Abstract
Nanosized extracellular vesicles derived from bacteria contain diverse cargo and transfer intercellular bioactive molecules to cells. Due to their favorable intercellular interactions, cell membrane-derived bacterial extracellular vesicles (BEVs) have great potential to become novel drug delivery platforms. In this review, we summarize the biogenesis mechanism and compositions of various BEVs. In addition, an overview of effective isolation and purification techniques of BEVs is provided. In particular, we focus on the application of BEVs as bioactive nanocarriers for drug delivery. Finally, we summarize the advances and challenges of BEVs after providing a comprehensive discussion in each section. We believe that a deeper understanding of BEVs will open new avenues for their exploitation in drug delivery applications.
Keywords: Bacterial extracellular vesicles, Bioactive nanocarriers, Isolation and purification, Drug delivery
Graphical abstract
Highlights
-
•
Bacterial extracellular vesicles (BEVs) are excellent nanomaterials as drug delivery systems.
-
•
The unique nanosized structures and biofunctions of BEVs are attractive for their use as nanomedicine platforms.
-
•
BEVs have been investigated as biotherapeutics due to their loading capacity, ease of modification and industrialization.
-
•
This review provides new insights of BEVs in drug delivery applications, discussing potential opportunities and challenges.
1. Introduction
As one of the main groups of organisms, bacteria have formed a close relationship with human health and disease, including sepsis, inflammatory bowel disease, arthritis, and tumors [[1], [2], [3], [4]]. The combination of synthetic biology technology with bacteria has been shown to improve characteristics such as chemotaxis and molecule secretion, thereby facilitating their clinical applications in a range of diseases [5]. Accordingly, in recent years, the application of engineered bacteria in drug delivery has received increasing interest [6]. Bacteria-based delivery systems have led to an ingenious synergy of interdisciplinary research for various biomedical applications [7,8]. However, the pathogenicity and immunogenicity of bacteria are major challenges for bacteria-based drug delivery systems. Although attenuated bacteria with the deletion of virulence genes improve the safety of bacteria-based delivery systems, the potential and unknown side effects remain a significant issue [9]. Recently, with the in-depth understanding of bacteria-based drug delivery systems, the use of bacterial extracellular vesicles (BEVs) for biomedical applications has become a new trend [10].
Bacteria release nanosized extracellular vesicles with diameters ranging from 20 to 400 nm, which carry a range of cargo, including lipopolysaccharide (LPS, also called endotoxin), peptidoglycan (PG), membranes, periplasmic and cytoplasmic proteins, toxins, and nucleic acids. BEVs affect a variety of biological processes, including virulence, horizontal gene transfer, phage infection, transport of cell metabolites, and bacterial-bacterial or bacterial-host interactions [11,12]. From a drug delivery perspective, BEVs are comparable to liposomes, given that both comprise biocompatible lipid bilayers [13]. Lipid-based nanocarriers offer a versatile platform for drug encapsulation, which has led to clinical translation of several formulations [14]. Similarly, cell membrane derived BEVs can transport various bioactive molecules to recipient cells, thereby changing the physiology of those cells. BEVs have been widely investigated as biotherapeutics due to their drug loading capacity, immunostimulatory capability, and ease of modification [15,16]. Such attractive properties have drawn a great deal of attention regarding the use of BEVs for drug delivery, which may overcome issues related to liposomes and other synthetic drug delivery systems.
In this review, we have extensively focused on the recent development of extracellular vesicles derived from gram-negative (G−) bacteria and gram-positive (G+) bacteria. Following a particular emphasis on the biogenesis and contents of BEVs, this review also describes the technology used for efficient isolation and purification of BEVs. Clearly understanding the biogenesis mechanism of BEVs and successfully obtaining pure BEVs are the most basic steps for their application as drug delivery systems. Moreover, we critically discuss the application of BEVs as drug delivery vehicles and as new therapeutics. With a comprehensive discussion in each section, we summarize the advances in BEVs and discuss the challenges and future directions.
2. Biogenesis and contents of bacterial extracellular vesicles
Based on their structure, morphology, and staining properties, bacteria are divided into G+ or G− types. G+ bacteria have thick cell walls with abundant PG, while G− bacteria have inner and outer membranes. The generation of extracellular vesicles is a spontaneous process that does not require energy consumption [17,18]. The differences in bacterial structure and physiology lead to distinct types of extracellular vesicles [19]. Moreover, the formation mechanisms and contents of BEVs are different between G− bacteria and G+ bacteria (Fig. 1).
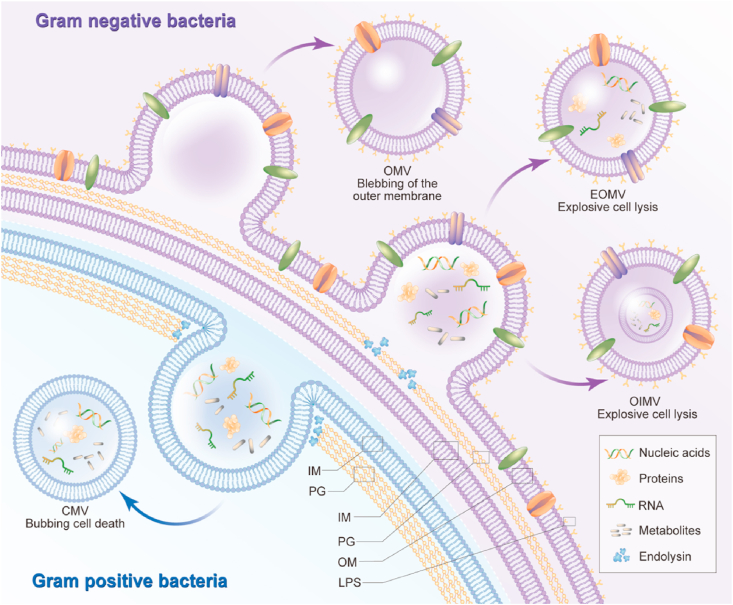
Fig. 1.
The biogenesis and composition of bacterial extracellular vesicles. The G− BEVs are generated by two main models, blebbing of the outer membrane and explosive cell lysis. The insertion of hydrophobic molecules or the instability of PG biosynthesis into the outer membrane cause blebbing of the outer membrane, which produces classic OMV. The weakness of PG layer by endolysin leads the inner membrane protrudes into the periplasm, which generate explosive EOMV or OIMV. On the other hand, the biogenesis mechanism of G+ BEVs release is bubbling cell death. The endolysin degrades the PG layer and triggers bubbling cell death in G+ bacteria and produces CMVs. The difference in contents of G+ and G− BEVs goes beyond the presence of LPS and includes other molecules, such as nucleic acids, proteins, lipids, and metabolites. LPS: lipopolysaccharide, OM: other membrane, PG: peptidoglycan, IM: Inner membrane.
2.1. Gram-negative bacteria
As with the history of mammalian extracellular vesicles (MEVs), BEVs have always been regarded as cellular debris from the decomposition of dead cells. The discovery that bacteria produce extracellular vesicles through metabolic activity and the fact that BEVs and MEVs have many similarities in structure and function strongly indicate that extracellular vesicles are released by living bacteria, especially G− bacteria [[20], [21], [22]]. G− bacteria are characterized by an outer membrane, cytoplasmic membrane, and periplasmic space [23]. The outer membrane contains an exterior leaflet of LPS and an interior leaflet of phospholipids. The cytoplasmic membrane, which serves as an electrochemical barrier, is composed of a typical phospholipid bilayer. The periplasmic space contains a PG layer, lipoproteins, and periplasmic proteins [24]. The reticulated PG layer in the periplasm imparts bacterial morphology and protects the cell from absolute pressure and osmotic changes.
It has been observed that all types of G− bacteria generate BEVs in different environments, such as biofilms, planktonic cultures, fresh and salt water, and inside eukaryotic cells and mammalian hosts [[25], [26], [27]]. Extracellular vesicles were first reported in G− bacteria in the 1960s [28,29]. In 1995, Kadurugamuwa et al. provided obvious evidence that the G− bacterium Pseudomonas aeruginosa releases extracellular vesicles into medium during normal growth [30]. Transmission electron microscopy (TEM) demonstrated that these extracellular vesicles were spherical bilayers with a diameter of 50–150 nm. Moreover, immunoelectron microscopy and western blot analysis showed that these extracellular vesicles contain LPS and DNA [30]. The extracellular vesicles derived from Escherichia coli DH5α had a diameter of 15–100 nm [31]. Additionally, a total of 141 proteins were identified by using a proteomics approach in DH5α-derived extracellular vesicles, which were highly enriched in outer membrane proteins but lacking in inner membrane proteins. Other G− bacteria such as Salmonella sp. [32], Shigella sp. [33], Campylobacter jejuni [34], Helicobacter pylori [35], Vibrio sp. [36], Borrelia burgdorferi [37], Neisseria sp. [38], and Acinetobacter baumannii [39] et al. have also been reported to produce extracellular vesicles.
The extracellular vesicles derived from G− bacteria are generated by two main models: blebbing of the outer membrane and explosive cell lysis [19]. The insertion of hydrophobic molecules or the instability of PG biosynthesis into the outer membrane cause blebbing of the outer membrane, which produces classical outer membrane vesicles (OMVs) (Fig. 1) [12,19]. Both genetic background and growth conditions strongly affect the production of BEVs. It has been reported that the use of polymyxin or gentamicin can influence lipid homeostasis to trigger cell outer membrane stress and generate OMVs through blebbing of the outer membrane [40,41]. Structurally, OMVs are spherical particles with a size of 20–250 nm, which consist of an inner leaflet of phospholipid and an outer leaflet of LPS [16,18,42,43]. In this case, the inner membrane is not damaged, and the components in the cytoplasm cannot access OMVs. Therefore, these OMVs contain abundant outer membrane proteins and lipids. Growth conditions such as the temperature, oxidation state, nutrient availability, quorum sensing, and envelope-targeting antibiotics, etc., also affect the contents of cargo in OMVs [44,45]. As growth conditions have significant influence on the BEV process, late-stage cells display maximum OMV yields [46]. However, unavoidable cell death in later stages causes contamination with cytosolic proteins and membrane components. Moreover, nutrient deficiency and waste accumulation at later life stages of cells affect the composition profile of BEVs [47,48]. Notably, many studies have shown that OMVs contain cytosolic and periplasmatic proteins, DNA and RNA [49,50]. The selectivity of OMV cargo indicates that biogenesis of extracellular vesicles is a deliberate process, rather than a random event. However, the biogenesis mechanism of cargo selection is still ambiguous. In addition, the fact that cytoplasmic contents are present in OMVs remains unexplained and needs further study [51].
Another important mechanism of G− BEVs is explosive cell lysis. The weakening of the PG layer by endolysin causes the inner membrane to protrude into the periplasm, which generates explosive outer-membrane vesicles (EOMVs) or outer-inner membrane vesicles (OIMVs) [19] (Fig. 1). Evidence has accumulated that ciprofloxacin can induce the cellular SOS response, which triggers the expression of endolysins and stimulates explosive cell lysis [52,53]. EOMVs have only an outer membrane, while OIMVs have two membrane bilayers (outer membrane and inner membrane). Both OIMVs and EOMVs randomly contain cytoplasmic components, including outer membrane proteins, cytoplasmic membrane proteins, chromosomal DNA, RNA, endolysins, virulence factors, hydrophobic molecules, and phages [54,55]. Notably, the models of blebbing of the outer membrane and explosive cell lysis are not independent. G− bacteria such as Shewanella vesiculosa M7 have been reported to produce both OMVs and OIMVs, and the composition ratio of these two types of vesicles is quite different [56]. The proportion of OIMVs in total extracellular vesicles ranges from 0.1% (S. vesiculosa M7 [56]) to 49% (Pseudoalteromonas marina [57]). These differences in the types of BEVs indicate that although the production of extracellular vesicles is a universal phenomenon, bacteria synthesize vesicles in different ways.
2.2. Gram-positive bacteria
In recent decades, research on G− BEVs has increased substantially; however, there has been little research on G+ BEVs. The existence of extracellular vesicles derived from G+ bacteria was not observed until 1990 [58], appearing nearly 30 years after similar reports regarding G− bacteria. Vesicle-like blisters were found on the surface of the G+ bacteria Bacillus spp, but no in-depth research has been conducted [58]. Structurally, the inner layer of G+ bacteria is composed of a single lipid membrane, and the outer layer is composed of a thick PG and lipoteichoic acid layer [59]. The main reason for the lack of interest in G+ BEVs is the thick cell wall, which may be a physical barrier to the release of extracellular vesicles. However, there is growing evidence that BEVs can reach the extracellular space through the thick cell wall [[60], [61], [62]].
