Abstract
The discovery of SARS-CoV-2 virus in the water bodies has been reported, and the risk of virus transmission to human via the water route due to poor wastewater management cannot be disregarded. The main source of the virus in water bodies is the sewage network systems which connects to the surface water. Wastewater-based epidemiology has been applied as an early surveillance tool to sense SARS-CoV-2 virus in the sewage network. This review discussed possible transmission routes of the SARS-CoV-2 virus and the challenges of the existing method in detecting the virus in wastewater. One significant challenge for the detection of the virus is that the high virus loading is diluted by the sheer volume of the wastewater. Hence, virus preconcentration from water samples prior to the application of virus assay is essential to accurately detect traceable virus loading. The preparation time, materials and conditions, virus type, recovery percentage, and various virus recovery techniques are comprehensively discussed in this review. The practicability of molecular methods such as Polymer-Chain-Reaction (PCR) for the detection of SARS-CoV-2 in wastewater will be revealed. The conventional virus detection techniques have several shortcomings and the potential of biosensors as an alternative is also considered. Biosensing techniques have also been proposed as an alternative to PCR and have reported detection limits of 10 pg/μl. This review serves to guide the reader on the future designs and development of highly sensitive, robust and, cost effective SARS-CoV-2 lab-on-a-chip biosensors for use in complex wastewater.
Keywords: Wastewater based epidemiology, SARS-CoV 2, Coronavirus, Biosensor, Polymer-chain-reaction, Wastewater
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a highly contagious virus with exceeding 200 million reported cases worldwide in 2021 [1]. Efforts have been carried out to identify the routes of virus transmission and the stability of the viruses in specific environments [[2], [3], [4]]. It is essential that the modes of transmission for the coronavirus are established and identified to facilitate rapid detection and prevention of the disease spread. The main route of transmission for SARS-CoV-2 is the exposure of human to droplet sprays or aerosols that could last for hours in the air [4]. Although unprecedented, there also exist a risk of this virus being transmitted through the faecal-oral route; as the virus has already been detected in the urine and faecal discharge of coronavirus disease 2019 (COVID-19) patients [[5], [6], [7]]. Interestingly, prior to the COVID-19 pandemic, waterborne transmission of coronaviruses have rarely been given attention to, since it was believed that the enveloped viruses are not capable of surviving in water for extended period of time [8]. However, as pre-cautionary, attention should be given to the detection of SARS-CoV-2 in water since there is a possibility of waterborne SARS-CoV-2 transmission to humans and animals.
The ongoing COVID-19 pandemic has brought adverse impact to the health, economy, and social well-being of the general population. Previous studies [9,10] have shown that these impacts are not equally distributed among the population with marginalized communities being more likely to suffer the brunt of the impact due to unsanitary housing conditions, overcrowding, insufficient testing, and ineffective outbreak management strategies that cause them to be more susceptible to diseases. Clinical and home-based test kits are expensive when periodic testing is required, and low-income families often forego medical tests in lieu of more pressing needs such as shelter and food. More efficient testing methods, such as mass testing, is needed to better monitor and prevent any outbreaks from occurring in densely populated areas and especially in marginalized communities.
Recently, wastewater-based epidemiology (WBE) has received attention as a potential surveillance tool to complement both clinical and home-based testing [11]. The basic principle of WBE is to collect biological samples excreted by the human bodies via the local sewage network of the housing area of interest. These biological samples can be used to infer the size of the shedding population and provide community-level health information [6]. WBE data is especially powerful as it can create an early warning system that can inform the authorities of a possible outbreak. WBE data is a non-intrusive method of gathering data and finding asymptomatic virus carriers that have not undergone testing. Circulation of the virus in a particular community can be detected early when there is a sudden increase in the water. Using this information, public health measures such as individual testing, isolation of cases, and adequate warning to the community can be carried out to control the outbreak. Recent studies performed in Europe [12], China [13], India [7] and Middle East [14] have demonstrated the potential of WBE methods as they were able to detect positive COVID-19 samples in wastewater about a week prior to the reporting of actual cases in the region.
Despite its apparent advantages, WBE is not widely applied in the community as its implementation is fraught with challenges. The two main difficulties pertaining to pathogen detection using WBE are: i) The sewage samples are often in the form of complex matrix that requires pre-treatment before the main pathogen can be detected; ii) Low concentrations of virus particles in samples that require the use of biosensors with low limits of detection (LOD). To overcome these challenges, polymerase chain reaction-based assays (PCR) are often employed because the viral nucleic acids can be amplified and detected rapidly in a single step [15]. Various modified PCR techniques such as digital PCR (dPCR) [16], generation sequencing [17], CRISPR [18] and loop-mediated isothermal amplification (LAMP) [19] have been reported to improve detection in the complex wastewater matrix. These PCR methods, however, are costly, tedious, prioritised for testing human samples during the peak of infections and require a long assay time, making them unsuitable for wastewater detection [15]. Biosensors on the other hand, provide rapid results, are cost-effective and allow parallel sample testing, making them more ideal for use to test for the virus in wastewater. However, the possible presence of proteins inhibitor in the complex wastewater may impede the operation of the biosensors.
To the best of our knowledge, this is the first review paper that associate virus preconcentration method to the development of biosensor for rapid detection of SARS-CoV-2 in wastewater. The aim of this review is to identify the transmission routes of SARS-CoV-2 in wastewater, possible interfering compounds to biosensors, and virus preconcentration. It is of paramount that the researchers have insights on the characteristics of the wastewater being studied and, comprehend the difficulties of measuring low shedding of virus in a large volume of wastewater. Hence, the objectives of this review paper are to: i) shed light on the characteristics of SARS-CoV-2, transmission route and contents of the sewage water environment containing the virus; ii) identify the virus recovery techniques prior to application of biosensor detection; and iii) discuss recent development of biosensors for SARS-CoV-2 detection. This review also highlights the pre-requisites and criteria in designing a robust biosensor for rapid detection of viruses extracted from polluted wastewater.
2. Research methodology
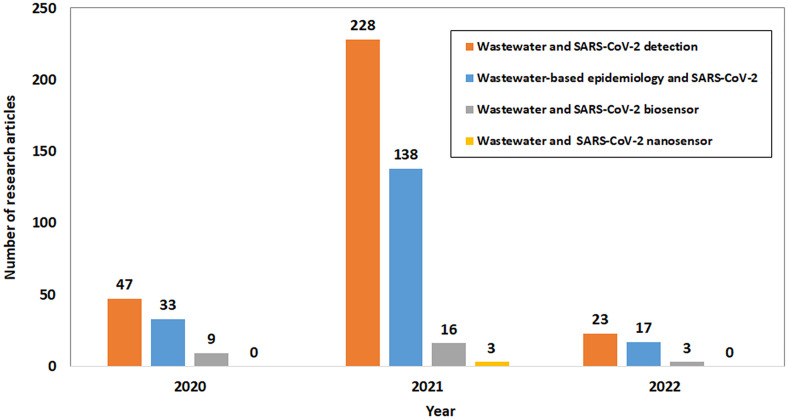
The outbreak of SARS-CoV-2 was found in late 2019, hence, the publication in WBE associated with SARS-CoV-2 are still limited. Based on the pre-selected terms, the metadata as shown in Fig. 1 on the quantity of research articles published associated with wastewater contamination by SARS-CoV-2 and detection was obtained from Science Direct between year 2020 to 2022. Initial search of research articles of relevant information related to this review were based on the search terms such as “wastewater and Covid-19 detection”, “wastewater and Covid-19 biosensor”, “wastewater and Covid-19 nanosensor”, and “wastewater-based epidemiology and Covid-19”. The search results from Science Direct are as shown in Fig. 1. After reviewing and shortlisting suitable articles, the information was collected and deliberated with suitable examples for the preparation of this review.
Fig. 1.
Number of research articles on the detection of SARS-CoV-2 in the wastewater.
Source: ScienceDirect.com, 2020 to 2022.
In the past 3 years, there were a total of 423 research articles on COVID-19 detection in wastewater reported (Fig. 1) which was searched using keywords “Wastewater” and “COVID-19 detection”. Most of these articles were based on the polymer chain reaction (PCR) techniques performed in the laboratory. The number of publications is comparatively low which indicates that the understanding of the contamination of the SARS-CoV-2 viruses in the wastewater sampled from sewage or wastewater plant is still scarce. Till date, the research for the in-situ detection of SARS-CoV-2 in wastewater using biosensor or nanosensor was found considerably less with a total published articles of 57 and 3, respectively. This indicates a vast opportunity and research gap to develop biosensor that could detect SARS-CoV-2 in complex wastewater on site. However, there are great challenges from translating the application of SARS-CoV-2 sensing technique applied for human samples to the application on wastewater samples. Various challenges had been identified and will be addressed in this review manuscript.
3. General morphology of SARS-CoV-2
The name “Coronavirus” stems from the appearance of crown-like thorns on the outer surface of the virus' structure [20]. Coronaviruses are enveloped and contain positive, single-stranded RNA with lengths ranging from 27 to 32 Kb, and with this length their genome is deemed to be one of the largest RNA viruses [20,21]. The viral particles are spherical with diameters between 60 and 140 nm with distinctive 9–12 nm-long spikes on the outer surface, volumes between 106 nm3 and mass of 103 MDa [22,23]. A coronavirus genome contains around 30,000 nucleotides and encodes four major proteins that are important for virus-host cell binding, virus morphogenesis and release of viral particles [21]. These proteins are the nucleocapsid (N), membrane (M), envelope (E) and spike (S) proteins [24]. The N-protein is a binding protein that is encoded with the virus that forms the RNA polymerase complex, which is crucial for the replication and transcription of the virus [25]. The M-protein is a positively-charged, basic transmembrane glycoprotein, and is the most abundant protein in the virus [24,25]. The E-protein is a small membrane protein that is mainly localised to the endoplasmic reticulum and Golgi body of the host cell [21]. Small portion of the E-protein is also integrated in the virion envelope [26]. The N-, M- and E-proteins are primary proteins for virus assembly and virus-like particle formation [21]. The S-protein, is a glycoprotein which decorates and are protruding on the outer surface of the virus, is responsible for early steps of viral infection that mediates virus entry into host cells [20].
The structure of SARS-CoV-2 has been well studied in several reports. On the surfaces of certain human cells, there are the angiotensin-converting enzyme 2 (ACE2) receptors which can attach to the S-protein of coronavirus [20,24]. These ACE2 receptors are dominantly expressed in lungs, oral mucosa and small intestine [27]. Upon entry of virus into the human host cell (also known as endocytosis), the S-protein complex rearranges itself to be able to merge into the host cell membrane [8]. Inside the host cell, the outer envelope of the virus dissociates and reveals the RNA genome, which will then attach to the ribosome and undergoes RNA translation to produce viral polymerase. The viral RNA genome also undergoes replication and transcription, which will generate more viral RNA and viral proteins and will ultimately form more virus cells that will be released outside the host cell [8]. It is worth noting that the ACE2 receptors are common in other mammalian species as well, and therefore, this explains why the coronaviruses have been observed in a wide range of animal hosts [8].
4. Transmission routes of SARS-CoV-2
4.1. The potential transmission routes of SARS-CoV-2 in the general environment
The survival and transmission of viral particles largely depend on the humidity, surrounding temperature, and contaminated surfaces [28]. These environmental factors are crucial for the viruses or pathogens to be viable outside the host; the period of viability of the virus in outer surroundings is dependent on the effects of various biotic and abiotic stresses on the viruses [29]. As mentioned, viral infections typically spread via aerosol production and direct contact between an infected patient and a healthy person. Aerosol transmission has been proven to be the main mode of transmission for SARS-CoV-2 [30], which involves generation of virus-infected aerosols or droplets from basic bodily mechanisms, voluntarily or involuntarily, such as talking, exhaling, coughing [31]. These mechanisms or actions can produce μm to mm-sized droplets and later evaporate to release suspended viral particles in the atmosphere [31]. For SARS-CoV-2, low humidity and temperature in closed environments are very conducive for droplet transmission, and small but loaded viral particles (> 5 μm) may travel up to 10 m from the infected host [32,33]. Virus transmission can also occur through fomites (inanimate objects/surfaces), and this transmission is believed to be quite dangerous since virus tends to have prolonged survival rate on plastic/metal surfaces compared to in air or droplets [34].
