Abstract
Thermal nociception involves the transmission of temperature-related noxious information from the periphery to the CNS and is a heritable trait that could predict transition to persistent pain. Rodent forward genetics complement human studies by controlling genetic complexity and environmental factors, analysis of end point tissue, and validation of variants on appropriate genetic backgrounds. Reduced complexity crosses between nearly identical inbred substrains with robust trait differences can greatly facilitate unbiased discovery of novel genes and variants. We found BALB/cByJ mice showed enhanced sensitivity on the 53.5°C hot plate and mechanical stimulation in the von Frey test compared to BALB/cJ mice and replicated decreased gross brain weight in BALB/cByJ versus BALB/cJ. We then identified a quantitative trait locus (QTL) on chromosome 13 for hot plate sensitivity (LOD = 10.7; p < 0.001; peak = 56 Mb) and a QTL for brain weight on chromosome 5 (LOD = 8.7; p < 0.001). Expression QTL mapping of brain tissues identified H2afy (56.07 Mb) as the top transcript with the strongest association at the hot plate locus (FDR = 0.0002) and spliceome analysis identified differential exon usage within H2afy associated with the same locus. Whole brain proteomics further supported decreased H2AFY expression could underlie enhanced hot plate sensitivity, and identified ACADS as a candidate for reduced brain weight. To summarize, a BALB/c reduced complexity cross combined with multiple-omics approaches facilitated identification of candidate genes underlying thermal nociception and brain weight. These substrains provide a powerful, reciprocal platform for future validation of candidate variants.
Keywords: Thermal nociception, hot plate, von frey, genetics, BALB/c, reduced complexity cross, brain weight, H2afy, macroH2A1
Introduction
Pain management poses large challenges for physicians globally and 11.2% of Americans report suffering from daily pain. 1 Pain is a known risk factor for several medical conditions including mood disorders, 2 suicidal behaviors, 3 and opioid misuse. 4 It is therefore vital to understand the genetic and neurobiological basis of pain traits to improve patient comfort and long-term health outcomes. Human familial studies of chronic pain have demonstrated heritability, 5 and twin studies have revealed shared genetic factors underlying several pain phenotypes. 6 Furthermore, human genome-wide association studies (GWAS) have identified loci associated with several modalities of chronic pain.7–10
Acute nociception is the transmission of nociceptive signals from peripheral nociceptors to the CNS that transmits the presence, type (chemical, mechanical, thermal), localization, and magnitude of the nociceptive stimulus.11–13 Acute nociception is critical for survival, as it alerts us to potential tissue damage and promotes avoidance. 14 Understanding the genetics and neurobiology of acute nociception is also important because its severity can often predict the transition to chronic pain.15,16
Thermal nociception refers to transmission of noxious information from the periphery to the CNS in response to heat- or cold-related stimuli and is highly heritable in mice (h2 = 0.59). 17 TRPV1, an important gene for thermal nociception, codes for transient receptor potential cation channel subfamily V member 1 (a.k.a., the capsaicin receptor)18–20 and has also has been implicated in inflammatory and neuropathic pain. 21 Furthermore, in humans single polymorphisms can reduce sensitivity to acute thermal pain and the severity of chronic pain, as is the case for the calcium channel CACNA2D3. 22 These examples demonstrate potential overlap in the genetic basis of acute and chronic pain. Given that increased acute pain sensitivity has been considered a risk factor for chronic pain,16,23–26 it is important to understand the genetics mediating the biology of thermal nociceptive sensitivity as it could provide insight into initiating factors underlying chronic pain progression.
Mice are an excellent model for discovery genetics of pain-related traits and offer several complementary advantages to human genetic studies. Most notably, mice allow for rigorous control of the experimental environment, detailed genetic knowledge of the subjects, the ability to control allelic frequency, the ability to sample the appropriate end point tissue, and the ability to identify and validate candidate causal variants within the same species on the most appropriate genetic backgrounds. Inbred mouse strains vary across several pain-associated phenotypes,17,27–29 genes underlying differences in pain-related phenotypes have been successfully mapped and validated in mouse populations, including Hydin, 30 Mc1r, 31 and in the case of Cacng2 have led to the identification of genetic risk factors in humans for post-surgical pain. 32
Individuals within an inbred strain are, for most intents and purposes, genetically identical. However, separation of founders and the establishment of new colonies can quickly lead to genetic drift and the independent fixation of spontaneous mutations that can significantly alter traits. Genetic drift leads to new genetic variation and presents unique opportunities to exploit the emergence phenotypic variance in the face of minimal genetic variance. 33 Rodent inbred substrains have nearly identical whole genome sequences and are much more closely related genetically than any two given classical inbred strains. For instance, C57BL/6J and C57BL/6NJ have approximately 30,000 SNPs + indels that distinguish them whereas most other classical inbred strains contain over five million SNPs + indels compared to the C57BL/6J reference genome. 34 This reduced genetic complexity can facilitate the identification of causal genes and variants influencing complex traits by reducing the density of genetic polymorphisms within a chromosomal interval and minimizing variant-variant interactions, given that a single major locus is typically identified for a given trait on an already nearly isogenic, segregating genetic background.35–37
We have had repeated success in using Reduced Complexity Crosses (RCC) between C57BL/6 substrains to map the genetic basis of complex traits ranging from binge-like eating 38 to thermal nociception as measured via hot plate nociceptive sensitivity 39 to methamphetamine stimulant sensitivity, 40 to name a few. Combining behavioral QTL with tissue-specific gene expression QTL analysis can facilitate identification of plausible candidate genes (e.g., Ryr1 for hot plate; Bryant et al., 2019) and even sometimes directly validate causal variants via CRISPR/Cas9 (e.g., Gabra2 for methamphetamine stimulant sensitivity). 40
As another example of substrains that can be used in a RCC, 36 the BALB/cJ (J) and BALB/cByJ (By) substrains of mice were separated in 1935 after the completion of backcrossing at generation F37 and have been maintained since then as separate inbred substrains. Over time, the fixation of spontaneous mutations and residually heterozygous loci, yielded approximately 8500 SNPs, insertions, and deletions that distinguish the substrains, comprising approximately a 500-fold reduction in genetic complexity compared to C57BL/6J versus most classical inbred strains.34,41 BALB/c substrains differ in several phenotypes that are of interest to neurobehavioral geneticists, including anxiety, social aggression, brain morphology, brain weight, immune response, reward and extinction learning, and gene expression27–29,42–46 (reviewed in Bryant et al.). 36 Thus, BALB/c substrains likely harbor a pool of readily identifiable causal variants underlying several behavioral traits relevant to a multitude of brain disorders.
In this study, we identified a robust difference between BALB/c substrains in thermal nociceptive sensitivity on the hot plate in the Bryant Lab at Boston University School of Medicine. We then expanded our assessment of pain-related phenotypes to include von Frey, cold plate, and Hargreaves assays which were conducted in the Young Lab at University of Kansas Medical Center. We also replicated the reduced brain weight in the BALB/cByJ substrain relative to BALB/cJ, 28 a phenotype that could potentially be associated with any number of neurobehavioral traits that differ between these substrains, 36 . Upon identifying robust trait differences between BALB/c substrains, we then generated an RCC between them to map QTLs underlying variable hot plate sensitivity and brain weight and triangulated on candidate gene identification using expression QTL mapping in multiple historically collected brain regions (striatum, hippocampus). Because there are more relevant CNS tissues involved in nociceptive transmission, we also performed transcriptome analysis via RNA-seq in spinal cord tissue and whole brain proteomic analysis between the parental substrains that allowed us to confirm differentially expressed genes identified from eQTL analysis at the protein level and provide further evidence for the candidacy of potential causal genetic factors.
Methods
Mice
All experiments were conducted in accordance with the National Institutes of Health Guidelines for the Use of Laboratory Animals (8th Ed.) 47 and were approved by the Institutional Animal Care and Use Committee at Boston University (BUSM) and University of Kansas Medical Center (KUMC). BALB/cJ and BALB/cByJ mice (7 weeks old) were purchased from The Jackson Laboratory (Bar Harbor, ME; #000651, #001026), housed 4/cage, and allowed 6 days to acclimate before testing. BALB/cJ x BALB/cByJ-F1 and -F2 mice were bred in house as described below and were tested between the ages of 60 and 130 days old. All mice were maintained on Teklad 18% protein diet (Envigo, Indiana; #2018) and a 12 h light/dark cycle.
Hot plate assay of thermal nociception (BUSM)
BALB/c substrains were originally phenotyped in a larger effort to characterize oxycodone responses in a large panel of inbred mouse strains, and therefore, one-half of the mice have prior exposure to a dosing regimen of oxycodone and related behavioral testing. Specifically, prior to hotplate testing, 64 BALB/cJ and 47 BALB/cByJ mice were tested for conditioned place preference using 1.25 mg/kg OXY (i.p.) as previously described. 48 Over the next 4 days, mice were administered once daily injections of 40 mg/kg (i.p.) OXY or SAL (10 mL/kg). On day 5 at 0700 h (1 h prior to testing), mice were moved into the testing room to acclimate, and water was removed from home cages. Mice were then placed on a 53.5°C hot plate (IITC Life Science Inc., Woodland Hills, CA USA) within a 15 cm diameter × 33 cm tall plastic cylinder. The latency to lick the hind paw or jump was recorded as a pain response and the mouse was returned to its home cage. Mice that did not respond in 60 s were removed, and a 60 s latency recorded. Two measures were taken, separated by 30 min and the average of the two baseline latencies was used for QTL mapping analysis. Water bottles were returned immediately after completion of testing.
von Frey test of mechanical sensation (KUMC)
Mice were placed in individual containment units resting on a mesh-floor testing table for a 15–20 min acclimation period. Following the acclimation, mechanical threshold testing was performed using graded monofilaments. Beginning with the 3.22 filament (0.16 g force), individual monofilaments were applied to the plantar surface of the hind paw and presence (lifting or flicking of the paw or vocalization in response to stimulus application) or absence of a withdrawal response was noted. Stimuli were presented using the Up-Down method. Mechanical Withdrawal Threshold (MWT) was defined as the force required to elicit a withdrawal response.
Hargreaves thermal analgesiometry (KUMC)
Thermal nociceptive thresholds were measured using a Paw Thermal Stimulator (University of California, San Diego). The heated glass floor was maintained at 30.0°C, corresponding to the latent paw temperature of a mouse. Mice were brought to the testing room and allowed to acclimate for 20 min before each experiment. Following the acclimation period, mice were placed in individual Plexiglas open-topped chambers on top of the Analgesiometer. The high-intensity light beam was directed at a hindpaw, and the latency to withdraw the hind paw was recorded with an automatic timer. Each mouse underwent three trials with a 3 min intertrial interval; values were averaged to determine the thermal withdrawal latency (TWL). A maximum stimulus latency of 20 s was used to prevent potential tissue damage.
Cold plate (KUMC)
Mice were brought to the testing facility and allowed to acclimate to the room for 30 min prior to testing. Mice were enclosed in a 4” × 8” clear, Plexiglas arena which allowed for free movement, resting atop a metal platform with digitally controlled temperature. The plate temperature began at ambient temperature and ramped down (24°C–0°C) at a rate of 10°C/min until the mouse exhibited a response. Once a response was observed, the experimenter discontinued the stimulus returning the plate to ambient temperature. Positive responses to unpleasant cold were recorded for rearing or lifting of paws off the surface of the plate in a coordinated fashion. Limits on maximum and minimum temperature settings were used to prevent tissue damage to the mouse. All mice exhibited a response within 5 min from trial initiation.
Whole brain weight dissections (BUSM)
Twenty four BALB/cJ and 24 BALB/cByJ mice (12 females and 12 males for each substrain) were trained and tested for drug-free and state-dependent CPP using 1.25 mg/kg OXY (i.p.) and procedures previously described. 48 Immediately after testing for Day 9 CPP, mice were sacrificed by rapid decapitation after the final day of testing. Brains were dissected from the skull (mice were 63 days old at the time of harvesting), olfactory bulbs trimmed, and brainstem was trimmed at the clearly demarcated pons-medulla boundary for consistency. Whole brains were collected and flash frozen while sitting in an aluminum foil boat placed into a bath of 100% ethanol cooled with dry ice, then placed in pre-weighed tubes, massed, and stored at −80°C for later proteomic analysis.
