Abstract
Coral reefs are declining due to anthropogenic disturbances, including climate change. Therefore, improving our understanding of coral ecosystems is vital, and the influence of bacteria on coral health has attracted particular interest. However, a gnotobiotic coral model that could enhance studies of coral–bacteria interactions is absent. To address this gap, we tested the ability of treatment with seven antibiotics for 3 weeks to deplete bacteria in Exaiptasia diaphana, a sea anemone widely used as a coral model. Digital droplet PCR (ddPCR) targeting anemone Ef1-α and bacterial 16S rRNA genes was used to quantify bacterial load, which was found to decrease six-fold. However, metabarcoding of bacterial 16S rRNA genes showed that alpha and beta diversity of the anemone-associated bacterial communities increased significantly. Therefore, gnotobiotic E. diaphana with simplified, uniform bacterial communities were not generated, with biofilm formation in the culture vessels most likely impeding efforts to eliminate bacteria. Despite this outcome, our work will inform future efforts to create a much needed gnotobiotic coral model.
Keywords: bacterial load, coral, ddPCR, Exaiptasia, gnotobiotic
Introduction
The sea anemone Exaiptasia diaphana (previously Aiptasia pallida [1, 2]) has become an important coral model as its intracellular symbiosis with photosynthetic algae of the family Symbiodiniaceae makes it useful for studying host–symbiont relationships [3, 4]. The breakdown of this relationship (i.e. bleaching) has been particularly well studied [5–9] due to an increase in the frequency of mass coral bleaching events linked to climate change [10].
E. diaphana’s ability to survive in a Symbiodiniaceae-free (i.e. aposymbiotic) state has clarified metabolic processes within cnidarians by separating the host and its algal symbiont to reveal the role of each in nutrient transfer [11–13] and their response to environmental stress [14–16]. However, studies that have investigated the relationship between E. diaphana and Symbiodiniaceae have often ignored the influence of bacteria on the holobiont, a functional entity comprising the host and all its microbial partners [17].
The bacterial component of the holobiont influences host health, for example through its involvement in nutrient cycling [18–21] and pathogen protection [22–24]. Therefore, removing bacteria from the holobiont represents an important next step in the elucidation of host and symbiont interdependence [25]. Although germ-free Symbiodiniaceae cells have been created [26], similar coral cultures or cell lines have not [25]. E. diaphana may be able to fill this gap.
There has been one report of germ-free E. diaphana, wherein anemones were exposed to two antibiotics to render them ‘aseptic’ [27]. Germ-free status was determined by culture methods and microscopy. However, as many bacteria cannot be cultured [28], and the extent to which an anemone can be screened by microscopy is limited, the germ-free status of the anemones is uncertain. In fact, creating germ-free E. diaphana might not be feasible as the anemones could require bacteria for normal host health and development, as in many other organisms [29–31]. Thus, E. diaphana that harbour reduced bacterial communities could represent a practical alternative to germ-free cultures. Strictly, these anemones would be described as gnotobiotic, that is, organisms with depleted microbial communities that are simple (i.e. possessing low individual, or alpha, diversity), uniform (i.e. possessing low inter-individual, or beta, diversity) and precisely defined [32].
In a recent step towards development of gnotobiotic E. diaphana, a method for bacterial depletion was reported [33, 34]. Depletion was achieved by exposure to four antibiotics with different mechanisms of action, and detection of bacteria in the treated anemones was by culture methods and PCR. However, depletion was only maintained with continuous treatment. In addition, bacterial load was not quantified, and the bacterial communities were not characterized, leaving the extent of the depletion and the uniformity and composition of the resulting bacterial communities unknown. Consequently, the efficacy of antibiotic approaches for generating gnotobiotic E. diaphana cultures remains unclear.
Here, we describe our efforts to produce gnotobiotic E. diaphana by exposure to antibiotics. Our aim was to determine whether anemone bacterial load could be reduced and the bacterial communities made uniform by antibiotic treatment. Further, by presenting our methods, we hope to assist other researchers seeking to create gnotobiotic E. diaphana, a resource that could help clarify the relationship between cnidarians and their bacterial associates.
Methods
Experimental set-up and antibiotic treatment
Clonal adult E. diaphana anemones (n=72; genotype AIMS2) were haphazardly selected from a single tank in the University of Melbourne (UoM) culture collection [35]. Individuals were transferred into single wells within sterile 12-well plates (CLS3513; Corning) where they were maintained in seawater reconstituted from Red Sea Salt (R11065) with reverse osmosis water at a salinity of ~34 p.p.t. The lidded plates were kept in clear, sterile plastic zip-lock bags to prevent contamination and were randomly positioned in a Hi-Point 740 incubator (Thermo Fisher) at 26 °C with lighting at ~33 µmol m–2 s–1 on a 12 h:12 h light–dark cycle. During a 1 week acclimation period the E. diaphana were fed twice with freshly hatched Artemia salina nauplii (Salt Creek, Premium GSL), and the water was changed three times (Table 1). After the acclimation period, antibiotic treatment and sampling commenced. All subsequent operations were performed using aseptic techniques. During the treatment period, water changes were performed with Red Sea Salt water sterilized by autoclaving (hereafter, ‘sRSS-water’). Treatment was scheduled to coincide with water changes to ensure the anemones were constantly exposed to antibiotics. Sampling was performed more frequently in the first week of treatment to track the impact of the antibiotics on the intact bacterial communities. When sampling coincided with treatment or water changes, samples were collected first.
Table 1.
E. diaphana maintenance, antibiotic treatment and sampling schedule
|
Monday |
Tuesday |
Wednesday |
Thursday |
Friday |
|---|---|---|---|---|
|
Transfer E. diaphana Hatch A. salina |
Feed E. diaphana |
Change water |
Hatch A. salina |
Change water Feed E. diaphana |
|
Day 0 Sampling Change water Treatment Hatch A. salina |
Day 1 Sampling Feed E. diaphana |
Day 2 Change water Treatment |
Day 3 Sampling Hatch A. salina |
Day 4 Change water Treatment Feed E. diaphana |
|
Day 7 Sampling Change water Treatment Hatch A. salina |
Day 8 Feed E. diaphana |
Day 9 Change water Treatment |
Day 10 Hatch A. salina |
Day 11 Change water Treatment Feed E. diaphana |
|
Day 14 Sampling Change water Treatment Hatch A. salina |
Day 15 Feed E. diaphana |
Day 16 Change water Treatment |
Day 17 Hatch A. salina |
Day 18 Change water Treatment Feed E. diaphana |
|
Day 21 Sampling |
|
|
|
|
On treatment days, half the E. diaphana were exposed to antibiotics selected for their different mechanisms of action and activity against Gram-positive or Gram-negative bacteria, and their previous use on cnidarians, Symbiodiniaceae, sponges and A. salina (Table 2). Maximum tolerable concentrations were determined in pre-treatment testing by exposing E. diaphana to increasing dilutions of the combined antibiotics until they maintained normal appearance, growth and feeding over an 18 day test period.
Table 2.
Antibiotics used to deplete bacteria in E. diaphana and A. salina
|
Antibiotic |
Concentration (µg ml–1) |
Target/mechanism of action |
Gram +/− activity |
References to prior use |
|---|---|---|---|---|
|
Carbenicillin |
25 |
dd-Transpeptidase/inhibits cell wall synthesis |
− |
[33, 97–100]* |
|
Chloramphenicol |
25 |
23S rRNA/inhibits protein synthesis |
+/− |
|
|
Nalidixic acid |
15 |
Gyrase/inhibits DNA replication |
− |
|
|
Neomycin |
10 |
30S rRNA assembly/inhibits protein synthesis |
+/− |
|
|
Polymyxin B |
10 |
Cell wall/increases cell wall permeability |
− |
|
|
Rifampicin |
10 |
RNA polymerase/inhibits transcription |
+ |
|
|
Streptomycin |
25 |
16S rRNA/inhibits protein synthesis |
+ |
*References to the use of Penicillin family antibiotics with the same mechanism of action.
