Abstract
Recently, Gagie et al. proposed a version of the FM-index, called the r-index, that can store thousands of human genomes on a commodity computer. Then Kuhnle et al. showed how to build the r-index efficiently via a technique called prefix-free parsing (PFP) and demonstrated its effectiveness for exact pattern matching. Exact pattern matching can be leveraged to support approximate pattern matching, but the r-index itself cannot support efficiently popular and important queries such as finding maximal exact matches (MEMs). To address this shortcoming, Bannai et al. introduced the concept of thresholds, and showed that storing them together with the r-index enables efficient MEM finding—but they did not say how to find those thresholds. We present a novel algorithm that applies PFP to build the r-index and find the thresholds simultaneously and in linear time and space with respect to the size of the prefix-free parse. Our implementation called can rapidly find MEMs between reads and large-sequence collections of highly repetitive sequences. Compared with other read aligners—PuffAligner, Bowtie2, BWA-MEM, and CHIC— MONI used 2–11 times less memory and was 2–32 times faster for index construction. Moreover, MONI was less than one thousandth the size of competing indexes for large collections of human chromosomes. Thus, MONI represents a major advance in our ability to perform MEM finding against very large collections of related references.
Keywords: r-index, run-length-encoded Burrows-Wheeler transform, thresholds, MEM-finding
1. INTRODUCTION
In the past couple of decades, the cost of genome sequencing has decreased at an amazing rate, resulting in more ambitious sequencing projects, including the 100K Genome Project (Turnbull et al., 2018) and the Vertebrate Genome Project (Rhie et al., 2021). Sequence read aligners—such as BWA (Li and Durbin, 2009), Bowtie (Langmead et al., 2009), and SOAP2 (Li et al., 2010)—have been fundamental methods for compiling and analyzing these and other data sets. Traditional read aligners build an index from a small number of reference genomes, find short exact matches between each read and the reference genome(s), and then extend these to find approximate matches for each entire read.
Maximal exact matches (MEMs), which are exact matches between a read R and genome G that cannot be extended to the left or right4, have been shown to be the most effective seeds for alignment of both short reads (Li, 2013) and long reads (Vyverman et al., 2015; Miclotte et al., 2016). Hence, any index used for unbiased alignment should efficiently support finding these MEMs and scale to indexing large numbers of genomes.
In recent years, we have come close to realizing such an index, but some gaps still remain. The FM-index consists of the Burrows-Wheeler transform (BWT) of the input text, a rank data structure over that BWT and the suffix array (SA) sampled at regular intervals. Mäkinen and Navarro (2007) showed how to store the BWT and rank data structure in space proportional to the number r of runs in the BWT, which tends to grow very slowly as we add genomes to a genomic database, and still quickly count how many times patterns occur in the text. Because the product of the size of the SA sample and the time to locate each of those occurrences are at least linear in the size of the input text, Mäkinen and Navarro's index is not a practical solution for alignment.
A decade later, Gagie et al. (2020a) showed how to sample entries of the SA such that locating queries is fast. The combination of their SA sample with Mäkinen and Navarro's rank data structure is called the r-index. However, Gagie et al. did not describe how to build the index—this was shown in a series of articles (Boucher et al., 2019; Kuhnle et al., 2020; Mun et al., 2020), which uses prefix-free parsing (PFP) to preprocess the data in such a way that allows for the BWT and SA samples to be computed from the compressed representation.
Exact pattern matching can be leveraged to support approximate pattern matching, by dividing patterns into pieces and searching for them separately, but the r-index itself cannot find MEMs. We cannot augment the r-index with the same auxiliary data structures that allow standard FM-indexes to find MEMs because, again, the product of their size and the query time grow linearly with the text.
To address this shortcoming of the r-index, Bannai et al. (2020) describe a variant of the r-index that supports MEM-finding. More specifically, it finds the matching statistics of a pattern with respect to the indexed text, from which we can easily compute the MEMs, using a two-pass process: first, working right to left, for each suffix of the query string, it finds a suffix of the text that matches for as long as possible; then, working left to right, it uses random access to the text to determine the length of those matches. We note that this computation of the matching statistics is enabled through the addition of a data structure that they refer to as thresholds. However, Bannai et al. did not say how to find those thresholds efficiently, and until we have a practical construction algorithm, we cannot say we really have a pangenomic index for MEM-finding.
In this article, we show how to use PFP to find the thresholds at the same time as we build the r-index. We refer to the final data structure as MONI, from the Finnish for “multi,” since our ultimate intention is to index and use multiple genomes as a reference, whereas other approaches are to pangenomic (Garrison et al., 2018; Li et al., 2020; Maarala et al., 2020) index models of genomic databases, but not the databases themselves. We compare MONI to PuffAligner (Almodaresi et al., 2021), Bowtie2 (Langmead and Salzberg, 2012), BWA-MEM (Li, 2013), and CHIC (Valenzuela and Mäkinen, 2017) using GRCh37 and haplotypes taken from The 1000 Genomes Project Consortium (2015), and the Salmonella genomes taken from GenomeTrakr (Stevens et al., 2017).
We show PuffAligner is between 1.7 and 4 slower for construction and uses between 3 and 12 times more memory for construction of the index for 32 or more haplotypes of chromosome 19. Bowtie2 is between 7 and 20 times slower for construction and uses between 2 and 15 times more memory for construction for these same data sets. BWA-MEM uses less memory but more time than Bowtie2 for construction. Only MONI and PuffAligner were capable of constructing an index for 1000 haplotypes of chromosome 19 in <24 hours; BWA-MEM and Bowtie2 surpassed this. Moreover, MONI used 21 GB of memory and 1.3 hours for construction of this index, whereas PuffAligner used over 260 GB of memory and 5.2 hours for construction. Finally, the size of the data structure of PuffAligner was 1114 times larger than that of MONI.
Similarly, we show that MONI required significantly less time for construction than Bowtie2 and BWA-MEM, and also used significantly less memory than PuffAligner for construction of indexes of 100 or more Salmonella genomes. Finally, we demonstrate the use of indexing a larger number of genomes by comparing MEM-finding with a single-reference genome, to that of 200 more genomes and show that we find MEMs (of length at least 75) for 4.6% more sequence reads.
We compare MONI with PuffAligner (Almodaresi et al., 2021), Bowtie2 (Langmead and Salzberg, 2012), BWA-MEM (Li, 2013), and VG (Garrison et al., 2018) using GRCh37 and from 10 to 200 complete genome haplotypes also taken from The 1000 Genomes Project Consortium (2015). We show that MONI and VG were the only tools able to build the index for 200 human genomes, in <48 hours, and using up to 756 GB of memory. In terms of wall clock time, PuffAligner is the fastest to index 10 and 20 genomes, MONI is the fastest to index 50 genomes, and VG is the fastest to index 100 and 200 genomes.
In terms of CPU time, MONI is always the fastest with a speedup that ranges from 5.92 to 1.16 with respect to the second fastest method. The peak memory usage of MONI is from 2.93 to 4.41 times larger than the best method. The final data structure size of MONI is at most 1.18 × times larger than the final data structure of VG, and from 2.2 to 12.35 times smaller than the third smallest method. We aligned 611,400,000 100-bp reads from Mallick et al. (2016) against the successfully built indexes and evaluated the query time. PuffAligner is always the fastest, followed by Bowtie2, BWA-MEM, MONI, and VG, while VG and MONI were the methods using the smallest amount of memory.