In 2009, Lee et al. provided the first definitive evidence that the G+ bacterium Staphylococcus aureus (S. aureus) could naturally produce extracellular vesicles [63]. In addition, proteomics characterization of S. aureus-derived extracellular vesicles has been performed, and many extracellular proteins have been identified. The diameter of extracellular vesicles derived from S. aureus ranges from 20 nm to 100 nm. Those of other G+ bacteria such as Streptomyces coelicolor- and Bacillus subtilis-derived extracellular vesicles have been visualized with TEM or scanning electron microscopy (SEM) [20,64]. As bacteria synthesize vesicles in different ways, the size of extracellular vesicles produced by Streptococcus pneumoniae, Staphylococcus spp., and Listeria monocytogenes ranges from 20 nm to 150 nm in diameter [[65], [66], [67]], while the diameter of extracellular vesicles derived from Bacillus spp., Streptomyces coelicolor and Clostridium perfringens ranges from 20 nm to 400 nm [20,60,64,68]. This variability in the size of these BEVs once again indicates that the generation of extracellular vesicles is a common but unique process.
Previously, three non-mutually exclusive hypotheses on the release mechanism of extracellular vehicles from thick cell walls in G+ bacteria have been proposed. 1) The extracellular carrier is first released from the cell membrane to generate turgor pressure, which triggers the release of extracellular vehicles from the cell wall. The cell wall thickness and pore size can regulate the size of extracellular vehicles to pass through the cell wall [69,70]. 2) The wall is loosened by protease released with extracellular vehicles, and the pore size is increased to facilitate extracellular vehicle release [63,71]. 3) Protein channels in the PG layer may release extracellular vehicles to the extracellular environment [69,70]. However, the biogenesis mechanism of G+ BEVs is now considered to be bubbling cell death [19]. Similarly, endolysin degrades the PG layer and triggers bubbling cell death in G+ bacteria, which generates cytoplasmic membrane vesicles (CMVs) (Fig. 1) [19]. Antibiotics that weaken the PG layer, such as β-lactams, have been shown to trigger CMV formation. In addition, other cell wall-damaging enzymes may have effects similar to those of endolysin. CMVs can carry a variety of cargo, including cytoplasmic membrane proteins, RNA, chromosomal DNA, endolysins, and virulence factors [11,42,[72], [73], [74]]. Interestingly, extracellular vesicles derived from G+ bacteria do not contain LPS, which is a compound that may cause an innate immune response [17]. However, the difference in the contents of G+ and G− BEVs goes beyond the presence of LPS and includes other molecules such as DNA, RNA, proteins, lipids, and metabolites, ultimately resulting in the different biological functions of these BEVs. Functionally, BEVs have been determined to have diverse roles, resulting in distinct host immune responses [11,16,17,19,75]. In general, it appears that bacteria can use extracellular vesicles as bioactive nanocarriers to communicate with other bacteria and hosts. Recent development of nanostructured BEVs for drug delivery and therapeutics will be introduced in detail in subsequent chapters.
3. Isolation and purification of bacterial extracellular vesicles
Here, effective BEV isolation and purification techniques are provided in this review (Fig. 2). After culturing for an appropriate amount of time, bacteria in the fermentation broth are removed by low-speed centrifugation (usually 2000×g ∼ 10,000×g) [31,63,65]. The supernatant is then filtered through a sterile filter (usually 0.22 μm or 0.45 μm, depending on the size of bacteria) to remove residual bacteria [76,77]. Subsequently, ultrafiltration membranes (usually 50 KDa ∼100 KDa) are required to concentrate BEVs and remove non-BEV-associated proteins. The ultrafiltration membrane technique is the commonly used method in the concentration step of BEVs. The choice of ultrafiltration membrane is critical for successful concentration. Regenerated cellulose membranes rather than polyether sulfone membranes are the first choice for dilute solutions. However, polyether sulfone membranes are preferred for high protein concentrations, such as porcine mucin for Akkermansia muciniphila (A. muciniphila) [17,78].
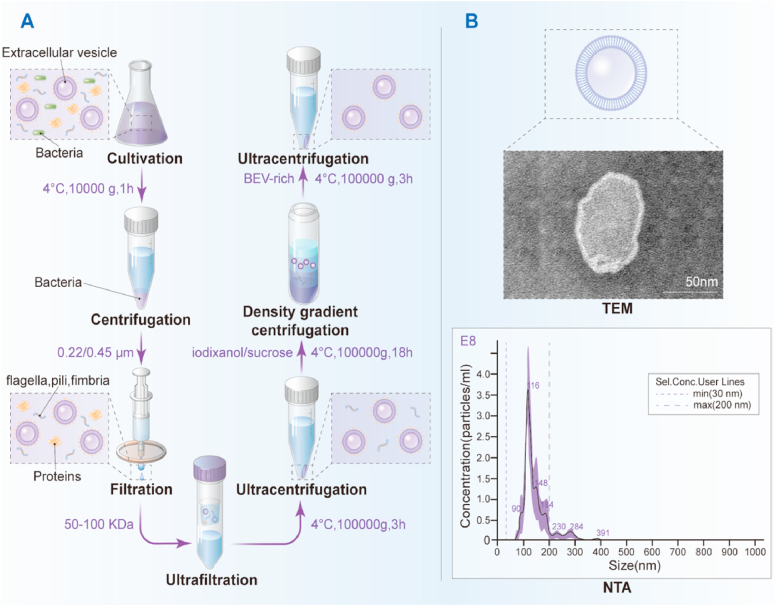
Fig. 2.
The isolation and purification of bacterial extracellular vesicles. (A) After cultured in appropriate time, the bacteria in the fermentation broth could be removed by low-speed centrifugation at 10, 000 g for 1 h. The supernatant is then filtered through a 0.22 μm sterile filter to remove residual bacteria. Subsequently, 100 KDa ultrafiltration membranes is required to concentrate BEVs and remove non-BEVs-associated proteins. The retentate is subjected to ultracentrifugation at 100, 000 g for 3 h to collect the non-purified BEVs, which could be further purified by density gradient centrifugation with iodixanol, and ultracentrifuged at 100, 000 g for 18 h. Finally, BEVs-rich fraction is diluted with PBS and collected by ultracentrifugation at 100, 000 g for 3 h. (B) TEM and NTA have been used to evaluate extracellular vesicles, as exemplified here by extracellular vesicles produced by Lactobacillus rhamnosus GG (scale bar = 50 nm).
Furthermore, the retentate is subjected to ultracentrifugation, which is the most conventional approach for extracellular vesicle isolation, to collect the BEVs [11,31,79]. However, unpurified BEVs can mix with bacteria flagella or large protein complexes, which may affect the subsequent experiments and lead to unexpected immunomodulatory effects [42]. Therefore, non-BEVs related materials are often separated by density gradient centrifugation, which is a more stringent form of ultracentrifugation [80]. Usually, density gradient centrifugation and ultracentrifugation are used in combination to obtain purified BEVs. The most frequently applied media for density gradient centrifugation are sucrose [31] and iodixanol [79]. Compared with other media, iodixanol has obvious advantages, and its solutions are isosmotic, protecting subtle membranous structures [81]. Unpurified BEVs are typically mixed with 60% w/v iodixanol and then stacked with 40% w/v, 20% w/v, and 10% w/v iodixanol [79,82,83]. BEVs migrate into an equilibrium position according to their density during ultracentrifugation. Then, the BEV-rich fraction is diluted to 50 mL PBS and centrifuged at 100,000×g ∼200,000×g to obtain BEV pellets [31,79]. Although gel filtration is another choice for BEV purification, this method requires special equipment and fillers, and it is not suitable for multiple sample operations at the same time.
After obtaining purified BEVs, their quantification and quality determination are vital steps for further application. TEM is a well-established and commonly used technique to demonstrate the presence of BEVs and to observe their sizes, shapes, and purity [84]. Moreover, dynamic light scattering (DLS) is usually used to measure the size distribution of extracellular vesicles in liquid suspension [79,[85], [86], [87]]. Liu et al. used both TEM and DLS to characterize the sizes and shapes of isolated extracellular vesicles derived from gut microbiota [79]. The TEM and DLS results showed that the extracellular vesicles derived from child gut microbiota, elderly gut microbiota, and A. muciniphila were spherical structures with diameters of nearly 200 nm. However, when DLS is used to measure smaller particles, the measured particle size has a large deviation from the actual size. To make up for the shortcomings of DLS, nanoparticle tracking analysis (NTA) technology was developed. NTA is another commonly used method to measure the size distribution and concentration of nanoparticles and has higher accuracy than DLS [88]. Therefore, it has become the most mainstream size measurement method in extracellular vesicles. Hu et al. applied both TEM and NTA to evaluate hybrid nanoparticles (liposomes and exosomes) [89]. The TEM and NTA results showed that the nanoparticles were spherical with a diameter of nearly 150 nm, accounting for 99.2% of the total amount [89]. Although the tunable resistive pulse sensing (TRPS) method is one option to measure the size distribution and concentration of extracellular vesicles, for small nanoparticles (usually <150 nm), NTA always detects more extracellular vesicles than TRPS [90].
4. Application of bacterial extracellular vesicles
Conventional synthetic nanomaterials, such as liposomes, metal-based nanoparticles, and polymers, have been widely used as drug carriers [91]. However, the simple conjugation of synthetic nanomaterials is not efficient in replicating intercellular interactions that facilitate nanoparticle trafficking and delivery [18]. Due to the ability to contain and deliver various bioactive molecules, nanosized BEVs have great potential as a new type of drug delivery carrier. Moreover, it has been demonstrated that BEVs can enter distant organs by systemic circulation through the disrupted tight junction under microbial disorder and inflammatory environment (Fig. 3) [92]. Here, we summarize the current applications of BEVs in gut, brain, bone, and tumor diseases (Fig. 4).
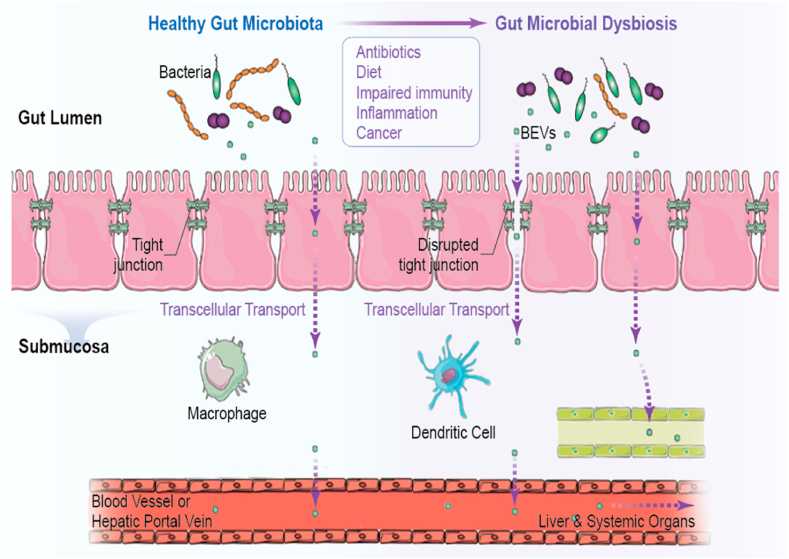
Fig. 3.
BEVs achieve access to systemic circulation through the disrupted tight junction under microbial disorder and inflammatory environment. Reprinted with permission [92]. Copyright 2020, Springer.
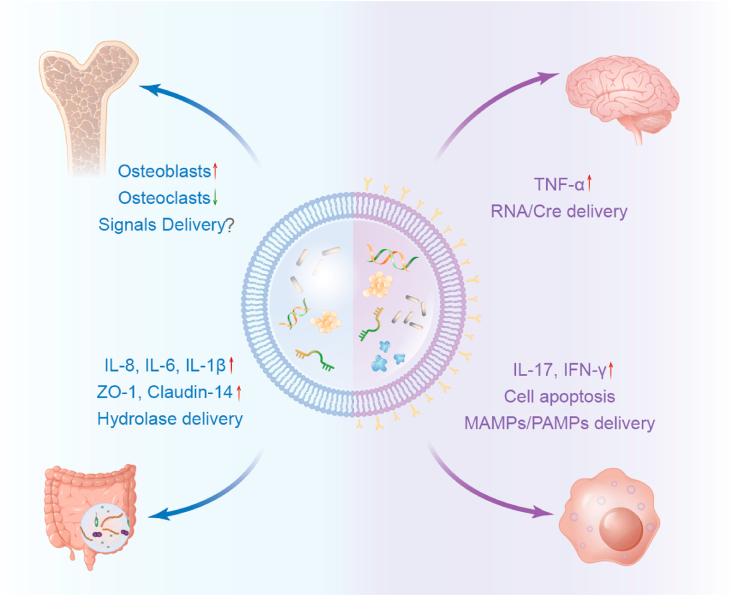
Fig. 4.