Control E. diaphana were fed with A. salina hatched in sRSS-water. Treated E. diaphana were fed with A. salina hatched in sRSS-water containing antibiotics at concentrations matching those used for E. diaphana.
Sampling and DNA extraction
On sampling days, six control and six treated E. diaphana were killed to measure changes in bacterial load and community composition. When collected, each anemone was gently passed two or three times through the tip of a sterile transfer pipette to remove loosely attached debris. Three 50 µl aliquots of a dense suspension of control and treated A. salina nauplii were also collected for bacterial load and community analyses. All samples were snap frozen and stored at –80 °C until processing. DNA was extracted from the bacterial analysis samples using a salting out protocol [36] modified according to Hartman et al. [37].
Bacterial load assessment (B/H ratio)
Bacterial load in E. diaphana and A. salina was quantified according to the number of bacterial gene copies to host gene copies in each DNA extract. The copy number data were obtained by digital droplet PCR (ddPCR) to allow calculation of the bacteria/host (B/H) cell ratio [38] for each sample. This approach has also been used to analyse bacterial load in insect samples with low mass and volume [39–41], with the use of a ratio accounting for differences in sample size. Primers targeting single-copy reference genes in E. diaphana and A. salina were used for host cell quantification (Table 3). The translation elongation factor 1 alpha gene (Ef1-α) was used for E. diaphana, and the beta actin gene (ß-actin) was used for A. salina. Primers targeting a conserved 98-nt sequence between the V2 and V3 regions of the bacterial 16S rRNA gene were used to estimate bacterial cell numbers as they produced small amplicons and no non-specific PCR product, which was essential for optimal ddPCR performance. No correction was made for 16S rRNA gene copy number, and hence the method is semi-quantitative.
Table 3.
Primers used in the present study to estimate host and bacterial cell numbers
|
Target (gene) |
Primer name |
Primer sequence |
Annealing temperature (°C) |
Product size (nt) |
References |
|---|---|---|---|---|---|
|
E. diaphana (Ef1-α) |
Ef1-α-fwd |
AGCACTGAGCCACCATACAG |
60 |
88 |
[107] |
|
Ef1-α -rev |
TTGGGTTATAGCCGGTCTTC |
60 |
[107] |
||
|
A. salina (ß-actin) |
art-actin-fwd |
GGTCGTGACTTGACGGACTATCT |
60 |
147 |
[108–111] |
|
art-actin-rev |
AGCGGTTGCCATTTCTTGTT |
60 |
[108–111] |
||
|
Universal bacteria (conserved inter V2–V3 16S rRNA gene region) |
259-fwd |
GGTAAHRGCYYACCAAG |
54 |
98 |
[112] |
|
357-rev |
CTGCTGCCTCCCGTAGGAG |
54 |
Reverse complement of ‘primer 1’ [113] |
Before performing ddPCR, DNA was restriction enzyme-digested to improve droplet encapsulation of DNA fragments and signal generation from low-concentration bacterial DNA [42]. Sample DNA was digested for 1.5 h at 37 °C in a volume of 20 µl comprising 7 µl sterile water, 2 µl of 10× restriction enzyme buffer, 10 µl DNA extract, and 1 µl (~20 U) HindIII (R3104S-HF; New England BioLabs). The digested DNA was then quantified by PicoGreen (P11496; Thermo Fisher) and diluted ≥1:4 to 10–20 ng µl–1 to create practical working concentrations and prevent PCR inhibition by the enzyme buffer.
ddPCRs for each DNA sample were prepared in an initial volume of 44 µl comprising 24 µl EvaGreen Supermix (QX200; Bio-Rad), sterile water and ~30 ng of digested DNA to ensure that DNA concentrations for each bacteria–host reaction pair were within the dynamic range of the ddPCR system. The mixture was then split into two 22 µl aliquots, one for host cell quantification (i.e. E. diaphana or A. salina) and one for bacteria. One microlitre each of the appropriate 5 µM forward and reverse primers (Table 3) was then added to each reaction aliquot, giving final primer concentrations of ~200 nM and volumes of 24 µl. From each 24 µl volume, 20 µl was loaded into a DG8 cartridge (1864008; Bio-Rad), followed by 70 µl of droplet generation oil for EvaGreen (1864005; Bio-Rad), and droplets were generated in a droplet-generator (QX200; Bio-Rad). A volume of 40 µl of generated droplets per reaction was then transferred to a 96-well plate and foil-sealed (1814040; Bio-Rad) with a thermal plate-sealer (PX1; Bio-Rad). One no-template control (NTC) reaction was included per plate. Thermal cycler settings were optimized according to Witte et al. [43]: one cycle at 95.0 °C for 5 min; 50 cycles at 95 °C for 1 min + 54 °C or 60 °C (see Table 3) for 2 min; one cycle at 4.0 °C for 5 min; one cycle at 90 °C for 5 min; 12 °C hold. All ramp rates were 1 °C s–1. Droplets were read on a Bio-Rad QX200 droplet reader, and fluorescence data were analysed in QuantaSoft v1.7.4.0917 (Tables S1 and S2, Fig. S1, available in the online version of this article).
Sample and data processing for metabarcoding analysis
In preparation for bacterial community analysis by metabarcoding, sample DNA was amplified by end-point PCR using primers with Illumina overhang sequences (underlined) targeting the V5–V6 regions of the 16S rRNA gene: 784F 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGGATTAGATACCCTGGTA-3′; 1061R 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCRRCACGAGCTGACGAC-3′ [44]. Triplicate PCRs were performed in 20 µl volumes comprising 1 µl DNA extract, 10 µl MyTaq HS Mix polymerase (Bioline), 0.5 µl of 10 µM 784F, 0.5 µl of 10 µM 1061R, and 8 µl MilliQ water. Thermal-cycler settings were: one cycle at 95.0 °C for 3 min; 30 cycles at 95.0, 55.0 and 72.0 °C for 15 s each; one cycle at 72 °C for 3 min; 12 °C hold. Each triplicate was pooled, then checked by 1% agarose gel electrophoresis. No-sample DNA extractions and no-template PCRs were performed to identify contaminants introduced during sample preparation. A volume of 25 µl of pooled PCR product from each sample was sent to the Ramaciotti Centre for Genomics (RCG), Sydney, Australia, for sequencing on a single Illumina MiSeq 2×250 bp run. RCG performed PCR product clean-up and normalization as part of library preparation prior to sequencing. The resulting Illumina MiSeq data were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA698456. Demultiplexed MiSeq reads were joined in QIIME2 v2020.8.0 [45]. Denoising, chimera filtering and trimming was performed using DADA2 within the QIIME2 environment [46]. Resulting amplicon sequence variants (ASVs) >289 nt were removed. Taxonomy was assigned to the remaining ASVs against the silva database (v132) trained with a naïve Bayes classifier [47–50]. Unidentified ASVs, and ASVs identified as mitochondrial or chloroplast sequences were removed.
