Abstract
Type 1 diabetes in children is heralded by a preclinical phase defined by circulating autoantibodies to pancreatic islet antigens. How islet autoimmunity is initiated and then progresses to clinical diabetes remains poorly understood. Only one study has reported gene expression in specific immune cells of children at risk associated with progression to islet autoimmunity. We analyzed gene expression with RNA sequencing in CD4+ and CD8+ T cells, natural killer (NK) cells, and B cells, and chromatin accessibility by assay for transposase-accessible chromatin sequencing (ATAC-seq) in CD4+ T cells, in five genetically at risk children with islet autoantibodies who progressed to diabetes over a median of 3 years (“progressors”) compared with five children matched for sex, age, and HLA-DR who had not progressed (“nonprogressors”). In progressors, differentially expressed genes (DEGs) were largely confined to CD4+ T cells and enriched for cytotoxicity-related genes/pathways. Several top-ranked DEGs were validated in a semi-independent cohort of 13 progressors and 11 nonprogressors. Flow cytometry confirmed that progression was associated with expansion of CD4+ cells with a cytotoxic phenotype. By ATAC-seq, progression was associated with reconfiguration of regulatory chromatin regions in CD4+ cells, some linked to differentially expressed cytotoxicity-related genes. Our findings suggest that cytotoxic CD4+ T cells play a role in promoting progression to type 1 diabetes.
Introduction
Type 1 diabetes (T1D) in children at genetic risk has a preclinical stage of asymptomatic pancreatic islet autoimmunity characterized by circulating autoantibodies to islet antigens. The rate of progression to clinical T1D varies, but most children with autoantibodies to more than one islet antigen become symptomatic within 10 years (1). While islet autoantibodies remain the most reliable marker of high risk for T1D, there is a clear need for additional biomarkers of preclinical disease and its natural history to improve prediction and facilitate potential prevention strategies. Epigenetic, gene expression, proteomic and metabolomic, serum analyte, and immune cell function studies have the potential to contribute new insights into disease pathogenesis and reveal biomarkers of disease progression. In the analysis of whole blood or peripheral blood mononuclear cells (PBMCs) investigators have identified genes and/or pathways that are altered in children at risk for T1D, including type 1–IFN-regulated genes (2,3) and genes linked to lymphocyte activation (4), natural killer (NK) cell–enriched transcriptional signatures (5), and a ubiquitin-dependent protein degradation pathway (6). However, gene expression in specific immune cells of children at risk has been the subject of only one study, by Kallionpää et al. (7), who identified transcriptional changes in T cells and PBMCs that preceded the appearance of islet autoantibodies. Gene expression alone provides limited insight into gene regulation, which can be deduced with analysis of chromatin structure and its cis- and trans-regulatory elements. We aimed to determine whether gene expression and regulation in specific immune cells could distinguish islet autoantibody–positive children who progressed most rapidly to clinical T1D.
Research Design and Methods
Study Cohort
Characteristics of the subjects in the primary discovery cohort and the validation cohort can be found in Supplementary Table 1. The discovery cohort comprised children and adolescents from one center (Melbourne, Australia) who had participated in A Randomised, Double-blind, Placebo-controlled Trial of Intranasal Insulin (440 IU) in Children and Young Adults at Risk of Type 1 Diabetes: Intranasal Insulin Trial II (INIT II) (clinical trial reg. no. NCT00336674, ClinicalTrials.gov). They had a first-degree relative with T1D and serum autoantibodies to at least two islet antigens, and all were HLA-DR3,4. Five (ages 5.0–13.8, median age 7 years) had progressed to clinical diabetes within 4 years (median 3.0 years) (“progressors”), and five (ages 7.1–13.8 years, median age 10.3 years) had not progressed over 3.1 years (median 2.9 years) (“nonprogressors”). The first blood sample analyzed was taken before treatment allocation in INIT II in the progressors. In both the discovery and validation cohorts, the “at clinical diagnosis” time point corresponds to the onset of clinical T1D in progressors; the criteria for the “earliest preclinical” time point were that samples were taken 1) closest to 3 years before clinical T1D and 2) before treatment allocation in INIT II. Nonprogressors were matched with progressors for sex and age at the time of collection of the earliest preclinical sample. The validation cohort, included for confirmation by quantitative RT-PCR (RT-qPCR) of the results of RNA-seq from the discovery cohort, comprised islet autoantibody–positive, HLA-DR3,4 subjects from several centers (Melbourne, Munich, and Adelaide, Australia) who had participated in INIT II or in an observational study of intestinal permeability (8). Samples for the validation cohort were collected over a period of 3.5 years up until diagnosis in the progressors. PBMCs were recovered from heparinized venous blood on a Ficoll-Hypaque density gradient and stored in liquid nitrogen. HLA-DR genotypes were analyzed as previously described (9). Subjects or parents/guardians gave written informed consent. INIT II was approved by the Human Research Ethics Committee (HREC) of Melbourne Health (2004.260) and the intestinal permeability study (9) by the HREC of the Women’s and Children’s Health Network, Adelaide (HREC/13/WCHN/29).
Islet Autoantibodies
Autoantibodies to insulin (IAA) were measured with a radiobinding assay as previously described (10) and autoantibodies to GAD65 (GADA), tyrosine phosphatase-like insulinoma antigen (IA2A), and zinc transporter 8 (COOH R/W dimer) (ZnT8A) with precipitation of 35S-methionine–labeled recombinant human proteins. Assays for IAA, GADA, IA2A, and ZnT8A scored 25%, 52%, 64%, and 52% for sensitivity and 98%, 100%, 100%, and 98% for specificity, respectively, in the Islet Autoantibody Standardization Program (IASP) 2016 serum exchange workshop. Positive cutoffs for autoantibodies had been defined as IAA ≥0.7 mU/L, GADA ≥5.0 units/mL, IA2A ≥3.0 units/mL, and ZnT8A ≥3.1 units/mL.
Cell Subset Isolation
Thawed PBMCs, ≥92% viable by acridine orange–ethidium bromide staining, were stained with anti-human αβ TCR (clone IP26, cat. no. 46-9986-42; eBioscience), anti-human CD4 (clone RPA-T4, cat. no. 555349; BD Pharmingen), anti-human CD45RA (clone 5H9, cat. no. 556626; BD Pharmingen), anti-human CD25 (clone M-A251, cat. no. 557741; BD Pharmingen), anti-human CD14 (clone 63D3, cat. no. 367104; BioLegend), anti-human CD16 (clone 3G8, cat. no. 557758; BD Pharmingen), anti-human HLA-DR (clone L243, cat. no. 48-9952-42; eBioscience), and anti-human CD19 (clone HIB19, cat. no. 302238; BioLegend). Naive CD4+ T cells (CD14−, CD16−, TCRαβ+, CD4+, CD45RA+, CD25−) and CD8+ T cells (CD14−, CD16−, TCRαβ+, CD4−, CD45RA+, CD25−), B cells (TCRαβ−, HLA-DR+, CD19+), and NK cells (CD14−, CD16+, CD56+) were flow sorted on a FACSAria (BD Biosciences).
