Abstract
The biosynthetic pathways and functions of ascaroside signaling molecules in the nematode Caenorhabditis elegans have been studied to better understand complex, integrative developmental decision-making. Although it is known that ascarosides play multiple roles in the development and behavior of nematode species other than C. elegans, these parallel pheromone systems have not been well-studied. Here, we show that ascarosides in the nematode Caenorhabditis briggsae are biosynthesized in the same manner as C. elegans and act to induce the alternative developmental pathway that generates the stress-resistant dauer lifestage. We show that ascr#2 is the primary component of crude dauer pheromone in C. briggsae; in contrast, C. elegans dauer pheromone relies on a combination of ascr#2, ascr#3, and several other components. We further demonstrate that Cbr-daf-22, like its C. elegans ortholog Cel-daf-22, is necessary to produce short-chain ascarosides. Moreover, Cbr-daf-22 and Cel-daf-22 mutants produce an ascaroside-independent metabolite that acts antagonistically to crude dauer pheromone and inhibits dauer formation.
Keywords: C. elegans, C. briggsae, ascarosides, dauer, daf-22, glo-1
Introduction
The nematode Caenorhabditis elegans has long been used as model organism not only for understanding general biological phenomena but also for the specific questions of nematode development and behavior. However, as we further explore this species, it is important to expand those findings to show they are applicable to other nematodes, beyond C. elegans.
The related nematode Caenorhabditis briggsae has previously been used to further investigate findings in C. elegans (Wang and Chamberlin 2002; Gupta et al. 2007; Hillier et al. 2007). Orthologous protein sequences are about 80% identical between C. elegans and C. briggsae, similar to the divergence between protein sequences of humans and mouse orthologs (78.5%) (Waterson et al. 2002; Stein et al. 2003). Caenorhabditisbriggsae shares many useful traits of C. elegans including selfing hermaphroditism, a fully annotated genome, optical transparency, and a similar and short lifecycle (Stein et al. 2003; Gupta et al. 2007). The growth conditions and methods for genetic and behavioral analysis are similar to those for C. elegans, and tools developed for C. elegans are relatively easy to modify for use in C. briggsae if they do not already exist as C. briggsae methods (Baird and Chamberlin 2006). In particular, the recent development of an easy and efficient CRISPR method in C. briggsae has made further investigation into the genetics and behavior of this species, both for itself and for comparison with C. elegans, much more feasible (Cohen and Sternberg 2019; Culp et al. 2020).
One of the most intriguing developmental questions in nematodes is the alternative development of larvae into the dauer diapause stage. This lifestage is thought to be vital to the survival of the species during times of inconsistent or nonexistent food supplies or other limiting conditions, and it is similar to the infective juvenile stage of many parasitic nematodes (Fodor et al. 1983). The dauer lifestage in C. elegans and C. briggsae is triggered by the confluence of both physical and chemical signals indicating the abundance of food, the concentration of conspecific nematodes in the local area, and the local amount of various dauer-inducing pheromones (Cassada and Russell 1975; Golden and Riddle 1982, 1984). Unlike C. elegans, the dauer lifestage in C. briggsae is not also triggered by high temperatures (Inoue et al. 2007). Genetic screens for dauer-constitutive and dauer-defective mutants have identified similar genes in both species in the few cases examined, suggesting that the dauer formation pathway is generally conserved between these species (Inoue et al. 2007).
Dauer pheromone was found to comprise various ascarosides, glycosides of the dideoxysugar ascarylose, named for the Ascaris parasitic nematodes in which they were first discovered (Jezyk and Fairbairn 1967; Jeong et al. 2005). Ascarosides are synthesized by nematodes throughout their lives and affect not only the developmental dauer decision, but also behaviors including mating, aggregation, and more (Edison 2009; Pungaliya et al. 2009; Srinivasan et al. 2012; Wharam et al. 2017). Nematodes across clades produce different ascaroside profiles indicating that many of these signals are species specific (Choe et al. 2012). A broad range of species responds to nematode-produced ascarosides including fungi (Hsueh et al. 2013) and plants (Manohar et al. 2020).
Ascarosides in C. elegans are made modularly, using building blocks derived from cellular waste products including the sugar ascarylose and fatty acid side chains; complexity is increased by modulating the length of the fatty acid-like side chain and attaching head or terminal groups scavenged from neurotransmitters, amino acids, and other readily available materials in the cell (von Reuss et al. 2012). Ascarosides require a shortened fatty acid side chain in order to be functional; this process is done through the peroxisomal beta-oxidation pathway comprising the 4 genes: Cel-acox-1, Cel-maoc-1, Cel-dhs-28, and Cel-daf-22 (Golden and Riddle 1985; Butcher et al. 2009; von Reuss et al. 2012). Mutants that lack a gene along the beta-oxidation pathway are unable to produce any of the short-chain ascarosides that affect behavior, although they are able to register exogenous ascaroside signals (Butcher et al. 2009). Similar to what is found in other Caenorhabditis species, C. briggsae has one-to-one orthologs of Cel-maoc-1 (Cbr-maoc-1), Cel-dhs-28 (Cbr-dhs-28), and Cel-daf-22 (Cbr-daf-22) (Harris et al. 2020). Caenorhabditisbriggsae also has orthologs of the several acox-1 genes (e.g. Cel-acox-1.1 and Cbr-acox-1.1) (WormBase WS282; Harris et al. 2020).
In C. elegans, further modification to the 4′ position of an ascaroside to form increasingly complex signals occurs in the gut granules (Panda et al. 2017). These are birefringent and autofluorescent lysosome-related organelles in the nematode intestine used to break down and recycle cellular waste (Hermann et al. 2005). Mutants that lack gut granules, such as Cel-glo-1, Cel-glo-3, or Cel-apb-3 cannot produce 4′-modified or some terminally modified ascarosides (Rabbitts et al. 2008; Panda et al. 2017; Le et al. 2020). Cbr-glo-1 is a one-to-one ortholog with its counterpart in C. elegans and has been shown to have a similar function; specifically, mutants lack both gut granules and the ability to form 4′-modified ascarosides (Le et al. 2020).