Data analyses
Data analyses were performed in R v4.0.3 [51], with differences considered significant at α=0.05 unless otherwise stated. The ddPCR count data were imported into R and the B/H ratios were calculated and plotted over time with the R package ggplot2 [52]. Overall differences in B/H were assessed by generalized least square (GLS) models with the R package nlme [53]. If the B/H data met homogeneity of variance [54] and normality criteria [55], Student’s t-test [56] was used to assess differences between samples, otherwise the Mann–Whitney U test [57] was used. Tabulated ASV counts, taxonomic and meta data were imported and converted into a phyloseq object for bacterial community analyses [58]. Rarefaction curves were generated with the R package vegan [59] to assess whether the metabarcoding samples had been sequenced sufficiently to capture species diversity. Putative contaminating ASVs were identified with the R package decontam [60] and removed. The ASV counts for each E. diaphana sample were multiplied by the corresponding B/H ratio ×103 to convert counts to 16S rRNA gene copies per host cell ×103 (hereafter, 16S/H × 103), thus producing absolute abundance values corrected for E. diaphana size differences [61, 62]. To identify samples with highly divergent bacterial compositions, the E. diaphana bacterial community data were visualized in a non-metric multidimensional scaling (nMDS) ordination based on Bray–Curtis dissimilarity [63], and a principal component analysis (PCA) ordination of centre log-ratio (CLR)-transformed data using vegan [59] and the R package mixOmics [64], respectively. Alpha diversity metrics for the E. diaphana bacterial communities were calculated in vegan [59] and plotted over time with ggplot2 after sub-sampling the data to 48 176 B/H-converted ASV counts per sample. Bacterial community richness was described by number of observed ASVs per E. diaphana sample. Community evenness was described using Simpson’s index [65]. General alpha diversity was described using Shannon’s index [66]. Overall differences in the alpha diversity metrics were assessed by GLS models and sample-wise differences were assessed using Mann–Whitney U tests, as above. Relationships between the untreated and treated E. diaphana bacterial communities at Day 0 and Day 21 were visualized in nMDS and PCA ordinations, as above, and differences between them were tested using generalized linear models (GLMs) in the R package mvabund [67]. Common and unique ASVs in the Day 21 control and treated E. diaphana were visualized in petal diagrams to compare the complexity of their bacterial communities. ASVs with an absolute abundance ≥1000 16S/H × 103 in the treated E. diaphana at all timepoints were identified to highlight antibiotic-tolerant bacteria [68]. The tolerant ASV abundances were plotted with ggplot2. To assess their possible origin or location, they were then compared to the A. salina bacterial community data from Maire et al. [69], which identified the bacteria associated with Symbiodiniaceae isolated from E. diaphana in the UoM culture collection.
Results
Changes in bacterial load (B/H ratio)
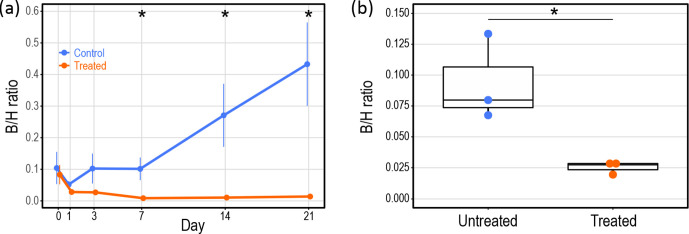
All antibiotic-treated E. diaphana survived until being killed and were phenotypically comparable to the control E. diaphana with respect to tentacle extension, mobility and feeding behaviour throughout the experiment. However, significant changes in B/H occurred in the E. diaphana according to treatment (GLS, χ 2=22.64, P<0.001) and time (GLS, χ 2=14.52, P<0.001) (Fig. 1a). The antibiotic-treated E. diaphana underwent a three-fold decrease in B/H from Day 0 to Day 1, and a significant six-fold decrease overall (Mann–Whitney, P=0.030). Despite the decrease, B/H in the treated E. diaphana remained above zero through to Day 21 indicating that bacteria were not completely eliminated. The B/H of the control E. diaphana also decreased from Day 0 to Day 1, but then recovered and underwent a four-fold increase from Day 7 to Day 21. Hatching the A. salina feedstock in the antibiotic solution significantly reduced its B/H 3.7-fold (Student’s t-test, P=0.029) (Fig. 1b). One treated Day 21 E. diaphana ddPCR sample (gt215) amplified poorly and was excluded from the B/H analysis. ddPCR counts for A. salina were also low, but these samples were retained as bacterial load in A. salina was not our primary focus.
Fig. 1.
Effect of antibiotic treatment on the bacterial load (B/H) of E. diaphana, and the A. salina feed stock. (a) Temporal change in B/H in E. diaphana. For each datapoint, n=5–6. Error bars±1 sem. (b) B/H in the untreated and treated A. salina feed stock, n=3. Asterisks indicate significant difference, α=0.05.
Metabarcoding data attributes
Sequencing produced 3747373 raw reads across the 72 E. diaphana and six A. salina samples (minimum 3825; mean 48043, maximum 88457 reads per sample). After merging, denoising and filtering, 2643644 reads remained (minimum 2404, mean 33891, maximum 55113 reads per sample) and 4628 ASVs were identified. Rarefaction curves for the E. diaphana samples plateaued, suggesting that sequencing had captured bacterial diversity (Fig. S2). Seven ASVs, which constituted 0.079 and 0.005% of the bacteria in the E. diaphana and A. salina respectively, were deemed contaminants by decontam and were removed from the analysis (Table S3). One E. diaphana sample (gt215) was removed because it could not be converted to absolute abundance as its B/H ratio could not be determined (see above). Two outlier E. diaphana samples were revealed in the nMDS (Fig. S3a) and PCA (Fig. S3b) ordinations and were also removed.
Changes in bacterial community composition
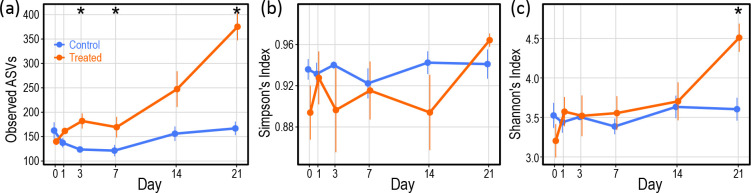
A significant change in the number of observed ASVs in the E. diaphana anemones (Fig. 2a) occurred according to treatment (GLS, χ 2=62.216, P<0.001) and time (GLS, χ 2=42.561, P<0.001). A difference between the control and treated E. diaphana emerged at Day 3 following an increase in observed ASVs in the treated E. diaphana and decrease in the controls (Mann–Whitney, P=0.002). However, the decrease in the controls was temporary and there was no significant difference between the number of ASVs observed in the controls at Day 21 compared to Day 0 (Mann–Whitney, P=1.000). In contrast, the number of ASVs observed in the treated E. diaphana at Day 21 was significantly higher than at Day 0 (Mann–Whitney, P=0.008).
Fig. 2.
Temporal change in alpha diversity in E. diaphana. (a) Number of observed ASVs; (b) Simpson’s index values; (c) Shannon’s index values. For each data point, n=5–6. Error bars±1 sem. Asterisks indicate significant differences, α=0.05.
A significant temporal change in evenness of the E. diaphana bacterial communities, measured according to Simpson’s evenness (Fig. 2b), was detected (GLS, χ 2=5.934, P=0.015). However, this was not supported by post-hoc tests for each timepoint, or in Day 0 versus Day 21 comparisons for each sample type. At the end of the treatment period, overall alpha diversity, measured according to Shannon’s index (Fig. 2c), was significantly higher in the bacterial communities of the treated E. diaphana anemones compared to the control E. diaphana anemones (Mann–Whitney, P=0.004).
