Abstract
TT virus (TTV) was first described in 1997 by representational difference analysis of sera from non-A to non-G posttransfusion hepatitis patients and hence intensively investigated as a possible addition to the list of hepatitis-inducing viruses. The TTV genome is a covalently closed single-stranded DNA of approximately 3.8 kb with a number of characteristics typical of animal circoviruses, especially the chicken anemia virus. TTV is genetically highly heterogeneous, which has led investigators to group isolates into numerous genotypes and subtypes and has limited the sensitivity of many PCR assays used for virus detection. The most remarkable feature of TTV is the extraordinarily high prevalence of chronic viremia in apparently healthy people, up to nearly 100% in some countries. The original hypothesis that it might be an important cause of cryptogenic hepatitis has not been borne out, although the possibility that it may produce liver damage under specific circumstances has not been excluded. The virus has not yet been etiologically linked to any other human disease. Thus, TTV should be considered an orphan virus.
The past few decades have witnessed formidable advances in the characterization of hepatotropic viruses of humans, with the discovery of five major hepatitis viruses (A to E) and the development of sensitive detection methods for each. However, a relatively small proportion of acute and chronic hepatitis cases still remain that cannot be ascribed to these viruses or to viruses (certain enteroviruses, adenoviruses, parvovirus B19, etc.) suspected of occasionally causing liver damage in the absence of overt multisystem clinical manifestations. While attempting to shed light on these forms, Nishizawa et al. in 1997 113 first identified TT virus (TTV) in Japanese patients who exhibited elevated alanine aminotransferase (ALT) levels following transfusions. The patients were negative for the A to E hepatitis viruses as well as for the GB virus C (also called hepatitis G virus due to never fully corroborated early claims that it might cause a form of non-A, non-E hepatitis). When investigated by the representational difference analysis approach developed by Lisitsyn et al. 84, which had already permitted the identification of GB virus C and herpesvirus 8, the sera of three such patients were found to contain a DNA sequence that lacked similarity to any sequence deposited in data banks. The presence of this new sequence, which was rapidly recognized as belonging to a novel viral agent, appeared to correlate with the development of hepatitis 113.
TTV owes its name to the initials of the patient in whom it was first identified. As noted by Nishizawa et al. 113, this acronym might also stand for transfusion-transmitted virus. However, this name would emphasize only one, and certainly not the most frequent, mode of this virus transmission (see below). In the few years elapsed since its discovery, TTV has been intensively studied to assess its molecular properties and whether it causes liver disease. This has led to recognition that the viremia is an extremely frequent occurrence in apparently healthy individuals worldwide, a feature so unusual among viruses that it has even been proposed that TTV might be a commensal virus. To the best of our knowledge, this concept had never been put forward in virology, although it is common wisdom for many other microorganisms (46; P. Simmonds, L. E. Prescott, C. Logue, F. Davidson, A. E. Thomas, and C. A. Ludlam, Letter, J. Infect. Dis. 180:1748–1749, 1999).
Knowledge on TTV is growing fast, but many fundamental aspects remain to be elucidated. This review should therefore be regarded as a stimulus to confront the many intellectual and technical challenges that the unprecedented features of this novel virus pose to the investigator more than as a state-of-the art appraisal that in any case will soon be outdated.
THE VIRUS AND ITS GENOME
TTV has been transmitted to chimpanzees and rhesus monkeys by intravenous inoculation of virus-positive human sera or fecal extracts 87, 106, 117, but there are no published attempts of experimental transmission to smaller animals. In addition, there is only one report dealing with growth in tissue culture 93. Thus, at present we do not know whether TTV will prove as difficult to propagate in the laboratory as many hepatitis-inducing viruses are. As noted by Nishizawa et al. 113, the chloroform-resistant, parvovirus-like agent identified by Bradley et al. 13 in chimpanzees inoculated with material derived from non-A, non-B hepatitis patient(s) prior to HCV characterization might have been TTV, and the same might be true for many viral particles of compatible size seen by electron microscopy in samples from primates inoculated with human materials by other investigators 148, 155, 188. However, to date TTV has not been visualized with certainty by electron microscopy.
The limited information currently available about the physical-chemical properties of TTV derives from studying plasma and, less commonly, fecal extracts from infected individuals. As determined by PCR analysis of infected serum passed through polycarbonate filters of decreasing pore sizes, the viral particle has a diameter of between 30 and 50 nm 106. The isopycnic density was found to be 1.31 to 1.35 g/ml in cesium chloride and 1.26 g/ml in sucrose gradients 106, 119. Because it remained unchanged following Tween 80 treatment, TTV has been deduced not to possess an external lipid envelope 119, a conclusion borne out also by the observation that TTV DNA is detectable in the bile of infected patients 173. As inferred from the prevalence rates of infection in hemophiliacs treated with different preparations of clotting-factor concentrates, virucidal treatments known to inactivate enveloped viruses, such as solvent-detergent and dry heat at 65°C for 96 h, appear to be poorly effective at destroying TTV infectivity. In contrast, immunoaffinity purification and more drastic heat treatments of clotting factors appear to be effective 17, 151, 160, 187. In the absence of more direct data, this information, together with what is known about virion structure, suggests that TTV may be at least as stable as parvoviruses 9.
The component of TTV currently best understood is its genome (Fig. 1 and 2). Early characterization showed that nucleic acids extracted from TTV were sensitive to DNase I and mung bean nuclease but resistant to RNase A and selected restriction enzymes, indicating that the viral genome is made up of single-stranded DNA 119. Initially, over 90% complete sequencing of the prototype Japanese isolate TA278 led to a proposal that the genome was linear 119. Subsequently, however, when sequencing of the viral genome was further extended, it was recognized that the two extremities were connected by a GC-rich stretch of about 100 nucleotides (nt), which in earlier studies had proven hard to amplify and sequence, to form a covalently closed circular molecule (Fig. 1). Hybridization and nuclease protection assays also showed that the virions encapsidate the minus strand 106.
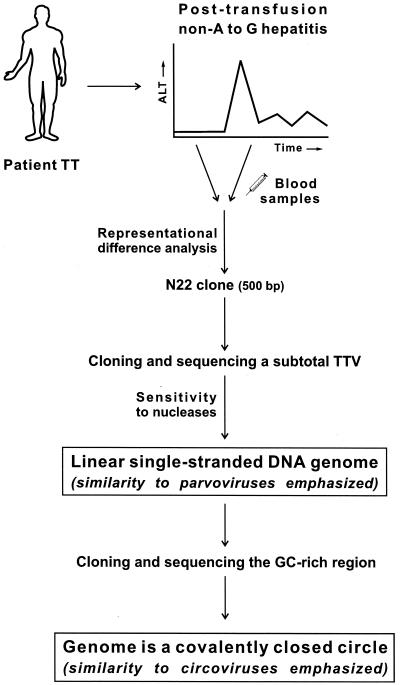
FIG. 1.
Strategy used for TTV discovery and characterization. Representational difference analysis was performed on paired sera from patient TT with posttransfusion non-A to -G hepatitis 113. By cloning and sequencing over 97% of the entire genome and testing its sensitivity to various nucleases (i.e., DNase I, RNase I, mung bean nuclease, and restriction endonuclease NdeI), the TTV genome was deemed to be single-stranded and linear DNA, similar to that of parvoviruses 119. By extending the genome toward its 5′ and 3′ ends, the genome was found to be circular, thus resembling that of circoviruses 101, 106.
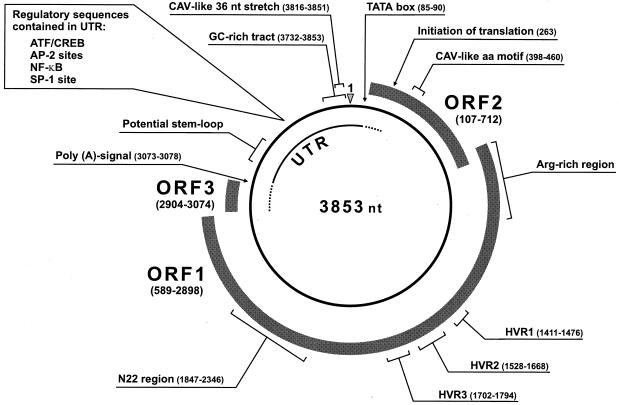
FIG. 2.
Genomic organization of TTV showing the circular, single-stranded DNA of negative polarity encapsidated into the virion. Nucleotide positions refer to the TA278 isolate. Shaded boxes indicate ORF1, ORF2, and ORF3, all present in the plus strand complementary to the virion DNA. ORF3 has been positioned downstream of ORF1, according to the structure in reference 29. ORF2 starts from the first ATG (positioned at nt 107), although, most probably, translation initiates at the second in-frame ATG (located at nt 263), which is conserved among all isolates. The dotted segments of the UTR identify potentially coding portions. The CAV-like amino acid motif and 36-nt stretch are indicated as in references 48 and 91, respectively. HVR are those in reference 114. The N22 region is the first tract of TTV DNA identified 113, which has been extensively used for molecular epidemiology surveys and genotyping. The potential stem-loop structures indicated in UTR are conserved among TTV isolates 53. The GC-rich tract also contains several potential stem-loop structures, which, however, vary considerably in shape among different isolates.
Currently, viruses of vertebrates with small single-stranded DNA genomes and no external lipoprotein envelopes are classified into either the Parvoviridae 9 or the Circoviridae 86 family. As is understandable for a virus of such recent recognition, the taxonomy of TTV is uncertain. Initially, it was noted that TTV had similarities to the parvoviruses 119, but more recently, our improved grasp of its molecular and biophysical structure discussed in detail by Hijikata et al. 53 has led us to emphasize its resemblance to circoviruses. These viruses are characterized by unsegmented circular genomes, isometric capsids that appear to be composed of a single structural protein (VP1) and may present well-defined peripheral protrusions 169, and lack of lipids and carbohydrates. Known animal circoviruses include the beak and feather disease virus of parrots and the porcine circovirus, both of which have a diameter of 15 to 20 nm and a genome of about 2 kb, and the chicken anemia virus (CAV), which stands a little apart because of its slightly larger capsid (23 to 25 nm) and genome (2.3 kb) and other distinguishing features 6, 111. Plant-infecting viruses that used to be classified together with animal circoviruses have recently been moved to a separate genus (Nanovirus). Animal circoviruses, which have very limited sequence homology among themselves and to TTV and no common antigenic determinants, circulate abundantly in their respective host species via a poorly understood means of transmission 31, 32. The genome of CAV, which resembles TTV more closely than the other members of the family, contains three major partially overlapping open reading frames (ORFs), named VP1, VP2, and VP3, which have the coding capacity for 52-, 24-, and 13-kDa proteins, respectively, and other smaller ORFs 63, 70. VP1 and VP2 have been reported to contain protective epitopes 77. Figure 3 shows the phylogenetic relationships of TTV to known animal circoviruses. It should be mentioned, however, that because the virion and genome sizes and other properties of TTV do not conform to those of known circoviruses, Mushahwar et al. 106 have suggested that TTV might represent the founding member of a new family of viruses, for which they tentatively propose the name Circinoviridae. Table 1 contrasts the characteristics of TTV with those of CAV and the human parvovirus B19. Most recently, Takahashi et al. 158 reported the existence of another new virus (designated TTV-like minivirus) with intermediate genomic properties between TTV and CAV and proposed that the three viruses be classified together into a new family, for which they coined the name Paracircoviridae. The relationships of SEN virus, another DNA virus recently detected in human blood 140, to these viruses are currently unknown since the sequences of the SEN virus have not been released.
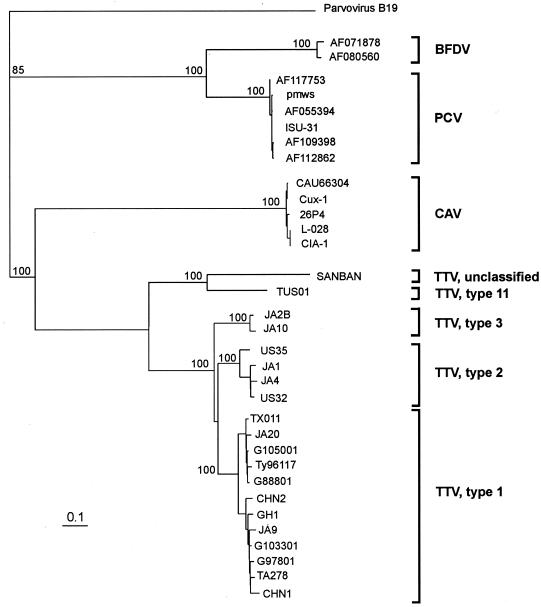
FIG. 3.
Phylogenetic relationships of TTV with the animal circoviruses CAV, porcine circovirus (PCV), and psittacine beak and feather disease virus (BFDV). The phylogenetic tree was constructed by comparing the entire deduced products of the ORFs that contain Rep protein motifs, namely, ORF1 (TTV), VP1 (CAV), ORF1 (PCV), V1 (BFDV), and VP1 (parvovirus B19), using the last as outgroup. TTV isolates are grouped based on N22 segment analysis 106, 120. The Fitch-Margoliash algorithm was used by randomizing the input order of sequences. Genetic distances were calculated by using the Poisson correction distance (PoissonP) as provided by DAMBE software (version 3.7.48) 182. Bootstrap values above 70 out of 100 are shown at the main branch points. The tree was drawn by using the Treeview program (version 1.5.2) 129. The bar indicates the number of amino acid substitutions per site. All sequences are available in the GenBank database and are indicated by their accession number or, whenever possible, isolate name.
TABLE 1.