In this study, we show that in C. briggsae, Cbr-daf-22 has a function similar to that of its C. elegans ortholog Cel-daf-22: both are essential for ascaroside biosynthesis. We combined our investigation of Cbr-daf-22 with the previously reported mutants of Cbr-glo-1 to characterize and compare the crude pheromones of AF16, Cbr-daf-22, and Cbr-glo-1. Through these experiments, we found that the major component of C. briggsae dauer-pheromone is ascr#2. We also found antidauer activity in the crude pheromones of both Cel-daf-22 and Cbr-daf-22. Investigation of the Cel-glo-1 and Cbr-glo-1 mutant pheromone, which lacks the additional information provided by 4′-modified ascarosides, was found to cause an irregular dauer response curve. Based on these findings, we postulate the existence of an antidauer compound, or class of compounds, that adds further complexity to the intricate and finely tuned system of ascaroside signaling in Caenorhabditis species.
Materials and methods
Caenorhabditis briggsae and C. elegans strains and strain maintenance
The Indian strain AF16 was used as a wild type for C. briggsae while the Bristol strain N2 was used as a wild type for C. elegans. See Table 1 for a list of strains used. All nematode strains were grown on NGM agar plates seeded with Escherichiacoli (OP50) at 20°C.
Table 1.
List of strains.
| Genotype | Strain | Source |
|---|---|---|
| C. elegans wild type | N2 | Brenner (1974) and CGC |
| Cbr-daf-22(sy1524) | PS8777 | This work |
| Cbr-daf-22(sy1525) | PS8778 | This work |
| Cbr-glo-1(sy1382) | PS8515 | Le et al. (2020) and Sternberg lab collection |
| C. briggsae wild type | AF16 | CGC |
| daf-22(ok693) | RB859 | The C. elegans Deletion Mutant Consortium (2012) and CGC |
| glo-1(zu391) | JJ1271 | Hermann et al (2005) and CGC |
CGC, Caenorhabditis Genetics Center.
Construction of Cbr-daf-22 mutants
Construction of the 2 Cbr-daf-22 mutants was done using the C. briggsae modifications of the universal STOP-IN cassette method as described in Wang et al. (2018) and Cohen and Sternberg (2019). The guide used was AATAGTGCATTAGACGATTG; the forward primer was ATGAGCCCAACCAAGCCAAA; and the reverse primer was CGGCTGGGTATGGAAGCTTT. Cbr-daf-22(sy1524) was a successful insertion of the universal STOP-IN cassette, while Cbr-daf-22 (sy1525), contains a 34 bp insertion from the Cbr-dpy-10 locus; both shown below with the flanking sequences underlined.
sy1524
GTGAATAGTGCATTAGACGAGGGAAGTTTGTCCAGAGCAGAGGTGACTAAGTGATAAGCTAGCTTGTGGACTAAAATATGCCG
sy1525
AAAGAGGCTGTGAATAGTGCTAAATCCGATTTGAAGACCTGTGACACACCGGTAGCTAGCTTATCACTTGTGGACTAAAATATGCCG
Liquid nematode culturing
Culturing began by chunking C. briggsae onto 10 cm NGM plates (each seeded with 800 µl of OP50 E. coli grown to stationary phase in Luria–Bertani broth) and incubated at 22°C. Once the food was consumed, each plate was then washed with 25 ml of S-complete medium into a 125 ml Erlenmeyer flask, and 1 ml of OP50 E. coli was added (E. coli cultures were grown to stationary phase in Luria–Bertani broth, pelleted and resuspended at 1 g wet mass per 1 ml M9 buffer), shaking at 220 rpm and 22°C. After 70 h, cultures were centrifuged at 1,000g for 1 min. After discarding supernatant, 24 ml H2O was added along with 6 ml bleach, 900 µl 10 M NaOH, and the mixture was shaken for 3 min to prepare eggs. Eggs were centrifuged at 1,000g, the supernatant was removed, and the egg pellet was washed with 25 ml M9 buffer twice and then suspended in a final volume of 5 ml M9 buffer in a 50 ml centrifuge tube. Eggs were counted and placed on a rocker and allowed to hatch as L1 larvae for 24 h at 22°C. A total of 75,000 L1 larvae were seeded in 25 ml cultures of S-complete with 1 ml of OP50 and incubated at 220 rpm and 22°C in a 125 ml Erlenmeyer flask. After 72 h, nematodes were spun at 1,000g for 5 min and spent medium was separated from nematode body pellet. Separated medium and nematode pellet were flash frozen over liquid nitrogen and then lyophilized. Two biological replicates were grown for each strain. Mutants were grown with parallel wild-type controls, and biological replicates were started on different days.
Dauer pheromone collection
Crude pheromones of AF16, Cbr-daf-22(sy1524), Cbr-glo-1(sy1382), N2, Cel-daf-22(ok693), and Cel-glo-1(zu391) were made using previously described methods (Golden and Riddle 1984; Schroeder and Flatt 2014). Briefly, 1 L of liquid nematode culture (4 flasks of each 250 ml S-complete and 8 ml of OP50 E. coli, grown in the same way as in the above liquid cultures) was grown until exhausted. The liquid culture supernatant was separated from the pellet, filtered, and then dried completely. The dried material was extracted using ethanol; the extract was then dried and redissolved in 1 ml of sterilized water.