Statistical assessment of normality, data normalization, and QTL model selection
Raw parental strain and F2 data were analyzed through ANOVA models to determine whether the residuals were distributed normally via Shapiro-Wilks test. To ensure the residuals are appropriately modeled to test normality we used an ANOVA to test for main effects of Sex, Prior Treatment, and Genotype in parental strain mice, and Sex, Prior Treatment, and Age for F2 mice. Factors with significant main effects were included in the untransformed ANOVA models and the residuals tested for normality with a Shapiro-Wilks test. When necessary, data were quantile-normalized using the orderNorm function from the BestNormalize R package. 49 In the case of F2 mice, normalized data were again tested for main effects of Sex, Prior Treatment, and Age and factors achieving significance were included in the QTL model as additive covariates.
F2 breeding and genotyping
BALB/cJ and BALB/cByJ mice (7 weeks old) were purchased from JAX and were first crossed to generate an F1 generation (J female x By male or By female x J male). F1 mice were then intercrossed, pairing mice so that each F2 offspring had a BALB/cJ and BALB/cByJ granddam and grandsire. F2 offspring were weaned at 21 days old, tails were collected for genotyping, and mice were housed 2–4 mice/cage. The first day of experimental testing began between 56 and 132 days old. The age range was larger than what we typically employ (50 days old to 100 days old) due to the COVID-19 shutdown. Breeder pairs were fed Teklad breeder diet (Envigo, Indiana; #7004), and F2 mice were fed Teklad 18% protein diet (Envigo, Indiana; #2018). Tails were harvested at the time of weaning and were shipped for DNA extraction and genotyping (Neogen GeneSeek Operations, Lincoln, NE, USA) using the miniMUGA array. 50 304 polymorphic markers on the miniMUGA array distinguished the BALB/cJ and BALB/cByJ progenitor strains (see QC below).
BALB/cJ x BALB/cByJ F2 hot plate and brain weight phenotyping
The 283 BALB/cJ x BALB/cByJ F2 mice that were phenotyped in this study were part of a larger F2 mapping project, and approximately one-half (152 mice) had previous exposure to oxycodone from the 9 day CPP paradigm described above, but were not administered the four 40 mg/kg OXY injections preceding hot plate as described for the parental substrains. Instead, F2 mice were tested for conditioned place preference over 9 days, 48 with OXY mice receiving 3 boluses of 1.25 mg/kg OXY on days 2, 4, and 9. On day 11, 2 days later, mice were tested in a red lit room on the elevated plus maze for 5 min. On day 12, mice were tested for baseline hot plate responses as described above. Three hour following hot plate testing, mice were sacrificed, and brains were dissected and weighed as described above.
QTL mapping
Quantitative trait locus mapping in the F2 cross was conducted using the R/qtl package. 51 Prior to mapping, QC measures were employed to ensure accurate genotypes. Markers with greater than a 5% no call rate were removed. Markers with allele frequencies that differed significantly from Mendellian inheritance in their ratios of homozygotes to homozygotes (J v By) and in heterozygotes to all homozygotes as determined by chi squared test were also removed. Using the countXO function within R/qtl, mice with aberrant crossover counts (>45 or <10) were also removed from analysis. Marker positions were transformed from Mb to sex-averaged cM using JAX Mouse Map Converter (http://cgd.jax.org/mousemapconverter/) prior to mapping. After QC there were 283 F2 animals and 216 polymorphic markers within the panel to conduct QTL mapping. The scanone function was used to compute Haley-Knott regression at each marker, and 1000 permutations were run to assign significance thresholds (p < 0.05), considering Sex and Prior Treatment (oxycodone, saline) as additive covariates. For significant QTLs, the bayesint function was used to calculate the Bayes credible interval and was conservatively expanded to the nearest genotyping markers and the percent phenotypic variance explained using the function fitqtl.
Whole genome sequencing and genotype calling of BALB/c substrains
We took advantage of data generated for other studies (BALB/c sequence: 41 ; M. Ferris, manuscript in prep) to access whole genome sequence of BALB/cJ and BALB/cByJ substrains. The BALB/cByJ sample was sequenced as paired-end (PE) 2 × 150 on an Illumina HiSeq. Reads were aligned to mouse reference (mm10) using BWA-MEM. We used GATK (v4.0.3.0) to identify variants segregating between BALB/cJ and BALB/cByJ within our QTL intervals. We further filtered these results based on GATK quality calls and to be consistent with expectations of inbred mouse strains (variants should be homozygous and not heterozygous). SNPs in our region were further annotated with SNPEff (v4.3t) to identify the functional consequences of these variants.
Parental strain spinal cord tissue collection (KUMC)
Eight BALB/cJ and 8 BALB/cByJ mice (4 males and 4 females per substrain) were euthanized with an overdose of isoflurane gas anesthesia, USP (Southmedic Inc, Ontario, Canada) in the Young Lab at University of Kansas Medical Center followed by transcardiac perfusion with ice cold Hanks Balance Salt Solution (1X HBSS) (Life Technologies Corporation, New York, USA). Spinal cords (L1–L5) were collected and submerged in RNA Stabilization Solution RNAlater TM Solution (Thermofisher) and stored at 4°C. RNAlater TM Solution was then removed, and tissue samples were stored at −80°C. Samples were subsequently shipped overnight on dry ice to the Bryant Lab where RNA was extracted.
RNA extractions, sequencing libraries, and read alignment
RNA was extracted in RNAlater-preserved tissue using Trizol (Qiagen), ethanol precipitation, filtering columns (Qiagen), DNAse digestion (Qiagen), and elution with RNAse and nucleotide free water 52 and diluted to 100 ng/uL. RNA library preparation (poly-A selection) and RNA-seq were conducted at the University of Chicago Genomics Facility on an Illumina NovaSEQ6000 using a NovaSEQ SP-100 bp flowcell/reagent cassette. We used the R/Bioconductor package “scruff” to conduct demultiplexing, read alignment, read counting, quality checking and data visualization (Wang et al., 2019). Reads were trimmed for quality using Trimmomatic. 53 Trimmed reads were then aligned to the mm10 mouse reference genome (Ensembl) to generate BAM files for alignment using STAR. 54 For differential gene analysis in the spinal cord, the featureCounts read summarization program was used to count reads mapping to the “exon” feature in a GTF file obtained from Ensembl (GRCm38). Genes without 10 reads per million in at least three samples were excluded from analysis using EdgeR, 55 and differential gene expression analysis of normalized counts was conducted using an appropriate design matrix, and reported using the topTable function.
BALB/cJ x BALB/cByJ F2 RNA expression QTL mapping
A subset of 64 BALB/cJ x BALB/cByJ F2 mice that were not tested on the hot plate were used for eQTL mapping and were 78–127 days old on day 1 of testing. Mice were trained in the CPP protocol with either saline (i.p.) or 1.25 mg/kg OXY (i.p.) 48 as described above and were sacrificed by rapid decapitation on the final day of experimental testing, 30 min after receiving either saline (i.p.) or OXY (1.25 mg/kg, i.p.). Using a brain matrix, striatal and hippocampal tissues were dissected. Striatal punches were harvested at bregma 1.5 to −0.5 mm and sampled with a 2 mm punch. Hippocampal tissues were dissected from bregma −0.5 to –2.5 using a sterile metal spatula to peel away the cortical layer. Samples were stored, extracted, sequenced, and prepared for analysis as described above. Count files were analyzed using R/MatrixEQTL 56 using the “linear cross” model and considered Sex, RNA extraction Batch (RNA extraction), and Prior Treatment (saline, oxycodone) as additive covariates.
Parental strain spinal cord alternative splicing analysis and exon-level expression QTL mapping
For analysis of F2 samples (striatum, hippocampus) and parental strains (spinal cord), spliceome analysis was conducted within the package R/ASpli. 57 For F2 striatum and hippocampus, the function gbcounts was used to extract to summarize reads from aligned BAMs to features. Features less than 50 bp and with fewer than 10 counts were excluded from analysis. These intron/exon counts were then analyzed using R/MatrixEQTL as described above. In parental strain spinal cord samples, the gbcounts function was used to summarize reads from aligned BAMs to features, and the gbDUreport and jDUreport functions were used to calculated differential feature and junction usage respectfully. Both functions evaluated the effect of Substrain with the inclusion of Sex as an additive covariate. Results were generated using splicingReport
Whole brain harvesting and tissue processing for proteomics
Eight BALB/cJ (4 females, 4 males) and 8 BALB/cByJ (4 females, 4 males) mouse brains were flash frozen and used for whole brain mass spectrometry analysis. Whole brains were homogenized in 5 mL of protein extraction buffer (100 mM Tris pH 8.5, 8 M Urea, 1 mM CaCl2, 10 mM TCEP, 40 mM Chloroacetamide) using a Polytron® PT 3100 tissue homogenizer at 20,000 r/min for 1 min. Protein extracts were sonicated with a Branson probe sonicator and were then quantified via Bradford assay. 300 μg of protein from each sample was diluted with 100 mM Tris, pH 8.5 buffer to lower the urea concentration to 1 M. Lysate proteins were then digested by the addition of trypsin (Pierce) at a 1:50 ratio (enzyme: protein, w/w) and incubated overnight at 37°C with shaking. Trypsin digestion was terminated with the addition of TFA to below pH 3 and the peptide digests were desalted via reversed-phase C18 columns (Sep-Pak, Waters) with a wash buffer of 0.1% TFA and elution buffer of 60% acetonitrile. The desalted peptides were then quantified with a Quantitative Colorimetric Peptide Assay (Pierce). Each sample comprising 100 μg peptides was TMT-labeled with TMTPro 16plex reagents (ThermoFisher, cat. # A44520) according to the manufacturer’s protocol. Labeled samples were combined and desalted on a C18 column prior to basic reversed-phase fractionation.
TMT-labeled peptides were fractionated via basic reversed-phase chromatography on the Agilent 1100 series HPLC instrument equipped with the XBridge Peptide BEH C18 column (130 Å, 3.5 μm, 4.6 mm × 250 mm, Waters Corporation). Prior to loading peptides, the C18 column was washed with 100% methanol and equilibrated with Buffer A (0.1% NH4OH and 2% acetonitrile). Peptides were injected via the autosampler and eluted from the column using a gradient of mobile phase A (2% acetonitrile, 0.1% NH4OH) to mobile phase B (98% acetonitrile, 0.1% NH4OH) over 48 min at a flow rate of 0.4 mL/min. The 48 fractions collected were orthogonally concatenated into 12 pooled fractions.
Mass spectrometry analysis
Approximately 2 μg of each multiplexed peptide fraction was resuspended in mobile phase A solvent (2% acetonitrile and 0.1% formic acid) to be analyzed on the Exploris 480 mass spectrometer equipped with FAIMS (ThermoFisher Scientific). The mass spectrometer was interfaced to the Easy-nLC 1200 HPLC system (ThermoFisher Scientific). Briefly, the peptides were first loaded onto a reversed-phase nanotrap column (Acclaim PepMap100 C18, 100 Å, 3 μm, 75 μm × 2 cm, ThermoScientific) in mobile phase A, and separated over an EASY-Spray column, (ES803 A, Thermo Scientific) using a gradient (6%–19% over 58 min, then 19%–36% over 34 min) of mobile phase B (0.1% formic acid, 80% acetonitrile) at a flow rate of 250 nL/min. The mass spectrometer was operated in positive ion mode with a capillary temperature of 275°C and a spray voltage of 2500 V. All data were acquired with the mass spectrometer operating in data dependent acquisition (DDA) mode, with FAIMS cycling through one of three compensation voltages (−50V, −57V, −64V) at each full scan. Precursor scans were acquired at a resolution of 60,000 FWHM with a maximum injection time of 120 milliseconds in the Orbitrap analyzer. The following 0.8 s were dedicated to fragmenting the most abundant ions at the same FAIMS compensation voltage, with charge states between 2 and 5, via HCD (NCE 33%) before analysis at a resolution of 45,000 FWHM with a maximum injection time of 60 ms.
