Abstract
Class B G protein-coupled receptors (GPCRs) are notoriously difficult to target by small molecules because their large orthosteric peptide-binding pocket embedded deep within the transmembrane domain limits the identification and development of non-peptide small molecule ligands. Using the parathyroid hormone type 1 receptor (PTHR) as a prototypic class B GPCR target, and a combination of molecular dynamics simulations and elastic network model-based methods, we demonstrate that PTHR druggability can be effectively addressed. Here we found a key mechanical site that modulates the collective dynamics of the receptor and used this ensemble of PTHR conformers to identify selective small molecules with strong negative allosteric and biased properties for PTHR signaling in cell and PTH actions in vivo. This study provides a computational pipeline to detect precise druggable sites and identify allosteric modulators of PTHR signaling that could be extended to GPCRs to expedite discoveries of small molecules as novel therapeutic candidates.
Introduction
G protein-coupled receptors (GPCRs) are the most abundant cell membrane receptors in the human genome for extracellular (EC) stimuli such as hormones and neurotransmitters, and the targets of ~35% of currently approved clinical drugs. While they are essential drug discovery targets, only 18% of the total estimated pool of >1000 human GPCRs are targeted1, with ~94% of drugs acting through class A GPCRs (rhodopsin-like receptors)2. The initial identification of a lead compound as a drug candidate is a critical step in the drug discovery process. Current strategy for GPCR drug discovery relies on screening large libraries, either via High-Throughput Screening (HTS) of selected drug-like molecule libraries, or a recent structure-guided in silico screening of virtual compound libraries and/or docking simulations to identify the best (highest affinity) fit to the target protein structure. The latter has experienced a substantial advancement with the recent development of an ultra-large make-on demand drug-like compound library3 and the first successful example of using this new tool4. Nonetheless, both approaches hold two substantial caveats: 1) due to the astronomical numbers (~1023-1060) of possible drug-like molecules5, only a limited number of compounds are selected to build libraries for testing in HTS assays, therefore results depend on subjective library selection that may not lead to successful lead compound discovery, and 2) virtual screening requires not only the high-resolution structure of the target receptor but also knowledge of one or more druggable sites in line with the conformational flexibility of the target, including allosteric sites to be selectively targeted for modulating selective interactions. These caveats may at least partially be overcome for class A GPCRs because: 1) many receptors of this class bind the endogenous ligands that are small molecules, such as amino acid derivatives or lipids, allowing for the design of compound libraries for HTS by selecting drug-like molecules with structures similar to those of the endogenous ligands, and 2) the orthosteric ligand binding sites of class A GPCRs are mostly compact pockets, making them well-suited docking sites in virtual screening approach. This partially explains the aforementioned success of drug discovery for class A GPCRs.
Of large interest is the development of orally available small molecule drugs for class B GPCRs (secretin-like receptors that bind moderately sized peptide hormones) to treat numerous metabolic disorders including diabetes, bone and mineral ion disorders, cardiovascular disease, neurodegeneration, and psychiatric disorders. The distinguishing structural feature of class B GPCRs is a large EC domain (ECD), which captures the endogenous peptide hormone ligands by their C-terminal part, leading to the insertion of the N-terminal portion of the peptide hormone deep into transmembrane (TMD) domain’s (TMD) EC cavity and subsequent receptor activation (reviewed in ref.6). Such hormone binding mode results in a highly extended endogenous ligand recognition and binding region spanning the entire ECD and reaching all the way down to the mid-region of the TMD. Undeniably, the highly extended endogenous ligand binding region coupled to the difficulty in perturbing large protein-protein interfaces by small organic molecules, constitute a major hurdle in the process of discovering non-peptidic small molecule drug candidates for class B GPCRs. Currently, ubrogepant –an orally administered calcitonin gene-related peptide receptor antagonist for the treatment of acute migraine– is the only U.S. Food and Drug Administration (FDA)-approved small molecule drug for a class B GPCR. Two other small molecule drug candidates have successfully completed phase 2 clinical trials: TTP273, orally available glucagon-like peptide 1 receptor (GLP1R) agonist for the treatment of type 2 diabetes mellitus–, and Verucerfont, orally available corticotrophin-releasing factor-1 (CRF-1) receptor small molecule antagonist for the treatment of post-traumatic stress disorder or alcoholism. The vast majority of the rest of the approved drugs targeting class B GPCRs are either native or modified endogenous peptides. However, the peptidic nature of a drug has several disadvantages including costs and low patient’s adherence to the therapy as they require frequent injections due to oral inaccessibility and short plasma half-lives. Furthermore, overshoot or undershoot of peptide injections can cause unwanted side-effects. Therefore, the identification of small molecules may lead to discovery of allosteric modulators, which may present particular advantages of low cost, oral accessibility, and potential to avoid side-effects as they can alter and/or fine-tune receptor signaling induced by endogenous ligands.
Here we investigate the druggability of the parathyroid hormone (PTH) type 1 receptor (PTHR), a prototypical class B GPCR and an essential target for the treatment of bone and mineral ion disorders7,8. By employing computational approaches including elastic network model (ENM)-based methods9-11, druggability simulations in the presence of drug-building fragments, pharmacophore modeling and virtual screening with experimental approaches, we identified druggable sites located within the EC vestibule of PTHR. Further comprehensive cell-based signaling analysis of one of these sites uncovered small molecule allosteric modulators of PTHR that reduce and bias subcellular location and duration of cAMP signaling in cultured cells and reduce calciotropic activities in mice in response to PTH, and thus makes these compounds promising pharmacological tools for development of therapeutics against hyperparathyroidism.
Results
Essential sites with potential allosteric effect.
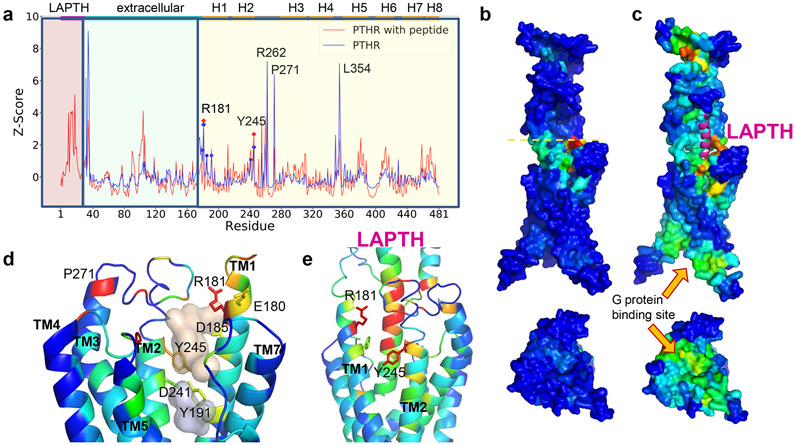
The recently resolved cryo-EM structure of PTHR (PDB id: 6NBF) in its fully activated state12 bound to a long-acting PTH (LA-PTH) and a dominant-negative Gs heterotrimer, reveal near-atomic details on intra- and intermolecular interaction networks that stabilize the signaling receptor conformation, and provides a structural framework for designing small molecule modulators of PTHR signaling. We used this cryo-EM PTHR structure in its ligand-removed form for modeling and simulations. We initially characterized the PTHR sites whose perturbation could elicit cooperative responses in the overall conformational dynamics of the PTHR, using a recently developed methodology, Essential Site Scanning Analysis (ESSA)11. ESSA identifies residues whose change in local density/packing, induced upon small molecule binding, exerts a global change in the conformational dynamics of the protein. As such, such sites are likely to serve as allosteric ligand/drug-binding sites, given their high potential to cooperatively alter the intrinsic dynamics of the target protein11. Here intrinsic dynamics is defined as the spectrum of modes of motion (i.e., thermal fluctuations) uniquely encoded by the protein fold under physiological conditions, analytically predicted by elastic network models (ENMs)13,14. ESSA probes the change in the frequency of the global/essential modes (i.e., the lowest frequency modes that usually underlie cooperative events or allosteric responses14 upon mimicking small molecule-binding near each residue, scanned over all residues. The peaks in figure 1a are such essential sites, color-coded diagrams in figure 1b and 1c. We distinguish residues R1811.33b, Y2452.72b and P271ECL1 (superscripts refer to Wootten’s class B GPCR nomenclature) in the EC-exposed vestibule lined by the transmembrane (TM) helices TM1 and TM2 (Fig. 1d) (prior to PTH binding). The same site also harbors two pockets detected by FPocket15, which further suggests that it might also be druggable (Fig. 1d and Extended Data Fig. 1). Notably, PTH- or LA-PTH-binding to this region (Fig. 1e) and ensuing dramatic change in the dispersion of essential sites (Fig. 1c) underscore the importance of ligand-binding to this site for eliciting an allosteric effect on distal sites, including the G protein binding epitope (see the cytoplasmic face in Fig. 1c bottom).
Figure 1. ESSA points to an extracellularly-exposed pocket as an essential site that can potentially alter the allosteric dynamics of PTHR upon ligand binding.
(a) Distribution of ESSA z-scores for PTHR active conformer in the presence (red curve) or absence (blue curve) of PTH. Peaks indicate the essential sites. Residue ranges of the PTH, EC domain and TM helices are indicated along the upper abscissa, also delimited by different shades in the graph. TM residues E180, R181, D185, Y191, D241 and Y245 on TM1 and TM2 exhibit z-score above the threshold value of 1.0 (without peptide) (see Methods). Among them, R181 and Y245 exhibit peaks in both structures. Other peaks correspond to loop regions. Thus, R181 and Y245 stand out as essential TM sites that can modulate the global dynamics.