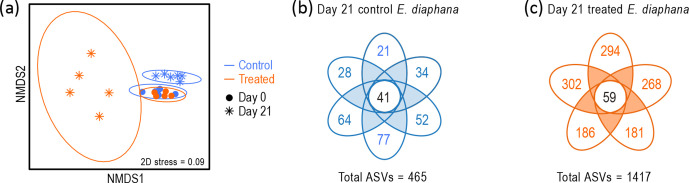
Grouping of Day 0 datapoints in nMDS (Fig. 3a) and PCA (Fig. S4b) ordinations of the E. diaphana bacterial community data suggested that the E. diaphana anemones were highly similar at the beginning of the experiment, and statistical analyses confirmed they were not significantly different (manyGLM, LRT=492, P=0.187). However, by Day 21, the bacterial communities of the control and treated E. diaphana anemones had become significantly different from their Day 0 counterparts (control: manyGLM, LRT=1126, P=0.003; treated: manyGLM, LRT=1 428, P=0.006) and each other (manyGLM, LRT=1725, P=0.001). Tight clustering of the Day 21 control E. diaphana datapoints suggested that despite undergoing compositional shifts, the bacterial communities of the control E. diaphana were still highly uniform after 21 days. In contrast, separation of the datapoints for the Day 21 treated E. diaphana indicated decreased uniformity. This may be partially explained by the increase in observed ASVs for the treated E. diaphana noted above. A survey of common and unique ASVs in the Day 21 control and treated E. diaphana explained this further by showing that, compared to the controls (Fig. 3b), each treated E. diaphana (Fig. 3c) harboured a high number of unique ASVs. Together, these data indicated an increase in bacterial beta diversity among the treated E. diaphana.
Fig. 3.
(a) nMDS ordination (Bray–Curtis dissimilarity) of bacterial communities in control and treated E. diaphana at Day 0 and Day 21 (n=5–6; see Fig. S4b for PCA ordination of the Day 0 and Day 21 data, and Fig. S5 for nMDS and PCA ordinations showing all timepoints). (b, c) Petal diagrams showing the number of common and unique ASVs in control (n=6) and treated (n=5) E. diaphana at Day 21.
Antibiotic-tolerant bacteria
Sixteen antibiotic-tolerant ASVs maintained absolute abundances of ≥1000 16S/H × 103 in the treated E. diaphana across all timepoints (Table 4; Fig. S6). Despite avoiding elimination, all tolerant ASVs declined in abundance from Day 0 to Day 21, with most declines being significant. Six of the tolerant ASVs were associated with the antibiotic-treated A. salina feedstock, and seven were associated with Symbiodiniaceae cells that were isolated from anemones in the UoM E. diaphana culture collection and washed to remove extracellular bacteria. All tolerant ASVs were associated with the control E. diaphana, indicating their ubiquity among the test anemones. Three tolerant ASVs were members of the genus Vibrio .
Table 4.
Summary of the antibiotic-tolerant ASVs (see Table S4 for full data)
Symbiodiniaceae associations are based on data from Maire et al. [69], with ‘Close’ and ‘Loose’ associates defined as ‘bacteria tightly attached to the algal cell’s exterior’ and ‘planktonic bacteria’ from the Symbiodiniaceae culture medium, respectively. P-values for the Day 0 vs. Day 21 comparisons were calculated using Mann–Whitney U tests (α=0.5).
|
Taxonomic classification |
Absolute abundance (16S/H × 103) |
Day 0 vs Day 21 P-value |
Present in A. salinas? |
Symbiodiniaceae association? |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Class |
Order |
Family |
Genus |
Day 0 |
Day 1 |
Day 3 |
Day 7 |
Day 14 |
Day 21 |
Intracellular |
Close |
Loose |
||
|
6928 |
6472 |
4745 |
1590 |
1417 |
1524 |
0.310 |
yes |
yes |
||||||
|
– |
– |
– |
20634 |
28507 |
9458 |
4674 |
2676 |
1061 |
0.008 |
|||||
|
4922 |
3712 |
5598 |
1908 |
3182 |
1204 |
0.016 |
yes |
|||||||
|
94123 |
56861 |
25879 |
11433 |
7282 |
6906 |
0.032 |
||||||||
|
Coxiellales |
43074 |
21771 |
15844 |
7171 |
78850 |
1903 |
0.008 |
|||||||
|
– |
15508 |
8025 |
11956 |
3753 |
4789 |
8869 |
0.095 |
yes |
yes |
|||||
|
10338 |
7333 |
6792 |
2439 |
1248 |
1238 |
0.032 |
yes |
yes |
yes |
yes |
||||
|
8616 |
26317 |
11092 |
1290 |
1002 |
1754 |
0.421 |
yes |
|||||||
|
– |
397755 |
67349 |
58628 |
10436 |
15986 |
3330 |
0.008 |
|||||||
|
SM2D12 |
– |
9099 |
9491 |
12574 |
3197 |
1861 |
1794 |
0.032 |
||||||
|
5299 |
1727 |
4680 |
1614 |
2682 |
3022 |
0.548 |
yes |
yes |
yes |
|||||
|
135153 |
5405 |
6342 |
2342 |
4440 |
2738 |
0.032 |
yes |
yes |
yes |
|||||
|
– |
56148 |
3157 |
6652 |
1988 |
7482 |
17836 |
0.222 |
yes |
||||||
|
32799 |
30630 |
37369 |
14014 |
20619 |
8801 |
0.016 |
yes |
yes |
yes |
yes |
||||
|
8025 |
6303 |
5786 |
1775 |
1185 |
1311 |
0.421 |
yes |
yes |
||||||
|
– |
28068 |
51153 |
58769 |
17803 |
10451 |
3742 |
0.032 |
|||||||
Discussion
Antibiotic treatment has been used previously to remove [27] or deplete [33] bacteria from E. diaphana, but with limited assessment of efficacy, and no information on changes in the bacterial communities. The present study sought to address these gaps by measuring changes in bacterial load and absolute abundance in antibiotic-treated E. diaphana. To our knowledge, describing bacterial communities in this way is novel in cnidarian research.
Antibiotic treatment did not completely eliminate bacteria
Although the treated E. diaphana and A. salina underwent 6- and 3.7-fold reductions in B/H, respectively, bacteria were not completely eliminated. It is possible that the ddPCR assay amplified DNA from dead bacteria as DNA can persist long after cell death [70, 71], particularly if cells are intact [72], causing bacteria to be overestimated. However, DNA in seawater aquaria has been shown to degrade to levels below detection by ddPCR in ≤94 h [73] and frequent water changes were performed in the present study, which would have removed free DNA and dead bacteria. Culture techniques could be used to test whether positive ddPCR signals originated from viable bacteria, but as previously noted, such analyses are not conclusive due to the uncultivability of many bacteria. Alternatively, viability staining [74] or RT-qPCR/ddPCR of bacterial mRNA [75] could be used to determine whether the remaining bacteria were alive.
Biofilm formation was evident in all wells of the culture plates (Fig. S7). The wells were not cleaned to avoid introducing bacteria or stressing the anemones, but the resulting biofilms undoubtedly impeded elimination of bacteria from the culture environment by shielding bacteria from the antibiotics [76].
The cnidarian surface mucus layer (SML) can be considered a host-associated biofilm [77, 78] that harbours generally transient bacterial communities distinct from the host tissue and surrounding seawater [79]. The SML protects the host by providing a physical barrier against pathogenic bacteria and can have selective antibiotic, but also antibiotic-inhibiting, properties [80]. Although our methods did not allow us to explore correlations between the E. diaphana SML and tolerant bacteria, it is possible that the SML assisted the survival of some bacteria.
Bacteria in the treated anemones may have also been protected by intracellular encapsulation or multicellular aggregation within E. diaphana tissue. For example, Palincsar et al. [81] found that E. diaphana exposed to high levels of chloramphenicol (125 mg ml–1) or streptomycin (25 mg ml–1) for 3 weeks reduced bacterial aggregates by only ~90 and ~50% respectively, thus emphasizing the challenge of using antibiotics to eliminate bacteria from E. diaphana.
