Abstract
Despite three decades of research to its name and increasing interest in immunotherapies that target it, LAG-3 remains an elusive co-inhibitory receptor in comparison to the well-established PD-1 and CTLA-4. As such, LAG-3 targeting therapies have yet to achieve the clinical success of therapies targeting other checkpoints. This could, in part, be attributed to the many unanswered questions that remain regarding LAG-3 biology. Of these, we address: (i) the function of the many LAG-3-ligand interactions, (ii) the hurdles that remain to acquire a high-resolution structure of LAG-3, (iii) the under-studied LAG-3 signal transduction mechanism, (iv) the elusive soluble form of LAG-3, (v) the implications of the lack of (significant) phenotype of LAG-3 knockout mice, (vi) the reports of LAG-3 expression on the epithelium, and (vii) the conflicting reports of LAG-3 expression (and potential contributions to pathology) in the brain. These mysteries which surround LAG-3 highlight how the ever-evolving study of its biology continues to reveal ever-increasing complexity in its role as an immune receptor. Importantly, answering the questions which shroud LAG-3 in mystery will allow the maximum therapeutic benefit of LAG-3 targeting immunotherapies in cancer, autoimmunity and beyond.
Keywords: LAG-3, sLAG-3, checkpoint targets, cancer immunotherapy, structural biology
Technical Box.
Surface plasmon resonance (SPR): A label-free technique used to determine receptor/ligand binding interactions and their strength, affinity (dissociation constant; KD), using purified protein samples. In SPR, one protein is immobilised to the chip surface (ligand) and potential binding partners (analytes) are flowed over the chip surface using a microfluidics system. Binding is detected in real-time due to a change in refractive index at the chip surface. Typical SPR experiments determine affinity through (i) steady-state analysis, where binding at steady state is measured across a dilution series of analyte, and/or (ii) kinetic analysis, whereby kinetic models are fitted to binding curves to determine on- and off-rates.
BioLayer interferometry (BLI): A similar label-free technique used to describe receptor-ligand binding interactions and determine affinities. In BLI, ligands are immobilised to a sensor probe surface which is immersed in analyte solution within a micro-well plate. Upon analyte binding, a change in the optical thickness of the probe surface is detected by a change in interference of optical white light which is reflected at the probe surface. BLI is less sensitive compared to SPR; however, higher-throughput is thus useful in searching for novel receptor/ligand interactions.
Affinity versus avidity: The term affinity (KD) describes the strength of binding between a receptor and ligand with a one-to-one binding mode. A lower measured KD value (units Molar) represents a higher affinity interaction. Many one-to-one protein/protein interactions bind in the micromolar (µM) range. Molecules such as antibodies, and LAG-3:Fc, are multimeric (IgG and LAG-3:Fc = dimeric) and, as a result, are able to engage multiple ligands. Consequently, multimeric molecules utilise avidity effects to more strongly engage ligands (typically with avidity values in the nanomolar to the picomolar range). Dimeric molecules can engage a second ligand species, thus prolonging off-rates. Avidity effects are dependent on the availability of multiple ligands on an attached surface (such as the cell surface or experimental chip or plate). It is important to remember that LAG-3:Fc has been artificially dimerised through the covalently linked Fc domain and thus the avidity effects are not representative of LAG-3 in a cellular context. Careful evaluation of experimental setup is required when evaluating the strength of LAG-3-ligand interactions using LAG-3:Fc.
Introduction
Lymphocyte activation gene 3 (LAG-3) is a single-pass transmembrane glycoprotein, which despite 30 years of research, continues to be elusive. The majority of LAG-3 research has focused on its function on conventional T cells, where it is generally understood to negatively regulate the immune response, for example, by inhibiting proliferation and reducing granzyme/cytokine production [1–3]. Its role in modulating the inhibitory function of regulatory T cells (Tregs) on the other hand is contentious, with conflicting evidence suggesting LAG-3 either limits or contributes to Treg inhibition [4–6]. Perhaps the next most recognised role of LAG-3 is its involvement in TLR-independent activation of plasmacytoid dendritic cells (pDC) [7]. LAG-3 expression has also been described on other immune cells, including B cells, NK (natural killer) cells and unconventional T cells; upon which the function of LAG-3 is less well understood [1,8–11].
Despite these significant knowledge gaps, LAG-3 is often referred to as the ‘next immune checkpoint’ target on lymphocytes [12–14]. The proposed targeting of LAG-3 in immunotherapies has taken many forms, including (i) the delivery of soluble dimeric LAG-3 as an adjuvant therapy [15], (ii) the antibody blockade of LAG-3 interactions with its ligand(s) in cancer which has also been combined with an anti-Programmed Cell Death Protein 1 (PD-1) targeting therapy [16], (iii) antibody-mediated depletion of LAG-3+ cells in autoimmunity [17] and (iv) modulation of LAG-3 expression through small molecule targeting of Glycogen synthase kinase-3 (GSK-3) in cancer [18]. Despite ongoing efforts, LAG-3 targeting therapies are yet to achieve the successes of PD-1 and cytotoxic T-lymphocyte associated protein 4 (CTLA-4) [4], despite being identified around the same time [19–21]. The targeting of LAG-3 in combination with other checkpoint therapies has been a focus of ongoing clinical trials – exemplified by a recent study that associated LAG-3 expression on peripheral CD8+ T cells with a poorer efficacy of checkpoint blockade in melanoma and urothelial carcinoma patients [22]. Here, we address seven mysteries of LAG-3 and discuss the hurdles that remain on the path to maximizing its therapeutic potential.
LAG-3 ligands: not just MHC-II?
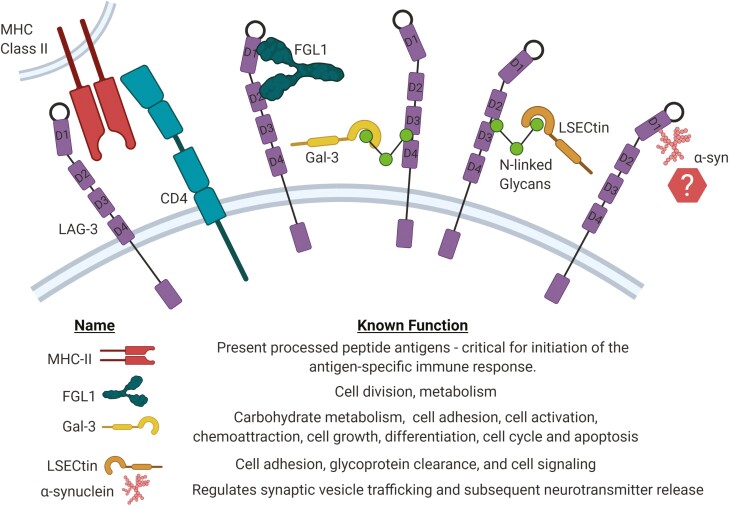
Upon first identification, parallels between LAG-3 gene structure and CD4 [23] led to an immediate investigation into major histocompatibility complex class II (MHC-II) as a potential ligand. LAG-3 expressing COS-7 cells engaged MHC-II bearing B lymphocytes which could be blocked by either anti-LAG-3 (clone 17B4) or anti-pan-HLA-II (clone D1.12) antibodies [24]. Today, however, multiple other binding partners have been proposed including liver and lymph node sinusoidal endothelial cell C-type lectin (LSECtin/CLEC4G), Galectin3 (Gal-3), Fibrinogen-like protein 1 (FGL1), and α-synuclein (α-syn) (Fig. 1) [25–27].
Figure 1.
Possible LAG-3 ligands and their functions; MHC-II, FGL1, Gal-3, LSECtin, and α-synuclein. The interaction between LAG-3 and MHC-II is believed to occur between the D1 loop of LAG-3 binding to a membrane-proximal site on MHC-II, similar to the defined MHC-II/CD4 binding site which is shown here for reference. FGL1 binds to LAG-3 at two sites in D1 and D2, while Gal-3 and LSECtin bind to N-linked glycans at glycosylation sites and α-syn has been shown to bind to the D1 domain of LAG-3.
MHC-II: the canonical ligand of LAG-3
Like the CD4 co-receptor, LAG-3 is located on chromosome 12 (12p13.1) and also has four extracellular immunoglobulin superfamily (IgSF) like domains (D1-4) (Fig. 2A and B). Molecular characterisation of LAG-3 binding to MHC-II molecules (and the discovery of novel ligands) has been hindered by difficulties in producing soluble LAG-3 protein. The formulation of LAG-3 as a soluble fusion protein, whereby D1-4 were fused to the fragment crystallisable (Fc) domain of IgG1 [28, 29], known as LAG-3:Fc, or as a pentabody [30], remains the only documented formulation for soluble LAG-3 protein production and have been used to confirm binding to MHC-II [29]; an interaction with greater avidity than equivalent MHC-II/CD4:Fc fusion dimers [28].
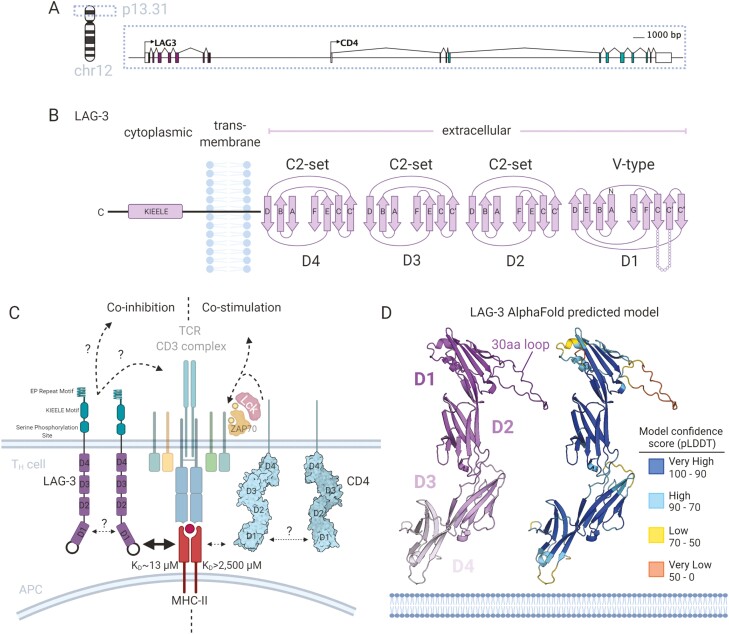
Figure 2.