Dauer assays
Dauer assays were run using the method described in Lee et al. (2017). The dauer plates were prepared by adding the desired amount of crude pheromone or ascaroside to a 35 × 10 mm plate before adding 2 ml of 2.5% agarose (NG agarose minus Bacto-Peptone). These plates were left to dry for 1 day. Prior to adding any nematodes, the plates were seeded with 10 µl of heat-killed OP50 (OP50 E. coli grown in Luria–Bertani broth, spun down, and heated at 97°C for 5 min) in the center of the plate. Ten adult nematodes were placed on the plate and allowed to lay eggs for 3 h. The adult nematodes were removed, and the plates were seeded with an additional 10 µl of OP50 on top of the eggs. The plates were stored in an incubator at 25.5°C for 24 h, after which the plates were quickly checked to confirm normal hatching and L1 development. After a further 24 h in the 25.5°C in the incubator (48 h after the start of the experiment) the total number of nematodes and dauer-stage nematodes were counted for each plate. To continue to ensure that the pheromones did not affect development, plates were kept for a further 24 h after the end of the experiment to check for expected continued development (i.e. that L3s became normal, egg-laying adults and that dauer animals remained in dauer).
Metabolite extraction
Lyophilized pellet and media samples were crushed and homogenized by shaking with 2.5 mm steel balls at 1,300 rpm for 3 min in 30 s pulses while chilled with liquid nitrogen (SPEX sample prep miniG 1600). Powdered media and pellet samples were extracted with 10 ml methanol in 50 ml centrifuge tubes, rocking overnight at 22°C. Extractions were pelleted at 5,000g for 10 min at 4°C, and supernatants were transferred to 20 ml glass scintillation vials. Samples were then dried in a SpeedVac (Thermo Fisher Scientific) vacuum concentrator. Dried materials were resuspended in 1 ml methanol and vortexed for 1 min. Samples were pelleted at 10,000g for 5 min and 22°C, and supernatants were transferred to 2 ml HPLC vials and dried in a SpeedVac vacuum concentrator. Samples were resuspended in 100 μl of methanol, transferred into 1.7-ml Eppendorf tubes, and centrifuged at 18,000g for 20 min at 4°C. Clarified extracts were transferred to HPLC vials and stored at −20°C until analysis was performed.
Mass spectrometric analysis
High-resolution LC–MS analysis was performed on a Thermo Fisher Scientific Vanquish Horizon UHPLC System coupled with a Thermo Q Exactive hybrid quadrupole-orbitrap high-resolution mass spectrometer equipped with a HESI ion source. One microliter of extract was injected and separated using a water-acetonitrile gradient on a Thermo Scientific Hypersil GOLD C18 column (150 mm × 2.1 mm 1.9 µm particle size 175 Å pore size, Thermo Scientific) and maintained at 40°C. Solvents were all purchased from Fisher Scientific as HPLC grade. Solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile. A/B gradient started at 1% B for 3 min, then from 1% to 100% B over 20 min, 100% for 5 min, then down to 1% B for 3 min. Mass spectrometer parameters: 3.5 kV spray voltage, 380°C capillary temperature, 300°C probe heater temperature, 60 sheath flow rate, 20 auxiliary flow rate, 2.0 spare gas; S-lens RF level 50.0, resolution 240,000, m/z range 150–1,000 m/z, AGC target 3e6. The instrument was calibrated with positive and negative ion calibration solutions (Thermo-Fisher) Pierce LTQ Velos ESI pos/neg calibration solutions. Peak areas were determined using Xcalibur 2.3 QualBrowser version 2.3.26 (Thermo Scientific) using a 5-ppm window around the m/z of interest.
Results
Strain construction and phenotype of Cbr-daf-22
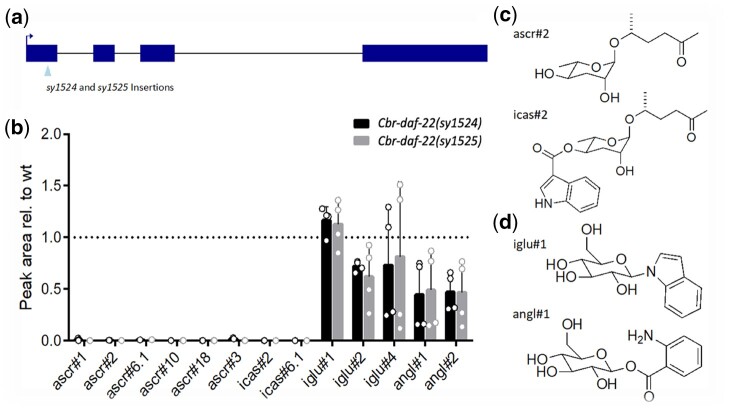
In previous studies, it has been shown that a Cbr-glo-1 loss-of-function mutant shares at least part of its phenotype with an orthologous Cel-glo-1 mutant: neither mutant is able to form gut granules (Hermann et al. 2005; Le et al. 2020). Concomitantly, a Cbr-glo-1 mutant is also unable to produce complex ascarosides that have been modified with a head group at the 4′ carbon positions (Le et al. 2020). To further explore the similarities between C. elegans and C. briggsae ascaroside formation, we made 2 mutant strains of Cbr-daf-22, ortholog of Cel-daf-22, which controls the last step in the C. elegans peroxisomal beta-oxidation pathway (Fig. 1a). These C. briggsae mutants are unable to form simple (e.g. ascr#2) and modular (e.g. icas#2) short-chain ascarosides (Fig. 1b), the phenotype associated with Cel-daf-22 mutant metabolomes in C. elegans (von Reuss et al. 2012). However, both Cbr-daf-22 and its ortholog Cel-daf-22 produce many other types of small molecules, including the previously identified small molecule classes of indole glucosides (iglu) and anthranilic acid glucosides (angl) (Fig. 1, b–d) (Coburn and Gems 2013; Stupp et al. 2013; Le et al. 2020).
