Abstract
Purpose:
Uveal melanoma (UM) is the most common eye cancer in adults. Approximately 50% of UM patients develop metastatic UM (mUM) in the liver, even after successful treatment of the primary lesions. mUM is refractory to current chemo- and immune-therapies, and most mUM patients die within a year. UM is characterized by gain-of-function mutations in GNAQ/GNA11, encoding Gαq proteins. We have recently shown that the Gαq-oncogenic signaling circuitry involves a non-canonical pathway distinct from the classical activation of PLCβ and MEK-ERK. GNAQ promotes the activation of YAP1, a key oncogenic driver, through focal adhesion kinase (FAK), thereby identifying FAK as a druggable signaling hub downstream from GNAQ. However, targeted therapies often activate compensatory resistance mechanisms leading to cancer relapse and treatment failure.
Experimental Design:
We performed a kinome-wide CRISPR-Cas9 sgRNA screen to identify synthetic lethal gene interactions that can be exploited therapeutically. Candidate adaptive resistance mechanisms were investigated by co-targeting strategies in UM and mUM in vitro and in vivo experimental systems.
Results:
sgRNAs targeting the PKC and MEK-ERK signaling pathways were significantly depleted after FAK inhibition, with ERK activation representing a predominant resistance mechanism. Pharmacological inhibition of MEK and FAK showed remarkable synergistic growth-inhibitory effects in UM cells and exerted cytotoxic effects leading to tumor collapse in UM xenograft and liver mUM models in vivo.
Conclusions:
Coupling the unique genetic landscape of UM with the power of unbiased genetic screens, our studies reveal that FAK and MEK-ERK co-targeting may provide a new network-based precision therapeutic strategy for mUM treatment.
Keywords: FAK, MEK, GNAQ, uveal melanoma, targeted therapy, precision medicine
Introduction
G protein-coupled receptors (GPCRs) are the largest family of cell surface proteins with over 800 members (1), and their dysregulation contributes to some of the most prevalent human diseases (2–4). GPCRs represent the largest family of targets for approved drugs. Strikingly, our recent analysis of human cancer genomes revealed that nearly 30% of human cancers present mutations in G proteins and GPCRs (5). In particular, uveal melanoma (UM) can be defined as a Gαq-driven malignancy. Indeed, Approximately 93% of UM lesions harbor activating mutations in GNAQ or GNA11, encoding for the alpha subunits Gαq and Gα11 of the heterotrimeric G protein respectively (6,7). An additional 4% harbor mutations in the Gαq-linked receptor CYSLTR2, also acting as a driver oncogene (8).
UM is diagnosed in about 2,500 adults in the United States every year, and is the most common primary cancer of the eye in adults and the second most common melanoma subtype after skin cutaneous melanoma (SKCM) (9). While the majority of early-stage UM lesions can be treated by irradiation or enucleation, approximately 50% of the patients will metastasize, primarily to the liver, within 5–10 years after diagnosis (10). Inactivating mutations or copy loss of the BAP1 gene, which is located on chromosome 3p21, are strongly associated with metastasis in UM patients (11), supporting that BAP1 functions as a metastasis suppressor (12). Most metastatic UM (mUM) patients are refractory to current chemotherapies and immune checkpoint blockade (13). Ultimately, the majority of advanced disease patients succumb within a year due to the suboptimal efficacy of these treatments, often combined with severe toxicities, underlying the high unmet medical need for new therapeutic strategies.
Most of the recent clinical research efforts in UM have focused on inhibiting the Gαq classical signaling pathway, PLCβ-PKC-ERK. MEK inhibitors (MEKi) selumetinib and trametinib have been extensively evaluated for mUM. Despite encouraging results in pre-clinical studies, MEK inhibition with these agents has been shown to have nearly no impact on the overall survival of mUM patients, as single-agent or when combined with chemotherapy (14–16). Our recent findings uncovered a non-canonical Gαq signaling pathway leading to the Rho-dependent activation of the Hippo/YAP pathway, which contributes to aberrant cancer cell growth (17,18). By further decoding this oncogenic signaling circuitry, we showed that the non-receptor focal adhesion kinase (FAK) is an integral node of this non-canonical Gαq pathway (19). Interestingly, FAK overexpression has already been associated with various cancer types including ovarian, head and neck, and breast cancers (20–22). Furthermore, multiple FAK inhibitors (FAKi) – defactinib (VS-6063), PF-562271, GSK2256098 and IN10018 – have been already tested in clinical trials, showing manageable toxicity profiles (23), and hence can be considered for mUM treatment. However, single-agent targeted therapies often activate compensatory mechanisms resulting in treatment resistance. This prompted us to perform a kinome-wide CRISPR-Cas9 sgRNA screen to identify synthetic lethal gene interactions in the context of FAKi that can be exploited therapeutically (24). Our study demonstrates that dual inhibition of MEK and FAK act synergistically to promote the conversion of cytostatic to cytotoxic inhibition of tumor growth, thereby identifying a new treatment option for mUM.
Materials and Methods
Reagents
DMEM, RPMI-1640 and antibiotic/antimycotic solution were purchased from Sigma-Aldrich. Turbofect, DMEM/F12 Glutamax, basic fibroblast growth factor (bFGF), epithelial growth factor (EGF), B-27 and N2 supplements were purchased from ThermoFisher Scientific. Fetal bovine serum (FBS, Lot 18H165) and penicillin/streptomycin from Sigma Aldrich, Polybrene from Millipore, Aquabluer from MultiTarget Pharmaceuticals.
VS-4718, VS-6063 and VS-6766 were provided by Verastem Oncology. All inhibitors used in this study were purchased from Selleck Chemicals (Houston, USA). Anhydrous DMSO, carboxymethyl cellulose (CMC), Polyethylene Glycol 400 (PEG400), Tween 80 were purchased from Sigma-Aldrich. Inhibitors were dissolved in DMSO to a stock concentration of 10 mM, aliquoted and stored at − 80 °C for in vitro experiments. For in vivo treatments, trametinib was prepared at a stock concentration 5mg/ml in DMSO and freshly diluted in PBS containing 5.2% PEG400 + 5.2% Tween 80 and a dose of 1mg/kg was administered once daily by intraperitoneal injection. VS-4718 was freshly diluted at 5 mg/ml in 0.5% carboxymethyl cellulose (CMC) (C5678, Sigma-Aldrich; St. Louis, MO) + 0.1% Tween 80 (P1754, Sigma Aldrich; St. Louis, MO) in sterile water (B. Braun Medical; Irvine, CA) and a dose of 10ml/kg was administered by oral gavage twice a day.
FAK, phospho-FAK, ERK, phospho-ERK, cleaved PARP1, cleaved Caspase 3, YAP, Cas9, and GAPDH antibodies were purchased from Cell Signaling. BAP1 and PARP1 were purchased from Santa Cruz Biotechnology. BrdU was purchased Abcam. All antibodies were diluted in 5% BSA at 1:500–1:1000 before use for immunoblotting and in 3% BSA at 1:100–1:400 for immunohistochemistry.
pHAGE PGK-GFP-IRES-LUC-W was from addgene (#46793).
Cell Lines
HEK 293T were cultured in High Glucose DMEM and supplemented with 10% FBS and antibiotic/antimycotic solution (100U penicillin, 0.1 mg/ml streptomycin and 0.25 μg/ml amphotericin B). UM cell lines were cultured in RPMI-1640 media supplemented with 10% FBS (92.1, OMM1.3, OMM1.5 and OMM1.5Cas9) or 20% FBS (MP41, MP46, MP38 and MM28) and antibiotic/antimycotic solution. All cell lines were tested for mycoplasma and proved to be mycoplasma free using the MycoAlert™ PLUS Mycoplasma Detection Kit (Lonza).
Sphere Formation Assay
Cells were seeded in 96-well ultra-low attachment culture dishes (Corning) at 100 cells/well with the indicated concentration of 10 nM trametinib, 1 uM VS-4718 or both. Medium consisted of serum-free DMEM/F12 Glutamax supplemented bFGF (20 ng/ml), EGF (20 ng/ml), B-27 (1:50 dilution), and N2 supplement (1:100 dilution). Three weeks after seeding the number of spheres in each well and their sizes were assessed by bright-field microscopy and quantify using ImageJ (25).