Molecular understanding of LAG-3 binding to MHC-II as a co-inhibitory competitor to CD4. (A) Gene structure and location of LAG-3 on chromosome 12 adjacent to CD4. (B) The predicted protein domain folds of LAG-3 containing four extracellular Ig-domains (D1–D4) of V-type (D1) and C2-set (D2–D4). LAG-3 has a single-pass transmembrane domain and a short cytoplasmic tail containing a unique KIEELE motif involved in signal transduction. (C) Model of LAG-3 co-inhibitory competition with CD4 whereby LAG-3 binds MHC-II at a higher affinity than the co-stimulatory CD4 and transducing inhibitory signalling through undescribed signalling pathways. (D) Model of the LAG-3 extracellular domain as predicted by the AlphaFold project. The LAG-3 model is shown as cartoon representation and coloured (left) by domain (colours indicated inset) and (right) model confidence score (pLDDT) with colours indicated by the inset legend table. pLDDT confidence levels and definitions are as described previously [31].
The LAG-3/MHC-II interface was subsequently attributed to a proline-rich 30 amino acid (a.a.) loop, not present in CD4, between the C and Cʹ β strands of the LAG-3 D1 domain, as the anti-LAG-3 (clone 17B4) antibody raised against the 30 a.a. loop blocked the interaction [24]. Later, LAG-3:Fc mutants were designed and tested for their ability to bind MHC-II molecules in a cell adhesion assay which again identified a cluster of residues at the base of the 30 a.a. loop which disrupted binding [32]. Interestingly, implication of the 30 a.a. loop in binding suggests a smaller binding surface area than the interaction between CD4 and MHC-II [33].
More recently, using LAG-3-EC (a pentabody of mLAG-3), enhanced binding to the mouse MHC-II molecules I-Ad, I-Ab, and I-Ag7 was observed when loaded with stable peptides compared to mutant peptides known to decrease MHC-II-peptide stability [30], which influenced the strength of inhibition through LAG-3 in the OT-II T cell receptor (TCR) system. Consequently, it was hypothesised that LAG-3 is selectively tuned to recognise MHC-II bearing high-affinity epitopes and mediate peripheral tolerance to high-affinity binding self-peptides. Although preferential binding of MHC-II allotypes has not been established, LAG-3:Fc was shown to bind a panel of HLA-DR, -DQ, and -DP molecules in a bead-based binding assay [34]. Thus, the known MHC-II ligands of LAG-3 were extended to 95 HLA-II molecules – all allotypes tested. Differences in affinity between LAG-3 and the HLA-II panel, however, were not examined.
LAG-3 has often been described as a high-affinity ligand for HLA-II owing to an apparent calculated affinity constant (KD) of 60 nM determined by titrated LAG-3:Fc binding to HLA-II expressing cells [28]. An accurate monovalent affinity of the LAG-3/HLA-II has been difficult to obtain however due to the difficulties in producing monomeric LAG-3 (see Technical box). Consequently, our lab has recently used surface plasmon resonance (SPR) (see Technical box) to directly measure binding between HLA-DR1 and LAG-3:Fc. Using titrated amounts of immobilised proteins on the sensor chip, we estimated the monovalent interaction between HLA-DR1 bearing a high-affinity peptide (Influenza A Haemagglutinin306–318) and LAG-3 to be in the low micromolar range (KD ≈ 13 µM) [35]. This affinity measurement is in a similar range as HLA-I binding co-inhibitory receptors, such as the ILT/LILRB family [36] but still significantly stronger affinity than that of HLA-II/CD4 which is estimated at KD = 2.5 mM [37]. These findings lend further support to the idea that LAG-3 can out-compete CD4 for HLA-II binding (Fig. 2C).
A new key ligand?
There are several anti-LAG-3 antibodies, with different MHC-II-blocking abilities. Mouse studies have largely used anti-mouse LAG-3 clone C9B7W, which binds to D2 of mouse LAG-3 (mLAG-3) and weakly attenuates the interaction between mLAG-3 and MHC-II [38]. Whereas many human studies have used clone 17B4, which binds to D1 (raised to the 30 a.a. loop), and strongly blocks binding to MHC-II [24]. Interestingly a mouse study that characterised blockade by three antibodies, which bound to either D1, D2, or D4 of LAG-3, found there was no difference in T cell activation between the D1 and D2 binding antibodies [38, 39]. This suggests that even partial blockade of the LAG-3/MHC-II interaction increases T cell activation. However, it is also possible that there exists another ligand for LAG-3, which may be strongly blocked by D2 binding antibodies, but not D1 binding antibodies.
In 2019, FGL1 was described as another ligand for LAG-3 in both human and mouse [27]. FGL1 was identified as a candidate ligand first using a semi-automatic gene expression and detection system. FGL1 shares a similar structure with fibrinogen beta and gamma but has no known role on platelets or in clot formation [40]. FGL1 is primarily secreted in low levels by hepatocytes and is thought to play a role in cell division and metabolism [41–45]. However, Wang et al. identified upregulation of FGL1 mRNA in some solid human tumours, despite being downregulated in others. The Cancer Genome Atlas (TCGA) indicates that FGL1 is upregulated in lung adenocarcinoma [27]. It is possible that the engagement of LAG-3 by FGL1 in the tumour micro-environment prevents an effective anti-cancer immune response.
Direct interaction was shown using BioLayer interferometry (BLI) analysis (see Technical box) using mFGL1 and mLAG-3:Fc, with a KD value of ~1.5 nM. This is an extremely high-affinity interaction (see Technical box), similar to an antibody that reflects multivalency of the mLAG-3:Fc to mFGL1. mLAG-3 mutagenesis studies by Wang et al., found mLAG-3 bound to the fibrinogen-like domain (FD) of mFGL1. Furthermore, the single point mutation (Y73F) in the Cʹ strand of mLAG-3 D1 domain, which was previously shown to disrupt MHC-II binding, did not prevent mFGL1 binding [27]. In addition, pre-incubation of LAG-3+ 293T cells with C9B7W, led to complete abrogation of mouse FGL1/LAG-3 binding. These data support the hypothesis that FGL1 is an additional ligand for LAG-3 which binds to a site distinct to MHC-II. It will be interesting to investigate whether dual treatment of anti-LAG-3 antibodies which strongly block both MHC-II and FGL1 improves anti-tumour immunity compared to monotherapy, or if there are no benefits from inhibiting both ligands.
LAG-3: more than just protein binding
LAG-3 is a glycosylated protein comprising N-linked glycans (branched glycans attached via a nitrogen atom of Asn residues at Asn-X-Ser/Thr motifs) [24, 46]. Lectins, glycan-binding proteins which include C-type lectin receptors (CLRs), siglecs, and galectins, are able to specifically interact with and select for these carbohydrate structures.
LSECtin, a member of the DC-SIGN family, is a Ca2+-dependent CLR shown to interact with mannose, N-acetyl glucosamine (GlcNAc), and fructose and is highly expressed in the liver [47]. Evidence that LSECTin limits T-cell activity was obtained using mouse models of acute viral infection [25, 48, 49]. To date, only a single study provides evidence that LSECTin can bind to N-glycans on LAG-3. Xu and colleagues, using SPR (see Technical box) and co-immunoprecipitation assays, showed that LSECtin was able to interact with mLAG-3, and this interaction could be blocked by treatment with C9B7W, which restored IFN-γ secretion [25]. Further studies are needed however to establish LSECTin as a physiological LAG-3 ligand in humans.
Galectins are small soluble proteins with one or two carbohydrate-binding domains specific for galactose-containing glycans [50]. Gal-3 (31 kDa) differs from other galectin family members having both a carbohydrate recognition domain and an oligomerisation domain that enables cross-linking of its binding targets. Gal-3 binds to highly branched N-glycans on extracellular matrix glycoproteins (such as CD29) and has been shown to regulate T-cell activation [51, 52]. Kouo et al., used co-immunoprecipitation assays to demonstrate that Gal-3 is bound to LAG-3 [26] and hypothesised that Gal-3 binding to LAG-3 forms cross-links resulting in an inhibitory signal and thus suppression of T cell function. A recent study using multiple myeloma patient bone marrow mononuclear cells demonstrated higher rates of proliferation when treated with an anti-Gal-3 antibody compared to a pan HLA-II antibody [53]. Characterising anti-LAG-3 epitopes and their capacity to block different LAG-3 ligands would help decipher the role of different LAG-3-ligand interactions and the consequences of their blockade on T cell function.
Intriguingly, LAG-3 expression has been identified in the brain and binding to α-syn, a presynaptic neuronal protein, has been postulated (Fig. 1). α-Syn does not have a defined structure, has been shown to localise to the presynaptic terminals, and is thought to be a modulator of synaptic transmission [54]. This interaction was investigated by Mao et al., using a mouse model of Parkinson’s disease (PD) [55]. Dual treatment with C97BW (D2 binding) and 410C9 (D3/D4 binding) mAb clones were shown to block α-syn pre-formed fibril (PFF) binding, endocytosis, and pathology. The potential role of LAG-3 in the brain and potential binding to α-syn is discussed in more detail in the section Is LAG-3 expressed in the brain?
Having multiple ligands, some of which have been shown to bind non-redundantly, may enable LAG-3 to modulate the immune response in different ways. The fact that there are both soluble and membrane-bound ligands adds another layer of complexity and control. In summary, there is still much work needed to understand the effects of LAG-3 engaging one or more of its ligands and which treatment strategy is most suited in different disease states.
What is the molecular structure of LAG-3?
Structural understanding of protein/protein or receptor/ligand interactions can be complementary in designing approaches for interfering with these interactions, such as in silico screening of candidate small molecules. So far, the structure of LAG-3 is unknown, so this above avenue has not yet opened. Single-pass transmembrane proteins such as LAG-3 are usually expressed for structural studies as truncated extracellular domains to remove (i) the hydrophobic transmembrane domain which acts as a barrier to solubility and (ii) the cytoplasmic signalling domain which is often highly mobile [56, 57].
Furthermore, heavily glycosylated proteins, such as LAG-3, are problematic for structural analysis due to the randomness or variability of glycosylation patterns [58]. So far, the expression of suitable LAG-3 protein for structural studies has proven challenging. Although LAG-3:Fc fusion proteins have been expressed and proven useful for LAG-3 ligand discovery, these fusion proteins are difficult to crystallise. Even cutting-edge technologies which open cryo-electron microscopy (cryo-EM) to smaller proteins – such as Volta phase plates [59, 60], and protein scaffolds [61] have yet to materialise more detailed structural analyses of LAG-3.
