.1.1.7) and does not contain del 69-70 mutation. The key feature of B.1.351 is E484K non-synonymous mutation in spike protein which alters RBD conformation. E484K was shown to reduce neutralization against numbers of sera [57]. Alongside E484K, there are several mutations like L18F, D80A, D215G, L242-244del, R246I, D614G, and A701V with K417N and N501Y, which also occurred in the RBD of spike protein (Fig. 1) [11, 58]. The variant gathered much attention through its capability to potentially limit therapeutic effectiveness, and also, there is evidence that it causes more severe disease [59]. K417N is also described to enhance antibody neutralizing capacities, but it has minimal impact on binding affinity to ACE2 [60]. K417 is a notable residue forming salt bridge interaction with the region in ACE2 called D30 and enhances the affinity of binding RBD to its target [48]. Given the mutations that occurred in the RBD of the Beta variant (N501Y, E484K, and K417T), the potential cross-species transmissibility of this clade was reflected by the study of Pan et al. in wild-type mouse infection. They indicated that B.1.351 could infect the mice directly with a high binding affinity to mouse ACE2 (mACE2), and also infected mice were able to infect other mice via close contact [61]. Additionally, N501Y alone may contribute to the increased infectivity in murine models [62], and other RBD mutations in the Beta variant have an additive effect in the virulence [63–65].
Gamma variant (P.1)
In mid-November 2020, a new variant was detected in Manaus, Brazil. The P.1 lineage contains three mutations localized in RBD of the spike protein: K417T, E484K, and N501Y (Fig. 1) [12]. The new variant, also known as the Gamma variant by WHO on May 31, 2021 [26], accounted for 42% of isolates found in Manaus and was also distinct enough in the manner of mutations to fit within the criteria of the new clade from B.1.1.28. The cluster of mutations laid within the mutational profile of P.1 comprises 17 substitutions and 3 deletions [12]. P.1 variant has some mutations in common with B.1.1.7 and B.1.351 variants by E484K, N501Y, and K417N, but they happened independently from each other, the process called convergent molecular adaptation [12]. Similar to the Beta variant, E484K, which is located in RBD, it can potentially create an obstacle in the path of targeting the spike protein through polyclonal antibodies in convalescent sera [66]. The panel of RBD mutations (K417T, E484K, and N501Y) in the Gamma variant transformed it into the clade with the cross-species transmission with virulence toward mice and roughly four times greater affinity to the mACE2 than the 614G mutant, the same phenomenon as the Beta variant [61, 64].
Delta variant (B.1.617.2)
Emerging from India in December 2020, B.1.617.2 led to an emergency situation by causing rapid outbreaks in more than 160 countries with over 1.3 million cases as of September 26, 2021, worldwide [13, 67, 68]. It is categorized as the subclade of B.1.617 variant with its sister variants of B.1.617.1 and B.1617.3. This lineage was labeled Delta variant by WHO on May 31, 2021, and assigned as a variant of concern (VOC) on May 11, 2021 [26]. Prominent spike mutations that took place in the Delta variant are L452R, T478K, P681R [69] along with T19R, T95I, G142D, del 156, del 157, R158G, D614G, and D950N [70] (Fig. 1). One study predicted that by acquiring K417N mutation in some isolates, RBD conformation was destabilized, and the local protein flexibility increased, which may be translated to increased binding affinity to ACE2 [71]. However, other studies demonstrated that K417N mutation attenuated the ACE-binding affinity, despite its immune evasion capabilities [60, 72, 73]. Using in silico method, L452R and T478K substitutions exhibited more positively charged RBD; thus, more binding affinity toward ACE2 and a more stable RBD-Spike complex are expected. This may partly explain the delta variant's higher transmissibility and an unexpected surge in cases in India peaking in May 2021 [71, 74]. For disease severity, a large cohort study illustrated that the patients with the Delta variant were more than two times at risk of hospitalization compared to the Alpha variant [75]. Together, these findings indicate that additional dangerous mutations may accumulate in this strain despite lacking E484Q. The appearance of the highly infectious Delta variant reflects that more dangerous and threatening variants may emerge as the infection rates elevate and bolds the necessity of widespread vaccination across the world.
Omicron variant (B.1.1.529)
A new variant with a high number of spike mutations was first identified in South Africa and Botswana in November 2021. While known by the Pango lineage of B.1.1.529, it elevated the concerns so high that WHO designated it as VOC and named it Omicron on November 26, 2021 [76]. With over 30 mutations in the spike and multiple ones in the ORFs and nucleocapsid (N), a possible new threat has been investigated since the early days, and as of December 22, 2021, it had been detected in all 6 WHO regions [77]. Spike mutations are in NTD: (A67V, 69-70 del, T95I, 142-144 del, Y145D, 211 del, L212I, and ins214EPE), RBD: (G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H), T547K, D614G, H655Y, N679K, P681H, and S2 subunit: (N764K, D796Y, N856K, Q954H, N969K, and L981F). Several notable mutations of Omicron are present in previous VOCs, including 69–70 del, K417N, T478K, N501Y, D614G, and P681H, while there are highly similar ones, such as E484A [78].
The mutations at the furin cleavage site are critical for evaluating the changes in transmissibility of different strains [79], while Omicron is accompanied by a collection of mutations at this site (H655Y, N679K, and P681H) (Fig. 1). Studies have shown that the Omicron transmissibility is remarkably superior to the Delta variant, possibly due to spike structural changes conferred by S371L, S373P, S375F, T478K, Q493R, Q498R, and N501Y mutations [80]. Also, the extended number of mutations in the RBD increased the positive electrostatic potential of the RBD interface, which interacts with the negatively charged surface of ACE2, therefore, yields to a higher affinity toward ACE2 and more infectivity [81]. However, there is speculation that the ACE2-binding affinity of Omicron RBD is similar to the Delta and Beta variants, suggesting that non-RBD mutations might play a role in increased transmissibility and enhanced cell entry of the Omicron [82].
Besides infectivity, the immune escape capability of Omicron has also been concerning since it may elevate reinfection rates and be less sensitive to neutralizing antibodies [83–85]. It was shown to evade the immunity conferred by previous VOC and VOI infections [84]. In one study, Omicron significantly impacted most of the neutralizing potency of therapeutic monoclonal antibodies (mAb), and nearly diminished the neutralization activity of BNT162b2 and AstraZeneca convalescent sera, 5 months after the two-dose vaccination. However, prior infection or a booster dose of BNT162b2, 8 months after the first dose, triggered strong immunity against Omicron [86]. Along with the immune escape of Omicron in other vaccines, the neutralization sensitivity decline of Omicron is considerably higher than previous VOCs [87]. Nevertheless, evidence of preserved CD8+ T-cell immunity against this variant opens the window of hope for current vaccines [88].
Early data regarding Omicron-related disease severity indicated that it probably leads to less severe disease than the Delta variant based on hospitalization and death rates; however, more evidence is required in more aspects of disease severity [89–91]. Omicron’s milder clinical manifestations versus the Delta might be related to its less efficient intracellular replication in TMPRSS2-rich cells, including lung cells [92].
Like the Beta and Gamma variant, Omicron may have acquired potential cross-species transmissibility due to Q493R mutation [93], which resembles Q493K mutation evolved in the study of SARS-CoV-2 adaptation in the mouse model [94].
Variants of interest
Lambda variant (C.37)
In August 2020, the earliest samples belonging to GISAID clade GR/452Q.V1 were detected in Peru [10]. It subsequently termed C.37 and quickly spread to Chile, the United States, Argentina, Ecuador [95]. On June 14, 2021, C.37 was assigned as a VOI and labeled Lambda variant by WHO [26]. It has now been detected in more than 37 countries, with over 7500 cases worldwide as of October 2021 [96]. Several notable mutations of spike and ORF1a have occurred in the Lambda variant, including seven amino acid deletion in the spike (del 246-252), G75V, T76I, L452Q, F490S, D614G, and T859N. The major alteration of ORF1a is del 3675-3677, which has also occurred in the Alpha, Beta, and Gamma variants [97].
Both L452Q and F490S are within the RBD of the spike; hence careful monitoring of the immune escaping potential of these mutations is required. L452Q is similar to L452R, which is present in Epsilon, Delta, and Iota variants and also associated with resistance to neutralizing antibodies [98, 99]. A study by Acevedo et al. on plasma samples from CoronaVac vaccines suggested that L452Q projects a similar immune escape pattern to L452R and reported higher infectivity and decreased neutralization for Lambda variant compared to wild type, Alpha, and Gamma variants [100]. Data on Lambda’s neutralization susceptibility to antibodies elicited by mRNA vaccines indicate that the variant is partially resistant to vaccine-elicited and convalescent sera neutralizing antibodies. The resistance was attributed to L452Q and F490S mutations, and the extent of the impact by L452Q was similar to that of L452R. In parallel to L452Q, F490S also caused a 2–3-fold decline in neutralizing activity of sera; however, the antibody titers remained sufficient to protect against infection caused by the Lambda variant [101]. Another feature of L452Q that resembles the L452R is in ACE2-binding affinity. It was documented that L452Q caused a three-fold increase in ACE2-binding affinity using pseudotyped virions, a pattern similar to N501Y and L452R that might contribute to higher transmission [101].
Mu variant (B.1.621)
The earliest document on the emergence of a novel variant belonging to B.1.621 lineage returns to January 2021 in Columbia. The variant is a descendent of B.1 clade and has been frequently observed later in Columbia during the third peak of the outbreak [102]. The highest prevalences of B.1621 variant have been reported in the United States, Columbia, Chile, Spain, and Mexico [103].
The variant was labeled with the Greek alphabet of Mu and subsequently classified as VOI by WHO on August 30, 2021 [26]. As of October 2021, over 11,000 cases in over 54 countries have been identified with lineages related to the Mu variant [104]. The accumulation of mutations shared with VOCs has raised concerns on potential more virulent disease and vaccine breakthrough infections. The spike mutational profile of the Mu variant includes T95I, Y144T, Y145S, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H, D950N [105]. Among spike mutations, E484K and N501Y have been associated with partial resistance to neutralizing and monoclonal antibodies, particularly E484K [64, 66, 106, 107]. Nevertheless, reports on the potency of vaccine-elicited neutralizing antibodies against the Mu variant indicate that current mRNA vaccines are still adequately neutralizing the variant [108, 109].
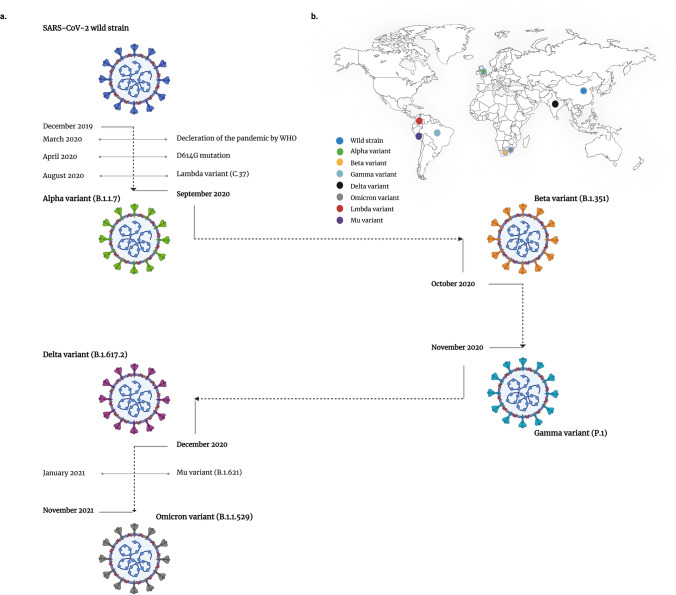
To get a better overview, the timeline of the emergence of VOCs and VOIs is depicted in Fig. 2.
Fig. 2.
a Timeline of the emergence of SARS-CoV-2 VOCs and VOIs. b Countries of origin for SARS-CoV-2 VOCs and VOIs
Miscellaneous variants
Since its appearance, SARS-CoV-2 has undergone many changes in its mutational profile with a diverse family of variants. Some of them arose independently from each other and contained concerning mutations. Nevertheless, some clades of minor concern reside in this family that may not greatly impact strategies coping with the virus, and some of them might not be present in current viral circulation, while others have alarming mutations with unknown biological significance. European variants of 20A.EU1 (also referred to as lineage B.1.177) and 20A.EU2 accounted for multiple epidemics across Europe in early summer 2020. 20A.EU1 attracted attention with spike mutation of A222V. Although A222V had slightly elevated viral titers than those without mutation by an average of 1.3-fold, it is not comparable to D614G for increasing fitness of virus [110]. The other variant of 20A.EU2 contained a notable mutation in the spike that was S477N and was previously discussed to be beneficial for viral entry [60]. Together, these variants had not reached the level of prevalence to illustrate a great effect on disease rates and shift in the distribution of confirmed cases [110].