Immunoblotting
UM cells were harvested at the indicated timepoints post-treatment with 10 nM trametinib and 1 μM VS-4718. Cell lysates were subjected to SDS/PAGE on 10% acrylamide gels and electroblotted to PVDF membranes. Blocking and primary and secondary antibody incubations of immunoblots were performed in Tris-buffered saline/Tween 20 [10 mM Tris (pH 7.4), 150 mM NaCl, and 0.1% Tween 20] supplemented with 5% (wt/vol) bovine serum albumin (BSA). The primary antibodies were used according to the manufacturers’ instructions. HRP-conjugated donkey anti-rabbit and anti-mouse IgGs were used at a dilution of 1:5,000, and immunoreactive bands were detected using enhanced chemiluminescence. Full blots are shown in Supplementary Fig. S4
Flow cytometry
Apoptosis was determined by detecting phosphatidylserine exposure on cell plasma membranes using the fluorescent dye FITC Annexin V (BD Biosciences, San Diego, CA, USA) according to the manufacturer’s protocol. Briefly, at the end of the treatment period, the cells (including floating cells) were harvested and washed twice with cold PBS. After resuspension in 100 μl 1X binding buffer, 5 μl FITC Annexin V was added and the cells were incubated for 15 min at room temperature in the dark. Finally, 400 μl 1X binding buffer was added to the cells and flow cytometric analysis was conducted. The experiments were conducted in triplicate.
Immunohistochemistry
For IHC, all tissue samples were processed and stained as previously described (26). Slides were scanned using a Zeiss Axioscan Z1 slide scanner equipped with a 20×/0.8 NA objective. All images analysis was performed using the QuPath software (27) to perform cell detection and quantify each marker as a percentage of cells stained.
Kinome-wide CRISPR screen
Generation and validation of stable Cas9 lines
For lentivirus-Cas9 production, 293T cells were plated in a poly-D-lysine–coated dish and, 16 hours later, transfected with 12 μg pLenti-CAS9-Blast, 8 μg psPAX2, 4 μg pCMV-VSV-G, using 32 μl TurboFect™ Transfection Reagent, and media was refreshed 6 hours post-transfection. 48 and 72h hours later, the virus-containing media was collected, filtered through a low protein binding filter unit (PVDF, 0.45um, Sigma-Aldrich) and stored at 4°C up to 5 days or at −80°C.
For lentivirus-Cas9 infection, OMM 1.5 cells were plated in a 6-well plate and, 16 hours later, transduced using 1 ml virus-containing media + 1 ml complete media + 10 μg/ml polybrene. The plate was centrifuged for 15 min at 1200 rpm. The same process was repeated after 24 hours and 48 hours later the cells were selected with blasticidin (10 μg/ml).
Cas9 editing efficiency was measured by quantifying the editing frequency of the safe-harbor locus AAVS1(TGCCTAACAGGAGGTGGGGGTTAGACCCAATATCAGGAGACTAGGAAGGAGGAGGCCTAAGGATGGGGCTTTTCTGTCACCAATCCTGTCCCTAGTGGCCCCACTGTGGGGTGGAGGGG) by next-generation sequencing using two separate sgRNA sequences (sgT1 and sgT2). Briefly, cells were transduced with either AAVS1 sgRNA and, after hygromycin-B selection, genomic DNA was isolated using the Qiagen DNeasy Blood and Tissue kit (Cat # 69504). The AAVS1 region was amplified, DNA gel-purified, and barcoded using the NEBNext Multiplex Oligos for Illumina kit (E7335) for multi-plex sequencing. Analysis of genome editing was performed using CRISPResso (28).
Pooled CRISPR library details and lentivirus preparation
In total, 3,052 unique sgRNAs targeting 763 human kinome genes for 4 guides per target were used for pooled CRISPR screens (Brunello Human Kinome CRISPR Knockout Library (29); Addgene Cat #75312). Lenti-X 293T cells were seeded in 15 cm plates and transfected the next day with: pooled CRISPR library DNA (21 ug), PAX2 (14 ug), and VSV-g (7 ug). Viral media was collected 48- and 72-hours post-transfection, pooled and concentrated using an ultracentrifugation protocol, and stored at −80°C in aliquots.
Infectious virus titer determination and transduction
Several aliquots of concentrated viral stock were used to infect separate 15 cm plates containing 4.8×106 OMM1.5-Cas9 cells that were seeded the day before. Non-infected plates for OMM 1.5 parental and Cas9 were also plated as antibiotic selection controls. 48 hours post-transduction, cells were treated with 2 ug/ml puromycin for 72 hours. After puromycin selection, complete cell death was achieved in uninfected control plates. The number of puromycin-resistant cells in each infected plate was counted using the Countess Automated Cell Counter to calculate the functional titer of the viral stock. Cells for screening were infected with a virus dose to achieve a MOI of 0.4–0.5 with sufficient cell numbers plated to obtain a screening depth of >1000 cells per sgRNA.
Screen set-up
4.8×106 cells were seeded into 15 cm plates a day before CRISPR library transduction. 48 hours post-transduction, cells were subject to puromycin selection (2 μg/ml; 72 hours). Cells were amplified and seeded into 15 cm plates (4×106 cells per plate) divided into two treatment arms: 3 replicate plates for either vehicle/DMSO or VS-4718 treatments. 4×106 cells from each individual plate were passaged into a new plate containing DMSO or 0.5 μM VS-4718 every 3–4 days for a total of 10 days of treatment. 5×106 cells were aliquoted from each plate at the beginning and end of the screen and stored at −80°C for sgRNA quantification.
sgRNA quantification
Genomic DNA was isolated using the Qiagen DNeasy Blood and Tissue kit (Cat # 69504) according to manufacturer’s protocol. DNA concentration was measured using a Qubit assay (ThermoFisher Scientific; Cat #Q33226). Amplification of sgRNAs for next-generation sequencing was performed according to the Broad Institute’s recommended protocol. NGS read counts were processed, aligned, and analyzed using PinAPL-Py (30).
Pre-screen evaluation of sgRNAs:
sgRNAs were quantified immediately prior to the start of the screen to evaluate CRISPR library representation: sgRNA representation was 99.5% and gene representation was 100%.
Post-screen evaluation of sgRNAs:
sgRNAs were quantified from each replicate plate at the end of the screen and analyzed to identify sgRNAs depleted in VS-4718 treated cells relative to DMSO treated cells.
Synergy determination
Dose-response curves and determination of IC50 values
Cells were seeded at a density of 5×103 to 1×104 cells/well in 96-well white plates. Eight different dilutions of each inhibitor were assayed in technical triplicates for 72 h in each experiment. Cell viability was measured with the Aquabluer Cell Viability Reagent on a Spark microplate reader (Tecan). Using the GraphPad Prism v8.2.0 software, the half-maximal inhibitor concentration values (GI50) were determined from the curve using the nonlinear log (inhibitor) vs response-variable slope (three parameters) equation. GI50 values were only determined for compounds which inhibited growth by more than 50%.
Synergy determination with the Chou-Talalay method
The Chou-Talalay method (31) was used to determine possible synergistic effects of selected kinase inhibitor combinations. Briefly, cells were seeded at a density of 5×103 to 1×104 cells/well in 96-well white plates (CulturePlate; PerkinElmer Inc.). Cells were treated with either single inhibitors or combinations thereof using eight different dilutions of each inhibitor and in technical triplicates. Cell viability was measured, after 72 h treatment, with the Aquabluer Cell Viability Reagent on a Spark microplate reader (Tecan). Combination Index (CI) values showing either synergy (< 1) or antagonism (> 1) were calculated using the following equation:
CI = (D)1/(Dx)1+(D)2/(Dx)2, where Dx equal the concentration of the tested substance used in the single treatment that was required to decrease the cell viability by x% and D equals the concentration of the tested substance 1 in combination with the concentration of the tested substance 2 that when combined decreased the cell number by x%.
Synergy determination with the Bliss delta score
The Bliss independence model(32) assumes a stochastic process in which two drugs elicit their effects independently, and the expected combination effect was calculated using the following equation: IAB = IA + IB – IA x IB, where IA and IB are the single agent inhibition levels at fixed concentrations. If the experimentally measured effect of the drug combination was equal to, higher than or lower than the expected effect (IAB), the combination was considered to be additive (ΔBliss =0), synergistic (<0) or antagonistic (>0), respectively.
Human Xenograft Tumor Models
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of University of California, San Diego with protocol S15195. Female 4- to 6-week-old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (SCID-NOD) mice were purchased from the UCSD in-house breeding program. Mice were injected subcutaneously in both flanks with either 2×106 or 2.5×106 92.1 or OMM1.3 cells, respectively. Mice were monitored twice weekly for tumor development. Tumor growth analysis was assessed as LW2/2, where L and W represent length and width of the tumor. 5 mg/ml VS-4718 was prepared in 0.5% carboxymethyl cellulose (CMC) (Sigma-Aldrich; St. Louis, MO) and 0.1% Tween 80 (Sigma-Aldrich) in sterile water. 0.1 mg/ml trametinib was prepared in 5.2% polyethylene glycol 400 (Sigma-Aldrich) and 5.3% Tween 80 (Sigma-Aldrich) in DPBS. Mice were administered 50 mg/kg VS-4718 (Verastem Oncology; Needham, MA) twice daily by oral gavage and 1 mg/kg Trametinib once daily by intra-peritoneal injection (IP); control group was treated with each vehicle. Mice were euthanized at the indicated time points and tumors were isolated for sequencing, histologic, and immunohistochemical evaluation. Results of mice experiments were expressed as mean ± SEM of a total of tumors analyzed.