The rest of this article is laid out as follows: in Section 2, we introduce some basic definitions, review of PFP, the definition of thresholds, and the computation of the matching statistics; in Section 3, we show how to compute the thresholds from the PFP; in Section 4, we present our experimental results; and in Section 5, we discuss future work. For the sake of brevity, we assume that readers are familiar with SAs, BWTs, wavelet trees, FM-indexes, and so on, and their use in bioinformatics. We refer the interested reader to Mäkinen et al. (2015) and Navarro (2016) for an in-depth discussion on these topics.
2. PRELIMINARIES
2.1. Basic definitions
A string of length is a sequence of n characters drawn from an alphabet of size . We denote the empty string as . Given two indices , we use to refer to the string if , while if . We refer to as a substring of S, as the j-th prefix of S, and as the i-th suffix of S. A substring T of S is called proper if . Hence, a proper suffix is a suffix that is not equal to the whole string, and a proper prefix is a prefix that is not equal to the whole string.
2.2. SA, ISA, and the longest common prefix
Given a text S, the suffix array (Manber and Myers, 1993) of S, denoted by , is the permutation of such that is the i-th lexicographically smaller suffix of S. The inverse SA is the inverse permutation of the SA, that is, .
We denote the length of the longest common prefix (LCP) of S and T as . And we define the LCP array of S as , the array storing the values of the LCP between two lexicographically consecutive suffixes of S, that is, and for all .
2.3. BWT, RLBWT, and LF-mapping
The BWT (Burrows and Wheeler, 1994) of the text S, denoted by , is a reversible permutation of the characters of S. It is the last column of the matrix of the sorted rotations of the text S, and can be computed from the SA of S as , where S is considered to be cyclic, that is, . The LF-mapping is a permutation on such that .
We represent the with its run-length-encoded representation , where r is the number of equal character runs of maximal size in the , for example, runs of A's and C's. We write for the character of the i-th run of the , and for its length.
When clear from the context, we remove the reference to the text S from the data structures, for example, we write instead of and instead of .
2.4. Matching statistics and thresholds
The matching statistics of a string R with respect to S are an array of pairs saying, for each position in R, the length of the longest substring starting at that position that occurs in S, and the starting position in S of one of its occurrences. They are useful in a variety of bioinformatic tasks (Mäkinen et al., 2015), including computing the MEMs of R with respect to S.
Definition 1. The matching statistics of R with respect to S are an array of pairs such that: (1) ; and (2) does not occur in S.
Suppose we have already computed and now want to compute . Furthermore, suppose we know the position q in the BWT of (or, equivalently, the position of in ). If , then we can set , and the position in the BWT of is .
Bannai et al. (2020) observed that, if , then we can choose as the position in S of either the last copy of that precedes , or the first copy of that follows , depending on which one of the suffixes of S following those copies of has a longer common prefix with . For simplicity, we ignore here cases in which or is undefined.
They also pointed out that, if we consider moving from immediately after to immediately before , then the length of the LCP of the suffix of S following and the suffix of S following is nonincreasing; the length of the LCP of the suffix of S following and the suffix of S following is nondecreasing. Therefore, we can choose a threshold in that interval between and —that is, between two consecutive runs of copies of —such that if is above that threshold, then we can choose as the position in S of , and we can otherwise choose as the position in S of .
Bannai et al. proposed storing a rank data structure over the of S, and so, we can compute , the entry at the beginning and ending of each run in the BWT and, for each consecutive pair of runs of the same character, the position of a threshold in the interval between them. With these, we can compute from right to left. We note they only said the thresholds exist and did not say how to find them efficiently; consequently, they did not give an implementation.
They also proposed storing a data structure supporting random access to S so that, once we have all the values, we can compute the values, from left to right: if we already have and now we want to compute ; since , we can find by comparing with character by character until we find a mismatch. The number of characters we compare is a telescoping sum over i, so we use random accesses in total. Figure 1 shows an example of the computation of the matching statistics using the algorithm of Bannai et al.
FIG. 1.
An illustration of the thresholds and matching statistics for identifying pattern R in the string S. Shown on the left is pattern R, the longest Prefix of the suffix of the pattern that occurs in S, its length in the matching statistics M, and the position of Prefix in the text. Shown on the right, continuing from left to right, are the , , the thresholds for the characters A, T, and $, the , and all rotations of S, with the LCP between each consecutive rotation highlighted in red. We note that in practice we build the and the entries at the beginning and end of each run in the . For example, we would store for the first run of T's in the . Moreover, the is just shown in red for illustrative purposes. The arrows illustrate the position in the , which each prefix corresponds to. BWT, Burrows/Wheeler transform; LCP, longest common prefix; SA, suffix array.
Examining Bannai et al.'s observations, we realized that we can choose the threshold between a consecutive pair of runs to be the position of the minimum LCP value in the interval between them. This allows us to build Bannai et al.'s index efficiently.
2.5. Prefix-free parsing
Kuhnle et al. (2020) showed how to compute the r-index (i.e., and the SA entries at the starting and ending positions of runs in ) using PFP. For PFP, we parse S into overlapping phrases by passing a sliding window of length w over it and inserting a phrase boundary whenever the Karp-Rabin hashing of the contents of the window is 0 modulo a parameter. This can be done efficiently using only sequential access to S, so it works well in external memory, and it can also be parallelized easily.
We call the substring contained in the window when we insert a phrase boundary a trigger string and we include it as both a suffix of the preceding phrase and a prefix of the next phrase. We treat S as cyclic, so the contents of the sliding window during the last w steps of the parsing are . It follows that: (1) each phrase starts with a trigger string, ends with a trigger string, and contains no other trigger strings; and (2) each character of S appears in exactly one phrase where it is not in the last w characters of that phrase.
The first property means no phrase suffix of length at least w is a proper prefix of any phrase suffix, which is the reason for PFP's name. This and the second property mean that, for any pair of characters and , we can compare lexicographically the suffixes starting at and , again viewing S as cyclic, by (1) finding the unique phrases containing and not in their last w characters, (2) comparing the suffixes of those phrases starting at and ; and (3) if those phrase suffixes are equal, comparing the suffixes of S starting at the next phrase boundaries.
Example 1. Let and . Let be the set of strings where the Karp-Rabin hash is 0 modulo a parameter, therefore the parsing is and the dictionary is , , , , . We note that in this example, the set of proper phrase suffixes of is , and and are the only one of length at least w.
2.5.1. Data structures on parse and dictionary
We let D be the dictionary of distinct phrases and let be the parse, viewed as a string of phrase identifiers. We define to be the text obtained by the concatenation of the phrases of the dictionary, that is, . With a slight abuse of the notation, we refer to as D when it is clear from the context. We build the SA of , the inverse SA of , the LCP array of , and a succinct range minimum query (RMQ) data structure on top of . Our last data structure we compute from D is a bitvector of length , which contains a 1 at the positions in that correspond to the beginning of a phrase. We also provide bD with and support, that is, and return the number of elements equal to q up to position i and select returns the position of the i-th occurrence of q where q is 0 or 1.