TTV, CAV, and human parvovirus B19 at a glancea
| Property | B19 | CAV | TTV |
|---|---|---|---|
| Virion structure | |||
| Shape | Unenveloped, isometric nucleocapsid; surface projections may be present | Unenveloped, isometric nucleocapsid; surface projections may be present | Parvovirus- and circovirus-like? |
| Size (nm) | 18–22b | 23–25c | 30–50 |
| Symmetry | Icosahedral | Icosahedral | Icosahedral |
| Physicochemical properties | |||
| CsCl buoyant density (g/ml) | 1.38–1.51b | 1.33–1.34c | 1.31–1.35 |
| Stability | Resistant to solvents, dry heat, and low pH | Resistant to solvents, dry heat, and low pH | Resistant to solvents, dry heat, and presumably low pH |
| Isoelectric point of the largest encoded proteind | 6.17 (VP1) | 11.02 (VP1) | 10.52 (ORF1) |
| Genome | |||
| Type | Single-stranded DNA | Single-stranded DNA | Single-stranded DNA |
| Size | ∼5.0 | ∼2.3de | ∼3.8 |
| Form | Linear, with inverted terminal repeats at both endsf | Covalently closed circle | Covalently closed circle |
| Polarity | Negative and positiveg | Negative | Negative |
| ORFs | ORF-S and ORF-L | VP1, VP2, VP3, and others smaller | ORF1, ORF2, and others smaller |
| Variability | Low | Low | High |
Genome Organization and Predicted Products
At the end of 1999, gene depositories contained the entire nucleotide sequences of 10 TTV isolates, which varied in length between 3,808 nt (SANBAN isolate) and 3,853 nt (isolates TA278 and JA20). Judging from the presence of well-conserved ORFs among the sequenced isolates, the genome of TTV is divided into a potentially coding region of ∼2.6 kb and an untranslated region (UTR) of ∼1.2 kb (Fig. 2). The former consists of two major potential protein-coding genes (ORF1 and ORF2) which are present in the plus strand complementary to the genomic DNA, are in different reading frames, and partially overlap. In the prototype TA278 sequence, ORF1 spans nt 589 to 2898 and ORF2 spans nt 107 to 712, which correspond to a coding capacity of 770 and 150 amino acids (aa), respectively. However, the lengths of the two ORFs may vary somewhat in individual isolates. Analysis of the sequences of 11 isolates 29 revealed the presence of an additional small ORF (designated ORF3) with coding capacity for 57 aa, located immediately downstream of ORF1 (nt 2904 to 3074 in the TA278 isolate) and well conserved in most isolates (70 to 100% identity). Other putative ORFs have been recognized in some isolates, but they might be nonfunctional, as suggested by their absence in most other isolates and additional molecular considerations 29, 101.
If the genomic organization of TTV is indeed analogous to that of CAV, ORF1 might encode the equivalent of the VP1 protein of CAV. The predicted translation product has a large proportion (approximately 40%) of arginine residues in the amino-terminal first 100 aa 119, thus resembling the N terminus of the capsid protein of CAV. Of note, similar hydrophobic domains are found in the core proteins of other viruses as well and are believed to mediate genome binding to the capsid and transportation to the cell nucleus 37, 149, 179, 185. The ORF1 product also contains a few potential asparagine-linked glycosylation sites that vary both in number and in location in individual isolates 53, 159. As noted by Hijikata et al. 53, such differences in glycosylation pattern might affect several biological properties of the resulting protein, including antigenic specificity. However, until the encoded products are characterized directly, it will be difficult to determine with certainty whether glycosylation actually occurs and what its functions are, if any. Also of interest is the observation that the ORF1 products of both TTV and CAV contain short amino acid motifs characteristic for the replication-associated proteins (Rep) that are common denominators of DNAs replicating by the rolling-circle mechanism 47, 78, 98. The other circoviruses also possess such motifs 94, 98.
ORF2 is believed to code for a nonstructural protein involved in viral replication. Due to the presence of two potential in-frame start codons and the length diversity among different isolates, the precise size of the protein encoded has been a matter of some uncertainty 119, but it is now generally accepted that translation starts at the second initiation triplet (Fig. 2), which would lead to a protein product of around 150 aa and whose size varies slightly in different isolates 53, 106, 120. Interestingly, as noted by Hijikata et al. 53, in spite of high heterogeneity among TTV isolates (see below), the segment encompassed between aa 46 and 66 of the ORF2 protein contains five positions conserved among all the isolates examined and also in CAV, thus further corroborating the relatedness of these two viruses.
The noncoding UTR of TTV genome contains the already mentioned G+C-rich (∼90%) segment as well as various regulatory sequences (Fig. 2). This segment also possesses multiple inverted repeats and hence has the potential to form several stem-loop structures 120. Another candidate stem-loop has been identified in the UTR just downstream of the polyadenylation site (Fig. 2). This was found to be conserved in all the sequences (including in the highly divergent SANBAN isolate) aligned by Hijikata et al. 53 with the exception of only one isolate (KC002-3′). The latter, however, presented “covariant mutations” that could maintain the base pairing necessary to form the stem-loop 53. Similar secondary structures are found in CAV and other circoviruses, albeit located in other regions 6. These structures might play a pivotal role in viral replication 120, but since they vary significantly in shape among different TTV isolates, their function in guiding viral replication awaits further verification 53, 101, 106, 120. Another similarity between the UTRs of TTV and CAV is the presence of a 36-nt stretch which has over 80% identity 101.
Replication
The mechanisms of TTV replication are currently unknown. A precise understanding of the replication strategies of related circoviruses might be helpful, but even these have not been studied in great detail. What is known about the replication strategies of animal circoviruses can be readily summarized. These viruses can be propagated in lymphoid cell lines and other cell types, where they replicate in the nucleus and appear to be dependent on cellular proteins expressed during the S phase of the cell cycle 31, 32. Transcription analysis of CAV has led to the identification of a single RNA transcript of 2.0 kb which might function as a polycistronic mRNA 131. In contrast, for the porcine circovirus, it has been demonstrated that two mRNAs are encoded by the minus strand and one mRNA, corresponding to the spliced transcript of ORF1, is encoded by the plus strand 95. This indicates that the transcriptional strategies of different circoviruses can vary considerably. The mechanism of DNA replication is not known with certainty, but there are molecular indications that circoviruses use a rolling-circle mechanism 94, 98, while the absence of motifs typical of known DNA polymerases in the encoded products suggests that the genome is replicated by a cellular enzyme. The latter circumstance is also suggested by the considerable degree of genetic conservation of circoviruses among isolates obtained worldwide (Fig. 3). Virus particles are assembled in the cytoplasm, where they can contribute to form clearly evident inclusions, and are presumably released by cell lysis. Interestingly, the VP3 protein of CAV (also known as apoptin) induces a p53-independent type of apoptosis that, being accelerated by the proto-oncogene Bcl-2 and being especially active in transformed cells, has been proposed as a potential antitumor agent 24, 25.
Regarding TTV, the existence of a conserved Rep protein motif in the genome might suggest that this virus also replicates via a rolling-circle mechanism in spite of the absence of the nonanucleotide that is conserved in many viruses that replicate in this manner 29. As discussed by Gilbert and Dressler 43, this mechanism is particularly well suited for replicating single-stranded DNAs. It should be noted, however, that the great genetic diversity of TTV (see below) has also led investigators to hypothesize that the viral DNA is replicated by a machinery with poor or no proofreading activity. That TTV might replicate through an RNA intermediate like the hepatitis B virus seems unlikely because no reverse transcriptase motif has been identified in its genome 114.
Genetic Heterogeneity
In contrast to most DNA viruses, TTV isolates exhibit a high level of genetic heterogeneity (Fig. 3). Comparison of the sequences available has revealed that the divergence is unevenly distributed throughout the genome. The UTR is comparatively well conserved (73% nucleotide identity between the two highly divergent isolates TA278 and SANBAN versus 57% for the entire genome) and contains several segments with greater than 90% identity, indicating that much variation is not tolerated in these regions 29, 53. In contrast, the translated portion presents an extraordinarily high degree of diversity. For example, the ORF1 of isolate SANBAN codifies for a 25-aa-shorter protein with only 32% amino acid identity relative to the prototype, TA278. The divergence is especially high in the central tract of ORF1, at least for subtype 1a (see below), where three hypervariable regions (HVRs) (HVR1, HVR2, and HVR3, of 22, 47, and 31 aa, respectively [Fig. 2]) have been defined that may contain multiple codon insertions or deletions and diverge up to 70% at the amino acid level 29, 53, 114, 159. Variability is lower in the arginine-rich amino-terminal portion and a few other short stretches, which might be indicative of constraints dictated by protein form and function.
More extensive investigations of the genetic diversity and phylogenetic relationships of TTV isolates have been carried out by sequencing amplicons obtained from the N22 segment (Fig. 1). A segment of approximately 220 nt in this region has been suggested as suitable for genotyping 119–121, although others have found that fragments longer than 500 nt appear to be the most useful for this purpose 29. Nomenclature and criteria used for grouping TTV isolates parallel those used for HCV classification, but, because of their higher heterogeneity, genotypes (designated by Arabic numbers) are separated by sequence differences of >30% or evolutionary distances (number of nucleotide substitutions per site) of >0.30 within the N22 segment, while subtypes (identified by lowercase letters) are separated by evolutionary distances of >0.15 119, 120. With the growing numbers of isolates characterized, appreciation of the genetic diversity of TTV has rapidly increased. So far, phylogenetic trees constructed on the N22 region have led investigators to subdivide TTV into up to 16 genotypes 120, 123 and several subtypes or to refrain from designating new types 56.
It is likely that the full extent of TTV diversity has yet to emerge for the following reasons: (i) the PCR protocols used in most studies published are suboptimal at demonstrating the entire variety of circulating TTV strains, and presumably with the improvement of methods the range of genetic divergence will become wider; and (ii) very recently, nonhuman primates were shown to harbor TTV strains indistinguishable from those of humans, as well as at least four TTV genotypes distinct from those detected in humans so far 2, 117, 143.
Related viruses such as CAV, porcine circovirus, and human parvovirus B19 are much less variable than TTV 30, 51, 106. Thus, the reasons for the vast genetic diversity of TTV are puzzling. According to Hijikata et al. 53, the only possible explanation is that TTV has existed for eons and drifted with time. However, other factors may have facilitated the expansion of TTV genetic diversity: (i) several domestic animal species harbor TTV or TTV-like viruses, most of which could not be distinguished from human TTV isolates in the genomic regions analyzed 79, 176, making cross-species transmission a likely possibility: the need to adapt to new hosts would represent an important drive to genetic change; (ii) as also discussed below, mixed infections by multiple genotypes are frequent 5, 10, 106, 143, 161, 165, 180, and hence, genetic recombination of human TTV between themselves and with related viruses of animals might also be an important multiplier of genetic diversity, as occurs in other viruses with circular single-stranded DNA genomes 42, 128: indeed, very recent analysis of a collection of full-length genomes showed that nearly half were of mosaic viruses 181; and (iii) the great ability of TTV to produce chronic productive infections characterized by conspicuous plasma viremia lasting many years (see below) may offer the host immune system the opportunity to exert a continuous pressure for virus evolution. As noted by Prescott et al. 138, other poorly pathogenic DNA viruses which produce long-lasting infections and have presumably coexisted with primates for millions of years (e.g., the papillomaviruses) also exhibit a vast diversity of genetic variants.
Information about the rate of TTV change in infected hosts is limited and confounded by the fact that patients frequently carry multiple viral strains. In a recent retrospective study of sequential serum samples, Nishizawa et al. 114 compared cloned PCR-amplified ORF1 sequences obtained from two patients over periods of 3 and 8 years and found that at the start of the study TTV circulated as a quasispecies of related sequences markedly divergent within the HVRs and that the quasispecies composition changed appreciably with time of infection, due to continued variation in such regions. In contrast, analysis of three subjects with acute, resolving TTV infections revealed a striking homogeneity of the HVR sequences. These findings were interpreted as suggesting that TTV HVRs are under strong pressure for change during persistence, presumably to escape immune surveillance, and might thus be functionally similar to the HVRs found in the envelopes of human immunodeficiency virus type 1 (HIV)-1 and hepatitis C virus (HCV). Another published report on intrahost TTV evolution is in disagreement with these findings. Ball et al. 5 studied the variability of TTV in three chronically infected individuals over periods up to 6 years. The extent of TTV genetic variation over the period examined was found to be quite modest, except in an individual who carried at least seven distinct TTV strains belonging to four different genotypes. Evidence for intrapatient TTV evolution over the time of persistence has also been detected by Irving et al. 59 but not by Biagini et al. 10, Gallian et al. 38, or Khudyakov et al. 74.
METHODS FOR DEMONSTRATING TTV INFECTION
The laboratory diagnosis of TTV infection is still rather primitive for many reasons: (i) no tissue culture system of sufficient sensitivity has been described that might be used for TTV isolation; (ii) the immune responses elicited by the virus are poorly understood, and, in particular, there are no easy-to-use serological methods, nor it is known whether antiviral antibodies may be of practical utility in the clinical virology laboratory; (iii) although viremia can be quite substantial, no methods have been reported that detect viral antigen in plasma; (iv) none of the many PCR formats assays used for demonstrating viral DNA have been validated for their ability to detect the entire spectrum of TTV variants; and (v) uncertainties about the clinical relevance of TTV infection have discouraged commercial companies from developing and marketing adequate reagents and diagnostic kits. While many of these aspects will be addressed below, here we discuss the molecular assays that have been used for TTV detection and characterization. Understanding their performances is crucial for a correct evaluation of many issues examined in the following sections.
Due to the great genetic variability of TTV, the choice of the viral DNA segment targeted for amplification has an enormous impact on PCR assay sensitivity 27, 62, 79, 157, possibly more so than for most viruses. For historical reasons, the N22 tract of ORF1 has been extensively targeted in nested or heminested PCR protocols. The primers designed on this region have been considerably improved over time, but nonetheless, they still may fail to amplify TTV isolates other than types 1 to 6, especially if the virus content in test samples is low (62, 73, 113, 119, 120; Simmonds et al., Letter). This limit has been solved only partly by using multiple sets of primers 26, 27, 29. Due to its higher conservation, the UTR of the TTV genome is much more suitable for primer design (Table 2). Indeed, generally UTR PCR has proven satisfactory at detecting most if not all the 16 TTV genotypes currently recognized (62, 79, 80; Simmonds et al., Letter) and, compared to ORF1 PCR, has greatly increased the rates of plasma or serum samples that gave a positive reaction to TTV DNA: from 23 to 92% in studies by Takahashi et al. 157, from 9 to 50% in studies by Irving et al. 59, and from 20 to 95% in studies by Itoh et al. 62. It has however been pointed out that even UTR-PCR assays can be biased toward the detection of type 1 isolates, unless the primers are carefully designed (29, 79, 123; A. S. Muerhoff, T. P. Leary, S. M. Desai, and I. K. Mushahwar, Letter, J. Infect. Dis. 180:1750, 1999). The lower limits of detection of the qualitative PCR assays discussed so far have generally been reported to be on the order of 102 to 103 genomes per ml of plasma, but such values should be regarded as valid only for the specific TTV genotype used for assay evaluation 5, 123, 173.
TABLE 2.
