Abstract
The chromatin environment plays a central role in regulating developmental gene expression in metazoans. Yet, the ancestral regulatory landscape of metazoan embryogenesis is unknown. Here, we generate chromatin accessibility profiles for six embryonic, plus larval and adult stages in the sponge Amphimedon queenslandica. These profiles are reproducible within stages, reflect histone modifications, and identify transcription factor (TF) binding sequence motifs predictive of cis-regulatory elements operating during embryogenesis in other metazoans, but not the unicellular relative Capsaspora. Motif analysis of chromatin accessibility profiles across Amphimedon embryogenesis identifies three major developmental periods. As in bilaterian embryogenesis, early development in Amphimedon involves activating and repressive chromatin in regions both proximal and distal to transcription start sites. Transcriptionally repressive elements (“silencers”) are prominent during late embryogenesis. They coincide with an increase in cis-regulatory regions harboring metazoan TF binding motifs, as well as an increase in the expression of metazoan-specific genes. Changes in chromatin state and gene expression in Amphimedon suggest the conservation of distal enhancers, dynamically silenced chromatin, and TF-DNA binding specificity in animal embryogenesis.
Embryogenesis occurs in most animals and includes fertilization, activation of the zygotic genome, cell proliferation, cell differentiation, and patterning (Kalinka and Tomancak 2012). Conserved transcription factors (TFs) and signaling pathways, Hedgehog, Notch, TGFB, and Wnt, underlie these processes across metazoan development (Carroll 2008; Levin et al. 2016). Despite this conservation, embryogenesis varies markedly between and within phyla, suggesting that changes in gene expression and regulation are the basis for animal body plan diversification (Wray 2003; Carroll 2008; Kalinka and Tomancak 2012).
The chromatin environment plays a key role in regulating complex developmental programs. Within this environment, cis-regulatory elements, including promoters and enhancers, orchestrate the precise gene expression patterns required for multicellular development. Via their interaction with trans-acting proteins, notably TFs, cis-regulatory elements modulate the chromatin accessibility landscape and define cell states, identities, and developmental fates (Zeitlinger 2020). Where promoters are primarily involved in initiating transcription, enhancers play an essential role in tuning expression in a spatiotemporal context (Long et al. 2016).
Sponges (poriferans) are widely considered one of the earliest branching extant animal phyla. Their body plan is simple: Sponges have no nervous system, muscle cells, or gut. Yet, their regulatory genome and gene repertoire is complex and animal-like. Amphimedon displays an extensive repertoire of noncoding elements, including microRNAs, long noncoding RNAs, and piwiRNAs (Grimson et al. 2008; Gaiti et al. 2017; Calcino et al. 2018). The larval and adult stages possess metazoan regulatory innovations, including distal regulatory elements and bivalent promoters (possessing both activation and repressive histone marks), both of which are not found in the unicellular relative Capsaspora (Bernstein et al. 2006; Bulger and Groudine 2011; Fernandez-Valverde and Degnan 2016; Sebé-Pedrós et al. 2016; Gaiti et al. 2017). Furthermore, despite the lack of primary sequence conservation and the absence of shared cell types, developmental enhancers in conserved microsyntenic regions in Amphimedon drive cell type–specific expression in developing vertebrates (Wong et al. 2020). This last discovery suggests a cis-regulatory grammar arose before the divergence of sponge and vertebrate lineages some 700 million years ago and has been maintained in conserved genomic regulatory blocks.
Advances in sequencing technology have enabled the mapping and characterization of the metazoan gene regulatory landscape during embryogenesis. Transcriptomes have been compared across the development of multiple divergent animal phyla (Levin et al. 2016), and post-translational histone modifications have also been profiled during development in several species (Bogdanović et al. 2012; Schwaiger et al. 2014; Daugherty et al. 2017; Gaiti et al. 2017; Jänes et al. 2018; Domcke et al. 2020; Floc'hlay et al. 2021). With the advent of transposase-accessible chromatin using sequencing (ATAC-seq) (Buenrostro et al. 2015), genome-wide profiling of chromatin accessibility across development can also be undertaken using small amounts of starting material to provide insights into the genomic environment where the transcriptional machinery operates (Daugherty et al. 2017; Sebé-Pedrós et al. 2018b; Esmaeili et al. 2020).
Hence, to study the chromatin dynamics of embryogenesis in an early-branching metazoan phylum, we profiled the chromatin accessibility of Amphimedon queenslandica across eight life stages. We interrogated differentially accessible regions across developmental stages to identify the collection of cis-regulatory motifs underpinning Amphimedon embryogenesis. We integrated chromatin structure and gene expression at matched life stages to characterize developmental dynamics and to infer the regulatory genome of early metazoans. Finally, we tested the ability of Amphimedon chromatin-accessible sequences to predict other species’ developmental cis-regulatory regions using a machine-learning framework.
Results
Dense chromatin accessibility landscapes across Amphimedon embryogenesis
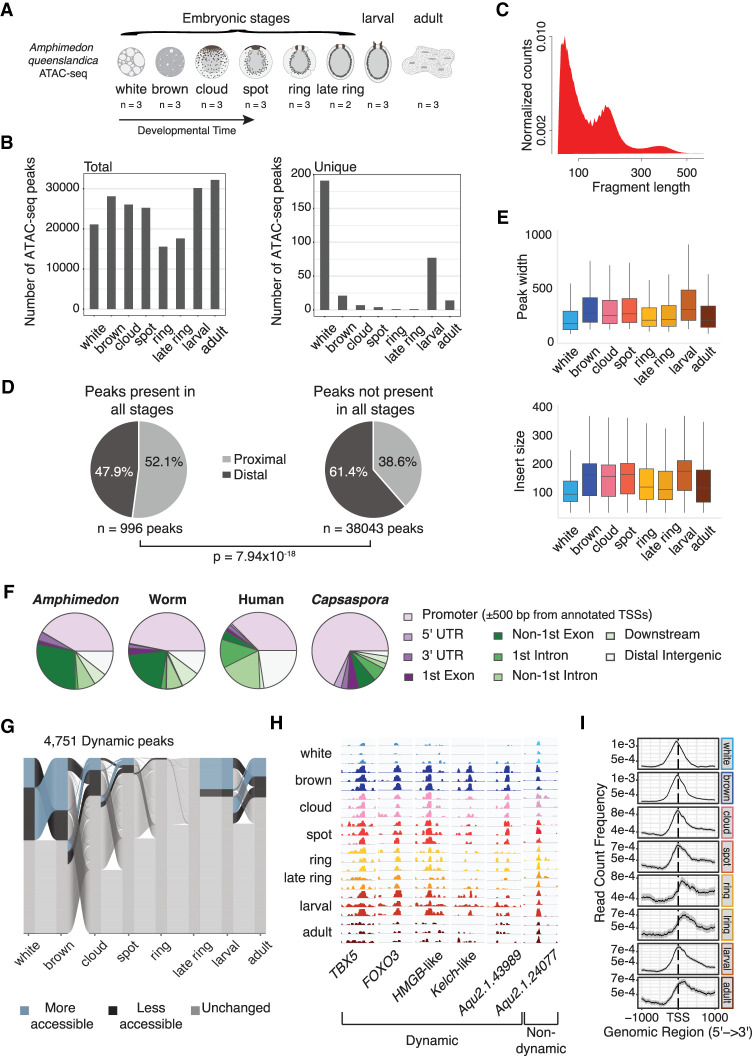
To investigate the genome-wide dynamics of chromatin accessibility during A. queenslandica embryogenesis, we collected individual animals from the following embryonic stages: white, brown, cloud, spot, ring, late ring embryonic stages; planktonic larval and sessile adult stages in triplicate (duplicate in late ring stage). We mapped transposase-accessible chromatin by short-read sequencing (ATAC-seq) across the eight life stages. Amphimedon is a viviparous sponge and embryonic stages occur throughout the year in brood chambers (Degnan et al. 2015). Embryogenesis is staged by the position and pattern of pigment cells in the embryo (Fig. 1A). Early cleavage stages are termed white-stage embryos. At this stage, blastomeres are irregular in size and shape and are mixed with maternal nurse cells. The transition to a two-layer embryo is called the brown stage, which is characterized by dispersed pigments. This is followed by the cloud stage, in which the pigment cells mark the anterior–posterior axis. Subsequently, pigment cells begin to concentrate at the posterior pole defining the spot and ring stages, each with their specific patterns of pigmentation. These stages are also characterized by the appearance of specific cell types.
Figure 1.
