Summary:
Objectives:
We aimed to identify genes associated with genetic generalized epilepsy (GGEs) by combining large cohorts enriched with individuals with a positive family history. Secondarily, we set out to compare the association of genes independently with familial and sporadic GGE.
Methods:
We performed a case-control whole exome sequencing study in unrelated individuals of European descent diagnosed with GGE (previously recruited and sequenced through multiple international collaborations) and ancestry-matched controls. The association of ultra-rare variants with epilepsy (URVs; in 18,834 protein coding genes) was examined in 1,928 individuals with GGE (vs. 8,578 controls), then separately in 945 individuals with familial GGE (vs. 8,626 controls), and finally in 1,005 individuals with sporadic GGE (vs. 8,621 controls). We additionally examined the association of URVs with familial and sporadic GGE in two gene sets important for inhibitory signaling (19 genes encoding GABAA receptors, 113 genes representing the GABAergic pathway).
Results:
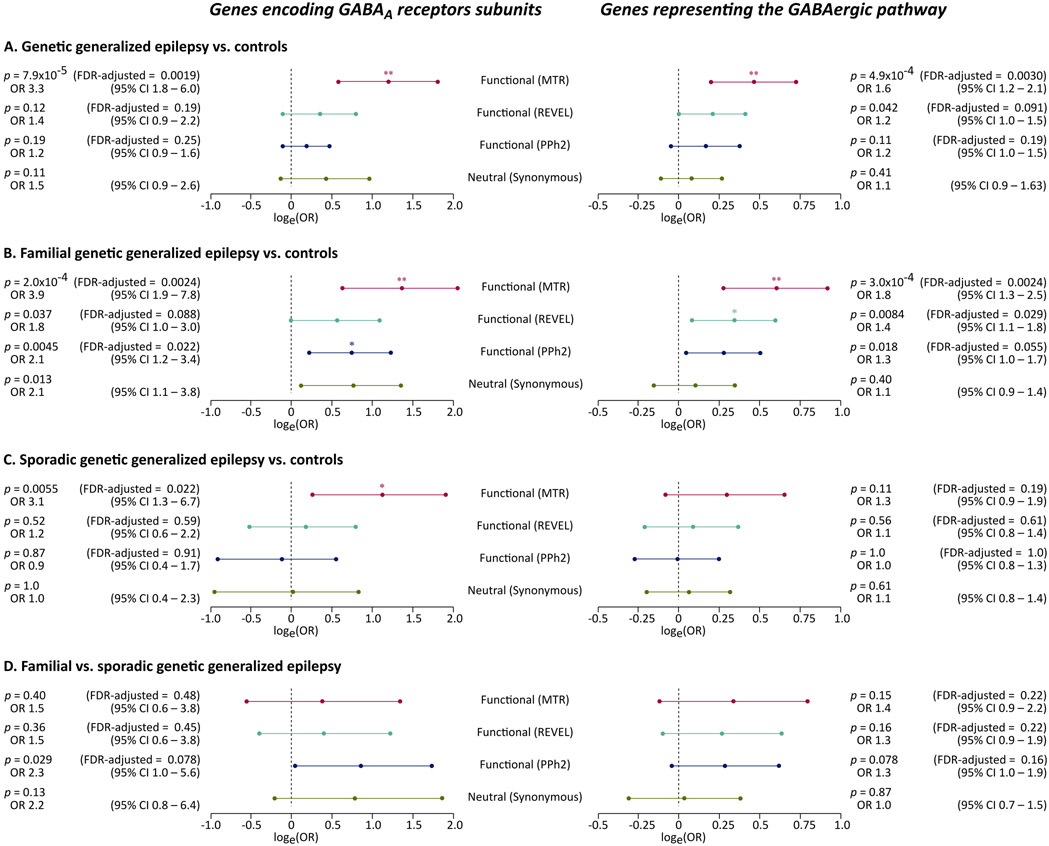
GABRG2 was associated with GGE (p = 1.8x10−5), approaching study-wide significance in familial GGE (p = 3.0x10−6), whereas no gene approached a significant association with sporadic GGE. Deleterious URVs in the most intolerant sub-genic regions in genes encoding GABAA receptors were associated with familial GGE (OR = 3.9, 95% CI = 1.9 – 7.8, FDR-adjusted p = 0.0024), whereas their association with sporadic GGE had marginally lower odds (OR = 3.1, 95% CI = 1.3 – 6.7, FDR-adjusted p = 0.022). URVs in GABAergic pathway genes were associated with familial GGE (OR = 1.8, 95% CI = 1.3 – 2.5, FDR-adjusted p = 0.0024) but not with sporadic GGE (OR = 1.3, 95% CI = 0.9 – 1.9, FDR-adjusted p = 0.19).
Significance:
URVs in GABRG2 are likely an important risk factor for familial GGE. The association of gene sets of GABAergic signaling with familial GGE is more prominent than with sporadic GGE.
Keywords: GGE, familial epilepsy, sporadic epilepsy, GABRG2, GABAA receptors
Introduction:
The genetic risk factors of generalized epilepsies have proven challenging to decipher despite evidence of its heritability from twin and family studies.1,2 Initial gene discovery, guided by linkage analysis, was performed in large families with autosomal dominant inheritance, but these cases proved rare3 and thus not necessarily representative of generalized epilepsies. Subsequently, both genome-wide association studies4,5 and rare variant association studies6-9 investigated increasingly larger cohorts of genetic generalized epilepsies (GGEs). These studies provided key insights into the heritability and genetic architecture of GGE, which seems to involve ultra-rare genetic variants,6,8,9 common variants,4,5,10 and copy number alterations.11-13 Repeat expansions have also been recently implicated in dominantly inherited Familial Adult Myoclonic Epilepsy syndromes.14-17
Prior large-scale sequencing studies of individuals with familial GGE failed to show statistically significant associations in single genes.6,7 Nonetheless, gene set burden analyses in these studies demonstrated that ultra-rare coding variants (URVs) in multiple phenotypically and biologically informed gene sets (e.g., dominant epilepsy and developmental epileptic and encephalopathy (DEE) genes, genes encoding GABAA receptors) are associated with an increased disease risk.6,7 These patterns were later replicated in independent case-control studies of predominantly sporadic GGE cases, which found few single genes approached study-wide significance despite much larger cohorts.8,9 A paradigm in which familial and sporadic epilepsy may have different genetic architectures has been previously established by our work on non-acquired focal epilepsies (NAFEs) demonstrating a markedly higher burden of URVs in familial compared to sporadic NAFE.6 This, however, was not investigated so far in GGE.