Molecular epidemiology of TTV viremia in the general population of different countries
| Country | % Positive by:
|
Genotypes identified | Reference(s) | |
|---|---|---|---|---|
| ORF1 PCR | UTR PCR | |||
| Africa | ||||
| Congo | 43–44 | 1, 2, ONTa | 26, 138 | |
| Egypt | 29 | 85 | 1, 2 | 1, 36 |
| Gambia | 86 | 1, 2, ONT | 138 | |
| Kenya | Unknown | 1–3, 8–10 | 121 | |
| Nigeria | 52 | 1, 2, 3, ONT | 138 | |
| Sudan | 7 | 2 | 138 | |
| Asia | ||||
| China | 5–11 | 1, 2 | 18 | |
| Japan | 10–58 | 70–93 | 1, 2, 3, 4–8, ONT | 119, 120, 157, 174 |
| Korea | 14 | 1, 2, 4 | 109 | |
| Mongolia | 43 | 1, 2, 4 | 71 | |
| Myanmar | 96 | 1, 2 | 1 | |
| Nepal | 82 | 1, 2, 4 | 1 | |
| Pakistan | 16 | 1, 2, 3 | 138 | |
| Saudi Arabia | 19 | 100 | 1 | 138 |
| Singapore | 98 | Simmonds et al., Letter | ||
| Taiwan | 10–53 | 55, 69 | ||
| Thailand | 7–36 | 1, 2, 3 | 135, 165 | |
| Europe | ||||
| Finland | 17 | 73 | Simmonds et al., Letter | |
| France | 5 | 1, 2, ONT | 11 | |
| Germany | 7–14 | 1, 2, 3, 4, ONT | 8, 146 | |
| Italy | 9–50 | 1, 2, 3, 4 | 90, 156 | |
| Spain | 14 | 1, 2, ONT | 44 | |
| The Netherlands | 72 | Simmonds et al., Letter | ||
| United Kingdom | 2–10 | 47–57 | 1, 2, 3, ONT | 138; Simmonds et al., Letter |
| North America | ||||
| United States | 1–11 | 1, 2, 11 | 15, 27 | |
| Oceania | ||||
| Papua New Guinea | 75 | 1, 3, ONT | 138 | |
| South America | ||||
| Bolivia | 82 | 1 | 1 | |
| Brazil | 20–62 | 1, 2, 3, ONT | 112, 138 | |
| Colombia | 16 | 1, 2, 4 | 164 | |
| Ecuador | 71 | 1, 2, ONT | 1 | |
ONT, others not typed.
As discussed at length below, it is likely that attempts to correlate TTV to specific diseases will depend on precise genetic characterization and quantification of the virus found in the patients under scrutiny. The TTV genotyping methods used to date include sequencing approximately 220 nt of the N22 segment of ORF1 gene, which reportedly permitted a satisfactory grouping of many genotypes 120; restriction fragment length polymorphism analysis within the same region, which has proven capable of characterizing types 1 to 6 with fair precision 90, 165; and genotype-specific PCR tests, which have been used to identify selected subtypes 52, 156.
Assays used for measuring the viral content of plasma and other clinical samples include semiquantitative end-point dilution PCR 5, 59, 90, a competitive PCR assay 16, and various real-time PCR protocols 58, 72, 103, 134. However, because molecular methods for measuring viral loads can perform very differently depending on the (sub)type of the virus being tested 82, 100, quantitative PCR methods will have to be designed and evaluated with even greater care than qualitative methods. In general, viral loads determined using UTR-based assays have provided viremia titers 10- to 100-fold higher than those found in ORF1-based assays 123, 157, and this might reflect the higher sensitivity of the former assays as well as the presence in the test samples of TTV (sub)types poorly amplified by the latter assays 123. In a recent study, Kato et al. 72 reported mean values of 3.45 ± 0.67 log copies per ml in Japanese blood donors and similar values in HCV-infected patients. The concomitant presence of multiple TTV (sub)types, a frequent occurrence in certain groups of patients 5, 106, 161, is yet another compounding factor which can significantly affect the reliability of quantitative (and genotyping) assays.
An additional issue that warrants investigation is the importance of examining clinical specimens other than blood in the diagnosis of TTV infection. TTV DNA has been detected in feces, saliva, nose swabs, throat swabs, and breast milk of viremic subjects and sometimes also in persons with no detectable viremia (60, 116, 144; S. Yokozaki, Y. Fukuda, I. Nakano, Y. Katano, M. Okamura, and M. Naruse, Letter, Blood 93:3569–3570, 1999).
EPIDEMIOLOGY
Presently, the only well-documented aspect of TTV epidemiology is the extremely high diffusion of active infection throughout the world. In spite of their relatively low sensitivity, early PCR assays detected TTV in the plasma of approximately one-third of the general population, although with considerable variations in different countries and in different investigations within the same country. In particular, prevalence rates tended to be moderate in the United States and northern Europe, intermediate in Asia, and high in Africa and South America (Table 2). Similarly, Matsumoto et al. 96 found that the prevalence rates for TTV were higher among Asians and African Americans than among Caucasians in a survey of sera from the area of Greater Washington and Chesapeake. Geographically segregated populations having limited contact with the rest of the world also exhibited high viremia rates 85, 139, 164. Early studies also demonstrated that infection was very frequent in the early months of life (26, 45, 55, 58, 115; M. Yamada-Osaki, R. Sumazaki, E. Noguchi, M. Shibasaki, and A. Matsui, Letter, Lancet 352:1309–1310, 1998), although in some surveys the viremia rates tended to increase with age and peaked in young adulthood or later in life (17, 26, 55, 90, 145, 174; Simmonds et al., Letter).
As suspected in several such studies, more recent epidemiological surveys employing PCR assays of improved sensitivity, based on either the UTR or the most highly conserved tracts of the coding portion of the viral genome, have shown that the prevalence rates found in early studies were a gross underestimate of TTV diffusion. In most countries surveyed, virus carriers are now known to range around 80%, with peaks in countries such as Japan, Myanmar, Saudi Arabia, and Singapore, where almost everyone was viremia positive (Table 2). These studies also suggest that geographical and age-related gradients of prevalence are much less pronounced than previously estimated 62, 123, 157.
Although it is assumed that TTV must be highly contagious, the modes of spread are poorly understood. Most screenings have found the highest detection rates amongst polytransfused, thalassemic, long-term hemodialysis patients, hemophiliacs treated with the nonvirally inactivated clotting-factor concentrates used prior to the HIV epidemics, and intravenous drug abusers; a positive correlation often existed between the quantity of blood or blood factors received and the TTV prevalence rate 12, 17, 38, 67, 76, 96, 115, 137, 160, 175, clearly indicating that TTV can behave as a transmissible blood-borne virus. Additional observations corroborating this conclusion were as follows: (i) TTV DNA was detected in commercial human plasma, clotting-factor concentrates, and intramuscular immunoglobulin (Ig) preparations (133, 151, 187; V. Gerolami, P. Halfon, H. Chambost, I. Thuret, and G. Halimi, Letter, Lancet 352:1309, 1998; R. Sumazaki, M. Yamada-Osaki, Y. Kajiwara, A. Shirahata, and A. Matsui, Letter, Lancet 352:1308–1309, 1998); (ii) in certain studies hepatitis B and C patients were coinfected with TTV more frequently than control groups were 26, 27, 38, 59, 73, 96, 162, 174; and (iii) individuals at risk for parenteral transmission often exhibited mixed infections by multiple TTV (sub)types 33, 106, 161.
On the other hand, from the very beginning it was also patent that TTV viremia rates in the general population were much too high to be explained solely in terms of apparent or inapparent blood-borne transmission. Feces of viremic individuals contain TTV 88, 116, 143, 144, 173, which is infectious in tissue culture 93 and nonhuman primates 87, suggesting the oral-fecal route as the most common mode of transmission. Undoubtedly, considering the high prevalence of virus carriers and the putative high resistance of TTV infectivity to physicochemical agents, even if TTV were shed in the stools only intermittently and/or at low titers, as existing data seem to indicate 116, 144, the extent of TTV contamination in the human environment is likely to be remarkably high. At present, however, there is no other evidence that the main means of TTV spread is enteric, except for a recent report showing that Brazilian women having anti-hepatitis A antibodies were TTV infected more frequently than those lacking such antibodies 145. Attempts should be made to monitor the levels of TTV contamination in drinking water and in food under different sanitation conditions and to evaluate whether TTV might be useful as a marker of contamination with viruses of fecal origin, similar to the use of Escherichia coli as a marker for enteric bacteria.
Mother-to-fetus transmission should be considered for a ubiquitous virus which is acquired early in life. Initial studies failed to detect TTV DNA in cord blood samples and in neonates born to infected mothers 55, 125, 186, favoring the idea that child infection was due to early postnatal exposure (26; Yokozaki et al., Letter). However, failure to find TTV in neonatal samples in these studies may have been due to pitfalls in the detection tests used, since it has subsequently been reported repeatedly that prepartum transmission of TTV not only occurs but may actually be very common 103, 145, 153. Viral sequences similar to those found in the corresponding maternal blood were detected in a number of newborns 26, and in a recent study, TTV DNA was detected in over half the cord blood samples examined 103. Furthermore, some cord blood samples had a TTV DNA content at least as high as that in the respective mother's blood, leaning in favor of transplacental transmission well before delivery 103.
Alternative modes of TTV spread have been only sparsely investigated. McDonald et al. 89 and Poovorawan et al. 135 reported that, in contrast to hepatitis B and C, the frequency of TTV infection in individuals at high risk for sexually transmitted diseases was not particularly high. Together with other findings 26, 55, 105, this suggests that sexual transmission plays an unimportant role, if any. Attempts to obtain evidence for household contact transmission have been inconclusive 68, 69, 153, although the detection of TTV in saliva, nasopharyngeal secretions 39, 60, 144, and skin and hair 127 might imply that it is also possible. Evidence suggestive of non-transfusion-associated nosocomial TTV infections has also been reported 96. Whether the TTV or TTV-like viruses recently reported to be widespread in large percentages (19 to 30%) of chickens, pigs, cows, and sheep 79 contribute to TTV epidemiology in humans is also unclear. More detailed analysis of these viruses will be needed to establish whether they might add to the enormous reservoir of TTV genomes existing in nature and contribute to human TTV evolution and diversification.
Several studies have also investigated the geographical distributions of TTV genotypes. Types 1 and 2 are very common worldwide, even if their prevalence rates vary substantially in different areas, whereas other types seem to be less frequent or restricted to specific geographical regions (Table 2). It is uncertain whether these data reflect real differences in genotype distribution or are instead biased by preferential amplification of selected genotypes by the PCR assays used. Whether the (sub)type frequency varies with the age of infected subjects and/or with the route of transmission, similarly to what was observed for HCV (reviewed in reference 7), also remains to be determined. Recently, Japanese hemophiliacs were found to harbor type 2 much more frequently than nonhemophiliacs were, suggesting the predominance of this type in the imported clotting factors they use 121, 187.
In conclusion, more extensive investigations with more standardized diagnostic tools are needed to obtain a coherent understanding of TTV epidemiology. It seems, however, that TTV spreads both vertically and horizontally. This factor and its outstanding ability to persist (see below) presumably represent the attributes that have permitted the extraordinary penetration of active TTV infection into human communities worldwide. It is noteworthy that the other human viruses that produce chronic infection with comparable prevalence rates (Epstein-Barr virus, cytomegalovirus, papillomaviruses, etc.) are mostly cell associated and circulate freely in plasma only briefly, if ever.
INTERACTIONS WITH INFECTED HOSTS
Evidence about the natural history of TTV infection is just beginning to accumulate. A well-recognized feature is that infection is usually chronic and characterized by the continued presence of abundant virus in the bloodstream. Longitudinal investigations by Lefrère et al. 81, Oguchi et al. 115, and other groups have clearly shown that TTV carriage tends to last many years; a patient was identified who had carried the virus for at least 22 years 96, thus suggesting lifelong persistence as a clear possibility. In addition, sequential blood samples obtained over prolonged periods were found to contain viral sequences ascribable to the TTV strain present at first sampling, although cases in which the initial strain appeared to be replaced by a different one were also observed 59. This is clear evidence that long-term TTV viremia usually results from genuine virus persistence and not from repeated reexposure to the virus.
Whether self-limited infections also exist is less well defined. Descriptions of viremias that seemingly spontaneously resolved after periods of a few weeks to several years are common in the literature 76, 81, 96, 115, 137, 160, 190, and experimentally infected chimpanzees also appeared to clear the virus after months of virus carriage 106. However, the possibility cannot be excluded that such findings reflect reduced viremia, resulting from decreased virus shedding into blood, increased virus clearance, or even viral DNA sequence modifications that render the virus undetectable by the detection tests used, rather than complete virus eradication. The possibility that TTV can become temporarily or permanently latent should also be considered 190. As pointed out by Okamoto et al. 118, it also remains to be determined whether TTV can behave as an episome or integrate into the host cell DNA, like other single-stranded DNA viruses 83. The only studies in this direction were carried out with patients with hepatocellular carcinomas and hematopoietic malignancies and provided no evidence that TTV was integrated 166, 184.
The sites and cell types where TTV undergoes primary amplification following entry into the body, as well as the definitive target tissues and organs from which fresh virus is continuously shed into the circulation, are unknown. Since related single-stranded DNA viruses require actively multiplying cells for productive replication, it seems likely that most, if not all, TTV is produced by cycling cells. So far, TTV DNA has been consistently detected in liver tissue and bile specimens at concentrations that frequently were 10 to 100 times higher than those in the corresponding plasma samples and in fecal extracts at lower concentrations 108, 116, 144, 173. Furthermore, in the study by Ukita et al. 173, complete concordance was observed between the TTV DNA sequences present in paired serum and bile samples and, in one patient whose stools were also virus positive, in fecal samples. Collectively, these findings strongly support the concept that the liver is a major site of TTV replication. This concept is supported by a recent report demonstrating circular double-stranded replicative forms of TTV DNA in the liver 124 and by hybridization findings showing TTV DNA within hepatocytes 19, 142.
Peripheral blood mononuclear cells (PBMC) but not red blood cells freshly harvested from infected individuals also harbor TTV DNA 93, 118, 125, 136, suggesting that the virus might replicate in lymphoid cells as well as in hepatocytes. Interestingly, the genotypes of plasma- and PBMC-associated virus sometimes differed in one study 118, suggesting a compartmentalization of TTV infection similar to what was observed for HIV-1 and HCV 91. However, whether the PBMC-associated virus resulted from active replication within these cells, from adsorbed and/or endocytized but nonreplicating virus, or from residual contaminating plasma was not established with certainty. Recently, Maggi et al. 93 found that the contents of TTV DNA in freshly harvested PBMC and purified B and T lymphocytes, monocytes, and polymorphonuclear leukocytes were uniformly low and tentatively ascribed the finding to the resting state of such cells. Indeed, cultures of phytohemagglutinin-stimulated TTV-negative PBMC could be productively infected and released substantial amounts of infectious TTV into the supernatant. That proliferating hematopoietic cells may indeed represent an important source of circulating TTV is suggested by the finding that baseline TTV viremia decreased to undetectable levels in patients given myelosuppressive treatments (cyclophosphamide and total body irradiation) for bone marrow transplantation 68 and other evidence 75, 122. Future studies on the permissiveness for TTV of different cell types in vivo and in vitro would greatly benefit from the development of suitable animal models of infection and of sensitive and reliable techniques for demonstrating positive-sense DNA and other intermediates of TTV replication.