Human Metastasis Tumor Model
Generation of stable GFP-Luc expressing 92.1
For lentivirus-GFP-Luc production, 293T cells were plated in a poly-D-lysine–coated 15cm dish and, 16 hours later, transfected with 30 μg pHAGE PGK-GFP-IRES-LUC-W, 3 μg VSV-G, 1.5 μg Tat1b, 1.5 μg Rev1b, 1.5 μg Gag/Pol using 25.2 μl p3000 buffer and 25.2 μl of Lipofectamine 3000 transfection reagent, and media was refreshed 16 hours post-transfection. 48 and 72h hours later, the virus-containing media was collected, filtered through a low protein binding filter unit (PVDF, 0.45um, Sigma-Aldrich) and stored at 4°C up to 5 days or at −80°C.
For lentivirus-Cas9 infection, 92.1 cells were plated in a 6-well plate and, 16 hours later, transduced using 1 ml virus-containing media + 1 ml complete media + 10 μg/ml polybrene. The plate was centrifuged for 15 min at 1200 rpm. GFP expression was validated by fluorescent microscopy.
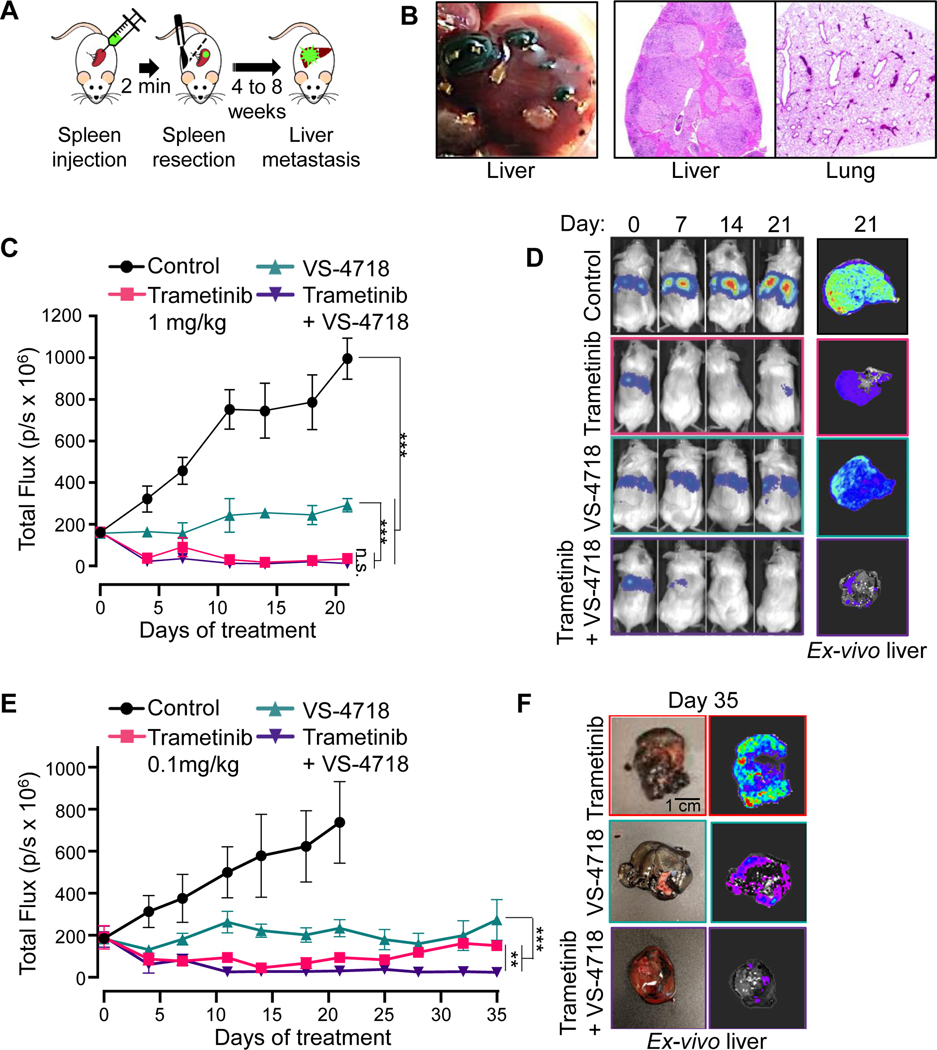
Splenic injection
Mice were injected with 1×106 92.1 GFP-Luc cells in the spleen, followed by removal of the spleen at 2 minutes post-injection. Tumor implantation by bioluminescence was assessed twice weekly by bioluminescence images captured using the In Vivo Imaging System (IVIS) Spectrum (Perkin Elmer, Santa Clara, CA). To this end, mice received an intraperitoneal injection of 200 mg/kg D-luciferin firefly potassium salt diluted in PBS 15 minutes before imaging (GoldBio, St Louis, MO). Total bioluminescence was determined upon subtracting the background from the region of interest (ROI). Vehicle, trametinib, VS-4718 or trametinib/VS-4718 were administered, starting 7 days post-surgery, with the described above dosing.
Statistical analysis.
GraphPad Prism version 8 for Windows (GraphPad Software, San Diego, CA) was used to perform data analyses, variation estimation and validation of test assumptions. Statistical analysis was performed using either a paired Student’s t test or one-way ANOVA.
Results
Identification of conditionally lethal drug targets of FAK inhibition.
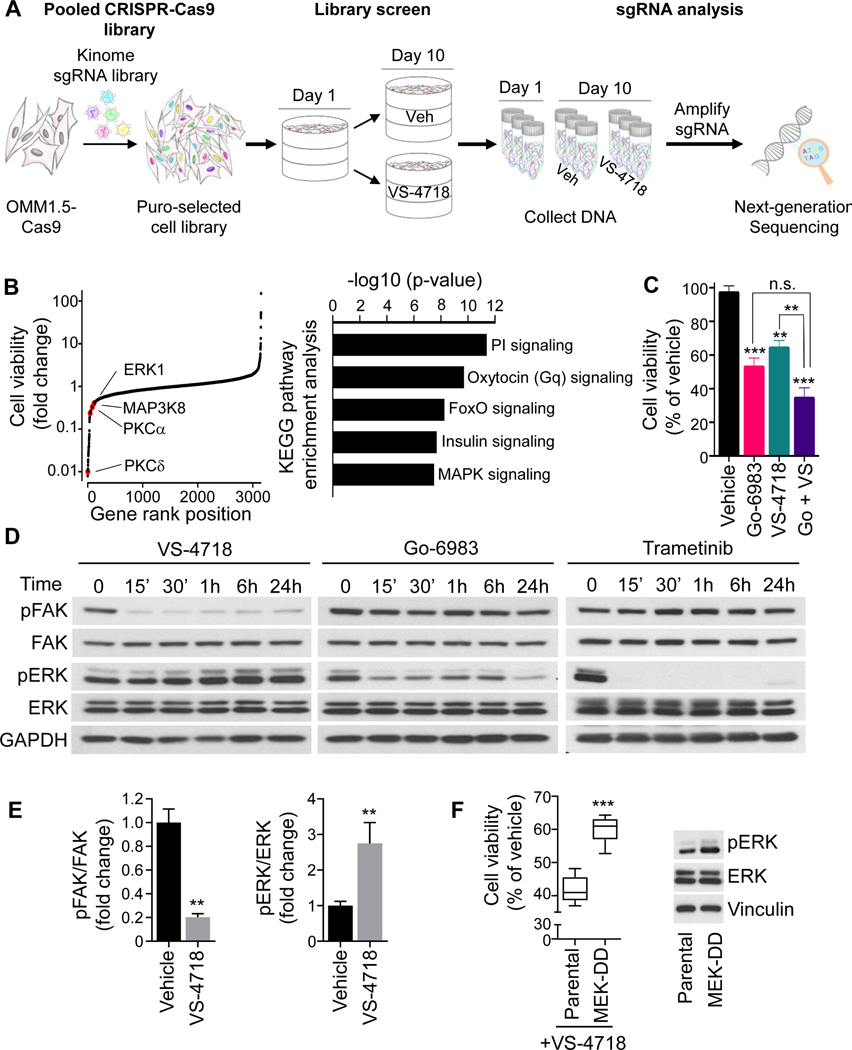
As the kinome has been the target of most drug discovery efforts, the identification of kinases whose activity are essential for UM survival in the context of FAK inhibition may facilitate the discovery of new co-targeting strategies. To perform an efficient loss-of-function screen, we used a two-vector CRISPR system, expressing a Cas9 transgene in representative UM cells (OMM1.5, originally derived from metastasis, hereby called OMM1.5Cas9), in which gene editing efficiency was validated (Fig. 1 and Supplementary Fig. S1A,B). OMM1.5Cas9 cells were infected with the Human Kinome Brunello pooled sgRNAs library (29), targeting 763 kinase genes and containing 3,052 unique sgRNAs along with 100 non-targeting controls, and subjected to puromycin selection. Surviving cells were treated with either selective FAKi (VS-4718) or vehicle for 10 days, followed by the collection of cellular DNA for gRNA analysis by NextGen sequencing (Fig. 1A). Depleted sgRNAs (dropouts) suggest that inhibition of their gene targets could sensitize cells to FAKi treatment. Our analysis of synthetic lethal interactors of FAKi revealed a significant enrichment of Gαq-PLC and MAPK components of the classical Gαqsignaling cascade (Fig. 1B and Supplementary Fig. S1C). Using the pan-PKC inhibitor, Go-6983 (33), we found that inhibition of the canonical PLCβ-PKC-ERK pathway decreased UM cell viability and that this inhibitory effect was further enhanced by combining Go-6983 with VS-4718 (Fig. 1C). However, PKC inhibition with Go-6983 was less effective in reducing active ERK (pERK) than the MEKi trametinib (Fig. 1D). This is aligned with prior studies supporting only partial pERK reduction after prolonged PKC inhibition (34), which may explain its very modest clinical activity in mUM (35). Thus, we focused on direct MEK inhibitors for further studies. Interestingly, inhibition of FAK by VS-4718 led to a gradual increase in pERK levels (Fig. 1D, E) and UM cells stably expressing the constitutive active MEK mutant (MEK-S218/222D, MEK-DD) were more resistant to VS-4718 (Fig. 1F), suggesting a possible compensatory mechanism that may contribute to drug resistance.