Previous research on transmission routes of SARS-CoV-2 have been mainly about transmissions via droplets and surface contaminations, however only a few studies have been focused on viral transmission in the water media [8,27,35,36]. The possibility of waterborne transmission of coronaviruses has been well debated since there are evidences on survival of coronaviruses in water bodies, especially in wastewater [24]. One confirmed route of viral contamination in water is by faecal matter of infected COVID-19 patients [8,36]. Reports of soil and groundwater contaminations from previous epidemics (from SARS-CoV and MERS-CoV) suggested that excretions of urine and faeces were the main contaminants [37,38]. Other than direct faecal elimination, vertical soil stacks may be contaminated with aerosols containing the virus upon toilet flushing; this happened during the 2002 SARS pandemic in a large community in Hong Kong, where a flawed sewage-disposal system led to aerosolization of infected wastewater, resulting in a cluster outbreak [39,40]. These findings suggest that the main pathway for virus contamination in water is via infected faecal matters existing in sewages and wastewater, and may infect our urban water cycle through untreated wastewater, leaked sewage pipes and surface water contamination [9]. Community with less hygienic sewage facilities or poor sanitation are potentially prone to the risk of waterborne transmission. Hence, the past studies [39,40] should be and have served as a basic guideline on what and where to look for when examining the virus contamination of water and the possibility of waterborne virus transmission.
4.2. Reported transmission route of SARS-CoV-2 from human to the water bodies
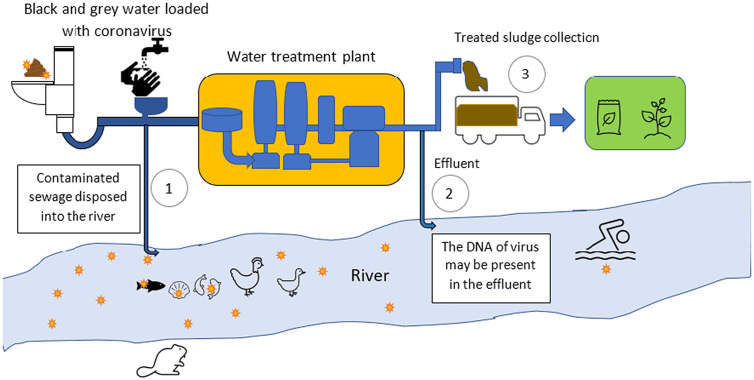
Since the beginning of the current pandemic, there are growing concerns over potential contamination of SARS-CoV-2 on surface water bodies, an area that is still not being well-studied. Well-known waterborne viruses such as Hepatitis A, Hepatitis E, adenovirus and rotavirus mainly affects people from developing countries, and these viruses normally spread in water environment through human faecal-oral contamination, and most of these viruses' outbreaks began and spread through the wastewater [8]. Also, certain viruses from aquatic lives (for example, from fish and crustaceans) are viable and temporarily virulent in both freshwater and wastewater [41]. Based on the studies of these waterborne viruses, it is also possible for SARS-CoV-2 to be virulent in water bodies, hence studies on virus existence and concentrations in untreated and under-treated wastewater is crucial in understanding the epidemiological risks of the infected surface waterways as shown in Fig. 2 [42]. Just like most waterborne viruses, the primary contamination of SARS-CoV-2 in wastewater is through viral shedding in patients' faecal matter, while other contaminations include hand washing, vomiting and urination [43,44]. Most viral infected water systems are caused by sewage discharges from hospitals and quarantine centres [43,45], but contamination of sewage pipes in urban areas (for example, residential areas) occupied with infected people is also possible [13,46]. Although this virus has not been detected in any drinking water source and water treatment plants to date, these water sources can still be contaminated within areas of non-point discharges, mainly in third-world countries where they normally urinate and defecate in open pit toilets and can possibly contaminate the soil and groundwater [7,47]. However, the virus loading in the sludge collected from water treatment plant can be reduced by treatment before disposal and anaerobic digestion in the earth [48].
Fig. 2.
The routes of transmission for coronavirus to the water bodies.
Since faecal contamination of SARS-CoV-2 is prominent in wastewater, studies of faeces of COVID-19 patients discharged into the sewage were reported [49,50]. In addition to the main symptom of respiratory problems, many COVID-19 patients have suffered from gastrointestinal symptoms such as diarrhoea, which is an initial indication of our digestive system being a potential route of infection [51]. In fact, it was suggested that these viruses can replicate in the human gastrointestinal tract and can infect the gastrointestinal glandular epithelial cells [52,53]. Approximately 2–10% confirmed COVID-19 cases were associated with diarrhoea, and many studies have reported the detection of SARS-CoV-2 RNA in stool samples of infected patients [51,54,55]. Interestingly, there are studied cases [53,56] of recovering patients obtaining negative respiratory swab tests but there was still presence of the viral particles in their stool samples. A study [53] reported that 53% of tested faecal samples (39 out of 73) were tested positive with SARS-CoV-2, and it was found later that 23% of recovering patients still had traces of the virus in their faeces, although no virus was detected in their respiratory systems. This indicates that the virus may still survive in the human body for a longer time, and some clinical studies reported that the virus is viable in human faeces between 7 and 33 days after a negative swab test [57,58]. High viral RNA antibodies in stool samples were present during the early stages of COVID-19 infection, with the highest presence in the first week of the infection [59]. The concentrations of the viral RNA in human faecal matter can be up to 108 copies per gram of faeces [59]. In addition to faeces, SARS-CoV-2 can also be observed in urine samples of infected patients. It was suggested that the virus is more stable in urine compared to faeces, and a few studies recorded around 105 to 108 RNA copies per litre found in patients' urine samples [3,51,60]. However, only an insignificant number of COVID-positive cases reported the detection of the virus in urine, hence clinical studies on this topic is very limited [3,61].
Although there are no official reports of COVID-19 patients being infected by sewage or drinking water, this virus still has the potential of waterborne transmission from untreated water since they can live on water surfaces for days. Extensive studies of the virus in sewage and untreated water have been done, and several studies [12,62,63] reported that this virus is viable up to 2 days in dechlorinated tap water in both hospital and domestic areas, and could endure low temperature of 4 °C for 14 days in wastewater. These data indicate the importance of wastewater and surface water monitoring, albeit there is still no human health risk related with virus exposure from environmental samples [42]. The studies on the challenges in the detection of SARS-CoV-2 in wastewater will be discussed in the next section.
5. Challenges of existing techniques for the detection of viruses in water
Virus detection in water media is critical for creating mitigation strategies and health and safety procedures [64,65], hence, rigorous studies on SARS-CoV-2 detection in various water media and wastewater have been reported. Additionally, the detection of virus is very useful for our health and government officials as an epidemiological indicator to determine the rate and magnitude of virus spreading, since the presence of virus in water media poses a huge risk as it can spread uncontrollably [66]. Hence, understanding the state-of-the-art on methods for detecting, quantifying, and determining virus infectivity in aqueous matrices is critical. It is also worth noting that the infectivity (ability to infect/transmit) of a virus is not directly related to its detection, thus differing analysis on determining the viral infectivity is also necessary [66]. Generally, the existing virus detection methods in water media are designed for detecting waterborne and non-enveloped viruses such as adenovirus and norovirus [67,68]. As mentioned, CoVs are protected with outer envelopes, so these viruses and other enveloped viruses have different physical and biochemical features from those traditional waterborne viruses, the typical detection methods will not yield the same recovery efficiency [69]. Therefore, many different methods for detection of virus have been studied for various infected water samples.
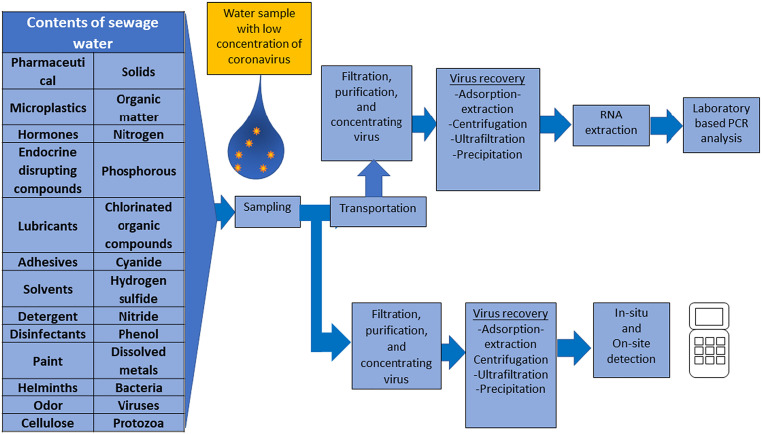
Unlike blood or saliva or blood samples, sewage water is a highly complex fluid, and the contents of sewage water are as indicated in Fig. 3 . Detecting the presence of virus in sewage water is not as straightforward as some may expect; there are processes involved prior to actual virus detection that are vital, such as sample preparation and pre-treatment of samples (Fig. 3). Proper preparation and/or pre-treatment of virus samples will increase the virus detection accuracy, and these parameters depend on the volume of virus sample, extraction/concentrated yield and sample purity [70]. It has been reported that the concentration of nucleic acids (for nucleic acid-based methods) and the type of the extraction procedures have a substantial impact on the overall results of the community composition, specificity and pathogen detection of the viruses [63]. Hence, pre-treatment, virus recovery and detection methods must be selected accordingly to achieve the best results. Concentrating the samples is by far the most common and effective pre-treatment method for virus detection, especially for aqueous samples [71]. Since untreated (unconcentrated) samples are taken from large water bodies, the virus concentration is initially very dilute, so concentrating water samples is important to increase the results yield above the detection limit [49,71]. Various approaches for concentrating viruses obtained from any water media have been developed; however, it should be noted that the majority of those concentration methods were established for non-enveloped enteric viruses which are generally waterborne [72]. The details of various methods for concentrating the viruses to increase the efficacy of detection would be discussed in the following section.
Fig. 3.
The workflow in detecting virus loaded in complex wastewater in the laboratory and using in-situ test instrument.
5.1. Virus recovery methods from water samples prior to virus detection
In addition to the sample processing, the origin of water samples from different environments can largely impact the virus detection accuracy. For example, flowing water bodies or influents usually have higher virus concentration compared to other water samples [72]. Also, small volumes of sludge and wastewater (< 100 mL) samples tend to have higher virus concentration compared to larger volumes of surface, recreational and drinking water (> 10 L) with much lower viral concentrations [72]. Furthermore, the presence of significant levels of heavy metals and organic acids in influent wastewater may impede virus detection that involves molecular methods or nucleic acid-based detections [73]. Untreated wastewater samples also have more suspended solids, greater concentration of organic matter and higher turbidity compared to other environmental water samples [73,74]. These are examples of environmental factors that have a huge impact on the detection accuracy, which can be improved by concentrating the samples. Although research on virus concentration methods have been broadly developed and established, there is still no definite method that is considered as reliable for all virus detection [75,76]. This is because a significant number of viral particles can still be lost after pre-treatment of the samples [75,76].