Antibiotic treatment increased bacterial diversity
If completely eliminating bacteria from E. diaphana is not possible, microbiologically standardized gnotobiotic cultures with low bacterial loads and diversity would still be highly valuable [82]. However, the treated anemones underwent significant increases in bacterial alpha and beta diversity despite reductions in B/H. The increase in bacterial richness in the treated anemones points to a large, diverse pool of bacteria within each anemone that were initially below detection. These bacteria were probably held in check by competition from more abundant bacteria but had higher antibiotic tolerance than those they superseded. The large number of unique ASVs detected in each treated anemone also suggests high bacterial variation between each anemone, which could complicate efforts to generate gnotobiotic E. diaphana with uniform bacterial communities.
Some bacteria tolerated antibiotic treatment
Six of the 16 antibiotic-tolerant ASVs identified in the treated E. diaphana were A. salina associates, thus implicating A. salina as the source, particularly as correlations between E. diaphana and A. salina feedstock microbiomes have been previously observed [83]. Seven tolerant ASVs were also associated with Symbiodiniaceae cells isolated from E. diaphana and washed to remove extracellular ASVs [69]. Therefore, these ASVs may have been located intracellularly within Symbiodiniaceae, which also reside within E. diaphana tissue. As this dual encapsulation probably aided the survival of the ASVs by reducing antibiotic exposure, bacterial depletion could be improved by using aposymbiotic anemones (i.e. free of algal symbionts), particularly as the number of Symbiodiniaceae harboured by each anemone (~127 cells mm–2 [84]) suggests they could account for a high fraction of bacterial load. However, the ability of E. diaphana to withstand both bleaching and antibiotic exposure would need to be tested. The dual encapsulation described above could also explain the survival of a tolerant Coxiella ASV, since members of this genus are obligate intracellular parasites [85]. Three tolerant ASVs were members of Vibrio , a genus often associated with microbiome dysbiosis and disease in cnidarians [86], that has many members in the marine environment known to possess antibiotic resistance [87]. The presence of bacteria that are not only tolerant but also antibiotic resistant may make generation of gnotobiotic E. diaphana by antibiotic treatment alone difficult. Four tolerant ASVs belonged to taxonomic groups ( Thalassobius , Saprospiraceae , Marinobacter and Oligoflexaceae ) with members previously identified as core E. diaphana associates [83]. Among these, the family Saprospiraceae is noteworthy as it contains species that are frequently found in plastic-associated marine biofilms, highlighting the role probably played by biofilms in the survival of the tolerant ASVs identified in our study [88–90].
Recommendations for improved bacterial depletion in E. diaphana
Based on our findings, we provide the following recommendations for future gnotobiotic E. diaphana work. First and foremost, the increase in B/H in the control anemones suggests that even under sterile conditions, maintaining vessel cleanliness is essential to remove biofilms and cellular debris that can harbour and protect bacteria. Indeed, since the present study was conducted, rearing E. diaphana under sterile conditions with regular cleaning has been shown to substantially reduce bacterial alpha diversity [91]. Second, due to the correlation between the E. diaphana and A. salina bacteria, proper feedstock sterilization is vital. This could be achieved by hatching the A. salina in higher antibiotic concentrations or by using chemical decapsulation as described by Sorgeloos et al. [92] and employed by Costa et al. [33]. However, care would be required not to introduce more antibiotics through feeding than could be tolerated by E. diaphana. Third, the survival of all treated E. diaphana suggests that prolonged antibiotic treatment is viable, and therefore bacterial load and diversity reduction via long-term exposure should be explored. Fourth, the recent discovery that Symbiodiniaceae contain intracellular bacteria suggests the use of aposymbiotic anemones could help reduce bacterial load and we encourage exploration of this approach. Finally, methods that address the increased bacterial beta diversity we observed in antibiotic-treated E. diaphana should be investigated. These include generating E. diaphana cultures from a single founder anemone, or alternatively from pedal lacerates or cell fragments (i.e. artificial lacerates) since smaller treatment subjects probably harbour fewer bacteria and few, or no, bacterial aggregates. Taken further, sterilization of fertilized eggs or larvae, as performed on other organisms [93–95], could provide the ideal path towards gnotobiotic E. diaphana, although closing the cycle of sexual reproduction in lab-reared E. diaphana has proven elusive [96]
Conclusion
Antibiotic exposure for 3 weeks significantly reduced the bacterial load of E. diaphana, but also increased the complexity and variability of the anemones’ bacterial communities, and hence they could not be defined as gnotobiotic. Extended treatment could improve bacterial depletion, providing culture vessels and food are sterile. However, the efficacy of antibiotic treatment could ultimately be limited by the diversity of native bacteria, some of which probably possess antibiotic tolerance or resistance. Therefore, using treatment subjects with naïve bacterial communities might be needed to create gnotobiotic E. diaphana which, if produced, would represent a substantial advance in cnidarian symbiosis research.
Supplementary Data
Funding information
This study was supported with funding from the Australian Research Council (grant ID: DP160101468) to M.v.O. and L.L.B. M.v.O acknowledges Australian Research Council Laureate Fellowship FL180100036.
Acknowledgement
The authors thank Dr Ruben Costa and Prof. Christian Voolstra from the King Abdullah University of Science and Technology (KAUST) for sharing a draft version of their gnotobiotic Aiptasia protocol. L.M.H. also thanks Ms Franca Casagranda from the University of Melbourne for her assistance in developing the ddPCR B/H assay.
Author contributions
L.M.H., L.L.B. and M.v.O. conceived the study. L.M.H. conducted the investigation, performed the formal analysis, and prepared the original draft. L.L.B. and M.v.O. supervised the study. L.M.H., L.L.B. and M.v.O. reviewed and edited the final manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ASV, amplicon sequence variant; B/H ratio, bacteria/host ratio; ddPCR, digital droplet PCR; GLM, generalized linear model; GLS, generalized least square; nMDS, non-metric multidimensional scaling; NTC, no-template control; PCA, principal component analysis; RCG, Ramaciotti Centre for Genomics; SML, surface mucus layer; sRSS-water, sterile Red Sea Salt-water; UoM, University of Melbourne.
Eight supplementary figures and four supplementary tables are available with the online version of this article.