Overview of Amphimedon cis-regulatory regions. (A) Amphimedon queenslandica developmental stages. The number of ATAC-seq libraries for every developmental stage is shown. (B) Total and unique number of ATAC-seq peaks by developmental stage. Numbers for each stage calculated using the arithmetic mean across replicates. A peak must have at least a normalized count above 10 in at least one stage. Summary plot across stages required peaks with a mean count per million above zero across all replicates for each stage. (C) Density plot of ATAC-seq fragment length (base pair). (D) Pie charts show the number of proximal (within 500 bp of the TSS) and distal peaks for (1) constitutively open peaks and (2) those not accessible in all developmental stages. χ2 test was used to compute P-value. Accessible peaks across all stages: over zero normalized counts across all libraries. Peaks with varying accessibility in all stages: normalized count over one in three or more libraries. (E) Boxplot of ATAC-seq peak width and insert size. Numbers for each stage calculated using the arithmetic mean across replicates. (F) Distribution of Amphimedon, C. elegans, human, and Capsaspora ATAC-seq peaks across genomic features (Buenrostro et al. 2013; Sebé-Pedrós et al. 2016; Daugherty et al. 2017). Promoter region is defined as a region within 500 bp of the TSS for all species. Downstream is defined as ≤300 bp of the end of a gene. (G) Alluvial plot shows peak dynamics across life stages for peaks that change between at least one life stage (n = 4751) (n = 808, 1422, 334, 202, 3, 0, 851, and 509 more-accessible peaks for white, brown, cloud, spot, ring, late ring, larval, and adult, respectively; n = 1422, 808, 693, 213, 48, 0, 202, and 563 less-accessible peaks for the same stages). Differential accessibility determined by beta-binomial model (Methods). (H) Genome browser view of read coverage at selected dynamically accessible regions (TBX5: Aqu2.1.27488; FOXO3: Aqu2.1.27411; HMGB-like: Aqu2.1.41331; Kelch-like: Aqu2.1.41157; Aqu2.1.43989) and a selected consistently accessible peak (Aqu2.1.24077) across all life stages. (I) Chromatin accessibility read density around the TSS (within 1 kb) by life stage. Peaks were used only if at least 50% of bases overlapped across biological replicates.
Across all life stages, we identified a total of 40,218 nonoverlapping ATAC-seq peaks (P < 1 × 10−5) (Supplemental Table S1). These spanned 32 Mb (20%) of the Amphimedon genome. The median number of peaks was 19,225; the average fraction of reads in peaks across libraries, 23% (Supplemental Table S2). Peak counts were highly correlated among biological replicates (Supplemental Figs. S1, S2). Overlaps to genome-wide chromatin states defined by histone marks (H3K4me3, H3K27ac, H3K27me3, H3K4me1, H3K36me3) and PolII binding in adults (Gaiti et al. 2017), revealed overall enrichment of the ATAC-seq peaks in the active parts of the sponge genome. ∼64% of consensus ATAC-seq peaks matched a nonquiescent region compared with the genome background (42%) (binomial test P = 2.1×10−322, OR > 2).
Of all stages, adults have the higher number of peaks and the highest number of expressed genes, which likely reflects the greater complexity of cell types in adulthood (Fig. 1B; Supplemental Table S3). On the other hand, the highest number of stage-specific peaks was found at the earliest cleavage white stage. We also observed that the typical periodicity of read fragment density, which reflects the regular positioning of nucleosomes, was not observed at this stage in any replicated individual (Fig. 1C; Supplemental Fig. S3). Further analysis suggests that this observation may reflect an abundance of maternal nurse cells, which undergo apoptosis in the early embryo (Eden et al. 2009; Degnan et al. 2015). We found genes proximal to white-stage-specific peaks showed a sixfold enrichment in apoptosis-related pathways (GO term “anoikis,” hypergeometric test, P = 1.3×10−4).
Although the Amphimedon genome is compact and gene dense, where ∼60% of the genome is genic, 61% ATAC-seq peaks were located more than ±500 bp from the transcriptional start sites (TSSs) of coding genes (Supplemental Fig. S4; Fernandez-Valverde and Degnan 2016). Examining distal peaks, we found that they were more dynamically regulated compared with TSS proximal peaks (χ2 test, P = 7.9×10−18, OR = 1.7) (Fig. 1D). This confirms prior results that distal regulatory elements tend to be cell type–specific, whereas promoters are more constitutively accessible across all cell types (Bulger and Groudine 2011; Klemm et al. 2019).
The genomic locations of where ATAC-seq peaks were located corresponded more closely between sponges, worms, flies, and humans than the unicellular organism Capsaspora, potentially reflecting a common metazoan genome organization. For example, ∼40% of metazoan developmental open-chromatin regions were at proximal regulatory regions (Fig. 1E; Supplemental Fig. S5; Sebé-Pedrós et al. 2016). Amphimedon peaks numbers were also comparable to the numbers of cis-regulatory elements identified in other metazoans during development, including Caenorhabditis elegans and zebrafish (Bogdanović et al. 2012; Daugherty et al. 2017; Jänes et al. 2018). Amphimedon ATAC-seq peak widths were also similar to those of the fruit fly and human, ranging between 260 and 538 bp (Fig. 1F; Supplemental Table S3; Sebé-Pedrós et al. 2016).
Despite the rapid evolution of regulatory sequences, we found 436 proximal and 772 distal Amphimedon peaks weakly mapped to the human genome with an overlap of ≥1 bp, potentially suggesting a small degree of regulatory conservation (BLASTN E-value <1 × 10−3). Based on human gene functional term annotation, association with the closest sponge gene revealed these aligned distal peaks were significantly enriched in environmental sensing terms (hypergeometric test, false-discovery rate [FDR] < 7 × 10−8) (Supplemental Table S4).
We next assessed differential chromatin accessibility between consecutive stages to interrogate chromatin dynamics during developmental transitions. We found 4751 peaks that are differentially accessible between consecutive life stage (Methods; Fig. 1G–H). The greatest change in accessibility occurred during early embryogenesis, during the transition between the white and brown stages, consistent with the high number of stage-specific peaks at the white stage (Fig. 1B,G). Focusing on the TSS, we next mapped the density of ATAC-seq reads upstream of and downstream from the TSS region for each stage of development. We found the highest level of accessible chromatin was located immediately before the TSS in the white stage but after the TSS in the ring, late ring, and adult stages (Fig. 1I; Supplemental Fig. S6). At downstream coding regions, overall chromatin accessibility was most reduced in the white stage, suggesting that transcription has not yet been initiated, despite accessible chromatin at the TSS. Based on this, we can also infer that the measured RNA at this stage is likely to be predominantly maternally deposited. An increase in downstream accessibility occurs as embryogenesis progresses, indicating increased transcription in the developing embryo following loss of the maternal nurse cells (Fig. 1H). Consistent with this, many genes that are accessible during early embryogenesis in Drosophila, zebrafish, mouse, and human are not transcribed until later development (Blythe and Wieschaus 2016; Lu et al. 2016; Wu et al. 2016; Pálfy et al. 2020).
Amphimedon embryogenesis involves transcriptionally activating and repressive chromatin
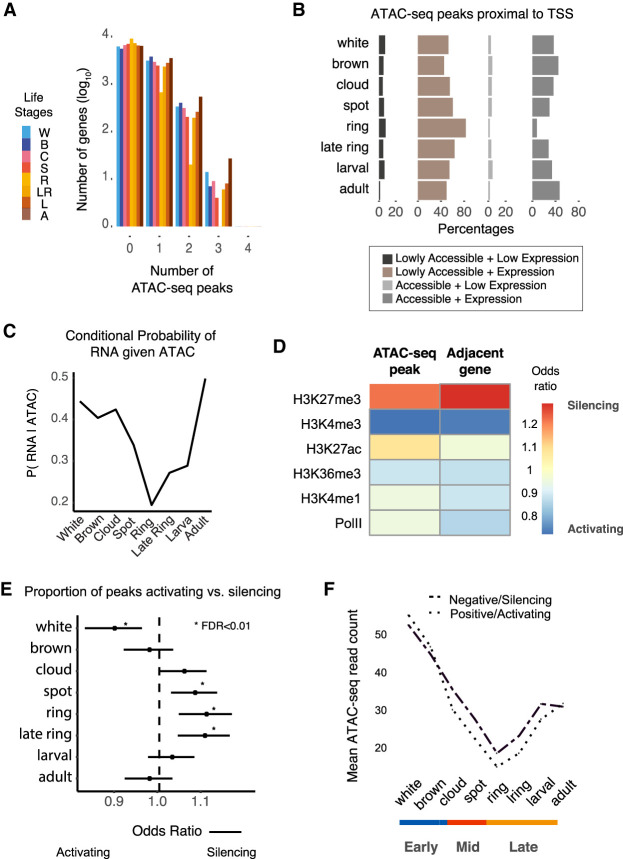
Developmental cell fate decisions involve the interplay between repressive and active interactions. TF and cis-regulatory elements frequently repress genes (Koenecke et al. 2017; Pang and Snyder 2020; Zeitlinger 2020). Hence, open chromatin regions can harbor cis-regulatory elements with either activating or repressive potential (Bernstein et al. 2006; Schoenfelder et al. 2018). To identify potential activating and repressive cis-regulatory elements, we integrated chromatin accessibility with gene expression data by leveraging a comprehensive set of CEL-Seq data for 61 individuals at matched Amphimedon life stages (Levin et al. 2016). Of the 10,766 expressed genes with a median count per million of 10 in at least one stage, 7451 genes (69%) were proximal to at least one ATAC-seq peak within 1 kb of the TSS (Fig. 2A,B). Genes involved in transcription and cell-to-cell communication tend to be adjacent to a higher number of chromatin-accessible regions (Supplemental Fig. S7).
Figure 2.