Finally, we build the SA of P, the inverse SA of P, and the array of P. This leads to the following result describing the total time and space needed for the construction from P and D. Kuhnle et al. (2020) showed that the construction of and from the dictionary and parse is linear in the size of the parse and dictionary. Moreover, the construction of the remaining structures is also linear (Navarro, 2016). Therefore, it follows that this data structure can be constructed in space and time since each individual structure can be constructed in time and space linear to the size of or P.
2.5.2. Computing the BWT
Suppose we lexicographically sort the distinct proper phrase suffixes of length at least w, and store the frequency of each such suffix in S. For each such phrase suffix , all the characters preceding occurrences of in S occur together in , and the starting position of the interval containing them is the total frequency in S of all such phrase suffixes lexicographically smaller than . It may be that is preceded by different characters in S, because is a suffix of more than one distinct phrase, but then those characters' order in is the same as the order of the phrases containing them in the BWT of P.
These observations allow us to compute from D and P without decompressing them. Kuhnle et al. (2020) showed that computing using PFP as a preprocessing step takes much less time and memory than computing it from S directly since D and P are significantly much smaller than S. The pseudocode for the construction algorithm of Kuhnle et al. is shown in Algorithm 1 in the Supplementary Data.
2.5.3. Random access to the SA
Now suppose, as shown by Boucher et al. (2021), we store a wavelet tree over the BWT of P, with the leaves labeled from left to right with the phrase identifiers in the colexicographic order of the phrases. For any phrase suffix , all the identifiers of phrases ending with appear consecutively in the wavelet tree; therefore, given an integer j and , with the wavelet tree we can perform a 3-side range selection query to find the jth phrase in the BWT of P that ends with . With some other auxiliary data structures whose sizes are proportional to the sizes of D and P, this lets us support random access to SA.
We emphasize that we keep the wavelet tree and auxiliary data structures only during the construction of the r-index or Bannai et al.'s index, and that once we discard them we lose random access to SA. If we want to find , we first determine (1) which such phrase suffix follows in S, and (2) the lexicographic rank j of the suffix of S preceded by , among all the suffixes of S prefixed by , from the cumulative frequencies of the proper phrase suffixes of length at least w. We then use the wavelet tree to find the jth phrase in the BWT of P that ends with . Finally, we use the SA of P to find that phrase's position in S and compute the position of in S.
3. COMPUTING THRESHOLDS, COMPUTING MATCHING STATISTICS, AND FINDING MEMs
In the previous section, we reviewed PFP. In this section, we augment the PFP-based algorithm to compute the and the samples to additionally compute the thresholds. Our description starts with our observation that the threshold positions correspond to the position of a minimum of the array in the interval between the two consecutive runs of the same character. This is our refinement of the definition. Next, we describe how to retrieve the thresholds. We show how to find MEMs from the matching statistics computed using Bannai et al.'s algorithm. Finally, we show implementation details on the threshold construction algorithm.
3.1. Redefining thresholds
Here, we show that Bannai et al.'s thresholds are equivalent to the positions of a minimum value in the interval between two consecutive runs of the same character. Given two suffixes p1 and p2 of S, we let q1 and q2 be their positions in the SA, that is, and . We recall that the length of the LCP between and can be computed as the minimum value of the array of S in the interval , assuming w.l.o.g. that . Let . This insight allows us to rewrite the definition of threshold in terms of values as follows. Given a text S, let and be two consecutive runs of the same character in . From the definition of thresholds, we want to find a position i between positions j and k, such that for all , is larger than or equal to , while for all , is larger than or equal to . This shows that position i is a threshold if the following holds:
assuming that if , that is, i is the position of a minimum value in . This can be summarized by the following observation.
Observation 1 Given text S, let and be two consecutive runs of the same character in . A position is a threshold if it corresponds to the minimum value in .
3.2. Computing thresholds
We can find positions of minima in intervals in the of S, similarly to how we can compute the SA samples, and thus compute Bannai et al.'s thresholds. If we want to find the position of a minimum in , we first check if and are followed in S by the same proper phrase suffix of length at least w. If they are not, we can find the position of the minimum from the array of the proper phrase suffixes of length at least w: since the suffixes of S following and are not prefixes of each other, their LCP is a proper prefix of both of them. The situation is more complicated when and are followed in S by the same proper phrase suffix of length at least w.
First, let us consider the simpler problem of finding the length of the LCP of the suffixes of S following and , using some more auxiliary data structures. From and we can find the phrases containing and in S. Using the inverse SA of P, we find the lexicographic rank of the suffixes of S starting at the next phrase boundaries after and , among all suffixes of S starting at phrase boundaries. Using a range-minimum data structure over all such suffixes of S, we find the length of the LCP of those two suffixes. Finally, we add , the length of the phrase suffixes after and minus the length of their overlaps with the next phrases.
The RMQ mentioned above gives us the position of a minimum in , which could be a much wider interval than . To see why, consider that each of the suffixes of S starting at one of the positions consists of followed by a suffix starting at one of the positions , but not all the suffixes starting at the latter positions are necessarily preceded by . We find the position of a minimum in by filtering out the positions in S that are not preceded by : we find the value t in such that is after the minimum in .
We can do this efficiently using the wavelet tree over the BWT of P: the position of a minimum in corresponds to a certain phrase boundary in S, and thus to the identifier in the BWT of P for the phrase preceding that phrase boundary; we are looking for the next identifier in the BWT of P for a phrase ending with , which we can find quickly because the phrase identifiers are assigned to the leaves of the wavelet tree in a colexicographic order.
3.3. Computing matching statistics and finding MEMs
Given a query string R, we compute the matching statistics of R from the thresholds using the algorithm of Bannai et al. given in Subsection 2.4. Next, it follows that we can find the MEMs for R by storing the nondecreasing values of the lengths of the matching statistics. This is summarized in the following lemma.
Lemma 1. Given input text and query string , let be the matching statistics of R against S. For all , is a maximal exact match of length in S if and only if and .
Proof. First, we show that if is a maximal exact match of length in S then and . Since is an MEM, then there exists a j such that and hence, . Moreover, is the length of the longest prefix of that occurs in S since does not occur in S, that is, . It also holds that does not occur in S. This implies that the length of the LCP of that occurs in S is smaller than , and therefore, .
Next, we prove the other direction. Given , by definition of matching statistics, there exists a j such that and does not occur in S. We also have that does not occur in S. Since then also , implying is a maximal exact match.
3.4. Implementation
We implemented using the bitvectors of the sdsl-lite library (Gog et al., 2014) and their rank and select supports. We used SACA-K (Nong, 2013) to lexicographically sort the parses, and gSACA-K (Louza et al., 2017) to compute the and array of the dictionary. We provide random access to the reference using the practical random access to SLPs of Gagie et al. (2020b) built on the grammar obtained using BigRePair (Gagie et al., 2019). We used the ksw2 (Suzuki and Kasahara, 2018; Li, 2018) library available at https://github.com/lh3/ksw2 to compute the local alignment.
3.4.1. Removing the wavelet tree
In Section 2, for the sake of explanation, we used a wavelet tree to provide random access to , but as shown by Kuhnle et al. (2020), to build , we only need sequential access to . To provide sequential access to , we store an array called inverted list that stores a list for each phrase in P of the sorted occurrences of the phrases in . When processing the proper phrase suffixes in the lexicographic order, if a proper phrase suffix is a suffix of more than one phrase, and is preceded by more than one character, we merge the lists of the occurrences of the phrases that contain in the of P using a heap. Since we want to find the phrases in the of P ending with in order, it is enough to scan the elements of the merged lists in an increasing order.