Wherever it replicates, TTV is shed into the bloodstream in relatively large quantities. Following transfusion of uninfected patients with contaminated blood, TTV became detectable in plasma in one to few weeks 76, 96. Studies have indicated that the TTV content in plasma is low compared to the content of other viruses that produce protracted plasma viremias, such as HIV-1 and HCV 5, 26, 59, 119; however more recent analyses by Pistello et al. 134, using a sensitive real-time PCR assay, demonstrated that plasma viremia loads vary widely, ranging between 103 and over 108 genomes per ml in different persons. Studies have also shown that in some individuals viremia levels fluctuated extensively while in others they remained essentially stable 5, 96. A clear understanding of the dynamics of plasma viremia will, however, need more detailed longitudinal studies of acutely and chronically infected individuals.
Elucidating the mechanisms by which TTV persists so efficiently in infected hosts represents an interesting challenge. As in most other persistent infections 3, a combination of virus and host factors is likely to be involved. For example, as observed for HBV and other viruses, the fact that children are exposed to TTV early in prenatal or postnatal life might greatly facilitate the establishment of persistence. Also, given the likely requirements of TTV replication discussed above, it is feasible that maintenance of persistence is made easy by the fact that only a fraction of potentially susceptible cells are fully permissive at any given time, as often seen in carrier culture models of persistence 97. It may also be that anti-TTV immune responses are not particularly vigorous or that the immune effectors produced are poorly efficient at controlling TTV infection.
While nothing is known about anti-TTV cell-mediated immune responses, what is currently known about humoral responses does not speak in favor of a powerful antiviral activity of anti-TTV antibody. Studies of anti-TTV immunity have been hampered by the lack of suitable viral antigens. The only published attempt to develop recombinant TTV antigens, a technology that has been extremely fruitful for many viruses, has yielded negative results. Recombinant 15-kDa proteins from the N22 segment of two TTV isolates proved completely unreactive in Western blotting against human sera, regardless of whether they were TTV DNA positive or negative 85. Tsuda et al. 171 used whole TTV from fecal extracts as the antigen and PCR to demonstrate anti-Ig-facilitated immunoprecipitation. Of 44 blood donors tested (38 TTV DNA negative and 6 positive), 12 were anti-TTV antibody positive, only 1 of whom was also viremia positive. These investigators also examined sequential serum samples from two patients with putative TTV-related posttransfusion hepatitis and observed the appearance of TTV-IgG immune complexes in the circulation followed by loss of detectable TTV from plasma. In one patient who could be monitored for long enough, anti-TTV antibodies persisted for at least 4 years. In partial agreement with the above findings, Nishizawa et al. 114 found that in chronically infected patients, but not in recently infected ones, a large proportion of circulating TTV was complexed with IgG, and Pisani et al. 133 found that commercial human Ig contained detectable TTV DNA if it had been prepared for intramuscular use but not if it had been freed of complement-activating aggregates for intravenous usage. Freer et al. (G. Freer, P. Rovero, and F. Maggi, unpublished data) found low levels of anti-TTV reactivity in sera from PCR-positive and -negative individuals by using an enzyme-linked immunosorbent assay exploiting two synthetic peptides deduced from well-conserved regions of ORF1. Interestingly, sera with high TTV loads were somewhat more reactive than those with low TTV loads, possibly suggesting that the peptides were representative of viral epitopes not exposed on the virion surface. Prior to the discovery of TTV, Tischer et al. 168 had detected antibodies that reacted with the porcine circovirus in over 20% of the human sera tested. As discussed by Okamoto et al. 121, seen in retrospect, these data might be indicative of the existence of cross-reactive antigens between porcine circovirus and TTV.
Taken together, the above findings show that TTV-infected hosts mount detectable antiviral antibodies which fail to eradicate the virus, at least in the great majority of cases. On the other hand, the existence of mixed infections by multiple TTV strains suggests that antiviral immunity is also unsuccessful at protecting against superinfections sustained by heterologous TTV (sub)types. As noted by Viazov et al. 177, the vast sequence diversity of the coding genomic regions has the potential to markedly affect TTV antigenic specificity. It remains to be determined whether antibodies are capable of modulating viremia levels and exerting a significant selective pressure for change on external TTV proteins, as postulated 114, 121.
CLINICAL SIGNIFICANCE
The temporal concomitance between viremia and ALT elevations observed in three of the five blood-transfused patients from whom TTV was originally demonstrated and other evidence led Nishizawa et al. 113 to propose TTV as a possible cause of some forms of acute and chronic hepatitis and fulminant liver failure that still remain etiologically unresolved. This was certainly instrumental in putting the new virus in the limelight, but subsequent findings have cast serious doubts on the correlation: (i) as extensively discussed above, TTV viremia is widely prevalent not only among patients with cryptogenic hepatitis but also, and at similar rates, in control groups with other forms of liver disease or no liver injury at all 8, 23, 35, 50, 66, 109, 170, 178; (ii) when hemophiliacs and other persons with or without risk of blood-borne infections were sorted into TTV DNA-positive and -negative groups, the two groups often showed similar ALT levels 8, 55, 85, 96, 109, 115, 130, 157, 160, 175; (iii) in several series of blood transfusion patients, there was no correlation between the acquisition of TTV infection and the development of hepatitis, and, in any case, the dynamics of ALT were unrelated to TTV viremia 44, 59, 81, 96, 115, 147; (iv) in several studies of patients with hepatitis B or C, no correlation was found between the severity of liver damage or responsiveness to alpha interferon therapy and concomitant TTV infection 8, 36, 44, 48, 66, 69, 96, 115, 126, 167; and (v) chimpanzees naturally or experimentally infected with TTV or TTV-like viruses showed no biochemical and histological signs of liver damage 106, 176. Furthermore, retrospective analyses of patients treated with alpha interferon with or without ribavirin for underlying HCV infection have shown that therapy may result in a generally transient disappearance of detectable TTV from blood, especially if baseline TTV viremia is low, but that this is not accompanied by modulation of ALT levels unless HCV is also cleared 4, 8, 16, 48. In addition, attempts to link TTV to hepatocellular carcinoma in patients positive or negative for HBV and HCV have given inconsistent results 107, 132, 156, 184.
Undoubtedly, the above observations rule out the notion that clinically evident liver disease is a frequent consequence of TTV infection. However, the evidence is not totally unequivocal, since data also exist that are compatible with the possibility that, in some cases, transient and mild abnormalities in liver enzyme levels are associated with TTV infection 33, 34, 56, 61, 65, 68, 81, 88, 110, 126. Likewise, in several series of HCV patients, coinfection with TTV appeared to be associated with increased severity of biochemical and histological parameters of liver damage 15, 22, 48, 56, 172, 175, 191. In a series of 26 patients with fulminant liver failure studied by Tanaka et al. 163, mortality was 100% in TTV-infected patients versus less than 50% in uninfected ones. Based on these findings and the apparent ability of TTV to replicate in the liver 19, 124, 142, several investigators consider it likely that TTV may cause occasional liver injury. In other words, TTV would be a candidate for cryptogenic hepatitis etiology not dissimilar from many other viruses, such as enteroviruses, adenoviruses, cytomegalovirus, Epstein-Barr virus, measles virus, rubella virus, and influenza virus, that are known to cause usually transient liver dysfunction of varying severity in minor subsets of infected patients. As in the above infections, the viral and host determinants that might determine or enhance the hepatopathogenicity of TTV are not known. It has been suggested that certain (sub)types or variants of TTV might be especially hepatotropic 62, 120, similarly to what was observed for certain enteroviruses and adenoviruses, or that TTV might cause disease only when activated by superinfection with other viruses 157. Alternatively, liver damage might become evident only when the extent of virus replication is above a certain threshold, due to large viral inocula or other factors. Pistello et al. 134 found that patients with cryptogenetic chronic hepatitis had mean viremia loads significantly higher than did patients with other pathologies. Finally, individuals exposed to TTV for the first time after loss of maternal antibody or reexposed to partially cross-reactive or non-cross-reactive TTV strains might be especially prone to liver damage.
Undoubtedly, the circumstances in which TTV was discovered have greatly influenced the direction of attempts to link it to clinical disease. Possibly for this reason, reported attempts to evaluate the role of acute or long-term TTV infection in the generation of extrahepatic illnesses are few and disparate. In a series of 49 consecutive mixed-cryoglobulinemia patients, TTV infection appeared to be neither a risk factor nor a cofactor 14. In ambulatory children, TTV DNA was most frequent among subjects with acute gastroenteritis 58. In another study, diabetic patients exhibited a 50% prevalence of viremia versus 5% in matched controls 38. No association was found with the Kawasaki syndrome 21 or with multiple sclerosis and other central nervous system disorders (F. Maggi, C. Fornai, M. L. Vatteroni, G. Siciliano, F. Menichetti, C. Tascini, S. Specter, M. Pistello, and M. Bendinelli, submitted for publication). Viremic hemophiliacs had increased levels of IgG and IgM in serum relative to TTV-negative hemophiliacs 187. Maggi et al. 90 found that infection rates in patients with psoriasis, rheumatoid arthritis, or systemic lupus erythematosus were not higher than in patients with miscellaneous diseases. In another study, viremic patients with rheumatoid arthritis had a greatly enhanced frequency of rheumatoid factor positivity 54. Attempts to correlate TTV with progression of HIV infection have generated conflicting reports 20, 141.
In conclusion, there are no clinical manifestations that have been unequivocally associated with TTV to date. Because of this and because active infection is highly prevalent among apparently healthy individuals, there have been repeated suggestions that TTV should be considered essentially devoid of pathogenic potential. Griffiths 46 and Simmonds et al. (Letter) have actually put forward the idea that TTV might represent part of the normal human microflora. While these are provocative speculations, it should be kept in mind that to date the only clinical entities that have been systematically investigated for a possible etiological correlation are some forms of liver disease. Thus, at this point, it is probably more appropriate (and stimulating) to keep TTV in the category of “orphan” viruses. Virology textbooks are full of examples of viruses that had to wait for years after discovery before being linked to disease(s): echoviruses, reoviruses (which still carry the term “orphan” in their acronym), adenoviruses, parvovirus B19, Epstein-Barr virus, and herpesviruses 6 and 7, to name a few. TTV might behave exactly as these viruses, in that only occasional infections might be sufficiently aggressive to become the cause of significant clinical disease. In this context, it is worth noting that until recently, the list of orphan viruses also included the porcine circovirus, a genetic variant of which is now known to cause a multisystemic wasting syndrome of piglets, characterized by lymphadenopathy, hepatitis, nephritis, pneumonia, myocarditis, and gastritis 28, 49, 99, 102.
Clearly, further possible etiological associations should be sought. Possible guidance in choosing the directions for clinical investigation may come from an improved understanding of the natural history of infection and from the diseases produced in their host species by related viruses of animals. In regard to the first criterion, the fact that TTV seems to circulate in blood complexed with IgG 114 should encourage investigations on glomerulonephritis, vasculitis, and other illnesses in which chronic immune complexemia has long been suspected as an important etiopathogenic mechanism. Chronic infection by HCV has already been associated with this kind of pathology (reviewed in reference 7). Also, if the observation that TTV replicates in lymphoid cells only when they are activated to proliferate 93 is confirmed, TTV might represent an ideal candidate agent for unexplained immunosuppressive syndromes. With regard to the diseases produced by related viruses, in addition to that just mentioned regarding the porcine circovirus, it is noteworthy that CAV causes a variety of pathologies including aplastic anemia, hemorrhage, lymphoid depletion, and increased mortality 64, 189. In any case, the large reservoir of TTV in healthy subjects demands that disease associations be investigated through carefully designed clinical studies. The significance of most investigations discussed in this section is, in fact, flawed by the small sizes of the case series, the use of TTV detection methods now known to be insensitive or to identify some (sub)types only, and the lack of proper TTV load measurements.
FUTURE DIRECTIONS
TTV was discovered hardly 3 years ago, and it is therefore not surprising that we still have only a nebulous understanding of its biological properties and pathogenic potential. The many gaps in our present comprehension of TTV have been discussed in detail throughout this review. Here, we will emphasize some issues that, in our opinion, should be addressed first in future investigations.
(i) Molecular systems capable of efficiently detecting and quantitating the full spectrum of existing TTV strains and serological IgG and IgM tests that permit the distinction between recent and remote infections should be developed and standardized. Without improved tools, the laboratory diagnosis and epidemiology of this ubiquitous infectious agent will remain as muddled as they are now.
(ii) Further full-length sequences of isolates worldwide should be obtained. This will improve our present TTV classifications and, in particular, help decide whether all of the viral isolates currently labeled TTV should be considered a single viral species or a genus comprising different species. Okamoto et al. 121 have already questioned whether genomes coding for proteins that differ by 60% or more in amino acid sequence can be attributed to a single viral species, and Hijikata et al. 53 have speculated that the SANBAN isolate might be representative of a distinct although related viral entity. Also, most recently, Khudyakov et al. 74 have proposed that TTV exists as a swarm of at least five closely related but different viral species.
(iii) Cell culture systems should be identified in which TTV replication can be readily investigated in vitro. Advancement in this area will be essential for understanding the relationships that TTV establishes with infected cells and for defining whether the remarkable ability of TTV to produce chronic, active infections in vivo is attributable mainly to the type of interaction established with infected cells or to an outstanding ability to resist the host antiviral defenses; producing large amounts of clean virus will also help in the development of reliable immunoassays.
(iv) The body sites and cell types where TTV replicates should be identified. This will permit a better glimpse at the natural history of infection and provide clues to targeting clinical research on possible TTV-induced pathologies.
(v) Clinical virology should focus on acute disorders that are temporally correlated with elevated levels of TTV viremia. Since it has been speculated that specific TTV strains or variants may exist that are tropic for distinct tissues and more virulent or pathogenic (120; P. Simmonds, F. Davidson, and L. M. Jarvis, Letter, Lancet 352:1310–1311, 1998), the infecting viruses should be molecularly characterized.