Interplay between transcription and proximal cis-regulatory elements. (A) Number of cis-regulatory peaks near Amphimedon genes (flanking 1 kb of the TSS). Number of genes was log10-transformed. (B) Percentage of chromatin-accessible/inaccessible peaks near expressed/unexpressed genes for each life stage (flanking 1 kb of the TSS) (expressed genes were defined as those with one or more median cpm in stage; accessible peaks were those with one or more median normalized counts in every stage). (C) Conditional probability of gene expression given proximal ATAC-seq accessibility. (D) Heatmap denotes the ratio between silencer and active sets of peaks and proximal genes overlapping adult Amphimedon histone marks and PolII binding sites. Peak and gene are considered active if there is a positive association between chromatin accessibility and gene expression across time. Silencers are defined as those negatively associated between ATAC-seq peak and gene. (E) Forest plot shows the proportion of peaks that are active versus repressive at each life stage. Fisher's exact tests are used to assess the significance of change relative to the total number of active and repressive peaks identified. Bars denote 95% confidence intervals. (F) Change in average chromatin accessibility read counts for active and silencer peaks.
To examine the stage-specific interplay between chromatin and transcription, we used conditional probability to examine the relationship between the presence/absence of gene expression and the presence/absence of accessible chromatin. Conditional analysis allowed us to account for peak number variation between stages owing to uneven sequencing depth. We found that the probability of gene expression, given the presence of a proximal ATAC-seq peak, was most reduced at ring stages (19% compared with 40%–50% in other stages) (Fig. 2C).
We next integrated chromatin accessibility and gene expression data to establish potential activating and repressive cis-regulatory elements. We took a two-step approach to identify regulatory pairs by associating peaks that were up to 1 kb from an active TSS. We used LASSO regression (Tibshirani 1996) to determine the most informative peaks and classified peaks into either repressive or activating based on the correlation of chromatin regions to the expression of genes across life stages. A cis-regulatory region with increased accessibility with increasing gene expression will positively associate (termed “activating”). On the other hand, a cis-regulatory element correlated to decreased expression would have a coefficient term below zero (termed “repressive,” i.e., increased accessibility at these regions corresponded to lower expression across time). Where only one peak was proximal to a gene, ordinary least squares regression was used. In total, 5254 peaks were positively associated with gene expression, and 3686 peaks were negatively correlated.
To assess whether these assignments of activating and repressive regions were biologically meaningful, we overlapped the regions to histone marks profiled in adult Amphimedon. Attesting to our overall ability to distinguish between cis-regulatory regions with opposing regulatory function, we found a high correspondence between inferred gene activity based on histone marks and peak classification. Active chromatin marks in adults, particularly H3K4me3, were highly enriched at positively correlated peaks and genes proximal to these regions. On the other hand, the repressive polycomb-mediated H3K27me3 mark, also profiled in adults, was enriched at negatively correlated peaks, consistent with H3K27me3 regions marking silencers (Fig. 2D; Cai et al. 2021). Linking this corroborating information to developmental stages, we saw a shift toward increased silencing at mid and late development, whereas activating peaks were most prevalent during early embryogenesis and adulthood (Fig. 2E,F). In line with this, we found increased accessibility at motifs of RE1-silencing transcription factor (REST) during late development (FDR < 0.01; Methods) (Supplemental Table S5), where regions bound by REST have been associated with the H3K27me3 mark during the differentiation of murine neuronal cells (Arnold et al. 2013).
In summary, by integrating chromatin accessibility and gene expression, our results suggest that transcriptional repression plays a crucial role in dynamically controlling Amphimedon developmental gene expression. The repressive chromatin marks, H3K9me3 and H3K27me3, are lacking in some unicellular organisms (Sebé-Pedrós et al. 2016), supporting the notion that transcriptional control of development through repressive elements is a critical component of multicellularity.
Human TF binding motifs separate developmental transitions in Amphimedon
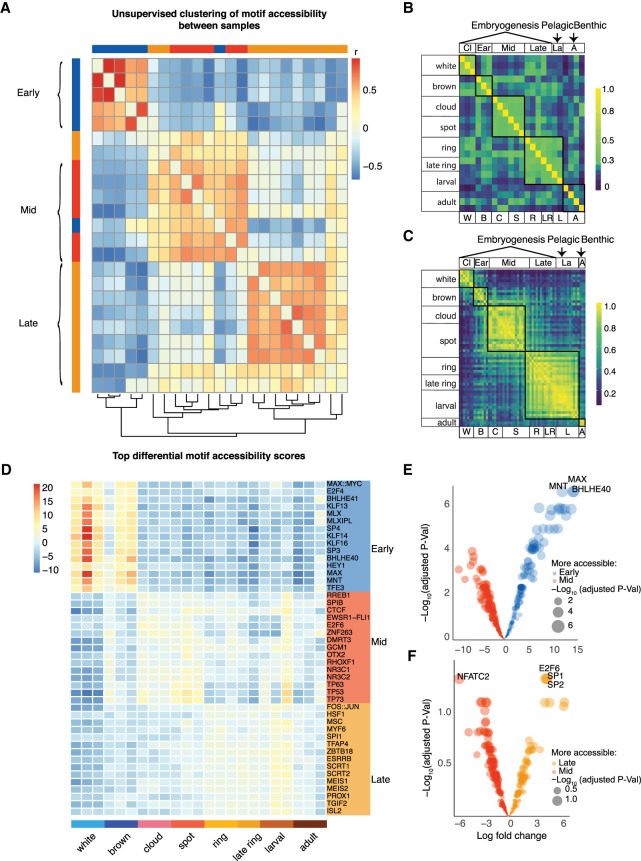
To elucidate the dynamic changes in DNA sequence associated with developmental gene expression, we used position-weighted matrices (PWMs) to search for TF binding motifs underlying accessible chromatin. In our use of PWMs, we leveraged the fact that TF gene families are highly conserved (Nitta et al. 2015; Kribelbauer et al. 2019), and used mammalian matrices to identify known motifs. To assess motif enrichment for each time point, we combined motif alignment scores and ATAC-seq counts to measure the accessibility of each motif for each library (n = 386 PWMs) (Methods; Supplemental Table S5). A set of background peaks matched for GC content and average accessibility was used to assess motif enrichment.
Unsupervised clustering of motif accessibility scores clustered the libraries into three major groups, recapitulating the groupings by developmental trajectory based on both peak counts and gene expression (Fig. 3A–D). We identified 85, 17, and 74 up-regulated differential accessible motifs at the early, mid, and late developmental stages, respectively (adjusted P-value < 0.05), with the greatest changes, both in terms of significance and total number of motifs, occurring in early embryogenesis (Fig. 3E,F). Top motifs at early embryonic stages were associated with TFs linked to stem and cancer cell states, including MAX, E2F4, and Kruppel like factors (KLFs). Motifs enriched during mid-embryogenesis included RREB1, tumor suppressor TP53, and glucocorticoid receptor NR3C1. Late development showed enrichment for the FOS::JUN dimer, MEIS, and ISL motifs. To explore regulatory motifs underlying accessibility dynamics that did not correspond to known motifs, we performed de novo searches for 8-mers and identified differentially enriched accessible regions between sponge developmental stages (Supplemental Table S6; Supplemental Fig. S8). We identified 32,896 enriched 8-mers across consensus peaks relative to a random background set of matched GC content and an average number of fragments across all stages (Methods). Of these, 12,523, 6080, and 7277 8-mers were differentially accessible between early, mid, and late development, respectively (FDR < 0.05). We searched for similarities of the top six 8-mers for each stage, ranked by statistical significance (n = 18 motifs), against JASPAR PWMs. Only six of these 18 8-mers showed discernible similarities to known PWMs (q-value < 0.5) (Supplemental Table S7).
Figure 3.
Motif analyses define three major Amphimedon developmental transitions. (A) Heatmap of Pearson correlation for each library based on TF motif deviation scores. Hierarchical clustering of rows and columns was performed (Methods). (B) Hierarchical clustered heatmap of Pearson correlation of the most variable 2000 peaks based on log10-transformed ATAC-seq read counts across life stages. (C) Hierarchical clustered heatmap of the most variable 1000 genes based on expression (counts per million) across life stage. (D) Heatmap of the top differentially enriched motifs (right) based on motif accessibility, where heatmap values represent motif deviation z-score (Methods). (E) Volcano plot of differentially enriched motifs between the early and mid stages. (F) Volcano plot of differentially enriched motifs between the mid and late stages. Each colored dot represents a motif, and the size of the dot is relative to the −log10(q-value). The x-axis is the log fold change. (Early) White and brown; (Mid) cloud, spot; and (Late) ring, late ring, and larval.
TFs are known to interact cooperatively or competitively in binding to DNA, and enhancers that harbor different TF binding sites show more activity than those with a single binding site (Zinzen et al. 2009; Smith et al. 2013). Hence, we examined the motif colocalization among the top 10 most differentially accessible JASPAR motifs at each Amphimedon developmental stage in a pairwise manner. Early highly significant colocalized TF motifs include MNT & SP4 and SP4 & BHLHE40 (z-score > 10) (Supplemental Table S8). Amphimedon orthologs for these genes include an ortholog to MNT, a member of the MYC/MAX/MAD network (Aqu2.1.41999), and two SP4-like orthologs (Aqu2.1.26963, Aqu2.1.26964). No one-to-one ortholog to human BHLHE40 has been identified, although the Amphimedon genome contains multiple bHLH genes, including ARNTL, which is a core component of the circadian clock in mammals that interacts with BHLHE40 (Aqu2.1.29954, Aqu2.1.05065).
