Abstract
Recent evolutionary studies reveal that microorganisms including yeasts and fungi are more closely related to mammals than was previously appreciated. Possibly as a consequence, many natural-product toxins that have antimicrobial activity are also toxic to mammalian cells. While this makes it difficult to discover antifungal agents without toxic side effects, it also has enabled detailed studies of drug action in simple genetic model systems. We review here studies on the antifungal actions of antineoplasmic agents. Topics covered include the mechanisms of action of inhibitors of topoisomerases I and II; the immunosuppressants rapamycin, cyclosporin A, and FK506; the phosphatidylinositol 3-kinase inhibitor wortmannin; the angiogenesis inhibitors fumagillin and ovalicin; the HSP90 inhibitor geldanamycin; and agents that inhibit sphingolipid metabolism. In general, these natural products inhibit target proteins conserved from microorganisms to humans. These studies highlight the potential of microorganisms as screening tools to elucidate the mechanisms of action of novel pharmacological agents with unique effects against specific mammalian cell types, including neoplastic cells. In addition, this analysis suggests that antineoplastic agents and derivatives might find novel indications in the treatment of fungal infections, for which few agents are presently available, toxicity remains a serious concern, and drug resistance is emerging.
Much of the success of medicine over the past century in preventing and treating infectious diseases is directly attributable to the introduction of vaccines, improvements in sanitation and water quality, and antibiotics. However, compared to antibiotics for bacterial infections, advances in the treatment of established viral or fungal infections have been slower, in large part because the target cell is either an infected human cell or a eukaryotic cell similar in structure and function to mammalian cells. The problems in treating viral and fungal infections are in many ways similar to those faced in developing treatments for cancer. Moreover, the growing problem of drug resistance in fungal infections (25, 72, 179, 270, 339, 340) and in cancer chemotherapy are similar and in many cases involve overexpression of multidrug resistance pumps (5, 335). We review here the antifungal activities of antineoplastic agents and propose that existing and candidate chemotherapy agents, which have known potential to increase the risk of fungal infections, can paradoxically represent an excellent resource for the discovery of novel antifungal targets and agents.
Deep-seated, invasive mycoses have never been more commonly reported than over the last decade (6, 14). From the endemic mycoses and Cryptococcus neoformans to the human colonizing yeast Candida albicans and the ubiquitous mold Aspergillus, invasive mycoses have become a modern medical problem in the treatment of neoplasms (226, 326, 338). Two factors have produced a major contribution to this epidemic of fungal infections. First, the human immunodeficiency virus (HIV) pandemic enlarged the worldwide immunosuppressed populations and provided large populations of at-risk individuals for both neoplasms and these secondary fungal pathogens. Second, modern cancer chemotherapy with its known immunosuppressive qualities such as profound neutropenia and the prolongation of life with broad-spectrum antibiotics and a myriad of invasive catheters have added to the numbers of immunosuppressed or at-risk pool of patients.
With this background, we review in detail a major risk factor for fungal infections: antineoplastic agents. However, we also focus on the effects of antineoplastic agents as direct antifungal agents or their use in the development of antifungal targets. There have been no clinical studies which suggest that any of the antineoplastic agents presently used in clinical practice have a positive impact on either preventing or treating fungal infections. On the other hand, many of the targets for these drugs have homologs in the eukaryotic fungal pathogens, and a careful examination of these classes of drugs may lead to the development of novel selective antifungal drugs. An increase in the incidence of invasive mycoses has brought about a significant expansion in the use of antifungal drugs in clinical practice and a need for new, broad-spectrum fungicidal agents that can be used prophylactically, empirically, and therapeutically in these immunosuppressed patients (103, 104, 121, 262). None of the presently available antifungal agents completely fill the medical need, since in some respect they have weaknesses in spectrum, potency, safety, or pharmacokinetics.
In addition, the development of both antifungal and antineoplastic agents can benefit from studies in model and pathogenic yeasts. These fungi have been valuable tools in our efforts to understand the mechanisms of action of certain drugs and identify the targets of these drugs and explore their unique actions in mammals and fungi. Remarkably, studies with the yeast Saccharomyces cerevisiae as a model system have provided invaluable insights into the actions of a diverse array of drugs and compounds with quite specific activities in both mammals and fungi. For instance, cytotoxic topoisomerase I inhibitors (camptothecin, topothecan, and irinothecan), immunosuppressants that block T-lymphocyte function (cyclosporin A, FK506, and rapamycin), the phosphatidylinositol (PI) kinase inhibitor wortmannin, the HSP90 inhibitor geldanamycin, steroid receptor antagonists including tamoxifen, and the angiogenesis inhibitors fumagillin and TNP-470 have been studied mechanistically in yeast. Furthermore, recent advances in genome sequencing, genome arrays, combinatorial chemistry, and the development of a novel yeast three-hybrid assay promise to further extend the utility of yeast as a drug discovery tool, both in the identification of candidate antifungal and antineoplastic agents and in the elucidation of their mechanisms of action. For many years, yeast has been touted as an ideal model eukaryotic cell; these recent findings reveal that yeast is a better model for mammalian cell biology than we might have ever dreamed.
TOPOISOMERASES AS TARGETS OF ANTINEOPLASTIC AND ANTIFUNGAL AGENTS
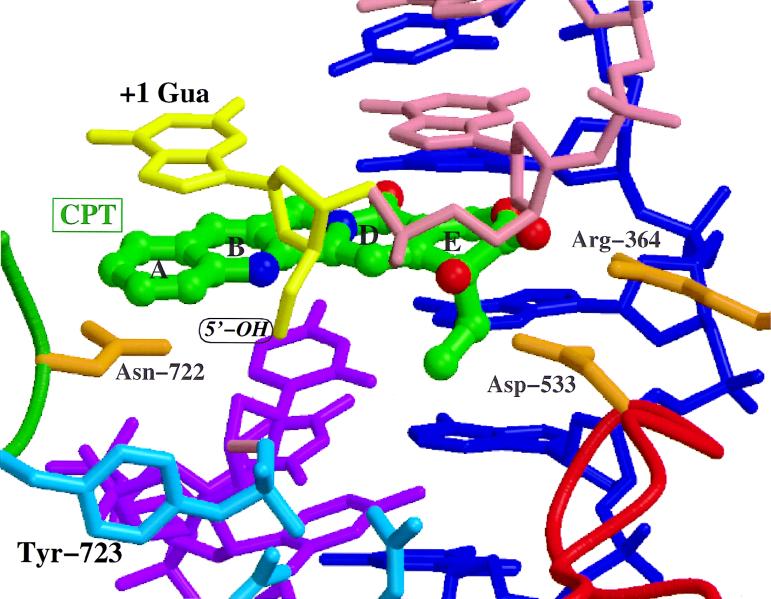
Topoisomerases are enzymes that control the topological state of DNA by introducing transient enzyme-bridged DNA breaks (single-strand DNA for type I and double-strand DNA for type II) that allow passage of DNA strands (330). Topoisomerase inhibitors stabilize the transient enzyme-DNA complexes, resulting in an inhibition of transcription and replication that ultimately leads to DNA damage and cell death (66, 195). Redinbo et al. recently solved the crystal structures of human topoisomerase I in both covalent and noncovalent complexes with DNA (279); a model for the interaction of the anticancer drug camptothecin with the human topoisomerase I-DNA covalent complex has been proposed (312) (Fig. 1).
FIG. 1.
Model of the human topoisomerase I-camptothecin interaction. A binding model of the three-dimensional structure of camptothecin (shown in green and labelled CPT) bound to the DNA-topoisomerase I complex is shown. Arg-364 and Asp-533 on the enzyme are hydrogen bonded to functional groups on the camptothecin E ring. A prominent feature of the model is that to accommodate camptothecin, the guanosine in the +1 position on the scissile DNA strand (labelled +1 Gua) is proposed to be flipped out of the DNA double helix. This could provide additional interactions, allowing the guanine ring to stack above the five-member ring system of the camptothecin molecule. This model will probably prove valuable in understanding the activity of camptothecin analogs and in developing novel analogs that specifically target human or fungal topoisomerase I enzymes. This figure is based on the X-ray structure of the DNA-topoisomerase I complex solved by Redinbo et al. (279) and was kindly provided by Matthew Redinbo and Wim Hol. It is important to note that this represents a binding model for the enzyme-drug-DNA complex and is not an experimentally determined binding mode for camptothecin association with the enzyme-DNA complex.
Significant progress has been made on the potential for molecular modeling and development of topoisomerase I-specific drugs. Furthermore, the importance of topoisomerase I as a drug target is emphasized by the development of two new antineoplastic agents, topotecan and irinotecan, which target topoisomerase I and are being used for antineoplastic chemotherapy against a variety of different neoplasms. The topoisomerase II inhibitor etoposide is also in clinical use as an antineoplastic agent. Several reviews have covered either type I or type II topoisomerase inhibitors as antitumor agents (18, 328, 329). The success of topoisomerase inhibitors in anticancer chemotherapy in humans underscores the potential of fungal topoisomerases as targets for development of novel antifungal compounds.
There has been substantial work on the structure and function of topoisomerase I and II in fungi. In the yeast S. cerevisiae, topoisomerase I and II serve separate functions important to cell transcription, replication, and recombination (111, 243). Topoisomerase II is essential for viability in S. cerevisiae (145), whereas topoisomerase I is not (111). Recent studies with the pathogenic basidiomycete Cryptococcus neoformans reveal that topoisomerase I is essential for viability (68). Previous studies with C. albicans (156) also suggest that topoisomerase I is essential in this common fungal pathogen. Because topoisomerase I is essential for viability in fungal pathogens, it is an ideal antifungal drug target.
Importantly, the fungal and mammalian topoisomerase I enzymes exhibit structural differences. The C. albicans and Cryptococcus neoformans topoisomerase I primary structures contain amino acid insertions not found in the mammalian enzyme (68, 317). These inserts are located in the linker domain of the fungal enzymes. For instance, in human topoisomerase I, the linker domain is 77 amino acids long, whereas in these two pathogenic fungi, the topoisomerase I linker domain consists of 155 amino acids. Redinbo et al. (279) have shown that human topoisomerase I is a multidomain enzyme that contains two conserved globular domains, the core and the COOH-terminal domains, which are essential for catalytic activity. Two other regions, the NH2-terminal and the linker domain, are not required for catalytic or relaxation activity (279). The functions of the fungal inserts are not yet known but could possibly be exploited in the development of fungus-specific topoisomerase I inhibitors.
The structure of human topoisomerase I is of significant medical importance because this enzyme is the unique target of the antineoplastic drug camptothecin and its derivatives topotecan and irinotecan. Previous studies demonstrated that C. albicans and human topoisomerase I are differentially inhibited by several compounds, including quinizarin, 5-HIAA (5-hydroxy-1H-indole-3-acetic acid), and A-3253 (96, 97). One issue that should be addressed is the ability of these topoisomerase I inhibitors to traverse the fungal cell membrane.
We have examined the in vitro activity of camptothecin and a series of camptothecin derivatives against the Cryptococcus neoformans topoisomerase I enzyme; several compounds are potent inhibitors of the fungal enzyme. While irinotecan and topotecan are not very potent inhibitors of the C. neoformans enzyme, some camptothecin derivatives are extremely active (67). Irinotecan and topotecan are 10-hydroxycamptothecin analogs. Topotecan has a dimethylaminomethyl hydroxychloride group substitution at the 9 position, whereas irinotecan has a carbamate ester at the 10-hydroxy group. Although these substitutions increase solubility, topotecan is a weaker topoisomerase I inhibitor than camptothecin whereas irinotecan is inactive against topoisomerase I in cancer cell lines. Similarly, both of these compounds are inactive against yeast cells (MIC > 500 μg/ml). Other analogs, such as the 10,11-methylenedioxy derivative and the corresponding glycinate esters, potentiate the topoisomerase I-inhibitory activity of camptothecin analogs both in cancer cell lines and in yeast cells. However, only the glycinate esters are active against yeast cells (MIC < 6.25 μg/ml). These findings support the notion that the addition of the glycinate ester increases the transport of the compound into the cell (67).
Topoisomerase II has been less extensively studied as an antifungal target. The antineoplastic topoisomerase II inhibitor etoposide has weak antifungal activity for Candida species, with MICs of 50 to 100 μg/ml (288). However, the fact that topoisomerase II is essential in yeast and fungi makes it a potentially important target for the development of future selective inhibitors and possibly new antifungal drugs.
DIRECT ANTIFUNGAL EFFECTS OF CYTOTOXIC AND ANTIMETABOLIC CHEMOTHERAPY AGENTS
The direct effects of standard cytotoxic and metabolic antineoplastic agents against fungi have been studied in vitro by several investigators. The MICs of methotrexate, cyclophosphamide, vincristine, bleomycin, doxorubicin, and mitomycin C for three yeast strains (C. albicans, C. tropicalis, C. kefyr) are between 500 and 1,500 μg/ml (108). In addition, bleomycin, carmustine, daunorubicin, doxorubicin, asparaginase, and 5-fluorouracil have MICs less than 100 μg/ml by in vitro susceptibility testing (261). In fact, these compounds were always more active against C. tropicalis than against C. glabrata and C. albicans, and both 5-fluorouracil and asparginase have MICs less than 1 μg/ml for C. tropicalis (288). However, bleomycin is toxic to yeasts in vitro but has no antifungal activity in an experimental murine candidiasis model (115).
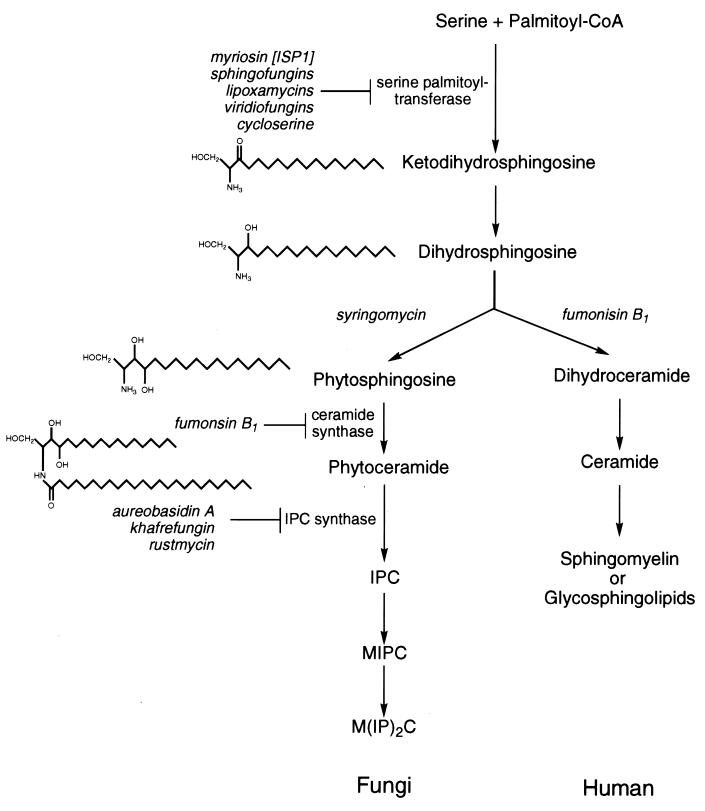
Direct antifungal activity of 5-fluorouracil against yeast is anticipated. Flucytosine is a known antifungal agent, in clinical use, which crosses the fungal cell wall and membrane and is converted by cytosine deaminase to 5-fluorouracil within the fungus. This cellular pyrimidine is then incorporated into fungal RNA in place of uracil, resulting in a block to protein synthesis (267). The ability to use the less toxic precursor of 5-fluorouracil and to target a unique fungal enzyme has allowed flucytosine to be developed as an antifungal agent. Drawbacks of flucytosine as an antifungal drug are the ease with which resistance develops when it is used as a single agent and the limited spectrum of antifungal activity; nonetheless, flucytosine demonstrates the potential to exploit differences in fungal and mammalian metabolism. Another pyrimidine target for fungi and cancer chemotherapy has been dihydroorotate dehydrogenase. A series of 4-phenoxyquinoline agricultural fungicides which target this enzyme in mitochondria have been developed (51, 103, 125).
Despite the success of agricultural and medical fungicides such as benomyl and griseofulvin on tubulin formation in fungi (152), the microtubule aggregation inhibited by vincristine and vinblastine (288) and the disaggregation of microtubules by paclitaxel appear to have little or no direct impact on fungal growth in vitro. In fact, paclitaxel antagonizes the activity of amphotericin B against fungi in vitro (230). On the other hand, depolymerization of microtubules by nocodazole produces a potent G2-M cell cycle arrest in S. cerevisiae. There appears to be a definite difference in potency or activity between antifungal and antineoplastic agents at this target, which is similar to differences in antitubule drugs in their activity against certain tumors and fungi. For instance, griseofulvin can inhibit dermatophytes both in vitro and in vivo but has no impact on growth or survival of pathogenic yeasts such as C. albicans.
Ornithine decarboxylase, the rate-limiting enzyme in polyamine biosynthesis, is a favorite target of anticancer chemotherapy (220). For instance, eflornithine, which is an irreversible suicide inhibitor of ornithine decarboxylase, can inhibit the growth of Cryptococcus neoformans (265).
Leptomycins A and B, produced by Streptomyces sp. strain ATS 1287, are metabolites with both antitumor and antifungal activity. Interestingly, leptomycin B has in vitro activity against the commonly drug-resistant zygomycetes such as Mucor spp. and Rhizopus spp. In Schizosaccharomyces pombe, leptomycin B inhibits cell division, producing elongated cells with morphologically altered nuclei and cell plates (128, 129, 175, 176). Leptomycin B inhibits the cell cycle in both the G1 and G2 phases in rat fibroblasts and S. pombe cells (357). Furthermore, investigators performed bioconversion screenings with several bacterial and fungal strains to reduce the cytotoxicity profile of leptomycin B. Although the antiproliferative effect could be reduced by bioconversion, none of the metabolites possessed greater activity than the parental leptomycin B compound (180).
The target of leptomycin B has recently been identified as the Crm1 protein, (242), which is a component of the protein nuclear export complex consisting of Crm1, RanGTP, and proteins containing a nuclear export signal (95, 102, 254, 352). As a consequence, leptomycin B has a variety of different effects on proteins that shuttle out of the nucleus. For example, Wolff and coworkers have shown that leptomycin B inhibits HIV-1 Rev protein translocation and therefore may be useful as a novel antiretroviral therapy (180, 344). In addition, nuclear export is required for the degradation of the p53 tumor suppressor protein by either Mdm2 or papillomavirus E6-mediated mechanisms; leptomycin B blocks the nuclear export and degradation of p53, and might therefore serve to stabilize this tumor suppressor protein in some neoplastic cell types (100). The nuclear export of several other proteins involved in neoplastic transformation, including steroid receptors and the abl tyrosine kinase, are similarly inhibited by leptomycin B (314, 320). Thus, further studies of leptomycin B as a novel inhibitor of nuclear export via its action on the conserved Crm1 protein are warranted, and leptomycin B may find indications as a novel antineoplastic or antimicrobial agent.
Glidobactins A, B, and C represent a complex of new antitumor antibiotics produced by Polyangium brachysporum. A variety of glidobactin analogs which exhibit even better in vitro anticancer and antifungal activity have been isolated (217). However, the therapeutic activity of the main component A of these analogs against systemic experimental fungal infections was found to be marginal (247).
STUDIES ON CISPLATIN COMPOUNDS IN MICROORGANISMS
Cisplatin is a DNA-damaging agent that causes DNA strand cross-links and which has revolutionized the treatment of some solid tumors, most prominently testicular carcinoma. The actions of cisplatin have been previously studied in the yeasts S. cerevisiae, Schizosaccharomyces pombe, and C. albicans. Cisplatin was originally studied as a potential antimicrobial drug (280) and, along with two related metal-complex agents (palladium), was found to be a particularly effective inhibitor of mammalian cell proliferation (80, 173, 345). Studies have shown that these compounds can inhibit C. albicans when used at concentrations between 100 and 180 μg/ml (234). However, these levels are higher than those which could be achieved for any significant period in an infected host. In C. albicans, pretreatment of cells with the antifungal agent amphotericin B or miconazole increases their sensitivity to cisplatin in vitro, suggesting that these compounds might have synergistic antifungal activity in patients being given concomitant treatment (286), but clinical studies have yet to confirm any positive relationship.
Cisplatin has been used in studies on drug mechanisms in yeast. Studies in S. cerevisiae and Schizosaccharomyces pombe have focused on the toxic effects of cisplatin in DNA repair mutants and the mechanisms of repair of cisplatin-induced DNA lesions. The SNM1 gene encodes a protein required for the repair of interstrand DNA cross-links induced by cisplatin and is transcriptionally induced in response to DNA damage by cisplatin or UV irradiation but not by methylating agents (346). Perego et al. examined the action of cisplatin and derivatives against a panel of 23 radiation-sensitive Schizosaccharomyces pombe rad mutants (260). Mutations in the RAD1 and RAD3 checkpoint genes and also in the RAD18 DNA repair gene conferred marked hypersensitivity (28- to 58-fold) to cisplatin, whereas mutations in 15 additional rad genes conferred more modest (5- to 20-fold) sensitivity. These findings suggest that human tumors with specific checkpoint or DNA repair defects might be uniquely sensitive to cisplatin in comparison to nonneoplastic cells and, similarly, that if compounds that inhibit such activities were available, these agents might sensitize additional cell types to cisplatin toxicity.
Recent studies have identified a novel high-mobility group (HMG) DNA binding protein, Ixr1, that plays a central role in cisplatin toxicity in S. cerevisiae. The Ixr1 protein was identified by its ability to bind with high affinity to intrastrand DNA cross-links produced by cisplatin (34). Mutants lacking Ixr1 are viable and modestly resistant to cisplatin (twofold) and accumulate only one-third as many cisplatin-DNA cross-links as an isogenic wild-type strain expressing Ixr1 does. Subsequent studies revealed that Ixr1 binds with 10-fold-higher affinity to DNA containing cisplatin adducts than to undamaged DNA and that the Ixr1-DNA complex covers approximately 15 bp of damaged DNA (219). These studies suggest that the Ixr1 protein, or other DNA binding proteins, plays a role in the cellular responses to cisplatin, and may either cause toxic DNA-adduct-protein lesions, block the repair of DNA cross-links by other repair enzymes, or assist in the recognition and repair of DNA cross-links. Thus, specific inhibitors of cisplatin adduct repair enzymes might find use as novel antineoplastic or antifungal agents.
EFFECTS OF DRUG COMBINATIONS (ANTINEOPLASTICS AND ANTIFUNGALS)
Ghannoum et al. (109) took the examination of antineoplastic and antifungal agents even further. For instance, they performed interactive studies on the combination of antifungal and antineoplastic agents. Several findings were noted: (i) inhibitory drug combinations whose MICs were lower than those of the respective agents alone were found; (ii) interactive effects between antineoplastic and antifungal agents may be very large; (iii) optimal combinations of drugs for yeast inhibition depend on both the relative and absolute concentrations of drugs in the mixture; and (iv) effective combinations were generally active against multiple fungal species, but the ratios of drugs may vary (109). In general, a polyene combined with an antineoplastic agent was the most potent combination against yeast growth. For example, the combination of amphotericin B and either methotrexate, mitomycin C, doxorubicin, or 5-fluorouracil had a positive interaction against yeast growth. This finding was partially explained by the membrane perturbation caused by amphotericin B, although it did not necessarily extend to another polyene, nystatin. While, as expected, 5-fluorouracil had the most positive antifungal interaction with amphotericin B, drugs such as cyclophosphamide and bleomycin antagonized the antifungal activity of the polyene (108).