Recently, a predicted model of LAG-3 was proposed by the AlphaFold project [62] whereby structure predictions for all human proteins were performed and made available [31] (Fig. 2D). Here, agreement with the IgSF domain arrangement first proposed in 1990 by Triebel et al. [23] (Fig. 2B) was similarly predicted. Likewise, the adjunct 30 a.a. extra loop between the C and Cʹ anti-parallel β-sheets of D1 was also observed in the prediction. This loop, however, exhibits very low positional confidence likely owing to sequence novelty and resultant lack of templates in the prediction model. As a result, a molecular understanding of how LAG-3 engages ligands remains unsolved and, at present, experimental structural data of LAG-3 is limited to a crude low-resolution envelope of LAG-3:Fc [63].
How does LAG-3 transduce a signal?
Understanding the signal processes which regulate LAG-3 expression and its functionality as an immune receptor is essential in describing the full breadth of LAG-3 activity and unlocking the potential for therapeutic targeting. The cytoplasmic tail of LAG-3 was shown to be indispensable to LAG-3 mediated inhibition of IL-2 production [64], highlighting that signal transduction through LAG-3 drives inhibitory function. The 54 amino acid cytoplasmic domain, however, is unique as it does not encode any of the classical inhibitory phosphatase binding motifs found amongst other immune-modulatory receptors e.g. immunoreceptor tyrosine-based inhibitory motifs (ITIMs). The sequence does, however, contain three characteristic features first highlighted through conservation between human and mouse LAG-3: (i) a potential serine phosphorylation motif (S454), (ii) a repetitive segment consisting of a glutamic acid-proline dipeptide (EP motif) and (iii) a unique KIEELE motif [64].
Of these three features, the KIEELE motif was initially identified as the key driver of LAG-3 signal transduction through truncation experiments whilst S454 was seemingly less important [64]. More recently, however, using an in vitro T cell activation system with LAG-3-MHC-II blocking mAbs, it was shown that loss of the KIEELE sequence did not affect the inhibitory function of LAG-3 [38]. Instead, T-cell activation was altered via two other distinct mechanisms. Firstly, inhibitory function was honed to an additional region proximal to the membrane termed the FxxL motif; implicating residues F475 and L478 in directly mediating IL-2 inhibition. Secondly, whilst LAG-3+ cells deficient for the EP motif (also termed EX repeat) initially demonstrated similar inhibitory capacity as full-length LAG-3, mutating LAG-3 to lack both the FxxL motif and truncating the EP motif instead completely abrogated inhibition and, in fact, rendered LAG-3 co-stimulatory [38]. This suggested that the membrane-proximal region containing the FxxL motif plays the greater inhibitory role [38]. The FxxL motif is more reminiscent of the ITIM motif YxxL and thus it is hypothesised that intracellular signalling factors may recruit to this motif [38]. Whilst unknown for FxxL, an intracellular factor has been identified for the EP motif which was shown to directly recruit the LAG-3-associated protein (LAP) [65]. The downstream signalling pathways associated with these motifs and the full repertoire of signalling molecules associated with LAG-3, however, remain unsolved.
Does soluble LAG-3 have a function?
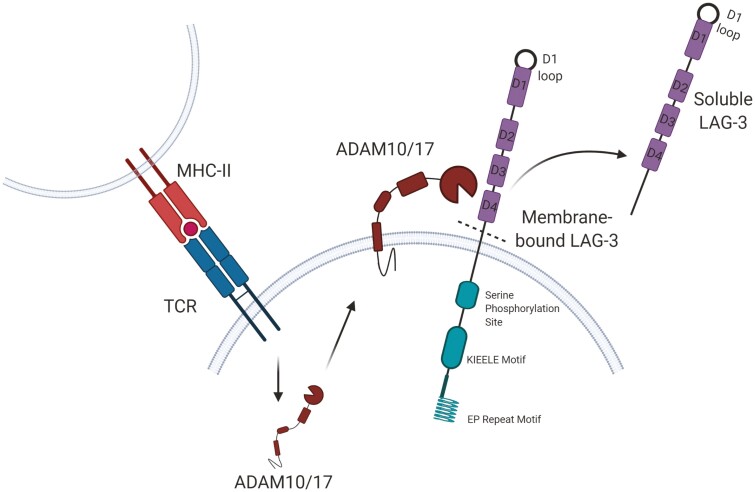
Cell surface LAG-3 is regulated, in part, via cleavage of the extracellular domain, resulting in a 52 kDa soluble form of LAG-3, known as soluble LAG-3 (sLAG-3). Cleavage is mediated by two TCR-induced metalloproteases, a dis-integrin and metalloproteinase domain-containing proteins 10 and 17 (ADAM10 and ADAM17) (Fig. 3) [66, 67]. Although sLAG-3 has been investigated in many chronic disease settings such as cancer and neurodegenerative diseases, no clear biological function has been identified [68–72]. It is unclear whether increased serum sLAG-3 levels reflect an increase in LAG-3 expression and turnover, or whether an increase in cleavage of cell surface LAG-3 plays a particular role in modulating its functions. Expression of sLAG-3 has been attributed, in vivo, to activated T cells [66] but also pDCs which produce potentially five-fold more sLAG-3 than activated T cells [9]. Given the expression of membrane-bound LAG-3 on both B cells [8] and NK cells [73], these cell types may also produce the soluble form.
Figure 3.
Cleavage of LAG-3 from the cell membrane. Soluble LAG-3 is produced when membrane-bound LAG-3 is cleaved by matrix metalloproteinases ADAM10 or ADAM17 between the D4 and transmembrane domains. The role of sLAG-3 is still unknown.
Soluble forms of other immune checkpoints
Evidence demonstrates that sLAG-3 is produced via cleavage from the cell surface or from alternatively spliced RNA [66, 67, 74]. Several co-inhibitory molecules such as PD-1, PD-L1, and CTLA-4 also have both membrane-bound and soluble forms [75]. Studies have suggested that soluble PD-1 mediates an immune-modulating effect in part by binding to membrane-bound PD-L1, obstructing the interaction of membrane PD-1 and PD-L1, counteracting the inhibitory effects of this interaction [76, 77]. On the other hand, soluble PD-L1 has also been shown to bind to PD-1 on the surface of T cells, inducing an inhibitory signal that prevents T cell activation and proliferation. Soluble CTLA-4, secreted during an immune response, has also demonstrated potent inhibitory properties [78, 79]. Whilst the contribution of these soluble forms to immunosuppression is not well understood, it is reasonable to postulate that sLAG-3 may perform a similar role.
Does cleavage of LAG-3 serve more than a regulatory function?
Li et al. showed that cleavage of LAG-3 from the cell surface increases T cell function yet sLAG-3 had no effect on antigen-driven T cell activation and proliferation [67]. Further to this, the same group showed that the half-life of passively transferred, purified sLAG-3 was less than four hours, and that sLAG-3 did not specifically bind to MHC-II or B cells. This led to the hypothesis that once LAG-3 is removed from the cell surface, it is degraded or secreted serving no further function [67]. In summary, it still needs to be determined whether sLAG-3 is purely a short-lived bi-product of T cell activation or whether it serves a specific immune-modulating function whether this is inhibitory or stimulatory as discussed below.
A role for sLAG-3 in prognosis?
Despite uncertainties regarding function, serum sLAG-3 has been investigated as a prognostic marker, particularly in cancer. In non-small cell lung cancer (NSCLC), serum sLAG-3 was inversely correlated with stage; levels were significantly higher in stages I–II NSCLC than in stages III–IV [68]. Additionally, high serum sLAG-3 level at diagnosis for breast cancer patients was associated with longer disease-free survival after a long follow-up [70]. When serum sLAG-3 levels were examined in patients with gastric cancer (GC), its expression correlated with TNM stage, depth of tumour invasion and degree of tumour differentiation in addition to positively correlating with IL-12 and IFN-γ levels [69]. This study also demonstrated that the administration of recombinant sLAG-3 in GC-bearing mice prolonged overall survival, increased survival rate and increased the CD8+ T cell response. The authors speculated that this was due to a greater expansion of antigen-specific CD8+ T cells [69]. Hence, in certain solid epithelial cancers, the presence of sLAG-3 is associated with a favourable prognosis. Despite identifying sLAG-3 levels in the sera of patients in these disease settings, identification of the cell types involved in producing sLAG-3 is lacking owing to the difficulties of tracing back to the cells responsible for this measured sLAG-3. However, in patients with chronic lymphocytic leukaemia (CLL), where sLAG-3 levels were higher in patients whose disease had progressed compared to those with stable CLL, sLAG-3 production was able to be attributed to the malignant B cells themselves [80]. Here, sLAG-3 was shown as a marker of tumour burden by tracking mRNA expression of the soluble isoform of LAG-3 (LAG-3V3) as opposed to the cleaved soluble form.
In another study, increased LAG-3 expression on CD4+ T cells was shown to correspond with high bacterial burden and active tuberculosis (TB) infection [81]. A follow-up study indicated that sLAG-3 levels increased during treatment, corresponding to treatment response and patients with only small increases in sLAG-3 levels during treatment demonstrated poor clinical outcome and/or failed to respond to their treatment [82]. These two studies taken together imply that during TB treatment, LAG-3 is cleaved from the cell surface at an increased rate, due to the increased activity of Th1 cells. Moreover, sLAG-3 levels in the plasma may act as a marker of treatment efficacy in TB.
