Significance
Histone-like protein from Escherichia coli strain U93 (HU) protein is the most abundant nucleoid-associated protein in bacteria, which plays a fundamental role in chromosomal compaction and organization. It is essential for most bacteria as well as Apicomplexans, thus an important target for the development of antimicrobial and antimalaria drugs. We report Gp46 as a phage protein HU inhibitor. It inhibits HU of Bacillus subtilis by occupying its DNA binding site, thus preventing chromosome segregation during cell division. As key residues for the interaction are highly conserved, Gp46 interacts with HUs of a broad range of pathogens, including many pathogenic bacteria and Apicomplexan parasites like Plasmodium falciparum. Thus, this cross-species property could benefit antibiotic and antimalaria drug development that targets HU.
Keywords: HU, bacteriophage, nucleoid, plasmodium, Gp46
Abstract
The architectural protein histone-like protein from Escherichia coli strain U93 (HU) is the most abundant bacterial DNA binding protein and highly conserved among bacteria and Apicomplexan parasites. It not only binds to double-stranded DNA (dsDNA) to maintain DNA stability but also, interacts with RNAs to regulate transcription and translation. Importantly, HU is essential to cell viability for many bacteria; hence, it is an important antibiotic target. Here, we report that Gp46 from bacteriophage SPO1 of Bacillus subtilis is an HU inhibitor whose expression prevents nucleoid segregation and causes filamentous morphology and growth defects in bacteria. We determined the solution structure of Gp46 and revealed a striking negatively charged surface. An NMR-derived structural model for the Gp46–HU complex shows that Gp46 occupies the DNA binding motif of the HU and therefore, occludes DNA binding, revealing a distinct strategy for HU inhibition. We identified the key residues responsible for the interaction that are conserved among HUs of bacteria and Apicomplexans, including clinically significant Mycobacterium tuberculosis, Acinetobacter baumannii, and Plasmodium falciparum, and confirm that Gp46 can also interact with these HUs. Our findings provide detailed insight into a mode of HU inhibition that provides a useful foundation for the development of antibacteria and antimalaria drugs.
The bacterial nucleoid is an “open” region of the cytoplasm containing genetic material that is not confined within a membrane envelop, in contrast to the eukaryotic nucleus (1). Furthermore, prokaryotic nucleoid-associated proteins (NAPs), which are responsible for organization of the nucleoid and the control of gene expression, are also distinct from eukaryotic histones (2). Bacterial NAPs are capable of not only binding to DNA but also, altering the quaternary structure of the DNA double helix by inducing wrapping or bending. Integration host factor, histone-like nucleoid-structuring protein, and histone-like protein from Escherichia coli strain U93 (HU) protein are among the best-known members of the NAP family (3, 4). HU, with ∼50,000 molecules per bacterial cell, is the most abundant and conserved NAP in bacteria (5, 6). It plays fundamental roles in chromosomal structuring and transcription/replication regulation as well as interacting with messenger RNA (mRNA) and noncoding RNA (7–9). HU binds to double-stranded DNA (dsDNA) with no sequence specificity, but it prefers specialized DNA structures and provides key architectural roles in DNA compaction (10).
HUs form homodimers in most bacteria but heterodimers in Enterobacteriaceae (9). In Enterobacteriaceae, including E. coli, HU dimers have two forms: a homodimer by HUα self-association (HUαα) or a heterodimer by HUα–HUβ (HUαβ) interactions. Deleting both HU genes in E. coli causes a deleterious filamentous morphology and poor proliferation rate but is not lethal. Interestingly, the composition of HU dimers is interchangeable; the HUαα homodimer is the dominant form in the lag and exponential growth phases, whereas the HUαβ heterodimer is present mostly in early and late stationary phases (11). In most other bacteria, including Mycobacterium tuberculosis, HUs exist as homodimers and are essential for bacterial survival (4, 12). Other than bacteria, orthologs of HU in Apicomplexans, including Plasmodium berghei and Toxoplasma gondii, are also indispensable (13, 14). As HUs are essential for the viability of many clinically significant pathogens and structurally distinctive from human histone proteins, they represent an interesting target for antibacterial and antimalaria drug development (13, 15, 16).
Bacteriophages are dedicated viruses for bacteria that specialize in utilizing host machineries for their own reproduction. To take over the host and counter bacterial resistance systems, such as restriction–modification (RM) and toxin–antitoxins (TA), phages produce multiple host inhibitors to hijack host metabolism and evade host defense. For example, E. coli phage T7 produces an RM inhibitor Ocr (17) and a TA inhibitor Gp4.5 (18) as well as the host RNA polymerase (RNAP) inhibitor Gp2 (19). Although many phage proteins that are used in hijacking their host have been described, a host HU inhibitor has not been reported to our knowledge (19–21). Lytic phage SPO1 of Bacillus subtilis has a complex genome encoding over 200 proteins, including a gene cluster called the host takeover module (HTM), which is transcribed during the initial host takeover event (22). The 24 HTM members are mostly early genes that are characteristically smaller than average size and possess promoters and ribosome binding sites for very high gene expression. Gene gp46 is one of the early genes locates in operon 3 of the HTM, neighboring another early gene with known function, gp44, which encodes a host RNAP inhibitor (23). While it has been shown that expressing plasmid-borne Gp46 attenuates the growth of B. subtilis, the underlying molecular mechanism remains unclear (24). As the protein products encoded by HTM are highly expressed during the early phage infection stage and most of them have been shown to inhibit host metabolism and redirect resources for phage production (25–27), gp46 is likely to play a role in a similar event.
Here, we report that Gp46 is an HU protein inhibitor. Overexpressing Gp46 prevents nucleoid segregation and causes cell filamentation in B. subtilis, which resembles the phenotype of HU-deleted E. coli. Using NMR spectroscopy (NMR), we have solved the solution structure of Gp46 and derived a model for the HU–Gp46 complex. Our model reveals that Gp46 occupies the DNA binding motif of the histone-like protein from E. coli strain U93 of B. subtilis (HBsu), utilizing the striking negatively charged surface to interact with the dsDNA binding surface of HU. This together with an important hydrophobic contact allows Gp46 to displace dsDNA from HBsu–dsDNA complex and form a highly stable HBsu–Gp46 complex. Moreover, key residues that are crucial for Gp46–HBsu interaction are highly conserved across many pathogenic bacteria and Apicomplexans, including Enterococcus faecalis, M. tuberculosis, Staphylococcus aureus, Acinetobacter baumannii, and Plasmodium falciparum. We also demonstrate that Gp46 can interact with HUs of these important pathogens; therefore, our work provides a potential platform for the development of antibiotics and antimalaria drugs.
Results
Gp46 Causes Host Filamentation and Prevents Chromosome Segregation.