Flow Cytometry
PBMCs were stained with different antibody panels (Supplementary Table 5). Intracellular staining was performed after fixation and permeabilization with use of the Foxp3/Transcription Factor Staining Buffer Set (eBioscience). Samples were acquired using with a Cytek Aurora flow cytometer and data analyzed with FlowJo 10.0.6 software (Tree Star, Inc.).
RNA Sequencing
RNA was isolated with the miRNeasy Micro Kit (QIAGEN) and reverse transcribed to cDNA with SuperScript II Reverse Transcriptase (Invitrogen), and libraries were prepared with the TruSeq Stranded Total RNA kit (Illumina) according to the manufacturer’s protocol. Libraries were sequenced on the Illumina NextSeq 500 platform to produce 2 × 75 paired-end reads. All samples were aligned to the human genome, build hg38, with the Rsubread aligner, version 1.32.2 (11). Fragments overlapping human genes were then summarized into counts with Rsubread’s featureCounts function. Genes were identified using the GENCODE annotation of the human genome (Human Release 35).
Chromosome Accessibility
The Omni-ATAC protocol (12) for transposase-accessible chromatin sequencing (ATAC-seq) was used to determine chromosome accessibility.
Differential Gene Expression Analysis
Differential expression was analyzed with edgeR v3.30.3 (13) and limma v3.44.3 (14) software packages. Prior to analysis, the XIST gene and genes unique to the Y chromosome were removed to avoid sex bias, together with all non-protein-coding genes. Each cell type was then analyzed independently, with comparison of progressors to nonprogressors at each time point and for the combined time points. For each cell type, genes were filtered with use of edgeR’s filterByExpr function with default parameters. For B cells, hemoglobin genes were also removed. Compositional differences between samples were then normalized with the trimmed mean of M values method in each case. Each cell type was then analyzed as described in Supplementary Material. Differential gene expression analysis of publicly available data was performed as described in Supplementary Material. For all analyses, genes with a false discovery rate <5% were classified as differentially expressed.
Quantitative RT-PCR
Total RNA from CD4+ T cells was isolated with a miRNeasy Micro Kit (QIAGEN) and 120 ng reversed transcribed with SuperScript III Reverse Transcriptase (Invitrogen). Quantitative real-time PCR (qPCR) was performed in duplicate with use of TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific) and predesigned TaqMan Gene Expression Assays (EOMES, Hs00172872_m1; GZMB, Hs00188051_m1; CST7, Hs00175361_m1; NKG7, Hs01120688_g1, DPYSL4, Hs00428780_m1; GZMH, Hs00277212_m1; GAPDH, Hs02786624_g1, XRRA1, Hs01566298_m1). The expression of any given gene was quantified relative to GAPDH expression with use of the 2-ΔCt method. RNA-sequencing (RNA-seq) data confirmed that the expression of GAPDH did not differ between progressors and nonprogressors across time.
Analysis of the Flow Cytometry and qPCR Data
Normality of data were confirmed with use of the ggqqplot and ggpubr R packages. Flow cytometry and qPCR gene expression data were each analyzed using linear mixed models with the function lmer from the lme4, version 1.1-27 R (15), on the log2-transformed data. The model included the progressor status * time point interaction term and was adjusted for age and batch and a random factor that accounts for the correlation between counts from the same donors across the time. The P value of the interaction term exceeded 0.05 in all the associations computed, indicating that the relationship between progression status and gene expression did not vary across time.
ATAC-seq Data Preprocessing and Peak Calling
Preprocessing of the ATAC-seq data was performed according to the methods of Reske et al., 2020 (16). (Details can be found in Supplementary Material.) Peaks were called on the merged BAM file using Model-based Analysis of ChIP-Seq (MACS2) (v2.1.0) callpeak (with parameters --nomodel, --format BAMPE, --qvalue 0.05). ATAC-seq reads overlapping the peaks were summarized using Rsubread’s featureCounts function. Lowly accessible peaks were filtered out if their average log2 count per million, computed with edgeR’s aveLogCPM function, was negative, resulting in a final set of 31,262 peaks. ATAC-seq was evaluated against standard parameters including the number of total reads, number of mapped reads, number of final usable reads, and fraction of reads in called peaks and transcription start site (TSS) scores (Supplementary Table 6).
Analysis of the Chromatin Accessibility
Differential accessibility analysis was undertaken using edgeR v3.32.1 (13) and limma v3.46.0 (14) software packages. The trimmed mean of M values method was applied to normalize compositional differences between libraries. A mean-dependent trend was fitted to the negative binomial dispersions with the estimateDisp function, and differential accessibility between all cell types was assessed with the likelihood ratio tests in edgeR. As with the differential expression analysis, linear models incorporated a correction for donor age effect. P values were adjusted for multiple testing with the Benjamini-Hochberg method. Peaks with a false discovery rate <5% were defined as differentially accessible regions (DARs).
Enrichment of TF Binding Motifs
We used the Hypergeometric Optimization of Motif EnRichment (HOMER) suite, v4.10 (17), to determine transcription factor (TF) binding motif enrichment within ATAC-seq peaks, using the findMotifsGenome.pl function (with parameters hg38 and –size given).
Footprinting Analysis
The TOBIAS package (18) was used to analyze footprinting signatures from the ATAC-seq data. Merged BAM files from progressors and nonprogressors were processed with ATACorrect, footprint scores calculated with FootprintScores, and differential binding analysis performed with BINDetect. TFs with absolute differential binding scores >0.075 and P values >0.0001 were defined as differentially bound.
Annotation of ATAC-seq Peaks
ATAC-seq peaks were annotated as described in Supplementary Material.
Visualization of Data
Plots were generated in R and ggplot. Multidimensional scaling (MDS) plots were constructed with limma’s plotMDS function applied to the filtered and normalized log2-counts per million values for each library. limma’s removeBatchEffect function was used to adjust for the effect of the age and batch in the data. ATAC-seq and RNA-seq coverage was plotted with the plotBedgraph function of the Sushi version 1.22.0 R package.
Data and Resource Availability
Processed RNA-seq and ATAC-seq data generated for this study are available from the National Center for Biotechnology Information Gene Expression Omnibus as GEO series GSE185191 (reviewer token: gxgnokoobrsjjab). Raw data are available on request, subject to approval by our institutional Data Access Committee (dataaccess@wehi.edu.au) to ensure preservation of subject confidentiality. The codes used for the analysis of RNA-seq data from CD4+ T cells are available at https://bioinf.wehi.edu.au/resources/BediagaT1D2021/.