(b–c) Same results as in a, illustrated by color-coded diagrams side (top) and cytoplasmic-facing (bottom) views. Panel b shows a global hinge site (yellow dashed line) that corresponds to the blue peaks in A. High-to-low scores are color-coded from red-to-blue. The peptide PTH is shown in magenta ribbon in C. A significant increase in sensitivity is observed at the G protein binding region in the presence of PTH.
(d) Two hydrophobic pockets (wheat and gray) determined by Fpocket, surrounded by five of the essential residues detected by ESSA (without the peptide). See also Extended Data Figure 1.
(e) Structural elements identified by ESSA to potentially alter the essential dynamics of the receptor, color coded by ESSA score from red (z-score > 3) to blue (z-score close to zero). The analysis was performed for PTH-bound PTHR structure in an active state (PDB id: 6nbf) after modeling the missing loops.
Druggability simulations and pharmacophore modeling.
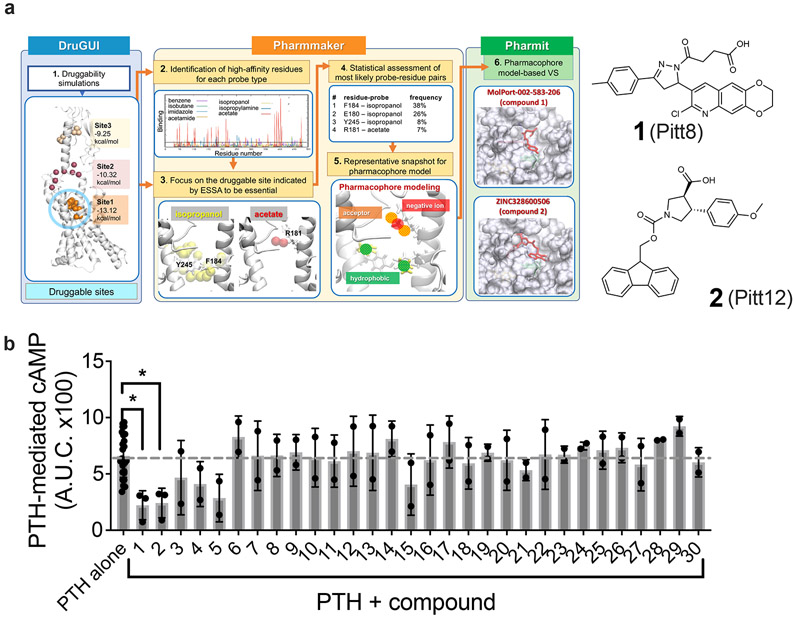
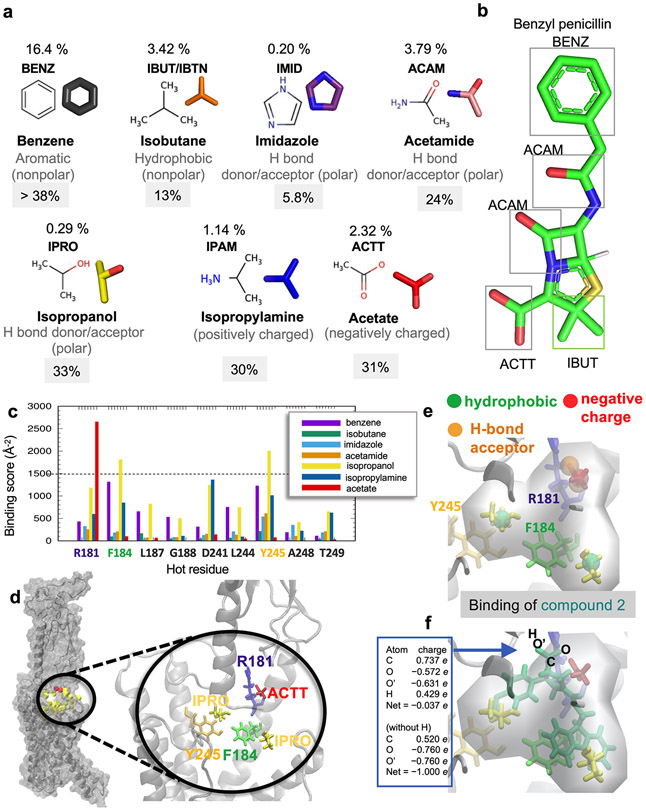
We next examined the druggability properties of PTHR and studied whether the essential sites identified above (Fig. 1d) were druggable. This was the Step 1 of a 6-step protocol we carried out in silico, schematically described in figure 2a. Druggability simulations are molecular dynamics (MD) simulations of the target protein in the presence of explicit water and probe molecules representative of drug-like fragments16. We used as probes benzene, isobutane, imidazole, acetamide, isopropanol, isopropylamine, and acetate –derived16 from statistical evaluation of chemical/functional groups most frequently observed in FDA-approved drugs such as penicillin (Extended Data Fig. 2a, b). Simulations revealed three druggable sites, Sites 1-3, distinguished by persistent and/or high concentration of probes (Fig. 2a left diagram). Site 1 corresponds to a wide-open pocket, which also includes the essential sites identified in figure 1d, mainly E1801.32b, R1811.33b, F1841.36b and Y2452.72b. We focused on that subsite within Site 1 as an essential site that is also druggable. This subsite exhibited the highest affinity for the probe molecules among all subsites in Site 1, and was therefore selected for pharmacophore modeling. Steps 2 and 3 in figure 2a (center diagrams) provide information on the type of probes that bind to Site 1, as well as an upper boundary for ligand-binding affinity at each site using our pharmacophore modeling tool Pharmmaker17. The propensity of binding different types of probes near Site 1 residues is further shown by color-coded bars in Extended Data figure 2c. A closeup view of the interaction between the most stable probes (one acetate and two isopropanol molecules) and amino acids R1811.33b, F1841.36b and Y2452.72b is presented in Extended Data figure 2d. We note that the subspace occupied by probe molecules in this selected subsite does not overlap with the space otherwise occupied by the PTH, so as to allow for possible binding of the small molecule to the PTH-bound or -unbound forms of PTHR. Steps 4 and 5 in figure 2a describe the construction of a pharmacophore model (PM) accounting for these interactions. To this end, residue-probe interactions are ranked based on their frequency of occurrence within Site 1, and MD snapshots that simultaneously exhibit multiple frequently-observed (entropically favored) interactions are used to construct the PMs. A snapshot showing the interaction between the most stable probes (one acetate and two isopropanols) and amino acids R1811.33b, F1841.36b and Y2452.72b is a corresponding PM are presented in Extended Data Fig. 2d-e.
Figure 2. Identification of druggable sites and drug-like small molecules targeting PTHR.
(a) Summary of a 6-step computational protocol toward identification of small molecule modulators of PTHR activity. The pipeline comprises three major components: druggability simulations using DruGUI (left; blue box); detection of a high-affinity essential site and pharmacophore modeling using Pharmmaker (middle; yellow box), and virtual screening (VS) of one or more pharmacophore models (PM) against libraries of small molecules using Pharmit (right, green box). See text for details. The figure illustrates how two hits, compounds 1 and 2 (thereafter designated as Pitt8 and Pitt12, shown on right) were derived using the PM deduced from DruGUI.
(b) Effect of computationally identified small molecules on PTH-induced cAMP production in HEK293 cells stably expressing the recombinant human PTHR. The bars graph represents the area under the curve (AUC) of cAMP time-courses (shown in Supplementary Figure 1) with 1 nM PTH and with or without 10 μM of compound (see Supplementary Tables 1 and 2). The dashed grey line depicts the average level of PTH alone induced cAMP and is shown for guidance. Error bars represent the mean values ± s.d. of N = 3 independent experiments for Pitt8 and Pitt12 (compounds 1 and 2), N = 2 for the rest compounds, and N = 25 for PTH alone. Only Pitt8 and Pitt12 significantly reduced PTH-induced cAMP generation (P values were determined by one-way ANOVA multiple comparisons test, and are 0.0268 for Pitt8, and 0.017 for Pitt12.
Virtual screening of library of small molecules.
In Step 6 we screened the PM against two libraries of small molecules, ZINC 18 and MolPort, using the Pharmit server 19. This procedure yielded an ensemble of compounds, 30 of which were selected as top-scoring hits (compounds 1-30, Supplementary Tables 1 and 2). Two hits, compounds 1 and 2 (hereafter renamed as Pitt8 and Pitt12, respectively), are shown in figure 2a (right diagrams). The computed binding properties of these compounds were further examined by docking simulations and energy minimization. Extended Data figure 2f illustrates the superposition of Pitt12 (ZINC328600506/MolPort 039-313-655) onto the PM. This shows how Pitt12 inserts into the Site 1 identified by both ESSA and druggability simulations, which suggests that it can potentially serve as an allosteric drug candidate, to be investigated next experimentally.
Selection of lead compounds.
We performed an initial bioactivity screening of the 30 top-ranking hit compounds identified (Supplementary Table 1) by the above computational protocol to assess their capacity to modulate the time courses of PTH-induced cAMP production by PTHR stably expressed in HEK-293 cells (Supplementary Fig. 1). Two of these molecules, Pitt8 and Pitt12, displayed a significant potential to elicit negative effects when co-applied with PTH (Fig. 2b). Notably, none of these molecules at concentration up to 10 μM exerted any effect on cAMP generation in the absence (Supplementary Fig. 2a, b) or before the addition of PTH (Supplementary Fig. 1) when cAMP was measured using either the multi-plate Glo-sensor or single cell FRET-based assays However, we noticed that compound concentrations that exceed 10 μM interfered with the multi-plate Glo-sensor cAMP assay before the addition of PTH and were not used in this study.