Fig. 1.
Caenorhabditis briggsae daf-22 mutant and phenotypes. a) Two Cbr-daf-22 strains made using the CRISPR/Cas9 triple stop knock-in method to insert the full 43 base pair triple-stop insert (sy1524) and a 34 base pair insert (sy1525) in the first exon. b) The metabolomes of both Cbr-daf-22 strains do not produce any functional short-chain ascarosides (e.g. ascr#2) or their 4′ modifications (e.g. icas#2), shown in (c). The Cbr-daf-22 strains are, however, still able to produce glucoside sugars modified at the 1′ position including indole glucosides (e.g. iglu#1) and anthranilic acid glucosides (e.g. angl#1) as shown in (d).
Caenorhabditis briggsae crude pheromone dauer assays
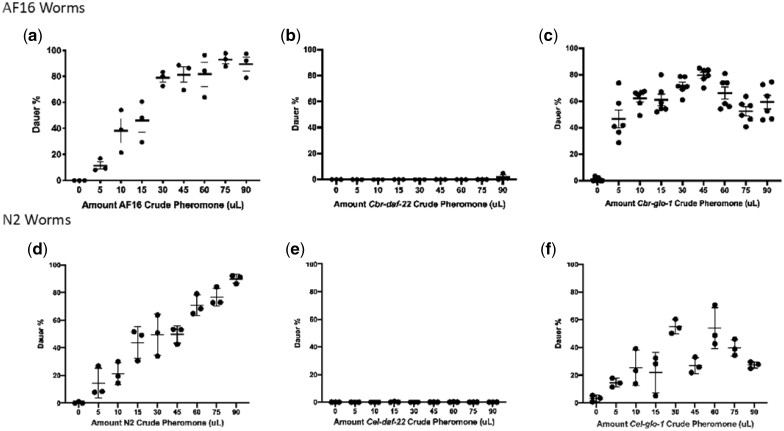
We performed a series of dauer assays using wild-type (AF16) nematodes to determine how the various innate crude pheromone preparations from Cbr-glo-1 and Cbr-daf-22 mutants would affect wild-type nematode dauer formation across a range of concentrations. Wild-type (AF16) crude pheromone exhibited an expected dose–response curve that showed a stable increase in the number of dauers as the amounts of pheromone increased (Fig. 2a). At high pheromone concentrations, almost all nematodes went into dauer. This assay was repeated using crude dauer pheromone from Cbr-daf-22 (Fig. 2b) and Cbr-glo-1 mutants (Fig. 2c). As hypothesized, due to its lack of innate short-chain ascarosides, Cbr-daf-22 crude dauer pheromone was unable to induce dauer formation at any concentration. Cbr-glo-1 crude pheromone appeared to induce dauer formation at fairly consistent but intermediate levels across a broad range of dosages. A Kolmogorov–Smirnov test showed that the dauer curve of Cbr-glo-1 pheromone is significantly different from that of AF16 (Supplementary Fig. 1a). Even the highest concentration tested was unable to replicate the dauer-inducing effectiveness of pheromone produced by the wild type. These data suggest that the dauer pheromone in C. briggsae are simple (i.e. unmodified) short-chain ascarosides.
Fig. 2.
Response to doses of crude pheromone in wild-type C. briggsae and C. elegans. a) The dauer curve of wild-type nematodes treated with crude pheromone prepared from wild-type nematodes creates a linear increase in dauer percentage with the increase in the amount of pheromone, approaching 100%. b) Cbr-daf-22(sy1524) crude pheromone produces no dauer-inducing effect on wild-type C. briggsae nematodes. c) Cbr-glo-1(sy1382) crude pheromone produces a dauer curve that remains stable despite changing amounts of dauer pheromone and never approaches 100%. d) When carried out in C. elegans, wild-type pheromone, e) Cel-daf-22 pheromone, and f) Cel-glo-1 pheromone produce very similar curves to their C. briggsae counterparts. Error bars indicate standard deviation.
These dauer assay experiments were also performed in C. elegans, testing the effects on wild-type nematodes of crude pheromone prepared from N2 wild type (Fig. 2d), Cel-daf-22 (Fig. 2e), and Cel-glo-1 (Fig. 2f). The same pattern occurs in these assays whereby the N2 pheromone has a typical dose–response curve, the Cel-daf-22 pheromone induces no dauers, and the Cel-glo-1 pheromone shows an inconsistent dauer induction effect relative to the amount added. Again, a Kolmogorov–Smirnov test showed that the dauer curve of Cel-glo-1 pheromone is significantly different from that of N2 (Supplementary Fig. 1b).
Isolation of main dauer pheromone component in C. briggsae
The response of Cbr-glo-1 mutants to doses of crude pheromone (Fig. 2c) indicates that at least 1 major component of the C. briggsae dauer-inducing pheromone must be a simple ascaroside since Cbr-glo-1 pheromone is able to induce dauer formation. As the C. briggsae metabolome primarily consists of ascr#2 and ascr#6.1 and their derivatives (Dong et al. 2016; von Reuss 2018), we were able to delineate the likely major dauer pheromone component to those 2 simple ascarosides. Caenorhabditisbriggsae has been shown to respond to C. elegans dauer pheromone (Fodor et al. 1983). C. elegans dauer pheromone contains ascr#2 (Butcher et al. 2007), making ascr#2 a leading candidate for the dominant dauer pheromone signal in C. briggsae.