Figure 1. Kinome-wide CRISPR screen for synthetic lethal interactors of FAKi.
(A) OMM 1.5 cells expressing Cas9 were infected with the Brunello Human Kinome CRISPR sgRNA KO library at a MOI of 0.3. After selection, cells were treated with vehicle or 0.5μM VS-4718 (FAKi) for 10 days. (B) Left, Cell viability represented as fold change in FAKi-treated cells compared to control. Highlighted significant hits represent synthetic lethal genes with FAKi treatment. Right, KEGG pathways analysis for the top depleted sgRNAs (n=200). (C) 92.1 cell viability after 72h treatment with vehicle, 1μM Go-6983 (PKCi), 1μM VS-4718 or combination of both. (D) Time-course analysis of FAK and ERK phosphorylation in 92.1 cells treated with VS-4718 (1μM), Go-6983 (1μM) or trametinib (MEKi, 10nM). (E) Quantification of pFAK/FAK and pERK/ERK ratios in 92.1 cells treated with 1μM VS-4718 or vehicle for 1h. (F) Left, Cell viability after 72h treatment with VS-4718 (1μM) in 92.1 cells expressing or not MEK-DD (S218/222D). Right, Immunoblot showing pERK levels in 92.1 cells expressing or not MEK-DD (S218/222D). (C, E and F) Data shown represent the mean ± SEM of three independent experiments. ***p<0.001; **p< 0.01; n.s. not significant.
Synergistic antiproliferative effect of MEK and FAK co-targeting in UM cells.
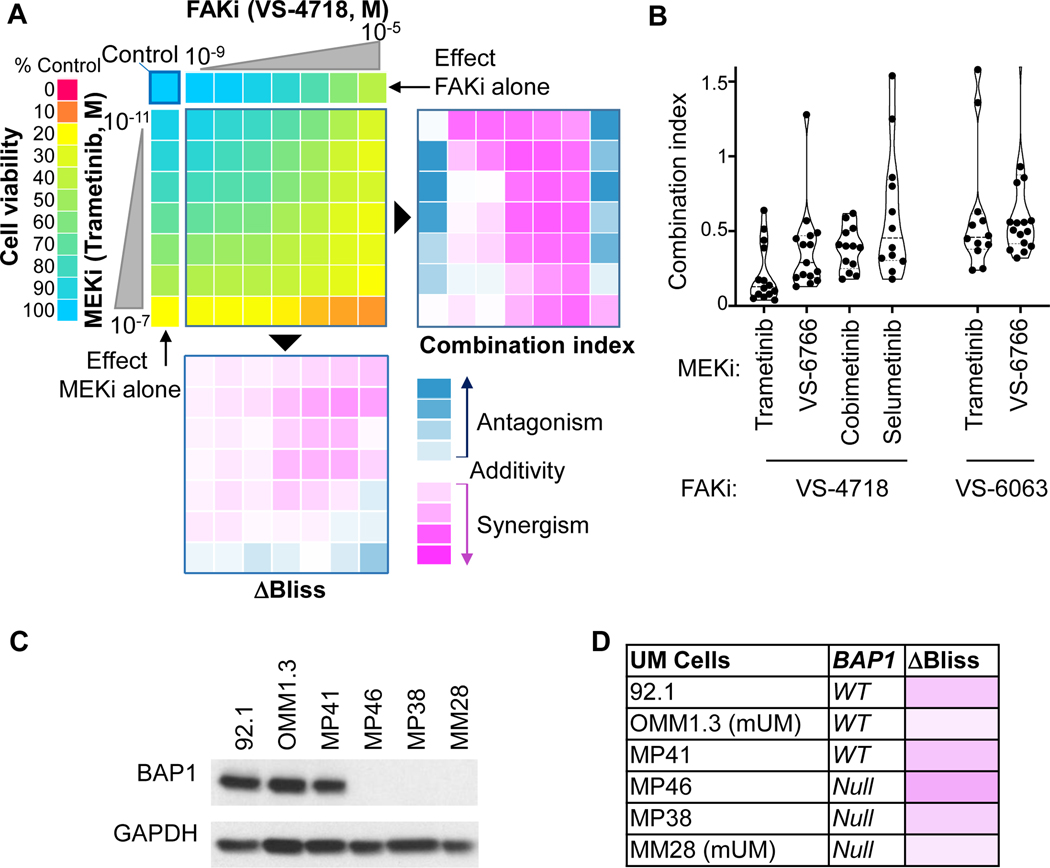
Co-targeting of MEK and FAK inhibits both canonical and non-canonical Gαq signaling pathways, and may provide a suitable drug combination for clinical application in mUM. We used the effect-based ΔBliss model and the dose-effect-based Chou-Talalay combination index (CI) to assess synergistic, additive, or antagonistic drug interaction in a given combination. We confirmed that trametinib (MEKi) and VS-4718 (FAKi) decrease UM cell growth as single agents. CI and ΔBliss scores both converged to demonstrate a synergistic interaction between trametinib and VS-4718 (Fig. 2A). Synergistic antiproliferative effects were observed using multiple clinically relevant MEKi (trametinib, cobimetinib, and selumetinib) as well as the second-generation RAF/MEK inhibitor VS-6766, combined with two different FAKi (VS-4718 and the clinically relevant defactinib). All combinations showed remarkable synergistic activity at relevant doses supporting a general drug-drug class pharmacodynamics interaction (Fig. 2B). Using a panel of GNAQ mutant UM cells lines with different expression levels of the metastasis suppressor protein BAP1 (12), we confirmed similar synergistic profiles in BAP1 wild-type or null cells (Fig. 2C & D) suggesting that this combination may also be active in mUM.
Figure 2. Synergy between FAKi and MEKi in UM and mUM cells.
(A) Left, 92.1 cell viability 72h after treatment. Right, Combination Index values (CI) determined using the Chou-Talalay method (CI<1 synergism, CI=1 additivity, CI>1 antagonism, scale from −2 to +2). Bottom, ΔBliss scores (score<0 synergism, score=0 additivity, score>0 antagonism, scale from −1 to +1). (B) CI at relevant doses (viability=50±5%) using various combinations of FAKi/MEKi. (C) Immunoblot depicting BAP1 levels in UM and mUM patient-derived cells. (D) Delta score (ΔBliss), assessing synergism between MEKi (trametinib, 10nM) and FAKi (VS-4718, 1μM) in a panel of UM and mUM cells with distinct BAP1 status.
MEKi/FAKi combination increases UM apoptotic cell death and reduces melanosphere formation.