The in vitro antifungal activity of camptothecin against yeast strains is greatly enhanced by mutations such as erg6 that cause defects in cell membranes (67, 244). Thus, the use of certain drugs in combination with others could alter the cell membrane and enhance the antineoplastic and antifungal activity of antitumor drugs. For example, the use of amphotericin B and cisplatin together results in an in vitro synergistic antineoplastic effect against both sensitive and resistant ovarian carcinoma cells and human colon cancer cells (10, 166). In combination assays, amphotericin B can potentiate the activity of a few antineoplastic agents against fungi (108), but the possible clinical relevance of this in vitro observation is not known.
RAPAMYCIN AND TORS IN THE YEAST, FUNGAL, AND MAMMALIAN CELL CYCLE
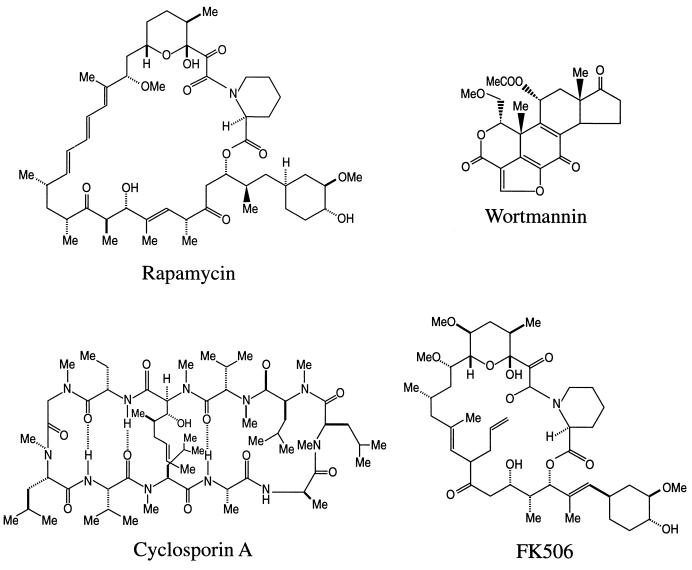
Rapamycin (Fig. 2) is a natural product with antimicrobial and immunosuppressant activities due to its ability to inhibit signal transduction cascades (reviewed in reference 44). Rapamycin is in late phase III clinical trials as an immunosuppressive drug for organ and bone marrow transplant recipients, and Food and Drug Administration approval is anticipated shortly. Rapamycin is also toxic to a number of tumor cell lines and thus may find additional indications as a novel chemotherapy agent. Finally, rapamycin is one of the most potent antifungal agents ever discovered, and nonimmunosuppressive analogs hold great promise as novel, potent, broad-spectrum antifungal agents (11, 324).
FIG. 2.
Structures of rapamycin, wortmannin, CsA, and FK506. The molecular structures of several natural products that inhibit signal transduction cascades are depicted. CsA is a cyclic peptide, rapamycin and FK506 are structurally related macrolide antibiotics, and wortmannin is related to the steroids.
In both yeast and mammalian cells, the action of rapamycin is mediated by its association with a highly conserved binding protein, the peptidylprolyl isomerase FKBP12 (140, 174, 301). Remarkably, yeast mutants lacking FKBP12 are viable and resistant to rapamycin toxicity, indicating that both the protein and the drug are required for rapamycin action and providing strong support for a model in which the FKBP12-rapamycin complex is the active in vivo agent. Overexpression of FKBP12 in mammalian cells increases sensitivity to rapamycin, and cell lines with reduced levels of FKBP12 are rapamycin resistant, providing evidence that FKBP12 is the conserved target of rapamycin action in yeast and mammals (28, 101).
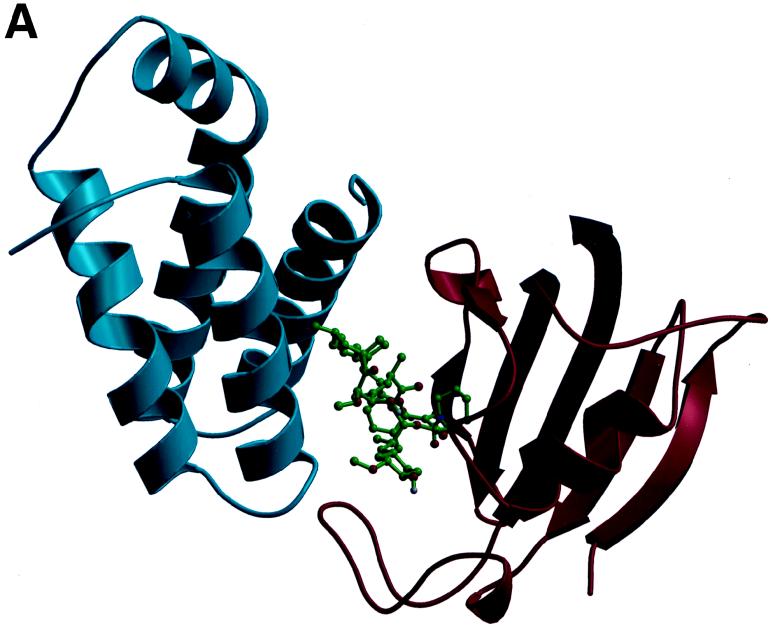
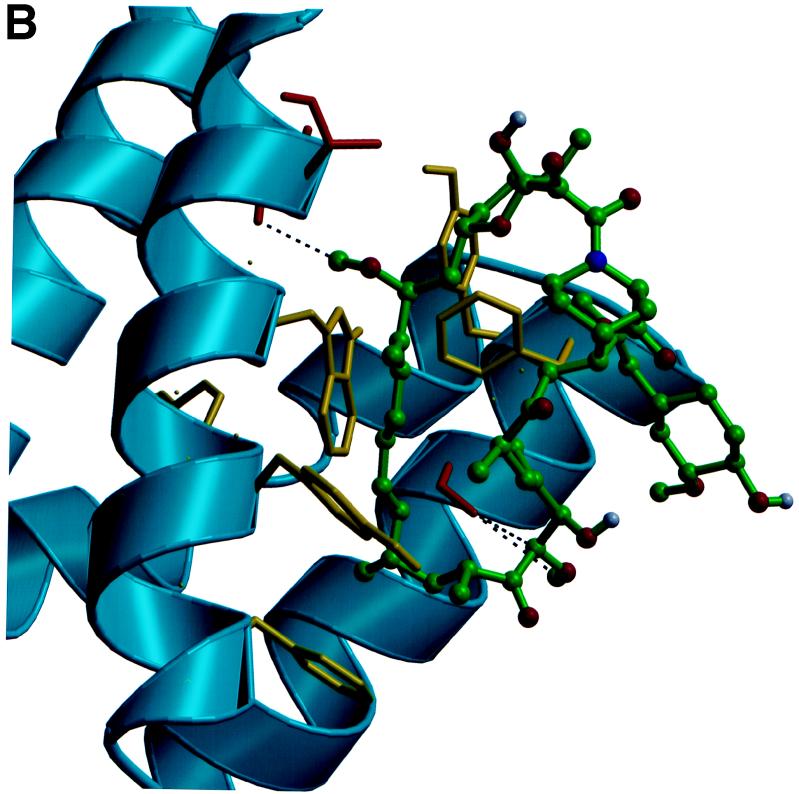
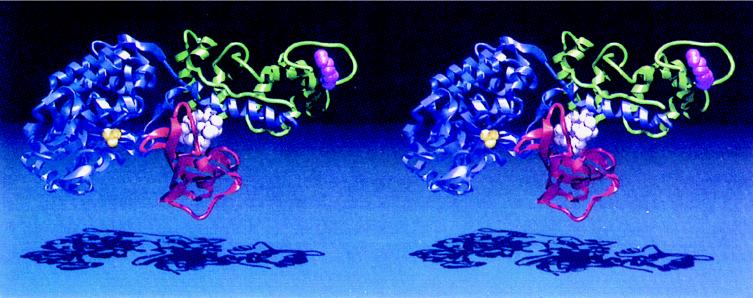
The targets of the toxic FKBP12-rapamycin complex are novel kinase homologs, the TOR1 and TOR2 proteins in yeast and the mTOR protein in mammals, which are conserved from yeast to humans. Genetic studies in yeast, in which rapamycin-resistant mutants were isolated, first implicated the TOR1 and TOR2 gene products as targets of the FKBP12-rapamycin complex (140). Analysis of the cloned TOR1 and TOR2 genes revealed the potential to encode large (∼280-kDa) proteins that have 67% overall identity (40, 141, 183). However, despite this remarkable similarity, TOR1 and TOR2 serve both shared and distinct functions. Deletion of the TOR1 gene confers only a modest growth defect under most conditions, whereas deletion of TOR2 is lethal. Subsequently, a mammalian TOR homolog (mTOR/FRAP/RAFT1/SEP/RAPT1) was identified by its ability to physically interact with FKBP12-rapamycin and was found to have ∼50% identity to the yeast TOR proteins (31, 52, 281, 282). The highest level of identity between the yeast and the mammalian TOR proteins is in the carboxy-terminal domain, which exhibits sequence identity to both lipid and protein kinases. The X-ray structure of FKBP12-rapamycin bound to a small portion of the mTOR protein (the FRB domain) has been solved (Fig. 3) (53). This structure revealed that rapamycin docks into a hydrophobic pocket on the surface of the TOR protein that has been highly conserved from yeast to humans. Few protein-protein contacts are apparent in the X-ray structure, but they should exist and play an important role in the complex, because rapamycin does not bind to the mTOR protein alone and thus FKBP12-TOR contacts should contribute to the ternary complex. Genetic studies had identified three residues that play a critical role in FKBP12-rapamycin binding to the yeast TOR1 and TOR2 proteins (205); these three residues, Ser1975, Trp2042, and Phe2049, are all conserved in the mammalian TOR protein and form the base and sides of the hydrophobic rapamycin binding pocket on mTOR (Fig. 3) (53).
FIG. 3.
Structures of the FKBP12-rapamycin-mTOR complex. (A) Structure of the ternary complex between the FKBP12-rapamycin binding domain on the FRAP/mTOR molecule (blue), rapamycin (green), and FKBP12 (red). (B) Detailed view of the FRB domain of FRAP/mTOR (blue) bound to rapamycin (green). Rapamycin binds into a hydrophobic pocket on the surface of TOR composed of the highly conserved residues Phe2108, Tyr2105, Trp2101, Phe2039, and Tyr2038 (side chains all in gold). These residues are conserved from yeast to humans and provide critical interactions with the inhibitor. This figure is based on the X-ray crystal structure of the FRB-rapamycin-FKBP12 complex solved by Choi et al. (54) and was kindly provided by Jon Clardy.
The TOR proteins are members of a ubiquitous family of signalling proteins, which includes the yeast cell cycle checkpoint and DNA repair proteins TEL1, MEC1, and RAD3 and the lipid kinases PIK1 and STT4, the Drosophila MEI41 protein, and the mammalian ataxia-telangiectasia (AT) protein ATM and its related protein ATR, the catalytic subunit of DNA-dependent protein kinase, and several phosphatidylinositol 3-kinases (PI3-kinases), PI4-kinases (for reviews, see references 162 and 359). In general, these proteins play conserved roles in transducing signals, either in response to exogenous stimuli such as growth factors or in response to endogenous cellular signals such as DNA damage and telomere length.
Rapamycin is also toxic to a number of pathogenic yeasts and fungi, including C. albicans, Cryptococcus neoformans, and Aspergillus fumigatus (59, 249, 347). In fact, rapamycin was originally discovered at Wyeth-Ayerst Pharmaceutical company in a screen for novel antifungal agents directed at C. albicans (304), and only much later was it appreciated to have potent and specific immunosuppressive activity, which is responsible for renewed interest in this compound. The FKBP12 homolog RBP1 has been identified and cloned in C. albicans, but studies demonstrating that this protein mediates rapamycin action in C. albicans have not yet been reported (89). Similarly, sequences derived from a TOR homolog are available in the systematic C. albicans genome-sequencing project, but no molecular studies have been published.
Rapamycin is also toxic to the opportunistic fungal pathogen Cryptococcus neoformans (249), and recently TOR1 and TOR2 homologs were identified and cloned based on homology to degenerate PCR (TOR1) and random sequencing of an expressed sequence tag (EST) database (TOR2) (59). Moreover, a C. neoformans FKBP12 homolog was cloned based on its ability to interact with the TOR1 FKBP12-rapamycin binding domain in a novel two-hybrid screen (59). Importantly, disruption of the FKBP12 encoding gene FRR1 by homologous recombination revealed that in C. neoformans, as in S. cerevisiae, mutants lacking FKBP12 are fully viable and resistant to rapamycin and to FK506. Spontaneous rapamycin-FK506-resistant mutants were also found to harbor FKBP12 mutations that prevent protein expression (59). Finally, a spontaneous rapamycin-resistant mutant was shown to have a mutation in the conserved serine residue in the FKBP12-rapamycin binding domain of TOR1 that is required for drug action in S. cerevisiae and mammalian cells (59). Taken together, these studies reveal that the antifungal activity of rapamycin is mediated via conserved complexes with FKBP12 and TOR homologs in C. neoformans. Furthermore, these studies suggest that nonimmunosuppressive rapamycin analogs have potential as novel antifungal agents.
One function of the TOR proteins that is shared by both S. cerevisiae TOR proteins and their mammalian homolog mTOR/FRAP/RAFT1 is required for signalling translational initiation and thereby cell cycle progression from G0 or G1 to S phase (13, 17). The second essential function of the yeast TOR2 protein is the control, via the RHO1 and RHO2 GTPases, of polarized distribution of the actin cytoskeleton during cell cycle progression (293, 294); however, it is not yet known whether this function is conserved in mammalian cells. The precise roles of the TOR proteins in either function are not yet well understood, and both the relevant substrates for TOR kinase activity and the upstream regulatory elements in these pathways are just beginning to be identified.
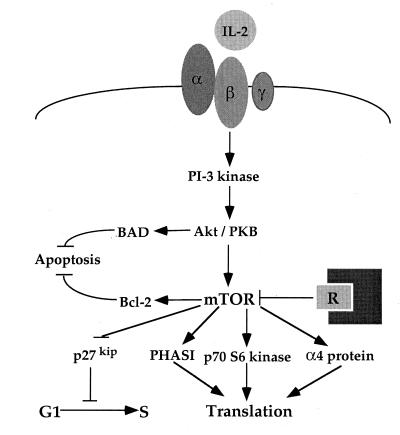
Genetic studies reveal that integrity of the TOR kinase domain is essential for TOR in vivo function in yeast and mammalian cells. Thus, TOR1 and TOR2 kinase-inactive mutants fail to complement tor1 or tor2 mutations in yeast (43, 360), overexpression of TOR kinase-inactive mutants is toxic in yeast (4, 360), and TOR kinase-inactive mutants are unable to function in a mammalian cell transfection assay involving TOR-dependent, rapamycin-sensitive activation of the p70 S6 kinase (32). The mammalian TOR protein (mTOR) is capable of autophosphorylation on serine residues, and this activity depends on the integrity of the kinase domain and is inhibited by both FKBP12-rapamycin and wortmannin (32, 36). Moreover, studies in which yeast TOR-mTOR chimeric proteins were expressed in wild-type or tor mutant S. cerevisiae strains revealed that the function of the kinase domain has been conserved between yeast and humans and that the mTOR kinase domain can regulate rapamycin-sensitive cell cycle progression in yeast cells (3). Earlier studies with rapamycin implicated the translational regulators p70 S6 kinase and PHAS-I as components functioning downstream of TOR (196, 327). Recently, mTOR was shown to phosphorylate PHAS-I and thereby mediate its dissociation from eukaryotic initiation factor 4E (eIF-4E) (35, 39) (Fig. 4). Dissociation of PHAS-I from eIF-4E is a crucial step toward activating translational initiation of certain mRNAs (reviewed in reference 188).
FIG. 4.
Rapamycin mechanism of action. Rapamycin inhibits a conserved signaling cascade that drives cell proliferation in response to interleukin-2 (IL-2) and other cytokines. The target of rapamycin kinase (TOR) functions to regulate the activities of the translational regulators PHAS-I and p70 S6 kinase. Signaling via the Tor cascade provides a proliferative signal that can in part prevent apoptosis. As a consequence, rapamycin promotes apoptosis under certain conditions.
Phosphorylation and activation of p70 S6 kinase is mitogen regulated and rapamycin sensitive (327). Interestingly, phosphorylation of p70 S6 kinase by mTOR in vitro at residues that are phosphorylated and rapamycin sensitive in vivo has been observed (39). Collectively, these observations support the view that in mammalian cells, TOR control of translational initiation occurs via phosphorylation of PHAS-I and p70 S6 kinase (Fig. 4). Although TOR action in control of translational initiation appears very similar in yeast and mammalian cells, the molecular TOR effectors do not seem to be conserved. No obvious p70 S6 kinase homologue is apparent in the completed yeast genome sequence. We have detected a robust kinase activity intrinsic to the yeast TOR1 protein by using PHAS-I as the substrate (4). Similar to the mTOR kinase, TOR1 protein kinase activity is blocked by active-site mutations and is inhibited by FKBP12-rapamycin and wortmannin (4).
Another possible target of the TOR kinase signaling cascade is the phosphatase regulator TAP42 in S. cerevisiae and its homolog, the α4 protein in mammals (Fig. 4). Overexpression of TAP42 suppresses mutations in the protein phosphatase 2A (PP2A) subunits SIT4, PPH21, and PPH22 (75). Interestingly, the TAP42 gene is essential, and studies of a conditional mutant reveal that TAP42, like TOR, is required for translation initiation (75). TAP42 forms a stable complex with SIT4 that is disrupted by either rapamycin or nutrient deprivation, and certain conditional alleles of TAP42 confer modest dominant rapamycin resistance in yeast, possibly by stabilizing the TAP42-PP2A complex initiation (75). Recent studies suggest that the TOR signaling pathway in yeast regulates the activity of a nutrient-regulated kinase, Npr1, that regulates the stability of nutrient permeases on the plasma membrane in response to nutrient availability, and that TAP42 may also participate in this regulatory cascade (292). Finally, the TAP42 homolog in mammalian cells, α4, is a known phosphoprotein that associates with PP2A, and the stability of the α4-PP2A complex is disrupted by rapamycin (150, 236). Further studies are required to define the role of TOR kinase activity in regulating PP2A signaling cascades that are conserved from S. cerevisiae to humans.
In both yeast and mammalian cells, the FKBP12-rapamycin complex inhibits G1-to-S cell cycle progression, but the mechanisms of cell cycle arrest are as yet unknown. In mammalian cells, rapamycin inhibits several cell cycle-linked events, including mitogen-induced activation of p70 S6 kinase (55, 184, 273), cyclin-dependent kinase activation, retinoblastoma protein phosphorylation (208), and downregulation of the cdk/cyclin inhibitor p27 kip1 (246) (Fig. 4). However, cells derived from mice in which the p27 gene has been disrupted by homologous recombination are only partially rapamycin resistant, indicating that rapamycin can inhibit cell cycle progression by p27-independent mechanisms (208). On the other hand, mutations in the yeast PP2A phosphatase subunit SIT4 cause a G1 cell cycle arrest similar to that due to rapamycin (88, 313), and this may result from inhibition of TOR-dependent TAP42-SIT4 activity by the FKBP12-rapamycin complex.
Growth factors such as cytokines and insulin trigger both mitogenic and antiapoptotic signals, providing a homeostatic balance between proliferation and cell death (276). In addition to its antiproliferative effects, rapamycin enhances apoptosis in mammalian cells in response to growth factor or serum deprivation, Fas activation (228), dexamethasone (151), and cisplatin (299), indicating that mTOR is a component of a signal transduction pathway dedicated to survival. One of the immediate events upon stimulation by growth factors, cytokines, or insulin is the activation of PI3-kinase (reviewed in reference 47). A prominent downstream target of PI3-kinase is the activation of the serine/threonine kinase Akt (also known as protein kinase B) (99). Recently, overexpression and activation of Akt was shown to rescue cells from Myc- or Fas-mediated apoptosis (136, 163). Among other responses, stimulation by interleukin-2 and epidermal growth factor results in the transcriptional induction of the transcription factors c-Fos, c-Jun, c-Myc, and the antiapoptotic protein bcl-2. Interestingly, rapamycin blocks the induction of bcl-2 but not of c-Fos, c-Jun, or c-Myc (231). Furthermore, evidence has been provided that mTOR kinase activation by insulin is mediated by the PI3-kinase Akt pathway (298). While there are multiple relevant targets for the survival signal of Akt, including direct phosphorylation and thereby inactivation of the apoptotic factor BAD, it is possible that the antiapoptotic effects of Akt are mediated at least in part by mTOR. Finally, rapamycin has recently been discovered to markedly inhibit the growth of several pediatric brain tumors, notably medulloblastomas; thus, rapamycin may find additional clinical use as an antitumour drug, possibly in combination with current chemotherapy regimens. Collectively, these observations suggest that the TOR proteins play important roles in cell cycle control and proliferation by pathways that have been conserved from yeast and fungal pathogens to humans.
WORTMANNIN AS AN INHIBITOR OF PI3, PI4, AND PI-RELATED KINASES
Wortmannin is a hydrophobic estrogen-related fungal metabolite of the fungus Talaromyces wortmanni (Fig. 2). The in vivo anti-inflammatory and immunosuppressive effects shown by wortmannin first suggested that it was a potent inhibitor of signal transduction pathways (reviewed in reference 321). Wortmannin blocks cellular responses emanating from stimulation of G-protein-coupled receptors. For example, wortmannin inhibits neutrophil activation by a variety of ligands (252) and histamine secretion by basophilic leukemia cells (354). In addition, wortmannin blocks insulin stimulation of glucose uptake in adipocytes (251).
Several lines of evidence suggested that the relevant molecular target of wortmannin in these effects is the lipid kinase that phosphorylates the 3 position of the phosphatidylinositol ring, PI3-kinase. Wortmannin blocks antigen- or insulin-dependent stimulation of PI3-kinase activity in rat basophils and adipocytes, respectively (251, 252). In mammalian cells, PI3-kinase is a heterodimer composed of a 110-kDa catalytic subunit and an 85-kDa regulatory subunit that interacts with other signal transduction elements via SH2 domains (160). The activity of PI3-kinase is potently inhibited in vivo and in vitro by wortmannin (251, 252, 311). Furthermore, by using anti-wortmannin antibodies and protease digestion, it was shown that wortmannin forms a covalent complex with an active-site residue of the 110-kDa PI3-kinase catalytic subunit, lysine 802 (351).
Although wortmannin potently inhibits the PI3-kinase with a 50% inhibitory concentration (IC50) of 5 nM, more recent studies have shown that it also inhibits PI4-kinases. A wortmannin-sensitive membrane-associated PI4-kinase was identified and cloned in mammalian cells (227, 239). Demethoxyviridin, a structural analog of wortmannin, inhibits an unidentified membrane-associated PI4-kinase from the fission yeast Schizosaccharomyces pombe (IC50 = 100 nM) (348). Interestingly, wortmannin is also toxic to the budding yeast S. cerevisiae (60). However, although wortmannin can inhibit the yeast PI3-kinase VPS34 in vitro at concentrations higher (IC50 = 3 μM) than those required to inhibit the mammalian PI3-kinase (308), mutant yeast cells lacking VPS34 are viable and remain wortmannin sensitive (60). This result suggests that wortmannin toxicity in yeast is mediated via another target. These observations led to the identification of a wortmannin target in yeast as the PI4-kinase STT4 (60). Thus, overexpression of STT4 in yeast rescues cells from wortmannin toxicity. Moreover, STT4 PI4-kinase activity in vitro is sensitive to 10 nM wortmannin. The second PI4-kinase in yeast, PIK1, is resistant to wortmannin inhibition (60).