Aiming to identify protein coding genes where URVs are significantly associated with an increased risk of generalized epilepsy, we performed a combined analysis of multiple cohorts of individuals with GGE and ancestry-matched controls. To improve the power of genetic discovery, we enriched our analysis with individuals with a positive family history of the disease, and also examined this subset of familial GGE separately. In additional analyses, we investigated individuals with sporadic GGE to understand if familial and sporadic GGE had different genetic architectures.
Methods:
Study design and participants:
In this case-control rare variant association study, we investigated the association of ultra-rare and rare genetic variants with epilepsy in individuals with a diagnosis of a GGE and matched controls of European descent. We jointly analyzed whole exome sequencing (WES) data from two independent datasets encompassing GGE patients previously studied by (1) the Epi4K Consortium and the Epilepsy Phenome/Genome Project6 (referred to hereafter, along with matched controls, as the first dataset) or (2) the Canadian Epilepsy Network and the EpiPGX, and EuroEPINOMICS-CoGIE Consortia7 (referred to, with their matched controls, as the second dataset). Control cohorts were obtained for the first dataset from local collections available at the Institute for Genomic Medicine9,18 (IGM) (New York, USA), and for the second dataset from controls available at the Luxembourg Centre for Systems Biology (LCSB) (Esch-sur-Alzette, Luxembourg) obtained from the database of Genotypes and Phenotypes19 or the Epi25 Collaborative.8 Ethical approvals from Institutional Review Boards and relevant Ethics Committees and written informed consent procedures were previously obtained and detailed elsewhere.6,7 The details of the recruitment or acquisition of analyzed case or control cohorts, diagnostic and inclusion criteria were also previously described.6-8 Here, we intended primarily to identify genes significantly increasing the risk of GGE by combining these cohorts. To that aim, we analyzed data from 2,203 affected individuals (1,214 from the first dataset and 989 from the second dataset; before quality control). Subsequently, we examined the strength of the association separately in 1,035 individuals (659 from the first dataset; 376 from the second dataset) with a positive family history of epilepsy. Afterwards, we went on to assess the remaining 1,168 individuals (555 from the first dataset; 613 from the second dataset) without a family history or with an unknown family history status.
Sequencing and quality control:
WES data generation for the case and control cohorts was previously described.6-8 In compliance with privacy regulations, the genotypes from the two datasets were processed in parallel at the IGM and the LCSB. A neural network predictive model was used to exclude individuals unlikely to be of a non-Finnish European descent. We removed one sample from each pair of duplicates/related individuals within each dataset and one sample from each pair of duplicates between the two datasets. We also performed quality control procedures to remove low quality samples/variants as well, to harmonize the coverage and call rate between the cases and controls within each dataset. Contingent on case-control matching, the final number of cases or controls included in each analysis (all, familial and sporadic GGEs analyses) differed slightly across analyses (see the results). The joint analysis strategy and the quality control procedures are outlined in Fig. S1 and detailed in the supplemental methods (see the Supplementary Material).
Variant annotations:
The analysis was limited to coding variants located in the exons of 18,834 protein coding genes from the consensus coding sequence20 (CCDS) release 20, extended with two bases on each side to accommodate canonical intronic splice sites. Variant effects were annotated using ClinEff21 v1.0c. Population allele frequencies were estimated from the Genome Aggregation Database22 (gnomAD r2.1) and DiscovEHR23 database v1. Since a portion of our control samples overlapped with gnomAD r2.1 exomes (see the Supplementary Material), gnomAD allele frequencies were based on gnomAD r2.1 genomes. Missense variants were further annotated with three in silico deleteriousness and intolerance scores (selected based on our previous work6,9): Polyphen2 (PPh2) Human Diversity based score,24 the Rare Exome Variant Ensemble Learner (REVEL) score25 and the Missense Tolerance Ratio (MTR) v1 score.26 The population allele frequencies and in silico missense deleteriousness and intolerance scores were annotated for the first dataset (and its matched controls) using the Analysis Tool for Annotated Variants (ATAV) platform18 and for the second dataset (and its matched controls) using Annovar27 and bcftools.28
Analysis models:
We defined three primary analysis models to examine the association of functional coding variation with GGE, based on a combination of three filtering criteria: minor allele frequency, variant types (effects), and in silico predictions (specifically for missense variants). We targeted URVs which we defined as those with a minor allele frequency (MAF) < 0.05% in our test datasets (internal MAF) and not seen in independent gnomAD & DiscovEHR population reference datasets (external MAF). Functional variants (i.e., presumed to affect the function of protein coding genes products) included those with predicted Loss-of-Function (pLoF: canonical splice-site, stop-gain & frameshift variants), in-frame insertions and deletions and missense variants. For each of the three models, missense variants were filtered further based on their expected (in silico) deleteriousness predicted using PPh2, REVEL or based on REVEL in combination MTR to capture the degree of sub-genic intolerance of the affected site. The latter approaches based on REVEL & MTR (i.e., analysis of deleterious variants identified with an ensemble method designed for rare variants in combination with sub-genic intolerance limiting) were recently shown to improve pathogenicity prediction in epilepsy and other disorders.9,26,29 A control model targeting synonymous URVs presumed to have a neutral effect was used to assess potential biases in cases vs. controls comparisons that are unlikely to be related to disease risk. We supplemented our primary analyses with additional secondary models to examine the association of (i) rare functional variants (defined as those with both internal and external MAFs lower than 0.1%) with and without URVs and (ii) pLoF variants without other types of functional variants (as these represent a class of high effect variants). Altogether, eight models were investigated (one control model, three primary models, and five secondary models) as summarized in Table 1.