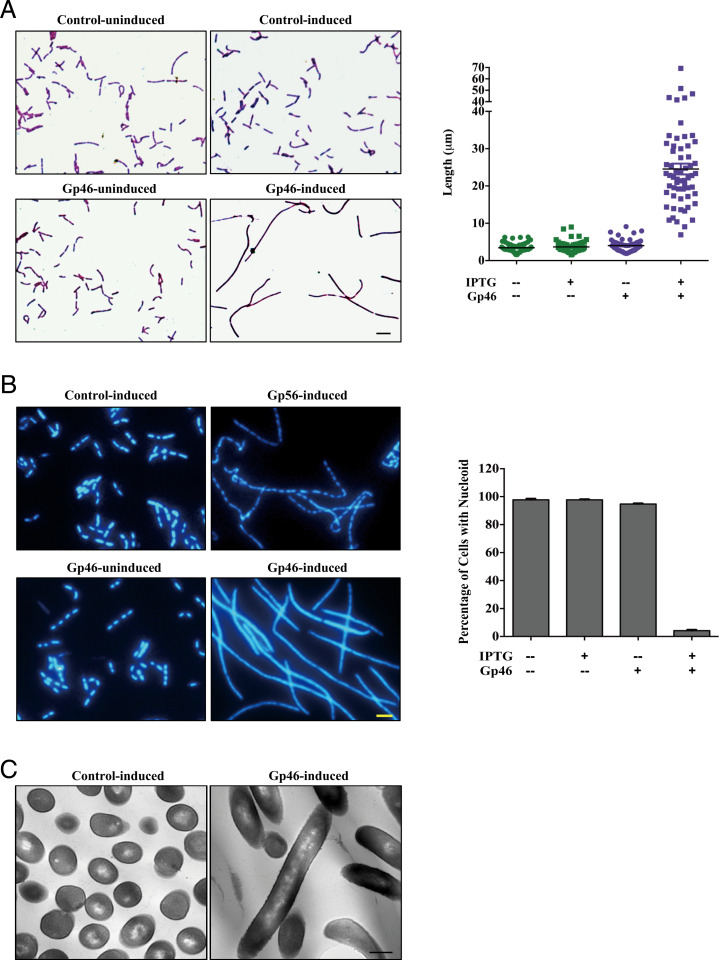
As Gp46 is reported as a host growth inhibitor, we verified the growth attenuation effect by expressing plasmid-borne gp46 in B. subtilis (strain 168) (SI Appendix, Fig. S1A). In addition to the expected growth defects, we observed an interesting morphological change for Gp46 expression Bacilli. In Gram staining, we found that the bacteria exhibited a filamentous morphology, and the average length of Gp46-expressing cells measured 24.5 ± 11.2 µm, which is over sixfold longer than that for control cells bearing the empty pHT01 plasmid (3.7 ± 1.6 µm) (Fig. 1A). These effects on cell growth and morphology are similar to another SPO1 protein Gp56, which is known to cause distinctive filamentation appearance of the bacteria (27). Gp56 inhibits cell division by interacting with FtsL but does not inhibit chromosome segregation with distinctive and well-defined repetitive nucleoids evident under DNA staining. To further investigate the cause of filamentation, we used the DAPI stain to examine the nucleoids and compared the phenotypes caused by Gp56 and Gp46. Strikingly, most nucleoids in Gp46-expressing cells display nondiscrete sizes, with the distribution of DNA expanded toward elongated cell walls, while the nucleoids of Gp56-expressing cells have a well-defined repetitive pattern throughout the bodies of the filaments (Fig. 1B). These results suggest that Gp46 may prevent normal chromosome segregation. To further explore this, we examined the distribution of Spo0J, which is an important indicator for normal chromosome segregation in B. subtilis, as it is concentrated into discrete foci during vegetative growth (28). However, Gp46 expression renders Spo0J–Green fluorescent protein (GFP) distributed evenly all over the cells (SI Appendix, Fig. S1 B and C), indicating unsuccessful chromosome segregation. To confirm our staining results, we used transmission electron microscopy (TEM) to examine the morphology of bacterial cells expressing Gp46. Under the microscope, we found that numerous Gp46-expressing bacteria display an elongated cell body with continuous and unseparated chromosomes (Fig. 1C). Taken together, these results indicate that Gp46 causes bacteria filamentation and prevents nucleoid separation.
Fig. 1.
Overexpressing Gp46 causes host filamentation and prevents nucleoid segregation. (A) Gram staining images of control (B. subtilis cells bearing empty pHT01 plasmid) and B. subtilis cells bearing pHT01–Gp46 plasmid before and after isopropyl ß-D-1-thiogalactopyranoside (IPTG) induction. The quantitation of the measured cell lengths of control and Gp46-expressing cells is displayed in the chart in Right; each point denotes the cell length from a single cell. (Scale bar: 5 µm.) (B) DAPI staining images of control and Gp56- or Gp46-expressing B. subtilis nucleoid. Quantitation of cells with segregated nucleoid from control and Gp46-expressing cells is displayed in the chart in Right. (Scale bar: 5 µm.) (C) TEM images of control and Gp46-expressing B. subtilis cells. (Scale bar: 500 nm.)
Gp46 Interacts with HBsu.
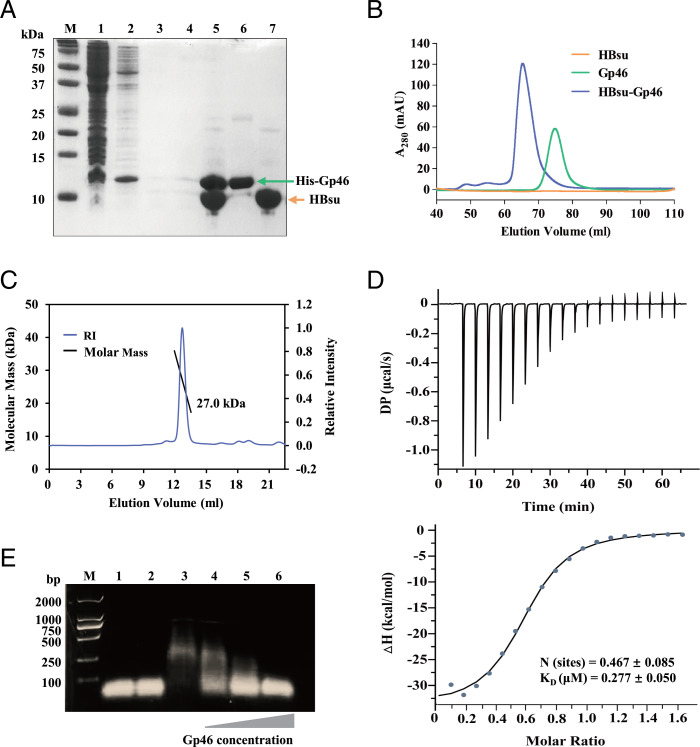
Interestingly, the distinctive phenotype caused by the Gp46 resembles that of the hup (coding HUα and HUβ proteins) null mutant of E. coli, which has an extreme filamentous morphology with a high rate of nucleoid loss (29). As the HU protein is one of the major NAPs that interacts with DNA and Gp46 is highly negatively charged (isoelectric point [pI] = 4.3), we speculated that Gp46 could displace DNA from HU, causing an hup knockout-like phenotype. To confirm the potential interaction, we performed a pull-down assay using column-immobilized Gp46 with cell lysate of B. subtilis and analyzed it by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 2A). The electrophoresis result revealed that an unknown protein with a molecular weight (MW) of ∼10 coeluted with Gp46 (Fig. 2 A, lane 5), suggesting that a complex is formed between Gp46 and the unknown protein. The identity of the bound protein was then confirmed by mass spectrometry to be HBsu (SI Appendix, Fig. S1D). The Gp46–HBsu complex is stable and could be separated by size exclusion chromatography (SEC) (Fig. 2B). The SEC profiles of Gp46, HBsu, and the HBsu–Gp46 complex indicate that the complex elutes at the volume corresponding to a molecule that is larger than Gp46. To determine the size of the complex, we used multiangle light scattering coupled with size exclusion chromatography (SEC-MALS) and determined the MW at 27.0 (Fig. 2C). As the MWs of Gp46 and HBsu (dimer) are 9.0 and 19.6, respectively, our results suggest that Gp46 forms a 1:2 complex with HBsu.
Fig. 2.