Prior to concentration of samples, samples also undergo initial pre-treatments that includes filtration, centrifugation, and settling time to remove suspended solids and debris [66]. Inactivation of virus can also be done by pasteurization at around 60 °C for over 1 h, this is to ensure safe handling of tested samples. As mentioned above, pre-treatments of the water samples should be chosen and done attentively since they can drastically alter the quantification of virus. The exclusion of viral particles adsorbed onto particulate in the pre-treatment process may decrease the detection accuracy [66]. As for the actual concentration methods, there are many ways to concentrate viral water samples, most common methods include adsorption extraction method, ultracentrifugation, ultrafiltration, and precipitation (Table1). These techniques have been widely tested on numerous enteric viruses and some CoVs in the past in the control process for concentrating the virus, however only limited studies were reported for SARS-CoV-2. Hence, this manuscript will put emphasis on reviewing these virus concentration methods as process control methods specifically on SARS-CoV-2.
-
a)
Adsorption extraction method
Virus adsorption extraction is a simple method begins with the acidification of samples to pH 4–6.9, followed by an addition of MgCl2. Subsequently, the pre-treated samples pass through electronegative membranes with a pore size of 0.45 μm. The electronegative retains the electronegative viral capsids [75]. The membrane is then inserted into a beating tube ready for RNA extraction. This method was demonstrated with relative high level of MHV recovery (65.7 ± 23%). Alternatively, aluminum-based adsorption-precipitation is a popular method to concentrate virus samples. This method requires the adjustment of sample pH to 6.0, shaking, extraction and the centrifugation [77]. Then, the pellet is resuspended in buffer solution. However, the virus recovery percentage (0.02–30%) derived from this method is lower compared to the use of electronegative membrane technique (Table 1 ).
-
b)
Centrifugation
Table 1.
Virus recovery using different methods for concentrating virus from wastewater.
| Method | Sample type | Materials/condition | Time | Virus | Virus recovery percentage | Ref |
|---|---|---|---|---|---|---|
| Adsorption extraction method | Wastewater Inoculated with virus |
|
< 30 min | Murine Hepatitis Virus (MHV) Seeded into wastewater |
26.7 ± 15.3% (Acidification of sample) 65.7 ± 23% (pre-treatment with MgCl2) |
[49,75,99] |
| Wastewater Inoculated with virus |
|
< 2 h | SARS-CoV2 Mengovirus (MgV) PEDV strain CV777 |
30.2 ± 17.7% 6.8 ± 4.8% <2% |
[91] | |
| Influent wastewater collected from wastewater plant |
|
< 2 h | MgV | 0.02–4.3% | [77,100] | |
| Biobanked influent water samples inoculated with viruses |
|
< 2 h | Porcine epidemic diarrhea virus (PEDV) strain CV777, enveloped virus Non-enveloped virus, Mengovirus (MgV) |
PEDV: 11 ± 3.5% (Influent water) 3.3 ± 1.6% (Effluent water) MgV: 11 ± 2.1% (Influent water) 6.2 ± 1.0%(Effluent water) |
[77] | |
| Ultracentrifugation | Influent wastewater collected from wastewater plant | Centrifugation 140,000 ×g for 2 h 20 min 120,000 ×g for 30 min 229,600 ×g for 1 h |
<5 h | Non-enveloped virus, MgV | 8.04–25.72% | [77] |
| Wastewater From water treatment plant |
150,000 ×g at 90 min | <2 h | SARS-CoV-2 | 12% | [35,80] | |
| Wastewater | Centrifugation 100,000 ×g for 1 h 12,000 ×g for 15 min |
< 2 h |
Murine Hepatitis Virus (MHV) Seeded into wastewater |
35.5 ± 12.1% | [76] | |
| Raw sewage water from sewage treatment plant | 100,000 ×g for 1 h 12,000 ×g for 15 min 100,000 ×g for 1 h |
< 2 h | Rotavirus (RV-A) Bacteriophasge (PP7) |
Range 34%–67% Mean: 47% |
[101] | |
| Ultrafiltration | Wastewater | 10, 30 and 100 kDa membrane filter PEG precipitation |
CoV | 33–42% | [81,82] | |
| Wastewater From water treatment plant seeded with viruses |
CP-Select (Concentration pipette ultrafiltration tips, InnovaPrep) Centricon® Plus 70 centrifugal ultrafiltration devices (CeUF) |
< 2 h | Bacteriophage MS2 Murine hepatitis coronavirus (MHV) |
CP-Select MS2: 27.72 ± 24.4% MHV: 7.51 ± 6.14% CeUF MS2: 26.34 ± 22.71% MHV: 24.07 ± 14.48% |
[81] | |
| Wastewater influent from water treatment plant | Centricon® Plus 70 centrifugal ultrafiltration devices (CeUF) | ~ 1 h | Mengovirus | 17.5–100.5% | [83] | |
| Wastewater | CP-Select (Concentration pipette ultrafiltration tips, InnovaPrep) Electronegative membrane (EM) |
< 1 h | Bovine Coronavirus |
CP-Select: 5.5 ± 2.1% EM: 4.8 ± 2.8% |
[102] | |
| Wastewater | Americon Ultra-15(30 K) Centricon plus-70 |
<1 h |
Murine Hepatitis Virus (MHV) Seeded into wastewater |
56.7 ± 32.3% 28.0 ± 9.1% |
[49,103] | |
| Precipitation | Wastewater Inoculated with virus |
10% PEG8000 2% NaCl w/v |
< 1 h | Murine Hepatitis Virus (MHV) Seeded into wastewater |
44.0 ± 27.7% | [84] |
| Grab wastewater Inoculated with virus |
PEG8000(20%) 25 mL of Tris Glycine-Beef Extract buffer (TGEB) NaCl 0.3 M |
Overnight incubation | SARS-CoV2 Mengovirus (MgV) PEDV strain CV777 |
52.8 ± 18.2% 11.1 ± 4.9% 2.6% |
[91] | |
| Grab wastewater from wastewater treatment plant | PEG8000 and NaCl2 | <24 h | F-phage | ~10% | [87] | |
| Grab wastewater from wastewater treatment plant | Pre-centrifugation PEG8000 and 10%NaCl2 |
Overnight incubation <24 h |
Bacteriophage MS2 Pseudomano Phage Ҩ6 |
27.5% -77.6% 29.8%–49.8% |
[88] |
Centrifugation is a very straightforward process, where the sample mixture can be separated/concentrated through high-speed spinning motion [78]. Ultracentrifugation is generally a rapid method for concentrating wastewater samples, and it is the most common concentration technique used. Centrifugation method is usually used on sludge samples, and the American Society for Testing Materials (ASTM) has established a protocol on pre-treatment of sludge samples (ASTM D449-19). This protocol promotes virus concentration onto sludge flocs by adsorption and subsequent elution [70]. Promotion of virus adsorption to the flocs to the optimum level is done by increasing the acidity of sample (to pH 3.5) by adding aluminium chloride (AlCl3) solution. After that, the mixture is centrifuged to separate the liquid phase from the solid phase. To desorb the viruses, the solids are rinsed with an eluent solution, and the eluate is centrifuged again. The pelleted solids are then discarded, and the supernatant is filtered to eliminate bacteria and other particles using a 0.22 m filter [78,79]. This protocol was tested on enteric viruses (for example, Rotavirus A, HAV, HAdV and NoV GII) recovered from primary sewage sludge samples, and it was found that only less than 7.5% of mean recovery was recorded for all viruses. This means that the concentration protocol was unable to increase the virus recovery, which hampered subsequent molecular-based detections [73]. This experiment involved adding inactivated SARS-CoV-2 into wastewater samples, and after ultracentrifugation the mean recovery was approximately 12% [80]. Despite the low mean recovery of targeted analyte from ultracentrifugation, these numbers are enough for amplification-based nucleic acid detection, whereas for more sensitive environmental detection and surveillance, further optimisation of pre-treatment methods and virus detection should be investigated [35].
-
c)
Ultrafiltration
Another common concentration method is ultrafiltration, which is a size-exclusion based method. Samples will be passed through membrane filters with different pore sizes that are smaller than the viral particles, and membrane filters with 10, 30 and 100 kDa weight cut-offs are typically used to concentrate CoVs in water samples [35]. Compared to centrifugation, there is no need for pH altering of samples and long precipitation time prior to ultrafiltration [81]. Fores et al. recently evaluated two different types of ultrafiltration devices (Centricon® and CP-Select™) on spiked viral wastewater, which can rapidly concentrate the viral samples. The size of filters used was 30 kDa to favour viral retention overall and avoid retention of smaller molecules that can act as enzymatic inhibitors. It was reported that the highest concentration of SARS-CoV-2 detected in supernatant, from nine tested viral samples, was 77%, and the remaining 23% was adsorbed to the solid fraction. Another study comparing different types of concentration methods were done on untreated wastewater spiked with SARS-CoV-2 [82]. The two methods were centrifugal ultrafiltration and polyethylene glycol (PEG) precipitation. For ultrafiltration using Vivaspin columns, the mean recovery rates were between 33 and 42.6%, while the PEG precipitation reported mean recovery rates of 59.4–63.7%. Although the ultrafiltration method yielded lower mean recovery rates, both methods are still deemed effective for concentrating enveloped viruses. A study of stability of virus RNA biomarkers in wastewater influent were done by prolonging the storage time and verifying the storage temperatures of different viruses of SARS-CoV-2, SARS-CoV-1 and norovirus GII [83]. From this study, they used mengovirus to determine the rate of virus recovery for ultrafiltration and reported recovery efficiencies from 17.5% to 100.5% in the supernatant fractions. It is very common for researchers to use different types of less harmful, non-pathogenic viruses to determine the mean recovery rates of concentration methods due to the stringent biosafety requirements for handling SARS-CoV-2, in which, this explains the limited studies that do not provide information for the percentage of virus recovery rates [84].
-
d)
Precipitation
Precipitation of viral samples is normally done by adding chemicals such as aluminium hydroxide Al(OH3) or polyethylene glycol (PEG) to induce the precipitation process [85]. This method is perhaps the most used concentration method since it does not need any advanced or fancy equipment. PEG precipitation is generally applied for proteins' precipitation since PEG can act as an “inert solvent sponge” that can break the solvation layer that surrounds the outer protein shell of a virus [86]. Without the solvation layers, the interactions between viruses will greatly increase, which will then precipitate to the bottom. This precipitation method was also suggested to be effective for concentrating enveloped viruses in surface water and wastewater samples [84,87,88]. A study by Kumar et al. suggested that detection of genetic material of SARS-CoV-2 can potentially be used in wastewater surveillance in India [7]. After precipitation, the concentrated viral samples were mixed with MS2 bacteriophages that act as a molecular process inhibition (MPC) control [72]. This step was taken to assess the efficiency of nucleic acid extraction and PCR inhibition, and it was found that the recovery of spiked MS2 in the tested samples had stable CT (threshold cycle) values (22.2–22.46), indicating efficient removal of wastewater matrix that may inhibit RNA extraction and detection. Note that samples are considered positive with SARS-CoV-2 with CT values below 40 [89]. Quantification of naturally occurring bacteriophages in viral wastewater samples is a practical indicator of efficient virus recovery. Another example, Hata et al. quantified the recovery efficiency of F-phages that are abundant in wastewater, and they reported a solid 45% geometric mean recovery of the F-phages, which can also indicate an efficient recovery for SARS-CoV-2 using PEG precipitation [87]. A mean recovery efficiency of >10% can be considered amply high and stable [72]. In addition to the use for precipitating agent for pathogens, Al(OH)3 has the ability to destabilise fine particulate particles (viruses and bacteria), and the viral particles will adsorb to the hydroxide and thus settle out as a precipitate [90]. Randazzo et al. employed the Al(OH)3 adsorption-precipitation to detect SARS-CoV-2 RNA in wastewater located in low prevalence area [77]. They validated this method using porcine coronavirus (Porcine Epidemic Diarrhoea Virus, PEDV) and mengovirus prior to applying it for SARS-CoV-2 detection and reported average recoveries of 11 ± 3.5% and 11 ± 2.1% in influent water samples for PEDV and mengovirus respectively. They repeated this study by comparing this method with PEG precipitation, in addition of using inactive (gamma-irradiated) SARS-CoV-2 [91]. With these precipitation methods, they managed to obtain mean recovery rates of 42.9 ± 9.5%, 27.5 ± 14.3% and 9.0 ± 2.2% for SARS-CoV-2, PEDV and mengovirus respectively.