Use of sLAG-3 in therapy
LAG-3 based therapy has employed the use of the LAG-3:Fc dimer, described previously, comprising the extracellular Ig domains of LAG-3 joined to the Fc section of an antibody [29]. Eftilagimod alpha (also known as LAG-3:Fc or IMP321) stimulates dendritic cells (DCs) via MHC-II. This results in an enhanced presentation of antigen to T cells and as such is classified as a member of the new class of adjuvant/antigen-presenting cell (APC) activators, use of which may result in a better response to vaccination [83–85]. During an early phase I study in 2007, patients co-injected with Eftilagimod alpha and the Hepatitis B vaccine (HBsAg) demonstrated a lower incidence of adverse events, faster and higher antibody responses, and an increased number of vaccine responders when compared to the group injected with the vaccine alone [83]. Eftilagimod alpha has been used as an adjuvant for treatment strategies in a range of phase I studies, including renal cell carcinoma, breast cancer, melanoma, NSCLC, and metastatic HNSCC – enhancing immune response and anti-tumour activity with little to no toxicity [70,83,86]. Additionally, a phase IIb clinic trial, that ended in December 2020, investigating Eftilagimod alpha with paclitaxel in patients with metastatic breast cancer presented with favourable results (https://www.globenewswire.com/news-release/2020/03/25/2005929/0/en/Immutep-Reports-Supportive-Efficacy-Data-from-the-Phase-IIb-AIPAC-Study-Overall-Survival-Data-Expected-in-Late-2020.html) [85]. There are multiple LAG-3 therapies in the clinic for a range of diseases, Table 1 examines those that are complete.
Table 1.
Clinical trials (complete, withdrawn, terminated, or suspended) that have involved the use of LAG-3 as a therapeutic either alone, in combination, or as an adjuvant therapy
| Identifier | Therapeutic (LAG-3 mechanism) | Disease | Phase | Participants | Status | Results |
|---|---|---|---|---|---|---|
| NCT00354861 | sLAG-3 (IMP321/eftilagimod alpha) an APC activator (sLAG-3:Fc) with hepatitis B antigen | Healthy volunteers (male) | 1 | 48 | Completed February 2006 | No study results have been reported |
| NCT00354263 | sLAG-3 (IMP321/eftilagimod alpha) alone or as an adjuvant to a reference flu antigen |
Healthy volunteers (male) | 1 | 60 | Completed February 2006 | No study results have been reported |
| NCT00351949 | sLAG-3 (IMP321/eftilagimod alpha) | Metastatic renal cell carcinoma | 1 | 24 | Completed October 2008 | No clinically significant adverse events were observed. Tumour growth reduced and PFS better in patients with higher doses [84] |
| NCT00349934 | sLAG-3 (IMP321/eftilagimod alpha) with paclitaxel (chemotherapy) | Metastatic breast cancer | 1 | 33 | Completed January 2010 | Increased number and activation of APC and percentage of NK and effector memory CD8 T cells. Clinical benefit observed in 90% of patients [87] |
| NCT00732082 | sLAG-3 (IMP321/eftilagimod alpha) with gemcitabine (chemotherapy) |
Pancreatic cancer | 1 | 18 | Terminated September 2012 | Company manufacturing study drug was unable to continue production |
| NCT00365937 | sLAG-3 (IMP321 with eight HLA-A2 restricted peptides |
Melanoma | 1/2 | 19 | Terminated December 2013 | New regulations of the peptides by the pharmaceutical company |
| NCT01308294 | sLAG-3 (IMP321/eftilagimod alpha) | Melanoma | 1/2a | 16 | Terminated April 2014 | Low enrolment rate. Side effects were mild to moderate. Specific CD4 T-cell responses found in all 16 patients. Conclude that vaccination with IMP321 is promising and safe and induces sustained immune responses [88] |
| NCT02195349 | Anti-LAG-3 (GSK2831781) IgG1 antibody-dependent cell cytotoxicity (ADCC) enhanced mAb – depleting | Healthy volunteers and patients with plaque psoriasis | 1 | 67 | Completed March 2018 | No safety or tolerability concern was identified following a single IV dose of GSK2831781 up to 5 mg/kg. Psoriasis disease activity improved compared to placebo [17] |
| NCT02676869 | sLAG-3 (IMP321/eftilagimod alpha) and pembrolizumab (Anti-PD-1) |
Metastatic melanoma | 1 | 24 | Completed December 2019 | No study results have been reported |
| NCT03965533 | Anti-LAG-3 (GSK2831781) Depleting antibody |
Healthy volunteers | 1 | 37 | Completed December 2019 | No study results have been reported |
| NCT03489369 | Anti-LAG-3 (Sym022) Fc-inert mAb -antagonistic |
Metastatic cancer, solid tumour or l ymphomas | 1 | 15 | Completed January 2020 | No study results have been reported |
With the information we have to date, it is clear that further work is urgently required to determine the physiological role of sLAG-3 and its utility as an immunotherapeutic agent. Importantly, the lack of progression of LAG-3 monotherapies, despite a relative lack of toxicity issues, may indicate a potential redundancy between co-inhibitory receptors (as explored further below). This also highlights the need to explore combination blockade further as a possible key to unlocking the potential of LAG-3 targeting therapies.
LAG-3 knockouts: where is the phenotype?
Perhaps indicative of the modest effect of LAG-3 blockade as a monotherapy in human cancers [89] and mouse tumour models [90, 91] complete knockout of LAG-3 in mice displayed normal immune function [10] – in stark contrast to the lymphoproliferative disease observed in CTLA-4 knockout mice [92]. First described by Miyazaki et al. [10], homozygous LAG-3 mutant mice appeared healthy, displayed normal T cell thymic development, and possessed typical numbers of peripheral CD4+ and CD8+ T cells with expected behaviours [10]. Subsequent studies showed that after 5 weeks of age, LAG-3−/− mice possessed approximately twice the number of αβ T cells with no impact on T cell phenotype or the ratio of T cell types (naïve, memory, or regulatory) [93]. LAG-3−/− mice also had higher numbers of other immune cells, including γδ T cells, NK cells, B cells, macrophages, and DCs [93]. Given that one of the key characteristics of LAG-3 is its ability to inhibit T cell proliferation, the increased number of T cells in LAG-3−/− mice is unsurprising [1, 4]. Accordingly, during adoptive transfer experiments, the genetic ablation of LAG-3 results in enhanced homeostatic expansion of LAG-3−/− T cells compared to wild-type (WT) T cells [93, 94]. This is in contrast to recent observations that CAR-T cells lacking LAG-3 were not functionally distinct to their LAG-3 expressing counterparts [95]. Despite this, LAG-3 along with PD-1 and TIM-3 was reported to define a subpopulation of T cells positively associated with clinical response in CAR-T therapy in patients with large B cell lymphoma [96].
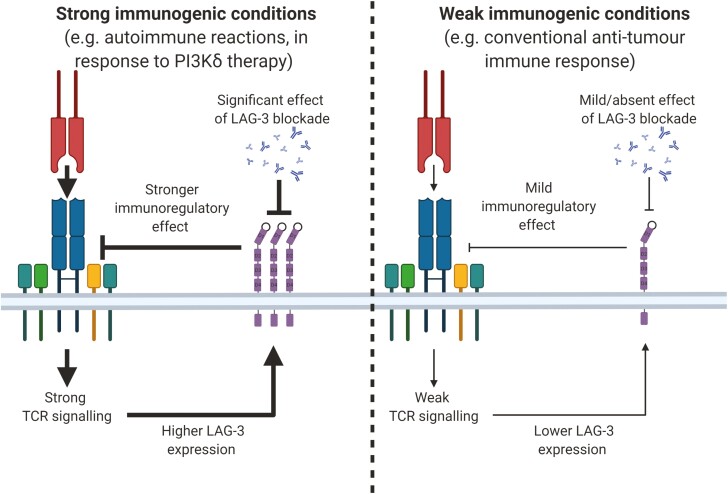
Further clues may also hide within recently made observations that LAG-3−/− mice express higher levels of the co-inhibitory receptors TIM-3, 2B4, and particularly PD-1 on T cells [97]. Similarly, in reverse, knockout of PD-1 results in an upregulation of LAG-3 on T cells, with this effect being more significant for CD8+ T cells [97]. These observations may point towards an underlying mechanism for the synergy observed between LAG-3 and PD-1 combinatorial blockade [90]. A recently proposed role of LAG-3 is that it functions as a rheostat of T-cell activation (Fig. 4). This is supported by the observation that the magnitude of LAG-3 expression is proportional to the affinity of a TCR for its pMHC-II complex [98–101]. In keeping with the rheostat role, the inhibitory potential of LAG-3 is proportional to the amount of LAG-3 on the cell surface [38]. It is noteworthy that LAG-3− mice injected with tumour cell lines exhibit minimal survival advantage when compared to WT mice [27, 90, 102]. Similarly, we have recently reported that LAG-3 blockade alone induced no survival advantage in mice with established 4T1 or MC38 tumours [91].
Figure 4.
LAG-3 as a rheostat of T-cell immune responses. Immunogenicity may be an important factor that should be considered when assessing the role of LAG-3 in regulating immune responses. Under low immunogenic conditions, LAG-3 expression is likely low and in turn, mild/no effects are observed upon LAG-3 blockade. Instead, during stronger immunogenic conditions during which the role of LAG-3 in immune regulation may be greater, it is possible that a more significant effect is observed upon blockade of LAG-3.
Nonetheless, when directly comparing LAG-3−/− T cells to WT T cells in adoptive transfer experiments, LAG-3−/− T cells display enhanced effector function compared to WT T cells during viral infections [10, 103, 104]. Although the phenotype of LAG-3−/− mice has been underwhelming, non-obese diabetic (NOD)-LAG-3−/− mice develop T-cell-driven autoimmune diabetes very rapidly, and LAG-3−/− mice are more prone to mercury-induced autoimmunity [105–107]. The subtle phenotype observed in LAG-3−/− mice is presumably the result of enhanced effector T-cell function, reduced Treg function, or a combination of both. Immunogenicity might be an important factor to consider in this context. This is exemplified by recent observations made in our lab, where LAG-3 blockade alone did not provide a survival advantage to mice bearing 4T1/MC38 tumours, likely due to low tumour immunogenicity resulting in the absence of an immune response requiring regulation. However, when mice were treated with a small-molecule inhibitor of the PI3Kδ subunit which reduced Treg function and allowed an immune response to develop in some mice, LAG-3 blockade displayed a strongly potentiating effect on anti-tumour immunity [91]. Perhaps, under weak immunogenic conditions (e.g. in the presence of a B16 melanoma [108]) there is no clear LAG-3−/− phenotype. However, under conditions where an immune response requires regulation (e.g. under pro-autoimmune conditions) a LAG-3−/− phenotype emerges.
Is LAG-3 expressed on epithelium?
Emerging studies have suggested that beyond expressing ligands for co-inhibitory receptors, tumour cells can also express co-inhibitory receptors such as CTLA-4 and PD-1 [109–112]. Interestingly, unlike on T cells, PD-1 signalling on melanoma cells was found to have pro-tumorigenic effects. These findings may reveal unforeseen benefits of checkpoint blockade immunotherapies.