We further examined receptor selectivity by testing the effect of Pitt8 and Pitt12 on cAMP production mediated by agonists for the β2-adrenergic receptor (β2AR) and the vasopressin type 2 receptor (V2R). These receptors were chosen for the following reasons: the β2AR expresses in many cell types including those expressing the PTHR, and can increase both PTHR signaling in cells by increasing the duration of cAMP production20, and PTHR function in vivo by increasing bone formation in mice in response to intermittent injection of PTH21; the V2R shares signaling mechanisms with the PTHR as they are both involved in the regulation of endosomal cAMP production by β-arrestin, G protein subunits Gβγ, and the retromer complex22 (Supplementary Fig. 3a). Co-application of Pitt8 or Pitt12 had no effect on β2AR-mediated cAMP production, and the relative selectivity of these compounds was further supported by their lack of effect on vasopressin-mediated cAMP (Supplementary Fig. 3b).
Next, we found that Pitt8 and Pitt12 compounds had similar negative modulation on cAMP responses mediated by the PTH-related peptide (PTHrP1-36), the N-terminal fragment of other endogenous PTHR agonist (Supplementary Fig. 3c). Notably, the modulation of PTH-induced cAMP generation by Pitt8 and Pitt12 was not an artifact of recombinant PTHR overexpression, as inhibitory effects were obtained for these compounds in rat primary osteoblasts and human RPTEC cell lines expressing native PTHR (Supplementary Fig. 4a, b); however, the lack of effect on the PTH-induced peak response of cAMP suggests possible differences between recombinant and native cell systems that might be due to PTHR genus (homo vs rattus) or presence or absence of proteins affecting PTHR signaling.
Pharmacological characterization of selected compounds.
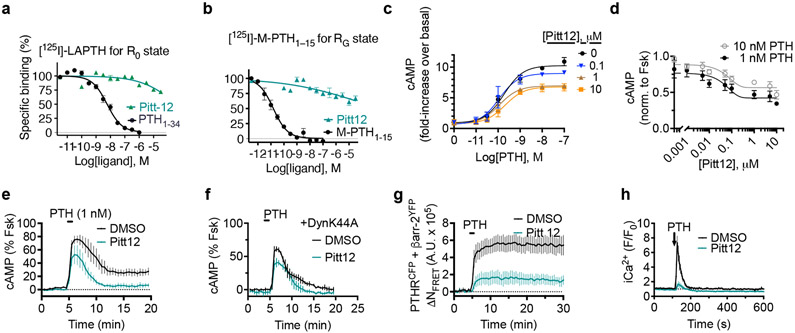
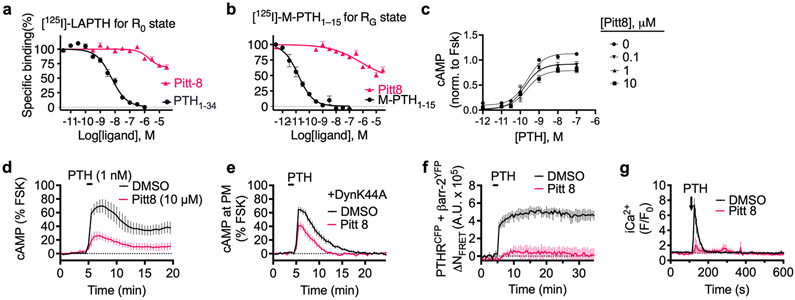
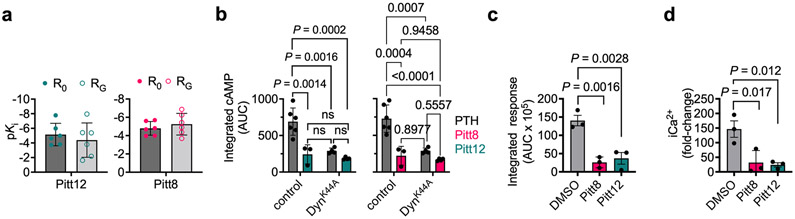
We next fully characterized Pitt8 and Pitt12 as lead negative modulators of PTH mediated PTHR signaling. Previous studies have established that PTHR can adopt at least two active conformational states, the R0 state that is preferentially stabilized by ligands that mediate endosomal cAMP production such as PTH and the long-acting PTH analog (LA-PTH)23-25. This R0 conformation is distinct from the classical G protein-dependent high-affinity receptor conformation, hereafter noted RG, which is preferentially stabilized by ligands such as PTHrP1-36 or M-PTH1–15 that do not engage endosomal cAMP signaling. To probe which conformational states the compounds are affecting and to determine their apparent affinity to PTHR, we performed competitive binding to the human PTHR (hPTHR) in R0 state (assayed in presence of GTPγS that irreversibly activates Gα subunits thus decoupling them from receptors) or RG state using PTHR-expressing cell membranes. Accordingly, [125I]-LAPTH or [125I]-M-PTH(1–15) were used as tracer radioligands for R0 or RG state, respectively. The inhibitory binding constants (Ki) of Pitt8 and Pitt12 were in the μM range (16.2 μM and 5.6 μM for Pitt8, and 7.1 μM and 40.7 μM for Pitt12 in R0 and RG states, respectively) with no significant differences in apparent affinity for a given conformational state of PTHR (Fig. 3a,b for Pitt12, Extended Data Fig. 3a,b for Pitt8, and Extended Data Fig. 4a).
Figure 3. Actions of Pitt12 on PTH signaling.
(a–b) Binding isotherms showing competitive inhibition of radio-labelled peptide ligand binding to PTHR by PTH or Pitt12, using plasma membrane extracts from HEK293 cells expressing recombinant PTHR. Inhibition of [125I]-LAPTH binding to G protein-independent conformational R0 state of PTHR (i.e., in the absence of G proteins) by PTH1-34 (black) or Pitt12 (teal) (a). Inhibition of [125I]-M-PTH1-15 binding to G protein-dependent RG conformational state of PTHR (i.e., in the presence of G proteins) by M-PTH1-15 (black) or Pitt12 (teal) (b). Mean ± s.e.m. of N = 6 experiments carried out in duplicate (c). (c, d) Concentration-response curves for cAMP production by PTH alone or together with Pitt12 (c), and effect of a range of Pitt12 concentrations on PTH mediated cAMP (d). Data are mean ± s.e.m. of N = 3 (c) and N = 5 (d) independent experiments. (e, f) Averaged cAMP time-courses following brief stimulation with 1 nM PTH without (Ctrl, black) or with 10 μM Pitt12 measured by FRET changes from HEK293 cells stably expressing PTHR and a FRET-based cAMP sensor EpacCFP/YFP in the absence (e) or presence of the dominant-negative dynamin mutant (DynK44A) tagged with RFP (f). Data are mean ± s.d. of N = 3 experiments. (g) Time course of β-arrestin 2 interaction with PTHR measured by FRET in HEK293 cells transiently expressing PTHRCFP and βarr-2YFP following brief stimulation with 10 nM PTH without (Ctrl) or with 10 μM Pitt12. Cells were continuously perfused with control buffer or PTH alone or together with compound Pitt12 (horizontal bar). Data are the mean ± s.e.m. of N = 3 experiments with n = 71 (DMSO) and n = 71 (Pitt12) cells examined. (h) Time courses of Ca2+ release in response to PTH (100 nM) with or without Pitt12 (10 μM) in live HEK-293 cells expressing recombinant PTHR. Data are the mean ± s.e.m. of N = 3 experiments with n = 42 (DMSO) and n = 33 (Pitt12) cells examined.
Both compounds act as negative allosteric modulators of PTHR signaling by decreasing the efficacy (Emax) of PTH to mediate cAMP increase without any significant effects on potency (Fig. 3c and Extended Data Fig. 3c). Saturable maximal effects are observed for both Pitt8 and Pitt12, which blocked ≈ 40% of the cAMP response mediated by saturating PTH concentrations (Fig. 3c,d).
We next sought to confirm that Pitt8- and Pitt12-mediated alterations in PTHR signaling occur at the level of the ligand-receptor complex by assessing their ability to modulate PTH-induced interaction of G-proteins with the activated receptor. Indeed, FRET-based time courses in cells expressing PTHR C-terminally fused to CFP (PTHRCFP) and mini-GS and -Gq fused to Venus (mGSvenus and mGqvenus, respectively) revealed that Pitt8 and Pitt12 cause a decrease in both the kinetics and magnitude of G proteins association with the receptor (Supplementary Fig. 5a,b).