The wavelet tree is also used to find the position of the minimum in a given interval, that is, to perform an RMQ. First, we store a variation of the array of P, we called , which we define as follows. We let . Next, for all , we let be equal to , where pi and are the positions in of the beginning of the lexicographically i-th and -th phrases of the parse. The array can be computed in -time with a slight modification of the algorithm of Kasai et al. (2001). Next, we build a succinct RMQ data structure from the .
Given the array, the following lemma shows how to compute the values:
Lemma 2. Given the PFP P of with dictionary D, for all , let and be the unique proper phrase suffixes of length at least w of the phrases that and belong to, and let p1 and p2 be the positions of the phrases that and belong to in . Assuming w.l.o.g. that , then
where .
Proof. First, we consider the case where . Since , where is the prefix-free set of proper phrase suffixes of length at least w, then . In the second case, that is, if , . We note that and are suffixes of S starting at phrase boundaries, and hence, their LCP can be computed using as follows. Let . We have to show that .
Since the phrases in P are represented by their lexicographic rank in D, the relative lexicographic rank of suffixes of P is the same as the relative lexicographic rank of their corresponding suffixes on S. Hence, the value of corresponds the minimum value in the interval between the positions in of the suffixes starting with phrases and . Thus, computing the minimum value in is equivalent to computing the minimum value in the interval between the positions in of the suffixes starting with phrases and . Then . We subtract w to the length because the last w character of is the same as the first w character of the phrase following , included in the values of .
We observe that, during the construction of the thresholds, we do not need to answer an arbitrary range of minimum queries, but only those between two equal-letter runs. In addition we are building the sequentially. Hence, while building the , we can store, for each character of the alphabet, the position of the minimum in the interval starting at the last occurrence of the character. We can clearly compute all the values of , while building the , however, this would require to visit, for each proper phrase suffix, all the occurrences of the phrases containing it, in the of P.
This can be avoided by noticing that if a proper phrase suffix is always preceded by the same character, the minimum value in the interval is in the position of the first suffix, because the previous suffix starts with a different proper phrase suffix. For the other values, we use the positions of the phrases in the of P that are computed using the inverted list, and we use the RMQ over the to compute the length of the LCP between two consecutive suffixes of S. We also note that the P, , and are used only during the construction of and the inverted list, and therefore are discarded after this construction.
Algorithm 2 in the Supplementary Data gives the pseudocode for building the thresholds.
4. EXPERIMENTS
We demonstrate the performance of through the following experiments: (1) comparison between our method and general data structures and algorithms that can calculate thresholds, (2) comparison between the size and time required to build the index of and that of competing read aligners, and (3) comparison between and competing read aligners with respect to their alignment performance.
The experiments were performed on a server with Intel(R) Xeon(R) CPU E5-2640 v4 @ 2.40 GHz with 40 cores and 756 GB of RAM running Ubuntu 16.04 (64bit, kernel 4.4.0). The compiler was g++ version 5.4.0 with -O3 -DNDEBUG -funroll-loops -msse4.2 options. The running time was found using the C ++ 11 high_resolution_clock facility, and memory usage was found with the malloc_count tool (https://github.com/bingmann/malloc_count) when available; otherwise, we used the maximum resident set size provided by/usr/bin/time. Where not specified, we refer to wall clock time as runtime. All experiments that either exceeded 24 hours or require more than 756 GB of RAM were omitted from further consideration, for example, chr19.1000 and salmonella.10,000 for gsacak and sdsl. MONI is publicly available at https://github.com/maxrossi91/moni.
4.1. Data sets
We used the following data for our experiments: Salmonella genomes taken from GenomeTrakr (Stevens et al. (2017) and sets of haplotypes from The 1000 Genomes Project Consortium (2015). In particular, we used collections of 50, 100, 500, , , and Salmonella genomes, where each collection is a superset of the previous. We denote these as salmonella.50,.., salmonella.10,000. We created a collection of chromosome 19 haplotypes using the bcftools consensus tool to integrate variant calls from the phase-3 callset into chromosome 19 of the GRCh37 reference.
We did this for sets of 1, 2, 4, 8, 16, 32, 64, 128, 256, 512, and distinct haplotypes, where each set is a superset of the previous. We denote these as chr19.1,.., chr19.1000. Lastly, we repeated this for the whole human genome and obtained sets of 1, 10, 20, 50, 100, and 200 distinct haplotypes, where each set is a superset of the previous. We denote these as HG, HG.10, HG.20, HG.50, HG.100, and HG.200. All DNA characters in the reference besides A, C, G, T, and N were removed from the sequences before construction.
4.2. Competing read aligners
We compare with Bowtie2 (Langmead and Salzberg, 2012) (v2.4.2) and BWA-MEM (Li and Durbin, 2009) (v0.7.17), and to more recent tools that have demonstrated efficient alignment to repetitive text, that is, PuffAligner (Almodaresi et al., 2021) (v1.0.0) and CHIC (Valenzuela et al., 2018) (v0.1). PuffAligner was released in 2020 and compared against deBGA (Liu et al., 2016), STAR (Dobin et al., 2013), and Bowtie2. We build Bowtie2, BWA-MEM, and PuffAligner using the default options, while we build CHIC using the relative Lempel-Ziv parsing method –lz-parsing-method = RLZ with as prefix length of text from which phrases in the parse can be sourced, and we fixed the maximum pattern length to be 100.
We tested CHIC with both BWA-MEM and Bowtie2 as kernel managers, which we denote as chic-bwa and chic-bowtie2. We run all methods using 32 threads, except the construction of the BWA-MEM index where multithreading is not supported.
4.3. Comparison with general data structures: threshold construction
We compare with other data structures that can compute thresholds. First, we compute the thresholds using the minimum value in each run of the , which we build using the construction algorithm of Prezza and Rosone (2019) that is available at https://github.com/nicolaprezza/rlbwt2lcp. We denote this method as bwt2lcp. Next, we compute the thresholds directly from the array computed using gSACA-K (Louza et al., 2017). We denote this method as gsacak. Both methods take as input the text S and provide as output the , the samples of at the beginning and at the end of a run, and the thresholds.
Hence, includes the construction of the PFP using the parsing algorithm of BigBWT (Boucher et al., 2019), while bwt2lcp includes the construction of the and the samples using BigBWT. In both cases, BigBWT is executed with 32 threads, window size , and parameter . We ran each algorithm five times for sets of chromosome 19 up to 64 distinct haplotypes.
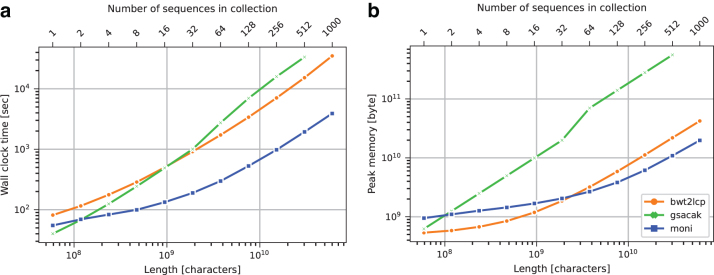
We compare , gsacak, and bwt2lcp with respect to the time and peak memory required to construct the thresholds using the Chromosome 19 data set. Figure 2 illustrates these values. We can observe that is fastest except for very small data sets, that is, chr19.1 and chr19.2, where gsacak is fastest. 's highest speedup is 8.9 × with respect to bwt2lcp on chr19.1000 and 17.3 × with respect to gsacak on chr19.512. bwt2lcp uses the least memory for collections up to 32 sequences, but uses the least for larger collections.