The application of bacterial extracellular vesicles in drug delivery. As a nano-scale bioactive material, BEVs deliver a variety of active molecules such as nucleic acids, protein, metabolisms etc. for the treatment of gut, brain, bone, and tumors.
4.1. Gut
The gut microbiota, which is the basis for regulating human health, actively promotes the functions of the intestine and the distal organs [93]. The intestinal microbiota and host cells have established a complex and dynamic relationship, also known as interkingdom crosstalk [93]. Epithelial cells from both the gastrointestinal tract, lung, cornea, and oral cavity are besieged by numerous pathogens, opportunistic pathogens, and probiotics. In an in vivo experiment, both Porphyromonas gingivalis cells and their vesicles increased IL-8 expression, but monocytes were more likely to be recruited by extracellular vesicles, indicating a vesicle-induced innate immune response in the epithelial system [94]. Moreover, excessive IL-8 production was observed in primary human bronchial and intestinal epithelial cells treated with P. aeruginosa and E. coli O157 extracellular vesicles [95,96]. The immunostimulatory functions of BEVs have also been appreciated in mouse models, represented by other cytokines including IL-1β, IL-6 and Ccl2. For example, mitochondrial apoptosis and NLRP3 inflammasome activation were induced by extracellular vesicles from Neisseria gonorrhoeae, E. coli and P. aeruginosa, thus activating and releasing IL-1β [97]. After injection of A. baumannii-derived extracellular vesicles, the expression of proinflammatory cytokines, including IL-6, macrophage inflammatory protein-1α, Ccl2 and Ccl3, was upregulated in mouse lung epithelial cells [98]. Despite this evidence about BEV-induced epithelial immune reactions, the specific mechanism and promising application are not fully understood. Under the threat of millions of microbiomes, how epithelial cells maintain the gastrointestinal barrier remains unknown.
There are two layers of mucus gel in vivo: an outer layer that is easy to remove by simple suctioning and an inner layer that is free of bacteria [99,100]. Muc2 mucin from the outer sparse layer not only provides an anchor point for bacterial adhesion but also acts as a source of carbon and energy or even an anoxic environment if necessary [101]. Pathogenic alterations or genetic defects such as Muc2−/− lead to direct contact between bacteria and epithelia and trigger an immune response, chronic inflammation or even cancerization [102]. BEVs from C. jejuni proteolytic activity in vitro were able to cleave both E-cadherin and occludin, thus disrupting both tight junctions and adherens junctions [103]. On the other hand, probiotics can help maintain the physiologic barrier via membrane proteins or BEVs. A special membrane protein named Amuc_1100 increased the expression level of tight junction proteins including occludin and claudin3 through TLR2 activation [104]. The extracellular vesicles from probiotic E. coli Nissle 1917 enhanced the intestinal mucosal barrier by upregulating the tight junction proteins ZO-1, ZO-2 and claudin-14 [105]. Moreover, these vesicles were shown to rescue intestinal epithelial barrier dysfunction induced by enteropathogenic E. coli [106]. The extracellular vesicles secreted by probiotic A. muciniphila improved tight junctions and gut permeability by activating AMP-activated protein kinase (AMPK) [107]. Although protein differences have been shown to influence AMPK activation, it has not been determined which specific protein of A. muciniphila extracellular vesicles is responsible. Moreover, proteins are not the only bioactive components of extracellular vesicles. It has been reported that the extracellular vesicles derived from B. thetaiotaomicron contains inositol polyphosphates phosphatases, which could promote polysaccharide digestion [108]. Similarly, another common intestinal colonizing bacteria, Bacteroides spp. can release extracellular vesicles that can deliver a variety of hydrolase to participate in the decomposition of glycans and mucins (Fig. 4) [109].
4.2. Cancer
4.2.1. Gut microbiome, BEVs and cancer development
The close relationship of gut microbiota and cancer in organs has become fully appreciated in recent decades [110]. To date, microbiome dysbiosis has been identified in colorectal cancer [111,112], breast cancer [113], lung cancer [114], liver cancer [115], and melanoma [116]. In addition, bacteria inside colorectal cancer travel along with tumor cells during liver metastasis, and metronidazole treatment reduces the Fusobacterium load and overall tumor growth [117]. However, it remains unknown what role BEVs play in the complex crosstalk between gut microbiota, intratumoral microflora and host cells, and their effects may be subtle and context dependent.
As illustrated in the last chapter, BEVs can gain access to systemic circulation through disrupted tight junctions under microbial disorders and an inflammatory environment (Fig. 3). Microbe-associated molecular patterns (MAMPs) can be delivered by BEVs, thus triggering various immune responses (Fig. 5). MAMPs are recognized by pattern recognition receptors (PRRs), which are key components of innate immunity (Fig. 5). Interestingly, extracellular vesicles from different bacteria lead to disparate host immune responses, depending mainly upon their relationship with host cells [92]. For example, extracellular vesicles derived from enterotoxigenic Bacteroides fragilis (ETBF) and nontoxigenic Bacteroides fragilis (NTBF) present different enzyme systems and biological functions, despite their genetic similarity. Compared with polysaccharide utilization enzymes in NTBF, many proteins contributing to stability, capability and virulence factor delivery were identified in ETBF [118].
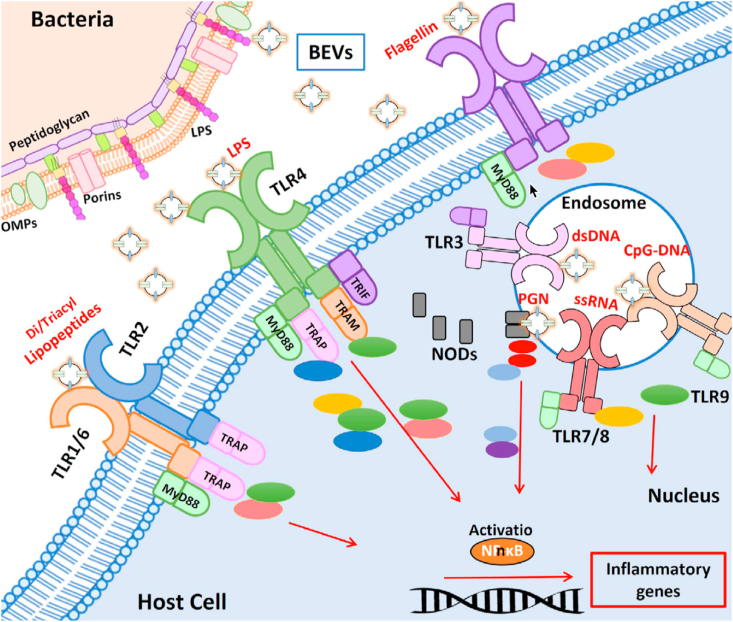
Fig. 5.
Recognition of BEV-associated molecular patterns by host immune receptors. TLR1/6, TLR2, TLR4, and TLR5 located at the host cell membrane, TLR3, TLR7/8, TLR9 located at the endosomal membranes. The downstream signaling pathways that lead to activation of NF-kB and inflammatory genes. Adaptor molecules: MyD88, TRAP, TRIF, TRAM. TLRs: toll-like receptors. Reprinted with permission [93]. Copyright 2021, Wiley-VCH GmbH.
4.2.2. BEVs and cancer therapy
Compared to live or weakened bacteria, BEVs have been thought to be safe, as they cannot be autonomously duplicated in vivo. BEVs exhibit great stability against temperature and carry multiple immunogenic membrane-associated or cytoplasm-associated components, and these intrinsic characteristics make them ideal candidates for both vaccines or immune modulators in novel cancer therapy [119]. Extracellular vesicles from LPS-depleted E. coli were reported to target cancer cells in vivo and reduce tumor burden by constant CXCL10 and interferon gamma (IFN-γ) production and antitumor immune responses [120]. Antigen-displaying BEVs were introduced to induce dendritic cell (DC) maturation and antigen presentation to T cells and yield tumor-specific antibodies like vaccines [121]. To expand the efficiency and application area of BEVs, Chen et al. combined melanoma cytomembranes, Salmonella vesicles and photothermal modules to suppress tumorigenesis [122].
BEV-derived nano-therapy has received wide attention as an antitumor vaccine or targeting vehicle [123]. The protein-rich membrane structure of BEVs makes them attractive biological components for hybrid nanoparticles when combined with traditional synthetic materials. Nanoparticles coated with S. aureus-derived membrane vesicles were shown to present homologous macrophage affinity, release antibiotics, and eliminate intracellular S. aureus efficiently [124]. BEV-coated gold nanoparticles demonstrated enhanced biostability and elevated IFN-γ and IL-17 production, indicating a faster immune response, which could be applied in cancer cell-targeted therapy [125]. Chemotherapeutic-loaded BEVs enabled increased doxorubicin infiltration and tumor cell apoptosis [126], indicating a promising method for targeted delivery. Furthermore, bioengineered BEVs with kinesin spindle protein (KSP) siRNA were able to target and kill cancer cells [127].
4.3. Brain
4.3.1. BEV-related neurologic abnormality
It is well recognized that bacteria-induced immune responses could be responsible for brain disorders, such as meningitis. Recent evidence has shown that BEVs not only activate the immune response via inherited components such as LPS and other antigens but also manipulate brain disorders through hereditary substance delivery [128]. BEVs from Alzheimer's disease (AD) patients were reported to affect blood-brain barrier (BBB) permeability and impair the learning memory ability of mice [129]. It has been confirmed that Paenalcaligenes hominis-derived extracellular vesicles can deliver cargo to the brain through the blood and vagus nerve and result in AD [130]. In addition, enterohemorrhagic E. coli (EHEC)-derived extracellular vesicles induce permeabilization of the mitochondrial membranes and trigger the mitochondrial apoptotic pathway in human brain microvascular endothelial cells; this result might suggest an underlying regulatory mechanism between the gut microbiota and central nervous system [131].
4.3.2. From gut to brain: BBB penetration
Various studies have identified BEVs as BBB breakers and have found that BEVs play a critical role in gut-brain communication [132]. Two papers recently published by Lee et al. introduced periodontopathogenic extracellular vesicles to deliver RNA cargo to intracranial immune cells such as macrophages and induce neuroinflammatory diseases [133,134]. For instance, BEVs from Aggregatibacter actinomycetemcomitans can travel across the BBB and successfully upregulate TNF-α expression. Due to this natural penetration ability, BEVs are valued as potential players in intracranial delivery. Nanogold and bacterial vesicle hybrid particles were introduced by Chen et al. [135] to augment chemoradiotherapy against glioblastoma and achieve longer survival of experimental mice. 4CMenB (Bexsero; GSK Biologicals), a vaccine against Neisseria meningitidis serogroup B (MenB), was introduced in 2013 and was confirmed to protect infants and children from MenB [136]. Porin protein A displays BEVs and is one of the four constituents in the vaccine [137,138], indicating enormous potential in both pharmacology and materials science. Recently, Bittel et al. visualized the process of transfer the microbial biomolecules (Cre/RNA) by BEVs in vivo via Cre/LoxP system (Fig. 4) [139].
4.4. Bone
The gut microbiome plays a critical role in bone metabolism during skeletal development and dynamic homeostasis [140]. Insulin-like growth factor 1 (IGF-1) was the first metabolite identified to be a messenger between intestine and bone, and antibiotic administration decreased serum IGF-1 levels and thus inhibited bone formation [141]. Recently, microbiota-derived short-chain fatty acids (SCFAs) were confirmed to connect the intestinal tract and bone metabolism [142]. Oral administration of lactulose increased SCFA production and attenuated ovariectomy-induced osteoporosis, indicating a close relationship between the gut and bone [143]. Claire et al. declared that warmth enhances bacterial polyamine biosynthesis, thus increasing bone strength [144]. Despite all these studies on the gut-bone axis, the role of BEVs in the close communication between the microbiota, gut epithelium and bone is still a mystery. According to the gut-bone axis and BEV behavior in other systems such as the brain and cancer, we hypothesize that BEVs participate in bone metabolism in both intestinal-mediated and bone-targeting manners, and we are currently conducting experiments to prove this hypothesis.