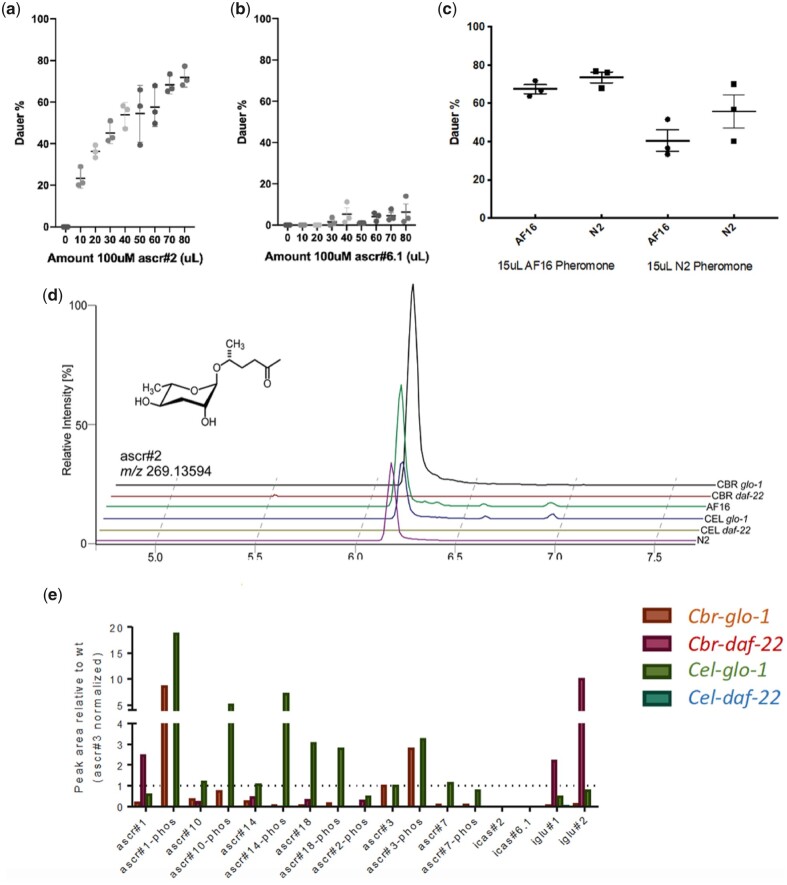
The dauer pheromone curve for ascr#2 demonstrated that it was the main dauer ascaroside for C. briggsae (Fig. 3a). Caenorhabditisbriggsae dauer ascarosides were active as low as the micromolar range, consistent with the general amounts of crude pheromone or purified single ascaroside needed to produce dauer formation effects in C. elegans (Srinivasan et al. 2012) (Supplementary Fig. 2). In contrast to the dauer-promoting effects of ascr#2, the other main ascaroside produced by C. briggsae, ascr#6.1, induced little or no dauer formation, even at high concentrations (Fig. 3b). In agreement with the metabolomics performed above (Fig. 1b), ascr#2 is present in all dauer-inducing crude pheromone tested and is not present in crude pheromone that is unable to induce dauer formation (Fig. 3c).
Fig. 3.
Ascarosides induce dauer formation. a) ascr#2, a main component of the C. briggsae metabolome, induces dauer formation increasingly as the number of ascaroside increases, and is a main component of dauer pheromone in C. briggsae. b) ascr#6.1, another major component of the C. briggsae metabolome, does not significantly induce dauer, even at high concentrations. c) ascr#2 is found in the crude pheromone of C. briggsae wild type, C. elegans wild type, Cbr-glo-1, and Cel-glo-1, all of which are able to induce dauer formation; ascr#2 is not found in the crude pheromone of Cbr-daf-22 or Cel-daf-22. d) Crude pheromone affects C. elegans and C. briggsae similarly except that wild-type C. briggsae pheromone appears to be slightly more potent than wild-type C. elegans pheromone; P< 0.05. e) Crude pheromone from Cbr-daf-22(sy1524) shows an increase in the production of modular glucosides compared to wild type, whereas Cbr-glo-1(sy1382) crude pheromone shows an increase in the production of phosphorylated ascarosides. Error bars indicate standard deviation.
Comparison of C. briggsae and C. elegans crude dauer pheromones shows that C. briggsae pheromone elicits a slightly higher rate of dauer formation in C. briggsae than the same amount of C. elegans pheromone (Fig. 3d). However, each pheromone affects the 2 species similarly.
The crude pheromone for Cbr-daf-22 shows an increase in indole glucosides relative to wild type; this is similar to what is found in the general Cbr-daf-22 metabolome (Fig. 3e). In contrast, Cbr-glo-1 and Cel-glo-1 crude pheromone both show an increase in phosphorylated ascarosides (Fig. 3e).