Table 1:
Overview of association analysis models.
| Models | Primary models | Secondary models | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | Ultra-rare functional variants | Rare functional variants | Loss of Function variants | ||||||
| Synonymous | PPh2 | REVEL | MTR | Rare variants + URVs |
Rare variants − URVs |
Rare variants + URVs |
Rare variants − URVs |
URVs only | |
| Minor Allele Frequencies | |||||||||
| Internal MAF | <0.0005 | <0.0005 | <0.0005 | <0.0005 | <0.001 | without URVs | <0.001 | without URVs | <0.0005 |
| DiscovEHR MAF | 0 | 0 | 0 | 0 | <0.001 | <0.001 | 0 | ||
| gnomAD r2 MAF | 0 | 0 | 0 | 0 | <0.001 | <0.001 | 0 | ||
| Classes of Variants | |||||||||
| ClinEff Effects | Synonymous | Functional | Functional | Functional | Functional | Functional | pLoF | pLoF | pLoF |
| Missense variants filters | |||||||||
| PPh2 prediction | - | "Probably" | - | - | "Probably" | "Probably" | - | - | - |
| REVEL score | - | - | ≥ 0.5 | ≥ 0.5 | - | - | - | - | - |
| MTR score | - | - | - | ≤ 0.78 | - | - | - | - | - |
MAF: Minor Allele Frequency. PPh2: Polyphen 2 Human Diversity based prediction. REVEL: Rare Exome Variant Ensemble Learner. MTR: Missense Tolerance Ratio score. pLoF: predicted loss-of-function variants. pLoF variants included stop-gain & stop-loss variants, frameshift insertions & deletions, and canonical splice-site variants. Functional variants included pLoF, in-frame insertions & deletions, and missense variants (the missense variants were filtered using PPh2, REVEL, and MTR predictions as indicated). MAF from gnomAD were based on the ‘genomes’ subset. The cut-offs for REVEL & MTR scores were based on Ref.8
Gene-level associations:
As adopted in our previous studies,6 we performed gene collapsing analyses by assigning a 1 or 0 indicator in a gene by sample matrix to indicate the presence or absence (respectively) of qualifying variants. Qualifying variants (QVs) were defined as variants matching the criteria for each analysis model in a given gene and study individual (assuming dominant inheritance). The collapsing analysis was performed separately in our two independent study datasets and a Cochran–Mantel–Haenszel exact test (CMH) was then used to quantify the gene-level association between case status and QV carrier status9 (by comparing the counts of cases and controls with QVs in the two datasets while accounting for cohort stratification). Separate comparisons were performed for all, familial and sporadic GGE cohorts, each against their ancestry-matched controls. We adopted a Bonferroni multiple testing correction for gene-level p values (α = 0.05) accounting for three phenotypic groups, three primary analysis models and 18,834 protein-coding genes with a study-wide significance cut-off of 2.9 x 10−7. The homogeneity in the observed odds between the two data sets was examined using Breslow-Day and Woolf tests. The genomic inflation factor (λ) was estimated as detailed in the supplements. The collapsing and subsequent joint statistical analyses were performed using ATAV18 or R data.table,30 R tidyverse,31 and R stats32 on R32 v3.3.
Gene set associations analyses:
We also studied two gene sets that are important for inhibitory signaling in which GGEs had previously shown an increased burden of deleterious URVs. This association was established in a subset of our current samples7 and was later validated in additional datasets.8 However, a stratified analysis based on family history was not performed in our previous work. Here, we examined the association of URVs in these gene sets with familial GGE (vs. controls), with sporadic GGE (vs. controls), and directly between individuals with familial GGE vs. those with a sporadic GGE. We complemented these comparisons with an analysis of all GGEs vs. controls (as a positive control). To measure the association, we did gene set collapsing analyses by collapsing QVs across all genes in the investigated gene set (i.e., a case/control was a carrier if they harbored a QV in any gene in the gene set) followed by CMH test. P values from the analyses of functional variants were adjusted for 24 multiple tests (four phenotypic comparisons, three URV analysis models, and two gene sets) using a Benjamini-Hochberg False Discovery Rate (FDR) procedure to maximize the power (as opposed to Bonferroni correction for Familywise Error Rate).
Results:
We studied the association of coding URVs with generalized epilepsy in a cohort of 1,928 unrelated individuals diagnosed with familial or sporadic GGE and 8,578 matched controls of European descent. We also performed separate association studies for individuals diagnosed with familial GGE (n = 945, studied against 8,626 matched controls) and individuals with a diagnosis of a sporadic GGE (n = 1,005, studied against 8,621 matched controls; all counts after quality control). As case-control matching on principal components was performed separately for each analysis, the total number of controls differed slightly between the analyses, and the total number of samples in the analysis of all individuals with GGE was slightly less than the total of familial and sporadic cases. The sample counts from the two study cohorts are detailed in the Supplementary Material (Table S1). We did not detect a prominent deviation of observed p values from expected p values in synonymous collapsing analysis (λ = 0.86 – 1.06, Fig. S5) indicating adequate population substructure matching between individuals with epilepsy (cases) and without epilepsy (controls).
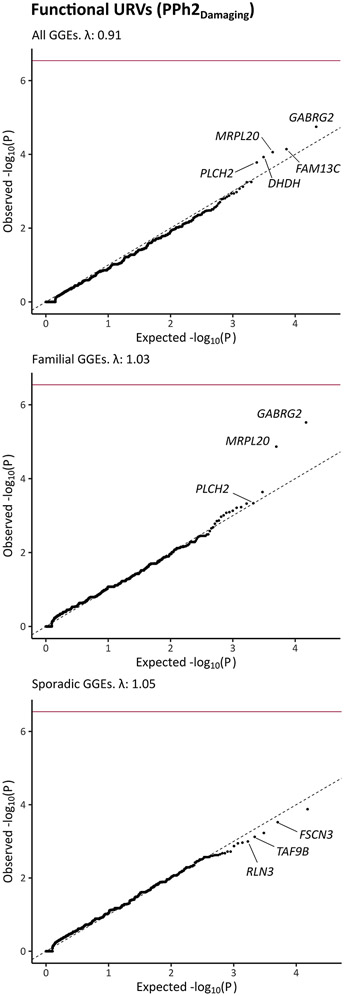
No single gene achieved study-wide significance. However, GABRG2 (Mendelian Inheritance in Man33 (MIM) gene number: 137164) was the top-ranked gene in the analysis of the combined GGE cohort, showing prominent association in two primary models; it had p value of 1.8x10−5 in the PPh2 model (examining the association of functional URVs while filtering missense variants based on a damaging Polyphen2 prediction; Fig. 1) and p value of 1.2x10−5 in the MTR model (combining sub-genic intolerance with REVEL; Fig. S7). Limiting the cases to individuals with a family history of epilepsy strengthened the association with GABRG2 in the PPh2 model (p = 3.0x10−6). Using REVEL combined with MTR did not outperform the PPh2 model in terms of significance in the analysis of familial GGEs (p = 1.4x10−5). Nonetheless, it maximized the separation between cases and controls, resulting in higher odds by preferentially filtering all GABRG2 variants seen in our control sets (Table 2, Tables S2 - S3).
Fig. 1: Association of ultra-rare variation in protein coding genes with genetic generalized epilepsy.