Results
A Cytotoxicity-Related Gene Expression Signature in CD4+ T Cells of Progressors
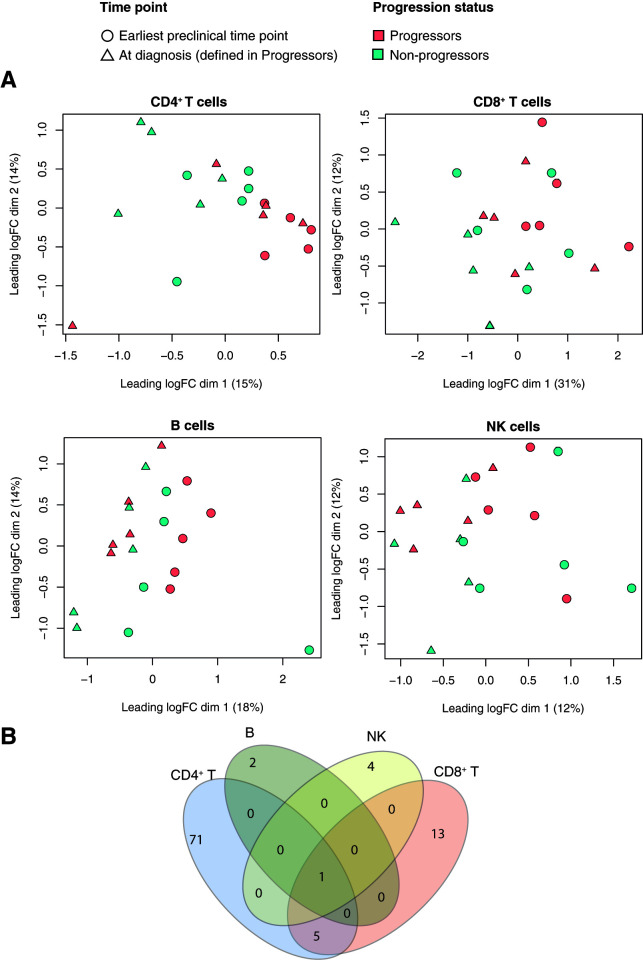
To define gene expression changes associated with progression from islet autoantibody positivity to clinical T1D, we performed RNA-seq analysis of CD4+ T cells, CD8+ T cells, NK cells, and B cells in a discovery cohort of 10 genetically at risk children and adolescents with autoantibodies to two or more islet antigens, 5 of whom progressed to clinical T1D (Supplementary Table 1A). Progressors and nonprogressors were matched for age, sex, and HLA (DR3,4). MDS of the gene expression data showed that samples clustered by progression status over dimension 1 in CD4+ T cells but not in CD8+ T, NK, or B cells (Fig. 1A). Distances correspond to the leading fold change, i.e., the average log2-fold change for the most divergent 500 genes between each pair of samples. For each cell type, we compared gene expression between progressors and nonprogressors. This yielded 77, 19, 5, and 3 differentially expressed genes (DEGs) in CD4+ T, CD8+ T, NK, and B cells, respectively (Fig. 1B and Supplementary Table 2). Six DEGs were shared between CD4+ and CD8+ T cells; four (GPD1L, NTPCR, MANBA, GLIS3) decreased, and two (XRRA1, PNMA8A) increased in progressors and only one (XRRA1) that increased in progressors was common to the four cell types (Fig. 1B). Pathway analysis of the upregulated DEGs in CD4+ T cells with the Gene Ontology (GO) database revealed enrichment for immune system–associated GO terms including “cytolysis” (GO:0019835), “cytolytic granule” (GO:0044194), “cell killing” (GO:0001906), “granzyme-mediated apoptotic signaling pathway” (GO:0008626), “cell activation” (GO:0001775), “immune effector process” (GO:0002252), and “leukocyte mediated immunity” (GO:0002443), among others. DEGs in the other cell types did not show enrichment for any pathway. To further characterize identified DEGs, we compared the CD4+ T cell signature with predefined modules of immune genes previously constructed, based on coexpression between relevant marker genes (cell surface and TF genes) and all other genes in whole blood and/or purified cells (19). Genes overexpressed in progressors were enriched for those in “cytotoxic” modules. These include the module of genes associated with EOMES (EOMES.mod), the master TF regulator of cytotoxicity in CD8+ and CD4+ T cells (20,21), as well as other modules that share significant numbers of genes with EOMES.mod (GZMA.mod, FASLG.mo, GNLY.mod, KLRD1.mod, NKG7.mod, PRF1.mod, GZMB.mod, KIR2DL1.mod, KIR2DS4.mod, NCAM1.mod, GZMM.mod, RUNX3.mod, CD160.mod, CD244.mod, and IL2RB.mod gene modules; hypergeometric P value <1E−03) (Fig. 2A [genes with asterisks in the heat map]). Moreover, upregulated DEGs in progressors were also enriched for target genes of EOMES (eomesodermin) (21) (hypergeometric P value = 0.0005, top-ranked 200 EOMES targets) (Fig. 2A, genes in boldface type in the heat map). Last, when we compared this signature to single-cell RNA profiles of different subsets of CD4+ T cells (central memory [CD45RA−, CCR7+], effector memory [CD45RA−, CCR7−], naive [CD45RA+, CCR7+], and T effector memory T cells reexpressing CD45RA, termed TEMRA cells [CD45RA+, CCR7−] [22]), we found that upregulated DEGs in CD4+ T cells from progressors were enriched for genes that characterize TEMRA cells (fry test P value < 4E-04) (Supplementary Fig. 1), which are known to have a cytotoxic phenotype.
Figure 1.
Transcriptional changes in four immune cell types associated with progression to clinical T1D. A: MDS plots of RNA-seq log2 counts per million values with samples colored by progression status and shaped by time point for CD4+ T, CD8+ T, NK, and B cells. The log2 counts per million values are corrected for the age and batch variables. B: Venn diagram showing the number of overlapping DEGs between progressors and nonprogressors in the cell types analyzed. logFC, log2-fold change. dim, dimension.
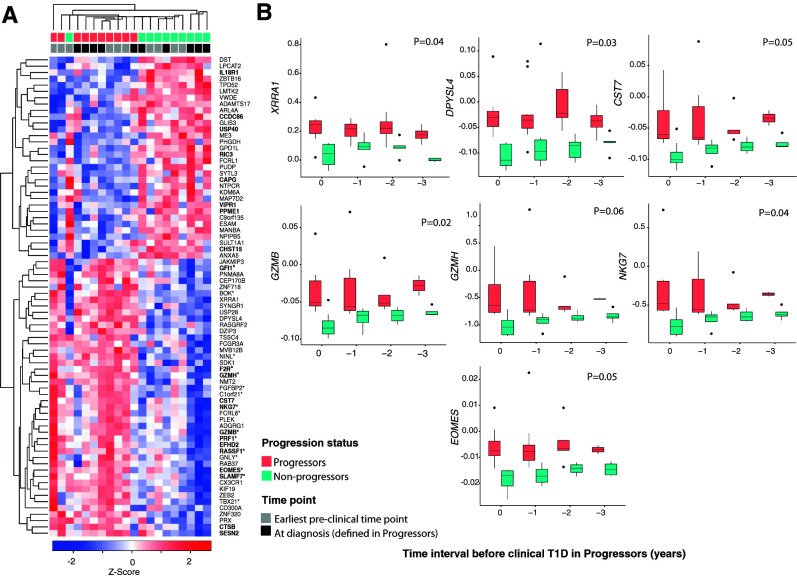
Figure 2.