Other concentration methods that have been investigated for detecting SARS-CoV-2 in wastewater include adsorption-extraction, electronegative membrane vortex (EMV) and skimmed milk flocculation and many more [71,84,88,[92], [93], [94]]. Although there are various types of concentration methods available, the effectiveness of these methods, especially for the detection of SARS-CoV-2 and other enveloped viruses in any water body, required further investigation and optimisation.
5.2. Feasibility of molecular based methods for the detection of SARS-CoV-2 detection in water
Virus detection in water bodies requires methods that are sensitive, rapid, cost-effective and high accuracy. By far, no specific method fulfils all these requirements, hence, more optimisation of the detection methods is required. Although many detection methods of pathogens in water bodies have been studied [105], most of them are established for non-enveloped enteric viruses that are structurally and chemically very different from enveloped CoVs, hence many studies on SARS-CoV-2 detection usually include modifications of the conventional methods. Before molecular analysis methods were popularized, cell culture methods were the gold standard for virus isolation since they give more accurate results [95]. Cell culture studies of different CoVs have been done on spiked wastewaters to determine the longevity and infectivity of the virus in different aqueous matrices [96]. These studies determined that the inactivation of CoVs greatly depends on the surrounding temperature, content of organic matter and presence of antagonistic bacteria [97]. Also, RNA of virus was found to be more persistent in sewage and different water bodies compared to the entire infectious virus themselves [98].
Currently, molecular methods, specifically polymerase chain reaction (PCR) are becoming more favoured due to their high sensitivity and specificity, and able to detect non-culturable viruses [72]. By PCR, a tiny sample of DNA can be rapidly replicated or amplified to billions of copies that are more than enough for qualitative and quantitative analyses of target study [66]. Because of high specificity of the technique, PCR has been regularly adopted for many years for detection of viruses in the environment, especially enteroviruses and hepatitis A virus (HAV) [[104], [105], [106]]. Although regularly used, PCR-based methods do have several disadvantages. It is important to note that these methods are highly susceptible to inhibition, and the inhibitory substances such as humic acids that are abundantly present in wastewater, are often inadvertently concentrated together with the target viruses [72]. Other known inhibitor of RNA are mostly organic compounds such as, urea, polysaccharides, sodium dodecyl sulphate, tannic acid, ethanol, urea, bile salts, melanin, collagen, myoglobin and haemoglobin [107]. These are the compounds commonly present in the wastewater (Fig. 2). Hence, the inhibitors will partially or totally decrease the sensitivity or giving false negative which is depending on the concentration of the inhibitor. In addition, PCR can only detect single virus type, and the sensitivity and specificity of the method is highly dependent on the efficiency of virus recovery [106].
The genetic material for CoVs and viruses in general is the ribonucleic acid (RNA), hence, to apply PCR, reverse-transcription (RT) from RNA to complementary DNA strand is compulsory. Till date, most of the SARS-CoV-2 detection in water bodies applies RT-PCR or RT-qPCR (q – quantitative) techniques [14,49,93]. For wastewater samples, the absolute limit of detection (ALOD) should be low since the concentration of viruses is usually diluted in water bodies. However, most RT-qPCR assays do not display ALOD data since these assays are designed to do rapid screening tests [6]. Hence, RT-qPCR assays that display <10 RNA copies per reaction could be useful for wastewater samples tests for SARS-CoV-2 [108,109]. Sequencing analysis is usually done to confirm the positive RT-qPCR signals in viral wastewater before the assay specificities have been confirmed against environmental samples. This is due to the design of current RT-qPCR assays that is first tailored made for clinical diagnosis instead of environmental surveillance. The study of sequence analysis for SARS-CoV-2 detection in wastewater has been done by sequencing after regular PCR or direct sequencing of the qPCR products [110,111]. To date, a standard assay for SARS-CoV-2 detection in water bodies has yet to be established; there are several studies that target different protein regions of SARS-CoV-2 for detection, such as the nucleocapsid N-protein, spike S-protein, envelope E-protein, membrane M-protein, ORF1ab and RNA-dependent RNA polymerase (RdRP), and employ different types of assays that contain different types of mentioned genes to determine the limit of detection of each assay with respective protein gene [22,49,102,108,[112], [113], [114]]. Among these screened encoding genes, ORF1ab is the most sensitive region for RNA amplification, although N- and E- genes coding can also be applied for SARS-CoV-2 detection [108,115]. Some of the established RT-PCR/qPCR assays for SARS-CoV-2 detection include CDC N1, N2, N3, E_Sarbeco, N_Sarbeco, and NIID_2019-nCOV_N assays. Buonerba et al. has written an in-depth review that summarizes these studies [66]. A study by Ahmed et al. employed the N_Sarbeco and NIID_2019-nCOV assays to study two different virus concentration methods, and recorded inconsistent results for both assays [49]. Even the positive samples were below RT-qPCR quantification level. Only N_Sarbeco assays provided two positive samples, indicating that this assay is more sensitive than the NIID_2019-nCOV assay to a certain extent. Medema et al. employed four different primer/probe sets; CDC N1-N3 and E_Sarbeco assays, to detect SAR-CoV-2 RNA in several wastewater samples in Netherlands [12,108]. All the wastewater samples tested with all four assays recorded positive results at 26–1800 gc/mL, taken on 25th March 2020. The CDC N1-N3 assays managed to yield similar quantitative results. Wu et al. studied the concentration of SARS-CoV-2 titers in wastewater using the three N assays, and reported that both N1 and N3 assays recorded positive for 3 out of 5 different samples each (CT < 40) while the N2 assay only recorded positive for only 1 out of 5 samples [116]. Sherchan et al. studied two different concentration methods for SARS-CoV-2 RNA in wastewater, and they used N1 and N2 assays to perform RT-qPCR tests [93]. Only 13% of the wastewater samples prepared with ultrafiltration were tested positive. Referring to some of the assay studies, La Rosa et al. tested the new assays for SARS-CoV-2 detection in influent sewage samples taken from several wastewater plants [117]. Out of the 50% samples that tested positive for SARS-CoV-2, they reported that the assay targeting for RdRP gene is more sensitive than the assay that targets the S-gene, although no quantitative analysis was provided. Kumar et al. reported the detection of ORF1ab, N- and S-genes in influent water samples that were tested with readily available RT-PCR kits (TaqPath™ COVID-19 RT-PCR Kit) [7]. The estimated maximum concentration of the SARS-CoV-2 genes was 3.5 × 102 copies/L. A study by Tanhaei et al. was conducted by testing wastewater samples in Iran [92]. They reported the detection of SARS-CoV-2 in 8 out of 10 effluent wastewater samples using RT-qPCR kits also detected ORF1ab and N-genes. The CT range values for ORF1ab and N-genes were 29.62–34.45 and 27.60–30.53, respectively.
Digital PCR (dPCR) is a newly emerging approach for SARS-CoV-2 detection and quantification. Compared to RT-qPCR, dPCR has been reported to have down to 10 times lower detection limit, indicating that dPCR is a more sensitive method [118]. Another version of dPCR is the droplet dCPR (ddPCR), and this method has reported a detection limit of 500 times lower than RT-qPCR, which is deemed suitable for detecting low concentrations of SARS-CoV-2 in wastewater in low COVID-19 prevalence area [118]. Other advantages of using dPCR/ ddPCR include less affected by PCR inhibitors and quantification without calibration curve [119,120]. Since dPCR does not rely on external calibration, it can be used to calibrate reference standards for qPCR [119]. Despite these advantages, only a few studies have been done to detect and quantify SARS-CoV-2 in water bodies. One notable study is a one-step ddPCR by Graham et al., and they found that this method is very sensitive in detecting SARS-CoV-2 in settled solids from wastewater due to the reduced effect of PCR inhibitors [50]. On the contrary, a study by D'Aoust et al. reported a superior quantification of SARS-CoV-2 using RT-qPCR compared to RT-ddPCR [121]. Several disadvantages of dPCR include lower specimen throughput and limited sample volume per reaction [119]. Another interesting PCR method is the integrated cell culture PCR (ICC-PCR), which, integrates the cell culture method into the PCR-based methods. ICC-PCR was found useful in differentiating infectious viruses from non-pathogenic viruses [122]. However, no reports have been published on SARS-CoV-2 detection by ICC-PCR.
6. Biosensors for the detection of coronavirus in wastewater
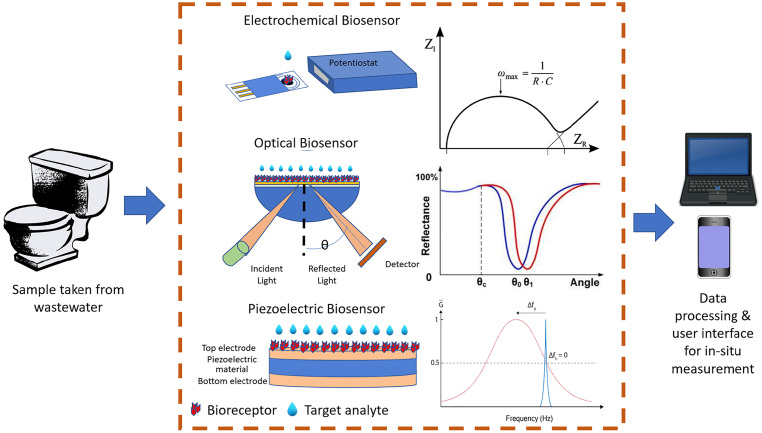
A biosensor typically consists of a biorecognition element (bioreceptor) and a physical transducer such as electrochemical, optical, and piezoelectric as shown in Fig. 4 . The bioreceptor could be antibodies, nucleic acids, enzymes, phage, or whole cells and microorganisms. Interactions between the bioreceptor and the target analyte are converted into electrical signals by a transducer. Readout circuits then process these signals and transmit them either into a computer or smart device, making it easily accessible to the user. The use of biosensors allows rapid detection of viruses, at low sample volumes and reagents, making it an ideal candidate for WBE systems that require a cost effective, yet continuous monitoring method of SARS-CoV-2 detection [123,124]. Correlation between wastewater viral load and the number of infected individuals can be made to infer the extent of the outbreak.
Fig. 4.
Existing biosensors (electrochemical [134], optical [135], and piezoelectric [136]) suitable for in-situ COVID-19 monitoring through WBE.