The quantile-quantile plots compare the observed p values (Cochran-Mantel-Haenszel exact test) and the expected p values (drawn from a uniform distribution) in analyses of 1,928 individuals genetic generalized epilepsies (GGEs) in comparison to 8,578 matched controls (top panel) as well as subsequent analyses of familial GGEs (middle panel; 945 cases and 8,626 controls) or sporadic GGEs (bottom panel; 1,005 cases and 8,621 controls). These analyses focused on functional ultra-rare variants (URVs; with minor allele frequencies < 0.05% in the test dataset & not seen in DiscovEHR/gnomAD) that were annotated as predicted Loss-of-Function variants (pLoF), damaging missense variants with Polyphen2 (PPh2), or in-frame insertions and deletions. Study-wide significance after Bonferroni correction (dark red line) was defined by a p value < 2.9 x 10−7. λ: Genomic inflation factor. Among the five top-ranked genes, genes that had a higher carrier frequency in cases vs. controls in both study datasets are labeled (not labeled among top-ranked: enriched in controls or in cases in one dataset only).
Table 2:
Top-ranked genes in the primary analyses of ultra-rare functional variants.
| Analysis | URVs | HGNC | Epilepsy gene |
Qualifying Cases | Qualifying Controls | OR (95% CI) |
P value (homogeneity) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1st Dataset |
2nd Dataset |
Both Datasets |
1st Dataset |
2nd Dataset |
Both Datasets |
||||||
| All GGEs | PPh2 | GABRG2 | yes | 7 (0.64%) | 3 (0.36%) | 10 (0.52%) | 3 (0.04%) | 1 (0.06%) | 4 (0.05%) | 12.1 (3.4 – 54.1) | 1.8 x 10−5 (0.54) |
| REVEL | 4 (0.36%) | 3 (0.36%) | 7 (0.36%) | 0 (0.00%) | 1 (0.06%) | 1 (0.01%) | 28.3 (3.4 – 1307.3) | 1.3 x 10−4 (0.15) | |||
| MTR | 4 (0.36%) | 3 (0.36%) | 7 (0.36%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | ∞ (6.1 – ∞) | 1.2 x 10−5 (0.53) | |||
| Familial GGEs | PPh2 | GABRG2 | yes | 6 (0.95%) | 2 (0.63.%) | 8 (0.85%) | 3 (0.04%) | 1 (0.06%) | 4 (0.05%) | 18.9 (5 – 86.5) | 3.0 x 10−6 (0.63) |
| REVEL | 3 (0.48%) | 2 (0.63%) | 5 (0.53%) | 0 (0.00%) | 1 (0.06%) | 1 (0.01%) | 40.6 (4.4 – 1934.3) | 1.0 x 10−4 (0.19) | |||
| MTR | 3 (0.48%) | 2 (0.63%) | 5 (0.53%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | ∞ (7.9 – ∞) | 1.4 x 10−5 (0.64) | |||
| Sporadic GGEs | PPh2 | FAM13C | - | 5 (0.81%) | 0 (0.00%) | 5 (0.50%) | 4 (0.05%) | 0 (0.00%) | 4 (0.05%) | 17.6 (3.8 – 89.0) | 1.3 x 10−4 (0.44) |
| REVEL | TNFRSF21 | - | 2 (0.47%) | 2 (0.39%) | 4 (0.40%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | ∞ (5.1 – ∞) | 2.3 x 10−4 (0.52) | |
| MTR | TRPV5 | - | 3 (0.70%) | 0 (0.00%) | 3 (0.30%) | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) | ∞ (5.8 – ∞) | 3.0 x 10−4 (0.18) | |
Odds Ratio (OR) and p values are given from a Cochran-Mantel-Haenszel exact test. No gene reached study-wide significance (p < of 2.9 × 10−7). The accompanying homogeneity p value indicates the lowest p value from Breslow-Day & Woolf tests for homogeneity of odds, where p values < 0.05 indicate significantly different odds between the two analysis datasets. CI: Confidence Interval. HGNC: HUGO (Human Genome Organization) Gene Nomenclature Committee gene names. QVs: qualifying variants. URVs: Ultra-rare Variants. See Table 1 for the details of the PPh2, REVEL & MTR analysis models.
The analysis of sporadic GGEs was generally unremarkable for GABRG2 (p = 0.15 – 0.015) and the top-ranked genes did not include biologically meaningful candidates (Table 2). Rare variant analyses (up to a MAF of 0.1%) resulted in the inclusion of additional GABRG2 variants exclusively in the control cohorts (Table S3). In general, secondary analyses of rare functional and pLoF variants neither captured significantly associated single genes nor strong novel candidates with biological relevance (Tables S4 - S5 and Figs. S8 - S9). Although not study-wide significant, GABRG2 achieved a higher rank than in our prior URVs analysis6 in 640 familial GGEs vs. 3,877 controls using an analysis model comparable to the current PPh2 model (rank 7, p = 9.2x10−4). Its rank was higher than that seen in two recent large-scale analyses from the Epi25 Collaborative8,9 in 3,108 GGEs vs. 8,436 controls (rank 3, p = 6.2 x 10−4) and 5,303 GGEs vs. 15,677 controls (rank = 37, p = 6.1 x 10−3).
Most URVs in GABRG2 were missense and four URVs were seen in individuals with absence seizures (Table S2). Eight variants were seen in the familial GGE cohort including two that were confirmed to be inherited: p.R177P, identified in a proband with Early Onset Absence Epilepsy (EOAE), was inherited from a parent with a similar phenotype, whereas p.Y213* was inherited from a parent not diagnosed with epilepsy (Fig. S10). We did not have sufficient data and samples to determine the allelic origin of the remaining variants (p.A160S, p.M199V, p.V252A, p.G413S, p.D450Y and p.N456S). Two other canonical splice donor variants (IVS2+2G>T and IVS6+2G>T) were seen in our sporadic GGE cohort. The predicted protein changes for these variants are based on the transcript NM_198904.
Few of these URVs were recurrent. p.M199V (familial GGE) was reported in a previous study,34 in which it segregated with a phenotype of Generalized Epilepsy with Febrile Seizures Plus (GEFS+) and also in an individual with NAFE in the first Epi25 Collaborative study.8 p.R177P (familial GGE) affected a codon for which a different change (p.R177G) was seen previously in a family with febrile seizures35 (FS). IVS2SD (sporadic GGE) was previously reported in ClinVar (ID: VCV001067627.1) as likely pathogenic for Childhood Absence Epilepsy (CAE) and FS (no details on family history). Last, IV6SD was previously associated with familial CAE and FS.36 The patient included here (sporadic GGE) had absence epilepsy but no history of FS. Sample overlap or relatedness to these previously reported individuals was not investigated genetically but it was considered unlikely based on our patients’ clinical and family histories.