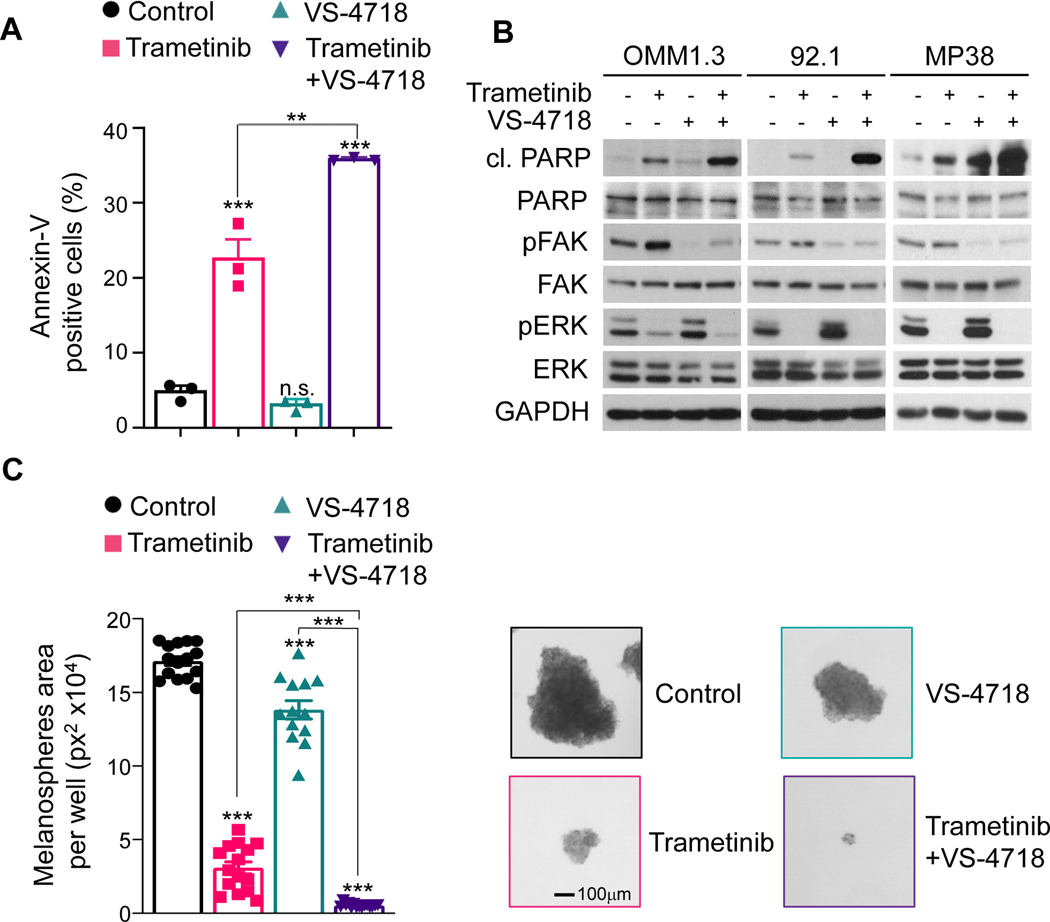
Several FAKi inhibitors have been evaluated in the clinic and demonstrated primarily a cytostatic effect as single agents (23). Given the high synergistic activity of MEKi and FAKi combination in vitro, we next evaluated the ability of trametinib and VS-4718 to induce cell death. As shown in Fig. 3A, FACS analysis suggests that tramenitib induces an increase in the fraction of apoptotic cells [Annexin V+], while VS-4718 has no apoptotic activity as a single agent. Nonetheless, when both drugs were combined, at a ratio of 1:100 based on our isobologram analysis (Supplementary Fig. S2), the apoptotic response was significantly increased.
Figure 3. MEKi/FAKi combination induces apoptosis and reduces UM melanosphere formation.
(A) FACS analysis of cells positive for Annexin V was used to assess the apoptotic response to trametinib (10 nM), VS-4718 (1 μM), and their combination after 24 h of treatment. (B) Immunoblot showing cleaved-PARP, pFAK (pY-397) and pERK levels upon treatment with vehicle, trametinib (10 nM), VS-4718 (1 μM) or trametinib+VS-4718 for 48 hours in UM cells. (C) Left, OMM1.3 melanospheres formation after treatment with vehicle (control), trametinib (10 nM), VS-4718 (1 μM) or trametinib+VS-4718 for 3 weeks. Right, Representative spheres (A and C) Data shown represent the mean ± SEM of three independent experiments. ***p<0.001; **p< 0.01; n.s. not significant.
One of the most common signaling cascades involved in apoptosis is the activation of a highly specialized family of cysteinyl-aspartate proteases (caspases) (36). Caspase-mediated cell death is achieved through the cleavage of multiple key proteins that are essential to cell survival. PARP-1 is one of the substrates of caspases and a well-established marker for apoptosis (37). Aligned with our prior results, cleaved-PARP levels were strongly increased by the combination of FAKi and MEKi (Fig. 3B), while single agents were less active. We then evaluated the ability of VS-4718 and trametinib to induce apoptosis in BAP1 null mUM cells. FAKi alone showed a minimal apoptotic response, and the combination of VS-4718 and trametinib resulted in increased cleaved-PARP compared to trametinib alone (Fig. 3B).
Cancer initiating cells are believed to play a central role in drug resistance and metastasis (38). A typical feature of these cells is their ability to form 3D-tumorspheres when cultured in suspension in a stem-cell medium. Interestingly, we found that the metastatic-derived OMM1.3 cells, but not 92.1 cells originating from a primary tumor, were able to form melanospheres. Using this model, we found that both VS-4718 and trametinib significantly reduced melanosphere formation as single agents when compared to vehicle (Fig. 3C) and this effect was further enhanced when both drugs were combined. Our results demonstrate that MEKi/FAKi combination acts by inhibiting cancer cell proliferation, including cancer initiating cells, and enhancing apoptotic cell death. This suggests that this multimodal targeted therapy could be effective on primary and metastatic UM.
Potent antitumoral and cytotoxic effects of MEKi/FAKi combination in UM xenograft and liver metastasis models.
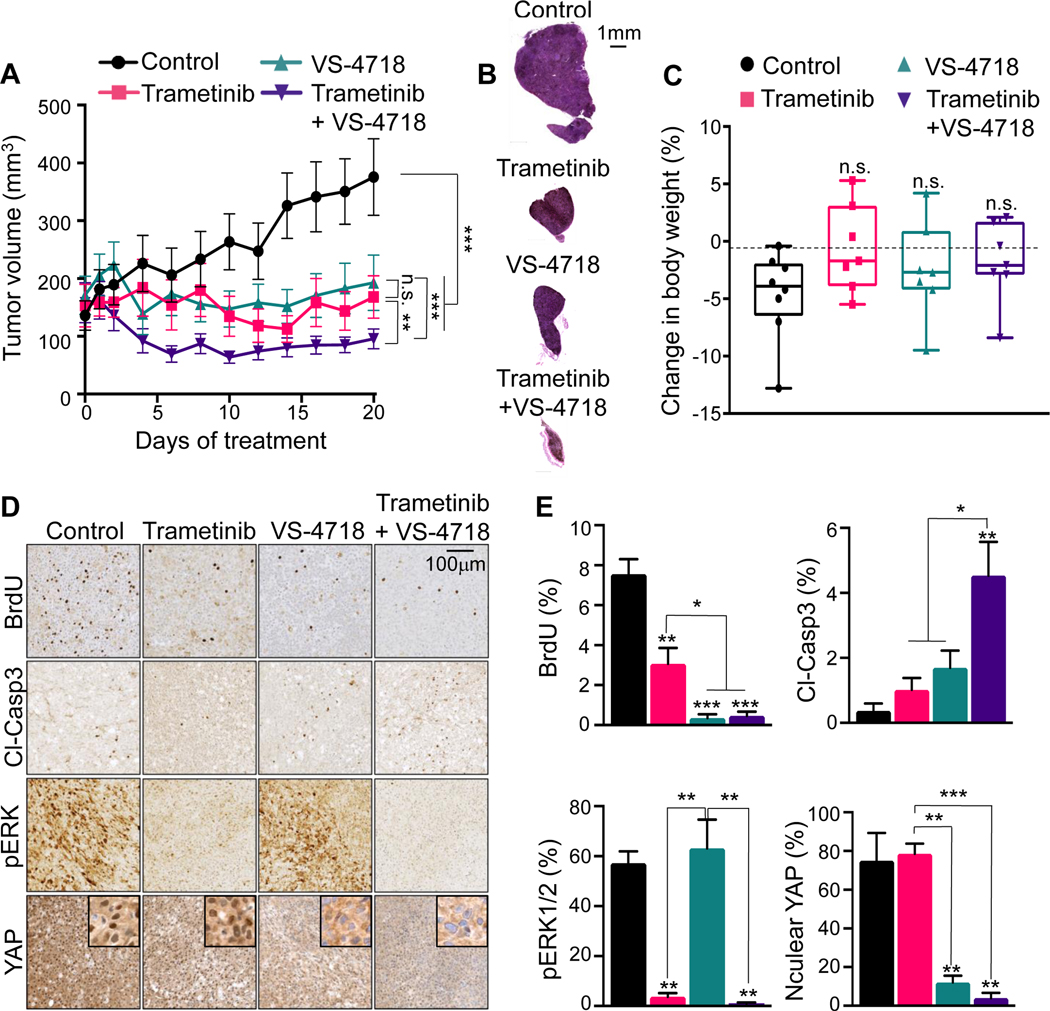
We used UM xenograft models to evaluate the anticancer activity of the MEKi/FAKi combination in vivo. Tumor-bearing mice were randomly divided into four groups: control, trametinib, VS-4718 and trametinib/VS-4718. In this regard, whereas VS-4718 has better pharmacokinetics than defactinib in mice, the latest is better able to inhibit tumor FAK activity with tolerability in cancer patients (39). Therefore, VS-4718 is used as a surrogate FAKi in murine pre-clinical models. As shown in Fig. 4A and B, in the 92.1 xenograft model, although trametinib and VS-4718 effectively induced tumor stasis as single agents, only the combination of trametinib with VS-4718 was able to induce tumor regression. No significant difference in body weight was observed between the control and any of the treated groups (Fig. 4C), suggesting that all treatments were well tolerated by the mice. These results were further confirmed using another UM cellular system harboring GNAQ mutations, OMM1.3, albeit in this case MEKi alone was quite potent (see Supplementary Fig. S3). We extended these studies in allograft experiments using the recently developed GNAQ-driven B2905 syngeneic mouse melanoma model (40). These cells are highly sensitive to the Gαq inhibitor FR900359, as we reported in a large panel UM cells (41), and to the combined inhibition of MEK and FAK (Supplementary Fig. 4). These cells exhibit limited responses to FAKi in vivo, likely reflecting the complexity of genetic alterations of these cells (40). MEKi limited tumor growth, but prolonged inhibition resulted in acquisition of resistance and rapid tumor regrowth, thereby compromising animal survival. Remarkably, the combination therapy was quite effective in promoting tumor regression, and no resistance was observed for the prolonged duration of the mouse experiments (>40 days).