(vi) An attentive eye should also be kept on possible long-term effects of chronic TTV carriage. Although the available evidence indicates that chronic productive infection does not represent a heavy burden for the body, evidence also indicates that much TTV circulates in blood in the form of immune complexes; it seems unlikely that this occurs with no pathological consequences for the kidneys and other preferential sites of immune complex deposition.
There is no doubt that the astonishingly high prevalence of TTV worldwide with minimal or no disease association is perplexing. Hopefully, investigating the many fundamental questions that remain to be answered will lead to the identification of hitherto unsuspected etiological links and, ultimately, perhaps reveal further avenues for significant public health improvements, for example in the field of transfusion and organ transplantation medicine.
ACKNOWLEDGMENT
We thank Ministero della Università e Ricerca Scientifica, Rome, Italy, for the financial support that allowed us to develop TTV research in our laboratories.
REFERENCES
- 1.Abe K, Inami T, Asano K, Miyoshi C, Masaki N, Hayashi S, Ishikawa K I, Takebe Y, Win K M, El-Zayadi A R, Han K H, Zhang D Y. TT virus infection is widespread in the general populations from different geographic regions. J Clin Microbiol. 1999;37:2703–2705. doi: 10.1128/jcm.37.8.2703-2705.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abe K, Inami T, Ishikawa K, Nakamura S, Goto S. TT virus infection in nonhuman primates and characterization of the viral genome: identification of simian TT virus isolates. J Virol. 2000;74:1549–1553. doi: 10.1128/jvi.74.3.1549-1553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed R, Morris L A, Knipe D M. Persistence of viruses. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincot-Raven; 1996. pp. 219–249. [Google Scholar]
- 4.Akahane Y, Sakamoto M, Miyazaki Y, Okada S, Inoue T, Ukita M, Okamoto H, Miyakawa Y, Mayumi M. Effect of interferon on a nonenveloped DNA virus (TT virus) associated with acute and chronic hepatitis of unknown etiology. J Med Virol. 1999;58:196–200. doi: 10.1002/(sici)1096-9071(199907)58:3<196::aid-jmv2>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 5.Ball J K, Curran R, Berridge S, Grabowska A M, Jameson C L, Thomson B J, Irving W L, Sharp P M. TT virus sequence heterogeneity in vivo: evidence for co-infection with multiple genetic types. J Gen Virol. 1999;80:1759–1768. doi: 10.1099/0022-1317-80-7-1759. [DOI] [PubMed] [Google Scholar]
- 6.Bassami M R, Berryman D, Wilcox G E, Raidal S R. Psittacine beak and feather disease virus nucleotide sequence analysis and its relationship to porcine circovirus, plant circoviruses, and chicken anaemia virus. Virology. 1998;249:453–459. doi: 10.1006/viro.1998.9324. [DOI] [PubMed] [Google Scholar]
- 7.Bendinelli M, Vatteroni M L, Maggi F, Pistello M. Hepatitis C virus: biology, pathogenesis, epidemiology, clinical description, and diagnosis. In: Specter S, editor. Viral hepatitis: diagnosis, therapy, and prevention. Totowa, N.J: Humana Press; 1999. pp. 65–127. [Google Scholar]
- 8.Berg T, Schreier E, Heuft H G, Hohne M, Bechstein W O, Leder K, Hopf U, Neuhaus P, Wiedenmann B. Occurrence of a novel DNA virus (TTV) infection in patients with liver diseases and its frequency in blood donors. J Med Virol. 1999;59:117–121. doi: 10.1002/(sici)1096-9071(199909)59:1<117::aid-jmv19>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 9.Berns K I. Parvoviridae: the virus and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincot-Raven; 1996. pp. 2173–2199. [Google Scholar]
- 10.Biagini P, Gallian P, Attoui H, Cantaloube J F, de Micco P, de Lamballerie X. Determination and phylogenetic analysis of partial sequences from TT virus isolates. J Gen Virol. 1999;80:419–424. doi: 10.1099/0022-1317-80-2-419. [DOI] [PubMed] [Google Scholar]
- 11.Biagini P, Gallian P, Cantaloube J F, De Micco P, de Lamballerie X. Presence of TT virus in French blood donors and intravenous drug users. J Hepatol. 1998;29:684–685. doi: 10.1016/s0168-8278(98)80167-0. [DOI] [PubMed] [Google Scholar]
- 12.Bonis P A L. TT virus. J Am Soc Nephrol. 1999;10:1828–1832. doi: 10.1681/ASN.V1081828. [DOI] [PubMed] [Google Scholar]
- 13.Bradley D W, Maynard J E, Popper H, Cook E H, Ebert J W, McCaustland K A, Schable C A, Fields H A. Post-transfusion non-A, non-B hepatitis: physicochemical properties of two distinct agents. J Infect Dis. 1983;148:254–265. doi: 10.1093/infdis/148.2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cacoub P, Halfon P, Musset L. Transfusion-transmitted virus and mixed cryoglobulinemia. Ann Intern Med. 1999;130:451–452. doi: 10.7326/0003-4819-130-5-199903020-00021. [DOI] [PubMed] [Google Scholar]
- 15.Charlton M, Adjei P, Poterucha J, Zein N, Moore B, Therneau T, Krom R, Wiesner R. TT-virus infection in North American blood donors, patients with fulminant hepatic failure, and cryptogenic cirrhosis. Hepatology. 1998;28:839–842. doi: 10.1002/hep.510280335. [DOI] [PubMed] [Google Scholar]
- 16.Chayama K, Kobayashi M, Tsubota A, Kobayashi M, Arase Y, Suzuki Y, Saitoh S, Murashima N, Ikeda K, Okamoto K, Hashimoto M, Matsuda M, Koike H, Kobayashi M, Kumada H. Susceptibility of TT virus to interferon therapy. J Gen Virol. 1999;80:631–634. doi: 10.1099/0022-1317-80-3-631. [DOI] [PubMed] [Google Scholar]
- 17.Chen B P, Rumi M G, Colombo M, Lin Y H, Ramaswamy L, Luna J, Liu J K, Prati D, Mannucci P M. TT virus is present in a high frequency of Italian hemophilic patients transfused with plasma-derived clotting factor concentrates. Blood. 1999;94:4333–4336. [PubMed] [Google Scholar]
- 18.Cheng J, Hada T, Fukui K, Ohno M, Hara N, Ohkawa T, Yokoyama Y, Imanishi H, Iijima H, Shimomura S, Yamamoto T, Amuro Y, Higashino K. Detection of TTV DNA in serum of patients with chronic liver disease and interferon efficacy. Hepatol Res. 1999;14:97–104. [Google Scholar]
- 19.Cheng J D, Hada T, Liu W D, Imanishi H, Iijima H, Shimomura S, Amuro Y, Kubota A, Higashino K. Investigation of TTV by in situ hybridization in patients with chronic hepatitis. Hepatol Res. 2000;18:43–53. doi: 10.1016/s1386-6346(99)00083-2. [DOI] [PubMed] [Google Scholar]
- 20.Christensen J K, Eugen-Olsen J, Sorensen M, Ullum H, Gjedde S B, Pedersen B K, Nielsen J O, Krogsgaard K. Prevalence and prognostic significance of infection with TT virus in patients infected with human immunodeficiency virus. J Infect Dis. 2000;181:1796–1799. doi: 10.1086/315440. [DOI] [PubMed] [Google Scholar]
- 21.Chua P K, Nerurkar V R, Yu Q, Woodward C L, Melish M E, Yanagihara R. Lack of association between Kawasaki syndrome and infection with parvovirus B19, human herpesvirus 8, TT virus, GB virus C/hepatitis G virus or Chlamidia pneumoniae. Pediatr Infect Dis J. 2000;19:477–479. doi: 10.1097/00006454-200005000-00019. [DOI] [PubMed] [Google Scholar]
- 22.Cleavinger P J, Persing D H, Li H, Moore S B, Charlton M R, Sievers C, Thernea T M, Zein N N. Prevalence of TTV virus infection in blood donors with elevated ALT in the absence of known hepatitis markers. Am J Gastroenterol. 2000;95:772–776. doi: 10.1111/j.1572-0241.2000.01820.x. [DOI] [PubMed] [Google Scholar]
- 23.Colombatto P, Brunetto M R, Kansopon J, Oliveri F, Maina A, Aragon U, Bortoli M L, Scatena F, Baicchi U, Houghton M, Bonino F, Weiner A J. High prevalence of G1 and G2 TT-virus infection in subjects with high and low blood exposure risk: identification of G4 isolates in Italy. J Hepatol. 1999;31:990–996. doi: 10.1016/s0168-8278(99)80310-9. [DOI] [PubMed] [Google Scholar]
- 24.Danen-Van Oorschot A A, Fisher D F, Grimbergen J M, Klein B, Zhuang S, Falkenburg J H, Backendorf C, Quax P H, van der Eb A J, Noteborn M H. Apoptin induces apoptosis in human transformed and malignant cells but not in normal cells. Proc Natl Acad Sci USA. 1997;94:5843–5847. doi: 10.1073/pnas.94.11.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danen-Van Oorschot A A, van der Eb A J, Noterborn M H. BCL-2 stimulates apoptin-induced apoptosis. Adv Exp Med Biol. 1999;457:245–249. doi: 10.1007/978-1-4615-4811-9_26. [DOI] [PubMed] [Google Scholar]
- 26.Davidson F, MacDonald D, Mokili J L K, Prescott L E, Graham S, Simmonds P. Early acquisition of TT virus (TTV) in an area endemic for TTV infection. J Infect Dis. 1999;179:1070–1076. doi: 10.1086/314730. [DOI] [PubMed] [Google Scholar]
- 27.Desai S M, Muerhoff A S, Leary T P, Erker J C, Simons J N, Chalmers M L, Birkenmeyer L G, Pilot-Matias T J, Mushahwar I K. Prevalence of TT virus infection in US blood donors and populations at risk for acquiring parenterally transmitted viruses. J Infect Dis. 1999;179:1242–1244. doi: 10.1086/314735. [DOI] [PubMed] [Google Scholar]
- 28.Ellis J, Krakowka S, Lairmore M, Haines D, Bratanich A, Clark E, Allan G, Konoby C, Hassard L, Meehan B, Martin K, Harding J, Kennedy S, McNeilly F. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J Vet Diagn Investig. 1999;11:3–14. doi: 10.1177/104063879901100101. [DOI] [PubMed] [Google Scholar]
- 29.Erker J C, Leary T P, Desai S M, Chalmers M L, Mushahwar I K. Analyses of TT virus full-length genomic sequences. J Gen Virol. 1999;80:1743–1750. doi: 10.1099/0022-1317-80-7-1743. [DOI] [PubMed] [Google Scholar]
- 30.Farkas T, Tanaka A, Kai K, Kanoe M. Cloning and sequencing of the genome of chicken anaemia virus (CAV) TK-5803 strain and comparison with other CAV strains. J Vet Med Sci. 1996;58:681–684. doi: 10.1292/jvms.58.681. [DOI] [PubMed] [Google Scholar]
- 31.Fenner F J, Gibbs E P J, Murphy F A, Rott R, Studdert M J, White D O. Veterinary virology. 2nd ed. San Diego, Calif: Academic Press, Inc.; 1993. [Google Scholar]
- 32.Fields B N. Virology. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincot-Raven; 1996. p. 2950. [Google Scholar]
- 33.Forns X, Hegerich P, Darnell A, Emerson S U, Purcell R H, Bukh J. High prevalence of TT virus (TTV) infection in patients on maintenance hemodialysis: frequent mixed infections with different genotypes and lack of evidence of associated liver disease. J Med Virol. 1999;59:313–317. [PubMed] [Google Scholar]
- 34.Fujiwara T, Iwata A, Iizuka H, Tanaka T, Okamoto H. Transfusion transmitted virus. Lancet. 1998;352:1310. doi: 10.1016/S0140-6736(05)70520-1. [DOI] [PubMed] [Google Scholar]
- 35.Fukuda Y, Nakano I, Katano Y, Kumada T, Hayashi K, Nakano S, Hayakawa T. TT virus (TTV) is not associated with acute sporadic hepatitis. Infection. 1999;27:125–128. doi: 10.1007/BF02560512. [DOI] [PubMed] [Google Scholar]
- 36.Gad A, Tanaka E, Orii K, Kafumi T, Serwah A E H, El-Sherif A, Nooman Z, Kiyosawa K. Clinical significance of TT virus infection in patients with chronic liver disease and volunteer blood donors in Egypt. J Med Virol. 2000;60:177–181. [PubMed] [Google Scholar]
- 37.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 38.Gallian P, Berland Y, Olmer M, Raccah D, De Micco P, Biagini P, Simon S, Bouchouareb D, Mourey C, Roubicek C, Touinssi M, Cantaloube J F, Dussol B, de Lamballerie X. TT virus infection in French hemodialysis patients: study of prevalence and risk factors. J Clin Microbiol. 1999;37:2538–2542. doi: 10.1128/jcm.37.8.2538-2542.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallian P, Biagini P, Zhong S, Touinssi M, Yeo W, Cantaloube J F, Attoui H, de Micco P, Johnson P J, de Lamballerie X. TT virus: a study of molecular epidemiology and transmission of genotypes 1, 2, and 3. J Clin Virol. 2000;17:43–49. doi: 10.1016/s1386-6532(00)00066-4. [DOI] [PubMed] [Google Scholar]
- 40.Gelderblom H, Kling S, Lurz R, Tischer I, von Bülow V. Morphological characterization of chicken anaemia agent (CAA) Arch Virol. 1989;109:115–120. doi: 10.1007/BF01310522. [DOI] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42.Gibbs M J, Weiller G F. Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc Natl Acad Sci USA. 1999;96:8022–8027. doi: 10.1073/pnas.96.14.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilbert W, Dressler D. DNA replication: the rolling circle model. Cold Spring Harbor Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- 44.Gimenez-Barcons M, Forns X, Ampurdanes S, Guilera M, Soler M, Soguero C, Sanchez-Fueyo A, Mas A, Bruix J, Sanchez-Tapias J M, Rodes J, Saiz J C. Infection with a novel human DNA virus (TTV) has no pathogenic significance in patients with liver diseases. J Hepatol. 1999;30:1028–1034. doi: 10.1016/s0168-8278(99)80256-6. [DOI] [PubMed] [Google Scholar]