Apart from GABRG2, there was little overlap between the leading associations in the recent analyses6,8,9 and this study (Table S6). CACNA1B [MIM: 601012], the top hit in our prior analysis6 (p = 1.7x10−5), showed a less prominent association than previously seen (rank 5 in the MTR model/familial GGEs; p = 0.00098). Our analysis also did not recapture two genes previously seen as top hits with suggestive association8,9 (CACNA1G [MIM: 604065] with p = 2.5x10−4 and SLC6A1 [MIM: 137165] with p = 2.1x10−6). GABRA1 [MIM: 137160] was among few shared top hits, achieving comparable ranks in all studies (rank 9 in the MTR model analysis of all GGEs with p = 0.0023; rank 8 in the 1st Epi25 Collaborative study8 with p = 0.0022; rank 9 in the 2nd study9 with p = 0.0013). Few other MIM genes previously suggested to increase the susceptibility to GGE were among the top hits (Tables S7 - S8). On the other hand, several MIM genes underlying dominant DEEs were among the top-ranked genes (Table S9), as expected from the known enrichment of such URVs in genes causing dominant DEEs in generalized epilepsies.6,8,9
The association of URVs in two gene sets important for GABAergic signaling (genes encoding GABAA receptors and GABAergic pathway genes) with the phenotype was not prominent in the analysis of deleterious URVs, whereas the incorporation of sub-genic intolerance in the definition of QVs improved the power9 and unraveled clear association signals in the analysis of all GGEs (Fig. 2A) and familial GGEs (Fig. 2B). It also aided the identification of an association between genes encoding GABAA receptors and sporadic GGEs, though weaker than that observed in comparisons of familial GGEs vs. controls (Fig. 2C). We did not detect an association between GABAergic pathway genes and sporadic GGE as expected from previous findings,8 possibly due to insufficient power or differences in the analysis models (Fig 2C). The outcomes of a direct comparison of 945 individuals with familial GGE vs. 1,005 individuals with sporadic GGE was unremarkable and also likely underpowered (Fig. 2D).
Fig. 2: Association of ultra-rare variation in genes encoding GABAA receptors with familial and sporadic genetic generalized epilepsy.
The forest plots show the association of ultra-rare deleterious and intolerant variants with the phenotype in analyses of 1,928 individuals with GGE vs. 8,578 controls (A), 945 individuals with familial GGE vs. 8,626 controls (B), 1,005 individuals with sporadic GGE vs. 8,621 controls (C), and a direct comparison of 945 individuals with familial GGE vs. 1,005 individuals with sporadic GGE (D). Four (primary and control) ultra-rare variant models are shown (y axis). The association in each analysis is displayed as the natural logarithm of stratified odds ratio from a Cochran-Mantel-Haenszel exact test (x axis). Errors bars indicated the logarithm of the 95% confidence intervals (CI). The corresponding odds ratios and associated p values, and False Discovery Rate (FDR) adjusted p values are displayed on the side. The tests for synonymous variants were not adjusted for multiple testing.
Discussion:
Here, we add to the evidence indicating that deleterious URVs in GABRG2 are a risk factor for generalized epilepsies, though this gene did not reach study-wide significance. Notably, this association appears to be driven by ultra-rare private variants rather than rare variants (possibly seen in external population controls). This work emphasizes the role of ultra-rare variation in less severe epilepsies and corroborates the association of coding variation in GABRG2 with familial GGE.6,8,9,37 The current analysis benefits from a higher number of individuals with familial GGE and a balanced distribution of familial and sporadic cases compared to recent large-scale analyses8,9 enriched for sporadic GGEs. Our attempts to integrate multiple cohorts from independent studies to achieve this larger sample size came with some limitations. Quality control and harmonization measures mandated the exclusion of putative qualifying variants in genes of interest. The restrictions in genotype sharing across study sites limited the possibilities to invoke analysis methods incorporating covariates to handle residual population stratification. Also, the use of phenotypic definitions and classifications from independent studies might have resulted inadvertently in minor inconsistencies in sample stratification across the familial and sporadic cohorts (which included individuals with unknown family history status).
Absence seizures, a seizure type that was prominent in earlier GABRG2 families featuring an overlap of GGE and GEFS+,38-40 was also predominant among individuals with QVs in GABRG2 in this cohort (Table S2). The phenotypes in individuals with a positive family history and their affected siblings or parents were mostly congruent (Fig. S10). The small number of affected individuals and the limited segregation analysis precluded reliable estimation of penetrance or heterogeneity. Although the segregation of GABRG2 variants could be studied only in two families, these showed that pathogenic variants could be inherited both from affected and non-affected parents. This is concordant with prior observations that penetrance was typically incomplete and that GABRG2-related GGE had complex inheritance; most inherited pathogenic GABRG2 variants had reduced penetrance sometimes with phenotypic heterogeneity whereas de novo variants were more prevalent in individuals with severe or developmental phenotypes (Table S10).
The lack of study-wide significance in rare variant association studies in GGE and the failure to reproduce multiple leading associations speak to the marked genetic heterogeneity of GGEs. The exact extent of the contribution of rare coding variation in GGE heritability is largely unknown. It remains, therefore, difficult to speculate on the interpretation of any negative findings, and on whether a further increase in statistical power might result in suggestive associations reaching significance. Using a similar study design to the one used to examine the current set (slightly exceeding 10,000 samples), we estimate that a total sample size exceeding 16,000 samples would be required to achieve study-wide significance in a gene with rates of qualifying variants similar to those observed in GABRG2. These carrier rates seem, however, to be an upper-bound estimate due to the multitude of familial GGEs included here; the sample size required is probably much larger when examining sporadic GGEs.8,9
Nonetheless, the observed association of GABRG2 with GGE further validates the outcomes of an analysis performed by the Epi25 Collaborative8 (albeit with partial overlap in datasets; see the Supplementary Material). The prominent difference in GABRG2 rank in a second iteration9 of the Epi25 study with an expanded sample size might be explained by the familial origin of GABRG2 variants, since both studies had considerably lower ratios of familial to sporadic GGEs (approximately 1:7 ratio). GABRG2 was also the lead association in a burden analysis of pLoF URVs (p value = 6.9x10−5) in a recent study investigating the exomes of 3,999 individuals with epilepsy (without further phenotypic sub-classification) vs. 277,586 controls from the UK Biobank.41 The different definitions of qualifying variants in these studies might also explain the variable outcomes. Our PPh2 analysis model is similar to the prior model we used to analyze a subset of our samples6 (thus allowing for comparisons of outcomes with the increase in sample size).