The inhibitory activity of wortmannin is not restricted to PI3- and PI4-kinases, and at higher concentrations wortmannin also inhibits several members of a novel family of PI-related protein kinases. These wortmannin-sensitive enzymes include the mammalian target of rapamycin mTOR (IC50 ≈ 200 nM [36]) and yeast TOR1 (IC50 ≈ 100 to 200 nM [4]), and also DNA damage control proteins including the human DNA-dependent protein kinase (DNA-PK) (IC50 = 200 nM [a more recent study reported 16 nM] [287]), the ataxia-telangiectaxia (AT) mutated (ATM) protein (IC50 = 150 nM), and the ATM- and Rad3-related protein ATR (IC50 = 1.8 μM) (132, 287).
Unreplicated and damaged DNAs are detected by checkpoints, which in turn trigger a cascade of cellular events including cell cycle arrest, DNA repair, and cell death or apoptosis (162, 233). The ATM, ATR, and DNA-PK proteins are components of the molecular machinery that senses and responds to DNA damage. Because DNA-PK is activated by binding to DNA double-strand ends, this kinase may play a proximal sensing role in initiating the checkpoint that responds to DNA damage. Consistent with this hypothesis, murine Scid (severe combined immunodeficiency) cells and the mutant Chinese hamster ovary cell line V-3, which are radiosensitive and defective in double-strand DNA break repair and VDJ recombination, also lack the DNA-PK catalytic subunit, whereas the murine XRCC5 mutant cell lines lack the Ku-80 component of DNA-PK (20, 26, 107, 169, 264, 278, 315). Moreover, the radiosensitive human cell line MO59J, which is defective in DNA double-strand break repair, also lacks the DNA-PK catalytic subunit (190). Interestingly, the human XRCC7 gene, to which the DNA-PK catalytic subunit maps, complements the defects in DNA double-strand break repair and VDJ recombination in murine Scid cell lines (305). Substrates and possible effectors of DNA-PK include transcription factors such as the tumor suppressor p53, SP1, Fos, Jun, Myc, serum response factor, and the C-terminal domain of RNA polymerase II (reviewed in reference 7).
The clinical disorders displayed by AT patients, who carry a mutated ATM gene, underscore the crucial role of DNA damage checkpoints in the maintenance of genomic stability. AT individuals suffer a progressive cerebellar degeneration, immunodeficiency, radiation hypersensitivity, a higher incidence of certain forms of cancer, premature aging, and death during the second or third decade of life (38). Cells bearing a mutated ATM gene are extremely sensitive to ionizing radiation (158) and are defective in the G1, S, and G2 cell cycle checkpoints as well as in the p53 DNA damage response pathway (161, 165).
Wortmannin radiosensitizes CHO-K1 cells at concentrations of 1 to 20 μM, which are not cytotoxic (27). Moreover, human tumor cell lines are sensitive to wortmannin treatment. The most wortmannin-sensitive cell lines are GC3 colon carcinoma, IGROV1 ovarian carcinoma, and CCRF-CEM leukemia cells, for which the IC50 is in the range of 0.3 to 2.1 μM (297). Other cancer cell lines, particularly breast carcinomas that overexpress the Her/neu or epidermal growth factor receptor, were refractory to wortmannin treatment (IC50 > 50 μM). The degree of sensitivity to wortmannin shown by these tumor cell lines may be a reflection of the oncogenic mechanisms operating in these different forms of cancer as well as of the different affinities with which wortmannin inhibits these mechanisms. Thus, enhanced proliferation resulting from perturbations in the response to extracellular signals such as growth factors or cytokines in which the PI3-kinase plays a role are likely to be more susceptible to wortmannin inhibition than perturbations in the mechanisms emanating from DNA damage responses mediated by the DNA-PK, ATM, and ATR kinases.
The resistance of cancer cells to DNA-damaging agents such as radiation is largely mediated by DNA repair processes. The ability of wortmannin to inhibit key DNA repair enzymes makes it a potential candidate for use in combination with ionizing radiation. In accord with this prediction, recent studies found that wortmannin potently inhibits DNA double-strand break repair and sensitizes cells to ionizing radiation (27). Furthermore, the concentration of drug required to render cells radiosensitive, 10 to 20 μM for 10% survival, is in the range at which ATM and DNA-PK are inhibited by wortmannin (27, 287). In summary, the cytotoxic and radiosensitizing activities of wortmannin highlight its potential as an antineoplastic drug. However, the multiple molecular targets inhibited by wortmannin raise caution about its immediate use in human subjects. Thus, use of wortmannin as an antineoplastic or antifungal agent awaits the development of wortmannin derivatives with higher specificity for individual targets in both eukaryotic cell types. Crystallographic studies of wortmannin bound to its target, as well as more knowledge of the oncogenic mechanisms operating in different types of cancer and pathogenic fungal species, should prove useful in developing wortmannin-based strategies for anticancer and antifungal chemotherapy treatments.
CALCINEURIN IS THE CONSERVED TARGET OF CYCLOSPORIN A AND FK506
FK506 (tacrolimus) and cyclosporin A (CsA) are natural products of soil microorganisms that have potent immunosuppressive and antifungal activity (Fig. 2). CsA and FK506 were discovered to be potent inhibitors of mixed lymphocyte responses (24, 168) and have been widely used to treat and prevent graft rejection and graft-versus-host disease in solid-organ and bone marrow transplant recipients. Furthermore, both drugs recently have been shown to play a role in reversing multidrug resistance in several types of cancer by inhibiting the efflux of anticancer drugs. The mechanisms of action of these immunosuppressants have been extensively studied in the ascomycetous yeast S. cerevisiae as well as in T cells (for reviews, see references 33, 45, 46, 139, and 296). FK506 and CsA diffuse into the cell and bind intracellular receptors known as immunophilins, which catalyze cis-trans-peptidylprolyl isomerization, a rate-limiting step in protein folding (for reviews, see references 76, 139, and 291). FK506 binds to the immunophilin FKBP12, whereas CsA binds to the protein cyclophilin A. Although FK506 binds to the FKBP12 active site and inhibits prolyl isomerase activity, this is not the mechanism of immunosuppression in T cells or the toxic action in S. cerevisiae. Instead, the FKBP12-FK506 protein-drug complexes target proteins required for signal transduction and cell growth. The target of the FKBP12-FK506 and cyclophilin A-CsA complexes in S. cerevisiae and in T cells is calcineurin, a Ca2+-calmodulin-regulated serine/threonine-specific protein phosphatase (29, 94, 199).
Calcineurin, also known as PP2B, is a heterodimer protein composed of catalytic (CnA) and regulatory (CnB) subunits (172). The catalytic subunit has a molecular mass of 60 kDa and contains a calmodulin binding domain. The regulatory subunit is a 19-kDa member of the EF-hand family and contains four Ca2+ binding loops (2). Calcineurin activation requires both Ca2+ and calmodulin binding. Calcineurin has been characterized from several different organisms including S. cerevisiae (62, 63, 201, 356), C. neoformans (249), Dictyostelium (64), Drosophila (124), mouse (167), and human (122, 123, 235).
The X-ray crystal structures of the calcineurin AB holoenzyme and of the inhibited calcineurin AB-FK506-FKBP12 ternary complex have been solved (119, 171) (Fig. 5). These studies reveal that the FKBP12-FK506 complex binds to a hydrophobic groove that lies between the CnA catalytic subunit and the CnB regulatory subunit, making contacts with both subunits of the heterodimeric enzyme (Fig. 5). These observations are in accord with previous genetic and biochemical studies which had revealed the importance of both the CnA (42) and CnB (191, 229) subunits in inhibition of calcineurin by FKBP12-FK506 and cyclophilin-CsA (Fig. 5).
FIG. 5.
Structure of the FKBP12-FK506-calcineurin complex. The ternary complex between CnAB, FK506, and FKBP12 is depicted. Calcineurin is a heterodimer, composed of the CnA catalytic subunit (blue) and the CnB regulatory subunit (green). A molecule of phosphate bound in the calcineurin active site is shown in yellow, and the N-terminal myristoyl group on CnB is shown in purple. FKBP12 (red) bound to FK506 (grey) binds in a hydrophobic groove composed of an extended α-helical arm of CnA and regions of CnB. Note that the FKBP12-FK506 inhibitor complex does not bind in the active site of calcineurin but probably inhibits by occluding the docking of large protein substrates to the phosphatase. Modified from reference 119 with permission of the publisher.
In T cells, calcineurin is activated in response to an increase in the level of intracellular Ca2+ that occurs upon stimulation of the antigen receptor pathway (reviewed in reference 44). Calcineurin is known to mediate T-cell activation by dephosphorylation of the NF-AT transcription factor, which unmasks its nuclear localization signal and thus promotes NF-AT translocation into the nucleus, where it induces a number of genes in response to cytokine activation (154, 245). Other proposed functions of calcineurin in mammalian cells include roles in apoptosis (355), neutrophil migration (144, 189), and other pathways regulated by Ca2+, such as neuronal growth (50, 90; for a review, see reference 143).
Why are CsA and FK506 specific for calcineurin, and why are calcineurin inhibitors relatively specific for T cells in a complex organism? First, while the CnA catalytic subunit is homologous to other protein phosphatases, the CnB subunit is unique to calcineurin. Because FKBP12-FK506 and cyclophilin A-CsA binding to calcineurin is mediated in part by the CnB subunit, this renders these inhibitors highly specific for calcineurin and not for other related phosphatases. Second, FK506 and CsA are relatively specific for T cells because T cells have lower levels of calcineurin than do other tissues, and thus lower concentrations of inhibitor are required to block T-cell function. In addition, some isoforms of the calcineurin target NF-AT are relatively T-cell specific.
The mechanism of action of CsA and FK506 has been conserved from T cells to the yeast S. cerevisiae. In a similar way to T cells, CsA and FK506 bind to their respective yeast immunophilins and target the yeast homolog of calcineurin. Two genes encoding the catalytic and one the regulatory subunit have been characterized (62, 63, 182, 201, 356). Vegetative growth of the majority of S. cerevisiae strains is not sensitive to CsA or FK506. Even though calcineurin is not required for vegetative growth of yeast, it is required for yeast survival after prolonged exposure to pheromone (62, 63, 342). Furthermore, calcineurin mutation or inhibition confers hypersensitivity to Li+, Na+, Mn2+, and other ions (86, 222, 240, 271). Calcineurin mediates the regulation of ion homeostasis and cell wall synthesis by activating the transcription factor Crz1/Tcn1, which in turn regulates the transcription of several genes that encode ion pumps or cell wall biosynthetic enzymes (including FKS2, PMR2, PMC1, and PMR1) (218, 309). Finally, a variety of mutations that render calcineurin essential for viability have been identified, and these strains are sensitive to growth inhibition by CsA and FK506 (29, 142, 259, 318).
CALCINEURIN IS THE TARGET OF CYCLOSPORIN A AND FK506 IN PATHOGENIC AND OTHER FUNGI
The immunosuppressants FK506 and CsA are toxic to several fungi, including Aspergillus fumigatus (135), Cryptococcus neoformans (249), Neurospora crassa (318), and Coccidioides immitis (170). The action of CsA and FK506 in these organisms is mediated via conserved complexes with cyclophilin A and FKBP12 homologs. Calcineurin appears to be the conserved target, and its inhibition mediates the toxic activity of these compounds. The genes encoding calcineurin have been cloned from several fungi. In the opportunistic pathogen C. neoformans, calcineurin is essential for growth at 37°C, in 5% CO2, or at alkaline pH, conditions which mimic the human host environment. As a consequence, calcineurin is required for C. neoformans virulence in an animal model of cryptococcal meningitis (249). As in S. cerevisiae, calcineurin controls Na+ and Li+ homeostasis in C. neoformans. On the other hand, calcineurin plays a role in the regulation of cell cycle in the pathogenic fungus Aspergillus nidulans, where it is essential for cell cycle progression at the G1-to-S-phase transition (277). Furthermore, calcineurin is involved in hyphal growth, and the regulatory subunit is required for normal vegetative growth in the ascomycete Neurospora crassa (177, 275). Calcineurin mutants of the fission yeast Schizosaccharomyces pombe are viable but have defects in cytokinesis, cell polarity, mating, spindle body positioning, and growth at 22°C (358). In summary, calcineurin plays an important role in normal growth, morphology, mating, and virulence in fungi.
Even though CsA and FK506 are toxic to pathogenic fungi such as C. neoformans, their immunosuppressive activity outweighs their antifungal potential because immunosuppression exacerbates fungal infection in animal models (263) and humans. However, nonimmunosuppressive CsA and FK506 analogs that retain some level of antifungal activity have been identified (249, 250). One such analog is the C-18-hydroxy C-21-ethyl derivative of FK506 (L-685,818), which is toxic to C. neoformans as a result of inhibition of calcineurin. Thus, drug analogs can capitalize on subtle structural differences between host and fungal targets to spare host immune function yet impair fungal growth. Because CsA and FK506 have been approved for use in transplant recipients, the development and implementation of analogs with a novel indication should be more rapid, given clinical and toxicity experience and approval of the parent compounds.
CYCLOSPORIN A AND FK506 ACTION ON MULTIDRUG RESISTANCE MECHANISMS
Multidrug resistance (MDR) is a generalized phenomenon in which cells develop resistance to chemically dissimilar compounds. One of the best-characterized mechanisms of MDR involves overexpression of ATP binding cassette (ABC) transporters. These conserved proteins function as drug efflux pumps and have been implicated in MDR in bacteria (8, 105, 268), fungi (12, 285), helminths (30), and human cancer cells (110, 112, 148, 193). In human cancer cells, P-glycoprotein (Pgp) is the best characterized ABC transporter and is responsible for resistance to standard cancer chemotherapy regimens (for reviews, see references 16, 106, 113, and 197). Pgp is a 170-kDa membrane protein that consists of two similar parts, each with an ATP binding site and six membrane binding domains (159). In vitro, MDR mediated by Pgp can be reversed by the binding of several pharmacological compounds to Pgp, thereby blocking efflux of anticancer drugs. CsA and FK506 are known to bind to and inhibit mammalian Pgp (9, 69, 98, 153, 232, 269, 283). Furthermore, CsA has been used as an MDR reversal agent for acute leukemia and has shown promising preliminary results (198, 215).
Since the onset of the AIDS epidemic, the frequency of fungal infections has increased dramatically and the isolation of azole-resistant yeast strains has become more common (5, 335). Thus, there is an ongoing need for more effective antifungal therapies with reduced toxicity and reduced emergence of antifungal drug resistance. Antifungal drug resistance is mediated partially through an MDR mechanism involving several ABC transporters, including the CDR1 and CDR2 proteins characterized from C. albicans (272, 284, 285). CsA, FK506, or analogs may inhibit these fungal ABC transporters involved in antifungal resistance, since FK506 inhibits the S. cerevisiae ABC transporter PDR5 (82). In addition, two nonimmunosuppressive CsA analogs that are potent inhibitors of mammalian MDR seem to have similar effects on lower eukaryotes, such as Toxoplasma gondii and Plasmodium falciparum; however, the binding of these analogs to mammalian or parasite Pgp has not yet been demonstrated (15, 302).
ANGIOGENESIS INHIBITORS: THE FUMAGILLIN TARGET METHIONINE AMINOPEPTIDASE 2 IS CONSERVED FROM YEAST TO HUMANS
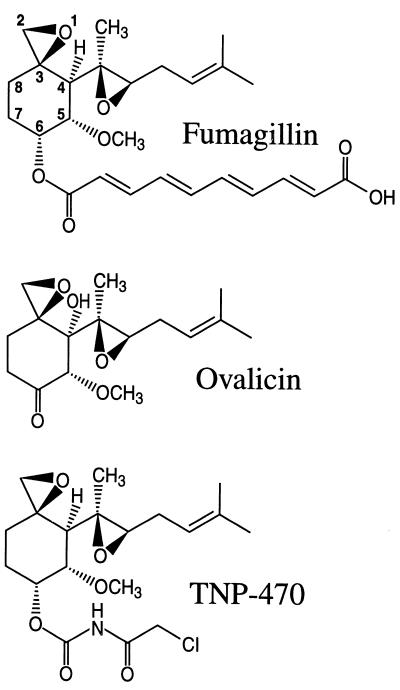
The development of clinically important tumors involves an initial neoplastic transformation of a mammalian cell and also requires the recruitment of new blood vessels (angiogenesis) to supply the tumor with oxygen and nutrients. Interestingly, angiogenesis occurs mainly at sites of new tissue growth, and tumors actively secrete growth factors that trigger angiogenesis. Over 20 years ago, Judah Folkman suggested that novel chemotherapy agents might target angiogenesis to restrain tumor growth (92, 93). Recently, several natural products have been shown to inhibit angiogenesis in in vitro culture and animal models, and they show promise in animal studies as chemotherapy agents (21, 149, 258). These include fumagillin, a natural product of the pathogenic fungus A. fumigatus, the fumagillin analog TNP-470 which is in clinical trials, and ovalicin (Fig. 6).
FIG. 6.
Structures of angiogenesis inhibitors. The structures of the natural-product angiogenesis inhibitors fumagillin and ovalicin and of the derivative TNP-470, currently in clinical trials, are depicted.
Although fumagillin potently inhibits angiogenesis, the target of fumagillin was only recently identified. Early studies revealed that fumagillin and its derivative TNP-470 inhibited vascularization and caused endothelial cells to arrest in the G1 phase of the cell cycle (1). Moreover, fumagillin inhibits early cell cycle events, including phosphorylation of the retinoblastoma protein, expression of cyclin E, and activity of the cyclin-dependent kinases cdk2 and cdk4. Recently, two groups took similar approaches to identify the target of fumagillin as a highly conserved enzyme, methionine aminopeptidase 2 (MetAP2) (117, 303). Both groups synthesized fumagillin affinity matrices or radiolabelled photo-cross-linking derivatives. These modified compounds were shown to retain the antiangiogenic activity of the parent fumagillin molecule against human vascular endothelial cells. Next, these affinity reagents were used to identify a 67-kDa fumagillin binding protein that was present in bovine brain or mouse embryo extracts. Binding to this 67-kDa protein was specific and was competed by unlabelled fumagillin. Amino acid sequence analysis by two approaches revealed sequences derived from a known protein, the enzyme MetAP2. Antisera to MetAP2 specifically recognized the 67-kDa fumagillin binding protein. Moreover, fumagillin derivatives and ovalicin bind covalently to MetAP2 and inhibit its activity with a tetrapeptide substrate in vitro. The abilities of a series of fumagillin and ovalicin analogs to inhibit MetAP2 enzymatic activity and endothelial cell proliferation were well correlated, providing additional evidence that MetAP2 is the relevant in vivo target (117). MetAP2 has a second activity, namely, the ability to inhibit phosphorylation of eIF-2α by the heme-regulated inhibitor kinase. This activity is not inhibited by fumagillin, indicating that it is the aminopeptidase activity of MetAP2 that is the target for inhibition by fumagillin (117). Finally, the immunosuppressive and antiangiogenic activities of fumagillin analogs are strongly correlated, which suggests a unique role of MetAP2 in lymphocytes (319).
Remarkably, genetic studies with S. cerevisiae demonstrated that the relevant in vivo target of fumagillin and ovalicin is MetAP2 (117, 303). In previous studies, two MetAP enzymes were identified in S. cerevisiae (192). The two enzymes are encoded by the MAP1 and MAP2 genes. Individual map1 or map2 mutant cells are viable, albeit slow growing, whereas map1 map2 double mutants are inviable, indicating that at least one isoform of the enzyme is essential for viability and that the two enzymes are at least partially redundant for an essential function (192). Most interestingly, MAP1 MAP2 wild-type yeast strains expressing both MetAP1 and MetAP2 are not at all sensitive to fumagillin whereas map1 mutants that lack the MetAP1 enzyme and express only the MetAP2 enzyme are sensitive to 50 nM fumagillin or ovalicin (117, 303). These studies demonstrate that fumagillin and ovalicin inhibit the MetAP2 enzyme in vivo and that MetAP2 is the relevant in vivo target for the cytotoxic effects of these angiogenesis inhibitors. Moreover, these genetic studies confirm that fumagillin and its derivatives do not inhibit the MetAP1 enzyme in vivo, because wild-type cells expressing both MetAP1 and MetAP2, or the map2 mutant cells expressing only MetAP1, were resistant to fumagillin. While the target(s) of MetAP2 in yeast or mammalian cells required for viability is not yet known, these studies provide a genetic approach to define important cellular targets of MetAP2 in vivo.
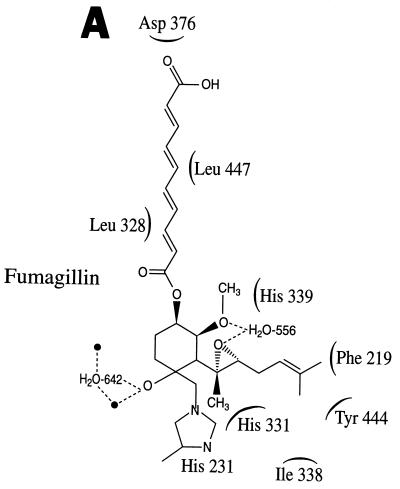
Fumagillin binds covalently to the MetAP2 enzyme, and recent biochemical studies and the X-ray crystal structure of the fumagillin-MetAP2 complex (Fig. 7) reveal that the ring epoxide of fumagillin becomes covalently cross-linked to an active-site histidine residue (118, 200, 207). Lowther et al. found that fumagillin covalently modifies a conserved active-site histidine residue in the distantly related Escherichia coli MetAP enzyme (207). Recent studies revealed that His231 in the human MetAP2 enzyme is covalently modified by reaction with the ring epoxide of fumagillin (118). Finally, the X-ray structure of the fumagillin-human MetAP2 complex has recently been solved at 1.8-Å resolution (200). This structure reveals that the inhibitor is covalently linked to His231 in the active site and via a rich network of interactions with the ring portion of fumagillin but has essentially no contacts to the extended chain of the inhibitor (Fig. 7). Modeling provides insights into the specificity of fumagillin as an inhibitor of MetAP2 but not of the closely related MetAP1 enzyme, in which the target active-site histidine residue is not close enough to covalently cross-link to the drug and in which the drug binding pocket is narrower than in MetAP2 (200). These studies provide a structural basis for further design of MetAP2 inhibitors with antiangiogenic and immunosuppressive activities.
FIG. 7.
Structure of the fumagillin-MetAP2 complex. (A) Schematic view of fumagillin bound to the active site of MetAP2. (B) Structure of the human MetAP2 enzyme (blue and red) and the bound molecule of fumagillin (gold). Notably, the active-site residue His231 is covalently linked to a reactive ring epoxide on fumagillin. This figure is based on the structure of the aminopeptidase-fumagillin complex solved by Liu et al. (200) and was generously provided by Maria C. Nonato and Jon Clardy.
In previous sections we described the use of yeast as a model system to analyze the functions of drugs that specifically target T lymphocytes of the mammalian immune system. Now these studies reveal that yeast can also be used to identify and study the cellular targets of inhibitors of angiogenesis, a process that is highly specific to mammalian systems but is also linked to pathogenic fungi through an understanding in yeast.