We further examined TF cooperatively and antagonism by calculating the correlation coefficient of motif pairs across life stages. In contrast to testing for motif co-occurrence above, this measured the correlation of motif accessibility across development. A positive correlation suggests the binding proteins are active at the same developmental stage even though the proteins may not directly interact at the same locus. A negative correlation coefficient suggests the binding proteins are enriched at different developmental times. As expected, for the top 10 most differentially accessible motifs at each stage, motif pairs were generally strongly positively correlated, suggesting their cognate TFs were active at the same stage and may cooperate in similar molecular processes (Supplemental Table S9). For example, YY1 and KLF13/14 motifs were both highly accessible during early embryogenesis (Pearson rho = 0.9). Incidentally, these motifs also co-occurred in the same peak, suggesting their cognate proteins may interact (OR = 2.30). In contrast, a strong negative correlation was apparent between YY1 and TP53 (Pearson rho = −0.7), where YY1 was most accessible during mid-embryogenesis when TP53 was low. Consistent with this, YY1 negatively regulates TP53 and has dual activator and repressor function in other animals (Sui et al. 2004).
In summary, we find chromatin-accessible regions harbor specific combinations of TF motifs in a developmental stage-dependent manner. These relationships in Amphimedon can be inferred using human TFs profiles, revealing a potential deep conservation of TF binding–DNA specificity.
Up-regulation of metazoan TFs during late embryogenesis
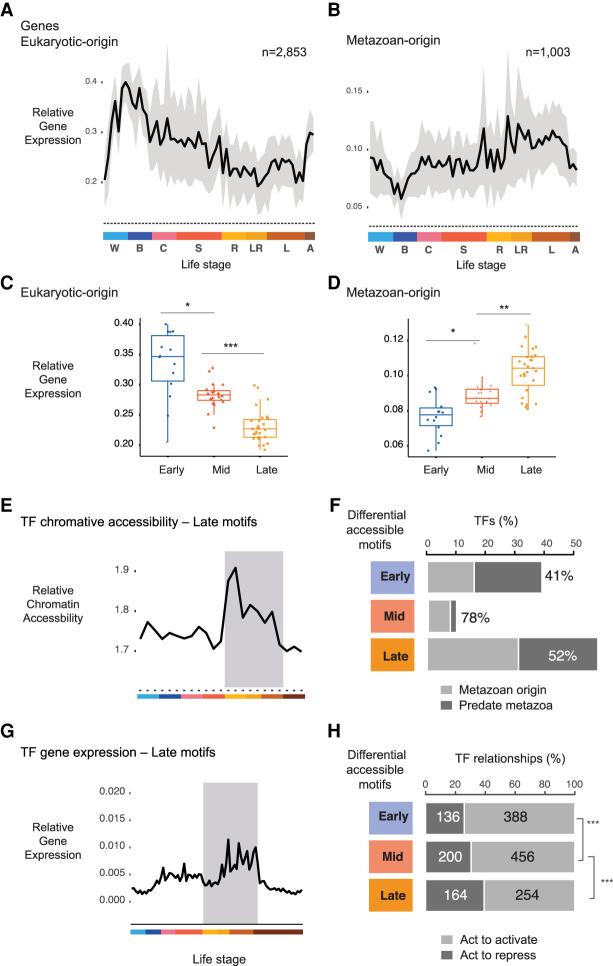
In zebrafish and fly, gene expression during development shows strong phylogenetic signatures corresponding to gene age (Domazet-Lošo and Tautz 2010). We asked whether a similar evolutionary pattern for gene expression and cis-regulatory regions exists in the sponge. To this end, we combined information on the phylogenetic age of Amphimedon genes with our time-series expression and chromatin accessibility data. We used 4967 expressed sponge genes that mapped to human using the TreeFam database; 2853 were of eukaryotic origin, 1003 of metazoan origin, and 1110 originated earlier at the opisthokonts (Li et al. 2006). Expression values were grouped by the age of the associated gene and normalized to total expression (Methods). Genes of eukaryotic-origin initiated early in Amphimedon development, whereas metazoan-specific genes dominated the expression profile as development progressed (Fig. 4A–D). A similar increase in the expression of metazoan-specific genes as development progressed has been reported in fly and zebrafish (Domazet-Lošo and Tautz 2010).
Figure 4.
Metazoan TF motifs are enriched in late development. (A) Relative expression values for sponge genes that can be traced to a eukaryotic ancestor. Gray area denotes 95% CI as determined by bootstrapping. Color denotes life stages. (B) Relative expression values for Amphimedon genes traced to the metazoan stem. Relative gene expression of transcription factors of eukaryotic origin (C) and metazoan origin (D) with binding motifs enriched in early, mid, and late Amphimedon development. (*) P < 0.01, (**) P < 0.001, and (***) P < 0.0001 (Mann–Whitney U test). (E) Relative chromatin accessibility values at TFs whose binding motifs are enriched in late Amphimedon development. Color denotes life stages as in A and B. (F) Bar plots show the number of TFs whose motifs are differentially enriched for each stage, grouped to whether the TF originated in metazoan stem versus those that predate metazoan (based on the TreeFam Amphimedon vs. human comparison). Percentage denotes TFs from the metazoan stem. (G) Relative gene expression of transcription factors with binding motifs enriched in late Amphimedon development. Color denotes life stages as in A and B. (H) Bar plots depict the number of activating versus repressive genes based on human database TRRUST (Han et al. 2018). Genes for each stage are TFs associated to stage through differential motif analyses. Numbers denote the number of unique interactions found for that gene in the activatory or repressive category. (***) P-value significance from Fisher's exact tests: early versus late, P = 2 × 10−5; mid versus late, P = 4 × 10−3.
Next, in a similar manner, we tested whether this trajectory is reflected by chromatin accessibility. Here, high scores implied increased accessibility at the promoters of younger genes, whereas low values reflected the accessibility at promoters of more ancient genes. Consistent with gene expression, the cis-regulatory elements of metazoan genes became increasingly accessible during late development (Fig. 4E). Late developmental peaks were highly enriched for motifs from metazoan genes but not genes whose origin predate metazoans (eukaryotic or opisthokont; Fisher's exact test P = 2 × 10−8, OR = 4.6) (Fig. 4F). In contrast, early developmental peaks were enriched for motifs of ancient/eukaryote-specific genes (OR = 1.9) (Fig. 4F). Reassuringly, TFs linked to motifs we previously identified at late developmental peaks were also more highly expressed during late development than in other stages (Fig. 4G). We further sought to determine whether there were differences in regulatory mechanisms among the TFs of differentially accessible binding motifs across development. We used TRRUST, a curated database of gene–gene interactions, to infer the mode of action (either activating or repressive) between the TFs that were associated with the 15 most differentially accessible motifs for each developmental stage and their associated gene targets (Han et al. 2018). Over 50% of the peaks containing these motifs can be associated with an expressed gene (within 1 kb of the TSS). TFs of motifs enriched in late development were significantly more likely to show a repressive mode of action than early and mid-stage TFs (Fisher's exact test, P = 6.4 × 10−5, OR = 1.6) (Fig. 4H).
Taken together, we identified increased metazoan gene expression and chromatin accessibility at promoters as embryogenesis progresses. Motifs at chromatin-accessible regions during late development corresponded to highly expressed metazoan TFs enriched for repressive function.
Amphimedon cis-regulatory motif composition distinguishes developmental cis-regulatory elements
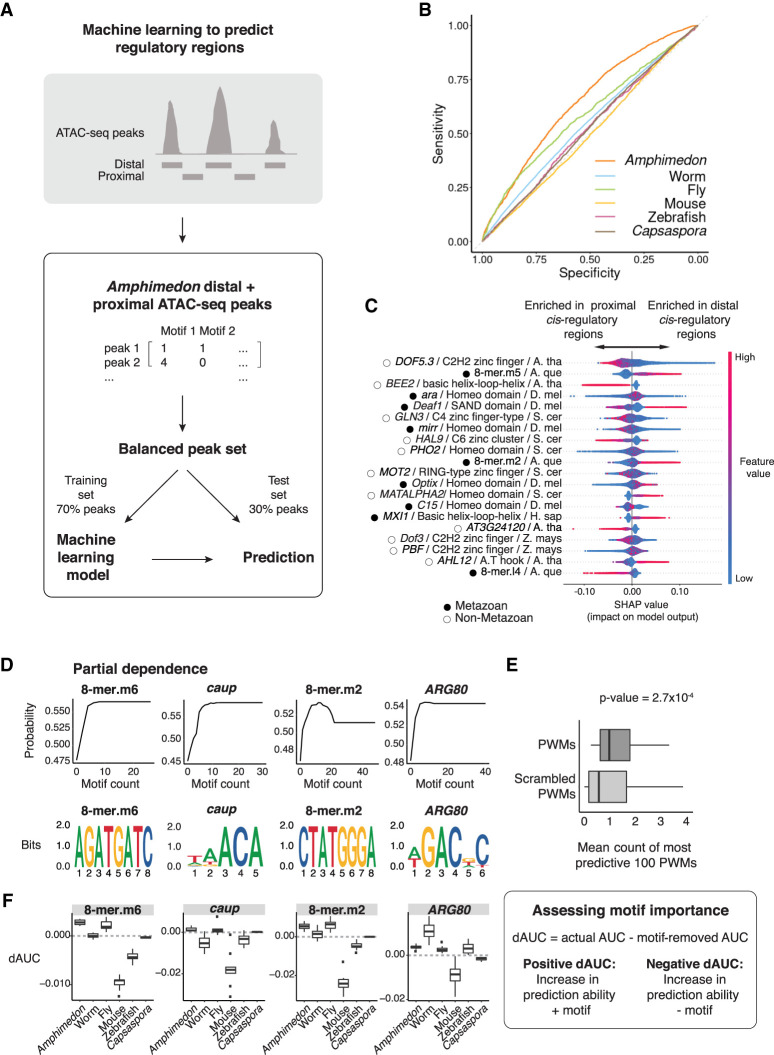
To further dissect the sequence basis for our chromatin-accessible regions, we used a machine learning framework to test whether TF binding sequence motifs could distinguish between proximal and distal Amphimedon peaks (Methods; Fig. 5A; Supplemental Fig. S9). We generated 10 balanced data sets for model training and testing, where for each data set, a matrix was constructed with rows as ATAC-seq peaks and columns as JASPAR CORE PWMs and the 18 developmental stage–enriched de novo 8-mers identified above. The matrix was populated using motif counts and used as input to the XGBoost gradient boosted decision trees algorithm (Methods). Results showed accuracies of ∼0.6 in classifying between proximal and distal Amphimedon peaks (distal defined as >1 kb upstream of TSS) (Fig. 5B; Supplemental Table S10). Motif importance scores were highly correlated among the 10 randomly subsampled balanced sets, suggesting robustness to peak selection (Supplemental Tables S11, S12). Proximal and distal regions showed representative differences, reflecting sequence differences between promoter and distal regulatory regions (Fig. 5C). Promoters were best explained by plant TF motifs, and these were often associated with environmental sensing. For example, the g-box motif (CACTG) is present at light responsive genes (Fig. 5C; Shen et al. 2008). In contrast, metazoan TF motifs (e.g., Optix, Deaf1, MXI1) were only predictive at distal regions, further supporting the idea that distal enhancers are metazoan specific (Fig. 5C; Sebé-Pedrós et al. 2016).