The latest research has reported that specific BEVs from the gut microbiome travel across the gut mucosal barrier and arrive at the marrow space. For example, BEVs isolated from A. muciniphila were found to reach the marrow space in 1 h via oral, rectal, and intravenous administration. The study identifies that the gut bacterium A. muciniphila mediates the antiosteoporotic effects of the gut microbiota of children and presents a novel mechanism underlying the exchange of signals between the gut microbiota and host bone [79]. Moreover, it has been reported that extracellular vesicles from A. muciniphila promoted osteoblasts and inhibited osteoclasts (Fig. 4) [79]. Based on rapid industrial production, bone-targeting engineering techniques reported for exosomes could be translated to BEVs to generate novel nanocarriers. For instance, stromal cell-derived factor 1 (SDF1) is mainly provided by bone marrow stromal cells (BMSCs) and brings CXCR4-positive cells back to bone marrow, including hematopoietic stem cells (HSCs) [145] and breast cancer cells [146]. Our group constructed a bone-targeting hybrid exosome-based nanocarrier system for bone microenvironment regulation [89].
5. Advantages and challenges
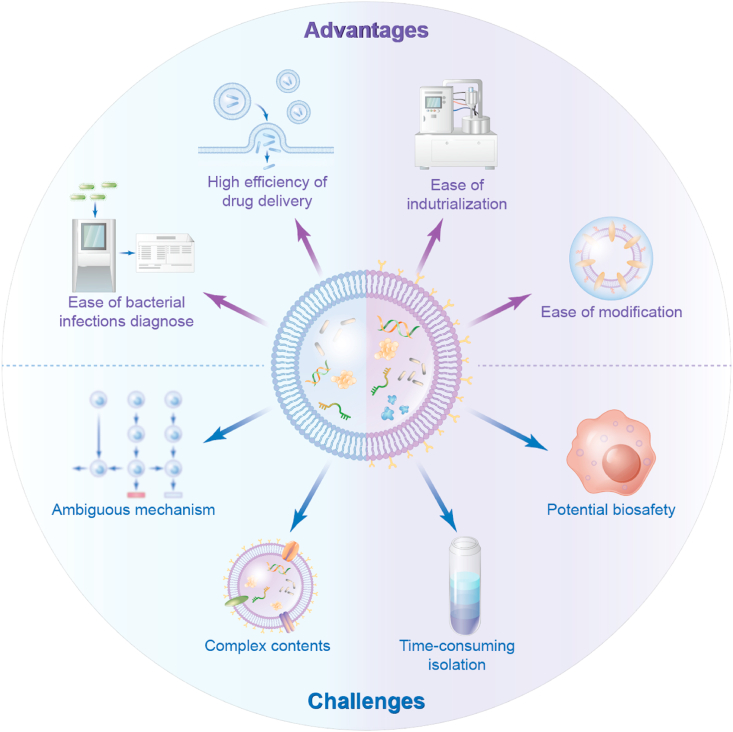
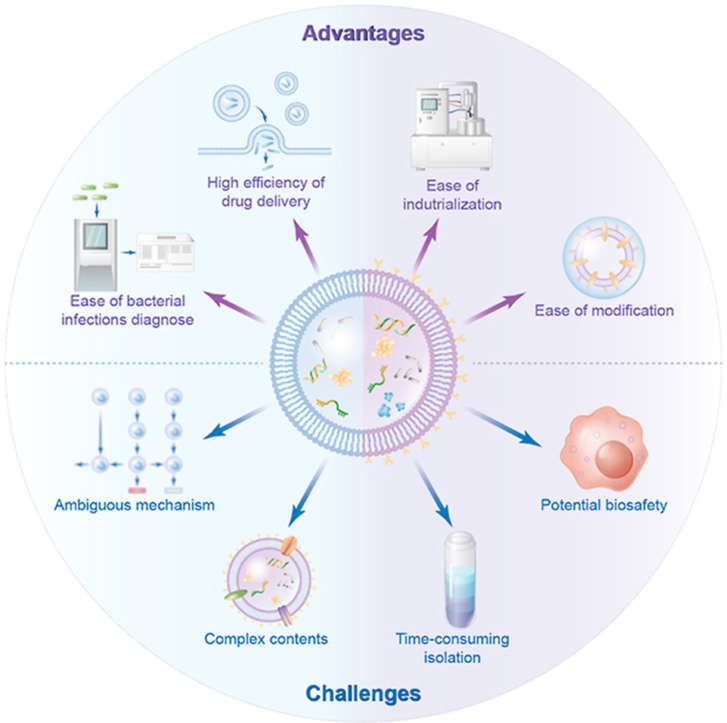
Bacteria and humans have formed a close and inseparable relationship during the long history of evolution. The extracellular vesicles secreted by bacteria offer various ingenious synergies for biomedical applications. Due to their proven immunomodulatory properties, BEVs have been used as viable tools for drug delivery, vaccine development, disease diagnosis, etc. Although the study of BEVs is less developed than that of eukaryotic extracellular vesicles (such as exosomes), the number of BEV studies has continuously increased in recent years [17]. Here, we summarize the advantages and challenges associated with BEVs (Fig. 6).
Fig. 6.
The advantages and challenges of bacterial extracellular vesicles. BEVs display many desirable qualities such as easy of industrialization, high efficiency of drug delivery, ease of modification, and ease of bacterial infections diagnose. On the other side, there are still significant challenges for the clinical translation of BEVs as delivery systems or therapeutics, including potential biosafety, complex contents, time-consuming isolation, ambiguous mechanism.
5.1. Advantages
5.1.1. Ease of industrialization
Compared with eukaryotic cells, bacteria show remarkable advantages including rapid proliferation abilities, mature high cell density culture techniques, and easy gene editing methods [147], which have the opportunity to produce more extracellular vesicles. Moreover, bacterial fermentation has always been a more viable production process because it offers the advantages of cost effectiveness and scalability [148,149]. Therefore, bacterial-derived extracellular vesicles have greater opportunities for industrialization. For example, Liu and colleagues were granted a national invention patent on the anti-osteoporotic effect of probiotic A. muciniphila and its extracellular vesicles [79], which lays the foundation for the future industrial application of probiotics or their functional extracellular vesicles to prevent and treat osteoporosis.
5.1.2. High efficiency of drug delivery
As nanosized materials, BEVs can enhance the efficiency of drug uptake and delivery. In addition, BEVs present great biostability, which can ensure efficient drug delivery. BEVs can protect bioactive molecules from degradation, which enhances the stability of cargo and enables delivery of cargo under functional conditions to target cells [150]. For example, RNA, an unstable bioactive molecule, cannot survive without shelter during systemic circulation. However, bacterial RNA has been found in the brains of patients with AD [151,152], suggesting that BEVs deliver RNA throughout the microbiota-gut-brain axis.
5.1.3. Ease of modification
Moreover, the loading of bioactive molecules in BEVs can occur in vivo or in vitro. In vivo, molecules of interest can be loaded in the generated extracellular vesicles when cells are treated with these molecules. For example, the strain P. aeruginosa PAO1 produced extracellular vesicles containing gentamicin when it was cultured with gentamicin [153]. This method may be used to produce extracellular vesicles carrying the compound of interest. In vitro, advances in genetic engineering have enabled the use of BEVs to deliver specific drugs and agents as a strategy to circumvent biocompatibility and large-scale production issues associated with synthetic nanomaterials [154]. Purified extracellular vesicles could also be encapsulated with different types of compounds through electroporation. Gujrati et al. developed a simple electroporation method to load siRNA into BEVs and showed that the established protocol was effective [155]. Similarly, Ayed et al. established a novel drug delivery system by loading BEVs with gold nanoparticles via electroporation [156]. The capacitance on the membrane creates pores in the lipid bilayer of BEVs, which allows any nanoparticle smaller than 10 nm to enter the vesicles. Subsequently, the closure of the BEV membrane captures the nanoparticles as cargo.
5.1.4. Ease of bacterial infections diagnose
BEVs can be used for the diagnosis of certain bacterial infections [157]. Since BEVs spread with intestinal microorganisms and are easily found in body fluids, diagnosis based on extracellular vesicles has significant advantages. Therefore, BEVs provide new insights into the links between microorganisms and the health status of the host. In addition, BEVs are a promising candidate for vaccine development against bacterial infections [34,83,84,95,158]. One of the advantages of using BEVs for vaccination is their ability to present native antigens, thus inducing effective immune responses.
5.2. Challenges
5.2.1. Ambiguous mechanism
Although BEVs have been proven to be a new drug delivery system for targeted drug delivery [18], there are still many areas for exploration of BEVs. For example, the mechanism of BEV internalization by host cells remains ambiguous. Several internalization pathways such as clathrin-mediated endocytosis [84,159], micropinocytosis [16], non-clathrin-mediated endocytosis [160], and membrane fusion [161] have been found. It is necessary to address which receptors mediate the internalization of BEVs. In addition, the mechanism of packaging these components in BEVs is not yet fully understood [17]. Is a molecule specific to certain strains and extracellular vesicles? Is it specific to certain environmental conditions? These questions remain to be elucidated in the future.
5.2.2. Potential biosafety
BEVs, especially the extracellular vesicles derived from G− bacteria, have been shown to contain many virulence factors, such as LPS and virulent proteins [32,84]. Therefore, regarding safety issues, caution should be exercised when BEVs are used as drug delivery nanocarriers because LPS could cause an innate immune response [97]. This immunotoxicity of BEVs will be a challenging aspect in drug delivery applications. For G− bacteria, LPS is one of the most abundant components of BEVs and is displayed on the outer surface of these vesicles. There are also mutant G− bacteria that do not contain LPS, such as E. coli EMKV15. Alternatively, the antibiotic polymyxin B can be used to neutralize LPS [162]. In principle, extracellular vesicles derived from G+ bacteria, which do not contain LPS and thus are generally less toxic than G− bacteria-derived extracellular vesicles, may be a better choice in drug delivery [119].
5.2.3. Time-consuming isolation
Although an overview of BEV isolation and purification techniques has been provided, the methods are still complicated and time-consuming. A timesaving, cost-effective, and efficient isolation method from a wide range of biological matrices needs to be proposed. Moreover, the existing techniques still cannot successfully separate BEVs, and prokaryotic extracellular vesicles exist together in the same sample. The human body is inhabited by approximately 1013 microbes composing a multicomplex system, which is strongly involved in the regulation and maintenance of homeostasis. BEVs can be found in several human biofluids (such as blood, plasma, and urine) and can be used as tools for bacterial detection and identification [163]. In addition, the specific (surface) markers present on BEVs from different sources are still unknown. Vanaja et al. used OmpF, an outer membrane protein of E. coli, as a marker for E. coli-derived extracellular vesicles [84]. The separation of BEVs and prokaryotic extracellular vesicles may be one of the most promising methods to discover specific markers and reveal the pathophysiological mechanisms of diseases.
5.2.4. Complex contents
Although BEVs are rich in a variety of effective bioactive and functional molecules, the composition is still complex and ambiguous. Notably, the contents and sizes of BEVs can change drastically when growth conditions or strains change [164]. In addition, the mechanism of packaging these components into BEVs is not yet fully understood and needs to be addressed in the future. A better characterization of these extracellular vesicles will increase our understanding of the possible functions and applications. With the development of multiomics technology, the composition of extracellular vesicles will be clearly identified.
6. Concluding remarks and outlook
We have shown that both G+ and G− bacteria can produce BEVs. Bacteria have two types of vesicle formation mechanisms, namely, membrane blebbing and endolysin-triggered cell lysis. Both mechanisms generate various extracellular vesicles, such as OMVs, OIMVs, EOMVs, and CMVs. The difference in the contents of G+ and G− BEVs goes beyond the presence of LPS and includes other molecules, such as nucleic acids, proteins, lipids, and metabolites.
Isolation and purification of BEVs are essential steps for subsequent applications. Moreover, the lack of standard methods of isolation and purification and the lack of well-defined identification of individual markers present on BEVs are obstacles to the progress of BEV research [165]. At present, information on achieving a high production of BEVs from fermentation broth including a complex medium composition and metabolites is limited. Here, we provide an effective protocol for BEV isolation and purification.