FIG. 2.
Chromosome 19 data set threshold construction running time (a) and peak memory (b).
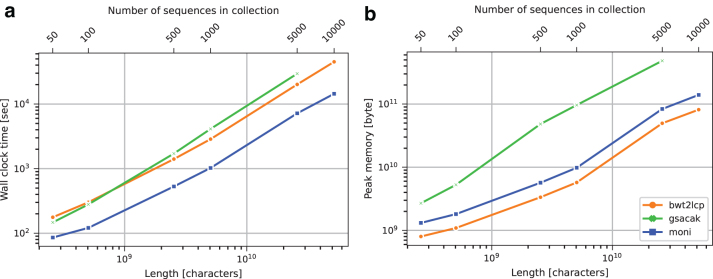
On the Salmonella data set, in Figure 3 we report that is always the fastest with the highest speedup of 4.1 × with respect to gsacak on salmonella.5000 and 3.1 × with respect to bwt2lcp on salmonella.10,000. On the contrary, bwt2lcp always uses less memory than and gsacak. The high memory consumption of on salmonella is mainly due to the size of the dictionary for salmonella, about 16 times larger than the dictionary for chromosome 19.
FIG. 3.
Salmonella data set threshold construction running time (a) and peak memory usage (b).
4.4. MEM-finding in a pangenomic index
Next, we evaluated the degree to which indexing more genomes allowed MONI to find longer MEMs, and thus, more reliable anchors for alignment. We used to build the thresholds for the GRCh37 reference genome (HG), and for GRCh37 and randomly selected haplotypes from The 1000 Genomes Project phase-3 callset, which we denote as HG.i for . Next, we computed the MEMs for 611,400,000 100-bp reads from Mallick et al. (2016) (accession no. ERR1019034_1). Table 1 shows the number of reads having an MEM of length at least 25, 50, and 75.
Table 1.
The Number of Reads Containing a Maximal Exact Match of Minimum Length 25, 50, and 75 for the Reference Genome, 10, 20, 50, 100, and 200 Haplotypes
| MEM | HG |
HG.10 |
HG.20 |
HG.50 |
HG.100 |
HG.200 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of reads | No. of reads | reads | No. of reads | reads | No. of reads | reads | No. of reads | reads | No. of reads | reads | |
| 25 | 411,665,608 | 412,292,276 | 626,668 | 412,383,562 | 91,286 | 412,580,107 | 196,545 | 412,678,277 | 98,170 | 412,818,387 | 140,110 |
| 50 | 309,876,128 | 311,825,333 | 1,949,205 | 311,986,530 | 161,197 | 312,172,012 | 185,482 | 312,298,469 | 126,457 | 312,460,499 | 162,030 |
| 75 | 253,953,551 | 264,406,220 | 10,452,669 | 264,941,235 | 535,015 | 265,311,230 | 369,995 | 265,510,475 | 199,245 | 265,770,839 | 260,364 |
We give the total number of reads that have an MEM (“No. of Reads”) and the number of additional reads that have an MEM (“Reads”). We note that Reads compare the number of MEMs of the current to the next-previous set, that is, HG.100–HG.50.
MEM, maximal exact match.
For larger collections, we also measured the number of additional reads having an MEM of each length with respect to the next-smaller collection (“+Reads”). For example, MONI was able to find over an additional 10,452,669 MEMs of length at least 75 when using the HG.10 collection compared with using HG, an increase of 4.12%. This demonstrates the utility of indexing a set of reference genomes rather than a single genome: reads tend to have longer MEMs, corresponding to longer uninterrupted stretches of genetically identical sequence between the read and a closely related reference in the set.
4.5. Comparison with read aligners: construction space and time
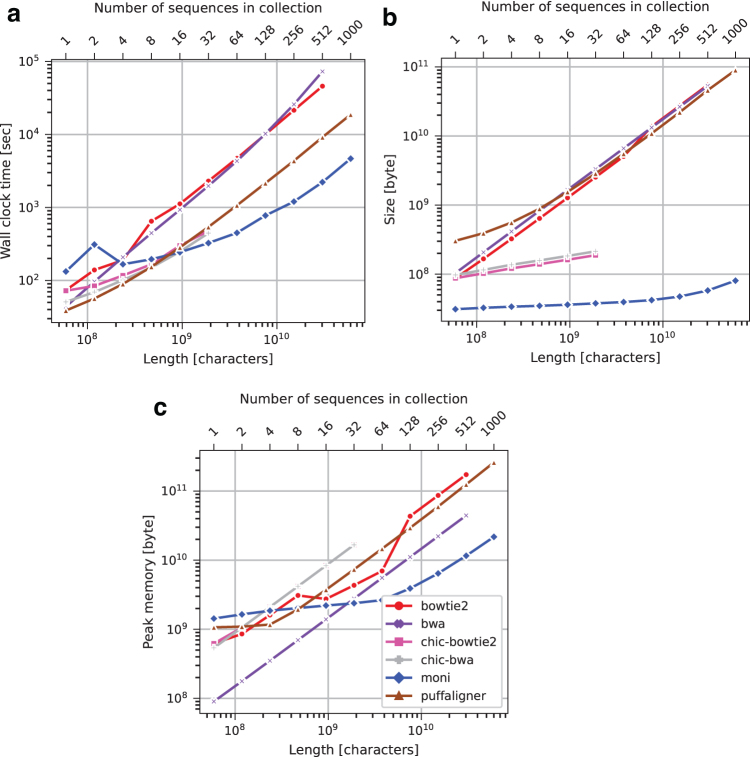
We compare with competing read aligners with respect to the time used for construction, peak memory used for construction, and size of the resulting data structure on disk. Figures 4 and 5 illustrate these comparisons for the human chromosome 19 and Salmonella data sets, respectively. For chromosome 19, is faster than Bowtie2, BWA-MEM, and PuffAligner for 16 or more copies of chromosome 19.
FIG. 4.
Chromosome 19 data set index construction running time (a), index size (b), and peak memory usage (c).