Gp46 interacts with HBsu. (A) SDS-PAGE shows the pull-down experiment using column-immobilized Gp46 and cell lysate of B. subtilis. M: Marker. (Lane 1) Flow through after applying cell lysate of B. subtilis. (Lane 2) Elute after applying 10 column volumes of binding buffer. (Lane 3) First elute after applying 20 column volumes of wash buffer. (Lane 4) Second elute after applying 20 column volumes of wash buffer. (Lane 5) Elute after applying 1 column volume of elution buffer. (Lane 6) Recombinantly expressed His–Gp46. (Lane 7) Recombinantly expressed HBsu. (B) The overlay of gel filtration chromatography profiles of HBsu (orange), Gp46 (green), and HBsu–Gp46 complex (blue). A280: absorbance at 280 nm. Note that HBsu does not contain any Trp, Tyr, or Cys; thus, it is not visible under ultraviolet spectrophotometry. (C) The absolute molar mass of the HBsu–Gp46 complex was determined at about 27.0 kDa by SEC-MALS. (D) ITC result for the Gp46 and HBsu interaction in a buffer containing 20 mM Tris, 150 mM NaCl, and 0.2 mM tris(2-carboxyethyl)phosphine (TCEP) (pH 7.5). The KD values for the interaction were calculated to be 0.277 ± 0.050 μM. DP: differential power. (E) Image of an agarose gel showing EMSA to demonstrate competition between Gp46 and dsDNA for HBsu. (Lane 1) dsDNA (40 µM). (Lane 2) dsDNA (40 µM) + Gp46 (40 µM). (Lane 3) dsDNA (40 µM) + HBsu (40 µM). (Lane 4) dsDNA (40 µM) + HBsu (40 µM) + Gp46 (20 µM). (Lane 5) dsDNA (40 µM) + HBsu (40 µM) + Gp46 (40 µM). (Lane 6) dsDNA (40 µM) + HBsu (40 µM) + Gp46 (80 µM).
Gp46 Replaces dsDNA from HBsu.
HU–DNA interaction is well characterized, and the affinity (KD) of the interaction has been determined to be 0.370 ± 0.021 µM using S. aureus histone-like protein from E. coli strain U93 (Sa_HU) in a buffer containing 20 mM trisaminomethane (Tris) and 150 mM NaCl (pH 7.5) (30). We calculated the KD for HBsu–DNA using isothermal titration calorimetry (ITC) under the same condition and compared it with that of the HBsu–Gp46 interaction. While the determined affinity of HBsu–DNA (0.359 ± 0.038 µM) (SI Appendix, Fig. S2A) is comparable with Sa_HU–DNA (0.370 ± 0.021 µM), the affinity of HBsu–Gp46 (0.277 ± 0.050 µM) is slightly higher (Fig. 2D). With the slight preference over dsDNA for HBsu and the abundance of Gp46 in a phage-infected cell, it should be able to displace DNA from the HBsu–DNA complex. To verify this, we performed electrophoretic mobility shift assays (EMSAs) using agarose gel. The HBsu–DNA complex is clearly visible as the migration profile on the gel is very different from that of dsDNA (Fig. 2E). With the increasing amount of added Gp46 into the HBsu–DNA mixture, the migration profile of the mixture increasingly resembles that of dsDNA, suggesting the gradual displacement of DNA from the HBsu–DNA complex. At one molar equivalent to the HBsu dimer, Gp46 almost completely replaced dsDNA, and the migration profile of the mixture is nearly identical to that of the dsDNA. This is further confirmed by a complementary EMSA using native polyacrylamide gel, in which the HBsu–dsDNA–Gp46 migration profile resembles that of HBsu–Gp46 rather than HBsu–dsDNA (SI Appendix, Fig. S2B).
Gp46 is very acidic, which raises the notion that DNA charge mimicry could play a role in its interaction with HBsu. To test this hypothesis, we explored the electrostatic nature of the interaction by performing a pull-down assay in a buffer containing 20 mM Tris, 300 mM NaCl, and 0.2 mM TCEP (pH 7.5; high-salt buffer), which would weaken the interaction. Under high-salt conditions, the pull-down assay failed to show stable complex formation between HBsu and Gp46 (SI Appendix, Fig. S2C). In addition, SEC shows no shift in the elution volume of the HBsu–Gp46 mixture, indicating no complex formation under the high-salt condition (SI Appendix, Fig. S2D). While ITC failed to quantify the interaction, we determined the KD to be 5.87 ± 0.11 µM by surface plasmon resonance (SPR), indicating much weaker affinity in the high-salt buffer (SI Appendix, Fig. S2E). Thus, our data suggest that an electrostatic contribution plays an important role in the complex formation and Gp46 displays negatively charged surface properties similar to dsDNA. As the HBsu–Gp46 interaction is highly dependent on salt concentration and ITC, SPR, and EMSA were performed at different salt concentrations, the results of these interaction experiments suggest that Gp46 is preferentially bound over DNA by HBsu under a variety of in vitro conditions, therefore providing support for the in vivo assays.
Gp46 Interacts with HU via Its DNA Binding Surface and a Key Hydrophobic Contact.
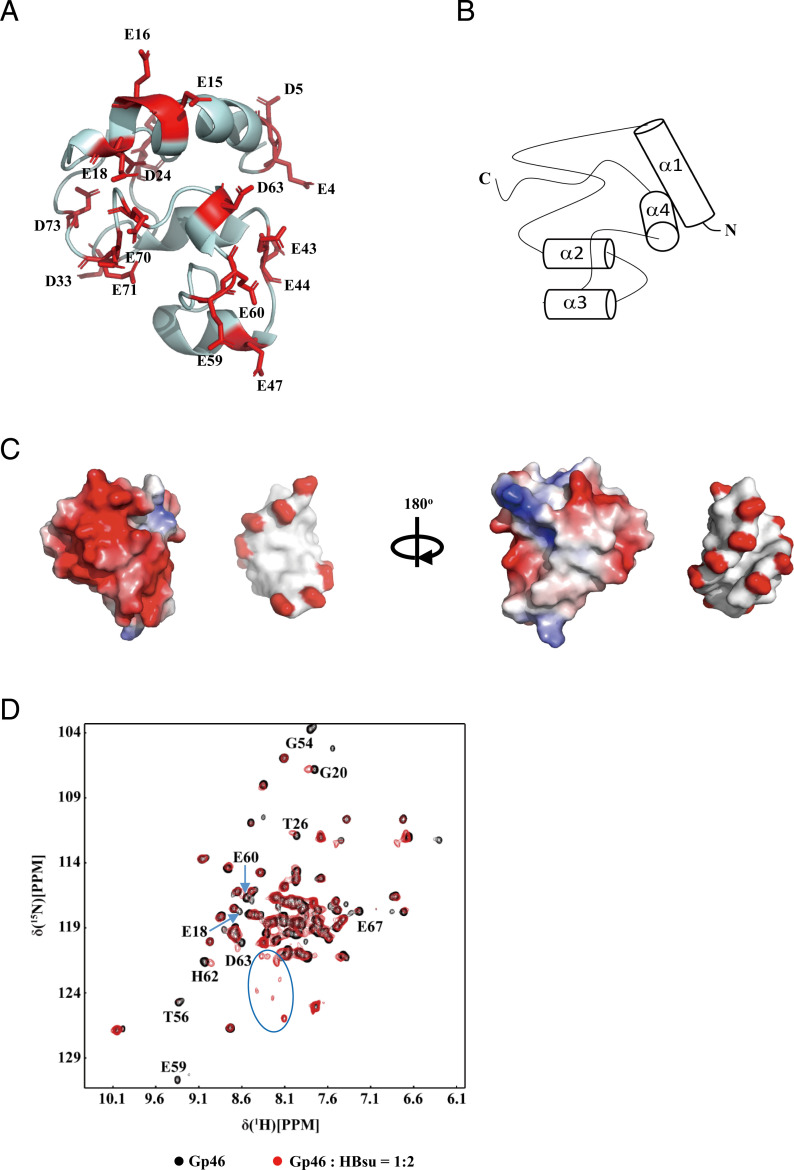
To map the HU interaction on Gp46, we expressed recombinant Gp46 in E. coli and determined the solution structure of Gp46 (Protein Data Bank [PDB] ID code 7by7 and Biological Magnetic Resonance Bank [BMRB] ID code 27767) by using standard multidimensional NMR experiments in a buffer containing 300 mM NaCl (31). The structure reveals that Gp46 has a globular fold with exposed negatively charged residues (Asp and Glu) localized on one side of the protein, while the rest of the surface is mainly uncharged (Fig. 3 A and B).