Figure 4. MEKi/FAKi combination UM growth in in vivo xenograft mouse models.
(A) Changes in 92.1 xenograft tumor volume in mice treated with vehicle (Control), trametinib 1 mg/kg, VS-4718 50 mg/kg or trametinib+VS-4718. (B) H&E staining of representative xenograft tumor sections after 20 days of treatment. (C) Difference in mice body weight between day 0 and day 20 of the indicated treatment in 92.1 xenografts mice. Box and whiskers plot with minimum and maximum whiskers (7 mice/group). (D) 92.1 tumor bearing mice were treated with vehicle (Control), trametinib 1 mg/kg, VS-4718 50 mg/kg or trametinib+VS-4718 for 20 days. Representative IHC staining tumor sections for BrdU, cleaved-Caspase3 (cl-Casp3), pERK and YAP. Scale bar is 100 μm and insets are 50 μm wide. (E) Quantification of the IHC stained tumor sections. (A and E) Data are mean±SEM (7 mice/group). *p<0.05; **p<0.01; ***p<0.001; n.s. not significant.
We next evaluated MEK/ERK and FAK/YAP pathway activities in the UM xenograft tumor specimens by immunohistochemistry (Fig. 4D and E). As expected, pERK1/2 was strongly decreased only in the trametinib and trametinib/VS-4718 treated groups. We monitored YAP nuclear exclusion as a readout for YAP inactivation upon FAKi treatment (18), and observed decreased nuclear YAP in the VS-4718 and trametinib/VS-4718 treatment groups exclusively. The percentage of proliferating BrdU positive cells was significantly decreased in all treated groups, with the VS-4718 and trametinib/VS-4718 treatment groups showing the most significant inhibition. Using cleaved-Caspase3 (cl-Casp3) as a marker for apoptosis, we detected a significant increase of cl-Casp3 in the trametinib/VS-4718 group compared to the single agent groups, suggesting that the MEKi/FAKi combination induces a switch from cytostatic to cytotoxic activity, consistent with the increased cl-PARP levels observed in vitro.
To assess the efficacy of the MEKi/FAKi combination treatment on a murine system that better represents the advanced disease, we developed a preclinical liver metastasis model. Luciferase/GFP expressing 92.1 cells (92.1-Luc) were injected into the spleen and allowed hematogenous dissemination followed by complete splenic resection (Fig. 5A). Mice were sacrificed 8-weeks post-injection and necropsies showed macroscopic hepatic metastases with no sign of dissemination to any other organs (Fig. 5B), confirming the strong liver tropism of UM cells. Remarkably, when tumor bearing mice were treated with trametinib, VS-4718 or trametinib/VS-4718, the metastatic burden was reduced by MEKi and FAKi treatment and their combination (Fig. 5C, D). FAKi alone was cytostatic, whereas MEKi and MEKi/FAKi combination induced tumor regression. However, detailed analysis showed that the residual disease in the trametinib alone treated group represented 24.2±0.6 % of the initial tumor compared to 7.5±0,2% in the combination treated group (p=0.0002), achieving none versus 40% complete responses, respectively (Fig. 5E). Resistance to trametinib in patients has led to the failure of this agent in UM clinical trial. To further investigate resistance to trametinib as a single agent or as part as a combination with FAKi in our metastatic model, we adjusted the dose of trametinib to reach maximum plasma levels similar to those achieved in humans at the recommended therapeutic dose, 2 mg/day, which is approximately 22 ng/ml upon repeated dosing (42). Mice treated with 0.1 mg/kg trametinib, achieving approximately 20 ng/ml (43) become resistant to treatment with a progressive increase of metastatic burden, whereas tumor burden remains barely undetectable in mice treated with the combination. Taken together, these findings indicate that treatment with trametinib combined with VS-4718 inhibited UM tumor growth in subcutaneous and liver metastasis models.
Figure 5. MEKi/FAKi combination reduces UM cells growth in an in vivo liver metastasis model.
(A) Schematic of the hematogenous dissemination model for UM liver metastasis using 92.1 GFP-Luc cells. (B) Left, Macroscopic view of liver metastasis 8 weeks post-splenic injection. Right, H&E staining of liver and lung. (C) Hepatic tumor burden tracked by IVIS imaging after injection of 92.1 UM cells in SCID/NOD mice treated with vehicle (Control), trametinib 1 mg/kg, VS-4718 50 mg/kg or both. Data are mean±SEM (6 mice/group). ***p<0.001; n.s. not significant. (D) Representative mice treated with vehicle (Control), trametinib 1 mg/kg, VS-4718 50 mg/kg or both, at the indicated days of treatment, and representative ex-vivo imaging of the liver obtained at day 21. (E) Hepatic tumor burden tracked by IVIS imaging after injection of 92.1 UM cells in SCID/NOD mice treated with vehicle (Control), trametinib 0.1 mg/kg, VS-4718 50 mg/kg or both. Data are mean±SEM (5 mice/group). ***p<0.001; **p<0.01. (F) Representative ex-vivo imaging of the liver from mice treated for 35 days with trametinib 0.1 mg/kg, VS-4718 50 mg/kg or both.
Discussion
The limited responses of UM to immunotherapies (40) and the lack of an FDA-approved standard of care for mUM patients poses an urgent unmet medical need that necessitates the development of new targeted treatment options. Most clinical studies in UM have focused on canonical kinases activated downstream from Gq/G11 with MEK as the major therapeutic target (44). MAPK/ERK pathway inhibitors have been proven to be effective treatments in various cancer types, but their effectiveness is often short-lived, with resistance developing often after the start of treatment (45–48). UM was no exception and despite encouraging results in an initial phase-II trial, with the MEK inhibitor selumetinib (AZD6244) showing an improved progression-free survival (PFS) compared to dacarbazine or temozolomide (15). The subsequent phase III double-blind trial failed to show improvement in PFS and overall survival (OS) with selumetinib + dacarbazine compared to dacarbazine alone (16). Here, we found that adaptive activation of MEK-ERK represents a compensatory resistance mechanism to FAK inhibition, and that in turn concomitant targeting FAK and MEK-ERK converts the cytostatic effects of FAKi into cytotoxic, resulting in UM cell death. These findings suggest that dual inhibition of MEK and FAK may represent a promising therapeutic option for advanced and metastatic UM patients.