We subsequently determined the actions of small molecules on location bias in PTHR signaling. Biased signaling is a dominant theme in GPCR research, in part, because it has major implications for pharmaceutical development. For the PTHR, the concept is particularly germane, given that the receptor can mediate diverse modes of cAMP signaling, including transient signaling at the plasma membrane or sustained signaling from within endosomes. Such altered modes of signaling likely contribute to the diversity of biological processes regulated by the PTHR, which include the maintenance of calcium homeostasis, bone remodeling, cell differentiation in developing tissues, and the control of renal phosphate excretion. Intriguingly, Pitt8 and Pitt12 exerted selective reduction of endosomal cAMP production with a moderate alteration of plasma membrane cAMP when cells are treated with 1 nM PTH. We found that blockade of receptor internalization by expression of a dominant-negative dynamin mutant (DynK44A) prevented significantly sustained cAMP responses induced by PTH alone (Fig. 3e,f and Extended Data Fig. 4b, black curves) as previously reported20,26,27. Those induced by PTH when coapplied with Pitt8 or Pitt12 were not significantly affected in the presence of DynK44A (Fig. 3e,f for Pitt12, Extended Data Fig. 3d,e for Pitt8, and Extended Data Fig. 4b). Consistent with the failure of PTH to engage endosomal cAMP in the presence of the selected small molecules, FRET experiments in cells expressing PTHRCFP and β-arrestin-2 fused to YFP (βarr2YFP) showed that PTHR coupling to βarr2 was markedly impaired (Fig. 3g, Extended Data Fig. 3f, and Extended Data Fig. 4c). In addition, Pitt8 and Pitt12 induced noticeable reduction of PTH-mediated release of stored intracellular Ca2+ (Fig. 3h, Extended Data Fig. 3g, and Extended Data Fig. 4d), suggesting that the compounds strongly impair Gq activation, a result that was corroborated by the reduced Gq coupling to PTHR (Supplementary Fig. 5b). Notably, such observed reduction in Gq activation and βarr2 coupling to PTHR is consistent with our recent results supporting the requirement of Gq activation-dependent biosynthesis of phosphatidylinositol (3,4,5)-triphosphate for PTHR–βarrestin assembly in response to PTH28.
These data suggest that small molecules Pitt8 and Pitt12 can bias the location of cAMP production mediated by PTH, a key feature given that duration and location of PTHR-mediated cAMP responses lead to distinct physiological outcomes, and thus make these compounds promising pharmacological tools for potential therapeutical development.
Refinement and validation of the Pitt12 binding site.
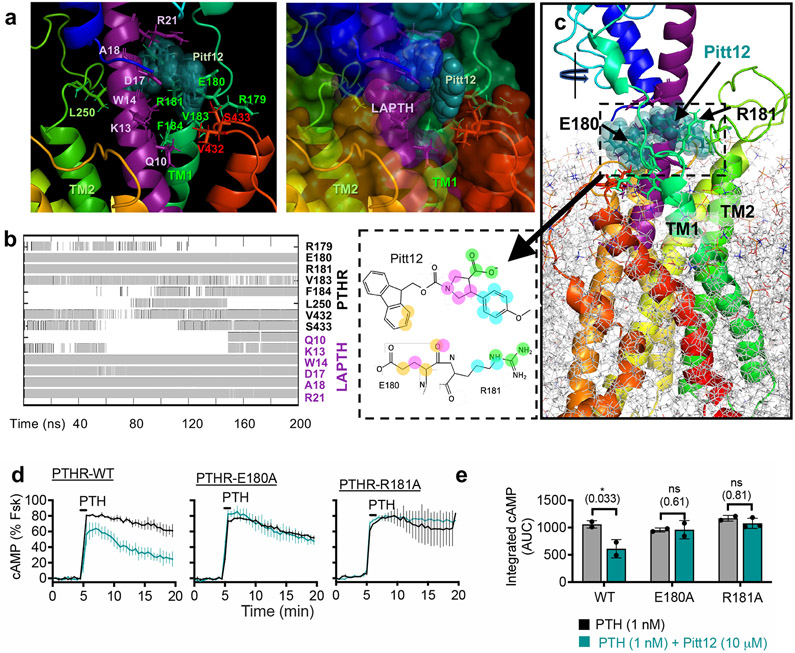
Our initial druggability simulations were performed using ligand- and membrane-free PTHR form. To refine the binding pose of Pitt12 in a biologically relevant context and to get insight into mechanism of its action, we performed additional all-atom 200 ns MD simulations using the active state, LA-PTH–bound PTHR structure12 with receptor embedded in membrane. Results of MD simulations showed predictive interactions (Fig. 4a) and time-evolution of Pitt12 engagement with the peptide ligand and PTHR residues (Fig. 4b) as well as the overall view of the simulation system and bound Pitt12 (Fig. 4c). Pitt12 binds to ligand peptide residues facing TM1 and spanning the peptide’s mid-region Q10–R21 (Fig. 4a), overall bridging the LA-PTH and PTHR interaction. Notably, Pitt12 makes persistent contacts with PTHR residues E1801.32b and R1811.33b, identified in the initial druggability simulations (see Supplementary Movie 1). To experimentally validate this prediction, we generated receptor mutants with individual replacement of E1801.32b and R1811.33b to alanines (E180A, R181A), and assessed their sensitivity to Pitt12 modulation of PTH-induced cAMP signaling (Fig. 4d, e). While each individual mutation did not alter receptor bioactivity, the mutant receptors lost their ability to respond to negative modulation by Pitt12 suggesting that PTHR residues E1801.32b and R1811.33b are critical for the negative allosteric action of Pitt12.
Figure 4. Refinement and validation of Pitt12 binding site in PTHR.
(a–c) Results from MD simulations (200 ns) of Pitt12 binding to active state, LA-PTH–bound PTHR (PDB 6NBF)12 embedded in membrane, showing predictive interactions (a), and time-evolution of Pitt12 engagement with the peptide ligand and PTHR residues (b), as well as the overall view of the simulation system and bound Pitt12 (c). Interaction is defined as existence of any heavy atom contact pairs between PTHR (or LA-PTH) and Pitt12 within distance < 4.0 Å. Inset of panel c shows 2D representations of closely interacting units, where same color circles (orange, magenta, cyan, or green) indicate interacting pairs of atoms between Pitt12 and E180/R181. As the initial pose of Pitt12- and LA-PTH-bound PTHR, we used the cryo-EM structure of LA-PTH-bound PTHR (PDB id: 6NBF), into which we inserted Pitt12 upon structural alignment with our snapshot from druggability simulations (which was adopted in the construction of the pharmacophore model as well as in virtual screening).
(d, e) Experimental validation of Pitt12 binding site in PTHR. PTH (1 nM)-induced cAMP generation time-courses (d) and corresponding integrated cAMP responses (e) from wild-type and receptor mutants carrying individual E180A or R181A mutation, in presence or absence of 10 μM Pitt12. Mean ± s.e.m. of N = 2 or 3 experiments with n = 39 (PTH) and n = 45 (PTH+Pitt12) cells examined for PTHR-WT, n = 46 (PTH) and n = 50 (PTH+Pitt12) cells examined for PTHR-E180A, and n = 40 (PTH) and n = 57 (PTH+Pitt12) cells examined for PTHR-R181A. P values were assessed by two-tailed Student’s t-test.
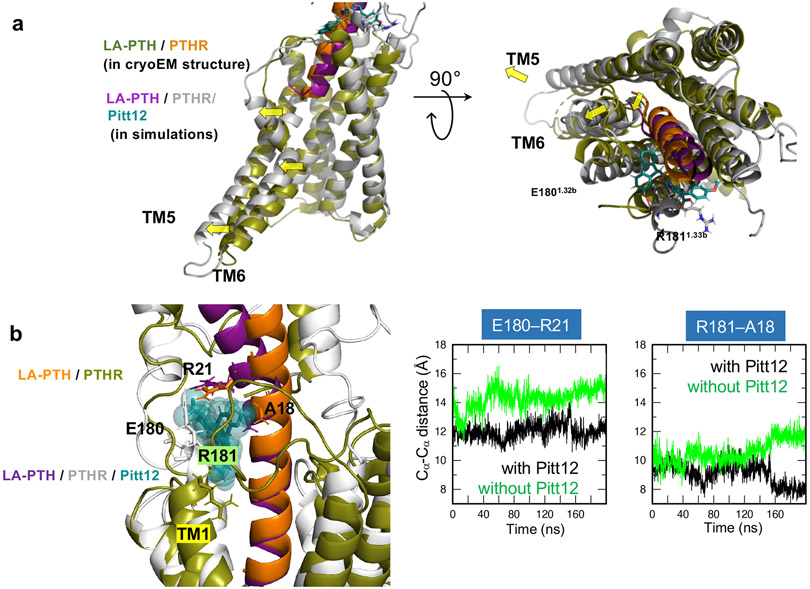
An intriguing observation predicted by these simulations is that Pitt12 binding alters the mode of LA-PTH interaction with PTHR, in particular shifting the position of peptide’s N-terminal tip, which leads to outward displacement of TM5 and TM6 helices (Fig. 5a). These conformational changes likely disfavor G-protein coupling consistent with negative allosteric action of Pitt12 in time course FRET recordings of PTHR interaction with Gs or Gq (Supplementary Fig. 5).
Figure 5. Conformational change of PTHR due to Pitt12 binding.
(a) Predicted Pitt12 binding-induced conformational changes in PTHR. N-terminal tip of LA-PTH pushes extracellular tip of TM6 further outwards leading to outward displacement of TM5 and TM6 helices.
(b) Comparison of the conformations stabilized in the MD runs performed for LA-PTH/PTHR in the presence and absence of Pitt12. Left, the superimposed final (t = 200 ns) snapshots from the two runs. Essential residues coordinating Pitt12 are shown in stick representation and labeled. Right, the time evolution of Cα-Cα distances between E180 of PTHR and R21 of LA-PTH (left panel) and between R181 of PTHR and A18 of LA-PTH (right panel) are presented, for the runs conducted in the presence (black) or in the absence of Pitt12 (green). The space between PTHR and LA-PTH is wider in the absence of Pitt12.
We also performed control MD simulations of the LA-PTH/PTHR in the absence of Pitt12 and compared with those performed for LA-PTH/PTHR in the presence of Pitt12 (Fig. 5b). The simulations showed that in the absence of Pitt12, LA-PTH has higher flexibility, based on the larger fluctuations in intermolecular distances (Fig. 5b). Precisely, we observed that the binding space between PTHR and LA-PTH residue pairs (E180–R21 and R181–A18) became tighter and more stable in the presence of Pitt12.