Epsilon variant (B.1.427/B.1.429)
First discovered in an isolate from Southern California in July 2020, CAL.20C was responsible for an outbreak in California initiated in October 2020 that later spread to other countries [111]. The Pango lineage of this variant is B.1.427/B.1.429 and harbors non-synonymous L452R mutation while accounting for more than half of the cases in the US by early 2021 [98]. This variant was designated Epsilon variant by WHO on May 31, 2021, and subsequently deescalated from VOI to the Variant Under Monitoring (VUM) on July 6, 2021, due to lack of evidence for epidemiological impact [26] and recognized by five notable mutations that include: ORF1a: 14205V, ORF1b: D1183Y; S: S13I; W152C; L452R [111].
L452R occurred at one of the RBD motifs replacing amino acid leucine with arginine. The residues in this area have direct contact with ACE2, and by substitution arginine, the changes in charge interactions made it easier for RBD to fit with its receptor and improve the affinity, thus defining its more transmissibility [66, 112]. There are mounting concerns about how contagious and lethal this variant may be in normal and immunocompromised populations. According to one study conducted by colleagues at the University of California, San Francisco (UCSF), sequencing 2172 viral genomes across California state revealed that 466 (21.5%) genomes belonged to the B.1.427/B.1.429 variant [98]. The viral RNA loads found in nasal/nasopharyngeal swabs from samples infected with B.1.427/B.1.429 were roughly twice as high as other examinees, and the observed resistance to neutralizing antibodies conferred by prior infection and vaccination were 4.0 to 6.7-fold and 2-fold, respectively [98].
Eta variant (B.1.525)
This lineage arose first in December 2020 and spread across Europe, North America, Asia, and Africa, with the most prevalence in Denmark and UK. The variant labeled Eta by WHO on May 31, 2021 [26] holds some worrisome mutations shared with clades of concern B.1.351 and B.1.1.7, including E484K and del 69–70, and also mutations like Q677H, F888L in spike protein which can pose a threat to vaccine development and therapeutics according to cov-lineages.org [113, 114].
Iota variant (B.1.526)
First appeared in samples dating back to November 2020, a new lineage of the virus has been identified using a software tool named Variant Database (VDB) novel variant termed B.1.526, and by WHO Iota on May 31, 2021 [26] and as of February 2021, the so-called variant was present in 25% of viral genome sequencing isolates from New York. The finding was based on recognizing clusters with shared mutations in the spike [115]. The novel clade harbors mutations in common with previous types, including L5F, T95I, N501Y, E484K, or S477N, D253G, D614G, and A701V [115, 116]. The viral isolates are divided into two major branches containing either E484K, or S477N [115]. E484K substitution similar to that of Beta (B.1.351) and Gamma (P.1) and N501Y as of Alpha (B.1.1.7) have previously been discussed to benefit virus with higher binding affinity to cells by altering RBD conformation. The worrisome mutations in New York’s lineage are S477N and D253G. S477 is adjacent to RBD and has been investigated in European outbreaks several times. The resulting modest increase in binding affinity with ACE2 could explain the higher infectivity of this mutation [60, 117]. The pattern of mutations and prevalence of changes in S477 with multiple lineages containing the mutation might be the consequence of immune pressure from host immune responses [110, 115]. D253G has a different feature as it occurred in the supersite or N5 loop of NTD and contributes to one of the most interactions with neutralizing antibodies [107, 118]. The presence of A701V has been observed previously in B.1.351 lineage and placed adjacent to the S2 cleavage site of the nearby protomer of the spike trimer [11].
Variants of B.1.617.1/Kappa variant and B.1.617.3
First cases of distinct clade from other circulating variants around India were detected in October 2020, and WHO announced it as a variant of interest on April 4th [26]. The variant was labeled by Pango lineage B.1.617, and as of 6 May 2021, it was identified in 28 countries, predominantly India and the united kingdom [119]. The variant encompasses three sub-lineages of B.1.617.1 or Kappa, B.1.617.2 or Delta (now considered as VOC) that both labeled Greek letters by WHO on May 31, 2021 [26], and B.1.617.3 of which contained similar mutations and appeared in close intervals. Concerning mutations regarding the variant are the spike mutations of E484Q, L452R in RBD, and P681R in the furin cleavage site, which appears to be the outcome of convergent evolution. Due to locating at the furin cleavage site, P681R may enable the virus to fuse with the host cell membrane more efficiently thus, heightens viral load and improves viral transmissibility [69, 120]. E484Q and L452R are present in other variants and reported to weaken the antibody-mediated responses to the viral infection [64].
The notable mutations of the spike that occurred in B.1.617.1 are G142D, E154K, L452R, E484Q, D614G, P681R, and Q1071H [69, 121]. However, E484Q is not present in B.1.617.2, and besides common mutations with its sister lineage B.1.617.1, new substitutions were detected in this sublineage, including R158G, T478K, and D950N. Of B.1.617.3, the major differences are the presence of E484Q and the absence of R158G, with the remaining mutations being in common [122].
Cluster V (Y453F)
In November 2020, 240 cases of COVID-19 were reported related to mink farms. The new variant, in most cases, had Y453F mutation in the RBD of spike protein [123]. This potentially allowed the virus to penetrate the cells more easily resulted in altered tropism. However, in 11 cases, 3 additional mutations in a spike also located in RBD were found (del69-70, I629V, M1229I). This variant is termed Cluster V and a reduction in antibody-mediated response was detected for an average of 3.58 folds (1–13.5) [124]. These findings indicate that there could be a potential impact on antibody-mediated immunity provided by SARS-CoV-2 infection or vaccines [123]. The evolution of SARS-CoV-2 in minks has concerned health care workers, and many countries have raised surveillance levels, and the selective slaughter of minks was implemented. Since November 19th, 2020, cluster V has not been observed and may have become extinct [125].
B.1.2 lineage
20G: S677H Robin 1 was first identified in the US isolates from southern states in October 2020; the variant harbors important Q677P spike mutation. Similar to 677H, 677P showed equal significance as the place of substitution near the S1/S2 cleavage site may impose increased spread and could be the sign of simultaneous convergent evolution [126–128].
Consequences of evading immune responses
As SARS-CoV-2 keeps mutating over time, the host immunity and monoclonal antibodies against novel variants spike glycoprotein may be less effective [129]. However, new data suggest that the combination of multiple antibodies with different affinities may be helpful in antiviral strategies [130].
RBD of S protein is the primary target for neutralizing antibodies by being immunodominant and holding 90% of serum neutralizing activity [131]. However, it does not indicate that all escape mutations are necessarily at the RBD domain, which contacts antibodies. Several notable mutations occur at residues that are not located in RBD or antibody-mediated regions [132].
There are four main classes of neutralizing antibodies with distinct patterns of targeting the epitopes. Class 1 antibodies block ACE2 and only interact with the upstate of RBD, and the VH3-53 gene segment encodes these antibodies. Similar to class 1, class 2 antibodies block ACE2, but in addition to binding to more open structure RBDs, they can contact RBDs with more compact conformations and adjacent RBDs with longer loops. The majority of neutralizing antibodies comprise classes 1 and 2. Class 3 and 4 recognize outside of ACE2 sites and interact with RBD with less significance [107, 130].
Alongside RBD, at the S1 subunit of spike protein, the N-terminal domain (NTD) has also been impacted by escape mutations. There are three loops involved in NTD, with all of them belonging to one immunodominant region of NTD: N-1, N-3, N-5 loops [107]. In terms of vulnerability to neutralizing antibodies, some mutations enabled the virus to evade the immune system more efficiently, known as escape mutations. On the other hand, there are subsets of mutations conferring more susceptibility to neutralizing antibodies.
D614G mutation has been present in most current variants since its emergence early in the pandemic. Although the substitution did not occur at RBD, it favored the virus with higher transmissibility and spike density due to the allosteric effect [9, 41]. In D614, two conformations of spike with their RBD state “1-up” and “3-down” exist [133]. The ratio of the two states substantially differs in D614 and 614G variants. With cryo-electron microscopy data, the “1-up” state significantly outnumbered the “3-down” state in 614G (89% versus 18%) compared to D614, which two states had a roughly same population (54% “3-down” versus 46% “1-up”) [37, 134]. Alteration of “1-up” RBD state population in 614G shifts S protein to ACE2-competing state [134], and by more open conformation site it makes RBD more prone to neutralizing antibodies through extra exposing RBD residues [28].
In B.1.1.7 lineage, despite its rapid growth over 40 countries and bearing 8 spike mutations with key changes, data revealed a modest reduction in neutralization with sera following the first dose of mRNA vaccine (BNT162b2) administration. A similar outcome was observed using a panel of convalescent sera against pseudovirus with eight amino acid changes in spike protein found in B.1.1.7 clade. The most pronounced effect on neutralization was reported from samples containing monoclonal antibodies (mAbs) against the NTD domain (nine out of ten) of the spike. In contrast, there was no reduction in neutralization activity in “out of RBD” regions by mAbs [106].
The key mutations impacting neutralization include N501Y, del 69–70, and P681H. N501Y substitution features a critical amino acid change due to its location in RBD and has also been described in newer variants. With 20 human vaccine-elicited sera by BNT162b2 mRNA vaccine trial against 501Y, there was no indication of diminished neutralizing activity against N501Y spike mutation [135, 136]. There is a slight reduction in sensitivity of B.1.1.7 clade harboring these mutational profiles against neutralizing mAbs and convalescent sera [136]. There is no evidence of a higher reinfection rate related to B.1.1.7 compared to previous strains [137]. However, when N501Y and E484K are both present in the spike as detected in samples isolated from the number of B.1.1.7-infected patients and in all of B.1.351 viral sequences, it resulted in significant loss of neutralization (19 out of 31) against both convalescent and sera obtained after the first and second dose of mRNA vaccination [106].
Findings indicate that E484K, present in B.1.351 and P.1 clades, could potentially impair the neutralization response elicited by both vaccine and convalescent sera but not thoroughly reduce the effect [106, 138]. Marked mutations in B.1.351 South African lineage comprise N501Y, E484K, and K417N in RBD and a cluster in NTD (e.g., del 242–244, R2461) [11, 136]. The magnitude of the drop in neutralization of sera was the highest when these RBD triple mutants coexist in the viral sample, with 27% of total sera samples losing all their activity against RBD with these three substitutions [107]. For polyclonal neutralization assays, a significant decrease in neutralization was observed in the presence of E484K and N501Y simultaneously, not individually; thus, B.1.351 may compromise immune responses greatly [64]. K417N alone is reported to abolish some of the most potent mAbs relating to VH3-53/66 germline (class 1 and 2) and recently approved therapeutic mAbs like LY-CoV016 [136, 139]. However, K417N shows increased sensitivity to polyclonal antibodies [64]. There are some reports regarding alarmingly high resistance of P.1 clade to both convalescent (3.4-fold) and vaccine sera (3.8–4.8-fold) for the original virus [140]. Regarding Y453F and N439K, which occurred in Cluster V, data suggest that they potentially exhibit partial resistance to some mAbs but not to a great extent; therefore, this variant may not be the clade concern [66, 141, 142]. The other mutations with the evidence of impacting neutralizing antibodies targeting RBD are E484Q and L452R, with L452R presenting in both Indian variants of B.1.617 and California’s B.1.427/B.1.429 lineage [99, 120]. However, data from 1 study of 12 patients with B.1.617 from India indicated that convalescent sera from patients contracted with COVID-19 and vaccinee sera of BBV152 (Covaxin) provided neutralization against B.1.617 [121]. A study by Planas et al. [143] revealed that delta variant (B.1.617.2) strongly impacted the natural immunity against the previous viral strains, which reflected in four times lower antibody titers in convalescent sera compared to the Alpha variant, and also some monoclonal antibodies were lost their potency to neutralize this variant [143, 144]. It is also consistent with the reduced effectiveness of both single and two doses regimen of BNT162b2 (35.6–88% effectiveness rate) and ChAdOx1 nCoV-19 (30–67% effectiveness rate) vaccines against highly infectious delta variant compared to alpha variant [145].
Ultimately, the most threatening mutation to immune evasion is E484K, shared by both Beta (B.1.351) and Gamma (P.1) variants. Recent studies demonstrate compelling evidence that E484K noticeably diminished sera neutralization activity from class 1 and 2 mAbs up to 10 folds [66]. The amino acid change at the immunodominant sidechain of RBD alters the charges and electrostatic interactions needed for antibody binding to the RBD epitope [146]. Therefore, the large drop in neutralization activity of sera but not infectivity conferred by notable E484K mutation strongly projects that both Beta (B.1.351) and Gamma (P.1) variants could pose a threat to the efficacy of neutralizing antibodies and vaccines targeting highly immunogenic epitopes [64].