What is the function of the MetAP2 enzyme in vascular endothelial cell proliferation and angiogenesis? While the specific function of MetAP2 required for angiogenesis has not yet been elucidated, there are several very interesting possible models. First, the primary function of this enzyme is to remove the initiator methionine at the amino terminus of proteins. This posttranslational processing event is required for myristoylation, a lipid modification to glycine residues that functions in protein targeting and stability. Proteins that are known to be N myristoylated include the catalytic subunit of cyclic AMP-dependent protein kinase, the Gα subunits of heterotrimeric G proteins, the regulatory B subunit of the protein phosphatase calcineurin, oncogenic protein kinases such as src, and the ADP-ribosylation factor that regulates protein synthesis. N-myristoyltransferase is known to be essential in S. cerevisiae and Cryptococcus neoformans (203), and differences between the substrate specificity of the C. neoformans NMT1 enzyme and the mammalian enzyme have been demonstrated, which could be exploited in the development of novel antifungal agents. Another function of N-terminal processing is to reveal the adjacent amino acid which then regulates protein stability by the N-end rule, in which following cleavage of the N-terminal methionine, the now penultimate amino acid residue regulates protein stability (323). Inhibition of MetAP2 would be predicted to stabilize proteins that are normally short-lived because of N-end rule-mediated degradation, and this might perturb endothelial-cell proliferation and angiogenesis. Third, it is possible that one or more target proteins are nonfunctional when the amino-terminal methionine is simply not removed by MetAP1 or MetAP2. Finally, it is worth noting that the tissue distributions of MetAP1 and MetAP2 have not yet been determined, and endothelial cells might have relatively low levels of MetAP2 compared to other tissues, rendering them more uniquely susceptible to inhibition by fumagillin. Similar arguments have been proposed to explain, in part, the marked specificity of CsA and FK506 for T cells that have relatively lower levels of their common target protein, calcineurin (see above).
Fumagillin does not exhibit broad-spectrum antimicrobial activity, possibly because many organisms express both the MetAP2 fumagillin-sensitive target and the fumagillin-resistant MetAP1 enzyme. However, one early report revealed that fumagillin is toxic to a fungal disease of honeybees that is due to Nosema (164). More recently, the fumagillin analog TNP-470, which is in clinical trials as an angiogenesis inhibitor, has been shown to have potent activity against microsporidia, which are obligate intracellular parasites that cause diarrhea and wasting syndrome in AIDS and other immunocompromised patients (58). These findings suggest that further analysis of fumagillin and its analogs as novel antimicrobial agents, possibly with a highly selective spectrum of action, would be warranted.
HSP90-DEPENDENT STEROID RECEPTORS AND ONCOGENIC KINASES STUDIED IN YEAST: MECHANISMS OF ACTION OF GELDANAMYCIN AND ESTROGEN RECEPTOR ANTAGONISTS
The chaperone complex associated with the 90-kDa heat shock protein (HSP90) serves a dual function in the cell. First, HSP90 and associated partner proteins interact with heat-damaged proteins following heat shock and either promote refolding by inhibiting aggregation or target severely damaged proteins for degradation (295). Second, under normal cellular conditions, the HSP90 chaperone complex interacts with several proteins that require HSP90 for activity, including steroid receptors and oncogenic kinases such as src. For example, HSP90 binding maintains steroid receptors in a state competent to bind hormone. The HSP90 complexes consist of several proteins, including HSP70, p60, p48, p23, and a large immunophilin, which can be either cyclophilin 40, FKBP52, or FKBP54. Remarkably, the HSP90 chaperone complex has been conserved from the yeast S. cerevisiae to humans, and thus yeast genetic approaches can be used to elucidate the functions of the HSP90 chaperone complex with target proteins. Importantly, the HSP90-target complex is targeted by several different drugs with antineoplastic or antifungal activity, including geldanamycin (HSP90), CsA (cyclophilin 40), FK506 and rapamycin (FKBP52/54), and steroid receptor agonists and antagonists such as tamoxifen (estrogen receptor). For recent reviews of HSP90 functions and structure, see references 41 and 289.
Previous studies have focused on the heterologous expression of mammalian steroid receptors and the oncogenic kinase src in S. cerevisiae (49, 91, 209, 225, 266). Such studies have demonstrated that a variety of different steroid receptors can be expressed in a functional form and that ligand binding results in transcriptional activation of corresponding target reporter genes. These systems provide a powerful assay of steroid receptor function. Mutations that reduce the cellular levels of Hsp90 compromise steroid receptor function, providing direct evidence that Hsp90 is required for steroid receptor function (266). Moreover, Hsp90-associated proteins, including Hsp70, p60/Sti1, and cyclophilin 40, are conserved in yeast (48, 78, 241) and are required both for steroid receptor function and for the stability of oncogenic kinases (49, 79).
Recent studies revealed that the novel antiproliferative agent geldanamycin inhibits HSP90. Geldanamycin is a benzoquinone ansamycin natural product of Streptomyces hygroscopicus subsp. geldanus that inhibits the proliferation of a wide variety of tumor cell lines. Initial findings revealed that geldanamycin inhibits the activity of several different protein kinases, but subsequent studies revealed this to be an indirect effect of HSP90 inhibition (336). The X-ray crystal structure of an HSP90-geldanamycin complex reveals that the drug binds in the N-terminal ATP binding site that has recently been defined on HSP90 (Fig. 8) (116, 274, 310). Geldanamycin binding inhibits HSP90-dependent ATPase activity (256). Geldanamycin also prevents binding of the p23 cochaperone to HSP90 and inhibits the release of substrates (22, 85). Yeast mutants lacking the yeast p23 homolog SBA1 are viable, and steroid receptor signaling is still sensitive to geldanamycin in sba1 mutant cells, indicating that inhibition of p23 binding is not the only mechanism by which geldanamycin inhibits HSP90 functions (22, 85). Yeast mutants lacking the cyclophilin 40 homolog CPR7 are uniquely sensitive to reductions in HSP90 levels (79). We recently discovered that yeast cpr7 mutants are hypersensitive to growth inhibition by geldanamycin (78). These findings suggest that further studies with yeast will provide a facile route to analyze inhibition of HSP90 by geldanamycin in vivo.
FIG. 8.
The HSP90 chaperone complex is targeted by geldanamycin. (A) Schematic view of geldanamycin bound in the ATPase active site of HSP90. (B) Stereoview of the geldanamycin-HSP90 active-site complex, with HSP90 shown in blue, side chains shown in purple, and geldanamycin shown in white and yellow. Interactions between geldanamycin and HSP90 are indicated by green dotted lines. (C) Space-filling model of the complex between geldanamycin, which snugly docks into the ATPase binding site on the surface of HSP90 (blue). Modified from reference 310 with permission of the publisher and kindly provided by Nikola Pavletich.
The use of the estrogen receptor antagonists tamoxifen and raloxifene in breast cancer treatment and prevention has generated considerable enthusiasm (253). Further studies of their novel antagonist and partial agonist actions on the estrogen receptors in different tissues are of paramount importance. A central question in the field is how a single agent, such as tamoxifen, can have beneficial antiestrogenic activities in the breast yet undesired estrogenic action in the endometrium. On the other hand, raloxifene exhibits antiestrogenic action in both tissues. There are several possible explanations. First, a second estrogen receptor has been recently discovered (181), and differing actions on the α and β forms of the estrogen receptor could provide an explanation for these disparate actions (255). Second, the estrogen receptors are associated with different adapter proteins in different tissues, or at different estrogen response elements in the genome (353), and these accessory proteins or sites of action could be responsible for differences in the actions of estrogen receptor ligands.
Studies with yeast provide an important model for steroid receptor functions and interactions with antagonists such as antiestrogens. For example, a variety of different steroid hormone receptors, including the α form of the estrogen receptor, have been reconstituted in yeast cells (221). These types of yeast model systems have been used to (i) define atypical estrogen response elements from the genome (65), (ii) study the actions of agonists and antagonists of the receptor (300), (iii) analyze the structure and function of the hormone binding domain of the human estrogen receptor (84, 349), (iv) demonstrate that the antiestrogen agent ICI 164,384 has no estrogen agonist activity with the α estrogen receptor expressed in yeast (19), and (v) isolate and analyze the functional status of the wild-type and mutant human estrogen receptors present in breast carcinoma cells, revealing that receptor variants with altered activity may be responsible for resistance to tamoxifen (322). These studies underscore the potential of yeast as a model system for further study of novel estrogen receptor ligands and promise to revolutionize the treatment and prevention of breast cancer with reduced undesirable side effects in other tissues. Finally, it is worth noting that with prolonged therapy, many breast carcinoma cells either develop resistance to or become dependent upon tamoxifen for proliferation. Additional antagonists, such as raloxifene, may be particularly useful in treating tamoxifen-resistant or -dependent tumors.
Studies have also examined the possible effects of FK506 on the activity of steroid receptors. In this case, the power of yeast as a model system is revealed by studies that exclude, rather than demonstrate, interesting models of drug action. Previous studies revealed that FK506 could potentiate gene activation by the progesterone receptor expressed in S. cerevisiae (316). An FK506 analog that poorly inhibits calcineurin (15-O-desmethyl-FK520) failed to enhance progesterone signaling in yeast, leading the authors to suggest that calcineurin might be required to dephosphorylate and activate the progesterone receptor (316). However, FK506 continued to enhance signaling even in yeast mutant strains lacking FKBP12 (316), in which FK506 has no effects on calcineurin activity or function (29). The authors also suggested that FK506 might potentiate progesterone signaling by acting on a yeast homolog of the HSP90-associated FKBP52 or FKBP54 protein (316), but now that the yeast genome has been sequenced, it is clear that there are no FKBP52 or FKBP54 homologs in yeast. What, then, is the target for FK506 action in this system? Studies by Yamamoto and colleagues have identified an ABC transporter MDR homolog in yeast, LEM1, which pumps steroid hormones out of the cell. FK506 is known to inhibit mammalian MDR pumps and also turns out to inhibit the yeast LEM1 steroid pump (178). Hence, FK506 effectively increases the intracellular concentrations of exogenously added steroids by inhibiting steroid export by LEM1. Thus, the effects of FK506 on progesterone receptor function in yeast are indirect and result from a relative increase in intracellular steroid levels, an effect mediated by LEM1 and not by calcineurin, FKBP12, or an FKBP52-FKBP54 homolog.
N-MYRISTOYLTRANSFERASE: NOVEL TARGET FOR CHEMOTHERAPEUTIC AND ANTIFUNGAL DRUGS
The enzyme N-myristoyltransferase (NMT) modifies proteins at the N-terminal end with lipophilic myristic acid groups, a modification that is involved in both membrane targeting and protein stability. The enzyme is conserved from microorganisms to humans and is known to be essential for viability in S. cerevisiae, C. albicans, and Cryptococcus neoformans (81, 203). Several oncoproteins are known to be myristoylated in mammalian cells, including the src and yes oncogenic tyrosine kinases; however, despite the potential for clinical benefit, inhibitors of NMT as antineoplastic agents remain to be fully explored (for a review, see reference 87).
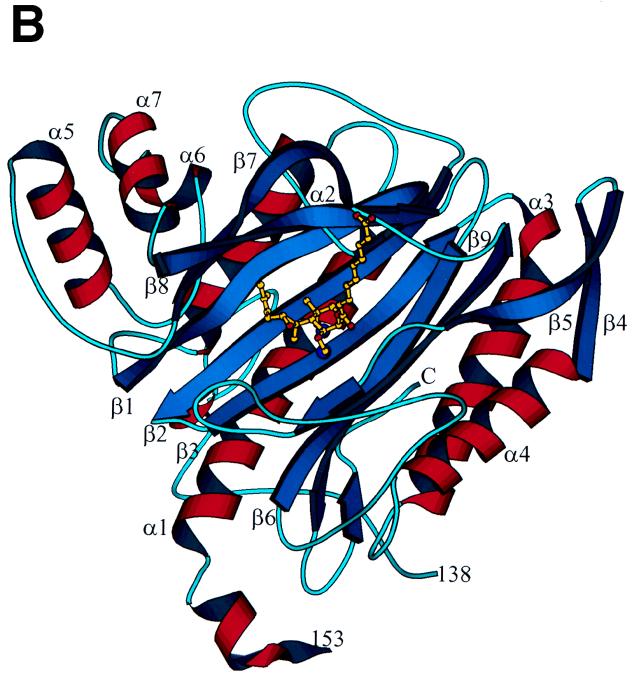
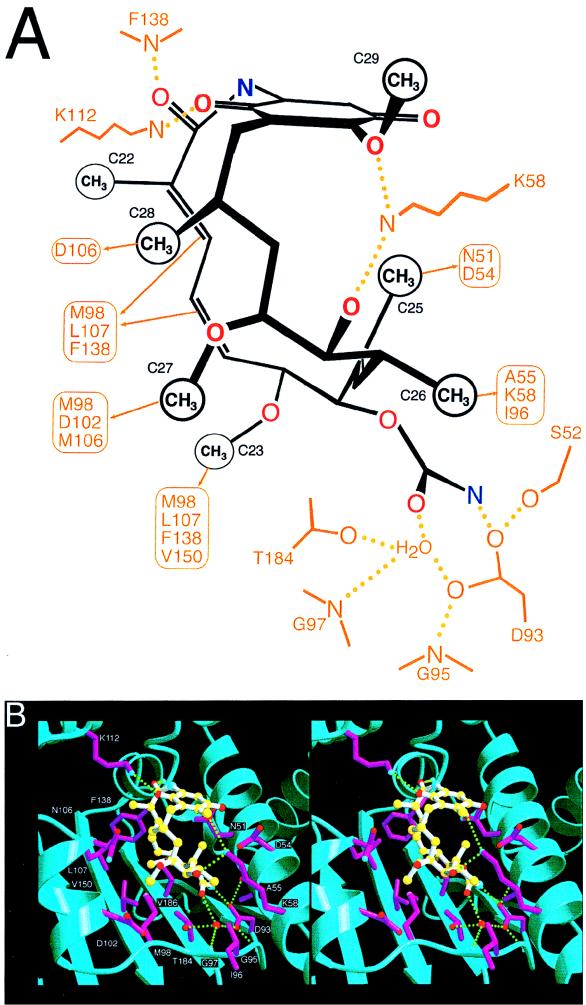
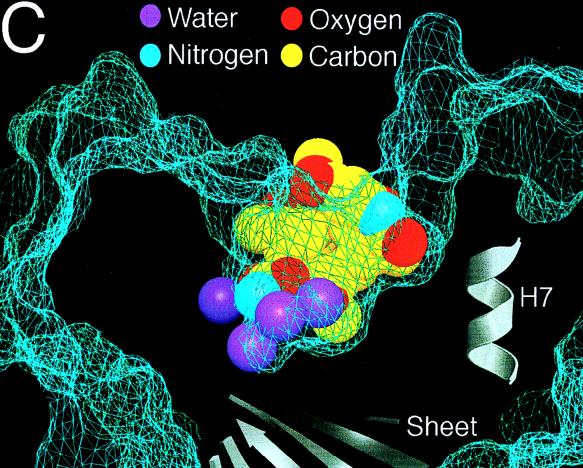
On the other hand, great progress has been made recently in the development of novel NMT inhibitors with potential as antifungal agents. Pioneering studies by Gordon and coworkers identified the NMT genes in S. cerevisiae, C. albicans, and Cryptococcus neoformans and demonstrated that these enzymes are essential for viability (81, 157, 203, 204). Hence, inhibitors would be expected to have fungicidal activity, which may be important in the development of newer broad-spectrum antifungal agents in severely immunosuppressed patients. An important recent advance is the solution of a high-resolution X-ray crystal structure of C. albicans NMT (333).
Lodge, Gordon, Sikorski and coworkers identified important differences in the substrate specificity of the fungal and mammalian NMT, suggesting that inhibitors with relative specificity for the fungal enzyme could be obtained (204). Starting with the octapeptide inhibitor ALYASKLS, a series of imidazole-substituted serine-lysine dipeptide amides were designed as candidate specific inhibitors of C. albicans NMT (71). Further structure-function optimization led to the synthesis of a series of tripeptide-based inhibitors that are markedly selective for the fungal rather than the human NMT (560- to 2,200-fold) and which have antifungal activity against C. albicans at ∼50 μM (71). Recently, nonpeptide derivatized versions of these inhibitors that retain antifungal activity and selectivity for the fungal rather than the human NMT have been synthesized (202), and trials of these compounds in animal models of fungal infection should be forthcoming. Finally, other investigators have been exploring the potential of myristic acid analogs as inhibitors of NMT as novel antifungal agents (257).
These studies on the design of selective inhibitors of fungal but not human NMT raise an important question in antifungal drug discovery, namely, when is there enough difference between the fungal and mammalian enzymes that such selective activity can be achieved? This is now a central question in the development of novel antifungal agents, and one bias has been to target only fungus-specific enzymes, such as 1,3-β-glucan synthase. In our opinion, this disregards a wealth of excellent antifungal targets that differ significantly from their mammalian homologs. Indeed, recent studies on the identification of novel and quite specific inhibitors of the mammalian enzyme cyclooxygenase 2 (COX2) are an important case in point. Aspirin and other nonsteroidal anti-inflammatory drugs inhibit both COX2 to yield their therapeutic benefits (analgesia, antipyresis, antithrombosis, and anti-inflammation), but they also inhibit cyclooxygenase 1 (COX1) in the gastric mucosa to cause undesired ulcerogenic side effects. However, a number of nonsteroidal anti-inflammatory drugs differ in their relative potency against COX1 and COX2, and this initially suggested that isoform-specific inhibitors could be obtained. This is in fact the case; the COX2-specific inhibitor celecoxib has just received Food and Drug Administration approval, and a number of other compounds are nearing approval. What is important about this example is that COX1 and COX2 differ by only a single amino acid in the active site, and this sequence divergence is apparently sufficient to achieve selective inhibition. A second case in point is the HIV-1 protease, which is sensitive to a variety of different protease inhibitors, yet single-amino-acid substitutions in the active site/drug binding pocket permit it to retain its enzyme activity but render the strains drug resistant. Clearly, a quite modest sequence difference should be fully sufficient for specific targeting of fungal rather than host enzymes. These differences can be further exploited and modeled since crystal structures are available for a series of these potential targets (Fig. 1, 3, 5, 7, and 8).
INHIBITORS OF SPHINGOLIPID METABOLISM AS ANTIFUNGAL AGENTS
Sphingolipids are ubiquitous and essential components of eukaryotic plasma membranes (127, 131). The basic structure and metabolism of these lipids are conserved in fungal and human cells, with some differences which could be exploited for the development of antifungal drugs. Correspondingly, compounds such as ceramide trigger apoptosis, and thus agonists of sphingolipid signaling may find use as anticancer chemotherapeutic agents. Enzymes in the sphingolipid metabolic pathways are targets of an array of natural compounds produced by many microorganisms (103, 223). We review sphingolipid metabolism to illustrate how sphingolipid metabolism in fungi is similar to and yet different from that in animals, and we suggest a strategy to use the inhibitors of the enzymes in sphingolipid metabolism as novel antifungal drugs.
The first half of the de novo synthesis pathway producing sphingoid bases and ceramides is essentially conserved among fungi, plants, and animals, while the second half of the pathway involves the more complex sphingolipids and is dissimilar among fungi, plants, and animals (73) (Fig. 9). The conserved first half starts with the rate-limiting step in de novo synthesis involving condensation of l-serine and palmitoyl coenzyme A to produce 3-ketodihydrosphingosine (3-ketosphinganine), which is subsequently converted to dihydrosphingosine (sphinganine). In animal cells, dihydrosphingosine is N acylated to form dihydroceramide (N-acyl sphinganine) and then desaturated to ceramide (N-acyl sphingosine), while in fungi and plants, it is either hydroxylated to phytosphingosine (5-hydroxysphinganine) and then N acylated to phytoceramide (N-acyl-5-hydroxysphinganine) or first N acylated to dihydroceramide and then hydroxylated to phytoceramide. Phytoceramide in fungi and ceramide in humans are the basic backbones upon which the more complex sphingolipids are built.
FIG. 9.
Sphingolipid metabolic cascades are conserved between yeast and humans. Differences in the de novo synthesis of sphingolipids between human and fungi are shown. Inhibitors of specific enzymes are shown in italics.
In animals, the phosphocholine moiety of phosphatidylcholine is transferred to ceramide to yield sphingomyelin (a phosphosphingolipid) or added to a variety of sugar moieties to become one of more than a few hundred different types of glycosphingolipids. In fungi, phytoceramide is sequentially added to phosphorylated sugar moieties to become inositolphosphoceramide (IPC), mannose inositolphosphoceramide (MIPC), and mannose diinositolphosphoceramide [M(IP)2C]. Analysis of sphingolipids from S. cerevisiae, C. albicans, and Cryptococcus neoformans suggests that sphingolipid metabolism is conserved among many fungi (325, 332).
Sphingolipids make up only a small portion of membrane phospholipids and yet are structurally diverse and essential. This suggests that sphingolipids may be involved in biological regulation or signal transduction rather than as structural components of the plasma membrane. Biological phenomena involving sphingolipids or metabolites, such as ceramide, sphingosine, and sphingosine 1-phosphate, are numerous and include apoptosis, cell cycle regulation, cellular senescence, differentiation, calcium homeostasis, and tumor progression (130). As an example, extracellular stimuli such as Fas, tumor necrosis factor alpha, ionizing radiation, and chemotherapeutic agents activate sphingomyelinase, which hydrolyzes sphingomyelin to ceramide and phosphocholine. Ceramide induces pleiotropic biological responses, including apoptosis, which could be clinically useful in cancer treatment strategies (130, 248). The proapoptotic function of ceramide was suggested to be counteracted by antiapoptotic function of sphingosine 1-phosphate, a further metabolite of ceramide (61, 307).
S. cerevisiae has been the target of extensive studies of sphingolipid metabolism and signal transduction (73). In S. cerevisiae, heat stress activates serine palmitoyltransferase, a key sphingolipid biosynthetic enzyme, leading to a marked increase in sphingoid bases including dihydrosphingosine and phytosphingosine (74, 155). Recently, it was shown that the serine palmitoyltransferase is also activated by diverse stresses including nutrient starvation and DNA damage, resulting in increased levels of phytosphingosine (56). This, in turn, leads to the degradation of nutrient permeases necessary for growth (56).
Serine palmitoyltransferase is the enzyme catalyzing the committed step in sphingolipid biosynthesis (Fig. 9). In S. cerevisiae, serine palmitoyltransferase is composed of the LCB1 and the LCB2 gene products (37, 237). Inhibitors of this enzyme block the production of all sphingolipids, which, because these are essential lipids, is lethal. Many inhibitors of this pathway, including myriosin (223), sphingofungins (361), lipoxamycins (210, 334), viridiofungins (212), and cycloserine (337), have been reported. Except for cycloserine, all of these are natural compounds and show very potent antifungal activity in the nano- to picomolar concentration range. However, since the serine palmitoyltransferase is conserved in both function and amino acid sequence from fungi to humans, these agents also inhibit the mammalian enzyme to various degrees.
Dihydrosphingosine desaturase converts dihydrosphingosine to dihydroceramide in humans and dihydrosphingosine to phytosphingosine in fungi (Fig. 9). In S. cerevisiae, this enzyme is encoded by the SYR2 gene (120, 126), and a mutation in this gene renders yeast resistant to syringomycin, a cyclic lipodepsipeptide produced by Pseudomonas syringae pv. syringae (57, 306). At present, it is not known if syringomycin has specific inhibitory effects on the fungal enzyme compared to the mammalian enzyme.
Ceramide synthase acylates dihydrosphingosine and phytosphingosine to produce dihydroceramide and phytoceramide, respectively (Fig. 9). Fumonisin B1 and australifungins inhibit this enzyme activity. Fumonisin B1 was shown to be active against both fungal and mammalian enzymes (223, 224), but it has relatively low cytotoxicity against S. cerevisiae (350). Australifungin is isolated from the fermentation extracts of Sporomiella australis and shows antifungal activity against Candida spp., Cryptococcus neoformans, and Aspergillus spp. (211); it has not yet been determined if australifungin also inhibits the mammalian enzyme.