Selected biosensors that employ these three mechanisms (bioreceptor-analytes interactions, signals conversion and digital readout) and are commonly used for the detection of pollutants and infectious diseases in WBE are listed in Table 2 . The suggested advantages and disadvantages of each method can be considered for the development of COVID-19 WBE-based in-situ monitoring. For WBE applications, in which samples are a complex matrix and may be contaminated, electrochemical sensors perform better as a detection tool as compared to the optical methods which have poorer LOD [125]. Other advantages of electrochemical biosensors are fast response times, low cost, and ability to be miniaturized with other portable devices [126]. However, purification process and virus concentration methods as discussed earlier on may improve the LOD of electrochemical biosensors. Optical detection methods such colorimetry and fluorescence are more suitable for discrete, pure and clean samples as these techniques are easily disrupted by the coloured and turbid samples of the wastewater [125]. Higher end optical techniques such as surface plasmon resonance (SPR) [127,128] and resonant mirror [129,130] have been reported to be sensitive and selective enough to detect virus detection in turbid samples. Functionalized metal nanomaterials such as gold nanoparticles can be used in both colorimetry [131] and Raman spectroscopy [132] to further enhance the sensitivities of these biosensors. While optical biosensors show a lot of potential in terms of sensitivities, these high-end systems are expensive both to purchase and maintain, making it less practical to be implemented in low-income communities. From the latest literature, it is interesting to note that Kumar et al. 2021 [133] reported an improved electrochemical DNA sensor using printed circuit board (PCB) electrodes integrated with portable PCR instruments for SARS-CoV-2 wastewater-based monitoring. This finding can be the way forward for the incorporation of electrochemical sensor for WBE SARS-CoV-2 monitoring by overcoming the limitations related to pre-treatment and concentration methods. In addition, it is important to note that the possible interference from the compounds present in the complex wastewater needs to be considered when interpretating the results obtained using these techniques.
Table 2.
Selected biosensors of various signal detection methods that are commonly used in WBE to detect various pathogens and infectious diseases.
| Signal detection methods | Advantages | Limitations | Infectious diseases / pathogens | Ref |
|---|---|---|---|---|
| Electrochemical (Voltammetric, Amperometric, Potentiometric) |
|
Potentiometric has issues related to immobilization, poor linear range, low reproducibility potential |
|
[[144], [145], [146]] |
| Optical (Fluorescence, Luminescence, Colorimetry) |
|
Complex, High cost, Bulky equipment (not portable), relatively high LOD |
|
[[147], [148], [149]] |
| Piezoelectric |
|
Long incubation times, difficulties in regenerating crystal surfaces, multiple washing and drying steps, difficult to immobilize the antibodies on quartz crystal |
|
[139,140] |
| Microfluidics |
|
Requires in-depth study on functional materials to simplify device, not miniaturize enough |
|
[141,150,151] |
| Paper-Based Devices |
|
Challenge to deal with complex WBE matrix |
|
[[152], [153], [154]] |
Another promising detection method for SARS-CoV-2 in WBE is the piezoelectric sensors. A piezoelectric biosensor is a mini weighing scale that detects changes mass on top of the sensor by measuring resonant frequency changes in the crystal. The mass change is associated with the interaction of bioreceptors immobilized on the crystal surface and biomolecules from the samples [137]. Due to its high sensitivities, simplicity and rapid detection, the piezoelectric-based biosensor has been employed in large screening operations such as HIV, Influenza and SARS. However, as reported by Mohsen et al. [138], only few piezoelectric sensors have been reported for wastewater-based detection [139,140]. This is probably due to difficulties in immobilization of antibodies on the crystal surface [138].
A comprehensive COVID-19 WBE system requires sensing, readout and sample or fluid handling. All these lab processes can be miniaturized into a single ‘micro total analysis system’ (uTAS) or lab-on-chip (LOC) device using microfluidic channels and reaction chambers. LOC devices provide advantages of low fluid volume consumption (less reagents, samples, and waste), which also result in faster analysis and response times. Multiple biosensing techniques such as PCR, electrochemistry, optical and loop mediated isothermal amplification (LAMP) for DNA can be integrated in a single device and for virus detection [141]. These microfluidic devices have been reported to reliably and precisely quantify antibodies and protein biomarkers [142]; detect pollutants such as copper, nitrite, aromatic amines, sulphide and ammonium [143] in WBE systems. Most importantly, uTAS are cost-effective, easily deployed on-site and can be placed in hostels or low-cost flats, which are the target areas of these WBE systems.
While in-situ WBE biosensors have been widely tested for infectious diseases such as Hepatitis A [[144], [145], [146]], commercially available COVID-19 biosensors have not yet been used in WBE. Most WBE testing rely of PCR data [50,121]. Promising development has been shown in this direction where a low-cost in-situ COVID-19 sensor in WBE has been successfully developed by a team of researchers from the University of Strathclyde, Glasgow, and the Indian Institute of Technology (IIT), Bombay [133]. The fabricated electrochemical-based biosensor utilizes easy-to-clean printed circuit board electrodes which enables reusability and, therefore, have a long shelf life, making the test even more cost-effective. The sensors are used to detect SARS-CoV-2 virus using samples collected from a sewage treatment plant based in Mumbai, India. Another significant in-situ COVID-19 WBE biosensor development is paper-based biosensor as it offers simple detection process where all processes required for nucleic acid testing include extraction, enrichment, purification, elution, amplification, and visual detection can be integrated onto an inexpensive paper through wax printing [155]. The testing process involves simple methods such as paper-folding and naked eye detection. Hydrophilic/hydrophobic microchannel networks are also easily placed on paper-based devices for detection of various pollutants/pathogens making them a simple, low cost and portable solution for in-situ monitoring when coupled with smartphone image processing [156]. An example of such system was a paper analytical device (μPAD) that detects norovirus in field water samples using antibody-conjugated fluorescent submicron particles [152]. Park and Jeong, 2015 [153] showed another application of a multichannel, cellulose-based chromatographic paper sensor used for E. coli detection.
Microfluidic paper-based devices combined with various sensing platforms including isothermal amplification, LAMP, PCR and thermal lysis have also been considered as the future candidate for in-situ SARS-CoV-2 detection in WBE as reported by [157]. While biosensors have lower sensitivities than PCR, nanomaterials such as nanoparticles, nanorods, nanowires, carbon nanotubes and nanocomposites can also be incorporated in biosensors to improve the sensors' sensitivity and LOD to counteract the low concentration of virus particles' in WBE. An example of a paper-based microfluidic device has been developed by Cranfield University researchers who reported test results in less than one hour through the detection of nucleic acids specific to COVID-19 [158]. This biosensor was developed as part of the UK National Wastewater Epidemiology Surveillance Programme (N-WESP) to develop an early COVID-19 detection system by testing samples from wastewater treatment plants. In more recent development, these researchers have discussed the feasibility of coupling the biosensors with mobile health technology creating a rapid, comprehensive, internet-of-thing (IOT) system that provides early warning, allowing effective public health interventions to manage the epidemic [159].
In summary, biosensors coupled with fluidics have the potential to be used as a rapid, real-time, low-cost, and portable solution for in-situ SARS-CoV-2 detection in WBE. Combination of various detection technologies (optical, electrochemical, PCR), with functional nanomaterials, LAMP and low-cost microfluidics may pave the way forward for rapid in-situ SARS-CoV-2 monitoring in WBE. Coupled with data analytics, this system can be an early indicator of the onset of a COVID-19 outbreak and can be used by public health officials to warn the spread of the disease and employ appropriate interventions.
7. Challenges and outlook
Improving the efficiency of COVID-19 virus detection and an improvement of the quantification techniques in the highly complex wastewater are still challenging. Although various molecular techniques are extensively utilized for detecting viruses in wastewater, it should be ensured that a wider range of viruses and analysis of the viability of viruses are available. To implement countermeasures and mitigate the public health outbreak risk, the application of biomarkers (pathogens, chemicals, and metabolites) is immensely important. Two major challenges of biomarkers are high sensitivity and low detection limits. Biosensors with nanobiotechnology with exact recognition features have a specific affinity towards target pathogens [160]. Nanobiotechnology-assisted sensors are advantageous due to their low cost, fast detection, and facile application. Thus, the biosensors can feasibly be used for real-time field monitoring in wastewater. Table 3 summarizes the possible application and challenges of various biosensor detection techniques.
Table 3.
Promising applications and challenges of various biosensor detection techniques for WBE.
| Detection technique | Applications in WBE | Challenges | Ref |
|---|---|---|---|
| Indirect (PCR, CRISP, and genome sensing) | Commonly used technique to detect nucleic acids; Precise and sensitive detection of SARS-Cov-2 RNA; examine complex wastewater or biofluids. | Require trained personnel otherwise false negative; specialized equipment to interpret results for disease circulation; human health risks; inconsistency of sample strains vs. reference. | [[163], [164], [165], [166]] |
| Surface-enhanced Raman scattering (SERS) based biosensors (liquid, paper-based, microfluidic, magnetic) | Highly sensitive, low cost, rapid; detect at environmentally relatable concentrations; wastewater monitoring due to active SERS substrates; handheld systems appropriate for field analysis. | SERS substrate heterogeneity; weak SERS signals; require additional data analysis; reproducibility; challenge in detecting in complex wastewater at nano concentrations. | [167,168] |
| Electrochemical and field-effect transistor (FET) sensors | Simple lab on a chip integration; detect at environmentally relatable concentrations; portable; compatible for on-site analysis; simple operation. | Non-specific adsorption of interfering molecules and electrochemical signals in unstable physiological conditions in operation for the complex wastewater analysis. | [169,170] |
| Spectroelectrochemical (SEC) sensors | Capable of detecting single-molecule; feasible in using in complex wastewater media as complementary data allows to resolve overlapping signals coming from interfering molecules. | Device reproducibility with EC-SERS substrates; complexity in data analysis and interpretation; appropriate design needed for on-site analysis. | [171,172] |
As the COVID-19 pandemic has caused total disruption of public health, early detection and diagnosis are necessary. To maximize the benefits of WBE, improved development and monitoring of biosensors have great potential for real-time implementation. However, there are several challenges to optimizing the methods with improved efficacy, flexibility, and functionality in the application of complex systems of wastewater consisting of various types of biomarkers. Stability of the biomaterials and nanomaterials needed for the design of the sensor should be guaranteed in every circumstances. Reproducible and reliable techniques should be developed. Standardized analytical methods are required to detect the analytes. To overcome the drawbacks of existing diagnostic methods, robust approaches with widespread availability, low cost, rapid testing response and consistency are essential to detect SARS-CoV-2 viruses in the complex wastewater. A full risk assessment for the personnel is essential due to the potential risks of infection from exposure to live coronavirus.
The reproducibility, repeatability, and reliability of COVID-19 WBE biosensors can be determined by comparing the wastewater surveillance data utilizing the developed biosensors with the PCR-based sensors as previously reported by Xiao et al. [161]. PCR-based WBE data was compared to the gold standard clinical surveillance data by considering significant quantitative metrics including time lag and transfer function between wastewater and clinical reporting. Another significant finding is that by coupling WBE with targeted clinical testing, presence of COVID-19 cases in the community can be isolated and used to control potential outbreaks. This method was implemented in University of Arizona to detect the presence of COVID-19 in dormitories upon student re-entry to campus [162]. Detection of viral DNA in wastewater samples were followed by clinical testing in affected dorms, allowing identification, isolation, and containment of potential outbreaks. Similar data comparison can be done for COVID-19 WBE biosensors, only with faster turnaround times as biosensors can produce faster results compared to PCR. Community sewage biosensors coupled with clinical testing is a promising early-warning tool to detect the presence of COVID-19 presence in high density areas.
8. Conclusion
Although aerosol transmission of SARS-CoV-2 is well described, waterborne transmission by the human excreta (faecal matter and urine) is less understood. It was reported that the sewage sludge typically contains high virus load compared to the sewage runoff. In fact, the viruses have been found surviving in aquatic species and hence, can be virulent in the surface water or untreated wastewater. Unlike human biological samples, the wastewater consists of various substances that differ in composition depending on the sewage system of interest. This pose a significant challenges to the development of the biosensor for virus detection. The polar and non-polar chemicals found in the wastewater samples may interfere and influence the measurement of the biosensor. Hence, pre-treatment procedures such as purification, extraction and concentration of viruses are essential and should be carefully selected to ensure a good detection limit for SARS-CoV-2. The protocol and equipment for these pre-requisite preparations must simple, portable and have good efficacy for onsite test. Adsorption-extraction, ultrafiltration, ultracentrifugation, and precipitation are common techniques used for virus recovery. Among these techniques, ultrafiltration and precipitation techniques seem to be delivering reasonable virus recovery percentage, acceptable preparation time and portability. PCR based molecular techniques targeting the S-proteins, nucleocapsids, and ACE-2 receptors for virus detection are tedious and highly skill dependent. Alternatively, a combination of various detection techniques (optical, electrochemical, PCR), with functional nanomaterials, LAMP and low-cost microfluidics have great potential to be implemented as a rapid SARS-CoV-2 biosensor. However, the accuracy, specificity and sensitivity of these sensors to detect SARS-CoV-2 in a complex water system is still lacking and should be improved.