LAG-3 expression has been described on lung cancer cell lines and fresh surgical lung cancer specimens using immunohistochemistry [113]. The results from this study are yet to be corroborated by other laboratories. Two separate studies, one assessing LAG-3 expression on 55 NSCLC cell lines and the other addressing glioblastoma, did not find LAG-3 expression on tumour cells [114, 115]. To add to the opacity surrounding LAG-3 expression on tumour cells, a recent abstract (American Society of Clinical Oncology Meeting 2020) reported expression of LAG-3 on tumour cells from B cell lymphomas as well as a range of epithelial tumours including lung, breast, and ovarian cancers [116]. Furthermore, assessment of RNAseq databases of cancer cell lines like the ‘Expression Atlas’ [117] suggests that various cancer cell lines express at least medium levels (11–1000 transcripts per million) of LAG-3 RNA transcripts, including leukaemia (e.g. TALL-1), lymphoma (e.g. A3-KAW), neuroblastoma (e.g. KP-N-YN) glioma (KALS-1), multiple myeloma (e.g. KMS-20), small cell lung carcinoma (e.g. NCI-H2171) and pancreatic carcinoma cell lines (e.g. KP4). It is important, however, to replicate these observations at the protein level before conclusions can be drawn.
Nonetheless, with these conflicting reports in mind, it is unclear what role, if any, LAG-3 plays if expressed on tumour cells. A possible scenario is one in which LAG-3 expression results in pro-tumorigenic effects in a similar fashion to PD-1 [111]. To learn more, further studies are needed which examine LAG-3 expression in situ on resected tumours and its association with prognosis.
Is LAG-3 expressed in the brain?
LAG-3 is potentially expressed in the central nervous system (CNS), initially shown to be expressed in neuronal culture cells [55]. Its exact location remains to be clearly defined and may include neurons and supportive cells such as microglia [118]. Evidence also suggests that LAG-3 is expressed in gliomas with an active inflammatory microenvironment [115, 119, 120]; the modulation of which is being investigated in a current clinical trial (ClinicalTrials.gov Identifier: NCT02658981).
It is emerging that proteins with fundamentally important roles within the immune system are expressed in the CNS where their roles are distinct. For instance, there is evidence that MHC-I molecules help CNS development and plasticity [121]. LAG-3 binding to α-syn has been implicated in Parkinson’s disease (PD) pathogenesis. The cell to cell spread of misfolded α-syn appears to contribute to a group of diseases, including PD, known as α-synucleinopathies [55]. Misfolded pre-formed fibrils (PFFs) of α-syn (rather than monomers) are suggested to be a ligand for LAG-3 where binding leads to endocytosis of the complex, possibly facilitating their cell to cell spread [55, 122–125]. Using the recombinant PFFs [55] and human A53T α-syn models [126], both of which drive α-synucleinopathies in mice, treatment with anti-LAG-3 antibodies or LAG-3−/− knockout resulted in reduced pathology [55, 126]. Further to this, in a recent study investigating the mechanism of pathological α-syn spread, the alkaline surface of the D1 domain of LAG-3 was shown to bind to the acidic C-terminus of α-syn [127]. Serum sLAG-3 levels in PD patients were significantly higher compared to healthy controls [128, 129], indicating that this interaction between LAG-3 and α-syn could provide a possible target for the development of therapeutics designed to slow the progression of α-synucleinopathies.
In another study investigating regional atrophy in PD, LAG-3, and the RAS-related protein, Rab5A were identified as two predictive candidate genes [130]. Interestingly, the previously discussed study identified that LAG-3 facilitated endocytosis of α-syn PFFs and subsequent co-localisation with the early endosomal marker Rab5A [55]. This group hypothesised that LAG-3 and Rab5A control regional propagation of α-syn following initiation of disease in certain brain regions and that α-synucleinopathies spread in stereotypical patterns from one brain region to another [130]. These two studies linking LAG-3 and Rab5A with α-syn aggregation and transfer could be an important starting point in determining whether these proteins are involved in PD initiation and progression.
In contrast, a recent publication by Emmenegger et al. found no evidence of LAG-3 in neuronal cell lines, neuronal stem cell-derived cultures nor human brain samples; assaying for expression using multiple anti-LAG-3 antibodies [131]. Marginal LAG-3 expression above background was, however, observed in mouse microglia at the RNA level. It was suggested that the association between LAG-3 and PFFs was due to the promiscuous binding of the PFFs rather than a specific interaction between LAG-3 and α-syn, given α-syn also bound other proteins such as CD4 and tau. It should be noted that Emmenegger et al. did not observe the increased survival and delayed α-syn PFF formation in LAG-3 knockout mice reported by Mao et al. [55]. Thus, LAG-3 expression in the brain and the implications of the LAG-3 α-syn axis in PD is contentious and the subject of ongoing discussion; further work is required to resolve these conflicting data.
Conclusion
It is clear that several uncharted territories exist on the map of LAG-3. These will require further exploration if we are to fully exploit LAG-3 as a therapeutic target. We summarise these by raising the following questions:
Do different LAG-3 ligands perform distinct roles, and if so, which LAG-3-ligand interactions should be targeted in each scenario?
Will developing techniques finally enable the acquisition of the LAG-3 structure? If obtained, what light would this shed on LAG-3 interactions with its ligands or on targeting strategies? Study of LAG-3 via cryo-EM may be achievable as the size barrier is lowered, however, engineering of novel LAG-3 constructs would be beneficial to both cryo-EM and X-ray crystallography approaches.
Why does LAG-3 utilise multiple signalling motifs that are not observed in other immune inhibitory receptors? The complexity and uniqueness of LAG-3 signalling leave this fundamental aspect of LAG-3 poorly understood and consequently more difficult to target. Improving our understanding of LAG-3 signalling may provide further downstream targets for therapeutic modulation.
Beyond arising as a result of the regulation of LAG-3 expression, does sLAG-3 possess a function that is yet to be identified, and if so, what implications might this have for LAG-3 targeting therapies?
Might the LAG-3−/− phenotype hold more subtle traits that have not yet been observed and could this phenotype hold the key to understanding the synergy between LAG-3 and PD-1 combinatorial blockade?
Can tumours ectopically express LAG-3, and if so, what implications might this have for LAG-3 targeting therapies? Definition of LAG-3 expression and purpose on tumour cells might reveal a novel insight about LAG-3 function.
Is LAG-3 expressed in the brain and if so, on what cells is it expressed? Is it there by chance or as a result of evolutionary motives? It is important to determine whether LAG-3 could be a novel target for the treatment of conditions like PD.
Despite these mysteries, clinical trials targeting LAG-3 remain ongoing. Antibodies aimed at blocking the LAG-3/MHC-II interaction are being tested in five active cancer immunotherapy clinical trials (NCT03365791, NCT03499899, NCT02460224, NCT02061761, NCT02658981) and 15 more are currently recruiting at the time of publication [19]. In addition, an anti-PD-1/LAG-3 bispecific antibody trial is also recruiting (NCT04140500) based on data that antibody targeting of PD-1 and LAG-3 synergistically inhibited tumour growth in vivo [90]. So much remains to be answered in the exciting field of LAG-3 biology, and perhaps these empirical experiments framed in clinical trials will offer further key insights: time will tell.
Acknowledgements
Figures were created with Biorender.com. The authors would like to thank the reviewers for their helpful suggested additions to the manuscript. The Editor-in-Chief, Tim Elliott, and handling editor, Adriana Bonomo, would like to thank the following reviewers, Elizabeth Mann and Martin Bonamino, for their contribution to the publication of this article.
Glossary
Abbreviations
- α-syn
α-synucelin
- µM
Micro molar
- a.a
Amino acid
- ADAM10/17
A dis-integrin and metalloproteinase domain-containing protein 10/17
- APC
Antigen presenting cells
- ATPase
adenosine triphosphatase
- BLI
BioLayer interferometry
- Ca2+
Calcium
- CAR-T cell
Chimeric antigen receptor T cell
- CEACAM
Carcinoembryonic antigen-related cell adhesion molecule 1
- CLRs
C-type lectin receptors
- CML
Chronic myelogenous leukemia
- CNS
Central nervous system
- Cryo-EM
Cryo electron microscopy
- CTLA-4
Cytotoxic T-lymphocyte associated protein 4
- D1/2/3/4
Domain 1/2/3/4
- DCs
Dendritic cells
- DC-SIGN
Dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin
- DSS
Dextran sulphate sodium
- Fab
Fragment antigen binding
- Fc
Fragment crystallisable
- FD
Fibrinogen-like domain
- FGL1
Fibrinogen-like protein 1
- Gal-3
Galectin 3
- GC
Gastric cancer
- GFP
Green fluorescent protein
- GlcNAc
N-acetyl glucosamine
- GnTI
N-acetylglucosaminyltransferase I
- GSK3
Glycogen synthase kinase-3
- HLA
Human leukocyte antigen
- hLAG-3
Human LAG-3
- HMGB1
High mobility group protein B1
- HNSCC
Head and neck squamous cell carcinoma
- IFNγ
Interferon gamma
- Ig
Immunoglobulin
- IL-2,-12
Interleukin-2, -12
- KD
Affinity constant
- kDa
Kilo dalton
- LAG-3
Lymphocyte activation gene 3
- LSECtin
Liver and lymph node sinusoidal endothelial cell C-type lectin
- mAb
monoclonal antibody
- MHC-II
Major histocompatibility complex class II
- mLAG-3
Mouse LAG-3
- mRNA
Messenger ribonucleic acid
- MS
Multiple sclerosis
- Na+/K+
Sodium/potassium
- NK
Natural killer
- NOD
Non-obese diabetic
- NSCLC
Non-small cell lung cancer
- OPCs
Oligodendrocyte precursor cells
- PD
Parkinson’s disease
- PD-1
Programmed cell death protein 1
- PD-L1
Programmed cell death protein ligand 1
- pDC
Plasmacytoid dendritic cell
- PFF
Pre-formed fibrils
- PI3K
Phosphoinositide 3-kinase
- pMHC-II
MHC-II protein
- PPI
Protein–protein interactions
- PtdSer
Phosphatidylserine
- Rab5A
RAS-related protein 5A
- sLAG-3
Soluble LAG-3
- SPR
Surface plasmon resonance
- TB
Tuberculosis
- TCGA
The cancer genome atlas
- TCR
T cell receptor
- TIM-3
T cell immunoglobulin and mucin-domain containing-3
- TNFα
Tumour necrosis factor alpha
- TNM
Tumour, nodes, metastasis
- Treg
T regulatory cells
- WT
Wild type
Funding
S.E.A.B. supported by the Wales Cancer Research Centre (517190). L.C. supported by a GW4 BIOMED MRC-DTP PhD studentship (MR/N0137941/1). B.J.M. is supported by a Sêr Cymru award from the Welsh Assembly Government. G.H.M. is supported by Immutep SAS. A.G. and A.M.G. supported by CRUK (C16731/A21200), Cancer Research Wales and the Wellcome Trust (209213/Z/17/Z).