In vivo action of Pitt12.
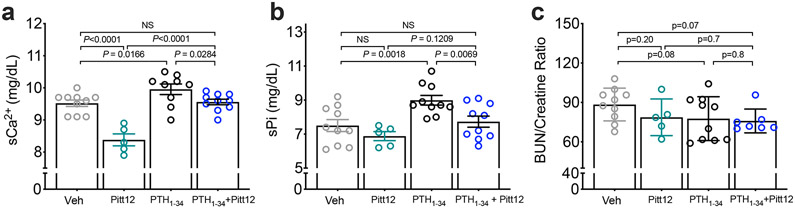
We selected Pitt12 to examine its in vivo action. As shown previously29,30, injections of a pharmacological dose of PTH1–34 into animals or humans results in acute increase in serum levels Ca2+ (sCa2+) and phosphate (sPi). We thus examined the ability of of Pitt12 to modulate these responses to endogenous and exogenous PTH. Four groups of C57BL/6 mice were randomized and injected subcutaneously with either vehicle, Pitt12 (20 μmol/kg body wt), or PTH1-34 (40 μg/kg body wt) alone or in combination and serum parameters were measured 3h after injections. Pitt12 alone significantly reduced sCa2+ levels and produced a trend of reducing sPi levels, presumably due to its inhibitory action against endogenous PTH (Fig. 6a, b). Consistent with this result, abilities of PTH1-34 injection to increase sCa2+ and sPi were significantly (P < 0.05) reduced by co-injection of Pitt12. The lack of significant changes in BUN (Blood Urine Nitrogen)/creatine ratio indicate normal kidney function and hydration status (Fig. 6c). Overall, our findings support antagonizing actions of Pitt12 against PTHR-mediated effects on mineral homeostasis, potentially by acting as a negative allosteric regulator of PTHR signaling.
Figure 6. In vivo action of Pitt12.
(a–c) Serum Ca2+ (sCa2+) and phosphate (sPi) levels, as well as BUN/creatine ratio were measured 3 hrs after injections of vehicle (Veh), Pitt12 (20 μmole/kg), PTH1-34 (40 μg/kg), or Pitt12+PTH1-34. Mean ± s.e.m. of N = 10 (veh), 5 (Pitt12), 9 (PTH1-34) and 10 (PTH1-34+Pitt12) mice for sCa2+, and N = 10 (veh), 5 (Pitt12), 10 (PTH1-34) and 10 (PTH1-34+Pitt12) mice for sPi, and N = 10 (veh), 5 (Pitt12), 9 (PTH1-34) and 7 (PTH1-34+Pitt12) mice for BUN/Creatine ratio. P values were assessed by one-way ANOVA with Dunnett test. *P values are indicated.
Discussion
PTH is vital for maintaining normal Ca2+, phosphate, and vitamin D levels in the body (blood and circulation fluid), and bone turnover. These biological hallmarks are collectively mediated via heterotrimeric Gs and Gq proteins activated in response to PTHR, a class B GPCR mainly expressed in bone and kidney cells. Disturbances in the maintenance of normal mineral-ions due to over-secretion of PTH by the parathyroid glands and PTHR hyperactivity result in hyperparathyroidism, a primary cause of kidney stones and a trigger of osteoporosis (loss of bone mass), two detrimental and costly endocrine diseases affecting the public health. Current pharmaceutical therapies for the treatment of PTH hypersecretion, including calcimimetics (such as cinacalcet, an allosteric activator of the calcium-sensing receptor), calcitriol (active form of Vitamin D) and its analogs to lower serum PTH level, are limited by patients’ intolerance, gastrointestinal toxicity, and hypocalcemia also associated with the risk of parathyroidectomy (surgical removal of parathyroid glands). The negative modulation of PTH-induced PTHR signaling is thus a desirable property for drug candidates investigated to treat bone and mineral diseases linked to PTHR hyperactivity, either intrinsic or caused by PTH hypersecretion.
To overcome the limitations of PTH efficacy on skeletal health and address the important gap in the treatment of PTHR hyperactivity, several attempts have been previously made to discover orally available small molecule PTHR ligands leading to identification of three molecules, AH-396031, SW10632 and PCO37133. The former two molecules were characterized as weak agonist (AH-3960) and weak antagonist (SW106), both acting as orthosteric PTHR ligands. However, their studies were discontinued due to too weak affinities and efficacies. The third molecule (PCO371) is currently in phase 1 clinical trial as a potential orally available drug candidate for hypoparathyroidism treatment34. It is characterized as an orthosteric agonist of PTHR. Our identified molecules are unique by their mode of action: they bind to an allosteric site on the extracellular vestibule of the PTHR and negatively modulate receptor signaling induced by PTH in the cytoplasm. As negative modulators, they are candidates to hyperparathyroidism treatment, and as allosteric molecules they have a key advantage over orthosteric ligands in that they modulate endogenous PTH actions and their effects are saturable.
The drug discovery process comprises five major stages, starting with 1) disease target identification and validation, followed by 2) lead compound discovery, 3) compound optimization, and 4) preclinical and clinical development, and culminating with 5) a new drug approvement by the regulatory agencies (reviewed in35). Our structure-guided computational approach tackled stage 2 by the identification of allosteric druggable sites in PTHR and small molecule compounds that act as negative allosteric modulators of PTHR signaling. When tested in vivo, Pitt12 caused significant reduction in the effects of PTH on serum Ca2+ and Pi levels in mice, showing that Pitt12 can act as a negative modulator of PTHR function in vivo. These small molecules are suitable lead compounds for further optimization and development into potential drug candidates for treatment of hyperparathyroidism.
While future studies are needed to complete the next stages in the drug discovery process including an ADME pharmacokinetics study and testing the compounds for their selectivity to PTHR against a large number of GPCRs using for example the PRESTO-TANGO assay36, our computational pipeline for identifying specific compounds that can modulate PTHR structure, dynamics and function complemented by in vitro and in vivo experiments provides a comprehensive protocol including the detection of druggable sites and the construction of pharmacophore models. Our study suggests that this approach could be extended to other GPCRs, in particular to class B peptide hormone receptors, and serve as a novel approach for the detection of allosteric druggable sites in GPCRs as well as identification of novel small molecules with therapeutic potential against a plethora of diseases.
Methods
Essential site scanning analysis.
ESSA analysis is based on modeling the structure as a Gaussian network model11,37 (GNM). We used the default cutoff distance of 10 Å for defining the inter-residue contact topology using the ProDy application programming interface (API)38. GNM yields a spectrum of N-1 nonzero modes of motions for a protein of N residues, where each mode is characterized by a frequency and a mode shape described by the respective eigenvalues and eigenvectors of the network connectivity matrix. To mimic the ‘crowding’ induced upon ligand binding, the local density around each residue was increased by adding extra nodes at the positions of heavy atoms other than its α-carbons. The resulting perturbation in the vibrational spectrum of the protein was quantified as a residue-specific z-score (or ESSA score) by evaluating the mean percent shift in the frequency of the ten lowest frequency (also called global) modes for each residue.
Druggability simulations and analyses.
Druggability simulations were performed for the resolved PTHR (PDB id: 6NBF) in the presence of the probes shown in Extended Data figure 2a using the all-atom MD simulation package NAMD39 with the CHARMM36 force field for proteins40, the TIP3P water model41, and the CGenFF force field42 for the probes. The trajectories were analyzed using the DruGUI module16 of ProDy38,43. The target proteins used for simulations are PTHR (PDB id: 6NBF) and GLP-1R (PDB id: 6VCB). Six independent runs of 40 ns were performed, as recommended. All MD snapshots were superposed onto the reference PDB structure using Cα-atoms and a cubic grid-based representation of the space, with the grid edge size was set to 0.5 Å. Probe molecules having non-hydrogen atoms within 4.0 Å from protein atoms were considered to interact with the protein. For each probe type, the individual occupancy of grids was calculated using their centroids. We evaluated the occupancy of each probe for a given voxel and quantified binding energies with the inverse Boltzmann relation. Briefly, each probe molecule was localized to a particular voxel using the centroid of each molecule based on heavy atoms. The ratio of the actual occupancy of each probe in each voxel on the protein surface (ni) relative to that in solution (n0) was used to calculate the binding free energy using ∆Gi = −RT ln(ni/n0) where R is the gas constant and T is the absolute temperature (in K). The resulting binding free energy map identifies interaction spots with low energy for one or more probe types. High occupancy voxels, called hot spots, within a distance of 5.6 Å were merged and druggable sites were defined by clusters of at least six such hot spots. Their maximal binding affinities were calculated as the sum of binding energies of their hot spots, allowing us to rank the druggable sites. Representative druggable sites for PTHR are shown in blue box in figure 2a. The highest affinity site (Site 1) was further analyzed further to build a PM.
Pharmacophore modeling.