References
- 1.ICZN Opinion 2404 (Case 3633) – Dysactis pallida Agassiz in Verrill, 1864 (currently Aiptasia pallida; Cnidaria, Anthozoa, Hexacorallia, Actiniaria): precedence over Aiptasia diaphana (Rapp, 1829), Aiptasia tagetes (Duchassaing de Fombressin & Michelotti, 1864), Aiptasia mimosa (Duchassaing de Fombressin & Michelotti, 1864) and Aiptasia inula (Duchassaing de Fombressin & Michelotti, 1864) not approved. Bull Zool Nomencl. 2017;74:130. doi: 10.21805/bzn.v74.a034. [DOI] [Google Scholar]
- 2.Grajales A, Rodríguez E. Morphological revision of the genus Aiptasia and the family Aiptasiidae (Cnidaria, Actiniaria, Metridioidea) Zootaxa. 2014;3826:55–100. doi: 10.11646/zootaxa.3826.1.2. [DOI] [PubMed] [Google Scholar]
- 3.Voolstra CR. A journey into the wild of the cnidarian model system Aiptasia and its symbionts. Mol Ecol. 2013;22:4366–4368. doi: 10.1111/mec.12464. [DOI] [PubMed] [Google Scholar]
- 4.Weis VM, Davy SK, Hoegh-Guldberg O, Rodriguez-Lanetty M, Pringle JR. Cell biology in model systems as the key to understanding corals. Trends Ecol Evol. 2008;23:369–376. doi: 10.1016/j.tree.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Gates RD, Baghdasarian G, Muscatine L. Temperature stress causes host cell detachment in symbiotic cnidarians: implications for coral bleaching. Biol Bull. 1992;182:324–332. doi: 10.2307/1542252. [DOI] [PubMed] [Google Scholar]
- 6.Goulet TL, Cook CB, Goulet D. Effect of short-term exposure to elevated temperatures and light levels on photosynthesis of different host-symbiont combinations in the Aiptasia pallida/Symbiodinium symbiosis . Limnol Oceanogr. 2005;50:1490–1498. doi: 10.4319/lo.2005.50.5.1490. [DOI] [Google Scholar]
- 7.Núñez-Pons L, Bertocci I, Baghdasarian G. Symbiont dynamics during thermal acclimation using cnidarian-dinoflagellate model holobionts. Mar Environ Res. 2017;130:303–314. doi: 10.1016/j.marenvres.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Perez SF, Cook CB, Brooks WR. The role of symbiotic dinoflagellates in the temperature-induced bleaching response of the subtropical sea anemone Aiptasia pallida . J Exp Mar Biol Ecol. 2001;256:1–14. doi: 10.1016/s0022-0981(00)00282-3. [DOI] [PubMed] [Google Scholar]
- 9.Tolleter D, Seneca FO, DeNofrio JC, Krediet CJ, Palumbi SR, et al. Coral bleaching independent of photosynthetic activity. Curr Biol. 2013;23:1782–1786. doi: 10.1016/j.cub.2013.07.041. [DOI] [PubMed] [Google Scholar]
- 10.Hughes TP, Anderson KD, Connolly SR, Heron SF, Kerry JT, et al. Spatial and temporal patterns of mass bleaching of corals in the Anthropocene . Science. 2018;359:80–83. doi: 10.1126/science.aan8048. [DOI] [PubMed] [Google Scholar]
- 11.Oakley CA, Ameismeier MF, Peng L, Weis VM, Grossman AR, et al. Symbiosis induces widespread changes in the proteome of the model cnidarian Aiptasia . Cell Microbiol. 2016;18:1009–1023. doi: 10.1111/cmi.12564. [DOI] [PubMed] [Google Scholar]
- 12.Lehnert EM, Mouchka ME, Burriesci MS, Gallo ND, Schwarz JA, et al. Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3. 2014;4:277–295. doi: 10.1534/g3.113.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medrano E, Merselis DG, Bellantuono AJ, Rodriguez-Lanetty M. Proteomic basis of symbiosis: a heterologous partner fails to duplicate homologous colonization in a novel cnidarian- symbiodiniaceae mutualism. Front Microbiol. 2019;10:1153. doi: 10.3389/fmicb.2019.01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthews JL, Crowder CM, Oakley CA, Lutz A, Roessner U, et al. Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc Natl Acad Sci U S A. 2017;114:13194–13199. doi: 10.1073/pnas.1710733114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burriesci MS, Raab TK, Pringle JR. Evidence that glucose is the major transferred metabolite in dinoflagellate-cnidarian symbiosis. J Exp Biol. 2012;215:3467–3477. doi: 10.1242/jeb.070946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bieri T, Onishi M, Xiang T, Grossman AR, Pringle JR. Relative contributions of various cellular mechanisms to loss of algae during cnidarian bleachingContributions of Various Cellular Mechanisms to Loss of Algae during Cnidarian Bleaching. PLoS One. 2016;11:e0152693. doi: 10.1371/journal.pone.0152693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margulis L. In: Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. Margulis L, Fester R, editors. Cambridge: MIT Press; 1991. Symbiogenesis and symbionticism; pp. 1–14. [PubMed] [Google Scholar]
- 18.Raina J-B, Tapiolas D, Willis BL, Bourne DG. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol. 2009;75:3492–3501. doi: 10.1128/AEM.02567-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas S, Burdett H, Temperton B, Wick R, Snelling D, et al. Evidence for phosphonate usage in the coral holobiont. ISME J. 2010;4:459–461. doi: 10.1038/ismej.2009.129. [DOI] [PubMed] [Google Scholar]
- 20.Ceh J, Kilburn MR, Cliff JB, Raina J-B, van Keulen M, et al. Nutrient cycling in early coral life stages: Pocillopora damicornis larvae provide their algal symbiont (Symbiodinium) with nitrogen acquired from bacterial associates. Ecol Evol. 2013;3:2393–2400. doi: 10.1002/ece3.642. [DOI] [Google Scholar]
- 21.Lesser M, Falcón L, Rodríguez-Román A, Enríquez S, Hoegh-Guldberg O, et al. Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral Montastraea cavernosa. Mar Ecol Prog Ser. 2007;346:143–152. doi: 10.3354/meps07008. [DOI] [Google Scholar]
- 22.Brown T, Rodriguez-Lanetty M. Defending against pathogens - immunological priming and its molecular basis in a sea anemone, cnidarian. Sci Rep. 2015;5:17425. doi: 10.1038/srep17425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krediet CJ, Ritchie KB, Paul VJ, Teplitski M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc Biol Sci. 2013;280:20122328. doi: 10.1098/rspb.2012.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao-Jones J, Ritchie KB, Jones LE, Ellner SP. How microbial community composition regulates coral disease development. PLoS Biol. 2010;8:e1000345. doi: 10.1371/journal.pbio.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohwer F, Kelley S. In: Coral Health and Disease. Rosenberg E, Loya Y, editors. Berlin: Springer; 2004. Culture-independent analyses of coral-associated microbes; pp. 265–277. [Google Scholar]
- 26.Xiang T, Hambleton EA, DeNofrio JC, Pringle JR, Grossman AR. Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity(1) J Phycol. 2013;49:447–458. doi: 10.1111/jpy.12055. [DOI] [PubMed] [Google Scholar]
- 27.Wang JT, Douglas AE. Essential amino acid synthesis and nitrogen recycling in an alga-invertebrate symbiosis. Mar Biol. 1999;135:219–222. doi: 10.1007/s002270050619. [DOI] [Google Scholar]
- 28.Lewis WH, Tahon G, Geesink P, Sousa DZ, Ettema TJG. Innovations to culturing the uncultured microbial majority. Nat Rev Microbiol. 2021;19:225–240. doi: 10.1038/s41579-020-00458-8. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez-Perez G, Hicks AL, Tekieli TM, Radens CM, Williams BL, et al. Maternal antibiotic treatment impacts development of the neonatal intestinal microbiome and antiviral immunity. J Immunol. 2016;196:3768–3779. doi: 10.4049/jimmunol.1502322. [DOI] [PubMed] [Google Scholar]
- 30.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Björkholm B, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brune A. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol. 2014;12:168–180. doi: 10.1038/nrmicro3182. [DOI] [PubMed] [Google Scholar]
- 32.Basic M, Bleich A. Gnotobiotics: Past, present and future. Lab Anim. 2019;53:232–243. doi: 10.1177/0023677219836715. [DOI] [PubMed] [Google Scholar]
- 33.Costa RM, Cárdenas A, Voolstra C. Protocol for bacterial depletion of Aiptasia anemones - towards the generation of gnotobiotic/germ-free cnidarian host animals. protocols.io. 2019. [DOI]