Transcriptional changes in CD4+ T cells associated with progression to clinical T1D. A: Heat map of genes differentially expressed in CD4+ T cells, determined by RNA-seq, between progressors (n = 5) and nonprogressors (n = 5) in the discovery cohort at the earliest preclinical and at clinical diagnosis time points. The z score below the heat map represents the number of SD units a specific data point is from the mean (derived from the entire set of genes expressed in CD4+ T cells). Red and blue indicate increased and decreased expression, respectively. Samples and genes were arranged according to the ward.D2 clustering method with the function coolmap in the limma package. Genes with an asterisk denote cytotoxicity-related genes (19), while those in boldface type are EOMES target genes (21). Samples are color coded by progression status and time point. B: Box plots showing corrected (for age and batch) expression by RT-qPCR of DEGs over time in the validation cohort (n = 24). The x-axis shows time intervals measured in years from time 0 (clinical T1D in progressors). Number of samples tested in each time point is as follows: time 0, 9 nonprogressors and 8 progressors; time −1, 7 nonprogressors and 10 progressors; time −2, 6 nonprogressors and 8 progressors; and time −3, 5 nonprogressors and 4 progressors. Shown are the median (central horizontal line), interquartile range (boxes), values of the upper and lower quartiles (whiskers), and outliers beyond 1.5 interquartile range (filled circles). Statistical comparisons were performed using linear mixed models and included the progressor status * time point interaction term and adjustment for age, batch, and donor identifier as a random factor.
To compare gene expression changes in preclinical and clinical T1D, we performed differential expression analysis of samples drawn a median of 3 years before (“earliest preclinical” T1D) and at the time of clinical diagnosis (“at clinical diagnosis”) in progressors. At the earliest preclinical time point, 37 and 2 DEGs were identified in CD4+ and CD8+ T cells, respectively, (Supplementary Table 3), of which 11 (CX3CR1, XRRA1, GZMH, DPYSL4, NTPCR, NMT2, JAKMIP3, GPD1, SYNGR1, EFHD2, and BOK) and two (XRRA1, ROBO1), respectively, remained differentially expressed at clinical diagnosis (Supplementary Table 4). The latter therefore represent potential disease progression biomarkers.
It is important to note that the changes in gene expression we observed were not related to treatment in INIT II. Gene expression in samples taken both at the earliest preclinical and clinical diagnosis time points did not differ between subjects in the placebo and nasal insulin arms of INIT II (Supplementary Fig. 2).
Validation of the Cytotoxicity-Related Gene Signature in Progressors
By RT-qPCR of RNA from CD4+ T cells, we validated the RNA-seq findings in a semi-independent cohort of genetically at risk children with autoantibodies to two or more islet antigens (13 progressors and 11 nonprogressors, including the 5 progressors and 5 nonprogressors from the discovery cohort) (Supplementary Table 1B), sampled at diagnosis and 1 to ≥4 years before diagnosis. Genes selected for validation were XRRA1 and DPYSL4 (shared DEGs between earliest preclinical and at clinical diagnosis time points) and CST7, GZMB, GZMH, NKG7, and EOMES (cytotoxicity-related DEGs). All were confirmed as differentially expressed in progressors compared with nonprogressors over the period of observation (Fig. 2B).
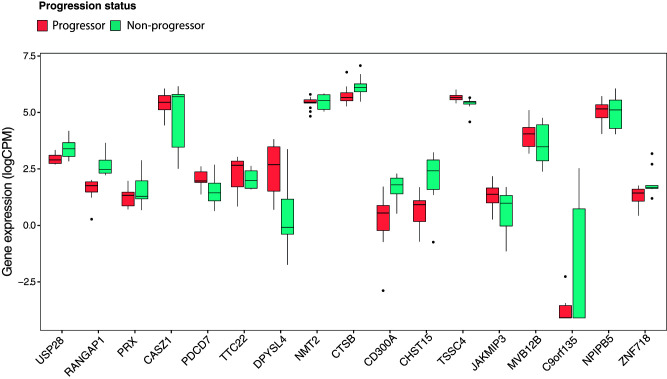
Subjects in the discovery and validation cohorts had all seroconverted. To investigate whether genes differentially expressed in progressors might be altered before the appearance of islet autoantibodies we took advantage of the RNA-seq data of Kallionpää et al. (7) (accession no. EGAS00001004071), from PBMCs and T cells of 14 children with HLA DR and DQ risk alleles for T1D, followed from 3 months to 3 years of age, one-half of whom seroconverted and progressed to clinical T1D. In reanalyzing their data set with our informatic pipelines (see research design and methods), we observed that 20% (n = 17) of our progression-associated genes were differentially expressed when progressors were compared with nonprogressors before seroconversion (Fig. 3). This suggests that the genes that predispose to islet autoimmunity may not necessarily be the same as those that promote subsequent progression to clinical T1D. The overlapping genes included CASZ1 and CD300A. CASZ1, which was downregulated in progressors in our data set, is a conserved zinc-finger TF reported to restrain the expression of some of the genes that drive Th1 lineage commitment in the mouse (23). CD300A, which was upregulated in progressors, is a member of the CD300 glycoprotein family of leukocyte surface proteins on Th1 cells that upregulate EOMES after stimulation (24). Down- and upregulation of CASZ1 and CD300A, respectively, could therefore favor Th1 lineage differentiation during disease progression.
Figure 3.
Differential expression of genes in progressors in relation to seroconversion. A: Box plots showing normalized (log2 counts per million [logCPM]) expression values by RNA-seq in total CD4+ T cells from the Kallionpää et al. (7) data set (n = 14). DEGs plotted represent genes differentially expressed between progressors and nonprogressors to T1D both before and after seroconversion. Preseroconversion DEGs were defined using data from Kallionpää et al. (7), while postseroconversion DEGs were from the current study. Shown are the median (central horizontal line), interquartile range (boxes), values of the upper and lower quartiles (whiskers), and outliers beyond 1.5 interquartile range (circles).