Compared to PPh2 filtering, GABRG2 URVs had higher odds of association with GGE when missense variants were filtered using REVEL, in line with REVEL’s higher performance in discriminating pathogenic and benign rare variants.25 Additional filtering on sub-genic intolerance (MTR) increased the odds further, consistent with recent findings suggesting that sub-genic intolerance filtering is particularly effective for analyses geared towards specificity as opposed to sensitivity.9,29 REVEL and MTR cut-offs similar to those utilized in the most extensive and recent rare variant association study on epilepsy were used. Different values for these filters maximize the separation of benign and pathogenic variants in different types of epilepsy.9 However, most functionally validated GABRG2 variants previously implicated in epilepsy fit one or more of the qualifying variant models we used, indicating good recall of disease-related variants with the current parameters (Table S10).
The recurrence of the same GABRG2 variants in individuals with different types of epilepsy (GEFS+, DEE, GGE & NAFE; Table S10), as well as in familial and sporadic GGEs with overlapping phenotypes, underscores a considerable genetic overlap and possibly a complex inheritance. Although we found the most substantial contribution from deleterious variants not seen in gnomAD and DiscovEHR databases, a small contribution from rare variants or variants with benign predictions to this complex genetic predisposition cannot be ruled out (for instance, p.N79S previously identified in individuals with GGE or NAFE causes subtle functional alterations 8,42-44 and is seen in 3 individuals in gnomAD release 3.1.2). The GABRG2 locus was recently found to be associated with febrile seizures,45 highlighting the role of common variants in a phenotype that was prominent in earlier families with an increased susceptibility to GGE and GEFS+ linked to rare GABRG2 variants.38-40
Prior burden analyses also revealed the presence of shared patterns of risk determinants between severe epilepsies (DEE) and common epilepsies (GGE & NAFE) in gene sets that are key for inhibitory signaling.7,8 A former analysis (in 3,108 individuals with GGE) did not capture a considerable change in URVs burden in genes encoding GABAA receptors or GABAergic pathway genes upon the exclusion of a relatively small subset (n = 380) of familial samples.8 Conversely, we found a more prominent association between ultra-rare coding variation in GABAergic pathway genes and familial GGE in comparison to its association with sporadic GGE, albeit not demonstrable in direct (familial vs. sporadic) comparisons. Direct comparisons with sufficient power could help confirm the subtle differences in risk profiles.
In summary, we show that URVs in GABRG2 are potentially an important risk factor for GGE, though not reaching study-wide significance. The association of URVs in genes representing the GABAergic pathway is likely more prominent in familial GGE than in sporadic GGE. Future work on epilepsy cohorts enriched with familial cases, extending the analysis to additional types of genetic variation (e.g., alterations in copy numbers and repeats, rare intronic and regulatory variants, and common risk alleles), could further our understanding of the genetic heterogeneity in GGE and the evidently complex inheritance.
Supplementary Material
Key points:
Although not study-wide significant, GABRG2 is likely an important risk gene for genetic generalized epilepsy (GGE).
Compared to controls, ultra-rare coding variants (URVs) in GABRG2 are seen more frequently in individuals with a familial GGE than a sporadic GGE.
Similarly, the association of URVs in GABAergic pathway genes is more prominent with familial GGE than with sporadic GGE.
Acknowledgments:
We thank the individuals who participated in CENet, Epi4K, EP/GP, EpiPGX, and EuroEPINOMICS-CoGIE studies. This work was supported by the Research Unit FOR-2715 of the German Research Foundation and the Fond Nationale de la Recherche of Luxembourg (DFG/FNR grants INTER/DFG/17/11583046 and Le1030/16-1) and foundation ‘no epilep’ (to HL). MK was support by the German Academic Exchange Service (DAAD program number 57214224; doctoral grant to MK). JEM is supported by the National Institutes of Health (TL1TR001875). PM obtained FNR funding as part of the National Centre of Excellence in Research on Parkinson’s disease (NCER-PD, FNR11264123). CENet was funded by joint funding from Genome Canada and Genome Quebec. The Epi4K consortium and EP/GP were supported by grants from the National Institute of Neurological Disorders and Stroke and Epilepsy-Research UK. The EpiPGX projects were supported by the European Commission Sixth and Seventh Framework Programmes. The EuroEPINOMICS project was supported by the European Science Foundation through contributing national funding agencies. We are thankful to the Epi25 Collaborative for providing access to their control cohort of Italian individuals that was of great help to match the southern European samples. This work used sequence data available through dbGaP under the accession numbers phs000806 and phs000572 or the European Nucleotide Archive under the accession number PRJEB20726. The full acknowledgement statement is provided in the supplements.
Appendix
Canadian Epilepsy Network (CENet): Simon Girard, Claudia Moreau, Véronique Cloutier, Caroline Meloche, Micheline Gravel, Dang K. Nguyen, Anne Lortie, Richard Desbiens, Cécile Cieuta-Walti, and Patrick Cossette.
Epi4K Consortium: Andrew S. Allen, Susannah T. Bellows, Samuel F. Berkovic, Joshua Bridgers, Rosemary Burgess, Gianpiero L. Cavalleri, Seo-Kyung Chung, Patrick Cossette, Norman Delanty, Dennis Dlugos, Michael P. Epstein, Catharine Freyer, David B. Goldstein, Erin L. Heinzen, Michael S. Hildebrand, Michael R. Johnson, Ruben Kuzniecky, Daniel H. Lowenstein, Anthony G. Marson, Caroline Mebane, Heather C. Mefford, Terence J. O’Brien, Ruth Ottman, Steven Petrou, Slavé Petrovski, William O. Pickrell, Annapurna Poduri, Rodney A. Radtke, Mark I. Rees, Brigid M. Regan, Zhong Ren, Ingrid E. Scheffer, Graeme J. Sills, Rhys H. Thomas, and Quanli Wang.