Figure 5.
A machine learning model trained on Amphimedon motifs distinguishes between proximal versus distal cis-regulatory elements. (A) An extreme gradient boosting (XGB) machine was used on a balanced data set of Amphimedon distal and proximal cis-regulatory regions. Seventy percent of peaks was used to train an XGB model. Motif counts for each peak were used to predict distal versus proximal cis-regulatory regions in the Amphimedon test sets (30% of peaks) and other species data sets. Distal was defined as >1 kb upstream of the TSS. (B) Receiver operating characteristic (ROC) curve of distal versus proximal cis-regulatory regions prediction in Amphimedon. (C) Relative importance score (SHAP) of most predictive known motifs and 8-mers for Amphimedon distal versus proximal cis-regulatory regions. Motifs are ordered according to their importance. Every dot in the model represents a peak used for the training of the XGB model. The SHAP values (x-axis) show the impact on the prediction. Color reflects the motif count in the peak. The name, class, and species of motifs are indicated: (S. cer) S. cerevisiae, (A. tha) A. thaliana, (D. mel) D. melanogaster, (A. que) A. queenslandica, (Z. mays) Z. mays, (H. sap) H. sapiens. Metazoan and nonmetazoan TFs are indicated with black-filled and black-outlined circles, respectively. (D, top row) Partial dependence of top four most predictive PWMs of distal cis-regulatory regions (compared with genomic background), showing the relationship between the number of instances of the motifs and the probability of a region being an actual ATAC-seq peak. (Bottom row) Sequence logos of the motifs shown in D, top row. (E) Boxplots of the mean number of instances of the top 100 most predictive actual PWMs (dark gray; compared with genomic background) and permuted PWMs (light gray), Mann–Whitney U-test P-value shown (estimate = 0.41). Outliers removed from the plot and defined as values smaller than 1Q − 1.5 × IQR or bigger than 3Q + 1.5 × IQR, where “1Q” is the 1st quartile, “3Q” represents the 3rd quartile, and IQR (interquartile range) is the difference between the 3Q and 1Q. (F) Schematic of the process to calculate dAUC as a measurement of the effect of individual motifs on distal cis-regulatory regions prediction ability (right); dAUC values for the motifs shown in D (left).
Furthermore, motifs of key developmental metazoan TFs were significantly overrepresented at cis-regulatory regions compared with a genome-wide background, including GATA-type, homeodomain, and Forkhead domain factors (Supplemental Table S13). Among TF families, the TALE homeodomain superclass was overrepresented at cis-regulatory regions (Fisher's exact test, FDR = 4 × 10−2) (Supplemental Fig. S10), including the MEIS1 motif, which was differentially accessible throughout late Amphimedon embryogenesis (AmqTALE, Aqu2.1.41527) (Larroux et al. 2008).
Finally, we investigated whether models trained on Amphimedon regions could classify similar developmental regulatory elements in other species, including in worm, fruit fly, mouse, zebrafish, and nonmetazoan unicellular organism, Capsaspora (Bogdanović et al. 2012; Sebé-Pedrós et al. 2016; Daugherty et al. 2017; Pijuan-Sala et al. 2020; Floc'hlay et al. 2021). Negative sets were constructed controlling for cis-regulatory region size for each species’ genome (Supplemental Fig. S11). Again, motif importance scores were highly correlated among models trained on data subsets (Supplemental Tables S14–S17). Models trained on motifs identified at Amphimedon regulatory regions could weakly predict both proximal and distal developmental cis-regulatory regions from the respective genome background in worm, mouse, zebrafish, and fruit fly, but not Capsaspora (area under ROC: 0.6–0.7 metazoan, 0.5 for Capsaspora) (Supplemental Figs. S12, S13; Supplemental Tables S18, S19). Partial dependence (PD) plots reveal the relationship between motifs enrichment and their effect on the model's output (Fig. 5D). Comparing the number of JASPAR motifs per regulatory peak to scrambled PWMs showed that actual motifs were significantly more abundant than motifs identified using scrambled PWMs, suggesting that PWMs identified biologically important information content, independent of overall motif nucleotide composition (Mann–Whitney U test, P < 2.7 × 10−4; top 100 most informative PWMs) (Fig. 5E; Supplemental Fig. S14). Normalizing across all data sets to adjust for species-specific differences in peak widths did not change the overall findings (Methods; Supplemental Fig. S15). Systematically removing each motif and recalculating prediction scores for each species revealed the most informative motifs to distinguish distal Amphimedon cis-regulatory regions from background were also informative of regulatory elements in other species (Fig. 5F; Supplemental Fig. S16).
Taken together, the number and type of TF motifs could distinguish between proximal and distal Amphimedon regulatory regions. Metazoan cis-regulatory regions across several species can be identified using a machine learning model trained on sponge cis-regulatory regions. Although Amphimedon regulatory regions possessed motifs of TFs key to metazoan development, some of the most informative motifs at ATAC-seq regions originate from nonmetazoan species.
Discussion
We mapped the cis-regulatory dynamics of Amphimedon embryogenesis using ATAC-seq to assay accessible chromatin in individual embryos, larvae, and adults. Chromatin accessibility was dynamically regulated across sponge embryogenesis, and changes in accessibility were concordant with changes in gene expression. Despite the compactness of the Amphimedon genome, the regulatory landscape was dominated by distal (>500 bp from the TSS) open chromatin regions. We identified transcriptionally repressive elements indicative of repressive elements (“silencers”), which are characteristic of development in other animals although less well studied compared with elements that activate transcription (Liu et al. 2016; Koenecke et al. 2017; Pang and Snyder 2020). Confirming a key role of gene repression to early development, we established that Amphimedon repressive elements were increased in activity during late embryogenesis, coinciding with the expression of metazoan-specific genes as well as increased cell complexity. Consistent with this, distal regulatory elements and the repressive histone mark H3K27me3 are not found in the unicellular organism Capsaspora and thus appear to be animal-specific innovations (Sebé-Pedrós et al. 2016).
Despite the rapid evolution of cis-regulatory elements, the repertoire of TF binding motifs at Amphimedon regulatory regions showed clear similarities to bilaterian animals. Sponge developmental transitions were well described by human PWMs, despite many human TFs lacking one-to-one orthologs in Amphimedon. In line with this, studies of TFs from the same structural family have been shown to recognize similar DNA sequences despite evolutionary divergence; the expanded repertoire of TFs in humans compared with fruit flies does not appear to have generally produced new TF specificities (Nitta et al. 2015; Kribelbauer et al. 2019). Thus, our finding supports the notion that structural constrains may limit both TF family diversity and binding specificity and, consequently, the evolution of markedly different TF binding sites. Yet, PWMs of 40 bilaterian TFs were not found in Amphimedon (Supplemental Table S20), raising the possibility that new TF-DNA specificities may have played a role in the emergence of bilaterian gene regulatory networks. However, we cannot rule out that changes to the arrangement and context of motifs (i.e., sequence grammar) may be sufficient mechanisms for cellular and phenotypic innovations.
Along this line, we showed that a machine learning model trained with the genome-wide collection of Amphimedon TF binding motifs (i.e., a bag-of-motif) at ATAC-seq peaks could distinguish accessible chromatin regions in other metazoans but not in the unicellular Capsaspora. The findings suggest a potential divergence in the overall composition of regulatory sequences that can be traced back to the evolutionary divergence of metazoans and nonanimals. However, a genome-wide model cannot capture specificities of individual enhancer–promoter connections in a locus-specific manner. This is an inherent limitation, which may, in part, explain why we did not observe stronger predictions.
Notably, some of the most informative sequences in discriminating between ATAC-seq regions and background were found enriched in the genome-wide background. This is likely due to mechanisms, such as DNA repair, that differ in efficiency between active and closed chromatin, resulting in mutational biases.