A PM for Site 1 was built using Pharmmaker17 (see yellow box in Fig. 2a). We identified the residues involved in high affinity interactions with each probe type (Step 2), and also identified highly druggable spots for each probe type (shown in spheres in the figure) within Site 1 (Step3). Residue-probe interactions were rank-ordered based on their frequency of occurrence within Site 1 at the druggable spots in multiple runs (entropic contribution, Step 4). In Steps 3 and 4 in Fig. 2a, we displayed focusing on high affinity residues and probe types we used for PM although we considered all the high affinity residues and probe types at Site 1. Snapshots that exhibited the highest number of the most frequently occurring interaction pairs (e.g., F1841.36b-isopropanol, Y2452.72b-isopropanol, and R1811.33b-acetate) were selected as template to construct PMs (Step 5). The PM was screened against ZINC and MolPort libraries using Pharmit19 (Step 6). MolPort contains a dataset of 110,832,826 conformers corresponding to 7,719,859 compounds. The Zinc library of drug-like fragments that we screened contained 123,399,574 conformers of 13,190,317 compounds. Further refinement of Pitt12 shown in figure 4a-c and figure 5a-b was performed using all-atom MD. In conformity with the acetate group used in constructing the PM and in virtual screens, the carboxylate group of Pitt12 is deprotonated (see Extended Data figure 2f). We built a system of LA-PTH–bound PTHR (PDB id 6NBF) embedded in POPC lipid membrane and employed simulation steps, using CHARMM-GUI bilayer builder44. MD simulation of 200 ns was performed using the package NAMD39 with the CHARMM36m force field39,45. All-atom MD simulations for LA-PTH/PTHR in the absence of Pitt12 (Fig. 5b) was performed using the same setup as that in the presence of Pitt12. Data visualization was performed using PyMOL 1.8.6. (Schrödinger L. The PyMOL Molecular Graphics System, Version 1.8.4.2. (2016)).
Cell culture.
Cell culture reagents were obtained from Corning (CellGro). Human embryonic kidney (HEK293; ATCC, Georgetown, DC) cells stably expressing the recombinant human PTHR were grown in selection medium (DMEM, 5% FBS, penicillin/streptomycin 5%, 500 μg/ml neomycin) at 37°C in a humidified atmosphere containing 5% CO2. Primary calvarial osteoblast (Ob) cells were isolated and cultured as previously described20; human renal proximal tubule epithelial cells (RPTEC, ATCC Georgetown, DC) were grown in DMEM/F12 supplemented with 5 pm triiodo-L-thyronine, 10 ng/ml recombinant human epidermal growth factor, 25 ng/ml prostaglandin E1, 3.5 μg/ml ascorbic acid, 1 mg/ml insulin, 0.55 mg/ml transferrin, 0.5 μg/ml sodium selenite, 25 ng/ml hydrocortisone) plus 1% penicillin and streptomycin. For transient expression, cells were seeded on glass coverslips coated with poly-D-lysine in six-well plates and cultured for 24 hours prior transfection with the appropriate cDNAs using Fugene-6 (Promega) or Lipofectamine 3000 (Life Technologies) for 48-72 h before experiments.
Peptides and small molecules.
PTH1–34 and PTHrP1–36 were purchased from Bachem. The peptide was resuspended in 10 mM acetic acid to make 1 mM stock solution. Forskolin (#344270) was purchased from EMD-Millipore. Small molecules were purchased from MolPort. Compound purity was >90% for all studied compounds.
Receptor mutagenesis.
We used Genewiz service to generate Human influenza hemagglutinin (HA)-tagged human PTHR mutants carrying E180A (HA-PTHRE180A) and R181A (HA-PTHRR181A), individual mutations.
Time-course measurements of cAMP production, and PTHR recruitment of β-arrestin in single live cells.
Cyclic AMP was assessed using FRET-based assays. Cells were transiently transfected with the FRET-based biosensors, Epac1-CFP/YFP for measuring cAMP and PTHR-CFP with βarr2-YFP for measuring arrestin recruitment. For inter-molecular FRET experiments, we optimized expression conditions to ensure the expression of fluorescently labeled proteins was similar in examined cells by performing experiments in cells displaying comparable fluorescence levels. Measurements were performed and analyzed as previously described46. In brief, cells plated on poly-D-lysine coated glass coverslips were mounted in Attofluor cell chambers (Life Technologies), maintained in Hepes buffer containing 150 mM NaCl, 20 mM Hepes, 2.5 mM KCl and 0.1–10 mM CaCl2, 0.1% BSA, pH 7.4, and transferred on the Nikon Ti-E equipped with an oil immersion 40X N.A 1.30 Plan Apo objective and a moving stage (Nikon Corporation). CFP and YFP were excited using a mercury lamp. Fluorescence emissions were filtered using a 480 ± 20 nm (CFP) and 535 ± 15 nm (YFP) filter set and collected simultaneously with a LUCAS EMCCD camera (Andor Technology) using a DualView 2 (Photometrics) with a beam splitter dichroic long pass of 505 nm. Fluorescence data were extracted from single cell using Nikon Element Software (Nikon Corporation). The FRET ratio for single cells was calculated and corrected as previously described46. Individual cells were perfused with buffer or with the ligand for the time indicated by the horizontal bar.
Concentration-response curves for cAMP.
HEK293 cells stably expressing human PTH type 1 receptor (hPTHR) and pGloSensor™-22R were seeded in white 96-well plates coated with poly-D lysine at a density of 1.5 × 104 cells/well. Cells were maintained at 37°C and 5% CO2 overnight. For the assay, cell media was replaced by 80 μl equilibration Locke buffer (NaCl 154 mM, KCl 5.6 mM, CaCl2 1.3 mM, MgCl2 1 mM, NaHCO3 3.6 mM, glucose 5.6 mM, HEPES 20mM, pH 7.4) containing 450 μg/ml D-luciferin (Biotium Cat:10101, Lot: 14L0408) and 0.5 mM IBMX (Cayman chemical, item No.13347, CAS No. 28822-58-4), and were incubated 45 min at 37°C for loading with luciferase. Before drug addition, background time luminescence was recorded at 560 nm in a SpectraMax Paradigm Multiple-Mode microplate reader (Molecular Devices) for 5 min. The plates were removed from the instrument, a range of PTH1–34 concentrations with or without small molecule compounds were added at different concentrations, and luminescence was again recorded in kinetic mode for 45 min. Peak luminescence or the area-under-curve (auc) of the time course of luminescence response for each PTH1-34 concentration were determined and used to generate concentration-response curves. Data were normalized to the response mediated by of 100 μM forskolin. Curves were fit using GraphPad Prism 8.0 to determine potency (EC50) and maximum efficacy (Emax) of the PTH1–34 response. Data are reported in Supplementary Table 1 as mean ± s.d. of at least N = 2–3 independent experiments, each done in duplicate.
Intracellular Ca2+ measurements.
Changes in intracellular calcium [Ca2+]i were measured by loading cells with the live cell calcium sensor Fluo-4 according to manufacturer’s directions (Molecular Probes, Carlsbad, CA). Cells were imaged at 2 s intervals using an inverted epifluorescence microscope as previously described47.
Radioligand receptor binding.
Competitive binding to the human PTHR (hPTHR) in G protein-dependent (RG) or G protein-independent (R0, assayed in presence of 10 μM GTPγS) states was assessed using membranes prepared from previously described48 HEK-293-derived GP-2.3 cells (stable expression of hPTHR) and either [125I]-M-PTH(1–15) or [125I]-LAPTH as tracer radioligands25. In brief, reactions (230 μl) were assembled in 96-well vacuum filtration plates (MultiScreen; 0.65 μM, Durapore HV, Millipore Corp., Millford, MA) and contained membrane assay buffer (20 mM HEPES pH 7.4, 0.1 M NaCl, 3 mM MgSO4, 20% glycerol, 3 mg/ml bovine serum albumin (VWR #97068), 1X protease inhibitor cocktail (SIGMA cat# P8340) radioligand (~25,000 cpm/well), various concentrations of unlabeled ligand, and membranes (80 μg/ml), added last to start the reactions. Reactions were incubated at room temperature for 90 minutes, and terminated by vacuum filtration followed by two rinses of the filters with membrane assay buffer; the filters were then detached, and counted for gamma irradiation. Nonspecific binding was determined using 0.5 μM PTH(1–34). Specific bound radioactivity was plotted vs ligand concentration using a four-parameter sigmoidal dose-response equation. The Ki values were calculated using Cheng-Prusoff equation49.
Photometric FRET recordings of G-protein interactions.
FRET experiments were performed as previously described46. In brief, cells grown on glass coverslips were maintained in buffer A (137 mM NaCl, 5 mM KCl, 1 mM MgCl2, 20 mM HEPES, pH 7.4) at room temperature and placed on a Zeiss inverted microscope (Axiovert 200) equipped with an oil immersion X100 objective and a dual emission photometric system (Till Photonics, Germany). Cells were excited with light from a polychrome V (Till Photonics). To minimize photobleaching, the illumination time was set to 5–15 ms applied with a frequency between 1 and 20 Hz. Individual cells were perfused with buffer, PTH1–34 alone or in combination with small molecule for the time indicated by the horizontal bar. The emission fluorescence intensities were determined at 535 ± 15 and 480 ± 20 nm (beam splitter dichroic long-pass (DCLP) 505 nm) upon excitation at 436 ± 10 nm (DCLP 460 nm) and were corrected for the spillover of CFP into the 535-nm channel, the spillover of YFP into the 480-nm channel, and the direct YFP excitation to give a corrected FRET emission ratio FCFP/FYFP. Changes in fluorescence emissions due to photobleaching were systematically subtracted.
Mouse Studies.