The immune evasion capability of the Omicron dramatically reduced sensitivity to the neutralizing antibodies. One study found that Omicron had 8.4-fold lower neutralizing titters than D614G mutant when tested against convalescent sera for original strain infection [84]. The main culprit for this escape is likely the synergistic effect of RBD mutations found in Alpha, Beta, Gamma, and Delta, including K417N, E484A (similar to E484K), T478K, and N501Y [58, 64, 147]. Besides, E484A and Y505H are in antibody-interfering locations that may lead to antibody resistance [148], and many new mutations at RBD could reduce neutralizing sensitivity due to conformational change [80, 82]. Despite the impacted humoral response, T-cell immunity by CD8+ cells is likely preserved against this variant, implying that vaccines may still be effective [88].
Current vaccine efficacy
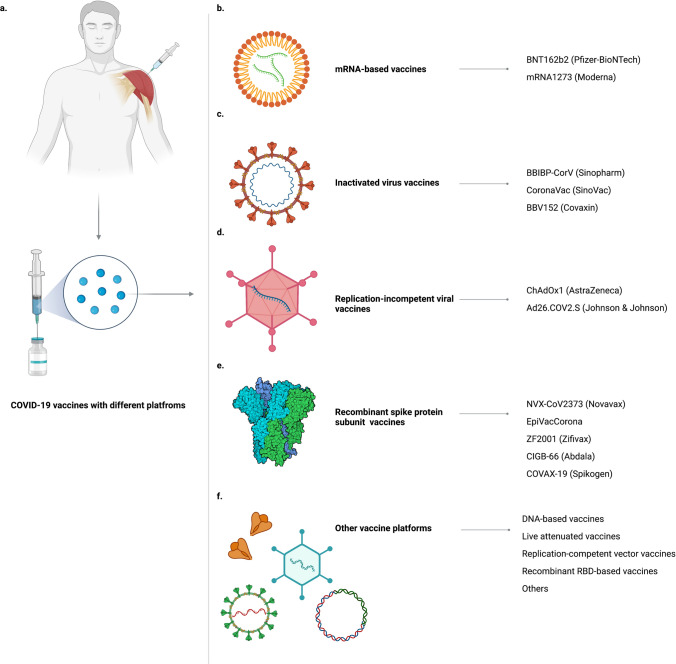
Since the outbreak of novel coronavirus, the race for vaccine developments by large multinational companies has taken at unprecedented and lightning speed. Many vaccine candidates have established robust clinical trials with efficacy studies. According to WHO, as of September 2021, there are 121 vaccines at clinical development and 194 at the preclinical stage [149]. Different ranges of platforms for vaccine production are used, including RNA-based, viral vector, inactivated viral particle, to name a few. The cutting-edge technology used for manufacturing RNA-based vaccines has captured much attention, since it can deliver new therapeutic approaches with more flexibility in a seemingly short period. Two vaccines initiatives using new RNA-based technology are Pfizer/ BioNTech BNT162b2 and Moderna’s mRNA-1273. Other vaccines of different platforms are ChAdOx1 AstraZeneca (AZD1222) in the form of an adenoviral-vector vaccine, CoronaVac (Sinovac COVID-19 vaccine), BBIBP-CorV (Sinopharm COVID-19 vaccine), and Covaxin (BBV152) inactivated-virus-based vaccines, Janssen JNJ-78436735 non-replicating viral vector, and some other developed vaccines with similar platforms. As of December 2021, 31 COVID-19 vaccines have been approved in at least 1 country worldwide, with 10 issued by WHO emergency use listing (EUL) [150]. The major vaccine platforms include mRNA-based, inactivated virus-based, replication incompetent viral vector, and recombinant spike protein subunit vaccines (Fig. 3).
Fig. 3.
a COVID-19 vaccines with different platforms. b mRNA-based vaccines. c Inactivated-virus vaccines. d Replication-incompetent viral vector vaccines. e Recombinant spike protein subunit vaccines. f Other vaccine platforms
Pfizer-BioNTech BNT162b2 vaccine comprises lipid nanoparticle encircling mRNA intended to deliver the information needed to produce the spike protein of SARS-CoV-2 [151], because the primary target for most SARS-CoV-2 vaccine candidates is the virus’s spike protein [152]. Following phase 3 clinical trial for BNT162b2, data indicated that the vaccine efficacy rate are 95% in individuals who had or did not have prior exposure to the virus [153]. Studies performed in the United States and Germany revealed a robust immune response with high titers of neutralizing antibodies and T-cell defense after administrating two 30 µg doses of BNT162b2 [154]. Even a single dose of vaccine in a community-tested study was shown to prevent 46% of catching the infection, 74% of hospitalizations, and 72% of death after 14–20 days [155].
Regarding the neutralizing potency of antibodies elicited by BNT162b2 on Alpha, Beta, and Gamma variants, a study using recombinant viruses representing these lineages revealed that vaccinee sera with full immunization effectively neutralized all three recombinant variants. However, neutralization of Beta variant was lower to Alpha and Gamma variants despite being robust, meaning vaccine is still protective against these variants [156]. Similarly, to determine neutralizing antibody activity from BNT162b2 vaccination against Alpha, Beta, and Delta variants, Wall et al. found that neutralizing antibody titers reduced 5.8-fold against Delta, 4.9-fold against Beta, and 2.6-fold against Alpha variants relative to the wild type [157].
In respect to immunity persistence conferred by BNT162b2, evidence from a large retrospective cohort study in the US presented the effectiveness of 97% (95CI: 95–99) against infection of non-Delta variants after the first month of full vaccination that declined to 67% (95CI: 45-80) after 4–5 months. For the Delta variant, the results were 93% (95CI: 85–97) and 53% (95CI: 39–65), respectively. Also, the vaccine effectiveness against hospital admission was 93% (95CI: 84–96) for the Delta variant. Together, these data provide strong support for mass vaccination since the vaccine is still highly protective, and waning immunity with time is probably the culprit for effectiveness decline rather than Delta variant immune escape [158].
Consistent with these findings, a test-negative case–control study by Bernal et al. evaluated the partial and full vaccination effectiveness of BNT162b2 against the Alpha and Delta variants. One dose of the vaccine showed the effectiveness of 35.6% (95CI: 22.7–46.4) against the Delta variant for symptomatic disease, and for two doses, the estimate was 88% (95CI: 85.3–90.1). The results for the Alpha variant were 47.5% (95CI: 41.6–52.8) and 93.7% (95CI: 91.6–95.3), respectively [145]. Regarding the highly transmissible variant of Omicron, vaccine effectiveness after the first month of primary inoculation was significantly lower than the Delta in the Danish Cohort study and was reported to be 55.2% (95CI: 23.5–73.7) [159]. Even though it largely escapes the immunity by two-dose vaccination with BNT162b2 [160, 161], multiple studies suggest that the booster dose could improve the protection against this variant [162, 163].
On February 25th, 2021, Pfizer and BioNTech announced that they are considering booster shots to participants drawn from phase 1 study, 6–12 months after receiving their two-dose regimen to evaluate the effectiveness of the third dose against new evolving variants [164]. The booster shot has been evaluated safe and efficacious at the rate of 95.6% (95CI: 89.3–98.6) against the symptomatic disease in 10,000 individuals over 16 years of age [165]. Moreover, in December 2021, Food and Drug Administration (FDA) extended the authorization of the booster dose of BNT162b2 to individuals over 16 years of age that completed primary vaccination with FDA-authorized or approved COVID-19 vaccines [166].
Moderna’s 1273 mRNA vaccine projected promising anti-SARS-CoV-2 immune response in phase 1 of clinical trials by analyzing antibody responses with enzyme-link immunosorbent assay (ELISA) following injection of three different dosages of vaccine to three groups of healthy adults between the age of 18 and 55 years, 28 days apart [167]. Also, preliminary results for the 1273 mRNA phase 2 trial with two-dose regimens of 50 and 100 µg of the vaccine were indicated to be effective with the significant immune response [168]. Phase 3 consisted of a double-blinded, randomized, and placebo-controlled study with 30,420 volunteers randomly assigned into two groups. Most of the participants received two doses of vaccine, and results were conclusive that the vaccine is effective at the rate of 94.1% (p < 0.001) [169]. Single-dose administration of the vaccine in individuals with prior exposure to the virus seems to be as effective or more beneficial than two-dose regimen in terms of the level of protection [170].
Data on whether 1273 mRNA can trigger an adequate immune response to tackle new variants or not were analyzed using a pseudovirus-based model bearing a range of spike mutations present in new emerging variants. The test was performed based on mutations from D614G, B.1.1.7, B.1.351, 20E(EU1), 20A.EU2, N439K-D614G, and cluster V lineages. The drop in neutralization against both full and partial subset of mutations in B.1.1.7 by serum from participants in phase 1 was insignificant. On the other hand, assays for the partial panel of mutations related to B.1.351 resulted in 2.4 folds, and for the whole set, 6.4 folds of reduction in neutralization activity elicited by the vaccine. Notably, the serum samples' concentration neutralized the pseudotype virus was low; therefore, the vaccine’s efficacy against B.1.351 is not conclusive and needs further investigation. Neutralizing activity of vaccine against 20E (EU1), 20A.EU2, N439K-D614G, and cluster V clades equal the D614G isolate. Thus, the vaccine coverage for these variants remains no place for concern [171]. Likewise, B.1.351, P.1, and each strain carrying these E484K, K417N/T, or N501Y substitutions together have also been demonstrated with the pseudotyped virus to compromise the efficacy of mRNA vaccines like Moderna’s 1273 mRNA vaccine [64].
To further evaluate neutralizing capacity elicited by full immunization with mRNA-1273, a study by Choi et al. using recombinant spike-pseudotyped variants showed that neutralizing titers against the Alpha variant were minimally different from those of the D614G mutant. Neutralization titers for other variants, including Delta and Gamma, were compromised by the decline range from 2.1-fold to 8.4-fold, with the greatest observed in the Beta variant. Nevertheless, the study implied that all variants remained susceptible to neutralizing antibodies induced by mRNA-1273 [172]. The greatest dip in vaccine neutralization potency was observed in Omicron with 49–84-fold less sensitivity to neutralizing antibodies than the D614G variant, 4 weeks after primary vaccination [173]. In parallel, vaccine efficacy dropped to 36.7% (95CI: -69.9-76.4) in a cohort study in Denmark [159]; however, similar to BNT162b2, a booster dose of vaccine could confer proper response against this variant [173].
The company announced its plan in February 2021 to address SARS-CoV-2 variants of concern, including a variant-specific booster shot named mRNA-1273.351 and two other similar approaches [122]. In November 2021, the FDA authorized a single booster dose of mRNA-1273 for individuals over 18 years of age who completed primary vaccination of authorized or approved COVID-19 vaccines [174].
The next vaccine we are dealing with is ChAdOx1 AstraZeneca (AZD1222) vaccine. It is fundamentally different from the two previous vaccines as it is an adenovirus vector vaccine. With phase 1/2 trial (registered at clinicaltrial.gov NCT04324606) and phase 2/3 (NCT04400838), the vaccine is gaining more upcoming data, including its better tolerance in older adults [175]. Large-scale randomized and controlled phase 3 carried in UK, Brazil and South Africa between April 23 and November 4 showed an overall efficacy of 70.4%. The 23,848 participants were divided into two groups in which one group received a half dose of vaccine at the first shot and the original dose for the second, and the other group received the original dose both times. The efficacy for the group who received the half dose first was 90% and for the two original dose groups was 62.1% [176]. Even single-dose administration of vaccine was reported to confer good immunity against the virus for 90 days, and if the interval between first and second shot is prolonged to 12 weeks instead of 6 weeks, it can boost antibody responses [176]. The vaccine potency to deliver a sufficient immune response against B.1.1.7 is equal to other clades based on a recent trial [177]. However, it projected disappointing results against B.1.351 and did not confer immunity against mild and moderate disease cases due to B.1.351. The safety efficacy was tested with two-dose regimens of vaccine or placebo with the interval of 21–35 days. Both pseudotyped and live virus neutralization assays showed the efficacy of 10.4% against the Beta (B.1.351) variant [178]. An exploratory analysis of ChAdOx1 efficacy at phase 3 of the trial in Brazil depicted that after 2 doses, the efficacy against the Gamma variant for the symptomatic disease was 64% (95CI: 2-87). The study also pointed to the maintained protection of ChAdOx1 against the Gamma variant despite a dip in efficacy compared to other variants. However, the conclusion was unpowered due to the limited cases and wide confidence interval [179]. On ChAdOx1 effectiveness against Delta variant, Bernal et al. [145] estimated one dose effectiveness against the symptomatic disease of 30.7% (95CI: 25.2–35.7) and 67% (95CI: 61.3–71.8) for two doses. Data around vaccine neutralization activity against the Omicron variant exhibited minimal activity after 5 months of two-dose immunization [86]. Also, no efficacy was yielded by the heterologous regimen of BNT162b2/ChAdOx1 against this mutant; however, a significant increase in neutralization titers was observed in the BNT162b2-boosted heterologous regimen [83].