Certain overrepresented 8-mers in regulatory chromatin regions showed a superior ability to distinguish developmental enhancers between metazoans compared with known motifs, suggesting that there may be additional yet uncharacterized sequences for determining genome accessibility during development (Fig. 5C; Supplemental Tables S11, S14, S16). Because of their low sequence similarity to previously characterized motifs, these sequences may influence other determinants of chromatin accessibility rather than bind canonical TFs. For example, motifs influencing DNA shape, nucleosome positioning, and DNA methylation make significant contributions to enhancer grammar (Barozzi et al. 2014; Domcke et al. 2015; Levo et al. 2015; Soufi et al. 2015).
In conclusion, by profiling the chromatin landscape in Amphimedon, a representative of an early-diverging metazoan lineage, we show that the ancestral regulatory landscape of metazoan embryonic development consisted of dynamic and abundant distal nongenic regions that contain shared developmental cis-regulatory motifs. Given conserved TF-DNA specificity, we suggest these TF motifs cooperatively modulate gene expression networks that are reused and rewired in the evolutionary acquisition of morphological diversity in metazoans. However, how these cis-regulatory elements diverge in sequence and in function and contribute to new gene regulatory networks and higher-order phenotype remain to be fully investigated. We anticipate that advances in single-cell multimodal data and statistical/machine learning will provide new insights by tracing how gene regulatory processes have evolved during cell and developmental evolution.
Methods
Temporal profiles of Amphimedon chromatin accessibility
Adult A. queenslandica were collected from Shark Bay, Heron Island, Great Barrier Reef, Queensland, Australia (23°26′37.92′′ S, 151°55′8.81′′ E) under Marine Parks permit number G16/38120.1 Sponges were transferred to a closed aquaria system at the University of Queensland, Brisbane, Australia, and maintained as previously described (Degnan et al. 2008). Adult brood chambers were dissected, and embryos were staged according to the method of Adamska et al. (2007). Larvae naturally released from sponges were collected before dissecting brood chambers (Leys et al. 2008). Cell suspensions from adult sponges were generated by cutting away externally facing tissue, avoiding any potential contaminating tissues, and washing three times in 0.2 µm filtered artificial seawater (FSW). Cleaned adult tissue was transferred to 0.2 µm filtered Ca+ and Mg+ free artificial seawater (CMFSW) and passed through a 20-µm nylon mesh to generate a single-cell suspension (Sebé-Pedrós et al. 2018a).
Dissected individual embryos were placed in CMFSW and loaded into a P1000 pipette tip, which had been cut and fitted with sealed 60-µm mesh at the end. This allowed a small volume of CMFSW to be used to push the embryo through the mesh using a hand-held pipette and the resulting cell suspension to be collected into a 1.7-mL centrifuge tube. To collect any tissue stuck on the mesh, the filter tip was then placed in 1× trypsin (Sigma-Aldrich)/CMFSW solution and placed for 10 min at 37°C. This was washed twice with CMFSW and added to the initial cell suspension. The above procedure was performed with larvae and the resulting cell suspensions manually counted with a hemocytometer following trypan blue exclusion staining. A single larva was estimated to have approximately 35,000 cells. Cell suspensions from adult tissues were also counted using trypan blue staining, and an equivalent number of cells was used to make adult libraries.
Cell suspensions from individual embryos and adult samples were centrifuged at 500g for 5 min, and ATAC-seq libraries were made according to the method of Buenrostro et al. (2013). Libraries using the unique primers were amplified for fewer than seven additional cycles. The quality of each individual library was assessed using a Bioanalyzer high-sensitivity DNA analysis kit (Agilent). Equal volumes of libraries from three individual replicates for each stage were then pooled and run again on the Bioanalyser. Because libraries appeared to have a large concentration of free primer, we incorporated an additional purification step by adding a 1.8× concentration of AMPure beads (Beckman Coulter), washed twice with 70% ethanol, and eluted in 40 µL water. A 30 nM pooled library was prepared for sequencing.
The ATAC-seq library pool was quantified on the Agilent Bioanalyzer with the high-sensitivity DNA kit (Agilent Technologies 4067-4626). The pool is assessed by qPCR using the KAPA library quantification kit–Illumina/Universal (KAPA Biosystems KK4824) combined with the Applied Biosystems ViiA 7 real-time PCR instrument. Pool QC and sequencing were performed at the Institute for Molecular Bioscience Sequencing Facility (University of Queensland) using the Illumina NextSeq 500 (NextSeq control software v2.0.2/Real-Time Analysis v2.4.11). The library pool was diluted and denatured according to the standard NextSeq protocol and sequenced to generate paired-end 76-bp reads using a 150 cycle NextSeq 500/550 high output reagent kit v2 (catalog no. FC-404-2002). After sequencing, FASTQ files were generated using bcl2fastq2 (v2.17; Illumina).
ATAC-seq read alignment and peak calling
Each ATAC-seq library was sequenced to a mean depth of approximately 12 million reads. Reads were aligned using Bowtie 2 (v2.3.0) (Langmead and Salzberg 2012) to the Aqu1 genome with Aqu2.1 gene annotations (Fernandez-Valverde et al. 2015). Correctly mapped paired reads and those above a MAPQ value of 10 were retained using SAMtools “view” (v1.6) (Li et al. 2009) for peak calling. Peak calling was performed using MACS2 (Zhang et al. 2008, 2) using the “callpeak” command, a genome size of 1.2 × 10−8, and in -BAMPE mode where insert sizes of read pairs were inferred using alignment results. A set of consensus variable-sized but nonoverlapping peaks was created for downstream analysis. Peaks counts reflect the number reads mapping at each peak. To remove technical compositional biases that can manifest from variable read coverage, we scaled each library based on the expected mean counts before analysis. Normalization for composition biases was performed using size factors calculated with the genomic mean for each peak across all libraries using the R package “DESeq2” (Love et al. 2014; R Core Team 2020). To select the most robust peaks for downstream analysis, we only kept those peaks with 10 or more normalized counts in at least three libraries. BEDTools (v2.28.0) was used to calculate FRiP scores (Quinlan and Hall 2010). Irreproducible discovery rates (IDRs), were calculated following ENCODE (Li et al. 2011). We used conditional probabilities to understand the relationship between expression and chromatin accessibility to account for potential technical differences in peak number between stages. We used the arithmetic mean to average the number of counts across biological replicates for each life stage for summary figures associating peaks to gene expression.
Differential accessibility analysis
We performed differential accessibility analyses of peaks using R packages “edgeR” and “voom/limma” (Robinson et al. 2010; Ritchie et al. 2015; R Core Team 2020). Normalization for composition biases was performed using factors calculated as described above. Peaks with low counts were removed from the analysis by filtering out peaks with less than one normalized count per million reads mapped in less than four samples; our design matrix tests for differences between adjacent life stages. The “voom” function was used to calculate precision weights to remove heteroscedasticity from the data. Counts were modelled by “lmFit” using empirical Bayes moderation to improve the precision of peak variability. A log fold change cut-off of one and FDR < 0.05 were used to define significantly changed peaks.
Integrative correlative analysis with gene expression
To understand how accessible chromatin and transcription were linked during development, we associated genes with proximal ATAC-seq peaks located in the vicinity of TSSs (1 kb bases upstream and downstream). In linear regression across life stages, we use ATAC-seq peaks as independent variables and gene expression data as the dependent variable to identify activating and repressive cis-regulatory regions. If multiple peaks are adjacent to a TSS (within 10 kb), a LASSO regression (Tibshirani 1996) was performed to determine the most informative peak using the R package “glmnet.” Otherwise, ordinary least squares regression was used. CEL-Seq and ATAC-seq data were averaged by arithmetic mean among stages and log10 + 1 transformed. The direction of association was used to class peak–gene relationship as either activating or repressive, with a positive coefficient indicative of activating and a negative coefficient suggestive of the binding of factors repressing expression.
Gene evolutionary history analysis
To examine the connection between development and gene phylogeny, we partitioned expressed genes (at least three CEL-Seq libraries with count >10) by their estimated evolutionary age using TreeFam (v9) (Li et al. 2006), where gene orthology was assigned by phylogenetic analysis. Genes were placed into the following phylogenetic clades: Eukaryota, Opisthokont, and Metazoan, reflecting the oldest clade that founders of the gene can be traced to, where possible based on orthology assignment. For each phylogenetic group and across developmental data sets, we calculate a relative measure of gene expression by taking the sum of all counts of genes classified to the same evolutionary origin and then normalizing this by the sum of total expression values across all genes. This value was bootstrapped 500 times to generate a confidence interval.
where g represents the genes (i) in the evolutionary clade and n represents all the genes (i) expressed in the library.