To test the impact of Pitt12 and PTH (PTH1–34) (Bachem, Torrance, CA, Cat# H-5460) on serum Ca2+ (sCa) and phosphate (sPi) parameters, 3-month-old male C57BL/6J (C57/B6) mice (Jackson Laboratory, Stock No: 000664) were ear-tagged for identification, randomly assigned to groups, and injected with the drugs (40 μg/kg bogy weight for PTH, 20 μmole/kg for Pitt12). Pitt12 was injected 15 minutes before PTH1–34. In this study, blood was sampled via retroorbital route under isoflurane anesthesia 3 hrs after the drug injection. sCa2+ and sPi levels were assessed by an ACE Axcel bioanalyzer (Alfa Wassermann, West Coldwell, NJ)50. All mice were kept in a climate-controlled room (22°C; 45-54% relative humidity) with a 12-hour light/12-hour dark cycle. Water and standard chow (1.3% calcium and 1.03% phosphate) were given ad libitum. The animal experiments were performed according to procedures approved by the Institutional Animal Care and Use Committee of the San Francisco Department of Veteran Affairs Medical Center (Protocol numbers: 18-017). Mice were acclimated with local environment for 2 weeks before randomization and the experimental procedure. Animal numbers were estimated by a power analysis using the following parameters: standard deviation= 15% based our prior studies testing the effects of PTH1–34, two-sided test, p value=0.05, and power of the test=0.8. Additional 20% of mice were included to account for potential fatality due to the drug treatments. The experiments were repeated with 2 different batches of mice that were acquired 2 months apart.
Statistical analysis.
Data were processed using Excel 2013 (Microsoft Corp., Redmond, WA) and GraphPad Prism 7.0 or 8.0 (GraphPad Software, La Jolla California USA). Data are expressed as mean ± s.e.m. or s.d. Binding or cAMP data from concentration–response assays were analyzed by using a sigmoidal dose–response model with variable slope. Paired data sets were statistically compared by using Student’s t test (two-tailed) assuming unequal variances for the two sets. Statistical analyses were performed using two-way ANOVA for in vivo data.
Extended Data
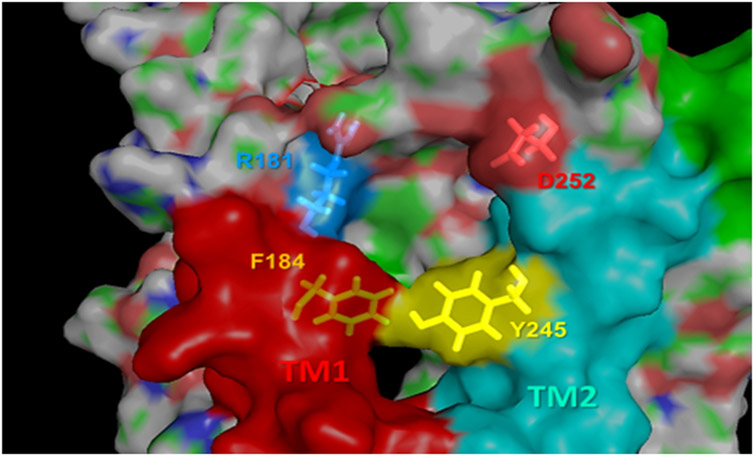
Extended Data Fig. 1. Close-up view of the pocket at the EC vestibule.
The pocket is lined by TM1 (red) and TM2 (cyan). Essential residues R1811.33b and Y2452.72b, whose interaction with ligands is predicted to affect the global dynamics of the overall PTHR, are shown in yellow sticks. Two additional residues, D252ECL1 and F1841.36b are also shown, which may help attract or coordinate the ligands through electrostatic interactions. The role of F184 in complementing R1811.33b and Y2452.72b to line a druggable pocket is further corroborated by druggability simulations.
Extended Data Fig. 2. Probe molecules used in druggability simulations.
(a) Seven probe molecules used in PTHR druggability simulations. The structure of each probe is shown along with its 4-letter acronym, its name and main chemical properties. Also shown is the percentage of drugs containing those fragments based on the SMILES data of 2,453 approved drugs in DrugBank. (b) An example of a common drug, benzyl-penicillin, containing four types of fragments represented by the probe molecules. (c) Binding score profile for PTHR residues near Site 1 (labeled along the abscissa), evaluated for different probe molecules (bars in different colors). The binding score is defined as Σ (1/dki)2 where k is frame/snapshot index and dki is corresponding distance between the probe and the residue i summed over all snapshots n (12,000 compiled from six independent runs), provided that they make atom-atom contacts of dki < 4.0 Å. Residues with score above 1500/Å2 (dashed line) are selected as high affinity residues for each probe type: R1811.33b for acetate, and F1841.36b and Y2452.72b for isopropanol. (d) Close-up view of site 1 preferentially sampled by three probes (two isopropanol and an acetic acid, shown in yellow and red sticks respectively), and associated residues R1811.33b, F1841.36b and Y2452.72b (right). (e, f) Construction of pharmacophore model (PM) composed of a hydrogen bond acceptor, a negatively charged region, and two hydrophobic sites (spheres), based on the preferential positions observed in (c), and overlay of a hit compound Pitt12 (aquamarine sticks, extracted from the ZINC database) and the PM. The box on the left in panel (f) shows the partial charges for the carboxylate group atoms O, C, O’, and H in Pitt12 in its protonated and deprotonated forms. In line with the use of acetate probe molecule to construct the PM, we used the deprotonated form in further analyses and simulations.
Extended Data Fig. 3. Actions of Pitt8 on PTH signaling.
(a, b) Binding isotherms showing competitive inhibition of radio-labeled peptide ligand binding to PTHR by PTH or Pitt8, using plasma membrane extracts from HEK293 cells expressing recombinant PTHR. Inhibition of [125I]-LAPTH binding to G protein-independent conformational R0 state of PTHR (i.e., in the absence of G proteins) by PTH1-34 (black) or Pitt8 (pink) (a). Inhibition of [125I]-M-PTH1-15 binding to G protein-dependent RG conformational state of PTHR (i.e., in the presence of G proteins) by M-PTH1-15 (black) or Pitt8 (pink). Mean ± s.d. of N = 6 experiments. (c) Concentration-response curves for cAMP production by PTH alone or together with Pitt8. Data are mean ± s.e.m. of N = 3 independent experiments. (d, e) Averaged cAMP time-courses following brief stimulation with 1 nM PTH without (Ctrl, black) or with 10 μM Pitt12 measured by FRET changes from HEK293 cells stably expressing PTHR and a FRET-based cAMP sensor EpacCFP/YFP in the absence (d) or presence of the dominant-negative dynamin mutant (DynK44A) tagged with RFP (e). (f) Time course of β-arrestin 2 interaction with PTHR measured by FRET in HEK293 cells transiently expressing PTHRCFP and βarr-2YFP following brief stimulation with 10 nM PTH without (Ctrl) or with 10 μM Pitt8. Cells were continuously perfused with control buffer or PTH alone or together with Pitt8 (horizontal bar). Data are the mean ± s.e.m. of N = 3 experiments with n = 52 (DMSO) and 47 (Pitt8) cells examined. (i) Time courses of Ca2+ release in response to PTH (100 nM) with or without Pitt8 (10 μM) in live HEK-293 cells expressing recombinant PTHR. Data are the mean ± s.e.m. of N = 3 experiments with n = 32 (DMSO) and 32 (Pitt8) cells examined.
Extended Data Fig. 4. Statistics for integrated responses from Figure 3 and ED Figure 3.
(a) pKi values for Pitt12 for the RG state were: −10.77 ± 0.15 (M-PTH1-15) and −4.39 ± 0.96 (Pitt12); for the R0 state: −8.26 ± 0.14 (PTH1-34) and −5.15 ± 0.63. pKi values for Pitt8 the RG state were: −10.77 ± 0.15 (M-PTH1-15) and −5.25 ± 0.48 (Pitt8); for the R0 state: −8.26 ± 0.14 (PTH1-34) and −4.79 ± 0.30 (Pitt8). Mean ± s.d. of N = 6 experiments. (b) Quantitation of cAMP responses by measuring the area under the curve (A.U.C.) from 0 to 20 min for figure 3e,f, and ED-figure 3d,e. Data are mean ± s.d. of N = 6 (PTH), 3 (Pitt12), and 3 (Pitt8) experiments for control, and N = 4 (PTH), 4 (Pitt12), and 4 (Pitt8) experiments for DynK44A. NS, not significant, **P < 0.002 and ***P < 0.0002 by two-way ANOVA with Tukey-Kramer post-hoc test. (c, d) Data are from figure 3h and ED-figure 3g. Mean ± s.e.m. of N = 3 independent experiments. P values were assessed by two-tailed Student’s t-test with *P<0.05, **P<0.005.
Supplementary Material
Acknowledgment
Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Disease (NIDDK), the National Institute of General Medical Sciences (NIGMS), the National Institute on Drug Abuse (NIDA), and the National Center for Advancing Translational Sciences (NCATS) of the US National Institutes of Health (NIH) under grant Awards Number R01-DK116780 (to J.-P.V.), R01-DK122259 (to W.C. and J-P.V.), P41 GM103712 and R01 GM139297 (to I.B. and P.D.), P01-DK011794 (to T.J.G.), and by the University of Pittsburgh Clinical and Translational Science Institute (NIH UL1TR001857).
Footnotes
Competing Interests
I.S., J.Y.L., B.K., I.B., and J.-P.V. acknowledge potential competing financial interests. No conflicts of interest were disclosed by other authors.
Data Availability
Source of datasets analyzed during this study for figures (Fig. 1-6, Extended Data Fig. 2-4, and Supplementary Fig. 1-5) are provided with this paper. Materials and associated protocols will be made available to all qualified investigators from the corresponding authors upon reasonable request.