IPC synthase catalyzes the addition of inositol phosphate to phytoceramide, which is the first fungus-specific step in sphingolipid biosynthesis (238). Therefore, this enzyme is an ideal target of antifungal drugs. There are several natural compounds that inhibit the activity of this enzyme, including aureobasidin A, khafrefungin, and rustmycin (Fig. 10). These are potent inhibitors of the enzyme, with IC50s in the subnanomolar range. Aureobasidin A is a cyclic depsipeptide isolated from Aureobasidium pullulans and has been reported to show antifungal activity against C. albicans, Aspergillus spp., and S. cerevisiae (83, 134, 137, 238). A number of novel aureobasidin analogs with enhanced antifungal activity against C. albicans have been recently reported (185). In addition, aureobasidin is exported from fungal and mammalian cells by MDR pumps, and some novel aureobasidin analogs with increased activity against mammalian MDR pumps have been described and may prove useful in treating MDR tumors (186). Khafrefungin is an aldonic acid ester linked to a C22 modified alkyl chain and shows fungicidal activity against C. albicans and Cryptococcus neoformans (214). Notably, khafrefungin does not inhibit the synthesis of mammalian sphingolipids including sphingomyelin. Rustmycin is a 14-member macrolide from an actinomycete (Fig. 10); it has especially potent fungicidal activity against Cryptococcus neoformans and was used in a mouse model, where it effectively reduced infection with this yeast (213).
FIG. 10.
Inhibitors of sphingolipid biosynthesis. The structures of three inhibitors of IPC synthase, aureobasidin A (a cyclic peptide), khafrefungin, and rustmicin, are shown.
That fungal sphingolipids are essential and differ in some respects from human sphingolipid metabolism makes these an attractive target for antifungal drugs. Because some parts of the metabolic pathways are more highly conserved than others, the enzymes which are more dissimilar may represent better targets. In this regard, the IPC synthase is the enzyme catalyzing the first step of fungus-specific sphingolipid synthesis, and therefore it may be an ideal target for antifungal drug development. Several antifungal drugs have been isolated from microbial extracts which were screened for inhibitory effects on IPC synthesis, and more extensive screening could identify additional inhibitors for preclinical testing in animal models of fungal infection.
YEAST AS A MODEL TO IDENTIFY NOVEL TOXINS WITH CHEMOTHERAPEUTIC POTENTIAL AGAINST FUNGI AND TUMORS
Recent advances in genomic sequencing, analysis of gene expression with genome arrays, combinatorial chemistry, and novel screening approaches promise to revolutionize drug discovery. Lee Hartwell and Steve Friend have recently pioneered a novel approach in which S. cerevisiae is used to discover compounds with novel chemotherapeutic potential (133, 331). The strategy involves the construction of a panel of isogenic yeast strains with mutations in genes encoding proteins and enzymes involved in DNA replication, recombination, repair, and cell cycle checkpoint function. Importantly, these mutants are viable but are more sensitive to DNA damage or agents that perturb the cell cycle than are the isogenic wild-type cells. Next, a panel of natural and synthetic compounds in the National Cancer Institute collection of compounds was screened for increased toxicity against one or more mutant strains compared to wild-type cells. A number of especially promising compounds have been identified by this approach. Next, these compounds were screened for activity against a panel of 60 human tumor cell lines. In several cases, compounds with activity against a restricted subset of the cell lines were identified. The hypothesis is that tumor cells, like the mutant yeast strains, may have mutations in DNA repair or cell cycle checkpoint functions that render them uniquely sensitive to a particular toxin compared to wild-type cells. Thus, the very mutations that originally contribute to the development of neoplastic potential can be used to target the tumor cell for destruction by novel agents. The great facility with which the yeast genome can be manipulated by homologous recombination and the speed of simple growth assays suggest that this type of approach could be quite robust. An additional adaptation might be to include combinatorial chemistry approaches on microbeads that can be used to assay for yeast growth inhibitors in simple plating assays (23, 147).
Sequencing of the S. cerevisiae genome has been recently completed, and two different whole-genome expression analysis approaches have been developed. The first involves binding of complete open reading frames for the ∼6,000 yeast genes as an ordered array on the surface of a glass slide (70, 187, 290). The array is then hybridized to fluorescently labelled cDNA derived from mRNA of cells grown under different conditions. The second approach involves the generation of high-density whole-genome arrays by chemical synthesis of short oligonucleotides on a scaffold matrix (146, 341, 343).
Genome arrays to monitor genome-wide expression patterns are now being exploited for drug discovery. As an example, Marton et al. recently analyzed the changes in gene expression in yeast cells that had been treated with the calcineurin inhibitor FK506 and compared these cells to mutant cells in which the genes encoding the calcineurin catalytic subunits had been deleted (216). While many of the changes in gene expression pattern were similar in cells lacking calcineurin and in cells with inhibited calcineurin, interestingly FK506 had additional effects on gene expression that were not attributable to calcineurin or FKBP12 inhibition. Importantly, many genes involved in amino acid biosynthesis were induced by FK506, and this induction required the transcription factor GCN4, which is known to be involved in activating the expression of amino acid biosynthetic genes in response to amino acid starvation (216). Earlier findings had revealed that FK506 inhibits amino acid transport in S. cerevisiae and that this drug effect is independent of calcineurin and the four known FKBP proteins in yeast (77, 138, 206).
A second recent example of the potential of combinatorial chemistry and whole-genome expression analysis is the development of a novel inhibitor of the human cdk2 cyclin-dependent kinase and the related S. cerevisiae Cdc28 kinase (114). Starting with novel trisubstituted purine analogs of the inhibitors olomoucine and roscovitine, Gray et al. (114) used combinatorial chemistry to develop more specific and potent inhibitors of the human cdk2 kinase. Subsequently, the X-ray structure of the complex between one analog, purvalanol B, and cdk2 was solved, revealing the molecular details of inhibitor binding to the cdk2 ATP binding site. Interestingly, this inhibitor also has potent activity against the S. cerevisiae CDC28 kinase and the related PHO85 kinase. Using a drug-sensitized yeast strain with mutations in the ERG6 gene, which is required for the final biosynthetic step leading to the membrane sterol ergosterol and two genes encoding MDR pump homologs (PDR5 and SNQ2), Gray et al. observed inhibition of yeast growth with an IC50 of 30 μM (114). Examination of the pattern of gene expression by whole-genome arrays revealed similar, but not identical, alterations in gene expression by inhibitor compared to cdc28 or pho85 mutations (114).
One final novel approach that has been developed has great promise to exploit S. cerevisiae for use in the discovery of novel drug targets. A novel variation on the yeast two-hybrid system, called the three-hybrid assay, has been developed to identify novel drug targets (194). The three-hybrid assay has several components. First, the DNA binding domain of the GAL4 transcription factor is fused to the glucocorticoid receptor (the first hybrid). Next, the transcriptional activation domain of the GAL4 protein is fused to a random library of cDNA sequences from a human cell line or organism of interest (the second hybrid). Finally, the third hybrid molecule is a hybrid drug in which glucocorticoid has been fused to a drug of interest, such as FK506. In this three-hybrid system, the ability of the hybrid steroid-drug conjugate to mediate the binding of the two isolated halves of the GAL4 transcription factor is then detected by monitoring the expression of reporter genes fused to a promoter regulated by the GAL4 transcription factor (Fig. 11). When the hybrid drug can bind to both halves of the GAL4 transcription factor fusion proteins, reporter gene expression is induced. For example, when the steroid-FK506 conjugate was added to yeast cells expressing the GAL4-DNA binding domain-steroid receptor fusion protein and transformed with a library of human T-cell cDNA clones fused to the GAL4 activation domain, library isolates in which the GAL4 activation domain had been fused to the human FK506 binding protein FKBP12 were identified. While the full promise of this three-hybrid system has yet to be realized in the identification of a novel, unknown drug target, clearly there are a multitude of possible applications in the identification of proteins which interact with small molecules that can be tethered to a steroid and can penetrate yeast cells.
FIG. 11.
Schematic view of the yeast three-hybrid assay for drug target discovery. The two-hybrid system for detecting protein-protein interactions was modified to develop a genetic method to identify drug targets. The GAL4 DNA binding domain was fused to a steroid receptor ligand binding domain. The GAL4 activation domain was fused to a library. The third hybrid is a conjugate drug in which dexamethosone (A) is conjugated to a second ligand (B). This hybrid drug is added to cells, and reporter gene expression is monitored to identify clones from the library that encode proteins that interact with the ligand of interest.
Finally, with the use of the yeast genome sequence and functional genomics, it is possible to identify all genes in S. cerevisiae essential for viability. These essential genes could then become targets for candidate chemical library screens. In fact, with human and yeast databases, unique essential genes could be identified by an electronic Southern analysis and those unique to fungi or mammalian cells could be used as a focus for antifungal or anticancer drug discovery.
CONCLUSION
In contrast to bacterial infections, cancer and viral and fungal infections are particularly difficult to treat and target with novel drugs because they all result from maladies caused by eukaryotic cells similar to or modified from the host cells. However, many of the same strategies that are used in the design of novel cancer chemotherapy agents can be brought to bear on the development of antiviral and antifungal agents. This is most apparent in the recent development of several novel antiviral agents (zidovudine, didanosine, and zalcitibine) by using nucleotide analogs that had been originally developed as candidate anticancer chemotherapy agents. Similar approaches can be used in the development of novel antifungal drugs. Although mammals have diverged from yeasts and fungi over approximately one billion years of evolution, recent studies underscore that much of the basic cellular machinery and signaling pathways are remarkably highly conserved and that yeasts and fungi are excellent model systems to understand the mechanisms of action and identify the targets of antineoplastic drugs. Finally, there is a rich collection of natural products produced by microorganisms that remains to be explored for novel chemotherapeutic potential. Microorganisms probably evolved to produce these natural products as antimicrobial agents, yet these compounds often have quite unique and useful actions in mammals via conserved target proteins, and these activities remain to be exploited for therapeutic benefit.
ACKNOWLEDGMENTS
Our studies described herein were supported, in part, by K01 award CA77075 from the NCI to Maria E. Cardenas; by R01 grants AI39115, AI41937, and AI42159 from NIAID; and by program project grant P01 AI44975 from NIAID to the Duke University Mycology Research Unit. Joseph Heitman is a Burroughs Wellcome Scholar in Molecular Pathogenic Mycology and an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Abe J, Zhou W, Takuwa N, Taguchi J, Kurokawa K, Kumada M, Takuwa Y. A fumagillin derivative angiogenesis inhibitor, AGM-1470, inhibits activation of cyclin-dependent kinases and phosphorylation of retinoblastoma gene product but not protein tyrosyl phosphorylation or protooncogene expression in vascular endothelial cells. Cancer Res. 1994;54:3407–3412. [PubMed] [Google Scholar]
- 2.Aitken A, Klee C B, Cohen P. The structure of the B subunit of calcineurin. Eur J Biochem. 1984;139:663–671. doi: 10.1111/j.1432-1033.1984.tb08055.x. [DOI] [PubMed] [Google Scholar]
- 3.Alarcon C M, Cardenas M E, Heitman J. Mammalian RAFT1 kinase domain provides rapamycin-sensitive TOR function in yeast. Genes Dev. 1996;10:279–288. doi: 10.1101/gad.10.3.279. [DOI] [PubMed] [Google Scholar]
- 4.Alarcon C M, Heitman J, Cardenas M E. Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast. Mol Biol Cell. 1999;10:2531–2546. doi: 10.1091/mbc.10.8.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander B D, Perfect J R. Antifungal resistance trends towards the year 2000. Implications for therapy and new approaches. Drugs. 1997;54:657–678. doi: 10.2165/00003495-199754050-00002. [DOI] [PubMed] [Google Scholar]
- 6.Anaissie E J. Opportunistic mycoses in the immunocompromised host: experience at a cancer center and review. Clin Infect Dis. 1992;14:S43–53. doi: 10.1093/clinids/14.supplement_1.s43. [DOI] [PubMed] [Google Scholar]
- 7.Anderson C W, Lees-Miller S P. The nuclear serine/threonine protein kinase DNA-PK. Crit Rev Eukaryotic Gene Expression. 1992;2:283–314. [PubMed] [Google Scholar]
- 8.Andreana A, Gollapudi S, Gupta S. Salmonella typhimurium induces expression of P glycoprotein (multidrug resistance 1 gene product) in a promonocytic cell line chronically infected with human immunodeficiency virus type 1. J Infect Dis. 1994;169:760–765. doi: 10.1093/infdis/169.4.760. [DOI] [PubMed] [Google Scholar]
- 9.Arceci R J, Stieglitz K, Bierer B E. Immunosuppressants FK506 and rapamycin function as reversal agents of the multidrug resistance phenotype. Blood. 1992;80:1528–1536. [PubMed] [Google Scholar]
- 10.Assem M, Bonvalot S, Beltramo J L, Garrido C, Dimanche-Boitrel M T, Genne P, Rebibou J M, Caillot D, Chauffert B. Deleterious effect of serum proteins on the amphotericin B-induced potentiation of cisplatin in human colon cancer cells. Br J Cancer. 1994;70:631–635. doi: 10.1038/bjc.1994.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker H, Sidorowicz A, Sehgal S N, Venzina C. Rapamycin (AY-22,989), a new antifungal antibiotic. III. In vitro and in vivo evaluation. J Antibiot. 1978;31:539–545. doi: 10.7164/antibiotics.31.539. [DOI] [PubMed] [Google Scholar]
- 12.Balzi E, Goffeau A. Yeast multidrug resistance: the PDR network. J Bioenerg Biomembr. 1995;27:71–76. doi: 10.1007/BF02110333. [DOI] [PubMed] [Google Scholar]
- 13.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck-Sague C M, Jarvis W R. Secular trends in the epidemiology of nosocomical fungal injections in the United States, 1980–1990. National Nosocomial Infections Surveillance System. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 15.Bell A, Wernli B, Franklin R M. Roles of peptidyl-prolyl cis-trans isomerase and calcineurin in the mechanisms of antimalarial action of cyclosporin A, FK506, and rapamycin. Biochem Pharmacol. 1994;48:495–503. doi: 10.1016/0006-2952(94)90279-8. [DOI] [PubMed] [Google Scholar]
- 16.Bellamy W T, Dalton W S. Multidrug resistance in the laboratory and clinic. Adv Clin Chem. 1994;31:1–61. doi: 10.1016/s0065-2423(08)60332-7. [DOI] [PubMed] [Google Scholar]
- 17.Beretta L, Gingras A-C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 18.Berger J M, Wang J C. Recent developments in DNA topoisomerase II structure and mechanism. Curr Opin Struct Biol. 1996;6:84–90. doi: 10.1016/s0959-440x(96)80099-6. [DOI] [PubMed] [Google Scholar]
- 19.Berry M, Metzger D, Chambon P. Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J. 1990;9:2811–2818. doi: 10.1002/j.1460-2075.1990.tb07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blunt T, Finnie N J, Taccioli G E, Smith G C, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 21.Boehm T, Folkman J, Browder T, O’Reilly M S. Antiangiogenic therapy of experimental cancer does not induce acquired drug resistance. Nature. 1997;390:404–407. doi: 10.1038/37126. [DOI] [PubMed] [Google Scholar]
- 22.Bohen S P. Genetic and biochemical analysis of p23 and ansamycin antibiotics in the function of Hsp90-dependent signaling proteins. Mol Cell Biol. 1998;18:3330–3339. doi: 10.1128/mcb.18.6.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borchardt A, Liberles S D, Biggar S R, Crabtree G R, Schreiber S L. Small molecule-dependent genetic selection in stochastic nanodroplets as a means of detecting protein-ligand interactions on a large scale. Chem Biol. 1997;4:961–968. doi: 10.1016/s1074-5521(97)90304-5. [DOI] [PubMed] [Google Scholar]
- 24.Borel J F. Comparative study of in vitro and in vivo drug effects on cell-mediated cytotoxicity. Immunology. 1976;31:631–641. [PMC free article] [PubMed] [Google Scholar]
- 25.Boschman C R, Bodnar U R, Tornatore M A, Obias A A, Noskin G A, Englund K, Postelnick M A, Suriano T, Peterson L R. Thirteen-year evolution of azole resistance in yeast isolates and prevalence of resistant strains carried by cancer patients at a large medical center. Antimicrob Agents Chemother. 1998;42:734–738. doi: 10.1128/aac.42.4.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boubnov N V, Hall K T, Wills Z, Lee S E, He D M, Benjamin D M, Pulaski C R, Band H, Reeves W, Hendrickson E A. Complementation of the ionizing radiation sensitivity, DNA end binding, and V(D)J recombination defects of double-strand break repair mutants by the p86 Ku autoantigen. Proc Natl Acad Sci USA. 1995;92:890–894. doi: 10.1073/pnas.92.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boulton S, Kyle S, Yalcintepe L, Durkacz B W. Wortmannin is a potent inhibitor of DNA double strand break but not single strand break repair in Chinese hamster ovary cells. Carcinogenesis. 1996;17:2285–2290. doi: 10.1093/carcin/17.11.2285. [DOI] [PubMed] [Google Scholar]
- 28.Bram R J, Hung D T, Martin P K, Schreiber S L, Crabtree G R. Identification of the immunophilins capable of mediating inhibition of signal transduction by cyclosporin A and FK506: roles of calcineurin binding and cellular location. Mol Cell Biol. 1993;13:4760–4769. doi: 10.1128/mcb.13.8.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breuder T, Hemenway C S, Movva N R, Cardenas M E, Heitman J. Calcineurin is essential in cyclosporin A- and FK506-sensitive yeast strains. Proc Natl Acad Sci USA. 1994;91:5372–5376. doi: 10.1073/pnas.91.12.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broeks A, Gerrard B, Allikmets R, Dean M, Plasterk R H. Homologues of the human multidrug resistance genes MRP and MDR contribute to heavy metal resistance in the soil nematode Caenorhabditis elegans. EMBO J. 1996;15:6132–6143. [PMC free article] [PubMed] [Google Scholar]
- 31.Brown E J, Albers M W, Shin T B, Ichikawa K, Keith C T, Lane W S, Schreiber S L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–759. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 32.Brown E J, Beal P A, Keith C T, Chen J, Shin T B, Schreiber S L. Control of p70 S6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 33.Brown E J, Schreiber S L. A signaling pathway to translational control. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 34.Brown S J, Kellett P J, Lippard S J. Ixr1, a yeast protein that binds to platinated DNA and confers sensitivity to cisplatin. Science. 1993;261:603–605. doi: 10.1126/science.8342024. [DOI] [PubMed] [Google Scholar]
- 35.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi J, Houghton P J, Lawrence J J C, Abraham R T. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 36.Brunn G J, Williams J, Sabers C, Wiederrecht G, John J, Lawrence C, Abraham R T. Direct inhibition of the signal functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J. 1996;15:5256–5267. [PMC free article] [PubMed] [Google Scholar]
- 37.Buede R, Rinker-Schaffer C, Pinto W J, Lester R L, Dickson R C. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J Bacteriol. 1991;173:4325–4332. doi: 10.1128/jb.173.14.4325-4332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bundey S. Clinical and genetic features of ataxia-telangiectasia. Int J Radiat Biol. 1994;66(Suppl.):23–29. [PubMed] [Google Scholar]
- 39.Burnett P E, Barrow R K, Cohen N A, Snyder S H, Sabatini D M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cafferkey R, Young P R, McLaughlin M M, Bergsma D J, Koltin Y, Sathe G M, Faucette L, Eng W-K, Johnson R K, Livi G P. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol Cell Biol. 1993;13:6012–6023. doi: 10.1128/mcb.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caplan A J. Yeast molecular chaperones and the mechanism of steroid hormone action. Trends Endocrinol Metab. 1997;8:271–276. doi: 10.1016/s1043-2760(97)00079-9. [DOI] [PubMed] [Google Scholar]
- 42.Cardenas M, Muir R S, Breuder T, Heitman J. Targets of immunophilin-immunosuppressant complexes are distinct highly conserved regions of calcineurin A. EMBO J. 1995;14:2772–2783. doi: 10.1002/j.1460-2075.1995.tb07277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cardenas M E, Heitman J. FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cardenas M E, Lorenz M, Hemenway C, Heitman J. Yeast as model-T cells. Perspect Drug Discovery Design. 1994;2:103–126. [Google Scholar]
- 45.Cardenas M E, Sanfridson A, Cutler N S, Heitman J. Signal-transduction cascades as targets for therapeutic intervention by natural products. Trends Biotechnol. 1998;16:427–433. doi: 10.1016/s0167-7799(98)01239-6. [DOI] [PubMed] [Google Scholar]
- 46.Cardenas M E, Zhu D, Heitman J. Molecular mechanisms of immunosuppression by cyclosporine, FK506, and rapamycin. Curr Opin Nephrol Hypertens. 1995;4:472–477. doi: 10.1097/00041552-199511000-00002. [DOI] [PubMed] [Google Scholar]
- 47.Carpenter C L, Cantley L C. Phosphoinositide kinases. Curr Opin Cell Biol. 1996;8:153–158. doi: 10.1016/s0955-0674(96)80060-3. [DOI] [PubMed] [Google Scholar]
- 48.Chang H-C J, Lindquist S. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J Biol Chem. 1994;269:24983–24988. [PubMed] [Google Scholar]
- 49.Chang H-C J, Nathan D F, Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol Cell Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang H Y, Takei K, Sydor A M, Born T, Rusnak F, Jay D G. Asymmetric retraction of growth cone filopodia following focal inactivation of calcineurin. Nature. 1995;376:686–690. doi: 10.1038/376686a0. [DOI] [PubMed] [Google Scholar]
- 51.Chen S F, Perrilla F W, Behrens D L, Papp L M. Inhibitor of dehydroorotate dihydrogenase activity by brequinar sodium. Cancer Res. 1992;52:3521–3527. [PubMed] [Google Scholar]
- 52.Chiu M I, Katz H, Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci USA. 1994;91:12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi G H, Chen B, Nuss D L. Virus-mediated or transgenic suppression of a G-protein α subunit and attenuation of fungal virulence. Proc Natl Acad Sci USA. 1995;92:305–309. doi: 10.1073/pnas.92.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi J, Chen J, Schreiber S L, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 55.Chung J, Kuo C J, Crabtree G R, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 Kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 56.Chung, N., C. Mao, G. M. Jenkins, Y. A. Hannun, J. Heitman, and L. M. Obeid. De novo synthesis of sphingolipids is required for stress-induced, ubiquitin-dependent degradation of nutrient permeases in Saccharomyces cerevisiae. Submitted for publication.