- 34.Costa RM, Cárdenas A, Loussert-Fonta C, Toullec G, Meibom A, et al. Surface Topography, bacterial carrying capacity, and the prospect of microbiome manipulation in the sea anemone coral model Aiptasia . Front Microbiol. 2021;12:492. doi: 10.3389/fmicb.2021.637834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dungan AM, Hartman LM, Tortorelli G, Belderok R, Lamb AM, et al. Exaiptasia diaphana from the great barrier reef: a valuable resource for coral symbiosis research. Symbiosis. 2020;80:195–206. doi: 10.1007/s13199-020-00665-0. [DOI] [Google Scholar]
- 36.Wilson K, Li Y, Whan V, Lehnert S, Byrne K, et al. Genetic mapping of the black tiger shrimp Penaeus monodon with amplified fragment length polymorphism. Aquaculture. 2002;204:297–309. doi: 10.1016/S0044-8486(01)00842-0. [DOI] [Google Scholar]
- 37.Hartman LM, van Oppen MJH, Blackall LL. The effect of thermal stress on the bacterial microbiome of Exaiptasia diaphana . Microorganisms. 2019;8:20. doi: 10.3390/microorganisms8010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deb R, Nair A, Agashe D. Host dietary specialization and neutral assembly shape gut bacterial communities of wild dragonflies. PeerJ. 2019;7:e8058. doi: 10.7717/peerj.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhukova M, Sapountzis P, Schiøtt M, Boomsma JJ. Diversity and transmission of gut bacteria in Atta and Acromyrmex leaf-cutting ants during development. Front Microbiol. 2017;8:1942. doi: 10.3389/fmicb.2017.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Catterson JH, Khericha M, Dyson MC, Vincent AJ, Callard R, et al. Short-term, intermittent fasting induces long-lasting gut health andTerm, Intermittent Fasting Induces Long-Lasting Gut Health and TOR-independent lifespan extensionIndependent Lifespan Extension. Curr Biol. 2018;28:1714–1724. doi: 10.1016/j.cub.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong L, Meng Y, Wang J, Liu Y. Evaluation of droplet digital PCR for characterizing plasmid reference material used for quantifying ammonia oxidizers and denitrifiers. Anal Bioanal Chem. 2014;406:1701–1712. doi: 10.1007/s00216-013-7546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witte AK, Mester P, Fister S, Witte M, Schoder D, et al. A systematic investigation of parameters influencing droplet rain in the Listeria monocytogenes prfA assay - reduction of ambiguous results in ddPCR. PLoS One. 2016;11:e0168179. doi: 10.1371/journal.pone.0168179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. doi: 10.5555/1953048.2078195. [DOI] [Google Scholar]
- 49.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Core Team 2020 R: a language and environment for statisitical computing. version 4.0.3. 2020. http://www.R-project.org
- 52.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. 2016. https://ggplot2.tidyverse.org
- 53.Pinheiro J, Bates D, DebRoy S, Sarkar D. 2020 nlme: linear and nonlinear mixed effects models. R package. version 3.1-152. 2020. https://CRAN.R-project.org/package=nlme
- 54.Levene H. In: Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling. Olkin I, Ghurye SG, Hoeffding W, Madow WG, Mann HB, editors. Menlo Park: Stanford University Press; 1960. Robust tests for equality of variances; pp. 278–292. [Google Scholar]
- 55.Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. doi: 10.1093/biomet/52.3-4.591. [DOI] [Google Scholar]
- 56.Student The probable error of a mean. Biometrika. 1908;6:1. doi: 10.2307/2331554. [DOI] [Google Scholar]
- 57.Whitney J. Testing for differences with the nonparametric Mann-Whitney U test. J Wound Ostomy Continence Nurs. 1997;24:12. doi: 10.1016/s1071-5754(97)90044-9. [DOI] [PubMed] [Google Scholar]
- 58.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, et al. 2020 vegan: community ecology package. R package. version 2.5-7. 2020. https://CRAN.R-project.org/package=vegan
- 60.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vandeputte D, Kathagen G, D’hoe K, Vieira-Silva S, Valles-Colomer M, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551:507–511. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 62.Jian C, Luukkonen P, Yki-Järvinen H, Salonen A, Korpela K. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS One. 2020;15:e0227285. doi: 10.1371/journal.pone.0227285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bray JR, Curtis JT. An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr. 1957;27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 64.Rohart F, Gautier B, Singh A, Lê Cao K-A. mixOmics: an R package for omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simpson EH. Measurement of dDiversity. Nature. 1949;163:688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 66.Shannon CE, Weaver W. The Mathematical Theory of Communication. Champaign, IL: University of Illinois Press; 1949. [Google Scholar]
- 67.Wang Y, Naumann U, Wright ST, Warton DI. mvabund - an R package for model-based analysis of multivariate abundance data. Methods Ecol Evol. 2012;3:471–474. doi: 10.1111/j.2041-210X.2012.00190.x. [DOI] [Google Scholar]
- 68.Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 69.Maire J, Girvan SK, Barkla SE, Perez-Gonzalez A, Suggett DJ, et al. Intracellular bacteria are common and taxonomically diverse in cultured and in hospite algal endosymbionts of coral reefs. ISME J. 2021;15:2028–2042. doi: 10.1038/s41396-021-00902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young G, Turner S, Davies JK, Sundqvist G, Figdor D. Bacterial DNA persists for extended periods after cell death. J Endod. 2007;33:1417–1420. doi: 10.1016/j.joen.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Salo WL, Aufderheide AC, Buikstra J, Holcomb TA. Identification of Mycobacterium tuberculosis DNA in a pre-Columbian Peruvian mummy. Proc Natl Acad Sci U S A. 1994;91:2091–2094. doi: 10.1073/pnas.91.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brundin M, Figdor D, Roth C, Davies JK, Sundqvist G, et al. Persistence of dead-cell bacterial DNA in ex vivo root canals and influence of nucleases on DNA decay in vitro. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2010;110:789–794. doi: 10.1016/j.tripleo.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 73.Wood SA, Biessy L, Latchford JL, Zaiko A, von Ammon U, et al. Release and degradation of environmental DNA and RNA in a marine system. Sci Total Environ. 2020;704:135314. doi: 10.1016/j.scitotenv.2019.135314. [DOI] [PubMed] [Google Scholar]
- 74.Emerson JB, Adams RI, Román CMB, Brooks B, Coil DA, et al. Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome. 2017;5:86. doi: 10.1186/s40168-017-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Magalhães AP, França Â, Pereira MO, Cerca N. RNA-based qPCR as a tool to quantify and to characterize dual-species biofilms. Sci Rep. 2019;9:13639. doi: 10.1038/s41598-019-50094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 77.Rivera-Ortega J, Thomé PE. Contrasting antibacterial capabilities of the surface mucus layer from three symbiotic cnidarians. Front Mar Sci. 2018;5:392. doi: 10.3389/fmars.2018.00392. [DOI] [Google Scholar]
- 78.Sweet MJ, Croquer A, Bythell JC. Development of bacterial biofilms on artificial corals in comparison to surface-associated microbes of hard corals. PLoS One. 2011;6:e21195. doi: 10.1371/journal.pone.0021195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sweet MJ, Croquer A, Bythell JC. Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs. 2010;30:39–52. doi: 10.1007/s00338-010-0695-1. [DOI] [Google Scholar]
- 80.Ritchie KB. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser. 2006;322:1–14. doi: 10.3354/meps322001. [DOI] [Google Scholar]