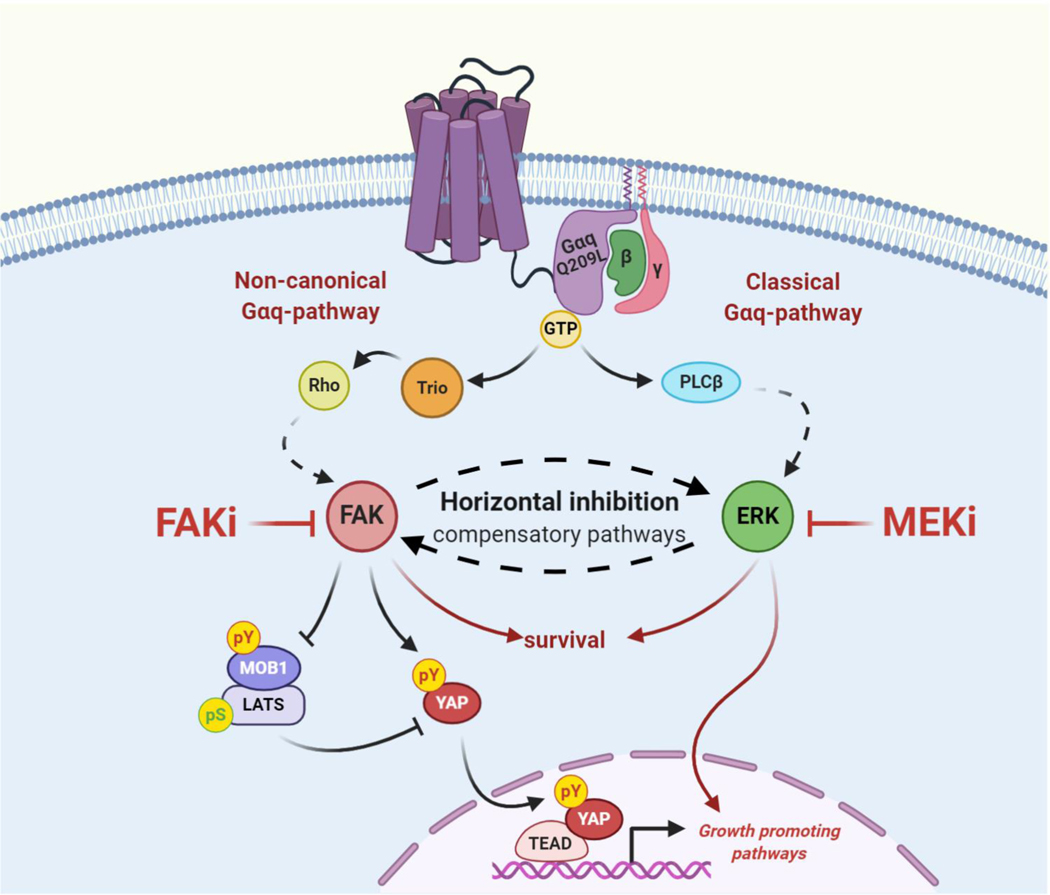
Our previous studies revealed that Gαq triggers the activation of the FAK/Hippo-YAP pathway, which represents a major driver in UM progression (18,19). Given that, we aimed at identifying synthetic lethal interactors that could potentiate FAKi efficiency and simultaneously reduce the risk of developing drug resistance (24), which is often observed in single-drug therapy. Our kinome-wide CRISPR/Cas9 screen unveiled potent synthetic lethality between FAKi and the MEK/ERK pathway, uncovering the therapeutic potential of a co-targeting strategy. This idea is reinforced by a recent study demonstrating that treatment of UM cells with the MEKi trametinib increased YAP activity and cell proliferation (49), and our findings that FAK/Hippo-YAP pathway inhibition leads to a gradual increase of pERK. These results suggest that both Gαq-activated pathways, FAK/Hippo-YAP and MEK/ERK, can be part of compensatory feed-back processes, and that horizontal inhibition of FAK and MEK might be necessary in order to achieve tumor regression, and prevent treatment resistance and tumor relapse (Fig. 6). In addition, concomitant inhibition of FAK and MEK triggers the activation of apoptotic cell death programs, whose full elucidation may facilitate the identification of mechanistic biomarkers predicting therapeutic response. Of interest, most UM and mUM models used in our studies are driven by the GNAQ oncogene. However, based on its high similarity with the GNA11 oncogene, our findings are also expected to be applicable to GNA11 mutant UM lesions (6,7), which should be nonetheless tested experimentally.
Figure 6. Horizontal inhibition of compensatory pathways in UM using MEKi/FAKi combination.
The cartoon depicts the proposed pathways by which active GNAQ mutant controls cell proliferation in UM cells. Horizontal inhibition of FAK and MEK likely acts by disabling growth promoting pathways regulated by YAP while concomitantly targeting parallel converging core survival mechanisms, thereby resulting in mUM regression.
Several potent FAKis have been tested in the clinic in multiple cancer types, showing target inhibition and manageable toxicity profiles. Indeed, based on our findings, a Phase 1 clinical trial using IN10018 as a potent FAKi has been recently initiated in mUM (NCT04109456). The use of FAKis as single agents may act primarily as cytostatic, suggesting the possibility of identifying suitable combination strategies to reach a greater clinical efficacy. Our study supports the clinical potential of co-targeting FAK and its sensitizing pathway, MEK/ERK, as a precision therapy approach in GNAQ-driven UM, achieving tumor regression. In this regard, the FAKi defactinib has been recently combined with a RAF/MEKi (VS-6766) in multiple cancer types (NCT03875820). Through use of an intermittent dosing schedule, the defactinib/VS-6766 combination has shown a manageable safety profile with initial clinical activity in early clinical trial results to date (50). Altogether, we believe our study presents a strong potential for clinical translation as a multimodal signal-transduction-based precision therapy for mUM.
Supplementary Material
Statement of significance.
Most metastatic uveal melanoma (mUM) patients are refractory to chemotherapies and immune checkpoint blockade, leading to patient death within a year of diagnosis. To date, there are no effective treatment options for mUM, highlighting an urgent need for novel therapeutic strategies. Uveal melanoma (UM) is characterized by gain-of-function mutations in GNAQ/GNA11, encoding Gαq proteins. Our recent studies identified focal adhesion kinase (FAK) as an integral component of the oncogenic Gαq signaling circuitry in UM, and the clinical benefits of targeting FAK in mUM are already under current investigation. However, single-agent targeted therapies often activate adaptive mechanisms resulting in drug resistance and treatment failure. Taking advantage of the unique genetic landscape of UM and the use of unbiased genetic screens, our studies reveal that horizontal inhibition of FAK and the adaptive activation of MEK-ERK results in UM cell death and tumor regression, thereby providing a novel multimodal precision therapy for mUM.
Acknowledgements
pHAGE PGK-GFP-IRES-LUC-W was a generous gift from Dr. Schoenberger (La Jolla Institute for Immunology). We thank Dr. Alfredo Molinolo for insightful discussion and Dr. Helen T. Michael for characterization of mouse melanoma cells. We thank the La Jolla Institute Microscopy and Histology Core Facility for their support with the IHC data presented in this manuscript. We thank the Moores Cancer Center Microscopy Core for the in vivo imaging support. The cartoon was created with BioRender.com.
Additional information: This work was supported, in whole or in part, by National Institutes of Health Grant R33CA225291 and U54CA209891 to JSG and R01CA196278 and R01CA182635 to AEA. JSP was supported by a fellowship from the Canadian Institute for Health Research. KF was supported by the National Science Foundation Graduate Research Fellowship Program (DGE-1650112). SL was supported by funding from AIRC and from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 800924. NA was supported by the National Science Foundation Graduate Research Fellowship Program (DGE-1650112). Verastem Oncology provided defactinib, VS-4718, and VS-6766.
Footnotes
Conflict of interest disclosure statement: Silvia Coma and Jonathan A. Pachter are employees and stockholders of Verastem Oncology (Needham, MA). Andrew E. Aplin has ownership interest in patent number 9880150. J. Silvio Gutkind is a member of the scientific advisory board of Oncoceutics, Vividion Therapeutics, and Domain Therapeutics, and interested part of a patent filed by UCSD, number 16/824639.