References
- 1.Insel PA et al. GPCRomics: An Approach to Discover GPCR Drug Targets. Trends Pharmacol Sci 40, 378–387, doi: 10.1016/j.tips.2019.04.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pandy-Szekeres G et al. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res 46, D440–D446, doi: 10.1093/nar/gkx1109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyu J et al. Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229, doi: 10.1038/s41586-019-0917-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein RM et al. Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature 579, 609–614, doi: 10.1038/s41586-020-2027-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basith S et al. Exploring G Protein-Coupled Receptors (GPCRs) Ligand Space via Cheminformatics Approaches: Impact on Rational Drug Design. Front Pharmacol 9, 128, doi: 10.3389/fphar.2018.00128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutkeviciute I & Vilardaga JP Structural insights into emergent signaling modes of G protein-coupled receptors. The Journal of biological chemistry 295, 11626–11642, doi: 10.1074/jbc.REV120.009348 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardella TJ & Vilardaga JP International Union of Basic and Clinical Pharmacology. XCIII. The parathyroid hormone receptors--family B G protein-coupled receptors. Pharmacol Rev 67, 310–337, doi: 10.1124/pr.114.009464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheloha RW, Gellman SH, Vilardaga JP & Gardella TJ PTH receptor-1 signalling-mechanistic insights and therapeutic prospects. Nat Rev Endocrinol 11, 712–724, doi: 10.1038/nrendo.2015.139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurkcuoglu Z, Findik D, Akten ED & Doruker P How an Inhibitor Bound to Subunit Interface Alters Triosephosphate Isomerase Dynamics. Biophys J 109, 1169–1178, doi: 10.1016/j.bpj.2015.06.031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y et al. Intrinsic dynamics is evolutionarily optimized to enable allosteric behavior. Curr Opin Struct Biol 62, 14–21, doi: 10.1016/j.sbi.2019.11.002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaynak BT, Bahar I & Doruker P Essential site scanning analysis: A new approach for detecting sites that modulate the dispersion of protein global motions. Comput Struct Biotechnol J 18, 1577–1586, doi: 10.1016/j.csbj.2020.06.020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao LH et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science 364, 148–153, doi: 10.1126/science.aav7942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahar I JR, Dill K Protein Actions: Principles & Modeling. (Garland Sciebce, Taylor & Francis Group, New York, NY, 2017). [Google Scholar]
- 14.Zhang Y et al. Intrinsic dynamics is evolutionarily optimized to enable allosteric behavior. Curr Opin Struct Biol 62, 14–21, doi: 10.1016/j.sbi.2019.11.002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Guilloux V, Schmidtke P & Tuffery P Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics 10, 168, doi: 10.1186/1471-2105-10-168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakan A, Nevins N, Lakdawala AS & Bahar I Druggability Assessment of Allosteric Proteins by Dynamics Simulations in the Presence of Probe Molecules. J Chem Theory Comput 8, 2435–2447, doi: 10.1021/ct300117j (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JY, Krieger JM, Li H & Bahar I Pharmmaker: Pharmacophore modeling and hit identification based on druggability simulations. Protein Sci, doi: 10.1002/pro.3732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sterling T & Irwin JJ ZINC 15--Ligand Discovery for Everyone. J Chem Inf Model 55, 2324–2337, doi: 10.1021/acs.jcim.5b00559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sunseri J & Koes DR Pharmit: interactive exploration of chemical space. Nucleic Acids Res 44, W442–448, doi: 10.1093/nar/gkw287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jean-Alphonse FG et al. beta2-adrenergic receptor control of endosomal PTH receptor signaling via Gbetagamma. Nature chemical biology 13, 259–261, doi: 10.1038/nchembio.2267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanyu R et al. Anabolic action of parathyroid hormone regulated by the beta2-adrenergic receptor. Proceedings of the National Academy of Sciences of the United States of America 109, 7433–7438, doi: 10.1073/pnas.1109036109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feinstein TN et al. Noncanonical control of vasopressin receptor type 2 signaling by retromer and arrestin. The Journal of biological chemistry 288, 27849–27860, doi: 10.1074/jbc.M112.445098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda A et al. Critical role of parathyroid hormone (PTH) receptor-1 phosphorylation in regulating acute responses to PTH. Proc Natl Acad Sci U S A 110, 5864–5869, doi: 10.1073/pnas.1301674110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okazaki M et al. Prolonged signaling at the parathyroid hormone receptor by peptide ligands targeted to a specific receptor conformation. Proceedings of the National Academy of Sciences of the United States of America 105, 16525–16530, doi: 10.1073/pnas.0808750105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dean T, Vilardaga JP, Potts JT Jr. & Gardella TJ Altered selectivity of parathyroid hormone (PTH) and PTH-related protein (PTHrP) for distinct conformations of the PTH/PTHrP receptor. Molecular endocrinology 22, 156–166, doi: 10.1210/me.2007-0274 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrandon S et al. Sustained cyclic AMP production by parathyroid hormone receptor endocytosis. Nature chemical biology 5, 734–742, doi: 10.1038/nchembio.206 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White AD et al. Ca(2+) allostery in PTH-receptor signaling. Proceedings of the National Academy of Sciences of the United States of America 116, 3294–3299, doi: 10.1073/pnas.1814670116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White AD et al. Gq/11-dependent regulation of endosomal cAMP generation by parathyroid hormone class B GPCR. Proceedings of the National Academy of Sciences of the United States of America 117, 7455–7460, doi: 10.1073/pnas.1918158117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burnett-Bowie SM, Henao MP, Dere ME, Lee H & Leder BZ Effects of hPTH(1-34) infusion on circulating serum phosphate, 1,25-dihydroxyvitamin D, and FGF23 levels in healthy men. J Bone Miner Res 24, 1681–1685, doi: 10.1359/jbmr.090406 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Z et al. Calcium-Sensing Receptors in Chondrocytes and Osteoblasts Are Required for Callus Maturation and Fracture Healing in Mice. J Bone Miner Res 35, 143–154, doi: 10.1002/jbmr.3864 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rickard DJ et al. Intermittent treatment with parathyroid hormone (PTH) as well as a non-peptide small molecule agonist of the PTH1 receptor inhibits adipocyte differentiation in human bone marrow stromal cells. Bone 39, 1361–1372, doi: 10.1016/j.bone.2006.06.010 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Carter PH et al. Discovery of a small molecule antagonist of the parathyroid hormone receptor by using an N-terminal parathyroid hormone peptide probe. Proceedings of the National Academy of Sciences of the United States of America 104, 6846–6851, doi: 10.1073/pnas.0605125104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura T et al. Identification of an orally active small-molecule PTHR1 agonist for the treatment of hypoparathyroidism. Nat Commun 7, 13384, doi: 10.1038/ncomms13384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishimura Y et al. Lead Optimization and Avoidance of Reactive Metabolite Leading to PCO371, a Potent, Selective, and Orally Available Human Parathyroid Hormone Receptor 1 (hPTHR1) Agonist. J Med Chem 63, 5089–5099, doi: 10.1021/acs.jmedchem.9b01743 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Hughes JP, Rees S, Kalindjian SB & Philpott KL Principles of early drug discovery. Br J Pharmacol 162, 1239–1249, doi: 10.1111/j.1476-5381.2010.01127.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kroeze WK et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol 22, 362–369, doi: 10.1038/nsmb.3014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 37.Bahar I, Atilgan AR & Erman B Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des 2, 173–181, doi: 10.1016/S1359-0278(97)00024-2 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Bakan A, Meireles LM & Bahar I ProDy: protein dynamics inferred from theory and experiments. Bioinformatics 27, 1575–1577, doi: 10.1093/bioinformatics/btr168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips JC et al. Scalable molecular dynamics with NAMD. J Comput Chem 26, 1781–1802, doi: 10.1002/jcc.20289 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Best RB et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J Chem Theory Comput 8, 3257–3273, doi: 10.1021/ct300400x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen WL CJ, Madura JD, Impey RW, Klein ML Comparison of simple potential functions for simulating liquid water. J Chem Phys 79, 926–935, doi: 10.1063/1.445869 (1983). [DOI] [Google Scholar]
- 42.Vanommeslaeghe K & MacKerell AD Jr. CHARMM additive and polarizable force fields for biophysics and computer-aided drug design. Biochim Biophys Acta 1850, 861–871, doi: 10.1016/j.bbagen.2014.08.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakan A et al. Evol and ProDy for bridging protein sequence evolution and structural dynamics. Bioinformatics 30, 2681–2683, doi: 10.1093/bioinformatics/btu336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J Chem Theory Comput 12, 405–413, doi: 10.1021/acs.jctc.5b00935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang J et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods 14, 71–73, doi: 10.1038/nmeth.4067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gidon A et al. Endosomal GPCR signaling turned off by negative feedback actions of PKA and v-ATPase. Nature chemical biology 10, 707–709, doi: 10.1038/nchembio.1589 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feinstein TN et al. Retromer terminates the generation of cAMP by internalized PTH receptors. Nature chemical biology 7, 278–284, doi: 10.1038/nchembio.545 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheloha RW, Maeda A, Dean T, Gardella TJ & Gellman SH Backbone modification of a polypeptide drug alters duration of action in vivo. Nat Biotechnol 32, 653–655, doi: 10.1038/nbt.2920 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng Y & Prusoff WH Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22, 3099–3108, doi: 10.1016/0006-2952(73)90196-2 (1973). [DOI] [PubMed] [Google Scholar]
- 50.McElderry JD et al. Tracking circadian rhythms of bone mineral deposition in murine calvarial organ cultures. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research 28, 1846–1854, doi: 10.1002/jbmr.1924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source of datasets analyzed during this study for figures (Fig. 1-6, Extended Data Fig. 2-4, and Supplementary Fig. 1-5) are provided with this paper. Materials and associated protocols will be made available to all qualified investigators from the corresponding authors upon reasonable request.