Janssen JNJ-78436735 or Ad26.COV2.S vaccine is an adenovirus serotype 26 vaccine that encodes the full-length spike protein of SARS-CoV-2. In randomly assigned, placebo-controlled phase 1 and 2 trials of the vaccine, 29 days following the first vaccine dose, 90% of participants were detected with neutralizing antibody in their sera, and after 57 days, all the participants showed even increased titers of neutralizing antibody [180]. In phase 3 of the study, 43,783 participants over the age of 18 were examined 14 and 28 days after single-dose administration, and data revealed that the vaccine are effective at preventing moderate-to-severe forms of the disease at the rate of 66.9% (95CI: 59.0–73.4) at least 14 days after administration and 66.1% (95CI: 55.0–74.8) at least 28 days after receiving the vaccine. It was also effective for preventing hospitalization, with the efficacy of 93.1% (95CI: 72.2–99.2) and 100% (95CI: 74.3–100) in a similar timeline. The data also suggest that the vaccine efficacy reached 72% (95CI: 58.2–81.7) in the United States and 64% (95CI: 41.2–78.7) in South Africa against moderate-to-severe disease, with at least 28 days after receiving the vaccine. [181]. With the predominancy of the Beta variant at the phase 3 trial in South Africa, the vaccine efficacy remained high at preventing moderate-to-severe disease and hospitalization [181].
A study by Barouch et al., evaluated the antibody responses against VOC in the subjects receiving Ad26.COV2.S vaccine in 29 and 239 days after inoculation. The greatest impact was reported against the Beta variant on day 29, with over 13-fold reduction in neutralizing antibody titers than wild strain; however, the decrease was moderate with over 3-fold on day 239. Other variants, including Alpha, Gamma, and Delta, had higher titers and showed increased neutralization titers over time, suggesting a durable immunity induced by Ad26.COV2.S vaccine [182]. However, Ad26.COV2.S showed almost no antiviral activity against the Omicron variant in the pseudovirus assay study [93].
As of December 2021, among 137 SARS-CoV-2 vaccine candidates in the clinical studies, protein subunit (PS) vaccines account for 36% of all platforms [183]. Currently, 11 PS vaccines have been approved by at least one country [150]. Here, we discuss Novavax NVX-CoV2373, Zifivax ZF2001, EpiVacCorona, Abdala CIGB-66, and COVAX-19.
Novavax NVX-CoV2373 is a developing vaccine consisting of recombinant spike protein with Matrix-M1 adjuvant, which is encapsulated by the nanoparticle. Preliminary data with post hoc analysis from phase 3 clinical trials from 15,000 individuals in the UK showed that by full immunization, it is 96.4% (95CI: 73.8–99.4) effective at preventing the symptomatic disease caused by non-alpha variants, predominantly the wild type; however, it was efficacious at the rate of 86.3% (95CI: 71.3–93.5) against the Alpha variant’s symptomatic disease. The overall efficacy for protecting against SARS-CoV-2 infection was 89.7% (95CI: 80.2–94.6) [184]. Similar to other vaccines, B.1.351 has also impacted NVX-CoV2373 efficacy. The trial conducted in South Africa resulted in 60.1% (95CI: 19.9–80.1) efficacy in HIV-negative subjects and 49.4% (95CI: 6.1–72.8) for all participants where the majority of the cases (25 of 27) were involved with B.1.351 (Table 1) [185, 186]. Regarding the Omicron and Delta variants, Novavax announced cross-reactive immune responses for primary two-dose and strong immune response with the third dose after 6 months of the second dose [187].
Table 1.
Summary of SARS-CoV-2 novel variants of concern and current vaccine efficacy
| Lineage name | Mutations related to spike protein | First detected | Transmissibility/disease severity | Vaccines’ efficacy/effectiveness |
|---|---|---|---|---|
| B.1.1.7 (Alpha) | 69-70 del, 144 del, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | September 2020, UK | Increased transmissibility and risk for serious illness [55, 59, 224] |
The majority of current vaccines displayed decent coverage against all outcomes [225] For asymptomatic infection, AZD1222 was reported to have a moderate loss of efficacy [177] CoronaVac showed a moderate loss in neutralization capacity, and NVX-CoV2373 showed a mild decline in efficacy against symptomatic disease [184, 206] |
| P.1 (Gamma) | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | November 2020, Brazil | Increased transmissibility and risk for serious illness [12, 59, 224] |
Scarce data on the impact [225] mRNA-1273 and BNT162b2 exhibited a partial reduction in neutralization but provided robust protection against all forms of the disease [140, 226, 227] AZD1222 has mildly lost efficacy against symptomatic disease and Ad26.COV2.S showed mild to moderate loss of neutralization capacity [179, 182] CoronaVac has significantly lost neutralization potency [208] |
| B.1.351 (Beta) | L18F, D80A, D215G, L242-244 del, R246I, K417N, E484K, N501Y, D614G, A701V | October 2020, South Africa | Increased transmissibility and risk for serious illness [11, 59, 224] |
Protection against severe illness reserved [225] Protection against symptomatic disease attenuated [225] AZD1222 and NVX-CoV2373 exhibited considerable efficacy loss against symptomatic disease, and CoronaVac showed a significant decline in neutralization capacity [178, 185, 206] Ad26.COV2.S, mRNA-1273, and BNT162b2 showed moderate loss in neutralization [182, 226, 228, 229] |
| B.1.617.2 (Delta) | T19R, T95I, G142D, 156-157 del, R158G, L452R, T478K, D614G, P681R, D950N | December 2020, India | Increased transmissibility and risk for serious illness [74, 75, 224] |
Protection against severe illness reserved [225] Possible reduction in protection against infection and symptomatic disease [225, 230] Neutralizing potency of BNT162b2, mRNA-1273, and Covaxin have been impacted mildly. AZD1222 and BNT162b2 showed a mild reduction in efficacy against symptomatic disease [121, 144, 145] Ad26.COV2.S and CoronaVac significantly lost the neutralization capacity [206, 231] |
| B.1.1.529 (Omicron) | A67V, del69-70, T95I, 142-144 del, Y145D, 211 del, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F | November 2021, South Africa and Botswana | Increased transmissibility and probable less risk for serious illness [80, 81, 89–91] |
All studied vaccines have been impacted as this variant showed severe immune evasion and elevated risk of reinfection [77] Booster doses of studied vaccines enhanced neutralization titers against this variant [83, 162, 163, 187, 192, 200] |
Zifivax vaccine, also named ZF2001, is a three-dose dimeric RBD-based vaccine developed by Anhui Zhifei Longcom Biopharmaceutical. In phases 1 and 2 of randomized placebo-controlled trials in China, both two-dose and three-dose regimens have been evaluated for safety and immunogenicity. The interval between intramuscular doses was 30 days in either two- or three-dose regimen of 25 or 50 µg groups and the placebo group. In phase 2, on a two-dose schedule, the seroconversion rate for the 25 µg group was 76% and 72% for the 50 µg group, 14 days after the second dose. The results for the three-dose regimen were 97% and 93% for the two groups, respectively, and the three-dose 25 µg regimen vaccine was reported to be safe and immunogenic [188]. The vaccine is currently in phase 3 that has extended outside China, including Ecuador, Indonesia, Pakistan, and Uzbekistan. The neutralization capacity of ZF2001 was evaluated against 4 VOCs (Alpha, Beta, Gamma, and Delta) in the study of Zhao et al. The results indicated that sera neutralization activity was retained against all four VOCs despite the decrease in neutralizing titers [189]. These findings are consistent with another study that examined the neutralizing titers of ZF2001 vaccinee sera against the Beta variant. The study exhibited no evidence of immune escape by Beta variant against ZF2001 even though the titers declined 1.59-fold compared to the wild strain [190]. Against the Omicron variant, neutralizing antibody titer of three-dose (0–1–5) ZF2001 was reduced 3.1-fold compared to the prototype virus, and neutralizing antibodies were found in all study participants. Thus, it induced proper immunity against this heavily mutated variant [191]. The results of the third-dose vaccination by ZF2001 in individuals with primary vaccination of inactivated vaccines also showed that it could boost the neutralization titers against the Omicron [192].
EpiVacCorona or Aurora-CoV is another protein subunit vaccine developed by Russia's Vektor center of virology and biotechnology. It is based on the synthetic immunogenic peptide representing viral spike antigens conjugated to a protein carrier with aluminum hydroxide adjuvant. It is administered intramuscularly in two doses, 14 days apart, and was reported to be safe and immunogenic in the preclinical study [193]. Phases 1 and 2 of the trial were conducted in Russia and demonstrated the seroconversion rate of 100% in participants after 42 days from the first injection [194]. Phase 3 was initiated in November 2020 with 3000 participants to evaluate the vaccine’s efficacy (NCT04780035); however, no data on this phase, along with the vaccine’s neutralizing potency against the VOCs, has been published.
Two other protein subunit vaccines approved by at least one country are CIGB-66 (monomeric RBD-based three-dose vaccine developed by the center for genetic engineering and biotechnology, Cuba) and COVAX-19 (recombinant protein subunit two-dose vaccine developed by CinnaGen institute, Iran, and Vaxine company, Australia). These vaccines exhibited antigenicity and safety in preclinical studies [195, 196]; however, for more detailed data around vaccine efficacy and protection against VOCs, results of clinical trials are pending.
Regarding inactivated-virus-based COVID-19 vaccines, results of the clinical trials highlight the safety and high immunogenicity of these vaccines. BBIBP-CorV, a two-dose regimen vaccine developed by Sinopharm’s Beijing institute of biological products (BIBP), was safe and well-tolerated alongside inducing a proper humoral response in all participants at phases 1 and 2 of its trial [197]. Phase 3 of a large randomized double-blinded controlled trial in Bahrain and the United Arab Emirates demonstrated that the BBIBP-CorV effectively prevents 78.2% (95CI 64.8–86.3%) against symptomatic disease and 100% against severe disease 14 days following the injection of the second dose [198].
For assessing the antibody responses conferred by BBIBP-CorV against novel variants, a study by Jeewandara et al. comprehensively evaluated neutralizing antibody levels against RBD of the Alpha (B.1.1.7), Beta (B.1.351), Delta (B.1.617.2), and wild strains. Although the vaccine exhibited high seroconversion rates among participants, it was shown that the Alpha, Beta, and Delta variants had impacted anti-RBD antibody levels compared to the wild strain with the reduction of 1.1-fold, 10-fold, and 1.38-fold in titers, respectively [199]. Also, the vaccine is considered to confer immunity against the Beta and Delta variants to the same extent as natural infection [199]. For assessing protection against the Omicron variant, a study from China found that two-dose vaccination with BBIBP-CorV provides a weak immune response 8 months after the prime vaccination. However, a booster dose enhanced the neutralizing antibody titers against the Omicron despite showing partial loss of sensitivity to neutralization provided by booster [200].
CoronaVac (Sinovac COVID-19 vaccine) is another inactivated whole-virion vaccine that presented promising results in experimental and real-world situations. The seroconversion of neutralizing antibodies against SARS-CoV-2 in phases 1 and 2 of its trial in China varied by different doses of 3 and 6 µg and vaccine schedules of 14 and 28 days after receiving the first dose. Phase 2 showed a higher immune response with the seroconversion of over 90% at both 3 and 6 µg groups [201]. The CoronaVac was evaluated as a safe, highly tolerable, and protective vaccine by inducing humoral responses against in both 18–59 and over 60 years [201, 202].
A phase 3 trial of the CoronaVac was conducted in Turkey, with the primary outcome of preventing symptomatic diseases at least 14 days following the second dose. The results indicated the vaccine efficacy of 83.5% (95CI: 65.4–92.1) and prevented hospitalization and death by 100% [203]. Also, phase 3 of the trial in Brazil on healthcare professionals with the interval of 14 days between two doses revealed 50.7% (95CI: 36.0–62.0) efficacy against symptomatic disease, 83.7% (95CI: 58.0–93.7) against cases that required medical assistance, and 100% (95CI: 56.4–100.0) against severe disease [204]. A real-world study involving 10.2 million subjects in Chile demonstrated the effectiveness of full immunization with CoronaVac against symptomatic disease by 65.9% (95CI: 65.2–66.6), 87.5% (95CI: 86.7–88.2) against hospitalization, 90.3% (95CI: 89.1–91.4) against ICU admission, and 86.3% (95CI: 84.5–87.9) against death [205].
The impact of variants on CoronaVac efficacy or neutralizing antibody titers has been reported for Alpha, Beta, Gamma, and Delta variants. A recent study found that CoronaVac elicits lower neutralizing antibody titers against Alpha, Beta, and Delta variants than natural infection [206]. An estimate by a negative case–control study on healthcare workers in Manaus, Brazil, with the dominance of Gamma variant in late 2020, showed the efficacy of 49.6% (95CI: 11.3–71.4) 14 days following single-dose and paradoxically 36.8% (95CI: 54.9–74.2) for two doses against symptomatic disease [207]. Consistently, a significant reduction in the neutralizing capacity of CoronaVac-elicited antibodies has been reported for the Gamma variant compared to a B.1 lineage isolate [208]. For the Delta variant, a study from China evaluated the efficacy of single-dose and complete vaccination, and the adjusted efficacy of full immunization against pneumonia was 69.5% (95CI: 42.8–96.3), and it was 100% efficacious at preventing severe disease [209]. Against the Omicron variant, CoronaVac conferred no detectable neutralizing antibodies 56 days after the first administration of prime vaccination in the recipient of a study in Hong Kong [160].