Bibliography
- 1.Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 2003;63(6):1256–72 doi 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 2.Schöneberg T, Schulz A, Biebermann H, Hermsdorf T, Römpler H, Sangkuhl K. Mutant G-protein-coupled receptors as a cause of human diseases. Pharmacology & therapeutics 2004;104(3):173–206 doi 10.1016/j.pharmthera.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature 2009;459(7245):356–63 doi 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon MI, Strathmann MP, Gautam N. Diversity of G proteins in signal transduction. Science 1991;252(5007):802–8 doi 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 5.Wu V, Yeerna H, Nohata N, Chiou J, Harismendy O, Raimondi F, et al. Illuminating the Onco-GPCRome: Novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J Biol Chem 2019;294(29):11062–86 doi 10.1074/jbc.REV119.005601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009;457(7229):599–602 doi nature07586 [pii] 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med 2010;363(23):2191–9 doi 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore AR, Ceraudo E, Sher JJ, Guan Y, Shoushtari AN, Chang MT, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet 2016;48(6):675–80 doi 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aronow ME, Topham AK, Singh AD. Uveal Melanoma: 5-Year Update on Incidence, Treatment, and Survival (SEER 1973–2013). Ocul Oncol Pathol 2018;4(3):145–51 doi 10.1159/000480640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvajal RD, Schwartz GK, Tezel T, Marr B, Francis JH, Nathan PD. Metastatic disease from uveal melanoma: treatment options and future prospects. Br J Ophthalmol 2017;101(1):38–44 doi 10.1136/bjophthalmol-2016-309034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shain AH, Bagger MM, Yu R, Chang D, Liu S, Vemula S, et al. The genetic evolution of metastatic uveal melanoma. Nat Genet 2019;51(7):1123–30 doi 10.1038/s41588-019-0440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Afshar AR, Damato BE, Stewart JM, Zablotska LB, Roy R, Olshen AB, et al. Next-Generation Sequencing of Uveal Melanoma for Detection of Genetic Alterations Predicting Metastasis. Transl Vis Sci Technol 2019;8(2):18 doi 10.1167/tvst.8.2.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komatsubara KM, Carvajal RD. Immunotherapy for the Treatment of Uveal Melanoma: Current Status and Emerging Therapies. Curr Oncol Rep 2017;19(7):45 doi 10.1007/s11912-017-0606-5. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Wu Q, Depeille P, Chen P, Thornton S, Kalirai H, et al. RasGRP3 Mediates MAPK Pathway Activation in GNAQ Mutant Uveal Melanoma. Cancer Cell 2017;31(5):685–96 e6 doi 10.1016/j.ccell.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvajal RD, Sosman JA, Quevedo JF, Milhem MM, Joshua AM, Kudchadkar RR, et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: a randomized clinical trial. Jama 2014;311(23):2397–405 doi 10.1001/jama.2014.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvajal RD, Piperno-Neumann S, Kapiteijn E, Chapman PB, Frank S, Joshua AM, et al. Selumetinib in Combination With Dacarbazine in Patients With Metastatic Uveal Melanoma: A Phase III, Multicenter, Randomized Trial (SUMIT). Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2018;36(12):1232–9 doi 10.1200/JCO.2017.74.1090. [DOI] [PubMed] [Google Scholar]
- 17.Vaque JP, Dorsam RT, Feng X, Iglesias-Bartolome R, Forsthoefel DJ, Chen Q, et al. A genome-wide RNAi screen reveals a Trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol Cell 2013;49(1):94–108 doi 10.1016/j.molcel.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell 2014;25(6):831–45 doi 10.1016/j.ccr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng X, Arang N, Rigiracciolo DC, Lee JS, Yeerna H, Wang Z, et al. A Platform of Synthetic Lethal Gene Interaction Networks Reveals that the GNAQ Uveal Melanoma Oncogene Controls the Hippo Pathway through FAK. Cancer Cell 2019;35(3):457–72 e5 doi 10.1016/j.ccell.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sood AK, Coffin JE, Schneider GB, Fletcher MS, DeYoung BR, Gruman LM, et al. Biological significance of focal adhesion kinase in ovarian cancer: role in migration and invasion. Am J Pathol 2004;165(4):1087–95 doi 10.1016/S0002-9440(10)63370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canel M, Secades P, Rodrigo JP, Cabanillas R, Herrero A, Suarez C, et al. Overexpression of focal adhesion kinase in head and neck squamous cell carcinoma is independent of fak gene copy number. Clin Cancer Res 2006;12(11 Pt 1):3272–9 doi 10.1158/1078-0432.CCR-05-1583. [DOI] [PubMed] [Google Scholar]
- 22.Luo M, Guan JL. Focal adhesion kinase: a prominent determinant in breast cancer initiation, progression and metastasis. Cancer Lett 2010;289(2):127–39 doi 10.1016/j.canlet.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohanty A, Pharaon RR, Nam A, Salgia S, Kulkarni P, Massarelli E. FAK-targeted and combination therapies for the treatment of cancer: an overview of phase I and II clinical trials. Expert Opin Investig Drugs 2020;29(4):399–409 doi 10.1080/13543784.2020.1740680. [DOI] [PubMed] [Google Scholar]
- 24.Lee JS, Das A, Jerby-Arnon L, Arafeh R, Auslander N, Davidson M, et al. Harnessing synthetic lethality to predict the response to cancer treatment. Nat Commun 2018;9(1):2546 doi 10.1038/s41467-018-04647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods 2012;9(7):671–5 doi 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madera D, Vitale-Cross L, Martin D, Schneider A, Molinolo AA, Gangane N, et al. Prevention of tumor growth driven by PIK3CA and HPV oncogenes by targeting mTOR signaling with metformin in oral squamous carcinomas expressing OCT3. Cancer Prev Res (Phila) 2015;8(3):197–207 doi 10.1158/1940-6207.CAPR-14-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 2017;7(1):16878 doi 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nature biotechnology 2019;37(3):224–6 doi 10.1038/s41587-019-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 2018;9(1):5416 doi 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spahn PN, Bath T, Weiss RJ, Kim J, Esko JD, Lewis NE, et al. PinAPL-Py: A comprehensive web-application for the analysis of CRISPR/Cas9 screens. Sci Rep 2017;7(1):15854 doi 10.1038/s41598-017-16193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010;70(2):440–6 doi 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 32.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacological reviews 2006;58(3):621–81 doi 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 33.Wu-Zhang AX, Newton AC. Protein kinase C pharmacology: refining the toolbox. Biochem J 2013;452(2):195–209 doi 10.1042/BJ20130220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, Wu Q, Tan L, Porter D, Jager MJ, Emery C, et al. Combined PKC and MEK inhibition in uveal melanoma with GNAQ and GNA11 mutations. Oncogene 2014;33(39):4724–34 doi 10.1038/onc.2013.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piperno-Neumann S, Larkin J, Carvajal RD, Luke JJ, Schwartz GK, Hodi FS, et al. Genomic Profiling of Metastatic Uveal Melanoma and Clinical Results of a Phase I Study of the Protein Kinase C Inhibitor AEB071. Molecular cancer therapeutics 2020;19(4):1031–9 doi 10.1158/1535-7163.MCT-19-0098. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene 2008;27(48):6194–206 doi 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- 37.Chaitanya GV, Steven AJ, Babu PP. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun Signal 2010;8:31 doi 10.1186/1478-811X-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res 2005;65(20):9328–37 doi 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 39.Fennell DA, Baas P, Taylor P, Nowak AK, Gilligan D, Nakano T, et al. Maintenance Defactinib Versus Placebo After First-Line Chemotherapy in Patients With Merlin-Stratified Pleural Mesothelioma: COMMAND-A Double-Blind, Randomized, Phase II Study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2019;37(10):790–8 doi 10.1200/JCO.2018.79.0543. [DOI] [PubMed] [Google Scholar]
- 40.Pérez-Guijarro E, Yang HH, Araya RE, El Meskini R, Michael HT, Vodnala SK, et al. Multimodel preclinical platform predicts clinical response of melanoma to immunotherapy. Nat Med 2020;26(5):781–91 doi 10.1038/s41591-020-0818-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Annala S, Feng X, Shridhar N, Eryilmaz F, Patt J, Yang J, et al. Direct targeting of Galphaq and Galpha11 oncoproteins in cancer cells. Science signaling 2019;12(573) doi 10.1126/scisignal.aau5948. [DOI] [PubMed] [Google Scholar]
- 42.Drugs@FDA. Trametinib (Mekinist) NDA 20114/0. https://wwwaccessdatafdagov/drugsatfda_docs/nda/2013/204114Orig1s000ClinPharmR.pdf 2013.
- 43.Drugs@FDA. Trametinib (Mekinist) NDA 204114Orig1s000. https://wwwaccessdatafdagov/drugsatfda_docs/nda/2013/204114Orig1s000PharmR.pdf 2013.
- 44.Croce M, Ferrini S, Pfeffer U, Gangemi R. Targeted Therapy of Uveal Melanoma: Recent Failures and New Perspectives. Cancers (Basel) 2019;11(6) doi 10.3390/cancers11060846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welsh SJ, Rizos H, Scolyer RA, Long GV. Resistance to combination BRAF and MEK inhibition in metastatic melanoma: Where to next? Eur J Cancer 2016;62:76–85 doi 10.1016/j.ejca.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60–00 trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2010;28(3):466–74 doi 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 47.Lauchle JO, Kim D, Le DT, Akagi K, Crone M, Krisman K, et al. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature 2009;461(7262):411–4 doi 10.1038/nature08279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov 2014;4(1):61–8 doi 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faião-Flores F, Emmons MF, Durante MA, Kinose F, Saha B, Fang B, et al. HDAC Inhibition Enhances the. Clin Cancer Res 2019;25(18):5686–701 doi 10.1158/1078-0432.CCR-18-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinde R, Terbuch A, Little M, Caldwell R, Kurup R, Riisnaes R, et al. Phase I study of the combination of a RAF-MEK inhibitor CH5126766 and FAK inhibitor defactinib in an intermittent dosing schedule with expansions in KRAS mutant cancers. 2020 June 22–24; Philadelphia (PA). AACR; 2020. Abstract nr CT143. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.