All acquired MS/MS spectra were searched against the complete SwissProt mouse proteome (downloaded on 2020-10-20) using MaxQuant (Version 1.6.7.0), which integrates the Andromeda search engine. TMT reporter ion quantification was performed using MaxQuant with default settings. Briefly, enzyme specificity was set to trypsin and up to two missed cleavages were allowed. Cysteine carbamidomethylation was specified as fixed modification whereas oxidation of methionine and N-terminal protein acetylation were set as variable modifications. Precursor ions were searched with a maximum mass deviation of 4.5 ppm and fragment ions with a maximum mass deviation of 20 ppm. Peptide and protein identifications were filtered at 1% FDR using the target-decoy database search strategy. 58 Proteins that could not be differentiated based on MS/MS spectra alone were grouped to protein groups (default MaxQuant settings). The MaxQuant output file designated “proteinGroups” was used for data normalization and other statistical analysis using the Omics Notebook analysis pipeline. 59
Results
BALB/cByJ mice are more sensitive compared to BALB/cJ in thermal nociception on the hot plate test and in mechanical stimulation on the von Frey test
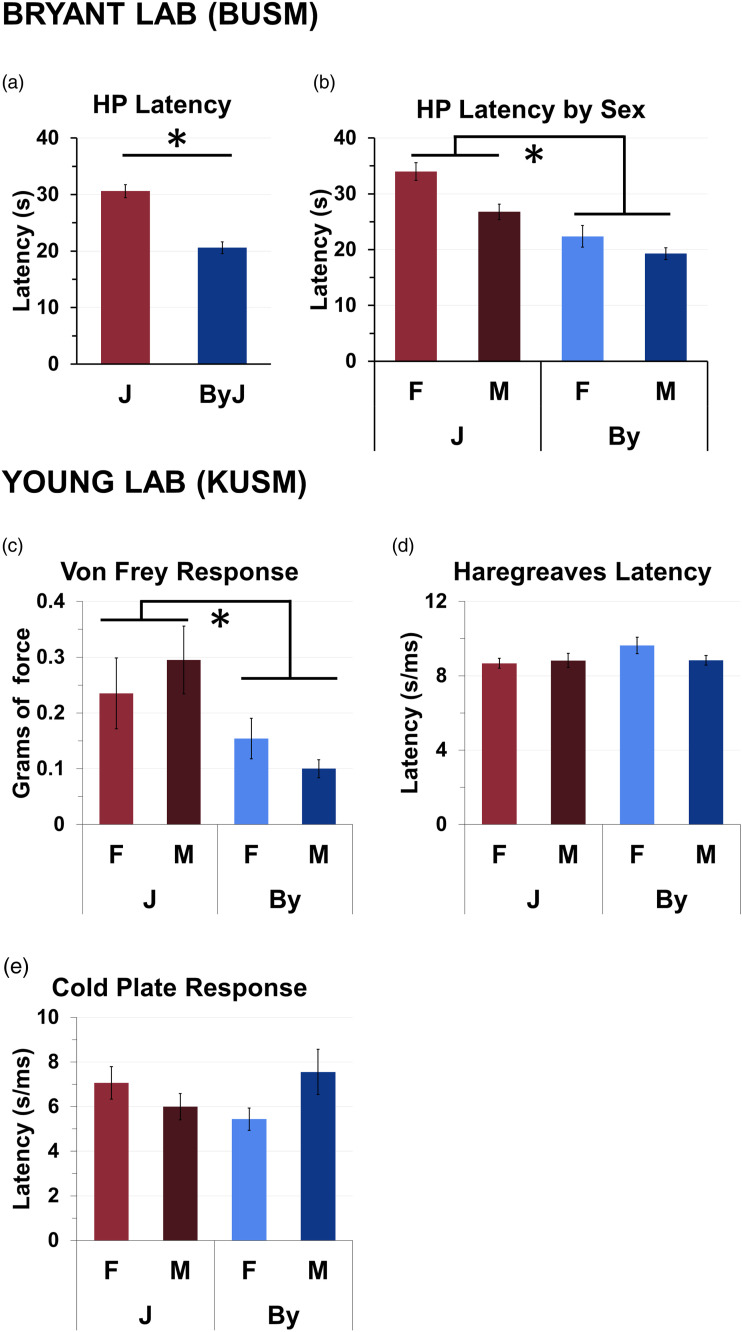
The distribution of the residuals for hot plate latencies deviated significantly from normality (W = 0.94, p = 5.98e−5). To facilitate interpretation, we plotted the raw latencies in the main results. Three-way ANOVA of normalized latencies indicated that there was no effect of prior Treatment (saline, oxycodone) or interaction of Treatment with Substrain or Sex on hot plate latencies (p’s > 0.30). Therefore, we removed Treatment from the ANOVA model (a breakdown of the hot plate data by treatment is provided in Supplementary Figure 1(a)). Two-way ANOVA (Substrain, Sex) indicated a decreased hot plate threshold in the By substrain (Substrain effect: F (1, 107) = 54.45, p =3.61e-11; Figure 1(a)). There was also a main effect of Sex [F (1, 107) = 13.1, p = 4.53e−4], with females showing higher latencies (Figure 1(b)) but no Substrain x Sex interaction F (1,107) = 0.78; p = 0.38, Figure 1(b)). For the Von Frey threshold, By also showed a lower latency (Substrain effect: F (1, 31) = 8.19, p < 0.008; Figure 1(c)), but not in the Hargreaves test (Substrain Effect: F (1, 31) = 1.95, p <0.173; Figure 1(d)) or cold plate test (F (1, 31) = 0.05, p <0.82; Figure 1(e)).
Figure 1.
BALB/cByJ mice are more sensitive to thermal nociception and mechanical stimulation compared to BALB/cJ mice. (a): By mice showed increased hot plate (53.5°C) sensitivity as indicated via a significant reduction in hind paw lick latencies compared to J (p = 3.61e−11). (b): Female mice showed increased hot plate latencies (p = 4.53e−4). There was no significant Sex x Strain interaction (p = 0.38). (c): By mice were more sensitive to mechanical stimulation than J mice as indicated via a reduced force threshold (g) to initiate a withdrawal response (p = 0.008). (d,e): There was no effect of Substrain in the Hargreaves test (p = 0.173) or cold plate (p = 0.82).
BALB/cByJ mice show decreased brain weight compared to BALB/cJ
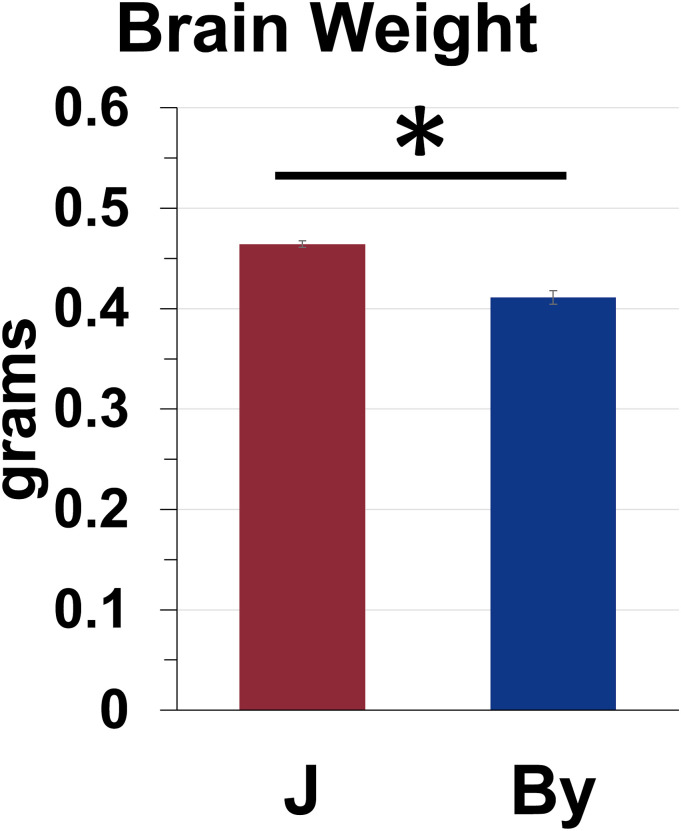
Residuals for brain weight deviated significantly from normality (W = 0.81, p = 2.6e−6). To facilitate interpretation, we plotted the raw brain weight (g) in the main results. Three-way ANOVA of the normalized brain weight data indicated no effect of prior Treatment (saline, oxycodone) or interaction of Treatment with Substrain or Sex on hot plate latencies (p’s > 0.22). Furthermore, there was no effect of Sex or interactions of Sex with Substrain (p’s > 0.22). Therefore, we removed Treatment and Sex from the statistical model (Supplementary Figure 1(b) shows brain weight broken down by prior Treatment). A follow-up simple main effect test revealed that By brains weighed less than J brains (p < 0.0001; Figure 2).
Figure 2.
BALB/cByJ brains weigh less than BALB/cJ brains. Simple main effect test: p < 0.0001.
Normalization of F2 data and ANOVA analysis of F2 mouse hot plate latencies (s) and brain weight (g) to identify covariates for QTL analysis
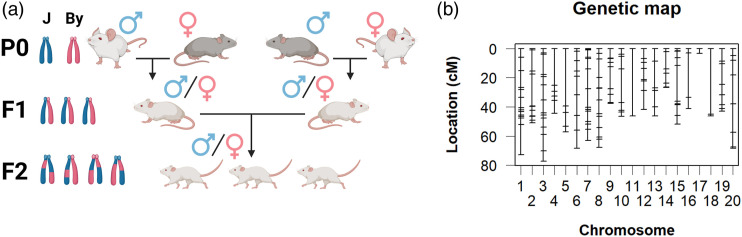
The breeding scheme for generating F2 mice is shown in Figure 3(a). BALB/cJ and BALB/cByJ mice were reciprocally crossed (J females x By males; J males x By females) generate F1 mice and then intercrossed so that each F2 offspring had both a BALB/cJ and BALB/cByJ granddam and grandsire. A total of 216 polymorphic markers were included in QTL analysis (Figure 3(b)). Neither primary outcome measure was normally distributed (hot plate latency: W = 0.92, p = 8.87e−11; brain weight: W = 0.94, p = 5.98e−5); thus, these phenotypes were quantile normalized. We then ran a 3-way ANOVA (Sex, Prior Treatment, and Age) to inform QTL model selection. Regarding hot plate, there was a significant effect of prior OXY treatment (Prior Treatment effect: F (1, 275) = 8.08, p = 0.003), and Sex (F (1, 275) = 5.59, p = 0.015), but not of Age (F (1, 275)=1.45, p = 0.23). For this reason, both Sex and Treatment were included as covariates in the QTL model for hot plate latencies. In examining normalized brain weight, a three-way ANOVA indicated a main effect of prior OXY treatment (Treatment effect: F (1, 247) = 36.27, p = 6.18e−9), an effect of Age (Age effect: F (1, 247) = 18.23, p = 2.79e−5), and a nearly significant effect of Sex (Sex effect: F (1, 247) = 3.476, p = 0.063). Therefore, these three covariates were included in the QTL model for normalized brain weight.
Figure 3.
Breeding scheme and genotyping marker panel for the BALB/c reduced complexity cross. (a): Breeding scheme used to produce F2 mice. Each F2 mouse has a grandsire and granddame of each parental strain. Created with BioRender.com (b): Genetic map of the 216 miniMUGA markers passing QC and used for QTL mapping.