Cbr-daf-22 pheromone has anti-dauer activity
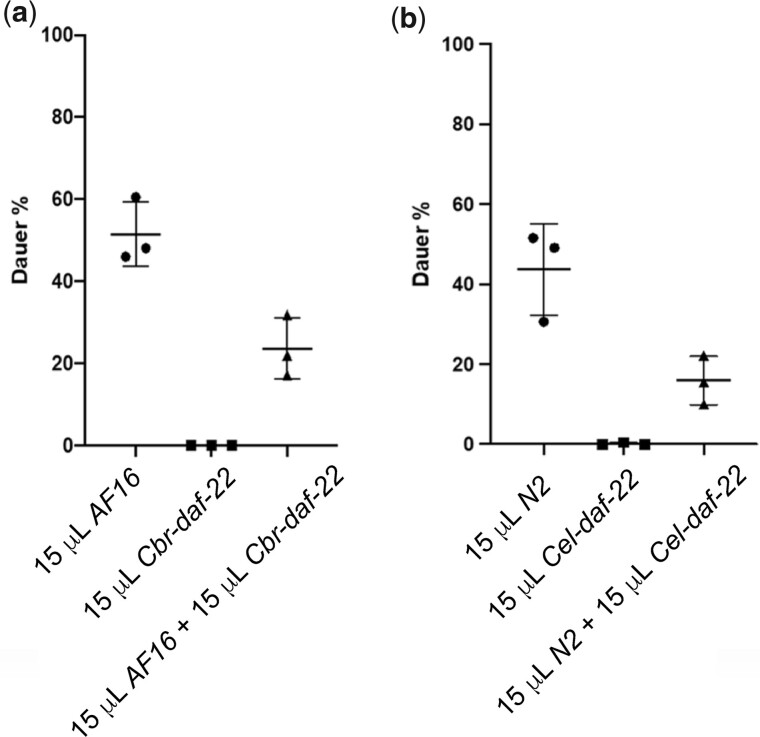
As shown in Fig. 3b, we found that ascr#6.1 had essentially no effect on dauer formation in C. briggsae. However, at high concentrations, ascr#6.1 does induce a small fraction of animals to become dauer larvae. When this was compared to the overall scarcity of dauer formation in response to Cbr-daf-22 mutant dauer pheromone at all concentrations the Cbr-daf-22 mutant pheromone appeared to indicate an actively antidauer effect. To test for such an antidauer effect, we combined the crude pheromones of C. briggsae wild type and Cbr-daf-22 to see if such a combination was additive or antagonistic. We found dauer formation was significantly depressed whenever Cbr-daf-22 pheromone was present (Fig. 4a). When this experiment was performed in C. elegans using pheromone produced from the corresponding C. elegans strains, the same phenomenon was observed (Fig. 4b). When AF16 and N2 nematodes encountered each other’s daf-22 pheromone, the same results—no dauer formation—were seen (Supplementary Fig. 3).
Fig. 4.
Combinations of crude pheromone are not additive. a) When combined with wild-type C. briggsae crude pheromone, Cbr-daf-22 mutant crude pheromone actively suppresses dauer formation in C. briggsae wild-type hermaphrodites. b) The same suppression occurs when wild-type C. elegans crude pheromone is combined with Cel-daf-22 mutant crude pheromone to treat C. elegans wild-type hermaphrodites. Error bars indicate standard deviation; P> 0.05.
Discussion
Comparison of ascaroside formation pathways in C. briggsae and C. elegans
As C. briggsae and C. elegans are closely related evolutionarily, so too are the basic pathways with which they create their myriad of ascarosides, many which are shown to be nematode-specific communication molecules (reviewed by von Reuss 2018). The C. briggsae one-to-one orthologs of Cel-daf-22 (Cbr-daf-22) and Cel-glo-1 (Cbr-glo-1) have been shown to be physiologically and metabolomically equivalent (this work; Le et al. 2020).
Cel-daf-22 and Cbr-daf-22 mutants cannot create the biologically relevant short-chain ascarosides used by both nematode species. However, they are still able to produce indole glucosides (iglu) and anthranilic acid glucosides (angl) at levels comparable to, or slightly lower than wild type. Functionally, the absence of short-chain ascarosides in Cel-daf-22 and Cbr-daf-22 crude pheromone causes a complete lack of dauer formation under the same conditions where increased amounts of C. briggsae or C. elegans wild-type crude pheromone, respectively, causes a linear increase in dauer formation.
Cel-glo-1 and Cbr-glo-1 mutants have previously been shown to lack gut granules and the subsequent ability to form 4′-modified ascarosides (Panda et al. 2017; Le et al. 2020). Functionally, this lack appears to also create an imbalance in the fine-tuned system of dauer-formation communication signaling and causes Cbr-glo-1 and Cel-glo-1 mutant crude pheromone to have unusual dose response curves where the amount of dauer formation appears to be independent of the amount of pheromone encountered. As phosphorylated ascarosides are upregulated in both Cel-glo-1 and Cbr-glo-1 (Fig. 3e), they are possible candidates for this effect.
With these similarities of the main ascaroside-forming pathways, we can infer that other major components of the ascaroside formation pathways, such as the remainder of the beta-oxidation pathway, are likely to be similarly conserved. We also expect to see further conservation in the major components of ascaroside formation between C. elegans and other nematode species, especially when their ascaroside profiles overlap.
Isolation of primary dauer pheromone component in C. briggsae
Caenorhabditis briggsae releases fewer types of known ascarosides than C. elegans and, subsequently, has seemingly fewer behaviors regulated by these ascarosides (Dong et al. 2016). The pathways that are conserved between the 2 species indicate their importance to those behaviors.
We confirmed that ascr#2 is the main component of dauer pheromone in C. briggsae. Thus, ascr#2 acts functionally the same in both C. briggsae and C. elegans. Combined with the conserved ascaroside formation pathways, this finding indicates that it may be an ascaroside originally used by the evolutionary ancestor of both species. However, the 2 species then diverge in other dauer-inducing ascaroside signals such as ascr#3, which is another major component of C. elegans dauer pheromone (Butcher et al. 2007), but which is absent in the C. briggsae metabolome.
Caenorhabditis elegans and C. briggsae not only are closely related in the elegans group of the Caenorhabditis genus, but also have overlapping habitats (Cutter et al. 2006). The 2 species have diverged climatically and temporally with C. briggsae preferring warmer climates and seasons than C. elegans (Félix and Duveau 2012). But this divergence likely did not change their underlying communication needs when it comes to basic dauer-ascaroside messaging. Overlying yet distinct ascaroside profiles may also indicate that, in the wild, their pheromone signaling may be mutually beneficial to both species using broad-strokes signals such as ascr#2. However, we know that indole-modified ascaroside biosynthesis and subsequent behavior is highly species dependent (Dong et al. 2016). Further refinement using species-specific signaling would allow the nematodes to give the best adaptive advantages to their own brethren. These different signals also would allow the nematodes to ignore signals that indicate imperfect conditions for other species when conditions for their own species (temperature proclivities, for example) are ideal.
It has been reported that an indole-modified version ascr#2, icas#2, acts as a sex pheromone in C. briggsae (Dong et al. 2016). The unmodified pheromone may be used as an indication of negative reproductive conditions and thus a dauer-promoting signal while the further modification indicates positive reproductive conditions.