Fig. 3.
NMR studies of Gp46. (A) The structure of Gp46 with all negatively charged residues on the front labeled (PDB ID code 7by7). The residues include E4, D5, E15, E16, E18, D24, D33, E43, E44, E47, E59, E60, E67, E70, and E71. (B) The topology of the fold acquired by Gp46. (C) Comparison of the surface charge distribution of Gp46 (as illustrated in A) and dsDNA. The Gp46 electrostatic potential surface is colored from red to blue representing the electrostatic potential. The DNA surface is colored according to atomic charge in Pymol. The two images are displayed in 180° rotation along the z axis. (D) Overlay of two-dimensional 1H-15N heteronuclear single quantum coherence (HSQC) spectra of Gp46 (black) and Gp46–HBsu complex (only Gp46 15N labeled; red) recorded at 300 K in a buffer containing 20 mM Tris, 150 mM NaCl, and 0.2 mM TCEP (pH 7.5). Some untraceable peaks in the bound form are indicated in the blue circle, and traceable peaks are labeled. PPM: parts per million.
The structure of Gp46 consists of four simple helices; however, structural homology searches using both the Dali server (32) and PDBefold (33) reveal no statistically significant similarity to other known proteins. A BlastP search shows that it shares sequence similarity with proteins only from a handful of Bacillus phages, including bacteriophage CampHawk, SIOphi, BSP9, phiNIT1, and Grass (SI Appendix, Fig. S3A). Among them, CampHawk is another SPO1-like phage belonging to the subfamily Spounavirinae; SIOphi belongs to subfamily Bastillevirinae, whereas BSP9, phiNIT1, and Grass fall into subfamily Nitunavirus. Gp46 represents a class of proteins currently only found in phages.
It is conceivable that the negative surface of Gp46 mimics the charge distribution of the phosphate backbone of dsDNA with an overall size similar to approximately five base pairs in length (Fig. 3C). Proteins that mimic DNA have already been described (34), with some strictly imitating the electrostatic surface of dsDNA, while others provide a more diffuse negatively charged surface and complement this with other specific interactions. A notable example includes DMP12 from Neisseria, which is a small monomeric protein that has a molecular shape and charge distribution similar dsDNA and forms a 1:2 complex with the HU dimer (35). Analogous to this, Gp46 could potentially be described as a member of this family of DNA mimic proteins, although its electrostatic surface is less of a strict DNA mimic. Although distinct from Gp46, another DNA mimic protein of note is the white spot syndrome virus ICP11, which is a dimeric protein that binds histones via mimicking the electrostatic properties of DNA and disrupting nucleosomes (36).
While the attempts to crystallize the HBsu–Gp46 complex have thus far failed, we sought to derive an NMR model. To assist in determining the interface of the interaction, we utilized our fully assigned NMR spectrum of Gp46 and performed NMR titrations using 15N-labeled Gp46 and unlabeled HBsu under both high- and low-salt conditions. As mixing Gp46 with HBsu in low-salt buffers induces precipitation during the NMR experiments, we mixed an excess amount of Gp46 with HBsu in the NMR buffer containing 300 mM NaCl and dialyzed into the buffer containing 150 mM NaCl used in ITC (low-salt buffer). Excess Gp46 precipitated out during the dialysis, leaving a homogenous sample of the HBsu–Gp46 complex (as illustrated in Fig. 2 B and C). In the spectra of the complex under lower-salt conditions, many peaks in the HBsu-bound Gp46 spectrum experience both chemical shift perturbation and significant peak broadening compared with free Gp46 (Fig. 3D), notably the negatively charged residues E18, E59, E60, D63, and E67. There are multiple peaks in the center of the spectrum, many of which are also negatively charged residues that also experience significant spectral perturbations but were not assignable due to peak overlap. In general, the chemical shift perturbations are small, and the significant broadening of peaks is the major effect in the complex. This is likely due to exchange within the complex, as Gp46 binds HU in a 1:2 stoichiometry and could switch interactional interfaces between monomers within the HU dimer. We, therefore, chose to use the extent of peak broadening as a key indicator of the interaction interface.
Under high-salt buffer, peak broadening is also the main observation in the titration (SI Appendix, Fig. S3B). Importantly, the peaks that were affected in the low-salt buffer titration are also among the most broadened peaks in the high-salt buffer, indicating that the underlying mechanism remains same, albeit with a reduced affinity. We plotted the change in peak intensity against residue number and mapped the residues that had >60% intensity reduction on the structure of Gp46 (SI Appendix, Fig. S3C). As expected, most of these residues (E15, E18, E59, E60, E67, and E70) have negatively charged side chains and locate on the one side of Gp46 (SI Appendix, Fig. S3D).
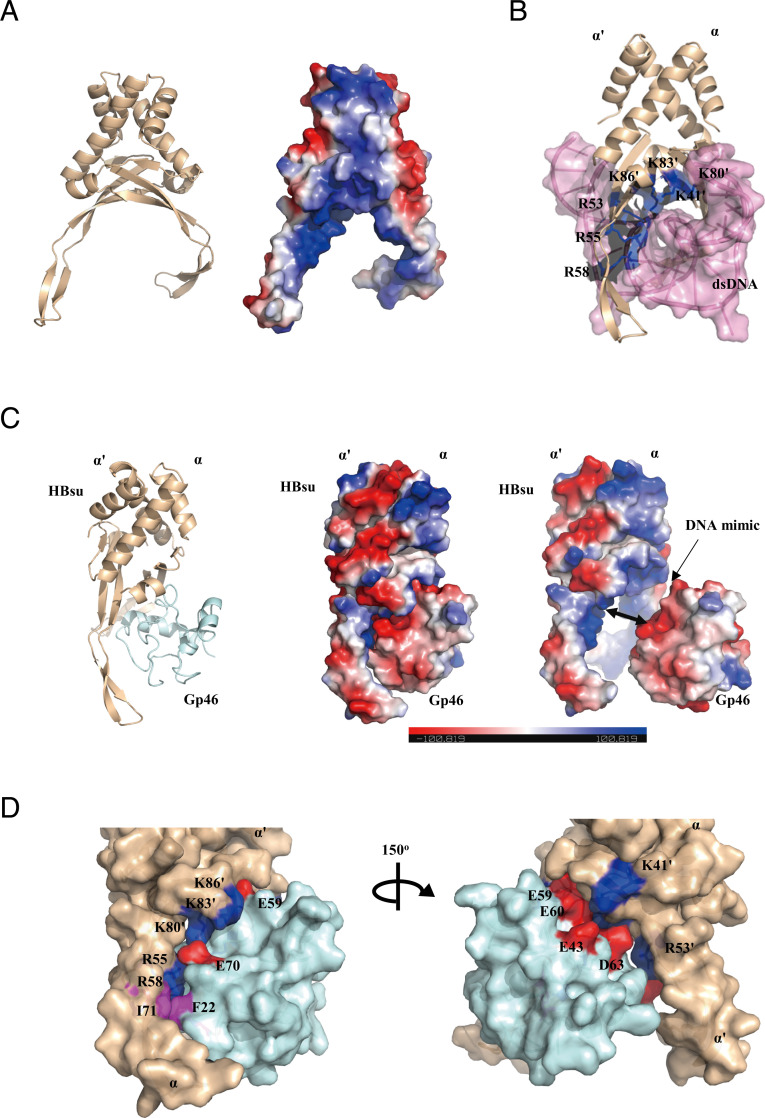
As the interface on Gp46 overlaps with negatively charged residues that may reflect DNA surface charge, we built a structural model using Gp46 NMR data and the identified DNA binding residues of HBsu. We first built a model of HBsu using the structure of the HU–dsDNA complex of S. aureus (Sa_HU; PDB ID code 4qju) as template using SWISS-MODEL (37) (Fig. 4A). As the DNA binding motifs are highly conserved among HU proteins, the residues involved are mostly identical across the species (30) (Fig. 4B). HBsu forms a symmetrical homodimer consisting of α- and α’-subunits, and its interaction with dsDNA involves both subunits. R53, R55, and R58 of the α-subunit and residues K41', K80', K83', and K86' of the α’-subunit are involved in DNA binding. Similarly, residues K41, K80, K83, and K86 of the α-subunit and R53', R55', and R58' of the α'-subunit can also bind dsDNA at the opposite side.