- 45.Goto K, Sugiyama K, Terabe K, Mizutani F, Wada Y. Detection rates of TT virus among children who visited a general hospital in Japan. J Med Virol. 1999;57:405–407. doi: 10.1002/(sici)1096-9071(199904)57:4<405::aid-jmv13>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 46.Griffiths P. Time to consider the concept of a commensal virus? Rev Med Virol. 1999;9:73–74. doi: 10.1002/(sici)1099-1654(199904/06)9:2<73::aid-rmv254>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 47.Hafner G J, Stafford M R, Wolter L C, Harding R H, Dale J L. Nicking and joining activity of banana bunchy top virus replication protein in vitro. J Gen Virol. 1997;78:1795–1799. doi: 10.1099/0022-1317-78-7-1795. [DOI] [PubMed] [Google Scholar]
- 48.Hagiwara H, Hayashi N, Mita E, Oshita M, Kobayashi I, Iio S, Hiramatsu N, Sasaki Y, Kasahara A, Kakinuma K, Yamauchi T, Fusamoto H. Influence of transfusion-transmitted virus infection on the clinical features and response to interferon therapy in Japanese patients with chronic hepatitis C. J Viral Hepatol. 1999;6:463–469. doi: 10.1046/j.1365-2893.1999.00183.x. [DOI] [PubMed] [Google Scholar]
- 49.Hamel A L, Lin L L, Nayar G P. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol. 1998;72:5262–5267. doi: 10.1128/jvi.72.6.5262-5267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He C, Nomura F, Yukimasa N, Itoga S, Yamada-Osaki M, Sumazaki R, Nakai T. Transfusion-transmitted virus infection in China: prevalence in blood donors and in patients with liver diseases. J Gastroenterol Hepatol. 1999;14:899–903. doi: 10.1046/j.1440-1746.1999.01970.x. [DOI] [PubMed] [Google Scholar]
- 51.Hemanuer A, von Poblotzki A, Gigler A, Cassinotti P, Siegl G, Wolf H, Modrow S. Sequence variability among different parvovirus B19 isolates. J Gen Virol. 1996;77:1781–1785. doi: 10.1099/0022-1317-77-8-1781. [DOI] [PubMed] [Google Scholar]
- 52.Hijikata M, Iwata K, Ohta Y, Nakao K, Matsumoto M, Matsumoto H, Kanai K, Baba K, Samokhvalov E I, Mishiro S. Genotypes of TT virus (TTV) compared between liver disease patients and healthy individuals using a new PCR system capable of differentiating 1a and 1b types from others. Arch Virol. 1999;144:2345–2354. doi: 10.1007/s007050050648. [DOI] [PubMed] [Google Scholar]
- 53.Hijikata M, Takahashi K, Mishiro S. Complete circular DNA genome of a TT virus variant (isolate name SANBAN) and 44 partial ORF2 sequences implicating a great degree of diversity beyond genotypes. Virology. 1999;260:17–22. doi: 10.1006/viro.1999.9797. [DOI] [PubMed] [Google Scholar]
- 54.Hirata D, Kaneko N, Iwamoto M, Yoshio T, Okazaki H, Mimori A, Masuyama J, Minota S. Infection with an unenveloped DNA virus (TTV) associated with non-A to G hepatitis in patients with rheumatoid arthritis. Br J Rheumatol. 1998;37:1361–1362. doi: 10.1093/rheumatology/37.12.1361. [DOI] [PubMed] [Google Scholar]
- 55.Hsieh S Y, Wu Y H, Ho Y P, Tsao K C, Yeh C T, Liaw Y F. High prevalence of TT virus infection in healthy children and adults and in patients with liver disease in Taiwan. J Clin Microbiol. 1999;37:1829–1831. doi: 10.1128/jcm.37.6.1829-1831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ikeda H, Takasu M, Inoue K, Okamoto H, Miyakawa Y, Mayumi M. Infection with an unenveloped DNA virus (TTV) in patients with acute or chronic liver disease of unknown etiology and in those positive for hepatitis C virus RNA. J Hepatol. 1999;30:205–212. doi: 10.1016/s0168-8278(99)80063-4. [DOI] [PubMed] [Google Scholar]
- 57.Inami T, Konomi N, Arakawa Y, Abe K. High prevalence of TT virus DNA in human saliva and semen. J Clin Microbiol. 2000;38:2407–2408. doi: 10.1128/jcm.38.6.2407-2408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iriyama M, Kimura H, Nishizawa K, Yoshioka K, Wakita T, Nishimura N, Shibata M, Ozaki T, Morishima T. The prevalence of TT virus (TTV) infection and its relationship to hepatitis in children. Med Microbiol Immunol. 1999;188:83–89. doi: 10.1007/s004300050109. [DOI] [PubMed] [Google Scholar]
- 59.Irving W L, Ball J K, Berridge S, Curran R, Grabowska A M, Jameson C L, Neal K R, Ryder S D, Thomson B J for the Trent HCV study group. TT virus infection in patients with hepatitis C: frequency, persistence, and sequence heterogeneity. J Infect Dis. 1999;180:27–34. doi: 10.1086/314825. [DOI] [PubMed] [Google Scholar]
- 60.Ishikawa T, Hamano Y, Okamoto H. Frequent detection of TT virus in throat swabs of pediatric patients. Infection. 1999;27:298. [PubMed] [Google Scholar]
- 61.Itoh K, Hirakawa K, Okamoto H, Ukita M, Tanaka H, Sawada N, Tsuda F, Miyakawa Y, Mayumi M. Infection by an unenveloped DNA virus associated with non-A to -G hepatitis in Japanese blood donors with or without elevated ALT levels. Transfusion. 1999;39:522–526. doi: 10.1046/j.1537-2995.1999.39050522.x. [DOI] [PubMed] [Google Scholar]
- 62.Itoh K, Takahashi M, Ukita M, Nishizawa T, Okamoto H. Influence of primers on the detection of TT virus DNA by polymerase chain reaction. J Infect Dis. 1999;180:1750–1751. doi: 10.1086/315107. [DOI] [PubMed] [Google Scholar]
- 63.Iwata N, Fujino M, Tuchiya K, Iwata A, Otaki Y, Ueda S. Development of an enzyme-linked immunosorbent assay using recombinant chicken anemia virus proteins expressed in a baculovirus vector system. J Vet Med Sci. 1998;60:175–180. doi: 10.1292/jvms.60.175. [DOI] [PubMed] [Google Scholar]
- 64.Jeurissen S H M, Wagenaar F, Pol J M A, van der Eb A J, Noteborn M H M. Chicken anaemia virus causes apoptosis of thymocytes after in vivo infection and of cell lines after in vitro infection. J Virol. 1992;66:7383–7388. doi: 10.1128/jvi.66.12.7383-7388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang R L, Lu Q, Luo K X, Tan F M. A prospective study of transfusion-transmitted virus transmission by blood transfusion. J Viral Hepatol. 1999;6:49–51. doi: 10.1046/j.1365-2893.1999.6120133.x. [DOI] [PubMed] [Google Scholar]
- 66.Kanda T, Yokosuka O, Ikeuchi T, Seta T, Kawai S, Imazeki F, Saisho H. The role of TT virus infection in acute viral hepatitis. Hepatology. 1999;29:1905–1908. doi: 10.1002/hep.510290613. [DOI] [PubMed] [Google Scholar]
- 67.Kanda Y, Chiba S, Tanaka Y, Kami M, Saito T, Asai T, Izutsu K, Yuji K, Ogawa S, Honda H, Mitani K, Yazaki Y, Hirai H. TT virus in frequently transfused patients. Am J Med. 1999;106:116–117. doi: 10.1016/s0002-9343(98)00366-0. [DOI] [PubMed] [Google Scholar]
- 68.Kanda Y, Tanaka Y, Kami M, Saito T, Asai T, Izutsu K, Yuji K, Ogawa S, Honda H, Mitani K, Chiba S, Yazaki Y, Hirai H. TT virus in bone marrow transplant recipients. Blood. 1999;93:2485–2490. [PubMed] [Google Scholar]
- 69.Kao J H, Chen W, Hsiang S C, Chen P J, Lai M Y, Chen D S. Prevalence and implication of TT virus infection: minimal role in patients with non-A-E hepatitis in Taiwan. J Med Virol. 1999;59:307–312. doi: 10.1002/(sici)1096-9071(199911)59:3<307::aid-jmv8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 70.Kato A, Fujino M, Nakamura T, Ishihama A, Otaki Y. Gene organization of chicken anemia virus. Virology. 1995;209:480–488. doi: 10.1006/viro.1995.1280. [DOI] [PubMed] [Google Scholar]
- 71.Kato H, Mizokami M, Nakano T, Kondo Y, Dashnyam B, Oyunsuren T, Ueda R. High prevalence and phylogenetic analysis of TT-virus infection in Mongolia. Virus Res. 1999;60:171–179. doi: 10.1016/s0168-1702(99)00016-7. [DOI] [PubMed] [Google Scholar]
- 72.Kato T, Mizokami M, Mukaide M, Orito E, Ohno T, Nakano T, Tanaka Y, Kato H, Sugauchi F, Ueda R, Hirashima N, Shimamatsu K, Kage M, Kojiro M. Development of a TT virus DNA quantification system using real-time detection PCR. J Clin Microbiol. 2000;38:94–98. doi: 10.1128/jcm.38.1.94-98.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kato T, Mizokami M, Orito E, Nakano T, Tanaka Y, Ueda R, Hirashima N, Iijima Y, Kato T, Sugauchi F, Mukaide M, Shimamatsu K, Kage M, Kojiro M. High prevalence of TT virus infection in Japanese patients with liver diseases and in blood donors. J Hepatol. 1999;31:221–227. doi: 10.1016/s0168-8278(99)80217-7. [DOI] [PubMed] [Google Scholar]
- 74.Khudyakov Y E, Cong M-E, Nichols B, Reed D, Dou X-G, Viazov S O, Chang J, Fried M W, Williams I, Bower W, Lambert S, Purdy M, Roggendorf M, Fields H A. Sequence heterogeneity of TT virus and closely related viruses. J Virol. 2000;74:2990–3000. doi: 10.1128/jvi.74.7.2990-3000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kikuchi K, Miyakawa H, Abe K, Kako M, Katayama K, Fukushi S, Mishiro S. Indirect evidence of TTV replication in bone marrow cells, but not in hepatocytes, of a subacute hepatitis/aplastic anemia patient. J Med Virol. 2000;61:165–170. [PubMed] [Google Scholar]
- 76.Kobayashi M, Chayama K, Arase Y, Kobayashi M, Tsubota A, Suzuki Y, Koida I, Saitoh S, Murashima N, Ikeda K, Koike H, Hashimoto M, Kobayashi M, Kumada H. Prevalence of TT virus before and after blood transfusion in patients with chronic liver disease treated surgically for hepatocellular carcinoma. J Gastroenterol Hepatol. 1999;14:358–363. doi: 10.1046/j.1440-1746.1999.01860.x. [DOI] [PubMed] [Google Scholar]
- 77.Koch G, van Roozelaar D J, Verschueren C A, van der Eb A J, Noteborn M H. Immunogenic and protective properties of chicken anaemia virus proteins expressed by baculovirus. Vaccine. 1995;13:763–770. doi: 10.1016/0264-410x(94)00034-k. [DOI] [PubMed] [Google Scholar]
- 78.Koonin E V, Ilyina T V. Computer-assisted dissection of rolling circle DNA replication. Biosystems. 1993;30:241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- 79.Leary T P, Erker J C, Chalmers M L, Desai S M, Mushahwar I K. Improved detection systems for TT virus reveal high prevalence in humans, non-human primates and farm animals. J Gen Virol. 1999;80:2115–2120. doi: 10.1099/0022-1317-80-8-2115. [DOI] [PubMed] [Google Scholar]
- 80.Leary T P, Erker J C, Chalmers M L, Desai S M, Mushahwar I K. Optimized PCR assay for the detection of TT virus. J Virol Methods. 1999;82:109–112. doi: 10.1016/s0166-0934(99)00100-7. [DOI] [PubMed] [Google Scholar]
- 81.Lefrère J J, Roudot-Thoraval F, Lefrere F, Kahfer A, Mariotti M, Lerable J, Thauvin M, Lefevre G, Rouger P, Girot R. Natural history of the TT virus infection through follow-up of TTV DNA-positive multiple-transfused patients. Blood. 1999;95:347–351. [PubMed] [Google Scholar]
- 82.Lin H J, Pedneault L, Hollinger F B. Intra-assay performance characteristics of five assays for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:835–839. doi: 10.1128/jcm.36.3.835-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Linden R M, Ward P, Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1996;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lisitsyn N, Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 85.Lo S Y, Peng K F, Ma H C, Yu J H, Li Y H, Lin H H, Lua A C, Lee M L. Prevalence of TT virus DNA in eastern Taiwan aborigenes. J Med Virol. 1999;59:198–203. doi: 10.1002/(sici)1096-9071(199910)59:2<198::aid-jmv12>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 86.Lukert P D, de Boer G F, Dale J L, Keese P, McNulty M S, Randers J W, Tisher I. Family Circoviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. pp. 166–168. [Google Scholar]
- 87.Luo K X, Liang W F, He H T, Yang S C, Wang Y, Xiao H, Liu D X, Zhang L. Experimental infection of nonenveloped DNA virus (TTV) in Rhesus monkey. J Med Virol. 2000;61:159–164. doi: 10.1002/(sici)1096-9071(200005)61:1<159::aid-jmv26>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 88.Luo K X, Zhang L, Wang S S, Nie J, Yang S C, Liu D X, Liang W F, He H T, Lu Q. An outbreak of enterically transmitted non-A, non-E viral hepatitis. J Viral Hepatol. 1999;6:59–64. doi: 10.1046/j.1365-2893.1999.6120132.x. [DOI] [PubMed] [Google Scholar]
- 89.MacDonald D M, Scott G R, Clutterbuck D, Simmonds P. Infrequent detection of TT virus infection in intravenous drug users, prostitutes, and homosexual men. J Infect Dis. 1999;179:686–689. doi: 10.1086/314642. [DOI] [PubMed] [Google Scholar]
- 90.Maggi F, Fornai C, Morrica A, Casula F, Vatteroni M L, Marchi S, Ciccorossi P, Riente L, Pistello M, Bendinelli M. High prevalence of TT virus viremia in Italian patients, regardless of age, clinical diagnosis, and previous interferon treatment. J Infect Dis. 1999;180:838–842. doi: 10.1086/314961. [DOI] [PubMed] [Google Scholar]
- 91.Maggi F, Fornai C, Morrica A, Vatteroni M L, Giorgi M, Marchi S, Ciccorossi P, Bendinelli M, Pistello M. Divergent evolution of hepatitis C virus in liver and peripheral blood mononuclear cells of infected patients. J Med Virol. 1999;57:57–63. [PubMed] [Google Scholar]
- 92.Reference deleted.