A BALB/c reduced complexity cross identifies a genome-wide significant QTL on chromosome 13 for baseline thermal nociceptive sensitivity on the hot plate test
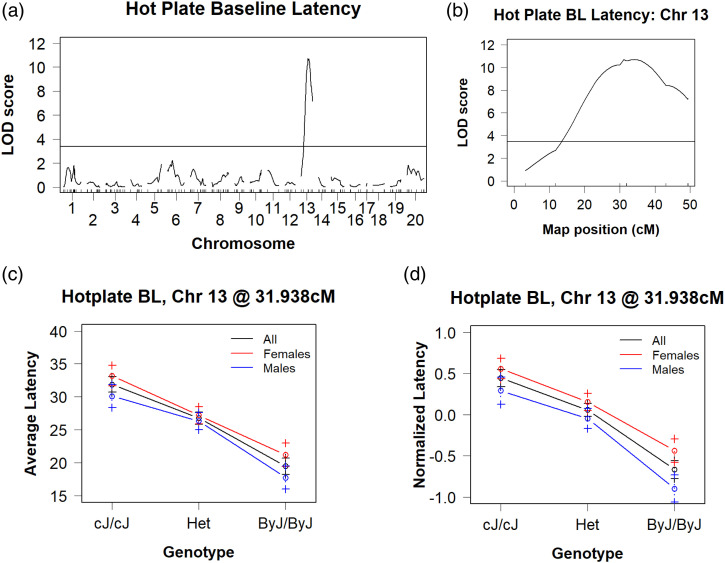
We identified a genome-wide significant QTL on chromosome 13 for normalized baseline hot plate latency (s) that peaked at 31.1 cM (58.7 Mb) and explained 15.7% of the phenotypic variance (LOD= 10.7; p < 0.001; Bayes: 53–73 Mb; Table 1, Figures 4(a) and (b)). The effect plot of the peak-associated marker (SBR132392332; 59.81 Mb), in particular when stratified by Sex, recapitulated the latencies of the parental substrains Figures 4(c) and (d)). We also stratified by Prior Treatment and illustrate qualitatively similar effects of Genotype on hot plate latency but with the also small, additive effect of prior Treatment (Supplementary Figures 2(a) and (b)). Note we still obtained genome-wide significant results even if we just ran the mice with prior SAL treatment and remove the mice with a prior history of oxycodone treatment from the analysis (Supplementary Figure 2(c)). Thus, prior Treatment had no effect on our ability to detect this hot plate QTL. Table 2 summarizes the results for the chromosome 13 QTL. We identified 42 protein coding genes with assigned polymorphisms that were contained in the Bayes interval. A complete list of all variants contained within the QTL interval can be found in Supplementary Table 1. Table 3 documents three candidate genes that we highlighted based on potentially highly disruptive mutations (i.e., mutations within the gene region, but not annotated as downstream, upstream, intronic, or intergenic) within the Bayesian interval.
Table 1.
Spinal cord intron/exon usage analysis in parental strains identified differential feature usage of H2afy Significant associations between genetic feature usage in the spinal cord of parental strain mice and substrain, considering sex as an additive covariate (FDR <0.05).
| Gene | Location (bp) | start | length | Element | Feature | logFC | p value | FDR |
|---|---|---|---|---|---|---|---|---|
| Dnah17 | 11:118021723–118130634 | 118023498 | 1096 | I | Intron 80 | 1.020851 | 2.19E-82 | 4.59E-77 |
| H2afy | 13:56073619–56136361 | 56084196 | 90 | E | Exon 7 | 0.373669 | 8.27E-10 | 8.65E-05 |
| Glp1r | 17:30901817–30940791 | 30936265 | 222 | E | Exon 13 | 0.858303 | 2.68E-07 | 0.014034 |
| Glp1r | 17:30901817–30940791 | 30936487 | 169 | E | Exon 13 | 0.891179 | 2.68E-07 | 0.014034 |
| H2afy | 13:56073619–56136361 | 56074393 | 8727 | I | Intron 8 | 0.368271 | 7.93E-07 | 0.033166 |
Figure 4.
A BALB/c reduced complexity cross identifies a genome-wide significant QTL on chromosome 13 for thermal nociceptive sensitivity on the hot plate. (a): We identified a single genome-wide significant peak on chromosome 13 (LOD = 10.71, p < 0.001; Bayes: 53–73 Mb) that explained 16% of the phenotypic variance. (b): Chromosome 13 QTL plot, with a peak association at 31 cM (59 Mb). (c): Effect plot of hot plate latencies at the peak-associated marker (sex-combined, females-only, males-only) recapitulates both the degree and magnitude of the original parental substrain difference shown in Figure 1. (d): Effect plot of normalized hot plate latencies at the peak-associated marker (sex-combined, females-only, males-only).
Table 2.
Summary of QTLs for thermal nociceptive sensitivity on the hot plate and brain weight in a BALB/c reduced complexity cross.
| N | Phenotype | Model | Chr | Peak (cM) | Peak (Mb) | LOD | p val | Bayesian interval (cM) | Bayesian interval (Mb) | 1.5 LOD Drop (cM) | 1.5 LOD Drop (Mb) | % Var explained |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 283 | Norm HP BL | Sex, Tx Add | 13 | 31.1 | 58.7 | 10.71 | <0.001 | 27–39 | 53–73 | 24–42 | 45–81 | 15.68 |
| 256 | Norm Br Wt | Sex, Tx, Age Add | 5 | 58.6 | 118.36 | 8.73 | <0.001 | 54–61 | 110–120 | 51–61 | 104–120 | 12.37 |
Table 3.
Positional candidate genes for thermal nociceptive sensitivity on the hot plate and brain weight. Salient mutations annotated to genes within the QTLs on chromosomes 13 (hot plate) and 5 (brain weight). Mutations were selected if they were annotated to splice regions, missense mutations, 5′UTR, 3′UTR, disruptive in-frame insertions, and frameshift mutations. Positions based on GRCm38/mm10 assembly of mouse genome. Functional annotation of genes derived from GO function terms.
| QTL | Loc (cM) | Loc (Mb) | Gene | Mutations | GO biological processes terms |
|---|---|---|---|---|---|
| Ch13: HP | 29.14 | 54.79 | Eif4e1b | 3'UTR | No entries found |
| 30.07 | 56.08 | H2afy | Intergenic, Splice region (2), downstream | Chromatin silencing, negative regulation of gene expression | |
| 34.67 | 67.47 | Zfp874b | 3'UTR | Regulation of transcription by RNA polymerase II | |
| 34.76 | 67.67 | Zfp738 | 3' UTR, downstream, intron (3) | Regulation of transcription by RNA polymerase II | |
| Chr 5: BrWt | 56.08 | 115.55 | Pxn | 3'UTR | Cytoskeleton organization, regulation of cell shape |
| 62.60 | 122.80 | Anapc5 | 3'UTR | Anaphase promoting complex, cell cycle, cell division | |
| 63.00 | 123.45 | Mlxip | 3'UTR | Regulation of transcription by RNA polymerase II | |
| 63.02 | 123.49 | Il31 | Disruptive inframe insertion | Acute inflammatory response to antigenic stimulus | |
| 68.30 | 129.86 | Sumf2 | 3'UTR | No entries found | |
| 83.22 | 144.21 | Tecpr1 | Missense | Autophagosome maturation |
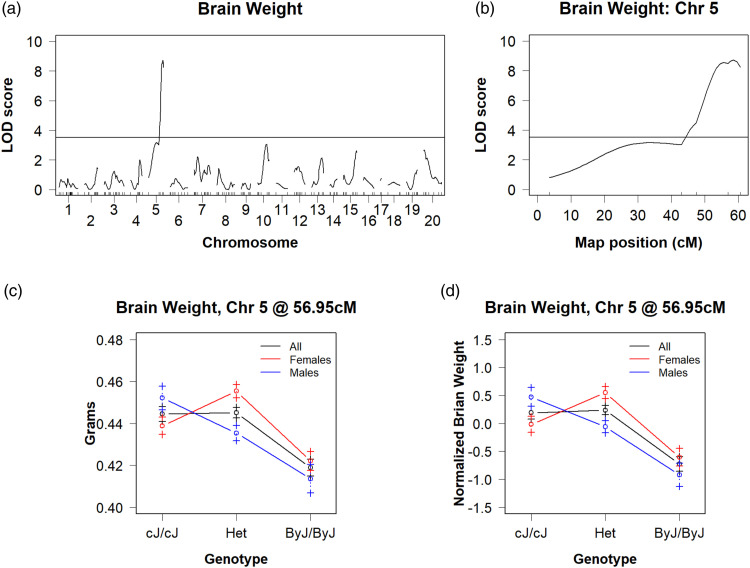
A BALB/c reduced complexity cross identifies a genome-wide significant QTL on chromosome 5 for brain weight
We identified a single genome-wide significant peak for normalized whole brain weight on chromosome 5 that peaked at 58.6 cM and explained 12% of the phenotypic variance (118.36 Mb, LOD = 8.73; p < 0.001; Bayes = 111 mB – End of Chromosome; Figures 5(a) and (b)). The effect plot of the peak-associated marker (SBJ054695332; 117.38 Mb) precisely recapitulated the results of the parental substrains (Figures 5(c) and (d)). The same chromosome 5 QTL for brain weight was also identified for the unnormalized brain weight (Supplementary Table 3). There are 70 polymorphic protein-coding genes within the QTL interval (Supplementary Table 2). Six of these genes are highlighted in Table 4 as candidate genes based on the presence of potentially highly disruptive mutations as defined above. A complete list of variants within the chromosome 5 QTL is provided in Supplementary Table 2.
Figure 5.
A BALB/c reduced complexity cross identifies a genome-wide significant QTL on chromosome 5 for brain weight (a): Genome-wide significant QTLs were identified on chromosome 5 (LOD = 8.73, p < 0.001; 12% of the variance explained). (b): QTL plot for the chromosome 5 QTL that peaks at 59 cM (118 Mb). (c): Chromosome 5 effect plot at the peak-associated marker recapitulates both the direction and magnitude of the original parental substrain difference shown in Figure 2. (d): Chromosome 5 effect plot at the peak-associated marker for normalized brain weight recapitulates observed trend in non-normalized data.
Table 4.
Striatal and hippocampal cis-eQTL transcripts for chromosome 13 and 5 QTLs. All cis-eQTLs on chromosome 13 and chromosome 5 in both the hippocampus and striatum (unadjusted p < 0.001).