A Cytotoxic CD4+ T Cell Phenotype in Progressors
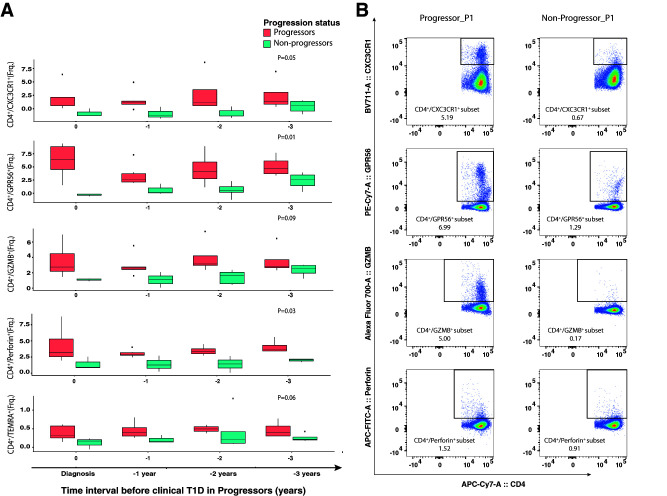
Because progression to clinical T1D was associated with differential expression of cytotoxicity-related genes, we sought to confirm their protein expression in CD4+ T cells. Antibodies were available for four proteins of differentially expressed cytotoxicity-related genes (Supplementary Table 5). Additional phenotype-defining markers included CD4, CD8, CCR7, CD28, CD62L, and CD45RA (Supplementary Table 5). Progressors had a higher frequency of CX3CR1+, GPR56+, GZMB+, and perforin+ CD4+ T cells than nonprogressors over the period of observation (Fig. 4). CD4+ TEMRA cells were also present at higher frequency at all time points in progressors (Fig. 4) and displayed higher expression of cytotoxicity-related proteins in progressors (Supplementary Fig. 3). Analogous to RNA expression, the cytotoxicity-related proteins were not differentially expressed between progressors and nonprogressors in CD8+ T cells. Thus, progression to T1D was associated with expansion of CD4+ T cells with a cytotoxic phenotype.
Figure 4.
Expression of cytotoxicity-related proteins in CD4+ T cells during progression to clinical T1D. A: Box plots showing the frequency (Frq.) of CD4+/CXC3CR1+, CD4+/GPR56+, CD4+/GZMB+, and CD4+/perforin+ measured by flow cytometry in progressors (n = 5) and nonprogressors (n = 5) over the period of observation. The x-axis shows time intervals measured in years from time 0 (clinical T1D in progressors). Shown are median (central horizontal line), interquartile range (boxes), values of the upper and lower quartiles (whiskers), and outliers beyond 1.5 interquartile range (circles). Statistical comparisons were performed using linear mixed models that included the progressor status * time point interaction term and were adjusted for age, batch, and donor identifier as a random factor. B: Flow cytometry data for CD4 vs. CX3CR1, CD4 vs. GPR56, CD4 vs. GZMB, and CD4 vs. perforin (after gating on CD3+ and CD4+ T cells) from a representative progressor and nonprogressor. APC, allophycocyanin; PE, phycoerythrin.
Changes in Chromatin Accessibility in Progressors
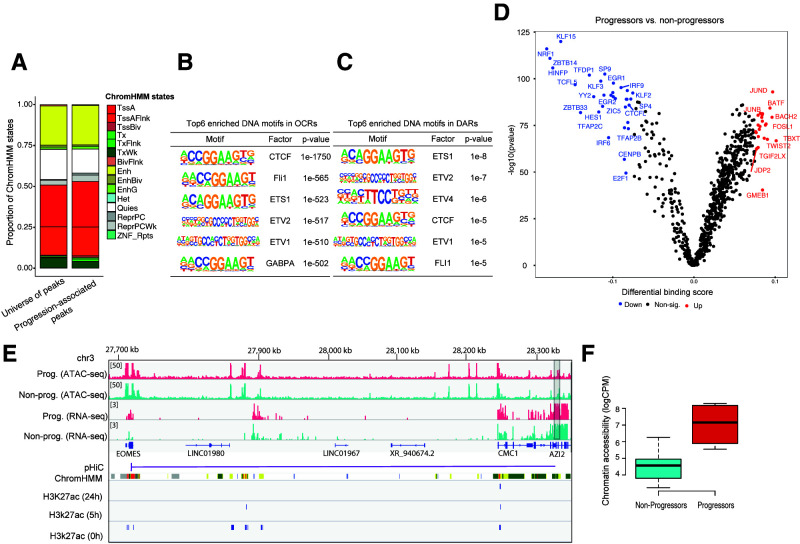
In samples taken at the earliest preclinical time point, we used ATAC-seq to profile chromatin accessibility and identify regulatory elements associated with the transcriptional changes we observed in CD4+ T cells. As far as we are aware, this is the first chromatin accessibility mapping in immune cells from individuals with preclinical T1D. The quality of the ATAC-seq data was demonstrated by the fragment size distribution and clear nucleosomal periodicity, as well as by the strong enrichment of ATAC-seq reads around TSS and by other standard parameters (Supplementary Table 6). Annotation of the ATAC-seq peaks using the ChromHMM states (25) from the Roadmap Epigenomics Project, as described in Supplementary Material, revealed that these open chromatin regions (OCRs) were heavily enriched for regulatory regions, with 43% of the OCRs at predicted active TSS or flanking TSS regions (compared with 1.3% genome wide) and 24% at predicted enhancers (compared with 2.7% genome wide) (Fig. 5A). OCRs were also enriched for TF binding sites (TFBS) recognized by TFs involved in regulation of chromatin architecture (CTCF, CTCFL) and T cell function (ETS1, FLI1, ERG, GABPA, EHF, ELK1, ELK4, RUNX, EOMES, and T-bet, among others) (Fig. 5B and Supplementary Table 7A). Of the 1,004 T1D single nucleotide polymorphisms (SNPs) in the GWAS catalog (Supplementary Material), 191 were located in OCRs, which is more than expected (binomial test, P < 2.2 10 × 10−6).
Figure 5.
Chromatin accessibility changes in CD4+ T cells associated with progression to clinical T1D. A: Bar plots showing the distribution of predicted chromatin states (using the ChromHMM states from the Roadmap Epigenomics Project) in both the complete set of OCRs and progression-associated DARs. BivFlnk, flanking bivalent TSS/enhancer; Enh, enhancers; EnhBiv, bivalent enhancer; EnhG, genic enhancers; Het, heterochromatin; Non-sig, nonsignificant; TssA, active TSS; TssAFlnk, flanking active TSS; TssBiv, bivalent/poised TSS; Tx, strong transcription; TxFlnk, transcription at gene 50 and 30; TxWk, weak transcription; Quies, quiescent/low; ReprPC, repressed polycomb; ReprPCWk, weak repressed polycomb; ZNF/Rpts, ZNF genes and repeats. B: Top six enriched DNA motifs in OCRs. C: Top six enriched DNA motifs in DARs. D: Pairwise comparison of TF activity between progressors and nonprogressors. The volcano plot shows the differential binding score against the −log10 (P value) (from TOBIAS) of all analyzed TF motifs, where each dot represents one motif. TFs with increased occupancy in progressors are labeled in red, while those with decreased occupancy are labeled in blue. E: Normalized read coverage plots of ATAC-seq and RNA-seq libraries around EOMES loci in both progressors (Prog.) (red) and nonprogressors (Non-prog.) (turquoise). The DAR is indicated by a gray rectangle at the right-hand side. pHiC shows promoter-enhancer interactions in primary CD4+ T cells predicted by pHiC (26) overlapping the DAR. ChromHMM shows ChromHMM tracks from Roadmap Epigenomics Project for “Primary T helper naive cells from peripheral blood” (E038). F: Box plot showing normalized (log2 counts per million [logCPM]) chromatin accessibility data (nonadjusted for age) in DAR linked to EOMES.