Covaxin (BBV152), developed by Bharat Biotech, harbors inactivated whole-virion technology similar to BBIBP-CorV and CoronaVac. Also, it contains a Toll-like receptor 7/8 agonist molecule as an adjuvant [210]. In phase 1 of the randomized controlled trial on 375 participants with three vaccine formulations administered 2 weeks apart, the seroconversion rates in all groups were over 80%, and in two groups, 16 participants showed proper T-cell responses. Results of phase 1 indicated the safety, tolerability, and immunogenicity of the vaccine [210]. In phase 2, with a schedule of 4 weeks between doses, seroconversion rates and T-cell responses were more robust than phase 1, at days 56 and 42 after the first injection, respectively. Also, the follow-up of samples at phase 1, 3 months after receiving the second dose, showed that neutralizing antibodies remained elevated in all participants [211].
Phase 3 of the trial analyzed the efficacy, immunogenicity, and safety of BBV152 on healthy adults 18–98 years old, 14 days after the second dose. The vaccine presented the efficacy of 63.6% (95CI: 29.0–82.4) against asymptomatic disease and 93.4% (95CI: 57.1–99.8) against severe disease. The study also evaluated the protection of BBV152 against the Delta variant (B.1.617.2) by 65.2% (95CI: 33.1–83.0) [212]. Data on vaccine efficacy against Alpha, Beta, and Delta variants indicate that BBV152 could effectively neutralize the Alpha variant [213], and despite impacting the neutralization titer by the Beta and Delta variants, the vaccine still protects against two variants [214, 215].
Conclusion
Large-scale genomic data and surveillances of the SARS-CoV-2 genome mainly contribute to spike glycoprotein. This region of the virus is the key to entering and invading the host cells, and it is a major target for neutralizing antibodies. Emerging new variants of SARS-CoV-2 have risen transmissibility and overall fitness and encouraged scientists worldwide to come up with new strategies against viral infection. This review covered the most notable variants and their mutations, enabling them to attach to the host cells faster and more efficiently. Large-scale genomic data and surveillances of the SARS-CoV-2 genome mainly contribute to spike glycoprotein. This viral component is the key to entering and invading the host cells and a central target for neutralizing antibodies. Emerging new variants of SARS-CoV-2 have risen transmissibility and overall fitness and encouraged scientists worldwide to develop new strategies against viral infection. This review covered the most notable variants and their mutations, enabling them to attach to the host cells faster and more efficiently. Furthermore, we pointed to the efficacy and neutralizing potency of currently developed vaccines against the concerning clades. The evolutionary pathway of the virus has led to the evasion of host immune response by emerging new mutants. Escape mutations could also ignite breakthrough infections and threaten global vaccination efforts. Nevertheless, current vaccines are still highly effective at preventing hospitalization and death by COVID-19.
While the virus keeps constantly mutating as it replicates in the large population, novel changes to the spike protein impact protection conferred by the vaccines, as seen in the variants of concern, especially Omicron. Thus, new approaches for developing universal vaccines seem sensible to tackle future variants by targeting shared features of coronaviruses. The ideal and desirable properties of universal coronavirus vaccines and critical research questions are reflected by Morens et al. Indeed, animal and clinical studies should aim to build vaccines with broad and long-lasting immunity against SARS-CoV-2 and optimally against all coronaviruses [216].
The past influenza pandemics and current HIV pandemic could hint us to prioritize addressing technical and real-world challenges of universal vaccines. Lessons from the universal vaccine for rapidly mutating influenza virus suggest that targeting conserved epitopes with sufficient immunogenicity and cross-reactivity may be a possible approach despite difficulties in development [217]. Candidates might be among proteins pivotal for coronaviral replication, of which mRNA codes could be added to the current mRNA-based vaccines [218]. Therefore, more profound knowledge around SARS-CoV-2 replication and related pathways are essential for accelerating universal vaccines.
Recent advancements in gene-editing tools, including CRISPER engineering, enabled scientists to propose a versatile phage-based vaccine platform that triggers robust humoral and T-cell responses against diverse epitopes of SARS-CoV-2 antigens [219]. This new delivery route for antigens is easy to update according to emerging variants. Another way the phage-based vaccines can be adapted is to insert a mixture of conserved epitopes into the phage [220]; therefore, exploring preserved immunogenic epitopes of SARS-CoV-2 and novel routes for exposing antigens potentially helps develop more protective vaccines. The concept of targeting conserved epitopes could also help prepare universal vaccines against other rapidly mutating viruses, including HIV-1 [221–223].
Collectively, the behavior of the coronaviruses signals us to shed light on shared characteristics and detailed replication pathways of this family. In parallel, we should gain a broader understanding of how mutations might affect the endemic transition of the SARS-CoV-2 pandemic as a possible scenario since the emergence of a highly contagious but less lethal variant of Omicron. To be optimistic about the future of the current pandemic and prevent the next ones, one game-changing prevention measure for global collaboration could be focusing on mass vaccination programs and the insight for robust universal coronavirus vaccines.
Acknowledgements
Not applicable.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- ELISA
Enzyme-linked immunosorbent assay
- ERGIC
Endoplasmic reticulum–Golgi intermediate compartment
- HIV
Human immunodeficiency virus
- ICU
Intensive care unit,
- MAb
Monoclonal antibody
- mACE2
Murine angiotensin- converting enzyme 2
- MERS-CoV
Middle East Respiratory Syndrome Coronavirus
- Mpro
Main protease, mRNA: Messenger ribonucleic acid
- COVID-19
Coronavirus disease 2019
- NTD
N-terminal domain
- ORF
Open reading frame
- PLpro
Papain-like protease
- RBD
Receptor-binding domain
- RdRp
RNA-dependent RNA polymerase
- RNA
Ribonucleic acid
- RNP
Ribonucleoprotein
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- TMPRSS2
Transmembrane serine protease 2
- UCSF
University of California San Francisco
- VDB
Variant database
- VLP
Virus-like particle
- WHO
World health organization
Author contributions
AR was the major contributor in writing and editing the manuscript. RD contributed in writing and preparing. FK carefully revised the manuscript. SA assisted in the literature search and editing. RJ framed the concept of the study and supervised the whole process. All authors have read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Amirmasoud Rayati Damavandi, Email: am-rayatid@student.tums.ac.ir.
Razieh Dowran, Email: raziehdowran@yahoo.com.
Sarah Al Sharif, Email: salshar@gmu.edu.
Fatah Kashanchi, Email: fkashanc@gmu.edu.
Reza Jafari, Email: Jafari.reza@umsu.ac.ir.
References
- 1.Weiss SR, Leibowitz JL. Coronavirus pathogenesis. Adv Virus Res. 2011;81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet (Lond, Engl) 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO COVID-19 Dashboard (2020) Geneva: World Health Organization, 2020. https://covid19.who.int/. Accessed 31 Dec 2021
- 9.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–27.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, Maceil A, et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage—United States, December 29, 2020-January 12, 2021. MMWR Morbid Mortal Weekly Rep. 2021;70(3):95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020;2020:5. [Google Scholar]
- 12.Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DDS, Mishra S, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Sci (N Y, NY). 2021;372(6544):815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Threat Assessment Brief: Emergence of SARS-CoV-2 B.1.617 variants in India and situation in the EU/EEA. 2021 . https://www.ecdc.europa.eu/en/publications-data/threat-assessment-emergence-sars-cov-2-b1617-variants. Accessed 30 Sep 2021
- 14.Ferrareze PAG, Franceschi VB, Mayer AM, Caldana GD, Zimerman RA, Thompson CE. E484K as an innovative phylogenetic event for viral evolution: genomic analysis of the E484K spike mutation in SARS-CoV-2 lineages from Brazil. Infect Genet Evol. 2021;93:104941. doi: 10.1016/j.meegid.2021.104941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Nyodu R, Maurya VK, Saxena SK (2020) Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2). Coronavirus Disease 2019 (COVID-19) 2020:23–31
- 16.Finkel Y, Mizrahi O, Nachshon A, Weingarten-Gabbay S, Morgenstern D, Yahalom-Ronen Y, et al. The coding capacity of SARS-CoV-2. Nature. 2021;589(7840):125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- 17.McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang S, Yang M, Hong Z, Zhang L, Huang Z, Chen X, et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmaceut Sin B. 2020;10(7):1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A, Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat Struct Mol Biol. 2020;27(12):1202–1208. doi: 10.1038/s41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boson B, Legros V, Zhou B, Siret E, Mathieu C, Cosset FL, et al. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J Biol Chem. 2020;296:100111. doi: 10.1074/jbc.RA120.016175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grubaugh ND, Petrone ME, Holmes EC. We shouldn't worry when a virus mutates during disease outbreaks. Nat Microbiol. 2020;5(4):529–530. doi: 10.1038/s41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sevajol M, Subissi L, Decroly E, Canard B, Imbert I. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niesen MJM, Anand P, Silvert E, Suratekar R, Pawlowski C, Ghosh P, et al. COVID-19 vaccines dampen genomic diversity of SARS-CoV-2: unvaccinated patients exhibit more antigenic mutational variance. medRxiv. 2021;2021:8. [Google Scholar]
- 24.Kemp SA, Collier DA, Datir RP, Ferreira I, Gayed S, Jahun A, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592(7853):277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.WHO announces simple, easy-to-say labels for SARS-CoV-2 Variants of Interest and Concern (2021) https://www.who.int/news/item/31-05-2021-who-announces-simple-easy-to-say-labels-for-sars-cov-2-variants-of-interest-and-concern. Accessed 27 Sep 2021
- 26.Tracking SARS-CoV-2 variants (2021) https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Accessed 27 Sep 2021
- 27.SARS-CoV-2 Variant Classifications and Definitions (2021) https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html#anchor_1632154493691. Accessed 27 Sep 2021
- 28.Mansbach RA, Chakraborty S, Nguyen K, Montefiori DC, Korber B, Gnanakaran S. The SARS-CoV-2 Spike variant D614G favors an open conformational state. Sci Adv. 2021;7:16. doi: 10.1126/sciadv.abf3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17(7):765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson CB, Zhang L, Farzan M, Choe H. Functional importance of the D614G mutation in the SARS-CoV-2 spike protein. Biochem Biophys Res Commun. 2021;538:108–115. doi: 10.1016/j.bbrc.2020.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann M, Kleine-Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78(4):779–84.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang IC, Bosch BJ, Li F, Li W, Lee KH, Ghiran S, et al. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J Biol Chem. 2006;281(6):3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gierer S, Bertram S, Kaup F, Wrensch F, Heurich A, Krämer-Kühl A, et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol. 2013;87(10):5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu T, Luo S, Libby P, Shi GP. Cathepsin L-selective inhibitors: a potentially promising treatment for COVID-19 patients. Pharmacol Therapeut. 2020;213:107587. doi: 10.1016/j.pharmthera.2020.107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao MM, Yang WL, Yang FY, Zhang L, Huang WJ, Hou W, et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct Target Ther. 2021;6(1):134. doi: 10.1038/s41392-021-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gobeil SM, Janowska K, McDowell S, Mansouri K, Parks R, Manne K, et al. D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 2021;34(2):108630. doi: 10.1016/j.celrep.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohammad A, Alshawaf E, Marafie SK, Abu-Farha M, Abubaker J, Al-Mulla F. Higher binding affinity of furin for SARS-CoV-2 spike (S) protein D614G mutant could be associated with higher SARS-CoV-2 infectivity. Int J Infect Dis. 2021;103:611–616. doi: 10.1016/j.ijid.2020.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharyya C, Das C, Ghosh A, Singh AK, Mukherjee S, Majumder PP, et al. SARS-CoV-2 mutation 614G creates an elastase cleavage site enhancing its spread in high AAT-deficient regions. Infect Genet Evol. 2021;90:104760. doi: 10.1016/j.meegid.2021.104760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniloski Z, Jordan TX, Ilmain JK, Guo X, Bhabha G, tenOever BR, et al. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. Elife. 2021;2021:10. doi: 10.7554/eLife.65365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11(1):6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Cai Y, Xiao T, Lu J, Peng H, Sterling SM, et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science (N Y, NY) 2021;372(6541):525–530. doi: 10.1126/science.abf2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pachetti M, Marini B, Benedetti F, Giudici F, Mauro E, Storici P, et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18(1):179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Groves DC, Rowland-Jones SL, Angyal A. The D614G mutations in the SARS-CoV-2 spike protein: Implications for viral infectivity, disease severity and vaccine design. Biochem Biophys Res Commun. 2021;538:104–107. doi: 10.1016/j.bbrc.2020.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A et al (2020) Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations,Written on behalf of COVID-19 Genomics Consortium UK. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563