Author contribution
SAZ: Investigation, writing - original draft, CFS: conceptualization, methodology, funding acquisition, supervision, writing - reviewing & editing, NS: writing - original draft. ANN: writing - original draft. RAR: writing - original draft, MAK: formal analysis. GPL: data curation. KST: investigation, visualization, writing-review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by the Malaysia Ministry of Higher Education; Research Excellence Consortium Grant Scheme (KKP) or KPM-Special Grant RMK-10 with reference number of JPT(BPKI)1000/016/018/25(54) or KKP Vot. No. K343.
References
- 1.W.H. Organization Coronavirus Disease 2019 (COVID-19) Situation Report. 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---27-july-2021 (Accessed on 1st August 2021)
- 2.Eslami H., Jalili M. The role of environmental factors to transmission of SARS-CoV-2 (COVID-19) AMB Express. 2020;10(1) doi: 10.1186/s13568-020-01028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiktorczyk-Kapischke N., Grudlewska-Buda K., Wałecka-Zacharska E., Kwiecińska-Piróg J., Radtke L., Gospodarek-Komkowska E., Skowron K. SARS-CoV-2 in the environment—non-droplet spreading routes. Sci. Total Environ. 2021;770:85–94. doi: 10.1016/j.scitotenv.2021.145260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al Huraimel K., Alhosani M., Kunhabdulla S., Stietiya M.H. SARS-CoV-2 in the environment: modes of transmission, early detection and potential role of pollutions. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140946. (140946-140946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. (115899-115899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. (139076-139076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. (141326-141326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohapatra S., Menon N.G., Mohapatra G., Pisharody L., Pattnaik A., Menon N.G., Bhukya P.L., Srivastava M., et al. The novel SARS-CoV-2 pandemic: Possible environmental transmission, detection, persistence and fate during wastewater and water treatment. Sci. Total Environ. 2021;765 doi: 10.1016/j.scitotenv.2020.142746. (142746-142746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul D., Kolar P., Hall S.G. A review of the impact of environmental factors on the fate and transport of coronaviruses in aqueous environments. npj Clean Water. 2021;4(1) [Google Scholar]
- 10.Bertrand I., Challant J., Jeulin H., Hartard C., Mathieu L., Lopez S., Schvoerer E., Courtois S., et al. Epidemiological surveillance of SARS-CoV-2 by genome quantification in wastewater applied to a city in the northeast of France: comparison of ultrafiltration- and protein precipitation-based methods. Int. J. Hyg. Environ. Health. 2021;233(January) doi: 10.1016/j.ijheh.2021.113692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguiar-Oliveira M.D.L., Campos A., Matos A.R., Rigotto C., Sotero-Martins A., Teixeira P.F.P., Siqueira M.M. Wastewater-based epidemiology (Wbe) and viral detection in polluted surface water: a valuable tool for covid-19 surveillance—a brief review. Int. J. Env. Res. Public Health. 2020;17(24):1–19. doi: 10.3390/ijerph17249251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 13.Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., et al. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140445. (140445-140445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albastaki A., Naji M., Lootah R., Almeheiri R., Almulla H., Almarri I., Alreyami A., Aden A., et al. First confirmed detection of SARS-COV-2 in untreated municipal and aircraft wastewater in Dubai, UAE: the use of wastewater based epidemiology as an early warning tool to monitor the prevalence of COVID-19. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.143350. (143350-143350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pilevar M., Kim K.T., Lee W.H. Recent advances in biosensors for detecting viruses in water and wastewater. J. Hazard. Mater. 2021;410 doi: 10.1016/j.jhazmat.2020.124656. (124656-124656) [DOI] [PubMed] [Google Scholar]
- 16.Rački N., Dreo T., Gutierrez-Aguirre I., Blejec A., Ravnikar M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods. 2014;10(1):42. doi: 10.1186/s13007-014-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrie B., Youdan J., Barden R., Kasprzyk-Hordern B. New framework to diagnose the direct disposal of prescribed drugs in wastewater - a case study of the antidepressant fluoxetine. Environ. Sci. Technol. Lett. 2016;50(7):3781–3789. doi: 10.1021/acs.est.6b00291. [DOI] [PubMed] [Google Scholar]
- 18.Broughton J.P., Deng X., Yu G. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin X., Huang X., Urmann K., Xie X., Hoffmann M.R. Digital loop-mediated isothermal amplification on a commercial membrane. ACS Sensors. 2019;4(1):242–249. doi: 10.1021/acssensors.8b01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittal A., Manjunath K., Ranjan R.K., Kaushik S., Kumar S., Verma V. COVID-19 pandemic: insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Path. 2020;16(8) doi: 10.1371/journal.ppat.1008762. (e1008762-e1008762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boopathi S., Poma A.B., Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2020;39(9):3409–3418. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whelan J., Editors M.W.M., Walker J.M. 2015. Mitochondria IN Series Editor. [Google Scholar]
- 26.Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panchal D., Prakash O., Bobde P., Pal S. SARS-CoV-2: sewage surveillance as an early warning system and challenges in developing countries. Environ. Sci. Pollut. Res. 2021;28(18):22221–22240. doi: 10.1007/s11356-021-13170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt A., Weber O., Wolff M.H. Birkhäuser Basel; Birkhäuser, Basel: 2005. Coronaviruses with special emphasis on first insights concerning SARS. [Google Scholar]
- 30.Liu Y.C., Kuo R.L., Shih S.R. COVID-19: The first documented coronavirus pandemic in history. Biomed. J. 2020;43(4):328–333. doi: 10.1016/j.bj.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chattopadhyay S., Taft S. U.S. Environmental Protection Agency; 2018. Exposure Pathways to High-consequence Pathogens in the Wastewater Collection and Treatment Systems. August. (96-96) [Google Scholar]
- 32.Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105730. (105730-105730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Audi A., AlIbrahim M., Kaddoura M., Hijazi G., Yassine H.M., Zaraket H. Seasonality of respiratory viral infections: will COVID-19 follow suit? Front. Public Health. 2020;8 doi: 10.3389/fpubh.2020.567184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020;104(3):246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao L., Zhu W., Shi J., Xu T., Qu G., Zhou W., Yu X.F., Zhang X., et al. Detection of coronavirus in environmental surveillance and risk monitoring for pandemic control. Chem. Soc. Rev. 2021;50(6):3656–3676. doi: 10.1039/d0cs00595a. [DOI] [PubMed] [Google Scholar]
- 36.Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5(4):335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao H., Yao H., Yang S., Li L. From SARS to MERS: evidence and speculation. Front. Med. 2016;10(4):377–382. doi: 10.1007/s11684-016-0466-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan J.F.W., Lau S.K.P., To K.K.W., Cheng V.C.C., Woo P.C.Y., Yue K.Y. Middle East Respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watts J. Report details lessons from SARS outbreak. Lancet. 2003;362(9391) doi: 10.1016/S0140-6736(03)14561-8. (1207-1207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu I.T.S., Li Y., Wong T.W., Tam W., Chan A.T., Lee J.H.W., Leung D.Y.C., Ho T. Evidence of airborne transmission of the wevere acute respiratory syndrome virus. N. Engl. J. Med. 2004;350(17):1731–1739. doi: 10.1056/NEJMoa032867. [DOI] [PubMed] [Google Scholar]
- 41.Oidtmann B., Dixon P., Way K., Joiner C., Bayley A.E. Risk of waterborne virus spread – review of survival of relevant fish and crustacean viruses in the aquatic environment and implications for control measures. Rev. Aquac. 2018;10(3):641–669. [Google Scholar]
- 42.Bandala E.R., Kruger B.R., Cesarino I., Leao A.L., Wijesiri B., Goonetilleke A. Impacts of COVID-19 pandemic on the wastewater pathway into surface water: a review. Sci. Total Environ. 2021;774:45586. doi: 10.1016/j.scitotenv.2021.145586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adelodun B., Ajibade F.O., Ibrahim R.G., Bakare H.O., Choi K.S. Snowballing transmission of COVID-19 (SARS-CoV-2) through wastewater: any sustainable preventive measures to curtail the scourge in low-income countries? Sci. Total Environ. 2020;742 doi: 10.1016/j.scitotenv.2020.140680. (140680-140680) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nomoto H., Ishikane M., Katagiri D., Kinoshita N., Nagashima M., Sadamasu K., Yoshimura K., Ohmagari N. Cautious handling of urine from moderate to severe COVID-19 patients. Am. J. Infect. Control. 2020;48(8):969–971. doi: 10.1016/j.ajic.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nabi G., Khan S. Risk of COVID-19 pneumonia in aquatic mammals. Environ. Res. 2020;188 doi: 10.1016/j.envres.2020.109732. (109732-109732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gandhi M., Yokoe D.S., Havlir D.V. The COVID-19 Reader. 2020. Asymptomatic transmission, the Achilles’ heel of current strategies to control COVID-19; pp. 36–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. (140444-140444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amoah I.D., Kumari S., Bux F. Coronaviruses in wastewater processes: source, fate and potential risks. Environ. Int. 2020;143:105962. doi: 10.1016/j.envint.2020.105962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. (138764-138764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., et al. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55(1):488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- 51.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leung W.K., To K.F., Chan P.K.S. Enteric involvement of severe acute respiratory syndrome. Gastroenterology. 2003;5085(03):1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. (e3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X., Zheng J., Guo L., Yao H., Wang L., Xia X.D., Zhang W. Fecal viral shedding in COVID-19 patients: clinical significance, viral load dynamics and survival analysis. Virus Res. 2020;289(August) doi: 10.1016/j.virusres.2020.198147. (198147-198147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiehao C., Jin X., Daojiong L., Zhi Y., Lei X., Zhenghai Q., Yuehua Z., Hua Z., et al. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020;71(6):1547–1551. doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morone G., Palomba A., Iosa M., Caporaso T., De Angelis D., Venturiero V., Savo A., Coiro P., et al. Incidence and persistence of viral shedding in COVID-19 post-acute patients with negativized pharyngeal swab: a systematic review. Front. Med. 2020;7(562) doi: 10.3389/fmed.2020.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92(7):833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 59.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 60.Jeong H.W., Kim S.M., Kim H.S., Kim Y.I., Kim J.H., Cho J.Y., Kim S.H., Kang H., et al. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020;26(11):1520–1524. doi: 10.1016/j.cmi.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frithiof R., Bergqvist A., Järhult J.D., Lipcsey M., Hultström M. Presence of SARS-CoV-2 in urine is rare and not associated with acute kidney injury in critically ill COVID-19 patients. Crit. Care. 2020;24(1):4–6. doi: 10.1186/s13054-020-03302-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tran H.N., Le G.T., Nguyen D.T., Juang R.S., Rinklebe J., Bhatnagar A., Lima E.C., Iqbal H.M.N., et al. SARS-CoV-2 coronavirus in water and wastewater: a critical review about presence and concern. Environ. Res. 2021;193 doi: 10.1016/j.envres.2020.110265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hjelmsø M.H., Hellmér M., Fernandez-Cassi X., Timoneda N., Lukjancenko O., Seidel M., Elsässer D., Aarestrup F.M., et al. Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS One. 2017;12(1):1–17. doi: 10.1371/journal.pone.0170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xagoraraki I., O'Brien E. 2020. Women in Water Quality. [Google Scholar]