| Marker | Gene | |||||||
|---|---|---|---|---|---|---|---|---|
| Brain Region | chr | pos (cM) | pos (Mb) | ID | Start (cM) | Start (Mb) | p value | FDR |
| Striatum | Chromosome 13 | 30.06 | 55.96 | H2afy | 30.07 | 56.07 | 9E-09 | 0.000221 |
| 30.06 | 55.96 | Ddx4 | 63.83 | 112.60 | 5.5E-05 | 0.435334 | ||
| 31.88 | 59.81 | H2afy | 30.07 | 56.07 | 4.53E-06 | 0.060237 | ||
| 31.88 | 59.81 | Msh3 | 47.58 | 92.21 | 0.000188 | 0.835602 | ||
| 31.94 | 60.21 | H2afy | 30.07 | 56.07 | 4.53E-06 | 0.060237 | ||
| 31.94 | 60.21 | Msh3 | 47.58 | 92.21 | 0.000188 | 0.835602 | ||
| 43.11 | 81.56 | H2afy | 30.07 | 56.07 | 2.55E-05 | 0.238048 | ||
| 43.11 | 81.56 | Msh3 | 47.58 | 92.21 | 5.93E-05 | 0.439217 | ||
| 49.36 | 95.03 | Msh3 | 47.58 | 92.21 | 3.88E-06 | 0.052652 | ||
| 49.36 | 95.03 | H2afy | 30.07 | 56.07 | 0.000262 | 0.94804 | ||
| 49.36 | 95.03 | Ccl28 | 67.52 | 119.62 | 0.000919 | 1 | ||
| 49.43 | 95.21 | Msh3 | 47.58 | 92.21 | 2.56E-05 | 0.238638 | ||
| 49.43 | 95.21 | H2afy | 30.07 | 56.07 | 0.000361 | 1 | ||
| 49.43 | 95.21 | Ccl28 | 67.52 | 119.62 | 0.000859 | 1 | ||
| 49.43 | 95.21 | Zfp953 | 34.61 | 67.34 | 0.000976 | 1 | ||
| Chr 5 | 44.06 | 83.69 | Ankrd61 | 81.21 | 143.89 | 0.000112 | 0.613657 | |
| 44.06 | 83.69 | Rfc3 | 85.59 | 151.64 | 0.000473 | 1 | ||
| 50.44 | 95.87 | Rfc3 | 85.59 | 151.64 | 0.000187 | 0.835099 | ||
| 50.44 | 95.87 | Ankrd61 | 81.21 | 143.89 | 0.000279 | 0.961505 | ||
| 65.59 | 117.38 | Ankrd61 | 81.21 | 143.89 | 0.000232 | 0.918532 | ||
| Hippocampus | Chr 13 | 30.06 | 55.96 | H2afy | 30.07 | 56.07 | 0.000436 | 0.503944 |
| 43.11 | 81.56 | Msh3 | 47.58 | 92.21 | 0.000107 | 0.462576 | ||
| 49.36 | 95.03 | Msh3 | 47.58 | 92.21 | 2.06E-05 | 0.308925 | ||
| 49.43 | 95.21 | Msh3 | 47.58 | 92.21 | 6.28E-05 | 0.434747 | ||
| 49.43 | 95.21 | Ccl28 | 67.52 | 119.62 | 0.000793 | 0.532853 | ||
| Chromosome 5 | 43.01 | 83.69 | Pf4 | 44.68 | 90.77 | 0.00054 | 0.513003 | |
| 47.44 | 95.87 | Pf4 | 44.68 | 90.77 | 0.000224 | 0.483621 | ||
| 47.44 | 95.87 | Wdr95 | 89.19 | 149.53 | 0.000253 | 0.483621 | ||
| 47.44 | 95.87 | Hopx | 41.48 | 77.09 | 0.000446 | 0.503944 | ||
| 47.44 | 95.87 | Hs3st1 | 21.11 | 39.61 | 0.000486 | 0.508738 | ||
| 47.44 | 95.87 | Wdfy3 | 48.94 | 101.83 | 0.000609 | 0.517917 | ||
| 56.95 | 117.38 | Hopx | 41.48 | 77.09 | 8.4E-05 | 0.443042 | ||
| 56.95 | 117.38 | Pus1 | 53.71 | 110.77 | 0.000909 | 0.539018 | ||
| 56.95 | 117.38 | Dnah10 | 63.77 | 124.73 | 0.000979 | 0.546122 | ||
| 60.62 | 120.43 | Dnah10 | 63.77 | 124.73 | 0.000329 | 0.493306 | ||
Cis-eQTL analysis of the hot plate QTL on chromosome 13 identifies H2afy as a positional and functional candidate gene underlying thermal nociceptive sensitivity
The RNA-seq data have been uploaded to the NCBI Gene Expression Omnibus (GEO) and are publicly available (GSE #196302, GSE #196334, and GSE #196352). Table 4 shows a comprehensive list of transcripts with significant cis-eQTLs on chromosome 13 within the striatum and hippocampus with an unadjusted cut-off of p < 0.001. For striatal tissue, we identified six protein coding genes with cis-eQTLs. Of these six genes, only H2afy contains any known polymorphisms, including two splice region variants. Only two genes were associated with the peak marker for the hot plate QTL, including H2afy (p = 4.53e−6, FDR = 0.06) and Msh3 (p = 1.88e−4, FDR = 0.84). For hippocampal tissue, we identified three protein coding cis-eQTL transcripts: Msh3, H2afy, and Ccl28. Of these, only Msh3 and H2afy are located within the Bayesian interval of the HP QTL, and only H2afy contains an assigned polymorphism. Thus, H2afy is a high-priority positional and functional candidate gene underlying thermal nociceptive sensitivity on the hot plate assay. Complete lists of cis-eQTLs with p < 0.001 within the striatum and hippocampus are provided in Supplementary Tables 3 and 4.
Cis-eQTL analysis of the brain weight QTL on chromosome 5
Table 4 shows a comprehensive list of chromosome 5 eQTL transcripts within the striatum and hippocampus with p < 0.001. For the striatum, we identified two transcripts with cis-eQTLs, only one of which, Ankrd61, was associated with the peak marker for the brain weight QTL. For the hippocampus, 7 transcripts with cis-eQTLs were identified, 3 of which were associated with the peak marker for the brain weight QTL, including Hopx, Pus1, and Dnah10. Interestingly, for this chromosome 5 locus, there was no overlap in cis-eQTLs between the striatum and hippocampus (Table 4).
Exon/Intron-level eQTL analysis of hot plate and brain weight QTLs
With regard to the hot plate QTL, we observed significant intron/exon level cis-eQTLs for the candidate gene H2afy at the peak marker for the hot plate QTL (SBR132392332) in both the hippocampus and striatum. These associations (FDR <0.05) are shown in Table 5. In both regions, we detected differential usage of introns 5, 6, and 7, as well as exon 6. With regard to the brain weight QTL, we observed significant intron/exon level cis-eQTLs for the candidate gene Acads at the peak brain weight marker (SBJ054695332) in both the hippocampus and striatum. For the hippocampus, we detected differential usage of introns 7 and 8, and for the striatum, we detected differential usage of introns 7 and 8 as well as exon 9. A complete list of intron/exon level cis-eQTL analysis is provided in Supplementary Tables 5 and 6.
Table 5.
Striatal and hippocampal intron/exon-level eQTLs at peak behavioral markers for chromosome 13 and 5 candidate genes. Significant associations between feature usage of candidate genes and top associated behavioral markers (FDR <0.05).
| QTL | Region | SNP | Name | Location (bp) | Element | Feature | Start (Mb) | Length (bp) | p value | FDR |
|---|---|---|---|---|---|---|---|---|---|---|
| Chr 13 Hotplate | Hippocampus | SBR132392332 | H2afy | 13:56073619–56136361 | I | Intron 5 | 56090059 | 5497 | 1.21E-09 | 0.0003 |
| SBR132392332 | H2afy | 13:56073619–56136361 | I | Intron 6 | 56088343 | 1415 | 3.20E-09 | 0.00072 | ||
| SBR132392332 | H2afy | 13:56073619–56136361 | I | Intron 6 | 56084522 | 3730 | 5.28E-09 | 0.00112 | ||
| SBR132392332 | H2afy | 13:56073619–56136361 | I | Intron 7 | 56083295 | 901 | 1.86E-08 | 0.00317 | ||
| SBR132392332 | H2afy | 13:56073619–56136361 | E | Exon 6 | 56089858 | 201 | 1.15E-07 | 0.0148 | ||
| Striatum | SBR132392332 | H2afy | 13:56073619–56136361 | I | Intron 7 | 56083295 | 901 | 1.95E-16 | 9.46E-11 | |
| SBR132392332 | H2afy | 13:56073619–56136361 | I | Intron 6 | 56088343 | 1415 | 3.88E-15 | 1.69E-09 | ||
| SBR132392332 | H2afy | 13:56073619–56136361 | I | Intron 6 | 56084522 | 3730 | 1.11E-14 | 3.89E-09 | ||
| SBR132392332 | H2afy | 13:56073619–56136361 | E | Exon 6 | 56088252 | 91 | 2.31E-12 | 3.72E-07 | ||
| SBR132392332 | H2afy | 13:56073619–56136361 | I | Intron 5 | 56090059 | 5497 | 9.55E-12 | 1.42E-06 | ||
| SBR132392332 | H2afy | 13:56073619–56136361 | I | Intron 5 | 56095642 | 482 | 2.44E-10 | 2.35E-05 | ||
| SBR132392332 | H2afy | 13:56073619–56136361 | E | Exon 6 | 56089858 | 201 | 1.57E-09 | 0.00013 | ||
| Chr 5 Brain Weight | Hipp | SBJ054695332 | Acads | 5:115110299–115119346 | I | Intron 8 | 115111151 | 168 | 1.92E-10 | 5.74E-05 |
| SBJ054695332 | Acads | 5:115110299–115119346 | I | Intron 7 | 115111415 | 222 | 2.27E-08 | 0.00376 | ||
| Striatum | SBJ054695332 | Acads | 5:115110299–115119346 | E | Exon 9 | 115111094 | 57 | 1.46E-09 | 0.00012 | |
| SBJ054695332 | Acads | 5:115110299–115119346 | I | Intron 8 | 115111151 | 168 | 5.10E-09 | 0.00038 | ||
| SBJ054695332 | Acads | 5:115110299–115119346 | I | Intron 7 | 115111415 | 222 | 1.22E-07 | 0.00675 |
Transcriptome analysis of spinal cord between BALB/c substrains
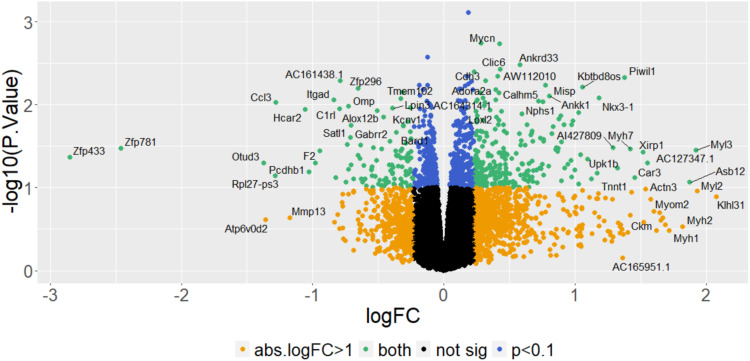
RNA-seq in hippocampus and striatum were historical datasets and are not the most relevant brain regions for pain-related traits such as thermal nociception. Therefore, we turned to differential gene expression analysis of the spinal cord between the parental substrains. Spinal cord tissue was collected from parental substrains at KUMC following pain testing as previously described above. The results are shown as a volcano plot in Figure 6. In considering genes with an absolute log2 fold change greater than 0.32 (absolute fold-change > 1.25), we observe a total of 109 differentially expressed genes at an unadjusted p < 0.05 and 245 genes at an unadjusted p < 0.1 (Supplementary Table 7). Of these 245 genes, none were located within our Chr 13 hot plate QTL. Interestingly, we found a larger number of differentially expressed genes in the males-only analysis (1366 genes, unadjusted p < 0.05; Supplementary Table 8) compared to females (375 genes, unadjusted p < 0.05; Supplementary Table 9). Notably, H2afy was differentially expressed in the spinal cord of male mice (logF = 0.5, adjP = 0.008) but not female mice (logFC = −0.2, adjP = 0.995). Of these same 245 genes, five genes are located within the chromosome 5 brain weight QTL, including Dao (114.1 Mb; 55.9 cM), Oas2 (120.9 Mb; 60.6 cM), Hcar2 (124 Mb, 63.2 cM), Piwil1 (128.8 Mb, 67.9 cM), and Wbscr25 (135 Mb; 74.9 cM).
Figure 6.
Spinal cord transcriptome analysis of differential expression in BALB/c substrains (a): Volcano plot shows the distribution of p values versus log2 fold changes in gene expression, controlling for Sex.
Spinal cord alternative splicing analysis
To deepen our RNA-seq analysis we considered intron and exon usage in the spinal cord of BALB/c substrains to understand differential splicing occurring between substrains. Aspli analysis revealed three genes with differentially used features (FDR <0.05), Dnah17, H2afy, and Glpr1. Within H2afy, exon 7 and intron 8 were differentially used between strains, with exon 7 reaching the highest significance (FDR = 8.63e−5). We did not detect differential exon usage for any genes within our chromosome 5 brain weight QTL. All differentially used features can be found in Supplementary Table 10.