Fig. 4.
The Gp46–HBsu complex model. (A) Homology modeling of HBsu by SWISS-MODEL. (B) Homology modeling of HBsu–DNA complex using the Sa_HU–DNA complex structure (PDB ID code 4qju), in which HBsu is colored in wheat and dsDNA is colored in pink. Residues colored in blue are R53, R55, and R58 of α-subunit, and K41', K80', K83', and K86' of α'-subunit of HBsu are responsible for the HBsu–DNA interaction. (C, Left) HBsu complex built by HADDOCK in which HBsu is colored in wheat and Gp46 is colored in cyan. (C, Center) The surface charge distribution of the complex. (C, Right) The surface charge distribution in an open complex. Positively charged residues are colored in blue, and negatively charged residues are colored in red. (D) A 150° rotation display of the HBsu–Gp46 complex model interface: Residues involved in the electrostatic interaction include multiple positively charged Lys and Arg (colored in blue) of HBsu and negatively charged Glu and Asp of Gp46 (colored in red). Hydrophobic residues involved in the interaction include I71 of HBsu and F22 of Gp46 (colored in magenta).
Using the NMR data in low-salt buffer and DNA binding residues of HBsu, we built a model of the Gp46–HBsu complex using High Ambiguity Driven protein–protein DOCKing (HADDOCK) (38) (Fig. 4C). The surface charge distributions and surface geometries of both interfaces complement well. The model shows that Gp46 binds to the HBsu homodimer in such an orientation that it could approach HBsu from either the front or back side of the DNA binding motif to block DNA binding. A notable hydrophobic contract (F22Gp46:I71HBsu) is also formed at the interface, which complements the electrostatic interaction (Fig. 4D).
Critical Residues for the Gp46–HBsu Interaction.
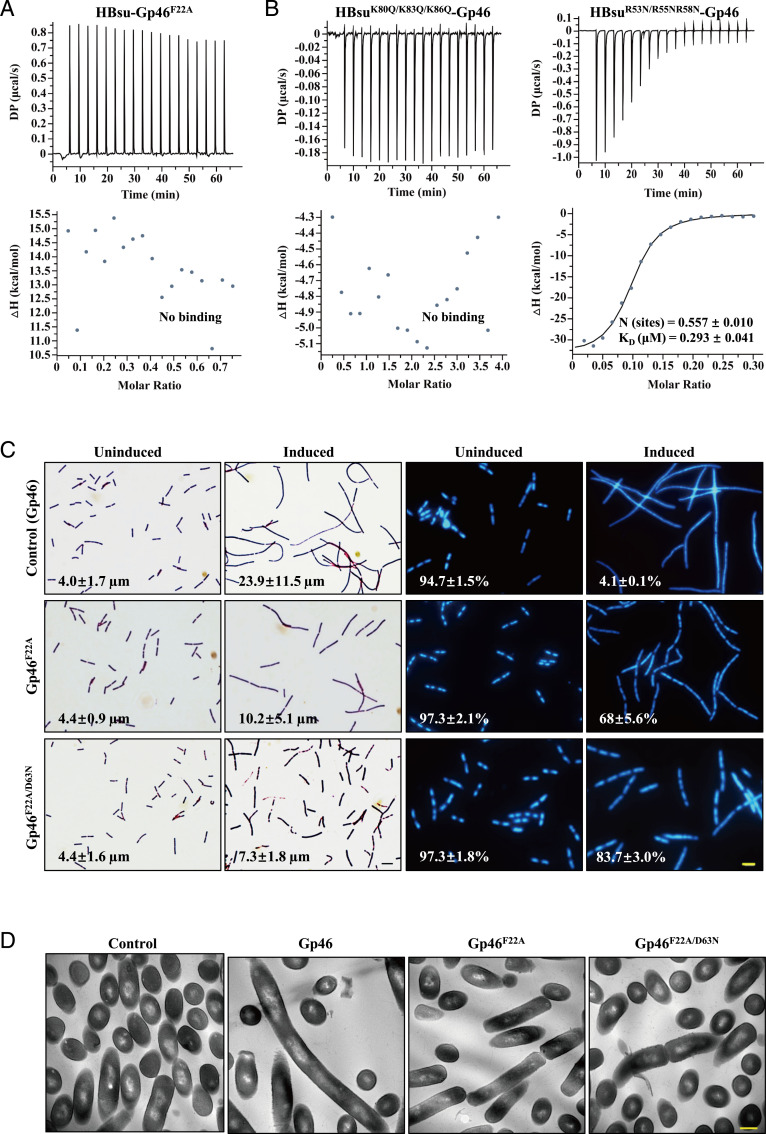
To validate the conclusions from our structural model, we mutated interfacial residues in the model and examined the effects with both in vitro and in vivo assays. First, we made mutations to disrupt the putative hydrophobic interaction between F22Gp46:I71HBsu. Gp46F22A fails to show an interaction with wild-type (WT) HBsu, and similarly, HBsuI71A interacts with WT Gp46 at much weaker affinity (KD = 6.41 ± 3.54 μM) (Fig. 5A and SI Appendix, Fig. S4A).
Fig. 5.
Key residues in the HBsu–Gp46 interaction. (A and B) ITC results for interactions between Gp46 and HBsu mutants as indicated. (C, Left) Gram staining images of Gp46- or Gp46 mutant–expressing B. subtilis cells. Values in the lower left corners represent the length of bacterial cells (average ± SD). (Scale bar: 5 μm.) (C, Right) DAPI staining images of Gp46- or Gp46 mutant–expressing B. subtilis nucleoid. Values in the lower left corners represent the percentage of cells with segregated nucleoid. (Scale bar: 5 μm.) (D) TEM images of Gp46- or Gp46 mutant–expressing B. subtilis cells. (Scale bar: 500 nm.)
In HBsu, the positive charged surface is localized in two clusters. The R53, R55, and R58 cluster is on the flexible “β-arms,” and the K41, K80, K83, and K86 cluster resides in the “pit” between the two arms. To evaluate if these charges are important for interaction with Gp46, we constructed two triple-charge neutralizing mutants, HBsuK80Q/K83Q/K86Q and HBsuR53N/R55NR58N, representing the charge neutralization on the pit and arm, respectively. Interestingly, only HUK80Q/K83Q/K86Q showed an abolished interaction, suggesting that only the electrostatic interaction in the pit residues is functionally important to the Gp46–HBsu interaction (Fig. 5B). In the arm region, the hydrophobic interaction between I71HBsu:F22Gp46 plays a more dominant role. In the structural model of the complex, D63 resides in the center of the charged patch of Gp46, and this complements the pit of HBsu. While charge-swapping mutant Gp46D63R loses its fully folded state, Gp46D63N shows a weaken interaction with HBsu as ITC (SI Appendix, Fig. S4B). Furthermore, Gp46D63N/F22A failed to show interaction with HBsu (SI Appendix, Fig. S4C). In conclusion, the mutagenesis data are consistent with our model of the complex, in which both the hydrophobic interaction in the arm and the electrostatic interaction in the pit contribute to complex formation.