- 65.Naddeo V., Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. Water Res. 2020;6(5):1213–1216. [Google Scholar]
- 66.Buonerba A., Corpuz M.V.A., Ballesteros F., Choo K.H., Hasan S.W., Korshin G.V., Belgiorno V., Barceló D., et al. Coronavirus in water media: analysis, fate, disinfection and epidemiological applications. J. Hazard. Mater. 2021;415 doi: 10.1016/j.jhazmat.2021.125580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bosch A., Guix S., Sano D., Pintó R.M. New tools for the study and direct surveillance of viral pathogens in water. Curr. Opin. Biotechnol. 2008;19(3):295–301. doi: 10.1016/j.copbio.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.La Rosa G., Fratini M., Libera S. Della, Iaconelli M., Muscillo M. Emerging and potentially emerging viruses in water environments. Ann. Ist. Super. Sanita. 2012;48(4):397–406. doi: 10.4415/ANN_12_04_07. [DOI] [PubMed] [Google Scholar]
- 69.Carducci A., Federigi I., Dasheng L., Julian T., Marco R.V. Making waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115907. (115907-115907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Corpuz M.V.A., Buonerba A., Vigliotta G., Zarra T., Ballesteros F., Campiglia P., Belgiorno V., Korshin G., et al. Viruses in wastewater: occurrence, abundance and detection methods. Sci. Total Environ. 2020;745 doi: 10.1016/j.scitotenv.2020.140910. (140910-140910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rusiñol M., Martínez-Puchol S., Forés E., Itarte M., Girones R., Bofill-Mas S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Health. 2020;17:21–28. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 73.Prado T., de Castro Bruni A., Barbosa M.R.F., Garcia S.C., de Jesus Melo A.M., Sato M.I.Z. Performance of wastewater reclamation systems in enteric virus removal. Sci. Total Environ. 2019;678:33–42. doi: 10.1016/j.scitotenv.2019.04.435. [DOI] [PubMed] [Google Scholar]
- 74.Falman J.C., Fagnant-Sperati C.S., Kossik A.L., Boyle D.S., Meschke J.S. Evaluation of secondary concentration methods for poliovirus detection in wastewater. Food Environ. Virol. 2019;11(1):20–31. doi: 10.1007/s12560-018-09364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bofill-mas S., Rusiñol M. Recent trends on methods for the concentration of viruses from water samples. Curr. Opin. Environ. Sci. Health. 2020;16:7–13. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. (144216-144216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pepper I., Gerba C., Gentry T. In: Environmental Microbiology. Third edition. Pepper I.L., Gerba C.P., Gentry T.J., editors. Academic Press; San Diego: 2015. Preface; p. xvii. [Google Scholar]
- 79.Xagoraraki I., Yin Z., Svambayev Z. Fate of viruses in water systems. J. Environ. Eng. 2014;140(7) (04014020-04014020) [Google Scholar]
- 80.Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., Insaf T., Kilaru P., et al. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X. 2021;11:100100. doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Forés E., Bofill-Mas S., Itarte M., Martínez-Puchol S., Hundesa A., Calvo M., Borrego C.M., Corominas L.L., et al. Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater. Sci. Total Environ. 2021;768:144786. doi: 10.1016/j.scitotenv.2020.144786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dumke R., Barron M.D.L.C., Oertel R., Helm B., Kallies R., Berendonk T.U., Dalpke A. Evaluation of two methods to concentrate SARS-CoV-2 from untreatedwastewater. Pathogens. 2021;10(2):1–7. doi: 10.3390/pathogens10020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hokajärvi A.M., Rytkönen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.M., Kankaanpää A., Gunnar T., et al. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021;770:145274. doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739:139960. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pérez-Cataluña A., Cuevas-Ferrando E., Randazzo W., Falcó I., Allende A., Sánchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758:143870. doi: 10.1016/j.scitotenv.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Atha K.C., Ingham K.C. Analysis in Terms of Excluded Volume. 1981. Mechanism of precipitation of proteins by polyethylene glycols; pp. 12108–12117. 256 23. [PubMed] [Google Scholar]
- 87.Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143578. (143578-143578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Torii S., Furumai H., Katayama H. Science of the Total Environment Applicability of polyethylene glycol precipitation followed by acid guanidinium thiocyanate-phenol-chloroform extraction for the detection of SARS-CoV-2 RNA from municipal wastewater. Sci. Total Environ. 2021;756 doi: 10.1016/j.scitotenv.2020.143067. (143067-143067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.W. World Health Organization . 2020. COVID-19 Coronavirus Real Time PCR Kit For In Vitro Diagnostic Use Only. (28-28) [Google Scholar]
- 90.Matsui Y., Matsushita T., Sakuma S., Gojo T., Mamiya T., Suzuoki H., Inoue T. Virus inactivation in aluminum and polyaluminum coagulation. Environ. Sci. Technol. 2003;37(22):5175–5180. doi: 10.1021/es0343003. [DOI] [PubMed] [Google Scholar]
- 91.Pérez-Cataluña A., Cuevas-Ferrando E., Randazzo W., Falcó I., Allende A., Sánchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758:143870. doi: 10.1016/j.scitotenv.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tanhaei M., Mohebbi S.R., Hosseini S.M., Rafieepoor M., Kazemian S., Ghaemi A., Shamloei S., Mirjalali H., et al. The first detection of SARS-CoV-2 RNA in the wastewater of Tehran, Iran. Environ. Sci. Pollut. Res. 2021;28(29):38629–38636. doi: 10.1007/s11356-021-13393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. (140621-140621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barril P.A., Pianciola L.A., Mazzeo M., Ousset M.J., Jaureguiberry M.V., Alessandrello M., Sánchez G., Oteiza J.M. Evaluation of viral concentration methods for SARS-CoV-2 recovery from wastewaters. Sci. Total Environ. 2021;756:144105. doi: 10.1016/j.scitotenv.2020.144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hematian A., Sadeghifard N., Mohebi R. Traditional and modern cell culture in virus diagnosis. Osong Public Health Res. Perspect. 2016;7(2):77–82. doi: 10.1016/j.phrp.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Giacobbo A., Rodrigues M.A.S., Zoppas Ferreira J., Bernardes A.M., de Pinho M.N. A critical review on SARS-CoV-2 infectivity in water and wastewater. What do we know? Sci. Total Environ. 2021;774 doi: 10.1016/j.scitotenv.2021.145721. (145721-145721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1(1):10–14. [Google Scholar]
- 98.Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(12):937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ibrahim Y., Ouda M., Kadadou D., Banat F., Naddeo V., Alsafar H., Yousef A.F., Barceló D., et al. Detection and removal of waterborne enteric viruses from wastewater: a comprehensive review. J. Environ. Chem. Eng. 2021;9(4):105613. [Google Scholar]
- 100.Salvo M., Moller A., Alvareda E., Gamazo P., Colina R., Victoria M. Evaluation of low-cost viral concentration methods in wastewaters: implications for SARS-CoV-2 pandemic surveillances. J. Virol. Methods. 2021;297 doi: 10.1016/j.jviromet.2021.114249. (114249-114249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fumian T.M., Leite J.P., Castello A.A., Gaggero A., Caillou M.S., Miagostovich M.P. Detection of rotavirus A in sewage samples using multiplex qPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J. Virol. Methods. 2010;170(1-2):42–46. doi: 10.1016/j.jviromet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 102.Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., et al. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. (116296-116296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739(139960):1–9. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gantzer C., Senouci S., Maul A., Levi Y., Schwartzbrod L. Enterovirus genomes in wastewater: concentration on glass wool and glass powder and detection by RT-PCR. J. Virol. Methods. 1997;65(2):265–271. doi: 10.1016/s0166-0934(97)02193-9. [DOI] [PubMed] [Google Scholar]
- 105.Egger D., Pasamontes L., Ostermayer M., Bienz K. Reverse transcription multiplex PCR for differentiation between polio- and enteroviruses from clinical and environmental samples. J. Clin. Microbiol. 1995;33(6):1442–1447. doi: 10.1128/jcm.33.6.1442-1447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lahrich S., Laghrib F., Farahi A., Bakasse M., Saqrane S., El Mhammedi M.A. Review on the contamination of wastewater by COVID-19 virus: impact and treatment. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.142325. (142325-142325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors – occurrence, properties and removal. J. Appl. Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 108.Corman V., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., et al. Detection of 2019-nCoV by RT-PCR. Euro. Surveill. 2020;25(3):1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., et al. Development of genetic diagnostic methods for detection for novel coronavirus 2019(nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020;73(4):304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 110.Nemudryi A., Nemudraia A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R. Vol. 3. 2020. Temporal Detection and Phylogenetic Assessment of Sars-Cov-2 in Municipal Wastewater; pp. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., et al. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2021;27(5):1–11. doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., et al. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751:141750. doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jung Y., Park G.S., Moon J.H., Ku K., Beak S.H., Kim S., Park E.C., Park D., et al. Comparative Analysis of Primer-Probe Sets for RT-qPCR of COVID-19 Causative Virus (SARS-CoV-2) ACS Infect Dis. 2020;6(9):2513–2523. doi: 10.1021/acsinfecdis.0c00464. [DOI] [PubMed] [Google Scholar]
- 116.Wu F., Zhang J., Xiao A., Gu X., Lee L., Armas F., Kauffman K. Crossm SARS-CoV-2 titers in wastewater are higher than expected. mSystems. 2020;5(4):1–9. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. (139652-139652) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Yang Y., Zhang Q., et al. ddPCR : a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020;9(1):1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.J. Kuypers, K.R. Jerome, Applications of Digital PCR for Clinical Microbiology, (1098-660X (Electronic)). [DOI] [PMC free article] [PubMed]