- 93.Maggi, F., C. Fornai, L. Zaccaro, A. Morrica, M. L. Vatteroni, P. Isola, S. Marchi, S. Ricchiuti, M. Pistello, and M. Bendinelli. TT virus (TTV) loads associated with different peripheral blood cell types and evidence for TTV replication in activated mononuclear cells. J. Med. Virol., in press. [DOI] [PubMed]
- 94.Mankertz A, Persson F, Mankertz J, Blaess G, Buhk H J. Mapping and characterization of the origin of DNA replication of porcine circovirus. J Virol. 1997;71:2562–2566. doi: 10.1128/jvi.71.3.2562-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mankertz J, Buhk H J, Blaess G, Mankertz A. Transcription analysis of porcine circovirus (PCV) Virus Genes. 1998;16:267–276. doi: 10.1023/a:1008022521329. [DOI] [PubMed] [Google Scholar]
- 96.Matsumoto A, Yeo A E T, Shih J W K, Tanaka E, Kiyosawa K, Alter H J. Transfusion-associated TT virus infection and its relationship to liver disease. Hepatology. 1999;30:283–288. doi: 10.1002/hep.510300118. [DOI] [PubMed] [Google Scholar]
- 97.Matteucci D, Paglianti M, Giangregorio A M, Capobianchi M R, Dianzani F, Bendinelli M. Group B coxsackieviruses readily establish persistent infections in human lymphoid cell lines. J Virol. 1985;56:651–654. doi: 10.1128/jvi.56.2.651-654.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Meehan B M, Creelan J L, McNulty M S, Todd D. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J Gen Virol. 1997;78:221–227. doi: 10.1099/0022-1317-78-1-221. [DOI] [PubMed] [Google Scholar]
- 99.Meehan B M, McNeilly F, Todd D, Kennedy S, Jewhurst V A, Ellis J A, Hassard L E, Clark E G, Haines D M, Allan G M. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol. 1998;79:2171–2179. doi: 10.1099/0022-1317-79-9-2171. [DOI] [PubMed] [Google Scholar]
- 100.Mellor J, Hawkins A, Simmonds P. Genotype dependence of hepatitis C virus load measurement in commercially available quantitative assays. J Clin Microbiol. 1999;37:2525–2532. doi: 10.1128/jcm.37.8.2525-2532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miyata H, Tsunoda H, Kazi A, Yamada A, Khan M A, Murakami J, Kamahora T, Shiraki K, Hino S. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J Virol. 1999;73:3582–3586. doi: 10.1128/jvi.73.5.3582-3586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morozov I, Sirinarumitr T, Sorden S D, Halbur P G, Morgan M K, Yoon K J, Paul P S. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Clin Microbiol. 1998;36:2535–2541. doi: 10.1128/jcm.36.9.2535-2541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morrica A, Maggi F, Vatteroni M L, Fornai C, Pistello M, Ciccorossi P, Grassi E, Gennazzani A, Bendinelli M. TT virus: evidence for transplacental transmission. J Infect Dis. 2000;181:803–804. doi: 10.1086/315296. [DOI] [PubMed] [Google Scholar]
- 104.Reference deleted.
- 105.Mulyanto M, Hijikata M, Matsushita M, Ingkokusmo G, Widjaya A, Sumarsidi D, Kanai K, Ohta Y, Mishiro S. TT virus (TTV) genotypes in native and non-native prostitutes of Irian Jaya, Indonesia: implication for non-occupational transmission. Arch Virol. 2000;145:63–72. doi: 10.1007/s007050050005. [DOI] [PubMed] [Google Scholar]
- 106.Mushahwar I K, Erker J C, Muerhoff A S, Leary T P, Simons J N, Birkenmeyer L G, Chalmers M L, Pilot-Matias T J, Desai S M. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA. 1999;96:3177–3182. doi: 10.1073/pnas.96.6.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nakagawa N, Ikoma J, Ishihara T, Yasui N, Fujita N, Iwasa M, Kaito M, Watanabe S, Adachi Y. High prevalence of transfusion-transmitted virus among patients with non-B, non-C hepatocellular carcinoma. Cancer. 1999;86:1437–1440. [PubMed] [Google Scholar]
- 108.Nakagawa N, Ikoma J, Ishihara T, Yasui-Kawamura N, Fujita N, Iwasa M, Kaito M, Watanabe S, Adachi Y. Biliary excretion of TT virus (TTV) J Med Virol. 2000;61:462–467. doi: 10.1002/1096-9071(200008)61:4<462::aid-jmv8>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 109.Nakano T, Park Y M, Mizokami M, Choi J Y, Orito E, Ohno T, Kato T, Kondo Y, Tanaka Y, Kato H, Kato T, Kim B S. TT virus infection among blood donors and patients with non-B, non-C liver diseases in Korea. J Hepatol. 1999;30:389–393. doi: 10.1016/s0168-8278(99)80095-6. [DOI] [PubMed] [Google Scholar]
- 110.Naoumov N V, Petrova E P, Thomas M G, Williams R. Presence of a newly described human DNA virus (TTV) in patients with liver disease. Lancet. 1998;352:195–197. doi: 10.1016/S0140-6736(98)04069-0. [DOI] [PubMed] [Google Scholar]
- 111.Niagro F D, Forsthoefel A N, Lawther R P, Kamalanathan L, Ritchie B W, Latimer K S, Lukert P D. Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch Virol. 1998;143:1723–1744. doi: 10.1007/s007050050412. [DOI] [PubMed] [Google Scholar]
- 112.Niel C, de Oliveira J M, Ross R S, Gomes S A, Roggendorf M, Viazov S. High prevalence of TT virus infection in Brazilian blood donors. J Med Virol. 1999;57:259–263. [PubMed] [Google Scholar]
- 113.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 114.Nishizawa T, Okamoto H, Tsuda F, Aikawa T, Sugai Y, Konishi K, Akahane Y, Ukita M, Tanaka T, Miyakawa Y, Mayumi M. Quasispecies of TT virus (TTV) with sequence divergence in hypervariable regions of the capsid protein in chronic TTV infection. J Virol. 1999;73:9604–9608. doi: 10.1128/jvi.73.11.9604-9608.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Oguchi T, Tanaka E, Orii K, Kobayashi M, Hora K, Kiyosawa K. Transmission of and liver injury by TT virus in patients on maintenance hemodialysis. J Gastroenterol. 1999;34:234–240. doi: 10.1007/s005350050249. [DOI] [PubMed] [Google Scholar]
- 116.Okamoto H, Akahane Y, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, Mayumi M. Fecal excretion of a nonenveloped DNA virus (TTV) associated with posttransfusion non A-G hepatitis. J Med Virol. 1998;56:128–132. [PubMed] [Google Scholar]
- 117.Okamoto H, Fukuda M, Tawara A, Nishizawa T, Itoh Y, Hayasaka I, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Species-specific TT viruses and cross-species infection in nonhuman primates. J Virol. 2000;74:1132–1139. doi: 10.1128/jvi.74.3.1132-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Okamoto H, Kato N, Iizuka H, Tsuda F, Miyakawa Y, Mayumi M. Distinct genotypes of a nonenveloped DNA virus associated with posttransfusion non-A to G hepatitis (TT virus) in plasma and peripheral blood mononuclear cells. J Med Virol. 1999;57:252–258. doi: 10.1002/(sici)1096-9071(199903)57:3<252::aid-jmv7>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 119.Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, Miyakawa Y, Mayumi M. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol Res. 1998;10:1–16. [Google Scholar]
- 120.Okamoto H, Nishizawa T, Ukita M. A novel unenveloped DNA virus (TT virus) associated with acute and chronic non-A to G hepatitis. Intervirology. 1999;42:196–204. doi: 10.1159/000024961. [DOI] [PubMed] [Google Scholar]
- 121.Okamoto H, Nishizawa T, Ukita M, Takahashi M, Fukuda M, Iizuka H, Miyakawa Y, Mayumi M. The entire sequence of a TT virus isolate from the United States (TUS01): comparison with reported isolates and phylogenetic analysis. Virology. 1999;259:437–448. doi: 10.1006/viro.1999.9769. [DOI] [PubMed] [Google Scholar]
- 122.Okamoto H, Takahashi M, Nishizawa T, Tawara A, Sugai Y, Sai T, Tanaka T, Tsuda F. Replicative forms of TT virus DNA in bone marrow cells. Biochem Biophys Res Commun. 2000;270:657–662. doi: 10.1006/bbrc.2000.2481. [DOI] [PubMed] [Google Scholar]
- 123.Okamoto H, Takahashi M, Nishizawa T, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, Mayumi M. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology. 1999;259:428–436. doi: 10.1006/viro.1999.9770. [DOI] [PubMed] [Google Scholar]
- 124.Okamoto H, Ukita M, Nishizawa T, Kishimoto J, Hoshi Y, Mizuo H, Tanaka T, Miyakawa Y, Mayumi M. Circular double-stranded forms of TT virus DNA in the liver. J Virol. 2000;74:5161–5167. doi: 10.1128/jvi.74.11.5161-5167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Okamura A, Yoshioka M, Kubota M, Kikuta H, Ishiko H, Kobayashi K. Detection of a novel DNA virus (TTV) sequence in peripheral blood mononuclear cells. J Med Virol. 1999;58:174–177. doi: 10.1002/(sici)1096-9071(199906)58:2<174::aid-jmv12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 126.Orii K, Tanaka E, Umemura T, Rokuhara A, Iijima A, Yoshizawa K, Imai H, Kiyosawa K. Prevalence and disease association of TT virus infection in Japanese patients with viral hepatitis. Hepatol Res. 1999;14:161–170. [Google Scholar]
- 127.Osiowy C, Sauder C. Detection of TT virus in human hair and skin. Hepatol Res. 2000;16:155–162. [Google Scholar]
- 128.Padidam M, Sawyer S, Fauquet C M. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- 129.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computer. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 130.Parquet M C, Yatsuhashi H, Koga M, Inoue O, Matsumoto T, Hamada R, Igarashi A, Yano M. Prevalence and clinical characteristics of TT virus (TTV) in patients with sporadic acute hepatitis of unknown etiology. J Hepatol. 1999;31:985–989. doi: 10.1016/s0168-8278(99)80309-2. [DOI] [PubMed] [Google Scholar]
- 131.Phenix K V, Meehan B M, Todd D, McNulty M S. Transcriptional analysis and genome expression of chicken anaemia virus. J Gen Virol. 1994;75:905–909. doi: 10.1099/0022-1317-75-4-905. [DOI] [PubMed] [Google Scholar]
- 132.Pineau P, Meddeb M, Raselli R, Qin L-X, Terris B, Tang Z-Y, Tiollais P, Mazzaferro V, Dejean A. Effect of TT virus infection on hepatocellular carcinoma development: results of a Euro-Asian survey. J Infect Dis. 2000;181:1138–1142. doi: 10.1086/315321. [DOI] [PubMed] [Google Scholar]
- 133.Pisani G, Cristiano K, Wirz M, Bisso G, Beneduce F, Morace G, Rapicetta M, Gentili G. Prevalence of TT virus in plasma pools and blood products. Br J Haematol. 1999;106:431–435. doi: 10.1046/j.1365-2141.1999.01568.x. [DOI] [PubMed] [Google Scholar]
- 134.Pistello, M., A. Morrica, F. Maggi, M. L. Vatteroni, G. Freer, C. Fornai, F. Casula, S. Marchi, P. Ciccorossi, P. Rovero, and M. Bendinelli. TT Virus levels in the plasma of infected individuals with different hepatic and extrahepatic pathologies. J. Med. Virol., in press. [PubMed]
- 135.Poovorawan Y, Theamboonlers A, Jantaradsamee P, Kaew-in N, Hirsch P, Tangkitvanich P. Hepatitis TT virus infection in high-risk groups. Infection. 1998;26:335–358. doi: 10.1007/BF02770835. [DOI] [PubMed] [Google Scholar]
- 136.Poovorawan Y, Theamboonlers A, Vimolket T, Jantaradsamee P, Kaew-in N, Hirsch P. Detection of TTV in peripheral blood mononuclear cells of intravenous drug users. Tohoku J Exp Med. 1999;188:47–54. doi: 10.1620/tjem.188.47. [DOI] [PubMed] [Google Scholar]
- 137.Prati D, Lin Y H, De Mattei C, Liu J K, Farma E, Ramaswamy L, Zanolla A, Lee H, Rebulla P, Allain J P, Sirchia G, Chen B. A prospective study on TT virus infection in transfusion-dependent patients with β-thalassemia. Blood. 1999;93:1502–1505. [PubMed] [Google Scholar]
- 138.Prescott L E, MacDonald D M, Davidson F, Mokili J, Pritchard D I, Arnot D E, Riley E M, Greenwood B M, Hamid S, Saeed A A, McClure M O, Smith D B, Simmonds P. Sequence diversity of TT virus in geographically dispersed human populations. J Gen Virol. 1999;80:1751–1758. doi: 10.1099/0022-1317-80-7-1751. [DOI] [PubMed] [Google Scholar]
- 139.Prescott L E, Simmonds P. Global distribution of transfusion-transmitted virus. N Engl J Med. 1998;339:776–777. doi: 10.1056/NEJM199809103391118. [DOI] [PubMed] [Google Scholar]
- 140.Primi D, Sottini A. Identification and characterization of SEN virus, a family of novel DNA viruses. Antiviral Ther. 2000;5:G7. [Google Scholar]
- 141.Puig-Basagoiti F, Cabana M, Guilera M, Giménez-Barcons M, Sirera G, Tural C, Clotet B, Sànchez-Tapias J-M, Rodés J, Saiz J-C, Martìnez M-A. Prevalence and route of transmission of infection with a novel DNA virus (TTV), hepatitis C virus, and hepatitis G virus in patients infected with HIV. J Acquired Immune Defic Syndr. 2000;23:89–94. doi: 10.1097/00126334-200001010-00012. [DOI] [PubMed] [Google Scholar]
- 142.Rodriguez-Inigo E, Casqueiro M, Bartolome J, Ortiz-Movilla N, Lopez-Alchorocho J M, Herrero M, Manzarbeitia F, Oliva H, Carreno V. Detection of TT virus DNA in liver biopsies by in situ hybridization. Am J Pathol. 2000;156:1227–1234. doi: 10.1016/S0002-9440(10)64993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Romeo R, Hegerich P, Emerson S U, Colombo M, Purcell R H, Bukh J. High prevalence of TT virus (TTV) in naive chimpanzees and in hepatitis C virus-infected humans: frequent mixed infections and identification of new TTV genotypes in chimpanzees. J Gen Virol. 2000;81:1001–1007. doi: 10.1099/0022-1317-81-4-1001. [DOI] [PubMed] [Google Scholar]
- 144.Ross R S, Viazov S, Runde V, Schaefer U W, Roggendorf M. Detection of TT virus DNA in specimens other than blood. J Clin Virol. 1999;13:181–184. doi: 10.1016/s1386-6532(99)00015-3. [DOI] [PubMed] [Google Scholar]
- 145.Saback F L, Gomes S A, de Paula V S, da Silva R R S, Lewis-Ximenez L L, Niel C. Age-specific prevalence and transmission of TT virus. J Med Virol. 1999;59:318–322. doi: 10.1002/(sici)1096-9071(199911)59:3<318::aid-jmv10>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 146.Schroter M, Feucht H H, Zollner B, Knodler B, Schafer P, Fischer L, Laufs R. Prevalence of TTV viremia among healthy subjects and individuals at risk for parenterally transmitted diseases in Germany. Hepatol Res. 1999;13:205–211. doi: 10.1046/j.1365-2893.2002.00390.x. [DOI] [PubMed] [Google Scholar]
- 147.Shang D, Lin Y H, Rigopoulou I, Chen B, Alexander G J M, Allain J-P. Detection of TT virus DNA in patients with liver disease and recipients of liver transplant. J Med Virol. 2000;61:455–461. doi: 10.1002/1096-9071(200008)61:4<455::aid-jmv7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 148.Shimizu Y K, Feinstone S M, Purcell R H, Alter H J, London W T. Non-A, non-B hepatitis: ultrastructural evidence for two agents in experimentally infected chimpanzees. Science. 1979;205:197–200. doi: 10.1126/science.451589. [DOI] [PubMed] [Google Scholar]
- 149.Shoya Y, Kobayashi T, Koda T, Ikuta K, Kakinuma M, Kishi M. Two proline-rich nuclear localization signals in the amino- and carboxyl-terminal regions of the Borna disease virus phosphoprotein. J Virol. 1998;72:9755–9762. doi: 10.1128/jvi.72.12.9755-9762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Reference deleted.