Epilepsy Phenome/Genome Project (EP/GP): Brian K. Alldredge, Dina Amrom, Eva Andermann, Jocelyn F. Bautista, Samuel F. Berkovic, Judith Bluvstein, Alex Boro, Gregory D. Cascino, Damian Consalvo, Patricia Crumrine, Orrin Devinsky, Dennis Dlugos, Michael P. Epstein, Miguel Fiol, Nathan B. Fountain, Jacqueline French, Catharine Freyer, Daniel Friedman, Eric B. Geller, Tracy Glauser, Simon Glynn, Kevin Haas, Sheryl R. Haut, Sucheta Joshi, Andres Kanner, Heidi E. Kirsch, Robert C. Knowlton, Eric H. Kossoff, Ruben Kuzniecky, Daniel H. Lowenstein, Paul V. Motika, Edward J. Novotny, Ruth Ottman, Juliann M. Paolicchi, Jack M. Parent, Kristen Park, Annapurna Poduri, Lynette G. Sadleir, Ingrid E. Scheffer, Renée A. Shellhaas, Elliott H. Sherr, Jerry J. Shih, Shlomo Shinnar, Rani K. Singh, Joseph Sirven, Michael C. Smith, Joseph Sullivan, Liu Lin Thio, Anu Venkat, Eileen P. G. Vining, Gretchen K. Von Allmen, Judith L. Weisenberg, Peter Widdess-Walsh, and Melodie R. Winawer.
EpiPGX Consortium: Stefan Wolking, Sarah Peter, Felicitas Becker, Yvonne G. Weber, Sarah Weckhuysen, Rikke S. Møller, Marina Nikanorova, Hiltrud Muhle, Andreja Avbersek, Costin Leu, Pasquale Striano, Antonio Gambardella, Sarah R. Langley, Martin Krenn, Karl M. Klein, Bianca Berghuis, Andrea Jorgensen, Pauls Auce, Ben Francis, Anja C. M. Sonsma, Josemir W. Sander, Fritz Zimprich, Chantal Depondt, Michael R. Johnson, Anthony G. Marson, Graeme J. Sills, Wolfram S. Kunz, Gianpiero L. Cavalleri, Norman Delanty, Federico Zara, Roland Krause, Holger Lerche, and Sanjay M. Sisodiya.
EuroEPINOMICS-CoGIE Consortium: Patrick May, Julian Schubert, Dheeraj R. Bobbili, Felicitas Becker, Stefan Wolking, Stéphanie Baulac, Janine Altmüller, Pasquale Striano, Hande Caglayan, Auli Siren, Kate Everett, Rikke S. Møller, Marina Nikanorova, Hiltrud Muhle, Ingo Helbig, Wolfram S. Kunz, Yvonne G. Weber, Sarah Weckhuysen, Sanjay M. Sisodiya, Rima Nabbout, Thomas Sander, Eric LeGuern, Bobby P. C. Koeleman, Anna-Elina Lehesjoki, Peter Nürnberg, Federico Zara, Roland Krause, and Holger Lerche.
Footnotes
Disclosure of Conflicts of Interest: RSD is a paid consultant of AstraZeneca. MK, JEM, KES, DRB, and PM have no conflicts of interest related to this article to disclose. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines. The funding agencies had no role in study design; in the collection, analysis, and interpretation of data; and in the writing and the decision to submit the paper for publication.
- Supplementary Material: Supplemental methods, results, acknowledgements and authors’ information (PDF file).
- Additional supporting data (the complete results of the presented gene-level association) are publicly available: Koko et al. (2021), “Supplements to the article: Association of ultra-rare coding variants with genetic generalized epilepsy”, Mendeley Data, V1, https://doi.org/10.17632/5p556jtwt9.1.
References:
- 1.Berkovic SF, Howell RA, Hay DA, Hopper JL. Epilepsies in twins: Genetics of the major epilepsy syndromes. Ann Neurol. 1998;43(4):435–45. 10.1002/ana.410430405. [DOI] [PubMed] [Google Scholar]
- 2.Peljto AL, Barker-Cummings C, Vasoli VM, Leibson CL, Hauser WA, Buchhalter JR, et al. Familial risk of epilepsy: a population-based study. Brain. 2014;137(3):795–805. 10.1093/brain/awt368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poduri A, Lowenstein D. Epilepsy genetics—past, present, and future. Curr Opin Genet Dev. 2011;21(3):325–32. 10.1016/j.gde.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The International League Against Epilepsy Consortium on Complex Epilepsies. Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol. 2014;13(9):893–903. 10.1016/S1474-4422(14)70171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The International League Against Epilepsy Consortium on Complex Epilepsies. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat Commun. 2018;9(1):5269. 10.1038/s41467-018-07524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Epi4K Consortium and the Epilepsy Phenome/Genome Project. Ultra-rare genetic variation in common epilepsies: a case-control sequencing study. Lancet Neurol. 2017;16(2):135–43. 10.1016/S1474-4422(16)30359-3. [DOI] [PubMed] [Google Scholar]
- 7.May P, Girard S, Harrer M, Bobbili DR, Schubert J, Wolking S, et al. Rare coding variants in genes encoding GABAA receptors in genetic generalised epilepsies: an exome-based case-control study. Lancet Neurol. 2018;17(8):699–708. 10.1016/S1474-4422(18)30215-1. [DOI] [PubMed] [Google Scholar]
- 8.The Epi25 Collaborative. Ultra-Rare Genetic Variation in the Epilepsies: A Whole-Exome Sequencing Study of 17,606 Individuals. Am J Hum Genet. 2019;105(2):267–82. 10.1016/j.ajhg.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Epi25 Collaborative. Sub-genic intolerance, ClinVar, and the epilepsies: A whole-exome sequencing study of 29,165 individuals. Am J Hum Genet. 2021;108(6):965–82. 10.1016/j.ajhg.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leu C, Stevelink R, Smith AW, Goleva SB, Kanai M, Ferguson L, et al. Polygenic burden in focal and generalized epilepsies. Brain. 2019;142(11):3473–81. 10.1093/brain/awz292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Kovel CGF, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133(1):23–32. 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, et al. Genome-Wide Copy Number Variation in Epilepsy: Novel Susceptibility Loci in Idiopathic Generalized and Focal Epilepsies. PLoS Genet. 2010;6(5):e1000962. 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niestroj L-M, Perez-Palma E, Howrigan DP, Zhou Y, Cheng F, Saarentaus E, et al. Epilepsy subtype-specific copy number burden observed in a genome-wide study of 17 458 subjects. Brain. 2020;143(7):2106–18. 10.1093/brain/awaa171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishiura H, Doi K, Mitsui J, Yoshimura J, Matsukawa MK, Fujiyama A, et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat Genet. 2018;50(4):581–90. 10.1038/s41588-018-0067-2. [DOI] [PubMed] [Google Scholar]