BEVs have been exploited as a new drug delivery platform. As the gene editing technology has been widely used in the bacteria, the overexpression or knockout of certain genes can influence the secretion and contents of extracellular vesicles. We hope that this review will raise the awareness that the drug delivery applications of BEVs are far beyond treating the gut, the brain, bone, and tumors. The intestinal microbiota contributes to the metabolic health of the human host [3,4,166]. Therefore, we hypothesize that these intestinal microbiota-derived extracellular vesicles may be involved in the metabolism of various tissues and result in different health statuses.
BEVs display many desirable qualities such as ease of industrialization, highly efficient drug delivery, ease of modification, and ease of bacterial infections diagnosis. On the other hand, there are still significant challenges for the clinical translation of BEVs as delivery systems or therapeutics, including potential biosafety, complicated and time-consuming isolation, complex and ambiguous contents, and mechanism (Fig. 6). While research on BEVs is in the early stage, their unique nanosized structure and biofunctions make them promising platforms in nanomedicine.
Authorship contribution statement
Han Liu, Qin Zhang, and Sicheng Wang contributed equally to this work. Han Liu, Qin Zhang, and Sicheng Wang drafted and wrote the manuscript of this review. Weizong Weng, Yingying Jing, and Jiacan Su guided and revised the manuscript of this review.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Key R&D Program of China (2018YFC2001500); National Natural Science Foundation of China (NSFC) Key Research Program in Aging (91749204); National Natural Science Foundation of China (82172098, 81771491, 81972254).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Weizong Weng, Email: drwengweizong@163.com.
Yingying Jing, Email: jingy4172@shu.edu.cn.
Jiacan Su, Email: jiacansu@smmu.edu.cn.
Abbreviations Used
- Lipopolysaccharide
LPS
- Gram-negative
G-
- Gram-positive
G+
- Bacteria extracellular vesicles
BEVs
- Mammalian extracellular vesicles
MEVs
- peptidoglycan
PG
- Outer membrane vesicles
OMVs
- Transmission electron microscopy
TEM
- Dynamic light scattering
DLS
- Nanoparticle tracking analysis
NTA
- Tunable resistive pulse sensing
TRPS
- Alzheimer's disease
AD
- AMP-activated protein kinase
AMPK
- Microbe-associated molecular patterns
MAMPs
- Pattern recognition receptors
PRRs
- Toxigenic Bacteroides fragilis
ETBF
- Nontoxigenic Bacteroides fragilis
NTBF
- Small interfering RNA
siRNA
- Insulin-like growth factor 1
IGF-1
References
- 1.Nguyen P.Q., Courchesne N.D., Duraj-Thatte A., Praveschotinunt P., Joshi N.S. Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 2018;30(19) doi: 10.1002/adma.201704847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nejman D., Livyatan I., Fuks G., Gavert N., Zwang Y., Geller L.T., Rotter-Maskowitz A., Weiser R., Mallel G., Gigi E., Meltser A., Douglas G.M., Kamer I., Gopalakrishnan V., Dadosh T., Levin-Zaidman S., Avnet S., Atlan T., Cooper Z.A., Arora R., Cogdill A.P., Khan M.A.W., Ologun G., Bussi Y., Weinberger A., Lotan-Pompan M., Golani O., Perry G., Rokah M., Bahar-Shany K., Rozeman E.A., Blank C.U., Ronai A., Shaoul R., Amit A., Dorfman T., Kremer R., Cohen Z.R., Harnof S., Siegal T., Yehuda-Shnaidman E., Gal-Yam E.N., Shapira H., Baldini N., Langille M.G.I., Ben-Nun A., Kaufman B., Nissan A., Golan T., Dadiani M., Levanon K., Bar J., Yust-Katz S., Barshack I., Peeper D.S., Raz D.J., Segal E., Wargo J.A., Sandbank J., Shental N., Straussman R. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Y., Pedersen O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021;19(1):55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 4.Zmora N., Suez J., Elinav E. You are what you eat: diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019;16(1):35–56. doi: 10.1038/s41575-018-0061-2. [DOI] [PubMed] [Google Scholar]
- 5.Li S., Jiang W., Zheng C., Shao D., Liu Y., Huang S., Han J., Ding J., Tao Y., Li M. Oral delivery of bacteria: basic principles and biomedical applications. J. Contr. Release. 2020;327:801–833. doi: 10.1016/j.jconrel.2020.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Zhou S. Synthetic biology: bacteria synchronized for drug delivery. Nature. 2016;536(7614):33–34. doi: 10.1038/nature18915. [DOI] [PubMed] [Google Scholar]
- 7.Song W., Anselmo A.C., Huang L. Nanotechnology intervention of the microbiome for cancer therapy. Nat. Nanotechnol. 2019;14(12):1093–1103. doi: 10.1038/s41565-019-0589-5. [DOI] [PubMed] [Google Scholar]
- 8.Rodrigo-Navarro A., Sankaran S., Dalby M.J., del Campo A., Salmeron-Sanchez M. Engineered living biomaterials. Nat. Rev. Mater. 2021;6:1175–1190. [Google Scholar]
- 9.Low K.B., Ittensohn M., Le T., Platt J., Sodi S., Amoss M., Ash O., Carmichael E., Chakraborty A., Fischer J., Lin S.L., Luo X., Miller S.I., Zheng L., King I., Pawelek J.M., Bermudes D. Lipid A mutant Salmonella with suppressed virulence and TNF alpha induction retain tumor-targeting in vivo. Nat. Biotechnol. 1999;17(1):37–41. doi: 10.1038/5205. [DOI] [PubMed] [Google Scholar]
- 10.Li Z., Wang Y., Liu J., Rawding P., Bu J., Hong S., Hu Q. Chemically and biologically engineered bacteria-based delivery systems for emerging diagnosis and advanced therapy. Adv. Mater. 2021 doi: 10.1002/adma.202102580. [DOI] [PubMed] [Google Scholar]
- 11.Brown L., Wolf J.M., Prados-Rosales R., Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015;13(10):620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwechheimer C., Kuehn M.J. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 2015;13(10):605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsharkasy O.M., Nordin J.Z., Hagey D.W., de Jong O.G., Schiffelers R.M., Andaloussi S.E., Vader P. Extracellular vesicles as drug delivery systems: why and how? Adv. Drug Deliv. Rev. 2020;159:332–343. doi: 10.1016/j.addr.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Herrmann I.K., Wood M.J.A., Fuhrmann G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021;16(7):748–759. doi: 10.1038/s41565-021-00931-2. [DOI] [PubMed] [Google Scholar]
- 15.Chen Q., Bai H., Wu W., Huang G., Li Y., Wu M., Tang G., Ping Y. Bioengineering bacterial vesicle-coated polymeric nanomedicine for enhanced cancer immunotherapy and metastasis prevention. Nano Lett. 2020;20(1):11–21. doi: 10.1021/acs.nanolett.9b02182. [DOI] [PubMed] [Google Scholar]
- 16.Kaparakis-Liaskos M., Ferrero R.L. Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol. 2015;15(6):375–387. doi: 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- 17.Ñahui Palomino R.A., Vanpouille C., Costantini P.E., Margolis L. Microbiota-host communications: bacterial extracellular vesicles as a common language. PLoS Pathog. 2021;17(5) doi: 10.1371/journal.ppat.1009508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S., Gao J., Wang Z. Outer membrane vesicles for vaccination and targeted drug delivery. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019;11(2):e1523. doi: 10.1002/wnan.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toyofuku M., Nomura N., Eberl L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019;17(1):13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 20.Brown L., Kessler A., Cabezas-Sanchez P., Luque-Garcia J.L., Casadevall A. Extracellular vesicles produced by the Gram-positive bacterium Bacillus subtilis are disrupted by the lipopeptide surfactin. Mol. Microbiol. 2014;93(1):183–198. doi: 10.1111/mmi.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Defourny K.A.Y., Smid E.J., Abee T. Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front. Microbiol. 2018;9:1502. doi: 10.3389/fmicb.2018.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao S., Klein M.I., Heim K.P., Fan Y., Bitoun J.P., Ahn S.J., Burne R.A., Koo H., Brady L.J., Wen Z.T. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J. Bacteriol. 2014;196(13):2355–2366. doi: 10.1128/JB.01493-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silhavy T.J., Kahne D., Walker S. The bacterial cell envelope. Cold Spring Harbor Perspect. Biol. 2010;2(5):a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kojer K., Riemer J. Balancing oxidative protein folding: the influences of reducing pathways on disulfide bond formation. Biochim. Biophys. Acta. 2014;1844(8):1383–1390. doi: 10.1016/j.bbapap.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Hickey C.A., Kuhn K.A., Donermeyer D.L., Porter N.T., Jin C., Cameron E.A., Jung H., Kaiko G.E., Wegorzewska M., Malvin N.P., Glowacki R.W., Hansson G.C., Allen P.M., Martens E.C., Stappenbeck T.S. Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe. 2015;17(5):672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beveridge T.J. Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1999;181(16):4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biller S.J., Schubotz F., Roggensack S.E., Thompson A.W., Summons R.E., Chisholm S.W. Bacterial vesicles in marine ecosystems. Science. 2014;343(6167):183–186. doi: 10.1126/science.1243457. [DOI] [PubMed] [Google Scholar]
- 28.Knox K.W., Vesk M., Work E. Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol. 1966;92(4):1206–1217. doi: 10.1128/jb.92.4.1206-1217.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Work E., Knox K.W., Vesk M. The chemistry and electron microscopy of an extracellular lipopolysaccharide from Escherichia coli. Ann. N. Y. Acad. Sci. 1966;133(2):438–449. doi: 10.1111/j.1749-6632.1966.tb52382.x. [DOI] [PubMed] [Google Scholar]
- 30.Kadurugamuwa J.L., Beveridge T.J. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 1995;177(14):3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee E.Y., Bang J.Y., Park G.W., Choi D.S., Kang J.S., Kim H.J., Park K.S., Lee J.O., Kim Y.K., Kwon K.H., Kim K.P., Gho Y.S. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7(17):3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- 32.Elhenawy W., Bording-Jorgensen M., Valguarnera E., Haurat M.F., Wine E., Feldman M.F. LPS remodeling triggers formation of outer membrane vesicles in Salmonella. mBio. 2016;7(4) doi: 10.1128/mBio.00940-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadurugamuwa J.L., Beveridge T.J. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other Gram-negative bacteria. Microbiology. 1999;145(Pt 8):2051–2060. doi: 10.1099/13500872-145-8-2051. [DOI] [PubMed] [Google Scholar]
- 34.Elmi A., Watson E., Sandu P., Gundogdu O., Mills D.C., Inglis N.F., Manson E., Imrie L., Bajaj-Elliott M., Wren B.W., Smith D.G., Dorrell N. Campylobacter jejuni outer membrane vesicles play an important role in bacterial interactions with human intestinal epithelial cells. Infect. Immun. 2012;80(12):4089–4098. doi: 10.1128/IAI.00161-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner L., Praszkier J., Hutton M.L., Steer D., Ramm G., Kaparakis-Liaskos M., Ferrero R.L. Increased outer membrane vesicle formation in a Helicobacter pylori tolB mutant. Helicobacter. 2015;20(4):269–283. doi: 10.1111/hel.12196. [DOI] [PubMed] [Google Scholar]
- 36.Chatterjee D., Chaudhuri K. Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett. 2011;585(9):1357–1362. doi: 10.1016/j.febslet.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 37.Shoberg R.J., Thomas D.D. Specific adherence of Borrelia burgdorferi extracellular vesicles to human endothelial cells in culture. Infect. Immun. 1993;61(9):3892–3900. doi: 10.1128/iai.61.9.3892-3900.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettit R.K., Judd R.C. The interaction of naturally elaborated blebs from serum-susceptible and serum-resistant strains of Neisseria gonorrhoeae with normal human serum. Mol. Microbiol. 1992;6(6):729–734. doi: 10.1111/j.1365-2958.1992.tb01522.x. [DOI] [PubMed] [Google Scholar]