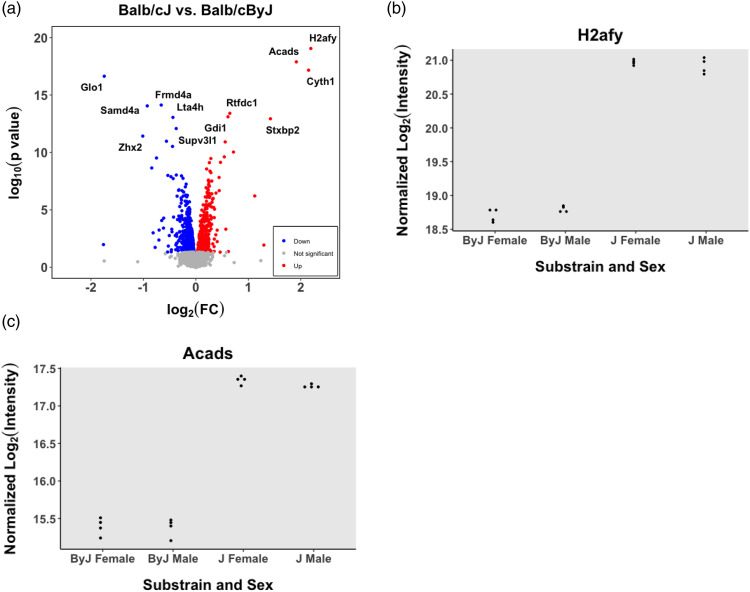
Whole brain proteomic results
Analysis of BALB/c substrain whole brain homogenates of saline and oxycodone pretreated parental substrains via mass spectrometry revealed 1375 differentially expressed genes across strain with an unadjusted p < 0.05. These genes are listed in Supplementary Table 6. There were only nine genes that were differentially expressed at the protein level within our chromosome 13 hot plate QTL interval (Bayesian), including Drd1, Arl10, Nop16, Cltb, Gprin1, H2afy, Tgfbi, Spock1, and Adcy2 (Figure 7). Of these genes Drd1, H2afy, Spock1, and Adcy2 contain at least one annotated variant. Forty genes within the chromosome 5 brain weight QTL interval were found on this list, with eight of these genes containing at least one annotated polymorphism (Rnft2, Aldh2, P2rx7, Anapc5, Smuf2, Dnaaf5, Cyth3, Tecpr1). Notably, Acads was identified as the gene with both the third largest change in protein expression, and second highest p value (logFC = 1.91, adjP = 5.7e−12). All differentially expressed proteins (unadjusted p < 0.05) are included in Supplementary Table 11.
Figure 7.
Whole brain proteomic analysis of differential gene expression between BALB/c substrains. (a): A volcano plot showing the distribution of differentially expressed proteins in whole brain homogenate (J-By expression), controlling for Sex. H2afy is located in the upper right hand corner. (b): Normalized intensity for H2AFY, a protein coded by the candidate gene H2afy for hot plate sensitivity, in J versus By mice (adjusted p = 7.79e−16). (c): Normalized intensity for ACADS, a protein coded by the candidate gene Acads for brain weight, in J versus By mice (adjusted p = 5.7e−12).
Discussion
We identified robust differences in baseline hot plate thermal nociception and mechanical stimulation between BALB/c substrains (Figure 1). In both cases, By mice were more sensitive to stimulation than J mice. We did not observe substrain differences in behavioral sensitivity in the related Hargreaves thermal nociceptive assay nor in the cold plate test, suggesting the genetic factor(s) influencing hot plate and von Frey sensitivity are distinct from these other measures but perhaps could share a common genetic influence within this genetic cross. Alternatively, enhanced sensitivity on the hot plate and von Frey in By mice could be explained by separate genetic variants. We identified a major genome-wide significant QTL for hot plate latency on chromosome 13 spanning ∼20 Mb that explained 15% of the phenotypic variance and recapitulated the BALB/c substrain phenotypic difference (Figure 4, Table 1). Cis-eQTL analysis of two historically processed brain regions (striatum and hippocampus) identified 6 cis-eQTL transcripts within the chromosome 13 interval, including two overlapping genes, Msh3 and H2afy (Table 4 and Table 5). We also replicated in the decrease in brain weight in By versus J28,60 and mapped this trait to a single locus on chromosome 5 that recapitulated the parental substrain difference (Figure 5, Table 1). and identified a differentially expressed protein as a plausible candidate gene within this region (Acads).
H2afy is clearly the most salient positional and functional candidate gene underlying the chromosome 13 QTL for hot plate sensitivity. In whole brain homogenate, H2AFY protein showed the greatest fold-change in expression and was the most significant differentially expressed protein out of all detectable proteins. In striatal and hippocampal eQTL analysis, the most significant associated marker for H2afy expression is one of the three annotated polymorphisms within the gene itself, suggesting that one or more of these H2afy variants could cause decreased mRNA and protein expression. In support, two of these H2afy variants are located within a predicted splice site at the boundary of intron 6 and exon 7 and are separated by only 4 bp. Differential usage of intron 6 in both striatum and hippocampus and differential usage of exon 7 in spinal cord further supported these variants as causal for changes in gene expression.
Surprisingly, we found ∼3.5-fold fewer genes to be differentially expressed in the spinal cord of females versus males (p < 0.05) and notably H2afy was not differentially expressed in the female spinal cord, despite this gene being differentially expressed in both sexes at the mRNA level in multiple brain tissues and at the protein level in whole brain tissue. RNA quality and abundance and similar overall read counts were similar females compared to males, which fails to support technical explanations for these results. Thus, we hypothesize that if H2afy is the causal gene underlying hot plate sensitivity, it is likely working through additional components of the so-called “pain matrix” (e.g., the brainstem, thalamus, anterior cingulate cortex, insula). In support, H2afy is ubiquitously expressed throughout the mouse brain. 61 H2afy, aka Macroh2a1, is a histone involved in transcriptional suppression and X chromosome inactivation 62 that is depleted in actively transcribing chromatin 63 and binds to autosomal chromatin. 64 Chronic pain is associated with global changes in chromatin accessibility. 65 Furthermore, changes in histone acetylation can alter pain phenotypes 66 and inhibition of histone deacetylase genes can reduce neuropathic 67 and inflammatory hyperalgesia. 68
A recent study using a mouse knockout model of H2afy did not find any significant difference in hotplate latencies. 69 However, this study was conducted on a C57BL/6 genetic background, and genetic background is known to influence the detection and direction of behavioral phenotypes in knockout mice. 70 Interestingly, the H2AFY knockout study found increased forced swim mobility, increased social interaction and investigation, and increased reaction to acoustic startle amplitude in H2afy knockout mice. 69 When comparing the By substrain (hypoexpression of H2afy), to the J substrain studies have observed increased forced swim mobility, decreased social aggression, and increased acoustic startle amplitude. 29 These set of observations raise the interesting possibility that the chromosome 13 QTL containing H2afy alters stress responsivity and emotional/affective-like neurobehavioral processes that in turn influence acute nociception and that all these behaviors are mediated by a shared genetic factor—H2afy. We therefore hypothesize that disruption of H2afy transcription through inappropriate splicing and subsequently reduced H2AFY protein in the By substrain perturbs normal transcriptional repression/chromosome accessibility at the genomic level, leading to an transcriptome profile influencing nociceptive neurotransmission and behavior.
BALB/cJ and BALB/cByJ mice differ in gross brain morphology, including corpus colossi length71,72 and brain weight.28,60 In agreement with these studies, we observed decreased whole brain weight in By mice and by extension, we discovered an important contribution to this literature by identifying a genome-wide significant QTL on chromosome 5 explaining 12% of the trait variance. Acads is a strong candidate gene within this region and codes for acyl-CoA-dehydrogenase short chain, a mitochondrial associated protein essential for fatty acid oxidation. This protein was the second most differentially expressed of all the detected proteins within our proteomic data set, with By mice showing reduced expression (pAdj = 5.7e−15, logFC = 1.91). Importantly, By mice harbor a private 278 bp deletion within Acads, resulting in deletion of exon 2 and intron 2 and a partial deletion of, intron 1 and exon 3. 73 This mutation creates two abnormal RNA transcripts harboring early stop codons, and a subsequent lack of ACADS protein. 73 The lack of a significant cis-eQTL for Acads could potentially be explained by a tissue-specific transcriptional difference or by overall transcript levels not being as robustly affected by this deletion compared to protein. In support, exon-level expression QTL analysis shows significantly different intron/exon usage associated with our peak brain weight QTL marker, suggesting that aberrant transcripts are leading to an abundance of non-functional ACADS mRNA.
In baseline cardiac tissues, By mice showed a ∼65% decrease in Acads transcripts compared to J mice, 73 but comparisons between mutant BALB/cByJ and non-mutant substrain BALB/cBy (Jackson Laboratory, #000650) did not reveal differences in Acads whole brain RNA expression. 74 Previous QTL mapping in NZB and NZW mice identified this same region of chromosome 5 underlying variation in plasma HDL quantity, and a coding mutation within a conserved region of Acads was proposed to comprise the mechanism. 75 BALB/cByJ mice showed increased plasma HDL compared to J and this phenotype segregates with the 278 bp deletion within Acads (MGI:3029768) that results in loss of function in By mice. In humans, mutations in ACADS that cause a loss of function lead to short-chain acyl-CoA dehydrogenase deficiency (SCADD) and are associated with microcephaly, developmental delays, epilepsy, and behavioral disorders.76,77 We therefore hypothesize that interruption of mitochondrial fatty acid metabolism through a loss of function mutation in Acads could lead to reduced brain weight. A comparison of NZW and NZO brain weights would provide corroborating evidence that reduced Acads protein underlies differences in brain weight. Whichever the causal gene(s)/variants(s) for reduced brain weight, the chromosome 5 QTL that we identified is likely to contribute to multiple neurobehavioral phenotypes previously reported in BALB/c substrains.
Our study has several limitations. Firstly, our QTL analysis of hot plate sensitivity does not address the question of whether or not the chromosome 13 QTL contributes to other pain-associated phenotypes, including mechanical sensitivity on the von Frey test. Furthermore, our eQTL analysis was not conducted in the most pain-relevant tissues. Given tissue-specific transcriptional and translational differences, especially within the CNS, this raises the possibility that tissue-specific eQTLs or DEGs in pain-relevant regions were missed. In the case of H2afy, confirmation of differential whole brain protein expression helps to mitigate this concern. When considering brain weight, we do not yet understand whether the observed differences are due to localized or global morphological, structural, and neurochemical changes. There is a body of literature demonstrating specific brain regions are disrupted between J and By substrains (i.e., corpus callosum morphology; Fairless et al. 78 ), and a more comprehensive understanding of these phenotypes could help facilitate candidate gene selection for future validation.
These findings leverage a systems genetics approach to identify the genetic loci and candidate genetic factors influencing pain-associated phenotypes and brain morphology. By leveraging the reduced genetic complexity of substrains with large phenotypic differences and combining eQTL, transcriptomic, and proteomic differential gene expression analysis, we identified a small set of candidate genes for these phenotypes. In considering the possibility that H2afy is a causal gene underlying variance in thermal nociception, changes in chromatin accessibility could lead to a widespread transcriptomic profile in nociceptors and/or various neuronal levels of nociceptive transmission (spinal cord, brainstem, thalamus, cortex) that affects multiple pain phenotypes. Layering epigenomic measurements on top of bulk RNA-seq and single cell RNA-seq assessment as it relates to chromatin in J and By mice could shed light on what genes lie downstream of H2afy that could contribute to the quantitative trait mechanism(s) underlying difference in nociceptive sensitivity. It is also interesting to note that exon-level eQTL analysis suggests that variants within H2afy cause the decrease in RNA abundance, and if H2afy and that the two closely localized splice site variants mutations could comprise the quantitative trait variants underlying H2afy expression and possibly hot plate sensitivity. Ultimately, causal gene validation via in vivo editing to both induce and “correct” the candidate variants will be necessary to provide causal validation. 40
Supplemental Material
Supplemental Material, sj-pdf-1-mpx-10.1177_17448069221079540 for Genetic basis of thermal nociceptive sensitivity and brain weight in a BALB/c reduced complexity cross by Jacob A Beierle, Emily J Yao, Stanley I Goldstein, Julia L Scotellaro, Katherine D Sena, Colton A Linnertz, Adam B Willits, Leena Kader, Erin E Young, Gary Peltz, Andrew Emili, Martin T Ferris and Camron D Bryant in Molecular Pain
Supplemental Material, sj-pdf-2-mpx-10.1177_17448069221079540 for Genetic basis of thermal nociceptive sensitivity and brain weight in a BALB/c reduced complexity cross by Jacob A Beierle, Emily J Yao, Stanley I Goldstein, Julia L Scotellaro, Katherine D Sena, Colton A Linnertz, Adam B Willits, Leena Kader, Erin E Young, Gary Peltz, Andrew Emili, Martin T Ferris and Camron D Bryant in Molecular Pain
Acknowledgments
Behavioral phenotyping was conducted in collaboration with the University of Kansas Medical Center Preclinical Models Core– Kansas Intellectual and Developmental Disabilities Research Center (NIH U54 HD 090216). RNA-seq was conducted at the University of Chicago Genomics Core. AE acknowledges generous startup funding from Boston University to the Center for Network Systems Biology which supported this study.