- 46.Polydorides S, Archontis G. Computational optimization of the SARS-CoV-2 receptor-binding-motif affinity for human ACE2. Biophys J. 2021;2021:1. doi: 10.1016/j.bpj.2021.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ortuso F, Mercatelli D, Guzzi PH, Giorgi FM. Structural genetics of circulating variants affecting the SARS-CoV-2 spike/human ACE2 complex. J Biomol Struct Dyn. 2021;2021:1–11. doi: 10.1080/07391102.2021.1886175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 49.Villoutreix BO, Calvez V, Marcelin AG, Khatib AM. In Silico Investigation of the New UK (B.1.1.7) and South African (501Y.V2) SARS-CoV-2 variants with a focus at the ACE2-spike RBD interface. Int J Mol Sci. 2021;22:4. doi: 10.3390/ijms22041695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson BA, Xie X, Bailey AL, Kalveram B, Lokugamage KG, Muruato A, et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591(7849):293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christian HCI, John N, Paul EO, Idowu O (2020) Detection of SARS-CoV-2 P681H Spike Protein Variant in Nigeria 2020. https://virological.org/t/detection-of-sars-cov-2-p681h-spike-protein-variant-in-nigeria/567
- 52.Kemp S, Datir R, Collier D. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion ΔH69/ΔV70. bioRxiv. 2020;2020:1. [Google Scholar]
- 53.Kemp SA, Meng B, Ferriera IATM, Datir R, Harvey WT, Papa G, et al. Recurrent emergence and transmission of a SARS-CoV-2 spike deletion H69/V70. bioRxiv. 2021;2020:12. [Google Scholar]
- 54.Chan CEZ, Seah SGK, Chye DH, Massey S, Torres M, Lim APC, et al. The Fc-mediated effector functions of a potent SARS-CoV-2 neutralizing antibody, SC31, isolated from an early convalescent COVID-19 patient, are essential for the optimal therapeutic efficacy of the antibody. bioRxiv. 2020;2020:107. doi: 10.1371/journal.pone.0253487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Sci (N Y, NY). 2021;372(6538):3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davies NG, Jarvis CI, Edmunds WJ, Jewell NP, Diaz-Ordaz K, Keogh RH. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593(7858):270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592(7855):616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. bioRxiv. 2021;2021:428137. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 59.Funk T, Pharris A, Spiteri G, Bundle N, Melidou A, Carr M, et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro Surveill. 2021;26:16. doi: 10.2807/1560-7917.ES.2021.26.16.2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan T, Chen R, He X, Yuan Y, Deng X, Li R, et al. Infection of wild-type mice by SARS-CoV-2 B.1.351 variant indicates a possible novel cross-species transmission route. Signal Transduct Target Therapy. 2021;6(1):420. doi: 10.1038/s41392-021-00848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu H, Chen Q, Yang G, He L, Fan H, Deng YQ, et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science (N Y, NY) 2020;369(6511):1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang R, Zhang Q, Ge J, Ren W, Zhang R, Lan J, et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54(7):1611–21.e5. doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Q, Nie J, Wu J, Zhang L, Ding R, Wang H, et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184(9):2362-71.e9. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Q, Huang XY, Sun MX, Li RT, Gu H, Tian Y, et al. Transient acquisition of cross-species infectivity during the evolution of SARS-CoV-2. Natl Sci Rev. 2021;8(11):nwab167. doi: 10.1093/nsr/nwab167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3):463–76.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tracking of Variants (2021) VOC Delta GK (B.1.617.2+AY*.) first detected in India]. https://www.gisaid.org/hcov19-variants/. Accessed 26 Sep 2021
- 68.Delta Variant Report (2021) https://outbreak.info/situation-reports/delta. Accessed 26 Sep 2021
- 69.Singh J, Rahman SA, Ehtesham NZ, Hira S, Hasnain SE. SARS-CoV-2 variants of concern are emerging in India. Nat Med. 2021;27(7):1131–1133. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- 70.Variant: 21A (Delta) (2021) https://covariants.org/variants/21A.Delta
- 71.Pascarella S, Ciccozzi M, Zella D, Bianchi M, Benetti F, Benvenuto D, et al. SARS-CoV-2 B.1.617 Indian variants: are electrostatic potential changes responsible for a higher transmission rate? J Med Virol. 2021;397:952. doi: 10.1002/jmv.27210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laffeber C, de Koning K, Kanaar R, Lebbink JHG. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. J Mol Biol. 2021;433(15):167058. doi: 10.1016/j.jmb.2021.167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yuan M, Huang D, Lee CD, Wu NC, Jackson AM, Zhu X, et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Sci (N Y, NY) 2021;373(6556):818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dhar MS, Marwal R, Radhakrishnan VS, Ponnusamy K, Jolly B, Bhoyar RC, et al. Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. medRxiv. 2021;2021:8076. doi: 10.1126/science.abj9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Twohig KA, Nyberg T, Zaidi A, Thelwall S, Sinnathamby MA, Aliabadi S, et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis. 2021;2021:1. doi: 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.World Health Organization (2021) Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. Geneva, Switzerland: World Health Organization. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern. Accessed 20 Dec 2021
- 77.Enhancing Readiness for Omicron (B.1.1.529): technical brief and priority actions for member states (2021) https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states. Accessed 27 Dec 2021
- 78.Hodcroft EB (2021) CoVariants: variants: Variant: 21K (Omicron) also known as BA.1 2021. https://covariants.org/variants/21K.Omicron. Accessed 20 Dec 2021
- 79.Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, et al. The furin cleavage site of SARS-CoV-2 spike protein is a key determinant for transmission due to enhanced replication in airway cells. bioRxiv. 2020;2020:311. [Google Scholar]
- 80.Kumar S, Thambiraja TS, Karuppanan K, Subramaniam G. Omicron and delta variant of SARS-CoV-2: a comparative computational study of spike protein. J Med Virol. 2021;2021:1. doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- 81.Pascarella S, Ciccozzi M, Bianchi M, Benvenuto D, Cauda R, Cassone A. The electrostatic potential of the omicron variant spike is higher than in delta and delta-plus variants: a hint to higher transmissibility? J Med Virol. 2021;2021:1. doi: 10.1002/jmv.27528. [DOI] [PubMed] [Google Scholar]
- 82.Zhang X, Wu S, Wu B, Yang Q, Chen A, Li Y, et al. SARS-CoV-2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct Target Ther. 2021;6(1):430. doi: 10.1038/s41392-021-00852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilhelm A, Widera M, Grikscheit K, Toptan T, Schenk B, Pallas C, et al. Reduced neutralization of SARS-CoV-2 omicron variant by vaccine sera and monoclonal antibodies. medRxiv. 2021;2021:7432. [Google Scholar]
- 84.Wang Y, Zhang L, Li Q, Liang Z, Li T, Liu S, et al. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg Microbes Infect. 2022;11(1):1–5. doi: 10.1080/22221751.2022.2140608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pulliam JRC, van Schalkwyk C, Govender N, von Gottberg A, Cohen C, Groome MJ, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv. 2021;2021:68. doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, et al (2022) Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 602(7898):671–5. [DOI] [PubMed]
- 87.Aggarwal A, Stella AO, Walker G, Akerman A, Milogiannakis V, Brilot F, et al (2021) SARS-CoV-2 Omicron: evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. medRxiv [Preprint]. 2021 Dec 15:2021:2021.12.14.21267772. 10.1101/2021.12.14.21267772
- 88.Redd AD, Nardin A, Kared H, Bloch EM, Abel B, Pekosz A, et al (2021) Minimal cross-over between mutations associated with Omicron variant of SARS-CoV-2 and CD8+ T cell epitopes identified in COVID-19 convalescent individuals. bioRxiv [Preprint]. 2021 Dec 9:2021.12.06.471446. 10.1101/2021.12.06.471446 [DOI] [PMC free article] [PubMed]
- 89.Madhi SA, Kwatra G, Myers JE, Jassat W, Dhar N, Mukendi CK, et al (2022) Population Immunity and Covid-19 Severity with Omicron Variant in South Africa. N Engl J Med. 2022 Feb 23. 10.1056/NEJMoa2119658. Epub ahead of print [DOI] [PMC free article] [PubMed]
- 90.Wolter N, Jassat W, Walaza S, Welch R, Moultrie H, Groome M, et al (2022) Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet (London, England).399(10323):437–46. [DOI] [PMC free article] [PubMed]
- 91.Mahase E. Covid-19: Hospital admission 50–70% less likely with omicron than delta, but transmission a major concern. BMJ (Clin Res Ed) 2021;375:3151. doi: 10.1136/bmj.n3151. [DOI] [PubMed] [Google Scholar]
- 92.Zhao H, Lu L, Peng Z, Chen LL, Meng X, Zhang C, et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect. 2021;2021:1–18. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, et al (2022) Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 602(7898):664–70. [DOI] [PMC free article] [PubMed]
- 94.Leist SR, Dinnon KH, 3rd, Schäfer A, Tse LV, Okuda K, Hou YJ, et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183(4):1070–85.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Romero PE, Dávila-Barclay A, Salvatierra G, González L, Cuicapuza D, Solís L, et al. The emergence of SARS-CoV-2 variant lambda (C.37) in South America. Microbiology Spectrum. 2021;9:e0078921. doi: 10.1128/Spectrum.00789-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.C.37 Lineage Report (2021) https://outbreak.info/situation-reports?pango=C.37. Accessed 18 Oct 2021
- 97.Novel sublineage within B.1.1.1 currently expanding in Peru and Chile, with a convergent deletion in the ORF1a gene (Δ3675–3677) and a novel deletion in the Spike gene (Δ246–252, G75V, T76I, L452Q, F490S, T859N) (2021) https://virological.org/t/novel-sublineage-within-b-1-1-1-currently-expanding-in-peru-and-chile-with-a-convergent-deletion-in-the-orf1a-gene-3675-3677-and-a-novel-deletion-in-the-spike-gene-246-252-g75v-t76i-l452q-f490s-t859n/685. Accesssed 18 Oct 2021
- 98.Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184(13):3426–37.e8. doi: 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McCallum M, Bassi J, De Marco A, Chen A, Walls AC, Di Iulio J, et al (2021) SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 373(6555):648–54. [DOI] [PMC free article] [PubMed]
- 100.Acevedo ML, Alonso-Palomares L, Bustamante A, Gaggero A, Paredes F, Cortés CP, et al (2021) Infectivity and immune escape of the new SARS-CoV-2 variant of interest Lambda. medRxiv [Preprint]. 2021 Jul 1:2021:2021.06.28.21259673. 10.1101/2021.06.28.21259673
- 101.Tada T, Zhou H, Dcosta BM, Samanovic MI, Mulligan MJ, Landau NR (2021) SARS-CoV-2 lambda variant remains susceptible to neutralization by mRNA vaccine-elicited antibodies and convalescent serum. bioRxiv [Preprint]. 2021 Jul 3:2021.07.02.450959. doi: 10.1101/2021.07.02.450959
- 102.Laiton-Donato K, Franco-Muñoz C, Álvarez-Díaz DA, Ruiz-Moreno HA, Usme-Ciro JA, Andrés Prada D, et al. Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. MedRxiv. 2021;26:2100008. doi: 10.1016/j.meegid.2021.105038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lineage B.1.621 (2021) https://cov-lineages.org/lineage.html?lineage=B.1.621. Accessed 18 Oct 2021
- 104.Mu Variant Report (2021) https://outbreak.info/situation-reports/mu. Accessed 18 Oct 2021
- 105.Variant: 21H (Mu) (2021) https://covariants.org/variants/21H.Mu. Accessed 18 Oct 2021