Motif analysis
Motif analyses using both known and de novo sequence motif were performed using the R package “chromVAR” (v3.11) (Schep et al. 2017; R Core Team 2020). The consensus set of nonoverlapping peaks was used. Known human motifs from JASPAR 2016 database (Mathelier et al. 2016) were used with the default cut-off of motif calling of 5 × 10−5. For each library, an accessibility score was calculated for each motif. This is the dot product across peaks of the motif score—calculated by the match of the motif PWM and the peak accessibility score—derived from the normalized read count. Technical biases, including GC content and average background accessibility, were controlled using matched sampled peaks of similar properties using the “getBackgroundPeaks” function in chromVAR. A deviation score was computed, reflecting the accessibility of peaks with that motif by subtracting the expectation based on background (Schep et al. 2017). Comparison of the deviation score between three broad developmental stages (early, mid, and late) used moderated t-tests with FDR P-value adjustments for multiple testing (Benjamini and Hochberg 1995). Using chromVAR, we also characterized de novo sequence motifs, k-mers of length 8, within chromatin-accessible regions. Deviation scores for k-mers are computed and compared between developmental stages using the “differentialDeviations” function and a two-sided t-test with FDR adjustments. The top 10 most-deviated JASPAR motifs and top six k-mers for each developmental stage were used for downstream analyses. The TF coaccessibility analysis was performed using the “getAnnotationSynergy” function in the chromVAR package, which assigns a z-score to each motif pairing, reflecting variability in peak accessibility of peaks containing both motifs compared with a background sample containing only one of the two motifs. To further elucidate the relationship between motifs at accessible peaks, we examined peaks where the motif pairs did not colocate. To do this, we calculated the Pearson correlation coefficient of the normalized accessibility (deviation score) across samples for each pairwise comparison, using the function “getAnnotationCorrelation.”
Machine learning model
The extreme gradient boosting (XGBoost) machine learning algorithm using sequential decision trees (Chen and Guestrin 2016) was trained to distinguish between (1) proximal and distal peaks and (2) actual peaks versus randomly sampled nonoverlapping regions (distal peaks were separated based on >1 kb upstream of the TSS, and genome background peaks were matched for peak width). To select balanced data sets we used (1) 10,000 distal Amphimedon peaks versus 10,000 proximal Amphimedon peaks, and (2) 10,000 Amphimedon peaks versus 10,000 nonoverlapping background peaks that were sampled from the Amphimedon genome using the function “shuffle” in BEDTools (v2.27.1; -noOverlapping and -excl, the last option is used to avoid selecting background peaks that overlap with the actual cis-regulatory regions). Input into the algorithm is a matrix of counts of motif instances for every peak, where the positive and background sets were coded in a binary manner (distal ATAC-seq peaks = 1, proximal ATAC-seq peaks = 0, actual ATAC-seq peaks = 1, random regions = 0). Motif counts are determined using the annotatePeaks.pl function from HOMER (v4.11) (Heinz et al. 2010).
The Amphimedon data matrix was then split into a training data set and a test data set (70% and 30% of the peaks, respectively) using R package “rsample” (version 0.0.5) (https://CRAN.R-project.org/package=rsample). Nonvariable columns were removed from the training data. We train the XGB model using the Amphimedon training data set using the following parameters: eta = 0.01, max_depth = 6, nround = 60,000, subsample = 0.5, nfold = 10, colsample_bytree = 0.5, objective = “reg:squarederror,” and early_stopping_rounds = 50 (function “xgboost” from R package “xgboost” v0.90.0.2) (https://rdrr.io/cran/xgboost), where “eta” is the learning rate, “max_depth” is the maximum depth of a tree, “nround” represent the maximum number of boosting iterations, “subsample” is the subsample ratio of training instances, “nfold” is the number of random partitions of the training data, and “colsample_bytree” is the ratio of columns sampled when each tree is created. The XGB model is trained to minimize the “objective” function, and it will stop if this value does not decrease in “early_stopping_rounds” rounds.
Convergence has been achieved when error did not decrease after 50 iterations. Probabilities were calculated using function “predict” (type = “response”) in the R package “stats” (v3.6.1). Predictions of cis-regulatory regions from other species (worm, fly, mouse, zebrafish, and Capsaspora) were performed in a similar way. To account for differences in peak widths across species, we trained an XGB model normalizing for peak widths by dividing each count by the peak size (base pair) and then multiplying by 10,000. ROC curves were generated using the function “roc” from R package “pROC” (v1.16.2) (Sing et al. 2005). A threshold of 0.5 was used to transform the raw predicted probabilities into predicted classes to calculate accuracy. SHAP values were calculated using the count matrix used to train the XGB model and the model produced by xgboost (Lundberg et al. 2020).
Zebrafish (Danio rerio) distal regulatory elements were defined by ChIP-seq peaks of H3K4me1 excluding regions overlapping H3K4me3 regions and the TSS. The data spans four developmental stages: dome (1878 peaks), 80% epiboly (23,748 peaks), 24 hpf (23,419 peaks), and 48 hpf (15,388 peaks) (Bogdanović et al. 2012). We used Drosophila melanogaster ATAC-seq data from three developmental stages (2–4, 6–8, 10–12 h post egg-laying) (Floc'hlay et al. 2021). Consensus peaks more than 20 reads were used (7241 peaks in total). Consensus ATAC-seq peaks from Capsaspora owczarzaki profiled from three developmental stages (filopodiated amoeba, aggregative multicellular stage, and cystic stage; 11,927 peaks) were used (Sebé-Pedrós et al. 2016). Mouse and worm cis-regulatory regions were from Pijuan-Sala et al. (2020) and Daugherty et al. (2017), respectively. Worm ATAC-seq data spans three developmental stages (early embryo, larval stage 3, and young adult; n = 55,432 total unique peaks). Mouse scATAC-seq peaks were restricted to those accessible in at least 5% of the cells profiled (corresponding to mouse embryos at 8.25 d post fertilization) (Pijuan-Sala et al. 2020). Background peaks for worm, fly, zebrafish, mouse, and Capsaspora selected like in sponge (option -chrom).
Data access
All sequence data generated in this study have been submitted to ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-10203.
Supplementary Material
Acknowledgments
We thank Ozren Bogdanovic for helpful discussions. P.C-P. was supported by a University of New South Wales International Postgraduate Award PhD scholarship. S.M.D. was supported by Australian Research Council (ARC) funding DP170102353. B.M.D. was supported by ARC funding DP160100573 and FL110100044. E.S.W. was supported by Advanced Queensland Maternity Fund, ARC funding DE160100755 and DP200100250.
Author contributions: E.S.W. and B.M.D. conceived of the study. B.M.D., S.M.D., E.S.W., and K.R. planned the experiments. K.R. performed the experiments. E.S.W. designed the analyses. E.S.W. and P.C-P. performed the analyses. E.S.W. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275864.121.
Competing interest statement
The authors declare no competing interests.
References
- Adamska M, Degnan SM, Green KM, Adamski M, Craigie A, Larroux C, Degnan BM. 2007. Wnt and TGF-β expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. PLoS One 2: e1031. 10.1371/journal.pone.0001031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold P, Schöler A, Pachkov M, Balwierz PJ, Jørgensen H, Stadler MB, van Nimwegen E, Schübeler D. 2013. Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting. Genome Res 23: 60–73. 10.1101/gr.142661.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. 2014. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell 54: 844–857. 10.1016/j.molcel.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Blythe SA, Wieschaus EF. 2016. Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. eLife 5: e20148. 10.7554/eLife.20148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanović O, Fernandez-Miñán A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen S, Veenstra GJC, Gómez-Skarmeta JL. 2012. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res 22: 2043–2053. 10.1101/gr.134833.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J, Wu B, Chang H, Greenleaf W. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21.29.1–21.29.9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339. 10.1016/j.cell.2011.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Zhang Y, Loh YP, Tng JQ, Lim MC, Cao Z, Raju A, Lieberman Aiden E, Li S, Manikandan L, et al. 2021. H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. Nat Commun 12: 719. 10.1038/s41467-021-20940-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcino AD, Fernandez-Valverde SL, Taft RJ, Degnan BM. 2018. Diverse RNA interference strategies in early-branching metazoans. BMC Evol Biol 18: 160. 10.1186/s12862-018-1274-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. 10.1016/j.cell.2008.06.030 [DOI] [PubMed] [Google Scholar]
- Chen T, Guestrin C. 2016. XGBoost: a scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 785–794. Association for Computing Machinery, New York. 10.1145/2939672.2939785 [DOI] [Google Scholar]
- Daugherty AC, Yeo RW, Buenrostro JD, Greenleaf WJ, Kundaje A, Brunet A. 2017. Chromatin accessibility dynamics reveal novel functional enhancers in C. elegans. Genome Res 27: 2096–2107. 10.1101/gr.226233.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan BM, Adamska M, Craigie A, Degnan SM, Fahey B, Gauthier M, Hooper JNA, Larroux C, Leys SP, Lovas E, et al. 2008. The demosponge Amphimedon queenslandica: reconstructing the ancestral metazoan genome and deciphering the origin of animal multicellularity. Cold Spring Harb Protoc 2008: pdb.emo108. 10.1101/pdb.emo108 [DOI] [PubMed] [Google Scholar]
- Degnan BM, Adamska M, Richards GS, Larroux C, Leininger S, Bergum B, Calcino A, Taylor K, Nakanishi N, Degnan SM. 2015. Porifera. In Evolutionary developmental biology of invertebrates 1: introduction, non-bilateria, Acoelomorpha, Xenoturbellida, Chaetognatha (ed. Wanninger A), pp. 65–106. Springer, Vienna. [Google Scholar]