- 81.Palincsar EE, Jones WR, Palincsar JS, Glogowski MA, Mastro JL. Bacterial Aggregates within the epidermis of the sea anemone Aiptasia pallida . The Biological Bulletin. 1989;177:130–140. doi: 10.2307/1541840. [DOI] [Google Scholar]
- 82.Gordon HA, Pesti L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol Rev. 1972;35:390–429. doi: 10.1128/br.35.4.390-429.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hartman LM, van Oppen MJH, Blackall LL. Microbiota characterization of Exaiptasia diaphana from the Great Barrier Reef. Anim Microbiome. 2020;2:10. doi: 10.1186/s42523-020-00029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tortorelli G, Belderok R, Davy SK, McFadden GI, van Oppen MJH. Host Genotypic effect on algal symbiosis establishment in the coral model, the anemone Exaiptasia diaphana, from the Great Barrier Reef. Front Mar Sci. 2020;6:833. doi: 10.3389/fmars.2019.00833. [DOI] [Google Scholar]
- 85.Voth DE, Heinzen RA. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii . Cell Microbiol. 2007;9:829–840. doi: 10.1111/j.1462-5822.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- 86.Munn CB. The role of Vibrios in diseases of corals. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.VE-0006-2014. [DOI] [PubMed] [Google Scholar]
- 87.Loo K, Letchumanan V, Law JW, Pusparajah P, Goh B, et al. Incidence of antibiotic resistance in Vibrio spp. Rev Aquacult. 2020;12:2590–2608. doi: 10.1111/raq.12460. [DOI] [Google Scholar]
- 88.Zettler ER, Mincer TJ, Amaral-Zettler LA. Life in the “Plastisphere”: microbial communities on plastic marine debris. Environ Sci Technol. 2013;47:7137–7146. doi: 10.1021/es401288x. [DOI] [PubMed] [Google Scholar]
- 89.Oberbeckmann S, Osborn AM, Duhaime MB. Microbes on a bottle: substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLoS One. 2016;11:e0159289. doi: 10.1371/journal.pone.0159289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kirstein IV, Wichels A, Gullans E, Krohne G, Gerdts G. The Plastisphere - uncovering tightly attached plastic “specific” microorganisms. PLoS One. 2019;14:e0215859. doi: 10.1371/journal.pone.0215859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dungan AM, van Oppen MJH, Blackall LL. Short-term exposure to sterile seawater reduces bacterial community diversity in the sea anemone, Exaiptasia diaphana . Front Mar Sci. 2021;7:599314. doi: 10.3389/fmars.2020.599314. [DOI] [Google Scholar]
- 92.Sorgeloos P, Bossuyt E, Laviña E, Baeza-Mesa M, Persoone G. Decapsulation of Artemia cysts: a simple technique for the improvement of the use of brine shrimp in aquaculture. Aquaculture. 1977;12:311–315. doi: 10.1016/0044-8486(77)90209-5. [DOI] [Google Scholar]
- 93.Forberg T, Milligan-Myhre K. In: Gnotobiotics. Schoeb TR, Eaton KA, editors. London: Academic Press; 2017. Gnotobiotic fish as models to study host-microbe interactions; pp. 369–383. [DOI] [Google Scholar]
- 94.Ridley EV, Wong ACN, Douglas AE. Microbe-dependent and nonspecific effects of procedures to eliminate the resident microbiota from Drosophila melanogaster . Appl Environ Microbiol. 2013;79:3209–3214. doi: 10.1128/AEM.00206-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leigh BA, Liberti A, Dishaw LJ. Generation of germ-free Ciona intestinalis for studies of gut-microbe interactions. Front Microbiol. 2016;7:2092. doi: 10.3389/fmicb.2016.02092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grawunder D, Hambleton EA, Bucher M, Wolfowicz I, Bechtoldt N, et al. Induction of gametogenesis in the cnidarian endosymbiosis model Aiptasia sp. Sci Rep. 2015;5:15677. doi: 10.1038/srep15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reyes-Bermudez A, Miller DJ. In vitro culture of cells derived from larvae of the staghorn coral Acropora millepora. Coral Reefs. 2009;28:859–864. doi: 10.1007/s00338-009-0527-3. [DOI] [Google Scholar]
- 98.Sweet MJ, Croquer A, Bythell JC. Experimental antibiotic treatment identifies potential pathogens of white band disease in the endangered Caribbean coral Acropora cervicornis . Proc Biol Sci. 2014;281:20140094. doi: 10.1098/rspb.2014.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Soffer N, Gibbs PDL, Baker AC. In: Proceedings — 11th International Coral Reef Symposium Fort Lauderdale. Riegl B, Dodge RE, editors. Davie, FL: Nova Southeastern University National Coral Reef Institute; 2008. Practical applications of contaminant-free Symbiodinium cultures grown on solid media; pp. 159–163. [Google Scholar]
- 100.Weiland-Bräuer N, Neulinger SC, Pinnow N, Künzel S, Baines JF, et al. Composition of bacterial communities associated with Aurelia aurita changes with compartment, life stage, and population. Appl Environ Microbiol. 2015;81:6038–6052. doi: 10.1128/AEM.01601-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rahat M, Dimentman C. Cultivation of bacteria-free Hydra viridis: missing budding factor in nonsymbiotic hydra. Science. 1982;216:67–68. doi: 10.1126/science.7063873. [DOI] [PubMed] [Google Scholar]
- 102.Richardson C, Hill M, Marks C, Runyen-Janecky L, Hill A. Experimental manipulation of sponge/bacterial symbiont community composition with antibiotics: sponge cell aggregates as a unique tool to study animal/microorganism symbiosis. FEMS Microbiol Ecol. 2012;81:407–418. doi: 10.1111/j.1574-6941.2012.01365.x. [DOI] [PubMed] [Google Scholar]
- 103.Mills E, Shechtman K, Loya Y, Rosenberg E. Bacteria appear to play important roles in both causing and preventing the bleaching of the coral Oculina patagonica . Mar Ecol Prog Ser. 2013;489:155–162. doi: 10.3354/meps10391. [DOI] [Google Scholar]
- 104.Polne-Fuller M. A novel technique for preparation of axenic cultures of Symbiodinium (Pyrrophyta) through selective digestion by amoebae. J Phycol. 1991;27:552–554. doi: 10.1111/j.0022-3646.1991.00552.x. [DOI] [Google Scholar]
- 105.Glasl B, Herndl GJ, Frade PR. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 2016;10:2280–2292. doi: 10.1038/ismej.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.D’Agostino A. In: Proceedings — 1st Conference on Culture of Marine Invertebrate Animals Greenport. Smith WL, Chanley MH, editors. Boston, MA: Springer; 1975. Antibiotics in cultures of invertebrates; pp. 109–133. [DOI] [Google Scholar]
- 107.Hawkins TD, Hagemeyer JCG, Hoadley KD, Marsh AG, Warner ME. Partitioning of respiration in an animal-algal symbiosis: implications for different aerobic capacity between Symbiodinium spp. Front Physiol. 2016;7:128. doi: 10.3389/fphys.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jiang L, Hou L, Zou X, Zhang R, Wang J, et al. Cloning and expression analysis of p26 gene in Artemia sinica . Acta Biochim Biophys Sin. 2007;39:351–358. doi: 10.1111/j.1745-7270.2007.00287.x. [DOI] [PubMed] [Google Scholar]
- 109.Niu Y, Defoirdt T, Baruah K, Van de Wiele T, Dong S, et al. Bacillus sp. LT3 improves the survival of gnotobiotic brine shrimp (Artemia franciscana) larvae challenged with Vibrio campbellii by enhancing the innate immune response and by decreasing the activity of shrimp-associated vibrios. Vet Microbiol. 2014;173:279–288. doi: 10.1016/j.vetmic.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 110.Valverde EJ, Labella AM, Borrego JJ, Castro D. Artemia spp., a susceptible host and vector for lymphocystis disease virus. Viruses. 2019;11:E506. doi: 10.3390/v11060506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Korea Polar Research Institute The whole genome shotgun sequencing project of Artemia franciscana . 2021. [ June 12; 2021 ]. https://antagen.kopri.re.kr/project/genome_info_iframe.php?Code=AF01 accessed.
- 112.Wang Y, Qian P-. Y. In: Encyclopedia of Metagenomics. Nelson KE, editor. New York: Springer; 2013. Conserved regions in 16S ribosome RNA sequences and primer design for studies of environmental microbes. [Google Scholar]
- 113.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.