Conflicts of interest
The authors declare no conflicts of interest.
Author contributions
S.E.A.B., L.C., G.H.M., A.M.G., and A.G. conceptualisation. A.M.G. and A.G. supervision. S.E.A.B., L.C., G.H.M., and B.J.M. writing. A.M.G. and A.G. editing and proofreading.
Data availability
There are no new data associated with this article.
References
- 1. Durham NM, Nirschl CJ, Jackson CMet al. . Lymphocyte activation gene 3 (LAG-3) modulates the ability of CD4 T-cells to be suppressed in vivo. PLoS One 2014;9:e109080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen N, Liu Y, Guo Yet al. . Lymphocyte activation gene 3 negatively regulates the function of intrahepatic hepatitis C virus-specific CD8+ T cells. J Gastroenterol Hepatol 2015;30:1788–95. [DOI] [PubMed] [Google Scholar]
- 3. Workman CJ, Rice DS, Dugger KJet al. . Phenotypic analysis of the murine CD4-related glycoprotein, CD223 (LAG-3). Eur J Immunol [Internet] 2002;32(8):2255. [DOI] [PubMed] [Google Scholar]
- 4. Zhang Q, Chikina M, Szymczak-Workman ALet al. . LAG3 limits regulatory T cell proliferation and function in autoimmune diabetes. Sci Immunol [Internet]. 2017. Mar 31;2(9):eaah4569. Available from: http://immunology.sciencemag.org/lookup/doi/10.1126/sciimmunol.aah4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang CT, Workman CJ, Flies Det al. . Role of LAG-3 in regulatory T cells. Immunity 2004;21:503–13. [DOI] [PubMed] [Google Scholar]
- 6. Camisaschi C, Casati C, Rini Fet al. . LAG-3 expression defines a subset of CD4(+)CD25(high)Foxp3(+) regulatory T cells that are expanded at tumor sites. J Immunol 2010;184:6545–51. [DOI] [PubMed] [Google Scholar]
- 7. Camisaschi C, De Filippo A, Beretta Vet al. . Alternative activation of human plasmacytoid DCs in vitro and in melanoma lesions: involvement of LAG-3. J Invest Dermatol 2014;134:1893–902. [DOI] [PubMed] [Google Scholar]
- 8. Kisielow M, Kisielow J, Capoferri-Sollami Get al. . Expression of lymphocyte activation gene 3 (LAG-3) on B cells is induced by T cells. Eur J Immunol 2005;35:2081–8. [DOI] [PubMed] [Google Scholar]
- 9. Workman CJ, Wang Y, El Kasmi KCet al. . LAG-3 regulates plasmacytoid dendritic cell homeostasis. J Immunol 2009;182:1885–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miyazaki T, Dierich A, Benoist Cet al. . Independent modes of natural killing distinguished in mice lacking Lag3. Science 1996;272:405–8. [DOI] [PubMed] [Google Scholar]
- 11. Shaler CR, Choi J, Rudak PTet al. . MAIT cells launch a rapid, robust and distinct hyperinflammatory response to bacterial superantigens and quickly acquire an anergic phenotype that impedes their cognate antimicrobial function: defining a novel mechanism of superantigen-induced immunopathology and immunosuppression. PLoS Biol 2017;15:e2001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderson AC, Joller N, Kuchroo VK.. Lag-3, Tim-3, and TIGIT: co-inhibitory receptors with specialized functions in immune regulation. Immunity 2016;44:989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maruhashi T, Sugiura D, Okazaki IM, Okazaki T.. LAG-3: from molecular functions to clinical applications. J Immunother Cancer 2020;8(2):e001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lecocq Q, Keyaerts M, Devoogdt N, Breckpot K.. The next-generation immune checkpoint LAG-3 and its therapeutic potential in oncology: third time’s a charm. Int J Mol Sci 2020;22(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Atkinson V, Khattak A, Haydon Aet al. . Eftilagimod alpha, a soluble lymphocyte activation gene-3 (LAG-3) protein plus pembrolizumab in patients with metastatic melanoma. J Immunother Cancer [Internet] 2020;8(2):e001681. Available from: https://jitc.bmj.com/content/jitc/8/2/e001681.full.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin C-C, Garralda E, Schöffski Pet al. . 387 A phase II, multicenter study of the safety and efficacy of LAG525 in combination with spartalizumab in patients with advanced malignancies. J Immunother Cancer [Internet] 2020;8(Suppl 3):A235–A235. Available from: https://jitc.bmj.com/content/jitc/8/Suppl_3/A235.full.pdf [Google Scholar]
- 17. Ellis J, Marks DJB, Srinivasan Net al. . Depletion of LAG-3+ T cells translated to pharmacology and improvement in psoriasis disease activity: a phase I randomized study of mAb GSK2831781. Clin Pharmacol Ther [Internet] 2021;109(5):1293–303. Available from: https://ascpt.onlinelibrary.wiley.com/doi/abs/10.1002/cpt.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rudd CE, Chanthong K, Taylor A.. Small molecule inhibition of glycogen synthase kinase 3 (GSK-3) specifically inhibits the transcription of inhibitory co-receptor LAG-3 for enhanced anti-tumor immunity. SSRN Electron J [Internet] 2019;30(7):2075–82.e4. Available from: 10.1016/j.celrep.2020.01.076 [DOI] [PubMed] [Google Scholar]
- 19. Qin S, Xu L, Yi Met al. . Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4. Mol Cancer 2019;18:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brunet JF, Denizot F, Luciani MFet al. . A new member of the immunoglobulin superfamily–CTLA-4. Nature 1987;328:267–70. [DOI] [PubMed] [Google Scholar]
- 21. Ishida Y, Agata Y, Shibahara Ket al. . Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. Embo J 1992;11:3887–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shen R, Postow MA, Adamow Met al. . LAG-3 expression on peripheral blood cells identifies patients with poorer outcomes after immune checkpoint blockade. Sci Transl Med [Internet] 2021;13(608):eabf5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Triebel F, Jitsukawa S, Baixeras Eet al. . LAG-3, a novel lymphocyte activation gene closely related to CD4. J Exp Med 1990;171:1393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baixeras E, Huard B, Miossec Cet al. . Characterization of the lymphocyte activation gene 3-encoded protein. A new ligand for human leukocyte antigen class II antigens. J Exp Med 1992;176:327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu F, Liu J, Liu Det al. . LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T-cell responses. Cancer Res 2014;74:3418–28. [DOI] [PubMed] [Google Scholar]
- 26. Kouo T, Huang L, Pucsek ABet al. . Galectin-3 shapes antitumor immune responses by suppressing CD8+ T cells via LAG-3 and inhibiting expansion of plasmacytoid dendritic cells. Cancer Immunol Res 2015;3:412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang J, Sanmamed MF, Datar Iet al. . Fibrinogen-like protein 1 is a major immune inhibitory ligand of LAG-3. Cell 2019;176:334–347.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huard B, Prigent P, Tournier Met al. . CD4/major histocompatibility complex class II interaction analyzed with CD4 − and lymphocyte activation gene-3 (LAG-3)-Ig fusion proteins. Eur J Immunol 1995;25:2718–21. [DOI] [PubMed] [Google Scholar]
- 29. Huard B, Prigent P, Pagès Fet al. . T cell major histocompatibility complex class II molecules down-regulate CD4+ T cell clone responses following LAG-3 binding. Eur J Immunol 1996;26:1180–6. [DOI] [PubMed] [Google Scholar]
- 30. Maruhashi T, Okazaki IM, Sugiura Det al. . LAG-3 inhibits the activation of CD4+ T cells that recognize stable pMHCII through its conformation-dependent recognition of pMHCII. Nat Immunol 2018;19:1415–26. [DOI] [PubMed] [Google Scholar]
- 31. Tunyasuvunakool K, Adler J, Wu Zet al. . Highly accurate protein structure prediction for the human proteome. Nature 2021;596:590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huard B, Mastrangeli R, Prigent Pet al. . Characterization of the major histocompatibility complex class II binding site on LAG-3 protein. Proc Natl Acad Sci U S A 1997;94:5744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yin Y, Wang XX, Mariuzza RA.. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc Natl Acad Sci U S A 2012;109:5405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Niehrs A, Garcia-Beltran WF, Norman PJet al. . A subset of HLA-DP molecules serve as ligands for the natural cytotoxicity receptor NKp44. Nat Immunol 2019;20:1129–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. MacLachlan BJ, Mason GH, Greenshields-Watson Aet al. . Molecular characterization of HLA class II binding to the LAG-3 T cell co-inhibitory receptor. Eur J Immunol 2021;51:331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shiroishi M, Tsumoto K, Amano Ket al. . Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci U S A 2003;100:8856–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jönsson P, Southcombe JH, Santos AMet al. . Remarkably low affinity of CD4/peptide-major histocompatibility complex class II protein interactions. Proc Natl Acad Sci U S A 2016;113:5682–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maeda TK, Sugiura D, Okazaki IMet al. . Atypical motifs in the cytoplasmic region of the inhibitory immune co-receptor LAG-3 inhibit T cell activation. J Biol Chem 2019;294:6017–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cemerski S, Zhao S, Chenard Met al. . T cell activation and anti-tumor efficacy of anti-LAG-3 antibodies is independent of LAG-3 – MHCII blocking capacity. J Immunother Cancer [Internet] 2015;3(Suppl 2):P183–P183. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4649371/ [Google Scholar]
- 40. Yamamoto T, Gotoh M, Kitajima Met al. . Thymosin beta-4 expression is correlated with metastatic capacity of colorectal carcinomas. Biochem Biophys Res Commun 1993;193:706–10. [DOI] [PubMed] [Google Scholar]
- 41. Hara H, Yoshimura H, Uchida Set al. . Molecular cloning and functional expression analysis of a cDNA for human hepassocin, a liver-specific protein with hepatocyte mitogenic activity. Biochim Biophys Acta 2001;1520:45–53. [DOI] [PubMed] [Google Scholar]