Antidauer effects of Cbr-daf-22 and Cel-daf-22 dauer pheromone
Although the absence of short-chain ascarosides and their dauer-inducing signals staves off dauer-formation under favorable developmental conditions, the almost complete lack of dauer formation in Cbr-daf-22 and Cel-daf-22 samples indicated something more than a simple absence of dauer ascarosides. Subsequently, we found that Cbr-daf-22 and Cel-daf-22 pheromone actively depresses dauer formation, even when in the presence of dauer-promoting crude pheromone from wild-type nematodes.
Our findings indicate that Cbr-daf-22 and Cel-daf-22 metabolomes contain either a specific antidauer compound or a compound that acts as a receptor antagonist. Potential candidates for either process include the angl or iglu families of glycosides, as these small molecules are highly abundant in Cbr-daf-22 and Cel-daf-22 metabolomes. The iglu family of glycosides is especially interesting as a potential antidauer signal as previous studies have shown these small molecules represent detoxification products of indole derived from E. coli, the primary food source of lab-cultured nematodes (Stupp et al. 2013). It would also help explain the idea that if there is some food, there will never be 100% dauer even at extremely high concentrations of dauer pheromone (Golden and Riddle 1984). As the Cel-daf-22 and Cbr-daf-22 pheromones both are able to suppress dauer in both C. briggsae and C. elegans, the antidauer pheromone for both species is hypothesized to contain at least 1 identical compound.
In addition to food, population density, and known dauer pheromones, it is clear that the dauer-decision system is further adjusted through the use of additional pheromone signaling. Thus, the dauer decision is not simply made under unfavorable conditions opposite to the standard L3 reproductive stage—instead there is an active antidauer signal that adds to the complexity of the finely tuned dauer-decision process in C. elegans and C. briggsae.
Data availability
Strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at G3 online.
Supplementary Material
Acknowledgments
The authors thank all members of the Sternberg and Schroeder labs especially Mengyi Cao, Hillel Schwartz, Heenam Park, Jessica Sun, and Katherine Norton for their input, assistance, and helpful comments throughout the project. They thank the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440) for some C. elegans strains used in these experiments, WormBase for sequences, and Tsui-Fen Chou for Cas9 protein.
Funding
This work was supported by the National Science Foundation Graduate Research Fellowship under Grant DGE 1745301 to SMC, the National Institutes of Health through Grants UF1NS111697 and R24OD023041 to PWS, and the National Institutes of Health through Grant R01GM113692 to FCS. FCS is also a Faculty Scholar of the Howard Hughes Medical Institute.
Conflicts of interest
None declared.
Literature cited
- Baird SE, Chamberlin HM.. Caenorhabditis briggsae Methods. WormBook; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA, Fujita M, Schroeder FC, Clardy J.. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat Chem Biol. 2007;3(7):420–422. doi: 10.1038/nchembio.2007.3. [DOI] [PubMed] [Google Scholar]
- Butcher RA, Ragains JR, Li W, Ruvkun G, Clardy J, Mak HY.. Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc Natl Acad Sci USA. 2009;106(6):1875–1879. doi: 10.1073/pnas.0810338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada RC, Russell RL.. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol. 1975;46(2):326–342. doi: 10.1016/0012-1606(75)90109-8. [DOI] [PubMed] [Google Scholar]
- Choe A, von Reuss SH, Kogan D, Gasser RB, Platzer EG, Schroeder FC, Sternberg PW.. Ascaroside signaling is widely conserved among nematodes. Curr Biol. 2012;22(9):772–780. doi: 10.1016/j.cub.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn C, Gems D.. The mysterious case of the C. elegans gut granule: death fluorescence, anthranilic acid, and the kynurenine pathway. Front Genet. 2013;4:151. doi: 10.3389/fgene.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Sternberg PW.. Genome editing of Caenorhabditis briggsae using CRISPR/Cas9 co-conversion marker dpy-10. MicroPubl Biol. 2019. doi: 10.17912/micropub.biology.000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp E, Richman C, Sharanya D, Jhaveri N, van den Berg W, Gupta BP.. Genome editing in the nematode Caenorhabditis briggsae using the CRISPR/Cas9 system. Biol Methods Protoc. 2020;5(1):bpaa003. doi: 10.1093/biomethods/bpaa003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Félix M-A, Barrière A, Charlesworth D.. Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics. 2006;173(4):2021–2031. doi: 10.1534/genetics.106.058651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Dolke F, von Reuss SH.. Selective MS screening reveals a sex pheromone in Caenorhabditis briggsae and species-specificity in indole ascaroside signaling. Org Biomol Chem. 2016;14(30):7217–7225. doi: 10.1039/C6OB01230B. [DOI] [PubMed] [Google Scholar]
- Edison AS. Caenorhabditis elegans pheromones regulate multiple complex behaviors. Curr Opin Neurobiol. 2009;19(4):378–388. doi: 10.1016/j.conb.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M-A, Duveau F.. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 2012;10(1):59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor A, Riddle DL, Nelson FK, Golden JW.. Comparison of a new wild-type Caenorhabditis briggsae with laboratory strains of C. briggsae and C. elegans. Nematol. 1983;29(2):203–216. doi: 10.1163/187529283X00456. [DOI] [Google Scholar]
- Golden JW, Riddle DL.. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science. 1982;218(4572):578–580. doi: 10.1126/science.6896933. [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL.. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol. 1984;102(2):368–378. doi: 10.1016/0012-1606(84)90201-X. [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL.. A gene affecting production of the Caenorhabditis elegans dauer-inducing pheromone. Mol Gen Genet. 1985;198(3):534–536. doi: 10.1007/bf00332953. [DOI] [PubMed] [Google Scholar]