- 39.Rumbo C., Fernández-Moreira E., Merino M., Poza M., Mendez J.A., Soares N.C., Mosquera A., Chaves F., Bou G. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011;55(7):3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadurugamuwa J.L., Clarke A.J., Beveridge T.J. Surface action of gentamicin on Pseudomonas aeruginosa. J. Bacteriol. 1993;175(18):5798–5805. doi: 10.1128/jb.175.18.5798-5805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roier S., Zingl F.G., Cakar F., Durakovic S., Kohl P., Eichmann T.O., Klug L., Gadermaier B., Weinzerl K., Prassl R., Lass A., Daum G., Reidl J., Feldman M.F., Schild S. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 2016;7:10515. doi: 10.1038/ncomms10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kulp A., Kuehn M.J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mashburn-Warren L.M., Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 2006;61(4):839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- 44.Orench-Rivera N., Kuehn M.J. Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol. 2016;18(11):1525–1536. doi: 10.1111/cmi.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toyofuku M., Zhou S., Sawada I., Takaya N., Uchiyama H., Nomura N. Membrane vesicle formation is associated with pyocin production under denitrifying conditions in Pseudomonas aeruginosaPAO1. Environ. Microbiol. 2014;16(9):2927–2938. doi: 10.1111/1462-2920.12260. [DOI] [PubMed] [Google Scholar]
- 46.Klimentová J., Stulík J. Methods of isolation and purification of outer membrane vesicles from Gram-negative bacteria. Microbiol. Res. 2015;170:1–9. doi: 10.1016/j.micres.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Tashiro Y., Ichikawa S., Shimizu M., Toyofuku M., Takaya N., Nakajima-Kambe T., Uchiyama H., Nomura N. Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2010;76(11):3732–3739. doi: 10.1128/AEM.02794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCaig W.D., Koller A., Thanassi D.G. Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J. Bacteriol. 2013;195(6):1120–1132. doi: 10.1128/JB.02007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sjöström A.E., Sandblad L., Uhlin B.E., Wai S.N. Membrane vesicle-mediated release of bacterial RNA. Sci. Rep. 2015;5:15329. doi: 10.1038/srep15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bitto N.J., Chapman R., Pidot S., Costin A., Lo C., Choi J., D'Cruze T., Reynolds E.C., Dashper S.G., Turnbull L., Whitchurch C.B., Stinear T.P., Stacey K.J., Ferrero R.L. Bacterial membrane vesicles transport their DNA cargo into host cells. Sci. Rep. 2017;7(1):7072. doi: 10.1038/s41598-017-07288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renelli M., Matias V., Lo R.Y., Beveridge T.J. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology (Read.) 2004;150(Pt 7):2161–2169. doi: 10.1099/mic.0.26841-0. [DOI] [PubMed] [Google Scholar]
- 52.Turnbull L., Toyofuku M., Hynen A.L., Kurosawa M., Pessi G., Petty N.K., Osvath S.R., Cárcamo-Oyarce G., Gloag E.S., Shimoni R., Omasits U., Ito S., Yap X., Monahan L.G., Cavaliere R., Ahrens C.H., Charles I.G., Nomura N., Eberl L., Whitchurch C.B. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 2016;7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bauwens A., Kunsmann L., Karch H., Mellmann A., Bielaszewska M. Antibiotic-mediated modulations of outer membrane vesicles in Enterohemorrhagic Escherichia coli O104:H4 and O157:H7. Antimicrob. Agents Chemother. 2017;61(9) doi: 10.1128/AAC.00937-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pérez-Cruz C., Delgado L., López-Iglesias C., Mercade E. Outer-inner membrane vesicles naturally secreted by Gram-negative pathogenic bacteria. PLoS One. 2015;10(1) doi: 10.1371/journal.pone.0116896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J., Azam F., Zhang S. Outer membrane vesicles containing signalling molecules and active hydrolytic enzymes released by a coral pathogen Vibrio shilonii AK1. Environ. Microbiol. 2016;18(11):3850–3866. doi: 10.1111/1462-2920.13344. [DOI] [PubMed] [Google Scholar]
- 56.Pérez-Cruz C., Carrión O., Delgado L., Martinez G., López-Iglesias C., Mercade E. New type of outer membrane vesicle produced by the Gram-negative bacterium Shewanella vesiculosa M7T: implications for DNA content. Appl. Environ. Microbiol. 2013;79(6):1874–1881. doi: 10.1128/AEM.03657-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hagemann S., Stöger L., Kappelmann M., Hassl I., Ellinger A., Velimirov B. DNA-bearing membrane vesicles produced by Ahrensia kielensis and Pseudoalteromonas marina. J. Basic Microbiol. 2014;54(10):1062–1072. doi: 10.1002/jobm.201300376. [DOI] [PubMed] [Google Scholar]
- 58.Dorward D.W., Garon C.F. DNA is packaged within membrane-derived vesicles of Gram-negative but not Gram-positive bacteria. Appl. Environ. Microbiol. 1990;56(6):1960–1962. doi: 10.1128/aem.56.6.1960-1962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shockman G.D., Barrett J.F. Structure, function, and assembly of cell walls of Gram-positive bacteria. Annu. Rev. Microbiol. 1983;37:501–527. doi: 10.1146/annurev.mi.37.100183.002441. [DOI] [PubMed] [Google Scholar]
- 60.Rivera J., Cordero R.J., Nakouzi A.S., Frases S., Nicola A., Casadevall A. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. U. S. A. 2010;107(44):19002–19007. doi: 10.1073/pnas.1008843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prados-Rosales R., Baena A., Martinez L.R., Luque-Garcia J., Kalscheuer R., Veeraraghavan U., Camara C., Nosanchuk J.D., Besra G.S., Chen B., Jimenez J., Glatman-Freedman A., Jacobs W.R., Jr., Porcelli S.A., Casadevall A. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J. Clin. Invest. 2011;121(4):1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hong S.W., Kim M.R., Lee E.Y., Kim J.H., Kim Y.S., Jeon S.G., Yang J.M., Lee B.J., Pyun B.Y., Gho Y.S., Kim Y.K. Extracellular vesicles derived from Staphylococcus aureus induce atopic dermatitis-like skin inflammation. Allergy. 2011;66(3):351–359. doi: 10.1111/j.1398-9995.2010.02483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee E.Y., Choi D.Y., Kim D.K., Kim J.W., Park J.O., Kim S., Kim S.H., Desiderio D.M., Kim Y.K., Kim K.P., Gho Y.S. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics. 2009;9(24):5425–5436. doi: 10.1002/pmic.200900338. [DOI] [PubMed] [Google Scholar]
- 64.Schrempf H., Koebsch I., Walter S., Engelhardt H., Meschke H. Extracellular Streptomyces vesicles: amphorae for survival and defence. Microb Biotechnol. 2011;4(2):286–299. doi: 10.1111/j.1751-7915.2011.00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee E.Y., Choi D.S., Kim K.P., Gho Y.S. Proteomics in Gram-negative bacterial outer membrane vesicles. Mass Spectrom. Rev. 2008;27(6):535–555. doi: 10.1002/mas.20175. [DOI] [PubMed] [Google Scholar]
- 66.Lee J.H., Choi C.W., Lee T., Kim S.I., Lee J.C., Shin J.H. Transcription factor σB plays an important role in the production of extracellular membrane-derived vesicles in Listeria monocytogenes. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0073196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olaya-Abril A., Prados-Rosales R., McConnell M.J., Martín-Peña R., González-Reyes J.A., Jiménez-Munguía I., Gómez-Gascón L., Fernández J., Luque-García J.L., García-Lidón C., Estévez H., Pachón J., Obando I., Casadevall A., Pirofski L.A., Rodríguez-Ortega M.J. Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J. Proteonomics. 2014;106:46–60. doi: 10.1016/j.jprot.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 68.Jiang Y., Kong Q., Roland K.L., Curtiss R., 3rd Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int J Med Microbiol. 2014;304(3–4):431–443. doi: 10.1016/j.ijmm.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vallejo M.C., Nakayasu E.S., Longo L.V., Ganiko L., Lopes F.G., Matsuo A.L., Almeida I.C., Puccia R. Lipidomic analysis of extracellular vesicles from the pathogenic phase of Paracoccidioides brasiliensis. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0039463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodrigues M.L., Nakayasu E.S., Oliveira D.L., Nimrichter L., Nosanchuk J.D., Almeida I.C., Casadevall A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell. 2008;7(1):58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Albuquerque P.C., Nakayasu E.S., Rodrigues M.L., Frases S., Casadevall A., Zancope-Oliveira R.M., Almeida I.C., Nosanchuk J.D. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell Microbiol. 2008;10(8):1695–1710. doi: 10.1111/j.1462-5822.2008.01160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Burdett I.D., Murray R.G. Electron microscope study of septum formation in Escherichia coli strains B and B-r during synchronous growth. J. Bacteriol. 1974;119(3):1039–1056. doi: 10.1128/jb.119.3.1039-1056.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aschtgen M.S., Lynch J.B., Koch E., Schwartzman J., McFall-Ngai M., Ruby E. Rotation of Vibrio fischeri flagella produces outer membrane vesicles that induce host development. J. Bacteriol. 2016;198(16):2156–2165. doi: 10.1128/JB.00101-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aschtgen M.S., Wetzel K., Goldman W., McFall-Ngai M., Ruby E. Vibrio fischeri-derived outer membrane vesicles trigger host development. Cell Microbiol. 2016;18(4):488–499. doi: 10.1111/cmi.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dhital S., Deo P., Stuart I., Naderer T. Bacterial outer membrane vesicles and host cell death signaling. Trends Microbiol. 2021;29(12):1106–1116. doi: 10.1016/j.tim.2021.04.003. [DOI] [PubMed] [Google Scholar]
- 76.Prados-Rosales R., Brown L., Casadevall A., Montalvo-Quirós S., Luque-Garcia J.L. Isolation and identification of membrane vesicle-associated proteins in Gram-positive bacteria and mycobacteria. MethodsX. 2014;1:124–129. doi: 10.1016/j.mex.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chutkan H., Macdonald I., Manning A., Kuehn M.J. Quantitative and qualitative preparations of bacterial outer membrane vesicles. Methods Mol. Biol. 2013;966:259–272. doi: 10.1007/978-1-62703-245-2_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rubin D., Christy C. Selecting the right ultrafiltration membrane for biopharmaceutical applications. Pharmaceut. Technol. Eur. 2002;14(12):39–40+43. [Google Scholar]
- 79.Liu J.H., Chen C.Y., Liu Z.Z., Luo Z.W., Rao S.S., Jin L., Wan T.F., Yue T., Tan Y.J., Yin H., Yang F., Huang F.Y., Guo J., Wang Y.Y., Xia K., Cao J., Wang Z.X., Hong C.G., Luo M.J., Hu X.K., Liu Y.W., Du W., Luo J., Hu Y., Zhang Y., Huang J., Li H.M., Wu B., Liu H.M., Chen T.H., Qian Y.X., Li Y.Y., Feng S.K., Chen Y., Qi L.Y., Xu R., Tang S.Y., Xie H. Extracellular vesicles from child gut microbiota enter into bone to preserve bone mass and strength. Adv. Sci. 2021;8(9):2004831. doi: 10.1002/advs.202004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Graham J., Ford T., Rickwood D. The preparation of subcellular organelles from mouse liver in self-generated gradients of iodixanol. Anal. Biochem. 1994;220(2):367–373. doi: 10.1006/abio.1994.1351. [DOI] [PubMed] [Google Scholar]
- 81.Ford T., Graham J., Rickwood D. Iodixanol: a nonionic iso-osmotic centrifugation medium for the formation of self-generated gradients. Anal. Biochem. 1994;220(2):360–366. doi: 10.1006/abio.1994.1350. [DOI] [PubMed] [Google Scholar]