- 57.Cliften P, Wang Y, Mochizuki D, Miyakawa T, Wangspa R, Hughes J. SYR2, a gene necessary for syringomycin growth inhibition of Saccharomyces cerevisiae. Microbiology. 1996;142:477–484. doi: 10.1099/13500872-142-3-477. [DOI] [PubMed] [Google Scholar]
- 58.Coyle C, Kent M, Tanowitz H B, Wittner M, Weiss L M. TNP-470 is an effective antimicrosporidial agent. J Infect Dis. 1998;177:515–518. doi: 10.1086/517390. [DOI] [PubMed] [Google Scholar]
- 59.Cruz M C, Cavallo L M, Görlach J M, Perfect J R, Cardenas M E, Heitman J. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol. 1999;19:4101–4112. doi: 10.1128/mcb.19.6.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cutler N S, Heitman J, Cardenas M E. STT4 is an essential phosphatidylinositol 4-kinase that is a target of wortmannin in Saccharomyces cerevisiae. J Biol Chem. 1997;272:27671–27677. doi: 10.1074/jbc.272.44.27671. [DOI] [PubMed] [Google Scholar]
- 61.Cuvillier O, Pirianov G, Kleuser B, Vanek P G, Coso O A, Gutkind S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature. 1996;381:800–803. doi: 10.1038/381800a0. [DOI] [PubMed] [Google Scholar]
- 62.Cyert M S, Kunisawa R, Kaim D, Thorner J. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc Natl Acad Sci USA. 1991;88:7376–7380. doi: 10.1073/pnas.88.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cyert M S, Thorner J. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol Cell Biol. 1992;12:3460–3469. doi: 10.1128/mcb.12.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dammann H, Hellstern S, Husain Q, Mutzel R. Primary structure, expression and developmental regulation of a Dictyostelium calcineurin A homologue. Eur J Biochem. 1996;238:391–399. doi: 10.1111/j.1432-1033.1996.0391z.x. [DOI] [PubMed] [Google Scholar]
- 65.Dana S L, Hoener P A, Wheeler D A, Lawrence C B, McDonnell D P. Novel estrogen response elements identified by genetic selection in yeast are differentially responsive to estrogens and antiestrogens in mammalian cells. Mol Endo. 1994;8:1193–1207. doi: 10.1210/mend.8.9.7838152. [DOI] [PubMed] [Google Scholar]
- 66.D’Arpa P, Liu L F. Topoisomerase-targeting antitumor drugs. Biochim Biophys Acta. 1989;989:163–177. doi: 10.1016/0304-419x(89)90041-3. [DOI] [PubMed] [Google Scholar]
- 67.Del Poeta, M., S. Chen, D. V. Hoff, C. C. Dykstra, M. C. Wani, G. Manikumar, J. Heitman, M. E. Wall, and J. R. Perfect. Structure-activity relationships of the topoisomerase I inhibitors camptothecin, nitidine and their derivatives in fungal and cancer cells. Submitted for publication.
- 68.Del Poeta M, Toffaletti D L, Rude T H, Dykstra C C, Heitman J, Perfect J R. Topoisomerase I is essential in Cryptococcus neoformans: role in pathobiology and as an antifungal target. Genetics. 1999;152:167–178. doi: 10.1093/genetics/152.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Demeule M, Wenger R M, Beliveau R. Molecular interactions of cyclosporin A with P-glycoprotein. Photolabeling with cyclosporin derivatives. J Biol Chem. 1997;272:6647–6652. doi: 10.1074/jbc.272.10.6647. [DOI] [PubMed] [Google Scholar]
- 70.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 71.Devadas B, Freeman S K, Zupec M E, Lu H F, Nagarajan S R, Kishore N S, Lodge J K, Kuneman D W, McWherter C A, Vinjamoori D V, Getman D P, Gordon J I, Sikorski J A. Design and synthesis of novel imidazole-substituted dipeptide amides as potent and selective inhibitors of Candida albicans myristoylCoA:protein N-myristoyltransferase and identification of related tripeptide inhibitors with mechanism-based antifungal activity. J Med Chem. 1997;40:2609–2625. doi: 10.1021/jm970094w. [DOI] [PubMed] [Google Scholar]
- 72.Dick J D, Merz W G, Saral R. Incidence of polyene-resistant yeasts recovered from clinical specimens. Antimicrob Agents Chemother. 1980;18:158–163. doi: 10.1128/aac.18.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dickson R C. Sphingolipid functions in Saccharomyces cerevisiae: Comparison to mammals. Annu Rev Biochem. 1998;67:27–48. doi: 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- 74.Dickson R C, Nagiec E E, Skrzypek M, Tillman P, Wells G B, Lester R L. Sphingolipids are potential heat stress signals in Saccharomyces. J Biol Chem. 1997;272:30196–30200. doi: 10.1074/jbc.272.48.30196. [DOI] [PubMed] [Google Scholar]
- 75.Di Como C J, Arndt K T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 76.Dolinski K, Heitman J. Peptidyl-prolyl isomerases (PPIases) In: Tooze S, editor. Guidebook to molecular chaperones and protein folding catalysts. Oxford, United Kingdom: Oxford University Press; 1997. pp. 359–369. [Google Scholar]
- 77.Dolinski K, Muir R S, Cardenas M E, Heitman J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dolinski K J, Cardenas M E, Heitman J. CNS1 encodes an essential p60/Sti1 homolog in Saccharomyces cerevisiae that suppresses cyclophilin 40 mutations and interacts with Hsp90. Mol Cell Biol. 1998;18:7344–7352. doi: 10.1128/mcb.18.12.7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duina A A, Chang H C, Marsh J A, Lindquist S, Gaber R F. A cyclophilin function in Hsp-90 dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 80.Durant J R. Cisplatin: a clinical overview. In: Prestayko A W, Crooke S T, Carter S K, editors. Cisplatin: current status and new developments. New York, N.Y: Academic Press, Inc.; 1980. pp. 317–321. [Google Scholar]
- 81.Duronio R J, Towler D A, Heuckeroth R O, Gordon J I. Disruption of the yeast N-myristoyl transferase gene causes recessive lethality. Science. 1989;243:796–800. doi: 10.1126/science.2644694. [DOI] [PubMed] [Google Scholar]
- 82.Egner R, Rosenthal F E, Kralli A, Sanglard D, Kuchler K. Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol Biol Cell. 1998;9:523–543. doi: 10.1091/mbc.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Endo M, Takesako K, Kato I, Yamaguchi H. Fungicidal action of aureobasidin A, a cyclic depsipeptide antifungal antibiotic, against Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1997;41:672–676. doi: 10.1128/aac.41.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eng F C, Lee H S, Ferrara J, Willson T M, White J H. Probing the structure and function of the estrogen receptor ligand binding domain by analysis of mutants with altered transactivation characteristics. Mol Cell Biol. 1997;17:4644–4653. doi: 10.1128/mcb.17.8.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fang Y, Fliss A E, Rao J, Caplan A J. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Farcasanu I C, Hirata D, Tsuchiya E, Nishiyama F, Miyakawa T. Protein phosphatase 2B of Saccharomyces cerevisiae is required for tolerance to manganese, in blocking the entry of ions into the cells. Eur J Biochem. 1995;232:712–717. [PubMed] [Google Scholar]
- 87.Felsted R L, Glover C J, Hartman K. Protein N-myristoylation as a chemotherapeutic target for cancer. J Natl Cancer Inst. 1995;87:1571–1573. doi: 10.1093/jnci/87.21.1571. [DOI] [PubMed] [Google Scholar]
- 88.Fernandez-Sarabia M J, Sutton A, Zhong T, Arndt K T. SIT4 protein phosphatase is required for the normal accumulation of SWI4, CLN1, CLN2, and HCS26 RNAs during late G1. Genes Dev. 1992;6:2417–2428. doi: 10.1101/gad.6.12a.2417. [DOI] [PubMed] [Google Scholar]
- 89.Ferrara A, Cafferkey R, Livi G P. Cloning and sequence analysis of a rapamycin-binding protein-encoding gene (RBP1) from Candida albicans. Gene. 1992;113:125–127. doi: 10.1016/0378-1119(92)90679-j. [DOI] [PubMed] [Google Scholar]
- 90.Ferreira A, Kincaid R, Kosik K S. Calcineurin is associated with the cytoskeleton of cultured neurons and has a role in the acquisition of polarity. Mol Biol Cell. 1993;4:1225–1238. doi: 10.1091/mbc.4.12.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fliss A E, Fang Y, Boschelli F, Caplan A J. Differential in vivo regulation of steroid hormone receptor activation by Cdc37p. Mol Biol Cell. 1997;8:2501–2509. doi: 10.1091/mbc.8.12.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Folkman J. Tumor angiogenesis. Adv Cancer Res. 1974;19:331–358. doi: 10.1016/s0065-230x(08)60058-5. [DOI] [PubMed] [Google Scholar]
- 93.Folkman J. The vascularization of tumors. Sci Am. 1976;234:58–64. doi: 10.1038/scientificamerican0576-58. [DOI] [PubMed] [Google Scholar]
- 94.Foor F, Parent S A, Morin N, Dahl A M, Ramadan N, Chrebet G, Bostian K A, Nielsen J B. Calcineurin mediates inhibition by FK506 and cyclosporin of recovery from α-factor arrest in yeast. Nature. 1992;360:682–684. doi: 10.1038/360682a0. [DOI] [PubMed] [Google Scholar]
- 95.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 96.Fostel J, Montgomery D. Identification of the aminocatechol A-3253 as an in vitro poison of DNA topoisomerase I from Candida albicans. Antimicrob Agents Chemother. 1995;39:586–592. doi: 10.1128/AAC.39.3.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fostel J, Montgomery D, Lartey P. Comparison of responses of DNA topoisomerase I from Candida albicans and human cells to four new agents which stimulate topoisomerase-dependent DNA nicking. FEMS Microbiol Lett. 1996;138:105–111. doi: 10.1111/j.1574-6968.1996.tb08142.x. [DOI] [PubMed] [Google Scholar]
- 98.Foxwell B M, Mackie A, Ling V, Ryffel B. Identification of the multidrug resistance-related P-glycoprotein as a cyclosporine binding protein. Mol Pharmacol. 1989;36:543–546. [PubMed] [Google Scholar]
- 99.Franke E K, Luban J. Cyclophilin and Gag in HIV-1 replication and pathogenesis. In: Andrieu J-M, Lu W, editors. Cell activation and apoptosis in HIV Infection. New York, N.Y: Plenum Press; 1995. pp. 217–228. [DOI] [PubMed] [Google Scholar]
- 100.Freedman D A, Levine A J. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fruman D A, Wood M A, Gjerston C K, Katz H R, Burakoff S J, Bierer B E. FU506 binding protein 12 mediates sensitivity to both FU506 and rapamycin in murine mast cells. Eur J Immunol. 1995;25:563–571. doi: 10.1002/eji.1830250239. [DOI] [PubMed] [Google Scholar]
- 102.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 103.Georgopapadakou N H. Antifungals: mechanism of action and resistance, established and novel drugs. Curr Opin Microbiol. 1998;1:547–557. doi: 10.1016/s1369-5274(98)80087-8. [DOI] [PubMed] [Google Scholar]
- 104.Georgopapadakou N H, Walsh T J. Human mycoses: drugs and targets for emerging pathogens. Science. 1994;264:371–373. doi: 10.1126/science.8153622. [DOI] [PubMed] [Google Scholar]
- 105.Gerlach J H, Endicott J A, Juranka P F, Henderson G, Sarangi F, Deuchars K L, Ling V. Homology between P-glycoprotein and a bacterial haemolysin transport protein suggests a model for multidrug resistance. Nature. 1986;324:485–489. doi: 10.1038/324485a0. [DOI] [PubMed] [Google Scholar]
- 106.Germann U A, Pastan I, Gottesman M M. P-glycoproteins: mediators of multidrug resistance. Semin Cell Biol. 1993;4:63–76. doi: 10.1006/scel.1993.1008. [DOI] [PubMed] [Google Scholar]
- 107.Getts R C, Stamato T D. Absence of a Ku-like DNA end binding activity in the xrs double-strand DNA repair-deficient mutant. J Biol Chem. 1994;269:15981–15984. [PubMed] [Google Scholar]
- 108.Ghannoum M A, Abu-Elteen K H, Motawy M S, Abu-Hatab M A, Ibrahim A S, Criddle R S. Combinations of antifungal and antineoplastic drugs with interactive effects on inhibition of yeast growth. Chemotherapy. 1990;36:308–320. doi: 10.1159/000238782. [DOI] [PubMed] [Google Scholar]
- 109.Ghannoum M A, Motawy M S, Abu-Hatab M A, Abu-Elteen K A, Criddle R S. Interactive effect of antifungal and antineoplastic agents on yeasts commonly prevalent in cancer patients. Antimicrob Agents Chemother. 1989;33:726–730. doi: 10.1128/aac.33.5.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goldstein L J, Gottesman M M, Pastan I. Expression of the MDR1 gene in human cancers. Cancer Treat Res. 1991;57:101–119. doi: 10.1007/978-1-4615-3872-1_5. [DOI] [PubMed] [Google Scholar]
- 111.Goto T, Wang J C. Cloning of yeast TOP1, the gene encoding DNA topoisomerase I and DNA topoisomerase II. Proc Natl Acad Sci USA. 1985;82:7178–7182. doi: 10.1073/pnas.82.21.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gottesman M M, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 113.Gottesman M M, Pastan I, Ambudkar S V. P-glycoprotein and multidrug resistance. Curr Opin Genet Dev. 1996;6:610–617. doi: 10.1016/s0959-437x(96)80091-8. [DOI] [PubMed] [Google Scholar]
- 114.Gray N S, Wodicka L, Thunnissen A-M W H, Norman T C, Kwon S, Espinoza F H, Morgan D O, Barnes G, LeClerc S, Meijer L, Kim S-H, Lockhart D J, Schultz P G. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 115.Graybill J R, Bocanegra R, Fothergill A, Rinalid M G. Bleomycin therapy of experimental disseminated candidiasis in mice. Antimicrob Agents Chemother. 1996;40:816–818. doi: 10.1128/aac.40.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grenert J P, Sullivan W P, Fadden P, Haystead T A J, Clark J, Mimnaugh E, Krutzsch H, Ochel H J, Schulte T W, Sausville E, Neckers L M, Toft D O. The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J Biol Chem. 1997;272:23843–23850. doi: 10.1074/jbc.272.38.23843. [DOI] [PubMed] [Google Scholar]
- 117.Griffith E C, Su Z, Turk B E, Chen S, Chang Y-H, Wu Z, Biemann K, Liu J O. Methionine aminopeptidase (type 2) is the common target for angiogenesis inhibitors AGM-1470 and ovalicin. Chem Biol. 1997;4:461–471. doi: 10.1016/s1074-5521(97)90198-8. [DOI] [PubMed] [Google Scholar]
- 118.Griffith E C, Su Z, Niwayama S, Ramsay C A, Chang Y H, Liu J O. Molecular recognition of angiogenesis inhibitors fumagillin and ovalicin by methionine aminopeptidase 2. Proc Natl Acad Sci USA. 1998;95:15183–15188. doi: 10.1073/pnas.95.26.15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Griffith J P, Kim J L, Kim E E, Sintchak M D, Thomson J A, Fitzgibbon M J, Felming M A, Caron P R, Hsiao K, Navia M A. X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 120.Grilley M M, Stock S D, Dickson R C, Lester R L, Takemoto J Y. Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J Biol Chem. 1998;273:11062–11068. doi: 10.1074/jbc.273.18.11062. [DOI] [PubMed] [Google Scholar]
- 121.Groll A H, Piscitelli S C, Walsh T J. Clinical pharmacology of systemic antifungal agents: a comprehensive review of agents in clinical use, current investigational compounds, and putative targets for antifungal drug development. Adv Pharmacol. 1998;44:343–500. doi: 10.1016/s1054-3589(08)60129-5. [DOI] [PubMed] [Google Scholar]
- 122.Guerini D, Klee C B. Cloning of human calcineurin A: evidence for two isozymes and identification of a polyproline structural domain. Proc Natl Acad Sci USA. 1989;86:9183–9187. doi: 10.1073/pnas.86.23.9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guerini D, Krinks M H, Sikela J M, Hahn W E, Klee C B. Isolation and sequence of a cDNA clone for human calcineurin B, the Ca2+-binding subunit of the Ca2+/calmodulin-stimulated protein phosphatase. DNA. 1989;8:675–682. doi: 10.1089/dna.1.1989.8.675. [DOI] [PubMed] [Google Scholar]
- 124.Guerini D, Montell C, Klee C B. Molecular cloning and characterization of the genes encoding the two subunits of Drosophila melanogaster calcineurin. J Biol Chem. 1992;267:22542–22549. [PubMed] [Google Scholar]
- 125.Gustafson G, Davis G, Waldron C, Smith A, Henry M. Identification of a new antifungal target site through a dual biochemical and molecular-genetics approach. Curr Genet. 1996;30:159–165. doi: 10.1007/s002940050115. [DOI] [PubMed] [Google Scholar]
- 126.Haak D, Gable K, Beeler T, Dunn T. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J Biol Chem. 1997;272:29704–29710. doi: 10.1074/jbc.272.47.29704. [DOI] [PubMed] [Google Scholar]
- 127.Hakomori S. Chemistry of glycosphingolipids. In: Kanfer J N, Hakomori S, editors. Handbook of lipid research. 3. Sphingolipid biochemistry. New York, N.Y: Plenum Publishing Co.; 1983. pp. 1–150. [Google Scholar]
- 128.Hamamoto T, Seto H, Beppu T. Leptomycins A and B, new antifungal antibiotics. II. Structure elucidation. J Antibiot (Tokyo) 1983;36:646–650. doi: 10.7164/antibiotics.36.646. [DOI] [PubMed] [Google Scholar]
- 129.Hamamoto T, Uozumi T, Beppu T. Leptomycins A and B, new antifungal antibiotics. III. Mode of action of leptomycin B on Schizosaccharomyces pombe. J Antibiot (Tokyo) 1985;38:1573–1580. doi: 10.7164/antibiotics.38.1573. [DOI] [PubMed] [Google Scholar]
- 130.Hannun Y A. Functions of ceramide in coordinating cellular responses to stress. Science. 1996;274:1855–1859. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- 131.Hannun Y A, Bell R M. Functions of sphingolipids and sphingolipid breakdown products in cellular regulation. Science. 1989;243:500–507. doi: 10.1126/science.2643164. [DOI] [PubMed] [Google Scholar]
- 132.Hartley K O, Gell D, Smith G C M, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 133.Hartwell L H, Szankasi P, Roberts C J, Murray A W, Friend S H. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 134.Hashida-Okado T, Ogawa A, Endo M, Yasumoto R, Takesako K, Kato I. AUR1, a novel gene conferring aureobasidin resistance on Saccharomyces cerevisiae: a study of defective morphologies in Aur1p-depleted cells. Mol Gen Genet. 1996;251:236–244. doi: 10.1007/BF02172923. [DOI] [PubMed] [Google Scholar]
- 135.Hatanaka H, Kino T, Miyata S, Inamura N, Kuroda A, Goto T, Tanaka H, Okuhara M. FR-900520 and FR-900523, novel immunosuppressants isolated from a Streptomyces. II. Fermentation, isolation and physico-chemical and biological characteristics. J Antibiot. 1988;41:1592–1601. doi: 10.7164/antibiotics.41.1592. [DOI] [PubMed] [Google Scholar]
- 136.Häusler P, Papoff G, Eramo A, Reif K, Cantrell D A, Ruberti G. Protection of CD95-mediated apoptosis by activation of phosphatidylinositide 3-kinase and protein kinase B. Eur J Immunol. 1998;28:57–69. doi: 10.1002/(SICI)1521-4141(199801)28:01<57::AID-IMMU57>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 137.Heidler S A, Radding J A. The AUR1 gene in Saccharomyces cerevisiae encodes dominant resistance to the antifungal agent aureobasidin A ( LY295337) Antimicrob Agents Chemother. 1995;39:2765–2769. doi: 10.1128/aac.39.12.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Heitman J, Koller A, Kunz J, Henriquez R, Schmidt A, Movva N R, Hall M N. The immunosuppressant FK506 inhibits amino acid import in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5010–5019. doi: 10.1128/mcb.13.8.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Heitman J, Movva N R, Hall M N. Proline isomerases at the crossroads of protein folding, signal transduction, and immunosuppression. New Biol. 1992;4:448–460. [PubMed] [Google Scholar]
- 140.Heitman J, Movva N R, Hall M N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 141.Helliwell S B, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall M N. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell. 1994;5:105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hemenway C S, Dolinski K, Cardenas M E, Hiller M A, Jones E W, Heitman J. vph6 mutants of Saccharomyces cerevisiae require calcineurin for growth and are defective in vacuolar H+-ATPase assembly. Genetics. 1995;141:833–844. doi: 10.1093/genetics/141.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hemenway C S, Heitman J. Calcineurin structure, function, and inhibition. Cell Biochem Biophys. 1999;30:115–151. doi: 10.1007/BF02737887. [DOI] [PubMed] [Google Scholar]
- 144.Hendey B, Lawson M, Marcantonio E E, Maxfield F R. Intracellular calcium and calcineurin regulate neutrophil motility on vitronectin through a receptor identified by antibodies to integrins alphav and beta3. Blood. 1996;87:2038–2048. [PubMed] [Google Scholar]
- 145.Holm C, Goto T, Wang J C, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 146.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 147.Huang J, Schreiber S L. A yeast genetic system for selecting small molecule inhibitors of protein-protein interactions in nanodroplets. Proc Natl Acad Sci USA. 1997;94:13396–13401. doi: 10.1073/pnas.94.25.13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ichikawa M, Yoshimura A, Furukawa T, Sumizawa T, Nakazima Y, Akiyama S. Glycosylation of P-glycoprotein in a multidrug-resistant KB cell line, and in the human tissues. Biochim Biophys Acta. 1991;1073:309–315. doi: 10.1016/0304-4165(91)90136-5. [DOI] [PubMed] [Google Scholar]
- 149.Ingber D, Fujita T, Kishimoto S, Sudo K, Kanamaru T, Brem H, Folkman J. Synthetic analogues of fumagillin that inhibit angiogenesis and suppress tumour growth. Nature. 1990;348:555–557. doi: 10.1038/348555a0. [DOI] [PubMed] [Google Scholar]
- 150.Inui S, Sanjo H, Maeda K, Yamamoto H, Miyamoto E, Sakaguchi N. Ig receptor binding protein 1 (alpha4) is associated with a rapamycin-sensitive signal transduction in lymphocytes through direct binding to the catalytic subunit of protein phosphatase 2A. Blood. 1998;92:539–546. [PubMed] [Google Scholar]
- 151.Ishizuka T, Sakata N, Johnson G L, Gelfand E W, Terada N. Rapamycin potentiates dexamethasone-induced apoptosis and inhibits JNK activity in lymphoblastoid cells. Biochem Biophys Res Commun. 1997;230:386–391. doi: 10.1006/bbrc.1996.5967. [DOI] [PubMed] [Google Scholar]
- 152.Iwasaki S. Antimitotic agents: chemistry and recognition of tubulin molecule. Med Res Rev. 1993;13:183–198. doi: 10.1002/med.2610130205. [DOI] [PubMed] [Google Scholar]
- 153.Jachez B, Boesch D, Grassberger M A, Loor F. Reversion of the P-glycoprotein-mediated multidrug resistance of cancer cells by FK-506 derivatives. Anti-Cancer Drugs. 1993;4:223–229. doi: 10.1097/00001813-199304000-00015. [DOI] [PubMed] [Google Scholar]
- 154.Jain J, McCaffrey P G, Miner Z, Kerppola T K, Lambert J N, Verdine G L, Curran T, Rao A. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 155.Jenkins G M, Richards A, Wahl T, Mao C, Obeid L, Hannun Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J Biol Chem. 1997;272:32566–32572. doi: 10.1074/jbc.272.51.32566. [DOI] [PubMed] [Google Scholar]
- 156.Jiang W, Gerhold D, Kmiec E B, Hauser M, Becker J M, Koltin Y. The topoisomerase I gene from Candida albicans. Microbiology. 1997;143:377–386. doi: 10.1099/00221287-143-2-377. [DOI] [PubMed] [Google Scholar]
- 157.Johnson D R, Cok S J, Feldmann H, Gordon J I. Suppressors of nmt1-181, a conditional lethal allele of the Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase gene, reveal proteins involved in regulating protein N-myristoylation. Proc Natl Acad Sci USA. 1994;91:10158–10162. doi: 10.1073/pnas.91.21.10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jorgensen T J, Shiloh Y. The ATM gene and the radiobiology of ataxia telangiectasia. Int J Radiat Biol. 1996;69:527–537. doi: 10.1080/095530096145535. [DOI] [PubMed] [Google Scholar]