- 151.Simmonds P, Davidson F, Lycett C, Prescott L E, MacDonald D M, Ellender J, Yap P L, Ludlam C A, Haydon G H, Gillon J, Jarvis L M. Detection of a novel DNA virus (TTV) in blood donors and blood products. Lancet. 1998;352:191–195. doi: 10.1016/s0140-6736(98)03056-6. [DOI] [PubMed] [Google Scholar]
- 152.Reference deleted.
- 153.Sugiyama K, Goto K, Ando T, Mizutani F, Terabe K, Kawabe Y, Wada Y. Route of TT virus infection in children. J Med Virol. 1999;59:204–207. doi: 10.1002/(sici)1096-9071(199910)59:2<204::aid-jmv13>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 154.Reference deleted.
- 155.Tabor E, Snoy P, Jackson D R, Schaff Z, Blatt P M, Gerety R J. Additional evidence for more than one agent of human non-A, non-B hepatitis. Transmission and passage studies in chimpanzees. Transfusion. 1984;24:224–230. doi: 10.1046/j.1537-2995.1984.24384225027.x. [DOI] [PubMed] [Google Scholar]
- 156.Tagger A, Donato F, Ribero M L, Binelli G, Gelatti U, Portera G, Albertini A, Fasola M, Chiesa R, Nardi G the Brescia HCC study. A case-control study on a novel DNA virus (TT virus) infection and hepatocellular carcinoma. Hepatology. 1999;30:294–299. doi: 10.1002/hep.510300115. [DOI] [PubMed] [Google Scholar]
- 157.Takahashi K, Hoshino H, Ohta Y, Yoshida N, Mishiro S. Very high prevalence of TT virus (TTV) infection in general population of Japan revealed by a new set of PCR primers. Hepatol Res. 1998;12:233–239. [Google Scholar]
- 158.Takahashi K, Iwasa Y, Hijikata M, Mishiro S. Identification of a new human DNA virus (TTV-like minivirus, TLMV) intermediately related to TT virus and chicken anemia virus. Arch Virol. 2000;145:979–993. doi: 10.1007/s007050050689. [DOI] [PubMed] [Google Scholar]
- 159.Takahashi K, Ohta Y, Mishiro S. Partial ∼2.4 Kb sequences of TT virus (TTV) genome from eight Japanese isolates: diagnostic and phylogenetic implications. Hepatol Res. 1998;12:111–120. [Google Scholar]
- 160.Takayama S, Miura T, Matsuo S, Taki M, Sugii S. Prevalence and persistence of a novel DNA TT virus (TTV) infection in Japanese haemophiliacs. Br J Haematol. 1999;104:626–629. doi: 10.1046/j.1365-2141.1999.01207.x. [DOI] [PubMed] [Google Scholar]
- 161.Takayama S, Yamazaki S, Matsuo S, Sugii S. Multiple infection of TT virus (TTV) with different genotypes in Japanese hemophiliacs. Biochem Biophys Res Commun. 1999;256:208–211. doi: 10.1006/bbrc.1999.0270. [DOI] [PubMed] [Google Scholar]
- 162.Tanaka H, Okamoto H, Luengrojanakul P, Chainuvati T, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Infection with an unenveloped DNA virus (TTV) associated with posttransfusion non-A to G hepatitis in hepatitis patients and healthy blood donors in Thailand. J Med Virol. 1999;56:234–238. doi: 10.1002/(sici)1096-9071(199811)56:3<234::aid-jmv10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 163.Tanaka M, Nishiguchi S, Tanaka T, Enomoto M, Takeda T, Shiomi S, Kuroki T, Otani S. Prevalence of TT virus in patients with fulminant hepatic failure in Japan. J Gastroenterol. 1999;34:589–593. doi: 10.1007/s005350050377. [DOI] [PubMed] [Google Scholar]
- 164.Tanaka Y, Mizokami M, Orito E, Nakano T, Kato T, Ding X, Ohno T, Ueda R, Sonoda S, Tajima K, Miura T, Hayami M. A new genotype of TT virus (TTV) infection among Colombian native indians. J Med Virol. 1999;57:264–268. doi: 10.1002/(sici)1096-9071(199903)57:3<264::aid-jmv9>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 165.Tanaka Y, Mizokami M, Orito E, Ohno T, Nakano T, Kato T, Kato H, Mukaide M, Park Y M, Kim B S, Ueda R. New genotypes of TT virus (TTV) and a genotyping assay based on restriction fragment length polymorphism. FEBS Lett. 1998;437:201–206. doi: 10.1016/s0014-5793(98)01231-9. [DOI] [PubMed] [Google Scholar]
- 166.Tanaka Y, Mizokami M, Orito E, Ohno T, Nakano T, Kato T, Iida S, Ueda R. Lack of integrated TT virus (TTV) genomes into cellular DNA in infected human hematopoietic cells. Leuk Lymphoma. 2000;38:411–417. doi: 10.3109/10428190009087033. [DOI] [PubMed] [Google Scholar]
- 167.Tangkijvanich P, Theamboonlers A, Hirsch P, Kullavanijaya P, Suwangool P, Poovorawan Y. TT virus infection in chronic liver disease. Hepatogastroenterology. 1999;46:1053–1058. [PubMed] [Google Scholar]
- 168.Tischer I, Bode L, Apodaca J, Timm H, Peters D, Rasch R, Pociuli S, Gerike E. Presence of antibodies reacting with porcine circovirus in sera of humans, mice, and cattle. Arch Virol. 1995;140:1427–1439. doi: 10.1007/BF01322669. [DOI] [PubMed] [Google Scholar]
- 169.Todd D, Creelan J L, Mackie D P, Rixon F, McNulty M S. Purification and biochemical characterization of chicken anaemia agent. J Gen Virol. 1990;71:819–823. doi: 10.1099/0022-1317-71-4-819. [DOI] [PubMed] [Google Scholar]
- 170.Toniutto P, Fabris C, Falleti E, Lombardelli T, Gasparini V, Barillari G, Biffoni F, Pirisi M. Evidence against a direct role played by transfusion-transmitted virus infection in causing hepatic or hematologic manifestations. Blood. 1999;93:2426–2427. [PubMed] [Google Scholar]
- 171.Tsuda F, Okamoto H, Ukita M, Tanaka T, Akahane Y, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. Determination of antibodies to TT virus (TTV) and application to blood donors and patients with post-transfusion non-A to G hepatitis in Japan. J Virol Methods. 1999;77:199–206. doi: 10.1016/s0166-0934(98)00154-2. [DOI] [PubMed] [Google Scholar]
- 172.Tuveri R, Jaffredo F, Lunel F, Nalpa B, Pol S, Feray C, Marcellin P, Thibault V, Delagneau J F, Opolon P, Scarpa B, Brechot C, Thiers V. Impact of TT virus infection in acute and chronic, viral- and non viral-related liver diseases. J Hepatol. 2000;33:121–127. doi: 10.1016/s0168-8278(00)80168-3. [DOI] [PubMed] [Google Scholar]
- 173.Ukita M, Okamoto H, Kato N, Miyakawa Y, Mayumi M. Excretion into bile of a novel unenveloped DNA virus (TT virus) associated with acute and chronic non-A–G hepatitis. J Infect Dis. 1999;179:1245–1248. doi: 10.1086/314716. [DOI] [PubMed] [Google Scholar]
- 174.Umemura T, Tanaka E, Ota M, Orii K, Yoshizawa K, Imai H, Sodeyama T, Kiyosawa K. TT virus infection in an area of high-endemicity for hepatitis C. Hepatol Res. 1999;13:212–220. [PubMed] [Google Scholar]
- 175.Utsunomiya S, Yoshioka K, Wakita T, Seno H, Takagi K, Ishigami M, Yano M, Watanabe K, Kobayashi M, Watanabe K, Kishimoto H, Kakumu S. TT virus infection in hemodialysis patients. Am J Gastroenterol. 1999;94:3567–3570. doi: 10.1111/j.1572-0241.1999.01647.x. [DOI] [PubMed] [Google Scholar]
- 176.Verschoor E J, Langenhuijzen S, Heeney J L. TT viruses (TTV) of non-human primates and their relationship to the human TTV genotypes. J Gen Virol. 1999;80:2491–2499. doi: 10.1099/0022-1317-80-9-2491. [DOI] [PubMed] [Google Scholar]
- 177.Viazov S, Ross R S, Niel C, de Oliveira J M, Varenholz C, Da Villa G, Roggendorf M. Sequence variability in the putative coding region of TT virus: evidence for two rather than several major types. J Gen Virol. 1998;79:3085–3089. doi: 10.1099/0022-1317-79-12-3085. [DOI] [PubMed] [Google Scholar]
- 178.Viazov S, Ross R S, Varenholz C, Lange R, Holtmann M, Niel C, Roggendorf M. Lack of evidence for an association between TTV infection and severe liver disease. J Clin Virol. 1998;11:183–187. doi: 10.1016/s0928-0197(98)00060-9. [DOI] [PubMed] [Google Scholar]
- 179.Vihinen-Ranta M, Kakkola L, Kalela A, Vilja P, Vuento M. Characterization of a nuclear localization signal of canine parvovirus capsid proteins. Eur J Biochem. 1997;250:389–394. doi: 10.1111/j.1432-1033.1997.0389a.x. [DOI] [PubMed] [Google Scholar]
- 180.White P A, Li Z, Zhai X, Marinos G, Rawlinson W D. Mixed viral infection identified using heteroduplex mobility analysis (HMA) Virology. 2000;271:382–389. doi: 10.1006/viro.2000.0323. [DOI] [PubMed] [Google Scholar]
- 181.Worobey M. Extensive homologous recombination among widely divergent TT viruses. J Virol. 2000;74:7666–7670. doi: 10.1128/jvi.74.16.7666-7670.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xia X. Data analysis in molecular biology and evolution. Boston, Mass: Kluwer Academic Publishers; 2000. [Google Scholar]
- 183.Reference deleted.
- 184.Yamamoto T, Kajino K, Ogawa M, Gotoh I, Matsuoka S, Suzuki K, Moriyama M, Okubo H, Kudo M, Arakawa Y, Hino O. Hepatocellular carcinomas infected with the novel TT DNA virus lack viral integration. Biochem Biophys Res Commun. 1998;251:339–343. doi: 10.1006/bbrc.1998.9420. [DOI] [PubMed] [Google Scholar]
- 185.Yi C K, Bansal O B, Hong M L, Chatterjee S, Roy P. Sequences within the VP6 molecule of bluetongue virus that determine cytoplasmic and nuclear targeting of the protein. J Virol. 1996;70:4778–4782. doi: 10.1128/jvi.70.7.4778-4782.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Reference deleted.
- 187.Yokozaki S, Toyoda H, Nakano I, Katano Y, Ebata M, Fukuda Y, Takamatsu J, Saito H, Hayakawa T. Infection with TT virus, a novel transfusion-transmissible DNA virus, in haemophiliacs and in blood products. Br J Haematol. 1999;105:1114–1119. doi: 10.1046/j.1365-2141.1999.01452.x. [DOI] [PubMed] [Google Scholar]
- 188.Yoshizawa H, Itoh Y, Iwakiri S, Kitajima K, Tanaka A, Nojiri T, Miyakawa Y, Mayumi M. Demonstration of two different types of non-A, non-B hepatitis by reinjection and cross-challenge studies in chimpanzees. Gastroenterology. 1981;81:107–113. [PubMed] [Google Scholar]
- 189.Yuasa N, Taniguchi T, Yoshida I. Isolation and some properties of an agent inducing anaemia in chicks. Avian Dis. 1979;23:366–385. [Google Scholar]
- 190.Yuki N, Kato M, Masuzawa M, Ishida H, Inoue T, Tabata T, Matsushita Y, Kishimoto H, Sasaki Y, Hayashi N, Hori M. Clinical implications of coinfection with a novel DNA virus (TTV) in hepatitis C virus carriers on maintenance hemodialysis. J Med Virol. 1999;59:431–436. doi: 10.1002/(sici)1096-9071(199912)59:4<431::aid-jmv3>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 191.Zein N N, Arslan M, Li H, Charlton M R, Gross J B, Jr, Poterucha J J, Therneau T M, Kolbert C P, Persing D H. Clinical significance of TT virus infection in patients with chronic hepatitis C. Am J Gastroenterol. 1999;94:3020–3027. doi: 10.1111/j.1572-0241.1999.01457.x. [DOI] [PubMed] [Google Scholar]