We then applied key mutations to our in vivo assays. In the bacterial growth assay, both Gp46D63 and Gp46F22A expressions had a smaller impact on growth attenuation compared with the WT (SI Appendix, Fig. S4D). In Gram and DAPI staining experiments, bacteria expressing Gp46D63N or Gp46F22A exhibited less severe cell filamentation and nucleoid deformation (Fig. 5C and SI Appendix, Fig. S5A). Under TEM, Gp46F22A expressing B. subtilis showed some signs of chromosome segregation and cell division. Notably, the double-mutant Gp46F22A/D63N expressing B. subtilis showed a near-normal growth rate (SI Appendix, Fig. S4D). Furthermore, Gp46F22A/D63N expressing B. subtilis displayed near-WT morphology in Gram and DAPI staining and further improved chromosome segregation and cell division under TEM (Fig. 5 C and D and SI Appendix, Fig. S5). Finally, to ensure that overexpression of Gp46 does not overpower HU with large excess copies in the bacterial cell, we estimated that there were ∼90,000 molecules of Gp46 in one bacterial cell, which is comparable with the number (50,000) for HU (SI Appendix, Fig. S6). In summary, our in vivo mutagenesis experiments are in full agreement with conclusions from the structural and biochemical studies.
Gp46 Interacts with Several Homodimeric HUs of Clinically Significant Pathogens In Vitro.
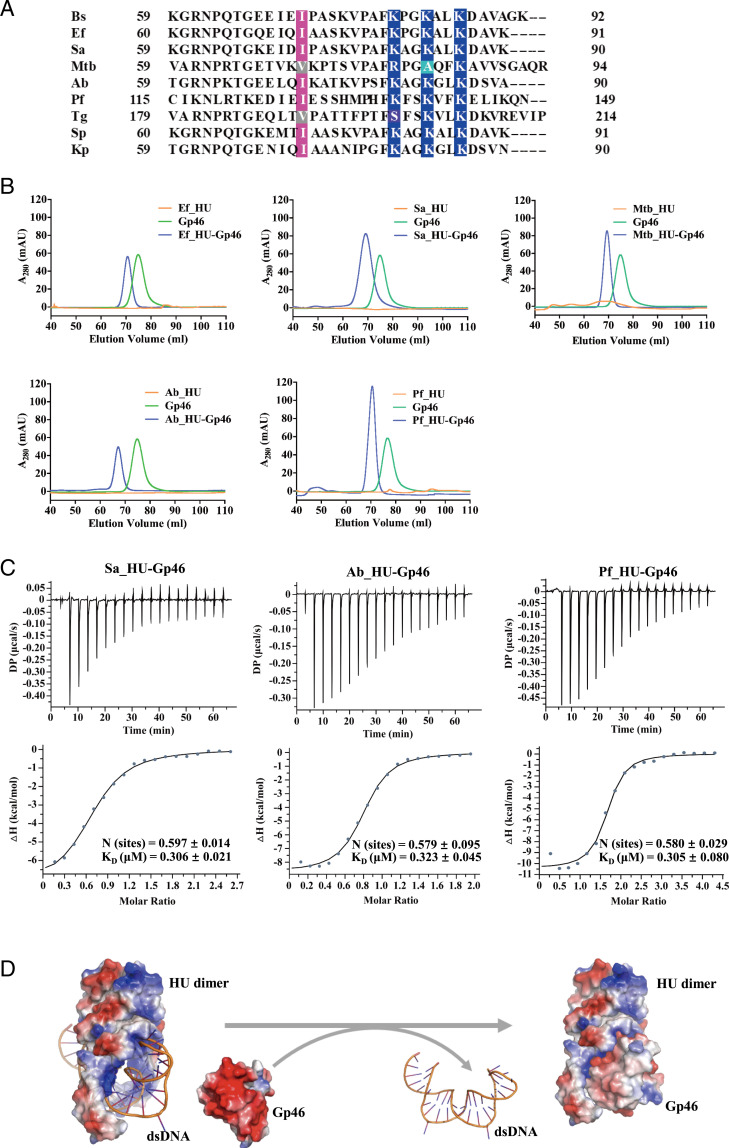
As HU proteins are highly conserved among bacteria and all HU structures deposited in PDB adopt the same fold, we speculated that Gp46 should also interact with other HUs. We selected HUs from some of the most clinically significant pathogens, including A. baumannii, E. faecalis, M. tuberculosis, S. aureus, P. falciparum, T. gondii, Streptococcus pneumoniae, and Klebsiella pneumoniae, and performed sequence alignment (Fig. 6A). Strikingly, the key residues for HBsu identified in our study are highly conserved among these HUs, indicating a high possibility of interaction with Gp46. To verify our hypothesis, we expressed and purified some of these HUs and checked the complex formation between Gp46 with various HUs. In gel filtration experiments, Gp46 forms stable complexes with HUs of A. baumannii, E. faecalis, M. tuberculosis, S. aureus, and P. falciparum (Fig. 6B). We also determined the binding affinities of three of the Gp46–HU interactions at ∼0.3 µM, which is comparable with the Gp46–HBsu interaction (Fig. 6C). As the key residues are highly conserved among these HUs, we anticipate that Gp46 is capable of binding most if not all homodimeric HUs.
Fig. 6.
Gp46 forms a complex with HUs of a range of pathogens. (A) Sequence alignment of different HU homologs: B. subtilis (Bs), A. baumannii (Ab), E. faecalis (Ef), K. pneumoniae (Kp), M. tuberculosis (Mtb), P. falciparum (Pf), S. aureus (Sa), S. pneumoniae (Sp), and T. gondii (Tg). Positively charged key residues Lys and Arg are colored in blue, and conserved Ile are colored in magenta. Mtb_HU contains a conserved Val (colored in grey) at residue 71 and a charge neutralizing Ala at residue 83 (colored in light blue). (B) The overlay of gel filtration chromatography profiles for different HU–Gp46 interactions: HU (orange), Gp46 (green), and HU–Gp46 complex (blue). (C) The affinities of Sa_HU–Gp46, Ab_HU–Gp46, and Pf_HU–Gp46 interaction calculated by ITC. (D) The proposed model for the Gp46–HU–DNA interaction.
Thus, we show that Gp46 can interact with HUs from a range of clinically significant pathogens. In our proposed model, Gp46 deactivates HU by blocking the DNA binding motifs and preventing HU–DNA complex formation (Fig. 6D). As the HU–DNA interaction is important to cell viability for many bacteria, Gp46 is a broad-range antimicrobial agent, and its mode of action may inspire drug design against HU.
Discussion
HU proteins are highly conserved, and HU is an essential NAP for many bacteria and Apicomplexa. The structure of HU adopts β-saddle fold, in which its DNA binding motifs consist of extended two β-arms and a central pit that lies between. Residues 54 to 76 form the mobile arms, which are poorly defined in many HU structures, and residues at the bottom of the saddle form the pit and contribute to HU dimerization. As HU is indispensable for many pathogenic bacteria and Apicomplexans, it represents an interesting target for therapeutic approaches. In a recent study, small molecule HU inhibitors targeting the pit region were shown to be effective against M. tuberculosis (16). Furthermore, using structure-based drug design, inhibitors targeting the α-helical domain (the saddle) of HU have also shown activity in Spiroplasma melliferum. However, sequence alignment shows that the overall sequence conservation within the pit region and α-helical domain is low; therefore, these targeted inhibitors may work on a limited range of bacteria.