- 159.Juranka P F, Zastawny R L, Ling V. P-glycoprotein: multidrug-resistance and a superfamily of membrane-associated transport proteins. FASEB J. 1989;3:2583–2592. doi: 10.1096/fasebj.3.14.2574119. [DOI] [PubMed] [Google Scholar]
- 160.Kapeller R, Cantley L C. Phosphatidylinositol 3-kinase. Bioessays. 1994;16:565–576. doi: 10.1002/bies.950160810. [DOI] [PubMed] [Google Scholar]
- 161.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 162.Keith C T, Schreiber S L. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 163.Kennedy S G, Wagner A J, Conzen S D, Jordán J, Bellacosa A, Tsichlis P N, Hay N. The PI 3-kinase/Akt signaling pathway delivers and anti-apoptotic signal. Genes Dev. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- 164.Ketznelson H, Jamieson C A. Control of Nosema disease of honey bees with fumagillin. Science. 1952;115:70–71. doi: 10.1126/science.115.2977.70. [DOI] [PubMed] [Google Scholar]
- 165.Khanna K K, Beamish H, Yan J, Hobson K, Williams R, Dunn I, Lavin M F. Nature of G1/S cell cycle checkpoint defect in ataxia-telangiectasia. Oncogene. 1995;11:609–618. [PubMed] [Google Scholar]
- 166.Kikkawa F, Kojima M, Oguchi H, Maeda O, Ishikawa H, Tamakoshi K, Mizuno K, Kawai M, Suganuma N, Tomoda Y. Potentiating effect of amphotericin B on five platinum anticancer drugs in human cis-diamminedichloroplatinum (II) sensitive and resistant ovarian carcinoma cells. Anticancer Res. 1993;13:891–896. [PubMed] [Google Scholar]
- 167.Kincaid R L, Giri P R, Higuchi S, Tamura J, Dixon S C, Marietta C A, Amorese D A, Martin B M. Cloning and characterization of molecular isoforms of the catalytic subunit of calcineurin using nonisotopic methods. J Biol Chem. 1990;265:11312–11319. [PubMed] [Google Scholar]
- 168.Kino T, Hatanaka H, Hashimoto M, Nishiyama M, Goto T, Okuhara M, Kohsaka M, Aoki H, Imanaka H. FK-506, a novel immunosuppressant isolated from a Streptomyces. I. Fermentation, isolation, physico-chemical and biological characteristics. J Antibiot. 1987;40:1249–1255. doi: 10.7164/antibiotics.40.1249. [DOI] [PubMed] [Google Scholar]
- 169.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 170.Kirkland T N, Fierer J. Cyclosporin A inhibits Coccidioides immitis in vitro and in vivo. Antimicrob Agents Chemother. 1983;24:921–924. doi: 10.1128/aac.24.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kissinger C R, Parge H E, Knighton D R, Lewis C T, Pelletier L A, Tempczyk A, Kalish V J, Tucker K D, Showalter R E, Moomaw E W, Gastinel L N, Habuka N, Chen X, Maldonado F, Barker J E, Bacquet R, Villafranca J E. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 172.Klee C B, Draetta G F, Hubbard M J. Calcineurin. Adv Enzymol Relat Areas Mol Biol. 1988;61:149–209. doi: 10.1002/9780470123072.ch4. [DOI] [PubMed] [Google Scholar]
- 173.Kociba R J, Sleight S D, Rosenberg B. Inhibition of Dunning ascitic leukemia and Walker 256 carcinosarcoma with cis-diamminedichloroplatinum (NCS-119875) Cancer Chemother Rep. 1970;54:325–328. [PubMed] [Google Scholar]
- 174.Koltin Y, Faucette L, Bergsma D J, Levy M A, Cafferkey R, Koser P L, Johnson R K, Livi G P. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidylprolyl cis-trans isomerase related to human FK506-binding protein. Mol Cell Biol. 1991;11:1718–1723. doi: 10.1128/mcb.11.3.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Komiyama K, Okada K, Hirokawa Y, Masuda K, Tomisaka S, Umezawa I. Antitumor activity of a new antibiotic, kazusamycin. J Antibiot (Tokyo) 1985;38:224–249. doi: 10.7164/antibiotics.38.224. [DOI] [PubMed] [Google Scholar]
- 176.Komiyama K, Okada K, Tomisaka S, Umezawa I, Hamamoto T, Beppu T. Antitumor activity of leptomycin B. J Antibiot (Tokyo) 1985;38:427–429. doi: 10.7164/antibiotics.38.427. [DOI] [PubMed] [Google Scholar]
- 177.Kothe G O, Free S J. Calcineurin subunit B is required for normal vegetative growth in Neurospora crassa. Fungal Genet Biol. 1998;23:248–258. doi: 10.1006/fgbi.1998.1037. [DOI] [PubMed] [Google Scholar]
- 178.Kralli A, Bohen S P, Yamamoto K R. LEM1, an ATP-binding-cassette transporter, selectively modulates the biological potency of steroid hormones. Proc Natl Acad Sci USA. 1995;92:4701–4705. doi: 10.1073/pnas.92.10.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Krcmery V, Jr, Oravcova E, Spanik S, Mrazova-Studena M, Trupl J, Kunova A, Stopkova-Grey K, Kukuckova E, Krupova I, Demitrovicova A, Kralovicova K. Nosocomial breakthrough fungaemia during antifungal prophylaxis or empirical antifungal therapy in 41 cancer patients receiving antineoplastic chemotherapy: analysis of aetiology risk factors and outcome. J Antimicrob Chemother. 1998;41:373–380. doi: 10.1093/jac/41.3.373. [DOI] [PubMed] [Google Scholar]
- 180.Kuhnt M, Bitsch F, Ponelle M, Sanglier J J, Wang Y, Wolff B. Microbial conversion products of leptomycin B. Appl Environ Microbiol. 1998;64:714–720. doi: 10.1128/aem.64.2.714-720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kuiper G G, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson J A. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kuno T, Tanaka H, Mukai H, Chang C-D, Hiraga K, Miyakawa T, Tanaka C. cDNA cloning of a calcineurin B homolog in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1991;180:1159–1163. doi: 10.1016/s0006-291x(05)81188-x. [DOI] [PubMed] [Google Scholar]
- 183.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva N R, Hall M N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 184.Kuo C J, Chung J, Fiorentino D F, Flanagan W M, Blenis J, Crabtree G R. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature. 1992;358:70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- 185.Kurome T, Inoue T, Takesako K, Kato I. Syntheses of antifungal aureobasidin A analogs with alkyl chains for structure-activity relationship. J Antibiot (Tokyo) 1998;51:359–367. doi: 10.7164/antibiotics.51.359. [DOI] [PubMed] [Google Scholar]
- 186.Kurome T, Takesako K, Kato I. Aureobasidins as new inhibitors of P-glycoprotein in multidrug resistant tumor cells. J Antibiot (Tokyo) 1998;51:353–358. doi: 10.7164/antibiotics.51.353. [DOI] [PubMed] [Google Scholar]
- 187.Lashkari D A, DeRisi J L, McCusker J H, Namath A F, Gentile C, Hwang S Y, Brown P O, Davis R W. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci USA. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lawrence J C, Jr, Abraham R T. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem Sci. 1997;22:345–349. doi: 10.1016/s0968-0004(97)01101-8. [DOI] [PubMed] [Google Scholar]
- 189.Lawson M A, Maxfield F R. Ca2+- and calcineurin-dependent recycling of an integrin to the front of migrating neutrophils. Nature. 1995;377:75–79. doi: 10.1038/377075a0. [DOI] [PubMed] [Google Scholar]
- 190.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M, Day I R S, Barron G M, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 191.Li W, Handschumacher R E. Specific interaction of the cyclophilin-cyclosporin complex with the B subunit of calcineurin. J Biol Chem. 1993;268:14040–14044. [PubMed] [Google Scholar]
- 192.Li X, Chang Y H. Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc Natl Acad Sci USA. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Licht T, Pastan I, Gottesman M, Herrmann F. P-glycoprotein-mediated multidrug resistance in normal and neoplastic hematopoietic cells. Ann Hematol. 1994;69:159–171. doi: 10.1007/BF02215949. [DOI] [PubMed] [Google Scholar]
- 194.Licitra E J, Liu J O. A three-hybrid system for detecting small ligand-protein receptor interactions. Proc Natl Acad Sci USA. 1996;93:12817–12821. doi: 10.1073/pnas.93.23.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lima C D, Mondragon A. Mechanism of type II DNA topoisomerases: a tale of two gates. Structure. 1994;2:559–560. doi: 10.1016/s0969-2126(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 196.Lin T A, Kong X, Saltiel A R, Blackshear P J, Lawrence J C. Control of PHAS-I by insulin in 3T3-L1 adipocytes. Synthesis, degradation, and phosphorylation by a rapamycin-sensitive and mitogen-activated protein kinase-independent pathway. J Biol Chem. 1995;270:18531–18538. doi: 10.1074/jbc.270.31.18531. [DOI] [PubMed] [Google Scholar]
- 197.Ling V. P-glycoprotein and resistance to anticancer drugs. Cancer. 1992;69:2603–2609. doi: 10.1002/1097-0142(19920515)69:10<2603::aid-cncr2820691034>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 198.List A F, Spier C, Greer J, Wolff S, Hutter J, Dorr R, Salmon S, Futscher B, Baier M, Dalton W. Phase I/II trial of cyclosporine as a chemotherapy-resistance modifier in acute leukemia. J Clin Oncol. 1993;11:1652–1660. doi: 10.1200/JCO.1993.11.9.1652. [DOI] [PubMed] [Google Scholar]
- 199.Liu J, Farmer J D, Lane W S, Friedman J, Weissman I, Schreiber S L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 200.Liu S, Widom J, Kemp C W, Crews C M, Clardy J. Structure of human methionine aminopeptidase-2 complexed with fumagillin. Science. 1998;282:1324–1327. doi: 10.1126/science.282.5392.1324. [DOI] [PubMed] [Google Scholar]
- 201.Liu Y, Ishii S, Tokai M, Tsutsumi H, Ohki O, Akade R, Tanaka K, Tsuchiya E, Fukui S, Miyakawa T. The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin-binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol Gen Genet. 1991;227:52–59. doi: 10.1007/BF00260706. [DOI] [PubMed] [Google Scholar]
- 202.Lodge J K, Jackson-Machelski E, Higgins M, McWherter C A, Sikorski J A, Devadas B, Gordon J I. Genetic and biochemical studies establish that the fungicidal effect of a fully depeptidized inhibitor of Cryptococcus neoformans myristoyl-CoA:protein N-myristoyltransferase (Nmt) is Nmt-dependent. J Biol Chem. 1998;273:12482–12491. doi: 10.1074/jbc.273.20.12482. [DOI] [PubMed] [Google Scholar]
- 203.Lodge J K, Jackson-Machelski E, Toffaletti D L, Perfect J R, Gordon J I. Targeted gene replacement demonstrates that myristoyl-CoA:protein N-myristoyltransferase is essential for viability of Cryptococcus neoformans. Proc Natl Acad Sci USA. 1994;91:12008–12012. doi: 10.1073/pnas.91.25.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lodge J K, Johnson R L, Weinberg R A, Gordon J I. Comparison of myristoyl-CoA:protein N-myristoyltransferases from three pathogenic fungi: Cryptococcus neoformans, Histoplasma capsulatum, and Candida albicans. J Biol Chem. 1994;269:2996–3009. [PubMed] [Google Scholar]
- 205.Lorenz M C, Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- 206.Lorenz M C, Muir R S, Lim E, McElver J, Weber S C, Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- 207.Lowther W T, McMillen D A, Orville A M, Matthews B W. The anti-angiogenic agent fumagillin covalently modifies a conserved active-site histidine in the Escherichia coli methionine aminopeptidase. Proc Natl Acad Sci USA. 1998;95:12153–12157. doi: 10.1073/pnas.95.21.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Luo Y, Marx S O, Kiyokawa H, Koff A, Massague J, Marks A R. Rapamycin resistance tied to defective regulation of p27kip1. Mol Cell Biol. 1996;16:6744–6751. doi: 10.1128/mcb.16.12.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mak P, McDonnell D P, Weigel N L, Schrader W T, O’Malley B W. Expression of functional chicken oviduct progesterone receptors in yeast (Saccharomyces cerevisiae) J Biol Chem. 1989;284:21613–21618. [PubMed] [Google Scholar]
- 210.Mandala S M, Frommer B R, Thornton R A, Kurtz M B, Young N M, Cabello M A, Liesch J M, Smith J L, Horn W S. Inhibition of serine palmitoyl-transferase activity by lipoxamycin. J Antibiot. 1994;47:376–379. doi: 10.7164/antibiotics.47.376. [DOI] [PubMed] [Google Scholar]
- 211.Mandala S M, Thornton R A, Frommer B R, Curotto J E, Rozdilsky W, Kurtz M B, Giacobbe R A, Bills G F, Cabello M A, Martin I, et al. The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J Antibiot (Tokyo) 1995;48:349–356. doi: 10.7164/antibiotics.48.349. [DOI] [PubMed] [Google Scholar]
- 212.Mandala S M, Thornton R A, Frommer B R, Dreikorn S, Kurtz M B. Viridiofungins, novel inhibitors of sphingolipid synthesis. J Antibiot (Tokyo) 1997;50:339–343. doi: 10.7164/antibiotics.50.339. [DOI] [PubMed] [Google Scholar]
- 213.Mandala S M, Thornton R A, Milligan J, Rosenbach M, Garcia-Calvo M, Harris G, Abruzzo G K, Flattery A M, Gill C J, Bartizal K, Kurtz M B. Rustmicin, a potent antifungal agent, inhibits sphingolipid synthesis at inositol phosphoceramide synthase. J Biol Chem. 1998;273:14942–14949. doi: 10.1074/jbc.273.24.14942. [DOI] [PubMed] [Google Scholar]
- 214.Mandala S M, Thornton R A, Rosenbach M, Milligan J, Garcia-Calvo M, Kurtz M B. Khafrefungin, a novel inhibitor of sphingolipid synthesis. J Biol Chem. 1997;272:32709–32714. doi: 10.1074/jbc.272.51.32709. [DOI] [PubMed] [Google Scholar]
- 215.Marie J P, Bastie J N, Coloma F, Suberville A M F, Delmer A, Rio B, Delmas-Marsalet B, Leroux G, Casassus P, Baumelou E, et al. Cyclosporin A as a modifier agent in the salvage treatment of acute leukemia (AL) Leukemia. 1993;7:821–824. [PubMed] [Google Scholar]
- 216.Marton J J, DeRisi J L, Bennett H A, Iyer V R, Meyer M R, Roberts C J, Stoughton R, Burchard J, Slade D, Dai H, Bassett J D E, Hartwell L H, Brown P O, Friend S H. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 217.Masahisa O, Numata K, Nishiyama Y, Kamei H, Konishi M, Oki T, Kawaguchi H. Chemical modification of the antitumor antibiotic glidobactin. J Antibiot (Tokyo) 1998;41:1812–1822. doi: 10.7164/antibiotics.41.1812. [DOI] [PubMed] [Google Scholar]
- 218.Matheos D P, Kingsbury T J, Ahsan U S, Cunningham K W. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 1997;11:3445–3458. doi: 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.McA’Nulty M M, Whitehead J P, Lippard S J. Binding of Ixr1, a yeast HMG-domain protein, to cisplatin-DNA adducts in vitro and in vivo. Biochemistry. 1996;35:6089–6099. doi: 10.1021/bi952877u. [DOI] [PubMed] [Google Scholar]
- 220.McCann P P, Pegg A E. Ornithine decarboxylase as an enzyme target for therapy. Pharmacol Ther. 1992;54:195–215. doi: 10.1016/0163-7258(92)90032-u. [DOI] [PubMed] [Google Scholar]
- 221.McDonnell D P, Nawaz Z, Densmore C, Weigel N L, Pham T A, Clark J H, O’Malley B W. High level expression of biologically active estrogen receptor in Saccharomyces cerevisiae. J Steroid Biochem Mol Biol. 1991;39:291–297. doi: 10.1016/0960-0760(91)90038-7. [DOI] [PubMed] [Google Scholar]
- 222.Mendoza I, Rubio F, Rodriquez-Navarro A, Pardo J M. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- 223.Merrill A H, Jr, Wang E, Gilchrist D G, Riley R T. Fumonisins and other inhibitors of de novo sphingolipid biosynthesis. Adv Lipid Res. 1993;26:215–234. [PubMed] [Google Scholar]
- 224.Merrill A H, Jr, Wang E, Vales T R, Smith E R, Schroeder J J, Menaldino DS, Alexander C, Crane H M, Xia J, Liotta D C, Meredith F I, Riley R T. Fumonisin toxicity and sphingolipid biosynthesis. Adv Exp Med Biol. 1996;392:297–306. doi: 10.1007/978-1-4899-1379-1_25. [DOI] [PubMed] [Google Scholar]
- 225.Metzger D, White J H, Chambon P. The human oestrogen receptor functions in yeast. Nature. 1988;334:31–36. doi: 10.1038/334031a0. [DOI] [PubMed] [Google Scholar]
- 226.Meunier F. Targeting fungi: a challenge. Am J Med. 1995;99:60S–67S. doi: 10.1016/s0002-9343(99)80291-5. [DOI] [PubMed] [Google Scholar]
- 227.Meyers R, Cantley L C. Cloning and characterization of a wortmannin-sensitive human phosphatidylinositol 4-kinase. J Biol Chem. 1997;272:4384–4390. doi: 10.1074/jbc.272.7.4384. [DOI] [PubMed] [Google Scholar]
- 228.Migita K, Eguchi K, Ichinose Y, Kawabe Y, Tsukada T, Aoyagi T, Nagataki S. Effects of rapamycin on apoptosis of rheumatoid synovial cells. Clin Exp Immunol. 1997;108:199–203. doi: 10.1046/j.1365-2249.1997.d01-1002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Milan D, Griffith J, Su M, Price E R, McKeon F. The latch region of calcineurin B is involved in both immunosuppressant-immunophilin complex docking and phosphatase activation. Cell. 1994;79:437–447. doi: 10.1016/0092-8674(94)90253-4. [DOI] [PubMed] [Google Scholar]
- 230.Mishra S K, Ramani R, Yankov L K, Sharma A, Trevaskiss T, Misra R, Pandey R C. Program and abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1998. Paclitaxel interference with the antifungal activities of amphotericin B, abstr. J-153; p. 496. [Google Scholar]
- 231.Miyazaki T, Liu Z-J, Kawahara A, Minami Y, Yamada K, Tsujimoto Y, Barsoumian E L, Perlmutter R M, Taniguchi T. Three distinct IL-2 signaling pathways mediated by bcl-2, c-myc, and lck cooperate in hematopoietic cell proliferation. Cell. 1995;81:223–231. doi: 10.1016/0092-8674(95)90332-1. [DOI] [PubMed] [Google Scholar]
- 232.Mizuno K, Furuhashi Y, Misawa T, Iwata M, Kawai M, Kikkawa F, Kano T, Tomoda Y. Modulation of multidrug resistance by immunosuppressive agents: cyclosporin analogues, FK506 and mizoribine. Anticancer Res. 1992;12:21–26. [PubMed] [Google Scholar]
- 233.Morgan S E, Kastan M B. p53 and ATM: cell cycle, cell death and cancer. Found Cancer Res. 1997;71:1–25. doi: 10.1016/s0065-230x(08)60095-0. [DOI] [PubMed] [Google Scholar]
- 234.Moussa N M, Ghannoum M A, Whittaker P A, el-Ezaby M S, Quraman S. Effects of cisplatin and two novel palladium complexes on Candida albicans. Microbios. 1990;62:252–253. [PubMed] [Google Scholar]
- 235.Muramatsu T, Kincaid R L. Molecular cloning of a full-length cDNA encoding the catalytic subunit of human calmodulin-dependent protein phosphatase (calcineurin A alpha) Biochim Biophys Acta. 1993;1178:117–120. doi: 10.1016/0167-4889(93)90117-8. [DOI] [PubMed] [Google Scholar]
- 236.Murata K, Wu J, Brautigan D L. B cell receptor-associated protein alpha4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc Natl Acad Sci USA. 1997;94:10624–10629. doi: 10.1073/pnas.94.20.10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nagiec M M, Baltisberger J A, Wells G B, Lester R L, Dickson R C. The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc Natl Acad Sci USA. 1994;91:7899–7902. doi: 10.1073/pnas.91.17.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nagiec M M, Nagiec E E, Baltisberger J A, Wells G B, Lester R L, Dickson R C. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- 239.Nakagawa T, Goto K, Kondo H. Cloning, expression, and localization of 230-kDa phosphatidylinositol 4-kinase. J Biol Chem. 1996;271:12088–12094. doi: 10.1074/jbc.271.20.12088. [DOI] [PubMed] [Google Scholar]
- 240.Nakamura T, Liu Y, Hirata D, Namba H, Harada S-I, Hirokawa T, Miyakawa T. Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Nicolet C M, Craig E A. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3638–3646. doi: 10.1128/mcb.9.9.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 243.Nitiss J L. Roles of DNA topoisomerases in chromosomal replication and segregation. Adv Pharmacol. 1994;29A:103–134. doi: 10.1016/s1054-3589(08)60542-6. [DOI] [PubMed] [Google Scholar]
- 244.Nitiss J L. Using yeast to study resistance topoisomerase II-targeting drugs. Cancer Chemother Pharmacol. 1994;34(Suppl.):S6–13. doi: 10.1007/BF00684857. [DOI] [PubMed] [Google Scholar]
- 245.Northrop J P, Ullman K S, Crabtree G R. Characterization of nuclear and cytoplasmic components of the lymphoid-specific nuclear factor of activated T cells (NF-AT) complex. J Biol Chem. 1993;268:2917–2923. [PubMed] [Google Scholar]
- 246.Nourse J, Firpo E, Flanagan W M, Coats S, Polyak K, Lee M-H, Massague J, Crabtree G R, Roberts J M. Interleukin-2-mediated elimination of the p27Kip1 cyclin-dependent kinase inhibitor prevented by rapamycin. Nature. 1994;372:570–573. doi: 10.1038/372570a0. [DOI] [PubMed] [Google Scholar]
- 247.Numata K, Murakami T, Oka M, Yamamoto H, Hatori M, Miyaki T, Oki T, Kawaguchi H. Enhanced production of the minor components of glidobactins in Polyangium brachysporum. J Antibiot (Tokyo) 1988;41:1358–1365. doi: 10.7164/antibiotics.41.1358. [DOI] [PubMed] [Google Scholar]
- 248.Obeid L M, Linardic C M, Karolak L A, Hannun Y A. Programmed cell death induced by ceramide. Science. 1993;259:1769–1771. doi: 10.1126/science.8456305. [DOI] [PubMed] [Google Scholar]
- 249.Odom A, Muir S, Lim E, Toffaletti D L, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Odom A, Poeta M D, Perfect J, Heitman J. The immunosuppressant FK506 and its nonimmunosuppressive analog L-685,818 are toxic to Cryptococcus neoformans by inhibition of a common target protein. Antimicrob Agents Chemother. 1997;41:156–161. doi: 10.1128/aac.41.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Okada T, Kawano Y, Sakakibara T, Hazeki O, Ui M. Essential role of phosphatidylinositol 3-kinase in insulin-induced glucose transport and antilipolysis in rat adipocytes. J Biol Chem. 1994;269:3568–3573. [PubMed] [Google Scholar]
- 252.Okada T, Sakuma L, Fukui Y, Hazeki O, Ui M. Blockage of chemotactic peptide-induced stimulation of neutrophils by wortmannin as a result of selective inhibition of phosphatidylinositol 3-kinase. J Biol Chem. 1994;269:3563–3567. [PubMed] [Google Scholar]
- 253.Osborne C K. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 254.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 255.Paech K, Webb P, Kuiper G G, Nilsson S, Gustafsson J, Kushner P J, Scanlan T S. Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 256.Panaretou B, Prodromou C, Roe S M, O’Brien R, Ladbury J E, Piper P W, Pearl L H. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Parang K, Knaus E E, Wiebe L I, Sardari S, Daneshtablab M, Csizmadia F. Synthesis and antifungal activities of myristic acid analogs. Arch Pharm. 1996;329:475–482. doi: 10.1002/ardp.19963291102. [DOI] [PubMed] [Google Scholar]
- 258.Parangi S, O’Reilly M, Christofori G, Holmgren L, Grosfeld J, Folkman J, Hanahan D. Antiangiogenic therapy of transgenic mice impairs de novo tumor growth. Proc Natl Acad Sci USA. 1996;93:2002–2007. doi: 10.1073/pnas.93.5.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Parent S A, Nielsen J B, Morin N, Chrebet G, Ramadan N, Dahl A M, Hsu M-J, Bostian K A, Foor F. Calcineurin-dependent growth of an FK506- and CsA-hypersensitive mutant of Saccharomyces cerevisiae. J Gen Microbiol. 1993;139:2973–2984. doi: 10.1099/00221287-139-12-2973. [DOI] [PubMed] [Google Scholar]
- 260.Perego P, Zunino F, Carenini N, Giuliani F, Spinelli S, Howell S B. Sensitivity of cisplatin and platinum-containing compounds of Schizosaccharomyces pombe rad mutants. Mol Pharmacol. 1998;54:213–219. doi: 10.1124/mol.54.1.213. [DOI] [PubMed] [Google Scholar]
- 261.Perfect, J. R. Unpublished results.