In comparing progressors and nonprogressors, we identified a total of 211 DARs in progressors (Supplementary Table 8), the majority of which (191 of 211) increased in accessibility. Annotation of the DARs for ChromHMM states and SNPs revealed modest enrichment for active promoters (binomial test, P value < 0.05) (Fig. 4B), while no further enrichment was observed for GWAS SNPs with respect to the OCRs. Results of analysis of the DARs using HOMER showed enrichment of TFBS for ETS1, ETV2, ETV4, CTCF, ETV1, FlI1, GABPA, and ERG (Fig. 5C and Supplementary Table 8B [q value <0.05]), while no enrichment was observed for EOMES or T-bet. With TF binding analysis using TOBIAS we identified 79 differentially bound TFs in progressors including JUND, JUNB, BACH2, and BATF (up) and NRF1, KLF15, EGR1, and IRF9 (down) (Fig. 5D). EOMES and T-bet did not show differential binding between progressors and nonprogressors.
To focus on potential regulatory functions for DEGs, we selected DARs within 250 kb of a TSS from a DEG. Of the 211 DARs, four spanned the TSS of SLAMF7, CAPG, GNLY, and CX3CR1. By using promoter-enhancer interactions previously identified in CD4+ T cells by promoter-capture Hi-C (pHiC) (26), we confirmed two of these interactions, with SLAMF7 and CAPG (Supplementary Fig. 4), and identified a new one, namely, EOMES (Fig. 5). Interestingly, these three potential “promoter-interacting” DARs also overlapped enhancers predicted by ChromHMM or the H3K27ac histone marks (27) (Fig. 5 and Supplementary Fig. 4). Together, these findings indicate that progression to T1D is associated with a modest reconfiguration of the chromatin at regulatory regions and that some DARs are linked to cytotoxicity-related DEGs.
Discussion
We characterized gene expression by RNA-seq in naive CD4+ and CD8+ T cells, NK cells, and B cells from islet autoantibody–positive, HLA-DR3,4 children 1–4 years before and at the time of clinical diagnosis in five progressors and five age- and sex-matched nonprogressors. Changes in gene expression across time and between progressors and nonprogressors were most apparent in CD4+ T cells. DEGs were validated by RT-qPCR of RNA from CD4+ T cells of 24 multiple islet autoantibody–positive HLA-DR3,4 children also followed over time. Previous genome-wide studies investigating T1D-specific gene expression in immune cells have mostly been restricted to bulk whole blood and PBMCs in both clinical (28–30) and preclinical (2–5,31) disease. The interpretation of such studies is confounded by cellular heterogeneity within PBMCs and genetic heterogeneity between individuals. The exceptions are the studies of Irvine et al. (32) in monocytes, Kallionpää et al. (7) in total CD4+ and CD8+ T cells, Vecchio et al. (33) in neutrophils, and Terrazzano et al. (34), Ferraro et al. (35), and Jailwala et al. (36) in different types of regulatory CD4+ T cells. Other studies have used single-cell transcriptomics to characterize different immune cells in at risk or recently diagnosed individuals (7,37–39). However, among these bulk and single-cell studies, only Kallionpää et al. (7) investigated longitudinal T cell transcriptomes in genetically at risk individuals before and after seroconversion. In reanalyzing the data of Kallionpää et al., we observed that only a minority of DEGs associated with progression after seroconversion were also detected in their preseroconversion data set. Moreover, differential expression analysis of our data sets separately at the preclinical and clinical stages of T1D revealed that only 11 of the 37 genes at the earliest preclinical time point were also differentially expressed at clinical diagnosis. This suggests that genes that predispose to islet autoimmunity may not necessarily be the same as those that promote subsequent progression to clinical T1D, with the caveat that we and Kallionpää et al. (7) studied different subjects and T cell subtypes. Furthermore, whether the changes we observed in gene expression are “intrinsic” to progressors remains unclear. Further studies of much larger numbers of children followed from birth will be required to discern the relative contributions of genes and environment to differential gene expression and rates of progression to seroconversion and then clinical T1D.
The CD4+ T cell gene signature in progressors was enriched for cytotoxicity-related genes GZMH, GZMB, GZMA, NKG7, FCRL6, SLAMF7, CX3CR1, and PRF1, among others. The progression signature was also enriched for previously reported EOMES-associated transcriptional profiles (19). Furthermore, the T-box family TFs, T-bet, and EOMES, master regulators of cytotoxicity in both CD8+ and CD4+ T cells, bind to the promoters of GZMB, NKG7, PRF1, EOMES, EFHD2, F2R, and SLAMF7 (20,21). These cytotoxicity-related genes were strongly upregulated in progressors, suggesting that the transcriptional reprograming of EOMES and TBX21 during progression to T1D might mediate the observed upregulation of cytotoxicity-related genes. Moreover, flow cytometric profiling showed that progression to T1D was associated with an increase in the frequency of CD4+ T cells expressing cytotoxic markers. Expansion of circulating EOMES+ CD4+ cytotoxic T cells was observed in multiple sclerosis during progression from relapsing-remitting to secondary progressive disease, and the pathologic significance of these cells was inferred by their presence in the brain lesion (40). The 1858T PTPN22 gene variant, linked to several autoimmune diseases including T1D (41), has been associated with a higher frequency of EOMES+ CD4+ T cells (42). Furthermore, EOMES risk variants themselves have been associated with the autoimmune diseases rheumatoid arthritis (43) and multiple sclerosis (44). Altogether our findings indicate that progression from islet autoantibody positivity to clinical T1D is associated with an expansion of cytotoxic CD4+ T cells that appears to be driven by EOMES.
CD4+ T cells with cytotoxic potential have been described for several decades (45,46). They circulate at very low numbers in healthy humans but are markedly expanded in the context of chronic infection, particularly viral (45–47), within tumors (48), and in autoimmune diseases including rheumatoid arthritis (49) and multiple sclerosis (50–52). These studies, with some exceptions (47,48), defined cytotoxic phenotypes, not cytolytic function. This is a limitation of the current study as well. However, based on the expression of mediators of cytotoxicity, granzyme B and perforin, it is reasonable to assume that more rapid progression to clinical T1D is reflected by expansion of CD4+ T cells that are cytolytic. Human cytotoxic CD4+ T cells are reported to be enriched in TEMRA cells, defined as CD3+CD4+CD45RA+CCR7− (22), believed to be induced by chronic antigen stimulation (53). An increase in CD4+ and CD8+ T cells with the TEMRA phenotype was reported in individuals with long-standing T1D and attributed potentially to chronic stimulation by persisting virus or by autoantigens (54). The antigen specificity of cytotoxic CD4+ T cells in T1D will be important to address in future studies. Our findings suggest that their induction may occur early in the development of T1D, in which case their monitoring may help identify children at high risk of progression to clinical T1D.