Antibiotic resistance, regarded as one of the biggest medical challenges along with cancer, is a growing threat to global health (39, 40). Bacteriophages are the natural killers of bacteria, including many antibiotic resistance strains (41). The rejuvenation of bacteriophage therapy has been successfully applied to many otherwise untreatable patients (42, 43). Other than the direct application of phage for therapeutic purposes, the strategies that phages employ to inhibit the host could also inspire the discovery of new methods for treating microbial infection (44). Inhibiting host DNA polymerase, RNAP, and σ-factors are among the reported hijacking strategies used by the phages to take over host nucleic acid metabolism (45–47); however, a phage-encoded HU inhibitor has until now not been reported. Bacteriophage SPO1 is the obligate phage of bacterium B. subtilis and possesses a complex genome encoding over 200 proteins, with 24 proteins in the HTM (25). Eight growth inhibitors have been identified within the HTM, including Gp56, which interacts with FtsL and the RNAP binding Gp44 (23, 48). Gp46 is also one of the HTM-encoded host inhibitors and has no sequence similarity to other proteins with known function.
Here, we report the function of Gp46 as an HU inhibitor that causes host growth attenuation and cell filamentation and prevents chromosome segregation. Our solution structure of Gp46 and model for its complex with HU reveal that it occupies the DNA binding motif of the HU and prevents the critical HU–DNA interaction. The “unprotected” dsDNA would provide an opportunity for other phage factors to access the nucleoid, such as organization of the early viral DNA replication machinery as seen in another B. subtilis phage Φ29 (49). We identify the key residues for the Gp46–HU complex and demonstrate that their mutagenesis abrogates the interaction and Gp46 function in vivo. Complementary electrostatic surfaces in the pit of the HBsu dimer and Gp46 and a key hydrophobic interaction between I71HBsu:F22Gp46 on the β-arm are crucial for HBsu–Gp46 complex formation. The nature of the Gp46–HU interaction is reminiscent of that for the Neisseria conserved hypothetical protein DMP12, which is described as a DNA mimic that binds to histone-like HU proteins. Unlike Gp46, the interaction is several folds weaker that the dsDNA–HU interaction, which led to the notion that DMP12 is a chaperone for HU dimers that stabilizes flexible regions during HU reorganization within the nucleoid. It was also suggested that DMP12 may play a role in manipulating the expression of the bacterial genome to favor replication (50). In contrast, Gp46 possesses a comparable and even slightly higher affinity for HU than dsDNA. Thus, it is a strong HU competitor that is able to displace DNA from the HU complex, disrupt nucleoid formation, and inhibit bacterial replication. This action is more akin to ICP11 from the mammalian white spot syndrome virus, which sequesters histones via mimicking the electrostatic properties of DNA, disrupting nucleosomes and causing cell death (36).
We also demonstrate that Gp46 can form complexes with a wide range of homodimeric HUs due to the sequence conservation of these key residues. Thus, engineering of therapeutic phages to carry a gp46 gene would benefit from its broad inhibitory ability regardless of host specificity. Meanwhile, the molecular interaction mechanism between Gp46 and HU serves as a model for the development of small molecule– or peptide-based inhibitors for a broad range of bacterial pathogens. Last but not the least, the above strategies may also be applied in developing treatments for malaria and potentially, other apicomplexan-related diseases.
Materials and Methods
Bacterial Growth Curve and Calculation of Gp46 Expression.
B. subtilis (strain 168) was used in this study for Gp46 expression and phenotyping. Details of the B. subtilis growth curve and calculation of Gp46 expression in B. subtilis are described in SI Appendix.
Gram Staining and Nucleoid Staining.
Gram staining was performed as previously reported (51). The length of each cell was measured using ImageJ software from random fields. For nucleoid staining, bacterial cells were first fixed with 4% (vol/vol) paraformaldehyde for 10 min followed by 0.05% (vol/vol) Triton X-100 permeabilization for 1 h with occasional agitation. Cells were then stained with 10 μg/mL DAPI for 10 min before being mounted on slides for photography using a Zeiss Axio Scope A1. For SPO0J–GFP imaging, cells transformed with the indicated constructs were induced with or without 1 mM IPTG for 4 h and mounted on slides before images were taken with the Zeiss Axio Scope A1 microscope.
TEM.
Sample preparation and electron microscopy are described in SI Appendix, Transmission Electron Microscopy (TEM).
Protein Expression and Purification.
SPO1 genes 46 and 56 and bacterial HU proteins are expressed in E.coli, and details about protein expression and purification are described in SI Appendix, Protein Expression in E. coli and Purification.
Pull-Down Assays.
Pull-down assays were performed using Nikel nitrilotriacetic acid (Ni-NTA) beads (Qiagen) in tandem with binding buffer (50 mM KH2PO4, 50 mM NaCl, 0.2 mM TCEP, 10 mM imidazole [pH 7.5]), wash buffer, and elution buffer as described previously (20). B. subtilis strain 168 (ATCC no. 23857) was used in the pull-down assay. Four liters of B. subtilis culture was harvested at optical density at 600 nm (OD600) ∼ 1.2 by centrifugation at 5,000 rpm at 4 °C for 10 min, resuspended in 100 mL of the binding buffer with one tablet of ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor mixture (cOmplete), and lysed by ultrasonication. The supernatant of cell lysate was filtered with a 0.2-µm filter before loading onto a benchtop His-trap column with His–Gp46 immobilized. The column was then subsequently washed with 10 volumes of binding buffer, 2 × 10 volumes of wash buffer, and 2 volumes of elution buffer.
Gp46–HBsu Interaction.
SEC-MALS, ITC, and SPR were employed to characterize Gp46–HBsu interactions (SI Appendix, Gp46–HBsu Interaction).
EMSA.
The EMSA assays were performed with dsDNA (the sequence is listed in SI Appendix, Table S1), Gp46, and HU in a buffer consisting of 20 mM Tris, 150 mM NaCl, and 0.2 mM TCEP (pH 7.5). The reaction mixtures containing different reagents as illustrated in Fig. 2E were topped up with the buffer to give a final volume of 20 μL and incubated for 30 min at 4 °C. The total binding solutions were loaded onto 1% agarose gels in 0.5× 45 mM Tris-borate and 1 mM EDTA buffer.
For EMSA performed in a native polyacrylamide gel, 10 μL of HBsu (0.1 μM), dsDNA (0.1 μM), and Gp46 (0.1 μM) were premixed as indicated. The mixtures were incubated on ice for 20 min before being loaded on a 10% native polyacrylamide polyacrylamide gel electrophoresis (PAGE). Electrophoresis was run at 100 V for 100 min on ice with a running buffer containing 3.03% (wt/vol) Tris-base and 14.4% glycine (wt/vol; pH 8.8). All EMSAs were performed in duplicate, and results show representative gels.
NMR Structure Determination.
NMR structure was carried out as previously described in ref. 52. and further described in SI Appendix, NMR Structure Determination. The structural statistics are shown in SI Appendix, Table S2.
NMR Titration.
NMR titrations were performed in either the high-salt buffer (300 mM NaCl, 50 mM KH2PO4, 0.2 mM TCEP [pH 6.5]) or the low-salt buffer (20 mM Tris, 150 mM NaCl, 0.2 mM TCEP [pH 7.5]). Unlabeled HBsu protein was added to 15N-labeled Gp46 (0.5 mM) according to the stoichiometric ratio indicated to perform NMR titration.
Mutagenesis.
All mutants carrying amino acid substitutions were obtained using the Q5 Site-Directed Mutagenesis Kit (NEB), and primers were designed using the online NEBaseChanger (SI Appendix, Table S1).
HADDOCK Calculation.
The HBsu structure generated by homology modeling using Sa_HU (PDB ID code 4qju) and the solution structure of Gp46 were used to build a model of a 1:1 complex of Gp46 bound to the HBsu dimer using HADDOCK (v2.2). Further description can be found in SI Appendix, HADDOCK Calculation.