Author Contributions: JAB, EJY, JLS, and KDS conducted behavioral phenotyping at Boston University. EEY, ABW, and LK conducted behavioral phenotyping and tissue collection at KUMC. Martin Ferris and Colton Linnertz provided BALB/cByJ whole genome sequence data and technical advice during QTL analysis. AE and SIG conducted proteomic analysis. JAB and CDB wrote the article, and it was reviewed and edited by EEY, GP, AE, and MTF. All other statistical analysis and bench work not mentioned was conducted by JAB.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the NIH/NIDA U01DA050243 (C.D.B.), R01DA039168 (C.D.B.), U01DA044399 (G.P.), P20GM103418 (D.E.W.), T32GM008541 (D.H. Farb), and by the Burroughs Welcome Fund Transformative Training Program in Addiction Science Grant 1011479.
Supplemental Material: Supplemental material for this article is available online.
ORCID iD
Jacob A Beierle https://orcid.org/0000-0001-6517-7614
References
- 1.Nahin RL. Estimates of pain prevalence and severity in adults: United States, 2012. J Pain 2015; 16: 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Heer EW, Ten Have M, van Marwijk HWJ, Dekker J, de Graaf R, Beekman ATF, van der Feltz-Cornelis CM. Pain as a risk factor for common mental disorders. Results from the Netherlands mental health survey and incidence study-2: a longitudinal, population-based study. Pain 2018; 159: 712–718. [DOI] [PubMed] [Google Scholar]
- 3.Ratcliffe GE, Enns MW, Belik S-L, Sareen J. Chronic pain conditions and suicidal ideation and suicide attempts: an epidemiologic perspective. Clin J Pain 2008; 24: 204–210. [DOI] [PubMed] [Google Scholar]
- 4.Stumbo SP, Yarborough BJH, McCarty D, Weisner C., Green C. A. Patient-reported pathways to opioid use disorders and pain-related barriers to treatment engagement. J Subst Abuse Treat 2017; 73: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hocking LJ, Generation Scotland. Morris AD, Dominiczak AF, Porteous DJ, Smith BH. Heritability of chronic pain in 2195 extended families. Eur J Pain 2012; 16: 1053–1063. [DOI] [PubMed] [Google Scholar]
- 6.Vehof J, Zavos HMS, Lachance G, Hammond CJ, Williams FMK. Shared genetic factors underlie chronic pain syndromes. Pain 2014; 155: 1562–1568. [DOI] [PubMed] [Google Scholar]
- 7.Johnston KJA, Adams MJ, Nicholl BI, Ward J, Strawbridge RJ, Ferguson A, McIntosh AM, Bailey MES, Smith DJ. Genome-wide association study of multisite chronic pain in UK Biobank. Plos Genet 2019; 15: e1008164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H, Ramsay E, Lee H, Wahl S, Dionne RA. Genome-wide association study of acute post-surgical pain in humans. Pharmacogenomics 2009; 10: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters MJ, Broer L, Willemen HL, Eiriksdottir G, Hocking LJ, Holliday KL, Horan MA, Meulenbelt I, Neogi T, Popham M, Schmidt CO, Soni A, Valdes AM, Amin N, Dennison EM, Eijkelkamp N, Harris TB, Hart DJ, Hofman A, Huygen FJ, Jameson KA, Jones GT, Launer LJ, Kerkhof HJ, de Kruijf M, McBeth J, Kloppenburg M, Ollier WE, Oostra B, Payton A, Rivadeneira F, Smith BH, Smith AV, Stolk L, Teumer A, Thomson W, Uitterlinden AG, Wang K, van Wingerden SH, Arden NK, Cooper C, Felson D, Gudnason V, Macfarlane GJ, Pendleton N, Slagboom PE, Spector TD, Völzke H, Kavelaars A, van Duijn CM, Williams FM, van Meurs JB. Genome-wide association study meta-analysis of chronic widespread pain: evidence for involvement of the 5p15.2 region. Ann Rheum Dis 2013; 72: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suri P, Palmer MR, Tsepilov YA, Freidin MB, Boer CG, Yau MS, Evans DS, Gelemanovic A, Bartz TM, Nethander M, Arbeeva L, Karssen L, Neogi T, Campbell A, Mellstrom D, Ohlsson C, Marshall LM, Orwoll E, Uitterlinden A, Rotter JI, Lauc G, Psaty BM, Karlsson MK, Lane NE, Jarvik GP, Polasek O, Hochberg M, Jordan JM, Van Meurs JBJ, Jackson R, Nielson CM, Mitchell BD, Smith BH, Hayward C, Smith NL, Aulchenko YS, Williams FMK. Genome-wide meta-analysis of 158,000 individuals of European ancestry identifies three loci associated with chronic back pain. PLoS Genet 2018; 14: e1007601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell 2009; 139: 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ralston HJ. Pain and the primate thalamus. Prog Brain Res 2005; 149: 1–10. [DOI] [PubMed] [Google Scholar]
- 13.Willis WD. The somatosensory system, with emphasis on structures important for pain. Brain Res Rev 2007; 55: 297–313. [DOI] [PubMed] [Google Scholar]
- 14.Tracey WD. Nociception. Curr Biol 2017; 27: R129–R133. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen CS, Staud R, Price DD. Individual differences in pain sensitivity: measurement, causation, and consequences. J Pain 2009; 10: 231–237. [DOI] [PubMed] [Google Scholar]
- 16.Perkins FM, Kehlet H. Chronic pain as an outcome of surgery. Anesthesiology 2000; 93: 1123–1133. [DOI] [PubMed] [Google Scholar]
- 17.Mogil JS, Wilson SG, Bon K, Eun Lee S, Chung K, Raber P, Pieper JO, Hain HS, Belknap JK, Hubert L, Elmer GI, Mo Chung J, Devor M. Heritability of nociception I: responses of 11 inbred mouse strains on 12 measures of nociception. Pain 1999; 80: 67–82. [DOI] [PubMed] [Google Scholar]
- 18.Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 1997; 389: 816–824. [DOI] [PubMed] [Google Scholar]
- 19.Caterina MJ, Rosen TA, Tominaga M, Brake AJ, Julius D. A capsaicin-receptor homologue with a high threshold for noxious heat. Nature 1999; 398: 436–441. [DOI] [PubMed] [Google Scholar]
- 20.Caterina MJ, Leffler A, Malmberg AB, Martin WJ, Trafton J, Petersen-Zeitz KR, Koltzenburg M, Basbaum AI, Julius D. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science 2000; 288: 306–313. [DOI] [PubMed] [Google Scholar]
- 21.Marrone MC, Morabito A, Giustizieri M, Chiurchiù V, Leuti A, Mattioli M, Marinelli S, Riganti L, Lombardi M, Murana E, Totaro A, Piomelli D, Ragozzino D, Oddi S, Maccarrone M, Verderio C, Marinelli S. TRPV1 channels are critical brain inflammation detectors and neuropathic pain biomarkers in mice. Nat Commun 2017; 8: 15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, Griffin RS, Belfer I, Dai F, Smith SB, Diatchenko L, Gupta V, Xia C, Amann S, Kreitz S, Heindl-Erdmann C, Wolz S, Ly CV, Arora S, Sarangi R, Dan D, Novatchkova M, Rosenzweig M, Gibson DG, Truong D, Schramek D, Zoranovic T, Cronin SJF, Angjeli B, Brune K, Dietzl G, Maixner W, Meixner A, Thomas W, Pospisilik JA, Alenius M, Kress M, Subramaniam S, Garrity PA, Bellen HJ, Woolf CJ, Penninger JM. A Genome-wide drosophila screen for heat nociception identifies α2δ3 as an evolutionarily conserved pain gene. Cell 2010; 143: 628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Althaus A, Arránz Becker O, Neugebauer E. Distinguishing between pain intensity and pain resolution: using acute post-surgical pain trajectories to predict chronic post-surgical pain. Eur J Pain Lond Engl 2014; 18: 513–521. [DOI] [PubMed] [Google Scholar]
- 24.Blichfeldt-Eckhardt MR, ϕrding H, Andersen C, Licht PB, Toft P. Early visceral pain predicts chronic pain after laparoscopic cholecystectomy. Pain 2014; 155: 2400–2407. [DOI] [PubMed] [Google Scholar]
- 25.Hickey OT, Burke SM, Hafeez P, Mudrakouski AL, Hayes ID, Shorten GD. Severity of acute pain after breast surgery is associated with the likelihood of subsequently developing persistent pain. Clin J Pain 2010; 26: 556–560. [DOI] [PubMed] [Google Scholar]
- 26.Tasmuth T, Kataja M, Blomqvist C, von Smitten K, Kalso E. Treatment-related factors predisposing to chronic pain in patients with breast cancer--a multivariate approach. Acta Oncol Stockh Swed 1997; 36: 625–630. [DOI] [PubMed] [Google Scholar]
- 27.Hilakivi LA, Lister RG. Comparison between BALB/cJ and BALB/cByJ mice in tests of social behavior and resident-intruder aggression. Aggress Behav 1989; 15: 273–280. [Google Scholar]
- 28.Sittig LJ, Jeong C, Tixier E, Davis J, Barrios-Camacho CM, Palmer AA. Phenotypic instability between the near isogenic substrains BALB/cJ and BALB/cByJ. Mamm Genome 2014; 25: 564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velez L, Sokoloff G, Miczek KA, Palmer AA, Dulawa SC. Differences in aggressive behavior and DNA copy number variants between BALB/cJ and BALB/cByJ substrains. Behav Genet 2010; 40: 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Recla JM, Robledo RF, Gatti DM, Bult CJ, Churchill GA, Chesler EJ. Precise genetic mapping and integrative bioinformatics in diversity outbred mice reveals hydin as a novel pain gene. Mamm Genome 2014; 25: 211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mogil JS. Melanocortin-1 receptor gene variants affect pain and -opioid analgesia in mice and humans. J Med Genet 2005; 42: 583–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nissenbaum J. From mouse to humans: discovery of the CACNG2 pain susceptibility gene. Clin Genet 2012; 82: 311–320. [DOI] [PubMed] [Google Scholar]
- 33.Bryant CD. The blessings and curses of C57BL/6 substrains in mouse genetic studies. Ann N Y Acad Sci 2011; 1245: 31–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan X, Nellåker C, Goodstadt L, Nicod J, Bhomra A, Hernandez-Pliego P, Whitley H, Cleak J, Dutton R, Janowitz D, Mott R, Adams DJ, Flint J. Sequence-based characterization of structural variation in the mouse genome. Nature 2011; 477: 326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bryant CD, Ferris MT, De Villena FPM, Damaj MI, Kumar V, Mulligan MK. Chapter 8 - reduced complexity cross design for behavioral genetics. In: Gerlai RT. (ed). Molecular-genetic and statistical techniques for behavioral and neural research. Cambridge, MA: Academic Press, 2018, pp. 165–190. DOI: 10.1016/B978-0-12-804078-2.00008-8 [DOI] [Google Scholar]