To investigate whether progression from islet autoimmunity to clinical T1D is associated with reconfiguration of the chromatin landscape, particularly around EOMES and T-bet DNA motifs, we analyzed genome-wide chromatin accessibility in CD4+ T cells. In progressors compared with nonprogressors, 211 regions were differentially accessible, clearly distinguishing the two groups. DARs were enriched for promoter-TSS regions and could be linked to differential expression of EOMES and CAPG with use of publicly available data on enhancer-promoter interactions defined by capture-Hi-C in human CD4+ T cells. DARs were also enriched for TFBS involved in T cell function, namely, CTCF, ETS1, and ETV2, but not EOMES or T-bet. Likewise, neither EOMES nor T-bet showed differential binding between progressors and nonprogressors. This suggests that EOMES and T-bet might contribute to transcriptomic reprograming by virtue of changes in their expression rather than alterations in their chromatin assembly, as previously reported for cytotoxic CD8+ T cells (55).
A strength of the current study is that the children were relatively homogeneous in terms of age, ethnicity, and HLA class II type. Non-HLA-related genotypic heterogeneity is likely to influence the rate of disease progression, but its genome-wide analysis would require a much larger sample size. Furthermore, phenotypic variability, e.g., in BMI, ethnicity, age at onset, first islet autoantibody, suggests genetic variation in pathways to β-cell destruction. Therefore, a further caveat is that our findings may not be broadly applicable and must be further validated in larger cohorts of at risk children with more diverse genetic backgrounds. Nevertheless, it is not unreasonable to suggest that the cytotoxic signature we identified in CD4+ T cells may be a final common feature associated with β-cell destruction.
In summary, we demonstrate that CD4+ T cells with a cytotoxicity-related phenotype distinguish children positive for multiple autoantibodies who progress more rapidly to clinical T1D. We also provide the first map of chromatin accessibility in immune cells of islet autoantibody–positive children as a resource for exploring the transcriptomic and epigenomic basis of T1D.
Article Information
Acknowledgments. The authors thank Dr. Stephen Wilcox (Genomics Hub, Walter and Eliza Hall Institute of Medical Research [WEHI], Parkville, Victoria, Australia) for assistance with RNA-seq, Kelly Watson and Peter Colman (Endocrine Laboratory, The Royal Melbourne Hospital, Victoria, Australia) for islet autoantibody assays, and Riitta Lahesmaa and Mikael Knip (Turku Bioscience Centre, University of Turku and bo Akademi University, Turku, Finland) for facilitating access to the data of Kallionpää et al (7).
Funding. This research was supported by JDRF Australia (JDRFA)/National Health and Medical Research Council of Australia (NHMRC) Centre of Research Excellence for the Protection of Pancreatic Beta Cells (1078106), JDRFA Australian Type 1 Diabetes Clinical Research Network, JDRFA/The Leona M. and Harry B. Helmsley Charitable Trust (3-SRA-2019-899-M-N), and JDRF International (1-SRA-2018-543-S-B). Additional support was provided by an NHMRC Program Grant (LCH APP1150425) and NHMRC Research Fellowships (to G.K.S and L.C.H). The work was made possible through Victorian State Government Operational Infrastructure Support Program and NHMRC Research Institute Infrastructure Support Scheme.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. N.G.B. and L.C.H. conceptualized, designed, and led the study and wrote and edited the manuscript. N.G.B., A.L.G., J.C., G.K.S., and L.C.H. analyzed data. G.K.S. provided bioinformatic guidance and significant edits to the manuscript. L.C.H., J.J.C., J.E.H., and A.-G.Z. provided cell samples. G.N., E.B.-S., and N.L.S. provided technical support. J.M.W. commented on experimental approaches and edited the manuscript. All authors read and commented on the manuscript. L.C.H. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form online at the 17th Immunology of Diabetes Society Congress, Beijing, China, 15–18 October 2020.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.17185898.
References
- 1. Ziegler AG, Rewers M, Simell O, et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 2013;309:2473–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kallionpää H, Elo LL, Laajala E, et al. Innate immune activity is detected prior to seroconversion in children with HLA-conferred type 1 diabetes susceptibility. Diabetes 2014;63:2402–2414 [DOI] [PubMed] [Google Scholar]
- 3. Reynier F, Pachot A, Paye M, et al. Specific gene expression signature associated with development of autoimmune type-I diabetes using whole-blood microarray analysis. Genes Immun 2010;11:269–278 [DOI] [PubMed] [Google Scholar]
- 4. Jin Y, Sharma A, Bai S, et al. Risk of type 1 diabetes progression in islet autoantibody-positive children can be further stratified using expression patterns of multiple genes implicated in peripheral blood lymphocyte activation and function. Diabetes 2014;63:2506–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xhonneux LP, Knight O, Lernmark Å, et al. Transcriptional networks in at-risk individuals identify signatures of type 1 diabetes progression. Sci Transl Med 2021;13:eabd5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mehdi AM, Hamilton-Williams EE, Cristino A, et al. A peripheral blood transcriptomic signature predicts autoantibody development in infants at risk of type 1 diabetes. JCI Insight 2018;3:e98212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kallionpää H, Somani J, Tuomela S, et al. Early detection of peripheral blood cell signature in children developing β-cell autoimmunity at a young age. Diabetes 2019;68:2024–2034 [DOI] [PubMed] [Google Scholar]
- 8. Harbison JE, Roth-Schulze AJ, Giles LC, et al. Gut microbiome dysbiosis and increased intestinal permeability in children with islet autoimmunity and type 1 diabetes: a prospective cohort study. Pediatr Diabetes 2019;20:574–583 [DOI] [PubMed] [Google Scholar]
- 9. Nguyen C, Varney MD, Harrison LC, Morahan G. Definition of high-risk type 1 diabetes HLA-DR and HLA-DQ types using only three single nucleotide polymorphisms. Diabetes 2013;62:2135–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colman PG, Steele C, Couper JJ, et al. Islet autoimmunity in infants with a type I diabetic relative is common but is frequently restricted to one autoantibody. Diabetologia 2000;43:203–209 [DOI] [PubMed] [Google Scholar]
- 11. Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 2019;47:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corces MR, Trevino AE, Hamilton EG, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 2017;14:959–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw 2015;67:1–48 [Google Scholar]