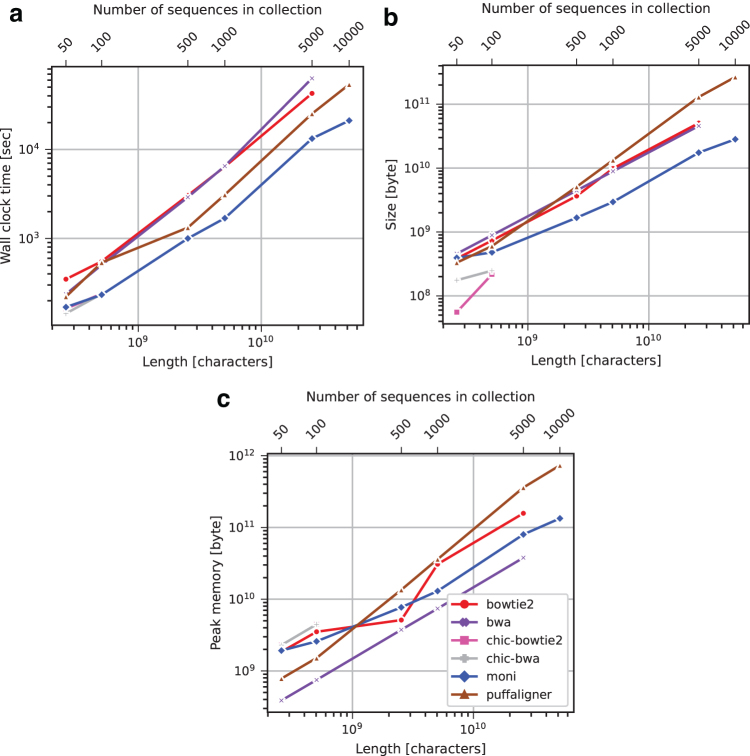
FIG. 5.
Salmonella data set index construction running time (a), index size (b), and peak memory usage (c).
In particular, for 16 or more copies of chromosome 19, is between 1.6 and 4 times faster than PuffAligner, 3.8 and 32.8 times faster than BWA-MEM, and 4.6 and 20.6 times faster than Bowtie2. Bowtie2 and BWA-MEM were only faster than on chr19.1 and chr19.2. For small input (i.e., chr19.1 to chr19.8), there was negligible difference between all the methods, that is, <200 CPU seconds. Bowtie2 and BWA-MEM are not shown for chr19.1000 in Figure 4 because they required over 24 hours for construction. has lower peak memory usage compared with BWA-MEM for more than 32 copies of chromosome 19, with Bowtie2 for more than 8 copies of chromosome 19, and with PuffAligner when the number of copies for chromosome 19 exceeded 8. Bowtie2 used between 1.2 and 14 times more memory than , BWA-MEM used between 1.1 and 3.8 times more, and PuffAligner used between 1.7 and 12 times more.
For small input (i.e., chr19.1, chr19.2, and chr19.8), there was negligible difference between the methods (i.e., <1 GB). In addition, 's data structure was the smallest for all experiments using chromosome 19. The index of Bowtie2, BWA-MEM, and PuffAligner was between 2.8 and 945, between 3.3 and 913, and between 9.8 and 1114 times larger than ours. PuffAligner consistently had the largest index. Although chic-bwa and chic-bowtie had competitive construction time and produced smaller indexes compared with Bowtie2, BWA-MEM, and PuffAligner, they required more memory and produced larger indexes compared with . Moreover, the chic-based methods were unable to index more than 32 copies of chromosome 19; after this point it truncated the sequences.
Results for Salmonella are similar to those for chromosome 19. was faster and used less memory for construction compared with PuffAligner, Bowtie2, and BWA-MEM for all sets of salmonella greater than 50; the one exception is salmonella.500, where Bowtie2 used ∼2 GB less memory for construction. Our final data structure had a consistently smaller disk footprint compared with indexes for BWA-MEM, Bowtie2, and PuffAligner for 100 or more strains of Salmonella. For salmonella.50, the difference in size between , PuffAligner, Bowtie2, and BWA-MEM was negligible.
Although chic-bwa and chic-bowtie were competitive with respect to construction time and size, they truncated the sequences after there were more than 100 strains of Salmonella. Bowtie2 and BWA-MEM are not shown for salmonella.10,000 in Figure 5 because they required over 24 hours for construction.
In summary, was the most efficient with respect to construction time and memory usage for 32 or more copies of chromosome 19. In general, PuffAligner had faster construction time than Bowtie2 and BWA-MEM, but had higher peak memory usage than BWA-MEM. PuffAligner and Bowtie2 had comparable peak memory usage. BWA-MEM had the most competitive peak memory usage to , but had the longest construction time for larger inputs, that is, for 128 or more chromosome 19 haplotypes and 1000 or more strains of Salmonella.
4.6. Comparison with short read aligners: human pangenome
We attempted to index the HG, HG.10, HG.20, HG.50, HG.100, and HG.200 collections using , BWA-MEM, Bowtie2, PuffAligner, and VG. We recorded the wall clock time, CPU time, and the maximum resident set size using/usr/bin/time. All tools that used >48 hours of wall clock time or exceeded a peak memory footprint of 756 GB were omitted from further consideration. BWA-MEM required >48 hours of wall clock time to index HG.20, Bowtie2 required >48 hours of wall clock time to index HG.50, and PuffAligner required >756 GB of main memory to index HG.200.
BWA-MEM, Bowtie2, PuffAligner, and VG all report alignments in SAM format. Although is not a full-featured aligner, we implemented a dynamic programming algorithm to extend MEMs as a fairer comparison. joins MEMs as follows: (1) we compute matching statistics and MEMs for a given read using our index, (2) if the read has an MEM of length at least 25, we extract the MEM plus the 100-bp regions on either side from the corresponding reference, and last, (3) we perform Smith–Waterman alignment (Smith and Waterman, 1981) using a match score of 2, a mismatch penalty of 2, a gap open penalty of 5, and a gap extension penalty of 2. We consider aligning a read with a Smith–Waterman score greater than , where m is the length of the read.
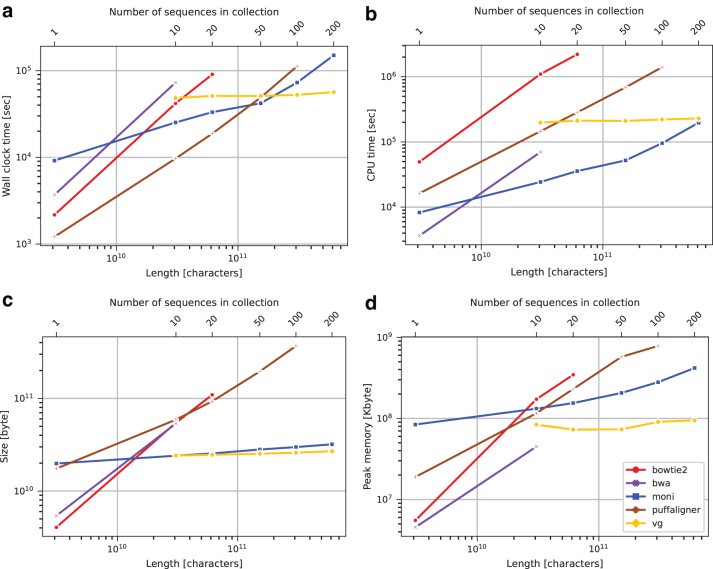
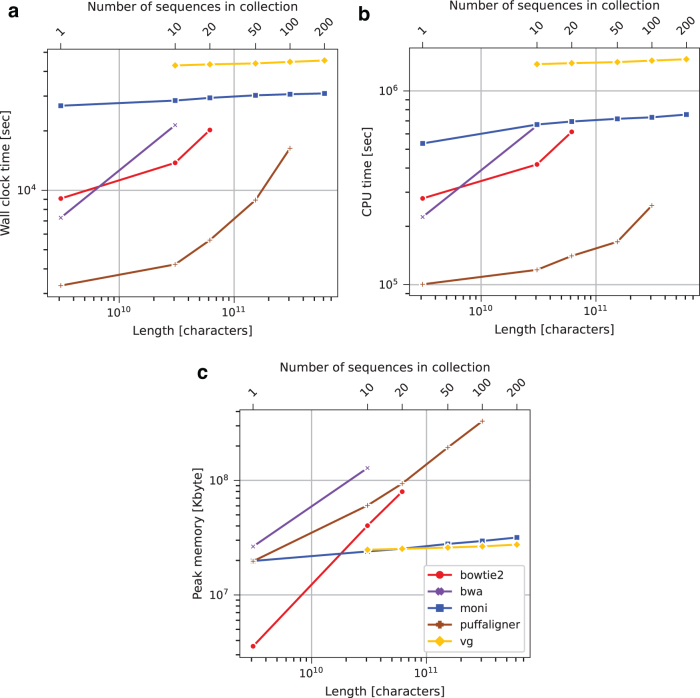
Using the aligners and collections for which we could successfully build an index, we aligned the 611,400,000 reads from Mallick et al. (2016) and measured the wall clock time, the CPU time, and the peak memory footprint required for alignment (Fig. 6). All tools were run using 32 simultaneous threads. All methods successfully built indexes for HG.10. was the fastest tool to build the index when considering the CPU time (7 hours), and the second fastest when considering the wall clock time (6 hours and 45 minutes), after PuffAligner.