- 42. Liu Z, Ukomadu C.. Fibrinogen-like protein 1, a hepatocyte derived protein is an acute phase reactant. Biochem Biophys Res Commun 2008;365:729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yan J, Ying H, Gu Fet al. . Cloning and characterization of a mouse liver-specific gene mfrep-1, up-regulated in liver regeneration. Cell Res [Internet] 2002;12(5–6):353–61. Available from: http://europepmc.org/abstract/MED/12528893 [DOI] [PubMed] [Google Scholar]
- 44. Li CY, Cao CZ, Xu WXet al. . Recombinant human hepassocin stimulates proliferation of hepatocytes in vivo and improves survival in rats with fulminant hepatic failure. Gut 2010;59:817–26. [DOI] [PubMed] [Google Scholar]
- 45. Kim MS, Pinto SM, Getnet Det al. . A draft map of the human proteome. Nature 2014;509:575–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gagneux P, Varki A.. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology 1999;9:747–55. [DOI] [PubMed] [Google Scholar]
- 47. Liu W, Tang L, Zhang Get al. . Characterization of a novel C-type lectin-like gene, LSECtin: demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node. J Biol Chem 2004;279:18748–58. [DOI] [PubMed] [Google Scholar]
- 48. Tang L, Yang J, Liu Wet al. . Liver sinusoidal endothelial cell lectin, LSECtin, negatively regulates hepatic T-cell immune response. Gastroenterology 2009;137:1498–508.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu B, Wang M, Wang Xet al. . Liver sinusoidal endothelial cell lectin inhibits CTL-dependent virus clearance in mouse models of viral hepatitis. J Immunol 2013;190:4185–95. [DOI] [PubMed] [Google Scholar]
- 50. Thiemann S, Baum LG.. Galectins and immune responses-just how do they do those things they do? Annu Rev Immunol 2016;34:243–64. [DOI] [PubMed] [Google Scholar]
- 51. Chen HY, Fermin A, Vardhana Set al. . Galectin-3 negatively regulates TCR-mediated CD4+ T-cell activation at the immunological synapse. Proc Natl Acad Sci U S A 2009;106:14496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gilson RC, Gunasinghe SD, Johannes Let al. . Galectin-3 modulation of T-cell activation: mechanisms of membrane remodelling. Prog Lipid Res 2019;76:101010. [DOI] [PubMed] [Google Scholar]
- 53. Bae J, Accardi F, Hideshima Tet al. . Targeting LAG3/GAL-3 to overcome immunosuppression and enhance anti-tumor immune responses in multiple myeloma. Leukemia [Internet] 2022;36:138–54. Available from: 10.1038/s41375-021-01301-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stefanis L. α-Synuclein in Parkinson’s disease. Cold Spring Harb Perspect Med 2012;2:a009399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mao X, Ou MT, Karuppagounder SSet al. . Pathological α-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science 2016;353(6307):aah3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dong D, Zheng L, Lin Jet al. . Structural basis of assembly of the human T cell receptor-CD3 complex. Nature 2019;573:546–52. [DOI] [PubMed] [Google Scholar]
- 57. Wray V, Mertins D, Kiess Met al. . Solution structure of the cytoplasmic domain of the human CD4 glycoprotein by CD and 1H NMR spectroscopy: implications for biological functions. Biochemistry 1998;37:8527–38. [DOI] [PubMed] [Google Scholar]
- 58. Chang VT, Crispin M, Aricescu ARet al. . Glycoprotein structural genomics: solving the glycosylation problem. Structure 2007;15:267–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Khoshouei M, Radjainia M, Baumeister Wet al. . Cryo-EM structure of haemoglobin at 3.2 Å determined with the Volta phase plate. Nat Commun 2017;8:16099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fan X, Wang J, Zhang Xet al. . Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution. Nat Commun 2019;10:2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yao Q, Weaver SJ, Mock JYet al. . Fusion of DARPin to aldolase enables visualization of small protein by cryo-EM. Structure 2019;27:1148–1155.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jumper J, Evans R, Pritzel Aet al. . Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Maclachlan B. Molecular Characterisation of CD4+ T Cell Responses to Tumour Antigens. Vol. Thesis, Cardiff:Cardiff University, 2016. [Google Scholar]
- 64. Workman CJ, Dugger KJ, Vignali DA.. Cutting edge: molecular analysis of the negative regulatory function of lymphocyte activation gene-3. J Immunol 2002;169:5392–5. [DOI] [PubMed] [Google Scholar]
- 65. Iouzalen N, Andreae S, Hannier Set al. . LAP, a lymphocyte activation gene-3 (LAG-3)-associated protein that binds to a repeated EP motif in the intracellular region of LAG-3, may participate in the down-regulation of the CD3/TCR activation pathway. Eur J Immunol 2001;31:2885–91. [DOI] [PubMed] [Google Scholar]
- 66. Li N, Workman CJ, Martin SMet al. . Biochemical analysis of the regulatory T cell protein lymphocyte activation gene-3 (LAG-3; CD223). J Immunol 2004;173:6806–12. [DOI] [PubMed] [Google Scholar]
- 67. Li N, Wang Y, Forbes Ket al. . Metalloproteases regulate T-cell proliferation and effector function via LAG-3. Embo J 2007;26:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. He Y, Cao J, Zhao Cet al. . TIM-3, a promising target for cancer immunotherapy. Onco Targets Ther 2018;11:7005–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li N, Jilisihan B, Wang Wet al. . Soluble LAG3 acts as a potential prognostic marker of gastric cancer and its positive correlation with CD8+T cell frequency and secretion of IL-12 and INF-γ in peripheral blood. Cancer Biomark 2018;23:341–51. [DOI] [PubMed] [Google Scholar]
- 70. Triebel F, Hacene K, Pichon MF.. A soluble lymphocyte activation gene-3 (sLAG-3) protein as a prognostic factor in human breast cancer expressing estrogen or progesterone receptors. Cancer Lett 2006;235:147–53. [DOI] [PubMed] [Google Scholar]
- 71. Guo W, Zhou M, Qiu Jet al. . Association of LAG3 genetic variation with an increased risk of PD in Chinese female population. J Neuroinflammation 2019;16:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Angelopoulou E, Paudel YN, Villa Cet al. . Lymphocyte-activation gene 3 (LAG3) protein as a possible therapeutic target for Parkinson’s disease: molecular mechanisms connecting neuroinflammation to α-synuclein spreading pathology. Biology (Basel) [Internet] 2020;9(4):86. Available from: https://pubmed.ncbi.nlm.nih.gov/32340360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Workman CJ, Rice DS, Dugger KJet al. . Phenotypic analysis of the murine CD4-related glycoprotein, CD223 (LAG-3). Eur J Immunol 2002;32:2255–63. [DOI] [PubMed] [Google Scholar]
- 74. Triebel F. LAG-3: a regulator of T-cell and DC responses and its use in therapeutic vaccination. Trends Immunol 2003;24:619–22. [DOI] [PubMed] [Google Scholar]
- 75. Zhu X, Lang J.. Soluble PD-1 and PD-L1: predictive and prognostic significance in cancer. Oncotarget 2017;8:97671–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xiao H, Huang B, Yuan Yet al. . Soluble PD-1 facilitates 4-1BBL-triggered antitumor immunity against murine H22 hepatocarcinoma in vivo. Clin Cancer Res 2007;13:1823–30. [DOI] [PubMed] [Google Scholar]
- 77. Gu D, Ao X, Yang Yet al. . Soluble immune checkpoints in cancer: production, function and biological significance. J Immunother Cancer 2018;6:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li Y, Xiao Y, Su Met al. . Role of soluble programmed death-1 (sPD-1) and sPD-ligand 1 in patients with cystic echinococcosis. Exp Ther Med 2016;11:251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ward FJ, Dahal LN, Wijesekera SKet al. . The soluble isoform of CTLA-4 as a regulator of T-cell responses. Eur J Immunol 2013;43:1274–85. [DOI] [PubMed] [Google Scholar]
- 80. Shapiro M, Herishanu Y, Katz BZet al. . Lymphocyte activation gene 3: a novel therapeutic target in chronic lymphocytic leukemia. Haematologica 2017;102:874–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Phillips BL, Mehra S, Ahsan MHet al. . LAG3 expression in active Mycobacterium tuberculosis infections. Am J Pathol 2015;185:820–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lienhardt C, Azzurri A, Amedei Aet al. . Active tuberculosis in Africa is associated with reduced Th1 and increased Th2 activity in vivo. Eur J Immunol 2002;32:1605–13. [DOI] [PubMed] [Google Scholar]
- 83. Brignone C, Grygar C, Marcu Met al. . IMP321 (sLAG-3), an immunopotentiator for T cell responses against a HBsAg antigen in healthy adults: a single blind randomised controlled phase I study. J Immune Based Ther Vaccines 2007;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Brignone C, Escudier B, Grygar Cet al. . A phase I pharmacokinetic and biological correlative study of IMP321, a novel MHC class II agonist, in patients with advanced renal cell carcinoma. Clin Cancer Res 2009;15:6225–31. [DOI] [PubMed] [Google Scholar]
- 85. Dirix L, Triebel F.. AIPAC: a phase IIb study of eftilagimod alpha (IMP321 or LAG-3Ig) added to weekly paclitaxel in patients with metastatic breast cancer. Future Oncol 2019;15:1963–73. [DOI] [PubMed] [Google Scholar]
- 86. Felip E, Doger B, Majem Met al. . Initial results from a phase II study (TACTI-002) in metastatic non-small cell lung or head and neck carcinoma patients receiving eftilagimod alpha (soluble LAG-3 protein) and pembrolizumab . J Clin Oncol [Internet] 2020;38(15_suppl):3100. Available from: https://ascopubs.org/doi/abs/10.1200/JCO.2020.38.15_suppl.3100 [Google Scholar]
- 87. Brignone C, Gutierrez M, Mefti Fet al. . First-line chemoimmunotherapy in metastatic breast carcinoma: combination of paclitaxel and IMP321 (LAG-3Ig) enhances immune responses and antitumor activity. J Transl Med 2010;8:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Legat A, Maby-El Hajjami H, Baumgaertner Pet al. . Vaccination with LAG-3Ig (IMP321) and peptides induces specific CD4 and CD8 T-cell responses in metastatic melanoma patients–report of a phase I/IIa clinical trial. Clin Cancer Res 2016;22:1330–40. [DOI] [PubMed] [Google Scholar]