- 82.Manabe T., Kato M., Ueno T., Kawasaki K. Flagella proteins contribute to the production of outer membrane vesicles from Escherichia coli W3110. Biochem. Biophys. Res. Commun. 2013;441(1):151–156. doi: 10.1016/j.bbrc.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 83.Jang K.S., Sweredoski M.J., Graham R.L., Hess S., Clemons W.M., Jr. Comprehensive proteomic profiling of outer membrane vesicles from Campylobacter jejuni. J. Proteonomics. 2014;98:90–98. doi: 10.1016/j.jprot.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vanaja S.K., Russo A.J., Behl B., Banerjee I., Yankova M., Deshmukh S.D., Rathinam V.A.K. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell. 2016;165(5):1106–1119. doi: 10.1016/j.cell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lawrie A.S., Albanyan A., Cardigan R.A., Mackie I.J., Harrison P. Microparticle sizing by dynamic light scattering in fresh-frozen plasma. Vox Sang. 2009;96(3):206–212. doi: 10.1111/j.1423-0410.2008.01151.x. [DOI] [PubMed] [Google Scholar]
- 86.Xu Y., Nakane N., Maurer-Spurej E. Novel test for microparticles in platelet-rich plasma and platelet concentrates using dynamic light scattering. Transfusion. 2011;51(2):363–370. doi: 10.1111/j.1537-2995.2010.02819.x. [DOI] [PubMed] [Google Scholar]
- 87.Sitar S., Kejžar A., Pahovnik D., Kogej K., Tušek-Žnidarič M., Lenassi M., Žagar E. Size characterization and quantification of exosomes by asymmetrical-flow field-flow fractionation. Anal. Chem. 2015;87(18):9225–9233. doi: 10.1021/acs.analchem.5b01636. [DOI] [PubMed] [Google Scholar]
- 88.Gardiner C., Ferreira Y.J., Dragovic R.A., Redman C.W., Sargent I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu Y., Li X., Zhang Q., Gu Z., Luo Y., Guo J., Wang X., Jing Y., Chen X., Su J. Exosome-guided bone targeted delivery of Antagomir-188 as an anabolic therapy for bone loss. Bioact Mater. 2021;6(9):2905–2913. doi: 10.1016/j.bioactmat.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Akers J.C., Ramakrishnan V., Nolan J.P., Duggan E., Fu C.C., Hochberg F.H., Chen C.C., Carter B.S. Comparative analysis of technologies for quantifying extracellular vesicles (EVs) in clinical cerebrospinal fluids (CSF) PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0149866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoo J.W., Irvine D.J., Discher D.E., Mitragotri S. Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat. Rev. Drug Discov. 2011;10(7):521–535. doi: 10.1038/nrd3499. [DOI] [PubMed] [Google Scholar]
- 92.Chronopoulos A., Kalluri R. Emerging role of bacterial extracellular vesicles in cancer. Oncogene. 2020;39(46):6951–6960. doi: 10.1038/s41388-020-01509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Díaz-Garrido N., Badia J., Baldomà L. Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J. Extracell. Vesicles. 2021;10(13) doi: 10.1002/jev2.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ho M.H., Guo Z.M., Chunga J., Goodwin J.S., Xie H. Characterization of innate immune responses of human endothelial cells induced by Porphyromonas gingivalis and their derived outer membrane vesicles. Front Cell Infect Microbiol. 2016;6:139. doi: 10.3389/fcimb.2016.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bauman S.J., Kuehn M.J. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microb. Infect. 2006;8(9–10):2400–2408. doi: 10.1016/j.micinf.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bielaszewska M., Marejková M., Bauwens A., Kunsmann-Prokscha L., Mellmann A., Karch H. Enterohemorrhagic Escherichia coli O157 outer membrane vesicles induce interleukin 8 production in human intestinal epithelial cells by signaling via Toll-like receptors TLR4 and TLR5 and activation of the nuclear factor NF-κB. Int J Med Microbiol. 2018;308(7):882–889. doi: 10.1016/j.ijmm.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 97.Deo P., Chow S.H., Han M.L., Speir M., Huang C., Schittenhelm R.B., Dhital S., Emery J., Li J., Kile B.T., Vince J.E., Lawlor K.E., Naderer T. Mitochondrial dysfunction caused by outer membrane vesicles from Gram-negative bacteria activates intrinsic apoptosis and inflammation. Nat Microbiol. 2020;5(11):1418–1427. doi: 10.1038/s41564-020-0773-2. [DOI] [PubMed] [Google Scholar]
- 98.Jun S.H., Lee J.H., Kim B.R., Kim S.I., Park T.I., Lee J.C., Lee Y.C. Acinetobacter baumannii outer membrane vesicles elicit a potent innate immune response via membrane proteins. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Johansson M.E., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U. S. A. 2008;105(39):15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Atuma C., Strugala V., Allen A., Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;280(5):G922–G929. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- 101.Bäckhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 102.Hansson G.C. Mucins and the microbiome. Annu. Rev. Biochem. 2020;89:769–793. doi: 10.1146/annurev-biochem-011520-105053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Elmi A., Nasher F., Jagatia H., Gundogdu O., Bajaj-Elliott M., Wren B., Dorrell N. Campylobacter jejuni outer membrane vesicle-associated proteolytic activity promotes bacterial invasion by mediating cleavage of intestinal epithelial cell E-cadherin and occludin. Cell Microbiol. 2016;18(4):561–572. doi: 10.1111/cmi.12534. [DOI] [PubMed] [Google Scholar]
- 104.Plovier H., Everard A., Druart C., Depommier C., Van Hul M., Geurts L., Chilloux J., Ottman N., Duparc T., Lichtenstein L., Myridakis A., Delzenne N.M., Klievink J., Bhattacharjee A., van der Ark K.C., Aalvink S., Martinez L.O., Dumas M.E., Maiter D., Loumaye A., Hermans M.P., Thissen J.P., Belzer C., de Vos W.M., Cani P.D. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017;23(1):107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 105.Alvarez C.S., Badia J., Bosch M., Giménez R., Baldomà L. Outer Membrane Vesicles and soluble factors released by probiotic Escherichia coli Nissle 1917 and commensal ecor63 enhance barrier function by regulating expression of tight junction proteins in intestinal epithelial cells. Front. Microbiol. 2016;7:1981. doi: 10.3389/fmicb.2016.01981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alvarez C.S., Giménez R., Cañas M.A., Vera R., Díaz-Garrido N., Badia J., Baldomà L. Extracellular vesicles and soluble factors secreted by Escherichia coli Nissle 1917 and ECOR63 protect against enteropathogenic E. coli-induced intestinal epithelial barrier dysfunction. BMC Microbiol. 2019;19(1):166. doi: 10.1186/s12866-019-1534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chelakkot C., Choi Y., Kim D.K., Park H.T., Ghim J., Kwon Y., Jeon J., Kim M.S., Jee Y.K., Gho Y.S., Park H.S., Kim Y.K., Ryu S.H. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018;50(2) doi: 10.1038/emm.2017.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stentz R., Osborne S., Horn N., Li A.W., Hautefort I., Bongaerts R., Rouyer M., Bailey P., Shears S.B., Hemmings A.M., Brearley C.A., Carding S.R. A bacterial homolog of a eukaryotic inositol phosphate signaling enzyme mediates cross-kingdom dialog in the mammalian gut. Cell Rep. 2014;6(4):646–656. doi: 10.1016/j.celrep.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Elhenawy W., Debelyy M.O., Feldman M.F. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio. 2014;5(2):e00909–e00914. doi: 10.1128/mBio.00909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gopalakrishnan V., Helmink B.A., Spencer C.N., Reuben A., Wargo J.A. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33(4):570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Garrett W.S. The gut microbiota and colon cancer. Science. 2019;364(6446):1133–1135. doi: 10.1126/science.aaw2367. [DOI] [PubMed] [Google Scholar]
- 112.Song M., Chan A.T., Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158(2):322–340. doi: 10.1053/j.gastro.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parida S., Wu S., Siddharth S., Wang G., Muniraj N., Nagalingam A., Hum C., Mistriotis P., Hao H., Talbot C.C., Jr., Konstantopoulos K., Gabrielson K.L., Sears C.L., Sharma D. A procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and β-Catenin Axes. Cancer Discov. 2021;11(5):1138–1157. doi: 10.1158/2159-8290.CD-20-0537. [DOI] [PubMed] [Google Scholar]
- 114.Jin C., Lagoudas G.K., Zhao C., Bullman S., Bhutkar A., Hu B., Ameh S., Sandel D., Liang X.S., Mazzilli S., Whary M.T., Meyerson M., Germain R., Blainey P.C., Fox J.G., Jacks T. Commensal microbiota promote lung cancer development via γδ T cells. Cell. 2019;176(5):998–1013. doi: 10.1016/j.cell.2018.12.040. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ma C., Han M., Heinrich B., Fu Q., Zhang Q., Sandhu M., Agdashian D., Terabe M., Berzofsky J.A., Fako V., Ritz T., Longerich T., Theriot C.M., McCulloch J.A., Roy S., Yuan W., Thovarai V., Sen S.K., Ruchirawat M., Korangy F., Wang X.W., Trinchieri G., Greten T.F. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360(6391) doi: 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., Cogdill A.P., Zhao L., Hudgens C.W., Hutchinson D.S., Manzo T., Petaccia de Macedo M., Cotechini T., Kumar T., Chen W.S., Reddy S.M., Szczepaniak Sloane R., Galloway-Pena J., Jiang H., Chen P.L., Shpall E.J., Rezvani K., Alousi A.M., Chemaly R.F., Shelburne S., Vence L.M., Okhuysen P.C., Jensen V.B., Swennes A.G., McAllister F., Marcelo Riquelme Sanchez E., Zhang Y., Le Chatelier E., Zitvogel L., Pons N., Austin-Breneman J.L., Haydu L.E., Burton E.M., Gardner J.M., Sirmans E., Hu J., Lazar A.J., Tsujikawa T., Diab A., Tawbi H., Glitza I.C., Hwu W.J., Patel S.P., Woodman S.E., Amaria R.N., Davies M.A., Gershenwald J.E., Hwu P., Lee J.E., Zhang J., Coussens L.M., Cooper Z.A., Futreal P.A., Daniel C.R., Ajami N.J., Petrosino J.F., Tetzlaff M.T., Sharma P., Allison J.P., Jenq R.R., Wargo J.A. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bullman S., Pedamallu C.S., Sicinska E., Clancy T.E., Zhang X., Cai D., Neuberg D., Huang K., Guevara F., Nelson T., Chipashvili O., Hagan T., Walker M., Ramachandran A., Diosdado B., Serna G., Mulet N., Landolfi S., Ramon Y.C.S., Fasani R., Aguirre A.J., Ng K., Élez E., Ogino S., Tabernero J., Fuchs C.S., Hahn W.C., Nuciforo P., Meyerson M. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zakharzhevskaya N.B., Vanyushkina A.A., Altukhov I.A., Shavarda A.L., Butenko I.O., Rakitina D.V., Nikitina A.S., Manolov A.I., Egorova A.N., Kulikov E.E., Vishnyakov I.E., Fisunov G.Y., Govorun V.M. Outer membrane vesicles secreted by pathogenic and nonpathogenic Bacteroides fragilis represent different metabolic activities. Sci. Rep. 2017;7(1):5008. doi: 10.1038/s41598-017-05264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van der Pol L., Stork M., van der Ley P. Outer membrane vesicles as platform vaccine technology. Biotechnol. J. 2015;10(11):1689–1706. doi: 10.1002/biot.201400395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim O.Y., Park H.T., Dinh N.T.H., Choi S.J., Lee J., Kim J.H., Lee S.W., Gho Y.S. Bacterial outer membrane vesicles suppress tumor by interferon-γ-mediated antitumor response. Nat. Commun. 2017;8(1):626. doi: 10.1038/s41467-017-00729-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schetters S.T.T., Jong W.S.P., Horrevorts S.K., Kruijssen L.J.W., Engels S., Stolk D., Daleke-Schermerhorn M.H., Garcia-Vallejo J., Houben D., Unger W.W.J., den Haan J.M.M., Luirink J., van Kooyk Y. Outer membrane vesicles engineered to express membrane-bound antigen program dendritic cells for cross-presentation to CD8(+) T cells. Acta Biomater. 2019;91:248–257. doi: 10.1016/j.actbio.2019.04.033. [DOI] [PubMed] [Google Scholar]
- 122.Chen Q., Huang G., Wu W., Wang J., Hu J., Mao J., Chu P.K., Bai H., Tang G. A hybrid eukaryotic-prokaryotic nanoplatform with photothermal modality for enhanced antitumor vaccination. Adv. Mater. 2020;32(16) doi: 10.1002/adma.201908185. [DOI] [PubMed] [Google Scholar]
- 123.Zhuang Q., Xu J., Deng D., Chao T., Li J., Zhang R., Peng R., Liu Z. Bacteria-derived membrane vesicles to advance targeted photothermal tumor ablation. Biomaterials. 2021;268:120550. doi: 10.1016/j.biomaterials.2020.120550. [DOI] [PubMed] [Google Scholar]