- 16. Reske JJ, Wilson MR, Chandler RL. ATAC-seq normalization method can significantly affect differential accessibility analysis and interpretation. Epigenetics Chromatin 2020;13:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bentsen M, Goymann P, Schultheis H, et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat Commun 2020;11:4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Linsley PS, Speake C, Whalen E, Chaussabel D. Copy number loss of the interferon gene cluster in melanomas is linked to reduced T cell infiltrate and poor patient prognosis. PLoS One 2014;9:e109760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Juno JA, van Bockel D, Kent SJ, Kelleher AD, Zaunders JJ, Munier CM. Cytotoxic CD4 T cells-friend or foe during viral infection? Front Immunol 2017;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li J, He Y, Hao J, Ni L, Dong C. High levels of Eomes promote exhaustion of anti-tumor CD8+ T cells. Front Immunol 2018;9:2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patil VS, Madrigal A, Schmiedel BJ, et al. Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci Immunol 2018;3:eaan8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhaskaran N, Liu Z, Saravanamuthu SS, et al. Identification of Casz1 as a regulatory protein controlling T helper cell differentiation, inflammation, and immunity. Front Immunol 2018;9:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Borrego F. The CD300 molecules: an emerging family of regulators of the immune system. Blood 2013;121:1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 2012;9:215–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Javierre BM, Burren OS, Wilder SP, et al.; BLUEPRINT Consortium . Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 2016;167:1369–1384.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yukawa M, Jagannathan S, Vallabh S, et al. AP-1 activity induced by co-stimulation is required for chromatin opening during T cell activation. J Exp Med 2020;217:e20182009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Evangelista AF, Collares CVA, Xavier DJ, et al. Integrative analysis of the transcriptome profiles observed in type 1, type 2 and gestational diabetes mellitus reveals the role of inflammation. BMC Med Genomics 2014;7:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ferreira RC, Guo H, Coulson RM, et al. A type I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes 2014;63:2538–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaizer EC, Glaser CL, Chaussabel D, Banchereau J, Pascual V, White PC. Gene expression in peripheral blood mononuclear cells from children with diabetes. J Clin Endocrinol Metab 2007;92:3705–3711 [DOI] [PubMed] [Google Scholar]
- 31. Elo LL, Mykkänen J, Nikula T, et al. Early suppression of immune response pathways characterizes children with prediabetes in genome-wide gene expression profiling. J Autoimmun 2010;35:70–76 [DOI] [PubMed] [Google Scholar]
- 32. Irvine KM, Gallego P, An X, et al. Peripheral blood monocyte gene expression profile clinically stratifies patients with recent-onset type 1 diabetes. Diabetes 2012;61:1281–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vecchio F, Lo Buono N, Stabilini A, et al. Abnormal neutrophil signature in the blood and pancreas of presymptomatic and symptomatic type 1 diabetes. JCI Insight 2018;3:e122146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Terrazzano G, Bruzzaniti S, Rubino V, et al. T1D progression is associated with loss of CD3+CD56+ regulatory T cells that control CD8+ T cell effector functions. Nat Metab 2020;2:142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferraro A, D’Alise AM, Raj T, et al. Interindividual variation in human T regulatory cells. Proc Natl Acad Sci U S A 2014;111:E1111–E1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jailwala P, Waukau J, Glisic S, et al. Apoptosis of CD4+ CD25(high) T cells in type 1 diabetes may be partially mediated by IL-2 deprivation. PLoS One 2009;4:e6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ahmed R, Omidian Z, Giwa A, et al. A public BCR present in a unique dual-receptor-expressing lymphocyte from type 1 diabetes patients encodes a potent T cell autoantigen. Cell 2019;177:1583–1599.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cerosaletti K, Barahmand-Pour-Whitman F, Yang J, et al. Single-cell RNA sequencing reveals expanded clones of islet antigen-reactive CD4+ T cells in peripheral blood of subjects with type 1 diabetes. J Immunol 2017;199:323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heninger AK, Eugster A, Kuehn D, et al. A divergent population of autoantigen-responsive CD4+ T cells in infants prior to β cell autoimmunity. Sci Transl Med 2017;9:eaaf8848. [DOI] [PubMed] [Google Scholar]
- 40. Raveney BJE, Sato W, Takewaki D, et al. Involvement of cytotoxic Eomes-expressing CD4+ T cells in secondary progressive multiple sclerosis. Proc Natl Acad Sci U S A 2021;118:e2021818118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stanford SM, Bottini N. PTPN22: the archetypal non-HLA autoimmunity gene. Nat Rev Rheumatol 2014;10:602–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chemin K, Ramsköld D, Diaz-Gallo LM, et al. EOMES-positive CD4+ T cells are increased in PTPN22 (1858T) risk allele carriers. Eur J Immunol 2018;48:655–669 [DOI] [PubMed] [Google Scholar]
- 43. Okada Y, Wu D, Trynka G, et al. RACI consortium; GARNET consortium . Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014;506:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium 2, Stephen Sawcer, Garrett Hellenthal, Matti Pirinen, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011;476:214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Appay V. The physiological role of cytotoxic CD4(+) T-cells: the holy grail? Clin Exp Immunol 2004;138:10–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takeuchi A, Saito T. CD4 CTL, a cytotoxic subset of CD4+ T cells, their differentiation and function. Front Immunol 2017;8:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Appay V, Zaunders JJ, Papagno L, et al. Characterization of CD4(+) CTLs ex vivo. J Immunol 2002;168:5954–5958 [DOI] [PubMed] [Google Scholar]
- 48. Cachot A, Bilous M, Liu YC, et al. Tumor-specific cytolytic CD4 T cells mediate immunity against human cancer. Sci Adv 2021;7:eabe3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fasth AE, Cao D, van Vollenhoven R, Trollmo C, Malmström V. CD28nullCD4+ T cells--characterization of an effector memory T-cell population in patients with rheumatoid arthritis. Scand J Immunol 2004;60:199–208 [DOI] [PubMed] [Google Scholar]
- 50. Markovic-Plese S, Cortese I, Wandinger KP, McFarland HF, Martin R. CD4+CD28- costimulation-independent T cells in multiple sclerosis. J Clin Invest 2001;108:1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peeters LM, Vanheusden M, Somers V, et al. Cytotoxic CD4+ T cells drive multiple sclerosis progression. Front Immunol 2017;8:1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Raveney BJ, Oki S, Hohjoh H, et al. Eomesodermin-expressing T-helper cells are essential for chronic neuroinflammation. Nat Commun 2015;6:8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Harari A, Vallelian F, Pantaleo G. Phenotypic heterogeneity of antigen-specific CD4 T cells under different conditions of antigen persistence and antigen load. Eur J Immunol 2004;34:3525–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matteucci E, Ghimenti M, Di Beo S, Giampietro O. Altered proportions of naïve, central memory and terminally differentiated central memory subsets among CD4+ and CD8 + T cells expressing CD26 in patients with type 1 diabetes. J Clin Immunol 2011;31:977–984 [DOI] [PubMed] [Google Scholar]
- 55. Scott-Browne JP, López-Moyado IF, Trifari S, et al. Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity 2016;45:1327–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]