- 106.Collier DA, De Marco A, Ferreira I, Meng B, Datir RP, Walls AC, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593(7857):136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- 107.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27(4):622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 108.Bruxvoort KJ, Sy LS, Qian L, Ackerson BK, Luo Y, Lee GS, et al. Effectiveness of mRNA-1273 against Delta, Mu, and other emerging variants. medRxiv. 2021;385:585. doi: 10.1136/bmj-2021-068848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miyakawa K, Jeremiah SS, Kato H, Ryo A. Neutralizing efficacy of vaccines against the SARS-CoV-2 Mu variant. medRxiv. 2021;12:987. [Google Scholar]
- 110.Hodcroft EB, Zuber M, Nadeau S, Crawford KHD, Bloom JD, Veesler D, et al. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. medRxiv. 2020;2020:1. [Google Scholar]
- 111.Zhang W, Davis BD, Chen SS, Sincuir Martinez JM, Plummer JT, Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA. 2021;325(13):1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tchesnokova V, Kulasekara H, Larson L, Bowers V, Rechkina E, Kisiela D, et al (2021) Acquisition of the L452R Mutation in the ACE2-Binding Interface of Spike Protein Triggers Recent Massive Expansion of SARS-CoV-2 Variants. J Clin Microbiol. 59(11):e0092121. [DOI] [PMC free article] [PubMed]
- 113.Pereira F, Tosta S, Lima MM, Reboredo de Oliveira da Silva L, Nardy VB, Gómez MKA, et al (2021) Genomic surveillance activities unveil the introduction of the SARS-CoV-2 B.1.525 variant of interest in Brazil: Case report. J Med Virol. 93(9):5523–6. [DOI] [PMC free article] [PubMed]
- 114.B.1.525 2021 (2021) https://cov-lineages.org/global_report_B.1.525.html
- 115.West AP, Jr., Wertheim JO, Wang JC, Vasylyeva TI, Havens JL, Chowdhury MA, et al (2021) Detection and characterization of the SARS-CoV-2 lineage B.1.526 in New York. Nat Commun. 12(1):4886. [DOI] [PMC free article] [PubMed]
- 116.Annavajhala MK, Mohri H, Wang P, Nair M, Zucker JE, Sheng Z, et al (2021) Emergence and expansion of SARS-CoV-2 B.1.526 after identification in New York. Nature. 597(7878):703–8. [DOI] [PMC free article] [PubMed]
- 117.Chen J, Wang R, Wang M, Wei GW. Mutations strengthened SARS-CoV-2 infectivity. J Mol Biol. 2020;432(19):5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184(9):2332–47.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lineage B.1.617 (2021) https://cov-lineages.org/lineages/lineage_B.1.617.html
- 120.Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, et al. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv. 2021;94:e00127. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yadav PD, Sapkal GN, Abraham P, Ella R, Deshpande G, Patil DY, et al. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. Clin Infect Dis. 2021;74:366–368. doi: 10.1093/cid/ciab411. [DOI] [PubMed] [Google Scholar]
- 122.Selected characteristics of SARS-CoV-2 Variants of Interest (2021) https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html
- 123.Detection of new SARS-CoV-2 variants related to mink (2020) https://www.ecdc.europa.eu/sites/default/files/documents/RRA-SARS-CoV-2-in-mink-12-nov-2020.pdf
- 124.Lassaunière R FJ, Rasmussen M, Frische A, Polacek Strandh C, Rasmussen TB et al (2020) Working paper on SARS-CoV-2 spike mutations arising in Danish mink, their spread to humans and neutralization data [Pre-print] Copenhagen: Statens Serum Institut. https://files.ssi.dk/Mink-cluster-5-short-report_AFO2
- 125.Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Sci (N Y, NY) 2021;371(6525):172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hodcroft EB, Domman DB, Snyder DJ, Oguntuyo KY, Van Diest M, Densmore KH et al (2021) Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677. medRxiv 2021:2021.02.12.21251658
- 127.Variant: S:677H.Robin1 (2021) https://covariants.org/variants/S.Q677H.Robin1
- 128.Variant: S:677P.Pelican (2021) https://covariants.org/variants/S.Q677P.Pelican
- 129.Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 2021;29(1):44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barnes CO, Jette CA, Abernathy ME, Dam KA, Esswein SR, Gristick HB, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183(4):1024–42.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science (N Y, NY) 2021;371(6531):850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Weissman D, Alameh MG, de Silva T, Collini P, Hornsby H, Brown R, et al. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. 2021;29(1):23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, et al. Structural and functional analysis of the D614G SARS-CoV-2 Spike protein variant. Cell. 2020;183(3):739–51.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, et al (2021) Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med. 27(4):620–1. [DOI] [PubMed]
- 136.Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 137.Graham MS, Sudre CH, May A, Antonelli M, Murray B, Varsavsky T, et al. Changes in symptomatology, reinfection, and transmissibility associated with the SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Public Health. 2021;6(5):e335–e345. doi: 10.1016/S2468-2667(21)00055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jangra S, Ye C, Rathnasinghe R, Stadlbauer D, Krammer F, Simon V, et al. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021;2:283–284. doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Lambson BE, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Lancet Microbe. 2021;586:583. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 140.Wang P, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, et al. Increased resistance of SARS-CoV-2 variant P1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747–751.e414. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Sci (N Y, NY) 2020;369(6506):1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Thomson EC, Rosen LE, Shepherd JG, Spreafico R, da Silva FA, Wojcechowskyj JA, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184(5):1171–87.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al (2021) Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 596(7871):276–80. [DOI] [PubMed]
- 144.Tada T, Zhou H, Dcosta BM, Samanovic MI, Mulligan MJ, Landau NR (2021) The Spike Proteins of SARS-CoV-2 B.1.617 and B.1.618 Variants Identified in India Provide Partial Resistance to Vaccine-elicited and Therapeutic Monoclonal Antibodies. bioRxiv [Preprint]. 2021 May 16:2021.05.14.444076. 10.1101/2021.05.14.444076
- 145.Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, et al (2021) Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med. 385(7):585–94. [DOI] [PMC free article] [PubMed]
- 146.Andreano E, Piccini G, Licastro D, Casalino L, Johnson NV, Paciello I, et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020;5:237. doi: 10.1073/pnas.2103154118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kannan SR, Spratt AN, Sharma K, Chand HS, Byrareddy SN, Singh K. Omicron SARS-CoV-2 variant: unique features and their impact on pre-existing antibodies. J Autoimmunity. 2022;126:102779. doi: 10.1016/j.jaut.2021.102779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.COVID-19 vaccine tracker and landscape (2021) https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines. Accessed 27 Dec 2021
- 150.Covid-19 Vaccine Tracker, Approved Vaccines (2021) https://covid19.trackvaccines.org/vaccines/approved/. Accessed 31 Dec 2021
- 151.Pardi N, Tuyishime S, Muramatsu H, Kariko K, Mui BL, Tam YK, et al. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J Controll Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Callaway E. The race for coronavirus vaccines: a graphical guide. Nature. 2020;580(7805):576–577. doi: 10.1038/d41586-020-01221-y. [DOI] [PubMed] [Google Scholar]
- 153.Pfizer and BioNTech Conclude Phase 3 Study of COVID-19 Vaccine Candidate, Meeting All Primary Efficacy Endpoints (2020) https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-conclude-phase-3-study-covid-19-vaccine
- 154.Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, et al. Publisher correction: COVID-19 vaccine BNT162b1 elicits human antibody and T(H)1 T cell responses. Nature. 2021;590(7844):E17. doi: 10.1038/s41586-020-03102-w. [DOI] [PubMed] [Google Scholar]
- 155.Dagan N, Barda N, Kepten E, Miron O, Perchik S, Katz MA, et al. BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N Engl J Med. 2021;384(15):1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu Y, Liu J, Xia H, Zhang X, Fontes-Garfias CR, Swanson KA, et al. Neutralizing activity of BNT162b2-elicited serum. N Engl J Med. 2021;384(15):1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wall EC, Wu M, Harvey R, Kelly G, Warchal S, Sawyer C, et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet (London, England). 2021;397(10292):2331–2333. doi: 10.1016/S0140-6736(21)01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tartof SY, Slezak JM, Fischer H, Hong V, Ackerson BK, Ranasinghe ON, et al (2021) Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet (London, England). 398(10309):1407–16. [DOI] [PMC free article] [PubMed]
- 159.Hansen CH, Schelde AB, Moustsen-Helms IR, Emborg H-D, Krause TG, Mølbak K et al (2021) Vaccine effectiveness against SARS-CoV-2 infection with the Omicron or Delta variants following a two-dose or booster BNT162b2 or mRNA-1273 vaccination series: a Danish cohort study. medRxiv [Preprint]. 2021 Dec 23:2021.12.20.21267966. doi: 10.1101/2021.12.20.21267966
- 160.Lu L, Mok BW, Chen LL, Chan JM, Tsang OT, Lam BH et al (2021) Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis.2021 Dec 16:ciab1041. 10.1093/cid/ciab1041. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 161.Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H, et al (2022) Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 602(7898):654–6. [DOI] [PMC free article] [PubMed]
- 162.Garcia-Beltran WF, St Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, et al (2022) mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell.185(3):457–66.e4. [DOI] [PMC free article] [PubMed]
- 163.Schmidt F, Muecksch F, Weisblum Y, Da Silva J, Bednarski E, Cho A, et al (2022) Plasma Neutralization of the SARS-CoV-2 Omicron Variant. N Engl J Med. 386(6):599–601. [DOI] [PMC free article] [PubMed]
- 164.Pfizer and BioNTech Initiate a Study as Part of Broad Development Plan to Evaluate COVID-19 Booster and New Vaccine Variants 2021 [Available from: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-initiate-study-part-broad-development.
- 165.Pfizer and BioNTech Announce Phase 3 Trial Data Showing High Efficacy of a Booster Dose of Their COVID-19 Vaccine (2021) https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-announce-phase-3-trial-data-showing. Accessed 26 Dec 2021
- 166.Coronavirus (COVID-19) Update: FDA Expands Eligibility for Pfizer-BioNTech COVID-19 Booster Dose to 16- and 17-Year-Olds (2021) https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-expands-eligibility-pfizer-biontech-covid-19-booster-dose-16-and-17. Accessed 26 Dec 2021
- 167.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chu L, McPhee R, Huang W, Bennett H, Pajon R, Nestorova B, et al. A preliminary report of a randomized controlled phase 2 trial of the safety and immunogenicity of mRNA-1273 SARS-CoV-2 vaccine. Vaccine. 2021;39(20):2791–2799. doi: 10.1016/j.vaccine.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Krammer F, Srivastava K, Alshammary H, Amoako AA, Awawda MH, Beach KF, et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N Engl J Med. 2021;384(14):1372–1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu K, Werner AP, Koch M, Choi A, Narayanan E, Stewart-Jones GBE, et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N Engl J Med. 2021;384(15):1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Choi A, Koch M, Wu K, Dixon G, Oestreicher J, Legault H, et al. Serum neutralizing activity of mRNA-1273 against SARS-CoV-2 variants. J Virol. 2021;2021:Jvio31321. doi: 10.1128/JVI.01313-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pajon R, Doria-Rose NA, Shen X, Schmidt SD, O'Dell S, McDanal C, et al (2022) SARS-CoV-2 Omicron Variant Neutralization after mRNA-1273 Booster Vaccination. N Engl J Med. 2022 Jan 26:NEJMc2119912. 10.1056/NEJMc2119912. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 174.Coronavirus (COVID-19) Update: FDA Expands Eligibility for COVID-19 Vaccine Boosters (2021) https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-expands-eligibility-covid-19-vaccine-boosters. Accessed 26 Dec 2021
- 175.Ramasamy MN, Minassian AM, Ewer KJ, Flaxman AL, Folegatti PM, Owens DR, et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet (Lond, Engl) 2021;396(10267):1979–1993. doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet (Lond, Engl) 2021;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Emary KRW, Golubchik T, Aley PK, Ariani CV, Angus B, Bibi S, et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet (Lond, Engl) 2021;397(10282):1351–1362. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Madhi SA, Baillie V, Cutland CL, Voysey M, Koen AL, Fairlie L, et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384(20):1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Clemens SAC, Folegatti PM, Emary KRW, Weckx LY, Ratcliff J, Bibi S, et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 lineages circulating in Brazil. Nat Commun. 2021;12(1):5861. doi: 10.1038/s41467-021-25982-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sadoff J, Le Gars M, Shukarev G, Heerwegh D, Truyers C, de Groot AM, et al. Interim results of a phase 1–2a trial of Ad26COV2S Covid-19 vaccine. N Engl J Med. 2021;384(19):1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sadoff J, Gray G, Vandebosch A, Cárdenas V, Shukarev G, Grinsztejn B, et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N Engl J Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Barouch DH, Stephenson KE, Sadoff J, Yu J, Chang A, Gebre M, et al. Durable humoral and cellular immune responses following Ad26.COV2.S vaccination for COVID-19. MedRxiv. 2021;325:1535. doi: 10.1056/NEJMc2108829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.World Health Organization, COVID-19 vaccine tracker and landscape (2021) https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines. Accessed 24 Dec 2021