- 120.Deiana M., Mori A., Piubelli C., Scarso S., Favarato M., Pomari E. Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR. Sci. Rep. 2020;10(1):18764. doi: 10.1038/s41598-020-75958-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.D'Aoust P.M., Mercier E., Montpetit D., Jia J.J., Alexandrov I., Neault N., Baig A.T., Mayne J., et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188:116560. doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reynolds K.A., Gerba C.P., Pepper I.L. Detection of infectious enteroviruses by an integrated cell culture-PCR procedure. Appl. Environ. Microbiol. 1996;62(4):1424–1427. doi: 10.1128/aem.62.4.1424-1427.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mehrotra P. Biosensors and their applications-a review. J. Oral Biol. Craniofac. Res. 2016;6:153–159. doi: 10.1016/j.jobcr.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Su L., Jia W., Hou C., Lei Y. Microbial biosensors: a review. Biosens. Bioelectron. 2011;26:1788–1799. doi: 10.1016/j.bios.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 125.Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020;54(7):3733–3735. doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- 126.Yang Z., Kasprzyk-Hordern B., Frost C.G., Estrela P., Thomas K.V. Community sewage sensors for monitoring public health. Environ. Sci. Technol. 2015;49(10):5845–5846. doi: 10.1021/acs.est.5b01434. [DOI] [PubMed] [Google Scholar]
- 127.Guo B., Wen B., Cheng W., Zhou X., Duan X., Zhao M., Xia Q., Ding S. An enzyme-free and label-free surface plasmon resonance biosensor for ultrasensitive detection of fusion gene based on DNA self-assembly hydrogel with streptavidin encapsulation. Biosens. Bioelectron. 2018;112:120–126. doi: 10.1016/j.bios.2018.04.027. [DOI] [PubMed] [Google Scholar]
- 128.Qiu G., Ng S.P., Wu C.-M.L. Bimetallic Au-Ag alloy nanoislands for highly sensitive localized surface plasmon resonance biosensing. Sens. Actuat. B. 2018;265:459–467. [Google Scholar]
- 129.Daghestani H.N., Day B.W. Theory and applications of surface plasmon resonance, resonant mirror, resonant waveguide grating, and dual polarization interferometry biosensors. Sensors. 2010;10(11):9630–9646. doi: 10.3390/s101109630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kassal P., Steinberg M.D., Steinberg I.M. Wireless chemical sensors and biosensors: a review. Sens. Actuat. B. 2018;266:228–245. [Google Scholar]
- 131.Jyoti A., Tomar R.S., Shanker R. Nanosensors for the detection of pathogenic bacteria. Nanosci. Food Agric. 2016;1:129–150. [Google Scholar]
- 132.Ejeian F., Etedali P., Mansouri-Tehrani H.A., Soozanipour A., Low Z.X., Asadnia M., Taheri-Kafrani A., Razmjou A. Biosensors for wastewater monitoring: a review. Biosens. Bioelectron. 2018;118:66–79. doi: 10.1016/j.bios.2018.07.019. [DOI] [PubMed] [Google Scholar]
- 133.Kumar M.S., Nandeshwar R., Lad S.B., Megha K., Mangat M., Butterworth A., Knapp C.W., Knapp M., Hoskisson P.A., Corrigan D.K., Ward A.C., Kondabagil K., Tallur S. Electrochemical sensing of SARS-CoV-2 amplicons with PCB electrodes. Sens. Actuat. B. 2021;343:130169. [Google Scholar]
- 134.Lisdat F., Schäfer D. The use of electrochemical impedance spectroscopy for biosensing. Anal. Bioanal. Chem. 2008;391:1555. doi: 10.1007/s00216-008-1970-7. [DOI] [PubMed] [Google Scholar]
- 135.Guo T., González-Vila Á., Loyez M., Caucheteur C. Plasmonic optical fiber-grating immunosensing: a review. Sensors (Basel) 2017;17(12):2732. doi: 10.3390/s17122732. (PMID: 29186871; PMCID: PMC5751598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Songkhla S.N., Nakamoto T. Overview of quartz crystal microbalance behavior analysis and measurement. Chemosensors. 2021;9:350. doi: 10.3390/chemosensors9120350. [DOI] [Google Scholar]
- 137.Chee C.Y.K., Tong L., Steven G.P. A review on the modelling of piezoelectric sensors and actuators incorporated in intelligent structures. J. Intell. Mater. Syst. Struct. 1998;9(1):3–19. [Google Scholar]
- 138.Mohsen P., Kim K.T., Lee W.H. Recent advances in biosensors for detecting viruses in water and wastewater. J. Hazard. Mater. 2021;410:124656. doi: 10.1016/j.jhazmat.2020.124656. [DOI] [PubMed] [Google Scholar]
- 139.König B., Grätzel M. Detection of viruses and bacteria with piezoelectric immunosensors. Anal. Lett. 1993;26:1567–1585. [Google Scholar]
- 140.König B., Grätzel M. A piezoelectric immunosensor for hepatitis viruses. Anal. Chim. Acta. 1995;309:19–25. [Google Scholar]
- 141.Basiri A., Heidari A., Nadi M.F., Fallahy M.T.P., Nezamabadi S.S., Sedighi M., Saghazadeh A., Rezaei N. Microfluidic devices for detection of RNA viruses. Rev. Med. Virol. 2020:e2154. doi: 10.1002/rmv.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tayyab M., Sami M.A., Raji H., Mushnoori S., Javanmard M. Potential microfluidic devices for COVID-19 antibody detection at point-of-care (POC): a review. IEEE Sens. J. 2021;21(4):4007–4017. doi: 10.1109/JSEN.2020.3034892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Saez J., Catalan-Carrio R., Owens R.M., Desmonts L.D., Lopez F.B. Microfluidics and materials for smart water monitoring: a review. Anal. Chim. Acta. 2021:338392. doi: 10.1016/j.aca.2021.338392. [DOI] [PubMed] [Google Scholar]
- 144.Manzano M., Viezzi S., Mazerat S., Marks R.S., Vidic J. Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron. 2018;100:89–95. doi: 10.1016/j.bios.2017.08.043. [DOI] [PubMed] [Google Scholar]
- 145.Tang D., Tang J., Su B., Ren J., Chen G. Simultaneous determination of five-type hepatitis virus antigens in 5 min using an integrated automatic electrochemical immunosensor array. Biosens. Bioelectron. 2010;25:1658–1662. doi: 10.1016/j.bios.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 146.Liu F., Choi K.S., Park T.J., Lee S.Y., Seo T.S. Graphene-based electrochemical biosensor for pathogenic virus detection. Biochip J. 2011;5:123–128. [Google Scholar]
- 147.Yildirim N., Li D., Long F., Gu A.Z. Direct detection of adenovirus in environmental waste waters by portable optical fiber sensor platform. Sensors. 2013:1–5. [Google Scholar]
- 148.Prabowo B.A., Wang R.Y., Secario M.K., Ou P.-T., Alom A., Liu J.-J., Liu K.-C. Rapid detection and quantification of Enterovirus 71 by a portable surface plasmon resonance biosensor. Biosens. Bioelectron. 2017;92:186–191. doi: 10.1016/j.bios.2017.01.043. [DOI] [PubMed] [Google Scholar]
- 149.Kim M.W., Park H.-J., Park C.Y., Kim J.H., Cho C.H., Park J.P., Kailasa S.K., Lee C.-H., Park T.J. Fabrication of a paper strip for facile and rapid detection of bovine viral diarrhea virus via signal enhancement by copper polyhedral nanoshells. RSC Adv. 2020;10:29759–29764. doi: 10.1039/d0ra03677c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu Q., Zhang X., Yao Y., Jing W., Liu S., Sui G. A novel microfluidic module for rapid detection of airborne and waterborne pathogens. Sensor. Actuator. B Chem. 2018;258:1138–1145. [Google Scholar]
- 151.Balasubramanian A.K., Soni K.A., Beskok A., Pillai S.D. A microfluidic device for continuous capture and concentration of microorganisms from potable water. Lab. Chip. 2007;7:1315–1321. doi: 10.1039/b706559k. [DOI] [PubMed] [Google Scholar]
- 152.Chung S., Breshears L.E., Perea S., Morrison C.M., Betancourt W.Q., Reynolds K.A., Yoon J.-Y. Smartphone-based paper microfluidic particulometry of norovirus from environmental water samples at the single copy level. ACS Omega. 2019;4:11180–11188. doi: 10.1021/acsomega.9b00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Park T.S., Yoon J.Y. Smartphone detection of Escherichia coli from field water samples on paper microfluidics. IEEE Sens. J. 2015;15(3):1902–1907. [Google Scholar]
- 154.Reboud J., Xu G., Garrett A., Adriko M., Yang Z., Tukahebwa E.M., Rowell C., Cooper J.M. Paper-based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities. Proc. Natl. Acad. Sci. U. S. A. 2019;116(11):4834–4842. doi: 10.1073/pnas.1812296116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mao K., Yang Z., Zhang H., Li X., Cooper J.M. Paper-based nanosensors to evaluate community-wide illicit drug use for wastewater-based epidemiology. Water Res. 2021;189:116559. doi: 10.1016/j.watres.2020.116559. [DOI] [PubMed] [Google Scholar]
- 156.Piaskowski K., Świderska-Dąbrowska R., Kaleniecka A., Zarzycki P.K. Advances in the analysis of water and wastewater samples using various sensing protocols and microfluidic devices based on PAD and μTAS systems. J. AOAC Int. 2017;100(4):962–970. doi: 10.5740/jaoacint.17-0170. [DOI] [PubMed] [Google Scholar]
- 157.Mao K., Zhang H., Pan Y., Yang Z. Biosensors for wastewater-based epidemiology for monitoring public health. Water Res. 2021;191:116787. doi: 10.1016/j.watres.2020.116787. [DOI] [PubMed] [Google Scholar]
- 158.What are Biosensors? How do Biosensors Work and What are Their Applications in the Water Sector? Aug 2021. https://www.envirotech-online.com/article/water-wastewater/9/swig/what-are-biosensors-how-do-biosensors-work-and-what-are-their-applications-in-the-water-sector/2997 Accessed on 10th February 2022.
- 159.Mao K., Zhang H., Yang Z. The potential of an integrated biosensor system with mobile health and wastewater-based epidemiology (iBMW) for the prevention, surveillance, monitoring and intervention of the COVID-19 pandemic. Biosens. Bioelectron. 2020 doi: 10.1016/j.bios.2020.112617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rahman K.S.A., Wang W., Garg A., Maile-Moskowitz A., Vikesland P.J. Nanobiotechnology enabled approaches for wastewater based epidemiology. Trends Anal. Chem. 2021;143:116400. doi: 10.1016/j.trac.2021.116400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xiao A., Wu F., Bushman M., Zhang J., Imakaev M., Chai P.R., Duvallet C., Endo N., Erickson T.B., Armas F., Arnold B., Chen H., Chandra F., Ghaeli N., Gu X., Hanage W.P., Lee W.L., Matus M., McElroy K.A., Moniz K., Rhode S.F., Thompson J., Alm E.J. Metrics to relate COVID-19 wastewater data to clinical testing dynamics. Water Res. 2022;212:118070. doi: 10.1016/j.watres.2022.118070. ISSN 0043-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Brown K.M.P., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779:146408. doi: 10.1016/j.scitotenv.2021.146408. ISSN 0048-9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu X., Feng J., Zhang Q., Guo D., Zhang L., Suo T., Hu W., Guo M., Wang X., Huang Z., Xiong Y., Chen G., Chen Y., Lan K. Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg. Microb. Infect. 2020;9(1):1175–1179. doi: 10.1080/22221751.2020.1772679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Parida M., Sannarangaiah S., Dash P.K., Rao P.V., Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008;18(6):407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J.…Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jayamohan H., Lambert C.J., Sant H.J., Jafek A., Patel D., Feng H.…Gale B.K. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal. Bioanal. Chem. 2021;413:49–71. doi: 10.1007/s00216-020-02958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Restaino S.M., White I.M. A critical review of flexible and porous SERS sensors for analytical chemistry at the point-of-sample. Anal. Chim. Acta. 2019;1060:17–29. doi: 10.1016/j.aca.2018.11.057. [DOI] [PubMed] [Google Scholar]
- 168.Langer J., Aizpurua J., Alvarez-Puebla R.A., Auguié B., Baumberg J.J., Bazan G.C.…Liz-Marzán L.M. Present and future of surface-enhanced Raman scattering. ACS Nano. 2020;14(1):28–117. doi: 10.1021/acsnano.9b04224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chandran G.T., Li X.W., Ogata A., Penner R.M. Electrically transduced sensors based on nanomaterials (2012-2016) Anal. Chem. 2017;89(1):249–275. doi: 10.1021/acs.analchem.6b04687. [DOI] [PubMed] [Google Scholar]
- 170.Zhang A., Lieber C.M. Nano-bioelectronics. Chem. Rev. 2016;116(1):215–257. doi: 10.1021/acs.chemrev.5b00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Murgida D.H. In situ spectroelectrochemical investigations of electrode-confined electron-transferring proteins and redox enzymes. ACS Omega. 2021;6(5):3435–3446. doi: 10.1021/acsomega.0c05746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Garoz-Ruiz J., Perales-Rondon J.V., Heras A., Colina A. Spectroelectrochemical sensing: current trends and challenges. Electroanalysis. 2019;31:1254. [Google Scholar]