- 262.Perfect J R. Fungal virulence genes as targets for antifungal chemotherapy. Antimicrob Agents Chemother. 1996;40:1577–1583. doi: 10.1128/aac.40.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Perfect J R, Durack D T. Effects of cyclosporine in experimental cryptococcal meningitis. Infect Immun. 1985;50:22–26. doi: 10.1128/iai.50.1.22-26.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Peterson S R, Kurimasa A, Oshimura M, Dynan W S, Bradbury E M, Chen D J. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Pfaller M A, Riley A J, Gerarden T. Polyamine deprivation and growth inhibition of Cryptococcus neoformans by α-difluoromethyl ornithine and cyclohexylamine. Mycopathologia. 1990;112:27–32. doi: 10.1007/BF01795176. [DOI] [PubMed] [Google Scholar]
- 266.Picard D, Khursheed B, Garabedian M J, Fortin M G, Lindquist S, Yamamoto K R. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- 267.Polak A, Scholer H J. Mode of action of 5-fluorocytosine and mechanisms of resistance. Chemotherapy. 1975;21:113–130. doi: 10.1159/000221854. [DOI] [PubMed] [Google Scholar]
- 268.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant stains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 269.Pourtier-Manzanedo A, Boesch D, Loor F. FK-506 (fujimycin) reverses the multidrug resistance of tumor cells in vitro. Anti-Cancer Drugs. 1991;2:279–283. doi: 10.1097/00001813-199106000-00010. [DOI] [PubMed] [Google Scholar]
- 270.Powderly W G, Kobayashi G S, Herzig G P, Medoff G. Amphotericin B-resistant yeast infections in severely immunocompromised patients. Am J Med. 1988;84:826–832. doi: 10.1016/0002-9343(88)90059-9. [DOI] [PubMed] [Google Scholar]
- 271.Pozos T C, Sekler I, Cyert M S. The product of HUM1, a novel yeast gene, is required for vacuolar Ca2+/H+ exchange and is related to mammalian Na+/Ca2+ exchangers. Mol Cell Biol. 1996;16:3730–3741. doi: 10.1128/mcb.16.7.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Prasad R, Wergifosse P D, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- 273.Price D J, Grove J R, Calvo V, Avruch J, Bierer B E. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- 274.Prodromou C, Roe S M, O’Brien R, Ladbury J E, Piper P W, Pearl L H. Identification and structural characterisation of the ATP/ADP binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 275.Prokisch H, Yarden O, Dieminger M, Tropschug M, Barthelmess I B. Impairment of calcineurin function in Neurospora crassa reveals its essential role in hyphal growth, morphology and maintenance of the apical Ca2+ gradient. Mol Gen Genet. 1997;256:104–114. doi: 10.1007/s004380050551. [DOI] [PubMed] [Google Scholar]
- 276.Raff M C, Barres B A, Burne J F, Coles H S, Ishizaki Y, Jacobson M D. Programmed cell death and the control of cell survival: lessons from the nervous system. Science. 1993;262:695–700. doi: 10.1126/science.8235590. [DOI] [PubMed] [Google Scholar]
- 277.Rasmussen C, Garen C, Brining S, Kincaid R L, Means R L, Means A R. The calmodulin-dependent protein phosphatase catalytic subunit (calcineurin A) is an essential gene in Aspergillus nidulans. EMBO J. 1994;13:2545–2552. doi: 10.1002/j.1460-2075.1994.tb06544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Rathmell W K, Chu G. Involvement of the Ku autoantigen in the cellular response to DNA double-strand breaks. Proc Natl Acad Sci USA. 1994;91:7623–7627. doi: 10.1073/pnas.91.16.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Redinbo M R, Stewart L, Kuhn P, Champoux J J, Hol W G J. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 280.Rosemberg B, Renshaw E C, Camp L V, Hartwick J, Drobnick J. Pt-induced filamentous growth in Escherichia coli. J Bacteriol. 1967;93:716–721. doi: 10.1128/jb.93.2.716-721.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sabatini D M, Erdjument-Bromage H, Lui M, Tempst P, Snyder S H. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 282.Sabers C J, Martin M M, Brunn G J, Williams J M, Dumont F J, Wiederrecht G, Abraham R T. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 283.Saeki T, Ueda K, Tanigawara Y, Hori R, Komano T. Human P-glycoprotein transports cyclosporin A and FK506. J Biol Chem. 1993;268:6077–6080. [PubMed] [Google Scholar]
- 284.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 285.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Sarachek A, Henderson L A. Modification of responses of Candida albicans to cisplatin by membrane damaging antimycotic agents. Mycoses. 1991;34:177–182. doi: 10.1111/j.1439-0507.1991.tb00640.x. [DOI] [PubMed] [Google Scholar]
- 287.Sarkaria J N, Tibbetts R S, Busby E C, Kennedy A P, Hill D E, Abraham R T. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- 288.Savani D V, Perfect J R, Durack D T. Program and abstracts of the 26th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1986. Activity of anti-cancer drugs against yeast in vitro, abstr. 838; p. 252. [Google Scholar]
- 289.Scheibel T, Buchner J. The Hsp90 complex—a super-chaperone machine as a novel drug target. Biochem Pharmacol. 1998;56:675–682. doi: 10.1016/s0006-2952(98)00120-8. [DOI] [PubMed] [Google Scholar]
- 290.Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 291.Schmid F X. Prolyl isomerase—enzymatic catalysis of slow protein-folding reactions. Annu Rev Biophys Biomol Struct. 1993;22:123–143. doi: 10.1146/annurev.bb.22.060193.001011. [DOI] [PubMed] [Google Scholar]
- 292.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Schmidt A, Bickle M, Beck T, Hall M N. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- 294.Schmidt A, Kunz J, Hall M N. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc Natl Acad Sci USA. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Schneider C, Sepp-Lorenzino L, Nimmesgern E, Ouerfelli O, Danishefsky S, Rosen N, Hartl F U. Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc Natl Acad Sci USA. 1996;93:14536–14541. doi: 10.1073/pnas.93.25.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Schreiber S L, Crabtree G R. The mechanism of action of cyclosporin A and FK506. Immunol Today. 1992;13:136–142. doi: 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- 297.Schultz R M, Merriman R L, Andis S L, Bonjouklian R, Grindey G B, Rutherford P G, Gallegos A, Massey K, Powis G. In vitro and in vivo antitumor activity of the phosphatidylinositol-3-kinase inhibitor, wortmannin. Anticancer Res. 1995;15:1135–1140. [PubMed] [Google Scholar]
- 298.Scott P H, Brunn G J, Kohn A D, Roth R A, Lawrence J C. Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. Proc Natl Acad Sci USA. 1998;95:7772–7777. doi: 10.1073/pnas.95.13.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Shi Y, Frankel A, Radvanyi L G, Penn L Z, Miller R G, Mills G B. Rapamycin enhances apoptosis and increases sensitivity to cisplatin in vitro. Cancer Res. 1995;55:1982–1988. [PubMed] [Google Scholar]
- 300.Shiau S P, Glasebrook A, Hardikar S D, Yang N N, Hershberger C L. Activation of the human estrogen receptor by estrogenic and antiestrogenic compounds in Saccharomyces cerevisiae: a positive selection system. Gene. 1996;179:205–210. doi: 10.1016/s0378-1119(96)00345-9. [DOI] [PubMed] [Google Scholar]
- 301.Siekierka J J, Hung S H Y, Poe M, Lin C S, Sigal N H. A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature. 1989;341:755–757. doi: 10.1038/341755a0. [DOI] [PubMed] [Google Scholar]
- 302.Silverman J A, Hayes M L, Luft B J, Joiner K A. Characterization of anti-Toxoplasma activity of SDZ 215-918, a cyclosporin derivative lacking immunosuppressive and peptidylprolyl-isomerase-inhibiting activity: possible role of a P glycoprotein in Toxoplasma physiology. Antimicrob Agents Chemother. 1997;41:1859–1866. doi: 10.1128/aac.41.9.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Sin N, Meng L, Wang M Q W, Wen J J, Bornmann W G, Crews C M. The anti-angiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP-2. Proc Natl Acad Sci USA. 1997;94:6099–6103. doi: 10.1073/pnas.94.12.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Singh K, Sun S, Vezina C. Rapamycin (AY-22,989), a new antifungal antibiotic. IV. Mechanism of action. J Antibiot (Tokyo) 1979;32:630–645. doi: 10.7164/antibiotics.32.630. [DOI] [PubMed] [Google Scholar]
- 305.Sipley J D, Menninger J C, Hartley K O, Ward D C, Jackson S P, Anderson C W. Gene for the catalytic subunit of the human DNA-activated protein kinase maps to the site of the XRCC7 gene on chromosome 8. Proc Natl Acad Sci USA. 1995;92:7515–7519. doi: 10.1073/pnas.92.16.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Sorensen K N, Kim K H, Takemoto J Y. In vitro antifungal and fungicidal activities and erythrocyte toxicities of cyclic lipodepsinonapeptides produced by Pseudomonas syringae pv. syringae. Antimicrob Agents Chemother. 1996;40:2710–2713. doi: 10.1128/aac.40.12.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Spiegel S, Cuvillier O, Edsall L C, Kohama T, Menzeleev R, Olah Z, Pirianov G, Thomas D M, Tu Z, Van Brocklyn J R, Wang F. Sphingosine-1-phosphate in cell growth and cell death. Ann N Y Acad Sci. 1998;845:11–18. doi: 10.1111/j.1749-6632.1998.tb09658.x. [DOI] [PubMed] [Google Scholar]
- 308.Stack J H, Emr S D. Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI 3-kinase activities. J Biol Chem. 1994;269:31552–31562. [PubMed] [Google Scholar]
- 309.Stathopoulos A M, Cyert M S. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Stebbins C E, Russo A A, Schneider C, Rosen N, Hartl F U, Pavletich N P. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 311.Stephens L, Smrcka A, Cooke F T, Jackson T R, Sternweis P C, Hawkins P T. A novel phosphoinositide 3 kinase activity in myeloid-derived cells is activated by G protein beta gamma subunits. Cell. 1994;77:83–93. doi: 10.1016/0092-8674(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 312.Stewart L, Redinbo M R, Qiu X, Hol W G J, Champoux J J. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 313.Sutton A, Immanuel D, Arndt K T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Taagepera S, McDonald D, Loeb J E, Whitaker L L, McElroy A K, Wang J Y, Hope T J. Nuclear-cytoplasmic shuttling of C-ABL tyrosine kinase. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Taccioli G E, Gottlieb T M, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann A R, Alt F W, Jackson S P, Jeggo P A. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 316.Tai P-K K, Albers M W, McDonnell D P, Chang H, Schreiber S L, Faber L E. Potentiation of progesterone receptor-mediated transcription by the immunosuppressant FK506. Biochemistry. 1994;33:10666–10671. doi: 10.1021/bi00201a014. [DOI] [PubMed] [Google Scholar]
- 317.Taylor A, Giles K, Sarthy A V, McGonigal T, Fostel J. Identification of the gene encoding DNA topoisomerase I from Candida albicans. FEMS Microbiol Lett. 1996;138:113–121. doi: 10.1111/j.1574-6968.1996.tb08143.x. [DOI] [PubMed] [Google Scholar]
- 318.Tropschug M, Barthelmess I B, Neupert W. Sensitivity to cyclosporin A is mediated by cyclophilin in Neurospora crassa and Saccharomyces cerevisiae. Nature. 1989;342:953–955. doi: 10.1038/342953a0. [DOI] [PubMed] [Google Scholar]
- 319.Turk B E, Su Z, Liu J O. Synthetic analogues of TNP-470 and ovalicin reveal a common molecular basis for inhibition of angiogenesis and immunosuppression. Bioorg Med Chem. 1998;6:1163–1169. doi: 10.1016/s0968-0896(98)00078-9. [DOI] [PubMed] [Google Scholar]
- 320.Tyagi R K, Amazit L, Lescop P, Milgrom E, Guiochon-Mantel A. Mechanisms of progesterone receptor export from nuclei: role of nuclear localization signal, nuclear export signal, and ran guanosine triphosphate. Mol Endocrinol. 1998;12:1684–1695. doi: 10.1210/mend.12.11.0197. [DOI] [PubMed] [Google Scholar]
- 321.Ui M, Okada T, Hazeki K, Hazeki O. Wortmannin as a unique probe for an intracellular signalling protein, phosphoinositide 3-kinase. Trends Biochem Sci. 1995;20:303–307. doi: 10.1016/s0968-0004(00)89056-8. [DOI] [PubMed] [Google Scholar]
- 322.van Dijk M A, Floore A N, Kloppenborg K I, van ter Veer L J. A functional assay in yeast for the human estrogen receptor displays wild-type and variant estrogen receptor messenger RNAs present in breast carcinoma. Cancer Res. 1997;57:3478–3485. [PubMed] [Google Scholar]
- 323.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Vezina C, Kudelski A, Sehgal S N. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 325.Vincent V L, Klig L S. Unusual effect of myo-inositol on phospholipid biosynthesis in Cryptococcus neoformans. Microbiology. 1995;141:1829–1837. doi: 10.1099/13500872-141-8-1829. [DOI] [PubMed] [Google Scholar]
- 326.Viscoli C, Castagnola E, Machetti M. Antifungal treatment in patients with cancer. J Intern Med Suppl. 1997;740:89–94. [PubMed] [Google Scholar]
- 327.von Manteuffel S R, Gingras A-C, Ming X-F, Sonenburg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Wall M E, Wani M C, Nicholas A W, Manikumar G, Tele C, Moore L, Truesdale A, Leitner P, Besterman J M. Plant antitumor agents. 30. Synthesis and structure activity of novel camptothecin analogs. J Med Chem. 1993;36:2689–2700. doi: 10.1021/jm00070a013. [DOI] [PubMed] [Google Scholar]
- 329.Wang J C. DNA topoisomerases. New break for archaeal enzyme. Nature. 1997;386:329–331. doi: 10.1038/386329a0. [DOI] [PubMed] [Google Scholar]
- 330.Wang J C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971;55:523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 331.Weinstein J N, Myers T G, O’Connor P M, Friend S H, Fornace J A J, Kohn K W, Fojo T, Bates S E, Rubinstein L V, Anderson N L, Buolamwini J K, van Osdol W W, Monks A P, Scudiero D A, Sausville E A, Zaharevitz D W, Bunow B, Viswanadhan V N, Johnson G S, Wittes R E, Paull K D. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 332.Wells G B, Dickson R C, Lester R L. Isolation and composition of inositolphosphorylceramide-type sphingolipids of hyphal forms of Candida albicans. J Bacteriol. 1996;178:6223–6226. doi: 10.1128/jb.178.21.6223-6226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Weston S A, Camble R, Colls J, Rosenbrock G, Taylor I, Egerton M, Tucker A D, Tunnicliffe A, Mistry A, Mancia F, de la Fortelle E, Irwin J, Bricogne G, Pauptit R A. Crystal structure of the anti-fungal target N-myristoyl transferase. Nat Struct Biol. 1998;5:213–221. doi: 10.1038/nsb0398-213. [DOI] [PubMed] [Google Scholar]
- 334.Whaley H A, Sebek O K, Lewis C. Production, isolation, characterization, and evaluation of lipoxamycin, a new antifungal agent. Antimicrob Agents Chemother. 1970;10:455–461. [PubMed] [Google Scholar]
- 335.White T C, Marr K A, Bowden R A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Whitesell L, Mimnaugh E G, Costa B D, Myers C E, Neckers L M. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Williams R D, Sgoutas D S, Zaatari G S, Santoianni R A. Inhibition of serine palmitoyltransferase activity in rabbit aorta by l-cycloserine. J Lipid Res. 1987;28:1478–1481. [PubMed] [Google Scholar]
- 338.Wingard J R. Infections due to resistant Candida species in patients with cancer who are receiving chemotherapy. Clin Infect Dis. 1994;19(Suppl.):S49–53. doi: 10.1093/clinids/19.supplement_1.s49. [DOI] [PubMed] [Google Scholar]
- 339.Wingard J R, Merz W G, Rinaldi M G, Johnson T R, Karp J E, Saral R. Increase in Candida krusei infection among patients with bone marrow transplantation and neutropenia treated prophylactically with fluconazole. N Engl J Med. 1991;325:1274–1277. doi: 10.1056/NEJM199110313251803. [DOI] [PubMed] [Google Scholar]
- 340.Wingard J R, Merz W G, Rinaldi M G, Miller C B, Karp J E, Saral R. Association of Torulopsis glabrata infections with fluconazole prophylaxis in neutropenia bone marrow transplants. Antimicrob Agents Chemother. 1993;37:1847–1849. doi: 10.1128/aac.37.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Winzeler E A, Richards D R, Conway A R, Goldstein A L, Kalman S, McCullough M J, McCusker J H, Stevens D A, Wodicka L, Lockhart D J, Davis R W. Direct allelic variation scanning of the yeast genome. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- 342.Withee J L, Mulholland J, Jeng R, Cyert M S. An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol Biol Cell. 1997;8:263–277. doi: 10.1091/mbc.8.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 344.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 345.Wolpert-De Filippes M K. Antitumor activity of cisplatin analogues. In: Prestayko A W, Crooke S T, Carter S K, editors. Cisplatin: current status and new developments. New York, N.Y: Academic Press, Inc.; 1980. pp. 183–191. [Google Scholar]
- 346.Wolter R, Siede W, Brendel M. Regulation of SNM1, an inducible Saccharomyces cerevisiae gene required for repair of DNA cross-links. Mol Gen Genet. 1996;250:162–168. doi: 10.1007/BF02174175. [DOI] [PubMed] [Google Scholar]
- 347.Wong G K, Griffith S, Kojima I, Demain A L. Antifungal activities of rapamycin and its derivatives, prolylrapamycin, 32-desmethylrapamycin, and 32-desmethoxyrapamycin. J Antibiot (Tokyo) 1998;51:487–491. doi: 10.7164/antibiotics.51.487. [DOI] [PubMed] [Google Scholar]
- 348.Woscholski R, Kodaki T, McKinnon M, Waterfield M D, Parker P J. A comparison of demethoxyviridin and wortmannin as inhibitors of phosphatidylinositol 3-kinase. FEBS Lett. 1994;342:109–114. doi: 10.1016/0014-5793(94)80482-6. [DOI] [PubMed] [Google Scholar]
- 349.Wrenn C K, Katzenellenbogen B S. Structure-function analysis of the hormone binding domain of the human estrogen receptor by region-specific mutagenesis and phenotypic screening in yeast. J Biol Chem. 1993;268:24089–24098. [PubMed] [Google Scholar]
- 350.Wu W I, McDonough V M, Nickels J T, Jr, Ko J, Fischl A S, Vales T R, Merrill A H, Jr, Carman G M. Regulation of lipid biosynthesis in Saccharomyces cerevisiae by fumonisin B1. J Biol Chem. 1995;270:13171–13178. doi: 10.1074/jbc.270.22.13171. [DOI] [PubMed] [Google Scholar]
- 351.Wymann M P, Bulgarelli-Leva G, Zvelebil M J, Pirola L, Vanhaesebroeck B, Waterfield M D, Panayotou G. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol. 1996;16:1722–1733. doi: 10.1128/mcb.16.4.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Yang J, Bardes E S, Moore J D, Brennan J, Powers M A, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Yang N N, Venugopalan M, Hardikar S, Glasebrook A. Identification of an estrogen response element activated by metabolites of 17β-estradiol and raloxifene. Science. 1996;273:1222–1225. doi: 10.1126/science.273.5279.1222. [DOI] [PubMed] [Google Scholar]
- 354.Yano H, Nakanishi S, Kimura K, Hanai N, Saitoh Y, Fukui Y, Nonomura Y, Matsuda Y. Inhibition of histamine secretion by Wortmannin through the blockade of phosphatidylinositol 3-kinase in RBL-2H3 cells. J Biol Chem. 1993;268:25846–25856. [PubMed] [Google Scholar]
- 355.Yazdanbakhsh K, Choi J W, Li Y, Lau L F, Choi Y. Cyclosporin A blocks apoptosis by inhibiting the DNA binding activity of the transcription factor Nur77. Proc Natl Acad Sci USA. 1995;92:437–441. doi: 10.1073/pnas.92.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Ye R R, Bretscher A. Identification and molecular characterization of the calmodulin-binding subunit gene (CMP1) of protein phosphatase 2B from Saccharomyces cerevisiae. Eur J Biochem. 1992;204:713–723. doi: 10.1111/j.1432-1033.1992.tb16686.x. [DOI] [PubMed] [Google Scholar]
- 357.Yoshida M, Nishikawa M, Nishi K, Abe K, Horinouchi S, Beppu T. Effects of leptomycin B on the cell cycle of fibroblasts and fission yeast cells. Exp Cell Res. 1990;187:150–156. doi: 10.1016/0014-4827(90)90129-x. [DOI] [PubMed] [Google Scholar]
- 358.Yoshida T, Toda T, Yanagida M. A calcineurin-like gene ppb1+ in fission yeast: mutant defects in cytokinesis, cell polarity, mating and spindle pole body positioning. J Cell Sci. 1994;107:1725–1735. doi: 10.1242/jcs.107.7.1725. [DOI] [PubMed] [Google Scholar]
- 359.Zakian V A. ATM-related genes: what do they tell us about functions of the human gene? Cell. 1995;82:685–685. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- 360.Zheng X-F, Fiorentino D, Chen J, Crabtree G R, Schreiber S L. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 361.Zweerink M M, Edison A M, Wells G B, Pinto W, Lester R L. Characterization of a novel, potent, and specific inhibitor of serine palmitoyltransferase. J Biol Chem. 1992;267:25032–25038. [PubMed] [Google Scholar]