- 124.Gao F., Xu L., Yang B., Fan F., Yang L. Kill the real with the fake: eliminate intracellular Staphylococcus aureus using nanoparticle coated with its extracellular vesicle membrane as active-targeting drug carrier. ACS Infect. Dis. 2019;5(2):218–227. doi: 10.1021/acsinfecdis.8b00212. [DOI] [PubMed] [Google Scholar]
- 125.Gao W., Fang R.H., Thamphiwatana S., Luk B.T., Li J., Angsantikul P., Zhang Q., Hu C.M., Zhang L. Modulating antibacterial immunity via bacterial membrane-coated nanoparticles. Nano Lett. 2015;15(2):1403–1409. doi: 10.1021/nl504798g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sagnella S.M., Trieu J., Brahmbhatt H., MacDiarmid J.A., MacMillan A., Whan R.M., Fife C.M., McCarroll J.A., Gifford A.J., Ziegler D.S., Kavallaris M. Targeted doxorubicin-loaded bacterially derived nano-cells for the treatment of neuroblastoma. Mol. Cancer Therapeut. 2018;17(5):1012–1023. doi: 10.1158/1535-7163.MCT-17-0738. [DOI] [PubMed] [Google Scholar]
- 127.Gujrati V., Kim S., Kim S.H., Min J.J., Choy H.E., Kim S.C., Jon S. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano. 2014;8(2):1525–1537. doi: 10.1021/nn405724x. [DOI] [PubMed] [Google Scholar]
- 128.Koeppen K., Hampton T.H., Jarek M., Scharfe M., Gerber S.A., Mielcarz D.W., Demers E.G., Dolben E.L., Hammond J.H., Hogan D.A., Stanton B.A. A Novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog. 2016;12(6) doi: 10.1371/journal.ppat.1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wei S., Peng W., Mai Y., Li K., Wei W., Hu L., Zhu S., Zhou H., Jie W., Wei Z., Kang C., Li R., Liu Z., Zhao B., Cai Z. Outer membrane vesicles enhance tau phosphorylation and contribute to cognitive impairment. J. Cell. Physiol. 2020;235(5):4843–4855. doi: 10.1002/jcp.29362. [DOI] [PubMed] [Google Scholar]
- 130.Lee K.E., Kim J.K., Han S.K., Lee D.Y., Lee H.J., Yim S.V., Kim D.H. The extracellular vesicle of gut microbial Paenalcaligenes hominis is a risk factor for vagus nerve-mediated cognitive impairment. Microbiome. 2020;8(1):107. doi: 10.1186/s40168-020-00881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bielaszewska M., Rüter C., Kunsmann L., Greune L., Bauwens A., Zhang W., Kuczius T., Kim K.S., Mellmann A., Schmidt M.A., Karch H. Enterohemorrhagic Escherichia coli hemolysin employs outer membrane vesicles to target mitochondria and cause endothelial and epithelial apoptosis. PLoS Pathog. 2013;9(12) doi: 10.1371/journal.ppat.1003797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cuesta C.M., Guerri C., Ureña J., Pascual M. Role of microbiota-derived extracellular vesicles in gut-brain communication. Int. J. Mol. Sci. 2021;22(8) doi: 10.3390/ijms22084235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Han E.C., Choi S.Y., Lee Y., Park J.W., Hong S.H., Lee H.J. Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF-α production in human macrophages and cross the blood-brain barrier in mice. Faseb. J. 2019;33(12):13412–13422. doi: 10.1096/fj.201901575R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ha J.Y., Choi S.Y., Lee J.H., Hong S.H., Lee H.J. Delivery of periodontopathogenic extracellular vesicles to brain monocytes and microglial IL-6 promotion by RNA cargo. Front Mol Biosci. 2020;7:596366. doi: 10.3389/fmolb.2020.596366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen M.H., Liu T.Y., Chen Y.C., Chen M.H. Combining augmented radiotherapy and immunotherapy through a nano-gold and bacterial outer-membrane vesicle complex for the treatment of glioblastoma. Nanomaterials. 2021;11(7) doi: 10.3390/nano11071661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Marshall H.S., McMillan M., Koehler A.P., Lawrence A., Sullivan T.R., MacLennan J.M., Maiden M.C.J., Ladhani S.N., Ramsay M.E., Trotter C., Borrow R., Finn A., Kahler C.M., Whelan J., Vadivelu K., Richmond P. Meningococcal B vaccine and meningococcal carriage in adolescents in Australia. N. Engl. J. Med. 2020;382(4):318–327. doi: 10.1056/NEJMoa1900236. [DOI] [PubMed] [Google Scholar]
- 137.Rodrigues F.M.P., Marlow R., Simões M.J., Danon L., Ladhani S., Finn A. Association of use of a Meningococcus group B vaccine with group B Invasive meningococcal disease among children in Portugal. JAMA. 2020;324(21):2187–2194. doi: 10.1001/jama.2020.20449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Harrison L.H., Stephens D.S. Good news and bad news - 4CMenB vaccine for group B Neisseria meningitidis. N. Engl. J. Med. 2020;382(4):376–378. doi: 10.1056/NEJMe1916440. [DOI] [PubMed] [Google Scholar]
- 139.Bittel M., Reichert P., Sarfati I., Dressel A., Leikam S., Uderhardt S., Stolzer I., Phu T.A., Ng M., Vu N.K., Tenzer S., Distler U., Wirtz S., Rothhammer V., Neurath M.F., Raffai R.L., Günther C., Momma S. Visualizing transfer of microbial biomolecules by outer membrane vesicles in microbe-host-communication in vivo. J. Extracell. Vesicles. 2021;10(12) doi: 10.1002/jev2.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zaiss M.M., Jones R.M., Schett G., Pacifici R. The gut-bone axis: how bacterial metabolites bridge the distance. J. Clin. Invest. 2019;129(8):3018–3028. doi: 10.1172/JCI128521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yan J., Herzog J.W., Tsang K., Brennan C.A., Bower M.A., Garrett W.S., Sartor B.R., Aliprantis A.O., Charles J.F. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc. Natl. Acad. Sci. U. S. A. 2016;113(47):E7554–e7563. doi: 10.1073/pnas.1607235113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lucas S., Omata Y., Hofmann J., Böttcher M., Iljazovic A., Sarter K., Albrecht O., Schulz O., Krishnacoumar B., Krönke G., Herrmann M., Mougiakakos D., Strowig T., Schett G., Zaiss M.M. Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat. Commun. 2018;9(1):55. doi: 10.1038/s41467-017-02490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen X., Zhang Z., Hu Y., Cui J., Zhi X., Li X., Jiang H., Wang Y., Gu Z., Qiu Z., Dong X., Li Y., Su J. Lactulose suppresses osteoclastogenesis and ameliorates estrogen deficiency-induced bone loss in mice. Aging Dis. 2020;11(3):629–641. doi: 10.14336/AD.2019.0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chevalier C., Kieser S., Çolakoğlu M., Hadadi N., Brun J., Rigo D., Suárez-Zamorano N., Spiljar M., Fabbiano S., Busse B., Ivanišević J., Macpherson A., Bonnet N., Trajkovski M. Warmth prevents bone loss through the gut microbiota. Cell Metabol. 2020;32(4):575–590. doi: 10.1016/j.cmet.2020.08.012. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zehentmeier S., Pereira J.P. Cell circuits and niches controlling B cell development. Immunol. Rev. 2019;289(1):142–157. doi: 10.1111/imr.12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Müller A., Homey B., Soto H., Ge N., Catron D., Buchanan M.E., McClanahan T., Murphy E., Yuan W., Wagner S.N., Barrera J.L., Mohar A., Verástegui E., Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 147.Yang D., Park S.Y., Park Y.S., Eun H., Lee S.Y. Metabolic engineering of Escherichia coli for natural product biosynthesis. Trends Biotechnol. 2020;38(7):745–765. doi: 10.1016/j.tibtech.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 148.Liu H., Hou Y., Wang Y., Li Z. Enhancement of sulfur conversion rate in the production of L-Cysteine by engineered Escherichia coli. J. Agric. Food Chem. 2020;68(1):250–257. doi: 10.1021/acs.jafc.9b06330. [DOI] [PubMed] [Google Scholar]
- 149.Liu H., Wang Y., Hou Y., Li Z. Fitness of chassis cells and metabolic pathways for L-cysteine overproduction in Escherichia coli. J. Agric. Food Chem. 2020;68(50):14928–14937. doi: 10.1021/acs.jafc.0c06134. [DOI] [PubMed] [Google Scholar]
- 150.Vader P., Mol E.A., Pasterkamp G., Schiffelers R.M. Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 2016;106(Pt A):148–156. doi: 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 151.Emery D.C., Shoemark D.K., Batstone T.E., Waterfall C.M., Coghill J.A., Cerajewska T.L., Davies M., West N.X., Allen S.J. 16S rRNA next generation sequencing analysis shows bacteria in Alzheimer's post-mortem brain. Front. Aging Neurosci. 2017;9:195. doi: 10.3389/fnagi.2017.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Haas-Neill S., Forsythe P. A budding relationship: bacterial extracellular vesicles in the microbiota-gut-brain axis. Int. J. Mol. Sci. 2020;21(23) doi: 10.3390/ijms21238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Allan N.D., Beveridge T.J. Gentamicin delivery to Burkholderia cepacia group IIIa strains via membrane vesicles from Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 2003;47(9):2962–2965. doi: 10.1128/AAC.47.9.2962-2965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gujrati V., Prakash J., Malekzadeh-Najafabadi J., Stiel A., Klemm U., Mettenleiter G., Aichler M., Walch A., Ntziachristos V. Bioengineered bacterial vesicles as biological nano-heaters for optoacoustic imaging. Nat. Commun. 2019;10(1):1114. doi: 10.1038/s41467-019-09034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gujrati V.B., Jon S. Bioengineered bacterial outer membrane vesicles: what is their potential in cancer therapy? Nanomedicine. 2014;9(7):933–935. doi: 10.2217/nnm.14.56. [DOI] [PubMed] [Google Scholar]
- 156.Ayed Z., Cuvillier L., Dobhal G., Goreham R.V. Electroporation of outer membrane vesicles derived from Pseudomonas aeruginosa with gold nanoparticles. SN Appl. Sci. 2019;1(12):1600. [Google Scholar]
- 157.Lee Y., Park J.Y., Lee E.H., Yang J., Jeong B.R., Kim Y.K., Seoh J.Y., Lee S., Han P.L., Kim E.J. Rapid assessment of microbiota changes in individuals with autism spectrum disorder using bacteria-derived membrane vesicles in urine. Exp Neurobiol. 2017;26(5):307–317. doi: 10.5607/en.2017.26.5.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Peng Y., Yin S., Wang M. Extracellular vesicles of bacteria as potential targets for immune interventions. Hum. Vaccines Immunother. 2021;17(3):897–903. doi: 10.1080/21645515.2020.1799667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Parker H., Chitcholtan K., Hampton M.B., Keenan J.I. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect. Immun. 2010;78(12):5054–5061. doi: 10.1128/IAI.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Furuta N., Takeuchi H., Amano A. Entry of Porphyromonas gingivalis outer membrane vesicles into epithelial cells causes cellular functional impairment. Infect. Immun. 2009;77(11):4761–4770. doi: 10.1128/IAI.00841-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bomberger J.M., Maceachran D.P., Coutermarsh B.A., Ye S., O'Toole G.A., Stanton B.A. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5(4) doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wai S.N., Lindmark B., Söderblom T., Takade A., Westermark M., Oscarsson J., Jass J., Richter-Dahlfors A., Mizunoe Y., Uhlin B.E. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115(1):25–35. doi: 10.1016/s0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- 163.Ricci V., Carcione D., Messina S., Colombo G.I., D'Alessandra Y. Circulating 16S RNA in biofluids: extracellular vesicles as mirrors of human microbiome? Int. J. Mol. Sci. 2020;21(23) doi: 10.3390/ijms21238959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Taboada H., Meneses N., Dunn M.F., Vargas-Lagunas C., Buchs N., Castro-Mondragón J.A., Heller M., Encarnación S. Proteins in the periplasmic space and outer membrane vesicles of Rhizobium etli CE3 grown in minimal medium are largely distinct and change with growth phase. Microbiology (Read.) 2019;165(6):638–650. doi: 10.1099/mic.0.000720. [DOI] [PubMed] [Google Scholar]
- 165.Macia L., Nanan R., Hosseini-Beheshti E., Grau G.E. Host-and microbiota-derived extracellular vesicles, immune function, and disease development. Int. J. Mol. Sci. 2019;21(1) doi: 10.3390/ijms21010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.DeJong E.N., Surette M.G., Bowdish D.M.E. The gut microbiota and unhealthy aging: disentangling cause from consequence. Cell Host Microbe. 2020;28(2):180–189. doi: 10.1016/j.chom.2020.07.013. [DOI] [PubMed] [Google Scholar]