Supplementary Material
Acknowledgments
This project was supported by National Key R&D Program of China Grant 2021YFA1301200 and 2021YFA1301201, National Natural Science Foundation of China Grant 81871662, and Key Research and Development Plan of Shaanxi Province Grants 2021ZDLSF01-10 and 2019ZDLSF03-02.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116278119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Dillon S. C., Dorman C. J., Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Browning D. F., Grainger D. C., Busby S. J., Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 13, 773–780 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Azam T. A., Ishihama A., Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 274, 33105–33113 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Oberto J., Nabti S., Jooste V., Mignot H., Rouviere-Yaniv J., The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One 4, e4367 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali Azam T., Iwata A., Nishimura A., Ueda S., Ishihama A., Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181, 6361–6370 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dame R. T., The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 56, 858–870 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Balandina A., Claret L., Hengge-Aronis R., Rouviere-Yaniv J., The Escherichia coli histone-like protein HU regulates rpoS translation. Mol. Microbiol. 39, 1069–1079 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Balandina A., Kamashev D., Rouviere-Yaniv J., The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J. Biol. Chem. 277, 27622–27628 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Pettijohn D. E., Histone-like proteins and bacterial chromosome structure. J. Biol. Chem. 263, 12793–12796 (1988). [PubMed] [Google Scholar]
- 10.Kamashev D., et al. , Comparison of histone-like HU protein DNA-binding properties and HU/IHF protein sequence alignment. PLoS One 12, e0188037 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammel M., et al. , HU multimerization shift controls nucleoid compaction. Sci. Adv. 2, e1600650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohavy O., et al. , Identification of a novel mycobacterial histone H1 homologue (HupB) as an antigenic target of pANCA monoclonal antibody and serum immunoglobulin A from patients with Crohn’s disease. Infect. Immun. 67, 6510–6517 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki N., et al. , The Plasmodium HU homolog, which binds the plastid DNA sequence-independent manner, is essential for the parasite’s survival. FEBS Lett. 583, 1446–1450 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Reiff S. B., Vaishnava S., Striepen B., The HU protein is important for apicoplast genome maintenance and inheritance in Toxoplasma gondii. Eukaryot. Cell 11, 905–915 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao J. H., et al. , Acetylome of Acinetobacter baumannii SK17 reveals a highly-conserved modification of histone-like protein HU. Front. Mol. Biosci. 4, 77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhowmick T., et al. , Targeting Mycobacterium tuberculosis nucleoid-associated protein HU with structure-based inhibitors. Nat. Commun. 5, 4124 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Walkinshaw M. D., et al. , Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell 9, 187–194 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Sberro H., et al. , Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning. Mol. Cell 50, 136–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cámara B., et al. , T7 phage protein Gp2 inhibits the Escherichia coli RNA polymerase by antagonizing stable DNA strand separation near the transcription start site. Proc. Natl. Acad. Sci. U.S.A. 107, 2247–2252 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabib-Salazar A., et al. , T7 phage factor required for managing RpoS in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 115, E5353–E5362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B., et al. , Bacteriophage Twort protein Gp168 is a β-clamp inhibitor by occupying the DNA sliding channel. Nucleic Acids Res. 49, 11367–11378 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart C. R., et al. , Genes and regulatory sites of the “host-takeover module” in the terminal redundancy of Bacillus subtilis bacteriophage SPO1. Virology 246, 329–340 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Wang Z., et al. , A bacteriophage DNA mimic protein employs a non-specific strategy to inhibit the bacterial RNA polymerase. Front. Microbiol. 12, 692512 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulvenna N., et al. , Xenogeneic modulation of the ClpCP protease of Bacillus subtilis by a phage-encoded adaptor-like protein. J. Biol. Chem. 294, 17501–17511 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart C. R., et al. , The genome of Bacillus subtilis bacteriophage SPO1. J. Mol. Biol. 388, 48–70 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampath A., Stewart C. R., Roles of genes 44, 50, and 51 in regulating gene expression and host takeover during infection of Bacillus subtilis by bacteriophage SPO1. J. Bacteriol. 186, 1785–1792 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart C. R., Deery W. J., Egan E. S., Myles B., Petti A. A., The product of SPO1 gene 56 inhibits host cell division during infection of Bacillus subtilis by bacteriophage SPO1. Virology 447, 249–253 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Ireton K., Gunther N. W. IV, Grossman A. D., spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176, 5320–5329 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huisman O., et al. , Multiple defects in Escherichia coli mutants lacking HU protein. J. Bacteriol. 171, 3704–3712 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D. H., et al. , β-Arm flexibility of HU from Staphylococcus aureus dictates the DNA-binding and recognition mechanism. Acta Crystallogr. D Biol. Crystallogr. 70, 3273–3289 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Zhang P., et al. , 1H, 13C and 15N NMR assignments of Bacillus subtilis bacteriophage SPO1 protein Gp46. Biomol. NMR Assign. 13, 245–247 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Holm L., Benchmarking fold detection by DaliLite v.5. Bioinformatics 35, 5326–5327 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Krissinel E., Henrick K., Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Wang H. C., Ho C. H., Hsu K. C., Yang J. M., Wang A. H., DNA mimic proteins: Functions, structures, and bioinformatic analysis. Biochemistry 53, 2865–2874 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Zhang K., et al. , Systemic expression, purification, and initial structural characterization of bacteriophage T4 proteins without known structure homologs. Front. Microbiol. 12, 674415 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H. C., et al. , White spot syndrome virus protein ICP11: A histone-binding DNA mimic that disrupts nucleosome assembly. Proc. Natl. Acad. Sci. U.S.A. 105, 20758–20763 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dominguez C., Boelens R., Bonvin A. M., HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Ventola C. L., The antibiotic resistance crisis. Part 1: Causes and threats. P&T 40, 277–283 (2015). [PMC free article] [PubMed] [Google Scholar]
- 40.Lushniak B. D., Antibiotic resistance: A public health crisis. Public Health Rep. 129, 314–316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golkar Z., Bagasra O., Pace D. G., Bacteriophage therapy: A potential solution for the antibiotic resistance crisis. J. Infect. Dev. Ctries. 8, 129–136 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Dedrick R. M., et al. , Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan Y., et al. , Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim B. O., et al. , Phage-derived antibacterials: Harnessing the simplicity, plasticity, and diversity of phages. Viruses 11, 268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.James E., et al. , Structural and mechanistic basis for the inhibition of Escherichia coli RNA polymerase by T7 Gp2. Mol. Cell 47, 755–766 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yano S. T., Rothman-Denes L. B., A phage-encoded inhibitor of Escherichia coli DNA replication targets the DNA polymerase clamp loader. Mol. Microbiol. 79, 1325–1338 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Tabib-Salazar A., et al. , Full shut-off of Escherichia coli RNA-polymerase by T7 phage requires a small phage-encoded DNA-binding protein. Nucleic Acids Res. 45, 7697–7707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhambhani A., et al. , Bacteriophage SP01 gene product 56 inhibits Bacillus subtilis cell division by interacting with FtsL and disrupting Pbp2B and FtsW recruitment. J. Bacteriol. 203, e00463-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muñoz-Espín D., Holguera I., Ballesteros-Plaza D., Carballido-López R., Salas M., Viral terminal protein directs early organization of phage DNA replication at the bacterial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 107, 16548–16553 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H. C., Wu M. L., Ko T. P., Wang A. H. J., Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein. Nucleic Acids Res. 41, 5127–5138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moyes R. B., Reynolds J., Breakwell D. P., Differential staining of bacteria: Gram stain. Curr. Protoc. Microbiol. Appendix 3, Appendix 3C (2009). [DOI] [PubMed] [Google Scholar]
- 52.Wang Z., et al. , Resonance assignments of bacteriophage SPO1 Gp49 protein. Biomol. NMR Assign. 14, 111–114 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.