- Gupta BP, Johnsen R, Chen N.. Genomics and Biology of the Nematode Caenorhabditis briggsae. WormBook; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Cho J, Davis P, Gao S, Grove CA, Kishore R, et al. WormBase: a modern model organism information resource. Nucleic Acids Res. 2020;48(D1):D762–D767. doi: 10.1093/nar/gkz920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, Fonarev P, Grant BD, Priess JR.. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16(7):3273–3288. doi: 10.1091/mbc.e05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, Miller RD, Baird SE, Chinwalla A, Fulton LA, Koboldt DC, Waterston RH.. Comparison of C. elegans and C. briggsae geneome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 2007;5(7):e167. doi: 10.1371/journal.pbio.0050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y-P, Mahanti P, Schroeder FC, Sternberg PW.. Nematode-trapping fungi eavesdrop on nematode pheromones. Curr Biol. 2013;23(1):83–86. doi: 10.1016/j.cub.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Ailion M, Poon SH, Kim HK, Thomas JH, Sternberg PW.. Genetic analysis of dauer formation in Caenorhabditis briggsae. Genetics. 2007;177(2):809–818. doi: 10.1534/genetics.107.078857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong P-Y, Jung M, Yim Y-H, Kim H, Park M, Hong E, Lee W, Kim YH, Kim K, Paik Y-K.. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature. 2005;433(7025):541–545. doi: 10.1038/nature03201. [DOI] [PubMed] [Google Scholar]
- Jezyk PF, Fairbairn D.. Ascarosides and ascaroside esters in Ascaris lumbricoides (Nematoda). Comp Biochem Physiol. 1967;23(3):691–705. doi: 10.1016/0010-406X(67)90334-9. [DOI] [PubMed] [Google Scholar]
- Le HH, Wrobel CJJ, Cohen SM, Yu J, Park H, Helf MJ, Curtis BJ, Kruempel JC, Rodrigues PR, Hu PJ, et al. Modular metabolite assembly in Caenorhabditis elegans depends on carboxylesterases and formation of lysosome related organelles. eLife. 2020;9:e61886. doi: 10.7554/eLife.61886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shih P-Y, Schaedel ON, Quintero-Cadena P, Rogers AK, Sternberg PW.. FMRFamide-like peptides expand the behavioral repertoire of a densely connected nervous system. Proc Natl Acad Sci USA. 2017;114(50):E10726–E10735. doi: 10.1073/pnas.1710374114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar M, Tenjo-Castano F, Chen S, Zhang YK, Kumari A, Williamson VM, Wang X, Klessig DF, Schroeder FC.. Plant metabolism of nematode pheromones mediates plant-nematode interactions. Nat Commun. 2020;11(1):208. doi: 10.1038/s41467-019-14104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda O, Akagi AE, Artyukhin AB, Judkins JC, Le HH, Mahanti P, Cohen SM, Sternberg PW, Schroeder FC.. Biosynthesis of modular ascarosides in C. elegans. Angew Chem Int Ed Engl. 2017;56(17):4729–4733. doi: 10.1002/anie.201700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg PW, Schroeder FC.. A shortcut to identifying small molecules signals that regulate behavior and development in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106(19):7708–7713. doi: 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts BM, Ciotti MK, Miller NE, Kramer M, Lawrenson AL, Levitte S, Kremer S, Kwan E, Weis AM, Hermann GJ.. glo-3, a novel Caenorhabditis elegans gene, is required for lysosome-related organelle biogenesis. Genetics. 2008;180(2):857–871. doi: 10.1534/genetics.108.093534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder NE, Flatt KM.. In vivo imaging of dauer-specific neuronal remodeling in C. elegans. J Vis Exp. 2014;(91):e51834. doi: 10.3791/51834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J, von Reuss SH, Bose N, Zaslaver A, Mahanti P, Ho MC, O'Doherty OG, Edison AS, Sternberg PW, Schroeder FC.. A modular library of small molecule signals regulates social behaviors in Caenorhabditis elegans. PLoS Biol. 2012;10(1):e1001237. doi: 10.1371/journal.pbio.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1(2):e45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp GS, von Reuss SH, Izrayelit Y, Ajredini R, Schroeder FC, Edison AS.. Chemical detoxification of small molecules by C. elegans. ACS Chem Biol. 2013;8(2):309–313. doi: 10.1021/cb300520u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Deletion Mutant Consortium. Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda). 2012;2(11):1415–1425. doi: 10.1534/g3.112.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Reuss SH. Exploring modular glycolipids involved in nematode chemical communication. Chimia (Aarau). 2018;72(5):297–303. doi: 10.2533/chimia.2018.297. [DOI] [PubMed] [Google Scholar]
- von Reuss SH, Bose N, Srinivasan J, Yim JJ, Judkins JC, Sternberg PW, Schroeder FC.. Comparative metabolomics reveals biogenesis of ascarosides, a modular library of small-molecule signals in C. elegans. J Am Chem Soc. 2012;134(3):1817–1824. doi: 10.1021/ja210202y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Park H, Liu J, Sternberg PW.. An efficient genome editing strategy to generate putative null mutants in Caenorhabditis elegans using CRISPR/Cas9. G3 (Bethesda). 2018;8(11):3607–3616. doi: 10.1534/g3.118.200662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chamberlin HM.. Multiple regulatory changes contribute to the evolution of the Caenorhabditis lin-48 ovo gene. Genes Dev. 2002;16(18):2345–2349. doi: 10.1101/gad.996302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. ; Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wharam B, Weldon L, Viney M.. Pheromone modulates two phenotypically plastic traits – adult reproduction and larval diapause – in the nematode Caenorhabditis elegans. BMC Evol Biol. 2017;17(1):197. doi: 10.1186/s12862-017-1033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at G3 online.