- 15.Florian RT, Kraft F, Leitão E, Kaya S, Klebe S, Magnin E, et al. Unstable TTTTA/TTTCA expansions in MARCH6 are associated with Familial Adult Myoclonic Epilepsy type 3. Nat Commun. 2019;10(1):4919. 10.1038/s41467-019-12763-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corbett MA, Kroes T, Veneziano L, Bennett MF, Florian R, Schneider AL, et al. Intronic ATTTC repeat expansions in STARD7 in familial adult myoclonic epilepsy linked to chromosome 2. Nat Commun. 2019;10(1):4920. 10.1038/s41467-019-12671-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeetong P, Pongpanich M, Srichomthong C, Assawapitaksakul A, Shotelersuk V, Tantirukdham N, et al. TTTCA repeat insertions in an intron of YEATS2 in benign adult familial myoclonic epilepsy type 4. Brain. 2019;142(11):3360–6. 10.1093/brain/awz267. [DOI] [PubMed] [Google Scholar]
- 18.Ren Z, Povysil G, Hostyk JA, Cui H, Bhardwaj N, Goldstein DB. ATAV: a comprehensive platform for population-scale genomic analyses. BMC Bioinformatics. 2021;22(1):149. 10.1186/s12859-021-04071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, et al. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42(D1):D975–9. 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pujar S, O’Leary NA, Farrell CM, Loveland JE, Mudge JM, Wallin C, et al. Consensus coding sequence (CCDS) database: a standardized set of human and mouse protein-coding regions supported by expert curation. Nucleic Acids Res. 2018;46(D1):D221–8. 10.1093/nar/gkx1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, snpEff. Fly (Austin). 2012;6(2):80–92. 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–43. 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dewey FE, Murray MF, Overton JD, Habegger L, Leader JB, Fetterolf SN, et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354(6319):aaf6814. 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 24.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet. 2016;99(4):877–85. 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Traynelis J, Silk M, Wang Q, Berkovic SF, Liu L, Ascher DB, et al. Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res. 2017;27(10):1715–29. 10.1101/gr.226589.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164–e164. 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Dhindsa RS, Carss K, Harper AR, Nag A, Tachmazidou I, et al. Rare variant contribution to human disease in 281,104 UK Biobank exomes. Nature. 2021;597:527–532. 10.1038/s41586-021-03855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dowle, Srinivasan. data.table: Extension of `data.fram`. 2019. Available from: https://CRAN.R-project.org/package=data.table. [Google Scholar]
- 31.Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4(43):1686. 10.21105/joss.01686. [DOI] [Google Scholar]
- 32.R Core Team. R: A language and environment for statistical computing. 2017. Available from: https://www.R-project.org/
- 33.Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD), 2021. Accessed: 01-09-2021. Available from: https://omim.org/.
- 34.Boillot M, Morin-Brureau M, Picard F, Weckhuysen S, Lambrecq V, Minetti C, et al. Novel GABRG2 mutations cause familial febrile seizures. Neurol Genet. 2015;1(4):e35. 10.1212/NXG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Audenaert D, Schwartz E, Claeys KG, Claes L, Deprez L, Suls A, et al. A novel GABRG2 mutation associated with febrile seizures. Neurology. 2006;67(4):687–90. 10.1212/01.wnl.0000230145.73496.a2. [DOI] [PubMed] [Google Scholar]
- 36.Kananura C, Haug K, Sander T, Runge U, Gu W, Hallmann K, et al. A Splice-Site Mutation in GABRG2 Associated With Childhood Absence Epilepsy and Febrile Convulsions. Arch Neurol. 2002;59(7):1137. 10.1001/archneur.59.7.1137. [DOI] [PubMed] [Google Scholar]
- 37.Bennett CA, Petrovski S, Oliver KL, Berkovic SF. ExACtly zero or once: A clinically helpful guide to assessing genetic variants in mild epilepsies. Neurol Genet. 2017;3(4):e163. 10.1212/NXG.0000000000000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baulac S, Huberfeld G, Gourfinkel-An I, Mitropoulou G, Beranger A, Prud’homme J-F, et al. First genetic evidence of GABAA receptor dysfunction in epilepsy: a mutation in the γ2-subunit gene. Nat Genet. 2001;28(1):46–8. 10.1038/ng0501-46. [DOI] [PubMed] [Google Scholar]
- 39.Wallace RH, Marini C, Petrou S, Harkin LA, Bowser DN, Panchal RG, et al. Mutant GABAA receptor γ2-subunit in childhood absence epilepsy and febrile seizures. Nat Genet. 2001;28(1):49–52. 10.1038/ng0501-49. [DOI] [PubMed] [Google Scholar]
- 40.Marini C, Harkin LA, Wallace RH, Mulley JC, Scheffer IE, Berkovic SF. Childhood absence epilepsy and febrile seizures: a family with a GABAA receptor mutation. Brain. 2003;126(1):230–40. 10.1093/brain/awg018. [DOI] [PubMed] [Google Scholar]
- 41.Karczewski KJ, Solomonson M, Chao KR, Goodrich JK, Tiao G, Lu W, et al. Systematic single-variant and gene-based association testing of 3,700 phenotypes in 281,850 UK Biobank exomes. medRxiv. 2021. 10.1101/2021.06.19.21259117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi X, Huang M-C, Ishii A, Yoshida S, Okada M, Morita K, et al. Mutational analysis of GABRG2 in a Japanese cohort with childhood epilepsies. J Hum Genet. 2010;55(6):375–8. 10.1038/jhg.2010.47. [DOI] [PubMed] [Google Scholar]
- 43.Migita K, Yamada J, Nikaido Y, Shi X, Kaneko S, Hirose S, et al. Properties of a novel GABAA receptor γ2 subunit mutation associated with seizures. J Pharmacol Sci. 2013;121(1):84–7. 10.1254/jphs.12222sc. [DOI] [PubMed] [Google Scholar]
- 44.Huang X, Hernandez CC, Hu N, Macdonald RL. Three epilepsy-associated GABRG2 missense mutations at the γ+/β- interface disrupt GABAA receptor assembly and trafficking by similar mechanisms but to different extents. Neurobiol Dis. 2014;68:167–79. 10.1016/j.nbd.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skotte L, Fadista J, Bybjerg-Grauholm J, Appadurai V, Hildebrand MS, Hansen TF, et al. Genome-wide association study of febrile seizures identifies seven new loci implicating fever response and neuronal excitability genes. medRxiv. 2020. 10.1101/2020.11.18.20233916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.