- 184.Heath PT, Galiza EP, Baxter DN, Boffito M, Browne D, Burns F, et al. Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. N Engl J Med. 2021;385(13):1172–1183. doi: 10.1056/NEJMoa2107659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shinde V, Bhikha S, Hoosain Z, Archary M, Bhorat Q, Fairlie L, et al. Efficacy of NVX-CoV2373 Covid-19 vaccine against the B.1.351 Variant. N Engl J Med. 2021;384(20):1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mahase E. Covid-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ (Clin Res Ed). 2021;372:296. doi: 10.1136/bmj.n296. [DOI] [PubMed] [Google Scholar]
- 187.Novavax Announces Initial Omicron Cross-Reactivity Data from COVID-19 Vaccine Booster and Adolescent Studies (2021) https://ir.novavax.com/2021-12-22-Novavax-Announces-Initial-Omicron-Cross-Reactivity-Data-from-COVID-19-Vaccine-Booster-and-Adolescent-Studiess. Accessed 27 Dec 2021
- 188.Yang S, Li Y, Dai L, Wang J, He P, Li C, et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect Dis. 2021;21(8):1107–1119. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhao X, Zheng A, Li D, Zhang R, Sun H, Wang Q, et al. Neutralisation of ZF2001-elicited antisera to SARS-CoV-2 variants. Lancet Microbe. 2021;2(10):e494. doi: 10.1016/S2666-5247(21)00217-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Huang B, Dai L, Wang H, Hu Z, Yang X, Tan W, et al. Serum sample neutralisation of BBIBP-CorV and ZF2001 vaccines to SARS-CoV-2 501Y.V2. Lancet Microbe. 2021;2(7):e285. doi: 10.1016/S2666-5247(21)00082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhao X, Li D, Ruan W, Zhang R, Zheng A, Qiao S et al (2021) Reduced sera neutralization to Omicron SARS-CoV-2 by both inactivated and protein subunit vaccines and the convalescents. bioRxiv [Preprint]. 2021 Dec 20:2021.12.16.472391. 10.1101/2021.12.16.472391
- 192.Ai J, Zhang H, Zhang Y, Lin K, Zhang Y, Wu J, et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg Microbes Infect. 2021;2021:1–24. doi: 10.1080/22221751.2021.2022440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ryzhikov AB, Ryzhikov EA, Bogryantseva MP, Danilenko ED, Imatdinov IR, Nechaeva EA, et al. Immunogenicity and protectivity of the peptide vaccine against SARS-CoV-2. Ann RAMS. 2021;76(1):5–19. doi: 10.15690/vramn1528. [DOI] [Google Scholar]
- 194.Ryzhikov AB, Ryzhikov EA, Bogryantseva MP, Usova SV, Danilenko ED, Nechaeva EA, et al. A single blind, placebo-controlled randomized study of the safety, reactogenicity and immunogenicity of the “EpiVacCorona” vaccine for the prevention of COVID-19, in volunteers aged 18–60 years (Phase I-II) Russ J Infect Immunity. 2021;11(1):283–296. doi: 10.15789/2220-7619-ASB-1699. [DOI] [Google Scholar]
- 195.Limonta-Fernández M, Chinea-Santiago G, Martín-Dunn AM, Gonzalez-Roche D, Bequet-Romero M, Marquez-Perera G, et al. The SARS-CoV-2 receptor-binding domain expressed in Pichia pastoris as a candidate vaccine antigen. medRxiv. 2021;11:1. doi: 10.1016/j.nbt.2022.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li L, Honda-Okubo Y, Huang Y, Jang H, Carlock MA, Baldwin J, et al. Immunisation of ferrets and mice with recombinant SARS-CoV-2 spike protein formulated with Advax-SM adjuvant protects against COVID-19 infection. Vaccine. 2021;39(40):5940–5953. doi: 10.1016/j.vaccine.2021.07.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Xia S, Zhang Y, Wang Y, Wang H, Yang Y, Gao GF, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect Dis. 2021;21(1):39–51. doi: 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Al Kaabi N, Zhang Y, Xia S, Yang Y, Al Qahtani MM, Abdulrazzaq N, et al. Effect of 2 inactivated SARS-CoV-2 vaccines on symptomatic COVID-19 infection in adults: a randomized clinical trial. JAMA. 2021;326(1):35–45. doi: 10.1001/jama.2021.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jeewandara C, Aberathna IS, Pushpakumara PD, Kamaladasa A, Guruge D, Jayathilaka D et al (2021) Persistence of antibody and T cell responses to the Sinopharm/BBIBP-CorV vaccine in Sri Lankan individuals. medRxiv [Preprint]. 2021 Oct 18:2021.10.14.21265030. doi: 10.1101/2021.10.14.21265030.
- 200.Yu X, Wei D, Xu W, Li Y, Li X, Zhang X-x, et al. Reduced sensitivity of SARS-CoV-2 Omicron variant to booster-enhanced neutralization. medRxiv. 2021;398:10316. [Google Scholar]
- 201.Zhang Y, Zeng G, Pan H, Li C, Hu Y, Chu K, et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect Dis. 2021;21(2):181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wu Z, Hu Y, Xu M, Chen Z, Yang W, Jiang Z, et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine (CoronaVac) in healthy adults aged 60 years and older: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect Dis. 2021;21(6):803–812. doi: 10.1016/S1473-3099(20)30987-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tanriover MD, Doğanay HL, Akova M, Güner HR, Azap A, Akhan S, et al. Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet (Lond, Engl) 2021;398(10296):213–222. doi: 10.1016/S0140-6736(21)01429-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Palacios R, Batista AP, Albuquerque CSN, Patiño EG, Santos JdP, Conde MTRP, et al (2021) Efficacy and Safety of a COVID-19 Inactivated Vaccine in Healthcare Professionals in Brazil: The PROFISCOV Study. SSRN [Preprint]. 2021 Apr 14. doi: 10.2139/ssrn.3822780.
- 205.Jara A, Undurraga EA, González C, Paredes F, Fontecilla T, Jara G, et al. Effectiveness of an Inactivated SARS-CoV-2 Vaccine in Chile. N Engl J Med. 2021;385(10):875–884. doi: 10.1056/NEJMoa2107715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Vacharathit V, Aiewsakun P, Manopwisedjaroen S, Srisaowakarn C, Laopanupong T, Ludowyke N, et al. CoronaVac induces lower neutralising activity against variants of concern than natural infection. Lancet Infect Dis. 2021;21(10):1352–1354. doi: 10.1016/S1473-3099(21)00568-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hitchings MDT, Ranzani OT, Torres MSS, de Oliveira SB, Almiron M, Said R, et al (2021) Effectiveness of CoronaVac among healthcare workers in the setting of high SARS-CoV-2 Gamma variant transmission in Manaus, Brazil: A test-negative case-control study. Lancet Reg Health Am 1:100025. [DOI] [PMC free article] [PubMed]
- 208.Souza WM, Amorim MR, Sesti-Costa R, Coimbra LD, Brunetti NS, Toledo-Teixeira DA, et al. Neutralisation of SARS-CoV-2 lineage P.1 by antibodies elicited through natural SARS-CoV-2 infection or vaccination with an inactivated SARS-CoV-2 vaccine: an immunological study. Lancet Microbe. 2021;2(10):e527–e535. doi: 10.1016/S2666-5247(21)00129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kang M, Yi Y, Li Y, Sun L, Deng A, Hu T, et al (2022) Effectiveness of Inactivated COVID-19 Vaccines Against Illness Caused by the B.1.617.2 (Delta) Variant During an Outbreak in Guangdong, China : A Cohort Study. Ann Intern Med. 2022 Feb 1:M21-3509. doi: 10.7326/M21-3509. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 210.Ella R, Vadrevu KM, Jogdand H, Prasad S, Reddy S, Sarangi V, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: a double-blind, randomised, phase 1 trial. Lancet Infect Dis. 2021;21(5):637–646. doi: 10.1016/S1473-3099(20)30942-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ella R, Reddy S, Jogdand H, Sarangi V, Ganneru B, Prasad S, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: interim results from a double-blind, randomised, multicentre, phase 2 trial, and 3-month follow-up of a double-blind, randomised phase 1 trial. Lancet Infect Dis. 2021;21(7):950–961. doi: 10.1016/S1473-3099(21)00070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ella R, Reddy S, Blackwelder W, Potdar V, Yadav P, Sarangi V, et al. Efficacy, safety, and lot to lot immunogenicity of an inactivated SARS-CoV-2 vaccine (BBV152): a, double-blind, randomised, controlled phase 3 trial. medRxiv. 2021;24:102298. doi: 10.1016/S0140-6736(21)02000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sapkal GN, Yadav PD, Ella R, Deshpande GR, Sahay RR, Gupta N, et al. Inactivated COVID-19 vaccine BBV152/COVAXIN effectively neutralizes recently emerged B.1.1.7 variant of SARS-CoV-2. J Travel Med. 2021;28:4. doi: 10.1093/jtm/taab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yadav PD, Sapkal GN, Ella R, Sahay RR, Nyayanit DA, Patil DY, et al. Neutralization of Beta and Delta variant with sera of COVID-19 recovered cases and vaccinees of inactivated COVID-19 vaccine BBV152/Covaxin. J Travel Med. 2021;28:7. doi: 10.1093/jtm/taab104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yadav PD, Sahay RR, Sapkal G, Nyayanit D, Shete AM, Deshpande G, et al. Comparable neutralization of SARS-CoV-2 Delta AY.1 and Delta with individuals sera vaccinated with BBV152. J Travel Med. 2021;384:2212. doi: 10.1093/jtm/taab154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Morens DM, Taubenberger JK, Fauci AS (2022) Universal Coronavirus Vaccines - An Urgent Need. N Engl J Med. 386(4):297–9. [DOI] [PMC free article] [PubMed]
- 217.Giurgea LT, Han A, Memoli MJ. Universal coronavirus vaccines: the time to start is now. NPJ Vaccines. 2020;5(1):43. doi: 10.1038/s41541-020-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Le Page M. Pre-existing immunity to covid-19 hints at universal coronavirus vaccine. New Sci. 2021;252(3361):19. doi: 10.1016/S0262-4079(21)02058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhu J, Ananthaswamy N, Jain S, Batra H, Tang WC, Lewry DA, et al. A universal bacteriophage T4 nanoparticle platform to design multiplex SARS-CoV-2 vaccine candidates by CRISPR engineering. Sci Adv. 2021;7(37):eabh1547. doi: 10.1126/sciadv.abh1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Markosian C, Staquicini DI, Dogra P, Dodero-Rojas E, Tang FHF, Smith TL, et al (2021) Apropos of Universal Epitope Discovery for COVID-19 Vaccines: A Framework for Targeted Phage Display-Based Delivery and Integration of New Evaluation Tools. bioRxiv [Preprint]. 2021 Aug 30:2021.08.30.458222. doi: 10.1101/2021.08.30.458222.
- 221.Lucchese G, Stufano A, Kanduc D. Searching for an effective, safe and universal anti-HIV vaccine: finding the answer in just one short peptide. Self/nonself. 2011;2(1):49–54. doi: 10.4161/self.2.1.14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Goldstein G, Chicca JJ., 2nd A universal anti-HIV-1 Tat epitope vaccine that is fully synthetic and self-adjuvanting. Vaccine. 2010;28(4):1008–1014. doi: 10.1016/j.vaccine.2009.10.129. [DOI] [PubMed] [Google Scholar]
- 223.Andresen BS, Vinner L, Tang S, Bragstad K, Kronborg G, Gerstoft J, et al. Characterization of near full-length genomes of HIV type 1 strains in Denmark: basis for a universal therapeutic vaccine. AIDS Res Hum Retroviruses. 2007;23(11):1442–1448. doi: 10.1089/aid.2007.0111. [DOI] [PubMed] [Google Scholar]
- 224.Paredes MI, Lunn SM, Famulare M, Frisbie LA, Painter I, Burstein R, et al (2022) Associations between SARS-CoV-2 variants and risk of COVID-19 hospitalization among confirmed cases in Washington State: a retrospective cohort study. medRxiv [Preprint]. 2022 Feb 16:2021.09.29.21264272. 10.1101/2021.09.29.21264272. [DOI] [PMC free article] [PubMed]
- 225.World Health Organization (2021) COVID-19 weekly epidemiological update 2021. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20211019_weekly_epi_update_62.pdf?sfvrsn=f0a4a5fe_27&download=true. Accessed 19 Oct 2021
- 226.Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, et al. SARS-CoV-2 variants B1351 and P1 escape from neutralizing antibodies. Cell. 2021;184(9):2384-93.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2372–83.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Tada T, Dcosta BM, Samanovic-Golden M, Herati RS, Cornelius A, Mulligan MJ, et al. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv. 2021;10:14031. [Google Scholar]
- 229.Wu K, Werner AP, Moliva JI, Koch M, Choi A, Stewart-Jones GBE, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021;586:567. [Google Scholar]
- 230.Nasreen S, Chung H, He S, Brown KA, Gubbay JB, Buchan SA, et al (2021) Effectiveness of mRNA and ChAdOx1 COVID-19 vaccines against symptomatic SARS-CoV-2 infection and severe outcomes with variants of concern in Ontario. medRxiv [Preprint]. 2021 Sep 30:2021.06.28.21259420. doi: 10.1101/2021.06.28.21259420 [DOI] [PubMed]
- 231.Tada T, Zhou H, Samanovic MI, Dcosta BM, Cornelius A, Mulligan MJ, et al. Comparison of neutralizing antibody titers elicited by mRNA and adenoviral vector vaccine against SARS-CoV-2 variants. BioRxiv. 2021;384:2187. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.