FIG. 6.
Human genome data set index construction wall clock time (a), CPU time (b), index size (c), and peak memory usage (d). CPU, central processing unit.
The tools produced indexes ranging in size from 24.06 to 58.91 GB. 's index was the smallest. Peak memory footprint varied from 44.92 to 172.49 GB, with having the fourth smallest (131.83 GB). On queries, BWA-MEM's large footprint (128.22 GB) is likely due to the fact that it was run with 32 simultaneous threads. Wall clock times required ranged from >1 to <12 hours, with being the second slowest (7 hours and 54 minutes). The CPU time required ranged from >1 to >15 days, with being the second slowest (7 days and 18 hours). For the HG.100 collection, only , PuffAligner, and VG were able to build an index, with VG being the fastest to build the index in terms of wall clock time (14 hours and 36 minutes), and was the second fastest (20 hours and 15 minutes). was the fastest when considering the CPU time (1 day and 2 hours).
VG used the smallest amount of peak memory (90.65 GB) and a final data structure size (25.93 GB). used 3.08 times more peak memory (206.25 GB), but the final data structure size is only 3.8 GB larger (29.80 GB). PuffAligner used the largest amount of both peak memory (782.78 GB) and final data structure size (368.22 GB). As shown in Figure 7, PuffAligner performed queries the fastest with respect to both wall time (4 hours and 32 minutes) and CPU time (2 day and 23 hours), but used the largest amount of peak memory (329.18 GB), while was the second fastest on both wall clock (8 hours and 31 minutes) and CPU time (8 days and 10 hours), and used the second smallest amount of memory (29.57 GB). VG was the slowest on both wall clock time (12 hours and 25 minutes) and CPU time (16 days and 14 hours), but used the smallest amount of peak memory (26.56 GB).
FIG. 7.
Human genome query wall clock time (a), CPU time (b), and peak memory usage (c).
For the HG.200 collection, only and VG were able to build an index, with VG being the fastest in terms of wall clock time (15 hours and 41 minutes), and being the fastest in terms of CPU time (2 days and 6 hours). VG used the smallest amount of peak memory (94.65 GB) and had the smallest final data structure size (26.88 GB). On queries, was the fastest (8 hours and 35 minutes), while VG used the smallest amount of peak memory (27.84 GB).
We note that running time heavily depends on the total length of the input reference, while the running time of VG depends on the size of the VCF file representing the multiple aligned sequences. This explains the differences in space and time growth between and VG.
Furthermore, VG requires multiple aligned genomes in input, while does not. Hence, would be able to index also nonmultiple-aligned genomes, for example, a set of long-read assemblies.
4.7. Comparison with read aligners: alignment analysis
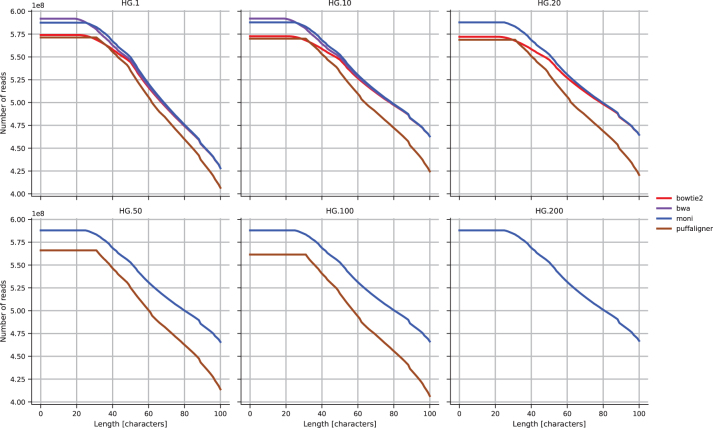
We analyze the results of the alignment of the reads from Mallick et al. (2016) computed in the previous section. In Figure 8 we report, for each length , the cumulative number of reads with the longest match of length at least i that is aligned. We computed the length of the longest match from the CIGAR string and the MD:Z field of the SAM file. The MD:Z field of the SAM file is not available for VG, and hence, it was not possible to include it in this analysis.
FIG. 8.
Human genome data set cumulative number of reads with the longest match of length at least i, for all .
We observe that is the tool having always more reads with a longest match of length at least 26. BWA-MEM has a very similar trend, with having more reads with a longest match from 0 to 25. Bowtie2 has a larger gap with respect to and BWA-MEM, for reads with longest matches from 0 to 50. PuffAligner has always fewer longest matches of length at least 40, than all the other tools. There is an evident increase in the number of reads with longer matches when moving from HG to HG.10.
By increasing the number of genomes from HG.10 to HG.20 in the reference, the curve for and Bowtie2 increases, while for PuffAligner decreases. This trend is preserved increasing even more the number of genomes in the reference. This also demonstrates the importance of indexing a population of genomes, rather than a single genome.
5. CONCLUSION
We described , a new indexing method and software tool for finding MEMs between sequencing reads and large collections of reference sequences with minimal memory footprint and index size. While it is not a full-featured aligner—for example, lacking the ability to compute mapping qualities— represents a major advance in our ability to perform MEM finding against large collections of references. proved to be competitive with graph-based pangenomic indexes such as VG. This is promising toward the possibility to perform MEM finding on long-read assemblies. The next step is to thoroughly investigate how to extend these MEMs to full approximate alignments in a manner that is both efficient and accurate.
As explained in this article, our method hinges on a novel way of computing Bannai et al.'s thresholds with the PFP while simultaneously building the r-index. We conclude by noting that there are possible uses of thresholds beyond sequence alignment and these warrant further investigation. For example, as a by-product of our construction, it is possible to compute the array of the text, which has practical applications in bioinformatics [i.e., single nucleotide polymorphism (SNP) finding (Prezza et al., 2019)].
Supplementary Material
ACKNOWLEDGMENT
The authors thank Nicola Prezza for the code of rlbwt2lcp.