- 36.Bryant CD, Smith DJ, Kantak KM, Nowak TS, Williams RW, Damaj MI, Redei EE, Chen H, Mulligan MK. Facilitating complex trait analysis via reduced complexity crosses. Trends Genet 2020; 36: 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar V, Kim K, Joseph C, Kourrich S, Yoo S-H, Huang HC, Vitaterna MH, Pardo-Manuel de Villena F, Churchill G, Bonci A, Takahashi JS. C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science 2013; 342: 1508–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirkpatrick SL, Goldberg LR, Yazdani N, Babbs RK, Wu J, Reed ER, Jenkins DF, Bolgioni AF, Landaverde KI, Luttik KP, Mitchell KS, Kumar V, Johnson WE, Mulligan MK, Cottone P, Bryant CD. Cytoplasmic FMR1-interacting protein 2 is a major genetic factor underlying binge eating. Biol Psychiatry 2017; 81: 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryant CD, Bagdas D, Goldberg LR, Khalefa T, Reed ER, Kirkpatrick SL, Kelliher JC, Chen MM, Johnson WE, Mulligan MK, Imad Damaj M. C57BL/6 substrain differences in inflammatory and neuropathic nociception and genetic mapping of a major quantitative trait locus underlying acute thermal nociception. Mol Pain 2019; 15: 1744806918825046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldberg LR, Yao EJ, Kelliher JC, Reed ER, Cox JW, Parks C, Kirkpatrick SL, Beierle JA, Chen MM, Johnson WE, Homanics GE, Williams RW, Bryant CD, Mulligan MK. A quantitative trait variant in gabra2 underlies increased methamphetamine stimulant sensitivity. Genes Brain Behav 2021: 20: 450337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellåker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assunção JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 2011; 477: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dam SA, Jager A, Oomen CA, Buitelaar JK, Arias-Vasquez A, Glennon JC. Inhibitory control in BALB/c mice sub-strains during extinction learning. Eur Neuropsychopharmacol 2019; 29: 509–518. [DOI] [PubMed] [Google Scholar]
- 43.Jager A, Dam SA, Van Der Mierden S, Oomen CA, Arias-Vasquez A, Buitelaar JK, Kozicz T, Glennon JC. Modulation of cognitive flexibility by reward and punishment in BALB/cJ and BALB/cByJ mice. Behav Brain Res 2020; 378: 112294. [DOI] [PubMed] [Google Scholar]
- 44.Perincheri S, Dingle RWC, Peterson ML, Spear BT. Hereditary persistence of -fetoprotein and H19 expression in liver of BALB/cJ mice is due to a retrovirus insertion in the Zhx2 gene. Proc Natl Acad Sci 2005; 102: 396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poyntz HC, Jones A, Jauregui R, Young W, Gestin A, Mooney A, Lamiable O, Altermann E, Schmidt A, Gasser O, Weyrich L, Jolly CJ, Linterman MA, Gros GL, Hawkins ED, Forbes-Blom E. Genetic regulation of antibody responsiveness to immunization in substrains of BALB/c mice. Immunol Cel Biol 2019; 97: 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner JK, McAllister MM, Xu JL, Tapping RI. The resistance of BALB/cJ mice to yersinia pestis maps to the major histocompatibility complex of chromosome 17. Infect Immun 2008; 76: 4092–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.National Research Council (US) . Committee for the update of the guide for the care and use of laboratory animals. Guide for the care and use of laboratory animals. Washington, DC: National Academies Press (US), 2011. [PubMed] [Google Scholar]
- 48.Kirkpatrick SL, Bryant CD. Behavioral architecture of opioid reward and aversion in C57BL/6 substrains. Front Behav Neurosci 2015; 8: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peterson RA. Finding optimal normalizing transformations via best normalize. R J 2021; 13: 310. [Google Scholar]
- 50.Sigmon JS, Blanchard MW, Baric RS, Bell TA, Brennan J, Brockmann GA, Burks AW, Calabrese JM, Caron KM, Cheney RE, Ciavatta D, Conlon F, Darr DB, Faber J, Franklin C, Gershon TR, Gralinski L, Gu B, Gaines CH, Hagan RS, Heimsath EG, Heise MT, Hock P, Ideraabdullah F, Jennette JC, Kafri T, Kashfeen A, Kulis M, Kumar V, Linnertz C, Livraghi-Butrico A, Lloyd KCK, Lutz C, Lynch RM, Magnuson T, Matsushima GK, McMullan R, Miller DR, Mohlke KL, Moy SS, Murphy CEY, Najarian M, O’Brien L, Palmer AA, Philpot BD, Randell SH, Reinholdt L, Ren Y, Rockwood S, Rogala AR, Saraswatula A, Sassetti CM, Schisler JC, Schoenrock SA, Shaw GD, Shorter JR, Smith CM, St Pierre CL, Tarantino LM, Threadgill DW, Valdar W, Vilen BJ, Wardwell K, Whitmire JK, Williams L, Zylka MJ, Ferris MT, McMillan L, Manuel de Villena FP. Content and performance of the MiniMUGA genotyping array: a new tool to improve rigor and reproducibility in mouse research. Genetics 2020; 216: 905–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003; 19(7): 889–890, doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 52.Yazdani N, Parker CC, Shen Y, Reed ER, Guido MA, Kole LA, Kirkpatrick SL, Lim JE, Sokoloff G, Cheng R, Johnson WE, Palmer AA, Bryant CD. Hnrnph1 is a quantitative trait gene for methamphetamine sensitivity. PLoS Genet 2015; 11(12): e1005713, doi: 10.1371/journal.pgen.1005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30(15): 2114–2120, doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29(1): 15–21, doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 2012; 28: 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mancini E, Rabinovich A, Iserte J, Yanovsky M, Chernomoretz A. ASpli: integrative analysis of splicing landscapes through RNA-Seq assays. Bioinformatics 2021; 37: 2609–2616. [DOI] [PubMed] [Google Scholar]
- 58.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 2007; 4: 207–214. [DOI] [PubMed] [Google Scholar]
- 59.Blum BC, Emili A. Omics notebook: robust, reproducible and flexible automated multiomics exploratory analysis and reporting. Bioinforma Adv 2021; 1: vbab024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fairless AH, Dow HC, Kreibich AS, Torre M, Kuruvilla M, Gordon E, Morton EA, Tan J, Berrettini WH, Li H, Abel T, Brodkin ES. Sociability and brain development in BALB/cJ and C57BL/6J mice. Behav Brain Res 2012; 228: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ng L, Bernard A, Lau C, Overly CC, Dong H-W, Kuan C, Pathak S, Sunkin SM, Dang C, Bohland JW, Bokil H, Mitra PP, Puelles L, Hohmann J, Anderson DJ, Lein ES, Jones AR, Hawrylycz M. An anatomic gene expression atlas of the adult mouse brain. Nat Neurosci 2009; 12: 356–362. [DOI] [PubMed] [Google Scholar]
- 62.Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 1998; 393: 599–601. [DOI] [PubMed] [Google Scholar]
- 63.Changolkar LN, Pehrson JR. macroH2A1 histone variants are depleted on active genes but concentrated on the inactive X chromosome. Mol Cel Biol 2006; 26: 4410–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev 2010; 24: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stephens KE, Zhou W, Renfro Z, Ji Z, Ji H, Guan Y, Taverna SD. Global gene expression and chromatin accessibility of the peripheral nervous system in animal models of persistent pain. J Neuroinflammation 2021; 18: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tran L, Schulkin J, Ligon CO, Greenwood-Van Meerveld B. Epigenetic modulation of chronic anxiety and pain by histone deacetylation. Mol Psychiatry 2015; 20: 1219–1231. [DOI] [PubMed] [Google Scholar]
- 67.Denk F, Huang W, Sidders B, Bithell A, Crow M, Grist J, Sharma S, Ziemek D, Rice ASC, Buckley NJ, McMahon SB. HDAC inhibitors attenuate the development of hypersensitivity in models of neuropathic pain. Pain 2013; 154: 1668–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bai G, Wei D, Zou S, Ren K, Dubner R. Inhibition of class II histone deacetylases in the spinal cord attenuates inflammatory hyperalgesia. Mol Pain 2010; 6: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chiodi V, Domenici MR, Biagini T, De Simone R, Tartaglione AM, Di Rosa M, Lo Re O, Mazza T, Micale V, Vinciguerra M. Systemic depletion of histone macroH2A1.1 boosts hippocampal synaptic plasticity and social behavior in mice. FASEB J 2021; 35: e21793. [DOI] [PubMed] [Google Scholar]
- 70.Sittig LJ, Carbonetto P, Engel KA, Krauss KS, Barrios-Camacho CM, Palmer AA. Genetic background limits generalizability of genotype-phenotype relationships. Neuron 2016; 91: 1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wahlsten D. Deficiency of the corpus callosum: incomplete penetrance and substrain differentiation in BALB/c Mice. J Neurogenet 1989; 5: 61–76. [DOI] [PubMed] [Google Scholar]
- 72.Wahlsten D. Genetic and developmental defects of the mouse corpus callosum. Experientia 1989; 45: 828–838. [DOI] [PubMed] [Google Scholar]
- 73.Hinsdale ME, Kelly CL, Wood PA. Null allele at Bcd-1 locus in BALB/cByJ mice is due to a deletion in the short-chain Acyl-CoA dehydrogenase gene and results in missplicing of mRNA. Genomics 1993; 16: 605–611. [DOI] [PubMed] [Google Scholar]
- 74.Tafti M, Petit B, Chollet D, Neidhart E, de Bilbao F, Kiss JZ, Wood PA, Franken P. Deficiency in short-chain fatty acid β-oxidation affects theta oscillations during sleep. Nat Genet 2003; 34: 320–325. [DOI] [PubMed] [Google Scholar]
- 75.Su Z, Leduc MS, Korstanje R, Paigen B. Untangling HDL quantitative trait loci on mouse chromosome 5 and identifying scarb1 and acads as the underlying genes. J Lipid Res 2010; 51: 2706–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kılıç M, Şenel S, Karaer K, Ceylaner S. Microcephaly and developmental delay caused by short-chain acyl-coa dehydrogenase deficiency. Turk J Pediatr 2017; 59: 708. [DOI] [PubMed] [Google Scholar]
- 77.van Maldegem BT, Wanders RJA, Wijburg FA. Clinical aspects of short-chain acyl-CoA dehydrogenase deficiency. J Inherit Metab Dis 2010; 33: 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fairless AH, Dow HC, Toledo MM, Malkus KA, Edelmann M, Li H, Talbot K, Arnold SE, Abel T, Brodkin ES. Low sociability is associated with reduced size of the corpus callosum in the BALB/cJ inbred mouse strain. Brain Res 2008; 1230: 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-pdf-1-mpx-10.1177_17448069221079540 for Genetic basis of thermal nociceptive sensitivity and brain weight in a BALB/c reduced complexity cross by Jacob A Beierle, Emily J Yao, Stanley I Goldstein, Julia L Scotellaro, Katherine D Sena, Colton A Linnertz, Adam B Willits, Leena Kader, Erin E Young, Gary Peltz, Andrew Emili, Martin T Ferris and Camron D Bryant in Molecular Pain
Supplemental Material, sj-pdf-2-mpx-10.1177_17448069221079540 for Genetic basis of thermal nociceptive sensitivity and brain weight in a BALB/c reduced complexity cross by Jacob A Beierle, Emily J Yao, Stanley I Goldstein, Julia L Scotellaro, Katherine D Sena, Colton A Linnertz, Adam B Willits, Leena Kader, Erin E Young, Gary Peltz, Andrew Emili, Martin T Ferris and Camron D Bryant in Molecular Pain