- 89. Papadopoulos KP, Lakhani NJ, Johnson MLet al. . First-in-human study of REGN3767 (R3767), a human LAG-3 monoclonal antibody (mAb), ± cemiplimab in patients (pts) with advanced malignancies. J Clin Oncol [Internet] 2019;37(15_suppl):2508. Available from: https://ascopubs.org/doi/abs/10.1200/JCO.2019.37.15_suppl.2508 [Google Scholar]
- 90. Woo SR, Turnis ME, Goldberg MVet al. . Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res 2012;72:917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lauder SN, Smart K, Kersemans Vet al. . Enhanced antitumor immunity through sequential targeting of PI3Kδ and LAG3. J Immunother Cancer 2020;8(2):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tivol EA, Borriello F, Schweitzer ANet al. . Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity 1995;3:541–7. [DOI] [PubMed] [Google Scholar]
- 93. Workman CJ, Vignali DA.. Negative regulation of T cell homeostasis by lymphocyte activation gene-3 (CD223). J Immunol 2005;174:688–95. [DOI] [PubMed] [Google Scholar]
- 94. Grosso JF, Kelleher CC, Harris TJet al. . LAG-3 regulates CD8+ T cell accumulation and effector function in murine self- and tumor-tolerance systems. J Clin Invest 2007;117:3383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhang Y, Zhang X, Cheng Cet al. . CRISPR-Cas9 mediated LAG-3 disruption in CAR-T cells. Front Med 2017;11:554–62. [DOI] [PubMed] [Google Scholar]
- 96. Galon J, Scholler N, Perbost Ret al. . Tumor microenvironment associated with increased pretreatment density of activated PD-1+ LAG-3+/− TIM-3− CD8+ T cells facilitates clinical response to axicabtagene ciloleucel (axi-cel) in patients (pts) with large B-cell lymphoma. J Clin Oncol [Internet] 2020;38(15_suppl):3022. Available from: https://ascopubs.org/doi/abs/10.1200/JCO.2020.38.15_suppl.3022 [Google Scholar]
- 97. Huang RY, Francois A, McGray ARet al. . Compensatory upregulation of PD-1, LAG-3, and CTLA-4 limits the efficacy of single-agent checkpoint blockade in metastatic ovarian cancer. Oncoimmunology 2017;6:e1249561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zahm CD, Colluru VT, McNeel DG.. Vaccination with high-affinity epitopes impairs antitumor efficacy by increasing PD-1 expression on CD8+ T cells. Cancer Immunol Res 2017;5:630–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sprouse ML, Shevchenko I, Scavuzzo MAet al. . Cutting edge: low-affinity TCRs support regulatory T cell function in autoimmunity. J Immunol 2018;200:909–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Han C, Sim SJ, Kim SHet al. . Desensitized chimeric antigen receptor T cells selectively recognize target cells with enhanced antigen expression. Nat Commun 2018;9:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Colluru VT, Zahm CD, McNeel DG.. Mini-intronic plasmid vaccination elicits tolerant LAG3+ CD8+ T cells and inferior antitumor responses. Oncoimmunology 2016;5:e1223002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Huang RY, Eppolito C, Lele Set al. . LAG3 and PD1 co-inhibitory molecules collaborate to limit CD8+ T cell signaling and dampen antitumor immunity in a murine ovarian cancer model. Oncotarget 2015;6:27359–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Richter K, Agnellini P, Oxenius A.. On the role of the inhibitory receptor LAG-3 in acute and chronic LCMV infection. Int Immunol 2010;22:13–23. [DOI] [PubMed] [Google Scholar]
- 104. Cook KD, Whitmire JK.. LAG-3 confers a competitive disadvantage upon antiviral CD8+ T cell responses. J Immunol 2016;197:119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Okazaki T, Okazaki IM, Wang Jet al. . PD-1 and LAG-3 inhibitory co-receptors act synergistically to prevent autoimmunity in mice. J Exp Med 2011;208:395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bettini M, Szymczak-Workman AL, Forbes Ket al. . Cutting edge: accelerated autoimmune diabetes in the absence of LAG-3. J Immunol 2011;187:3493–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Jha V, Workman CJ, McGaha TLet al. . Lymphocyte activation gene-3 (LAG-3) negatively regulates environmentally-induced autoimmunity. Plos One 2014;9:e104484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Turk MJ, Guevara-Patiño JA, Rizzuto GAet al. . Concomitant tumor immunity to a poorly immunogenic melanoma is prevented by regulatory T cells. J Exp Med 2004;200:771–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yao H, Wang H, Li Cet al. . Cancer cell-intrinsic PD-1 and implications in combinatorial immunotherapy. Front Immunol 2018;9:1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Contardi E, Palmisano GL, Tazzari PLet al. . CTLA-4 is constitutively expressed on tumor cells and can trigger apoptosis upon ligand interaction. Int J Cancer 2005;117:538–50. [DOI] [PubMed] [Google Scholar]
- 111. Kleffel S, Posch C, Barthel SRet al. . Melanoma cell-intrinsic PD-1 receptor functions promote tumor growth. Cell 2015;162:1242–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Li H, Li X, Liu Set al. . Programmed cell death-1 (PD-1) checkpoint blockade in combination with a mammalian target of rapamycin inhibitor restrains hepatocellular carcinoma growth induced by hepatoma cell-intrinsic PD-1. Hepatology 2017;66:1920–33. [DOI] [PubMed] [Google Scholar]
- 113. Ma C, Sun X, Shen Det al. . Ectopic expression of LAG-3 in non-small-cell lung cancer cells and its clinical significance. J Clin Lab Anal 2020;34:e23244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. He Y, Yu H, Rozeboom Let al. . LAG-3 protein expression in non-small cell lung cancer and its relationship with PD-1/PD-L1 and tumor-infiltrating lymphocytes. J Thorac Oncol 2017;12:814–23. [DOI] [PubMed] [Google Scholar]
- 115. Harris-Bookman S, Mathios D, Martin AMet al. . Expression of LAG-3 and efficacy of combination treatment with anti-LAG-3 and anti-PD-1 monoclonal antibodies in glioblastoma. Int J Cancer 2018;143:3201–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Chen F, Sherwood T, De Costa Aet al. . Immunohistochemistry analyses of LAG-3 expression across different tumor types and co-expression with PD-1. J Clin Oncol [Internet] 2020;38(15 suppl):e15086–e15086. Available from: https://ascopubs.org/doi/abs/10.1200/JCO.2020.38.15_suppl.e15086 [Google Scholar]
- 117. Papatheodorou I, Moreno P, Manning Jet al. . Expression atlas update: from tissues to single cells. Nucleic Acids Res 2020;48:D77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chen VS. Immune Mediators of Central Nervous System Demyelination and Remyelination. University of North Carolina, 2008. [Google Scholar]
- 119. Mair M, Kiesel B, Feldmann Ket al. . Lymphocyte-activation gene 3 (LAG-3) expression in the inflammatory microenvironment of glioma. J Clin Oncol [Internet] 2020;38(15 suppl):2553. Available from: https://ascopubs.org/doi/abs/10.1200/JCO.2020.38.15_suppl.2553 [Google Scholar]
- 120. Mair MJ, Kiesel B, Feldmann Ket al. . LAG-3 expression in the inflammatory microenvironment of glioma. J Neurooncol 2021;152:533–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Huh GS, Boulanger LM, Du Het al. . Functional requirement for class I MHC in CNS development and plasticity. Science 2000;290(5499):2155–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Birol M, Wojcik SP, Miranker AD, Rhoades E.. Identification of N-linked glycans as specific mediators of neuronal uptake of acetylated α-synuclein. PLoS Biol [Internet] 2019;17(6):e3000318. Available from: 10.1371/journal.pbio.3000318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Musila JM, Rhoades E.. Structural evaluation of aromatic residues in α-syn and their role in glycan binding and cellular uptake. Biophys J 2019;116(3):494a. [Google Scholar]
- 124. Acosta K, Rhoades E.. Characterizing alpha-synuclein binding to glycans. Biophys J [Internet] 2018;114(3):77a. Available from: 10.1016/j.bpj.2017.11.467 [DOI] [Google Scholar]
- 125. Bieri G, Gitler AD, Brahic M.. Internalization, axonal transport and release of fibrillar forms of alpha-synuclein. Neurobiol Dis 2018;109:219–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Gu H, Yang X, Mao Xet al. . Lymphocyte activation gene 3 (Lag3) contributes to α-synucleinopathy in α-synuclein transgenic mice. Front Cell Neurosci 2021;15:656426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zhang S, Liu Y-Q, Jia Cet al. . Mechanistic basis for receptor-mediated pathological α-synuclein fibril cell-to-cell transmission in Parkinson’s disease. Proc Natl Acad Sci [Internet] 2021;118(26):e2011196118. Available from: https://www.pnas.org/content/pnas/118/26/e2011196118.full.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Roy A, Choudhury S, Banerjee Ret al. . Soluble LAG-3 and toll-interacting protein: novel upstream neuro-inflammatory markers in Parkinson’s disease. Parkinsonism Relat Disord [Internet] 2021;91:121–3. Available from: https://www.sciencedirect.com/science/article/pii/S1353802021003485 [DOI] [PubMed] [Google Scholar]
- 129. Cui SS, Du JJ, Liu SHet al. . Serum soluble lymphocyte activation gene-3 as a diagnostic biomarker in Parkinson’s disease: a pilot multicenter study. Mov Disord 2019;34:138–41. [DOI] [PubMed] [Google Scholar]
- 130. Freeze B, Acosta D, Pandya Set al. . Regional expression of genes mediating trans-synaptic alpha-synuclein transfer predicts regional atrophy in Parkinson disease. Neuroimage Clin 2018;18:456–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Emmenegger M, De Cecco E, Hruska-Plochan Met al. . LAG3 is not expressed in human and murine neurons and does not modulate α-synucleinopathies. EMBO Mol Med 2021;13:e14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no new data associated with this article.