AUTHORS' CONTRIBUTIONS
M.R., T.G., and C.B. conceptualized the idea and developed the algorithmic contributions of this work. M.R. implemented the tool. M.R. and M.O. conducted the experiments. M.R., M.O., C.B., and B.L. assisted and oversaw the experiments and implementation. All authors contributed to the writing of this article.
AUTHOR DISCLOSURE STATEMENT
The authors declare they have no competing financial interests.
FUNDING INFORMATION
M.R., M.O., T.G., B.L., and C.B. are funded by the National Science Foundation NSF IIBR (grant no. 2029552) and the National Institutes of Health (NIH) NIAID (grant no. HG011392). M.R., M.O., and C.B. are funded by NSF IIS (grant no. 1618814) and NIH NIAID (grant no. R01AI141810). T.G. is funded by the NSERC Discovery Grant (grant no. RGPIN-07185-2020).
SUPPLEMENTARY MATERIAL
Formally, given a genome and read , of length is a MEM of R in G if occurs in G and and are not substrings of G.
REFERENCES
- Almodaresi, F., Zakeri, M., and Patro, R.. 2021. Puffaligner: An efficient and accurate aligner based on the pufferfish index. Bioinformatics. Online ahead of print, DOI: 10.1093/bioinformatics/btab408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai, H., Gagie, T., and Tomohiro, I.. 2020. Refining the r-index. Theor. Comput. Sci. 812, 96–108. [Google Scholar]
- Boucher, C., Cvacho, O., Gagie, T., et al. 2021. PFP compressed suffix trees. In: 2021 Proceedings of the Symposium on Algorithm Engineering and Experiments (ALENEX), 60–72. [DOI] [PMC free article] [PubMed]
- Boucher, C., Gagie, T., Kuhnle, A., et al. 2019. Prefix-free parsing for building big BWTs. Algorithms Mol. Biol. 14, 13:1–13:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows, M., and Wheeler, D.J.. 1994. A block sorting lossless data compression algorithm. Technical Report 124, Digital Equipment Corporation. [Google Scholar]
- Dobin, A., Davis, C.A., Schlesinger, F., et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, J., and Heun, V.. 2011. Space-efficient preprocessing schemes for range minimum queries on static arrays. SIAM J. Comput. 40, 465–492. [Google Scholar]
- Gagie, T., Navarro, G., and Prezza, N.. 2020a. Fully functional suffix trees and optimal text searching in BWT-runs bounded space. J. ACM. 67, 2:1–2:54. [Google Scholar]
- Gagie, T., Tomohiro, I., Manzini, G., et al. 2019. Rpair: Rescaling RePair with Rsync. In: Proceedings of the 26th International Symposium on String Processing and Information Retrieval (SPIRE). 35–44.
- Gagie, T., Tomohiro, I., Manzini, G., et al. 2020b. Practical random access to SLP-compressed texts. In: Proceedings of the 27th International Symposium on String Processing and Information Retrieval (SPIRE). 221–231.
- Garrison, E., Sirén, J., Novak, A.M., et al. 2018. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat. Biotechnol. 36, 875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gog, S., Beller, T., Moffat, A., et al. 2014. From theory to practice: Plug and play with succinct data structures. In: Proceedings of the 13th International Symposium on Experimental Algorithms (SEA). 326–337.
- Kasai, T., Lee, G., Arimura, H., et al. 2001. Linear-time longest-common-prefix computation in suffix arrays and its applications. In: Proceedings of the 12th Annual Symposium on Combinatorial Pattern Matching (CPM). 181–192.
- Kuhnle, A., Mun, T., Boucher, C., et al. 2020. Efficient construction of a complete index for pan-genomics read alignment. J. Comput. Biol. 27, 500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg, S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., Trapnell, C., Pop, M., et al. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. [Google Scholar]
- Li, H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34, 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and Durbin, R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler Transform. Bioinformatics. 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Feng, X., and Chu, C.. 2020. The design and construction of reference pangenome graphs with minigraph. Genome Biol. 21, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., Zhu, H., Ruan, J., et al. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Guo, H., Brudno, M., et al. 2016. deBGA: read alignment with de Bruijn graph-based seed and extension. Bioinformatics. 32, 3224–3232. [DOI] [PubMed] [Google Scholar]
- Louza, F.A., Gog, S., and Telles, G.P.. 2017. Inducing enhanced suffix arrays for string collections. Theor. Comput. Sci. 678, 22–39. [Google Scholar]
- Maarala, A.I., Arasalo, O., Valenzuela, D., et al. 2020. Scalable reference genome assembly from compressed pan-genome index with spark. In: Proceedings of the 9th International Conference on Big Data (BIGDATA), 68–84.
- Mäkinen, V., Belazzougui, D., Cunial, F., et al. 2015. Genome-Scale Algorithm Design: Biological Sequence Analysis in the Era of High-Throughput Sequencing. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- Mäkinen, V., and Navarro, G.. 2007. Rank and select revisited and extended. Theor. Comput. Sci. 387, 332–347. [Google Scholar]
- Mallick, S., Li, H., Lipson, M., et al. 2016. The Simons genome diversity project: 300 genomes from 142 diverse populations. Nature. 538, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manber, U., and Myers, G.W.. 1993. Suffix arrays: a new method for on-line string searches. SIAM J. Comput. 22, 935–948. [Google Scholar]
- Miclotte, G., Heydari, M., Demeester, P., et al. 2016. Jabba: hybrid error correction for long sequencing reads. Algorithms Mol. Biol. 11, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun, T., Kuhnle, A., Boucher, C., et al. 2020. Matching reads to many genomes with the r-index. J. Comput. Biol. 27, 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, G. 2016. Compact Data Structures—A Practical Approach. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- Nong, G. 2013. Practical linear-time O(1)-workspace suffix sorting for constant alphabets. ACM Trans. Inform. Syst. 31, 15. [Google Scholar]
- Prezza, N., Pisanti, N., Sciortino, M., et al. 2019. SNPs detection by eBWT positional clustering. Algorithms Mol. Biol. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prezza, N., and Rosone, G.. 2019. Space-efficient computation of the LCP array from the Burrows-Wheeler transform. In: Proceedings of the 30th Annual Symposium on Combinatorial Pattern Matching (CPM). 7, 1–7:18. [Google Scholar]
- Rhie, A., McCarthy, S.A., Fedrigo, O., et al. 2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 592, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.F., and Waterman, M.S.. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197. [DOI] [PubMed] [Google Scholar]
- Stevens, E.L., Timme, R., Brown, E.W., et al. 2017. The public health impact of a publically available, environmental database of microbial genomes. Front. Microbiol. 8, 808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., and Kasahara, M.. 2018. Introducing difference recurrence relations for faster semi-global alignment of long sequences. BMC Bioinform. 19, 33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature. 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull, C., Scott, R.H., Thomas, E., et al. 2018. The 100,000 genomes project: bringing whole genome sequencing to the nhs. Br. Med. J. 361. [DOI] [PubMed] [Google Scholar]
- Valenzuela, D., and Mäkinen, V.. 2017. CHIC: a short read aligner for pan-genomic references. bioRxiv. [Google Scholar]
- Valenzuela, D., Norri, T., Välimäki, N., et al. 2018. Towards pan-genome read alignment to improve variation calling. BMC Genomics. 19, 123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyverman, M., De Baets, B., Fack, V., et al. 2015. A long fragment aligner called ALFALFA. BMC Bioinform. 16, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.