- Domazet-Lošo T, Tautz D. 2010. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468: 815–818. 10.1038/nature09632 [DOI] [PubMed] [Google Scholar]
- Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schübeler D. 2015. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 528: 575–579. 10.1038/nature16462 [DOI] [PubMed] [Google Scholar]
- Domcke S, Hill AJ, Daza RM, Cao J, O'Day DR, Pliner HA, Aldinger KA, Pokholok D, Zhang F, Milbank JH, et al. 2020. A human cell atlas of fetal chromatin accessibility. Science 370: eaba7612. 10.1126/science.aba7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeili M, Blythe SA, Tobias JW, Zhang K, Yang J, Klein PS. 2020. Chromatin accessibility and histone acetylation in the regulation of competence in early development. Dev Biol 462: 20–35. 10.1016/j.ydbio.2020.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Valverde SL, Degnan BM. 2016. Bilaterian-like promoters in the highly compact Amphimedon queenslandica genome. Sci Rep 6: 22496. 10.1038/srep22496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Valverde SL, Calcino AD, Degnan BM. 2015. Deep developmental transcriptome sequencing uncovers numerous new genes and enhances gene annotation in the sponge Amphimedon queenslandica. BMC Genomics 16: 387. 10.1186/s12864-015-1588-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floc'hlay S, Wong ES, Zhao B, Viales RR, Thomas-Chollier M, Thieffry D, Garfield DA, Furlong EEM. 2021. Cis-acting variation is common across regulatory layers but is often buffered during embryonic development. Genome Res 31: 211–224. 10.1101/gr.266338.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiti F, Jindrich K, Fernandez-Valverde SL, Roper KE, Degnan BM, Tanurdžić M. 2017. Landscape of histone modifications in a sponge reveals the origin of animal cis-regulatory complexity. eLife 6: e22194. 10.7554/eLife.22194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. 2008. Early origins and evolution of microRNAs and piwi-interacting RNAs in animals. Nature 455: 1193–1197. 10.1038/nature07415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Cho J-W, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, et al. 2018. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res 46: D380–D386. 10.1093/nar/gkx1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jänes J, Dong Y, Schoof M, Serizay J, Appert A, Cerrato C, Woodbury C, Chen R, Gemma C, Huang N, et al. 2018. Chromatin accessibility dynamics across C. elegans development and ageing. eLife 7: e37344. 10.7554/eLife.37344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinka AT, Tomancak P. 2012. The evolution of early animal embryos: conservation or divergence? Trends Ecol Evol 27: 385–393. 10.1016/j.tree.2012.03.007 [DOI] [PubMed] [Google Scholar]
- Klemm SL, Shipony Z, Greenleaf WJ. 2019. Chromatin accessibility and the regulatory epigenome. Nature Reviews Genetics 20: 207–220. 10.1038/s41576-018-0089-8 [DOI] [PubMed] [Google Scholar]
- Koenecke N, Johnston J, He Q, Meier S, Zeitlinger J. 2017. Drosophila poised enhancers are generated during tissue patterning with the help of repression. Genome Res 27: 64–74. 10.1101/gr.209486.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kribelbauer JF, Rastogi C, Bussemaker HJ, Mann RS. 2019. Low-affinity binding sites and the transcription factor specificity paradox in eukaryotes. Annu Rev Cell Dev Biol 35: 357–379. 10.1146/annurev-cellbio-100617-062719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larroux C, Luke GN, Koopman P, Rokhsar DS, Shimeld SM, Degnan BM. 2008. Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol 25: 980–996. 10.1093/molbev/msn047 [DOI] [PubMed] [Google Scholar]
- Levin M, Anavy L, Cole AG, Winter E, Mostov N, Khair S, Senderovich N, Kovalev E, Silver DH, Feder M, et al. 2016. The mid-developmental transition and the evolution of animal body plans. Nature 531: 637–641. 10.1038/nature16994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levo M, Zalckvar E, Sharon E, Dantas Machado AC, Kalma Y, Lotam-Pompan M, Weinberger A, Yakhini Z, Rohs R, Segal E. 2015. Unraveling determinants of transcription factor binding outside the core binding site. Genome Res 25: 1018–1029. 10.1101/gr.185033.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leys SP, Larroux C, Gauthier M, Adamska M, Fahey B, Richards GS, Degnan SM, Degnan BM. 2008. Isolation of Amphimedon developmental material. Cold Spring Harb Protoc 2008: pdb.prot5095. 10.1101/pdb.prot5095 [DOI] [PubMed] [Google Scholar]
- Li H, Coghlan A, Ruan J, Coin LJ, Hériché J-K, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, et al. 2006. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res 34: D572–D580. 10.1093/nar/gkj118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brown JB, Huang H, Bickel PJ. 2011. Measuring reproducibility of high-throughput experiments. Ann Appl Stat 5: 1752–1779. 10.1214/11-AOAS466 [DOI] [Google Scholar]
- Liu X, Wang C, Liu W, Li J, Li C, Kou X, Chen J, Zhao Y, Gao H, Wang H, et al. 2016. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537: 558–562. 10.1038/nature19362 [DOI] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J. 2016. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167: 1170–1187. 10.1016/j.cell.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, Zhang Y. 2016. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 165: 1375–1388. 10.1016/j.cell.2016.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, Nair B, Katz R, Himmelfarb J, Bansal N, Lee S-I. 2020. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell 2: 56–67. 10.1038/s42256-019-0138-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen C, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al. 2016. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44: D110–D115. 10.1093/nar/gkv1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta KR, Jolma A, Yin Y, Morgunova E, Kivioja T, Akhtar J, Hens K, Toivonen J, Deplancke B, Furlong EEM, et al. 2015. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. eLife 4: e04837. 10.7554/eLife.04837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pálfy M, Schulze G, Valen E, Vastenhouw NL. 2020. Chromatin accessibility established by Pou5f3, Sox19b and nanog primes genes for activity during zebrafish genome activation. PLoS Genet 16: e1008546. 10.1371/journal.pgen.1008546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B, Snyder MP. 2020. Systematic identification of silencers in human cells. Nat Genet 52: 254–263. 10.1038/s41588-020-0578-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijuan-Sala B, Wilson NK, Xia J, Hou X, Hannah RL, Kinston S, Calero-Nieto FJ, Poirion O, Preissl S, Liu F, et al. 2020. Single-cell chromatin accessibility maps reveal regulatory programs driving early mouse organogenesis. Nat Cell Biol 22: 487–497. 10.1038/s41556-020-0489-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. 2017. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 14: 975–978. 10.1038/nmeth.4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Mifsud B, Senner CE, Todd CD, Chrysanthou S, Darbo E, Hemberger M, Branco MR. 2018. Divergent wiring of repressive and active chromatin interactions between mouse embryonic and trophoblast lineages. Nat Commun 9: 4189. 10.1038/s41467-018-06666-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Schönauer A, Rendeiro AF, Pribitzer C, Schauer A, Gilles AF, Schinko JB, Renfer E, Fredman D, Technau U. 2014. Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res 24: 639–650. 10.1101/gr.162529.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Ballaré C, Parra-Acero H, Chiva C, Tena JJ, Sabidó E, Gómez-Skarmeta JL, Di Croce L, Ruiz-Trillo I. 2016. The dynamic regulatory genome of Capsaspora and the origin of animal multicellularity. Cell 165: 1224–1237. 10.1016/j.cell.2016.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Chomsky E, Pang K, Lara-Astiaso D, Gaiti F, Mukamel Z, Amit I, Hejnol A, Degnan BM, Tanay A. 2018a. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat Ecol Evol 2: 1176–1188. 10.1038/s41559-018-0575-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Saudemont B, Chomsky E, Plessier F, Mailhé M-P, Renno J, Loe-Mie Y, Lifshitz A, Mukamel Z, Schmutz S, et al. 2018b. Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell 173: 1520–1534.e20. 10.1016/j.cell.2018.05.019 [DOI] [PubMed] [Google Scholar]
- Shen H, Cao K, Wang X. 2008. AtbZIP16 and AtbZIP68, two new members of GBFs, can interact with other G group bZIPs in Arabidopsis thaliana. BMB Rep 41: 132–138. 10.5483/BMBRep.2008.41.2.132 [DOI] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. 2005. ROCR: visualizing classifier performance in R. Bioinformatics 21: 3940–3941. 10.1093/bioinformatics/bti623 [DOI] [PubMed] [Google Scholar]
- Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, Ovcharenko I, Ahituv N. 2013. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet 45: 1021–1028. 10.1038/ng.2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. 2015. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161: 555–568. 10.1016/j.cell.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui G, Affar EB, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, et al. 2004. Yin Yang 1 is a negative regulator of p53. Cell 117: 859–872. 10.1016/j.cell.2004.06.004 [DOI] [PubMed] [Google Scholar]
- Tibshirani R. 1996. Regression shrinkage and selection via the lasso. J R Stat Soc Series B Stat Methodol 58: 267–288. 10.1111/j.2517-6161.1996.tb02080.x [DOI] [Google Scholar]
- Wong ES, Zheng D, Tan SZ, Bower NI, Garside V, Vanwalleghem G, Gaiti F, Scott E, Hogan BM, Kikuchi K, et al. 2020. Deep conservation of the enhancer regulatory code in animals. Science 370: eaax8137. 10.1126/science.aax8137 [DOI] [PubMed] [Google Scholar]
- Wray GA. 2003. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20: 1377–1419. 10.1093/molbev/msg140 [DOI] [PubMed] [Google Scholar]
- Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, et al. 2016. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534: 652–657. 10.1038/nature18606 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J. 2020. Seven myths of how transcription factors read the cis-regulatory code. Curr Opin Syst Biol 23: 22–31. 10.1016/j.coisb.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EEM. 2009. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462: 65–70. 10.1038/nature08531 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.